Summary
Lyme disease is on the rise. Caused by a spirochete Borreliella burgdorferi, it affects an estimated 500,000 people in the US alone. The antibiotics currently used to treat Lyme disease are broad-spectrum, damage the microbiome and select for resistance in non-target bacteria. We therefore sought to identify a compound acting selectively against B. burgdorferi. A screen of soil microorganisms revealed a compound highly selective against spirochetes, including B. burgdorferi. Unexpectedly, this compound was determined to be hygromycin A, a known antimicrobial produced by Streptomyces hygroscopicus. Hygromycin A targets the ribosomes and is taken up by B. burgdorferi, explaining its selectivity. Hygromycin A cleared the B. burgdorferi infection in mice, including animals that ingested the compound in a bait, and was less disruptive to the fecal microbiome than clinically relevant antibiotics. This selective antibiotic holds the promise of providing a better therapeutic for Lyme disease, and eradicating it in the environment.
Keywords: Lyme disease, antibiotic, microbiome, transport, B. burgdorferi, Spirochetes
Graphical Abstract
One sentence summary:
Hygromycin A selectively accumulates in Borreliella burgdorferi and eradicates Lyme disease in a mouse model of infection
In Brief:
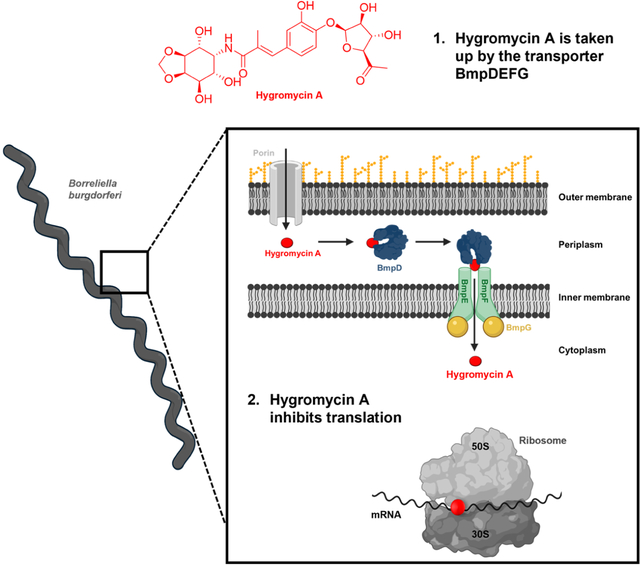
The use of broad-spectrum antibiotics to treat specific pathogens can damage the host microbiome and contribute to antibiotic resistance. Hygromycin A is selectively taken up through a nucleoside transporter specific to spirochete bacteria providing a highly selective antibiotic for spirochete infections such as Lyme disease.
Introduction
The incidence and geographic range of Lyme disease caused by Borreliella burgdorferi (formerly Borrelia) have been increasing due to a variety of factors, including expansion of the habitat range of the tick vector, increasing intersection of human domiciles and animal hosts of ticks, and longer seasonal activity due to climate change (Steere et al., 2016).
Current estimates from the Centers of Disease Control suggest that there are almost half a million cases of Lyme disease in the U.S. per year (Schwartz et al., 2021). The cost of Lyme disease to the U.S. economy has been estimated at approximately $3 billion in direct costs (Adrion et al., 2015), with even higher losses due to lost work hours. Acute infection is notable for a characteristic rash called erythema migrans, which starts at the site of inoculation by the tick. From there, the bacteria disseminate quickly to other skin sites, the heart, and the peripheral and central nervous system causing carditis, radiculitis/nerve palsies, and meningitis. If not treated during the early phase of infection, late symptoms of infection with B. burgdorferi include arthritis and neurological issues.
Related species of Borreliella causing Lyme disease include B. garinii and B. afzelii which are found predominantly in Eurasia. Although the disease is largely similar between the different Borreliella species, there is an increased incidence of arthritis with B. burgdorferi while B. garinii and B. afzelii are more closely linked to neurologic disease and a skin manifestation called acrodermatitis atrophicans, respectively.
The acute disease is treated with broad-spectrum antibiotics such as doxycycline, amoxicillin and ceftriaxone. Treatment with broad-spectrum compounds comes at a considerable cost to the patient, disrupting the gut microbiome and selecting for resistance in off-target bacteria (Modi et al., 2014; Willing et al., 2011). The microbiome shapes the immune system during development and contributes to maintaining a healthy gastrointestinal (GI) tract and preventing cardiovascular, mental health and autoimmune diseases (Belkaid and Hand, 2014; Grenham et al., 2011; Kasselman et al., 2018; Romano et al., 2015; Valles-Colomer et al., 2019; Vatanen et al., 2018; Zhang et al., 2021). Development of narrow-spectrum antibiotics is therefore highly desirable. With this in mind, we sought to identify compounds that act selectively against B. burgdorferi.
Most classes of antibiotics currently in use have been discovered from soil actinomycetes (Lewis, 2020). These have been heavily overmined, and more recently novel compounds are coming from other genera (Imai et al., 2019; Ling et al., 2015). However, previous searches for active compounds from actinomycetes have focused on broad-spectrum compounds, and narrow-spectrum antibiotics might have been overlooked. We recently reported several novel compounds from actinomycetes acting selectively against Mycobacterium tuberculosis, including lassomycin (Gavrish et al., 2014) that targets the unique ClpC1, P1, P2 protease and amycobactin (Quigley et al., 2020) that inhibits SecY of the protein export system.
We reasoned that compounds acting selectively against spirochetes may have evolved in nature, and are produced by actinomycetes. A selective screen against B. burgdorferi uncovered hygromycin A, a known compound with poor activity against a variety of bacterial species. Hygromycin A proved to be highly active, and selective against spirochetes. The compound acts by binding to a conserved region in the 23S rRNA. We find that hygromycin A is smuggled into B. burgdorferi by a nucleoside transporter, explaining its selectivity. Hygromycin A effectively clears an infection in mice by oral administration, and had a minimal effect on the gut microbiome. These properties make hygromycin A an attractive candidate for developing an antibiotic for targeted treatment of Lyme disease.
Results
We developed a selective screen for actinomycete extracts against B. burgdorferi using S. aureus as a counter-screen (Fig. 1A). This small screen of 452 actinomycetes yielded 24 hits, 6 of which were selective against B. burgdorferi. One of the hits showed potent activity against B. burgdorferi and good selectivity, and came from Streptomyces hygroscopicus, a common and well-studied species of Streptomyces. S. hygroscopicus is a prolific producer of secondary metabolites, a surprisingly large number of which are important commercial products. S. hygroscopicus makes rapamycin, an immunosuppressant; milbemycin, a major antihelmintic drug; bialaphos, a widely used herbicide; geldanamycin, an inhibitor of Hsp90, in development as an anti-cancer agent; and antimicrobials hygromycin B, an aminoglycoside antibiotic targeting the ribosome, used in cell culture media; nigericin, an H+/Na+ antiporter, growth promoter; and validamycin, an inhibitor of trehalase used to treat fungal pathogens in agriculture.
Figure 1. A screen for a selective anti-Borreliella compound.
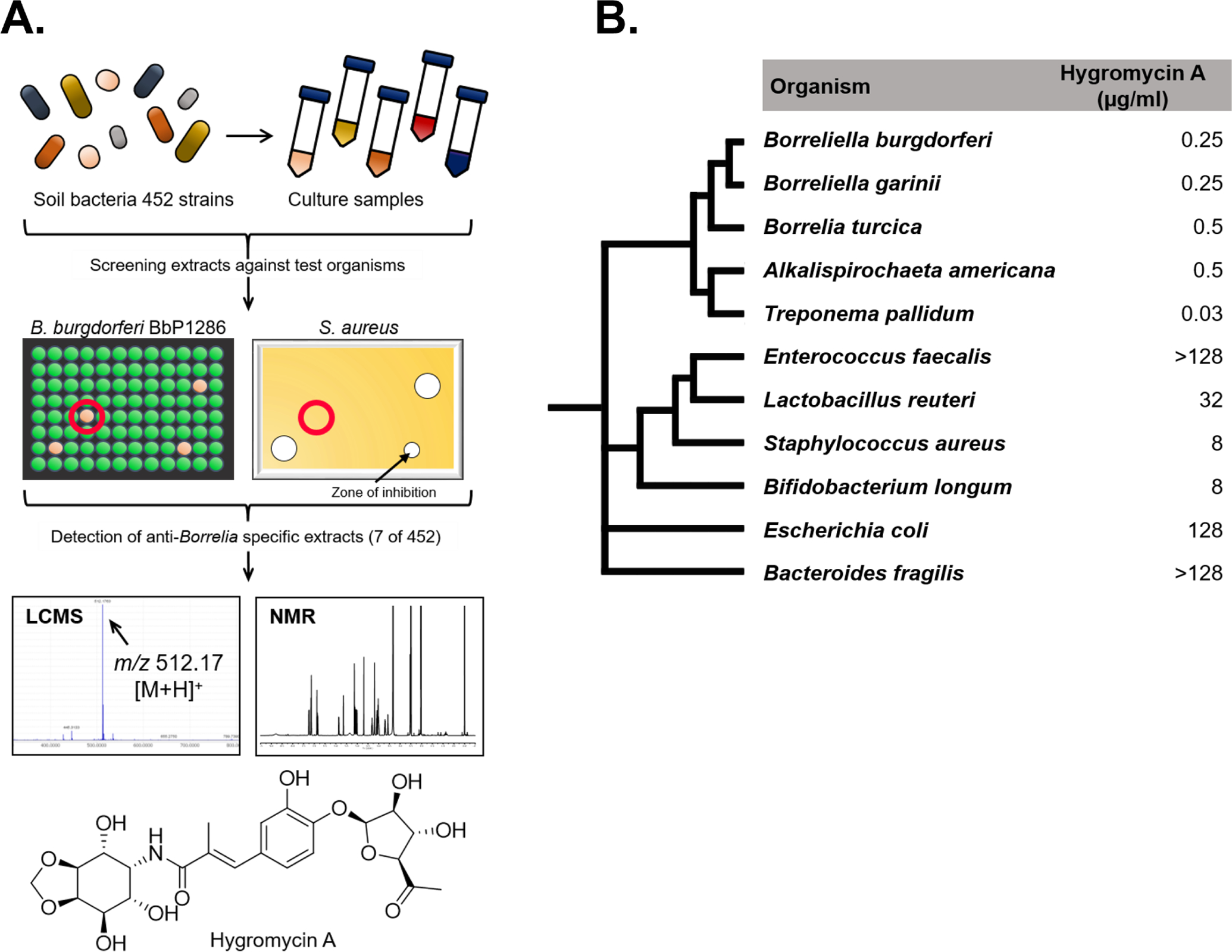
A. Actinomycete strains were fermented in liquid culture. Concentrated culture supernatants were screened against B. burgdorferi Bb1286 expressing GFP, and against S. aureus HG003. Extracts showing specific activity against B. burgdorferi were fractionated by HPLC, and their activity was determined by bioassay. After LCMS and NMR analysis, the active compound from one of the producers was determined to be hygromycin A. B. MIC of hygromycin A against spirochaetes, human pathogens and commensals. Phylogenetic tree shows similarities between strains.
An extract of S. hygroscopicus was separated into 40 fractions using high-performance liquid chromatography (HPLC). The fractions were tested for growth inhibition of B. burgdorferi, resulting in the isolation of a pure compound with a molecular mass of 511 Da (Fig. 1A). The structure was determined by a combination of MS and NMR analysis (Fig. S1, S2, Table S1, S2). Surprisingly, a search of AntiBase database revealed the identity of this compound to hygromycin A, a known antibiotic (Fig. 1A). This was unexpected, given the selectivity of the compound against B. burgdorferi. Hygromycin A was discovered in 1953 by a group from Eli Lilly (Pittenger et al., 1953). The compound contains a modified cinnamic acid flanked by a furanose sugar and aminocyclitol (not to be confused with hygromycin B, an unrelated aminoglycoside). Hygromycin A was isolated from S. hygroscopicus, including strains from Selman Waksman’s collection. We examined the spectrum of activity of hygromycin A in detail and found that it is highly active against spirochetes (Fig. 1b, Table 1). The class Spirochaetia consists of 4 defined orders (Brachyspirales, Brevinematales, Leptospirales, Spirochaetales). Hygromycin A was previously found to be active against Brachyspira hyodysenteriae with an MIC of 1.56 μg/ml (previously called Serpulina hyodysenteriae or Treponema hyodysenteriae) that belongs to the order Brachyspirales (Hayashi et al., 1997). In our study, we find that hygromycin A has the highest activity against Treponema pallidum, the causative agent of syphilis, with a minimal inhibitory concentration (MIC) of 0.03 μg/ml. The compound is equally active against B. burgdorferi and other species responsible for Lyme disease in Eurasia – B. afzelii, B. garinii and B. bavariensis as well as against Borrelia miyamotoi, a spirochete that causes tick-borne relapsing fever (Breuner et al., 2018), with an MIC of 0.25 μg/ml. It is also active against environmental spirochetes such as Alkalispirochaeta americana. All these species belong to the order Spirochaetales that displayed higher sensitivity towards hygromycin A in comparison to B. andersonii, L. biflexa and L. interrogans; species that belong to the orders Brevinematales and Leptospirales, respectively (Table 1). Activity against S. aureus, which was used in the counter-screen, is 32 to 64 times lower as compared to B. burgdorferi. Importantly, the compound is relatively ineffective against both Gram-positive and Gram-negative gut symbionts (Figure1B, Table 1). Conveniently, production of hygromycin A by S. hygroscopicus is high, 120 mg/L in the strain KLE3558 isolated from soil. This is probably why the compound was originally discovered, despite its poor activity against non-spirochetes. Hygromycin A is bactericidal against B. burgdorferi, with a minimal bactericidal concentration (MBC) of 1 μg/ml (at 4× MIC). We also tested hygromycin B against B. burgdorferi and as expected found it to be fairly inactive, with an MIC of 125 μg/ml.
Table 1.
