Abstract
The severe acute respiratory syndrome (SARS-CoV-2), a newly emerging of coronavirus, continues to infect humans in the absence of a viable treatment. Neutralizing antibodies that disrupt the interaction of RBD and ACE2 has been under the spotlight as a way of developing the COVID-19 treatment. Some animals, such as llamas, manufacture heavy-chain antibodies that have a single variable domain (VHH) instead of two variable domains (VH/VL) as opposed to typical antibodies. Nanobodies are antigen-specific, single-domain, changeable segments of camelid heavy chain-only antibodies that are recombinantly produced. These types of antibodies exhibit a wide range of strong physical and chemical properties, like high solubility, and stability. The VHH's high-affinity attachment to the receptor-binding domain (RBD) allowed the neutralization of SARS-CoV-2. To tackle COVID-19, some nanobodies are being developed against SARS-CoV-2, some of which have been recently included in clinical trials. Nanobody therapy may be useful in managing the COVID-19 pandemic as a potent and low-cost treatment. This paper describes the application of nanobodies as a new class of recombinant antibodies in COVID-19 treatment.
Keywords: Single-domain antibodies, Nanobody, COVID-19, SARS-CoV-2
Introduction
From December 2019, the new coronavirus disease 2019 (COVID-19) has spread quickly around the world. Being reported in over 200 countries, it has claimed a large number of lives [1, 2]. As there is no effective medication for the severe acute respiratory syndrome (SARS-CoV-2), treating COVID-19 patients, particularly those with severe pneumonia, is challenging [3]. Neutralizing antibodies are critical for SARS-CoV-2 immunity as well as COVID-19 prevention and treatment [4]. Although monoclonal antibody-based therapy can be useful for those with mild COVID-19 symptoms, it requires exceedingly large doses, typically a few grams intravenously [5, 6]. Moreover, they cannot be produced fast or economically, and are unable to target several epitopes at the same time [7]. Camelid antibodies with a single variable domain (also known as nanobodies/VHH) are a practical alternative, as they are tiny in size (13 to 15 kDa), and have excellent solubility, and stability. They are particularly well suited for pulmonary administration [8, 9]. VHH's high-affinity compete with the receptor-binding domain (RBD) of viral spike, enabling SARS-CoV-2 neutralization. Therefore, the S1 RBD has become a prominent target for vaccine research [10, 11]. The first stage of a viral life cycle is entering a host cell [12, 13]. The interaction of the RBD of the viral spike with the cellular receptor, known as angiotensin-converting enzyme 2 (ACE2), is required for SARS-CoV-2 cell entry [14, 15]. Besides, Nbs that target chemokines and cytokines can be customized to modify inflammation responses, which could be conducive to COVID-19 patients' recovery [16]. In this review, we elaborate on the use of nanobodies, a new type of recombinant antibody, as a treatment for COVID-19.
SARS-CoV-2 characteristics
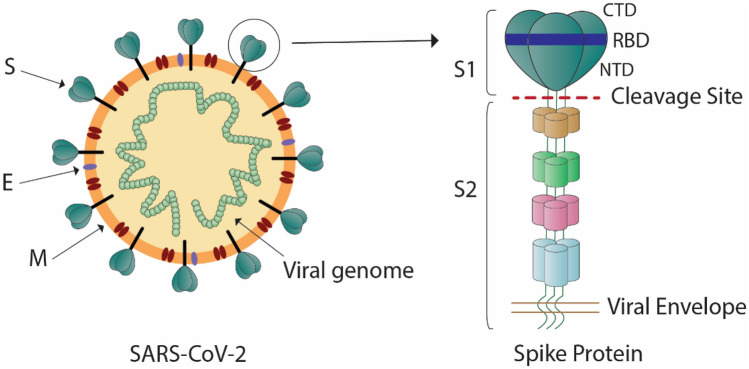
An unknown respiratory tract infection began in Wuhan, China, in the autumn of 2019. Based on the patients’ symptoms, clinicians diagnosed virus-induced pneumonia [17]. According to genomic sequencing, this pneumonia was caused by a novel coronavirus [18]. The international committee on taxonomy of viruses (ICTV) named this unexpected coronavirus SARS-CoV-2 [19]. As the seventh coronavirus, SARS-CoV-2 is able to infect humans [20]. The SARS-CoV-2 is an enveloped, non-segmented, positive-sense single-stranded RNA virus with a genomic size of about 30 kb (22), which encodes both structural and non-structural proteins. The non-structural proteins such as Papain-like protease, 3C‑like serine protease (3CL-protease), RNA-dependent RNA polymerase, Helicase, Endoribonuclease, and the structural proteins are spike glycoprotein (S), an envelope protein (E), membrane protein (M), and nucleocapsid protein (N). The coronavirus spiral nucleocapsid is covered by phospholipid bilayers with M/E proteins, and the trimmers of S protein are found on the virus particle’s surface [21].
The S protein of coronaviruses is a trimeric class I viral fusion protein. It consists of two subunits: S1 (in the amino-terminal) that contains RBD, and S2 (in the carboxy-terminal) that induces membrane fusion (Fig. 1) [22]. The RBD of the SARS-CoV-2 S protein mediates cell entry by attaching to human angiotensin-converting enzyme 2 (ACE2). The ACE2 is a transmembrane protein placed on the epithelial cells of the nasal mucosa, lungs, heart, kidneys, stomach, bladder, and intestine [23]. The dissociation of S1 from ACE2 is induced by receptor contact, causing the transition of S2 from a metastable pre-fusion mode to a stable post-fusion mode, which is essential for membrane fusion [24]. The S protein can be a target of antibody-mediated neutralization due to its crucial function, and the characterization of the pre-fusion S structure can provide useful information for vaccine formulation and development [24, 25].
Fig. 1.
The SARS-CoV-2 spike structure. S spike, E envelope, M membrane, RBD receptor-binding domain, CTD C-terminal domain, NTD N-terminal domain. The SARS-CoV-2 surface protein Spike (S) is a structural glycoprotein. It comprises S1 and S2 subunits. The RBD region is located in the CTD part of the S1 and mediates ACE2 binding. The S protein is a possible therapeutic target since it promotes virus entry into target cells in response to ACE2 receptor interaction
Virus mutations and variants are major impediments to the control of the SARS-CoV-2 pandemic and the development of an effective vaccine. The coronavirus will probably mutate during the replication phase, which will alter the virus's behavior as the genomes of RNA viruses are intrinsically unstable [26]. The mutation E484K, called an escape mutation, takes place in diverse variants identified in Brazilian (B.1.1.28), South African (B.1.351), and UK (B.1.1.7). The E484K mutation identified in the RBD is associated with the ability to evade neutralizing antibodies and the body's immunological responses. This mutation can influence vaccine efficacy. The E484K mutation increases the number of serum antibodies needed to protect cells from infection [27, 28]. Another mutation in the spike is the N501Y, which in the case of SARS-CoV-2, appeared in Brazilian, South African, and United Kingdom variants [29]. The SARS-CoV-2 RBD, with the N501Y mutation, is linked to the improved receptor binding specificity and the virus growth speed. The SARS-CoV-2 with N501Y mutation was tested in the mouse model and increased infectivity and virulence were reported [29]. Several mutations in the Spike's S subunits have been reported in India, including the D614G. In addition, G1124V in the Spike (S2 subunit) protein has been recognized as a nonsynonymous mutation [30]. SARS-CoV-2 features can be altered by any of these spike mutations.
