Abstract
CRISPR and Cas proteins, often referred to as CRISPR/Cas, are the components of a bacterial genome editing system that can be used to perturb genes in cells and tissues. A classic application is to use CRISPR/Cas to generate genetic loss-of-function. When performed at large scale and combined with deep sequencing techniques, CRISPR-based perturbations can be performed in a high throughput setting to screen many candidate genomic elements for their roles in a phenotype of interest. Here, we discuss major considerations in the design, execution, and analysis of CRISPR screens. We focus on CRISPR knockout screens but also review adaptations to the CRISPR/Cas system that highlight the versatility of the system to make other types of experimental genetic changes as well. We also discuss examples of CRISPR genetic screens in investigative dermatology and how they may be used to answer key scientific questions in the field.
INTRODUCTION
The human genome contains over 20,000 protein-coding genes, and their disruption underlies many diseases, underscoring the need to comprehensively understand their biological functions. One approach to studying gene functions and biological phenotypes is to perform a genetic screen, which aims for systematic functional interrogation of many candidate elements in a single experiment (Doench, 2018; Ford et al., 2019; Sanjana, 2017; Schuster et al., 2019). In a typical cell culture–based screen, systematic loss-of-function of a set of candidates is applied to identify elements contributing to a phenotype of interest. RNA interference (RNAi) and transposon-based technologies have been used successfully, but since their development, CRISPR/Cas-based tools have become a preferred method for genetic screens (Doench, 2018; Ford et al., 2019; Guitart et al., 2016; Schuster et al., 2019).
GENE KNOCKOUT USING CRISPR IN POOLED HIGH-THROUGHPUT SCREENS
CRISPR-based screens demonstrate improved versatility, efficacy, and lower off-target effects compared with approaches such as RNAi (Ford et al., 2019; Guitart et al., 2016; Schuster et al., 2019). For a comprehensive description of the fundamentals of CRISPR-mediated genome editing, we refer to a previous Research Techniques Made Simple article (Guitart et al., 2016). In brief, the bacterial Cas enzyme (usually Cas9) is guided to a genomic DNA target by a single guide RNA (sgRNA), an approximately 20-nucleotide sequence that specifies the genomic target, such as a protein-coding gene. Once present at the target, Cas9 catalyzes a double-strand DNA (dsDNA) break. Cells repair the dsDNA break, most commonly by nonhomologous end joining (NHEJ). Because NHEJ is error-prone, small insertions or deletions (indels) are introduced during repair, which leads to frameshifts and/or a premature stop codon at the target that result in loss-of-function. The classical CRISPR-based nuclease approach is therefore also referred to as CRISPR knockout (CRISPR-ko).
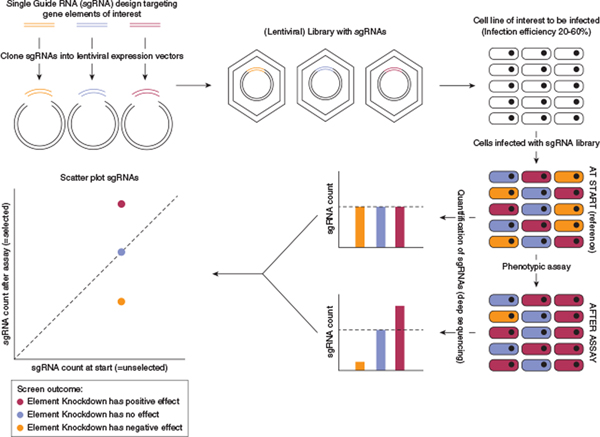
By using one sgRNA, CRISPR-ko of a single target can be achieved. However, by using multiple sgRNAs designed to target distinct genes, the investigator can generate a sgRNA library, which allows high-throughput genetic screens (Figure 1). Construction of a library containing sgRNAs against all protein-coding genes enables a genome-wide screen, whereas a smaller library containing sgRNAs against preselected genes enables targeted assessment of a specific gene set. During a CRISPR screen (Figure 1), the sgRNA library is introduced into a cell population in a manner such that each cell receives only one sgRNA. As a result, each cell within the bulk population undergoes a single knockout event, but the targeted elements differ between cells. After subjecting the CRISPR-ko cells to assays that enable positive or negative selection for a phenotype of interest, the effect of a gene knockout can be quantitated by assessing the relative enrichment or depletion of the causative sgRNA compared with its abundance in the starting population (Figures 1 and 2).
Figure 1. Overview of the key steps during a typical CRISPR-ko screen.
