Abstract
Hypoxia-inducible factor 1α (HIF-1α) functions as a transcription factor that is activated by decreased cellular oxygen concentrations to induce expression of a network of genes involved in angiogenesis, erythropoiesis, and glucose homeostasis. Here we demonstrate that two members of the SRC-1/p160 family of transcriptional coactivators harboring histone acetyltransferase activity, SRC-1 and transcription intermediary factor 2 (TIF2), are able to interact with HIF-1α and enhance its transactivation potential in a hypoxia-dependent manner. HIF-1α contains within its C terminus two transactivation domains. The hypoxia-inducible activity of both these domains was enhanced by either SRC-1 or the CREB-binding protein (CBP)/p300 coactivator. Moreover, at limiting concentrations, SRC-1 produced this effect in synergy with CBP. Interestingly, this effect was strongly potentiated by the redox regulatory protein Ref-1, a dual-function protein harboring DNA repair endonuclease and cysteine reducing activities. These data indicate that all three proteins, CBP, SRC-1, and Ref-1, are important components of the hypoxia signaling pathway and have a common function in regulation of HIF-1α function in hypoxic cells. Given the absence of cysteine residues in one of the Ref-1-regulated transactivation domains of HIF-1α, it is thus possible that Ref-1 functions in hypoxic cells by targeting critical steps in the recruitment of the CBP–SRC-1 coactivator complex.
Reduced tissue oxygen tension levels (hypoxia) play a major role in the regulation of many physiological and pathological processes (for reviews, see references 8 and 55). Adaptive physiological responses to hypoxia include induced expression of genes encoding erythropoietin and vascular endothelial growth factor and activation of genes involved in glucose transport and metabolism (reviewed in references 8 and 55). Moreover, hypoxia is a critical determinant of the pathogenesis of many disorders, including tumor angiogenesis, ischemic heart disease, and stroke. Under hypoxic conditions, the diverse target genes described above are all transcriptionally up-regulated by the transcription factor hypoxia-inducible factor 1 (HIF-1).
HIF-1 is a heterodimeric complex of two basic helix-loop-helix (bHLH)/Per-Arnt-Sim domain (PAS) proteins, HIF-1α and Arnt (54). The HIF-1α protein is rapidly degraded in normoxic cells by the ubiquitin-proteasome pathway but is stabilized under hypoxic conditions (20, 21, 25, 27, 44) and shows hypoxia-induced import into the nucleus (26), thus allowing heterodimerization with the nuclear factor Arnt (12). The active HIF-1 heterodimer binds DNA at conserved hypoxia response elements (HREs) to activate transcription of target genes (55). The mechanism of hypoxia-dependent activation of HIF-1α function is not yet understood. We and others have recently demonstrated that the mechanism of signal transduction by HIF-1α is a multistep process since nuclear translocation of the protein per se is not sufficient for transcriptional activation by HIF-1α (26). In addition, within the nucleus HIF-1α requires hypoxia-dependent activation to enable it to recruit the CREB-binding protein (CBP)/p300 coactivator protein (26), which has previously been shown to play a role in activation of the erythropoietin and vascular endothelial growth factor genes in response to hypoxia (5).
CBP and p300 are ubiquitous, evolutionarily conserved, nuclear phosphoproteins that function, at least in part, by linking several different signal-responsive transcriptional activators to the basal transcription apparatus (28, 29, 32). It has been shown that CBP/p300 can form a complex with SRC-1, a protein originally identified as a coactivator of steroid hormone receptors (15, 28, 42, 60). SRC-1 belongs to a family of 160-kDa proteins that interact with several members of the nuclear receptor family in a hormone-dependent manner and thereby enhance transcriptional activation (50). To date, several structurally and functionally related proteins have been identified, including transcription intermidiary factor 2 (TIF2) (52) (also known as GRIP1 [17]), p/CIP (49) (also known as ACTR [10]), RAC3 (33), AIB1 (2), and TRAM-1 (48). The finding that the p160 coactivators can bind to other type of coactivators, such as CBP/p300 and p/CAF (10, 28, 60), suggests that p160 proteins play an important role in bridging the interactions of receptor activation functions with the basal transcription machinery to stimulate transactivation. Moreover, both the CBP/p300 and SRC-1/p160 classes of coactivators possess intrinsic histone acetyltransferase (HAT) activity which may contribute to locally remodel chromatin structures for better accessibility of the transcription machinery to DNA (6, 10, 41, 47).
The mechanism underlying hypoxia-dependent transcriptional activation by HIF-1α remains unclear. Understanding how functional activity of HIF-1α is triggered by low oxygen tension and how the activated protein is able to mediate transcriptional activation is necessary to elucidate the mechanism of signal transduction by HIF-1α. In this study, we demonstrate that two p160 proteins, SRC-1 and TIF2, interact with HIF-1α to enhance its hypoxia-dependent transactivation function. We also show that SRC-1 and CBP/p300 synergistically enhance HIF-1α-mediated transcriptional regulation under hypoxic conditions. Moreover, the redox-dependent enzyme Ref-1 appears to be critical in linking the effects of the two coactivator proteins CBP/p300 and SRC-1 to HIF-1α.
MATERIALS AND METHODS
Plasmids and fusion proteins.
Plasmids containing full-length HIF-1α (pCMV4/HIF-1α), full-length Arnt (pCMV4/Arnt), and the GAL4-driven luciferase reporter gene have been previously described (14, 26). The Rc/RSV-mCBP-HA construct expressing full-length mouse CBP was obtained from R. H. Goodman (Vollum Institute, Portland, Oreg.), and plasmids pCMVβ-p300-HA and pCMVβ-p300Δ1254-1376 were a kind gift from D. M. Livingston (Dana-Farber Cancer Institute and Harvard Medical School, Boston, Mass.). Plasmids containing human full-length SRC-1 and SRC-1ΔPAS cDNAs in pSG5 vector (42) were kindly provided by B. W. O'Malley (Baylor College of Medicine, Houston, Tex.). pSG5/SRC-1 M1234 (16) was obtained from M. G. Parker (Imperial Cancer Research Fund, London, United Kingdom). pCMV5/Ref-1 was produced by inserting the 1.45-kb EcoRI fragment of pBS/Ref-1 (57) (kindly provided by T. Curran, St. Jude's Children Hospital, Memphis, Tenn.) into the EcoRI site of pCMV5. pSG5/TIF2 was a kind gift from P. Chambon (Institut de Génétique et de Biologie Moléculaire et Cellulaire, Strasbourg, France). pT81/HRE-luc contains three tandem copies of the erythropoietin HRE in front of the herpes simplex thymidine kinase promoter and the luciferase gene (P. Coumailleau, P. Carrero, and L. Poellinger, unpublished data).
GAL4 DNA binding domain (DBD) fusion proteins encoding HIF-1α subfragments (corresponding to amino acids 1 to 826, 1 to 813, 526 to 642, 526 to 826, and 776 to 826) were subcloned into the pCMX expression vector (51). The green fluorescent protein (GFP) fusion protein containing full-length HIF-1α has been described elsewhere (26). pGFP/Ref-1 was produced by inserting the BamHI-SpeI fragment of pCRII/Ref-1 into the BamHI-NheI sites of pGFP. A GAL4 DBD fusion protein carrying the ΔbHLH/HIF-1α sequence (residues 71 to 826) was generated by amplifying the equivalent DNA sequence by PCR using Pfu DNA polymerase (Stratagene) together with primer pairs carrying KpnI or BamHI ends. The resulting product was inserted in frame to KpnI-BamHI sites of pCMX. GAL4/HIF 776–813 was generated by cleaving the C-terminal 13 amino acids from GAL4/HIF 776–826 by PstI digestion. GAL4/HIF 531–584 was produced by inserting the XhoI-BamHI fragment of CMV4/HIF-1α 531–584 (obtained from T. Pereira) into the XhoI-BamHI sites of pCMX. Nucleotide sequences were confirmed by sequencing. All GAL4 fusion constructs show similar levels of expression under hypoxic conditions (27). A glutathione S-transferase (GST)–HIF-1α fusion protein was generated by excision of full-length HIF-1α from a corresponding bacterial GST fusion expression vector (a kind gift from I. Pongratz) and insertion in frame into the BglII-XhoI sites of the pBC expression vector (9).
Cell culture.
COS7 cells (from the American Type Culture Collection) were routinely maintained in high-glucose Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal calf serum (FCS), 100 IU of penicillin per ml, 100 μg of streptomycin sulfate per ml, 2 mM l-glutamine, and 1× minimal essential medium containing nonessential amino acids. The human embryonic kidney cell line (293 cells) was cultured in 1:1 mixture of DMEM and F-12 medium supplemented with 10 and 5% FCS, respectively, penicillin-streptomycin sulfate, and nonessential amino acids as described above. All media and growth factors were purchased from Life Technologies, Inc.
Transfection and transient expression assays.
One day before transfection, cells were seeded in six-well plates, and the medium was changed to OPTI-MEM medium lacking phenol red (Life Technologies) before transfection. A DNA mixture containing 0.5 μg of luciferase reporter, 0.2 μg of a combination of expression vectors for HIF-1α and Arnt, 0.75 to 1.5 μg of expression vectors for different coactivators, and 0.2 μg of internal control plasmid pRSV-AF together with carrier DNA pCMV4 was prepared and mixed with Lipofectamine (Life Technologies) according to the manufacturer's recommendations. DNA precipitates were allowed to form at room temperature for 30 min and added to the culture. After 6 h of transfection, the medium was replaced with DMEM–10% FCS. Hypoxic induction was achieved by incubation of the cells with 1% O2, or 21% O2 for normoxia. After 36 h, aliquots of medium were collected and analyzed for alkaline phosphatase activity, and cells were harvested and extracts were analyzed for luciferase activity. Alkaline phosphatase activity was used to correct for differences in transfection efficiency. All transfections were done in duplicate; results are presented as mean ± standard error (SE).
For analysis of nuclear translocation of HIF-1α in living cells, we transiently expressed GFP-tagged full-length HIF-1α and pSG5/TIF2, pSG5/SRC-1, or pSG5/SRC-1 M1234 in COS7 cells as described previously (26). Briefly, a plasmid mixture containing 6 μg of the expression vector for GFP–HIF-1α, 5 μg of pSG5/TIF2, pSG5/SRC-1, or pSG5/SRC-1 M1234 plasmid when indicated, and carrier DNA pGEM-3Z to keep the total amount of DNA constant at 11 μg was mixed with 20 μl of TransIT-LT1 reagent (Panvera Corp., Madison, Wis.) and added to the culture. After 6 h of incubation, the medium was replaced as described above, and cells were induced 24 h later with the hypoxia-mimicking agent 2,2′-dipyridyl (100 μM) or treated with vehicle only. Two hours after treatment, the medium was withdrawn and cells were refed fresh DMEM containing 10% FCS. Visualization of intracellular trafficking of GFP-tagged proteins was performed as described elsewhere (26).
For protein-protein interaction assays, COS7 cells were transfected by using Lipofectamine (Life Technologies) with 10 μg of the GST–HIF-1α expression vector and pCMV4 as carrier DNA. After 6 h of transfection, the medium was replaced with DMEM–10% FCS. Hypoxic induction was achieved by incubation of the cells with the hypoxia mimic CoCl2 (100 μM) or with vehicle only. After 24 h, cells were harvested in ice-cold phosphate-buffered saline (PBS). Whole-cell extracts were prepared in buffer containing 0.4 M KCl, 20 mM HEPES (pH 7.4), 1 mM dithiothreitol, and 20% glycerol. The protein content of cell extracts was determined by a colorimetric method (Bio-Rad).
