Abstract
Methylation of cytosines in the CpG dinucleotide is generally associated with transcriptional repression in mammalian cells, and recent findings implicate histone deacetylation in methylation-mediated repression. Analyses of histone acetylation in in vitro-methylated transfected plasmids support this model; however, little is known about the relationships among de novo DNA methylation, transcriptional repression, and histone acetylation state. To examine these relationships in vivo, we have developed a novel approach that permits the isolation and expansion of cells harboring expressing or silent retroviruses. MEL cells were infected with a Moloney murine leukemia virus encoding the green fluorescent protein (GFP), and single-copy, silent proviral clones were treated weekly with the histone deacetylase inhibitor trichostatin A or the DNA methylation inhibitor 5-azacytidine. Expression was monitored concurrently by flow cytometry, allowing for repeated phenotypic analysis over time, and proviral methylation was determined by Southern blotting and bisulfite methylation mapping. Shortly after infection, proviral expression was inducible and the reporter gene and proviral enhancer showed a low density of methylation. Over time, the efficacy of drug induction diminished, coincident with the accumulation of methyl-CpGs across the provirus. Bisulfite analysis of cells in which 5-azacytidine treatment induced GFP expression revealed measurable but incomplete demethylation of the provirus. Repression could be overcome in late-passage clones only by pretreatment with 5-azacytidine followed by trichostatin A, suggesting that partial demethylation reestablishes the trichostatin-inducible state. These experiments reveal the presence of a silencing mechanism which acts on densely methylated DNA and appears to function independently of histone deacetylase activity.
Transcriptional repression of heterologous genetic elements such as proviruses is often observed concomitantly with their integration and packaging into chromatin in the host cell genome (57). Transcriptional activity is negatively influenced in cis by deacetylation of nucleosomal histones (4, 54, 56) and methylation of cytosines (22, 29, 45, 55), particularly in the dinucleotide 5′CpG (cytosine-guanine). Transfection of in vitro-methylated constructs reveals a clear relationship between methylation density and transcriptional repression (6, 27). While the repression associated with methylated DNA is known to require a chromatin template (8, 31) and involve a condensed chromatin structure (32), the mechanism underlying this alteration remained unresolved until recently. The description of a biochemical association between the transcriptional-repression domain (TRD/MRD) of the methyl-CpG (mCpG)-binding protein MeCP2 and a corepressor complex containing histone deacetylases (HDACs) suggests that mCpGs suppress transcription by recruiting HDACs which deacetylate chromatin in cis (30, 43). Chromatin immunoprecipitation experiments lend further support to this model: in contrast to an unmethylated reporter gene, stable transfection of an identical but in vitro-methylated construct reveals no preferential association with acetylated nucleosomal fractions (17). However, failure of the extremely potent histone deacetylase inhibitor trichostatin A (TSA) (60) to fully reactivate TRD/MRD-mediated repression (30, 43), as well as a report of repression mediated by the MeCP2 methyl-CpG binding domain (MBD) alone (34) suggests that MeCP2-mediated repression may also involve acetylation-independent mechanisms. Furthermore, the recent description of several novel proteins with MBDs demonstrates that additional factors may play a role in methylation-mediated repression (15, 24).
Experiments involving in vitro methylation and a stable episomal reporter demonstrate that transcriptional activity and induction with the nonspecific HDAC inhibitor sodium butyrate are inversely dependent on methylation density (27). Similarly, a mutant FMR1 gene harboring an expanded, hypermethylated CGG repeat is refractory to TSA-mediated transcriptional induction (14). Recently, Cameron et al. (10) showed that several densely methylated endogenous genes could be reactivated only by treatment with 5-azacytidine (5-azaC) followed by TSA and proposed, based on these results, that CpG methylation and histone deacetylation act synergistically to silence CpG island-containing genes. While TSA has been shown to induce the expression of integrated viruses (11, 56), the relationships among methylation density, histone deacetylation, and expression of proviral sequences have yet to be addressed. In this study, we used a green fluorescent protein (GFP)-based retroviral reporter system (1) to assess the dynamic relationship between proviral induction, de novo methylation, and histone acetylation. In contrast to conventional drug selection, this fluorescence-activated cell sorter (FACS)-based assay permits the isolation and expansion of expressing or nonexpressing subpopulations for further analysis. Furthermore, while transfection or microinjection studies of in vitro-methylated DNA provide a static picture of the influence of DNA methylation (6, 27, 42), the proviral template characterized here is integrated in the host genome and progressively methylated de novo. Consequently, we believe that this system more accurately reflects the native process of mCpG-associated transcriptional repression of foreign genetic elements (52).
We generated MEL cell clones with single-copy proviral integrants in which expression could be induced by TSA. At weekly intervals after infection, subcultures of these clones were treated with TSA or the cytidine analog 5-azaC (22), which inhibits DNA methylation, and proviral expression was monitored by FACS analysis. Proviral methylation status was simultaneously measured by Southern analysis and the bisulfite sequencing method. Surprisingly, in the majority of clones analyzed, a progressive diminution of drug-inducible expression was observed coincident with an increase in proviral methylation. Under the conditions used, the provirus was only partially demethylated in the presence of 5-azaC, yielding a methylation density comparable to that seen in earlier-passage TSA-inducible cells. In late-passage clones, the provirus was densely methylated and transcriptional repression was relieved by pretreatment with 5-azaC followed by TSA, but not with either drug alone, results consistent with those recently reported for several hypermethylated endogenous genes (10). These experiments reveal the dynamic nature of DNA methylation and associated transcriptional repression and suggest that while repression of nascent provirus bearing a low level of methylation is mediated primarily by HDAC activity, late-passage highly methylated proviral DNA appears to be repressed by an HDAC-independent mechanism.
MATERIALS AND METHODS
Retroviral infections.
To generate a GFP gene optimized for flow cytometric analysis, a “humanized” GFP gene (hGFP) (optimized for translational efficiency in mammalian cells [23]) was mutated to improve stability and fluorogenic properties (reference 1 and data not shown), yielding hGFP-Bex1. This GFP variant was cloned between the NcoI and BamHI sites of the Moloney murine leukemia virus (M-MuLV) vector MFG (16), yielding the retroviral vector MFG-hGFP. High-titer MFG-hGFP retrovirus was generated by calcium phosphate transfection of Phoenix A retroviral producer cells as described previously (25). Retroviral supernatant was added to 5 × 104 MEL (745-A) cells cultured in log phase in 2 ml of growth medium (RPMI 1640 medium supplemented with 10% bovine calf serum, 100 U of penicillin per ml, 0.05 mM streptomycin, and 2 mM glutamine). To maximize infection efficiency (3), the cell suspension-retroviral supernatant mix was supplemented with 5 μg of Polybrene per ml and centrifuged at 2,000 rpm for 1 h at room temperature. The cells were harvested 7 h later, washed, resuspended in Polybrene-free growth medium, and cultured under standard laboratory conditions. The MEL MFG-hGFP clones were derived as described in the legend to Fig. 1. Cells were maintained in log-phase growth for all experiments.
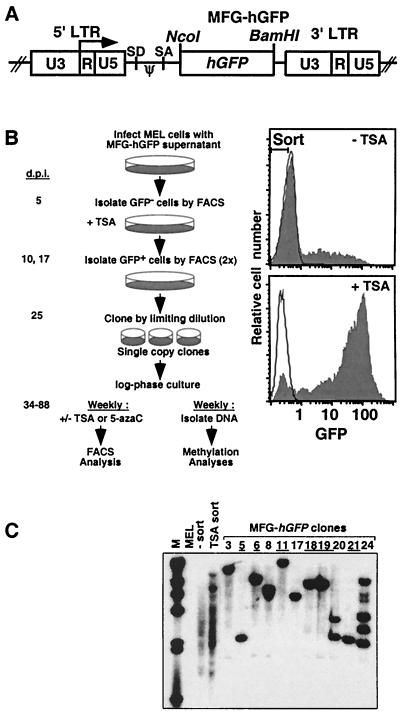
FIG. 1.
