Influenza A viruses contain sialic acid (SIA) receptor‐binding hemagglutinin (HA) and receptor‐destroying neuraminidase (NA). The balance between HA and NA is adjusted to the SIA receptor repertoire of the host species. Besides a conserved catalytic site, NA carries a 2nd SIA‐binding site (2SBS). The 2SBS enhances the activity of the catalytic site and affects the HA‐NA balance. Conservation or loss of the 2SBS is associated with (changes in) host tropism.
Keywords: hemagglutinin, host range, influenza A virus, neuraminidase, second sialic acid‐binding site, sialic acid
Abstract
Influenza A viruses (IAVs) are a major cause of human respiratory tract infections and cause significant disease and mortality. Human IAVs originate from animal viruses that breached the host species barrier. IAV particles contain sialoglycan receptor‐binding hemagglutinin (HA) and receptor‐destroying neuraminidase (NA) in their envelope. When IAV crosses the species barrier, the functional balance between HA and NA needs to be adjusted to the sialoglycan repertoire of the novel host species. Relatively little is known about the role of NA in host adaptation in contrast to the extensively studied HA. NA prevents virion aggregation and facilitates release of (newly assembled) virions from cell surfaces and from decoy receptors abundantly present in mucus and cell glycocalyx. In addition to a highly conserved catalytic site, NA carries a second sialic acid‐binding site (2SBS). The 2SBS preferentially binds α2,3‐linked sialic acids and enhances activity of the neighboring catalytic site by bringing/keeping multivalent substrates in close contact with this site. In this way, the 2SBS contributes to the HA‐NA balance of virus particles and affects virus replication. The 2SBS is highly conserved in all NA subtypes of avian IAVs, with some notable exceptions associated with changes in the receptor‐binding specificity of HA and host tropism. Conservation of the 2SBS is invariably lost in human (pandemic) viruses and in several other viruses adapted to mammalian host species. Preservation or loss of the 2SBS is likely to be an important factor of the viral host range.
Abbreviations
- 2SBS
second sialic acid‐binding site
- BLI
biolayer interferometry
- H1–16
hemagglutinin subtypes 1–16
- H1N1pdm09
pandemic influenza A virus H1N1 from 2009
- HA
hemagglutinin
- IAV
influenza A virus
- N1–9
neuraminidase subtypes 1–9
- NA
neuraminidase
- Neu5Ac
N‐acetylneuraminic acid
- Neu5Gc
N‐glycolylneuraminic acid
- SIA
sialic acid
- STD‐NMR
saturation‐transfer difference nuclear magnetic resonance
Introduction
Sialoglycans are omnipresent at the surface of every cell type and therefore appear attractive attachment/entry receptors for viruses. These glycans contain terminal sialic acids (SIAs), derivatives of the nine‐carbon monosaccharide neuraminic acid (reviewed by Ref. [1]). Members of several virus families initiate cell infection by binding to cell surface sialoglycans, such as influenza viruses, coronaviruses, picornaviruses, and paramyxoviruses [2, 3]. Binding to sialoglycans comes at a cost, however, as the sheer abundance of these glycans makes it inherently difficult for viruses to bind their bona fide receptors as required for cell entry, and on the other hand to be released from cells at the end of the infection cycle. To solve this problem, some enveloped viruses not only contain glycan receptor‐binding properties, but also receptor‐destroying activity, carried in either a separate glycoprotein or an enzymatic domain genetically fused to the glycan‐binding protein. These include neuraminidase (NA) proteins of influenza A and influenza B viruses (IAV and IBVs), hemagglutinin‐esterase fusion proteins of influenza C and influenza D viruses, hemagglutinin–esterase proteins of some coronaviruses (embecoviruses), and hemagglutinin–neuraminidase proteins of some paramyxoviruses.
Cleavage of sialoglycans has originally been recognized as being required for release of newly assembled virions and to prevent virion aggregation [4], but it is increasingly being appreciated that it also enables virion mobility through mucus layers containing heavily sialylated mucin decoy receptors (reviewed by Ref. [5]). A functional balance between low‐affinity binding of individual attachment proteins to sialoglycans, in combination with glycan‐destroying activity, is thought to enable these virions to move through environments with high concentrations of sialoglycans in search for their cell surface entry receptors.
Influenza A viruses are zoonotic pathogens that infect a broad range of host species, including humans, wild birds, poultry, pigs, horses and marine mammals [6, 7]. Aquatic birds are the natural host reservoir, from which IAVs can jump to other host species. In humans, IAVs cause seasonal epidemics resulting in major public health problems and a huge economic burden [8]. Occasional pandemics of influenza are caused by animal IAVs that managed to cross the host species barrier and adapt to humans [9]. In little more than 100 years, four of such pandemics have been recorded: H1N1 Spanish pandemic in 1918, H2N2 Asian pandemic in 1957, H3N2 Hong Kong pandemic in 1968, and H1N1 pandemic in 2009 (H1N1pdm09) [9, 10]. The H1N1 Spanish pandemic, which stands as the single most fatal event in human history, killed an estimated 50 million or more people. The other pandemics were less severe, resulting in much lower casualties. In postpandemic years, the pandemic viruses establish themselves in the human population and cause seasonal influenza, thereby usually replacing previous human IAV subtypes.
Influenza A virus particles contain two major surface glycoproteins: hemagglutinin (HA) and NA. HA, of which 16 subtypes exist in aquatic birds (H1–16), is responsible for virus binding to SIA receptors on the cell surface and mediates virus–cell membrane fusion after endocytosis (for recent reviews, see Ref. [11, 12]). The receptor‐destroying enzyme NA, of which nine subtypes are known in wild waterfowl (N1–9), cleaves SIA from glycosylated proteins and lipids. Influenza A‐like viruses (H17N10 and H18N11) that have been found in bats neither bind nor cleave SIAs (reviewed by Ref. [13]). As HA and NA have apparent opposite functions and work in concert, a balance between HA binding and NA cleavage is needed for efficient viral replication [5, 14, 15, 16]. For example, substitutions in NA, conferring resistance to antiviral drugs, often result in reduced NA activity altering the HA‐NA balance [17, 18, 19]. The balance is subsequently restored by selecting for compensatory substitutions in NA and/or HA that enhance NA activity [20, 21, 22] or decrease HA‐binding affinity [23].
The HA‐NA balance of IAVs is probably highly adapted to the specific sialoglycan host repertoire and represents an important host range determinant. When viruses cross the species barrier and encounter a novel sialoglycan repertoire in mucus and on the epithelial cell surface, the HA‐NA balance of these viruses needs to be readjusted to allow efficient replication and intra‐ and inter‐host spread. When avian IAVs cross the species barrier and adapt to humans, this host switch is accompanied by a change in the receptor‐binding preference of HA from SIAs connected to the penultimate galactose via α2,3‐linkage (avian‐type receptors) to SIAs connected by α2,6‐linkage (human‐type receptors) [24, 25, 26]. The substitutions in the receptor‐binding site of HA that cause this alteration have been extensively studied and characterized for different HA subtypes (for reviews, see Refs [27, 28, 29]). Compared to the well‐documented adaptive changes in the HA, much less is known about the role of NA in host adaptation. Here, we review the available literature on a 2nd SIA‐binding site (2SBS) in NA, which is increasingly being recognized as an important determinant of NA enzymatic activity, HA‐NA balance, virus replication, and host tropism.
Neuraminidase is a type II glycoprotein, which forms tetramers of four identical polypeptides. Each protomer of about 470 amino acids harbors four distinct domains: the N‐terminal cytoplasmic tail, the transmembrane region, the thin stalk, and the catalytic head (Fig. 1) (reviewed by Refs [30, 31]). Crystal structures of the box‐shaped NA head domain have been determined for all IAV NA subtypes ([32] and references therein). Each protomer forms a six‐bladed propeller‐like structure, with each blade having four antiparallel β‐sheets that are stabilized by disulfide bonds and connected by loops of variable length. Each head domain contains a catalytic site with highly conserved residues that directly contact SIA and framework residues that keep the catalytic site in place. Close to the catalytic site, NA contains a 2SBS (also referred to as hemadsorption site) with sialoglycan‐binding properties (Fig. 1) [33, 34].
Fig. 1.
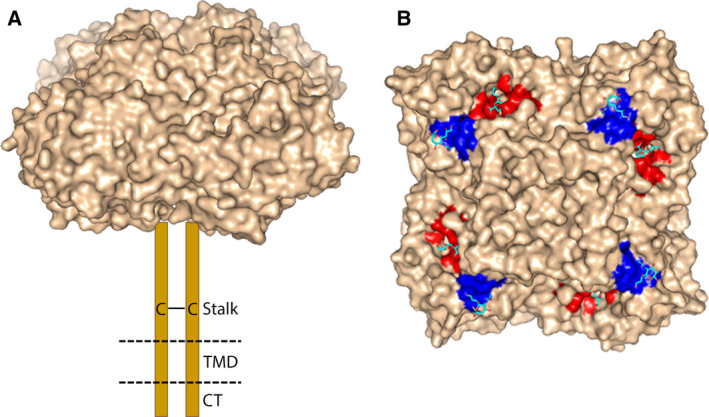
NA structure. (A) Schematic representation of NA structure. NA forms a homo‐tetramer of four identical subunits. Each subunit contains a cytoplasmic tail (CT), transmembrane domain (TMD), and stalk and head domain (PDB ID: 1W20). Cys residues present in the stalk domain result in the formation of two disulfide‐bonded protomers (C—C) (for reviews on the structure, see Refs [30, 31]). (B) Crystal structure of N6 NA head domain (PDB ID: 1W20) complexed with sialic acid shown in surface representation. The catalytic site and 2SBS are colored red and blue, respectively. Two SIA molecules bound to each NA monomer are shown as sticks (oxygen in red; nitrogen in blue; carbon in cyan). The figures were made using pymol (DeLano Scientific, San Carlos, CA, USA) http://www.csb.yale.edu/userguides/graphics/pymol/index.html.
