Abstract
SARS-CoV-2 infections display tremendous interindividual variability, ranging from asymptomatic infections to life-threatening disease. Inborn errors of, and autoantibodies directed against, type I interferons (IFNs) account for about 20% of critical COVID-19 cases among SARS-CoV-2-infected individuals. By contrast, the genetic and immunological determinants of resistance to infection per se remain unknown. Following the discovery that autosomal recessive deficiency in the DARC chemokine receptor confers resistance to Plasmodium vivax, autosomal recessive deficiencies of chemokine receptor 5 (CCR5) and the enzyme FUT2 were shown to underlie resistance to HIV-1 and noroviruses, respectively. Along the same lines, we propose a strategy for identifying, recruiting, and genetically analyzing individuals who are naturally resistant to SARS-CoV-2 infection.
Subject terms: Viral infection, Infection
In this Perspective, Spaan and colleagues propose a strategy for identifying, recruiting, and genetically analyzing individuals who are naturally resistant to SARS-CoV-2 infection.
Main
The COVID-19 pandemic has reminded us that infections are unique among diseases in their potential to rapidly cause massive morbidity and mortality worldwide. Throughout history, infectious diseases have imposed strong selection pressures on humans1–3. In particular, viral pandemics, including ones caused by coronaviruses, have occurred repeatedly over the last century, and probably throughout human history4–7. Clinical variability in response to infection, viral or otherwise, can be explained, at least in some individuals, by human genetic factors8. The introduction of SARS-CoV-2 to a naive population, on a global scale, has provided yet another demonstration of the remarkable clinical variability between individuals in the course of infection, ranging from asymptomatic infections to life-threatening disease9–11. Our understanding of the pathophysiology of life-threatening COVID-19 has progressed considerably since the disease was first described in December 2019 (refs. 12,13), but we still know very little about the human genetic and immunological basis of inborn resistance to SARS-CoV-2. Mean secondary attack rates for SARS-CoV-2 infections can reach up to 70% in specific households14,15, and a number of families have been reported in which all the members except one of the spouses are infected16, suggesting that some highly exposed individuals may be resistant to infection with this virus. Here, we review examples of genetically determined susceptibility to severe outcomes of two infectious diseases—tuberculosis (TB) and COVID-19—while covering in greater depth the three known cases of inborn resistance to infections. We then consider candidate genes directly relevant to resistance to SARS-CoV-2 infection. Finally, we propose a strategy for recruiting and genetically analyzing individuals who are naturally resistant to infection with the virus. Above all, we advocate for further studies to develop our understanding of the causal mechanisms of inborn resistance to SARS-CoV-2 infection and provide a framework for the use of this knowledge for therapeutic purposes.
Inborn susceptibility to life-threatening infectious diseases
Human evolution has been marked by microorganisms that are sufficiently pathogenic to exert selective pressure on genes crucial for host defense2. One of the deadliest scourges of human health is TB, which has caused an estimated one billion deaths in Europe over the past two millennia17. Paradoxically, less than 10% of humans infected with Mycobacterium tuberculosis develop TB. Since the turn of the twentieth century, the contribution of human genetics to TB pathogenesis has been deciphered through classic genetics and experimental studies18,19. More recently, rare inborn errors of immunity (IEIs), including autosomal recessive interleukin-12 receptor β1 (IL12RB1)20,21 and tyrosine kinase 2 (TYK2) deficiencies22, in particular, have been identified in a few people with TB. The broader relevance of this finding was shown when the analysis was expanded to more common variants, revealing that homozygosity for the TYK2(P1104A) polymorphism was associated with a high risk of developing TB17,23. p.P1104A homozygosity disrupts the capacity of TYK2 to mediate IL-23-dependent IFN-γ immunity to mycobacteria23. Its minor allele frequency is highest among Europeans17. An analysis of ancient DNA showed that the frequency of TYK2(P1104A) has strongly decreased over the last 2,000 years in Europe owing to strong negative selection, concomitant with the high TB burden in Europe24.
