Abstract
Chromatin conformation capture (3C) methods and fluorescent in situ hybridization (FISH) microscopy have been used to investigate the spatial organization of the genome. Although powerful, both techniques have limitations. Hi-C is challenging for low cell numbers and requires very deep sequencing to achieve its high resolution. In contrast, FISH can be done on small cell numbers and capture rare cell populations, but typically targets pairs of loci at a lower resolution. Here we detail a protocol for optical reconstruction of chromatin architecture (ORCA), a microscopy approach to trace the 3D DNA path within the nuclei of fixed tissues and cultured cells with a genomic resolution as fine as 2 kb and a throughput of ~10,000 cells per experiment. ORCA can identify structural features with comparable resolution to Hi-C while providing single-cell resolution and multimodal measurements characteristic of microscopy. We describe how to use this DNA labeling in parallel with multiplexed labeling of dozens of RNAs to relate chromatin structure and gene expression in the same cells. Oligopaint probe design, primary probe making, sample collection, cryosectioning and RNA/DNA primary probe hybridization can be completed in 1.5 weeks, while automated RNA/DNA barcode hybridization and RNA/DNA imaging typically takes 2–6 d for data collection and 2–7 d for the automated steps of image analysis.
Introduction
Technical challenges have limited the application of Hi-C methods in complex tissues, where key biological processes are believed to be regulated by chromatin structure. The challenges arise because achieving high resolution with these sequencing-based assays requires: (i) relatively large numbers of homogenous cells, limiting application to rare-cell populations; (ii) dissociation of cells, leading to loss of spatial context; and (iii) nuclei extraction, leading to loss of RNA transcripts, which are the readouts of enhancer function. Microscopy studies using fluorescent in situ hybridization (FISH) have revealed various cell type–specific enhancer-promoter interactions in tissues1–4, but largely achieve lower resolution than sequencing methods5. In addition, FISH is limited by the number of fluorescent channels available and, thus, allows only for observation of a targeted handful of loci. Super-resolution imaging using stochastic optical reconstruction microscopy (STORM)6,7 provides further detail on the spatial structure of domains ranging from several kilobases to several mega-bases2,8–11. However, the globular structures resolved via STORM cannot be used to distinguish sequence features, such as enhancer and promoter, or genomic positions.
Both sequencing and imaging approaches typically sacrifice analysis of RNA transcripts when measuring DNA contacts, which limits the ability to study how chromatin structure and transcription correlate at the cellular scale. Several studies have relied on correlating these properties at the bulk level by using separate assays, such as Hi-C for mapping cis-interactions and RNA-seq for characterizing the transcriptional state12–23. However, at the single-cell level, both structure and expression vary substantially from cell to cell24. Thus, we developed a method that reveals high-resolution single-cell chromatin structure for thousands of individual cells within cryosectioned tissue, which we have termed ‘optical reconstruction of chromatin architecture’ (ORCA)25 (Fig. 1). A key feature of our approach is its compatibility with multiplexed RNA labeling of both mature and nascent transcripts25–32, allowing the interaction of transcription and genome structure to be measured at the single-cell level. The protocols we describe here for DNA and multiplexed RNA labeling can be performed separately or sequentially, providing both measurements within the same individual cells.
Fig. 1 |. Optical reconstruction of chromatin architecture (ORCA) approach.
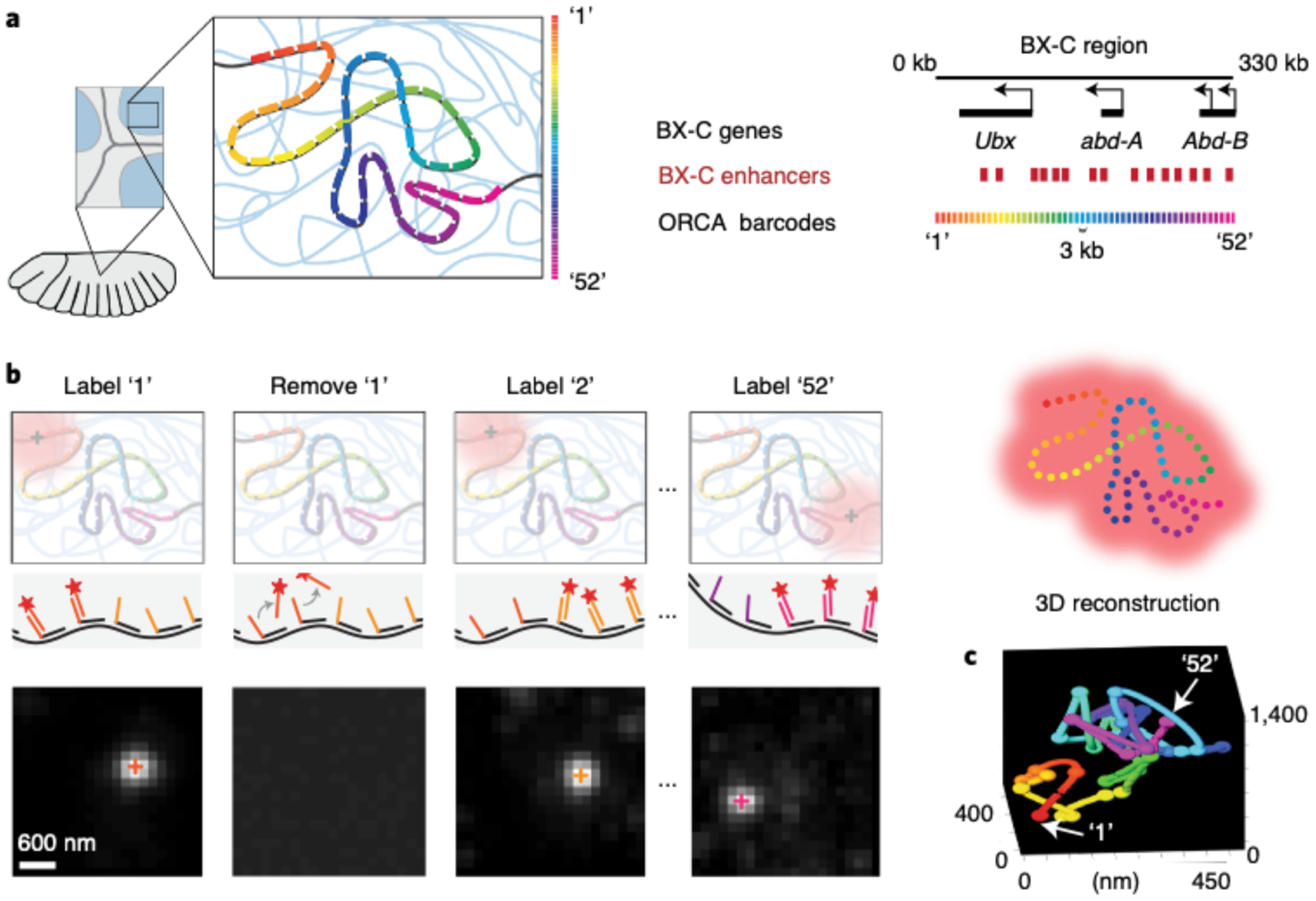
This figure was adapted from our previously published work13. a, In a cryosectioned Drosophila embryo, the bithorax complex (BX-C) region is tiled with primary probes. These probes partition the region of interest into 52 3-kb genomic segments and each segment contains a unique barcode. b, ORCA relies on sequential hybridization rounds to label and image every barcode through the process of adding a complementary readout oligo bound to a fluorophore, removing the readout probe after imaging it, and repeating this process for all 52 barcodes. The top represents a schematic of this process and the bottom illustrates example data from imaged barcodes. The 3D centroid of each labeled barcode, seen as a white spot, is shown as ‘+’ symbols on the spots. c, 3D positions of the labeled barcodes are seen as spheres and are pseudocoloured. Arrows are pointing at the first (‘1’) and last (‘52’) labeled barcodes. The spheres are linked together to generate a 3D polymer for a single cell.
Overview of the multiplex RNA labeling and DNA tracing procedure
Both multiplex RNA labeling and DNA tracing with ORCA begin with designing an Oligopaint library33–35 (Fig. 2, Step 1A). For DNA probes, first select a genomic region of interest and an intended step size (e.g., 10 or 2 kb). Using the corresponding reference genome (e.g., dm3, mm10) and a list of abundant repetitive sequences to exclude, select ≥20 primary probes with a length of 40 bp per genomic step to hybridize onto the genome. Although prior multiplex single-molecule RNA FISH protocols have relied on shorter hybridization regions26–28,30–32, in our hands, longer sequences have provided more robust detection. When selecting primary probes, make sure to avoid repetitive regions and oligonucleotide properties known to reduce hybridization specificity. Examples of such off-target lists are provided at https://github.com/BoettigerLab/ORCA-public/tree/master/probe-building. Selection of oligos targeting a genomic region while respecting filter conditions is a routine filtering task, which can be facilitated for rapid probe construction through recently described algorithms9,25,34,36,37.
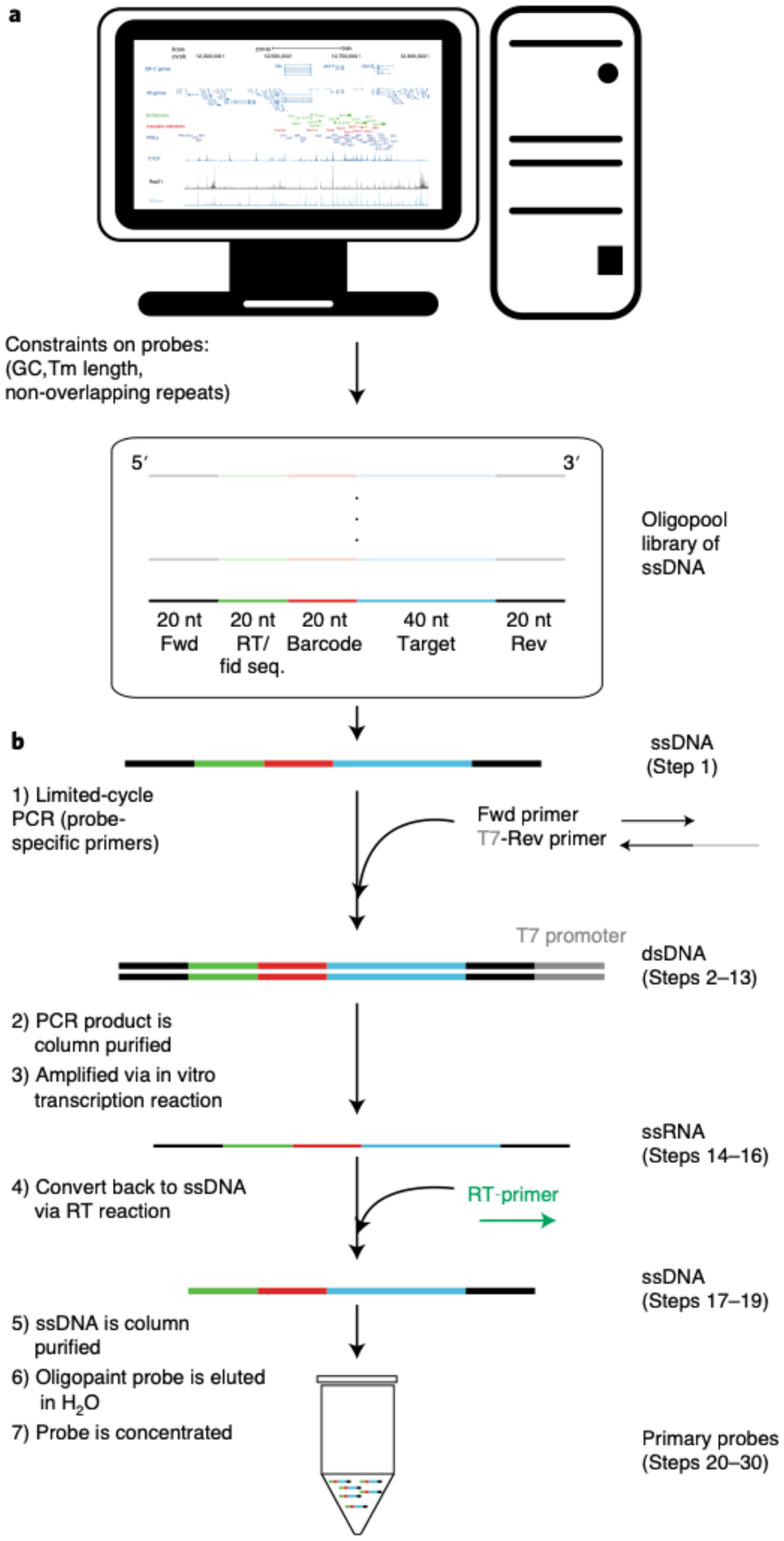
Fig. 2 |. Schematic overview of ORCA and RNA labeling experimental process and computational analysis.
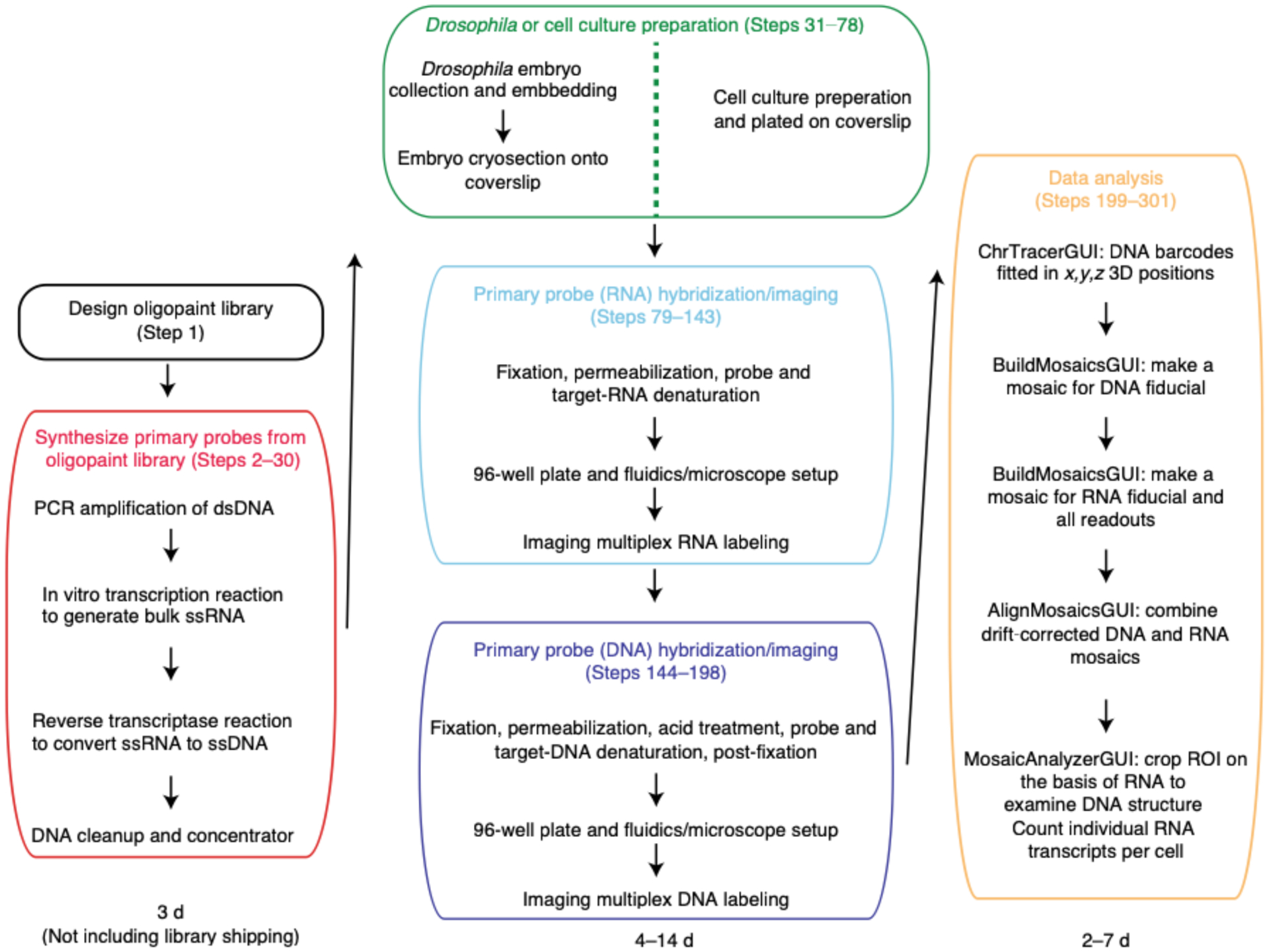
Designing the Oligopaint library (boxed in black) takes less than a day and the library is typically shipped from IDT within a month of ordering. Synthesizing the primary probes (boxed in red) from the library is a two day process that involves turning double-stranded DNA (dsDNA) into single-stranded RNA (ssRNA), and then to single-stranded DNA (ssDNA). Sample preparation length (boxed in green) differs between embryos and cell culture. Embryo collection, embedding and cryosectioning takes two days. Adding cultured cells onto a poly-lysine charged coverslip and fixing the cells takes less than 30 minutes. Hybridizing RNA primary probe (boxed in cyan) can be done right after sample preparation. After the overnight hybridization step, the sample is ready to be imaged. Imaging acquisition (boxed in cyan) time depends upon the number of FOVs chosen and the number of barcodes in a region of interest. For example, if one were to image 18 FOVs and 10 RNAs for mESCs, this process would take ~9.5 hrs as imaging each RNA for 18 FOVs takes ~55 min. Typically for embryos, imaging ~50 FOVs and 29 RNAs takes 1.5 days. Hybridizing DNA primary probes (boxed in purple) is a 1.5 days process as there is an overnight step. Again, the imaging duration (boxed in purple) depends upon the number of FOVs and barcodes in a given experiment. For example, imaging a single barcode for 30 FOVs takes ~44 minutes each and an experiment with 55 barcodes will last approximately 1 day and 16 hrs. Another example is an experiment where one images 16 barcodes for 15 FOVs takes ~10 hrs. Thus, the span of time one could take to prepare a sample and image it (middle column) can be between 4–14 days. The duration of image analysis (boxed in yellow) depends upon the size of the data acquired. If the user wants to only look at DNA data, ChrTracer3 analysis can take approximately 1 day for a dataset for 75 barcodes and 34 FOVs in cell culture. Note, the number of nuclei imaged in 1 FOV for mESCs can range between 300–500 while the number of nuclei in 1 FOV for embryos is ~1600. Thus, analyzing mESCs data through ChrTracer3 will be quicker compared to embryos. Typically, analyzing embryo data through ChrTracer3 can range between 1–4 days depending upon the number of FOVs selected. If the user chooses to use all provided MATLAB software for DNA and RNA analysis (boxed in yellow), the process can take 4–8 days.
For DNA probes, if the target genomic region overlaps with transcribed regions, we recommend designing probes that match the sense strand, to avoid hybridization to RNA (Step 1B). Although all RNA probes should be anti-sense to the RNA transcript, for nascent RNA probes, we recommend targeting 5′ ends of the introns, because these are the first to be transcribed and will appear closer to the promoter firing event, which is especially important for long genes. After selecting the 40mer targeting sequences, these need to be concatenated with a 20mer barcode sequence and a common 20mer ‘fiducial-binding’ sequence. Whereas the barcode sequence is unique for each RNA species or, for DNA, each genomic step size, the fiducial-binding sequence is the same for all probes. Finally, unique primer sequences added to either end allow for amplification and multiplexing of different DNA/RNA probe templates in the same commercial oligonucleotide pool. RNA and DNA probesets should be given distinct primers so that they may be synthesized separately. Combining probesets into the same oligonucleotide pool is useful for reducing costs (most vendors sell fixed increments of unique oligos in a complex pool and have decreasing costs for larger pools). Example primer sequences, 384 unique barcode sequences and scripts to aid in design of the oligonucleotide pools are provided at https://github.com/BoettigerLab/ORCA-public/tree/master/probe-building.
Because the unique barcodes are sequentially imaged for a given genomic region of interest, the following types of oligos must be generated: (i) oligos that bind to the barcode for imaging and (ii) oligos that remove any previous barcode signal. These oligos are referred to as the ‘readout’ probe and the ‘strand-displacement’ oligo, respectively (see https://github.com/BoettigerLab/ORCA-public/tree/master/probe-building for readout and strand-displacement sequences). The 30mer strand-displacement oligos are designed first, and the 20mer readout sequences are designed afterward to be complementary to these sequences. When a strand-displacement oligo is introduced, it will bind to its complementary readout sequence, which will result in the removal of the readout probe from the barcode. In addition, strand-displacement sequences should not bind to the genome and should not form hairpin structures.
Primary probes are synthesized (Fig. 2, Steps 2–30) from the oligopool of probe templates by using non-saturating PCR to isolate and amplify the sequences, and are further amplified to high concentration probes through in vitro transcription. A reverse transcription (RT) reaction converts the RNA product to single-stranded DNA (ssDNA). After a final cleanup, DNA can be stored at 4 °C or frozen until needed for hybridization. This protocol produces a high concentration of ssDNA products while requiring only small-volume reactions38, which lowers costs (Supplementary Data 1).
Because thin samples allow for better detection of fluorescent oligos, which bind to the readout probes, fixed Drosophila embryos (Steps 31–49) should first be cryosectioned at 6–10 μm. These tissue sections are adhered to positively charged cover glasses before labeling (Fig. 2, Steps 50–75). Cultured cells may be grown on cover glasses or plated on cover glasses from suspended cells and fixed (Steps 76–78).
For combining RNA with DNA measurements, RNA labeling and imaging (Steps 79–143) are conducted before DNA labeling (Steps 144–166) because the latter involves genomic DNA denaturation (Fig. 2). Performing RNA FISH first prevents cross-talk of RNA probes binding their genomic DNA complements. RNA labeling begins with a brief membrane permeabilization followed by probe and target RNA denaturation on a hot plate. Heat denaturing facilitates probe binding and alleviates the need for protease treatments to remove proteins that can mask RNA binding39. The sample is hybridized overnight in an enclosed humidified chamber. Before imaging, a 96-well plate containing fluorescently labeled readout probes that bind to unique barcodes of the primary probe is placed into our homebuilt robotic fluidics system (Steps 95–103 and 167). The first well contains a Cy3-labeled fiducial oligo, which binds to the fiducial sequence in all primary probes. In addition, all wells—with the exception of the first one—contain the strand-displacement oligo that will remove the previous readout oligo. The sample is mounted in a flow chamber (Steps 104–113 and 168) connected to a robotic fluidics system for sequential readout probe hybridizations (see Supplementary Data 2 for information regarding the cost of each round of readout probe hybridization). Once the flow chamber is secured to a motorized stage on the microscope, the fields of view (FOVs) are manually chosen for imaging. Both sequential readout hybridization and barcode imaging are performed using automated software (Steps 114–143; Supplementary Data 3 has details of imaging time). See the Troubleshooting table (for Step 142) for our recommendations on validating RNA labeling.
After imaging of RNA species, the sample is removed from the microscope, and genomic DNA is hybridized with DNA probes (Fig. 2, Steps 144–166). The sample may be treated with RNase after RNA imaging and before DNA hybridization to avoid cross-hybridization; however, we find this is not essential if only sense-strand probes are made. Although DNA labeling is similar to the procedure for RNA labeling, it has an additional and crucial genomic DNA denaturation step, which is induced by addition of HCl after permeabilization and before heating. In addition, after overnight incubation, the sample is post-fixed in glutaraldehyde solution for further stabilization of 3D genome structure. The FOVs selected for imaging are the same as those that were previously imaged for RNA species. Sequential readout hybridization and barcode imaging are also performed using the automated software (Steps 169–198). See Supplementary Data 3 for details on how long it takes to complete an imaging experiment.
Additional counterstains, such as DAPI, nuclear lamina labeling or other antibody staining, may be added at any point after adhering the tissue/cells onto the coverslip, as long as the associated fluorescent probes do not overlap spectrally with those being used for imaging the readout probes. The efficiency of antibody labeling after RNA and/or DNA sample preparation and labeling is antigen specific; some antigens denature during the hybridization process. In general, adding and imaging counterstains before RNA/DNA labeling will provide the best signal, whereas imaging in parallel with the fiducial label (Steps 143 and 198) will provide the best registration.
The raw images are analyzed using our freely available graphical user interface (GUI)–based tools written in MATLAB, which use an ‘Experimental Table’ to distinguish between fiducial and readout data images (Fig. 2, Steps 199–233). Images are first corrected for x-y stage drift, which occurs between imaging rounds because of microscope stage movement, using Cy3 fiducial oligo data. The fiducial signal is used for registration purposes. Individual nuclei are cropped using supervised, automated segmentation and then further corrected for sub-pixel drift in the 3D image volume using fiducial data. Then, the images of each DNA barcode are fitted in x, y and z using a 3D-Gaussian spot-fitting algorithm. The nanoscale 3D positions of each barcode are recorded in a data table. This process occurs for every labeled nucleus throughout all FOVs. The degree of unspecific background signal is sample and probe set dependent. It is recommended that the user manually inspect the data and the quality of fits for a randomly selected subset of labeled chromosomes to evaluate the potential for fitting errors. We provide a GUI system with instructions to facilitate this process (see ChrTracer3—image analysis of DNA structure in the Procedure). See Supplementary Data 3 for details about the time it takes to complete the analysis of the raw images.
Mosaics are created for both RNA and DNA data where FOV-to-FOV drift is corrected using fiducial-labeled nuclei that overlap between adjacent FOVs (Fig. 2, Steps 235–255). The RNA and DNA mosaics are aligned on top of one another using the Cy3 fiducial oligo signal and the autofluorescence of the sample (Fig. 2, Steps 256–275). Lastly, once RNA and DNA information has been compiled into a single mosaic, regions of interest can be manually selected to analyze chromatin structure for a subset of cells, and RNA transcripts can be counted (Fig. 2, Steps 276–301). The user may also wish to consult various detailed protocols on RNA transcript counting28,32,40–45.
We recommend that first-time users start with a downscaled experiment when learning the approach. For example, we recommend the use of adherent cells before using cryosectioned tissue samples. In addition, users can first start with a smaller number of barcodes when targeting RNA species and/or DNA—such as 5 for RNAs and 20 for DNA—when adopting the method, and then scale up to more complex experiments (e.g., 50–100 barcodes for DNA). For practical time considerations, we typically aim for ≤100 total barcodes in a DNA labeling experiment (Supplementary Data 3). For DNA labeling, the number of barcodes informs the choice of resolution—e.g., a 1-Mb domain can be imaged at 10-kb resolution with 100 barcodes, or a 200-kb domain at 2-kb resolution. To simplify the experiment, the user should image only 5–10 FOVs. With these recommendations, the user should take 1.5 d to synthesize primary probes, 3.5 d to hybridize and image RNA and DNA and <3 d to analyze the images. Thus, this starting experiment should take ~1 week. It is also advisable to try RNA and DNA labeling as separate experiments when learning the method before combining both on the same sample. Lastly, we recommend that users attempt their first experiment from our validated primary probe list found here: https://bit.ly/3fdwBx0. We have included fasta files of primary probes targeting RNA and DNA that have worked previously in Drosophila embryos and mammalian cultured cells25,46,47.
Comparison to other methods
Bulk methods for analysis of chromatin structure, including chromosome conformation capture methods such as Hi-C (Fig. 3a) or ligation-free alternatives such as split-pool recognition of interactions by tag extension48 and genome architecture mapping49, have the ability to provide a genome-wide view of chromosome organization50. However, these methods do not capture variation at the single-cell level. In contrast, ORCA provides access to high-resolution structural information from single cells for targeted genomic regions (Fig. 3b). In addition, because ORCA is compatible with RNA labeling, we have the ability to measure important co-localized features, such as transcript levels and cell position within a tissue, as well as the ability to directly connect these cell identity features with genome structure. Although single-cell Hi-C (scHi-C) combines whole genome coverage with cellular resolution51–56, it loses access to these important cellular features that may drive variation in structure (Fig. 3b). In addition, scHi-C provides significantly lower genomic resolution of chromatin features compared to Hi-C or ORCA (Fig. 3a,b). This limitation arises not only from limited sample recovery, which might be improved, but also from inherent limitations from ligation-based proximity assays. In the dense nuclear environment, multiple sequences are probably within ligation distance of each restriction site, yet only one pair may ligate, intrinsically losing multi-way contact information and reducing the resolution of the data. Restraint-based modeling has been used to augment the resolution of scHi-C52,55 but requires additional assumptions about the typical physical extent of the chromatin, which ORCA does not.
Fig. 3 |. Comparison between ORCA and other methods.
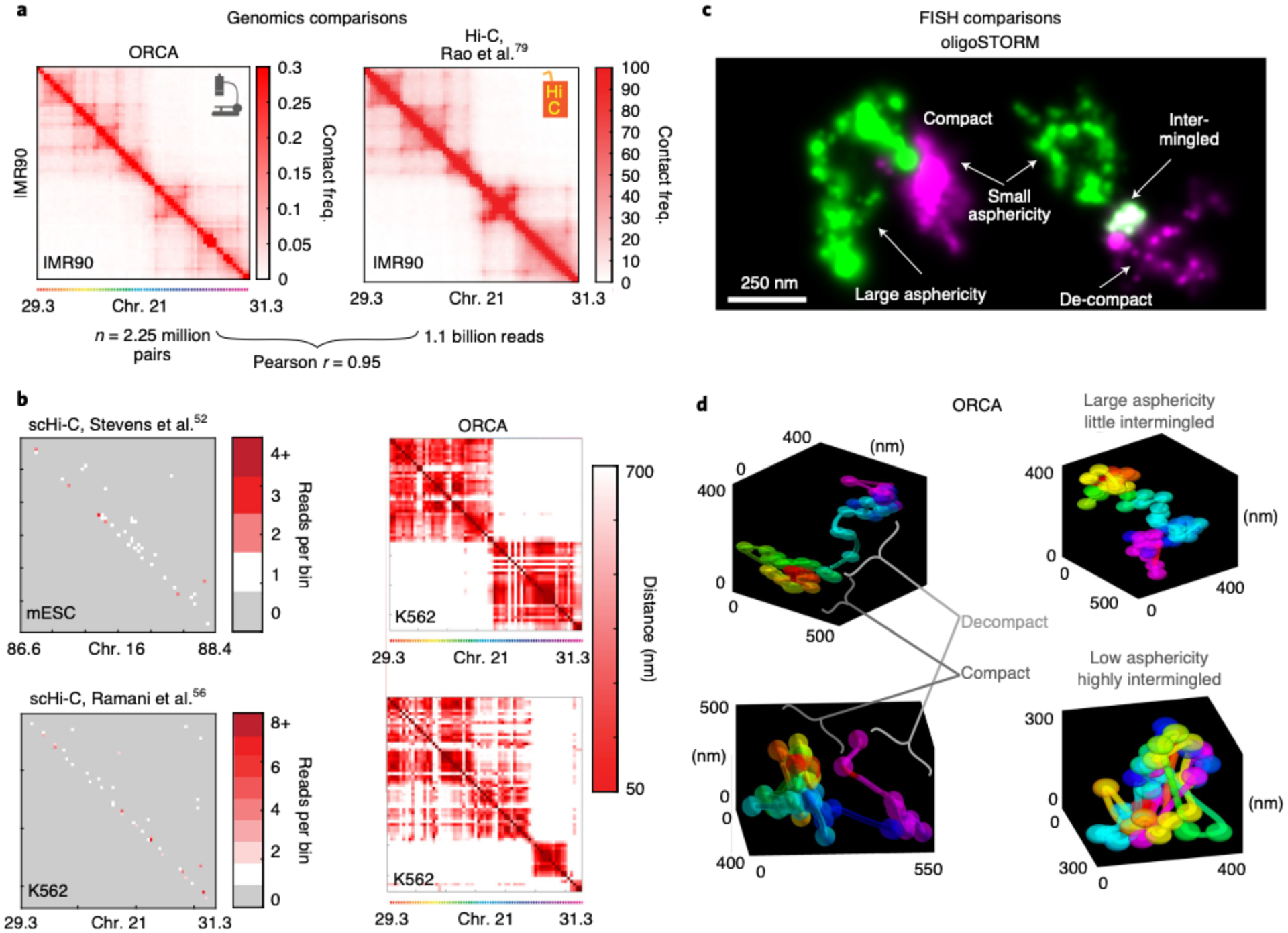
a, Comparison of ORCA generated average contact frequency map46 to a Hi-C generated map79, for a 2 Mb region from IMR90 human fibroblasts. b, Comparison of maps from individual cells imaged by ORCA46 or by single cell Hi-C52,56 for syntenic genomic intervals for the indicated cell lines. For the scHi-C data, the cell with the most total reads in the dataset is shown. Missed detection events are grey in the scHi-C maps and appear as white stripes in the ORCA maps. Please refer to Step 234 in the Troubleshooting table for potential reasons why these stripes are present in the single-cell ORCA distance maps. c, oligoSTORM resolves chromatin structural ‘shape properties’ below the diffraction limit, such as asphericity and compaction. (Left) 150 kb H3K4me2-rich chromatin (green) and ~150kb of adjacent H3K27me3 rich chromatin (magenta), (chr3R:12.65–12.95 Mb). (Right) Two adjacent regions of H3K4me2-rich chromatin in green and magenta (chr2R:19.7–19.9 Mb), images from Drosophila KC167 cells9. d, ORCA allows both the 3D features identified from STORM to be quantified and provides additional information about the path of the chromatin fiber. Examples are from BX-C (chr3R:12.5–12.8 Mb) in different cells of a Drosophila embryo, which are part of our previously published data25.
Compared to using traditional super-resolution approaches to image Oligopaint-labeled chromatin, such as sequential STORM6,46,57 or DNA-PAINT8,58,59 (Fig. 3c), ORCA provides a 3D polymer path rather than a ‘cloud’ of points arising from single-fluorophore localization events (Fig. 3d)38. The steps in this polymer path represent the centroid of multiple fluorophores, all concentrated within a small region of the nucleus (e.g., a specific 2-kb genomic segment), rather than the centroid of individual fluorophores detected through STORM. Adjacent ‘steps’ along the genome are resolved in physical space at better than diffraction limit because they are sequentially detected, much like the sequential detection of adjacent fluorophores in STORM and DNA-PAINT. Moreover, because detection events are not stochastic, each detection event contains information about genomic sequence identity as well as the physical location (Fig. 3c,d).
Because sequential hybridization is much slower than stochastic switching of fluorophores, it takes much longer to reconstruct the image of a domain (days compared to minutes). However, ORCA has the capability to image an abundant number of cells because of its compatibility with large illumination volumes and large detectors. Thus, ORCA allows for the processing of chromosome paths from many cells in parallel, enabling us to access much larger cell numbers than we have with STORM9,25. Data from both ORCA and STORM/DNA-PAINT can resolve ‘shape-properties’, such as density/compaction, eccentricity and the overlap of domains (Fig. 3c)2,9–11,20,60. Mapping sequence-specific features, such as topologically associated domains (TADs) or enhancer-promoter loops, requires the sequence specificity provided by the sequential labeling of barcodes. Notably, sequential labeling of barcodes has also been used in conjunction with STORM46,57, providing access to shape-properties within each barcode and genomic resolution at the scale of the barcoding (typically, 30–5,000 kb). Individual barcodes can be imaged much faster in ORCA because movies of blinking fluorophores are not required. In contrast, STORM images require ~100-fold more images to ensure sparse localization9,35,46,57, which increases the challenges and costs for data storage and analysis.
In ORCA, the resolution is limited by the genomic step size and precision with which the barcode label can be imaged and aligned with the rest of the domain. We have successfully imaged tiles with step sizes as fine as 2 kb and as early as blastoderm stage Drosophila nuclei. Because of our strand-displacement strategy to remove the signal from the previously imaged barcode, rather than purely photobleaching, we can re-label the same barcode. By re-imaging a subset of barcodes at the end of the ORCA experiment, it is also possible to measure the localization precision and resolution of the experiment. We find the resolution typically to be ~20–35 nm in x/y and ~40–60 nm in z25, similar to our results from the centroids of features measured with STORM46. Overall, ORCA allows for faster imaging per genomic step but measures only the centroid of this step. The approach achieves super-resolution only when the barcodes are adjacently tiling short genomic regions (e.g., kilobases, not 100s of kilobases), and when drift and any chromatic aberrations have also been corrected to the nanoscale. Alternatively, larger probes spanning larger gaps of DNA have been effectively used to map chromosome structure at large length scales, with correspondingly reduced resolution, including entire chromosomes with sequential labeling61 or in separate multicolor experiments spanning a significant portion of the genome62.
Many of these advantages of ORCA are captured by a conceptually similar approach developed in parallel called ‘high- throughput, high-resolution, high-coverage microscopy-based technology’ (Hi-M)63,64. This approach covered larger genomic distances with fewer probes (targeting ~21 genomic loci) by using variable-sized gaps between the probes. Avoiding these variable gaps in ORCA achieves a higher resolution view of the chromatin path that is more robust to the stochastic missed detection events. Regular genomic spacing between barcodes rather than variable spacing also facilitates interpretation and direct comparison with Hi-C data. In addition, the use of strand-displacement probes in ORCA, in place of bleaching or fluorophore-cleavage in other sequential FISH studies27,28,34,61,63,65,66, allows individual signals to be repeatedly extinguished and then relabeled.