CRISPR sgRNA libraries can be ordered commercially, obtained through public repositories (e.g., Addgene), or custom-designed. For custom CRISPR libraries, pooled sgRNAs can be ordered as a DNA oligonucleotide pool. After PCR amplification, the library is cloned into a delivery vector (e.g., a lentiviral vector). Following packaging into lentiviral particles, the CRISPR library is infected into target cells at an infection efficiency of 20–60% to maximize the percentage of cells transduced with a single sgRNA. After infection, cells are selected with an antibiotic to deplete noninfected cells. A subset of cells is then collected as reference (at start, reference and/or unselected). An assay is then applied to select for cells displaying a desired phenotype, and cells are harvested at the endpoint and optionally at intermediate timepoints. Genomic DNA of both reference and endpoint cells is isolated and primers flanking the sgRNAs are deep sequenced from bulk DNA to measure the abundance of each sgRNA (sgRNA count) at each timepoint. sgRNA abundances can be visualized by plotting the sgRNA counts pre- and post-screen. Negative control sgRNAs (without biological targets) should appear around the dotted line, representing no change. CRISPR-ko, CRISPR knockout; sgRNA, single guide RNA.
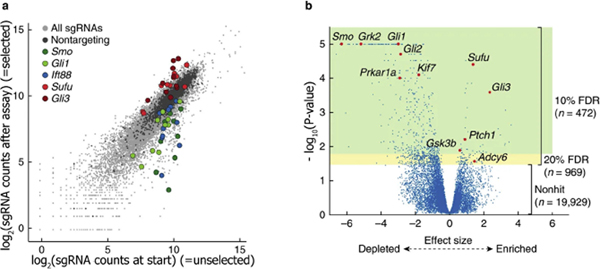
Figure 2. Example of output of a published CRISPR-ko screen.
(a) Scatter plot of a CRISPR screen aiming to systematically identify genes essential for Hedgehog signaling (Breslow et al., 2018). A transcriptional reporter assay allowed selection of cells in which the Hedgehog signaling pathway is active. The plot shows the abundance of sgRNAs (10 per target) at the start of the screen (x-axis; reference population) and after selection for cells with an active Hedgehog signaling pathway (y-axis). sgRNAs targeting selected genes are highlighted, similar colors indicate different sgRNAs targeting the same gene. (b) Volcano plot showing the effect size (x-axis) and P-values (y-axis) as calculated by the Cas9 high throughput likelihood estimator (casTLE) algorithm for this screen (Breslow et al., 2018). Select Hedgehog signaling pathway components are highlighted. Genes with P-value cut-offs corresponding to 10% FDR are highlighted in green, and those corresponding to a 20% FDR are in yellow. FDR, false discovery rate; sgRNA, single guide RNA. Reprinted with permission from Springer-Nature.
Here, we provide an overview of the key steps in designing a CRISPR-ko screen. Furthermore, we review adaptations to the CRISPR/Cas system that extend the genomic targets that can be studied, discuss examples of CRISPR genetic screens in investigative dermatology, and provide examples of prominent CRISPR screens from other research fields.
Overview of the methodology
The design of a pooled genomic CRISPR-ko screen (Figure 1) is characterized by four key steps (Ford et al., 2019), each with specific considerations (Table 1) influencing practical execution and screen results.
Table 1.
Key Considerations When Performing a CRISPR-ko Screen
| Screen Phase | Considerations | Advantages/Disadvantages |
|---|---|---|
|
I. Targets What genes will be studied? |
Choice of library | • Genome-scale libraries have the benefit of being comprehensive but can be resource-intensive. Predesigned genome-wide CRISPR libraries are available • Targeted libraries focus on class of elements (kinases, transcription factors, etc.) or can be custom-designed and generated by the investigator |
|
II. Model What cells should be used? |
Cas9 expression | • Primary cells require delivery of both Cas9 and sgRNA, which may be technically challenging • Stably expressing Cas9 cell lines provide uniform high expression of Cas9 |
|
III. Assay How are cells screened? |
Phenotype | • A classical screen results in positive or negative selection from a selective pressure (e.g., exposure to a drug) • Combining CRISPR screens with other genetic tools such as reporter cell lines can facilitate screening for diverse phenotypes |
|
IV. Analysis How are screen results evaluated? |
Measuring screen outputs | • Changes in sgRNA abundance, measured by next-generation sequencing, are a classical output of CRISPR-ko screens • A variety of validated CRISPR screen analysis pipelines are publicly available |
Abbreviations: CRISPR-ko, CRISPR knockout; sgRNA, single guide RNA.