With the advent of the COVID-19 pandemic, specific IEIs were shown to have a role in defining susceptibility to severe COVID-19. The COVID Human Genetic Effort (http://www.covidhge.com) reported 23 critically ill people with IEIs at 8 loci governing TLR3- and IRF7-dependent type I IFN induction and amplification13. Remarkably, four unrelated and previously healthy adults had autosomal recessive IRF7 or IFNAR1 deficiency. Although rare, the individuals with IEIs demonstrate that type I IFN immunity is indispensable for the control of SARS-CoV-2 infection. This finding led to the subsequent discovery, also by the consortium, of pre-existing neutralizing autoantibodies against type I IFNs as a phenocopy of type I IFN-related IEIs12. Subsequent studies in independent cohorts confirmed the presence of neutralizing autoantibodies against type I IFNs in more than 10% of people with severe COVID-19 (refs. 25–30). More recently, the consortium found that autoantibodies neutralizing lower, more physiological concentrations of type I IFNs account for about 20% of patients older than 70 years with critical pneumonia31. Moreover, the consortium also reported that about 1% of male patients younger than 60 years of age with critical pneumonia have X-recessive TLR7 deficiency32. Surprisingly, the individuals with IEIs identified and those with autoantibodies had not displayed any particular susceptibility to other severe infectious diseases before exposure to SARS-CoV-2. This finding is consistent with the smaller amounts of type I IFNs induced by SARS-CoV-2 than by seasonal influenza virus, for example33. However, type I IFN autoantibodies have been shown to underlie a third of adverse reactions to the live attenuated yellow fever virus vaccine34. Collectively, these examples illustrate how the genetic elucidation of an immunological deficit in a few rare individuals can indicate a mechanism that is disrupted by other causes in many more people.
Inborn resistance to infection upon exposure
An individual’s genetically determined protection against an infectious disease is the mirror image of genetically determined susceptibility to life-threatening disease. The term ‘protective’ is applied to a given locus when the allele associated with a lower risk of disease is the least frequent, alternative allele. Far fewer genetic studies on infectious diseases have focused on protective alleles than on susceptibility to infection, whether monogenic or polygenic. In the early 1950s, Anthony Allison showed that the HbS sickle-cell trait is maintained at high frequency in African areas where malaria is endemic, owing to a heterozygous advantage1 of the allele for providing protection against severe Plasmodium falciparum infections35. Other examples of protection against poor infection outcomes include the occurrence of specific HLA class I alleles in long-term nonprogressing HIV-1-infected individuals36, and the role of a type III interferon (IFNL3-IFNL4) haplotype in viral clearance following infection with hepatitis C virus (HCV)37,38. These alleles confer protection against severe disease in infected people, but not against contraction of the infection itself.
The genetic determinism of resistance to infection has been even less studied than that of protection against poor infection outcomes, and study has always been from a monogenic angle. Only three mechanisms of Mendelian resistance to infection have been identified to date. In the 1970s, Louis Miller discovered that the absence of the Duffy antigen on erythrocytes prevented these cells from becoming infected with Plasmodium vivax39,40. The molecular genetic basis of this autosomal recessive resistance trait was not determined until the 1990s. The causal variant affects the GATA-1 binding site in the DARC promoter, selectively preventing gene transcription in erythroid cells41. At about the same time, autosomal recessive CCR5 deficiency was found to confer resistance to infection with HIV-1 (refs. 42–44). The most common loss-of-function mutation in CCR5 is a 32-base-pair deletion with a minor allele frequency of 10% in the European population. Finally, autosomal recessive FUT2 deficiency was discovered to confer resistance to gastrointestinal infections with noroviruses45. As for DARC and the P. vivax Duffy binding protein, and CCR5 and the HIV-1 gp120–gp41 heterodimer, FUT2 expression is required for binding of the norovirus VPg capsid. It is probably no coincidence that these examples of Mendelian resistance to infection are complete deficiencies of receptors or coreceptors exploited by the pathogen as a means of entering cells. The genetic mechanisms of protection against severe infectious outcomes and those underlying resistance to infection itself are both subject to positive selection, as they provide a survival advantage46.
Candidate SARS-CoV-2 resistance genes
The proportion of humans naturally resistant to SARS-CoV-2 infection is unknown, but a number of candidate genes potentially involved in human inborn resistance to SARS-CoV-2 infection have emerged from several lines of evidence. One is the ABO locus, which was identified in genome-wide association studies (GWAS)47,48. Although initial data on the impact of blood group on COVID-19 severity were inconsistent, a recent meta-analysis of nearly 50,000 people from 46 studies confirmed an effect of this locus on susceptibility to infection49. The protective effect of the O allele, however, is small, with an odds ratio of ~0.90. Although no unified mechanism of resistance has yet been proposed50, ABO blood groups may play a direct role in infection by serving as coreceptors for SARS-CoV-2 (ref. 47). Pandemic-associated pernio (chilblain) is a rare manifestation in individuals exposed to SARS-CoV-2 that could provide insight into mechanisms of resistance to infection51,52. Pandemic-associated pernio (‘COVID toes’) mimics the skin lesions of familial chilblain lupus and Aicardi–Goutières syndrome, monogenic disorders caused by mutations leading to an upregulation of type I IFN signaling53. Most people with pernio remain seronegative, but the presence of the SARS-CoV-2 spike protein has been demonstrated in skin biopsy specimens, and a robust local type I IFN response has also been observed, suggesting early clearance of the virus54. These observations imply the presence of infection, and, thus, the absence of natural resistance to infection. Nevertheless, by understanding the pathophysiology of this phenomenon, we may be able to shed light on host mechanisms restricting viral replication and promoting resilience upon SARS-CoV-2 infection.