Assays of GST protein-protein interaction.
The analysis of interactions between SRC-1 and HIF-1α bound to glutathione-Sepharose was carried out by the method of Chatton et al. (9), with minor modifications. Approximately 250 μg of whole-cell extracts from COS7 cells transiently expressing GST–HIF-1α was incubated with [35S]methionine-labeled in vitro-translated SRC-1 protein in 250 μl of a low-stringency buffer (50 mM Tris-HCl [pH 7.8], 0.1% NP-40, 250 mM NaCl). The complexes were immobilized on 60 μl of a 50% suspension of glutathione-Sepharose beads (Pharmacia Biotech). After washing, the radiolabeled proteins were eluted, separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE), and detected by fluorography.
Bacterially expressed GST–Ref-1 fusion protein (1 μg) or GST (0.5 μg) was incubated with 45 μg of protein A-Sepharose CL-4B (Pharmacia Biotech) in 250 μl of PBS with 0.4 μg of GST antibody for 1 h at 4°C. After washing, the beads were incubated with [35S]methionine-labeled in vitro-translated proteins in PBS containing 3% bovine serum albumin and 1 mM dithiothreitol, with or without 5 mM diamide, at room temperature for 30 min. The beads were then washed five times, and bound proteins were eluted, separated by SDS-PAGE, and visualized by autoradiography.
RESULTS
TIF2 enhances HIF-1α activity and interacts with HIF-1α in a hypoxia-dependent manner in mammalian cells.
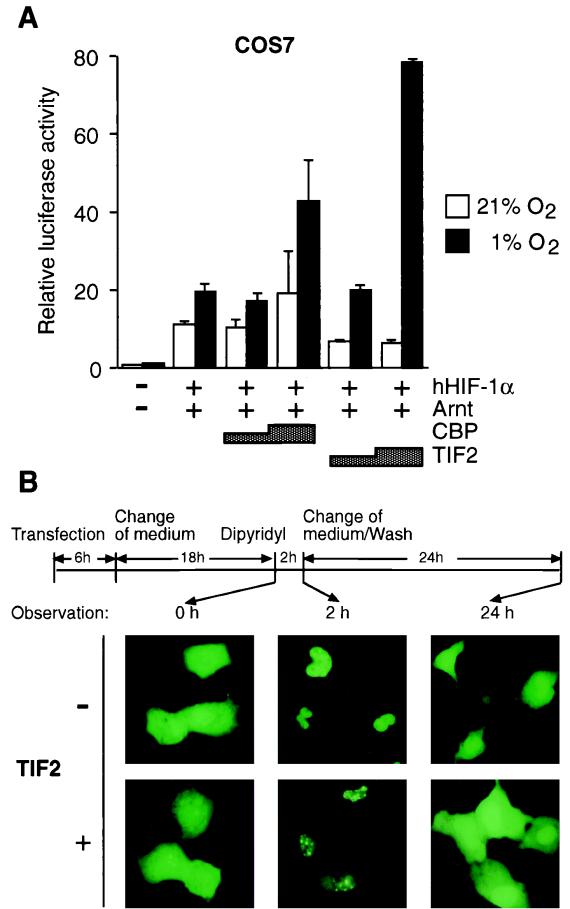
To gain a better understanding of the mechanism of transcriptional activation by HIF-1α, we tested the effect of transient expression of the transcriptional coactivator TIF2 on the functional activity of HIF-1α. TIF2 belongs to the p160 family of coactivators which have been shown to be required for nuclear receptor-mediated transcriptional activation (17, 48, 52). Transient transfection experiments were initially performed with COS7 cells. As shown in Fig. 1A, the reporter gene activity was not significantly altered by hypoxic treatment (1% O2) in the absence of coexpressed proteins, most probably due to the low levels of endogenous HIF-1 activity in COS7 cells (unpublished observations). Upon transient coexpression of HIF-1α and Arnt, pT81/HRE-luc reporter gene activity was stimulated around 10-fold under normoxic conditions, possibly due to a small but detectable pool of HIF-1α localized in the nucleus under these conditions (26). This reporter gene activity showed a modest (∼1.5-fold) increase by hypoxia (Fig. 1A). However, as expected (5, 26), ectopic expression of CBP further enhanced hypoxia-dependent activation by HIF-1α in a dose-dependent manner compared with hypoxia-stimulated activity in the absence of exogenous coactivators (two- to threefold). Although expression of CBP slightly stimulated HIF-1α activity already at normoxia, its overall activity was almost entirely dependent on the exposure to hypoxia as has been described previously (5, 26). The effect was most likely limited to threefold due to the large quantities of endogenous CBP already present in the cell prior to transfection. Coexpression of HIF-1α together with TIF2 resulted in potent (around 12-fold) activation of the reporter gene over the levels observed at normoxia, thereby establishing that both TIF2 and CBP support transcription by the HIF-1α–Arnt complex (Fig. 1A). In conclusion, TIF2 which was originally identified as a coactivator supporting hormone-dependent transcriptional activation by members of the steroid hormone receptor family, also functions as a conditionally regulated coactivator in hypoxia-dependent transcriptional activation by the HIF-1α–Arnt complex.
FIG. 1.
TIF2 enhances the HIF-1α-mediated transactivation function and interacts with HIF-1α in vivo. (A) COS7 cells were transiently cotransfected as described in Materials and Methods with 0.5 μg of pT81/HRE-luc reporter construct, hHIF1-α expression vector (pCMV4/HIF-1α; 0.2 μg), Arnt expression vector (pCMV4/Arnt; 0.2 μg), and 0.75 to 1.5 μg of either TIF2 (pSG5/TIF2) or CBP (Rc/RSV-CBP-HA), as indicated. The total amount of DNA was kept constant by the addition of parental pCMV4 when appropriate. Following transfection cells were incubated either under normoxic (21% O2) or hypoxic (1% O2) conditions. Luciferase activities were normalized for transfection efficiency by cotransfection of alkaline phosphatase-expressing pRSV-AF. Data are presented as luciferase activity relative to cells transfected with pCMV4 and reporter gene only and incubated at normoxia. Values represent the mean ± SE of two independent experiments. (B) Intranuclear redistribution of the GFP-HIF-1α fusion protein in the presence of TIF2. GFP–HIF-1α was transfected into COS7 cells in the absence or presence of exogenous TIF2 (pSG5/TIF2). After 24 h of expression, either 100 μM 2,2′-dipyridyl or vehicle (H2O) was added to the culture medium and incubated for 2 h before observation. Subsequently, 2,2′-dipyridyl was withdrawn by washing the cells and changing the medium, and the cells were thereafter incubated for an additional 24 h. Cells were observed microscopically at different time points as indicated by arrows. Photographs were taken with a Zeiss fluorescence microscope.
In response to hypoxia, HIF-1α is imported to the nucleus and shows a strictly nuclear localization (26). To characterize the mechanism of TIF2-dependent enhancement of transcriptional activation by HIF-1α in living cells, we next examined whether overexpression of TIF2 would affect the intracellular localization of HIF-1α. To this end, we transiently transfected COS7 cells with an expression vector encoding an in-frame fusion of GFP with full-length HIF-1α. As shown previously (26), fluorescence of this GFP–HIF-1α construct was uniformly distributed throughout the cell under normoxic conditions, and treatment of the transfected cells with the hypoxia-mimicking agent 2,2′-dipyridyl for 2 h led to a complete nuclear accumulation of the GFP–HIF-1α protein (26) (Fig. 1B). Transient expression of the TIF2 coactivator protein in the presence of GFP–HIF-1α had no effect on either the intensity or the subcellular distribution of fluorescence activity at normoxia. However, upon exposure of cells coexpressing GFP–HIF-1α and TIF2 to 2,2′-dipyridyl, GFP–HIF-1α-dependent fluorescence activity was detected in dot-like structures throughout the nucleus (Fig. 1B). It has previously been described that TIF2 is a nuclear protein mainly associated with such dot-like discrete bodies within the nucleus (52). These observations strongly suggest that TIF2 induced relocalization of GFP–HIF-1α within the nucleus, indicating that GFP–HIF-1α and TIF2 interacted with the one another in vivo and that the dot-like fluorescence pattern might represent transcriptionally active chromatin.
We have previously observed that return of cells to normoxia following exposure to hypoxia or withdrawal of hypoxia-mimicking chemicals results in an export of the nuclear pool of HIF-1α (26). To address whether the interaction between GFP–HIF-1α and TIF2 in vivo would be dependent on the hypoxic stimulus, we therefore initially exposed the transfected cells to 2,2′-dipyridyl and then further incubated them at normal levels of atmospheric oxygen tension after withdrawal of 2,2′-dipyridyl by washing of the cells in medium (see schematic representation in Fig. 1B). As expected (26), after 24 h of incubation under normoxic conditions, we observed a distribution of GFP–HIF-1α in both the nuclear and cytoplasmic compartments that was indistinguishable from the distribution of fluorescence activity prior to exposure to the hypoxic signal (Fig. 1B). Transient expression of TIF2 did not induce any quantitative or qualitative differences in the export of GFP–HIF-1α (Fig. 1B), demonstrating that the hypoxia-induced nuclear relocalization of HIF-1α by TIF2 could be reversed following withdrawal of the hypoxic signal.
Regulation of the hypoxia-inducible transactivation function of HIF-1α by SRC-1.
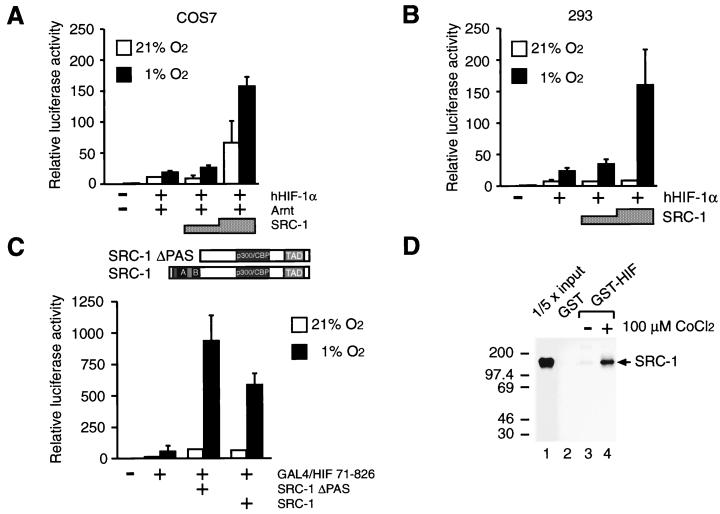
As outlined above, TIF2 belongs to a growing family of the p160 family of coactivators including SRC-1. TIF2 and SRC-1 appear to have similar activities as coactivators to enhance the transcriptional potential of many members of the ligand-activated nuclear hormone receptors (18, 28, 42, 52). SRC-1 has been demonstrated to directly and constitutively interact with CBP (28, 60). Moreover, in analogy to CBP (6, 41), SRC-1 has been shown to possess intrinsic HAT activity (10, 47). Against this background we also tested the ability of SRC-1 to modulate HIF-1α-dependent transcriptional activation of the minimal HRE-driven reporter gene. In transient transfection experiments using COS7 cells, HRE-dependent reporter gene activity was stimulated from two- to eightfold in a dose-dependent manner by SRC-1, compared with hypoxia-stimulated activity in the absence of exogenous coactivators (Fig. 2A). Transient expression of SRC-1 also enhanced the transcriptional activity of human HIF-1α (hHIF-1α) under normoxic conditions, albeit to a lesser extent (around threefold [Fig. 2A]). Thus, SRC-1 and TIF2 have similar properties in enhancing the transcriptional potential of hHIF-1α. To determine whether regulation of HIF-1α function by members of the p160 family of coactivators was cell type specific, we next carried out similar transfection experiments with the human 293 embryonic kidney cell line. In these experiments we observed that both SRC-1 (Fig. 2B) and TIF2 (data not shown) can significantly enhance hHIF-1α-dependent transcriptional activation in 293 cells.