The ability to relabel the same genomic region at different times throughout the experiment (or repeat the entire experiment on the same cell if desired) provides a valuable control for whether sample chromatin structure changes during imaging and allows for accurate quantification of the resolution. Note that an initial photobleaching step before fluorescent labeling is still helpful to reduce auto-fluorescent background, and a rapid photobleaching between imaging different barcodes helps suppress the return of autofluorescence without extinguishing readout signal. The use of readout sequences instead of directly labeled barcodes25,46 dramatically reduces the experimental cost for ORCA compared to Hi-M. Similar readouts or ‘bridges’ have also been used in STORM57. In contrast to the approach described here using 6-μm cryosections, the Hi-M protocol demonstrated robust detection in the epithelial nuclei of whole Drosophila embryos (~10-μm layer around an ~100-μm embryo), overcoming the substantial background and signal aberrations associated with thicker samples.
Simultaneous labeling of DNA and RNA is historically difficult because of different hybridization conditions required for labeling67,68 and the potential for RNA probes to also bind the complementary DNA sequences from which they were transcribed. Some prior protocols have addressed these challenges by compromising hybridization conditions to find a good balance to allow labeling of abundant RNAs (e.g., XIST, a highly expressed noncoding RNA involved in X-silencing) and repetitive DNA33,67,69. Other groups cross-link RNA probes with amines or antibodies before DNA hybridization or imaging, which also works best with abundant transcripts63,64,68.
Our approach circumvents the compromise in simultaneous RNA-DNA labeling through sequential rounds of labeling and imaging, similar to early multi-label methods70. RNA is labeled first under hybridization conditions optimal for RNA labeling that do not denature the DNA helix and expose it to hybridization. Multiple RNA species can be labeled at once and distinguished through unique barcodes25 or combinations of barcodes28. After the imaging is complete, the sample is then removed from the microscope, and probes are hybridized to the DNA in an optimal DNA hybridization condition. Distinct barcodes are used with DNA and RNA labeling. The use of fiducial markers and automated alignment software minimizes the registration problem that made earlier implementations of a sequential imaging approach challenging70. Successful avoidance of cross-talk is most easily assessed with intronic RNA probes targeting nascent transcripts and when the sample contains distinct cell populations that do and do not express the target gene. Bright puncta should be detected only in a subset of the cells that express the target, and none of the cells that do not (Extended Data Fig. 1). DNA probes target the sense strand to avoid potential hybridization to RNA targets, which can be validated with RNase treatment.
Limitations
Labeling of DNA with primary probes requires denaturation of the DNA double helix. To achieve robust probe hybridization, DNA must be sufficiently denatured through separate treatments with acid, formamide and heat. These conditions also denature nuclear proteins. Consequently, at the molecular scale, chemical structures have been disrupted. It is not well understood to what extent structural features at the super-resolution scales measured by ORCA (tens to hundreds of nanometers) are also disrupted by these denaturing conditions, because we cannot yet directly image and measure the structural features seen in ORCA without denaturation. Notably, some features that can be measured by other methods, such as the boundaries of TADs and the relative pairwise contact frequencies among domains, agree quantitatively with non-denaturing methods such as Hi-C25,46. Importantly, denaturation is carried out in a densely cross-linked protein gel created by fixation before perturbation. Fixation will limit the mobility of the denatured proteins and DNA strands, and as long as they remain confined within the resolution limit of imaging, this perturbation will not significantly affect the nanoscale measurement of chromatin structure. Emerging alternatives to acid, formamide and heat, such as Cas9-mediated DNA targeting71 or the use of exonucleases to remove one strand16, may reduce the degree of perturbation.
We look forward to further improvements in this area. Ideally, such methods should be tested for sequences and chromatin state bias9,25. Furthermore, existing and emerging alternative methods should be tested for their effect on the kilobase-scale chromatin path by measuring it before and after treatment. This requires the ability for reliable super-resolution imaging of the path of DNA before and after denaturation, which to our knowledge has not been achieved, although some creative proxies have been used72. Improvements in correlative light and electron microscopy and in super-resolution live imaging will probably improve our understanding of these perturbations and the ways they can be minimized in the future.
Denaturation creates several additional limitations to ORCA. If DAPI and/or fluorescent dyes are introduced to the sample before ORCA, the signal intensity of these additional labels is notably reduced after ORCA. Because our protocol involves acid and heat treatment of the sample, many antigens may unfold and reduce or disrupt recognition by antibodies (although some chromatin-associated proteins are compatible with DNA FISH). Examples include geminin, a cell cycle marker46; fibrillarin, a nucleolar protein; and SC35, a splicing factor associated with nuclear speckles73.
Failure to accurately correct drift, contamination of readout probes or improper dilution and mixing of readout oligos or strand-displacement oligos can lead to artifacts in the data. With proper controls and experience, such artifacts can be readily detected, as shown in the Troubleshooting table.
Raw data require considerable storage. For example, a single experiment with ~100 FOVs (each ~200 × 200 μm) using 100 barcodes and three barcode replicates requires >16 TB of storage (Supplementary Data 4). Large storage systems can be a challenge to maintain, and storage costs are easily in the range of two orders of magnitude greater than the cost of the chemical reagents. As with the analysis of the raw imaging data from Illumina sequencing, processed data result in significant compression, providing an avenue by which storage costs might be reduced in the future by limiting retention of image data after processing. Read/write operations on large datasets are also the primary limitation to the speed of data analysis. Consequently, processing data from external hard drives connected by USB to the analysis computer should be avoided, because USB achieves a slower read-write speed than internal serial ATA (SATA) or external high-speed network attached storage systems.
Another important limitation is the detection efficiency for individual barcodes, which affects both RNA and DNA labeling. Because of the stochastic nature of probe binding and photon emission, among other effects, individual genomic steps along the ORCA trajectory are not detected in some cells. For the barcodes provided in our GitHub repository (https://github.com/BoettigerLab/ORCA-public/tree/master/probe-building), we find that the efficiency is largely uniform across the sequences used, and shows no systematic variation with chromatin state or cell type in the domains that we examined (Extended Data Fig. 2). Typically, detection efficiency per barcode is in the 60–90% range, with the highest efficiency observed for larger step sizes and lower efficiency for smaller step sizes, as expected. This dropout rate has a minor effect on the number of cells one needs to analyze in a population to compute cell type–specific contact frequencies and identify cell type–specific boundaries, but does not otherwise impede these measurements. Similarly, the three-way or four-way interaction frequencies can be reliably measured with this barcode efficiency range46. Care must be taken in the analysis that the missing data are excluded and not erroneously treated as a non-contact event. The analysis routines discussed below in the Procedure (see also Code availability) implement handling of missing data. An exciting future direction will be the development of improved approaches to impute some of the missed barcode positions from the collected data. Particularly for small genomic step sizes, there is a high correlation in the 3D position of adjacent sequences, which could be used to resolve otherwise ambiguous detection events that are discarded and counted as missed in the current approach.
Applications
The current ORCA protocol is suitable for both cryosectioned tissues and cultured cells, including adherent and suspension cell lines and mouse embryonic stem cell spheroids25,46.
General considerations
This protocol details the approach originally described in our earlier report25, in which we use ORCA to image the 3D DNA structure in nuclei of cryosectioned Drosophila embryos in combination with multiplexed RNA labeling. We include several updates reflecting recent improvements. These include adjustments for reading multiple barcodes per hybridization with spectrally distinct fluorophores and a wholly rebuilt pipeline for processing mosaic data made from overlays of multiple FOVs. We provide resources to guide users to build a robotic fluidics system to assist with sequential labeling of barcodes and a list of parts to replicate the optics system that we used in these experiments (Tables 1 and 2 and Supplementary Data 5). Manual pipetting can substitute for robotic pipetting and may be preferable for troubleshooting or for users with a small number of barcodes. We provide step-by-step instructions and recommendations for both wet laboratory and computational procedures. Our protocol is written for researchers with experience in microscopy and minimal computational skills. We provide MATLAB scripts that will be necessary to process all raw data (https://github.com/BoettigerLab/ORCA-public) along with a basic walkthrough of how to use this software. Sample data for testing the software are provided at https://bit.ly/2S6eCjk.
Table 1 |.
Details of the components used for the homebuilt fluidics system
| Supplier | Part number | Quantity | Unit | Cost ($) | Description |
|---|---|---|---|---|---|
| Hamilton | 7750–16 | 1 | 6/pk | 71.00 | 25-gauge, Kel-F Hub needle, custom length (5 inches), point style 3, 6/pk |
| Gilson | F155006 | 1 | Each | 2,438.00 | Peristaltic pump |
| Gilson | 361832 | 1 | Each | 436.00 | 361832 361832: 508 interface module, 110–220 V |
| Gilson | F117933 | 1 | Pk-12 | 41.00 | PVC manifold tubing, 0.38-mm i.d. (0.015 inches), orange/green collars |
| Bioptechs | 03060319–2-NH | 1 | Set | 2,686.86 | FCS2 chamber flow cell |
| Bioptechs | 060319-2-2611 | 1 | Each | 970.26 | Zeiss (K), Marzhauser, ASI, Ludl & prior adapter |
| Bioptechs | 1907–750 | 0 | Set-5 | 37.32 | Blank gasket cut using die# 443792, 0.75-mm thick—5/pack—1907–250 |
| Bioptechs | 40-1313-03192 | 1 | 500 pk | 522.00 | 40-mm coverslips |
| Amazon | N/A | 1 | Each | 22.95 | Keyspan by Tripp Lite USA-19HS high-speed USB serial adapter, PC, MAC, supports Cisco Break Sequence |
| Amazon | N/A | 1 | Each | 9.99 | Anker ultra slim 4-port USB 3.0 data hub |
| Amazon | N/A | 1 | Each | 3.45 | StarTech 6-feet straight through serial cable - DB9 F/F (MXT100FF) |
| Amazon | N/A | 3 | Each | 5.79 | AmazonBasics USB 3.0 extension cable—a-male to a-female—9.8 feet |
| Amazon | N/A | 1 | 2-pack | 8.99 | CableCreation (2-pack) 3 feet 18 AWG universal power cord for NEMA 5–15P to IEC320C13 cable, 0.915M/black |
| McMaster | 5583k52 | 0 | Each | 212.00 | Abrasion-resistant white ETFE tubing, 0.020 inch i.d., 1/16 inch o.d., 50 feet |
| eBay | CNC3040T | 1 | Each | 445.01 | 3 aXIS USB router engraver engraving drilling milling machine, 300 × 400 mm |
AWG, American wire gauge; N/A, not applicable; NEMA, National Electrical Manufacturers Association; pk, pack; PVC, polyvinyl chloride.
Table 2 |.
Overview of components used for our homebuilt microscope
| Item | Cost ($) |
|---|---|
| Optics table and setup supplies | 14,163.80 |
| Lasers (405, 488, 560, 647 and 750 nm) | 82,796.99 |
| Laser optics for combing beams | 10,534.50 |
| CMOS camera | 19,475.00 |
| Microscope body and objectives | 20,737.20 |
| Microscope stage | 30,852.20 |
| AOTF | 5,911.46 |
| Focus lock | 7,063.08 |
| Laser-proof enclosure | 691.27 |
CMOS, complementary metal oxide semiconductor. Further information, including supplier(s), part number(s), quantity and units, can be found in Supplementary Data 5.
Fluidics setup
Typically, an ORCA experiment can last for 2–3 d and require hundreds of fluid exchanges to the sample; thus, we built a homebuilt robotic system to handle the fluidic exchanges (Fig. 4). The fluidics system consists of a Bioptechs FCS2 fluidics chamber (which we refer to as the sample flow chamber), a Gilson Minipuls 3 peristaltic pump and a commercially available computer numerical control (CNC) mill, which is modified into a robotic pipette to support fluidics operations (Fig. 4). We developed a set of open source Python libraries that communicate with the USB controller to replace the commercial CNC software, allowing easy custom control and future development (additional details are included in the fluidics Github repository (https://github.com/BoettigerLab/fluidics-control; see Code availability). All necessary buffers, including the 2x SSC (bleach buffer) and the 30% (vol/vol) formamide in 2x SSC (wash buffer), 0.5 mg/ml glucose oxidase, 40 μg/ml catalase and 10% (wt/vol) glucose in 2x SSC (imaging buffer)74,75, and the 96-well plate holder are mounted on the CNC platform (Fig. 4). The mill motor was replaced with a hypodermic needle (Hamilton, 7750–16), which is used for extracting specific readout probes and strand-displacement oligos from each well of the 96-well plate (per hybridization round) and from buffer stock solutions for the intermediate steps. Together, these solutions are individually flowed using an ethylene tetrafluoroethylene (ETFE) plastic tubing that allows solutions to flow over the FCS2 chamber by using a peristaltic pump. The timing and flow rate are programmed by using our custom software written in Python (‘Kilroy’), which is adapted from ref.28. Components of the fluidic system are listed in Table 1.
Fig. 4 |. Fluidics system layout.
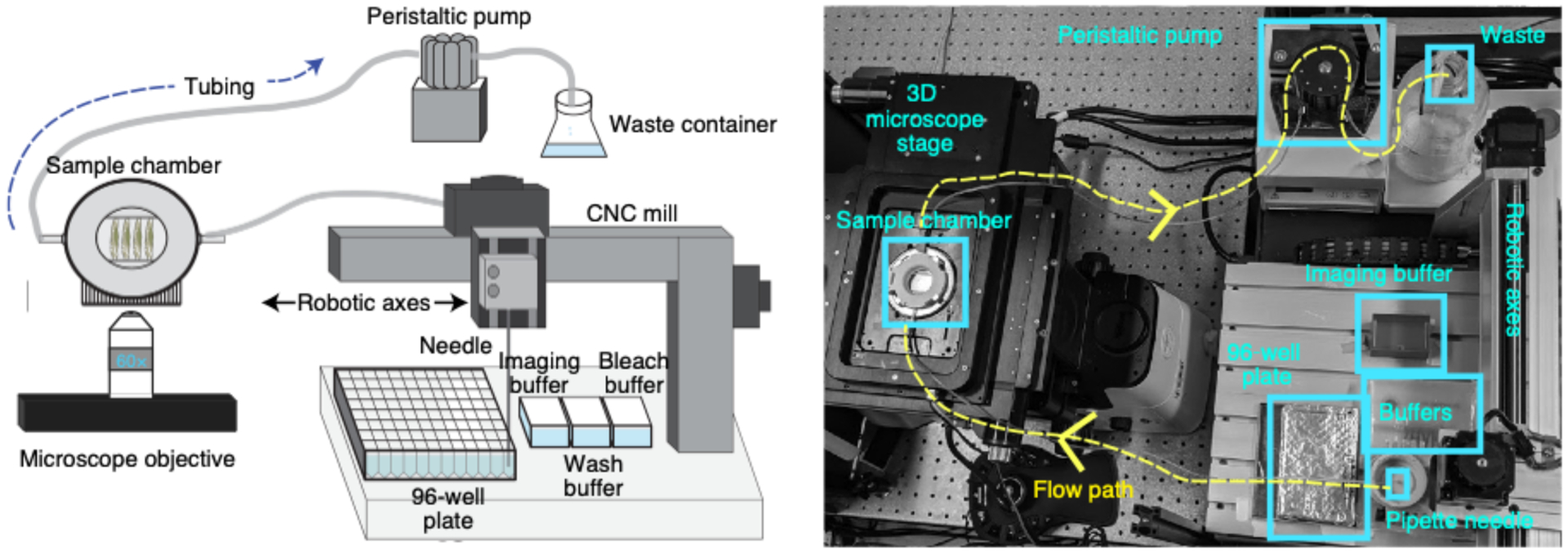
Schematic (left) and a photograph (right) of the homebuilt robotic system used to hybridize readout probes onto barcodes, and exchange bleach, wash, and imaging buffers onto the sample. Components of the fluidic system are labeled, and further information about these parts can be found in Table 1.
Microscope setup
Single-molecule FISH requires a high-resolution, low-noise imaging system to obtain high-quality data. Essential components include a low-noise, high-sensitivity camera for single-molecule imaging (e.g., Hamamatsu’s ORCA-FLASH4.0); high-intensity, controllable laser illumination, with corresponding dichroic mirrors; and an auto-focusing system, to actively correct for focus drift and reset the focus after each hybridization round or stage movement (Table 2 and Supplementary Data 5). The layout of the components used for our homebuilt microscope is shown schematically in Fig. 5a. We anticipate that various commercial systems could also be adapted for this imaging process, because similar components are available on many systems intended for single-molecule imaging and particle tracking. We have not currently tested the complete protocol on commercial instruments. Some adjustments to the software to allow integration of the automated hybridization steps with the imaging steps will be required, a feature we have not found on most commercial systems currently in use, but something we anticipate will be more readily available in the future. Adjustment of automated focusing systems to be compatible with the range of buffers used in this protocol may also be required.
Fig. 5 |. Homebuilt microscope.
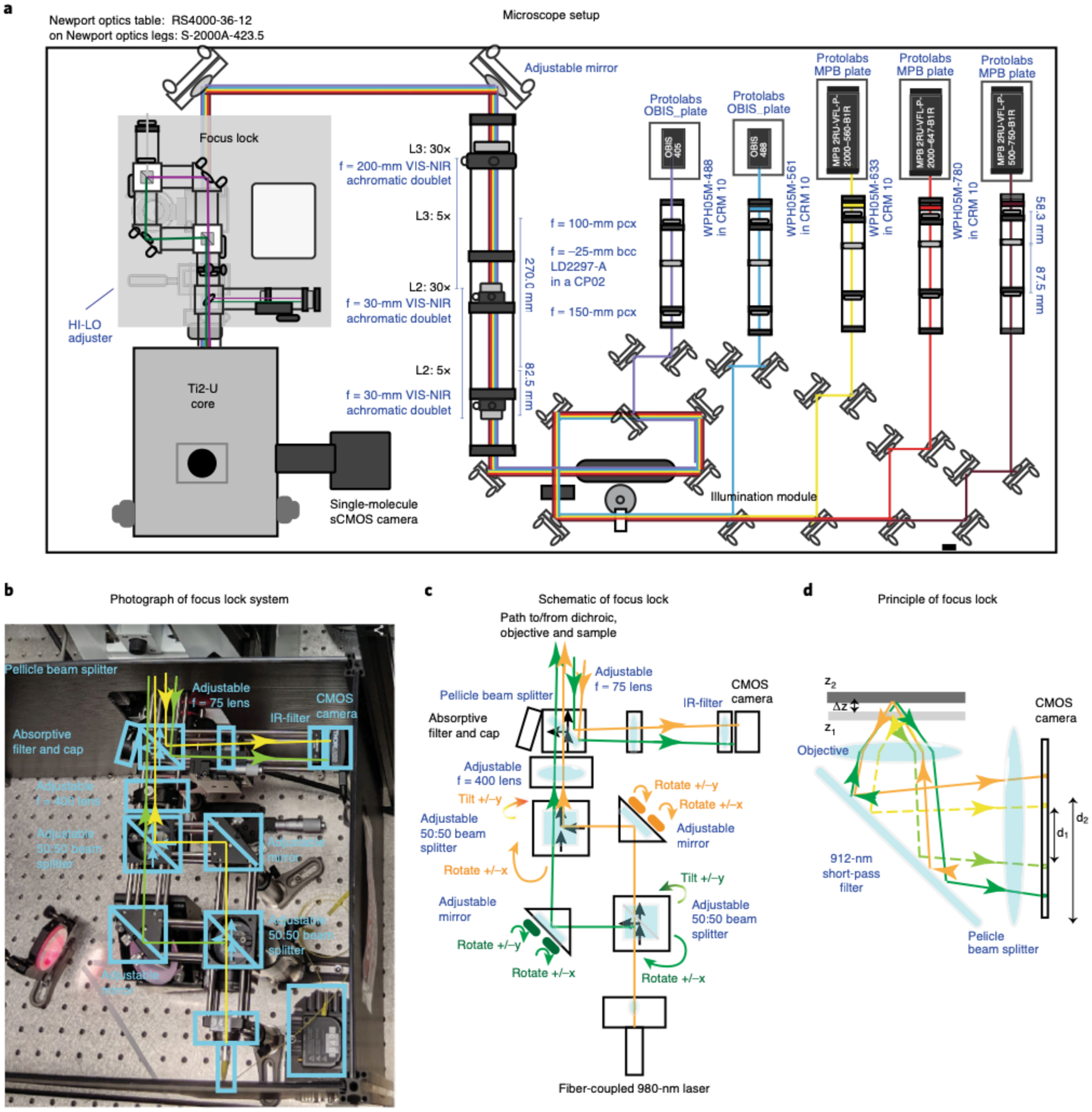
a, Schematic layout of the custom-built microscope with components labeled (information about the parts can be found in Supplementary Data 5). The 405-nm, 488-nm, 560-nm, 647-nm, and 750-nm laser paths are illustrated. The laser beams were combined using dichroics and directed based on custom polychroic mirrors placed on top of a high-performance optical table. b, Photograph of the custom Focus Lock system showing the parts used to properly align the beam path. The names of the key optical components are labeled. The beam path for the two 980-nm beams are illustrated in yellow and green. For simplicity, unused branches at the beam splitters are not shown. c, Schematic illustration of the beam path for the Focus Lock. All optical components are indicated. The green and orange paths trace the split 980-nm laser beam. The controls for separately adjusting the two beam paths are indicated in matching colors. d, Schematic illustration of the operating mechanism of the Focus Lock: how a small displacement in z is projected into a large displacement in the separation of the reflected laser beam. Dotted line and solid line show alternate paths for two different focal planes. Closed-loop active feedback is achieved with a precision piezo-z stage to maintain the distance between the two reflected beams.
Lasers
Several high-power solid-state lasers provide the illumination for the system. These are combined into a common beampath with dichroic mirrors and passed through an adjustable, custom beam expander as shown in Fig. 5a (‘illumination module’). The 2-W 561-nm laser is primarily used for imaging the fiducial signal. The 2-W 647-nm and 500-mW 750-nm lasers are used for imaging barcodes. Lower power (<200 mW) 488-nm and 405-nm lasers are used for imaging of additional counterstains, such as antibody-labeled nuclear lamina and DAPI, respectively, although the 488-nm laser can be used for imaging the fiducial signal. Although fiducial and barcode imaging are not restricted to a particular laser, it is not recommended to use the 488-nm and 405-nm wavelengths for imaging ORCA barcodes, because high-frequency lasers can photocleave DNA. Laser illumination is provided as a highly inclined and laminated optical setup76,77 to reduce out-of-focus background. An acoustic optical tunable filter (AOTF) is used to selectively deflect the laser beams to enable rapid switching and power tuning of the individual beams (Fig. 5a). A standard Nikon Ti2-U microscope core is used to mount the dichroic filters, tube lens and high-resolution piezo stage (Fig. 5a, ‘Ti2-U core’).
Camera
We tested camera pixel sizes from ~100 nm to 150 nm with comparable results. For samples with a good signal-to-noise ratio, the smaller pixel size provides a slight boost in localization precision while somewhat decreasing overall throughput due to a smaller total FOV. Typical photon counts per pixel for labeled foci are in the 300–5,000 range, with a signal over background of 1.5–10, depending on the label size and fluorophore. The analysis software is robust to this substantial variation in absolute signal strength and signal-to-noise ratio, the effects of which are recorded in the computed uncertainty estimates.
Motorized stages enable automation
The use of fiducial signals in the experiment provides ~25-nm accuracy in x, y drift correction25, minimizing also the need for a highly accurate mechanical x, y stage. This could be used to reduce cost in the future by purchasing a lower cost x, y platform. Z-scanning is conducted in 100-nm steps by using a fast piezo stage, which is also used for active axial drift correction as described below. The resulting 3D point spread function will be computationally processed to identify its centroid in 3D in downstream analyses (Steps 221 and 233). We found that comparable results could be obtained with either a piezo stage or piezo objective positioner. Additional details of the parameters of image acquisition can be found in the parameter files at https://bit.ly/2FlYQOc (this is a link to the ‘Settings’ folder of the mini dataset that we provide) and the documentation included for the microscope software discussed below.
Focus lock assembly and calibration
To maintain focus during imaging acquisition and between successive rounds of barcode labeling and x-y–stage movement, we built a custom focus lock system, modified from earlier work7 (Fig. 5b–d). An infrared (980-nm) laser beam is split into two paths with a beam-splitter cube. Both paths are directed to the sample and reflected off the coverslip at a converging angle (Fig. 5d). The reflected beam is directed back by a dichroic mirror and directed to a small CMOS camera by using a pellicle beam splitter. This beam splitter allows the original incident beam to reach the sample and the returning beam to reach the CMOS without any change in the wavelength. The 50% loss of power at each pass is not an obstacle because directly imaging the reflected laser provides plenty of power. The pellicle beam splitter reduces secondary reflections that can otherwise interfere with the focus lock.
The position at which the reflected beam strikes the camera is dependent on the beam path length (and thus focal position), which is robustly read out in the separation between where the two beams hit the camera (Fig. 5d). A simple algorithm computes this separation and activates a piezo stage if the difference is greater or less than the value set by the lock. Because of the rapid response of the piezo stage and sensitivity of the beam position to nanoscale displacements, this closed feedback loop provides robust stabilization across the ~10-μm focal range of the instrument. To align or adjust the lock, each beam passes through two mechanically controlled reflective surfaces (one mirror and one right angle surface in a beam splitter), allowing both beams to be independently adjusted (Fig. 5c). A 400-mm lens focuses the beams onto the backplane of the objective, and a 75-mm lens controls the angle at which the beams intersect the camera (Fig. 5c).
Microscope software
Lasers and the camera are controlled by using the ‘storm-control’ software package written by Hazen Babcock and Xiaowei Zhuang’s laboratory78, with modifications (https://github.com/BoettigerLab/storm-control). This software provides an intuitive GUI control system and Python wrappers to integrate and control many widely used optical components, including various motorized stages, piezo stages, AOTFs, laser systems and cameras. The software also supports timing integration and communicates with the fluidics control software. Directions for basic installation and troubleshooting are included in the software. The program named ‘Hal’ allows interactive control of the microscope, including moving the stage, setting the laser power, changing the image acquisition parameters, such as exposure time or z-scan depth, and recording images. The program named ‘Steve’ directs Hal to record images in tiled mosaic, which is useful for reconstructing a larger field of view. The program ‘Kilroy’ controls the fluidics system, including choosing which well/buffer the needle goes to and when and at what speed the pump works. The program named ‘Dave’ passes pre-written image acquisition protocols to Hal and prewritten fluidics control programs to Kilroy. Detailed instructions for using the software to set up an automated experiment are provided in the protocol below (Steps 117–120 and 125–142).
Considerations about the laboratory facilities
All the steps in our protocol can be done in a standard laboratory. We advise you to have an RNase-free table to generate the primary probes. We also advise having a dedicated microscope-fluidics system in a separate room.
Materials
Biological materials
Canton-S Drosophila wild-type strain (Stanford Fly Facility)
IMR90 cell line (American Type Culture Collection, cat. no. CCL-186; RRID: CVCL_0347)
K562 cell line (American Type Culture Collection, cat. no. CCL-243; RRID: CVCL_0004)
Reagents
Probe synthesis
Oligopaint library (CustomArray; see Procedure Step 1 for instructions on how to design the probes targeting RNA and/or DNA)
Ultrapure H2O
Phusion High-Fidelity PCR Master Mix with HF buffer (New England BioLabs, M0531L)
Forward primer (Integrated DNA Technology (IDT); https://github.com/BoettigerLab/ORCA-public/tree/master/probe-building/Oligos/FwdPrimers.fasta)
Reverse primer (IDT; https://github.com/BoettigerLab/ORCA-public/tree/master/probe-building/Oligos/RevPrimers.fasta)
EvaGreen, 20x in water (Fisher Scientific, cat. no. NC0521178)
LE agarose (GeneMate, cat. no. E-3120–500)
Borax (Sigma-Aldrich, cat. no. 221732)
6x Orange DNA loading dye (Thermo Fisher Scientific, cat. no. R0631)
DNA Clean & Concentrator (DCC)-5 kit (Zymo Research, cat. no. D4013)
NTP buffer mix (HiScribe T7 Quick High Yield RNA synthesis kit (New England BioLabs, cat. no. E2050S))
T7 RNA polymerase mix (from HiScribe kit, cat. no. E2050S)
RNasin Plus RNase inhibitor (Fisher Scientific, cat. no. PRN2615)
dNTP (Thermo Fisher, cat. no. N8080260)
RT buffer (Maxima H Minus kit (Thermo Fisher Scientific, cat. no. EP0753))
Unlabeled RT-primer (IDT; sequence: catcaacgccacgatcagct)
Maxima H Minus reverse transcriptase (Thermo Fisher Scientific, cat. no. EP0753)
RNasin Plus RNase inhibitor (Promega, cat. no. N2611)
EDTA, 0.5 M (Fisher, cat. no. AM9262)
NaOH, 1 M (Fisher Scientific, cat. no. BP-359–500)
Ethanol, 100% (vol/vol)
DCC-25 kit (Zymo Research, cat. no. D4006)
DCC-5 kit (Zymo Research, cat. no. D4004)
Oligo-binding buffer (Zymo Research, cat. no. D4060-1-40)
Urea PAGE gel
Urea (Fisher Scientific, U15–3)
Acrylamide:bisacrylamide (19:1) (Fisher Scientific, cat. no. BP1406–1)
Tris-Borate-EDTA (TBE), 10x solution (Fisher Scientific, cat. no. BP1333–1)
ddH2O
Ammonium persulfate (APS), (Sigma, cat. no. A3678)
Tetramethylethylenediamine (TEMED), (Fisher Scientific, cat. no. EC50325ML)
GeneRuler Ultra Low Range DNA Ladder, ready-to-use (Thermo Fisher, cat. no. SM1213)
Urea-loading buffer (Thermo Fisher, cat. no. LC6876)
GelGreen Nucleic Acid Gel Stain, 10,000x in water (Biotium, cat. no. 41005)
Apple juice–agar plate
Apple juice (Amazon, Tropicana apple juice, 10 ounces, 24 count)
Methyl P-hydroxybenzoate (MP Biomedicals, LLC, cat. no. ICN10234101)
Sucrose (Research Products International, cat. no. S24060–1000.0)
Bacto agar (BD Biosciences, cat. no. 214010)
ddH2O
Drosophila egg laying and collection
Yeast (Amazon, Fleischmann’s yeast for bread machines, 4-ounce jars (pack of 2))
Apple juice–agar plate
Commercial Clorox bleach (Fisher Scientific, cat. no. 50371500) !CAUTION Bleach is toxic. It should be handled with gloves and under a fume hood and should be discarded with care and according to your institution’s environmental health and safety rules.
ddH2O
Heptane (Fisher Scientific, cat. no. H3601)
Formaldehyde aqueous solution, 32% (wt/vol) (Fisher Scientific, cat. no. 50980494) !CAUTION Formaldehyde is toxic on inhalation and with skin contact. It should be handled with gloves and under a fume hood. It should be discarded with care and according to your institution’s environmental health and safety rules.
EGTA, 0.5 M (Bio-World, cat. no. NC0997810)
PBS, 10x, pH 7.4 (Fisher Scientific, cat. no. 70011044)
Methanol, 100% (vol/vol) (JT Baker, cat. no. 9070–01) !CAUTION Methanol is both toxic and volatile. Methanol should be handled with gloves and under a fume hood. It should be discarded with care and according to your institution’s environmental health and safety rules.
Drosophila embryo embedding and cryosectioning
Methanol (JT Baker, cat. no. 9070–01) !CAUTION Methanol is both toxic and volatile. Methanol should be handled with gloves and under a fume hood. It should be discarded with care and according to your institution’s environmental health and safety rules.
PBS, 10x, pH 7.4 (Fisher Scientific, cat. no. 70011044)
Sucrose (Research Products International, cat. no. S24060–1000.0)
Cryomold, 15-mm × 15-mm × 5-mm Tissue-Tek Cryomold (VWR, cat. no. 25608–924 (PK))
Optical cryogenic temperature compound (OCT) (Fisher Scientific, cat no. 23-730-571)
Gelatin (JT Baker, cat. no. 2124–01)
Chromium potassium sulfate (Fisher Scientific, cat. no. C337–500)
Gasket 2 discardable template (homemade)
Cell culture fixation preparation
Poly-lysine (Sigma, cat. no. A003E)
Formaldehyde aqueous solution, 32% (wt/vol) (Fisher Scientific, cat. no. 50980494) !CAUTION Formaldehyde is toxic on inhalation and with skin contact. It should be handled with gloves and under a fume hood. Formaldehyde should be discarded with care and according to your institution’s environmental health and safety rules.
PBS, 10x, pH 7.4 (Fisher Scientific, cat. no. 70011044)
Hybridization of RNA or DNA probe
Ultrapure H2O
Formaldehyde aqueous solution, 32% (wt/vol) (Fisher Scientific, cat. no. 50980494) !CAUTION Formaldehyde is toxic on inhalation and with skin contact. It should be handled with gloves and under a fume hood. It should be discarded with care and according to your institution’s environmental health and safety rules.
Triton-X (Acros Organics, cat. no. AC32737-1000)
SSC, 20x (Fisher Scientific, cat. no. 15-557-044)
Formamide, deionized (Millipore, cat. no. S4117) !CAUTION Formamide is toxic and can be absorbed through the skin. It should be handled with gloves and under a fume hood. It should be discarded with care and according to your institution’s environmental health and safety rules.
Tween-20 (Fisher Scientific, cat. no. AAJ20605AP)
RNase A solution, DNase and protease-free (10 mg/ml) (Fisher Scientific, cat. no. EN0531)
Dextran sulfate, 50% solution (vol/vol) (Millipore, cat. no. S4030)
Sigmacote siliconizing reagent for glass and other surfaces (Sigma-Aldrich, cat. no. SL2–100ML)
Glutaraldehyde (GA) 50% aqueous solution (wt/vol) (Fisher Scientific, cat. no. 50-262-17) !CAUTION GA is toxic. It should be handled with gloves and under a fume hood. It should be discarded with care and according to your institution’s environmental health and safety rules.
Preparing a 96-well plate for hybridization
▲CRITICAL For example probe and primer sequences, see Primers and probes in Equipment.
Readout probes, 10 μM (IDT)
Strand-displacement probes, 30 μM (IDT)
Fluorescently labeled oligos (IDT)
Tris, pH 8.0 (Fisher Scientific, cat. no. AM9855G)
EDTA, 0.5 M (Fisher Scientific, cat. no. AM9262)
Tris-EDTA (TE) buffer, 1x
Ethylene carbonate (EC), 98% (Sigma, cat. no. E26258–3KG)
Imaging buffer
Catalase (Fisher Scientific, cat. no. ICN10040250)
Glucose oxidase (Sigma, cat. no. G2133–250KU)
Glucose (Sigma, cat. no. G8270–1KG)
ddH2O
SSC, 20x (Fisher Scientific, cat. no. 15-557-044)
Mineral oil, pure (Fisher Scientific, cat. no. AC415080010)
Bleach buffer
SSC, 20x (Fisher Scientific, cat. no. 15-557-044)
ddH2O
Wash buffer
Formamide, deionized (Millipore, cat. no. S4117) !CAUTION Formamide is toxic and can be absorbed through the skin. It should be handled with gloves and under a fume hood. It should be discarded with care and according to your institution’s environmental health and safety rules.
SSC, 20x (Fisher Scientific, cat. no. 15-557-044)
ddH2O
Equipment
−20 °C, −80 °C freezer
4 °C refrigerator
96-well plate (Thermo Fisher Scientific, cat. no. AB-0765)
100-mm × 15-mm Petri dishes (Fisher Scientific, cat. no. FB0875712)
Blade, cryosectioning blade (Fisher Scientific, cat. no. 22–030530)
Blade, standard razor blade
Carbon dioxide tank
Countess cell counting chamber slides (Fisher Scientific, cat. no. C10228)
Centrifuge (Fisher Scientific, cat. no. 75-002-431)
Cover glass, 18 × 18 mm (Fisher Scientific, cat. no. 12–542-AP)
Coverslips, 40-mm glass coverslips (Bioptechs, cat. no. 40-131-03193)
Cryomold, 15-mm × 15-mm × 5-mm Tissue-Tek Cryomold (VWR, cat. no. 25608–924 (PK))
Cryostat machine (Leica, CM3050 S)
Dissecting microscope
DNA SpeedVac (Thermo Fisher Scientific, serial no. L23B-542783-LB)
Dry ice (large block size)
Embryo collection cage (Flystuff, cat. no. 9–101)
Embryo collection basket with mesh (Flystuff, cat. no. 46–102)
Eppendorf tube, 1.7 ml
Falcon conical centrifuge tubes (15 and 50 ml)
Flow chamber (FCS2 Chamber Flow Cell) (Bioptechs, SKU 03060319–2-NH)
Gasket 1 and gasket 2 (blank gasket, 0.75-mm thickness, pack of 5) (Bioptechs, cat. no. 1907–750)
Gasket 2 discardable template (homemade)
Gel imager
Heat block
Hybridization chamber (made from an empty pipette tip box filled with water)
Kimtech Kimwipes, small and large
Magnetic stir bar
Miicroaqueduct slide (Bioptechs, SKU 130119–5)
Microwave
Mini-PROTEAN tetra vertical electrophoresis cell, two-gel, for 0.75-mm-thick handcast gels; includes two casting frames, 10-well combs, five short plates and five spacer plates, with TGX Stain-Free FastCast acrylamide starter kit, 10% (Bio-Rad, 1658002FC)
Mini table shaker (Thermo Scientific, cat. no. 11-676-681)
NanoDrop spectrophotometer
Paintbrush
PCR strip tubes
Plastic Petri dishes (for apple juice plates, 100 mm × 15 mm (Fisherbrand, cat. no. FB0875712); for holding glass coverslips, 60 × 15 mm (Fisherbrand, cat. no. 08772F))
Real-time PCR machine
Scintillation vials (Fisher Scientific, cat. no. 03-341-25E)
Specimen disk (Fisher Scientific, cat. no. NC0626750)
Storage CryoBox (Amazon, BioCision BCS-206MC TruCool hinged CryoBox, 81-place, multi-color (pack of 5))
Superfrost Plus microscope slide (Fisher Scientific, cat. no. 22-037-246)
Vortex
Water bath
Primers and probes
We have provided a list of barcode, readout, strand-displacement and primer sequences at https://github.com/BoettigerLab/ORCA-public/tree/master/probe-building. In this same folder, we have also included the repetitive sequences from various model organisms and a script called DemoProbe-Synthesis.m that will aid new users in building their own primary probes.
