SUMMARY
Many organisms evolved strategies to survive desiccation. Plant seeds protect dehydrated embryos from various stressors and can lay dormant for millennia. Hydration is the key trigger to initiate germination, but the mechanism by which seeds sense water remains unresolved. We identified an uncharacterized Arabidopsis thaliana prion-like protein we named FLOE1, which phase separates upon hydration and allows the embryo to sense water stress. We demonstrate that biophysical states of FLOE1 condensates modulate its biological function in vivo in suppressing seed germination under unfavorable environments. We find intragenic, intraspecific, and interspecific natural variation in FLOE1 expression and phase separation and show that intragenic variation is associated with adaptive germination strategies in natural populations. This combination of molecular, organismal, and ecological studies uncovers FLOE1 as a tunable environmental sensor with direct implications for the design of drought resistant crops, in the face of climate change.
Keywords: Phase separation, Prion-like, Intrinsically Disordered Proteins, Seed germination, Water stress, Salt stress, Biomolecular condensate, Bet-hedging, Water sensing, Seed desiccation, Adaptation
Graphical Abstract
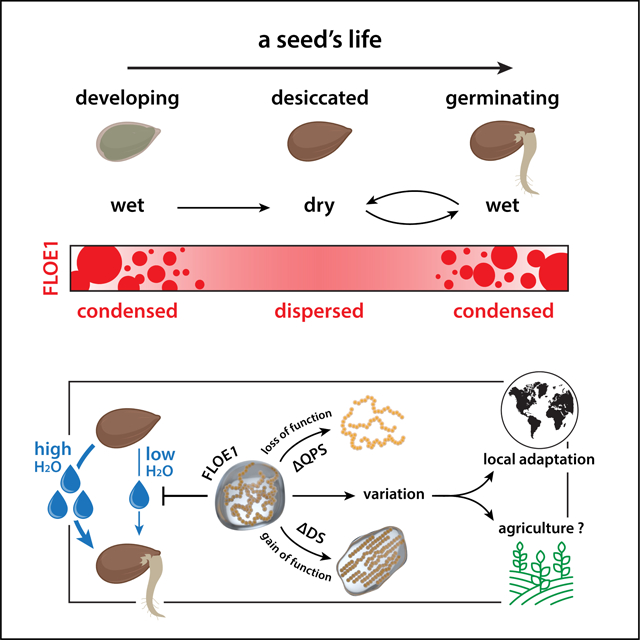
In Brief:
A previously uncharacterized protein, FLOE1, functions in seeds as a water potential sensor and can undergo reversible phase separation.
INTRODUCTION
Although water is essential for life, numerous organisms developed ways to withstand severe water stress or have quiescent desiccated life stages (Boothby and Pielak, 2017; Esbelin et al., 2018; Giarola et al., 2017). Among the organisms that have come up with such extreme adaptation are the seed plants. Seeds are specialized propagation vectors that can mature to a quiescent, desiccated state, allowing them to remain viable in harsh conditions anywhere from a few years to millennia (Sallon et al., 2020; Sano et al., 2016; Yashina et al., 2012). They can survive extreme desiccation by accumulating protective molecules that profoundly change their cellular biophysical properties (Buitink and Leprince, 2008; Leprince and Buitink, 2015). Upon water uptake, called imbibition, seeds undergo a cascade of biochemical and mechanical events and resume cellular activities (Nonogaki et al., 2018; Rajjou et al., 2012). Seeds can endure multiple hydration-dehydration cycles while remaining viable and desiccation tolerant (Bai et al., 2012). However, once committed to germination, seeds lose this ability (Rajjou et al., 2012). Thus, poor timing of germination can severely limit seedling survival (Kranner et al., 2010). Despite the fundamental importance of germination control, the molecular details that underpin this decision remain elusive.
Limited water availability dramatically alters protein solubility and induces protein aggregation. To protect their cytoplasmic components, seeds have an arsenal of protective mechanisms. These include the upregulation of osmoprotectants (ElSayed et al., 2014) and chaperones, and a cytoplasmic liquid-to-glass transition during seed maturation (Buitink and Leprince, 2008; Leprince and Buitink, 2015). Given the complex biophysical changes proteins must endure under such extreme conditions, we investigated how seed proteins might have adapted to deal with them. We used transcriptomic data to identify the Arabidopsis thaliana seed-enriched proteome. Interestingly, we found an enrichment for protein disorder, among which were several prion-like proteins. These types of proteins have been previously connected to protein phase separation, a physical process that allows cells to compartmentalize biomolecules into membraneless biomolecular condensates (Boeynaems et al., 2018; Shin and Brangwynne, 2017). While this process has been recently implicated in plant biology (Chakrabortee et al., 2016; Fang et al., 2019; Jung et al., 2020; Powers et al., 2019; van Dop et al., 2020; Zhang et al., 2020), it remains unclear how phase separation could play a role in seeds , especially given the challenges regarding protein aggregation under water-limiting conditions.
Here we report the identification of an uncharacterized prion-like protein, FLOE1, which specifically phase separates upon seed imbibition. Through phenotypic and transcriptomic analyses of mutant lines, we provide evidence that the protein allows the embryo to sense water potential and attenuates germination under unfavorable conditions. FLOE1 undergoes phase separation and functions to regulate germination under conditions of water stress. We dissected the molecular grammar of FLOE1 and created a set of mutants that span the material state spectrum. Using these to change FLOE1’s biophysical state resulted in altered seed germination, providing evidence for the functional importance of a biomolecular condensate’s material properties in vivo in a multicellular organism. Finally, we uncovered a vast family of FLOE homologs across green plants. We show that natural sequence variation drives differences in FLOE phase behavior suggesting that FLOE variation might regulate seed germination in the wild.
RESULTS
FLOE1 is an uncharacterized seed-specific prion-like protein
To find seed-enriched proteins, we re-analyzed publicly available A. thaliana transcriptomics data and found 449 protein-coding genes that are expressed at higher levels in dry seeds compared to other tissues (Fig. 1A, Table S1) (Austin et al., 2016; Schmid et al., 2005). These proteins had a different amino acid compositional profile (Fig. 1B) and were enriched for regions of structural disorder compared to the rest of the proteome (Fig. 1C). Intrinsically disordered proteins (IDPs) have emerged as key players in cell biology that, among many roles, orchestrate how cells organize themselves and their contingent biochemical reactions into discrete membraneless compartments by a process called liquid-liquid phase separation (LLPS) (Boeynaems et al., 2018; Shin and Brangwynne, 2017). A subset of IDPs harbors a class of domains known as prion-like domains (PrLD), and we identified 14 proteins with PrLDs in the seed-enriched proteins (Fig. 1D). PrLDs were first identified in fungal prions and later shown to drive reversible protein phase separation in diverse eukaryotic species (Alberti et al., 2009). In yeast, PrLDs give rise to phenotypic diversity via a proposed bet-hedging strategy to help cope with a fluctuating environment (Halfmann et al., 2012). All but one of these PrLD-containing seed-enriched proteins had annotated functions or domains related to nucleic acid metabolism. The one that did not, AT4G28300, was an uncharacterized plant-specific protein, which we named FLOE1, inspired by the second movement of Glassworks by Philip Glass as well as the definition of floe being ‘a sheet of floating ice’, which is a phase-separated body of water.
Figure 1: FLOE1 is an uncharacterized seed protein that undergoes biomolecular condensation in a hydration-dependent manner.
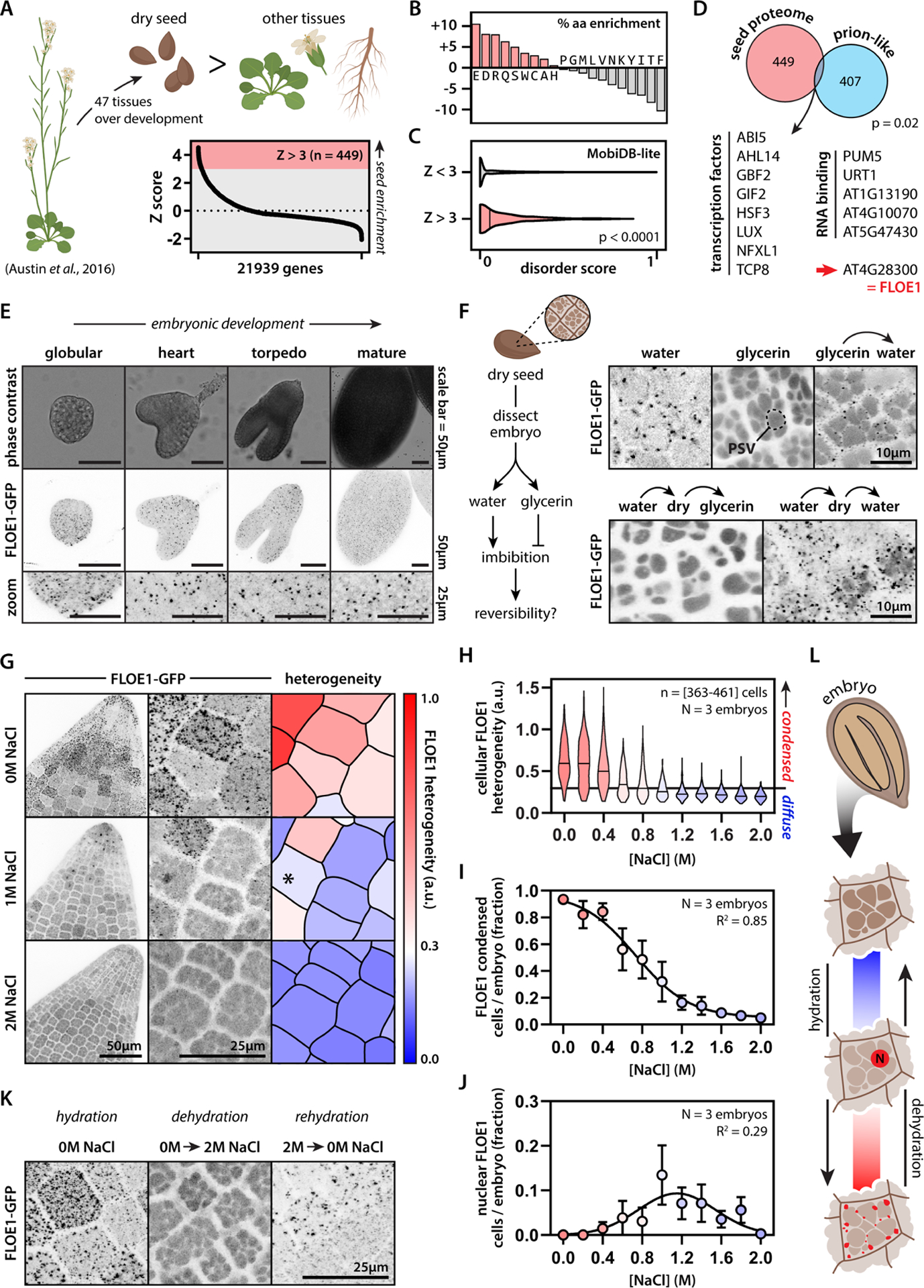
(A) Identification of genes enriched in dry Arabidopsis seeds. (B–C) The seed proteome is enriched for specific amino acids (B) and intrinsic disorder (C). Mann-Whitney test. (D) The seed proteome is enriched for prion-like proteins. Binomial test. AT4G28300 is an uncharacterized prion-like protein, which we named FLOE1. (E) FLOE1p:FLOE1-GFP is expressed during embryonic development and forms condensates. (F) FLOE1-GFP forms condensates in embryos dissected from dry seed in a hydration-dependent and reversible manner. Embryonic cotyledons are shown. PSV denotes autofluorescent protein storage vacuoles that are more prominent in the dry state than in the hydrated state (see Fig. S1E). (G) Cell-to-cell variation in subcellular FLOE1-GFP heterogeneity in response to salt. Radicles are shown. * denotes nuclear localization. (H) Quantification of cellular FLOE1 heterogeneity as a function of salt concentration. Black line denotes the 95th percentile of the 2M NaCl heterogeneity distribution. (I) Quantification of the percentage of cells per radicle that show FLOE1 condensation as a function of salt concentration. Mean ± SEM. Four-parameter dose-response fit. (J) Quantification of the percentage of cells per radicle that show FLOE1 nuclear localization as a function of salt concentration. Mean ± SEM. Gaussian fit. (K) FLOE1-GFP condensation exhibits reversibility between high and no salt treatment. Radicles are shown. (L) Scheme highlighting different FLOE1 behaviors upon imbibition. Fluorescence microscopy images are maximum projections. GFP signal is displayed as an inverted gray scale.
FLOE1 undergoes hydration-dependent phase separation
FLOE1 accumulates during embryo development and its expression peaks in the mature desiccated state (Fig. S1A–B). We generated transgenic A. thaliana lines expressing FLOE1’s genomic region—from its predicted promoter to its last coding codon—fused to GFP. FLOE1 formed cytoplasmic condensates during embryonic development (Fig. 1E, Fig. S1C) and in embryos dissected in water from dry seeds (Fig. 1F, Fig. S1D). However, when we dissected dry seeds in glycerin instead of water (to mimic the desiccated environment), FLOE1 did not form condensates and was diffusely localized (Fig. 1F, Fig. S1E–F). When we transferred these embryos from glycerin to water, FLOE1 condensates spontaneously appeared (Fig. 1F) and were fully reversible with repeated hydration-dehydration cycles (Fig. 1F, Fig. S1K). Pre-treating seeds with the translation inhibitor cycloheximide did not affect the formation of FLOE1 condensates, indicating that they are distinct from stress granules and processing bodies (Gutierrez-Beltran et al., 2015), and that their emergence did not depend on FLOE1 translation upon imbibition (Fig. S1G). In line with this, FLOE1 condensates appeared rapidly upon hydration (in less than a minute), arguing against the involvement of biosynthetic processes and for a rapid biophysical response to water. To directly test whether FLOE1 forms condensates in response to changes in water potential, we dissected embryos in solutions of varying concentrations of salt, mannitol, or sorbitol (Fig. 1G–I, Fig. S1H–I). High concentrations of salt resembled dry conditions–embryos lacked visible FLOE1 condensates (Fig. 1G–I). Lowering the salt concentration resulted in a gradual emergence of condensates, which was highly variable at the cell-to-cell (Fig. 1G–H) and tissue levels (Fig. 1I). In intermediate concentrations, we observed a small number of cells with apparent nuclear localization of FLOE1 (Fig. 1J, Fig. S1J), suggesting this could be a behavior associated with early steps of imbibition, before the majority of the protein condenses in the cytoplasm. Like our observations with repeated hydration-dehydration cycles, FLOE1 condensation was also reversible by moving seeds back and forth between solutions of high or no salt (Fig. 1K, Fig. S1K). Thus, FLOE1 forms cytoplasmic condensates in response to changes in water potential in vivo and these are fully reversible (Fig. 1L).
