Abstract
Realistic lesion generation is a useful tool for system evaluation and optimization. In this work, we investigate a data-driven approach for categorical lung lesion generation using public lung CT databases. We propose a generative adversarial network with a Wasserstein discrimination and gradient penalty to stabilize training. We further included conditional inputs such that the network can generate user-specified lesion categories. Novel to our network, we directly incorporated radiomic features in an intermediate supervision step to encourage similar textures between generated and real lesions. We evaluated the network using lung lesions from the Lung Image Database Consortium (LIDC) database. The lesions are divided into two categories: solid vs. non-solid. We performed quantitative evaluation of network performance base on four criteria: 1) overfitting in terms of structural and morphological similarity to the training data, 2) diversity of generated lesions in terms of similarity to other generated data, 3) similarity to real lesions in terms of distribution of example radiomics features, and 4) conditional consistency in terms of classification accuracy using a classifier trained on the training lesions. We imposed a quantitative threshold for similarity based on visual inspection. The percentage of non-solid and solid lesions that satisfy low overfitting and high diversity is 96.9% and 88.6% of non-solid and solid lesions respectively. The distribution of example radiomics features are similar in the generated and real lesions indicated by a low Kullback–Leibler divergence score. Classification accuracy for the generated lesions are comparable with that for the real lesions. The proposed network is a promising approach for data-driven generation of realistic lung lesions.
Keywords: lesion generation, virtual clinical trial, deep learning, generative adversarial network
1. INTRODUCTION
Realistic lesion generation is a useful tool for system evaluation and optimization. Generated lesions can serve as realistic imaging tasks for task-base image quality assessment, as well as targets in virtual clinical trials. Virtual clinical trial (VCT) in particular has been receiving increasing attention as an efficient and cost-effective tool for the preclinical assessment of imaging systems. Within the VCT workflow, realistic lesion generation is a critical element. Not only should the lesions capture realistic morphology and texture, but should also systematically represent different categories of lesion types and diagnostic features.
Traditionally, lesion generation has mostly relied on procedural approaches. In this work, we propose a data-driven approach which leverages the increasing availability of large-scale patient databases and enables rich feature discovery unconstrained by a pre-selected set of basis functions. In particular, we develop a novel network architecture based on generative adversarial network (GAN). We directly incorporated radiomics features in the training process via an intermeidate supervision step. We additionally combined several state-of-the-art elements to improve stability and convergence of the network. We implemented the network to conditionally generate solid and non-solid lung lesions, and presented quantitative evaluation of network performance.
2. METHOD
2.1. Network Architecture
As illustrated in Fig.1, the network employs a GAN architecture where the generator G learns a mapping from samples from a prior distribution z′ ~ p(z′) to the real distribution x′ ~ p(x′). We included the following extensions to a basic GAN:
Figure 1:
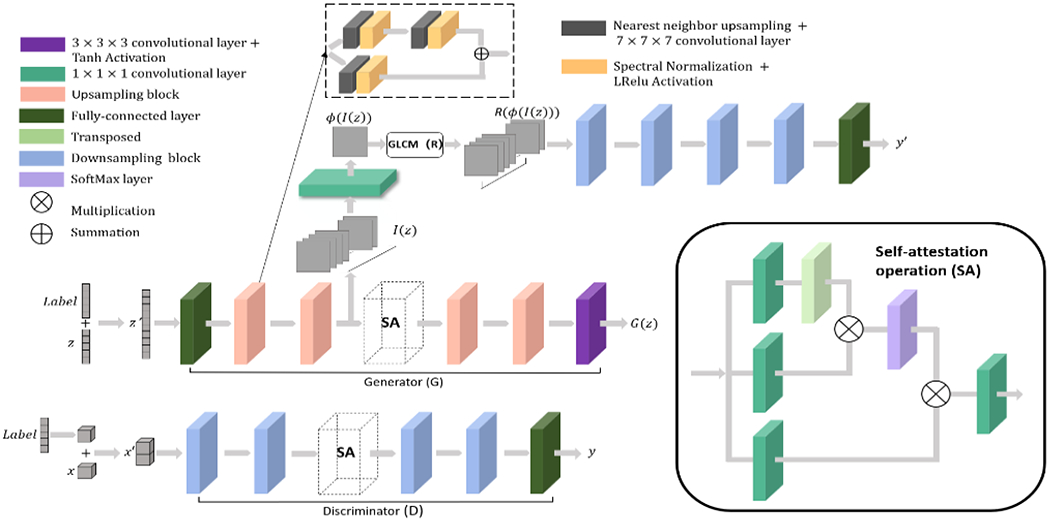
Network structure: The GAN network proposed adopts a Wasserstein discriminator with gradient penalty. Self-attention layers are included in both the generator and discriminator to model long-range dependencies in the image. The radiomics supervision network is included after the first half of the generator to encourage texture similarity between generated and real lesions. Input conditions are encoded in a one-hot label vector and appended to the inputs of both the generator and discriminator. Upsampling and down-sampling ratios are 2. We adopted the RMSprop optimizer with an initial learning rate of 0.0001. The learning rate decays to 0.9 of its value every 20 epochs. Batch size is 8 and a total of 1200 epochs were used in training.
