Summary
Climate change makes plant‐parasitic nematodes (PPN) an increasing threat to commercial crops. PPN can be managed sustainably by the biocontrol fungus Pochonia chlamydosporia (Pc). Chitosan generated from chitin deacetylation enhances PPN parasitism by Pc. In this work, we investigate the molecular mechanisms of Pc for chitosan resistance and root‐knot nematode (RKN) parasitism, using transcriptomics. Chitosan and RKN modify the expression of Pc genes, mainly those involved in oxidation–reduction processes. Both agents significantly modify the expression of genes associated to 113 GO terms and 180 Pc genes. Genes encoding putative glycoproteins (Pc adhesives) to nematode eggshell, as well as genes involved in redox, carbohydrate and lipid metabolism trigger the response to chitosan. We identify genes expressed in both the parasitic and endophytic phases of the Pc lifecycle; these include proteases, chitosanases and transcription factors. Using the Pathogen—Host Interaction database (PHI‐base), our previous RNA‐seq data and RT‐PCR of Pc colonizing banana we have investigated genes expressed both in the parasitic and endophytic phases of Pc lifecycle.
Introduction
Root‐knot nematodes (RKN) are a persistent problem in fruit and vegetable crops (Ralmi and Khandaker, 2016). Biological control is used to reduce and avoid the use of toxic chemical nematicides and fumigants, introducing non‐harmful organisms for plants that can manage pests and diseases in a sustainable way (Mankau, 1980).
Pochonia chlamydosporia (=Metacordyceps chlamydosporia) (Goddard) Zare and Gams (Pc) is a nematophagous fungus used for biocontrol of RKN (Meloidogyne spp.) (Forghani and Hajihassani, 2020), cyst nematodes (Heterodera spp. and Globodera spp.) (Willcox and Tribe, 1974; Manzanilla‐Lopez et al., 2011) and false RKN (Nacobbus spp.) (Flores‐Camacho et al., 2008). Pc is distributed worldwide and may also adopt saprotrophic and endophytic lifestyles (Bordallo et al., 2002; Maciá‐Vicente et al., 2009; Manzanilla‐López et al., 2011; Zavala‐Gonzalez et al., 2017).
Chitosan is a linear polymer of β‐(1‐4)‐linked N‐acetyl‐2‐amino‐2‐deoxy‐d‐glucose (acetylated) and 2‐amino‐2‐deoxy‐d‐glucose (deacetylated) (Kaur and Dhillon, 2014). This polymer is an elicitor of plant defences (Benhamou and Thériault, 1992; Lafontaine and Benhamou, 1996; Yin et al., 2016; Suarez‐Fernandez et al., 2020) and has antifungal activity (Shih et al., 2019), inhibiting or killing fungal pathogens. Chitosan also promotes the growth of resistant fungi such as Pc and entomopathogenic fungi (Palma‐Guerrero et al., 2007). The molecular mechanisms that determine whether a fungus is resistant or sensitive to chitosan remain to be determined. Pc is resistant to chitosan and can use it as a nutrient source (Palma‐Guerrero et al., 2010). Chitosan‐resistant fungi produce valuable bioproducts from chitosan degradation due to their chitinases and chitosanases (Kaczmarek et al., 2019). The Pc genome encodes a high number of chitosanases that are induced during nematode egg parasitism (Aranda‐Martinez et al., 2016). Chitosan improves efficiency in reducing nematode pests by nematophagous fungi (Escudero et al., 2017; Mwaheb et al., 2017). Therefore, combining Pc and chitosan could be a good strategy to manage PPN infections in plants.
Global unbiased transcriptomic analyses are a useful tool for determining genes involved in the response of fungi to elicitors such as chitosan (Zhang et al., 2020). These analyses also show which genes are involved in biological processes, such as pathogenicity to nematodes (Balestrini et al., 2019). The activation of specific genes can trigger the transition from endophytism to parasitism and vice versa in fungi (Fesel and Zuccaro, 2016; Zhang et al., 2018). In view of the multitrophic interaction of chitosan and Pc with plants and PPN, it is relevant to study genes in common between endophytism and pathogenicity. This could reveal which genes are key for interacting with other organisms.
This work aims to analyse the molecular mechanisms that are activated in Pc when interacting with chitosan, RKN or both as well as to unravel which mechanisms would increase RKN parasitism by Pc. These mechanisms could explain what makes a fungus resistant or sensitive to chitosan. In this work we focus on identifying which genes are shared in both lifestyles: RKN parasitism and plant root endophytism.
Results
Chitosan and RKN eggs modify Pc gene expression
Twelve samples were processed (four treatments per triplicate), with average total read bases (total reads × read length) of 17 894 300 929 yielding approximately 100 GB of transcriptomic raw data. When using DESeq2 in a genome‐guided analysis to Pc123 genome 39 009 transcripts in 20 502 loci were found. In addition, 5746, 710 and 6595 genes were expressed in Pc treated with chitosan (PcQ), Pc treated with nematodes (PcRKN) and Pc treated with chitosan and nematode eggs (PcRKNQ) respectively (all data are available in Supplementary Information File). Considering all treatments together, 80 upregulated and 99 downregulated genes can be found with a threshold of ±2 in log2 fold change values (Fig. 1; Tables 1, 2, 3). In PcQ there are 136 differentially expressed genes (DEG, up and downregulated), in PcRKNQ 75 DEG, while in PcRKN only 10. Therefore, chitosan is a stronger modulator of Pc gene expression than RKN eggs are. Two genes encoding proteins (RZR63781.1, ribonuclease H‐like protein and RZR63940.1, ankyrin repeat protein) are downregulated in all treatments. These results are supported by qRT‐PCR (Supplementary Fig. 1).
Fig 1.
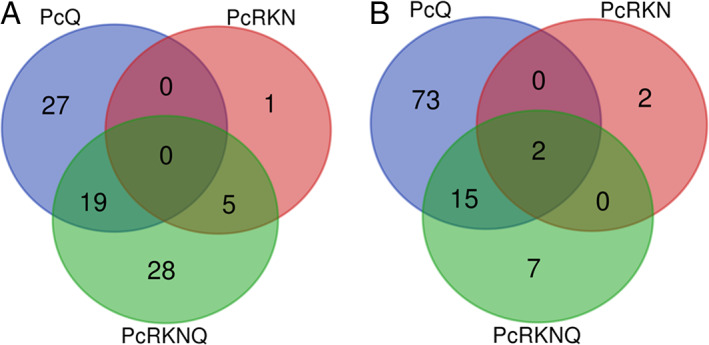
Chitosan and RKN modify P. chlamydosporia gene expression. Venn diagrams show a total of 80 upregulated (A) and 99 downregulated (B) genes when a threshold of ±2 in log2 fold change is set and adjusted P‐value is taken into account (44.7% vs. 55.3%). Treatments: PcQ, P. chlamydosporia with chitosan (1 mg·ml−1); PcRKN, P. chlamydosporia with M. javanica eggs (1 egg·μl−1); PcRKNQ, P. chlamydosporia with M. javanica eggs (1 egg·μl−1) and chitosan (1 mg ml−1). All treatments were applied for 4 days.
Table 1.
Differentially expressed genes in PcQ treatment.
| GenBank accession | Sequence description | Log2 fold change | Adj. P‐value |
|---|---|---|---|
| RZR66026.1 | Secreted aspartic proteinase precursor | 8.155 | 6.36E‐13 |
| RZR63431.1 | Peptidase S8/S53, subtilisin/kexin/sedolisin | 8.029 | 5.38E‐12 |
| RZR59126.1 | Hypothetical protein I1G_00009219 | 7.792 | 3.35E‐04 |
| RZR63795.1 | Glycoside hydrolase family 75 | 6.745 | 3.92E‐07 |
| RZR63158.1 | Peptidase A1 | 6.520 | 3.68E‐11 |
| RZR61856.1 | Glucokinase | 6.489 | 1.27E‐09 |
| RZR62208.1 | Peptidase A4 family protein | 6.016 | 3.61E‐11 |
| RZR61845.1 | Glycoside hydrolase family 75 protein | 5.858 | 5.70E‐05 |
| RZR66080.1 | CipC1 protein, concanamycin induced protein C | 5.738 | NA |
| RZR62018.1 | Metallo‐endopeptidase | 5.064 | 4.21E‐05 |
| RZR61625.1 | Cytochrome P450 ClCP1 | 5.053 | 7.12E‐06 |
| RZR61472.1 | Oligopeptide transporter OPT‐like protein | 4.947 | 6.16E‐04 |
| RZR70313.1 | Chitosanase CSN1 | 4.664 | 2.15E‐06 |
| RZR61846.1 | Major facilitator superfamily domain, general substrate transporter | 4.641 | 8.25E‐08 |
| RZR65223.1 | WSC domain‐containing protein | 4.559 | 1.09E‐04 |
| RZR68359.1 | Nucleotide‐binding, alpha‐beta plait | 4.427 | 5.32E‐07 |
| RZR60329.1 | Carboxyl‐terminal proteinase | 3.907 | 5.36E‐04 |
| RZR61987.1 | Tripeptidyl‐peptidase 1 precursor | 3.851 | 6.55E‐06 |
| RZR62940.1 | Glycoside hydrolase family 75 | 3.801 | 9.84E‐04 |
| RZR66604.1 | Hypothetical protein VFPPC_00169 | 3.368 | 2.91E‐03 |
| RZR68451.1 | Maltose permease | 3.141 | 5.70E‐05 |
| RZR62148.1 | P‐loop containing nucleoside triphosphate hydrolase protein | 3.132 | 4.50E‐03 |
| RZR69895.1 | Putative polyketide synthase | 2.968 | 4.94E‐03 |
| RZR70338.1 | Glycoside hydrolase, subgroup, catalytic core | 2.916 | 4.59E‐03 |
| RZR66611.1 | Hypothetical protein I1G_00003511 | 2.630 | 1.15E‐02 |
| RZR66588.1 | AtmA protein | 2.622 | 2.81E‐07 |
| RZR70289.1 | Extracellular soluble lytic transglycosylase | 2.599 | 7.85E‐06 |
| RZR60800.1 | Hypothetical protein VFPPC_03295 | 2.575 | 2.39E‐03 |
| RZR64639.1 | Hypothetical protein I1G_00010772 | 2.561 | 2.28E‐04 |