We have included a short list of primary probe sequences that have been tested to work robustly in Drosophila embryos and cultured cells: https://bit.ly/3fdwBx0. This could be used as a positive control by laboratories that are adopting our method.
Example dataset
For a single experiment, our microscope data can easily be >2 TB. We provide a mini dataset to help users become comfortable analyzing raw microscope data through our MATLAB GUIs: https://bit.ly/2S6eCjk. This 337-GB mini dataset contains RNA and DNA images for a single 10–12-h Drosophila embryo (imaged in four FOVs). There are five folders in this dataset, including ‘Settings’, ‘Mosaics’, ‘RNA_Expt’, ‘DNA_Expt’ and ‘ORCAMapsPolymerOutput’. The ‘Settings’ folder contains all the files that were used for this automated experiment. We provide the embryo mosaic that was imaged in brightfield to select for FOVs before RNA imaging in the ‘Mosaic’ folder. The ‘RNA_Expt’ and ‘DNA_Expt’ folders contain separate ‘Readout_XXX’ folders, which hold the raw microscope images (called ConvZscan_XX.dax) and maximum projection fiducial/data images (the latter were generated by the GUIs in downstream analysis). In addition, both the ‘RNA_Expt’ and ‘DNA_Expt’ folders also have an ‘Analysis’ folder where one can find data tables, text files and images that were generated through ChrTracer3, BuildMosaicsGUI, AlignMosaicsGUI and MosaicAnalyzer. The ‘ORCAMapsPolymerOutput’ folder contains a MATLAB script (ORCA_MapsAndPolymers.m) that takes the ChrTracer3 outputs to produce the images (EPS and PNG images) found here. This script generates the following: (i) a heatmap of the fraction that two labeled barcodes fail to pair with one another (WT_BXC3kb_NaN_Fraction.eps), (ii) a distance map of the total population of cells (WT_BXC3kb_DistanceMap.eps), (iii) a contact frequency map of the total population of cells (WT_BXC3kb_ContactFreq.eps) and (iv) 3D polymers for single cells (you have the option to scroll through all the polymers in the data and select images to save by interacting with the command window).
Software
MATLAB
Python
The ‘storm-control’ software controls both the lasers and the camera and is available at https://github.com/BoettigerLab/storm-control.
The software for the fluidics handling is available at https://github.com/BoettigerLab/fluidics-control.
Software for processing the raw data, including the GUIs used throughout the procedure and some demo analysis scripts, is available at https://github.com/BoettigerLab/ORCA-public.
Reagent setup
Apple juice–agar plate preparation
Add 0.6 g of methyl P-hydroxybenzoate and 10.0 g of sucrose into 250 ml of apple juice. Bring to boil and then let it cool to 65 °C.
Add 9.0 g of Bacto agar to 300 ml of H2O. Bring to boil and then let it cool to 65 °C.
Once cooled, mix both the apple juice and agar solutions.
Pour into 100-mm × 15-mm Petri dishes. Petri dish sizes can vary depending on the size of the collection cage.
Store in 4 °C for 2 months. Older plates can get dried out, and flies will not lay as many eggs.
Yeast paste
Mix 1 g of yeast with 1 ml of ddH2O. If needed, add more water to make the yeast paste a peanut butter–like consistency. Store in 4 °C for 2 weeks. Make new yeast paste if it starts smelling like beer.
Borax solution
Dissolve 1 g of anhydrous sodium tetraborate in 1 liter of dH2O. Store at room temperature indefinitely.
2% (wt/vol) agarose borax gel
Dissolve 2 g of agarose in 100 ml of borax solution by boiling for 30 s. Prepare freshly.
NaOH-EDTA solution (1:1 (vol/vol) 0.5 M EDTA and 1 M NaOH)
Mix 1 ml of 0.5 M EDTA and 1 ml of 1 M NaOH. Store at room temperature (20–22 °C) indefinitely.
Gel for urea PAGE (15% (wt/vol) urea TBE gel) (two 0.75-mm gels)
Weigh out 6 g of urea in a 100-ml beaker.
Add 3.5 ml of 40% (wt/vol) acrylamide (19:1) Bis, 2.5 ml of 5x TBE and 1.5 ml of ultrapure H2O.
Add a magnetic stir bar.
Stir and heat gently on low for 5 min, until urea is dissolved. Let cool.
Add 20 μl of TEMED and 40 μl of 10% (wt/vol) APS. Mix thoroughly.
Cast the polyacrylamide gel into 0.75-mm-thick spacers. ▲CRITICAL Polymerization will happen in a few minutes after adding APS. If the gel polymerizes before you can pour the gels, use less APS next time. Allow gels to fully polymerize before removing them from the casting unit. Use the excess gel as a test for when polymerization is complete. If the gel does not polymerize within 1 h, repeat and use 2x APS. ▲CRITICAL Unused gels may be stored for a few days to a week wrapped in Saran wrap with a moist paper towel backing. These can last longer if kept cold; however, sometimes 4 °C causes the urea to crash out of solution, and, thus, room temperature may be better.
1x TBE
Mix 100 ml of 10x TBE and 900 ml of ddH2O. Store at room temperature indefinitely.
1x GelGreen in 1x TBE
Mix 2 μl of 10,000x GelGreen and 20 ml of 1x TBE. ▲CRITICAL Make fresh each time.
1x PBS
Mix 5 ml of 10x PBS and 45 ml of ddH2O. Store at room temperature indefinitely.
2x SSC (bleaching buffer)
Mix 5 ml of 20x SSC and 45 ml of ddH2O. Store at room temperature indefinitely.
8% (vol/vol) formaldehyde + 50 mM EGTA in 1x PBS
Combine 10 ml of 32% (vol/vol) formaldehyde, 4 ml of 0.5 M EGTA, 4 ml of 10x PBS and 22 ml of ddH2O. Store under the hood for 1 month. !CAUTION Formaldehyde is toxic on inhalation and with skin contact. It should be handled with gloves and under a fume hood. It should be discarded with care and according to your institution’s environmental health and safety rules.
50% (vol/vol) methanol in 1x PBS
Mix 5 ml of 1x PBS and 5 ml of 100% (vol/vol) methanol. !CAUTION Methanol is both toxic and volatile. Methanol should be handled with gloves and under a fume hood. It should be discarded with care and according to your institution’s environmental health and safety rules. ▲CRITICAL Make fresh each time.
30% sucrose (wt/vol) in 1x PBS
Add 15 g of sucrose to 5 ml of 10x PBS.
Add 30 ml of H2O and leave in a water bath at 60 °C to dissolve the sucrose.
Once the sucrose has dissolved, add water until the final solution is 50 ml.
Store at 4 °C for 1–3 months.
Chromium gelatin
Add 750 mg of gelatin to 145 ml of ultrapure H2O. Heat in a water bath/microwave to dissolve.
Let it cool down at room temperature.
While cooling, thoroughly mix 75 mg of chromium potassium sulfate in 5 ml of ultrapure H2O. Vortex to dissolve the chromium potassium sulfate.
Once the gelatin solution has cooled, add the chromium potassium sulfate solution from the previous step to the gelatin solution.
Store at 4 °C for 1–3 months.
4% (vol/vol) formaldehyde in 1x PBS
Combine 6.25 ml of 32% (vol/vol) formaldehyde, 5 ml of 10x PBS and 38.75 ml of ultrapure H2O. Store under the hood for 1 month. !CAUTION Formaldehyde is toxic on inhalation and with skin contact. It should be handled with gloves and under a fume hood. It should be discarded with care and according to your institution’s environmental health and safety rules.
RNase A solution (10 μg/ml) in 1x PBS
Combine 50 μl of RNase A solution (10 mg/ml) and 5 ml of 10x PBS. Add water until the final solution is 50 ml. Store at room temperature indefinitely.
Hybridization no. 1 (0.1% (vol/vol) Tween-20, 50% (vol/vol) formamide and 2x SSC)
Combine 5 ml of 20x SSC, 25 ml of formamide and 50 μl of Tween-20. Add water until the final solution is 50 ml. Store at 4 °C indefinitely. !CAUTION Formamide is toxic and can be absorbed through the skin. It should be handled with gloves and under a fume hood. It should be discarded with care and according to your institution’s environmental health and safety rules.
Hybridization no. 2 (50% (vol/vol) formamide, 2x SSC, 0.1% (vol/vol) Tween-20 and 10% (vol/vol) dextran sulfate)
Combine 1 ml of 20x SSC, 5 ml of formamide, 10 μl of Tween-20 and 2 ml of dextran sulfate 50% (wt/vol) aqueous solution. Add water until the final solution is 10 ml. Store at 4 °C indefinitely. !CAUTION Formamide is toxic and can be absorbed through the skin. It should be handled with gloves and under a fume hood. It should be discarded with care and according to your institution’s environmental health and safety rules.
Post-fix buffer (8% (vol/vol) formaldehyde and 2% (vol/vol) GA in 1x PBS)
Combine 10 ml of 32% (vol/vol) formaldehyde, 1.6 ml of 50% (wt/vol) GA, 4 ml of 10x PBS and 24.4 ml of ultrapure H2O. Store under the hood for 1 month. !CAUTION Formaldehyde is toxic on inhalation and with skin contact. It should be handled with gloves and under a fume hood. It should be discarded with care and according to your institution’s environmental health and safety rules.
Imaging buffer (0.5 mg/ml glucose oxidase, 40 μg/ml catalase and 10% (vol/vol) glucose in 2x SSC)
In a 1.5-μl Eppendorf tube, weigh out 10 mg of glucose oxidase and dissolve in 500 μl of 2x SSC by vortexing it.
Add 100 μl of catalase solution (20 mg/ml stock) and mix it by pipetting up and down three times.
In a 50-ml conical tube, add 8 ml of 50% (wt/vol) glucose in H2O.
Add 4 ml of 20x SSC.
Add all 600 μl of enzyme mixture (from the Eppendorf tube) into the conical tube.
Fill to 40 ml with ultrapure H2O.
Overlay solution with 1 inch of mineral oil to prevent oxygen from the air dissolving back into the buffer.
Store at room temperature under oil for ≤4–5 d. ▲CRITICAL Make fresh each time. If you are doing an RNA+DNA experiment within a 4–5-d span, a batch of fresh imaging buffer should be enough for both RNA and DNA imaging. Use a pH strip to check if the imaging buffer pH is <4 at any point. If the pH is <4, do not use the imaging buffer, because it can be harmful to your sample.
25% (vol/vol) EC in 2x SSC
▲CRITICAL EC is used as a low-toxicity, low-cost, high-stability replacement for formamide. Like formamide, EC partially destabilizes hydrogen bonds, increasing the stringency of hybridization without heating. It is a solid at room temperature.
Heat EC to 42 °C in a water bath to liquefy.
Add 20 ml of 20x SSC to a bottle.
Add 130 ml of ultrapure H2O.
Add 50 ml of liquified EC. EC is added last to avoid crystallization.
Store at room temperature for 2 months.
Wash buffer (30% (vol/vol) formamide in 2x SSC)
Combine 50 ml of 20x SSC, 150 ml of formamide and 300 ml of ultrapure H2O. Store at room temperature for 2 months. !CAUTION Formamide is toxic and can be absorbed through the skin. It should be handled with gloves and under a fume hood. It should be discarded with care and according to your institution’s environmental health and safety rules.
1x TE buffer
Combine 2 ml of Tris (pH 8.0), 400 μl of 0.5 M EDTA (pH 8.0) and 197.6 ml of ultrapure H2O. Store at room temperature indefinitely.
Readout probes and strand-displacement oligo preparation for a single well in a 96-well plate
▲CRITICAL The plate should be freshly made. To each well of a 96-well plate, add the following:
600 μl of 25% (vol/vol) EC in 2x SSC
0.11 μM Cy5-labeled imaging oligo (0.33 μl of 200 μM stock)
0.233 μM Cy3-labeled fiducial oligo (0.7 μl of 200 μM stock). ▲CRITICAL STEP Add the fiducial label only to the first well.
0.1-μM readout probe (6 μl of 10 μM stock)
0.3-μM strand-displacement oligo (6 μl of 30 μM stock). ▲CRITICAL There is no need to add the strand-displacement oligo for the first well.
Procedure
Oligopaint library design ● Timing 1 d
-
1See option A or B for designing probes that target DNA or RNA, respectively, and Fig. 6a for a schematic of the probe design. Some users may be interested in designing only DNA, only RNA or both. Familiarity with a basic scripting language such as Python, MATLAB or R is recommended. These languages all have basic genomics/bioinformatics toolboxes that may be useful. We recommend that the user is familiar with elementary bioinformatics operations, such as sequence alignment and melting temperature calculations. Numerous published approaches are available for accomplishing these steps9,28,33–37,65.
- Designing probes targeting DNA
-
Generate a FASTA file containing the sequence for regions of interest.Make sure that the sequence selected is sense to avoid cross-talk with RNA, or use RNase to remove RNA signal before labeling DNA (as described in Step 152).
- Take the FASTA file of the target DNA region and partition it into even blocks of sequences at the chosen step size (e.g., 2 kb).
- For each block, pick non-overlapping 40-nt sequences. These 40-nt sequences must have (i) a melting temperature >65 °C, (ii) a G/C range of 20–80%, (iii) a homology <12 nt to any other barcode in the pool and (iv) a homology <14 nt to repetitive elements in the genome.
- Choose a minimum of 20 of the 40-nt sequences with the same barcode sequence to label a single genomic step size.
- Assign a common unique barcode sequence (20 nt) to all 40-nt sequences that are within the same block.
- Assign a common fiducial 20-nt fiducial probe–binding sequence (catcaacgccacgatcagct) to all of the 60-nt sequences (40-nt sequence + 20-nt unique barcode). The fiducial probe–binding sequence is also the RT sequence that will be used for downstream probe synthesis.
- Assign a unique forward and reverse primer to all the sequences, to allow a region of interest to be isolated from the Oligopaint library.
-
- Designing probes targeting RNA
- Select genes and isoforms. There is no strict limit to the number of genes that may be probed in an experiment. However, including additional genes of minor interest may increase the background signal and decrease the signal-to-noise ratio in the experiment; thus, we recommend exercising some restraint. We typically chose 10–100.
- Assemble a FASTA file of all genes/isoforms. For stable RNA expression, use the spliced mRNA sequence for the isoform of interest. For nascent RNA, use ≥1 kb of intron, ideally the most 5′ of the isoform’s intronic sequence.
-
For each gene/isoform, select non-overlapping 40-nt sequences and assign a unique barcode sequence. These 40-nt sequences must have (i) a melting temperature >65 °C, (ii) a G/C range of 20–80%, (iii) a homology <12 nt to any other barcode in the pool and (iv) a homology <14 nt to highly expressed protein-coding genes (e.g., ribosomal proteins) and RNAs (rRNA and tRNA).▲CRITICAL STEP Make sure that the sequence selected is anti-sense to the target RNA.
- Choose a minimum of 20 of the 40-nt sequences with the same barcode sequence to label a single RNA species.
- Assign a common fiducial 20-nt fiducial probe–binding sequence (catcaacgccacgatcagct) to the 60-nt sequences (40-nt sequence + 20-nt unique barcode).
-
Assign a unique forward and reverse primer to all the sequences, to allow a region of interest to be isolated from the Oligopaint library.■PAUSE POINT The Oligopaint library from Twist Biosciences or Genscript typically ships at 30–100-ng/μl final concentration and should be stored at −20 °C. Working stocks for probe amplification may be diluted to 1–2 ng/μl in TE buffer and stored at −20 °C.
Fig. 6 |. Primary probe synthesis.
a, Computational design of the Oligopaint library. The region of interest is partitioned into genomic segments (e.g. 2 kb or 10 kb) and each segment is assigned a minimum of twenty 40-nt target sequences. The following are taken into consideration for the oligonucleotides: GC content melting temperature, homology to barcode sequences, and repetitive sequences. Our MATLAB scripts take into account these constraints and tile the oligonucleotides across a genomic region of interest in a uniform manner. Each primary probeset from the oligopool has a unique forward (Fwd) and reverse (Rev) index primer sequence and a 20-nt reverse transcription (RT) sequence (catcaacgccacgatcagct) that will be used for probe synthesis. Additionally, the RT sequence serves as the fiducial probe binding sequence (Fid seq.), which will be used for drift-correction after data collection. b, Schematic of the primary probe synthesis. Unique primers allow for the amplification specific primary probes via limited-cycle PCR. Following the PCR, the double-stranded DNA (dsDNA) undergoes an in vitro transcription reaction to generate a bulk number of single-stranded RNAs (ssRNAs), which are converted back to single stranded DNA via a reverse transcription reaction. The DNA is then column purified, and the primary probes are concentrated.
Primary probe synthesis from the oligopool ● Timing 2 d
▲CRITICAL When synthesizing a new primary probe, we recommend making a primary probe from an Oligopaint library that has previously worked to serve as a positive control and for troubleshooting purposes. We have provided a list of validated primary probes here: https://bit.ly/3fdwBx0. Figure 6b illustrates the process of primary probe synthesis.
-
2Amplify primary probes from a diluted Oligopaint library in 1x TE (~1 ng/μl) via limited-cycle PCR. Add the following into a PCR tube and prepare on ice:
Reagent Quantity (μl) Final concentration ddH20 15 - Phusion High Fidelity PCR Master Mix with HF buffer (2×) 25 1× Forward primer (10 μM) 2.5 500 nM Reverse primer with T7 promoter sequence (10 μM) 2.5 500 nM Diluted Oligopaint library (−1 ng/μl) 2.5 −50 pg/μl EvaGreen dye (20×) 2.5 1× Total 50 - -
3Run reactions on a quantitative real-time PCR machine with the settings listed below. Stop the reaction for each tube while it is still in the exponential growth phase and before it reaches saturation. This should take 15–32 cycles.
Cycle no. Denature Anneal Extend no. 1 Imagea Extend no. 2 1 98 °C, 3 min - - - - 2–32 98 °C, 5 s 65 °C, 10 s 72 °C, 10 s Image 72 °C, 8 s aThis is the step where the quantitative PCR machine images each well to see how much double-stranded DNA (dsDNA) is detected by EvaGreen Dye labeling.▲CRITICAL STEP Be sure to stop the reaction only at ‘Extend no. 2’. Stopping during the annealing phase will result in cross-annealed hybrids.
?TROUBLESHOOTING
-
4
(Optional) Validate the PCR reaction by running 2 μl of sample on a 2% (wt/vol) agarose borax gel. A satisfying result will feature a single band, typically at 120–160 bp, depending on the length of the library.
-
5
In a 1.5-ml tube, add 350 μl of Zymo DNA binding buffer to ~50 μl of PCR product.
-
6
Transfer to a Zymo DCC-5 column and spin at >10,000g for 30 s at room temperature.
-
7
Add 200 μl of DNA wash buffer to the column and spin at >10,000g for 30 s.
-
8
Repeat Step 7.
-
9
Discard flow-through by aspirating the collection tube dry.
-
10
Spin at >10,000g for 1 min to remove the residual buffer.
-
11
Transfer the Zymo DCC-5 column to a clean 1.5-ml tube.
-
12
Pipette 11 μl of ultrapure H2O onto the membrane of the column and allow it to sit on the membrane at room temperature for 1 min.
-
13
Centrifuge the tube at >10,000g for 30 s to collect the DNA.
■PAUSE POINT The reaction can be stored at 4 °C indefinitely.
-
14Prepare the following in an RNase-free environment.
Reagent Quantity (μl) PCR product (Step 13) 10 NTP buffer mix (20 mM each NTP) 10 T7 RNA polymerase mixture 2 RNAse inhibitor 1 Total 23 -
15
Incubate at 37 °C for 4–16 h. This process will turn the dsDNA to single-stranded RNA (ssRNA).
■PAUSE POINT The reaction can be stored at 4 °C for ≤1 week.
-
16
At this point, you have ssRNA. There should be >2 μmol of ssRNA from the T7 reaction above. Purification of RNA is not necessary here; however, purification is needed for quantification purposes. We do not perform any RNA quantification, because it is not necessary. Doing so would increase the opportunity for RNA degradation by increased handling and removing the RNAse inhibitor. Moreover, probe synthesis is not sensitive to the yield of the PCR, and, thus, there is no need to over-amplify any of the products. More details of this can be found in Boettiger and Murphy38.
Use RT to generate ssDNA probes from the ssRNA. Add the following into the same PCR tube from the previous step (containing ~20 μl of T7 reaction) for the RT reaction. Note that the 200 μM Cy3-labeled RT-primer can be used during the reverse transcriptase step to fluorescently label all primary probes. However, to be cost efficient, we choose to not use the fluorophore primer for this step.Reagent Quantity (μl) T7 reaction 20 dNTP (10 mM) 10 RT buffer (5×) 2 Labeled or unlabeled RT-primer (200 (μM) 3 Maxima H Minus RT enzyme 1 RNasin RNase inhibitor 4 Total 40 ▲CRITICAL STEP Use an RNase inhibitor engineered to work at 50 °C. RNasin does work at this temperature, but most do not.
-
17
Incubate the sample at 50 °C for 1 h.
-
18
Digest the remaining RNA by adding 25 μl of NaOH-EDTA solution to the product and heating it at 95 °C for 10 min.
-
19
Recommended: Test the ssDNA reaction product by running 2 μl out on a 15% (wt/vol) urea PAGE gel. 15% (wt/vol) Mini-PROTEAN TBE-urea precast gels (Bio-Rad, 4565054) can be bought or made (see Reagent setup). Pre-run gel for 10 min in a water bath at 60 °C and at 120 V to equilibrate the gel temperature. Mix 2 μl of reaction product with loading buffer. Use the GeneRuler Ultra Low Range DNA Ladder and mix it with the loading buffer. Load the gel, place the gel in a water bath (at 60 °C) and run the gel for 50 min at 120 V. Gently extract the gel from the casing; the gel can be extracted while it is still warm. Incubate in 1x GelGreen in 1x TBE for 10 min. Image the gel and check for a clear single band at ~100 nt.
!CAUTION When running the gel, make sure to not submerge electrodes into the water bath, because this may cause an electroduction hazard.
?TROUBLESHOOTING
-
20
Add 150 μl of oligo-binding buffer to a clean 1.5-ml Eppendorf tube.
-
21
Add NaOH-treated RT product (75 μl) to the Eppendorf tube and mix by pipetting.
-
22
Add 600 μl of 100% ethanol (vol/vol) to the Eppendorf tube and mix.
-
23
Transfer to a Zymo DCC-25 column, spin at ≥10,000g for 30 s and discard the wash-through. The maximum capacity of the column is 800 μl. Larger volumes may be split over two spins.
-
24
Add 750 μl of DNA wash buffer.
-
25
Spin at >10,000g for 30 s and discard the flow-through.
-
26
Spin at ≥10,000g for 30 s to remove excess buffer. Discard any flow-through.
-
27
Transfer the column to a clean 1.5-ml Eppendorf tube.
-
28
Elute the primary probes in 50 μl of ultrapure H2O.
-
29
Measure the final primary probe concentration by using a NanoDrop.
-
30
Record the concentration into a document/notebook. An expected yield for the final concentration is 1–2 μmol of ssDNA.
■PAUSE POINT Primary probes can be stored at −20 °C indefinitely.
?TROUBLESHOOTING
Drosophila egg laying in collection cage ● Timing 1.5 d (Timing is based on the time that it takes to collect 10–12-h embryos. This process can be shorter or longer depending on the desired developmental stage.)
-
31
Spread fresh yeast in the middle of a fresh apple juice–agar plate to encourage egg laying. The smell of yeast paste stimulates egg production.
-
32
Use carbon dioxide to knock out the flies.
-
33
Transfer both female and male flies into a collection cage and seal the cage with the apple juice–agar plate (Fig. 7a).
-
34
Let the flies acclimate to the collection cage for 1 d. Female flies may internally hold their fertilized eggs. Consequently, embryos start developing within the female. The first eggs laid in the collection cage may be slightly more developed than desired. After 1 d within the collection cage, the fresh yeast paste induces females to lay eggs shortly after fertilization.
-
35
On the next day, put the apple juice–agar plate in the −20 °C freezer for 1–2 h (embryos will not survive this temperature) and then discard it in the trash. Spread new yeast paste onto a new apple juice–agar plate and place it in the cage for 2 h. Afterward, remove the apple juice–agar plate from the cage.
-
36
Leave the plate at room temperature until it reaches the desired developmental stage.
?TROUBLESHOOTING
Fig. 7 |. Sample preparation for primary probe hybridization.
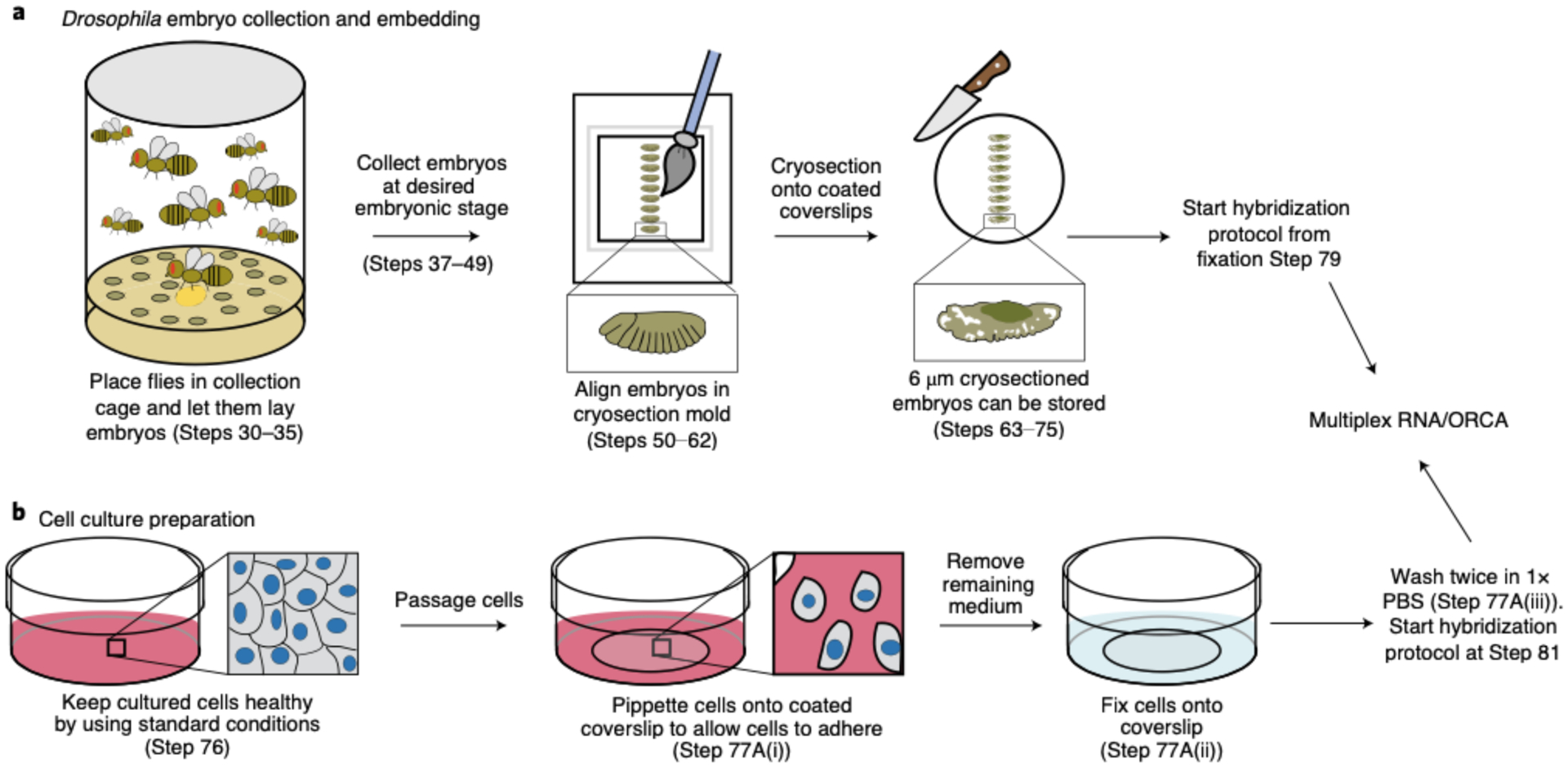
a, Flies are placed in a collection cage containing an apple juice agar plate with yeast paste. After a day of acclimation, a new plate is placed in the collection cage and flies lay eggs for two hours. The plate is then removed and eggs are allowed to develop until the desired embryonic stage. Embryos are collected using a standard collection protocol. Next, embryos are embedded onto a cryomold. During this process any remaining sucrose is removed and a paintbrush is used to align embryos along the anterior-posterior axis. After freezing with dry ice, embryos are cryosectioned at 6 μm sections onto chromium-gelatin coated 40-mm coverslips. Embryo sections can be immediately hybridized or stored. b, Schematic of the general process of cell culture preparation. Cells are maintained healthy using standard cell culture protocols. During passaging, cells are placed onto a poly-lysine coated glass coverslip and fixed with 4% formaldehyde for ten minutes and then washed twice in 1x PBS. Continue primary probe hybridization at Step 81 of section ‘Hybridization with RNA primary probes’ or Step 149 of section ‘Hybridization with DNA primary probes’ if you want to target RNA or DNA, respectively.
Drosophila embryo collection ● Timing 40 min
-
37
Use a Kimwipe to gently remove any remaining yeast from the apple juice–agar plate.
-
38
Take the mesh gauze fabric and screw it into the lid of the collection basket to act as a mesh barrier for embryo collection.
-
39
Pour bleach on the agar plate until the plate is fully covered to remove the embryonic chorion.
!CAUTION Bleach is toxic. It should be handled with gloves and under a fume hood. It should be discarded with care and according to your institution’s environmental health and safety rules.
-
40
Swirl the agar plate with bleach for 1 min. If embryos do not detach from the plate during the swirling, use a clean paintbrush to physically detach them from the plate.
-
41
Pour the embryos into a collection basket and rinse with water for 1 min.
-
42
In the hood, unscrew the collection tube and dip the gauze fabric embryo-side down into a scintillation vial filled with 5 ml of heptane and 5 ml of 8% (vol/vol) formaldehyde + 50 mM EGTA in 1x PBS.
!CAUTION Formaldehyde is toxic on inhalation and with skin contact. It should be handled with gloves and under a fume hood. It should be discarded with care and according to your institution’s environmental health and safety rules.
-
43
Use a paintbrush to transfer any embryos stuck in the gauze fabric and/or rim of the collection tube into the scintillation vial.
-
44
Take the scintillation vial out of the hood and place it on the shaker table for 25 min at 400 rpm. Both the heptane and the embryos’ vitelline membrane are hydrophobic. Thus, the heptane will be able to go through the vitelline membrane and allow the formaldehyde to enter the embryo for fixation during this step.
-
45
The formaldehyde mixture will be on the bottom, and the heptane will be on the top layer of the vial. The embryos should float between the two layers. In the hood, pipette off the bottom formaldehyde mixture and discard it in the hazardous waste.
!CAUTION Formaldehyde is toxic on inhalation and with skin contact. It should be handled with gloves and under a fume hood. It should be discarded with care and according to your institution’s environmental health and safety rules.
▲CRITICAL STEP Do not pipette up any embryos, because they are sticky and will get stuck inside the pipette tip. If embryos do get stuck inside the pipette tip, you can attempt to use a paintbrush to rescue them and place them back into the vial.
-
46
Add 5 ml of 100% (vol/vol) methanol and vortex for 3 min. Embryos should sink to the bottom of the vial.
!CAUTION Methanol is both toxic and volatile. Methanol should be handled with gloves and under a fume hood. It should be discarded with care and according to your institution’s environmental health and safety rules.
?TROUBLESHOOTING
-
47
Use a razor blade to cut 1/4 inch off the end of a 1,000-μl pipette tip and pipette embryos into a clean 1.7-ml Eppendorf tube.
-
48
Wash the embryos three times with 1 ml of 100% (vol/vol) methanol.
!CAUTION Methanol is both toxic and volatile. Methanol should be handled with gloves and under a fume hood. It should be discarded with care and according to your institution’s environmental health and safety rules.
-
49
Discard used heptane and methanol into flammable waste.
■PAUSE POINT Embryos can be stored in 100% (vol/vol) methanol in a −20 °C freezer for several months.
Embedding Drosophila embryos for cryosectioning ● Timing 2 h 35 min
-
50
Carefully pipette out the 1 ml of 100% (vol/vol) methanol from the Eppendorf tube containing the collected embryos. Do not pipette up any of the embryos.
!CAUTION Methanol is both toxic and volatile. Methanol should be handled with gloves and under a fume hood. It should be discarded with care and according to your institution’s environmental health and safety rules.
-
51
Wash embryos with 50% (vol/vol) methanol in 1x PBS for 5 min in a mini table shaker at 250 rpm.
-
52
Wash two times quickly with 500 μl of 1x PBS.
-
53
Wash with 1x PBS and leave in the mini table shaker for 10 min at 250 rpm.
-
54
Remove the 1x PBS and add 500 μl of 30% (wt/vol) sucrose in 1x PBS.
-
55
Leave embryos to sink in sucrose for 2 h in the 4 °C refrigerator.
-
56
Take a Cryomold and use a disposable gasket 2 as a stencil to outline the middle of the Cryomold.
This is the area where embryos will be embedded.
▲CRITICAL STEP If you are using the Bioptechs flow chamber, remember that there is a limited area of the slide that can be imaged because of the size of the hole in gasket 2 (see Extended Data Fig. 3a). Any cells/tissues placed underneath the plastic portion of gasket 2 cannot be imaged. For Drosophila embryos, we highly recommend taking this into account when embedding embryos into the Cryomold and later when cryosectioning them onto the coated coverslip. Failure to do so can result in embryos being completely covered by the plastic portion of gasket 2, and you will not image them.
-
57
Use a razor blade to cut 1/4 inch off the end of a 200-μl pipette tip and pipette up 25–50 μl of embryos from the bottom of the Eppendorf tube.
-
58
Pipette out the embryos within the marked outline of the Cryomold.
-
59
Under a dissection microscope, use a thin clean paintbrush to move the embryos inside the marked area of the Cryomold (Fig. 7a).
-
60
Remove the rest of the remaining sucrose in the Cryomold by using a pipette and/or a Kimwipe to wick out the sucrose.
▲CRITICAL STEP Excess liquid during this step will disrupt tissue morphology during the cryosectioning process. We have found that failure to remove most liquid makes the Drosophila cryosectioned sample look like ‘Swiss cheese’, because the sample appears to be covered with holes.
-
61
Under a dissection scope, use a paintbrush to orient each embryo along its anterior to posterior axis. In addition, use the paintbrush to make sure that embryos are embedded in a monolayer and not mounted on top of each other.
▲CRITICAL STEP This step should be done quickly (within 30 s to 1 min maximum) to make sure that embryos do not completely dry out.
-
62
Fill the Cryomold with OCT compound and place the Cryomold on top of flat dry ice for 10 min.
■PAUSE POINT Embryos in a Cryomold can be stored for ≤1 year in the −80 °C freezer. We have not attempted to RNA/DNA-label cryosection embryos past 1 year of storage.
Preparing slides for cryosectioning ● Timing 1 h
-
63
Place a large Kimwipe on top of the working bench. Glass coverslips will be placed on top of the large Kimwipe to keep them clean.
-
64
Take a round glass coverslip and wipe down both sides with methanol. Leave it to dry.
-
65
Mark an outline of gasket 2 onto the coverslip by using an alcohol-resistant pen (an example of a marked coverslip can be seen in Extended Data Fig. 3a). The marked side of the coverslip will be the back of the slide and will not make contact with the sample. We find that the alcohol-resistant pen does not harm the sample in any way.
-
66
Pipette 500 μl of chromium gelatin over the gasket 2–marked area on the front of the coverslip.
-
67
Leave the chromium gelatin solution on the coverslip for 40 min.
-
68
Afterward, completely aspirate the chromium gelatin off the coverslip.
■PAUSE POINT Coverslips can be used immediately or stored for ≤1 week at 4 °C. To store the coverslips, take a piece of foam (typically one that comes within laboratory purchase packages) and cut it to fit within a storage CryoBox. Next, take a blade and cut the foam to make slits large enough to place the coverslips. Make sure that the coverslips do not touch one another when placing them within the slits in the foam. 20–25 coverslips can fit in a storage CryoBox.
?TROUBLESHOOTING
Embryo cryosectioning ● Timing 1 h
-
69
On the cryostat machine, set the temperature for both the chamber and the object to −21 °C.
-
70
Place the OCT Cryomold containing the embryos inside the cryostat chamber for ≥15 min before cryosectioning. Also place the paintbrush and blades inside the chamber to equilibrate with the chamber’s temperature.
-
71
Carefully, pop out the frozen OCT with the sample from the plastic Cryomold. Make sure that the top side (the side that is closest to the embryos) of the sample is facing toward you. Add a layer of fresh OCT onto the specimen disk. It is easier adding the OCT outside the chamber so that it does not freeze right away. Place the sample onto the specimen disk inside the chamber. Flatten the sample onto the OCT-filled specimen disk by using the ‘pressing tool’ found within the cryostat machine until everything has solidified.
-
72
Place the specimen disk into the specimen head and orient the Cryomold.
