ABSTRACT
The interferon-induced transmembrane (IFITM) gene family performs multiple functions in immunity, including inhibition of virus entry into cells. The IFITM repertoire varies widely between species and consists of protein-coding genes and pseudogenes. The selective forces driving pseudogenization within gene families are rarely understood. In this issue, the human pseudogene IFITM4P is characterized as a virus-induced, long noncoding RNA that contributes to restriction of influenza A virus by regulating mRNA levels of IFITM1, IFITM2, and IFITM3.
KEYWORDS: IFITM, interferons, lncRNA, miRNA, pseudogene, retrogene
TEXT
Innate immunity plays a critical role in the initial detection and restriction of virus infections at the level of individual cells. Upon pathogen invasion, pattern recognition receptors, such as retinoic acid-inducible gene I (RIG-I), recognize pathogen-associated molecular patterns (PAMPs) and initiate a cascade of signaling that leads to the production of type I interferons (IFN-α and IFN-β) (1). These cytokine mediators interact with specific cellular receptors and activate the transcription of a distinct set of genes known as IFN-stimulated genes (ISGs) that collectively confer an antiviral state to cells (2). Regulation of ISG functions by transcriptional and posttranscriptional mechanisms is a blossoming area of research at the interface of immunology and virology. In this issue of the Journal of Virology, Xiao et al. (3) report that the interferon-inducible transmembrane (IFITM) proteins are regulated by a long noncoding RNA (lncRNA) encoded by a pseudogene known as IFITM4P.
IFITM proteins are a group of small transmembrane proteins known to regulate various cellular processes, including virus entry into cells (4). The human genome contains five IFITM genes (IFITM1, IFITM2, IFITM3, IFITM5, and IFITM10). IFITM1-3 are upregulated by interferons and are the family members ascribed with immune functions (5). IFITM1-3 broadly inhibit infection by diverse enveloped viruses, including orthomyxoviruses, coronaviruses, flaviviruses, filoviruses, paramyxoviruses, and retroviruses (5, 6). All three immune-related IFITM proteins have been shown to remodel cellular membranes in a manner that disfavors virus-cell fusion. The functions of IFITM3 are best characterized. IFITM3 dimers increase membrane lipid order (rigidity) (7) and curvature (8) in order to disfavor virus-cell membrane fusion (9) and traffic endocytosed virions toward lysosomes for degradation (10, 11). IFITM3 exhibits the most potent antiviral activity against most viruses studied in cell culture, and it is required for control of influenza A virus in vivo (12–14). As components of the cell’s first line of antiviral defense, there is much interest in the spatiotemporal regulation of IFITM proteins. Most human tissues contain cell types that express one or more IFITM proteins constitutively (15), including tissue-resident memory T cells (16, 17), respiratory epithelial cells (13), and hematopoietic stem cell progenitors (18–20). Beyond the level of transcription, posttranslational modifications play important roles in the stability and subcellular localization of IFITM proteins (21). For example, the E3 ligase NEDD4 ubiquitinates IFITM2 and IFITM3 and promotes their degradation in lysosomes (21–23). On the other hand, there is relatively little known about pathways that regulate the fate of IFITM mRNAs.
In addition to the five protein-coding IFITM genes found in humans, there are multiple IFITM-like loci, which are designated pseudogenes due to apparent truncation of the open reading frame (ORF) and/or lack of an intron. At least 12 IFITM-derived pseudogenes, including IFITM4P, exist in the human genome, but functional information for their biological roles is lacking. Xiao et al. (3) used a cDNA microarray approach to identify RNAs that are up- or downregulated in lung epithelial cells during influenza A virus infection and chose to focus on the RNA encoded by IFITM4P due to its relatedness to the antiviral IFITM protein family. They showed that IFITM4P expression was induced 10 to 14 h following infection by multiple strains of influenza A virus and was also upregulated by another RNA virus (Sendai virus) and a DNA virus (herpes simplex virus 1). This observation was maintained in many, but not all, additional cell lines tested. Due to the authors’ inability to detect a protein product, the IFITM4P mRNA (which is 300 bp long) was established as an lncRNA with potential functional roles played during virus infections. The authors went on to show that IFITM4P expression was triggered by purified viral RNA or poly(I·C), which represent the natural and synthetic ligand, respectively, of the pathogen recognition receptor RIG-I. Accordingly, the induction of IFITM4P by influenza A virus was lost in lung epithelial cells in which RIG-I was depleted. Furthermore, type I interferon treatment elevated IFITM4P levels, indicating that it may be an ISG. These findings suggested that IFITM4P may contribute to the antiviral state activated in response to virus infection.
