Abstract
Atomic-resolution protein structural models are prerequisites for many downstream activities like structure-function studies or structure-based drug discovery. Unfortunately, this data is often unavailable for some of the most interesting and therapeutically important proteins. Thus, computational tools for building native-like structural models from less-than-ideal experimental data are needed. To this end, interaction homology exploits the character, strength and loci of the sets of interactions that define a structure. Each residue type has its own limited set of backbone angle-dependent interaction motifs, as defined by their environments. In this work, we characterize the interactions of serine, cysteine and S-bridged cysteine in terms of 3D hydropathic environment maps. As a result, we explore several intriguing questions. Are the environments different between the isosteric serine and cysteine residues? Do some environments promote the formation of cystine S–S bonds? With the increasing availability of structural data for water-insoluble membrane proteins, are there environmental differences for these residues between soluble and membrane proteins? The environments surrounding serine and cysteine residues are dramatically different: serine residues are about 50% solvent exposed, while cysteines are only 10% exposed; the latter are more involved in hydrophobic interactions although there are backbone angle-dependent differences. Our analysis suggests that one driving force for –S–S– bond formation is a rather substantial increase in burial and hydrophobic interactions in cystines. Serine and cysteine become less and more, respectively, solvent-exposed in membrane proteins. 3D hydropathic environment maps are an evolving structure analysis tool showing promise as elements in a new protein structure prediction paradigm.
Keywords: Serine, Cysteine, Cystine, Hydropathic interactions, Soluble proteins, Membrane proteins, Hydrophobic effect
Graphical abstract
Highlights
-
•
Ser and Cys residues have unique sets of 3D hydropathic interaction environments.
-
•
Ser is more solvent exposed and usually engages in strong hydrogen bonds.
-
•
Cys is likely to be buried and found mostly in hydrophobic interaction environments.
-
•
Cys –S–S– bridge formation is driven by further burial in hydrophobic environments.
-
•
Ser residues in membrane proteins are less solvent exposed while Cys are more.
1. Introduction
In 1957 and in 1960, Sir John Kendrew and Max Perutz solved the first structures for myoglobin and hemoglobin, respectively (Kendrew et al., 1960; Perutz et al., 1960). These extraordinary achievements awarded them the Nobel Prize in chemistry in 1962. Their structural models revealed the complexity of protein 3D structure, including the α helices and β pleats predicted by Linus Pauling in 1951 (Pauling and Corey, 1951; Pauling et al., 1951). Although Pauling never solved a protein crystal structure, he discovered the fundamental structural motifs of proteins. Perhaps most importantly, he proposed that protein structure should arise from a repetition of stable motifs (Edison, 2001), i.e., the existence of biological polymers. Therefore, it is crucial to investigate the basic units of the protein framework, a knowledge that could unwind its biochemical mechanism and provide aid to many relevant biological questions. At the heart of a protein blueprint, there are 20 different amino acids with a surprising array of distinctive characteristics encoded in their sidechains. The geometry (Robson and Suzuki, 1976; Shapovalov and Dunbrack, 2011) and chemistry (Bywater, 2018, Lodish et al., 2000) of these residues represent the raw material of a complex mosaic of details organized into a biologically meaningful whole (Richardson, 1981). A single variation in one of these themes can alter the functionality of a protein, as well as its structure.
The properties of each of the 20 amino acid residues are likely to be influenced by the local environment surrounding them: the protein core, cofactors, water, or the lipid bilayer. In fact, the specific identity of each amino acid residue is directed by its environment as the result of evolution. We previously described how, regardless of the localization in the protein, amino acid hydropathic environments will cluster into limited sets of three-dimensional microenvironments, each possessing a unique system of interactions (Ahmed et al., 2015; Ahmed et al., 2019; AL Mughram et al., 2021; Herrington and Kellogg, 2021). Our core HINT model classifies interactions in terms of four classes: favorable polar (e.g., hydrogen bond, acid-base), unfavorable polar (acid-acid, base-base, repulsive Coulombic), favorable hydrophobic (hydrophobic-hydrophobic, hydrophobic packing, π-π stacking), and unfavorable hydrophobic (hydrophobic-polar, desolvation) (Kellogg and Abraham, 2000; Kellogg et al., 1991; Sarkar and Kellogg, 2010).
This work will focus on the similarity and dissimilarity of two amino acid residues: serine (SER) and cysteine (CYS). Many intriguing questions can be posed concerning these two residues. The first is: are the environments different between these isosteric residues, or, to turn it around, why has Nature settled on a serine vs. a cysteine in a particular locus? Clearly, one reason is cysteine's ability to form CYS-CYS disulfide bridges, a major structural feature of proteins, which then suggests a second question: are there discernible environmental differences around cysteines that bridge and those that do not? Third, with the increasing availability of structural data for water-insoluble membrane proteins, another question is: are there environmental differences for CYS and SER residues between soluble and membrane proteins?
1.1. Serine vs. cysteine
Serine is among the most frequently found amino acids in proteins due to the six codons (9.37% of the genetic code) encoding it, whereas cysteine is only coded by two (3.12%). Current thinking is that cysteine is a later addition to the genetic code, accumulating in complex organisms, ranging from 0.50% in some bacteria to 2.26% in mammals (Brooks and Fresco, 2002; Foden et al., 2020; Liu et al., 2010; Miseta and Csutora, 2000; Trifonov, 2000). In a broader sense, it has been known for some time that a relationship exists between the physicochemical properties of the amino acids and specific aspects of their codon positions and identities (Sjostrom and Wold, 1985).
Serine and cysteine would seemingly be the two most similar amino acids. The difference of only a sulfur atom replacing an oxygen yields only minor differences in bond lengths and angles, and the result is that these two residues are isosteric and perhaps isostructural. However, serine, by nature, is highly polar owing to its sidechain hydroxyl, with a log10Po/w of around −5. While the sidechain is electrically neutral, this functional group creates an electric dipole giving a partial charge and has dual characteristics of a hydrogen bond donor and acceptor. This feature suggests that SER has a high probability of being on the protein surface and having high solvent accessibility (Tien et al., 2013).
Serine's congener, CYS, has a larger bag of tricks: it is both more hydrophobic (sidechain log10Po/w ∼ −4), but also more acidic with a pKa < 9 (vs. SER's 13) (Hofer et al., 2020). Its slightly polar nature is due to the partial negative charge from the four unpaired electrons in the outer shell of sulfur. Compared with the other polar residues, cysteine is a very poor hydrogen donor or acceptor but can form weak to intermediately strong interactions. There are some suggestions that the sulfur σ∗ orbital can interact with π electron clouds (Beno et al., 2015; Zhou et al., 2009). The weak polarity of cysteine is also evidenced by its buriedness compared to the other polar residues, which is, in this case, probably due to its thiol being very reactive and easily oxidized (Marino and Gladyshev, 2010).
Serine is often phosphorylated, glycosylated or N-acetylated. These post-translational modifications are crucial for organic life (Barzkar et al., 2021), and is a precursor in the biosynthesis of other amino acids like glycine, cysteine, and D-serine (Murtas et al., 2020). SER is also essential in the production of phosphoglycerides, glycerides, sphingolipids and phosphatidylserine (Hirabayashi and Furuya, 2008; Kent, 1995). Serine residues are also found in the active site (as a nucleophile) with histidine and aspartic acid (i.e., the catalytic triad) of serine protease, the most abundant type of endopeptidase, in which they perform several functions (Di Cera, 2009). Although cysteine is the least abundant residue, it still plays essential physiological roles. For example, it is found in the active site of cysteine (thiol) protease, an endopeptidase similar to serine protease in which the cysteine residue takes the role of the nucleophile. Cysteine and histidine are the most frequent residues to coordinate to metals, and are often found stabilizing protein structure elements such as zinc fingers (Klug and Schwabe, 1995), or involved in catalytic reactions by acting as a redox switch (Klomsiri et al., 2011; Kroncke and Klotz, 2009). Like its counterpart, cysteine is also subject to reversible post-translational modifications, e.g., participating in S-nitrosylation and glutathionylation or forming sulfenic acid and most interestingly disulfide bonds (Duan et al., 2017). Other functionalizations include thioether bonds with lipid residues that anchor the protein to the membrane (i.e., prenylation) (Casey and Seabra, 1996; Paulsen and Carroll, 2013), and disulfide bonds with endogenous hydrogen sulfide, a potent signal molecule (Marino and Gladyshev, 2011). Also, cysteine is an especially well-known target for covalent (irreversible) inhibition (Singh et al., 2011; Hallenbeck et al., 2017, Long and Aye, 2017; Heppner, 2021).
As described above, there are many similarities and differences between serine and cysteine. There have been many studies focusing on one or the other of these two residues, but surprisingly few rationalizing their differences, and none to our knowledge exploring the differences between their environments within proteins. We believe that our unique approach of sampling interaction environments with 3D maps could shed light on the nature and differences in serine and cysteine structural and physicochemical properties, and perhaps yield insight into their differences on a functional basis.
1.2. The mechanism of cysteine bridging
When two cysteines in close proximity are under oxidizing conditions, they can covalently bond, losing two protons and two electrons, resulting in a disulfide bond, sometimes called cystine. Enzymes often catalyze this reaction in the endoplasmic reticulum. However, the propensity of two cysteines to dimerize is due to the size of the sulfur atom; its large volume can better stabilize the negative charge of the transition state (Thornton, 1981). Disulfides have a prominent and unique role in stabilizing a protein structure, and they are typically found in proteins excreted in harsh, e.g., extracellular, environments. The bridge not only contributes to protein stability, primarily by decreasing the entropy of the unfolded state, but also protects the highly reactive thiol group, thus avoiding cysteine side reactions. The formation of the disulfide bond is also involved in the sensing and signaling of oxidative stress intracellularly; vice versa, the reduction of the bond may serve as an effector functioning extracellularly (Bhattacharyya et al., 2004; Bulaj, 2005; Dombkowski et al., 2014; Fass, 2012; Moomaw et al., 1995; Petersen et al., 1999).
Understanding the forces driving cysteine bridging has been a major computational goal for many years. A wide variety of methodologies have been used, particularly molecular dynamics and related tools, with a number of parameters (Marino and Gladyshev, 2012; Manteca et al., 2017; Qin et al., 2015). While all of these studies took into consideration the influence of the local environment, we believe that our 3D map tools will probe these interaction environments in a different way, and help explain the difference between a free cysteine and one participating in a covalent bond. In particular, the HINT model is very sensitive to small differences in hydrophobicity.
1.3. Membrane proteins
Membrane proteins (MPs) play a central role in many cellular functions (Coskun and Simons, 2011). They represent ∼20% of the human proteome, and they act as enzymes, transporters, ion channels, and receptors (Wallin and von Heijne, 1998). The efficiency of these activities has been proven to be mainly modulated by specific interactions between MPs and their local lipid environments (Guo, 2020). Despite intensive research, the thermodynamics driving this process remains fairly ambiguous. The barrier to understanding is likely the hydrophobic nature of transmembrane segments of proteins and lipid bilayers (MacCallum et al., 2007; McIntosh and Simon, 2007; White, 2007; Wolfenden, 2007). At an amino acid level, the change in the properties, e.g., electrostatic potential, pressure, pH and dielectric constant, of the membrane result in a bias for a particular amino acid to be located in a different part of the protein. Because of the different environments, even the rotamers of these amino acids could differ if compared with the soluble proteins (Ramachandran et al., 1963; Ramachandran and Sasisekharan, 1968). Therefore, our questions arise: a) Are serine, cysteine and cystine disulfide bridges in membrane proteins fundamentally different from those in soluble proteins? b) Are these differences manifested in the hydropathic interaction environments surrounding these residues? c) If these environments are different, are there new principles of protein structure analysis and prediction to be discerned?
1.4. 3D interaction homology
To explore these phenomena, we characterized the residue environments by assembling a database of backbone-angle-dependent 3D maps that fully describe the sets of preferred conformations and interaction environments surrounding each. We have already analyzed other residue types, tyrosine (Ahmed et al., 2015), alanine (Ahmed et al., 2019), the aromatic residues (AL Mughram et al., 2021) and aspartic acid, glutamic acid and histidine (Herrington and Kellogg, 2021) with this approach. These reports revealed significant insight about protein structure, largely due to the initially unexpected fact that there were commonalities in these maps independent of the identity of the specific molecular species in the environment. Thus, the maps – sometimes many thousands – could be clustered into limited sets of distinct and informative environmental interaction motifs. These maps encode interaction types, interaction strengths and the 3D midpoint of the interacting atoms. Furthermore, the maps record π-π and donor-π interactions (AL Mughram et al., 2021), and are responsive to local and global pH effects.
In this contribution, we are using this suite of 3D map-based structural analysis tools to explore three fundamental structural properties of serine and cysteine. First, we will mine our protein structure database to isolate and model with 3D maps the environments surrounding SER and CYS residues, and identify similarities and differences between the two residues. We will show that the seemingly small structural and physicochemical differences between SER and CYS produce significantly divergent environments in proteins. Second, we will also extract bridging cysteine residues (in this work "CYX") from the structure database to examine the potential differences in environments that may support bridge formation. To simulate the pre-bridge environments, we broke the bridges and protonated both cysteines to create a new data set called "CYZ". In our analyses, the hydropathic environmental differences between CYS and CYZ are surprisingly subtle, with CYZ being somewhat more likely found in polar environments. Third, we extracted serine and cysteine 3D hydropathic interaction datasets from a structural collection of membrane proteins that were artificially "solvated" with lipids and subjected to dynamics. These sets, "SERm", "CYSm" and "CYXm", are ideal for comparing to soluble protein hydropathic interaction environments, and several notable differences will be described.
