SUMMARY
Viruses are intracellular parasites that subvert the functions of their host cells to accomplish their infection cycle. The endoplasmic reticulum (ER)-residing chaperone proteins are central for the achievement of different steps of the viral cycle, from entry and replication to assembly and exit. The most abundant ER chaperones are GRP78 (78-kDa glucose-regulated protein), GRP94 (94-kDa glucose-regulated protein), the carbohydrate or lectin-like chaperones calnexin (CNX) and calreticulin (CRT), the protein disulfide isomerases (PDIs), and the DNAJ chaperones. This review will focus on the pleiotropic roles of ER chaperones during viral infection. We will cover their essential role in the folding and quality control of viral proteins, notably viral glycoproteins which play a major role in host cell infection. We will also describe how viruses co-opt ER chaperones at various steps of their infectious cycle but also in order to evade immune responses and avoid apoptosis. Finally, we will discuss the different molecules targeting these chaperones and the perspectives in the development of broad-spectrum antiviral drugs.
KEYWORDS: ER chaperone, GRP78, GRP94, calnexin, calreticulin, protein disulfide isomerase, DNAJ, viral infection
INTRODUCTION
As obligate intracellular parasites, viruses have evolved to subvert the functions of their host cells to replicate. Despite divergences between the different groups of the Baltimore classification, the viral infectious cycle is classically subdivided into the following steps: entry, translation, and genome replication, assembly of progeny viruses, and exit. Entry is itself made up of a sequence of events: attachment to the host cell, penetration, cytoplasmic trafficking, and uncoating, after which the genome can be released. Along their cycle, viruses can potentially use all the cellular organelles. Among these, the endoplasmic reticulum (ER) appears critical for the achievement of different steps of the infectious cycle, from entry and replication to assembly and exit, depending on the virus family (reviewed in reference 1). The ER is in charge of the synthesis of a large fraction of cellular proteins, whether cytosolic, membrane, or secreted (reviewed in reference 2). It also ensures the folding of proteins and their posttranslational modifications, including glycosylation, disulfide bond formation, and proline isomerization. The presence of an efficient quality control system referred to as ERAD (ER-associated degradation) ensures the degradation of misfolded proteins by directing them to the proteasome. Viruses exploit the ER machineries for synthesis and maturation of viral proteins. This has been especially well studied when it comes to the glycoproteins of enveloped viruses (3). Moreover, some viruses, such as flaviviruses, coronaviruses, polioviruses, and rotaviruses, exploit the ER’s large and dynamic membrane to replicate or even assemble (1, 4). Finally, the ER may also be used for virus entry, as shown for polyomaviruses, nonenveloped viruses that hijack the ERAD for entry into the cytosol (reviewed in reference 5).
Viral infection as well as ER stress can saturate the ERAD and lead to the accumulation of unfolded and misfolded proteins in the ER lumen. Some viruses can also express viroporins, pore-forming proteins acting as ion channels on the ER membrane which alter ER calcium homeostasis (6). Cells cope with ER stress by activating the unfolded protein response (UPR). The UPR is a complex network of signaling pathways that coordinate the adaptive response to ER stress. It consists of three pathways, each engaged by activation of one specific signal transducer: inositol-requiring protein 1 (IRE1), protein kinase RNA (PKR)-like ER kinase (PERK), or activating transcription factor 6 (ATF6). These pathways work in concert to decrease the load of unfolded proteins in the ER by simultaneously decreasing protein synthesis, increasing the folding capacity of the ER, increasing ERAD, and modifying the intracellular protein trafficking and secretory pathways. Failure of the UPR leads to apoptotic cell death. The UPR can therefore affect the progression of viral replication. However, many viruses hijack the UPR to upregulate their replication by overriding the inhibition of translation and preventing apoptosis. A considerable number of publications have reported that viruses induce ER stress and at the same time modulate the UPR (7–35).
The ER functions, including protein synthesis, folding, transport, quality control, UPR, and regulation of calcium homeostasis, are tightly controlled by ER-residing chaperone proteins. The main ER chaperones are glucose-regulated chaperones GRP78 (78-kDa glucose-regulated protein), GRP94 (94-kDa glucose-regulated protein), the carbohydrate or lectin-like chaperones calnexin (CNX) and calreticulin (CRT), the protein disulfide isomerases (PDIs), and the DNAJ chaperones. They are dedicated to the co- and posttranslational folding of polypeptides. They can detect and bind unfolded and misfolded proteins and constitute the essential recognition components for the degradation machinery (36). They also have the ability to protect proteins from stress-induced denaturation, misfolding, and aggregation and to assist the UPR and can regulate apoptotic signaling pathways as well as immune responses. In addition, most ER chaperones contribute to calcium buffering in the ER lumen by binding calcium (37). Some of them can migrate to the cell surface, where they also play important roles in cell survival/apoptosis as well as in immune responses (38).
This review will focus on the role of these ER chaperones in productive viral infection. We will analyze their role in the folding and quality control of viral proteins, in various steps of the infectious cycle, and how they are exploited by viruses to avoid apoptosis of the host cell as well as immune responses. The sometimes-overlapping activities of ER chaperones, their pleiotropic roles, and the complexity of their network make them difficult cellular components to target therapeutically. However, there are some interesting results that have been achieved so far in the field of broad-spectrum antiviral strategies which we will discuss in the present review.
GLUCOSE-REGULATED PROTEINS GRP78 AND GRP94 IN VIRAL INFECTION
GRP78 (also called BiP [binding immunoglobulin protein] or HSPA5) and GRP94 (also called Gp96 or HSPC4) are the most abundant ER chaperones.
GRP78
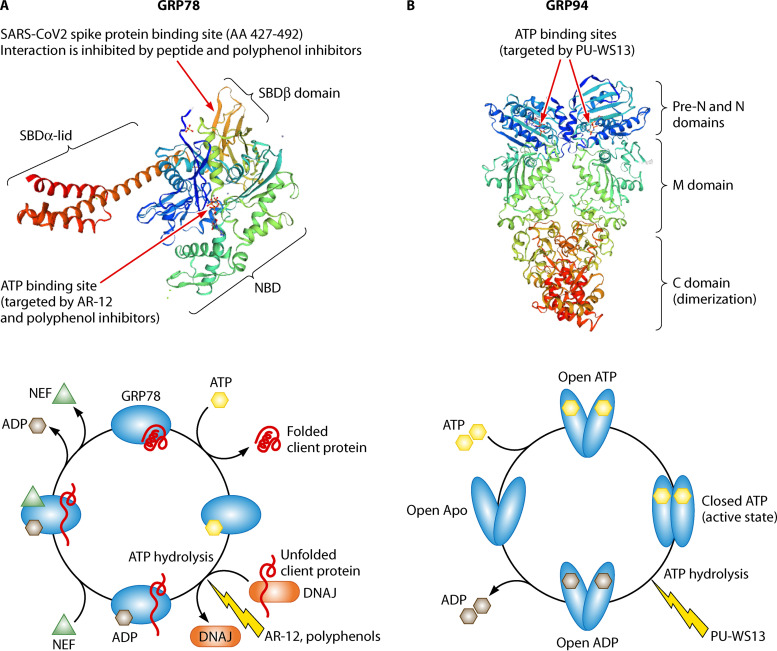
GRP78 is considered the master regulator of ER functions. This member of the HSP70 family is comprised of an N-terminal nucleotide binding domain (NBD) endowed with ATPase activity and a C-terminal substrate binding domain (SBD) bridged by a linker. In mammalian cells, GRP78 is assisted by HSP40/DNAJ cochaperones for binding of the protein to be chaperoned (often called client protein) and hydrolysis of ATP to ADP and by nucleotide exchange factors (NEFs) and GRP170 for the release of ADP and the client (39) (Fig. 1A). GRP78 binds nascent proteins as they transit to the lumen of the ER, ensuring de novo polypeptide folding, their subsequent assembly, and transport. GRP78 also protects proteins from stress-induced misfolding and aggregation. In addition, GRP78 can target misfolded proteins for degradation by guiding them to the retrotranslocation machinery at the ER membrane, where they are ubiquitinated before being delivered to the proteasome (reviewed in reference 40). Finally, GRP78 is also a calcium binding protein involved in maintaining calcium homeostasis (37). This chaperone is considered a major regulator of the UPR, acting as an ER stress sensor. Under normal conditions, it binds to the three transmembrane UPR transducers, IRE1α, PERK, and ATF6, keeping them in an inactive conformation. Upon ER stress, GRP78’s higher affinity for exposed hydrophobic residues on unfolded proteins causes it to release the effectors that can therefore initiate UPR. At the same time, because of its antiapoptotic properties, GRP78 can block ER stress-induced cell death. Indeed, GRP78 can bind and repress some effectors of the apoptotic signaling pathway, such as caspase-7 and -12 and BIK (Bcl-2-interacting killer), a BH3-only protein of the bcl-2 family localized on the outer membrane of the ER (41–43). Furthermore, under stress conditions, GRP78 can be exposed at the cell surface (38), where it assists membrane proteins in transducing signal.
FIG 1.
Glucose-regulated ER chaperone structures. (A) GRP78. Upper panel, structure in the ATP-bound state (SMTL ID 5e84). NBD, nucleotide binding domain; SBD, substrate binding domain. Arrows indicate the two sites which have been targeted by inhibitors, i.e., the ATP binding site for ATPase inhibitors (ATP mimicry) and the site of interaction (SBDβ) with the glycoprotein S of SARS-CoV-2 (205). Lower panel, GRP78 folding cycle. Involvement of cochaperones (DNAJ, NEF) and inhibitors are shown. (B) GRP94. Upper panel, structure in the ATP-bound state (homodimer, SMTL ID 5uls.1). PU-WS13 targets the ATP binding site (109). Lower panel, GRP94 cycle. Exactly where in the cycle the client proteins are bound and released is unclear.
GRP78 is by far the most studied ER chaperone for its role in the progression of viral infectious cycles and the modulation of cell survival and of immune responses. In addition, an increased expression of GRP78 (reflecting ER stress) has often been reported in cells infected either in vitro or in vivo with DNA or RNA viruses or in cells transfected with a wide range of viral proteins (7).
Role in the virus life cycle.
(i) Virus entry.
Although GRP78 is mainly expressed in the ER, it can also be found on the cell surface. GRP78 levels at the cell surface are usually low but upregulated under ER stress conditions, including when caused by virus infection (38). An increase in cell surface GRP78 has also been observed in uninfected bystander cells in the proximity of neurons infected with dengue virus (DENV) (44). This suggests intercellular communication of ER stress. Viruses use specific cell surface receptors to attach to target cells, and this first interaction may be followed by a second one before penetration into the cell. Since GRP78 can interact with a number of viral envelope proteins, it can act as a coreceptor for virus entry (Fig. 2). The identification of GRP78 as a coreceptor was first described for coxsackie A9 virus (CVA9) by Triantafilou et al. in 2002 (45). They showed that CVA9, which utilizes integrin αvβ3 as a receptor, requires GRP78 as a coreceptor molecule, whereas major histocompatibility complex class I (MHC-I) molecules associated with GRP78 serve as the internalization pathway of this virus. They further showed that the αvβ3 receptors along with GRP78 and MHC-I accessory molecules are present within lipid rafts at the plasma membrane (46). Borna disease virus (BDV), a highly neurotropic virus, may also use GRP78 as a coreceptor (47). GRP78 can interact with domain III of envelope E proteins of Japanese encephalitis virus (JEV), DENV, and Zika virus (ZIKV) (48–51), the E protein playing a key role in receptor binding during virus entry. Wu et al. (52) showed that JEV could use GRP78 present in lipid rafts for efficient cell surface attachment. GRP78 was found to be expressed at the plasma membrane of Neuro2a (a murine primary neuronal cell line) and Huh-7 (a human epithelial cell line) cells. In these, the presence of an antagonist antibody targeting the N-terminal domain of GRP78 (amino acids [aa] 1 to 100) or depletion of GRP78 significantly inhibited JEV entry (49), supporting GRP78’s role in virus uptake. JEV uses GRP78 as a coreceptor for entry, while heparan sulfate proteoglycans (HSPGs) and glycosaminoglycans (GAGs) serve as attachment factors (53). Finally, GRP78 was reported to comigrate with JEV particles in sucrose density gradients, and those mature viruses that do not cofractionate with GRP78 showed a significant decrease in infectivity (52). However, a more recent study did not confirm these results (49). This discrepancy could be explained by differences in the methodologies of the two studies.
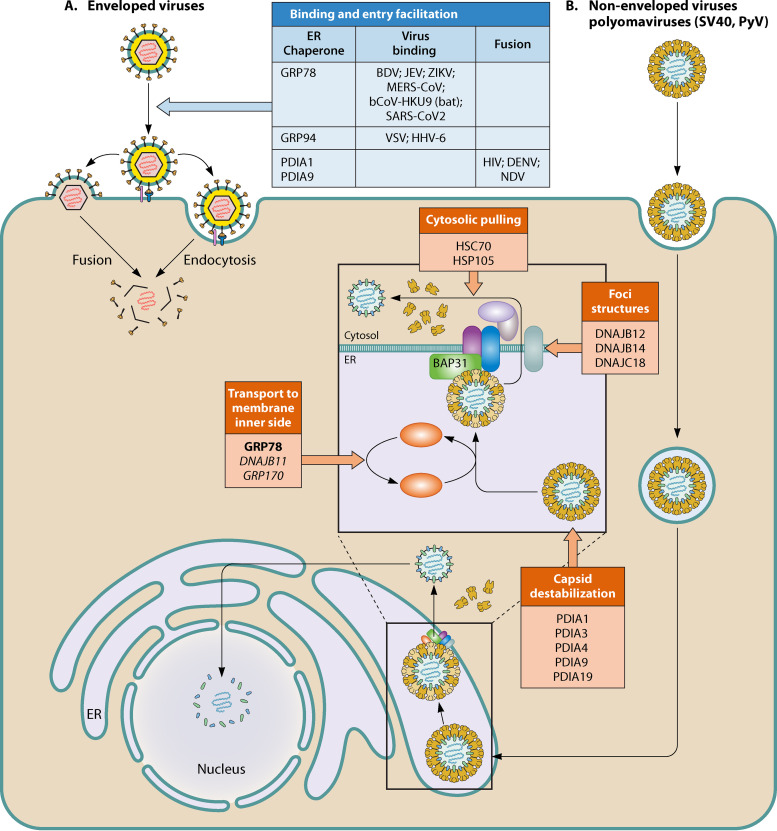
FIG 2.
Summary of the role of ER chaperones in viral entry of enveloped (A) and nonenveloped (B) viruses. (A) Enveloped viruses. ER chaperones involved either in viral attachment as coreceptors or through specific client proteins or in fusion. (B) Nonenveloped viruses, for example, polyomaviruses. PDIs, GRP78, and DNAJ ER chaperones work in conjunction with cytosolic chaperones to allow cytosolic entry of polyomaviruses after trafficking to the ER following receptor-mediated endocytosis. Inside the ER, PDIs first destabilize the capsid, exposing the hydrophobic VP2 and VP3 proteins. This allows GRP78 binding and transport to the ER membrane, where the capsid binds to the B cell receptor-associated protein 31 (BAP31) and is inserted. Various transmembrane DNAJs contribute to forming foci at the membrane, where the inserted capsids are extracted into the cytosol by HSC70 and HSP105.
Recently, Khongwichit et al. reported that the N-terminal domain of GRP78 also plays an important role in ZIKV internalization (50). As for DENV, GRP78 was first reported to act as an entry coreceptor in HepG2 (human liver) cells (54). However, another study in HEK 293T cells concluded that GRP78 had no role in DENV virus entry, although it was important during viral antigen production (44).
Besides CVA9, BDV, JEV, and ZIKV, several betacoronaviruses, RNA viruses that attach and enter cells via their envelope spike protein (S), have been shown to use GRP78 as a coreceptor. Chu et al. (55) identified membrane-associated GRP78 as an additional binding target of the Middle East respiratory syndrome coronavirus (MERS-CoV) spike, the main cell receptor being dipeptidyl peptidase 4 (DPP4). They showed that GRP78 could not render nonpermissive cells susceptible to MERS-CoV infection on its own but could facilitate MERS-CoV entry into permissive cells by increasing virus attachment. Moreover, GRP78 could also interact with the spike protein of a bat coronavirus, HKU9 (bCoV-HKU9), and facilitate its attachment to the host cell surface. Angiotensin-converting enzyme 2 (ACE2) is the main receptor for the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) responsible for the COVID-19 pandemic. However, Ibrahim et al. (56) hypothesized that the spike protein S of SARS-CoV-2 could also bind to GRP78, as observed for MERS-CoV. Using in silico models, they proposed that residues 480 to 488 (located in the receptor binding domain) of the spike protein could interact with the substrate binding domain (SBD) of GRP78. They raised the possibility that GRP78 could be a facilitator for SARS-CoV-2 viral entry. Using several in vitro and ex vivo models, Aguiar et al. (57) demonstrated the presence of GRP78 in the respiratory mucosa. They suggested that alternative receptors for SARS-CoV-2 could exist and facilitate initial host cell infection. The different viruses for which GRP78 facilitates entry are summarized in Fig. 2.
To summarize, these different studies show a role for membrane-bound GRP78 as a coreceptor to facilitate viral entry after attachment. The GRP78 domains involved have been reported for SARS-CoV-2 (SBD), JEV, and ZIKV (N-terminal domain [aa 1 to 100]). Whether inhibitors of the ATPase activity of GRP78 could inhibit these interactions deserves to be explored. In any case, monoclonal antibodies targeting the regions involved may represent relevant approaches to inhibit GRP78 as a host cell coreceptor for these viruses.
(ii) Viral protein folding, virus assembly, and production.
