Abstract
Mutations in PTEN-induced Kinase 1 (PINK1) leads to early-onset autosomal recessive Parkinson’s disease in humans. In healthy neurons, full-length PINK1 (fPINK1) is post-translationally cleaved into different lower molecular weight forms, and cleaved PINK1 (cPINK1) gets shuttled to the cytosolic compartments to support extra-mitochondrial functions. While numerous studies have exemplified the role of mitochondrially-localized PINK1 in modulating mitophagy in oxidatively-stressed neurons, little is known regarding the physiological role of cPINK1 in healthy neurons. We have previously shown that cPINK1, but not fPINK1, modulates neurite outgrowth and the maintenance of dendritic arbors by activating downstream Protein Kinase A (PKA) signaling in healthy neurons (Das Banerjee et al., 2017). However, the molecular mechanisms by which cPINK1 promotes neurite outgrowth remain to be elucidated. In this report, we show that cPINK1 supports neuronal development by modulating the expression and extracellular release of BDNF. Consistent with this role, we observed a progressive increase in the level of endogenous cPINK1 but not fPINK1 during pre-natal and post-natal development of mouse brains and during development in primary cortical neurons. In cultured primary neurons, pharmacological activation of endogenous PINK1 leads to enhanced downstream PKA activity, subsequent activation of the PKA-modulated transcription factor CREB, increased intracellular production and extracellular release of BDNF, and enhanced activation of the BDNF receptor TRKβ. Mechanistically, cPINK1-mediated increased dendrite complexity requires the binding of extracellular BDNF to TRKβ. In summary, our data support a physiological role of cPINK1 in stimulating neuronal development by activating the PKA-CREB-BDNF signaling axis in a feedforward loop.
Keywords: Parkinson’s Disease, Dendrite, Synaptic Plasticity, PKA-CREB signaling, PINK1 KO
Introduction
Parkinson’s disease (PD) is a chronic, progressive neurodegenerative disease caused by loss of dopaminergic (DA) neurons in the substantia nigra pars compacta (SNpc) of the midbrain. PD patients primarily suffer from motor symptoms, including bradykinesia, rigidity, and loss of posture/balance. However, a spectrum of non-motor symptoms, including cognitive decline, dominates the pathological outcome during advanced stages of PD (Gatt et al., 2016; Huang et al., 2019). Cognitive decline in PD ranges from mild cognitive impairment to PD dementia (PDD) (Aarsland and Kurz, 2010; Goldman et al., 2018). The primary pathology exhibited in PDD, besides DA neuronal degeneration, is the presence of Lewy body pathology coupled with the loss of structural plasticity in cortical and limbic structures (Hurtig et al., 2000; Apaydin et al., 2002; Bereczki et al., 2017). Impaired structural plasticity in DA neurons in the form of synaptic loss and retracted dendrites, disrupts voltage-gated ion channels, and reduces the propagation of action potentials, culminating in skewed cortico-limbic circuitry (McGregor and Nelson, 2019).
Autosomal recessive mutations in PINK1, a serine/threonine (ser/thr) kinase, causes early-onset PD in humans (Valente et al., 2004). PINK1 is targeted to the mitochondria for post-translational processing via the N-terminal region that contains a mitochondrial targeting sequence (MTS). In healthy mitochondria, PINK1 is imported, truncated, and released as cleaved-PINK1(cPINK1), which is subsequently targeted for proteasome-mediated degradation or retro-translocation into the cytosol (Lin and Kang, 2008). cPINK1 resides in both cytosolic and mitochondrial compartments. While the majority of cPINK1 resides in the cytosol compartment, a lesser fraction of cPINK1 is localized to the mitochondria (Dagda et al., 2014). In contrast, upon mitochondrial insult, PINK1 accumulates at the outer mitochondrial membrane (OMM) and initiates the recruitment of Parkin to depolarized mitochondria where it ubiquitylates multiple mitochondrial substrates to trigger mitophagy (Dagda et al., 2009; Vives-Bauza et al., 2010; Kondapalli et al., 2012; Lazarou et al., 2012). Although PINK1 function is predominantly associated with mitophagy, there is growing evidence to support a broader role of cPINK1 in mitochondrial retrograde signaling (Dagda et al., 2014; Das Banerjee et al., 2017). Mitochondrial retrograde signaling through mROS, NADPH oxidase, cardiolipin, tricarboxylic acid cycle metabolites, and processed proteins govern essential physiological functions in cells including transcriptional regulation, epigenetic modification, immunological response, apoptosis, mitochondrial biogenesis, antioxidant responses, and cell survival (Chandel et al., 1998; Haque et al., 2008; Murata et al., 2011; Ellis et al., 2013; Ko et al., 2016).
BDNF is an essential growth factor that modulates critical neuronal functions, including development of neuronal circuits, modulating neuronal survival, enhancing the arborization of dendrites in developing neurons, and the formation and maturation of dendritic spines (Bramham and Messaoudi, 2005; Numakawa et al., 2010). BDNF is produced initially as a precursor form, a pro-BDNF (30–35 kDa) before it is proteolytically cleaved into a mature BDNF form (12–13 kDa) (Mowla et al., 2001) that can bind to its cognate neurotrophin receptor Trkβ. PKA is a cAMP-dependent master regulator of neuronal processes, including neuronal development, synaptic plasticity, and dopamine synthesis (Nishi et al., 2011). When bound to cAMP, the catalytic subunits of PKA are released to catalyze the phosphorylation of the PKA-modulated transcription factor CREB. Phosphorylated CREB (p-CREB) subsequently translocates from the cytosol to the nucleus to bind to cyclic AMP response elements (CRE) in CRE-regulated genes to promote the transcription of several proteins involved in neuronal plasticity, including BDNF, tyrosine hydroxylase, cytoskeletal proteins (e.g., MAP2B) and other neuronal survival genes (e.g., cfos) (West et al., 2002; Lewis-Tuffin et al., 2004; Nam et al., 2015). Enhanced BDNF levels directly contribute to synaptic strength by increasing the availability of PSD95 at the postsynaptic sites (Hu et al., 2011).
While our published studies suggest that cPINK1, not the mitochondrial pool of PINK1, promotes anterograde trafficking to enhance the level of mitochondrial content in dendrites by activating downstream PKA signaling in the mitochondrion. However, PINK1-mediated enhancement of PKA activity at the mitochondrion alone is insufficient to promote dendrite outgrowth. Therefore, an investigation into the molecular mechanisms by which PINK1 stimulates dendritic outgrowth is imminent. Here, we report how endogenous cPINK1 stimulates neuronal development by modulating BDNF signaling via downstream activation of PKA in healthy neurons.
Materials and Methods
Experimental animals:
All in vivo studies were performed following the National Institutes of Health Office of Laboratory Animal Welfare Policy and Institutional Animal Care and Use Committee (IACUC) at the University of Nevada, Reno (protocol #0572). In brief, C57Bl/6J (Charles Rivers Laboratory, Reno, NV, USA) and PINK1-knockout (PINK1-KO) (Catalog#: B6.129S4-Pink1tm1Shn/J, RRID:IMSR_JAX:017946, Jackson Laboratories, Bar Harbor, ME) mice were maintained (2–4 in number/cage) under controlled temperature 25–26 °C, 12/12 h light/dark cycle, a humidity of 30%–70% (mean 40%) with food and water ad libitum. No physical enrichment was provided. Cages were changed once per week with minimal handling. To verify the genotype of animals (Suppl. Fig. 1A), the genomic DNA was extracted from tail snips, and the tissue was digested with proteinase K as previously published (Bonaparte et al., 2013). After verifying that the quality and yield of genomic DNA were acceptable, PCR was performed using primers (WT R: ACTGCCACACTCAGTCCTTG; Common F: GCACTACAGCGAACTGCATC; Mutant R: GCCAGAGGCCACTTGTGTAG) used for corroborating the genotype of PINK1-KO and WT mice. Equal number of male and female mice were incorporated into the respective experimental groups.
Culturing primary neurons and SH-SY5Y cells:
Primary cortical neurons (PCNs) were prepared from 14–15 day C57BL/6 and PINK1-KO mouse embryos as previously described (Das Banerjee et al., 2017). Briefly, the embryos were individually harvested and placed into separate wells containing dissection media [DMEM (Thermo Fisher Scientific, Waltham, MA), 5% FBS (Sigma-Aldrich, St. Louis, MO), and 0.5mM Glutamax (Thermo Fisher Scientific, Waltham, MA)]. The cortices were micro-dissected from the embryos and placed into ice-cold plating media [Neurobasal media (Thermo Fisher Scientific, Waltham, MA), 2% FBS, 2% B27 (Thermo Fisher Scientific, Waltham, MA), 0.5mM Glutamax, 25μM Glutamic acid (Sigma-Aldrich, St. Louis, MO), 100 U/ml penicillin/streptomycin (Thermo Fisher Scientific, Waltham, MA)] and mechanically dissociated with repeated pipetting with a 1mL pipettor. The cell density was determined using a hemocytometer, and subsequently, primary cells were plated to poly-L-Lysine (P4832; Thermo Fisher Scientific, Waltham, MA) coated sterile cell culture plates [Lab-Tek™ IV Chamber Slides (0.5ml media/well)- 2 X 106 cells/mL; 12 well plate (0.5ml media/well)- 2 X 106 cells/mL; 6 well plate (2ml media/well)- 3.5 X 105cells/mL] in prewarmed plating media. The plates containing PCNs were maintained at 37°C, 5% CO2 / 95% humidity in a cell culture incubator. Approximately, 1/3rd of the medium was then changed every three days and replaced by fresh serum-free maintenance media [Neurobasal media (Thermo Fisher Scientific, Waltham, MA), 2% B27 (Thermo Fisher Scientific, Waltham, MA), 0.5mM Glutamax (Thermo Fisher Scientific, Waltham, MA)]. Primary hippocampal neurons were prepared from 15–16 day C57BL/6 and PINK1-KO mouse embryos and further processed as previously described (Merrill et al., 2011a).
Parental SH-SY5Y neuroblastoma cells were grown in culture media (DMEM, 0.5mM Glutamax, and 10% FBS) in cell culture plates [Lab-Tek™ IV Chamber Slides (0.5ml media/well)- 6 X 104 cells/mL; 6 well plate (2ml media/well)- 1 X 105 cells/ml] maintained at 37°C, 5% CO2 / 95% humidity in a cell culture incubator. For specific experiments, SH-SY5Y cells were differentiated with 10μM retinoic acid (RA) for up to three days (Sigma-Aldrich, St. Louis, MO). All experiments requiring the use of ATTC approved cell lines and treatment/transfection of cells with pharmacological compounds, reagents, and recombinant DNA was performed per a Memorandum of Understanding and Agreement on the Use of Biological Agents and Recombinant DNA (MOUA #B2016–06, expiration date: 03–15-2022) that was approved by the University of Nevada, Reno Institutional Board Committee.
Treatment of cells with compounds:
Cells were treated with following drugs: PINK1 activator- kinetin (50μM) [K3253 (Sigma-Aldrich, St. Louis, MO)], PKA activator-forskolin (10μM) [344273 (Sigma-Aldrich, St. Louis, MO)], human recombinant BDNF (20ng/ml) [B3795 (Sigma-Aldrich, St. Louis, MO)], Mitochondrial uncoupler- FCCP (20μM) [C920 (Sigma-Aldrich, St. Louis, MO)], 500μM cAMP [A9501 (Sigma-Aldrich, St. Louis, MO)], TRKβ receptor antagonist- ANA12 (1μM) [4781 (Tocris Bioscience, Bristol, UK)], MEK inhibitor- UO126 (5μM) [1144 (Tocris Bioscience, Bristol, UK)], PI3K inhibitor- LY294002 (25μM) [1130 (Tocris Bioscience, Bristol, UK)], PKA inhibitor- H89 (10μM) [B1427 (Sigma-Aldrich, St. Louis, MO)].
Plasmids used for transfection in primary neurons and SH-SY5Y cells:
C-terminally fused PINK1 (PINK1-GFP) and C-terminally Flag-tagged PINK1 (PINK1–3xFlag) were purchased from Genecopoeia (Rockville, MD) and characterized as previously published (Dagda et al., 2014). The DNA plasmids that encode GFP targeted to mitochondria (OMM-GFP), C-terminally GFP tagged inhibitor of PKA targeted to the OMM (Mito-PKI-GFP) were provided by Dr. Stefan Strack (University of Iowa, Department of Pharmacology) (Merrill et al., 2011b). A pathogenic PD-associated mutant of full-length PINK1 (G309D) was generated in the PINK1-GFP vector (pReceiver M03)(Kessler et al., 2005). An N-terminal truncation mutant of human PINK1 cDNA lacking the residues 1–111 (Haque et al., 2008) was generated via PCR amplification from the full-length wild-type PINK1–3X-Flag pReceiver M03 vector full-length sequence and the resulting amplicons were ligated upstream of GFP in the pEGFP-N1 vector (ΔN111-PINK1-GFP). Based on subcellular fractionization/Western blotting assays, ΔN111-PINK1-GFP was observed to be predominantly localized to the cytosolic compartments with a minor fraction anchored to the OMM as previously published (Dagda et al., 2014). A catalytic kinase-inactive construct of ΔN111-PINK1(K219M) was generated from ΔN111-PINK1 in the pReceiver M03 construct by using the QuickChange Site-Directed Mutagenesis kit (Stratagene, San Diego, CA) and was characterized for lack of neuroprotective and dendrite-growth promoting activity as previously published (Dagda et al., 2014). Wild type and K219M versions of PINK1-V5 were generously provided by Dr. Anurag Tandon (University of Toronto)(Petit et al., 2005). Cells were transfected with the DNA plasmids listed above by using Lipofectamine 2000 as previously described but with the following modifications (Dagda et al., 2014; Das Banerjee et al., 2017). For primary neurons (cortical and hippocampal ) grown in twelve well tissue culture plates and SH-SY5Y cells grown in Lab-Tek™ IV Chamber Slides, 1μg of DNA plasmid was diluted in OPTIMEM media (Thermo Fisher Scientific, Waltham, MA) and mixed with Lipofectamine- at a final concentration of 1%. Cells were subsequently treated with combined and diluted DNA: Lipofectamine mixture. 24h following transfection, up to 2/3rd of the media was changed with pre-warmed plating/cell culture media.