| RZR59556.1 | Solid‐state culture specific ATP‐grasp domain protein | 2.530 | 4.53E‐03 |
| RZR68085.1 | Sugar transporter family protein | 2.526 | 3.84E‐03 |
| RZR64446.1 | Cytochrome P450 6A1 | 2.469 | 1.28E‐03 |
| RZR67153.1 | Hypothetical protein I1G_00011003 | 2.454 | 1.85E‐02 |
| RZR65901.1 | l‐amino‐acid oxidase | 2.447 | 1.39E‐02 |
| RZR64948.1 | Fungal chitosanase | 2.411 | 4.53E‐03 |
| RZR69865.1 | Cytochrome P450 oxidoreductase | 2.383 | 2.99E‐03 |
| RZR63081.1 | Glycoside hydrolase family 2 protein | 2.379 | 4.99E‐03 |
| RZR63277.1 | l‐ascorbate oxidase | 2.333 | 9.16E‐03 |
| RZR63926.1 | Lactonase, 7‐bladed beta‐propeller domain‐containing protein | 2.280 | 1.76E‐02 |
| RZR63275.1 | Cytochrome b561, eukaryote | 2.169 | 2.16E‐03 |
| RZR69240.1 | APSES transcription factor | 2.140 | 7.08E‐03 |
| RZR64143.1 | Flavin‐binding monooxygenase‐like family protein | 2.104 | 2.06E‐02 |
| RZR59143.1 | Hypothetical protein I1G_00011200 | 2.094 | 1.37E‐02 |
| RZR63080.1 | Nuclear distribution protein pac‐1a | 2.049 | 1.31E‐02 |
| RZR69181.1 | Hypothetical protein I1G_00010741 | 2.034 | 1.36E‐03 |
| RZR67141.1 | Killer toxin, Kp4/SMK‐like, core | 2.008 | 1.02E‐02 |
| RZR65937.1 | Nitrate reductase (NADH) | −2.024 | 2.37E‐02 |
| RZR60029.1 | Aldo/keto reductase | −2.034 | 8.26E‐03 |
| RZR63940.1 | Ankyrin repeat protein | −2.040 | 1.36E‐03 |
| RZR67896.1 | MFS transporter, SP family, general alpha glucoside:H+ symporter | −2.051 | 4.65E‐03 |
| RZR66454.1 | Hypothetical protein I1G_00004264 | −2.055 | 9.55E‐03 |
| RZR65015.1 | Nitrate reductase‐like protein | −2.056 | 4.99E‐03 |
| RZR69659.1 | Integral membrane protein | −2.057 | 5.70E‐05 |
| RZR66259.1 | Cytochrome P450 | −2.068 | 2.61E‐03 |
| RZR62313.1 | Hypothetical protein I1G_00005303 | −2.097 | 1.24E‐02 |
| RZR67230.1 | NADP‐dependent alcohol dehydrogenase C | −2.103 | 7.72E‐03 |
| RZR64929.1 | FMN‐dependent alpha‐hydroxy acid dehydrogenase | −2.104 | 1.08E‐02 |
| RZR59541.1 | Hypothetical protein I1G_00010383 | −2.107 | 2.21E‐02 |
| RZR66613.1 | Oxidoreductase | −2.114 | 7.04E‐03 |
| RZR67320.1 | Related to double substrate‐specificity short chain dehydrogenase/reductase 2 | −2.149 | 2.72E‐02 |
| RZR65483.1 | MFS transporter | −2.157 | 1.89E‐02 |
| RZR62047.1 | Cell surface flocculin, putative | −2.178 | 1.24E‐02 |
| RZR61511.1 | Reductase | −2.181 | 2.16E‐03 |
| RZR59404.1 | Transcription factor | −2.183 | 1.03E‐02 |
| RZR66789.1 | Predicted protein | −2.185 | 2.18E‐02 |
| RZR66794.1 | Hypothetical protein I1G_00009684 | −2.193 | 2.55E‐02 |
| RZR67810.1 | l‐isoaspartate O‐methyltransferase | −2.220 | 9.55E‐03 |
| RZR61034.1 | Thioredoxin domain‐containing protein | −2.232 | 2.91E‐03 |
| RZR64973.1 | NA | −2.234 | 0.01 |
| RZR70090.1 | Zinc transporter protein | −2.240 | 2.35E‐02 |
| RZR66702.1 | QI74 protein | −2.258 | 2.49E‐02 |
| RZR70243.1 | MUS38‐like protein | −2.274 | 4.08E‐04 |
| RZR59616.1 | Oxidoreductase | −2.275 | 1.15E‐02 |
| RZR62422.1 | Related to short‐chain alcohol dehydrogenase | −2.277 | 4.10E‐03 |
| RZR66328.1 | S‐(hydroxymethyl)glutathione dehydrogenase | −2.283 | 9.35E‐03 |
| RZR70238.1 | Predicted protein | −2.283 | 1.36E‐03 |
| RZR62423.1 | Catalase A | −2.301 | 9.61E‐03 |
| RZR64023.1 | Hypothetical protein I1G_00011265 | −2.304 | 1.35E‐02 |
| RZR64512.1 | MIP transporter | −2.324 | 1.32E‐02 |
| RZR67229.1 | 3‐dehydroshikimate dehydratase protein | −2.328 | 6.62E‐03 |
| RZR67902.1 | Alpha/beta hydrolase domain‐containing protein | −2.340 | 1.87E‐02 |
| RZR59365.1 | SUR7 protein | −2.359 | 6.50E‐03 |
| RZR65734.1 | Related to diacylglycerol pyrophosphate phosphatase DPP1 | −2.380 | 2.56E‐03 |
| RZR70207.1 | Major facilitator superfamily domain, general substrate transporter | −2.386 | NA |
| RZR67256.1 | Oligosaccharide translocation protein RFT1 | −2.412 | 6.87E‐03 |
| RZR60041.1 | Ribonuclease H‐like protein | −2.448 | 1.88E‐02 |
| RZR67257.1 | Putative phosphatidylinositol phosphate kinase | −2.470 | 3.98E‐04 |
| RZR69040.1 | Double‐stranded RNA binding motif domain‐containing protein | −2.471 | 2.39E‐03 |
| RZR68713.1 | BTB domain transcription factor | −2.478 | 1.69E‐03 |
| RZR65017.1 | Short‐chain dehydrogenase/reductase family protein | −2.486 | 5.58E‐03 |
| RZR66003.1 | Potassium channel | −2.496 | 1.40E‐03 |
| RZR65013.1 | 3‐oxoacyl‐(acyl‐carrier‐protein) reductase | −2.522 | 1.04E‐02 |
| RZR67228.1 | 6‐phosphogluconate dehydrogenase, decarboxylating | −2.539 | 5.23E‐03 |
| RZR65468.1 | Hypothetical protein I1G_00002210 | −2.550 | 2.91E‐03 |
| RZR67249.1 | Hypothetical protein I1G_00007967 | −2.594 | 1.34E‐04 |
| RZR65014.1 | Lactamase_B domain‐containing protein | −2.619 | 1.40E‐02 |
| RZR65332.1 | Hypothetical protein I1G_00001300 | −2.625 | 1.65E‐04 |
| RZR62987.1 | C6 transcription factor | −2.667 | 1.20E‐02 |
| RZR66334.1 | Protein kinase domain protein | −2.711 | 6.50E‐03 |
| RZR64692.1 | Hypothetical protein I1G_00007179 | −2.717 | 3.78E‐03 |
| RZR64363.1 | ATP synthase protein 9 (Lipid‐binding protein) | −2.745 | 4.58E‐03 |
| RZR67237.1 | Choline and nitrogen mustard permease | −2.795 | 5.61E‐04 |
| RZR61033.1 | Glucose repressible protein Grg1 | −2.810 | 8.46E‐07 |
| RZR69039.1 | DUF1929 multi‐domain protein | −2.824 | 1.18E‐03 |
| RZR69535.1 | NAD(P)‐binding domain protein | −2.844 | 8.91E‐04 |
| RZR66687.1 | l‐amino acid oxidase | −2.848 | 1.64E‐02 |
| RZR64977.1 | Related to molybdopterin biosynthesis protein moeA | −2.849 | 4.99E‐03 |
| RZR69169.1 | Transcriptional regulatory protein GAL4 | −2.866 | 3.21E‐03 |
| RZR67231.1 | d‐xylulose 5‐phosphate/d‐fructose 6‐phosphate phosphoketolase | −2.874 | 2.56E‐03 |
| RZR66951.1 | Hypothetical protein I1G_00006786 | −2.876 | 1.26E‐02 |
| RZR60802.1 | Siderophore iron transporter mirB | −3.029 | 1.50E‐02 |
| RZR60798.1 | Alcohol acetyltransferase | −3.046 | 1.50E‐02 |
| RZR63453.1 | Protein bli‐3 | −3.067 | 1.92E‐03 |
| RZR66605.1 | Beta‐lactamase‐like protein | −3.069 | 5.20E‐04 |
| RZR62135.1 | MFS transporter | −3.134 | 5.81E‐04 |
| RZR68164.1 | MARVEL‐like domain protein | −3.165 | 5.35E‐04 |
| RZR61718.1 | Ctr copper transporter | −3.188 | 1.39E‐02 |
| RZR59602.1 | 30 kDa heat shock protein | −3.333 | 1.42E‐04 |
| RZR60799.1 | Transferase family protein | −3.417 | 1.21E‐02 |
| RZR67450.1 | Histone acetylase complex subunit | −3.523 | 3.28E‐03 |
| RZR65938.1 | HHE domain containing protein | −3.575 | 1.31E‐03 |
| RZR62210.1 | Major allergen Asp f 2‐like protein | −4.322 | 3.45E‐05 |
| RZR62374.1 | Ctr copper transporter family protein | −4.413 | 7.43E‐03 |
| RZR63781.1 | Ribonuclease H‐like protein | −4.450 | 8.42E‐04 |
| RZR63947.1 | Cycloheximide resistance protein | −4.655 | 3.53E‐05 |
| RZR68445.1 | FAD binding domain protein | −5.118 | 5.70E‐05 |
| RZR60696.1 | Siderophore iron transporter | −5.628 | 3.78E‐03 |
| RZR61719.1 | Ferric‐chelate reductase | −5.658 | 4.11E‐03 |
| RZR66686.1 | Glutamyl‐tRNA(Gln) amidotransferase | −6.049 | 4.79E‐04 |
| RZR63736.1 | Monocarboxylate permease‐like protein | −6.675 | 1.61E‐03 |
| RZR61425.1 | Hypothetical protein I1G_00001446 | −6.766 | 1.26E‐06 |
| RZR66909.1 | Aldo/keto reductase | −9.653 | 9.66E‐11 |
| RZR60251.1 | Cysteine synthase B | −10.592 | 3.46E‐04 |
| RZR60252.1 | MFS drug transporter | −10.824 | 2.28E‐04 |
| RZR69808.1 | Symbiotic chitinase | −21.784 | 1.45E‐11 |
| RZR58608.1 | Hypothetical protein I1G_00008738 | −26.011 | 2.10E‐07 |
Log2 fold change value for upregulated genes >2. Log2 fold change value for downregulated genes < −2.
Table 2.
Differentially expressed genes in PcRKN treatment.
| GenBank accession | Sequence description | Log2 fold change | Adj. P‐value |
|---|---|---|---|
| RZR69242.1 | Floculation protein FLO1 | 7.501 | 5.75E‐11 |
| RZR67544.1 | Isochorismatase family protein | 4.199 | 6.31E‐04 |
| RZR68026.1 | Putative som1 protein | 3.157 | 3.60E‐04 |
| RZR64799.1 | Floculation protein FLO1 | 3.078 | 4.35E‐06 |
| RZR59618.1 | Hypothetical protein I1G_00011582 | 2.634 | 2.30E‐03 |
| RZR64511.1 | CRAL/TRIO domain protein | 2.099 | 1.85E‐03 |
| RZR63940.1 | Ankyrin repeat protein | −2.148 | 1.35E‐03 |
| RZR64281.1 | Zinc finger, C2H2‐like protein | −2.605 | 1.09E‐04 |
| RZR63781.1 | Ribonuclease H‐like protein | −2.729 | 7.32E‐03 |
| RZR64322.1 | Hypothetical protein I1G_00000198 | −2.934 | 1.35E‐03 |
Log2 fold change value for upregulated genes >2. Log2 fold change value for downregulated genes < −2.
Table 3.