-
73
Set the cryostat machine to cut 6-μm sections.
▲CRITICAL STEP We do not use the Trimming option on the cryostat machine; instead, we only use the Section option and set the cutting size at 6 μm from the start because of the size of the Drosophila embryo. Keep in mind that Drosophila embryonic tissue is not visible in the Cryomold, unlike a mouse brain that is visibly pink in the Cryomold. Section away at the Cryomold and check whether you are cutting through the sample by transferring the cut slices onto a Superfrost Plus microscope slide. Then look at the slide under a dissecting scope to check if any tissue is being transferred onto the slide. Once sectioning is confirmed, start transferring the tissue sections onto the center of the coated glass coverslip (from Step 68), within the templated region.
-
74
Once you have started sectioning tissue, section through the whole sample and transfer the tissue onto the round glass 40-mm coverslips (Fig. 7a). Typically, 20–25 glass coverslips can be used during one cryosectioning session for Drosophila embryos.
-
75
Once done, thoroughly clean up the cryostat machine and workspace.
■PAUSE POINT The cryosectioned sample can be used immediately for RNA/DNA hybridization or stored for ≤0.5 years at −80 °C. We have not tested our approach on samples stored for longer.
Cell culture fixation preparation ● Timing 20 min
-
76
Use standardized cell culture protocols to culture cells and make sure that cells are healthy.
-
77Fix the cells. Follow option A for adherent cells (e.g., IMR90s) or option B for suspended cells (e.g., K562s). We typically use a 60-mm Petri dish as a container for the 40-mm cover glass during the fixation procedure.
- For adherent cells
- Plate cells on a 40-mm coated cover glass (e.g., poly-lysine) at the desired confluency and allow cells to adhere (typically overnight).
-
Fix cells to a coated cover glass by removing growth medium and covering the cells in 4% (vol/vol) formaldehyde in 1x PBS for 10 min (Fig. 7b).!CAUTION Formaldehyde is toxic on inhalation and with skin contact. It should be handled with gloves and under a fume hood. Formaldehyde should be discarded with care and according to your institution’s environmental health and safety rules.▲CRITICAL STEP 2–4 ml is sufficient to cover the cover glass in a 60-mm Petri dish.
- Remove the fixation buffer and wash twice with 1x PBS. Pour 1x PBS onto the Petri dish, avoiding pouring it directly on top of the cells, and use enough 1x PBS to cover the entire cover glass.
- For suspended cells
- Coat the 40-mm cover glass with poly-lysine. One may choose to just coat the cover glass area that will be imaged.
- Aspirate poly-lysine before plating cells.
- Spin down 2–10 ml of cells in a 15-ml centrifuge tube at 1,200 rpm and room temperature.
- (Optional) You can count the number of cells at this point to plate the desired amount onto the plate by using the Countess cell counting chamber slides. For K562 cells, we found that using ~500 μl of poly-lysine is enough to coat the center of the glass slide, and spotting ~500,000–1 million cells provides a dense monolayer of cells.
- Remove most of the supernatant, leaving ~100 μl in the tube.
- Add cells in a droplet to the poly-lysine–coated cover glass.
- Spin cells at 3,000 rpm by placing the 60-mm Petri dish holding the cover glass in a swinging bucket centrifuge at maximum acceleration for 90 s. Holding the pulse spin for 90 s may provide better acceleration.
-
Fix cells to the cover glass by adding 2–4 ml of 4% (vol/vol) formaldehyde in 1x PBS for 10 min (after spinning, most of the liquid will have moved to the sides of the plate) (Fig. 7b).!CAUTION Formaldehyde is toxic on inhalation and with skin contact. It should be handled with gloves and under a fume hood. Formaldehyde should be discarded with care and according to your institution’s environmental health and safety rules.
- Remove the fixation buffer and wash twice with 1x PBS. Pour 1x PBS onto the Petri dish, avoiding pouring it directly on top of the cells, and use enough 1x PBS to cover the entire cover glass.
-
78
(Optional) If you will be labeling RNA and DNA, draw a shape (e.g., star) on the coverslip by using a pipette tip. This will scrape off cells from the coverslip, but it will also create a shape that can be used as a spatial orientation indicator when manually selecting for the same FOVs in RNA and DNA imaging.
■PAUSE POINT Fixed cultured cells can be used immediately for RNA/DNA hybridization or stored for up to 2 weeks at 4 °C. We have not tested our approach on samples stored for longer. For storing, we recommend leaving the slide covered in 1x PBS inside the 60-mm Petri dish and placing it in a humid chamber. To make sure that the 1x PBS does not evaporate and dry out the slide, make sure that the Petri dish is completely sealed with Parafilm.
Hybridization with RNA primary probes ● Timing 1.5 d
▲CRITICAL If you are interested only in targeting DNA, and not RNA, skip to Step 147 for cryosectioned tissue or Step 149 for cultured cells.
▲CRITICAL If you are working with cell culture, skip to Step 81 to permeabilize cells. From Step 81 onward, the protocol is the same for cultured cells and tissue.
▲CRITICAL In regard to the liquid volume used for 4% (vol/vol) formaldehyde in 1x PBS, 1x PBS, 0.5% (vol/vol) Triton-X in 1x PBS, hybridization no. 1, hybridization no. 2 and 2x SSC, we pour the solutions onto the Petri dish. Pour enough of the solution onto the Petri dish to cover the entire round cover glass. Make sure to not pour directly on top of the cells/tissues, because this could cause the sample to peel away.
-
79
Fix the cryosectioned embryo sample with 4% (vol/vol) formaldehyde in 1x PBS for 10 min. We typically use a 60-mm Petri dish as a container for the 40-mm cover glass during the hybridization procedure.
!CAUTION Formaldehyde is toxic on inhalation and with skin contact. It should be handled with gloves and under a fume hood and should be discarded with care and according to your institution’s environmental health and safety rules.
-
80
Remove the fixation buffer and wash three times with 1x PBS.
-
81
Permeabilize the sample by adding 0.5% (vol/vol) Triton-X in 1x PBS for 10 min.
-
82
Wash three times in 1x PBS.
-
83
Incubate in hybridization no. 1 for 35 min.
-
84
During the 35-min wait, set the heat block to 90 °C. Place a hybridization chamber (this can be an empty pipette tip box with water added into the pipette tip holes) in a 42 °C incubator and allow it to equilibrate. Take an 18 × 18-mm cover glass and clean it with 100% (vol/vol) methanol. Cover the 18 × 18-mm cover glass in Sigmacote and allow it to air dry in the hood. After the 35-min wait, rinse the remaining Sigmacote from the 18 × 18-mm cover glass with ultrapure H2O and dry it with a Kimwipe.
-
85
Remove hybridization no. 1.
-
86
Add the hybridization mixture (1.5 μl of primary probe from a 500-ng/μl stock in 30 μl of hybridization no. 2) onto the sample while avoiding introducing air bubbles.
-
87
Place the Sigmacote-treated 18 × 18-mm cover glass on top of the hybridization mixture to spread the primary probe.
-
88
Set the time of the heat denaturation step for tissue or cultured cells, respectively. For tissue/embryos, place the Petri dish on top of the heat block at 90 °C for 10 min. The timing can be shortened to 5 min, but we generally observe better embryo labeling with 10-min heat denaturation. For cultured cells, place the Petri dish on top of the heat block at 90 °C for 3 min. We find that 3 min is sufficient for labeling.
-
89
Hybridize the sample inside the hybridization chamber and leave it overnight in a 42 °C incubator.
■PAUSE POINT The sample can be left at 42 °C for ≤24 h. We typically allow the sample to stay in an enclosed humid chamber overnight at 42 °C, but find that 6 h is generally sufficient for some signal. However, we generally recommend leaving the sample incubating overnight for a strong RNA labeling signal.
-
90
The following day, pre-warm 2x SSC solution to 42 °C.
-
91
Add 2x SSC to the sample and return the sample to the 42 °C incubator for 10 min.
-
92
Carefully remove the 18 × 18-mm cover glass from the sample by using forceps.
-
93
Wash in 2x SSC by incubating for 10 min at 42 °C.
-
94
Rinse once in 2x SSC at room temperature.
■PAUSE POINT The sample may be stored in 2x SSC at 4 °C for ≤1 month.
96-well plate preparation for robotic hybridization ● Timing 20 min
-
95
Our readout probes and strand-displacement probes are divided into aliquots in 1x TE at 200 μl in PCR tube stripes and are organized in separate 96-well PCR tube racks. The final concentrations of readout probes and strand-displacement probes are 10 and 30 μM, respectively.
-
96
Take out a clean 96-well plate for your experiment with well capacity >600 μl.
-
97
Calculate how much 25% (vol/vol) EC in 2x SSC and Cy5imaging oligo, which will bind to readout probes, will be needed for the size of your experiment. Each well will contain 600 μl of 25% (vol/vol) EC in 2x SSC with 0.33 μl of 200 μM Cy5 imaging oligo (Fig. 8a). Make a master mixture of the amount of dye and buffer needed for your experiment.
-
98
Use a multichannel pipette to add 600 μl of 25% (vol/vol) EC in 2x SSC and Cy5-labeled oligo in your wells.
-
99
In the first well, add 0.7 μl of 100 μM Cy3 imaging oligo, which will bind to the fiducial sequences for all primary probes. The Cy3 signal will be very bright, and, thus, this oligo needs to be added only for the first well (Fig. 8a).
-
100
In your notebook, design a layout for this experiment. Organize the order of the readout probes and make sure that you will add the appropriate strand-displacement oligo to remove the previous signal (Fig. 8b).
-
101
Follow your designed layout and add 6 μl of 10 μM readout probes to your 96-well plate by using either a multichannel pipette or a regular pipette.
▲CRITICAL STEP Be sure to check your pipette tips to ensure accurate fluid transfer. If tips are not properly attached to the multichannel pipette, they will not aspirate the correct volume. This could lead to artifacts in the data.
-
102
Follow your designed layout and add 6 μl of 30-μM strand-displacement oligo to your 96-well plate by using either a multichannel pipette or a regular pipette.
-
103
Use a multichannel pipette and 300-μl pipette tips to properly mix the contents in each well.
▲CRITICAL STEP Make sure to use fresh pipette tips when mixing to not cross-contaminate your wells. Insufficient mixing of individual wells will lead to significantly reduced hybridization efficiency of individual readouts across the sample.
Fig. 8 |. Oligo setup for barcode labeling.
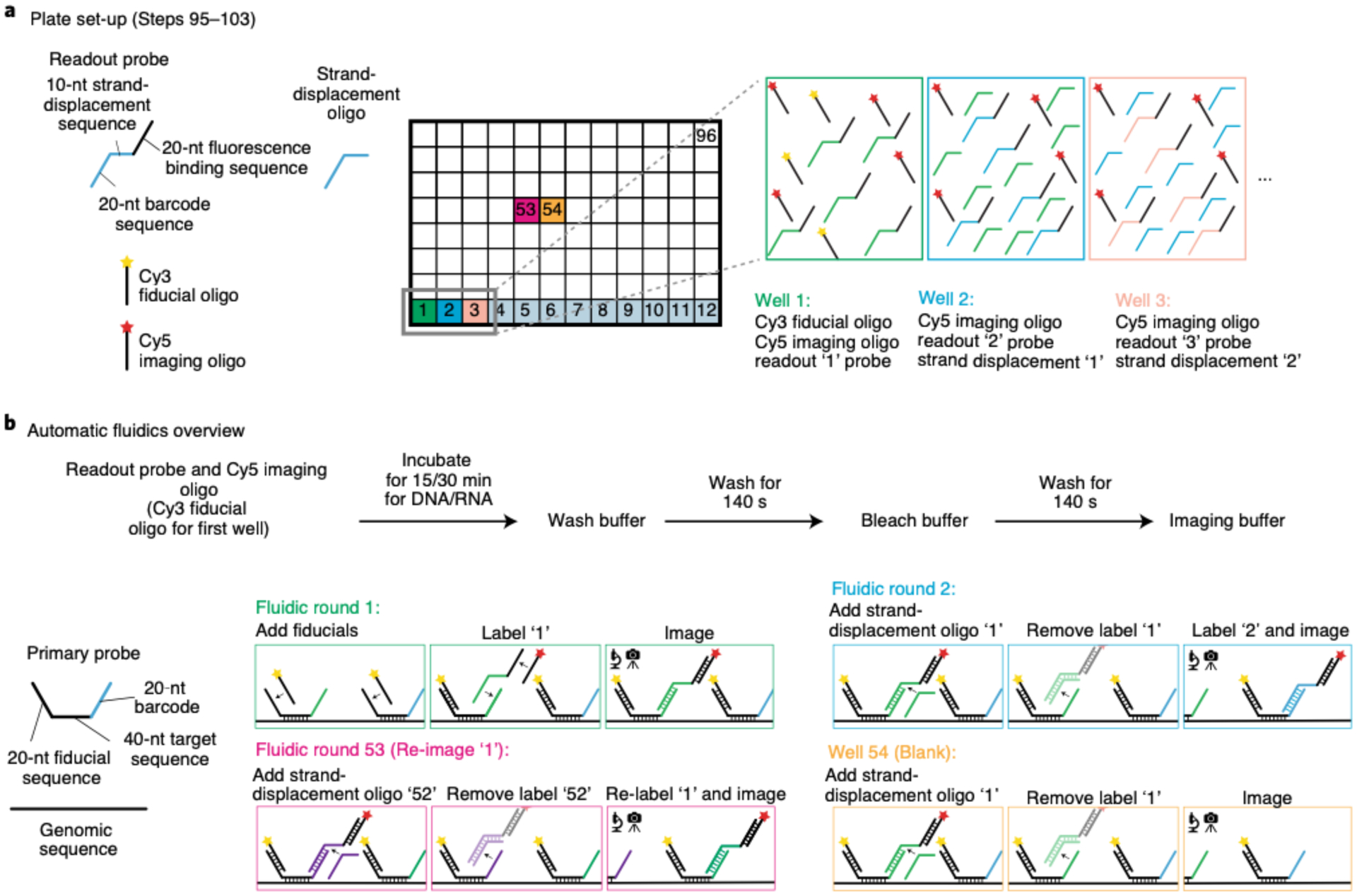
a, Readout probe, strand displacement oligos, Cy3 fiducial oligo, and Cy5 imaging oligo are pipetted into a 96-well plate containing 600 μL of 25% (vol/vol) EC in 2x SSC. As all primary probes tiling the region of interest contain a 20-nt fiducial sequence, the Cy3 fiducial oligo only needs to be added to the first well of the 96-well plate. The Cy5 imaging oligo will be used to acquire readout signals during automated imaging. Each readout probe contains a 20-nt barcode sequence to bind onto the barcode of the primary probe, a 10-nt strand displacement sequence for the removal of the previous readout probe, and a 20-nt sequence to bind a Cy5 imaging oligo. Add the appropriate readout probe and strand displacement oligo to each well at the proper concentration. b, The flow chart illustrates the timing for each automated fluidic round. Each primary probe contains a 40-nt DNA target sequence, a 20-nt fiducial sequence, and a 20-nt barcode to bind the readout oligo. Barcoded genomic segments contain a minimum of 20 primary probes. Schematic of data acquisition sequence: in fluidic round 1, the Cy3 fiducial oligos bind to the primary probes, which will be imaged during each round of readout data collection. Readout oligo ‘1’ binds its target barcode ‘1’ and the Cy5 imaging oligo. Barcode ‘1’ is imaged simultaneously with the fiducial Cy3 signal; in fluidic round 2, readout probe ‘1’ is displaced by its corresponding strand displacement oligo; additionally, readout probe ‘2’ binds with target barcode ‘2’ and the Cy5 imaging oligo. Both steps in fluidic round 2 are important for sequential imaging. Fluidic round 53 illustrates the re-labeling and imaging of barcode ‘1’ after the end of the experiment. During analysis, we use the 3D position of the re-labeled barcode ‘1’ to quantify the localization precision against its original measurements. Fluidic round 54 is an example of how one could test the efficiency of a strand displacement oligo. In this example, strand displacement oligo ‘1’ is introduced to remove the readout probe ‘1’ signal. If the signal from readout probe ‘1’ is completely gone (i.e. the Cy5 channel should appear blank), then the strand displacement oligo is working properly. However, if there is Cy5 signal, it is recommended to dilute a fresh strand displacement oligo.
Mounting the sample in the flow chamber ● Timing 5 min
▲CRITICAL For step-by-step photos, refer to Extended Data Fig. 3.
-
104
Fit the circular rubber gasket 1 that has the two holes over the two metal aqueduct ports on the plastic chamber (Extended Data Fig. 3b).
-
105
Take the sample (from Step 94) out of the Petri dish and aspirate off excess 2x SSC buffer from the coverslip.
-
106
Carefully place gasket 2 on top of the coverslip (Extended Data Fig. 3c). Make sure that the tissue sample is within the open area of gasket 2 and not underneath it.
-
107
Place the microaqueduct slide on top of gasket 2 (Extended Data Fig. 3d). Make sure that the microaqueduct slide wells are perpendicular to gasket 2 to avoid formation of bubbles when flowing fluid across the chamber and sample.
-
108
Place the plastic chamber top upside down on a bench.
-
109
Place the sandwiched coverslip with tissue sample, gasket 2 and microaqueduct slide on top of the circular rubber gasket 1 (Extended Data Fig. 3e). Make sure to properly align the two holes on the microaqueduct slide to the metal aqueduct ports.
▲CRITICAL STEP Improper alignment may cause flow problems, such as no flow, for the automated fluidics system.
-
110
Place the metal mount on top of the plastic chamber containing the materials from the previous step by aligning the clamps and holes (Extended Data Fig. 3f).
-
111
Clasp the plastic chamber top against the metal chamber bottom to hold everything together, and then very carefully invert the chamber (Extended Data Fig. 3g).
▲CRITICAL STEP Misalignment of the microaqueduct slide relative to the metal aqueduct ports will lead to fluid flow issues downstream and potential damaging of the microaqueduct slide. Misalignment of the coverslip within the chamber may cause the circular coverslip to crack when the chamber is sealed tight.
-
112
Move the metal clamps into place and tighten by using the metal collar. To tighten, move the clamps in a counterclockwise manner (Extended Data Fig. 3h).
▲CRITICAL STEP Make sure to tighten the chamber with the clamp as much as possible, because a loose chamber can result in air being trapped in the chamber, which would lead to inadequate flow or the introduction of bubbles. Tightening the chamber too much can also lead to the coverslip and/or the microaqueduct slide cracking.
-
113
Using a pipette, manually flow in 200 μl of 2x SSC into the chamber (from the connect ‘in’ tube) to check whether the sample mounting was successful. The flow should be bubble free and without resistance through the chamber.
Fluidics and microscope setup for RNA multiplex labeling ● Timing 1 h
-
114
With a Kimwipe, gently clean the bottom of the coverslip with 100% (vol/vol) methanol.
-
115
Add immersion oil to the 60x objective and mount the flow chamber firmly onto the microscope stage (Fig. 9a).
▲CRITICAL STEP Failure to tightly secure the stage will cause major stage drifts throughout the experiment, and the worst case is that you could lose all your FOVs.
-
116
Connect the ‘In’ and ‘Out’ fluidics tubes to the flow chamber.
-
117
Create an ‘Experiment’ folder on the data drive and include an appropriate folder name containing the data and a short description of experimental sample(s) (e.g., E:\2017-11-30_WT_10–12hrs_(HoxRNA)_(BXC3kb)).
-
118
Create a ‘Settings’ folder inside the Experiment folder to save the fluidics protocol in the next two steps and later the microscope camera software parameter settings.
-
119
Open the fluidics system software (our software is called ‘Kilroy’).
-
120
Load the RNA fluidics protocol into Kilroy (Fig. 9b). This protocol can be copied and pasted from a previous experiment. This xml file contains information pertaining to the x, y and z position of the modified robotic pipette to draw out fluid from the 96-well plate, bleach buffer reservoir (2x SSC), wash buffer reservoir (30% (vol/vol) formamide in 2x SSC) and imaging buffer chamber. In addition, the Kilroy xml file has protocols specifying the flow rate (0.6 ml/min) and time for each solution. In our example dataset (https://bit.ly/2S6eCjk), users will find that within the Settings folder, we have two Kilroy files: (i) Kilroy_15mins.xml and (ii) Kilroy_30mins.xml. We use the latter file for the RNA multiplex labeling because we see that a longer hybridization time of the readout probe to the barcode results in a better RNA signal.
!CAUTION Formamide is toxic and can be absorbed through the skin. It should be handled with gloves and under a fume hood. It should be discarded with care and according to your institution’s environmental health and safety rules.
▲CRITICAL STEP Depending on the length of the tubing from the fluidics system to the sample on the microscope, you may have to adjust the flow timing in the fluidics protocol. To validate the flow timing, with the flow chamber connected, or the tubes disconnected from the chamber but connected with a thin plastic tube, introduce a small bubble into the needle and time its rate of travel through the clear tubing. A small bubble can be generated by using Kilroy and making the needle move up and down while fluid is flowing through the system. Small bubbles generally do not disrupt the sample, but extensive exposure to air can lead to evaporation and disrupt morphology and labeling, so it should be avoided when possible.
▲CRITICAL STEP For an RNA experiment, make sure that the readout probe, strand-displacement oligo and fluorescent oligos sit on the sample for 30 min in the fluidics protocol.
-
121
Place the 96-well plate containing readout probes, oligo dye(s) and strand-displacement oligos on the flow system.
-
122
Fill up the bleach buffer and wash buffer reservoirs.
▲CRITICAL STEP We use Parafilm to cover our reservoirs to prevent particles from falling into them. In addition, we replace the buffers at least every 1–2 months because of potential evaporation that might change the concentration of our buffers.
▲CRITICAL STEP Low levels of buffers can be detrimental to the experiment. If levels are too low, the needle will not be able to draw any fluid. Instead, the needle will draw air, which damages the tissue and creates pressure inside the tube, making the flow slower.
-
123
Through the fluidics system software, flow bleach buffer to make sure the flow is behaving normally. It is recommended to check if the flow is normal before recording a mosaic of your sample or taking any images.
▲CRITICAL STEP Troubleshooting the flow might require you to disconnect the ‘In’ and ‘Out’ tubes from the flow chamber, and this action can cause the flow chamber to move. Thus, all stage positions would change because of this movement.
-
124
Turn on the microscope main-power switch, which provides power to the camera, the stages, the timing hardware and the AOTF. Also turn on the lasers for the focus lock and for both fiducial and data channel(s), and the power for the fluidics system. Be sure to follow all laser operation procedures for your system.
-
125
Open the microscope camera software (our software is called ‘Hal’).
-
126
Load parameter files into the microscope camera software (Fig. 9c). These xml files include information about the number of lasers that will be used, which lasers will be used, the order that the lasers will illuminate, the number of frames that will be taken and the Z-scan step size. In our example dataset (https://bit.ly/2S6eCjk), users will find that within the Settings folder, we have two parameter files: (i) ParsBleach_647.xml and (ii) ParsZscan_5um_647_561.xml. The former file will be used for the initial 30-frame photobleaching of the sample and then for quick 3-frame photobleaching in between readout probe hybridization rounds. This is done with the 647-nm laser at maximum power (19,000 mW). The latter parameter file is used for sequential imaging using the 561-nm and 647-nm lasers at 100-nm z-step sizes for a 10-micron scan through the sample. We typically set the AOTF to use the 561-nm laser at 10% power of the 200 mW at the laser head and the 647-nm laser at 23% power of the 19,000 mW at the laser head.
-
127
Create an ‘RNA_Experiment’ folder inside the data folder to save raw data images, which are formatted as dax files.
-
128
In the microscope camera software, get the sample in focus in brightfield and activate the focus lock. Set the focus lock to Z-scan.
-
129
Create a ‘Mosaics’ folder inside the Experiment folder.
-
130
Open the mosaic software (ours is called ‘Steve’), and using the microscope camera software, record a mosaic of your sample in brightfield (Fig. 9d). Make sure to uncheck ‘Run Shutters’ in the microscope camera software when imaging the mosaic. If you have already added in the dye oligos, you can choose to record the mosaic in either the fiducial channel or the readout channel. This is helpful, for example, if you are interested in imaging embryos that have positive expression for a gene of interest.
-
131
Select RNA FOVs (Fig. 9d). A box will appear around the area that will be imaged.
▲CRITICAL STEP Make sure there is a bit of an overlap between FOVs within an embryo. This is essential for accurate drift correction to reconstruct the mosaic in downstream analysis.
-
132
Save RNA FOVs as a .txt document in the Settings folder (e.g., RNA_FOVs.txt). This document contains the x and y stage position of each FOV.
-
133
Save the mosaic in the Mosaic folder (e.g., RNA_mosaic.msc).
-
134Create an executable file (ours is called ‘Dave Recipe’), which is an xml file (Fig. 9e). Copy a previous executable file into the Settings folder and modify it for the current experiment. The purpose of the executable file is to dictate the order of the fluidics and imaging steps of the entire experiment. This file makes it possible for the experiment to be automated, as it will control both the fluidics system and microscope. This master file contains information including the directories where files will be saved, the fluidics protocol, the z position of where the sample is in focus, the laser parameters and the number of frames it will take during image acquisition. In our example dataset (https://bit.ly/2S6eCjk), within the Settings folder, we have provided the Dave Recipe we used for RNA imaging (dave_Hox_RNA_Expt.xml). Use the checklist below to verily that all the parameters are correct.
Dave Recipe checklist - Check that the directory paths are updated
- Check that the fluidics commands are correct
- Check that the parameter files are correct for the lasers needed
- Check that your executable file follows a similar pattern as the following: (i) bleach the data channel by using the laser that corresponds to the channel (e.g., 647 nm laser) for 30 frames to reduce tissue autofluorescence, (ii) hybridize the readout probe and fluorescent oligo(s) from the well plate, (iii) record a movie of readout and fiducial data, (iv) bleach the data channel for three frames and (v) repeat steps ii-iv. Remember to include strand displacement oligos to remove the previous readout signal
- Check that the ‘lock target’ number in the executable file matches that of the ‘offset value’ that can be found in the microscope camera software when the sample is in focus. This is important for proper z-scanning of the entire sample
-
135
Once everything has been checked, save the executable file in the Settings folder.
-
136
Generate empty folders in RNA_Experment in which to save data (i.e., Bleaching, Read_100, Read_101, etc).
-
137
Open the executable software (ours is called ‘Dave’).
-
138
Load the executable file onto the software (Fig. 9f).
-
139
Once the file has loaded, a window will appear where you will be asked to select the RNA FOV positions txt document (e.g., RNA_positions.txt).
-
140
You will be prompted to save the executable xml file that records the settings used in the experiment.
-
141
Click the ‘Validate’ button, which will check whether the proper parameter files are loaded, whether the fluidics system is active and that the target folders exist.
-
142
Run the experiment. We recommend that after bleaching the data channel for 30 frames and flowing in the contents from the first well (readout probe, readout fluorescent oligo and fiducial fluorescent oligo), you adjust the laser intensities for the experiment on the basis of the staining of the fiducial and readout probes. You might also want to adjust the focus lock if your entire sample is not getting imaged in Z.
■PAUSE POINT Typically, an RNA experiment with 30 readout probes will take 1.5 d to complete. Be sure to occasionally check on the microscope and fluidics to make sure that the experiment is running smoothly, and replenish buffers if necessary.
?TROUBLESHOOTING
-
143
(Optional) If you are not labeling DNA and wish to add and image a counterstain, such as DAPI or an antibody, you can add it and label it here. If you do want to add an antibody, make sure that the secondary fluorescent antibody does not overlap with a fluorescent channel that is being used to image the fiducial and readout probes. You can add the counterstain(s) in a well(s) from the 96-well plate and write a fluidics protocol to control for fluidics timing and washing in Kilroy. The timing should reflect that of a normal DAPI/antibody staining protocol. Refer to the fluidics protocol for a better understanding of how to control for fluidics timing. Keep in mind that you can choose to add additional counterstains at any point of the protocol. We chose to make it an optional step here, because adding a counterstain after RNA imaging is best for registration purposes during the analysis.
Fig. 9 |. Microscope and fluidics setup for an automated experiment.
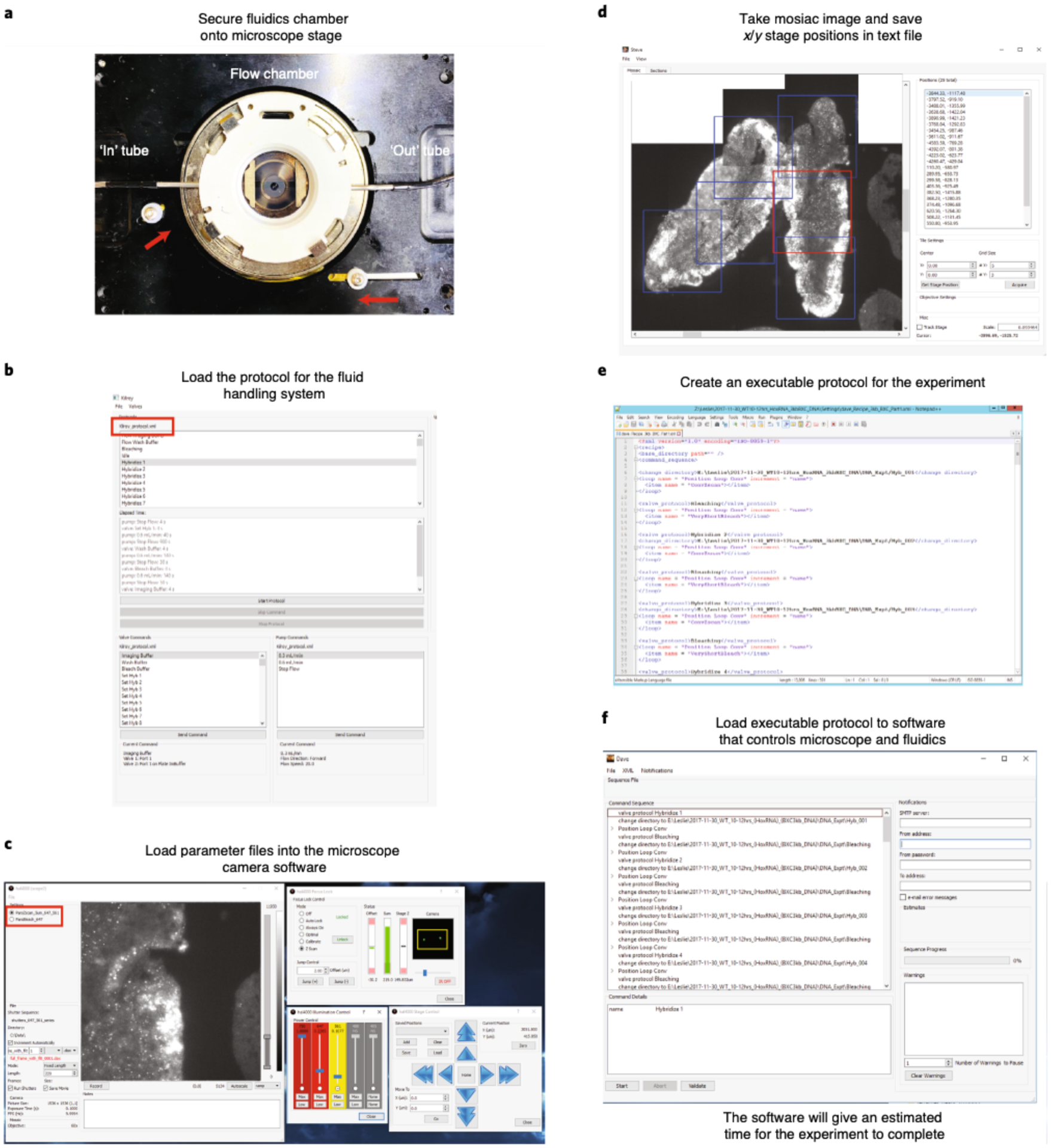
a, Tightly secure the flow chamber onto the microscope stage. Knobs on the stage have to be pushed in toward the flow chamber (shown with red arrows). Connect the ‘in’ and ‘out’ tubes (shown in white arrows) to the flow chamber. b, Open the fluidics system software and load the fluidics protocol. Examples of the fluidic Kilroy protocols are provided in the ‘Settings’ folder of the mini dataset (https://bit.ly/2S6eCjk), which are named Kilroy_15mins.xml and Kilroy_30mins.xml. The name of the loaded protocol is shown in the red box. Run Bleach Buffer through your sample to make sure that the flow is working properly. c, Open the microscope camera software and load the laser parameter files. Examples of the fluidic laser parameter files are provided in the ‘Settings’ folder of the mini dataset, which are named ParsBleach_647.xml and ParsZscan_5um_647_561.xml. The name of the parameters loaded is shown in the red box. Adjust the focus lock for your sample, which can be done in brightfield. The sample is in focus when you see two bright spots on the IR camera as shown inside the yellow box. In this example, we have already flown in the Cy3 fiducial oligo onto the cryosectioned Drosophila embryos that have been hybridized with RNA probes targeting 29 RNA transcripts. The 561-nm (Cy3) laser is used to visualize the fiducial signal for RNA primary probes. d, Using both the microscope camera software and the mosaic software, image a mosaic of your sample and manually record FOVs. Here we show a mosaic taken in the Cy3 channel. Example of a mosaic is in the ‘Mosaic’ folder of the mini dataset, which is named RNA_Brightfield_X.stv. Boxes represent the chosen FOVs where we purposely chose overlapping FOVs for downstream analysis. The red box indicates the currently selected FOV (which defaults to the last selected) The positions of these FOVs are saved and an example of these txt files (RNA_positions.txt and DNA_positions.txt) can be found in the ‘Settings’ folder of the mini dataset. e, The executable protocol can be copied and modified from a previous experiment. Here we show an example of a protocol. We have various examples of these protocols (‘dave.X.xml’) in the ‘Settings’ folder of the mini dataset. f, Open the Executable software and load in protocol from (e). Click ‘Validate’ to make sure that all parameter files are loaded onto their appropriate software and that the target folders, where data will be saved, exist. Afterward, click ‘Start’ to run your experiment.
Hybridization with DNA primary probes ● Timing 1.5 d
▲CRITICAL If you are working with cell culture and did not previously label RNA, start at Step 149 to permeabilize cells. The fixation step was already done in Cell culture fixation preparation.
▲CRITICAL In regard to the liquid volume used for 4% (vol/vol) formaldehyde in 1x PBS, 1x PBS, 0.5% (vol/vol) Triton-X in 1x PBS, 0.1 M HCl, RNase solution, hybridization no. 1, hybridization no. 2, 2x SSC and 2% (vol/vol) GA, 8% (vol/vol) formaldehyde in 1x PBS, we pour the solutions onto the Petri dish. Pour enough of the solution onto the Petri dish to cover the entire round cover glass. Make sure to not pour directly on top of the cells/tissues, because this could cause the sample to peel away.
-
144
If hybridizing DNA probes after imaging RNA, carefully take the coverslip with the sample off the flow chamber. The coverslip will most likely be stuck with the microaqueduct slide and gasket 2. To avoid cracking the coverslip, place the sandwiched coverslip, microaqueduct slide and gasket 2 in a 60-mm Petri dish and fill it with 2x SSC.
-
145
Put the Petri dish inside the 42 °C incubator for 30 min. The microaqueduct slide should separate from the cover glass on its own. Alternatively, you can immediately separate the cover glass and microaqueduct slide by using a pipette tip and sliding the end of the pipette tip between the cover glass and microaqueduct slide. However, we find that this alternative route leads to a higher probability of breaking the cover glass.
-
146
Place the round cover glass in the 60-mm Petri dish and remove the 2x SSC.
■PAUSE POINT If you want to store the RNA-labeled sample and do the DNA labeling at a later time, add 2x SSC to the sample. Seal the Petri dish with Parafilm to make sure that the 2x SSC does not evaporate and leave your slide dry. Store the Petri dish in a humid chamber and put it in the 4 °C refrigerator. In our experience, it is fine to do DNA labeling after the sample has been in the refrigerator for 1.5 weeks. We do not have any experience with samples stored for a longer period of time.
-
147
Fix the sample with 4% (vol/vol) formaldehyde in 1x PBS for 10 min.
-
148
Remove the fixation buffer and wash three times with 1x PBS.
-
149
Permeabilize the sample by adding 0.5% (vol/vol) Triton-X in 1x PBS for 10 min.
-
150
Wash three times in 1x PBS.
-
151
Add 0.1 M HCl for 5 min.
-
152
Wash three times in 1x PBS. Afterward, it is optional to incubate the sample for 1 h at 37 °C in RNase A solution to remove any RNA signal and then wash three times in 1x PBS.
-
153
Incubate in hybridization no. 1 for 35 min.
■PAUSE POINT Coverslips can be stored for ≤1 week at 4 °C in hybridization no. 1. Alternatively, you can wash three times in 2x SSC and store it at 4 °C.
-
154
During the 35-min wait, set the heat block to 90 °C. Place a hybridization chamber (this can be an empty pipette tip box with water added into the pipette tip holes) in a 42 °C incubator and allow it to equilibrate. Take an 18 × 18-mm cover glass and clean it with 100% (vol/vol) methanol. Cover the 18 × 18-mm cover glass in Sigmacote and allow it to air dry in the hood. After the 35-min wait, rinse the remaining Sigmacote from the 18 × 18-mm cover glass with ultrapure H2O and dry it with a Kimwipe.
-
155
Remove hybridization no. 1.
-
156
Add the hybridization mixture (1.5 μl of primary probe from a 500-ng/μl stock in 30 μl of hybridization no. 2) onto the sample while avoiding introducing air bubbles.
-
157
Place the Sigmacote-treated 18 × 18-mm cover glass on top of the hybridization mixture to spread the primary probe.