Minimum inhibitory concentration (MIC) of hygromycin A and clinically relevant antibiotics against spirochaetes, human pathogens and commensals. MIC was determined by microdilution assay as described in material and methods. MIC of doxycycline and ceftriaxone against T. pallidum from (Edmondson et al., 2020; Norris et al., 2001).
| Concentration (μg/ml) | ||||
|---|---|---|---|---|
| Organism and genotype | Hygromycin A | Doxycycline | Amoxicillin | Ceftriaxone |
| Spirochaetes | ||||
| Treponema pallidum | 0.03 | 0.1d | N/A | 0.0007e |
| Borreliella burgdorferi P1286a | 0.12 | 0.25 | 0.12 | 0.01 |
| Borreliella afzelii a | 0.25 | 0.12 | 0.01 | <0.01 |
| Borreliella bavariensis a | 0.25 | 0.12 | 0.06 | 0.01 |
| Borreliella burgdorferi 297a | 0.25 | 0.25 | 0.25 | 0.01 |
| Borreliella burgdorferi B31a | 0.25 | 0.25 | 0.25 | 0.01 |
| Borreliella burgdorferi N40a | 0.25 | 0.25 | 0.12 | 0.01 |
| Borreliella garinii a | 0.25 | 0.12 | 0.12 | 0.01 |
| Borreliella miyamotoi a | 0.25 | 0.12 | 0.12 | 0.01 |
| Borrelia turcica DSM 16138a | 0.5 | 0.25 | 0.25 | 0.06 |
| Alkalispirochaeta americana DSM 14872b | 0.5 | N/A | N/A | N/A |
| Brevinema andersonii ATCC 43811a | 2 | 0.12 | 0.12 | 0.01 |
| Leptospira biflexa ATCC 23582 | 4 | 2 | 8 | 2 |
| Leptospira interrogans ATCC BAA-1198 | 4 | N/A | N/A | N/A |
| Opportunistic pathogens | ||||
| Fusobacterium nucleatum KLE 2428b,c | 1 | 0.12 | 2 | 0.5 |
| Clostridium tertium KLE 2303b,c | 8 | 0.03 | 32 | 1 |
| Clostridium perfringens KLE 2326b,c | 8 | 0.25 | 0.12 | <0.01 |
| Staphylococcus aureus HG003 | 8 | 0.12 | 1 | 8 |
| Escherichia coli W0153 | 16 | 0.12 | 4 | 0.01 |
| Staphylococcus epidermidis KLE 2378b,c | 32 | 0.5 | >128 | 1 |
| Shigella sonnei ATCC 25931b | 64 | 4 | 2 | 0.25 |
| Escherichia coli MG1655 | 128 | 2 | 16 | 0.06 |
| Pseudomonas aeruginosa PAO1 | >128 | 32 | >128 | 64 |
| Salmonella typhimurium LT2 ATCC 19585b | >128 | 8 | 16 | 0.25 |
| Gut commensal bacteria | ||||
| Streptococcus salivarius KLE 2370b,c | 2 | 0.12 | 0.06 | 0.03 |
| Streptococcus parasanguinis KLE 2375b,c | 4 | 4 | 64 | 0.06 |
| Bifidobacterium longum ATCC BAA-999b | 8 | 0.25 | 1 | 4 |
| Bacteroides nordii KLE 2369b,c | 8 | 0.25 | 32 | 1 |
| Bacteroides cellulosilyticus KLE 2342b,c | 8 | 2 | 128 | 128 |
| Streptococcus sanguinis KLE 2374b,c | 16 | 0.25 | 2 | 0.12 |
| Parabacteroides merdae KLE 2238b,c | 16 | 0.06 | 1 | 0.25 |
| Lactobacillus reuteri ATCC 23272b | 32 | 8 | 4 | 32 |
| Bacteroides fragilis ATCC 25285b | 32 | 0.5 | 16 | 2 |
| Blautia producta ATCC 27340b | 64 | 2 | 1 | 0.5 |
| Bacteroides ovatus KLE 2306b,c | 64 | 0.01 | 0.5 | 0.02 |
| Bacteroides vulgatus KLE 2381b,c | 64 | 1 | 128 | 128 |
| Bacteroides eggerthii KLE 2382b,c | 64 | 0.01 | 0.5 | N/A |
| Enterococcus faecalis KLE 2341b,c | >128 | 0.5 | 0.5 | >128 |
| Enterobacter cloacae ATCC 13047b | >128 | 8 | >128 | 64 |
| Bacteroides xylanisolvens KLE 2305b,c | >128 | 0.5 | 32 | 64 |
N/A, not available.
Cultivated under microaerophilic conditions
Cultivated under anaerobic conditions
Human stool isolate, KLE laboratory collection
Selectivity of hygromycin A against spirochetes is puzzling, since it targets the conserved peptidyl transferase center (PTC) of the bacterial ribosome (Guerrero and Modolell, 1980). To confirm the effect of hygromycin A on protein synthesis, we used a strain of B. burgdorferi that ectopically expresses GFP (Whetstine et al., 2009). Cells were treated with hygromycin A or a known protein synthesis inhibitor (spectinomycin), and GFP production was then induced with anhydrotetracycline. Cells with impaired protein synthesis are expected to remain non-fluorescent. Uninduced cells, non-treated cells or cells treated with the cell wall-acting antibiotic amoxicillin served as control. GFP signal was then measured for 100,000 cells by Flow cytometry (Figure 2A). Cells treated with hygromycin A had a dramatically lower level of GFP as compared to a control, showing that hygromycin A indeed inhibits protein synthesis. The structure of hygromycin A bound to the Thermus thermophilus 70S ribosome shows the interaction of the compound with the conserved 23S rRNA nucleotides that form the core of the peptidyl transferase center (Polikanov et al., 2015). The identity of these nucleotides is the same in Borreliella, based on our analysis of the genome sequence of B. burgdorferi (Figure 2B, C). Nevertheless, we initially considered the possibility that hygromycin A could have a much higher affinity to B. burgdorferi ribosomes. We first tested the action of hygromycin A on ribosomes of E. coli. During in vitro translation of the model ORFs, the complete arrest of translation was observed even at the lowest concentration of antibiotic (0.6 μM), which roughly approximates the concentration of active ribosomes in the reaction mixture (Figure 2D). Under these conditions, essentially all the ribosomes were captured at the start codons of the ORFs, indicating binding of hygromycin A to the ribosome with a very high affinity (Kd <10 nM). Since it is not possible to improve on a 1:1 binding, this result suggests that the basis for selective action of hygromycin A against spirochetes lies elsewhere. We therefore considered that the transport of hygromycin A into spirochetes might be responsible for its selectivity.
Figure 2. The mechanism of action of Hygromycin A.
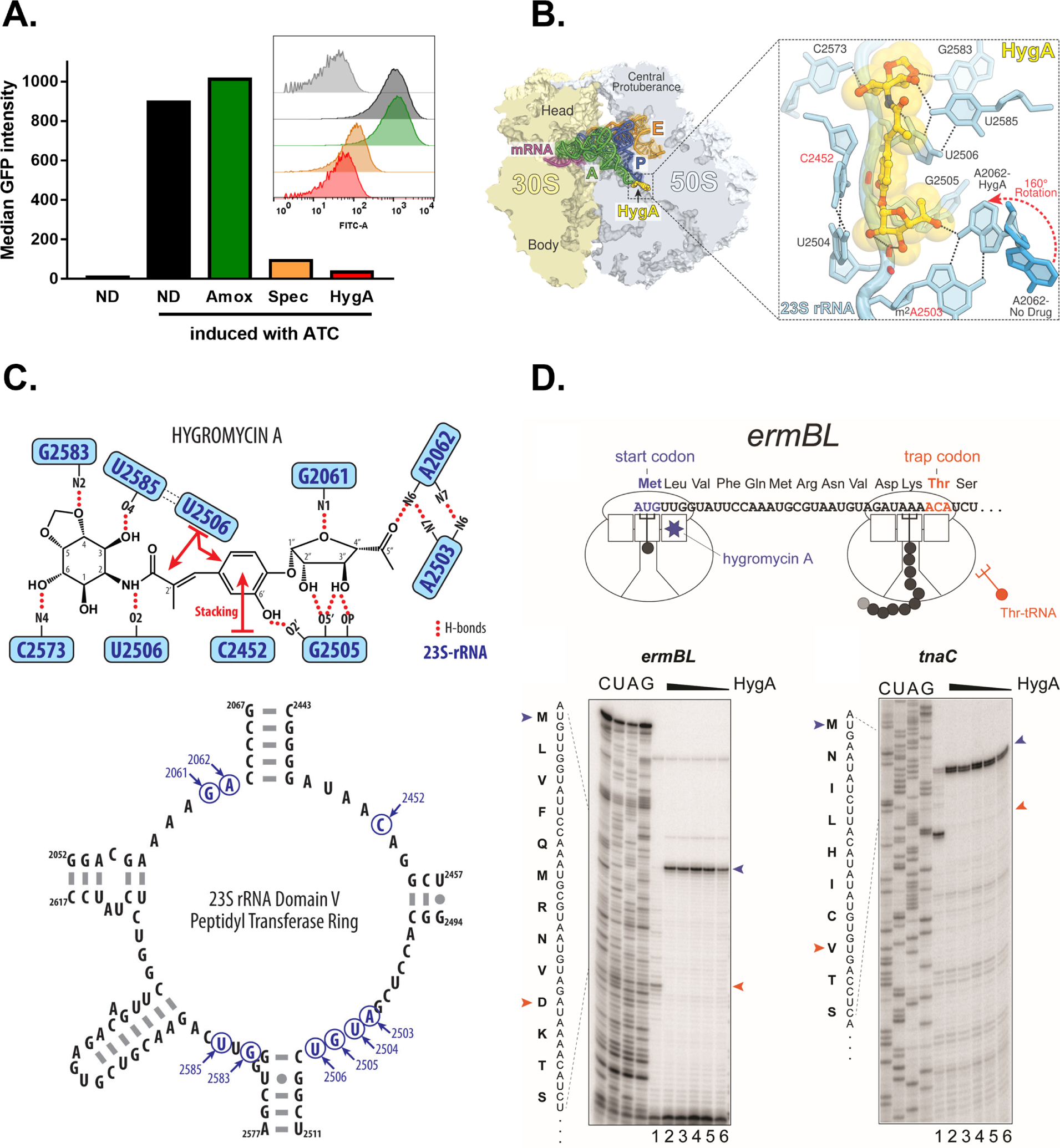
A. Measurement of protein synthesis inhibition using the GFP inducible (5 μg/ml anhydrotetraycycline, ATC) B. burgdorferi strain pCRW53 and flow cytometry. The median fluorescent intensity of 105 analyzed cells is shown. Insert shows a histogram of FITC-A (GFP) distribution. B. Overview of the hygromycin A binding site in the T. thermophilus 70S ribosome viewed as a cross- cut section through the ribosome. Insert shows close-up view of hygromycin A bound in the PTC. The E. coli nucleotide numbering is used throughout. H-bond interactions are indicated with dashed lines. Note that by forming an H-bond with the base of nucleotide A2062 of the 23S rRNA (light blue), hygromycin A causes characteristic rotation of this nucleotide by approximately 160 degrees to form Hoogsteen base-pair with the m2A2503 of the 23S rRNA (red dashed arrow). The unrotated conformation of A2062 observed in the absence of the drug is shown in blue (PDB entry 4Y4P) (Polikanov et al., 2015). C. Schematic representation of the interactions of hygromycin A with the ribosome shown in panel B. Potential H-bond interactions are indicated with dashed lines, stacking interactions are shown with red arrows (upper panel). Secondary structure of the peptidyl transferase ring of the domain V of the 23S rRNA from Gram-negative bacterium Thermus thermophilus (lower panel). Nucleotides involved in HygA coordination are highlighted in blue and are conservative between T. thermophilus, E. coli, and B. burgdorferi. D. Toe-printing analysis of the HygA-induced translation arrest at the start codons of the ORFs. Cartoon representation of the toe-printing experiment illustrated with the ermBL gene (upper panel). Binding of hygromycin A (HygA) to the ribosomal A site arrests translation at the start codon of the gene.
Similar to Gram-negative bacteria, spirochetes have an outer membrane that serves as an additional penetration barrier, which, however lacks lipopolysaccharide (LPS) and is not as efficient a barrier as that of other species such as E. coli. In E. coli, LPS on the surface of the outer membrane forms a hydrophilic network that restricts penetration of large or hydrophobic compounds. Substances that leak through the barrier are extruded by trans-envelope multidrug-resistant pumps (MDR)(Zgurskaya and Nikaido, 2000). We therefore compared penetration of hygromycin A into B. burgdorferi with that of wild type and “hyper-porinated” E. coli ΔtolC-Pore. This E. coli strain carries a modified, inducible FhuA siderophore receptor that forms a large pore in the outer membrane (Krishnamoorthy et al., 2016). As a result, the outer membrane is structurally intact, but has lost its barrier function. E. coli ΔtolC-Pore also carries a deletion in TolC, which encodes a channel serving as an exit portal for MDR substrates, further increasing the permeability of this strain.
To measure penetration of hygromycin A, B. burgdorferi and E. coli cells were incubated with the antibiotic, samples were withdrawn, rapidly filtered, lysed, and intracellular concentration of the compound was determined by mass spectrometry analysis of material separated by liquid chromatography (LC/MS) (Figure S3). There was little penetration of the compound into E. coli (Figure 3A). Importantly, there was no difference in MIC between cells with an intact outer membrane and those expressing the FhuA pore, suggesting that active efflux pumps and the inner membrane of E. coli are the barriers for hygromycin A (Table S3, Figure S4A). In agreement with this, efflux-deficient E. coli ΔtolC cells accumulate higher levels of hygromycin A (Figure 3C, middle panel). By contrast, accumulation of hygromycin A into cells of B. burgdorferi was rapid and substantial (note that the relatively high levels of antibiotic in this experiment were used since a dense culture of cells is required to measure the level of intracellular compound). No significant difference in the levels of accumulated hygromycin A could be seen after a short 1 minute incubation and a long incubation for 40 minutes. Apparently, hygromycin A is smuggled into B. burgdorferi by a transporter that is unique to spirochetes and is responsible for the selectivity of the compound against this group of microorganisms.
Figure 3. The basis for hygromycin A selectivity.
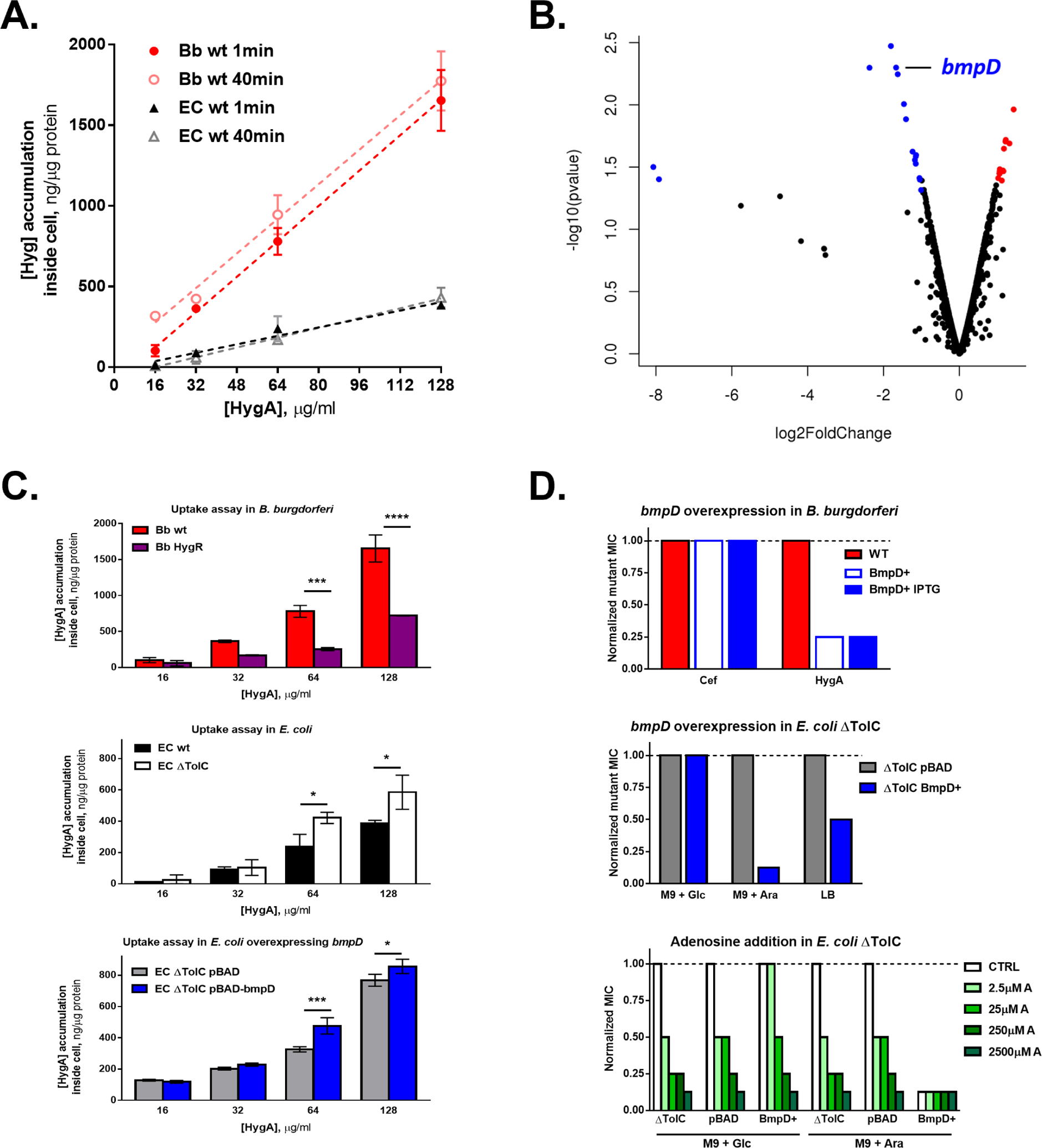
A. B. burgdorferi and E. coli cells were incubated at indicated concentrations of hygromycin A for 1 min or 40 min and intracellular accumulation was quantified by UHPLC/MS. Mean ± SD, N = 2. Linear regression is shown in dashed lines. Slopes are significantly different between B. burgdorferi and E. coli (p< 0.0001). B. RNAseq was performed on the B. burgdorferi hygromycin A resistant mutant in the presence (4μg/ml) or absence of hygromycin A. The volcano plot shows differential expression of genes in the presence of Hygromycin A relative to no drug control. Blue dots indicate statistically significant lower abundance, red dots indicate statistically significant higher abundance and black dots indicate that abundance is not significantly different. C. B. burgdorferi and E. coli cells were incubated at indicated concentrations of hygromycin A for 1 min and intracellular accumulation was quantified by UHPLC/MS. Mean ± SD, N = 2. (****=p< 0.0001, ***=p< 0.001, *=p< 0.05) D. Hygromycin A MIC fold-change of B. burgdorferi overexpressing bmpD as compared to B. burgdorferi wild-type (upper panel), of E. coli dTolC overexpressing bmpD as compared to E. coli dTolC under repressing (M9 + Glucose), inducing (M9 + Arabinose) and regular LB growth conditions (middle panel) and with addition of adenosine (A, lower panel).
