Abstract
Telomere length control is influenced by several factors, including telomerase, the components of telomeric chromatin structure, and the conventional replication machinery. Although known components of the replication machinery can influence telomere length equilibrium, little is known about why mutations in certain replication proteins cause dramatic telomere lengthening. To investigate the cause of telomere elongation in cdc17/pol1 (DNA polymerase α) mutants, we examined telomeric chromatin, as measured by its ability to repress transcription on telomere-proximal genes, and telomeric DNA end structures in pol1-17 mutants. pol1-17 mutants with elongated telomeres show a dramatic loss of the repression of telomere-proximal genes, or telomeric silencing. In addition, cdc17/pol1 mutants grown under telomere-elongating conditions exhibit significant increases in single-stranded character in telomeric DNA but not at internal sequences. The single strandedness is manifested as a terminal extension of the G-rich strand (G tails) that can occur independently of telomerase, suggesting that cdc17/pol1 mutants exhibit defects in telomeric lagging-strand synthesis. Interestingly, the loss of telomeric silencing and the increase in the sizes of the G tails at the telomeres temporally coincide and occur before any detectable telomere lengthening is observed. Moreover, the G tails observed in cdc17/pol1 mutants incubated at the semipermissive temperature appear only when the cells pass through S phase and are processed by the time cells reach G1. These results suggest that lagging-strand synthesis is coordinated with telomerase-mediated telomere maintenance to ensure proper telomere length control.
Telomeres, consisting of simple, tandem DNA repeats and associated proteins, are located at the ends of linear eukaryotic chromosomes, where they ensure chromosome stability and integrity and facilitate the completion of DNA replication (reviewed in references 7, 25, and 61). Telomeres contribute to the overall stability of the genome by protecting chromosomes from the exonucleolytic degradation and end-to-end fusions that are often associated with broken chromosome ends (40). In addition to functioning as protective caps on chromosome ends, telomeres contribute to the faithful completion of DNA replication because of the unique mechanism of telomere synthesis. Due to the incomplete replication of chromosome ends that occurs because DNA polymerases require an RNA primer, there remains a terminal single-stranded gap that cannot be filled by conventional polymerases (57). Without a mechanism to balance this loss of DNA sequence, telomere sequences would be progressively lost until chromosomes become unstable, causing cell death. Since telomeres are replicated by telomerase, which does not require an RNA primer or a DNA template, telomere synthesis prevents the gradual loss of DNA sequence that would otherwise occur during each round of replication (reviewed in references 7, 25, and 61). Telomerase, telomeric chromatin structure, and the replication complex all contribute to the maintenance of an equilibrium telomere length, which is important for ensuring proper chromosome function.
Telomerase plays an obvious role in maintaining telomere length; it is a ribonucleoprotein complex that adds newly synthesized telomeric sequences onto the ends of the 5′-to-3′ (G-rich) strands after the completion of bulk DNA synthesis. This addition of telomeric DNA is followed by fill-in synthesis of the complementary C-rich strand by the conventional replication machinery (25). Telomerase is an RNA-directed DNA polymerase, or reverse transcriptase (17, 35), that uses its RNA component as a template for the addition of new telomeric sequences onto chromosome ends. Yeast strains carrying a deletion of the TLC1 gene, which encodes the RNA component of telomerase, show progressive telomere shortening, which ultimately leads to a loss of cell viability (53). A similar phenotype is caused by mutations in the catalytic subunit of telomerase, encoded by the EST2 gene (17, 33, 35). The inexorable telomere shortening caused by perturbations in telomerase components indicates that telomerase function is vital for telomere length maintenance.
Many reports suggest that alterations in chromatin structure at telomeres can also affect telomere length equilibrium. A unique telomeric chromatin structure called the telosome is formed by a complex of telomere-binding proteins and telomeric DNA in Saccharomyces cerevisiae (60). Examination of the sensitivity of telomeric chromatin to nucleases revealed that the telosome is protected by a complex of proteins, including Rap1p, which is different from the nucleosomal complexes that protect the subtelomeric regions (60). Genes that are artificially inserted adjacent to this telosome are subject to reduced transcriptional expression, known as the telomere position effect (TPE) or telomeric silencing (23). Mutations in several chromatin components in yeast, such as Rap1p, Sir2p, Sir3p, and Sir4p, cause the loss of TPE (2, 30). Some of these mutations also cause telomere length changes (30, 37), suggesting that chromatin components are important for controlling telomere length. In particular, it has recently been shown that the number of Rap1p-binding sites is directly correlated with telomere length (39, 49).
Components of the replication machinery also appear to be involved in the control of telomere length. In 1985, Carson and Hartwell first determined that mutations in the structural gene for DNA polymerase α, encoded by the CDC17/POL1 gene, cause telomeres to lengthen by several kilobases compared to telomeres in wild-type strains (15). Our later studies revealed that telomere lengthening can also result from mutations in the gene encoding the large subunit of replication factor C, CDC44/RFC1 (1). Strains carrying temperature-sensitive mutations in either CDC17/POL1 or CDC44/RFC1 have slightly elongated telomeres at permissive temperature but display greatly elongated telomeres after growth at the semipermissive temperature (1, 15). Furthermore, telomere lengthening in these mutants is correlated with defects in DNA replication (1). However, other mutations affecting DNA replication do not show this effect. These observations suggest that Cdc17p/Pol1p and Cdc44p/Rfc1p may play a functional role in telomere length control.
To determine why mutations in certain DNA replication proteins cause telomeres to lengthen, we examined the possibility that telomere elongation in cdc17/pol1 mutants is caused by structural defects at the telomeres. Since a mutant polymerase is likely to cause defects in the DNA itself, we hypothesized that cdc17/pol1 mutants have DNA defects that perturb the overall structure of telomeric chromatin. The consequence of this perturbation may be an increased accessibility of telomerase to the telomere substrates or an alteration in the regulation of telomerase. This study reveals that temperature-sensitive DNA polymerase α mutants grown at the semipermissive temperature, at which telomere elongation occurs, show a striking reduction in TPE compared to wild-type cells. This loss of TPE is correlated with a significant increase in the amount of single-stranded DNA (ssDNA) character at the telomeres but not at internal sequences. Furthermore, the generation of excess telomeric ssDNA, or G tails, can occur in the absence of telomerase but is still cell cycle regulated. These results suggest that the telomere elongation in DNA polymerase α mutants may be caused by alterations in telomeric chromatin structure that derive from defective fill-in synthesis on the lagging (C-rich) strand during S phase.
MATERIALS AND METHODS
Yeast strains and media.
Yeast strains used in this study are listed in Table 1. The URA3 gene was placed at chromosome VII-L telomeres in strains CH2377 and CH2378 by transforming strains CH2146 and CH2147 (1), respectively, with the EcoRI-SalI fragment of pVII-L URA3-TEL (kindly provided by D. Gottschling), as described previously (23). Southern hybridization analysis was used to confirm the integration of the URA3 gene at the telomere. The ura3 gene was deleted in strains CH2514 and CH2515 by replacing the entire ura3 coding region with the Escherichia coli kan gene (55) in strains CH2146 and CH2148 (1), respectively; integrants were selected on yeast extract-peptone (YEP) agar containing Geneticin (200 mg/liter; Gibco-BRL) (55). Insertion of the kan gene at the deleted ura3 locus was confirmed both by PCR analysis and by the absence of a ura3 RNA transcript on Northern blots. Additionally, the URA3 gene was placed at chromosome VII-L telomeres in strains CH2514 and CH2515, as described above. Strain RWY120 (cdc17-1 tlc1Δ) was generated by crossing strain CH2248 (1) with RWY12, a tlc1 deletion strain. As a cdc17-1 control, strain RWY120 (cdc17-1 tlc1Δ) carrying the TLC1 gene on a CEN plasmid (pAZ1), was used (5). The entire coding region of the BAR1 gene was replaced by the E. coli kan gene in strain CH2146 to yield strain RWY121. The bar1 deletion fragment was generated by using plasmid pRS400 (11) and primers Bar1f (5′-CGAGGTTCGCGATTATTAACACCATTACGTCTTTAACAAAAGATTGTACGTAGAGTGCAC-3′) and Bar1r (5′-TCGTGACAGTTTCTGTTATGAGCTGTCTTATGAGTAGGCCGCTGTGCGGTATTTCACACCG-3′).
TABLE 1.
