Abstract
The newly identified p53 homolog p73 mimics the transcriptional function of p53. We have investigated the regulation of p73's transcriptional activity by p300/CREB binding protein (CBP). p73-p300 complexes were identified in HeLa cell extracts by cofractionation and coimmunoprecipitation assays. The p73-p300 interaction was confirmed in vitro by glutathione S-transferase–protein association assays and in vivo by coimmunoprecipitating the overexpressed p300 and p73 in human p53-free small-cell lung carcinoma H1299 or osteosarcoma Saos-2 cells. The N terminus but not the N-terminal truncation of p73 bound to the CH1 domain (amino acids [aa] 350 to 450) of p300/CBP. Accordingly, this p73 N-terminal deletion was unable to activate transcription or to induce apoptosis. Overexpression of either p300 or CBP stimulated transcription mediated by p73 but not its N-terminally deleted mutant in vivo. The N-terminal fragment from aa 19 to 597, but not the truncated fragment from aa 242 to 1700 of p300, reduced p73-mediated transcription markedly. p73-dependent transcription or apoptosis was partially impaired in either p300- or CBP-deficient human breast carcinoma MCF-7 or H1299 cells, suggesting that both coactivators mediate transcription by p73 in cells. These results demonstrate that the N terminus of p73 directly interacts with the N-terminal CH1 domain of p300/CBP to activate transcription.
Transcriptional activation of class II genes by RNA polymerase II (RNAPII) involves protein-protein interactions between transcriptional activators and their coactivators (50). One extensively studied group of coactivators is the p300/CREB binding protein (CBP) family (72). p300 and CBP are distinct proteins encoded by two different genes and were initially identified independently (13, 18, 81). However, these two proteins not only share significant homology in their functional domains but also mediate transcription by binding to similar sets of transcriptional activators (72). Both can be negatively regulated by the 12S form of adenovirus E1A (2, 4, 56). Thus, these proteins are often referred to as p300/CBP. There are a variety of p300/CBP-interacting transcriptional activators, including CREB (44), c-Jun (46), ATF-2 (39), nuclear receptors (11, 34, 85), MyoD (87), SREBP-2 (64), and YY1 (47), suggesting that p300/CBP may play an integrating role in different cellular events mediated by these activators, such as cell proliferation, differentiation, and signaling (70, 72, 86). Interestingly, p300/CBP also binds to the p53 transcription activator and stimulates p53-dependent transcriptional activity in vivo (3, 26, 51), indicating that this group of coactivators also participate in the p53 response pathway.
The transactivation activity of the p53 tumor suppressor protein is important for regulating cell growth and apoptosis in response to various cellular stress signals (23, 42, 48, 49). This activity is contributed by three main domains of this protein: the N-terminal transactivation domain (amino acids [aa] 1 to 45) (21, 67), the central sequence-specific DNA binding domain (aa 113 to 290) (6, 12, 66, 80), and the C-terminal regulatory region (30). The N terminus contacts several transcriptional regulators, including components of the RNAPII transcriptional machinery, such as TAFII31 and TAFII70 (54, 75), and the coactivators p300 and CBP (3, 26, 51). Phosphorylation of this domain at Ser15 regulates the activity and stability of p53 in response to DNA damage (71, 73). The central sequence-specific DNA binding domain recognizes and binds to a specific consensus sequence with two copies of the 10-mer 5′-RRRC(A/T)(T/A)GYYY-3′ (19), which has been identified in many p53 target genes (76). The products of these genes, including p21waf1 (17, 20), MDM2 (82), Gadd45 (36), BAX-1 (59), IGF-BP3 (10), and 14-3-3ς (29), act as downstream effectors of the p53-mediated growth arrest or apoptosis. The C terminus regulates p53 transcriptional activity (30). Posttranslational modification of this region, such as DNA damage-responsive phosphorylation (35, 55) or acetylation (25, 53, 68), leads to activation of p53's DNA binding and transcriptional activities. Because of its crucial importance in cell growth regulation, the transcriptional activity is conserved among p53 homologs.
Several p53-like proteins have been reported recently. One gene, designated the p73 gene, has been isolated and found to encode two spliced polypeptides, p73α and p73β. p73β (499 aa), possessing a unique pentamer at its extreme C terminus, is 137 aa shorter than p73α (636 aa) (33). The p73 protein resembles p53 in both sequence (approximately 60% identity with p53 in the central domain and 29% identity in the N terminus) and function (31, 33). Like p53, p73 transactivates p53 target genes in vivo and causes apoptosis and growth suppression (31, 33). Although p73 was expressed monoallelically in neuroblastoma, its tumor suppression function remains uncertain, because only the wild-type form has been identified in all tumors or tumor cell lines tested (33). Another group of p53 homologs, p51/p63 (65, 69, 77, 83), was also identified and found to share 55 to 65% homology with p53 in the central domain. These p53 homologs can also suppress cell growth, induce apoptosis, and transactivate p53-responsive genes (65, 83), although it is unclear whether they suppress tumor growth. Finally, two additional p53-like activities have been identified, p53-competing protein from mouse (9) and NBP (non-p53 response element [p53RE] binding protein) from human (90) cell lines, although their identity remains to be clarified. Thus, the transcriptional function is well conserved in the p53 family.
Identification of multiple p53 homologs suggests that these proteins have distinct roles during embryogenesis and development or in response to different cellular signals. In fact, two recent p63 knockout studies demonstrated that p63, in contrast to p53 (16), is essential for limb and epidermal morphogenesis (58, 84). Also, unlike p53, p73 was not induced by some DNA damage signals (33), suggesting a distinct pathway for this protein. Indeed, p73 has recently been shown to be activated through c-Abl-mediated tyrosine phosphorylation in response to DNA damage caused by cisplatin or γ but not UV irradiation (1, 22, 89). Because of the lower level of homology between p53 and p73 in the N and C termini (33), it would be interesting to learn whether these transcriptional activators interact with the same set of coactivators, such as p300/CBP, or with the same domains of these coactivators. It is clear that different domains of p300/CBP mediate transcription and thus signaling by different transcriptional activators (72). Hence, identifying p73-interacting proteins or domains of the proteins would provide clues for the potential signaling of p73. In attempt to address this issue, we have investigated whether p300 and CBP regulate p73-dependent function. We found that p300/CBP bound to p73 both in vitro and in vivo and that it enhanced p73-dependent transcription. Functional mapping revealed that unlike p53 (3, 26, 51; X. Zeng and H. Lu, unpublished data), p73 through its the N terminus utilized the N-terminal CH1 domain (aa 390 to 450) of p300/CBP for transcriptional activation and apoptosis. Consistent with this observation, p73 functions were found to be impaired to different degrees in p300- and CBP-deficient cells. Thus, this study provides evidence that p73 interacts with the N-terminal domain of p300/CBP to execute its transcriptional function.
MATERIALS AND METHODS
Plasmids and antibodies.
The pCDNA3-HA-p73α and pCDNA3-HA-p73β plasmids were obtained from William G. Kaelin, Jr. (Dana-Farber Cancer Institute, Boston, Mass.). pCDNA3-Flag-p300 or CBP plasmids were constructed. pCMV-p300-Ha was obtained from David Livingston (Dana-Farber Cancer Institute). pGSTCBP1(aa 390-790) and pGSTCBP3(aa1990-2441) were obtained from Robert G. Roeder (Rockefeller University, New York, N.Y.). pGST-p300(aa 1571-2414) was a gift from Yang Shi (Harvard Medical School, Boston, Mass.). pCDNA3-ΔN-p73α was constructed by PCR-directed mutagenesis using pCDNA3-HA-p73α as a template. pCNA3-GFP, encoding green fluorescent protein (GFP), was a gift from Moshe Oren (Weizmann Institute, Rehovot, Israel). pCDNA3-Flag-p73β was constructed by putting a Flag epitope in front of this insert. pEGFP-C1 was purchased from GIBCO-BRL. The monoclonal anti-p73 antibodies ER15, recognizing both p73α and p73β, and ER13, recognizing only p73α, were generously provided by William G. Kaelin, Jr. (57). Polyclonal anti-CBP antibodies recognizing both p300 and CBP were raised against the N-terminal region of CBP from aa 350 to 550. Monoclonal anti-p300 antibodies against the C-terminal domain of p300 were purchased from Upstate Biotechnology. Polyclonal anti-p53 antibodies were purchased from Santa Cruz Biotechnology. A monoclonal anti-p73 antibody against the C terminus of this protein was purchased from Oncogene.
Buffers.
Lysis buffer consisted of 50 mM Tris-HCl (pH 8.0), 0.5% NP-40, 1 mM EDTA, 150 mM NaCl, and 1 mM phenylmethylsulfonyl fluoride. SNNTE buffer contained 50 mM Tris-HCl (pH 7.4), 5 mM EDTA, 1% NP-40, 500 mM NaCl, and 5% sucrose. Radioimmunoprecipitation assay (RIPA) buffer consisted of 50 mM Tris-HCl (pH 7.4), 150 mM NaCl, 1% Triton X-100, 0.1% sodium dodecyl sulfate (SDS), and 1% (wt/vol) sodium deoxycholate. Buffer C 100 (BC100) included 20 mM Tris-HCl (pH 7.9), 0.1 mM EDTA, 10% glycerol, 100 mM KCl, 4 mM MgCl2, 0.2 mM phenylmethylsulfonyl fluoride, 1 mM dithiothreitol (DTT), and 0.25 μg of pepstatin A/ml.
Cell culture.
Human cervical carcinoma HeLa, human osteosarcoma Saos-2, and small-cell lung carcinoma H1299 cells were cultured in Dulbecco's modified Eagle medium supplemented with 5 to 10% fetal bovine serum, 50 U of penicillin/ml, and 0.1 mg of streptomycin/ml at 37°C in a 5% CO2 atmosphere. p300- or CBP-deficient H1299 or human breast carcinoma MCF-7 cell lines were cultured in the same medium except that G418 (0.4 μg/ml) was added as a selection marker as previously described (88).
Construction and preparation of GST-p300 and GST-p73 fusion proteins.
The glutathione S-transferase (GST)–p300 fusion protein expression plasmids were constructed as previously described (87). The GST-p73 fusion protein expression vectors were made by inserting the PCR-generated fragments of p73 into pGEX-KT (Pharmacia) at the EcoRI sites. The orientation and sequence of each p73 fragment were confirmed by sequencing and Western blotting (WB). These GST fusion proteins were expressed in and purified from bacteria. The GST-p73 (aa 411 to 636) fusion protein was also used as an antigen for generation of polyclonal anti-p73 antibodies as described above.