2. Results and discussion
2.1. Structural data
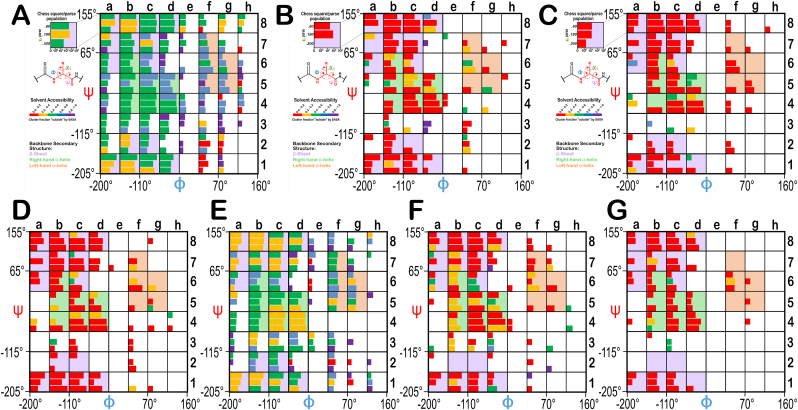
From the dataset of 2703 soluble protein structures used in our previous studies (Ahmed et al., 2015; Ahmed et al., 2019; AL Mughram et al., 2021), we extracted 46,869 SER, 5500 CYS and 5237 CYX residues. From the dataset suggested by Grazhdankin et al. (2020) we selected 369 membrane protein structures from the MemProtMD database (Newport et al., 2019) with 23,791 serines, 4265 cysteines and 962 S–S bridged cysteines residues, which we are naming SERm, CYSm and CYXm, respectively. The process of binning and parsing our residues dataset is well-documented and thoroughly explained in earlier publications (Ahmed et al., 2015; Ahmed et al., 2019; AL Mughram et al., 2021). Briefly, the Ramachandran plot was modified by superimposing an 8 by 8 chessboard, shifted such that each grid square contains higher-density populations rather than splitting them between two chess squares, with dimensions of 45° by 45° in φ – ψ (see Fig. 1) (Ramachandran et al., 1963; Ramachandran and Sasisekharan, 1968). The grid squares were named as a1 through h8. Each residue was then classified into its square by its backbone φ and ψ angle, superimposed onto the center of that chess square (Ahmed et al., 2015), and further parsed by their χ1 angles into three groups corresponding to those typically observed in rotamer libraries: ∼60°, ∼180°, and ∼300°. While this parsing is not per se necessary, it does provide increased computational efficiency although the map-based clustering (vide infra) generally identified this low level of detail nearly flawlessly (Ahmed et al., 2015). From here on, the chess square names will be given in bold italics, e.g., b1, c5, etc. The χ1 parses will be denoted by the suffixes .60, .180, and .300.
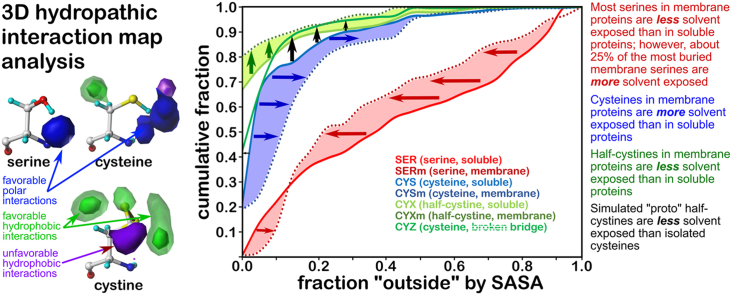
Fig. 1.
Ramachandran chessboard displaying the chess square/parse population for each residue type. The Ramachandran φ vs ψ plot is rendered into sixty-four 45°by 45° (π/4 by π/4) chess squares. The (χ1) parse (∼60°, ∼180°, ∼300°) populations are represented in log10 scale with the lengths of the colored bars. Their colors reflect the average weighted fraction outside or solvent-exposed, that is, “foutside” a measure of solvent accessibility (see text for definition). The φ vs ψ regions associated with β-pleat, α-helix, and left-hand α-helix secondary structure motifs are shaded in light purple, light green, and light orange, respectively. A) serine in soluble proteins (SER), B) cysteine in soluble proteins (CYS), C) cysteine with intact disulfide bridge in soluble proteins (CYX), D) cysteine with “broken” disulfide bridge in soluble proteins (CYZ), E) serine in membrane proteins (SERm), F) cysteine in membrane proteins (CYSm), and G) cysteine with intact disulfide bridge in membrane proteins (CYXm). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
The occupancies of the chess square/parses range from 0 to 3255 (d4.300) for SER, to 3711 (d4.300) for SERm, to 823 (d4.300) for CYS, to 1388 (d4.300) for CYSm, to 606 (c8.300) for CYX and to 124 (c8.300) for CYXm. The broken bridge cysteine, CYZ, has an occupancy of 684 in d4.300. For SER, 119 (of 192) chess square/parses contain 5 or more residues. For SERm, CYS, CYSm, CYX, CYXm and CYZ, 105, 56, 52, 47, 33 and 51 chess square/parses, respectively, contain 5 or more residues. Table S2 (Supporting Information) displays the occupancies in the Ramachandran chessboards for these seven residues. Any parses with less than 5 members were not clustered. To simplify nomenclature in this article, we use a numerical scheme wherein the sequential number of that residue in its chess square/parse is its name. Thus, cysteine 100 in chess square a1.60 is the 100th cysteine in that chess square/parse combination. Supporting Information Table S1 decodes this nomenclature in terms of the actual pdbid, residue number and chain ID for each amino acid residue included in this study. Clusters (vide infra) will be named for the residue closest to its centroid or exemplar and will be given in bold numerals.
As in our previous work, we will be focusing the discussion on only four of the chess squares, sampling three of the common secondary structure elements β-pleat with b1, right-hand α-helix with c5 and d5 and left-hand α-helix with f6. The c5, d5 pair allows us to compare independently-calculated map and environment data between chess squares within the same right-hand α-helix structural motif region. However, all numeric data for all chess squares is available as Supporting Information Tables S4–S10.
2.2. Calculation and clustering of hydropathic environments
Using methods we previously reported, we used the HINT force field and score model to evaluate interatomic interactions (Kellogg et al., 1991; Kellogg and Abraham, 2000; Sarkar and Kellogg, 2010). This forcefield is derived from log Po/w (for 1-octanol and water solute transfer), a measure of the free energy of interaction, and a term related to the solvent-accessible surface area. HINT has shown the ability to estimate ΔΔG for ligand-protein, protein-protein, and other complexes in various systems, such that a change of ∼500 HINT score units = −1 kcal mol−1 (Burnett et al., 2001; Cozzini et al., 2004). Using atom-atom interaction scores applied to three-dimensional Gaussians at the midpoint between the atoms, maps were constructed by sampling within rectangular boxes large enough to contain each of the studied residue types along with their interacting atoms (see Methods). For this report, we are most interested in the interactions made between the residue sidechains and the remainder of the protein. Our maps categorize interactions in "quartets" of four distinct types: favorable polar, unfavorable polar, favorable hydrophobic, and unfavorable hydrophobic.
As stated above, a major hypothesis in our studies is that, despite there being thousands of each residue type in our dataset, there will be only a relatively small number of unique 3D hydropathic interaction environments that are specific to the residue's chemical properties. We refer to this as the hydropathic valence, i.e., the constellation of interactions made by the residues. These interactions are characterized by their type, strength and geometry, but not necessarily molecular or residue identity. To extract the information encoded in the 3D hydropathic interaction maps, we applied a map-map similarity metric to score two maps m and n (see Methods) (Ahmed et al., 2015). After loading the similarities for each m-n map pair in each chess square/parse into square matrices, we performed statistical analysis by clustering these matrices with k-means within the R programming environment (Hartigan and Wong, 1979; R Development Core Team, 2013). We set a maximum number of 6 clusters per chess square/parse for SER and CYS in the soluble proteins dataset, which was sufficient for capturing the diversity of residue environments. We expected a greater diversity for these residues in the membrane protein dataset due to the additional possibility of interactions with the lipid bilayer. Thus, we set a maximum of 8 clusters for SERm and CYSm. Finally, both CYX and CYXm were allowed to cluster with up to 10 clusters to compensate for the flexibility of the disulfide bond. Table S2 (SI) sets out the numbers of clusters found for each chess square/parse of the seven residue-type datasets in this study.
If clustering failed to reach this predetermined maximum, the optimum number of clusters for a chess square/parse was generally chosen as the most stable solution available. An average set of maps was then calculated for each cluster using a Gaussian weighting scheme (Euclidian distance from the cluster's centroid) reported earlier (Ahmed et al., 2015). Using the same scheme, the average molecular structure of the residue for the cluster's members was also calculated. Residue and atomistic RMSDs and average χ1 angles were calculated as additional evaluation metrics to validate the map-based clustering.
2.3. Solvent-accessible surface area (SASA) and foutside
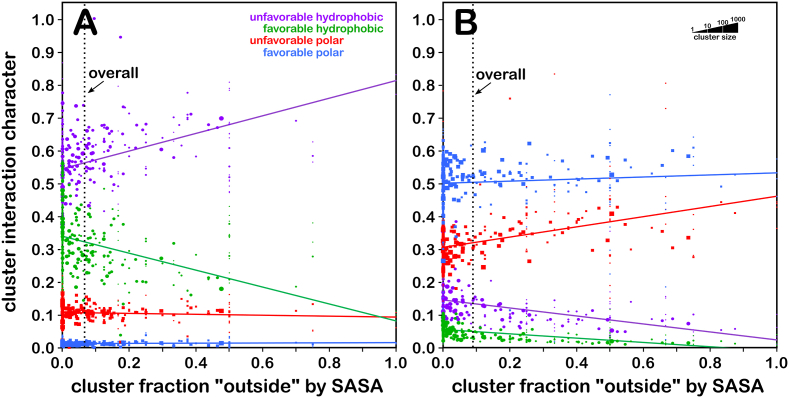
As previously described (AL Mughram et al., 2021), we combined our modified Ramachandran plot, based on the residue backbones, with the solvent-accessible surface area (SASA) of each residue, which is dependent not only on the backbone but also on the χ angles. It can be seen that the solvent exposure is correlated with the backbone protein structure (Fig. 1). The residue SASAs were estimated using default parameters in the online software GETAREA (Fraczkiewicz and Braun, 1998), which utilizes a rolling-ball algorithm with a 1.4 Å water probe. We adapted the "In/Out" output based on the ratio between the calculated sidechain SASA and reference random-coil values relative to the Gly-X-Gly (Src, 77.4 Å2 for SER, 102.3 Å2 for CYS). With ratios less than 0.2, the residue is "In" (buried); greater than 0.5, the residue is "Out" (exposed); between 0.2 and 0.5 is "indeterminate". Our metric "foutside" averages these descriptors recast as 0.0, 1.0, and 0.5, respectively, for a cluster parse or chess square. The foutside values for each parse are illustrated in Fig. 1 with the color of the bars. The parse populations are represented by the bar's lengths.
As expected, these parameters show, overall, that the exposure by residue type follows this trend: CYX < CYS ∼ CYZ < SER; we will discuss the membrane protein residues later. The Ramachandran plot for SER (Fig. 1A) shows the lowest foutside in the β-pleat region with most parses averaging in the range of 0.4–0.6 (green) with a significant number of parses between 0.2 and 0.4 (yellow). Whereas, in the right-handed α-helix region, most parses indicate foutside in the 0.4 to 0.6 range, with a few more exposed parses in the c5 and d5 chess squares in the 0.6–0.8 range (blue). Among the secondary structures, the left-handed α-helix region is the most exposed with an overall foutside of 0.6–0.8. For cysteine (Fig. 1B), the data suggest that this residue likes to be buried, foutside < 0.2 (red), and to be in the β-pleat and right-handed α-helix regions, with very few showing up in the left-handed helix. The foutside trends for CYX (Fig. 1C) suggest even more buriedness, with only the a1.300, b4.60, d1.60, d4.60, d5.60 and d5.180 parses showing foutside > 0.2, but these also are less populated. Finally, in CYZ (Fig. 1D), the disulfide bond reduction recasts the populations relative to CYX such that they are more similar to CYS in terms of buriedness. The SASA and foutside values for all residues in this study are included in the Supporting Information Tables S3–S10.
2.4. Serine and cysteine in soluble proteins
Our intent is to exploit these clustered average maps to highlight the structural roles of the like CB methylene and the unlike OG hydroxyl and SG sulfhydryl groups of SER and CYS, respectively, as reflected in their hydropathic environments. Because the role of the environments could potentially stabilize the ionization of cysteine, particularly in response to changes in pH, we evaluated this effect by determining the pH titration curve of the CYS residues in our data set, and developed methodology to tune the molecular models to particular pH values. A further key environmental factor that we calculated for our data is the residue-level solvent accessibility.
2.4.1. Hydropathic interaction maps of serine
The chessboard schema we use to bin residues by their secondary structures has previously revealed dramatic differences between sidechain map sets in β-pleat, right-hand and left-hand α-helix conformations (Ahmed et al., 2015; Ahmed et al., 2019; AL Mughram et al., 2021) The additional binning by χ1 parse also – not surprisingly – affects the maps. We are focusing here, as in previous reports, the analyses on four particular chess squares, b1, c5, d5 and f6, to survey the environments from each of the three Ramachandran secondary structural motifs.
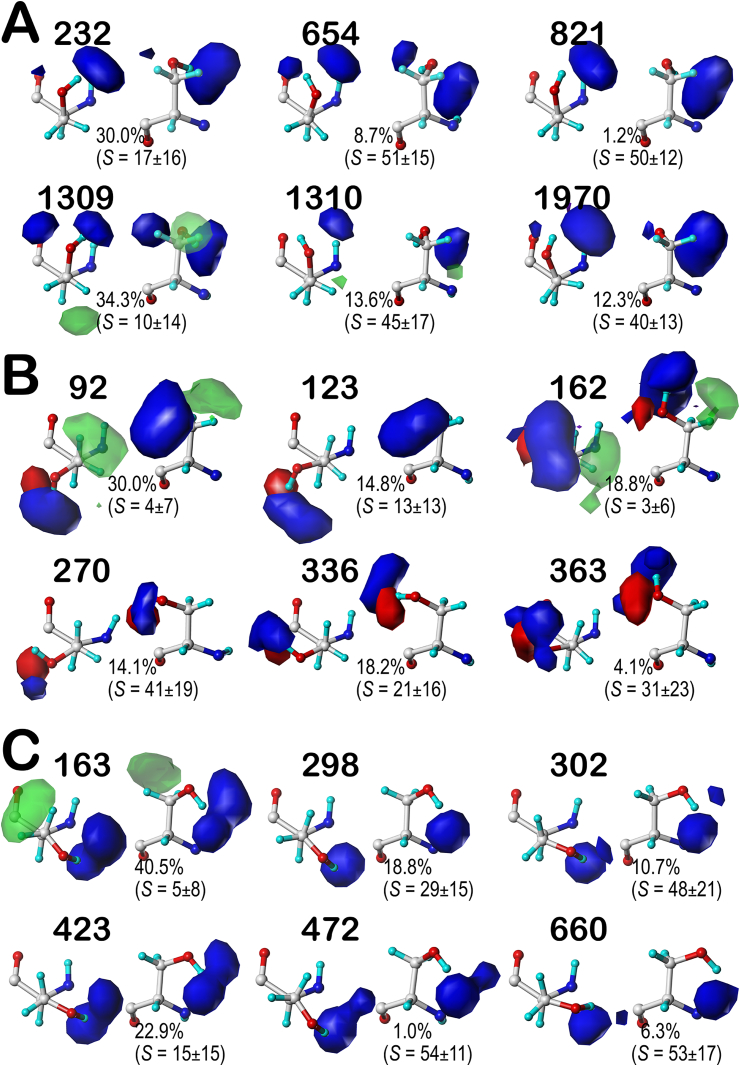
Serine's hydroxyl group has the characteristic of being both a donor and an acceptor (with its two oxygen lone pairs). We expect to see two things: 1) a plethora of maps indicating strong favorable and perhaps unfavorable polar interactions localized around the hydroxyl end of the side chain and 2) strong evidence for SER residues to be highly solvent accessible. The latter is due to the high presence of serine residues on protein exteriors, where they can form hydrogen bonds with water molecules or participate in post-translational modification. For brevity, we will only discuss the averaged map contour plots for the SER sidechain clusters of chess square b1 for the .60, .180, and .300 parses (Fig. 2). Two views are shown for each map to help visualization: the left element of each pair is rotated such that the x-axis points to the right. The z-axis (the CA-CB bond) points up, while the second orientation (a rotation around the x-axis) brings the z-axis to the front. The maps are superimposed on the exemplar structure for the map. The contour levels chosen for all map pairs are identical to allow visual comparisons of relative interaction strengths: favorable polar (blue, +24); unfavorable polar (red, −24); favorable hydrophobic (green, +6); and unfavorable hydrophobic (purple, −12). In some maps, to illustrate the presence of weak hydrophobic interactions, contours at +3 were also plotted in translucent green. It should be noted that the displayed contours are showing interactions, and favorable polar interaction contours may arise from serine hydroxyls acting as either a donor or an acceptor with an appropriate complement. Noted on each is the cluster name (the ordinal residue number of the cluster's exemplar), the percentile contribution of each cluster to the chess square/parse and the average solvent-accessible surface areas (S) calculated with GETAREA (Fraczkiewicz and Braun, 1998).