The antiviral functions of IFITM4P were revealed by assessing the effect of IFITM4P knockdown or overexpression on influenza A virus, which promoted or inhibited infection, respectively. Importantly, levels of IFITM4P were inconsequential for infection by Sendai virus, which was previously shown to be insensitive to restriction by IFITM1-3 (24). These results indicated that IFITM4P may function similarly to the known antiviral effectors IFITM1-3. Alternatively, since IFITM4P is an lncRNA, it may exhibit indirect antiviral activity by regulating the expression of IFITM1-3. Indeed, Xiao et al. (3) show that knockdown of IFITM4P resulted in reduced levels of IFITM3 protein, which was accompanied by reduced levels of mRNA corresponding to IFITM3, IFITM2, and IFITM1. Conversely, overexpression of IFITM4P resulted in increased levels of IFITM1, IFITM2, and IFITM3 mRNAs. These data suggested that IFITM4P lncRNA promotes the stability of IFITM mRNAs and provided an explanation for its observed anti-Influenza activity.
The fact that IFITM4P lncRNA stabilized mRNAs from the related IFITM1-3 raised the possibility that it interferes with a process that negatively regulates these mRNAs. The authors used prediction software to identify microRNAs (miRNAs) capable of engaging IFITM4P lncRNA and identified miR-24-3p as one that is functionally capable of decreasing levels of IFITM4P as well as mRNA levels of IFITM1-3. Mutagenesis of IFITM4P or IFITM1-3 mRNAs at nucleotides complementary to miR-24-3p rendered the transcripts resistant to miR-24-3p-mediated suppression.
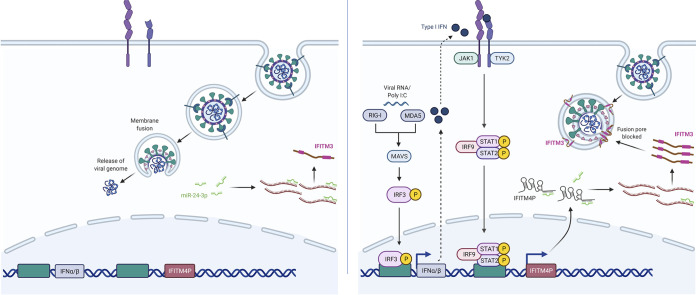
Therefore, IFITM4P promotes the production of IFITM proteins by interfering with the silencing activity of miRNAs targeting IFITM transcripts. This mechanism of action led the authors to designate IFITM4P as a competing endogenous RNA (ceRNA). By acting as a decoy for miR-24-3p, IFITM4P augments the abundance of IFITM1-3 mRNAs available for translation (Fig. 1). Furthermore, the regulatory relationship between IFITM4P and IFITM1-3 mRNAs is bidirectional—increased levels of IFITM1-3 mRNAs also augment the levels of IFITM4P, and this is due to the shared miR-24-3p-binding sequence they possess.
FIG 1.
IFITM4P-mediated regulation of innate immunity to curb virus infection. (Left panel) The entry pathway exhibited by influenza A virus and other endocytic viruses is shown. Following internalization by endocytosis, membrane fusion between virus and endosome enables release of the viral ribonucleoprotein complex and initiation of virus replication. Binding of miR-24-3p to IFITM1-3 mRNAs suppresses the production of IFITM1-3 proteins. (Right panel) Viral RNA or poly(I·C) is recognized by RIG-I or MDA5 and initiates the production of type I interferons (IFNs). IFN binds to its cognate receptor in an autocrine fashion (and in a paracrine fashion [not shown]), leading to induction of interferon-stimulated genes and IFITM4P. IFITM4P encodes a long noncoding RNA that acts as a decoy for miR-24-3p, releasing IFITM1-3 mRNAs from a suppressed state and augmenting IFITM1-3 protein levels. Elevated IFITM proteins, particularly IFITM3, inhibit formation of the fusion pore during virus-cell membrane fusion. Figure created with BioRender.
