ABSTRACT
Trypanosoma brucei subspecies cause African sleeping sickness in humans, an infection that is commonly fatal if not treated, and available therapies are limited. Previous studies have shown that heat shock protein 90 (Hsp90) inhibitors have potent and vivid activity against bloodstream-form trypanosomes. Hsp90s are phylogenetically conserved and essential catalysts that function at the crux of cell biology, where they ensure the proper folding of proteins and their assembly into multicomponent complexes. To assess the specificity of Hsp90 inhibitors and further define the role of Hsp90s in African trypanosomes, we used RNA interference (RNAi) to knock down cytosolic and mitochondrial Hsp90s (HSP83 and HSP84, respectively). Loss of either protein led to cell death, but the phenotypes were distinctly different. Depletion of cytosolic HSP83 closely mimicked the consequences of chemically depleting Hsp90 activity with inhibitor 17-AAG. In these cells, cytokinesis was severely disrupted, and segregation of the kinetoplast (the massive mitochondrial DNA structure unique to this family of eukaryotic pathogens) was impaired, leading to cells with abnormal kinetoplast DNA (kDNA) structures. Quite differently, knockdown of mitochondrial HSP84 did not impair cytokinesis but halted the initiation of new kDNA synthesis, generating cells without kDNA. These findings highlight the central role of Hsp90s in chaperoning cell cycle regulators in trypanosomes, reveal their unique function in kinetoplast replication, and reinforce their specificity and value as drug targets.
KEYWORDS: Trypanosoma brucei, Hsp90, RNAi, 17-AAG, HSP83, HSP84, kDNA, cytokinesis, kinetoplast, topoisomerase
TEXT
The ancient divergent eukaryote Trypanosoma brucei, transmitted by the tsetse fly, causes human African trypanosomiasis (sleeping sickness) (1). The life cycle of this and related kinetoplastid pathogens entails differentiation between bloodstream and insect forms and proliferative division within these stages. Bloodstream-form T. brucei, the subject of this report, is an extracellular parasite that progresses to the central nervous system (CNS) in late-stage infection, causing a meningoencephalitis that is fatal if not treated (2). While investigating the therapeutic potential of repurposing heat shock protein 90 (Hsp90) inhibitors, we found that geldanamycin analogues such as 17-allylamino-17-demethoxygeldanamycin (17-AAG) potently cause rapid and severe morphologic abnormalities in trypanosomes and cure infected mice (3). The current studies were designed to illuminate the role of Hsp90s in the proliferative division of disease-causing bloodstream forms and to corroborate the putative target of Hsp90 inhibitors in trypanosomes.
Essential for cell survival, Hsp90s are phylogenetically ubiquitous protein chaperones. Assisted by multiple cochaperones, they ensure correct maturation and localization of client proteins, themselves mediators of critical regulatory pathways (4, 5). This central and wide-reaching role makes them attractive drug targets (6, 7). Hsp90s have an N-terminal Bergerat-fold ATP binding and hydrolysis domain, and their C termini contain motifs that interact with cochaperones and clients. Several forms of Hsp90 exist in mammalian cells. HSP90α and HSP90β differ in transcriptional regulation, and their protein products are abundant in cytosol (4). The two organellar Hsp90s, TRAP-1/HSP75 in mitochondria and HSP90B1/GRP94 in the endoplasmic reticulum, have 34 to 49% identity (respectively) to cytosolic HSP90, suggesting unique and specialized roles (8, 9). In T. brucei, cytosolic Hsp90 is expressed from 10 tandem genes that differ by just a few nucleotides (Data Set S1 in the supplemental material) and is termed HSP83 based on size (10–12). HSP83 is 60% identical to human HSP90 and has the required Bergerat fold (13). In T. brucei, at least three genes in addition to HSP83 likely encode Hsp90 proteins (11). Genome-wide RNA interference (RNAi) surveys identified HSP83 as essential in T. brucei (14), but, to our knowledge, there are no reports, for any Hsp90, of the phenotypic consequences of knockdown on cell proliferation of bloodstream parasites.
T. brucei has several unique structural features and a complex cell cycle (Fig. 1 and Fig. 2A) (15). A single attached flagellum runs the length of the elongated cell body and is nucleated by a basal body. Connected to these structures via a tripartite attachment complex is the kinetoplast, the dense network of mitochondrial DNA (kinetoplast DNA [kDNA]) unique to trypanosomes and related kinetoplastids (16). kDNA is situated at the posterior of the cell, within the single mitochondrion, as a compact disc visible by fluorescence microscopy. When isolated, it expands to reveal a network consisting of thousands of interlocked DNA circles. Several dozen 20- to 30-kb maxicircles contain protein-coding genes, but some 90% of network DNA comprises 1-kb minicircles that encode guide RNAs for maxicircle transcript editing (17). During proliferation, cellular structures replicate in an ordered fashion, following which a cleavage furrow, originating at the anterior of the cell, ingresses and divides the daughter cells (18). These primitive eukaryotes lack tight checkpoints, so small disruptions to this process are catastrophic. RNAi knockdown of candidate proteins has provided valuable insights into the details of trypanosome cell cycle regulation (15, 16, 19).
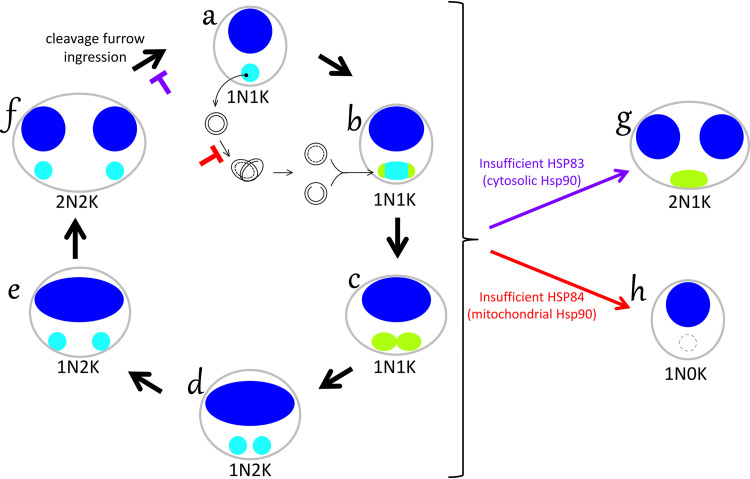
FIG 1.
Nuclear and mitochondrial DNA replication in trypanosomes and lesions from loss of Hsp90s. In control cells, replication of the nuclear (dark blue) and mitochondrial (light blue) genomes is tightly linked, and mitochondrial kinetoplast DNA (kDNA) is fully replicated and scissioned before the double-size nucleus divides (clockwise, forms a to f). This schematic summarizes results of three experimental methods, including DAPI staining of intact cells (blue), examination of isolated network-free kDNA minicircles (black forms between forms a and b, which reside within the mitochondrion, not, as depicted for clarity, outside the cell), and TdT labeling of isolated kDNA networks to detect newly replicated minicircles (green, depicted as if observed within the cell). Nonreplicating cells have a single nucleus and a single kinetoplast (1N1K, form a). To replicate kDNA, individual covalently closed template minicircles are serially decatenated from the network and proceed via theta structure to produce two daughters. As daughters with residual nicks and gaps are reattached at the poles (green areas, form b), the network grows and is remodeled into a dumbbell shape (form c). Minicircle nascent-strand discontinuities are covalently closed only after all circles have replicated at the time of scission (1N2K, forms c and d). Kinetoplasts in 1N2K cells are increasingly separated from one another (forms d and e) prior to nuclear mitosis (2N2K, form f), which is followed by rapid cleavage furrow ingression. Chemical or genetic depletion of cytosolic HSP83 activity inhibits cytokinesis (purple block), manifested by the accumulation of 2N2K cells and cells with incomplete cleavage furrows. The accompanying disruption of network remodeling and scission yields a large population of abnormal 2N1K cells (form g) that have undergone nuclear but not kinetoplast segregation. Distinctly different from this pattern, genetic depletion of mitochondrial HSP84 halts the initiation of kDNA minicircle synthesis (red block), without affecting nuclear replication, mitosis, or cytokinesis, to yield 1N0K cells (form h).