What are nanobodies
Nanobodies represent a relatively new type of recombinant antibody The Camelidae members, which include ancient species like dromedaries and camels as well as more recent species such as llamas and alpacas, can produce non-conventional antibodies besides conventional antibodies [31, 32]. The conventional (classical) antibodies produced by all mammals have a heterotetrameric structure with two heavy and two light chains, but non-conventional antibodies are only composed of two heavy chains and have a single variable domain (VHH, k15kDa), which is the antigen-binding region (Fig. 2a and b) [33]. Heavy-chain antibodies (HcAbs) are the third generation of antibodies that provided a new approach to the creation of therapeutic antibodies [32, 34]. Even though the lack of a light chain variable domain (VL) may seem disadvantageous in terms of antigen binding, nanobodies have evolved to compensate, generating characteristics that improve stability [33]. These antibodies' variable parts contain intriguing features compared to other antibodies including higher solubility, small size, greater resistance to denaturation, stability in high temperatures, and high/low pH, cost-effective production, high specificity, low immunogenicity, ease of manipulation, and identification of variable epitopes [35, 36]. Furthermore, tissue penetration and extravasation are better in nanobodies than in classical monoclonal antibodies, which is of great therapeutic value [37].
Fig. 2.
Nanobodies neutralize the SARS-CoV-2. FC Fragment crystallizable, Fab fragment antigen-binding, VH Heavy chain variable domain, VL Light chain variable domain, CH heavy chain constant region, CL Light chain constant region, ACE2 angiotensin-converting enzyme 2. SARS-CoV-2 neutralization was made possible by inhibiting the target cell entrance. VHHs bind to the spike RBD and prevent the ACE2 engagement with RBD
The overall-architecture framework regions (FR1/2/3/4) and complementarity determining regions (CDR1/2/3) of nanobodies are the same as VHs. The CDR3 is a key factor in antigen recognition and specificity and its length are significantly greater in nanobodies than in human VH domains. The CDR1 and CDR2 are proteins that contribute to the binding strength [38]. However, human immunogenic reactions could be triggered by nanobodies in humans. Thus, for therapeutic purposes, humanization protocols have been developed to identify the aminoacid sequences in the framework regions that correspond to their human heavy-chain variable domain counterparts [39, 40]. The first adult medication developed based on the single-domain antibody (sdAb) was approved in November 2018 by the European Medicines Agency (EMA) and the US Food and Drug Administration (FDA) [38, 41]. The antiviral effects of nanobodies against respiratory viruses such as coronavirus have been demonstrated in a host of investigations [42]. The SARS-CoV-2 spike protein has been used to immunize some animals and discover nanobodies that interact with the virus's receptor-binding region [43].
Nanobodies against SARS-CoV-2
There are some effective diagnostic procedures for the diagnosis of SARS-CoV-2 infection, but no therapy that can interfere with SARS-CoV-2 replication has been identified so far [44]. Antibody treatment is one of the different methods for controlling COVID-19 infection [35]. The therapeutic potential of nanobodies against coronaviruses has been established in recent studies [45–47]. A primary target for anti-SARS-CoV-2 neutralizing antibodies involves blocking the interaction of the SARS-CoV-2 S1 protein with ACE2 [46–48]. The S1 RBD has been the main target of vaccine development, as SARS-CoV-2 can be neutralized by VHH's high-affinity competition with the spike RBD (Fig. 2c) [10, 11]. Several techniques are currently used to identify nanobodies that neutralize SARS-CoV-2, including llama immunization, the phage display of a naive llama nanobody library or humanized synthetic nanobody library, as well as the yeast surface display of synthetic nanobodies [23, 39, 40, 43, 49–54]. The developed nanobodies effectively neutralized both SARS-CoV-2 pseudovirus and live virus [55]. Recently, researchers in the United States have isolated nanobodies that bind to the receptor-binding domain of the SARS-CoV-2 spike protein and prevent the interaction of spike protein with ACE2 [50]. Furthermore, the results of an in vitro analysis suggest two nanobodies (H11-D4 and H11-H4) with high affinity to spike RBD protein that can prevent the spike's attachment to ACE2 [40]. In another study, Hanke et al. discovered Ty1, a SARS-CoV-2 RBD-specific nanobody that effectively neutralizes the virus. The Ty1 nanobody has several practical advantages, including high-yield production in bacteria, low cost, and scalability. As suggested in the literature, future research needs to explore a range of techniques to improve Ty1's potency and efficacy [54]. Three VHHs, H11-D4, H11-H4, and Ty1, have been identified to target the SARS-CoV-2 spike RBD and disrupt its interaction with ACE2 [56, 57].
Nanobodies have special biophysical characteristics, such as thermostability and tiny size, which enable their aerosolized administration. They can be readily nebulized and inhaled straight into the lungs by an inhaler [8, 58]. One of the advantages of inhalable nanobodies is antibodies like PiN-21 and Nb11-59, which can successfully target deep and local lung tissues such as terminal alveoli that share a border with alveolar cells rich in ACE2 receptors [34, 59]. Thus, inhalation therapy can be used to administer nanobody therapeutics [8], aside from other routes of delivery such as intravenous, intramuscular, or subcutaneous treatment [50].
Synthetic nanobodies (or sybodies)
Traditional nanobodies are extracted from immunized camelids, but creating libraries of synthetic nanobodies (sybodies) is a new technique for fast drug development that produces highly selective binders with neutralizing potentials in a short time [60]. In this context, another option is the chimeric nanobodies-Fc (in which the variable area of nanobody is bonded to Fc of human immunoglobulin). The synthetic nanobodies are tiny, aerosolizable and heat stable, which makes them a feasible option for COVID-19 prevention and therapy [60].
Stefan et al. designed a huge synthetic VHH library (3.18 × 1010), finding over 50 VHH candidates that can bind to SARS-2 [61]. Schoof et al. (2020) identified a panel of Nbs that can bind to various epitopes on Spike from a synthetic Nbs library. These Nbs are divided into two groups. Class I binds directly to the RBD and competes with the ACE2 receptor on human cell surfaces. Class II, identified as a different binding site, contributes to the alteration of the RBD's structural conformation, preventing it from the recognition of the ACE2 receptor [52]. Researchers have identified a new synthetic Nb called SR31. Stronger binding affinities and neutralizing activities of this synthetic nanobody have been reported in fusion with additional sybodies. Additionally, to reinforce affinity and efficacy, it can be coupled with monoclonal antibodies or other antibody fragments [62]. In the context of synthetic nanobodies, a recent study has used a mix of ribosome display and phage display. Accordingly, it managed to develop 99 distinct sybodies against the SARS-CoV-2 S-RBD from three libraries, which neutralized SARS-CoV-2 pseudo-viruses efficiently and selectively [63]. Synthetic or naive llama nanobodies including IE2, 2F2, 3F11, 4D8, 5F8, H11-D4, and H11-H4 can block RBD–ACE2 binding, and neutralize the pseudo-typed and live SARS-CoV-2 infections under in vitro conditions through SARS-CoV-2 RBD-targeting [39, 40]. Custodio et al. reported that sybody 23 (Sb23) neutralized SARS-CoV-2 spike pseudovirus by competing with ACE2 binding. This type of synthetic nanobody, displaying a high affinity for the recombinant RBD and the prefusion spike glycoprotein, has strong neutralizing activities [64]. However, synthetic nanobodies have several limitations. As monomers, they often lack the high binding affinity required for therapeutic use. In this way, multivalent or multi-paratopic nanobodies might offer a faster way to exploit avidity in order to improve affinity and effectiveness [65, 66].
