Abstract
Background
Synovial inflammation is associated with pain severity in patients with knee osteoarthritis (OA). The aim here was to determine in a population with knee OA, whether synovial tissue from areas associated with pain exhibited different synovial fibroblast subsets, compared to synovial tissue from sites not associated with pain. A further aim was to compare differences between early and end-stage disease synovial fibroblast subsets.
Methods
Patients with early knee OA (n = 29) and end-stage knee OA (n = 22) were recruited. Patient reported pain was recorded by questionnaire and using an anatomical knee pain map. Proton density fat suppressed MRI axial and sagittal sequences were analysed and scored for synovitis. Synovial tissue was obtained from the medial and lateral parapatellar and suprapatellar sites. Fibroblast single cell RNA sequencing was performed using Chromium 10X and analysed using Seurat. Transcriptomes were functionally characterised using Ingenuity Pathway Analysis and the effect of fibroblast secretome on neuronal growth assessed using rat DRGN.
Findings
Parapatellar synovitis was significantly associated with the pattern of patient-reported pain in knee OA patients. Synovial tissue from sites of patient-reported pain exhibited a differential transcriptomic phenotype, with distinct synovial fibroblast subsets in early OA and end-stage OA. Functional pathway analysis revealed that synovial tissue and fibroblast subsets from painful sites promoted fibrosis, inflammation and the growth and activity of neurons. The secretome of fibroblasts from early OA painful sites induced greater survival and neurite outgrowth in dissociated adult rodent dorsal root ganglion neurons.
Interpretation
Sites of patient-reported pain in knee OA exhibit a different synovial tissue phenotype and distinct synovial fibroblast subsets. Further interrogation of these fibroblast pathotypes will increase our understanding of the role of synovitis in OA joint pain and provide a rationale for the therapeutic targeting of fibroblast subsets to alleviate pain in patients.
Funding
This study was funded by Versus Arthritis, UK (21530; 21812)
Keywords: scRNAseq, Osteoarthritis, Synovial fibroblasts, Obesity, Inflammation
Research in context.
Evidence before this study
Despite the significant impact on quality of life, effective alleviation of joint pain in OA remains elusive and generic analgesics provide minimal relief and are associated with significant side effects. Understanding the molecular basis for OA joint pain is important in seeking to develop targeted and more efficacious analgesic therapies.
Added value of this study
Our study is the first to report a differential transcriptome and distinct synovial fibroblast subsets in parapatellar synovial issue from sites of patient-reported pain in knee OA, with distinct differences in the predominance of clusters between OA disease stage and the presence of pain.
Implications of all the available evidence
Our study has implications for understanding the role of the synovial membrane in OA pain and disease progression. The observation that painful synovial sites in early OA patients exhibit a distinct subset of synovial fibroblasts with a gene signature and secretome that promotes the formation and development of neurites provides a rationale for the therapeutic targeting of fibroblast subsets to alleviate pain in OA patients.
Alt-text: Unlabelled box
1. Introduction
Joint pain in osteoarthritis (OA) is a leading cause of disability and shortening of adult-working life [1]. Despite the significant impact on quality of life, effective alleviation of joint pain remains elusive since generic analgesics provide minimal relief and are associated with significant side effects when taken chronically [2]. Understanding the molecular basis for OA joint pain is important in seeking to develop targeted and more efficacious analgesic therapies.
Inflammation of the synovial membrane, termed synovitis, is strongly associated with pain severity in patients with knee OA [3]. Synovitis presents early in the disease course, when radiographic signs of cartilage damage is minimal, and is typified by hyperplasia of synovial fibroblasts, which are a major source of the increased pro-inflammatory cytokines present within the OA synovial joint [4,5], and increased infiltration of activated immune cells [6,7]. OA pain is generated via nociceptive peripheral input from the tissues of the OA joint, including the synovium. These afferent nerve fibres and nociceptors can be activated and sensitised by pro-inflammatory cytokines present in the OA synovial fluid including TNF-α, IL-1β, CCL2 (MCP1) and IL-6 [8], the concentrations of which correlate with patient-reported pain [9]. Indeed, compared to healthy synovial fluid, synovial fluid from patients with knee OA increases the excitability of sensory neurones [10].
Notably, it was recently reported that pain in knee OA patients was associated with distinct patterns of synovitis, particularly in regions around the patellar [11]. Furthermore, single-cell RNA sequencing (scRNA-seq) has identified the existence of different subset populations of synovial fibroblasts in patients with rheumatoid arthritis (RA) [12], which differentially drive inflammation [13]. Therefore, the aim of this study was firstly to determine in a population with either early or end-stage knee OA the relationship between patterns of patient-reported knee pain, pain severity and the anatomical site and degree of synovitis. Secondly, to determine whether sites of patient-reported pain exhibit synovial tissue with a different transcriptomic phenotype containing different cellular subsets of synovial fibroblasts, compared to sites of no pain.
2. Methods
2.1. Patient recruitment, pain scoring and tissue collection
Following ethical approval (NRES 17/SC/0456), 51 patients were recruited who either had early knee OA (n = 29) or end-stage knee OA (n = 22) and were scheduled for elective knee arthroscopy or arthroplasty at either The Royal Orthopaedic Hospital NHS Foundation Trust (Birmingham, UK) or Russell's Hall Hospital, Dudley, UK (Table 1). Prior to surgery, patients completed EQ-5D and Oxford Knee Score (OKS) questionnaires, a visual analog scale (VAS) to record pain severity and a patient-reported knee pain map [11], to record anatomical sites of most and least pain. Intra-operatively, synovial fluid was aspirated and synovial tissue collected from four anatomical sites, namely the inferior and superior patellar sites from both lateral and medial sides of the joint.
Table 1.
Anthropometric data and patient reported outcome measures.
| All Patients | Early OA | End-stage OA | p-value | ||
|---|---|---|---|---|---|
| Agea | 61 ± 1.8 | 54 ± 2.2 | 69 ± 2.0 | < 0.001 | |
| Gender (male:female) | 25:26 | 12:17 | 13:9 | ||
| Height (cm)a | 169 ± 1.5 | 169 ± 2.0 | 169 ± 2.3 | 0.92 | |
| Weight (kg)a | 92 ± 2.8 | 96 ± 4.0 | 85 ± 3.5 | < 0.05 | |
| BMIa | 31.9 ± 0.8 | 33.6 ± 1.2 | 29.6 ± 0.9 | < 0.05 | |
| Waist circumference (cm)a | 102 ± 2 | 102 ± 3 | 102 ± 3 | 0.86 | |
| Hip circumference (cm)a | 113 ± 1.9 | 115 ± 2.8 | 110 ± 2.2 | 0.25 | |
| W:Ha | 0.91 ± 0.01 | 0.89 ± 0.02 | 0.92 ± 0.02 | 0.15 | |
| Oxford Knee Score (0-48)b | Median | 18.5 (13.8–37) | 24 (15.5–29.5) | 16 (11.8–37) | < 0.05 |
| Oxford Knee Score Category c | Severe (0–19) | 53% (27) | 41% (12) | 68% (15) | < 0.05 |
| Moderate (20–29) | 31% (16) | 34% (10) | 27% (6) | < 0.05 | |
| Mild (30–39) | 16% (8) | 24% (7) | 5% (1) | < 0.05 | |
| Normal (40–48) | - | - | - | - | |
| EQ5D summative | 8.6 ± 0.20 | 8.4 ± 0.26 | 8.9 ± 0.32 | 0.43 | |
P-values represents the comparison between early OA and end-stage OA patients.
values represent mean ± SEM for All patients (n = 51), early OA patients (n = 29) and end-stage OA patients (n = 22).
values represent median (interquartile range).
values represent % (number) of patients in each category.
2.2. Synovitis scoring using MRIs
Pre-surgical PDFS (proton density fat suppressed) images were scored for synovitis at 11 different sites, namely the medial parapatellar recess, lateral parapatellar recess, suprapatellar, infrapatellar, intercondylar, adjacent to PCL (posterior cruciate ligament), adjacent to ACL (anterior cruciate ligament), medial perimeniscal, lateral perimeniscal, within Baker's cyst and presence or absence of loose bodies according to the protocol of Guermazi et al [14]. These were performed by a Consultant Musculoskeletal Radiologist and research fellow.