S. cerevisiae strains used in this study
| Strain | Genotype | Source or reference |
|---|---|---|
| CH557a | MATα ade6 ade2 ura1 lys2 tyr1 gal1 his7 cdc28 | 26 |
| CH2146b | MATa ura3 trp1-289 ade2-101 tyr1 gal2 can1 his pol1-17 | 1 |
| CH2147b | MATα ura3 trp1-289 ade2-101 tyr1 gal2 can1 his POL1 | 1 |
| CH2148b | MATa ura3 trp1-289 ade2-101 tyr1 gal2 can1 his POL1 | 1 |
| CH2248c | MATα ura1 leu2 his7 ura3-52 cdc17-1 | 1 |
| CH2377d | MATa ura3 trp1-289 ade2-101 tyr1 gal2 can1 his pol1-17 URA3/VII-L-TEL | This study |
| CH2378d | MATα ura3 trp1-289 ade2-101 tyr1 gal2 can1 his POL1 URA3/VII-L-TEL | This study |
| CH2493de | MATa ura3 trp1-289 ade2-101 tyr1 gal2 can1 his pol1-17 | This study |
| CH2494de | MATa ura3 trp1-289 ade2-101 tyr1 gal2 can1 his POLI | This study |
| CH2514d | MATa ura3Δ::KAN trp1-289 ade2-101 tyr1 gal2 can1 his pol1-17 URA3/VII-L-TEL | This study |
| CH2515d | MATa ura3Δ::KAN trp1-289 ade2-101 tyr1 gal2 can1 his POL1 URA3/VII-L-TEL | This study |
| RWY12 | MATa ura3-52 lys2-801 ade2-101 trp1-Δ63 his3-Δ200 leu2-Δ1 VR-ADE2-TEL tlc1Δ::LEU2 | 53 |
| RWY120 | MATa ura3-52 lys2-801 leu2 cdc17-1 tlc1Δ::LEU2 | This study |
| RWY121d | MATa ura3 trp1-289 ade2-101 try1 gal2 can1 his pol1-17 bar1Δ::KAN | This study |
This strain has an A364A genetic background.
These strains are equivalent to strains SS111 or SS111-17 (4); they carry an unidentified his auxotrophy.
This strain is a mixture of S288C and A364A genetic backgrounds.
These strains are derivatives of strains SS111 or SS111-17 (14); they carry an unidentified his auxotrophy.
These strains carry the E. coli kan gene in an unidentified location.
Strains were grown at the specified temperatures and in standard yeast extract-peptone-dextrose (YEPD) or synthetic minimal medium supplemented with amino acids (52). For TPE assays, synthetic minimal medium was supplemented with 0.02 mg of uracil per ml, 0.02 mg of adenine per ml, 0.02 mg of histidine per ml, 0.03 mg of tyrosine per ml, and 0.02 mg of tryptophan per ml to generate synthetic complete (SC) plates. 5-Fluoroorotic acid (5FOA) plates consisted of synthetic minimal medium supplemented with 0.75 mg of 5FOA (American Biorganics Inc.) per liter, 0.05 mg of uracil per ml, 0.02 mg of adenine per ml, 0.02 mg of histidine per ml, 0.03 mg of tyrosine per ml, and 0.02 mg of tryptophan per ml.
Telomeric silencing.
To examine telomeric silencing in pol1-17 mutant cells with elongated telomeres, strains CH2378 (POL1) and CH2377 (pol1-17) were grown to stationary phase (approximately 10 generations of growth) at the permissive temperature (24°C) and diluted 1:1,000 into fresh YEPD and samples were incubated at 24 or 30°C. After the cultures reached stationary phase, each was diluted 1:1,000 once again and then incubated at the previous temperature. This process was repeated twice to allow mutant cells to grow for approximately 40 generations at 30°C so as to allow telomeres to elongate (15). TPE was assayed in these cultures by the method described by Gottschling et al. (23). Cell density was determined for the final stationary cultures, and the cells from each culture were plated onto SC plates and incubated at the previous temperatures. After 5 days of growth at 24°C or 4 days of growth at 30°C, colonies were picked from the SC plates, resuspended in 1.0 ml of double-distilled H2O, and plated at a density of 300 cells per plate onto YEPD, SC, SC−uracil, and 5FOA plates. In addition, mutant cells were plated onto 5FOA plates at densities of 3,000 cells and 106 cells per plate. Since 5FOA is a toxic analog of uracil, uptake of 5FOA is lethal for URA3 cells whereas ura3 cells are viable in the presence of 5FOA (8). Thus, the percent silencing is calculated as the ratio of the number of cells capable of forming colonies on 5FOA to the total number of colony-forming cells on complete medium.
Northern blot analysis.
Total RNA was isolated by a phenol-freeze method (51). Standard techniques were used for gel electrophoresis, RNA transfer, and hybridization of RNA samples (20 μg/lane) (56); Quick-Hyb hybridization solution (Stratagene) was used for the hybridizations of the RNA immobilized onto Hybond-N nylon membrane (Amersham). Expression of HML in strains CH2493 (pol1-17) and CH2494 (POL1) was examined with a PCR-generated 746-bp fragment containing the entire Yα segment of the α cassette, which recognizes both the α1 and α2 genes from the HML locus (4); the PCR primers used were 5′-CTCATCTGTGATTTGTGGAT-3′ and 5′-AAGTAGTCCCATATTCCGTG-3′. A 571-bp fragment containing the entire ACT1 coding sequence (generated by PCR with primers 5′-CGATTTGGCCGGTAGAGATT-3′ and 5′-TAGATGGACCACTTTCGTCG-3′) was used to control for RNA loading. The probes were labeled with [α-32P]ATP by random priming (DECAprime; Ambion). For analysis of telomeric URA3 expression, strains CH2514 (pol1-17) and CH2515 (POL1) were grown to early logarithmic phase at 24°C, cultures were split, and aliquots were shifted to 24 or 30°C for 2 or 5 h. The URA3 probe, which was labeled as described above, is a 911-bp NdeI-NsiI DNA fragment of pCH1099, containing the entire coding region of the URA3 gene. Quantitation was performed with a Molecular Dynamics STORM PhosphorImager and ImageQuaNT software.
Responsiveness to α-factor.
Responsiveness to α-factor was examined by using single-cell shmoo assays (20). Strain CH2494 (POL1) was grown at 24 or 30°C, strain CH2493 [pol1-17(short)] was grown at 24°C, and strain CH2493 [pol1-17(long)] was grown at 30°C. The strains were grown to logarithmic phase at the indicated temperatures, and α-factor was added as described previously (20). Aliquots of cells were collected at zero time and 3 h after the addition of α-factor and fixed with formaldehyde. For each strain, the cell morphology of individual formaldehyde-fixed cells was examined to quantitate the number of shmooed, unbudded, budded, and dumbbell-shaped cells. On average, 98% of pol1 cells at 30°C, 84% of pol1-17(short) cells at 24°C, and 94% of pol1-17(long) cells at 30°C formed shmoos following α-factor treatment.
In-gel nondenaturing hybridization.
Strains CH2378 (POL1) and CH2377 [pol1-17(short)] were grown to early logarithmic phase at 23°C and shifted to 30°C. At each hourly time point, genomic DNA was isolated, digested with XhoI, and subjected to gel electrophoresis under nondenaturing conditions as previously described (18). In-gel hybridization analysis of single-stranded telomeric DNA was performed with a single-stranded 22-mer CA oligonucleotide probe or a 22-mer single-stranded oligonucleotide TG probe (18). The Y′-specific probe was generated as described previously (18). To confirm that the lanes in the nondenaturing gels were loaded equally, nondenaturing gels were denatured, neutralized, and reprobed with a telomeric or Y′ probe under regular conditions.
Cell cycle synchrony.
To produce a synchronously dividing culture, a pol1-17 bar1Δ strain (RWY121) growing exponentially at 23°C in YEPD was initially treated with 0.2 μM α-factor to arrest the cells in G1. After 11 h at 23°C, the culture was split into two portions: one portion remained at 23°C for an additional 1 h, and the other portion was shifted to 30°C for 1 h to ensure that the cells were at 30°C before the removal of α-factor. The cells remained at these temperatures for the remainder of the experiment. To release cells into the cell cycle, the 23 and 30°C portions were each split into two aliquots; α-factor was retained in one aliquot as a control, and it was removed from the other aliquot by centrifuging and washing the cells and reincubating them in YEPD without α-factor and containing pronase (100 μg/ml). Following the removal of α-factor, samples of cells were collected at the times indicated in Fig. 7 and processed for DNA and fluorescence-activated cell sorting (FACS) analysis. The appearance of telomeric ssDNA was examined by in-gel nondenaturing hybridization (described above), and cell cycle progression was assessed by FACS as described previously (24).