In vitro interactome studies have identified additional candidate host genes supporting the viral life cycle. Early in the pandemic, it was discovered that SARS-CoV-2 infection is dependent on the ACE2 receptor for cell entry and the serine protease TMPRSS2 for spike protein priming55–58. Indeed, a rare variant located close to ACE2 was found, by GWAS, to confer protection against SARS-CoV-2 infection, possibly by decreasing ACE2 expression59. Furthermore, although their impact on infection is unknown, some human ACE2 polymorphisms bind the SARS-CoV-2 spike protein with different affinities in vitro60. In a genome-wide CRISPR knockout screen for infection with SARS-CoV-2 and other coronaviruses, TMEM41B was identified as a requirement for permissive infection with the virus61. TMEM41B is an endoplasmic reticulum transmembrane protein that is also required by flaviviruses62. Its impact on SARS-CoV-2 infection remains to be established, but an allele common in East and South Asians has been shown to be associated with a lower capacity to support flavivirus infection in vitro62. Like genome-wide CRISPR knockout screens, affinity purification-mass spectrometry on human proteins interacting with SARS-CoV-2 has yielded an extensive protein interaction map63,64. Functional assessments of this interactome have resulted in its translation into a catalog of essential host factors required for SARS-CoV-2 infection65. Although no human studies linking the SARS-CoV-2 interactome to susceptibility to infection have yet been published, the genes concerned—along with the loci identified by GWAS—can be regarded as candidates for the identification of inborn variants conferring resistance to infection.
Genetic and immunological strategies
There are two key challenges in the search for individuals naturally resistant to SARS-CoV-2 infection. First, demonstrating an absence of infection poses a diagnostic hurdle. PCR-based molecular diagnostic approaches using respiratory specimens provide only snapshot information. Serology is useful for assessing the occurrence of prior infections for many viral infections, but some individuals remain seronegative despite infection with SARS-CoV-2 (refs. 66,67). Pre-existing crossreactive T-cell-mediated immunity as a result of prior infections with other coronaviruses might contribute to a resilient response upon infection with SARS-CoV-2 (refs. 68–71). At the same time, T cell responses to SARS-CoV-2-specific antigens could provide a sensitive and specific marker for the qualitative assessment of prior infection with SARS-CoV-2 (ref. 68). A second challenge lies in the probability of virus transmission. The likelihood of infection is influenced by both the duration and intensity of exposure to an infected individual, and the intrinsic transmission characteristics of the pathogen. The basic reproduction number (R0, the average number of secondary infections produced by a typical case of an infection in a population where everyone is susceptible) of SARS-CoV-2 is between 2.5 and 5.0, on average72–74. However, coronaviruses are known to be transmitted during superspreader events with very high secondary attack rates75. Identifying these events, other large-scale outbreaks, and households in which one or very few individuals remained uninfected14–16 would be of particular interest for the study of inborn variants conferring resistance to SARS-CoV-2.
When testing the hypothesis that monogenic inborn variants of immunity confer natural resistance to SARS-CoV-2 infection, we apply a four-step strategy to overcome diagnostic limitations and uncertainties about exposure (Fig. 1). We first focus on uninfected household contacts of people with symptomatic COVID-19 (score of 3 or higher on the World Health Organization’s clinical progression scale76). We then consider individuals exposed to an index case without personal protection equipment, for at least 1 hour per day, and during the first 3–5 days of symptoms in the index case. Priority is given to the study of serodiscordant spouses and sleeping partners. We subsequently enroll individuals with a negative PCR result when tested plus negative serological results obtained 4 weeks after exposure. Finally, we assess SARS-CoV-2-specific T cell responses in the candidate resistant individuals and compare their responses with those of SARS-CoV-2-infected individuals. We differentiate T cell responses induced by vaccination from those provoked by natural infection. Study participants lacking a SARS-CoV-2-reactive T cell response will be analyzed by whole-exome/genome sequencing. The results will be compared with those for SARS-CoV-2-infected controls, with the aim of identifying rare or common variants with a strong effect on resistance to infection11–13,77. Finally, as in studies of IEIs78, the genetic findings will be validated experimentally, including with cells from the study participants, to dissect the mechanisms of resistance at the molecular, cellular, tissue, immunological, and whole-organism levels (Fig. 1).
Fig. 1. A global effort to dissect the human genetic basis of resistance to SARS-CoV-2 infection.