2.1.1. Wassertein discriminator with gradient penalty
To improve training stability, we implemented a Wasserstein GAN where the discriminator D computes the Wasserstein-1 distance y between the synthetic samples G(z′) and the real samples x′. We additionally included a gradient penalty1 in the objective function to constrain the discriminator to be K-Lipschitz. The objective function is given by:
| (1) |
where γ is the penalty coefficient empirically chosen to be 10, and is a random mixture uniformly sampled from the pairs of real and synthetic images.
2.1.2. Conditional GAN
To enable lesion generation in different categories, we included lesion category (in this work, solid vs non-solid) as a one-hot encoded label vector.2 The label vector is concatenated with input z to the generator, and input x to the discriminator.
2.1.3. Self-attention layers
To model long-range dependencies across image regions, we implement one SA layer3 in both the generator and the discriminator. The 3D SA layer performs the following operation:
| (2) |
where ys is the output and xs is the input of the SA layer, S indicates Softmax operation. Wf, Wθ, Wϕ and Wg are trainable 1x1x1 convolution layers. γ is a trainable weight for the non-local responses.
2.1.4. Intermediate supervision using radiomics features
Novel to our network, we directly included radiomics features in an intermediate supervision step. Intermediate supervision was originally proposed for convolutional neural networks to speed up convergence and alleviate vanishing gradient.4 As shown in Fig.1, the intermediate supervision network effectively serves as a discriminator for the first half of the generator to encourage similar textures between the generated and real lesions. The objective function for this block is given by:
| (3) |
where is the radiomics feature, chosen here as the homogeneity feature computed from the gray-level co-occurance matrix (GLCM-Homongeneity), computed for a 1-pixel offset at 13 angles (i.e., all adjacent pixel neighbors in 3D); is the radiomics computed from a randomly chosen pairs of real lesions and intermediate outputs. For each iteration, training is divided into two stages, first for the radiomics supervision network according to the objective function in Eq.3, and second for the complete generator and discriminator using the objective function in Eq.1.
2.2. Training Data and Preprocessing
For training data, we used segmented 3D lung lesions from the Lung Image Database Consortium (LIDC) database.5 For this work, we aim to conditionally generate lesions of different textures. Therefore, we used the “Texture” ratings provided in the LIDC and divided all lesions into two categories: non-solid (by aggregating Texture ratings 1 to 3), and solid (Texture ratings 4 and 5). We excluded lesions smaller than 8x8x8, since clinical assessment of textures are only performed for larger lesions. Each lesion volume is centered and resized to 32x32x32 by cubic interpolation. For training, all voxel values are normalized between −1 and 1. A common normalization scheme is used for the entire dataset so that Hounsfield numbers can be recovered post training.
2.3. Performance Evaluation
We evaluate four aspects of network performance: overfitting, diversity of generated lesions, statistical similarity between generated and real lesions, and conditional consistency of generated lesion categories. A total of 640 lesions were generated for performance evaluation.
Overfitting
We first analyze whether generated lesions are as a result of overfitting to the training data. Each generated lesion was compared with each training lesion in terms of their shape similarity (in terms of the Dice score) and structural similarity (in terms of the Multi-scale Structural Similarity Index, MS-SSIM). An empirical threshold was chosen for each score based on visual inspection to exclude lesions deemed too similar to the training lesions.
Diversity
GANs are known to suffer from mode collapse where different noise inputs produce similar outputs. To assess the extent of mode collapse, the remaining lesions were analyzed for diversity, i.e., whether generated lesions are unique. Each generated lesion was compared with every other generated lesion in terms of the Dice score and MS-SSIM. A threshold was similarly chosen to select unique lesions.
Statistical similarity to real lesions
Lesion passing the first two tests were then evaluated on whether they are drawn from the same statistical distribution as the real lesions. We computed the distributions of several radiomics features (GLCM-homogeneity, contrast, energy,) of generated lesions and compared with those for real lesions. Similarity between the distributions were assessed using the Kullback–Leibler divergence(KL).