The MFG-hGFP vector and strategy for generating TSA-inducible MEL clones. (A) The 720-bp hGFP gene was inserted downstream of the splice acceptor site (SA) in the MFG retroviral vector. Transcriptional regulation of hGFP is mediated by the 5′ LTR promoter-enhancer (ψ, packaging signal; SD, splice donor). (B) MEL cells were infected with MFG-hGFP as described in Materials and Methods. At 5 days p.i. (d.p.i.), 5 × 104 GFP-negative cells were sorted from the infected population (Top filled histogram, with sort gate). The sorted negative cells were treated with TSA at 9 days p.i., and GFP-positive cells were sorted from this population on day 10. The majority of cells in this pool of TSA-inducible cells became silent by day 16, at which time the cells were treated again with TSA (bottom filled histogram), and GFP-positive cells were sorted on day 17. These TSA-responsive cells were cloned by limiting dilution at 25 days p.i. Of 24 clones analyzed, 10 showed very low to undetectable levels of spontaneous GFP expression. Beginning at 34 days p.i., subcultures of six of these clones (clones 5, 6, 11, 18, 19, and 21) were harvested weekly for analysis of TSA and 5-azaC inducibility, as measured by FACS, and for methylation status, as measured by Southern blotting and bisulfite analysis. Unfilled histograms show the fluorescence distributions of uninfected MEL cells. (C) The proviral copy number was determined by Southern blotting with an hGFP probe. Each of the clones chosen for further analysis (underlined) harbors a single MFG-hGFP provirus at a unique integration site (lanes: MEL, uninfected cells; − sort, infected MEL cells sorted 5 days p.i.; TSA sort, infected pool immediately prior to cloning).
TSA and 5-azaC treatment, FACS analysis, and cell sorting.
Preliminary experiments with TSA revealed that both cytotoxicity and proviral expression increase in a dose-dependent manner (data not shown). In all experiments, 100 nM TSA was used, a concentration which yielded maximal induction while minimizing cytotoxicity; >90% of cells were viable as measured by propidium iodide (PI) staining. For each experiment, aliquots of TSA (Wako Pure Chemical Industries, Ltd.) (dissolved at 5 mg/ml in methanol and stored at −20°C) were thawed, diluted in fresh growth medium, and immediately added to MEL cells. Aliquots of 5-azaC (Sigma) (1.2 mg/ml in water) were added at a final concentration of 5 μM (58). Cells were harvested 24 h after addition of TSA and/or 5-azaC, unless otherwise indicated. Prior to analysis, single-cell suspensions were centrifuged at 1,200 × g for 5 min and washed with staining medium (phosphate-buffered saline supplemented with 3% [vol/vol] fetal calf serum). Cells were resuspended in staining medium supplemented with propidium iodide (PI) (1 μg/ml, final concentration) for live-dead discrimination. FACS analyses and cell sorting were carried out on FACSCalibur and FACS Vantage (Becton Dickinson) cytometers, respectively, both equipped with the standard fluorescein filter set. Multiparameter data were analyzed by using the FlowJo analysis package (Tree Star Inc.). Data on a minimum of 10,000 live cells were collected for each sample. For cell sorting, electronic gates were established to exclude dead cells on the basis of light scatter and PI fluorescence.
Southern blot hybridization and methylation blotting.
Preparation of high-molecular-weight genomic DNA, restriction digests, membrane transfers, and preparation of the DNA probe were performed by standard methods (50). The GFP probe used for Southern hybridization was generated by digestion of the MFG-hGFP plasmid with NcoI and BamHI, yielding a restriction fragment including the 720-bp hGFP gene. Single-copy integrants were identified by digestion of genomic DNA with BamHI, which cuts once in the MFG-hGFP provirus, followed by Southern blot analysis with the GFP probe. The methylation status of the hGFP gene was determined by digestion of 20 μg of genomic DNA with BamHI and the methylation-sensitive enzyme HpaII or, as a control, the methylation-insensitive isoschizomer MspI. The methylation status of the 5′ long terminal repeat (LTR) was determined by digestion with BamHI and the methylation-sensitive enzyme BssHII. The restriction fragments were quantitated by PhosphorImager (Molecular Dynamics) analysis, which revealed a weak nonspecific band that comigrated with the BssHII-BamHI fragment. To correct for the contribution of this nonspecific band in the quantitative analyses, the fraction of comigrating DNA in the BamHI-alone lane was subtracted from the total DNA value and the fraction of DNA resistant to BssHII digestion in each sample was divided by the resulting value. The nonspecific band represented a maximum of 10% of the DNA in each lane.
Sodium bisulfite treatment.
The bisulfite conversion was carried out, with minor modifications, by the method developed by Clark et al. (13). Genomic DNA was linearized with BamHI, phenol-chloroform extracted, and ethanol precipitated. For the time course study, 100 ng of digested DNA was mixed with 2 μg of tRNA as carrier and denatured by adding freshly prepared NaOH to a final concentration of 0.3 M in a 20-μl reaction volume and incubating the mixture at 42°C for 30 min. For the cell-sorting experiment, 5,000 sorted viable cells were resuspended in a proteinase K (40 μg/ml, final concentration)–sodium dodecyl sulfate (1.6%, final concentration) solution and denatured as for the genomic DNA. Fresh solutions of sodium bisulfite (Sigma), adjusted to pH 5 with NaOH, and hydroquinone (Sigma) were prepared and added to the denatured DNA at final concentrations of 3.4 M and 1 mM, respectively, in a final volume of 100 μl. DNA solutions were gently mixed, overlaid with mineral oil, and incubated at 55°C for 8 to 16 h. Unreacted bisulfite was removed by spin column chromatography (MicroSpin sephacryl S-200 HR; Amersham) as recommended by the manufacturer. Purified DNA samples (equilibrated in Tris-EDTA [TE] buffer) were mixed with NaOH at a final concentration of 0.3 M and incubated at 37°C for 20 min. The desulfonated samples were neutralized on an S-200 HR column, and the flowthrough fraction (∼100 μl) containing the converted DNA was stored at −20°C.
PCR amplification, cloning, and sequence analysis.
Reaction mixtures containing 5 μl of bisulfite-treated DNA (50 μl final vol) were subjected to 25 to 32 amplification cycles with a GeneAmp PCR system 9700 (Perkin-Elmer), with denaturation at 94°C, annealing at 49 to 56°C and extension at 72°C. Nested or seminested amplification was performed with 2 μl of product from the first round in a 50-μl reaction mixture. Primers were designed to favor the amplification of bisulfite-converted DNA. If the template strand included a CpG, degeneracy was incorporated into the primer at the nucleotide position corresponding to the cytosine such that no bias for amplification of methylated template was introduced. For analysis of the methylation state of the plus strand of the M-MuLV 5′ LTR, the primers used in the first round were +25+ (TAGGTTTGGTAAGTTAGTTTAAGTAAYGTT) (where Y is thymosine or cytidine) and +1080− (TAAAAAAATAATAACAAACTAACCCRAAC) (where R is adenosine or guanosine) and the primers used in the second round were +58+ (TTGTAAGGTATGGAAAAATATATAATTG) and +665− (TAAATTACTAACCAACTTACCTCCCRATAA). For analysis of the methylation state of the plus strand of the GFP gene, the primers used in the first round were +1870+ (ATTATTTTTTAGATTGTTATGGTGAGTAAGGG) and +2300− (CTCAAGCTTATAATTATACTCCAACTTATACCCCA) and the primers used in the second round were +1903+ (GAGGAGTTGTTTATYGGGGTGGTGTTT) and +2300−. Amplification products were directly purified by using the Qiaquick PCR purification kit (Qiagen) or subjected to electrophoresis in 1.5% Tris-acetate-EDTA (TAE) agarose gels and purified by using the Qiaquick gel extraction kit (Qiagen). The purified amplification products were ligated into plasmid p-GEM-T Easy by using the p-GEM-T Easy vector system (Promega) and transformed into competent Escherichia coli XL-1 Blue by the CaCl2 method. Colonies were screened on the basis of 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) staining, and plasmid DNA was prepared by the alkali lysis method. Plasmid clones harboring inserts of the appropriate size, as measured by restriction analysis, were amplified with the SP6 promoter primer and the ABI PRISM Dye Terminator cycle sequencing ready-reaction kit and sequenced with an ABI PRISM 377 DNA sequencer (Perkin-Elmer). Sequence analyses were conducted with the Sequencher sequence analysis package (Gene Codes Corporation), and sequence data from bisulfite-treated DNA was compared to the vector sequence generated with nonconverted template DNA. Each plasmid clone represents a single “allelic” variant of the original clonal population. The sample mean value of mCpGs for each genomic sample was determined by summing the number of mCpGs determined for each allele and dividing by the total number of alleles sequenced. The standard error of the mean (SEM) is given for each allelic population. Statistical significance was determined by using the unpaired two-sample t test assuming unequal variance. Two-tailed P values are presented.