Fig. 2.
Hydropathic interaction maps illustrating the Gaussian-weighted average clustered SER sidechain environments for the b1 chess square. A) 60° parse; B) 180° parse; C) 300° parse. Two views are shown for each map: left) the CA-CB z-axis points out of the page, right) the CA-CB axis points up. The x-axes of both views point right and the y-axis points up on the left and back on the right. The blue contours represent favorable polar interactions between the hydroxyl and neighboring residues; red contours are unfavorable polar interactions; green contours are favorable hydrophobic-hydrophobic interactions between the methylene and neighbors; purple contours are unfavorable hydrophobic-polar interactions. Translucent green contours, when present, are plotted at one-half the map density of the solid green contours. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
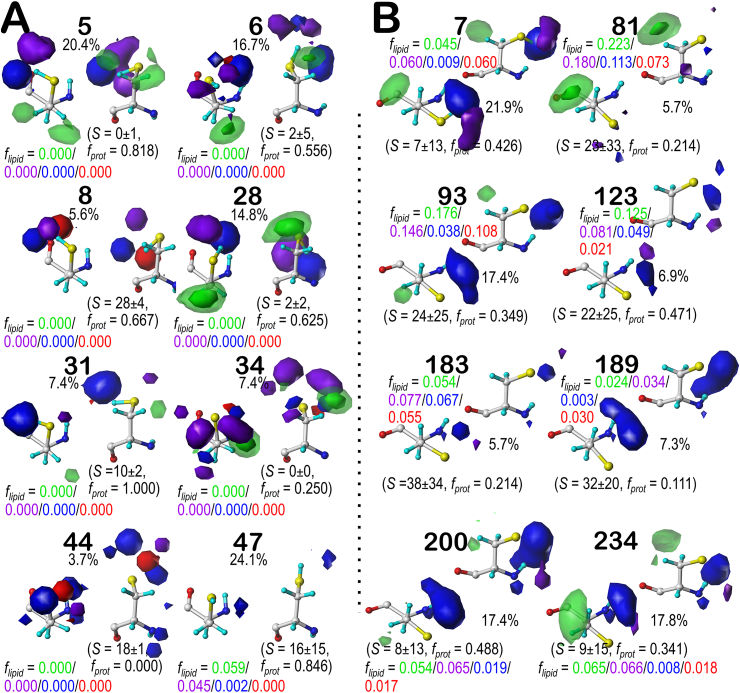
The b1 chess square appears to be, comparatively, the least solvent-exposed of the four we report on here, and collectively contains 3519 (7.5%) maps of the overall SERs in our dataset. Of these, two-thirds are in the .60 parse (Fig. 2A), with the other third split between the .180 (13.3%, Figs. 2B) and .300 (20.0%, Fig. 2C) parses. With these high populations, all three parses were successfully clustered into 6 unique cluster environment map sets. Most interactions are positive polar, which is expected, given the chemical nature of SER. These are the prominent blue contours near the hydroxyl group that signifies hydrogen bonds between this group and its environment. Somewhat surprisingly, the b1.60 parse (Fig. 2A) – the most populated group – is over-clustered: the average inter-cluster similarity is 0.9284; three of the clusters (654, 821 and 1970) appear very similar in the maps, have similar average S values, and have map-map similarities of between 0.9661 and 0.9791. Cluster 1309 is the most different (average similarity with the other maps = 0.8776), as it shows favorable hydrophobic interactions (green contours) near the CB atom and is in a buried environment (S ≤ 10 Å2).
In the other two b1 parses, .180 (Figs. 2B) and .300 (Fig. 2C), similar features can be observed, but the blatantly high inter-cluster similarities of b1.60 are not as evident in either of these cases. In fact, the average similarities are 0.7957 and 0.8538, respectively. Clearly, the b1.180 map data (Fig. 2B) present quite differently than either of the others: the S values are quite a bit smaller, indicating more buriedness, and this conformation produces unfavorable polar interactions (red contours) because the more restricted space makes it difficult to satisfy both the donor and acceptor properties of the hydroxyl. The b1.300 map data (Fig. 2C) is largely comparable to the b1.60 data.
Our description of the three map sets for b1 above will serve as guidelines for viewing and interpretation of the other map sets for c5, d5 and f6, which are available as Supporting Information Fig. S1 (for SER c5.60, c5.180 and c5.300), S2 (for SER d5.60, d5.180 and d5.300) and S3 (for SER f6.60, f6.180 and f6.300). Contour maps for these three chess squares show broadly similar map profiles to the previously discussed chess square. The average similarities for the chess square/parses are 0.9293, 0.8298 and 0.8921 (c5: .60, .180 and .300, respectively); 0.9217, 0.7661 and 0.8217 (d5); and 0.8263, 0.8116 and 0.8818 (f6). These are consistent with the b1 average similarities. Another observation is that serines have larger solvent accessibility (S) in α-helix conformations. This is manifested with clusters that appear to be largely or completely void of interactions, e.g., Fig. S1B (c5.180), clusters 12 and 109. Such clusters represent scenarios where the serine sidechains interact with water molecules, i.e., on the surface, but these water molecules are not explicit in the molecular models.
In summary, the examination of the maps illustrates our rationale for treating the data this way: 1) each map appears to be a backbone-dependent representation of a unique collection of interactions made by serine, with respect to type, strength and spatial location; and 2) low SASA cases where SER shows a few hydrophobic interactions in addition to hydrogen bonding can be differentiated by map-based clustering, whereas high SASA cases with few interactions of any type are less interpretable but still informative.
2.4.2. pKa and the ionization state of cysteine residues
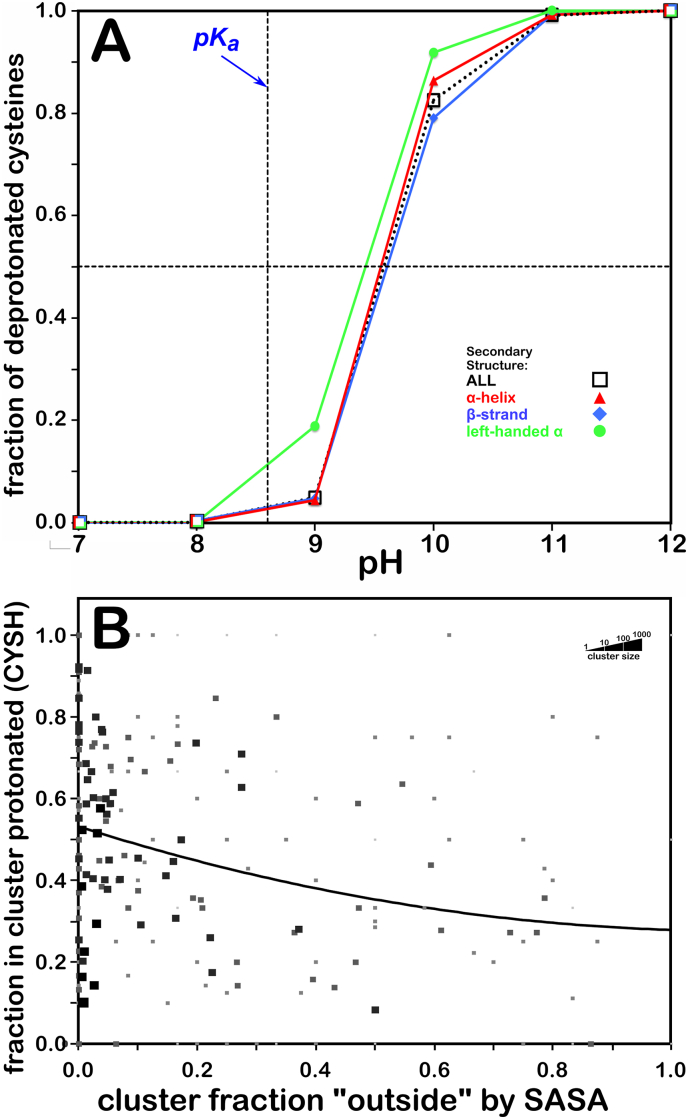
We recognized that a key feature of cysteine that distinguishes it from serine is that its pKa is within an accessible range. Thus, we were interested in knowing how the CYS environments affect its ionization state. The computational titration algorithm we reported in early publications (Kellogg and Abraham, 2000; Fornabaio et al., 2003, Kellogg et al., 2004), and recently optimized for our study of aspartic acid, glutamic acid and histidine (Herrington and Kellogg, 2021), was also applied here to evaluate the effect of pH on CYS ionization states in each protein local environment represented by our dataset. We calculated the total fraction of cysteines expected to be protonated at pHs 7 through 12 in increments of 1 pH unit, as shown in the titration curve of Fig. 3A. Overall, the titration curve is centered at pH 9.5814 (a value that we are calling pH50), which is nearly 1.0 pH units higher than the experimental pKa for this residue obtained using small peptides (Bulaj et al., 1998). It is interesting that there is an apparent backbone conformation effect on protonation: cysteines in the left-hand α-helix conformation are more easily ionized than those in the right-hand α or the β-pleat regions. Our model calculates the free energy required for the deprotonation in an environment where heavy atoms are held static, and thus does not account for any local conformational flexibility. Also, the algorithm does not take into consideration the presence of metals or water of solvation not explicit in the models. Therefore, a number of potential interactions between the –SH/-S- functional groups and neighboring chemical moieties may not be present in our pKa prediction. In our previous application of this algorithm, for aspartic acid, glutamic acid and histidine (Herrington and Kellogg, 2021), we found good agreement with experiment (∼0.5 pH units) for the two acids, but poorer agreement for histidine. In that case, and for cysteine as well, the experimental data is sparser, and many of the experimental cases involve metal coordination. The goal here, however, was to create a methodology such that the actual ionization state for each cysteine in our data set could be modeled prior to calculating its hydropathic interaction map. As a secondary benefit, we have the means to "tune" these environments, molecular models and maps with respect to pH.
Fig. 3.
A) Titration curves of CYS residue by secondary structure. The native pKa for cysteine is indicated. B) Protonation of cysteine as a function of solvent accessibility. Each marker on the plots represents a cluster in the data set. The size and gray shade of the marker represents the population of the clusters in a logarithmic-like scheme, i.e., clusters with fewer members are depicted with smaller, lighter gray squares. The fit lines are weighted by these populations. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
In Fig. 3B, we explore the relationship between solvent accessibility and ionization state. The fprot metric represents the fraction of residues protonated in a cluster, parse, chess square, etc. Here, it was calculated for each residue/map cluster at the pH50 protonation level. We plotted these values as a function of the corresponding cluster's foutside. The marker sizes and shading are related to the cluster populations. Note that the fit curve is not statistically meaningful; it serves only as a visualization guide. Clearly, cysteine likes to be buried: most cluster/parses shown in Fig. 1B indicate foutside < 0.2 and there is a much higher density of points on the low foutside portion of Fig. 3B. While there does not seem to be much of an overall trend in this plot, that assessment belies the information content within each cluster composing this plot.
2.4.3. Hydropathic interaction maps of cysteine
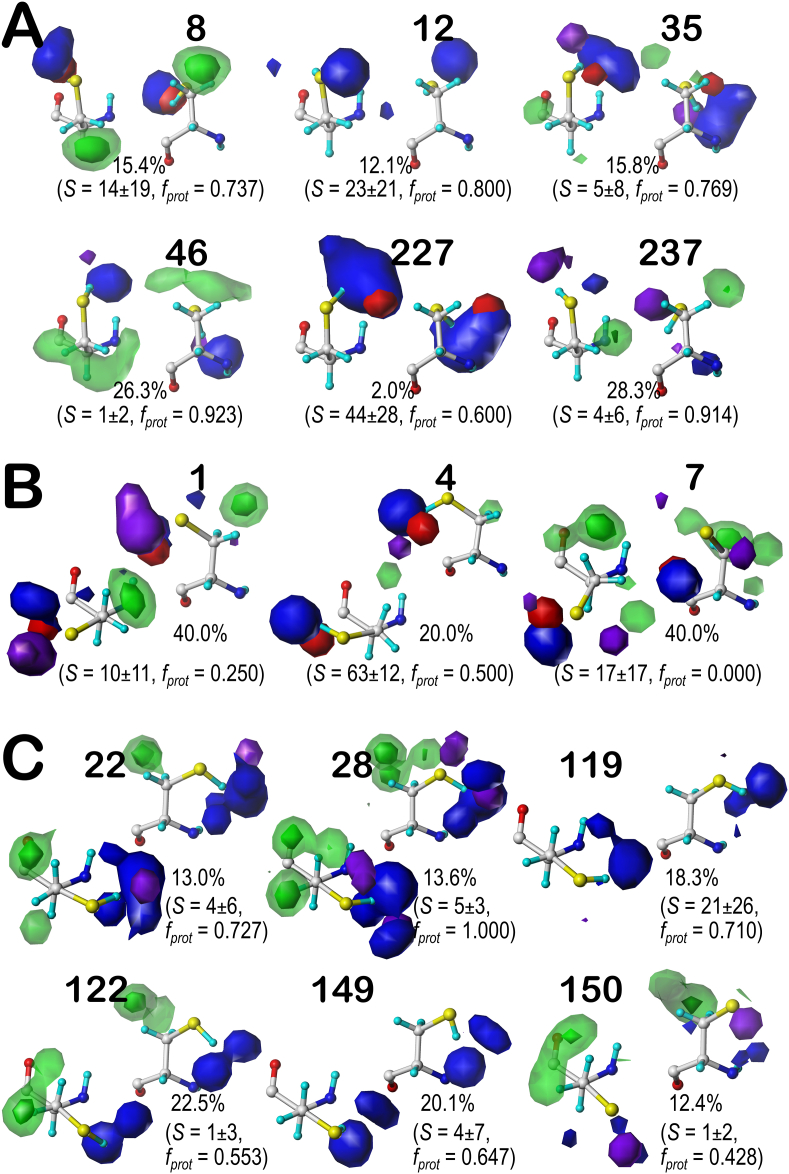
We performed complete studies for CYS at pH 7, 9, and 11, and finally at the pH at which half of the CYS residues were protonated, i.e., pH50. However, we only constructed and present here the visual map contour displays at that latter value, as we believed this pH would best illustrate the diversity of maps in protonated and deprotonated cases. Considering cysteine's more modest polar character and its higher tendency to be buried, we should expect to see more of the CYS maps with favorable and unfavorable hydrophobic interactions localized around the sulfhydryl group. The contour levels chosen for CYS are identical to those used for SER. Now added to each map is the fraction of the members of that cluster that are protonated (fprot). Again, as the displayed contours are showing interactions, cases where the CYS is deprotonated (i.e., an H-bond acceptor) interacting with a donor may be indistinguishable from cases where the CYS is protonated (donor) interacting with an acceptor. Our description of CYS maps (see Fig. 4) will focus on the b1 chess square (like above for SER) in which they were binned into their .60, .180, and .300 parses; similarly, b1 is the least solvent exposed of the group we are discussing and accounts for 7.7% of all CYS residues in our dataset.