Conditional consistency
Lastly, we assess whether the lesions were correctly generated to their respective categories. To highlight the effect of the labels, we generated lesions belonging to both categories using the same noise realization by only changing the label vector. We deploy MATLAB library to trained a L1 soft-margin Support Vector Machine (SVM) to classify the training lesions as solid vs. non-solid based on 38 radiomics features, including features from GLCM, Gray-Level Run-Length Matrix (GLRLM), Gray-Level Size Zone Matrix (GLSZM) and three more global features: Skewness, Variance and Kurtosis. The SVM is trained with radial basis function kernel and optimized by quadratic programming. It is applied to the generated lesions to test whether they can be correctly categorized.
3. RESULT
Eight example generated lesions in each category is shown in Fig.2. Quantitative analysis results in Sec.2.3 is summarized in Table 1.
Figure 2:
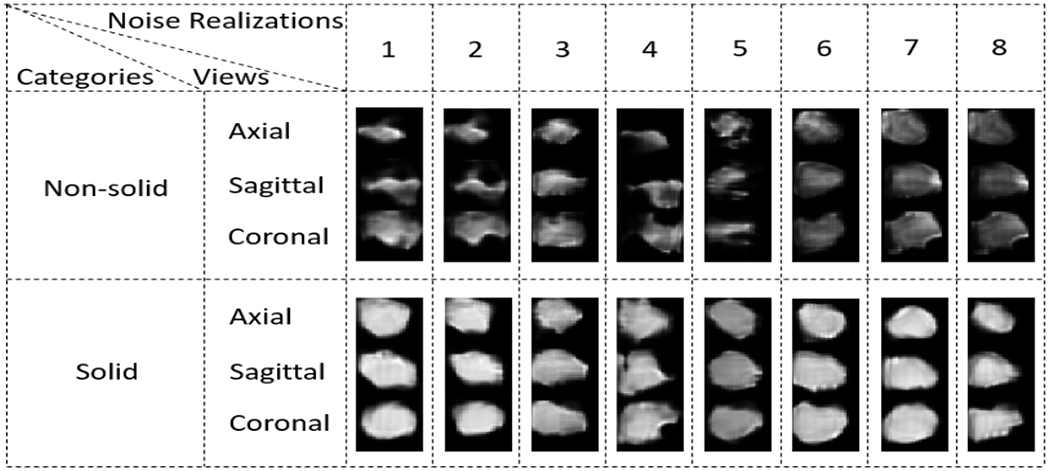
Example lesions from both non-solid and solid categories.
Table 1:
Quantitative analysis: Table shows statistics for the overfitting analysis, the diversity analysis, the conditional consistency analysis and the overall statistical consistency. And a percentage of the high-quality synthetic lesion is presented.
| Overfitting Statistics | Diversity Analysis | Condition Analysis | Statistical Analysis | High-quality lesion (%) | |||||
|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||
| MS-SSIM | Dice | New lesion (%) | MS-SSIM | Dice | Unique lesion (%) | Match expected condition (%) | Empirical KL divergence | ||
| Non-solid | 0.75 | 0.62 | 100 | 0.91 | 0.81 | 96.9 | 94.8 | 2.05 | 92.7 |
| Solid | 0.78 | 0.75 | 100 | 0.92 | 0.87 | 88.6 | 92.7 | 5.92 | 84.2 |
Overfitting
The distribution of Dice score and MS-SSIM for solid and non-solid lesions are shown in Fig.4. From visual inspection, the thresholds separating similar and dissimilar lesions are set to 0.9567 for the MS-SSIM and 0.9175 for the Dice score, where the justification is shown in Fig.3. Out of 640 total lesions, 100% of non-solid lesions and solid lesions satisfy both criteria simultaneously and are therefore considered sufficiently different from the training data.
Figure 4:
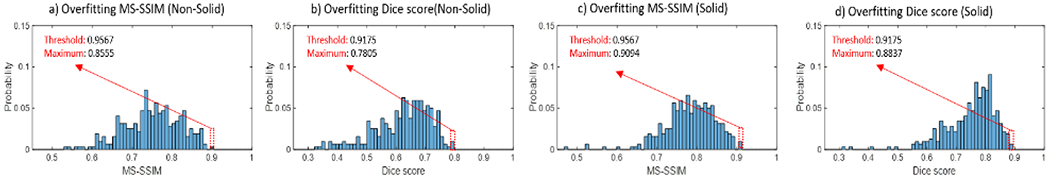
Overfitting: The maximum MS-SSIM and Dice score between generated lesions and each real lesions are shown for both the solid and non-solid categories. The red dotted bars indicate the location of the threshold below which lesions are considered dissimilar.