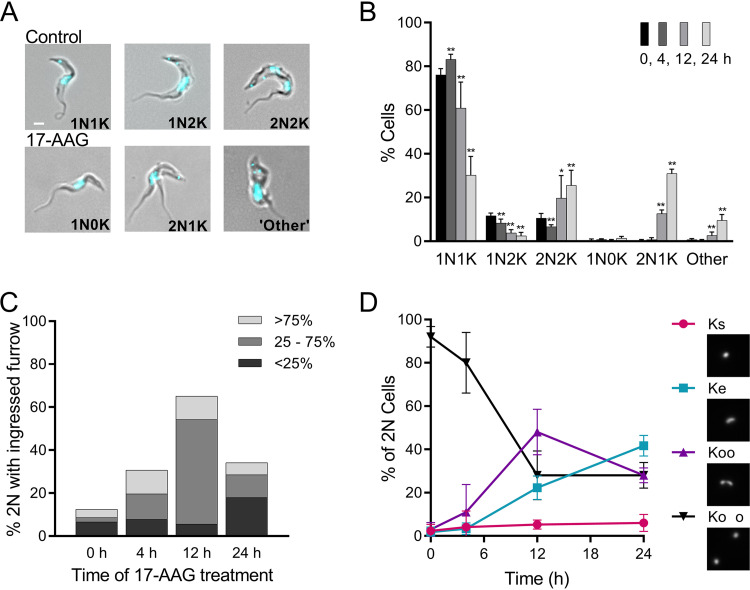
FIG 2.
Effects of increasing exposure to 17-AAG on kinetoplast segregation and cleavage furrow ingression. (A) Merged phase-contrast and DAPI fluorescence images of untreated controls and trypanosomes treated with 100 nM 17-AAG. In controls, the cell cycle progresses from 1 nucleus (N) and 1 kinetoplast (K) (1N1K) to 1 nucleus and 2 kinetoplasts (1N2K), followed by 2 nuclei and 2 kinetoplasts (2N2K) before cell division (Fig. 1). With 17-AAG treatment, abnormal nuclear and kinetoplast configurations occur, including 1N0K, 2N1K, or multiple nuclei and/or kinetoplasts (other). White bar, 1 μm. (B) Nuclear and kinetoplast content of trypanosomes over time with 100 nM 17-AAG treatment. Data are mean ± SD of ≥3 independent experiments; n = 3 to 11 at each point. *, P < 0.05; **, P < 0.01; t test, two-tailed equal variance to 0-h control. (C) Impact of 100 nM 17-AAG on cleavage furrow ingression. Cells were sampled at indicated times after start of 17-AAG treatment and examined for 2N cells with visible cleavage furrows (>50 cells scored per time point). Populations were subdivided by extent of furrow ingression. (D) Effect of 100 nM 17-AAG on kinetoplast separation. 2N cells in timed samples stained with DAPI were examined by fluorescence microscopy and scored as having the expected two fully separated kinetoplasts (Ko o, black; Fig. 1f), or as being abnormal, including those with directly adjacent kinetoplasts (Koo, purple), a single elongated kinetoplast (Ke, blue), or a single kinetoplast (Ks, red). Insets, example images of network morphologies. Mean ± SD of ≥3 independent experiments; >175 2N cells for each time point.
We further examined defects in proliferative growth that result from treatment of bloodstream trypanosomes with Hsp90 inhibitor 17-AAG and compared these lesions with the phenotype of genetic knockdown of cytosolic HSP83 or of putative mitochondrial Hsp90, with particular focus on kDNA as a sensitive and readily evident in situ reporter of disorders in DNA synthesis, mitosis, and cytokinesis.
RESULTS
Chemical inhibition of Hsp90 causes severe defects in kinetoplast replication and cytokinesis.
Bloodstream-form trypanosomes treated with classic Hsp90 inhibitor 17-AAG at 100 nM (90% effective concentration [EC90]) ceased growing within hours of exposure (Fig. S1 in the supplemental material) and, as we previously reported, had severe cell cycle defects apparent by microscopy (3). In control cells, 76% have a single nucleus and kinetoplast (1N1K; nonreplicating or early replication), 12% have divided kinetoplasts but single nucleus (1N2K; midreplication), and 11% have both kinetoplasts and nuclei clearly separated (2N2K; late replication) (Fig. 1 and Fig. 2A and B). Rapid ingression of the cleavage furrow follows and was not commonly observed in controls (0 h) (Fig. 2C). By 24 h, 17-AAG markedly decreased 1N1K and 1N2K populations and quintupled the percentage of cells with two nuclei (Fig. 2B). Over time, an increasing proportion of 2N cells, and by 24 h, 70% of them, contained abnormally proximal daughter kinetoplasts or a single elongated kinetoplast (Fig. 2D). These aberrant cells reflect a failure of mitochondrial kDNA to complete replication before nuclear mitosis. As cells with greater than 2N were not observed, nuclear mitosis apparently halted after the initial division.
Anomalies in cleavage furrow ingression occurred early after drug exposure: within 4 h, the proportion of 2N cells with visible furrows tripled (Fig. 2C); at 12 h, 65% had partially ingressed furrows, with most halted midway along the cell length; and by 24 h, as cells began to die, the visible furrows were mainly short. The presence of an ingressed furrow and abnormal kinetoplast(s) were not strictly linked (Fig. S2).
17-AAG treatment prevents network remodeling.
Replication of kinetoplasts is a fascinating and complex process unique to trypanosomatids (17). As a solution to the substantial topological challenges posed by a network of interlocked circles, individual minicircles are serially decatenated and replicated as “free minicircles” (Fig. 1). Covalently closed network-free template circles proceed via theta structure intermediates to produce daughters with residual nicks or gaps that migrate to replication complexes at antipodal sides of the network and are reattached to the network prior to their covalent closure. The pool of network-free replication intermediates can be evaluated by electrophoresis in agarose gels containing ethidium bromide. 17-AAG caused a rapid (by 2 h) decline in theta and early nicked forms, followed by a growing population of abnormal linearized minicircles (Fig. 3A; Fig. S3A). The linears were covalently bound to protein, a hallmark of topoisomerase dysfunction (Fig. S3B) (20). Linearized maxicircles and total cell content of mini- and maxicircle DNA also gradually increased (Fig. S3C and D).
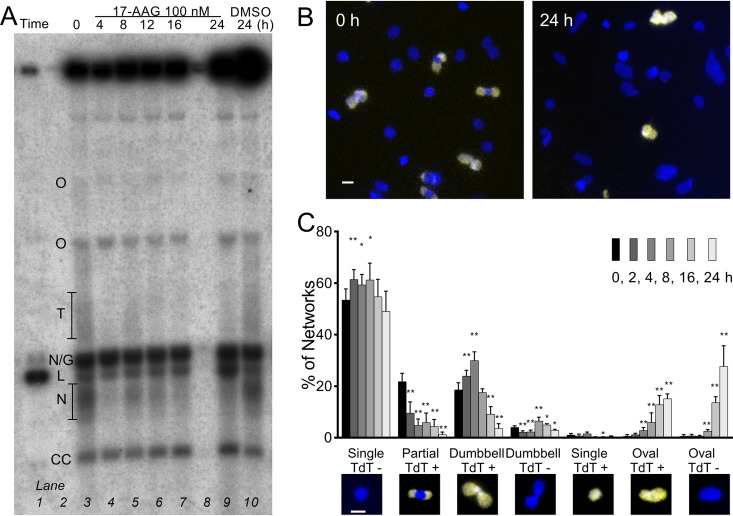
FIG 3.