In order to identify a possible transporter, we selected for mutants resistant to hygromycin A. To do so, we first used a standard approach to obtain resistant mutants by embedding a large number of cells (108–109) in semi-solid BSK medium containing various concentrations of hygromycin A. Colonies growing on hygromycin A containing plates were then picked, grown in liquid medium without antibiotic, and plated again on medium with a higher concentration of compound. We also used an evolutionary selection by serial passaging in increasing concentrations of hygromycin A in liquid cultures. For both approaches, we used B. burgdorferi B31 wild-type, a B. burgdorferi hyper-mutator strain with a deletion in mutS, a fast-growing Borrelia species (B. turcica), a pooled transposon mutant library of B. burgdorferi and EMS-mutagenized B. burgdorferi. For the evolutionary approach, cultures growing at the highest hygromycin A concentration were reinoculated in liquid medium without antibiotic and then challenged with higher concentrations of the compound. This approach however produced slow-growing cells. It is known that frequent passaging of B. burgdorferi under laboratory conditions leads to plasmid loss (Grimm et al., 2003). In addition, most resistant isolates were unstable and lost resistance after growth in the absence of hygromycin A. Combined, this very large effort yielded only two mutants: B. turcica KLEX1 and B. burgdorferi B31 KLEX2 with stable hygromycin A resistance, MICs of 8 μg/ml and 4 μg/ml, respectively, which is a 16-fold increase as compared to their wild type parent strains. The two hygromycin A resistant mutants did not have any growth defect. The genomes of the mutants were sequenced and the mutations were analyzed. KLEX1 and KLEX2 isolates carried the same two mutations in the gene coding for the 23S rRNA, the known target of hygromycin A (2629G>T and 2618G>T, B. turcica and B. burgdorferi nucleotide numbering, respectively, Table S4, S5). B. burgdorferi has two copies of 23S rRNA (Schwartz et al., 1992), equivalent to two targets, which will diminish the probability of resistance development (Silver, 2007). Given that the conserved target is not responsible for the selectivity of hygromycin A, we considered that additional mutations in the resistant strains might lead to a decrease in the transport of the compound. Measuring hygromycin A penetration showed that uptake of the compound was severely impaired in the B. burgdorferi KLEX2 mutant (Figure 3C, upper panel), but not in the B. turcica KLEX1 mutant (Figure S4B).
The B. burgdorferi KLEX2 mutant accumulated a total of 21 polymorphisms, but no non-synonymous mutations were found in genes known to be involved in transport (Table S5). Only one of the genes (BB 0336) contained a frameshift mutation adjacent to an operon involved in oligopeptide transport (OppABCDF). If hygromycin A was using the oligopeptide transporter for uptake, then a mutation in BB0336 could lead to downregulation of OppABCDF expression and antibiotic resistance. However, hygromycin A susceptibility was not changed in a B. burgdorferi strain with a knockout in oppDF. This shows that the oligopeptide transport system of B. burgdorferi is not involved in hygromycin A uptake. We next considered that one or more of the 21 SNPs in the KLEX2 mutant might lead to downregulation of expression of a hygromycin A transporter. Such downregulation could then be detected by transcriptome analysis. By comparing the transcriptome of wild type B. burgdorferi with KLEX2, we observed the absence of reads for many transcripts in the resistant mutant, which is unsurprising, because B. burgdorferi tends to lose plasmids when serially passaged in the lab. We then compared the transcriptome of KLEX2 treated with hygromycin A at 1x MIC to that of untreated cells (Table S6). Notably, the transcriptome of hygromycin A treated cells showed a 3.18-fold decrease in the expression of bmpD, a periplasmic substrate-binding protein of an ABC-type purine nucleoside transporter composed of two transmembrane proteins (BB0678 and BB0679) and the ATP-binding protein BB0677 (subsequently referred to as BmpDEFG) (Åstrand et al., 2019; Cuellar et al., 2020) (Figure 3b). B. burgdorferi lacks enzymes required for de novo purine synthesis, and BmpDEFG is essential for nucleoside uptake. No other genes involved in transport were significantly differentially regulated.
Since a decrease in expression of BmpD correlated with resistance to hygromycin A, we expected that overexpressing this protein will produce an opposite effect, increasing susceptibility to the antibiotic. We cloned bmpD under the control of an IPTG-inducible promoter using the B. burgdorferi expression vector pJSB275m (Blevins et al., 2007) and assessed the MIC of this overexpression strain. Overexpression of bmpD in B. burgdorferi resulted in a four-fold reduction in hygromycin A MIC (Figure 3d, upper panel), while the MIC for ceftriaxone remained unchanged. This effect was IPTG-independent, likely due to the leakiness of the promoter in the construct. Overexpression of BmpD had no effect on the MIC of ceftriaxone, a cell wall-acting antibiotic. We then examined the ability of BmpD to affect hygromycin A resistance in a heterologous system, using E. coli as a host. To this end, bmpD was cloned into the pBAD30 expression vector under an arabinose promoter and transformed into E. coli ΔtolC (carrying a deletion in the TolC component of MDR pumps, to avoid effects from efflux of hygromycin A). Inducing expression of BmpD resulted in an 8-fold reduction of E. coli hygromycin A MIC (Figure 3d, middle panel), in good agreement with a similar result obtained with B. burgdorferi. Next, we measured penetration of hygromycin A into E. coli carrying recombinant BmpD. Expression of the protein increased penetration of hygromycin A (Figure 3c, lower panel), suggesting that nucleoside transporters contribute to the uptake of hygromycin A. E. coli nucleoside transporters NupC, NupG and YegT are induced by adenosine (Xie et al., 2004). Addition of adenosine to E. coli resulted in a concentration-dependent decrease in MIC up to 8-fold (Figure 3d, lower panel). When bmpD was induced, addition of adenosine did not result in a further decrease of MIC, suggesting that either the uptake of hygromycin A, the target engagement or both are saturated under these conditions.
Given the high potency and selectivity of hygromycin A against Borreliella, we sought to determine its potential as a therapeutic. Testing hygromycin A against a number of human cell lines showed no cytotoxicity (EC50 >512), giving an impressive in vitro therapeutic index of >2,000 (Table S7). Next, we tested hygromycin A in a mouse model of acute Lyme disease (Hodzic et al., 2003). Mice are infected with B. burgdorferi N40 by subcutaneous injection, after which the pathogen propagates and spreads from the site of injection. Three weeks after inoculation, hygromycin A was delivered by intraperitoneal (IP) injection as an aqueous solution. Therapy in the mouse model aims to emulate treatment of humans, and antibiotics are administered twice a day for five days. In this model, the pathogen burden is determined by culturing skin samples in a liquid medium, and performing quantitative PCR of the pathogen 16S rRNA (Ornstein and Barbour, 2006). Culturing in liquid is a stringent test of efficacy, because even one surviving cell of the pathogen will result in growth. After five days of treatment, hygromycin A cleared the infection, similarly to the ceftriaxone control (Table 2).
Table 2.
The efficacy of hygromycin A and control antibiotics against a murine infection with B. burgdorferi. C3H mice or Peromyscus leucopus were infected subcutaneously with B. burgdorferi N40 or B. burgdorferi B31, respectively, and after 3 weeks were treated for 5 days with hygromycin A via oral gavage, IP injection, or by bait, with ceftriaxone via IP injection, with amoxicillin via oral gavage, or with doxycycline via oral gavage or by bait. All mice were treated twice a day and the total daily dose (mg/kg/day) is indicated. After treatment, the presence of B. burgdorferi cells was detected by dark-field microscopy from culture of a whole ear in BSK-II media. The percentage of mice from which cultures were positive is reported.
| Mouse | Route of Administration | Compound | Daily Dose (mg/kg/day) | Culture positive (%) |
|---|---|---|---|---|
| C3H | IP. | Saline | 0 | 100 |
| Hygromycin A | 100 | 0 | ||
| Ceftriaxone | 312 | 0 | ||
| C3H | Oral gavage | Hygromycin A | 50 | 50 |
| Hygromycin A | 100 | 0 | ||
| Hygromycin A | 140 | 0 | ||
| Hygromycin A | 500 | 0 | ||
| Amoxicillin | 200 | 0 | ||
| Doxycycline | 100 | 0 | ||
| C3H | Bait | Hygromycin A | 100 | 0 |
| Doxycycline | 200 | 0 | ||
| Peromyscus leucopus | Oral gavage | Hygromycin A | 100 | 100 |
| Hygromycin A | 200 | 0 |
I.P. = Intraperitoneal injection
We also tested oral administration of hygromycin A and found similar efficacy. There were no indications of toxicity even at very high doses, suggesting the compound is safe.
Broad-spectrum antibiotics can disrupt the gut microbiome (Ferrer et al., 2017). Since hygromycin A is selective against spirochetes and is inactive against many intestinal bacterial isolates (Table 1), we examined microbiome changes that accompany treatment of C3H mice with this compound. Mice were infected with B. burgdorferi N40, treated with clinically relevant doses of hygromycin A, ceftriaxone, or amoxicillin, and the microbiome composition was analyzed by sequencing 16S rDNA. Ampicillin and amoxicillin caused significant changes in the composition of the microbiota (Figure 4C, D, S5). Notably, there was a prominent bloom of pathogenic Enterococcus, resulting in 98% of total microbiome abundance in some of the animals. Amoxicillin also produced a bloom of Bacteroides in some of the mice, apparently due to the presence of resistant bacteria. By contrast, the changes produced by hygromycin A were milder, with a prominent increase in symbiotic Lactobacillus and Lactococcus. Given the low activity of hygromycin A against gut bacteria, the reason for this observed shift is presently unknown.
Figure 4: Animal efficacy of hygromycin A.
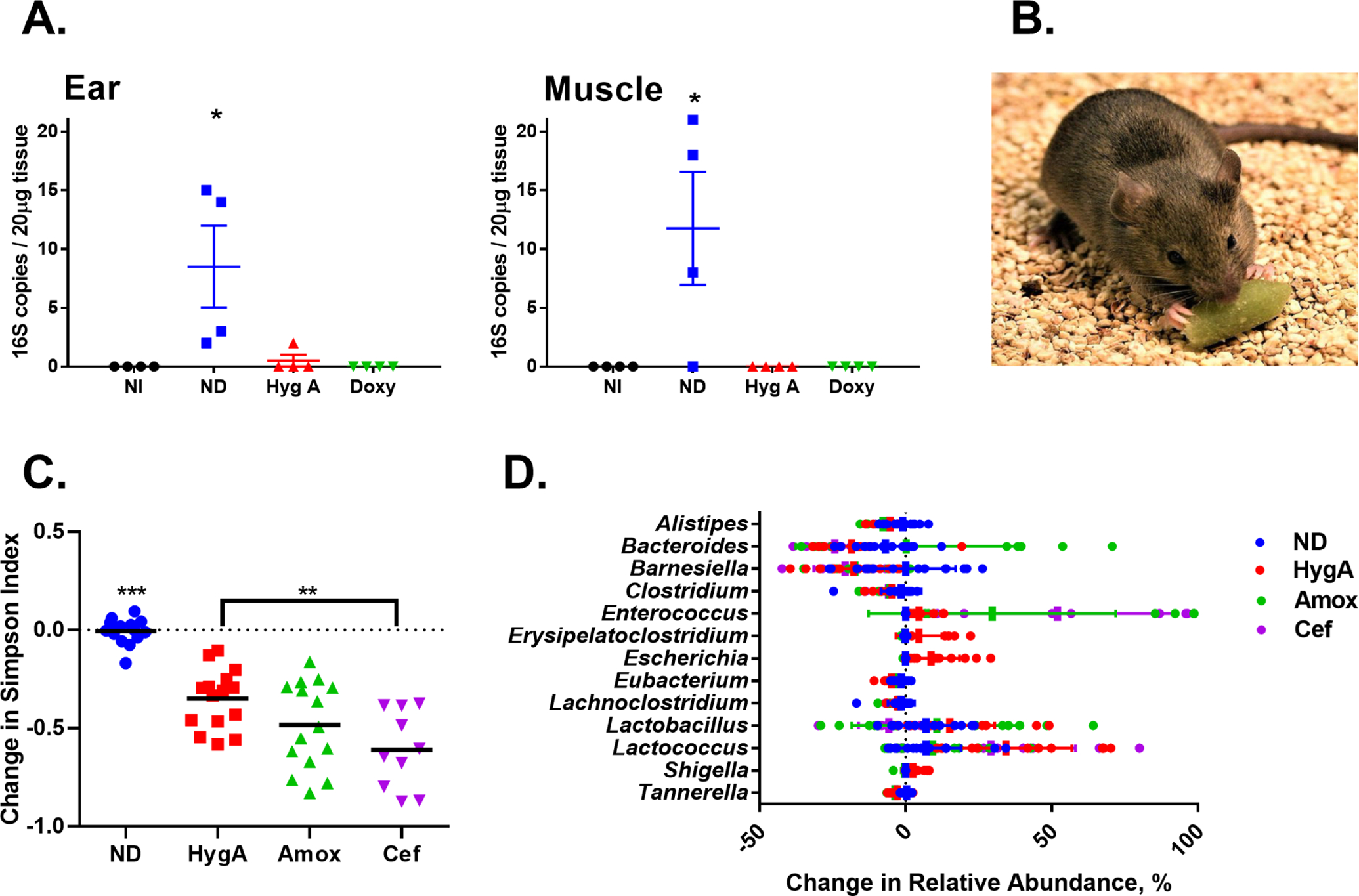
A. Quantification of B. burgdorferi in tissues (ear, muscle) of animals that ingested baits. The amount of 16S rRNA was converted into cell count as described in Materials and Methods. Infected mice were given hygromycin A bait (HygA) or doxycycline bait (Doxy); infected non-treated group (ND) and non-infected group (NI) were given drug-free bait for 5 days. B. Photograph of a C3H mouse eating a bait. C. The change in alpha diversity based on the Simpson index of the murine fecal microbiome from before to after treatment with hygromycin A (HygA) (per os), amoxicillin (Amox) (per os), or ceftriaxone (Cef) (subcutaneous). Mice were infected with B. burgdorferi N40 and treated twice a day for 5 days or were untreated (ND). Stool was collected before and after treatment and sequenced for the 16S rRNA gene and the alpha diversity was calculated using the Simpson Index metric. Each point represents the change in the Simpson index from before to after treatment for an individual mouse across three individual experiments. Bars represent the mean. Statistical significance was calculated using a one-way ANOVA with Tukey’s multiple comparisons test (**=p<0.01, ***=p<0.001). D. The change in relative abundance (%) of the most abundant genera in the murine gut microbiome from before to after treatment with hygromycin A (per os), amoxicillin (per os), or ceftriaxone (subcutaneous). Mice were infected with B. burgdorferi N40 and treated twice a day for 5 days or were untreated. Stool was collected before and after treatment and was sequenced for the 16S rRNA gene. Each point represents the change in the relative abundance of the respective genus for an individual mouse across three individual experiments. Bars represent the mean.