2.3. RNA sequencing analysis
Total RNA was extracted from snap-frozen whole synovium tissue from painful and non-painful samples from n = 6 patients with early OA and n = 6 patients with end-stage OA (Supplementary Table 1) using Trizol (Life Technologies, UK) and DNase treated (Qiagen DNase kit). RNA integrity Number (RIN) values were > 7 (Agilent Bioanalyser). Library preparation and RNA-sequencing was performed by Genomics Facility at University of Birmingham using QuantSeq 3’ kit (Lexogen, Austria) and sequenced on Illumina's NextSeq 500. RNA-sequencing data was imported to the Galaxy web platform (usegalaxy.org) [15] and aligned with HISAT2. Aligned BAM files where imported into Qlucore Omics Explorer v3.6 (Qlucore AB, Lund, Sweden) and differential gene expression analysis performed following TMM (Trimmed Mean of M values) normalisation. Data was filtered to identify differentially expressed genes (DEGs) using two-group statistical comparison for > 1.5 fold change and p < 0.05. Sequence data is available through the GEO database under accession number GSE176223.
2.4. Single-cell RNA sequencing analysis
Synovial tissue from painful and non-painful sites of n = 4 early OA patients and from painful sites of n=4 end-stage OA patients (Supplementary Table 1) was diced and cultured in growth media (RPMI 1640 containing 10% FCS, 1% penicillin/streptomycin, 5% L-glutamine, 5% sodium pyruvate and 5% non-essential amino acids (Sigma Aldrich, Dorset, UK)) for primary synovial fibroblasts outgrowth over 3–7 days as previously described [16]. scRNA-seq (Chromium 10X, Genomics Facility, University of Birmingham) was performed on a total of ∼4247 cells (representing ∼353 cells per patient sample). In brief, individual cells were partitioned into nanolitre-scale Gel Bead-In-Emulsions (GEMs) such that reverse transcription produced specific bar-coded cDNA for each cell. Following cDNA amplification, single cell 3’ cDNA libraries were constructed, pooled and sequenced using the Nextseq 500 (Illumina) platform. Sequencing data was analysed using Seurat v2.3.4 (https://satijalab.org/seurat/). Individual datasets were merged and low quality cells or doublets were filtered by excluding cells that expressed > 7,100 genes/cell and > 15% mitochondrial gene expression. 4180 cells remained post quality control (QC). Data was subjected to global-scale normalisation and log-transformed. Technical noise was accounted for by scaling data based on nUMI and % mitochondrial genes. Highly variable genes were identified and used to perform principal component analysis (PCA). Using the top 15 PCs, t-distributed stochastic neighbour embedding (t-SNE) analysis was performed to cluster the cells, with a resolution of 0.6. Monocle v2.9.0 was used to analyse pseudotime trajectories (http://monocle-bio.sourceforge.net/). Genes were selected based on differentially expressed genes that were expressed by at least 10 cells and with q < 0.01. Using the DDRTree method, the dimensionality of the data was reduced and cells were ordered in pseudotime. Sequence data is available through the GEO database under accession number GSE176308.
2.5. Pathway analysis
Differentially expressed genes from each cluster (> 1.5 fold change, p < 0.05) were analysed using Ingenuity Pathway Analysis (IPA; Qiagen, UK) to determine the significance of the association with a particular pathway/function. Fisher's exact test was used to calculate a p-value of the association between the genes and the pathway or function. The identification of upstream regulators by IPA utilised information derived from the IPA Knowledge Base, which is the largest knowledge base of its kind, including modelled relationships between proteins, genes, complexes, cells, tissues, drugs, pathways, and diseases. The IPA knowledge base includes information from curated literature, internally curated knowledge, and a wide variety of trusted 3rd party sources and databases (including NCBI databases-EntrezGene, RefSeq, OMIM disease associations, targets and pharmacological relevance of FDA-approved and clinical trial drugs, clinical biomarkers, Gene Ontology annotations, a normal gene expression body atlas for over 30 tissues and the NCI-60 panel of cancer cell lines, microRNA-mRNA target databases, GWAS databases, HumanCyc metabolic pathways and reactions. For the prediction of upstream regulators, a p-value and z-score was computed based on the significant overlap between genes in the dataset and known targets regulated by the transcriptional regulator.
2.6. Isolation of adult rat dorsal root ganglion neurons
Primary dorsal root ganglion neurons (DRGN) from 6–8 week old Sprague-Dawley rats (Charles River, UK) were dissociated and cultured in Neurobasal-A supplemented media (NBA) in 8-well chamber slides, as we have previously described [17]. After 24 h, the media was replaced with fresh NBA media containing either conditioned media from fibroblasts from non-painful or painful sites (n = 4 patients), or control medium (diluted 1:4) and cultured for a further 24 h. Cell soma and neurites (n = 4 wells/condition) were immunostained with an anti-βIII tubulin antibody (Sigma-Aldrich), imaged using an Axioplan 2 epi-fluorescent microscope equipped with an AxioCam HRc and controlled through Axiovision Software (Carl Zeiss, UK). Quantification of DRGN survival, the longest DRGN neurite length and % DRGN with neurites was assessed by an investigator masked to the treatment conditions, as described by us previously [17]. Briefly, each 8-well chamber slide was divided into 9 quadrants and random images of each quadrant were used to measure the longest DRGN neurite (at least 144 DRGN/condition) using the measurement function in Axiovision Software, % DRGN was determined in 36 images of each condition by counting DRGN with neurites versus those without neurites whilst DRGN survival was determined in each quadrant (36 images/condition) by counting the number of βIII-tubulin+ DRGN.
2.7. Statistical analysis
Data were analysed using Graphpad Prism v7. Specific tests performed are detailed within the figure legends, with statistical significance determined to be p < 0.05. To avoid bias the investigators were blinded during data analysis. OA patient synovial values of pro-inflammatory cytokines and synovial fibroblast RNA sequencing data from previously published articles were used to calculate sample sizes [4,5,13].
2.8. Role of the funding source
The funder (Versus Arthritis) had no involvement in the study or any role in the writing of the manuscript or the decision to submit it for publication.
2.9. Ethics statement
Ethical approval was granted by the UK National Research Ethics Committee (NRES 17/SC/0456). Consent was provided by all patients.
3. Results
3.1. Patient reported pain and synovitis in patients with early and end-stage knee OA
Based on pre-operative OKS, the majority of patients were characterised as having either moderate or severe OA. However, as expected, end-stage OA patients (median OKS= 16) were characterised as having significantly (p < 0.05) more severe arthritis, compared to early OA patients (median OKS = 24) (Table 1). End-stage OA patients were also on average older, and exhibited lower body weight and BMI than early stage OA patients (Table 1).
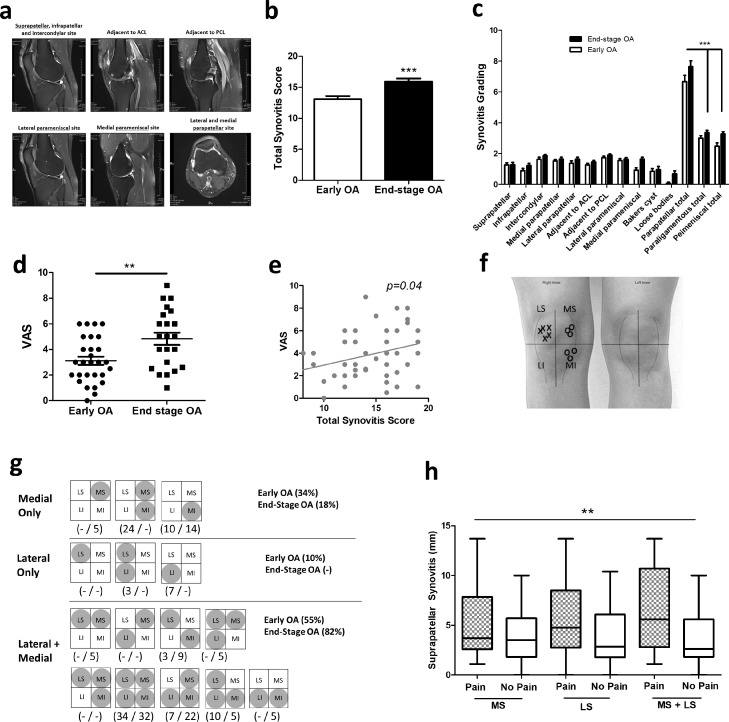
Summing the total synovitis scores from MRI analysis at 11 anatomical locations (Fig. 1a, b), patients with end-stage OA exhibited significantly greater total synovitis (15.91 ± 0.51), compared to early stage OA patients (13.07 ± 0.52). Indeed, 95% of end-stage OA patients were designated as having severe synovitis (score > 13), compared to only 48% of early OA patients (Table 2). Comparing synovitis across the different anatomical sites revealed that in both early and end-stage OA patients the majority of synovitis was in the parapatellar region, with significantly greater synovitis in parapatellar sites (p < 0.001), compared to either paraligamentous or parameniscal sites (Fig. 1c). Furthermore, total parapatellar synovitis was significantly greater (p < 0.05) in end-stage OA patients (7.64 ± 0.39), compared to early OA patients (6.67 ± 0.42) (Table 2).
Fig. 1.