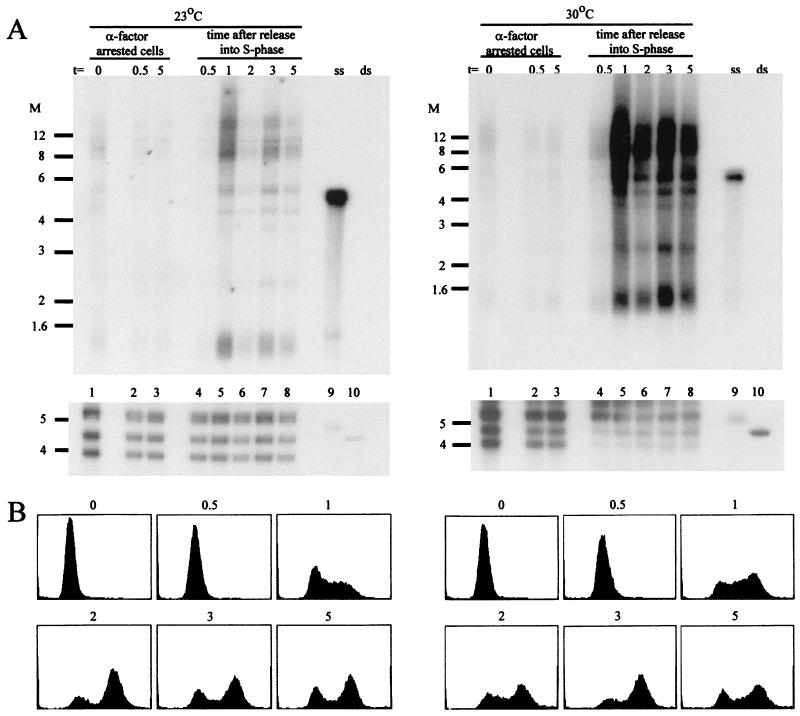
FIG. 7.
The appearance of telomeric ssDNA occurs during S phase in pol1-17 cells at 30°C. (A) In-gel hybridization of DNA derived from pol1-17 cells arrested in G1 (lanes 1 to 3) and released into a synchronous cell cycle (lanes 4 to 8) at 23°C (left panels) and 30°C (right panels). Exponentially growing cells of strain RWY121 (pol1-17 bar1Δ) were treated with α-factor for 11 h at 23°C to arrest the cells in the G1 phase. The culture was then split into two portions, which were incubated at 23 or 30°C for the remainder of the experiment. The portions were split into two aliquots after 1 h (t = 0, lanes 1), and the cells of one aliquot were kept in G1 with α-factor (lanes 2 and 3) while the other aliquot was released into the cell cycle by α-factor removal (lanes 4 to 8). Samples of cells were collected from all four aliquots at 0.5 h (lanes 2 and 4), 1 h (lanes 5), 2 h (lanes 6), 3 h (lanes 7), and 5 h (lanes 3 and 8) after t = 0. These aliquots were used for DNA isolation and analysis of the DNA end structures by nondenaturing in-gel hybridization (as described in the legend to Fig. 3) (A); they also were used for FACS analysis of DNA content (B). Hybridization under denaturing conditions demonstrates that all lanes were approximately evenly loaded (lower panels in panel A). Lanes 9 and 10 contain control single-stranded and double-stranded TG1–3 DNA, respectively. Note that for the cultures remaining in G1, only the DNA isolated after 0.5 and 5 h is shown. The DNA isolated from cells harvested at the time points in between those two times yielded indistinguishable very weak hybridization in the nondenaturing gels (data not shown). Similarly, only the FACS profiles of cells at t = 0 and cells collected after the release into the cell cycle are shown; the profiles of the cells remaining at G1 were indistinguishable from those at t = 0 (data not shown). pol1-17 cells released into the cell cycle at 30°C display an excess of ssDNA that initially appears only when the cells enter S phase. Molecular weight (M) markers (in thousands) are indicated on the left of each panel.
RESULTS
TPE is greatly reduced in pol1-17 mutants with elongated telomeres.
To determine if cdc17/pol1 mutants exhibit alterations in telomeric chromatin structure, we examined the extent of telomeric silencing in pol1-17 cells with elongated telomeres. Telomeric silencing reflects the ability of the telosome to repress the transcription of telomere-proximal genes (23), and alterations in the protein components of telomeric chromatin can cause the derepression of genes adjacent to the telomere (16, 27, 41, 44). Thus, an alteration in telomeric chromatin structure could be reflected by a loss of telomeric silencing in pol1-17 cells with fully elongated telomeres. Since telomere elongation occurs progressively in pol1 mutants, pol1-17 cultures must be grown for many generations at the semipermissive temperature (30°C) for dramatic telomere lengthening to occur. In these studies, pol1-17 mutants were grown initially at 24°C, where telomeres are only slightly elongated [hereafter called pol1-17(short)] and then shifted to 30°C. After approximately 40 generations of growth at 30°C, telomeres are fully elongated [hereafter called pol1-17(long)]. The ability to silence a telomere-proximal URA3 gene was assessed by determining if POL1, pol1-17(short), and pol1-17(long) cells bearing URA3 at the telomere are capable of growth in the presence of 5FOA. Telomeric silencing of URA3 allows cells to grow on 5FOA plates, while loss of telomeric repression prevents cell growth on 5FOA. Thus, the level of telomeric silencing is determined by comparing the number of viable colonies on 5FOA with the total number of colony-forming cells on complete medium (23).
pol1-17(long) cells displayed a striking loss of telomeric silencing (Table 2). In POL1 colonies at 24 and 30°C and pol1-17(short) colonies, 85 to 90% of the cells were capable of telomeric silencing (Table 2). In sharp contrast, pol1-17(long) cells showed a dramatic decrease in the repression of the telomeric URA3 gene (URA3 was silenced in only 3.5 in 105 cells). Interestingly, the loss of TPE in pol1-17 mutants at 30°C was also observed in pol1-17 cultures that had been incubated at the semipermissive temperature for only 10 generations of growth (8.3 in 104 cells were silenced), well before telomeres were fully elongated (data not shown). These results suggest that the elongating telomeres in pol1-17 mutants have an altered chromatin structure that prevents the repression of telomere-proximal genes.
TABLE 2.
Loss of telomeric silencing in pol1-17 strains at 30°Ca
| Genotype | Conditions | Fraction that is 5FOARb |
|---|---|---|
| POL1 | 24°C | 0.65 |
| POL1 | 30°C | 0.86 |
| pol1-17(short) | 24°C | 0.76 |
| pol1-17 | 30°C, 10 generations | 8.3 × 10−4 |
| pol1-17(long) | 30°C | 3.5 × 10−5 |
TPE was examined in strains CH2378 (POL1) grown in YEPD liquid at 24 or 30°C, CH2377 [pol1-17(short)] grown at 24°C, CH2377(Pol1-17) grown at 30°C for 10 generations, and CH2377 [pol1-17(long)] grown at 30°C.
The percent TPE is calculated as a ratio of the number of cells capable of forming colonies on 5FOA to the total number of colony-forming cells on rich medium. The average of two independent trials is presented.
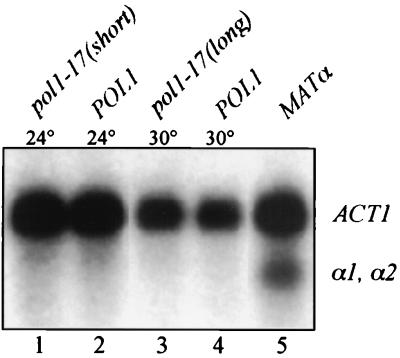
To determine if the loss of TPE is due to a specific defect at the telomeres rather than a generalized defect along the entire length of the chromosome, we examined the level of silencing at the cryptic mating-type HML locus. Since telomeric silencing requires many of the same proteins that are required for silencing at HML (2, 13), loss of silencing at both locations would reflect a nonspecific silencing defect. We examined the level of silencing at the HML locus by determining whether the HML α1 and α2 mating-type information genes are transcribed. Total RNA was isolated from POL1, pol1-17(short), and pol1-17(long) strains and then subjected to Northern hybridization with a probe that recognizes both the α1 and α2 transcripts, which comigrate on an agarose gel (28, 46) (Fig. 1). Although transcripts were easily visualized in a control MATα strain in which α1 and α2 are expressed (Fig. 1, lane 5), no hybridization to α1 and α2 transcripts was detected in RNA from POL1 (lanes 2 and 4), pol1-17(short) (lane 1), or pol1-17(long) (lane 3) cells. These data are consistent with results obtained from single-cell shmoo assays to examine the repression of the silent mating loci (see Materials and Methods) (data not shown). These results suggest that the HML locus remains silenced in pol1-17 mutants with elongated telomeres.
FIG. 1.
Loss of silencing in pol1-17 mutants does not occur at HML. Total RNA was isolated from strains CH2494 (MATaPOL1) at 24 and 30°C, CH2493 [MATapol1-17(short)], and CH2493 [MATapol1-17(long)]. Expression of MATα information from the HML locus was examined by Northern hybridization analysis of MATa strains with a probe that recognizes both the α1 and α2 transcripts, which comigrate on an agarose gel. Expression of the α1 and α2 transcripts is repressed to the same extent in POL1 cells (lanes 2 and 4), pol1-17(short) cells (lane 1), and pol1-17(long) cells (lane 3). As a positive control, RNA also was isolated from strain CH557 (MATα POL1) (lane 5). To monitor RNA loading in each lane, actin mRNA levels were measured by hybridization with an ACT1 probe. A longer exposure of this gel also fails to show any α1 or α2 transcripts in pol1-17(long) cells (lane 3).
Thus, there is a cis-acting defect associated with the telomeres that causes the loss of silencing specifically at the telomeres. This defect appears after less than 10 generations of growth, well before the telomeres are fully elongated (data not shown).