Antibodies:
The following primary antibodies were used for immunocytochemistry: BDNF (1:100 in 2.5% BSA & TBST; Abcam Cat# ab108319, RRID:AB_10862052), CREB (1:200 in 2.5% BSA & TBST; Cell Signaling Technology Cat# 9197, RRID:AB_331277), Phospho-CREB (Ser133)(1:100 in 2.5% BSA & TBST; Cell Signaling Technology Cat# 9198, RRID:AB_2561044), PSD 95 (K28/43)(1:100 in 2.5% BSA & TBST; Antibodies Incorporated Cat# 75–028, RRID:AB_2292909), Synaptophysin (1:200 in 3% BSA & TBST; Abcam Cat# ab14692, RRID:AB_301417), GFP(1:200 in 3% BSA & TBST; Rockland Cat# 600–101-215, RRID:AB_218182), MAP2A-MAP2B (1:200 in 3% BSA & TBST; Millipore Cat# MAB378, RRID:AB_11214935). The following primary antibodies were used for Western blotting analysis of the following proteins of interest: PINK1 (1:500 in 5% milk & TBST; Novus Cat# BC100–494, RRID:AB_10127658), β-Tubulin (1:1000 in 5% BSA & TBST; Abcam Cat# ab6046, RRID:AB_2210370), BDNF (1:1000 in 5% BSA & TBST; Abcam Cat# ab108319, RRID:AB_10862052), CREB (1:500 in 5% BSA & TBST; Cell Signaling Technology Cat# 9197, RRID:AB_331277), Phospho-CREB (Ser133)(1:750 in 3% BSA & TBST; Cell Signaling Technology Cat# 9198, RRID:AB_2561044), PSD 95 (K28/43)(1:1000 in 3% BSA & TBST; Antibodies Incorporated Cat# 75–028, RRID:AB_2292909), Synaptophysin (1:1000 in 3% BSA & TBST; Abcam Cat# ab14692, RRID:AB_301417), GFP(1:1000 in 3% BSA & TBST; Rockland Cat# 600–101-215, RRID:AB_218182), MAP2A-MAP2B (1:750 in 3% BSA & TBST; Millipore Cat# MAB378, RRID:AB_11214935), TRKβ (1:250; 5% Milk & TBST; Abcam Cat# ab18987, RRID:AB_444716), Phospho-TRKβ (Tyr515) (1:250 in 5% BSA & TBST; Thermo Fisher Scientific Cat# PA5–38076, RRID:AB_2554679), PKA C (pT197) 1:1000; 3% BSA; (Cell Signaling Technology Cat# 4781, RRID:AB_2300165), LC3 B (1:1000, 3% BSA, Thermo Fisher Scientific Cat# PA1–46286, RRID:AB_2234770).
Immunohistochemistry:
Mouse brain sections were prepared for immunohistochemistry, as previously described (Das Banerjee et al., 2017). Briefly, mice were anesthetized by 5% isoflurane induction and transcardially perfused at a rate of 2ml/min with 30 ml ice-cold PBS followed by 4% ice-cold PFA. The mice were then euthanized by decapitation with the whole brain quickly removed and fixed in 4% paraformaldehyde (PFA) at 4°C for 48h and followed by sequential incubation in 10%, 20%, and 30% sucrose at 4°C overnight. Fixed mouse brains were frozen in OCT mounting media at −80°C. Midbrain sections (10 μm) were prepared from frozen blocks using a cryostat (CM1510-S, Leica Microsystems Inc, Germany), and sections were applied to pre-coated slides (Superfrost Plus GoldMicroscope Slides, Fisher Scientific, Hampton, NH) and stored in −20 °C until further use. Mid-brain sections were air-dried, rinsed with PBS, and re-fixed with 4% PFA for 1h. Post-fixated brain sections were blocked in 5% bovine serum albumin [BSA, (Sigma-Aldrich, St. Louis, MO)], PBS, and 0.1% Triton X-100 (PBST) for 1 h at room temperature (RT). The midbrain sections were incubated in primary antibodies for 24h at 4°C, washed four times in PBST (10 min per wash), followed by incubation in secondary antibodies for 2h at RT. Brain sections were subsequently mounted in ProLong™ Gold Antifade mounting media (Thermo Fisher Scientific, Waltham, MA) in order to preserve the brain sections. A cover slip was placed on the slide and imaged using an EVOS-FL Cell Imaging System (Thermo Fisher Scientific, Waltham, MA) equipped with EVOS Light cubes specific for GFP (Ex/Em of 470/510), RFP (Ex/Em of 531/593) and Cy5 (Ex/Em of 655/794), at a magnification of 20X (0.45NA) or 40X (0.60NA). The captured images were analyzed using NIH ImageJ software version 1.44 (Bethesda, MD). 10–15 sections from midbrain derived from 4–5 animals per genotype (WT & PINK KO) were immunostained for TH and BDNF to count the number of BDNF positive dopaminergic neurons per epifluorescence field. Up to 200–300 BDNF co-stained with TH neurons were analyzed per genotype. Importantly, the complete SN was analyzed including the SN pars compacta, SN pars reticulata and SN pars lateralis as a validated method to assess for neurodegeneration of midbrain dopamine neurons in PD models as previously published (Grigoruta et al., 2020) and excluding the ventral tegmental area (VTA).
Immunocytochemistry:
PFA-fixed primary cortical and hippocampal neurons were immunolabeled as previously described (Das Banerjee et al., 2017), but with the following minor modifications. Briefly, dissociated cultured primary neurons were fixed in 4% PFA and then permeabilized with PBST for 15 min. Subsequently, cells were blocked in 2.5% BSA for 1h, incubated in primary antibodies overnight at 4°C, washed four times in PBST (5–10 min/wash), and subsequently incubated in applicable secondary antibody for 1h at RT. Cells were then counter-stained with 4′,6-diamidino-2-phenylindole (DAPI) at a concentration of 1.25μg/ml). The cells were imaged by EVOS-FL Cell Imaging System (Thermo Fisher Scientific, Waltham, MA), ImageXpress Nano automated microscope (Molecular Devices, San Jose, CA) and FluoView 1000 laser scanning confocal microscope (Olympus America Inc., Melville, NY).
Western blotting:
Cells were harvested and lysed on ice using lysis buffer [50 mM Tris-HCl, 50 mM -GP,1mM EGTA, 1 mM EDTA, 1% Triton X, 10 mM NaF, 1 mM dithiothreitol, 0.5 mM NaVO3, 1X protease inhibitor cocktail (Roche, Basel, Switzerland)]. Cell lysates were then centrifuged for 15 min at 13,000 x g, and the resulting supernatants were transferred to sterile 1.5 mL microcentrifuge tubes. The protein amount in cell lysates was determined by employing the Pierce BCA Protein Assay Kit per manufacturer’s instructions (Thermo Fisher Scientific, Waltham, MA). Sample buffer [5% Mercaptoethanol in 4X Laemmli buffer (Bio-Rad Laboratories, Hercules, CA)] was added to each of the cell lysate samples and boiled for 5 min at 90°C. Proteins were then resolved in SDS/PAGE electrophoresis using polyacrylamide gels composed of acrylamide percentages, per the molecular weight of the proteins of interest to be analyzed (7.5%, 10%, 12.5 %). The separated proteins were transferred electrophoretically onto a PVDF membrane using a semi-dry transfer system (BioRad Laboratories, Hercules, Ca) at 250 mA for 45 min. The PVDF membrane was then blocked in TBST [20 mM Tris, pH 7.5, 150 mM NaCl, 0.05% Tween 20 (v/v)] containing 5% skimmed milk or with 2–5% BSA for 2h at RT with gentle shaking on an orbital rocker. The PVDF membranes were then incubated overnight at 4 °C with primary antibodies. Following incubation with primary antibodies, the PVDF membranes were rinsed in TBST and subsequently incubated with the respective horseradish peroxidase (HRP)-conjugated secondary antibodies for 2 h at RT. Chemiluminescence substrate kit- SuperSignal™ Western Blotting Substrate was used to detect immunoreactive proteins and were visualized by a detector [ChemiDoc™MP Imaging System (170–8280) Bio-Rad Laboratories, Hercules, CA]. Densitometric analysis of immunoreactive bands in technical replicates of each protein marker of interest was performed by using NIH ImageJ (version 1.44–1.50i) software (NIH, Bethesda, MD) as previously published (Das Banerjee et al., 2017). The integrated density for each immunoreactive band of interest was normalized to β-tubulin.
ELISA:
To analyze the effects of pharmacologically activating PINK1 (by treating PCNs with kinetin) on the extracellular release of the mature form of BDNF at various time points, 2ml of cell culture medium was collected from untreated or 50μM kinetin treated PCNs and examined for the presence of released BDNF by ELISA. In brief, to prevent the degradation of BDNF, the collected medium was treated with a cell culture specific protease inhibitor cocktail (P1860, Sigma-Aldrich, St. Louis, MO) at a final concentration of (100x). ELISA was performed by employing the DuoSet ELISA development system as per the manufacturer’s instructions (DY248, R&D Systems, Minneapolis, MN). Briefly, polystyrene flat bottom 96 well plates (Thermo Fisher Scientific, Waltham, MA) were coated with 100μl monoclonal capture antibody (mouse anti-human/mouse BDNF; R&D Systems, Minneapolis, MN) and incubated at 4°C overnight. The following day, the plates were blocked with 1% BSA (R&D Systems, Minneapolis, MN) in PBS (pH 7.2–7.4) for 2 h, subsequently incubated with the samples of cell culture media for 1 h. after which the plate was incubated with biotinylated secondary antibody (R&D Systems, Minneapolis, MN), respectively. Following the incubation with biotinylated secondary antibodies, the amount of released BDNF in each well was calorimetrically detected by incubating the plate with streptavidin, horseradish peroxidase conjugate in the presence of 3,3’,5,5’- tetramethylbenzidine substrate (Sigma-Aldrich, St. Louis, MO) for 2 h. The colorimetric reaction was then stopped by incubating the plate with 2 N sulphuric acid, and the amount of color formation was read at an optical density (OD) of 450nm by using an M3 SpectraMax plate reader (Molecular Devices, San Jose, CA). The ELISA experiments were conducted in technical replicates. To determine the absolute amount of BDNF detected for each experimental condition, a standard curve for BDNF was prepared by using exogenous human recombinant BDNF standards provided by the manufacturer. The abundance of BDNF in experimental groups were normalized to Control (DMSO treated cells) and the results were expressed as a percentage fold change of BDNF from the standard curve.
Image acquisition and analysis:
To analyze for dendrite length, MAP2B immunolabeled primary neurons were imaged by using an EVOS-FL Cell Imaging System (technical capabilities described above) by capturing at least 10–15 representative regions of interest containing at least ten cells per well at a magnification of 20X. Neurite lengths were analyzed by using the NIH Image J plug-in program ‘NeuronJ’ (Erik Meijering, Biomedical Imaging Group Rotterdam, Netherlands) as previously described (Dagda et al. 2014). The experimentalist performing imaging was blinded to the experimental groups. Neurite length was determined in at least 25–30 neurons per experiment. For analysis of the mean fluorescent intensity and counting the number of punctae of the postsynaptic marker PSD95 and presynaptic marker synaptophysin in neurons, fluorescent images were collected by using a Fluoview1000 laser‐scanning confocal microscope running Fluoview FV10‐ASW (Olympus Corporation, Tokyo, Japan), by using a 60X oil objective [(1.42 numerical aperture) excitation/emission filter: 488 nm/510 nm; 561 nm/592 nm] and by employing a digital zoom factor of (2X). Given that dendrites and axons can traverse multiple plains of view and show variable thickness in dissociated culture primary neurons, 3D reconstructions were performed by confocal microscopy by acquiring stacks of fluorescent images by using the following parameters: Z‐steps of 0.15–2 μm, which yielded a volume of approximately 24 μm thick (range 11.5–36 μm). 3D projections were prepared from the fluorescent image stacks by employing the maximum intensity projection algorithm using ImageJ. In order to analyze the number of BDNF puncta, immunolabeled PCNs cultured on 96 well tissue culture plates were imaged at 40X by employing an ImageXpress Nano automated microscope (Molecular Devices, San Jose, CA) equipped with DAPI, FITC, and CY5 filter sets. The ImageXpress Nano automated microscope captured up to twelve regions of interest at predetermined positions for each well for up to 8–12 wells per experimental condition. It is worth noting that a single field of view at a magnification of 40X was able to capture up to 120 cells for each experimental condition, which facilitated statistical analysis in a relevant number of cells. Automated analysis of BDNF puncta per cell was analyzed using the Cell Scoring analysis module of the MetaXPress software at the following configurations (Approx. minimum width=0.5μm. Approx. max width=2μm, intensity above local background=200 gray levels).
Statistical analysis:
Unless indicated otherwise, results are expressed as mean ± SEM from three independent experiments. Data were analyzed by Student’s t-test (two-tailed) for pairwise comparisons, whereas multiple group comparisons were made by performing a one-way ANOVA followed by Bonferroni-corrected Tukey’s test and Dunnett’s multiple comparison test, using the GraphPad Prism software (version 6.0). P-values less than 0.05 were considered statistically significant.