The relationship between synovitis and patient-reported knee pain in early knee OA and end-stage knee OA
(a) Representative proton-density MRI scans of OA patient knee joint (axial, coronal and sagittal scans), showing effusive synovitis (white/greying regions) across different anatomical sites. (b) Total synovitis score in early OA patients (n = 29) and end-stage OA patients (n = 22). ***= p < 0.001 (unpaired t-test), significantly different between early and end-stage OA. (c) Synovitis grading at each of 11 anatomical sites in early OA patients (n = 29) and end-stage OA patients (n = 22). ***= p < 0.001 (1-way ANOVA with Tukey's multiple comparison post-hoc test), significantly different between parapatellar compared to either paraligamentous or parameniscal. (d) Patient reported pain severity by VAS in early OA and end-stage OA patients. **= p < 0.01 (Mann Whitney test), significantly different between early and end-stage OA. (e) Correlation between OA patient reported pain severity (VAS) and total synovitis score. (f) Representative example of a completed patient-reported knee pain map. Crosses (x) represent sites of patient-reported pain, circles (o) represent sites of patient-reported no pain. LS and MS refer to lateral and medial supraparapatellar respectively. LI and MI refer to lateral and medial infraparapatellar respectively. (g) Different patterns of knee pain reported in early OA (n = 29 patients) and end-stage OA patients (n = 22). Greyed circles within either LS, MS, LI or MI represent sites of patient reported pain. (h) Supraparapatellar synovitis (mm) in patients who report pain in suprapatellar compartments (medial vs lateral vs medial+lateral), compared to those who report no pain. **=p < 0.01 (2-way ANOVA), significantly different between pain and no pain.
Table 2.
MRI synovitis grades in early and end-stage knee OA patients.
| All patients | Early OA | End-stage OA | |
|---|---|---|---|
| Supra parapatellar | 1.27 ± 0.11 | 1.26 ± 0.15 | 1.27 ± 0.16 |
| Infra parapatellar | 1.04 ± 0.11 | 0.89 ± 0.15 | 1.23 ± 0.15 |
| Intercondylar | 1.74 ± 0.08 | 1.63 ± 0.13 | 1.86 ± 0.07 |
| Medial parapatellar | 1.57 ± 0.08 | 1.52 ± 0.10 | 1.64 ± 0.12 |
| Lateral parapatellar | 1.49 ± 0.10 | 1.37 ± 0.15 | 1.64 ± 0.12 |
| Adjacent to ACL | 1.35 ± 0.08 | 1.26 ± 0.11 | 1.46 ± 0.11 |
| Adjacent to PCL | 1.82 ± 0.06 | 1.74 ± 0.10 | 1.91 ± 0.06 |
| Lateral parameniscal | 1.59 ± 0.08 | 1.56 ± 0.11 | 1.64 ± 0.11 |
| Medial parameniscal | 1.25 ± 0.11 | 0.93 ± 0.15 | 1.65 ± 0.12 |
| Bakers cysts | 0.89 ± 0.14 | 0.85 ± 0.18 | 0.95 ± 0.21 |
| Loose bodies | 0.35 ± 0.11 | 0.07 ± 0.07 | 0.68 ± 0.20 |
| Parapatellar total | 7.10 ± 0.29 ‡ | 6.67 ± 0.42 ‡ | 7.64 ± 0.39 ‡* |
| Paraligamentous total | 3.16 ± 0.11 | 3.00 ± 0.16 | 3.36 ± 0.14 |
| Parameniscal total | 2.84 ± 0.15 | 2.48 ± 0.22 | 3.27 ± 0.15 |
| Total | 14.35 ± 0.42 | 13.07 ± 0.52 | 15.91 ± 0.51 ψ |
| % Normal (0–4) | - | - | - |
| % Mild (5–8) | 2% | 4% | - |
| % Moderate (9–11) | 29% | 48% | 5% |
| % Severe (> 13) | 69% | 48% | 95% |
Values represent synovitis grade (0, 1 or 2) mean ± SEM for All patients (n = 51), early OA patients (n = 29) and end-stage OA patients (n = 22).
= p < 0.001, significantly different from paraligamentous total and parameniscal total.
= p < 0.01, significantly different between early and end-stage OA.
= p < 0.001, significantly different between early and end-stage OA.
Examining the relationship between synovitis and patient reported knee pain we found that end-stage OA patients reported significantly (p < 0.01) greater pain severity (VAS = 4.8 ± 0.49) than patients with early OA (VAS = 3.1 ± 0.32) (Fig. 1d). Furthermore, VAS was significantly correlated (p < 0.05) to total synovitis scores (Fig. 1e). However, assessment of the patient-reported anatomical knee pain maps showed considerable heterogeneity in the pattern of patient reported pain in both early OA and end-stage OA patients (Fig. 1f, g).
Notably, the majority of either early OA or end-stage OA patients (70%) reported pain that was localised with at least 1 compartment (i.e. medial or lateral supra or infraparapatellar) being pain-free (Fig. 1g). In early OA, 43% of patients report pain that is either localised to the medial or lateral side of the joint, in contrast to only 18% of end-stage OA patients. Indeed, a higher proportion of end-stage OA patients (82% vs 55%) reported pain involving both lateral and medial compartments (Fig. 1g). Examining the relationship between patient-reported pain maps and synovitis we found a highly significant relationship between synovitis and the pattern of patient-reported knee pain (p < 0.01). In particular, supraparapatellar synovitis was found to be significantly greater in patients who reported pain that involved supraparapatellar sites (either medial or lateral), compared to patients who reported no pain in supraparapatellar compartments (Fig. 1h).
3.2. RNA sequencing analysis of synovium from sites of patient-reported pain and no pain in early and end-stage OA
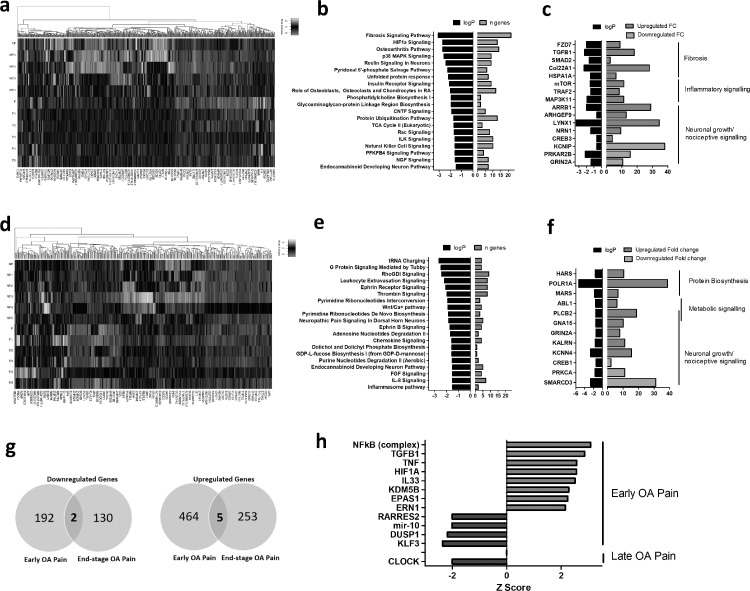
Having found that synovitis was predominantly located at parapatellar sites and that the pattern of knee pain is related to synovitis we next examined the transcriptome of parapatellar synovial tissue from patient-matched painful and non-painful sites from early OA patients (n = 6) or end-stage OA patients (n = 6) by RNAseq. Comparing pain-associated with no-pain synovial tissue in early OA patients, a total of 667 genes were differentially expressed (> 1.5 fold change, p < 0.05) of which 469 were upregulated and 198 were downregulated genes in pain-associated synovium. This included 567 protein coding genes, 68 long non-coding RNAs, 29 pseudogenes, 1 vault RNA and 2 miscRNAs (Fig. 2a; Supplementary Tables 2 & 3). Functional pathway analysis (ingenuity Pathway Analysis) of these differentially expressed genes revealed that the most significant canonical pathways were “Fibrosis Signalling”, “HIF1α Signalling”, “Osteoarthritis Pathway” and “p38 MAPK Signalling”. Furthermore, amongst the top 20 most significant canonical pathways were several known pain-mediator pathways, namely “Reelin Signalling in Neurones”, “Endocannabinoid Developing Neuron Pathway” and “NGF Signalling” (Fig. 2b). In line with the pathway analysis, the most differentially expressed genes included genes that mediate fibrosis, inflammatory signalling and neuronal growth/nociceptive signalling (Fig. 2c).
Fig. 2.