RESULTS
TSA- and 5-azaC-induced proviral expression decreases with time in culture.
Infection of MEL cells with the M-MuLV-based vector MFG-hGFP (Fig. 1A) yielded a subpopulation of infected cells harboring silenced proviral integrants (Fig. 1B and C, lane 3), reflecting the presence of integration sites which do not support expression (26, 28). These GFP-negative cells were sorted and, to isolate the subpopulation of proviral integrants inducible with TSA, subjected to two consecutive rounds of TSA treatment and FACS sorting of GFP-positive cells. The TSA-inducible cells were cloned by limiting dilution and screened for proviral copy number by Southern blotting (Fig. 1C). Six single-copy TSA-inducible proviral clones (clones 5, 6, 11, 18, 19, and 21) were serially passaged in log-phase growth. At weekly intervals, subcultures of each clone were treated with TSA or the methylation inhibitor 5-azaC, previously shown to demethylate and activate M-MuLV proviral integrants (26, 44, 53). After 24 h of treatment, the cells were harvested and analyzed for GFP expression by FACS. Untreated cells were also harvested for analysis of proviral DNA methylation (Fig. 1B).
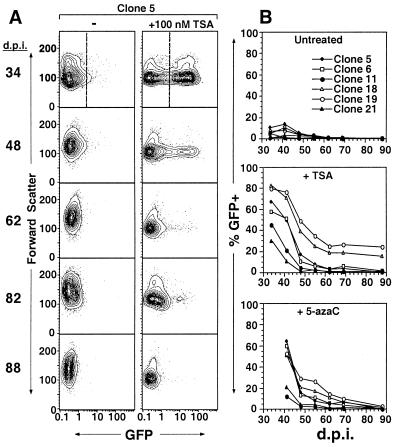
FACS analysis revealed a progressive diminution of TSA and 5-azaC induction with time in culture. Figure 2A shows a representative clone (clone 5) in which TSA induced GFP expression in more than 60% of viable cells 34 days postinfection (p.i.) while fewer than 2% responded 88 days p.i. A summary of the data collected for each clone reveals a clear time-dependent decrease in both spontaneous and inducible GFP expression, with TSA and 5-azaC inducibility being greatly diminished or entirely extinguished by day 88 p.i. (Fig. 2B). A similar diminution of responsiveness was detected when the clones were treated with sodium butyrate (reference 48 and data not shown). Comparison of the TSA and 5-azaC results revealed a clear correlation between TSA and 5-azaC responsiveness; for clones 5, 6, 18 and 19, a higher percentage of cells responded to either drug than did the cells of clones 11 and 21. This correlation reveals that the same position effect may influence the rate at which proviruses become refractory to TSA- and 5-azaC-mediated induction.
FIG. 2.
Induction of GFP expression by TSA and 5-azaC declines over time. MEL MFG-hGFP clones were generated as described in the legend to Fig. 1 and cultured in log phase. Aliquots were removed weekly and treated with 100 nM TSA or 5 μM 5-azaC for 24 h. Electronic gating in the PI staining and forward- and side-scatter channels was applied to exclude dead cells. The induction experiments were initiated 34 days p.i. (d.p.i.) because of the time required to derive TSA-responsive clones. (A) TSA-treated and untreated samples from clone 5 assayed on successive days p.i., displayed as 5% probability contour plots of GFP fluorescence versus forward scatter. Note that in early-passage cultures, a small subpopulation of untreated cells spontaneously express low levels of GFP. (B) At each time point, an electronic gate in the GFP channel was applied (e.g., the dashed line on day 34 in panel A), as established by gating on uninfected MEL cells (data not shown), such that >99% of viable cells were excluded. The percentages of viable GFP-positive cells in the absence of drug or in the presence of TSA or 5-azaC at successive time points following retroviral infection are shown.
Diminution of drug-induced expression is strongly correlated with de novo methylation of the GFP gene and the 5′ LTR.
TSA and 5-azaC are believed to induce expression by counteracting the repressive affect of methylation, the former by inhibiting associated HDAC activity (17, 30, 43) and the latter by inhibiting methylation of newly synthesized DNA (22, 44). To study the relationship between de novo methylation and transcriptional induction in the clones characterized above, the methylation state of the provirus was studied in detail. Genomic DNA isolated weekly from untreated subcultures was subjected to Southern blotting, which allows rapid analysis of multiple samples but is limited to methylation-sensitive restriction enzyme sites, and bisulfite genomic sequencing, which allows the detection of any methylated cytosine.
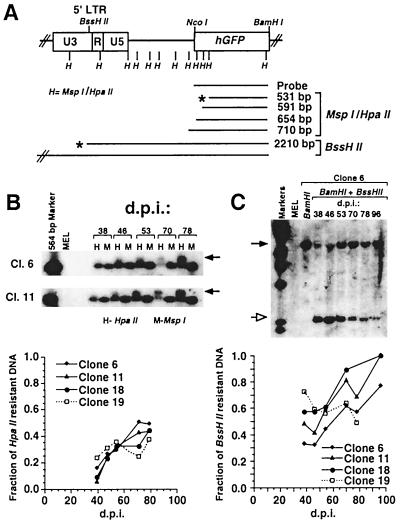
Preliminary analysis of de novo methylation of the hGFP gene was carried out with the methylation-sensitive enzyme HpaII (Fig. 3A). Digestion and Southern blotting of genomic DNA isolated during the course of the TSA and 5-azaC induction experiments revealed that the fraction of methylated HpaII sites within the hGFP gene was initially low but increased with time in three of the four clones (clones 6, 11, and 18) analyzed (Fig. 3B, graph). Clone 19, which appears to be methylated at a lower rate, also maintained the highest residual TSA and 5-azaC responsiveness (Fig. 2B). To characterize the methylation density of the hGFP coding region at higher resolution, genomic DNA isolated 39 and 78 days p.i. was analyzed by bisulfite analysis (13, 19). Under the appropriate conditions, sodium bisulfite catalyzes the conversion of cytosine to uracil whereas 5-methylcytosine is unreactive. PCR with strand-specific primers yields a product in which uracil residues are amplified as thymine while 5-methylcytosine residues are amplified as cytosine. By cloning and sequencing these PCR products, the methylation status of all cytosines in the template strand can be determined.
FIG. 3.
Change in methylation state of the hGFP gene and the proviral LTR over time. (A) The MspI-HpaII (H) and BssHII sites in the MFG-hGFP provirus are shown in relation to the 5′ LTR and hGFP gene. The schematic diagram also shows the expected fragments if digestion with HpaII or BssHII is not inhibited (∗) or is partially inhibited by methylation. Total genomic DNA was prepared weekly from the untreated MEL MFG-hGFP clones. Digested fragments were separated by electrophoresis on a 0.7% agarose gel, Southern blotted, and hybridized with the hGFP probe. (B) Genomic DNA from clones 6, 11, 18, and 19 was digested with BamHI and either HpaII (H) or its methylation-insensitive isoschizomer, MspI (M). Southern blots of clones 6 and 11 are shown. Methylation of the MspI-HpaII sites in the GFP coding region yields a slower-migrating fragment (arrow) in the HpaII digest lanes. PhosphorImager analysis was performed to quantitate the DNA in the HpaII-resistant (slower-migrating) and -sensitive bands. The fraction of the HpaII-resistant band (no slower-migrating bands were present) over the sum of the HpaII-resistant and -sensitive bands reflects the degree of methylation within the hGFP gene. For each sample, this value is plotted against the day p.i. (d.p.i.) on which genomic DNA was harvested (Fig. 1). (C) Analysis of the methylation state of the LTR with the methylation-sensitive enzyme BssHII, which cuts once in the LTR at the 3′ end of the U3 region. Genomic DNA from MFG-hGFP clones 6, 11, 18, and 19 was digested with BamHI alone or in combination with BssHII. Southern blots of clones 11 and 18 are shown. Digestion at the BamHI and BssHII sites yields a 2,210-bp fragment (open arrow). The presence of a methyl group at the BssHII site yields a clone-specific fragment of the same size as that in the BamHI-alone lane (solid arrow). The fraction of DNA resistant to BssHII digestion was determined and plotted as described for the MspI-HpaII digests.
