Abstract
A variant polyadenylation signal, which is conserved and employed by mammalian hepadnaviruses, has a sequence resembling that of the TATA box. We report here that this composite box manifests all the promoter characteristics. It binds effectively TATA-binding protein with TFIIB and TFIIA in a synergistic manner. This capacity, however, is lost when the box is converted to a canonical and simple poly(A) signal. Furthermore, we show that it has promoter activity and supports transcription of reporter genes preferentially in liver-derived cells, a characteristic behavior of the hepatitis B virus (HBV) promoters. In addition, we show that the HBV noncanonical poly(A) signal supports transcription initiation from the viral genome, suggesting that it is a genuine promoter, possibly of the polymerase/reverse transcriptase gene. Finally, we found that this deviant poly(A) signal is crucial for HBV replication since a viral mutant with a canonical poly(A) box is impaired in replication. Our data, therefore, raise the interesting and novel possibility that a composite poly(A) box might have a dual function. At the level of DNA it functions as a promoter to initiate transcription, whereas at the level of RNA it serves as a poly(A) signal to process RNA. An interesting outcome of this strategy of gene expression is that it provides a novel mechanism for the synthesis of an approximately genome length transcript.
The 3′ end of the eukaryotic mRNA is polyadenylated by a reaction that involves site-specific endonucleolytic cleavage. The AAUAAA sequence, the important polyadenylation signal, is located about 15 to 30 nucleotides upstream of the cleavage site. Some variation of this signal is tolerated, although it often results in diminished processing efficiency (39, 43). Transcripts that contain the deviant UAUAAA poly(A) signal are processed much less efficiently (about 17%). In fact, at the DNA level the deviant sequence (TATAAA) resembles a TATA box more than a canonical poly(A) signal (AATAAA). Interestingly, in spite of its remarkable inefficiency some viruses tend to prefer this deviant poly(A) signal. These include all the mammal hepadnaviruses (33), the figwart mosaic virus (34), and Epstein-Barr virus (40). Particularly puzzling is the fact that this deviant box is conserved among the different members of mammalian hepadnaviruses, raising an interesting possibility that it has a unique but yet-unidentified role.
Hepatitis B virus (HBV) is the prototype of the hepadnaviruses. This enveloped DNA virus has a very small 3.2-kb genome replicating via reverse transcription and is primarily hepatotropic. The genome contains four partially overlapping open reading frames (ORFs), each translated from a specific viral transcript. The largest two viral transcripts known are the 3.5-kb precore mRNA (pcRNA) and the 3.4-kb pregenomic mRNA (pgRNA). pgRNA encodes the core (HBcAg) protein and possibly the viral polymerase/reverse transcriptase (Pol). pgRNA has a third function in viral replication, which is to serve as a template for the reverse transcripts. Two additional known transcripts are the 2.3- to 2.1-kb mRNAs, which encode the S, PreS1, and PreS2 viral surface antigens. The last known transcript is the 0.7-kb mRNA encoding the regulatory X protein (pX). pX has transcription coactivation activity (14–17, 25) and is an effector of cellular signaling (3, 9, 23, 26, 28, 41).
HBV transcription is regulated by the cellular transcriptional activators that are preferentially found in liver cells (8, 12, 20, 29, 30, 38). The viral genome contains multiple promoters; each regulates the synthesis of a distinct transcript, all of which are processed at a single poly(A) signal. Except for the promoter of the 2.3-kb transcript, none of the viral promoters contains a classical TATA box (37). The juxtaposed pc- and pgRNA promoters contain a number of AT-rich boxes that bind recombinant TATA-binding protein (TBP) (6). By employing recombinant general transcription factors (GTFs), we attempted to characterize the functional and cryptic TATA boxes of the different HBV promoters. Unexpectedly, we found that the deviant poly(A) signal of the virus binds GTFs effectively in a manner characteristic of a promoter. Furthermore, this box has promoter activity and supports the transcription of reporter genes. Our data, therefore, describe an interesting composite poly(A) box with dual roles. At the level of DNA it functions as a promoter to initiate transcription, whereas at the level of RNA it serves as a poly(A) signal to process RNA.
MATERIALS AND METHODS
Cell culture.
HepG2, SK-Hep1, and Huh7 cells were maintained in Dulbecco's modified Eagle's minimal essential medium (GIBCO Laboratories) containing penicillin (100 U/ml) and streptomycin (100 μg/ml), supplemented with 8% fetal calf serum (GIBCO Laboratories). Transfection was carried out by the CaPi method as previously described (16). Cells were seeded 8 to 12 h prior to transfection at about 60% confluence and were transfected as indicated in each figure. When necessary, pGEM3 plasmid was added at various concentrations to reach total amounts of 6 and 20 μg of DNA per 6- or 10-cm-diameter plate, respectively.
Plasmid constructions.
For construction of simian virus 40 (SV40) enhancer/P(A)S/TATA reporter plasmid, the StyI-BglII DNA fragment from the HBV genome (subtype adw), containing the HBV poly(A) signal, either wild type (wt) or mutant, was cloned via HindIII and BamHI sites into the pBluescript plasmid (Stratagene). A KpnI-SacI fragment of this plasmid harboring this HBV fragment with the flanking polylinker sequence was cloned to the KpnI-XhoI sites in the pGL2-Enhancer vector (Promega). The pGL2 plasmid containing the SV40 enhancer with the SV40 early promoter served as the positive control; the same plasmid containing the SV40 enhancer alone served as the negative control.
For construction of the G5/P(A)S/TATA reporter plasmids, the G5 luciferase plasmid containing five repeats of the UASGal synthetic enhancer element was used. The XbaI fragment of this plasmid was replaced with an XbaI fragment of pGL3 (Promega) containing the luciferase gene. The SmaI-BglII DNA fragments from the SV40 enhancer/P(A)S/TATA reporter plasmid, containing either the wt or mutant sequence, were cloned downstream of G5 but upstream of the luciferase gene.
RNA analysis.