Loss of TPE occurs rapidly in pol1-17 mutants upon shift to 30°C.
Because TPE appears to be lost before telomeres fully elongate, it is possible that structural alterations at the telomere are the cause of telomere lengthening in cdc17/pol1 cells. To more closely examine the kinetics of loss of TPE compared to the kinetics of increase in telomere length, we used Northern analysis to examine telomeric URA3 expression immediately after shifting pol1-17 and POL1 cultures to the semipermissive temperature (30°C). To confirm that the results from the Northern experiments reflect the same physiological state as the TPE experiments, we first examined the levels of URA3 mRNA in pol1-17(long) cells that exhibit a loss of telomeric silencing (Fig. 2A). Northern analysis indicated that the telomeric URA3 gene was strongly expressed in pol1-17(long) cells (Fig. 2A, lane 2) whereas URA3 expression was undetectable in pol1-17(short) cells (lane 1) and POL1 cells grown at either 24 or 30°C (lanes 3 and 4, respectively). Thus, Northern analysis and TPE experiments yielded consistent results.
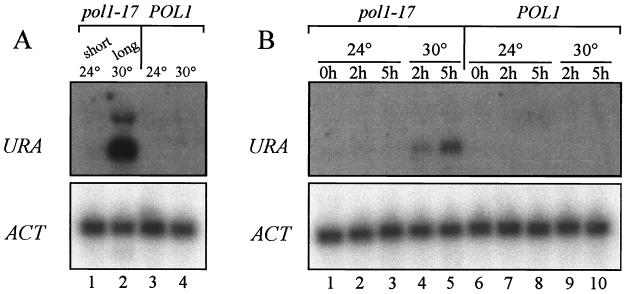
FIG. 2.
A telomere-proximal URA3 gene is rapidly derepressed in pol1-17 mutants upon the shift to 30°C. (A) Telomeric URA3 expression was examined in strains CH2514 [pol1-17(short)] (lane 1), CH2514 [pol1-17(long)] (lane 2), and CH2515 (POL1) grown at either 24°C (lane 3) or 30°C (lane 4). pol1-17(long) cells exhibit a dramatic loss of telomeric URA3 repression. (B) Strains CH2514 [pol1-17(short)] and CH2515 (POL1) were grown to early log phase at 24°C, and aliquots were shifted to 24 or 30°C for 2 or 5 h. URA3 derepression in pol1-17 cells occurs by 2 to 5 h after the shift to 30°C (compare lane 1 with lanes 4 and 5). For each sample in panels A and B, RNA was isolated and the expression of a telomere-proximal URA3 gene was examined by Northern hybridization with a URA3 probe. To monitor RNA loading in each lane, actin transcript levels were measured by hybridization with an ACT1 probe.
To determine if telomeric URA3 repression is lost before or after telomere elongation occurs in pol1-17 mutants, cultures of POL1 and pol1-17(short) cells were grown to early logarithmic phase at 24°C, the cultures were split, and aliquots were shifted to 30 or 24°C for 2 or 5 h. At each time point, RNA was isolated to measure URA3 transcript levels. Northern hybridization of RNA from temperature-shifted strains revealed that derepression of the telomeric URA3 gene in pol1-17 strains was detectable by 2 h after the shift to 30°C (Fig. 2B, lane 4); a higher level of expression was detected by 5 h after the shift (lane 5). These data suggest that TPE is lost progressively in pol1-17 strains upon the shift to 30°C. Southern hybridization to genomic DNA revealed that telomere lengthening in pol1-17 mutants was first detectable by 5 to 6 h after the shift to 30°C (see Fig. 4B, compare the telomeric DNA smear in lanes 2 and 3; see Fig. 3B, compare lane 1 with lanes 2 to 6). Since telomeric URA3 expression occurred before any detectable changes in telomere length in pol1-17 cells, alterations in telomeric chromatin structure, as measured by URA3 derepression, may be a cause of telomere elongation. Of course, we cannot rule out the possibility that changes in TPE occur gradually as telomeres lengthen. Regardless of the kinetics, however, it is clear that telomere lengthening is accompanied by alterations of chromatin structure, as evidenced by the loss of TPE.
FIG. 4.
The increase in the level of telomeric ssDNA in pol1-17 cells is reversible. Strain CH2377 [pol1-17(short)], grown to early log phase at 23°C, was shifted to 30°C, and aliquots of cells were collected after 1 and 5 h of growth. Subsequently, the culture of pol1-17 cells was shifted back down to 23°C and aliquots again were collected after 1 and 5 h of growth. DNA isolated at each time point was digested with XhoI and analyzed as described in the legend to Fig. 3. (A) Nondenaturing in-gel hybridization with a CA-oligonucleotide probe reveals that the ssDNA that appears at 30°C (lanes 2 and 3) is lost immediately after the cells are shifted back to 23°C (lanes 4 and 5). (B) Stripping, denaturing, and reprobing of the native gel in panel A with a Y′ probe indicates that the lanes are approximately evenly loaded. Lanes 6, 7, and 8 contain control double-stranded TG1–3 DNA, single-stranded TG1–3 DNA, and Y′ DNA, respectively. Because a Y′ probe was used for this hybridization, the pattern of bands after denaturation differs in appearance from those in other figures. The smear of Y′-containing telomeric DNA is indicated by an arrow in panel B. Molecular weight (MW) markers (in thousands) are indicated in the middle.
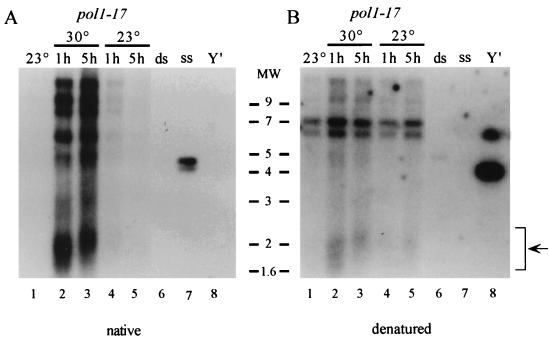
FIG. 3.
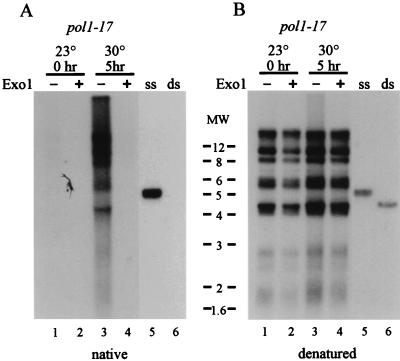
pol1-17 mutants quickly exhibit single-stranded character at the telomeres upon the shift to 30°C. Strains CH2377 (pol1-17) and CH2378 (POL1) were grown to early log phase at 23°C, and aliquots were shifted to 30°C for various times. DNA isolated at each time point was digested with XhoI, which releases terminal telomeric fragments on Y′-containing telomeres, and analyzed by nondenaturing in-gel hybridization (A) and denaturing hybridization (B) with a CA oligonucleotide probe. Hybridization to high-molecular-weight DNA represents non-Y′-containing telomeres. ssDNA on the native gel is observed by 1 h after the shift to 30°C (lane 2), and its level increases by 2 h after the shift to 30°C (lane 3). Stripping and reprobing the gel under denaturing conditions demonstrates that the lanes were approximately evenly loaded. Lanes 15 and 16 contain control double-stranded and single-stranded TG1–3 DNA, respectively. The smear of Y′-containing telomeric DNA from pol1-17 cells is indicated by an arrow in panel B. Molecular weight markers (in thousands) are indicated on the left.
pol1-17 mutants rapidly exhibit increased single-stranded character at the telomeres upon shift to 30°C.
To explain the alteration in chromatin structure in pol1-17 mutants, we reasoned that a DNA polymerase mutant is likely to display defects in the telomeric DNA itself. Increased single-stranded character at the telomeres recently has been observed to result from mutations in the CDC13 and Ku genes, which encode proteins that affect telomere metabolism (22, 24). Furthermore, strains with the RAD27 gene deleted also display increased single strandedness at telomeres (45). To determine if pol1 mutants exhibit increased single strandedness at the telomeres at times when telomeric URA3 derepression is observed, we examined telomeric DNA by a nondenaturing in-gel hybridization technique, in which DNA is hybridized within an agarose gel without denaturation of the DNA (18). The presence of ssDNA in pol1-17(short) cells initially grown at 23°C and then shifted to 30°C for various times was examined (Fig. 3). Interestingly, single-strandedness in pol1-17 strains grown at 30°C was detectable by 1 h after the shift to 30°C (Fig. 3A, lane 2) and increased even further by 2 h after the temperature shift (lane 3).
To determine if the single strandedness that appears at 30°C is reversible, a logarithmically growing culture of pol1-17 cells was shifted from 23 to 30°C and DNA was isolated after 1 and 5 h of growth; the culture was then shifted back to 23°C, and DNA was again collected after 1 and 5 h of growth. Examination of ssDNA in the collected samples reveals that the ssDNA signal that appeared at 30°C (Fig. 4A, lanes 2 and 3) was mostly lost after the cells were returned to 23°C for just 1 h of growth (lane 4); even less signal was present after 5 h of growth at 23°C (lane 5). Thus, the increase in single-stranded character that appears rapidly upon the shift to 30°C is lost just as quickly when the cells are shifted back to 23°C. This result indicates that the DNA defects generated at the telomere in pol1-17 cells at 30°C are reversible.