Differentially expressed genes in PcQRKN treatment.
| GenBank accession | Sequence description | Log2 fold change | Adj. P‐value |
|---|---|---|---|
| RZR63795.1 | Glycoside hydrolase family 75 | 9.036 | 5.45E‐15 |
| RZR69242.1 | Floculation protein FLO1 | 8.287 | 1.43E‐17 |
| RZR61856.1 | Glucokinase | 8.287 | 5.87E‐18 |
| RZR61845.1 | Glycoside hydrolase family 75 protein | 8.263 | 1.50E‐10 |
| RZR62940.1 | Glycoside hydrolase family 75 | 7.067 | 8.19E‐11 |
| RZR61846.1 | Major facilitator superfamily domain, general substrate transporter | 6.368 | 2.80E‐17 |
| RZR70395.1 | Acid phosphatase | 5.598 | 2.36E‐04 |
| RZR65223.1 | WSC domain‐containing protein | 5.234 | 4.47E‐07 |
| RZR70313.1 | Chitosanase CSN1 | 5.180 | 1.53E‐09 |
| RZR62842.1 | FAD‐dependent monooxygenase | 5.073 | 6.19E‐05 |
| RZR63081.1 | Glycoside hydrolase family 2 protein | 4.696 | 6.21E‐10 |
| RZR68451.1 | Maltose permease | 4.380 | 1.44E‐10 |
| RZR68026.1 | Putative som1 protein | 4.259 | 1.37E‐08 |
| RZR64799.1 | Floculation protein FLO1 | 4.011 | 1.31E‐12 |
| RZR70420.1 | Floculation protein FLO1 | 3.419 | 2.62E‐03 |
| RZR66026.1 | Secreted aspartic proteinase precursor | 3.373 | 4.89E‐04 |
| RZR64948.1 | Fungal chitosanase | 3.260 | 1.31E‐05 |
| RZR61625.1 | Cytochrome P450 ClCP1 | 3.111 | 7.79E‐04 |
| RZR68359.1 | Nucleotide‐binding, alpha‐beta plait | 3.097 | 5.56E‐05 |
| RZR70289.1 | Extracellular soluble lytic transglycosylase | 3.002 | 1.26E‐09 |
| RZR68649.1 | Glycoside hydrolase family 75 | 2.988 | 1.91E‐03 |
| RZR59618.1 | Hypothetical protein I1G_00011582 | 2.929 | 7.41E‐05 |
| RZR70338.1 | Glycoside hydrolase, subgroup, catalytic core | 2.862 | 2.22E‐03 |
| RZR64159.1 | Major facilitator superfamily domain, general substrate transporter | 2.790 | 2.64E‐03 |
| RZR64864.1 | Hydrophobic surface binding protein A domain‐containing protein | 2.686 | 3.46E‐03 |
| RZR59400.1 | Alpha‐l‐rhamnosidase A | 2.672 | 7.69E‐03 |
| RZR68019.1 | Glycoside hydrolase, family 29 | 2.660 | 5.33E‐03 |
| RZR59412.1 | Cell wall protein | 2.629 | 5.88E‐03 |
| RZR65779.1 | Aromatic‐ring hydroxylase‐like protein | 2.565 | 7.38E‐03 |
| RZR66703.1 | Major facilitator superfamily domain, general substrate transporter | 2.552 | 3.44E‐03 |
| RZR70254.1 | Thioredoxin‐like protein | 2.543 | 3.53E‐04 |
| RZR64866.1 | Antigenic cell wall galactomannoprotein | 2.518 | 5.77E‐03 |
| RZR66682.1 | Cell wall protein | 2.456 | 5.88E‐03 |
| RZR67148.1 | Purple acid phosphatase‐lik | 2.455 | 1.21E‐04 |
| RZR59293.1 | General substrate transporter | 2.450 | 5.47E‐03 |
| RZR63431.1 | Peptidase S8/S53, subtilisin/kexin/sedolisin | 2.405 | 7.09E‐03 |
| RZR61740.1 | Extracellular serine‐rich protein | 2.381 | 3.98E‐03 |
| RZR60307.1 | Cytochrome P450 | 2.368 | 5.67E‐04 |
| RZR69862.1 | Cell wall galactomannoprotein | 2.363 | 7.81E‐03 |
| RZR66588.1 | AtmA protein | 2.350 | 3.02E‐07 |
| RZR62045.1 | EF‐hand calcium‐binding domain‐containing protein | 2.198 | 5.47E‐03 |
| RZR62051.1 | Lipase 5 | 2.182 | 7.81E‐03 |
| RZR65709.1 | Exo‐beta‐d‐glucosaminidase | 2.180 | 6.40E‐04 |
| RZR62208.1 | Peptidase A4 family protein | 2.165 | 5.88E‐03 |
| RZR59617.1 | Galactose oxidase | 2.145 | 1.27E‐07 |
| RZR64865.1 | Hydrophobic surface binding protein A domain‐containing protein | 2.141 | 1.01E‐02 |
| RZR65777.1 | Related to glutathione S‐transferase GST‐6.0 | 2.112 | 1.30E‐02 |
| RZR64511.1 | CRAL/TRIO domain protein | 2.096 | 3.05E‐04 |
| RZR62042.1 | Helix–loop–helix DNA‐binding protein | 2.090 | 6.40E‐03 |
| RZR67153.1 | Hypothetical protein I1G_00011003 | 2.068 | 1.51E‐02 |
| RZR62046.1 | Polyketide synthase | 2.054 | 7.69E‐03 |
| RZR61204.1 | Cell wall protein | 2.037 | 1.19E‐02 |
| RZR66605.1 | Beta‐lactamase‐like protein | −2.001 | 8.76E‐03 |
| RZR67311.1 | Phosphoglycerate mutase family protein | −2.028 | 3.07E‐03 |
| RZR67249.1 | Hypothetical protein I1G_00007967 | −2.046 | 1.32E‐03 |
| RZR66003.1 | Potassium channel | −2.047 | 3.80E‐03 |
| RZR65332.1 | Hypothetical protein I1G_00001300 | −2.075 | 1.46E‐03 |
| RZR58491.1 | Hypothetical protein I1G_00003346 | −2.088 | 6.40E‐03 |
| RZR66328.1 | S‐(hydroxymethyl)glutathione dehydrogenase | −2.090 | 7.81E‐03 |
| RZR59616.1 | Oxidoreductase | −2.097 | 8.76E‐03 |
| RZR69857.1 | Protein SERAC1 | −2.104 | 1.23E‐03 |
| RZR61435.1 | Repetitive proline‐rich cell wall protein | −2.130 | 5.56E‐05 |
| RZR61534.1 | Trehalose synthase (Ccg‐9) | −2.148 | 9.96E‐10 |
| RZR68989.1 | Zn(2)‐C6 fungal‐type DNA‐binding domain protein | −2.149 | 8.76E‐03 |
| RZR61511.1 | Reductase | −2.153 | 5.91E‐04 |
| RZR63940.1 | Ankyrin repeat protein | −2.212 | 5.71E‐05 |
| RZR63259.1 | Hypothetical protein I1G_00011648 | −2.240 | 6.80E‐04 |
| RZR69039.1 | DUF1929 multidomain protein | −2.336 | 2.91E‐03 |
| RZR63453.1 | Protein bli‐3 | −2.365 | 5.70E‐03 |
| RZR61033.1 | Glucose repressible protein Grg1 | −2.502 | 1.10E‐06 |
| RZR68164.1 | MARVEL‐like domain protein | −2.566 | 1.73E‐03 |
| RZR66334.1 | Protein kinase domain protein | −2.677 | 2.64E‐03 |
| RZR62135.1 | MFS transporter | −2.845 | 4.60E‐04 |
| RZR59365.1 | SUR7 protein | −2.876 | 2.57E‐04 |
| RZR63781.1 | Ribonuclease H‐like protein | −2.944 | 3.80E‐03 |
| RZR66909.1 | Aldo/keto reductase | −3.348 | 2.91E‐03 |
Chitosan favours oxidation–reduction and associated processes in Pc
We assessed the sets of differentially expressed Pc genes for enrichment in Gene Ontology (GO) terms (Fig. 2). We considered terms from all three GO domains (Biological Processes, BP; Molecular Function, MF; Cellular Component, CC) for upregulated (Fig. 2A) and downregulated genes (Fig. 2B). For upregulated genes, ‘oxidation–reduction’ (GO:0055114) and ‘polysaccharide catabolism’ (GO:0000272) are the most enriched BP in chitosan treatments. The importance of oxidation–reduction and polysaccharide catabolism is also reflected in MF (e.g. ‘oxidoreductase activity acting on paired donors, with incorporation or reduction of molecular oxygen’ (GO:0016705) and ‘monooxygenase activity’ (GO:0004497) for oxidation–reduction processes; and ‘chitosanase activity’ (GO:0004568) for polysaccharide metabolism). Proteolysis (reflected in MF in ‘aspartic‐type endopeptidase’ (GO:0004190) and ‘endopeptidase’ (GO:0004175) activities) is also an enriched GO term in chitosan treatments. Transmembrane transport is a BP enriched in all treatments. ‘Carbohydrate transport’ (GO:0008643) and ‘carbohydrate derivative metabolic process’ (GO:1901135) are also enriched GO terms in chitosan treatments. Taken together, this would indicate a high chitosan turnover by Pc.
Fig 2.
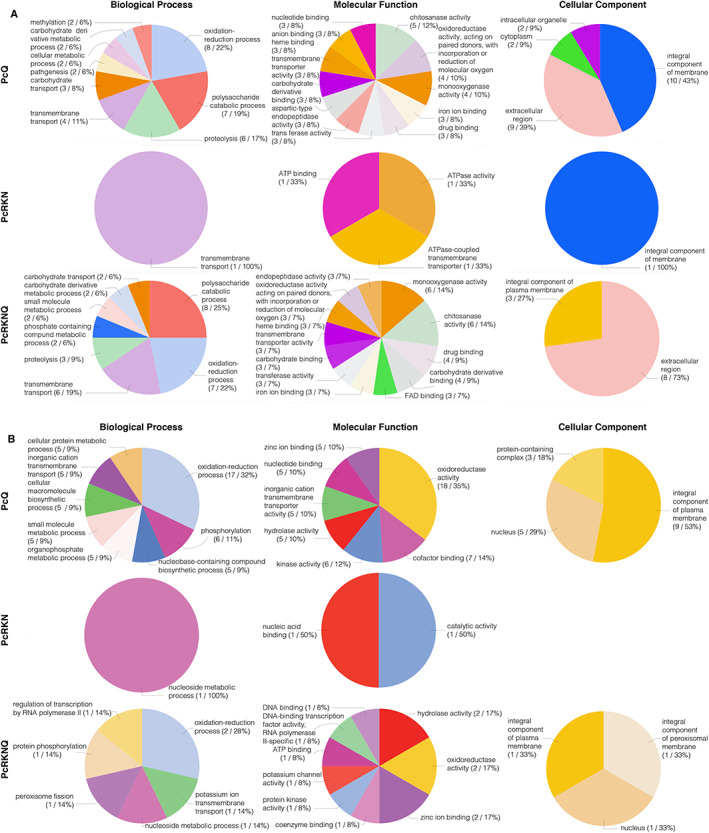
Chitosan favours redox processes in Pc. Gene ontology annotation of differentially expressed Pc genes with nematode eggs and chitosan using a threshold of ±2 in log2 fold change value.
A. Upregulated genes,
B. downregulated genes. Treatments: PcQ, P. chlamydosporia with chitosan (1 mg ml−1); PcRKN, P. chlamydosporia with M. javanica eggs (1 egg μl−1); PcRKNQ, P. chlamydosporia with M. javanica eggs (1 egg μl−1) and chitosan (1 mg ml−1). All treatments were applied for 4 days.