Fig. 4.
Hydropathic interaction maps illustrating the Gaussian-weighted average clustered CYS sidechain environments for the b1 chess square. A) 60° parse; B) 180° parse; C) 300° parse. See caption for Fig. 2.
Cysteine tells a more intriguing story than serine, although some of the points made for SER apply here, as well. For example, the bulk of the interactions made with the sulfhydryl group, like those with the hydroxyl of SER, are of the positive polar type. One aspect of the CYS maps that we expected to see was much more significant hydrophobicity than the SER one. The maps show favorable hydrophobic interactions (green contours) generally localized around the CB in about two-thirds of the b1 cases, and a number of unfavorable hydrophobic interactions (purple contours) signifying mismatches between the sulfhydryl and neighboring hydrophobic groups, e.g., cluster 35 (Fig. 4A), or between the methylene and neighboring polar groups, e.g., cluster 7 (Fig. 4B). The unfavorable hydrophobic interactions are somewhat more evident in the clusters with deprotonated CYS such as 1 and 7 in b1.180 and 150 in b1.300, likely because CYS is more solvent buried than SER, and deprotonated CYS− is more polar than CYSH. Maps for the other selected chess squares (c5, d5 and f6) are available as Supporting information (Figs. S4–S6). We calculated the intracluster similarities for the CYS map set: for b1 these ranged between 0.7963 and 0.8418, for c5 between 0.7232 and 0.8309, for d5 they are around 0.777, and for f6 ranging between 0.7756 and 0.9064. This result demonstrates how much more diverse the CYS maps are compared to those of SER, where we saw numerous cases of similarity >0.90.
2.4.4. Serine and cysteine: environments and roles
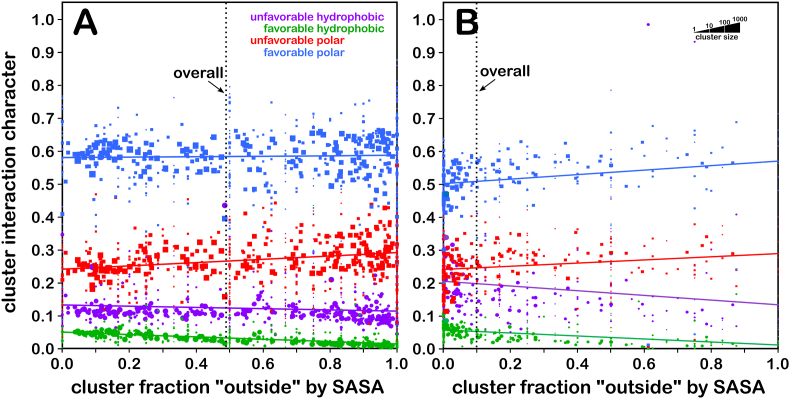
We calculated the fractional environmental characters of each cluster by summing and normalizing the values of the grid points in the four interaction type maps for all cluster members (see Methods). The non-normalized data can be found in Supporting Information Tables S4 and S5. Fig. 5 show plots of these descriptors as functions of foutside for SER (5A) and CYS (5B). While it can be seen that the interaction environments for serines are more polar than for cysteines, a lot of that can be attributed to its higher solvent exposure.
Fig. 5.
Hydropathic Interaction Character for SER and CYS Clusters. A) interaction character for serine; B) interaction character for cysteine. The green markers and line represent favorable hydrophobic interactions, purple represents unfavorable hydrophobic, blue represents favorable polar and red represents unfavorable polar. Each data point plots interaction character (summed from grid points as described in the text) as a function of foutside for a map environment cluster. The sizes of the markers are log10-scaled by the number of members of the cluster. The fit lines (y = ax + b) are the result of weighted least squares analyses as described in the text. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
There are clearly fundamental and substantial electronic differences between these two more or less isosteric residues that simply cannot be explained without applying quantum chemistry. However, from a structural viewpoint, the 3D interaction maps and the associated properties of the underlying molecular structures tell a rather complete story of the roles for SER and CYS in protein structure. Their small differences in hydrophobicity are responsible for a larger and even dramatic difference in buriedness. The biological accessibility of cysteine's pKa is an additional factor of structure and key to many biological processes (Bulaj et al., 1998; Jensen et al., 2009; Zeida et al., 2014a). CYS has been shown to be a later entrant in the genetic code; its reactivity and functional role in active sites appears to be responsible for its distribution and abundance in proteins(Marino and Gladyshev, 2010). This fairly facile ionization also enables cysteine's most important special ability: to form –S–S– bridges.
2.5. Cysteine disulfide bridges
When two adjacent cysteines are under oxidizing conditions, they can covalently bond, losing their two hydrogens, resulting in a disulfide bond. The cystine is less polar than the free thiol group explaining why cystines are usually found in the hydrophobic core (Fig. 1C) rather than the hydrophilic surface. To facilitate our understanding of the underlying energetics of the interactions in the cystine (disulfide bridged cysteine) microenvironments, we tackled the problem into two parts: 1) for the (intact bridge) CYX, where we wanted to evaluate the environment of the cystine –S–S–, we truncated the cystine as a pseudo-reside composed of its CA, CB, SG, SG' and CB' and removing CA' from consideration; and 2) we broke the disulfide bond adding HG to each sulfur and energy minimized the resulting proteins to build models that simulate the environmental conditions before bridge formation. We refer to this residue dataset as CYZ. An important point is that a cystine actually has five dihedral angles (χ1, χ2, χ3, χ2', χ1'), and our truncated CYX model incorporates the first three of these. We opted to bin the CYX only by its χ1, as before for SER and CYX (and now CYZ), but compensated for the additional flexibility by increasing the maximum allowed clusters to 10 (instead of 6). The alternative approach of two-level parsing (χ1 + χ2) could not be supported by the unfortunately sparse CYX dataset.
2.5.1. Deconstruction of the CYS93A-CYS116A cystine in 1M8N
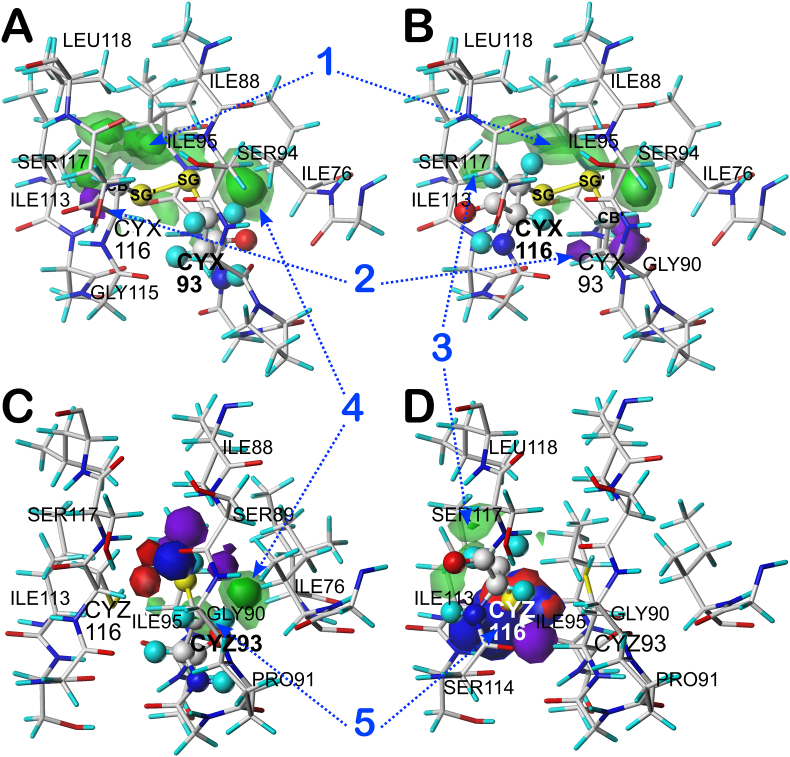
To illustrate our approach, we will discuss a single cystine example from our dataset. The hydropathic environment surrounding a CYX in the highly active antifreeze protein from Choristoneura fumigerana (CfAFP) was chosen as a model. Its 9 kDa isoform CfAFP501 was crystallized at 2.45 Å (PDB id 1M8N) (Zeida et al., 2014b) in a highly regular β-helical structure, including a disulfide bond between CYS93 and CYS116, both in its A chain. The first corresponding CYX is, in our nomenclature, residue 67 of the c8.180 chess square/parse (β-sheet region of the Ramachandran plot) and is a member of cluster 223. The second corresponding CYX is residue 53 of b1.300 and a member (and exemplar) of cluster 53. As is evinced from Fig. 6A, this map – from the CYS93A perspective – is dominated by strong favorable hydrophobic interactions between the disulfide bond and its microenvironment ILE76, ILE88, GLY90, ILE95, ILE113 and LEU118. Also seen are unfavorable hydrophobic interaction likely caused by interactions between CYS93's CB' with CYS116's and GLY115’s backbones. Fig. 6B illustrates the environment from the CYS116A perspective. With this orientation what is captured are strong favorable hydrophobic interactions between the CYX and ILE76, ILE88, ILE113, GLY115 and LEU118 with other clear unfavorable hydrophobic interactions with the backbones of CYS93 and GLY90.
Fig. 6.
Deconstruction of the CYS93A-CYS116A Cystine in 1M8N. A) Hydropathic environment map from the perspective of CYS93. Here, the CYX construct (sphere display) includes SG’ and CB’, which are the SG and CB of CYS116; B) Hydropathic environment map from the perspective of CYS116. Here, the CYX construct includes the SG and CB of CYS93 as SG’ and CB’; C) Hydropathic environment map from the perspective of the CYZ93 construct (sphere display), which is a simulation of the pre-bridging cysteine residue; and D) Hydropathic environment map from the perspective of the CYZ116 construct. The contours are colored: purple – unfavorable hydrophobic, green – favorable hydrophobic, red – unfavorable polar, blue – favorable polar. Notes: 1 – The CYX environments are dominated by strong hydrophobic interactions, are largely symmetric and lastly, as expected, are complementary; 2 – unfavorable hydrophobic interactions near the CB’ atoms of both constructs are likely truncation artifacts; 3 - the environments surrounding CB in both CYX116 and CYZ116 are very similar and largely favorable hydrophobic; 4 - the environments surrounding CB in CYX93 and CYZ93 are also very similar; and 5 – the large favorable and unfavorable polar interactions around the two CYZ constructs, as they transition to the highly favorable interaction environment depicted by (1) is a very dramatic indication of the hydrophobic effect as a key factor driving cystine bridge formation. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
After deleting the S–S bond of this cystine, adding the hydrogens and performing the energy minimization described in Methods, the backbone φ and ψ and the χ1 angles for CYX93A and CYX116A changed. The minimization caused the two residues to be parsed into different chess squares of the β-sheet region of the Ramachandran plot and reformed the χ1 and χ2 angles of both. CYX(Z)93A is now residue 51 in the b7.180 chess square/parse of CYZ (cluster 9) whereas CYX(Z)116A is 232 in the c1.300 (cluster 31). The CYZ data set was subjected to the same computational titration protocol as CYS (above): in this case the pH50 was 9.496 (see Fig. S7). All CYZ maps were calculated at this pH. From Fig. 6C, CYZ93 is making favorable hydrophobic interactions with ILE76, GLY90, PRO91 and GLY92 and favorable polar interactions with SER89 and, of course, CYZ116. These interactions are counterbalanced, if not dominated, by unfavorable hydrophobic and polar interactions with ILE88, GLY90, CYZ116 and SER117. Similarly (Fig. 6D), CYZ116 has favorable hydrophobic interactions with ILE113 and LEU118, favorable polar with CYZ93 and SER114, and unfavorable interactions with ILE95, SER114, GLY115 and SER117.
2.5.2. CYX and CYZ hydropathic interaction maps
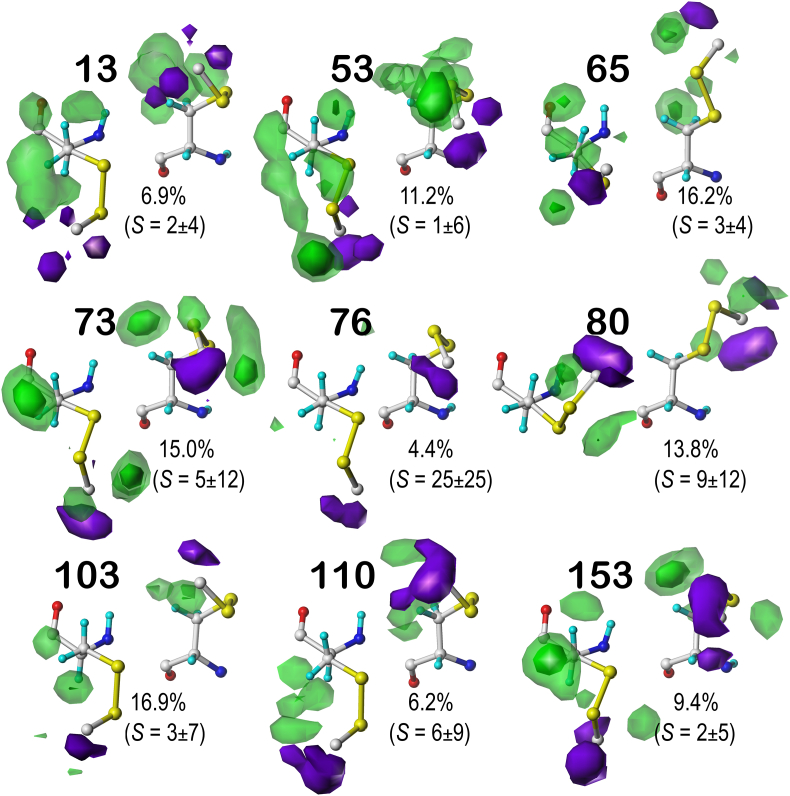
With this orientation to what is captured by the CYX maps, it can be seen (Fig. 7) that the interactions described above are common in all 9 clusters of b1.300: strong hydrophobic interactions around the disulfide bond, unfavorable hydrophobic interactions associated with the CYX CB and the backbone of its bonded CYX, and only a few, generally small, favorable polar interactions. However, note the diversity of conformations in χ2 and χ3 exhibited by the molecules underlying these maps. Although there are 5500 CYX residues in our dataset, comprising 2750 cystines, once binned into their chess squares and χ1 parses, only a handful of such bins were populated sufficiently for robust clustering at our conservative target of 10 clusters per bin (see Supporting Information Tables S2 and S3). The numerically data describing the clusters and their memberships, properties, etc. are in Table S6. A few additional CYX contoured map sets are available in Fig. S8 for b1.60, and S9 for c5.60, c5.180 and c5.300.