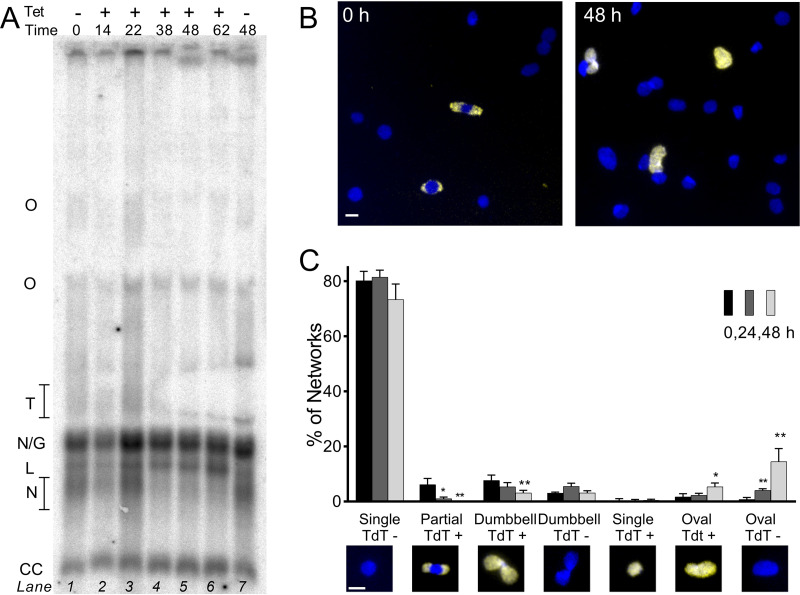
17-AAG interferes with network remodeling. (A) Southern blot of free minicircles isolated from 100 nM 17-AAG-treated trypanosomes and separated in ethidium agarose. Lane 1, linear minicircle marker; lanes 3 to 7 and 9, increasing duration of 17-AAG; lane 10, controls treated with DMSO for 24 h. Lanes loaded by equal cell equivalents. O, oligomers of two or more interlocked circles; T, theta structures; N/G, mature nicked or gapped daughters; L, linears, N, immature nicked daughters; CC, covalently closed template circles. (B) Kinetoplasts isolated from cells treated for 0 or 24 h with 100 nM 17-AAG and then labeled by TdT with fluorescent dUTP to reveal discontinuities in replicated minicircles (yellow) and stained with DAPI (blue). White bar, 1 μm. (C) Morphology and TdT labeling of networks isolated from trypanosomes treated for increasing intervals with 100 nM 17-AAG. Insets, example images of network morphologies; blue, DAPI; yellow, TdT-incorporated dUTP. White bar, 1 μm. Values are means ± SD from independent experiments; at each point, n = 3 to 9. *, P < 0.05; **, P < 0.01 t test, two-tailed equal variance to 0-h control.
Reattachment of nicked and gapped daughters normally leads to an oval network that is increasingly populated at the poles with these incompletely replicated circles and is progressively remodeled to a dumbbell shape (Fig. 1b and c). In the final stage, when all template circles have been replicated, residual discontinuities in the network-bound daughter circles are fully repaired, and the network (now comprised entirely of covalently closed minicircles) divides. Kinetoplasts isolated from treated cells were spread on slides, and residual gaps in network-bound replicating circles were detected by terminal deoxynucleotidyl transferase (TdT) incorporation of fluorescent dUTP (Fig. 3B). In networks from untreated trypanosomes, half were nonreplicating (unit-size round networks that do not label), and the balance was divided between replicating pole-labeled ovals and dumbbells (Fig. 3C). By just 2 h of 17-AAG exposure, significant changes were apparent, which were dynamic over time. Normal replicating pole-labeled oval and dumbbell networks decreased, and there was a dramatic rise in abnormal populations of ovals. These evolved from diffusely labeled to unlabeled as gaps in the incorrectly positioned daughters were repaired (Fig. 3C). The abnormal double-sized networks were nevertheless still intricately catenated, consistent with the accumulation of elongated single kinetoplasts within cells (Fig. 2D). In the face of ongoing DNA synthesis, 17-AAG thus retarded the remodeling of growing networks. Inability to reshape and scission the network likely reflects improper topoisomerase activities and, perhaps, incorrect localization of the usually polar replication machinery (19, 21).
RNAi of HSP83 causes severe growth defect.
To compare chemical inhibition with genetic knockdown of the cytosolic Hsp90, an RNAi stem-loop construct was generated against HSP83 and stably integrated into single-marker bloodstream-line (SMB) trypanosomes. The 10-tandem copies of HSP83 are 99.7% identical; thus, a single 487-base RNAi sequence was chosen that was sufficient for all (Data Set S1) and, to minimize off-target effects, had a less than 19-base match with any other trypanosome gene (22–24). Three independent rounds of transfection generated 11 clones. By just 24 h after initiation of RNAi, 9 of 11 clones displayed a severe growth defect (Fig. 4A). Notably, although onset of this defect was rapid, cell death and decline in numbers took longer, a pattern reminiscent of that for 17-AAG (Fig. S1). Reductions in mRNA and protein levels were moderate: Northern blot confirmed a 57% (n = 2) decline in HSP83 mRNA (Fig. 4B; Fig. S4A), and Western blots probed with anti-human Hsp90 revealed a band at 80 to 90 kDa that decreased 54% ± 16% (mean ± SD; n = 4) by 24 h (Fig. 4C; Fig. S4B).
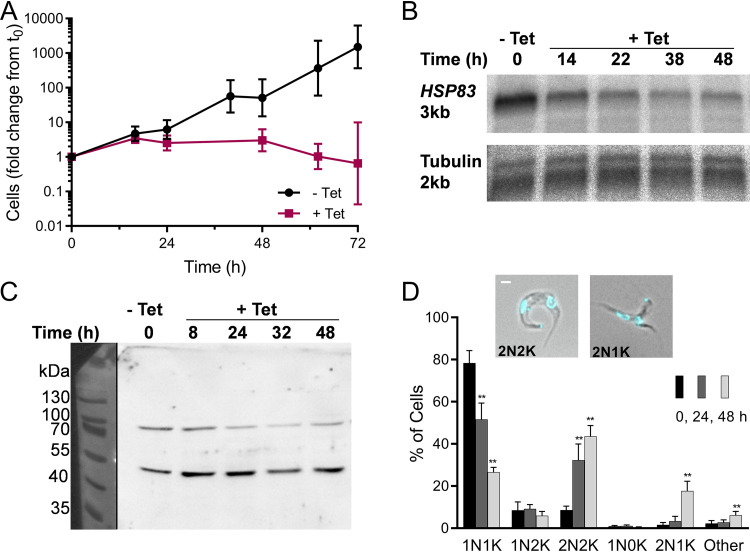
FIG 4.
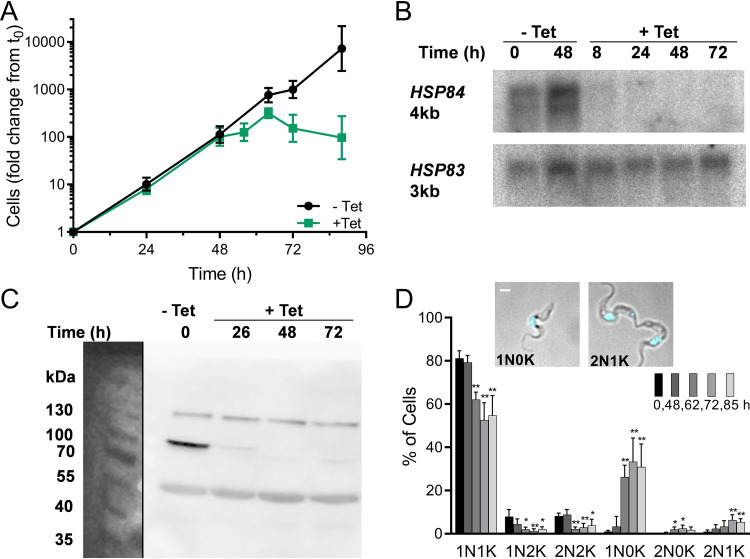
HSP83 RNAi causes severe growth defect and cytokinesis inhibition. (A) Growth of trypanosomes engineered to express, under tetracycline (Tet) control, stem-loop HSP83 RNA. Values are from 23 growth curves representing 9 clones. The clone used for follow-up work is represented by six independent inductions. Geometric mean ± SD; n = 3 to 21 at each point. (B) Northern blots of RNA from cells before and after induction of HSP83 RNAi, probed for HSP83 mRNA, and reprobed for tubulin mRNA and size markers; loaded with 700 ng RNA. Approximate lengths calculated from simultaneous size markers (not shown). Quantitation is given in Fig. S4A in the supplemental material. (C) Western blot of proteins from trypanosomes induced for RNAi of HSP83, probed with anti-human Hsp90 to reveal HSP83 (83 kDa) and an unknown cross-reacting band (50 kDa). Loaded by cell equivalents; quantitation given in Fig. S4B. (D) Nuclear and kinetoplast contents of trypanosome populations at increasing intervals after HSP83 RNAi induction. Inset, examples of abnormal 2N2K and 2N1K trypanosomes after 24 and 48 h of HSP83 RNAi, respectively (merged phase-contrast and DAPI fluorescent images). White bar, 1 μm. Data are means ± SD from independent experiments; n = 3 to 6 at each time point. **, P < 0.01; t test, two-tailed equal variance to 0-h control.