Total RNA was extracted from transfected cells by TRI REAGENT (MRC, Inc.) and treated with RNase-free DNase I (Boehringer) for 15 min at 37°C. The RNA quality and quantity were monitored by measuring UV absorption and by ethidium bromide staining. For Northern blot analysis 10 to 20 μg of total RNA per sample was separated on a 1% formaldehyde-agarose gel and blotted to a Hybond-N nylon membrane (Amersham). Radioactive probes were prepared by random priming with a full-length HBV DNA and [α-32P]dCTP (Amersham; 3,000 Ci/mmol). About 106 cpm (10 ng of DNA) of labeled DNA per ml of hybridization buffer was used. After hybridization the membrane was washed for 60 min at 65°C in a 0.1 × SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)–0.1% sodium dodecyl sulfate buffer and exposed to an X-ray film for autoradiography. Densitometry was performed by a Fujix Bas 2500 phosphorimager (Fuji).
For the RNase protection assay, total RNA from transfected cells was extracted by TRI REAGENT and analyzed as previously reported (10). For this analysis a single-stranded RNA probe was generated by using T7 RNA polymerase to transcribe an antisense probe labeled with [α-32P]UTP (Amersham).
The primer extension analysis was performed with poly(A)-containing RNA by using as a 32P-end-labeled primer the single-stranded synthetic oligonucleotide 5′-ACTCTAAGGCTTCTCGATAC-3′ (nucleotides 2014 to 2034 in the viral genome considering the unique EcoRI site as 1). The reactions were conducted as previously described (10).
Preparation of recombinant proteins.
Recombinant TBP, TFIIB, and TFIIA proteins were prepared in Escherichia coli as previously reported (17).
Protein-DNA interaction assays.
The electrophoretic mobility shift assays (EMSA) were performed as described previously (5, 27). The composition of the binding buffer was 10 mM HEPES-KOH (pH 7.9), 4 mM MgCl2, 0.1 mM EDTA, 5 mM (NH4)2SO4, 2% (wt/vol) polyethylene glycol, 8% (vol/vol) glycerol, 10 μM Zn acetate, 0.025% NP-40, 50 to 100 mM KCl, 0.14 mg of poly(dC-dG)/ml, bovine serum albumin (100 μg/ml), and 2 mM dithiothreitol. As a DNA probe the HBV StyI-BglII 102-bp fragment containing the wt poly(A) signal or the mutated one was used. The fragment was labeled by a fill-in reaction. For each assay 5 to 10 ng of DNA (about 5,000 cpm) was used. The binding reaction was carried out at 30°C for 30 min, and the complexes were separated through nondenaturing 4 to 5% polyacrylamide gels. The running buffer contained 25 mM Tris (pH 7.9), 100 mM glycine, 1 mM EDTA, 0.025% NP-40, 1 mM β-mercaptoethanol, 3% glycerol, 1.5 mM MgCl2, and 10 μM ZnOAc.
For DNase I footprinting of the core promoter, a 283-bp (StuI-BglII) DNA fragment was subcloned in pGEM3Z (Promega) and was digested and labeled at the unique EcoRI site of the plasmid polylinker region. The minus strand of the DNA fragment was labeled by a fill-in reaction and used for DNase footprinting according to our published protocols (29). For DNase I footprinting of the poly(A) signal of HBV the fragment used for EMSA was employed.
Virus replication assays.
HepG2 cells were transfected with equivalent amounts of plasmid HBV DNA, either wt or mutant, harvested after 5 and 7 days, and analyzed for the presence of replicative HBV DNA intermediates by a published protocol (31).
To assay for endogenous DNA polymerase activity in viral particles the culture medium of the transfected cells was collected after 7 days, virions were harvested and treated, and an endogenous DNA polymerase assay was performed according to the published protocols (21, 42).
RESULTS
Analysis of the HBV pc- and pgRNA promoter region.
To characterize the functional HBV pol-II promoters, we took the advantage of the fact that their core box binds GTFs in a defined manner (5, 27). TFIIB and TFIIA are the first GTFs to associate with the TBP during formation of a transcription initiation complex on RNA pol-II promoters. It has been reported that both TFIIB and TFIIA have the intrinsic ability to directly increase the affinity of TBP to the TATA box (19). Also, TFIIB was reported to directly interact with DNA immediately upstream of the TBP binding region (24).
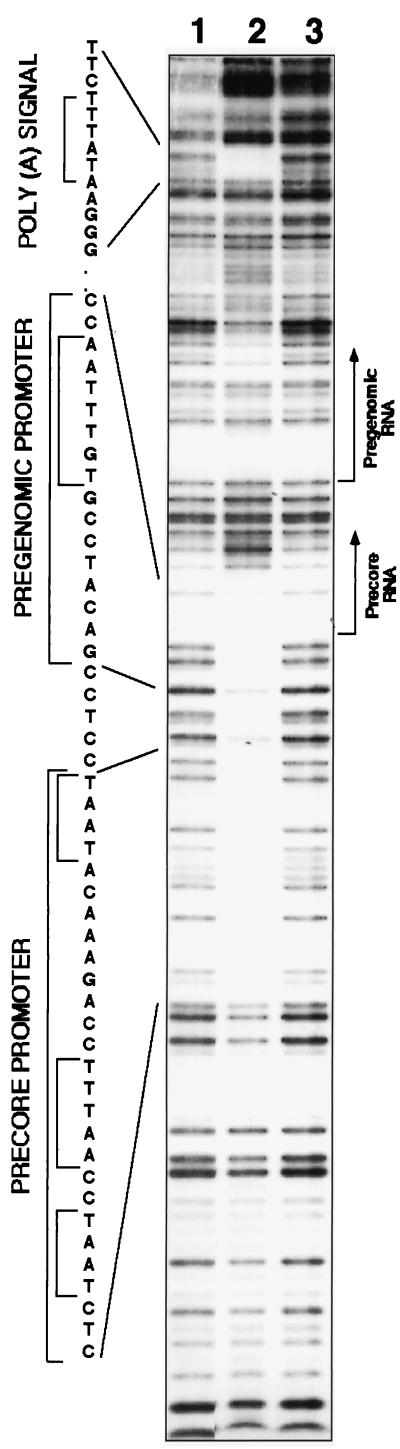
To characterize the structure of the HBV pc- and pgRNA promoter region, we conducted DNase I footprinting assays with recombinant TBP, TFIIA, and TFIIB proteins. An extended protected region at the expected DNA sequence was obtained, suggesting that the employed recombinant GTFs bind the pc- and pgRNA promoter(s) at multiple and possibly overlapping sites (Fig. 1, lane 2). This region does not contain a canonical TATA box, but three AT-rich regions that were reported to bind recombinant TBP were found (6). Unexpectedly, however, an additional footprinted region was detected at the region that contains the viral poly(A) site. The HBV poly(A) signal [P(A)S] has a deviant TATAAA sequence that resembles that of a TATA box. We refer to this composite box as P(A)S-TATA.