The presence of RKN represses nucleoside metabolic process (e.g. reflected in MF by ‘nucleic acid binding’ (GO:0003676), ‘catalytic activity’ (GO:0003824) and ‘DNA binding’ (GO:0003677)). These GO terms are enriched in both PcRKN and PcRKNQ treatments. ‘Oxidation–reduction process’ (GO:0055114) is the most enriched GO term for downregulated genes in chitosan treatments (reflected in MF by ‘oxidoreductase’ (GO:0016491) and ‘hydrolase’ (GO:0016787) activities). ‘Plasma membrane’ (GO:0005886) is the most enriched CC GO term for downregulated genes associated with chitosan treatments. PcRKN does not display any GO term associated with downregulated genes linked to CC GO domain.
Chitosan and RKN significantly modify the expression of genes associated with energy, lipid and chitosan catabolism and proteolysis
Statistical analyses of GO terms associated with Pc DEG show 113 GOs enriched in all treatments (Supplementary Table 1). These can be classified into nine clusters according to their behaviour with chitosan (Fig. 3). Pc genes included in these significantly enriched GO terms are shown in Supplementary Table 2. Behaviour of the individual GO term is shown in Supplementary Fig. 2. Chitosan promotes the expression of Pc genes associated with 38 GO terms (Fig. 3A) when nematodes are absent. Twenty‐four of them are related to the cell cycle, 12 to protein synthesis and modification and two to sugar metabolism. In the absence of nematodes, chitosan represses Pc genes involved in metal transport and redox metabolism GO terms (Fig. 3B and C; Supplementary Table 1). Nematode eggs reverse this behaviour. Conversely, chitosan induces genes associated with energy GO terms (Fig. 3D). Chitosan also induces the expression of genes associated with lipid metabolism GO terms (specially sphingomyelin (GO:0004767, GO:0006684 and GO:0006685) metabolism; Fig. 3E). Chitosan increases expression of genes in GO terms related to chitin and chitosan degradation (‘chitosanase activity’ (GO:0016977) and ‘exo‐1,4‐beta‐d‐glucosaminidase’ activity (GO:0052761)). This is further enhanced when RKN eggs are present (Fig. 3F). ‘Phosphate pentose shunt’ (GO:0006098) and ‘NADPH regeneration’ (GO:0006740) (Fig. 3G) are not affected by chitosan. Both are repressed by the presence of nematodes. ‘Metallocarboxypeptidase activity’ (GO:0004181) is overexpressed with chitosan in the absence of nematode eggs (Fig. 3H). ‘Structural constituent of cell wall’ (GO:0005199) is induced with chitosan (Fig. 3I).
Fig 3.
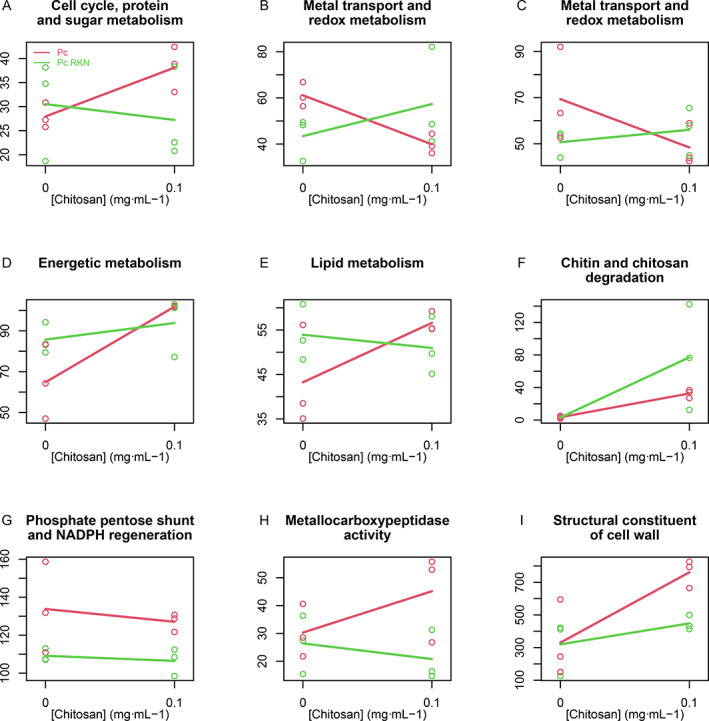
Chitosan and RKN modify the expression of genes associated to 113 GO terms. A cluster of the median profile of 113 GO terms analysis generated nine groups. These GOs are enriched and show a significant behaviour.
A. Cluster related to cell cycle and associated processes.
B. Redox and transport.
C. Redox and transport metabolism.
D. Includes GO terms related to the previous step to energy production.
E. Lipid metabolism.
F. All GO terms related to chitin/chitosan degradation.
G. Phosphate pentose shunt and NADPH regeneration GO terms.
H. This cluster is represented by one GO term: metallocarboxypeptidase activity.
I. Associated with cell wall–associated processes. Legend is shown in Fig. 3A.
Chitosan and RKN significantly modify Pc gene expression dynamics
In response to nematodes and chitosan, 180 Pc genes cluster into nine significant unique gene trends (Fig. 4, Supplementary Table 3). Individual gene behaviour is shown in Supplementary Fig. 3. Chitosan activates the expression of the genes associated with clusters 1–3 (Fig. 4A, B and C). Cluster 1 includes, among others, genes encoding proteins related with reactive oxygen species (ROS) metabolism, such as cytochrome P450 ClCP1 (RZR61625.1) and thioredoxin‐like protein (RZR70254.1). ROS metabolisms play a key role in fungal response to chitosan inducing oxidative metabolism in chitosan‐sensitive fungi such as Neurospora crassa (Lopez‐Moya et al., 2016). In Cluster 2, the presence of nematodes mitigates the increase in gene expression. This can be confirmed in Tables 1 and 3. Genes encoding peptidases (RZR60329.1, RZR61987.1, RZR62018.1, RZR62208.1, RZR62805.1, RZR63158.1, RZR63431.1, RZR66026.1 and RZR69491.1) exhibit this behaviour. RKN and chitosan induce gene expression in Cluster 3. This cluster includes genes encoding chitosanases (RZR61845.1, RZR62940.1, RZR63795.1, RZR64948.1 and RZR70313.1), adhesives (FLO1; RZR64799.1 and RZR69242.1) and sugar catabolism proteins (RZR61856.1, RZR63081.1, RZR65709.1 and RZR66181.1). Differential expression of genes in this cluster may explain why chitosan increases RKN egg parasitism by Pc, since chitosanases and polysaccharide degrading enzymes are involved in RKN egg parasitism (Aranda‐Martinez et al., 2016) and chitosan assimilation. Genes in Clusters 4–6 (Fig. 4D, E and F) are repressed with chitosan when nematodes are absent. When nematodes are present chitosan does not modify gene expression dynamics. Cluster 7 (Fig. 4G) includes genes repressed by chitosan regardless of the presence of nematodes. Some membrane transporters show this trend. Nematode eggs increase expression of genes in Cluster 8 (Fig. 4H), but chitosan represses them. The most significant encodes a salicylate hydroxylase (RZR59707.1). Nematode eggs and chitosan induce the expression of genes in Cluster 9 (Fig. 4I). Genes encoding the following proteins belong to this cluster: RZR65223.1 WSC domain‐containing protein, RZR69865.1 Cytochrome P450, RZR64444.1 GMC oxidoreductase, RZR64079.1 3‐beta hydroxysteroid dehydrogenase/isomerase. These genes would play a crucial role for RKN egg parasitism by Pc with chitosan.
Fig 4.
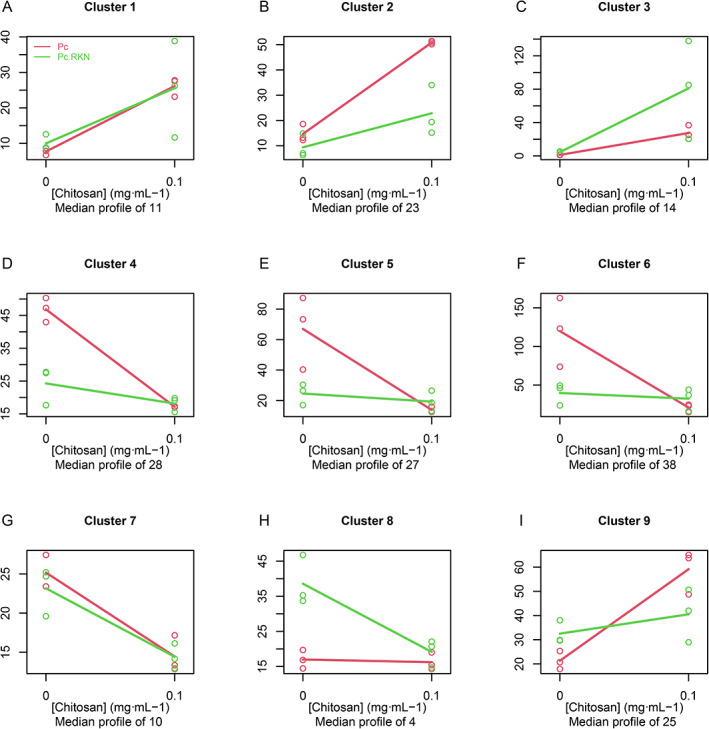
Chitosan and RKN significantly modify the expression of 180 genes. A median profile of 180 genes clustered in nine groups according to their trends with chitosan. Legend is shown in Fig. 4A.
Eight Pc genes are expressed in endophytism, pathogenicity and response to chitosan
To investigate Pc genes shared between endophytism and parasitism, we combined PHI‐base (Urban et al., 2020; www.phi-base.org), Pc upregulated genes found in this work (RKN parasitism and chitosan metabolism) and Pc genes expressed during Barley colonization (Larriba et al., 2014). The intersection between these datasets includes eight genes encoding the following proteins (Fig. 5): RZR66026.1 (secreted aspartic proteinase precursor, overexpressed in PcQ and PcRKNQ), RZR62042.1 (helix–loop–helix DNA‐binding protein, overexpressed in PcRKNQ), RZR63158.1 (peptidase A1, overexpressed in PcQ), RZR64159.1 (major facilitator superfamily domain, general substrate transporter, overexpressed in PcRKNQ), RZR62046.1 (polyketide synthase (PKS), overexpressed in PcRKNQ), RZR61625.1 (cytochrome P450 ClCP1, overexpressed in PcQ and PcRKNQ), RZR69240.1 (APSES transcription factor, overexpressed in PcQ) and RZR61845.1 (glycoside hydrolase family 75 protein (chitosanase) overexpressed in PcQ and PcRKNQ). We have investigated the expression of these genes in banana plants colonized by Pc. Banana plants modify the expression of these Pc genes (Supplementary Fig. 4). Both barley and banana are monocots, and the former has been widely used for endophytism studies (Maciá‐Vicente et al., 2009; Murphy et al., 2014a; Murphy et al., 2014b; Larriba et al., 2015).
Fig 5.
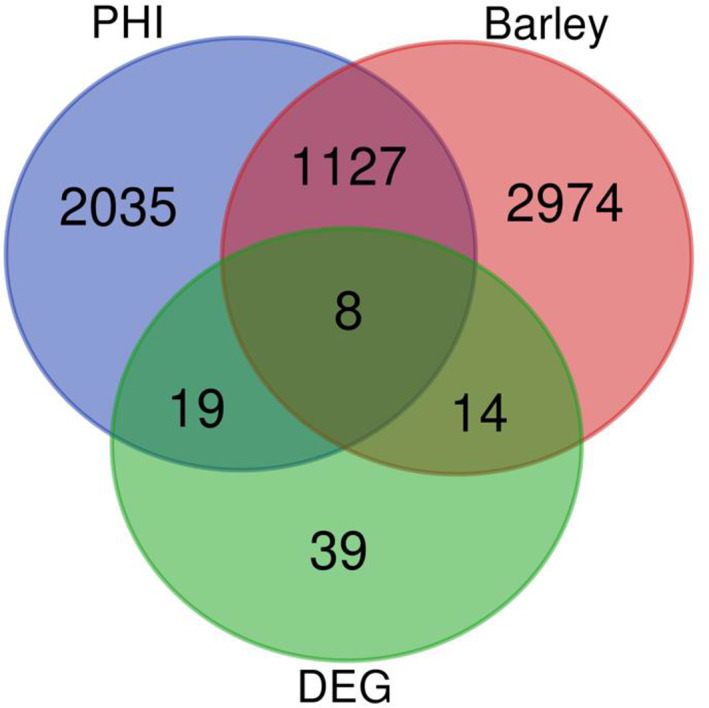
Eight Pc genes are common to endophytism, pathogenicity and chitosan response. Venn diagram showing the intersection of genes expressed when Pc colonizes barley (Larriba et al., 2014), PHI database data and differentially expressed genes (DEG; log2 fold change >2) in this study. Eight genes are found common to all studies.