Fig. 7.
Hydropathic interaction maps illustrating the Gaussian-weighted average clustered CYX sidechain environments for the b1.300 chess square/parse. See caption for Fig. 2.
Maps for the CYZ environmental constructs are generally analogous in appearance to those of CYS, and are not shown. There are a handful of map-map similarities (CYS-to-CYZ) around 0.9 in most of the chess square χ1 parses. Numerical data for CYZ is available in Table S7, and is also similar to that of CYS (Table S5).
2.5.3. CYX environments and insight into cysteine bridging
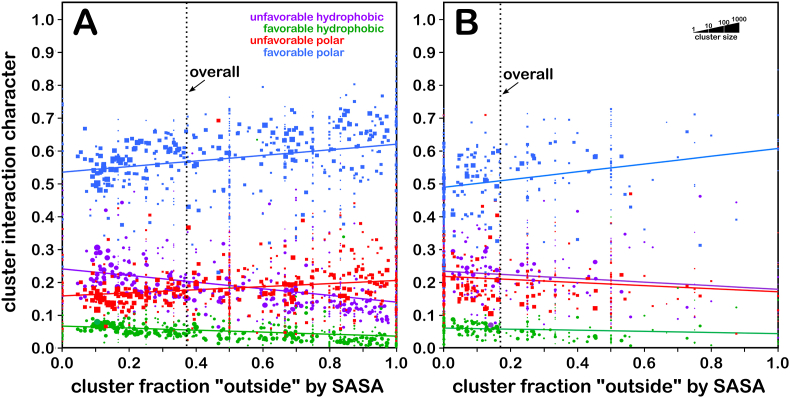
Similar to Fig. 5 above, Fig. 8 shows plots of the hydropathic character as functions of foutside for the CYX (A) and CYZ (B) constructs (see also Tables S6 and S7). The interaction environment for CYX (Fig. 8A) is dramatically different than any of the other three – most notably, the major interaction class contribution is unfavorable hydrophobic, followed by favorable hydrophobic, and virtually no favorable polar interactions. Also obvious is that CYX residues are only very rarely solvent-exposed. On the other hand, CYZ data (Fig. 8B) are remarkably similar to CYS data (Fig. 5B), with CYZ showing somewhat less favorable and unfavorable hydrophobic characters than CYS. As noted above, we created the CYZ data set by double protonation and energy minimization of the resulting models. Clearly a molecular dynamics protocol would have been preferable, but with over 2700 structures in-play, MD was not practical. We expected a larger difference in pH50 than observed, i.e., ionization of a CYZ construct would be much easier with a readily available –S–H from its formerly bonded cystine partner. However, while that was not observed – pH50 for CYZ is only 0.09 less than pH50 for CYS – poorly optimized structures would be probably much easier to ionize, which is additional evidence that our models are probably reliable.
Fig. 8.
Hydropathic Interaction Character for CYX and CYZ Clusters. A) interaction character for CYX (cysteine, intact bridge); B) interaction character for CYZ (cysteine, broken bridge). See Fig. 5 caption.
The small difference in pH50 was surprising for another reason: we had expected that differences in the bridged CYX-CYX environment, when wound back to a pair of CYS residues in close proximity (as in our CYZ construct models) would suggest obvious deprotonation of one of the CYZs (Poole, 2015). Paired cysteines cannot react spontaneously, but they required oxidants or enzyme to catalyze the dimerization of the two thiol groups (Fass and Thorpe, 2018). For example, protein disulfide isomerase (PDI) acts via the thiol-disulfide exchange mechanism, a process in which a disulfide bond donates to a substrate and in turn becomes reduced. This process is pH-dependent, because it affects the deprotonation of the cysteine thiol group to form the active thiolate, which is required for starting the nucleophilic attack on the cysteine in the active site of PDI (Hatahet and Ruddock, 2009). Despite huge advances in understanding cystine formation, the precise mechanism via which correct dimerization is achieved remains not fully understood (Robinson and Bulleid, 2020). The larger atomic volume of sulfur better stabilizes the negative charge of the transition state (Zeida et al., 2014a). Certainly, ionization remains a contributing factor, and even the small pH50 change we calculated corresponds to pKas that increase the fraction of CYS− by about 25%. However, another factor, which is clearly shown in Fig. 8A, is that a key driving force for the formation of the disulfide bridge is the hydrophobic effect concomitant with better burial (Cadenas and Packer, 2010). Proximal cysteines prefer to form a bridge at the expense of their hydrogens to reduce their conformational entropy, while increasing the system's entropy. Another advantage is that the reactivity of free thiol groups is ameliorated by burial.
2.6. Are there environmental differences between soluble and membrane SER and CYS?
2.6.1. Membrane proteins dataset
Using as starting point the snapshots of coarse-grained molecular dynamics simulations created and catalogued in the MemProtMD database by Newport, Sansom, and Stansfeld, we calculated the hydropathic maps of SER, CYS and CYX (that we are calling SERm, CYSm and CYZm) from 369 such proteins in the presence of the reported artificial lipid set based on dipalmytoylphosphatidylcholine (DPPC) (Newport et al., 2019; Stansfeld et al., 2015). In our soluble protein data set, SER, CYS and CYX accounted for 6.21%, 0.73% and 0.69% of residues; in the membrane protein set, 6.05%, 1.09% and 0.24% of the residues are SERm, CYSm, and CYXm, respectively. The major difference seems to be that proportionately fewer cysteines form cystine bridges in the membrane proteins. While we are accounting for some degree of membrane protein interactions with this approach, this dataset still has significant limitations due to the absence of native lipids. Biological membranes are typically crowded, and their lipids compositions vary in terms of composition character. It is probable that the local properties in the lipid bilayer are different region by region (Engelman, 2005; MacCallum and Tieleman, 2011). Biological membranes are thus more complicated than the single-component lipid bilayer simulated within the MemProtMD database, but this analysis is still helpful in conceptualizing the local hydropathic environments and roles for the individual amino acids in membranes.
We calculated the maps using the same conditions as reported above and in Methods for the soluble protein data set. We treated the DPPC as a residue, and this new interaction set was accounted for in the final maps. A complete summary of all the data for the membrane protein dataset can be found in Tables S1-S3 and S8-S10.
2.6.2. Solvent-accessible surface area (SASA) and foutside
With SASA calculations for the soluble proteins, we can safely equate accessibility with water; in the case of membrane proteins, accessibility could be either with lipid or water. The GETAREA algorithm currently has no scope to differentiate these two cases. Thus, some care must be taken in evaluating these results. Fig. 1E, F and G show the Ramachandran plot superposed with foutside for SERm, CYSm, and CYXm. As noted above, the fractions of serine and cysteine residues are consistent between soluble and membrane proteins. However, the latter is structurally confined in space by the lipid environment, causing them to follow different conformational principles than in globular proteins (MacCallum and Tieleman, 2011). They fall into two classes: those α-helical and those considered to be β-barrel proteins. This behavior can explain the increased percentage of SER and CYS in the Ramachandran α-helix region of membrane proteins (62%–73%) than in the water-soluble proteins (44%–47%). However, the notion that these fractions are fluxional due to the evolving availability of membrane protein structural data should not be ignored. Another aspect that is quite clear in Fig. 1E is the increased buriedness of SERm, particularly in the helical regions. This may be explained by serine protecting its hydroxyl group from the lipid environment by instead making intra- or inter-helical hydrogen bonds in the membrane protein's core. The opposite behavior is seen for CYSm, as this residue seems to prefer more exposure than CYS to the solvent. For CYXm, the small amount of data available compared to CYX (<1:5) does not allow for many confident comparisons, but in general, it seems that CYXm exposure is slightly higher than CYX.
2.6.3. Clustering and hydropathic environment maps for SERm, CYSm, and CYXm
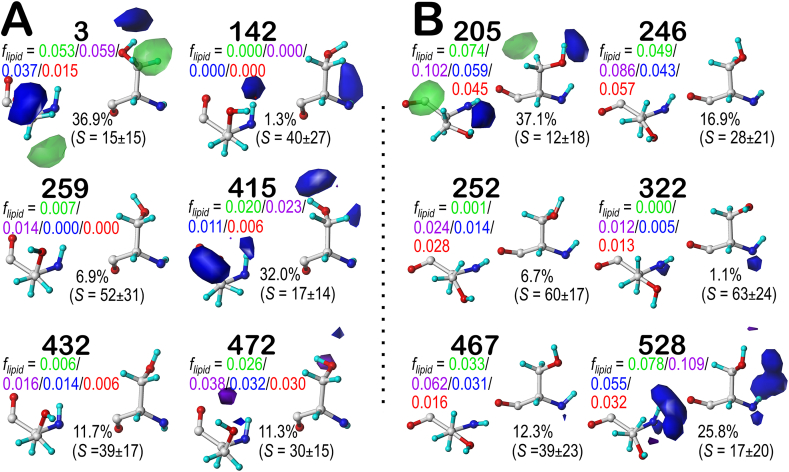
To explore the role of lipid interactions in the hydropathic environment of CYSm and SERm, we calculated 3D interaction maps. To ensure we account for the expected additional interaction profiles, we increased the maximum number of clusters to 8 (instead of 6) for CYSm. We also introduced a new metric, flipid, representing the fractional environmental character of interaction types between each residue and lipid-related to all interactions made by that residue in the resulting clusters. The contour levels chosen for CYSm and SERm are identical to those used for SER and CYS. Our description of SERm and CYSm maps will focus on the b1.60 and c5.300 chess square/parses. These were chosen to highlight the differences in sidechain burial between SERm and CYSm by secondary structure. The SERm and CYSm b1 chess squares are no longer the most buried, and they account for only 3.23% and 2.25% of the total residues, slightly more than one-third of that seen in the soluble proteins. The c5 chess squares have 8.71% and 7.95% of the SERm and CYSm populations, respectively, closer to the count in their soluble counterparts. During the following discussion, recall that highly solvent-exposed sidechains do not necessarily mean that they are in water environments, but may be in lipid. To distinguish these two situations, we can correlate the SASA values with the flipid descriptors. High SASA and flipid values mean that the residue or, in this case, cluster is interacting with, i.e., exposed to, the lipid bilayer. In contrast, high SASA and low flipid indicate that the cluster is water exposed.
As shown in Fig. 9A, we can see that clusters 259 and 432 in SERm b1.60 do not show any strong interactions between these sets of residues and their environments. This pair may be an example of over-clustering this parse, but including cluster 142, defines a collection (20%) of cases where the hydroxyl group is solvent-accessible, based on high SASA (39–52 Å2) and very low flipid values displayed on the maps. In contrast, cluster 472 shows a moderate SASA and intermediate flipids. The hydroxyl group is likely making unfavorable hydrophobic interactions with the phospholipid tail, supported by the small purple contours (unfavorable hydrophobic) near the OG-HG. The two lower SASA clusters, 3 and 415, while mostly buried, suggest that their CB methylenes are lipid accessible, to a larger degree for 3. The c5.300 maps for SERm are displayed in Fig. 9B. The most interesting observation is that 62% of the residues have a SASA lower than 20 Å2 compared to the only 25% in SER, confirming that serine in membrane proteins likes to be buried, especially in the helical secondary structure motifs. Interpretation of these maps parallels rather closely the b1.60 maps: while mostly buried, the methylenes of 205 and 528 are interacting with the phospholipid tail; 247 and 467 have moderate SASAs and are interacting with the lipids likely through both the CB and OG-HG; and 252 and 322 are likely solvent (water) exposed with relatively high SASAs (≥60 Å2). While the SER maps (Figs. 2A and S1C) and SERm maps (Fig. 9) illustrate similar features, they are also quantitatively similar: several map pairs have map-map similarities larger than 0.9, e.g., SER cluster 1309 and SERm cluster 3 in b1.60 (0.9266). It remains to be determined whether these correspondences are only a consequence of over-clustering serine environments or indicative of these environments being truly indistinct. See Supporting Information Fig. S10 (SERm d5.300 maps), Fig. S11 (f6.300 maps), and Table S8 (SERm data summary).
Fig. 9.
Hydropathic interaction maps illustrating the Gaussian-weighted average clustered SERm (serine, membrane protein) sidechain environments. A) b1.60 chess square/parse; B) c5.300 chess square parse. flipid values are the fraction of all interaction scores arising from residue-to-lipid interactions in the molecular models by type (green-favorable hydrophobic, purple-unfavorable hydrophobic, blue-favorable polar, red-unfavorable polar) See also caption for Fig. 2. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
With CYSm possessing a pH50 of 9.4506, it is ∼35% more likely to be protonated at a given pH than CYS. To emphasize diversity, CYSm maps were calculated at pH50. As illustrated in Fig. 10A, excepting 47, which appears to be somewhat exposed to the lipid bilayer, all clusters have a flipid value of 0, emphasizing that they are very buried (clusters 5, 6, 28, and 31) or modestly water exposed (8 and 44). The overall buriedness of b1.60 residues in CYSm is the same as in the water-soluble b1.60 (Fig. 4A). CYSm tends to form more unfavorable hydrophobic interactions than CYS and makes stronger favorable hydrophobic interactions. The relatively rare clusters 8 and 44 display unfavorable polar interactions (red contours), indicating structural errors in the small number of structures contributing to these clusters. The differences in buriedness for the c5.300 chess square/parse in CYSm vs. CYS are dramatic. For the latter (Fig. S4C), only 8% were solvent-exposed (SASA >20 Å2), but in CYSm, 43% are exposed. Also, all c5.300 maps indicate interactions with the lipid, e.g., cluster 81 with its strong favorable hydrophobic interactions, likely with the lipid tail. Cluster-cluster similarities between the CYS and CYSm clusters are far more supportive of them having distinct hydropathic environments. With the exception of similarities involving a likely to be "extra" and unneeded cluster in CYS c5.300 (218, Fig. S4C), there are none greater than 0.9, and quite a few less than 0.7. See also Supporting Information Fig. S12 (CYSm d5.300 maps), Fig. S13 (CYSm f6.300 maps) and Table S9 (CYSm data summary).