Multivalent nanobodies
In certain cases, SARS-CoV-2 undergoes rapid mutations, which seem to have avoided the antibody response [67–69]. The developing variations, B.1.1.7, B.1.351, and P.1 (first reported in the UK, South Africa, and Brazil, respectively) have undermined the effectiveness of serum in COVID-19 patients and immunotherapies approved for emergency use [70–72]. Multivalent nanobodies or variable domains of heavy-chain Abs have been developed by investigating the precise architectures of SARS-CoV-2 epitopes and binding modalities to the S protein of the virus, as well as fusing virus to the cell membrane through the S protein [43]. An in-silico technique that leads to the fusion of VHHs to Fc domains is currently utilized to create multi-specific antibodies with elevated avidity and affinity as well as enhanced S/ACE2 blocking [49]. The mutations of SARS-CoV-2 variant are countered by multivalent nanobodies via two mechanisms: amplified avidity for the binding domain of ACE2 and detection of preserved epitopes, which are chiefly not accessible to human antibodies [69]. Moreover, the serum half-life could be improved by the oligomerization of nanobodies [73]. In the S1 RBD and S/ACE2 inhibition of SARS-CoV-2, the bi-specific VHH-Fc antibodies are significantly more effective than monoclonal VHH-Fcs [49]. Ma et al. reported the isolation of seven anti-RBD Nbs from alpacas immunized with SARS-CoV-2 RBD. In their analysis, combining two Nbs with different epitopes led to the creation of two hetero-bivalent Nbs with high affinity, producing antibodies with significant SARS-CoV-2 neutralizing efficacy [11]. In line with these findings, the heterodimer nanobody Nb91-Nb3-hFc with an IC50 of 1.54 nM displayed the highest RBD-binding affinity and neutralizing activity against SARS-CoV-2 pseudo-viruses. By limiting the interaction of spike protein with ACE2 through RBD targeting, the neutralizing nanobodies were able to drastically reduce SARS-CoV-2 pseudo-virus infection in the host HEK293T-ACE2 cells [47]. Pymm et al. used four bivalent nanobodies to neutralized SARS-CoV-2 (WNbFc 2, WNbFc7, WNbFc 15, and WNbFc 36). They discovered that nanobody cocktails containing two noncompeting nanobodies could prevent ACE2 engagement with RBD variations commonly found in human populations and neutralize both wild-types of SARS-CoV-2 and the N501Y D614G RBD variant at low quantities [4]. Another study suggested that tri-specific VHH-Fc antibodies as promising therapeutics for COVID-19 treatment and prevention. These tri-specific VHH-Fcs were found to be particularly effective in binding to SARS-CoV-2 S1 RBD and blocking S/ACE2, while inhibiting the infection of human target cells by a SARS-CoV-2 pseudo-virus [44].
Researchers developed a VH-phage library and targeted the binding interface of the SARS-CoV-2 Spike receptor-binding domain's angiotensin-converting enzyme 2 (ACE2) (Spike-RBD). They discovered VH binders for two non-overlapping epitopes, which were combined into multivalent and biparatopic forms. Compared to stand-alone VH domains, they demonstrated an increased affinity for spike (up to 600-fold) and neutralization efficacy (up to 1400-fold) on pseudo-typed SARS-CoV-2 virus [66]. Koenig et al. engineered multivalent nanobodies using more than 100 neutralizing activities of monovalent nanobodies, which targeted the receptor-binding domain of the SARS-CoV-2 spike protein. The biparatopic nanobody fusions inhibited the scape of viral mutants. By targeting two distinct epitopes, these multivalent nanobodies prevented SARS-CoV-2 infection and inhibited mutational escape [43].
A recent study by Zupancic et al. has suggested that a hexavalent VHH-72 nanobody with excellent stability, solubility, and non-specific binding was successfully attached to spike proteins in SARS-CoV-2 variants with high transmission (B.1.1.7 and B.1.351) and neutralizes them effectively [74]. Another study discovered three new bispecific nanobodies (Nb15-Fc, Nb22-Fc, and Nb31-Fc) that could significantly inhibit the wild-type and variations of SARS-CoV-2, such as circulating forms, as was the case in mutant viruses found in the UK and South Africa with the N501Y mutation [75]. Sun et al. reported that SARS-CoV-2 neutralizing Nbs can be grouped into three epitope classes. Class I includes some of the most effective SARS-CoV-2 neutralizing Nbs discovered so far. However, a single point mutation (E484K/Q) found in Gamma (P.1), Beta (B.1.351), or Kappa (B.1.617.1) variations can prevent class I Nbs from binding to RBD. Class II Nbs are designed to target conserved epitopes resistant to existing variants of concern (VOCs) and are expected to resist mutation. Class III Nbs can bind to formerly unidentified high-affinity epitopes with strong affinities, such as the Nb17 epitope, which is likely inaccessible to conventional antibodies [76]. Thus, nanobodies can be a potential tool to neutralize SARS-CoV-2 variants, even if new mutations continue to develop. Table 1 outlines a summary of similarities and differences in published nanobodies.
Table 1.
Summary of similarities and differences in published nanobodies
| Nanobody | Method | Neutralizing pseudovirus (IC50) | Affinity to RBD | Function | References |
|---|---|---|---|---|---|
| VHH-E | Immunized phage display library | 60 nM | 1.86 nM | Blocking RBD-ACE2 interaction/prevent the emergence of viral escape mutants | [90] |
| Nb11-59 | Immunized phage display library | 36.7 nM | 21 nM | Blocking RBD-ACE2 interaction/high binding activity to the RBD | [34] |
| Sybody (n3021) | Ribosome and phage display | – | 0.63 nM | Blocking RBD-ACE2 interaction/binding to the full-length SARS-CoV-2 spike protein | [91] |
| Sybody (MR3) | Ribosome and phage display | 40 nM | 24.22 nM | Blocking RBD-ACE2 interaction | [92] |
| Nb6 | yeast surface-displayed library | 2 uM | 210 nM | Blocking RBD-ACE2 interaction/Binding Spike in a fully inactive conformation with its receptor binding domains | [93] |
| H11-D4 | naive llama single-domain antibody library | – | 39 nM | Blocking RBD-ACE2 interaction/Binding to all three RBDs in the spike trimer | [94] |
| H11-H4 | naive llama single-domain antibody library | – | 12 nM | Blocking RBD-ACE2 interaction/Binding to all three RBDs in the spike trimer | [94] |
| Ty1 | Immunized phage display library | 54 nM | 5–10 nM | Blocking RBD-ACE2 interaction/Binding to the RBD with high affinity | [95] |
| Nbs 89 | Immune library and MS proteomic | 0.133 nM | 108 pM | Blocking RBD-ACE2 interaction/inhibit viral infection | [96] |
| WNb | Immunized phage display library | – | ≤ 80 nM | Blocking RBD-ACE2 interaction/neutralize both wildtype SARS-CoV-2 and the N501Y D614G variant | [4] |
Nanobodies modulate inflammation
COVID-19 hyper-inflammation, one of the uncontrolled systemic inflammations and a key process in acute respiratory distress syndrome (ARDS), is also known as a cytokine storm. It takes place when copious numbers of pro-inflammatory cytokines (e.g. TNF, IL-1, and IL-6) and chemokines (e.g. CXCL-10) are released [77–79]. SARS-CoV-2 S-interacting proteins expressed in myeloid cells can operate as signaling receptors to activate particular hyperinflammatory responses. Also, they play a key role in COVID-19 immunopathogenesis and immunological dysregulation. Researchers manufactured a bispecific anti-spike nanobody, A8-G11-Fc, which inhibited both ACE2-mediated infection and myeloid receptor-mediated proinflammatory responses [80].