-
158
Time the heat denaturation step for tissue or cultured cells, respectively, as follows. For tissue/embryos, place the Petri dish on top of the heat block at 90 °C for 10 min. The timing can be shortened to 5 min, but we generally observe better embryo labeling with 10-min heat denaturation. For cultured cells, place the Petri dish on top of the heat block at 90 °C for 3 min. We find that 3 min is sufficient for decent labeling.
-
159
Hybridize the sample inside the hybridization chamber and leave it overnight in a 42 °C incubator.
■PAUSE POINT The sample can be left at 42 °C for ≤24 h.
-
160
The following day, pre-warm 2x SSC solution to 42 °C.
-
161
Add 2x SSC to the sample and return the sample to the 42 °C incubator for 10 min.
-
162
Carefully remove the 18 × 18-mm cover glass from the sample by using forceps.
-
163
Wash in 2x SSC and incubate for 10 min at 42 °C.
-
164
Wash in 2x SSC at room temperature.
-
165
Add post-fix buffer (2% (vol/vol) GA, 8% (vol/vol) formaldehyde in 1x PBS) for 1 h.
!CAUTION Both GA and formaldehyde are toxic and should be handled with gloves and under a fume hood. This buffer should be discarded with care and according to your institution’s environmental health and safety rules.
-
166
Wash three times in 2x SSC at room temperature.
■PAUSE POINT The sample may be stored in 2x SSC at 4 °C for ≤2 months.
96-well plate preparation for robotic hybridization ● Timing 20 min
-
167
Follow the same instructions as Steps 95–103.
Mounting the sample in the flow chamber ● Timing 5 min
-
168
Follow the same instructions as Steps 104–113.
Fluidics and microscope setup for DNA labeling ● Timing 1 h
-
169
With a Kimwipe, gently clean the bottom of the coverslip with 100% (vol/vol) methanol.
-
170
Add immersion oil to the objective and mount the flow chamber firmly onto the microscope stage (Fig. 9a).
-
171
Connect the ‘In’ and ‘Out’ fluidics tubes to the flow chamber.
-
172
Create a Settings folder inside the Experiment folder, which was created earlier for the RNA experiment, to save the fluidics protocol in the next two steps and later the microscope camera software parameter settings. If you did not run an RNA experiment earlier, create an Experiment folder on the data drive and include an appropriate folder name containing the data and a short description of the experimental sample(s) (e.g., E:\2017-11-30_WT_10–12hrs_(BXC3kb)). Create a Settings folder inside this folder.
-
173
Open the fluidics system software (our software is called ‘Kilroy’).
-
174
Load the fluidics protocol xml file onto Kilroy (Fig. 9b). This xml file contains information pertaining to the x, y and z positions of the modified robotic pipette to draw out fluid from the 96-well plate, bleach buffer reservoir (2x SSC), wash buffer reservoir (30% (vol/vol) formamide in 2x SSC) and imaging buffer chamber. In addition, the Kilroy xml file has protocols specifying the flow rate (0.6 ml/min) and time for each solution. In our example dataset (https://bit.ly/2S6eCjk), users will find that within the Settings folder, we have two Kilroy files: (i) Kilroy_15mins.xml and (ii) Kilroy_30mins.xml. For a DNA experiment, make sure that the readout probe, strand-displacement oligo and fluorescent oligos sit on the sample for 15 min and thus use the Kilroy_15mins.xlml protocol.
!CAUTION Formamide is toxic and can be absorbed through the skin. It should be handled with gloves and under a fume hood. This solution should be discarded with care and according to your institution’s environmental health and safety rules.
▲CRITICAL STEP Depending on the length of the tubing from the fluidics system to the sample on the microscope, you may have to adjust the flow timing in the fluidics protocol. To validate the flow timing, with the flow chamber connected, or the tubes disconnected from the chamber but connected with a thin plastic tube, introduce a small bubble into the needle and time its rate of travel through the clear tubing. A small bubble can be generated by using Kilroy and making the needle move up and down while fluid is flowing through the system. Small bubbles generally do not disrupt the sample, but extensive exposure to air can lead to evaporation and disrupt morphology and labeling, so it should be avoided when possible.
-
175
Place the 96-well plate containing readout probes, oligo dye(s) and strand-displacement oligos on the flow system.
-
176
Fill up the bleach buffer and wash buffer reservoirs.
▲CRITICAL STEP Low levels of buffers can be detrimental to the experiment. If levels are too low, the needle will not be able to draw any fluid. Instead, the needle will draw air, which damages the tissue and creates pressure inside the tube, making the flow slower.
-
177
Through the fluidics system software, flow bleach buffer (2× SSC) to make sure the flow is behaving normally. It is recommended to check if the flow is normal before recording a mosaic of your sample or taking any images.
▲CRITICAL STEP Troubleshooting the flow might require you to disconnect the ‘In’ and ‘Out’ tubes from the flow chamber, and this action can cause the flow chamber to move. Thus, all stage positions would change because of this movement.
-
178
Turn on the microscope main-power switch, which provides power to the camera, the stages, the timing hardware and the AOTF. Also turn on the lasers for the focus lock and for both fiducial and data channel(s), and the power for the fluidics system. Be sure to follow all laser operation procedures for your system.
-
179
Open the microscope camera software (our software is called ‘Hal’).
-
180
Load parameter files into the microscope camera software (Fig. 9c). These xml files include information about the number of lasers that will be used, which lasers will be used, the order that the lasers will illuminate, the number of frames that will be taken and the Z-scan step size. In our example dataset (https://bit.ly/2S6eCjk), users will find that within the Settings folder, we have two parameter files: (i) ParsBleach_647.xml and (ii) ParsZscan_5um_647_561.xml. The former file will be used for the initial 30-frame photobleaching of the sample and then for quick 3-frame photobleaching in between readout probe hybridization rounds. This is done with the 647-nm laser at maximum power (19,000 mW). The latter parameter file is used for sequential imaging using the 561-nm and 647-nm lasers at 100-nm z-step sizes for a 10-micron scan through the sample. We typically set the AOTF to use the 561-nm laser at 10% power of the 200 mW at the laser head and the 647-nm laser at 23% power of the 19,000 mW at the laser head.
-
181
Create a ‘DNA_Experiment’ folder inside the data folder to save raw data images, which are formatted as dax files.
-
182
In the microscope camera software, get the sample in focus in brightfield and activate the focus lock. Set the focus lock to Z-scan.
-
183
Open the mosaic software in two separate windows (our mosaic software is called ‘Steve’). You will need two mosaics for the next two steps.
-
184
In the first mosaic software window, load the previous RNA mosaic (e.g., RNA_mosaic.msc) and RNA FOVs (e.g., RNA_FOVs.txt).
-
185
In the second mosaic software window, use the microscope camera software to record a mosaic of your sample in brightfield (Fig. 9d). Make sure to uncheck ‘Run Shutters’ in the microscope camera software when imaging the mosaic.
-
186
Manually select DNA FOVs that cover the same area as the RNA FOVs. Use the RNA mosaic and recorded RNA FOVs to help pick new DNA FOVs. We use the shape of the cryosectioned embryo to help us during this realignment process because each embryo has a unique shape. For cultured cells, after plating cells onto the 40-mm glass coverslip, we find it useful to draw a shape (star, hexagon, heart, etc.) on the coverslip by using a pipette tip. This action will scrape off cells from the coverslip, but it will also create a shape that can be used as a spatial orientation indicator when realigning. Do not worry about attempting to manually select DNA FOVs that look identical to the RNA FOVs. The downstream analysis will take care of rotational changes between the datasets. It is also not necessary to choose the DNA FOVs in the same order as the RNA FOVs.
-
187
Save DNA FOVs as a text file in the Settings folder (e.g., DNA_FOVs.txt).
-
188
Save the mosaic in the Mosaic folder (DNA_mosaic.msc).
-
189Create an executable file (ours is called ‘Dave Recipe’), which is an xml file (Fig. 9e). Copy a previous executable file into the Settings folder and modify it for the current experiment. The purpose of the executable file is to dictate the order of the fluidics and imaging steps of the entire experiment. This file makes it possible for the experiment to be automated as it will control both the fluidics system and the microscope. This master file contains information including the directories where files will be saved, the fluidics protocol, the z position of where the sample is in focus, the laser parameters and the number of frames that will be taken during image acquisition. In our example dataset (https://bit.ly/2S6eCjk), within the Settings folder, we have provided the Dave Recipes we used for DNA imaging (dave_Recipe_3kb_BXC_Part1.xml and dave_Recipe_3kb_BXC_Part2.xml). We have two recipes because the first one was used to image the first 24 barcodes and the second was used to image the remaining barcodes (barcodes 25–52 and control repeat post-experiment). Use the checklist below to verify that the Dave Recipe file has been modified correctly.
Dave Recipe checklist - Check that the directory paths are updated
- Check that the fluidics commands are correct
- Check that the parameter files are correct for the lasers needed
- Check that your executable file follows a similar pattern as the following: (i) bleach the data channel by using the laser that corresponds to the channel (e.g., 647-nm laser) for 30 frames to reduce tissue autofluorescence, (ii) hybridize the readout probe and fluorescent oligo(s) from the well plate, (iii) record a movie of readout and fiducial data, (iv) bleach the data channel for three frames and (v) repeat steps ii iv. Remember to include strand-displacement oligos to remove the previous readout signal
- Check that the ‘lock target’ number in the executable file matches that of the ‘offset value’ that can be found in the microscope camera software when the sample is in focus. This is important for proper z-scanning of the entire sample
-
190
Once everything has been checked, save the Executable File in the Settings folder.
-
191
Generate empty folders in DNA_Experment to save data in (i.e. Bleaching, Read_001, Read_002, etc).
-
192
Open the Executable software (ours is called Dave).
-
193
Load the Executable File onto the software (Fig. 9f).
-
194
Once loaded, a window will appear where you will be asked to select the DNA FOVs positions txt document (e.g. DNA_positions.txt).
-
195
You will be prompted to save the Executable XML file that records the settings used in the experiment.
-
196
Click the Validate button, which will check whether the proper parameter files are loaded and that the target folders exist.
-
197
Run experiment. We recommend that after bleaching the data channel for 30 frames and flowing in the contents from the first well (readout probe, readout fluorescent oligo, and fiducial fluorescent oligo), you adjust the laser intensities for the experiment based on the staining of the fiducial and readout probe.
■PAUSE POINT Typically a DNA experiment with ~50 readout probes will take 2–3 days to complete. Be sure to occasionally check on the microscope and fluidics to make sure that the experiment is running smoothly.
?TROUBLESHOOTING
-
198
(Optional) If you wish to add and image a counterstain after the DNA labeling, such as DAPI or an antibody, you can add it and label it here. If you do want to add an antibody, make sure that the secondary fluorescent antibody does not overlap with a fluorescent channel that is being used to image the fiducial and readout probes. You can add the counterstain(s) in a well(s) from the 96-well plate and write a fluidics protocol to control for fluidics timing and washing in Kilroy. The timing should reflect that of a normal DAPI/antibody staining protocol. Refer to the fluidics protocol for a better understanding of how to control for fluidics timing. Keep in mind that you can choose to add additional counterstains at any point of the protocol. We chose to make it an optional step here as adding a counterstain after RNA/DNA imaging is best for registration purposes during the analysis.
Image pre-processing ● Timing 5 hours
-
199
Transfer images from the microscope computer to a remote host with greater computer memory size.
-
200
Within the RNA_Expt folder, create an.xlsx table that describes the organization of your data: folder name (e.g. Read_001), Readout, Data type, Readout number, Channels, Channel Layout (one of these should contain fiducial data), Buffer frames, Total Frames, and RNA Names. Example can be seen in Supplementary Data 6. Save the.xlsx table as RNAtable.xlsx. Do not use commas in the “rnaName” column. The system uses a comma separated list to organize the channel names and will get confused.
-
201
Within the DNA_Expt folder, create an.xlsx table that describes the organization of your data: folder name (e.g. Read_001), Readout, Data type, Readout number, Channels, Channel Layout (one of these should contain fiducial data), Buffer frames, Total Frames. Example can be seen in Supplementary Data 7. Save the.xlsx table as ExperimentLayout.xlsx.
ChrTracer3—image analysis of DNA structure ● Timing 1–4 days
▲CRITICAL ChrTracer3 software was developed for analysis of raw DNA labeled images. As an input, it takes an.xlsx table containing information and folder names of the DNA experiment. As an output, it returns tab delimited.txt files with drift-corrected x, y, z positions for all labeled barcodes. These can be used directly to calculate the nm scale distances between all pairs of labeled loci. The current version of the software as of this writing is ChrTracer3. The GitHub repository (https://github.com/BoettigerLab/ORCA-public) will contain the latest public release of the software, which can be found in ‘ORCA-public/GUIs’.
-
202
Make a new folder within DNA_Experiment called ‘Analysis’ (e.g. E:\2017-11-30_WT_10–12hrs_(HoxRNA)_(BXC3kb)\DNA_Expt\Analysis\). ChrTracer3 outputs will be saved in this folder.
-
203
Start MATLAB (currently running MATLAB 2018b).
-
204
Launch ChrTracer3 () in a MATLAB terminal to begin labeled chromatin analysis. A GUI labeled “ChrTracer3” should pop up.
-
205
Load file names and create max-project data by adding ‘Ex. Table’ and ‘Save Folder’ information onto the GUI (Fig. 10a). This step will parse your experiment table, which includes information about the channels used for your readout and fiducial data. This step will create max projections along the z-axis for all fiducial data for each FOV. The projections will later be used for x-y drift registration. (Z-drift is largely handled by the live-feedback auto-focus system. Nanoscale z-drift is further corrected in the ChrTracer3 GUI). The fiducial max projections will be saved within its respective readout folder (e.g. Read_001) within the DNA_Experiment folder. Look at the table below for input information.
Make sure to add ‘.xlsx’ to the end of your input for the Ex. Table. Also, add a ‘\’ to the end of your input directory for the ‘Save Folder’. Not doing so will cause errors to the software.Directory path input Ex. (Experiment) Table Type in the directory containing the ExperimentLayout.xIsx (e.g. E:\2017-11-30_WT_10-12hrs_(HoxRNA)_(BXC3kb)\DNA_Expt\ExperimentlLayout.xlsx) Save Folder Type in the directory targeting the Analysis folder (e.g. E:\2017-11-30_WT_10-12hrs_(HoxRNA)_(BXC3kb)\DNA_Expt\Analysis\) -
206
Click ‘Load Ex. Table’. This step takes a long time depending on the number of FOVs and readouts within the experiment. For example, this loading step takes 44 minutes for just 4 FOVs and 57 barcodes.
-
207
Click ‘Next Step’.
-
208
Click ‘Fix Global Drift’ to compute corrections for x-y stage drift (Fig. 10b). We recommend trying the default values for all parameters for this step to start. This step aligns images from sequential hybridizations to one another to correct for stage-drift and sample-drift between imaging rounds, using the fiducial image data specified in the reference table. Alignment is done by maximizing the cross-correlation between the fiducial images. For speed, a coarse alignment is performed first using a downsampled version of the image, then a fine-scale alignment is performed using only a subset of the image, once the two are brought approximately in register (see Fig. 10b). All output data are saved in the ‘Analysis’ folder. Stage drift information for individual readouts within each FOV will be saved in a.csv file (e.g. fov001_registeredData). An overlay of the corrected Fiducial is also saved as a png image. This step takes a while depending on the number of FOVs and readouts within the experiment. For example, this step takes 10 minutes to correct the drift for 4 FOVs with 57 barcodes.
-
209
(Optional) If you choose to change any of the default parameters, click ‘Step Pars’. Table 3 lists the parameters you can change.
?TROUBLESHOOTING
-
210
Click ‘Validate Drift Fix’ to see the result of the x-y drift corrections (Fig. 10c). A new window will display the overlay of the drift corrected Fiducials for all FOVs. The final image shows a multicolor overlay, where each channel is assigned a unique step on the color wheel. If all channels are aligned correctly, the resulting image should be white. Click ‘Next’ on the new window to scan through each FOV. This step will save the parameters chosen for this step in a.csv file (e.g. ParsValidateDriftFix.csv).
-
211(Optional) If you choose to change any of the default parameters, click ‘Step Pars’. The following are some of the parameters you can change:
Parameter name Default value Function verbose true Results will be printed to the command terminal recordValidation true Results of your selection will be saved and printed to the command terminal at the end selectFOVs inf If you do not want to Validate Drift Fix for all FOVs, you can specify particular FOVs here (e.g. 1, 2, 3, 10, 11, 12) -
212
On the new window, if a FOV is not properly drift corrected (the spots are not white and instead there are multiple colors for a single spot), then click ‘Bad’ (marked in a red box in Fig. 10c). This will record the number of the FOV that was not properly drift corrected.
?TROUBLESHOOTING
-
213
Click ‘Select Spots’. You will manually select a single Fiducial spot to examine how well the readouts labeled (Fig. 10d). A new window will display an image of the overlay of the Fiducial for FOV 1. The parameters for this step will be saved as a.csv file (e.g. ParsSelectSpots.csv). We will later return to this step to further validate our parameters, but for now choose one spot.
-
214(Optional) If you choose to change any of the default parameters, click ‘Step Pars’. The following are the parameters you can change:
Parameter name Default value Function selectFOVs 1 If you do not want to select a fiducial spot in FOV 1, which is the default, you can specify particular FOVs here imcontrastLow 0 Corresponds to the contrast to use when rendering an image. This fraction of the dimmest pixels will be set to zero/black imcontrastHigh 0.999 Pixels brighter than this fraction will be saturated -
215
In the new window, use the zoom-in tool to zoom in on an area with bright white spots on the image.
-
216
Use the data tips tool (marked in a red box in Fig. 10d) to dick on a bright white spot that is well isolated from other spots. Once a spot is selected, then press the Return button on your keyboard to record the position of the spot. The command window will display “Position Captured”.
-
217
Click ‘Next Step’.
-
218
Click ‘Crop Spot’ to see your individually labeled readouts and fiducials (Fig. 10e). Four windows will display the x-y and x-z max projection for each individual readout probe and the fiducial signal for the selected spot. Two windows will display the overlay of the max projected fiducial and readout in both x-y and x-z. The last window will display the intensity signal for each readout probe. This step will save the parameters for this step as a.csv file (e.g. ParsCropSpots.csv) in the ‘Analysis’ folder. This step might take a few minutes.
-
219(Optional) if you choose to change any of the default parameters, click ‘Step Pars’. The following is one of the parameters you can change if you see that your spot is too large to completely crop:
Parameter name Default value Function boxWidth 18 Corresponds to the width in pixels that will be used to crop your selected spot. This can be increased if your spot is large in terms of genomic size or perhaps your genomic region of interest is very decondensed and requires a larger size to crop -
220
Click ‘Next Step’.
-
221
Click ‘Fit Spot’ to find the centroid of the fiducial and readouts using a 3D-Gaussian spot-fitting algorithm (Fig. 10e). This step first uses the fiducial data from each hybridization to correct for nanoscale drift. This is accomplished by upsampling the cropped fiducial images and then computing the x, y, and z shifts which maximize the 3D cross-correlation between the cropped image and a reference image (by default the first fiducial image). After 3D finescale drift correction, this step uses 3D-Gaussian fitting to identify to sub-pixel precision the center of each of the spots in the data channel(s) for each hybridization. The windows with x-y and x-z max projections for readouts will now display a yellow circle around the detected readout spots. The max projected fiducial image will display a plus symbol on the center of the fiducial spot. The parameters for this step will be saved as a.csv file (e.g. ParsFitSpots.csv) in the ‘Analysis’ folder.
-
222
(Optional) If you choose to change any of the default parameters, click ‘Step Pars’. The parameters you can change are listed in Table 4.
?TROUBLESHOOTING
-
223
Repeat Steps 213–222 for at least three fiducial spots to see whether the parameters for Fit Spots are detecting the readout spots well. To repeat these steps, click the ‘Back Step’ button twice to return to the ‘Select Spots’ step. After selecting a new fiducial spot in the ‘Select a Single Spot’ step, click ‘Next Step’. This will prompt a message asking you if you want to overwrite the existing data in ParsSelectSpots.csv. A similar message appears after the ‘Crop Spot’ and ‘Fit Spot’ steps. It does not hurt to overwrite these steps.
-
224
Click ‘Next Step’.
-
225
Click ‘Select All Spots’ to select all fiducial spots for all FOVs (Fig. 10f). This will open a new window with an image of the overlay of the Fiducial for FOV 1. Additionally, a new GUI named ChrTracer3_SpotSelector will pop up. This GUI will be used to select all fiducial spots for each FOV.
-
226
On ChrTracer3_SpotSelector GUI, click ‘Find Spots’.
-
227
(Optional) If too few or too many spots are chosen for a FOV, go to ‘Parameters’ and modify ‘autoSelectThreshold’ prior to changing any other parameter (Table 5).
-
228
If the threshold was changed in Step 227, click ‘Find Spots’ again.
-
229If you are still missing spots or have too many spots after changing the threshold, use these optional tools that can be found on the ChrTracer3_SpotSelector to help select spots:
ChrTracer3_SpotSelector Tools Function Add by Click A cursor tool will allow you to manually select fiducial spots that might have been missed Remove by Click A cursor tool will allow you to manually select fiducial spots that are not real spots. This could be background noise or in the case of Drosophila embryos, e.g. signal from the embryonic yolk which does not represent real fiducial spots Remove in Area This is a useful bounding polygon feature that will allow you to select a region of interest, such as embryonic yolk, that might not have any real fiducial spots Remove IDs Each spot has a given ID and you can type the spot ID below Remove ID and then click this button to remove desired spots Source Data By default, spot selection is based on the fiducial from readout 1. You can choose to change spot selection to the data channel instead and can choose which readout to visualize. We typically do not change this parameter, but include it here in case there is too much background in the fiducial channel -
230
Once you are satisfied with the number of spots in a FOV, click ‘Save Spots’. This step will save a.csv file containing the x and y coordinates for all spots chosen in this step. (e.g. Fov001_SelectSpots.csv). The GUI will automatically go to the next FOV.
-
231
Continue this process to select all spots for all FOVs. A warning sign will appear when you have reached the last FOV.
-
232
Click ‘Next Step’ on the ChrTracer3 GUI.
-
233Click ‘Fit All’ to automatically detect the 3D centroids (Fig. 10g). This will find the centroid of each readout probe for all fiducial spots for all FOVs and save its 3D position. All information is stored in a.csv file (e.g. Fov001_AllFits.csv) in the ‘Analysis’ folder (Fig. 10h). An example is shown in Supplementary Data 8. This step might take 1–3 days depending on how many readouts were labeled and how many FOVs were recorded. For example, this step takes 44 minutes for 4 FOVs that contain a total of 4,268 labeled nuclei, with 57 barcodes. We typically do not change any of the parameters for ‘Fit All’, but include them here nevertheless:
Parameter name Default value Function numParallel 10 Number of parallel pool of workers overwrite No By default, the output files (e.g Fov001_AllFits.csv) will not get overwritten. Change to ‘Yes’ if you want to overwrite these files checkOffset false Request software to align images along the z-dimension (axial) using the offset files saveData false Saves a compressed version of the data that contains only the cropped regions of the image. See function Readlmage5D for details about this file format
Fig. 10 |. ChrTracer3 GUI.
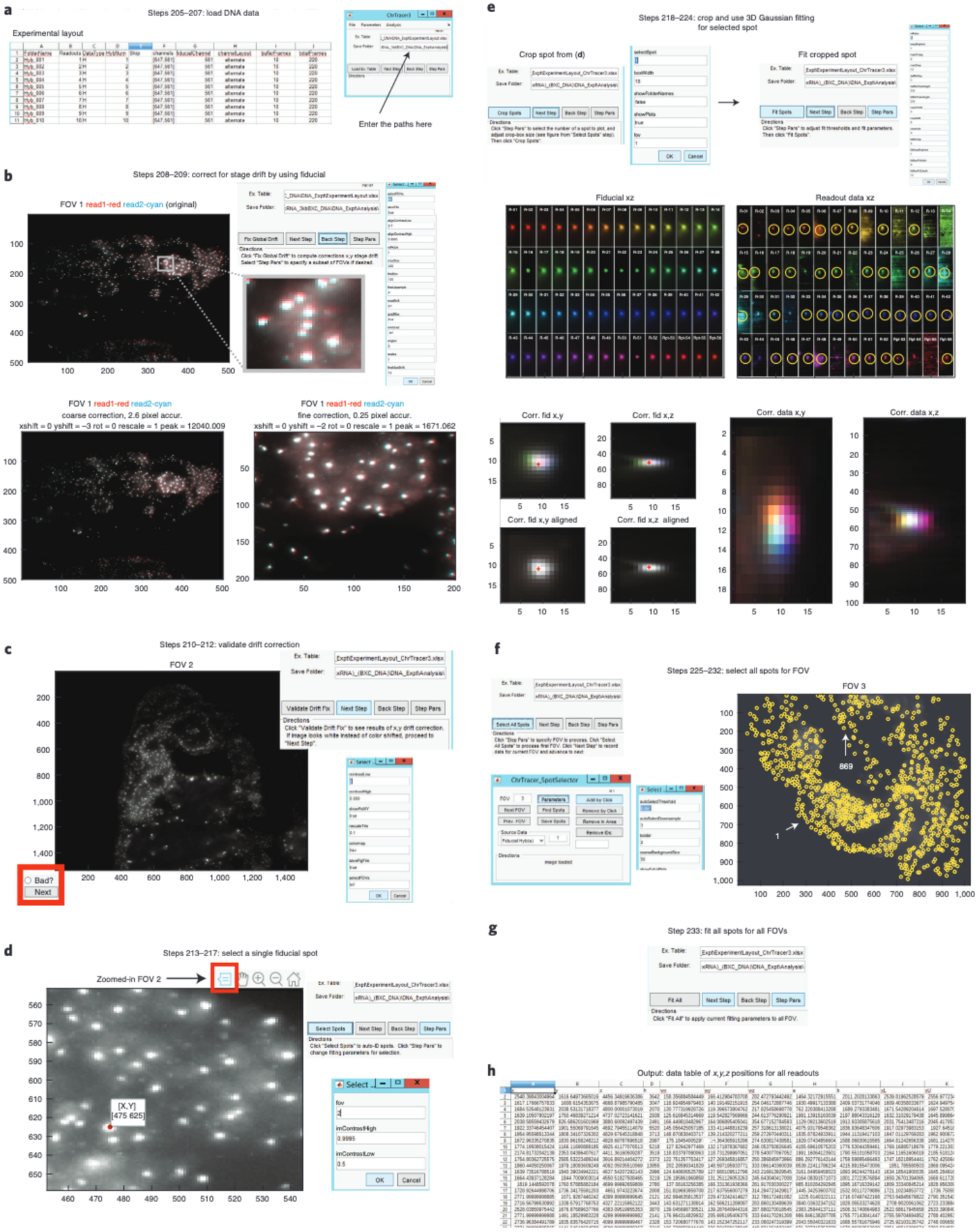
a, In ‘Ex. Table’, enter the directory path for your ExperimentLayout.xlsx file. In ‘Save Folder’, enter the directory path for your ‘Analysis’ folder. Click ‘Load Ex. Table’. b, Click ‘Fix Global Drift’ to correct for x-y stage drift between imaging rounds. A new window appears showing the drift correction process. Here we show the alignment of the fiducial that was imaged simultaneously with readout ‘1’ and readout ‘2’ for FOV 1. The Original image displays the drift that occurs between imaging rounds and this is further highlighted in the zoomed-in Original image. Coarse and fine-scale alignments are performed for drift-correction. The fine-scale alignment further corrects for drift by focusing on the brightest pixels from the down sampled version of the image. c, Validate the drift correction from (b) by looking at the multicolor fiducial overlay of all FOVs. Here is an example of an FOV that was properly aligned. Note that all the fiducial spots in the image are white. If the FOV was or was not properly corrected, click ‘Next’ or ‘Bad?’ (highlighted in a red box), respectively. d, Zoom into the fiducial overlay of an FOV and use the data tips tool (shown in a red box) to select a fiducial spot. e, Cropping spot from (d) will display images of x-y and x-z max projections of fiducial and readout data. The yellow circles show the readout spots detected based on the parameters chosen for the ‘Fit Spot’ step. The centroids of all fiducial and readout spots are detected using the 3D Gaussian algorithm. Centroid of the corrected (corr.) overlay fiducial spots are shown with “+”. Corrected overlay or readout data are also shown for x-y and x-z. Note how the readout spots do not overlay like fiducial, which represents the readout spots’ unique 3D spatial positions. f, Use the ChrTracer3 SpotSelector GUI to select all fiducial spots for all FOVs. All selected fiducial spots are shown within yellow circles. In this example, FOV 3 has a total of 869 fiducial spots. Typically, a unique number appears next to each fiducial spot. To simplify the figure, we have omitted these numbers. Instead, we use white arrows to highlight fiducial spots ‘1’ and ‘869’. g, Click ‘Fit All’ to automatically fit the centroid of each readout probe for all fiducial spots. h, Example of data table output that stores all 3D nanoscale positions for every readout probe.
Table 3 |.
Parameters for the ‘Fix Global Drift’ step in ChrTracer3
| Parameter name | Default value | Function |
|---|---|---|
| selectFOVs | inf | You can choose to correct stage drift for specific FOVs by specifying them under SelectFOVs. If inf (infinite) all FOVs will be used |
| saveFits | true | Will save the coarse alignments |
| alignContrastLow | 0.7 | Auto contrast to enhance feature matching between images. Pixels dimmer than this fraction of the brightness histogram will be set to zero |
| alignContrastHigh | 0.9995 | Auto contrast to enhance feature matching between images. Pixels brighter than this fraction of the brightness histogram will be saturated |
| refReadout | 1 | As default, all fiducials are aligned with the Fiducial imaged simultaneously with Readout 1. If you want, you can choose to align all Fiducial images with any readout. For example, if you rather align Fiducials based on the Fiducial image taken simultaneously with Readout 2, change refReadout from ‘1’ to ‘2’ |
| maxSize | 400 | All images will be downsampled to this size in terms of pixels for the coarse registration, prior to a second round of alignment on a subset of the image, at higher resolution. Set equal to image size or larger (‘inf’ is allowed) to do single step alignment |
| fineBox | 100 | Corresponds to the 100×100 pixel box size from the coarse alignment that will be used for fine alignment between fiducials from all FOVs. The fine alignment will use the brightest area of your image for this step |
| fineUpsample | 1 | The fine scale alignment can be performed to sub-pixel accuracy by upsampling each image. Values between ‘1’ (no upsampling) to ‘6’ (1/6 pixel accuracy) are recommended |
| maxShift | inf | This allows for shifts between fiducial images to occur during the coarse alignment without any shift restrictions |
| gradMax | true | Align where the gradient of the cross-correlation is largest (i.e. where the correlation strength changes the fastest, rather than where it is maximum). This typically performs better than maximizing the cross-correlation when aligning punctile data with varying brightness |
| minGrad | -inf | Optionally, select a minimum positive value for the gradient difference to accept the fine scale alignment. If this minimum improvement is not met, the maximum of the 2D cross-correlation function will be used instead of the gradient |
| angles | 0 | This refers to the angle of rotation potentially needed to drift correction. No angles need to be corrected here as the sample has not moved in a rotational manner during the experiment |
| scales | 1 | Isotropic expansion or contraction of image around the centroid. Typically not used here |
| fineMaxShift | 10 | Refers to the max shift in pixels that can be used for the fine alignment |
| fineAngles | 0 | If upsampling was used, the image can be corrected to sub-pixel resolution by scanning angles here. Typically not used here |
| fineScales | 1 | If upsampling was used, isotropic expansion can be corrected to sub-pixel accuracy by changing the scale here. Typically not used here |
| fineCenter | [0 0] | The computer will choose the best x-y position to do the fine alignment based on the pixel intensity of the fiducial spots. Optionally, this can be set to focus the fine-alignment on a particular pixel |
| showplot | true | This allows you to see Fig. 10b. It is not recommended to change this as visual inspection is always useful. However, if you decide to change this to ‘false’, you will be able to use MATLAB freely without the figure continuously appearing on your screen |
| fastDisplay | true | Downsample the overlay images displayed on the screen to the size specified by ‘displayWidth’. If the ‘maxSize’ is larger than the ‘displayWidth’, fastDisplay as ‘true’ will increase the run speed |
| displayWidth | 500 | If you wish to increase or decrease the display resolution of Fig. 10b, you can increase the size here and vice versa |
| showExtraPlot | false | Display the 2D correlation and gradient of the 2D correlation in addition to showing the alignment. Only recommended for troubleshooting |
| minFinelmprovement | inf | Optionally, require the absolute improvement in the correlation-coefficient of the fine alignment to be at least this much better than the correlation-coefficient of the coarse alignment. If this condition is not met, the coarse alignment values will be used and a warning message ‘fineAlign failed’ will be printed |
| showCorrAlign | true | This allows you to visualize the overlay of the corrected Fiducial for the latest readout corrected FOV |
Table 4 |.
Parameters for the ‘Fit Spot’ step in ChrTracer3
| Parameter name | Default value | Function |
|---|---|---|
| refHybe | 1 | Determines which hybridization round’s fiducial data is used as the reference for a final fine-scale 3D drift correction |
| keepsBrightest | 1 | Corresponds to the number of data spot(s) per readout that the computer will try to fit using the 3D Gaussian algorithm |
| maxXYstep | 8 | This is the number of pixels away from the center of the fiducial that can be considered a valid localization for the data |
| maxZstep | 10 | This is the number of z-sections away from the center of the fiducial signal that can be considered a valid localization for the data |
| datMinSep | 3 | If keepBrightest is more than 1, only readout spots within this number of pixels from one another will be considered to be in the same cell |
| datMinPeakHeight | 500 | This sets the minimum readout signal intensity value which is considered a valid localization |
| fidMinPeakHeight | 200 | This sets the minimum fiducial signal intensity value that is considered a valid image for alignment |
| maxXYdrift | 4 | The maximum number of pixels in XY that the algorithm is allowed to shift the image during the registration step |
| maxZdrift | 6 | Same as above, but for the z-drift |
| upsample | 4 | The correlation based alignment will upsample the fiducial data by this amount prior to registration. So ‘4’ corresponds to a registration accuracy of ¼ a voxel width |
| fidMinSep | 5 | If fidKeepBrightest is more than 1, Fiducial spots within this number of pixels from one another will be considered to be in the same cell |
| fidKeepsBrightest | 1 | This determines the maximum number of distinct fiducial spots the algorithm will try to detect per crop area. It will further attempt to find a data spot within the search range of each fiducial spot |
| fidMaxFitWidth | 8 | The Gaussian fitting will crop to a box of this width in pixels in x, y around the brightest pixel prior to fitting. This prevents stray background signals from dragging the fit off peak. This is used only for determining the centroid of the fiducial signal |
| fidMaxFitZdepth | 12 | This parameter is like the previous, but for the z-pixels (z-steps) |
| datMaxFitWidth | 8 | The Gaussian fitting will crop to a box of this width in pixels in x, y around the brightest pixel prior to fitting. This prevents stray background signals from dragging the fit off peak |
| datMaxFitZdepth | 12 | This parameter is like the previous, but for the z-pixels (z-steps). Separate control is allowed since z-step size may be different than x-y pixel size |
| datMinHBratio | 1.2 | The minimum ratio between the peak brightness and the background. This parameter affects how ‘spot like’ the signal needs to be counted. Decrease it to make fitting less stringent and increase the number to make fitting more stringent |
| datMinAHratio | 0.25 | The minimum ratio between the fitted amplitude of the Gaussian and the peak brightness. This parameter affects how ‘spot like’ the signal needs to be counted. Decrease it to make fitting less stringent and increase the number to make fitting more stringent |
| datMaxUncert | 2 | The maximum uncertainty in pixels for a spot to be recorded. Increase it to make fitting less stringent and decrease the number to make fitting more stringent |
| nmXYpix | 154 | This is the x-y pixel size in nanometers |
| nmZpix | 100 | This is the z-step between frames in the z-stack in nanometers (the z pixel size) |
| showPlots | true | The SpotFinder will plot the results of the fit as a tile of multicolor spots when finished |
Table 5 |.
Parameters for the ChrTracer3_SpotSelector GUI
| Parameter name | Default value | Function |
|---|---|---|
| autoSelectThreshold | 0.997 | If you want more fiducial spots, lower the threshold number and if there are too many spots then increase the threshold |
| autoSelectDownsample | 3 | Centroids are called on a downsampled image. If the spots are large, try increasing this value. You may need to lower autoSelectThreshold to compensate |
| coarseBackgroundSize | 50 | Image will be downsampled to this pixel size to estimate the illumination profile. “0” deactivates coarseBackground subtraction |
| densityFilterNeibs | 0 | Sets a threshold for the number of neighbors a spot may have within the cutoff distance set by densityFilterMinDist. Extra spots will be removed. A value 0 decativates this filter |
| densityFilterMinDist | 30 | Distance in pixels for the filter above |
| laplaceFilter | false | Uses a laplacian filter to correct brightness differences set to ‘true’ |
| edgeThresh | 0 | Uses an edge detection filter to correct brightness differences with a threshold set here. Selection of 0 deactivates this filter |
| edgeSigma | 1.2 | Controls the width of the edge detection filter above |
| imContrastHigh | 0.9995 | Pixels brighter than this fraction will be saturated |
| imContrastLow | 0.5 | Corresponds to the contrast to use when rendering an image. This fraction of the dimmest pixels will be set to zero/black |
| displayContrastHigh | 0.999 | Changes the contrast in the display image only, does not saturate the image used to call spots |
| displayContrastLow | 0.5 | Changes the contrast in the display image only, does not saturate the image used to call spots, pixels below this fraction will be set to zero/black |
| backgroundCorrect | none | Choose between ‘none’, ‘file’, or ‘median’. Decide if illumination should be flattened. If you recorded dax files of a blank illumination, select ‘file’. If you have many fields of view with randomly positioned data, you may use ‘median’ to estimate the illumination bias from the data. To skip background correction, select 'none' |
| border | 2 | Spots within this number of pixels from the edge will be removed |
| removeEdgeStack | 20 | If this many spots are observed in the same pixel row or column, they will all be removed. This helps correct edge effects in some images |
| showExtraPlots | false | If true, the software will display the results of any of the filters selected above, and wait for any key-press before proceeding to show the result of the next filter. For troubleshooting only |
Analyzing ORCA output ● Timing 1 h
-
234
In our Mini Dataset, we provide examples of the output of ChrTracer3, which are the ‘FovX_AllFits.csv’ files (see https://bit.ly/3j442Us). We also provide a MATLAB script, called ORCA_MapsAndPolymers.m, for generating Distance maps, contact maps, and single-cell 3D polymers (see https://bit.ly/3k5hj0p which will direct your to a folder called ‘ORCAMapsPolymer-Output’). The ORCA_MapsAndPolymers.m script has step by step instructions in the comments to aid with analyzing the output tables. While we primarily use MATLAB to analyze our data, other programming languages can be used for this step. Looking at the generated distance maps can be very useful for detecting potential issues during the ORCA barcode labeling.
