Abstract
The neu (c-erbB-2) proto-oncogene encodes a tyrosine kinase receptor that is overexpressed in 20 to 30% of human breast tumors. Herein, cyclin D1 protein levels were increased in mammary tumors induced by overexpression of wild-type Neu or activating mutants of Neu in transgenic mice and in MCF7 cells overexpressing transforming Neu. Analyses of 12 Neu mutants in MCF7 cells indicated important roles for specific C-terminal autophosphorylation sites and the extracellular domain in cyclin D1 promoter activation. Induction of cyclin D1 by NeuT involved Ras, Rac, Rho, extracellular signal-regulated kinase, c-Jun N-terminal kinase, and p38, but not phosphatidylinositol 3-kinase. NeuT induction of the cyclin D1 promoter required the E2F and Sp1 DNA binding sites and was inhibited by dominant negative E2F-1 or DP-1. Neu-induced transformation was inhibited by a cyclin D1 antisense or dominant negative E2F-1 construct in Rat-1 cells. Growth of NeuT-transformed mammary adenocarcinoma cells in nude mice was blocked by the cyclin D1 antisense construct. These results demonstrate that E2F-1 mediates a Neu-signaling cascade to cyclin D1 and identify cyclin D1 as a critical downstream target of neu-induced transformation.
The neu (c-erbB-2, HER-2) proto-oncogene encodes a receptor tyrosine kinase that is a member of a growth factor receptor family, which includes the epidermal growth factor (EGF) receptor (ErbB-1), ErbB-3, and ErbB-4. neu is overexpressed in 20 to 30% of human breast tumors (64). Both Neu and the EGF receptor stimulate proliferation of breast cancer cells, and overexpression of these two proteins correlates with progression of human breast cancer and poor patient prognosis (28, 31, 47). A substitution point mutation at residue 664 (Val→Glu) in the transmembrane domain of rat Neu (referred to as NeuT) encodes an activated transforming tyrosine kinase (7). Overexpression of either wild-type Neu or NeuT in transgenic mice under the control of the murine mammary tumor virus (MMTV) long terminal repeat induces mammary adenocarcinoma with high frequency (25, 41). Several independent transgenic strains bearing the identical MMTV-neuT transgene developed synchronous, multifocal mammary tumors involving all mammary glands (24), providing strong evidence that activated neu requires few if any additional genetic events to transform the epithelial cell.
In mammary tumors of mice transgenic for the wild-type Neu receptor (MMTV-neu mice), the receptor's intrinsic tyrosine kinase activity was increased in association with in-frame somatic mutations of the transgene (61). Introduction of these extracellular domain deletion (ECD) mutations into the wild-type Neu cDNA enhanced neu transforming potential (61). Transgenic mice expressing these Neu deletion mutants in the mammary gland (MMTV-NDL mice) developed multifocal mammary adenocarcinomas with high frequency and shorter latency compared with mice transgenic for the wild-type neu. In primary human breast tumors, a splice variant of ErbB-2 encoding a similar ECD deletion which can also transform Rat-1 fibroblasts has been detected, suggesting that such activating mutants of ErbB-2 may similarly have a critical role in human breast cancer induction and progression (62).
The mitogenic activity of the ErbB family, induced by ligand addition, is initiated through the formation of heterodimeric and homodimeric receptor signaling complexes (49). ErbB-2 is capable of heterodimerizing with other family members, and induction of ErbB-2 tyrosine phosphorylation stimulates mitogenesis (21, 49). Overexpression of ErbB-2 results in autophosphorylation and induction of signaling pathways involving Ras and c-Src (27, 43, 51). Mitogenic intracellular signaling mediators induced through ErbB proteins include the p42 and p44 extracellular signal-regulated kinases (ERKs) (5), the stress-activated protein kinases (49), and a wortmannin-sensitive phosphoinositide 3-OH kinase (PI 3-kinase) activity (68). The mitogenic and transforming potential of Neu is well documented and raises the possibility that Neu directly affects components of the cell cycle regulatory apparatus implicated in cellular transformation.
The functional inactivation of the retinoblastoma tumor suppressor protein (pRB) is a common finding in many tumor types (74). Phosphorylation and inactivation of pRB by cyclin-dependent kinase (Cdk) holoenzymes, consisting of D-type cyclins and the catalytic subunits including Cdk4 or Cdk6, block the growth suppressive function of pRB (reviewed in references 57 and 74). Cyclin D1 overexpression in cultured cells hyperphosphorylates pRB (54), and in cells containing pRB, the abundance of cyclin D1 is rate limiting in progression through the first gap (G1) phase of the cell cycle. Aberrant overexpression of cyclin D1 is associated with breast cancer formation, with cyclin D1 mRNA overexpressed in 70 to 100% of breast tumor cell lines (32, 56) and the majority of breast cancers (75). Targeted overexpression of cyclin D1 induced mammary adenocarcinoma (69), and transgenic mice lacking both cyclin D1 alleles failed to develop normal mammary glands (59). These studies, though consistent with a role for cyclin D1 in both oncogenesis and breast development, also demonstrate the limited utility of the cyclin D1−/− animals for analysis of cyclin D1's role in mammary tumorigenesis.
Cyclin D1 abundance is induced transcriptionally, and the protein is degraded rapidly upon the withdrawal of growth factors via the proteasome pathway (20). It has been proposed that induction of cyclin D1 by growth factors and oncogenes may contribute to the transformed phenotype (reviewed in references 57 and 74). Cyclin D1 is induced by several proteins involved in proliferative signaling and transformation, including Ras (1), Rac (76), Src (33), STATs (11, 38), and simian virus 40 (SV40) small t antigen (71). Further, cyclin D1 collaborates with Ras and Myc in tumor formation (10). Transformation by Ras in certain cell types requires the GTPase Rac1 and RhoA (52, 53). Rac induced cyclin D1 (29, 76), while a cyclin D1 antisense construct reduced Ras-induced colony-forming activity (35), suggesting that cyclin D1 is a downstream target of Ras- and perhaps Rac-induced transformation. In the present study, we examined the role of cyclin D1 in neu-induced transformation and identified the intracellular signaling pathway by which NeuT induces cyclin D1.
MATERIALS AND METHODS
Cyclin D1 immune-complex assays.
Cyclin D1 immunoprecipitation kinase assays were performed as previously described (33, 72). Tissues from MMTV-neu and MMTV-NDL transgenic mice (25, 62) were Dounce homogenized in lysis buffer (150 mM NaCl, 50 mM HEPES pH 7.2, 1 mM EDTA, 1 mM EGTA, 1 mM dithiothreitol, 0.1% Tween 20, 0.1 mM phenylmethylsulfonyl fluoride, 2.5 μg of leupeptin per ml, 0.1 mM sodium orthovanadate [Sigma, St. Louis, Mo.]) at 4°C. Lysates (100 μg) were precipitated with protein A-agarose beads precoated with the cyclin D1 antibody DCS-11 (NeoMarkers, Fremont, Calif.). Phosphorylated proteins were separated by electrophoresis and quantified after exposure to autoradiographic film (Labscientific, Inc., Livingston, N.J.) by densitometry using ImageQuant version 1.11 (Molecular Dynamics, Sunnyvale, Calif.).
Western blots.
The abundance of cyclin D1 and Neu proteins in 50 μg of lysate was determined by Western analysis as previously described (33, 72), using a cyclin D1 antibody (DCS-6; NeoMarkers), a c-Neu antibody (Ab-3; Oncogene Research Products, Cambridge, Mass.), a keratin-8 antibody (M20; ICN Biomedicals, Inc., Aurora, Ohio), an α-tubulin antibody (5H1) (13), and a guanine nucleotide dissociation inhibitor (GDI) antibody (a generous gift from Perry Bickel, Washington University, St. Louis, Mo.) (55).
Immunohistochemistry.
Immunostaining of the mammary tissue from seven transgenic animals was performed as previously described (33). In each tumor, 500 cells were scored for nuclear cyclin D1 staining. Tissues were fixed in 4% paraformaldehyde, blocked in paraffin, sectioned at 5 μm, and stained with hematoxylin and eosin or used for immunohistochemistry. Cyclin D1 was detected by using antibody DCS-6 with the Vectastain ABC system (Vector Laboratories, Burlingame, Calif.).
Construction of reporter and expression vectors.
The human cyclin D1 promoter reporter constructions, the c-fos promoter, the (UAS)5E1BTATALUC reporter, and the PALUC reporter, which contains 7 kb of the human cyclin A promoter (1, 33, 70), were previously described. The E2F site of the cyclin D1 promoter was mutated from TTTGGCGCC to TTTcttGaC (mutated bases are in lowercase) in the context of the −163 bp fragment, using PCR to form −163E2FmtCD1LUC. The serum response element from the c-fos promoter from −332 to −277 was linked to the minimal TATA region of the E4 promoter and cloned into the reporter pA3LUC.
The expression vectors encoding Neu (pJ4ΩNeuN and pSV2NeuN), NeuT (pJ4ΩNeuT and pSV2NeuT), the ECD mutants of Neu (8142, 8340, 8342, and 8567) (61), the carboxy-terminal deletion of NeuT (ΔCT), and the ΔCT mutants pLSV P1, P1F, P2,3, P4, P5, and Y1253F (9) were previously described. RSV (Rous sarcoma virus)-RasN17, RSV-RasL61, RSV-RasL61S186 (1), pEXV3N19Rho, pEXV3N17Rac, and the dominant negative MEK1 plasmid pEXVMEKC (MEKAla-218/Ala-222) (52, 71), and c-Jun N-terminal kinase (JNK) inhibitor JIP-1 (JNK-interacting protein 1) (18, 33) were previously described. The cDNAs encoding N17Rac and N19Rho were cloned into the tetracycline-regulated vector pBPSTR-1 (46). The human cyclin D1 cDNA antisense construct from the tetracycline-regulated plasmid pUHD10.3 CD1AS (shown to reduce cyclin D1 protein levels in rat H19-7 cells [79]) was recloned into pBPSTR-1 to form pBPSTR-1CD1AS.
The p16INK4a in vitro expression plasmid was a gift from L. Zhu. The vectors pCMV-E2F-1, pCMV-DC-E2F-1 E132, pCMV-E2F-1-Y411C, pCMV-HA-DP-1, and pCMV-HA-DP-1Δ103-126 were previously described (70, 80). The Sp1, Sp3, and E2F-1 activation domains were linked to the GAL4 DNA binding domain to form GAL4-Sp1(83-621) (63), GAL4-Sp3(1-382), GAL4–E2F-1(368-437), and GAL4–E2F-1(Δ413-417), which is defective in regulation by pRB. GAL4-PAG236 encodes the constitutively active transactivation domain.
Reporter assays and cell culture.
Cell culture, transfections, and luciferase assays were performed as previously described (1). MCF7 cells were maintained in Dulbecco modified Eagle medium (DMEM) with 10% (vol/vol) calf serum and 1% penicillin-streptomycin. Rat-1, MDA-MB-231, MDA-MB-453, HBL-100, and BT-483 cells were grown in DMEM-Ham's F12 (1:1) with 10% calf serum and 1% penicillin-streptomycin. The MCF7 stable cell line −1745CD1LUC was established by selection with G418 in the presence of pcDNA3. The NIH 3T3 stable cell lines DHFR/G8 (overexpressing wild-type neu), B104-1-1, and neuΔC-1 have been described elsewhere (78). The NAFA cell line from the MMTV-neuT mouse (41) was grown in DMEM with 10% fetal bovine serum and 1% penicillin-streptomycin.