The significance of the single strandedness in pol1-17 mutants grown at 30°C was investigated by examining the nature of the ssDNA. To determine if single strandedness is specific to telomeric DNA or is a general chromosomal defect, nondenaturing in-gel hybridization was performed with probes to DNA located internally on the chromosome. Although hybridization to control ssDNA was observed, no hybridization occurred with probes to Y′ DNA or rDNA (data not shown), suggesting that single strandedness is not detectable in these regions of the chromosome. Furthermore, a TG oligonucleotide probe failed to hybridize with non-denatured DNA, which suggests that the increased single strandedness is due to defects on the C-rich lagging strand (data not shown). In addition, to determine if the single strandedness is terminal, the ssDNA in pol1-17 mutants was treated with E. coli exonuclease I (ExoI), an ssDNA-specific exonuclease that degrades DNA from the 3′ end (32). ExoI treatment resulted in loss of hybridization of a CA probe to pol1-17 telomeric DNA (Fig. 5A, lane 4), suggesting that the single strandedness is located in terminal sequences rather than being caused by the presence of internal gaps or nicks. Finally, these single-stranded G-strand extensions occur in a cell cycle-regulated fashion in pol1-17 cells, even at 30°C (see below). Although we cannot exclude the possibility that they differ from ssDNA extensions that occur at the telomeres in wild-type cells, they share the defining characteristics of G tails. Thus, we will refer to them as G tails.
FIG. 5.
Telomeric ssDNA in pol1-17 cells is removed by ExoI. CH2377 [pol1-17(short)] was grown to early log phase at 23°C and shifted to 30°C for 5 h. DNA was isolated before and after the temperature shift and incubated in the presence (lanes 2 and 4) or absence (lanes 1 and 3) of ExoI. The DNA was digested with XhoI and examined by nondenaturing in-gel hybridization (A) and denaturing hybridization (B) with a CA-oligonucleotide probe, as described in the legend to Fig. 3. Telomeric ssDNA in pol1-17 cells is lost following treatment with ExoI (compare lanes 3 and 4 in panel A). Lanes 5 and 6 contain control single-stranded and double-stranded TG1–3 DNA, respectively. Molecular weight (MW) markers (in thousands) are indicated in the middle.
In summary, pol1-17 mutants show telomeric G tails even before the appearance of detectable telomere elongation beyond the length of telomeres in pol1-17 cells grown at 23°C (Fig. 3A and B, compare lanes 1 and 2). Furthermore, the ssDNA is telomere specific and appears to be due to a loss of DNA on the C-rich, lagging strand. This rapid appearance of excessive G tails is temporally consistent with the loss of repression of the telomeric URA3 gene expression that begins to occur by 2 to 5 h after the shift to the semipermissive temperature. These data suggest that alterations in telomeric chromatin structure could be the cause of telomere elongation in pol1-17 mutants.
The appearance of telomeric ssDNA in cdc17-1 strains can occur even in the absence of telomerase.
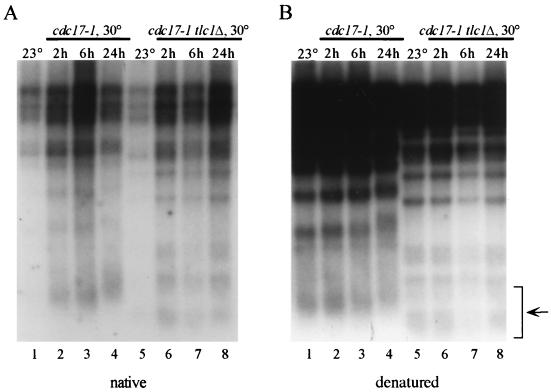
The failure to detect hybridization with a TG probe and the absence of telomeric ssDNA in pol1-17 mutants following ExoI treatment indicate that the single strandedness is due to a terminal absence of DNA on the C-rich, lagging strand. To determine if the appearance of these excess G tails requires elongation of the G-rich, leading strand by telomerase, we examined the amount of single-stranded character in cdc17-1 tlc1Δ strains, which lack telomerase activity. As expected, cdc17-1 strains shifted to 30°C showed increased single strandedness at the telomeres (Fig. 6A, lanes 2 to 4) as well as telomere elongation (Fig. 6B, compare the telomeric DNA smear in lanes 1 and 4). Interestingly, cdc17-1 tlc1Δ strains shifted to 30°C for either 2, 6, or 24 h also exhibited single strandedness (Fig. 6A, lanes 6 to 8), although their telomeres did not elongate (Fig. 6B, compare lane 5 with lanes 6 to 8). The presence of ssDNA in cdc17-1 tlc1Δ strains suggests that the terminal ssDNA is caused by a specific loss of telomeric DNA on the G-rich strand in a telomerase-independent manner. These observations are consistent with the hypothesis that cdc17/pol1 mutants are defective in telomeric lagging-strand synthesis, which would predict that the ssDNA is generated as a result of an S-phase defect caused by the mutant polymerase.
FIG. 6.
ssDNA tails occur in the absence of telomerase, but the telomeres do not elongate. DNA was isolated from strain RWY120 (cdc17-1 tlc1Δ) cells or from strain RWY120 cells carrying a plasmid-borne copy of TLC1 after growth at 23°C (lanes 1 and 5) and after the 23°C culture had been shifted to 30°C for 2 h (lanes 2 and 6), 6 h (lanes 3 and 7), or 24 h (lanes 4 and 8) of growth. (A) Nondenaturing in-gel hybridization with a CA-oligonucleotide probe indicates that the telomeric ssDNA that is present in cdc17-1 cells at 30°C (lanes 2 to 4) is produced even in the absence of telomerase (lanes 6 to 8). (B) Denaturing hybridization with a CA-oligonucleotide probe of the gel in panel A demonstrates that the lanes are approximately evenly loaded. Note that the telomeres are substantially shorter in the cdc17-1 tlc1Δ strain than in the cdc17-1 strain; telomere shortening occurs during the 30 generations of growth at 23°C that is necessary to produce the cdc17-1 tlc1Δ strain (compare telomeric DNA smears [arrow]).
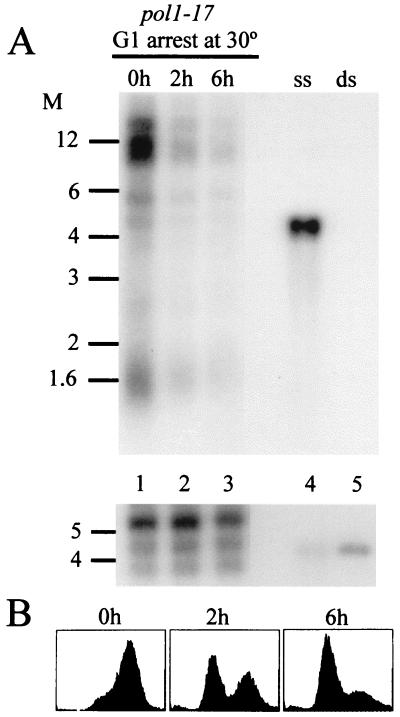
To determine whether the excess telomeric ssDNA in pol1-17 mutants is generated during S phase, we examined the appearance of G tails in synchronized cultures of pol1-17 cells as they moved through S phase. pol1-17 cells grown at 23°C were arrested with α-factor to synchronize the cells in G1. The culture was split into four aliquots, which were incubated at 23 or 30°C, with or without α-factor (see Materials and Methods for details). Examination of isolated DNA by nondenaturing in-gel hybridization (Fig. 7A) revealed that G1-arrested pol1-17 cells at 23 or 30°C failed to exhibit ssDNA (Fig. 7A). In contrast, pol1-17 cells released from α-factor arrest at 23°C (Fig. 7A, left panels) exhibited a peak of ssDNA 1 h after release from arrest (Fig. 7A, lane 5), when the cells were in S phase (Fig. 7B). Furthermore, the level of telomeric ssDNA decreased when the cells exited S phase (Fig. 7A, lane 6, and Fig. 7B) and increased again slightly as the culture became asynchronous (Fig. 7A, lanes 7 and 8, and Fig. 7B). The timing of the appearance of the telomeric ssDNA corresponded to earlier evidence that telomeric G tails are generated only during S phase (58, 59). pol1-17 cells released from α-factor arrest at 30°C displayed a striking excess of ssDNA (Fig. 7A, right panels), which initially appeared when the cells entered S phase (Fig. 7A, lane 5, and Fig. 7B). Notably, more signal for ssDNA was observed in pol1-17 cells at 30 than at 23°C, even as the cultures became asynchronous (Fig. 7A and B, compare 1 and 5 h), and a similarly elevated signal for ssDNA was present 24 h later (data not shown). Taken together, these results suggest that the telomeric ssDNA generated in pol1-17 mutants at 30°C occurred as a result of an S-phase-specific defect, such as defective telomeric lagging-strand synthesis.