Preparation and fractionation of HeLa cell nuclear extracts.
Nuclear extracts were prepared from HeLa cells as described elsewhere (14). Briefly, 30-ml aliquots of nuclear extracts containing 10 mg of protein/ml (∼5 × 109 HeLa cells) were fractionated through a phosphocellulose (P11) column as described previously (54). Proteins were eluted by stepwise washes with 0.1, 0.3, 0.5, and 1.0 M KCl-containing buffer C.
Coimmunoprecipitation and WB analyses.
Fifty-microliter aliquots of the 0.3 M protein fractions from the P11 column were incubated at 4°C for 4 h with 30 μl of protein A-agarose and 1 μg of antibodies specifically against p300 (monoclonal) and p73 (polyclonal). The beads were washed as described above. Bound proteins were subjected to SDS-polyacrylamide gel electrophoresis (PAGE) and analyzed by WB as described elsewhere (54), using the same antibodies, preimmune serum, or antibody 419 as a control, and detected by enhanced chemiluminescence (Amersham).
Purification of Flag-p300 and Flag-CBP.
Flag-p300 and Flag-CBP were purified from a baculovirus expression system by immunoaffinity chromatography (27, 54) and used for protein association assays described below.
Transfection and coimmunoprecipitation.
H1299 cells (three 60-mm-diameter plates) were transfected with 3 μg of the parental pCDNA3, p73β-HA, or p73α-HA expression plasmid by means of LipofectAmine (GIBCO-BRL). Six hours after transfection, fresh Dulbecco's modified Eagle medium was added to the plates to replace the medium containing plasmids. Cells were harvested 48 h posttransfection. Cell lysates were prepared as described previously (55). Lysates (1 mg of proteins) were precleared with 35 μl of protein A-agarose (50% slurry) and then incubated for 3 h at 4°C with fresh protein A-beads (35 μl) and 2 μg of monoclonal antibody ER15, 421, or 419. The beads were loaded directly onto a SDS-acrylamide gel after vigorously washes twice with lysis buffer, twice with SNNTE, and once with RIPA buffer. The coimmunoprecipitated proteins were detected by WB using ER15 or polyclonal anti-CBP antibodies (detecting both p300 and CBP).
Metabolic labeling of cells.
H1299 cells (50% confluent) were transfected with plasmids as indicated in the legend to Fig. 1, using LipofectAmine (Promega) as described above. At 32 h posttransfection, cells were metabolically labeled with [35S]Met (1 mCi/60-mm-diameter dish) for 8 h and then harvested for immunoprecipitation; 2 million cpm was used for each immunoprecipitation reaction, carried out as described above.
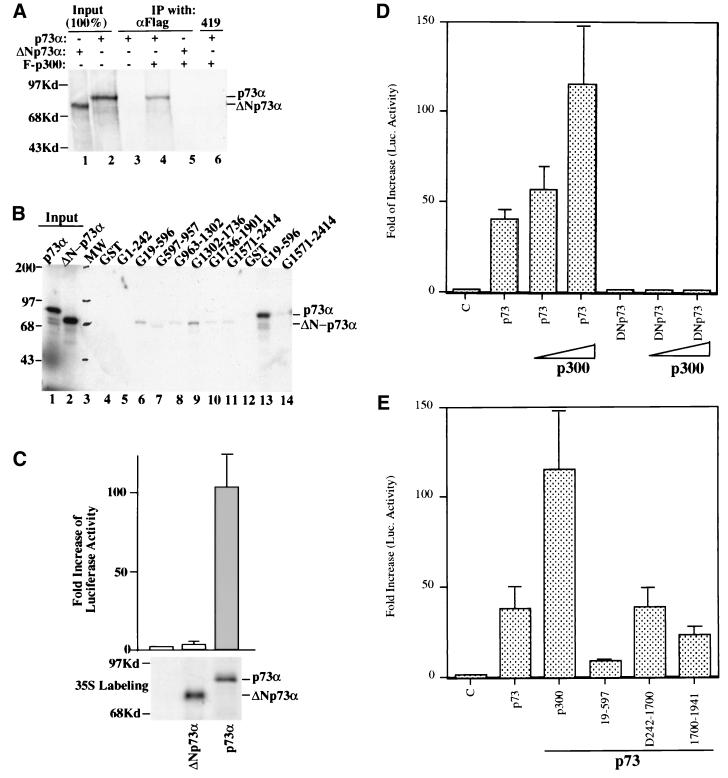
FIG. 1.
p73 forms a complex with p300 in vivo. (A) WB analysis of the protein fractions from P11 and DE-52 columns. Twenty-five-microliter aliquots of the fractions as indicated on the top were directly loaded onto an SDS–10% gel for WB analysis with polyclonal anti-CBP antibodies after probing with the monoclonal anti-p73α antibody ER13. NE, nuclear extract; FT, flowthrough; ΦS, protein samples containing the NBP activity from a phenol-Superose column (90), used as a negative control. (B) Immunoprecipitation followed by WB analysis of the p73-p300 complex. Fifty microliters (approximately 20 μg of protein) of the 0.3 M wash fraction of a P11 column was incubated with antibodies as indicated on top. Immunoprecipitation (IP)-WB analysis was carried out as described in Materials and Methods. The membrane was probed with ER15 and anti-p300 antibodies, respectively. 419 is a monoclonal antibody specific for the SV40 T antigen, used as a negative control (also for panel C). PreI, preimmune serum. (C) Immunoprecipitation of the p73-p300 complexes after transient transfection. H1299 cells (50% confluent in a 60-mm-diameter dish) were transfected with plasmids encoding Flag-p300 (F-p300) and/or HA-p73α as indicated at the top; 32 h posttransfection, cells were metabolically labeled with [35S]methionine for 8 h and harvested for immunoprecipitation. p300-p73α complexes were analyzed by immunoprecipitation using monoclonal anti-p73α (αp73α) or anti-Flag (αFlag) antibodies, as indicated. The asterisk denotes the nonspecific bands brought down by mouse immunoglobulin G, as they appeared equally in all lanes. Here and in other figures, positions of molecular weight markers (in kilodaltons) are indicated at the left. The fast-migrating band of the p300 doublet may represent a shortened large fragment of Flag-p300. The result was obtained after a 2-day exposure to X-ray film. The tiny scratch above the Flag-p300 band on lane 2 was a nonspecific spot caused by film exposure. (D) p73, when overexpressed, binds to endogenous p300 or CBP. H1299 cells (6 × 105/60-mm-diameter dish) were transfected with vectors (3 μg) encoding no insert, p73α, or p73β, as indicated, and harvested for immunoprecipitation with ER15 (lanes 1 to 3) or 421 (lane 4), followed by WB with ER15 and polyclonal anti-CBP antibodies. Each lane represents the result from three dishes of cells (with 30-min exposure to X-ray film).
Cotransfection and CAT assay.
Transfections were carried out as described above with H1299 and Saos-2 cells. Totals of 15 and 4 μg of plasmid DNA were used for 100- and 60-mm-diameter plates, respectively. At 48 h after transfection, the transfected cells were harvested for chloramphenicol acetyltransferase (CAT) assays as described previously (82).
Transient transfection and luciferase assay.
H1299 cells (60% confluent in a 12-well plate) were transfected with a pCMV-β-galactoside reporter plasmid (0.2 μg) and a luciferase reporter plasmid (0.2 μg) driven by two copies of the p53RE motif derived from the MDM2 promoter (82), together with a combination of different plasmids (total plasmid DNA = 1 μg/well) as indicated in Fig. 5 and 7, using GenePORTER (Gene Therapy Systems, Inc., San Diego, Calif.). At 48 h posttransfection, cells were harvested for luciferase assays as described previously (41). Luciferase activity was normalized by a factor of β-galactosidase activity tested in the same assay.
FIG. 5.
The N-terminal deletion mutant of p73α is defective in p300 binding and transcriptional activation. (A) The N-terminally deleted mutant of p73α does not bind to p300. Three microliters of in vitro-translated and 35S-labeled wild-type or N-terminally deleted mutant p73α (lacking aa 1 to 56), as indicated at the top, were incubated with Flag-p300 (200 ng). Bound proteins were coimmunoprecipitated by anti-Flag antibodies and detected by autoradiography. (B) The N-terminally deleted mutant of p73α does not bind to the N-terminal domain of p300 in vitro. Three microliters of in vitro-translated and 35S-labeled wild-type or N-terminally deleted mutant p73α were incubated with different GST-p300 fusion proteins as indicated at the top. Bound proteins were analyzed by SDS-PAGE and detected by autoradiography. Lanes 1 and 2, 100% input of wild-type (p73α) and mutant (ΔN-p73α) proteins, respectively; lane 3, molecular weight markers; lanes 4 through 12, results with N-terminally deleted mutant p73α; lanes 13 and 14, positive controls with wild-type p73α. (C) The N-terminally deleted mutant of p73α is inactive in transcriptional activation. H1299 cells (105/60-mm-diameter plate) were transfected with plasmids (total, 3 μg of DNA) encoding no gene, p73α, or ΔN-p73α (1 μg) together with a luciferase reporter plasmid (0.2 μg) driven by the p53RE motif derived from the p21 promoter and a β-galactosidase reporter plasmid (0.2 μg) driven by a cytomegalovirus promoter; 48 h posttransfection, cells were harvested for luciferase (Luc.) and β-galactosidase assays. Luciferase activity was normalized by a factor of the internal β-galactosidase activity. Fold increase in luciferase activity was calculated and is presented in the graph (each column is from three independent assays, and bars show standard deviations). The bottom panel shows expression of p73α or ΔN-p73α detected by immunoprecipitation with anti-p73 antibodies (Santa Cruz), followed by autoradiography, after in vivo [35S]Met metabolic labeling of the transfected cells. (D) ΔN-p73α lacks transcriptional activity and its potential to cooperate with p300 in transcription. Transient transfection-luciferase assays similar to that in Fig. 5C were carried out except that H1299 cells grown to 60% confluence in 12-well plates were transfected with total plasmid DNA of 1 μg (per well) including 0.1 μg of the luciferase reporter plasmid, 0.1 μg of β-galactosidase reporter, 0.1 μg of p73α or ΔN-p73α alone, and 0.35 or 0.7 μg of p300, as indicated. Each column represents a result from triplicate assays. (E) The CH1-containing fragment of p300 inhibits p73α-mediated transcription in vivo. H1299 cells were transfected with a p73α expression plasmid (0.1 μg) in the presence or absence of a plasmid (0.7 μg) encoding p300 or a deletion mutant of p300 (one including the fragment from aa 19 to 597, one without aa 242 to 1700, and one with deletion of aa 1700 to 1941) as described for panel D.