?TROUBLESHOOTING
Build Mosaics: Drift-correct DNA and RNA mosaics ● Timing 1–8 h automated (step 237), 1–2 h interactive (steps 238–255)
▲CRITICAL The BuildMosaicsGUI should be run separately for the RNA and DNA experiment. For DNA, the output of ChrTracer3 will be used as inputs for BuildMosaicsGUI, including the max projection fiducial/readout images and drift correction information. The output of the BuildMosiacsGUI for a DNA experiment is a.xml file containing FOV to FOV drift corrected information. Using this file ensures that overlapping FOVs are correctly positioned relative to each other (most useful for tissue level analyses). If you only have DNA data, and do not care about spatial positioning of cells (e.g. tissue culture cells), Steps 235 – 301 are unnecessary.
For RNA, an.xlsx table containing information about each readout in the RNA experiment is provided as an input. The output of the BuildMosaicsGUI for an RNA experiment is a Tif mosaic for each RNA where overlapping FOVs are positioned correctly relative to each other.
-
235
Start MATLAB (currently running MATLAB 2018b).
-
236
Launch BuildMosaicsGUI () in a MATLAB terminal to begin mosaic analysis. A GUI labeled “Build Mosaics GUI” should pop up.
-
237
Load file names and max-project data (Fig. 11a). Click ‘Parameters’.
This step will parse the experiment table created in ‘Image pre-processing’ to find the necessary data, and create max projections along the z-axis for all channels as a preliminary step for x-y drift registration. Note, creating max-projections of terabytes of data takes a long time. Data outputs will be saved in the ‘Analysis’ folder so that this step runs faster if repeated in the future. Make sure to add ‘.xlsx’ to the end of your input for the ‘chnTableFile’ in the parameters below. Not doing so will cause errors to the software. The following are parameter options that need to be changed differently for both DNA and RNA data: -
238
Click ‘Run Step’. This may take a while depending on how many FOVs were selected for the experiment.
-
239
Click ‘Next Step’.
-
240
Click ‘Run Step’ to correct channel-to-channel drift (Fig. 11b). We recommend trying the default values for all parameters. This step aligns images from sequential hybridizations to one another to correct for stage-drift and sample-drift between imaging rounds, using the fiducial image data specified in the experimental table. This same function with the same parameters is called by BuildMosaicsGUI and ChrTracer3. If DNA/ORCA alignment has already been performed, the software can detect it by the presence of the fov*_regData.csv files for each FOV, and will offer to skip this step. A final image shows a multicolor overlay, where each channel is assigned a unique step on the color wheel. If all channels are aligned correctly, the resulting image should be white (Fig. 11b). Stray points that appear in only a subset of channels will show up in the corresponding color. Gradual drift will also appear as a rainbow, projecting in the direction of the drift along the “hsv” colormap. Stage drift information for individual readouts will be saved in a.csv file, (e.g. fov001_registeredData) and for each FOV. An overlay of the corrected Fiducial is also saved as a png image.
-
241
(Optional) If you choose to change any of the default parameters, click ‘Parameters’. The parameters you can change are listed in Table 8.
-
242
Click ‘Next Step’.
-
243
Click ‘Run Step’ to create an initial mosaic in preparation for FOV-to-FOV-drift correction (Fig. 11c). A new figure should appear and display a mosaic of all tiles. It is recommended this is done with all FOVs, and using the fiducial data from the first hybridization. However, you may select any round or any data type in the ‘parameters’.
-
244
(Optional) If you choose to change any of the default parameters, click ‘Parameters’. The parameters you can change are listed in Table 9.
-
245
(Optional) A new GUI will appear called ‘Mosaic Viewer App’ (Fig. 11c). If you want to view the outline of the tiles of the mosaic, on the new GUI click ‘Show Boxes’ (shown in a green rectangle). The tiles and their corresponding names will appear on mosaic (see top right corner of Fig. 11c).
-
246
(Optional) The Mosaic Viewer App GUI allows you to further examine the stage drift between adjacent FOVs. First, zoom into the mosaic using the figure toolbar and in the new GUI, click ‘Zoom In Here’. This will allow you to zoom into an area of the mosaic at high resolution. Next, click ‘Select Tile’ (shown in a red rectangle) and use the cross-hairs to select a tile on the zoomed-in mosaic. One tile will turn red and the other will turn cyan. If you further zoom in using the figure toolbar, you will see the stage drift between FOVs (see bottom of Fig. 11c). Spots that should be overlapping are in close proximity to each other.
-
247
Click ‘Next Step’.
-
248
Click ‘Run Step’ to manually adjust tile positions in mosaic (Fig. 11d). The computer will use the fiducial data within the ~10% overlapping edge pixels between adjacent FOVs to correct the offset. However, if the fiducial spots in the overlapping FOV region remain drifted, follow the instructions on the GUI’s screen to manually shift a FOV using the keyboard.
This step aligns the different FOVs, to correct for stage-drift and inaccuracies in stage-positioning within a single imaging round. Because of this drift, the hardware reported coordinates of two partially overlapping fields of view will not match in the overlapping region to nanoscale accuracy. However, with sufficient overlap (typically 10% of the image area), this difference can be corrected to create smooth, unbroken images of the tissue. We refer to the different FOVs in the mosaic as ‘tiles’ in the mosaic image. First the software determines the order in which tiles should be placed. The first tile placed determines the new reference coordinates -- all other tiles placed in the mosaic must be positioned accurately with respect to this tile, so they must overlap with it, or with tiles placed after it that overlap the first tile. If there are disconnected regions each of multiple tiles, the tiles for each region may be placed independently. Tile order is determined by examining the tile of images to identify which tiles have overlap, and to what extent the image data (above background signal) in the tile also overlaps.
Having determined the number of connected regions, the algorithm starts in the largest connected region with the single tile that has the most overlap, and proceeds to place all its overlapping fellows in order from most to least overlap, and spirals out from there until all tiles in the first connected region are placed. Tile placement occurs in a supervised automated manner, with maximum cross-correlation (or maximum gradient of the cross-correlation) being used to determine overlap. If drift is large, or the sample quite sparse, some tiles may fail to have sufficient identifiable overlapping features to register on, even though their stage coordinates overlap. In such cases, maximum correlation will suggest an erroneous position. We therefore require the user to validate each automatically determined tile position. If there is insufficient overlap, the user may postpone placing a tile instead, and it is added to the back of the stack. Typically, placing subsequent tiles provides enough image data to constrain the later placement of this tile (and if no features constrain its placement, then the absolute coordinates are not essential). The user may also use the key-pad to manually adjust tile positions which have been inaccurately placed by the automatic algorithm.
This step will save the coordinates of the tile positions in a regFOV.csv file in the ‘Analysis’ folder. This file will be used to construct the final mosaic in Step 252. Additionally, regFOV.csv is used to construct both the DNA and RNA mosaics in AlignMosaics GUI () below (steps 258 and 261 for DNA and RNA, respectively).
-
249
(Optional) If you choose to change any of the default parameters, click ‘Parameters’. The parameters you can change are listed in Table 10.
-
250
(Optional) Once done aligning FOVs, you’ll be given the option ‘Select individual tiles to validate alignment’. If you wish to validate the alignment, click ‘Run’. This will prompt the same Mosaic Viewer App described in Step 246, where you can click ‘Select Tile’ and then click an area of the mosaic to see how well your adjacent FOVs aligned (see the bottom right comer of Fig. 11d). After selecting a tile, if needed, follow the instructions on Mosaic Viewer App to manually move the FOV tile. You will be able to use your keyboard again to further adjust an FOV.
-
251
Click ‘Next Step’.
-
252
For RNA data only, click ‘Run step’ to inspect multicolor RNA mosaic (Fig. 11e).
If you are processing DNA/ORCA data, skip Steps 252–255. The overlay of all data channels for DNA is rather uninteresting at the mosaic scale as all spots overlap. For DNA/ORCA only data, ChrTracer3 can be run prior to the MosaicBuilder and the output super-resolution traces from ChrTracer3 can be imported later in the mosaic analyzer.
This step loads the max projections of all the data channels and arranges them in the mosaic based on the FOV positions registered in Step 248 and the channel positions registered in Step 240. The data are shown as a multicolor projection. The “Ncolor” display tool allows the user to select which channels to display (left panel), what colors to display them as, and allows for the contrast to be adjusted (select channel to adjust using the right panel) -- all controlled with intuitive buttons and sliders, (see Fig. 11e). The channel names are determined from the experiment layout Excel file. Hold down the “ctrl” key to look at multiple channels. Click ‘Edit Colormap’ on Ncolor App to change RGB values (ranging from 0 to 1) for gene(s) of interest.
To provide accelerated viewing of large mosaics, the GUI automatically downsamples the data before plotting (current defaults are set to a 2400 ×2400 pixel image, which means on most monitors no detail is lost). Using the built in MATLAB zoom tool will expand the region selected, but will not increase the resolution. After zooming into a select region, click ‘Zoom in Here’ in the Ncolor App to show this part of the mosaic at higher resolution (up to full image size). To zoom in on a different region, first select “Zoom Out” to restore the full mosaic.
Note, the Ncolor adjustments only affect the display. The full resolution, unsaturated image is stored in memory. It is not necessary to interact with the Ncolor popup in order to proceed to the export Tif step. The parameter editor lets you choose which FOVs and channels (hybs) to view and change the default contrast. Typically no parameter adjustment is needed for this step, nevertheless the parameters and their functions are listed in Table 11. Defaults for hybs automatically changes to ‘inf’, and dataType changes to ‘all’.
-
253
Click ‘Next Step’.
-
254
Click ‘Run Step’ to save Data for RNA Mosaic (Fig. 11f).
A Tif file for every channel is saved. To avoid data loss, the images are not contrasted and some contrast adjustment in later viewing may be needed if the image appears mostly black. Tif compression avoids writing extra bits to cover portions of the mosaic not occupied by data. These images may be read by later tools described here (e.g. AlignMosaicsGUI, MosaicAnalyzer) or by third party applications such as ImageJ.
-
255(Optional) If you choose to change the default parameters, click ‘Parameters’. The following are the only two parameters you can change:
Parameter name Default value Function saveName Mosaic The Tif file will be saved in the ‘Analysis’ folder, along with the tile positions and tile filenames needed to reconstruct the mosaic from the max projected image files. Channel names are also saved as a txt file for easy reference. The default file name is ‘Mosaic’, but you can change it to any other name if you want saveFolder By default, the Tif file will be saved in the folder that was specified in the first step of BuildMosaicsGUI. However, you can change it to a new path directory here
Fig. 11 |. BuildMosaics GUI.
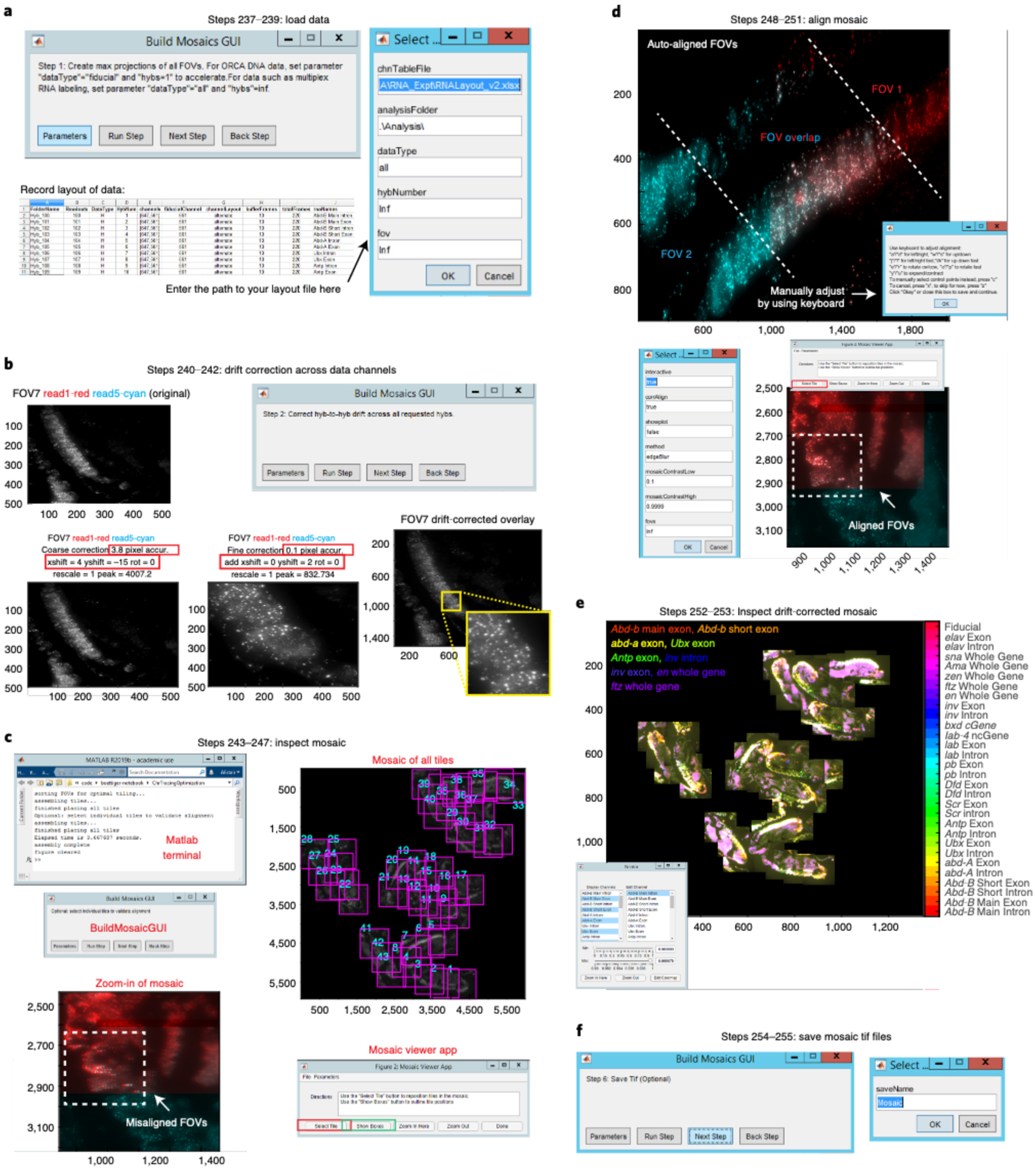
a, Load the experimental table and enter the appropriate parameters in ‘datatype’ and ‘hybNumber’ depending on if you are building a mosaic for RNA or DNA data. b, This step corrects for stage drifts across the experiment by using the fiducial data. In this example, fiducial data that was taken alongside readout ‘1’ and readout ‘5’ in FOV 7 is used for alignment. After coarse and fine alignment, an overlay of the corrected fiducial is displayed for FOV 7. c, View the mosaic of the fiducial from your first readout in the experimental table prior to FOV-to-FOV-drift correction. Click ‘Show Boxes’ (green rectangle) to view the position of all FOVs in the mosaic. Click ‘Select a tile’ (red rectangle) in the Mosaic Viewer App to see that FOVs are misaligned in the mosaic at this step. This is obvious by observing the non-overlapping red and cyan spots between overlapping FOVs. d, The computer uses an automatic algorithm to attempt to correct for FOV-to-FOV stage-drift by using the fiducial spots that overlap two FOVs. Manually check to see whether red and cyan spots overlap one another. If they do not, then read the instructions on the new window to manually adjust the FOVs by using your keyboard. After FOV-to-FOV drift correction, a new window will display a drift-corrected mosaic. You can click ‘Select a tile’ in the Mosaic Viewer App to see that FOVs are now aligned as the spots overlay with one another. e, Mosaic of all max projected readout channels. Here we are only showing 9 out of 29 RNAs. Use the Ncolor GUI to view gene(s) of interest. You can view multiple genes by holding onto the ‘Control’ button on your keyboard and clicking on different genes. Changing the Min/Max pixel intensity using the slider for gene(s) of interest will help with visualization. If you want to view a portion of the mosaic at high resolution, zoom-into the mosaic and then click ‘Zoom in Here’ in the Ncolor GUI. Click ‘Edit Colormap’ to change RGB values for gene(s) of interest. A list of RBG values that range from 0 to 1 will appear and the genes are in the same order as seen in Ncolor GUI. f, Save Tif files for every channel in your ‘Analysis’ folder.
Table 6 |.
Loading parameters for DNA data in BuildMosaicsGUI
| Parameter name | Default value | Change | Function value to |
|---|---|---|---|
| chnTableFile | Enter the path to your experiment layout file here (e.g. E:\2017-11-30_WT_10-12hrs_(HoxRNA)_(BXC3kb)\DNA\ExperimentLayout.xlsx) | ||
| analysisFolder | .\Analysis\ | Default value “.\Analysis\” is recommended. Saved alignment results and mosaics will be placed in the Analysis folder. “.\” notation saves data in whichever folder the chnTableFile is located, inside the newly created folder ‘Analysis’. There should already be an ‘Analysis’ folder created if ChrTracer3 ran before this step. No changes need to be made here | |
| dataType | all | fiducial | This specifies which data to create max projections of, fiducial, readout data, or all. The default is ‘all’, which corresponds to both the fiducial and readout channel. For creating a DNA/ORCA mosaic, type in 'fiducial' since you will only need to know the nanoscale positions of fiducial spots and do not need to create a mosaic for individual DNA readouts. If ChrTracer3 was already used in this dataset, it would have already created max-projections and this step will go by quickly as BuildMosaicsGUI will only need to load already created max projected fiducial data |
| hybNumber | inf | 1 | To create a DNA mosaic, only the fiducial data from readout 1 is needed. Thus, change the default ‘inf’ to ‘1’ to specify to use data only from readout 1 |
| fov | inf | The default is ‘inf’ to process data from all FOVs in the folder. This default value is recommended to use. It is optional to select a subset of FOVs (e.g. “[2:10,15:35]”) |
Table 7 |.
Loading parameters for RNA data in BuildMosaicsGUI
| Parameter name | Default value | Function |
|---|---|---|
| chnTableFile | Enter the path to your experiment layout file here (e.g. E:\20l7-11-30_WT_10-12hrs_(HoxRNA)_(BXC3kb)\RNA\RNAtable.xlsx) | |
| analysisFolder | .\Analysis\ | Saved alignment results and mosaics will be placed in the Analysis folder. “.\” notation saves data in whichever folder the chnTableFile is located, inside the newly created folder ‘Analysis’ |
| dataType | all | This specifies which data to create max projections of, fiducial, readout data, or all. The default is ‘all’, which corresponds to both the fiducial and readout channel. For creating a RNA mosaic, do not change the default setting. ‘All’ is necessary for loading RNA data as it will take the fiducial information for drift correction and the readout data to display RNA information. Depending on how many RNAs you labeled and the number of FOVs recorded, this step will take a while |
| hybNumber | inf | Do not change the default ‘inf’ as this will use fiducial data from all RNA readouts for drift correction |
| fov | inf | The default is ‘inf’ to process data from all FOVs in the folder. This default value is recommended to use. It is optional to select a subset of FOVs (e.g. “[2:10,15:35]”) |
Table 8 |.
Parameters for channel-to-channel drift-correction in BuildMosaicsGUI
| Parameter name | Default value | Function |
|---|---|---|
| alignContrastLow | 0.7 | This fraction of the dimmest pixels will be ignored for the alignment |
| alignContrastHigh | 0.9995 | Pixels brighter than this fraction of the brightest pixels will be saturated for the alignment |
| refHybe | 1 | The fiducial data from this hybridization round (e.g. ‘1’) will be used as the alignment reference |
| maxSize | 400 | For the coarse alignment step, the images larger than this size will be downsampled so the average dimension is this size |
| fineBox | 200 | Fine scale alignment extends this far in pixels around the center point |
| fineUpsample | 1 | Data will be upsampled by this factor to improve accuracy of fine alignment |
| maxShift | inf | The maximum number of pixels the coarse alignment may shift the image |
| gradMax | true | Boolean indicates whether or not to use the gradient of the correlation plot instead of the correlation values |
| minGrad | -inf | Minimum slope observed in the correlation map to accept a shift. To use any shift, keep default of “-inf” |
| fineMaxShift | 10 | Maximum number of pixels the fine alignment may shift the image |
| fineCenter | [0 0] | Fine scale alignment is centered at this point. If left at [0,0], the brightest region (not brightest pixel) of the image will be selected |
| showplot | true | Show plots during imaging |
| fastdisplay | true | Boolean, indicating whether display should be accelerated by downsampling. Typically limited by screen resolution; if you aren’t pausing and zooming in on the data, this should not affect quality |
| displayWidth | 500 | Image size to downsample to for fast display |
| showExtraPlot | false | As the default, correlation maps will not be displayed. Can be changed to ‘true’ to show the correlation maps, which can be useful for troubleshooting |
| minFineImprovement | 0 | If the correlation coefficient does not improve by this amount upon fine-scale alignment, reject the fine scale alignment |
| showCorrAlign | true | Shows images of before alignment, after coarse alignment, and after fine alignment. It is not recommended to change this parameter as keeping it ‘true’ allows you to visually check if the alignment is happening correctly |
Table 9 |.
Parameters for FOV-to-FOV drift-correction in BuildMosaicsGUI
| Parameter name | Default value | Function |
|---|---|---|
| backgroundCorrect | none | Parameter options are ‘none’, ‘file’, and ‘median’. Decide if illumination should be flattened. If you recorded dax files of a blank illumination, select ‘file’. If you have many fields of view with randomly positioned data, you may use ‘median’ to estimate the illumination bias from the data. To skip background correction, keep the default ‘none’ |
| method | edgeBlur | Keeping the default ‘edgeBlur’ is recommended. This setting uses a linear gradient of overlapping images regions to stitch borders. This is typically best for avoiding artificial edges. Other options include ‘mean’ - average the overlap, ‘sum’ - add the overlap, ‘last’ - use the last image and cover over what was imaged before, ‘first’ - use the first image and ignore later data |
| mosaicContrastLow | 0.1 | Contrast to use when rendering images. This fraction of the dimmest pixels will be set to zero/black |
| mosaicContrastHigh | 0.9999 | Contrast to use when rendering an image. This fraction of the brightest pixels in the image will be saturated |
| fovs | inf | “inf” default value is recommended to use all FOV in the folder. Optionally, select a subset (e.g. “[2:10,15:35]”) |
| hybs | 1 | Hybridization data that will be displayed in the mosaic. By default, we show data from readout ‘1’ |
| dataType | fiducial | Data to show for alignment mosaic |
| transpose | true | The default is ‘true’, which gives you the option to transpose tiles in creating mosaic |
| scope | autoDetect | Depending on the camera orientation, individual tiles may be flipped across the left-right and up-down axis to properly render the mosaic. In our case, the camera orientation is automatically detected based on the pixel size of the raw images. Outside users will have to configure this setting in the MATLAB code to make sure that the tiles are oriented correctly. Our code within the Github link (https://github.com/Boettigerlab/ORCA-public) is commented in BuildMosaicsGUI.m to make sure the user knows where to make the appropriate changes based on their camera orientation |
| trimBorder | 10 | Remove the number of specified pixels around the border - sometimes our cameras have a bright line of pixels at the edge of the image, for reasons currently unclear. This removes them |
Table 10 |.
Parameters for manually adjusting FOV tiles in BuildMosaicsGUI
| Parameter name | Default value | Function |
|---|---|---|
| interactive | true | Boolean, set to ‘true’ to run the alignment tool in interactive mode or not. We recommend using the default setting. Set to false to skip alignment |
| corrAlign | true | Boolean, set to ‘true’ perform correlation based alignment. Set to false to skip alignment |
| showplot | false | Set to ‘true’ to show additional alignment plots; recommended only for troubleshooting alignments |
| method | edgeBlur | Uses a linear gradient of overlapping images to stitch borders. This is typically best for avoiding artificial edges |
| corrAlignLow | 0.8 | Controls the lower bound for image contrasting used during automated alignment. This fraction of pixels will be set to 0/black, so the alignment is driven by the data and not the background |
| corrAlignHigh | 0.999 | Controls the upper bound for image contrasting used during automated alignment. Pixels brighter than this fraction of all pixels will be saturated |
| mosaicContrastLow | 0.1 | Contrast to use when rendering image. This fraction of the dimmest pixels will be set to zero/black |
| mosaicContrastHigh | 0.9999 | Contrast to use when rendering an image. This fraction of the brightest pixels in the image will be saturated |
| fovs | inf | Use all FOVs in the folder. Optionally, select a subset (e.g. “[2:10,15:35]”) |
Table 11 |.
Parameters for generating the RNA mosaic in BuildMosaicsGUI
| Parameter name | Default value | Function |
|---|---|---|
| backgroundCorrect | none | Parameter options are ‘none’, ‘file’, and ‘median’. Decide if illumination should be flattened. If you recorded dax files of a blank illumination, select ‘file’. If you have many fields of view with randomly positioned data, you may use ‘median’ to estimate the illumination bias from the data. To skip background correction, keep the default ‘none’ |
| method | edgeBlur | Keeping the default ‘edgeBlur’ recommended. Uses a linear gradient of overlapping images regions to stitch borders. This is typically best for avoiding artificial edges. Other options include ‘mean’ - average the overlap, ‘sum’ - add the overlap, ‘last’ - use the last image and cover over what was imaged before, ‘first’ - use the first image and ignore later data |
| mosaicContrastLow | 0.1 | Contrast to use when rendering images. This fraction of the dimmest pixels will be set to zero/black |
| mosaicContrastHigh | 0.9999 | Contrast to use when rendering an image. This fraction of the brightest pixels in the image will be saturated |
| fovs | inf | “inf” default value is recommended to use all FOV in the folder. Optionally, select a subset (e.g. “[2:10,15:35]”) |
| hybs | inf | Hybridization data that will be displayed in the mosaic |
| dataType | all | Fiducial and readout data will be displayed in the mosaic |
| transpose | true | The default is ‘true’, which gives you the option to transpose tiles in creating mosaic |
| scope | autoDetect | Depending on the camera orientation, individual tiles may be flipped across the left-right and up-down axis to properly render the mosaic. In our case, the camera orientation is auto detected based on the pixel size of the raw images. Outside users will have to configure this setting in the MATLAB code to make sure that the tiles are oriented correctly. Our code within the Github link (https://github.com/BoettigerLab/ORCA-public) is commented in BuildMosaicsGUI.m to make sure the user knows where to make the appropriate changes based on their camera orientation |
| trimBorder | 10 | Remove the number of specified pixels around the border - sometimes our cameras have a bright line of pixels at the edge of the image, for reasons currently unclear. This removes them |
Align Mosaics: Combining drift-corrected RNA and DNA ● Timing 1–3 h
▲CRITICAL AlignMosaicsGUI takes as input the FOV to FOV drift-corrected RNA and DNA information from BuildMosiacsGUI. Each RNA FOV will be aligned on top of a DNA FOV. AlignMosiacsGUI will output a table containing the upper left x-y position and rotational information of each RNA FOV relative to the DNA FOVs. This GUI will also output a.txt file with every RNA name and RNA/DNA fiducial. Lastly, this GUI will save a combined RNA and DNA mosaic.
-
256
Start MATLAB.
-
257
Launch AlignMosaicsGUI () in a MATLAB terminal to begin mosaic analysis. A GUI labeled “Align Mosaics GUI” should pop up.
-
258
Load DNA mosaic (Fig. 12a) by clicking ‘Parameters’. Make sure to change chnTableFile for the appropriate DNA Excel file. The other parameters (Table 12) are optional to change.
Only fiducial data is necessary for DNA mosaics. The mosaic is loaded from the raw dax files and uses the FOV-to-FOV alignment specified by BuildMosaicsGUI () in Step 248.
-
259
Click ‘Run Step’. This step should be quick because it is only loading data and no longer creating max-projections as previously done in other GUIs. A new window will display the DNA mosaic.
-
260
Click ‘Next Step’.
-
261
Load RNA mosaic (Fig. 12b) by clicking ‘Parameters’. Make sure to change chnTableFile for the appropriate RNA Excel file. Hybs and dataType are the only parameters that are changed by default in this step. These and the remaining parameters and their functions are listed in Table 13.
For RNA data, you will need both fiducial and readout data for the complete RNA mosaic. The mosaic is loaded from the raw dax files and uses the FOV-to-FOV alignment specified by BuildMosaicsGUI () in Step 248.
-
262
Click ‘Run Step’. This step should be quick because it is only loading data and no longer creating max-projections as previously done in other GUIs. A new window will display the RNA mosaic.
-
263
Click ‘Next Step’.
-
264
Click ‘Run Step’ to coarse align DNA and RNA mosaics (Fig. 12d). This step will take a few minutes. It creates a rough alignment of the DNA and RNA mosaics at the whole mosaic scale. A new window will appear where you can see the alignment between mosaics happening. The alignment is based on the fiducials from DNA and RNA mosaics, the sample edge, and autofluorescence of the sample. The red and cyan spots represent the DNA and RNA fiducials, respectively. This step will correct for rotational variation between datasets, which is introduced when removing and mounting the sample from and onto the microscope, respectively, between RNA and DNA experiments. Note, the sample does not rotate further due to potential swelling or peeling of the tissue during the imaging process (more information on tissue shrinking/expanding at Step 267) The parameters used for this step will be saved in the RNA_Experiment ‘Analysis’ folder as a coarseAlign.csv file.
-
265
(Optional) Table 14 lists parameter options that can be changed in order to help the coarse alignment if needed. However, we find that we usually do not need to change these parameters.
-
266
Click ‘Next Step’.
-
267
Click ‘Run Step’ for fine alignment of DNA and RNA mosaics (Fig. 12d). This will start the interactive process of aligning DNA and RNA FOVs. Follow the directions on the window that appears and, if needed, move FOVs using your keypad. If FOVs do not overlap, press ‘z’ on your keypad and then click ‘OK’. If you skipped non-overlapping FOVS, after you are done going through all the FOVs, repeat this step by clicking ‘Run Step’ again. A new window will appear saying “Found X existing docking positions”. Click ‘Skip’ and the GUI will do the fine alignment for the FOVs that were previously skipped. Often it does a better job at aligning these skipped FOVs the second time around since you have already aligned the majority of RNA-DNA FOVs.
Because the two mosaics separately correct the different stage drift, the images from the two mosaics won’t necessarily align to pixel/single-DNA spot accuracy. For this, we conduct a fine-scale alignment and redock each RNA tile into the DNA mosaic directly.
As the RNA fiducial contains data with intron labels, which are transcribed from and tethered to the same DNA sequence labeled by DNA fiducial probes, these two generally align well, but are not literally images of the same subject. These spots align well due to the fact that nascent RNA is tethered to DNA. A single FOV will contain 100–1000 DNA spots, corresponding to the genomic region labeled with ORCA. If the user has labeled introns from genes within this region, the cells within this FOV will be transcriptionally active and have RNA spots in almost the exact location (although not precisely) as the DNA spots. If only 10 out of 100 of the DNA spots also have RNA spots, this is more than enough to compute the alignment for the FOV (at least 2 spots are needed for alignment and we typically have more than 10). Moreover, if the genomic region contains multiple genes and these introns are labeled, there is an even higher fraction of DNA spots that will have corresponding matches to the RNA fiducial channel.
The outline of tissue or cells can also facilitate this alignment. Manual inspection will greatly facilitate the alignment, and we launch an interactive GUI for supervised alignment. We find that the fine alignment usually does a great job on its own and we usually do not need to assist this process. A new GUI will pop up with instructions on how to manually align each FOV, which will appear as new images, using the keyboard. The red and cyan images represent DNA and RNA fiducial data, respectively.
Additionally, we find that a sample may shrink (very slightly <1–2%) after a long imaging experiment (4–5 days of imaging). In this case, the parameter in Step 268 called ‘scales’ handles the shrinking and expansion that might have occurred. We emphasize that we only need at least 2 spots where RNA and RNA labels overlap for the fine alignment and we usually have at least 10 spots where they overlap. Thus, we do not have difficulties aligning RNA to DNA even when the sample has slightly shrunken/expanded.
-
268
(Optional) Table 15 lists parameter options that can be changed in order to help the fine alignment if needed.
-
269
Click ‘Next Step’.
-
270
Click ‘Run Step’ to validate fine alignment (Fig. 12e). This will generate an image of the fine aligned DNA and RNA mosaic for visual inspection. Once you click ‘Run Step’, a message on Align Mosaics GUI will appear with directions, “Zoom in and validate alignment”.
?TROUBLESHOOTING
-
271It is optional to change the following parameters for ‘Validate fine alignment’, although we typically do not change them:
Parameter name Default value Function redContrastLow 0.1 For the red (DNA) channel, this fraction of the dimmest pixels will be set to zero/black redContrastHigh 0.9999 For the red (DNA) channel, pixels brighter than this fraction will be saturated cyanContrastLow 0.1 For the cyan (RNA) channel, this fraction of the dimmest pixels will be set to zero/black cyanContrastHigh 0.999 For the cyan (RNA) channel, pixels brighter than this fraction will be saturated -
272
Click ‘Next Step’.
-
273
Click ‘Run Step’ to assemble mosaic (Fig. 12f). Using the rotations computed from the RNA fiducial images, all of the RNA data images need to be rotated, aligned, and then the borders need to be properly merged where each tile overlaps. This step will create a table specifying the upper left x-y position and rotation theta to place each RNA tile into reference with the DNA tiles and this information will be saved in a uls2to1.csv file in the RNA_Experiment ‘Analysis’ folder. Additionally, this step will save a text file of the names of the RNA channels specified in RNAtable.csv in the ‘Analysis’ folder. However, this new text file will also include the fiducial channel for both DNA and RNA in that list. (Example of list: fiducial1-DNA, Abd-B Main Exon, Abd-B Main Intron, abd-A Exon, abd-A Intron, Ubx Exon, Ubx Intron, fiducial2-RNA). No parameters need to be changed for this step.
-
274
Click ‘Next Step’.
-
275Click ‘Run Step’ to save alignment data (Fig. 12g). This step will save Tif images of the new mosaic coordinates for all channels, including DNA, RNA fiducials channels, and every RNA readout channel. We call this our ComboMosaic. The reason that each channel is saved as a separate Tif file is due to the 4GB size constraint of Tif files. All Tif files will be saved in the RNA_Experiment ‘Analysis’ folder. If you choose to change any of the two parameters below, make sure to click ‘Run Step’ afterward.
Parameter name Default value Change value to Function saveName ComboMosaic By default, ‘ComboMosaic’ will be a common name given to all Tif files for every RNA readout channel. You can change it to any other name that you want saveFolder Value does not need to be changed as this will automatically save your files in the RNA ‘Analysis’ folder. You can choose to change the directory path here
Fig. 12 |. AlignMosaics GUI.