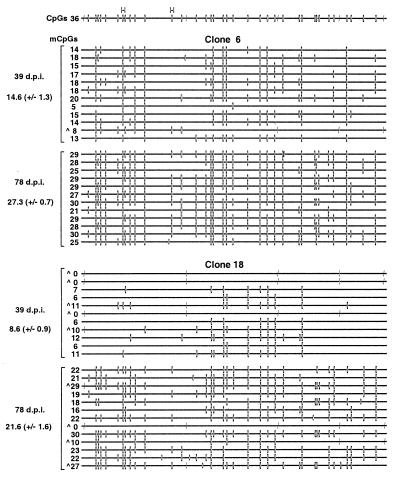
We generated primers to amplify a 457-bp fragment at the 5′ end of the 720-bp hGFP gene. Since the hGFP gene has a very high density of CpGs (see Discussion), the resulting “allelic” amplification product yielded information on a total of 127 cytosines, 36 of which were in the context of CpG. Two of these CpGs were within the HpaII sites characterized in the Southern analyses. Genomic DNA isolated from clones 6, 11, and 18 on days 39 and 78 p.i. was bisulfite treated, amplified, and subcloned. Analysis of the sequenced alleles revealed that the density of mCpGs increased significantly for each clonal population (P < 0.001; Student's t test). The sample mean increased from 14.6 ± 1.3 to 27.3 ± 0.7 mCpGs for clone 6 (excluding strands in which only CpNpG methylation was detected; see Discussion) (Fig. 4A), from 19.9 ± 0.8 to 30.3 ± 0.8 for clone 11 (data not shown), and from 8.6 ± 0.9 to 21.6 ± 1.6 for clone 18 (Fig. 4B). This trend confirmed that seen by Southern analysis and was consistent with the previous observation that the de novo methylation rate is dependent on the site of integration (26). While specific CpGs may be preferentially methylated within a clonal population, no sites were consistently hypermethylated in a clone-to-clone comparison. Taken together with the data in Fig. 2, these results reveal that during the time when the provirus becomes refractory to TSA induction, the methylation density increases significantly, suggesting the involvement of an as yet uncharacterized methylation density-dependent, histone deacetylation-independent mechanism of transcriptional repression that acts on proviruses as well as endogenous genes (10).
FIG. 4.
The mCpG density of the hGFP gene increases with time in culture. Genomic DNA isolated from clones 6 (A) and 18 (B) at 39 and 78 days p.i. (d.p.i.) was bisulfite treated and amplified in a seminested reaction with the GFP-specific primer pairs +1870+ plus +2300− and +1903+ plus +2300− in the first and second rounds, respectively. Amplified products were ligated, cloned, and sequenced as described in Materials and Methods. For each genomic sample, at least two amplification and ligation reactions were carried out. A map of the amplified region of the hGFP gene is presented at the top of the figure, including the potentially methylated CpG (–□–) and CCa/tGG (–|) sites, as well as the HpaII (H) sites analyzed in the Southern analyses. For each allele, mCpG (–□–) and methylated CC(a/t)GG (–|) sequences are shown. Presumed nonconverted cytosines (−C−) (cytosines present in the bisulfite-treated DNA but not originally in the context of CpG or CpNpG) are also displayed. The number of mCpGs inferred for each allele and the sample mean ± SEM of each genomic isolate are presented on the left of the figure. Alleles with CC(a/t)GG methylation (∧) are also labeled.
The bisulfite analysis also revealed non-CpG cytosine methylation in the hGFP gene, particularly of the internal cytosine in the sequence context CC(a/t)GG (Fig. 4), consistent with the previous description of CpNpG methylation in mammalian cells (12). This modification was often detected on strands lacking conventional CpG methylation. CC(a/t)GG is the recognition site of the E. coli Dcm methylase. Maintenance of the original methylation pattern of the plasmid transfected into the Phoenix A retroviral producer cells is unlikely, since the retroviral genome passes through an RNA intermediate. Furthermore, since the genomic DNA is bisulfite treated prior to transformation into the dcm+ E. coli XL-1 Blue strain, contamination is not a possibility at this step in the procedure. However, plasmid DNA contamination of the genomic DNA prior to bisulfite conversion remains a formal possibility. We think that this explanation is unlikely for two reasons. First, in several alleles, CpG methylation, which is unique to mammalian cells, coexists with CC(a/t)GG methylation on the same molecule of DNA. Second, adenine methylation in the sequence GATC, the recognition site of E. coli Dam methylase, was not detected: digestion of MFG-hGFP plasmid DNA isolated from the dam+ E. coli XL-1 Blue strain with the methylation-sensitive enzyme MboI showed complete inhibition of digestion, while Southern blotting of MboI-digested clone 18 genomic DNA isolated 39 days p.i. revealed complete cutting (data not shown). Interestingly, CC(a/t)GG methylation was detected primarily in early-passage cells, suggesting that CpNpG methylation may be an early event in proviral silencing.
Several groups have observed that methylation of the promoter region is particularly effective in repressing expression (9, 33), while others report that methylation density, regardless of localization, is the critical factor in methylation-mediated repression (6, 27). To study the methylation state of the MFG-hGFP promoter, we used the methylation-sensitive enzyme BssHII, which cuts once in the proviral LTR (Fig. 3A) just upstream of the TATA box. As with the HpaII analyses, BssHII digestion revealed that methylation increased over time for clones 6, 11, and 18 but remained unchanged or decreased for clone 19 (Fig. 3C). For each clone, this site was significantly more methylated at each time point than the HpaII sites analyzed in the hGFP gene, suggesting that de novo methylation of the promoter may occur at a higher rate.
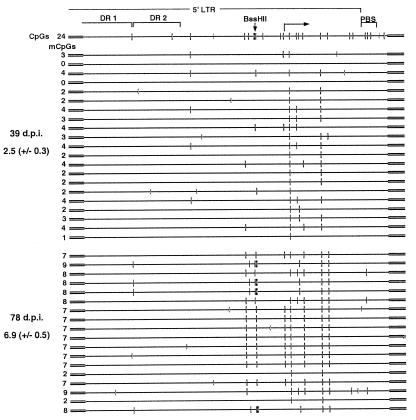
To study the methylation state of the enhancer/promoter region at higher resolution, primers were designed to amplify a 565-bp fragment, including most of the 5′ LTR and the primer binding site. The M-MuLV enhancer is made up of two direct repeats, which include binding sites for a number of transcription factors (21) and which, like many enhancer elements, include few CpGs. In contrast, the downstream promoter region contains a cluster of CpGs, including the BssHII site. Bisulfite analysis of clone 6 reveals that the mean number of mCpGs in the 5′ LTR increased significantly (P < 0.001), from 2.5 ± 0.3 at 39 days p.i. to 6.9 ± 0.5 at 78 days p.i. (Fig. 5). In contrast to the direct-repeat region, which remained hypomethylated, the promoter region became hypermethylated relative to flanking DNA.
FIG. 5.
The mCpG density of the LTR increases with time in culture. Clone 6 genomic DNA isolated at 39 and 78 days p.i. (d.p.i.) was bisulfite treated and amplified in a nested reaction with the vector-specific primers +25+ and +1080− in the first round and the 5′ LTR-specific primers +58+ and +665− in the second round. A map of the 5′ LTR is presented at the top of the figure, including the potentially methylated CpG (–□–) and CCa/tGG (–|) sites, the locations of the direct repeats (DR) of the tandem enhancer, the transcription start site, the primer binding site (PBS), and the BssHII site analyzed in the Southern analyses. mCpGs (–□–) and nonconverted cytosines (−C−) are shown for each sequenced allele. The number of mCpGs inferred for each allele and the sample mean ± SEM are presented on the left of the figure.
GFP expression in late-passage clones can be induced only by sequential treatment with 5-azaC followed by TSA.