FIG. 7.
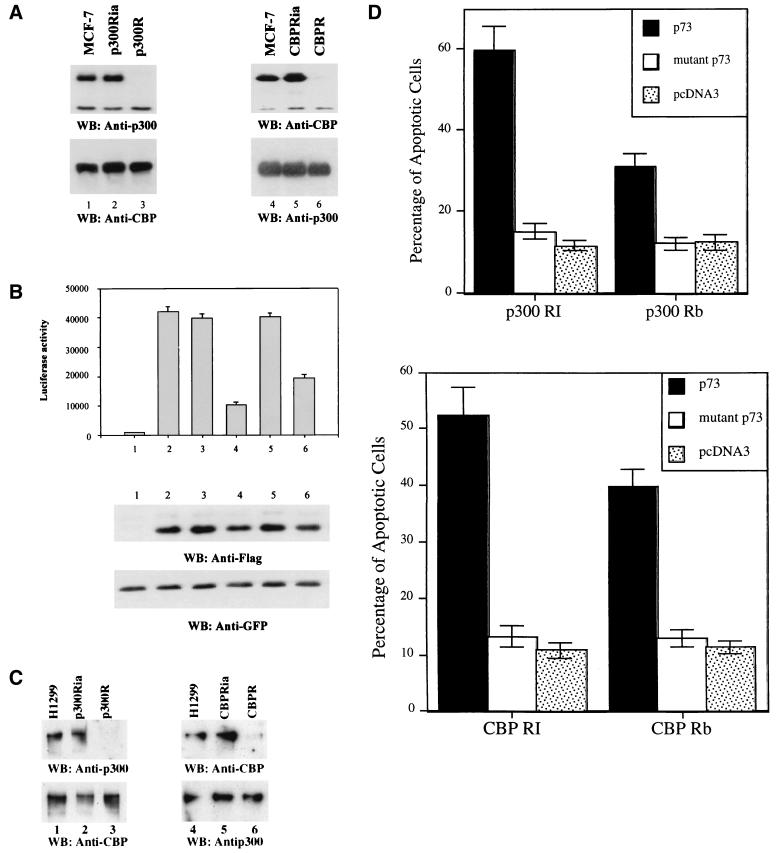
p73 transcription and apoptosis are impaired in p300- or CBP-deficient cells. p300- or CBP-deficient MCF-7 or H1299 cells were generated as described previously (88). (A) Cell lysates prepared from the indicated MCF cell lines were analyzed by WB with anti-p300 (RW109; Upstate Biotechnology) or anti-CBP antibodies. (B) The pCDNA3-Flag control vector or Flag-p73β was cotransfected with a luciferase reporter driven by an MDM2 promoter-derived p53RE motif into MCF-7 (columns 1 and 2), p300Ria (column 3), p300R (column 4), CBPRia (column 5), and CBPR (column 6) cells. pEGFP-C1 was included as transfection control (bottom panel). Luciferase activity was measured with a normalized protein concentration 24 h posttransfection. Cell lysates were also analyzed by WB with anti-Flag to detect Flag-p73β (middle panel) and anti-GFP antibodies (bottom panel). (C) WB analysis of p300 or CBP in p300- or CBP-deficient H1299 cell lines. (D) Apoptotic analysis of H1299 cell lines after transfection with plasmids as indicated, using the same approaches as described for Fig. 6. p300 RI, p300 RB, CBP RI, and CBP Rb stand for the p300Ria, p300R, CBPRia, and CBPR cell lines. p73 or mutant p73 denotes the wild-type p73α or N-terminally deleted p73α. The result for the parental H1299 cells was similar to that for the p300Ria or CBPRia cell lines; thus, results for only the latter are presented here as a control.
GST fusion protein association assay.
The fusion proteins were expressed in Escherichia coli and purified on a glutathione (GSH)-Sepharose 12B column. Protein-protein association assays were conducted as reported previously (54), using fusion protein-containing beads. The purified and in vitro-translated, 35S-labeled p53 protein or in vitro-translated, 35S-labeled p73α and p73β proteins generated by using TNT kits (Promega) were incubated with the GSH-Sepharose 4B beads (50% slurry) containing approximately 400 ng of GST-p300, GST-CBP, and GST, respectively. One hour after incubation at room temperature, the mixtures were washed once in BC100 containing 0.1% NP-40, twice in SNNTE, and once in RIPA buffer. Bound proteins were analyzed on an SDS–10% gel and detected by WB using the anti-p53 monoclonal antibody 421 to detect p53-GST-CBP interactions and autoradiography to detect interactions of p53 or p73 with GST-CBP or GST-p300. Similar assays were also conducted to map the p300 or CBP binding site on p73, using GST-p73 fusion proteins as described elsewhere (91).
Apoptotic analysis.
The analysis was carried out as previously described (91). H1299 or p300- or CBP-deficient cells (105/35-mm-diameter dish) were transfected with a plasmid encoding GFP (0.85 μg of DNA/dish) together with combinations of expression plasmids (total plasmid DNA = 2 μg/dish) as indicated in Fig. 6. Transfected cells in cultures were analyzed under a fluorescence microscope and identified by the presence of green fluorescence. Apoptotic cells were identified by their rounded and shrunken morphology in contrast to the spread-out appearance of nonapoptotic H1299 cells, counted on a blind basis, and presented as a percentage of the total population of fluorescent cells (see Fig. 6).
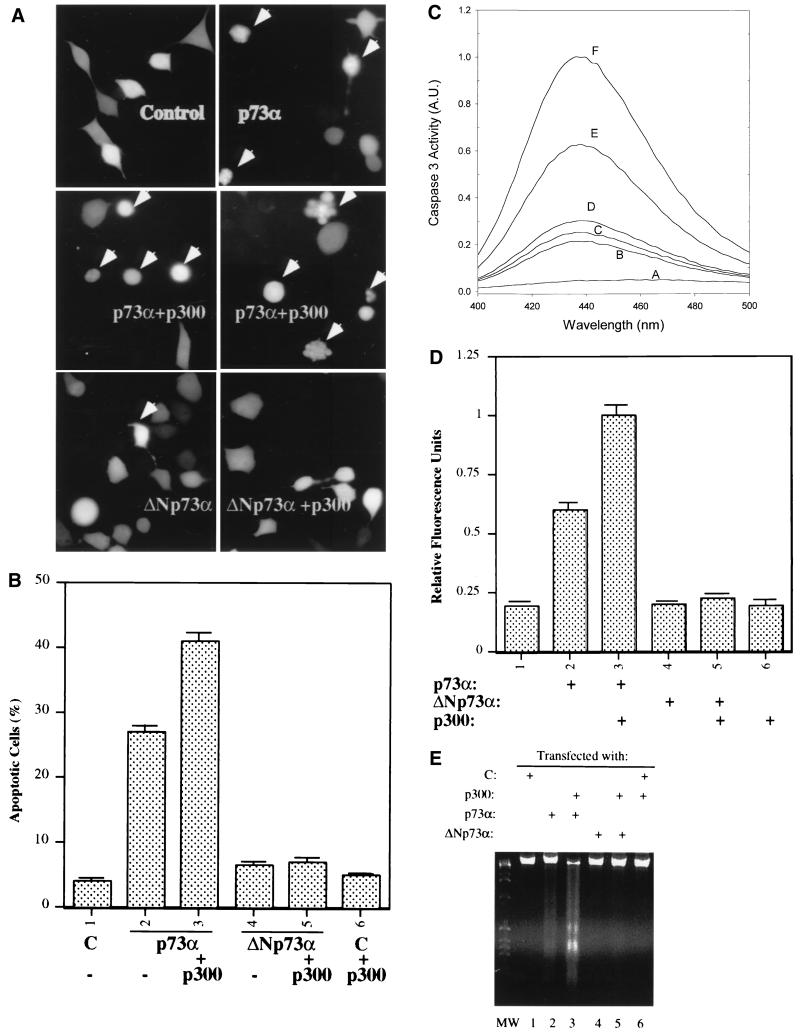
FIG. 6.
p300 enhances p73-induced apoptosis. (A and B) The N-terminal deletion of p73 lacks apoptosis-promoting potential. As indicated, plasmids encoding no protein as a control (C; 3 μg), p73α (1.0 μg), ΔNp73α (1.0 μg), and/or p300 (2.0 μg), together with the pGFP plasmid (200 ng), were introduced into H1299 cells (105/60-mm-diameter dish) using LipofectAmine (GIBCO-BRL); 32 h posttransfection, cells (100) expressing GFP were counted under a fluorescence microscope using a blind approach, with morphologically round cells (arrows) identified as apoptotic (A). (B) Statistical data (with standard error indicated by a bar above each column) from three independent assays. (C and D) Caspase 3 activation by wild-type but not N-terminus-deleted p73α is enhanced by p300. Transient transfection as described for panel A was conducted. Cells were harvested for caspase 3 activity analysis as described in Materials and Methods. (C) Representative result of the fluorescence graph of AMC released from Ac-DEVD-AMC substrates for one set of experiments with plasmids encoding no protein (curve B), ΔNp73α (curve C), ΔNp73α plus p300 (curve D), p73α (curve E), or p73α plus p300 (curve F). Curve A is a water control. Caspase 3 activity is presented in arbitrary units (A.U.) after normalization by a factor of 105. (D) Statistical values for caspase 3 activity in triplicate assays. (E) DNA fragmentation analysis of p73-induced apoptosis in H1299 cells was conducted as described in Materials and Methods; 10 μg of DNA was analyzed by electrophoresis on 2% agarose. Each lane represents the result for three 60-mm-diameter plates of cells. MW, molecular weight markers.
Caspase 3 assays.
H1299 or p300- or CBP-deficient H1299 cells (60% confluent) were transiently transfected with a plasmid expressing p73α or a p73α mutant lacking the N-terminal aa 1 to 56 (ΔNp73α). Cells were harvested and lysed 34 h posttransfection for caspase 3 assays according to a protocol provided by PharMingen. The caspase 3 reaction mix contained 20 μM Ac–DEVD–7-amino-4-methyl coumarin (AMC) substrate for caspase 3 (PharMingen), 150 μg of lysates, and 500 μl of protease buffer (20 mM HEPES [pH 7.5], 10% glycerol, 2 mM DTT freshly added); the reaction was conducted at 37°C for 60 min. AMC released from Ac-DEVD-AMC substrates was measured with a spectrofluorometer (Photon Technology International) with an excitation wavelength of 380 nm and an emission wavelength range of 400 to 500 nm.