The use of monoclonal antibodies to target and neutralize proinflammatory cytokines provides a potential therapeutic approach for inflammatory disorders. Nanobodies offer novel tools to modulate inflammatory responses in COVID-19 patients [81]. By targeting chemokines and cytokines, they can be adjusted to control inflammation responses, which contributes to COVID-19 patients' recovery [16]. For example, the isolation of anti-CXCL10 polyclonal HcAbs has been conducted to create specific Nbs, which can selectively target CXCL10 for in vivo therapies [82]. TNF-alpha is a critical cytokine and TNF-blocking sdAbs have been reported in a llama immunized with human and mouse antigens. It has been shown to be more effective than TNF-blocking antibodies like Infliximab and Adalimumab in TNF neutralization [83]. Furthermore, antigen presenting through antigen presentation cells (APCs) enhances antibody formation, CD4+ T cell activation, and CD8+ T cell responses, all of which help boost immunological responses. APCs contain high levels of MHC-II products, integrins (CD11b), and scavenger receptors (CD36). VHHs targeting these molecules improved immune responses in dendritic cells (DCs), induced humoral immunity, and detected inflammation in order to cure or prevent SARS-CoV-2 infection [16, 84].
According to the evidence, nanobodies can act as an ion-channel blocking agent. P2X7 is a ligand-gated ion channel expressed by lymphocytes and monocytes. This channel triggers a pro-inflammatory signaling cascade when detecting adenosine 5′-triphosphate produced by injured cells. It involves the production of pro-inflammatory cytokines like interleukin-1beta (IL-1β), IL-18, and IL-33 [85–87]. This ion channel is a potential therapeutic target in inflammatory diseases [88]. A variety of nanobodies capable of regulating the function of human and mouse P2X7 have been identified [87]. For example, the nanobody Dano1 has exhibited complete effectiveness in blocking human P2X7 [89]. This nanobody was found to be 1000 times more effective than small molecule inhibitors of P2X7 at blocking ATP-induced IL-1b release from monocytes [87]. In this way, nanobodies pave the way for novel experimental and therapeutic immunomodulation approaches.
Conclusion
Tiny, stable, and easy to make, the Nbs show great potentials as COVID-19 therapeutic proteins. To develop suitable nanobodies against SARS-CoV-2, the bulk of studies have employed the spike protein, especially RBD domains. Given that nanobodies can be utilized as an inhaler, it seems that the produced nanobodies can be applied to inhibit the infection of the lungs by the virus. Developing methods with effective nanobodies, high-expression yield and reasonable costs is crucial to control the COVID-19 pandemic.
Data availability
The data used to support the findings of this study are included in the article.
Declarations
Conflict of interest
The authors declare that there is no conflict of interests.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Consent for publication
All of the authors consent for publication.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Arghavan Zebardast, Email: azebardast@razi.tums.ac.ir.
Tayebeh latifi, Email: t.latifi1992@gmail.com, Email: tlatifi@razi.tums.ac.ir.
References
- 1.Ksiazek TG, Erdman D, Goldsmith CS, Zaki SR, Peret T, Emery S, Tong S, Urbani C, Comer JA, Lim W. A novel coronavirus associated with severe acute respiratory syndrome. N Engl J Med. 2003;348:1953–1966. doi: 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]
- 2.Zaki AM, Van Boheemen S, Bestebroer TM, Osterhaus AD, Fouchier RA. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N Engl J Med. 2012;367:1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- 3.Raza SS, Khan MA. Mesenchymal stem cells: a new front emerge in COVID19 treatment: mesenchymal stem cells therapy for SARS-CoV2 viral infection. Cytotherapy. 2020 doi: 10.1016/j.jcyt.2020.07.002. [DOI] [Google Scholar]
- 4.Pymm P, Adair A, Chan L-J, Cooney JP, Mordant FL, Allison CC, Lopez E, Haycroft ER, O’Neill MT, Tan LL. Nanobody cocktails potently neutralize SARS-CoV-2 D614G N501Y variant and protect mice. Proc Natl Acad Sci. 2021;118:19. doi: 10.1073/pnas.2101918118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen P, Nirula A, Heller B, Gottlieb RL, Boscia J, Morris J, Huhn G, Cardona J, Mocherla B, Stosor V. SARS-CoV-2 neutralizing antibody LY-CoV555 in outpatients with Covid-19. N Engl J Med. 2021;384:229–237. doi: 10.1056/NEJMoa2029849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weinreich DM, Sivapalasingam S, Norton T, Ali S, Gao H, Bhore R, Musser BJ, Soo Y, Rofail D, Im J. REGN-COV2, a neutralizing antibody cocktail, in outpatients with Covid-19. N Engl J Med. 2021;384:238–251. doi: 10.1056/NEJMoa2035002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Iwasaki A, Yang Y. The potential danger of suboptimal antibody responses in COVID-19. Nat Rev Immunol. 2020;20:339–341. doi: 10.1038/s41577-020-0321-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Van Heeke G, Allosery K, De Brabandere V, De Smedt T, Detalle L, de Fougerolles A. Nanobodies® as inhaled biotherapeutics for lung diseases. Pharmacol Ther. 2017;169:47–56. doi: 10.1016/j.pharmthera.2016.06.012. [DOI] [PubMed] [Google Scholar]
- 9.Larios Mora A, Detalle L, Gallup JM, Van Geelen A, Stohr T, Duprez L, Ackermann MR, editors. Delivery of ALX-0171 by inhalation greatly reduces respiratory syncytial virus disease in newborn lambs. London: Taylor & Francis; 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Respaud R, Vecellio L, Diot P, Heuzé-Vourc’h N. Nebulization as a delivery method for mAbs in respiratory diseases. Expert Opin Drug Deliv. 2015;12:1027–1039. doi: 10.1517/17425247.2015.999039. [DOI] [PubMed] [Google Scholar]