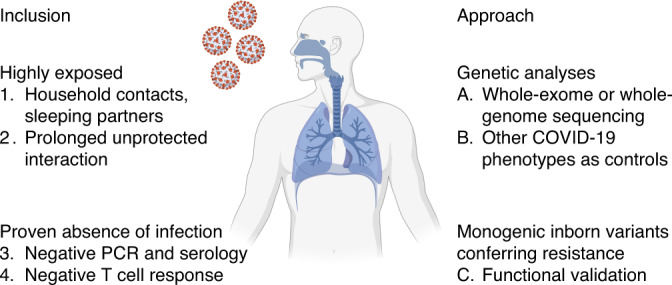
Inclusion criteria, and approach for the identification and validation of inborn variants conferring resistance to SARS-CoV-2 infection (http://www.covidhge.com). Created with BioRender.com.
Concluding remarks
Historical examples of inborn resistance to infection with other pathogens provide a road map for testing the hypothesis of monogenic inborn resistance to infection with SARS-CoV-2. Some more common inborn variants of resistance in candidate genes may have relatively small effects. However, we also aim to identify candidate genes with potentially rare variants and a large effect size. These variants are of particular interest for two reasons. First, they can provide a deep understanding of the essential biological pathways involved in infection with SARS-CoV-2. Second, they will allow for the development of innovative therapeutic interventions to prevent or treat SARS-CoV-2 infection in others. The proof-of-principle for this second reason of interest has been provided by CCR5 and its antagonist maraviroc, which is used for the treatment of HIV-1 infections in specific settings79. In addition, transplantation of CCR5-deficient bone marrow has been successfully applied to cure HIV infection in a few people80,81. No specific drug effective against COVID-19 has been discovered since the start of the pandemic. Lessons learned from experiments of nature could potentially guide us toward such specific treatments for COVID-19. We have already enrolled more than 400 individuals meeting the criteria for inclusion in a dedicated resistance study cohort. The collaborative enrollment of study participants is continuing (http://www.covidhge.com), and subjects from all over the world are welcome.
Acknowledgements
The Laboratory of Human Genetics of Infectious Diseases is supported by the National Institutes of Health (NIH) (R01AI088364), the National Center for Advancing Translational Sciences (NCATS), NIH Clinical and Translational Science Award (CTSA) program (UL1TR001866), a Fast Grant from Emergent Ventures, Mercatus Center at George Mason University, the Yale Center for Mendelian Genomics and the GSP Coordinating Center funded by the National Human Genome Research Institute (NHGRI) (UM1HG006504 and U24HG008956), the Fisher Center for Alzheimer’s Research Foundation, the Meyer Foundation, the French National Research Agency (ANR) under the Investments for the Future program (ANR-10-IAHU-01), the Integrative Biology of Emerging Infectious Diseases Laboratory of Excellence (ANR-10-LABX-62-IBEID), the French Foundation for Medical Research (FRM) (EQU201903007798), the FRM and ANR GENCOVID project (ANR-20-COVI-0003), ANRS-COV05, the Fondation du Souffle, the Square Foundation, Grandir - Fonds de solidarité pour l’enfance, the SCOR Corporate Foundation for Science, the Howard Hughes Medical Institute, the Rockefeller University, the St. Giles Foundation, Institut National de la Santé et de la Recherche Médicale (INSERM), and the University of Paris. E.A. is supported by research grants from the European Commission’s Horizon 2020 research and innovation program (IMMUNAID, grant no. 779295, CURE, grant no. 767015 and TO_AITION grant no. 848146) and the Hellenic Foundation for Research and Innovation (INTERFLU, no. 1574). C.O.F. is supported in part by the Science Foundation Ireland COVID-19 Program. G.N. is supported by a grant awarded to Regione Lazio (Research Group Projects 2020) no. A0375-2020-36663, GecoBiomark. A.P. is supported in part by the Horizon 2020 program under grant no. 824110 (EasiGenomics grant no. COVID-19/PID12342) and the CERCA Program/Generalitat de Catalunya. H.S. is supported in part by the Intramural Research Program of the National Institute of Allergy and Infectious Diseases, National Institutes of Health. A.S. is supported in part by the European Union’s Horizon 2020 research and innovation program (Marie Sklodowska-Curie grant no. 789645).
Competing interests
The authors declare no competing interests.
Footnotes
Peer review information Nature Immunology thanks the anonymous reviewers for their contribution to the peer review of this work. Primary Handling editor: Zoltan Fehervari, in collaboration with the Nature Immunology team.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
A list of authors and their affiliations appears at the end of the paper.
Change history
11/24/2021
A Correction to this paper has been published: 10.1038/s41590-021-01096-9
Contributor Information
Evangelos Andreakos, Email: vandreakos@bioacademy.gr.