Pc acidic peptidases modify their expression when banana plants are present. Secreted aspartic proteinase precursor gene is induced, while the A1 peptidase gene is slightly repressed. A chitosanase gene is expressed whenever chitosan is present. The fungus does not modify the expression of this gene when it grows endophytically on bananas, but the maximum expression occurs in the ‘artificial rhizosphere’ (liquid surrounding roots) with chitosan. One Pc PKS protein, involved in the secondary metabolism of the fungus, is slightly induced by the presence of the plant. Secondary metabolism plays an essential role during plant–host interaction (Macheleidt et al., 2016). Endophytism in Pc also modifies the expression of transcription factors. Helix–loop–helix DNA binding protein is overexpressed in all treatments. Cytochrome P450 ClCP1 is related to oxidative metabolism, which is also involved in this process.
Discussion
In this work, we have carried out a transcriptomic study, using RNA‐seq, to determine Pc genes involved in the response to chitosan and RKN parasitism. We have found that chitosan on its own modifies the expression of more Pc genes than RKN does. This may be because chitosan permeabilizes plasma membrane in fungi triggering the activation of reactive oxygen species (ROS) and cell death (Lopez‐Moya et al., 2019). Besides, chitosan solutions reach all fungal cells, whereas nematodes are in contact with only specific parts (mainly appressoria) of the fungus. Chitosan enhances redox processes, proteolysis and carbohydrate metabolism in Pc. Redox metabolism is the most affected process by chitosan. To this respect, the gene encoding CipC1 (concanamycin induced protein C), a ROS‐related protein, is the most induced by chitosan. This protein has also been found induced in citrus fruits infected by the fungal pathogen Penicillium digitatum (López‐Pérez et al., 2015). CipC1 is involved in hyphal branching and upregulated in Laccaria bicolor in response to Pseudomonas fluorescens (Deveau et al., 2007). This gene could be overexpressed in Pc to overcome chitosan‐induced ROS. Other genes overexpressed in chitosan‐induced ROS are Cytochromes P450 and b561, l‐ascorbate oxidase and aldo/keto reductase. This enhanced activation of redox metabolism–related genes connects resistance with chitosan‐sensitive fungi (Jaime et al., 2012; Lopez‐Moya et al., 2016). Pc has to fight plasma membrane oxidation generated by chitosan‐induced ROS. Chitosan permeabilizes the plasma membrane of fungi in an energy‐dependent manner (Palma‐Guerrero et al., 2009). In our work, Pc activates energy and lipid metabolism genes with chitosan. This was previously found on chitosan‐sensitive yeasts and filamentous fungi (Jaime et al., 2012; Lopez‐Moya et al., 2015; Lopez‐Moya et al., 2016). Lipid oxidation generates oxylipins (Gabbs et al., 2015), which can modify fungal morphology, promoting appressoria differentiation (Niu et al., 2020). We then may speculate that Pc oxylipins could be involved in chitosan induction of Pc appressoria found previously (Escudero et al., 2016). ROS increase in the cell could lead to Pc methylation (Wu and Ni, 2015), a highly represented GO term in chitosan treatments. This indicates chitosan may also trigger Pc epigenetic mechanisms (Razin and Cedar, 1991; Phillips, 2008). ROS also causes damage to proteins, probably activating protease induction (Schieber and Chandel, 2014). To this respect, chitosan induces Pc genes encoding all families of peptidases. In our study, chitosan enhances Pc subtilisin S8/S53 expression. This protease may help the fungus to adapt to the ecological niches, facilitating nutrition (Li et al., 2017). Pc subtilisins have been detected in both the parasitic (Escudero et al., 2016) and endophytic (Lopez‐Llorca et al., 2010) phases of the fungal lifestyle. Peptidases A1 and A4 (fungal family G1), involved in plant parasitism and host adaptation (Kirshnan et al., 2018), are also upregulated in Pc with chitosan. Finally, Pc metallo‐endopeptidases are also upregulated with chitosan. This wide range of biological processes and molecular functions affected by chitosan indicates the effort made by the cellular machinery of Pc to overcome stress. Further work should quantify the response of individual Pc cells against chitosan, since antibiotics generate dead, dying and resistant active and inactive cells (Bamford et al., 2017).
Pc has mechanisms to efficiently degrade chitosan and thus reduce its damage (mainly caused by ROS) on cells. Pc overexpresses chitosanases (GH75) with chitosan, degrading it into monosaccharides. One WSC domain‐containing protein, a stress responsory related to carbohydrate binding (Tong et al., 2016; Oide et al., 2019), is also upregulated with chitosan. Genes coding for membrane transporters, such as the major facilitator superfamily domain, general substrate transporter, oligopeptide transporter OPT‐like protein, maltose permease and sugar transporter family protein, are also overexpressed with chitosan. They may be involved in the assimilation of monosaccharides generated upon chitosan degradation by Pc. Neurospora crassa, a chitosan‐sensitive fungus, also shows the activation of monosaccharide transport genes (Lopez‐Moya et al., 2016) upon chitosan treatment but not as many as Pc. This may be a further reason to explain why Pc is more resistant to chitosan than N. crassa. Overexpression of Pc glucokinase in the presence of chitosan is probably related to the final catabolism of this polymer (Maitra and Lobo, 1983). Other glycoside hydrolases (GH2 and 3) are also overexpressed in chitosan treatments. This may indicate the high metabolic potential of Pc to degrade and assimilate chitosan.
RKN eggs stimulate Pc to overexpress rather than repress genes. RKN also induce gene expression of nematode‐trapping fungi, such as Arthrobotrys conoides (Pandit et al., 2017), indicating the activation of parasitic pathways. In this work, the addition of chitosan to nematode eggs displays the highest values of Pc gene expression in all treatments tested. This shows that chitosan is a strong elicitor of genes potentially involved in RKN egg parasitism by Pc. In cluster analyses, genes encoding proteins for adhesion (FLO1), chitosan and sugar degradation (GH2, GH3, GH75), membrane transport (MFS‐transporters) and carbohydrate metabolism (glucokinase) are overexpressed in chitosan and RKN treatments. In a previous study (Lin et al., 2018), RKN eggs in Minimal Medium (MM) were found to induce Pc adhesives (CFEM), GHs enzymes and proteases. In our GO cluster analysis, we have found GO terms related to chitin/chitosan degradation with an enhanced expression upon addition of nematode eggs and chitosan. Genes that share this behaviour may explain why RKN eggs parasitism by Pc increases in the presence of chitosan. The trend pattern (increase or decrease in expression by adding chitosan to treatments) is best observed in a gene rather than in GO cluster analyses. This is because GOs share genes and when some have opposing trends, the average GO behaviour does not show up in the cluster analysis. Pc deploys its machinery to putatively attach to RKN eggshell by binding peptides, lipids and carbohydrates. FLO1 proteins are flocculation proteins present in yeasts, related to adhesion to hyphae (Moreno‐García et al., 2018). FLO1 is a mannose‐binding glycoprotein, which could be a determinant for hyphal adhesion to the nematode eggshell. A CRAL/TRIO domain protein is upregulated when Pc is in contact with RKN. This domain is related to the binding to small lipophilic molecules (Panagabko et al., 2003). It could also be involved to the attachment to the eggshell lipid layer (Johnston and Dennis, 2012). We hypothesize that once the fungus is attached to the eggshell, its degradation begins by transforming the RKN egg chitin layer (Johnston and Dennis, 2012) to chitosan using chitindeacetylases (Aranda‐Martinez et al., 2016) and degrading the resulting chitosan mainly using chitosanases (GH75). Other glycosyl hydrolases (GH2 and GH3) may also contribute to degradation. Sugars may be introduced to the cells of the fungus through transporters (MFS). All molecular processes involved in egg parasitism are enhanced by the addition of chitosan. The increase in ROS could explain this behaviour.
Pc can act both as an endophyte (Bordallo et al., 2002) or RKN egg parasite (Lopez‐Llorca et al., 2002). In this work, we have explored the common ‘gene toolbox’ involved in endophytism (Larriba et al., 2014), pathogenicity (PHI‐base) and response to chitosan. We have found eight candidate genes, among them proteases, chitosanases, redox‐related proteins and transcription factors.
Proteases modify their expression in endophytic processes. This is related to parasitism in other fungi (Druzhinina et al., 2012) and to endophytism in insect pathogenic fungi (Moonjely et al., 2016). In works of gene expression during endophytism, it has been found that Pc overexpresses ribosomal proteins, proteases, secreted proteins and heat shock proteins, among others probably related to transitions in the lifestyle of the fungus (Pentimone et al., 2019). The fact that chitosan increases the secretion of Pc proteases could explain why the fungus colonizes the plant efficiently when chitosan is present. Pc does not modify the expression of GH75 when it grows endophytically on bananas, but the maximum expression of this gene occurs in the rhizosphere with chitosan. Endophytic fungi have enzymes that modify chitin and chitosan (Govinda Rajulu et al., 2011; Venkatachalam et al., 2015). This could mean that the fungus enhances its chitin‐ and chitosanolytic metabolism in order to start plant root colonization. Chitosan and its derivatives are highly related to redox metabolism (Sarangapani et al., 2018; Ivanova and Yaneva, 2020), as we commented before. Cytochrome P450 ClCP1 is also involved in oxidative metabolism (Korzekwa, 2014), and it is expressed in endophytes (Chadha et al., 2018). This is consistent with studies which prove that root colonization processes activate ROS (Segal and Wilson, 2018). One Pc PKS protein is slightly induced by the presence of the plant. PKS are expressed in biocontrol fungus such as Clonostachys rosea during fungal–fungal interactions (Fatema et al., 2018) and virulence events (Tsai et al., 1998). This gene is also related to melatonin biosynthesis (Knapp et al., 2018). Melatonin is a precursor of plant hormones (Arnao and Hernández‐Ruiz, 2018). It has been shown that endophytic organisms secrete melatonin and derivatives during root colonization (Jiao et al., 2016). This could mean that Pc is secreting metabolites homologous to plant hormones in order to facilitate plant root colonization. This could be one of the reasons why Pc increases banana plant growth and development (Mingot‐Ureta et al., 2020). Finally, endophytism in Pc modifies the expression of transcription factors. Helix–loop–helix DNA binding protein is overexpressed in all treatments. Related to previous results, Pc seems to be activating metabolic routes in order to colonize the plant properly hiding from plant defences.