RNAi of HSP83 impairs cytokinesis and kinetoplast remodeling.
Cell cycle progression after RNAi induction was examined by microscopy of DAPI (4′,6-diamidino-2-phenylindole)-stained cells. Over 48 h, HSP83 RNAi caused a substantial cytokinesis defect with a 5-fold increase in 2N2K cells and accumulation of an abnormal 2N1K population (Fig. 4D). The free minicircle profile revealed a progressive increase in linearized minicircles (Fig. 5A; Fig. S5). Census of isolated networks by shape and TdT labeling disclosed a loss of normal replicating forms, and new populations of abnormal ovals, both labeled and unlabeled (Fig. 5B and C), confirming lesions in remodeling and scission. The similarity of this pattern to that from inhibition by 17-AAG is striking. As seen with 17-AAG, nuclear division did not reoccur following the initial mitosis to 2N; differently, however, the number of 2N cells containing a visible cleavage furrow did not significantly increase (Fig. S6). Dissimilarity in cleavage furrow retardation by inhibitor versus knockdown techniques may reflect the difference in their times to onset of action (hours versus days) or in the extent of HSP83 depletion, which is almost certainly more severe with 17-AAG.
FIG 5.
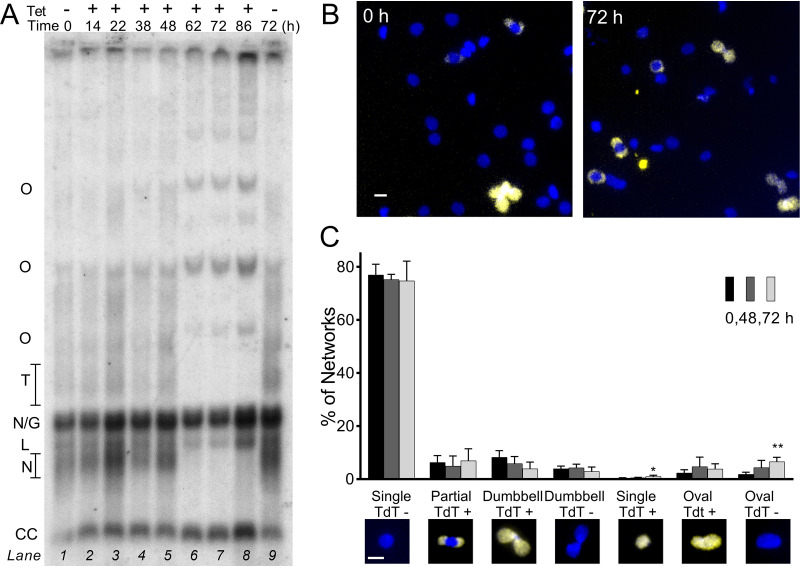
HSP83 RNAi interferes with network remodeling. (A) Southern blot of free minicircles isolated from HSP83 RNAi-induced trypanosomes and separated in ethidium agarose. Lane 1, uninduced cells at T0h; lanes 2 to 6, increasing hours of RNAi; lane 7, uninduced cells at T48h. Lanes loaded by equal cell equivalents. O, oligomers of two or more interlocked circles; T, theta structures; N/G, mature nicked or gapped daughters; L, linears, N, immature nicked daughters; CC, covalently closed template circles (blot without artifact in theta region given in Fig. S5 in the supplemental material). (B) Kinetoplast networks isolated from cells uninduced (0 h) or induced (48 h) for HSP83 RNAi, and then labeled with fluorescent dUTP to reveal discontinuities in replicated minicircles (yellow) and stained with DAPI (blue). White bar, 1 μm. (C) Kinetoplast network morphology and TdT labeling after increasing durations of HSP83 RNAi. Insets, example images of network morphologies; blue, DAPI; yellow, TdT-incorporated dUTP. White bar, 1 μm. Data are means ± SD from independent experiments; n = 3 to 4 at each point. *, P < 0.05; **, P < 0.01; t test, two-tailed equal variance to 0-h control.
RNAi of mitochondrial HSP84 causes delayed growth defect.
We speculated there may be a mitochondrial Hsp90 that contributes to kinetoplast regulation and perhaps to 17-AAG action. BLAST search against the T. brucei genome with TRAP-1, the human mitochondrial Hsp90, identified Tb427tmp.02.0250 (Tb927.11.2650 reference strain) as a homologue with 62% positive and 43% identical matches in amino acid sequence. Tb427tmp.02.0250, annotated as putative heat shock protein 84, contains a mitochondrial targeting sequence and has previously been identified in the trypanosome mitochondrial proteome (25). We refer to this gene as HSP84. For RNAi, a 592-base sequence of HSP84 (<19-base overlap with any other gene) was cloned into a stem-loop construct and transfected into cells. Three independent rounds gave 12 clones, all displaying similar growth defect after 48 h (Fig. 6A). Northern blot confirmed loss of HSP84, but not HSP83, mRNA (Fig. 6B; densitometry in Fig. S7A). Commercial antibodies to human TRAP-1 did not detect any correct-sized bands in Western blotting, so an endogenous copy of HSP84 in an RNAi line was C-terminally tagged with cMyc. Four of six tested transformants retained the growth defect, and anti-cMyc antibodies recognized an 80- to 90-kDa protein band that disappeared by 48 h (Fig. 6C; densitometry in Fig. S7B).
FIG 6.
HSP84 RNAi causes growth defect and kinetoplast loss. (A) Growth of trypanosomes engineered to express double-stranded HSP84 RNA, with or without induction (±Tet). Values are from 21 growth curves representing 12 clones; the clone used in follow-up work is represented by 6 independent inductions. Values are geometric means ± SD; n = 3 to 19 at each point. (B) Northern blots of RNA isolated from cells induced to express HSP84 RNAi, probed for HSP84 mRNA, and then reprobed for HSP83 mRNA. Approximate lengths calculated from simultaneous size markers (not shown). Loaded with 700 ng RNA. Quantitation given in Fig. S7A. (C) Western blot of cMyc-tagged HSP84 (87 kDa) and cross-reacting bands (130 kDa, 60 kDa) after induction of RNAi. Probed with anti-cMyc; loaded by cell equivalents. Quantitation given in Fig. S7B. (D) Nuclear and kinetoplast counts after increasing duration of HSP84 RNAi. Insets, examples of abnormal trypanosomes after 72 h of HSP84 RNAi (merged phase-contrast and DAPI fluorescent images). White bar, 1 μm. Data are from independent experiments; n = 3 to 5 at each time point. *, P < 0.05; **, P < 0.01 t test, two-tailed equal variance to 0-h control.
RNAi of mitochondrial HSP84 results in loss of kinetoplasts.