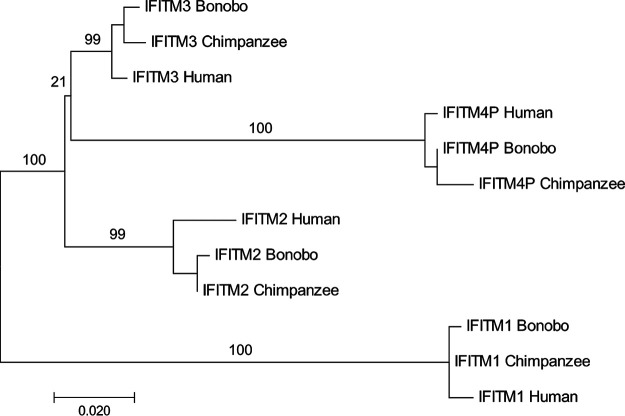
Given the importance of IFITM4P in the regulation of antiviral immunity, we decided to investigate the structure, origin, and evolutionary conservation of this gene to better understand its functions and to assess similarities to other IFITM-derived pseudogenes. lncRNAs are generally poorly conserved among various species relative to other well-studied noncoding RNAs, such as miRNAs and small nucleolar RNAs (snoRNAs) (25–27), implying that lncRNAs may perform species-specific functions. A previous publication stated that IFITM4P is found in mice (28). However, our genomics analyses in Ensembl (ensembl.org) found IFITM4P orthologs in 31 of 90 eutherian mammal species, including some rodent and bat species, but no such gene was identified in the mouse genome. Mammalian IFITM4P orthologs exhibited various degrees of sequence conservation, with IFITM4P from chimpanzees, bonobos, and pig-tailed macaques most closely resembling the human gene. Elsewhere, including in other primate species, the IFITM4P locus has diverged or decayed. Phylogenetic analysis suggests that IFITM4P may have descended from IFITM3 (Fig. 2). Overall, IFITM4P may act as a functional lncRNA in some, but not all, mammalian species. Furthermore, its role as a ceRNA impacting antiviral immunity may be confined to a select number of primates.
FIG 2.
Phylogenetic reconstruction of IFITM4P, IFITM1, IFITM2, and IFITM3 in great apes. Using MEGA7, open reading frames were aligned and gaps were removed such that a total of 335 nucleotides per sequence were obtained. Phylogenetic analysis was inferred by maximum likelihood, and the consensus tree with the highest log likelihood is shown. Initial trees for the heuristic search were obtained automatically by applying neighbor-joining and BioNJ algorithms to a matrix of pairwise distances estimated using the maximum composite likelihood (MCL) approach, and the topology with the superior log likelihood value was selected. The tree branches are drawn to scale, with branch lengths measured in number of substitutions per site. Confidence levels were determined using the bootstrapping method (500 iterations), and the numbers above branches indicate the percentage of trees in which the associated taxa clustered together.
The sequence architecture of IFITM4P indicates that it is a processed pseudogene because it lacks introns and contains a poly(A) tail in its 3′ untranslated region (UTR) (Table 1). Processed pseudogenes, also known as retrogenes, are genes produced from mRNA transcripts that have been spliced, reverse transcribed into DNA by an endogenous retroelement, and integrated into a new genomic location (29). During pseudogenization, enhancers and promoter elements that were once required for gene expression are lost but may be reacquired following sequence evolution and natural selection. In that light, in contrast to the parental locus IFITM3, we did not detect a canonical interferon response element (ISRE) motif upstream of IFITM4P. This raises the possibility that the interferon inducibility of IFITM4P observed by Xiao et al. (3) is actually the result of interferon-induced IFITM1, IFITM2, and/or IFITM3 mRNA transcripts binding to miR-24-3p, releasing IFITM4P from a silenced state.
TABLE 1.