FIG. 2.
SRC-1 stimulates HIF-1α activity in a hypoxia-dependent manner and interacts in vitro with HIF-1α. COS7 (A) and human embryonic kidney 293 (B) cells were cotransfected with pT81/HRE-luc (0.5 μg), 0.2 μg of pCMV4/HIF-1α (hHIF1-α), 0.2 μg of pCMV4/Arnt (Arnt), and 0.75 to 1.5 μg of SRC-1 (pSG5/SRC-1), as indicated. Six hours after transfection, cells were exposed to either 21 or 1% O2 for 36 h before harvest. Luciferase values are presented as relative luciferase activity as described in the legend to Fig. 1. The results of three independent experiments performed in duplicate ± SE are shown. (C) The SRC-1 PAS domain is not required for functional interaction with HIF-1α. (Top) Schematic representation of full-length SRC-1 and SRC-1ΔPAS. (Bottom) pGAL4/HIF 71-826 was cotransfected into COS7 cells together with a reporter plasmid expressing the luciferase gene driven by the thymidine kinase minimal promoter under the control of five copies of GAL4 binding sites and 1.5 μg of an expression vector encoding either full-length SRC-1 or a deletion mutant of SRC-1, SRC-1ΔPAS, lacking the PAS domain. Cells were exposed to 21 or 1% O2 for 36 h before harvest and reporter gene assays. Luciferase values are presented as relative luciferase activity as described in the legend to Fig. 1. The results of two independent experiments performed in duplicate ± SE are shown. (D) In vitro interaction between SRC-1 and HIF-1α. COS7 cells were transfected with 10 μg of the expression plasmid pGST-HIF-1α (GST-HIF) or empty GST expression vector (GST). Cells were exposed to either 100 μM CoCl2 or vehicle (H2O) for 24 h. Cell extracts were prepared and incubated with [35S]methionine-labeled in vitro-translated SRC-1 protein. The complexes were immobilized on glutathione-agarose beads for 2 h and eluted with the sample buffer by boiling. The eluted material was analyzed by SDS-PAGE (5% gel) and visualized by fluorography. Lane 1 represents one-fifth of the amount of [35S]methionine-labeled SRC-1 used in the binding reactions. Positions of molecular mass markers are shown on the left in kilodaltons.
Interestingly, in analogy to HIF-1α, the p160 family of coactivators belongs to the larger family of bHLH/PAS factors (schematically represented in Fig. 2C). SRC-1 was originally cloned as an N-terminally truncated fragment (42), here termed SRC-1ΔPAS (Fig. 2C), lacking the bHLH and PAS motifs. Both the bHLH and PAS domains represent potent protein-protein interaction interfaces (34, 35) and are critical for dimerization between HIF-1α and Arnt (14, 54). To investigate the role of the bHLH and PAS motifs of SRC-1 in hypoxia-dependent transactivation by HIF-1α, we examined the effect of overexpression of SRC-1ΔPAS or full-length SRC-1 on the transcriptional activity of a fusion protein, GAL4/HIF 71-826, containing the GAL4 DBD fused in frame to a fragment of hHIF-1α spanning amino acids 71 to 826. Hypoxia-dependent activation of this construct was monitored by using a GAL4-responsive reporter gene. As shown in Fig. 2C, the chimeric protein mediated ∼4-fold hypoxia-dependent activation of reporter gene activity. In agreement with the effect of SRC-1 on transcriptional activation mediated by the HIF-1α–Arnt complex, coexpression of full-length SRC-1 coactivator further enhanced the activity of GAL4/HIF 71-826 about 12-fold under hypoxic conditions, resulting in ∼45-fold stimulation of the reporter gene over the levels observed at normoxia (Fig. 2C). SRC-1 showed no detectable effect on the transcriptional activity by the GAL4 DBD alone (data not shown). These results demonstrate that SRC-1 targets the HIF-1α protein. Next, we examined the effect of the SRC-1ΔPAS mutant on GAL4/HIF 71-826-mediated transactivation. This truncated coactivator fragment showed activity very similar to if not more potent than that of full-length SRC-1 in enhancing the hypoxia-dependent activation response (Fig. 2C). Thus, in excellent agreement with the high activity of SRC-1ΔPAS in enhancing steroid hormone receptor-dependent transcriptional regulation (42), the bHLH and PAS motifs of SRC-1 were not important to support HIF-1α function.
The present experiments indicated the functional importance of both CBP and SRC-1 for enhancing the transcriptional potential by HIF-1α in hypoxic cells. CBP has previously been demonstrated to physically interact with HIF-1α (5). To investigate whether SRC-1 was able to interact with HIF-1α, we performed GST precipitation assays using cell extracts prepared from COS7 cells transiently expressing GST-tagged HIF-1α in the presence and absence of 100 μM CoCl2, a chemical known to mimic hypoxic induction of target gene expression and to activate HIF-1α (8, 55). This material was subsequently incubated with in vitro-translated, 35S-labeled SRC-1. As shown in Fig. 2D, in the presence of CoCl2, 35S-labeled SRC-1 was specifically retained by the GST–HIF-1α fusion protein immobilized on glutathione-Sepharose beads (lane 4), whereas we observed no significant binding to GST–HIF-1α extracted from nontreated cells (lane 3) or to GST alone (lane 2). Thus, SRC-1 interacted with HIF-1α in a hypoxia-dependent manner in vitro.
Role of LXXLL motifs in HIF-1α–SRC-1 interaction.
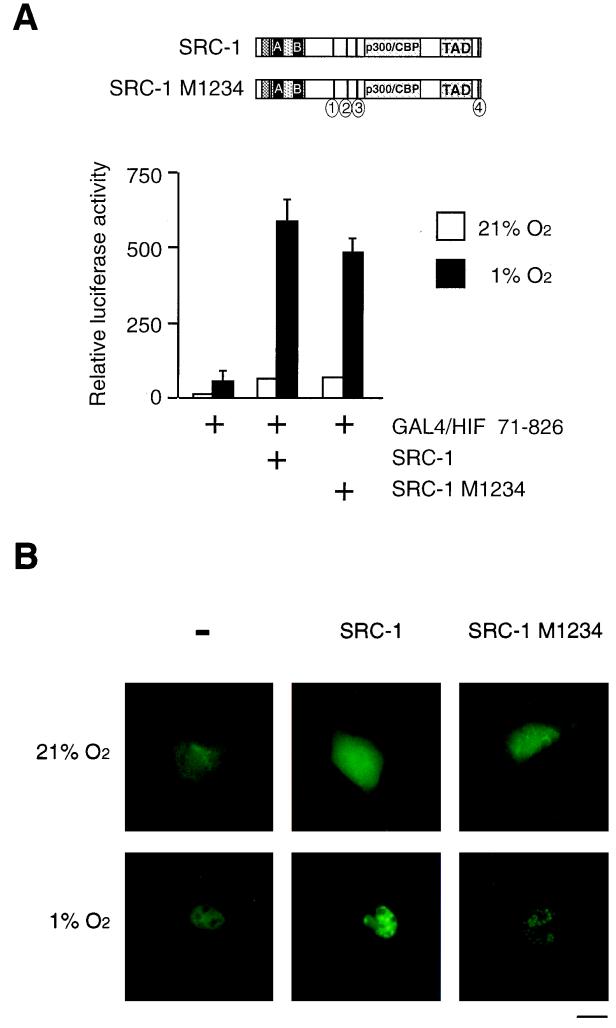
SRC-1 contains four copies of the short sequence motif LXXLL. Three of these motifs were identified in the central domain of the protein, and the fourth motif was found in the eight most C-terminal amino acids of human SRC-1 (16). The LXXLL motifs have been shown to be necessary to mediate the binding of SRC-1 to ligand-occupied nuclear receptors (16, 38, 56). To investigate the role of the LXXLL motifs of SRC-1 in hypoxia-dependent transactivation by HIF-1α, we compared the activities of wild-type SRC-1 and SRC-1 M1234, a mutant protein in which the conserved leucine couplets were changed to alanine at residues 636/7, 693/4, 752/3, and 1438/9 (16) in transient transfection experiments. As shown in Fig. 3A, the mutant protein in which all four functional binding motifs are disabled enhanced the activity of GAL4/HIF 71-826 to the same extent as the wild-type SRC-1 under hypoxic conditions. Moreover, transient expression of either wild-type SRC-1 or SRC-1 M1234 together with GFP–HIF-1α protein in COS7 cells had no effect on the subcellular distribution of fluorescence activity at normoxia. Under hypoxic conditions, SRC-1 induced relocalization of GFP-HIF-1α from a uniform nuclear distribution to discrete dot-like structures within the nucleus (Fig. 3B). This effect was also produced by the mutant SRC-1 M1234 protein (Fig. 3B), indicating that both proteins interact with HIF-1α in a hypoxia-dependent manner in vivo. Taken together, these results suggest that in contrast to nuclear receptor signaling, the LXXLL motif is not required for HIF-1α–SRC-1 interaction.
FIG. 3.
LXXLL motifs of SRC-1 are not required for HIF-1α-mediated transactivation function and interaction with HIF-1α in vivo. (A) (Top) Schematic representation of full-length SRC-1 and mutant SRC-1 M1234. Black bars represent the approximate locations of the LXXLL motifs in the linear SRC-1 sequence; circles with numbers indicate sites of mutation of LXXLL motifs in which conserved leucine residues are replaced by alanines. (Bottom) pGAL4/HIF 71-826 was cotransfected into COS7 cells together with a GAL4-responsive reporter plasmid and 1.5 μg of either full-length SRC-1 or mutated SRC-1, SRC-1 M1234 expression vectors. Cells were exposed to 21 or 1% O2 for 36 h before harvest and reporter gene assays. Luciferase values are presented as relative luciferase activity as described in the legend to Fig. 1. The results of two independent experiments performed in duplicate ± SE are shown. (B) Intranuclear redistribution of the GFP–HIF-1α fusion protein in the presence of SRC-1 and SRC-1 M1234. pGFP-HIF-1α was transfected into COS7 cells with or without SRC-1 or SRC-1 M1234 as indicated. After 24 h of expression, either 100 μM 2,2′-dipyridyl or vehicle (H2O) was added to the culture medium and incubated for 2 h. Cells were observed using a fluorescence microscope. Photographs of several cells for each condition were taken and representative cells are shown. Bar = 10 μm.
Definition of the functional domains of HIF-1α which interact with the coactivators CBP and SRC-1.