FIG. 1.
DNase I footprinting analysis of the HBV core promoter region. An HBV DNA fragment (StuI-BglII), was end labeled and was incubated either with TBP, TFIIB, and TFIIA in a binding reaction (lane 2) or alone (lanes 1 and 3) and subjected to DNase I footprinting. The sequence of the protected regions, as determined by the comigrated products of the Maxam-Gilbert sequencing reaction, is shown at the left. Also, indicated are the TA-rich boxes that were previously reported to bind recombinant TBP (6). At the right the initiation sites of the pc- and pgRNA are shown; the arrows indicate the transcription direction.
The HBV P(A)S-TATA composite box binds GTFs.
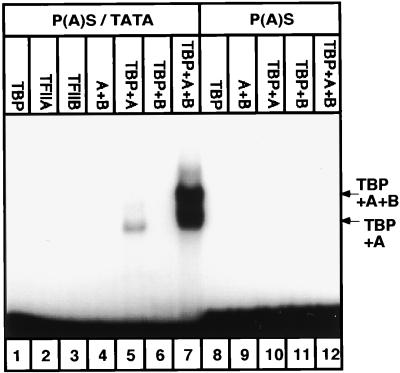
To further characterize the capacity of the HBV P(A)S-TATA composite box to bind GTFs in a sequence-specific manner, we introduced a point mutation to convert it from a composite to a simple canonical poly(A) signal. Binding of the recombinant TBP, TFIIA, and TFIIB was analyzed by EMSA (Fig. 2). A suboptimal TBP concentration was used to show synergism in DNA-binding activity between TBP and TFIIA and TFIIB (19). Under these conditions P(A)S-TATA does not bind TBP efficiently (lane 1) but binds the mixture of the three GTF proteins (lane 7). A much weaker complex is seen in the absence of TFIIB (lane 5), but no DNA binding was detected in the absence of TBP (lanes 2 to 4). Thus, the observed DNA-GTF binding activity is fully TBP dependent and the joining of TFIIB to the shifted complex (lane 7) seems to be TFIIA dependent. This stepwise assembly behavior is in accordance with the reported GTF binding activity (5, 27). Interestingly, the point mutant with the simple and canonical poly(A) signal [P(A)S] no longer binds GTFs (Fig. 2, lanes 8 to 12). We therefore concluded that the HBV composite P(A)S-TATA box binds GTFs in a manner characteristic of a genuine TATA box and that this binding activity is completely dependent on the composite nature of this box.
FIG. 2.
wt HBV P(A)S-TATA binds the basal transcription factors in a manner characteristic of a promoter. Two DNA fragments (102 bp) which span from the StyI site to the BglII site, harboring the HBV P(A)S-TATA sequence (TATAAA) (lanes 1 to 7), or the canonical P(A)S sequence (AATAAA; lanes 8 to 12) were subjected to EMSA with purified recombinant basal transcription factors. The amounts of the different basal transcription factors in the reaction mixture were as follows: 125 pg of TBP, 2 ng of TFIIA, and 10 ng of TFIIB. The protein mixtures in the different reactions are indicated.
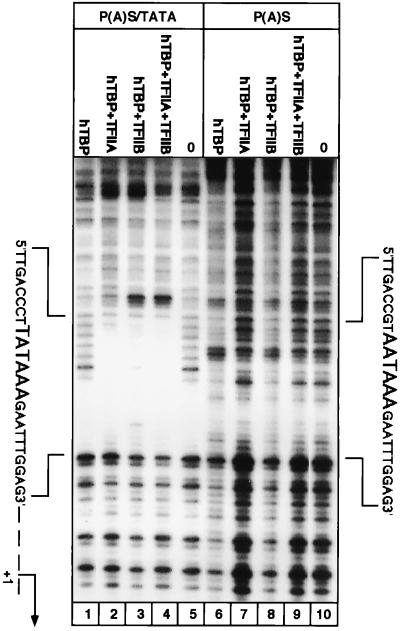
To further substantiate the above findings and to delineate the GTF-interacting regions, DNase I footprinting was employed. An HBV 102-bp fragment (StyI-BglII) containing the P(A)S-TATA box was end labeled and incubated with recombinant GTFs. Here again, a suboptimal concentration of the recombinant human TBP was used to minimize its direct DNA interaction (Fig. 3, lane 1). The addition of TFIIA resulted in the appearance of a protected region over the P(A)S-TATA sequence (lane 2). When TBP was incubated with TFIIB, this region was protected to a lesser extent but a strong DNase I-hypersensitive site at the 5′ end of the protected region was evident (lane 3). This is expected, given the recent finding that TFIIB directly binds DNA at the 5′ end of the TATA box region (24). Significantly, a fully protected region with a hypersensitive site was detected when all the three proteins were coincubated (lane 4). Under similar conditions, a mutant DNA fragment bearing the simple and canonical poly(A) box displayed very poor GTF binding activity (lanes 6 to 10). We concluded that the cryptic TATA box embedded within the composite P(A)S-TATA sequence binds the GTFs in a manner genuinely characteristic of a promoter. Furthermore, the position of the TFIIB-dependent, DNase I-hypersensitive site defined the correct orientation of this box. Thus, the deviant and composite poly(A) signal of the mammal hepadnaviruses binds GTFs at the level of DNA and, given its documented function, it must bind the poly(A)-processing proteins at the RNA level.
FIG. 3.
DNase I footprinting analysis of the novel TATA box in the HBV genome. The examined DNA fragments and the binding conditions are identical to those used for EMSA in Fig. 2, except that 2 ng of TBP, 4 ng of TFIIA, and 10 ng of TFIIB were used. The different probes and the different protein mixtures in each reaction are indicated above the lanes. Lanes 0, no protein added. The sequences of the protected region of the P(A)S-TATA (left) and of the unprotected region of the P(A)S sequence (right) are shown. hTBP, human TBP.
The composite P(A)S-TATA box has promoter activity.
