Abstract
Hus1 is one of six checkpoint Rad proteins required for all Schizosaccharomyces pombe DNA integrity checkpoints. MYC-tagged Hus1 reveals four discrete forms. The main form, Hus1-B, participates in a protein complex with Rad9 and Rad1, consistent with reports that Rad1-Hus1 immunoprecipitation is dependent on the rad9+ locus. A small proportion of Hus1-B is intrinsically phosphorylated in undamaged cells and more becomes phosphorylated after irradiation. Hus1-B phosphorylation is not increased in cells blocked in early S phase with hydroxyurea unless exposure is prolonged. The Rad1–Rad9–Hus1-B complex is readily detectable, but upon cofractionation of soluble extracts, the majority of each protein is not present in this complex. Indirect immunofluorescence demonstrates that Hus1 is nuclear and that this localization depends on Rad17. We show that Rad17 defines a distinct protein complex in soluble extracts that is separate from Rad1, Rad9, and Hus1. However, two-hybrid interaction, in vitro association and in vivo overexpression experiments suggest a transient interaction between Rad1 and Rad17.
DNA integrity checkpoint pathways arrest progression through the cell cycle when replication is perturbed (replication checkpoint) or the DNA is damaged (DNA damage checkpoint) (23, 50). In the fission yeast Schizosaccharomyces pombe, the checkpoint Rad proteins (Rad1, Rad3, Rad9, Rad17, Rad26, and Hus1) are required for both the replication checkpoint and the DNA damage checkpoint (1, 8, 14, 40, 41). Downstream of the checkpoint Rad proteins are the Cds1 and Chk1 kinases, which directly impinge on the cell cycle machinery (5, 17, 35, 39, 52). Cds1 is activated when DNA replication is blocked by hydroxyurea (HU; which inhibits ribonucleotide reductase) or when DNA is damaged during S phase (29, 33). Chk1 is phosphorylated in response to DNA damage induced in late S or G2 (31, 48, 49). Phosphorylation of Cds1 and Chk1 is dependent on the kinase domain of Rad3, a large protein kinase with a lipid kinase motif similar to that first identified in DNA-dependent protein kinases (4, 22, 29, 31). While phosphorylation of Cds1 correlates with its activation (29), the phosphorylation of Chk1 has not been associated with increased Chk1 kinase activity.
Five of the six checkpoint Rad proteins, and both of the downstream kinases Chk1 and Cds1, are structurally and functionally conserved through evolution (Table 1). Based on the observation that null mutations in any one of the six checkpoint rad genes (but not the downstream kinases Chk1 and Cds1) result in similar loss-of-all-checkpoint phenotypes, it was suggested that Rad3 is the central element of a “guardian complex,” which can monitor stalled replication complexes and sense DNA damage (7). Biochemical data for the S. pombe, Saccharomyces cerevisiae, and Homo sapiens systems indicate that Rad1, Rad9, and Hus1 participate in a single protein complex (26, 27, 44, 47). Both Rad1 and Hus1 have been reported to be structurally related to PCNA (a DNA sliding clamp required for DNA replication and repair) (2, 46). In addition, Rad1 has a potential nuclease domain, and the purified human protein (Rad1Hs) (throughout, superscripts denote the organisms of origin: H. sapiens [Hs], S. pombe [Sp], or S. cerevisiae [Sc]) and an Ustilago maydis homolog, Rec1, have been reported to possess nuclease activity in vitro (37, 38, 45). Recent evidence indicates that the concept of a single guardian complex may be a simplification. A biochemical association has been observed between Rad3 and Rad26 (12). Rad1, Rad9, Hus1, and Rad17 are not stably associated with the Rad3-Rad26 complex (≈750 kDa) in soluble extracts (12). We show here that Rad1, Rad9, and Hus1 form a protein complex of approximately 450 kDa that is distinct from the Rad3-Rad26 complex and that a third protein complex contains Rad17. This is consistent with previous reports that upon overexpression and immunoprecipitation, the human and S. cerevisiae homologs of Rad1, Rad9, and Hus1 associated with each other but not with the Rad17 homologs (26, 47). Rad17Sp/Hs (Rad24Sc) is related to all five subunits of replication factor C (RFC) (18), a protein complex required for loading of PCNA onto DNA. The reported association of Rad24Sc (Rad17Sp) with Rfc5, a small subunit of RFC (43), suggests that Rad17 forms an RFC-like protein complex.
TABLE 1.
Checkpoint proteins of S. pombe, S. cerevisiae, and H. sapiens
| Checkpoint protein
| |||
|---|---|---|---|
| S. pombe | S. cerevisiae | H. sapiens | Sequence motif |
| Rad1 | Rad17 | Rad1 | PCNA-like, nuclease |
| Rad3 | Mec1 | ATR | Lipid kinase domain |
| Rad9 | Ddc1 | Rad9 | PCNA-like |
| Rad17 | Rad24 | Rad17 | RFC-related |
| Rad26 | ? | ? | Coiled-coiled domain |
| Hus1 | Mec3 | Hus1 | PCNA-like |
| Chk1 | Chk1 | Chk1 | Serine/threonine kinase |
| Cds1 | Rad53 | Chk2 | Serine/threonine kinase |
Since Rad26, which is associated with Rad3, is phosphorylated in response to DNA damage in the absence of the remaining checkpoint proteins (12), we have proposed that the Rad3-Rad26 complex is able to recognize damaged chromatin directly. To investigate the roles of Hus1, Rad9, and Rad1 in S. pombe, the hus1, rad1, and rad9 genes were genomically epitope tagged and antibodies were generated against Rad9 and Rad1. Hus1-MYC was found to exist in four distinct forms of 75 kDa (Hus1-A), 65 kDa (Hus1-B), 60 kDa (Hus1-C), and 35 kDa (Hus1-D), respectively. The most abundant form becomes hyperphosphorylated in a checkpoint-dependent manner in response to DNA damage and associates with Rad1 and Rad9 in a soluble protein complex. Indirect immunofluorescence microscopy reveals that Hus1 and Rad9 are nuclear and that their localization is dependent on the presence of Rad17. Two-hybrid data, in vitro binding experiments, and coimmunoprecipitation after overexpression show that Rad17 can interact with Rad1, although this interaction is not detected in soluble extracts. These data suggest that the checkpoint Rad proteins associate in different protein complexes. How they interact in vivo to generate a checkpoint signal is still not clear.
MATERIALS AND METHODS
Genetics and cell biology techniques.
Strains were constructed by standard genetic techniques (32). The protocols for checkpoint measurements, cell scoring, and irradiation are described in reference 13. Indirect immunofluorescence microscopy was performed according to a protocol previously described (19). The following changes were made: cells were fixed in 3.7% formaldehyde for 10 min in the culture medium at 29°C, the anti-MYC antibody was diluted 1:200, and fixed cells were incubated for 6 h with a 1:100 dilution of the secondary fluorescein isothiocyanate-conjugated antibody.
PCR-based gene tagging.
Using a PCR-based method, genes were C-terminally tagged in S. pombe wild-type cells (501; leu1-32 ura4-D18 ade6-704 h−) with 3 hemagglutinin (HA) or 13 MYC epitopes as previously described (3).
Two-hybrid assay.
S. cerevisiae Y190 (21) was transformed with Gal4 DNA binding domain (DB) and activation domain (AD) plasmids. Transformants were assayed for β-galactosidase activity by streaking or plating cells on selective synthetic complete medium, incubating the cells for 1 to 2 days at 30°C, and transferring them to nitrocellulose filters. After 10 s in liquid nitrogen, filters were transferred onto Whatman 3MM paper soaked with buffer Z (60 mM Na2HPO4, 40 mM NaH2PO4, 10 mM KCl, 1 mM MgSO4, 40 mM 2-mercaptoethanol) containing 1 mg of 5-bromo-4-chloro-3-indolyl-β-d-galactoside (X-Gal) per ml at 30°C. The time elapsed before appearance of blue color was recorded, and the inverse of this time was used as a measure of enzyme activity.