GRP78 has been shown to colocalize and interact with many viral proteins from both DNA and RNA viruses, mainly glycoproteins that are essential for the formation of the viral envelopes (Fig. 3). Examples include hemagglutinin (HA) (58, 59) and neuraminidase (NA) (60) of influenza virus, F protein of respiratory syncytial virus (RSV) (61), L protein of hepatitis B virus (HBV) (62), and E envelope proteins of Chikungunya virus (CHIKV) (8) and of several flaviviruses, such as DENV (48, 63), JEV (52), ZIKV (50, 51), and hepatitis C virus (HCV) (64, 65). The role of GRP78 in the folding of viral envelope glycoproteins has been studied using influenza virus HA as a model. The HA transmembrane glycoprotein is synthetized as an 84-kDa precursor (HA0) in the ER and then undergoes rapid initial folding and trimerization before being transported to the Golgi complex. Hurtley et al. (58) reported that both spontaneously misfolded HA0 (about 5 to 10% of newly synthetized molecules in their model) and aberrant HA0 molecules synthesized in the presence of tunicamycin, a drug that inhibits N-linked glycosylation, rapidly self-aggregated and associated with GRP78 in a permanent noncovalent way. The complexes were retained in the ER and slowly degraded. HA folding seemed to be independent of GRP78 but dependent on the lectin-like chaperones CNX/CRT, suggesting that GRP78 is more likely involved in misfolded protein degradation than in folding of nascent HA. Molinari and Helenius (59) proposed that glycoproteins such as HA that have glycans close to the NH2 terminus enter the CNX/CRT pathway directly without prior binding to GRP78, whereas glycoproteins like the Semliki Forest virus E1 and vesicular stomatitis virus (VSV) glycoproteins, which have C-terminal glycans, first associate with GRP78. In a similar manner, GRP78 and CNX/CRT were shown to interact with the E1 and E2 envelope glycoproteins of HCV. E1 and E2 proteins form a heterodimer before leaving the ER compartment. During their folding process, a significant amount of these proteins form aggregates that considerably decrease the yield of heterodimer assembly. GRP78 has been shown to interact preferentially with misfolded aggregates, leading to nonproductive folding, whereas CNX was involved in proper folding of E1-E2 (64). In fact, it is now known that HSP70 family proteins, including GRP78, are recruited to protein aggregates by cochaperones of the DNAJ family, which recognize sequences predicted to have high aggregation potential (66–68).
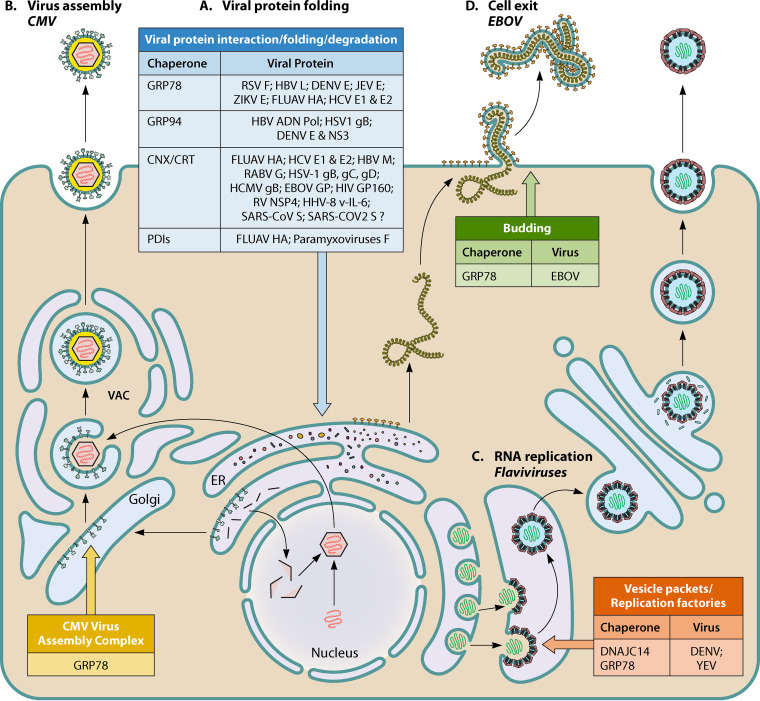
FIG 3.
Summary of the roles of ER chaperones in viral protein folding, virus assembly, RNA replication, and cell exit. (A) Viral protein folding. ER chaperones reported to interact with, fold, or degrade the listed viral proteins. FLUAV, influenza A virus; RABV, rabies virus; RV, rotavirus. (B) Virus assembly. GRP78 contributes to the structure and function of the viral assembly compartment (VAC) of CMV. (C) RNA replication. ER chaperones involved in replication factories of flaviviruses. (D) Cell exit. GRP78 is involved in EBOV cell exit.
The importance of GRP78 in the production of viral particles was demonstrated for flaviviruses, RNA viruses that specifically replicate and assemble at the ER membrane. Using GRP78 knockout (KO) cells infected with DENV, Limjindaporn et al. (48) observed a 90% decrease in the production of infectious viral particles. In the same way, Wati et al. (44) showed that inactivation of GRP78 by cleaving it with the bacterial subtilase cytotoxin (SubAB) decreased DENV release by 10- to 100-fold without affecting viral RNA levels. As for ZIKV, GRP78 knockdown by a small interfering RNA (siRNA) induced a significant decrease in viral titer and in viral genome copies (50, 51). Royle et al. (51) reported that siRNA-mediated depletion of GRP78 decreased ZIKV protein synthesis and viral titers and altered the localization of viral replication factories (Fig. 3). It has been suggested that GRP78 may be involved in JEV assembly as GRP78 knockdown impaired virus production and GRP78 colocalized in round perinuclear structures along with E envelope proteins (52). Interestingly, the E protein has been shown to play an important role in the formation of flavivirus assembly units (69). However, the precise role of GRP78 and of other potential ER chaperones in this step of the cycle remains unknown.
Finally, an indirect role of GRP78 in flavivirus production has been proposed. Jitobaom et al. (63) reported an interaction between GRP78, the host cell’s voltage-dependent anion channel (VDAC), and the viral E protein of DENV. DENV infection induced the translocation of VDAC from the outer membrane of mitochondria toward the ER. Silencing of VDAC decreased viral protein production, demonstrating the importance of this relocalization. Similar results were obtained for JEV in insect cells (70). GRP78 has also been involved in rotavirus morphogenesis, another virus assembling in the ER (71). Finally, a specific role for GRP78 in virus assembly has been reported for human cytomegalovirus (HCMV), a DNA virus from the Herpesviridae family whose virion assembly does not occur in the ER but in a cytoplasmic compartment located in the perinuclear area (Fig. 3). Buchkovich et al. (72) showed that the immediate early protein IE1-72 of cytomegalovirus upregulates GRP78 gene transcription and protein levels. In infected cells, GRP78 was found localized in the viral assembly compartment, where it interacts with the HCMV virion tegument proteins pp28 and TRS1, which play a role in viral assembly (73, 74). Depletion of GRP78 by SubAB or short hairpin RNA (shRNA) blocked virion assembly and disorganized the assembly compartment (73), suggesting that GRP78 contributes to the structure and function of this compartment.
(iii) Progeny virion release.
The last step in a successful viral life cycle is the release of progeny virions. Few data are available concerning the role of GRP78 in this stage. Reid et al. (75) used epigallocatechin gallate (EGCG) to inhibit the GRP78 ATPase activity and show that GRP78 is essential for Ebola virus (EBOV) infection (Fig. 3). Furthermore, in vitro and in vivo gene targeting of GRP78 impaired viral replication and protected animals in a lethal infection model. Researchers used cells transfected with a GRP78 targeting siRNA to show that cellular release of VP40 (an EBOV protein required for viral particle release) virus-like particles required a functional GRP78. The mechanism was not determined in this study; however, GRP78 had been previously shown to interact with VP40 (76).
In summary, even though many studies have reported the interaction and colocalization of GRP78 with several viral proteins as well as a decrease in virus production upon GRP78 depletion, few have deciphered the precise mechanisms involved. Little is especially known on how the chaperone contributes to the assembly of viruses that replicate and assemble in the ER. The contribution of GRP78, in particular when membrane bound or secreted by ER stress, to viral egress is also unknown.
Role in cell survival/apoptosis.
In the context of cancer, GRP78 is a well-known prosurvival protein which prevents apoptosis and induces cell proliferation, including during ER stress (77). However, Mukherjee et al. (18) proposed that upregulation of GRP78 in JEV-infected neural/stem progenitor cells was essential in mediating ER stress-induced apoptosis. In their study, knockdown of GRP78 using an siRNA decreased both viral load and caspase-3 activation. On the other hand, BHK-21 cells with persistent JEV infection resist virus-induced apoptosis through constant upregulation of GRP78 expression, resulting in the complete depletion of the C/EBP homologous protein (CHOP). That could explain in part the development of persistent JEV infection in mammalian cells (78). In the case of HBV, Shu et al. (79) recently demonstrated that GRP78 was upregulated by HBV in hepatocytes, and this inhibited HBV transcription and replication and played a crucial role in the survival of hepatocytes by maintaining mild ER stress. They suggested that HBV may sacrifice part of its replication in order to establish a persistent infection through induction of GRP78 and concluded that targeting GRP78 may be an interesting therapeutic strategy against chronic HBV infection and the associated hepatocellular carcinoma. Upregulation of GRP78 by HBV may result from the induction of ER stress or from direct activation of GRP78’s promoter by HBV proteins (80).
In the case of human immunodeficiency virus 1 (HIV-1), Lopez et al. (81) reported differences in cell survival or apoptosis of U87-MG astrocytoma cells depending on whether the cells were infected by the B or C strains. The authors concluded that this could explain differences in neuropathogenesis induced by the two clades (82). They showed that the HIV-1 envelope glycoprotein gp120 from clade B induced proliferation of astrocytoma cells that relied on the expression of GRP78. gp120 from HIV-1 clade C, on the other hand, induced proapoptotic markers. GRP78 seemed to be a key player in HIV-1 clade B and C neuropathogenic discrepancies, in accordance with the fact that GRP78 was reported to be upregulated in HIV‐positive cortical tissue from autopsied neurocognitively impaired individuals (83).
Role in immune responses.
GRP78 has been reported to have either a pro- or antiviral effect, depending on its role in immune responses, as summarized in Table 1. Inhibition of cell surface expression of MHC-I molecules by HCMV is a well-known mechanism of cellular escape from recognition by CD8+ cytotoxic T lymphocytes (CTLs). This is due to the degradation by the proteasome of the heavy chain of MHC-I during the early phase of infection, mediated by the HCMV US2 and US11 gene products. GRP78 was reported to contribute to this process by binding to MHC-I in a US2-dependent manner, consistent with its role in aberrant ER protein elimination (84, 85). On the other hand, GRP78 has been shown to have an antiviral effect via the innate immune response, especially the interferon (IFN) type I response. Ma et al. (86) showed that GRP78 was one of the most significantly upregulated proteins induced by HBV replication in HepAD38 and HepG2 cells as well as in HBV-infected human liver biopsy specimens, whereas its amount decreased in the liver of chronic hepatitis B patients after effective anti-HBV treatment. Knockdown of GRP78 induced an increase in HBV virions in HepG2 cells, whereas its overexpression led to HBV suppression that correlated with the activation of the IFN-β1-mediated 2′,5′-oligoadenylate synthetase and RNase L signaling pathway. In the case of HCV, it has been reported that GRP78 contributes to the innate immune response in hepatocytes through TLR3 and IRF3 (interferon regulatory factor 3) (87). The authors showed that TLR3-dependent induction of IFN-stimulated genes and chemokines was compromised by depletion of GRP78. Moreover, expression of GRP78 correlated with that of RANTES (regulated upon activation, normal T-cell expressed and secreted) and CXCL10, two inflammatory chemokines frequently elevated in HCV-infected livers. Of note, as with HBV, GRP78 was significantly upregulated in livers from patients with chronic hepatitis C. Finally, Richardson et al. (88) reported that GRP78 interacts with and stabilizes IFI6 (IFN alpha [IFN-α]-inducible protein 6), an antiviral protein which prophylactically protects uninfected cells by preventing the formation of single-membrane invaginations of the ER caused by some flaviviruses, including yellow fever virus (YFV), West Nile virus (WNV), DENV, and ZIKV. This was not the case with HCV, which replicates in double-membrane vesicles that protrude outwards from the ER. They proposed that GRP78 facilitates the proper folding and/or localization of IFI6 at the ER membrane and thus regulates the IFN response to these flaviviruses (88). Thus, their findings suggest a model in which the IFN response is armed with membrane-targeted effectors that block the establishment of the ER microenvironment required for viral replication. So far, IFI6 is the only such effector reported to be stabilized by GRP78.
TABLE 1.
Role of ER chaperones in immune responsesa
| Chaperone | Antiviral immune response | Immune escape |
|---|---|---|
| GRP78 | IFN response: 2′5′-oligoadenylate synthetase and RNase L activation (HBV) (86); TLR3-mediated IRF-3 activation (stabilizes p-IRF3 in endolysosomes) (HCV) (87); IFI-6 stabilization (prevents the formation of replication factories) (DENV, YFV, ZIKV, WNV) (88) | Contributes to MHC class I heavy chain degradation mediated by US2 protein (HCMV) (GRP78-CNX-CRT-US2 complex) (84, 85) |
| GRP94 | APC peptide cross-presentation: extracellular GRP94 proposed as an adjuvant for vaccination | Potential TGF-β activation by chaperoning GARP and integrins; suggested role in immunosuppression induced by E2 protein (membrane GRP94, HCV) (105, 120, 122, 123) |
| CNX | Contributes to MHC class I heavy chain degradation mediated by US2 protein (HCMV) (see above) (84) and E5 protein (HPV) (148); contributes to CD1d degradation mediated by E5 protein (HPV) (149) | |
| CRT | Immunogenic cell death (membrane CRT, many viruses) (151) | Contributes to MHC class I heavy chain degradation mediated by US2 (HCMV) (84); decreases IFN-α production and activity (HBV) (150) |
| PDIs | Proposed to improve HA vaccine immunogenicity in mice (PDIA3, FLUAV) (177) | Contribute to MHC class I heavy chain degradation mediated by US2 (HCMV) as a component of the signal peptide peptidase-mediated ERAD (PDIA1, PDIA3) (174); inhibit TNF-α production by monocytes (PDIA3, DENV) (175) |
| DNAJC3 (P58IPK) | Regulates PKR, leading to a decrease in eIF2a phosphorylation and IRF3-mediated IFN type I and III production (FLUAV [189]; coronaviruses? [191]) |
FLUAV, influenza A virus.
GRP94
GRP94 belongs to the HSP90 family. Like the other HSP90s, GRP94 contains three major domains, the N-terminal domain (N), which is linked to the middle domain (M) via a charged linker, and the C-terminal (C) domain. The N domain is the site of ATP binding, the M domain is involved in ATP hydrolysis, and the C domain is required for homodimerization (Fig. 1B). In GRP94, the N domain is preceded by a pre-N domain that is not conserved in the other HSP90s and has been identified as essential for client protein maturation and ATPase activity regulation (89). The M domain is a calcium binding domain. GRP94 is a chaperone involved in folding and/or assembly of membrane and secreted proteins, many of them being involved in immune responses. It also contributes to calcium buffering in the ER and to targeting misfolded proteins to ERAD (90).
Role in the virus life cycle.
(i) Virus entry.
GRP94 has been shown to be involved in cell entry for two viruses, vesicular stomatitis virus (VSV) and human herpesvirus 6 (HHV-6) (Fig. 2). VSV causes vesicular stomatitis in domestic animals. Although the disease is usually mild in humans, VSV has gained interest because of significant biomedical potential. Indeed, it is being developed into an oncolytic agent for cancer therapy. Bloor et al. (91) showed that GRP94 was essential for infection with VSV, as viruses bearing the envelope protein G are unable to enter GRP94-deficient cells. They showed that the ATPase activity of GRP94 is necessary, suggesting that GRP94 is required because it chaperones a cellular protein(s) essential for viral entry. Finkelshtein et al. (92) further reported that these cellular proteins are members of the low-density lipoprotein (LDL) receptor family (93) that serve as cellular receptors for VSV. As an integrin chaperone, GRP94 is expected to be necessary for integrin-mediated cell attachment or entry of several enveloped viruses (e.g., HIV, WNV, JEV, ZIKV, HSV, or probably SARS-CoV2). In addition, naked viruses (e.g., HAV, HEV, adenoviruses) have been shown to be packaged into extracellular vesicles by infected cells at a prelytic stage. These vesicles, expressing integrins and/or integrin ligands on their surface, may facilitate integrin-mediated entry in target cells (94). Concerning HHV-6, a herpesvirus belonging to the Betaherpesvirus subfamily, the cellular receptors on T cells are CD46 and CD134 for HHV-6A and HHV-6B, respectively. Prusty et al. (95) first reported that GRP94 interacts with HHV-6A virions and contributes to HHV-6A attachment to target cells during infection of the HSB-2 cell line. They also found that HHV-6A infection promotes GRP94 expression at the cell surface. Ma et al. (96) found that the viral glycoprotein gQ1 from both HHV-6A and HHV-6B interacts with GRP94 and that depletion of GRP94, competition with soluble GRP94, or its blockade with an antibody inhibited HHV-6 infection. They also found that cell surface expression of GRP94 was induced as early as 5 min postinfection. How GRP94 functions together with CD46 or CD134 during cell entry remains unclear.
GRP94 could also act early in the viral entry process by regulating cellular proteases responsible for the cleavage of viral envelope glycoproteins involved in cell entry. Indeed, whereas binding to the host cell receptor/coreceptor is an essential first step in establishing infection, the proteolytic activation step is often critical for viral fusion with the host cell membranes (97). This is notably the case for coronaviruses, whose envelope spike protein S needs to be activated by proteases (98). The S proteins of HKU1, MERS-CoV, and SARS-CoV-2, but not SARS-CoV, contain a furin-like cleavage site, and cleavage by furin at the S1/S2 site is essential for SARS-CoV-2 entry into human lung cells (99–102). Cell surface TMPRSS and lysosomal cathepsins have also been shown to be involved in further cleavage of the S protein of SARS-CoV-2 (101, 103). Furin preactivation allows SARS-CoV-2 to be less dependent on target cells. GRP94 has been shown to interact with and to regulate furin in HEK293F cells, and inhibition of GRP94 with an siRNA suppresses intracellular trafficking of furin as well as its proteolytic activity on the zymogen of the metalloproteinase ADAMTS9 at the cell membrane (104). We recently found that GRP94 directly interacts with cathepsin L (105). These reported data suggest that GRP94 activity could potentially impact viral entry by modulating furin and cathepsins.
In conclusion, GRP94 acts as a coreceptor interacting with HHV-6 glycoprotein gQ1 to facilitate entry of HHV-6 after attachment, but, conversely to GRP78, the region of GRP94 involved remains unknown. On the other hand, GRP94 has been shown to chaperone cellular receptors (LDLR). In this case, the ATPase activity is necessary and thus could be targeted by specific inhibitors (see below). While its involvement as a chaperone of integrins has not been reported, it should be noted that integrins are receptors of several enveloped viruses and may play a role in the entry of naked viruses enclosed in extracellular vesicles. Lastly, GRP94, through its association with the proteases cathepsin L and furin, might be involved in the cleavage of envelope proteins of several highly pathogenic viruses. This, however, has yet to be reported. All together, these findings strengthen the interest of GRP94 as a broad-spectrum antiviral target.
(ii) Viral protein folding and virus production.