Results
The endogenous level of cleaved PINK1 coincides with markers of neuronal development
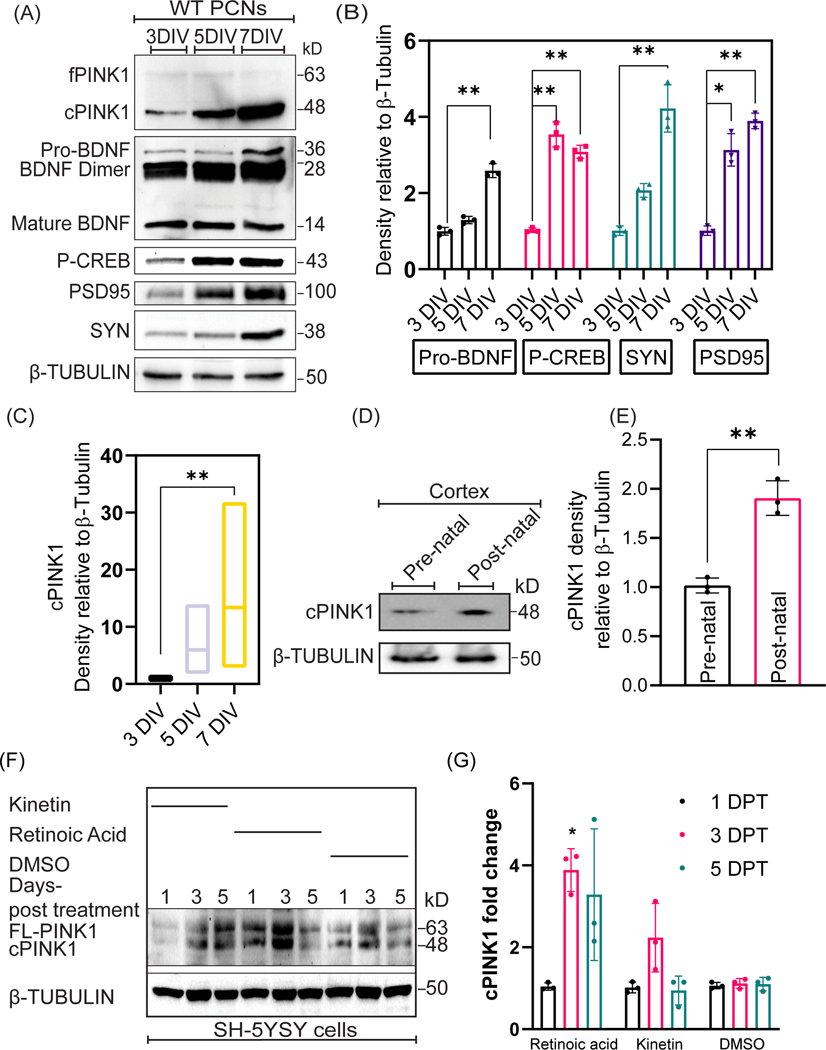
BDNF is a neuropeptide that governs differentiation, development, protection, and maintenance of neurons (Kowiański et al., 2018). Given that both cPINK1 and BDNF mediate dendrite outgrowth in developing neurons by eliciting downstream PKA (Das Banerjee et al., 2017; Kowiański et al., 2018), we hypothesized that the cleaved pool of PINK1 (cPINK1: 54 and 48 kDa) promotes dendrite outgrowth via BDNF by upregulating PKA signaling. To address this hypothesis, we first examined whether the expression of endogenous PINK1 is developmentally regulated in-vivo and in cultured PCNs. The expression profile of the different post-translationally processed forms of PINK1 throughout neuronal development was analyzed by immunoblotting the cell lysates collected from 3, 5, and 7 days in-vitro (DIV) PCNs. While the level of expression of endogenous full-length PINK1 (fPINK1, ~66kDa) was low and invariable throughout neuronal development (3–7DIV), a significant and progressive increase in cleaved PINK1 (cPINK1, ~48kDa) (F=8, N=3, p=0.0396), expression was observed throughout neuronal development of PCNs (Fig. 1A & 1C). Invariably, we also observed an enhanced expression of cPINK1 (F=5, N=3, p-value=0.0013) in the post-natal mouse cortex when compared to the pre-natal mouse cortex (Fig. 1D & 1E). Given the serendipitous finding that cPINK1 expression levels are significantly increased in response to neuronal development relative to fPINK1, we wanted to assess the specificity of the anti-human PINK1 antibody, as well as the abundance and distribution of endogenous cPINK1, by performing a battery of immunocytochemical and biochemical assays as further described below. By immunocytochemistry, we observed that endogenous PINK1 is localized throughout the dendrites and the soma of paraformaldehyde fixed PCNs. The immunoreactivity detected by the anti-human PINK1 antibody is specific for PINK1 as PCNs transfected with siRNAs specific for PINK1 significantly reduced this immunoreactivity by Immunocytochemistry (Suppl. Fig. 2A). Furthermore, Western blot assays revealed that endogenous cPINK1 can be immunodepleted by using an anti-PINK1 peptide (Suppl. Fig. 2B), suggesting that the anti-PINK1 antibody used in our assays specifically detects cPINK1. In accordance with prior studies (Das Banerjee et al., 2017), we observed that cPINK1 is localized to both cytosolic and mitochondrial fraction with over more than 75% of PINK1 residing in the cytosol suggesting that PINK1 predominantly resides in the cytosol whereas a minor fraction is bound to mitochondria (Suppl. Fig. 3). Neuronal maturation involves the formation of functional synapses, that occurs between 5–7 days in-vitro (Lesuisse and Martin, 2002). Consistently, our Western blotting data shows that PCNs show increased expression of synaptic markers such as synaptophysin (F=5, N=3, p-value=0.0041) and PSD95 (F=5, N=3, p-value=<0.0001) at 7 DIV (Fig. 1A & 1B). The neurotrophic factor BDNF stimulates neuronal development in part by activating the PKA holoenzyme, which subsequently phosphorylates the transcription factor CREB on serine133 to elicit the transcription of genes that are critical for neuronal development and survival (Delghandi et al., 2005). Our Western blot data show that the endogenous level of BDNF (F=5, N=3, p-value=0.0020) and phosphorylation of p-CREB (Ser133) (F=5, N=3, p value=0.0011) (Fig. 1A & 1B) progressively increased during neuronal development in-vitro.
Figure 1. The level of cleaved PINK1 coincides with markers of neuronal development.
A. Representative Western blotting for the indicated proteins and phosphoprotein in cell lysates extracted from 3, 5, and 7 DIV WT PCNs. B, C. Densitometric analysis of the Western blotting data shown in A on the level of Pro-BDNF, Phospho-CREB, Synaptophysin, PSD95, and cPINK1 expression levels in 3, 5, and 7 DIV PCNs. Values were normalized to β-Tubulin. Ordinary one-way ANOVA and post-hoc analysis using Dunnett’s multiple comparison test and Sidak’s multiple comparisons test were used for statistical analysis. Bars denote the average ± SEM and are representative of three independent experiments. D. Representative Western blotting of cPINK1 in tissue lysates extracted from pre-natal and post-natal embryo cortices. E. Densitometric analysis of the Western blotting data shown in D cPINK1 expression levels in pre-natal and post-natal embryo cortices. Values were normalized to β-Tubulin. An unpaired t-test was used for statistical analysis. Bars denote the average ± SEM and are representative of two independent experiments (N=4 mice per genotype). F. Representative Western blotting for the indicated proteins in lysates extracted at differential time-points from DMSO, kinetin, and RA treated SH-5YSY cells. G. Densitometric analysis of the Western blotting data shown in F pertaining to cPINK1 expression levels at 1, 3, 5 days post-treatment with DMSO, kinetin, and RA treated SH-5YSY cells. Values were normalized to β-Tubulin. Ordinary one-way ANOVA and post-hoc analysis using Tukey’s multiple comparisons test were used for statistical analysis. Bars denote the average ± SEM and are representative of three independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001.
To investigate the possible role of PINK1 in neuronal differentiation, we analyzed the effects of pharmacologically induced differentiation of SH-SY5Y neuroblastoma cells, a widely used human midbrain DA cell line used to study neuronal development and neurodegeneration in the context of PD (Xicoy et al., 2017), on the endogenous levels of fPINK1 and cPINK1. In a similar manner to our in-vivo data, the level of endogenous cPINK1 is significantly increased in SH-SY5Y neuroblastoma cells treated with RA, a well-characterized inducer of neuronal differentiation, or with exposure to kinetin, a pharmacological activator of endogenous PINK1 (Fig 1F & 1G). In summary, these results suggest that the expression profile of cPINK1 coincides with neuronal development and differentiation, presumably by activating downstream PKA signaling and the level of BDNF.
PINK1 modulates BDNF levels
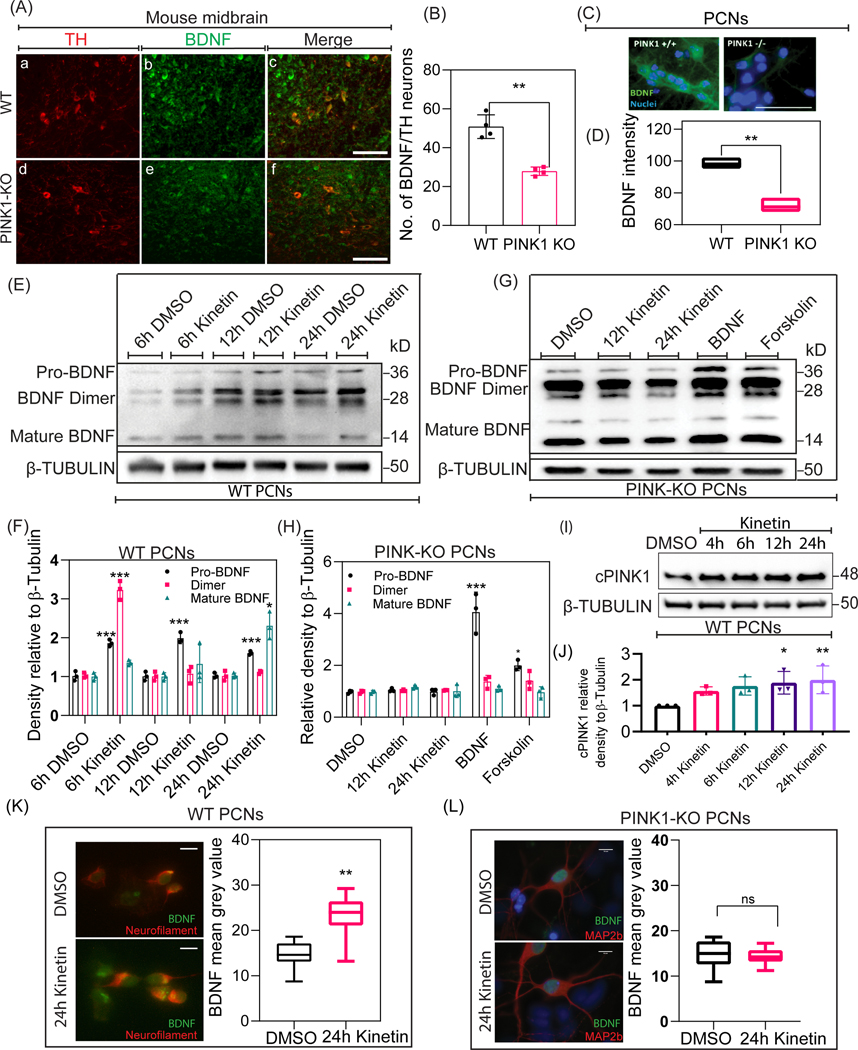
PINK1-KO mice exhibit reduced dopamine release, a retraction of dendrites, and mitochondrial dysfunction, resulting in neuronal plasticity abnormalities in the cortico-striatal pathway in the absence of overt neuronal death (Kitada et al., 2007a; Das Banerjee et al., 2017). Given that the level of endogenous PINK1 is developmentally modulated and coincides with the level of BDNF (Fig. 1A), we hypothesized that PINK1 regulates neuronal development by modulating the level of BDNF in a PKA-dependent manner. To address this hypothesis, we first measured endogenous BDNF levels by performing immunohistochemical analyses of midbrain sections derived from 10-month-old PINK1-KO and WT mice. Midbrain sections from PINK1-KO mice showed significantly reduced intracellular levels of BDNF (Fig. 2A & 2B) compared to midbrain sections from 10-month-old WT mice. Furthermore, consistent with our immunohistochemical analysis of BDNF levels, Western blot analysis revealed that the overall endogenous levels of immature and mature forms of BDNF are reduced in the cortex of 10 month old PINK1-KO mice relative to wild-type mice (Supp. Fig. 1B). It is worth mentioning that the mean number of TH neurons per field across multiple sections did not changed significantly (Suppl. Fig. 2C) in the midbrain in WT vs. PINK1-KO consistent with previous observations that 10-month-old PINK1-KO mice do not show significant loss of dopaminergic neurons (Kitada et al., 2007b). Additionally, immunocytochemical studies revealed a significant decrease in the endogenous level of intracellular BDNF in 5DIV PINK1-KO PCNs compared to WT PCNs (Fig. 2C & 2D). The precursor form of BDNF (~32kDa) is proteolytically cleaved to its mature form (~14kDa), which binds to its cognate receptor TRKβ to activate a multitude of downstream signaling pathways to regulate neuronal differentiation and survival. Hence, given that a lack of PINK1 is associated with the reduced endogenous level of intracellular BDNF, we surmised that a reduction in the intracellular level of BDNF delays neuronal development in PINK1-deficient PCNs. To address this hypothesis, we pharmacologically activated endogenous PINK1 by exposing PCNs to kinetin, a plant hormone, that is metabolized intracellularly into kinetin triphosphate (KTP) which acts as an ATP analog and specifically binds to the ATP-binding pocket of PINK1 to stimulate its kinase activity (Hertz et al., 2013). Western blotting analysis of cell lysates derived from PCNs exposed to kinetin (50μM) showed that pharmacological activation of endogenous PINK1 led to a significant time-dependent increase in the endogenous level of cPINK1 (F=14, N=3, 12h p-value= 0.0175; 24h p value= 0.0069) (Fig. 2I). Moreover, Western blotting data showed that exposure to kinetin enhanced the level of the mature form of BDNF (~14kDa) compared to untreated WT PCNs. Specifically, exposure of WT PCNs to kinetin significantly elevated the levels of the immature (28 and 36kDa) forms of BDNF by 6h (F=18, N=3, p-value=<0.0001), 12h (F=18, p-value=<0.0001) and 24h (F=18, p-value=0.0002) post-treatment compared to PCNs treated with DMSO (control). Likewise, the endogenous level of the mature form of BDNF (~14kDa) was significantly elevated after 24h kinetin treatment (F=18, N=3, p-value=0.0230) compared to DSMO treated control (Fig. 2E & 2F). In contrast, exposure of PINK1-deficient PCNs with kinetin failed to upregulate the expression of either the immature or mature forms of BDNF, indicating that an increased level of intracellular BDNF throughout neuronal development occurs in a PINK1-dependent manner. As expected, and consistent with the model that PKA regulates BDNF levels, treating PINK1-KO PCNs with human recombinant BDNF (F=14, N=3, p-value=<0.0001) or with forskolin (F=14, N=3, p-value= 0.0183), a pharmacological activator of PKA, for 24 h were able to increase the endogenous level of immature, but not the mature forms of BDNF (Fig. 2G & 2H). Moreover, immunocytochemical analysis of BDNF showed that exposure of 5 DIV PCNs to kinetin for 24 h led to a significant increase in the level of BDNF in the soma (F=29, N=30, p-value= 0.0194) compared to DMSO (Control)-treated PCNs (Fig. 2K). In contrast, no significant changes in the level of intracellular BDNF were observed in PINK1-deficient PCNs treated for 24 h with kinetin compared to DMSO (Control)-treated PCNs (Fig. 2L) suggesting that kinetin-induced increase in the level of intracellular BDNF requires endogenous PINK1. For the first time, our compiled data so far suggest that PINK1 modulates the level of BDNF in a kinase activity-dependent manner.