Good efficacy, safety, oral availability and selectivity of hygromycin A open an additional intriguing possibility of eradicating Lyme disease by targeting its environmental reservoir. In a previously published study, a bait containing doxycycline was spread in a limited area, and cleared the infection in 87% of mice and 94% of Ixodes ticks (Dolan et al., 2011). This is far above levels required to reduce infection below the transmission threshold - the percentage of infected ticks and mice needed to sustain the infectious cycle in the wild (Lou et al., 2014; Mount et al., 1997). Doxycycline however is an essential part of our shrinking antibiotic arsenal, and spreading it on large territory is unfeasible due to the risk of selecting for resistant microorganisms. Hygromycin A, with its limited activity against non-spirochetal organisms would make an ideal reservoir targeted antibiotic against B. burgdorferi. We therefore tested the efficacy of hygromycin A to clear B. burgdorferi by incorporating it into mouse baits. Consuming baits with hygromycin A efficiently cleared B. burgdorferi infection in the animals (Figure 4 A, B).
Discussion
The rise in antibiotic resistance, and the important role the microbiome plays in human health suggest a transition from traditional broad-spectrum to selectively acting compounds. Here we report the rediscovery of hygromycin A as a selective and efficient antibiotic acting against B. burgdorferi, the causative agent of Lyme disease. We identified hygromycin A in a differential screen of soil actinomycetes against B. burgdorferi, counter screening against S. aureus. Produced by S. hygroscopicus, hygromycin A is an old, abandoned antibiotic, with poor activity against both Gram-positive and Gram-negative bacteria. The one exception has been Brachyspira (formerly Treponema)(Omura et al., 1987), and for a while hygromycin A was evaluated as a potential treatment for pig dysentery caused by this pathogen. While efficacy was established, apparently, the economics of this application were not favorable due to low activity against other pathogens, and the compound was largely forgotten. Testing a variety of bacteria showed that hygromycin A is highly potent against spirochetes, with MIC around 0.2 μg/ml. Apparently, hygromycin A evolved in actinomycetes as a weapon against environmental spirochetes.
Given the poor activity of hygromycin A against most bacteria, we examined its effect on the mouse microbiome. By comparison to conventional broad-spectrum antibiotics used to treat Lyme disease, changes following hygromycin A administration were mild (Figure 4C, D). A particularly troublesome consequence of therapy with broad-spectrum antibiotics is a dramatic expansion of drug-resistant mutants of opportunistic pathogens such as Enterococci that take over the population. This both harms the microbiome and contributes to the spread of drug resistance. Such expansion was not observed with hygromycin A.
The mechanism of selectivity of hygromycin A is puzzling since the compound inhibits protein synthesis by binding to the ribosomal 23S rRNA in the catalytic peptidyl transferase center. This functional part of the ribosome is highly conserved among bacteria, including B. burgdorferi. If action against the target cannot be responsible for the observed selectivity, the remaining logical possibility is transport. Tracking penetration by LC/MS showed rapid accumulation of hygromycin A into B. burgdorferi, but not E. coli. Transcriptome analysis of a resistant mutant showed a down-regulation of BmpD, the periplasmic substrate-binding protein of an ABC-type transporter (BmpDEFG) of purine nucleosides. Ectopic overexpression of BmpD had the opposite effect, increasing susceptibility to hygromycin A. BmpD is conserved in the Borreliella and Borrelia genera and T. pallidum has a PnrA nucleoside transporter homologous to BmpD (Åstrand et al., 2019; Cuellar et al., 2020), explaining selectivity of hygromycin A. In B. burgdorferi, BmpDEFG is likely essential, since the spirochetes are auxotrophic for purines and this is the only transporter that brings them into the cell (Cuellar et al., 2020). This is why we did not observe selection for high- frequency null mutants in BmpDEFG conferring resistance to hygromycin A.
Compounds acting against selective bacterial groups are well-documented, and it appears that their number reflects the effort expended to identify them, rather than their incidence in nature. A considerable effort to find antibiotics acting against M. tuberculosis resulted in the discovery of cyclomarin, lassomycin and kitamycobactin targeting the essential mycobacterial protease ClpP1P2C1 (Gavrish et al., 2014; Quigley et al., 2020; Schmitt et al., 2011); griselimycin, targeting DNA polymerase (Kling et al., 2015); amycobactin, targeting the SecY protein export component of mycobacteria (Quigley et al., 2020), and others. A screen against Pseudomonas aeruginosa led to the discovery of pacidamycins that target MraY, a conserved essential enzyme of the peptidoglycan biosynthetic pathway. Pacidamycins appeared to be selectively taken up by an oligopeptide transporter of P. aeruginosa (Mistry et al., 2013). This is similar to hygromycin uptake, however, the pseudomonas transporter is non-essential and resistant mutants appear with high frequency. In a majority of known cases, selectivity is determined by the target. Selectivity determined by an essential transporter we report in this study significantly increases the repertoire of possible underlying mechanisms, and with it, increases the possibility of identifying additional compounds targeting a narrow taxonomic group of pathogens. We may expect a growing number of such selective antibiotics focused on pathogens and sparing the microbiome.
Hygromycin A has a number of features that makes it an attractive candidate for treating Lyme disease. Apart from sparing the microbiome, resistance development is very low, and only occurs upon multiple passages. There is no detectable cytotoxicity against human cells, and we were unable to detect toxicity upon oral administration even at very high doses, exceeding MIC 2,500 fold. This is probably a consequence of poor penetration through membranes of this highly water-soluble compound (LogP −0.66) that requires a transporter to smuggle it to its target in spirochetes. By the same token, hygromycin A is unlikely to reach a homologous 23S rRNA in mitochondrial ribosomes. Hygromycin A was able to clear a B. burgdorferi infection in mice upon oral administration. Oral availability is a desirable property for an antibiotic, especially for compounds that need to be administered over lengthy periods of time. Acute Lyme disease is usually treated with a 2-week (Stupica et al., 2012) course of doxycycline, which is not particularly short, but Lyme arthritis that develops if the infection is not diagnosed in time requires up to 3 months of antibiotic therapy. Apart from treating acute infection, antibiotics are also given prophylactically in case of a tick bite to prevent possible infection. Such prophylaxis may become more routine if it involves an antibiotic acting selectively against the pathogen with fewer side-effects.
We find that hygromycin A is also highly potent against another important human pathogen, T. pallidum, the spirochete causing syphilis. T. pallidum is becoming increasingly resistant to current therapies (Tien et al., 2020), and hygromycin A may be a candidate for treating this sexually transmitted disease.
Acute Lyme infection, even if treated in time, produces a poorly-understood post-Lyme disease in about 10–20% of patients (Aucott, 2015; Aucott et al., 2013; Marques, 2008; Rebman and Aucott, 2020; Rebman et al., 2017). This condition is characterized by fatigue, muscle pain, a ‘foggy’ mind and other symptoms. The significance of this debilitating disease has been recently brought into focus by a set of very similar symptoms in patients with “Long COVID” (Aucott and Rebman, 2021). There are a number of objective markers that are associated with post-Lyme disease, such as elevated microglial activation(Coughlin et al., 2018), a decrease of the CD57 lymphocyte subset (Stricker et al., 2002; Stricker and Winger, 2001), and increased levels of the chemokine CCL19 (Aucott et al., 2016), and the cytokine IL-23 (Strle et al., 2014), and, interestingly, a characteristic shift in the microbiome composition (Morrissette et al., 2020). Whether this change in the microbiome of post-Lyme patients contributes to the disease is unclear, but having therapeutics that avoid harming the microbiome may be particularly desirable in the case of Lyme disease.
Lyme disease is on the rise and in many locations, limits our ability to enjoy outdoor activities. While vaccination can protect those that receive the vaccine, it does not get to the heart of the problem and requires continued health care expenditures. A more permanent solution would require eradicating the source of the disease. We show that baits containing hygromycin A clear B. burgdorferi infection in mice, the principal host of the pathogen. This opens the attractive possibility of environmental eradication of this troublesome pathogen.
Limitations of the study
The aim of this study was to identify a compound that acts selectively against B. burgdorferi, and can be developed into a dedicated therapeutic to treat Lyme disease. A differential screen of soil isolates led to the identification of hygromycin A that selectively kills spirochetes. We then examined the mechanism of selectivity, and the potential of hygromycin A as a therapeutic. Hygromycin A is known to bind to the conserved ribosomal 23S rRNA, and we therefore searched for an alternative explanation for its selective action - a possible transporter unique to spirochetes that could smuggle the compound into the cell. Transcriptome analysis showed that in the presence of hygromycin, expression of BmpD, a periplasmic binding protein of the spirochete nucleoside transporter, is downregulated. Subsequent experiments indicated that the BmpD transport system is involved in bringing the compound into the cell, explaining the nature of selectivity. At present, we do not know how hygromycin causes a decrease in the expression of BmpD. We also assume that the BmpD transport system is essential, based on indirect evidence, such as the auxotrophy of spirochetes for nucleosides, and the lack of transposon insertions in this locus. This would explain why we failed to obtain high-frequency null mutants in BmpD resistant to hygromycin A. However, direct evidence for the essentiality of BmpD to spirochetes is lacking. Regarding the therapeutic potential of hygromycin A, we show that the compound is efficacious in a mouse model of Lyme disease by both parenteral and oral administration, and there were no signs of toxicity at fairly high doses. The compound also shows no cytotoxicity. This indicates that the compound may be safe and efficacious. At the same time, these are only the first steps in the preclinical evaluation of a compound, which will require a detailed pharamcokinetic/pharamacodynamic, metabolism and toxicology evaluation before it enters into clinical trials. The preclinical work will show whether hygromycin A, as most natural product antibiotics that became drugs, can be developed in its native form, or will require medicinal chemistry optimization to improve its properties.
STAR Methods
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Kim Lewis (k.lewis@northeastern.edu).
Materials Availability
Strains generated in this study are available from the Lead Contact upon request.
Data and Code Availability
Data have been deposited at https://data.mendeley.com/datasets/wxcfg58zg3/draft?a=94a40a19-499f-4d95-8e9f-8a5d890ffa35 and are publicly available as of the date of publication. DOIs are listed in the key resources table. This paper does not report original code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Bacterial Strains
Actinomycete strains were isolated as follows: soil was collected, mixed with calcium carbonate and enriched for spores by dry heat at 70°C for 30 minutes. Cells were then serially diluted ten-fold and plated on oatmeal agar medium. After 7 days of incubation, plates were examined under a dissecting microscope and colonies with characteristic Actinomycete morphology were re-streaked to fresh plates. After 7 days of incubation, biomass was scraped and re-suspended in TSB with 15% glycerol. Stocks were stored at −80°C. Several Actinomycete strains were purchased from the ARS Culture Collection (NRRL). B. burgdorferi sensu stricto strain B31 (clone 5A19) was kindly provided by Monica Embers at the Tulane National Primate Research Center (Purser and Norris, 2000).
B. burgdorferi sensu stricto strain B31 5A4 NP1 GFP+ (BbP1286) strain was kindly provided by Melissa Caimano at the University of Conecticut Health Center, School of Medicine (Iyer et al., 2015).B. burgdorferi sensu stricto strain N40 (clone D10E9), B. afzelii PKo serotype 2, B. garinii PBr serotype 3 and B. bavariensis Pbi serotype 4 were kindly provided by John Leong at Tufts-New England Medical Center Hospital (Lin et al., 2014). B. turcica strain IST7 (DSM 16138) and Alkalispirochaeta americana strain ASpG1 (DSM 14872) were purchased from DSMZ. B. burgdorferi (strain 297; ATCC 53899), Brevinema andersonii (ATCC 43811), Leptospira interrogans serovar Copenhageni (ATCC BAA-1198) and Leptospira biflexa Patoc 1 (ATCC 23582) were purchased from ATCC.
B. burgdorferi (strains B31, BbP1286, N40 and 297), B. afzelii, B. garinii, B. bavariensis, B. miyamotoi, B. turcica and B. andersonii cultures were grown in Barbour-Stoenner-Kelly II (BSKII) medium in a microaerophilic chamber (34°C, 3% O2, 5% CO2) as described previously for B. burgdorferi (Wu et al., 2018).
In the case of B. burgdorferi BbP1286 expressing GFP under the control of flab promoter50 μg/mL gentamicin and 100 μg/mL kanamycin were added as described previously (Blevins et al., 2007).
Alkalispirochaeta americana (DSM 14872) was grown in DSMZ media 1165 as described previously (Hoover et al., 2003) at pH 9.4 in an anaerobic chamber (37°C, 5% H2, 10% CO2, and 85% N2). L. biflexa and L. interrogans were grown in ATCC medium 1470 without agar (modified Leptospira medium) containing per liter, 0.3 g Peptone (BD 211677), 0.2 g beef extract, 0.5 g sodium chloride, 10% v/v rabbit serum and 0.0012 g hemin adjusted to pH 7.4. Cultures were grown aerobically and static at 30°C.
Treponema pallidum subspecies pallidum Nichols, initially isolated from the cerebrospinal fluid of a neurosyphilis patient in Baltimore, Maryland, U.S.A. in 1912, was a gift to SJN from J. N. Miller at the UCLA Geffen School of Medicine. In vitro cultivation of T. pallidum has been described in detail by (Edmondson et al., 2018). Briefly, T. pallidum is grown in co-culture with rabbit epithelial cells (Sf1Ep (NBL-11),(ATCC CCL-68TM)). Stocks of Sf1EP cells were between passage 36 and 40 and were maintained in Sf1Ep medium consisting of Eagle’s MEM with non-essential amino acids, L-glutamine, sodium pyruvate, and 10% heat-inactivated FBS (48) at 37°C in air with 5% CO2. One day prior to initiation of antibiotic testing, Sf1Ep cells were seeded into tissue culture-treated 6-well cluster plates at 0.5×105 per well. T. pallidum cultivation medium (TpCM-2) was prepared the day before experiment initiation and pre-equilibrated in a BBL™ GasPak™ jar in which a vacuum was drawn five times (house vacuum, ~12–18 um Hg). The jar was refilled with 5% CO2:95% N2 four times and a final time with 1.5% O2:5% CO2:93.5% N2. The medium was then incubated overnight in a tri-gas incubator (ThermoFisher Forma Model 3130) maintained at 34°C and 1.5% O2:5% CO2:93.5% N2 (hereafter referred to as the low oxygen incubator). All subsequent steps in the incubation of T. pallidum cultures were carried out under these conditions.