- 11.Ma H, Zeng W, Meng X, Huang X, Yang Y, Zhao D, Zhou P, Wang X, Zhao C, Sun Y. Potent neutralization of sars-cov-2 by hetero-bivalent alpaca nanobodies targeting the spike receptor-binding domain. J Virol. 2021;95:e02438–e2520. doi: 10.1128/JVI.02438-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li F. Structure, function, and evolution of coronavirus spike proteins. Annu Rev Virol. 2016;3:237–261. doi: 10.1146/annurev-virology-110615-042301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bosch BJ, Van der Zee R, De Haan CA, Rottier PJ. The coronavirus spike protein is a class I virus fusion protein: structural and functional characterization of the fusion core complex. J Virol. 2003;77:8801–8811. doi: 10.1128/JVI.77.16.8801-8811.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoffmann M, Kleine-Weber H, Schroeder S, Krüger N, Herrler T, Erichsen S, Schiergens TS, Herrler G, Wu N-H, Nitsche A. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang Q, Zhang Y, Wu L, Niu S, Song C, Zhang Z, Lu G, Qiao C, Hu Y, Yuen K-Y. Structural and functional basis of SARS-CoV-2 entry by using human ACE2. Cell. 2020;181:894–904. doi: 10.1016/j.cell.2020.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bessalah S, Jebahi S, Mejri N, Salhi I, Khorchani T, Hammadi M. Perspective on therapeutic and diagnostic potential of camel nanobodies for coronavirus disease-19 (COVID-19) 3 Biotech. 2021;11:1–14. doi: 10.1007/s13205-021-02647-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li Q, Guan X, Wu P, Wang X, Zhou L, Tong Y, Ren R, Leung KS, Lau EH, Wong JY. Early transmission dynamics in Wuhan, China, of novel coronavirus–infected pneumonia. New Engl J Med. 2020;382:1199–1207. doi: 10.1056/NEJMoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, Zhao X, Huang B, Shi W, Lu R. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382(8):727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gorbalenya AE, Baker SC, Baric R, Groot RJd, Drosten C, Gulyaeva AA, Haagmans BL, Lauber C, Leontovich AM, Neuman BW. Severe acute respiratory syndrome-related coronavirus: the species and its viruses–a statement of the Coronavirus Study Group. bioRxiv. 2020 doi: 10.1101/2020.02.07.937862. [DOI] [Google Scholar]
- 20.Covid C, Team R, COVID C, Team R, COVID C, Team R, Bialek S, Boundy E, Bowen V, Chow N Severe outcomes among patients with coronavirus disease 2019 (COVID-19)—United States, February 12–March 16, 2020. Morb Mortal Wkly Rep. 2020;69:343. doi: 10.15585/mmwr.mm6912e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pal M, Berhanu G, Desalegn C, Kandi V. Severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2): an update. Cureus. 2020;12:3. doi: 10.7759/cureus.7423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yadav R, Chaudhary JK, Jain N, Chaudhary PK, Khanra S, Dhamija P, Sharma A, Kumar A, Handu S. Role of structural and non-structural proteins and therapeutic targets of SARS-CoV-2 for COVID-19. Cells. 2021;10:821. doi: 10.3390/cells10040821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wrapp D, Wang N, Corbett KS, Goldsmith JA, Hsieh C-L, Abiona O, Graham BS, McLellan JS. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Donoghue M, Hsieh F, Baronas E, Godbout K, Gosselin M, Stagliano N, Donovan M, Woolf B, Robison K, Jeyaseelan R. A novel angiotensin-converting enzyme–related carboxypeptidase (ACE2) converts angiotensin I to angiotensin 1–9. Circ Res. 2000;87:e1–e9. doi: 10.1161/01.res.87.5.e1. [DOI] [PubMed] [Google Scholar]
- 25.Gui M, Song W, Zhou H, Xu J, Chen S, Xiang Y, Wang X. Cryo-electron microscopy structures of the SARS-CoV spike glycoprotein reveal a prerequisite conformational state for receptor binding. Cell Res. 2017;27:119–129. doi: 10.1038/cr.2016.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen J, Gao K, Wang R, Wei G-W. Prediction and mitigation of mutation threats to COVID-19 vaccines and antibody therapies. Chem Sci. 2021;12:6929–6948. doi: 10.1039/d1sc01203g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wise J. Covid-19: the E484K mutation and the risks it poses. Br Med J. 2021 doi: 10.1136/bmj.n359. [DOI] [PubMed] [Google Scholar]
- 28.Collier DA, De Marco A, Ferreira IA, Meng B, Datir R, Walls AC, Bassi J, Pinto D, Fregni CS, Bianchi S. SARS-CoV-2 B. 1.1. 7 escape from mRNA vaccine-elicited neutralizing antibodies. MedRxiv. 2021;593:136–141. doi: 10.1038/s41586-021-03412-7. [DOI] [PubMed] [Google Scholar]
- 29.Mahase E. Covid-19: what new variants are emerging and how are they being investigated? Br Med J. 2021 doi: 10.1136/bmj.n158. [DOI] [PubMed] [Google Scholar]
- 30.Maitra A, Sarkar MC, Raheja H, Biswas NK, Chakraborti S, Singh AK, Ghosh S, Sarkar S, Patra S, Mondal RK. Mutations in SARS-CoV-2 viral RNA identified in Eastern India: possible implications for the ongoing outbreak in India and impact on viral structure and host susceptibility. J Biosci. 2020;45:1–18. doi: 10.1007/s12038-020-00046-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zare H, Rajabibazl M, Rasooli I, Ebrahimizadeh W, Bakherad H, Ardakani LS, Gargari SLM. Production of nanobodies against prostate-specific membrane antigen (PSMA) recognizing LnCaP cells. Int J Biol Markers. 2014;29:169–179. doi: 10.5301/jbm.5000063. [DOI] [PubMed] [Google Scholar]
- 32.Hamers-Casterman C, Atarhouch T, Sa M, Robinson G, Hammers C, Songa EB, Bendahman N, Hammers R. Naturally occurring antibodies devoid of light chains. Nature. 1993;363:446–448. doi: 10.1038/363446a0. [DOI] [PubMed] [Google Scholar]
- 33.Yang EY, Shah K. Nanobodies: next generation of cancer diagnostics and therapeutics. Front Oncol. 2020;10:1182. doi: 10.3389/fonc.2020.01182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gai J, Ma L, Li G, Zhu M, Qiao P, Li X, Zhang H, Zhang Y, Chen Y, Ji W. A potent neutralizing nanobody against SARS-CoV-2 with inhaled delivery potential. MedComm. 2021;2:101–113. doi: 10.1002/mco2.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zare H, Aghamollaei H, Hosseindokht M, Heiat M, Razei A, Bakherad H. Nanobodies, the potent agents to detect and treat the Coronavirus infections: a systematic review. Mol Cell Probes. 2021;55:101692. doi: 10.1016/j.mcp.2020.101692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hu Y, Liu C, Muyldermans S. Nanobody-based delivery systems for diagnosis and targeted tumor therapy. Front Immunol. 2017;8:1442. doi: 10.3389/fimmu.2017.01442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yu S, Xiong G, Zhao S, Tang Y, Tang H, Wang K, Liu H, Lan K, Bi X, Duan S. Nanobodies targeting immune checkpoint molecules for tumor immunotherapy and immunoimaging. Int J Mol Med. 2021;47:444–454. doi: 10.3892/ijmm.2020.4817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jovčevska I, Muyldermans S. The therapeutic potential of nanobodies. BioDrugs. 2020;34:11–26. doi: 10.1007/s40259-019-00392-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chi X, Liu X, Wang C, Zhang X, Li X, Hou J, Ren L, Jin Q, Wang J, Yang W. Humanized single domain antibodies neutralize SARS-CoV-2 by targeting the spike receptor binding domain. Nat Commun. 2020;11:1–7. doi: 10.1038/s41467-020-18387-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huo J, Le Bas A, Ruza RR, Duyvesteyn HM, Mikolajek H, Malinauskas T, Tan TK, Rijal P, Dumoux M, Ward PN. Neutralizing nanobodies bind SARS-CoV-2 spike RBD and block interaction with ACE2. Nat Struct Mol Biol. 2020;27:846–854. doi: 10.1038/s41594-020-0469-6. [DOI] [PubMed] [Google Scholar]