Fig. 10.
Hydropathic interaction maps illustrating the Gaussian-weighted average clustered CYSm (cysteine, membrane protein) sidechain environments. A) b1.60 chess square/parse; B) c5.300 chess square parse. See captions for Fig. 2, Fig. 9.
Unfortunately, we do not have enough structural data to perform full analyses on CYXm. Disulfide bridges are more often found in shorter proteins (<200 residues) (Bosnjak et al., 2014) than those we had available for this study, or are generally not found as membrane proteins. Thus, only three of the chess square/parses contained more than 100 residues. The data we did collect and calculate is available in Table S10.
2.6.4. The triumph of subtlety?
In keeping with their relative positions in the periodic table, oxygen and sulfur have – at first look – similar properties, but their differences in electronegativity, atomic radii and sulfur’s ability to utilize its d shell, yield significantly different chemistries. While serine and cysteine have similar structures, their differences in atomic properties are magnified in residue properties. These differences are accentuated by environments – serine in membrane proteins overall has a notably higher tendency to be buried (foutside ∼35%) than in soluble proteins (∼50%), while cysteine shows the opposite trend (membrane proteins, foutside ∼18%; soluble proteins, ∼10%). The associated character plots are displayed as Fig. 11 (see also Tables S8 and S9). Comparing Fig. 5, Fig. 11, one can see that there is modestly higher favorable hydrophobic character in the membrane proteins for both SER and CYS, more unfavorable hydrophobic (especially for SERm), but less unfavorable polar. These data, combined with the flipid analysis (examining interactions between the SER and CYS residues and the artificial DPPC lipid set in the MemProtMD models) (Newport et al., 2019) is filling in the pieces of an understanding of how interaction environment affects these residues. For example, some highly solvent exposed SERm and CYSm residues are interacting favorably with the lipids (Fig. 11A and B, lower right corners). Nevertheless, despite their different protein structures and their varied structural roles, most measurable differences between the membrane and soluble protein serine and cysteine residues are quite subtle. One flaw in our analyses is that we need more structural data, particularly for the disulfide cystine. Also, as membrane protein structure solution is an emerging field, it is difficult to say whether the structures solved to date are representative, although multiple technological and methodological developments have led to the deposition of 5045 membrane protein structures in the Protein Data Bank with 1286 unique structures according to White’s mpstruc database (https://blanco.biomol.uci.edu/mpstruc/) as of June 2021.
Fig. 11.
Hydropathic Interaction Character for SERm and CYSm Clusters. A) interaction character for SERm (serine, membrane protein); B) interaction character for CYSm (cysteine, membrane protein). See Fig. 5 caption.
3. Summary and conclusions
The hydropathic environment of each residue can be mapped in terms of its backbone and influenced by its surroundings, like water exposure, protein core, and membrane bilayer (Ahmed et al., 2015; Ahmed et al., 2019; AL Mughram et al., 2021) We analyzed and mapped the interaction environments of more than 85,000 amino acid residues (serine, cysteine, and cystine) in a diverse collection of globular and a separate collection of integral membrane protein structures. With these analyses, we have produced backbone-dependent libraries of sidechain rotamers and their 3D residue interaction preferences encoded in maps by type, strength and position. Accordingly, the residue maps for serine and cysteine contained similar polar features representing hydrogen bonding, etc., but additional favorable and unfavorable hydrophobic interactions were often observed in the latter. One additional aspect of this study is that we explored the ionization of cysteine by applying a computational tool that adjusts the ionization states of cysteine and other residues like aspartatic acid, glutamic acid and histidine (Herrington and Kellogg, 2021) based on their specific environments and solution pH. Our backbone-dependent rotamer library and map set incorporates this feature as well. The maps are critically dependent on the pH at which they are calculated, and they can be pH-tuned.
Although serine and cysteine are isosteric, their slight difference in hydrophobicity significantly impacts their solvent exposure. One explanation is that Nature has applied a negative selection to remove cysteine residues from the protein surface due to their relatively high reactivity, which is, for example, responsible for formation of cystine. We explored this phenomenon by evaluating the 3D hydropahic interaction environment maps for cystine and a broken-bridge construct of two protonated cysteines. The residues of the latter construct were not significantly easier to ionize than other cysteines. However, their environments and that of the intact-bridge cystine strongly suggest that a key driving force for bridge formation is that it parallels one of the classical dogmas for protein folding, i.e., the hydrophobic effect. The environments surrounding disulfide bridges are somewhat surprisingly dominated by both favorable and unfavorable hydrophobic interactions, rather than the polar interactions observed in non-bridging cysteine environments. Further refinement of these data may lead to a strategy for identifying cysteines and cystines that can be targeted by selective covalent inhibitors (Long and Aye, 2017; Heppner, 2021).
Lastly, we applied our analyses to a comparison of serine and cysteine in soluble and membrane protein environments. We did not know what to expect, i.e., whether our serine and cysteine rotamer/map sets would be applicable for the membrane proteins. In some respects, there are substantial differences: first, while the frequencies of serine and cysteine remain more or less consistent in these protein sets, the number of cysteines involved in –S–S– (cystine) bridging drops dramatically; second, there are notable shifts in the frequency of finding serine and cysteine from the β-pleat to the α-helix motifs, although it is unknown whether that is universal or a consequence of the structures solved to-date; and third, most importantly, there is a shift in the exposure of these two residues with serine becoming more buried trying to protect its hydroxyl group from the lipid environment, while cysteine instead becomes more exposed and willing to make favorable interaction with lipids. These analyses using DPPC as a stand-in for the true lipid environment are snapshots of membrane protein structure, but do carry some caveats. The experimental structure determination of membrane proteins are considerably more difficult than those that are water-soluble, and despite remarkable advances in the structural elucidation of isolated membrane proteins, obtaining atomic resolution remains challenging: most structures are currently in the resolution range of 3.0–5.0 Å. Newer technologies with higher resolution, e.g., cryo-electron microscopy, are becoming increasingly available. However, regardless of how the structures are obtained, extraction of membrane proteins in their active, folded form is fraught with difficulties. Membrane protein folding and the resulting activity require the presence of the native lipid environment, which is often corrupted by the detergents used for extraction(Guo, 2020). The recent developments of detergent-free systems (Gulamhussein et al., 2020; Guo, 2021; Kroeck et al., 2020; Lee et al., 2016; Marconnet et al., 2020; Qiu et al., 2018; Simon et al., 2018; Yang et al., 2021) allows co-extraction and stabilization of membrane proteins and their associated lipids in their near-to-native conformation. This increases the environmental diversity of the lipid bilayer, enhancing our understanding of protein structure, and the critical roles of lipids as designed by Nature.
Our long-term goal is to develop detailed understanding of the full set of amino acid residues through calculation of 3D hydropathic interaction maps. In this report we described serine and cysteine in terms of these maps. This complements our earlier reports on alanine (Ahmed et al., 2019), phenylalanine, tyrosine and tryptophan (AL Mughram et al., 2021) and aspartic acid, glutamic acid and histidine (Herrington and Kellogg, 2021). With the full set of these maps, we envision a scheme for protein structure building and prediction; with what we have learned about cysteine, we can very likely anticipate in our predictions the formation (or not) of disulfide bridges, because we have shown that the 3D hydropathic environments, solvent exposure and ionization propensities are different. Our paradigm for structure model building and refinement will clearly benefit from further exploration of membrane protein structures. However, the disadvantages of the artificial DPPC environment in our data set compared to the true native lipid environments are unknown.
4. Methods
4.1. Datasets
From a collection of 2703 randomly selected proteins from the RCSB Protein Data Bank, using only structures containing no ligand or cofactor, we extracted all SER, CYS, and CYX residues from each structure, excluding N- and C-terminal residues. For these structures, we have previously described our selection criteria (Ahmed et al., 2015). Similarly, we extracted the same residue types from 369 membrane protein structures in the Grazhdankin et al. dataset (2020), which is a subset of the MemProtMD database (Newport et al., 2019) of preoriented membrane proteins. Water molecules, ions and lipids more than 6 Å away from the protein were removed and missing hydrogen atoms were added to all heavy atoms of all structures based on their hybridization states and their positions were subjected to conjugate gradient minimizations in Sybyl X.2.1 (Tripos, St. Louis, MO, USA). A similar procedure was followed to create the CYZ (cysteine, broken bridge) dataset: after removal of the S–S bond from all bridged (CYX) cystines in our soluble protein dataset, protons were added each thiolate, and the resulting models were energy minimized. Clearly, a molecular dynamics-powered procedure would have been preferable, but with over 2700 structures, that was impractical.
4.2. Alignment calculations
We overlayed an 8 by 8 "chessboard", where each "chess square" has dimensions of 45° by 45° in φ (phi) – ψ (psi) space, on the standard Ramachandran plot. The grid of the board was shifted by −20° and −25° in the φ and ψ directions, respectively, to more closely align higher-density regions of the plot with the chessboard system. The φ, ψ, and χ angles were calculated for every residue in our dataset to bin each residue within its proper chess square with respect to its φ and ψ angles. All residues were further classified by their χ1 angles into three parse groups: group A, (0° ≤ χ1 < 120°), group B (120° ≤ χ1 < 240°), and group C (240° ≤ χ1 < 360°), which we are calling .60, .180, and .300, respectively. Table S1 contains all information for each residue of each type in our dataset, including their chess squares, parses, PDB IDs, φ, ψ and ω torsion angles and atom numbers for the backbone atoms and CB of each residue.
A single model residue of each type was constructed at the center of each chess square with characteristic φ and ψ angles for that centroid. The CA of the protein residue’s backbone was placed at the origin with the CA-CB oriented along the z-axis and the CA-HA bond oriented into the -y, -z quadrant of the yz-plane. All residues of each type were aligned to this model, and rotation and translation matrices were calculated by least-squares fitting of the residue constituent atoms to the model. This effectively shifted coordinates of every protein structure to align the residue of interest with the centroid within a common frame and ensures that all calculated maps and environments are attributable to a residue's interactions and not misalignments in backbone structure. The average root-mean square distances (RMSDs) for superimpositions of backbone atoms in each chess square are close to 0.15 Å, indicating that errors arising from aligning residue backbones to the centroid model (based on the CA-CB bond) are minimal. The models for CYX (cysteine, intact bridge) residues were created differently: in addition to its N, CA, C, O, CB and SG atoms (and all bonded hydrogens), the atoms SG’ and CB’ from its –S–S– bonded cysteine were included.
4.3. HINT scoring function
The HINT forcefield (Kellogg et al., 1991; Kellogg and Abraham, 2000; Sarkar and Kellogg, 2010) was used for all scoring of interactions between protein atoms. HINT relies on atom-focused parameters, namely the hydrophobic atom constant (a1) and a value for solvent-accessible surface area (SASA, Si) for atom i. Generally speaking, ai > 0 for hydrophobic atoms and ai < 0 for polar atoms.
Si is greater for more solvent-exposed external atoms. The interaction score between atoms i and j is calculated by:
| bij = ai Si aj Sj Tij e-r + Lij |
where r is the distance in angstroms between atoms i and j. Tij is equivalent to −1, 0, or 1 to account for acidic, basic, etc. character of atoms involved and assign the proper sign to the interaction score. Finally, Lij implements the Lennard-Jones potential function (Kellogg and Abraham, 2000). bij > 0 for favorable interactions, such as Lewis acid-base and hydrophobic-hydrophobic interactions, while bij < 0 for unfavorable interactions, including hydrophobic-polar or Lewis base-base interactions.
4.4. Computational titration of Ionizable cysteine residues
To determine the optimal ionization state of each cysteine, we adapted an algorithm that we reported previously for improving protein-ligand models for scoring (Fornabaio et al., 2003; Spyrakis et al., 2004). Our algorithm scores all possible ionization states of a model residue with other residues in its environment. Here, we optimized the ionization states of cysteine by first calculating the normal (environment-free) cost for ionizations of thiol group using published data (pKa = 8.66) (Bulaj et al., 1998) and applying the Henderson-Hasselbalch equation. For CYS, at pH 7, log ([S−]/[SH]) = 8.66–7.00, which is an equilibrium constant that can be converted to a ΔG of −2.26 kcal mol−1. Using the previously reported relation that −1 kcal mol−1 ≈ 500 HINT score units (Kellogg et al., 1991; Kellogg and Abraham, 2000), the energy cost in HINT score units for deprotonating cysteine at pH 7, in the absence of local pH effects is 1132.
The second term, calculated in varying protonation states, also as a HINT score, measures the effects of the local environment around the residue. This assessment of the environment scores the interactions of the residue in question in each accessible protonation state, with those in its neighborhood. These scores determine the protonation state of the residue. We examined the ionized (thiolate, S−) and neutral states with protonation at the sulfur atom (SH). If the HINT score was 50 or more (∼0.1 kcal mol−1) than the starting case, the residue's molecular model was replaced with the (protonated or deprotonated) trial model for that case. All further calculations at that pH were performed with the resulting optimized residue structure and coordinates.
4.5. HINT basis interaction maps
Each residue with its CA-CB bond along the z-axis, was placed within a three-dimensional box large enough to accommodate the structure of a residue, plus an additional 5 Å on each dimension. These boxes, based on residue type, are as follows: SER, −8.0 Å ≤ x ≤ 8.0 Å; −8.0 Å ≤ y ≤ 8.0 Å; −7.5 Å ≤ z ≤ 9.0 Å, (37,026 points, 4224 Å3); and CYS/CYX, −8.5 Å ≤ x ≤ 8.5 Å; −8.5 Å ≤ y ≤ 8.5 Å; −7.5 Å ≤ z ≤ 9.0 Å, (45,325 points, 4769 Å3); all with a point spacing of 0.5 Å. As described previously (Ahmed et al., 2015), HINT was used to calculate an interaction grid representing the 3D interaction space surrounding a residue of interest. In short, these maps interpret sums of pairwise HINT scores into 3D map objects indicating position, intensity, and type of interaction between atoms of the residue and those close in proximity (Kellogg and Abraham, 2000; Sarkar and Kellogg, 2010). Each grid point for a map was calculated, according to:
| ρxyz = ∑ bij exp (–[(x – xij)2 + (y – yij)2 + (z – zij)2] / σ) |
where ρxyz is the map interaction score at coordinates (x, y, z), xij, yij and zij are coordinates of the midpoint of the vector between atoms i and j, and σ is the width of the Gaussian map peak, 0.5 for our purposes. Map data were calculated for sidechain atoms of all SER, CYS, CYX, CYZ, CYSm, SERm and CYXm residues with individual maps for the four interaction classes: favorable/unfavorable polar and favorable/unfavorable hydrophobic.