FLOE1 attenuates germination under water stress
We next asked whether FLOE1 functions in germination. Lines carrying the knockout allele floe1-1 did not show any obvious developmental defects, and floe1-1 seeds had the same size, shape, and weight as wildtype seeds (Fig. S2A). floe1-1 seeds germinated indistinguishably from wildtype seeds under standard conditions (Fig. S2B) but, intriguingly, had higher germination levels under conditions of water deprivation induced by salt (Fig. 2A, Fig. S2C) or mannitol (Fig. S2C). We confirmed that these phenotypes were caused by mutations in FLOE1 using independent lines carrying CRISPR-Cas9 deletion alleles and floe1-1 lines complemented with the wildtype allele (Fig. S2C–F). Germination during stressful environmental conditions is risky for a plant and can reduce fitness. Indeed, both wildtype and floe1-1 seedlings displayed developmental defects or eventually died under these conditions (Fig. S2G), whereas ungerminated seeds retained their full germination potential upon stress alleviation (Fig. 2B), in line with bet-hedging strategies utilized by stressed seeds (Gremer and Venable, 2014; Johnston and Bassel, 2018; Villa Martin et al., 2019). To define the role of FLOE1 condensates in stress-mediated attenuation of germination, we first asked whether the condensates would reform upon stress alleviation. Whereas ungerminated salt-stressed seeds were largely devoid of FLOE1 condensates, even after 15 days of incubation, alleviating salt stress robustly induced their appearance (Fig. 2C, Fig. S2H). Moreover, the extent of FLOE1 phase separation correlated with germination levels (Fig. S3). Importantly, in contrast to the hydration experiments we performed on naked embryos by submersion (Fig. 1F–K), we performed these experiments by incubating intact seeds on agar plates with the indicated salt concentrations–representing more physiologically relevant conditions (Fig. S3D). These results show that FLOE1 phase separates in vivo and that this event coincides with the onset of germination. Moreover, under water limitation, FLOE1 attenuates germination, thereby enabling a better chance of survival (Fig. 2D).
Figure 2: FLOE1 attenuates germination under water stress.
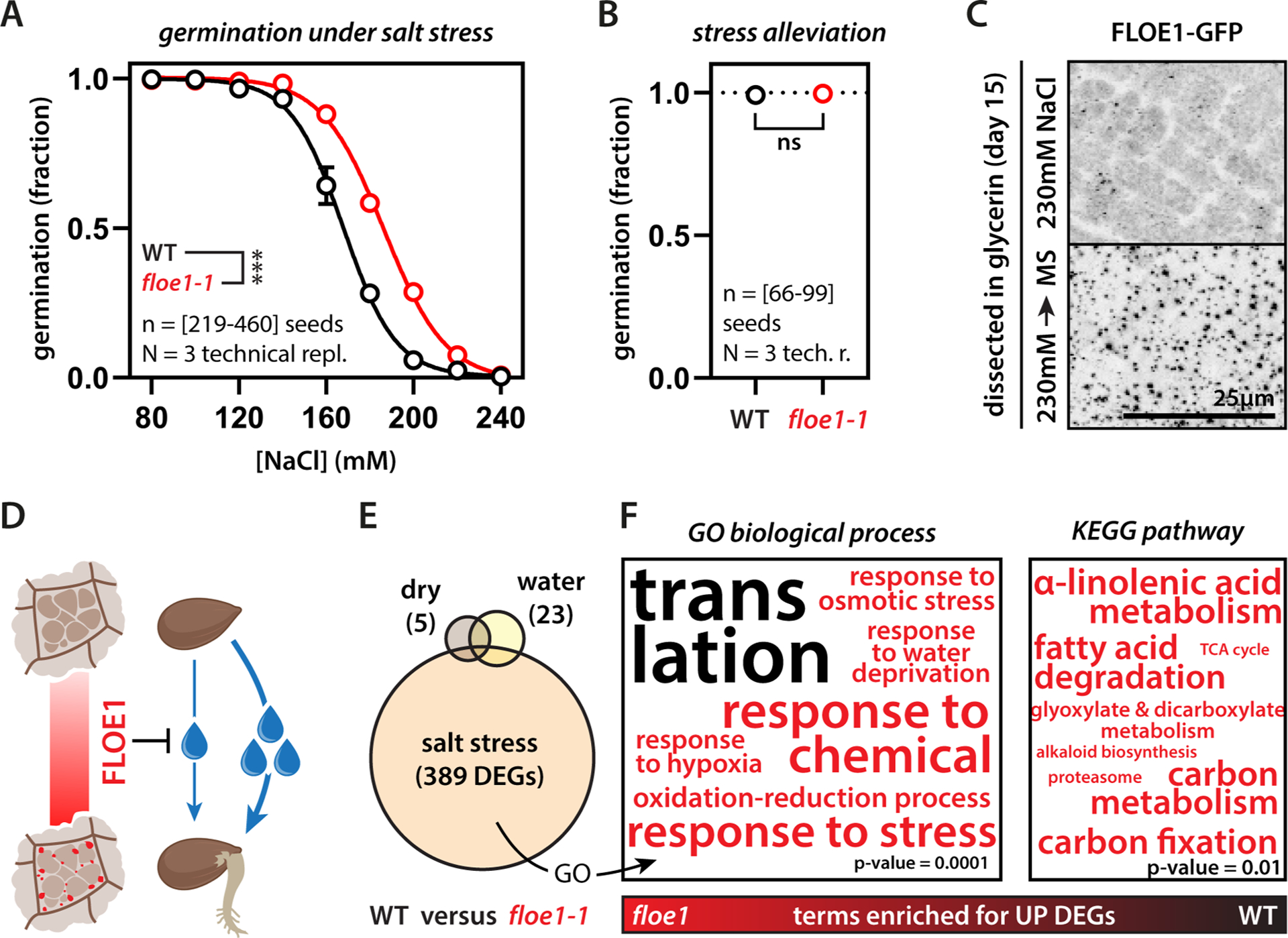
(A) floe1-1 seeds show higher germination levels under salt stress. Two-way ANOVA, four-parameter dose-response fit, *** p-value < 0.001. Representative of three independent experiments. ΔIC50 is 18.5mM. (B) Seeds retain full germination potential under standard conditions after a 15-day 230mM salt stress treatment. Representative of three independent experiments. (C) Condensates are largely absent in ungerminated seeds after 15 days of incubation under salt stress. FLOE1 condensates appear within two hours after transfer to standard conditions (MS medium). Maximum projection images of radicle cells. GFP signal is displayed as an inverted gray scale. (D) Scheme highlighting the potential function of FLOE1 in attenuating germination when water potential is low. Droplets indicate water availability. (E) floe1-1 seeds show high numbers of differentially expressed genes (DEGs) after imbibition under salt stress, as opposed to unimbibed (dry) and normally imbibed (water) seeds. Imbibition was performed on MS medium by first stratifying for 5 days followed by 4h incubation in a growth cabinet. (F) floe1-1 seeds upregulate stress response genes and genes implicated in metabolism compared to wildtype seeds, and have relatively lower expression of genes involved in ribosomal biogenesis. The only KEGG pathway enriched for the WT was “ribosome” (p-value = 3.88E-17, not shown). See also Table S2. Font size correlates to −log10 (p-value). P-values at bottom-right for scale.
To place FLOE1’s role in the context of other germination pathways, we performed transcriptomics on wildtype and floe1-1 plants. RNA-seq analysis suggests that FLOE1 functions upstream of key germination pathways. floe1-1 seeds had only few changes in gene expression compared to wildtype seeds in both their desiccated state and under normal imbibition. But under salt stress, loss of FLOE1 caused a significant change in the transcriptome (Fig. 2E). In line with the observation that floe1-1 seeds germinate more under water stress, their salt stress-responsive transcriptome was marked by an upregulation of metabolic genes, consistent with promoting germination, and stress-response genes, as the seeds exit their stress-tolerant desiccated state (Fig. 2F, Fig. S2I, Table S2). Interestingly, floe1-1 seeds showed relatively lower transcript levels of genes involved in ribosome biogenesis compared to wildtype seeds under salt stress (Fig. 2F, Fig. S2I). We attribute the apparent downregulation of these housekeeping genes in floe1-1 seeds under salt stress to the initiation of gene expression upon germination. Finally, since the ratio of the phytohormones abscisic acid (ABA) and gibberellin (GA) is a major regulator of germination under unfavorable conditions, including salinity (Shu et al., 2017), we explored whether FLOE1 is involved in this hormonal regulation. Disruption of the ABA/GA balance through supplementation of ABA or inhibition of GA synthesis did not induce any germination differences between floe1-1 and wildtype seeds (Fig. S2J–K). Together, these results suggest that FLOE1 regulates germination through a distinct process that involves the direct sensing of water potential.
Synergistic and opposing molecular forces regulate FLOE1’s phase behavior
Numerous yeast proteins undergo oligomerization or phase separation upon stress-induced quiescence (Munder et al., 2016) but FLOE1 undergoes biomolecular condensation upon release from the quiescent state. To define the mechanism by which FLOE1 undergoes this switch, we dissected the molecular grammar underlying this behavior. We first expressed and purified MBP-tagged FLOE1 protein from E. coli to determine if it can undergo phase separation in isolation. Upon cleavage of the MBP tag, FLOE1 spontaneously demixed to form droplets (Fig. 3A, Fig. S4A–B) that enriched FLOE1 but excluded MBP (Fig. S4C), similar to the behavior of other phase-separating proteins (Burke et al., 2015). To test if FLOE1 can phase separate in the range of physiological protein concentrations, we set out to estimate the local protein concentration of FLOE1 in vivo and performed dilution series in vitro. By calculating the volume of the cytoplasmic fraction of A. thaliana embryos (see STAR Methods), we found that FLOE1’s in vivo concentration is within the same range it can phase separate in vitro (Fig. S4D, Fig. S5A–G, Movie S1, Table S3). Because FLOE1 can phase separate in isolation, we next defined which protein domains drive phase separation. FLOE1 harbors a predicted coiled-coil domain and a conserved plant-specific domain of unknown function (DUF1421) (Fig. 3B). Disorder prediction algorithms identified another predicted folded region and two different disordered regions, one enriched for aspartic acid and serine (DS-rich) and the other enriched for glutamine, proline, and serine (QPS-rich). We heterologously expressed FLOE1 in two orthogonal systems, tobacco leaves (Fig. 3C, Fig. S1L–M, Movie S2) and the human osteosarcoma cell line U2OS (Fig. 3D). In these two systems, like in A. thaliana (Fig. 1), FLOE1 formed spherical condensates, providing independent platforms for interrogating the molecular drivers of condensation. We deleted each domain of FLOE1 and assayed the impact on cytoplasmic condensation (Fig. 3C–E). In both tobacco and human cells, mutants lacking either the short coiled-coil domain or DUF1421 behaved identically to the wildtype protein (Fig. 3C–E). In contrast, deletion of the other domains altered FLOE1 condensation (Fig. 3C–E). Deletion of the predicted folded domain, which we refer to as the nucleation domain, abolished cytoplasmic condensation, resulting in a fraction of the protein redistributing to the nucleus. Folded oligomerization domains play important roles in nucleating phase separation of several IDPs (Boeynaems et al., 2018). Indeed, expression of chimeric fusion proteins revealed that this domain is sufficient to nucleate phase separation of the FLOE1 PrLD (i.e., QPS) and the PrLD derived from the human FUS protein, which has been extensively studied for its phase separation behavior (Patel et al., 2015) (Fig. 4A). All-atom simulations and homology-based modeling suggest that the nucleation domain adopts a trimeric coiled coil conformation (Fig. S4E), providing the multivalent interactions required for nucleating protein phase separation.
Figure 3: Molecular dissection of FLOE1 phase separation.
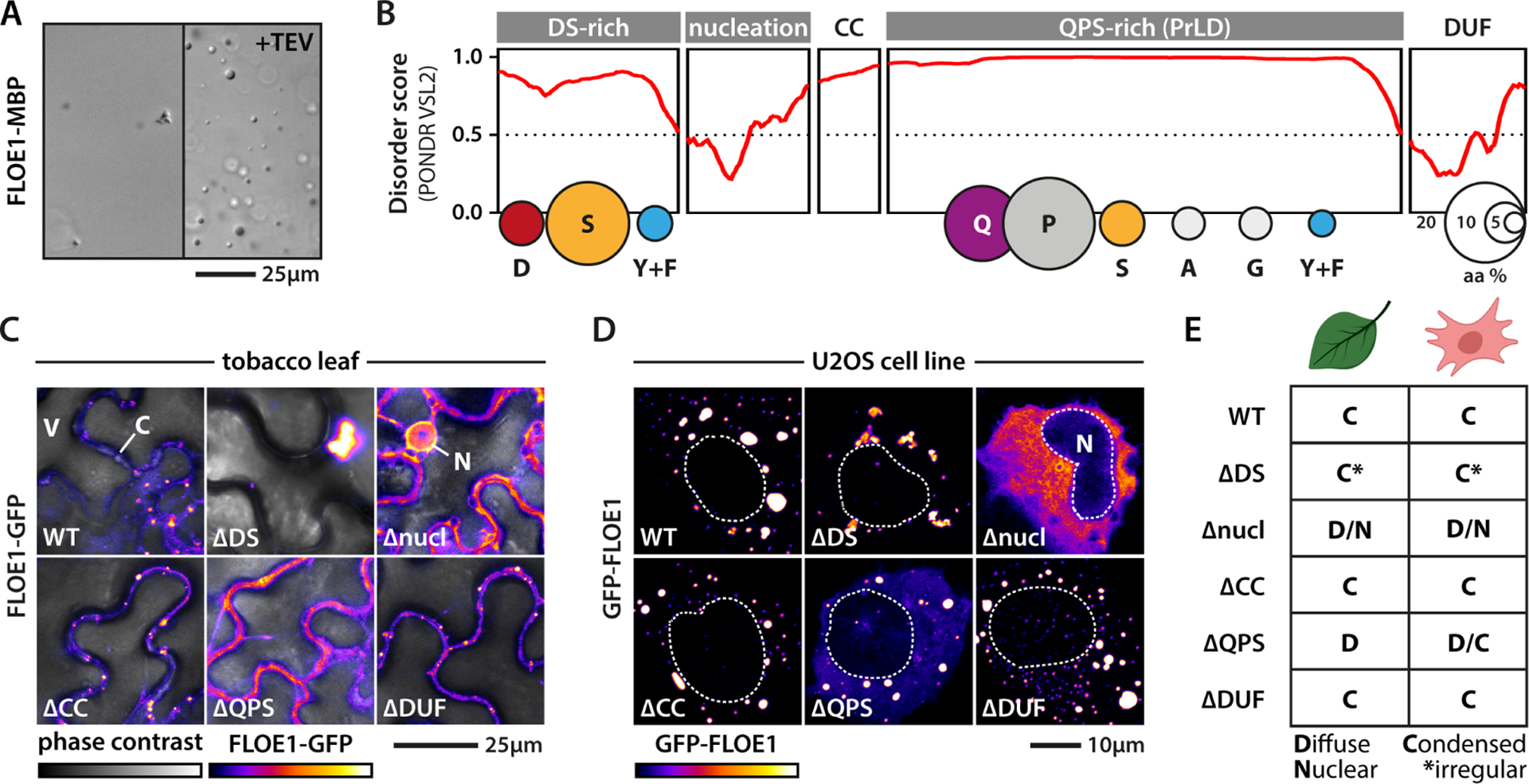
(A) Recombinant MBP-FLOE1 phase separates in the test tube upon MBP cleavage with TEV protease. Irregular small aggregates can be seen pre-cleavage highlighting FLOE1 aggregation-propensity. DIC imaging. (B) FLOE1 domain structure. CC = predicted coiled coil, DUF = DUF1421. Balloon plots show amino acid composition of the disordered domains. (C–D) Expression of FLOE1 domain deletion mutants in tobacco epidermal pavement cells (C) and human U2OS cells (D). V = vacuole, C = cytoplasm, N = nucleus. (E) Summary of FLOE1 behavior in tobacco and human cells. Fluorescence microscopy images are single optical sections. GFP signal is displayed as a false-colored intensity scale.
Figure 4: Molecular dissection of FLOE1 phase separation.