András N. Spaan, Email: aspaan@rockefeller.edu
COVID Human Genetic Effort:
Paul Bastard, Catherine M. Biggs, Benedetta Bigio, Bertrand Boisson, Alexandre Bolze, Anastasiia Bondarenko, Petter Brodin, Samya Chakravorty, John Christodoulou, Aurelié Cobat, Antonio Condino-Neto, Stefan N. Constantinescu, Hagit Baris Feldman, Jacques Fellay, Carlos Flores, Rabih Halwani, Emmanuelle Jouanguy, Yu-Lung Lau, Isabelle Meyts, Trine H. Mogensen, Satoshi Okada, Keisuke Okamoto, Tayfun Ozcelik, Qiang Pan-Hammarström, Rebeca Pérez de Diego, Anna M. Planas, Anne Puel, Lluis Quintana-Murci, Laurent Renia, Igor Resnick, Anna Sediva, Anna Shcherbina, Ondrej Slaby, Ivan Tancevski, Stuart E. Turvey, K. M. Furkan Uddin, Diederik van de Beek, Mayana Zatz, Pawel Zawadzki, and Shen-Ying Zhang
References
- 1.Haldane JBS. Disease and evolution. Ric. Sci. 1949;19:66–76. [Google Scholar]
- 2.Quintana-Murci L. Human immunology through the lens of evolutionary genetics. Cell. 2019;177:184–199. doi: 10.1016/j.cell.2019.02.033. [DOI] [PubMed] [Google Scholar]
- 3.Casanova JL, Abel L. Inborn errors of immunity to infection: the rule rather than the exception. J. Exp. Med. 2005;202:197–201. doi: 10.1084/jem.20050854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Morens DM, Fauci AS. Emerging pandemic diseases: how we got to COVID-19. Cell. 2020;182:1077–1092. doi: 10.1016/j.cell.2020.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Enard D, Cai L, Gwennap C, Petrov DA. Viruses are a dominant driver of protein adaptation in mammals. Elife. 2016;5:e12469. doi: 10.7554/eLife.12469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Enard D, Petrov DA. Evidence that RNA viruses drove adaptive introgression between Neanderthals and modern humans. Cell. 2018;175:360–371.e313. doi: 10.1016/j.cell.2018.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Souilmi Y, et al. An ancient viral epidemic involving host coronavirus interacting genes more than 20,000 years ago in East Asia. Curr. Biol. 2021;31:3504–3514. doi: 10.1016/j.cub.2021.05.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Casanova JL, Abel L. Lethal infectious diseases as inborn errors of immunity: toward a synthesis of the germ and genetic theories. Annu Rev. Pathol. 2021;16:23–50. doi: 10.1146/annurev-pathol-031920-101429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gandhi RT, Lynch JB, del Rio C. Mild or moderate COVID-19. N. Engl. J. Med. 2020;383:1757–1766. doi: 10.1056/NEJMcp2009249. [DOI] [PubMed] [Google Scholar]
- 10.Lurie MB, Abramson S, Heppleston AG. On the response of genetically resistant and susceptible rabbits to the quantitative inhalation of human type tubercle bacilli and the nature of resistance to tuberculosis. J. Exp. Med. 1952;95:119–134. doi: 10.1084/jem.95.2.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Casanova JL, Su HC. A global effort to define the human genetics of protective immunity to SARS-CoV-2 infection. Cell. 2020;181:1194–1199. doi: 10.1016/j.cell.2020.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bastard P, et al. Autoantibodies against type I IFNs in patients with life-threatening COVID-19. Science. 2020;370:eabd4585. doi: 10.1126/science.abd4585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang Q, et al. Inborn errors of type I IFN immunity in patients with life-threatening COVID-19. Science. 2020;370:eabd4570. doi: 10.1126/science.abd4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cerami, C. et al. Household transmission of SARS-CoV-2 in the United States: living density, viral load, and disproportionate impact on communities of color. Clin. Infect. Dis.10.1093/cid/ciab701 (2021). [DOI] [PMC free article] [PubMed]
- 15.Madewell ZJ, Yang Y, Longini IM, Jr., Halloran ME, Dean NE. Household transmission of SARS-CoV-2: a systematic review and meta-analysis. JAMA Netw. Open. 2020;3:e2031756. doi: 10.1001/jamanetworkopen.2020.31756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reukers, D. F. M. et al. High infection secondary attack rates of SARS-CoV-2 in Dutch households revealed by dense sampling. 10.1093/cid/ciab237 (2021). [DOI] [PMC free article] [PubMed]
- 17.Kerner G, et al. Homozygosity for TYK2 P1104A underlies tuberculosis in about 1% of patients in a cohort of European ancestry. Proc. Natl Acad. Sci. USA. 2019;116:10430–10434. doi: 10.1073/pnas.1903561116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Abel L, et al. Genetics of human susceptibility to active and latent tuberculosis: present knowledge and future perspectives. Lancet Infect. Dis. 2018;18:e64–e75. doi: 10.1016/S1473-3099(17)30623-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Boisson-Dupuis S. The monogenic basis of human tuberculosis. Hum. Genet. 2020;139:1001–1009. doi: 10.1007/s00439-020-02126-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Altare F, et al. Interleukin-12 receptor β1 deficiency in a patient with abdominal tuberculosis. J. Infect. Dis. 2001;184:231–236. doi: 10.1086/321999. [DOI] [PubMed] [Google Scholar]