GRP94’s role in viral protein folding has been shown for the HBV polymerase by Kim et al. (106). They showed that GRP94 was associated with HBV polymerase and was necessary for the latter’s production in an active conformation in the human liver HepG2 cell line. As reported for GRP78 (59, 64), GRP94 plays a role in protein quality control by eliminating misfolded viral proteins that may compromise viral replication. Indeed, Ramakrishnan et al. (107) reported that GRP94 forms a stable association with mutated glycoprotein B of herpes simplex virus 1 (HSV-1) but not with the fully processed viral protein. Rothan et al. identified GRP94 as essential in the DENV and ZIKV infectious cycles (108). They showed that GRP94 knockout inhibited DENV2 replication in Huh-7 cells by decreasing viral protein (envelope and NS3) synthesis and final virus production. Moreover, PU-WS13, a specific inhibitor of GPR94 (109), decreased DENV2 and ZIKV viral titers. These antiviral activities were observed for multiple DENV serotypes and ZIKV strains. The authors propose that GRP94 could be involved in protein folding and/or delivering misfolded proteins to the ERAD, thus avoiding accumulation of misfolded proteins that could interfere with viral replication. In support of this hypothesis, they showed that GRP94 was a critical component of the HrD1 ubiquitin ligase complex that has an essential role in flavivirus replication, although the mechanism is not currently understood. The different viruses for which GRP94 facilitates viral protein folding are summarized in Fig. 3.
Role in immune responses.
GRP94 is a master chaperone for immune responses, and, like GRP78, it has a dual role. It increases inflammatory and immune responses as it acts as a molecular chaperone for most TLRs, particularly TLR7, and for most integrins (110, 111). Also, secreted GRP94 initiates inflammatory conditions in the absence of other activators by activating NF-κB and the NLRP3 inflammasome in antigen-presenting cells (APCs) (112). GRP94 also favors cross-presentation of antigenic peptides by APCs (113). In addition, our team has shown that extracellular GRP94 can interact with C3, the key protein of the complement system (114). We have also recently shown that GRP94 was present at the cell surface of M2 but not M1 macrophages. In these cells, functional GRP94 was secreted under ER stress conditions, and this correlated with a switch to a proinflammatory profile which was dependent on activation of the UPR through IRE1α. Under these conditions, GRP94 is cosecreted with C3 and may facilitate its cleavage by cathepsin L. We also showed that GRP94 interacts directly with cathepsin L (105). Finally, GRP94 controls regulatory T cells by chaperoning GARP (glycoprotein A repetitions predominant), the docking receptor for transforming growth factor beta (TGF-β) (115) and the integrins αvβ6 and β8 involved in its release.
Although GRP94 has been involved in these numerous immune responses, when it comes to viral infections, it has mostly been studied for its adjuvant properties. This is a consequence of its ability to bind intracellular viral peptides and promote their cross-presentation by MHC class I molecules in professional APCs. This in turn activates peptide-specific CTL responses (Table 1). CD91, a major endocytosis receptor, has been proposed to be essential for GRP94 binding and endocytosis by APCs (113). However, it was later demonstrated that CD91 was dispensable and that fluid-phase, rather than receptor-mediated, mechanisms predominate during GRP94 peptide internalization and entry into the cross-presentation pathway (116). In addition, this report established heparin sulfate proteoglycans as cell surface GRP94 binding sites. The use of GRP94 as an adjuvant has been proposed for different viral vaccines. For example, Ju et al. showed that GRP94 elicited a cross-protective CD8+ T cell response against the conserved internal viral nucleoprotein NP (117). This cross-protection was proven effective against challenges with a heterologous influenza virus in mice. It has also been reported that immunization of transgenic HBV-replicating mice with a combined formulation of HBV surface (HBs) and core (HBc) antigens along with GRP94 induced robust antiviral T cells and antibodies as well as a decrease in regulatory T cells (Tregs) (118). This correlated with a decrease in serum HBs Ag and HBV DNA levels, suggesting a therapeutic effect. A significant enhancement in cellular responses against HBc Ag and in infiltration of CD8+ T cells in transgenic mouse livers was observed with GRP94 treatment. Selinger et al. also proposed the use of GRP94 as an adjuvant for vaccination against HIV (119).
GRP94’s immunosuppressive role has not been reported in viral infections. Nevertheless, although its own role has not been deciphered, cell surface migration of GRP94 has been associated with increased TGF-β production in a hepatocyte cell line following transfection with the hepatitis C E2 envelope protein (120). The authors of the study showed that this resulted from the degradation by E2 of AIMP1/p43 (amino-acyl-tRNA synthetase-interacting multifunctional protein 1/p43), which is a protein that retains GRP94 in the ER (121) and negatively regulates TGF-β signaling (122). More specifically, E2 inhibited the interaction between GRP78 and AIMP1/p43 in which GRP78 stabilizes AIMP1/43. Of note, in the same study, GRP94 migration to the cell surface was also observed when E2 protein was expressed in a B lymphocyte cell line. Moreover, Kwon et al. (123) analyzed the interactions of HCV E2 with human monocyte-derived macrophages and showed that it induced a polarization of macrophages toward an M2-like anti-inflammatory phenotype, which is consistent with the increase in TGF-β production by hepatocytes. Although they did not study GRP94 specifically, we can hypothesize that E2 also induces AIMP1 degradation and GRP94 migration to the cell membrane in macrophages, consistent with our findings in which we showed that membrane GRP94 is a signature of M2 macrophages (105).
The potential effects of GRP94 in immune escape are summarized in Table 1.
OTHER ER CHAPERONES
The Carbohydrate or Lectin-Like Chaperones Calnexin and Calreticulin
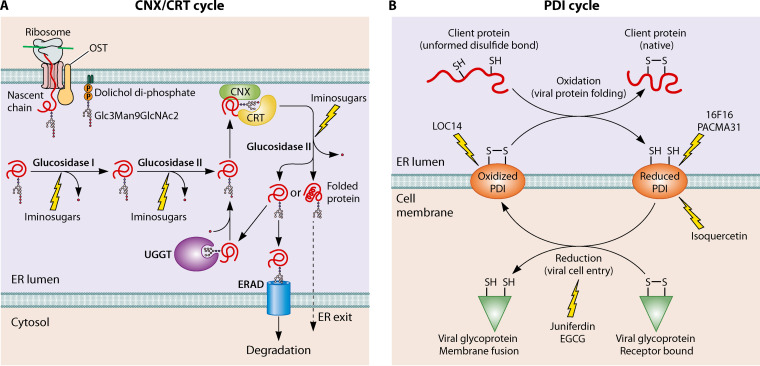
The transmembrane protein calnexin (CNX) and its soluble paralog, calreticulin (CRT), are chaperones specifically located in the ER to ensure the proper folding of glycoproteins (reviewed in reference 124) (Fig. 4A). N-linked glycoproteins are first cotranslationally modified at the luminal face of the ER through the addition of one or more precursor glycans. Interaction of the polypeptide chain with CNX/CRT takes place after the sequential cleavage of the two terminal glucose residues by glucosidases I and II. If a protein is still misfolded, it is reglucosylated and rebinds to CNX/CRT. This is referred to as the CNX/CRT folding cycle. Proteins that fail to emerge from the CNX/CRT folding cycle are targeted for ERAD. CNX/CRT also play a major role in calcium buffering in the ER (37).
FIG 4.
CNX/CRT and PDI cycles. (A) CNX/CRT cycle. N-linked glycoproteins are cotranslationally modified at the luminal face of the ER through the addition of one or more precursor glycans in the form of Glc3Man9GlcNAc2 by the oligosaccharyl transferase complex (OST). Interaction of the nascent polypeptide chain with CNX/CRT takes place after sequential cleavage of the two terminal glucose residues by glucosidases I and II. Folded glycoproteins are transported to the Golgi compartment following release of the third and last terminal glucose by glucosidase II. If a protein is misfolded, it is reglucosylated by UDP-glucose:glycoprotein glucosyltransferase (UGGT) to allow its rebinding to CNX/CRT. This is referred to as the CNX/CRT folding cycle. Proteins that fail to emerge from the CNX/CRT folding cycle in a folded state are targeted for ERAD. The inhibitors so far described of the CNX/CRT cycle target glucosidases I and II. (B) PDI cycle. Oxidized PDIs catalyze disulfide bond formation of nascent peptides in the ER (upper part). At the cell membrane, PDIs act primarily as reductants of viral glycoproteins and induce conformational changes allowing fusion (lower part). Arrows indicate the sites which have been targeted by inhibitors.
Role in the virus life cycle.
Glycoproteins are one of the major components of human-pathogenic viruses. They are key actors of enveloped virus infectivity, as they allow virus attachment to host cell receptors, fusion of the viral envelope with the cell membrane, and viral release. Glycoproteins also play a role in immune evasion either as secreted decoys or because they mimic host glycans (glycan shield) (125). CNX and CRT, which have similar oligosaccharide preferences (126), are essential in the folding of all viral glycoproteins as long as they have an N-linked glycan. These include envelope proteins of several viruses, such as influenza virus (HA and NA) (59, 127–129), HBV (M protein) (130), rabies virus (G protein) (131), HSV-1 (gB, gC, and gD) (132), HCMV (gB) (133), Hantaan virus (Gn, Gc) (134), EBOV (GP) (135), and HIV-1 (gp160) (136, 137). This is also true for nonenveloped virus glycoproteins such as the nonstructural protein NSP4 of rotaviruses, which is synthesized as an ER transmembrane protein and plays an important role in morphogenesis and pathogenesis (138), and the viral interleukin 6 produced by human herpesvirus 8 (v-IL-6) (139) (Fig. 3). The binding of CNX to glycoproteins has been analyzed by Hammond et al. (127) in a study using influenza A virus (FLUAV) HA as a model. They showed the high affinity of CNX for proteins with monoglucosylated oligosaccharides. Tatu et al. (128) identified several intermediates in the conformational maturation process of HA. CNX can associate with the incompletely oxidized, monomeric forms of HA. Once fully oxidized, these intermediates dissociate from CNX and can assemble into mature homotrimers. The cotranslational maturation process of FLUAV NA has been deciphered by Wang et al. (129). They showed that NA formed intramolecular disulfide bonds prematurely and aggregated when CNX and CRT were absent. Sequential binding with, first, CNX and then CRT is required for efficient and accurate maturation. Inhibition of the interaction between CNX and the M protein of HBV blocked the secretion of viral envelopes of HBV (130). A role for CNX along with CRT and GRP78 in the folding and assembly of HCV glycoprotein E1-E2 heterodimer complexes has been reported (64, 140). CRT and GRP78 interact preferentially with aggregates, whereas CNX associates with monomeric forms of HCV glycoproteins or noncovalent complexes (140). As with the knockdown of GRP78, CNX or CRT knockdown by siRNAs decreased the production of infectious dengue virions by about 50% (48). The authors furthermore show that the interaction of these three chaperones with the DENV E protein plays an important role in virion production. Concerning HIV-1, it has been suggested that it may be stabilized by binding of CNX to both the glycan and the peptide regions of gp160, as the α-glucosidase inhibitor castanospermine failed to inhibit the interaction and to affect the infectivity of virions. Furthermore, Jennelle et al. reported an increased risk of developing atherosclerosis in HIV patients (141). They showed that this involved CNX and the HIV Nef (negative regulatory factor) accessory protein. The interaction between the two disrupted the interaction of CNX with the cholesterol efflux pump ABCA1 (ATP-binding cassette transporter A1) but increased CNX’s affinity for HIV-1 gp160. 1-[(7-oxo-7H-benz[de]anthracene-3-yl)amino]anthraquinone was discovered to block the Nef-CNX interaction, partially restoring ABCA1 activity in HIV-infected cells and reducing foam cell formation in a culture of HIV-infected macrophages (142). CNX was demonstrated to also control the maturation of the spike protein S of SARS-CoV, and in the presence of α-glucosidase inhibitors, the produced virions showed significantly lower infectivity than those produced in the absence of those compounds (143). Williams and Goddard-Borger (144) suggested that the S protein of SARS-CoV-2 is also likely folded by CNX and proposed alpha-glucosidase inhibitors as antivirals for evaluation. Finally, Zhao et al., using a CRISPR screening of a porcine single guide RNA (sgRNA) library, identified CRT as a major host factor for JEV replication (145).
Role in immune responses.
CNX and CRT have been shown to influence antigen presentation to CD8 T cells by controlling the assembly of MHC class I molecules and their cell surface expression (reviewed in references 146 and 147). Several viruses target the interaction between CNX and MHC class I molecules to escape CD8-mediated immune responses (Table 1). For example, Gruener et al. (148) showed that the E5 protein of human papillomaviruses forms a ternary complex with CNX and the heavy chain of HLA-I and causes retention of HLA-I molecules in the ER. Of note, E5 also targets CD1d to the cytosolic proteolytic pathway by inhibiting CNX-mediated CD1d folding and trafficking, resulting in the abrogation of CD1d-mediated production of interleukin-12 that may help HPV-infected cells evade immunological surveillance (149). Oresic and Tortorella (84) showed that CMV US2-mediated destruction of class I heavy chains implicated CNX and CRT besides GRP78, showing that the same chaperones involved in folding/maturation of MHC class I molecules are also quality control sensors that target misfolded proteins for degradation.
Concerning CRT, HBV upregulates its expression, resulting in increased viral replication. Indeed, CRT suppressed the production of endogenous IFN-α by reducing the nuclear translocation of IFN regulatory factor 7 (150). Furthermore, it also suppressed the antiviral activity of IFN-α by inhibiting the phosphorylation of STAT1 and decreasing the expression of two IFN-α downstream effectors, protein kinase R and 2′,5′-oligoadenylate synthetase (150).
Finally, CRT can be exposed at the cell surface during ER stress, especially on preapoptotic cells, where it acts as an eat-me signal favoring immunogenic phagocytosis. CRT translocation to the plasma membrane depends on ER stress, especially PERK and the phosphorylation of eIF2α, and on the apoptotic process involving caspase-8 activation, BAP31 cleavage, and Bak/Bax activation. Viruses have developed strategies (inhibition of eIF2α kinases, inhibition of the caspase-8 pathway) that result in inhibition of CRT exposure, thereby avoiding immunogenic cell death (reviewed in reference 151). Cell surface CRT has been considered a key determinant and a biomarker of immunogenic cell death and is used to assess the efficacy of oncolytic viruses (152). The effects of CNX and CRT in antiviral immune responses are summarized in Table 1.
Protein Disulfide Isomerases
The protein disulfide isomerases (PDIs) form a family of redox enzymes that also have a chaperone activity. They have thioredoxin-like (TRX) domains which assist in the oxidative folding and disulfide bond rearrangement of peptides. Oxidized PDIs catalyze disulfide bond formation of nascent peptides in the ER (Fig. 4B). The reoxidation of PDI is due primarily to the endoplasmic reticulum oxireductin 1 (ERO1), which uses flavin adenine dinucleotide (FAD) as a cofactor to transfer electrons on molecular oxygen, thereby generating hydrogen peroxide (67).
There are over 20 members in the PDI family, distinguished by the number and organization of their TRX domains (153).
Role in the virus life cycle.
(i) Virus entry.
In addition to their abundance in the ER, PDIs can also be found at the plasma membrane. In contrast to their predominantly oxidizing activity in the ER, surface-associated PDIs act primarily as reductants (154). PDIs have been much studied for their role in cell entry of enveloped viruses, in particular the membrane fusion step (Fig. 2). Virus-cell membrane fusion is the result of an active virus-cell interaction that induces conformational changes within the envelope. For some viruses, such as influenza virus, these conformational changes are induced by exposure to an acidic milieu during the early steps of infection. For other viruses, they are mediated at neutral pH by cell surface-associated oxidoreductases which reduce disulfide bridges in viral envelope glycoproteins (155). For example, HIV-1 entry requires attachment of the gp120 subunit of the viral envelope to its primary receptor, CD4. This interaction induces a structural rearrangement in gp120 that allows binding to the coreceptor, either CCR5 or CXCR4. Binding of gp120 to the coreceptor results in conformational changes in gp41 that allow insertion of the fusion peptide into the cell membrane and then fusion. PDIA1 antibodies as well as the chemical inhibitors of PDIs, DTNB [5,5′-dithiobis(2-nitrobenzoic acid] and bacitracin, have been shown to inhibit HIV cell entry by preventing the conformational changes in gp120 and gp41, thus demonstrating the importance of thiol/disulfide exchanges during the first steps of HIV infection (156–159). In fact, three different oxidoreductases have been involved, PDIA1, thioredoxin-1 (Trx1), and glutaredoxin-1 (Grx1) (160, 161). Ou and Silver (161), using an siRNA against PDIA1, showed that thioredoxin had a greater effect on HIV-1 envelope-mediated fusion than PDIA1 and was able to reduce disulfide bonds in gp120 and increase disulfide bond reduction in CD4 in the presence of HIV-1 gp120. PDIA1 as well as PDIA19 are also required for membrane fusion directed by protein F of Newcastle disease virus, an avian paramyxovirus that may occasionally infect professionals in contact with infected birds (162). Inhibitors of PDIs also inhibit the fusion function of the CHIKV E1 protein (163). Another mechanism has been described in the case of DENV. Cell surface PDIA1 was shown to colocalize with and activate β1 and β3 integrins (164), and its blockade inhibited DENV entry into endothelial cells. Finally, PDIs have also been shown to be involved in cell entry of nonenveloped viruses such as polyomaviruses, rotaviruses, and human astroviruses. Polyomaviruses enter target cells by receptor-mediated endocytosis and then traffic to the ER, where they are uncoated before the genome is translocated into the nucleus. It was shown that PDIA9 extrudes the viral VP1 C-terminal extremity to initiate ER membrane penetration of the murine polyomavirus (PyV), used as a model to study polyomavirus cell entry (165). The same team further demonstrated that the PDI family members PDIA1, PDIA3, and PDIA4 collaborate with PDIA9, forming a network that facilitates polyomavirus infection (166). In fact, this network also includes GRP78 and DNAJ chaperones (see below), which play an essential role in the last step of polyomavirus entry in the cytosol (Fig. 2). PDIs have also been shown to be essential for rotavirus cell entry (165) and to be required for astrovirus type 8 uncoating during cell entry (PDIA4) (167).
(ii) Viral protein folding and virus production.