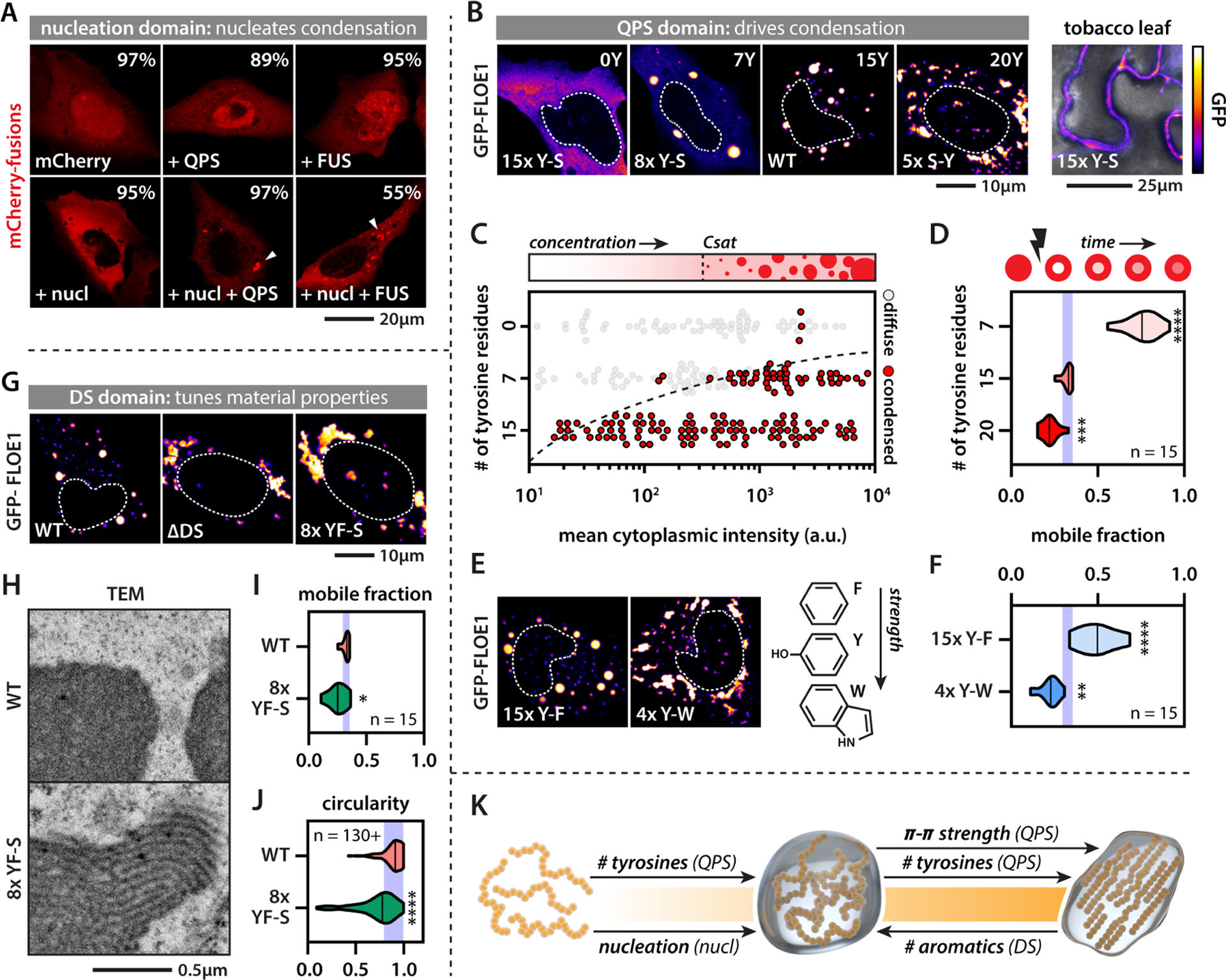
(A) Chimeric proteins containing both the FLOE1 nucleation domain and the PrLDs from FLOE1 (QPS) or the human FUS protein form cytoplasmic condensates. Percentages display number of cells lacking or containing condensates. Average of three experiments. Arrowheads point at cytoplasmic condensates. (B) The number of QPS tyrosine residues alters FLOE1 phase separation in human and tobacco cells. (C) FLOE1 phase diagram as a function of concentration and number of QPS tyrosines. (D) Number of QPS tyrosines affects intracondensate FLOE1 dynamics. Mobile fraction as assayed by FRAP is shown. One-way ANOVA. (E–F) QPS tyrosine-phenylalanine and tyrosine-tryptophan substitutions alter condensate morphology (E) and intracondensate dynamics compared to WT (F). One-way ANOVA. (G) DS deletion or DS tyrosine/phenylalanine-serine substitutions alter condensate morphology. (H) TEM shows that mutant DS FLOE1 condensates have filamentous substructure that is absent in the WT. U2OS cells. (I) DS tyrosine/phenylalanine-serine substitutions alter intracondensate dynamics. Student’s t-test. (J) DS tyrosine/phenylalanine-serine substitutions alter condensate morphology. Mann-Whitney. (K) Scheme summarizing synergistic and opposing roles of FLOE1 domains on the material property spectrum. * p-value < 0.05, ** p-value < 0.01, *** p-value < 0.001, **** p-value < 0.0001. Purple band denotes WT mean ± SD (D, F, I, J). Fluorescence microscopy images are single optical sections. GFP signal is displayed as a false-colored intensity scale (scale in panel B).
In line with their role in driving phase separation of other prion-like proteins, deletion of the QPS PrLD reduced condensate formation in cells (Fig. 3C–E) and in the test tube (Fig. S4F). Consistent with the emerging sticker-spacer framework for PrLDs (Martin et al., 2020; Wang et al., 2018), the QPS PrLD has regularly spaced aromatic tyrosine residues along its sequence that may act as attractive stickers. Substituting tyrosine residues for serines (Y-S) decreased condensate formation in both human and tobacco cells in a dose-dependent manner (Fig. 4B, Table S3). By mapping out a phase diagram (Fig. 4C) and probing the molecular dynamics using fluorescence recovery after photobleaching (Fig. 4D) of Y-S and S-Y mutants, we confirmed that the number of tyrosines determines both the saturation and gelation concentrations of FLOE1 condensates, consistent with what has been shown for other PrLDs (Martin et al., 2020). These findings provide further evidence that FLOE1 condensates form via LLPS, and increasing its multivalency drives gelation into more solid-like irregular assemblies. While changing the number of stickers can drive a liquid-to-gel transition, altering sticker strength may also alter the gelation concentration. Substituting tyrosines for weaker (phenylalanine) or stronger (tryptophan) aromatic residues affected both condensate morphology and intracondensate dynamics in a predictable manner, in which increasing the stickiness of the QPS PrLD induced gelation of FLOE1 (Fig. 4E–F, Table S3).
Surprisingly, removing the N-terminal DS domain also induced FLOE1 gelation (Fig. 3C–E, Fig. 4G). Serine substitution of aromatic residues in the DS domain had a similar effect as DS deletion (Fig. 4G), suggesting that the aromatic residues in the DS and QPS domains have opposing functions. Intriguingly, electron microscopy revealed that the gel-like condensates formed via DS-8xY/F-S (8 phenylalanine and tyrosine residues changed to serine in the DS region) had a regular striated substructure and were strikingly distinct from the homogeneous wildtype condensates (Fig. 4H, Fig. S5I–M, Movie S3), consistent with their reduced dynamics and change in morphology (Fig. 4I–J). This striated substructure is reminiscent of synaptonemal complexes that have also been proposed to form through phase separation of a coiled coil protein (Rog et al., 2017). Thus, synergistic and opposing molecular forces tightly regulate FLOE1’s biophysical phase behavior, and changing this balance allows its properties to be experimentally toggled between dilute, liquid droplet, and solid gel states (Fig. 4K).
FLOE1’s condensate material properties modulate its function in germination
To directly test whether FLOE1’s role in seed germination is a function of its biophysical state, we generated A. thaliana lines carrying wildtype or different FLOE1 domain deletion mutants in the null background (Fig. 5A–B). These mutants behaved consistently in A. thaliana embryos as they did in human and tobacco cells (Fig. 3C–E). The ΔQPS mutant was unable to phase separate upon imbibition (Fig. 5B), whereas the ΔDUF mutant formed condensates that were indistinguishable from the wildtype (Fig. 5B–D). In contrast, the ΔDS mutant formed condensates that were larger than those formed by the wildtype protein (Fig. 5B–D), consistent with what we observed in tobacco and human cells. The ΔDS mutant has lost its hydration-dependency and seems to exist in a constitutively condensed state (Fig. 5E). This is important because now we have a way to eliminate the reversibility of FLOE1 condensate formation. We assayed germination levels under salt stress and found that removing the DUF domain resulted in phenotypes that were indistinguishable from those of knockout lines complemented with wildtype FLOE1 (Fig. 5F, Fig. S6A–D). However, removing the QPS domain resulted in phenotypes indistinguishable from those of knockout mutants (Fig. 5F, Fig. S6A–D), suggesting that this domain is required for FLOE1 function. Surprisingly, removing the DS domain resulted in greatly enhanced germination levels under salt stress, surpassing even that of the null mutant, indicating that ΔDS likely functions as a gain-of-function mutation (Fig. 5F, Fig. S6A–D). These data provide evidence that the reversibility of FLOE1 condensate formation is important for its function, but it remains possible that the ΔQPS and ΔDS mutations have additional effects on FLOE1 function independent of phase separation (see Discussion).
Figure 5: FLOE1 condensate material properties regulate its role in seed germination under salt stress.
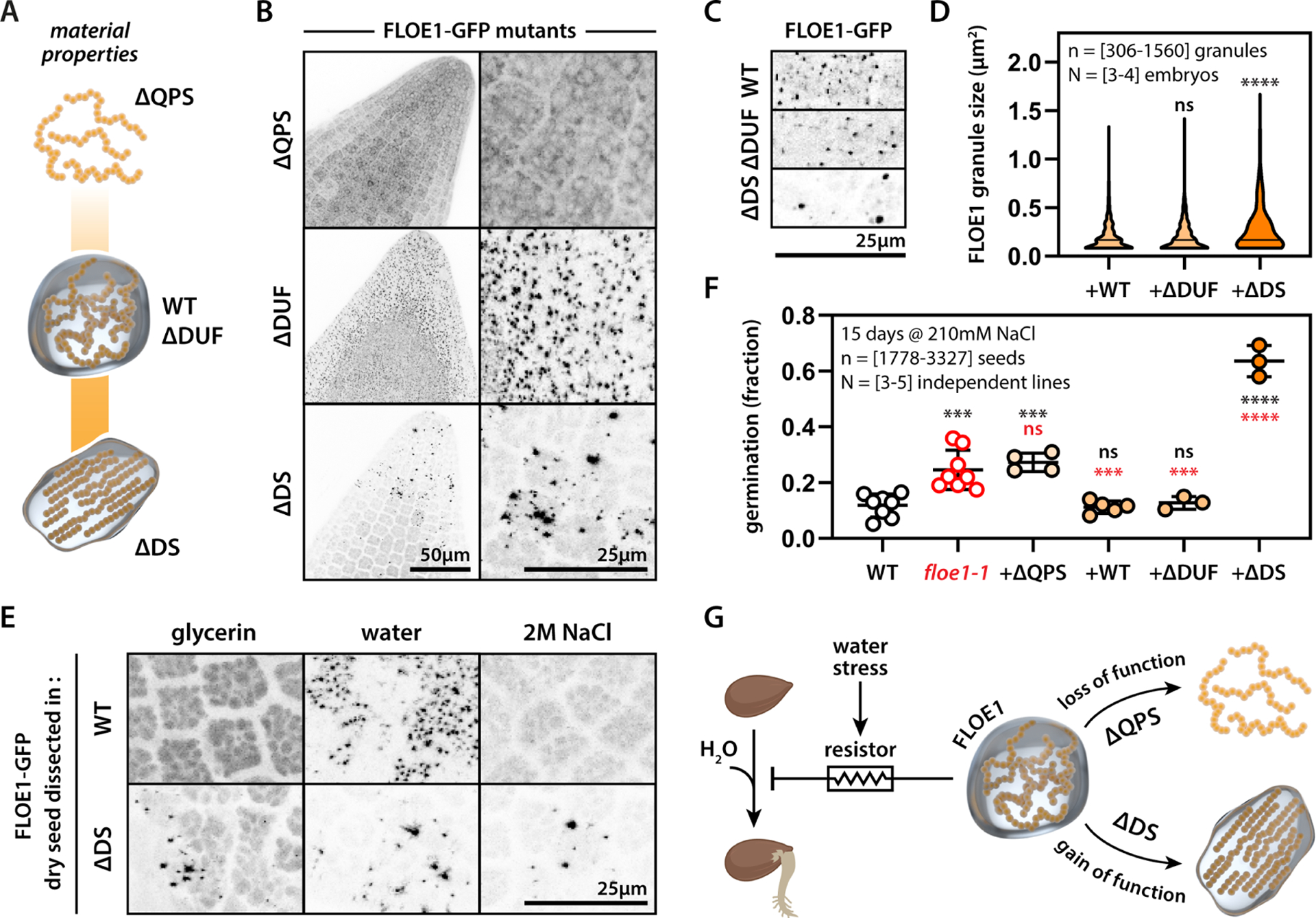
(A) Scheme highlighting position of tested FLOE61 mutants on the material properties spectrum. (B) Representative images of floe1-1 mutants complemented with ΔQPS, ΔDUF and ΔDS forms of FLOE1 upon dissection in water. Maximum projection images from embryo radicles. (C) Close-up images of WT and mutant FLOE1 condensates. Single optical sections from embryo radicles. (D) Quantification of FLOE1 condensate size. One-way ANOVA. (E) ΔDS FLOE1 condensates are not dependent on hydration. Maximum projection images from embryo radicles. (F) Germination levels of WT, floe1-1 and complemented lines. One-way ANOVA. Representative of three independent experiments. (G) Scheme highlighting FLOE1’s role in regulating germination and the effect of mutants with altered material properties. * p-value < 0.05, ** p-value < 0.01, *** p-value < 0.001, **** p-value < 0.0001. GFP signal is displayed as an inverted gray scale.
Assaying the transcriptome of ΔDS seeds showed large changes in gene expression compared to wildtype complemented lines (Fig. S6F–G, Table S2). In contrast to floe1-1 seeds, where gene expression changes were limited to salt stress (Fig. 2E), in ΔDS seeds, we observed significant changes in both dry and non-stress imbibition conditions (Fig. S6F–G), suggesting the possibility of non-stress related phenotypes. Indeed, ΔDS seeds displayed faster germination rates under standard conditions (Fig. S6D–E). These findings indicate that switching the material properties of a biomolecular condensate can have broad phenotypic effects that can impact organismal fitness in a variety of environmental conditions (Fig. 5G).