In transient expression studies, cells were transfected by calcium phosphate precipitation, the medium was changed after 6 h, and luciferase activity was determined after another 24 h. The effect of an expression vector was compared with that of an equal amount of empty vector. Treatments with PD098059 (10 to 20 μM), SB203580 (10 to 20 μM), wortmannin (20 to 100 nM), and rapamycin (100 pM to 50 nM) were performed for 24 h and compared with dimethyl sulfoxide (DMSO) vehicle treatment. Luciferase content was measured during the initial 10 s of the reaction with an AutoLumat LB953 (EG&G Berthold), and the values were expressed in arbitrary light units (72). Statistical analyses were performed by using the Mann-Whitney U test, with significant differences established as P < 0.05.
Oligodeoxyribonucleotides.
The oligonucleotide sequences used in electrophoretic mobility shift assays (EMSAs) were as follows: for the adenovirus E2F site, 5′ GCC GTC CAG TTT CGC GCC CTT TCT CAA ATT TAA GCA GCT CGA; for the cyclin D1 E2F site, 5′ TCC CGG CGT TTG GCG CCC GCG CCC; for the cyclin D1 Sp1 site (−130 to −99), 5′ TCC CCC TGC GCC CGC CCC CGC CCC CCT CCC GC; and for the consensus Sp1 site, 5′ ATT CGA TCG GGG CGG GGC GAG C. For the mutant cyclin D1 E2F oligonucleotide, the wild-type TTT GGC GCC CG sequence was changed to cga Gct GCC CG. The sequence of the primer used to mutate the cyclin D1 E2F site (TTTGGCGCC) was 5′ GGT ACC TCG CTG CTC CCG GCG TTT ctt gaC CGC G. (Mutated bases are shown in lowercase.) The primers used for PCR in chromatin immunoprecipitation (CHIP) assays were −349CD1 (5′ CTC CAC CTC ACC CCC TAA AT) and −112CD1 (5′ GGG GGC GGG CGC AGG GGG A). PCR primers for neu from the NAFA cell tumors were 5′ CGG AAC CCA CAT CAG GCC and 5′ TTT CCT GCA GCC TAC GC.
EMSAs.
EMSAs using the Sp1 sequence with MCF7 nuclear extracts were performed as described previously (1, 72). Extracts were incubated in a mixture containing 20 mM HEPES (pH 7.9), 80 mM KCl, 5 mM MgCl2, 0.2 mM dithiothreitol, 2% Ficoll, 5% glycerol, 0.1 mM EDTA, and 500 ng of poly(dI-dC) on ice for 30 min. 32P-labeled oligonucleotides (100 fmol, 50,000 cpm) were added to the reaction mix and incubated at room temperature for 30 min. The protein-DNA complexes were electrophoresed through a 5% polyacrylamide gel with 0.5× TBE (0.5× TBE is 0.045 M Tris-borate plus 0.001 M EDTA) buffer and 2.5% glycerol. The gels were dried and exposed to autoradiographic film.
EMSAs using the E2F sequences were performed essentially as previously described (66). Sf9 cells were infected with equal amounts (PFU) of baculoviruses encoding E2F-1, E2F-4, DP-1, or DP-2. Equal amounts of 32P-labeled probes (10,000 to 20,000 cpm) were incubated with equal volumes of extracts from baculovirus-infected Sf9 cells. Antisera to E2F and DP proteins (Santa Cruz Biotechnology, Inc., Santa Cruz, Calif.) were used to confirm the components of each protein-DNA complex (not shown). To determine relative binding activities of the individual complexes, radioactivity in each complex was calculated with a phosphorimager (Packard Instant Imager), and the counts in the cyclin D1 complexes were normalized to the value for the corresponding adenovirus E2F complexes. The mean of two independent experiments was used for each complex.
CHIP.
The CHIP method was described elsewhere (65). Briefly, 2 × 107 MCF7 cells were fixed by addition of formaldehyde to the tissue culture media (final concentration, 1%). Isolated chromatin was sonicated to an average length of 0.5 to 1 kb and treated with 1 μg of rabbit anti-E2F-1 antibody (C-20; Santa Cruz Biotechnology) or control rabbit immunoglobulin G (IgG) for 16 h at 4°C. The complexes were immunoprecipitated with 10 μl of blocked Staph A cells and washed with immunoprecipitation buffer (100 mM Tris-Cl [pH 9.0], 500 mM LiCl, 1% NP-40, 1% deoxycholate). After elution and reversal of cross-links, DNA was isolated and analyzed by PCR; 10 ng of −1745CD1LUC plasmid DNA, sterile water, and immunoprecipitation buffer were included as controls. PCR products were visualized on a 2% agarose gel with ethidium bromide.
Transformation assays.
Transformation assays were conducted exactly as previously described (16). Rat-1 cells (105/60-mm-diameter dish) were transfected by calcium phosphate precipitation, with a 45-s glycerol shock performed after 5 to 8 h. After 3 days, the cells were trypsinized and passed into 100-mm-diameter dishes. The medium was changed twice weekly for 3 weeks. For colony counting, cells were washed twice with phosphate-buffered saline (PBS), fixed (10 min, 10% acetic acid), and stained (10 min, 0.4% crystal violet in 10% ethanol). The dishes were rinsed, inverted, and dried at room temperature.
Tumorigenicity in immunodeficient mice.
Nude mice injections were performed as described previously (23). Male, 6- to 8-week-old nude mice (strain BALB/cAnNCr-nuBR) were obtained from the National Cancer Institute. NAFA cells were transfected by calcium phosphate precipitation with plasmid pMACS4.1 (Miltenyi Biotec Inc., Auburn, Calif.) and either pBPSTR-1 or pBPSTR-1CD1AS. Transfected cells were enriched by magnetic bead-activated cell sorting (autoMACS; Miltenyi Biotec) (6) according to the manufacturer's protocol. Transfected cells were washed twice, suspended in 0.1 ml of PBS, and injected subcutaneously. For each mouse, the left flank was injected with NAFA cells transfected with cyclin D1 antisense, while the right flank received NAFA cells transfected with the control vector. Thus, each mouse received two injections of at least 3 × 105 cells. Tumor growth localized to the site of injection was monitored for 3 to 5 weeks, at which time mice were sacrificed. Tumors that formed were measured, excised, and analyzed for presence of neuT by PCR, expression of NeuT by Western blotting (not shown), and histopathology. Injections into nude mice were performed on four separate occasions, with similar results.
RESULTS
Neu induces cyclin D1 abundance in the mammary gland tumors of transgenic mice and stable cell lines.
To examine a possible role for Neu in regulating cyclin D1, we assessed stable NIH 3T3 cell lines overexpressing wild-type or transforming neu (shown schematically in Fig. 1A). Stable overexpression of wild-type neu in NIH 3T3 cells did not induce the transformed phenotype. A stable NIH 3T3 cell line (B104-1-1) expressing NeuT exhibited a transformed phenotype associated with Shc tyrosine phosphorylation and Shc-Grb2 complex formation (78). The NIH 3T3 stable cell line neuΔC-1 contains an internal carboxy-terminal deletion (amino acids 1007 to 1248) of NeuT which removes most of the Neu autophosphorylation sites but maintains transforming potential. In the transformed cell lines B104-1-1 (Fig. 1A, lane 3) and neuΔC-1 (Fig. 1A, lane 2), cyclin D1 levels were increased 7- and 6.5-fold, respectively, compared with the cells expressing wild-type Neu (Fig. 1A, lane 1).
FIG. 1.
Cyclin D1 protein levels are induced by Neu. (A) Western blot analysis of NIH 3T3 cell lines DHFR/G8 (expresses wild-type Neu [NeuWt]), B104-1-1 (expresses NeuT), and neuΔC-1 (contains a carboxy-terminal deletion of NeuT). α-Tubulin is shown as a protein loading control. Right, schematic representation of the Neu mutants showing the signal peptide (SP), cysteine-rich domains (CRD), transmembrane domain (TM), tyrosine kinase (TK), and carboxy terminus (CT). Within the CT are shown the five autophosphorylation sites (P1 to P5). The mutation within the transmembrane domain (Glu664) is shown. (B) Cyclin D1 protein levels were assessed in human breast cancer cell lines that have amplification of neu (MDA-MB-453 and BT-483) compared with cells with wild-type neu (MDA-MB-231 and HBL-100). Cells were deprived of serum (0.5% serum) for 24 h and then refed serum (10% serum) for 0, 4, or 8 h. Neu protein levels are indicated, with GDI blotting for protein loading control. (C) Western blot for endogenous cyclin D1 in MCF7 cells transfected with the NeuT expression vector, with comparison made to transfection of the empty expression vector cassette. (D) Mammary tumors of MMTV-neu and MMTV-NDL transgenic animals were analyzed for cyclin D1 protein levels by Western blotting (lower panel). Cyclin D1 immune-complex assays were conducted with the cyclin D1-specific antibody DCS-11. Phosphorylation of the GST-pRB substrate is indicated by the arrow (upper panel). NBE, normal breast epithelium. (E) The relative cyclin D1 protein levels and kinase activity for each tumor (MMTV-neu in blue and MMTV-NDL in red). Fold induction is shown in comparison with the mean derived from assays of three normal mammary glands, indicated by dashed lines. (F) Representative cyclin D1 immunohistochemical staining of MMTV-neu mammary gland tumors, with positive tumor cells appearing brown (top, yellow arrow) and negative cells appearing blue (red arrow). Normal mammary gland from the same animal demonstrated little nuclear cyclin D1 positivity (bottom).
Because neu has been found to be amplified in 20 to 30% of human breast tumors (64), we compared cyclin D1 levels in human breast carcinoma cell lines with and without amplification of neu. The cell lines MDA-MB-453 and BT-483, which harbor amplification of neu, exhibited higher cyclin D1 protein levels under serum deprivation and stimulation conditions compared with MDA-MB-231 cells and HBL-100 cells, which contain wild-type levels of neu (Fig. 1B). Neu levels were readily detectable in the neu-amplified cells lines MDA-MB-453 and BT-483. GDI levels were determined as a control for equivalent amounts of loaded protein (55).
To determine whether NeuT was also capable of inducing cyclin D1 protein levels in mammary epithelial cells, the human breast cancer cell line MCF7 was used. MCF7 cells are useful for determining Neu signaling mechanisms because Neu levels in MCF7 cells are similar to levels in normal breast tissue (27). In MCF7 cells, cyclin D1 protein levels are induced by estrogen, serum, and EGF (83). Immunoneutralization and antisense experiments have demonstrated that in MCF7 cells, the abundance of cyclin D1 is rate limiting in G1 phase progression (8, 42). MCF7 cells were transfected with the NeuT plasmid, and Western blotting was performed. Cyclin D1 protein levels were induced fivefold in the NeuT-transfected cells compared with cells transfected with the empty expression vector cassette (Fig. 1C). Blotting for α-tubulin confirmed equal protein loading in both lanes (Fig. 1C).
To determine if Neu induces cyclin D1 levels in vivo, cyclin D1 protein levels were assessed in the mammary tumor tissue from 18 independent MMTV-neu (wild-type Neu-expressing) (25) and MMTV-NDL (Neu deletion mutants-expressing) (62) transgenic mice, and comparison was made with normal mammary gland tissue from three nontransgenic animals (Fig. 1D). Equal amounts of protein loading were confirmed by Ponceau S staining and α-tubulin abundance (not shown). Cyclin D1 protein levels were increased up to 12.9-fold in all tumor samples examined except one (Fig. 1E). Cyclin D1 kinase (CD1K) activity was also assessed in each tumor, using a cyclin D1 immunoprecipitation kinase assay and glutathione S-transferase (GST)–pRB as the substrate (70, 72). The phosphorylated pRB band was dependent on the addition of pRB substrate and was inhibited by the addition of p16INK4a protein (not shown), consistent with the specificity of the kinase assays (12). Equal amounts of total protein were assessed in the assay, with comparison made between the activity generated by mammary tumor tissue from the transgenic mice and the mean mammary gland tissue CD1K activity from three nontransgenic animals (Fig. 1D). CD1K activity was increased in each tumor examined, from 1.5- to 17.7-fold (Fig. 1E, MMTV-neu [blue] and MMTV-NDL [red]).