The NA catalytic site is highly conserved among all IAVs. Regardless whether NA is derived from an avian or a human virus, it cleaves α2,3‐linked sialosides more efficiently than α2,6‐linked sialosides. Human viruses, however, are generally more efficient in cleaving α2,6‐linked SIAs than avian viruses [35, 36, 37]. For example, N2 protein of human H2N2 and H3N2 viruses evolved an increased ability to cleave α2,6‐linked SIAs, presumably to match the binding preference of HA. Increased cleavage of α2,6‐linked SIAs was, however, not yet observed in the early pandemic H2N2 viruses [35]. The dual cleavage specificity of NA, that is, the ability to cleave both α2,3‐ and α2,6‐linked SIAs, appears to contrast the apparent clear receptor preference of HA of human viruses for α2,6 sialylated glycans and of avian viruses for α2,3 sialylated glycans (for reviews on HA receptor specificity, see Refs [28, 38]).
In contrast to the NA catalytic site, the stalk domain is subject to host species adaptation. Stalk deletions in several NA subtypes are regarded as a poultry adaptation of IAVs derived from aquatic birds [39, 40]. Experimental studies showed that stalk truncations confer increased virulence in chickens [41] and interestingly also in mice [42]. All human pandemic and seasonal IAVs contain full‐length stalk domains. The length of the stalk domain has been shown to affect NA enzymatic activity of virus particles on multivalent receptor surfaces [39, 43]. Deletions in the stalk result in lower enzymatic activity presumably by reducing access of the NA catalytic site to sialoglycan substrates [44]. The role of the stalk domain in IAV replication and host tropism has been reviewed in detail elsewhere [40, 45]. Besides the stalk domain, also the 2SBS displays host species adaptation. While the 2SBS is highly conserved in most avian IAVs, it is invariably lost in human IAVs [37, 46, 47, 48], suggesting that the 2SBS plays an important role in the adaptation of IAV to humans as is discussed in more detail below.
Structure and conservation of the 2SBS
Hemagglutinating activity of NA was first discovered in 1984 for N9 NA [34]. In contrast to the NA catalytic activity, the hemadsorption activity of NA was not inhibited by an NA inhibitor targeting the NA catalytic site. Amino acid residues responsible for hemadsorption were identified by sequencing monoclonal antibody escape mutants of N9 NA that had lost this activity [49]. These effects led to the conclusion that enzymatic and hemadsorption activities of NA are associated with two separate sites on the head domain of N9. Hemagglutinating activity was acquired upon transfer of these amino acids by site‐directed mutagenesis to an hemadsorption‐negative N2 NA [50]. Structural evidence for the role of 2SBS in the hemadsorption activity of NA was obtained in 1997, by solving the crystal structure of N9 NA in complex with SIAs bound to both the catalytic site and the 2SBS [33]. While the catalytic site forms a deep pocket, the 2SBS forms a shallow pocket (Fig. 2). Three peptide loops, 370 loop (residues 366–373), 400 loop (residues 399–404), and 430 loop (residues 430–433), form the 2SBS and contain SIA contact residues: S367, S370, S372, N400, W403, and K432 (N2 numbering is used; for an alignment, see Ref. [30]). These residues directly interact with SIA via hydrophobic interactions (W403) and via the formation of hydrogen bonds (other residues). Until now, direct binding of SIA to the 2SBS has been demonstrated using X‐ray crystallography for N2, N5, N6, and N9 NAs [32, 33, 51, 52] (Fig. 2) and by saturation‐transfer difference nuclear magnetic resonance (STD‐NMR) for N1 NA [53]. Similar residues form direct contacts between the 2SBS and SIA for the different NAs as determined by X‐ray crystallography, with exception of the residue at position 432, which forms a direct hydrogen bond with SIA only in N9 NA (Fig. 2). Residue 432 is, however, hydrogen bonded to SIA via a water molecule in N2 and N5 NAs, while in N6 NA rotation of the side chain of K432 can readily allow formation of a hydrogen bond directly or via water. SIAs in the 2SBS and catalytic site adopt the chair and boat conformation, respectively [32]. SIA bound to the receptor‐binding site of HA adopts the chair conformation as well [27]. There is no direct interaction between the penultimate galactose residue and the 2SBS, which is different from HA receptor binding where the galactose and N‐acetylglucosamine residues in the sialoglycan also interact with the HA receptor‐binding site [27, 32].
Fig. 2.
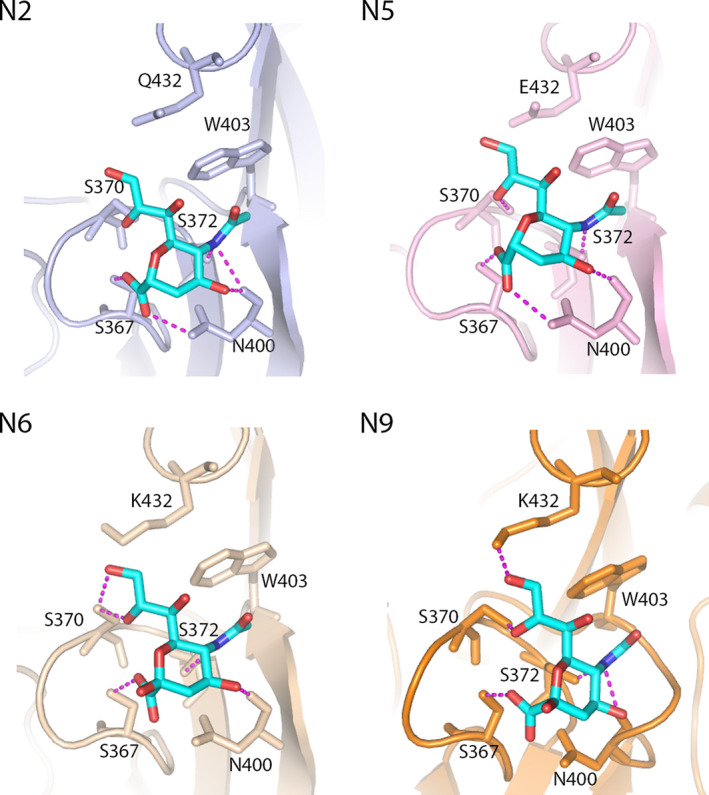
Interaction of 2SBS with Neu5Ac. The complexes of Neu5Ac with the 2SBS of neuraminidases N2 (PDB ID: 4H53; [51]), N5 (PDB ID: 4QN5; [32]), N6 (PDB ID: 1W20 [52]), and N9 (PDB ID: 1MWE; [33]) are shown. Neu5Ac is shown as sticks (oxygen in red; nitrogen in blue; carbon in cyan). The numbering of residues in the 2SBS that contact Neu5Ac is indicated. Hydrogen bonds are shown as dashed magenta lines. W403 forms van der Waals contacts with the N‐acetyl group of Neu5Ac. The figures were made by using pymol.
To visualize the conservation of the 2SBS of IAVs derived from different host species, we generated sequence logos of the three loops that constitute the 2SBS. The SIA contact residues in the 370 and 400 loops are highly conserved in avian IAVs regardless of the NA subtype (Fig. 3), with exception of residues at position 400 in N1 NA and 370 in N3 NA. The K432 SIA contact residue in the 430 loop of N9 is conserved in some, but not in other NA subtypes. Substitution of K432 in N1 [37] or N9 [49] NA negatively affected sialoglycan binding via the 2SBS. However, as avian N2 proteins carrying Q432 have a functional 2SBS [47, 48], a K at position 432 is not absolutely required for sialoglycan binding. Of note, N1 and N9 NA display some variations in residues, which do not form direct contacts with SIA (positions 369 and 401) but may nevertheless affect the receptor‐binding properties of NA [37, 54]. Although receptor‐binding activity of 2SBS has only been demonstrated for N1, N2, N5, N6, and N9 NAs, all other NA subtypes, with the exception of N3 NA, display a high conservation of the SIA contact residues in the 370 and 400 loops in avian viruses, indicating that a functional 2SBS is a highly conserved feature in most avian viruses.
Fig. 3.

Sequence logos of the 2SBS of avian N1–9 NA. Sequence logos were created for the three loops in the 2SBS: 370 loop (residues 366 to 373), 400 loop (residues 399 to 404), and 430 loop (residues 430 to 433) with the WebLogo program (https://weblogo.berkeley.edu/logo.cgi) [55] using all sequences available for avian N1 to N9 NAs in Influenza Research Database (https://www.fludb.org/). For N2 NA, H9N2 viruses were excluded from the analysis. The overall height of the stack represents the sequence conservation at that position, while the height of the symbols within the stack indicates the relative frequency of each amino acid at that position. Asterisks indicate SIA contact residues. Six residues interacting with SIA (SIA contact residues) are labeled with black asterisks.