Figure 2. PINK1 modulates BDNF levels in neurons.
A, B. Representative epifluorescence images and quantification of BDNF (green) immunoreactivity in coronal sections of WT and PINK1-KO midbrains. Scale bar represents 50μm. An unpaired t-test was used for statistical analysis. Bars denote the average ± SEM and are representative of three independent experiments (N=4 mice per genotype; 10–15 brain sections analyzed per genotype). C, D. Representative epifluorescence images and quantification of BDNF (green) immunoreactivity in WT and PINK1-KO PCNs. Nuclei were counterstained with DAPI (blue). Scale bar represents 75μm. An unpaired t-test was used for statistical analysis. Bars denote the average ± SEM and are representative of three independent experiments (N=30; 10 neurons per genotype per experiment). E. Representative Western blotting of BDNF expression in cell lysates extracted at differential time-points from DMSO and kinetin treated WT PCNs. F. Densitometric analysis of BDNF expression levels after 6h DMSO, 6h kinetin, 12h DMSO, 12h kinetin, 24h DMSO, and 24h kinetin treatments in WT PCNs. Values were normalized to β-Tubulin. Ordinary one-way ANOVA and post-hoc analysis using Tukey’s multiple comparisons test were used for statistical analysis. Bars denote the average ± SEM and are representative of three independent experiments. G. Representative Western blotting of BDNF expression in cell lysates extracted at differential time-points from DMSO, kinetin, Human recombinant BDNF, and forskolin treated PINK1-KO PCNs. H. Densitometric analysis of BDNF expression levels after DMSO, 12h kinetin, 24h kinetin, Human recombinant BDNF, and forskolin treatments in PINK1-KO PCNs. Values were normalized to β-Tubulin. Ordinary one-way ANOVA and post-hoc analysis using Tukey’s multiple comparisons test were used for statistical analysis. Bars denote the average ± SEM and are representative of three independent experiments. I. Representative Western blotting of cPINK1 expression in lysates extracted at differential time-points from DMSO and kinetin treated WT PCNs. J. Densitometric analysis of the Western blotting data shown in I of cPINK1 expression in DMSO, 4h kinetin, 8h kinetin, 12h kinetin, and 24h kinetin treated WT PCNs. Values were normalized to β-Tubulin. Ordinary one-way ANOVA and post-hoc analysis using Tukey’s multiple comparisons test were used for statistical analysis. Bars denote the average ± SEM and are representative of three independent experiments. K, L. Representative epifluorescence images and quantification of BDNF (green) immunoreactivity in WT and PINK1-KO PCNs. An unpaired t-test was used for statistical analysis. Scale bar represents 10μm Bars denote the average ± SEM and are representative of three independent experiments (N=30; 10 neurons per genotype per experiment). *P < 0.05, **P < 0.01, ***P < 0.001.
Cleaved but not full- length PINK1 modulates BDNF-mediated signaling
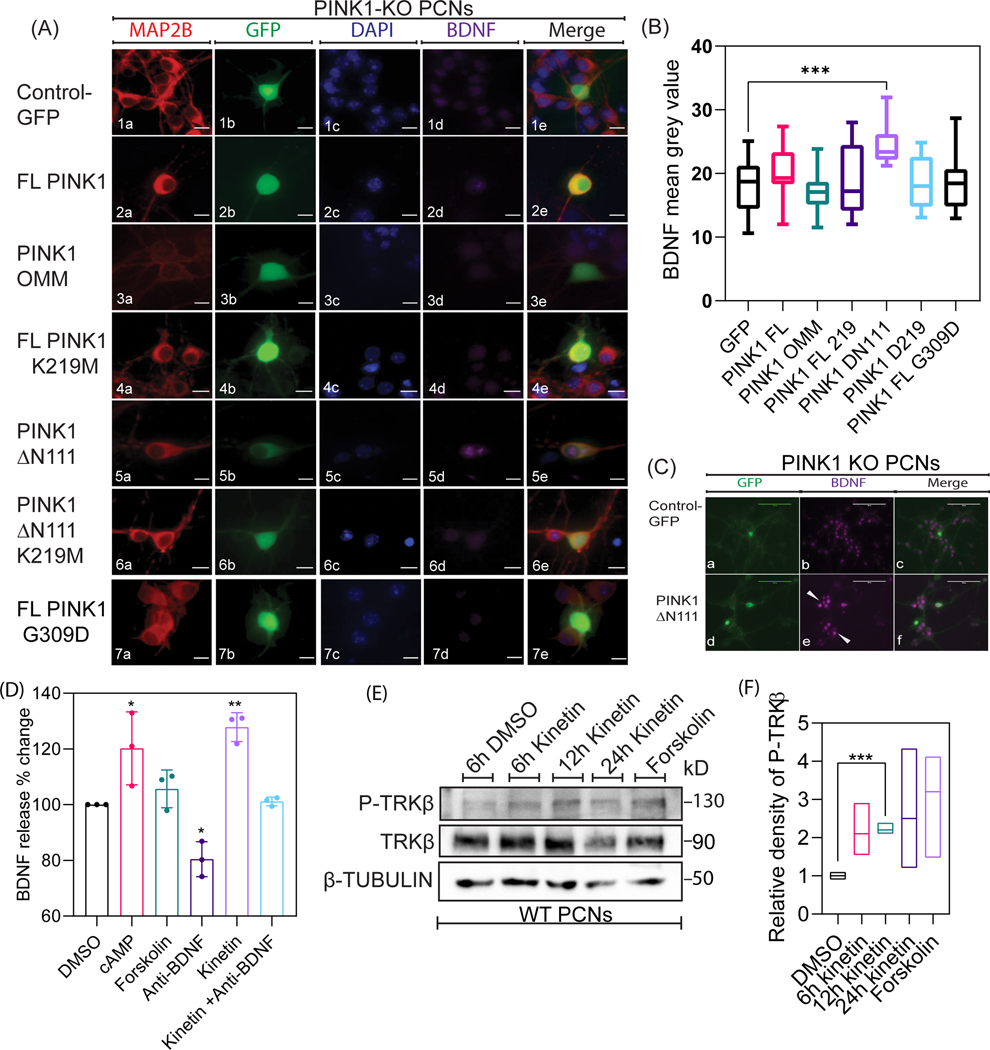
While a significant number of published research findings have increased our understanding of the functional roles of fPINK1 in the context of mitophagy, the physiological role of cPINK1 remains obscure. Specifically, cPINK1, which lacks the 111 amino acid-long N-terminal domain that includes its putative MTS sequence, is predominantly localized in the cytosol and to some extent to the OMM to exert extra-mitochondrial functions including enhancing anterograde trafficking of mitochondria to the distal region of dendrites, promoting neurite outgrowth and upregulating PI-3 kinase signaling to enhance neuronal survival (Haque et al., 2008; Das Banerjee et al., 2017; Furlong et al., 2019). Therefore, given the existence of mitochondrial and cytosolic pools of PINK1 (Suppl. Fig. 3), we transiently transfected PCNs with a series of GFP tagged constructs that included GFP as control, wild-type PINK1 (FL-PINK1), chimeric constructs designed to target PINK1 to the OMM via a TOM20 leader sequence (PINK1-OMM-GFP), catalytically inactive mutant (PINK1-K219M-GFP), ΔN111-PINK1-GFP to restrict PINK1 to the cytosol as it lacks the N-terminal MTS or the kinase dead ΔN111-PINK1-K219M-GFP and PD-associated PINK1 mutant (PINK1-G309D-GFP). Two days following lipofectamine-mediated transfection of DNA plasmids, we analyzed the level of intracellular BDNF via immunocytochemistry in PFA-fixed PCNs. While PINK1-deficient PCNs showed more than a 25% reduction in the intracellular level of endogenous BDNF (Fig. 2C & 2D), transiently expressing Δ111N-PINK1-GFP, but not a kinase dead version of Δ111N-PINK1 (K219M) significantly increased (F=149, N=30, p= 0.0006) the level of intracellular BDNF expression in PINK1-deficient PCNs compared to PINK1-deficient PCNs transiently expressing GFP or OMM-PINK1-GFP (Fig. 3A & 3B). In addition, we observed a non-significant increase in the intracellular level of endogenous BDNF expression in PCNs transiently expressing FL-PINK1-GFP compared to PCNs expressing GFP. In contrast, PINK1-KO PCNs transiently expressing the PD-associated pathogenic mutation K219M (FL-PINK1- K219M), which reduces its kinase activity, were unable to significantly increase the level of intracellular BDNF compared to PCNs transiently expressing GFP control. Overall, our immunocytochemistry data suggest that cPINK1, but not fPINK1, which is predominantly localized to mitochondria, modulates the level of intracellular BDNF in a kinase-dependent manner.
Figure 3. Cleaved but not full-length PINK1 modulates BDNF mediated signaling in neurons.
A. Representative epifluorescence images of MAP2B, GFP, and BDNF immunostaining in PINK1 KO PCNs transiently transfected with plasmids coding for Control (GFP), wild type full-length PINK1 (f-PINK1-GFP), OMM-anchored PINK1 (OMM-PINK1-GFP), catalytically inactive mutant (FL-PINK1-K219M-GFP), N-terminal cleaved PINK1 localized to the cytosol (ΔN111-PINK1-GFP, and an N-terminal cleaved kinase dead PINK1 localized to the cytosol) (ΔN111-PINK1-K219M-GFP), and PD associated PINK1 mutant (PINK1-G309D-GFP).. Nuclei were visualized by counterstaining with DAPI (blue). Scale bar represents 10μm. The arrow shows enhanced BDNF expression (magenta) in PINK1-KO PCNs transfected with ΔN111-PINK1-GFP relative to non-transfected PCNs. B. Quantification of mean BDNF expression in PINK1 KO PCNs transiently transfected with plasmids coding for Control (GFP), FL-PINK1, OMM-PINK1-GFP, FL-PINK1_K219M, PINK1-ΔN111-GFP, PINK1- ΔN111-K219M-GFP, and ΔN111-PINK1-G309D-GFP. Ordinary one-way ANOVA and post-hoc analysis using Sidak’s multiple comparisons test were used for statistical analysis. Bars denote the average ± SEM and are representative of three independent experiments. C. Representative epifluorescence images of GFP and BDNF immunostaining in PINK1 KO PCNs transiently transfected with a plasmid coding for ΔN111-PINK1-GFP. Scale bar represents 5μm. The arrow shows enhanced BDNF expression (magenta) in neurons adjacent to ΔN111-PINK1-GFP transfected PINK1 KO PCN. D. Graphical representation of BDNF expression using ELISA quantified from the media collected from WT PCNs transiently treated with DMSO, cAMP, Forskolin, Anti-BDNF antibody, 24h kinetin, and Anti-BDNF antibody+ 24h kinetin. Ordinary one-way ANOVA and post-hoc analysis using Sidak’s multiple comparisons test were used for statistical analysis. Bars denote the average ± SEM and are representative of three independent experiments. E. Representative Western blotting of Phospho-TRKβ and TRKβ expression in cell lysates extracted at differential time-points from DMSO, kinetin, and forskolin treated WT PCNs. H, Densitometric analysis of BDNF expression levels after DMSO, kinetin, and forskolin treated WT PCNs. Values were normalized to β-Tubulin. Ordinary one-way ANOVA and post-hoc analysis using Tukey’s multiple comparisons test were used for statistical analysis. Bars denote the average ± SEM and are representative of three independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001.
Furthermore, we observed an unexpected phenomenon. An increase in the level of BDNF expression was observed in the non-transfected PINK1-KO PCNs adjacent to Δ111N-PINK1-GFP expressing PINK1-KO PCNs when compared to GFP transfected PINK1-KO PCNs (Fig. 3C), raising the possibility that cPINK1 modulates the level of BDNF via an undescribed, non-cell-autonomous manner. Given that the mature form of BDNF is released to the extracellular space to bind to TRKβ leading to a PKA-mediated increase in the intracellular level of BDNF in a feedforward manner, we surmised pharmacological stimulating endogenous PINK1 by treating PCNs with kinetin will result in a significant increase in the extracellular release of endogenous BDNF to mediate dendritic development in developing neurons. To address this hypothesis, we performed ELISA to measure the level of extracellular BDNF in the medium from WT PCNs treated with a 24 h dose of vehicle control DMSO or with kinetin (50μM) for 24 h. Indeed, we observed a significant increase in the extracellular level of BDNF in the medium from WT PCNs treated with kinetin (F=14, N=3 p-value= 0.0017) or with cAMP for 24h. (F=14, N=3, p-value=0.0185). Conversely, treating WT PCNs with a BDNF-blocking antibody abolished (F=14, N=3, p-value= 0.0229) the ability of PINK1 to enhance the release of BDNF into the medium (Fig. 3D). Additionally, Western blotting data showed that exposure of PCNs to kinetin enhanced the activation of the TRKβ compared to untreated WT PCNs (Fig. 3E). Specifically, 12 h kinetin administration significantly (F=14, N=3, p-value=0.009) increased the phosphorylation of TRKβ (Tyr515) receptor when compared to DMSO treated control in WT PCNs (Fig. 3F). Thus far, our compiled data suggest that the cleaved, but not the mitochondrial pool of PINK1, modulates the intracellular level of BDNF, and is released to the extracellular space to influence neuronal development in a non-cell-autonomous manner, at least as shown in cultured PCNs.