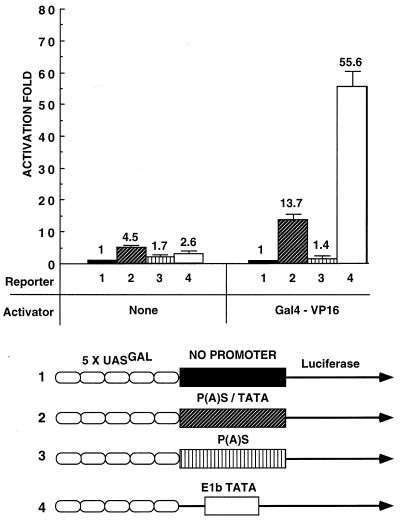
To investigate the functional significance of the ability of the composite P(A)S-TATA box to bind GTFs, we employed the luciferase reporter system. Each of the examined HBV DNA fragments was inserted into a promoterless plasmid at the 5′ end of the luciferase reporter gene. The reporter plasmid also contained an artificial enhancer composed of five copies of the yeast GAL4 binding site (UASGal). An empty vector without a TATA box and a vector with the adenovirus E1b TATA box were used as negative and positive controls, respectively. The reporter plasmids were cotransfected together with a plasmid expressing the VP16 acidic activation domain fused to the GAL4 DNA-binding domain. A significant transcription activation was obtained by the Gal4VP16 activator and the reporter plasmid containing either the wt HBV fragment (13.7-fold) or the E1b TATA box (55.6-fold), suggesting that the P(A)S-TATA box has intrinsic promoter activity (Fig. 4). In contrast the HBV mutated-DNA fragment with the simple poly(A) sequence was inactive. We concluded that the embedded TATA box in the composite HBV P(A)S-TATA signal has promoter activity. The activity of this box, however, is fourfold weaker than that of the canonical E1b TATA box promoter.
FIG. 4.
The HBV composite P(A)S-TATA box exerts promoter activity in the context of a reporter plasmid. HepG2 cells were transfected with 1 μg of each of the four reporter plasmids schematically described at the bottom and with 50 ng of DNA of the Gal4VP16 activator plasmid per 6-cm-diameter plate. Luciferase activity was measured 48 h posttransfection. To calculate the fold activation, the activity of each reporter was divided by that obtained with reporter 1. The experiments were performed in triplicate, and the average values and standard deviations are shown.
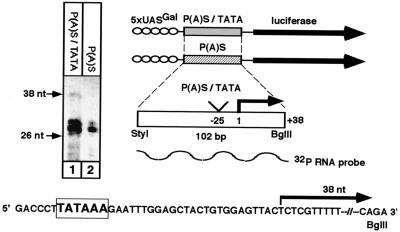
To confirm that the activation of the reporter plasmids is regulated on the level of transcription, we performed RNase mapping analysis. The Huh7 hepatocytes were transiently cotransfected with the reporter and the Gal4VP16 activator plasmids. RNA was extracted and subjected to an RNase protection reaction. As a probe an antisense 32P-RNA containing the sequence of the putative promoter region was used. A specific band with an approximate length of 38 nucleotides was detected with the composite but not with the simple P(A)S reporter plasmid (Fig. 5). This band is a likely candidate to represent the 5′ ends of the transcripts that are initiated by the P(A)S-TATA promoter element. The exact 5′ end of the protected band is mapped at the CT sequence (this was also confirmed by primer extension analysis [see below]). The obtained lower-molecular-weight bands are the products of the RNase digestion at the “breathing” region of five consecutive Ts (see the sequence in Fig. 5). These data, together with the results of the luciferase assays, suggest that the composite P(A)S-TATA element can function as a promoter, at least in the context of a reporter plasmid.
FIG. 5.
Analysis by RNase mapping of the 5′ portion of the RNA molecules produced by the reporter plasmids. Huh7 cells were cotransfected with 0.2 μg of DNA of the activator plasmid Gal4VP16, together with 2 μg of DNA of the luciferase reporter plasmids with either the composite P(A)S-TATA or the simple P(A)S sequence, per 10-cm-diameter plate. Sixty hours posttransfection, total RNA was extracted and RNase mapping was performed with a 32P-labeled antisense RNA probe. The bands with lower molecular weights are likely to be the products of RNase digestion at the breathing region with five consecutive Ts.
Cell type preference of the P(A)S-TATA promoter element.
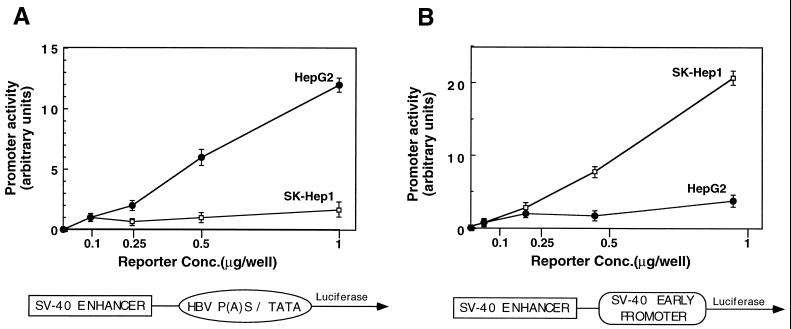
Previously, we have reported that one of the characteristic features of the HBV promoters is that under the SV40 enhancer they display greater activity in the liver-derived cell lines than the SV40 promoter/enhancer unit (18). To determine the possible cell-specific promoter activity of the composite P(A)S-TATA element, we constructed a reporter plasmid that contains the SV40 enhancer upstream of the composite HBV box and performed transient transfection experiments. Both highly and poorly differentiated hepatoma cell lines HepG2 and SK-Hep1, respectively, were transfected. All the reporter plasmids displayed dose-dependent activity regardless of the cell origin. However, the reporter plasmid containing the P(A)S-TATA promoter under the SV40 enhancer was more active in the highly differentiated HepG2 cell line (Fig. 6A) than in the SK-Hep1 cell line. In contrast, and in agreement with our previous findings (18), a reverse picture was seen with the control SV40 promoter/enhancer reporter plasmid (Fig. 6B). Thus, similar to the other HBV promoters, the novel P(A)S-TATA promoter is more active in the liver-derived differentiated cells.
FIG. 6.
Preferential promoter activity of the P(A)S-TATA box in highly differentiated hepatocytes. SK-Hep1 cells and HepG2 cells, poorly and highly differentiated hepatocytes, respectively, were transfected with increasing amounts of the luciferase reporter plasmids (0.1 to 1.0 ng of DNA per 2-cm-diameter plate). The SV40 enhancer/HBV P(A)S-TATA (A) and the SV40 enhancer/promoter (B) reporter plasmids were used. Luciferase activity was measured 48 h posttransfection. The experiments were performed in triplicate, and the average values and standard deviations are shown. Conc., concentration.