E. coli WO153 was kindly provided by Lucy Ling, Novobiotic Pharmaceuticals (Ling et al., 2015). Shigella sonnei (ATCC 25931), Salmonella enterica Typhimurium LT2 (ATCC 19585), Enterobacter cloacae (ATCC 13047), Bifidobacterium longum (ATCC BAA-999), Lactobacillus reuteri (ATCC 23272), Blautia producta (ATCC 27340) and Bacteroides fragilis (ATCC 25285) strains were purchased from ATCC. Bacteria from laboratory KLE collection were isolated as described previously (Imai et al., 2019). S. aureus HG003, E. coli W0153, E. coli MG1655 and P. aeruginosa PO1 were grown aerobically in cation-adjusted Mueller-Hinton broth (MHIIB), all other bacteria and members of the gut flora were grown in an anaerobic chamber (5% H2, 10% CO2, and 85% N2) in Brain Heart Infusion (BHI) broth, supplemented with 0.5% Yeast Extract, 50mM MOPS buffer, 0.1% cysteine hydrochloride and 15 μg/ml hemin (BHI-YMCH).
Cell lines
The cell lines used were FaDu pharynx squamous cell carcinoma (ATCC HTB-43), HepG2 liver hepatocellular carcinoma (ATCC HB-8065), and HEK293-RFP human embryonic kidney red fluorescent protein tagged (GenTarget SC007) cells. All cells were cultured in Eagle’s Minimum Essential Medium supplemented with 10% fetal bovine serum at 37°C.
Mice
Wild type 6 weeks old female C3H mice (Charles River Laboratories) were used for all animal experiments. All animal experiments were conducted according to protocols that were approved by the Institute of Animal Care and Usage Committee (IACUC) at Northeastern University (Approval #16 -−0619R). Mice were randomly assigned to experimental groups.
METHOD DETAILS
Minimum Inhibitory Concentration (MIC)
The microbroth dilution Minimum Inhibitory Concentration (MIC) method was used to quantitatively measure the in vitro antibacterial activity of hygromycin A against bacterial strains. Anti-Borreliella MIC assays were performed as described previously (Wu et al., 2018). Briefly, cultures were grown to stationary phase and diluted 1:10 (1:100 for L. biflexa) in a 96-well plate containing Hygromycin A diluted serially 2 fold. Plates with Borrelia cultures were incubated in a microaerophilic chamber (Coy hypoxic O2 control glove box) for 7 days (34°C, 3% O2, 5% CO2) and scored visually for a pink to yellow color change of the phenol red present in the medium. Plates with A. americana and Leptospira cultures were grown for 4 days under anaerobic conditions and 6 days under aerobic conditions respectively, and scored visually for change in color imparted by resazurin (0.001 g/L) present in the medium. The lowest concentration of antibiotic that prevented color change was interpreted as the MIC. The samples were also observed by dark field microscopy to count the number of spirochetes and results corroborated the visual MIC score.
Three hours prior to the start of a T. pallidum MIC experiment, the medium in the 6-well plates containing Sf1EP cells was removed, and the plates were rinsed with the pre-equilibrated TpCM-2 to remove traces of Sf1Ep medium and the medium replaced with 4 ml of the TpCM-2. The antibiotic to be tested was then added to each well to obtain the indicated antibiotic concentrations. Plates were then pre-equilibrated in the GasPak jar as described above and then transferred to the low oxygen incubator. Antibiotic sensitivity testing was initiated by inoculation of cultures with 3.3–3.5 × 106 T. pallidum trypsinized from an actively growing culture. Sf1Ep cultures containing antibiotics were briefly removed from the incubator, inoculated with T. pallidum, pre-equilibrated, and then returned to the low oxygen incubator. Three biological replicates were used for each antibiotic concentration. Following seven days of culture the T. pallidum were trypsinized to remove them from the Sf1Ep cells (Edmondson et al., 2018) and quantitated by darkfield microscopy using Helber counting chambers with Thoma rulings (Hawksley, Lancing, Sussex, UK). Each culture was counted at least twice using this method. Motility of each organism was also assessed. The MIC was calculated by interpolating the yields relative to the inoculum, with the point at which the yield = inoculum representing the MIC. Hygromycin A spectrum of activity against a panel of lab strains, pathobionts and symbionts was determined as described previously (Imai et al., 2019). Briefly, aerobic lab strains (i.e. S. aureus HG003, E. coli W0153 and MG1655, P. aeruginosa PAO1) from liquid cultures were diluted into the assay plate to achieve 5 × 105 CFU/mL; pathobionts and symbionts from anaerobic cultures were diluted 100 fold. Assay plates were prepared by 2-fold dilution of compound across the plate and included a positive growth control. After incubating the aerobes at 37 °C for 16–20 hours and the anaerobes in an anaerobic chamber for 24–48 hours, the MIC was determined as the lowest concentration of compound that inhibits growth of the bacteria as detected by the unaided eye. All MIC assays were repeated at least in triplicate.
Minimum bactericidal concentration (MBC)
The MBC of hygromycin A against B. bugdorferi was determined in 1.5 ml centrifuge tubes. Exponential phase B31 cultures (107/ml) were incubated with compounds at 1x, 2x, 4x and 8x MIC for 5 days in the microaerophilic chamber. Cells were washed 3 times with BSK-II medium, serially diluted and mixed in semi-agar plates (Sharma et al., 2015). All the plates were incubated in the microaerophilic chamber for 20 days before counting. The lowest concentration that killed more than 3 logs of cells was determined as MBC.
Cytotoxicity
A microplate Alamar Blue assay (MABA/resazurin) was used to determine the cytotoxicity of Hygromycin A. The cell lines used were FaDu pharynx squamous cell carcinoma (ATCC HTB-43), HepG2 liver hepatocellular carcinoma (ATCC HB-8065), and HEK293-RFP human embryonic kidney red fluorescent protein tagged (GenTarget SC007) cells, all cultured in Eagle’s Minimum Essential Medium supplemented with 10% fetal bovine serum. Exponentially growing cells were seeded into a 96-well, flat bottom, tissue culture treated plate (Corning) and incubated at 37°C with 5% CO2. After 24 hours, the medium was aspirated and replaced with fresh medium containing test compounds (2 μL of a two-fold serial dilution in water to 98 μL of media). After 72 hours of incubation at 37°C with 5% CO2, resazurin (Acros Organics) was added to each well to a final concentration of 0.15 mM. After three hours, the A544 and A590 were measured using a BioTek Synergy H1 microplate reader to determine cell viability. Experiments were performed with biological replicates.
Screening of anti-Borreliella specific compounds
Actinomycetes were inoculated into 50 mL Falcon tubes containing 10 mL modified R5 (MR5) medium with 0.1% vitamin supplement (ATCC® MD-VS™), and incubated at 28°C with shaking (200 rpm). After 10 days of incubation, 1 mL samples were collected and centrifuged (12,000×g, 10 min) to remove the cells. Culture supernatants (750 μl) were transferred into 96 deep well plates, and dried by centrifugal evaporator. The dried samples were resuspended in 50 μL of water, and yielded 15 times concentrated samples as compared to culture supernatant. Activity of culture extracts were evaluated against B. burgdorferi BbP1286 and counter-screened against S. aureus HG003. Activity against B. burgdorferi BbP1286 was evaluated by measuring GFP fluorescence by plate reader (Synergy™ H1, BioTek Instruments) with 485/528 nm wavelength. B. burgdorferi BbP1286 was cultured and incubated as described above. After 5 days of growth, the culture was diluted 1:100 into BSKII with 50 μg/mL gentamicin and 100 μg/mL kanamycin. In a 96-well plate, 200 μL of the diluted B. burgdorferi BbP1286 culture as added to 2 μL of extract. The plate was incubated in a microaerophilic chamber for 7 days. A reduction of 80% of the GFP signal compared to cell only control was considered positive anti-Borrelia activity. For the counter-screening, S. aureus exponential phase culture (OD600 of 0.1–0.9) was diluted to OD600 of 0.03, and evenly plated onto MHIIA plates. Concentrated samples from the actinomycetes cultures (3 μl) were spotted directly onto the bacterial lawn, and anti-Staphylococcus activity was evaluated based on the presence of a zone of inhibition.
Initial isolation of anti-Borreliella compound
S. hygroscopicus was inoculated in a 250 mL flask containing 40 mL MR5 medium with 0.1% vitamin supplement, and incubated for 7 days at 28°C with shaking (200 rpm). A culture sample (1 mL) was separated into fractions using semi-preparative high-performance liquid chromatography (HPLC) with C18 reverse-phase column (Ultra C18 5 μm Column 250 × 10 mm, Restek) and eluted at a flow rate of 5 ml/min. The HPLC apparatus included a Shimadzu HPLC system equipped with an SPD-M20A diode array detector (SHIMADZU Co. Ltd., Japan). The solvent and conditions used were 0 to 5 min of 5% acetonitrile (ACN) that contained 0.1% formic acid (FA), 5 to 30 min of a linear gradient of 5 to 25% ACN that contained 0.1% FA, 30 to 31 min of a linear gradient of 25 to 100% ACN that contained 0.1% FA, and 31 to 40 min of 100% ACN. Culture sample was fractionated every 1 min, and 40 fractions were generated. Each fraction was subjected to bioassay against B. burgdorferi, and activity was observed in samples from retention time 20–21 min.
Scale up production of hygromycin A
S. hygroscopicus was inoculated in a 2 L Erlenmeyer flask containing 1 L MR5 medium with 0.1% vitamin supplement and incubated at 28°C with shaking (200 rpm). After 10–14 days of cultivation, the culture was centrifuged, and the cell pellet was removed. The supernatant was treated with XAD16N resin (20–60 mesh, Sigma-Aldrich) to bind the active compound, and incubated overnight with agitation. After discarding supernatant, the active fraction containing hygromycin A was eluted from XAD16N resin with 1L 100% methanol. The methanol extract was dried using a rotary evaporator, and the sample was dissolved in MilliQ water. The sample was then subjected to preparative HPLC with a C18 reverse-phase column [Luna® 5 μm C18(2) 100 Å, LC Column 250 × 21.2 mm, Phenomenex] and eluted at a flow rate of 10 ml/min. The solvent and conditions used were 0 to 5 min of 7% ACN that contained 0.1% FA and 5 to 43 min of a linear gradient of 7 to 15.5% ACN that contained 0.1% FA. Active compound was eluted as single peak at retention time 40 min (HPLC peak at 215nm with purity of approximately 90%).
MS and structure elucidation
The HRMS and molecular formula of the active compound were determined by LC-MS/MS analysis on an LTQ Orbitrap XL™ Hybrid Ion Trap-Orbitrap Mass Spectrometer (Thermo Scientific) in positive ion mode coupled with an UltiMate 3000 RSLCnano System chromatography (Dionex). The HPLC purified active fraction was prepared in a concentration of 100 μg/mL in water with 0.1 % (v/v) formic acid. The solution was separated at 0.2 μL/min on a capillary column (150 mm by ID 75 μm) packed with C18 2.5μm resin (XSelect® CSH C18) under water:acetonitrile containing 0.1% (v/v) formic acid starting with 98:2 for 5 minutes followed by 50:50 for 20 minutes. The ion peak in positive mode m/z 512.1763 for C23H30NO12 [M+H]+ (calculated 512.1768 for C23H30NO12) was determined by full-scan high resolution electrospray ionization mass spectroscopy analysis, revealing C23H29NO12 as the molecular formula (Fig. S1). The compound of molecular mass of 511.17 Da determined by mass spectrometry was a match to hygromycin A in commercially available databases (AntiBase, Wiley). For further structural elucidation, 1H, 13C, and various two-dimensional NMR techniques, including 1H-1H correlation spectroscopy (COSY), heteronuclear multiple bond correlation (HMBC), and heteronuclear single quantum correlation (HSQC) were recorded on a 500 MHz on a Varian Inova spectrometer. For comparison with the previously reported hygromycin A data, 1H and 13C NMR spectra were recorded in CD3OD on 400 MHz Varian Mercury spectrometer (Fig. S2). Chemical shifts were in accordance with those previously reported(Lo et al., 2012; Yoshida et al., 1986). 1H NMR (500 MHz, DMSO-d6) δ 8.79 (br s, 1H), 7.25 (d, J = 9.1 Hz, 1H), 7.15 (d, J = 8.5, 1H), 7.14 (s, 1H), 6.88 (d, J = 2.1 Hz, 1H), 6.85 (dd, J = 8.5 and 2.1 Hz, 1H), 5.86 (m, 1H), 5.65 (d, J = 4.2 Hz, 1H), 5.36 (br s, 1H), 5.13 (s, 1H), 5.11 (d, J = 4.3 Hz, 1H), 5.06 (d, J = 5.8 Hz, 1H), 5.02 (d, J = 5.7 Hz, 1H), 4.69 (s, 1H), 4.31 (ddd, J = 9.0, 6.0, and 3.0 Hz, 1H), 4.22–4.18 (m, 2H), 4.09 (dd, J = 6.4 and 3.9 Hz, 1H), 4.04−3.98 (m, 3H), 3.70 (m, 1H), 3.57 (dd, J = 7.1 and 3.8 Hz, 1H), 2.03 (d, J = 1.3 Hz, 3H), 2.02 (s, 3H). (400 MHz, CD3OD) δ 7.22 (s, 1H), 7.18 (d, J = 8.4 Hz, 1H), 6.89 (s, 1H), 6.84 (d, J = 8.4 Hz, 1H), 5.58 (d, J = 4.1 Hz, 1H), 5.19 (s, 1H), 4.75 (s, 1H), 4.46 (dd, J = 6.1 and 2.4 Hz, 1H), 4.31 (t, J = 6.6 Hz, 1H), 4.24 (d, J = 6.2 Hz, 1H), 4.15 (m, 4H), 3.93 (t, J = 6.8 Hz, 1H), 3.76 (br s, 1H), 2.08 (s, 3H), 2.07 (s, 3H).
13C NMR (125 MHz, DMSO-d6) δ 207.6, 169, 146.5, 144, 132.1, 131.2, 130.7, 121, 116.6, 115.7, 101.3, 94.1, 87.2, 77.1, 77, 76.5, 76, 69.9, 69.4, 69.4, 49.2, 25.9, 14.5. (100 MHz, CD3OD) δ 210.1, 172.7, 148.4, 146.1, 135.1, 133.0, 132.4, 122.7, 118.5, 117.9, 103.8, 96.3, 88.6, 78.7, 78.3, 78.3, 77.7, 72.7, 71.7, 71.4, 50.4, 26.3, 14.7.
Measurement of protein synthesis inhibition
Exponential cultures of the GFP-inducible B. burgdorferi strain pCRW53 (Whetstine et al., 2009) were treated with antibiotics at 2x MIC overnight (amoxicillin 0.12ug/ml, spectinomycin 4ug/ml, hygromycin A 0.5ug/ml). Cultures were then induced with 5μg/ml anhydrotetracycline overnight. No-drug cultures and uninduced cultures were used as controls. Cells were washed and analyzed using a BD FACSAria II flow cytometer with a 70-μm nozzle. B. burgdorferi cells were gated by size using forward scatter (FSC) and side scatter (SSC). GFP fluorescence (FITC-A) was acquired for 100,000 cells.