The sequence logos of the 2SBS of N1 NA indicate that, while the SIA contact residues are highly conserved in N1 NAs of avian viruses, this conservation is invariably lost for both the seasonal (prior 2009) and the H1N1pdm09 viruses (Fig. 4) [37, 47]. Based on Brownian dynamic simulation, it was concluded that some of the key features of the 2SBS are retained in N1 NA of H1N1pdm09, although this NA displayed a 16‐fold reduced k on compared with N1 NA from an avian virus [56]. In agreement herewith, STD‐NMR analyses indicated that the 2SBS of NA of seasonal H1N1 virus (isolated prior to the 2009 pandemic) and of H1N1pdm09 bound very weakly if at all to α2,3‐sialyllactose as compared with readily detectable binding of the 2SBS of an avian H5N1 virus [53].
Fig. 4.
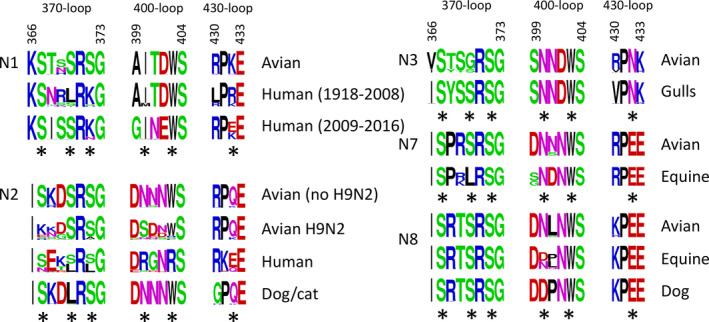
Sequence logos of the 2SBS of NA proteins from viruses that infect different species. Sequence logos were generated for the three loops that constitute the 2SBS similarly as described in the legend to this figure. All NA sequences available for viruses infecting different species from the Influenza Research Database (https://www.fludb.org/) were used with the indicated limitations. For N1 NA of human viruses, sequences were limited to those of seasonal H1N1 IAVs isolated prior to 2009 (Human (1918–2009)) and swine‐origin human H1N1pdm09 IAVs isolated between years 2009 and 2016 (Human (2009–2016)). For N2 NA of human viruses, sequences were limited to sequences prior to 2012. N3 NA avian sequences include sequences from gull viruses. SIA contact residues are labeled with black asterisks.
Just as in N1 NA of human viruses, the 2SBS of human H2N2 and H3N2 viruses essentially invariably contains substitutions. The NA of the H2N2/1957 pandemic influenza virus originated from an avian precursor. The large majority of virus strains that circulated during the first year of the pandemic contained substitutions in one of the contact residues of 2SBS, which markedly reduced hemadsorption activity [47]. Additional substitutions in both contact and noncontact residues of 2SBS were acquired during subsequent seasonal circulation of H2N2 viruses in humans and after transmission of this NA via reassortment to a novel pandemic H3N2 virus in 1968 (Fig. 4) [46, 47, 48]. The lack of conservation in the 2SBS of both N1 and N2 NAs of human viruses indicates that a 2SBS with substitutions represents a marker of influenza virus adaptation to humans.
The 2SBS is highly conserved in N2 of avian HXN2 viruses, with the exception of H9N2 viruses, which display a large variability in contact and noncontact residues [57, 58, 59, 60] (Fig. 4). H9N2 viruses transmitted and adapted from aquatic birds to gallinaceous poultry (chicken, turkey, and quail) in Asia in early 1990s and since then became endemic in farmed poultry across Asia, Middle East, and North and West Africa [61]. Transmission to poultry was accompanied by substitutions in 2SBS of NA, among them S367K and N400S [60].
H3N2 viruses of dogs originated from avian H3N2 viruses and can now transmit between dogs and from dogs to cats [62, 63]. The change in host range of these H3N2 viruses was accompanied by a S370L substitution in the 370 loop of the 2SBS (Fig. 4). Equine H7N7 and H3N8 viruses also contain substitutions in the 2SBS compared with their avian counterparts. All equine H7N7 viruses contain substitution S370L, while approximately half of the equine H3N8 viruses contain N400D. The conservation of some of the noncontact residues is also less strict in the equine viruses. All H3N8 dog viruses, which originate from equine H3N8 viruses, contain D400 in the 2SBS. Most N3 NAs of avian viruses do not have a strict conservation of the SIA contact residue S370 and rather contain G370. S370 is, however, strictly conserved in H16N3 gull viruses and in a specific lineage of H7N3 chicken viruses. Although founder effects cannot be excluded, the obvious correlation between structure of the 2SBS and the virus–host species is indicative of an important role for the 2SBS in host range of IAVs. More detailed phylogenetic and functional analysis of substitutions in the 2SBS in relation to IAV host range is warranted.
Receptor‐binding properties and contribution to NA catalytic activity
In early studies, the receptor‐binding specificity of the 2SBS was studied by analyzing the interaction of red blood cells with cell surface‐expressed NA. Hemadsorption by NA could be observed with erythrocytes from chicken and human origin, but not with equine, bovine, or swine erythrocytes, suggesting that these latter erythrocytes do not contain sufficient amounts of sialoglycans that are recognized by the 2SBS [46]. Equine, bovine, and swine erythrocytes abundantly display N‐glycolylneuraminic acid (Neu5Gc), in addition to N‐acetylneuraminic acid (Neu5Ac). Neu5Gc is absent in humans and most avian species [1], suggesting that recognition of Neu5Gc‐containing sialoglycans by the 2SBS would play no role in these species. In other studies, erythrocytes that had been desialylated, followed by linkage‐type specific re‐sialylation using α2,3‐ or α2,6‐sialyltransferases, were used to show that the 2SBS of N1 and N2 NAs of avian viruses bound to both α2,3‐ and α2,6‐linked SIAs [47, 64]. The 2SBS bound α2,3‐sialyllactose in the presence of oseltamivir carboxylate, which efficiently competed with binding of this sialoglycan to the NA catalytic site [53]. This finding of preferred binding of NA inhibitors to the catalytic site over the 2SBS was supported by the Brownian dynamic simulations [65]. In contrast to the observed dual specificity when using resialylated red blood cells in combination with cell‐expressed NA [47, 64], cell‐derived vesicles containing N1 or N2 NA proteins were shown, by using biolayer interferometry (BLI), to exclusively bind via 2SBS to synthetic glycans containing α2,3‐linked SIAs [37, 48]. Also, N9, as shown by glycan array analysis, prefers binding via its 2SBS to α2,3‐linked sialosides, particularly bi‐ and triantennary glycans with multiple LacNAc repeats, whereas weak or no binding to α2,6‐linked sialosides was observed [54]. This discrepancy in the data on receptor‐binding preference of the 2SBS is probably explained by the different methods used to analyze the receptor specificity of the 2SBS. For example, a higher receptor density on erythrocytes compared with the BLI sensor surface might allow for 2SBS binding of α2,6‐linked SIAs. In addition, binding to resialylated erythrocytes might be affected by prior incomplete desialylation.
Interestingly, receptor binding via the 2SBS of avian N9 NA appeared more efficient than that of avian N1 NA, as hemagglutination in the presence of oseltamivir carboxylate was observed for nanoparticles multivalently displaying recombinant N9 NA, but not N1 NA [37]. Despite that, glycan receptor binding, including hemagglutination, could be demonstrated for N1 NA by displaying the protein in vesicles or virus‐like particles, resulting in increased multivalency [37, 66]. The presence of I400 in N1 NA might explain the reduced receptor binding of this NA compared with N9 (Fig. 2) [37]. Using STD‐NMR, binding of α2,3‐sialyllactose via the 2SBS was observed for avian N1 NA, which was much stronger than for N1 NA of H1N1pdm09 [53]. Substitutions in all three loops of the 2SBS were shown to affect the receptor‐binding properties of NA. These include substitutions of SIA contact residues in both the 370 loop (S367N for N2 and N9; S370L for N1, N2, and N9; and S372Y for N9) [47, 48, 49, 64, 67] and the 400 loop (N400S for N2, N400K for N9, and W403R for N2) [47, 49], as well as some noncontact residues (N369H for N1; A369D and T401A for N9) [37, 49, 54]. Also, substitutions of residue K432, which is a direct SIA contact residue in N9 (Fig. 2), were shown to abolish hemagglutination by N1 NA (substitution K432E; [37]) and reduce activity of N9 NA (substitution K432N; [49]).