Variation in the FLOE1 DS domain regulates variation in phase behavior, seed dormancy and germination in natural populations
To investigate the role of the material properties of FLOE1 in an ecological and evolutionary context, we first examined FLOE1’s intragenic variation in phase behavior, and explored variation in expression level and seed dormancy across hundreds of A. thaliana ecotypes. FLOE1 encodes two isoforms from alternatively spliced transcripts (6A). Besides the full-length isoform (FLOE1.1), which is the dominant form in all ecotypes (Fig. S7A), FLOE1 encodes a shorter splice isoform that lacks the majority of the DS domain (6A). This shorter isoform (FLOE1.2) forms larger condensates (Fig. 6B, Fig. S6H) that can recruit the longer isoform (Fig. 6C). We next asked whether this intragenic variation in material properties of FLOE1.1 and FLOE1.2, which, based on our mutant studies, are associated with attenuating and promoting germination, respectively, could tune germination in natural populations. To explore this question, we generated a set of floe1-1 lines complemented with different expression levels of FLOE1, and found that FLOE1 transgene expression inversely correlated with germination levels under salt stress (Fig. S2D, Fig. S6B–C). This provided an opportunity to look for evidence for such a tuning mechanism in wild populations, and to see if FLOE1 isoforms elicited differential effects in nature. We took advantage of publicly available transcriptomic data for a wide array of A. thaliana ecotypes (1001 Genomes Consortium, 2016), with a final set of 478 ecotypes (Table S4), and several published phenotypic studies (Martinez-Berdeja et al., 2020; Togninalli et al., 2020). Consistent with our findings from manipulating gene expression levels and FLOE1 mutants, seed dormancy across ecotypes was positively correlated with FLOE1.1 expression, but not with FLOE1.2 expression (Fig. S7B). Since FLOE1.1 is the dominant isoform (Fig. S7A), this observation is in line with our results showing that FLOE1 dose-dependently attenuates germination in lab strains (Fig. S2D). By contrast, FLOE1.2 expression across ecotypes was strongly correlated with promoting germination, whereas this correlation was less pronounced for FLOE1.1 expression (Fig. S7C–F). Given that FLOE1.2 seems to mimic the phase behavior of our ΔDS mutant, the increased germination levels coinciding with its increased expression are consistent with the gain-of-function phenotype of ΔDS seeds (Fig. 5F, Fig. S6B,E). While these correlations corroborate our experimental data, they do not address whether the expression of FLOE1 isoforms is tuned to the specific ecological niche, and whether this variation could drive phenotypic adaptation in wild populations. To explore this question, we examined the isoform expression across ecotypes with climate data from their respective GPS coordinates of collection. In accordance with the hypothesis that FLOE1 expression is tuned by local environmental conditions, its expression was correlated with precipitation and temperature (Fig. S7G–L). Surprisingly, FLOE1.1/1.2 expression was positively correlated with summer precipitation, while negatively correlated with temperature, indicating this dormancy/germination switch may be a mechanism more prevalent in colder regions of the distribution. Since bet-hedging strategies are often employed by organisms to cope with changing environments, we asked how expression of the isoforms behaved in the context of variability in precipitation by calculating the coefficient of variation of July rainfall at the geographic locations of origin for a time-series from 1958–2017 (Abatzoglou et al., 2018). Strikingly, FLOE1.2 (putative promoter of germination) expression negatively correlated with the yearly variation in precipitation, whereas this was far less pronounced for FLOE1.1 (putative attenuator of germination) (Fig. S7M–N), indicating that natural populations living in environments with unpredictable precipitation express lower levels of FLOE1.2. This suggests that expression of FLOE1.2 to control germination/dormancy may not be an advantageous strategy for plants living in environments with unpredictable rainfall such as the Mediterranean basin, where temperature may be a more reliable cue to fine-tune dormancy, as shown by other studies of temperature sensing germination delayers (Exposito-Alonso, 2020). In summary, natural variation in the DS domain between FLOE1 isoforms may fine-tune FLOE1 function in seed dormancy and germination across ecotypes for local adaptation.
Figure 6: The DS-rich domain drives variation in condensate properties of FLOE1 isoforms and paralogs.
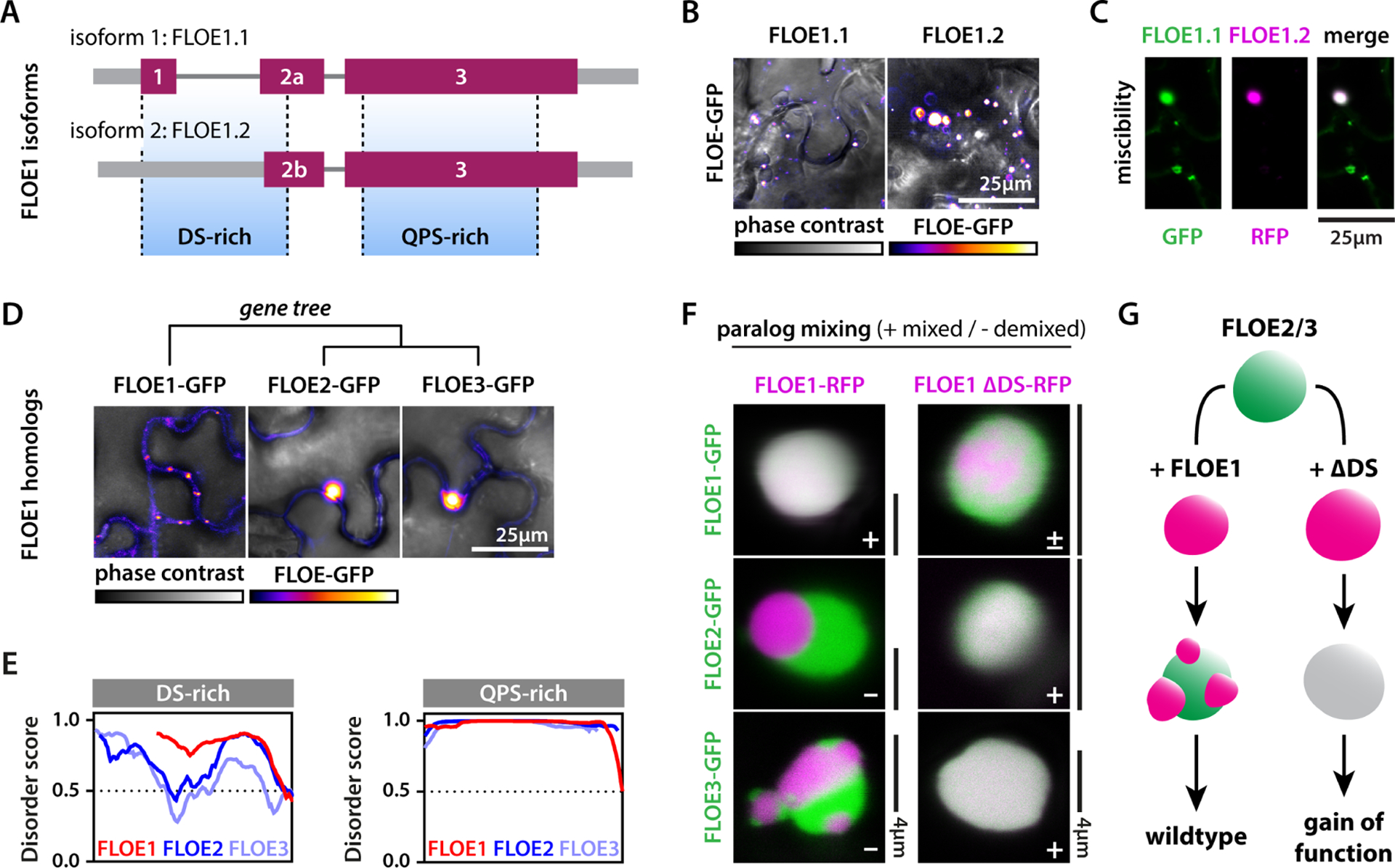
(A–B) A. thaliana has two FLOE1 isoforms. FLOE1.2 is missing most of the DS-rich region (A) and forms larger condensates than FLOE1.1 in tobacco leaves (B). (C) The large FLOE1.2 condensates recruit FLOE1.1. (D) FLOE1 has two A. thaliana paralogs that form larger condensates in tobacco leaves. (E) Disorder plots show strong length- and disorder variation between FLOE1 and FLOE2/3 in their DS-rich domains, but not in their QPS-rich domains. (F) FLOE1 condensates do not mix with FLOE2 and FLOE3 condensates. Deletion of the FLOE1 DS-rich domain partially disrupts mixing with wildtype FLOE1, but drives uniform mixing with FLOE2/3 condensates. (G) Scheme highlighting the switch-like role of the FLOE1 DS-domain in condensate mixing and the corresponding phenotypes. All images are single optical sections of tobacco epidermal pavement cells. (B, D) GFP signal is displayed as false-colored intensity scale (scale in panel D). (C, F) GFP and RFP signal are false-colored green and magenta.
Variation in the DS domain resides not only intragenically, but also intergenically. The A. thaliana genome also encodes two FLOE1 paralogs, which we named FLOE2 (AT5G14540) and FLOE3 (AT3G01560). When expressed in tobacco cells, FLOE2 and FLOE3 also formed larger condensates compared to FLOE1.1 (Fig. 6D, Fig. S6H). FLOE2 and FLOE3 have expanded DS regions with lower overall structural disorder, while their QPS domains were very similar to FLOE1’s (Fig. 6E). To see if FLOE2 and FLOE3 could recruit FLOE1.1 similar to FLOE1.2, we coexpressed them in tobacco. Surprisingly, coexpression of FLOE1 with FLOE2 or FLOE3 resulted in the formation of demixed condensates, illustrating immiscibility of FLOE1 with its paralogs (Fig. 6F, Fig. S6I). Such complex topologies are common among biomolecular condensates, and have been linked to differences in surface tension or viscosity between the different phases (Boeynaems et al., 2019; Feric et al., 2016). Since the DS domain is a key tuner of FLOE1 material properties, we wondered how deletion of this domain would impact condensate mixing. Surprisingly, while FLOE1/ΔDS FLOE1 condensates showed weak ΔDS substructures, consistent with the more gel-like nature of this mutant, combining the ΔDS FLOE1 mutant with FLOE2 or FLOE3 condensates lead to complete condensate mixing (Fig. 6F). Since the ΔDS FLOE1 mutant had such a pronounced phenotype compared to floe1-1 lines in promoting germination under salt stress, we hypothesize that the aberrant interaction of ΔDS FLOE1 with endogenous FLOE2/3 paralogs may be in part driving its gain-of-function phenotype (Fig. 6G).
Wide variety in FLOE phase behavior across the plant kingdom
To assess whether phase separation is conserved in the FLOE family, we explored the diversity of this protein family in nature. We found FLOE homologs in all green plant lineages, even in those preceding seed evolution (Fig. 7A–B, Fig. S6J). Phylogenetic analysis revealed the emergence of two major clades, FLOE1-like and FLOE2-like. While the latter clade includes the algal lineage, the FLOE1-like clade is restricted to seed plants. Both clades show conserved variation in the length of the two disordered domains (Fig. 7C). By testing a set of FLOE homologs across the plant kingdom, we find wide phenotypic variation in phase separation (Fig. 7D, Fig. S6K) that involves differences in condensate sizes and nuclear localization (Fig. 3C). These findings indicate there exists a wealth of natural sequence variation that might give rise to these different behaviors. Given our observations linking the variation in FLOE1 isoforms and paralogs in A. thaliana to phenotypic variation in both engineered and natural strains, this wide diversity of FLOE paralogs with differential phase separation behaviors suggests the possibility that this gene family could be a driver of functional phenotypic variation across the plant lineage.
Figure 7: Natural sequence variation tunes FLOE phase separation.
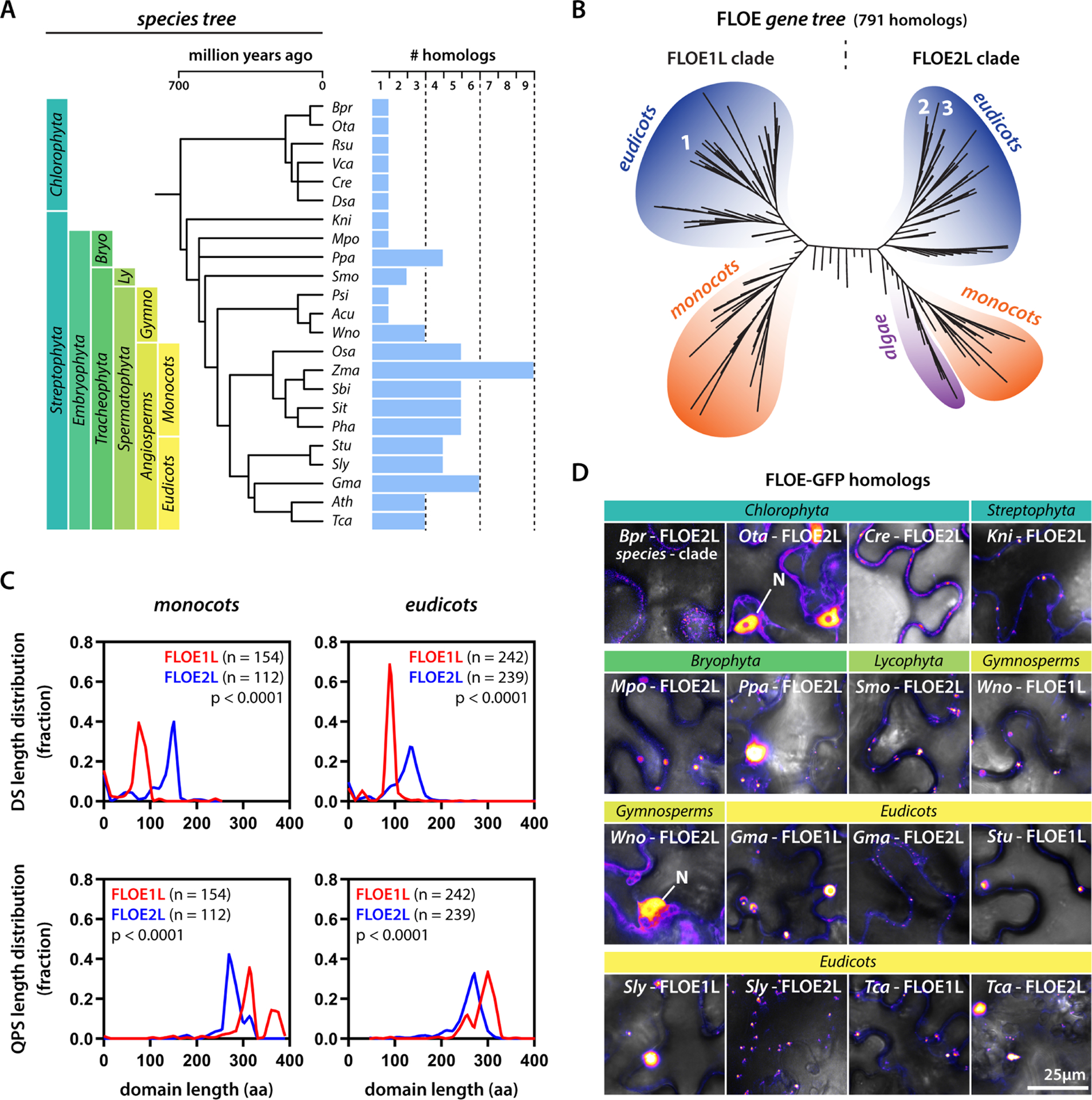
(A) A species tree of the plant kingdom with example species and their number of FLOE homologs. (B) Gene tree of FLOE homologs. Numbers correspond to FLOE1, FLOE2, and FLOE3. (C) Distribution of DS and QPS length differences between the FLOE1-like (FLOE1L) and FLOE2-like (FLOE2L) clades among monocots and eudicots. Mann-Whitney. (D) Examples of FLOE homologs from across the plant kingdom. N denotes nuclear localization. Single optical sections of tobacco epidermal pavement cells. GFP signal is displayed as false-colored intensity scale. Full species names for (B,D) in Fig. S6J. Each panel indicates the specific clade of the homolog (FLOE1L or FLOE2L).
DISCUSSION
Phase separation is emerging as a universal mechanism to explain how cells compartmentalize biomolecules. Despite advances involving the widespread identification of novel biomolecular condensates and elucidating their underlying molecular grammar, many questions remain. Chief among these is whether protein phase separation is truly functional for cellular physiology or whether it is an unintended consequence of protein evolution (Martin and Holehouse, 2020). Recent work in yeast shows that phase separation of prion-like and related proteins is important for their function (Franzmann et al., 2018; Riback et al., 2017), suggesting that these protein domains may indeed have evolved to drive protein condensation. Interestingly, amyloid-based aggregation of the same prion-like proteins has also been shown to confer fitness advantages to yeast under certain environmental conditions (Halfmann et al., 2012). This observation indicates that the material properties of biomolecular condensates may be important in regulating protein function. Yet, this picture is less clear for multicellular organisms, especially since aggregation of several of these phase separation-prone proteins is implicated in human disease (King et al., 2012; Ramaswami et al., 2013; Scheckel and Aguzzi, 2018). The question therefore still remains whether this process is truly functional in multicellular eukaryotes. Emerging evidence suggests the functionality of protein condensates in plants (Chakrabortee et al., 2016; Fang et al., 2019; Jung et al., 2020; Powers et al., 2019; van Dop et al., 2020; Zhang et al., 2020) and flies (Bakthavachalu et al., 2018). However, in vivo evidence for a functional role of the emergent properties of phase separation is lacking. Here we demonstrate that conformational switches between liquid and solid-like states of FLOE1 can drive functional phenotypic variability via bet-hedging in a multicellular organism, similar to the prion-based bet-hedging strategies described in yeast (Halfmann et al., 2012).