Properties of human IFITM pseudogenes
| Gene name | Ensembl ID | Genomic location | ISRE | Poly(A) tail | ORF length relative to IFITM3 (aa) |
|---|---|---|---|---|---|
| IFITM3P1 | ENSG00000236562 | Chr4(+) | No | Yes | 22–133 |
| IFITM3P2 | ENSG00000223722 | Chr12(+) | Yes | Yes | 1–133 |
| IFITM3P3 | ENSG00000196114 | Chr6(−) | Yes | Yes | 22–45 |
| IFITM3P4 | ENSG00000230191 | Chr7(+) | Yes | Yes | 1–42 |
| IFITM3P5 | ENSG00000238168 | Chr12(+) | Yes | Yes | 22–147 |
| IFITMP6 | ENSG00000258352 | Chr12(−) | No | Yes | 1–97 |
| IFITM3P7 | ENSG00000233419 | Chr1(−) | No | Yes | 1–24 |
| IFITM3P8 | ENSG00000271377 | Chr8(+) | No | Yes | 1–31 |
| IFITM3P9 | ENSG00000271134 | Chr2(−) | Yes | Yes | 1–69 |
| IFITM4P | ENSG00000235821 | Chr6(−) | No | Yes | 1–113 |
| IFITM8P | ENSG00000215096 | Chr8(+) | Yes | Yes | 1–131 |
| IFITM9P | ENSG00000213275 | Chr11(−) | Yes | Yes | 1–20 |
lncRNAs have previously been found to participate in cellular functions by acting as decoys, guides, or scaffolds, but the demonstration that IFITM4P regulates the expression of IFITM3 indicates that pseudogenes can be functionally selected to regulate the parental loci from which they were derived. We and others previously found that IFITM3 has undergone recurrent gene duplication in a lineage-specific manner (23, 30). As such, the number of IFITM genes carried by vertebrates varies widely by species. Gene duplication and subsequent sequence divergence at the protein level can alter the subcellular localization of antiviral effectors or provide new functional modalities, a process known as neofunctionalization. However, this description of IFITM4P as a ceRNA reveals that RNA-based functions may also be a selective driving force for the rampant proliferation of IFITM genes in vertebrates.
The human genome contains at least 12 IFITM-derived pseudogenes, including IFITM4P (Table 1). It is possible that other IFITM pseudogenes share a complementary or redundant function with IFITM4P, such that multiple lncRNAs act on IFITM mRNAs simultaneously to further amplify mRNA stability and boost the antiviral protection conferred to cells. Alternatively, other IFITM pseudogenes may perform roles that are distinct from that of IFITM4P. Several contain an ISRE in their promoter, suggesting that they are interferon inducible and that this capacity may be the result of natural selection. In addition, four of them (IFITM3P1, IFITM3P2, IFITM3P5, and IFITM8P) contain intact open reading frames (with start and stop codons) that are as long as IFITM3, hinting that they may perform functions at the protein level (see Fig. S1 in the supplemental material). Apart from IFITM4P, the functional potential of these loci remains uncharacterized. However, a recent study on severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2)-infected patients suggested that a potent and early induction of ISGs across immune cell subtypes was associated with successful containment of virus and avoidance of COVID-19 (31). This protective gene signature included the likes of IFITM3P1, IFITM3P2, IFITM3P3, IFITM3P6, and IFITM3P9. Therefore, studies addressing the antiviral activities of IFITM pseudogenes against SARS-CoV-2 and other viruses are warranted. Particularly, it would be interesting to assess whether they perform indirect (lncRNA-based) or direct (protein-based) antiviral activities.
Prior to the demonstration that it bound to IFITM4P, miR-24-3p was known to target other cellular transcripts. miR-24-3p has been shown to play an antiviral role against influenza A virus by negatively regulating the expression of cellular furin, a protease required for the cleavage and activation of the viral fusion protein (32). In addition, miR-24-3p may downregulate neuropilin-1, which promotes the internalization of SARS-CoV-2 into cells (33, 34). These targets of miR-24-3p suggest that high levels of IFITM4P, through its actions as a ceRNA, may also promote infection by respiratory viruses. Therefore, the overall impact of IFITM4P on virus infections may result from the net effect of its antiviral and proviral effects.
The positive regulation of IFITM1-3 via the ceRNA activity of IFITM4P has implications beyond antiviral immunity. IFITM proteins are known to be upregulated in various cancers, and at least in some cases, they are believed to play a direct role in promoting tumorigenic phenotypes, such as cell proliferation, migration, and invasion (35). miR-24-3p expression is also associated with cancers and was previously reported to influence cell proliferation and migration by regulating SOX7 and ING1 (36–38). By competing for miR-24-3p binding, IFITM4P represents a previously unrecognized link (lnc!) between IFITM proteins and miR-24-3p and may impact cancer pathogenesis through multiple pathways. Detailing the functional crossroads between tumorigenesis and the antiviral state of cells is an important priority for biomedical research.