Uhlendorff et al. [47] showed that a single amino acid substitution, which reduced binding activity of the 2SBS, had no effect on desialylation of monovalent sialosides, but significantly reduced SIA cleavage from polyvalent natural and synthetic glycoconjugates containing multiple copies of SIA. These experiments demonstrated that binding of the SIA‐containing macromolecules to 2SBS facilitates their desialylation by the catalytic site. These findings were later confirmed and expanded in the studies on different NAs and sialoglycan substrates [37, 48, 54, 66, 67]. Thus, 2SBS‐positive N9 NA displayed a lower K m value for fetuin, but not for monovalent 2′‐(4‐methylumbelliferyl)‐α‐d‐N‐acetylneuraminic acid, compared with a 2SBS‐negative N9 NA variant, while their k cat values were similar [67]. Substitutions in all three receptor‐binding loops of the 2SBS of N1, N2, and N9 NAs were shown to affect cleavage of multivalent substrates [37, 48, 54]. In agreement herewith, glycan array analysis showed that glycans that were efficiently bound by the N9 2SBS were also cleaved efficiently [54]. Cleavage of synthetic glycans immobilized on BLI sensors by N1 and N2 NAs carrying a functional 2SBS was observed for α2,3‐linked but not α2,6‐linked sialoglycans, corresponding with their binding preference for α2,3‐linked glycans [37, 48]. Likewise, cleavage of SIAs from fetuin, which contains both α2,3‐ and α2,6‐linked sialosides, was generally much more affected by disruption of the 2SBS than cleavage of SIAs from transferrin, which only contains α2,6‐linked sialosides [37, 48, 54, 66]. Nevertheless, cleavage of some multivalent synthetic and natural substrates only containing α2,6‐linked sialosides was affected by substitutions in the 2SBS [37, 47, 48, 54, 66], suggesting that the 2SBS binds to some extent to these human‐type receptors. Collectively, these studies indicate that amino acid substitutions in the 2SBS affect NA catalytic activity by changing substrate binding via the 2SBS rather than by directly affecting the structure and function of the adjacent catalytic site.
Enhancement of NA catalytic activity by the 2SBS may be explained by a ‘bind and trans‐cleave’ model [47, 54], in which the 2SBS recruits and keeps sialosides in close proximity of the catalytic site (Fig. 5A). More recently, an alternative model was proposed (Fig. 5B) [68], in which the SIA receptors first bind to the 2SBS before being transferred to the catalytic site, referred to as a ‘bind and transfer’ mechanism. This latter model is based on the Brownian dynamic simulations, which indicate that substrates bind faster to the 2SBS of avian NA than to the catalytic site [56, 69], and on chlorine anion distribution and the projection of the electrostatic potential onto the NA surface, which may indicate the existence of a path between the 2SBS and the catalytic site wide enough to allow movement of negatively charged SIAs [68]. Even though the 2SBS prefers binding to α2,3‐linked SIAs, cleavage of α2,6‐linked SIAs by NA may be enhanced to a similar extent by a functional 2SBS as cleavage of α2,3‐linked SIAs, as long as the multivalent substrate (e.g., fetuin) also displays α2,3‐linked sialosides [48]. This result fits well with the ‘bind and trans‐cleave’ model but is more difficult to reconcile with the ‘bind and transfer’ model. In this latter model, enhanced cleavage of substrates would only be expected for those glycans that are efficiently bound by the 2SBS. Furthermore, the ‘bind and transfer’ model is not consistent with the lack of an effect of the 2SBS on cleavage of monovalent sialosides [47, 48]. Further studies are needed to clarify the mechanism behind 2SBS‐mediated enhancement of the NA catalytic activity. Of note, enhancement of catalytic efficiency of glycoside hydrolases, by combining catalytic and glycan‐binding domains, appears to be a common theme that is observed for most eukaryotic and bacterial NAs [70] and for other viral receptor‐destroying enzymes, such as the hemagglutinin–esterase protein of embecoviruses [71, 72] and hemagglutinin–neuraminidase protein of paramyxoviruses [73, 74].
Fig. 5.
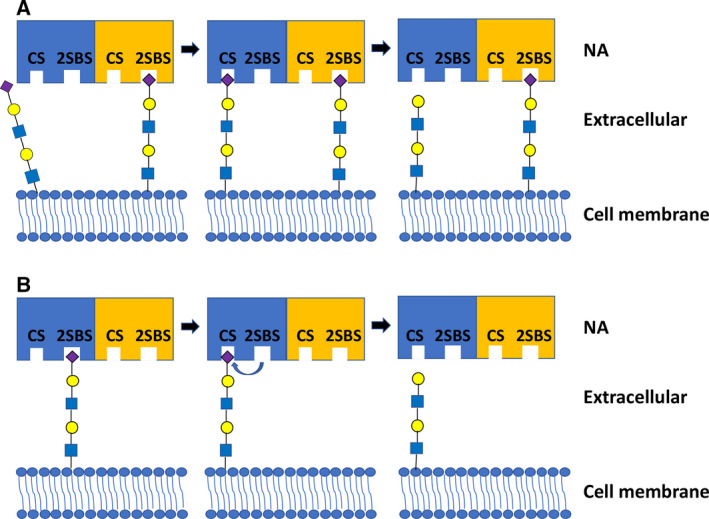
Proposed models of the enhanced cleavage activity induced by the 2SBS. (A) ‘Bind and trans‐cleave’ model [47, 54]: binding of the 2SBS to SIA residues presented on multivalent substrates brings adjacent SIA closer to the catalytic site (CS) in the same or another protomer, resulting in a higher cleavage efficiency. (B) ‘Bind and transfer’ model [68]: SIA receptors first bind to the 2SBS, after which they are transferred to the CS, resulting in enhanced cleavage of SIA by CS. Blue and yellow rectangles represent NAs. Purple diamond: Neu5Ac; yellow circle: galactose; green circle: mannose; blue square: N‐acetylglucosamine.
Importance of the 2SBS for the HA‐NA receptor balance
Uhlendorff et al. [47] showed that a functional 2SBS contributes to catalytic activity of both isolated NA molecules and NA in the context of virions. Recombinant variants of pandemic virus A/Hong Kong/1/68 (H3N2) that only differed in their 2SBS displayed identical hemagglutination activity but differed in the NA‐driven disaggregation of the erythrocytes. Thus, virus with a nonfunctional 2SBS destroyed receptors on red blood cells more slowly than did its 2SBS‐positive counterpart. Recent application of BLI‐based assays to study virus–receptor interactions [75] enabled a more detailed analysis of the contribution of the 2SBS to the HA‐NA receptor balance. BLI enables analysis of virus binding by determining the initial binding rate of virus binding to a receptor‐coated surface [75], or by analyzing the receptor density at which half‐maximum binding occurs [76]. These assays are usually performed in the presence of inhibitors of the NA catalytic site. When NA activity is not or no longer inhibited, the latter by washout of the inhibitor, virus binding is followed by NA‐driven virion mobility on the sensor surface, which finally results in virion self‐elution when the receptor density is too low to sustain virion association [75]. The rate, at which viruses are released from the sensor surface, reflects the HA‐NA receptor balance, as it depends on the interplay between HA binding to the immobilized receptors and NA cleavage of these receptors.
Using BLI, the 2SBS was shown to contribute to receptor binding of virus, the effect being dependent on properties of both viral HA and sialic acid‐containing receptor molecules [48, 67]. A functional 2SBS was shown to enhance H3N2 virion binding to synthetic glycans carrying α2,3‐linked but not α2,6‐linked SIAs, in agreement with the binding preference of the 2SBS. When combined with an H3 HA that preferentially bound to α2,6‐linked SIAs, the 2SBS of N2 NA also enhanced binding to glycoprotein receptors LAMP1, carrying mostly N‐glycans, and glycophorin A, carrying mainly O‐linked sugars. Enhanced binding to these glycoproteins was not observed when N2 NA was combined with an H3 HA that preferentially bound to α2,3‐linked SIAs [48]. The presence of a functional 2SBS was also shown to result in a lower receptor density, at which half‐maximum virion binding was observed [67]. The 2SBS is also a critical determinant of the HA‐NA receptor balance of virus particles, as analyzed by BLI. 2SBS‐positive viruses (H3N2 [48], H5N1, and H1N1 [66]) display faster self‐elution from surfaces containing LAMP1 or glycophorin A. Of note, the contribution of the 2SBS to virion self‐elution is larger when NA is combined with an HA that binds these receptors more strongly. Substitutions that decreased HA receptor‐binding avidity also decreased the contribution of the 2SBS to virion self‐elution [66]. Apparently, a weaker‐binding HA does not require a 2SBS‐positive NA with a higher enzymatic activity for efficient self‐elution from the receptor‐coated surface.
Disruption of the 2SBS affects virus replication in vitro in an HA‐ and cell type‐dependent manner. Several studies show that the loss of a functional 2SBS negatively affected virus replication when NA was combined with HAs preferring binding to α2,3‐linked sialoglycans (H1 [37], H2 [46], H3 [48], H5 [37, 66]). A recombinant virus, in which a 2SBS‐negative N1 NA was combined with H5 HA, rapidly reverted to a functional 2SBS, indicating that the virus with the restored 2SBS has replicative advantage in cell culture [37, 66]. In contrast, a recombinant virus harboring the same NA and H1 HA of the laboratory‐adapted strain A/PR8/8/34, which displays a lower receptor‐binding avidity than H5 HA, stably maintained the debilitating substitution in the 2SBS [37, 66]. In agreement herewith, self‐elution of the virus with the weak‐binding H1 HA from sialoglycoproteins was much less inhibited by disruption of the 2SBS, than that of the virus with the strong binding H5 HA [66].