4.6. Calculation of map-map correlation metrics
The calculation of map-map correlations, i.e., comparison of two maps, m and n, are based on:
| if |Gt|/F > 1.0, At = (Gt/|Gt|) log10 (|Gt|/F); else, At = 0 |
where each raw map data point (Gt, for point at index t) is transformed to log10 space and normalized with a predefined floor value, F = 1.0. Calculational methods defining the similarity between maps m and n, defined as D(m,n) was calculated as described previously in detail (Ahmed et al., 2015). Also, all correlation calculations were performed with in-house GPU-powered programs that exploit the inherent parallelism of our methods, especially for calculating maps and similarity matrices.
4.7. Clustering and validation
We utilized the freely available R programming language and environment to perform our clustering analysis on the pairwise map similarity matrices calculated above.(R Development Core Team, 2013) We determined that for our purposes, out of a number of different clustering methods, the k-means method was most reliable (Ahmed et al., 2015). We opted to set a uniform maximum number of clusters of 6 for each chess square-parse combination for SER, CYS and CYZ; 8 for SERm and CYSm; 10 for CYX and CYXm. This allows for significant map diversity and facilitates inter-chess square/inter-residue comparisons. A limitation of the k-means clustering is that it does not form singleton clusters, so we developed protocols to optionally recover them by reconstructing the cluster solutions possessing missing singletons. Any chess square-parse with four or fewer maps was not subjected to clustering, but, was instead averaged to create what is, effectively, a 1-cluster case.
4.8. Average map, RMSD, and solvent-accessible surface area calculations
Careful consideration must be given to calculation of average maps. First, to avoid a phenomeon that we described as "brown mapping" (Ahmed et al., 2015), only maps sharing high similarity should be combined. Second, the average maps are calculated by Gaussian weighting (w) the contribution of each map with respect to its Euclidean distance from the cluster centroid, given by:
| w = exp [–(d2/σ2)] |
where d is the map's distance from the centroid and σ = dmax/8, which is the average of all maximum distances across all clusters in the chess square. This weighting ensures that maps closer to the centroid contribute more significantly to the average map of the cluster, whereas taking a flat average of all map data would overweight the importance of maps further from the centroid, of which there are more. While a formal definition exists for "exemplar" in affinity propagation clustering, for our purposes, it represents the residue datum closest to the centroid of each cluster output by the k-means algorithm.
RMSDs (root-mean square distances) for each residue type were calculated by weighted averaging, as above, all atomic positions from all residues in a cluster to construct one average residue structure. For each non-hydrogen atom, an RMSD was calculated from the average structure, and then all atomic values were averaged to obtain the reported RMSD for the cluster.
We calculated SASAs for all residue sidechains using the GETAREA algorithm (Fraczkiewicz and Braun, 1998) and its default settings. The protein coordinates in PDB files were submitted as input. Also, from GETAREA's "In/Out" parameter, we created a new metric "foutside" to represent the buriedness of the set of residues in a cluster, parse, chess square, etc. by recasting "In" as 0.0, "Out" as 1.0 and "indeterminant" as 0.5, and averaging the set.
4.9. Calculation of map character and lipid-specific interactions
Map characters were calculated for each residue map from the grid points values in the associated map quartets – favorable and unfavorable hydrophobic and polar. For each, the grid point values (v) were summed [Σvhydro(−), Σvhydro(+), Σvpolar(−), Σvpolar(+)], and analyzed. The fractional interaction character of each residue’s environment was calculated as these sums normalized by the sum of all interactions, e.g., fhydro(+) = |Σ vhydro(+)|/{ |Σ vhydro(−)| + |Σ vhydro(+)| + |Σ vpolar(−)| + |Σ vpolar(+)| }. In this work, these values were averaged on a cluster-by-cluster basis, but the individual data are available in Supporting Information Tables S4–S10.
In order to assess the contribution of the artificial lipids set to the residue environments in the membrane proteins set (SERm, CYSm, CYZm), we created a DPPC “residue” for incorporation into the HINT partition dictionary, i.e., we assigned both hydropathic atom constants (ai) and solvent-accessible surface areas (Si) for each DPPC atom (vide supra). These atoms were thus explicitly included in map calculations for the residues in membrane proteins. Then, for each residue, the ratios of interaction scores involving the lipid atoms to all interaction scores, for each interaction type, were calculated. These are reported as flipid values (fLipHyd(−), fLipHyd(+), fLipPol(−), fLipPol(+)) and averaged on a cluster-by-cluster basis (see Tables S8–S10).
Credit author statement
Claudio Catalano: Conceptualization, Methodology, Investigation, Formal Analysis, Writing – Original Draft, Visualization; Mohammed H. AL Mughram: Methodology, Investigation, Formal Analysis, Visualization; Youzhong Guo: Writing – Review & Editing, Supervision, Funding Acquisition; Glen E. Kellogg: Conceptualization, Methodology, Software, Data Curation, Writing – Original Draft, Editing and Review, Supervision, Funding Acquisition.
Data availability
All numerical data generated in this work is provided in Supporting Information. Contact the author (glen.kellogg@vcu.edu) for access to other results from the study.
Declaration of competing interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests: 1910 Genetics, Inc. provided support for preliminary studies but had no role in the reporting of data or derived conclusions.
Acknowledgments
We acknowledge the motivation for continuing this project given to us by numerous proposal and manuscript reviewers who have critiqued our past work. Further, Dr. J. Néel Scarsdale provided advice and insight into protein structure. Preliminary studies for this research were partially funded by 1910 Genetics, Cambridge, Massachusetts. We are also grateful to Jen Nwankwo of 1910 Genetics for her enthusiasm.
Handling Editor: Chandra shekhar verma
Footnotes
Glen E. Kellogg reports financial support was provided by 1910 Genetics Inc.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.crstbi.2021.09.002.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- Ahmed M.H., Koparde V.N., Safo M.K., Scarsdale J.N., Kellogg G.E. 3D interaction homology: the structurally known rotamers of tyrosine derive from a surprisingly limited set of information-rich hydropathic interaction environments described by maps. Proteins. 2015;83:1118–1136. doi: 10.1002/prot.24813. [DOI] [PubMed] [Google Scholar]
- Ahmed M.H., Catalano C., Portillo S.C., Safo M.K., Neel Scarsdale J., Kellogg G.E. 3D interaction homology: the hydropathic interaction environments of even alanine are diverse and provide novel structural insight. J. Struct. Biol. 2019;207:183–198. doi: 10.1016/j.jsb.2019.05.007. [DOI] [PubMed] [Google Scholar]
- AL Mughram M.H., Catalano C., Bowry J.P., Safo M.K., Scarsdale J.N., Kellogg G.E. 3D interaction homology: hydropathic analyses of the “π–cation” and “π–π” interaction motifs in phenylalanine, tyrosine, and tryptophan residues. J. Chem. Inf. Model. 2021;61:2937–2956. doi: 10.1021/acs.jcim.1c00235. [DOI] [PubMed] [Google Scholar]
- Barzkar N., Khan Z., Jahromi S.T., Pourmozaffar S., Gozari M., Nahavandi R. A critical review on marine serine protease and its inhibitors: a new wave of drugs? Int. J. Biol. Macromol. 2021;170:674–687. doi: 10.1016/j.ijbiomac.2020.12.134. [DOI] [PubMed] [Google Scholar]
- Beno B.R., Yeung K.S., Bartberger M.D., Pennington L.D., Meanwell N.A. A survey of the role of noncovalent sulfur interactions in drug design. J. Med. Chem. 2015;58:4383–4438. doi: 10.1021/jm501853m. [DOI] [PubMed] [Google Scholar]
- Bhattacharyya R., Pal D., Chakrabarti P. Disulfide bonds, their stereospecific environment and conservation in protein structures. Protein Eng. Des. Sel. 2004;17:795–808. doi: 10.1093/protein/gzh093. [DOI] [PubMed] [Google Scholar]
- Bosnjak I., Bojovic V., Segvic-Bubic T., Bielen A. Occurrence of protein disulfide bonds in different domains of life: a comparison of proteins from the Protein Data Bank. Protein Eng. Des. Sel. 2014;27:65–72. doi: 10.1093/protein/gzt063. [DOI] [PubMed] [Google Scholar]
- Brooks D.J., Fresco J.R. Increased frequency of cysteine, tyrosine, and phenylalanine residues since the last universal ancestor. Mol. Cell. Proteomics. 2002;1:125–131. doi: 10.1074/mcp.m100001-mcp200. [DOI] [PubMed] [Google Scholar]
- Bulaj G. Formation of disulfide bonds in proteins and peptides. Biotechnol. Adv. 2005;23:87–92. doi: 10.1016/j.biotechadv.2004.09.002. [DOI] [PubMed] [Google Scholar]
- Bulaj G., Kortemme T., Goldenberg D.P. Ionization-reactivity relationships for cysteine thiols in polypeptides. Biochemistry. 1998;37:8965–8972. doi: 10.1021/bi973101r. [DOI] [PubMed] [Google Scholar]
- Burnett J.C., Botti P., Abraham D.J., Kellogg G.E. Computationally accessible method for estimating free energy changes resulting from site-specific mutations of biomolecules: systematic model building and structural/hydropathic analysis of deoxy and oxy hemoglobins. Proteins. 2001;42:355–377. doi: 10.1002/1097-0134(20010215)42:3<355::aid-prot60>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- Bywater R.P. Why twenty amino acid residue types suffice(d) to support all living systems. PloS One. 2018;13 doi: 10.1371/journal.pone.0204883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadenas E., Packer L. Thiol redox transitions in cell signaling, Pt A: chemistry and biochemistry of low molecular weight and protein thiols. Methods Enzymol. 2010;473 [Google Scholar]
- Casey P.J., Seabra M.C. Protein prenyltransferases. J. Biol. Chem. 1996;271:5289–5292. doi: 10.1074/jbc.271.10.5289. [DOI] [PubMed] [Google Scholar]
- Di Cera E. Serine proteases. IUBMB Life. 2009;61:510–515. doi: 10.1002/iub.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coskun U., Simons K. Cell membranes: the lipid perspective. Structure. 2011;19:1543–1548. doi: 10.1016/j.str.2011.10.010. [DOI] [PubMed] [Google Scholar]
- Cozzini P., Fornabaio M., Marabotti A., Abraham D.J., Kellogg G.E., Mozzarelli A. Free energy of ligand binding to protein: evaluation of the contribution of water molecules by computational methods. Curr. Med. Chem. 2004;11:3093–3118. doi: 10.2174/0929867043363929. [DOI] [PubMed] [Google Scholar]
- Dombkowski A.A., Sultana K.Z., Craig D.B. Protein disulfide engineering. FEBS Lett. 2014;588:206–212. doi: 10.1016/j.febslet.2013.11.024. [DOI] [PubMed] [Google Scholar]
- Duan J.C., Gaffrey M.J., Qian W.J. Quantitative proteomic characterization of redox-dependent post-translational modifications on protein cysteines. Mol. Biosyst. 2017;13:816–829. doi: 10.1039/c6mb00861e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edison A.S. Linus Pauling and the planar peptide bond. Nat. Struct. Biol. 2001;8:201–202. doi: 10.1038/84921. [DOI] [PubMed] [Google Scholar]
- Engelman D.M. Membranes are more mosaic than fluid. Nature. 2005;438:578–580. doi: 10.1038/nature04394. [DOI] [PubMed] [Google Scholar]
- Fass D. Disulfide bonding in protein biophysics. Annu. Rev. Biophys. 2012;41:63–79. doi: 10.1146/annurev-biophys-050511-102321. [DOI] [PubMed] [Google Scholar]
- Fass D., Thorpe C. Chemistry and enzymology of disulfide cross-linking in proteins. Chem. Rev. 2018;118:1169–1198. doi: 10.1021/acs.chemrev.7b00123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foden C.S., Islam S., Fernandez-Garcia C., Maugeri L., Sheppard T.D., Powner M.W. Prebiotic synthesis of cysteine peptides that catalyze peptide ligation in neutral water. Science. 2020;370:865–869. doi: 10.1126/science.abd5680. [DOI] [PubMed] [Google Scholar]
- Fornabaio M., Cozzini P., Mozzarelli A., Abraham D.J., Kellogg G.E. Simple, intuitive calculations of free energy of binding for protein-ligand complexes. 2. Computational titration and pH effects in molecular models of neuraminidase-inhibitor complexes. J. Med. Chem. 2003;46:4487–4500. doi: 10.1021/jm0302593. [DOI] [PubMed] [Google Scholar]
- Fraczkiewicz R., Braun W. Exact and efficient analytical calculation of the accessible surface areas and their gradients for macromolecules. J. Comput. Chem. 1998;19:319–333. [Google Scholar]
- Grazhdankin E., Stepniewski M., Xhaard H. Modeling membrane proteins: the importance of cysteine amino-acids. J. Struct. Biol. 2020;209:107400. doi: 10.1016/j.jsb.2019.10.002. [DOI] [PubMed] [Google Scholar]
- Gulamhussein A.A., Uddin R., Tighe B.J., Poyner D.R., Rothnie A.J. A comparison of SMA (styrene maleic acid) and DIBMA (di-isobutylene maleic acid) for membrane protein purification. Biochim. Biophys. Acta. 2020;1862:183281. doi: 10.1016/j.bbamem.2020.183281. [DOI] [PubMed] [Google Scholar]
- Guo Y. Be cautious with crystal structures of membrane proteins or complexes prepared in detergents. Crystals. 2020;10:86. doi: 10.3390/cryst10020086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y. Detergent-free systems for structural studies of membrane proteins. Biochem. Soc. Trans. 2021 doi: 10.1042/BST20201080. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallenbeck K.K., Turner D.M., Renslo A.R., Arkin M.R. Targeting non-catalytic cysteine residues through structure-guided drug discovery. Curr. Top. Med. Chem. 2017;17:4–15. doi: 10.2174/1568026616666160719163839. 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartigan J.A., Wong M.A. Algorithm AS 136: a k-means clustering algorithm. J. R. Stat. Soc., C: Appl. Stat. 1979;28:100–108. [Google Scholar]
- Hatahet F., Ruddock L.W. Protein disulfide isomerase: a critical evaluation of its function in disulfide bond formation. Antioxid. Redox Signal. 2009;11:2807–2850. doi: 10.1089/ars.2009.2466. [DOI] [PubMed] [Google Scholar]