The composite P(A)S-TATA box is an active promoter in the context of the viral genome.
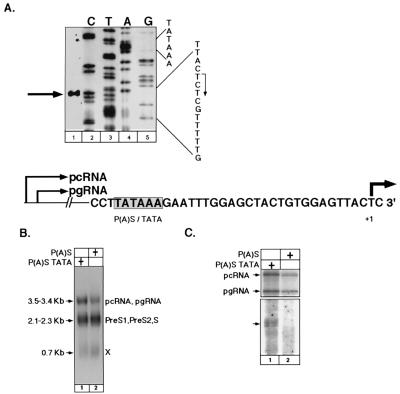
Having demonstrated that the HBV composite P(A)S-TATA box is active in GTF binding and in supporting transcription, we next investigated its possible promoter activity in the context of the viral genome. HepG2 permissive cells were transfected with HBV DNA, and RNA was extracted and subjected, along with an end-labeled 32P-labeled oligonucleotide primer, to primer extension reactions. The products were resolved in a denaturing polyacrylamide-urea gel along with that of the dideoxy-sequencing reaction. Interestingly, a band that mapped exactly at the expected site of transcription initiation programmed by the P(A)S-TATA box was detected (Fig. 7A). We concluded, therefore, that this composite box has TATA box activity in the context of the intact viral genome.
FIG. 7.
The composite P(A)S-TATA box is an active promoter in the context of the viral genome, as shown by primer extension analysis. HepG2 cells were transfected with 20 μg of a head-to-tail dimer of HBV DNA, RNA was extracted, and 3 μg of poly(A) RNA was incubated with an end-labeled 32P-oligonucleotide primer in primer extension reactions. The products were separated in a denaturing polyacrylamide-urea gel along with that of the dideoxy-sequencing reactions (A). The initiation sites are indicated (+1), with an arrow to show the transcription direction. To show that this initiation point is regulated by the composite poly(A) box, wt [P(A)S-TATA] DNA and a point mutant HBV DNA with a simple poly(A) signal [P(A)S] were used to transfect cells. Total RNA (10 μg) of each transfected plate was used for Northern blotting and hybridization with a 32P-HBV DNA probe (B). The known HBV RNA species and their calculated sizes are indicated. Poly(A) RNA (3 μg) from each of the transfected cells was used for primer extension reactions (C). The 5′ ends of the pc- and pgRNA are shown in the upper radiogram, and those of the novel transcript, initiating about 25 bases downstream of the composite poly(A) signal, are shown in the lower radiogram. The upper and lower radiograms were exposed for different times. Note the sharp reduction in the level of this RNA in the HBV mutant, while pc- and pgRNA levels were moderately affected (70% of wt).
The fact that the exact initiation site was also detected in the context of a reporter plasmid (Fig. 5) strongly suggests that a common promoter core element, i.e., a P(A)S-TATA box, was utilized in both cases. To further substantiate this possibility, an HBV DNA mutant that contained a simple poly(A) signal instead of the composite one was generated. The constructed HBV plasmids were transfected in the HepG2 cells, and RNA was extracted and analyzed. Northern blot analysis revealed that the expected HBV transcripts are produced by both wt and mutant HBV (Fig. 7B), although in the latter case the levels of pc- and pgRNA are lower (about 70%). Thus, under the employed conditions, the gene expression program of the HBV mutant was not dramatically changed. Next the RNA samples were subjected to primer extension analysis. This analysis revealed that both wt and mutant HBV DNA genomes utilize the same transcription initiation sites for the synthesis of pc- and pgRNA (Fig. 7C, top), but, as expected, the level of the mutant's transcripts was lower (about 70% of the wt). In sharp contrast the level of the novel transcript was undetectable when the mutant template was used (Fig. 7C, bottom). The fact that a simple poly(A) box cannot support the production of the novel transcript suggests that the composite HBV P(A)S-TATA box is a functional TATA box.
The deviant poly(A) signal is instrumental in HBV replication.
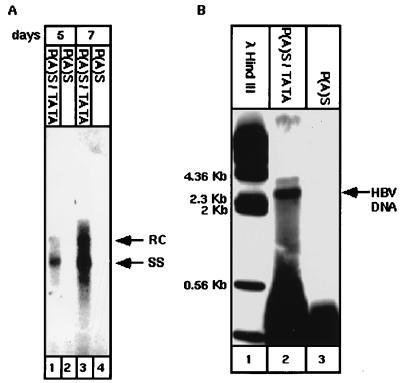
Having demonstrated that an HBV mutant bearing the canonical poly(A) signal is capable of programming the synthesis of the major viral transcripts, we next asked whether this virus is replication competent. Cells were transfected and analyzed after 5 and 7 days for the presence of the replicative HBV DNA intermediates. Both wt and canonical poly(A)-containing viruses produced equal amounts of HBsAg (data not shown); however, replicative HBV DNA intermediates were detected only when cells were transfected with wt HBV DNA (Fig. 8A). As has been reported previously (31), HBV DNA is accumulated after transfection, with the late appearance of the relaxed circular form (Fig. 8A, lane 3). These data clearly suggest that the composite poly(A) signal with the embedded TATA box is essential for virus replication. A similar conclusion was drawn from an experiment whereby the virions in the culture medium were assayed for DNA polymerase activity (Fig. 8B). Virions with active DNA polymerase were found only in the culture of wt-transfected cells, as measured by their capacity to incorporate [α-32P]dCTP into the viral genome.
FIG. 8.
The composite P(A)S-TATA is instrumental in viral replication. Cells were transfected with the wt and the mutant plasmids as for Fig. 7 and were harvested after 5 and 7 days. The extracted DNA was subjected to 1% agarose gel electrophoresis, Southern blotted, and hybridized with a 32P-labeled HBV DNA probe to determine the level of relaxed circular (RC) forms and single-stranded (SS) HBV DNA replicative intermediates (A). Viral particles were collected from culture media and assayed for their endogenous polymerase activity by measuring [α-32P]dATP incorporation into the viral genome (B).