Also, the preference of HA for α2,3‐linked or to α2,6‐linked SIAs affects the contribution of the 2SBS to virus replication. An H3N2 virus with preferred binding to α2,3‐linked SIAs displayed reduced self‐elution from sialylated glycoproteins and reduced replication upon loss of a functional 2SBS. In contrast, the absence of a functional 2SBS in N2 NA enhanced virus replication when combined with H3 from the 1968 pandemic virus, which prefers binding to α2,6‐linked SIAs [48]. In this latter case, reduced virus replication and fuzzy plaques (resulting from the presence of many noninfected cells), in the presence of a functional 2SBS, correlated with increased catalytic activity and thus virion self‐elution from sialoglycoproteins in the BLI assay. These results suggest that high NA activity combined with low HA‐binding avidity shifts the HA‐NA receptor balance toward virus dissociation from the cell surface before it can enter the cells. Replication in MDCK cells of a recombinant H3N9 virus‐carrying HA from a recent seasonal human H3N2 virus was enhanced by the presence of a functional 2SBS in N9. No difference in virus propagation was observed, however, on MDCK cells with increased levels of α2,6‐linked sialoglycans [67]. Possibly, a functional 2SBS can enhance binding to cells when insufficient numbers of receptors are present. In summary, an optimal HA‐NA balance appears to exist for virus replication in vitro, which is significantly influenced by the absence or presence of a functional 2SBS. The importance of a functional 2SBS in NA depends on HA‐binding affinity for the receptor repertoire on the surface of target cells.
So far, only one study investigated potential effects of a functional 2SBS on virus replication in vivo [46]. No difference in virus replication in ducks was reported for viruses with or without a 2SBS, although replication in vitro was reduced in the absence of the 2SBS. In this study, recombinant viruses were used containing HA and NA derived from different viruses. This may have resulted in a mismatched HA‐NA combination, in which the presence of the 2SBS might be of minor influence. Alternatively, the virus acquired compensatory substitutions in HA (discussed below) or a functional 2SBS may not be required for efficient replication, but rather for transmission between ducks. Obviously, additional studies are needed to demonstrate the importance of the 2SBS for IAV replication and transmission in vivo.
In view of the interplay between HA and NA, it is not surprising that substitutions in the 2SBS often go hand in hand with substitutions in HA that affect its receptor‐binding properties. This, as mentioned above, holds true for the human N1‐ and N2‐containing viruses, in which NAs with a disrupted 2SBS (Fig. 4) are associated with HAs that prefer binding to human‐type receptors. Likewise, substitutions in the 2SBS in canine H3N2 and H3N8 viruses is for both subtypes correlated with a W222L substitution in the receptor‐binding site of H3 [77, 78]. Canine H3 proteins predominantly bind to α2,3‐linked sialosides, but display higher binding avidity to fucosylated and sulfated glycan receptors, and glycans with Neu5Gc moieties, compared with precursor avian and equine HAs lacking this substitution [77, 78, 79]. Similarly, equine H7N7 viruses not only carried NA with a disrupted 2SBS, but also contained HA that preferentially bound to α2,3‐linked Neu5Gc, in contrast to avian H7‐containing viruses which prefer binding to Neu5Ac‐containing sialoglycans [80, 81]. H16 subtype viruses that predominantly circulate in gulls differ by their receptor‐binding properties from avian viruses of other species [82]. This feature correlates with differences in the 2SBS at position 370 between gull H16N3 viruses and N3 NA of other avian viruses.
Two scenarios can be envisioned for the emergence of IAVs with substitutions in the receptor‐binding sites of both HA and NA. The first scenario is that mutations in HA precede and drive mutations in NA. Thus, changes obtained in HA as an adaptation to the sialoglycan repertoire of a novel host species may be followed by compensatory substitutions in the 2SBS of NA. Such NA mutations may also serve to escape inhibitory molecules that may be present in the novel host species, for example, in saliva [83]. An alternative scenario is that substitutions in the 2SBS of NA may precede or drive compensatory mutations in HA, thereby facilitating a host tropism jump. Often, it is unclear which protein changed first due to a lack of sequencing data. In the case of the 1918 pandemic H1N1 human virus, only a single NA sequence is available from 1918 (A/Brevig Mission/1/1918; [84]). NA of this virus does not contain substitutions at any of the six SIA contact residues compared with the avian consensus sequence shown in Fig. 4. HA of this virus contains D190 and D225 [85] that result in reduced binding to α2,3‐linked and increased binding to α2,6‐linked SIAs, which may suggest that substitutions in HA preceded those in the 2SBS of NA. The 1918 NA does display, however, reduced hemadsorption activity compared with two avian NAs [47], which might be related to substitution R430Q compared with the avian consensus sequence. The next available H1N1 NA sequences of human viruses come from the 1930s and do contain substitutions of SIA contact residues. Passaging of these latter historical isolates may have resulted, however, in acquisition of egg/cell culture‐derived adaptive substitutions. For early pandemic H2N2 viruses, the situation is also complicated by laboratory passaging of historical isolates. H2N2 viruses with and without Q226L and G228S substitutions in HA, associated with a switch in receptor‐binding preference, are both found in combination with NAs with and without a functional 2SBS, making determination of the order of mutations challenging.
For some viruses, phylogenetic analysis indicates that the disruption of the 2SBS preceded mutations that changed the receptor‐binding properties of HA. Thus, the substitution of T401A in NA of novel H7N9 viruses, known to decrease receptor binding via the 2SBS and NA catalytic activity, preceded the Q226L substitution in H7 HA [54] that results in decreased binding to avian‐type and increased binding to human‐type receptors [29, 86, 87]. Also for H9N2 viruses, phylogenetic analysis indicates that alteration of the 2SBS preceded substitutions in HA at positions 183, 190, and 226, including Q226L [66], that are known or expected to affect receptor binding [60, 88, 89]. For position 190, it cannot be excluded, however, that this substitution occurred simultaneously with disruption of the 2SBS, as only viruses with both mutations have been isolated [60]. The plausibility of a scenario, in which substitutions in the 2SBS drive acquisition of substitutions in HA that alter its receptor‐binding properties, was recently experimentally shown for H5N1 virus [66]. Passaging H5N1 viruses with a disrupted 2SBS in cell culture rapidly resulted in the selection of substitutions in NA that restored the 2SBS and/or in HA that reduced binding to avian‐type receptors to compensate for the lowered NA activity. Of note, one of these mutations in HA (resulting in substitution S223N) concomitantly increased binding to human‐type receptors, similarly as observed for H7N9 and H9N2 field viruses. It is tempting to speculate that also for H7N9 and H9N2 viruses, substitutions that increased binding to human‐type receptors were selected for their negative effect on binding to avian‐type receptors in order to restore the HA‐NA balance after loss of a functional 2SBS. Clearly, more research on the interplay between the receptor‐binding properties of HA and NA is desirable.
Outlook
A complex interplay exists between receptor‐binding and receptor‐destroying activities of IAV virions, which needs to be adapted to the sialoglycan repertoire of the virus–host species and thus represents an important determinant of viral host range. As emphasized by this review, the precarious HA‐NA receptor balance that is required for efficient viral replication includes not only the receptor‐destroying activity of NA per se, but also the receptor‐binding properties of its 2SBS. This binding may affect virion binding to a receptor‐containing surface but is also of critical importance for the catalytic activity of the viral NA. Likewise, receptor‐binding via HA was also shown to contribute to the catalytic activity of virions [75, 90]. To add to this complexity, receptor binding by the catalytic site of NA may also contribute to the initial binding rate of virions, which appears more pronounced for low activity NA [75, 91, 92]. The HA‐NA receptor balance needs adjustment when IAVs infect a novel host species, but it may also be under selective pressure to change in the postadaptation stage owing to antigenic drift. Antigenic drift in HA has been shown to be accompanied with altered receptor‐binding properties [93, 94, 95, 96, 97, 98]. Likewise, antigenic drift in NA, for example, resulting from substitutions in the 2SBS, may also alter NA catalytic activity. Contribution of substitutions in the 2SBS to antigenic drift in humans has been demonstrated [49, 99, 100, 101, 102, 103]. Recently developed assays based on live imaging [104, 105] and BLI analyses [75] enable researchers to study the evolution of the HA‐NA receptor balance of IAVs, and the contribution of the 2SBS thereto, in detail. Such kinetic assays are needed to understand the full complexity of IAV evolution, including links between HA‐NA receptor balance, host tropism, antigenic drift, and the zoonotic potential of IAVs.
Conflict of interest
The authors declare no conflict of interest.
Author contributions
WD and CAMH wrote the manuscript. FJMK, EV, and MM provided critical feedback and contributed to the final manuscript.
PEER REVIEW
The peer review history for this article is available at https://publons.com/publon/10.1111/febs.15668.
[Correction added on 24 March 2021, after first online publication: URL for peer review history has been corrected.]