The most studied PDI, PDIA3, has been shown to be required for efficient folding of some viral proteins such as FLUAV HA (168) and the F protein of paramyxoviruses (169) (Fig. 3). Roberson et al. showed that influenza virus increased PDIA3 levels in primary lung epithelial cells and that PDIA3 knockout decreased virus production and cell apoptosis (170). The increase and importance of PDIA3 in influenza infection have been recently confirmed in vivo by Chamberlain et al. (171). Furthermore, the deletion of PDIA3 in lung epithelial cells in mice induced a decrease in viral production as well as in inflammatory cytokine levels in the bronchoalveolar lavage fluid. PDIA1 and PDIA4 may also be involved (172). In the case of paramyxoviruses, the inhibition of PDIA3 by tizoxanide, a metabolite of the antiprotozoal/antimicrobial nitazoxanide, induced the formation of F protein aggregates and inhibited its trafficking to the plasma membrane (169).
Role in immune responses.
As with calreticulin, PDIA1 and PDIA3 associate with the TAP-tapasin-MHC class I complex to form the peptide loading complex necessary for antigen presentation through MHC class I (173). In the case of HCMV infection, PDIA1 is involved in the dissociation of MHC class I from the US2 protein, which leads to MHC class I dislocation to the cytosol before degradation by the proteasome (Table 1). Knockdown of PDIA1 by an siRNA was shown to inhibit the degradation of MHC class I molecules induced by the US2 protein (174).
Mishra et al. (175) reported that increased levels of PDIA3 expression in DENV-infected THP1 cells favor viral replication by suppressing TNF-α production (Table 1). Of note, antibodies against DENV nonstructural protein 1 (NS1) cross-react with PDIA1 on the surface of platelets, causing inhibition of platelet aggregation, which may contribute to the pathogenesis of dengue disease (176).
Finally, the capacity of PDI to stabilize proteins could be exploited to enhance their immunogenicity. This is how Wu et al. (177) improved immunization against H3N2 and H1N1 in mice. In their study, they showed that immunizing mice with H3 HA from 293T cells overexpressing PDIA3 was more effective in protecting the mice than H3 produced in conventional or PDIA3-depleted 293T cells (Table 1).
DNAJ Chaperones
DNAJ proteins, also called HSP40, are characterized by the presence of a conserved 70-amino-acid domain called the J-domain (related to the Escherichia coli DNAJ chaperone). They have been categorized into three subfamilies (A, B, and C). Some of these chaperones are localized in the ER, either at the membrane or in the lumen (178). One ER luminal member, DNAJC3 (also named P58IPK), is of particular interest in viral infections due to its specific role in host cell defense (179). Indeed, P58IPK acts as a repressor of protein kinase R (PKR), a kinase activated by double-stranded RNA (dsRNA) which phosphorylates eiF2α, resulting in protein translation blockage (180). P58IPK also transiently inhibits PERK, the latter being usually activated during ER stress to protect cells (181). Moreover, ER stress activates P58IPK gene transcription.
Role in the virus life cycle.
(i) Virus entry.
The role of DNAJs in cell entry has been studied in detail for simian virus 40 (SV40), another polyomavirus also used as a model (reviewed in references 5 and 182). These studies demonstrated the corecruitment of HSP40/DNAJ chaperones (Fig. 2). The exposition of hydrophobic regions of VP2 and VP3 in the ER lumen results from interaction of the SV40 capsid with PDIs (PDIA1, PDIA3, and PDIA19). GRP78 then associates with the hydrophobic capsids, maintaining these in a soluble state until their integration in the ER membrane. Then, SV40 reorganizes the ER membrane to form penetration sites (called foci) where the virus enters the cytosol. This process involves several ER proteins, including DNAJ chaperones B12, B14, and C18. DNAJB12, DNAJB14, and DNAJC18 chaperones then recruit a cytosolic chaperone complex, including HSC70 and HSP105, that extracts SV40 from the membrane to bring the virus into the cytosol (Fig. 2). This process involving several PDIs and several DNAJ chaperones represents an interesting example of the coordination of ER and cytosolic chaperones in viral entry of a nonenveloped virus, a key step of the viral life cycle. On the basis that most of these chaperones are members of the ERAD machinery and that, during ERAD, misfolded ER proteins are recognized and ejected into the cytosol, it has been proposed that SV40 and PyV exploit ERAD during their life cycle (5). Moreover, the authors suggested that papillomaviruses, which also traffic from the cell surface to the ER, might also use the ERAD machinery to access the cytosol.
(ii) Viral replication.
Flavivirus infection induces ER membrane structures termed vesicle packets (VPs), which house the viral replication complex (RC) and form replication factories (Fig. 3). DNAJ chaperones have been involved in protein folding and RNA replication of DENV and YFV. Concerning DENV, it has been shown that the ER chaperone DNAJB11 participates in the virus replication complexes and associates with viral RNA (178). Yi et al. showed, using the YFV model, that DNAJC14 is recruited to and interacts with viral NS proteins within the ER membrane, in detergent-resistant domains, via its transmembrane or membrane binding domain. Clustering of these domains facilitates protein interactions and RC formation (183, 184). Furthermore, DNAJC14 was shown to regulate the cleavage of the single polyprotein synthesized on ER membranes at the NS3/4A site (180). This is important to ensure virus replication, NS4A being possibly involved in the induction of the membrane curvature required for the formation of the VPs (185).
Role in apoptosis.
Cardiomyocyte apoptosis is a specific feature of coxsackievirus B3 (CVB3)-induced myocarditis. Zhang et al. (186) showed that CVB3 infection activates the three UPR pathways and induces ER stress and caspase-12 (187). They further reported, using a P58IPK siRNA and a CVB3‐infected cell line expressing a dominant negative ATF6a, that P58IPK suppresses CVB3-induced apoptosis through selective activation of the PI3K/Akt pathway that requires activation of ATF6a (188).
Role in immune responses.
An important strategy of the host to block viral replication is to inhibit translation of viral mRNAs by activating the eIF2α transcription factor. Two kinases, PKR and PERK, are involved in eIF2α phosphorylation, PKR being activated by dsRNA and PERK by ER stress. Both are regulated by P58IPK. Another crucial host pathway which is activated by PKR is IRF3-mediated IFN Ι and III production. P58IPK was originally identified in FLUAV-infected cells (180). Goodman et al. (189), using P58IPK KO murine fibroblasts and cells lacking PKR or PERK, reported that P58IPK positively regulates FLUAV protein synthesis and infection via a PKR-mediated mechanism (Table 1). They further showed in vivo (190) that P58IPK also functions as a regulator of the host innate immune response, as infection of P58IPK KO mice with FLUAV resulted in increased lung pathology, immune cell apoptosis, PKR activation, and mortality. This demonstrates the complexity of the co-optation of host cell proteins by viruses. Indeed, P58IPK benefits both the host through the innate immune response and the virus through increased viral protein synthesis. Interestingly, coronaviruses which inhibit IFNs have been shown to inhibit the PKR-IRF3 pathway (191); however, their impact on P58IPK has not yet been reported.
INHIBITORS OF ER CHAPERONES AS ANTIVIRAL THERAPEUTIC AGENTS
Drugs targeting ER chaperones have been studied with the aim to propose broad-spectrum and resistance-refractory antiviral molecules, especially against severe viral diseases for which no therapy or vaccine is yet available. Most are also studied as anticancer agents. All of the ER chaperones described in this review except DNAJs have inhibitors that have been studied for their antiviral potential, as summarized in Table 2.
TABLE 2.
ER chaperone inhibitors
| ER chaperone targeted | Inhibitor | Mechanism of action | Targeted virus | Development stage |
|---|---|---|---|---|
| GRP78 | AR-12 (OSU-03012) (celecoxib derivative kinase inhibitor) | Competitive inhibition of ATPase; GRP78 decrease; other mechanisms GRP78 independent? | Lassa, Marburg, and Nipah viruses, EBOV, coxsackievirus B4, adenovirus 5, HAV, HBV, HCV, CHIKV, mumps, rubella, HCMV, Junin, HIV, SARS-CoV-2 | In vitro (195, 196, 201, 203) |
| ZIKV, DENV | In vivo (197, 198) | |||
| EGCG (polyphenol) | Competitive inhibition of ATPase; inhibition of the GRP78-spike protein S interaction | SARS-CoV2 | In silico (205) | |
| EBOV | In vivo (75) | |||
| Other polyphenols: homoeriodictyol, isorhamnetin, curcumin | Competitive inhibition of ATPase; inhibition of the GRP78-spike protein interaction | SARS-CoV-2 | In silico (205) | |
| Peptides | Inhibition of the GRP78-spike protein interaction | SARS-CoV-2 | In silico (205) | |
| GRP94 | PU-WS13 | Competitive inhibition of ATPase | DENV, ZIKV | In vitro (108) |
| CNX/CRT (ER α-glucosidases) | 1-Deoxynojirimycin (DNJ) derivatives (monocyclic): UV-4, IHVR-19029, IHVR-17028 | Competitive inhibition | Flaviviruses | In vitro, in vivo, clinical trials (210, 214) |
| Influenza virus | In vivo (210) | |||
| EBOV and Marburg virus | In vivo (211, 214) | |||
| SARS-CoV, CoV NL63, SARS-CoV-2 | In vitro (216, 217) | |||
| Miglitol, miglustat | SARS-CoV-2 | Approved in other diseases, proposed for SARS-CoV-2 (144) | ||
| Indolizidine derivatives (bicyclic): celgosivir/castanospermine) | Competitive inhibition | SARS-CoV-2 | In vitro (217) | |
| ZIKV | In vitro (213) | |||
| HCV, DENV, HIV | Clinical trials (209) | |||
| PDIs | Juniferdin | Reductase activity inhibition | FLUAV, FLUBV | In vitro (172) |
| PACMA 31 | Covalent binding with active cysteines | FLUAV, FLUBV | In vitro (172) | |
| 16F16 | Covalent binding with active cysteines | FLUAV, FLUBV, human astroviruses 1 and 8 | In vitro (172); in vitro (167) | |
| Isoquercetin | Constrains PDI conformation | FLUAV, FLUBV | In vitro (172) | |
| EGCG | Reductase activity inhibition | FLUAV, FLUBV | In vitro (172) | |
| Bacitracin | Competitive enzymatic activity inhibition; nonspecific effects ? | DENV Rotavirus |
In vitro (221) In vitro (165) |
|
| Origamicin | HCV | In vitro (222) | ||
| Compound 147 | Covalent binding; polypharmacological effects beyond PDI targeting | DENV, ZIKV | In vitro (224) | |
| PDIA1, PDIA3 | LOC14 | Oxidizes PDI | FLUAV | In vitro (171) |
| PDIA3 | Tizoxanide | Noncompetitive enzymatic activity inhibition | Paramyxovirus, FLUAV, FLUVB | In vitro (169, 172) |
GRP78 Inhibitors
GRP78 inhibitors are numerous and include natural products, synthetic molecules, specific peptides, and monoclonal antibodies (reviewed in reference 192). Two of them, AR-12 (OSU-03012) and epigallocatechin-3-O-gallate (EGCG), have been evaluated in vitro or in preclinical models for their antiviral potential. Both molecules inhibit the ATPase activity of GRP78 (193) (Fig. 1A). AR-12 has also been reported to decrease GRP78 expression by inducing its degradation (194). However, these molecules have multiple effects, including effects on other chaperones (195), making it difficult to link their antiviral activity to GRP78 specifically. AR-12 is a celecoxib derivative kinase inhibitor that does not inhibit cyclooxygenase activity. It has been evaluated in lymphoma in a phase I clinical trial (ClinicalTrials registration no. NCT00978523). AR-12 inhibits replication of several viruses requiring cellular kinases in vitro (Lassa, Ebola, Marburg, and Nipah viruses [196]). It also inhibits replication of DENV and ZIKV, two flaviviruses that are known to require GRP78 and activate the PI3K/AKT pathway (197–199). AR-12 has also been shown to protect mice in murine models of DENV (198) and ZIKV infection (197). A decrease in AKT activity was reported in both cases, whereas a decrease in GRP78 activity was observed only with DENV (198). Yang et al. (200) developed two AR-12 derivatives, P12-23 and P12-24, which showed a 10-fold improvement in efficacy and selectivity indices compared to AR-12, against three flaviviruses, DENV, JEV, and ZIKV (50% inhibitory concentrations [IC50] for AR-12, P12-23, and P12-24: 660, 70, and 52 nM for DENV, respectively; 510, 53, and 56 nM for JEV; 1,373, 130, and 119 nM for ZIKV). However, the mechanism of action proposed seemed to be independent of GRP78. Booth et al. (195, 201) showed that AR-12 associated with sildenafil, a phosphodiesterase 5 inhibitor, decreased the expression of the coxsackie and adenovirus receptor (CAR) and of several other receptors, namely, NPC1 and TIM1 (Ebola, Marburg, and hepatitis A virus), LAMP1 (Lassa fever virus), NTCP1 (hepatitis B virus), and CD81 (HCV). Moreover AR-12 combined with sildenafil reduced infectivity and replication of serotype 5 adenovirus and coxsackie B4 virus. This inhibition was abolished by overexpression of GRP78. The production of CHIKV, HCMV, HIV, EBOV, and influenza, mumps, rubella, and Junin viruses was also reduced. However, Park et al. did not observe any antiviral activity against FLUAV (202). Rayner et al. demonstrated that AR-12 suppresses the ability of SARS-CoV-2 to produce the viral spike protein S and to generate infectious virions in cell culture (203). Other potential GRP78 inhibitors have been proposed as anti-SARS-CoV-2 agents through an in silico screening approach: phytoestrogens and estrogens (204) as well as polyphenols (EGCG, homoeriodictyol, isorhamnetin, and curcumin) and peptides (205). The five peptides and the polyphenols selected by Allam et al. (205) could interfere with the interaction of SARS-CoV-2 with its target cells by blocking the interaction of the GRP78 cellular receptor (substrate binding domain) with the S protein. In addition, the polyphenols also interact with the ATPase domain of GRP78 (Fig. 1A). More recently, using a humanized anti-GRP78 monoclonal antibody (hMAb159) selected for its safe clinical profile in preclinical models to treat lung epithelial cells, researchers have demonstrated a decrease in cell surface GRP78 and ACE2 expression, as well as decreased spike-driven SARS-CoV-2 viral entry and infection in vitro (206). As a strategy against EBOV, Reid et al. (75) used EGCG as well as a phosphorodiamidate morpholino oligomer targeting GRP78 (an antisense DNA nucleotide analog) in vitro and in a murine model. Although siRNAs of GRP78 have been used in vitro as a tool to decipher its role in infection as previously mentioned, they have not been further explored for a therapeutic use.
GRP94 Inhibitors
Among GRP94 inhibitors developed as potential anticancer agents, PU-WS13 is a purine-based one that occupies the ATP binding site, blocking GRP94’s ATPase activity (Fig. 1B). This compound was reported to decrease DENV2 and ZIKV viral titers as well as the expression of DENV2 Env and NS3 proteins in Huh7 cells (108). The EC50 (half-maximal effective concentrations) for both viruses were about 25 nm. The antiviral activity of PU-WS13 was confirmed for the four DENV serotypes and for different strains of ZIKV as well as in different cell lines. Virus replication inhibition was demonstrated to occur at postentry steps of the viral life cycle (108).
Inhibitors of the CNX/CRT Cycle
Iminosugars are glucomimetics possessing a nitrogen in place of the endocyclic oxygen atom and act as ER α-glucosidase inhibitors. They compete with endogenous substrates for the enzymes α-glucosidases I and II that are responsible for controlling entry into the CNX/CRT cycle described in Fig. 4A and subsequent folding of glycoproteins. Given the importance of glycoproteins in virus pathogenesis, inhibition of the glycoprotein folding cycle by using glucosidase I and II inhibitors constitutes a promising approach in the development of broad-spectrum antiviral molecules (207). Interestingly, Sadat et al. (208) reported that cells from patients genetically deficient in the gene encoding ER α-glucosidase I are naturally resistant to multiple enveloped viruses. Two families of α-glucosidase inhibitors, 1-deoxynojirimycin (DNJ)-derived (monocyclic) and indolizidine compounds (bicyclic, celgosivir/castanospermine, the active form), have been reported to be active in vitro against many enveloped viruses, including DENV, JEV, and ZIKV (IC50 range, 40 nM to 20 μM, depending on the virus, the compound, and the cell type), EBOV and Marburg virus (IC50 range, 6 to 33 and 8 to 48 μM for EBOV and Marburg virus, respectively, depending on the compound), and influenza virus. These compounds have also been shown to be active in vivo in animal models of DENV, JEV, EBOV, influenza virus, and Marburg virus (reviewed in reference 209; see also references 210 and 211). UV-4 iminosugar [N-(9-methoxynonyl)-1-deoxynojirimycin] has been described to present a low risk for selection of drug-resistant influenza A (H1N1 and H3N2 subtypes) and B mutant viruses in vitro and in vivo (212). The iminosugars deoxynojirimycin, castanospermine, and celgosivir tested against ZIKV in vitro were able to reduce viral RNA and infectious virus titers in Vero and CHME3 cells with no effect on cellular apoptosis and antiviral responses (213). To increase their efficacy against EBOV and YFV infections, Ma et al. (214) proposed to use them in association with other antiviral molecules. A combination of the ER α-glucosidase inhibitor IHVR-19029 with favipiravir improved the antiviral efficacy against EBOV and YFV in vitro and in vivo in a murine model of EBOV infection, whereas previous results had shown inconsistent results (211). As key glycosylated host receptors could be also affected, Miller et al. (215) investigated whether iminosugar treatment of primary macrophages with N-butyl-1-DNJ (NB-DNJ) and N-(9-methoxynonyl)-1-DNJ (MON-DNJ) affected the expression and functionality of 11 host receptors, including attachment/entry and immune response receptors. They showed that treatment did not affect expression of the receptors examined on its own but reversed the DENV-mediated downregulation of IFN-γ receptor expression and signaling. Zhao et al. (216) demonstrated that the iminosugar IHVR-17028 altered the N-linked glycan structure of ACE2 but not its expression on the cell surface or its binding to the SARS-CoV spike glycoprotein. However, it impaired viral entry via a post-receptor-binding mechanism. Finally, Clarke et al. (217) showed that celgosivir prevented SARS-CoV-2-induced cell death and reduced viral replication and S protein levels in a dose-dependent manner in Vero E6 cells. Castanospermine was also able to inhibit SARS-CoV-2. Moreover, three ER glucosidase inhibitors, miglitol (N-hydroxyethyl-1-deoxynojirimycin), miglustat (N-butyl-deoxynojirimycin [NB-DNJ]), and celgosivir were suggested as potential therapeutic drugs against SARS-CoV-2 infection by Williams and Goddard-Borger (144). Of interest, miglitol and miglustat are already approved drugs that are used in long-term treatments of type 2 diabetes and type 1 Gaucher’s disease, respectively. Concerning viral infections, three candidate iminosugars have advanced to clinical trials to evaluate their safety and efficacy against viral infections: celgosivir (HCV, DENV, and HIV), NB-DNJ (HIV), and UV-4B (healthy subjects). Results, although not yet conclusive, are so far encouraging and indicate good tolerance (209).