Because the subcellular distribution of cyclin D1 varies with cell cycle progression and the nuclear location of cyclin D1 is important for its ability to inactivate pRB (19, 37), we assessed the distribution of cyclin D1 within the tumors by immunohistochemistry. Immunostaining of mammary tumors from both MMTV-neu and MMTV-NDL transgenic animals demonstrated increased nuclear abundance of cyclin D1 in the adenocarcinoma (Fig. 1F, top) compared with normal mammary tissue from the same animal (Fig. 1F, bottom), shown at higher magnification to demonstrate the absence of cyclin D1 staining. Analyses were performed on separate tumors from seven animals. The mean percentage of cells positively staining for nuclear cyclin D1 in the mammary gland tumors was 55% (range, 10 to 85%) compared with 2 to 5% nuclear staining in adjacent normal mammary epithelium.
NeuT activates the cyclin D1 promoter in MCF7 cells.
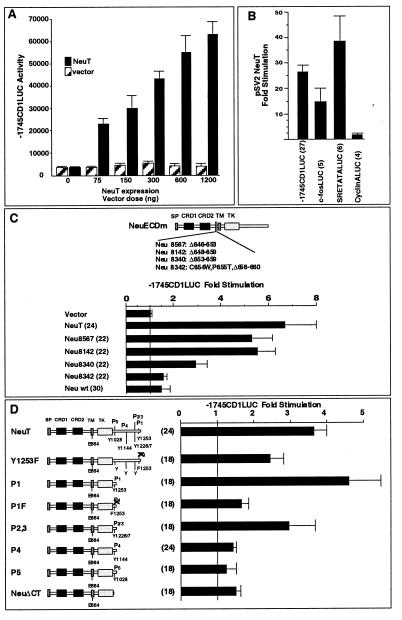
To determine whether the cyclin D1 gene is a direct transcriptional target of Neu, the human cyclin D1 promoter linked to a luciferase reporter was examined in MCF7 cells. Overexpression of NeuT induced cyclin D1 promoter activity in a dose-dependent manner (Fig. 2A). Wild-type Neu induced the cyclin D1 promoter (1.4-fold [not shown]). The induction was observed whether wild-type Neu was driven from the SV40 or pJ4Ω expression vector. The cyclin D1 promoter was induced a mean of 26-fold by NeuT when expressed from the SV40 promoter, which is highly active in MCF7 cells (Fig. 2B). To examine the specificity of the NeuT induction of cyclin D1, we examined several other gene promoters, including those for the c-fos gene and the cyclin A gene. The c-fos promoter was induced 14-fold, and the serum response element of the c-fos promoter was induced 39-fold. In contrast, the cyclin A promoter was not induced by NeuT in MCF7 cells (Fig. 2B).
FIG. 2.
Neu stimulates the cyclin D1 promoter in MCF7 cells. (A) The −1745CD1LUC reporter was transfected with increasing amounts of the Neu expression vector (pSV2NeuT) into MCF7 cells. Luciferase activity (relative light units) is shown with the activity induced by equal amounts of control vector cassette. (B) Neu induces the cyclin D1 promoter but not the cyclin A promoter. Cotransfection experiments were conducted with plasmids for the cyclin D1, c-fos, and cyclin A promoters linked to the luciferase reporter gene. The c-fos serum response element (SRE) linked to the minimal TATA box was also assessed. Induction by pSV2NeuT is shown. (C and D) Expression vectors encoding different neu mutants, previously described for their transforming ability, were assessed for their effects on cyclin D1 promoter activity in MCF7 cells. Data are shown as the mean ± standard error of the mean of the number of experiments shown in parentheses. wt, wild type.
The specificity of the promoter induction by NeuT was examined further, through driving expression of NeuT from a distinct promoter (pJ4Ω NeuT). This promoter is less active in MCF7 cells; however, the cyclin D1 promoter was induced 6.7-fold (Fig. 2C), compared with 1.5-fold induction by wild-type Neu. To determine whether the transforming Neu ECD mutants found in the MMTV-neu mice (61) could directly activate the cyclin D1 promoter, cotransfection experiments were conducted with MCF7 cells. The most transforming mutant (Neu 8142) induced the cyclin D1 promoter 5.4-fold (Fig. 2C). For each of the ECD mutants, the magnitude of cyclin D1 promoter induction corresponded well with the previously published transforming ability of these mutants in Rat-1 cells (61).
The carboxy-terminal domain of Neu includes five autophosphorylation sites. The autophosphorylation capacity of Neu is linked to its transforming capacity in focus-forming assays in fibroblasts and in the formation of tumors in athymic mice (9). The mutants examined in Fig. 2D included a carboxy-terminal deletion mutant (ΔCT) and a series of mutants in which the carboxy-terminal 12 amino acids of Neu were linked to the ΔCT, to serve as a functional docking site for SH2-containing proteins, with phosphorylation sites sequentially added. The mutant P1 includes the carboxy-terminal Neu phosphorylation site (tyrosine 1253). In this series of mutants, the cyclin D1 promoter was induced 3.6-fold by NeuT and 4.4-fold by the P1 mutant. The P1F mutant, which has a substitution of the terminal tyrosine for phenylalanine, was poorly transforming in Rat-1 cells and in athymic mice and was defective in activation of the cyclin D1 promoter. The induction of cyclin D1 by P1F was 37% of the value for P1 (Fig. 2D), and in athymic mice its transforming capacity was 25% of that of P1 (9). Mutation of tyrosine 1253 to phenylalanine (Y1253F) in the context of NeuT reduced transforming ability to 17% (9). This mutant induced the cyclin D1 promoter 50% compared with P1.
Previous studies had suggested that additional phosphorylation sites within the carboxy terminus contribute to the transforming capacity of NeuT. We assessed whether additional phosphorylation sites contribute to the induction of cyclin D1. The P2/3 mutant induced the cyclin D1 promoter 2.9-fold. The P4 and P5 mutants had displayed 2 to 3% transforming ability in NIH 3T3 cells and induced the cyclin D1 promoter 1.4- and 1.2-fold, respectively (Fig. 2D). Thus, full induction of the cyclin D1 promoter could be induced through the same carboxy-terminal phosphorylation events as previously shown to correlate with Neu's transforming ability.
Intracellular signaling pathways governing neuT induction of cyclin D1.
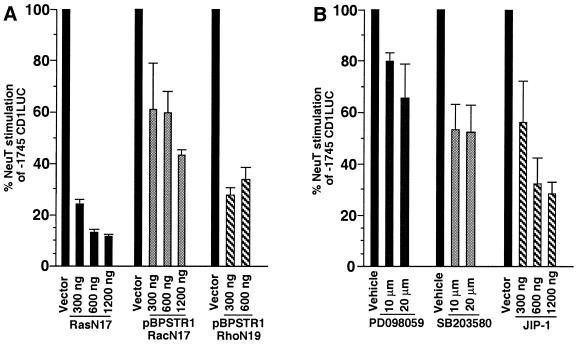
Due to its potent transforming capacity, which correlated with its induction of the cyclin D1 promoter, we focused on NeuT to determine the signaling pathway governing cyclin D1 regulation downstream of activated Neu. In previous studies, neutralizing Ras antibodies inhibited Neu-induced DNA synthesis (9). Ras is known to induce cyclin D1 (1, 38). We therefore assessed whether NeuT induction of cyclin D1 required Ras. NeuT-induced promoter activity was reduced 80 to 90% by N17Ras (Fig. 3A). The inhibition by N17Ras of NeuT induction of the cyclin D1 promoter was dose dependent and was observed with similar concentrations of N17Ras as shown previously to inhibit JNK activation by EGF (40). The RasL61S186 mutant, which is incapable of inserting in the plasma membrane, did not affect activity of cyclin D1 in the presence of NeuT (not shown).
FIG. 3.
Effects of inhibitors on NeuT induction of cyclin D1. To examine the intracellular signaling pathways involved in Neu induction of the cyclin D1 promoter, the −1745CD1LUC reporter was introduced into MCF7 cells with the NeuT expression vector. (A) Cotransfection experiments were conducted using increasing amounts of dominant negative expression vector, and the inhibition of Neu-induced promoter activation is shown as percent activity. Comparison was made between the effect of the dominant negative mutants for N17Ras, N17Rac, or N19Rho and equal amounts of empty expression vector cassette. (B) The chemical inhibitors of the MEK/ERK pathway (PD098059) (n = 8) and the p38 pathway (SB203580) (n = 8) were added to the culture medium and compared with equal volumes of DMSO vehicle. The expression vector encoding the inhibitor of JNK signaling (JIP-1) (n = 4) for each concentration of plasmid is shown compared with equal amounts of empty expression vector cassette. Inhibition is significant at P < 0.05.
The pathways activated by Ras diverge selectively through activation of distinct monomeric GTPases, with Rac1 and Rho activating distinct pathways (67). We therefore assessed the individual contributions of Rac and Rho in NeuT-mediated induction of the cyclin D1 promoter in MCF7 cells. RacN17 inhibited NeuT-induced cyclin D1 promoter activity 40 to 50% (P < 0.05) (Fig. 3A). Similar results were found whether the dominant negative expression was driven by the pBPSTR-1 tetracycline-regulated expression system (Fig. 3A) or the pEXV3 promoter (not shown). Overexpression of the dominant negative Rho (pEXVN19Rho) reduced NeuT induction of cyclin D1 60 to 70% (Fig. 3A). The dominant negative expression plasmids did not inhibit the promoter driving the NeuT expression vector at any of the concentrations used in the experiments (not shown), suggesting that the effect was not due to an indirect effect on NeuT expression. In contrast with the inhibition of −1745CD1LUC by pEXVN19Rho, activating mutants of Rho (RhoAL63) further induced the cyclin D1 promoter (not shown). In addition, plasmids pEXVN19Rho and pEXVN17Rac inhibited RasL61 induction of the collagenase AP-1 site reporter (p3TPLUX) (71) in a dose-dependent manner in MCF7 cells (not shown), consistent with previous studies of these same mutant plasmids performed in fibroblasts and HeLa cells (40, 52, 71).
NeuT induced ERK activity in cultured cells (9), and induction of the cyclin D1 promoter in MCF7 cells by another oncogenic tyrosine kinase, pp60v-src, involved the ERK, p38, and JNK pathways (33). The MEK inhibitor PD098059, which inhibits induction of the MEK/ERK pathway (3), reduced the NeuT induction of the cyclin D1 promoter by 30% compared with the DMSO control (n = 8, P < 0.05) (Fig. 3B). The p38 mitogen-activated protein kinase (MAPK) pathway chemical inhibitor SB203580 reduced the NeuT induction of the cyclin D1 promoter by 40 to 50% (n = 8, P < 0.05) at concentrations previously shown to inhibit p38 MAPK activity (Fig. 3B). Finally, introduction of JIP-1 (18), which sequesters JNK in the cytoplasm and therefore inhibits its activity, reduced NeuT-induced cyclin D1 promoter activation by 70% (n = 4, P < 0.05) (Fig. 3B).