Acknowledgements
The work of the authors was funded by the Chinese Scholarship Council, file number 201603250057 (WD), Mizutani Foundation, reference number 180094 (CAMdH), and the Deutsche Forschungsgemeinschaft (German Research Foundation), project number 197785619‐SFB 1021 (MM). The funders had no role in study design, data collection, and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Varki A, Schnaar RL & Schauer R (2015) Sialic acids and other nonulosonic acids. In Essentials of Glycobiology (rd, Varki A, Cummings RD, Esko JD, Stanley P, Hart GW, Aebi M, Darvill AG, Kinoshita T, Packer NH, Prestegard JH et al., eds), pp. 179–195, Cold Spring Harbor, New York, NY. [PubMed] [Google Scholar]
- 2. Matrosovich M, Herrler G & Klenk HD (2015) Sialic acid receptors of viruses. In SialoGlyco Chemistry and Biology II: Tools and Techniques to Identify and Capture Sialoglycans (Gerardy‐Schahn R, Delannoy P & von Itzstein M, eds), pp. 1–28. Springer International Publishing, Cham. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Thompson AJ, de Vries RP & Paulson JC (2019) Virus recognition of glycan receptors. Curr Opin Virol 34, 117–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Palese P & Compans RW (1976) Inhibition of influenza virus replication in tissue culture by 2‐deoxy‐2,3‐dehydro‐N‐trifluoroacetylneuraminic acid (FANA): mechanism of action. J Gen Virol 33, 159–163. [DOI] [PubMed] [Google Scholar]
- 5. de Vries E, Du W, Guo H & de Haan CAM (2020) Influenza A virus hemagglutinin‐neuraminidase‐receptor balance: preserving virus motility. Trends Microbiol 28, 57–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Short KR, Richard M, Verhagen JH, van Riel D, Schrauwen EJ, van den Brand JM, Mänz B, Bodewes R & Herfst S (2015) One health, multiple challenges: the inter‐species transmission of influenza A virus. One Health 1, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Yoon SW, Webby RJ & Webster RG (2014) Evolution and ecology of influenza A viruses. Curr Top Microbiol Immunol 385, 359–375. [DOI] [PubMed] [Google Scholar]
- 8. Krammer F, Smith GJD, Fouchier RAM, Peiris M, Kedzierska K, Doherty PC, Palese P, Shaw ML, Treanor J, Webster RG et al. (2018) Influenza. Nat Rev Dis Primers 4, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Taubenberger JK & Kash JC (2010) Influenza virus evolution, host adaptation, and pandemic formation. Cell Host Microbe 7, 440–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kilbourne ED (2006) Influenza pandemics of the 20th century. Emerg Infect Dis 12, 9–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gamblin SJ, Vachieri SG, Xiong X, Zhang J, Martin SR & Skehel JJ (2020) Hemagglutinin structure and activities. Cold Spring Harb Perspect Med. Epub ahead of print: a038638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wu NC & Wilson IA (2020) Influenza hemagglutinin structures and antibody recognition. Cold Spring Harb Perspect Med 10, a038778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Brunotte L, Beer M, Horie M & Schwemmle M (2016) Chiropteran influenza viruses: flu from bats or a relic from the past? Curr Opin Virol 16, 114–119. [DOI] [PubMed] [Google Scholar]
- 14. Gaymard A, Le Briand N, Frobert E, Lina B & Escuret V (2016) Functional balance between neuraminidase and haemagglutinin in influenza viruses. Clin Microbiol Infect 22, 975–983. [DOI] [PubMed] [Google Scholar]
- 15. Wagner R, Matrosovich M & Klenk H‐D (2002) Functional balance between haemagglutinin and neuraminidase in influenza virus infections. Rev Med Virol 12, 159–166. [DOI] [PubMed] [Google Scholar]
- 16. Byrd‐Leotis L, Cummings RD & Steinhauer DA (2017) The interplay between the host receptor and influenza virus hemagglutinin and neuraminidase. Int J Mol Sci 18, 1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Carr J, Ives J, Kelly L, Lambkin R, Oxford J, Mendel D, Tai L & Roberts N (2002) Influenza virus carrying neuraminidase with reduced sensitivity to oseltamivir carboxylate has altered properties in vitro and is compromised for infectivity and replicative ability in vivo. Antiviral Res 54, 79–88. [DOI] [PubMed] [Google Scholar]
- 18. Ives JAL, Carr JA, Mendel DB, Tai CY, Lambkin R, Kelly L, Oxford JS, Hayden FG & Roberts NA (2002) The H274Y mutation in the influenza A/H1N1 neuraminidase active site following oseltamivir phosphate treatment leave virus severely compromised both in vitro and in vivo. Antiviral Res 55, 307–317. [DOI] [PubMed] [Google Scholar]
- 19. Marjuki H, Mishin VP, Chesnokov AP, De La Cruz JA, Davis CT, Villanueva JM, Fry AM & Gubareva LV (2015) Neuraminidase mutations conferring resistance to oseltamivir in influenza A(H7N9) viruses. J Virol 89, 5419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bloom JD, Gong LI & Baltimore D (2010) Permissive secondary mutations enable the evolution of influenza oseltamivir resistance. Science 328, 1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Butler J, Hooper KA, Petrie S, Lee R, Maurer‐Stroh S, Reh L, Guarnaccia T, Baas C, Xue L, Vitesnik S et al. (2014) Estimating the fitness advantage conferred by permissive neuraminidase mutations in recent oseltamivir‐resistant A(H1N1)pdm09 influenza viruses. PLoS Pathog 10, e1004065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Duan S, Govorkova EA, Bahl J, Zaraket H, Baranovich T, Seiler P, Prevost K, Webster RG & Webby RJ (2014) Epistatic interactions between neuraminidase mutations facilitated the emergence of the oseltamivir‐resistant H1N1 influenza viruses. Nat Commun 5, 5029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ginting TE, Shinya K, Kyan Y, Makino A, Matsumoto N, Kaneda S & Kawaoka Y (2012) Amino acid changes in hemagglutinin contribute to the replication of oseltamivir‐resistant H1N1 influenza viruses. J Virol 86, 121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Connor RJ, Kawaoka Y, Webster RG & Paulson JC (1994) Receptor specificity in human, avian, and equine H2 and H3 influenza virus isolates. Virology 205, 17–23. [DOI] [PubMed] [Google Scholar]
- 25. Matrosovich M, Tuzikov A, Bovin N, Gambaryan A, Klimov A, Castrucci MR, Donatelli I & Kawaoka Y (2000) Early alterations of the receptor‐binding properties of H1, H2, and H3 avian influenza virus hemagglutinins after their introduction into mammals. J Virol 74, 8502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Rogers GN & Paulson JC (1983) Receptor determinants of human and animal influenza virus isolates: differences in receptor specificity of the H3 hemagglutinin based on species of origin. Virology 127, 361–373. [DOI] [PubMed] [Google Scholar]
- 27. Shi Y, Wu Y, Zhang W, Qi J & Gao GF (2014) Enabling the ‘host jump’: structural determinants of receptor‐binding specificity in influenza A viruses. Nat Rev Microbiol 12, 822–831. [DOI] [PubMed] [Google Scholar]
- 28. de Graaf M & Fouchier RAM (2014) Role of receptor binding specificity in influenza A virus transmission and pathogenesis. EMBO J 33, 823–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. de Vries RP, Peng W, Grant OC, Thompson AJ, Zhu X, Bouwman KM, de la Pena ATT, van Breemen MJ, Ambepitiya Wickramasinghe IN, de Haan CAM et al. (2017) Three mutations switch H7N9 influenza to human‐type receptor specificity. PLoS Pathog 13, e1006390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Air GM (2012) Influenza neuraminidase. Influenza Other Respir Viruses 6, 245–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. McAuley JL, Gilbertson BP, Trifkovic S, Brown LE & McKimm‐Breschkin JL (2019) Influenza virus neuraminidase structure and functions. Front Microbiol 10, 39. 10.3389/fmicb.2019.00039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sun X, Li Q, Wu Y, Wang M, Liu Y, Qi J, Vavricka CJ & Gao GF (2014) Structure of influenza virus N7: the last piece of the neuraminidase “jigsaw” puzzle. J Virol 88, 9197–9207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Varghese JN, Colman PM, van Donkelaar A, Blick TJ, Sahasrabudhe A & McKimm‐Breschkin JL (1997) Structural evidence for a second sialic acid binding site in avian influenza virus neuraminidases. Proc Natl Acad Sci USA 94, 11808–11812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Laver WG, Colman PM, Webster RG, Hinshaw VS & Air GM (1984) Influenza virus neuraminidase with hemagglutinin activity. Virology 137, 314–323. [DOI] [PubMed] [Google Scholar]
- 35. Baum LG & Paulson JC (1991) The N2 neuraminidase of human influenza virus has acquired a substrate specificity complementary to the hemagglutinin receptor specificity. Virology 180, 10–15. [DOI] [PubMed] [Google Scholar]