Total cell extracts.
Logarithmically growing cells (108) were harvested, washed in buffer A (1× phosphate-buffered saline, 50 mM NaF, 10 mM NaN3), washed in 20% (wt/vol) trichloroacetic acid (TCA), and resuspended in 200 μl of 20% TCA. After cells were broken with glass beads, 400 μl of 5% (wt/vol) TCA was added, and the cell homogenate was spun (4,000 rpm; Heraeus centrifuge) into a new test tube. Following centrifugation at 14,000 for 5 min, the supernatant was discarded and the pellet was resuspended in 200 μl of loading buffer (250 mM Tris-HCl [pH 8], 2% sodium dodecyl sulfate [SDS], 5% glycerol, 5% β-mercaptoethanol, 0.1% bromphenol blue). Boiled samples were analyzed by SDS-polyacrylamide gel electrophoresis (PAGE) (8% gel).
Liquid nitrogen extracts and size fractionation.
Logarithmically growing cells (2 × 109) were harvested, washed in buffer A, washed in buffer B (50 mM NaH2PO4-Na2HPO4 [pH 7], 150 mM NaCl, 0.1% IGEPAL CA-630 [Sigma], 10% glycerol, 0.5 mM dithiothreitol, 5 mM EGTA, 60 mM β-glycerophosphate, 0.1 mM NaF, 10 μg of aprotinin per ml, 10 μg of leupeptin per ml, 2.5 mM p-aminobenzamidine, 1.0 mM phenylmethylsulfonyl fluoride, 1 mM orthovanadate), and resuspended in 1 ml of buffer B. Cell suspension was dripped into a mortar filled with liquid nitrogen, and frozen cells were homogenized using a pestle. Liquid nitrogen was continuously added during the homogenization. Frozen cells were transferred to a test tube and allowed to thaw. The homogenate was precleared by centrifugation for 3 h at 4°C (Sorvall AH-650 rotor, 43,000 rpm, 170,000 × g). The protein concentration was measured using the Bradford assay.
A Superdex 200 HR 10/30 (Pharmacia) column was equilibrated and run in buffer C (50 mM NaH2PO4-Na2HPO4 [pH 7], 150 mM NaCl, 0.1% IGEPAL CA-630 [Sigma], 10% glycerol, 0.5 mM dithiothreitol, 5 mM EGTA, 60 mM β-glycerophosphate, 0.1 mM NaF). A sample of 200 μl (1.5 mg of protein) was injected, and 500-μl fractions were collected. The column was calibrated using a low- and high-molecular-weight gel filtration calibration kit (Pharmacia). The elution profile of the size standards was not affected by the presence of crude extract. When fraction 10 was rerun, the tagged protein peaked again at approximately 450 kDa.
Immunoprecipitation and phosphatase treatment.
Immunoprecipitation and phosphatase treatment were performed as previously described (29). Anti-Rad1 antibody G5 was affinity purified (20). When the anti-Rad1 antibody was used for Western blotting, proteins were transferred in 10 mM 3-(cyclohexylamino)-1-propanesulfonic acid (CAPS)–NaOH (pH 11) buffer. Either 0.5 mg of protein of soluble liquid nitrogen extract or 200 to 400 μl of a fraction from the gel filtration column was diluted to 0.5 ml with buffer B, antibodies prebound to protein G beads were added, and the mixture was incubated for 1 to 2 h at 4°C.
Antibodies specific for Rad9 and Rad1.
The rad9 cDNA was cloned into plasmid pGEX-4T1 (Pharmacia), and glutathione S-transferase fusion protein was expressed in Escherichia coli DH5α. The purification method is described in reference 15. Between 200 and 500 μg of soluble protein was used for immunization of rabbits. The anti-Rad9 antibody was affinity purified (20).
The rad1 cDNA was cloned into pET16b (Novagen), and the His-tagged protein was purified under denaturing conditions according to instructions. Purified protein was SDS-PAGE purified, and approximately 200 μg was used for rabbit immunization.
RESULTS
Hus1Sp exists in four distinct forms.
Previous studies have reported that Hus1 is a 33-kDa protein (28) which is phosphorylated in response to DNA damage and HU treatment in a checkpoint-dependent manner (27). Like wild-type cells, hus1-MYC cells (C-terminally tagged with 13 MYC epitopes) arrest progression through the cell cycle upon ionizing radiation and HU treatment (data not shown) (Fig. 1E). Analysis of Hus1-MYC prepared from total cell extracts revealed that hus1-MYC cells contain at least four distinct forms of Hus1: Hus1-A (75 kDa), Hus1-B (65 kDa), Hus1-C (60 kDa), and Hus1-D (35 kDa) (Fig. 1A). This was surprising, because the MYC tag was expected to reveal only one protein with a size of approximately 60 kDa (the tag adds 30 kDa). Since the extracts were prepared under denaturing conditions, it is unlikely that the smaller forms of Hus1 result from degradation in the extract.
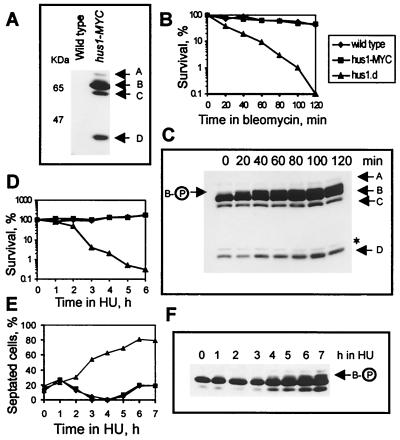
FIG. 1.
Hus1-MYC exists in four forms. (A) Total cell extracts were prepared under denaturing conditions from wild-type and hus1-MYC cells and probed with anti-MYC antibody. The arrows mark the four forms of Hus1-MYC. (B) Wild-type cells, hus1-MYC cells, and hus1.d cells were incubated for 120 min in bleomycin-containing medium (40 μg/ml), and 500 cells were plated at the indicated time points to analyze cell viability. (C) At the same time points, 108 cells were removed and total cell extracts were probed with anti-MYC antibody. The asterisk marks an unspecific band. (D) Wild-type cells, hus1-MYC cells, and hus1.d cells were incubated in the presence of 10 mM HU, and cells were plated at the indicated time points to analyze cell viability. (E) The same strains were incubated in 10 mM HU for 7 h, and samples were fixed in methanol to monitor cell cycle arrest; (F) 108 cells were collected to prepare total extracts. The arrow indicates phosphorylated Hus1-B. Checkpoint-deficient hus1.d cultures show a strong increase in septated cells, because they enter a catastrophic mitosis in the presence of HU, in which the septum cuts through the nucleus.
To further test the functionality of the hus1-MYC allele, cells were treated with bleomycin, the radiomimetic drug previously used to characterize Hus1 phosphorylation (27). While checkpoint-deficient hus1.d cells lost viability within 120 min, hus1-MYC cells behaved like wild-type cells, remaining viable during the course of the experiment (Fig. 1B). In response to DNA damage, Hus1-B was the only form of Hus1 that showed a mobility shift (Fig. 1C). Consistent with the previous report (27), the mobility shift of Hus1-B was dependent on the integrity of the checkpoint Rad proteins and could be reversed by phosphatase treatment (data not shown). In the absence of DNA damage, Hus1-B was found to run as a diffuse band on SDS-PAGE (Fig. 1A and C), suggesting a low level of intrinsic phosphorylation.