Figure 3:
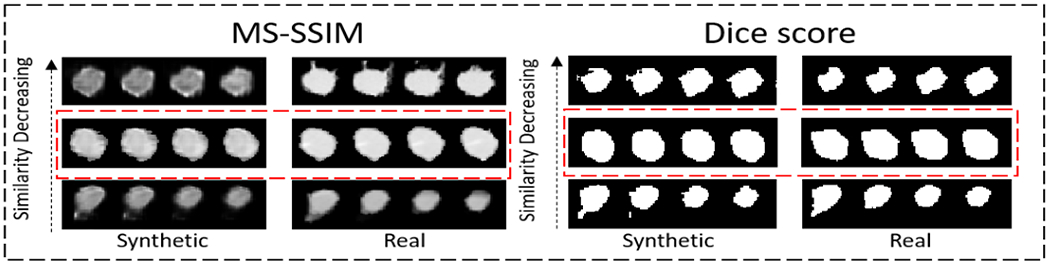
Quantitative thresholds for the MS-SSIM metric and Dice score were chosen based on visual inspection. Example lesions that, are considered dissimilar (below the threshold, top row, and at the threshold, middle row) and similar (above the threshold, bottom row) are plotted. In both (a) and (b), The left column shows four axial slices through a generated lesion volume, while the right column shows the same for a real lesion.
Diversity
The distribution of Dice score and MS-SSIM for solid and non-solid lesions are shown in Fig.5. Using the same thresholds determined previously, 96.9% non-solid lesions and 88.6% solid lesions are considered unique. We observed lower diversity in solid lesions, which indicates a partial mode collapse (the network produces limited varieties of samples) for solid lesion generation. This is likely due to insufficient data for solid lesions: in empirical experiments, we tend to see more mode collapse with a small dataset with relative large variance (see Fig. 7). This behavior can be mitigated with more training data.
Figure 5:
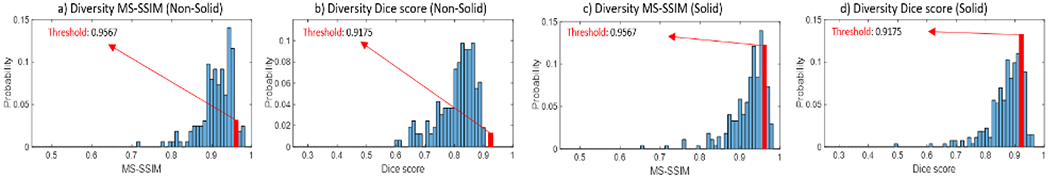
Diversity: The maximum MS-SSIM find Dice score between each generated lesions and every other generated lesions are shown for both the solid and non-solid categories. Red bars indicates the thresholds below which lesions are considered unique.
Figure 7:
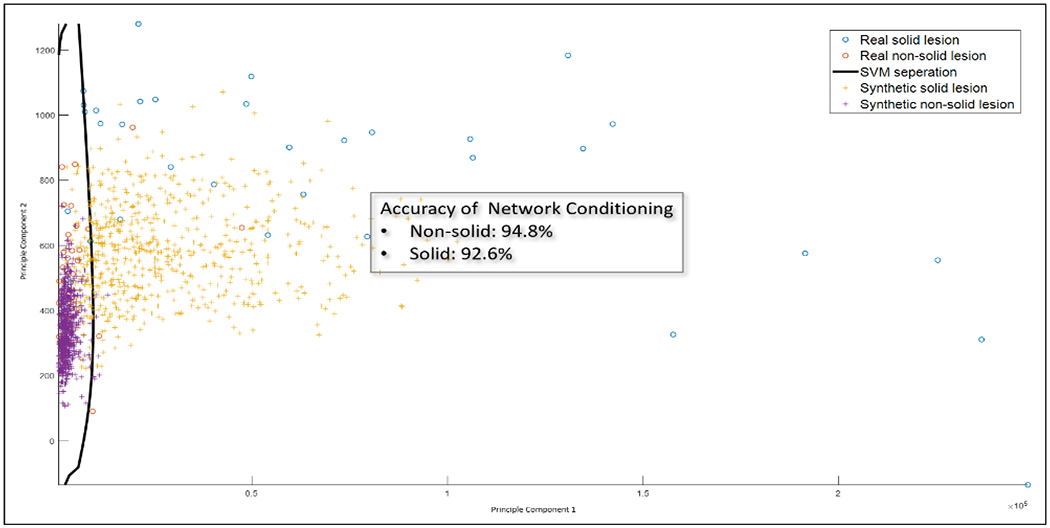
Conditional consistency: The SVM classifier trained to classify solid vs non-solid lesions. The classifier were trained on real lesions and applied to generated lesions. The classification accuracy is comparable between real and generated lesions, indicating the conditional network could produce lesions belong to distinct categories.