Pharmacological activation of PINK1 promotes the activation of CREB
Given that PINK1 upregulates downstream cytosolic and mitochondrial PKA signaling and the fact that PKA partly modulates the level of endogenous BNDF, we next sought to determine the extent to which pharmacological activation of PINK1 in PCNs promotes the phosphorylation status of CREB, a substrate of PKA (Dagda et al., 2011; Dagda et al., 2014; Das Banerjee et al., 2017; Wang et al., 2018a). To this end, we performed immunofluorescent analysis on the PKA-mediated phosphorylation of CREB in PCNs treated with kinetin by double labeling PFA-fixed cells for p-CREB (Ser133) and MAP2B. We observed that the subcellular localization of p-CREB (Ser133) was significantly shifted from the cytosol to the nuclei of PCNs treated with kinetin for 6 h (F=14, N=30, p-value=0.0078) compared to DMSO (Control)-treated PCNs. At the same time, we did not observe a significant change in total CREB between kinetin treated and untreated PCNs (Fig. 4A–4D). To further corroborate our immunocytochemical data, we examined the phosphorylation status of CREB in PCNs treated with kinetin in a time-dependent manner by Western blotting. Indeed, consistent with our immunofluorescence data shown in figure 4(A–D), Western blotting data shows that the total protein level of CREB remained unchanged in response to kinetin treatment. On the other hand, the level of p-CREB (Ser133) significantly increased in PCNs treated with kinetin for 24h (F=14, N=3, p-value = 0.0064) (Fig. 4E & 4F). Also, it is worth noting that sustained levels of p-CREB (Ser133) could be attributed to latency in kinetin’s conversion to kinetin triphosphate (Hertz et al., 2013). In contrast, no changes in the level of CREB and p-CREB (Ser133) were observed in PINK1-deficient PCNs treated for 6, 12, and 24h with kinetin compared to DMSO (Control)-treated PCNs (Suppl. Fig. 4) suggesting that kinetin-induced increased phosphorylation of CREB requires endogenous PINK1.
Figure 4. Pharmacological activation of PINK1 promotes the activation of CREB.
A, C. Representative epifluorescence images of Phospho-CREB, CREB, and MAP2B immunostaining in WT PCNs treated with DMSO (Control) and 24h kinetin. Nuclei were visualized by counterstaining with DAPI. Scale bar represents 5μm. The arrow points to enhanced Phospho-CREB expression (red) in WT PCNs treated with kinetin compared to untreated PCNs. B, D. Quantification of Phospho-CREB expression levels in WT PCNs treated with DMSO (Control) and with 24h treatment with kinetin. An unpaired t-test was used for statistical analysis. Bars denote the average ± SEM and are representative of three independent experiments (N=30; 10 neurons per genotype per experiment). E. Representative Western blotting for the indicated proteins in cell lysates extracted at different time-points from DMSO, 4h kinetin, 8h kinetin, 12h kinetin, and 24h kinetin treated WT PCNs. F. Densitometric analysis of Phospho-CREB and CREB expression levels in from DMSO, 4h kinetin, 8h kinetin, 12h kinetin, and 24h kinetin treated WT PCNs. Values were normalized to β-Tubulin. Ordinary one-way ANOVA and post-hoc analysis using Dunnett’s multiple comparison test were used for statistical analysis. Bars denote the average ± SEM and are representative of three independent experiments.
Pharmacological activation of PINK1 supports neuronal plasticity
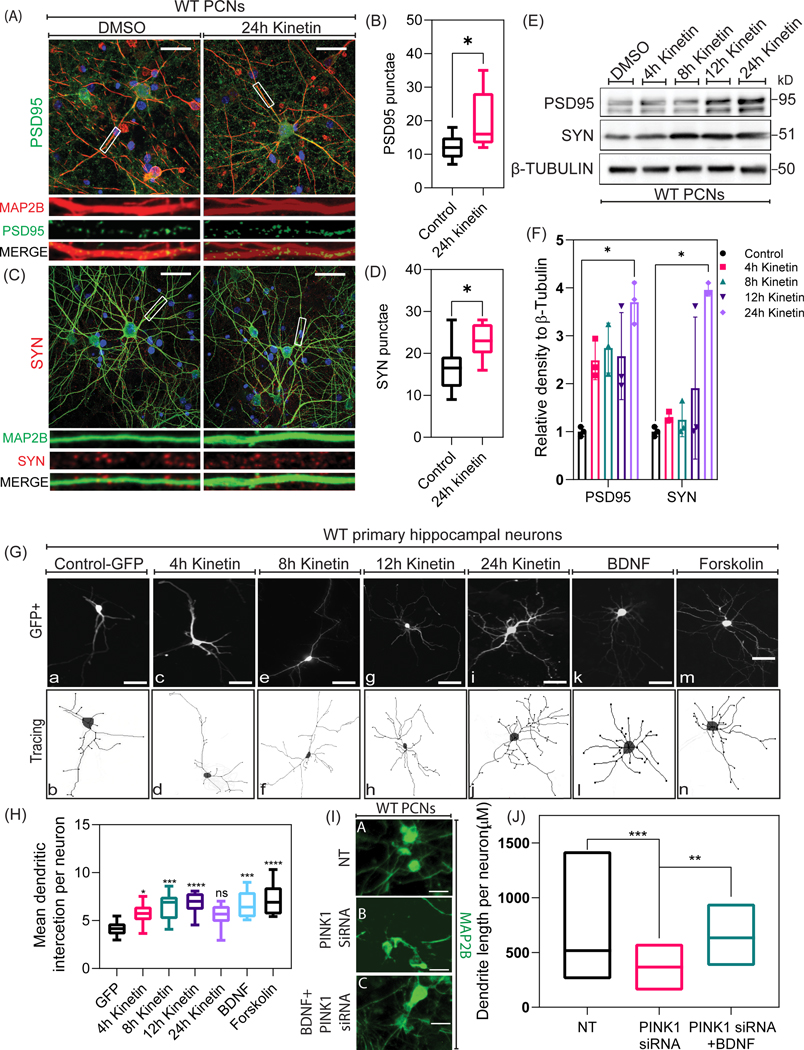
The developmental and pro-survival effects of BDNF in neurons have been well characterized (Genheden et al., 2015; Kowiański et al., 2018). Besides, it is worth noting that neuronal development in dissociated primary neurons physiologically resembles in-vivo neuronal development (Arlotta et al., 2005), including neurite outgrowth and neuronal maturation. Given that BDNF is a modulator of dendrite development and complexity, we then investigated the effects of pharmacological enhancement of PINK1 activity on synaptic connectivity in PCNs. By using confocal microscopy, we analyzed the amount of PSD95 puncta within MAP2B-positive dendrites in WT PCNs treated with kinetin at 24 h. Indeed, we observed a significant increase in the localization (F=34, N=30, p-value=0.0125) of PSD95 puncta within the dendrites of PCNs treated with kinetin compared to DMSO (Control) (Fig. 5A & 5B) but not in PINK1-deficient PCNs (Suppl. Fig. 5) suggesting that kinetin-mediated increase in PSD95 puncta requires endogenous PINK1. On the other hand, PINK1-KO PCNs were unable to respond to kinetin-stimulated increase in the number of PSD95 puncta within MAP2B-positive dendrites (Suppl. Fig. 5), suggesting that BDNF-enhanced differentiation of PCNs requires endogenous PINK1. Similarly, the level of the presynaptic marker synaptophysin was significantly increased near the dendrites (F=37, N=30, p-value=0.0220) in WT PNCs following the pharmacological activation of PINK1, suggesting that PINK1 mediates synaptogenesis in a kinase-dependent manner (Fig. 5C & 5D). To corroborate our immunocytochemical data biochemically, we performed Western blotting analysis of PSD95 and synaptophysin levels in cell lysates derived from PCNs treated at various time points with kinetin. Indeed, a significant time-dependent increase in the level of PSD95 (F=14, N=3, p-value=0.0134) and of synaptophysin (F=14, N=3, p-value= 0.0482) was observed in response to kinetin treatment at 24 h compared to DMSO (Control) treated WT PCNs (Fig. 5E & 5F) but not PINK1 KO PCN’s (Suppl. Fig. 5).
Figure 5. Pharmacological activation of PINK1 promotes neuronal plasticity.
A. Representative epifluorescence merged images of PSD-95 (green) immunostaining in WT PCNs treated with DMSO (Control) and 24h kinetin. Nuclei were visualized by counterstaining with DAPI (blue). Magnified images of PSD-95 (green) punctae on dendrites (MAP2b-red). Scale bar represents 5μm. B. Quantification of the endogenous level of PSD-95 in WT PCNs treated with DMSO (Control) and 24h kinetin. An unpaired t-test was used for statistical analysis. Bars denote the average ± SEM and are representative of three independent experiments (N=30; 10 neurons per genotype per experiment). C. Representative merged epifluorescence images of Synaptophysin (Red) immunostaining in WT PCNs treated with DMSO (Control) and 24h kinetin and co-immunolabeled for MAP2B. The nucleus is counterstained by DAPI (blue). Magnified images of Synaptophysin (Red) punctae around dendrites (MAP2b-green). Scale bar represents 5μm. D. Quantification of Synaptophysin level in WT PCNs treated with DMSO (Control) and 24h kinetin. An unpaired t-test was used for statistical analysis. Bars denote the average ± SEM and are representative of three independent experiments (N=30; 10 neurons per genotype per experiment). E. Representative Western blotting for PSD95 and synaptophysin in cell lysates extracted at differential time-points from DMSO (Control), 4h kinetin, 8h kinetin, 12h kinetin, and 24h kinetin treated WT PCNs. F. Densitometry analysis of the level of endogenous PSD95 and synaptophysin in DMSO (Control), 4h kinetin, 8h kinetin, 12h kinetin, and 24h kinetin treated WT PCNs. Values were normalized to β-Tubulin. Ordinary one-way ANOVA and post-hoc analysis using Dunnett’s multiple comparison test were used for statistical analysis. Bars denote the average ± SEM and are representative of three independent experiments. G. Representative epifluorescence images and traces of GFP immunostained WT PCNs transiently transfected with plasmids coding for Control (GFP) and treated with DMSO (Control), 4h kinetin, 8h kinetin, 12h kinetin, 24h kinetin, BDNF, and forskolin. Scale bar represents 10μm. H. Quantification of mean dendrite intersections in WT primary hippocampal neurons treated with DMSO (Control), 4h kinetin, 8h kinetin, 12h kinetin, 24h kinetin, BDNF, and forskolin. Ordinary one-way ANOVA and post-hoc analysis using Dunnett’s multiple comparison test were used for statistical analysis. Bars denote the average ± SEM and are representative of three independent experiments (N=30; 10 neurons per genotype per experiment). I.Representative epifluorescence images of MAP2b immunostaining in WT PCNs transfected with NT siRNA, PINK1 siRNA alone, and PINK1 siRNA-pretreated with Human recombinant BDNF. J. Quantification of mean dendrite length in WT PCNs treated with NTsiRNA, PINK1 siRNA alone, and PINK1siRNA- pretreated with human recombinant BDNF. Ordinary one-way ANOVA and post-hoc analysis using Dunnett’s multiple comparison test were used for statistical analysis. Bars denote the average ± SEM and are representative of three independent experiments (N=30; 10 neurons per genotype per experiment).*P < 0.05, **P < 0.01, ***P < 0.001.
Compared to PCNs, primary hippocampal neurons express a higher level of BDNF and exhibit a more heightened dendrite development in response to BDNF treatment, as evidenced by an increase in mean dendrite length and dendrite complexity (Tolwani et al., 2002; Zagrebelsky et al., 2018). To further evaluate the effect of pharmacologically activating endogenous PINK1 on dendrite development, primary hippocampal neurons derived from WT 14–15 days mouse embryos were transiently transfected with GFP to facilitate the visualization of dendritic arbors by confocal microscopy, and dendrite complexity was analyzed by Sholl analysis. In brief, Sholl analysis of reconstructed images of GFP-transfected primary hippocampal neurons suggests that hippocampal neurons treated with kinetin showed a higher degree of complexity of dendritic trees, when compared to untreated hippocampal neurons (Fig. 5G & 5H). Specifically, the number of branch points significantly increased at 8h, and 12h following treatment of WT primary hippocampal neurons with kinetin, when compared to DMSO treated primary hippocampal neurons. Additionally, treating primary hippocampal neurons with exogenous human recombinant BDNF or with forskolin had a similar effect as treating WT primary hippocampal neurons with kinetin for 12h. To further corroborate that endogenous PINK1 enhances dendritic complexity via BDNF, we examined the effect of siRNA-mediated knockdown of endogenous PINK1 on dendrite length in the presence or absence of exogenous recombinant human BDNF. We observed that PCNs transfected with PINK1 siRNA for three days exhibited significantly reduced dendrite length compared to WT PCNs transfected with non-targeting (NT) control siRNA (Fig. 5I & 5J). Conversely, treating PINK1 siRNA-expressing PCNs with exogenous recombinant human BDNF for 24h, significantly reversed the supplementary increase in dendrite shortening induced by siRNA-mediated knockdown of endogenous PINK1. Collectively, our data suggest that pharmacological activation of endogenous PINK1 or increased level of PINK1 leads to downstream activation of PKA to promote neuronal plasticity through a mechanism that involves the binding of BDNF to TRKβ.