To analyze Hus1 phosphorylation in response to HU treatment, hus1-MYC cells were incubated in HU-containing medium. As seen with wild-type cells, hus1-MYC cells remained viable (Fig. 1D) and arrested progression through the cell cycle (Fig. 1E). In contrast to the previous report (27), Hus1-B was not phosphorylated after incubation for 3 h in the presence of 10 mM HU (at which point all cells are arrested in S phase). Phosphorylation of Hus1-B became evident after 4 h when cells started to pass through S phase (Fig. 1 E and F). Total extracts prepared from HU-treated cells showed an increase in the amount of all four forms of Hus1-MYC (Fig. 1F and data not shown). Taken together, these data strongly suggest that the C-terminal extension does not interfere with the checkpoint or survival function(s) of Hus1 and argue that the four forms of Hus1-MYC are unlikely to be artifacts resulting from the addition of a C-terminal epitope tag.
Hus1-B forms a protein complex with Rad1 and Rad9.
Previous studies have indicated that Hus1Sp/Hs (Mec3Sc), Rad1Sp/Hs (Rad17Sc), and Rad9Sp/Hs (Ddc1Sc) are part of a single protein complex (26, 27, 44, 47). We have used size exclusion chromatography to determine the native size of Hus1-MYC. Soluble extracts loaded onto the column contained approximately 90% of the total amount of Hus1-MYC forms, including phosphorylated Hus1-B (not shown). The majority of Hus1-B (65 kDa) eluted from the column in fractions 9 to 12 (Fig. 2A). The size marker ferritin (440 kDa) eluted in fractions 10 to 12, peaking at fraction 11. This suggests that the majority of Hus1-B is associated with a protein complex of approximately 450 kDa. However, a significant amount of Hus1-B was also found in fractions in which thyroglobulin (669 kDa) and catalase (232 kDa) eluted during calibration of the column (Fig. 2A). The low abundance of Hus1-B present in fraction 17, where the monomeric form is predicted to elute, indicates that Hus1-B exists mainly in association with other proteins. The other forms of Hus1-MYC eluted from the column with different predicted molecular weights (Fig. 3A and data not shown).
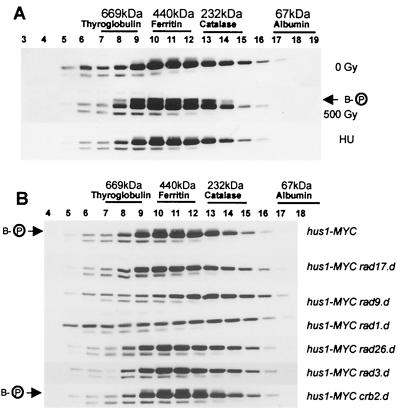
FIG. 2.
Hus1-B forms a protein complex which is sensitive to loss of Rad1 and Rad9. (A) Soluble extracts were prepared from untreated hus1-MYC cells (0 Gy) and from hus1-MYC cells irradiated with 500 Gy or treated with 10 mM HU for 3.5 h and subjected to size exclusion chromatography. (B) Soluble extracts were prepared from hus1-MYC cells deleted for rad17, rad9, rad1, rad26, rad3, or crb2/rhp9 and analyzed by size exclusion chromatography. All Western blots were probed with anti-MYC antibodies. The arrows indicate phosphorylated Hus1-B.
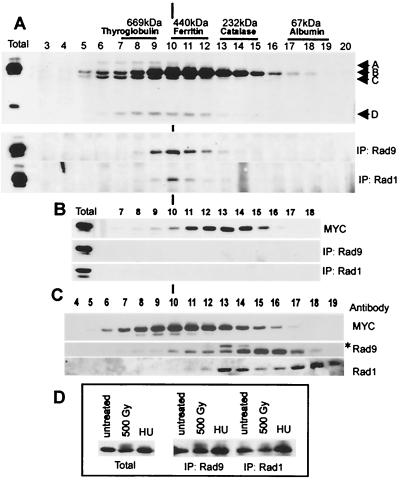
FIG. 3.
Hus1-B interacts with Rad1 and Rad9 in a rad9-dependent manner, but most of Rad1 and Rad9 do not coelute with Hus1-B. (A) A soluble protein extract prepared from untreated hus1-MYC cells was subjected to size exclusion chromatography. After removal of 50 μl for Western analysis (top; Western blot probed with anti-MYC antibodies, total = 30 μg of starting extract), half of each fraction was incubated with the Rad9-specific antibody (middle) and the other half was incubated with the Rad1-specific antibody (bottom). Precipitated Hus1-MYC was visualized using the anti-MYC antibody. Immunoprecipitations (IP) were performed in the presence of saturating antibody concentrations. The total remaining material after incubation with both antibodies was largely unchanged, indicating that both antibodies precipitate only a small proportion of Hus1-B. (B) The same experiment was performed using an extract prepared from hus1-MYC rad9.d cells (total = 30 μg of starting extract). (C) Fractions obtained from untreated hus1-MYC cells were analyzed directly using the anti-MYC antibody (top), the Rad9-specific antibody (middle), and the Rad1-specific antibody (bottom). The asterisk marks an unspecific band. (D) Soluble protein extracts were prepared from untreated hus1-MYC cells and from hus1-MYC cells irradiated with a dose of 500 Gy or arrested in 10 mM HU for 3.5 h. Left, 30 μg of total starting protein; middle, Hus1-B precipitated with the Rad9-specific antibody from 500 μg of total protein; right, Hus1-B precipitated with the Rad1-specific antibody from 500 μg of total protein. Hus1-B was visualized using the anti-MYC antibody.
Irradiation of hus1-MYC cells with 500 Gy of ionizing radiation clearly induced hyperphosphorylation of Hus1-B in fractions 8 to 14 (Fig. 2A). When cells were arrested early in S phase after an incubation of 3.5 h in 10 mM HU, no increase in Hus1-B phosphorylation above the intrinsic level was observed (Fig. 2A). Neither DNA damage nor replication arrest caused a change in the elution profile of Hus1, suggesting that the association of Hus1-B with other proteins is independent of checkpoint activation.
To analyze the involvement of the other checkpoint Rad proteins in the Hus1 complexes, extracts were prepared from hus1-MYC cells deleted for rad1, rad3, rad9, rad17, rad26, or crb2/rhp9 and subjected to size exclusion chromatography. The Rad3-Rad26 complex has previously been shown to elute in fractions 6 to 8 (12). Deletion of either Rad3 (280 kDa) or Rad26 (69 kDa) did not result in loss of any Hus1-B from fractions 6 to 8 or result in changes in any of the other fractions (Fig. 2B and data not shown). This suggests that no Hus1 is bound to the soluble Rad3-Rad26 complex. Deletion of Rad17 or Crb2/Rhp9 similarly did not change the Hus1 profiles during size exclusion. Only the absence of Rad9 and Rad1 caused a change in the elution profile but not in the overall abundance of Hus1-B. This was largely confined to fractions 9 to 12 (Fig. 2B). Interestingly, while deletion of rad9 resulted in redistribution of Hus1-B to smaller sizes, deletion of rad1 had the opposite effect, seeming to increase the intensity of Hus1-B in fractions containing larger sizes.
Intrinsically phosphorylated Hus1-B was mainly present in fractions 9 to 13 (Fig. 2B). This intrinsic phosphorylation is compromised by deletion of any of the checkpoint rad genes but not by the deletion of crb2/rhp9, which is thought to function downstream of the checkpoint Rad proteins and upstream of Chk1 (42, 51).
Most of Rad1 and Rad9 are in fractions that do not contain the Rad1–Rad9–Hus1-B complex.