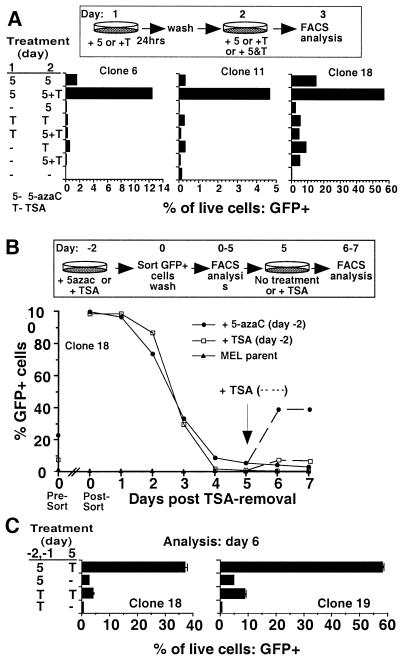
If the increased density of proviral mCpGs is responsible for the progressive diminution of TSA responsiveness, the efficacy of TSA treatment should be reestablished by reducing proviral methylation. Late-passage clones (with near-complete extinction of TSA inducibility) were pretreated with 5-azaC for 24 h and then treated with TSA and 5-azaC for 24 h. Pretreatment with 5-azaC increased the inductive potential of TSA ∼10-fold over that of TSA alone (Fig. 6A). This order of addition is required for the synergistic effect: pretreatment with TSA, or treatment with TSA and 5-azaC together for only 24 h, does not potentiate induction. As with treatment of early-passage clones with either drug alone, the degree of induction was dependent on the integration site and the fraction of cells induced within each clonal population, greatest for clone 18 and least for clone 11, was inversely related to the extent of methylation of each clone.
FIG. 6.
Pretreatment with 5-azaC potentiates induction of proviral expression by TSA. (A) MEL MFG-hGFP clones 6, 11, and 18 (101 days p.i.) were treated with 100 nM TSA (T) or 5μM 5-azaC (5) for 24 h, washed, and treated for a further 24 h either with the same drug or with both TSA and 5-azaC (5+T). At 24 h after the second treatment, the cells were harvested and prepared for flow cytometry. Results for control untreated cells, as well as samples treated only on the second day, are also presented. (B) MEL MFG-hGFP clone 18 cells (108 days p.i.) were treated with 100 nM TSA or 5 μM 5-azaC for 48 h. The percentage of GFP-positive cells prior to sorting is shown on the y axis (Pre-Sort day 0). Viable GFP-positive cells were sorted into complete medium and immediately reanalyzed by FACS (Post-Sort day 0) to confirm sorting purity. Sorted cells were washed in fresh complete medium and replated. Aliquots of the sorted population were removed daily for FACS analysis. Aliquots of the sorted cultures were treated with TSA on day 5 postsorting and analyzed for GFP expression on days 6 and 7 post-sorting (dotted lines). Each point represents the percentage of viable cells that were scored as GFP positive. Analyses of MEL parental cells are included on each day of analysis. (C) Treatment and sorting as described in panel B was repeated, and triplicate subcultures of sorted clone 18 and 19 cells were established. Subcultures were re-treated on day 5 and analyzed on day 6. Each bar represents the mean percentage of viable cells scored as GFP positive. Error bars show the standard deviation. For all samples, dead cells were excluded from the analyses on the basis of PI staining and forward- and side-scatter parameters.
If 5-azaC pretreatment potentiates induction by reducing the methylation density of proviral DNA, then, given the low rate of de novo methylation, the effect of 5-azaC should persist for some time after the drug is removed. Late-passage clone 18 cells were treated for 2 days either with 5-azaC or with TSA, and GFP-expressing cells were sorted to generate homogeneous populations of 5-azaC- or TSA-responsive cells. Sorted cells were washed, placed in culture, and monitored daily for GFP expression. Regardless of the inducing agent applied, retroviral expression is rapidly silenced. However, upon retreatment of the cultures with TSA 5 days postsorting (by which time >90% of the cells are silenced), only the cells originally treated with 5-azaC were effectively induced (Fig. 6B). Repetition of the experiment with sorted clone 18 and 19 cells showed the same effect (Fig. 6C). For both clones, the potentiating effect of 5-azaC was still apparent 11 days postsorting, indicating that alteration of the provirus mediated by 5-azaC extends well beyond the time when the silent state is reestablished (data not shown).
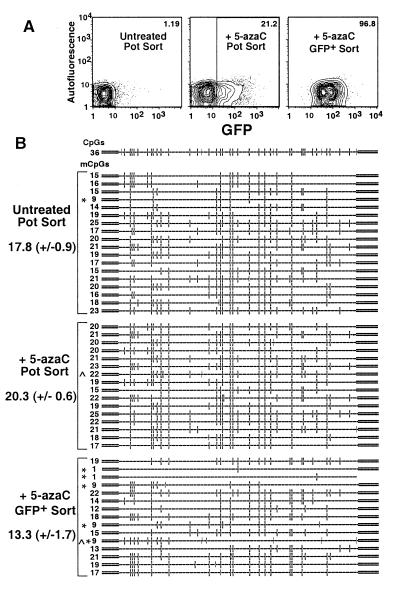
The results described above are consistent with a model in which proviral expression becomes refractory to TSA induction as a result of the accumulation of mCpGs in late-passage cells and in which 5-azaC treatment of these cells reduces proviral methylation density sufficiently to allow TSA induction. To explicitly test whether 5-azaC treatment under the conditions used inhibits maintenance methyltransferase activity and thus demethylates the provirus, subcultures of clone 18 cells at 48 days p.i. were treated with 5 μM 5-azaC for 48 h. Untreated and treated cells were sorted on the basis of viability alone, and treated viable GFP-positive cells were also sorted (Fig. 7A). Extracts of each sorted subpopulation were bisulfite treated and amplified with the hGFP primer set. In the absence of 5-azaC treatment, a mean of 17.8 ± 0.9 mCpGs were present in this region of the hGFP gene, within range of the predicted value from the initial time course experiment (Fig. 4A). No decrease in the number of mCpGs (20.3 ± 0.6) was detected in the cell population sorted on the basis of viability alone, indicating that in the majority of 5-azaC-treated cells, the hGFP gene is not demethylated (Fig. 7B). Given that GFP expression was induced in only 21% of these cells, this result is not surprising. In contrast, the 5-azaC-treated population sorted on the basis of GFP expression showed a significant decrease in hGFP methylation (P = 0.027), to a mean of 13.3 ± 1.7 mCpGs. The decrease in the number of mCpGs resulted primarily from the appearance of several alleles with very low levels of methylation, from 1 to 9 mCpGs/molecule (Fig. 7B). Given the duration of 5-azaC treatment in this experiment, the presence of alleles with higher mCpG density in the GFP-positive population is not surprising: since 5-azaC inhibits methylation during DNA replication, only the nascent strand in daughter cells is unmethylated. Such hemimethylated DNA is a poor substrate for the MeCP complexes (38, 41) and thus may adopt a chromatin state permissive for expression. The bisulfite sequencing assay yielded data on only one strand and therefore cannot be used to reveal the methylation status of complementary strands. Given that only 1% of the cells were GFP positive in the untreated sample, we conclude that 5-azaC-mediated proviral induction is strongly correlated with decreased local methylation density. Similar results were found for clone 6 (data not shown), and, as with the time course study, CC(a/t)GG methylation was also detected in the 5-azaC-treated samples. These results are consistent with the hypothesis that 5-azaC treatment of late-passage clones potentiates TSA-mediated GFP expression by reducing proviral methylation density and help to explain why the efficacy of 5-azaC treatment diminishes with time; the duration of treatment and concentration of drug used are not sufficient to fully demethylate the provirus.
FIG. 7.
Induction of GFP expression by 5-azaC treatment is correlated with partial demethylation of the hGFP gene. Subcultures of clone 18 cells (48 days p.i.) were treated with 5 μM 5-azaC for 48 h. Subsequently, mock-treated and 5-azaC-treated cells were FACS sorted on the basis of PI staining alone (Pot Sort), and 5-azaC-treated, viable GFP-positive cells were also sorted. Each sorted sample was immediately lysed and subjected to bisulfite conversion. (A) For each population, 5% probability plots are shown. The electronic gate used to sort GFP-positive cells from the 5-azaC-treated population is also shown. The percentage of GFP-positive cells in each population is shown at the top of each plot. Amplification, cloning, and sequencing were conducted as described in the legend to Fig. 4 and Materials and Methods. (B) Map of the amplified region of the hGFP gene, including the potentially methylated CpG (–□–) and CCa/tGG (–|) sites. For each allele, mCpG (–□–), methylated CC(a/t)GG sequences (–|), and presumed nonconverted cytosines (−C−) are shown. The number of mCpGs inferred for each allele and the sample mean ± SEM of each sorted population is presented on the left of the figure. Alleles with relatively low levels of methylation (∗) (<10 mCpGs) and/or CC(a/t)GG methylation (∧) are labeled accordingly.