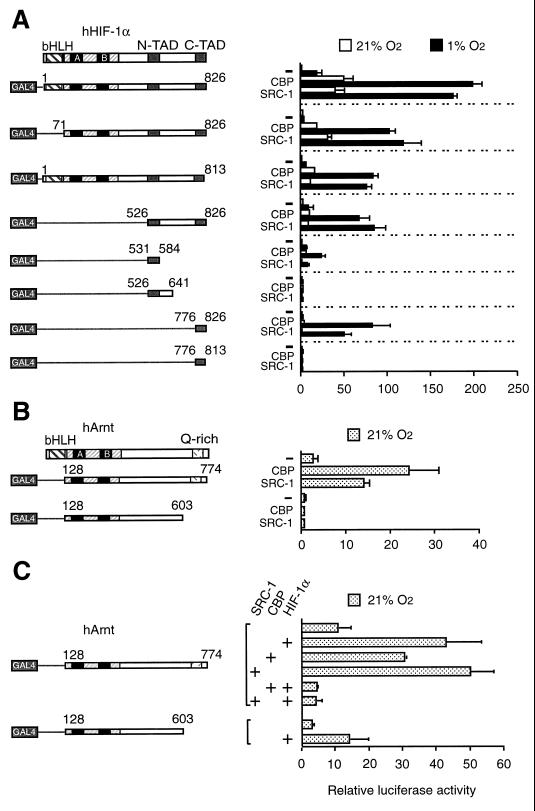
To identify HIF-1α structures which could serve as interaction surfaces with the SRC-1 and CBP coactivator proteins, we constructed a series of GAL4 DBD fusion proteins containing HIF-1α fragments of various lengths (as schematically represented in Fig. 4A). These constructs were transiently transfected into COS7 cells together with a GAL4-responsive reporter gene. Fusion of full-length HIF-1α to the GAL4 DBD produced significant hypoxia-dependent induction of reporter gene activity in the absence of ectopically expressed coactivator proteins (26) (Fig. 4A). Coexpression of either CBP or SRC-1 strongly (around 8- to 10-fold) enhanced the transcriptional potential of HIF-1α in hypoxic cells. These coactivators also increased to a lesser extent the transcriptional activity of GAL4/HIF 1-826 under normoxic conditions. Neither CBP nor SRC-1 enhanced the transcriptional activity of the GAL4 DBD alone (data not shown). GAL4/HIF 71-826, which lacks the bHLH domain of HIF-1α, induced relative luciferase activity about 3.5-fold over background levels (Fig. 4A). In the presence of coexpressed CBP or SRC-1, luciferase activity was induced under hypoxic conditions by GAL4/HIF 71-826 to a 30- or 34-fold-higher level, respectively, in comparison with the activity observed at hypoxia in the absence of exogenous coactivators.
FIG. 4.
Definition of HIF-1α structures which are regulated by the CBP and SRC-1 coactivators. (A) Different subregions of HIF-1α were fused to the GAL4 DBD and transfected into COS7 cells together with a GAL4-responsive reporter plasmid in the absence or presence of 1.5 μg of either CBP or SRC-1 expression plasmid. Cells were exposed to 21 or 1% O2 for 36 h before harvest. N-TAD and C-TAD, N- and C-terminal transactivation domains. (B) The C-terminal transactivation domain of Arnt mediates transcriptional activation by CBP and SRC-1. COS7 cells were cotransfected with the indicated GAL4-Arnt fusion proteins and 1.5 μg of CBP or SRC-1 expression vectors together with a GAL4-responsive reporter plasmid. Six hours after transfection, cells were exposed to either 21 or 1% O2 for 36 h before harvest and reporter gene assays. (C) HIF-1α prevents Arnt from functionally interacting with CBP and SRC-1. GAL4/Arnt 128-774 and a deletion mutant (GAL4/Arnt 128-603) were transfected into COS7 cells together with 0.2 μg of hHIF-1α expression vector and 1.5 μg of CBP and SRC-1 expression vectors, as indicated. Luciferase activity was measured following 36 h of exposure to either 21 or 1% O2. Normalized reporter gene activities are expressed relative to that of nonfusion GAL4 in normoxia. The results of three independent experiments performed in duplicate ± SE are shown.
We next examined the effect of CBP and SRC-1 on the activity of GAL4 fusion proteins containing sequences lying C terminal to the DNA binding and dimerization domains of HIF-1α. A hypoxia-inducible activation response was produced by GAL4/HIF 526-826 alone, in excellent agreement with the presence within this HIF-1α fragment of two distinct transactivation domains, labeled N-TAD and C-TAD in Fig. 4A, which function in a hypoxia-dependent fashion when fused to the GAL4 DBD (23, 43). Upon transient expression of CBP or SRC-1, the transcriptional activity of GAL4/HIF 526-826 was greatly (about seven- to ninefold) stimulated, demonstrating that these coactivators target the C terminus of HIF-1α. Moreover, these results further establish that the enhancing effect of both CBP and SRC-1 on HIF-1α function is independent of the presence of the bHLH and PAS domains of HIF-1α.
To further investigate whether one individual or both transcriptional activation domains of HIF-1α were targeted by the coactivators, we next deleted residues 813 to 826, a region that is contained within the C-terminal transactivation domain of HIF-1α (23, 43) and has been identified as a point of interaction with CBP (5, 7). Interestingly, GAL4/HIF 1-813 maintained a hypoxia-inducible response that was significantly enhanced (14- to 13-fold) by overexpression of CBP and SRC-1 (Fig. 4A), indicating that the hypoxia-dependent function of the N-terminal transactivation domain of HIF-1α may also be regulated by the coactivators. Against this background, we were interested in examining the regulatory properties of the individual transactivation domains of HIF-1α. As shown in Fig. 4A, GAL4/HIF 531-584, a chimeric protein spanning the N-terminal transactivation domain of HIF-1α (21, 23), showed hypoxia-dependent induction of reporter gene activity, albeit with a lower potency than the constructs spanning both transactivation domains. In the presence of CBP, however, an 11-fold stimulation of reporter gene activity was observed in hypoxic cells, whereas overexpression of SRC-1 only slightly (1.5-fold) enhanced the activity of GAL4/HIF 531-584 under identical conditions. We next examined the transactivation capacity of GAL4/HIF 526-641, which harbors both the N-terminal transactivation domain and a structure which has been proposed to function as an inhibitory sequence (23). In comparison with GAL4/HIF 531-584, the transcriptional activity of this chimeric protein was greatly reduced and showed complete abrogation of the effect of CBP and SRC-1 on its transactivation function (Fig. 4A), indicating either that an inhibitory region that may serve to modulate HIF-1α function under certain as yet unidentified conditions is contained between residues 584 and 641 (23) or, alternatively, that the fusion of the N-terminal transactivation domain to this sequence produces a malfolded domain when expressed outside the context of the full-length protein. In comparison, a fusion protein containing the carboxy-terminal transactivation domain of HIF-1α, GAL4/HIF 776-826, produced a very modest (about twofold) hypoxia-dependent induction response. However, its transcriptional activity was dramatically enhanced in the presence of either CBP (29-fold increase) or SRC-1 (17-fold increase). Upon deletion of the 13 most carboxy-terminal amino acids, both basal activity and inducible properties of GAL4/HIF 776-813 were completely abrogated independently of the absence or presence of coactivators (Fig. 4A). Taken together, these results suggest that (i) CBP mediates the hypoxia-inducible transcriptional activation by HIF-1α by targeting both of the transactivation domains of HIF-1α and (ii) the 13 most C-terminal amino acids of HIF-1α are crucial for stimulation of the activity of the C-terminal transactivation domain by CBP or SRC-1. However, in the context of the full-length protein, this C-terminal portion plays only a minor part in determining hypoxia-dependent functional activity in the presence of either CBP or SRC-1.
Since Arnt, the functional partner factor of HIF-1α, has recently been shown to interact with CBP/p300 (30), we also examined whether SRC-1 might functionally interact with Arnt. In normoxic cells, Arnt functions as a constitutively active transcription factor on E-box-driven promoters (1, 46). We transiently expressed in COS7 cells Arnt fused to the GAL4 DBD together with either CBP or SRC-1. In these experiments, we observed that constitutive activation of the reporter gene by GAL4/Arnt 128-774 was potently further enhanced by coexpression of CBP or SRC-1 (Fig. 4B). Deletion of the C-terminal Q-rich transactivation domain of Arnt (schematically represented in Fig. 4B) completely abolished the effect of these coactivators on transcription by GAL4/Arnt 128-603 (Fig. 4B). These observations suggest that the subunits of the HIF-1 heterodimer, HIF-1α and Arnt, interact independently with CBP and SRC-1. Interestingly, whereas Arnt interacted constitutively with CBP and SRC-1, the functional communication of the HIF-1α-Arnt heterodimeric complex with either CBP or SRC-1 was hypoxia dependent. Thus, dimerization with HIF-1α appears to impose hypoxia-dependent regulation on the ability of Arnt to functionally interact with these coactivator proteins. To further examine this question, we analyzed the effect of overexpression of HIF-1α on the activity of GAL4-Arnt fusion proteins at normoxia. Cotransfection of COS7 cells with HIF-1α and GAL4/Arnt 128-774 resulted in ∼4-fold activation of the reporter gene compared to cells transfected with the GAL4 fusion protein alone (Fig. 4C), probably due to the presence of small amounts of HIF-1α in the nucleus already at normoxia (26). However, constitutive activation of the reporter gene was completely abolished by coexpression of HIF-1α together with CBP or SRC-1 (Fig. 4C). This effect was indistinguishable from the one observed with GAL4/Arnt 128-603 in the presence of either of the coactivators (Fig. 4B). This deletion mutant, however, when cotransfected with HIF-1α resulted in further ∼5-fold stimulation of reporter gene activity (Fig. 4C), in agreement with its ability to functionally interact with HIF-1α via the PAS domain (14, 54). These results are consistent with the model that dimerization with HIF-1α impairs the ability of Arnt to functionally interact with CBP or SRC-1. However, the mechanism of this putative negative regulatory effect of HIF-1α needs to be further investigated.
SRC-1 and CBP act synergistically to enhance HIF-1α-dependent transcription.
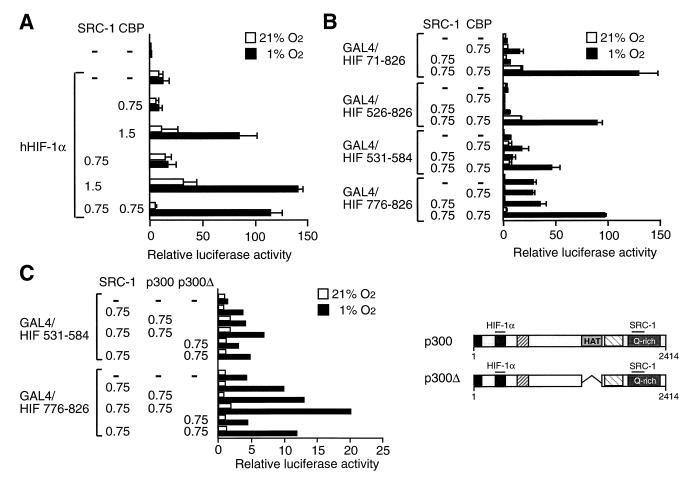
The ability of either CBP or SRC-1 to enhance the transcriptional activity of HIF-1α prompted us to investigate the effect of coexpression of both coactivators on HIF-1α function. We initially investigated the effects of transient expression of SRC-1 and CBP individually and in combination with one another on hypoxia-dependent activation by HIF-1α of a minimal HRE-driven reporter gene. As observed in Fig. 2A and B, the effect of SRC-1 on HIF-1α-dependent activation of the reporter gene was strictly dose dependent in COS7 and 293 cells, respectively. For instance, at a low concentration of SRC-1 expression vector (0.75 μg), no significant effect on HIF-1α-dependent activation of the reporter gene was detected (Fig. 2A, 2B, and 5A). In a similar fashion, an identically low concentration of CBP expression vector produced no enhancement of the transactivation function of HIF-1α (Fig. 5A). Under all of these conditions, poor hypoxia-dependent inducibility of the reporter gene was observed. However, a potent, hypoxia-dependent activation response could be reconstituted upon transient coexpression of SRC-1 and CBP at doses which yielded no significant effect individually (Fig. 5A), indicating strong synergy between these two coactivators in enhancing HIF-1α function.