Measurement of Hygromcin A affinity to ribosome
The ermBL template for toeprinting was generated by PCR. The resulting templates contained T7 promoter, ribosome binding site, the coding sequence and the binding site of the toeprinting primer. The ermBL ORF was generated by crossover 4-primer PCR using the primers ermB3-F (TAATACGACTCACTATAGGGCTTAAGTATAAGGAGGAAAAAATATGT TGGTATTCCAAATGCGTAATGTAGATAAAACATCTAC), ermB3-R (GGTTATAATGAATTTTGCTTATTAACGATAGAATTCTATCACTTATTTCAAAATAGTAGATGT TTTATCTACATTACG), T7 TAATACGACTCACTATAGGG and NV1 GGTTATAATGAATTTTGCTTATTAAC. The tnaC ORF was generated by PCR amplifying the tnaC gene from the pGF2500 plasmid (Martinez et al., 2014) using the primers T7-tnaC2 (TAATACGACTCACTATAGGGAGTTTTATAAGGAGGAAAACATATGAA TATCTTACATATATGTG) and R-44 (AGCGGATAACAATTTCACACAGGA). The toe-printing analysis of drug-dependent ribosome stalling was carried out as described (Orelle et al., 2013) numbering check. Briefly, the DNA templates (0.1 pmol) were transcribed and translated in a total volume of 5 μL of PURExpress (New England Biolabs, cat # E6800) reactions containing 10 pmol of E. coli ribosomes. Samples were incubated for 30 min at 37 °C, followed by addition of the [32 P]-labelled NV1 toe-printing primer (Vazquez-Laslop et al., 2008). The primer was extended by reverse transcriptase for 10 min and the reaction products were analyzed in sequencing gels. Gels were exposed overnight to the phosphorimager screens and scanned on Typhoon phosphorimager (GE).
The HygA-free ribosomes that escaped translation arrest at the start codon would be arrested at the downstream ‘trap’ codon because the presence of the Thr-tRNA synthetase inhibitor, borrelidin, depletes the system from Thr-tRNA.
(B, C) Toeprinting analysis of ribosome stalling during translation of the (B) ermBL gene or (C) tnaC gene in the absence (lane 1) or the presence of decreasing concentrations of HygA. HygA was present in the reactions at the following concentrations: lane 2: 50 μM, lane 3: 5.5 μM, lane 4: 1.9 μM, lane 6: 0.6 μM. Blue arrowheads mark the toeprint bands representing ribosomes stalled at the start codon. In both templates, the ribosomes could be trapped when Thr codon enters the A site (orange arrowhead) due to the presence of 50 μM of borrelidin in all the samples.
Isolation of hygromycin A resistant mutants
Plating was used to obtain CFU counts. BSK 1.5x medium was prepared as by Samuels (Samuels, 1995). B. burgdorferi B31 cells (6× 107) were plated onto semi-solid BSKII medium containing 2x, 4x, and 8x MIC hygromycin A. After a 4-week incubation, 12 colonies were isolated from the 2x MIC plate. The colonies were picked and used to inoculate liquid BSKII medium without hygromycin A to eliminate the effect of transient resistance. This strain was once again challenged with Hygromycin A as described above. After this passage strains were regrown in liquid BSKII without hygromycin A, and the MIC of hygromycin A was checked.
Whole Genome Sequencing
Genomic DNA from KLEx1 and KLEx2 was extracted using the QIAgen DNeasy Blood and Tissue Kit per the manufacturer’s instructions. DNA samples were sent to Omega Bioservices for library prep and sequencing using the Illumina Mi-Seq platform. We used Geneious to map parental Borreliella strains to reference genomes from the National Center for Biotechnology Information, and then to map resistant mutants to parental strains. Using Geneious, we also mapped polymorphisms between resistant mutants and parental strains.
RNAseq
The (resistant at 16x MIC HygA, MIC = 0.25ug/ml, 16x MIC= 4ug/ml) was grown to late exponential cultures in 500ml BSK-II, in the presence or absence of 4μg/ml hygromycin A. Treated and untreated cultures grew comparably and reached same cell density (as confirmed by dark-field microscopy). Cells were washed 4x in cold PBS, treated with RNA protect (QIAGEN) according to manufacturer’s instructions, and resuspended in TRIzol (Ambion). RNA was isolated B. burgdorferi hygromyin A resistant mutant KLEx2 using the RNA isolation kit from QIAGEN following the manufactures protocol. Genewiz (South Plainfileld, NJ) carried out further steps including quality control, DNAse treatment, rRNA depletion, RNA fragmentation, library preparation, and Illumina HiSeq2×150bp sequencing. Data analysis was performed by Genewiz by trimming and mapping reads to assess differential gene expression.
Construction of B. burgdorferi overexpressing bmpD
bmpD was amplified from B. burgdorferi B31 genome with primers NdeI_bmpD (gcgCATATGTTAAAAAAAGTTTATTATTTTTTAATTTTTTTATTTATTGTTGC) and bmpD_XhoI (ggcCTCGAGTTAATTTTCCATTTGCAAAACAAAGTTATCATAAGATACCTTGTC), introducing NdeI and XhoI restriction sites to the 5’ and 3’ end. The B. burgdorferi shuttle vector pJSB275 (Blevins et al., 2007; Groshong et al., 2012) – containing a codon-optimized lacI repressor transcribed from the PflaB promoter and an IPTG-inducible T5 promoter derived from pQE30 – was modified to introduce NdeI and XhoI restriction sites, yielding pJSB275m. The digested PCR fragment was then ligated into pJSB275m backbone and transformed into E. coli DH5α. E. coli transformants were selected with 50 μg/ml spectinomycin. The resulting shuttle vector pJSB275m_bmpD was verified by sequencing with primers pJSB275_up2 (ACCCGGAATAAGCAGTCAAG), pJSB275_down (GCTGCCTTACAAGCCTCTAC) and bmpD-internal_F (CAGGGCTTTCTGGTATAGGG). 30μg of ethanol precipitated plasmid pJSB275m_bmpD was transformed into electrocompetent B. burgdorferi cells as previously described (Hyde et al., 2011). Transformants were selected with kanamycin (100 μg/ml) and streptomycin (50 μg/ml) by semisolid plating. To quantify growth, plasmid pJSB275m_bmpD was transformed into a B. burgdorferi strain constitutively expressing GFP; Bb1286 (Caimano et al., 2015). Bb1286 was generated from B. burgdorferi strain B31 5A4 NP1 (bbe02 disrupted with PflgB-KanR (Kawabata et al., 2004) and carries a PflaB_gfp PflgB-aacC1 cassette inserted into the endogenous cp26 plasmid. Bb1286 transformants carrying the bmpD overexpression plasmid were confirmed by PCR and sequencing using primers pJSB275_up2, pJSB275_down and bmpD-internal_F. Plasmid content of transformants was verified by multiplex PCR as previously described (Bunikis et al., 2011). BmpD overexpression was confirmed by RT-PCR as previously described, using primers bmpD_RT_F (GGATACTTTGCGTCGAAGGC) and bmpD_RT_R (TGCATACTTAGCACCAGCTTCA) and normalizing to the levels of recA (Bockenstedt et al., 2006).
Construction of E. coli overexpressing bmpD
bmpD was amplified from B. burgdorferi B31 genome with primers SalI_bmpD (gatcGTCGACAAGCAAGGAGGATATTTTTATGTTAAAAAAAG) and bmpD_SbfI (gagaCCTGCAGGTTAATTTTCCATTTGCAAAACAAAGTTATCATAAGATACC), introducing SalI and SbfI restriction sites to the 5’ and 3’ end. The digested PCR fragment was then ligated into pBAD30 (Guzman et al., 1995) backbone and transformed into E. coli DH5α. E. coli transformants were selected with 50 μg/ml ampicillin. Transformants carrying pBAD30_bmpD were verified by sequencing with primers pBAD-F (ATGCCATAGCATTTTTATCC) and pBAD-R (AGTTTATGGCGGGCGTCCTG). Plasmid pBAD30_bmpD was then transformed into E. coli ΔTolC (Baba et al., 2006) by heat shock. Transformants were selected with kanamycin (50 μg/ml) and ampicillin (50 μg/ml) and verified by sequencing with primers mentioned above.
MIC fold-change and adenosine addition experiment
To investigate the effect of bmpD overexpression in B. burgdorferi on the efficacy of hygromycin A, Bb1286 + pJSB275m_bmpD was grown to early stationary phase in BSK-II medium containing kanamycin (100μg/ml) and streptomycin (50μg/ml). Cultures were diluted 1:10 in fresh BSK-II and divided into two; 1mM IPTG was added to one culture, while the other culture remained untreated. Hygromycin A MIC was determined by standard microbroth dilution method, as described previously. Inhibition of growth was measured via GFP fluorescence by a microplate reader (emission 528nm and excitation 485nm). Ceftriaxone was used as a control antibiotic. The MIC of the bmpD-overexpressing B. burgdorferi strain Bb1286 + pJSB275m_bmpD was normalized to the MIC of the wild-type B. burgdorferi strain (Bb1286) for each antibiotic. No growth difference was detected between B1286 + pJSB275m_bmpD with or without 1mM IPTG or as compared to B1286 wild-type, as assessed by daily GFP reads over the course of eight days.
To investigate the effect of bmpD overexpression in E. coli on the efficacy of hygromycin A, E. coli ΔTolC + pBAD30_bmpD was grown to mid exponential phase in M9 minimal medium with 0.2% glycerol as the sole carbon source and then shifted to either 0.2% L-arabinose containing medium for inducing conditions or to 0.2% D-Glucose containing medium for repressing conditions. E. coli ΔTolC carrying the empty plasmid pBAD30 was used as a control. Hygromycin A MIC was determined by standard microbroth dilution method, as described previously. Hygromycin A MIC was also assessed in rich LB medium and in the presence of different concentrations of adenosine (2.5–2500 μM) added to minimal M9 medium with inducing arabinose or repressing glucose conditions. Inhibition of growth was measured via optical density at 600bm by a microplate reader. The hygromycin A MIC of the bmpD overexpressing E. coli strain ΔTolC + pBAD30_bmpD and of the empty plasmid carrying E. coli strain ΔTolC + pBAD30 was normalized to the hygromycin A MIC of wild-type E. coli ΔTolC for each medium. No growth difference was detected between the strains in minimal medium under repressing or inducing conditions, as compared to the wild-type strain, or by addition of indicated concentrations of adenosine, as assessed by measuring optical density at 600nm.
Intracellular accumulation of hygromycin A
B. burgdorferi and the antibiotic hypersusceptible E. coli strain ΔTolC-Pore (Zgurskaya and Nikaido, 2000) were grown in BSKII and MOPS-M9 (pH 7.2) media, respectively. At OD600= 0.1–0.2 for E. coli and =0.01–0.02 for B. burgdorferi, 50 mL of cell cultures were pelleted by centrifugation at room temperature (RT). Cells were washed twice in MOPS-M9 and concentrated 20-fold. Cells were incubated with 16, 32, 64 and 128 μg/mL of antibiotics and after 1 and 40 min incubation at room temperature, 100 μL cell aliquots were collected by vacuum filtration onto 1.0 μm Glass Fiber Type B filters. Filters were washed twice with 10 mM Tris-HCl (pH 8.0), dried and placed into 100% methanol at −80°C for at least 10 min. Intracellular material was extracted by water bath sonication for 1 min. Cell and filter debris were separated by ultracentrifugation at 100,000×g for 8 min at 16°C and the pellet re-extracted with 80% methanol in water by sonication for 15 min followed by ultracentrifugation at the same conditions. Supernatants from two extractions were combined, evaporated to dryness under a vacuum and resuspended in 60 μL of 100% methanol. For compound quantification, 5 μL of solution was analyzed in triplicates. The calibration curve for hygromycin was generated in the same experiment by mixing antibiotic at increasing concentrations with the sonicated E. coli cell extracts (Supplemental Fig 3.).
An Agilent 1290 Infinity II ultrahigh-pressure liquid chromatography (UHPLC) system and 6545 quadrupole/time-of-flight (Q/TOF) system (Agilent Technologies) were used to quantify hygromycin. A Zorbax Rapid Resolution High Definition column (RRHD, 2.1×50mm, 1.8 μm) was used for the separation with a flow rate of 0.65 mL/min. The initial concentration of 5% MS grade Acetonitrile was maintained for 1 min, followed by a linear gradient to 80% over 3 min, and then by 100% over 1.1 min which was maintained for an additional 1.2 min. HPLC solvent mixtures contained 0.1% HPLC grade formic acid (SigmaAldrich) to improve ionization efficiency. MS parameters were as follows: gas temperature, 325°C; capillary voltage, 4000V; fragmentor voltage, 175V; m/z range, 50–1100; detector signal acquisition rate, 4GHz; and spectrum storage rate, 2s−1. MassHunter qualitative and quantitative analysis B8.0 was used to quantify the hygromycin A concentration using the calibration curve.
Hygromycin A was mixed with clarified E. coli cell lysates and analyzed by UHPLC/MS. At each concentration, the compound was injected in triplicates. The data were fit into a linear dependence and were used to calculate intracellular levels of antibiotic.
For hygromycin A accumulation in E. coli ΔtolC(pBAD30) and E. coli ΔtolC(pBAD30bmpD) cells, Rapidfire RF365 (Agilent technologies) coupled to a quadrupole time of flight (Q/TOF) 6545 mass spectrometer (Agilent technologies) was used. E. coli cells were grown in MOPS-M9 medium supplemented with 0.2% glycerol overnight, induced with 0.2% arabinose for 1 hour and concentrated 10-fold. The uptake was measured at increasing external hygromycin A concentrations (16–128 μg/ml). Intracellular material was extracted in two steps, (i) water and (ii) 50% methanol, by 96-well sonication for 2 min twice. Samples were aspirated during 500 ms into a 20 uL loop, then absorbed to a solid phase extraction C18 cartridge. Samples were washed for 3 seconds with 5 mM ammonium formate with a flow rate of 1.5 mL/min, then eluted from the cartridge to the mass spectrometer with 100% methanol containing 0.1% formic acid at a flow rate of 1.00 mL/min for 6 seconds. The system is then re-equilibrated with 5 mM of ammonium formate for 1 second. MSMS transition of 511.17 to 177.09 was used to quantify hygromycin A. MS parameters were as follows: gas temperature, 325°C; capillary voltage, 4000V; fragmentor voltage, 175V; m/z range, 50–1100; detector signal acquisition rate, 4GHz; collision energy 22 V and spectrum storage rate, 6.s−1. MassHunter quantitative analysis B8.0 was used to quantify hygromycin A concentration using the calibration curve.
Mouse infection model.