In conclusion, Pc modifies its gene expression to parasite RKN eggs, colonize plant roots or resist chitosan. When Pc is growing with chitosan, it activates metabolic pathways to avoid chitosan‐induced ROS damage. Pc enzymes capable of degrading it into sugars. Pc sugar carriers introduce them into the cell for catabolism. This process increases ROS content, and Pc activates lipid metabolism. Metal membrane transporters putatively carry out these reduction and oxidation reactions. During this process genes related to plant endophytism or nematode egg parasitism are activated, which could indicate that, in combination with the real stimulus, chitosan could be a non‐toxic additive to increase plant colonization by Pc and sustainably reduce plant‐parasitic nematodes, such as RKN, in banana and other agroecosystems.
Experimental procedures
Fungi, chitosan, nematodes and plants
Pochonia chlamydosporia var. chlamydosporia (=Metacordyceps chlamydosporia var. chlamydosporium) isolate 123 (Pc) (ATCC No. MYA‐4875; CECT No. 20929) was used in this study. Pc was obtained from Heterodera avenae–infected eggs (Olivares and López‐Llorca, 2002). Chitosan T8 (70 kDa and 80.5% deacetylation degree) was obtained from Marine Bioproducts GmbH (Bremerhaven, Germany). Chitosan solutions were prepared as described in Palma‐Guerrero et al. (2010). Meloidogyne javanica was a kind gift from Dr. Caridad Ros (IMIDA, Murcia, Spain). It was maintained in tomato susceptible plants (Solanum lycopersicum Mill cv. Marglobe). RKN egg masses were hand‐picked from infected tomato roots and surface‐sterilized with 1% sodium hypochlorite as in McClure et al. (1973). Meloidogyne javanica eggs in all developmental stages were used for experiments. One‐month‐old in vitro banana plantlets (Musa acuminata cv. Dwarf Cavendish) were purchased from Cultesa S.A. (Tacoronte, Canary Islands, Spain).
Pc, chitosan and RKN: RNA‐seq experimental design
Pc conidia (final concentration 106 conidia·ml−1) were inoculated into 100 ml flasks each containing 20 ml Czapek Dox broth medium (Ward et al., 2012). Flasks were incubated at 25°C with shaking at 120 rpm. After 5 days, mycelia were recovered by filtration through Miracloth (Calbiochem) and washed twice with sterile distilled water. Pc mycelia (ca. 0.2 g) were inoculated axenically into 100 ml flasks each containing 20 ml MM (Aranda‐Martinez et al., 2016) and amended with either: (i) chitosan (PcQ) (0.1 mg ml−1 final concentration), (ii) surface‐sterilized RKN eggs (PcRKN) (1 egg μl−1 final concentration), or (iii) both (PcRKNQ). Controls consisted of Pc mycelium growing in MM (Pc). All treatments were carried out in triplicate. Flasks were incubated for 4 days as before. Samples were then filtered through Miracloth, frozen in liquid N2, lyophilized and stored at −80°C until used. The experiment was performed three times.
RNA extraction and RNA‐seq performing
Total RNA extractions were performed using TRIzol reagent (Life Tech), following the manufacturer's protocol. The quality of all RNA samples was determined using a bioanalyzer (Agilent 2100 Bioanalyzer System) to confirm it was adequate for RNA‐seq analysis (Supplementary Table 4). Three replicates per treatment were then selected at random to perform RNA‐seq analysis. cDNA synthesis, library construction and Illumina sequencing were carried out by Macrogen (Seoul, South Korea). TruSeq Stranded mRNA LT Sample Prep Kit (Illumina) was used as Library Kit and TruSeq Stranded mRNA Sample Preparation Guide, Part #15031047 Rev. E as Library Protocol. The reagent used was NovaSeq 6000 S4 Reagent Kit (Illumina) and sequencing protocol NovaSeq 6000 System User Guide Document #1000000019358 v02.
Bioinformatic analyses
Raw reads were trimmed and filtered with Trimmomatic (Bolger et al., 2014) to remove adapters with up to two mismatches that had a palindrome read alignment accuracy of 30 and a sequence match accuracy of 10. Leading and trailing low‐quality or N bases (<3) were removed by using a 4‐base‐wide sliding window. Where average base quality was low (<15) reads were trimmed and any short reads (<80 bp) were removed. The quality of the reads was then checked with FASTQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/).
Reads were quantified using Salmon mapping against the P. chlamydospora 123 genome (GenBank accession: GCA_000411695.2) using a wrapper script (align_and_estimate_abundance.pl) from the Trinity software package (Grabherr et al., 2011). DEG were determined with the Bioconductor package DESeq2 (Love et al., 2014) using the likelihood ratio test. Genes were filtered so only those with at least three samples with >10 counts were analysed. P values were adjusted using the Benjamini–Hochberg (BH) correction (Benjamini and Hochberg, 1995). Genes were considered differentially expressed if they had a log2 fold change of ±2 and a BH‐FDR‐adjusted P‐value of ≤0.05. This process selected genes whose expression varied the most with treatments with respect to the control and characterized the main genes involved in the response of Pc to chitosan and RKN.
A consensus set of transcripts was functionally annotated with GO terms using Blast2GO (http://www.blast2go.com/b2ghome) (Edgar et al., 2002). Protein sequences were annotated using the InterPro (http://www.ebi.ac.uk/interpro) and KEGG databases (http://www.genome.jp/kegg/pathway.html) in OmicsBox (BioBam, Spain; http://www.biobam.com/omicsbox). Significant differential gene expression between treatments was analysed using maSigPro R package (Nueda et al., 2014). And significant differential GOs were obtained with maSigFun, an adaptation of maSigPro for dealing with groups of genes, as in Lopez‐Moya et al. (2016).
Pc‐Chitosan‐Musa : bioassays
Pc (106 conidia·ml−1) was inoculated in Cz liquid medium and incubated for 5 days as described above. Thirty‐six banana plantlets were placed individually in Magenta Boxes™ (Sigma) each containing 50 ml of MM supplemented or without chitosan (final concentration 0.1 mg ml−1). Half of the plants were inoculated with 0.2 g Pc mycelium and half were left uninoculated. Plants were maintained at 24°C, 60% relative humidity and 16:8 h light/darkness photoperiod, with 100 rpm shaking, for 4 days. Control replicates of 0.2 g of mycelium were inoculated in 100 ml flasks each containing 20 ml MM supplemented or not with chitosan (final concentration 0.1 mg ml−1). Controls with and without chitosan were made by triplicate to obtain replicability among the samples. To extract RNA, three plant roots from the same treatment were collected for each extraction. In this way, three replicates were obtained per treatment, each replicate with three whole roots from three different plants. RNA was extracted as described above. Final treatments were: Pc (Pc mycelium in MM), PcQ (Pc mycelium in MM amended with 0.1 mg ml−1 chitosan), PcB (Pc mycelium growing in MM close to banana roots), PcBQ (Pc mycelium growing in MM amended with 0.1 mg ml−1 chitosan close to Banana roots), BPc (banana roots colonized by Pc) and BPcQ (Banana roots colonized by Pc in medium amended with 0.1 mg ml−1 chitosan). To identify potential genes related to endophytism and pathogenicity, a whole‐genome blast search was conducted against the Pathogen–Host Interaction database v. 4.9 (PHI‐base; www.phi-base.org; Urban et al., 2020). These candidate genes were intersected with expressed genes when Pc colonizes Barley (Larriba et al., 2014) and upregulated genes (log2 fold change values ≧2) in all treatments in this work (data available in Supplementary Information File). Selected genes were evaluated by qRT‐PCR in banana treatments.
qRT‐PCR
RNA was treated twice with DNase (Turbo DNA‐free, Ambion) to remove any remnants of DNA in the samples. Then, cDNA was obtained using NZY First Strand cDNA Synthesis Kit (NZYtech). Finally, qRT‐PCRs were performed in a StepOnePlus™ Real‐Time PCR System machine, using SYBR Green with ROX (Roche) and a ΔΔCt methodology. qRT‐PCR analyses included three biological replicates with three technical replicates each. ∆∆Ct method was used to calculate the relative fold gene expression of samples and statistical analyses were performed using ANOVA in GraphPad Prism 7.0 Software (www.graphpad.com). Primers used for qRT‐PCRs are shown in Supplementary Table 5. Pochonia chlamydosporia allantoate permease (RZR69578.1; Rosso et al., 2014), glyceraldehyde‐3‐phosphate dehydrogenase (RZR61537.1; Escudero et al., 2016) and ß‐tubulin (RZR65128.1; Ward et al., 2012) were used as housekeeping genes.
Statistical analyses and figures
Statistical analysis for differential gene expression was determined using DESeq2 (Love et al., 2014). Statistics for gene set enrichment analysis were performed using OmicsBox (BioBam, Spain) and figures generated using GraphPad Prism version 7.00 for Mac, GraphPad Software, La Jolla, California, USA, (www.graphpad.com).
Author Contributions
L.V.L.‐L. conceived the original screening and research plans and supervised original writing; F.L.‐M. supervised the experiments and provided technical assistance; M.S.‐F. performed biological experiments and original writing; C.S. and D.J.S. performed bioinformatic analyses; M.J.N. performed analyses with maSigPro; all authors completed the writing; M.S.‐F. agrees to serve as the author responsible for contact and ensures communication.
Supporting information
Appendix S1. Supporting Information.
Supplementary Fig. 1. Confirmation of data replicability by qRT‐PCR. A, q‐PCR analysis. B, RNA‐seq log2 fold change data.
Supplementary Fig. 2. Individual trends of 113 GO terms included in clusters in Fig. 3.
Supplementary Fig. 3. Individual trends of 180 genes included in clusters in Fig. 4.
Supplementary Fig. 4. Pc gene expression of 8 selected genes when the fungus colonizes banana roots. Treatments: Pc (Pc mycelium in MM), PcQ (Pc mycelium in MM amended with 0.1 mg·mL−1 chitosan), PcB (Pc mycelium growing in MM close to banana roots), PcBQ (Pc mycelium growing in MM amended with 0.1 mg·mL−1 chitosan close to banana roots), BPc (banana roots colonized by Pc) and BPcQ (banana roots colonized by Pc in medium amended with 0.1 mg·mL−1 chitosan).
Supplementary Fig. 5. Workflow of the steps followed for the analysis and obtaining log2 fold change values from the raw data.
Supplementary Table 1. Statistical analyses of 113 GO enriched Terms from Fig. 3.
Supplementary Table 2. 113 GO enriched Terms from Fig. 3: ID, Description and genes included.
Supplementary Table 3. Classification and statistics of the 180 genes represented in clusters in Fig. 4.
Supplementary Table 4. Quality of RNA extracted. All samples were sent to Macrogen to perform RNA‐seq analyses.
Supplementary Table 5. Primers used in all gene expression analyses.
Acknowledgements
This work was supported by H2020 MUSA 727624 European Project. The authors would like to thank components of the Laboratory of Plant Pathology at the University of Alicante for their technical support.