Gene set to study and sgRNA library design
A first step in designing a CRISPR screen is to define the set of genes to study. The number of elements included in the screen determine the size, complexity, and cost of the experiment. A genome-wide CRISPR screen has the advantage of being comprehensive and avoids pretest selection bias. In addition, several validated genome-wide CRISPR sgRNA libraries are publicly available and can save the investigator from the task of designing and building their own library. Addgene is a nonprofit repository that distributes predesigned CRISPR libraries (www.addgene.org/crispr/libraries/).
Two prominent examples of genome-wide CRISPR-ko libraries are the Genome-scale CRISPR knockout (GeCKO) (Sanjana et al., 2014; Shalem et al., 2014) and Brunello (Doench et al., 2016) libraries, which both target all protein-coding genes in the human genome. These libraries can be ordered as pooled plasmids or directly as ready-to-use lentiviral particles. Within investigative dermatology, a genome-wide CRISPR-ko screen using the GeCKO library was used to identify genes whose loss is involved in resistance to the therapeutic cancer drug vemurafenib (Shalem et al., 2014).
A genome-wide screen can be both labor- and resource-intensive, and a more focused screen may be appropriate for scientific objectives where reasonable filters can be applied to narrow down the screening candidate list. For instance, RNA sequencing data can be used to identify only the set of genes that are expressed in the condition or cell type of interest. Another approach is to focus on a certain class of elements, such as transcription factors, kinases, or RNA binding proteins (Doench, 2018). Each of these approaches results in a more directed and manageable screening strategy but may require a custom-designed CRISPR library. For example, a targeted CRISPR library was used to screen for kinases that have a role in IL-17–mediated inflammatory signaling in primary keratinocytes (Slivka et al., 2019).
The overall sgRNA library size is principally determined by the number of candidate genes and the number of sgRNAs per target. Each sgRNA varies in its knockout effectiveness and target specificity. If multiple different sgRNAs targeting the same gene lead to consistent outcomes, the confidence of the finding increases. Therefore, including multiple sgRNAs per target improves the sensitivity and specificity of a CRISPR-ko screen (Doench et al., 2016). Predesigned and validated genome-wide human and mouse CRISPR libraries typically include >3–4 independent sgRNAs per gene (Doench, 2018). Online tools can assist in the design of effective sgRNAs (Table 1 in Doench, 2018), or the investigator can select specific sgRNAs from previously designed libraries. Additionally, the sgRNA library should contain negative and positive controls. Negative controls are typically nontargeting sgRNAs whose sequences do not match any sites in the genome. These nontargeting sgRNAs can be used to assess neutral variations in sgRNA abundance in the screen (Figure 2). Positive controls are sgRNAs that target essential (housekeeping) genes such as ribosomal or proteasomal subunits. These positive controls should be depleted in CRISPR-ko screens and serve as benchmarks to judge the confidence of the screen. The library can be synthesized by commercial vendors as an oligonucleotide pool and cloned into target vectors (e.g., lentiviral plasmids).
Cells of interest and CRISPR library delivery
CRISPR screens can be performed in primary cells, such as keratinocytes, melanocytes, and fibroblasts (Fenini et al., 2018; Slivka et al., 2019; Sun et al., 2015). However, primary cells can be difficult to transfect, generate lower gene-editing efficiency, or may have cell limitations that are incompatible with long-term library screens (Ford et al., 2019). For these reasons, use of transformed or immortalized cell lines, such as 293T or HeLa cells, are sometimes favored for their technical tractability.
CRISPR-based genome editing requires two components, the Cas9 protein and the sgRNA, which contains both a scaffold and a target-specific spacer sequence. There are several options to deliver these components into cells, each with their specific advantages and disadvantages (Table 2 in Ford et al., 2019). Some delivery methods can give rise to undesired effects, such as cytotoxicity or innate immunity responses (Kim et al., 2018). Therefore, choice and optimization of the preferred delivery method should be evaluated for the cell type of choice.
For many cell types, the preferred delivery is by utilizing lentivirus, which can stably integrate into the genome of the host and express Cas9 and RNA components. One important technical consideration is to determine how Cas9 is introduced into cells. Simultaneous delivery of both Cas9 and sgRNAs is a simple, one-step approach but can create variability in Cas9 protein expression among cells. Variability of Cas9 protein levels affects CRISPR-ko efficiency. An alternative approach is to establish or purchase cell lines stably expressing Cas9, such as from ATCC (www.atcc.org).