All animal experiments were conducted according to protocols that were approved by the Institute of Animal Care and Usage Committee (IACUC) at Northeastern University (Approval #16 -0619). Wild type female C3H mice (Charles River Laboratories) were infected with 105 B. burgdorferi N40 cells by subcutaneous injection, with no fewer than 3 animals per treatment group. Ear punches were collected from each animal after 2 weeks of infection and cultured into BSK-II media to confirm infection. The infection was established for 3 weeks, and then animals were dosed twice a day for 5 days with saline or ceftriaxone (156 mg/kg) by intraperitoneal injection, Hygromycin A (25, 50, 70 and 250 mg/kg) by oral gavage and 100 mg/kg by intraperitoneal injection.
qRT-PCR for in vivo B. burgdorferi quantification
The mouse and B. burgdorferi RNA from infected tissue was extracted using RNAeasy Mini Kit (Qiagen). cDNA was synthesized using High-Capacity cDNA Reverse Transcription Kits (Applied Biosysterms). Real -time PCR was performed with primers targeting the 16S rDNA gene (F: 5’- GGT CAA GAC TGA CGC TGA GTC and R: 5`GGC TTA GAA CTA ACG CTG) using SYBR green Supermix (Bio -RAD). B. burgdorferi N40 copy number was normalized to cDNA standard curve synthesized from a known copy number of B. burgdorferi cells.
Bait experiment
To prepare the mouse bait, 2 g of Bait formulation 3 (Food source), 0.5 g freshly ground peanut butter (Whole foods), 35 μl of green food coloring mix (Whole foods) were added into a 60 mm Petri dish. Boiling water (4 mL) was added and mixed with a spatula. The mixture was then dispensed into wells of a 96 well plate with a spatula such that about 2/3 of the well was full. Antibiotics or vehicle were added at appropriate volume and mixed with a pipette tip. Added volume was kept below 60 μL per well. The bait was allowed to air dry/solidify for a few hours and the prepared baits were stored at 4°C until use.
After one day of fasting, the animals were given bait once a day for 5 days containing Hygromycin A (200 mg/kg) or Doxycycline (100 mg/kg). Control groups included uninfected and infected animals, both given untreated bait. To ensure that the bait would be consumed, mice were housed individually with their mouse chow withheld during treatment. The doses used in mice were matched to the pharmacokinetic profile of humans given 1 g ceftriaxone every 12 hours (Sai Life Sciences Ltd, India) and 100 mg doxycycline every 12 hours (Bockenstedt et al., 2002; Crandon et al., 2010).
Animals were sacrificed 3 days after completion of antibiotic treatment and skin (whole ear), heart, and quadricep muscle were collected. Skin was used for liquid culture, and RNA extraction. All other tissues were used for RNA extraction. The growth of B. burgdorferi in 10 day liquid cultures was detected by dark field microscopy with a 100x objective.
Efficacy of Hygromycin A in Peromyscus mice
Efficacy studies of hygromycin A in eradicating B. burgdorferi from infected mice were performed by infecting Peromyscus leucopus mice by subcutaneous inoculation with 106 B. burgdorferi B31. After 3 weeks, mice were administered hygromycin A by oral gavage at either 50 mg/kg or 100 mg/kg twice daily for 5 days. Ear punches were taken each day for culture in BSKII media containing rifampicin, phosphomycin and amphotericin B while the mice were treated with hygromycin A. An additional ear punch culture was performed on Day 7. Mice were sacrificed on Day 11 and the heart, ankle, ear and inoculation skin site were taken for culture. Cultures were monitored by darkfield microscopy for 4 weeks or until positive.
Impact of Hygromycin A on the murine fecal microbiome
Wild type female C3H mice (Charles River Laboratories) were infected with 105 B. burgdorferi N40 cells by subcutaneous injection. The infection was established for 3 weeks, and then animals were dosed twice a day for 5 days with ceftriaxone (156 mg/kg) (Sigma) by subcutaneous injection, Amoxicillin (100 mg/kg) (Goldbio) by oral gavage, or Hygromycin A (50 mg/kg) by oral gavage, or were left untreated (n=4–5 per group per experiment, 3 experiments). Fecal pellets were collected 3 days before and 3 days after antibiotic treatment and were stored at −80°C in PBS. Sequencing of a stool pellet from each mouse before and after treatment was performed by MR DNA (www.mrdnalab.com, Shallowater, TX, USA) on an Ion Torrent PGM. Using PCR primers 515F (GTGCCAGCMGCCGCGGTAA) and 806R (GGACTACVSGGGTATCTAAT), the V4 variable region of the 16SrRNA gene was amplified in a single-step 30 cycle PCR with the HotStarTaq Plus Master Mix Kit (Qiagen, USA). The following conditions were used: 94°C for 3 minutes and 30 cycles of 94°C for 30 seconds, 53°C for 40 seconds and 72°C for 1 minute, then a final elongation step for 5 minutes at 72°C. These data were analyzed with a proprietary analysis pipeline (MR DNA, Shallowater, TX, USA). Barcodes and primers and then sequences <150 bp were removed. Further, sequences with homopolymer runs exceeding 6 bp and sequences with ambiguous calls were removed. The sequences were then denoised, OTUs were generated, and chimeras were removed; OTUs were clustered at 3% divergence. Taxonomic classification was performed using BLASTn against a database derived from RDPII (http://rdp.cme.msu.edu) and NCBI (www.ncbi.nlm.nih.gov). The change in relative abundance of the most abundant genera from before to after treatment of each individual mouse was reported. Further, raw sequences were processed in Qiita (Gonzalez et al., 2018) and were demultiplexed, trimmed to 100 bp, and closed reference OTU picking was performed against the Greengenes database (McDonald et al., 2012) at 97% similarity. The alpha diversity based on the Simpson index was calculated in Qiita. The change in the Simpson index was calculated for the fecal microbiome of each mouse from before to after treatment. Statistical significance was calculated using a one-way ANOVA with Tukey’s multiple comparisons test (p <0.05).
QUANTIFICATION AND STATISTICAL ANALYSIS
GraphPad Prism was used for statistical analysis. All of the statistical details for each experiment are described in the figure legends. A P-value ≤ 0.05 was considered statistically significant.
ADDITIONAL RESOURCES
The authors have not generated external sites linked to this study.
Supplementary Material
Fig. S1. High-resolution mass spectra (HRMS) showing a peak at m/z 512.1763 which corresponds to an [M + H]+ of formula C23H30NO12, consistent with the theoretical monoisotopic mass of hygromycin A 512.1768 [M + H]+, Δ0.97 ppm. Related to Figure 1.
Fig. S2. 1D and 2D NMR spectra of hygromycin A. (A) 13C (125 MHz) and 1H (500 MHz) NMR spectra in DMSO-d1, (B) 1H-1H COSY, 1H-13C HSQC, 1H-13C HMBC spectra (500 MHz) in DMSO-d6, and hygromycin A structure with key 2D NMR correlations, (C) 13C (100 MHz) and 1H (400 MHz) NMR spectra in CD3OD. Related to Figure 1.
Fig. S3. Calibration curve and the range of detection of hygromycin A. Hygromycin A was mixed with clarified E. coli cell lysates and analyzed by UHPLC/MS. At each concentration, the compound was injected in triplicates. The data were fit into a linear dependence and were used to calculate intracellular levels of antibiotic. Related to Figure 3.
Fig S4. A. Intracellular accumulation of hygromycin A in E. coli ΔtolC. Accumulation of hygromycin A was quantified in cells with the intact outer membrane and in hyper-porinated cells expressing FhuA siderophore receptor that forms a large pore in the outer membrane. Cells were incubated at indicated concentrations of hygromycin A for 40 min and intracellular accumulation was quantified by UHPLC/MS. Mean ± SD, N = 2. B. Intracellular accumulation of hygromycin A in B. turcica and the B. turcica hygromycin A resistant mutant, KLEx1. Cells were incubated at indicated concentrations of hygromycin A for 1 min and intracellular accumulation was quantified by UHPLC/MS. Mean ± SD, N = 6. Note: For 16ug/ml Hygromycin A addition, hygromycin A accumulation was below the limit of quantification. Related to Figure 3.
Fig S5. The fecal microbiome composition from before to after treatment in mice infected with B. burgdorferi N40 and treated twice daily for five days with hygromycin A, amoxicillin or ceftriaxone. The fecal microbiome was sequenced for the 16S rRNA gene and the average relative abundance of each genus was calculated and averaged for each group (n=4–5). Three individual experiments are represented (A–C). Related to Figure 4.
Highlights section:
A selective screen against B. burgdorferi led to the rediscovery of hygromycin A.
The mechanism of selectivity is puzzling since hygromycin A targets the ribosome.
Hygromycin A is smuggled into spirochetes by the conserved transporter BmpDEFG.
Hygromycin A is efficacious in a mouse model without disturbing the microbiome.
Acknowledgments
The authors thank Dr. Alexander Mankin and Dr. Nora Vázquez-Laslop for helpful discussions. This work was supported by The Steven and Alexandra Cohen Foundation, The Global Lyme Alliance, by NIH grant R01AI122286 to KL and LH, and NIH grant RO1AI132836 to HIZ.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interests
K.L. serves on the scientific advisory board of Flightpath.
References
- Adrion ER, Aucott J, Lemke KW, and Weiner JP (2015). Health care costs, utilization and patterns of care following Lyme disease. PLoS One 10, e0116767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Åstrand M, Cuellar J, Hytönen J, and Salminen TA (2019). Predicting the ligand-binding properties of Borrelia burgdorferi s.s. Bmp proteins in light of the conserved features of related Borrelia proteins. J Theor Biol 462, 97–108. [DOI] [PubMed] [Google Scholar]
- Aucott JN (2015). Posttreatment Lyme disease syndrome. Infect Dis Clin North Am 29, 309–323. [DOI] [PubMed] [Google Scholar]
- Aucott JN, Crowder LA, and Kortte KB (2013). Development of a foundation for a case definition of post-treatment Lyme disease syndrome. International journal of infectious diseases : IJID : official publication of the International Society for Infectious Diseases 17, e443–449. [DOI] [PubMed] [Google Scholar]
- Aucott JN, and Rebman AW (2021). Long-haul COVID: heed the lessons from other infection-triggered illnesses. Lancet 397, 967–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aucott JN, Soloski MJ, Rebman AW, Crowder LA, Lahey LJ, Wagner CA, Robinson WH, and Bechtold KT (2016). CCL19 as a chemokine risk factor for posttreatment Lyme disease syndrome: a arospective clinical cohort study. Clin Vaccine Immunol 23, 757–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, and Mori H (2006). Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Molecular systems biology 2, 2006.0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belkaid Y, and Hand TW (2014). Role of the microbiota in immunity and inflammation. Cell 157, 121–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blevins JS, Revel AT, Smith AH, Bachlani GN, and Norgard MV (2007). Adaptation of a luciferase gene reporter and lac expression system to Borrelia burgdorferi. Appl Environ Microbiol 73, 1501–1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bockenstedt LK, Liu N, Schwartz I, and Fish D (2006). MyD88 deficiency enhances acquisition and transmission of Borrelia burgdorferi by Ixodes scapularis ticks. Infect Immun 74, 2154–2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bockenstedt LK, Mao J, Hodzic E, Barthold SW, and Fish D (2002). Detection of attenuated, noninfectious spirochetes in Borrelia burgdorferi-infected mice after antibiotic treatment. J Infect Dis 186, 1430–1437. [DOI] [PubMed] [Google Scholar]
- Breuner NE, Hojgaard A, Replogle AJ, Boegler KA, and Eisen L (2018). Transmission of the relapsing fever spirochete, Borrelia miyamotoi, by single transovarially-infected larval Ixodes scapularis ticks. Ticks Tick Borne Dis 9, 1464–1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunikis I, Kutschan-Bunikis S, Bonde M, and Bergström S (2011). Multiplex PCR as a tool for validating plasmid content of Borrelia burgdorferi. J Microbiol Methods 86, 243–247. [DOI] [PubMed] [Google Scholar]
- Caimano MJ, Dunham-Ems S, Allard AM, Cassera MB, Kenedy M, and Radolf JD (2015). Cyclic di-GMP modulates gene expression in Lyme disease spirochetes at the tick-mammal interface to promote spirochete survival during the blood meal and tick-to-mammal transmission. Infect Immun 83, 3043–3060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coughlin JM, Yang T, Rebman AW, Bechtold KT, Du Y, Mathews WB, Lesniak WG, Mihm EA, Frey SM, Marshall ES, et al. (2018). Imaging glial activation in patients with post-treatment Lyme disease symptoms: a pilot study using [11C]DPA-713 PET. J Neuroinflammation 15, 346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crandon JL, Kuti JL, and Nicolau DP (2010). Comparative efficacies of human simulated exposures of telavancin and vancomycin against methicillin-resistant Staphylococcus aureus with a range of vancomycin MICs in a murine pneumonia model. Antimicrob Agents Chemother 54, 5115–5119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuellar J, Åstrand M, Elovaara H, Pietikäinen A, Sirén S, Liljeblad A, Guédez G, Salminen TA, and Hytönen J (2020). Structural and biomolecular analyses of Borrelia burgdorferi BmpD reveal a substrate-binding protein of an ABC-type nucleoside transporter family. Infect Immun 88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolan MC, Schulze TL, Jordan RA, Dietrich G, Schulze CJ, Hojgaard A, Ullmann AJ, Sackal C, Zeidner NS, and Piesman J (2011). Elimination of Borrelia burgdorferi and Anaplasma phagocytophilum in rodent reservoirs and Ixodes scapularis ticks using a doxycycline hyclate-laden bait. Am J Trop Med Hyg 85, 1114–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edmondson DG, Hu B, and Norris SJ (2018). Long-term in vitro culture of the syphilis spirochete Treponema pallidum subsp. pallidum. MBio 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edmondson DG, Wormser GP, and Norris SJ (2020). In vitro susceptibility of Treponema pallidum subsp. pallidum to doxycycline. Antimicrob Agents Chemother 64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrer M, Méndez-García C, Rojo D, Barbas C, and Moya A (2017). Antibiotic use and microbiome function. Biochem Pharmacol 134, 114–126. [DOI] [PubMed] [Google Scholar]
- Gavrish E, Sit CS, Cao S, Kandror O, Spoering A, Peoples A, Ling L, Fetterman A, Hughes D, Bissell A, et al. (2014). Lassomycin, a ribosomally synthesized cyclic peptide, kills Mycobacterium tuberculosis by targeting the ATP-dependent protease ClpC1P1P2. Chem Biol 21, 509–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez A, Navas-Molina JA, Kosciolek T, McDonald D, Vazquez-Baeza Y, Ackermann G, DeReus J, Janssen S, Swafford AD, Orchanian SB, et al. (2018). Qiita: rapid, web-enabled microbiome meta-analysis. Nat Methods 15, 796–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grenham S, Clarke G, Cryan JF, and Dinan TG (2011). Brain-gut-microbe communication in health and disease. Front Physiol 2, 94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm D, Elias AF, Tilly K, and Rosa PA (2003). Plasmid stability during in vitro propagation of Borrelia burgdorferi assessed at a clonal level. Infect Immun 71, 3138–3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groshong AM, Gibbons NE, Yang XF, and Blevins JS (2012). Rrp2, a prokaryotic enhancer-like binding protein, is essential for viability of Borrelia burgdorferi. J Bacteriol 194, 3336–3342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrero MD, and Modolell J (1980). Hygromycin A, a novel inhibitor of ribosomal peptidyltransferase. Eur J Biochem 107, 409–414. [DOI] [PubMed] [Google Scholar]