RNA-sequencing of synovial tissue from painful and non-painful sites in early OA and end-stage knee OA patients
(a) Heatmap of differentially expressed genes (FC > 1.5) in synovial tissue from painful sites, compared to non-painful sites in patients with early OA (n = 6). (b) Most significant canonical signalling pathways associated with the differential transcriptome of early OA painful synovial tissue. (c) LogP and fold-change values of upregulated and downregulated genes in early OA painful synovial tissue grouped into functional categories. (d) Heatmap of differentially expressed genes (FC> 1.5, p < 0.05) in synovial tissue from painful sites, compared to non-painful sites in patients with end-stage OA (n = 6). (e) Most significant canonical signalling pathways associated with the differential transcriptome of end-stage OA painful synovial tissue, as identified using IPA software. (f) LogP and fold-change values of upregulated and downregulated genes in end-stage OA painful synovial tissue grouped into functional categories. (g) Venn diagram showing the overlap and total numbers of significantly upregulated (> 1.5 FC) and down-regulated (< 1.5 FC) in painful synovial tissue in early OA and end-stage OA. (h) Z-scores of the identified upstream regulators of the transcriptome of painful synovial tissue in early OA and in end-stage OA. Positive z-scores (red bars) represent a predicted “activated” upstream regulator, negative z-scores (blue bars) represent a predicted “inhibited” upstream regulator.
Comparing pain-associated with no-pain synovial tissue in end-stage OA patients, a total of 390 genes were differentially expressed (> 1.5 fold change, p < 0.05) of which 258 were upregulated and 132 were downregulated genes in pain-associated synovium. This included 331 protein coding genes, 38 long non-coding RNAs, 18 pseudogenes, and 3 miscRNAs (Fig. 2d; Supplementary Tables 4 & 5). Functional pathway analysis of these differentially expressed genes revealed that the most significant canonical pathways were “tRNA Charging”, “G-protein Signalling Mediated by Tubby”, “RhoGDI Signalling” and “Leukocyte Extravasation Signalling”. Furthermore, similarly to the early OA pain synovium, amongst the top 20 most significant canonical pathways were several known pain-mediator pathways, namely “Neuropathic Pain in Dorsal Root Horn Neurons”, “Endocannabinoid Developing Neuron Pathway”, “Ephrin Receptor Signalling and “Ephrin B Signalling” (Fig. 2e). In line with the pathway analysis, the most differentially expressed genes included genes that mediate protein biosynthesis, metabolic signalling and neuronal growth/nociceptive signalling (Fig. 2f). Of interest, only a small number of genes were differentially expressed in both early OA pain and end-stage OA (Fig. 2g), which could indicate that different pathways mediate pain in early stage disease compared to late stage disease. To further investigate these findings we then performed an upstream analysis to identify any activated or inhibited upstream regulators of the pain-associated transcriptomes. In early OA pain, NFκB, TGFB, TNF, HIF1A, IL-33, KDM5B, EPSA1 and ERN were identified as activated upstream regulators, whilst RARRES2, miR10, DUSP1 and KLF3 were identified as inhibited upstream regulators. In end-stage OA pain, CLOCK was identified as an inhibited upstream regulator (Fig. 2h). To evaluate whether these differences could be due to differences in anatomical site rather than the patient-reported painful/non-painful designation, we re-analysed the RNA sequencing data to compare synovial tissue from medial, lateral, infra or supra parapatellar sites of the same patient-reported pain designation i.e either all painful or all non-painful. Although a number of genes were found to be differentially expressed in synovium from different anatomical sites, the the pathway analysis did not reveal that these genes mediated fibrotic, inflammatory or neuronal growth/nociceptive signalling pathways (Supplementary Fig. 1).
3.3. Single-cell RNA sequencing identifies differences in synovial fibroblast subsets between early and end-stage knee OA and between painful and non-painful synovial sites
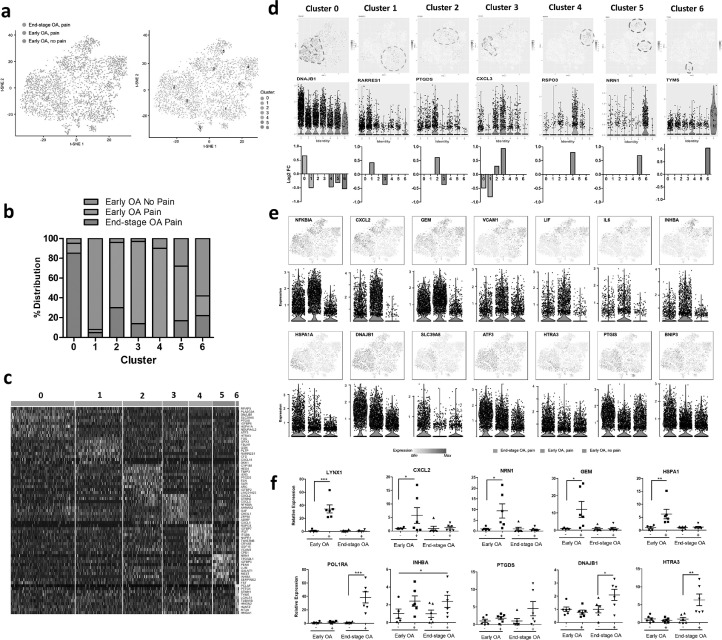
Having found differences in the transcriptome of synovial tissue from patient-reported pain sites we next examined whether synovial fibroblasts from pain-associated synovial tissue exhibited different cellular subsets. To this end, synovial fibroblasts from early OA pain and non-pain sites (n = 4), and from end-stage OA pain sites (n = 4) were subjected to scRNA-seq.
PCA and t-SNE analysis revealed seven distinct subsets of synovial fibroblasts (clusters 0–6) with clear differences between early OA and end-stage disease, as well as between painful and non-painful sites in early OA disease (Fig. 3a). Specifically, synovial fibroblasts isolated from end-stage OA patients were predominantly observed in cluster 0. In contrast, in early OA, fibroblasts from non-painful regions were predominantly observed in cluster 1 and cluster 6, whereas those fibroblasts isolated from painful regions were predominant in clusters 2–5 (Fig. 3b). The cells in each cluster showed distinct gene expression patterns, as visualised by the expression heatmap (Fig. 3c; Supplementary Table 6). t-SNE and violin plots featuring gene expression were then used to visualise gene expression signatures within each specific cluster 0–6 (Supplementary Figs. 2–8) and within patient cohorts. For example, expression of DNAJB1 was significantly associated with Cluster 0, a cluster predominant in fibroblasts from painful sites in end-stage. Expression of RARRES1 and TYPMS were significantly associated with Cluster 1 and Cluster 6 respectively, clusters predominant in fibroblasts from non-painful sites in early OA. Expression of PTGDS (Cluster 2), CXCL3 (Cluster 3), RSPO3 (Cluster 4), and NRN1 (Cluster 5) were all significantly associated with specific fibroblasts clusters predominantly found in early OA painful synovial sites (Fig. 3d). Furthermore, analysis of genes associated with particular patient cohorts, rather than any one specific cluster, revealed that fibroblasts from early OA painful synovial sites exhibited high gene expression of NFKBIA, CXCL2, GEM, VCAM1, LIF, IL-6 and INHBA, whilst fibroblasts from end-stage OA painful synovial sites were characterised by high expression of HSPA1A, DNAJB1, SLC39A8, HTRA3, ATF3, PTGIS and BNIP3 (Fig. 3e; Supplementary Tables 7 & 8). Examining the bulk expression of these genes in early OA and end-stage OA synovial tissue from patient-reported painful and non-painful joint sites we found that CXCL2, GEM, NRN1 and HSPA1A were significantly upregulated in early OA painful synovium, whilst INHBA was significantly increased in painful synovium irrespective of disease stage. In addition, RSPO3 exhibited on average greater expression in early OA painful synovium, whereas CXCL3, PTGDS, DNAJB1 and HTRA were on average more highly expressed in end-stage OA synovium (Fig. 3f).
Fig. 3.
Single cell RNA-sequencing identifies distinct synovial fibroblasts subsets in the synovium from painful and non-painful sites in early OA and in end stage OA patients. (a) T-distributed stochastic neighbour embedding (t-SNE) plots of scRNAseq of synovial fibroblasts showing the separation between fibroblasts from painful and non-painful synovial sites in early OA patients, and between early and end-stage OA painful sites, with identification of 7 fibroblast subsets based on transcript profile. In total, scRNAseq was performed on synovial fibroblasts from painful and non-painful synovial patient-matched sites of n=4 early OA patients, and on synovial fibroblasts from painful sites of n = 4 end-stage OA patients. (b) Percentage distribution of the 7 different subsets according to either early OA pain, early OA no pain or end-stage OA pain. (c) Heatmap showing the z-score average gene signature expression of the top 10 most differentially expressed genes within each of the 7 synovial fibroblast clusters. (d) FeaturePlots displaying expression of subset specific markers for each of the 7 subsets (clusters 0-6) on the t-SNE map with violin plots below showing the expression levels of these markers (y-axis) across the different subsets (x-axis). (e) FeaturePlots displaying expression of sample specific markers (identified by performing differential expression analysis on the non-parametric Wilcoxon rank sum test) on the t-SNE map with violin plots below showing the expression levels of these markers (y-axis) across the different sample cohorts, early OA pain, early OA no-pain or end-stage OA pain (x-axis). (f) Bulk expression of specific synovial fibroblast genes in OA synovial tissue from either early OA patient matched painful (+) and non-painful (-) sites (n = 6) or end-stage OA patient-matched painful (+) and non-painful (-) sites (n = 6). *= p < 0.05, **= p < 0.01, ***= p < 0.001 significantly different between painful and non-painful synovial sites as determined by ANOVA with Tukey post-hoc test.