DNA fragmentation assays.
Transfection of H1299 cells was conducted as described above. Floating or attached cells were harvested separately for DNA isolation 38 h posttransfection. Since few floating cells were found in the control, mutant p73-, or p300-alone-transfected dishes, DNA isolated from attached cells was used as a control. DNA fragments were detected by using a DNA ladder detection kit purchased from BioSource International, Inc., Camarillo, Calif.
RESULTS
Association of p73 with p300/CBP in vivo.
During purification of NBP, a recently identified p53-like transcriptional activity from HeLa cell nuclear extracts (90), we detected a significant amount of p73α protein by WB with anti-p73 antibodies in the 0.3 M wash fraction from a phosphocellulose column (Fig. 1A, lane 3 [90]). This finding prompted an examination of whether p73 cofractionates with other cellular proteins that may be important for its function. Given that p53 interacts with p300/CBP (3, 26, 51), the same membrane was then probed with polyclonal anti-CBP antibodies, which detect both p300 and CBP (4, 56). p300 and CBP were enriched in the same fraction (Fig. 1A, lane 3), implying that p73 may form a complex with p300/CBP. To examine this possibility, proteins in the 0.3 M fraction of the P11 column were coimmunoprecipitated with monoclonal anti-p300 or anti-p73α antibodies and detected by WB with antibodies against p73 and p300. As shown in Fig. 1B, the p73 protein was specifically coprecipitated by the monoclonal anti-p300 antibody or vice versa (lanes 2 and 3). As a control, the monoclonal anti-simian virus 40 (SV40) T-antigen antibody 419 or preimmune serum did not pull down any proteins detectable by either anti-p73α or anti-p300 antibodies (lanes 1 and 4). This result suggests that p73 can form a complex with p300.
To further confirm the p73-p300 interaction in vivo, H1299 cells containing no detectable endogenous p73 were transfected with mammalian expression plasmids encoding hemagglutinin (HA)-p73α or HA-p73β and/or Flag-p300. The cells were harvested after metabolic labeling for immunoprecipitation-autoradiograpic analyses as described in Materials and Methods. A representative result is shown in Fig. 1C for HA-p73α and Flag-p300. Flag-p300 was coimmunoprecipitated with p73α by the monoclonal anti-p73α antibody (Fig. 1C, lane 3), and the reverse was also true with the monoclonal anti-Flag antibody (lane 6). This occurred apparently when these proteins were coexpressed in the cells, as this complex was not found when either protein alone was introduced into the cells (compare lanes 1, 2, 4, and 5 with lanes 3 and 7). This interaction was also specific for these two antibodies, as it was not detected by a monoclonal antibody against SV40 large T antigen (lane 7). A fast-migrating band below the Flag-p300 protein (lanes 3, 4, and 6) could be a shortened large fragment of the exogenous Flag-p300 protein. These results were repeated using immunoprecipitation-WB after transient transfection with the same sets of plasmids; the p300-p73β complex was also detected by this method (data not shown). Moreover, the endogenous p300 or CBP was coimmunoprecipitated with exogenous HA-p73α or HA-p73β by monoclonal anti-p73 antibody ER15 when these transcriptional factors were overexpressed in H1299 cells (Fig. 1D, lanes 2 and 3). Since the polyclonal anti-CBP antibody recognizes both p300 and CBP, the doublet detected by this antibody may present both of the p300 and CBP polypeptides (lanes 2 and 3). The interaction between p300/CBP and p73 was specific, as it was not observed when anti-p53 antibodies were used (lane 4) or in the absence of p73 (lane 1). Taken together, these results clearly demonstrate that p73 can associate with p300 or CBP in cells.
p73 binds to the N-terminal domain of p300 or CBP.
Because p53 was previously reported to bind to the C-terminal domain (aa 1990 to 2441) of p300/CBP (26), we tested whether p73 targets the same region, using GST protein association assays. Equal amounts (1 μg) of GST-p300 fusion proteins coupled to Sepharose 4B resin were incubated with in vitro-translated, [35S]Met-labeled p73α or p73β. Bound proteins were analyzed by SDS-PAGE and detected by autoradiography. As shown in Fig. 2A, both p73α and p73β bound specifically to the p300 N-terminal region from aa 19 to 596 (19-596 domain) but not to the other regions of p300 (two top panels). When GST-CBP fusion proteins were incubated with radioactively labeled p73, both α and β forms of p73 were also observed to interact with the CBP N terminus, as shown by a representative result in Fig. 2B for p73β. As CBP fragments encompassing aa 1 to 450 (lane 2) and 390 to 790 (lane 3) retained an equivalent amount of p73, the p73 binding site in CBP (or p300) can be ascribed to the region from aa 390 to 450. Indeed, a CBP fragment of aa 350 to 450 was able to bind to p73 (unpublished data). The faster-migrating p73 band in lane 3 of Fig. 2B was due to its comigration with the GST-CBP 390-790 fusion protein on an SDS-gel. As a control, GST alone did not retain a significant amount of the p73 proteins (Fig. 2), nor did the C-terminal domains of CBP or p300 (Fig. 2). These results suggest that both p73α and p73β physically bind to the first C/H domain (potential zinc finger domain) of p300 and CBP. In contrast, p53 bound to both the N- and C-terminal domains of p300/CBP (Fig. 2A, middle panel). Consistent with a previous report (24), the binding of p53 to the N-terminal CH1 region of p300/CBP appears to be not important for its transcription activity, as a transcriptionally inactive 22/23 mutant of p53 also bound to this domain (Zeng and Lu, unpublished data). The binding of p73 to the domain is not due to an adherent property of this region, as it was not the case for TAFII31 (Fig. 2A, bottom panel), consistent with a recent report (62). Thus, p73 is associated with the N-terminal CH1 domain of p300/CBP.
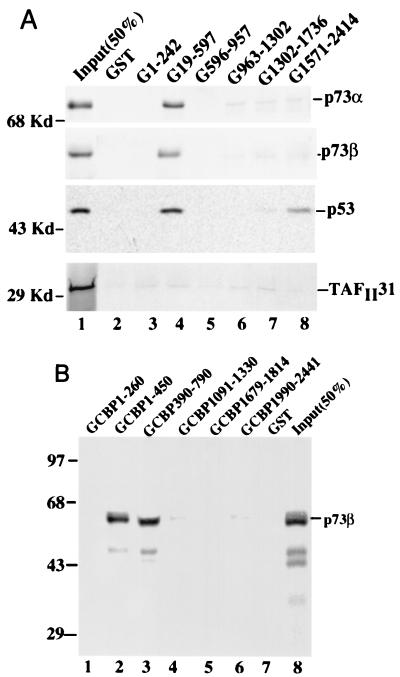
FIG. 2.
p73 and p53 bind to different domains of p300 or CBP. (A) Protein association analyses with in vitro-translated, 35S-labeled p53, p73α, p73β, and TAFII31. Equal amounts (3,000 cpm) for each of the in vitro-translated, 35S-labeled proteins (indicated on the right) were incubated with 30 μl of GSH-Sepharose 12B beads (50% slurry) bound by GST or GST-p300 deletion mutants (indicated at the top); 50% of the input was directly loaded onto the gel as a control (lane 1). (B) p73β binds to the first C/H domain of CBP. A GST pull-down assay as described above was carried out but with GST-beads containing a series of CBP deletion mutants as indicated; 50% of the p73 input was directly loaded onto the gel (lane 8). These results were reproducible; the same results were obtained in experiments with GST-CBP fusion proteins and p73α (Zeng and Lu, unpublished data).
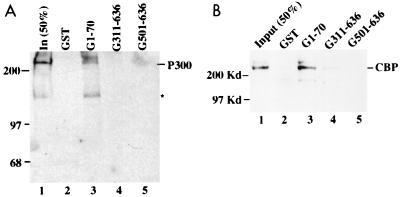
p300/CBP binds to the N-terminal domain of p73.
To map the p300/CBP binding site in the p73 protein, we carried out a similar set of GST pull-down assays using GST p73 fusion proteins. Bound p300 or CBP was analyzed by SDS-PAGE, followed by WB with antibodies against either p300 or CBP. Either p300 (Fig. 3A) or CBP (Fig. 3B) was retained by the GST-p73 fusion protein containing only the N-terminal domain (aa 1 to 70) but not by the C-terminal region of p73α (aa 311 to 636) (compare lanes 3 with lanes 4 and 5). p300 or CBP did not bind to the GST protein alone (lanes 2). Thus, p300 or CBP appears to recognize the N-terminal domain of p73, which shares a 29% identity with the p53 N-terminal 1-58 domain.
FIG. 3.
p300 or CBP binds to the N-terminal domain of p73. One hundred nanograms of partially purified p300 (A) or CBP (B) was incubated with different GST-p73 deletion mutants as indicated (1 μg). The bound p300 or CBP protein was detected by WB with anti-p300 or anti-CBP antibodies; 50% of the input was directly loaded onto the SDS–7% gel. The asterisk indicates degraded p300. These results were reproducible.
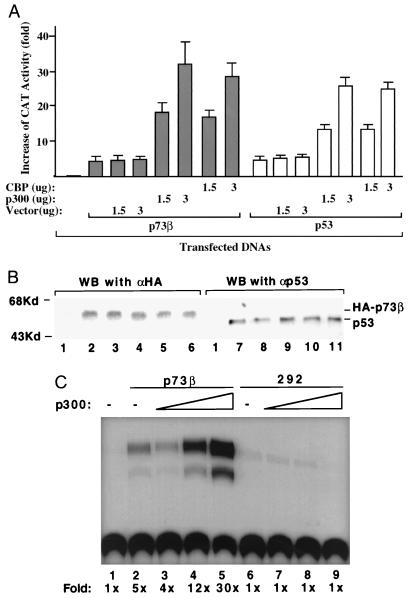
p300/CBP stimulates p73-dependent transcription in vivo.