Most library vectors include antibiotic resistance and/or fluorescence markers, allowing for selection of successfully infected cells. When using lentivirus to deliver the sgRNA library, viral titers and infection efficiencies should be determined. CRISPR libraries should be infected at low infection efficiencies to maximize the number of cells receiving a single sgRNA. This results in a single perturbation event per cell, an assumption that underlies accurate screening analysis and identification of candidates (Figure 1). In general, infection efficiencies of 20–60% are recommended (Doench, 2018; Ford et al., 2019).
It is essential to determine the total number of cells needed to perform the CRISPR screen. A 1× representation indicates that the number of cells infected matches the number of sgRNAs. For a reliable CRISPR screen, sgRNA representation of 300–1000× assures that all screening sgRNAs are present in the cell population (Doench, 2018; Ford et al., 2019; Schuster et al., 2019; Yau and Rana, 2018). A CRISPR screen with 50,000 sgRNAs and an infection efficiency of 40% requires a starting number of 125,000 cells to achieve approximately 1× representation after selection (125,000 × 0.40 = 50,000) and 125 million cells for 1,000× representation. To maintain library representation, >50 million cells need to be propagated throughout the screen. Factoring in biological and technical replicates, it is easy to envision how a screen can become an intensive effort. These prescreen planning steps should be performed to accurately estimate the resources (lentivirus, cells, culture space, plasticware, culture media, etc.) that will be needed.
Choosing the phenotype assay
The next step in a CRISPR screen is to choose or design an assay that provides a basis for positive or negative selection of cells in the screened population. As every sgRNA inflicts a genetic perturbation, the response of each genetic perturbation occurs within the bulk population of cells, and the effects are ultimately identified by changes in sgRNA abundance (Figure 1).
Most CRISPR screens are combined with assays that exert a selective stress on cell fitness. Cells with lower fitness decrease in abundance. Relative differences in cell fitness can be accelerated by applying the desired selective pressure, such as a drug or UVR. Although common, CRISPR screens are not just limited to cell fitness assays. Other groups have conducted CRISPR screens on cells engineered with a fluorescence reporter that activates upon triggering a desired phenotype (e.g., expression of cytokines) and combined their screen with FACS. Selection for a desired phenotype might require developing a novel assay. For instance, to select for cells with an active Hedgehog signaling pathway, researchers created an assay in which active Hedgehog signaling confers resistance to the antibiotic blasticidin, which allowed for their selection (Figure 2 in [Breslow et al., 2018]). Such a FACS-based phenotypic assay has also been applied in a CRISPR-ko screen in primary keratinocytes to screen for kinases affecting the expression of the cytokine IL-8 (Slivka et al., 2019).
Measuring and quantifying screen output
In the case of lentiviral-based screening, the sgRNA sequence delivered to a cell is integrated into the genome and serves as a unique identifier for that cell. sgRNAs that target genes involved in the phenotype of interest will be either enriched or depleted after the screen (Figures 1 and 2). To measure changes in sgRNA abundance, sgRNA sequences are amplified from genomic DNA isolated before and after the screen. Using primers flanking sgRNA sequences in bulk genomic DNA, deep sequencing is performed to assess sgRNA abundances (Yau and Rana, 2018).
Conceptually, a positive screen hit for a gene will result in multiple independent sgRNA abundances changing concordantly (Figures 1 and 2). A detailed review of CRISPR screen analysis is beyond the scope of this review, but many web-based and command-line analysis tools are available (Box 3 in [Schuster et al., 2019]).
LIMITATIONS, APPLICATIONS, AND FUTURE DIRECTIONS
CRISPR-ko screens can identify novel roles for genes contributing to a phenotype. However, even after a well-designed and executed screen, it is important to validate screen hits using an alternative knockdown method, such as RNAi, and/or by using complementary functional experiments. Ongoing innovations are expanding the application of CRISPR/Cas genetic screens to primary cells, tissue, and even in vivo models (Chow and Chen, 2018).
Although this review focuses on CRISPR-ko screens that classically target protein-coding genes, it is worthwhile to note that other genome elements can be studied as well. CRISPR screens have been used to study enhancers (Korkmaz et al., 2016) and microRNAs (Kurata and Lin, 2018). Additionally, the versatility of CRISPR/Cas has increased dramatically by re-engineering Cas proteins, such as the catalytically dead Cas9 protein (dCas9). By fusing dCas9 to different effector domains (Table 2), the variety of genomic elements that can be interrogated (especially those not reliably perturbed by small indels) can be expanded. As a result, CRISPR has been applied to study the role of long noncoding RNAs (Liu et al., 2017). Recently, we performed a CRISPR screen using dCas9 to identify long noncoding RNAs contributing to epidermis formation (Cai et al., 2020).