- 21.Boisson-Dupuis S, et al. IL-12Rβ1 deficiency in two of fifty children with severe tuberculosis from Iran, Morocco, and Turkey. PLoS ONE. 2011;6:e18524. doi: 10.1371/journal.pone.0018524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kreins AY, et al. Human TYK2 deficiency: mycobacterial and viral infections without hyper-IgE syndrome. J. Exp. Med. 2015;212:1641–1662. doi: 10.1084/jem.20140280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boisson-Dupuis S, et al. Tuberculosis and impaired IL-23-dependent IFN-γ immunity in humans homozygous for a common TYK2 missense variant. Sci. Immunol. 2018;3:eaau8714. doi: 10.1126/sciimmunol.aau8714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kerner G, et al. Human ancient DNA analyses reveal the high burden of tuberculosis in Europeans over the last 2,000 years. Am. J. Hum. Genet. 2021;108:517–524. doi: 10.1016/j.ajhg.2021.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.de Prost N, et al. Plasma exchange to rescue patients with autoantibodies against type I interferons and life-threatening COVID-19 pneumonia. J. Clin. Immunol. 2021;41:536–544. doi: 10.1007/s10875-021-00994-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Koning R, et al. Autoantibodies against type I interferons are associated with multi-organ failure in COVID-19 patients. Intensive Care Med. 2021;47:704–706. doi: 10.1007/s00134-021-06392-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Troya J, et al. Neutralizing autoantibodies to type I IFNs in >10% of patients with severe COVID-19 pneumonia hospitalized in Madrid, Spain. J. Clin. Immunol. 2021;41:914–922. doi: 10.1007/s10875-021-01036-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van der Wijst, M. G. P. et al. Type I interferon autoantibodies are associated with systemic immune alterations in patients with COVID-19. Sci Transl Med, eabh2624 (2021). [DOI] [PMC free article] [PubMed]
- 29.Vazquez SE, et al. Neutralizing autoantibodies to type I interferons in COVID-19 convalescent donor plasma. J. Clin. Immunol. 2021;41:1169–1171. doi: 10.1007/s10875-021-01060-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bastard P, et al. Preexisting autoantibodies to type I IFNs underlie critical COVID-19 pneumonia in patients with APS-1. J. Exp. Med. 2021;218:e2021055. doi: 10.1084/jem.20210554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bastard, P. et al. Autoantibodies neutralizing type I IFNs are present in ~4% of uninfected individuals over 70 years old and account for ~20% of COVID-19 deaths. Sci. Immunol.10.1126/sciimmunol.abl4340 (2021). [DOI] [PMC free article] [PubMed]
- 32.Asano, T. et al. X-linked recessive TLR7 deficiency in ~1% of men under 60 years old with life-threatening COVID-19. Sci. Immunol.10.1126/sciimmunol.abl4348 (2021). [DOI] [PMC free article] [PubMed]
- 33.Galani IE, et al. Untuned antiviral immunity in COVID-19 revealed by temporal type I/III interferon patterns and flu comparison. Nat. Immunol. 2021;22:32–40. doi: 10.1038/s41590-020-00840-x. [DOI] [PubMed] [Google Scholar]
- 34.Bastard P, et al. Auto-antibodies to type I IFNs can underlie adverse reactions to yellow fever live attenuated vaccine. J. Exp. Med. 2021;218:e20202486. doi: 10.1084/jem.20202486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Allison AC. Protection afforded by sickle-cell trait against subtertian malareal infection. Br. Med. J. 1954;1:290–294. doi: 10.1136/bmj.1.4857.290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Migueles SA, et al. HLA B*5701 is highly associated with restriction of virus replication in a subgroup of HIV-infected long term nonprogressors. Proc. Natl Acad. Sci. USA. 2000;97:2709–2714. doi: 10.1073/pnas.050567397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ge D, et al. Genetic variation in IL28B predicts hepatitis C treatment-induced viral clearance. Nature. 2009;461:399–401. doi: 10.1038/nature08309. [DOI] [PubMed] [Google Scholar]
- 38.Thomas DL, et al. Genetic variation in IL28B and spontaneous clearance of hepatitis C virus. Nature. 2009;461:798–801. doi: 10.1038/nature08463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Miller LH, Mason SJ, Dvorak JA, McGinniss MH, Rothman IK. Erythrocyte receptors for (Plasmodium knowlesi) malaria: Duffy blood group determinants. Science. 1975;189:561–563. doi: 10.1126/science.1145213. [DOI] [PubMed] [Google Scholar]