FIG. 5.
CBP and SRC-1 cooperatively enhance HIF-1α-mediated transcriptional activation. (A) COS7 cells were transiently cotransfected with pCMV4/HIF-1α and 0.75 to 1.5 μg of SRC-1 and/or 0.75 to 1.5 μg of CBP expression plasmids together with a hypoxia-responsive reporter gene (pT81/HRE-luc) and subsequently exposed to 21 or 1% O2. Luciferase values are presented as relative luciferase activity as described in the legend to Fig. 1. The results of three independent experiments performed in duplicate ± SE are shown. (B) Cooperative activation by CBP and SRC-1 of GAL4–HIF-1α fusion proteins. COS7 cells were cotransfected with GAL4–HIF-1α fusion constructs together with a GAL4-responsive reporter plasmid in the absence or presence of CBP and/or SRC-1, as indicated (in micrograms). Cells were exposed to 21 or 1% O2 before harvest. After normalization for transfection efficiency using alkaline phosphatase activity, reporter gene activities are expressed as relative to that of GAL4 in normoxia. The results of two independent experiments performed in duplicate ± SE are shown. (C) p300 protein stimulates the activity of the two minimal transactivation domains of HIF-1α in a hypoxia-dependent manner. (Right) Schematic representation of full-length p300 (pCMVβ-p300-HA) and p300Δ (pCMVβ-p300Δ1254-1376). (Left) The two minimal transactivation domains of HIF-1α were fused to GAL4 DBD and transfected into COS7 cells together with a GAL4-responsive reporter plasmid in the absence or presence of p300, p300Δ, and/or SRC-1 expression vectors, as indicated. Cells were exposed to 21 or 1% O2 before harvest. Normalized reporter gene activities are expressed as relative to that of GAL4 in normoxia. The results of a representative experiment performed in duplicate are shown.
To further substantiate this synergistic effect, we next examined the transactivation potential of a number of GAL4 DBD fusion proteins containing distinct subregions of HIF-1α. Low concentrations of SRC-1 and CBP produced modest stimulation of the transcriptional activity by GAL4/HIF 71-826, spanning HIF-1α lacking the N-terminal bHLH domain (Fig. 5B). In contrast, coexpression of SRC-1 and CBP generated strong enhancement of the GAL4/HIF 71-826-mediated activation response in hypoxic cells. This effect was more than additive relative to the effects detected in the presence of CBP or SRC-1 alone, and it was also observed with the GAL4/HIF 526-826 fusion protein, which contains both N-terminal and C-terminal transactivation domains of HIF-1α (Fig. 5B). Moreover, coexpression of low concentrations of SRC-1 and CBP potently stimulated hypoxia-inducible transcriptional activation by the individual transactivation domains of HIF-1α contained within GAL4/HIF 531-584 and GAL4/HIF 776-826, respectively. These results are in excellent agreement with the data demonstrating that the two transactivation domains of HIF-1α independently can functionally interact with either SRC-1 or CBP (Fig. 4A). These data also are consistent with the notion that CBP and SRC-1 constitutively interact with one another (28, 60) and, in analogy to the mechanism of transcriptional activation by members of the steroid hormone receptor family (45), that recruitment of both coactivators may be necessary to trigger the full activity of HIF-1α in hypoxic cells. It remains to be established whether the recruitment of both of these classes of coactivators to HIF-1α occurs simultaneously or in a temporally regulated fashion. Furthermore, it is unclear whether CBP, SRC-1, or both coactivators provide the physical contact points with the transactivation domains of HIF-1α upon hypoxic activation. Interestingly, the functional interaction between HIF-1α and the two coactivator proteins was synergistic when both transactivation domains of HIF-1α were contained within the analyzed GAL4–HIF-1α fusion proteins. In contrast, GAL4 fusion proteins spanning the isolated transactivation domains showed an additive mode of regulation in the presence of both SRC-1 and CBP, strongly suggesting that synergistic regulation by these two coactivators may require the presence and integrity of both transactivation domains of HIF-1α.
CBP and p300 are closely related proteins that exhibit strong sequence similarity and similar functions with respect to their roles as coactivators (3, 4, 11, 36). In analogy to CBP, p300 enhanced in the presence of SRC-1 hypoxia-dependent transcriptional activation of reporter genes by GAL4 fusion proteins containing either one of the two individual transactivation domains of HIF-1α (Fig. 5C). As observed in experiments using CBP (Fig. 5B), the CBP/p300 and SRC-1 classes of coactivators produced together rather modest (3- to 4-fold) hypoxic activation of the N-terminal transactivation domain located between residues 531 and 584 of HIF-1α (Fig. 5C), whereas in the presence of SRC-1, CBP or p300 more potently (around 11-fold) stimulated hypoxic activation by the C-terminal transactivation domain of HIF-1α (Fig. 5C).
Although the mechanism of action of the CBP/p300 class of coactivators remains largely unclear, recent observations have suggested that these proteins contribute to transcriptional regulation not only by acting as simple adaptors between DNA binding factors and transcription initiation factors but also by harboring intrinsic HAT activity, linking their effect to regulation of chromatin structure and/or function (6, 31, 41). To investigate the potential role of the acetyltransferase activity of p300 in supporting HIF-1α-dependent transcription, we transfected COS7 cells with a p300 deletion mutant, p300ΔHAT, lacking the HAT domain which is centrally located in the protein (schematically represented in the right panel of Fig. 5C). In the absence of SRC-1, transient expression of p300ΔHAT resulted in only very moderate (twofold) stimulation of the hypoxia-dependent activation response produced by GAL4/HIF 531-584, containing the N-terminal transactivation domain of HIF-1α, whereas it did not produce any significant effect on the hypoxia-induced transcriptional activity of GAL4/HIF 776-826, spanning the minimal C-terminal transactivation domain (Fig. 5C). In the presence of SRC-1, p300ΔHAT enhanced reporter gene activation, most notably when coexpressed in hypoxic cells together with GAL4/HIF 776-826, containing the C-terminal transactivation domain of HIF-1α. However, p300ΔHAT was about twofold less potent than wild-type p300 in producing this response (Fig. 5C), indicating that the acetyltransferase catalytic activity of p300 may contribute to trigger the full activity of the HIF-1α-mediated transcriptional activation response. However, given the observed synergy between CBP/p300 and SRC-1 in enhancing activation potency by HIF-1α, and the fact that SRC-1 also harbors intrinsic HAT activity (47), the effect of the deletion of the HAT domain of p300 may be masked by corresponding activities of CBP-associated proteins, possibly that of SRC-1 itself.
Ref-1 potentiates HIF-1α function in the presence of CBP and SRC-1.
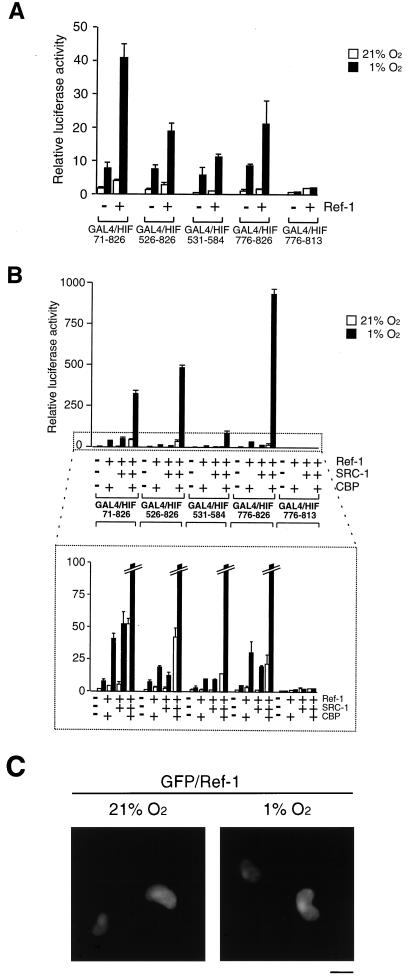
The nuclear redox regulator Ref-1 is known to stabilize the DNA binding activity of AP-1 by reduction of a conserved cysteine residue of Fos and Jun. Ref-1 is a bifunctional enzyme: it harbors both redox and endonuclease DNA repair activities (57) and has been implicated in up-regulation of HIF-1α-dependent induction of gene expression under hypoxic conditions (13, 20). Against this background, we wanted to further investigate the effect of Ref-1 on HIF-1α-dependent transcriptional activation and to examine its effect on the functional interaction of HIF-1α with the coactivators CBP and SRC-1. To investigate whether HIF-1α is a target of regulation by Ref-1, we initially used the different GAL4 DBD fusion proteins containing distinct subfragments of HIF-1α. As shown in Fig. 6A, transient overexpression of Ref-1 markedly potentiated hypoxic induction of reporter gene expression by the fusion proteins GAL4/HIF 71-826 and GAL4/HIF 526-826, spanning both transactivation domains of HIF-1α. The transcriptional activation function of GAL4/HIF 531-584, containing the N-terminal transactivation domain of HIF-1α, was only very moderately but significantly stimulated by Ref-1 in hypoxic cells, whereas Ref-1 produced more potent regulation of hypoxia-inducible transactivation by GAL4/HIF 776-826, containing the C-terminal transactivation domain (Fig. 6A).
FIG. 6.
Ref-1 enhances HIF-1α function. (A) COS7 cells were cotransfected with different GAL4–HIF-1α fusion constructs together with a GAL4-responsive reporter plasmid in the absence or presence of 1.5 μg of Ref-1 (pCMV5/Ref-1), as indicated. Cells were exposed to 21 or 1% O2 before harvest. After normalization for transfection efficiency using alkaline phosphatase activity, reporter gene activities were expressed as relative to that of GAL4 in normoxia. The results of two independent experiments performed in duplicate ± SE are shown. (B) Ref-1 potentiates CBP and SRC-1 activation of HIF-1α. The same GAL4– HIF-1α fusion proteins as shown in panel A were cotransfected into COS7 cells together with a GAL4-responsive reporter plasmid in the absence or presence of different combinations of Ref-1 (0.75 μg), CBP (0.75 μg), and/or SRC-1 (0.75 μg) expression vector, as indicated. The bottom panel shows an enlargement of the area marked with dots. (C) Effect of hypoxia treatment on subcellular localization of Ref-1. COS7 cells grown on coverslips were transiently transfected with 3 μg of pGFP/Ref-1. After 6 h of incubation, the medium was changed to fresh DMEM supplemented with 10% FCS and incubated for 24 h. Cells were then exposed to 21 or 1% O2 for 4 h. After being washed three times with PBS, cells were fixed with 4% paraformaldehyde in PBS for 2 h at room temperature, subsequently washed three times with PBS, and mounted. Cells were observed with a fluorescence microscope. Representative cells are shown. Bar = 10 μm.