The views expressed in this article do not necessarily reflect the views of the journal or of ASM.
Footnotes
Supplemental material is available online only.
Contributor Information
Alex A. Compton, Email: alex.compton@nih.gov.
Bryan R. G. Williams, Hudson Institute of Medical Research
REFERENCES
- 1.Mogensen TH. 2009. Pathogen recognition and inflammatory signaling in innate immune defenses. Clin Microbiol Rev 22:240–273. 10.1128/CMR.00046-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akira S, Uematsu S, Takeuchi O. 2006. Pathogen recognition and innate immunity. Cell 124:783–801. 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 3.Xiao M, Chen Y, Wang S, Liu S, Rai KR, Chen B, Li F, Li Y, Maarouf M, Chen J-L. 2021. Long noncoding RNA IFITM4P regulates host antiviral responses by acting as a competing endogenous RNA. J Virol 95:e00277-21. 10.1128/JVI.00277-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shi G, Schwartz O, Compton AA. 2017. More than meets the I: the diverse antiviral and cellular functions of interferon-induced transmembrane proteins. Retrovirology 14:53. 10.1186/s12977-017-0377-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brass AL, Huang IC, Benita Y, John SP, Krishnan MN, Feeley EM, Ryan BJ, Weyer JL, van der Weyden L, Fikrig E, Adams DJ, Xavier RJ, Farzan M, Elledge SJ. 2009. The IFITM proteins mediate cellular resistance to influenza A H1N1 virus, West Nile virus, and dengue virus. Cell 139:1243–1254. 10.1016/j.cell.2009.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bailey CC, Zhong G, Huang IC, Farzan M. 2014. IFITM-family proteins: the cell's first line of antiviral defense. Annu Rev Virol 1:261–283. 10.1146/annurev-virology-031413-085537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li K, Markosyan RM, Zheng Y-M, Golfetto O, Bungart B, Li M, Ding S, He Y, Liang C, Lee JC, Gratton E, Cohen FS, Liu S-L. 2013. IFITM proteins restrict viral membrane hemifusion. PLoS Pathog 9:e1003124. 10.1371/journal.ppat.1003124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guo X, Steinkühler J, Marin M, Li X, Lu W, Dimova R, Melikyan GB. 2021. Interferon-induced transmembrane protein 3 blocks fusion of diverse enveloped viruses by altering mechanical properties of cell membranes. ACS Nano 15:8155–8170. 10.1021/acsnano.0c10567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rahman K, Coomer CA, Majdoul S, Ding SY, Padilla-Parra S, Compton AA. 2020. Homology-guided identification of a conserved motif linking the antiviral functions of IFITM3 to its oligomeric state. eLife 9:e58537. 10.7554/eLife.58537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Spence JS, He R, Hoffmann H-H, Das T, Thinon E, Rice CM, Peng T, Chandran K, Hang HC. 2019. IFITM3 directly engages and shuttles incoming virus particles to lysosomes. Nat Chem Biol 15:259–268. 10.1038/s41589-018-0213-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Suddala KC, Lee CC, Meraner P, Marin M, Markosyan RM, Desai TM, Cohen FS, Brass AL, Melikyan GB. 2019. Interferon-induced transmembrane protein 3 blocks fusion of sensitive but not resistant viruses by partitioning into virus-carrying endosomes. PLoS Pathog 15:e1007532-35. 10.1371/journal.ppat.1007532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zani A, Yount JS. 2018. Antiviral protection by IFITM3 in vivo. Curr Clin Microbiol Rep 5:229–237. 10.1007/s40588-018-0103-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bailey CC, Huang IC, Kam C, Farzan M. 2012. Ifitm3 limits the severity of acute influenza in mice. PLoS Pathog 8:e1002909. 10.1371/journal.ppat.1002909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Coomer CA, Rahman K, Compton AA. 2021. CD225 proteins: a family portrait of fusion regulators. Trends Genet 37:406–410. 10.1016/j.tig.2021.01.004. [DOI] [PubMed] [Google Scholar]