To determine if the telomeric ssDNA structures disappear following the termination of S phase, the persistence of ssDNA was examined in cells that were arrested in the G1 phase. Strain RWY121 (pol1-17) was grown exponentially at 30°C to allow the formation of ssDNA at the telomeres. The cultures then were treated with α-factor to arrest the cells in G1; the DNA isolated from these cells was examined for the presence of ssDNA by nondenaturing in-gel hybridization. As shown in Fig. 8, the ssDNA disappeared when the cells were arrested in G1 for 2 or 6 h (Fig. 8A, compare lanes 2 and 3 with lane 1). This result suggests that although excessive G tails form during S phase in pol1-17 mutants grown at 30°C, the tails are still processed to normal chromosomal end structures, with no detectable G tails, by the time the cells reach the G1 phase. Thus, telomeric ssDNA is abundant in pol1-17 mutants only during a restricted part of the cell cycle.
FIG. 8.
The telomeric ssDNA structures generated in S phase can be processed to normal end structures in pol1-17-cells even at 30°C. Strain RWY121 (pol1-17 bar1Δ) was grown exponentially at 23°C and then shifted to 30°C overnight to allow production of telomeric ssDNA (0h, lane 1). The 30°C culture was then arrested in G1 with 600 μM α-factor for 2 h (lane 2) or 6 h (lane 3). (A) Nondenaturing in-gel hybridization to DNA derived from the cells at the individual time points (top panel) and the same gel hybridized after denaturing the DNA (bottom panel) as in Fig. 3. (B) FACS profiles of the same cells used in panel A. Note that pol1-17 cells growing at the semipermissive temperature always show a predominance of cells with a 2C DNA content (1). Cell- cycle arrest in the G1 phase of this culture was also monitored by measuring cell morphology (shmooed and single cells versus budded cells) at each time point to confirm that >85% of the cells were arrested in G1 after 6 h of α-factor treatment (data not shown). The ssDNA tails generated in pol1-17 cells at 30°C are processed to normal end structures by the time the cells reach the G1 phase. Molecular weight (M) markers (in thousands) are indicated on the left of each panel.
DISCUSSION
These studies demonstrate that cdc17/pol1 mutants exhibit alterations in telomeric chromatin structure at temperatures at which their telomeres also elongate. pol1-17 mutants with elongated telomeres display a dramatic loss of telomeric silencing, or TPE (Table 2). Interestingly, Northern hybridization analysis suggests that derepression of a telomeric URA3 gene in pol1-17 mutants may occur before detectable telomere lengthening (Fig. 2 to 4). Furthermore, cdc17/pol1 strains exhibit a striking and reversible increase in ssDNA character at the telomeres prior to telomere elongation (Fig. 5). This ssDNA is generated during S phase and occurs specifically at telomeres due to a loss of DNA on the C-rich strand (Fig. 5 and 7). Thus, cdc17/pol1 mutants appear to be defective in telomeric lagging-strand synthesis. Taken together, these results suggest that telomere elongation in cdc17/pol1 mutants is caused by alterations in telomere structure.
The loss of TPE in cdc17/pol1 mutants suggests that the protein complexes required for TPE are not properly assembled. One possible explanation for the improper assembly is that the elongated telomeres in pol1 mutants titrate a limiting silencing component, such as Sir3p (38, 50), to the ends of the extended telomeres and away from the telomeric URA3 gene. However, an examination of silencing at HML, which requires several proteins that are also required for TPE (3, 13), reveals that the HML locus is silenced to the same extent in POL1 strains and pol1-17 strains with fully elongated telomeres (Fig. 1 and data not shown). Furthermore, this interpretation is inconsistent with the kinetics of TPE loss in these cells: TPE is lost before detectable telomere lengthening occurs (Fig. 2 to 4). Thus, it appears unlikely that the loss of TPE in cdc17/pol1 cells is due to the titration of a limiting silencing component to the telomere ends. This interpretation is also consistent with the finding that wild-type cells with artificially elongated telomeres can exhibit even greater levels of telomeric silencing than wild-type cells with normal-length telomeres (31).
A more reasonable explanation for the loss of TPE is that cdc17/pol1 mutants suffer an alteration in the composition of telomeric proteins that are required for telomeric silencing and telomere length regulation. This hypothesis is consistent with observations of a number of other mutants. For example, some mutations affecting telomeric chromatin components, such as Rap1p, Sir2p, Sir3p, and Sir4p, cause dramatic reductions in the levels of telomeric silencing (2, 30). rap1t alleles cause telomere lengthening while also causing a derepression of telomere-proximal genes (30). Additionally, mutations in the genes encoding the yeast Ku proteins, which are believed to function in the regulation of the telomeric DNA end structure, also cause a loss of telomeric silencing and a loss of telomere length regulation (9, 24, 42). Thus, the absence of one or more important structural or regulatory components on the telomeres of cdc17/pol1 mutants could account for the loss of telomeric silencing.
The rapidity with which G tails appear in cdc17/pol1 mutants at 30°C makes it likely that the formation of telomeric ssDNA is the primary structural defect that causes telomere elongation and the loss of telomeric silencing. This defect appears to be telomere specific, rather than being a general DNA replication defect, because increased single strandedness occurs specifically at the telomeres and not at rDNA or Y′ DNA. The mutant polymerase may be defective in its ability to synthesize repetitive, simple DNA sequences like those found at telomeres. Because ssDNA appears before detectable telomere lengthening, telomere elongation may occur as a result of this alteration in telomeric DNA structure, possibly by affecting telomeric protein binding. This hypothesis is consistent with previous studies reporting that alterations in telomeric DNA can affect telomere length control. For example, in the yeast Kluyveromyces lactis, explosive telomere elongation results from mutations in the telomerase RNA template that perturb the sequence of newly synthesized telomeric DNA, thereby altering the binding sites for the telomere-binding proteins (29). According to this hypothesis, incomplete telomeric DNA synthesis in cdc17/pol1 mutants may trigger an altered regulation of telomerase and, as a consequence, the telomere length “set point” of the cells.
The excess telomeric ssDNA generated in cdc17/pol1 mutants appears to be caused by a specific loss of DNA sequence on the C-rich strand that can occur independently of telomerase. An economical explanation for these results is that the telomeric ssDNA observed in cdc17/pol1 mutants at 30°C is caused by a failure of the mutant polymerase to adequately synthesize the C-rich, lagging strand at the telomeres. In support of this hypothesis, the generation of excess telomeric ssDNA appears to occur specifically during S phase in cdc17/pol1 mutants. This observation is consistent with the recent demonstration that the generation of telomeric G tails requires not only that the cells pass through the S phase but also that the replication machinery reach the telomere (19). Of course, at least for cdc17/pol1 mutants, a lagging-strand defect is probably not the only explanation for the appearance of the ssDNA tails. While both cdc17-1 tlc1Δ and cdc17-1 strains exhibit ssDNA tails immediately upon the shift to 30°C, the telomeres in cdc17-1 strains become longer rather than shorter. Thus, we hypothesize that the G tails in the cdc17-1 cells may be generated by a combination of defective lagging-strand synthesis and excessive elongation by telomerase. Given that the hybridization signals to the ssDNA in the cdc17 and cdc17/tlc1Δ strains are not significantly different, the contribution of telomerase to the single-stranded tails as a whole may be minor. This hypothesis is consistent with the finding that the transient G-rich tails observed on DNA derived from tlc1Δ cells yield hybridization signals that are indistinguishable from those obtained with DNA derived from wild-type cells (18).
As mentioned above, the telomeric ssDNA structures in cdc17/pol1 cells may perturb telomere length control by affecting the regulation of telomerase activity. The presence of the ssDNA tails may cause excessive elongation by acting as good substrates for telomerase. However, the prolonged presence of telomerase substrates alone is probably not sufficient to cause telomere lengthening. Mutations in the Ku proteins, which are suggested to be important for telomere function and telomere length control (9, 24, 42), cause increased telomeric ssDNA character, loss of telomeric silencing, and loss of telomere length control (9, 24, 42). Interestingly, the telomeres in yeast Ku mutants shorten, rather than lengthen, despite the persistent presence of G tails throughout the cell cycle in these mutants (10, 24, 47, 48).