Given the striking potentiation of HIF-1α function by the coactivators CBP and SRC-1, we next examined the effect of Ref-1 on transcriptional activation by the GAL4–HIF-1α fusion proteins in combination with CBP, SRC-1, or both proteins. Following transient expression of Ref-1 together with either CBP or SRC-1, hypoxia-inducible transcriptional activation by the tested GAL4–HIF-1α fusion proteins was not significantly altered in comparison to the results obtained in the presence of the fusion proteins and Ref-1 alone (Fig. 6B). Remarkably, however, the hypoxia-inducible transcriptional potencies of all fusion proteins containing either both or one of the HIF-1α transactivation domains were dramatically (10- to 53-fold) enhanced by coexpression of Ref-1 in the presence of CBP and SRC-1 (Fig. 6B). Under these conditions, the fusion proteins spanning the N- or C-terminal transactivation domains produced 12- or 53-fold hypoxia-dependent activation responses, respectively, whereas the fusion protein containing the C-terminal transactivation domain lacking the CBP interaction interface, GAL4/HIF 776-813, showed no regulation by Ref-1 and the coactivators. Thus, these data indicate that these two coactivators may have been limiting under these conditions for the Ref-1-mediated effect on HIF-1α-dependent transcription. Moreover, these results clearly demonstrate that both the N-terminal and C-terminal transactivation domains of HIF-1α are targets of regulation by Ref-1, implying that Ref-1 together with the CBP and SRC-1 classes of transcriptional coactivators plays a key role in regulation of HIF-1α function. Moreover, this is the first example of a transactivation domain representing a target of regulation by Ref-1; conversely, these data represent the first example of a noncovalent protein modifier of the HIF-1α transactivation domains identified in mammalian cells.
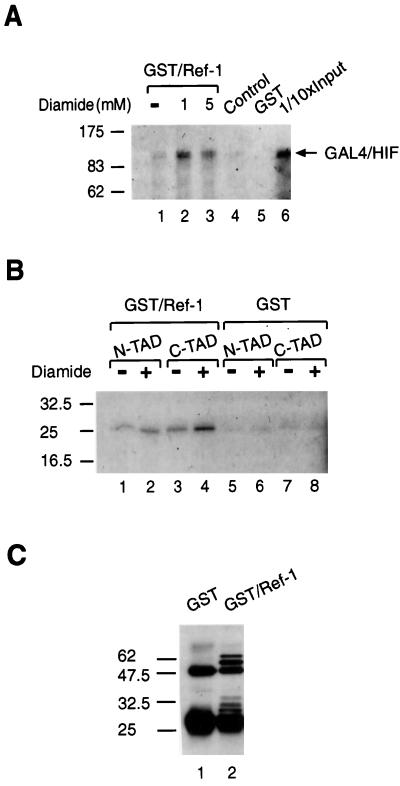
The precise mechanism by which Ref-1 activates HIF-1α is not known. Interestingly, a GFP-tagged Ref-1 fusion protein was exclusively localized in the nucleus both under normoxic and hypoxic conditions (Fig. 6C), indicating that nuclear translocation of HIF-1α is required for functional interaction with Ref-1. This redox regulator protein has been shown to form complexes with Jun in vitro (58). To further investigate whether Ref-1 could interact with HIF-1α, we tried to trap the interaction by using a cross-linking reagent such as diamide, which oxidizes cysteine sulfhydryls to disulfides. A series of experiments was performed with GST-tagged purified Ref-1 (Fig. 7). [35S]methionine-labeled in vitro-translated GAL4/HIF 1-826 was incubated with GST–Ref-1 in the presence or absence of diamide (Fig. 7A). GAL4/HIF 1-826 was found to weakly bind GST–Ref-1 (Fig. 7A, lane 1), and the cross-linking agent stabilized this interaction between Ref-1 and GAL4-HIF (compare lanes 2 and 3). No binding of [35S]methionine-labeled GAL4-HIF was observed when it was incubated with the anti-GST affinity gel alone in the absence of GST–Ref-1 (Fig. 7A, control lane 4) or when it was incubated with purified GST (Fig. 7A, lane 5), indicating that there was no significant background binding.
FIG. 7.
Ref-1 interacts with HIF-1α in vitro. [35S]methionine-labeled in vitro-translated GAL4/HIF 1-826 (A) and GAL4/HIF 531-584 (N-TAD) or GAL4/HIF 776-826 (C-TAD) (B) were incubated for 30 min at room temperature with recombinant GST/Ref-1 or GST in the presence or absence of diamide as indicated. Bound proteins were eluted in SDS sample buffer, run on SDS–7.5% (A) and 12.5% (B) polyacrylamide gels, and visualized by fluorography. Lane 6 in panel A represents 1/10 of the amount of the [35S]methionine-labeled HIF-1α protein used in the binding reactions; lane 4 represents [35S]methionine-labeled HIF-1α incubated with anti-GST affinity gel alone in the absence of GST/Ref-1. (C) Anti-GST Western blot. One-tenth input of proteins used in GST pull-down assays followed by elution and SDS-PAGE was analyzed by Western blotting using anti-GST antibodies. Positions of molecular mass markers are shown on the left in kilodaltons.
Next, we tried a series of pull-down experiments with [35S]methionine-labeled in vitro-translated GAL4/HIF 776-826 and GAL4/HIF 531-584. As shown in Fig. 7B, GAL4/HIF 531-584 bound GST–Ref-1, and no effect of diamide was observed (lanes 1 and 2), consistent with the absence of cysteine residues in the N-terminal transactivation domain of HIF-1α. An interaction between GAL4/HIF 726-826 and GST–Ref-1 was also detected, and this interaction was enhanced in the presence of diamide (Fig. 7B, lanes 3 and 4). In excellent agreement with this observation, the C-terminal transactivation domain contains two cysteine residues. Low levels of background binding of the labeled proteins to GST were observed in the presence or absence of diamide (Fig. 7B, lanes 5 to 8). Immunoblot analysis verified that the input concentrations of GST–Ref-1 and GST alone were similar (Fig. 7C).
The experiments above established that Ref-1 potentiates the effect of both the CBP and SRC-1 coactivators on hypoxia-inducible promoter activation by HIF-1α. Moreover, Ref-1 functionally and physically interacted with the distinct N- and C-terminal transactivation domains of HIF-1α. However, only the C-terminal and not the N-terminal transactivation domain of HIF-1α contains cysteine residues through which Ref-1 regulation occurs (57). These data indicate that an auxiliary factor, possibly a coactivator, may mediate interaction of Ref-1 with at least the N-terminal transactivation domain of HIF-1α.
DISCUSSION
In this report we demonstrate that two members of the p160 family, SRC-1 and TIF2, are able to interact with HIF-1α in a hypoxia-dependent manner and enhance its hypoxia-inducible transactivation potential. Moreover, low concentrations of SRC-1 can produce this effect in synergy with CBP, and importantly, this effect is greatly potentiated by the redox regulatory protein Ref-1, indicating that these three proteins are important components of the hypoxia signaling pathway.
Conditional recruitment of the SRC-1 and CBP classes of coactivators to HIF-1α is an important step in the hypoxia signaling pathway.
TIF2 and SRC-1 are related proteins that have originally been identified as coactivators of nuclear hormone receptors and are expressed in most tissues (17–19, 28, 42, 52, 53). The two proteins could have redundant functions, or they could serve as coactivators for different or overlapping subsets of transcription factors. Our results indicate that both SRC-1 and TIF2 are able to functionally interact in very similar manners with HIF-1α, a protein belonging to the bHLH/PAS family of transcription factors. In fact, SRC-1 and TIF2 are members of the same family of factors harboring a bHLH/PAS motif in their N termini. Whereas both the HLH (35) and PAS (34) motifs are potent dimerization interfaces mediating, for instance, HIF-1α–Arnt heterodimerization (14, 54), the N-terminal bHLH/PAS motif of SRC-1 is not important for functional interaction with HIF-1α. In agreement with this observation, the bHLH/PAS region of SRC-1 is irrelevant for enhancing steroid hormone receptor-dependent transcription or mediating physical interaction with the receptors (28, 42, 60). In the case of various members of these receptors, a conserved helix (helix 12) of the ligand binding domain is establishing a physical contact with the SRC-1 class of coactivators, and in turn, there are multiple motifs within the SRC-1 family of proteins mediating this interaction with receptors (19, 24, 38, 53, 56). These interaction interfaces appear to be characterized by the integrity of the short signature motif LXXLL, where L is leucine and X is any amino acid (16). Interestingly, our results indicate that an SRC-1 mutant protein with mutated LXXLL motifs that is unable to support estrogen receptor-dependent transcription (16) functionally interacts with HIF-1α. Thus, HIF-1α may employ a mechanism of coactivator recruitment that is different from that of steroid receptors. In a similar fashion, the interaction between SRC-1 and the p50 subunit of NF-κB occurs via a region that does not harbor an LXXLL motif (39). It will now be interesting to identify the structural motif(s) of SRC-1 which mediates interaction with the hypoxia-activated form of HIF-1α.
Here we have identified the two transactivation domains localized in the C terminus of HIF-1α as targets of regulation by SRC-1. These two functional domains of HIF-1α are contained within 54- or 38-residue-long stretches of amino acids. Interestingly, the identical regions of HIF-1α were also targeted for regulation by CBP, Ref-1, and, most notably, a combination of Ref-1 together with SRC-1 and CBP. This striking interdigitation in regulatory potential between these three proteins indicates a common link in their mechanisms of action. SRC-1 and CBP constitutively interact with one another (15, 28, 60), and both proteins appear to potentiate steroid hormone receptor-mediated transactivation as a complex (reviewed in reference 50). Furthermore, both proteins contain intrinsic HAT activity (6, 41, 47). It is thus reasonable to expect functional redundancy within the CBP–SRC-1 complex with regard to acetylation activity. Consistent with this notion, we observed only partial reduction of the effect on HIF-1α-mediated transcriptional activation by using the p300ΔHAT mutant lacking the domain harboring HAT activity (37) but, as schematically represented in Fig. 5C, maintaining intact HIF-1α and SRC-1 interaction domains.
It is unclear whether the transactivation domain of HIF-1α preferentially interacts with any specific component of the CBP–SRC-1 complex. It has been shown that CBP interacts with HIF-1α via its first cysteine/histidine-rich region (CH1 [5]). In contrast, interaction of CBP with Arnt has been reported to be mediated by the CREB-binding site of CBP (30). These observations suggest that HIF-1α and Arnt within the hypoxia-activated heterodimeric complex may interact independently with two distinct regions of CBP/p300. Interestingly, it has been reported that it is not possible to detect any interaction between CBP and Arnt in an in vitro protein-protein interaction assay, whereas the interaction was demonstrated using an in vivo assay (30). These data indicate that the interaction could be mediated or strengthened by a factor(s) missing in the in vitro system (5, 30). For instance, in analogy to the mechanism of coactivator assembly on nuclear receptors, SRC-1 is a plausible candidate to facilitate this interaction.
Regulation of HIF-1α function by the redox regulatory protein Ref-1.