- 15.Uhlén M, Fagerberg L, Hallström BM, Lindskog C, Oksvold P, Mardinoglu A, Sivertsson Å, Kampf C, Sjöstedt E, Asplund A, Olsson I, Edlund K, Lundberg E, Navani S, Szigyarto CA-K, Odeberg J, Djureinovic D, Takanen JO, Hober S, Alm T, Edqvist P-H, Berling H, Tegel H, Mulder J, Rockberg J, Nilsson P, Schwenk JM, Hamsten M, von Feilitzen K, Forsberg M, Persson L, Johansson F, Zwahlen M, von Heijne G, Nielsen J, Pontén F. 2015. Proteomics. Tissue-based map of the human proteome. Science 347:1260419. 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 16.Wakim LM, Gupta N, Mintern JD, Villadangos JA. 2013. Enhanced survival of lung tissue-resident memory CD8(+) T cells during infection with influenza virus due to selective expression of IFITM3. Nat Immunol 14:238–245. 10.1038/ni.2525. [DOI] [PubMed] [Google Scholar]
- 17.Bedford JG, O'Keeffe M, Reading PC, Wakim LM. 2019. Rapid interferon independent expression of IFITM3 following T cell activation protects cells from influenza virus infection. PLoS One 14:e0210132. 10.1371/journal.pone.0210132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu X, Dao Thi VL, Huang Y, Billerbeck E, Saha D, Hoffmann HH, Wang Y, Silva LAV, Sarbanes S, Sun T, Andrus L, Yu Y, Quirk C, Li M, MacDonald MR, Schneider WM, An X, Rosenberg BR, Rice CM. 2018. Intrinsic immunity shapes viral resistance of stem cells. Cell 172:423–438.e25. 10.1016/j.cell.2017.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ozog S, Timberlake ND, Hermann K, Garijo O, Haworth KG, Shi G, Glinkerman CM, Schefter LE, D'Souza S, Simpson E, Sghia-Hughes G, Carillo RR, Boger DL, Kiem HP, Slukvin I, Ryu BY, Sorrentino BP, Adair JE, Snyder SA, Compton AA, Torbett BE. 2019. Resveratrol trimer enhances gene delivery to hematopoietic stem cells by reducing antiviral restriction at endosomes. Blood 134:1298–1311. 10.1182/blood.2019000040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shi G, Ozog S, Torbett BE, Compton AA. 2018. mTOR inhibitors lower an intrinsic barrier to virus infection mediated by IFITM3. Proc Natl Acad Sci USA 115:E10069–E10078. 10.1073/pnas.1811892115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chesarino NM, McMichael TM, Yount JS. 2014. Regulation of the trafficking and antiviral activity of IFITM3 by post-translational modifications. Future Microbiol 9:1151–1163. 10.2217/fmb.14.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chesarino NM, McMichael TM, Yount JS. 2015. E3 ubiquitin ligase NEDD4 promotes influenza virus infection by decreasing levels of the antiviral protein IFITM3. PLoS Pathog 11:e1005095. 10.1371/journal.ppat.1005095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Compton AA, Roy N, Porrot F, Billet A, Casartelli N, Yount JS, Liang C, Schwartz O. 2016. Natural mutations in IFITM3 modulate post-translational regulation and toggle antiviral specificity. EMBO Rep 17:1657–1671. 10.15252/embr.201642771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hach JC, McMichael T, Chesarino NM, Yount JS. 2013. Palmitoylation on conserved and nonconserved cysteines of murine IFITM1 regulates its stability and anti-influenza A virus activity. J Virol 87:9923–9927. 10.1128/JVI.00621-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, Guernec G, Martin D, Merkel A, Knowles DG, Lagarde J, Veeravalli L, Ruan X, Ruan Y, Lassmann T, Carninci P, Brown JB, Lipovich L, Gonzalez JM, Thomas M, Davis CA, Shiekhattar R, Gingeras TR, Hubbard TJ, Notredame C, Harrow J, Guigo R. 2012. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res 22:1775–1789. 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pang KC, Frith MC, Mattick JS. 2006. Rapid evolution of noncoding RNAs: lack of conservation does not mean lack of function. Trends Genet 22:1–5. 10.1016/j.tig.2005.10.003. [DOI] [PubMed] [Google Scholar]