Table 2.
CRISPR Toolset for Genetic Screens
| DNA Binding Protein | DNA Cleavage? | Effector | Mechanistic Result | Assess the Role of |
|---|---|---|---|---|
| Cas9 | Yes | None | Loss-of-function (knockout) | Protein-coding genes, miRNAs, enhancers, … |
| dCas9 | No | None | Transcriptional repression | All genes |
| KRAB (CRISPR interference) | Transcriptional repression | All genes | ||
| VP16, VP64 (CRISPR activation) | Transcriptional activation | All genes |
Abbreviation: dCas9, dead Cas9.
In the future, CRISPR screens could be applied to address other questions in investigative dermatology. Can we identify novel therapeutic targets in keratinocyte cancers, melanomas, and other genetic skin diseases? What are the noncoding genomic regions that contribute to skin disease? For questions like these, CRISPR screens offer a powerful alternative way for new discoveries.
Supplementary Material
SUMMARY POINTS.
Advantages
The versatility and programmability of CRISPR/Cas genome editing enables high throughput genetic screens.
CRISPR genetic screens enable a systematic evaluation of many genetic elements in a single experiment.
The availability of predesigned CRISPR libraries provides opportunities to quick-start a CRISPR screen using prevalidated single guide RNAs.
The wide variety of CRISPR toolsets enables the study of many classes of genetic element (proteins, microRNAs, noncoding genes, and enhancers).
Limitations
CRISPR screens can be labor- and resource-intensive.
Screen readouts might require the development of an assay that allows selection for a phenotype of interest.
CRISPR screen hits need to be validated by complementary, independent functional techniques.
MULTIPLE CHOICE QUESTIONS.
- What is the main reason to perform a high throughput genetic screen, such as with CRISPR?
- It allows you to measure the effect of a gene in many different conditions.
- It provides a systematic assessment of genotype-phenotype relations in a systematic manner.
- It can be executed in every cell line because of the endogenous expression of CRISPR/Cas9 in mammalian cells.
- What is the main outcome parameter of a CRISPR-mediated genetic screen?
- Changes in gene knockout frequencies.
- Changes in lentiviral multiplicity of infection.
- Changes in single guide RNA (sgRNA) abundance.
- Which of the following parameters most likely ensures only one genetic perturbation per cell?
- An infection efficiency of 100%.
- An infection efficiency of 50%.
- An infection efficiency of 0%.
- What directly contributes most to a higher statistical certainty of a CRISPR screen output?
- Increasing the number of independent sgRNAs per target.
- Amplifying the sgRNA library to high titer.
- Decreasing the number of cells in the screen.
- Which of the following most accurately indicates a CRISPR screen hit?
- Decreased abundance of multiple sgRNAs targeting the same genomic element.
- Decreased mRNA expression of a candidate gene at the end of the screen.
- Markedly increased gain of abundance of a single sgRNA targeting a candidate.
See online version of this article for a detailed explanation of correct answers.
DETAILED ANSWERS.
-
What is the main reason to perform a high throughput genetic screen, such as with CRISPR?
Answer: B. By using libraries of different sgRNAs, CRISPR screens allow direct perturbations of a large number of genes, which are tested in a phenotypic assay.
-
What is the main outcome parameter of a CRISPR-mediated genetic screen?
Answer: C. The sgRNA sequence delivered to a cell serves as a unique identifier for that cell and the gene that is targeted.
-
Which of the following parameters most likely ensures only one genetic perturbation per cell?
Answer: B. Infection efficiencies of 20–60% are recommended, which maximizes the number of cells receiving a single sgRNA.
-
What directly contributes most to a higher statistical certainty of a CRISPR screen output?
Answer: A. True positive screen hits should have multiple, independent sgRNAs targeting the same gene change consistently, which improves screen sensitivity and specificity.
-
Which of the following most accurately indicates a CRISPR screen hit?
Answer: A. CRISPR screens are assessed by sgRNA abundances and independent sgRNAs targeting the same gene that change consistently are true screen hits.
ACKNOWLEDGMENTS
The authors gratefully acknowledge support from the National Institute of Arthritis and Musculoskeletal and Skin Diseases (R03AR075844 and K08AR067853 to BKS), the Doris Duke Charitable Foundation (2019088 to BKS), and the National Eczema Association (NEA18-RG108 to AO and BKS). The authors also greatly acknowledge Dr. Alan O’Neill and Dr. Prashant Mali for their critical evaluation and improvements to this manuscript.