Interpretation of previous results led to the assumption that Hus1, Rad1, and Rad9 define a single protein complex. We raised antibodies against Rad1 and Rad9 and studied Hus1-MYC association with Rad1 and Rad9 by coimmunoprecipitation from different fractions. Hus1-B was the only form of Hus1-MYC that was precipitated by either of the antibodies, indicating that none of the other forms bind to Rad9 or Rad1 (Fig. 3A). Anti-Rad9 sera precipitated Hus1-B mainly from fractions 9 to 13. Anti-Rad1 sera precipitated Hus1-B from the same fractions, appearing to be most efficient in precipitating from fraction 10 (Fig. 3A).
Since deletion of rad9 is known to prevent immunoprecipitation between Rad1 and Hus1, the coimmunoprecipitation experiment was repeated using anti-Rad1 sera and anti-Rad9 sera on fractions obtained from hus1-MYC rad9.d cells. As expected, both antibodies failed to precipitate Hus1-B, although fractions 10 to 15 still contained a significant amount of Hus1-B (Fig. 3B). This is in agreement with the requirement of Rad9 for the association of Hus1 with Rad1 (27). These experiments define an approximately 450-kDa Rad1 (35 kDa)–Rad9 (50 kDa)–Hus1-B (65 kDa) protein complex. The size of this complex differs from an expected trimeric size of approximately 150 kDa, arguing that the Rad1–Rad9–Hus1-B protein complex could have an oligomeric structure under these conditions or may contain as yet unidentified proteins.
Next we determined the profile of Rad9 and Rad1 proteins in the size exclusion chromatography fractions from hus1-MYC cell extracts (Fig. 3C). Rad9 eluted at a peak of approximately 150 kDa, and only a small quantity was found in the fractions (9 to 12) from which Hus1 can be precipitated with anti-Rad9 sera. Rad1 eluted as two peaks, one of approximately 200 kDa (fractions 13 and 14) and another corresponding to a molecular mass of approximately 70 kDa (fractions 17 to 19). Again only small amounts were seen in the fractions from which Hus1-MYC can be precipitated with anti-Rad1 sera. Together these data suggest that the majority of Rad9 and Hus1 exist in protein complexes which do not contain Hus1.
Rad9 interacts with phosphorylated and unphosphorylated Hus1-B.
Since fractions 9 to 12 contain phosphorylated Hus1-B, unphosphorylated Hus1-B, and the Rad1–Rad9–Hus1-B complex, Hus1-B was coimmunoprecipitated from total cell extracts prepared from untreated cells, cells irradiated with 500 Gy, and cells arrested for 3.5 h in 10 mM HU. Both Rad1 and Rad9 antibodies precipitated phosphorylated and unphosphorylated Hus1-B protein, although the Rad9-specific antibody appeared to be more efficient in precipitating the phosphorylated form (Fig. 3D). These data indicate that Rad9 and probably Rad1 bind to Hus1-B independently of its phosphorylation status.
Hus1-MYC is nuclear throughout the cell cycle, and nuclear localization requires the presence of Rad17.
The involvement of Hus1 in DNA structure-dependent checkpoints implies that Hus1 is a nuclear protein. Indirect immunofluorescence microscopy was used to investigate the cellular localization of Hus1-MYC. Incubation of hus1-MYC cells with anti-MYC-antibodies resulted in a strong nuclear signal (Fig. 4A). The presence of nuclear staining in G1/S-phase cells (binucleated cells with a septum) and in cells undergoing mitosis suggests that Hus1 is nuclear throughout the cell cycle. Neither ionizing radiation nor treatment with HU resulted in a detectable relocation of Hus1 (Fig. 4A).
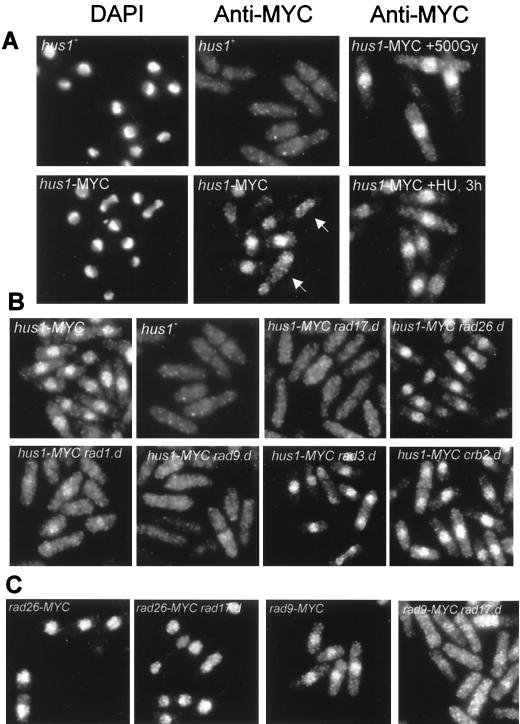
FIG. 4.
Rad17 is required for nuclear localization of Hus1 and Rad9. (A) Indirect immunofluorescence microscopy of wild-type cells (hus1+), untreated hus1-MYC cells, and hus1-MYC cells irradiated with a dose of 500 Gy or treated with HU for 3 h. 4′,6-Diamidino-2-phenylindole (DAPI) was used to stain the chromatin; anti-MYC antibodies were used to localize Hus1-MYC. The arrows mark a mitotic cell and a binucleated cell in G1/S phase. (B) Localization of Hus1-MYC in hus1-MYC cells deleted for rad1, rad3, rad9, rad17, rad26, and crb2/rhp9. (C) Localization of Rad26-MYC (left two panels) and Rad9-MYC (right two panels) in cells containing rad17+ and in cells deleted for rad17.
Rad3-dependent phosphorylation of Hus1 requires the function of Rad1, Rad9, Rad26, and Rad17 (reference 27 and data not shown). The requirement of Rad1 and Rad9 is in agreement with the finding that complex formation is a prerequisite for Hus1 phosphorylation (Fig. 2), and the requirement of Rad26 is consistent with the observation that Rad26 associates with Rad3 (12). As shown in Fig. 4B, Rad17 appears to be necessary to allow nuclear localization of Hus1. hus1-MYC rad17.d cells have lost the nuclear signal and showed a uniform staining throughout the cytosol, indicating that Rad17 is necessary to locate the Rad1–Rad9–Hus1-B complex in the nucleus. Nuclear localization of Hus1 was not affected by deletion of the rad3, rad26, or crb2/rhp9 genes. In agreement with the observation that the Hus1 complexes are disrupted in rad9.d cells (Fig. 2B), deletion of rad9 caused loss of the nuclear Hus1-MYC signal. Interestingly, hus1-MYC rad1.d cells still showed a small but significant level of nuclear hus1-MYC staining, suggesting that Rad1 association is not essential for all Hus1 to be localized to the nucleus (Fig. 4B).
To test the specificity of the Rad17 function in allowing nuclear import or retention of Hus1, we constructed rad17.d strains that carry either rad26 or rad9 C-terminally MYC tagged. Deletion of rad17 impaired the nuclear localization of Rad9-MYC but had no influence on nuclear localization of Rad26 (Fig. 4C).
Rad17 forms a third protein complex.
Rad3 and Rad26 are part of a protein complex that is distinct from the Rad1–Rad9–Hus1-B complex (12). Since none of these protein complexes appears to contain Rad17, size exclusion chromatography was used to analyze the native size of the previously characterized Rad17-MYC protein (18). As shown in Fig. 5A, Rad17-MYC eluted in fractions 9 to 13, containing proteins of approximately 450 kDa. While the Rad17-containing fractions are coincident with some of the Hus1-containing fractions, the elution pattern was not changed upon deletion of any of the other checkpoint rad genes, including hus1. This strongly implies that under these conditions, Rad17-MYC forms a protein complex distinct from the Rad3-Rad26 complex and the Rad1–Rad9–Hus1-B complex. Consistent with this interpretation, Rad9-specific antibodies and Rad26-specific antibodies failed to coimmunoprecipitate Rad17-MYC (data not shown). Rad17 elution profiles did not change in response to ionizing radiation or HU treatment.