- 36. Kobasa D, Kodihalli S, Luo M, Castrucci MR, Donatelli I, Suzuki Y, Suzuki T & Kawaoka Y (1999) Amino acid residues contributing to the substrate specificity of the influenza A virus neuraminidase. J Virol 73, 6743–6751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Du W, Dai M, Li Z, Boons G‐J, Peeters B, van Kuppeveld FJM, de Vries E & de Haan CAM (2018) Substrate binding by the second sialic acid‐binding site of influenza A virus N1 neuraminidase contributes to enzymatic activity. J Virol 92, e01243‐18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Gambaryan AS & Matrosovich MN (2015) What adaptive changes in hemagglutinin and neuraminidase are necessary for emergence of pandemic influenza virus from its avian precursor? Biochemistry (Mosc) 80, 872–880. [DOI] [PubMed] [Google Scholar]
- 39. Matrosovich M, Zhou N, Kawaoka Y & Webster R (1999) The surface glycoproteins of H5 influenza viruses isolated from humans, chickens, and wild aquatic birds have distinguishable properties. J Virol 73, 1146–1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Li J, Zu Dohna H, Cardona CJ, Miller J & Carpenter TE (2011) Emergence and genetic variation of neuraminidase stalk deletions in avian influenza viruses. PLoS One 6, e14722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Munier S, Larcher T, Cormier‐Aline F, Soubieux D, Su B, Guigand L, Labrosse B, Cherel Y, Quéré P, Marc D et al. (2010) A genetically engineered waterfowl influenza virus with a deletion in the stalk of the neuraminidase has increased virulence for chickens. J Virol 84, 940–952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zhou H, Yu Z, Hu Y, Tu J, Zou W, Peng Y, Zhu J, Li Y, Zhang A, Yu Z et al. (2009) The special neuraminidase stalk‐motif responsible for increased virulence and pathogenesis of H5N1 influenza A virus. PLoS One 4, e6277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Blumenkrantz D, Roberts KL, Shelton H, Lycett S & Barclay WS (2013) The short stalk length of highly pathogenic avian influenza H5N1 virus neuraminidase limits transmission of pandemic H1N1 virus in ferrets. J Virol 87, 10539–10551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Baigent SJ & McCauley JW (2001) Glycosylation of haemagglutinin and stalk‐length of neuraminidase combine to regulate the growth of avian influenza viruses in tissue culture. Virus Res 79, 177–185. [DOI] [PubMed] [Google Scholar]
- 45. Long JS, Benfield CT & Barclay WS (2015) One‐way trip: influenza virus' adaptation to gallinaceous poultry may limit its pandemic potential. BioEssays 37, 204–212. [DOI] [PubMed] [Google Scholar]
- 46. Kobasa D, Rodgers ME, Wells K & Kawaoka Y (1997) Neuraminidase hemadsorption activity, conserved in avian influenza A viruses, does not influence viral replication in ducks. J Virol 71, 6706–6713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Uhlendorff J, Matrosovich T, Klenk HD & Matrosovich M (2009) Functional significance of the hemadsorption activity of influenza virus neuraminidase and its alteration in pandemic viruses. Arch Virol 154, 945–957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Du W, Guo H, Nijman VS, Doedt J, van der Vries E, van der Lee J, Li Z, Boons GJ, van Kuppeveld FJM, de Vries E et al. (2019) The 2nd sialic acid‐binding site of influenza A virus neuraminidase is an important determinant of the hemagglutinin‐neuraminidase‐receptor balance. PLoS Pathog 15, e1007860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Webster RG, Air GM, Metzger DW, Colman PM, Varghese JN, Baker AT & Laver WG (1987) Antigenic structure and variation in an influenza virus N9 neuraminidase. J Virol 61, 2910–2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Nuss JM & Air GM (1991) Transfer of the hemagglutinin activity of influenza virus neuraminidase subtype N9 into an N2 neuraminidase background. Virology 183, 496–504. [DOI] [PubMed] [Google Scholar]
- 51. Vavricka CJ, Liu Y, Kiyota H, Sriwilaijaroen N, Qi J, Tanaka K, Wu Y, Li Q, Li Y, Yan J et al. (2013) Influenza neuraminidase operates via a nucleophilic mechanism and can be targeted by covalent inhibitors. Nat Commun 4, 1491. [DOI] [PubMed] [Google Scholar]
- 52. Rudino‐Pinera E, Tunnah P, Crennell SJ, Webster RG, Laver WG & Garman EF (2004) Structure of neuraminidase from English duck subtype N6 complexed with 30mM sialic acid (NANA, Neu5Ac), crystal soaked for 3 hours at 291 K. wwpdb.org, https://doi.org/10.2210/pdb1w20/pdb
- 53. Lai JCC, Garcia J‐M, Dyason JC, Böhm R, Madge PD, Rose FJ, Nicholls JM, Peiris JSM, Haselhorst T & von Itzstein M (2012) A secondary sialic acid binding site on influenza virus neuraminidase: fact or fiction? Angew Chem Int Ed 51, 2221–2224. [DOI] [PubMed] [Google Scholar]
- 54. Dai M, McBride R, Dortmans J, Peng W, Bakkers MJG, de Groot RJ, van Kuppeveld FJM, Paulson JC, de Vries E & de Haan CAM (2017) Mutation of the second sialic acid‐binding site, resulting in reduced neuraminidase activity, preceded the emergence of H7N9 influenza A virus. J Virol 91. e00049‐17. 10.1128/JVI.00049-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Crooks GE, Hon G, Chandonia J‐M & Brenner SE (2004) WebLogo: a sequence logo generator. Genome Res 14, 1188–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Sung JC, Van Wynsberghe AW, Amaro RE, Li WW & McCammon JA (2010) Role of secondary sialic acid binding sites in influenza N1 neuraminidase. J Am Chem Soc 132, 2883–2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Aamir UB, Wernery U, Ilyushina N & Webster RG (2007) Characterization of avian H9N2 influenza viruses from United Arab Emirates 2000 to 2003. Virology 361, 45–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Tombari W, Nsiri J, Larbi I, Guerin JL & Ghram A (2011) Genetic evolution of low pathogenicity H9N2 avian influenza viruses in Tunisia: acquisition of new mutations. Virol J 8, 467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Naguib MM, Arafa AS, Parvin R, Beer M, Vahlenkamp T & Harder TC (2017) Insights into genetic diversity and biological propensities of potentially zoonotic avian influenza H9N2 viruses circulating in Egypt. Virology 511, 165–174. [DOI] [PubMed] [Google Scholar]
- 60. Matrosovich MN, Krauss S & Webster RG (2001) H9N2 influenza A viruses from poultry in Asia have human virus‐like receptor specificity. Virology 281, 156–162. [DOI] [PubMed] [Google Scholar]
- 61. Carnaccini S & Perez DR (2020) H9 influenza viruses: an emerging challenge. Cold Spring Harb Perspect Med 10, a038588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Song D, Kang B, Lee C, Jung K, Ha G, Kang D, Park S, Park B & Oh J (2008) Transmission of avian influenza virus (H3N2) to dogs. Emerg Infect Dis 14, 741–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Li S, Shi Z, Jiao P, Zhang G, Zhong Z, Tian W, Long L‐P, Cai Z, Zhu X, Liao M et al. (2010) Avian‐origin H3N2 canine influenza A viruses in Southern China. Infect Genet Evol 10, 1286–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Hausmann J, Kretzschmar E, Garten W & Klenk H‐D (1995) N1 neuraminidase of influenza virus A/FPV/Rostock/34 has haemadsorbing activity. J Gen Virol 76, 1719–1728. [DOI] [PubMed] [Google Scholar]
- 65. Seitz C, Casalino L, Konecny R, Huber G, Amaro RE & McCammon JA (2020) Multiscale simulations examining glycan shield effects on drug binding to influenza neuraminidase. Biophys J 119, 2275–2289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Du W, Wolfert MA, Peeters B, van Kuppeveld FJM, Boons GJ, de Vries E & de Haan CAM (2020) Mutation of the second sialic acid‐binding site of influenza A virus neuraminidase drives compensatory mutations in hemagglutinin. PLoS Pathog 16, e1008816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Benton DJ, Wharton SA, Martin SR & McCauley JW (2017) Role of neuraminidase in influenza A(H7N9) virus receptor binding. J Virol 91, e02293‐16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Durrant JD, Kochanek SE, Casalino L, Ieong PU, Dommer AC & Amaro RE (2020) Mesoscale all‐atom influenza virus simulations suggest new substrate binding mechanism. ACS Cent Sci 6, 189–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Amaro RE, Ieong PU, Huber G, Dommer A, Steven AC, Bush RM, Durrant JD & Votapka LW (2018) A computational assay that explores the hemagglutinin/neuraminidase functional balance reveals the neuraminidase secondary site as a novel anti‐influenza target. ACS Cent Sci 4, 1570–1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Thobhani S, Ember B, Siriwardena A & Boons G‐J (2003) Multivalency and the mode of action of bacterial sialidases. J Am Chem Soc 125, 7154–7155. [DOI] [PubMed] [Google Scholar]
- 71. Langereis MA, Bakkers MJG, Deng L, Padler‐Karavani V, Vervoort SJ, Hulswit RJG, van Vliet ALW, Gerwig GJ, de Poot SAH, Boot W et al. (2015) Complexity and diversity of the mammalian sialome revealed by nidovirus virolectins. Cell Rep 11, 1966–1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Bakkers MJG, Zeng Q, Feitsma LJ, Hulswit RJG, Li Z, Westerbeke A, van Kuppeveld FJM, Boons G‐J, Langereis MA, Huizinga EG et al. (2016) Coronavirus receptor switch explained from the stereochemistry of protein–carbohydrate interactions and a single mutation. Proc Natl Acad Sci USA 113, E3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Bousse TL, Taylor G, Krishnamurthy S, Portner A, Samal SK & Takimoto T (2004) Biological significance of the second receptor binding site of Newcastle disease virus hemagglutinin‐neuraminidase protein. J Virol 78, 13351–13355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Streltsov VA, Pilling P, Barrett S & McKimm‐Breschkin JL (2015) Catalytic mechanism and novel receptor binding sites of human parainfluenza virus type 3 hemagglutinin‐neuraminidase (hPIV3 HN). Antiviral Res 123, 216–223. [DOI] [PubMed] [Google Scholar]