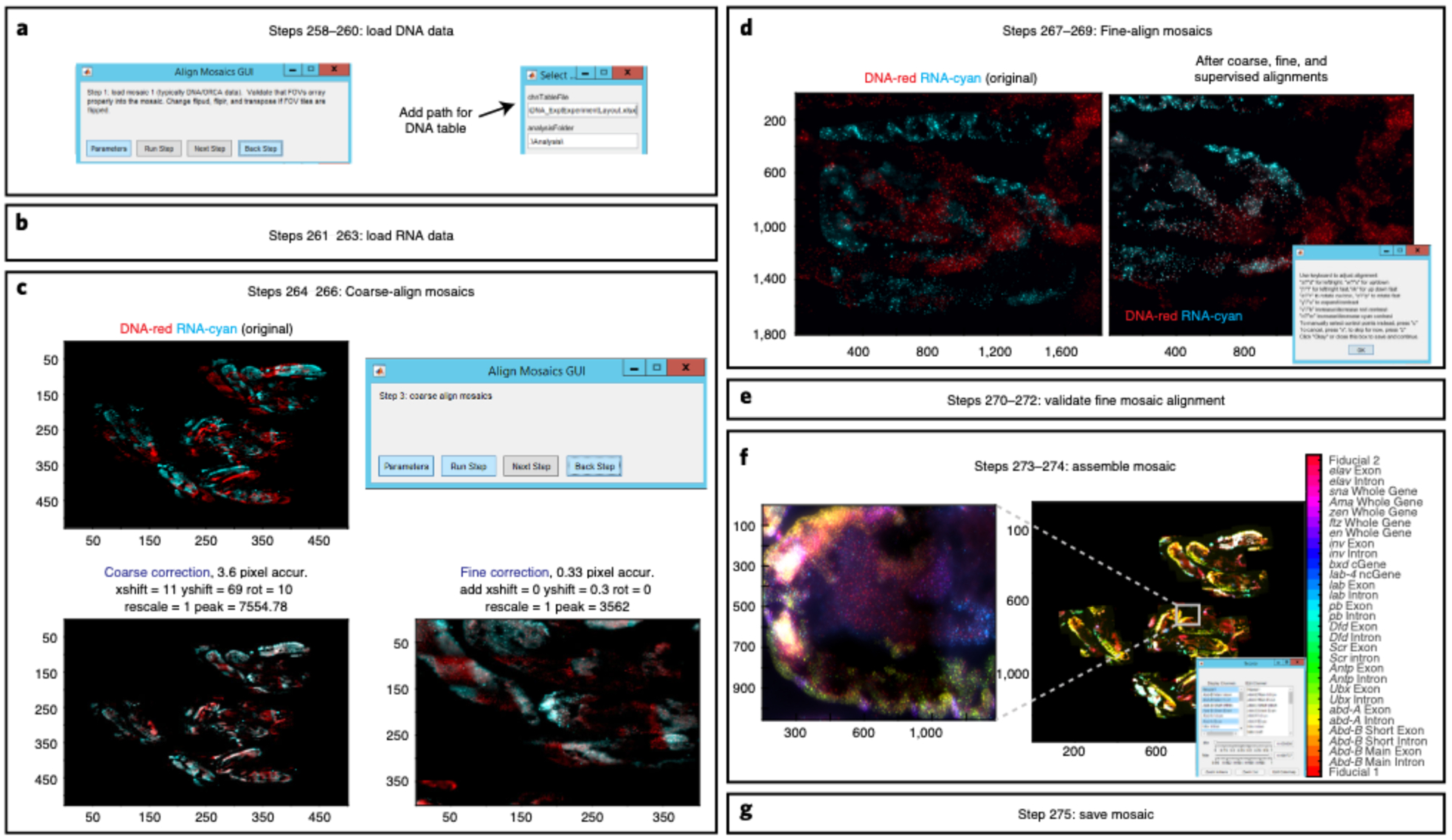
a, Load DNA data by entering the path of the experimental layout file. b, Load RNA data by entering the path of the RNA experimental layout file. c, Coarse and fine alignments of DNA and RNA mosaics based upon their fiducial data. d, Fine-scale alignment of RNA and DNA data at the FOV scale. Alignment relies upon fiducial from both datasets and the outline of the embryos. If the alignment could be further improved, follow the instructions on the new window and use your keyboard to make manual adjustments. e, A new window will appear and display drift-correct mosaic for visual inspection. f, This step will take into account all drift-corrected x-y and rotational changes made to the RNA and overlay each RNA channel on top of DNA fiducial data. You can inspect the mosaic by using the Ncolor GUI. g, Each channel will be saved with its new mosaic coordinates as a Tif image.
Table 12 |.
Parameters for loading DNA mosaic in AlignMosaicsGUI
| Parameter name | Default value | Function |
|---|---|---|
| chnTableFile | Enter Path To DNA Experimental Table: C:\ | Enter the path to your experiment layout file here (e.g. E:\2017-11-30_WT_10-12hrs_(HoxRNA)_(BXC3kb)\DNA\ExperimentLayout.xlsx) |
| analysisFolder | .\Analysis\ | The default value “.\Analysis\” is recommended. Saved alignment results and mosaics from previous ChrTracerGUI and BuildMosaicsGUI were placed in this folder |
| backgroundCorrect | none | Parameter options are ‘none’, ‘file’, and ‘median’. Decide if illumination should be flattened. If you recorded dax files of a blank illumination, select ‘file’. If you have many fields of view with randomly positioned data, you may use ‘median’ to estimate the illumination bias from the data. To skip background correction, keep the default ‘none’ |
| method | edgeBlur | Keeping the default ‘edgeBlur’ is recommended. This setting uses a linear gradient of overlapping images regions to stitch borders. This is typically best for avoiding artificial edges. Other options include ‘mean’ - average the overlap, ‘sum’ - add the overlap, ‘last’ - use the last image and cover over what was imaged before, ‘first’ - use the first image and ignore later data |
| mosaicContrastLow | 0.1 | Contrast to use when rendering images. This fraction of the dimmest pixels will be set to zero/black |
| mosaicContrastHigh | 0.9999 | Contrast to use when rendering an image. This fraction of the brightest pixels in the image will be saturated |
| fovs | inf | “inf” default value is recommended to use all FOV in the folder. Optionally, select a subset (e.g. “[2:10,15:35]”) |
| hybs | 1 | Hybridization data that will be displayed in the mosaic. By default, we show data from readout ‘1’ |
| data Type | fiducial | Show fiducial data for alignment mosaic |
| transpose | true | The default is ‘true’, which gives you the option to transpose tiles in creating mosaic |
| scope | autoDetect | Depending on the camera orientation, individual tiles may be flipped across the left-right and up-down axis to properly render the mosaic. In our case, the camera orientation is auto detected based on the pixel size of the raw images. Outside users will have to configure this setting in the MATLAB code to make sure that the tiles are oriented correctly. Our code within the Github link (https://github.com/BoettigerLab/ORCA-public) is commented in BuildMosaicsGUI.m to make sure the user knows where to make the appropriate changes based on their camera orientation |
| trimBorder | 10 | Remove the number of specified pixels around the border - sometimes our cameras have a bright line of pixels at the edge of the image, for reasons currently unclear. This removes them |
Table 13 |.
Parameters for loading RNA mosaic in AlignMosaicsGUI
| Parameter name | Default value | Function |
|---|---|---|
| chnTableFile | Enter Path To RNA Experimental Table: C:\ | Enter the path to your experiment layout file here (e.g. E:\2017-11-30_WT_10-12hrs_(HoxRNA)_(BXC3kb)\RNA\rnaTable.xlsx) |
| analysisFolder | .\Analysis\ | The default value “.\Analysis\” is recommended. Saved alignment results and mosaics from previous ChrTracerGUI and BuildMosaicsGUI were placed in this folder |
| backgroundCorrect | none | Parameter options are ‘none’, ‘file’, and ‘median’. Decide if illumination should be flattened. If you recorded dax files of a blank illumination, select ‘file’. If you have many fields of view with randomly positioned data, you may use ‘median’ to estimate the illumination bias from the data. To skip background correction, keep the default ‘none’ |
| method | edgeBlur | Keeping the default ‘edgeBlur’ is recommended. This setting uses a linear gradient of overlapping images regions to stitch borders. This is typically best for avoiding artificial edges. Other options include ‘mean’ - average the overlap, ‘sum’ - add the overlap, ‘last’ - use the last image and cover over what was imaged before, ‘first’ - use the first image and ignore later data |
| mosaicContrastLow | 0.1 | Contrast to use when rendering images. This fraction of the dimmest pixels will be set to zero/black |
| mosaicContrastHigh | 0.9999 | Contrast to use when rendering an image. This fraction of the brightest pixels in the image will be saturated |
| fovs | inf | “inf” default value is recommended to use all FOV in the folder. Optionally, select a subset (e.g. “[2:10,15:35]”) |
| hybs | inf | Hybridization data that will be displayed in the mosaic. All data from readouts taken throughout the RNA experiment will be shown |
| dataType | inf | Show both fiducial and readout data |
| transpose | true | The default is ‘true’, which gives you the option to transpose tiles in creating mosaic |
| scope | autoDetect | Depending on the camera orientation, individual tiles may be flipped across the left-right and up-down axis to properly render the mosaic. In our case, the camera orientation is auto detected based on the pixel size of the raw images. Outside users will have to configure this setting in the MATLAB code to make sure that the tiles are oriented correctly. Our code within the Github link (https://github.com/BoettigerLab/ORCA-public) is commented in BuildMosaicsGUI.m to make sure the user knows where to make the appropriate changes based on their camera orientation |
| trimBorder | 10 | Remove the number of specified pixels around the border - sometimes our cameras have a bright line of pixels at the edge of the image, for reasons currently unclear. This removes them |
Table 14 |.
Parameters for coarse alignment between DNA and RNA mosaics in AlignMosaicsGUI
| Parameter name | Default value | Function |
|---|---|---|
| angles | −12:0.5:12 | List of rotation angles to explore for mosaic alignment |
| downsample | 10 | Downsample the image this many fold for coarse alignment |
| maxSize | 400 | This is for correlation based analysis, the coarse alignment on the whole image is still done in a two step process. This is the maxSize for the first step |
| alignContrastHigh | 0.999 | Auto contrast to enhance feature matching between images. Pixels brighter than this fraction of the brightness histogram will be saturated |
| alignContrastLow | 0.9 | Auto contrast to enhance feature matching between images. Pixels dimmer than this fraction of the brightness histogram will be set to zero |
| maxShift | 300 | The maximum number of pixels the coarse alignment may shift the image |
| showPlot | true | Boolean, when set to ‘true’ will show progress image of the multistep alignment. The final result will always be shown |
| verbose | true | Boolean, when set to ‘true’ will show text in the terminal. The text will include information/instructions of the GUI’s current step |
Table 15 |.
Parameters for fine alignment between DNA and RNA mosaics in AlignMosaicsGUI
| Parameter name | Default value | Function |
|---|---|---|
| Interactive | true | Boolean, use the interactive GUI to guide adjust alignment |
| corrAlign | true | Boolean, use correlation based analysis |
| padAlign | 150 | Number of pixels with which to pad image prior to computing alignment |
| corrAlignHigh | 0.998 | Auto contrast to enhance feature matching between images. Pixels brighter than this fraction of the brightness histogram will be saturated |
| corrAlignLow | 0.65 | Auto contrast to enhance feature matching between images. Pixels dimmer than this fraction of the brightness histogram will be set to zero |
| angles | 0 | Rotations to try in docking already coarse rotated RNA images. Coarse correction in the previous step already took into account rotational changes and that is why the default is ‘0’ |
| scales | 1 | Isotropic expansion contraction of image around centroid |
| showplot | true | Show alignment plots |
| maxSize | 400 | All images will be downsampled to this size in terms of pixels for the coarse registration, prior to a second round of alignment on a subset of the image, at higher resolution. Set equal to image size or larger (‘inf’ is allowed) to do single step alignment |
| showExtraPlot | false | Change to ‘true’ to show extra plots for troubleshooting alignment |
| fineBox | 250 | Size of fineBox for the correlation based alignment |
| minFinelmprovement | 0.1 | Minimum improvement for the correlation based alignment |
MosaicAnalyzer ● Timing interactive tool to visualize and explore data, timing will vary.
▲CRITICAL MosaicAnalyzer software was developed for analysis of chromatin structure within specific cell types - which are defined based on RNA expression pattern- and for analysis of RNA transcript counts in individual cells. This GUI allows you to manually group cell types (such as Drosophila embryonic segments) containing ORCA data by using a polygon tool. As inputs, it takes the combined RNA/DNA mosaic built in AlignMosiacsGUI and ChrTracer3 data. As outputs, it returns four tables containing the following information: 1) The X_polygonTable.csv file contains the x and y positions for each comer of the manually drawn polygon/ROI in the table’s A and B column, respectively. Additionally, column C contains the ‘Tag’ that was given for that particular ROI. 2) The X_tableData.csv file contains ChrTracer3 information for the chromosomes that lie within the manually selected ROIs. Column number is variable, depending on the number of associated columns. The first 5 columns are always present: x, y, fov, s, and sidx. These respectively record the x/y position of the chromosome in the mosaic, the FOV number and spot number in that FOV from which the chromosome was originally analyzed in the ChrTracer3, and an ID number (sidx) which corresponds to its starting position in the AllFits table. Any ROI added in the MosaicAnalyzer will also generate a new column labeled by the associated tag. The column value (True/false, which is represented by 1/0) records whether or not this chromosome lies within that ROI. RNA counts in the cell associated with each chromosome may be added to this table using the transcript positions in the spotCountTable and the cell borders in the cellBorders table. This allows any combination of expression, physical position, and 3D chromatin structure to be assayed in downstream analysis. 3) X_cellBordersXY.csv contains the x and y positions of the xy polygon boundary coordinates for all cells. These coordinates are stored in two columns. Distinct cells are separated by NaNs and units are in pixels. 4) The X_spotCountTable.csv file contains the RNA spot information (this is for both single-molecule mRNA data and intron-RNA). In the table, the A and B columns correspond to the x and y position, respectively, of a single RNA spot. Column C includes information of the spot’s brightness represented by pixel intensity and column D includes the name of the RNA. All output files will get saved under the ‘Analysis’ folder for DNA_Expt
-
276
Start MATLAB.
-
277
Launch MosaicAnalyzer () in a MATLAB terminal. A GUI labeled “MosaicAnalyzer GUI” should pop up.
-
278
Load mosaic (Fig. 13a). This step will load the registered DNA and RNA Tif files. Under File (highlighted with a red box in Fig. 13a), click ‘Load Mosaic’ and find the directory where you saved your registered mosaics (these files are in ‘Analysis’ within the RNA_Expt folder). All the Tif file mosaics will have a common name, such as ComboMosaic. Click on one of these Tif files and then click ‘Open’. This will load all files that contain the word ComboMosaic (e.g. ComboMosaic_001). Once loaded, the ‘Ncolor’ GUI will appear with your RNA+DNA mosaic.
-
279
(Optional) Sometimes it is easier to view RNA exons/introns when you designate each exon and intron to a specific color. To change the color, click ‘Edit Colormap’ on ‘Ncolor’ GUI. A list of RBG values that range from 0 to 1 will appear and these are listed in the same order as the ‘Display Channels’ on the ‘Ncolor’ GUI. Change the RGB colors to help you visualize RNA patterns. For example, in Fig. 13a we changed the values of Abd-B Short Exon, abd-A Exon, and Ubx exon to [0 0 1], [0 1 0], and [1 0 0], respectively.
-
280
Load ChrTracer3 data (Fig. 13b). This step will load all ChrTracer3 FOVx_FitAll.xlsx files that were previously generated in Step 233 (these files are in ‘Analysis’ within the DNA_Expt folder). Under File (highlighted with a red box in Fig. 13b), click ‘Load CT Data’ and find the directory where you saved all ChrTracer3 outputs. Select all the FovX_AllFits.csv files and click ‘Open’. If you do not want to load a specific FovX_AllFits.csv file, perhaps due to large dye spots that overlap with the majority of your data spots, be sure to unselect it prior to loading all FovX_AllFits.csv files. Once the mosaic and ChrTracer3 outputs are loaded, you can use the MosaicAnalyzer GUI to explore your data.
-
281
(Optional) If you have previously run MosaicAnalyzer for your data and have saved ‘Annotations’, under ‘File’ click ‘Load Annotations’. Find the directory where you saved your previous annotations and select all desired tables (e.g. X_polygonTable.csv). Your loaded data will now appear under ‘Available Annotations’ to the right of the MosaicAnalyzer GUI (Fig. 13b).
-
282
Click on ‘Box FOVs’ to visualize the outline of all FOVs in the mosaic (Fig. 13b).
-
283
Click on ‘Show CT spots’ to visualize all the spots that you selected in ChrTracer3 (Fig. 13b). The following steps will allow you to assign ChrTracer3 spots to a specific group of cells, which will be used to examine cell-type specific structure downstream.
-
284
To manually crop individual embryos from the registered mosaic image click ‘Record Region of Interest (ROI)’ (Fig. 13c). Under MarkerTags, enter the names you wish to record (e.g. “Embryo1”, “Embryo2”, etc). Then click ‘Ok’.
-
285
A new GUI will appear with all the MarkerTags that you just specified. Choose which MarkerTag you wish to crop first and then click ‘New ROI’. A bounding polygon cursor will let you trace around your region of interest. Once you close the polygon, the shape of the polygon will change color and will display the MarkerTag name next to it. Once you are done tracing over every region of interest, go back to the GUI and click ‘Done’.
-
286
Click ‘Add Annotation to Table’ to add MarkerTags ROI information.
▲CRITICAL STEP If you click ‘Record ROI’ again before clicking ‘Add Annotation’, all the ROI manually selected will be overwritten.
-
287
To save your new annotations, to go File and click ‘Save Annotation’ to save table files containing the coordinates of the polygons (X_polygonTable.csv), the identity of the spots within the traced MarkerTags (X_tableData.csv), and number of RNA spots within a cell (X_spotCountTable.csv; this file should be empty for now but will be populated while saving during this step). Go to the directory where you want to save these annotations. Give these table files a common name (e.g WT_embryos) and then click ‘Save’.
-
288
To record embryonic segments within ROI embryos, repeat Steps 284–287 and make sure to specify MarkerTags with the appropriate name (e.g. “T3”, “A1”, “A2”, etc) (Fig. 13d). We recommend using the ‘Zoom in Here’ in the Ncolor App for a specific embryo before tracing its embryonic segments. Additionally, we highly recommend using the exonic expression pattern of BX-C genes (Ubx, abd-A, and Abd-B) and Engrailed (En) to correctly annotate embryonic segments. In WT, Ubx expression starts in segment T3. En marks the posterior compartment of each embryonic segment. If at any point of MosaicAnalyzer you decide to zoom out to view the whole mosaic again, first click ‘Zoom out’ and see if the mosaic looks normal or shifted. If the mosaic looks shifted to one side, close the mosaic (close Figure 100) and click any gene under Ncolor App. This will open a brand new mosaic figure.
-
289
To view the annotated ROI information, under ‘Available Annotations’, click on ‘ROIs’ and select your ROI(s) of interest (e.g. “A1”).
-
290You can view the distance map for a specific or all ROIs by going to ‘Analyze’ and clicking on ‘Distance Map’. The following parameters will appear:
Parameter name Default value Function ROIs all By default, a distance map for all ROIs that have been saved will be populated. You can choose to change this for a specific ROI (e.g. “A1”). If you choose to view multiple ROIs, make sure to not insert a space between each ROI (e.g. “A1,A2,A3”) figHandle 200 All figures are identified by a unique handle. To avoid overwriting currently active figures, select a unique number here badHybes After generating the distance map, you may notice that some barcodes have poor labeling (e.g. prominent stripes that are seen throughout the distance map). If so, enter the barcode number(s) that did not label well (e.g.16,26,39). Additionally, if you have previously generated distance maps for the total population of cells after running ChrTracer3, you might already know which barcode (s) labeled poorly. Enter those barcode numbers here In Fig. 13c, we show the distance map for the 330 kb BX-C genomic region generated via MosaicAnalyzer for A1 to A3 cells. In our example, we are showing the pairwise distances in nanometers between labeled barcodesThese repeated labeled barcodes are shown asspots - not to be confused with chromatin loops - and are displayed after barcode ‘52’. A slider is seen underneath the distance map and is used to control the maximum of the color bar, meaning the largest distance that can be displayed on the map.
-
291You can view the contact frequency map for a specific or all ROIs by going to ‘Analyze’ and clicking on ‘Contact Map’. The following parameters will appear:
Parameter name Default value Function ROIs all By default, a distance map for all ROIs that have been saved will be populated. You can choose to change this for a specific ROI (e.g. “A1”). If you choose to view multiple ROIs, make sure to not insert a space between each ROI (e.g. “A1,A2,A3”) threshold 150 This will display the fraction of cells in which the pairwise interactions are within 150 nm figHandle 200 ‘200’ is the default figure handle of the distance map (see the bottom-right corner of Fig. 13c). Having figures with unique figure handles allows you to access a specific figure, such as saving a specific figure. To avoid clearing a previously generated distance map, change the figHandle number to one that has not been previously used (e.g. 307) badHybes After generating the distance map, you may notice that some barcodes have poor labeling (e.g. prominent stripes that are seen throughout the distance map). If so, enter the barcode number(s) that did not label well (e.g. 16,26,39). Additionally, if you have previously generated distance maps for the total population of cells after running ChrTracer3, you might already know which barcode(s) labeled poorly. Enter those barcode numbers here -
292
Click ‘Segment Cells’ to segment all cells, which will use the ChrTracer3 fiducial spots as reference. This step will take a while. (Fig. 13d). Cell segmentation is sample dependent. Unique algorithms may be required for your specific sample type. The segmentation is saved as a two column text file, specifying the x,y coordinates of the borders of each cell, separated by NaNs. Cell borders computed by any other algorithms and formatted this way can be loaded into the MosaicAnalyzer software.
Additionally, following parameters can be changed:Parameter name Default value Function minSep 10 Spots within this number of pixels from one another will be considered to be in the same cell cellSize 25 This is the maximum radius which may be assigned to a cell during the segmentation -
293
Click ‘Add Annotation to Table’. Next, save the segmented cell information by going to ‘File’ and clicking ‘Save Annotation’. Choose a directory to record all segmented cells. Give the file the same common name as before and this will save an X_cellBorderXY.csv file.
-
294
To view the saved segmented cells, under ‘Available Annotations’, click ‘Cell edges’. The following steps record the number of RNA introns and/or single molecules (Fig. 13e).
-
295
Select an RNA channel of interest (e.g. abd-A Exon).
-
296
Zoom into the region where your RNA of interest is expressed and click ‘Count Spots’. If there are too few or too many spots for your RNA, change the threshold by using the threshold slider in the MosaicAnalyzer GUI (Fig. 13e).
-
297
Zoom out to view the whole mosaic (Fig. 13e).
-
298
Apply the RNA threshold globally by selecting ‘Count Spots” to count the number of RNAs in all cells (Fig. 13d).
-
299
Click ‘Add Annotation to Table’.
-
300
To Save Annotations for downstream applications, click ‘File’ and ‘Save Annotations’ to save RNA information into the X_spotCountTable.csv file.
-
301
To view the saved RNA spots, under ‘Available Annotations’, click ‘Spot counts’.
Fig. 13 |. MosaicAnalyzer GUI.
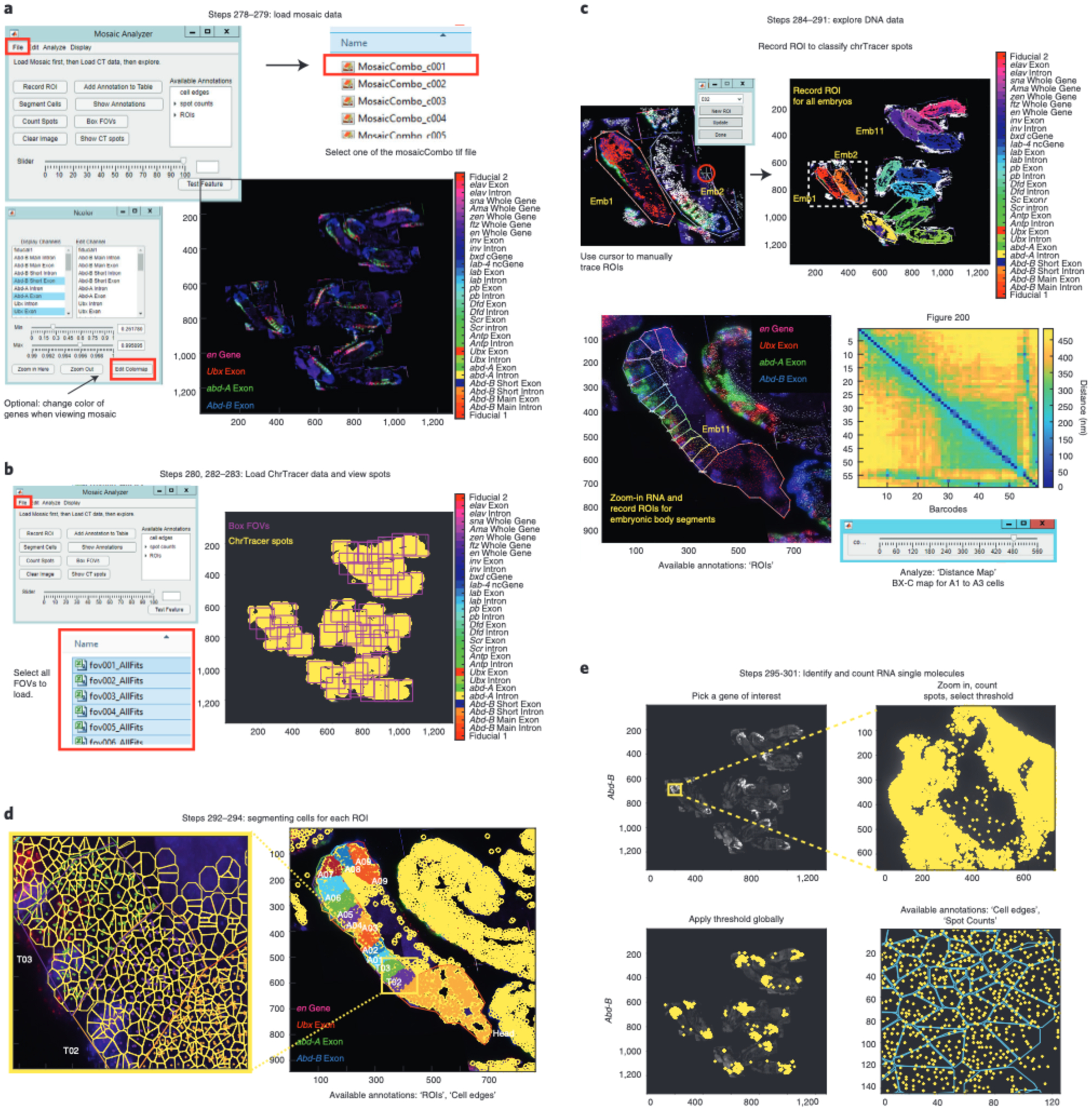
a, Load RNA-DNA registered mosaic by clicking on ‘File’ (highlighted with a red box) on the MosaicAnalyzer GUI and then clicking on ‘Load Mosaic (highlighted with a red box)’. Search for your directory path and open one of the ‘ComboMosaic_cX.tif’ files. If you have previously saved annotations, click ‘File’ and ‘Load Annotations’. The names of the annotations will appear on the right side of the MosaicAnalyzer GUI as shown here. In this example, RGB values for the BX-C exonic genes (Ubx in red, abd-A in green, and Abd-B in blue) are adjusted for visual purposes. b, To load ChrTracer3 processed tables, click ‘File’ and then ‘Load CT Data’. Find the directory path that contains all the ChrTracer outputs and click on all the ‘fovX_AllFits.xlsx’ files (highlighted with a red box). Once loaded, you can click ‘Show CT spots’ to see the position of all fiducial spots in the mosaic (shown as yellow outlined circles). You can also click ‘Box FOVs’ to see the outline of all FOVs. c, Click ‘Record ROI’ and use the bounding polygon cursor to trace around embryos. Note that all the fiducial data inside Embryo (Emb) 1 are red and match the outline of the traced embryo. Emb 2 is in the process of being traced and the polygon cursor is highlighted by the red circle. Each embryo is classified as a specific ROI and all are shown by different colors after tracing. For tracing embryonic segments, zoom into an embryo and adjust the Min/Max pixel intensity of the gene(s) of interest. Using ‘Record ROI’ trace around embryonic segment. Again, notice how fiducial data inside ROIs/embryonic segments change colors after being traced. Additionally, you can generate distance maps for the selected ROIs using MosaicAnalyzer. Map from A1, A2, and A3 cells from wild-type embryos 10–12 hpf, showing pairwise distances between all 52 barcodes that traced the 330-kb BX-C locus (chr3R:12.46–12.79 Mb (dm3)) at 3-kb resolution. The five loop-like structures that appear after barcode ‘52’ correspond to the barcodes that were re-labeled after the experiment. The slider under the distance map is used to select the largest distance shown on the map. d, Segment individual cells by clicking ‘Count Spots’. Display both ‘ROI polygons’ and ‘cell edges’ annotations to show the color coded individually segmented cells in each embryonic body segment. e, Example of identifying and counting Abd-B exonic molecules. Zoom-in to the posterior region of the embryo where Abd-B is expressed and adjust the threshold on the MosaicAnalyzer GUI slider to display the appropriate number of Abd-B exonic molecules (shown in yellow). Zoom-out and apply the threshold to the entire mosaic. Zoom into a region of the embryo to view Abd-B exonic spots by entering ‘Counted spots’ in ‘Show Annotations’. You can also display segmented cells (in cyan) on top of the exonic spots.
Troubleshooting
Troubleshooting advice can be found in Table 16.
Table 16 |.
Troubleshooting table
| Step(s) | Problem | Possible reason | Solution |
|---|---|---|---|
| 3 | Primary probes do not amplify during the run | Insufficient template DNA and/or primer | Add more Oligopaint library and/or primer to the reaction. Alternatively, dilute fresh primer |
| 19 | Substantial primer band is observed at the bottom of the PAGE gel | Primer dimer has formed | Use less forward and reverse primers next time |
| 30 | Concentration is below 500 ng/μl. | Incomplete elution | Concentrate the DNA using a DNA SpeedVac for 1–2 hrs. If the tube is completely dried after concentrating in the DNA SpeedVac, resuspend it in 25 μL ultrapure water |
| 36 | Flies have laid low numbers of eggs | Female flies could be old | Add virgin/young females to the collection cage as they lay more than older females |
| 46 | Embryos are floating between heptane-methanol layers after vortexing | Non-devitellined embryos can remain in the interphase | If you choose to collect all embryos, including non-devitellined embryos, add more methanol and continue vortexing until embryos sink to the methanol layer |
| 68 | Embryos cryosectioned onto glass coverslips coated with chromium-gelatin older than a month might peel away either during the primary probe hybridization and/or during imaging due to the constant flow of solutions within the flow chamber | Older chromium-gelatin may be less adhesive | Make fresh chromium-gelatin once a month |
| 142 | When testing a new RNA primary probe, the user is not sure if the probes labeled RNA or DNA | We recommend designing the RNA probes as antisense to avoid targeting DNA. However, some users might want to design probes differently and this might lead to cross-hybridizing to genomic DNA | We suggest the following 3 controls to make sure that probes are labeling RNA and not DNA: 1) Compare distinguishable populations of cells where one population should express the target RNAs and other should not, or at least should express it at a clearly distinct level. For example, Abd-B should never be expressed in the most anterior cells in the Drosophila embryo. 2) Compare distinguishable populations of cells labeled with primary probes targeting mature mRNAs. This should yield a clear difference in average fluorescence intensity of labeling seen at low magnification and in total spot counts seen at higher magnification. 3) Primary probes targeting mature mRNAs should provide multiple distinct puncta in the cytoplasm, whereas intronic-RNA and DNA primary probes should only label spots in the nuclei |
| 197 | High background during DNA imaging acquisition | Primary probes are antisense and could be binding to some RNA transcripts in the locus | Incubate the sample for 1 hr at 37°C in RNase solution in Step 152 |
| 209 | Fiducials do not align well in ‘Fix Global Drift’ | Larger than average stage drifts might have occurred during the imaging | Increase the size of fineBox. Keep in mind that increasing fineBox will slow down this step. You can also specify a particular region of the image to use instead of the default brightest area - see fineCenter |
| 209 | Fiducials do not align well in ‘Fix Global Drift’ | Sample has peeled over the course of the experiment, which makes it difficult for the computer to align fiducials at the edge of the sample | Specify that fine alignment would occur on an x-y position that has remained flat throughout the experiment |
| 212 | Drift correction failed for one or more FOVs as seen with multiple colors for a single spot | See possible troubleshooting reasons for step 209 | To correct the Bad FOVs, click ‘Back’ on the ChrTracer3 GUI. You will be back on the ‘Fix Global Drift’ step. Click on ‘Step Pars’ and type in the bad FOVs under SelectFOVs. Repeat Steps 208–212 for the Bad FOVs |
| 222 | General failure to detect the centroid of fiducial and readout signal | Fiducial data for the readout ‘1’ did not label well. By default ChrTracer uses this fiducial for the fine-scale drift correction when calculating for the 3D centroid of our spots | In ‘Step Pars’ change the refHybe to another readout, such as ‘2’, that has a fiducial signal that labeled well |
| 222 | Fit Spot’ is not detecting the readout spots | The readout spot might be further away, in terms of pixels, from the center of the fiducial. This could happen if the primary probe is labeling a large region of interest (~1 Mb) | In ‘Step Pars’, change the maxXYstep to increase the default search size in terms of X-Y pixels to fit the readout spot |
| 222 | Fit Spot’ is not detecting the readout spots | The readout spot might be further away, in terms of pixels, from the center of the fiducial. This could happen if the primary probe is labeling a large region of interest (~1 Mb) | In ‘Step Pars’, change the maxZstep to increase the default search size in terms of Z pixels to fit the readout spot |
| 222 | Fit Spot’ is not detecting the readout spots | The background level is very high, which can make it hard to detect readout spots | We find it is more robust to select spots based on the quality of the Gaussian fit, rather than a threshold, so this default value of datMinPeakHeight is intentionally chosen below the typical background. However, if you can visually see the readout probe against the high background, then you can choose to lower the default number. Don’t lower it too much or you might start picking up background noise |
| 222 | Fit Spot’ is not detecting the correct fiducial spots which will result in detecting erroneous readout spots | If the data has a lot of background in the fiducial channel, ‘Fit Spot’ might be choosing a fiducial spot that is actually background noise | In ‘Step Pars’, increase the threshold for fidMinPeakHeight. Remember that the fiducial signal is typically strong and, thus, you can easily increase the fidMinPeakHeight value to 750–1000 |
| 222 | Fit Spot’ is not detecting the readout spots | During the registration step, the algorithm allows the image to shift based on the default maxXYdrift. Frames might require larger shifts | The amount of shift is reported in the command terminal. If any frames are shifted over the max drift allowed, it is an indication that the alignment is not good. This may be acceptable for individual rounds but is generally an indication that more accurate drift correction was required during the Fix Global Drift step |
| 222 | Fit Spot’ is not detecting the readout spots | Same as above. Shifts between images might require a larger shift in Z | In ‘Step Pars’, increase the search size for maxZdrift in terms of Z pixels |
| 234 | Prominent stripes are seen across the distance map may indicate poor barcode labeling or noisy background | If the stripe suggests that the pairwise distance between a barcode is far away in nanoscale distance from all labeled barcodes, it may indicate that the readout probe failed to hybridize to the barcode | To further verify failure of labeling, you can calculate the detection efficiency for that particular barcode by using the FovX_AllFits.csv files generated from ChrTracer. You may need to re-dilute a fresh readout probe if failure of labeling occurred for a single readout. If it seems that many barcodes were not labeled throughout the experiment, this may suggest that something is wrong with your fluidics system. Perhaps there is salt build up from the 25% EC in 2× SSC or a tear in the tube that is causing the flow to be delayed. To determine if the stripes are real and not an artifact, perform a replicate of the experiment using newly diluted readout probes at the proper concentration |
| 234 | Prominent stripes are seen across the distance map may indicate poor barcode labeling or noisy background | If a stripe suggests that a single barcode is close in nanoscale distance to all labeled barcodes, it may indicate that the background noise was high when imaging the barcode | Repeat the experiment with a fresh sample and new buffers. You may need to re-dilute a fresh readout probe if the stripe occurred for a single readout. Alternatively, we have found that certain readout probes cause unspecific staining in our sample - perhaps due to the readout sequence complementing some other part of the organism’s genome - and these readout probes can create a stripe across the distance map. If this is the case, then we suggest dropping these readout probes when analyzing the distance maps. To determine if the stripes are real, perform a replicate of the experiment using newly diluted readout probes at the proper concentration |
| 234 | Erroneous stripe-like features in the distance maps | May be indicative of the strand displacement oligo not working properly | If you see a partial stripe (e.g. a stripe that starts at readout ‘25’ and continues) this means that the strand displacement oligo failed to remove a particular readout signal. To fix this, you can either re-dilute the strand-displacement oligo from the stock plate, or re-optimize the strand-displacement sequence |
| 234 | Erroneous loop-like features in the distance maps | May be indicative of contamination of the readout sequence by introducing two readout probes at the same time, due to pipetting error or cross-contamination between readouts. This might result in an unexpected loop between two barcodes, which would incorrectly suggest that these two barcodes are in close 3D proximity to one another | To confirm that a loop is real or not, we suggest using newly diluted readout sequences and repeating the experiment with a new sample |
Timing
Step 1, Oligopaints library design: 1 d
Steps 2–30, Primary probe synthesis: 2 d
Steps 31–62, Drosophila embryo collection and embedding: 1 d 15 h
Steps 63–75, Slide preparation and Drosophila embryo cryosectioning: 2 h
Steps 76–78, Cell culture fixation: 20 m
Steps 79–94, Hybridization with RNA primary probes: 1.5 d
Steps 95–113, 96-well plate preparation and mounting sample in flow chamber: 25 m
Steps 114–143, Microscope, fluidics setup, and RNA imaging: 1 d 13 h
Steps 144–166, Hybridization with DNA primary probes: 1.5 d
Steps 167–168, 96-well plate preparation and mounting sample in flow chamber: 25 m
Steps 169–198, Microscope, fluidics setup, and DNA imaging: 2–3 d
Steps 199–201, Image pre-processing: 5 h
Steps 202–234, ChrTracer3 and analyzing data: 1–4 d
Steps 235–255, Build Mosaics: 1–8 h automated (step 237), 1–2 h interactive (238–255)
Steps 256–275, Align Mosaics: 1–3 h
Steps 276–301, MosaicAnalyzer: interactive tool to visualize and explore data, timing will vary.
Anticipated results
Data from Fig. 14 was published in our previous work25. The RNA data was generated from 12 embryos (not all shown in the RNA mosaic in Fig. 14a), which were tiled together from 47 FOVs. Imaging 29 RNA transcripts, including both mRNA and nascent RNA, with unique barcodes takes approximately 1.5 days with our current protocol. Imaging RNAs takes longer compared to DNA because readout probes are hybridized to barcodes for 30 minutes, instead of 15 minutes for DNA, to acquire better RNA signals.