In addition to the effect of the presence of ssDNA, telomere elongation in cdc17/pol1 mutants may be caused by a loss of the coordination between the defective polymerase and telomerase. A similar phenomenon also has been observed in other systems. In Euplotes, for example, partial inhibition of C-rich strand synthesis by DNA polymerase α causes heterogeneous changes in the length of the G-rich strand that is synthesized by telomerase, suggesting that the functions of polymerase α and telomerase are coordinately regulated (21). We hypothesize that the activities causing C-strand degradation, and ultimately telomere shortening, may be regulated normally in cdc17/pol1 mutants, while the elongating activities (C-strand fill-in synthesis and telomerase) occur in an uncoordinated fashion. In this scenario, the generation of duplex telomeric DNA may function to directly or indirectly inhibit excessive telomere lengthening by telomerase. For example, the partial loss of duplex DNA at the telomeres in cdc17/pol1 mutants may create an unfavorable environment for the binding of the telomere-binding Rap1 protein; Rap1p requires duplex DNA for binding (6, 12, 36) and is central in the mechanism of telomere length control (39). Similarly, alterations in telomeric DNA structure in cdc17/pol1 mutants could affect the binding of Est1p or Cdc13p/Est4p, which are associated with the regulation of telomerase (34, 43, 54). Strikingly, cdc13-1ts and cdc17/pol1 mutants each exhibit telomere elongation at the semipermissive temperature (43) and display increases in telomeric ssDNA character (22). Thus, the telomeric DNA defects in cdc17/pol1 mutants grown at the semipermissive temperature may interfere with the binding or interactions of critical regulatory components. Consequently, cdc17/pol1 mutants could be incapable of establishing the proper telomeric context necessary for telomeric silencing and telomere length control.
Taken together, a failure of a mutant polymerase α to complete telomeric C-strand synthesis in yeast may directly prevent the formation of proper telomeric chromatin structure, which is important for telomere function and telomere length control. Future work is needed to more closely examine the composition of proteins present at telomeres in cdc17/pol1 cells grown at the permissive and semipermissive temperatures. Identification of the factors responsible for the differences observed on the telomeres in cdc17/pol1 cells exposed to these different conditions will contribute to a greater understanding of the elements required for telomere length regulation.
ACKNOWLEDGMENTS
A.A.M. and I.D. contributed equally to this work.
We thank Scott Oh in the Holm laboratory for providing technical assistance with the Northern analyses and Robert Lemire in the Wellinger laboratory for constructing and characterizing the cdc17 tlc1Δ strain. Thanks are also due to members of the Holm laboratory for many valuable discussions about this work.
This work was supported by a grant from the National Institutes of Health to C.H. (GM36510) and a grant from the Canadian Medical Research Council (MRC) to R.J.W. (MT12616). R.J.W. is a Chercheur-Boursier Senior of the FRSQ. A.A.M. was funded by an NRSA Minority Research Fellowship (GM18056). I.D. was supported by a studentship from the FCAR.
REFERENCES
- 1.Adams A K, Holm C. Specific DNA replication mutations affect telomere length in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:4614–4620. doi: 10.1128/mcb.16.9.4614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aparicio O M, Billington B L, Gottschling D E. Modifiers of position effect are shared between telomeric and silent mating-type loci in S. cerevisiae. Cell. 1991;66:1279–1287. doi: 10.1016/0092-8674(91)90049-5. [DOI] [PubMed] [Google Scholar]
- 3.Aparicio O M, Gottschling D E. Overcoming telomeric silencing: a trans-activator competes to establish gene expression in a cell cycle-dependent way. Genes Dev. 1994;8:1133–1146. doi: 10.1101/gad.8.10.1133. [DOI] [PubMed] [Google Scholar]
- 4.Astell C R, Ahlstrom-Jonasson L, Smith M. The sequence of the DNAs coding for the mating-type loci of Saccharomyces cerevisiae. Cell. 1981;27:15–23. doi: 10.1016/0092-8674(81)90356-1. [DOI] [PubMed] [Google Scholar]
- 5.Beeler T, Gable K, Zhao C, Dunn T. A novel protein, CSG2p, is required for Ca2+ regulation in Saccharomyces cerevisiae. J Biol Chem. 1994;269:7279–7284. [PubMed] [Google Scholar]
- 6.Berman J, Tachibana C Y, Tye B-K. Identification of a telomere-binding activity from yeast. Proc Natl Acad Sci USA. 1986;83:3713–3717. doi: 10.1073/pnas.83.11.3713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blackburn E H. Structure and function of telomeres. Nature. 1991;350:569–573. doi: 10.1038/350569a0. [DOI] [PubMed] [Google Scholar]
- 8.Boeke J D, Trueheart J, Natsoulis G, Fink G R. 5-Fluoroorotic acid as a selective agent in yeast molecular genetics. Methods Enzymol. 1987;154:164–175. doi: 10.1016/0076-6879(87)54076-9. [DOI] [PubMed] [Google Scholar]
- 9.Boulton S J, Jackson S P. Components of the Ku-dependent nonhomologous end-joining pathway are involved in telomeric length maintenance and telomeric silencing. EMBO J. 1998;6:1819–1828. doi: 10.1093/emboj/17.6.1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boulton S J, Jackson S P. Saccharomyces cerevisiae Ku70 potentiates illegitimate DNA double-strand bread repair and serves as a barrier to error-prone DNA repair pathways. EMBO J. 1996;15:5093–5103. [PMC free article] [PubMed] [Google Scholar]
- 11.Brachman C B, Davies A, Cost G J, Caputo E, Li J, Hieter P, Boeke J D. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast. 1998;14:115–132. doi: 10.1002/(SICI)1097-0061(19980130)14:2<115::AID-YEA204>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 12.Buchman A R, Kimmerly W J, Rine J, Kornberg R D. Two DNA-binding factors recognize specific sequences at silencers, upstream activating sequences, autonomously replicating sequences, and telomeres in Saccharomyces cerevisiae. Mol Cell Biol. 1988;8:210–225. doi: 10.1128/mcb.8.1.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Buck S W, Shore D. Action of a RAP1 carboxy-terminal silencing domain reveals an underlying competition between HMR and telomeres in yeast. Genes Dev. 1995;9:370–384. doi: 10.1101/gad.9.3.370. [DOI] [PubMed] [Google Scholar]
- 14.Budd M E, Wittrup K D, Bailey J E, Campbell J L. DNA polymerase I is required for premeiotic DNA replication and sporulation but not for X-ray repair in Saccharomyces cerevisiae. Mol Cell Biol. 1989;9:365–376. doi: 10.1128/mcb.9.2.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carson M J, Hartwell L. CDC17: an essential gene that prevents telomere elongation in yeast. Cell. 1985;42:249–257. doi: 10.1016/s0092-8674(85)80120-3. [DOI] [PubMed] [Google Scholar]
- 16.Cockell M, Palladino F, Laroche T, Kyrion G, Liu C, Lustig A J, Gasser S M. The carboxy termini of Sir4 and Rap1 affect Sir3 localization: evidence for a multicomponent complex required for yeast telomeric silencing. J Cell Biol. 1995;129:909–924. doi: 10.1083/jcb.129.4.909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Counter C M, Meyerson M, Eaton E N, Weinberg R A. The catalytic subunit of yeast telomerase. Proc Natl Acad Sci USA. 1997;94:9202–9207. doi: 10.1073/pnas.94.17.9202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dionne I, Wellinger R J. Cell-cycle-regulated generation of single-stranded G-rich DNA in the absence of telomerase. Proc Natl Acad Sci USA. 1996;93:13902–13907. doi: 10.1073/pnas.93.24.13902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dionne I, Wellinger R J. Processing of telomeric DNA ends requires the passage of a replication fork. Nucleic Acids Res. 1998;26:5365–5371. doi: 10.1093/nar/26.23.5365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Enomoto S, Berman J. Chromatin assembly factor I contributes to the maintenance, but not the re-establishment, of silencing at the yeast silent mating loci. Genes Dev. 1998;12:219–232. doi: 10.1101/gad.12.2.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fan X, Price C M. Coordinate regulation of G- and C-strand length during new telomere synthesis. Mol Biol Cell. 1997;8:2145–2155. doi: 10.1091/mbc.8.11.2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Garvik B, Carson M, Hartwell L. Single-stranded DNA arising at telomeres in cdc13 mutants may constitute a specific signal for the RAD9 checkpoint. Mol Cell Biol. 1995;15:6128–6138. doi: 10.1128/mcb.15.11.6128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gottschling D E, Aparicio O M, Billington B L, Zakian V A. Position effect at S. cerevisiae telomeres: reversible repression of Pol II transcription. Cell. 1990;63:751–762. doi: 10.1016/0092-8674(90)90141-z. [DOI] [PubMed] [Google Scholar]