Seed germination follows a bet-hedging strategy by spreading the risk of potentially lethal conditions, such as drought, across different seasons or years (Gremer and Venable, 2014; Johnston and Bassel, 2018; Villa Martin et al., 2019). While seeds withstand many stresses in their desiccated dormant state, seedlings are at the mercy of their environment and present the most vulnerable stage in a plant’s life. Therefore, precise timing of germination is crucial for plant fitness. Seeds should germinate under conditions that ensure the highest chance of seedling survival, yet a trade-off is that if they wait too long, they may miss the window of good conditions, increase their risk to be eaten or buried too deeply, or ultimately exhaust their viability. Importantly, rainfall cue sensing that drives seed behavior remains largely unresolved, and not only poses an interesting biological problem, but also an outstanding agricultural challenge of great societal importance (Baskin and Baskin, 2004; Johnston and Bassel, 2018).
To identify factors that could regulate seed timing and bet-hedging strategies, we investigated the seed-enriched proteome. These proteins were overrepresented for disorder and involved a set of prion-like proteins. Among them was an uncharacterized prion-like protein that we named FLOE1. Interestingly, FLOE1 formed reversible condensates in A. thaliana embryos that were hydration-dependent. Seed desiccation presents a peculiar biological challenge to proteins as they have to switch between a completely desiccated state and a ‘wet’ state that supports cellular biochemistry, all while preventing irreversible protein aggregation. FLOE1 undergoes hydration-dependent phase separation in plant seeds and it is likely that similar processes occur in a wide variety of organisms with quiescent desiccated life stages, including human pathogens, nematodes, and fungal spores (Boothby and Pielak, 2017; Esbelin et al., 2018; Giarola et al., 2017).
Given that water uptake is the key initiating event for germination, seeds must have evolved a water sensor that allows them to measure water potential. By characterizing FLOE1 knock-out and complemented lines, we provide evidence that FLOE1 attenuates germination under water-limiting conditions in a dose-dependent manner and that its condensation coincides with imbibition. Moreover, the extent of FLOE1 condensation is directly correlated with seed germination. An interesting observation is that despite germination being a binary decision for a seed, FLOE1 condensation displays large variation at the cellular and tissue levels. The link between cellular variability and organism-scale variability in seeds is still unclear (Mitchell et al., 2017), and future work should aim to elucidate how embryos integrate a distribution of cellular phenotypes within a tissue into an ON/OFF switch decision.
To test whether FLOE1’s biophysical state is important for its function as a germination regulator, we dissected its molecular grammar and interrogated mutants for their ability to rescue the floe1-1 phenotype. First, we found that FLOE1’s phase behavior is tuned by opposing forces exerted by two disordered domains. While the prion-like QPS domains drives condensation, its DS domain tunes the material state by fluidizing FLOE1 condensates. This information allowed us to engineer two key mutants: one that can no longer phase separate (ΔQPS) and one that presents solid-like condensates (ΔDS). Second, by complementing floe1-1 with these mutants, we find that the QPS domain is required for its function, whereas the ΔDS mutant acts in a gain-of-function manner leading to a dramatic germination phenotype. In the evolutionary game theory framework (Gremer and Venable, 2014; Johnston and Bassel, 2018; Villa Martin et al., 2019), the ΔDS mutant behaves like a “high-stakes gambler”—it perceives the risk of germination under stress (e.g., seedling dying) to be lower than the chance of there being a change in the environment (e.g., increased rainfall). Thus, our work shows that the emergent properties of a biomolecular condensate can tune fitness in vivo, by tuning bet-hedging strategies at a crucial step in a seed’s life (Fig. 5G).
The observation that variation in FLOE1 sequence and its expression level tune germination, make it a prime candidate as a seed bet-hedging gene. We made use of available transcriptomic, physiological, and climatologic data of A. thaliana ecotypes to test this hypothesis and found evidence that wild populations might use FLOE1 variation to tune their bet-hedging strategies according to their environment’s predictability. By analyzing the conservation of FLOE1, we find a vast repertoire of FLOE sequence diversity and phase separation behavior, suggesting the possibility that other species may also use this gene family to fine-tune germination as a local adaptation mechanism. Of note, the FLOE family precedes the origin of seed plants, and even in A. thaliana, FLOE1 paralogs—FLOE2 and FLOE3—show a broader tissue expression (Waese et al., 2017), pointing to potential functions beyond seed germination.
In conclusion, we have identified a putative plant water potential sensor that undergoes reversible hydration-dependent phase separation and regulates seed germination. Our findings have direct implications for not only cell biology and ecology, but also agriculture. By providing a set of guiding principles to engineer a novel biomolecular condensate (Hastings and Boeynaems, 2021), we hope to inspire the development of designer crops engineered to withstand the effects of climate change.
LIMITATIONS OF THE STUDY
The precise molecular function of FLOE1 remains unresolved. Our data are consistent with FLOE1 functioning at the transition between the desiccated state of the dry embryo and the reactivated hydrated state – when the protein undergoes reversible condensation. But there are alternative interpretations. It is possible that FLOE1 might have a function independent of phase separation. The ΔQPS mutant that we made to prevent condensation might disrupt this other FLOE1 function in the dispersed state independent of its ability to form a condensate. It is also formally possible that the ΔDS mutant, despite being in a constitutively condensed state, might have an enhanced function in a very minor dispersed fraction that we cannot readily detect. Future work will be needed to precisely define contributions of FLOE1 function in the disperse vs. condensed state. Because FLOE1 is dispersed in the dry state of the embryo, these analyses represent technical challenges and new experimental methods will be needed. Currently, the tools to investigate dry biological matter are limited. Perhaps techniques can be borrowed from the material science field to study the dry state.
Supplementary Material
Table S1 (Excel): The thaliana seed-enriched proteome, related to Figure 1
Table S2 (Excel): RNAseq analysis of floe1-1 versus Col-0 (WT), and of +ΔDS versus +wtFLOE1 complemented seeds, related to Figures 2 and 5.
Table S3 (Excel): Estimation of endogenous FLOE1 concentration, sequences of primers, constructs, CRISPR mutations and FLOE homologs, related to Figure 3 and STAR Methods.
Table S4 (Excel): A. thaliana ecotypes analysis, related to Figure 6.
Figure S1: Supplementary information on FLOE1 expression, localization and condensation, related to Figure 1. (A) RT-qPCR analysis of different developmental stages shows peak expression in mature dry seeds, and a decrease in expression upon imbibition. “Dark”, “green” and “yellow” refer to the maturation stages of the siliques (from younger to older), which roughly correspond to 4–7, 8–10 and 11–13 days post-anthesis (Mizzotti et al., 2018), and “imbibed” corresponds to seeds that were imbibed in sterile double-distilled water for 24 h. Col-0 (WT) plants were used. One-way ANOVA. **** p-value < 0.0001. Mean ± SD shown. Circles represent technical replicates. (B) Expression of FLOE1 in developing embryos detected by GUS staining in FLOE1p:FLOE1-GUS transgenic lines. (C) YFP-FLAG localizes diffusely with modest nuclear enrichment in Arabidopsis torpedo stage embryos without any condensates forming. (D) GFP localizes diffusely with modest nuclear enrichment in imbibed seed embryo radicles without any condensates forming. (E) Autofluorescence of protein storage vacuoles in non-transgenic control plants is much weaker in the hydrated state. (F) Dissection in glycerin does not alter the presence of FLOE1-GFP condensates throughout embryonic development before desiccation. For the mature stage, a zoom in of the radicle is shown. (G) Cycloheximide (CHX) treatment does not prevent FLOE1-GFP condensate formation in imbibed embryo radicles. (H–I) Incubation of FLOE1-GFP embryos in osmolyte solutions prevents FLOE1 condensate formation. Mannitol: Mann-Whitney. Sorbitol: One-way ANOVA Kruskal-Wallis. (J) Top panels: location of the nucleus among the protein storage vacuoles in a radicle region without FLOE1-GFP nuclear localization. Bottom panels: radicle region exhibiting FLOE1-GFP nuclear localization (denoted by red asterisks). (K) Cellular heterogeneity quantification for hydration (see Fig. 1F), dehydration (see Fig. 1F), and salt (see Fig. 1K) experiments. Mann-Whitney. **** p-value < 0.0001. a.u. = arbitrary units. Images in panel H are single optical sections, with GFP and DAPI signals displayed as inverted gray scales and false-coloring in the merged image, respectively green and blue. All other fluorescence microscopy images shown are maximum projections, with GFP signal displayed as inverted gray scales. (L) Both N- and C-terminal GFP fusions condense into cytoplasmic condensates. V denotes vacuole, C denotes cytoplasm. GFP signal is displayed as false-colored intensity scale. (M) Plasmolysis of tobacco leaves shows that FLOE1-GFP is located in the cytoplasm and not the cell wall. See also Movie S2 for evidence of cytoplasmic streaming of FLOE1-GFP condensates. GFP and propidium iodide (staining the plant cell wall) signals are displayed with green and magenta false coloring. All fluorescence microscopy images shown are single optical sections of epidermal pavement cells of tobacco leaves.
Figure S2: Supplementary data regarding FLOE1 function, related to Figure 2. (A) Absence of FLOE1 does not affect seed characteristics. Mann-Whitney. ns = not significant. Representative of three independent batches of seeds (B) Absence of FLOE1 does not affect germination under standard conditions. Mean ± SEM. Four-parameter dose-response fit. Two-way ANOVA with Šidák correction. Representative of three independent experiments. (C) Increased germination of floe1-1 knockout line under water stress is rescued by WT FLOE1 complementation (+WT). Mean ± SEM. One-way ANOVA. Representative of three independent experiments. (D) Various FLOE1 WT complemented lines in floe1-1 background with different expression levels, as assayed by RT-qPCR, show dose-dependent effect of FLOE1 on germination under salt stress. Mean ± SEM. Linear regression. (E) Two CRISPR-Cas9 FLOE1 mutant lines show enhanced germination under varying salt stress conditions. Mean ± SEM. Four-parameter dose-response fit. Two-way ANOVA. Representative of three independent experiments. (F) Four CRISPR-Cas9 FLOE1 mutants lines show enhanced germination under salt stress. Mean ± SEM. One-way ANOVA. Representative of three independent experiments. (G) Both WT and floe1-1 seedlings show similar developmental defects upon germination under salt stress. Root lengths were quantified 15 days after transfer of 4-day old seedlings from normal MS media to media supplemented with the indicated salt concentration. Mixed-effects model (modified two-way ANOVA). Representative of three independent experiments (H) Quantification of FLOE1 condensate formation upon alleviation from salt stress. Horizontal line indicates cut-off for FLOE1 condensation (see STAR Methods). Mann-Whitney. * p-value < 0.05, ** p-value < 0.01, *** p-value < 0.001, **** p-value < 0.0001. MS = Murashige and Skoog medium (standard conditions). a.u. = arbitrary units. (A–F) Biological replicates refer to seeds collected from different mother plants. (I) Molecular function GO ontology associated with differentially expressed genes in salt stressed floe1-1 seeds (See also Table S2). Font size correlates to −log10 (p-value). (J) Germination experiment in the presence of 2μM of the gibberellic acid (GA) inhibitor paclobutrazol shows no difference between wildtype and floe1-1 seeds. Mean ± SEM. Student’s t-test. Representative of three independent experiments. (K) Abscisic acid (ABA) (5μM) does not differentially affect wildtype of floe1-1 germination rates. Mean ± SEM. Four-parameter dose-response fit. Two-way ANOVA. Representative of three independent experiments.
Figure S3: FLOE1 condensation correlates with germination under water stress, related to Figure 2. (A) Quantification of cellular FLOE1 heterogeneity as a function of salt concentration. Black line denotes the 95th percentile of the 280 mM NaCl heterogeneity distribution. (B) Quantification of the percentage of cells per embryo that show FLOE1 condensation as a function of salt concentration. Mean ± SEM. Four-parameter dose-response fit. (C) Correlation between fraction of cells showing FLOE1 condensates and germination. Germination data is same as in Fig. 2A. Mean ± SEM. Four-parameter dose-response fit. (D) Schemes highlighting the experimental setups of the different FLOE1 condensation assays.
Figure S4: Recombinant FLOE1 experiments and in silico modelling, related to Figure 3. (A) Size exclusion chromatography of FLOE1-MBP. For more details about Peaks # 1 and 2, see STAR Methods. (B) Differential interference contrast (DIC) images of FLOE1-MBP before (left) and after (right) TEV cleavage. Some irregularly shaped aggregates can be seen in the starting material, but otherwise the sample has no droplets. After cleavage of the MBP protein tag by TEV, MBP is solubilized and FLOE1 forms spherical droplets indicative of liquid-liquid phase separation (LLPS). Scale bars are 10 μm. (C) SDS-PAGE confirms that TEV cleavage reaction was successful and shows the composition of the droplets observed in (B). Lane 1: FLOE1-MBP uncleaved construct (Peak #2 in (A)). Lane 2: Sample from Peak #2 following 20 hours of incubation with TEV. To see the composition of the droplets and ensure they are primarily composed of FLOE1, the products of this cleavage reaction were spun down for 10 minutes at 20,800 g and the supernatant was separated from the pellet. Lane 3: Pellet shows strong FLOE1 enrichment and very little MBP. Lane 4: Supernatant shows that both FLOE1 and MBP are present. The existence of a FLOE1 band in the supernatant indicates an equilibrium between a dilute and condensed FLOE1, as expected in LLPS. The strong MBP band indicates that these FLOE1 droplets selectively exclude MBP. Lane 5: TEV. (D) FLOE1 phase separation is observed even in the low μM range. (E) Predicted structure from the SWISS-MODEL homology prediction server. Side and top views are shown. A well-defined trimeric coiled-coil oligomeric state is strongly predicted. Inferred helicity from SWISS-MODEL prediction projected from N-to-C terminus and aligned with residue numbers. Predicted helicity from Phyre2 homology model prediction. Predicted helicity from PSI-PRED secondary structure prediction tool. Per-residue percentage helicity as obtained from all-atom simulations. Smoothened fit is shown in black. (F) Images are taken following overnight cleavage of 10 μM MBP-tagged constructs using TEV. Wildtype FLOE1 is found in condensates under a range of salt concentrations. The QPS mutant fails to phase separate and is only able to form aggregates, which are especially prevalent at high salt concentration.