Wortmannin, which blocks PI 3-kinase activation, added at concentrations from 20 to 100 nM (previously shown to inhibit PI 3-kinase but not PI 4-kinase, protein kinase A [PKA], PKC, or PKG [77]) did not affect cyclin D1 induction by NeuT (not shown), consistent with recent studies showing that the cyclin D1 promoter is not directly induced by the PI 3-kinase pathway (38). The pp70S6k pathway has been implicated in promoting cell cycle progression (50) and, like cyclin D1, is induced by activating Rac1 (15). Addition of rapamycin, which prevents induction of pp70S6k, from 100 pM to 20 nM did not reduce NeuT induction of cyclin D1 but caused an induction (60%, not shown), suggesting that the pp70S6k pathway may inhibit cyclin D1 in the presence of activating NeuT. These studies suggest that NeuT induction of cyclin D1 involves a Ras/Rac/Rho pathway in which ERK, JNK, and p38 MAPK but neither PI 3-kinase nor pp70S6k is a distal mediator.
Sp1 and E2F binding sites are required for full induction of the cyclin D1 promoter by NeuT.
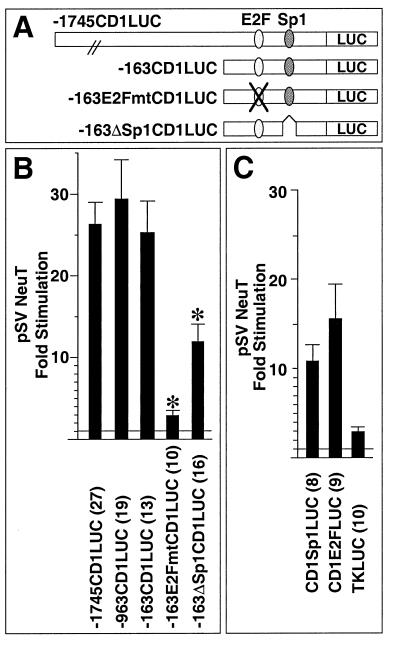
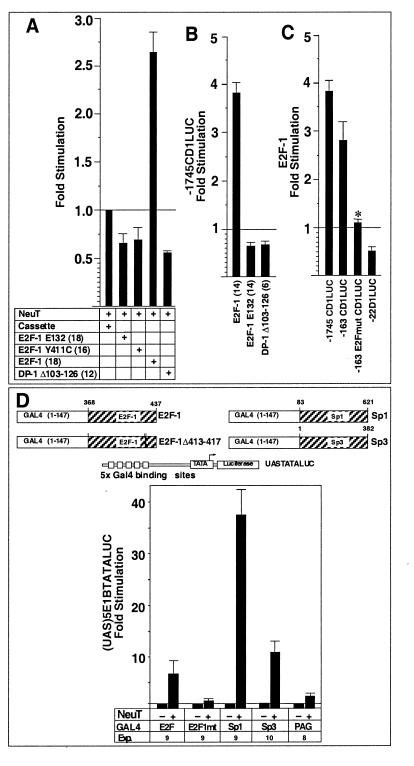
To determine the sequences of the cyclin D1 promoter required for full induction by NeuT, cotransfection experiments were conducted with a series of 5′ promoter deletion constructions. Deletion of the promoter from −1745 to −163 retained 25-fold induction by NeuT (compared to the vector control [Fig. 4B]). Within the proximal promoter, we had recently identified functional sequences resembling an E2F binding site and an Sp1 binding site (70) (Fig. 4A). Mutation of the E2F site in the context of the −163 bp promoter fragment (−163E2Fmt) reduced NeuT induction from 25- to 3-fold (Fig. 4B). Deletion of the Sp1 site (−163ΔSp-1) reduced NeuT induction from 25- to 13.6-fold, suggesting that the Sp1 site may also contribute to full induction of cyclin D1 by Neu (Fig. 4B). The cyclin D1 E2F and Sp1 sequences were linked to the minimal thymidine kinase (TK) promoter to form heterologous reporters and were examined for NeuT responsiveness. The cyclin D1 Sp1 site was induced 10.5-fold by NeuT, and the cyclin D1 E2F site reporter was induced 15.6-fold (Fig. 4C). In contrast, the TK promoter was not responsive to NeuT (Fig. 4C).
FIG. 4.
Promoter sequences involved in Neu induction of the cyclin D1 promoter. (A) Schematic representation of the cyclin D1 promoter, with the sequences homologous to E2F and Sp1 binding sites indicated. LUC, luciferase. (B) pSV2NeuT was transfected with cyclin D1 5′ promoter constructs into MCF7 cells. ∗ represents significant difference from the −163CD1LUC reporter for P < 0.05. (C) The heterologous constructions, encoding the E2F or Sp1 site of the cyclin D1 promoter linked to the minimal TK promoter, were transfected with pSV2NeuT into MCF7 cells. Comparison is made with the effect on the minimal TK reporter. In each case, fold stimulation reflects induction with NeuT compared with the empty expression vector cassette, with the number of experiments shown in parentheses.
E2F-1 and Sp1/Sp3 bind the cyclin D1 promoter in MCF7 cells.
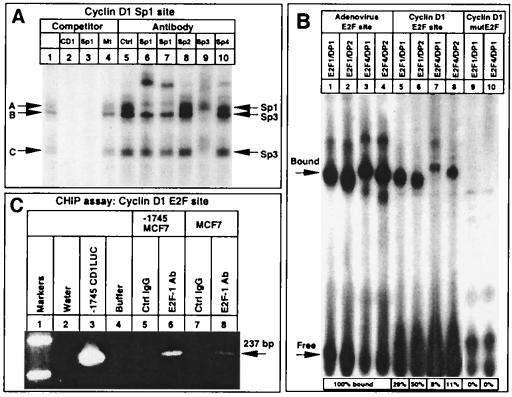
To determine which proteins bind the cyclin D1 Sp1 site in MCF7 cells, EMSAs were performed (Fig. 5A). The cyclin D1 Sp1 sequences formed three complexes with MCF7 nuclear extracts (Fig. 5A, lane 1, arrows A to C). These complexes were competed with 100-fold molar excess of the cold cognate oligonucleotide probe or a consensus Sp1 site probe (Fig. 5A, lanes 2 and 3) but not by an unrelated sequence (Fig. 5A, lane 4). Supershifting antibodies to members of the Sp family demonstrated that band A consisted of Sp1 (Fig. 5A, lanes 6 and 7). Bands B and C were inhibited with the addition of Sp3 antibody (Fig. 5A, lane 9). Control IgG (Fig. 5A, lane 5) or antibodies to Sp2 and Sp4 (Fig. 5A, lanes 8 and 10) had no effect. The inhibition of binding by the Sp3 antibody was not observed with other DNA sequences (not shown) and was associated with a partial supershift, consistent with the presence of Sp3 binding to this site.
FIG. 5.
Sp1/Sp3 and E2F-1 proteins bind the neuT-responsive elements of the cyclin D1 promoter. (A) The 32P-labeled cyclin D1 Sp1-like sequence was incubated with MCF7 cell nuclear extracts, and the effects of 100-fold excess of cognate competitor (lane 2), wild-type canonical Sp1 binding site competitor (lane 3), and an unrelated oligonucleotide competitor (lane 4) were determined. Specific antibodies to the Sp proteins or equal amounts of control IgG (lane 5) were added as indicated above the lanes (lanes 5 to 10). Arrows indicate the predicted proteins constituting the bands (A to C) identified through supershift or inhibition of DNA binding. (B) EMSA with extracts prepared from baculovirus-infected Sf9 cells. The 32P-labeled adenovirus E2F site (lanes 1 to 4) and wild-type (lanes 5 to 8) or mutant (lanes 9 to 10) cyclin D1 E2F sites were incubated with E2F and DP proteins as indicated above the lanes. Relative binding compared to the adenovirus E2F site for each E2F-DP complex is indicated below each lane. (C) CHIP assays were performed with the −1745 CD1LUC MCF7 cell line or wild-type MCF7 cells (lanes 5 to 8). PCR was performed with cyclin D1-specific primers on water (lane 2), control plasmid (lane 3), or immunoprecipitation buffer (lane 4) or after immunoprecipitation of formaldehyde cross-linked cell extracts with either IgG control (lanes 5 and 7) or E2F-1-specific antibody (lanes 6 and 8). The specific cyclin D1 promoter band is shown (arrow).
Previous studies had demonstrated that E2F-1, E2F-4, DP-1, and pRB family members bind the cyclin D1 E2F site (70). To compare the relative binding affinities of E2F-1 and E2F-4 to the cyclin D1 and adenovirus E2F sequences, EMSAs were performed with in vitro-prepared E2F and DP proteins (Fig. 5B). E2F-1 bound the cyclin D1 E2F sequence when incubated with either of its dimerization partners, DP-1 (Fig. 5B, lane 5) or DP-2 (Fig. 5B, lane 6). E2F-4 bound the cyclin D1 E2F site, albeit with less affinity than E2F-1 (Fig. 5B, lanes 7 and 8). Mutation of the cyclin D1 E2F sequence abolished binding of either E2F-1 or E2F-4 (Fig. 5B, lanes 9 and 10). The cyclin D1 E2F sequence had lower affinity for each E2F-DP complex compared with the adenoviral E2F sequence (Fig. 5B, compare lanes 5 to 8 with lanes 1 to 4, respectively; percentages of binding to the cyclin D1 E2F site compared to the adenoviral E2F site are given under lanes 5 to 8).
Because native chromatin structure may alter the ability of transcription factors to bind a specific DNA sequence, changes in E2F occupancy may be detected only with the use of higher-resolution analysis, such as in vivo footprinting (2). CHIP assays were performed to analyze E2F-1 binding to the cyclin D1 promoter in the context of native chromatin. The cyclin D1 promoter linked to the luciferase reporter gene was stably integrated into MCF7 cells. Analyses were performed of E2F-1 binding to the cyclin D1 promoter in the stable cell line (Fig. 5C). PCR amplification of chromatin cross-linked extracts immunoprecipitated with an E2F-1 specific antibody identified the product corresponding to the cyclin D1 E2F site and its surrounding genomic sequence (Fig. 5C, lane 6) (confirmed by sequence analysis [not shown]). In contrast, no amplification was observed in products of immunoprecipitations using equal amounts of control IgG (Fig. 5C, lane 5). CHIP analysis of wild-type MCF7 cells also identified E2F-1 binding to the endogenous cyclin D1 gene (Fig. 5C, lane 8). These studies demonstrate that E2F-1 binds to the cyclin D1 promoter in the context of its native chromatin structure in MCF7 cells.
Together, these results indicate that the DNA sequences within the cyclin D1 promoter that are sufficient for induction by NeuT in MCF7 cells are capable of binding Sp1, Sp3, and E2F-1.
Sp1, Sp3, and E2F-1 transactivation function is induced by NeuT.
To determine whether E2F-1 is critical for NeuT induction of cyclin D1 promoter activity, experiments were conducted with DNA-binding-defective mutant E2F-1 E132 or activation-defective mutant E2F-1 Y411C. E2F-1 E132 functions as a dominant inhibitor of E2F activity by dimerizing with DP proteins and thereby blocking the DNA binding of the active E2F-DP complex. E2F-1 Y411C binds competitively to E2F binding sites and thereby blocks activation by wild-type E2F-1. Overexpression of E2F-1 E132 or E2F-1 Y411C reduced the NeuT induction of cyclin D1 by 33 or 30%, respectively (Fig. 6A). Similarly, the DP-1 dominant negative (DP-1 Δ103-126) reduced NeuT induction of cyclin D1 by 43% (Fig. 6A). In contrast, overexpression of the wild-type E2F-1 further induced cyclin D1 promoter activity 2.6-fold (Fig. 6A). These results are consistent with a requirement for E2F-1 for optimal induction of cyclin D1 by NeuT.