Cell cycle progression of HSP84-deficient cells revealed abnormalities distinctively different from those after HSP83 knockdown or 17-AAG treatment. Normal 1N1K, 1N2K, and 2N2K forms fell slightly, but after 48 h (concurrent with the growth defect), distinctly abnormal 1N0K cells lacking a visible kinetoplast comprised 30% of the population (Fig. 6D). These likely arise from a kDNA replication defect unaccompanied by impaired cytokinesis, as aberrant 2N1K cells at abscission were clearly generating a 1N1K and 1N0K daughter (Fig. 6D). Also simultaneous with the growth defect were marked alterations in network-free minicircles. Replicating intermediates (theta and early nicked daughters) decreased and covalently closed template circles accumulated, collective evidence for a block in initiation of minicircle DNA synthesis, but not in nascent strand elongation or termination (Fig. 7A). Over time, multiple forms of abnormal interlocked circles (oligomers) and linearized circles appeared, both manifestations of impaired topoisomerase activity (20). Interestingly, TdT-labeled networks evidenced only minor change, with small increases in labeled and unlabeled ovals (Fig. 7B and C), which, in contrast, were the principal abnormality after 17-AAG or HSP83 RNAi.
FIG 7.
HSP84 RNAi interferes with minicircle replication. (A) Southern blot of free minicircles isolated from HSP84 RNAi-induced trypanosomes and separated into topological forms in ethidium agarose. Lane 1, uninduced cells at T0h; lanes 2 to 8, increasing duration of RNAi; lane 9, uninduced cells at T72. Lanes loaded by cell equivalents. O, oligomers of two or more interlocked circles; T, theta structures; N/G, mature nicked, or gapped daughters; L, linears, N, immature nicked daughters; CC, covalently closed template circles. (B) Networks isolated from cells uninduced (0 h) or induced (72 h) for HSP84 RNAi and then labeled with fluorescent dUTP to reveal discontinuities in replicated minicircles (yellow), and stained DAPI (blue). White bar, 1 μm. (C) Kinetoplast network morphology and TdT labeling with increasing durations of HSP84 RNAi. Insets, example images of network morphologies. Blue, DAPI; yellow, TdT-incorporated dUTP. White bar, 1 μm. Values are means ± SD from independent experiments; n = 3 to 5 at each time point, *, P < 0.05; **, P < 0.01; t test, two-tailed equal variance to 0-h control.
DISCUSSION
In these studies of pathogenic bloodstream-form African trypanosomes, RNAi knockdowns clearly illustrate the pivotal role of Hsp90s in orderly progression of the cell cycle. We focused on cytokinesis and kDNA replication, where the relatively rapid and vivid consequences of lost function are in keeping with the view that Hsp90 proteins are indispensable in eukaryotes and that they chaperone many key regulators of vital cell pathways (4). Furthermore, the essentiality of Hsp90s, the severity of cell cycle arrest and dysregulation on loss of function, and our previous demonstration that drugs that inhibit Hsp90 are curative in mice confirm that trypanosome Hsp90s are effective drug targets (3, 26). Finally, comparison of RNAi phenotypes and chemical inhibition indicates Hsp90 is the dominant (if not only) target of 17-AAG and that it is the cytosolic HSP83, not mitochondrial HSP84, that is inhibited.
After chemical or genetic depletion of cytosolic HSP83, early and prominent abnormalities occurred in cytokinesis (Fig. 2 and 4). In bloodstream trypanosomes, such errors often lead to cells with four or eight nuclei due to unchecked nuclear replication (27, 28); however, disruption of HSP83 led to accumulation of cells with just two nuclei (Fig. 1g), suggesting additional errors that preclude further nuclear replication, in keeping with an extensive clientele for HSP83. The order of events and mechanisms of cell cycle regulation in trypanosomes are subjects of active study. RNAi phenotypes have implicated several proteins in cleavage furrow initiation and ingression, including cytokinesis initiation factors regulated by polo-like kinase and aurora kinase, as well as nDBF-2-related kinases, TRACK, CDC34, and cytoskeletal-associated katanins and kinesins (29–35); they have also identified regulators of nuclear mitosis (e.g., aurora kinase, cyclin complex CRK12-CYC9) (36, 37). Polo-like kinase and aurora kinase are clients of mammalian Hsp90 (38–40); perhaps this dependence is conserved in trypanosomes and contributes to the observed defects. HSP83 should be considered in future research as a participant in the coordination of essential cell cycle proteins and events.
Kinetoplast DNA is a unique feature of trypanosomatids, and its carefully timed and regulated replication, tightly linked with that of the nucleus, differs markedly from that of mitochondrial DNA in mammalian host cells (16). 17-AAG caused failure in kinetoplast network remodeling and cleavage (Fig. 3C) and in the progressive separation of newly replicated networks within the cell (Fig. 2D). Importantly, these defects were closely mimicked by RNAi of HSP83 (Fig. 5C) but not of HSP84 (Fig. 7C), suggesting a requirement for cytosolic Hsp90 activity to chaperone kinetoplast replication proteins (many, if not all encoded in the nucleus) (15, 19) and, perhaps, to ensure their successful translocation from cytosol to mitochondrion. The potent action of 17-AAG in trypanosomes thus appears to be mediated largely by inhibition of cytosolic HSP83, with little to no contribution by HSP84, a conclusion in keeping with the reported inability of 17-AAG to access mammalian cell mitochondria (41).
RNAi knockdown of HSP84 revealed its critical role in kDNA replication and cell survival. Upon silencing, minicircle replication decreased, (de)catenation defects were pronounced, and linearized minicircles accumulated (Fig. 7A), all convincing evidence of topoisomerase dysfunction (42). Intriguingly, this pattern is essentially identical to that seen after trypanosomes are treated with DNA-binding antitrypanosomal drugs, including pentamidine, ethidium bromide, or isometamidium chloride, compounds that selectively poison mitochondrial—but not nuclear—type II topoisomerase activity (43) and that rapidly quench the initiation of minicircle DNA synthesis (Fig. S8 in the supplemental material). Interestingly, although initiation of new strand synthesis declines, the repair of existing discontinuities in newly replicated daughter circles continues (Fig. 7A). In keeping with a halt in minicircle initiation by HSP84 RNAi, by the time cell growth has stopped, nearly half the population is accounted for by 2N1K and 1N0K cells, the result of ongoing nuclear mitosis and cytokinesis despite defective kDNA synthesis (Fig. 1h). Mechanistically telling were late-replicating 2N1K cells in which the nascent daughters were clearly 1N1K and 1N0K (Fig. 6D, right inset). RNAi has clarified the role of several mitochondrial proteins in kDNA synthesis, including topoisomerase II (44), TbPif1 helicase (45), polymerases POL1B, POL1C, and POL1D (46), and minicircle replication factor 172 (MiRF172) (47). For these proteins, knockdown halted growth after 4 to 6 days, caused progressive shrinkage and loss of kDNA networks, and produced characteristic patterns of minicircle replication intermediates. Knockdown of HSP84 halted growth by just 2 days, caused an abrupt loss of kDNA networks, decreased the initiation of minicircle synthesis, and created a distinctively different pattern of free minicircle abnormalities. These findings resemble the phenotype of mitochondrial Hsp70 knockdown by RNAi (48). In mammalian cells, Hsp70 transfers a subset of client proteins to Hsp90 (5), which may likewise be the case in the trypanosome mitochondrion. Collectively, these findings suggest that the mitochondrial Hsp90, like its cytosolic homolog, has a matrix of client proteins, almost certainly several of which are directly involved in kDNA replication, and may include topoisomerase II, TbPif1, polymerases, and/or MiRF172.
Trypanosomes are particularly susceptible to cell cycle damage. They replicate rapidly and lack many of the checkpoints and repair pathways of higher eukaryotes. The observed defects in cytokinesis and kinetoplast replication (due to 17-AAG treatment or HSP83 RNAi) implicate regulators of these processes as sensitive clients of trypanosomal HSP83. We showed, we believe for the first time, that HSP84, the mitochondrial Hsp90, is also essential in the unique process of kinetoplast replication. The overarching regulation and pathway dynamics of cell cycle control in trypanosomes are key questions, and HSP83 and HSP84 both appear to play significant roles in these processes. Hsp90s in trypanosomes thus chaperone multiple clients and enable the function of cell cycle regulators. These findings further support Hsp90s as attractive targets for antitrypanosomal therapies.