- Heppner D. Structural insights into redox-active cysteine residues of the Src family kinases. Redox Biol. 2021;41:101934. doi: 10.1016/j.redox.2021.101934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrington N.B., Kellogg G.E. 2021. 3D Interaction Homology: Computational Titration of Aspartic Acid, Glutamic Acid and Histidine Can Create pH-Tunable Hydropathic Environment Maps. Manuscript submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirabayashi Y., Furuya S. Roles of L-serine and sphingolipid synthesis in brain development and neuronal survival. Prog. Lipid Res. 2008;47:188–203. doi: 10.1016/j.plipres.2008.01.003. [DOI] [PubMed] [Google Scholar]
- Hofer F., Kraml J., Kahler U., Kamenik A.S., Liedl K.R. Catalytic site pKa values of aspartic, cysteine, and serine proteases: constant pH MD simulations. J. Chem. Inf. Model. 2020;60:3030–3042. doi: 10.1021/acs.jcim.0c00190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen K.S., Hansen R.E., Winther J.R. Kinetic and thermodynamic aspects of cellular thiol-disulfide redox regulation. Antioxidants Redox Signal. 2009;11:1047–1058. doi: 10.1089/ars.2008.2297. [DOI] [PubMed] [Google Scholar]
- Kellogg G.E., Abraham D.J. Hydrophobicity: is LogP(o/w) more than the sum of its parts? Eur. J. Med. Chem. 2000;35:651–661. doi: 10.1016/s0223-5234(00)00167-7. [DOI] [PubMed] [Google Scholar]
- Kellogg G.E., Semus S.F., Abraham D.J. HINT: a new method of empirical hydrophobic field calculation for CoMFA. J. Comput. Aided Mol. Des. 1991;5:545–552. doi: 10.1007/BF00135313. [DOI] [PubMed] [Google Scholar]
- Kellogg G.E., Fornabaio M., Spyrakis F., Lodola A., Cozzini P., Mozzarelli A., Abraham D.J. Getting it right: modeling of pH, solvent and "nearly" everything else in virtual screening of biological targets. J. Mol. Graph. Model. 2004;22:479–486. doi: 10.1016/j.jmgm.2004.03.008. [DOI] [PubMed] [Google Scholar]
- Kendrew J.C., Dickerson R.E., Strandberg B.E., Hart R.G., Davies D.R., Phillips D.C., Shore V.C. Structure of myoglobin: a three-dimensional fourier synthesis at 2 Å resolution. Nature. 1960;185:422–427. doi: 10.1038/185422a0. [DOI] [PubMed] [Google Scholar]
- Kent C. Eukaryotic phospholipid biosynthesis. Annu. Rev. Biochem. 1995;64:315–343. doi: 10.1146/annurev.bi.64.070195.001531. [DOI] [PubMed] [Google Scholar]
- Klomsiri C., Karplus P.A., Poole L.B. Cysteine-based redox switches in enzymes. Antioxidants Redox Signal. 2011;14:1065–1077. doi: 10.1089/ars.2010.3376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klug A., Schwabe J.W.R. Protein motifs 5. Zinc fingers. Faseb. J. 1995;9:597–604. [PubMed] [Google Scholar]
- Kroeck K.G., Qiu W., Catalano C., Trinh T.K.H., Guo Y. Native cell membrane nanoparticles system for membrane protein-protein interaction analysis. J Vis Exp. 2020;10 doi: 10.3791/61298. 3791/61298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroncke K.D., Klotz L.O. Zinc fingers as biologic redox switches? Antioxidants Redox Signal. 2009;11:1015–1027. doi: 10.1089/ARS.2008.2269. [DOI] [PubMed] [Google Scholar]
- Lee S.C., Knowles T.J., Postis V.L.G., Jamshad M., Parslow R.A., Lin Y.P., Goldman A., Sridhar P., Overduin M., Muench S.P., Dafforn T.R. A method for detergent-free isolation of membrane proteins in their local lipid environment. Nat. Protoc. 2016;11:1149–1162. doi: 10.1038/nprot.2016.070. [DOI] [PubMed] [Google Scholar]
- Liu X.X., Zhang J.X., Ni F., Dong X., Han B.C., Han D.X., Ji Z.L., Zhao Y.F. Genome wide exploration of the origin and evolution of amino acids. BMC Evol. Biol. 2010;10 doi: 10.1186/1471-2148-10-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodish H., Berk A., Zipursky S.L., Matsudaira P., Baltimore D., Darnell J. fourth ed. W. H. Freeman; New York: 2000. Molecular Cell Biology. [Google Scholar]
- Long M.J.C., Aye Y. Privileged electrophile sensors: a resource for covalent drug development. Cell Chem. Biol. 2017;24:787–800. doi: 10.1016/j.chembiol.2017.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacCallum J.L., Tieleman D.P. Hydrophobicity scales: a thermodynamic looking glass into lipid-protein interactions. Trends Biochem. Sci. 2011;36:653–662. doi: 10.1016/j.tibs.2011.08.003. [DOI] [PubMed] [Google Scholar]
- MacCallum J.L., Bennett W.F.D., Tieleman D.P. Partitioning of amino acid side chains into lipid bilayers: results from computer simulations and comparison to experiment. J. Gen. Physiol. 2007;129:371–377. doi: 10.1085/jgp.200709745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manteca A., Alonso-Caballero A., Fertin M., Poly S., De Sancho D., Perez-Jimenez R. The influence of disulfide bonds on the mechanical stability of proteins is context dependent. J. Biol. Chem. 2017;292:13374–13380. doi: 10.1074/jbc.M117.784934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marconnet A., Michon B., Le Bon C., Giusti F., Tribet C., Zoonens M. Solubilization and stabilization of membrane proteins by cycloalkane-modified amphiphilic polymers. Biomacromolecules. 2020;21:3459–3467. doi: 10.1021/acs.biomac.0c00929. [DOI] [PubMed] [Google Scholar]
- Marino S.M., Gladyshev V.N. Cysteine function governs its conservation and degeneration and restricts its utilization on protein surfaces. J. Mol. Biol. 2010;404:902–916. doi: 10.1016/j.jmb.2010.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marino S.M., Gladyshev V.N. Redox biology: computational approaches to the investigation of functional cysteine residues. Antioxid. Redox Signal. 2011;15:135–146. doi: 10.1089/ars.2010.3561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marino S.M., Gladyshev V.N. Analysis and functional prediction of reactive cysteine residues. J. Biol. Chem. 2012;287:4419–4425. doi: 10.1074/jbc.R111.275578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntosh T.J., Simon S.A. Bilayers as protein solvents: role of bilayer structure and elastic properties. J. Gen. Physiol. 2007;130:225–227. doi: 10.1085/jgp.200709841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miseta A., Csutora P. Relationship between the occurrence of cysteine in proteins and the complexity of organisms. Mol. Biol. Evol. 2000;17:1232–1239. doi: 10.1093/oxfordjournals.molbev.a026406. [DOI] [PubMed] [Google Scholar]
- Moomaw J.F., Zhang F.L., Casey P.J. Isolation of protein prenyltransferases from bovine brain and baculovirus expression system. Meth. Enzymol. 1995;250:12–21. doi: 10.1016/0076-6879(95)50058-8. [DOI] [PubMed] [Google Scholar]
- Murtas G., Marcone G.L., Sacchi S., Pollegioni L. L-serine synthesis via the phosphorylated pathway in humans. Cell. Mol. Life Sci. 2020;77:5131–5148. doi: 10.1007/s00018-020-03574-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newport T.D., Sansom M.S.P., Stansfeld P.J. The MemProtMD database: a resource for membrane-embedded protein structures and their lipid interactions. Nucleic Acids Res. 2019;47:D390–D397. doi: 10.1093/nar/gky1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauling L., Corey R.B. Configurations of polypeptide chains with favored orientations around single bonds - 2 new pleated sheets. Proc. Natl. Acad. Sci. U.S.A. 1951;37:729–740. doi: 10.1073/pnas.37.11.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauling L., Corey R.B., Branson H.R. The structure of proteins: two hydrogen-bonded helical configurations of the polypeptide chain. Proc. Natl. Acad. Sci. U.S.A. 1951;37:205–211. doi: 10.1073/pnas.37.4.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulsen C.E., Carroll K.S. Cysteine-mediated redox signaling: chemistry, biology, and tools for discovery. Chem. Rev. 2013;113:4633–4679. doi: 10.1021/cr300163e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perutz M.F., Rossmann M.G., Cullis A.F., Muirhead H., Will G., North A.C.T. Structure of haemoglobin: a three-dimensional fourier synthesis at 5.5 Å resolution, obtained by X-ray analysis. Nature. 1960;185:416–422. doi: 10.1038/185416a0. [DOI] [PubMed] [Google Scholar]
- Petersen M.T.N., Johnson P.H., Petersen S.B. Amino acid neighbours and detailed conformational analysis of cysteines in proteins. Protein Eng. 1999;12:535–548. doi: 10.1093/protein/12.7.535. [DOI] [PubMed] [Google Scholar]
- Poole L.B. The basics of thiols and cysteines in redox biology and chemistry. Free Radic. Biol. Med. 2015;80:148–157. doi: 10.1016/j.freeradbiomed.2014.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin M., Wang W., Thirumalai D. Protein folding guides disulfide bond formation. Proc. Natl. Acad. Sci. U. S. A. 2015;112:11241–11246. doi: 10.1073/pnas.1503909112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu W., Fu Z., Xu G.G., Grassucci R.A., Zhang Y., Frank J., Hendrickson W.A., Guo Y. Structure and activity of lipid bilayer within a membrane-protein transporter. Proc. Natl. Acad. Sci. U.S.A. 2018;115:12985–12990. doi: 10.1073/pnas.1812526115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team . R Foundation for Statistical Computing; 2013. R: A Language and Environment for Statistical Computing. [Google Scholar]
- Ramachandran G.N., Sasisekharan V. In: Anfinsen C.B., Anson M.L., Edsall J.T., Richards F.M., editors. Vol. 23. Academic Press; 1968. Conformation of polypeptides and proteins; pp. 283–437. (Advances in Protein Chemistry). [DOI] [PubMed] [Google Scholar]
- Ramachandran G.N., Ramakrishnan C., Sasisekharan V. Stereochemistry of polypeptide chain configurations. J. Mol. Biol. 1963;7:95–99. doi: 10.1016/s0022-2836(63)80023-6. [DOI] [PubMed] [Google Scholar]
- Richardson J.S. The anatomy and taxonomy of protein structure. Adv. Protein Chem. 1981;34:167–339. doi: 10.1016/s0065-3233(08)60520-3. [DOI] [PubMed] [Google Scholar]
- Robinson P.J., Bulleid N.J. Mechanisms of disulfide bond formation in nascent polypeptides entering the secretory pathway. Cells. 2020;9:1994. doi: 10.3390/cells9091994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robson B., Suzuki E. Conformational properties of amino-acid residues in globular proteins. J. Mol. Biol. 1976;107:327–356. doi: 10.1016/s0022-2836(76)80008-3. [DOI] [PubMed] [Google Scholar]
- Sarkar A., Kellogg G.E. Hydrophobicity - shake flasks, protein folding and drug discovery. Curr. Top. Med. Chem. 2010;10:67–83. doi: 10.2174/156802610790232233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapovalov M.V., Dunbrack R.L. A smoothed backbone-dependent rotamer library for proteins derived from adaptive kernel density estimates and regressions. Structure. 2011;19:844–858. doi: 10.1016/j.str.2011.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon K.S., Pollock N.L., Lee S.C. Membrane protein nanoparticles: the shape of things to come. Biochem. Soc. Trans. 2018;46:1495–1504. doi: 10.1042/BST20180139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh J., Petter R.C., Baillie T.A., Whitty A. The resurgence of covalent drugs. Nat. Rev. Drug Discov. 2011;10:307–317. doi: 10.1038/nrd3410. [DOI] [PubMed] [Google Scholar]
- Sjostrom M., Wold S. A multivariate study of the relationship between the genetic-code and the physical-chemical properties of amino-acids. J. Mol. Evol. 1985;22:272–277. doi: 10.1007/BF02099756. [DOI] [PubMed] [Google Scholar]
- Spyrakis F., Fornabaio M., Cozzini P., Mozzarelli A., Abraham D.J., Kellogg G.E. Computational titration analysis of a multiprotic HIV-1 protease-ligand complex. J. Am. Chem. Soc. 2004;126:11764–11765. doi: 10.1021/ja0465754. [DOI] [PubMed] [Google Scholar]
- Stansfeld P.J., Goose J.E., Caffrey M., Carpenter E.P., Parker J.L., Newstead S., Sansom M.S.P. MemProtMD: automated insertion of membrane protein structures into explicit lipid membranes. Structure. 2015;23:1350–1361. doi: 10.1016/j.str.2015.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornton J.M. Disulfide bridges in globular-proteins. J. Mol. Biol. 1981;151:261–287. doi: 10.1016/0022-2836(81)90515-5. [DOI] [PubMed] [Google Scholar]
- Tien M.Z., Meyer A.G., Sydykova D.K., Spielman S.J., Wilke C.O. Maximum allowed solvent accessibilites of residues in proteins. PloS One. 2013;8 doi: 10.1371/journal.pone.0080635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trifonov E.N. Consensus temporal order of amino acids and evolution of the triplet code. Gene. 2000;261:139–151. doi: 10.1016/s0378-1119(00)00476-5. [DOI] [PubMed] [Google Scholar]
- Wallin E., von Heijne G. Genome-wide analysis of integral membrane proteins from eubacterial, archaean, and eukaryotic organisms. Protein Sci. 1998;7:1029–1038. doi: 10.1002/pro.5560070420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White S.H. Membrane protein insertion: the biology-physics nexus. J. Gen. Physiol. 2007;129:363–369. doi: 10.1085/jgp.200709741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfenden R. Experimental measures of amino acid hydrophobicity and the topology of transmembrane and globular proteins. J. Gen. Physiol. 2007;129:357–362. doi: 10.1085/jgp.200709743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L., Catalano C., Xu Y., Qiu W., Zhang D., McDermott A., Guo Y., Blount P. A native cell membrane nanoparticles system allows for high-quality functional proteoliposome reconstitution. BBA Advances. 2021;1:100011. doi: 10.1016/j.bbadva.2021.100011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeida A., Guardia C.M., Lichtig P., Perissinotti L.L., Defelipe L.A., Turjanski A., Radi R., Trujillo M., Estrin D.A. Thiol redox biochemistry: insights from computer simulations. Biophys. Rev. 2014;6:27–46. doi: 10.1007/s12551-013-0127-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeida A., Reyes A.M., Lebrero M.C.G., Radi R., Trujillo M., Estrin D.A. The extraordinary catalytic ability of peroxiredoxins: a combined experimental and QM/MM study on the fast thiol oxidation step. Chem. Commun. 2014;50:10070–10073. doi: 10.1039/c4cc02899f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Tian F.F., Lv F.L., Shang Z.C. Geometric characteristics of hydrogen bonds involving sulfur atoms in proteins. Proteins. 2009;76:151–163. doi: 10.1002/prot.22327. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All numerical data generated in this work is provided in Supporting Information. Contact the author (glen.kellogg@vcu.edu) for access to other results from the study.