PDI Inhibitors
Inhibition of the PDI cycle is yet another antiviral strategy targeting host ER chaperones (Fig. 4B). Juniferdin, PACMA 31, 16F16, isoquercetin, and epigallocatechin gallate (EGCG) were identified as PDI inhibitors and demonstrated to inhibit the replication of influenza A and B viruses in cells in the low micromolar range (172). The same study reported that siRNAs targeting PDIA1, PDIA3, and PDIA4 also inhibited the replication of influenza A and B viruses in vitro. 16F16 was also recently reported to inhibit human astrovirus types 1 and 8 (167). Also, LOC14, a reversible small molecule inhibitor of PDIA1 and PDIA3, had an efficient effect in decreasing viral burden and inflammation in primary tracheal epithelial cells infected with FLUAV. Moreover, the deficiency in disulfide bond formation after LOC14 treatment resulted in reduced mature HA levels. This suggests a possible inhibition of HA maturation in host cells (171). The folding of paramyxovirus protein F depends on ER-resident PDIA3. Tizoxanide, a metabolite of nitazoxanide, is a noncompetitive PDIA3 inhibitor that has been shown to inhibit paramyxovirus protein F folding and virus replication at the submicromolar range (169). Nitazoxanide is already used as an antiprotozoal/antimicrobial drug and has been shown to have antiviral activity against HBV, HCV, rotavirus, and influenza virus (218–220). The involvement of PDIA1 in HIV-1 and NDV has been described above. However, in the case of HIV-1, as other oxidoreductases are also involved, especially thioredoxin, specific PDI inhibition is not sufficient to block HIV entry (161). Rawarak et al. (221) reported that PDI inhibition by bacitracin, a mixture of bacterial peptides with PDI-inhibiting effects, decreased DENV virus production and the expression of viral proteins E and NS1 in the four DENV serotypes in a DENV-ADE in vitro model. This effect was observed in the early stages of infection. Another small molecule derived from abscisic acid, origamicin, was shown to inhibit HCV replication (222). Using a microarray mRNA profiling, the authors propose that besides the inhibition of disulfide bond formation in E1 and E2 glycoproteins, this molecule perturbates ER homeostasis through PDI inhibition, thus disrupting the portion of the HCV viral cycle which occurs in the ER. However, the exact mechanism must be elucidated. Finally, very recently, a small molecule regulator of ER proteostasis which binds to PDIs in an irreversible manner [compound 147, N-(2-hydroxy-5-methylphenyl)-3-phenylpropanamide] (223) has been reported to have a remarkable, although not very specific, antiviral effect on DENV (IC50, ∼1 μM) and ZIKV infection without inducing cell toxicity (224).
CONCLUDING REMARKS AND FUTURE DIRECTIONS
ER chaperones are essential to control the production of glycosylated proteins in the cell, including those necessary for the different stages of a viral infection and those playing a role in immune escape. They are also involved in various major functions at different steps of the infectious cycle. GRP78 and GRP94 facilitate the entry of many viruses, either on their own or through specific client proteins which are viral receptors or coreceptors. In the first case, it is not known if the ATPase activity of these chaperones is required. Further research is necessary to determine the role of GRP94 in protease activation of viral envelope proteins, a step that is required for cell entry of highly pathogenic viruses. GRP78, the most studied ER chaperone in viral infection, has also been shown to be co-opted at other steps of the life cycle. Furthermore, it is considered a crucial host factor in the biology of flaviviruses, which impact millions of people each year. However, much remains to be learned about the precise mechanisms involved. GRP94 is a master chaperone in immune responses, but although clinical work is relatively abundant when it comes to exploiting extracellular GRP94 in vaccine preparations, little is known about its role in antiviral immune responses, particularly in immunosuppression. Regarding other ER chaperones, CNX/CRT play a major role in viral infection as they are fundamental for viral glycoprotein folding. PDIs are involved in conformational changes allowing the fusion of enveloped viruses and in the entry of nonenveloped viruses, where they work together with DNAJ chaperones. Among the latter, DNAJC14 plays a singular role in the RNA replication of some flaviviruses. Another DNAJ chaperone, P58IPK (DNAJC3), impacts viral protein translation and IRF3 signaling by regulating PKR. ER chaperones and their interplay with other chaperones in viral immune escape, particularly through inhibition of type I and III IFNs, deserve to be studied in depth.
Different inhibitors of these chaperones already exist; most were initially identified through their anticancerous effect and have demonstrated their potential role as antiviral agents. Although some of them have already been evaluated as antiviral agents in clinical phases or in advanced preclinical studies, work is still needed in order to bring those molecules to the clinic. Monoclonal antibodies inhibiting membrane chaperones represent an interesting strategy to inhibit cell entry of viruses using these as a coreceptor. Also, inhibitors of DNAJ could be developed, particularly in order to inhibit RNA replication of a broad range of flaviviruses. Targeting ER chaperones is definitively a new avenue to consider in broad-spectrum antiviral therapy.
ACKOWLEDGMENTS
We thank the French Government grant managed by the French National Research Agency programs AAPG2021 SMART-PROGRESS and the Investissements d’Avenir with reference no. ANR-11-LABX-0021 (LabEX LipSTIC) and ISITE-BFC (contract no. ANR-15-IDEX-0003). We also thank the Conseil Régional de Bourgogne-Franche-Comté and the European Union program FEDER for their financial support.
Contributor Information
Evelyne Kohli, Email: evelyne.kohli@u-bourgogne.fr.
Carmen Garrido, Email: carmen.garrido@u-bourgogne.fr.
REFERENCES
- 1.Ravindran MS, Bagchi P, Cunningham CN, Tsai B. 2016. Opportunistic intruders: how viruses orchestrate ER functions to infect cells. Nat Rev Microbiol 14:407–420. 10.1038/nrmicro.2016.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Reid DW, Nicchitta CV. 2015. Diversity and selectivity in mRNA translation on the endoplasmic reticulum. Nat Rev Mol Cell Biol 16:221–231. 10.1038/nrm3958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Braakman I, van Anken E. 2000. Folding of viral envelope glycoproteins in the endoplasmic reticulum. Traffic 1:533–539. 10.1034/j.1600-0854.2000.010702.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Romero-Brey I, Bartenschlager R. 2016. Endoplasmic reticulum: the favorite intracellular niche for viral replication and assembly. Viruses 8:160. 10.3390/v8060160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dupzyk A, Tsai B. 2016. How polyomaviruses exploit the ERAD machinery to cause infection. Viruses 8:242. 10.3390/v8090242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fung TS, Torres J, Liu DX. 2015. The emerging roles of viroporins in ER stress response and autophagy induction during virus infection. Viruses 7:2834–2857. 10.3390/v7062749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li S, Kong L, Yu X. 2015. The expanding roles of endoplasmic reticulum stress in virus replication and pathogenesis. Crit Rev Microbiol 41:150–164. 10.3109/1040841X.2013.813899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Khongwichit S, Wikan N, Abere B, Thepparit C, Kuadkitkan A, Ubol S, Smith DR. 2016. Cell-type specific variation in the induction of ER stress and downstream events in chikungunya virus infection. Microb Pathog 101:104–118. 10.1016/j.micpath.2016.11.009. [DOI] [PubMed] [Google Scholar]
- 9.Liang JQ, Fang S, Yuan Q, Huang M, Chen RA, Fung TS, Liu DX. 2019. N-Linked glycosylation of the membrane protein ectodomain regulates infectious bronchitis virus-induced ER stress response, apoptosis and pathogenesis. Virology 531:48–56. 10.1016/j.virol.2019.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ma Y, Wang C, Xue M, Fu F, Zhang X, Li L, Yin L, Xu W, Feng L, Liu P. 2018. The coronavirus transmissible gastroenteritis virus evades the type I interferon response through IRE1alpha-mediated manipulation of the microRNA miR-30a-5p/SOCS1/3 axis. J Virol 92:e00728-18. 10.1128/JVI.00728-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Manning JT, Yun NE, Seregin AV, Koma T, Sattler RA, Ezeomah C, Huang C, de la Torre JC, Paessler S. 2020. The glycoprotein of the live-attenuated Junin virus vaccine strain induces endoplasmic reticulum stress and forms aggregates prior to degradation in the lysosome. J Virol 94:e01693-19. 10.1128/JVI.01693-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shi CS, Nabar NR, Huang NN, Kehrl JH. 2019. SARS-coronavirus open reading frame-8b triggers intracellular stress pathways and activates NLRP3 inflammasomes. Cell Death Discov 5:101. 10.1038/s41420-019-0181-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zou D, Xu J, Duan X, Xu X, Li P, Cheng L, Zheng L, Li X, Zhang Y, Wang X, Wu X, Shen Y, Yao X, Wei J, Yao L, Li L, Song B, Ma J, Liu X, Wu Z, Zhang H, Cao H. 2019. Porcine epidemic diarrhea virus ORF3 protein causes endoplasmic reticulum stress to facilitate autophagy. Vet Microbiol 235:209–219. 10.1016/j.vetmic.2019.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lim YX, Ng YL, Tam JP, Liu DX. 2016. Human coronaviruses: a review of virus-host interactions. Diseases 4:26. 10.3390/diseases4030026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guo X, Liu WL, Yang D, Shen ZQ, Qiu ZG, Jin M, Li JW. 2019. Hepatitis C virus infection induces endoplasmic reticulum stress and apoptosis in human fetal liver stem cells. J Pathol 248:155–163. 10.1002/path.5240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rios-Ocampo WA, Navas MC, Faber KN, Daemen T, Moshage H. 2019. The cellular stress response in hepatitis C virus infection: a balancing act to promote viral persistence and host cell survival. Virus Res 263:1–8. 10.1016/j.virusres.2018.12.013. [DOI] [PubMed] [Google Scholar]
- 17.Lewy TG, Grabowski JM, Bloom ME. 2017. BiP: master regulator of the unfolded protein response and crucial factor in flavivirus biology. Yale J Biol Med 90:291–300. [PMC free article] [PubMed] [Google Scholar]
- 18.Mukherjee S, Singh N, Sengupta N, Fatima M, Seth P, Mahadevan A, Shankar SK, Bhattacharyya A, Basu A. 2017. Japanese encephalitis virus induces human neural stem/progenitor cell death by elevating GRP78, PHB and hnRNPC through ER stress. Cell Death Dis 8:e2556. 10.1038/cddis.2016.394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Okamoto T, Suzuki T, Kusakabe S, Tokunaga M, Hirano J, Miyata Y, Matsuura Y. 2017. Regulation of apoptosis during flavivirus infection. Viruses 9:243. 10.3390/v9090243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang Q, Xin X, Wang T, Wan J, Ou Y, Yang Z, Yu Q, Zhu L, Guo Y, Wu Y, Ding Z, Zhang Y, Pan Z, Tang Y, Li S, Kong L. 2019. Japanese encephalitis virus induces apoptosis and encephalitis by activating the PERK pathway. J Virol 93:e00887-19. 10.1128/JVI.00887-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alfano C, Gladwyn-Ng I, Couderc T, Lecuit M, Nguyen L. 2019. The unfolded protein response: a key player in Zika virus-associated congenital microcephaly. Front Cell Neurosci 13:94. 10.3389/fncel.2019.00094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kolpikova EP, Tronco AR, Hartigh ABD, Jackson KJ, Iwawaki T, Fink SL. 2020. IRE1alpha promotes Zika virus infection via XBP1. Viruses 12:278. 10.3390/v12030278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mohd Ropidi MI, Khazali AS, Nor Rashid N, Yusof R. 2020. Endoplasmic reticulum: a focal point of Zika virus infection. J Biomed Sci 27:27. 10.1186/s12929-020-0618-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.He C, Qiu Y, Han P, Chen Y, Zhang L, Yuan Q, Zhang T, Cheng T, Yuan L, Huang C, Zhang S, Yin Z, Peng XE, Liang D, Lin X, Lin Y, Lin Z, Xia N. 2018. ER stress regulating protein phosphatase 2A-B56gamma, targeted by hepatitis B virus X protein, induces cell cycle arrest and apoptosis of hepatocytes. Cell Death Dis 9:762. 10.1038/s41419-018-0787-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li Y, Xia Y, Cheng X, Kleiner DE, Hewitt SM, Sproch J, Li T, Zhuang H, Liang TJ. 2019. Hepatitis B surface antigen activates unfolded protein response in forming ground glass hepatocytes of chronic hepatitis B. Viruses 11:386. 10.3390/v11040386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang P, Su C, Jiang Z, Zheng C. 2017. Herpes simplex virus 1 UL41 protein suppresses the IRE1/XBP1 signal pathway of the unfolded protein response via its RNase activity. J Virol 91:e02056-16. 10.1128/JVI.02056-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Johnston BP, McCormick C. 2019. Herpesviruses and the unfolded protein response. Viruses 12:17. 10.3390/v12010017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang LK, Wang B, Xin Q, Shang W, Shen S, Xiao G, Deng F, Wang H, Hu Z, Wang M. 2019. Quantitative proteomic analysis reveals unfolded-protein response involved in severe fever with thrombocytopenia syndrome virus infection. J Virol 93:e00308-19. 10.1128/JVI.00308-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mehrbod P, Ande SR, Alizadeh J, Rahimizadeh S, Shariati A, Malek H, Hashemi M, Glover KKM, Sher AA, Coombs KM, Ghavami S. 2019. The roles of apoptosis, autophagy and unfolded protein response in arbovirus, influenza virus, and HIV infections. Virulence 10:376–413. 10.1080/21505594.2019.1605803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Romeo MA, Masuelli L, Gaeta A, Nazzari C, Granato M, Gilardini Montani MS, Faggioni A, Cirone M. 2019. Impact of HHV-6A and HHV-6B lytic infection on autophagy and endoplasmic reticulum stress. J Gen Virol 100:89–98. 10.1099/jgv.0.001176. [DOI] [PubMed] [Google Scholar]
- 31.Luo XN, Yao HL, Song J, Song QQ, Shi BT, Xia D, Han J. 2018. Coxsackievirus B3 infection triggers autophagy through 3 pathways of endoplasmic reticulum stress. Biomed Environ Sci 31:867–875. 10.3967/bes2018.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reed M, Morris SH, Owczarczyk AB, Lukacs NW. 2015. Deficiency of autophagy protein Map1-LC3b mediates IL-17-dependent lung pathology during respiratory viral infection via ER stress-associated IL-1. Mucosal Immunol 8:1118–1130. 10.1038/mi.2015.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang L, Cheng W, Zhang Z. 2017. Respiratory syncytial virus infection accelerates lung fibrosis through the unfolded protein response in a bleomycin-induced pulmonary fibrosis animal model. Mol Med Rep 16:310–316. 10.3892/mmr.2017.6558. [DOI] [PubMed] [Google Scholar]
- 34.Duarte M, Vende P, Charpilienne A, Gratia M, Laroche C, Poncet D. 2019. Rotavirus infection alters splicing of the stress-related transcription factor XBP1. J Virol 93:e01739-18. 10.1128/JVI.01739-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rohde C, Becker S, Krahling V. 2019. Marburg virus regulates the IRE1/XBP1-dependent unfolded protein response to ensure efficient viral replication. Emerg Microbes Infect 8:1300–1313. 10.1080/22221751.2019.1659552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brodsky JL. 2007. The protective and destructive roles played by molecular chaperones during ERAD (endoplasmic-reticulum-associated degradation). Biochem J 404:353–363. 10.1042/BJ20061890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Prins D, Michalak M. 2011. Organellar calcium buffers. Cold Spring Harb Perspect Biol 3:a004069. 10.1101/cshperspect.a004069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ni M, Zhang Y, Lee AS. 2011. Beyond the endoplasmic reticulum: atypical GRP78 in cell viability, signalling and therapeutic targeting. Biochem J 434:181–188. 10.1042/BJ20101569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pobre KFR, Poet GJ, Hendershot LM. 2019. The endoplasmic reticulum (ER) chaperone BiP is a master regulator of ER functions: getting by with a little help from ERdj friends. J Biol Chem 294:2098–2108. 10.1074/jbc.REV118.002804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ibrahim IM, Abdelmalek DH, Elfiky AA. 2019. GRP78: a cell's response to stress. Life Sci 226:156–163. 10.1016/j.lfs.2019.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fu Y, Li J, Lee AS. 2007. GRP78/BiP inhibits endoplasmic reticulum BIK and protects human breast cancer cells against estrogen starvation-induced apoptosis. Cancer Res 67:3734–3740. 10.1158/0008-5472.CAN-06-4594. [DOI] [PubMed] [Google Scholar]
- 42.Reddy RK, Mao C, Baumeister P, Austin RC, Kaufman RJ, Lee AS. 2003. Endoplasmic reticulum chaperone protein GRP78 protects cells from apoptosis induced by topoisomerase inhibitors: role of ATP binding site in suppression of caspase-7 activation. J Biol Chem 278:20915–20924. 10.1074/jbc.M212328200. [DOI] [PubMed] [Google Scholar]