DISCUSSION
The infected cells characterized in this study contain a provirus that is silenced at the earliest time point analyzed, 5 days p.i. Bisulfite analysis of the hGFP gene in the subpopulation of MFG-hGFP infected cells not expressing GFP revealed that the provirus was methylated by day 11 p.i., the earliest time point studied (data not shown). The mean density of 2.6 ± 0.4 mCpGs/100 bp is within the range of the mCpG density at which the repressor MeCP2 effectively inhibits expression in vitro (>1 mCpG/100 bp) (42), indicating that the inductive effect of TSA may result from inhibition of HDAC-MeCP2 complexes associated with nascent mCpGs (17, 30, 43). Others have proposed that transcriptional repression precedes de novo methylation (20, 45), and we cannot rule out the possibility that HDACs are recruited to the nascent provirus by a mCpG-independent mechanism. For example, a sequence-specific DNA binding factor such as YY-1, which binds to a conserved region within the M-MuLV LTR (18) and is known to recruit an HDAC-associated complex (59), may play a role in repressing proviral expression. Alternatively, constitutively methylated flanking DNA may contribute to repression of the nascent provirus; for example, it has been reported that methylated DNA can silence an adjacent unmethylated promoter (31).
Regardless of the mechanism initiating transcriptional silencing, expression of the nascent provirus can be induced by treatment with TSA, consistent with several reports showing that inhibition of HDAC activity is sufficient to activate otherwise nonexpressing transgenes (11, 46, 56). However, in vitro methylation studies of an episomal reporter construct revealed that the efficacy of transcriptional induction by sodium butyrate is inversely related to methylation density (27), suggesting the presence of an HDAC-independent, methylation density-dependent repressive mechanism. We found that with increasing time in culture, the efficacy of TSA-mediated proviral induction diminished, to an extent dependent upon the proviral integration site. These results contrast with a previous report (11) of no diminution of TSA responsiveness of an rAAV/CMVlacZ reporter in HeLa cells. This discrepancy may be explained by the difference in enhancers or experimental conditions used. The concentration of TSA used in the study described in reference 11, 3,000 nM, yields significant cytotoxicity in MEL cells (M. Lorincz, unpublished observation).
The hGFP reporter and the E. coli lacZ gene have CpG densities of ∼9/100 bp. Although this CpG density is also comparable to that of endogenous genes, such as human ζ-globin (2), the majority of CpGs in hGFP were introduced during the codon optimization process (23) and thus may not reflect the normal distribution of CpGs in mammalian genes. The failure of TSA-mediated induction, however, is not peculiar to the hGFP reporter, since we observed a similar time-dependent diminution of drug induction when we replaced hGFP with the murine CD8α gene (4 CpGs/100 bp) in the MFG vector (data not shown). These observations have important implications for gene therapy protocols: given that TSA treatment yields only transitory expression of provirus methylated at low density (Fig. 6) and is completely ineffective in inducing the expression of highly methylated provirus, it is unlikely that TSA treatment to relieve suppression of therapeutic retroviral vectors will improve gene therapy for genetic diseases (11). Development of vectors which are refractory to de novo methylation should be a more efficacious approach (49).
To generate the TSA-inducible M-MuLV-hGFP MEL clones characterized in this study, infected cells were subject to TSA treatment and cell sorting on the basis of GFP expression. Subsequently, the clones were split weekly and only those subcultures to be screened for inducibility that week were treated with TSA or 5-azaC. Nevertheless, since TSA has pleiotropic effects on cells (60), we cannot rule out the possibility that the initial TSA treatment induced trans effects, such as alteration of the transcription factor milieu. We think that this explanation is unlikely, since infection of a late-passage (noninducible) MEL MFG-hGFP clone with MFG-CD8 yielded a subpopulation of cells expressing CD8 at levels comparable to those in MEL cells infected with MFG-CD8 alone (data not shown). That the complement of trans-acting factors necessary for expression from the M-MuLV enhancer are present and functional in these cells indicates that the progressive loss of responsiveness to TSA and 5-azaC results from a cis-mediated alteration of the provirus.
The bisulfite and Southern blot data reveals that the methylation density gradually increases in both the hGFP gene and the LTR with time in culture. However, the fraction of CpGs methylated in the hGFP gene (0.58 to 0.83) is clearly greater than that in the LTR (0.08 to 0.37) by day 88 p.i. At first impression, this observation contradicts that seen in the Southern analysis experiments, but closer inspection of the bisulfite data reveals that the 3′ HpaII site in the hGFP gene is hypomethylated compared with flanking CpGs and that the BssHII site in the LTR is hypermethylated compared to flanking CpGs. This data clearly illustrates that, given the heterogeneity in methylation of specific CpGs, the use of single sites to study methylation patterns can yield misleading results. The enhancer region, in particular, remains devoid of mCpGs, perhaps as a result of passive inhibition by transcription factor binding (40) or recruitment of a demethylating factor (39). Nevertheless, as previously shown for the simian virus 40 enhancer (6), the methylation-free M-MuLV enhancer is clearly insufficient to induce expression when the density of mCpGs in cis exceeds a certain threshold. In contrast to the enhancer, the promoter/cap site region became heavily methylated with time in culture. Several studies show that methylation of the promoter/preinitiation domain is sufficient to repress gene expression (47), perhaps by recruitment of methyl-binding proteins, such as MeCP1 (5, 51), which might inhibit the formation of the preinitiation complex (36). Such transcriptional repression may operate independently of HDAC activity, thus explaining the loss of TSA-mediated induction in late-passage cells. Unfortunately, given that the promoter region and the hGFP gene are de novo methylated concurrently, we cannot attribute the HDAC-independent repressive effect to methylation of a specific region in the provirus.
The GFP-based reporter system described here allowed a coordinated analysis of the transcriptional induction and methylation status of single proviral integrants at successive time points after infection. These experiments clearly revealed a diminution of TSA induction concomitant with the accumulation of proviral mCpGs during serial passage. Interestingly, de novo methylation continued to occur long after the provirus was silenced. Southern analysis of genomic DNA isolated from TSA-treated cultures revealed no reduction in proviral methylation in cells in which GFP expression was induced, suggesting that TSA does not act as a demethylating agent (reference 17 and data not shown). The observation that 5-azaC treatment only partially demethylates the provirus helps to explain why the efficacy of treatment with this drug also diminishes with time; residual methylation is probably sufficient to maintain MeCP2- and HDAC-mediated silencing in late-passage cells. Expression is inducible in such late-passage cells only by treatment with 5-azaC followed by TSA. These results are consistent with those recently reported by Cameron et al. (10), who found that several hypermethylated endogenous genes could be reactivated only by treatment with 5-azaC followed by TSA. Taken together, these experiments reveal that HDAC activity is the primary effector of repression mediated by low-density methylation whereas an as yet uncharacterized HDAC-independent mechanism predominates in the repression of highly methylated genes.
MBD proteins (24, 43), MeCP2-associated proteins such as mSin3 (35), or chromatin-specific proteins such as histone H1 (37) may be involved in such repression. MBD1 is a particularly intriguing candidate, since it lacks the MeCP2 TRD (24) required for binding to the Sin3-histone deacetylase complex. Interestingly, the MBD1 protein includes the CxxCxxC motif found in several mammalian proteins, including ALL-1/HRX, which is related to Drosophila trithorax (15), suggesting the potential involvement of the trithorax/polycomb protein groups in methylation-mediated repression. The MeCP1 complex may also be involved, since, in contrast to MeCP2 (38), it binds efficiently only to substrates containing an array of mCpGs (5, 41). The binding of MBD proteins may promote the recruitment of densely methylated provirus to a repressive nuclear compartment, such as centromeric heterochromatin foci (7). Whatever the mechanism, when a threshold of local methylation density is reached, ∼5 mCpGs/100 bp depending on the integration site, the provirus may become irreversibly silenced. Our time course experiments revealed the dynamic nature of methylation-mediated repression of a nascent provirus, with HDAC activity playing the predominant role early after infection and an HDAC-independent mechanism consolidating the silent state. Clearly, further characterization of methylation-mediated repression will require that mCpG density be taken into account.