Figure S5: Supplementary data regarding EM analyses, related to Figure 3. (A) 3D reconstruction and cellular annotation of A. thaliana embryo, used to calculate the number of cells and their volumes (see STAR Methods). (B–C) TEM image of an embryonic cell. N denotes nucleus, PSV denotes Protein Storage Vacuoles, and yellow shading highlights remainder of cytoplasm. (D) Quantification of cellular fraction excluding nuclei and PSVs. A total of 43 cells from 8 images were analyzed. (E–F) 3D-tomogram of an embryonic cell. Blue highlights lipid membranes and surface of lipid droplets (LD). See also Movie S1. (G) Quantification of cytoplasmic fraction excluding lipid droplets (LD) and membrane compartments (M) based on 7 tomograms from randomly selected regions. (H) By integrating cell count and volume estimates (A) with subcellular fraction estimates (B–G) we estimate that only 16.7% (68.77% * 24.6%) of the total cellular volume of embryos (~17,000,000 μm3) represents the true cytoplasmic fraction, which is estimated at ~2,840,000 μm3. (I–J) Correlative light-electron microscopy (CLEM) allows us to confirm the presence of GFP-FLOE1 in cytoplasmic condensates in transfected U2OS cells. (I) shows wildtype FLOE1 condensates, whereas (J) shows mutant DS domain (8xY/F–S) condensates. (K–L) Standard transmission electron microscopy of wildtype (K) and mutated DS domain (L) FLOE1 condensates. Zooms of (K–L) can be found in Fig. 4H. (L) Highlights filamentous substructure of mutated DS FLOE1 condensates. (M) 3D-tomography of DS(8xY/F-S) FLOE1 condensates. By looking onto the XY-plane of the tomogram one can observe a hexagonal arrangement of ‘dots’, which are the top view of the mutant FLOE1 filaments (small dashed lines). Upon tilting by 30° (around X-axis, ‘horizontal’ on page) the hexagonal symmetry of the dotted structure transforms into a line pattern, by superpositioning of the filaments. This observation demonstrates the hexagonal symmetry of the mutant FLOE1 filaments. The large dashed line indicates approximate boundary of the full condensates. Small dashed lines indicate a section of the condensate with filaments perpendicular to the tomographic section. See also Movie S3. Dimensions of tomographic volume: 2.5 × 2.5 × 0.2 μm.
Figure S6: Supplementary data regarding FLOE1 mutants and paralogs, related to Figures 5, 6 and 7. (A-B) Since FLOE1 is a dosage-dependent regulator of seed germination under water stress, we wanted to rule out that expression differences in the mutant lines would be responsible for the observed differences in their germination levels. (A) We assayed FLOE1 expression levels in dry seeds via RT-qPCR. (B) As shown before, there was a linear correlation between FLOE1 expression level and germination levels. floe1-1 lines complemented with the ΔDUF mutant followed a similar trend, confirming that the DUF domain deletion does not affect germination in our assays. floe1-1 lines complemented with the ΔDS mutant showed low levels of transgene expression according to RT-qPCR (A, Right Panel. One-way ANOVA. *** p-value < 0.001. Mean ± SEM.) which was consistent with the sparser localization of the protein in radicles (Fig. 5F). Yet, despite these low expression levels, the ΔDS complemented lines consistently induced extreme germination levels, which we never observed for floe1-1 or WT complemented lines. floe1-1 lines complemented with the ΔQPS mutant showed high levels of transgene expression according to RT-qPCR (A). Despite these high transgene levels, and robust protein expression in radicles (Fig. 5F), ΔQPS complemented lines had germination levels similar to the parental floe1-1 line, in stark contrast with WT complemented lines with higher relative expression, supporting the loss-of-function phenotype of this mutant and loss of dose dependency (B). B: Mean ± SEM. Germination data are representative of three independent experiments. (C) Correlation between qPCR-based expression measurements and protein concentrations as measured via GFP ELISA (see also Table S3). Color scheme is the same as in the previous panels. (D) All complemented lines are able to fully germinate under standard conditions (43.5h time point shown). Mean ± SEM. Representative of three independent experiments. (E) ΔDUF and ΔQPS complemented lines have similar germination rates as WT complemented lines. In contrast, ΔDS complemented lines show faster germination rates under standard conditions. Mean ± SEM. Two-way ANOVA. Biological replicates refer to seeds collected from different mother plants. Representative of two independent experiments. (F) ΔDS seeds show high numbers of differentially expressed genes (DEGs) compared to wildtype complemented seeds in all tested conditions. (G) ΔDS seeds upregulate stress response genes or genes implicated in ribosomal biogenesis depending on whether they are in a desiccated or active state. Font size correlates to −log10 (p-value). (H) Quantification of FLOE granule size in tobacco leaves. Isoform comparison: Mann-Whitney test. Paralog comparison: Kruskal-Wallis test. * p-value < 0.05, ** p-value < 0.01, **** p-value < 0.0001. (I) FLOE1 does not mix with FLOE2 and FLOE3, while FLOE2 and FLOE3 do mix. GFP and RFP signal false colored as green and magenta. (J) Species tree as in Fig. 7A with full species names. “Bryo”, “Ly” and “Gymno” refer to the bryophyte, lycophyte, and gymnosperm lineages. (K) Additional examples of FLOE homologs that condense upon expression in tobacco leaves. Belonging to the FLOE1L and FLOE2L clades are indicated. GFP signal displayed as false-colored intensity scale. All fluorescence microscopy images shown are single optical sections of epidermal pavement cells of tobacco leaves.
Figure S7: Ecological analysis of FLOE1 isoform expression, related to Figure 6. (A) FLOE1.1 is the dominant isoform across ecotypes. Mann-Whitney test. **** p-value < 0.0001. (B) Scatterplots of FLOE1 isoform expression show a positive correlation between FLOE1.1 expression and the dormancy trait DSDS50 (number of Days of Seed Dry Storage required to reach 50% germination) (Atwell et al., 2010), which is absent for FLOE1.2. Correlation was measured using Spearman’s rank correlation coefficient, which assesses the statistical dependence between the rankings of two variables. Fitted line is for visualization purposes and not meant to imply a linear relationship. (C–D) Scatterplots of FLOE1 isoform expression show a positive correlation between FLOE1.2 expression and germination at 10°C (Martinez-Berdeja et al., 2020), which is less pronounced for FLOE1.1. (E–F) Scatterplots of FLOE1 isoform expression show a positive correlation between FLOE1.2 expression and germination at 4°C (Martinez-Berdeja et al., 2020), which is less pronounced for FLOE1.1. Correlation was measured using Spearman’s rank correlation, which assesses the statistical dependence between the rankings of two variables. Fitted line is for visualization purposes and not meant to imply a linear relationship. TPM were log2 transformed and mean centered prior to plotting for better visualization. (G) Spearman’s rho correlation between normalized FLOE1.1 expression and average monthly precipitation (blue), maximum (red) and minimum (orange) temperature across 527 worldwide locations of A. thaliana ecotypes. Color of balloons indicates −log10(p-value). Balloons only shown for data points with significance at p < 0.01. (H–I) FLOE1.1 expression shows weak correlation with July precipitation or maximum temperature across locations. Fitted line is for visualization purposes and not meant to imply a linear relationship. (J) Spearman’s rho correlation between normalized FLOE1.2 expression and average monthly precipitation (blue) and maximum (red) and minimum (orange) temperature across 527 worldwide locations of A. thaliana ecotypes. Color of balloons indicates −log10(p-value). Balloons only shown for data points with significance at p < 0.01. (K–L) FLOE1.2 expression shows strong correlation with July precipitation or maximum temperature across locations. Fitted line is for visualization purposes and not meant to imply a linear relationship. (M) Correlation between FLOE1.1 and the coefficient of variation in inter-annual precipitation in the month of July between the years 1958–2017 (Spearman’s rho correlation was r = −0.2397752, p = 1.524E-9). (N) Correlation between FLOE1.2 and the coefficient of variation in inter-annual precipitation in the month of July between the years 1958–2017 (Spearman’s rho correlation was r = −0.2802625, p = 1.234E-12). Fitted lines are for visualization purposes and not meant to imply a linear relationship.
Movie S1: Electron tomographic reconstruction and segmentation of membrane-bound organelles (in blue) of an Arabidopsis thaliana embryo cell, related to Figure 3.
Movie S2: Cytoplasmic streaming of FLOE1-GFP granules in tobacco epidermal pavement cells, related to Figure 3.
Movie S3: 3D tomogram of DS(8xY/F-S) FLOE1 condensates in U2OS cells, related to Figure 4.
Dorone et al. (2021): HIGHLIGHTS.
FLOE1 is a plant-specific gene that regulates seed germination under water stress
FLOE1 undergoes reversible hydration-dependent phase separation
Ecological data suggests that FLOE1 variation is involved in local adaptation
FLOE1 can be engineered to tune seed germination with potential use for agriculture
ACKNOWLEDGEMENTS:
We thank Drs. M.B. Mudgett, D. Jarosz, H. Meyer, B. Schmidt, K. Lasker, C. Cuevas-Velasquez, K. Bradford, and members of the Gitler and Rhee laboratories as well as the Carnegie-Stanford Intrinsically Disordered Protein Scientific Interest Group (IDPSIG) for helpful discussion and suggestions. We are grateful to Drs. Z. Wang’s and T. Nakagawa’s labs for sharing reagents, G. Materassi-Shultz for growth facilities management, and Dr. N. Boruah for bioinformatics assistance. The computing for the ecological data was performed on the Memex and Calc clusters from the Carnegie Institution for Science. We thank the Stanford Neuroscience Microscopy Service for use of the core facility.
FUNDING:
This research was supported, in part, by the U.S. Department of Energy, Office of Science, Office of Biological and Environmental Research, Genomic Science Program grant nos. DE-SC0018277, DE-SC0008769, DE-SC0020366, and DE-SC0021286 and the U.S. National Science Foundation grants MCB-1617020 and IOS-1546838 to S.Y.R. Y.D. was supported, in part, by the Stanford Graduate Fellowship in Science and Engineering, Carnegie Institution for Science, and Brigitte Berthelemot. S.B. acknowledges an EMBO Long Term Fellowship. Work in the A.D.G. lab is supported by NIH (grant R35NS097263). Work in the M.S.O. lab is supported by NSF (MCB1614965) and NIH (1S10 OD026769-01). M.E.-A. and S.H. are supported by the Carnegie Institution for Science and Department of Energy (grant DE-SC0021286). Work in the S.S. lab is supported by NIH (grant R35GM137926). The Stanford Neuroscience Microscopy Service is supported by NIH (grant NS069375). Freeze-substitution experiments were performed on equipment acquired by FWO (grant AKUL/11/30).
Footnotes
Supplemental Information:
STAR Methods
Figs. S1 to S7
References
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DECLARATION OF INTERESTS: Y.D., S.B., A.D.G and S.Y.R: Inventors on a provisional patent application filed with the U.S. Patent and Trademark Office on August 7th 2020 by Leland Stanford University & Carnegie Institution for Science. Application number: 63063009. Aspect of the manuscript covered in the patent application: regulation of seed germination via FLOE1 modulation.
REFERENCES
- 1001 Genomes Consortium, 2016. 1,135 Genomes Reveal the Global Pattern of Polymorphism in Arabidopsis thaliana. Cell 166, 481–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abatzoglou JT, Dobrowski SZ, Parks SA, and Hegewisch KC (2018). TerraClimate, a high-resolution global dataset of monthly climate and climatic water balance from 1958–2015. Sci Data 5, 170191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afgan E, Baker D, Batut B, van den Beek M, Bouvier D, Cech M, Chilton J, Clements D, Coraor N, Gruning BA, et al. (2018). The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2018 update. Nucleic Acids Res 46, W537–W544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alberti S, Halfmann R, King O, Kapila A, and Lindquist S (2009). A systematic survey identifies prions and illuminates sequence features of prionogenic proteins. Cell 137, 146–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atwell S, Huang YS, Vilhjalmsson BJ, Willems G, Horton M, Li Y, Meng D, Platt A, Tarone AM, Hu TT, et al. (2010). Genome-wide association study of 107 phenotypes in Arabidopsis thaliana inbred lines. Nature 465, 627–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austin RS, Hiu S, Waese J, Ierullo M, Pasha A, Wang TT, Fan J, Foong C, Breit R, Desveaux D, et al. (2016). New BAR tools for mining expression data and exploring Cis-elements in Arabidopsis thaliana. Plant J 88, 490–504. [DOI] [PubMed] [Google Scholar]
- Bai B, Sikron N, Gendler T, Kazachkova Y, Barak S, Grafi G, Khozin-Goldberg I, and Fait A (2012). Ecotypic variability in the metabolic response of seeds to diurnal hydration-dehydration cycles and its relationship to seed vigor. Plant Cell Physiol 53, 38–52. [DOI] [PubMed] [Google Scholar]
- Bakthavachalu B, Huelsmeier J, Sudhakaran IP, Hillebrand J, Singh A, Petrauskas A, Thiagarajan D, Sankaranarayanan M, Mizoue L, Anderson EN, et al. (2018). RNP-Granule Assembly via Ataxin-2 Disordered Domains Is Required for Long-Term Memory and Neurodegeneration. Neuron 98, 754–766 e754. [DOI] [PubMed] [Google Scholar]
- Barbier de Reuille P, Routier-Kierzkowska AL, Kierzkowski D, Bassel GW, Schupbach T, Tauriello G, Bajpai N, Strauss S, Weber A, Kiss A, et al. (2015). MorphoGraphX: A platform for quantifying morphogenesis in 4D. Elife 4, 05864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baskin J, & Baskin C (2004). A classification system for seed dormancy. Seed Science Research, 14(1), 1–16. doi: 10.1079/SSR2003150 [DOI] [Google Scholar]
- Bassel GW, Stamm P, Mosca G, Barbier de Reuille P, Gibbs DJ, Winter R, Janka A, Holdsworth MJ, and Smith RS (2014). Mechanical constraints imposed by 3D cellular geometry and arrangement modulate growth patterns in the Arabidopsis embryo. Proc Natl Acad Sci U S A 111, 8685–8690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodenhofer U, Bonatesta E, Horejs-Kainrath C, and Hochreiter S (2015). msa: an R package for multiple sequence alignment. Bioinformatics 31, 3997–3999. [DOI] [PubMed] [Google Scholar]
- Boeynaems S, Alberti S, Fawzi NL, Mittag T, Polymenidou M, Rousseau F, Schymkowitz J, Shorter J, Wolozin B, Van Den Bosch L, et al. (2018). Protein Phase Separation: A New Phase in Cell Biology. Trends Cell Biol 28, 420–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeynaems S, De Decker M, Tompa P and Van Den Bosch L (2017). Arginine-rich Peptides Can Actively Mediate Liquid-liquid Phase Separation. Bio-protocol 7(17): e2525. DOI: 10.21769/BioProtoc.2525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boothby TC, Pielak GJ. Intrinsically Disordered Proteins and Desiccation Tolerance: Elucidating Functional and Mechanistic Underpinnings of Anhydrobiosis. Bioessays. 2017. November;39(11). doi: 10.1002/bies.201700119 [DOI] [PubMed] [Google Scholar]
- Bossi F, Fan J, Xiao J, Chandra L, Shen M, Dorone Y, Wagner D, and Rhee SY (2017). Systematic discovery of novel eukaryotic transcriptional regulators using sequence homology independent prediction. BMC Genomics 18, 480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray NL, Pimentel H, Melsted P, and Pachter L (2016). Near-optimal probabilistic RNA-seq quantification. Nat Biotechnol 34, 525–527. [DOI] [PubMed] [Google Scholar]
- Buitink J, and Leprince O (2008). Intracellular glasses and seed survival in the dry state. C R Biol 331, 788–795. [DOI] [PubMed] [Google Scholar]