FIG. 5.
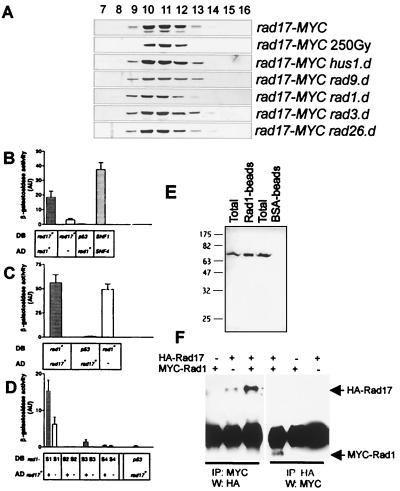
Rad17 forms a protein complex and interacts with Rad1. (A) Size exclusion chromatography of soluble extracts prepared from untreated rad17-MYC cells, rad17-MYC cells irradiated with a dose of 250 Gy, and rad17-MYC cells deleted for hus1, rad9, rad1, rad3, or rad26. Rad17-MYC was visualized using anti-MYC antibodies. (B) Two-hybrid interaction between Rad17 and Rad1. S. cerevisiae Y190 was cotransformed with a DB and an AD plasmid carrying inserts of wild-type or mutant genes or no insert, as indicated below each bar. Activity of the lacZ reporter was assayed with cell patches on nitrocellulose filters. Each bar represents six to nine independent measurements. Error bars indicate 95% confidence interval (AU, arbitrary units). rad17+ in DB vector and rad1+ in AD vector caused a significant increase in β-galactosidase activity. As a positive control, plasmids containing S. cerevisiae SNF1 and SNF4 genes were used. As negative control, the p53 gene was cotransformed with rad1. (C) Inverse orientation: rad1+ in DB vector and rad17+ in AD vector. This combination resulted in a strong background signal. (D) Activity of mutant rad1-S alleles in DB vector and rad17+ in AD vector. Together with AD-rad17+, DB-rad1-S1 and DB-rad1-S3 gave a signal that was above the background. (E) Whole-cell extracts from S. pombe cells overexpressing MYC-Rad17 were incubated with Rad1-coated or bovine serum albumin-coated beads. The washed beads were boiled in sample buffer, and MYC-Rad17 was visualized using anti-MYC antibodies after separation by SDS-PAGE on a 12.5% gel and Western blotting (Total, whole-cell lysate). (F) S. pombe cells were cotransformed with plasmids containing inserts encoding HA-Rad17, MYC-Rad1, or no insert, as indicated above each lane. After lysis, proteins were immunoprecipitated (IP) with anti-MYC or anti-HA antibodies. The precipitates were probed with the antibody that was not used for precipitation.
Rad17 and Rad1 interact in the two-hybrid system.
The Rad17-dependent nuclear localization of Hus1 and Rad9 suggests a potential interaction between the Rad17 complex and complexes containing Rad9 and/or Hus1-B. The yeast two-hybrid system was used to test interactions between Rad17, Rad1, Hus1, Rad9, Rad26, and Chk1. Full-length cDNAs encoding the corresponding genes were fused to both the Gal4 DB and the Gal4 AD. DB and AD constructs and empty vector controls were combined in the same cell, and β-galactosidase activity was assayed. The only reproducible positive result was obtained for Rad1 in the AD vector and Rad17 in the DB vector (Fig. 5B). When the cDNAs were switched between the DB vector and the AD vector, a strong signal was obtained from cells containing only DB-rad1 (Fig. 5C). Several site-specific mutations have previously been created in rad1 and observed to give a low background signal (reference 25 and data not shown) in the two-hybrid system. Therefore, the wild-type rad1 cDNA was substituted in the DB vector for mutated rad1-S cDNAs. When combined with AD-rad17, DB-rad1-S1 and DB-rad1-S3 cDNAs gave a signal that was substantially higher than the signal obtained from control cells harboring DB-rad1-S1 or DB-rad1-S3 and the empty AD plasmid (Fig. 5D). While Rad1 and Rad17 do not interact in soluble extracts, transient interactions can sometimes be detected by increasing the relative concentrations of the interacting partners. This was explored in two ways. First, HIS-Rad1 was purified from E. coli and coupled to Sepharose beads. A significant amount of MYC-Rad17 present in extracts of S. pombe cells overexpressing MYC-rad17 from the inducible nmt1 promoter of pREP41 was precipitated with Rad1-coupled beads but not with bovine serum albumin-coupled control beads (Fig. 5E). Second, both HA-Rad17 and MYC-Rad1 were overexpressed (pREP41 promoter) in the same cell and extracts were used for immunoprecipitation with either anti-MYC or anti-HA monoclonal antibodies. In both cases an interaction was evident by coimmunoprecipitation (Fig. 5F). Taken together, these data suggest that Rad17 and Rad1 can interact, but that the interaction may be unstable and/or transient in vivo at endogenous protein concentrations.
Hus1, Rad9, and Rad1 are all related to PCNA.
The reported findings that Rad17 contains RFC-like domains (18) and that Rad1 and Hus1 are both related to PCNA (2, 46) argue for a model in which Rad17 transiently interacts with the Rad1–Rad9–Hus1-B complex. Consistent with such a model, we found by using the generalized profile technique (6) that the PCNA relationship extends to Rad9Sp/Hs and its S. cerevisiae homolog Ddc1 (Fig. 6A). The alignment between PCNA, Rad1, and Hus1 spans the entire length of all three proteins, whereas the PCNA-like motifs found in Rad9Sp/Hs and Ddc1Sc are located in the N-terminal portion of these proteins. The conserved motifs are predominately found in the structural regions of PCNA and not in the loop domains which determine protein-protein interactions (24, 36). This suggests that the checkpoint proteins have a PCNA-like structure rather than similar interaction partners.
FIG. 6.
Rad9, Rad1, and Hus1 are all related to PCNA. Hus1Sp and Mec3Sc share a common domain. (A) Multiple alignment of PCNA, Rad1 (Rad17Sc), Hus1, and Rad9 (Ddc1Sc) from S. pombe (SP), S. cerevisiae (SC), and H. sapiens (HS). Identical residues are shown in white capital letters on black background, and conservative replacements are shown in black capital letters on grey background. (s, β-sheet; h, α-helix). (B) Multiple alignment of the N-terminal region of Hus1Sp, Hus1Hs, and Mec3Sc. Identical residues and conserved replacements are in bold letters and marked by asterisks.
Hus1Sp and Mec3Sc share a common domain.
The association of Mec3Sc with Ddc1Sc (Rad9Sp) and Rad17Sc (Rad1Sp) suggests that Mec3 could be related to Hus1Sp and possibly PCNA. However, no PCNA-like motifs were detected in Mec3 (reference 2 and data not shown). Application of the generalized profile technique (6) revealed instead the existence of an N-terminal domain of approximately 90 amino acids that is conserved between Hus1Sp, Hus1Hs, and Mec3Sc (Fig. 6B). Although the homology is restricted to this domain, this finding, combined with the similar protein interactions, supports a proposition that Mec3 and Hus1 can be considered orthologs.
DISCUSSION
Several points emerge from the analysis of Hus1-MYC. (i) Hus1-B, the most abundant form of Hus1 detected in tagged cell extracts, is phosphorylated after DNA damage but not during HU arrest. (ii) Hus1-B associates with Rad1 and Rad9 in a complex of approximately 450 kDa, larger than a simple trimeric complex. (iii) Rad1, Rad9, and Hus1 are all structurally related to PCNA, and Hus1 shares statistically significant similarity to Mec3, suggesting that Hus1 and Mec3 are orthologs. (iv) The majority of both Rad1 and Rad9 do not coelute with the Rad1–Rad9–Hus1-B complex, indicating that these proteins also form Hus1-independent protein complexes. (v) Rad17 forms a third protein complex, distinct from the Rad3-Rad26 complex and the Rad1–Rad9–Hus1-B complex. (vi) Rad17 is specifically required for nuclear localization of Hus1 and Rad9.