PINK1 activates downstream cytosolic-localized PKA to modulate BDNF signaling
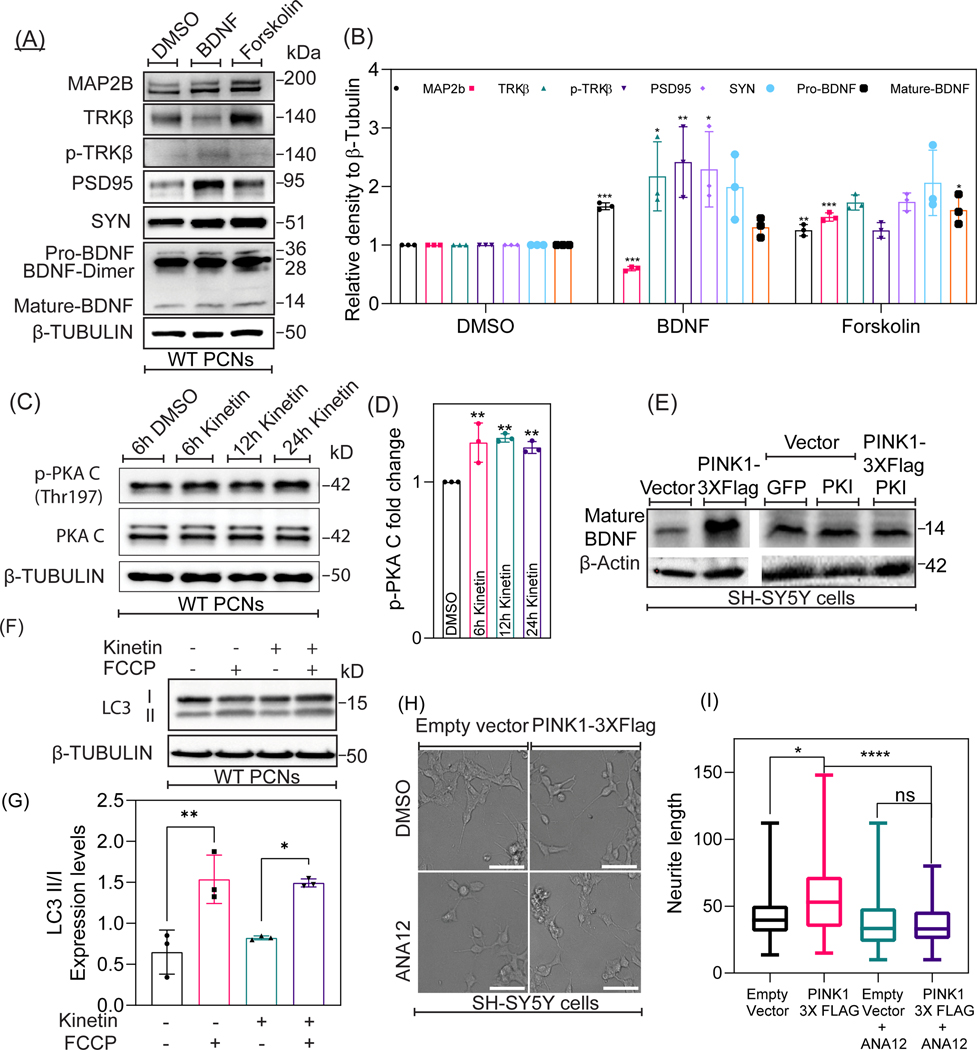
So far, our data suggest that pharmacological activation of endogenous PINK1 elicits PKA-mediated phosphorylation of CREB (Ser133) in developing neurons. Given that the canonical PKA-CREB-BDNF signaling pathway has been widely considered to promote neuronal development and plasticity, we construed that PINK1 acts as a modulator of BDNF signaling by specifically stimulating downstream PKA signaling in the cytosol. Consistent with other studies showing a role of BDNF and PKA in stimulating neuronal development and plasticity, Western blotting analysis of cell lysates derived from wild-type PCNs treated with exogenous human recombinant BDNF or with the pharmacological activator of PKA forskolin showed a significant increase in the level of various protein markers of neuronal plasticity and development including MAP2B (F=8, N=3, p-value=0.0062), PSD95 (F=8, N=3, p-value= 0.0051), and synaptophysin (F=8, N=3, p-value= 0.0314) compared to PCNs treated with DMSO (control) (Fig 6A & 6B). Besides, treating PCNs with forskolin for 24h, significantly increased the protein level of TRKβ (F=8, N=3, p-value= <0.0001) and of the cleaved, mature form of BDNF (F=8, N=3, p-value=0.0118) (Fig 6A & 6B). Given that transient overexpression of fPINK1 enhances downstream autocatalytic activation of PKA (Wang et al., 2018a), we next evaluated the extent to which pharmacological activation of PINK1 increases the autophosphorylation of the catalytic subunit of PKA (PKA/C) in kinetin treated PCNs. Indeed, Western blotting analysis shows that treating PCNs with kinetin led to a significant progressive increase in the autocatalytic-mediated phosphorylation of PKA-C (Thr197) across multiple time points, 6h (F=11, N=3, p-value =0.0045), 12h (F=11, N=3, p =0.0022) and 24h (F=11, N=3, p-value=0.0096) compared to DMSO treated control (Fig.6C & 6D). Total PKA-C levels remained unchanged in PCNs treated with kinetin throughout the same time points.
Figure 6. PINK1 activates downstream cytosolic-localized PKA to modulate BDNF signaling.
A. Representative Western blotting for the indicated proteins in lysates extracted at differential time-points from DMSO, BDNF, and forskolin treated WT PCNs. B. Densitometric analysis of the endogenous expression level of MAP2B, TRKβ, phospho-TRKβ, PSD95, Synaptophysin, and BDNF expression levels. Values were normalized to β-Tubulin. Ordinary one-way ANOVA and post-hoc analysis using Dunnett’s multiple comparison test were used for statistical analysis. Bars denote the average ± SEM and are representative of three independent experiments. C. Representative Western blotting of Phospho-PKA (cat Thr197 ) and PKA-C expression in cell lysates extracted from DMSO, 6h kinetin, 12h kinetin, and 24h kinetin treated WT PCNs. D. Densitometric analysis on the endogenous expression level of Phospho-PKA-C (Thr197) levels in DMSO, 6h kinetin, 12h kinetin, and 24h kinetin treated WT PCNs. Values were normalized to β-Tubulin. Ordinary one-way ANOVA and post-hoc analysis using Dunnett’s multiple comparison test were used for statistical analysis. Bars denote the average ± SEM and are representative of three independent experiments. E. Representative Western blotting for LC3 expression in lysates extracted from Control, FCCP treated, kinetin treated, and FCCP+kinetin treated WT PCNs. F. Densitometric quantification of the ratio of LCII to LCI ratio in Control, FCCP treated, kinetin treated, and FCCP+kinetin treated WT PCNs. Values were normalized to β-Tubulin. Ordinary one-way ANOVA and post-hoc analysis using Dunnett’s multiple comparison test were used for statistical analysis. Bars denote the average ± SEM and are representative of three independent experiments. G. Representative Western blotting of BDNF expression in cell lysates extracted from SH-SY5Y cells transfected with plasmids encoding for control (empty vector), PINK1–3XFlag, GFP alone, an inhibitor of Protein Kinase A (PKI) or co-transfected with PINK1–3XFlag and PKI. H. Representative bright-field images of SH-SY5Y cells transfected with plasmids coding for control (empty vector) and PINK1–3XFlag and treated with DMSO (control) and ANA12 (TRKβ antagonist). I. Quantification of mean neurite length in SH-SY5Y cells transfected with plasmids coding for control (empty vector) and PINK1–4XFlag and treated with DMSO (control) and ANA12. Ordinary one-way ANOVA and post-hoc analysis using Dunnett’s multiple comparison test were used for statistical analysis. Bars denote the average ± SEM and are representative of three independent experiments (N=30; 10 neurons per genotype per experiment). *P < 0.05, **P < 0.01, ***P < 0.001.
In the mitochondria, fPINK1 has been well-characterized to elicit the autophagy pathway to promote the clearance of oxidatively-damaged mitochondria in cells that are exposed to mitochondrial-targeted toxins or acutely treated with mitochondrial uncouplers (e.g., FCCP). We then sought to verify whether endogenous activation and increased intracellular level of cPINK1 via kinetin elicit autophagy in PCNs treated with FCCP. LC3, the human homolog of yeast ATG8, is a protein that governs the biogenesis and maturation of autophagosomes (AVs) and is a bona fide marker used to measure the intracellular levels of AVs by Western blotting (Levine and Klionsky, 2004). While LC3 is predominantly localized in the cytosol (termed LC3-I), LC3-I is conjugated to phosphatidylethanolamine through a series of conjugation reactions that allow it to bind to nascent AVs. The AV-bound form of LC3 is known as LC3-II. By performing Western blotting analysis of lysates derived from PCNs treated with FCCP in the presence or absence of kinetin, we examined the conversion of soluble LC3-I (15kDa) to AV-bound LC3-II, the lapidated form of LC3 which shows a retarded electrophoretic mobility relative to LC3-I (17kDa). Hence, an increase in the ratio of LC3-II to LC3-I is indicative of increased autophagy (Klionsky et al., 2016). In brief, while Western blotting analysis of LC3 suggests that a 4h FCCP treatment of PCNs significantly increased the ratio of LC3-II to LC3-I (F=11, N=3, p =0.0037), treating PCNs with kinetin alone did not increase the ratio of LC3 II to LC3-I nor did it enhanced FCCP-mediated increase in autophagy (Fig. 6F & 6G). In the absence of mitochondrial insult, our Western blotting data suggest that pharmacological activation of endogenous PINK1 kinase activity does not stimulate basal autophagy.
Given that PINK1 activates downstream PKA signaling (Dagda et al., 2014; Wang et al., 2018b) and increases the level of BDNF, we then sought to investigate the extent to which PINK1-mediated increase in BDNF requires PKA activity. SH-SY5Y cell line is frequently used in PD in vitro studies due to its catecholaminergic phenotype and higher transfection rate, enabling Western blotting analysis of markers of neuronal development in response to exogenous protein expression (Xicoy et al., 2017). Western blotting analysis of cell lysates derived from PINK1–3X Flag transfected SH-SY5Y cells showed that the level of the mature form of endogenous BDNF was increased compared to a stable cell line expressing an empty vector (Fig. 6E) which is consistent with the observation that pharmacological activation of endogenous PINK1 stimulates the extracellular release of BDNF (Fig. 3D). However, transiently expressing a GFP tagged form of the protein kinase inhibitor (PKI), an endogenous peptide that specifically inhibits PKA activity by binding to PKA/C (Cheng et al., 1986), blocked the ability of overexpressed PINK1 to increase the level of BDNF (Fig. 6E) suggesting that PINK1 requires PKA activity to enhance BDNF levels. We have previously reported that stable overexpression of full-length Flag-tagged human PINK1 (PINK1–3XFlag) in SH-SY5Y cells stimulated neurite outgrowth, as evidenced by a significant elongation of neurites when compared to the stable cell line that expressed an empty vector as a control (Dagda et al., 2014). Here, we sought to investigate the extent to which the PINK1-mediated increase in dendrite complexity requires the binding of extracellular BDNF to TRKβ. While naïve PINK1–3X-Flag cells showed a significant two-fold increase in the mean length of neurite-like extensions, treating PINK1–3XFlag SH-SY5Y cells with ANA-12 (45nM, 24h), a pharmacological inhibitor of TRKβ, blocked the ability of PINK1 in promoting neurite outgrowth (Fig. 6H & 6I).
In addition to PKA, it is worth noting that other ser/thr kinases can phosphorylate CREB at serine133 and increase the level of endogenous BDNF, including the MAPK/ERK pathway and the PI-3K/AKT. In order to rule out that PINK1 enhances the CREB-BDNF signaling pathway by activating other MAP kinases or PI-3K/AKT, we treated PCNs with kinetin to pharmacologically enhance the activity of PINK1 in the presence or absence of U0126, LY294002, and H89, pharmacological inhibitors of MAPK/ERK, PI3K/AKT, and PKA signaling pathways respectively. As expected, while immunofluorescent labeling showed a significant increase in the endogenous level BDNF in kinetin treated PCNs, pharmacological co-treatment with a 24 h dose of U0126, and LY294002, was unable to block the ability of kinetin in enhancing the level of BNDF. As expected, cotreating PCNs with H89 significantly reduced the ability of kinetin in enhancing the level of BNDF (Fig 7). Like PKA, CAMKII can also govern neuronal plasticity and differentiation. It is worth noting that we have not investigated CaMKII- dependent activation of CREB and possible correlation to PINK1-PKA signaling mechanisms, which needs to be further investigated in future studies. Collectively our results show that PINK1 regulates neuronal development by specifically activating PKA signaling resulting in augmented BDNF signaling.
Figure 7.
A. Representative epifluorescence images in WT PCNs immunostained for BDNF (green) and MAP2B (red) and treated with DMSO (Control), 24h kinetin, UO126, U)126+kinetin, LY294002, LY294002+kinetin, H89, and H89+kinetin. The nucleus is counterstained by DAPI (blue). Scale bar represents 5μM. B. Graphical representation of BDNF punctae quantification in WT PCNs treated with DMSO (Control), 24h kinetin, UO126 (MEK1/2 inhibitor), UO126+kinetin, LY294002, LY294002+kinetin, H89, and H89+kinetin. Ordinary one-way ANOVA and post-hoc analysis using Dunnett’s multiple comparison test were used for statistical analysis. Bars denote the average ± SEM and are representative of three independent experiments (N=30; 10 neurons per genotype per experiment). *P < 0.05, **P < 0.01, ***P < 0.001.
Discussion
While a majority of research endeavors have predominantly focused on functionally dissecting the pathological roles of fPINK1 in the context of PD-associated mutations of PINK1 and in in vitro and in vivo models of PD, there is limited knowledge regarding the physiological functions of cPINK1 in a healthy brain. We have previously shown that cPINK1, rather than fPINK1, promotes neurite outgrowth by activating downstream PKA signaling (Dagda et al., 2014; Das Banerjee et al., 2017). Beyond enhancing neurite outgrowth and modulating mitochondrial trafficking, here we report for the first time that endogenous cPINK1 governs neuronal development in healthy neurons through activating PKA mediated BDNF signaling.