3.4. Functional characterisation of synovial fibroblast clusters
To investigate the functional characteristics of these newly identified fibroblast populations we performed pathway analysis of the differentially expressed gene signatures for each fibroblast cluster (> 1.5 fold change, p < 0.05) to identify significantly associated canonical pathways, cellular functions and upstream regulators (Fig. 4a–c).
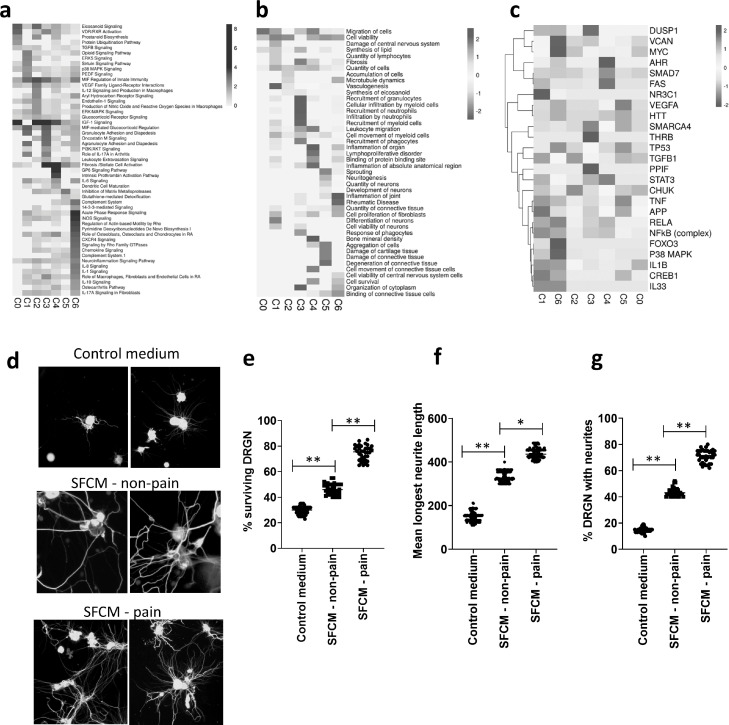
Fig. 4.
Functional characteristics of OA synovial fibroblasts clusters.
(a) Heatmap of canonical signalling pathways significantly associated with the transcriptome of each of the 7 synovial fibroblast subsets (C0-C6) as identified using IPA software, and coloured by logp value. (b) Heatmap of significant cellular functions for each of the 7 synovial fibroblast subsets (C0-C6) as identified using IPA software, and coloured by activation z-score with a positive z-score representing activation and a negative z-score representing inhibition. (c) Heatmap of identified upstream regulators for each of the 7 synovial fibroblast subsets (C0-C6) as identified using IPA software, and coloured by activation z-score with a positive z-score representing activation and a negative z-score representing inhibition. (d) Immunofluorescent images of rat DRGN stained with anti-βIII tubulin antibody (Sigma Aldrich; green) and after 4’,6-diamidino-2-phenylindole (DAPI; blue) nuclear stain after 24 h treatment with either fibroblast culture media diluted 1:4 in NBA-supplemented media (Control medium) or with synovial fibroblast conditioned media diluted 1:4 in NBA-supplemented media (SFCM) from either non-painful SFCM – non-pain) or painful (SFCM – pain). Stained DRGN (n = 4 per treatment group) were imaged using Axiovision Software (Carl Zeiss). (e) Quantification of the % surviving βIII-tubulin+ DRGN (36 images/condition) in control and SFCM – non pain and SFCM - pain treated DRGN 24 h after treatment. Quantification of neurite outgrowth in terms of (f) the longest DRGN neurite length (n = 144 DRGN/condition) and (g) % DRGN with neurites (36 images/condition) in control and SFCM – non-pain and SFCM – pain treated DRGN after 24 h treatment. *** = p < 0.001 and * = p < 0.05, one-way ANOVA with Dunnett's post hoc test.
Notably, the gene expression profile of fibroblasts in cluster 0 (predominantly representing end-stage OA pain) was significantly associated with “Eicosanoid Signalling” and “Prostanoid Biosynthesis”, both of which are purported to be contributors of OA pain, as well as “IGF-1 signalling”. The most significant cellular functions were “migration of cells” and “cell viability”, indicative of an activated fibroblast phenotype in synovial tissue from painful sites in end-stage OA.
Similar to the end-stage OA pain fibroblast cluster, the gene signatures of fibroblast subsets predominantly found in early OA painful synovial sites (Clusters 2–5) were also significantly associated with the canonical pathway “IGF-1 Signalling”. In addition, Cluster 2 was significantly associated with the canonical signalling pathways “MIF-regulation of immunity” and “eicosanoid signalling”, and characterised by the cellular functions “quantity of cells”, “accumulation of cells”, and “microtubule dynamics”. The gene signature of fibroblasts in cluster 3 was associated with the pathways “MIF-mediated glucocorticoid regulation”, “Agranulocyte Adhesion and Diapedesis”, “Granulocyte Adhesion and Diapedesis” and “Oncostatin M signalling”, and characterised by mediating innate immune cell recruitment, including the “recruitment of granulocytes”, “cellular infiltration of myeloid cells”, “cellular infiltration of leukocytes and ”recruitment of phagocytes”. Cluster 4 fibroblasts were associated with the canonical signalling pathways of “Fibrosis Signalling”, “GP6 signalling pathway and “Intrinsic Prothrombin Activation Pathway”, indicating a role in the collagen-induced activation and aggregation of platelets. The cellular functions most associated with Cluster 4 fibroblasts were “Inflammation of Organ” and “Inflammation of Anatomical Region”. Finally, Cluster 5 fibroblasts were significantly associated with activation of cellular functions involved in the development of neurons, including “neuritogenesis”, “sprouting”, “quantity of neurones” and “development of neurones”. Further, the most significantly associated canonical pathway was “inhibition of matrix metalloproteases”, a pathway that is known to mediate neural plasticity via the modification of the extracellular matrix.
Differentiating the fibroblast subsets that were predominantly found in early OA non-painful synovial sites (Clusters 1 and 6), it was found that the gene signature of Cluster 1 fibroblasts was associated with the canonical pathway “Role of IL-17A in RA” and the inhibition of the cellular functions of “Cell Proliferation of Fibroblasts”, “Differentiation of Neurones” and “Cellular Viability of Neurones”. Similarly, the gene signature of Cluster 6 was also associated with the inhibition of neuronal cell viability, with inhibition of the cellular function “Cell viability of CNS cells” and “Cell Survival”, whilst significant canonical pathways also included “Role of IL-17A in RA” and several to cytokine signalling pathways and the “Neuroinflammation Signalling” pathway.
To further interrogate the functional characterisation of these subsets we then performed an upstream analysis to identify any activated or inhibited upstream regulators of the individual clusters (Fig. 4c). Compared to the non-painful Clusters (Clusters 1 and 6), painful subsets showed increased activation of several known inflammatory upstream regulators including TNFα, IL-1β, NFκB, RELA and p38 MAPK. Furthermore, the anti-inflammatory glucocorticoid receptor NR3C1 was identified as an activated upstream regulator of the non-painful Cluster 1 fibroblast subset. In addition, TGFB1 was identified as an activated upstream regulator of the early OA pain Clusters 2 and 5, the mitochondrial enzyme PPIF as an activated regulator of Cluster 3, and the Aryl Hydrocarbon Receptor (AHR), SMAD7 and FAS identified as activated regulators of Cluster 4 (Fig. 4c).