- Burke KA, Janke AM, Rhine CL, and Fawzi NL (2015). Residue-by-Residue View of In Vitro FUS Granules that Bind the C-Terminal Domain of RNA Polymerase II. Mol Cell 60, 231–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabortee S, Kayatekin C, Newby GA, Mendillo ML, Lancaster A, and Lindquist S (2016). Luminidependens (LD) is an Arabidopsis protein with prion behavior. Proc Natl Acad Sci U S A 113, 6065–6070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ (2005). Floral dip: agrobacterium-mediated germ line transformation. Methods Mol Biol 286, 91–102. [DOI] [PubMed] [Google Scholar]
- Czechowski T, Stitt M, Altmann T, Udvardi MK, and Scheible WR (2005). Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol 139, 5–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davuluri RV, Sun H, Palaniswamy SK, Matthews N, Molina C, Kurtz M, and Grotewold E (2003). AGRIS: Arabidopsis gene regulatory information server, an information resource of Arabidopsis cis-regulatory elements and transcription factors. BMC Bioinformatics 4, 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekkers BJ, Willems L, Bassel GW, van Bolderen-Veldkamp RP, Ligterink W, Hilhorst HW, and Bentsink L (2012). Identification of reference genes for RT-qPCR expression analysis in Arabidopsis and tomato seeds. Plant Cell Physiol 53, 28–37. [DOI] [PubMed] [Google Scholar]
- Esbelin J, Santos T, and Hebraud M (2018). Desiccation: An environmental and food industry stress that bacteria commonly face. Food Microbiol 69, 82–88. [DOI] [PubMed] [Google Scholar]
- Fang X, Wang L, Ishikawa R, Li Y, Fiedler M, Liu F, Calder G, Rowan B, Weigel D, Li P, et al. (2019). Arabidopsis FLL2 promotes liquid-liquid phase separation of polyadenylation complexes. Nature 569, 265–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franzmann TM, Jahnel M, Pozniakovsky A, Mahamid J, Holehouse AS, Nuske E, Richter D, Baumeister W, Grill SW, Pappu RV, et al. (2018). Phase separation of a yeast prion protein promotes cellular fitness. Science. 2018 January 5;359(6371):eaao5654. doi: 10.1126/science.aao5654. [DOI] [PubMed] [Google Scholar]
- Giarola V, Hou Q, and Bartels D (2017). Angiosperm Plant Desiccation Tolerance: Hints from Transcriptomics and Genome Sequencing. Trends Plant Sci 22, 705–717. [DOI] [PubMed] [Google Scholar]
- Goodstein DM, Shu S, Howson R, Neupane R, Hayes RD, Fazo J, Mitros T, Dirks W, Hellsten U, Putnam N, et al. (2012). Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res 40, D1178–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gremer JR, and Venable DL (2014). Bet hedging in desert winter annual plants: optimal germination strategies in a variable environment. Ecol Lett 17, 380–387. [DOI] [PubMed] [Google Scholar]
- Gutierrez-Beltran E, Moschou PN, Smertenko AP, and Bozhkov PV (2015). Tudor staphylococcal nuclease links formation of stress granules and processing bodies with mRNA catabolism in Arabidopsis. Plant Cell 27, 926–943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halfmann R, Jarosz DF, Jones SK, Chang A, Lancaster AK, and Lindquist S (2012). Prions are a common mechanism for phenotypic inheritance in wild yeasts. Nature 482, 363–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hijmans RJ, Cameron S, Parra J, Jones P, Jarvis A, Richardson K, 2015. WorldClim-Global Climate Data. Free Climate Data for Ecological Modeling and GIS. [Google Scholar]
- Jefferson RA, Kavanagh TA, and Bevan MW (1987). GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6, 3901–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston IG, and Bassel GW (2018). Identification of a bet-hedging network motif generating noise in hormone concentrations and germination propensity in Arabidopsis. J R Soc Interface. 2018 April;15(141):20180042. doi: 10.1098/rsif.2018.0042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakatsu T, Huang SC, Jupe F, Sasaki E, Schmitz RJ, Urich MA, Castanon R, Nery JR, Barragan C, He Y, et al. (2016). Epigenomic Diversity in a Global Collection of Arabidopsis thaliana Accessions. Cell 166, 492–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley LA, Mezulis S, Yates CM, Wass MN, and Sternberg MJ (2015). The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc 10, 845–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King OD, Gitler AD, and Shorter J (2012). The tip of the iceberg: RNA-binding proteins with prion-like domains in neurodegenerative disease. Brain Res 1462, 61–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kranner I, Minibayeva FV, Beckett RP, and Seal CE (2010). What is stress? Concepts, definitions and applications in seed science. New Phytol 188, 655–673. [DOI] [PubMed] [Google Scholar]
- Kremer JR, Mastronarde DN, and McIntosh JR (1996). Computer visualization of three-dimensional image data using IMOD. J Struct Biol 116, 71–76. [DOI] [PubMed] [Google Scholar]
- Lancaster AK, Nutter-Upham A, Lindquist S, and King OD (2014). PLAAC: a web and command-line application to identify proteins with prion-like amino acid composition. Bioinformatics 30, 2501–2502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasker K, Boeynaems S, Lam V, Stainton E, Jacquemyn M, Daelemans D, Villa E, Holehouse AS, Gitler AD, Shapiro L (2021). A modular platform for engineering function of natural and synthetic biomolecular condensates. BioRxiv. doi: 10.1101/2021.02.03.429226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemoine F, Correia D, Lefort V, Doppelt-Azeroual O, Mareuil F, Cohen-Boulakia S, and Gascuel O (2019). NGPhylogeny.fr: new generation phylogenetic services for non-specialists. Nucleic Acids Res 47, W260–W265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leprince O, and Buitink J (2015). Introduction to desiccation biology: from old borders to new frontiers. Planta 242, 369–378. [DOI] [PubMed] [Google Scholar]
- Letunic I, and Bork P (2019). Interactive Tree Of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Res 47, W256–W259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, and Anders S (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin EW, Holehouse AS, Peran I, Farag M, Incicco JJ, Bremer A, Grace CR, Soranno A, Pappu RV, and Mittag T (2020). Valence and patterning of aromatic residues determine the phase behavior of prion-like domains. Science 367, 694–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Berdeja A, Stitzer MC, Taylor MA, Okada M, Ezcurra E, Runcie DE, and Schmitt J (2020). Functional variants of DOG1 control seed chilling responses and variation in seasonal life-history strategies in Arabidopsis thaliana. Proc Natl Acad Sci U S A 117, 2526–2534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastronarde DN (2005). Automated electron microscope tomography using robust prediction of specimen movements. J Struct Biol 152, 36–51. [DOI] [PubMed] [Google Scholar]
- McDonald KL, and Webb RI (2011). Freeze substitution in 3 hours or less. J Microsc 243, 227–233. [DOI] [PubMed] [Google Scholar]
- McGuffin LJ, Bryson K, and Jones DT (2000). The PSIPRED protein structure prediction server. Bioinformatics 16, 404–405. [DOI] [PubMed] [Google Scholar]
- Meng L, and Feldman L (2010). A rapid TRIzol-based two-step method for DNA-free RNA extraction from Arabidopsis siliques and dry seeds. Biotechnol J 5, 183–186. [DOI] [PubMed] [Google Scholar]
- Mizzotti C, Rotasperti L, Moretto M, Tadini L, Resentini F, Galliani BM, Galbiati M, Engelen K, Pesaresi P, and Masiero S (2018). Time-Course Transcriptome Analysis of Arabidopsis Siliques Discloses Genes Essential for Fruit Development and Maturation. Plant Physiol 178, 1249–1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munder MC, Midtvedt D, Franzmann T, Nuske E, Otto O, Herbig M, Ulbricht E, Muller P, Taubenberger A, Maharana S, et al. (2016). A pH-driven transition of the cytoplasm from a fluid- to a solid-like state promotes entry into dormancy. Elife. 2016 Mar 22;5:e09347. doi: 10.7554/eLife.09347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T, Kurose T, Hino T, Tanaka K, Kawamukai M, Niwa Y, Toyooka K, Matsuoka K, Jinbo T, and Kimura T (2007). Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J Biosci Bioeng 104, 34–41. [DOI] [PubMed] [Google Scholar]
- Nallamsetty S, Austin BP, Penrose KJ, and Waugh DS (2005). Gateway vectors for the production of combinatorially-tagged His6-MBP fusion proteins in the cytoplasm and periplasm of Escherichia coli. Protein Sci 14, 2964–2971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonogaki H, Chen F, and Bradford KJ (2018). Mechanisms and Genes Involved in Germination Sensu Stricto. In Annual Plant Reviews online, Roberts JA (Ed.). doi: 10.1002/9781119312994.apr0285 [DOI] [Google Scholar]
- Peddie CJ, Blight K, Wilson E, Melia C, Marrison J, Carzaniga R, Domart MC, O’Toole P, Larijani B, and Collinson LM (2014). Correlative and integrated light and electron microscopy of in-resin GFP fluorescence, used to localise diacylglycerol in mammalian cells. Ultramicroscopy 143, 3–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piovesan D, Tabaro F, Paladin L, Necci M, Micetic I, Camilloni C, Davey N, Dosztanyi Z, Meszaros B, Monzon AM, et al. (2018). MobiDB 3.0: more annotations for intrinsic disorder, conformational diversity and interactions in proteins. Nucleic Acids Res 46, D471–D476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers SK, Holehouse AS, Korasick DA, Schreiber KH, Clark NM, Jing H, Emenecker R, Han S, Tycksen E, Hwang I, et al. (2019). Nucleo-cytoplasmic Partitioning of ARF Proteins Controls Auxin Responses in Arabidopsis thaliana. Mol Cell 76, 177–190 e175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajjou L, Duval M, Gallardo K, Catusse J, Bally J, Job C, and Job D (2012). Seed germination and vigor. Annu Rev Plant Biol 63, 507–533. [DOI] [PubMed] [Google Scholar]
- Raudvere U, Kolberg L, Kuzmin I, Arak T, Adler P, Peterson H, and Vilo J (2019). g:Profiler: a web server for functional enrichment analysis and conversions of gene lists (2019 update). Nucleic Acids Res 47, W191–W198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riback JA, Katanski CD, Kear-Scott JL, Pilipenko EV, Rojek AE, Sosnick TR, and Drummond DA (2017). Stress-Triggered Phase Separation Is an Adaptive, Evolutionarily Tuned Response. Cell 168, 1028–1040 e1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roels J, Vernaillen F, Kremer A, Goncalves A, Aelterman J, Luong HQ, Goossens B, Philips W, Lippens S, and Saeys Y (2020). An interactive ImageJ plugin for semi-automated image denoising in electron microscopy. Nat Commun 11, 771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rog O, Kohler S, and Dernburg AF (2017). The synaptonemal complex has liquid crystalline properties and spatially regulates meiotic recombination factors. Elife. 2017 Jan 3;6:e21455. doi: 10.7554/eLife.21455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sano N, Rajjou L, North HM, Debeaujon I, Marion-Poll A, and Seo M (2016). Staying Alive: Molecular Aspects of Seed Longevity. Plant Cell Physiol 57, 660–674. [DOI] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat Methods 9, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid M, Davison TS, Henz SR, Pape UJ, Demar M, Vingron M, Scholkopf B, Weigel D, and Lohmann JU (2005). A gene expression map of Arabidopsis thaliana development. Nat Genet 37, 501–506. [DOI] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, and Eliceiri KW (2012). NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9, 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwember AR, and Bradford KJ (2010). Quantitative trait loci associated with longevity of lettuce seeds under conventional and controlled deterioration storage conditions. J Exp Bot 61, 4423–4436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin Y, and Brangwynne CP (2017). Liquid phase condensation in cell physiology and disease. Science. 2017 September 22;357(6357):eaaf4382. doi: 10.1126/science.aaf4382. [DOI] [PubMed] [Google Scholar]
- Steinert J, Schiml S, Fauser F, and Puchta H (2015). Highly efficient heritable plant genome engineering using Cas9 orthologues from Streptococcus thermophilus and Staphylococcus aureus. Plant J 84, 1295–1305. [DOI] [PubMed] [Google Scholar]
- Truernit E, Bauby H, Dubreucq B, Grandjean O, Runions J, Barthelemy J, and Palauqui JC (2008). High-resolution whole-mount imaging of three-dimensional tissue organization and gene expression enables the study of Phloem development and structure in Arabidopsis. Plant Cell 20, 1494–1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- UniProt C (2019). UniProt: a worldwide hub of protein knowledge. Nucleic Acids Res 47, D506–D515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dop M, Fiedler M, Mutte S, de Keijzer J, Olijslager L, Albrecht C, Liao CY, Janson ME, Bienz M, and Weijers D (2020). DIX Domain Polymerization Drives Assembly of Plant Cell Polarity Complexes. Cell 180, 427–439 e412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villa Martin P, Munoz MA, and Pigolotti S (2019). Bet-hedging strategies in expanding populations. PLoS Comput Biol 15, e1006529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitalis A, and Pappu RV (2009). ABSINTH: a new continuum solvation model for simulations of polypeptides in aqueous solutions. J Comput Chem 30, 673–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waese J, Fan J, Pasha A, Yu H, Fucile G, Shi R, Cumming M, Kelley LA, Sternberg MJ, Krishnakumar V, et al. (2017). ePlant: Visualizing and Exploring Multiple Levels of Data for Hypothesis Generation in Plant Biology. Plant Cell 29, 1806–1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walton J (1979). Lead aspartate, an en bloc contrast stain particularly useful for ultrastructural enzymology. J Histochem Cytochem 27, 1337–1342. [DOI] [PubMed] [Google Scholar]
- Wang J, Choi JM, Holehouse AS, Lee HO, Zhang X, Jahnel M, Maharana S, Lemaitre R, Pozniakovsky A, Drechsel D, et al. (2018). A Molecular Grammar Governing the Driving Forces for Phase Separation of Prion-like RNA Binding Proteins. Cell 174, 688–699 e616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse A, Bertoni M, Bienert S, Studer G, Tauriello G, Gumienny R, Heer FT, de Beer TAP, Rempfer C, Bordoli L, et al. (2018). SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res 46, W296–W303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao N, Cao DS, Zhu MF, and Xu QS (2015). protr/ProtrWeb: R package and web server for generating various numerical representation schemes of protein sequences. Bioinformatics 31, 1857–1859. [DOI] [PubMed] [Google Scholar]
- Xue B, Dunbrack RL, Williams RW, Dunker AK, and Uversky VN (2010). PONDR-FIT: a meta-predictor of intrinsically disordered amino acids. Biochim Biophys Acta 1804, 996–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yashina S, Gubin S, Maksimovich S, Yashina A, Gakhova E, and Gilichinsky D (2012). Regeneration of whole fertile plants from 30,000-y-old fruit tissue buried in Siberian permafrost. Proc Natl Acad Sci U S A 109, 4008–4013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Li Z, Chen N, Huang Y, and Huang S (2020). Phase separation of Arabidopsis EMB1579 controls transcription, mRNA splicing, and development. PLoS Biol 18, e3000782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Sarker U, Neumann G, and Ludewig U (2019). The LaCEP1 peptide modulates cluster root morphology in Lupinus albus. Physiol Plant 166, 525–537. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1 (Excel): The thaliana seed-enriched proteome, related to Figure 1
Table S2 (Excel): RNAseq analysis of floe1-1 versus Col-0 (WT), and of +ΔDS versus +wtFLOE1 complemented seeds, related to Figures 2 and 5.
Table S3 (Excel): Estimation of endogenous FLOE1 concentration, sequences of primers, constructs, CRISPR mutations and FLOE homologs, related to Figure 3 and STAR Methods.
Table S4 (Excel): A. thaliana ecotypes analysis, related to Figure 6.