MATERIALS AND METHODS
Cell culture and reagents.
All studies were conducted with bloodstream-form Trypanosoma brucei brucei maintained in log growth in HMI9 (phenol red-free [MediaTech]), 10% fetal bovine serum [FBS], 10% SerumPlus at 5% CO2 and 37°C. MiTat 1.2 strain 427 (George Cross) was used for 17-AAG studies; single-marker bloodstream line (SMB, ATCC), maintained with 2.5 μg/ml G418 (Gibco), was used for RNAi (49). 17-AAG (National Cancer Institute Developmental Therapeutics Program) was stored in dimethyl sulfoxide (DMSO) at −20°C. Tetracycline (Tet) solution was made fresh. Reagents were from MilliporeSigma unless noted.
DAPI staining and microscopy.
Trypanosomes (1 × 106) smeared on glass slides were air-dried, fixed in ice-cold methanol, and then coverslips were mounted in Fluoroshield (containing DAPI). Imaging with 100×/1.30 objective (Zeiss Axioskop) and Retiga EXi charge-coupled-device camera (QImaging) used iVision v4.0.13 (BioVision Technologies). Per slide, 100 minimum and average 250 cells were evaluated.
KDNA network isolation and TdT labeling.
Cells washed in Voorheis's modified phosphate-buffered saline (vPBS) (50) were lysed (5 mM Tris-HCl, 37.5 mM EDTA, pH 8.0, and 0.5% SDS; all Fisher Chemical), and lysates were digested with proteinase K (1 mg/ml) and then RNase A (0.1 mg/ml). Networks were pelleted (13,000 × g, 30 min) through 20% sucrose (Baker) in Tris-EDTA (10 mM Tris-HCl and 1 mM EDTA, pH 8.0), washed (Tris-EDTA), settled (1 h) on 0.01% polylysine-treated slides, labeled (terminal deoxynucleotidyl transferase [TdT; Roche] and 5 mM fluorescein-12-dUTP [Thermo Scientific]), stained with 5 μg/ml DAPI, and sealed using Vectashield (Vector). Per replicate, 100 minimum and average 1,030 networks were evaluated.
Southern blot analysis.
Protease- and RNase-digested lysates were loaded onto 20- by 24-cm agarose (Denville) gels (1.5% minicircles, 0.6% maxicircles) that were cast and run (70 mV, 17 h) in 90 mM Tris-borate (Baker), 2.5 mM EDTA, and 1 μg/ml ethidium bromide. Gels were processed in 0.25 N HCl (15 min), H2O (10 min), 0.6 M NaCl, 0.2 M NaOH (45 min), and 25 mM sodium phosphate (pH 6.5, 30 min, 2 times) and then blotted overnight (10× SSC is 1.5 M NaCl, 0.15 M sodium citrate) onto Hybond-N+ (GE Healthcare). Membranes were cross-linked (1,200 μJ UV twice), moistened (2× SSC), prehybridized (51), and then hybridized overnight (45°C) with [32P]dATP random primer (Invitrogen)-labeled probes (Klenow polymerase [NEB], 37°C, 1 h, purified through Sephadex G50). Templates were 1-kb minicircle DNA (20) or a 770-bp maxicircle fragment (amplified from whole-cell DNA; primers given in Table S1). Washed membranes (thrice, 1× SSC, 0.1% SDS) on PhosphorImager plates (Fuji) were scanned (Fuji BAS-2500 PhosphorImager; ImageGauge v3.45). Free minicircle profiles by Southern blots were, at minimum, in biological triplicate.
RNAi of trypanosome Hsp90s.
Full-length HSP83 (Tb10.26.1080/Tb927.10.10980) and putative mitochondrial Hsp90 (Tb427tmp.02.0250/Tb11.02.0250, here designated HSP84), identified via BLAST of human TRAP1 mRNA NM_016292.2 and NM_001272049.1 on TriTrypDb (11), were analyzed (RNAit; TrypanoFan [24]) to identify 487- and 592-bp segments of HSP83 and HSP84, respectively, suitable for RNAi (<19-base overlap with any other gene). These were amplified from whole-cell DNA, incorporating 5′ XbaI/XhoI and 3′ NdeI/AscI sites (primers given in Table S1 in the supplemental material) and then sequentially inserted into pLEW100v5X:pex11 (52) as inverted repeats. Constructs (sequence validated) were purified from Escherichia coli DH5α (Qiagen miniprep), linearized (NotI; NEB), and 10 μg electroporated into 107 SMB trypanosomes (Human T cell Nucleofector kit [Lonza] and Amaxa program X-001). Transformants selected in 2.5 μg/ml phleomycin (Invivogen) were induced with 100 ng/ml Tet (a concentration determined by serial dilution analysis to induce maximal response).
Tagging HSP84 with cMyc.
Fragments (400 bp) of 3′-end HSP84 (minus stop codon) and proximal 3′ untranslated region (UTR) were PCR amplified (primers given in Table S1) and then sequentially ligated in-frame into KpnI and XhoI-digested, then BamHI and SacI-digested (NEB), pMOTag33M vector encoding a 3× cMyc tag and modified for blasticidin resistance (53). The product was excised (KpnI, SacI) from purified constructs and electroporated into HSP84 RNAi clonal cells, as above. Transformants were selected with 5 μg/ml blasticidin; 6 of 43 were randomly chosen for clonal dilution.
Northern blot analysis.
Extracted RNA (Qiagen RNeasy Mini) was mixed (700 ng; NanoDrop) with formaldehyde loading dye (Invitrogen), heated (15 min, 65°C), cooled on ice, and loaded (12.5 μg/ml ethidium bromide) onto 6.6% formaldehyde, 1% agarose gels (5 V/cm, 3 h; 20 mM MOPS [morpholinepropanesulfonic acid], 8 mM sodium acetate, and 1 mM EDTA, pH 7.0). Gels in 1 M ammonium acetate were blotted overnight onto Hybond-N+. Membranes were cross-linked, probed (56°C), and imaged as for Southern blots. Probe templates were HSP83 and HSP84 fragments nonoverlapping with RNAi sequences or other genes (≤40 bp; primers given in Table S1) or α-tubulin (54). HSP83 RNAi Northern blots were in biological duplicate; HSP84 RNAi Northern blots were in biological triplicate.
Western blot analysis.
Lysates of cells boiled with SDS loading dye (5 to 10 min) were separated by SDS-PAGE (200 V, 45 min; 5% stacking, 8% running; Bio-Rad), transferred (100 V, 1 h, room temperature [RT]; Hybond-enhanced chemiluminescence [ECL]), blocked (5% nonfat dry milk, Tween-TBS); incubated (4°C, overnight) with rabbit polyclonal anti-human Hsp90 (1:5,000; catalog no. sc-7947; Santa Cruz) or anti-cMyc (1:500; catalog no. C3956; Sigma-Aldrich) in 5% bovine serum albumin (BSA) in Tween-Tris-buffered saline (TBS), washed, and then incubated with secondary antibody (1:30,000; goat-anti-rabbit horseradish peroxidase [HRP; catalog no. sc-2054; Santa Cruz], 5% BSA, 1 h, RT). Blots were developed (ECL Plus; catalog no. RPN2132, Amersham Biosciences) and imaged (Image Station 4000R Pro; Kodak, Carestream software). Western blots were, at minimum, in biological quadruplicate.
Densitometry.
Northern and Western blot images were quantified by mean gray value using ImageJ 1.50i. All bands in the same blot were circumscribed by equal-sized rectangles and their gray values measured. Background (mean gray values from rectangles taken just above or below bands) was subtracted. These measurements were repeated three times, and the mean value was taken for each band.
ACKNOWLEDGMENTS
We dedicate this paper to our colleague Paul Englund, who loved the intricacies of the trypanosome kinetoplast and whose presence is deeply missed.