Given that fibroblast clusters found predominantly in early OA painful synovial sites exhibited gene signatures that mediate cellular migration, inflammation and the development of neurons we next performed a cellular cross-talk assay utilising primary adult rat DRGN to examine the effect of the secretome of these cells on neuronal growth. To this end, we subjected DRGN to conditioned medium obtained from cultures of fibroblasts isolated from early OA painful and non-painful synovial sites. Incubation of DRGN with the fibroblast conditioned media from non-painful and painful sites for 24 h resulted in a significant (p < 0.0001) induction of DRGN survival (Fig. 4d and e) and neurite outgrowth (mean longest neurite length and % DRGN with neurites), compared to the DRGN incubated in control medium (Fig. 4d–g). Notably, both DRGN survival and neurite outgrowth (longest neurite length and % DRGN with neurites) were significantly increased in DRGN treated with conditioned medium from painful sites compared to those treated with fibroblast conditioned medium from non-painful sites (Fig. 4d–g). Analysis of the fibroblast secretome using an inflammatory proteomic array (Olink, Sweden) of patient matched conditioned media of fibroblasts from painful and non-painful sites (n = 4 patients) detected the presence of numerous neurotrophic factors including members of the CNTF family- IL-6, LIF and OSM, glial cell line-derived neurotrophic factor (GDNF), and other neurotrophins including VEGF, FGF21, CSF-1, IL-8 and HGF. Comparing differences between the fibroblast conditioned medial from painful and non-painful sites we did not find any individual factor to be significantly different. Instead, numerous factors were slightly elevated in the fibroblast conditioned media from painful sites, suggesting that an overall increase in the inflammatory mileu could be responsible for mediating the greater effect on neuronal growth and survival observed with fibroblasts from painful sites (Supplementary Fig. 9).
3.5. Transcriptional switch in fibroblast phenotype associated with OA progression and development of early OA pain
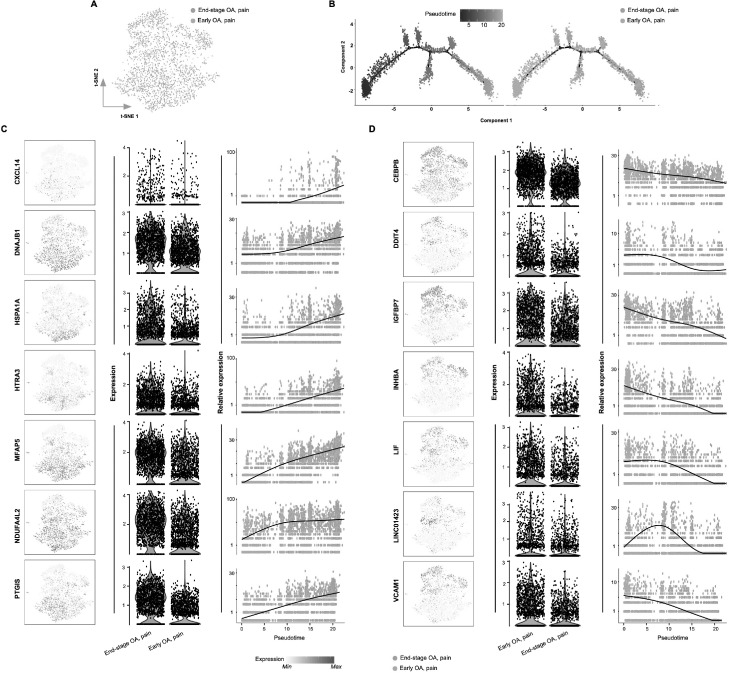
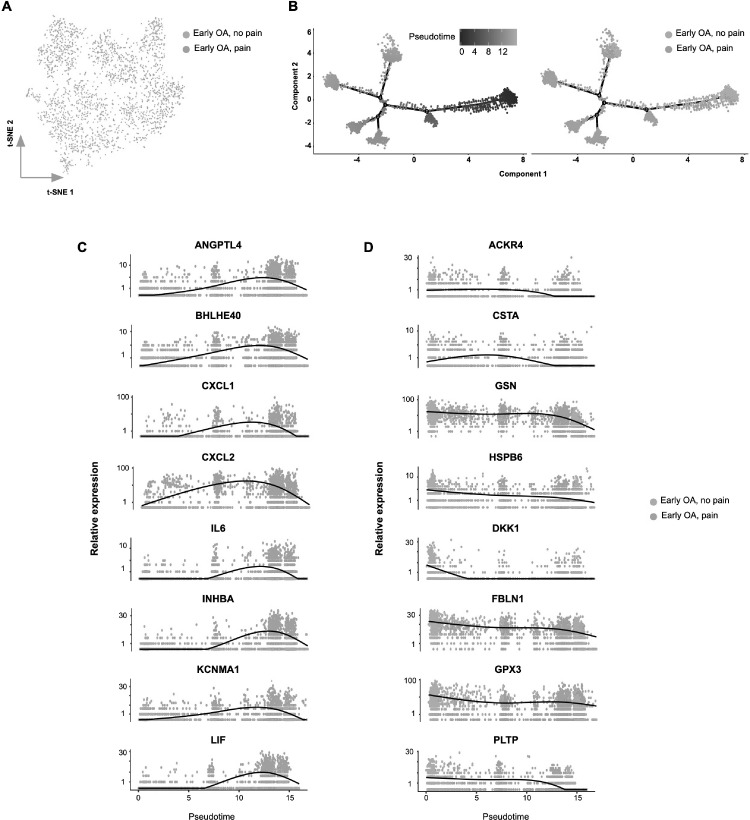
Having identified differences in fibroblast subsets between early OA pain and end-stage OA pain, we next constructed pseudotime trajectories (using Monocle) to investigate transcriptional changes in fibroblasts associated with OA disease progression. By analysing pseudotemporal expression dynamics, we observed a number of genes that were upregulated upon progression towards end-stage OA, namely CXCL14, DNAJB1, HSPA1A, HTRA3, MFAP5, NDUFA4L2 and PTGIS. We also observed a number of genes that were downregulated during OA progression, namely CEBPB, DDIT4, IGFBP7, INHBA, LIF, LINC01423 and VCAM1 (Fig. 5). Finally, we computationally ordered fibroblasts from early OA non-painful sites and early OA painful sites along pseudotime trajectories to investigate transcriptional changes during the transition from non-pain-associated towards pain-associated fibroblasts in early OA. Pseudotemporal expression dynamics revealed a number of genes that were upregulated genes with the transition towards pain-associated fibroblasts (ANGPTL4, BHLHE40, CXCL1, CXCL2, IL6, INHBA, KCNMA1 and LIF) and several genes that were down-regulated (ACKR4, CSTA, GSN, HSPB6, DKK1, FBLN1, GPX3 and PLTP) (Fig. 6).
Fig. 5.
Transcriptional switch in fibroblast phenotype associated with OA disease progression.
(a) t-SNE analysis of synovial fibroblast scRNAseq data showing the differential between fibroblasts from early OA painful sites and end-stage OA painful synovial sites. (b) Monocle pseudotime trajectory of the transition from early OA painful to end-stage OA painful synovial fibroblast transcriptome phenotype (left panel). The trajectory is overlaid with sample distribution (right panel). Cells are ordered in pseudotime based on differentially expressed genes (q-value < 0.01). Expression dynamics of upregulated genes (c) and downregulated genes (d) upon progression of synovial fibroblast phenotype towards end-stage OA overlaid with sample distribution. T-SNE map together with violin plots and featureplots showing the expression levels (y-axis) of the upregulated/downregulated genes for each sample (x-axis).
Fig. 6.
Transcriptional switch in fibroblast phenotype associated with development of pain in early OA.
(a) t-SNE analysis of synovial fibroblast scRNAseq data showing the differential between fibroblasts from early OA non-painful sites and painful synovial samples. (b) Monocle pseudotime trajectory of the transition from early OA non-painful to early OA painful synovial fibroblast transcriptome phenotype (left panel). The trajectory is overlaid with sample distribution (right panel). Cells are ordered in pseudotime based on differentially expressed genes (q-value < 0.01). Expression dynamics of upregulated genes (c) and downregulated genes (d) upon progression of synovial fibroblast phenotype towards early OA pain overlaid with sample distribution.
4. Discussion
This is the first study to compare patterns of patient-reported knee pain in early OA patients and end-stage OA patients and examine the relationship to synovitis and pain severity. This is also the first report of differences in synovial tissue phenotype and in synovial fibroblast subsets between painful and non-painful joint sites in patients with early OA or end-stage OA.
MRI analysis provided further evidence that synovitis is a key feature of knee OA pathology irrespective of disease stage, with all patients exhibiting significant synovitis, and the majority graded as having moderate to severe synovitis. Similar to the findings of de Lange-Brokaar et al [11], we also report a significant relationship between patterns of synovitis and patient-reported pain. Importantly, we show that although total synovitis score correlates with overall pain severity > 70% of knee early OA or end-stage OA patients reported pain that is to some degree localised, with 1 or more compartments being pain-free. Significantly, despite heterogeneity, the pattern of patient-reported knee pain was significantly associated with sites of synovitis. Indeed, patients who reported pain in either the medial or lateral supraparapatallar compartment exhibited significantly greater synovitis at this anatomical location, compared to those patients reporting no pain.