Statistical similarity to real lesions
The distributions of example radiomics features computed for the generated lesions are overlaid with those for the real lesions. For non-solid lesions, high similarity (indicated by low KL score) is observed for all four radiomics features. On the other hand, solid lesions have lower similarity, possibly attributed to the partial mode collapse mentioned previously.
Conditional consistency
For visualization purpose, we apply Principle Component Analysis (PCA) to the features and only plot the first two components, which together can explain 99.98% variance. The SVM classifier trained on the real lesions is capable of achieving 85.7% accuracy for non-solid lesions and 88.6% for solid lesions. When applied to the generated lesions, classification accuracy is 94.8% for non-solid lesions and 92.7% for solid lesions.
Combining all quantitative assessment above, the percentage of lesions that pass all criteria are 92.7% and 84.2% for non-solid and solid lesions, respectively.
4. CONCLUSION
In this work, we present a novel GAN with radiomics supervision for 3D lung lesion generation. The network structure has shown capability of conditionally generating user-specified lesion categories that exhibit low levels of overfitting and high intra-condition diversity. Ongoing and future work will focus on expanding lesion categories beyond texture to include features such as lesion shapes. We will further evaluate the generated lesions through an observer study involving radiologists.
Figure 6:
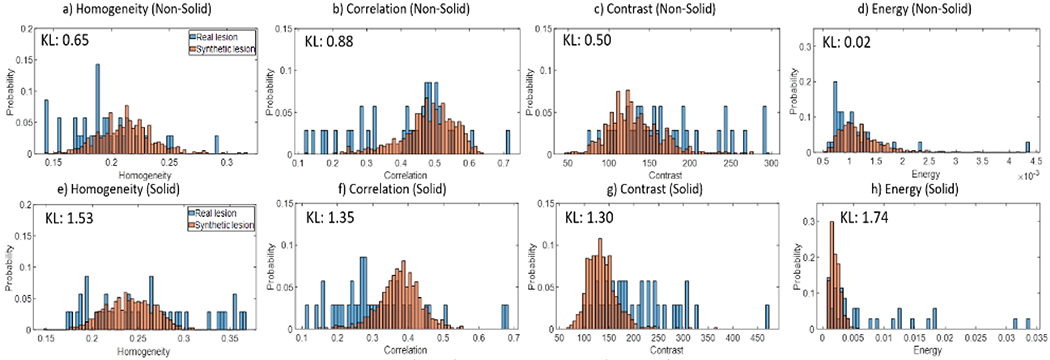
Example radiomics features for real (blue) and generated (orange) lesions are plotted for comparison. The histograms should have high similarity if lesion generation faithfully reproduce the range of texture features of real lesions. Generated non-solid lesions are able to achieve high similarity and low KL score with real lesions, while solid lesions perform worse, possibly due to partial mode collapse during training.
5. ACKNOWLEDGEMENT
This paper is supported, in part, by NIH grants R21CA219608 and R01CA249538.
REFERENCES
- [1].Gulrajani I, Ahmed F, Arjovsky M, Dumoulin V, and Courville A, “Improved training of wasserstein gans,” (2017). [Google Scholar]
- [2].Mirza M and Osindero S, “Conditional generative adversarial nets,” (2014). [Google Scholar]
- [3].Zhang H, Goodfellow I, Metaxas D, and Odena A, “Self-attention generative adversarial networks,” (2018). [Google Scholar]
- [4].Li C, Zia MZ, Tran Q-H, Yu X, Hager GD, and Chandraker M, “Deep supervision with intermediate concepts,” IEEE transactions on pattern analysis and machine intelligence 41(8), 1828–1843 (2018). [DOI] [PubMed] [Google Scholar]
- [5].Armato SG III, McLennan G, Bidaut L, McNitt-Gray MF, Meyer CR, Reeves AP, Zhao B, Aberle DR, Henschke CI, Hoffman EA, et al. , “The lung image database consortium (lidc) and image database resource initiative (idri): a completed reference database of lung nodules on ct scans,” Medical physics 38(2), 915–931 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]