We thank Emily Caton for assistance with microscopy and Rahul Bakshi and Beth Nenortas for thoughtfully reading and discussing the manuscript. 17-AAG was generously provided by the Open Chemical Repository of the Drug Synthesis and Chemistry Branch, Developmental Therapeutics Program, Division of Cancer Treatment and Diagnosis, NCI (http://dtp.cancer.gov).
This work was supported by National Institutes of Health [grant R01AI095453 to T.A.S.] and by Fulbright New Zealand and the Mustard Seed Foundation [to K.J.M.].
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Brun R, Blum J, Chappuis F, Burri C. 2010. Human African trypanosomiasis. Lancet 375:148–159. 10.1016/S0140-6736(09)60829-1. [DOI] [PubMed] [Google Scholar]
- 2.Kennedy PGE. 2019. Update on human African trypanosomiasis (sleeping sickness). J Neurol 266:2334–2337. 10.1007/s00415-019-09425-7. [DOI] [PubMed] [Google Scholar]
- 3.Meyer KJ, Shapiro TA. 2013. Potent antitrypanosomal activities of heat shock protein 90 inhibitors in vitro and in vivo. J Infect Dis 208:489–499. 10.1093/infdis/jit179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schopf FH, Biebl MM, Buchner J. 2017. The HSP90 chaperone machinery. Nat Rev Mol Cell Biol 18:345–360. 10.1038/nrm.2017.20. [DOI] [PubMed] [Google Scholar]
- 5.Radli M, Rudiger SGD. 2018. Dancing with the diva: Hsp90-client interactions. J Mol Biol 430:3029–3040. 10.1016/j.jmb.2018.05.026. [DOI] [PubMed] [Google Scholar]
- 6.Neckers L. 2002. Hsp90 inhibitors as novel cancer chemotherapeutic agents. Trends Mol Med 8:S55–61. 10.1016/s1471-4914(02)02316-x. [DOI] [PubMed] [Google Scholar]
- 7.Rochani AK, Singh M, Tatu U. 2013. Heat shock protein 90 inhibitors as broad spectrum anti-infectives. Curr Pharm Des 19:377–386. 10.2174/138161213804143608. [DOI] [PubMed] [Google Scholar]
- 8.Johnson JL. 2012. Evolution and function of diverse Hsp90 homologs and cochaperone proteins. Biochim Biophys Acta 1823:607–613. 10.1016/j.bbamcr.2011.09.020. [DOI] [PubMed] [Google Scholar]
- 9.Altieri DC, Stein GS, Lian JB, Languino LR. 2012. TRAP-1, the mitochondrial Hsp90. Biochim Biophys Acta 1823:767–773. 10.1016/j.bbamcr.2011.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jones C, Anderson S, Singha UK, Chaudhuri M. 2008. Protein phosphatase 5 is required for Hsp90 function during proteotoxic stresses in Trypanosoma brucei. Parasitol Res 102:835–844. 10.1007/s00436-007-0817-z. [DOI] [PubMed] [Google Scholar]
- 11.Aslett M, Aurrecoechea C, Berriman M, Brestelli J, Brunk BP, Carrington M, Depledge DP, Fischer S, Gajria B, Gao X, Gardner MJ, Gingle A, Grant G, Harb OS, Heiges M, Hertz-Fowler C, Houston R, Innamorato F, Iodice J, Kissinger JC, Kraemer E, Li W, Logan FJ, Miller JA, Mitra S, Myler PJ, Nayak V, Pennington C, Phan I, Pinney DF, Ramasamy G, Rogers MB, Roos DS, Ross C, Sivam D, Smith DF, Srinivasamoorthy G, Stoeckert CJ, Jr, Subramanian S, Thibodeau R, Tivey A, Treatman C, Velarde G, Wang H. 2010. TriTrypDB: a functional genomic resource for the Trypanosomatidae. Nucleic Acids Res 38:D457–D462. 10.1093/nar/gkp851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mottram JC, Murphy WJ, Agabian N. 1989. A transcriptional analysis of the Trypanosoma brucei hsp83 gene cluster. Mol Biochem Parasitol 37:115–127. 10.1016/0166-6851(89)90108-4. [DOI] [PubMed] [Google Scholar]
- 13.Pizarro JC, Hills T, Senisterra G, Wernimont AK, Mackenzie C, Norcross NR, Ferguson MA, Wyatt PG, Gilbert IH, Hui R. 2013. Exploring the Trypanosoma brucei Hsp83 potential as a target for structure guided drug design. PLoS Negl Trop Dis 7:e2492. 10.1371/journal.pntd.0002492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alsford S, Turner DJ, Obado SO, Sanchez-Flores A, Glover L, Berriman M, Hertz-Fowler C, Horn D. 2011. High-throughput phenotyping using parallel sequencing of RNA interference targets in the African trypanosome. Genome Res 21:915–924. 10.1101/gr.115089.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wheeler RJ, Gull K, Sunter JD. 2019. Coordination of the cell cycle in trypanosomes. Annu Rev Microbiol 73:133–154. 10.1146/annurev-micro-020518-115617. [DOI] [PubMed] [Google Scholar]
- 16.Schneider A, Ochsenreiter T. 2018. Failure is not an option - mitochondrial genome segregation in trypanosomes. J Cell Sci 131:jcs221820. 10.1242/jcs.221820. [DOI] [PubMed] [Google Scholar]
- 17.Jensen RE, Englund PT. 2012. Network news: the replication of kinetoplast DNA. Annu Rev Microbiol 66:473–491. 10.1146/annurev-micro-092611-150057. [DOI] [PubMed] [Google Scholar]
- 18.Hughes L, Borrett S, Towers K, Starborg T, Vaughan S. 2017. Patterns of organelle ontogeny through a cell cycle revealed by whole-cell reconstructions using 3D electron microscopy. J Cell Sci 130:637–647. 10.1242/jcs.198887. [DOI] [PubMed] [Google Scholar]
- 19.Mensa-Wilmot K, Hoffman B, Wiedeman J, Sullenberger C, Sharma A. 2019. Kinetoplast division factors in a trypanosome. Trends Parasitol 35:119–128. 10.1016/j.pt.2018.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shapiro TA, Klein VA, Englund PT. 1989. Drug-promoted cleavage of kinetoplast DNA minicircles. Evidence for type II topoisomerase activity in trypanosome mitochondria. J Biol Chem 264:4173–4178. 10.1016/S0021-9258(19)84979-7. [DOI] [PubMed] [Google Scholar]
- 21.Concepcion-Acevedo J, Miller JC, Boucher MJ, Klingbeil MM. 2018. Cell cycle localization dynamics of mitochondrial DNA polymerase IC in African trypanosomes. Mol Biol Cell 29:2540–2552. 10.1091/mbc.E18-02-0127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Djikeng A, Shi H, Tschudi C, Ullu E. 2001. RNA interference in Trypanosoma brucei: cloning of small interfering RNAs provides evidence for retroposon-derived 24–26-nucleotide RNAs. RNA 7:1522–1530. [PMC free article] [PubMed] [Google Scholar]
- 23.Durand-Dubief M, Kohl L, Bastin P. 2003. Efficiency and specificity of RNA interference generated by intra- and intermolecular double stranded RNA in Trypanosoma brucei. Mol Biochem Parasitol 129:11–21. 10.1016/s0166-6851(03)00071-9. [DOI] [PubMed] [Google Scholar]
- 24.Redmond S, Vadivelu J, Field MC. 2003. RNAit: an automated web-based tool for the selection of RNAi targets in Trypanosoma brucei. Mol Biochem Parasitol 128:115–118. 10.1016/s0166-6851(03)00045-8. [DOI] [PubMed] [Google Scholar]