Figure S1: Supplementary information on FLOE1 expression, localization and condensation, related to Figure 1. (A) RT-qPCR analysis of different developmental stages shows peak expression in mature dry seeds, and a decrease in expression upon imbibition. “Dark”, “green” and “yellow” refer to the maturation stages of the siliques (from younger to older), which roughly correspond to 4–7, 8–10 and 11–13 days post-anthesis (Mizzotti et al., 2018), and “imbibed” corresponds to seeds that were imbibed in sterile double-distilled water for 24 h. Col-0 (WT) plants were used. One-way ANOVA. **** p-value < 0.0001. Mean ± SD shown. Circles represent technical replicates. (B) Expression of FLOE1 in developing embryos detected by GUS staining in FLOE1p:FLOE1-GUS transgenic lines. (C) YFP-FLAG localizes diffusely with modest nuclear enrichment in Arabidopsis torpedo stage embryos without any condensates forming. (D) GFP localizes diffusely with modest nuclear enrichment in imbibed seed embryo radicles without any condensates forming. (E) Autofluorescence of protein storage vacuoles in non-transgenic control plants is much weaker in the hydrated state. (F) Dissection in glycerin does not alter the presence of FLOE1-GFP condensates throughout embryonic development before desiccation. For the mature stage, a zoom in of the radicle is shown. (G) Cycloheximide (CHX) treatment does not prevent FLOE1-GFP condensate formation in imbibed embryo radicles. (H–I) Incubation of FLOE1-GFP embryos in osmolyte solutions prevents FLOE1 condensate formation. Mannitol: Mann-Whitney. Sorbitol: One-way ANOVA Kruskal-Wallis. (J) Top panels: location of the nucleus among the protein storage vacuoles in a radicle region without FLOE1-GFP nuclear localization. Bottom panels: radicle region exhibiting FLOE1-GFP nuclear localization (denoted by red asterisks). (K) Cellular heterogeneity quantification for hydration (see Fig. 1F), dehydration (see Fig. 1F), and salt (see Fig. 1K) experiments. Mann-Whitney. **** p-value < 0.0001. a.u. = arbitrary units. Images in panel H are single optical sections, with GFP and DAPI signals displayed as inverted gray scales and false-coloring in the merged image, respectively green and blue. All other fluorescence microscopy images shown are maximum projections, with GFP signal displayed as inverted gray scales. (L) Both N- and C-terminal GFP fusions condense into cytoplasmic condensates. V denotes vacuole, C denotes cytoplasm. GFP signal is displayed as false-colored intensity scale. (M) Plasmolysis of tobacco leaves shows that FLOE1-GFP is located in the cytoplasm and not the cell wall. See also Movie S2 for evidence of cytoplasmic streaming of FLOE1-GFP condensates. GFP and propidium iodide (staining the plant cell wall) signals are displayed with green and magenta false coloring. All fluorescence microscopy images shown are single optical sections of epidermal pavement cells of tobacco leaves.
Figure S2: Supplementary data regarding FLOE1 function, related to Figure 2. (A) Absence of FLOE1 does not affect seed characteristics. Mann-Whitney. ns = not significant. Representative of three independent batches of seeds (B) Absence of FLOE1 does not affect germination under standard conditions. Mean ± SEM. Four-parameter dose-response fit. Two-way ANOVA with Šidák correction. Representative of three independent experiments. (C) Increased germination of floe1-1 knockout line under water stress is rescued by WT FLOE1 complementation (+WT). Mean ± SEM. One-way ANOVA. Representative of three independent experiments. (D) Various FLOE1 WT complemented lines in floe1-1 background with different expression levels, as assayed by RT-qPCR, show dose-dependent effect of FLOE1 on germination under salt stress. Mean ± SEM. Linear regression. (E) Two CRISPR-Cas9 FLOE1 mutant lines show enhanced germination under varying salt stress conditions. Mean ± SEM. Four-parameter dose-response fit. Two-way ANOVA. Representative of three independent experiments. (F) Four CRISPR-Cas9 FLOE1 mutants lines show enhanced germination under salt stress. Mean ± SEM. One-way ANOVA. Representative of three independent experiments. (G) Both WT and floe1-1 seedlings show similar developmental defects upon germination under salt stress. Root lengths were quantified 15 days after transfer of 4-day old seedlings from normal MS media to media supplemented with the indicated salt concentration. Mixed-effects model (modified two-way ANOVA). Representative of three independent experiments (H) Quantification of FLOE1 condensate formation upon alleviation from salt stress. Horizontal line indicates cut-off for FLOE1 condensation (see STAR Methods). Mann-Whitney. * p-value < 0.05, ** p-value < 0.01, *** p-value < 0.001, **** p-value < 0.0001. MS = Murashige and Skoog medium (standard conditions). a.u. = arbitrary units. (A–F) Biological replicates refer to seeds collected from different mother plants. (I) Molecular function GO ontology associated with differentially expressed genes in salt stressed floe1-1 seeds (See also Table S2). Font size correlates to −log10 (p-value). (J) Germination experiment in the presence of 2μM of the gibberellic acid (GA) inhibitor paclobutrazol shows no difference between wildtype and floe1-1 seeds. Mean ± SEM. Student’s t-test. Representative of three independent experiments. (K) Abscisic acid (ABA) (5μM) does not differentially affect wildtype of floe1-1 germination rates. Mean ± SEM. Four-parameter dose-response fit. Two-way ANOVA. Representative of three independent experiments.
Figure S3: FLOE1 condensation correlates with germination under water stress, related to Figure 2. (A) Quantification of cellular FLOE1 heterogeneity as a function of salt concentration. Black line denotes the 95th percentile of the 280 mM NaCl heterogeneity distribution. (B) Quantification of the percentage of cells per embryo that show FLOE1 condensation as a function of salt concentration. Mean ± SEM. Four-parameter dose-response fit. (C) Correlation between fraction of cells showing FLOE1 condensates and germination. Germination data is same as in Fig. 2A. Mean ± SEM. Four-parameter dose-response fit. (D) Schemes highlighting the experimental setups of the different FLOE1 condensation assays.
Figure S4: Recombinant FLOE1 experiments and in silico modelling, related to Figure 3. (A) Size exclusion chromatography of FLOE1-MBP. For more details about Peaks # 1 and 2, see STAR Methods. (B) Differential interference contrast (DIC) images of FLOE1-MBP before (left) and after (right) TEV cleavage. Some irregularly shaped aggregates can be seen in the starting material, but otherwise the sample has no droplets. After cleavage of the MBP protein tag by TEV, MBP is solubilized and FLOE1 forms spherical droplets indicative of liquid-liquid phase separation (LLPS). Scale bars are 10 μm. (C) SDS-PAGE confirms that TEV cleavage reaction was successful and shows the composition of the droplets observed in (B). Lane 1: FLOE1-MBP uncleaved construct (Peak #2 in (A)). Lane 2: Sample from Peak #2 following 20 hours of incubation with TEV. To see the composition of the droplets and ensure they are primarily composed of FLOE1, the products of this cleavage reaction were spun down for 10 minutes at 20,800 g and the supernatant was separated from the pellet. Lane 3: Pellet shows strong FLOE1 enrichment and very little MBP. Lane 4: Supernatant shows that both FLOE1 and MBP are present. The existence of a FLOE1 band in the supernatant indicates an equilibrium between a dilute and condensed FLOE1, as expected in LLPS. The strong MBP band indicates that these FLOE1 droplets selectively exclude MBP. Lane 5: TEV. (D) FLOE1 phase separation is observed even in the low μM range. (E) Predicted structure from the SWISS-MODEL homology prediction server. Side and top views are shown. A well-defined trimeric coiled-coil oligomeric state is strongly predicted. Inferred helicity from SWISS-MODEL prediction projected from N-to-C terminus and aligned with residue numbers. Predicted helicity from Phyre2 homology model prediction. Predicted helicity from PSI-PRED secondary structure prediction tool. Per-residue percentage helicity as obtained from all-atom simulations. Smoothened fit is shown in black. (F) Images are taken following overnight cleavage of 10 μM MBP-tagged constructs using TEV. Wildtype FLOE1 is found in condensates under a range of salt concentrations. The QPS mutant fails to phase separate and is only able to form aggregates, which are especially prevalent at high salt concentration.
Figure S5: Supplementary data regarding EM analyses, related to Figure 3. (A) 3D reconstruction and cellular annotation of A. thaliana embryo, used to calculate the number of cells and their volumes (see STAR Methods). (B–C) TEM image of an embryonic cell. N denotes nucleus, PSV denotes Protein Storage Vacuoles, and yellow shading highlights remainder of cytoplasm. (D) Quantification of cellular fraction excluding nuclei and PSVs. A total of 43 cells from 8 images were analyzed. (E–F) 3D-tomogram of an embryonic cell. Blue highlights lipid membranes and surface of lipid droplets (LD). See also Movie S1. (G) Quantification of cytoplasmic fraction excluding lipid droplets (LD) and membrane compartments (M) based on 7 tomograms from randomly selected regions. (H) By integrating cell count and volume estimates (A) with subcellular fraction estimates (B–G) we estimate that only 16.7% (68.77% * 24.6%) of the total cellular volume of embryos (~17,000,000 μm3) represents the true cytoplasmic fraction, which is estimated at ~2,840,000 μm3. (I–J) Correlative light-electron microscopy (CLEM) allows us to confirm the presence of GFP-FLOE1 in cytoplasmic condensates in transfected U2OS cells. (I) shows wildtype FLOE1 condensates, whereas (J) shows mutant DS domain (8xY/F–S) condensates. (K–L) Standard transmission electron microscopy of wildtype (K) and mutated DS domain (L) FLOE1 condensates. Zooms of (K–L) can be found in Fig. 4H. (L) Highlights filamentous substructure of mutated DS FLOE1 condensates. (M) 3D-tomography of DS(8xY/F-S) FLOE1 condensates. By looking onto the XY-plane of the tomogram one can observe a hexagonal arrangement of ‘dots’, which are the top view of the mutant FLOE1 filaments (small dashed lines). Upon tilting by 30° (around X-axis, ‘horizontal’ on page) the hexagonal symmetry of the dotted structure transforms into a line pattern, by superpositioning of the filaments. This observation demonstrates the hexagonal symmetry of the mutant FLOE1 filaments. The large dashed line indicates approximate boundary of the full condensates. Small dashed lines indicate a section of the condensate with filaments perpendicular to the tomographic section. See also Movie S3. Dimensions of tomographic volume: 2.5 × 2.5 × 0.2 μm.
Figure S6: Supplementary data regarding FLOE1 mutants and paralogs, related to Figures 5, 6 and 7. (A-B) Since FLOE1 is a dosage-dependent regulator of seed germination under water stress, we wanted to rule out that expression differences in the mutant lines would be responsible for the observed differences in their germination levels. (A) We assayed FLOE1 expression levels in dry seeds via RT-qPCR. (B) As shown before, there was a linear correlation between FLOE1 expression level and germination levels. floe1-1 lines complemented with the ΔDUF mutant followed a similar trend, confirming that the DUF domain deletion does not affect germination in our assays. floe1-1 lines complemented with the ΔDS mutant showed low levels of transgene expression according to RT-qPCR (A, Right Panel. One-way ANOVA. *** p-value < 0.001. Mean ± SEM.) which was consistent with the sparser localization of the protein in radicles (Fig. 5F). Yet, despite these low expression levels, the ΔDS complemented lines consistently induced extreme germination levels, which we never observed for floe1-1 or WT complemented lines. floe1-1 lines complemented with the ΔQPS mutant showed high levels of transgene expression according to RT-qPCR (A). Despite these high transgene levels, and robust protein expression in radicles (Fig. 5F), ΔQPS complemented lines had germination levels similar to the parental floe1-1 line, in stark contrast with WT complemented lines with higher relative expression, supporting the loss-of-function phenotype of this mutant and loss of dose dependency (B). B: Mean ± SEM. Germination data are representative of three independent experiments. (C) Correlation between qPCR-based expression measurements and protein concentrations as measured via GFP ELISA (see also Table S3). Color scheme is the same as in the previous panels. (D) All complemented lines are able to fully germinate under standard conditions (43.5h time point shown). Mean ± SEM. Representative of three independent experiments. (E) ΔDUF and ΔQPS complemented lines have similar germination rates as WT complemented lines. In contrast, ΔDS complemented lines show faster germination rates under standard conditions. Mean ± SEM. Two-way ANOVA. Biological replicates refer to seeds collected from different mother plants. Representative of two independent experiments. (F) ΔDS seeds show high numbers of differentially expressed genes (DEGs) compared to wildtype complemented seeds in all tested conditions. (G) ΔDS seeds upregulate stress response genes or genes implicated in ribosomal biogenesis depending on whether they are in a desiccated or active state. Font size correlates to −log10 (p-value). (H) Quantification of FLOE granule size in tobacco leaves. Isoform comparison: Mann-Whitney test. Paralog comparison: Kruskal-Wallis test. * p-value < 0.05, ** p-value < 0.01, **** p-value < 0.0001. (I) FLOE1 does not mix with FLOE2 and FLOE3, while FLOE2 and FLOE3 do mix. GFP and RFP signal false colored as green and magenta. (J) Species tree as in Fig. 7A with full species names. “Bryo”, “Ly” and “Gymno” refer to the bryophyte, lycophyte, and gymnosperm lineages. (K) Additional examples of FLOE homologs that condense upon expression in tobacco leaves. Belonging to the FLOE1L and FLOE2L clades are indicated. GFP signal displayed as false-colored intensity scale. All fluorescence microscopy images shown are single optical sections of epidermal pavement cells of tobacco leaves.
Figure S7: Ecological analysis of FLOE1 isoform expression, related to Figure 6. (A) FLOE1.1 is the dominant isoform across ecotypes. Mann-Whitney test. **** p-value < 0.0001. (B) Scatterplots of FLOE1 isoform expression show a positive correlation between FLOE1.1 expression and the dormancy trait DSDS50 (number of Days of Seed Dry Storage required to reach 50% germination) (Atwell et al., 2010), which is absent for FLOE1.2. Correlation was measured using Spearman’s rank correlation coefficient, which assesses the statistical dependence between the rankings of two variables. Fitted line is for visualization purposes and not meant to imply a linear relationship. (C–D) Scatterplots of FLOE1 isoform expression show a positive correlation between FLOE1.2 expression and germination at 10°C (Martinez-Berdeja et al., 2020), which is less pronounced for FLOE1.1. (E–F) Scatterplots of FLOE1 isoform expression show a positive correlation between FLOE1.2 expression and germination at 4°C (Martinez-Berdeja et al., 2020), which is less pronounced for FLOE1.1. Correlation was measured using Spearman’s rank correlation, which assesses the statistical dependence between the rankings of two variables. Fitted line is for visualization purposes and not meant to imply a linear relationship. TPM were log2 transformed and mean centered prior to plotting for better visualization. (G) Spearman’s rho correlation between normalized FLOE1.1 expression and average monthly precipitation (blue), maximum (red) and minimum (orange) temperature across 527 worldwide locations of A. thaliana ecotypes. Color of balloons indicates −log10(p-value). Balloons only shown for data points with significance at p < 0.01. (H–I) FLOE1.1 expression shows weak correlation with July precipitation or maximum temperature across locations. Fitted line is for visualization purposes and not meant to imply a linear relationship. (J) Spearman’s rho correlation between normalized FLOE1.2 expression and average monthly precipitation (blue) and maximum (red) and minimum (orange) temperature across 527 worldwide locations of A. thaliana ecotypes. Color of balloons indicates −log10(p-value). Balloons only shown for data points with significance at p < 0.01. (K–L) FLOE1.2 expression shows strong correlation with July precipitation or maximum temperature across locations. Fitted line is for visualization purposes and not meant to imply a linear relationship. (M) Correlation between FLOE1.1 and the coefficient of variation in inter-annual precipitation in the month of July between the years 1958–2017 (Spearman’s rho correlation was r = −0.2397752, p = 1.524E-9). (N) Correlation between FLOE1.2 and the coefficient of variation in inter-annual precipitation in the month of July between the years 1958–2017 (Spearman’s rho correlation was r = −0.2802625, p = 1.234E-12). Fitted lines are for visualization purposes and not meant to imply a linear relationship.
Movie S1: Electron tomographic reconstruction and segmentation of membrane-bound organelles (in blue) of an Arabidopsis thaliana embryo cell, related to Figure 3.
Movie S2: Cytoplasmic streaming of FLOE1-GFP granules in tobacco epidermal pavement cells, related to Figure 3.
Movie S3: 3D tomogram of DS(8xY/F-S) FLOE1 condensates in U2OS cells, related to Figure 4.