- 24.Gravel S, Larrivee M, Labrecque P, Wellinger R J. Yeast Ku as a regulator of chromosomal DNA end structure. Science. 1998;280:741–744. doi: 10.1126/science.280.5364.741. [DOI] [PubMed] [Google Scholar]
- 25.Greider C W. Telomere length regulation. Annu Rev Biochem. 1996;65:337–365. doi: 10.1146/annurev.bi.65.070196.002005. [DOI] [PubMed] [Google Scholar]
- 26.Hartwell L H, Mortimer R K, Culotti J, Culotti M. Genetic control of the cell division cycle in yeast: V. Genetic analysis of cdc mutants. Genetics. 1973;74:267–286. doi: 10.1093/genetics/74.2.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hecht A, Laroche T, Strahl-Bolsinger S, Gasser S M, Grunstein M. Histone H3 and H4 N-termini interact with SIR3 and SIR4 proteins: a molecular model for the formation of heterochromatin in yeast. Cell. 1995;80:583–592. doi: 10.1016/0092-8674(95)90512-x. [DOI] [PubMed] [Google Scholar]
- 28.Ivy J M, Klar A S, Hicks J B. Cloning and characterization of four SIR genes of Saccharomyces cerevisiae. Mol Cell Biol. 1986;6:688–702. doi: 10.1128/mcb.6.2.688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Krauskopf A, Blackburn E H. Control of telomere growth by interactions of RAP1 with the most distal telomeric repeats. Nature. 1996;383:354–357. doi: 10.1038/383354a0. [DOI] [PubMed] [Google Scholar]
- 30.Kyrion G, Boakye K A, Lustig A J. C-terminal truncation of RAP1 results in the deregulation of telomere size, stability and function in Saccharomyces cerevisiae. Mol Cell Biol. 1992;12:5159–5173. doi: 10.1128/mcb.12.11.5159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kyrion G, Liu K, Liu C, Lustig A J. RAP1 and telomere structure regulate telomere position effects in Saccharomyces cerevisiae. Genes Dev. 1993;7:1146–1159. doi: 10.1101/gad.7.7a.1146. [DOI] [PubMed] [Google Scholar]
- 32.Lehmann I R, Nussbaum A L. The deoxyribonucleases of Escherichia coli. V. On the specificity of exonuclease I (phosphodiesterase) J Biol Chem. 1964;239:2628–2634. [PubMed] [Google Scholar]
- 33.Lendvay T S, Morris D K, Sah J, Balasubramanian B, Lundblad V. Senescence mutants of Saccharomyces cerevisiae with a defect in telomere replication identify three additional EST genes. Genetics. 1996;144:1399–1412. doi: 10.1093/genetics/144.4.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lin J-J, Zakian V A. The Saccharomyces CDC13 protein is a single-strand TG1–3 telomeric DNA-binding protein in vitro that affects telomere behavior. Proc Natl Acad Sci USA. 1996;93:13760–13765. doi: 10.1073/pnas.93.24.13760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lingner J, Hughes T R, Shevchenko A, Mann M, Lundblad V, Cech T R. Reverse transcriptase motifs in the catalytic subunit of telomerase. Science. 1997;276:561–567. doi: 10.1126/science.276.5312.561. [DOI] [PubMed] [Google Scholar]
- 36.Longtine M S, Wilson N M, Petracek M E, Berman J. A yeast telomere binding activity binds to two related telomere sequence motifs and is indistinguishable from RAP1. Curr Genet. 1989;16:225–239. doi: 10.1007/BF00422108. [DOI] [PubMed] [Google Scholar]
- 37.Lustig A J, Kurtz S, Shore D. Involvement of the silencer and UAS binding protein RAP1 in regulation of telomere length. Science. 1990;250:549–553. doi: 10.1126/science.2237406. [DOI] [PubMed] [Google Scholar]
- 38.Lustig A J, Liu C, Zhang C, Hanish J P. Tethered Sir3p nucleates silencing at telomeres and internal loci in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:2483–2495. doi: 10.1128/mcb.16.5.2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Marcand S, Gilson E, Shore D. A protein-counting mechanism for telomere length regulation in yeast. Science. 1997;275:986–990. doi: 10.1126/science.275.5302.986. [DOI] [PubMed] [Google Scholar]
- 40.McClintock B. The stability of broken ends of chromosomes in Zea mays. Genetics. 1941;26:234–282. doi: 10.1093/genetics/26.2.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moretti P, Freeman K, Coodly L, Shore D. Evidence that a complex of SIR proteins interacts with the silencer and telomere-binding protein RAP1. Genes Dev. 1994;8:2257–2269. doi: 10.1101/gad.8.19.2257. [DOI] [PubMed] [Google Scholar]
- 42.Nugent C I, Bosco G, Ross L O, Evans S K, Salinger A P, Moore J K, Haber J E, Lundblad V. Telomere maintenance is dependent on activities required for end repair of double-strand breaks. Curr Biol. 1998;8:657–660. doi: 10.1016/s0960-9822(98)70253-2. [DOI] [PubMed] [Google Scholar]
- 43.Nugent C I, Hughes T R, Lue N F, Lundblad V. Cdc13p: a single-strand telomeric DNA-binding protein with a dual role in yeast telomere maintenance. Science. 1996;274:249–252. doi: 10.1126/science.274.5285.249. [DOI] [PubMed] [Google Scholar]
- 44.Palladino F, Laroche T, Gilson E, Axelrod A, Pillus L, Gasser S M. SIR3 and SIR4 proteins are required for the positioning and integrity of yeast telomeres. Cell. 1993;75:543–555. doi: 10.1016/0092-8674(93)90388-7. [DOI] [PubMed] [Google Scholar]
- 45.Parenteau J, Wellinger R J. Accumulation of single-stranded DNA and destabilization of telomeric repeats in yeast mutant strains carrying a deletion of RAD27. Mol Cell Biol. 1999;19:4143–4152. doi: 10.1128/mcb.19.6.4143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Park E-C, Szostak J W. Point mutations in the yeast histone H4 gene prevent silencing of the silent mating type locus HML. Mol Cell Biol. 1990;10:4932–4934. doi: 10.1128/mcb.10.9.4932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Polotnianka R M, Li J, Lustig A J. The yeast Ku heterodimer is essential for protection of the telomere against nucleolytic and recombinational activities. Curr Biol. 1998;8:831–834. doi: 10.1016/s0960-9822(98)70325-2. [DOI] [PubMed] [Google Scholar]
- 48.Porter S E, Greenwell P W, Ritchie K B, Petes T D. The DNA-binding protein Hdf1p (a putative Ku homologue) is required for maintaining normal telomere length in Saccharomyces cerevisiae. Nucleic Acids Res. 1996;24:582–585. doi: 10.1093/nar/24.4.582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ray A, Runge K W. The yeast telomere length counting machinery is sensitive to sequences at the telomere-nontelomere junction. Mol Cell Biol. 1999;19:31–45. doi: 10.1128/mcb.19.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Renauld H, Aparicio O M, Zierath P D, Billington B L, Chhablani S K. Silent domains are assembled continuously from the telomere and are defined by promoter distance and strength, and by SIR3 dosage. Genes Dev. 1993;7:1133–1145. doi: 10.1101/gad.7.7a.1133. [DOI] [PubMed] [Google Scholar]
- 51.Schmitt M E, Brown T A, Trumpower B L. A rapid and simple method for preparation of RNA from Saccharomyces cerevisiae. Nucleic Acids Res. 1990;18:3091–3092. doi: 10.1093/nar/18.10.3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sherman F, Fink G R, Hicks J B. Methods in yeast genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1986. [Google Scholar]
- 53.Singer M S, Gottschling D E. TLC1: template RNA component of Saccharomyces cerevisiae telomerase. Science. 1994;266:404–409. doi: 10.1126/science.7545955. [DOI] [PubMed] [Google Scholar]
- 54.Virta-Pearlman V, Morris D K, Lundblad V. Est1 has the properties of a single-stranded telomere end-binding protein. Genes Dev. 1996;10:3094–3104. doi: 10.1101/gad.10.24.3094. [DOI] [PubMed] [Google Scholar]
- 55.Wach A, Brachat A, Pohlmann R, Philippsen P. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast. 1994;10:1793–1808. doi: 10.1002/yea.320101310. [DOI] [PubMed] [Google Scholar]
- 56.Wahl G M, Meinkoth J L, Kimmel A R. Northern and Southern blots. Methods Enzymol. 1987;152:572–581. doi: 10.1016/0076-6879(87)52064-x. [DOI] [PubMed] [Google Scholar]
- 57.Watson J D. Origin of concatemeric T7 DNA. Nature New Biol. 1972;239:197–201. doi: 10.1038/newbio239197a0. [DOI] [PubMed] [Google Scholar]
- 58.Wellinger R J, Wolf A, Zakian V A. Origin activation and formation of single-stranded TG1–3 tails occur sequentially in last S phase on a yeast linear plasmid. Mol Cell Biol. 1993;13:4057–4065. doi: 10.1128/mcb.13.7.4057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wellinger R J, Wolf A J, Zakian V A. Saccharomyces telomeres acquire single-stranded TG1–3 tails late in S phase. Cell. 1993;72:51–60. doi: 10.1016/0092-8674(93)90049-v. [DOI] [PubMed] [Google Scholar]
- 60.Wright J H, Gottschling D E, Zakian V A. Saccharomyces telomeres assume a non-nucleosomal chromatin structure. Genes Dev. 1992;6:197–210. doi: 10.1101/gad.6.2.197. [DOI] [PubMed] [Google Scholar]
- 61.Zakian V A. Structure and function of telomeres. Annu Rev Genet. 1989;23:579–604. doi: 10.1146/annurev.ge.23.120189.003051. [DOI] [PubMed] [Google Scholar]