To determine the functional relevance of the p300/CBP-p73 interaction, we performed a set of transfection and CAT assays using a CAT reporter plasmid containing two copies of p53RE derived from the MDM2 promoter (82). A p53-deficient human osteosarcoma Saos-2 cell line was cotransfected with 100 ng of p73β, p73α, or p53 and increasing amounts of p300 or CBP. Since p73β and p73α displayed similar transcription activities in this assay, a representative result for only p73β in contrast with p53 is shown in Fig. 4A. The p73-dependent CAT activity in the presence of 3 μg of the p300 or CBP plasmid was ∼10-fold greater than that in the absence of plasmid (Fig. 4A, compare lane 2 with lanes 4 and 6). This stimulation was due to p300 or CBP, as it was not observed when a parental vector was used (Fig. 4A); also, p73 was equivalently expressed regardless of whether p300 or CBP was coexpressed (Fig. 4B, lanes 1 to 6). Moreover, transcriptional activity was specific for p73, as it disappeared when p73 mutant R292H, which is equivalent to the inactive p53 mutant R273H (31), was used in the assay (Fig. 4C). The transcriptional activity of p53 was enhanced, as expected, in the presence of exogenous p300 or CBP (Fig. 4A and B), consistent with previous studies (3, 26, 51). These results were also reproducible using H1299 cells devoid of the p53 protein (Zeng and Lu, unpublished data). We additionally observed that p73 or p53 requires a full-length p300 or CBP protein for synergistic activation of transcription, as p73 or p53 binding fragments of both proteins were unable to enhance p73- or p53-mediated transcription in vivo (Fig. 5E and data not shown). Thus, these studies demonstrate that the coactivators p300 and CBP not only bind to p73 but also enhance transcription mediated by this transcriptional activator.
FIG. 4.
p300 or CBP enhances the p73-dependent transcription in vivo. (A) p300 or CBP stimulates p73β- or p53-dependent CAT activity in transient transfection-CAT assays. Saos-2 cells (105/60-mm-diameter plate) were transfected with 0.2 μg of the BP100-CAT reporter vector containing two copies of p53RE derived from the MDM2 promoter (82), 100 ng of p73β or p53, together with 1.5 or 3 μg of p300- or CBP-expressing vector, respectively, as indicated. CAT assays were carried out 48 h posttransfection. Fold increase in CAT activity compared with that of the mock transfection is shown at the bottom. CAT activity from three experiments was quantified with a molecular imager (Bio-Rad) and presented as a graph. Vectors without an insert were used as controls. Bars denote fold increase in CAT activities, and standard deviations are indicated at the top of each bar. A similar result was obtained with p73α. (B) Expression of HA-p73β or p53, detected by immunoprecipitation with ER15 for HA-p73 or with 421 for p53, followed by WB with 12CA5 for HA-p73 or polyclonal anti-p53 antibodies. Lane 1, control. Lanes 2 to 6 correspond to columns 4 to 8 in panel A, and lanes 7 to 11 match columns 11 to 15 in panel A. Cells from three 60-mm-diameter dishes were collected for each lane. (C) p300 stimulates wild-type but not mutant p73α-dependent transcription. The assay described above was conducted with 100 ng of p73α or an inactive p73α mutant R292H. All experiments were repeated three times using Saos-2 and H1299 cells with similar results.
The N-terminally deleted mutant of p73 is defective in binding to the N terminus of p300 and stimulating transcription and apoptosis in vivo.
The interaction of the N terminus of p73 with p300/CBP suggests that the N terminus of p73 may serve as transactivation domain, though this domain shares a low degree homology (29% identity) with that of p53 (33). To test this possibility, we first examined the direct binding of the p73 N terminus to the N-terminal domain of p300. For this purpose, we generated ΔNp73α and analyzed its interaction with wild-type or mutant forms of p300 by in vitro coimmunoprecipitation and protein-protein association assays. Consistent with the results in Fig. 1 to 3 and our recent report (91), p73α was coimmunoprecipitated with Flag-p300 by anti-Flag antibodies (Fig. 5A, lane 4). However, ΔNp73α was unable to bind to p300 (compare lane 4 with lane 5), indicating that the N-terminal 1-56 domain directly interacts with p300. The interaction was specific, as no signals were immunoprecipitated by anti-Flag antibodies either in the absence of Flag-p300 or using polyclonal antibody 419 specifically against the SV40 T antigen (lane 3 or 6). This interaction was mediated by the N-terminal domain of p300, as binding of ΔNp73α to the N-terminal region (aa 19 to 596) of p300 was dramatically reduced compared with that of the wild-type p73 (Fig. 5B, compare lane 6 with lane 13). These results demonstrate that the N terminus of p73 directly binds to the N-terminal CH1-containing domain of p300.
Correlated with the results above, the N terminus of p73 is also essential for its transcriptional function, as ΔNp73α was inactive in transcriptional activation of a luciferase reporter gene driven by the p53RE motif derived from the p21 promoter (Fig. 5C; a luciferase assay was used, as it is convenient for statistical analysis). This defect was not due to lower expression of the p73 mutant, as both the wild-type and mutant p73 proteins were expressed equivalently (Fig. 5C, bottom panel). Moreover, p300 was unable to synergize transcriptional activation by this p73 mutant (Fig. 5D, compare left four columns with right three columns), likely due to loss of the interaction between these two proteins (Fig. 5A to C). To further test whether fragments of p300 can affect the transcriptional activity of p73α, mammalian expression plasmids encoding wild-type p300, the N-terminal p73 binding region (aa 19-596), a fragment lacking the p73 binding domain (deletion from aa 242 to 1700), and a fragment containing the acetylase-containing region (aa 1700-1941) (5, 63) were used for transient transfection-luciferase assays. Again, the result in Fig. 5E shows that p73-mediated transcription (p73 column) was significantly stimulated by p300 (p300 column). By contrast, the p73 binding fragment of p300 markedly inhibited p73-dependent transcription (19-597 column), suggesting that this fragment might prevent p73 from binding to p300/CBP (Fig. 2A and 5B), as this effect was not apparent when the other p300 deletion lacking the p73 binding domain was introduced into cells (Fig. 5E, D242-1700 column). A partial inhibitory effect on p73-mediated transcription was observed in the presence of the acetylase domain-containing fragment (1700-1941 column). These effects were not due to different p73 or p300 fragment levels expressed in the cells, as equivalent amounts of the proteins were revealed by immunoprecipitation-WB analysis (data not shown).
Transcriptional activity of p73 is believed to be responsible at least partially for apoptosis induced by this protein (31). Thus, the transcriptionally inactive p73 deletion mutant may also be defective in inducing apoptosis. To test this idea, the wild-type or N-terminally deleted form of p73 was introduced into H1299 cells together with a GFP expression vector. At 32 h posttransfection, apoptotic cells were identified by blind counting as rounded and shrunken under a fluorescence microscope (Fig. 6A). The percentage of apoptotic cells was calculated and presented in Fig. 6B. Approximately 4% of H1299 cells were undergoing apoptosis under normal conditions of cell culture. p73 when overexpressed caused a significant increase of apoptotic cells (26% apoptotic cells). p300 further enhanced p73-induced apoptosis (42% apoptotic cells) but had no effect on cells transfected with a control vector. By contrast, the N-terminally deleted mutant of p73 was unable to induce apoptosis, regardless of whether p300 was overexpressed. The apoptotic nature was ensured by a DNA ladder assay (Fig. 6E). To further confirm the apoptotic induction by p73 and p300, caspase 3 activity with the same set of experiments was detected by monitoring its cleavage substrates as described in Materials and Methods. A fluorescence graph of this activity is shown in Fig. 6C, indicating that the greater the emission at a wavelength of 435 nm is, the higher the caspase 3 activity is. For example, p73 expression induced caspase 3 significantly (curve E) and p300 further stimulated this activity (curve F), whereas either the N-terminally deleted p73 or p300 alone was without effect on this activity (curves C and D). The quantitation of three independent assays (Fig. 6D) again displays the same result as that in Fig. 6B, demonstrating that the N-terminal domain of p73 is not only crucial for its transcriptional activity but also essential for promoting apoptosis, and p300 can further enhance these functions.
After observing that the N-terminal domain of p73, unlike that of p53 (26), directly interacts with the N-terminal, instead of C-terminal, domain of p300/CBP (Fig. 2, 3, and 5), we also tested whether p53 binds to the N-terminal domain of p300/CBP and mediates transcription through this binding. We found that both wild-type and transcriptionally inactive 22/23 mutant forms of p53 were able to bind to the CH1 domain of p300/CBP, indicating that this binding may not be crucial for p53 transcriptional function (Zeng and Lu, unpublished data). This is consistent with a recent report showing that the binding of p53 with p300 through their N termini probably plays a role in a nontranscriptional function of the proteins (24).
p73-mediated transcription and apoptosis are impaired to different degrees in p300- and CBP-deficient cells.
It has been recently shown that p53 activation appears to require p300 but not CBP in response to DNA damage (88). Our results indicate that both p300 and CBP stimulate p73-mediated transcription (Fig. 4 and 5). To determine which of the coactivators is essential for p73-mediated transcription, cell lines expressing either active or inactive p300 or CBP ribozymes were used as described elsewhere (37, 88). In the active p300 or CBP ribozyme-expressing cells, p300 or CBP was hardly detectable by WB using anti-p300 or anti-CBP antibodies (Fig. 7A and C, lane p300R or CBPR), whereas these proteins were not affected in the inactive p300 or CBP ribozyme-expressing cells (lane p300Ria or CBPRia) compared with that in parental MCF-7 or H1299 cells (lane MCF-7 or H1299). p73β expression plasmids together with a luciferase reporter driven by the p53RE motif derived from the MDM2 promoter were transiently introduced into these cells as well as parental cells, and luciferase activity was measured (Fig. 7B). Consistent with the results above, luciferase activity was significantly elevated by p73 in the parental or inactive p300 or inactive CBP ribozyme-expressing MCF-7 cells (compare column 1 with columns 2, 3, and 5). By contrast, p73-dependent luciferase activity decreased by ∼76% in the p300-deficient cells (column 4) and by ∼53% in the CBP-deficient cells (column 6). The incomplete inhibition in either case was probably due to the existence of endogenous CBP or p300 (Fig. 7A, bottom panel). The reduction of p73-dependent transcription was not due to a difference in p73 expression, as p73 levels were equivalent in all cells transfected with Flag-p73 expression plasmids (Fig. 7B, middle panel). This result was also repeated with the p300- or CBP-deficient cell lines derived from H1299 (Zeng and Lu, unpublished data). The result suggests that both endogenous p300 and CBP proteins are utilized by p73 to execute transcription in cells, and lack of either of the coactivators impairs p73 transcriptional activity to a certain extent.
To further test whether p73 can induce apoptosis in the p300- or CBP-deficient cells, H1299 cells expressing either active or inactive p300 or CBP ribozymes were transfected with plasmids expressing no p73α, wild-type p73α, or N-terminally deleted p73α. Consistent with the result in Fig. 7A, p73-induced apoptosis was reduced markedly (∼60%) in the p300-deficient H1299 cells but to a lesser degree (∼37%) in the CBP-deficient H1299 cells (Fig. 7D). This difference may be explained by the possibility that the number of residual CBP molecules in CBPR cells is more than that of p300 in p300R cells (Fig. 7C); alternatively, p300 may be more critical than CBP in p73-induced apoptosis. Again, the N-terminally deleted p73 was unable to induce apoptosis in either cell line. These results suggest that p300 and probably CBP plays a role in p73-induced apoptosis.