Our analysis of synovial tissue from sites of patient-reported pain and no-pain in early knee OA and end-stage knee OA patients provides the first evidence linking a differential synovium transcriptome to knee OA joint pain. Analysis of early OA synovium demonstrated an exacerbated pro-fibrotic and pro-inflammatory synovial tissue transcriptomic phenotype at sites of patient-reported pain. Furthermore, intriguingly, the transcriptomes of synovial tissue from sites of patient-reported pain in both early OA and end-stage OA was shown to activate several neuronal growth and nociceptive signalling pathways, providing evidence that a differential synovium phenotype participates in mediating pain perception by promoting neuronal growth and sensitisation of afferent nociceptors. However, it should be noted that one limitation of the current study is that synovitis was determined by MRI only, and not confirmed by histopathology. Therefore, we cannot be sure whether all synovial tissue biopsies used for the transcriptomic analysis were similar in terms of histological grading of inflammation.
Our single cell analysis showed that the differential pain transcriptome of knee OA synovial tissue extends to the cellular level of the OA synovial fibroblast. In total, we determined the existence of 7 transcriptionally distinct synovial fibroblast subsets, with differences in their predominance between disease stage and presence of pain.
Changes in the synovium occur early in the disease course, often preceding cartilage damage [7]. Therefore, our observation of the existence of different populations of synovial fibroblasts in early OA has implications for understanding the role of the synovial membrane in OA progression. Previously, Zhang et al. [18] performed single cell analysis of RA fibroblast and end-stage OA fibroblast samples and identified four fibroblast subsets, with two subsets (SC-F1 and SC-F2) which were more prominent in the leukocyte-rich RA fibroblast samples, one subset (SC-F3) with markers evenly expressed in RA and OA fibroblast samples, and one subset (SC-F4) that was more prominent in OA samples. Interestingly, in contrasting our early OA fibroblast samples to our end-stage OA fibroblast samples, we found the cluster predominant in our end-stage OA fibroblast samples more closely resembled the clusters that Zhang et al found to be predominant in leukocyte-rich RA fibroblasts (SC-F1 and SC-F2), with greater expression of SERPINF1, VCAN, DCN, C3 and MFAP5 (SC-F1) and HLA-B, CXCL12, B2M and C1S (SC-F2). This could reflect the greater degree of synovitis we found in our end-stage OA patients, compared to our early OA patients, In contrast, our early OA clusters bore greater resemblance to the sublining SC-F3 and lining SC-F4 subsets, which Zhang et al. found to be more predominant in OA fibroblast samples, with greater expression of PDGFRB, ASPN, CADM1, OGN, VIM, ABHD2, AHNAK and VIM (SC-F3) and TIMP3, HTRA1, ITGB8, NTN4, PRG4, HBEGF, ERRF1Z (SC-F4).
Synovial fibroblasts from non-painful synovial sites of patients with early OA were mainly observed in a single cluster (Cluster 1) and formed majority of the cells within the smallest cluster (Cluster 6). In contrast, fibroblasts from the painful sites of these early OA patients were more diverse, being distributed into four distinct clusters (Clusters 2–5). However, in end-stage OA the majority of fibroblasts were present in one large cluster (Cluster 0), suggesting a shift with disease progression towards a more succinct OA-diseased fibroblast pathotype. Notably, the gene signatures of these end-stage OA fibroblasts were associated with eicosanoid and prostanoid signalling pathways, which are known mediators of inflammatory pain providing the rationale for NSAID analgesia [2]. End-stage OA fibroblasts were also characterised by high expression of the stress-response genes ATF3, HSPA1A and DNAJB1 (Supplementary Fig. 2). Interestingly, serum and synovial fluid concentrations of HSP70 (encoded by HSPA1A), correlate with knee OA severity [19] and are increased in synovial fibroblasts from RA-diseased joints [20]. End-stage OA pain fibroblasts also exhibited high expression of HTRA3, a serine proteinase implicated in the degradation of extracellular matrix [21], which could indicate that these cells also mediate cartilage destruction as disease progresses. Notably, the differential expression of HTRA3 in end-stage OA pain was observed in the bulk expression of synovial tissue.
In line with the bulk transcriptomic analysis of the synovial tissue, our finding that painful synovial tissue sites in early OA contain fibroblast subsets with high expression of pro-inflammatory mediators including CXCL1, CXCL2, IL-6 and LIF provides further evidence for the role of synovial inflammation in the development of early OA pain. CXCL1 enhances inflammatory pain via ERK activation, synaptic transmission and COX-2 expression in dorsal horn neurons [22], LIF alters neuropeptide content following axonal injury [23], and intrathecal administration of CXCL2 mediated hypersensitivity to thermal and mechanical stimuli in naïve mice [24]. Similarly, IL-6 is implicated in mediating inflammatory pain in the rodent antigen-induced arthritis model [25]. These data are further supported by our finding of increased KCNMA1 and decreased ACKR4 expression in fibroblast subsets from painful synovial sites in early OA patients. KCNMA1, a calcium-sensitive potassium ion channel, is involved in the regulation of pro-inflammatory and pro-invasive properties of synovial fibroblasts and its targeting has been suggested in the treatment of RA [26]. ACKR4, an atypical chemokine receptor, functions to scavenge chemokines and regulate leukocyte migration [27].
We also observed a reduction in GPX3 expression associated with both pain and OA disease progression. GPX3 protects against oxidative damage and decreased levels have been reported in OA cartilage [28]. Conceivably, the down-regulation of fibroblast GPX3 expression in the latter stages of OA we observe here could increase reactive oxygen species, which is a known driver of cartilage aggrecan proteoglycan degradation [29]. Our “non-painful” early OA synovial fibroblast subsets were also characterised by higher expression of GSN, coding for gelsolin, an actin binding protein. Experimental removal of GSN in synovial fibroblasts has been shown to cause synovial hyperplasia [30]. Based on our pseudotemporal expression dynamics it appears that fibroblast protective mechanisms against inflammation and oxidative damage are gradually lost during OA progression, whilst there is a simultaneous increase in expression of pro-inflammatory mediators.
One of the most notable findings from this study was the finding that painful synovial sites in early OA patients exhibit a distinct subset of synovial fibroblasts with a gene signature supporting the formation and development of neurites, in line with the transcriptome of the pain-associated synovial tissue. This specific fibroblast subset exhibited high expression of neuritin 1 and sulfatase 1, which are known promoters of neuronal survival, development and outgrowth [31,32]. Indeed, we demonstrated that the secretome of fibroblasts from painful sites promoted the greatest amounts of adult rodent DRGN survival and neurite outgrowth. This is the first time that the human OA synovial fibroblast secretome has been reported to induce survival and neurite outgrowth.
In summary, synovial tissue from sites of patient-reported pain in early OA and end-stage OA patients exhibits a differential transcriptomic phenotype with distinct synovial fibroblast subsets. Notably, subsets of fibroblasts from painful sites of early OA patients exhibit gene signatures that promote fibrosis, inflammation and neuronal growth and nociceptive signalling pathways and the secretome from these cells promoted neurite outgrowth. Further understanding of the precise cellular functions of these fibroblast subsets will help to identify new candidate targets and provide a rationale for the therapeutic targeting of specific fibroblast pathotypes to alleviate pain in OA.
5. Contributors
SWJ and MAL designed the study. SWJ, DEN, AB, AMH, SNW, RB and PH carried out experiments and data analysis. SWJ and DEN wrote the manuscript. SWJ, MAL, ZA and ETD carried out data interpretation and edited the manuscript. All authors approved the manuscript.
6. Data sharing statement
The data discussed in this publication have been deposited in NCBI's Gene Expression Omnibus and are accessible through GEO Series accession number GSE176223 and GSE176308.
Declaration of Competing Interest
SWJ declares grant funding from Versus Arthritis during the course of this study.
Acknowledgments
This study was funded by Versus Arthritis (21530; 21812). The funder had no involvement in the study or any role in the writing of the manuscript or the decision to submit it for publication. The corresponding author (Dr Simon Jones) confirms that authors were not precluded from accessing data in the study and they accept responsibility to submit for publication. The authors acknowledge all study participants, research staff at The Royal Orthopaedic Hospital NHS Foundation Trust and Russell's Hall Hospital, Dudley for obtaining consents and screening. This research made use of the Balena High Performance Computing (HPC) Service at the University of Bath.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.ebiom.2021.103618.