FIG. 6.
Induction of E2F-1, Sp1, and Sp3 transactivation by NeuT. The effects of wild-type E2F-1, the E132 mutant E2F-1, the Y411C mutant E2F-1, and the DP-1 dominant negative mutant on NeuT-induced (A) or basal (B) cyclin D1 promoter activity were assessed in MCF7 cells and compared with effects of the respective empty vectors. (C) The E2F-1 expression vector was transfected with cyclin D1 5′ promoter constructs into MCF7 cells. ∗ represents significant difference from the −163CD1LUC reporter for P < 0.05. (D) Schematic representation of the GAL4 constructs and the heterologous luciferase reporter containing five upstream activator binding sites for the GAL4 DNA binding domain. The reporter (UAS)5E1BTATALUC (2.4 μg) was transfected with expression vectors for GAL4–E2F-1, GAL4–E2F-1(Δ413-417) (the pRB-binding-defective E2F-1 mutant), GAL4-Sp1, GAL4-Sp3, PAG236, and either pSV2NeuT (600 ng) or empty expression vector cassette in MCF7 cells. Comparison was made between the effect of the NeuT expression vector and equal amounts of the parental vector. Data are mean fold induction ± standard error of the mean for the number of separate experiments indicated in parentheses.
As these results contrast with the inhibitory effect of E2F-1 on basal cyclin D1 promoter activity in trophoblastic cells and fibroblasts (70), we assessed whether E2F-1 has a cell-type-specific effect on basal activity of the −1745CD1LUC reporter in MCF7 cells. E2F-1 induced the cyclin D1 promoter in MCF7 cells 3.8-fold (Fig. 6B). Overexpression of either the E2F-1 E132 mutant or the DP-1 dominant negative inhibited the cyclin D1 promoter by 36 or 34%, respectively (Fig. 6B), whereas the E2F-1 Y411C mutant had no effect (not shown). To identify the DNA sequences required for induction of the cyclin D1 promoter by E2F-1, cotransfection experiments were conducted with the cyclin D1 promoter deletion constructs (Fig. 6C). The induction by E2F-1 was maintained in the −163 bp fragment but was lost upon point mutation of the E2F site (−163 E2FmutLUC), suggesting that the E2F site is the region required for optimal induction by E2F-1 overexpression (Fig. 6C). E2F-1 is therefore a positive regulator of cyclin D1 in MCF7 cells.
The results presented above indicated that NeuT induced the cyclin D1 promoter through an E2F binding site that bound E2F-1 and was required for E2F-1 induction in MCF7 cells and through an Sp1 binding site that bound Sp1 and Sp3. To understand how NeuT may induce cyclin D1 through the E2F-1 and Sp1/Sp3 binding sequences, we tested whether NeuT regulates E2F-1, Sp1, and Sp3 transactivation function. The transactivation domains of these proteins were linked to the GAL4 DNA binding domain. The NeuT-regulated activity of Sp1 and E2F-1 was examined in conjunction with a heterologous reporter construction, (UAS)5E1BTATALUC, consisting of multimeric GAL4 DNA binding sites linked to a luciferase reporter gene (Fig. 6D). E2F-1, Sp1, and Sp3 conveyed basal enhancer function in MCF7 cells, and overexpression of NeuT enhanced E2F-1 activity 6.3-fold. E2F-1 transactivation function is regulated by the relative abundance of pRB, and selected deletions within the carboxy-terminal pRB binding domain abolish transcriptional regulation by pRB (22). In contrast with GAL4–E2F-1, the carboxy-terminal E2F-1 mutant GAL4–E2F-1(Δ413-417) was not induced by NeuT. Overexpression of NeuT induced Sp1 activity 37.5-fold and Sp3 activity 11-fold. In contrast, the constitutively active GAL4 construct PAG236 was not induced by NeuT. Thus, NeuT induces transactivation function of the transcription factors binding to the neuT-responsive regions of the cyclin D1 promoter.
Cyclin D1 antisense blocks NeuT-induced transformation.
The findings that cyclin D1 levels were induced in neu-induced mammary gland tumors and that NeuT potently induced the cyclin D1 promoter raised the possibility that cyclin D1 is required for NeuT-induced transformation. Transformation assays were therefore performed with Rat-1 cells. NeuT (Fig. 7A) and RasL61 (not shown) both induced focus formation as previously described (61). Experiments were conducted to assess the involvement of cyclin D1 and several intracellular signaling proteins involved in mitogenic signaling in NeuT-induced transformation. Either a 1:1 or a 1:2 molar ratio of NeuT plasmid (2.5 μg) to antisense or dominant negative expression plasmid was used (Fig. 7). In previous studies of V12Ras-induced focus formation, the N17Rac plasmid was used in the ratio of 1:250 (V12Ras:N17Rac) for 70% inhibition of focus formation (53). Thus, our experiments used relatively small amounts of dominant negative or antisense expression plasmid in conjunction with the NeuT expression plasmid.
FIG. 7.
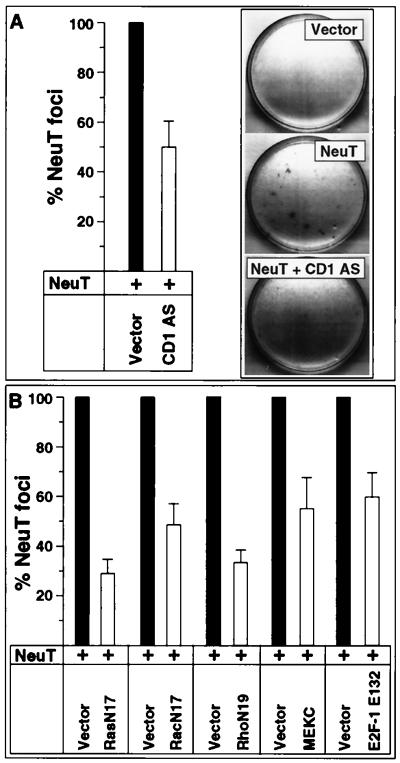
Cyclin D1 antisense inhibits neu-induced focus formation. Transformation assays were conducted with NeuT in Rat-1 cells. The activating NeuT mutant (2.5 μg) was introduced alone or in conjunction with one of the antisense or dominant negative expression plasmids listed. The cyclin D1 antisense (pBPSTR-1CD1AS) (A) and the dominant negative mutants for N17Ras, N17Rac, N19Rho, MEKC, and E2F-1 (E2F-1 E132) (2.5 or 5 μg) (B) were assessed in comparison to the empty expression vector cassette. The effect on transformation is shown for 1:1 and 1:2 molar ratios of NeuT vector to dominant negative or antisense plasmid. Panel A shows a representative assay with the effect of cyclin D1 antisense. The transformation induced by NeuT is shown as 100% in black bars throughout. The results are shown as percentage of transformation by NeuT for independent transformation assays compared with the effect of empty vector cassette (mean ± standard error of the mean).
We have previously used the full-length human cyclin D1 antisense under control of a tetracycline-regulated promoter to effectively reduce cyclin D1 protein levels in rat H19-7 cells (79). These experiments were conducted with the cyclin D1 antisense in Rat-1 cells in the tetracycline-regulated plasmid pBPSTR-1. The transformation experiments were conducted on eight separate occasions. In every experiment, the cyclin D1 antisense construct inhibited the number of foci induced by NeuT (Fig. 8A). Overexpression of the cyclin D1 antisense construct reduced NeuT-induced colony formation by a mean of 50% (Fig. 7A), and this effect was dose dependent.
FIG. 8.
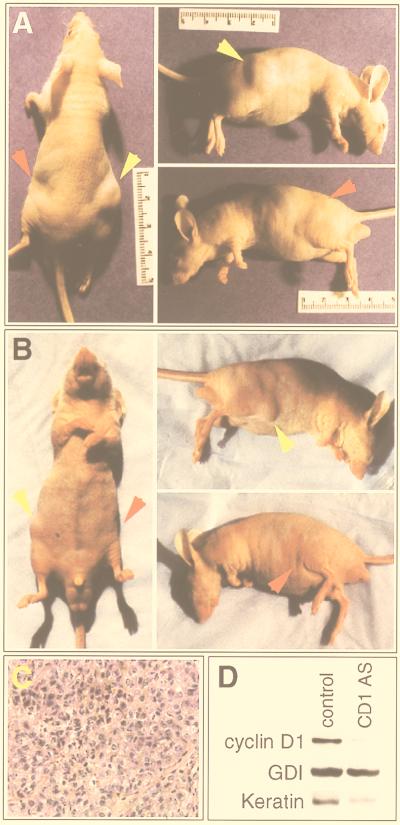
Cyclin D1 antisense inhibits growth of NeuT-transformed mammary epithelial cells in nude mice. Immunodeficient mice received subcutaneous injections into each flank with transfected NAFA cells in PBS. Sites injected with NAFA cells transfected with control vector (right flank, yellow arrow) showed development of tumors, whereas in the same animal, the left flank (red arrow), injected with NAFA cells transfected with cyclin D1 antisense, did not show the development of tumors. (A and B) Two examples of mice injected in both flanks; (C) hematoxylin and eosin staining of the right flank tumor of the mouse in panel A, demonstrating adenocarcinoma; (D) Western blot of implanted cells of the mouse in panel A, demonstrating reduced cyclin D1 protein levels in cells transfected with the cyclin D1 antisense (CD1 AS) compared with the control vector (control). GDI blotting confirmed equivalent protein loading, and keratin-8 blotting confirmed that the tissues were of epithelial origin.
Overexpression of the dominant negative N17Ras reduced NeuT-induced colony formation by 70% (Fig. 7B) in a dose-dependent manner. Rac (52) and Rho (53) were previously shown to convey an important component of V12Ras induced colony formation in Rat-1 cells. Therefore, we examined whether Rac and Rho pathways were involved in NeuT-induced transformation or whether these transforming pathways diverged. Dominant negative N17Rac reduced NeuT-induced Rat-1 focus formation by 50%, and N19Rho reduced colony formation by 67%. These results were seen whether the dominant negative mutants were expressed from pBPSTR-1 (Fig. 7B) or from the pEXV3 vector (not shown). In each of the seven separate experiments for each expression plasmid, inhibition of focus formation was observed. None of the dominant negative expression plasmids reduced activity of the promoter driving the NeuT expression plasmid (not shown), suggesting that the effect of each dominant negative was not mediated through an indirect effect inhibiting NeuT expression.
In previous studies, an interfering mutant of MAPK kinase 1 (MEKAla-218/Ala-222, MEKC) was shown to inhibit MAPK kinase activity (17). Rho is thought to preferentially regulate the MAPK pathway, and Rac is thought to preferentially regulate Jun kinase activity (67). To determine whether NeuT-induced transformation involved the MAPK pathway, the effect of MEKC was assessed. MEKC inhibited NeuT-induced transformation by 44% (Fig. 7B). The MEK inhibitor PD098059 also reduced NeuT-induced focus formation by 30% (mean of three determinations [not shown]).
Our studies above had found that a dominant negative mutant of E2F-1 was capable of inhibiting NeuT-induced cyclin D1 promoter activity, implicating E2F-1 as a downstream target of NeuT signaling. To examine the role of E2F-1 in NeuT-induced transformation, we coexpressed the E2F-1 E132 mutant with NeuT in Rat-1 cells. Compared with the empty expression vector, which had no effect on transformation, the E2F-1 E132 plasmid inhibited NeuT-induced transformation by 40% (Fig. 7B). Together, these results are consistent with a model in which NeuT-induced transformation involves Ras, Rac, Rho, MEK1, E2F-1, and cyclin D1 for full transformation.