Dysfunctional mitochondria, as evident by a significant decrease in ΔΨm, leads to the activation of canonical mitochondrial-PINK1 mediated mitophagy, a physiological pathway that is crucial for maintaining an equilibrium of a healthy pool of mitochondria to ensure ATP production, reduce detrimental ROS generation, calcium homeostasis, and allow for the biosynthesis of metabolites (Kubli and Gustafsson, 2012; Chandel, 2014; Soman et al., 2019). However, cPINK1, which is devoid of its MTS motif, is adequate to provide resistance to neurons against the toxic effects of the complex I inhibitors such as MPTP (Haque et al., 2008) and the proteasome inhibitor MG132 (Valente et al., 2004). The non-canonical roles of PINK1, attributed specifically to cPINK1, includes retrograde signaling to modulate neuronal development, govern mitochondrial trafficking within dendrites, promoting dendrite outgrowth and stabilizing dendrites against oxidative stress by activating downstream PKA signaling, protecting the soma of cells from proteomic stress, and stimulating pro-survival PI3K-AKT and PKA signaling (Hollville et al., 2014; Parganlija et al., 2014; Choi et al., 2016; Lee et al., 2017). In this report, we show that the increase in endogenous levels cPINK1 throughout neuronal development is associated with the upregulation of markers of neuronal plasticity such as BDNF, PSD95, and synaptophysin (Fig. 1A). Consistent with our study, other reports have also shown higher cPINK1 expression in different stages of neuronal development, and have reported defects in neuronal stem cell differentiation due to PINK1 deficiency (Choi et al., 2016; Agnihotri et al., 2017). It is worth noting that we also observed an increased level of endogenous PINK1 in RA-differentiated SH-SY5Y cells (Fig. 1F). We have previously shown that forced overexpression of cPINK1 promotes the differentiation of naïve SH-SY5Y cells by activating downstream PKA activity (Dagda et al., 2014). However, RA promotes the differentiation of naïve SH-SY5Y neuroblastoma cells in a PKC-dependent manner, suggesting that the induction of endogenous level of PINK1 is a phenomenon that occurs throughout neuronal development regardless of the signaling pathway activated to promote neuronal differentiation (PKA or PKC) (Kurie et al., 1993). For optimum cPINK1 function, a minimum amount of protein stability of PINK1 isoforms is necessary, and under basal conditions, cPINK1 is retained in the cytosol by the presence of Lys-63-linked ubiquitination or protected from proteolytic degradation by binding to chaperone proteins such as HSP90 and BAG5 (Lin and Kang, 2008; Wang et al., 2014; Lim et al., 2015). It is worth noting that PINK1 can be auto-phosphorylated at Ser228 and Ser402 to enhance its neuroprotective kinase activity, paving the way for therapeutic intervention by pharmacological activation of PINK1 (Kondapalli et al., 2012; Aerts et al., 2015).
Kinetin is a plant hormone that acts as a neo-substrate to PINK1. The metabolized form of kinetin (KTP) shows higher catalytic efficiency compared to the endogenous substrate of PINK1, ATP (Hertz et al., 2013). In this report, we observed an increased level of endogenous cPINK1 (Fig. 2I), but not fPINK1, following kinetin treatment in PCNs, suggesting stabilization of endogenous cPINK1 following kinetin treatment. This observation suggests that not only does KTP activates endogenous PINK1 to enhance its activity by binding to the ATP binding pocket of the ser/thr, but it also increases kinase activity by increasing the endogenous level of cPINK1, presumably as a mechanism to further enhance neuronal development in a feedforward through the PKA-BDNF signaling axis. Moreover, it is conceivable that the catalytic autophosphorylation of PINK1 initiated by kinetin could expedite the proteolytic processing of PINK1 to cleaved derivatives in healthy mitochondria, leading to enhanced protein stability, and subsequent increased endogenous levels of cPINK1.
A myriad of non-motor symptoms can manifest in advanced forms of PD, including cognitive decline, which can occur in over 50% of idiopathic PD cases (Aarsland and Kurz, 2010). Postmortem studies and experimental models of PD had highlighted the extent of synaptic loss in multiple brain regions, including SNpc and cerebral cortex before the loss of somatic compartments and ensued decrease in the number of neurons (Dauer and Przedborski, 2003; Wu et al., 2003; Reeve et al., 2018). Neurotrophins such as BDNF are the prime molecular modulators of synaptic plasticity, proper development and maintenance of dendrites, and regulating neuronal survival for many neuronal subpopulations, including SNpc neurons (Hyman et al., 1991; Leal et al., 2015). Consistent with a potential pathological role of reduced BDNF level in PD, BDNF null mice mostly die within two post-natal weeks and show extensive neuronal loss accompanied by the behavioral deficits in the context of poor coordination of movement and balance (Ernfors et al., 1994). Also, functional polymorphisms in BDNF (Val66Met) gene are linked with cognitive impairment in PD patients (Bialecka et al., 2014). Besides, a reduction in BDNF mRNA levels has been observed in the SNpc of the postmortem brain from PD patients (Howells et al., 2000). Consistent with that study, we observed diminished BDNF levels in the midbrain and PCN of 10-month-old PINK1-KO mice (Fig. 2A). Reduced BDNF levels in SNpc and serum are often associated with cognitive impairment in clinical and experimental conditions of PD (Costa et al., 2015; Wang et al., 2016; Sampaio et al., 2017). BDNF exerts its action through binding to its cognate receptors (TRKβ/p75NTR receptors), leading to downstream activation of several signaling pathways, including MAPK, PLC, NF-κB, and PI3K signaling pathways. Diminished BDNF-TRKβ signaling, as induced by downregulating the expression of TRKβ receptor, can lead to significant neurodegeneration of nigrostriatal DA neurons in the absence of toxic insult and enhances their sensitivity to MPTP (Baydyuk et al., 2011; Kang et al., 2017). Furthermore, it is worth noting that pharmacological agonists of TRKβ can significantly protect DA neurons from 6-OHDA and MPTP-mediated neurodegeneration in-vivo (Nie et al., 2015; Luo et al., 2016; Bhurtel et al., 2018) suggesting that increasing the level of BDNF via kinetin-mediated activation of PINK1 may be neuroprotective in models of PD. Also, we show that the reduction in the intracellular level of BDNF expression is rescued in PINK1-deficient PCNs by transient transfection of a plasmid encoding for human cPINK1, but not kinase-dead cPINK1 (Fig. 3A). Alternatively, kinetin-mediated activation of PINK1 leads to increased intracellular production and release of mature forms of BDNF in WT but not in PINK1-deficient PCNs (Fig 3D). While most of the clinical PINK1 mutations lead to reduced kinase activity, it is also worth noting that our data show that a PD-associated mutation of PINK1 (G309D) and catalytically inactive mutation (K219M), which impairs PINK1’s kinase activity, are unable to increase the intracellular level of BDNF in transiently transfected PCNs. This observation suggests that a substantial reduction in intracellular BDNF caused by kinase-impairing mutations in PINK1 may contribute to neurodegeneration in PD patients, due to decreased pro-survival input in SNpc and cortical neurons, two neuronal subpopulations targeted during the progression of PD.
In addition to initiating mitophagy, PINK1 directly or indirectly phosphorylates several non-conventional substrates including NDUFA10/ND42 subunit (Pogson et al., 2014), Bcl-xL (Arena et al., 2013), sequesters defective mitochondria via phosphorylation of Miro1 (Wang et al., 2011), sequestosome-1, and subsequent activation of Parkin independent aggresome-autophagy pathway (Gao et al., 2016). These observations indicate that the pro-survival activity of PINK1 can be distinct from mitophagy and likely dependent upon the ratio of proteolytically-cleaved forms of PINK1 to fPINK1. We had reported that loss of dendrites during PINK1 deficiency is accompanied by reduced PKA signaling in PINK1-deficient PCNs (Das Banerjee et al., 2017). A recent report suggested that dendrite arborization stimulated through PINK1 is dependent upon phosphorylation of PKAcat(pT197) (Wang et al., 2018a). Consistent with that study, here we show that endogenous PINK1 activation through kinetin supplementation phosphorylates the catalytic subunit of PKA PKAcat(pT197) to induce autocatalytic activation of the PKA holoenzyme (Fig. 6C). cAMP-dependent PKA activity triggered by Ca+2 is an essential biochemical pathway that promotes synaptic plasticity, regulates spine structure, and promotes long-term potentiation (Ohadi et al., 2019). On further inspection, we found that PINK1 activation prompts downstream phosphorylation of CREB at Ser133 (p-CREB) (Fig. 4), a post-translational event that leads to the transcription of cAMP-modulated gene products including BDNF, antiapoptotic protein Bcl2, and tyrosine-hydroxylase.
In conclusion, our data suggest that PINK1 initiates robust kinase-dependent activation of the PKA-CREB-BDNF signaling axis to promote neuronal development. Notably, we describe a novel signaling pathway activated through activation of cleaved PINK1, which promotes neuronal development via accelerating the maturation of presynaptic and postsynaptic compartments. This physiological role is distinct from the canonical functions ascribed to full length PINK1.
Supplementary Material
Significance statement.
The non-canonical role of PINK1 in regulating neuronal development is a classic example of mitochondrial retrograde signaling. In healthy neurons, proteases in mitochondria cleave fPINK1 to lower molecular weight forms (cPINK1) exert extra-mitochondrial functions. In our study, we observe significantly reduced BDNF levels in PINK1-deficient neurons, and pharmacological activation of endogenous PINK1 through kinetin restores the level of BDNF in PINK1-deficient neurons. Mechanistically, cPINK1 activates PKA-CREB-BDNF signaling pathway to stimulate neuronal plasticity. This novel role of PINK1 in stimulating BDNF signaling opens avenues for intervention in Parkinson’s disease patients with cognitive impairment.
Acknowledgments
Support: This project was supported by NIH grant R01 NS105783 and The William N. Pennington Foundation (Nevada, USA).
RRID:IMSR_JAX:017946, RRID:AB_10862052, RRID:AB_331277, RRID:AB_2561044, RRID:AB_2292909, RRID:AB_301417, RRID:AB_218182, RRID:AB_11214935, RRID:AB_10127658, RRID:AB_2210370, RRID:AB_10862052, RRID:AB_331277, RRID:AB_2561044, RRID:AB_2292909, RRID:AB_301417, RRID:AB_218182, RRID:AB_11214935, RRID:AB_444716, RRID:AB_2554679, RRID:AB_2300165, RRID:AB_2234770
Footnotes
CONFLICT OF INTEREST
The authors disclose no conflict of interest.
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding authors upon reasonable request.