Characterization of Hus1-MYC.
hus1-MYC cells are checkpoint proficient and resistant to treatment with bleomycin or HU (Fig. 1B and D). Four Hus1-MYC-specific isoforms were present in extracts, and the major form, Hus1-B, is hyperphosphorylated in response to DNA damage (Fig. 1C) and after prolonged HU treatment (Fig. 1F). In contrast with a previous report (27), Hus1 is not phosphorylated when cells are arrested in early S phase by HU (Fig. 1E and F). It is not clear if this is a result of the C-terminal MYC tag, or if it reflects differences in the experimental protocol. However, the C-terminal MYC tag does not interfere with DNA damage or S-phase checkpoint functions of Hus1.
The Rad1–Rad9–Hus1-B protein complex.
Size exclusion chromatography and subsequent coimmunoprecipitation demonstrate that Rad1, Rad9, and Hus1-B form a soluble protein complex and that this protein complex is undetectable upon loss of either Rad9 (Fig. 3B) or Rad1 (data not shown). This is consistent with the previous report that Hus1 and Rad1 interact in a rad9-dependent manner (27). It is also consistent with coimmunoprecipitation experiments showing that a similar protein complex exists in human cells and in S. cerevisiae (26, 44, 47). The Rad1–Rad9–Hus1-B complex eluted in fractions 8 to 14 (Fig. 3A), peaking with a profile similar to that for the size marker ferritin (440 kDa). Since the expected molecular mass of a trimeric Rad1 (35 kDa)–Rad9 (50 kDa)–Hus1-B (65 kDa) protein complex is approximately 150 kDa, this indicates that the trimeric complex either binds to other proteins or contains multiple Rad1, Rad9, and/or Hus1-B under these conditions. Since Rad1, Hus1, and Rad9 are all required for activation of Chk1 in response to DNA damage (48) and Rad3 can respond to DNA damage in the absence of rad1, rad9, and hus1 (12), these data suggest that the Rad1–Rad9–Hus1-B complex is required to transmit the checkpoint signal generated by the Rad3-Rad26 complex to Chk1.
However, several lines of evidence suggest that Rad1, Rad9, and Hus1-B form additional protein complexes. First, deletion of rad9 or rad1 did not affect Hus1-B fractionation as dramatically as would be expected if Hus1-B is present only in association with Rad9 and Rad1 (Fig. 2B). The majority of Rad9 is found in fractions 14 to 16 (∼200 kDa) and is not associated with Hus1-B, as judged by immunoprecipitation. The second line of evidence suggesting that Rad1, Rad9, and Hus1-B can form distinct protein complexes comes from our analysis of Rad1. Most of Rad1 (35 kDa) did not coelute with Hus1-B but showed two elution peaks corresponding to sizes of 200 and 70 kDa. Again, Hus1-B did not coimmunoprecipitate with anti-Rad1 sera (Fig. 3A) from the fractions containing the 200-kDa peak of Rad1 (fractions 13 and 14). This suggests that distinct checkpoint protein complexes may exist in soluble extracts. There is limited genetic evidence available, which supports distinct functions for different checkpoint protein complexes. Telomere shortening was evident in rad1.d, rad3.d, rad17.d, and rad26.d mutant cells, but telomeres were normal in either rad9.d or hus1.d mutants (9). Thus, a Rad1-containing complex that does not contain Rad9 and Hus1 could be required for correct telomere length maintenance. The dependency on Rad17 could be explained by lack of nuclear localization of this Rad1 complex. Another line of evidence comes from studies on UVDE (UV damage endonuclease). It has been reported that UVDE activity is constitutively high in rad9.d cells, but that in other checkpoint rad mutants and wild-type cells UVDE activity is found at similar levels only after DNA damage treatment (10, 11). Although these observations are contentious (34), possibly because cells were taken at different growth phases, they could suggest that a Rad9-containing complex acts independently of the other checkpoint Rad proteins in inducing UVDE activity. In this case, Rad9 seems to enter the nucleus independently of Rad17. Interestingly, in contrast to Hus1, some Rad9 is still seen in the nucleus of rad9-MYC rad17.d cells (Fig. 4C).
Rad17, which fractionates as a distinct complex, is required for nuclear localization of Hus1 and Rad9.
Rad3 and Rad26 are both part of a protein complex that elutes during size exclusion chromatography in fractions 6 to 8, and deletion of rad3 causes redistribution of Rad26 to smaller sizes (12). Since such redistribution was not observed for the proportion of Hus1 (Fig. 2B) eluting in fractions 6 to 8, it is unlikely that Hus1 associates with Rad3 in soluble extracts. Most of Rad17-MYC eluted in fractions 10 to 12 (Fig. 5A), indicating a native complex size of approximately 450 kDa, similar in size to the Rad1–Rad9–Hus1-B complex. However, while loss of Rad9 or Rad1 caused a reduction of the amount of Hus1-B in these fractions (Fig. 2B), loss of either Rad1, Rad9, or Hus1 did not change the elution profile of Rad17 (Fig. 5A) and loss of Rad17 did not change the elution profile of Hus1-B (Fig. 2B). The assumption that Rad17 forms a third protein complex that does not contain other checkpoint Rad proteins is consistent with the absence of Rad17Hs and Rad24Sc in the human Rad1-Rad9-Hus1 and S. cerevisiae Rad17-Ddc1-Mec3 complexes, respectively, when these proteins were co-overexpressed (26, 47).
Nonetheless, Rad17 is required for Rad3-dependent Hus1 phosphorylation (Fig. 2B) (27). To address possible physical interactions between these complexes, which might not be sufficiently stable to be detected in soluble extracts, we searched for potential interactions by using the two-hybrid method. We found significant evidence of an interaction between Rad1 and Rad17 and were also able to identify in vitro binding and in vivo coimmunoprecipitation of these proteins when they were overexpressed (Fig. 5B to F). Thus, Rad1 and Rad17 are capable of interacting. Together with the genetic and immunolocalization data (see below), this finding suggests that Rad1 and Rad17 are capable of transient and/or unstable interactions in vivo.
By examining the subcellular localization of Hus1, Rad9, and Rad26 using indirect immunofluorescence microscopy, we found that Rad9, Rad26, and Hus1 are all localized inside the nucleus (Fig. 4A and C). Furthermore, we also observed a Rad9-specific staining when nuclei were spread onto glass slides, often taken as evidence of chromatin association (T. Caspari and A. M. Carr, unpublished data). Nuclear localization of Rad9 and Hus1-B was compromised in rad17.d cells, while nuclear localization of Rad26 was not (Fig. 4C). It is not clear whether Rad17 is involved in nuclear import of Rad9 and Hus1-B or whether Rad17 is required to retain both proteins inside the nucleus. However, leptomycin B treatment, which is known to block Crm1-dependent nuclear export (16) and promote the accumulation of the Cdc25 mitotic inducer (30), did not result in accumulation of Hus1-MYC inside the nucleus of hus1-MYC rad17.d cells (data not shown). The presence of two nuclear localization motifs in Rad17 is consistent with an involvement of Rad17 in nuclear import of Rad9 and Hus1-B. However, the sequence similarities between Rad17 and RFC (18) and between Rad1, Rad9, Hus1, and PCNA (Fig. 6A) suggest that Rad17 may also be required to load Rad1, Rad9, and Hus1 onto DNA.
Conclusion.