ACKNOWLEDGMENTS
The GFP-Bex1 plasmid and Phoenix A retroviral producer cells were kind gifts of M. Anderson and G. Nolan, respectively. We thank Claire Francastel, Robert Eisenman, Steve Fiering, Reinhard Stoeger, Dan Cimbora, and the Martin and Groudine laboratories for helpful suggestions. We also thank the FHCRC Biotechnology and Flow Cytometry Shared Resource facilities for technical assistance and Kristy Seidel for advice on statistical analysis.
This work was supported by NIH grants to M.G. and D.I.K.M., who is a Scholar of the Leukemia Society of America, and fellowships from the NIH to M.C.L. and Deutsche Forschungsgemeinschaft to D.S.
REFERENCES
- 1.Anderson M T, Tjioe I M, Lorincz M C, Parks D R, Herzenberg L A, Nolan G P, Herzenberg L A. Simultaneous fluorescence-activated cell sorter analysis of two distinct transcriptional elements within a single cell using engineered green fluorescent proteins. Proc Natl Acad Sci USA. 1996;93:8508–8511. doi: 10.1073/pnas.93.16.8508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Antequera F, Bird A. Number of CpG islands and genes in human and mouse. Proc Natl Acad Sci USA. 1993;90:11995–11999. doi: 10.1073/pnas.90.24.11995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bahnson A B, Dunigan J T, Baysal B E, Mohney T, Atchison R W, Nimgaonkar M T, Ball E D, Barranger J A. Centrifugal enhancement of retroviral mediated gene transfer. J Virol Methods. 1995;54:131–143. doi: 10.1016/0166-0934(95)00035-s. [DOI] [PubMed] [Google Scholar]
- 4.Bartsch J, Truss M, Bode J, Beato M. Moderate increase in histone acetylation activates the mouse mammary tumor virus promoter and remodels its nucleosome structure. Proc Natl Acad Sci USA. 1996;93:10741–10746. doi: 10.1073/pnas.93.20.10741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boyes J, Bird A. DNA methylation inhibits transcription indirectly via a methyl-CpG binding protein. Cell. 1991;64:1123–1134. doi: 10.1016/0092-8674(91)90267-3. [DOI] [PubMed] [Google Scholar]
- 6.Boyes J, Bird A. Repression of genes by DNA methylation depends on CpG density and promoter strength: evidence for involvement of a methyl-CpG binding protein. EMBO J. 1992;11:327–333. doi: 10.1002/j.1460-2075.1992.tb05055.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brown K E, Guest S S, Smale S T, Hahm K, Merkenschlager M, Fisher A G. Association of transcriptionally silent genes with Ikaros complexes at centromeric heterochromatin. Cell. 1997;91:845–854. doi: 10.1016/s0092-8674(00)80472-9. [DOI] [PubMed] [Google Scholar]
- 8.Buschhausen G, Wittig B, Graessmann M, Graessmann A. Chromatin structure is required to block transcription of the methylated herpes simplex virus thymidine kinase gene. Proc Natl Acad Sci USA. 1987;84:1177–1181. doi: 10.1073/pnas.84.5.1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Busslinger M, Hurst J, Flavell R A. DNA methylation and the regulation of globin gene expression. Cell. 1983;34:197–206. doi: 10.1016/0092-8674(83)90150-2. [DOI] [PubMed] [Google Scholar]
- 10.Cameron E E, Bachman K E, Myohanen S, Herman J G, Baylin S B. Synergy of demethylation and histone deacetylase inhibition in the re-expression of genes silenced in cancer. Nat Genet. 1999;21:103–107. doi: 10.1038/5047. [DOI] [PubMed] [Google Scholar]
- 11.Chen W Y, Bailey E C, McCune S L, Dong J Y, Townes T M. Reactivation of silenced, virally transduced genes by inhibitors of histone deacetylase. Proc Natl Acad Sci USA. 1997;94:5798–5803. doi: 10.1073/pnas.94.11.5798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clark S J, Harrison J, Frommer M. CpNpG methylation in mammalian cells. Nat Genet. 1995;10:20–27. doi: 10.1038/ng0595-20. [DOI] [PubMed] [Google Scholar]
- 13.Clark S J, Harrison J, Paul C L, Frommer M. High sensitivity mapping of methylated cytosines. Nucleic Acids Res. 1994;22:2990–2997. doi: 10.1093/nar/22.15.2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Coffee B, Zhang F, Warren S T, Reines D. Acetylated histones are associated with FMR1 in normal but not fragile X-syndrome cells. Nat Genet. 1999;22:98–101. doi: 10.1038/8807. . (Erratum, 22:209.) [DOI] [PubMed] [Google Scholar]
- 15.Cross S H, Meehan R R, Nan X, Bird A. A component of the transcriptional repressor MeCP1 shares a motif with DNA methyltransferase and HRX proteins. Nat Genet. 1997;16:256–259. doi: 10.1038/ng0797-256. [DOI] [PubMed] [Google Scholar]
- 16.Dranoff G, Jaffee E, Lazenby A, Golumbek P, Levitsky H, Brose K, Jackson V, Hamada H, Pardoll D, Mulligan R C. Vaccination with irradiated tumor cells engineered to secrete murine granulocyte-macrophage colony-stimulating factor stimulates potent, specific, and long-lasting anti-tumor immunity. Proc Natl Acad Sci USA. 1993;90:3539–3543. doi: 10.1073/pnas.90.8.3539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eden S, Hashimshony T, Keshet I, Cedar H, Thorne A W. DNA methylation models histone acetylation. Nature. 1998;394:842. doi: 10.1038/29680. [DOI] [PubMed] [Google Scholar]
- 18.Flanagan J R, Becker K G, Ennist D L, Gleason S L, Driggers P H, Levi B Z, Appella E, Ozato K. Cloning of a negative transcription factor that binds to the upstream conserved region of Moloney murine leukemia virus. Mol Cell Biol. 1992;12:38–44. doi: 10.1128/mcb.12.1.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Frommer M, McDonald L E, Millar D S, Collis C M, Watt F, Grigg G W, Molloy P L, Paul C L. A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc Natl Acad Sci USA. 1992;89:1827–1831. doi: 10.1073/pnas.89.5.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gautsch J W, Wilson M C. Delayed de novo methylation in teratocarcinoma suggests additional tissue-specific mechanisms for controlling gene expression. Nature. 1983;301:32–37. doi: 10.1038/301032a0. [DOI] [PubMed] [Google Scholar]
- 21.Granger S W, Fan H. In vivo footprinting of the enhancer sequences in the upstream long terminal repeat of Moloney murine leukemia virus: differential binding of nuclear factors in different cell types. J Virol. 1998;72:8961–8970. doi: 10.1128/jvi.72.11.8961-8970.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Groudine M, Eisenman R, Weintraub H. Chromatin structure of endogenous retroviral genes and activation by an inhibitor of DNA methylation. Nature. 1981;292:311–317. doi: 10.1038/292311a0. [DOI] [PubMed] [Google Scholar]
- 23.Haas J, Park E C, Seed B. Codon usage limitation in the expression of HIV-1 envelope glycoprotein. Curr Biol. 1996;6:315–324. doi: 10.1016/s0960-9822(02)00482-7. [DOI] [PubMed] [Google Scholar]
- 24.Hendrich B, Bird A. Identification and characterization of a family of mammalian methyl-CpG binding proteins. Mol Cell Biol. 1998;18:6538–6547. doi: 10.1128/mcb.18.11.6538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hitoshi Y, Lorens J, Kitada S I, Fisher J, LaBarge M, Ring H Z, Francke U, Reed J C, Kinoshita S, Nolan G P. Toso, a cell surface, specific regulator of Fas-induced apoptosis in T cells. Immunity. 1998;8:461–471. doi: 10.1016/s1074-7613(00)80551-8. [DOI] [PubMed] [Google Scholar]
- 26.Hoeben R C, Migchielsen A A, van der Jagt R C, van Ormondt H, van der Eb A J. Inactivation of the Moloney murine leukemia virus long terminal repeat in murine fibroblast cell lines is associated with methylation and dependent on its chromosomal position. J Virol. 1991;65:904–912. doi: 10.1128/jvi.65.2.904-912.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hsieh C L. Dependence of transcriptional repression on CpG methylation density. Mol Cell Biol. 1994;14:5487–5494. doi: 10.1128/mcb.14.8.5487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Humphries E H, Allen R, Glover C. Clonal analysis of the integration and expression of endogenous avian retroviral DNA acquired by exogenous viral infection. J Virol. 1981;39:584–596. doi: 10.1128/jvi.39.2.584-596.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jahner D, Stuhlmann H, Stewart C L, Harbers K, Lohler J, Simon I, Jaenisch R. De novo methylation and expression of retroviral genomes during mouse embryogenesis. Nature. 1982;298:623–628. doi: 10.1038/298623a0. [DOI] [PubMed] [Google Scholar]