- 41.Morrison C. Nanobody approval gives domain antibodies a boost. Nat Rev Drug Discov. 2019;18:485–488. doi: 10.1038/d41573-019-00104-w. [DOI] [PubMed] [Google Scholar]
- 42.Ezzikouri S, Nourlil J, Tsukiyama-Kohara K, Kohara M, El Ossmani H, Windisch MP, Benjelloun S. Nanobodies: an unexplored opportunity to combat COVID-19. J Biomol Struct Dyn. 2020;1:1–3. doi: 10.1080/07391102.2020.1845801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Koenig P-A, Das H, Liu H, Kümmerer BM, Gohr FN, Jenster L-M, Schiffelers LD, Tesfamariam YM, Uchima M, Wuerth JD. Structure-guided multivalent nanobodies block SARS-CoV-2 infection and suppress mutational escape. Science. 2021;371:6530. doi: 10.1126/science.abe6230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dong J, Huang B, Wang B, Titong A, Kankanamalage SG, Jia Z, Wright M, Parthasarathy P, Liu Y. Development of humanized tri-specific nanobodies with potent neutralization for SARS-CoV-2. Sci Rep. 2020;10:1–12. doi: 10.1038/s41598-020-74761-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.He L, Tai W, Li J, Chen Y, Gao Y, Li J, Sun S, Zhou Y, Du L, Zhao G. Enhanced ability of oligomeric nanobodies targeting MERS coronavirus receptor-binding domain. Viruses. 2019;11:166. doi: 10.3390/v11020166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhao G, He L, Sun S, Qiu H, Tai W, Chen J, Li J, Chen Y, Guo Y, Wang Y. A novel nanobody targeting Middle East respiratory syndrome coronavirus (MERS-CoV) receptor-binding domain has potent cross-neutralizing activity and protective efficacy against MERS-CoV. J Virol. 2018;92:e00837-18. doi: 10.1128/JVI.00837-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lu Q, Zhang Z, Li H, Zhong K, Zhao Q, Wang Z, Wu Z, Yang D, Sun S, Yang N. Development of multivalent nanobodies blocking SARS-CoV-2 infection by targeting RBD of spike protein. J Nanobiotechnol. 2021;19:1–12. doi: 10.1186/s12951-021-00768-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shi R, Shan C, Duan X, Chen Z, Liu P, Song J, Song T, Bi X, Han C, Wu L. A human neutralizing antibody targets the receptor-binding site of SARS-CoV-2. Nature. 2020;584:120–124. doi: 10.1038/s41586-020-2381-y. [DOI] [PubMed] [Google Scholar]
- 49.Dong J, Huang B, Jia Z, Wang B, Gallolu Kankanamalage S, Titong A, Liu Y. Development of multi-specific humanized llama antibodies blocking SARS-CoV-2/ACE2 interaction with high affinity and avidity. Emerg Microb Infect. 2020;9:1034–1036. doi: 10.1080/22221751.2020.1768806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Esparza TJ, Martin NP, Anderson GP, Goldman ER, Brody DL. High affinity nanobodies block SARS-CoV-2 spike receptor binding domain interaction with human angiotensin converting enzyme. Sci Rep. 2020;10:1–13. doi: 10.1038/s41598-020-79036-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xiang Y, Nambulli S, Xiao Z, Liu H, Sang Z, Duprex WP, Schneidman-Duhovny D, Zhang C, Shi Y. Versatile and multivalent nanobodies efficiently neutralize SARS-CoV-2. Science. 2020;370:1479–1484. doi: 10.1126/science.abe4747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schoof M, Faust B, Saunders RA, Sangwan S, Rezelj V, Hoppe N, Boone M, Billesbølle CB, Zimanyi M, Deshpande I. An ultra-high affinity synthetic nanobody blocks SARS-CoV-2 infection by locking Spike into an inactive conformation. BioRxiv. 2020 doi: 10.1101/2020.08.08.238469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Custódio TF, Das H, Sheward DJ, Hanke L, Pazicky S, Pieprzyk J, Sorgenfrei M, Schroer MA, Gruzinov AY, Jeffries CM, Graewert MA, Svergun DI, Dobrev N, Remans K, Seeger MA, McInerney GM, Murrell B, Hällberg BM, Löw C. Selection, biophysical and structural analysis of synthetic nanobodies that effectively neutralize SARS-CoV-2. Nat Commun. 2020;11:5588. doi: 10.1038/s41467-020-19204-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hanke L, Perez LV, Sheward DJ, Das H, Schulte T, Moliner-Morro A, Corcoran M, Achour A, Hedestam GBK, Hällberg BM. An alpaca nanobody neutralizes SARS-CoV-2 by blocking receptor interaction. Nat Commun. 2020;11:1–9. doi: 10.1038/s41467-020-18174-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zupancic JM, Desai AA, Schardt JS, Pornnoppadol G, Makowski EK, Smith MD, Kennedy AA, de Mattos Barbosa MG, Cascalho M, Lanigan TM. Directed evolution of potent neutralizing nanobodies against SARS-CoV-2 using CDR-swapping mutagenesis. Cell Chem Biol. 2021;28:1379. doi: 10.1016/j.chembiol.2021.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rissiek B, Koch-Nolte F, Magnus T. Nanobodies as modulators of inflammation: potential applications for acute brain injury. Front Cell Neurosci. 2014;8:344. doi: 10.3389/fncel.2014.00344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sanaei M, Setayesh N, Sepehrizadeh Z, Mahdavi M, Yazdi MH. Nanobodies in human infections: prevention, detection, and treatment. Immunol Invest. 2020;49:875–896. doi: 10.1080/08820139.2019.1688828. [DOI] [PubMed] [Google Scholar]
- 58.Wrapp D, De Vlieger D, Corbett KS, Torres GM, Wang N, Van Breedam W, Roose K, van Schie L, COVID V-C, Team R Structural basis for potent neutralization of betacoronaviruses by single-domain camelid antibodies. Cell. 2020;181:1004–1015. doi: 10.1016/j.cell.2020.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nambulli S, Xiang Y, Tilston-Lunel NL, Rennick LJ, Sang Z, Klimstra WB, Reed DS, Crossland NA, Shi Y, Duprex PW. Inhalable Nanobody (PiN-21) prevents and treats SARS-CoV-2 infections in Syrian hamsters at ultra-low doses. bioRxiv. 2021;7:eabh3019. doi: 10.1126/sciadv.abh0319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Najmeddin A, Shapourabadi MB, Behdani M, Dorkoosh F. Nanobodies as powerful pulmonary targeted biotherapeutics against SARS-CoV-2, pharmaceutical point of view. Biochim Biophys Acta. 2021;18:129974. doi: 10.1016/j.bbagen.2021.129974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Stefan MA, Light YK, Schwedler JL, McIlroy PR, Courtney CM, Saada EA, Thatcher CE, Phillips AM, Bourguet FA, Mageeney CM. Development of potent and effective synthetic SARS-CoV-2 neutralizing nanobodies. bioRxiv. 2021 doi: 10.1080/19420862.2021.1958663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yao H, Cai H, Li T, Zhou B, Qin W, Lavillette D, Li D. A high-affinity RBD-targeting nanobody improves fusion partner’s potency against SARS-CoV-2. PLoS Pathog. 2021;17:e1009328. doi: 10.1371/journal.ppat.1009328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li T, Cai H, Yao H, Zhou B, Zhao Y, Qin W, Hutter CA, Lai Y, Bao J, Lan J (2020) Potent synthetic nanobodies against SARS-CoV-2 and molecular basis for neutralization. BioRxiv.