References
- Aarsland D, Kurz MW (2010) The epidemiology of dementia associated with Parkinson disease. J Neurol Sci 289:18–22. [DOI] [PubMed] [Google Scholar]
- Aerts L, Craessaerts K, De Strooper B, Morais VA (2015) PINK1 kinase catalytic activity is regulated by phosphorylation on serines 228 and 402. J Biol Chem 290:2798–2811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agnihotri SK, Shen R, Li J, Gao X, Bueler H (2017) Loss of PINK1 leads to metabolic deficits in adult neural stem cells and impedes differentiation of newborn neurons in the mouse hippocampus. Faseb j 31:2839–2853. [DOI] [PubMed] [Google Scholar]
- Arena G, Gelmetti V, Torosantucci L, Vignone D, Lamorte G, De Rosa P, Cilia E, Jonas EA, Valente EM (2013) PINK1 protects against cell death induced by mitochondrial depolarization, by phosphorylating Bcl-xL and impairing its pro-apoptotic cleavage. Cell Death Differ 20:920–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arlotta P, Molyneaux BJ, Chen J, Inoue J, Kominami R, Macklis JD (2005) Neuronal subtype-specific genes that control corticospinal motor neuron development in vivo. Neuron 45:207–221. [DOI] [PubMed] [Google Scholar]
- Baydyuk M, Nguyen MT, Xu B (2011) Chronic deprivation of TrkB signaling leads to selective late-onset nigrostriatal dopaminergic degeneration. Exp Neurol 228:118–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhurtel S, Katila N, Neupane S, Srivastav S, Park PH, Choi DY (2018) Methylene blue protects dopaminergic neurons against MPTP-induced neurotoxicity by upregulating brain-derived neurotrophic factor. Ann N Y Acad Sci 1431:58–71. [DOI] [PubMed] [Google Scholar]
- Bialecka M, Kurzawski M, Roszmann A, Robowski P, Sitek EJ, Honczarenko K, Mak M, Deptula-Jarosz M, Golab-Janowska M, Drozdzik M, Slawek J (2014) BDNF G196A (Val66Met) polymorphism associated with cognitive impairment in Parkinson’s disease. Neurosci Lett 561:86–90. [DOI] [PubMed] [Google Scholar]
- Bonaparte D, Cinelli P, Douni E, Herault Y, Maas M, Pakarinen P, Poutanen M, Lafuente MS, Scavizzi F (2013) FELASA guidelines for the refinement of methods for genotyping genetically-modified rodents: a report of the Federation of European Laboratory Animal Science Associations Working Group. Lab Anim 47:134–145. [DOI] [PubMed] [Google Scholar]
- Chandel NS (2014) Mitochondria as signaling organelles. BMC Biol 12:34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng HC, Kemp BE, Pearson RB, Smith AJ, Misconi L, Van Patten SM, Walsh DA (1986) A potent synthetic peptide inhibitor of the cAMP-dependent protein kinase. J Biol Chem 261:989–992. [PubMed] [Google Scholar]
- Choi I, Choi D-J, Yang H, Woo JH, Chang M-Y, Kim JY, Sun W, Park S-M, Jou I, Lee S-H, Joe E-H (2016) PINK1 expression increases during brain development and stem cell differentiation, and affects the development of GFAP-positive astrocytes. Molecular brain 9:5–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa A, Peppe A, Carlesimo GA, Zabberoni S, Scalici F, Caltagirone C, Angelucci F (2015) Brain-derived neurotrophic factor serum levels correlate with cognitive performance in Parkinson’s disease patients with mild cognitive impairment. Front Behav Neurosci 9:253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dagda RK, Gusdon AM, Pien I, Strack S, Green S, Li C, Van Houten B, Cherra SJ, Chu CT (2011) Mitochondrially localized PKA reverses mitochondrial pathology and dysfunction in a cellular model of Parkinson’s disease. Cell Death & Differentiation 18:1914–1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dagda RK, Pien I, Wang R, Zhu J, Wang KZ, Callio J, Banerjee TD, Dagda RY, Chu CT (2014) Beyond the mitochondrion: cytosolic PINK1 remodels dendrites through protein kinase A. J Neurochem 128:864–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das Banerjee T, Dagda RY, Dagda M, Chu CT, Rice M, Vazquez-Mayorga E, Dagda RK (2017) PINK1 regulates mitochondrial trafficking in dendrites of cortical neurons through mitochondrial PKA. J Neurochem 142:545–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dauer W, Przedborski S (2003) Parkinson’s disease: mechanisms and models. Neuron 39:889–909. [DOI] [PubMed] [Google Scholar]
- Delghandi MP, Johannessen M, Moens U (2005) The cAMP signalling pathway activates CREB through PKA, p38 and MSK1 in NIH 3T3 cells. Cellular Signalling 17:1343–1351. [DOI] [PubMed] [Google Scholar]
- Ernfors P, Lee KF, Jaenisch R (1994) Mice lacking brain-derived neurotrophic factor develop with sensory deficits. Nature 368:147–150. [DOI] [PubMed] [Google Scholar]
- Furlong RM, Lindsay A, Anderson KE, Hawkins PT, Sullivan AM, O’Neill C (2019) The Parkinson’s disease gene PINK1 activates Akt via PINK1 kinase-dependent regulation of the phospholipid PI(3,4,5)P(3). J Cell Sci 132. [DOI] [PubMed] [Google Scholar]
- Genheden M, Kenney JW, Johnston HE, Manousopoulou A, Garbis SD, Proud CG (2015) BDNF Stimulation of Protein Synthesis in Cortical Neurons Requires the MAP Kinase-Interacting Kinase MNK1. The Journal of Neuroscience 35:972–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigoruta M, Martinez-Martinez A, Dagda RY, Dagda RK (2020) Psychological Stress Phenocopies Brain Mitochondrial Dysfunction and Motor Deficits as Observed in a Parkinsonian Rat Model. Mol Neurobiol 57:1781–1798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haque ME, Thomas KJ, D’Souza C, Callaghan S, Kitada T, Slack RS, Fraser P, Cookson MR, Tandon A, Park DS (2008) Cytoplasmic Pink1 activity protects neurons from dopaminergic neurotoxin MPTP. Proc Natl Acad Sci U S A 105:1716–1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertz NT, Berthet A, Sos ML, Thorn KS, Burlingame AL, Nakamura K, Shokat KM (2013) A neo-substrate that amplifies catalytic activity of parkinson’s-disease-related kinase PINK1. Cell 154:737–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollville E, Carroll RG, Cullen SP, Martin SJ (2014) Bcl-2 family proteins participate in mitochondrial quality control by regulating Parkin/PINK1-dependent mitophagy. Mol Cell 55:451–466. [DOI] [PubMed] [Google Scholar]
- Howells DW, Porritt MJ, Wong JY, Batchelor PE, Kalnins R, Hughes AJ, Donnan GA (2000) Reduced BDNF mRNA expression in the Parkinson’s disease substantia nigra. Exp Neurol 166:127–135. [DOI] [PubMed] [Google Scholar]
- Hu X, Ballo L, Pietila L, Viesselmann C, Ballweg J, Lumbard D, Stevenson M, Merriam E, Dent EW (2011) BDNF-induced increase of PSD-95 in dendritic spines requires dynamic microtubule invasions. The Journal of neuroscience : the official journal of the Society for Neuroscience 31:15597–15603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyman C, Hofer M, Barde YA, Juhasz M, Yancopoulos GD, Squinto SP, Lindsay RM (1991) BDNF is a neurotrophic factor for dopaminergic neurons of the substantia nigra. Nature 350:230–232. [DOI] [PubMed] [Google Scholar]
- Kang SS, Zhang Z, Liu X, Manfredsson FP, Benskey MJ, Cao X, Xu J, Sun YE, Ye K (2017) TrkB neurotrophic activities are blocked by alpha-synuclein, triggering dopaminergic cell death in Parkinson’s disease. Proc Natl Acad Sci U S A 114:10773–10778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler KR, Hamscho N, Morales B, Menzel C, Barrero F, Vives F, Gispert S, Auburger G (2005) Dopaminergic function in a family with the PARK6 form of autosomal recessive Parkinson’s syndrome. J Neural Transm (Vienna) 112:1345–1353. [DOI] [PubMed] [Google Scholar]
- Kitada T, Pisani A, Porter DR, Yamaguchi H, Tscherter A, Martella G, Bonsi P, Zhang C, Pothos EN, Shen J (2007a) Impaired dopamine release and synaptic plasticity in the striatum of PINK1-deficient mice. Proc Natl Acad Sci U S A 104:11441–11446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitada T, Pisani A, Porter DR, Yamaguchi H, Tscherter A, Martella G, Bonsi P, Zhang C, Pothos EN, Shen J (2007b) Impaired dopamine release and synaptic plasticity in the striatum of PINK1-deficient mice. Proceedings of the National Academy of Sciences 104:11441–11446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky DJ et al. (2016) Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition). Autophagy 12:1–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondapalli C, Kazlauskaite A, Zhang N, Woodroof HI, Campbell DG, Gourlay R, Burchell L, Walden H, Macartney TJ, Deak M, Knebel A, Alessi DR, Muqit MM (2012) PINK1 is activated by mitochondrial membrane potential depolarization and stimulates Parkin E3 ligase activity by phosphorylating Serine 65. Open Biol 2:120080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowiański P, Lietzau G, Czuba E, Waśkow M, Steliga A, Moryś J (2018) BDNF: A Key Factor with Multipotent Impact on Brain Signaling and Synaptic Plasticity. Cell Mol Neurobiol 38:579–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubli DA, Gustafsson ÅB (2012) Mitochondria and mitophagy: the yin and yang of cell death control. Circulation research 111:1208–1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurie JM, Younes A, Miller WH Jr., Burchert M, Chiu CF, Kolesnick R, Dmitrovsky E (1993) Retinoic acid stimulates the protein kinase C pathway before activation of its beta-nuclear receptor during human teratocarcinoma differentiation. Biochim Biophys Acta 1179:203–207. [DOI] [PubMed] [Google Scholar]
- Leal G, Afonso PM, Salazar IL, Duarte CB (2015) Regulation of hippocampal synaptic plasticity by BDNF. Brain Res 1621:82–101. [DOI] [PubMed] [Google Scholar]
- Lee Y et al. (2017) PINK1 Primes Parkin-Mediated Ubiquitination of PARIS in Dopaminergic Neuronal Survival. Cell reports 18:918–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesuisse C, Martin LJ (2002) Immature and mature cortical neurons engage different apoptotic mechanisms involving caspase-3 and the mitogen-activated protein kinase pathway. J Cereb Blood Flow Metab 22:935–950. [DOI] [PubMed] [Google Scholar]
- Levine B, Klionsky DJ (2004) Development by self-digestion: molecular mechanisms and biological functions of autophagy. Dev Cell 6:463–477. [DOI] [PubMed] [Google Scholar]
- Lim GG, Chua DS, Basil AH, Chan HY, Chai C, Arumugam T, Lim KL (2015) Cytosolic PTEN-induced Putative Kinase 1 Is Stabilized by the NF-kappaB Pathway and Promotes Non-selective Mitophagy. J Biol Chem 290:16882–16893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin W, Kang UJ (2008) Characterization of PINK1 processing, stability, and subcellular localization. Journal of neurochemistry 106:464–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo D, Shi Y, Wang J, Lin Q, Sun Y, Ye K, Yan Q, Zhang H (2016) 7,8-dihydroxyflavone protects 6-OHDA and MPTP induced dopaminergic neurons degeneration through activation of TrkB in rodents. Neurosci Lett 620:43–49. [DOI] [PubMed] [Google Scholar]
- McGregor MM, Nelson AB (2019) Circuit Mechanisms of Parkinson’s Disease. Neuron 101:1042–1056. [DOI] [PubMed] [Google Scholar]
- Merrill RA, Dagda RK, Dickey AS, Cribbs JT, Green SH, Usachev YM, Strack S (2011a) Mechanism of neuroprotective mitochondrial remodeling by PKA/AKAP1. PLoS Biol 9:e1000612-e1000612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrill RA, Dagda RK, Dickey AS, Cribbs JT, Green SH, Usachev YM, Strack S (2011b) Mechanism of Neuroprotective Mitochondrial Remodeling by PKA/AKAP1. PLoS Biol 9:e1000612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie S, Xu Y, Chen G, Ma K, Han C, Guo Z, Zhang Z, Ye K, Cao X (2015) Small molecule TrkB agonist deoxygedunin protects nigrostriatal dopaminergic neurons from 6-OHDA and MPTP induced neurotoxicity in rodents. Neuropharmacology 99:448–458. [DOI] [PubMed] [Google Scholar]
- Ohadi D, Schmitt DL, Calabrese B, Halpain S, Zhang J, Rangamani P (2019) Computational Modeling Reveals Frequency Modulation of Calcium-cAMP/PKA Pathway in Dendritic Spines. Biophys J 117:1963–1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parganlija D, Klinkenberg M, Domínguez-Bautista J, Hetzel M, Gispert S, Chimi MA, Dröse S, Mai S, Brandt U, Auburger G, Jendrach M (2014) Loss of PINK1 impairs stress-induced autophagy and cell survival. PloS one 9:e95288-e95288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petit A, Kawarai T, Paitel E, Sanjo N, Maj M, Scheid M, Chen F, Gu Y, Hasegawa H, Salehi-Rad S, Wang L, Rogaeva E, Fraser P, Robinson B, St George-Hyslop P, Tandon A (2005) Wild-type PINK1 prevents basal and induced neuronal apoptosis, a protective effect abrogated by Parkinson disease-related mutations. J Biol Chem 280:34025–34032. [DOI] [PubMed] [Google Scholar]
- Pogson JH, Ivatt RM, Sanchez-Martinez A, Tufi R, Wilson E, Mortiboys H, Whitworth AJ (2014) The Complex I Subunit NDUFA10 Selectively Rescues Drosophila pink1 Mutants through a Mechanism Independent of Mitophagy. PLOS Genetics 10:e1004815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeve AK, Grady JP, Cosgrave EM, Bennison E, Chen C, Hepplewhite PD, Morris CM (2018) Mitochondrial dysfunction within the synapses of substantia nigra neurons in Parkinson’s disease. NPJ Parkinson’s disease 4:9–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampaio TB, Pinton S, da Rocha JT, Gai BM, Nogueira CW (2017) Involvement of BDNF/TrkB signaling in the effect of diphenyl diselenide on motor function in a Parkinson’s disease rat model. Eur J Pharmacol 795:28–35. [DOI] [PubMed] [Google Scholar]
- Soman SK, Bazala M, Keatinge M, Bandmann O, Kuznicki J (2019) Restriction of mitochondrial calcium overload by mcu inactivation renders a neuroprotective effect in zebrafish models of Parkinson’s disease. Biol Open 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolwani RJ, Buckmaster PS, Varma S, Cosgaya JM, Wu Y, Suri C, Shooter EM (2002) BDNF overexpression increases dendrite complexity in hippocampal dentate gyrus. Neuroscience 114:795–805. [DOI] [PubMed] [Google Scholar]
- Valente EM, Salvi S, Ialongo T, Marongiu R, Elia AE, Caputo V, Romito L, Albanese A, Dallapiccola B, Bentivoglio AR (2004) PINK1 mutations are associated with sporadic early-onset parkinsonism. Annals of Neurology 56:336–341. [DOI] [PubMed] [Google Scholar]
- Wang KZQ, Steer E, Otero PA, Bateman NW, Cheng MH, Scott AL, Wu C, Bahar I, Shih YT, Hsueh YP, Chu CT (2018a) PINK1 Interacts with VCP/p97 and Activates PKA to Promote NSFL1C/p47 Phosphorylation and Dendritic Arborization in Neurons. eNeuro 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang KZQ, Steer E, Otero PA, Bateman NW, Cheng MH, Scott AL, Wu C, Bahar I, Shih Y-T, Hsueh Y-P, Chu CT (2018b) PINK1 Interacts with VCP/p97 and Activates PKA to Promote NSFL1C/p47 Phosphorylation and Dendritic Arborization in Neurons. eNeuro 5:ENEURO.0466–0418.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Winter D, Ashrafi G, Schlehe J, Wong YL, Selkoe D, Rice S, Steen J, LaVoie MJ, Schwarz TL (2011) PINK1 and Parkin target Miro for phosphorylation and degradation to arrest mitochondrial motility. Cell 147:893–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Guo J, Fei E, Mu Y, He S, Che X, Tan J, Xia K, Zhang Z, Wang G, Tang B (2014) BAG5 protects against mitochondrial oxidative damage through regulating PINK1 degradation. PLoS One 9:e86276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Liu H, Zhang BS, Soares JC, Zhang XY (2016) Low BDNF is associated with cognitive impairments in patients with Parkinson’s disease. Parkinsonism Relat Disord 29:66–71. [DOI] [PubMed] [Google Scholar]
- Wu D-C, Teismann P, Tieu K, Vila M, Jackson-Lewis V, Ischiropoulos H, Przedborski S (2003) NADPH oxidase mediates oxidative stress in the 1-methyl-4-phenyl-1, 2, 3, 6-tetrahydropyridine model of Parkinson’s disease. Proceedings of the National Academy of Sciences 100:6145–6150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xicoy H, Wieringa B, Martens GJM (2017) The SH-SY5Y cell line in Parkinson’s disease research: a systematic review. Molecular Neurodegeneration 12:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zagrebelsky M, Gödecke N, Remus A, Korte M (2018) Cell type-specific effects of BDNF in modulating dendritic architecture of hippocampal neurons. Brain Struct Funct 223:3689–3709. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.