References
- Aranda‐Martinez, A. , Lenfant, N. , Escudero, N. , Zavala‐Gonzalez, E.A. , Henrissat, B. , and Lopez‐Llorca, L.V. (2016) CAZyme content of Pochonia chlamydosporia reflects that chitin and chitosan modification are involved in nematode parasitism: CAZome of Pochonia chlamydosporia . Environ Microbiol 18: 4200–4215. [DOI] [PubMed] [Google Scholar]
- Arnao, M.B. , and Hernández‐Ruiz, J. (2018) Melatonin and its relationship to plant hormones. Ann Bot 121: 195–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balestrini, R. , Rosso, L.C. , Veronico, P. , Melillo, M.T. , De Luca, F. , Fanelli, E. , et al. (2019) Transcriptomic responses to water deficit and nematode infection in mycorrhizal tomato roots. Front Microbiol 10: 1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bamford, R.A. , Smith, A. , Metz, J. , Glover, G. , Titball, R.W. , and Pagliara, S. (2017) Investigating the physiology of viable but non‐culturable bacteria by microfluidics and time‐lapse microscopy. BMC Biol 15: 121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benhamou, N. , and Thériault, G. (1992) Treatment with chitosan enhances resistance of tomato plants to the crown and root rot pathogen Fusarium oxysporum f. sp. radicis‐lycopersici. Physiol Mol Plant Path 41: 33–52. [Google Scholar]
- Benjamini, Y. , and Hochberg, Y. (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B Stat Methodol 57: 289–300. [Google Scholar]
- Bolger, A.M. , Lohse, M. , and Usadel, B. (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordallo, J.J. , Lopez‐Llorca, L.V. , Jansson, H.‐B. , Salinas, J. , Persmark, L. , and Asensio, L. (2002) Colonization of plant roots by egg‐parasitic and nematode‐trapping fungi. New Phytol 154: 491–499. [DOI] [PubMed] [Google Scholar]
- Chadha, S. , Mehetre, S.T. , Bansal, R. , Kuo, A. , Aerts, A. , Grigoriev, I.V. , et al. (2018) Genome‐wide analysis of cytochrome P450s of Trichoderma spp.: annotation and evolutionary relationships. Fungal Biol Biotechnol 5: 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deveau, A. , Palin, B. , Delaruelle, C. , Peter, M. , Kohler, A. , Pierrat, J.C. , et al. (2007) The mycorrhiza helper Pseudomonas fluorescens BBc6R8 has a specific priming effect on the growth, morphology and gene expression of the ectomycorrhizal fungus Laccaria bicolor S238N. New Phytol 175: 743–755. [DOI] [PubMed] [Google Scholar]
- Druzhinina, I.S. , Shelest, E. , and Kubicek, C.P. (2012) Novel traits of Trichoderma predicted through the analysis of its secretome. FEMS Microbiol Lett 337: 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar, R. , Domrachev, M. , and Lash, A.E. (2002) Gene expression omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res 30: 207–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escudero, N. , Ferreira, S.R. , Lopez‐Moya, F. , Naranjo‐Ortiz, M.A. , Marin‐Ortiz, A.I. , Thornton, C.R. , and Lopez‐Llorca, L.V. (2016) Chitosan enhances parasitism of Meloidogyne javanica eggs by the nematophagous fungus Pochonia chlamydosporia . Fungal Biol 120: 572–585. [DOI] [PubMed] [Google Scholar]
- Escudero, N. , Lopez‐Moya, F. , Ghahremani, Z. , Zavala‐Gonzalez, E.A. , Alaguero‐Cordovilla, A. , Ros‐Ibañez, C. , et al. (2017) Chitosan increases tomato root colonization by Pochonia chlamydosporia and their combination reduces root‐knot nematode damage. Front Plant Sci 8: 1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatema, U. , Broberg, A. , Jensen, D.F. , Karlsson, M. , and Dubey, M. (2018) Functional analysis of polyketide synthase genes in the biocontrol fungus Clonostachys rosea . Sci Rep 8: 15009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fesel, P.H. , and Zuccaro, A. (2016) Dissecting endophytic lifestyle along the parasitism/mutualism continuum in Arabidopsis . Curr Opin Microbiol 32: 103–112. [DOI] [PubMed] [Google Scholar]
- Flores‐Camacho, R. , Atkins, S.D. , Manzanilla‐López, R.H. , Prado‐Vera, I.C. , and Martínez‐Garza, A. (2008) Caracterización de Aislamientos Mexicanos de Pochonia chlamydosporia var. chlamydosporia (Goddard) Gams y Zare para el Control Biológico de Nacobbus aberrans (Thorne) Thorne y Allen. Rev Mex Fitopatol 26: 93–104. [Google Scholar]
- Forghani, F. , and Hajihassani, A. (2020) Recent advances in the development of environmentally benign treatments to control root‐knot nematodes. Front Plant Sci 11: 1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabbs, M. , Leng, S. , Devassy, J.G. , Monirujjaman, M. , and Aukema, H.M. (2015) Advances in our understanding of oxylipins derived from dietary PUFAs. Adv Nutr 6: 513–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govinda Rajulu, M.B. , Thirunavukkarasu, N. , Suryanarayanan, T.S. , Ravishankar, J.P. , El Gueddari, N.E. , and Moerschbacher, B.M. (2011) Chitinolytic enzymes from endophytic fungi. Fungal Divers 47: 43–53. [Google Scholar]
- Grabherr, M.G. , Haas, B.J. , Yassour, M. , Levin, J.Z. , Thompson, D.A. , Amit, I. , et al. (2011) Full‐length transcriptome assembly from RNA‐Seq data without a reference genome. Nat Biotechnol 29: 644–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanova, D.G. , and Yaneva, Z.L. (2020) Antioxidant properties and redox‐modulating activity of chitosan and its derivatives: biomaterials with application in cancer therapy. Biores Open Access 9: 64–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaime, M.D.L.A. , Lopez‐Llorca, L.V. , Conesa, A. , Lee, A.Y. , Proctor, M. , Heisler, L.E. , et al. (2012) Identification of yeast genes that confer resistance to chitosan oligosaccharide (COS) using chemogenomics. BMC Genomics 13: 267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao, J. , Ma, Y. , Chen, S. , Liu, C. , Song, Y. , Qin, Y. , et al. (2016) Melatonin‐producing endophytic bacteria from grapevine roots promote the abiotic stress‐induced production of endogenous melatonin in their hosts. Front Plant Sci 7: 1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston, W.L. , and Dennis, J.W. (2012) The eggshell in the C. elegans oocyte‐to‐embryo transition. Genesis 50: 333–349. [DOI] [PubMed] [Google Scholar]
- Kaczmarek, M.B. , Struszczyk‐Swita, K. , Li, X. , Szczęsna‐Antczak, M. , and Daroch, M. (2019) Enzymatic modifications of chitin, chitosan, and Chitooligosaccharides. Front Bioeng Biotechnol 7: 243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur, S. , and Dhillon, G.S. (2014) The versatile biopolymer chitosan: potential sources, evaluation of extraction methods and applications. Crit Rev Microbiol 40: 155–175. [DOI] [PubMed] [Google Scholar]
- Knapp, D.G. , Németh, J.B. , Barry, K. , Hainaut, M. , Henrissat, B. , Johnson, J. , et al. (2018) Comparative genomics provides insights into the lifestyle and reveals functional heterogeneity of dark septate endophytic fungi. Sci Rep 8: 6321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korzekwa, K. (2014) Enzyme kinetics of oxidative metabolism: cytochromes P450. Methods Mol Biol 1113: 149–166. [DOI] [PubMed] [Google Scholar]
- Krishnan, P. , Ma, X. , McDonald, B.A. , and Brunner, P.C. (2018) Widespread signatures of selection for secreted peptidases in a fungal plant pathogen. BMC Evol Biol 18: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafontaine, P.J. , and Benhamou, N. (1996) Chitosan treatment: an emerging strategy for enhancing resistance of greenhouse tomato plants to infection by Fusarium oxysporum f.sp. radicis‐lycopersici. Biocontrol Sci Technol 6: 111–124. [Google Scholar]
- Larriba, E. , Jaime, M.D.L.A. , Carbonell‐Caballero, J. , Conesa, A. , Dopazo, J. , Nislow, C. , et al. (2014) Sequencing and functional analysis of the genome of a nematode egg‐parasitic fungus, Pochonia chlamydosporia . Fungal Genetics Biol 65: 69–80. [DOI] [PubMed] [Google Scholar]
- Larriba, E. , Jaime, M.D.L.A. , Nislow, C. , Martín‐Nieto, J. , and Lopez‐Llorca, L.V. (2015) Endophytic colonization of barley (Hordeum vulgare) roots by the nematophagous fungus Pochonia chlamydosporia reveals plant growth promotion and a general defense and stress transcriptomic response. J Plant Res 128: 665–678. [DOI] [PubMed] [Google Scholar]
- Li, J. , Gu, F. , Wu, R. , Yang, J. , and Zhang, K.‐Q. (2017) Phylogenomic evolutionary surveys of subtilase superfamily genes in fungi. Sci Rep 7: 45456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, R. , Qin, F. , Shen, B. , Shi, Q. , Liu, C. , Zhang, X. , et al. (2018) Genome and secretome analysis of Pochonia chlamydosporia provide new insight into egg‐parasitic mechanisms. Sci Rep 8: 1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez‐Llorca, L.V. , Gómez‐Vidal, S. , Monfort, E. , Larriba, E. , Casado‐Vela, J. , Elortza, F. , et al. (2010) Expression of serine proteases in egg‐parasitic nematophagous fungi during barley root colonization. Fungal Genet Biol 47: 342–351. [DOI] [PubMed] [Google Scholar]
- Lopez‐Llorca, L.V. , Olivares‐Bernabeu, C. , Salinas, J. , Jansson, H.‐B. , and Kolattukudy, P.E. (2002) Pre‐penetration events in fungal parasitism of nematode eggs. Mycol Res 106: 499–506. [Google Scholar]
- Lopez‐Moya, F. , Colom‐Valiente, M.F. , Martinez‐Peinado, P. , Martinez‐Lopez, J.E. , Puelles, E. , Sempere‐Ortells, J.M. , and Lopez‐Llorca, L.V. (2015) Carbon and nitrogen limitation increase chitosan antifungal activity in Neurospora crassa and fungal human pathogens. Fungal Biol 119: 154–169. [DOI] [PubMed] [Google Scholar]
- Lopez‐Moya, F. , Kowbel, D. , Nueda, M.J. , Palma‐Guerrero, J. , Glass, N.L. , and Lopez‐Llorca, L.V. (2016) Neurospora crassa transcriptomics reveals oxidative stress and plasma membrane homeostasis biology genes as key targets in response to chitosan. Mol Biosyst 12: 391–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez‐Moya, F. , Suarez‐Fernandez, M. , and Lopez‐Llorca, L.V. (2019) Molecular mechanisms of chitosan interactions with fungi and plants. Int J Mol Sci 20: 332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López‐Pérez, M. , Ballester, A.‐R. , and González‐Candelas, L. (2015) Identification and functional analysis of Penicillium digitatum genes putatively involved in virulence towards citrus fruit. Mol Plant Pathol 16: 262–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love, M.I. , Huber, W. , and Anders, S. (2014) Moderated estimation of fold change and dispersion for RNA‐seq data with DESeq2. Genome Biol 15: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macheleidt, J. , Mattern, D.J. , Fischer, J. , Netzker, T. , Weber, J. , Schroeckh, V. , et al. (2016) Regulation and role of fungal secondary metabolites. Annu Rev Genet 50: 371–392. [DOI] [PubMed] [Google Scholar]
- Maciá‐Vicente, J.G. , Rosso, L.C. , Ciancio, A. , Jansson, H.‐B. , and Lopez‐Llorca, L.V. (2009) Colonisation of barley roots by endophytic Fusarium equiseti and Pochonia chlamydosporia: effects on plant growth and disease. Ann Appl Biol 155: 391–401. [Google Scholar]