- 64.Custódio TF, Das H, Sheward DJ, Hanke L, Pazicky S, Pieprzyk J, Sorgenfrei M, Schroer MA, Gruzinov AY, Jeffries CM. Selection, biophysical and structural analysis of synthetic nanobodies that effectively neutralize SARS-CoV-2. Nat Commun. 2020;11:1–11. doi: 10.1038/s41467-020-19204-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Palomo C, Mas V, Detalle L, Depla E, Cano O, Vázquez M, Stortelers C, Melero JA. Trivalency of a nanobody specific for the human respiratory syncytial virus fusion glycoprotein drastically enhances virus neutralization and impacts escape mutant selection. Antimicrob Agents Chemother. 2016;60:6498–6509. doi: 10.1128/AAC.00842-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bracken CJ, Lim SA, Solomon P, Rettko NJ, Nguyen DP, Zha BS, Schaefer K, Byrnes JR, Zhou J, Lui I. Bi-paratopic and multivalent VH domains block ACE2 binding and neutralize SARS-CoV-2. Nat Chem Biol. 2021;17:113–121. doi: 10.1038/s41589-020-00679-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhou Z, Wang X, Fu Y, Zhang X, Liu C. Letter to the editor: Neutralizing antibodies for the treatment of COVID-19. Acta Pharmaceutica Sinica B. 2021;11:304. doi: 10.1016/j.apsb.2020.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Greaney AJ, Loes AN, Crawford KH, Starr TN, Malone KD, Chu HY, Bloom JD. Comprehensive mapping of mutations in the SARS-CoV-2 receptor-binding domain that affect recognition by polyclonal human plasma antibodies. Cell Host Microbe. 2021;29:463–476. doi: 10.1016/j.chom.2021.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wibmer CK, Ayres F, Hermanus T, Madzivhandila M, Kgagudi P, Oosthuysen B, Lambson BE, De Oliveira T, Vermeulen M, Van der Berg K. SARS-CoV-2 501Y. V2 escapes neutralization by South African COVID-19 donor plasma. Nat Med. 2021;27:622–625. doi: 10.1038/s41591-021-01285-x. [DOI] [PubMed] [Google Scholar]
- 70.Wang P, Nair MS, Liu L, Iketani S, Luo Y, Guo Y, Wang M, Yu J, Zhang B, Kwong PD. Antibody resistance of SARS-CoV-2 variants B. 1.351 and B. 1.1. 7. Nature. 2021;593:130–135. doi: 10.1038/s41586-021-03398-2. [DOI] [PubMed] [Google Scholar]
- 71.Wang Z, Schmidt F, Weisblum Y, Muecksch F, Barnes CO, Finkin S, Schaefer-Babajew D, Cipolla M, Gaebler C, Lieberman JA. mRNA vaccine-elicited antibodies to SARS-CoV-2 and circulating variants. Nature. 2021;592:616–622. doi: 10.1038/s41586-021-03324-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wu K, Werner AP, Moliva JI, Koch M, Choi A, Stewart-Jones GB, Bennett H, Boyoglu-Barnum S, Shi W, Graham BS (2021) mRNA-1273 vaccine induces neutralizing antibodies against spike mutants from global SARS-CoV-2 variants. BioRxiv.
- 73.Bannas P, Hambach J, Koch-Nolte F. Nanobodies and nanobody-based human heavy chain antibodies as antitumor therapeutics. Front Immunol. 2017;8:1603. doi: 10.3389/fimmu.2017.01603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zupancic JM, Schardt JS, Desai AA, Makowski EK, Smith MD, Pornnoppadol G, de Mattos G, Barbosa M, Cascalho M, Lanigan TM, Tessier PM. Engineered multivalent nanobodies potently and broadly neutralize SARS-CoV-2 variants. Adv Therap. 2021;4:2100099. doi: 10.1002/adtp.202100099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wu X, Cheng L, Fu M, Huang B, Zhu L, Xu S, Shi H, Zhang D, Yuan H, Nawaz W (2021) A potent bispecific nanobody protects hACE2 mice against SARS-CoV-2 infection via intranasal administration [DOI] [PMC free article] [PubMed]
- 76.Sun D, Sang Z, Kim J, Xiang Y, Cohen T, Belford AK, Huet A, Conway JF, Sun J, Taylor D (2021) Potent neutralizing nanobodies resist convergent circulating variants of SARS-CoV-2 by targeting novel and conserved epitopes. bioRxiv [DOI] [PMC free article] [PubMed]
- 77.Bain W, Yang H, Shah FA, Suber T, Drohan C, Al-Yousif N, DeSensi RS, Bensen N, Schaefer C, Rosborough BR (2021) COVID-19 versus non-COVID ARDS: comparison of demographics, physiologic parameters, inflammatory biomarkers and clinical outcomes. Ann Am Thorac Soc [DOI] [PMC free article] [PubMed]
- 78.Bektas A, Schurman SH, Franceschi C, Ferrucci L. A public health perspective of aging: do hyper-inflammatory syndromes such as COVID-19, SARS, ARDS, cytokine storm syndrome, and post-ICU syndrome accelerate short-and long-term inflammaging? Immun Ageing. 2020;17:1–10. doi: 10.1186/s12979-020-00196-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Soy M, Keser G, Atagündüz P, Tabak F, Atagündüz I, Kayhan S. Cytokine storm in COVID-19: pathogenesis and overview of anti-inflammatory agents used in treatment. Clin Rheumatol. 2020;39:2085–2094. doi: 10.1007/s10067-020-05190-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lu Q, Liu J, Zhao S, Castro MFG, Laurent-Rolle M, Dong J, Ran X, Damani-Yokota P, Tang H, Karakousi T. SARS-CoV-2 exacerbates proinflammatory responses in myeloid cells through C-type lectin receptors and Tweety family member 2. Immunity. 2021;54:1304-19 e9. doi: 10.1016/j.immuni.2021.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Shi Y, Wang Y, Shao C, Huang J, Gan J, Huang X, Bucci E, Piacentini M, Ippolito G, Melino G. COVID-19 infection: the perspectives on immune responses. New York: Nature Publishing Group; 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sadeghian-Rizi T, Behdani M, Khanahmad H, Ghasemi-Dehkordi P, Sadeghi HM, Jahanian-Najafabadi A. Production of novel camelid anti-CXCL10 specific polyclonal antibodies and evaluation of their bioreactivity. Int J Pept Res Ther. 2019;25:535–540. [Google Scholar]