DISCUSSION
p300 and CBP have been shown to interact with a variety of transcriptional activators (72), including p53 (3, 26, 51), serving as a linker between these activators and the RNAPII transcriptional machinery. The study presented here further investigated whether this group of coactivators is also important for transcription mediated by the p53 homolog p73. This study demonstrates that p300 and CBP enhance p73-dependent transcription in vivo. Like p53, p73 interacted with p300 and CBP, as shown by in vitro protein-protein association assays (Fig. 2 and 3) or in vivo coimmunoprecipitation assays (Fig. 1). Unlike p53, which was originally found to bind to the C-terminal region of p300/CBP (3, 26, 51), the N-terminal domain of p73 bound to the N-terminal CH1-containing region of the coactivators (Fig. 2 and 5). Loss of this binding correlated with the defect in transcriptional activation by p73 (Fig. 5). Consistently, the N- but not the C-terminal fragments of p300, when overexpressed, inhibited p73-dependent transcription. By contrast, the C terminus of p300/CBP is crucial for transactivation by p53, as the transcriptionally inactive p53 mutant 22/23 (26, 52, 54) was still able to bind to the coactivator's N- but not C-terminal domain in vitro (data not shown; references 24 and 26). The difference between p73 and p53 in binding to distinct domains of p300/CBP was also observed in our recent study (91), in which MDM2, a cellular p53 inhibitor (60), selectively blocked the p73-p300 but not the p53-p300 interaction, as MDM2 itself bound to the p300 N terminus (aa 1 to 450) as well. However, binding of p53 to the N-terminal domain of p300/CBP was recently proposed to mediate MDM2-dependent p53 degradation (24), although the biological conditions under which p300 assists MDM2 to enhance p53 degradation are not known. Also, unlike p53, whose N-terminal domain (aa 1 to 45) was not absolutely essential for its apoptotic function (78), p73 appeared to require its N terminus for apoptosis (Fig. 6 and 7). This suggests that the transactivational activity of p73 may be closely related with its apoptosis function, although it remains possible that the N-terminal domain (aa 1 to 56) of p73 may induce apoptosis through a direct and transcription-independent pathway. Thus, these studies reveal that these two p53 family members share the same set of coactivators for their transactivational activities by targeting different domains of these partners.
It is interesting that p73 and p53 target different domains of p300/CBP (Fig. 2 and 5; references 24 and 26) for transactivation, despite the fact that the N termini of the two transcriptional activators are related (33). One interpretation for this difference could be that although p73 and p53 share an overall 29% identity in amino acid sequence in their N termini (33), the p73 N terminus is 16 residues longer than the p53 N terminus. This extra sequence in the middle of the p73 N terminus may cause some conformational variation that could, in turn, influence the manner by which p73 physically contacts p300 or CBP. This difference between p73 and p53 may serve to avoid a direct competition for the same set of coactivators between the activators of the same family within cells. Also, in line with the finding that p73 does not respond to some DNA-damaging reagents (33), this difference implies that p73 may be activated through a signaling pathway that requires the CH1 domain of p300/CBP, distinct from that for p53 activation.
One important question is whether p73 is also a substrate of the p300/CBP intrinsic acetylase, which acetylates histones (5, 63) and is vital for the biological function of these coactivators (32, 74). p53 was the first reported nonhistone protein and transcriptional activator shown to be acetylated by this family of coactivators (25). The acetylation of p53 by p300 or CBP stimulates its p53RE binding and transcriptional activities (25) in response to DNA damage (53, 68, 88). p73 requires a full-length p300 or CBP protein for synergistic activation of transcription (Fig. 5). This is reasonable because (i) the acetylase activity of p300/CBP, which maps to aa 1195-1810 (5, 63) of the coactivators, is required for transactivation in cells (25, 43, 79) and (ii) communication with the RNAPII transcriptional machinery important for p300/CBP functions is mediated through different regions of the coactivators (40, 61). Thus, it will be important to determine whether the p300/CBP-interacting p73 can be activated through acetylation as well.
It has not been resolved whether p300 and CBP contribute equivalently to transcriptional activation and thus cellular signaling mediated by different p300/CBP-interacting transcriptional activators in cells. One report showed that p300 and CBP can selectively stimulate transcription mediated by different human T-cell leukemia virus type 1 Tax mutants (7). Recently, p300 and CBP null mice have been generated by a gene knockout technique (86). The p300−/− or CBP−/− mice were embryonic lethal with severe proliferation defects, strongly demonstrating that these coactivators are essential for cell viability and development. Intriguingly, although retinoic acid receptor was reported to utilize both p300 and CBP as comediators (11, 34), retinoic acid receptor-dependent transcription was defective in the p300−/− but not in CBP−/− cells (86). This finding suggests that p300 and CBP may have some independent partners, which is consistent with the observation that p300 cannot compensate for the lack of CBP and vice versa (86). In agreement with this study is a recent report showing that p300 and CBP play distinct roles in retinoic acid-induced F9 cell phenotypes (38). p300 appears to be important for retinoic acid-induced differentiation, whereas CBP mediates retinoic acid-induced apoptosis (38). More recently, p53 was shown to be stabilized specifically by p300 but not CBP after ionizing irradiation using p300- or CBP-deficient MCF-7 cell lines (88). In contrast, using either the MCF-7 cell lines or p300- or CBP-deficient H1299 cells, we found that p73-mediated transcription was significantly impaired, though to different degrees, in the absence of either p300 or CBP (Fig. 7A and B). However, the inhibition of p73-mediated apoptosis was less dramatic in the CBP-deficient cells than in the p300-deficient cells (Fig. 7C and D). This difference suggests that p73-induced apoptosis may be regulated through mechanisms aside from its transcription activity and that p300 may be more important than CBP in regulating p73-dependent apoptosis. This question remains to be further investigated with the availability of cultured p300−/− and CBP−/− cell lines (86). Therefore, p300 and CBP may preferentially cooperate with p73 or p53 in mediating specific cellular processes.
In searching for p73 regulators (15, 91), it was found that p73 bound to p53's natural inhibitor MDM2 (60) and that this interaction reduced p73-mediated transcription in vivo. When overexpressed in cells, p73 was also able to induce the expression of MDM2 (91), which is a p53 target gene (82). Surprisingly, MDM2, though promoting p53 degradation (28, 45), failed to do so for the p73 protein (15, 91). Instead, it disrupted the interaction of p73 but not p53 with p300/CBP (91). These results indicate that MDM2 regulates p73 function through a mechanism distinct from that for p53 regulation, although p73 and p53 shares this MDM2 negative regulatory feedback loop. Recently, c-Abl was reported to up-regulate p73 but not p53 by phosphorylating its tyrosine residue 90 in response to γ irradiation or cisplatin treatment (1, 22, 89). This is also different from p53, which is activated by ATM (ataxia telangiectasia mutated) through serine 15 phosphorylation after γ irradiation (71, 73). Moreover, p73 does not appear to respond to UV-induced DNA damage (22, 33). Thus, despite the functional similarity between p73 and p53 in the downstream events (references 31, 33, and 91 and this study), p73 and p53 are regulated through different mechanisms in response to DNA damage signals. These differences may account for the distinct roles of p73 and p53 in cell growth and homeostasis. Identification of other upstream signals that regulate p73 functions is crucial for a better understanding its biological role.
ACKNOWLEDGMENTS
X. Li and A. Miller contributed equally to this study.
We thank William G. Kaelin, Tsu-Pan Yao, David Livingston, Kristen Walker, Hongwe Chen, Ronald Evans, Wei Gu, Robert Roeder, James Lundblad, Jean-Rene Cardinaux, Brian Elenbass, and Yang Shi for generously providing some of the reagents used in this study. We thank Steve Mansoor and David Farrens for assistance with fluorophotometer analysis, and we thank members in the laboratories of Hua Lu and Matt Thayer for discussion.
R. Goodman, W. Yuan, and R. P. S. Kwok were supported by NIH grants. This work was supported partly by grants to H. Lu from the American Cancer Society (RPG-98-191-01-CBE), NIH (R01 CA 79721), Medical Research Foundation of Oregon, Oregon Cancer Center, and Oregon division of the American Cancer Society.