- 43.Zhou H, Zhang Y, Fu Y, Chan L, Lee AS. 2011. Novel mechanism of anti-apoptotic function of 78-kDa glucose-regulated protein (GRP78): endocrine resistance factor in breast cancer, through release of B-cell lymphoma 2 (BCL-2) from BCL-2-interacting killer (BIK). J Biol Chem 286:25687–25696. 10.1074/jbc.M110.212944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wati S, Soo ML, Zilm P, Li P, Paton AW, Burrell CJ, Beard M, Carr JM. 2009. Dengue virus infection induces upregulation of GRP78, which acts to chaperone viral antigen production. J Virol 83:12871–12880. 10.1128/JVI.01419-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Triantafilou K, Fradelizi D, Wilson K, Triantafilou M. 2002. GRP78, a coreceptor for coxsackievirus A9, interacts with major histocompatibility complex class I molecules which mediate virus internalization. J Virol 76:633–643. 10.1128/jvi.76.2.633-643.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Triantafilou K, Triantafilou M. 2003. Lipid raft microdomains: key sites for Coxsackievirus A9 infectious cycle. Virology 317:128–135. 10.1016/j.virol.2003.08.036. [DOI] [PubMed] [Google Scholar]
- 47.Honda T, Horie M, Daito T, Ikuta K, Tomonaga K. 2009. Molecular chaperone BiP interacts with Borna disease virus glycoprotein at the cell surface. J Virol 83:12622–12625. 10.1128/JVI.01201-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Limjindaporn T, Wongwiwat W, Noisakran S, Srisawat C, Netsawang J, Puttikhunt C, Kasinrerk W, Avirutnan P, Thiemmeca S, Sriburi R, Sittisombut N, Malasit P, Yenchitsomanus PT. 2009. Interaction of dengue virus envelope protein with endoplasmic reticulum-resident chaperones facilitates dengue virus production. Biochem Biophys Res Commun 379:196–200. 10.1016/j.bbrc.2008.12.070. [DOI] [PubMed] [Google Scholar]
- 49.Nain M, Mukherjee S, Karmakar SP, Paton AW, Paton JC, Abdin MZ, Basu A, Kalia M, Vrati S. 2017. GRP78 is an important host factor for Japanese encephalitis virus entry and replication in mammalian cells. J Virol 91:e02274-16. 10.1128/JVI.02274-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Khongwichit S, Sornjai W, Jitobaom K, Greenwood M, Greenwood MP, Hitakarun A, Wikan N, Murphy D, Smith DR. 2021. A functional interaction between GRP78 and Zika virus E protein. Sci Rep 11:393. 10.1038/s41598-020-79803-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Royle J, Ramirez-Santana C, Akpunarlieva S, Donald CL, Gestuveo RJ, Anaya JM, Merits A, Burchmore R, Kohl A, Varjak M. 2020. Glucose-regulated protein 78 interacts with Zika virus envelope protein and contributes to a productive infection. Viruses 12:524. 10.3390/v12050524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wu YP, Chang CM, Hung CY, Tsai MC, Schuyler SC, Wang RY. 2011. Japanese encephalitis virus co-opts the ER-stress response protein GRP78 for viral infectivity. Virol J 8:128. 10.1186/1743-422X-8-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Su CM, Liao CL, Lee YL, Lin YL. 2001. Highly sulfated forms of heparin sulfate are involved in Japanese encephalitis virus infection. Virology 286:206–215. 10.1006/viro.2001.0986. [DOI] [PubMed] [Google Scholar]
- 54.Jindadamrongwech S, Thepparit C, Smith DR. 2004. Identification of GRP 78 (BiP) as a liver cell expressed receptor element for dengue virus serotype 2. Arch Virol 149:915–927. 10.1007/s00705-003-0263-x. [DOI] [PubMed] [Google Scholar]
- 55.Chu H, Chan CM, Zhang X, Wang Y, Yuan S, Zhou J, Au-Yeung RK, Sze KH, Yang D, Shuai H, Hou Y, Li C, Zhao X, Poon VK, Leung SP, Yeung ML, Yan J, Lu G, Jin DY, Gao GF, Chan JF, Yuen KY. 2018. Middle East respiratory syndrome coronavirus and bat coronavirus HKU9 both can utilize GRP78 for attachment onto host cells. J Biol Chem 293:11709–11726. 10.1074/jbc.RA118.001897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ibrahim IM, Abdelmalek DH, Elshahat ME, Elfiky AA. 2020. COVID-19 spike-host cell receptor GRP78 binding site prediction. J Infect 80:554–562. 10.1016/j.jinf.2020.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Aguiar JA, Tremblay BJ, Mansfield MJ, Woody O, Lobb B, Banerjee A, Chandiramohan A, Tiessen N, Cao Q, Dvorkin-Gheva A, Revill S, Miller MS, Carlsten C, Organ L, Joseph C, John A, Hanson P, Austin RC, McManus BM, Jenkins G, Mossman K, Ask K, Doxey AC, Hirota JA. 2020. Gene expression and in situ protein profiling of candidate SARS-CoV-2 receptors in human airway epithelial cells and lung tissue. Eur Respir J 56:2001123. 10.1183/13993003.01123-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hurtley SM, Bole DG, Hoover-Litty H, Helenius A, Copeland CS. 1989. Interactions of misfolded influenza virus hemagglutinin with binding protein (BiP). J Cell Biol 108:2117–2126. 10.1083/jcb.108.6.2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Molinari M, Helenius A. 2000. Chaperone selection during glycoprotein translocation into the endoplasmic reticulum. Science 288:331–333. 10.1126/science.288.5464.331. [DOI] [PubMed] [Google Scholar]
- 60.Hogue BG, Nayak DP. 1992. Synthesis and processing of the influenza virus neuraminidase, a type II transmembrane glycoprotein. Virology 188:510–517. 10.1016/0042-6822(92)90505-J. [DOI] [PubMed] [Google Scholar]
- 61.Anderson K, Stott EJ, Wertz GW. 1992. Intracellular processing of the human respiratory syncytial virus fusion glycoprotein: amino acid substitutions affecting folding, transport and cleavage. J Gen Virol 73:1177–1188. 10.1099/0022-1317-73-5-1177. [DOI] [PubMed] [Google Scholar]
- 62.Cho DY, Yang GH, Ryu CJ, Hong HJ. 2003. Molecular chaperone GRP78/BiP interacts with the large surface protein of hepatitis B virus in vitro and in vivo. J Virol 77:2784–2788. 10.1128/jvi.77.4.2784-2788.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jitobaom K, Tongluan N, Smith DR. 2016. Involvement of voltage-dependent anion channel (VDAC) in dengue infection. Sci Rep 6:35753. 10.1038/srep35753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Choukhi A, Ung S, Wychowski C, Dubuisson J. 1998. Involvement of endoplasmic reticulum chaperones in the folding of hepatitis C virus glycoproteins. J Virol 72:3851–3858. 10.1128/JVI.72.5.3851-3858.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Liberman E, Fong YL, Selby MJ, Choo QL, Cousens L, Houghton M, Yen TS. 1999. Activation of the grp78 and grp94 promoters by hepatitis C virus E2 envelope protein. J Virol 73:3718–3722. 10.1128/JVI.73.5.3718-3722.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Nillegoda NB, Wentink AS, Bukau B. 2018. Protein disaggregation in multicellular organisms. Trends Biochem Sci 43:285–300. 10.1016/j.tibs.2018.02.003. [DOI] [PubMed] [Google Scholar]
- 67.Zito E. 2015. ERO1: a protein disulfide oxidase and H2O2 producer. Free Radic Biol Med 83:299–304. 10.1016/j.freeradbiomed.2015.01.011. [DOI] [PubMed] [Google Scholar]
- 68.Behnke J, Mann MJ, Scruggs FL, Feige MJ, Hendershot LM. 2016. Members of the Hsp70 family recognize distinct types of sequences to execute ER quality control. Mol Cell 63:739–752. 10.1016/j.molcel.2016.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Barnard TR, Abram QH, Lin QF, Wang AB, Sagan SM. 2021. Molecular determinants of flavivirus virion assembly. Trends Biochem Sci 46:378–390. 10.1016/j.tibs.2020.12.007. [DOI] [PubMed] [Google Scholar]
- 70.Fongsaran C, Phaonakrop N, Roytrakul S, Thepparit C, Kuadkitkan A, Smith DR. 2014. Voltage dependent anion channel is redistributed during Japanese encephalitis virus infection of insect cells. ScientificWorldJournal 2014:976015. 10.1155/2014/976015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Maruri-Avidal L, Lopez S, Arias CF. 2008. Endoplasmic reticulum chaperones are involved in the morphogenesis of rotavirus infectious particles. J Virol 82:5368–5380. 10.1128/JVI.02751-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Buchkovich NJ, Yu Y, Pierciey FJ, Jr, Alwine JC. 2010. Human cytomegalovirus induces the endoplasmic reticulum chaperone BiP through increased transcription and activation of translation by using the BiP internal ribosome entry site. J Virol 84:11479–11486. 10.1128/JVI.01330-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Buchkovich NJ, Maguire TG, Paton AW, Paton JC, Alwine JC. 2009. The endoplasmic reticulum chaperone BiP/GRP78 is important in the structure and function of the human cytomegalovirus assembly compartment. J Virol 83:11421–11428. 10.1128/JVI.00762-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Buchkovich NJ, Maguire TG, Yu Y, Paton AW, Paton JC, Alwine JC. 2008. Human cytomegalovirus specifically controls the levels of the endoplasmic reticulum chaperone BiP/GRP78, which is required for virion assembly. J Virol 82:31–39. 10.1128/JVI.01881-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Reid SP, Shurtleff AC, Costantino JA, Tritsch SR, Retterer C, Spurgers KB, Bavari S. 2014. HSPA5 is an essential host factor for Ebola virus infection. Antiviral Res 109:171–174. 10.1016/j.antiviral.2014.07.004. [DOI] [PubMed] [Google Scholar]
- 76.Yamayoshi S, Noda T, Ebihara H, Goto H, Morikawa Y, Lukashevich IS, Neumann G, Feldmann H, Kawaoka Y. 2008. Ebola virus matrix protein VP40 uses the COPII transport system for its intracellular transport. Cell Host Microbe 3:168–177. 10.1016/j.chom.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lee AS. 2014. Glucose-regulated proteins in cancer: molecular mechanisms and therapeutic potential. Nat Rev Cancer 14:263–276. 10.1038/nrc3701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lyoo HR, Park SY, Kim JY, Jeong YS. 2015. Constant up-regulation of BiP/GRP78 expression prevents virus-induced apoptosis in BHK-21 cells with Japanese encephalitis virus persistent infection. Virol J 12:32. 10.1186/s12985-015-0269-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Shu W, Guo Z, Li L, Xiong Z, Wang Z, Yang Y, Li Y, He M, Gong R, Gao B. 2020. Regulation of molecular chaperone GRP78 by hepatitis B virus: control of viral replication and cell survival. Mol Cell Biol 40:e00475-19. 10.1128/MCB.00475-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Xu Z, Jensen G, Yen TS. 1997. Activation of hepatitis B virus S promoter by the viral large surface protein via induction of stress in the endoplasmic reticulum. J Virol 71:7387–7392. 10.1128/JVI.71.10.7387-7392.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lopez SN, Rodriguez-Valentin M, Rivera M, Rodriguez M, Babu M, Cubano LA, Xiong H, Wang G, Kucheryavykh L, Boukli NM. 2017. HIV-1 Gp120 clade B/C induces a GRP78 driven cytoprotective mechanism in astrocytoma. Oncotarget 8:68415–68438. 10.18632/oncotarget.19474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Constantino AA, Huang Y, Zhang H, Wood C, Zheng JC. 2011. HIV-1 clade B and C isolates exhibit differential replication: relevance to macrophage-mediated neurotoxicity. Neurotox Res 20:277–288. 10.1007/s12640-011-9241-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lindl KA, Akay C, Wang Y, White MG, Jordan-Sciutto KL. 2007. Expression of the endoplasmic reticulum stress response marker, BiP, in the central nervous system of HIV-positive individuals. Neuropathol Appl Neurobiol 33:658–669. 10.1111/j.1365-2990.2007.00866.x. [DOI] [PubMed] [Google Scholar]
- 84.Oresic K, Tortorella D. 2008. Endoplasmic reticulum chaperones participate in human cytomegalovirus US2-mediated degradation of class I major histocompatibility complex molecules. J Gen Virol 89:1122–1130. 10.1099/vir.0.83516-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Tirosh B, Iwakoshi NN, Lilley BN, Lee AH, Glimcher LH, Ploegh HL. 2005. Human cytomegalovirus protein US11 provokes an unfolded protein response that may facilitate the degradation of class I major histocompatibility complex products. J Virol 79:2768–2779. 10.1128/JVI.79.5.2768-2779.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ma Y, Yu J, Chan HL, Chen YC, Wang H, Chen Y, Chan CY, Go MY, Tsai SN, Ngai SM, To KF, Tong JH, He QY, Sung JJ, Kung HF, Cheng CH, He ML. 2009. Glucose-regulated protein 78 is an intracellular antiviral factor against hepatitis B virus. Mol Cell Proteomics 8:2582–2594. 10.1074/mcp.M900180-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wei D, Li NL, Zeng Y, Liu B, Kumthip K, Wang TT, Huo D, Ingels JF, Lu L, Shang J, Li K. 2016. The molecular chaperone GRP78 contributes to Toll-like receptor 3-mediated innate immune response to hepatitis C virus in hepatocytes. J Biol Chem 291:12294–12309. 10.1074/jbc.M115.711598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Richardson RB, Ohlson MB, Eitson JL, Kumar A, McDougal MB, Boys IN, Mar KB, De La Cruz-Rivera PC, Douglas C, Konopka G, Xing C, Schoggins JW. 2018. A CRISPR screen identifies IFI6 as an ER-resident interferon effector that blocks flavivirus replication. Nat Microbiol 3:1214–1223. 10.1038/s41564-018-0244-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Huck JD, Que NL, Hong F, Li Z, Gewirth DT. 2017. Structural and functional analysis of GRP94 in the closed state reveals an essential role for the pre-N domain and a potential client-binding site. Cell Rep 20:2800–2809. 10.1016/j.celrep.2017.08.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Christianson JC, Shaler TA, Tyler RE, Kopito RR. 2008. OS-9 and GRP94 deliver mutant alpha1-antitrypsin to the Hrd1-SEL1L ubiquitin ligase complex for ERAD. Nat Cell Biol 10:272–282. 10.1038/ncb1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Bloor S, Maelfait J, Krumbach R, Beyaert R, Randow F. 2010. Endoplasmic reticulum chaperone gp96 is essential for infection with vesicular stomatitis virus. Proc Natl Acad Sci USA 107:6970–6975. 10.1073/pnas.0908536107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Finkelshtein D, Werman A, Novick D, Barak S, Rubinstein M. 2013. LDL receptor and its family members serve as the cellular receptors for vesicular stomatitis virus. Proc Natl Acad Sci USA 110:7306–7311. 10.1073/pnas.1214441110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Weekes MP, Antrobus R, Talbot S, Hor S, Simecek N, Smith DL, Bloor S, Randow F, Lehner PJ. 2012. Proteomic plasma membrane profiling reveals an essential role for gp96 in the cell surface expression of LDLR family members, including the LDL receptor and LRP6. J Proteome Res 11:1475–1484. 10.1021/pr201135e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Nolte MA, Nolte-'t Hoen ENM, Margadant C. 2021. Integrins control vesicular trafficking; new tricks for old dogs. Trends Biochem Sci 46:124–137. 10.1016/j.tibs.2020.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Prusty BK, Siegl C, Gulve N, Mori Y, Rudel T. 2014. GP96 interacts with HHV-6 during viral entry and directs it for cellular degradation. PLoS One 9:e113962. 10.1371/journal.pone.0113962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ma J, Jia J, Jiang X, Xu M, Guo J, Tang T, Xu X, Wu Z, Hu B, Yao K, Li L, Tang H. 2020. gp96 is critical for both human herpesvirus 6A (HHV-6A) and HHV-6B infections. J Virol 94:e00311-20. 10.1128/JVI.00311-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Izaguirre G. 2019. The proteolytic regulation of virus cell entry by furin and other proprotein convertases. Viruses 11:837. 10.3390/v11090837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Millet JK, Whittaker GR. 2015. Host cell proteases: critical determinants of coronavirus tropism and pathogenesis. Virus Res 202:120–134. 10.1016/j.virusres.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Coutard B, Valle C, de Lamballerie X, Canard B, Seidah NG, Decroly E. 2020. The spike glycoprotein of the new coronavirus 2019-nCoV contains a furin-like cleavage site absent in CoV of the same clade. Antiviral Res 176:104742. 10.1016/j.antiviral.2020.104742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Hoffmann M, Kleine-Weber H, Pohlmann S. 2020. A multibasic cleavage site in the spike protein of SARS-CoV-2 is essential for infection of human lung cells. Mol Cell 78:779–784.e5. 10.1016/j.molcel.2020.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Hoffmann M, Kleine-Weber H, Schroeder S, Kruger N, Herrler T, Erichsen S, Schiergens TS, Herrler G, Wu NH, Nitsche A, Muller MA, Drosten C, Pohlmann S. 2020. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 181:271–280.e8. 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Shang J, Wan Y, Luo C, Ye G, Geng Q, Auerbach A, Li F. 2020. Cell entry mechanisms of SARS-CoV-2. Proc Natl Acad Sci USA 117:11727–11734. 10.1073/pnas.2003138117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Bestle D, Heindl MR, Limburg H, Van Lam van T, Pilgram O, Moulton H, Stein DA, Hardes K, Eickmann M, Dolnik O, Rohde C, Klenk HD, Garten W, Steinmetzer T, Bottcher-Friebertshauser E. 2020. TMPRSS2 and furin are both essential for proteolytic activation of SARS-CoV-2 in human airway cells. Life Sci Alliance 3:e202000786. 10.26508/lsa.202000786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Koo BH, Apte SS. 2010. Cell-surface processing of the metalloprotease pro-ADAMTS9 is influenced by the chaperone GRP94/gp96. J Biol Chem 285:197–205. 10.1074/jbc.M109.039677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Chaumonnot K, Masson S, Sikner H, Bouchard A, Baverel V, Bellaye PS, Collin B, Garrido C, Kohli E. 2021. The HSP GRP94 interacts with macrophage intracellular complement C3 and impacts M2 profile during ER stress. Cell Death Dis 12:114. 10.1038/s41419-020-03288-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kim SS, Shin HJ, Cho YH, Rho HM. 2000. Expression of stable hepatitis B viral polymerase associated with GRP94 in E. coli. Arch Virol 145:1305–1320. 10.1007/s007050070092. [DOI] [PubMed] [Google Scholar]