DISCUSSION
The HBV genome contains multiple promoters, each designed to support the production of a specific transcript. This type of gene regulation program, which rarely employs splicing to increase the RNA repertoire, is rather unique considering the small genome of the virus. In this report we describe the presence of an additional promoter that so far was overlooked, perhaps due to its strategic position. We provide biochemical as well as functional evidence that the HBV polyadenylation signal sustains a cryptic promoter activity. The HBV polyadenylation signal differs from the canonical one (AATAAA) by a single base change and contains the sequence TATAAA (the changed base is underlined). Such a change in the first nucleotide is very uncommon in vertebrate genomes (0.8% contain TATAAA and 98% contain AATAAA), and yet it is found in the genomes of all the hepadnaviruses that infect mammals. This deviant signal bears a cryptic TATA box and is therefore regarded as a composite element. By employing recombinant GTFs we could show that this cryptic TATA box binds TBP, TFIIA, and TFIIB in a manner characteristic of an authentic TATA box (19, 27). At a low TBP concentration no DNA-binding activity was seen, but this was changed by the addition of TFIIA and TFIIB. The former is sufficient to increase the DNA-binding activity of TBP (32, 45). The addition of TFIIB resulted in the appearance of a strong DNase I-hypersensitive site, as was reported by others (27). Recently, it was shown that TFIIB has a DNA-binding domain that interacts with a CG-rich sequence positioned immediately upstream of the TATA box (24). Interestingly, a similar sequence is found at the correct position next to the HBV composite P(A)S-TATA box (see Fig. 5 for the sequence). The fact that TFIIB induced the appearance of a strong DNase I-hypersensitive site is a good indication that it binds the TATA box upstream region. This behavior of TFIIB not only provides strong support for the HBV composite P(A)S-TATA box in sustaining promoter characteristics but also was helpful for mapping the orientation of this TATA box (27). It was reported that the TATA box can in fact act bidirectionally, and the involvement of auxiliary TFIIA and TFIIB is essential to determine the correct orientation (7). Based on these data we defined the TATA box orientation to be at the viral positive strand, where all the other viral promoters have been mapped.
Functional analysis in the context of a heterologous reporter system revealed that the HBV composite box has promoter activity. At the moment we do not know how this promoter is regulated, but our data tend to suggest that it is a weak one. Based on the primer extension analysis we have estimated that this promoter is about 20-fold weaker than the pc- and pgRNA promoters. The presence of a nearby repressor element could explain the weak activity of the promoter, but it also could be an intrinsic behavior of this bifunctional box. The fact that the embedded cryptic TATA box of the HBV composite P(A)S-TATA element was responsible for the promoter activity was confirmed by mutagenesis studies. A point mutation that changed the TATA sequence to AATA resulted in the lost of GTF binding activity, with concomitant inability to support transcription.
To show that this promoter is active in the context of the viral genome, we conducted DNA transfection experiments. In the absence of tissue culture infection this approach is widely used to study the viral life cycle (1, 36, 44). Under these conditions, by primer extension analysis we revealed a novel transcription initiation site next to the P(A)S-TATA box. Others have detected this very same initiation site in cell lines that stably express some of the viral transcripts (35). Thus, it is likely that this promoter is active under a wide range of conditions. However, to strengthen the biological significance of this finding, in vivo footprinting is required to confirm the occupancy of this box inside the cells. Although this is an important experiment, it cannot be performed with plasmid-transfected cells, and we must await the development of an infection system.
The HBV pgRNA is longer than the genome; therefore, for its synthesis the transcription machinery must ignore the poly(A) signal at the first round of transcription. The deviant nature of the HBV poly(A) signal might be important in this process (33). Indeed, in vitro studies designed to determine the relationship between the sequence of the poly(A) signal and its activity have classified the HBV type of poly(A) signal sequence (TATAAA) as a poor one because it supports RNA processing with only 17% efficiency (39). Although this model is appealing, in our experiments we see only about a twofold reduction in the level of pgRNA synthesis by a viral genome containing a canonical poly(A) signal compared to the wt level (Fig. 7A). This model is also inconsistent with the fact that duck HBV DNA, experiencing the same pattern of gene expression, contains a canonical poly(A) signal. Furthermore, the poly(A) signal of the retroviruses is often positioned at the R region, which is present at both the 5′ and 3′ ends. The former must be occluded, whereas the latter is efficiently utilized. This differential employment of the poly(A) signal of a given genome is not achieved by the presence of a deviant poly(A) box but rather by other mechanisms (reference 22 and the references therein). Therefore, a deviant poly(A) signal is not instrumental in this process but might have additional roles, such as functioning as a promoter.
A remarkable finding is that HBV DNA with a canonical poly(A) signal is replication defective despite its capacity to program, albeit with lower efficiency, the production of all the viral transcripts. This might suggest that a very essential activity is missing. The first relevant and functional AUG of the putative novel transcript is that of the viral Pol ORF. Pol is exceptional in the sense that it is the only viral protein whose mechanism of production remains obscure. It has been speculated that Pol is translated from the polycistronic pgRNA, but the underlying molecular mechanism is still unknown (for a review see reference 11). The present work raises the possibility that a novel HBV transcript regulates Pol production. This intriguing possibility was in part supported by genetic complementation analysis (data not shown). Mutation of the HBV P(A)S to a canonical one was accompanied by a single amino acid change encoded by the overlapping core ORF. The constructed HBV mutant was extremely inefficient in core protein accumulation, and, therefore, Pol was insufficient to rescue the HBV P(A)S. An optimal complementation, however, was achieved only when both the core and Pol were supplemented in trans (data not shown), consistent with the possibility that the novel transcript is responsible for Pol production.
Primer extension analysis revealed that the novel transcript is present at a much lower level (about 20-fold) than pc- and pgRNA. Also, the activity of its promoter in the context of the P(A)S-TATA reporter plasmids is lower than that of the E1b TATA box (Fig. 4). The poor activity of this novel promoter is in accordance with the assumption that only a single Pol polypeptide per virion is encapsidated (2), as opposed to the core protein, for which about 240 polypeptides are required (4). The low level of this putative transcript might be the reason why so far it has escaped detection by the conventional RNA analysis tools.
A composite poly(A) signal with dual functions might be a mechanism exclusively adopted by the viruses that have highly compact genome structures. However, an interplay between the poly(A) signal and transcription initiation was also found in Saccharomyces cerevisiae (13). It was found that a deletion in the GAL10 poly(A) signal resulted in complete inactivation of the GAL7 promoter, implying a pivotal role for the poly(A) site in the transcription of a downstream gene. Thus, the poly(A) signal is also involved in the termination to initiation switching. This documented case, together with our results, suggests the interesting possibility that the poly(A) signal may sustain additional functions in gene regulation beyond its well-characterized role in supporting correct processing of the RNA 3′ end.