Fig. 14 |. Data generated from a single RNA + ORCA labeling experiment.
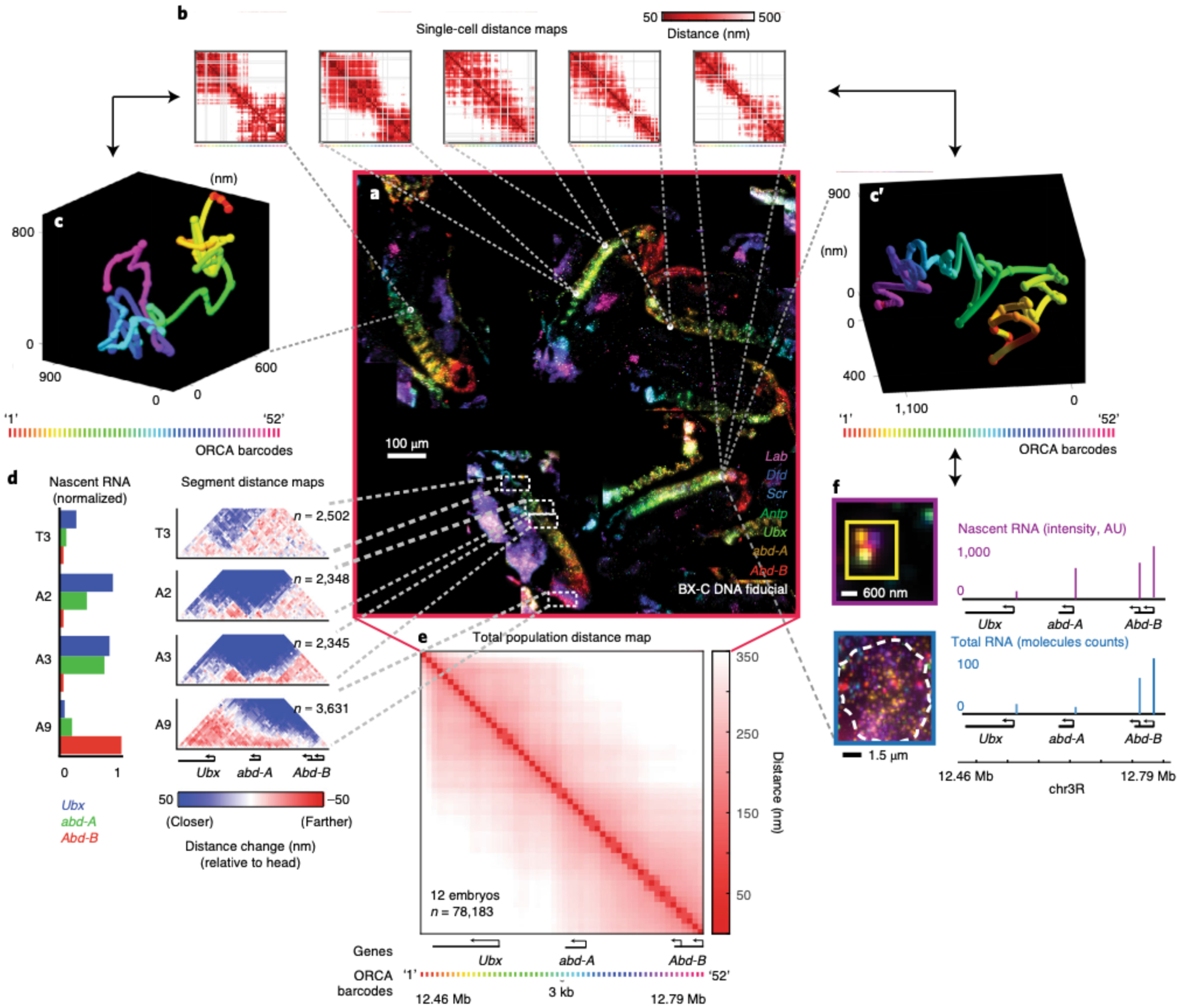
Overview of the multiple outputs that can be generated within an experiment. This 10–12 hours post fertilization (hpf) Drosophila embryo data was previously published13 and is the final result from the data analysis performed in Figs. 10–13. a, The mosaic in the center displays 7 mRNAs out of the 29 labeled RNA species that were used to identify specific embryonic body segments. mRNAs are shown for Lab (purple), Dfd (blue), Scr (cyan), Antp (green), Ubx (lime green), abd-A (orange), and Abd-B (red). Additionally, BX-C DNA fiducial spots are shown in white. ORCA was used to image the 330 kb BX-C locus (chr3R:12.46–12.79 Mb (dm3)) at 3 kb resolution. Because ORCA is compatible with RNA labeling in tissue, the user can use the data to investigate the relationship between chromatin structure and gene expression. These are some, not all, of the following ways the user can analyze the data: b, Single-cell distance maps from 5 individual cells, showing pairwise distances between labeled barcodes of the BX-C. c, c′, Example of two 3D traces of the BX-C chromatin path from single cells. The spheres on the polymer represent the 3D positions of the labeled barcodes. The spheres were linked together to generate a 3D polymer. d, As described in our previously published work25, we annotated body segments based on morphology and RNA expression pattern. The average nascent RNA intensity for the BX-C genes in body segments T3, A2, A3, and A9 are plotted on the left. These values are normalized based on the body segment with the highest RNA expression. To the right, body segment distance maps show the pairwise distances between the labeled BX-C barcodes. All maps are normalized relative to cells from head segments as these anterior cells have a compacted chromatin structure with no obvious TAD boundary partitioning. e, Average population distance map from all cells in 12 embryos. f, Individual RNA transcripts for each BX-C gene (note, Abd-B has two isoforms) per cell.
ORCA allows the user to examine chromatin structure in single cells (Fig. 14b), as polymer traces (Fig. 14c), in specific cell-types based on RNA expression patterns (Fig. 14d), and as a bulk population (Fig. 14e). The ORCA data from Fig. 14 was generated from the same 12 embryos (47 FOVs) previously imaged for RNA species. 52 unique barcodes were used to image the 330-kb bithorax complex (BX-C) region at 3 kb resolution. An additional 5 barcodes were relabeled at the end of the experiment to measure localization precision (relabeled barcodes are not displayed in the distance maps). Using our current protocol, the imaging time for a total of 57 barcodes and 47 FOVs is ~2 days. Within this time, we acquired a total of 5,358 3D fiducial and readout image stacks [(47 FOVs)*(57 barcodes * 2 fiducial/data images)] for analysis. A single FOV from a cryosectioned 10–12 hours post fertilization (hpf) Drosophila embryo contains ~1,600 nuclei, which allows for the imaging of ~78,000 nuclei from 12 embryos.
For the first time, we can visualize both chromatin structure at the single-cell level and count RNA transcripts within the same cell (Fig. 14f). Measurements of both RNA and DNA from a single experiment have generated much interest from biologists who are interested in examining gene regulation through chromatin structure. The image analysis can be performed using our software or can be performed using other microscopy image analysis software, such as Imaris.
Reporting Summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
We provide a mini dataset to help users become comfortable analyzing raw microscope data through our MATLAB GUIs: https://bit.ly/2S6eCjk. This 337 GB mini dataset contains RNA and DNA\ORCA images for a single embryo (4 FOVs) from Fig. 14. Images for an entire dataset are too large to append and, thus, image data is available upon request. All DNA paths from all cells are available upon request.
The ORCA data in Fig. 3 and Fig. 14 and the barcode detection efficiency data in Extended Data Fig. 2 are from our previous work25,46. The single-cell Hi-C and OligoSTORM data in Fig. 3 are from refs.52,56, (Single-cell Hi-C) and9 (OligoSTORM).
Code availability
The “storm-control” software controls both the lasers and the camera. This software is a fork of the “storm-control” system written by Hazen Babcock and the Zhuang lab, with modifications specific to our application included on our fork available at: https://github.com/BoettigerLab/storm-control. The software for the fluidics handling is available at: https://github.com/BoettigerLab/fluidics-control. Software, including the GUIs described above and some demo analysis scripts are available at https://github.com/BoettigerLab/ORCA-public. Demo data to test this software is also available (see ‘Data availability’. All software is open source and available under the CC-BY license. It may be freely edited, modified or shared. We ask only that users cite this work.
Extended Data
Extended Data Fig. 1 |. Distinct intronic RNA patterns in cryosectioned tissue.
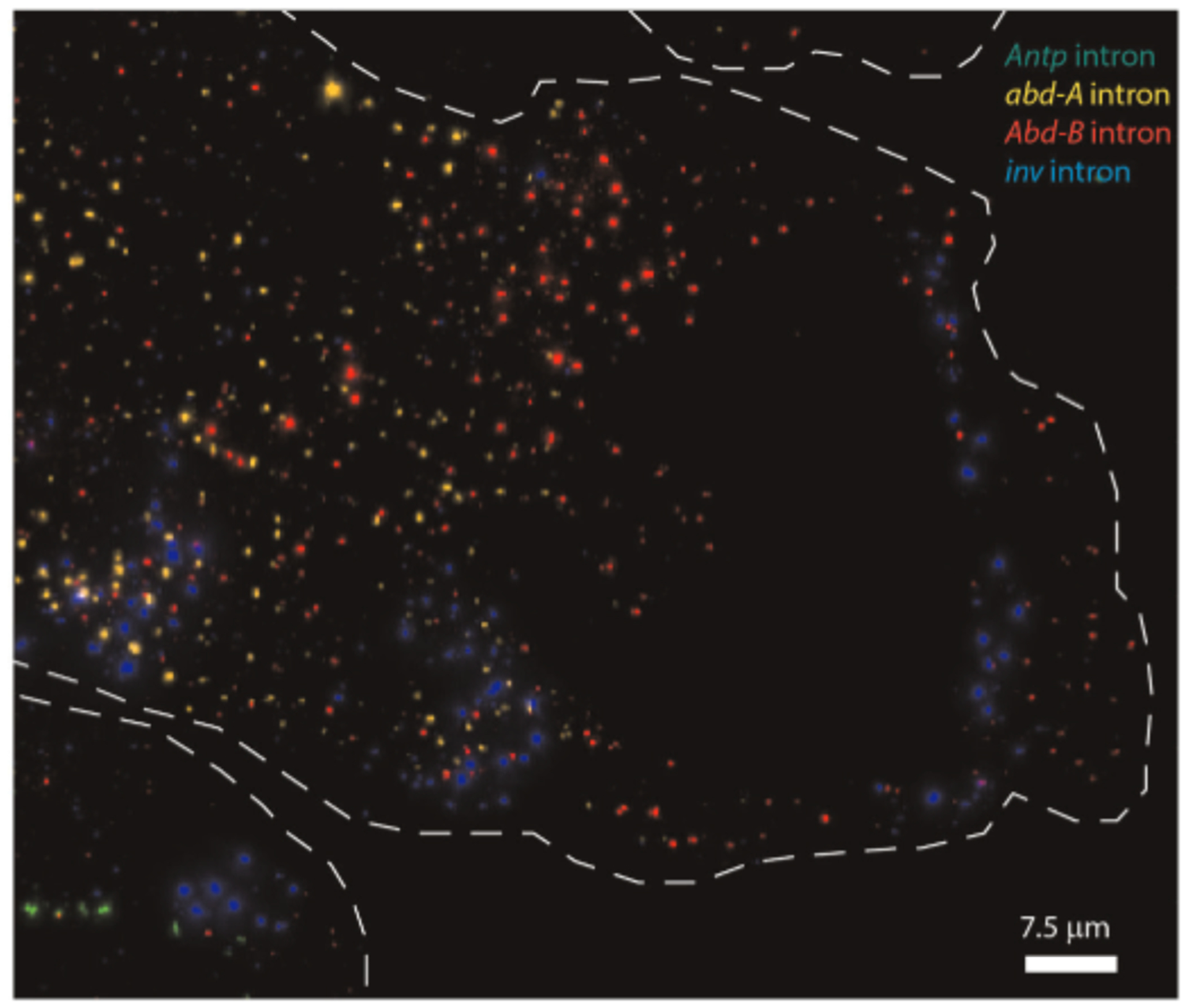
A zoomed-in image of the most posterior body segments of a 10–12 hpf Drosophila embryo. With multiplex RNA labeling, we targeted the following RNA introns: Antp (green), abd-A (yellow), Abd-B (red), inv (blue). Note that these introns have distinct RNA patterns.
Extended Data Fig. 2 |. Barcode detection efficiency.
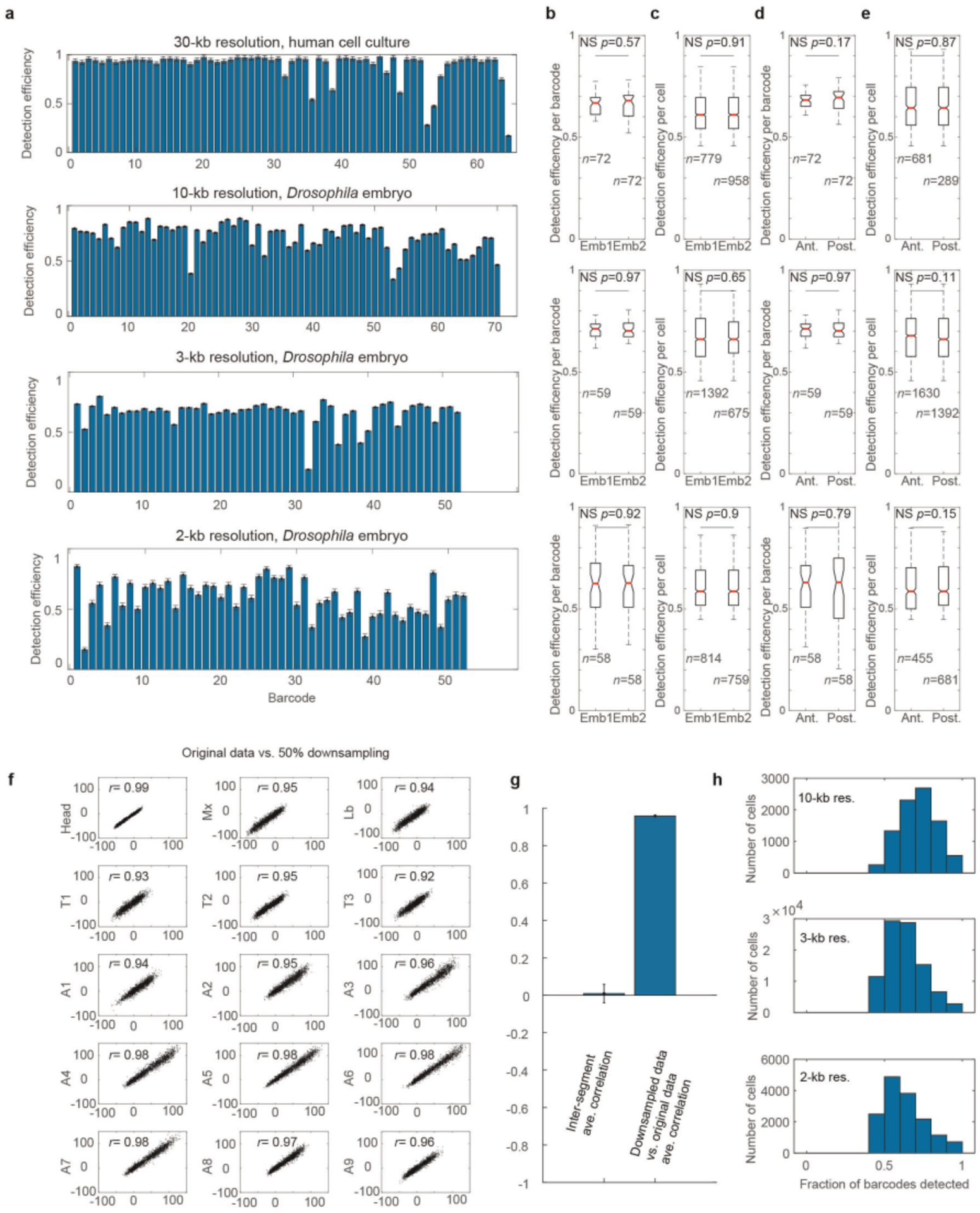
Figure modified from previous publication25. a, The average detection efficiency of each barcode is shown for probe sets with the resolution and cell type indicated. Error bars represent standard error of the mean, n = 1,979 cells, n = 8,801 cells. n = 94,300 cells, and n = 15,230 cells for the 30-kb, 10-kb, 3-kb, and 2-kb probesets, respectively. b, Box-and-whisker plots comparing the distribution of detection efficiencies across barcodes between two different embryos. N.S. indicates not statistically significant (two sided Wilcoxon test, p-values indicated). For the boxplots in (b-e), the red line marks the median, the upper and lower limits of the box mark the interquartile range, and the whiskers extend to the furthest datum within 1.5 times the interquartile range. Notches denote the 95% confidence interval around the median. Sample size (n) is indicated. c, Comparison of the distribution of detection efficiencies between cells of different embryos. d, Comparison of the detection efficiency across barcodes between anterior and posterior cells. e, Comparisons of the detection efficiency across all cells between anterior and posterior cells. Results are shown for the 10-kb, 3-kb and 2-kb probesets as indicated. f, Correlation between normalized distance data from different cell types and a modified dataset in which 50% of detected barcodes were removed at random to simulate a high missed detection rate. The correlation coefficients are indicated (Pearson), computed using all unique combinations of 52 barcodes. g, Bar graph comparing the mean correlation between matched segments in the original and downsampled data shown in (f) compared to the mean inter-segment correlation. The correlation coefficients are indicated (Pearson), computed using all unique combinations of 52 barcodes. All 15 matched segment pairs shown in f are plotted compared to all 210 inter-segment pairs. This illustrates that the difference between the substantially downsampled data and the original measurements are much more similar than the typical differences between segments, demonstrating the robustness of the segment-specific findings to missing data. Error bars denote standard error of the mean. h, Histogram showing the detection efficiency per cell for each probeset.
Extended Data Fig. 3 |. Setting up the flow chamber.
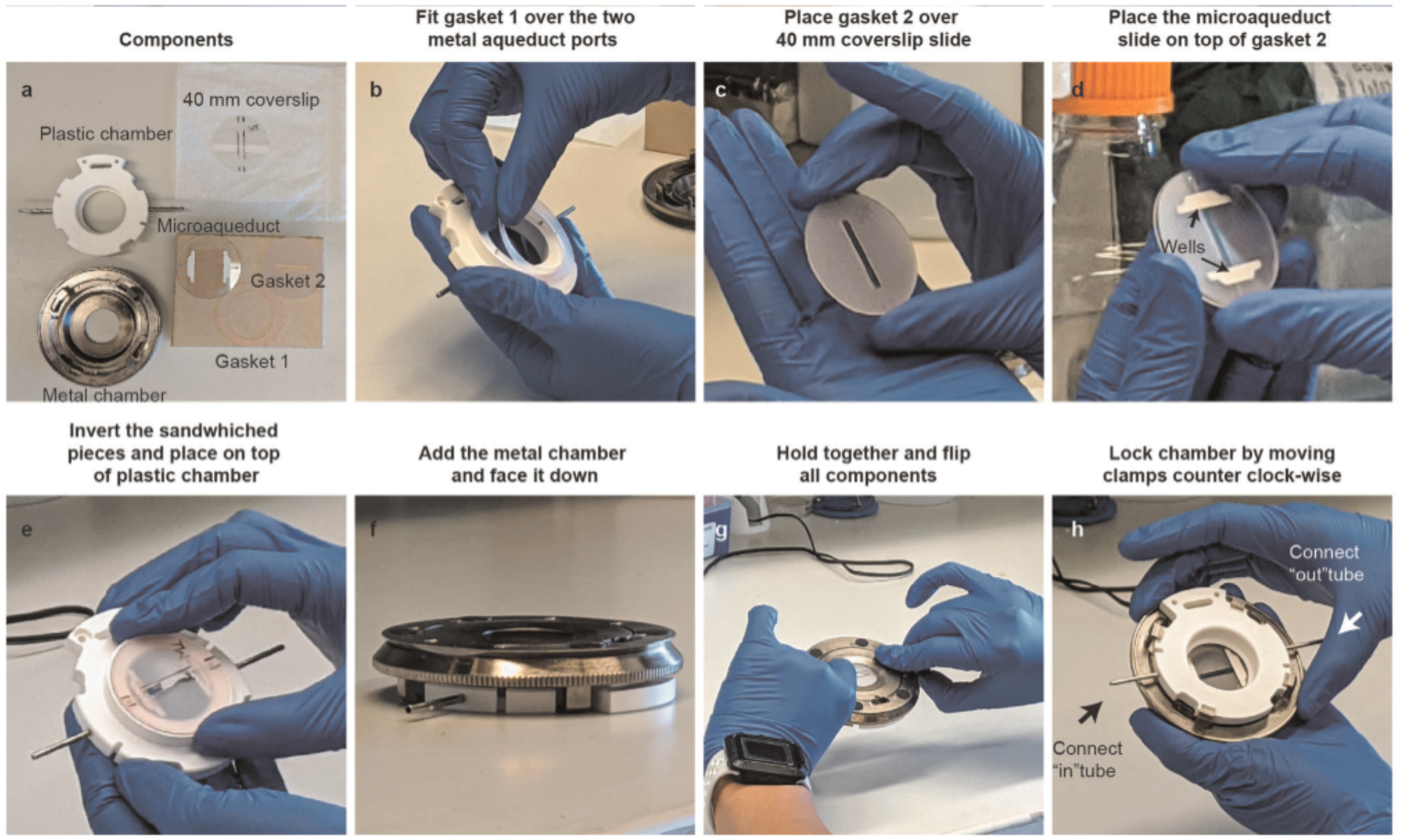
a, Layout of all the components that are needed to assemble the flow chamber. Before using, clean all components with a kimwipe that is slightly damp with ultra-pure water. Note that the 400 mm coverslip is marked with an alcohol-resistant pen. The marks indicate the location of the embryos, which are cryosectioned between the two lines in the center of the coverslip. This space is determined by the middle area of gasket 2. We also used the pen to label that our sample is WT. We find that labeling the slide does not harm the sample. b, Invert the plastic chamber. Then take gasket 1 and place its two holes over the two metal aqueduct ports of the plastic chamber. c, Carefully, place gasket 2 on top of the coverslip. You can use the pen labeled markers as an indicator of where to place the gasket. Make sure to not place gasket 2 directly on top of your sample. Any sample under the gasket will not be accessible for imaging. Also, make sure to not accidentally slide the gasket on top of your sample as this may result in your sample rubbing off the coverslip. d, Place the miroaqueuct glass slide on top of gasket 2. Carefully, place the microaqueduct slide so that its wells on the side (pointed by arrows) are perpendicular to gasket 2. e, Invert the sandwiched components - including the 40-mm coverslip, gasket 2, and the microaqueduct slide - and place the two holes of the microaqueduct slide on top of the plastic chamber’s aqueduct ports. f, Place the plastic chamber on a flat surface. Invert the metal mount and place it on top of the plastic chamber. When placing it on top, it is useful to make sure that the clamps are aligned to the plastic chamber holes as shown here. g, Invert all the components. During this flip, make sure to carefully hold onto both the metal chamber and plastic chamber. We find it useful to hold the metal part with our index/middle finger(s) and to have our thumbs holding onto the tubes of the plastic chamber. h, Tighten the metal clamps by moving the chamber counterclockwise to lock it. Make sure to tighten this as hard as you can, and without breaking the glass components inside. Afterward, connect the “In” and “Out” fluidics tube to the flow chamber’s metal tubes (shown with arrows).
Supplementary Material
Acknowledgements
We thank S. Murphy, T. Hafner, M. Park, and A. Rajpurkar for critical reading of the manuscript. This work was supported by the Searle Scholars Program, Burroughs Wellcome Careers at the Scientific Interface Award, Beckman Young Investigator Program, NIH New Innovator Award DP2 (DGM132935A), and the Packard Fellows Program (A.N.B.). L.J.M. was supported by the Howard Hughes Medical Institute (HHMI) Gilliam Fellowship. N.S.-A. was supported by a Stanford Graduate Fellowship.
Footnotes
Competing interests
The authors declare no competing interests.
Extended data is available for this paper at https://doi.org/10.1038/s41596-020-00478-x.
Supplementary information The online version contains supplementary material available at https://doi.org/10.1038/s41596-020-00478-x.
Reprints and permissions information is available at www.nature.com/reprints.
References
- 1.Amano T et al. Chromosomal dynamics at the Shh locus: limb bud-specific differential regulation of competence and active transcription. Dev. Cell 16, 47–57 (2009). [DOI] [PubMed] [Google Scholar]
- 2.Fabre PJ et al. Nanoscale spatial organization of the HoxD gene cluster in distinct transcriptional states. Proc. Natl Acad. Sci. USA 112, 13964–13969 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Williamson I, Lettice LA, Hill RE & Bickmore WA Shh and ZRS enhancer colocalisation is specific to the zone of polarising activity. Development 143, 2994–3001 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barbieri M et al. Active and poised promoter states drive folding of the extended HoxB locus in mouse embryonic stem cells. Nat. Struct. Mol. Biol 24, 515–524 (2017). [DOI] [PubMed] [Google Scholar]
- 5.Fraser J, Williamson I, Bickmore WA & Dostie J An overview of genome organization and how we got there: from FISH to Hi-C. Microbiol. Mol. Biol. Rev 79, 347–372 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rust MJ, Bates M & Zhuang X Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM). Nat. Methods 3, 793–795 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang B, Wang W, Bates M & Zhuang X Three-dimensional super-resolution imaging by stochastic optical reconstruction microscopy. Science 319, 810–813 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beliveau BJ et al. Single-molecule super-resolution imaging of chromosomes and in situ haplotype visualization using Oligopaint FISH probes. Nat. Commun 6, 7147 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boettiger AN et al. Super-resolution imaging reveals distinct chromatin folding for different epigenetic states. Nature 529, 418–422 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kundu S et al. Polycomb repressive complex 1 generates discrete compacted domains that change during differentiation. Mol. Cell 65, 432–446.e5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Szabo Q et al. TADs are 3D structural units of higher-order chromosome organization in Drosophila. Sci. Adv 4, eaar8082 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kieffer-Kwon K-R et al. Myc regulates chromatin decompaction and nuclear architecture during B cell activation. Mol Cell 67, 566–578.e10 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rao SSP et al. Cohesin loss eliminates all loop domains. Cell 171, 305–320.e24 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang P et al. Comparative analysis of three dimensional chromosomal architecture identifies a novel fetal hemoglobin regulatory element. Genes Devel. 31, 1704–1713 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hsu SC et al. The BET protein BRD2 cooperates with CTCF to enforce transcriptional and architectural boundaries. Mol. Cell 66, 102–116.e7 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brown JM et al. A tissue specific self-interacting chromatin domain forms independently of enhancer promoter interactions. Nat. Commun 9, 3849 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van Bemmel JG et al. The bipartite TAD organization of the X-inactivation center ensures opposing developmental regulation of Tsix and Xist. Nat. Genet 51, 1024–1034 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Williamson I et al. Developmentally regulated Shh expression is robust to TAD perturbations. Development 10.1242/dev.179523 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dixon JR et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485, 376–380 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nora EP et al. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature 485, 381–385 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nora EP et al. Targeted degradation of CTCF decouples local insulation of chromosome domains from genomic compartmentalization. Cell 169, 930–944.e22 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schwarzer W et al. Two independent modes of chromatin organization revealed by cohesin removal. Nature 551, 51–56 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Symmons O et al. The Shh topological domain facilitates the action of remote enhancers by reducing the effects of genomic distances. Dev. Cell 39, 529–543 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Finn EH & Misteli T Molecular basis and biological function of variability in spatial genome organization. Science 365, eaaw9498 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mateo LJ et al. Visualizing DNA folding and RNA in embryos at single-cell resolution. Nature 568, 49–54 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lubeck E & Cai L Single-cell systems biology by super-resolution imaging and combinatorial labeling. Nat. Methods 9, 743–748 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lubeck E, Coskun AF, Zhiyentayev T, Ahmad M & Cai L Single-cell in situ RNA profiling by sequential hybridization. Nat. Methods 11, 360–361 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen KH, Boettiger AN, Moffitt JR, Wang S & Zhuang X Spatially resolved, highly multiplexed RNA profiling in single cells. Science 348, 412 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fazal FM et al. Atlas of subcellular RNA localization revealed by APEX-Seq. Cell 178,473–490.e26 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Levsky JM, Shenoy SM, Pezo RC & Singer RH Single-cell gene expression profiling. Science 297, 836–840 (2002). [DOI] [PubMed] [Google Scholar]
- 31.Levesque MJ & Raj A Single-chromosome transcriptional profiling reveals chromosomal gene expression regulation. Nat. Methods 10, 246–248 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Raj A, van den Bogaard P, Rifkin SA, van Oudenaarden A & Tyagi S Imaging individual mRNA molecules using multiple singly labeled probes. Nat. Methods 5, 877–879 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Beliveau BJ et al. Versatile design and synthesis platform for visualizing genomes with Oligopaint FISH probes. Proc. Natl Acad. Sci. USA 109, 21301–21306 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moffitt JR et al. High-throughput single-cell gene-expression profiling with multiplexed error-robust fluorescence in situ hybridization. Proc. Natl Acad. Sci. USA 113, 11046–11051 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Beliveau BJ et al. In situ super resolution imaging of genomic DNA with OligoSTORM and OligoDNA-PAINT. Methods Mol. Biol 1663, 231–252 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Beliveau BJ et al. OligoMiner provides a rapid, flexible environment for the design of genome-scale oligonucleotide in situ hybridization probes. Proc. Natl Acad. Sci. USA 115, E2183–E2192 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gelali E et al. iFISH is a publically available resource enabling versatile DNA FISH to study genome architecture. Nat. Commun 10, 1636 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Boettiger A & Murphy S Advances in chromatin imaging at kilobase-scale resolution. Trends Genet. 36, 273–287 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Buxbaum AR, Wu B & Singer RH Single β-actin mRNA detection in neurons reveals a mechanism for regulating its translatability. Science 343, 419–422 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zenklusen D, Larson DR & Singer RH Single-RNA counting reveals alternative modes of gene expression in yeast. Nat. Struct. Mol. Biol 15, 1263–1271 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Paré A et al. Visualization of individual Scr mRNAs during Drosophila embryogenesis yields evidence for transcriptional bursting. Curr. Biol 19, 2037–2042 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Boettiger AN & Levine M Rapid transcription fosters coordinate snail expression in the Drosophila embryo. Cell Rep 3, 8–15 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Trcek T et al. Single-mRNA counting using fluorescent in situ hybridization in budding yeast. Nat. Protoc 7, 408–419 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rouhanifard SH et al. ClampFISH detects individual nucleic acid molecules using click chemistry-based amplification. Nat. Biotechnol 10.1038/nbt.4286 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lyubimova A et al. Single-molecule mRNA detection and counting in mammalian tissue. Nat. Protoc 8, 1743–1758 (2013). [DOI] [PubMed] [Google Scholar]
- 46.Bintu B et al. Super-resolution chromatin tracing reveals domains and cooperative interactions in single cells. Science 362, eaau1783 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Weber CM, Hafner A, Braun SMG, Boettiger AN & Crabtree GR mSWI/SNF promotes distal repression by titrating polycomb dosage. Preprint at bioRxiv 10.l101/2020.01.29.925586 (2020). [DOI] [Google Scholar]
- 48.Quinodoz SA et al. Higher-order inter-chromosomal hubs shape 3D genome organization in the nucleus. Cell 174, P744–757.E24 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Beagrie RA et al. Complex multi-enhancer contacts captured by genome architecture mapping. Nature 10.1038/nature21411 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McCord RP, Kaplan N & Giorgetti L Chromosome Conformation Capture and Beyond: Toward an Integrative View of Chromosome Structure and Function. Mol. Cell 77, 688–708 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nagano T et al. Single-cell Hi-C reveals cell-to-cell variability in chromosome structure. Nature 502, 59–64 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stevens TJ et al. 3D structures of individual mammalian genomes studied by single-cell Hi-C. Nature 544, 59–64 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Flyamer IM et al. Single-nucleus Hi-C reveals unique chromatin reorganization at oocyte-to-zygote transition. Nature 544, 110–114 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gassler J et al. A mechanism of cohesin-dependent loop extrusion organizes zygotic genome architecture. EMBO J. 36, 3600–3618 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tan L, Xing D, Chang C-H, Li H & Xie XS Three-dimensional genome structures of single diploid human cells. Science 361, 924–928 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ramani V et al. Massively multiplex single-cell Hi-C. Nat. Methods 14, 263–266 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nir G et al. Walking along chromosomes with super-resolution imaging, contact maps, and integrative modeling. PLoS Genet. 14, e1007872 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Beliveau BJ et al. In situ super-resolution imaging of genomic DNA with OligoSTORM and OligoDNA-PAINT. Methods Mol. Biol 1663, 231–252 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jungmann R et al. Multiplexed 3D cellular super-resolution imaging with DNA-PAINT and Exchange-PAINT. Nat. Methods 10.1038/nmeth.2835 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Georgieva M et al. Nanometer resolved single-molecule colocalization of nuclear factors by two-color super resolution microscopy imaging. Methods 105, 44–55 (2016). [DOI] [PubMed] [Google Scholar]
- 61.Wang S et al. Spatial organization of chromatin domains and compartments in single chromosomes. Science 353, 598–602 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Finn EH et al. Extensive heterogeneity and intrinsic variation in spatial genome organization. Cell 176, 1502–1515.e10 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cardozo Gizzi AM et al. Microscopy-based chromosome conformation capture enables simultaneous visualization of genome organization and transcription in intact organisms. Mol Cell 74, 212–222.e5 (2019). [DOI] [PubMed] [Google Scholar]
- 64.Cardozo Gizzi AM et al. Direct and simultaneous observation of transcription and chromosome architecture in single cells with Hi-M. Nat. Protoc 15, 840–876 (2020). [DOI] [PubMed] [Google Scholar]
- 65.Moffitt JR et al. High-performance multiplexed fluorescence in situ hybridization in culture and tissue with matrix imprinting and clearing. Proc. Natl Acad. Sci. USA 113, 14456–14461 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Eng C-HL et al. Transcriptome-scale super-resolved imaging in tissues by RNA seqFISH. Nature 10.1038/s41586-019-1049-y (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Magaraki A, Loda A, Gribnau J & Baarends WM Simultaneous RNA-DNA FISH in mouse pre-implantation embryos. Methods Mol. Biol 1861, 131–147 (2018). [DOI] [PubMed] [Google Scholar]
- 68.Lai L-T, Meng Z, Shao F & Zhang L-F in Long Non-Coding RNAs: Methods and Protocols (eds. Feng Y & Zhang L) 135–145 (Springer, 2016). [Google Scholar]
- 69.Lanzuolo C, Roure V, Dekker J, Bantignies F & Orlando V Polycomb response elements mediate the formation of chromosome higher-order structures in the bithorax complex. Nat. Cell Biol 9, 1167–1174 (2007). [DOI] [PubMed] [Google Scholar]
- 70.Lee JT, Strauss WM, Dausman JA & Jaenisch R A 450 kb transgene displays properties of the mammalian X-inactivation center. Cell 86, 83–94 (1996). [DOI] [PubMed] [Google Scholar]
- 71.Deng W, Shi X, Tjian R, Lionnet T & Singer R H. CASFISH: CRISPR/Cas9-mediated in situ labeling of genomic loci in fixed cells. Proc. Natl Acad. Sci. USA 10.1073/pnas.l515692112 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Solovei I et al. Spatial preservation of nuclear chromatin architecture during three-dimensional fluorescence in situ hybridization (3D-FISH). Exp. Cell Res 276, 10–23 (2002). [DOI] [PubMed] [Google Scholar]
- 73.Su J-H, Zheng P, Kinrot SS, Bintu B & Zhuang X Genome-scale imaging of the 3D organization and transcriptional activity of chromatin. Cell 10.1016/j.cell.2020.07.032 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rasnik I, McKinney SA & Ha T Nonblinking and long-lasting single-molecule fluorescence imaging. Nat. Methods 3, 891–893 (2006). [DOI] [PubMed] [Google Scholar]
- 75.Shi X, Lim J & Ha T Acidification of the oxygen scavenging system in single-molecule fluorescence studies: in situ sensing with a ratiometric dual emission probe. Anal. Chem 82, 6132–6138 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tokunaga M, Imamoto N & Sakata-Sogawa K Highly inclined thin illumination enables clear single-molecule imaging in cells. Nat. Methods 5, 159–161 (2008). [DOI] [PubMed] [Google Scholar]
- 77.Lim D, Ford TN, Chu KK & Mertz J Optically sectioned in vivo imaging with speckle illumination HiLo microscopy. J. Biomed. Opt 16, 016014 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Babcock H et al. ZhuangLab/storm-control: v2019.06.28 release. 10.5281/zenodo.3264857 (2019). [DOI]
- 79.Rao SSP et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 159, 1665–1680 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
We provide a mini dataset to help users become comfortable analyzing raw microscope data through our MATLAB GUIs: https://bit.ly/2S6eCjk. This 337 GB mini dataset contains RNA and DNA\ORCA images for a single embryo (4 FOVs) from Fig. 14. Images for an entire dataset are too large to append and, thus, image data is available upon request. All DNA paths from all cells are available upon request.
The ORCA data in Fig. 3 and Fig. 14 and the barcode detection efficiency data in Extended Data Fig. 2 are from our previous work25,46. The single-cell Hi-C and OligoSTORM data in Fig. 3 are from refs.52,56, (Single-cell Hi-C) and9 (OligoSTORM).