Appendix. Supplementary materials
References
- 1.Neogi T. The epidemiology and impact of pain in osteoarthritis. Osteoarthr Cartil. 2013;21:1145–1153. doi: 10.1016/j.joca.2013.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tonge D.P., Pearson M.J., Jones SW. The hallmarks of osteoarthritis and the potential to develop personalised disease-modifying pharmacological therapeutics. Osteoarthr Cartil. 2014;22:609–621. doi: 10.1016/j.joca.2014.03.004. [DOI] [PubMed] [Google Scholar]
- 3.Baker K., Grainger A., Niu J. Relation of synovitis to knee pain using contrast-enhanced MRIs. Ann Rheum Dis. 2010;69:1779–1783. doi: 10.1136/ard.2009.121426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pearson M.J., Herndler-Brandstetter D., Tariq M.A. IL-6 secretion in osteoarthritis patients is mediated by chondrocyte-synovial fibroblast cross-talk and is enhanced by obesity. Sci Rep. 2017;7:3451. doi: 10.1038/s41598-017-03759-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nanus D.E., Wijesinghe S.N., Pearson M.J. Regulation of the inflammatory synovial fibroblast phenotype by metastasis-associated lung adenocarcinoma transcript 1 long noncoding RNA in obese patients with osteoarthritis. Arthritis Rheumatol. 2020;72:609–619. doi: 10.1002/art.41158. [DOI] [PubMed] [Google Scholar]
- 6.Rhodes L.A., Conaghan P.G., Radjenovic A., Grainger A.J., Emery P., McGonagle D. Further evidence that a cartilage-pannus junction synovitis predilection is not a specific feature of rheumatoid arthritis. Ann Rheum Dis. 2005;64:1347–1349. doi: 10.1136/ard.2004.033688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Myers S.L., Brandt K.D., Ehlich J.W. Synovial inflammation in patients with early osteoarthritis of the knee. J Rheumatol. 1990;17:1662–1669. [PubMed] [Google Scholar]
- 8.Miller R.E., Miller R.J., Malfait AM. Osteoarthritis joint pain: the cytokine connection. Cytokine. 2014;70:185–193. doi: 10.1016/j.cyto.2014.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Orita S., Koshi T., Mitsuka T. Associations between proinflammatory cytokines in the synovial fluid and radiographic grading and pain-related scores in 47 consecutive patients with osteoarthritis of the knee. BMC Musculoskelet Disord. 2011;12:144. doi: 10.1186/1471-2474-12-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chakrabarti S., Jadon D.R., Bulmer D.C., Smith ESJ. Human osteoarthritic synovial fluid increases excitability of mouse dorsal root ganglion sensory neurons: an in-vitro translational model to study arthritic pain. Rheumatology (Oxford) 2020;59:662–667. doi: 10.1093/rheumatology/kez331. [DOI] [PubMed] [Google Scholar]
- 11.de Lange-Brokaar B.J., Ioan-Facsinay A., Yusuf E. Association of pain in knee osteoarthritis with distinct patterns of synovitis. Arthritis Rheumatol. 2015;67:733–740. doi: 10.1002/art.38965. [DOI] [PubMed] [Google Scholar]
- 12.Mizoguchi F., Slowikowski K., Wei K. Functionally distinct disease-associated fibroblast subsets in rheumatoid arthritis. Nat Commun. 2018;9:789. doi: 10.1038/s41467-018-02892-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Croft A.P., Campos J., Jansen K. Distinct fibroblast subsets drive inflammation and damage in arthritis. Nature. 2019;570:246–251. doi: 10.1038/s41586-019-1263-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guermazi A., Roemer F.W., Hayashi D. Assessment of synovitis with contrast-enhanced MRI using a whole-joint semiquantitative scoring system in people with, or at high risk of, knee osteoarthritis: the most study. Ann Rheum Dis. 2011;70:805–811. doi: 10.1136/ard.2010.139618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Afgan E., Baker D., Batut B. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2018 update. Nucleic Acids Res. 2018;46:W537–W544. doi: 10.1093/nar/gky379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Parsonage G., Falciani F., Burman A. Global gene expression profiles in fibroblasts from synovial, skin and lymphoid tissue reveals distinct cytokine and chemokine expression patterns. Thromb Haemost. 2003;90:688–697. doi: 10.1160/TH03-04-0208. [DOI] [PubMed] [Google Scholar]
- 17.Ahmed Z., Dent R.G., Suggate E.L. Disinhibition of neurotrophin-induced dorsal root ganglion cell neurite outgrowth on CNS myelin by siRNA-mediated knockdown of NgR, p75NTR and Rho-A. Mol Cell Neurosci. 2005;28:509–523. doi: 10.1016/j.mcn.2004.11.002. [DOI] [PubMed] [Google Scholar]
- 18.Zhang F., Wei K., Slowikowski K. Defining inflammatory cell states in rheumatoid arthritis joint synovial tissues by integrating single-cell transcriptomics and mass cytometry. Nat Immunol. 2019;20:928–942. doi: 10.1038/s41590-019-0378-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ngarmukos S., Scaramuzza S., Theerawattanapong N., Tanavalee A., Honsawek S. Circulating and synovial fluid heat shock protein 70 Are correlated with severity in knee osteoarthritis. Cartilage. 2020;11:323–328. doi: 10.1177/1947603518790075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nguyen T.T., Gehrmann M., Zlacka D. Heat shock protein 70 membrane expression on fibroblast-like synovial cells derived from synovial tissue of patients with rheumatoid and juvenile idiopathic arthritis. Scand J Rheumatol. 2006;35:447–453. doi: 10.1080/03009740600759621. [DOI] [PubMed] [Google Scholar]
- 21.Milner J.M., Patel A., Rowan AD. Emerging roles of serine proteinases in tissue turnover in arthritis. Arthritis Rheum. 2008;58:3644–3656. doi: 10.1002/art.24046. [DOI] [PubMed] [Google Scholar]
- 22.Cao D.L., Zhang Z.J., Xie R.G., Jiang B.C., Ji R.R., Gao YJ. Chemokine CXCL1 enhances inflammatory pain and increases NMDA receptor activity and COX-2 expression in spinal cord neurons via activation of CXCR2. Exp Neurol. 2014;261:328–336. doi: 10.1016/j.expneurol.2014.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sun Y., Zigmond RE. Involvement of leukemia inhibitory factor in the increases in galanin and vasoactive intestinal peptide mRNA and the decreases in neuropeptide Y and tyrosine hydroxylase mRNA in sympathetic neurons after axotomy. J Neurochem. 1996;67:1751–1760. doi: 10.1046/j.1471-4159.1996.67041751.x. [DOI] [PubMed] [Google Scholar]
- 24.Piotrowska A., Rojewska E., Pawlik K. Pharmacological blockade of spinal CXCL3/CXCR2 signaling by NVP CXCR2 20, a selective CXCR2 antagonist, reduces neuropathic pain following peripheral nerve injury. Front Immunol. 2019;10:2198. doi: 10.3389/fimmu.2019.02198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Boettger M.K., Leuchtweis J., Kummel D., Gajda M., Brauer R., Schaible HG. Differential effects of locally and systemically administered soluble glycoprotein 130 on pain and inflammation in experimental arthritis. Arthritis Res Ther. 2010;12:R140. doi: 10.1186/ar3079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hu X., Laragione T., Sun L. KCa1.1 potassium channels regulate key proinflammatory and invasive properties of fibroblast-like synoviocytes in rheumatoid arthritis. J Biol Chem. 2012;287:4014–4022. doi: 10.1074/jbc.M111.312264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thomson C.A., van de Pavert S.A., Stakenborg M. Expression of the atypical chemokine receptor ACKR4 identifies a novel population of intestinal submucosal fibroblasts that preferentially expresses endothelial cell regulators. J Immunol. 2018;201:215–229. doi: 10.4049/jimmunol.1700967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aigner T., Fundel K., Saas J. Large-scale gene expression profiling reveals major pathogenetic pathways of cartilage degeneration in osteoarthritis. Arthritis Rheum. 2006;54:3533–3544. doi: 10.1002/art.22174. [DOI] [PubMed] [Google Scholar]
- 29.Tiku M.L., Gupta S., Deshmukh DR. Aggrecan degradation in chondrocytes is mediated by reactive oxygen species and protected by antioxidants. Free Radic Res. 1999;30:395–405. doi: 10.1080/10715769900300431. [DOI] [PubMed] [Google Scholar]
- 30.Aidinis V., Carninci P., Armaka M. Cytoskeletal rearrangements in synovial fibroblasts as a novel pathophysiological determinant of modeled rheumatoid arthritis. PLoS Genet. 2005;1:e48. doi: 10.1371/journal.pgen.0010048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhou S., Zhou J. Neuritin, a neurotrophic factor in nervous system physiology. Curr Med Chem. 2014;21:1212–1219. doi: 10.2174/0929867321666131218093327. [DOI] [PubMed] [Google Scholar]
- 32.Kalus I., Rohn S., Puvirajesinghe T.M. Sulf1 and Sulf2 differentially modulate heparan sulfate proteoglycan sulfation during postnatal cerebellum development: evidence for neuroprotective and neurite outgrowth promoting functions. PLoS One. 2015;10 doi: 10.1371/journal.pone.0139853. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.