- 30.Jones P L, Veenstra G J, Wade P A, Vermaak D, Kass S U, Landsberger N, Strouboulis J, Wolffe A P. Methylated DNA and MeCP2 recruit histone deacetylase to repress transcription. Nat Genet. 1998;19:187–191. doi: 10.1038/561. [DOI] [PubMed] [Google Scholar]
- 31.Kass S U, Landsberger N, Wolffe A P. DNA methylation directs a time-dependent repression of transcription initiation. Curr Biol. 1997;7:157–165. doi: 10.1016/s0960-9822(97)70086-1. [DOI] [PubMed] [Google Scholar]
- 32.Keshet I, Lieman-Hurwitz J, Cedar H. DNA methylation affects the formation of active chromatin. Cell. 1986;44:535–543. doi: 10.1016/0092-8674(86)90263-1. [DOI] [PubMed] [Google Scholar]
- 33.Keshet I, Yisraeli J, Cedar H. Effect of regional DNA methylation on gene expression. Proc Natl Acad Sci USA. 1985;82:2560–2564. doi: 10.1073/pnas.82.9.2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kudo S. Methyl-CpG-binding protein MeCP2 represses Sp1-activated transcription of the human leukosialin gene when the promoter is methylated. Mol Cell Biol. 1998;18:5492–5499. doi: 10.1128/mcb.18.9.5492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Laherty C D, Yang W M, Sun J M, Davie J R, Seto E, Eisenman R N. Histone deacetylases associated with the mSin3 corepressor mediate mad transcriptional repression. Cell. 1997;89:349–356. doi: 10.1016/s0092-8674(00)80215-9. [DOI] [PubMed] [Google Scholar]
- 36.Levine A, Cantoni G L, Razin A. Methylation in the preinitiation domain suppresses gene transcription by an indirect mechanism. Proc Natl Acad Sci USA. 1992;89:10119–10123. doi: 10.1073/pnas.89.21.10119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Levine A, Yeivin A, Ben-Asher E, Aloni Y, Razin A. Histone H1-mediated inhibition of transcription initiation of methylated templates in vitro. J Biol Chem. 1993;268:21754–21759. [PubMed] [Google Scholar]
- 38.Lewis J D, Meehan R R, Henzel W J, Maurer F I, Jeppesen P, Klein F, Bird A. Purification, sequence, and cellular localization of a novel chromosomal protein that binds to methylated DNA. Cell. 1992;69:905–914. doi: 10.1016/0092-8674(92)90610-o. [DOI] [PubMed] [Google Scholar]
- 39.Lichtenstein M, Keini G, Cedar H, Bergman Y. B cell-specific demethylation: a novel role for the intronic kappa chain enhancer sequence. Cell. 1994;76:913–923. doi: 10.1016/0092-8674(94)90365-4. [DOI] [PubMed] [Google Scholar]
- 40.Matsuo K, Silke J, Georgiev O, Marti P, Giovannini N, Rungger D. An embryonic demethylation mechanism involving binding of transcription factors to replicating DNA. EMBO J. 1998;17:1446–1453. doi: 10.1093/emboj/17.5.1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Meehan R R, Lewis J D, McKay S, Kleiner E L, Bird A P. Identification of a mammalian protein that binds specifically to DNA containing methylated CpGs. Cell. 1989;58:499–507. doi: 10.1016/0092-8674(89)90430-3. [DOI] [PubMed] [Google Scholar]
- 42.Nan X, Campoy F J, Bird A. MeCP2 is a transcriptional repressor with abundant binding sites in genomic chromatin. Cell. 1997;88:471–481. doi: 10.1016/s0092-8674(00)81887-5. [DOI] [PubMed] [Google Scholar]
- 43.Nan X, Ng H H, Johnson C A, Laherty C D, Turner B M, Eisenman R N, Bird A. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature. 1998;393:386–389. doi: 10.1038/30764. [DOI] [PubMed] [Google Scholar]
- 44.Niwa O, Sugahara T. 5-Azacytidine induction of mouse endogenous type C virus and suppression of DNA methylation. Proc Natl Acad Sci USA. 1981;78:6290–6294. doi: 10.1073/pnas.78.10.6290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Niwa O, Yokota Y, Ishida H, Sugahara T. Independent mechanisms involved in suppression of the Moloney leukemia virus genome during differentiation of murine teratocarcinoma cells. Cell. 1983;32:1105–1113. doi: 10.1016/0092-8674(83)90294-5. [DOI] [PubMed] [Google Scholar]
- 46.Pikaart M J, Recillas-Targa F, Felsenfeld G. Loss of transcriptional activity of a transgene is accompanied by DNA methylation and histone deacetylation and is prevented by insulators. Genes Dev. 1998;12:2852–2862. doi: 10.1101/gad.12.18.2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Razin A, Cedar H. DNA methylation and gene expression. Microbiol Rev. 1991;55:451–458. doi: 10.1128/mr.55.3.451-458.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Riggs M G, Whittaker R G, Neumann J R, Ingram V M. n-Butyrate causes histone modification in HeLa and Friend erythroleukaemia cells. Nature. 1977;268:462–464. doi: 10.1038/268462a0. [DOI] [PubMed] [Google Scholar]
- 49.Robbins P B, Skelton D C, Yu X J, Halene S, Leonard E H, Kohn D B. Consistent, persistent expression from modified retroviral vectors in murine hematopoietic stem cells. Proc Natl Acad Sci USA. 1998;95:10182–10187. doi: 10.1073/pnas.95.17.10182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 51.Singal R, Ferris R, Little J A, Wang S Z, Ginder G D. Methylation of the minimal promoter of an embryonic globin gene silences transcription in primary erythroid cells. Proc Natl Acad Sci USA. 1997;94:13724–13729. doi: 10.1073/pnas.94.25.13724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Smith C L, Hager G L. Transcriptional regulation of mammalian genes in vivo. A tale of two templates. J Biol Chem. 1997;272:27493–27496. doi: 10.1074/jbc.272.44.27493. [DOI] [PubMed] [Google Scholar]
- 53.Stewart C L, Stuhlmann H, Jahner D, Jaenisch R. De novo methylation, expression, and infectivity of retroviral genomes introduced into embryonal carcinoma cells. Proc Natl Acad Sci USA. 1982;79:4098–4102. doi: 10.1073/pnas.79.13.4098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Struhl K. Histone acetylation and transcriptional regulatory mechanisms. Genes Dev. 1998;12:599–606. doi: 10.1101/gad.12.5.599. [DOI] [PubMed] [Google Scholar]
- 55.Stuhlmann H, Jahner D, Jaenisch R. Infectivity and methylation of retroviral genomes is correlated with expression in the animal. Cell. 1981;26:221–232. doi: 10.1016/0092-8674(81)90305-6. [DOI] [PubMed] [Google Scholar]
- 56.Van Lint C, Emiliani S, Ott M, Verdin E. Transcriptional activation and chromatin remodeling of the HIV-1 promoter in response to histone acetylation. EMBO J. 1996;15:1112–1120. [PMC free article] [PubMed] [Google Scholar]
- 57.Weintraub H. Assembly and propagation of repressed and depressed chromosomal states. Cell. 1985;42:705–711. doi: 10.1016/0092-8674(85)90267-3. [DOI] [PubMed] [Google Scholar]
- 58.Xu L, Yee J K, Wolff J A, Friedmann T. Factors affecting long-term stability of Moloney murine leukemia virus-based vectors. Virology. 1989;171:331–341. doi: 10.1016/0042-6822(89)90600-4. [DOI] [PubMed] [Google Scholar]
- 59.Yang W M, Inouye C, Zeng Y, Bearss D, Seto E. Transcriptional repression by YY1 is mediated by interaction with a mammalian homolog of the yeast global regulator RPD3. Proc Natl Acad Sci USA. 1996;93:12845–12850. doi: 10.1073/pnas.93.23.12845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yoshida M, Kijima M, Akita M, Beppu T. Potent and specific inhibition of mammalian histone deacetylase both in vivo and in vitro by trichostatin A. J Biol Chem. 1990;265:17174–17179. [PubMed] [Google Scholar]