- 40.Miller LH, Mason SJ, Clyde DF, McGinniss MH. The resistance factor to Plasmodium vivax in blacks. The Duffy-blood-group genotype, FyFy. N. Engl. J. Med. 1976;295:302–304. doi: 10.1056/NEJM197608052950602. [DOI] [PubMed] [Google Scholar]
- 41.Tournamille C, Colin Y, Cartron JP, Le Van Kim C. Disruption of a GATA motif in the Duffy gene promoter abolishes erythroid gene expression in Duffy-negative individuals. Nat. Genet. 1995;10:224–228. doi: 10.1038/ng0695-224. [DOI] [PubMed] [Google Scholar]
- 42.Dean M, et al. Genetic restriction of HIV-1 infection and progression to AIDS by a deletion allele of the CKR5 structural gene. Hemophilia Growth and Development Study, Multicenter AIDS Cohort Study, Multicenter Hemophilia Cohort Study, San Francisco City Cohort, ALIVE Study. Science. 1996;273:1856–1862. doi: 10.1126/science.273.5283.1856. [DOI] [PubMed] [Google Scholar]
- 43.Liu R, et al. Homozygous defect in HIV-1 coreceptor accounts for resistance of some multiply-exposed individuals to HIV-1 infection. Cell. 1996;86:367–377. doi: 10.1016/S0092-8674(00)80110-5. [DOI] [PubMed] [Google Scholar]
- 44.Samson M, et al. Resistance to HIV-1 infection in caucasian individuals bearing mutant alleles of the CCR-5 chemokine receptor gene. Nature. 1996;382:722–725. doi: 10.1038/382722a0. [DOI] [PubMed] [Google Scholar]
- 45.Lindesmith L, et al. Human susceptibility and resistance to Norwalk virus infection. Nat. Med. 2003;9:548–553. doi: 10.1038/nm860. [DOI] [PubMed] [Google Scholar]
- 46.Rausell A, et al. Common homozygosity for predicted loss-of-function variants reveals both redundant and advantageous effects of dispensable human genes. Proc. Natl Acad. Sci. USA. 2020;117:13626–13636. doi: 10.1073/pnas.1917993117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shelton JF, et al. Trans-ancestry analysis reveals genetic and nongenetic associations with COVID-19 susceptibility and severity. Nat. Genet. 2021;53:801–808. doi: 10.1038/s41588-021-00854-7. [DOI] [PubMed] [Google Scholar]
- 48.Ellinghaus D, et al. Genomewide association study of severe Covid-19 with respiratory failure. N. Engl. J. Med. 2020;383:1522–1534. doi: 10.1056/NEJMoa2020283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.COVID-19 Host Genetics Initiative. Mapping the human genetic architecture of COVID-19. 10.1038/s41586-021-03767-x (2021). [DOI] [PMC free article] [PubMed]
- 50.Zhang Y, Garner R, Salehi S, La Rocca M, Duncan D. Association between ABO blood types and coronavirus disease 2019 (COVID-19), genetic associations, and underlying molecular mechanisms: a literature review of 23 studies. Ann. Hematol. 2021;100:1123–1132. doi: 10.1007/s00277-021-04489-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Freeman EE, et al. Pernio-like skin lesions associated with COVID-19: a case series of 318 patients from 8 countries. J. Am. Acad. Dermatol. 2020;83:486–492. doi: 10.1016/j.jaad.2020.05.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tan SW, Tam YC, Oh CC. Skin manifestations of COVID-19: a worldwide review. JAAD Int. 2021;2:119–133. doi: 10.1016/j.jdin.2020.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Crow YJ, Manel N. Aicardi–Goutières syndrome and the type I interferonopathies. Nat. Rev. Immunol. 2015;15:429–440. doi: 10.1038/nri3850. [DOI] [PubMed] [Google Scholar]
- 54.Colmenero I, et al. SARS-CoV-2 endothelial infection causes COVID-19 chilblains: histopathological, immunohistochemical and ultrastructural study of seven paediatric cases. Br. J. Dermatol. 2020;183:729–737. doi: 10.1111/bjd.19327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hoffmann M, et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wei J, et al. Genome-wide CRISPR screens reveal host factors critical for SARS-CoV-2 infection. Cell. 2021;184:76–91. doi: 10.1016/j.cell.2020.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang R, et al. Genetic screens identify host factors for SARS-CoV-2 and common cold coronaviruses. Cell. 2021;184:106–119. doi: 10.1016/j.cell.2020.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Daniloski Z, et al. Identification of required host factors for SARS-CoV-2 infection in human. Cells Cell. 2021;184:92–105. doi: 10.1016/j.cell.2020.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Horowitz, J. E. et al. Genome-wide analysis in 756,646 individuals provides first genetic evidence that ACE2 expression influences COVID-19 risk and yields genetic risk scores predictive of severe disease. Preprint at medRxiv 10.1101/2020.12.14.20248176 (2021).