The data presented in this report suggest that the checkpoint Rad proteins form distinct protein complexes, which presumably interact in vivo to generate a checkpoint signal. Rad17 associates with yet unknown protein partners and is required to allow nuclear localization of Rad9 and Hus1. Rad9, Hus1, and Rad1 appear to form different protein complexes that might function downstream of the Rad3-Rad26 complex. Thus, the checkpoint Rad proteins seem to form a network of stable protein complexes rather than a single stable guardian complex. It will be interesting to determine whether these define a single chromatin-associated complex in vivo.
ACKNOWLEDGMENTS
We thank Per Uhrskov Christensen for reading the manuscript and helpful discussions.
T.C. was sponsored by fellowship Ca197/2-1 from the Deutsche Forschungsgemeinschaft (Bonn, Germany). A.M.C. and H.D.L. are supported by the MRC and Euratom contract F14PCT950010. P.S. acknowledges the Swedish Cancer Fund (grant 2163-B97-08XAC) and Swedish Radiation Protection Institute (grant 1092.98).
REFERENCES
- 1.Al-Khodairy F, Carr A M. DNA repair mutants defining G2 checkpoint pathways in Schizosaccharomyces pombe. EMBO J. 1992;11:1343–1350. doi: 10.1002/j.1460-2075.1992.tb05179.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aravind L, Walker D R, Koonin E V. Conserved domains in DNA repair proteins and evolution of repair systems. Nucleic Acids Res. 1999;27:1223–1242. doi: 10.1093/nar/27.5.1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bähler J, Wu J Q, Longtine M S, Shah N G, McKenzie A, Steever A B, Wach A, Philippsen P, Pringle J R. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast. 1998;14:943–951. doi: 10.1002/(SICI)1097-0061(199807)14:10<943::AID-YEA292>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 4.Bentley N J, Holtzman D A, Flaggs G, Keegan K S, DeMaggio A, Ford J C, Hoekstra M, Carr A M. The Schizosaccharomyces pombe rad3 checkpoint gene. EMBO J. 1996;15:6641–6651. [PMC free article] [PubMed] [Google Scholar]
- 5.Boddy M N, Furnari B, Mondesert O, Russell P. Replication checkpoint enforced by kinases Cds1 and Chk1. Science. 1998;280:909–912. doi: 10.1126/science.280.5365.909. [DOI] [PubMed] [Google Scholar]
- 6.Bucher P, Karplus K, Moeri N, Hofmann K. A flexible motif search technique based on generalized profiles. Comput Chem. 1996;20:3–23. doi: 10.1016/s0097-8485(96)80003-9. [DOI] [PubMed] [Google Scholar]
- 7.Carr A M. Control of cell cycle arrest by the Mec1sc/Rad3sp DNA structure checkpoint pathway. Curr Opin Genet Dev. 1997;7:93–98. doi: 10.1016/s0959-437x(97)80115-3. [DOI] [PubMed] [Google Scholar]
- 8.Caspari T, Carr A M. DNA structure checkpoint pathways in Schizosaccharomyces pombe. Biochimie. 1999;81:173–181. doi: 10.1016/s0300-9084(99)80050-9. [DOI] [PubMed] [Google Scholar]
- 9.Dahlen M, Olsson T, Kanter-Smoler G, Ramne A, Sunnerhagen P. Regulation of telomere length by checkpoint genes in Schizosaccharomyces pombe. Mol Biol Cell. 1998;9:611–621. doi: 10.1091/mbc.9.3.611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davey S, Han C S, Ramer S A, Klassen J C, Jacobson A, Eisenberger A, Hopkins K M, Lieberman H B, Freyer G A. Fission yeast rad12 regulates cell cycle checkpoint control and is homologous to the Bloom's syndrome disease gene. Mol Cell Biol. 1998;18:2721–2728. doi: 10.1128/mcb.18.5.2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Davey S, Nass M L, Ferrer J V, Sidik K, Eisenberger A, Mitchell D L, Freyer G A. The fission yeast UVDE DNA repair pathway is inducible. Nucleic Acids Res. 1997;25:1002–1008. doi: 10.1093/nar/25.5.1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Edwards R J, Bentley N J, Carr A M. A Rad3-Rad26 complex responds to DNA damage independently of other checkpoint proteins. Nat Cell Biol. 1999;1:393–398. doi: 10.1038/15623. [DOI] [PubMed] [Google Scholar]
- 13.Edwards R J, Carr A M. Analysis of radiation-sensitive mutants of fission yeast. Methods Enzymol. 1997;283:471–494. doi: 10.1016/s0076-6879(97)83038-8. [DOI] [PubMed] [Google Scholar]
- 14.Enoch T, Carr A M, Nurse P. Fission yeast genes involved in coupling mitosis to completion of DNA replication. Genes Dev. 1992;6:2035–2046. doi: 10.1101/gad.6.11.2035. [DOI] [PubMed] [Google Scholar]
- 15.Frangioni J V, Neel B G. Solubilization and purification of enzymatically active glutathione S-transferase (pGEX) fusion proteins. Anal Biochem. 1993;210:179–187. doi: 10.1006/abio.1993.1170. [DOI] [PubMed] [Google Scholar]
- 16.Fukuda M, Asano S, Nakamura T, Adachi M, Yoshida M, Yanagida M, Nishida E. CRM1 is responsible for intracellular transport mediated by the nuclear export signal. Nature. 1997;390:308–311. doi: 10.1038/36894. [DOI] [PubMed] [Google Scholar]
- 17.Furnari B, Rhind N, Russell P. Cdc25 mitotic inducer targeted by Chk1 DNA damage checkpoint kinase. Science. 1997;277:1495–1497. doi: 10.1126/science.277.5331.1495. [DOI] [PubMed] [Google Scholar]
- 18.Griffiths D J F, Barbet N C, McCready S, Lehmann A R, Carr A M. Fission yeast rad17: a homologue of budding yeast RAD24 that shares regions of sequence similarity with DNA polymerase accessory proteins. EMBO J. 1995;14:5812–5823. doi: 10.1002/j.1460-2075.1995.tb00269.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hagan I, Hayles J, Nurse P. Cloning and sequencing of the cyclin-related cdc13+ gene and a cytological study of its role in fission yeast mitosis. J Cell Sci. 1988;91:587–595. doi: 10.1242/jcs.91.4.587. [DOI] [PubMed] [Google Scholar]
- 20.Harlow E, Lane D. Antibodies: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1988. [Google Scholar]
- 21.Harper J W, Adami G R, Wei N, Keyomarsi K, Elledge S J. The p21 Cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell. 1993;75:805–816. doi: 10.1016/0092-8674(93)90499-g. [DOI] [PubMed] [Google Scholar]
- 22.Hartley K O, Gell D, Smith G C M, Zhang H, Divecha N, Connelly M A, Admon A, Lees-Miller S P, Anderson C W, Jackson S P. DNA-dependent protein kinase catalytic subunit: a relative of phosphatidylinositol 3-kinase and the ataxia telangiectasia gene product. Cell. 1995;82:849–856. doi: 10.1016/0092-8674(95)90482-4. [DOI] [PubMed] [Google Scholar]
- 23.Hartwell L H, Kastan M B. Cell cycle control and cancer. Science. 1994;266:1821–1828. doi: 10.1126/science.7997877. [DOI] [PubMed] [Google Scholar]