REFERENCES
- 1.Agami R, Blandino G, Oren M, Shaul Y. Interaction of c-Abl and p73α and their collaboration to induce apoptosis. Nature. 1999;399:809–813. doi: 10.1038/21697. [DOI] [PubMed] [Google Scholar]
- 2.Avantaggiati M L, Carbone M, Graessmann A, Nakatani Y, Howard B, Levine A C. The SV40 large T antigen and adenovirus E1a oncoproteins interact with distinct isoforms of the transcriptional co-activator, p300. EMBO J. 1996;15:2236–2248. [PMC free article] [PubMed] [Google Scholar]
- 3.Avantaggiati M L, Ogryzko V, Gardner K, Giordano A, Levine A S, Kelly K. Recruitment of p300/CBP in p53-dependent signal pathways. Cell. 1997;89:1175–1184. doi: 10.1016/s0092-8674(00)80304-9. [DOI] [PubMed] [Google Scholar]
- 4.Arany Z, Newsome D, Oldread E, Livingston D M, Eckner R. A family of transcriptional adaptor proteins targeted by the E1A oncoprotein. Nature. 1995;374:81–84. doi: 10.1038/374081a0. [DOI] [PubMed] [Google Scholar]
- 5.Bannister A J, Kouzarides T. The CBP co-activator is a histone acetyltransferase. Nature. 1996;384:641–643. doi: 10.1038/384641a0. [DOI] [PubMed] [Google Scholar]
- 6.Bargonetti J, Manfredi J J, Chen X, Marshak D R, Prives C. A proteolytic fragment from the central region of p53 has marked sequence-specific DNA-binding activity when generated from wild-type but not from oncogenic mutant p53 protein. Genes Dev. 1993;7:2565–2574. doi: 10.1101/gad.7.12b.2565. [DOI] [PubMed] [Google Scholar]
- 7.Bex F, Yin M J, Burny A, Gaynor R B. Differential transcriptional activation by human T-cell leukemia virus type 1 Tax mutants is mediated by distinct interactions with CREB binding protein and p300. Mol Cell Biol. 1998;18:2392–2405. doi: 10.1128/mcb.18.4.2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bhattacjarua S R, Eclmer, Grossman S, Oldread E, Arany Z, D'Andrea A, Livingston D M. Cooperation of Stat2 and p300/CBP in signaling induced by interferon-alpha. Nature. 1996;383:344–347. doi: 10.1038/383344a0. [DOI] [PubMed] [Google Scholar]
- 9.Bian J, Sun Y. P53CP, a putative p53 competing protein that specifically binds to the consensus p53 DNA binding sites: a third member of the p53 family? Proc Natl Acad Sci USA. 1997;94:14753–14758. doi: 10.1073/pnas.94.26.14753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Buckbinder L, Talbott R, Valesco-Miguel S, Takenaka I, Faha B, Seizinger B R, Kley N. Induction of the growth inhibitor IGF-binding protein 3 by p53. Nature. 1995;377:646–649. doi: 10.1038/377646a0. [DOI] [PubMed] [Google Scholar]
- 11.Chakravarti D, LaMorte V J, Nelson M C, Nakajima T, Schulman I G, Juguilon H, Monteminy M, Evans R M. Role of CBP/p300 in nuclear receptor signaling. Nature. 1996;383:99–103. doi: 10.1038/383099a0. [DOI] [PubMed] [Google Scholar]
- 12.Cho Y, Gorina S, Jeffrey P D, Pavietich N P. Crystal structure of a p53 tumor suppressor-DNA complex: understanding tumorigenic mutations. Science. 1994;265:346–355. doi: 10.1126/science.8023157. [DOI] [PubMed] [Google Scholar]
- 13.Chrivia J C, Kwok R P, Lamb N, Hagiwara M, Montminy M R, Goodman R H. Phosphorylated CREB binds specifically to the nuclear protein CBP. Nature. 1993;365:855–859. doi: 10.1038/365855a0. [DOI] [PubMed] [Google Scholar]
- 14.Dignam J D, Lebovitz R M, Roeder R G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dobbelstein M, Wienzek S, Konig C, Roth J. Inactivation of the p53-homologue p73 by the mdm2-oncoprotein. Oncogene. 1999;18:2101–2106. doi: 10.1038/sj.onc.1202512. [DOI] [PubMed] [Google Scholar]
- 16.Donehower L A, Harvey M, Slagle B L, McArthur M J, Montgomery C A, Jr, Butel J S, Bradley A. Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature. 1992;356:215–221. doi: 10.1038/356215a0. [DOI] [PubMed] [Google Scholar]
- 17.Dulic V, Kaufmann W K, Lees S J, Tisty T D, Lees E, Harper J W, Elledge S J, Reed S I. p53-dependent inhibition of cyclin-dependent kinase activities in human fibroblasts during radiation-induced G1 arrest. Cell. 1994;76:1013–1023. doi: 10.1016/0092-8674(94)90379-4. [DOI] [PubMed] [Google Scholar]
- 18.Eckner R, Ewen M E, Newsome D, Gerdes M, DeCaprio J A, Lawrence J B, Livingston D M. Molecular cloning and functional analysis of the adenovirus E1A-associated 300-kD protein (p300) reveals a protein with properties of a transcriptional adaptor. Genes Dev. 1994;7:869–884. doi: 10.1101/gad.8.8.869. [DOI] [PubMed] [Google Scholar]
- 19.El-Deiry W S, Kern S E, Pietenpol J A, Kinzler K W, Vogelstein B. Definition of a consensus binding site for p53. Nat Genet. 1992;1:45–49. doi: 10.1038/ng0492-45. [DOI] [PubMed] [Google Scholar]
- 20.El-Deiry W S, Tokino T, Velculescu V E, Levy D B, Parsons R, Trent J M, Lin D, Mercer W E, Kinzler K W, Vogelstein B. WAF1, a potential mediator of p53 tumor suppression. Cell. 1993;75:817–825. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- 21.Fields S, Jang S K. Presence of a potent transcription activating sequence in the p53 protein. Science. 1990;249:1046–1049. doi: 10.1126/science.2144363. [DOI] [PubMed] [Google Scholar]
- 22.Gong J G, Costanzo A, Yang H Q, Melino G, Kaelin W G, Jr, Levrero M, Wang J Y. The tyrosine kinase c-Abl regulates p73 in apoptotic response to cisplatin-induced DNA damage. Nature. 1999;399:806–809. doi: 10.1038/21690. [DOI] [PubMed] [Google Scholar]
- 23.Gottlieb T M, Oren M. p53 in growth control and neoplasia. Biochim Biophys Acta Gene Struct Expr. 1996;1287:77–102. doi: 10.1016/0304-419x(95)00019-c. [DOI] [PubMed] [Google Scholar]
- 24.Grossman S R, Perez M, Kung A L, Joseph M, Mansur C, Xiao Z, Kumar S, Howley P M, Livingston D M. p300/MDM2 complexes participate in MDM2-mediated p53 degradation. Mol Cell. 1998;2:405–415. doi: 10.1016/s1097-2765(00)80140-9. [DOI] [PubMed] [Google Scholar]
- 25.Gu W, Roeder R G. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell. 1997;90:595–606. doi: 10.1016/s0092-8674(00)80521-8. [DOI] [PubMed] [Google Scholar]
- 26.Gu W, Shi X L, Roeder R G. Synergistic activation of transcription by CBP and p53. Nature. 1997;387:819–823. doi: 10.1038/42972. [DOI] [PubMed] [Google Scholar]
- 27.Harlow E, Lane D. Antibodies: a laboratory manual. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1988. pp. 519–522. [Google Scholar]
- 28.Haupt Y, Maya R, Kazaza A, Oren M. Mdm2 promotes the rapid degradation of p53. Nature. 1997;387:296–299. doi: 10.1038/387296a0. [DOI] [PubMed] [Google Scholar]
- 29.Hermeking H, Lengauer C, Polyak K, He T, Zhang L, Thiagalingam S, Kinzler K W, Vogelstein B. 14-3-3ς is a p53-regulated inhibitor of G2/M progression. Mol Cell. 1997;1:3–11. doi: 10.1016/s1097-2765(00)80002-7. [DOI] [PubMed] [Google Scholar]
- 30.Hupp T R, Lane D P. Allosteric activation of latent p53 tetramers. Curr Biol. 1994;4:865–875. doi: 10.1016/s0960-9822(00)00195-0. [DOI] [PubMed] [Google Scholar]
- 31.Jost C A, Marin C M, Kaelin W G., Jr p73 is a human p53-related protein that can induce apoptosis. Nature. 1997;389:191–194. doi: 10.1038/38298. [DOI] [PubMed] [Google Scholar]
- 32.Kadonaga J T. Eukaryotic transcription: an interlaced network of transcription factors and chromatin-modifying machines. Cell. 1998;92:307–313. doi: 10.1016/s0092-8674(00)80924-1. [DOI] [PubMed] [Google Scholar]
- 33.Kaghad M, Bonnet H, Yang A, Creancier L, Biscan J, Valent A, Minty A, Chalon P, Lelias J, Dumont X, Ferrara P, McKeon F, Caput D. Monoallelically expressed gene related to p53 at 1p36, a region frequently deleted in neuroblastoma and other human cancers. Cell. 1997;90:809–819. doi: 10.1016/s0092-8674(00)80540-1. [DOI] [PubMed] [Google Scholar]
- 34.Kamei Y, Xu L, Heinzel T, Torchia J, Kurokawa R, Gloss B, Lin S, Heyman R A, Rose D W, Glass C K, Rosenfeld M G. A CBP integrator complex mediates transcriptional activation and AP-1 inhibition by nuclear receptors. Cell. 1996;85:403–414. doi: 10.1016/s0092-8674(00)81118-6. [DOI] [PubMed] [Google Scholar]
- 35.Kapoor M, Lozano G. Functional activation of p53 via phosphorylation following DNA damage by UV but not gamma radiation. Proc Natl Acad Sci USA. 1998;95:2834–2837. doi: 10.1073/pnas.95.6.2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kastan M B, Zhan Q, El-Deiry W S, Carrier F, Jacks T, Walsh W V, Plunkett B S, Vogelstein B, Fornace A J., Jr A mammalian cell cycle checkpoint pathway utilizing p53 and GADD45 is defective in ataxia-telangiectasia. Cell. 1992;71:587–597. doi: 10.1016/0092-8674(92)90593-2. [DOI] [PubMed] [Google Scholar]
- 37.Kawasaki H, Ohkawa J, Tanishige N, Yoshinari K, Murata T, Yokoyama K K, Taira K. Selection of the best target site for ribozyme-mediated cleavage within a fusion gene for adenovirus E1A-associated 300 kDa protein (p300) and luciferase. Nucleic Acids Res. 1996;24:3010–3016. doi: 10.1093/nar/24.15.3010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kawasaki H, Eckner R, Yao T-P, Taira K, Chiu R, Livingston D M, Yokoyama K K. Distinct roles of the co-activators p300 and CBP in retinoic-acid-induced F9-cell differentiation. Nature. 1998;393:284–289. doi: 10.1038/30538. [DOI] [PubMed] [Google Scholar]
- 39.Kawasaki H, Song J, Eckner R, Ugal H, Chiu R, Taira K, Shi Y, Jones N, Yokoyama K K. p300 and ATF-2 are components of the DRF complex, which regulates retinoic acid- and E1A-mediated transcription of the c-jun gene in F9 cells. Genes Dev. 1998;12:233–245. doi: 10.1101/gad.12.2.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kee B L, Arias J, Montminy M R. Adaptor-mediated recruitment of RNA polymerase II to a signal-dependent activator. J Biol Chem. 1996;271:2373–2375. doi: 10.1074/jbc.271.5.2373. [DOI] [PubMed] [Google Scholar]