Finally, the composite poly(A) signal, in the context of an episomal genome, can support the production of an approximately genome length transcript. Transcription initiation occurs about 20 to 30 bp downstream of the TATA box, and the stretch of A residues is usually added about 15 to 30 bases downstream of the poly(A) signal, where the initiation took place. At the moment we do not know what might be the role of a genome length transcript in the HBV life cycle. However, to our knowledge, in animal cells so far no mechanism for synthesis of such a transcript has been described; therefore, the significance of this finding might be general and relevant to different organisms.
ACKNOWLEDGMENT
N.P. and A.O. contributed equally to this work.
REFERENCES
- 1.Acs G, Sells M A, Purcell R H, Price P, Engle R, Shapiro M, Popper H. Hepatitis B virus produced by transfected Hep G2 cells causes hepatitis in chimpanzees. Proc Natl Acad Sci USA. 1987;84:4641–4644. doi: 10.1073/pnas.84.13.4641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bartenschlager R, Schaller H. Hepadnaviral assembly is initiated by polymerase binding to the encapsidation signal in the viral RNA genome. EMBO J. 1992;11:3413–3420. doi: 10.1002/j.1460-2075.1992.tb05420.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Benn J, Su F, Doria M, Schneider R J. Hepatitis B virus HBx protein induces transcription factor AP-1 by activation of extracellular signal-regulated and c-Jun N-terminal mitogen-activated protein kinases. J Virol. 1996;70:4978–4985. doi: 10.1128/jvi.70.8.4978-4985.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bottcher B, Wynne S A, Crowther R A. Determination of the fold of the core protein of hepatitis B virus by electron cryomicroscopy. Nature. 1997;385:88–91. doi: 10.1038/386088a0. [DOI] [PubMed] [Google Scholar]
- 5.Buratowski S, Hahn S, Guarente L, Sharp P A. Five intermediate complexes in transcription initiation by RNA polymerase II. Cell. 1989;56:549–561. doi: 10.1016/0092-8674(89)90578-3. [DOI] [PubMed] [Google Scholar]
- 6.Chen I H, Huang C J, Ting L P. Overlapping initiator and TATA box functions in the basal core promoter of hepatitis B virus. J Virol. 1995;69:3647–3657. doi: 10.1128/jvi.69.6.3647-3657.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cox J M, Hayward M M, Sanchez J F, Gegnas L D, van der Zee S, Dennis J H, Sigler P B, Schepartz A. Bidirectional binding of the TATA box binding protein to the TATA box. Proc Natl Acad Sci USA. 1997;94:13475–13480. doi: 10.1073/pnas.94.25.13475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dikstein R, Faktor O, Ben-Levy R, Shaul Y. Functional organization of the hepatitis B virus enhancer. Mol Cell Biol. 1990;10:3682–3689. doi: 10.1128/mcb.10.7.3683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Doria M, Klein N, Lucito R, Schneider R J. The hepatitis B virus HBx protein is a dual specificity cytoplasmic activator of Ras and nuclear activator of transcription factors. EMBO J. 1995;14:4747–4757. doi: 10.1002/j.1460-2075.1995.tb00156.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Faktor O, De-Medina T, Shaul Y. Regulation of hepatitis B virus S gene promoter in transfected cell lines. Virology. 1988;162:362–368. doi: 10.1016/0042-6822(88)90476-x. [DOI] [PubMed] [Google Scholar]
- 11.Ganem D. Hepadnaviridae and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 2703–2737. [Google Scholar]
- 12.Garcia A D, Ostapchuk P, Hearing P. Functional interaction of nuclear factors EF-C, HNF-4, and RXRα with hepatitis B virus enhancer I. J Virol. 1993;67:3940–3950. doi: 10.1128/jvi.67.7.3940-3950.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Greger I H, Proudfoot N J. Poly(A) signals control both transcriptional termination and initiation between the tandem GAL10 and GAL7 genes of Saccharomyces cerevisiae. EMBO J. 1998;17:4771–4779. doi: 10.1093/emboj/17.16.4771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haviv I, Matza Y, Shaul Y. pX, the HBV-encoded coactivator, suppresses the phenotypes of TBP and TAFII250 mutants. Genes Dev. 1998;12:1217–1226. doi: 10.1101/gad.12.8.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haviv I, Shamay M, Doitsh G, Shaul Y. Hepatitis B virus pX targets TFIIB in transcription coactivation. Mol Cell Biol. 1998;18:1562–1569. doi: 10.1128/mcb.18.3.1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Haviv I, Vaizel D, Shaul Y. The X protein of the hepatitis B virus coactivates acidic activation domains. Mol Cell Biol. 1995;15:1079–1085. doi: 10.1128/mcb.15.2.1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haviv I, Vaizel D, Shaul Y. pX, the HBV-encoded coactivator, interacts with components of the transcription machinery and stimulates transcription in a TAF-independent manner. EMBO J. 1996;15:3413–3420. [PMC free article] [PubMed] [Google Scholar]
- 18.Honigwachs J, Faktor O, Dikstein R, Shaul Y, Laub O. Liver-specific expression of hepatitis B virus is determined by the combined action of the core gene promoter and the enhancer. J Virol. 1989;63:919–924. doi: 10.1128/jvi.63.2.919-924.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Imbalzano A N, Zaret K S, Kingston R E. Transcription factor (TF) IIB and TFIIA can independently increase the affinity of the TATA-binding protein for DNA. J Biol Chem. 1994;269:8280–8286. [PubMed] [Google Scholar]
- 20.Johnson P F, Landschulz W H, Graves B J, McKnight S L. Identification of a rat liver nuclear protein that binds to the enhancer core element of three animal viruses. Genes Dev. 1987;1:133–146. doi: 10.1101/gad.1.2.133. [DOI] [PubMed] [Google Scholar]