What is the mechanism of hypoxia-inducible recruitment of the coactivators to HIF-1α? We and others have previously observed that hypoxia-dependent activation of HIF-1α function is a multistep mechanism including massive up-regulation of HIF-1α protein levels (20, 21, 25, 44) by inhibition of ubiquitination of HIF-1α (27), nuclear translocation (26), dimerization with the constitutively nuclear factor Arnt (14, 54), and recruitment of CBP (5, 26). Moreover, we have recently demonstrated that GAL4–HIF-1α fusion proteins which show constitutive nuclear compartmentalization due to the nuclear localization signal contained within the GAL4 DBD require the hypoxic signal for functional interaction with CBP (26). In analogy to the steroid receptor system, it is an attractive scenario that the hypoxic signal determines a conformational change in HIF-1α which is critical for recruitment of the coactivators. This model of conformational regulation of HIF-1α function is supported by the present experiments which show that the integrity of the C terminus of HIF-1α containing both transactivation domains is important for cooperative regulation by SRC-1 and CBP. Moreover, the transactivation domain of Arnt is also able to functionally interact with both CBP and SRC-1 (this study and reference 30). Thus, conformational regulation may also provide a mechanism of conditional coactivator assembly on the individual transactivation domains within the heterodimeric HIF-1α–Arnt complex. To understand the hypoxia signaling pathway, it is critical to identify what determines recruitment of the coactivators within the nucleus of hypoxic cells. It is possible that noncovalent modification of the C terminus of HIF-1α plays a role in determining this regulatory effect. Although this is a plausible mechanism, a protein kinase mediating such a modification has not yet been demonstrated. In the present study we observed that Ref-1 enhances the effects of SRC-1 and CBP on the transactivation potential of HIF-1α. Ref-1 is known to stimulate the DNA binding activity of a number of transcription factors, including Fos, Jun, and p53, by a redox-dependent mechanism (22, 57). The stimulatory effect on Fos and Jun is elicited by reduction of a conserved cysteine residue located in the DNA binding domain of each protein (reference 57 and references therein). What is the target of regulation by Ref-1 in the hypoxia signaling pathway? Intriguingly, only the C-terminal transactivation domain of HIF-1α contains cysteine residues, whereas the N-terminal one located between amino acid residues 531 and 584 lacks cysteines. Transient overexpression of Ref-1 alone resulted in significant stimulation of the hypoxia-inducible activity of the C-terminal transactivation domain of HIF-1α. Under identical conditions, Ref-1 produced only a subtle effect on the function of the N-terminal hypoxia-responsive transactivation domain, indicating preferential regulation of the C-terminal transactivation domain containing the cysteine residues. However, in combination with transient expression of both SRC-1 and CBP, Ref-1 dramatically stimulated the hypoxia-dependent activities of both transactivation domains. Moreover, Ref-1 protein interacts in vitro with both N- and C-terminal transactivation domains of HIF-1α. These data demonstrate that both domains are individually regulated by Ref-1, providing the first example of a transactivation domain as a target of Ref-1 function.
Given the absence of cysteine residues in the N-terminal transactivation domain of HIF-1α, the molecular target of Ref-1 in the hypoxia signaling pathway may not be restricted to HIF-1α itself. In line with this model, recent experiments have indicated hypoxia-inducible enhancement by Ref-1 on the ligand-dependent functional activities of either the retinoic acid receptor β2 or the dioxin receptor, a bHLH/PAS transcription factor (P. Carrero, K. Okamoto, and L. Poellinger, unpublished data). Thus, the effect of Ref-1 on CBP- and SRC-1-regulated transcriptional activities appears not to be restricted to HIF-1α but also detected in a hypoxia-dependent fashion among other conditionally regulated transcription factors (59). This suggests that Ref-1 may be involved in regulation of coactivator assembly on these conditionally regulated transcription factors by, for instance, affecting the stability of the coactivator complex tethered to the transactivation domains or by enhancing recruitment of the coactivators. We are currently investigating this possibility. To understand this mechanism of regulation, it will now be critical to examine whether cysteine residues within any of the components of the coactivator complex are targeted for regulation by Ref-1. If so, Ref-1 may represent under hypoxic conditions an important regulator of transcriptional activation processes that depend on inducible recruitment of the CBP–SRC-1 coactivator complex.
ACKNOWLEDGMENTS
We thank M. G. Parker for the SRC-1 M1234 construct. We also thank Anders Berkenstam and Yuichi Makino for stimulating discussions and helpful advice.
This study was supported by grants from the Swedish Medical Research Council, Pharmacia and Upjohn, Akiyama Foundation, and NOVARTIS Foundation (Japan) for the Promotion of Science.
REFERENCES
- 1.Antonsson C, Arulampalam V, Whitelaw M L, Pettersson S, Poellinger L. Constitutive function of the basic helix-loop-helix/PAS factor Arnt. Regulation of target promoters via the E box motif. J Biol Chem. 1995;270:13968–13972. doi: 10.1074/jbc.270.23.13968. [DOI] [PubMed] [Google Scholar]
- 2.Anzick S L, Kononen J, Walker R L, Azorsa D O, Tanner M M, Guan X-Y, Sauter G, Kallioniemi O-P, Trent J M, Meltzer P S. AIB1, a steroid receptor coactivator amplified in breast and ovarian cancer. Science. 1997;277:965–968. doi: 10.1126/science.277.5328.965. [DOI] [PubMed] [Google Scholar]
- 3.Arany Z, Sellers W R, Livingston D M, Eckner R. E1A-associated p300 and CREB-associated CBP belong to a conserved family of coactivators. Cell. 1994;77:799–800. doi: 10.1016/0092-8674(94)90127-9. [DOI] [PubMed] [Google Scholar]
- 4.Arany Z, Newsome D, Oldread E, Livingston D M, Eckner R. A family of transcriptional adaptor proteins targeted by the E1A oncoprotein. Nature. 1995;374:81–84. doi: 10.1038/374081a0. [DOI] [PubMed] [Google Scholar]
- 5.Arany Z, Huang L E, Eckner R, Bhattacharya S, Jiang C, Goldberg M A, Bunn H F, Livingston D M. An essential role for p300/CBP in the cellular response to hypoxia. Proc Natl Acad Sci USA. 1996;93:12969–12973. doi: 10.1073/pnas.93.23.12969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bannister A J, Kouzarides T. The CBP co-activator is a histone acetyltransferase. Nature. 1996;384:641–643. doi: 10.1038/384641a0. [DOI] [PubMed] [Google Scholar]
- 7.Bhattacharya S, Michels C L, Leung M-K, Arany Z P, Kung A L, Livingston D M. Functional role of p35srj, a novel p300/CBP binding protein, during transactivation by HIF-1. Genes Dev. 1999;13:64–75. doi: 10.1101/gad.13.1.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bunn H F, Poyton R O. Oxygen sensing and molecular adaptation to hypoxia. Physiol Rev. 1996;76:839–885. doi: 10.1152/physrev.1996.76.3.839. [DOI] [PubMed] [Google Scholar]
- 9.Chatton B, Bahr A, Acker J, Kedinger C. Eukaryotic GST fusion vector for the study of protein-protein associations in vivo: application to interaction of ATFa with Jun and Fos. BioTechniques. 1995;18:142–145. [PubMed] [Google Scholar]
- 10.Chen H, Lin R J, Schiltz R L, Chakravarti D, Nash A, Nagy L, Privalsky M L, Nakatani Y, Evans R M. Nuclear receptor coactivator ACTR is a novel histone acetyltransferase and forms a multimeric activation complex with P/CAF and CBP/p300. Cell. 1997;90:569–580. doi: 10.1016/s0092-8674(00)80516-4. [DOI] [PubMed] [Google Scholar]
- 11.Eckner R, Ewen M E, Newsome D, Gerdes M, DeCaprio J A, Lawrence J B, Livingston D M. Molecular cloning and functional analysis of the adenovirus E1A-associated 300-kD protein (p300) reveals a protein with properties of a transcriptional adaptor. Genes Dev. 1994;8:869–884. doi: 10.1101/gad.8.8.869. [DOI] [PubMed] [Google Scholar]
- 12.Eguchi H, Ikuta T, Tachibana T, Yoneda Y, Kawajiri K. A nuclear localization signal of human aryl hydrocarbon receptor nuclear translocator/hypoxia-inducible factor 1 is a novel bipartite type recognized by the two components of nuclear pore-targeting complex. J Biol Chem. 1997;272:17640–17647. doi: 10.1074/jbc.272.28.17640. [DOI] [PubMed] [Google Scholar]
- 13.Ema M, Hirota K, Mimura J, Abe H, Yodoi J, Sogawa K, Poellinger L, Fujii-Kuriyama Y. Molecular mechanisms of transcription activation by HLF and HIF-1α in response to hypoxia: their stabilization and redox signal-induced interaction with CBP/p300. EMBO J. 1999;18:1905–1914. doi: 10.1093/emboj/18.7.1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gradin K, McGuire J, Wenger R H, Kvietikova I, Whitelaw M L, Toftgård R, Tora L, Gassmann M, Poellinger L. Functional interference between hypoxia and dioxin signal transduction pathways: competition for recruitment of the Arnt transcription factor. Mol Cell Biol. 1996;16:5221–5231. doi: 10.1128/mcb.16.10.5221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hanstein B, Eckner R, DiRenzo J, Halachmi S, Liu H, Searcy B, Kurokawa R, Brown M. p300 is a component of an oestrogen receptor coactivator complex. Proc Natl Acad Sci USA. 1996;93:11540–11545. doi: 10.1073/pnas.93.21.11540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heery D M, Kalkhoven E, Hoare S, Parker M G. A signature motif in transcriptional co-activators mediates binding to nuclear receptors. Nature. 1997;387:733–736. doi: 10.1038/42750. [DOI] [PubMed] [Google Scholar]
- 17.Hong H, Kohli K, Trivedi A, Johnson D L, Stallcup M R. GRIP1, a novel mouse protein that serves as a transcriptional coactivator in yeast for the hormone binding domains of steroid receptors. Proc Natl Acad Sci USA. 1996;93:4948–4952. doi: 10.1073/pnas.93.10.4948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hong H, Kohli K, Garabedian M J, Stallcup M R. GRIP1, a transcriptional coactivator for the AF-2 transactivation domain of steroid, thyroid, retinoid, and vitamin D receptors. Mol Cell Biol. 1997;17:2735–2744. doi: 10.1128/mcb.17.5.2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hong H, Darimont B D, Ma H, Yang L, Yamamoto K R, Stallcup M R. An additional region of coactivator GRIP1 required for interaction with the hormone-binding domains of a subset of nuclear receptors. J Biol Chem. 1999;274:3496–3502. doi: 10.1074/jbc.274.6.3496. [DOI] [PubMed] [Google Scholar]