- 60.Suryamohan K, et al. Human ACE2 receptor polymorphisms and altered susceptibility to SARS-CoV-2. Commun. Biol. 2021;4:475. doi: 10.1038/s42003-021-02030-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schneider WM, et al. Genome-scale identification of SARS-CoV-2 and pan-coronavirus host factor networks. Cell. 2021;184:120–132. doi: 10.1016/j.cell.2020.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hoffmann HH, et al. TMEM41B is a pan-flavivirus host factor. Cell. 2021;184:133–148.e120. doi: 10.1016/j.cell.2020.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gordon DE, et al. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020;583:459–468. doi: 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gordon DE, et al. Comparative host–coronavirus protein interaction networks reveal pan-viral disease mechanisms. Science. 2020;370:eabe9403. doi: 10.1126/science.abe9403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hoffmann HH, et al. Functional interrogation of a SARS-CoV-2 host protein interactome identifies unique and shared coronavirus host factors. Cell Host Microbe. 2021;29:267–280. doi: 10.1016/j.chom.2020.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schwarzkopf, S. et al. Cellular immunity in COVID-19 convalescents with PCR-confirmed infection but with undetectable SARS-CoV-2-specific IgG. 10.3201/2701.203772 2021). [DOI] [PubMed]
- 67.Sekine T, et al. Robust T cell immunity in convalescent individuals with asymptomatic or mild COVID-19. Cell. 2020;183:158–168. doi: 10.1016/j.cell.2020.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sette A, Crotty S. Adaptive immunity to SARS-CoV-2 and COVID-19. Cell. 2021;184:861–880. doi: 10.1016/j.cell.2021.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mateus J, et al. Selective and cross-reactive SARS-CoV-2 T cell epitopes in unexposed humans. Science. 2020;370:89–94. doi: 10.1126/science.abd3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Braun J, et al. SARS-CoV-2-reactive T cells in healthy donors and patients with COVID-19. Nature. 2020;587:270–274. doi: 10.1038/s41586-020-2598-9. [DOI] [PubMed] [Google Scholar]
- 71.Le Bert N, et al. SARS-CoV-2-specific T cell immunity in cases of COVID-19 and SARS, and uninfected controls. Nature. 2020;584:457–462. doi: 10.1038/s41586-020-2550-z. [DOI] [PubMed] [Google Scholar]
- 72.Li Q, et al. Early transmission dynamics in Wuhan, China, of novel coronavirus-infected pneumonia. N. Engl. J. Med. 2020;382:1199–1207. doi: 10.1056/NEJMoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wu JT, Leung K, Leung GM. Nowcasting and forecasting the potential domestic and international spread of the 2019-nCoV outbreak originating in Wuhan, China: a modelling study. Lancet. 2020;395:689–697. doi: 10.1016/S0140-6736(20)30260-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sanche S, et al. High contagiousness and rapid spread of severe acute respiratory syndrome coronavirus 2. Emerg. Infect. Dis. 2020;26:1470–1477. doi: 10.3201/eid2607.200282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Al-Tawfiq JA, Rodriguez-Morales AJ. Super-spreading events and contribution to transmission of MERS, SARS, and SARS-CoV-2 (COVID-19) J. Hosp. Infect. 2020;105:111–112. doi: 10.1016/j.jhin.2020.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.WHO Working Group on the Clinical Characterisation and Management of COVID-19 infection. A minimal common outcome measure set for COVID-19 clinical research. Lancet Infect. Dis.20, e192–e197 (2020). [DOI] [PMC free article] [PubMed]
- 77.Sancho-Shimizu V, et al. SARS-CoV-2-related MIS-C: A key to the viral and genetic causes of Kawasaki disease? J. Exp. Med. 2021;218:e20210446. doi: 10.1084/jem.20210446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Meyts I, et al. Exome and genome sequencing for inborn errors of immunity. J. Allergy Clin. Immunol. 2016;138:957–969. doi: 10.1016/j.jaci.2016.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Thompson, M. A. et al. Primary care guidance for persons with human immunodeficiency virus: 2020 update by the HIV medicine association of the Infectious Diseases Society of America. 10.1093/cid/ciaa1391 (2020). [DOI] [PubMed]
- 80.Hütter G, et al. Long-term control of HIV by CCR5 Δ32/Δ32 stem-cell transplantation. N. Engl. J. Med. 2009;360:692–698. doi: 10.1056/NEJMoa0802905. [DOI] [PubMed] [Google Scholar]
- 81.Gupta RK, et al. HIV-1 remission following CCR5Δ32/Δ32 haematopoietic stem-cell transplantation. Nature. 2019;568:244–248. doi: 10.1038/s41586-019-1027-4. [DOI] [PMC free article] [PubMed] [Google Scholar]