- 24.Jonsson Z O, Hindges R, Hubscher U. Regulation of DNA replication and repair proteins through interaction with the front side of proliferating cell nuclear antigen. EMBO J. 1998;15:2412–2425. doi: 10.1093/emboj/17.8.2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kanter-Smoler G, Knudsen K E, Jimenez G, Sunnerhagen P, Subramani S. Separation of phenotypes in mutant alleles of the Schizosaccharomyces pombe cell-cycle checkpoint gene rad1+ Mol Biol Cell. 1995;6:1793–1805. doi: 10.1091/mbc.6.12.1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kondo T, Matsumoto K, Sugimoto K. Role of a complex containing Rad17, Mec3, and Ddc1 in the yeast DNA damage checkpoint pathway. Mol Cell Biol. 1999;19:1136–1143. doi: 10.1128/mcb.19.2.1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kostrub C, Knudsen K, Subramani S, Enoch T. Hus1p, a conserved fission yeast checkpoint protein, interacts with Rad1p and is phosphorylated in response to DNA damage. EMBO J. 1998;17:2055–2066. doi: 10.1093/emboj/17.7.2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kostrub C F, al-Khodairy F, Ghazizadeh H, Carr A M, Enoch T. Molecular analysis of hus1+, a fission yeast gene required for S-M and DNA damage checkpoints. Mol Gen Genet. 1997;254:389–399. doi: 10.1007/pl00008606. [DOI] [PubMed] [Google Scholar]
- 29.Lindsay H D, Griffiths D J F, Edwards R J, Christensen P U, Murray J M, Osman F, Walworth N, Carr A M. S-phase specific activation of Cds1 kinase defines a subpathway of the checkpoint response in Schizosaccharomyces pombe. Genes Dev. 1998;12:382–395. doi: 10.1101/gad.12.3.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lopez-Girona A, Furnari B, Mondesert O, Russell P. Nuclear localisation of Cdc25 is regulated by DNA damage and a 14-3-3 protein. Nature. 1999;397:172–175. doi: 10.1038/16488. [DOI] [PubMed] [Google Scholar]
- 31.Martinho R G, Lindsay H D, Flaggs G, DeMaggio A, Hoekstra M, Carr A M, Bentley N J. Analysis of Rad3 and Chk1 protein kinases defines different checkpoint responses. EMBO J. 1998;17:7239–7249. doi: 10.1093/emboj/17.24.7239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moreno S, Klar A, Nurse P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 1991;194:795–826. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- 33.Murakami H, Okayama H. A kinase from fission yeast responsible for blocking mitosis in S phase. Nature. 1995;374:817–819. doi: 10.1038/374817a0. [DOI] [PubMed] [Google Scholar]
- 34.Murray J M, Lindsay H D, Munday C A, Carr A M. Role of Schizosaccharomyces pombe RecQ homolog, recombination, and checkpoint genes in UV damage tolerance. Mol Cell Biol. 1997;17:6868–6875. doi: 10.1128/mcb.17.12.6868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.O'Connell M J, Raleigh J M, Verkade H M, Nurse P. Chk1 is a wee1 kinase in the G2 DNA damage checkpoint inhibiting cdc2 by Y15 phosphorylation. EMBO J. 1997;16:545–554. doi: 10.1093/emboj/16.3.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Oku T, Ikeda S, Sasaki H, Fukuda K, Morioka H, Ohtsuka E, Yoshikawa H, Tsurimoto T. Functional sites of human PCNA which interact with p21 (Cip1/Waf1), DNA polymerase delta and replication factor C. Genes Cells. 1998;3:357–369. doi: 10.1046/j.1365-2443.1998.00199.x. [DOI] [PubMed] [Google Scholar]
- 37.Onel K, Koff A, Bennett R L, Unrau P, Holloman W K. The REC1 gene of Ustilago maydis, which encodes a 3′-5′ exonuclease, couples DNA repair and completion of DNA synthesis to a mitotic checkpoint. Genetics. 1996;143:165–174. doi: 10.1093/genetics/143.1.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Parker A E, van de Weyer I, Laus M C, Oostveen I, Yon J, Verhasselt P, Luyten W H. A human homologue of the Schizosaccharomyces pombe rad1+ checkpoint gene encodes an exonuclease. J Biol Chem. 1998;273:18332–18339. doi: 10.1074/jbc.273.29.18332. [DOI] [PubMed] [Google Scholar]
- 39.Rhind N, Furnari B, Russell P. Cdc2 tyrosine phosphorylation is required for the DNA damage checkpoint in fission yeast. Genes Dev. 1997;11:504–511. doi: 10.1101/gad.11.4.504. [DOI] [PubMed] [Google Scholar]
- 40.Rhind N, Russell P. Mitotic DNA damage and replication checkpoints in yeast. Curr Opin Cell Biol. 1998;10:749–758. doi: 10.1016/s0955-0674(98)80118-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rowley R, Subramani S, Young P G. Checkpoint controls in Schizosaccharomyces pombe: rad1. EMBO J. 1992;11:1335–1342. doi: 10.1002/j.1460-2075.1992.tb05178.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Saka Y, Esashi F, Matsusaka T, Mochida S, Yanagida M. Damage and replication checkpoint control in fission yeast is ensured by interactions of Crb2, a protein with BRCT motif, with Cut5 and Chk1. Genes Dev. 1997;11:3387–3400. doi: 10.1101/gad.11.24.3387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shimomura T, Ando S, Matsumoto K, Sugimoto K. Functional and physical interaction between Rad24 and Rfc5 in the yeast checkpoint pathways. Mol Cell Biol. 1998;18:5485–5491. doi: 10.1128/mcb.18.9.5485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.St Onge R P, Udell C M, Casselman R, Davey S. The human G2 checkpoint control protein hRAD9 is a nuclear phosphoprotein that forms complexes with hRad1 and hHus1. Mol Biol Cell. 1999;10:1985–1995. doi: 10.1091/mbc.10.6.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thelen M P, Onel K, Holloman W K. The REC1 gene of Ustilago maydis involved in the cellular response to DNA damage encodes an exonuclease. J Biol Chem. 1994;269:747–754. [PubMed] [Google Scholar]
- 46.Thelen M P, Venclovas C, Fidelis K. A sliding clamp model for the Rad1 family of cell cycle checkpoint proteins. Cell. 1999;19:769–770. doi: 10.1016/s0092-8674(00)80587-5. [DOI] [PubMed] [Google Scholar]
- 47.Volkmer E, Karnitz L M. Human homologs of Schizosaccharomyces pombe Rad1, Hus1, and Rad9 form a DNA damage-responsive protein complex. J Biol Chem. 1999;274:567–570. doi: 10.1074/jbc.274.2.567. [DOI] [PubMed] [Google Scholar]
- 48.Walworth N, Bernards R. rad-dependent responses of the chk1-encoded protein kinase at the DNA damage checkpoint. Science. 1996;271:353–356. doi: 10.1126/science.271.5247.353. [DOI] [PubMed] [Google Scholar]
- 49.Walworth N, Davey S, Beach D. Fission yeast chk1 protein kinase links the rad checkpoint pathway to cdc2. Nature. 1993;363:368–371. doi: 10.1038/363368a0. [DOI] [PubMed] [Google Scholar]
- 50.Weinert T A, Hartwell L H. The RAD9 gene controls the cell cycle response to DNA damage in Saccharomyces cerevisiae. Science. 1988;241:317–322. doi: 10.1126/science.3291120. [DOI] [PubMed] [Google Scholar]
- 51.Willson J, Wilson S, Warr N, Watts F Z. Isolation and characterization of the Schizosaccharomyces pombe rhp9 gene: a gene required for the DNA damage checkpoint but not the replication checkpoint. Nucleic Acids Res. 1997;25:2138–2146. doi: 10.1093/nar/25.11.2138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zeng Y, Forbes K C, Wu Z, Moreno S, Piwnica-Worms H, Enoch T. Replication checkpoint requires phosphorylation of the phosphatase Cdc25 by Cds1 or Chk1. Nature. 1998;395:507–510. doi: 10.1038/26766. [DOI] [PubMed] [Google Scholar]