Cyclin D1 antisense blocks tumorigenesis of NeuT-transformed mammary cells in immunodeficient strains of mice.
As cyclin D1 antisense blocked NeuT-induced transformation in fibroblasts, we assessed its effect on growth of a mammary tumor cell line (NAFA) derived from an MMTV-neuT mouse (41). NAFA cells grow rapidly and have readily detectable levels of cyclin D1 by Western blot (not shown). NAFA cells were cotransfected with a truncated human CD4-encoding plasmid and either cyclin D1 antisense or empty vector (pBPSTR-1). Transfected cells were selected by magnetic bead-activated cell sorting (39, 60) and injected subcutaneously into the flanks of the same nude mouse (Fig. 8A and B), and tumor formation was assessed. Importantly, no tumors were observed at the sites of injection of the cyclin D1 antisense-transfected cells after 3 to 5 weeks. In contrast, the control vector-transfected NAFA cells injected into the contralateral flank developed histologically confirmed adenocarcinomas consisting of solid cords and nodules of neoplastic cells in each case (Fig. 8C). Tissues from the sites of implantation were assessed for cyclin D1 levels by Western blotting (Fig. 8D). Cyclin D1 levels were lower in cells transfected with the cyclin D1 antisense construct than in cells transfected with the control vector. Equivalent protein loading was confirmed by blotting for GDI, and the tissues were confirmed to be of epithelial origin by keratin-8 blotting. After correction for equivalent protein loading using GDI, cyclin D1 levels were determined to be reduced 90% in cells transfected with the cyclin D1 antisense. Four different mice were injected on different days, as independent experiments, with similar results. These results establish that downregulation of cyclin D1 is sufficient to abolish tumorigenicity in Neu-transformed cells.
DISCUSSION
Oncogenic activation of neu can occur through overexpression, point mutation, or deletion of the extracellular domain (7, 61). Similar to the murine MMTV-neu model of mammary tumorigenesis, in primary human breast cancers, the overexpression of ErbB-2 (64) and the recent identification of an in-frame deletion of a portion of the extracellular domain of ErbB-2 (62) suggest an important role for ErbB-2 in induction and progression of human breast tumors. The present studies identify for the first time the role of a rate-limiting component of the cell cycle in transformation by Neu in mammary adenocarcinoma cells in vivo. Cyclin D1 abundance and kinase activity were increased in mammary gland tumors from MMTV-neu and MMTV-NDL transgenic animals. The activating ECD mutations of Neu induced cyclin D1 promoter activity in MCF7 cells in a manner that corresponded well with their transforming capacity in Rat-1 cells (61). Cyclin D1 antisense inhibited neuT-induced transformation in a dose-dependent manner and abolished the growth of NeuT-transformed mammary cells in immunodeficient mice. These results suggest a critical role for cyclin D1 in proliferative and transforming signals downstream of oncogenic Neu.
In these studies, Ras, Rac, and Rho contributed to NeuT induction of cyclin D1 promoter activity and cellular transformation. Each of these small monomeric GTPases has been implicated in promoting cell cycle progression, and activating mutants of Ras and Rac1 have been shown to induce cyclin D1 (1, 76). The MEK inhibitor PD098059, the p38 inhibitor SB203850, and the JNK pathway inhibitor JIP-1 reduced induction of cyclin D1 by NeuT. These findings are consistent with our previous studies in which ERK directly induced cyclin D1, EGF induction of the cyclin D1 promoter involved a Ras/ERK pathway (1, 71), and the ERK, p38, and JNK pathways were involved in induction of the cyclin D1 promoter in MCF7 cells downstream of another oncogenic tyrosine kinase, pp60v-src (33).
These studies identify for the first time specific transcriptional targets activated by NeuT within the cyclin D1 promoter, indicating an important role for E2F-1 in both NeuT-induced transformation and cyclin D1 promoter activation. E2F-1 can function as both an oncogene and a tumor suppressor likely dependent on cellular context, although the molecular mechanisms governing these events is poorly understood (73). NeuT induced the cyclin D1 E2F site when linked to an heterologous promoter in MCF7 cells, whereas in previous studies performed in trophoblastic cells and mouse embryo fibroblasts (70), E2F-1 inhibited cyclin D1 abundance and promoter activity. Thus, E2F-1 conveys cell-type-specific effects on the cyclin D1 promoter likely related to additional E2F/DP family members or cofactors present within a given cell type. Recent studies have indicated that in vitro, distinct E2F sites have preferential affinities for members of the E2F family (66). The cyclin D1 E2F sequence resembles most closely the consensus sequence that was found to preferentially bind pRB–E2F-1–DP-1 complexes (66). Indeed, E2F-1 bound the cyclin D1 sequence with greater affinity than E2F-4 in these studies, and pRB is a component of complexes that can bind the cyclin D1 E2F site (reference 70 and data not shown). In MCF7 cells, CHIP assays demonstrated binding of E2F-1 specifically to the cyclin D1 promoter in the context of native chromatin. Further characterization of the additional proteins binding to the cyclin D1 promoter E2F site may provide greater insight into the mechanisms of neu-mediated transformation and is the focus of ongoing studies.
The E2F-1 transactivation domain linked to the GAL4 DNA binding domain was sufficient for induction by NeuT. Cyclin D1 overexpression leads to the induction of pRB phosphorylation and the release of free E2F-1 (74), which is capable of inducing promoter activity through either Sp1 or E2F sites (30). Consistent with a role for E2F-1 in NeuT induction of cyclin D1, the DNA-binding-defective and activation-defective mutants of E2F-1 and the dominant negative mutant of DP-1 inhibited NeuT induction of cyclin D1, and E2F-1 E132 inhibited NeuT-induced transformation. Because cyclin D1 overexpression can induce promoter activity through E2F sequences (82) and overexpression of pRB inhibits E2F-1 transactivation function (22), NeuT may sustain cyclin D1-mediated autoinduction through the E2F and Sp1 sites.
The Sp1 binding site of the cyclin D1 promoter was required for optimal induction by NeuT, and NeuT induced the Sp1 transactivation domain in MCF7 cells. Sequences resembling Sp1 binding sites contribute to the inducible expression of several growth factor-inducible genes (14, 44, 58). Sp1 binds to the promoters of Neu differentiation factor-inducible genes, including the acetylcholine receptor δ and ɛ subunits and neurotrophin-3 (4). Sp1 binds several intermediary proteins, some of which have been implicated in mitogenic signaling, including the E2F-1 and E2F-3 proteins (30). The Sp1 binding site of the α2-integrin gene is a site of inhibition by ErbB-2, suggesting that the inhibition of α2-integrin abundance may contribute to loosening of cell-cell contacts necessary for cell division (81). Together with the activation of cyclin D1, the induction of other growth factor-regulated genes through Sp1 binding sites and the inhibition of genes involved in cell contact may contribute to the proliferative and transforming phenotype induced by activated Neu.
As a common downstream target of several different mitogenic signaling pathways in breast cancer cells (36), cyclin D1 represents a logical target for inactivation by gene therapy. In addition to contributing to phosphorylation and inactivation of pRB, cyclin D1 binds other proteins, including proliferating cell nuclear antigen (34), a Myb-related protein (26), and the estrogen receptor (ER) (45). Neu is capable of phosphorylating the ER on tyrosine residues (48), and phosphorylation of the ER at tyrosine and/or serine residues has been associated with functional activation. The role of cyclin D1-associated proteins in neu-induced transformation remains to be determined. Cyclin D1 antisense constructs as well as the recently characterized dominant negative mutants of cyclin D1 (19) may provide important information about the requirement for cyclin D1 in neu-induced mammary gland tumor formation in vivo. Clearly, mammary tumorigenesis is a multistep process and may involve alterations in the expression of other tumor suppressor gene products. The abundance of the p16INK4a protein is frequently reduced in the MMTV-neu tumors, and overexpression of p16INK4a but not p19ARF can inhibit NeuT-induced focus formation in Rat-1 cells, suggesting that specific tumor suppressors may inhibit Neu-induced transforming pathways (M. D'Amico and R. G. Pestell, unpublished data). The roles of these additional tumor suppressor genes in Neu-induced transformation in mammary epithelial cells are the basis of ongoing studies.
ACKNOWLEDGMENTS
We are grateful to S. Cook, R. Davis, N. Dyson, G. Gill, M. Gilman, E. Harlow, W. Kaelin, J. Massague, F. McCormick, J. Nevins, S. Reeves, P. Farnham, J. Wells, and L. Zhu for plasmids, antibodies, and helpful discussions. We thank R. Russell for pathological assessment of tumors.
This work was supported in part by grants R29CA70897, RO1CA75503, and 5-P30-CA13330-26 (R.G.P.), CA536340 (J.M.H.), NIH training grant T32 DK 07513 (R.J.L.), and grant CA09475-12 (M.D.). W.J.M. is supported by research grants awarded by the Canadian Breast Cancer Initiative and is a recipient of an MRC of Canada Scientist award. R.G.P. is a recipient of the Irma T. Hirschl Award and an award from the Susan G. Komen Breast Cancer Foundation. Work conducted at the Albert Einstein College of Medicine was supported by Cancer Center Core National Institutes of Health grant 5-P30-CA13330-26 and the Mortimer Harrison Foundation (R.G.P.).