- Maitra, P.K. , and Lobo, Z. (1983) Genetics of yeast glucokinase. Genetics 105: 501–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mankau, R. (1980) Biological control of nematode pests by natural enemies. Annu Rev Phytopathol 18: 415–440. [Google Scholar]
- Manzanilla‐López, R.H. , Esteves, I. , Powers, S.J. , and Kerry, B.R. (2011) Effects of crop plants on abundance of Pochonia chlamydosporia and other fungal parasites of root‐knot and potato cyst nematodes. Ann Appl Biol 159: 118–129. [Google Scholar]
- McClure, M.A. , Kruk, T.H. , and Misaghi, I. (1973) A method for obtaining quantities of clean Meloidogyne eggs. J Nematol 5: 230. [PMC free article] [PubMed] [Google Scholar]
- Mingot‐Ureta, C. , Lopez‐Moya, F. , and Lopez‐Llorca, L.V. (2020) Isolates of the nematophagous fungus Pochonia chlamydosporia are endophytic in banana roots and promote plant growth. Agronomy 10: 1299. [Google Scholar]
- Moonjely, S. , Barelli, L. , and Bidochka, M.J. (2016) Insect pathogenic fungi as endophytes. Adv Genet 94: 107–135. [DOI] [PubMed] [Google Scholar]
- Moreno‐García, J. , Martín‐García, F.J. , Ogawa, M. , García‐Martínez, T. , Moreno, J. , Mauricio, J.C. , and Bisson, L.F. (2018) FLO1, FLO5 and FLO11 flocculation gene expression impacts Saccharomyces cerevisiae attachment to Penicillium chrysogenum in a co‐immobilization technique. Front Microbiol 9: 2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy, B.R. , Doohan, F.M. , and Hodkinson, T.R. (2014a) Fungal endophytes of barley roots. J Agric Sci 152: 602–615. [Google Scholar]
- Murphy, B.R. , Doohan, F.M. , and Hodkinson, T.R. (2014b) Yield increase induced by the fungal root endophyte Piriformospora indica in barley grown at low temperature is nutrient limited. Symbiosis 62: 29–39. [Google Scholar]
- Mwaheb, M.A.M.A. , Hussain, M. , Tian, J. , Zhang, X. , Hamid, M.I. , El‐Kassim, N.A. , et al. (2017) Synergetic suppression of soybean cyst nematodes by chitosan and Hirsutella minnesotensis via the assembly of the soybean rhizosphere microbial communities. Biol Control 115: 85–94. [Google Scholar]
- Niu, M. , Steffan, B.N. , Fischer, G.J. , Venkatesh, N. , Raffa, N.L. , Wettstein, M.A. , et al. (2020) Fungal oxylipins direct programmed developmental switches in filamentous fungi. Nat Commun 11: 5158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nueda, M.J. , Tarazona, S. , and Conesa, A. (2014) Next maSigPro: updating maSigPro bioconductor package for RNA‐seq time series. Bioinformatics 30: 2598–2602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oide, S. , Tanaka, Y. , Watanabe, A. , and Inui, M. (2019) Carbohydrate‐binding property of a cell wall integrity and stress response component (WSC) domain of an alcohol oxidase from the rice blast pathogen Pyricularia oryzae . Enzyme Microb Technol 125: 13–20. [DOI] [PubMed] [Google Scholar]
- Olivares, C.M. , and López‐Llorca, L.V. (2002) Fungal egg‐parasites of plant‐parasitic nematodes from Spanish soils. Rev Iberoam Micol 19: 104–110. [PubMed] [Google Scholar]
- Palma‐Guerrero, J. , Gómez‐Vidal, S. , Tikhonov, V.E. , Salinas, J. , Jansson, H.‐B. , and Lopez‐Llorca, L.V. (2010) Comparative analysis of extracellular proteins from Pochonia chlamydosporia grown with chitosan or chitin as main carbon and nitrogen sources. Enzyme Microb Technol 46: 568–574. [Google Scholar]
- Palma‐Guerrero, J. , Huang, I.‐C. , Jansson, H.‐B. , Salinas, J. , Lopez‐Llorca, L.V. , and Read, N.D. (2009) Chitosan permeabilizes the plasma membrane and kills cells of Neurospora crassa in an energy dependent manner. Fungal Genet Biol 46: 585–594. [DOI] [PubMed] [Google Scholar]
- Palma‐Guerrero, J. , Jansson, H.‐B. , Salinas, J. , and Lopez‐Llorca, L.V. (2007) Effect of chitosan on hyphal growth and spore germination of plant pathogenic and biocontrol fungi. J Appl Microbiol 104: 541–553. [DOI] [PubMed] [Google Scholar]
- Panagabko, C. , Morley, S. , Hernandez, M. , Cassolato, P. , Gordon, H. , Parsons, R. , et al. (2003) Ligand specificity in the CRAL‐TRIO protein family. Biochemistry 42: 6467–6474. [DOI] [PubMed] [Google Scholar]
- Pandit, R. , Patel, R. , Patel, N. , Bhatt, V. , Joshi, C. , Singh, P.K. , and Kunjadia, A. (2017) RNA‐Seq reveals the molecular mechanism of trapping and killing of root‐knot nematodes by nematode‐trapping fungi. World J Microbiol Biotechnol 33: 65. [DOI] [PubMed] [Google Scholar]
- Pentimone, I. , Colagiero, M. , Ferrara, M. , Nigro, F. , Rosso, L.C. , and Ciancio, A. (2019) Time‐dependent effects of Pochonia chlamydosporia endophytism on gene expression profiles of colonized tomato roots. Appl Microbiol Biotechnol 103: 8511–8527. [DOI] [PubMed] [Google Scholar]
- Phillips, T. (2008) The role of methylation in gene expression. Nat Educ 1: 116. [Google Scholar]
- Ralmi, N.H.A.A. , and Khandaker, M.M. (2016) Occurrence and control of root knot nematode in crops: a review. Aust J Crop Sci 10: 1649–1654. [Google Scholar]
- Razin, A. , and Cedar, H. (1991) DNA methylation and gene expression. Microbiol Rev 55: 451–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosso, L.C. , Colagiero, M. , Salatino, N. , and Ciancio, A. (2014) Observations on the effect of trophic conditions on Pochonia chlamydosporia gene expression. Ann Appl Biol 164: 232–243. [Google Scholar]
- Sarangapani, S. , Patil, A. , Ngeow, Y.K. , Elsa Mohan, R. , Asundi, A. , and Lang, M.J. (2018) Chitosan nanoparticles' functionality as redox active drugs through cytotoxicity, radical scavenging and cellular behaviour. Integr Biol (Camb) 10: 313–324. [DOI] [PubMed] [Google Scholar]
- Schieber, M. , and Chandel, N.S. (2014) ROS function in redox signaling and oxidative stress. Curr Biol 24: R453–R462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal, L.M. , and Wilson, R.A. (2018) Reactive oxygen species metabolism and plant‐fungal interactions. Fungal Gen Biol 110: 1–9. [DOI] [PubMed] [Google Scholar]
- Shih, P.‐Y. , Liao, Y.‐T. , Tseng, Y.‐K. , Deng, F.‐S. , and Lin, C.‐H. (2019) A potential antifungal effect of chitosan against Candida albicans is mediated via the inhibition of SAGA complex component expression and the subsequent alteration of cell surface integrity. Front Microbiol 10: 602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suarez‐Fernandez, M. , Marhuenda‐Egea, F.C. , Lopez‐Moya, F. , Arnao, M.B. , Cabrera‐Escribano, F. , Nueda, M.J. , et al. (2020) Chitosan induces plant hormones and defenses in tomato root exudates. Front Plant Sci 11: 572087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong, S.‐M. , Chen, Y. , Zhu, J. , Ying, S.‐H. , and Feng, M.‐G. (2016) Subcellular localization of five singular WSC domain‐containing proteins and their roles in Beauveria bassiana responses to stress cues and metal ions. Environ Microbiol Rep 8: 295–304. [DOI] [PubMed] [Google Scholar]
- Tsai, H.F. , Chang, Y.C. , Washburn, R.G. , Wheeler, M.H. , and Kwon‐Chung, K.J. (1998) The developmentally regulated alb1 gene of Aspergillus fumigatus: its role in modulation of conidial morphology and virulence. J Bacteriol 180: 3031–3038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urban, M. , Cuzick, A. , Seager, J. , Wood, V. , Rutherford, K. , Venkatesh, S.Y. , et al. (2020) PHI‐base: the pathogen‐host interactions database. Nucleic Acids Res 48: D613–D620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatachalam, A. , Govinda Rajulu, M. , Thirunavukkarasu, N. , and Suryanarayanan, T. (2015) Endophytic fungi of marine algae and seagrasses: a novel source of chitin modifying enzymes. Mycosphere 6: 345–355. [Google Scholar]
- Ward, E. , Kerry, B.R. , Manzanilla‐López, R.H. , Mutua, G. , Devonshire, J. , Kimenju, J. , and Hirsch, P.R. (2012) The Pochonia chlamydosporia serine protease gene vcp1 is subject to regulation by carbon, nitrogen and pH: implications for nematode biocontrol. PLoS One 7: e35657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willcox, J. , and Tribe, H.T. (1974) Fungal parasitism in cysts of Heterodera . Trans British Mycol Soc 62: 585–IN3. [Google Scholar]
- Wu, Q. , and Ni, X. (2015) ROS‐mediated DNA methylation pattern alterations in carcinogenesis. Curr Drug Targets 16: 13–19. [DOI] [PubMed] [Google Scholar]
- Yin, H. , Du, Y. , and Dong, Z. (2016) Chitin oligosaccharide and chitosan oligosaccharide: two similar but different plant elicitors. Front Plant Sci 7: 522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zavala‐Gonzalez, E.A. , Rodríguez‐Cazorla, E. , Escudero, N. , Aranda‐Martinez, A. , Martínez‐Laborda, A. , Ramírez‐Lepe, M. , et al. (2017) Arabidopsis thaliana root colonization by the nematophagous fungus Pochonia chlamydosporia is modulated by jasmonate signaling and leads to accelerated flowering and improved yield. New Phytol 213: 351–364. [DOI] [PubMed] [Google Scholar]
- Zhang, W. , Zhang, X. , Li, K. , Wang, C. , Cai, L. , Zhuang, W. , et al. (2018) Introgression and gene family contraction drive the evolution of lifestyle and host shifts of hypocrealean fungi. Mycology 9: 176–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Z. , Zhao, P. , Zhang, P. , Su, L. , Jia, H. , Wei, X. , et al. (2020) Integrative transcriptomics and metabolomics data exploring the effect of chitosan on postharvest grape resistance to Botrytis cinerea . Postharvest Biol Technol 167: 111248. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix S1. Supporting Information.
Supplementary Fig. 1. Confirmation of data replicability by qRT‐PCR. A, q‐PCR analysis. B, RNA‐seq log2 fold change data.
Supplementary Fig. 2. Individual trends of 113 GO terms included in clusters in Fig. 3.
Supplementary Fig. 3. Individual trends of 180 genes included in clusters in Fig. 4.
Supplementary Fig. 4. Pc gene expression of 8 selected genes when the fungus colonizes banana roots. Treatments: Pc (Pc mycelium in MM), PcQ (Pc mycelium in MM amended with 0.1 mg·mL−1 chitosan), PcB (Pc mycelium growing in MM close to banana roots), PcBQ (Pc mycelium growing in MM amended with 0.1 mg·mL−1 chitosan close to banana roots), BPc (banana roots colonized by Pc) and BPcQ (banana roots colonized by Pc in medium amended with 0.1 mg·mL−1 chitosan).
Supplementary Fig. 5. Workflow of the steps followed for the analysis and obtaining log2 fold change values from the raw data.
Supplementary Table 1. Statistical analyses of 113 GO enriched Terms from Fig. 3.
Supplementary Table 2. 113 GO enriched Terms from Fig. 3: ID, Description and genes included.
Supplementary Table 3. Classification and statistics of the 180 genes represented in clusters in Fig. 4.
Supplementary Table 4. Quality of RNA extracted. All samples were sent to Macrogen to perform RNA‐seq analyses.
Supplementary Table 5. Primers used in all gene expression analyses.