Abbreviations:
- CRISPR-ko
CRISPR knockout
- dCas9
dead Cas9
- dsDNA
double-strand DNA
- GeCKO
Genome-scale CRISPR knockout
- indel
insertion or deletion
- NHEJ
nonhomologous end joining
- RNAi
RNA interference
- sgRNA
single guide RNA
Footnotes
CONFLICT OF INTEREST
The authors state no conflict of interest.
SUPPLEMENTARY MATERIAL
Supplementary material is linked to this paper. Teaching slides are available as supplementary material.
REFERENCES
- Breslow DK, Hoogendoorn S, Kopp AR, Morgens DW, Vu BK, Kennedy MC, et al. A CRISPR-based screen for Hedgehog signaling provides insights into ciliary function and ciliopathies. Nat Genet 2018;50:460–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai P, Otten ABC, Cheng B, Ishii MA, Zhang W, Huang B, et al. A genome-wide long noncoding RNA CRISPRi screen identifies PRANCR as a novel regulator of epidermal homeostasis. Genome Res 2020;30:22–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow RD, Chen S. Cancer CRISPR screens in vivo. Trends Cancer 2018;4: 349–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doench JG. Am I ready for CRISPR? A user’s guide to genetic screens. Nat Rev Genet 2018;19:67–80. [DOI] [PubMed] [Google Scholar]
- Doench JG, Fusi N, Sullender M, Hegde M, Vaimberg EW, Donovan KF, et al. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat Biotechnol 2016;34:184–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenini G, Grossi S, Contassot E, Biedermann T, Reichmann E, French LE, et al. Genome editing of human primary keratinocytes by CRISPR/Cas9 reveals an essential role of the NLRP1 inflammasome in UVB sensing. J Invest Dermatol 2018;138:2644–52. [DOI] [PubMed] [Google Scholar]
- Ford K, McDonald D, Mali P. Functional genomics via CRISPR-Cas. J Mol Biol 2019;431:48–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guitart JR Jr, Johnson JL, Chien WW. Research techniques made simple: the application of CRISPR-Cas9 and genome editing in investigative dermatology. J Invest Dermatol 2016;136:e87–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Koo T, Jee HG, Cho HY, Lee G, Lim DG, et al. Crispr RNAs trigger innate immune responses in human cells. Genome Res 2018;28: 367–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korkmaz G, Lopes R, Ugalde AP, Nevedomskaya E, Han R, Myacheva K, et al. Functional genetic screens for enhancer elements in the human genome using CRISPR-Cas9. Nat Biotechnol 2016;34:192–8. [DOI] [PubMed] [Google Scholar]
- Kurata JS, Lin RJ. MicroRNA-focused CRISPR-Cas9 library screen reveals fitness-associated miRNAs. RNA 2018;24:966–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu SJ, Horlbeck MA, Cho SW, Birk HS, Malatesta M, He D, et al. CRISPRi-based genome-scale identification of functional long noncoding RNA loci in human cells. Science 2017;355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjana NE. Genome-scale CRISPR pooled screens. Anal Biochem 2017;532: 95–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjana NE, Shalem O, Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods 2014;11:783–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster A, Erasimus H, Fritah S, Nazarov PV, van Dyck E, Niclou SP, et al. RNAi/CRISPR screens: from a pool to a valid hit. Trends Biotechnol 2019;37:38–55. [DOI] [PubMed] [Google Scholar]
- Shalem O, Sanjana NE, Hartenian E, Shi X, Scott DA, Mikkelson T, et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science 2014;343:84–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slivka PF, Hsieh CL, Lipovsky A, Pratt SD, Locklear J, Namovic MT, et al. Small molecule and pooled CRISPR screens investigating IL17 signaling identify BRD2 as a novel contributor to keratinocyte inflammatory responses. ACS Chem Biol 2019;14:857–72. [DOI] [PubMed] [Google Scholar]
- Sun BK, Boxer LD, Ransohoff JD, Siprashvili Z, Qu K, Lopez-Pajares V, et al. CALML5 is a ZNF750- and TINCR-induced protein that binds stratifin to regulate epidermal differentiation. Genes Dev 2015;29: 2225–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yau EH, Rana TM. Next-generation sequencing of genome-wide CRISPR screens. Methods Mol Biol 2018;1712:203–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.