- 21.Junker M, Galle P, Schaller H. Expression and replication of the hepatitis B virus genome under foreign promoter control. Nucleic Acids Res. 1987;15:10117–10132. doi: 10.1093/nar/15.24.10117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Klasens B I, Das A T, Berkhout B. Inhibition of polyadenylation by stable RNA secondary structure. Nucleic Acids Res. 1998;26:1870–1876. doi: 10.1093/nar/26.8.1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Klein N P, Schneider R J. Activation of Src family kinases by hepatitis B virus HBx protein and coupled signaling to Ras. Mol Cell Biol. 1997;17:6427–6436. doi: 10.1128/mcb.17.11.6427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lagrange T, Kapanidis A N, Tang H, Reinberg D, Ebright R H. New core promoter element in RNA polymerase II-dependent transcription: sequence-specific DNA binding by transcription factor IIB. Genes Dev. 1998;12:34–44. doi: 10.1101/gad.12.1.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lin Y, Tang H, Nomura T, Dorjsuren D, Hayashi N, Wei W, Ohta T, Roeder R, Murakami S. The hepatitis B virus X protein is a co-activator of activated transcription that modulates the transcription machinery and distal binding activators. J Biol Chem. 1998;273:27097–27103. doi: 10.1074/jbc.273.42.27097. [DOI] [PubMed] [Google Scholar]
- 26.Luber B, Lauer U, Weiss L, Hohne M, Hofschneider P H, Kekule A S. The hepatitis B virus transactivator HBx causes elevation of diacylglycerol and activation of protein kinase C. Res Virol. 1993;144:311–321. doi: 10.1016/s0923-2516(06)80047-6. [DOI] [PubMed] [Google Scholar]
- 27.Maldonado E, Ha I, Cortes P, Weis L, Reinberg D. Factors involved in specific transcription by mammalian RNA polymerase II: role of transcription factors IIA, IID, and IIB during formation of a transcription-competent complex. Mol Cell Biol. 1990;10:6335–6347. doi: 10.1128/mcb.10.12.6335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Natoli G, Avantaggiati M L, Chirillo P, Puri P L, Ianni A, Balsano C, Levrero M. Ras- and Raf-dependent activation of c-jun transcriptional activity by the hepatitis B virus transactivator pX. Oncogene. 1994;9:2837–2843. [PubMed] [Google Scholar]
- 29.Ori A, Shaul Y. Hepatitis B virus enhancer binds and is activated by the hepatocyte nuclear factor 3. Virology. 1995;207:98–106. doi: 10.1006/viro.1995.1055. [DOI] [PubMed] [Google Scholar]
- 30.Patel N U, Jameel S, Isom H, Siddiqui A. Interactions between nuclear factors and the hepatitis B virus enhancer. J Virol. 1989;63:5293–5301. doi: 10.1128/jvi.63.12.5293-5301.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pugh J C, Yaginuma K, Koike K, Summers J. Duck hepatitis B virus (DHBV) particles produced by transient expression of DHBV DNA in a human hepatoma cell line are infectious in vitro. J Virol. 1988;62:3513–3516. doi: 10.1128/jvi.62.9.3513-3516.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Roeder R G. The role of general initiation factors in transcription by RNA polymerase II. Trends Biochem Sci. 1996;21:327–335. [PubMed] [Google Scholar]
- 33.Russnak R, Ganem D. Sequences 5′ to the polyadenylation signal mediate differential poly(A) site use in hepatitis B viruses. Genes Dev. 1990;4:764–776. doi: 10.1101/gad.4.5.764. [DOI] [PubMed] [Google Scholar]
- 34.Sanfacon H. Analysis of figwort mosaic virus (plant pararetrovirus) polyadenylation signal. Virology. 1994;198:39–49. doi: 10.1006/viro.1994.1006. [DOI] [PubMed] [Google Scholar]
- 35.Schranz P, Zentgraf H, Schroder C H. Integrated defective replication units of hepatitis B virus. Virus Genes. 1990;4:367–374. doi: 10.1007/BF00570031. [DOI] [PubMed] [Google Scholar]
- 36.Sells M A, Chen M L, Acs G. Production of hepatitis B virus particles in Hep G2 cells transfected with cloned hepatitis B virus DNA. Proc Natl Acad Sci USA. 1987;84:1005–1009. doi: 10.1073/pnas.84.4.1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shaul Y. Regulation of HBV transcription. In: McLachlan A, editor. Molecular biology of the hepatitis B virus. Boca Raton, Fla: CRC Press; 1991. pp. 193–211. [Google Scholar]
- 38.Shaul Y, Ben-Levy R. Multiple nuclear proteins in liver cells are bound to hepatitis B virus enhancer element and its upstream sequences. EMBO J. 1987;6:1913–1920. doi: 10.1002/j.1460-2075.1987.tb02451.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sheets M D, Ogg S C, Wickens M P. Point mutations in AAUAAA and the poly (A) addition site: effects on the accuracy and efficiency of cleavage and polyadenylation in vitro. Nucleic Acids Res. 1990;18:5799–5805. doi: 10.1093/nar/18.19.5799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Silver-Key S C, Pagano J S. A noncanonical poly(A) signal, UAUAAA, and flanking elements in Epstein-Barr virus DNA polymerase mRNA function in cleavage and polyadenylation assays. Virology. 1997;234:147–159. doi: 10.1006/viro.1997.8647. [DOI] [PubMed] [Google Scholar]
- 41.Su F, Schneider R J. Hepatitis B virus HBx protein activates transcription factor NF-κB by acting on multiple cytoplasmic inhibitors of rel-related proteins. J Virol. 1996;70:4558–4566. doi: 10.1128/jvi.70.7.4558-4566.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tuttleman J S, Pugh J C, Summers J W. In vitro experimental infection of primary duck hepatocyte cultures with duck hepatitis B virus. J Virol. 1986;58:17–25. doi: 10.1128/jvi.58.1.17-25.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wilusz J, Pettine S M, Shenk T. Functional analysis of point mutations in the AAUAAA motif of the SV40 late polyadenylation signal. Nucleic Acids Res. 1989;17:3899–3908. doi: 10.1093/nar/17.10.3899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yaginuma K, Shirakata Y, Kobayashi M, Koike K. Hepatitis B virus (HBV) particles are produced in a cell culture system by transient expression of transfected HBV DNA. Proc Natl Acad Sci USA. 1987;84:2678–2682. doi: 10.1073/pnas.84.9.2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yokomori K, Zeidler M P, Chen J L, Verrijzer C P, Mlodzik M, Tjian R. Drosophila TFIIA directs cooperative DNA binding with TBP and mediates transcriptional activation. Genes Dev. 1994;8:2313–2323. doi: 10.1101/gad.8.19.2313. [DOI] [PubMed] [Google Scholar]