- 27.Ma L, Bajic VB, Zhang Z. 2013. On the classification of long non-coding RNAs. RNA Biol 10:925–933. 10.4161/rna.24604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Diamond MS, Farzan M. 2013. The broad-spectrum antiviral functions of IFIT and IFITM proteins. Nat Rev Immunol 13:46–57. 10.1038/nri3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Harrison PM, Zheng D, Zhang Z, Carriero N, Gerstein M. 2005. Transcribed processed pseudogenes in the human genome: an intermediate form of expressed retrosequence lacking protein-coding ability. Nucleic Acids Res 33:2374–2383. 10.1093/nar/gki531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang Z, Liu J, Li M, Yang H, Zhang C. 2012. Evolutionary dynamics of the interferon-induced transmembrane gene family in vertebrates. PLoS One 7:e49265. 10.1371/journal.pone.0049265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pekayvaz K, Leunig A, Kaiser R, Brambs S, Joppich M, Janjic A, Popp O, Polewka V, Wange LE, Gold C, Kirchner M, Muenchhoff M, Hellmuth JC, Scherer C, Eser T, Deák F, Kuhl N, Linder A, Saar K, Tomas L, Schultz C, Enard W, Kroidl I, Geldmacher C, von Bergwelt-Baildon M, Keppler OT, Zimmer R, Mertins P, Hubner N, Hölscher M, Massberg S, Stark K, Nicolai L. 2021. Protective immune trajectories in early viral containment of non-pneumonic SARS-CoV-2 infection. bioRxiv. https://www.biorxiv.org/content/10.1101/2021.02.03.429351v1. [DOI] [PMC free article] [PubMed]
- 32.Loveday EK, Diederich S, Pasick J, Jean F. 2015. Human microRNA-24 modulates highly pathogenic avian-origin H5N1 influenza A virus infection in A549 cells by targeting secretory pathway furin. J Gen Virol 96:30–39. 10.1099/vir.0.068585-0. [DOI] [PubMed] [Google Scholar]
- 33.Mone P, Gambardella J, Wang X, Jankauskas SS, Matarese A, Santulli G. 2021. miR-24 targets SARS-CoV-2 co-factor Neuropilin-1 in human brain microvascular endothelial cells: insights for COVID-19 neurological manifestations. Res Square. https://www.researchsquare.com/article/rs-192099/v1. [DOI] [PMC free article] [PubMed]
- 34.Cantuti-Castelvetri L, Ojha R, Pedro LD, Djannatian M, Franz J, Kuivanen S, van der Meer F, Kallio K, Kaya T, Anastasina M, Smura T, Levanov L, Szirovicza L, Tobi A, Kallio-Kokko H, Osterlund P, Joensuu M, Meunier FA, Butcher SJ, Winkler MS, Mollenhauer B, Helenius A, Gokce O, Teesalu T, Hepojoki J, Vapalahti O, Stadelmann C, Balistreri G, Simons M. 2020. Neuropilin-1 facilitates SARS-CoV-2 cell entry and infectivity. Science 370:856–860. 10.1126/science.abd2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rajapaksa US, Jin C, Dong T. 2020. Malignancy and IFITM3: friend or foe? Front Oncol 10:593245. 10.3389/fonc.2020.593245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yan L, Ma J, Zhu Y, Zan J, Wang Z, Ling L, Li Q, Lv J, Qi S, Cao Y, Liu Y, Cao L, Zhang Y, Qi Z, Nie L. 2018. miR-24-3p promotes cell migration and proliferation in lung cancer by targeting SOX7. J Cell Biochem 119:3989–3998. 10.1002/jcb.26553. [DOI] [PubMed] [Google Scholar]
- 37.Gao Z, Zhou L, Hua S, Wu H, Luo L, Li L, Wang S, Liu Y, Zhou Z, Chen X. 2020. miR-24-3p promotes colon cancer progression by targeting ING1. Signal Transduct Target Ther 5:171. 10.1038/s41392-020-0206-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Khodadadi-Jamayran A, Akgol-Oksuz B, Afanasyeva Y, Heguy A, Thompson M, Ray K, Giro-Perafita A, Sanchez I, Wu X, Tripathy D, Zeleniuch-Jacquotte A, Tsirigos A, Esteva FJ. 2018. Prognostic role of elevated mir-24-3p in breast cancer and its association with the metastatic process. Oncotarget 9:12868–12878. 10.18632/oncotarget.24403. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1<br>. Download JVI.00680-21-s0001.pdf, PDF file, 0.09 MB (95.2KB, pdf)