- Guzman LM, Belin D, Carson MJ, and Beckwith J (1995). Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol 177, 4121–4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi SF, Norcia LJ, Seibel SB, and Silvia AM (1997). Structure-activity relationships of hygromycin A and its analogs: protein synthesis inhibition activity in a cell free system. J Antibiot (Tokyo) 50, 514–521. [DOI] [PubMed] [Google Scholar]
- Hodzic E, Feng S, Freet KJ, and Barthold SW (2003). Borrelia burgdorferi population dynamics and prototype gene expression during infection of immunocompetent and immunodeficient mice. Infect Immun 71, 5042–5055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoover RB, Pikuta EV, Bej AK, Marsic D, Whitman WB, Tang J, and Krader P (2003). Spirochaeta americana sp. nov., a new haloalkaliphilic, obligately anaerobic spirochaete isolated from soda Mono Lake in California. Int J Syst Evol Microbiol 53, 815–821. [DOI] [PubMed] [Google Scholar]
- Hyde JA, Weening EH, and Skare JT (2011). Genetic transformation of Borrelia burgdorferi. Curr Protoc Microbiol Chapter 12, Unit 12C.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai Y, Meyer KJ, Iinishi A, Favre-Godal Q, Green R, Manuse S, Caboni M, Mori M, Niles S, Ghiglieri M, et al. (2019). A new antibiotic selectively kills Gram-negative pathogens. Nature 576, 459–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer R, Caimano MJ, Luthra A, Axline D Jr., Corona A, Iacobas DA, Radolf JD, and Schwartz I (2015). Stage-specific global alterations in the transcriptomes of Lyme disease spirochetes during tick feeding and following mammalian host adaptation. Mol Microbiol 95, 509–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasselman LJ, Vernice NA, DeLeon J, and Reiss AB (2018). The gut microbiome and elevated cardiovascular risk in obesity and autoimmunity. Atherosclerosis 271, 203–213. [DOI] [PubMed] [Google Scholar]
- Kawabata H, Norris SJ, and Watanabe H (2004). BBE02 disruption mutants of Borrelia burgdorferi B31 have a highly transformable, infectious phenotype. Infect Immun 72, 7147–7154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kling A, Lukat P, Almeida DV, Bauer A, Fontaine E, Sordello S, Zaburannyi N, Herrmann J, Wenzel SC, König C, et al. (2015). Targeting DnaN for tuberculosis therapy using novel griselimycins. Science 348, 1106–1112. [DOI] [PubMed] [Google Scholar]
- Krishnamoorthy G, Wolloscheck D, Weeks JW, Croft C, Rybenkov VV, and Zgurskaya HI (2016). Breaking the permeability barrier of Escherichia coli by controlled hyperporination of the outer membrane. Antimicrob Agents Chemother 60, 7372–7381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis K (2020). The Science of Antibiotic Discovery. Cell 181, 29–45. [DOI] [PubMed] [Google Scholar]
- Lin YP, Benoit V, Yang X, Martinez-Herranz R, Pal U, and Leong JM (2014). Strain-specific variation of the decorin-binding adhesin DbpA influences the tissue tropism of the Lyme disease spirochete. PLoS pathogens 10, e1004238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling LL, Schneider T, Peoples AJ, Spoering AL, Engels I, Conlon BP, Mueller A, Schaberle TF, Hughes DE, Epstein S, et al. (2015). A new antibiotic kills pathogens without detectable resistance. Nature 517, 455–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo HJ, Chang YK, and Yan TH (2012). Chiral pool based efficient synthesis of the aminocyclitol core and furanoside of (−)-hygromycin A: formal total synthesis of (−)-hygromycin A. Org Lett 14, 5896–5899. [DOI] [PubMed] [Google Scholar]
- Lou Y, Wu J, and Wu X (2014). Impact of biodiversity and seasonality on Lyme-pathogen transmission. Theor Biol Med Model 11, 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marques A (2008). Chronic Lyme disease: a review. Infect Dis Clin North Am 22, 341–360, vii–viii. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez AK, Gordon E, Sengupta A, Shirole N, Klepacki D, Martinez-Garriga B, Brown LM, Benedik MJ, Yanofsky C, Mankin AS, et al. (2014). Interactions of the TnaC nascent peptide with rRNA in the exit tunnel enable the ribosome to respond to free tryptophan. Nucleic Acids Res 42, 1245–1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald D, Price MN, Goodrich J, Nawrocki EP, DeSantis TZ, Probst A, Andersen GL, Knight R, and Hugenholtz P (2012). An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J 6, 610–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mistry A, Warren MS, Cusick JK, Karkhoff-Schweizer RR, Lomovskaya O, and Schweizer HP (2013). High-level pacidamycin resistance in Pseudomonas aeruginosa is mediated by an Opp oligopeptide permease encoded by the opp-fabI operon. Antimicrob Agents Chemother 57, 5565–5571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modi SR, Collins JJ, and Relman DA (2014). Antibiotics and the gut microbiota. J Clin Invest 124, 4212–4218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrissette M, Pitt N, González A, Strandwitz P, Caboni M, Rebman AW, Knight R, D’Onofrio A, Aucott JN, Soloski MJ, et al. (2020). A distinct microbiome signature in posttreatment Lyme disease patients. mBio 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mount GA, Haile DG, and Daniels E (1997). Simulation of management strategies for the blacklegged tick (Acari: Ixodidae) and the Lyme disease spirochete, Borrelia burgdorferi. Journal of medical entomology 34, 672–683. [DOI] [PubMed] [Google Scholar]
- Norris SJ, Cox DL, and Weinstock GM (2001). Biology of Treponema pallidum: correlation of functional activities with genome sequence data. J Mol Microbiol Biotechnol 3, 37–62. [PubMed] [Google Scholar]
- Omura S, Nakagawa A, Fujimoto T, Saito K, Otoguro K, and Walsh JC (1987). Hygromycin A, an antitreponemal substance. I. Screening method and therapeutic effect for Treponema hyodysenteriae-caused infection in CF-1 mice. J Antibiot (Tokyo) 40, 1619–1626. [DOI] [PubMed] [Google Scholar]
- Orelle C, Szal T, Klepacki D, Shaw KJ, Vazquez-Laslop N, and Mankin AS (2013). Identifying the targets of aminoacyl-tRNA synthetase inhibitors by primer extension inhibition. Nucleic Acids Res 41, e144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ornstein K, and Barbour AG (2006). A reverse transcriptase-polymerase chain reaction assay of Borrelia burgdorferi 16S rRNA for highly sensitive quantification of pathogen load in a vector. Vector borne and zoonotic diseases 6, 103–112. [DOI] [PubMed] [Google Scholar]
- Pittenger RC, Wolfe RN, Hoehn MM, Marks PN, Daily WA, and Mc GJ (1953). Hygromycin. I. Preliminary studies on the production and biologic activity of a new antibiotic. Antibiot Chemother (Northfield) 3, 1268–1278. [PubMed] [Google Scholar]
- Polikanov YS, Starosta AL, Juette MF, Altman RB, Terry DS, Lu W, Burnett BJ, Dinos G, Reynolds KA, Blanchard SC, et al. (2015). Distinct tRNA accommodation intermediates observed on the ribosome with the antibiotics hygromycin A and A201A. Mol Cell 58, 832–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purser JE, and Norris SJ (2000). Correlation between plasmid content and infectivity in Borrelia burgdorferi. Proc Natl Acad Sci U S A 97, 13865–13870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quigley J, Peoples A, Sarybaeva A, Hughes D, Ghiglieri M, Achorn C, Desrosiers A, Felix C, Liang L, Malveira S, et al. (2020). Novel antimicrobials from uncultured bacteria acting against Mycobacterium tuberculosis. mBio 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebman AW, and Aucott JN (2020). Post-treatment Lyme disease as a model for persistent symptoms in Lyme disease. Front Med (Lausanne) 7, 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebman AW, Bechtold KT, Yang T, Mihm EA, Soloski MJ, Novak CB, and Aucott JN (2017). The clinical, symptom, and quality-of-life characterization of a well-defined group of patients with posttreatment Lyme disease syndrome. Front Med (Lausanne) 4, 224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romano KA, Vivas EI, Amador-Noguez D, and Rey FE (2015). Intestinal microbiota composition modulates choline bioavailability from diet and accumulation of the proatherogenic metabolite trimethylamine-N-oxide. mBio 6, e02481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuels DS (1995). Electrotransformation of the spirochete Borrelia burgdorferi. Methods Mol Biol 47, 253–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt EK, Riwanto M, Sambandamurthy V, Roggo S, Miault C, Zwingelstein C, Krastel P, Noble C, Beer D, Rao SP, et al. (2011). The natural product cyclomarin kills Mycobacterium tuberculosis by targeting the ClpC1 subunit of the caseinolytic protease. Angew Chem Int Ed Engl. [DOI] [PubMed] [Google Scholar]
- Schwartz AM, Kugeler KJ, Nelson CA, Marx GE, and Hinckley AF (2021). Use of commercial claims data for evaluating trends in Lyme disease diagnoses, United States, 2010–2018. Emerg Infect Dis 27, 499–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz JJ, Gazumyan A, and Schwartz I (1992). rRNA gene organization in the Lyme disease spirochete, Borrelia burgdorferi. Journal of bacteriology 174, 3757–3765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma B, Brown AV, Matluck NE, Hu LT, and Lewis K (2015). Borrelia burgdorferi, the causative agent of Lyme disease, forms drug-tolerant persister cells. Antimicrob Agents Chemother 59, 4616–4624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver LL (2007). Multi-targeting by monotherapeutic antibacterials. Nature Reviews Drug Discovery 6, 41–55. [DOI] [PubMed] [Google Scholar]
- Steere AC, Strle F, Wormser GP, Hu LT, Branda JA, Hovius JW, Li X, and Mead PS (2016). Lyme borreliosis. Nat Rev Dis Primers 2, 16090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stricker RB, Burrascano J, and Winger E (2002). Longterm decrease in the CD57 lymphocyte subset in a patient with chronic Lyme disease. Ann Agric Environ Med 9, 111–113. [PubMed] [Google Scholar]
- Stricker RB, and Winger EE (2001). Decreased CD57 lymphocyte subset in patients with chronic Lyme disease. Immunol Lett 76, 43–48. [DOI] [PubMed] [Google Scholar]
- Strle K, Stupica D, Drouin EE, Steere AC, and Strle F (2014). Elevated levels of IL-23 in a subset of patients with post-Lyme disease symptoms following erythema migrans. Clin Infect Dis 58, 372–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stupica D, Lusa L, Ruzić-Sabljić E, Cerar T, and Strle F (2012). Treatment of erythema migrans with doxycycline for 10 days versus 15 days. Clin Infect Dis 55, 343–350. [DOI] [PubMed] [Google Scholar]
- Tien V, Punjabi C, and Holubar MK (2020). Antimicrobial resistance in sexually transmitted infections. J Travel Med 27. [DOI] [PubMed] [Google Scholar]
- Valles-Colomer M, Falony G, Darzi Y, Tigchelaar EF, Wang J, Tito RY, Schiweck C, Kurilshikov A, Joossens M, Wijmenga C, et al. (2019). The neuroactive potential of the human gut microbiota in quality of life and depression. Nature microbiology 4, 623–632. [DOI] [PubMed] [Google Scholar]
- Vatanen T, Franzosa EA, Schwager R, Tripathi S, Arthur TD, Vehik K, Lernmark A, Hagopian WA, Rewers MJ, She JX, et al. (2018). The human gut microbiome in early-onset type 1 diabetes from the TEDDY study. Nature 562, 589–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez-Laslop N, Thum C, and Mankin AS (2008). Molecular mechanism of drug-dependent ribosome stalling. Mol Cell 30, 190–202. [DOI] [PubMed] [Google Scholar]
- Whetstine CR, Slusser JG, and Zückert WR (2009). Development of a single-plasmid-based regulatable gene expression system for Borrelia burgdorferi. Appl Environ Microbiol 75, 6553–6558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willing BP, Russell SL, and Finlay BB (2011). Shifting the balance: antibiotic effects on host-microbiota mutualism. Nat Rev Microbiol 9, 233–243. [DOI] [PubMed] [Google Scholar]
- Wu X, Sharma B, Niles S, O’Connor K, Schilling R, Matluck N, D’Onofrio A, Hu LT, and Lewis K (2018). Identifying vancomycin as an effective antibiotic for killing Borrelia burgdorferi. Antimicrob Agents Chemother 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie H, Patching SG, Gallagher MP, Litherland GJ, Brough AR, Venter H, Yao SY, Ng AM, Young JD, Herbert RB, et al. (2004). Purification and properties of the Escherichia coli nucleoside transporter NupG, a paradigm for a major facilitator transporter sub-family. Mol Membr Biol 21, 323–336. [DOI] [PubMed] [Google Scholar]
- Yoshida M, Takahashi E, Uozumi T, and Beppu T (1986). Hygromycin A and methoxyhygromycin, novel inhibitors of K88-antigen synthesis of enterotoxic Escherichia coli strain. Agr Biol Chem Tokyo 50, 143–149. [Google Scholar]
- Zgurskaya HI, and Nikaido H (2000). Multidrug resistance mechanisms: drug efflux across two membranes. Mol Microbiol 37, 219–225. [DOI] [PubMed] [Google Scholar]
- Zhang X, Borbet TC, Fallegger A, Wipperman MF, Blaser MJ, and Müller A (2021). An antibiotic-impacted microbiota compromises the development of colonic regulatory T cells and predisposes to dysregulated immune responses. mBio 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. High-resolution mass spectra (HRMS) showing a peak at m/z 512.1763 which corresponds to an [M + H]+ of formula C23H30NO12, consistent with the theoretical monoisotopic mass of hygromycin A 512.1768 [M + H]+, Δ0.97 ppm. Related to Figure 1.
Fig. S2. 1D and 2D NMR spectra of hygromycin A. (A) 13C (125 MHz) and 1H (500 MHz) NMR spectra in DMSO-d1, (B) 1H-1H COSY, 1H-13C HSQC, 1H-13C HMBC spectra (500 MHz) in DMSO-d6, and hygromycin A structure with key 2D NMR correlations, (C) 13C (100 MHz) and 1H (400 MHz) NMR spectra in CD3OD. Related to Figure 1.
Fig. S3. Calibration curve and the range of detection of hygromycin A. Hygromycin A was mixed with clarified E. coli cell lysates and analyzed by UHPLC/MS. At each concentration, the compound was injected in triplicates. The data were fit into a linear dependence and were used to calculate intracellular levels of antibiotic. Related to Figure 3.
Fig S4. A. Intracellular accumulation of hygromycin A in E. coli ΔtolC. Accumulation of hygromycin A was quantified in cells with the intact outer membrane and in hyper-porinated cells expressing FhuA siderophore receptor that forms a large pore in the outer membrane. Cells were incubated at indicated concentrations of hygromycin A for 40 min and intracellular accumulation was quantified by UHPLC/MS. Mean ± SD, N = 2. B. Intracellular accumulation of hygromycin A in B. turcica and the B. turcica hygromycin A resistant mutant, KLEx1. Cells were incubated at indicated concentrations of hygromycin A for 1 min and intracellular accumulation was quantified by UHPLC/MS. Mean ± SD, N = 6. Note: For 16ug/ml Hygromycin A addition, hygromycin A accumulation was below the limit of quantification. Related to Figure 3.
Fig S5. The fecal microbiome composition from before to after treatment in mice infected with B. burgdorferi N40 and treated twice daily for five days with hygromycin A, amoxicillin or ceftriaxone. The fecal microbiome was sequenced for the 16S rRNA gene and the average relative abundance of each genus was calculated and averaged for each group (n=4–5). Three individual experiments are represented (A–C). Related to Figure 4.
Data Availability Statement
Data have been deposited at https://data.mendeley.com/datasets/wxcfg58zg3/draft?a=94a40a19-499f-4d95-8e9f-8a5d890ffa35 and are publicly available as of the date of publication. DOIs are listed in the key resources table. This paper does not report original code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.