- 25.Panigrahi AK, Ogata Y, Zikova A, Anupama A, Dalley RA, Acestor N, Myler PJ, Stuart KD. 2009. A comprehensive analysis of Trypanosoma brucei mitochondrial proteome. Proteomics 9:434–450. 10.1002/pmic.200800477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meyer KJ, Caton E, Shapiro TA. 2018. Model system identifies kinetic driver of Hsp90 inhibitor activity against African trypanosomes and Plasmodium falciparum. Antimicrob Agents Chemother 62:e00056-18. 10.1128/AAC.00056-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hammarton TC. 2007. Cell cycle regulation in Trypanosoma brucei. Mol Biochem Parasitol 153:1–8. 10.1016/j.molbiopara.2007.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhou Q, Hu H, Li Z. 2014. New insights into the molecular mechanisms of mitosis and cytokinesis in trypanosomes. Int Rev Cell Mol Biol 308:127–166. 10.1016/B978-0-12-800097-7.00004-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hammarton TC, Lillico SG, Welburn SC, Mottram JC. 2005. Trypanosoma brucei MOB1 is required for accurate and efficient cytokinesis but not for exit from mitosis. Mol Microbiol 56:104–116. 10.1111/j.1365-2958.2005.04542.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rothberg KG, Burdette DL, Pfannstiel J, Jetton N, Singh R, Ruben L. 2006. The RACK1 homologue from Trypanosoma brucei is required for the onset and progression of cytokinesis. J Biol Chem 281:9781–9790. 10.1074/jbc.M600133200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rojas F, Koszela J, Bua J, Llorente B, Burchmore R, Auer M, Mottram JC, Tellez-Inon MT. 2017. The ubiquitin-conjugating enzyme CDC34 is essential for cytokinesis in contrast to putative subunits of a SCF complex in Trypanosoma brucei. PLoS Negl Trop Dis 11:e0005626. 10.1371/journal.pntd.0005626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hu L, Hu H, Li Z. 2012. A kinetoplastid-specific kinesin is required for cytokinesis and for maintenance of cell morphology in Trypanosoma brucei. Mol Microbiol 83:565–578. 10.1111/j.1365-2958.2011.07951.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Benz C, Clucas C, Mottram JC, Hammarton TC. 2012. Cytokinesis in bloodstream stage Trypanosoma brucei requires a family of katanins and spastin. PLoS One 7:e30367. 10.1371/journal.pone.0030367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang X, An T, Pham KTM, Lun ZR, Li Z. 2019. Functional analyses of cytokinesis regulators in bloodstream stage Trypanosoma brucei parasites identify functions and regulations specific to the life cycle stage. mSphere 4:e00199-19. 10.1128/mSphere.00199-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kurasawa Y, An T, Li Z. 2020. Polo-like kinase in trypanosomes: an odd member out of the Polo family. Open Biol 10:200189. 10.1098/rsob.200189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li Z, Umeyama T, Wang CC. 2009. The aurora kinase in Trypanosoma brucei plays distinctive roles in metaphase-anaphase transition and cytokinetic initiation. PLoS Pathog 5:e1000575. 10.1371/journal.ppat.1000575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Monnerat S, Almeida Costa CI, Forkert AC, Benz C, Hamilton A, Tetley L, Burchmore R, Novo C, Mottram JC, Hammarton TC. 2013. Identification and functional characterisation of CRK12:CYC9, a novel cyclin-dependent kinase (CDK)-cyclin complex in Trypanosoma brucei. PLoS One 8:e67327. 10.1371/journal.pone.0067327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.de Carcer G. 2004. Heat shock protein 90 regulates the metaphase-anaphase transition in a polo-like kinase-dependent manner. Cancer Res 64:5106–5112. 10.1158/0008-5472.can-03-2214. [DOI] [PubMed] [Google Scholar]
- 39.Hsieh YL, Tu HJ, Pan SL, Liou JP, Yang CR. 2019. Anti-metastatic activity of MPT0G211, a novel HDAC6 inhibitor, in human breast cancer cells in vitro and in vivo. Biochim Biophys Acta Mol Cell Res 1866:992–1003. 10.1016/j.bbamcr.2019.03.003. [DOI] [PubMed] [Google Scholar]
- 40.Wu Z, Gholami AM, Kuster B. 2012. Systematic identification of the HSP90 candidate regulated proteome. Mol Cell Proteomics 11:M111.016675. 10.1074/mcp.M111.016675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee C, Park HK, Jeong H, Lim J, Lee AJ, Cheon KY, Kim CS, Thomas AP, Bae B, Kim ND, Kim SH, Suh PG, Ryu JH, Kang BH. 2015. Development of a mitochondria-targeted Hsp90 inhibitor based on the crystal structures of human TRAP1. J Am Chem Soc 137:4358–4367. 10.1021/ja511893n. [DOI] [PubMed] [Google Scholar]
- 42.Burri C, Bodley AL, Shapiro TA. 1996. Topoisomerases in kinetoplastids. Parasitol Today 12:226–231. 10.1016/0169-4758(96)10017-x. [DOI] [PubMed] [Google Scholar]
- 43.Shapiro TA, Englund PT. 1990. Selective cleavage of kinetoplast DNA minicircles promoted by antitrypanosomal drugs. Proc Natl Acad Sci USA 87:950–954. 10.1073/pnas.87.3.950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang Z, Englund PT. 2001. RNA interference of a trypanosome topoisomerase II causes progressive loss of mitochondrial DNA. EMBO J 20:4674–4683. 10.1093/emboj/20.17.4674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu B, Yildirir G, Wang J, Tolun G, Griffith JD, Englund PT. 2010. TbPIF1, a Trypanosoma brucei mitochondrial DNA helicase, is essential for kinetoplast minicircle replication. J Biol Chem 285:7056–7066. 10.1074/jbc.M109.084038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bruhn DF, Sammartino MP, Klingbeil MM. 2011. Three mitochondrial DNA polymerases are essential for kinetoplast DNA replication and survival of bloodstream form Trypanosoma brucei. Eukaryot Cell 10:734–743. 10.1128/EC.05008-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Amodeo S, Jakob M, Ochsenreiter T. 2018. Characterization of the novel mitochondrial genome replication factor MiRF172 in Trypanosoma brucei. J Cell Sci 131:jcs211730. 10.1242/jcs.211730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Týč J, Klingbeil MM, Lukeš J. 2015. Mitochondrial heat shock protein machinery Hsp70/Hsp40 is indispensable for proper mitochondrial DNA maintenance and replication. mBio 6:e02425-14. 10.1128/mBio.02425-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wirtz E, Leal S, Ochatt C, Cross GA. 1999. A tightly regulated inducible expression system for conditional gene knock-outs and dominant-negative genetics in Trypanosoma brucei. Mol Biochem Parasitol 99:89–101. 10.1016/s0166-6851(99)00002-x. [DOI] [PubMed] [Google Scholar]
- 50.Field MC, Allen CL, Dhir V, Goulding D, Hall BS, Morgan GW, Veazey P, Engstler M. 2004. New approaches to the microscopic imaging of Trypanosoma brucei. Microsc Microanal 10:621–636. 10.1017/S1431927604040942. [DOI] [PubMed] [Google Scholar]
- 51.Church GM, Gilbert W. 1984. Genomic sequencing. Proc Natl Acad Sci USA 81:1991–1995. 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Silverman JS, Schwartz KJ, Hajduk SL, Bangs JD. 2011. Late endosomal Rab7 regulates lysosomal trafficking of endocytic but not biosynthetic cargo in Trypanosoma brucei. Mol Microbiol 82:664–678. 10.1111/j.1365-2958.2011.07842.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Oberholzer M, Morand S, Kunz S, Seebeck T. 2006. A vector series for rapid PCR-mediated C-terminal in situ tagging of Trypanosoma brucei genes. Mol Biochem Parasitol 145:117–120. 10.1016/j.molbiopara.2005.09.002. [DOI] [PubMed] [Google Scholar]
- 54.Povelones ML, Tiengwe C, Gluenz E, Gull K, Englund PT, Jensen RE. 2013. Mitochondrial shape and function in trypanosomes requires the outer membrane protein, TbLOK1. Mol Microbiol 87:713–729. 10.1111/mmi.12089. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material. Download AAC.00632-21-s0001.pdf, PDF file, 3.3 MB (3.3MB, pdf)