REFERENCES
- 1.Albanese C, Johnson J, Watanabe G, Eklund N, Vu D, Arnold A, Pestell R G. Transforming p21ras mutants and c-Ets-2 activate the cyclin D1 promoter through distinguishable regions. J Biol Chem. 1995;270:23589–23597. doi: 10.1074/jbc.270.40.23589. [DOI] [PubMed] [Google Scholar]
- 2.Alberts A S, Geneste O, Treisman R. Activation of SRF-regulated chromosomal templates by Rho-family GTPases requires a signal that also induces H4 hyperacetylation. Cell. 1998;92:475–487. doi: 10.1016/s0092-8674(00)80941-1. [DOI] [PubMed] [Google Scholar]
- 3.Alessi D R, Cuenda A, Cohen P, Dudley D T, Saltiel A R. PD 098059 is a specific inhibitor of the activation of mitogen-activated protein kinase kinase in vitro and in vivo. J Biol Chem. 1995;270:27489–27494. doi: 10.1074/jbc.270.46.27489. [DOI] [PubMed] [Google Scholar]
- 4.Alroy I, Soussan L, Seger R, Yarden Y. Neu differentiation factor stimulates phosphorylation and activation of the Sp1 transcription factor. Mol Cell Biol. 1999;19:1961–1972. doi: 10.1128/mcb.19.3.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Amundadottir L T, Leder P. Signal transduction pathways activated and required for mammary carcinogenesis in response to specific oncogenes. Oncogene. 1998;16:737–746. doi: 10.1038/sj.onc.1201829. [DOI] [PubMed] [Google Scholar]
- 6.Ashton A W, Watanabe G, Albanese C, Harrington E O, Ware J A, Pestell R G. Protein kinase Cδ inhibition of S-phase transition in capillary endothelial cells involves the cyclin dependent kinase inhibitor p27Kip1. J Biol Chem. 1999;274:20805–20811. doi: 10.1074/jbc.274.30.20805. [DOI] [PubMed] [Google Scholar]
- 7.Bargmann C I, Hung M C, Weinberg R A. Multiple independent activations of the neu oncogene by a point mutation altering the transmembrane domain of p185. Cell. 1986;45:649–657. doi: 10.1016/0092-8674(86)90779-8. [DOI] [PubMed] [Google Scholar]
- 8.Bartkova J, Lukas J, Muller H, Lutzhoft D, Strauss M, Bartek J. Cyclin D1 protein expression and function in human breast cancer. Int J Cancer. 1994;57:351–361. doi: 10.1002/ijc.2910570311. [DOI] [PubMed] [Google Scholar]
- 9.Ben-Levy R, Paterson H F, Marshall C J, Yarden Y. A single autophosphorylation site confers oncogenicity to the Neu/ErbB-2 receptor and enables coupling to the MAP-kinase pathway. EMBO J. 1994;13:3302–3311. doi: 10.1002/j.1460-2075.1994.tb06632.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bodrug S E, Warner B J, Bath M L, Lindeman G J, Harris A W, Adams J M. Cyclin D1 transgene impedes lymphocyte maturation and collaborates in lymphomagenesis with the myc gene. EMBO J. 1994;13:2124–2130. doi: 10.1002/j.1460-2075.1994.tb06488.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bromberg J F, Wrzeszczynska M H, Devgan G, Zhao Y, Albanese C, Pestell R G, Darnell J E J. Stat3 as an Oncogene. Cell. 1999;98:295–303. doi: 10.1016/s0092-8674(00)81959-5. [DOI] [PubMed] [Google Scholar]
- 12.Byeon I-J L, Li J, Ericson K, Selby T L, Tevelev A, Kim H-J, O'Maille P, Tsai M-D. Tumor suppressor p16Ink4a: determination of solution structure and analyses of its interaction with cyclin-dependent kinase 4. Mol Cell. 1998;1:421–431. doi: 10.1016/s1097-2765(00)80042-8. [DOI] [PubMed] [Google Scholar]
- 13.Caceres A, Binder L I, Payne M R, Bender P, Rebhun L, Steward O. Differential subcellular localization of tubulin and the microtubule-associated protein MAP2 in brain tissue as revealed by immunocytochemistry with monoclonal hybridoma antibodies. J Neurosci. 1983;4:394–410. doi: 10.1523/JNEUROSCI.04-02-00394.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen X, Azizkhan J C, Lee D C. The binding of transcription factor Sp1 to multiple sites is required for maximal expression from the rat transforming growth factor alpha promoter. Oncogene. 1992;7:1805–1815. [PubMed] [Google Scholar]
- 15.Chou M M, Blenis J. The 70 kDa S6 kinase complexes with and is activated by the Rho family G proteins Cdc42 and Rac1. Cell. 1996;85:573–583. doi: 10.1016/s0092-8674(00)81257-x. [DOI] [PubMed] [Google Scholar]
- 16.Clark G J, Cox A D, Graham S M, Der C J. Biological assays for Ras transformation. Methods Enzymol. 1995;255:395–412. doi: 10.1016/s0076-6879(95)55042-9. [DOI] [PubMed] [Google Scholar]
- 17.Cowley S, Paterson H, Kemp P, Marshall C J. Activation of MAP kinase kinase is necessary and sufficient for PC12 differentiation and for transformation of NIH 3T3 cells. Cell. 1994;77:841–852. doi: 10.1016/0092-8674(94)90133-3. [DOI] [PubMed] [Google Scholar]
- 18.Dickens M, Rogers J S, Cavanagh J, Raitano A, Xia Z, Halpern J R, Greenberg M E, Sawyers C L, Davis R J. A cytoplasmic inhibitor of the JNK signal transduction pathway. Science. 1997;277:693–696. doi: 10.1126/science.277.5326.693. [DOI] [PubMed] [Google Scholar]
- 19.Diehl J A, Sherr C J. A dominant negative cyclin D1 mutant prevents nuclear import of cyclin-dependent kinase 4 (CDK4) and its phosphorylation of CDK-activating kinase. Mol Cell Biol. 1997;17:7362–7374. doi: 10.1128/mcb.17.12.7362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Diehl J A, Zindy F, Sherr C J. Inhibition of cyclin D1 phosphorylation on threonine-286 prevents its rapid degradation via the ubiquitin-proteasome pathway. Genes Dev. 1997;11:957–972. doi: 10.1101/gad.11.8.957. [DOI] [PubMed] [Google Scholar]
- 21.Earp H S, Dawson T L, Li X, Yu H. Heterodimerization and functional interaction between EGF receptor family members: a new signaling paradigm with implications for breast cancer research. Breast Cancer Res Treat. 1995;35:115–132. doi: 10.1007/BF00694752. [DOI] [PubMed] [Google Scholar]
- 22.Flemington E K, Speck S H, Kaelin W G J. E2F-1-mediated transactivation is inhibited by complex formation with the retinoblastoma susceptibility gene product. Proc Natl Acad Sci USA. 1993;90:6914–6918. doi: 10.1073/pnas.90.15.6914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Galbiati F, Volonte D, Engelman J A, Watanabe G, Burk R, Pestell R G, Lisanti M P. Targeted Down-regulation of caveolin-1 is sufficient to drive cell transformation and activate the p42/p44 MAP kinase cascade. EMBO J. 1998;17:6633–6648. doi: 10.1093/emboj/17.22.6633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guy C T, Cardiff R D, Muller W J. Activated neu induces rapid tumor progression. J Biol Chem. 1996;271:7673–7678. doi: 10.1074/jbc.271.13.7673. [DOI] [PubMed] [Google Scholar]
- 25.Guy C T, Webster M A, Schaller M, Parson T J, Cardiff R D, Muller W J. Expression of the neu proto-oncogene in the mammary epithelium of transgenic mice induces metastatic disease. Proc Natl Acad Sci USA. 1992;89:10578–10582. doi: 10.1073/pnas.89.22.10578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hirai H, Sherr C. Interaction of D-type cyclins with a novel Myb-like transcription factor, DMP1. Mol Cell Biol. 1996;16:6457–6467. doi: 10.1128/mcb.16.11.6457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Janes P W, Daly R J, deFazio A, Sutherland R L. Activation of the Ras signalling pathway in human breast cancer cells overexpressing erbB-2. Oncogene. 1994;9:3601–3608. [PubMed] [Google Scholar]
- 28.Jardines L, Weiss M, Fowble B, Greene M. neu (c-erbB-2/HER2) and the epidermal growth factor receptor (EGFR) in breast cancer. Pathobiology. 1993;61:268–282. doi: 10.1159/000163805. [DOI] [PubMed] [Google Scholar]
- 29.Joyce D, Bouzahzah B, Fu M, D'Amico M, Albanese C, Steer J, Klein J U, Lee R J, Segal J D, Westwick J K, Der C J, Pestell R G. Integration of Rac-dependent regulation of cyclin D1 transcription through an NF-kB-dependent pathway. J Biol Chem. 1999;274:25245–25249. doi: 10.1074/jbc.274.36.25245. [DOI] [PubMed] [Google Scholar]
- 30.Karlseder J, Rotheneder H, Wintersberger E. Interaction of Sp1 with growth- and cell cycle-regulated transcription factor E2F. Mol Cell Biol. 1996;16:1659–1667. doi: 10.1128/mcb.16.4.1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lacroix H, Iglehart J D, Skinner M A, Kraus M H. Overexpression of erbB-2 or EGF-receptor proteins present in early stage mammary carcinoma is detected simultaneously in matched primary tumors and regional metastasis. Oncogene. 1989;4:145–151. [PubMed] [Google Scholar]
- 32.Lammie G A, Fantl V, Smith R, Schuuring E, Brookes S, Michalides R, Dickson C, Arnold A, Peters G. D11S287, a putative oncogene on chromosome 11q13, is amplified and expressed in squamous cell and mammary carcinomas and linked to BCL-1. Oncogene. 1991;6:439–444. [PubMed] [Google Scholar]
- 33.Lee R J, Albanese C, Stenger R J, Watanabe G, Inghirami G, Haines III G K, Webster M, Muller W J, Brugge J S, Davis R, Pestell R G. pp60v-src induction of cyclin D1 requires collaborative interactions between the ERK, p38 and Jun kinase pathways: a role for CREB and ATF-2 in pp60v-src signaling in breast cancer cells. J Biol Chem. 1999;274:7341–7350. doi: 10.1074/jbc.274.11.7341. [DOI] [PubMed] [Google Scholar]
- 34.Li R, Hannon G J, Beach D, Stillman B. Subcellular distribution of p21 and PCNA in normal and repair-deficient cells following DNA damage. Curr Biol. 1996;6:189–199. doi: 10.1016/s0960-9822(02)00452-9. [DOI] [PubMed] [Google Scholar]
- 35.Liu J-J, Chao J-R, Jiang M-C, Ng S-Y, Yen J J-Y, Yang-Yen H-F. Ras transformation results in an elevated level of cyclin D1 and acceleration of G1 progression in NIH 3T3 cells. Mol Cell Biol. 1995;15:3654–3663. doi: 10.1128/mcb.15.7.3654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lukas J, Bartkova J, Bartek J. Convergence of mitogenic signalling cascades from diverse classes of receptors at the cyclin D–cyclin-dependent kinase–pRb-controlled G1 checkpoint. Mol Cell Biol. 1996;16:6917–6925. doi: 10.1128/mcb.16.12.6917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lukas J, Bartkova J, Rohde M, Srauss M, Bartek J. Cyclin D1 is dispensable for G1 control in retinoblastoma gene-deficient cells independently of cdk4 activity. Mol Cell Biol. 1995;15:2600–2611. doi: 10.1128/mcb.15.5.2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Matsumura I, Kitamura T, Wakao H, Tanaka H, Hashimoto K, Albanese C, Downward J, Pestell R G, Kanakura Y. Transcriptional regulation of cyclin D1 promoter by STAT5: its involvement in cytokine-dependent growth of hematopoietic cells. EMBO J. 1999;18:101–111. doi: 10.1093/emboj/18.5.1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Miltenyi S, Muller W, Weichel W, Radbruch A. High gradient magnetic cell separation with MACS. Cytometry. 1990;11:231–238. doi: 10.1002/cyto.990110203. [DOI] [PubMed] [Google Scholar]
- 40.Minden A, Lin A, Claret F-X, Abo A, Karin M. Selective activation of the JNK signalling cascade and c-jun transcriptional activity by the small GTPases RAC and Cdc42Hs. Cell. 1995;81:1147–1157. doi: 10.1016/s0092-8674(05)80019-4. [DOI] [PubMed] [Google Scholar]
- 41.Muller W J, Sinn E, Pattengale P K, Wallace R, Leder P. Single step induction of mammary adenocarcinoma in transgenic mice bearing the activated c-neu oncogene. Cell. 1988;54:105–115. doi: 10.1016/0092-8674(88)90184-5. [DOI] [PubMed] [Google Scholar]