- 107.Ramakrishnan M, Tugizov S, Pereira L, Lee AS. 1995. Conformation-defective herpes simplex virus 1 glycoprotein B activates the promoter of the grp94 gene that codes for the 94-kD stress protein in the endoplasmic reticulum. DNA Cell Biol 14:373–384. 10.1089/dna.1995.14.373. [DOI] [PubMed] [Google Scholar]
- 108.Rothan HA, Zhong Y, Sanborn MA, Teoh TC, Ruan J, Yusof R, Hang J, Henderson MJ, Fang S. 2019. Small molecule grp94 inhibitors block dengue and Zika virus replication. Antiviral Res 171:104590. 10.1016/j.antiviral.2019.104590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Patel PD, Yan P, Seidler PM, Patel HJ, Sun W, Yang C, Que NS, Taldone T, Finotti P, Stephani RA, Gewirth DT, Chiosis G. 2013. Paralog-selective Hsp90 inhibitors define tumor-specific regulation of HER2. Nat Chem Biol 9:677–684. 10.1038/nchembio.1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Staron M, Yang Y, Liu B, Li J, Shen Y, Zuniga-Pflucker JC, Aguila HL, Goldschneider I, Li Z. 2010. gp96, an endoplasmic reticulum master chaperone for integrins and Toll-like receptors, selectively regulates early T and B lymphopoiesis. Blood 115:2380–2390. 10.1182/blood-2009-07-233031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Yang Y, Liu B, Dai J, Srivastava PK, Zammit DJ, Lefrancois L, Li Z. 2007. Heat shock protein gp96 is a master chaperone for toll-like receptors and is important in the innate function of macrophages. Immunity 26:215–226. 10.1016/j.immuni.2006.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Wang Y, Sedlacek AL, Pawaria S, Xu H, Scott MJ, Binder RJ. 2018. Cutting edge: the heat shock protein gp96 activates inflammasome-signaling platforms in APCs. J Immunol 201:2209–2214. 10.4049/jimmunol.1800505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Binder RJ, Srivastava PK. 2004. Essential role of CD91 in re-presentation of gp96-chaperoned peptides. Proc Natl Acad Sci USA 101:6128–6133. 10.1073/pnas.0308180101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Seignez A, Joly AL, Chaumonnot K, Hazoume A, Sanka M, Marcion G, Boudesco C, Hammann A, Seigneuric R, Jego G, Ducoroy P, Delarue P, Senet P, Castilla-Llorente C, Solary E, Durey MA, Rubio MT, Hermine O, Kohli E, Garrido C. 2017. Serum Gp96 is a chaperone of complement-C3 during graft-versus-host disease. JCI Insight 2:e90531. 10.1172/jci.insight.90531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Zhang Y, Wu BX, Metelli A, Thaxton JE, Hong F, Rachidi S, Ansa-Addo E, Sun S, Vasu C, Yang Y, Liu B, Li Z. 2015. GP96 is a GARP chaperone and controls regulatory T cell functions. J Clin Invest 125:859–869. 10.1172/JCI79014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Jockheck-Clark AR, Bowers EV, Totonchy MB, Neubauer J, Pizzo SV, Nicchitta CV. 2010. Re-examination of CD91 function in GRP94 (glycoprotein 96) surface binding, uptake, and peptide cross-presentation. J Immunol 185:6819–6830. 10.4049/jimmunol.1000448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Ju Y, Fan H, Liu J, Hu J, Li X, Li C, Chen L, Gao Q, Gao GF, Meng S. 2014. Heat shock protein gp96 adjuvant induces T cell responses and cross-protection to a split influenza vaccine. Vaccine 32:2703–2711. 10.1016/j.vaccine.2014.03.045. [DOI] [PubMed] [Google Scholar]
- 118.Wang S, Qiu L, Liu G, Li Y, Zhang X, Jin W, Gao GF, Kong X, Meng S. 2011. Heat shock protein gp96 enhances humoral and T cell responses, decreases Treg frequency and potentiates the anti-HBV activity in BALB/c and transgenic mice. Vaccine 29:6342–6351. 10.1016/j.vaccine.2011.05.008. [DOI] [PubMed] [Google Scholar]
- 119.Selinger C, Strbo N, Gonzalez L, Aicher L, Weiss JM, Law GL, Palermo RE, Vaccari M, Franchini G, Podack ER, Katze MG. 2014. Multiple low-dose challenges in a rhesus macaque AIDS vaccine trial result in an evolving host response that affects protective outcome. Clin Vaccine Immunol 21:1650–1660. 10.1128/CVI.00455-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Kim MS, Kim S, Myung H. 2014. Degradation of AIMP1/p43 induced by hepatitis C virus E2 leads to upregulation of TGF-beta signaling and increase in surface expression of gp96. PLoS One 9:e96302. 10.1371/journal.pone.0096302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Han JM, Park SG, Liu B, Park BJ, Kim JY, Jin CH, Song YW, Li Z, Kim S. 2007. Aminoacyl-tRNA synthetase-interacting multifunctional protein 1/p43 controls endoplasmic reticulum retention of heat shock protein gp96: its pathological implications in lupus-like autoimmune diseases. Am J Pathol 170:2042–2054. 10.2353/ajpath.2007.061266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Lee YS, Han JM, Son SH, Choi JW, Jeon EJ, Bae SC, Park YI, Kim S. 2008. AIMP1/p43 downregulates TGF-beta signaling via stabilization of smurf2. Biochem Biophys Res Commun 371:395–400. 10.1016/j.bbrc.2008.04.099. [DOI] [PubMed] [Google Scholar]
- 123.Kwon YC, Meyer K, Peng G, Chatterjee S, Hoft DF, Ray R. 2019. Hepatitis C virus E2 envelope glycoprotein induces an immunoregulatory phenotype in macrophages. Hepatology 69:1873–1884. 10.1002/hep.29843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Kozlov G, Gehring K. 2020. Calnexin cycle—structural features of the ER chaperone system. FEBS J 287:4322–4340. 10.1111/febs.15330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Watanabe Y, Bowden TA, Wilson IA, Crispin M. 2019. Exploitation of glycosylation in enveloped virus pathobiology. Biochim Biophys Acta Gen Subj 1863:1480–1497. 10.1016/j.bbagen.2019.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Peterson JR, Ora A, Van PN, Helenius A. 1995. Transient, lectin-like association of calreticulin with folding intermediates of cellular and viral glycoproteins. Mol Biol Cell 6:1173–1184. 10.1091/mbc.6.9.1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Hammond C, Braakman I, Helenius A. 1994. Role of N-linked oligosaccharide recognition, glucose trimming, and calnexin in glycoprotein folding and quality control. Proc Natl Acad Sci USA 91:913–917. 10.1073/pnas.91.3.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Tatu U, Hammond C, Helenius A. 1995. Folding and oligomerization of influenza hemagglutinin in the ER and the intermediate compartment. EMBO J 14:1340–1348. 10.1002/j.1460-2075.1995.tb07120.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Wang N, Glidden EJ, Murphy SR, Pearse BR, Hebert DN. 2008. The cotranslational maturation program for the type II membrane glycoprotein influenza neuraminidase. J Biol Chem 283:33826–33837. 10.1074/jbc.M806897200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Prange R, Werr M, Loffler-Mary H. 1999. Chaperones involved in hepatitis B virus morphogenesis. Biol Chem 380:305–314. 10.1515/BC.1999.042. [DOI] [PubMed] [Google Scholar]
- 131.Gaudin Y. 1997. Folding of rabies virus glycoprotein: epitope acquisition and interaction with endoplasmic reticulum chaperones. J Virol 71:3742–3750. 10.1128/JVI.71.5.3742-3750.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Yamashita Y, Yamada M, Daikoku T, Yamada H, Tadauchi A, Tsurumi T, Nishiyama Y. 1996. Calnexin associates with the precursors of glycoproteins B, C, and D of herpes simplex virus type 1. Virology 225:216–222. 10.1006/viro.1996.0590. [DOI] [PubMed] [Google Scholar]
- 133.Yamashita Y, Shimokata K, Mizuno S, Daikoku T, Tsurumi T, Nishiyama Y. 1996. Calnexin acts as a molecular chaperone during the folding of glycoprotein B of human cytomegalovirus. J Virol 70:2237–2246. 10.1128/JVI.70.4.2237-2246.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Shi X, Elliott RM. 2004. Analysis of N-linked glycosylation of Hantaan virus glycoproteins and the role of oligosaccharide side chains in protein folding and intracellular trafficking. J Virol 78:5414–5422. 10.1128/JVI.78.10.5414-5422.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Wang B, Wang Y, Frabutt DA, Zhang X, Yao X, Hu D, Zhang Z, Liu C, Zheng S, Xiang SH, Zheng YH. 2017. Mechanistic understanding of N-glycosylation in Ebola virus glycoprotein maturation and function. J Biol Chem 292:5860–5870. 10.1074/jbc.M116.768168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Land A, Braakman I. 2001. Folding of the human immunodeficiency virus type 1 envelope glycoprotein in the endoplasmic reticulum. Biochimie 83:783–790. 10.1016/S0300-9084(01)01314-1. [DOI] [PubMed] [Google Scholar]
- 137.Otteken A, Moss B. 1996. Calreticulin interacts with newly synthesized human immunodeficiency virus type 1 envelope glycoprotein, suggesting a chaperone function similar to that of calnexin. J Biol Chem 271:97–103. 10.1074/jbc.271.1.97. [DOI] [PubMed] [Google Scholar]
- 138.Mirazimi A, Nilsson M, Svensson L. 1998. The molecular chaperone calnexin interacts with the NSP4 enterotoxin of rotavirus in vivo and in vitro. J Virol 72:8705–8709. 10.1128/JVI.72.11.8705-8709.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Chen D, Choi YB, Sandford G, Nicholas J. 2009. Determinants of secretion and intracellular localization of human herpesvirus 8 interleukin-6. J Virol 83:6874–6882. 10.1128/JVI.02625-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Dubuisson J, Rice CM. 1996. Hepatitis C virus glycoprotein folding: disulfide bond formation and association with calnexin. J Virol 70:778–786. 10.1128/JVI.70.2.778-786.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Jennelle L, Hunegnaw R, Dubrovsky L, Pushkarsky T, Fitzgerald ML, Sviridov D, Popratiloff A, Brichacek B, Bukrinsky M. 2014. HIV-1 protein Nef inhibits activity of ATP-binding cassette transporter A1 by targeting endoplasmic reticulum chaperone calnexin. J Biol Chem 289:28870–28884. 10.1074/jbc.M114.583591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Hunegnaw R, Vassylyeva M, Dubrovsky L, Pushkarsky T, Sviridov D, Anashkina AA, Uren A, Brichacek B, Vassylyev DG, Adzhubei AA, Bukrinsky M. 2016. Interaction between HIV-1 Nef and calnexin: from modeling to small molecule inhibitors reversing HIV-induced lipid accumulation. Arterioscler Thromb Vasc Biol 36:1758–1771. 10.1161/ATVBAHA.116.307997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Fukushi M, Yoshinaka Y, Matsuoka Y, Hatakeyama S, Ishizaka Y, Kirikae T, Sasazuki T, Miyoshi-Akiyama T. 2012. Monitoring of S protein maturation in the endoplasmic reticulum by calnexin is important for the infectivity of severe acute respiratory syndrome coronavirus. J Virol 86:11745–11753. 10.1128/JVI.01250-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Williams SJ, Goddard-Borger ED. 2020. Alpha-glucosidase inhibitors as host-directed antiviral agents with potential for the treatment of COVID-19. Biochem Soc Trans 48:1287–1295. 10.1042/BST20200505. [DOI] [PubMed] [Google Scholar]
- 145.Zhao C, Liu H, Xiao T, Wang Z, Nie X, Li X, Qian P, Qin L, Han X, Zhang J, Ruan J, Zhu M, Miao YL, Zuo B, Yang K, Xie S, Zhao S. 2020. CRISPR screening of porcine sgRNA library identifies host factors associated with Japanese encephalitis virus replication. Nat Commun 11:5178. 10.1038/s41467-020-18936-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Cresswell P. 2005. Antigen processing and presentation. Immunol Rev 207:5–7. 10.1111/j.0105-2896.2005.00320.x. [DOI] [PubMed] [Google Scholar]
- 147.Raghavan M, Wijeyesakere SJ, Peters LR, Del Cid N. 2013. Calreticulin in the immune system: ins and outs. Trends Immunol 34:13–21. 10.1016/j.it.2012.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Gruener M, Bravo IG, Momburg F, Alonso A, Tomakidi P. 2007. The E5 protein of the human papillomavirus type 16 down-regulates HLA-I surface expression in calnexin-expressing but not in calnexin-deficient cells. Virol J 4:116. 10.1186/1743-422X-4-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Miura S, Kawana K, Schust DJ, Fujii T, Yokoyama T, Iwasawa Y, Nagamatsu T, Adachi K, Tomio A, Tomio K, Kojima S, Yasugi T, Kozuma S, Taketani Y. 2010. CD1d, a sentinel molecule bridging innate and adaptive immunity, is downregulated by the human papillomavirus (HPV) E5 protein: a possible mechanism for immune evasion by HPV. J Virol 84:11614–11623. 10.1128/JVI.01053-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Yue X, Wang H, Zhao F, Liu S, Wu J, Ren W, Zhu Y. 2012. Hepatitis B virus-induced calreticulin protein is involved in IFN resistance. J Immunol 189:279–286. 10.4049/jimmunol.1103405. [DOI] [PubMed] [Google Scholar]
- 151.Kepp O, Tesniere A, Zitvogel L, Kroemer G. 2009. The immunogenicity of tumor cell death. Curr Opin Oncol 21:71–76. 10.1097/CCO.0b013e32831bc375. [DOI] [PubMed] [Google Scholar]
- 152.Thomas S, Kuncheria L, Roulstone V, Kyula JN, Mansfield D, Bommareddy PK, Smith H, Kaufman HL, Harrington KJ, Coffin RS. 2019. Development of a new fusion-enhanced oncolytic immunotherapy platform based on herpes simplex virus type 1. J Immunother Cancer 7:214. 10.1186/s40425-019-0682-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Galligan JJ, Petersen DR. 2012. The human protein disulfide isomerase gene family. Hum Genomics 6:6. 10.1186/1479-7364-6-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Mathys L, Balzarini J. 2016. The role of cellular oxidoreductases in viral entry and virus infection-associated oxidative stress: potential therapeutic applications. Expert Opin Ther Targets 20:123–143. 10.1517/14728222.2015.1068760. [DOI] [PubMed] [Google Scholar]
- 155.Fenouillet E, Barbouche R, Jones IM. 2007. Cell entry by enveloped viruses: redox considerations for HIV and SARS-coronavirus. Antioxid Redox Signal 9:1009–1034. 10.1089/ars.2007.1639. [DOI] [PubMed] [Google Scholar]
- 156.Fenouillet E, Barbouche R, Courageot J, Miquelis R. 2001. The catalytic activity of protein disulfide isomerase is involved in human immunodeficiency virus envelope-mediated membrane fusion after CD4 cell binding. J Infect Dis 183:744–752. 10.1086/318823. [DOI] [PubMed] [Google Scholar]
- 157.Gallina A, Hanley TM, Mandel R, Trahey M, Broder CC, Viglianti GA, Ryser HJ. 2002. Inhibitors of protein-disulfide isomerase prevent cleavage of disulfide bonds in receptor-bound glycoprotein 120 and prevent HIV-1 entry. J Biol Chem 277:50579–50588. 10.1074/jbc.M204547200. [DOI] [PubMed] [Google Scholar]
- 158.Markovic I, Stantchev TS, Fields KH, Tiffany LJ, Tomic M, Weiss CD, Broder CC, Strebel K, Clouse KA. 2004. Thiol/disulfide exchange is a prerequisite for CXCR4-tropic HIV-1 envelope-mediated T-cell fusion during viral entry. Blood 103:1586–1594. 10.1182/blood-2003-05-1390. [DOI] [PubMed] [Google Scholar]
- 159.Ryser HJ, Levy EM, Mandel R, DiSciullo GJ. 1994. Inhibition of human immunodeficiency virus infection by agents that interfere with thiol-disulfide interchange upon virus-receptor interaction. Proc Natl Acad Sci USA 91:4559–4563. 10.1073/pnas.91.10.4559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Auwerx J, Isacsson O, Soderlund J, Balzarini J, Johansson M, Lundberg M. 2009. Human glutaredoxin-1 catalyzes the reduction of HIV-1 gp120 and CD4 disulfides and its inhibition reduces HIV-1 replication. Int J Biochem Cell Biol 41:1269–1275. 10.1016/j.biocel.2008.10.031. [DOI] [PubMed] [Google Scholar]
- 161.Ou W, Silver J. 2006. Role of protein disulfide isomerase and other thiol-reactive proteins in HIV-1 envelope protein-mediated fusion. Virology 350:406–417. 10.1016/j.virol.2006.01.041. [DOI] [PubMed] [Google Scholar]
- 162.Jain S, McGinnes LW, Morrison TG. 2008. Overexpression of thiol/disulfide isomerases enhances membrane fusion directed by the Newcastle disease virus fusion protein. J Virol 82:12039–12048. 10.1128/JVI.01406-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Langsjoen RM, Auguste AJ, Rossi SL, Roundy CM, Penate HN, Kastis M, Schnizlein MK, Le KC, Haller SL, Chen R, Watowich SJ, Weaver SC. 2017. Host oxidative folding pathways offer novel anti-chikungunya virus drug targets with broad spectrum potential. Antiviral Res 143:246–251. 10.1016/j.antiviral.2017.04.014. [DOI] [PubMed] [Google Scholar]
- 164.Wan SW, Lin CF, Lu YT, Lei HY, Anderson R, Lin YS. 2012. Endothelial cell surface expression of protein disulfide isomerase activates beta1 and beta3 integrins and facilitates dengue virus infection. J Cell Biochem 113:1681–1691. 10.1002/jcb.24037. [DOI] [PubMed] [Google Scholar]
- 165.Calderon MN, Guerrero CA, Acosta O, Lopez S, Arias CF. 2012. Inhibiting rotavirus infection by membrane-impermeant thiol/disulfide exchange blockers and antibodies against protein disulfide isomerase. Intervirology 55:451–464. 10.1159/000335262. [DOI] [PubMed] [Google Scholar]
- 166.Walczak CP, Tsai B. 2011. A PDI family network acts distinctly and coordinately with ERp29 to facilitate polyomavirus infection. J Virol 85:2386–2396. 10.1128/JVI.01855-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Aguilar-Hernandez N, Meyer L, Lopez S, DuBois RM, Arias CF. 2020. Protein disulfide isomerase A4 is involved in genome uncoating during human astrovirus cell entry. Viruses 13:53. 10.3390/v13010053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Solda T, Garbi N, Hammerling GJ, Molinari M. 2006. Consequences of ERp57 deletion on oxidative folding of obligate and facultative clients of the calnexin cycle. J Biol Chem 281:6219–6226. 10.1074/jbc.M513595200. [DOI] [PubMed] [Google Scholar]