- 20.Huang L E, Arany Z, Livingston D M, Bunn H F. Activation of hypoxia-inducible transcription factor depends primarily upon redox-sensitive stabilization of its α subunit. J Biol Chem. 1996;271:32253–32259. doi: 10.1074/jbc.271.50.32253. [DOI] [PubMed] [Google Scholar]
- 21.Huang L E, Gu J, Schau M, Bunn H F. Regulation of hypoxia-inducible factor 1α is mediated by an O2-dependent degradation domain via the ubiquitin-proteasome pathway. Proc Natl Acad Sci USA. 1998;95:7987–7992. doi: 10.1073/pnas.95.14.7987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jayaraman L, Murthy K G K, Zhu C, Curran T, Xanthoudakis S, Prives C. Identification of redox/repair protein Ref-1 as a potent activator of p53. Genes Dev. 1997;11:558–570. doi: 10.1101/gad.11.5.558. [DOI] [PubMed] [Google Scholar]
- 23.Jiang B-H, Zheng J Z, Leung S W, Roe R, Semenza G L. Transactivation and inhibitory domains of hypoxia-inducible factor 1α. J Biol Chem. 1997;272:19253–19260. doi: 10.1074/jbc.272.31.19253. [DOI] [PubMed] [Google Scholar]
- 24.Kalkhoven E, Valentine J E, Heery D M, Parker M G. Isoforms of steroid receptor co-activator 1 differ in their ability to potentiate transcription by the oestrogen receptor. EMBO J. 1998;17:232–243. doi: 10.1093/emboj/17.1.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kallio P J, Pongratz I, Gradin K, McGuire J, Poellinger L. Activation of hypoxia-inducible factor 1α: posttranscriptional regulation and conformational change by recruitment of the Arnt transcription factor. Proc Natl Acad Sci USA. 1997;94:5667–5672. doi: 10.1073/pnas.94.11.5667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kallio P J, Okamoto K, O'Brien S, Carrero P, Makino Y, Tanaka H, Poellinger L. Signal transduction in hypoxic cells: inducible nuclear translocation and recruitment of the CBP/p300 coactivator by the hypoxia-inducible factor-1α. EMBO J. 1998;17:6573–6586. doi: 10.1093/emboj/17.22.6573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kallio P J, Wilson W J, O'Brien S, Makino Y, Poellinger L. Regulation of the hypoxia-inducible transcription factor 1α by the ubiquitin-proteasome pathway. J Biol Chem. 1999;274:6519–6525. doi: 10.1074/jbc.274.10.6519. [DOI] [PubMed] [Google Scholar]
- 28.Kamei Y, Xu L, Heinzel T, Torchia J, Kurokawa R, Gloss B, Lin S-C, Heyman R A, Rose D W, Glass C K, Rosenfeld M G. A CBP integrator complex mediates transcriptional activation and AP-1 inhibition by nuclear receptors. Cell. 1996;85:403–414. doi: 10.1016/s0092-8674(00)81118-6. [DOI] [PubMed] [Google Scholar]
- 29.Kee B I, Arias J, Montminy M R. Adaptor-mediated recruitment of RNA polymerase II to a signal-dependent activator. J Biol Chem. 1996;271:2373–2375. doi: 10.1074/jbc.271.5.2373. [DOI] [PubMed] [Google Scholar]
- 30.Kobayashi A, Numayama-Tsuruta K, Sogawa K, Fujii-Kuriyama Y. CBP/p300 functions as a possible transcriptional coactivator of Ah receptor nuclear translocator (Arnt) J Biochem. 1997;122:703–710. doi: 10.1093/oxfordjournals.jbchem.a021812. [DOI] [PubMed] [Google Scholar]
- 31.Korzus E, Torchia J, Rose D W, Xu L, Kurokawa R, McInerney E M, Mullen T-M, Glass C K, Rosenfeld M G. Transcription factor-specific requirements for coactivators and their acetyltransferase functions. Science. 1998;279:703–707. doi: 10.1126/science.279.5351.703. [DOI] [PubMed] [Google Scholar]
- 32.Kwok R P S, Lundland J R, Chrivia J C, Richards J P, Bachinger H P, Brennan R G, Roberts S G E, Green M R, Goodman R H. Nuclear protein CBP is a coactivator for the transcription factor CREB. Nature. 1994;370:223–226. doi: 10.1038/370223a0. [DOI] [PubMed] [Google Scholar]
- 33.Li H, Gomes P J, Chen J D. RAC3, a steroid/nuclear receptor-associated coactivator that is related to SRC-1 and TIF2. Proc Natl Acad Sci USA. 1997;94:8479–8484. doi: 10.1073/pnas.94.16.8479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lindebro M C, Poellinger L, Whitelaw M L. Protein-protein interaction via PAS domains: role of the PAS domain in positive and negative regulation of the bHLH/PAS dioxin receptor-Arnt transcription factor complex. EMBO J. 1995;14:3528–3539. doi: 10.1002/j.1460-2075.1995.tb07359.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Littlewood T D, Evan G I. Transcription factors 2: helix-loop-helix. Protein Profile. 1995;2:621–702. [PubMed] [Google Scholar]
- 36.Lundblad J R, Kwok R P, Laurance M E, Harter M L, Goodman R H. Adenoviral E1A-associated protein p300 as a functional homologue of the transcriptional co-activator CBP. Nature. 1995;374:85–88. doi: 10.1038/374085a0. [DOI] [PubMed] [Google Scholar]
- 37.Martinez-Balbas M A, Bannister A J, Martin K, Haus-Seuffert P, Meisterernst M, Kouzarides T. The acetyltransferase activity of CBP stimulates transcription. EMBO J. 1998;17:2886–2893. doi: 10.1093/emboj/17.10.2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McInerney E M, Rose D W, Flynn S E, Westin S, Mullen T-M, Krones A, Inostroza J, Torchia J, Nolte R T, Assa-Munt N, Milburn M V, Glass C K, Rosenfeld M G. Determinants of coactivator LXXLL motif specificity in nuclear receptor transcriptional activation. Genes Dev. 1998;12:3357–3368. doi: 10.1101/gad.12.21.3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Na S Y, Lee S K, Han S J, Choi H S, Im S Y, Lee J W. Steroid receptor coactivator-1 interacts with the p50 subunit and activates nuclear factor kappaB-mediated transactivations. J Biol Chem. 1998;273:10831–10834. doi: 10.1074/jbc.273.18.10831. [DOI] [PubMed] [Google Scholar]
- 40.Nordeen S K. Luciferase reporter gene vectors for analysis of promoters and enhancers. BioTechniques. 1988;6:454–457. [PubMed] [Google Scholar]
- 41.Ogryzko V V, Schiltz R L, Russanova V, Howard B H, Nakatani Y. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell. 1996;87:953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- 42.Oñate S A, Tsai S Y, Tsai M J, O'Malley B W. Sequence and characterization of a coactivator for the steroid hormone receptor superfamily. Science. 1995;270:1354–1357. doi: 10.1126/science.270.5240.1354. [DOI] [PubMed] [Google Scholar]
- 43.Pugh C W, O'Rourke J F, Nagao M, Gleadle J M, Ratcliffe P J. Activation of hypoxia-inducible factor-1; definition of regulatory domains within the α subunit. J Biol Chem. 1997;272:11205–11214. doi: 10.1074/jbc.272.17.11205. [DOI] [PubMed] [Google Scholar]
- 44.Salceda S, Caro J. Hypoxia-inducible factor 1α (HIF-1α) protein is rapidly degraded by the ubiquitin-proteasome system under normoxic conditions. Its stabilization by hypoxia depends on redox-induced changes. J Biol Chem. 1997;272:22642–22647. doi: 10.1074/jbc.272.36.22642. [DOI] [PubMed] [Google Scholar]
- 45.Smith C L, Oñate S A, Tsai M-J, O'Malley B W. CREB binding protein acts synergistically with steroid receptor coactivator-1 to enhance steroid receptor-dependent transcription. Proc Natl Acad Sci USA. 1996;93:8884–8888. doi: 10.1073/pnas.93.17.8884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sogawa K, Nakano R, Kobayashi A, Kikuchi Y, Ohe N, Matsushita N, Fujii-Kuriyama Y. Possible function of Ah receptor nuclear translocator (Arnt) homodimer in transcriptional regulation. Proc Natl Acad Sci USA. 1995;92:1936–1940. doi: 10.1073/pnas.92.6.1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Spencer T E, Jenster G, Burcin M M, Allis C D, Zhou J X, Mizzen C A, McKenna N J, Oñate S A, Tsai S Y, Tsai M J, O'Malley B W. Steroid receptor coactivator-1 is a histone acetyltransferase. Nature. 1997;389:194–198. doi: 10.1038/38304. [DOI] [PubMed] [Google Scholar]
- 48.Takeshita A, Cardona G R, Koibuchi N, Suen C-S, Chin W W. TRAM-1, a novel 160-kDa thyroid hormone receptor activator molecule, exhibits distinct properties from steroid receptor coactivator-1. J Biol Chem. 1997;272:27629–27634. doi: 10.1074/jbc.272.44.27629. [DOI] [PubMed] [Google Scholar]
- 49.Torchia J, Rose D W, Inostroza J, Kamei Y, Westin S, Glass C K, Rosenfeld M G. The transcriptional co-activator p/CIP binds CBP and mediates nuclear-receptor function. Nature. 1997;387:677–684. doi: 10.1038/42652. [DOI] [PubMed] [Google Scholar]
- 50.Torchia J, Glass C, Rosenfeld M G. Co-activators and co-repressors in the integration of the transcriptional responses. Curr Opin Cell Biol. 1998;10:373–383. doi: 10.1016/s0955-0674(98)80014-8. [DOI] [PubMed] [Google Scholar]
- 51.Umesono K, Murakami K K, Thompson C C, Evans R M. Direct repeats as selective response elements for the thyroid hormone, retinoic acid and vitamin D3 receptors. Cell. 1991;65:1255–1266. doi: 10.1016/0092-8674(91)90020-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Voegel J J, Heine M J S, Zechel C, Chambon P, Gronemeyer H. TIF2, a 160 kDa transcriptional mediator for the ligand-dependent activation function AF-2 of nuclear receptors. EMBO J. 1996;15:3667–3675. [PMC free article] [PubMed] [Google Scholar]
- 53.Voegel J J, Heine M J S, Tini M, Vivat V, Chambon P, Gronemeyer H. The coactivator TIF2 contains three nuclear receptor-binding motifs and mediates transactivation through CBP binding-dependent and -independent pathways. EMBO J. 1998;17:507–519. doi: 10.1093/emboj/17.2.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang G L, Jiang B-H, Rue E A, Semenza G L. Hypoxia-inducible factor 1 is a basic-helix-loop-helix-PAS heterodimer regulated by cellular O2 tension. Proc Natl Acad Sci USA. 1995;92:5510–5514. doi: 10.1073/pnas.92.12.5510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wenger R H, Gassmann M. Oxygen(es) and the hypoxia-inducible factor-1. Biol Chem. 1997;378:609–616. [PubMed] [Google Scholar]
- 56.Westin S, Kurokawa R, Nolte R T, Wisely G B, McInerney E M, Rose D W, Milburn M V, Rosenfeld M G, Glass C K. Interactions controlling the assembly of nuclear-receptor heterodimers and co-activators. Nature. 1998;395:199–202. doi: 10.1038/26040. [DOI] [PubMed] [Google Scholar]
- 57.Xanthoudakis S, Miao G, Wang F, Pan Y C, Curran T. Redox activation of Fos-Jun DNA binding activity is mediated by a DNA repair enzyme. EMBO J. 1992;11:3323–3335. doi: 10.1002/j.1460-2075.1992.tb05411.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xanthoudakis S, Miao G G, Curran T. The redox and DNA-repair activities of Ref-1 are encoded by nonoverlapping domains. Proc Natl Acad Sci USA. 1994;91:23–27. doi: 10.1073/pnas.91.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yao K-S, Xanthoudakis S, Curran T, O'Dwyer P J. Activation of AP-1 and of a nuclear redox factor, Ref-1, in the response of HT29 colon cancer cells to hypoxia. Mol Cell Biol. 1994;14:5997–6003. doi: 10.1128/mcb.14.9.5997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yao T-P, Ku G, Zhou N, Scully R, Livingston D M. The nuclear hormone receptor coactivator SRC-1 is a specific target of p300. Proc Natl Acad Sci USA. 1996;93:10626–10631. doi: 10.1073/pnas.93.20.10626. [DOI] [PMC free article] [PubMed] [Google Scholar]