- 75. Guo H, Rabouw H, Slomp A, Dai M, van der Vegt F, van Lent JWM, McBride R, Paulson JC, de Groot RJ, van Kuppeveld FJM et al. (2018) Kinetic analysis of the influenza A virus HA/NA balance reveals contribution of NA to virus‐receptor binding and NA‐dependent rolling on receptor‐containing surfaces. PLoS Pathog 14, e1007233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Benton DJ, Martin SR, Wharton SA & McCauley JW (2015) Biophysical measurement of the balance of influenza a hemagglutinin and neuraminidase activities. J Biol Chem 290, 6516–6521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Wen F, Blackmon S, Olivier AK, Li L, Guan M, Sun H, Wang PG & Wan X‐F (2018) Mutation W222L at the receptor binding site of hemagglutinin could facilitate viral adaption from equine influenza A(H3N8) virus to dogs. J Virol 92, e01115‐18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Yang G, Li S, Blackmon S, Ye J, Bradley KC, Cooley J, Smith D, Hanson L, Cardona C, Steinhauer DA et al. (2013) Mutation tryptophan to leucine at position 222 of haemagglutinin could facilitate H3N2 influenza A virus infection in dogs. J Gen Virol 94, 2599–2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Collins PJ, Vachieri SG, Haire LF, Ogrodowicz RW, Martin SR, Walker PA, Xiong X, Gamblin SJ & Skehel JJ (2014) Recent evolution of equine influenza and the origin of canine influenza. Proc Natl Acad Sci USA 111, 11175–11180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Gambaryan AS, Matrosovich TY, Philipp J, Munster VJ, Fouchier RAM, Cattoli G, Capua I, Krauss SL, Webster RG, Banks J et al. (2012) Receptor‐binding profiles of H7 subtype influenza viruses in different host species. J Virol 86, 4370–4379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Broszeit F, Tzarum N, Zhu X, Nemanichvili N, Eggink D, Leenders T, Li Z, Liu L, Wolfert MA, Papanikolaou A et al. (2019) N‐glycolylneuraminic acid as a receptor for influenza A viruses. Cell Rep 27, 3284–3294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Gambaryan AS, Matrosovich TY, Boravleva EY, Lomakina NF, Yamnikova SS, Tuzikov AB, Pazynina GV, Bovin NV, Fouchier RAM, Klenk H‐D et al. (2018) Receptor‐binding properties of influenza viruses isolated from gulls. Virology 522, 37–45. [DOI] [PubMed] [Google Scholar]
- 83. Gilbertson B, Ng WC, Crawford S, McKimm‐Breschkin JL & Brown LE (2017) Mouse saliva inhibits transit of influenza virus to the lower respiratory tract by efficiently blocking influenza virus neuraminidase activity. J Virol 91, e00145‐17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Reid AH, Fanning TG, Janczewski TA & Taubenberger JK (2000) Characterization of the 1918 “Spanish” influenza virus neuraminidase gene. Proc Natl Acad Sci USA 97, 6785–6790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Reid AH, Fanning TG, Hultin JV & Taubenberger JK (1999) Origin and evolution of the 1918 “Spanish” influenza virus hemagglutinin gene. Proc Natl Acad Sci USA 96, 1651–1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Dortmans JCFM, Dekkers J, Wickramasinghe INA, Verheije MH, Rottier PJM, van Kuppeveld FJM, de Vries E & de Haan CAM (2013) Adaptation of novel H7N9 influenza A virus to human receptors. Sci Rep 3, 3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Shi Y, Zhang W, Wang F, Qi J, Wu Y, Song H, Gao F, Bi Y, Zhang Y, Fan Z et al. (2013) Structures and receptor binding of hemagglutinins from human‐infecting H7N9 influenza viruses. Science 342, 243–247. [DOI] [PubMed] [Google Scholar]
- 88. Neumann G & Kawaoka Y (2015) Transmission of influenza A viruses. Virology 479–480, 234–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Peacock THP, James J, Sealy JE & Iqbal M (2019) A global perspective on H9N2 avian influenza virus. Viruses 11, 620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Lai JCC, Karunarathna HMTK, Wong HH, Peiris JSM & Nicholls JM (2019) Neuraminidase activity and specificity of influenza A virus are influenced by haemagglutinin‐receptor binding. Emerg Microbes Infect 8, 327–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Lin YP, Gregory V, Collins P, Kloess J, Wharton S, Cattle N, Lackenby A, Daniels R & Hay A (2010) Neuraminidase receptor binding variants of human influenza A(H3N2) viruses resulting from substitution of aspartic acid 151 in the catalytic site: a role in virus attachment? J Virol 84, 6769–6781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Mögling R, Richard MJ, Vliet S, Beek R, Schrauwen EJA, Spronken MI, Rimmelzwaan GF & Fouchier RAM (2017) Neuraminidase‐mediated haemagglutination of recent human influenza A(H3N2) viruses is determined by arginine 150 flanking the neuraminidase catalytic site. J Gen Virol 98, 1274–1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Smith DJ, Lapedes AS, de Jong JC, Bestebroer TM, Rimmelzwaan GF, Osterhaus ADME & Fouchier RAM (2004) Mapping the antigenic and genetic evolution of influenza virus. Science 305, 371–376. [DOI] [PubMed] [Google Scholar]
- 94. Koel BF, Burke DF, Bestebroer TM, van der Vliet S, Zondag GCM, Vervaet G, Skepner E, Lewis NS, Spronken MIJ, Russell CA et al. (2013) Substitutions near the receptor binding site determine major antigenic change during influenza virus evolution. Science 342, 976–979. [DOI] [PubMed] [Google Scholar]
- 95. Lin YP, Xiong X, Wharton SA, Martin SR, Coombs PJ, Vachieri SG, Christodoulou E, Walker PA, Liu J, Skehel JJ et al. (2012) Evolution of the receptor binding properties of the influenza A(H3N2) hemagglutinin. Proc Natl Acad Sci USA 109, 21474–21479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Peng W, de Vries RP, Grant OC, Thompson AJ, McBride R, Tsogtbaatar B, Lee PS, Razi N, Wilson IA, Woods RJ et al. (2017) Recent H3N2 viruses have evolved specificity for extended, branched human‐type receptors, conferring potential for increased avidity. Cell Host Microbe 21, 23–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Gulati S, Smith DF, Cummings RD, Couch RB, Griesemer SB, St. George K, Webster RG & Air GM (2013) Human H3N2 influenza viruses isolated from 1968 to 2012 show varying preference for receptor substructures with no apparent consequences for disease or spread. PLoS One 8, e66325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Hensley SE, Das SR, Bailey AL, Schmidt LM, Hickman HD, Jayaraman A, Viswanathan K, Raman R, Sasisekharan R, Bennink JR et al. (2009) Hemagglutinin receptor binding avidity drives influenza A virus antigenic drift. Science 326, 734–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Jiang L, Fantoni G, Couzens L, Gao J, Plant E, Ye Z, Eichelberger MC & Wan H (2016) Comparative efficacy of monoclonal antibodies that bind to different epitopes of the 2009 pandemic H1N1 influenza virus neuraminidase. J Virol 90, 117–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Rijal P, Wang BB, Tan TK, Schimanski L, Janesch P, Dong T, McCauley JW, Daniels RS, Townsend AR & Huang K‐YA (2020) Broadly inhibiting antineuraminidase monoclonal antibodies induced by trivalent influenza vaccine and H7N9 infection in humans. J Virol 94, e01182‐19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Tulip WR, Varghese JN, Laver WG, Webster RG & Colman PM (1992) Refined crystal structure of the influenza virus N9 neuraminidase‐NC41 Fab complex. J Mol Biol 227, 122–148. [DOI] [PubMed] [Google Scholar]
- 102. Malby RL, Tulip WR, Harley VR, McKimm‐Breschkin JL, Laver WG, Webster RG & Colman PM (1994) The structure of a complex between the NC10 antibody and influenza virus neuraminidase and comparison with the overlapping binding site of the NC41 antibody. Structure 2, 733–746. [DOI] [PubMed] [Google Scholar]
- 103. Gao J, Couzens L, Burke DF, Wan H, Wilson P, Memoli MJ, Xu X, Harvey R, Wrammert J, Ahmed R et al. (2019) Antigenic drift of the influenza A(H1N1)pdm09 virus neuraminidase results in reduced effectiveness of A/California/7/2009 (H1N1pdm09)‐specific antibodies. mBio 10, e00307‐19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Sakai T, Nishimura SI, Naito T & Saito M (2017) Influenza A virus hemagglutinin and neuraminidase act as novel motile machinery. Sci Rep 7, 45043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Vahey MD & Fletcher DA (2019) Influenza A virus surface proteins are organized to help penetrate host mucus. eLife 8, e43764. [DOI] [PMC free article] [PubMed] [Google Scholar]