- 169.Piacentini S, La Frazia S, Riccio A, Pedersen JZ, Topai A, Nicolotti O, Rossignol JF, Santoro MG. 2018. Nitazoxanide inhibits paramyxovirus replication by targeting the fusion protein folding: role of glycoprotein-specific thiol oxidoreductase ERp57. Sci Rep 8:10425. 10.1038/s41598-018-28172-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Roberson EC, Tully JE, Guala AS, Reiss JN, Godburn KE, Pociask DA, Alcorn JF, Riches DW, Dienz O, Janssen-Heininger YM, Anathy V. 2012. Influenza induces endoplasmic reticulum stress, caspase-12-dependent apoptosis, and c-Jun N-terminal kinase-mediated transforming growth factor-beta release in lung epithelial cells. Am J Respir Cell Mol Biol 46:573–581. 10.1165/rcmb.2010-0460OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Chamberlain N, Korwin-Mihavics BR, Nakada EM, Bruno SR, Heppner DE, Chapman DG, Hoffman SM, van der Vliet A, Suratt BT, Dienz O, Alcorn JF, Anathy V. 2019. Lung epithelial protein disulfide isomerase A3 (PDIA3) plays an important role in influenza infection, inflammation, and airway mechanics. Redox Biol 22:101129. 10.1016/j.redox.2019.101129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Kim Y, Chang KO. 2018. Protein disulfide isomerases as potential therapeutic targets for influenza A and B viruses. Virus Res 247:26–33. 10.1016/j.virusres.2018.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Raghavan M, Del Cid N, Rizvi SM, Peters LR. 2008. MHC class I assembly: out and about. Trends Immunol 29:436–443. 10.1016/j.it.2008.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Lee SO, Cho K, Cho S, Kim I, Oh C, Ahn K. 2010. Protein disulphide isomerase is required for signal peptide peptidase-mediated protein degradation. EMBO J 29:363–375. 10.1038/emboj.2009.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Mishra KP, Shweta Diwaker D, Ganju L. 2012. Dengue virus infection induces upregulation of hn RNP-H and PDIA3 for its multiplication in the host cell. Virus Res 163:573–579. 10.1016/j.virusres.2011.12.010. [DOI] [PubMed] [Google Scholar]
- 176.Cheng HJ, Lei HY, Lin CF, Luo YH, Wan SW, Liu HS, Yeh TM, Lin YS. 2009. Anti-dengue virus nonstructural protein 1 antibodies recognize protein disulfide isomerase on platelets and inhibit platelet aggregation. Mol Immunol 47:398–406. 10.1016/j.molimm.2009.08.033. [DOI] [PubMed] [Google Scholar]
- 177.Wu J, Wang Y, Wei Y, Xu Z, Tan X, Wu Z, Zheng J, Liu GD, Cao Y, Xue C. 2020. Disulfide isomerase ERp57 improves the stability and immunogenicity of H3N2 influenza virus hemagglutinin. Virol J 17:55. 10.1186/s12985-020-01325-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Taguwa S, Maringer K, Li X, Bernal-Rubio D, Rauch JN, Gestwicki JE, Andino R, Fernandez-Sesma A, Frydman J. 2015. Defining Hsp70 subnetworks in dengue virus replication reveals key vulnerability in flavivirus infection. Cell 163:1108–1123. 10.1016/j.cell.2015.10.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Petrova K, Oyadomari S, Hendershot LM, Ron D. 2008. Regulated association of misfolded endoplasmic reticulum lumenal proteins with P58/DNAJc3. EMBO J 27:2862–2872. 10.1038/emboj.2008.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Lee TG, Tomita J, Hovanessian AG, Katze MG. 1990. Purification and partial characterization of a cellular inhibitor of the interferon-induced protein kinase of Mr 68,000 from influenza virus-infected cells. Proc Natl Acad Sci USA 87:6208–6212. 10.1073/pnas.87.16.6208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Yan W, Frank CL, Korth MJ, Sopher BL, Novoa I, Ron D, Katze MG. 2002. Control of PERK eIF2alpha kinase activity by the endoplasmic reticulum stress-induced molecular chaperone P58IPK. Proc Natl Acad Sci USA 99:15920–15925. 10.1073/pnas.252341799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Chen YJ, Bagchi P, Tsai B. 2020. ER functions are exploited by viruses to support distinct stages of their life cycle. Biochem Soc Trans 48:2173–2184. 10.1042/BST20200395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Yi Z, Sperzel L, Nurnberger C, Bredenbeek PJ, Lubick KJ, Best SM, Stoyanov CT, Law LM, Yuan Z, Rice CM, MacDonald MR. 2011. Identification and characterization of the host protein DNAJC14 as a broadly active flavivirus replication modulator. PLoS Pathog 7:e1001255. 10.1371/journal.ppat.1001255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Yi Z, Yuan Z, Rice CM, MacDonald MR. 2012. Flavivirus replication complex assembly revealed by DNAJC14 functional mapping. J Virol 86:11815–11832. 10.1128/JVI.01022-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Aktepe TE, Mackenzie JM. 2018. Shaping the flavivirus replication complex: it is curvaceous! Cell Microbiol 20:e12884. 10.1111/cmi.12884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Zhang HM, Ye X, Su Y, Yuan J, Liu Z, Stein DA, Yang D. 2010. Coxsackievirus B3 infection activates the unfolded protein response and induces apoptosis through downregulation of p58IPK and activation of CHOP and SREBP1. J Virol 84:8446–8459. 10.1128/JVI.01416-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Sharma K, Tripathi S, Ranjan P, Kumar P, Garten R, Deyde V, Katz JM, Cox NJ, Lal RB, Sambhara S, Lal SK. 2011. Influenza A virus nucleoprotein exploits Hsp40 to inhibit PKR activation. PLoS One 6:e20215. 10.1371/journal.pone.0020215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Zhang HM, Qiu Y, Ye X, Hemida MG, Hanson P, Yang D. 2014. P58(IPK) inhibits coxsackievirus-induced apoptosis via the PI3K/Akt pathway requiring activation of ATF6a and subsequent upregulation of mitofusin 2. Cell Microbiol 16:411–424. 10.1111/cmi.12229. [DOI] [PubMed] [Google Scholar]
- 189.Goodman AG, Smith JA, Balachandran S, Perwitasari O, Proll SC, Thomas MJ, Korth MJ, Barber GN, Schiff LA, Katze MG. 2007. The cellular protein P58IPK regulates influenza virus mRNA translation and replication through a PKR-mediated mechanism. J Virol 81:2221–2230. 10.1128/JVI.02151-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Goodman AG, Fornek JL, Medigeshi GR, Perrone LA, Peng X, Dyer MD, Proll SC, Knoblaugh SE, Carter VS, Korth MJ, Nelson JA, Tumpey TM, Katze MG. 2009. P58(IPK): a novel “CIHD” member of the host innate defense response against pathogenic virus infection. PLoS Pathog 5:e1000438. 10.1371/journal.ppat.1000438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Vabret N, Britton GJ, Gruber C, Hegde S, Kim J, Kuksin M, Levantovsky R, Malle L, Moreira A, Park MD, Pia L, Risson E, Saffern M, Salomé B, Esai Selvan M, Spindler MP, Tan J, van der Heide V, Gregory JK, Alexandropoulos K, Bhardwaj N, Brown BD, Greenbaum B, Gümüş ZH, Homann D, Horowitz A, Kamphorst AO, Curotto de Lafaille MA, Mehandru S, Merad M, Samstein RM, Sinai Immunology Review Project. 2020. Immunology of COVID-19: current state of the science. Immunity 52:910–941. 10.1016/j.immuni.2020.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Bailly C, Waring MJ. 2019. Pharmacological effectors of GRP78 chaperone in cancers. Biochem Pharmacol 163:269–278. 10.1016/j.bcp.2019.02.038. [DOI] [PubMed] [Google Scholar]
- 193.Bhattacharjee R, Devi A, Mishra S. 2015. Molecular docking and molecular dynamics studies reveal structural basis of inhibition and selectivity of inhibitors EGCG and OSU-03012 toward glucose regulated protein-78 (GRP78) overexpressed in glioblastoma. J Mol Model 21:272. 10.1007/s00894-015-2801-3. [DOI] [PubMed] [Google Scholar]
- 194.Booth L, Cazanave SC, Hamed HA, Yacoub A, Ogretmen B, Chen CS, Grant S, Dent P. 2012. OSU-03012 suppresses GRP78/BiP expression that causes PERK-dependent increases in tumor cell killing. Cancer Biol Ther 13:224–236. 10.4161/cbt.13.4.18877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Booth L, Roberts JL, Ecroyd H, Tritsch SR, Bavari S, Reid SP, Proniuk S, Zukiwski A, Jacob A, Sepulveda CS, Giovannoni F, Garcia CC, Damonte E, Gonzalez-Gallego J, Tunon MJ, Dent P. 2016. AR-12 inhibits multiple chaperones concomitant with stimulating autophagosome formation collectively preventing virus replication. J Cell Physiol 231:2286–2302. 10.1002/jcp.25431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Mohr EL, McMullan LK, Lo MK, Spengler JR, Bergeron E, Albarino CG, Shrivastava-Ranjan P, Chiang CF, Nichol ST, Spiropoulou CF, Flint M. 2015. Inhibitors of cellular kinases with broad-spectrum antiviral activity for hemorrhagic fever viruses. Antiviral Res 120:40–47. 10.1016/j.antiviral.2015.05.003. [DOI] [PubMed] [Google Scholar]
- 197.Chan JF, Zhu Z, Chu H, Yuan S, Chik KK, Chan CC, Poon VK, Yip CC, Zhang X, Tsang JO, Zou Z, Tee KM, Shuai H, Lu G, Yuen KY. 2018. The celecoxib derivative kinase inhibitor AR-12 (OSU-03012) inhibits Zika virus via down-regulation of the PI3K/Akt pathway and protects Zika virus-infected A129 mice: a host-targeting treatment strategy. Antiviral Res 160:38–47. 10.1016/j.antiviral.2018.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Chen HH, Chen CC, Lin YS, Chang PC, Lu ZY, Lin CF, Chen CL, Chang CP. 2017. AR-12 suppresses dengue virus replication by down-regulation of PI3K/AKT and GRP78. Antiviral Res 142:158–168. 10.1016/j.antiviral.2017.02.015. [DOI] [PubMed] [Google Scholar]
- 199.Hassandarvish P, Oo A, Jokar A, Zukiwski A, Proniuk S, Abu Bakar S, Zandi K. 2017. Exploring the in vitro potential of celecoxib derivative AR-12 as an effective antiviral compound against four dengue virus serotypes. J Antimicrob Chemother 72:2438–2442. 10.1093/jac/dkx191. [DOI] [PubMed] [Google Scholar]
- 200.Yang CF, Gopula B, Liang JJ, Li JK, Chen SY, Lee YL, Chen CS, Lin YL. 2018. Novel AR-12 derivatives, P12-23 and P12-34, inhibit flavivirus replication by blocking host de novo pyrimidine biosynthesis. Emerg Microbes Infect 7:187. 10.1038/s41426-018-0191-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Booth L, Roberts JL, Cash DR, Tavallai S, Jean S, Fidanza A, Cruz-Luna T, Siembiba P, Cycon KA, Cornelissen CN, Dent P. 2015. GRP78/BiP/HSPA5/Dna K is a universal therapeutic target for human disease. J Cell Physiol 230:1661–1676. 10.1002/jcp.24919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Park JG, Avila-Perez G, Nogales A, Blanco-Lobo P, de la Torre JC, Martinez-Sobrido L. 2020. Identification and characterization of novel compounds with broad-spectrum antiviral activity against influenza A and B viruses. J Virol 94:e02149-19. 10.1128/JVI.02149-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Rayner JO, Roberts RA, Kim J, Poklepovic A, Roberts JL, Booth L, Dent P. 2020. AR12 (OSU-03012) suppresses GRP78 expression and inhibits SARS-CoV-2 replication. Biochem Pharmacol 182:114227. 10.1016/j.bcp.2020.114227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Elfiky AA. 2020. Natural products may interfere with SARS-CoV-2 attachment to the host cell. J Biomol Struct Dyn 39:3194–3203. 10.1080/07391102.2020.1761881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Allam L, Ghrifi F, Mohammed H, El Hafidi N, El Jaoudi R, El Harti J, Lmimouni B, Belyamani L, Ibrahimi A. 2020. Targeting the GRP78-dependant SARS-CoV-2 cell entry by peptides and small molecules. Bioinform Biol Insights 14:1177932220965505. 10.1177/1177932220965505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Carlos AJ, Ha DP, Yeh DW, Van Krieken R, Tseng CC, Zhang P, Gill P, Machida K, Lee AS. 2021. The chaperone GRP78 is a host auxiliary factor for SARS-CoV-2 and GRP78 depleting antibody blocks viral entry and infection. J Biol Chem 296:100759. 10.1016/j.jbc.2021.100759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Alonzi DS, Scott KA, Dwek RA, Zitzmann N. 2017. Iminosugar antivirals: the therapeutic sweet spot. Biochem Soc Trans 45:571–582. 10.1042/BST20160182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Sadat MA, Moir S, Chun TW, Lusso P, Kaplan G, Wolfe L, Memoli MJ, He M, Vega H, Kim LJY, Huang Y, Hussein N, Nievas E, Mitchell R, Garofalo M, Louie A, Ireland DC, Grunes C, Cimbro R, Patel V, Holzapfel G, Salahuddin D, Bristol T, Adams D, Marciano BE, Hegde M, Li Y, Calvo KR, Stoddard J, Justement JS, Jacques J, Priel DAL, Murray D, Sun P, Kuhns DB, Boerkoel CF, Chiorini JA, Di Pasquale G, Verthelyi D, Rosenzweig SD. 2014. Glycosylation, hypogammaglobulinemia, and resistance to viral infections. N Engl J Med 370:1615–1625. 10.1056/NEJMoa1302846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Evans DeWald L, Starr C, Butters T, Treston A, Warfield KL. 2020. Iminosugars: a host-targeted approach to combat Flaviviridae infections. Antiviral Res 184:104881. 10.1016/j.antiviral.2020.104881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Warfield KL, Alonzi DS, Hill JC, Caputo AT, Roversi P, Kiappes JL, Sheets N, Duchars M, Dwek RA, Biggins J, Barnard D, Shresta S, Treston AM, Zitzmann N. 2020. Targeting endoplasmic reticulum alpha-glucosidase I with a single-dose iminosugar treatment protects against lethal influenza and dengue virus infections. J Med Chem 63:4205–4214. 10.1021/acs.jmedchem.0c00067. [DOI] [PubMed] [Google Scholar]
- 211.Warfield KL, Warren TK, Qiu X, Wells J, Mire CE, Geisbert JB, Stuthman KS, Garza NL, Van Tongeren SA, Shurtleff AC, Agans KN, Wong G, Callahan MV, Geisbert TW, Klose B, Ramstedt U, Treston AM. 2017. Assessment of the potential for host-targeted iminosugars UV-4 and UV-5 activity against filovirus infections in vitro and in vivo. Antiviral Res 138:22–31. 10.1016/j.antiviral.2016.11.019. [DOI] [PubMed] [Google Scholar]
- 212.Warfield KL, Schaaf KR, DeWald LE, Spurgers KB, Wang W, Stavale E, Mendenhall M, Shilts MH, Stockwell TB, Barnard DL, Ramstedt U, Das SR. 2019. Lack of selective resistance of influenza A virus in presence of host-targeted antiviral, UV-4B. Sci Rep 9:7484. 10.1038/s41598-019-43030-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Bhushan G, Lim L, Bird I, Chothe SK, Nissly RH, Kuchipudi SV. 2020. Iminosugars with endoplasmic reticulum alpha-glucosidase inhibitor activity inhibit ZIKV replication and reverse cytopathogenicity in vitro. Front Microbiol 11:531. 10.3389/fmicb.2020.00531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Ma J, Zhang X, Soloveva V, Warren T, Guo F, Wu S, Lu H, Guo J, Su Q, Shen H, Solon E, Comunale MA, Mehta A, Guo JT, Bavari S, Du Y, Block TM, Chang J. 2018. Enhancing the antiviral potency of ER alpha-glucosidase inhibitor IHVR-19029 against hemorrhagic fever viruses in vitro and in vivo. Antiviral Res 150:112–122. 10.1016/j.antiviral.2017.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Miller JL, Hill ML, Brun J, Pountain A, Sayce AC, Zitzmann N. 2019. Iminosugars counteract the downregulation of the interferon gamma receptor by dengue virus. Antiviral Res 170:104551. 10.1016/j.antiviral.2019.104551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Zhao X, Guo F, Comunale MA, Mehta A, Sehgal M, Jain P, Cuconati A, Lin H, Block TM, Chang J, Guo JT. 2015. Inhibition of endoplasmic reticulum-resident glucosidases impairs severe acute respiratory syndrome coronavirus and human coronavirus NL63 spike protein-mediated entry by altering the glycan processing of angiotensin I-converting enzyme 2. Antimicrob Agents Chemother 59:206–216. 10.1128/AAC.03999-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Clarke EC, Nofchissey RA, Ye C, Bradfute SB. 2020. The iminosugars celgosivir, castanospermine and UV-4 inhibit SARS-CoV-2 replication. Glycobiology 31:378–384. 10.1093/glycob/cwaa091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Korba BE, Montero AB, Farrar K, Gaye K, Mukerjee S, Ayers MS, Rossignol JF. 2008. Nitazoxanide, tizoxanide and other thiazolides are potent inhibitors of hepatitis B virus and hepatitis C virus replication. Antiviral Res 77:56–63. 10.1016/j.antiviral.2007.08.005. [DOI] [PubMed] [Google Scholar]
- 219.La Frazia S, Ciucci A, Arnoldi F, Coira M, Gianferretti P, Angelini M, Belardo G, Burrone OR, Rossignol JF, Santoro MG. 2013. Thiazolides, a new class of antiviral agents effective against rotavirus infection, target viral morphogenesis, inhibiting viroplasm formation. J Virol 87:11096–11106. 10.1128/JVI.01213-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Tilmanis D, van Baalen C, Oh DY, Rossignol JF, Hurt AC. 2017. The susceptibility of circulating human influenza viruses to tizoxanide, the active metabolite of nitazoxanide. Antiviral Res 147:142–148. 10.1016/j.antiviral.2017.10.002. [DOI] [PubMed] [Google Scholar]
- 221.Rawarak N, Suttitheptumrong A, Reamtong O, Boonnak K, Pattanakitsakul SN. 2019. Protein disulfide isomerase inhibitor suppresses viral replication and production during antibody-dependent enhancement of dengue virus infection in human monocytic cells. Viruses 11:155. 10.3390/v11020155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Ozcelik D, Seto A, Rakic B, Farzam A, Supek F, Pezacki JP. 2018. Gene expression profiling of endoplasmic reticulum stress in hepatitis C virus-containing cells treated with an inhibitor of protein disulfide isomerases. ACS Omega 3:17227–17235. 10.1021/acsomega.8b02676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Paxman R, Plate L, Blackwood EA, Glembotski C, Powers ET, Wiseman RL, Kelly JW. 2018. Pharmacologic ATF6 activating compounds are metabolically activated to selectively modify endoplasmic reticulum proteins. Elife 7:e37168. 10.7554/eLife.37168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Almasy KM, Davies JP, Lisy SM, Tirgar R, Tran SC, Plate L. 2021. Small-molecule endoplasmic reticulum proteostasis regulator acts as a broad-spectrum inhibitor of dengue and Zika virus infections. Proc Natl Acad Sci USA 118:e2012209118. 10.1073/pnas.2012209118. [DOI] [PMC free article] [PubMed] [Google Scholar]