- 83.Coppieters K, Dreier T, Silence K, Haard HD, Lauwereys M, Casteels P, Beirnaert E, Jonckheere H, Wiele CVD, Staelens L. Formatted anti–tumor necrosis factor α VHH proteins derived from camelids show superior potency and targeting to inflamed joints in a murine model of collagen-induced arthritis. Arthritis Rheum. 2006;54:1856–1866. doi: 10.1002/art.21827. [DOI] [PubMed] [Google Scholar]
- 84.Duarte JN, Cragnolini JJ, Swee LK, Bilate AM, Bader J, Ingram JR, Rashidfarrokhi A, Fang T, Schiepers A, Hanke L. Generation of immunity against pathogens via single-domain antibody–antigen constructs. J Immunol. 2016;197:4838–4847. doi: 10.4049/jimmunol.1600692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Danquah W, Meyer-Schwesinger C, Rissiek B, Pinto C, Serracant-Prat A, Amadi M, Iacenda D, Knop J-H, Hammel A, Bergmann P. Nanobodies that block gating of the P2X7 ion channel ameliorate inflammation. Sci Transl Med. 2016;8:366ra162-366ra162. doi: 10.1126/scitranslmed.aaf8463. [DOI] [PubMed] [Google Scholar]
- 86.Gu B, Zhang W, Bendall L, Chessell I, Buell G, Wiley J. Expression of P2X7 purinoceptors on human lymphocytes and monocytes: evidence for nonfunctional P2X7 receptors. Am J Physiol Cell Physiol. 2000;279:C1189–C1197. doi: 10.1152/ajpcell.2000.279.4.C1189. [DOI] [PubMed] [Google Scholar]
- 87.Koch-Nolte F, Eichhoff A, Pinto-Espinoza C, Schwarz N, Schäfer T, Menzel S, Haag F, Demeules M, Gondé H, Adriouch S. Novel biologics targeting the P2X7 ion channel. Curr Opin Pharmacol. 2019;47:110–118. doi: 10.1016/j.coph.2019.03.001. [DOI] [PubMed] [Google Scholar]
- 88.Burnstock G, Knight GE. The potential of P2X7 receptors as a therapeutic target, including inflammation and tumour progression. Purinergic Signal. 2018;14:1–18. doi: 10.1007/s11302-017-9593-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Stortelers C, Pinto-Espinoza C, Van Hoorick D, Koch-Nolte F. Modulating ion channel function with antibodies and nanobodies. Curr Opin Immunol. 2018;52:18–26. doi: 10.1016/j.coi.2018.02.003. [DOI] [PubMed] [Google Scholar]
- 90.Koenig P-A, Das H, Liu H, Kümmerer BM, Gohr FN, Jenster L-M, Schiffelers LDJ, Tesfamariam YM, Uchima M, Wuerth JD, Gatterdam K, Ruetalo N, Christensen MH, Fandrey CI, Normann S, Tödtmann JMP, Pritzl S, Hanke L, Boos J, Yuan M, Zhu X, Schmid-Burgk JL, Kato H, Schindler M, Wilson IA, Geyer M, Ludwig KU, Hällberg BM, Wu NC, Schmidt FI. Structure-guided multivalent nanobodies block SARS-CoV-2 infection and suppress mutational escape. Science. 2021;371:6230. doi: 10.1126/science.abe6230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Walter JD, Hutter CA, Zimmermann I, Earp J, Egloff P, Sorgenfrei M, Hürlimann LM, Gonda I, Meier G, Remm S (2020) Synthetic nanobodies targeting the SARS-CoV-2 receptor-binding domain. BioRxiv.
- 92.Li T, Cai H, Yao H, Zhou B, Zhao Y, Qin W, Hutter CAJ, Lai Y, Bao J, Lan J, Wong G, Seeger M, Lavillette D, Li D (2020) Potent synthetic nanobodies against SARS-CoV-2 and molecular basis for neutralization. bioRxiv.2020.06.09.143438
- 93.Schoof M, Faust B, Saunders RA, Sangwan S, Rezelj V, Hoppe N, Boone M, Billesbølle CB, Puchades C, Azumaya CM, Kratochvil HT, Zimanyi M, Deshpande I, Liang J, Dickinson S, Nguyen HC, Chio CM, Merz GE, Thompson MC, Diwanji D, Schaefer K, Anand AA, Dobzinski N, Zha BS, Simoneau CR, Leon K, White KM, Chio US, Gupta M, Jin M, Li F, Liu Y, Zhang K, Bulkley D, Sun M, Smith AM, Rizo AN, Moss F, Brilot AF, Pourmal S, Trenker R, Pospiech T, Gupta S, Barsi-Rhyne B, Belyy V, Barile-Hill AW, Nock S, Liu Y, Krogan NJ, Ralston CY, Swaney DL, García-Sastre A, Ott M, Vignuzzi M, Consortium QSB, Walter P, Manglik A An ultrapotent synthetic nanobody neutralizes SARS-CoV-2 by stabilizing inactive Spike. Science. 2020;370:1473–1479. doi: 10.1126/science.abe3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Huo J, Le Bas A, Ruza RR, Duyvesteyn HME, Mikolajek H, Malinauskas T, Tan TK, Rijal P, Dumoux M, Ward PN, Ren J, Zhou D, Harrison PJ, Weckener M, Clare DK, Vogirala VK, Radecke J, Moynié L, Zhao Y, Gilbert-Jaramillo J, Knight ML, Tree JA, Buttigieg KR, Coombes N, Elmore MJ, Carroll MW, Carrique L, Shah PNM, James W, Townsend AR, Stuart DI, Owens RJ, Naismith JH. Neutralizing nanobodies bind SARS-CoV-2 spike RBD and block interaction with ACE2. Nat Struct Mol Biol. 2020;27:846–854. doi: 10.1038/s41594-020-0469-6. [DOI] [PubMed] [Google Scholar]
- 95.Hanke L, Vidakovics Perez L, Sheward DJ, Das H, Schulte T, Moliner-Morro A, Corcoran M, Achour A, Karlsson Hedestam GB, Hällberg BM, Murrell B, McInerney GM. An alpaca nanobody neutralizes SARS-CoV-2 by blocking receptor interaction. Nat Commun. 2020;11:4420. doi: 10.1038/s41467-020-18174-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Xiang Y, Nambulli S, Xiao Z, Liu H, Sang Z, Duprex P, Schneidman-Duhovny D, Zhang C, Shi Y (2020) Versatile, Multivalent Nanobody Cocktails for Highly Efficient SARS-CoV-2 Neutralization. bioRxiv.2020.08.24.26433
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data used to support the findings of this study are included in the article.