- 42.Musgrove E A, Lee C S, Buckley M F, Sutherland R L. Cyclin D1 induction in breast cancer cells shortens G1 and is sufficient for cells arrested in G1 to complete the cell cycle. Proc Natl Acad Sci USA. 1994;91:8022–8026. doi: 10.1073/pnas.91.17.8022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Muthuswamy S K, Siegel P M, Dankort D L, Webster M A, Muller W J. Mammary tumors expressing the neu proto-oncogene possess elevated c-Src tyrosine kinase activity. Mol Cell Biol. 1994;14:735–743. doi: 10.1128/mcb.14.1.735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nehls M C, Grapilon M L, Brenner D A. NF-I/Sp1 switch elements regulate collagen alpha 1(I) gene expression. DNA Cell Biol. 1992;11:443–452. doi: 10.1089/dna.1992.11.443. [DOI] [PubMed] [Google Scholar]
- 45.Neuman E, Ladha M, Lin N, Upton T M, Miller S J, DiRenzon J, Pestell R G, Hinds P W, Dowdy S F, Brown M, Ewen M E. Cyclin D1 stimulation of estrogen receptor transcription independent of Cdk4 activation. Mol Cell Biol. 1997;17:5338–5347. doi: 10.1128/mcb.17.9.5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Paulus W, Baur I, Boyce F M, Breakefield X O, Reeves S A. Self-contained, tetracycline-regulated retroviral vector system for gene delivery to mammalian cells. J Virol. 1996;70:62–67. doi: 10.1128/jvi.70.1.62-67.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Peles E, Yarden Y. Neu and its ligands: from an oncogene to neural factors. Bioessays. 1993;15:815–824. doi: 10.1002/bies.950151207. [DOI] [PubMed] [Google Scholar]
- 48.Pietras R J, Arboleda J, Reese D M, Wongvipat N, Pegram M D, Ramos L, Gorman C M, Parker M G, Sliwkowski M X, Slamon D J. HER-2 tyrosine kinase pathway targets estrogen receptor and promotes hormone-independent growth in human breast cancer cells. Oncogene. 1995;10:2435–2446. [PubMed] [Google Scholar]
- 49.Pinkas-Kramarski R, Soussan L, Waterman H, Levkowitz G, Alroy I, Klapper L, Lavi S, Seger R, Ratzkin B J, Sela M, Yarden Y. Diversification of Neu differentiation factor and epidermal growth factor signaling by combinatorial receptor interactions. EMBO J. 1996;15:2452–2467. [PMC free article] [PubMed] [Google Scholar]
- 50.Proud C G. p70 S6 kinase: an enigma with variations. Trends Biochem Sci. 1996;21:181–185. [PubMed] [Google Scholar]
- 51.Qian X, Dougall W C, Fei Z, Greene M I. Intermolecular association and trans-phosphorylation of different neu-kinase forms permit SH2-dependent signaling and oncogenic transformation. Oncogene. 1995;10:211–219. [PubMed] [Google Scholar]
- 52.Qiu R G, Chen J, Kirn D, McCormick F, Symons M. An essential role for Rac in Ras transformation. Nature. 1995;374:457–459. doi: 10.1038/374457a0. [DOI] [PubMed] [Google Scholar]
- 53.Qiu R G, Chen J, McCormick F, Symons M. A role for Rho in Ras transformation. Proc Natl Acad Sci USA. 1995;92:11781–11785. doi: 10.1073/pnas.92.25.11781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Resnitzky D, Reed S I. Different roles for cyclins D1 and E in regulation of the G1-to-S transition. Mol Cell Biol. 1995;15:3463–3469. doi: 10.1128/mcb.15.7.3463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Scherer P E, Lewis R Y, Volonte D, Engelman J A, Galbiati F, Couet J, Kohtz D S, van Donselaar E, Peters P, Lisanti M P. Cell-type and tissue-specific expression of caveolin-2: caveolins 1 and 2 co-localize and form a stable hetero-oligomeric complex in vivo. J Biol Chem. 1997;272:29337–29346. doi: 10.1074/jbc.272.46.29337. [DOI] [PubMed] [Google Scholar]
- 56.Schuuring E, Verhoeven E, Mooi W J, Michalides R J. Identification and cloning of two overexpressed genes, U21B31/PRAD1 and EMS1, within the amplified chromosome 11q13 region in human carcinomas. Oncogene. 1992;7:355–361. [PubMed] [Google Scholar]
- 57.Sherr C J. Cancer cell cycles. Science. 1996;274:1672–1677. doi: 10.1126/science.274.5293.1672. [DOI] [PubMed] [Google Scholar]
- 58.Shin T H, Paterson A J, Grant J H 3, Meluch A A, Kudlow J E. 5-Azacytidine treatment of HA-A melanoma cells induces Sp1 activity and concomitant transforming growth factor alpha expression. Mol Cell Biol. 1992;12:3998–4006. doi: 10.1128/mcb.12.9.3998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sicinski P, Donaher J L, Parker S B, Li T, Fazeli A, Gardner H, Haslam S Z, Bronson R T, Elledge S J, Weinberg R A. Cyclin D1 provides a link between development and oncogenesis in the retina and breast. Cell. 1995;82:621–630. doi: 10.1016/0092-8674(95)90034-9. [DOI] [PubMed] [Google Scholar]
- 60.Siebenkotten G, Behrens-Jung U, Miltenyi S, Petry K, Radbruch A. Employing surface markers for the selection of transfected cells. In: Recktenwald D, Radbruch A, editors. Cell separation methods and applications. New York, N.Y: Marcel Dekker; 1998. pp. 271–281. [Google Scholar]
- 61.Siegel P M, Dankort D L, Hardy W R, Muller W J. Novel activating mutations in the neu proto-oncogene involved in induction of mammary tumors. Mol Cell Biol. 1994;14:7068–7077. doi: 10.1128/mcb.14.11.7068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Siegel P M, Ryan E D, Cardiff R D, Muller W J. Elevated expression of activated forms of Neu/ErbB-2 and ErbB-3 are involved in the induction of mammary tumors in transgenic mice: implications for human breast cancer. EMBO J. 1999;18:2149–2164. doi: 10.1093/emboj/18.8.2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sif S, Gilmore T D. Interaction of the v-Rel oncoprotein with cellular transcription factor Sp1. J Virol. 1994;68:7131–7138. doi: 10.1128/jvi.68.11.7131-7138.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Slamon D J, Clark G M, Wong S G, Levin W J, Ullrich A, McGuire W L. Human breast cancer: correlation of relapse and survival with amplification of the HER-2/neu oncogene. Science. 1987;235:177–182. doi: 10.1126/science.3798106. [DOI] [PubMed] [Google Scholar]
- 65.Solomon M J, Larsen P L, Varshavsky A. Mapping protein-DNA interactions in vivo with formaldehyde: evidence that histone H4 is retained on a highly transcribed gene. Cell. 1988;53:937–947. doi: 10.1016/s0092-8674(88)90469-2. [DOI] [PubMed] [Google Scholar]
- 66.Tao Y, Kassatly R F, Cress W D, Horowitz J M. Subunit composition determines E2F DNA-binding site specificity. Mol Cell Biol. 1997;17:6994–7007. doi: 10.1128/mcb.17.12.6994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Van Aelst L, D'Souza-Schorey C. Rho GTPases and signaling networks. Genes Dev. 1997;11:2295–2322. doi: 10.1101/gad.11.18.2295. [DOI] [PubMed] [Google Scholar]
- 68.Wallasch C, Weiss F U, Niederfellner G, Jallal B, Issing W, Ullrich A. Heregulin-dependent regulation of HER2/neu oncogenic signaling by heterodimerization with HER3. EMBO J. 1995;14:4267–4275. doi: 10.1002/j.1460-2075.1995.tb00101.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang T C, Cardiff R D, Zukerberg L, Lees E, Arnold A, Schmidt E V. Mammary hyperplasia and carcinoma in MMTV-cyclin D1 transgenic mice. Nature. 1994;369:669–671. doi: 10.1038/369669a0. [DOI] [PubMed] [Google Scholar]
- 70.Watanabe G, Albanese C, Lee R J, Reutens A, Vairo G, Henglein B, Pestell R G. Inhibition of cyclin D-kinase activity is associated with E2F-mediated inhibition of cyclin D1 promoter activity through E2F and Sp1. Mol Cell Biol. 1998;18:3212–3222. doi: 10.1128/mcb.18.6.3212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Watanabe G, Howe A, Lee R J, Albanese C, Shu I-W, Karnezis A, Zon L, Kyriakis J, Rundell K, Pestell R G. Induction of cyclin D1 by simian virus 40 small tumor antigen. Proc Natl Acad Sci USA. 1996;93:12861–12866. doi: 10.1073/pnas.93.23.12861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Watanabe G, Lee R J, Albanese C, Rainey W E, Batlle D, Pestell R G. Angiotensin II (AII) activation of cyclin D1-dependent kinase activity. J Biol Chem. 1996;271:22570–22577. doi: 10.1074/jbc.271.37.22570. [DOI] [PubMed] [Google Scholar]
- 73.Weinberg R A. E2F and cell proliferation: a world turned upside down. Cell. 1996;85:457–459. doi: 10.1016/s0092-8674(00)81244-1. [DOI] [PubMed] [Google Scholar]
- 74.Weinberg R A. The retinoblastoma protein and cell cycle control. Cell. 1995;81:323–330. doi: 10.1016/0092-8674(95)90385-2. [DOI] [PubMed] [Google Scholar]
- 75.Weinstat-Saslow D, Merino M J, Manrow R E, Lawrence J A, Bluth R F, Wittenbel K D, Simpson J F, Page D L, Steeg P S. Overexpression of cyclin D mRNA distinguishes invasive and in situ breast carcinomas from non-malignant lesions. Nat Med. 1995;1:1257–1259. doi: 10.1038/nm1295-1257. [DOI] [PubMed] [Google Scholar]
- 76.Westwick J K, Lambert Q T, Clark G J, Symons M, Van Aelst L, Pestell R G, Der C J. Rac regulation of transformation, gene expression, and actin organization by multiple, PAK-independent pathways. Mol Cell Biol. 1997;17:1324–1335. doi: 10.1128/mcb.17.3.1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wymann M P, Bulgarelli-Leva G, Zvelebil M J, Pirola L, Vanhaesebroeck B, Waterfield M D, Panayotou G. Wortmannin inactivates phosphoinositide 3-kinase by covalent modification of Lys-802, a residue involved in the phosphate transfer reaction. Mol Cell Biol. 1996;16:1722–1733. doi: 10.1128/mcb.16.4.1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Xie Y, Li K, Hung M-C. Tyrosine phosphorylation of Shc proteins and formation of Shc/Grb2 complex correlate to the transformation of NIH3T3 cells mediated by the point-mutation activated neu. Oncogene. 1995;10:2409–2413. [PubMed] [Google Scholar]
- 79.Xiong W, Pestell R G, Rosner M R. Role of cyclins in neuronal differentiation of immortalized hippocampal cells. Mol Cell Biol. 1997;17:6585–6597. doi: 10.1128/mcb.17.11.6585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Xu G, Livingston D M, Krek W. Multiple members of the E2F transcription factor family are the products of oncogenes. Proc Natl Acad Sci USA. 1995;92:1357–1361. doi: 10.1073/pnas.92.5.1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ye J, Xu R H, Taylor-Papadimitriou J, Pitha P M. Sp1 binding plays a critical role in Erb-B2- and v-ras-mediated downregulation of α2-integrin expression in human mammary epithelial cells. Mol Cell Biol. 1996;16:6178–6189. doi: 10.1128/mcb.16.11.6178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zerfass-Thome K, Schulze A, Zwerschke W, Vogt B, Helin K, Bartek J, Henglein B, Jansen-Durr P. p27KIP1 blocks cyclin E-dependent transactivation of cyclin A gene expression. Mol Cell Biol. 1997;17:407–415. doi: 10.1128/mcb.17.1.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zwijsen R M L, Klompmaker R, Wientjens E B H G M, Kristel P M P, van der Burg B, Michalides R J A M. Cyclin D1 triggers autonomous growth of breast cancer cells by governing cell cycle exit. Mol Cell Biol. 1996;16:2554–2560. doi: 10.1128/mcb.16.6.2554. [DOI] [PMC free article] [PubMed] [Google Scholar]