- 41.Keller D, Zeng X Y, Li X R, Kapoor M, Iordanov M S, Taya Y, Lozano G, Magun B, Lu H. The p38MAPK inhibitor SB203580 alleviates ultraviolet-induced phosphorylation at serine 389 but not serine 15 and activation of p53. Biochem Biophys Res Commun. 1999;261:464–471. doi: 10.1006/bbrc.1999.1023. [DOI] [PubMed] [Google Scholar]
- 42.Ko J L, Prives C. p53: puzzle and paradigm. Genes Dev. 1996;10:1054–1072. doi: 10.1101/gad.10.9.1054. [DOI] [PubMed] [Google Scholar]
- 43.Kuo M H, Zhou J, Jambeck P, Churchill M E, Allis C D. Histone acetyltransferase activity of yeast Gcn5p is required for the activation of target genes in vivo. Genes Dev. 1998;12:627–639. doi: 10.1101/gad.12.5.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kwok R P S, Lundblad J R, Chrivia J C, Richards J P, Bachinger H P, Brennan R G, Roberts S G E, Green M R, Goodman R H. Nuclear protein CBP is a coactivator for the transcription factor CREB. Nature. 1994;370:223–227. doi: 10.1038/370223a0. [DOI] [PubMed] [Google Scholar]
- 45.Kubbutat M H, Jones S N, Vousden K H. Regulation of p53 stability by mdm2. Nature. 1997;387:299–303. doi: 10.1038/387299a0. [DOI] [PubMed] [Google Scholar]
- 46.Lee J S, See R H, Deng T, Shi Y. Adenovirus E1A downregulates c-Jun- and JunB-mediated transcription by targeting their coactivator p300. Mol Cell Biol. 1996;16:4312–4326. doi: 10.1128/mcb.16.8.4312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lee J S, Zhang X, Shi Y. Relief of YY1 transcriptional repression by adenovirus E1A is mediated by E1A-associated protein p300. Genes Dev. 1995;9:1188–1198. doi: 10.1101/gad.9.10.1188. [DOI] [PubMed] [Google Scholar]
- 48.Levine A J. p53, the cellular gatekeeper for growth and division. Cell. 1997;88:323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- 49.Levine A J, Chang A, Dittmer D, Notterman D A, Silver A, Thorn K, Welsh D, Wu M. The p53 tumor suppressor gene. J Lab Clin Med. 1994;124:817–823. [PubMed] [Google Scholar]
- 50.Lewin B. Commitment and activation at pol II promoters: a tail of protein-protein interactions. Cell. 1990;61:1161–1164. doi: 10.1016/0092-8674(90)90675-5. [DOI] [PubMed] [Google Scholar]
- 51.Lill N L, Grossman S R, Ginsberg D, DeCaprio J, Livingston D M. Binding and modulation of p53 by p300/CBP coactivators. Nature. 1997;387:823–827. doi: 10.1038/42981. [DOI] [PubMed] [Google Scholar]
- 52.Lin J, Chen J, Elenbaas B, Levine A J. Several hydrophobic amino acids in the p53 amino-terminal domain are required for transcriptional activation, binding to MDM2 and the adenovirus 5 E1B 55-kd protein. Genes Dev. 1994;8:1235–1246. doi: 10.1101/gad.8.10.1235. [DOI] [PubMed] [Google Scholar]
- 53.Liu L, Scolnick D M, Trievel R C, Zhang H B, Marmorstein R, Halazonetis T D, Berger S L. p53 sites acetylated in vitro by PCAF and p300 are acetylated in vivo in response to DNA damage. Mol Cell Biol. 1999;19:1202–1209. doi: 10.1128/mcb.19.2.1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lu H, Levine A J. Human TAF-31 is a transcriptional coactivator of the p53 protein. Proc Natl Acad Sci USA. 1995;92:5154–5158. doi: 10.1073/pnas.92.11.5154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lu H, Taya Y, Ikeda M, Levine A J. Phosphorylation of p53 at serine 389 is responsive uniquely to UV- but not to gamma- and etoposide-induced DNA damage. Proc Natl Acad Sci USA. 1998;95:6399–6402. doi: 10.1073/pnas.95.11.6399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lundblad J R, Kwok R P S, Laurance M E, Harter M L, Goodman R H. Adenoviral E1A-associated protein p300 as a functional homologue of the transcriptional coactivator CBP. Nature. 1995;374:85–88. doi: 10.1038/374085a0. [DOI] [PubMed] [Google Scholar]
- 57.Marin M C, Jost C A, Irwin M S, DeCaprio J A, Caput D, Kaelin W G., Jr Viral oncoproteins discriminate between p53 and the p53 homolog p73. Mol Cell Biol. 1998;18:6316–6324. doi: 10.1128/mcb.18.11.6316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mills A A, Zheng B, Wang X J, Vogel H, Roop D R, Bradley A. p63 is a p53 homologue required for limb and epidermal morphogenesis. Nature. 1999;398:708–713. doi: 10.1038/19531. [DOI] [PubMed] [Google Scholar]
- 59.Miyashita T, Reed J C. Tumor suppressor p53 is a direct transcriptional activator of the human bax gene. Cell. 1995;80:293–299. doi: 10.1016/0092-8674(95)90412-3. [DOI] [PubMed] [Google Scholar]
- 60.Momand J, Zambetti G P, Olson D C, George D, Levine A J. The mdm-2 oncogene product forms a complex with the p53 protein and inhibits p53-mediated transactivation. Cell. 1992;69:1237–1245. doi: 10.1016/0092-8674(92)90644-r. [DOI] [PubMed] [Google Scholar]
- 61.Nakajima T, Uchida C, Anderson S F, Lee C G, Hurwitz J, Parvin J D, Montminy M R. RNA helicase A mediates association of CBP with RNA polymerase II. Cell. 1997;90:1107–1112. doi: 10.1016/s0092-8674(00)80376-1. [DOI] [PubMed] [Google Scholar]
- 62.Ogryzko V V, Kotani T, Zhang X, Schiltz R L, Howard T, Yang X, Howard B H, Qin Y, Nakatani Y. Histone-like TAFs within the PCAF histone acetylase complex. Cell. 1998;94:35–44. doi: 10.1016/s0092-8674(00)81219-2. [DOI] [PubMed] [Google Scholar]
- 63.Ogryzko V V, Schiltz R L, Russanova V, Howard B H, Nakatani Y. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell. 1996;87:953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- 64.Oliner J D, Anderson J M, Hansen S K, Zhou S, Tjian R. SREBP transcriptional activity is mediated through an interaction with the CREB-binding protein. Genes Dev. 1996;10:2903–2911. doi: 10.1101/gad.10.22.2903. [DOI] [PubMed] [Google Scholar]
- 65.Osada M, Ohba M, Kawahara C, Ishioka C, Kanamaru R, Katoh I, Ikawa Y, Nimura Y, Nakagawara A, Obinata M, Ikawa S. Cloning and functional analysis of human p51, which structurally and functionally resembles p53. Nat Med. 1998;4:839–843. doi: 10.1038/nm0798-839. [DOI] [PubMed] [Google Scholar]
- 66.Pavletich N P, Chambers K A, Pabo C O. The DNA binding domain of p53 contains the four conserved regions and the major mutation hotspots. Genes Dev. 1993;7:2556–2564. doi: 10.1101/gad.7.12b.2556. [DOI] [PubMed] [Google Scholar]
- 67.Raycroft L, Wu H, Lozano G. Transcriptional activation by wild-type but not transforming mutants of the p53 anti-oncogene. Science. 1990;249:1049–1051. doi: 10.1126/science.2144364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sakaguchi K, Herrera J E, Saito S, Miki T, Bustin M, Vassilev A, Anderson C W, Appella E. DNA damage activates p53 through a phosphorylation-acetylation cascade. Genes Dev. 1997;12:2831–2841. doi: 10.1101/gad.12.18.2831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Schmale H, Bamberger C. A novel protein with strong homology to the tumor suppressor p53. Oncogene. 1997;15:1363–1367. doi: 10.1038/sj.onc.1201500. [DOI] [PubMed] [Google Scholar]
- 70.Shi Y, Mello C. A CBP/p300 homolog specifies multiple differentiation pathways in Caenorhabditis elegans. Genes Dev. 1998;12:943–955. doi: 10.1101/gad.12.7.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Shieh S, Ikeda M, Taya Y, Prives C. DNA damage-induced phosphorylation of p53 alleviates inhibition by MDM2. Cell. 1997;91:325–334. doi: 10.1016/s0092-8674(00)80416-x. [DOI] [PubMed] [Google Scholar]
- 72.Shikama N, Lyon L, La Thangue N B. The p300/CBP family: integrating signals with transcription factors and chromatin. Trends Cell Biol. 1997;7:230–236. doi: 10.1016/S0962-8924(97)01048-9. [DOI] [PubMed] [Google Scholar]
- 73.Siliciano J D, Canman C E, Taya Y, Sakaguchi K, Appella E, Kastan M B. DNA damage induces phosphorylation of the amino terminus of p53. Genes Dev. 1998;11:3471–3481. doi: 10.1101/gad.11.24.3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Struhl K. Histone acetylation and transcriptional regulatory mechanisms. Genes Dev. 1998;19:599–606. doi: 10.1101/gad.12.5.599. [DOI] [PubMed] [Google Scholar]
- 75.Thut C J, Chen J L, Klemin R, Tjian R. P53 transcriptional activation mediated by coactivators TAFII40 and TAFII60. Science. 1995;267:100–104. doi: 10.1126/science.7809597. [DOI] [PubMed] [Google Scholar]
- 76.Tokino T, Thiagalingam S, El-Deiry W S, Waldman T, Kinzler K W, Vogelstein B. p53 tagged sites from human genomic DNA. Hum Mol Genet. 1994;3:1537–1542. doi: 10.1093/hmg/3.9.1537. [DOI] [PubMed] [Google Scholar]
- 77.Trink B, Okami K, Wu L, Sriuranpong V, Jen J, Sidransky D. A new human p53 homologue. Nat Med. 1998;4:747–748. doi: 10.1038/nm0798-747. [DOI] [PubMed] [Google Scholar]
- 78.Walker K K, Levine A J. Identification of a novel p53 functional domain that is necessary for efficient growth suppression. Proc Natl Acad Sci USA. 1996;93:15335–15340. doi: 10.1073/pnas.93.26.15335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wang L, Liu L, Berger S L. Critical residues for histone acetylation by Gcn5, functioning in Ada and SAGA complexes, are also required for transcriptional function in vivo. Genes Dev. 1998;12:640–653. doi: 10.1101/gad.12.5.640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wang Y, Reed M, Wang P, Stenger J E, Mayr G, Anderson M E, Schwedes J F, Tegtmeyer P. p53 domains: identification and characterization of two autonomous DNA-binding regions. Genes Dev. 1993;7:2575–2586. doi: 10.1101/gad.7.12b.2575. [DOI] [PubMed] [Google Scholar]
- 81.Whyte P, Williamson N M, Harlow E. Cellular targets for transformation by the adenovirus E1A proteins. Cell. 1989;56:67–75. doi: 10.1016/0092-8674(89)90984-7. [DOI] [PubMed] [Google Scholar]
- 82.Wu X, Bayle J H, Olson D, Levine A J. The p53-mdm-2 autoregulatory feedback loop. Genes Dev. 1993;7:1126–1132. doi: 10.1101/gad.7.7a.1126. [DOI] [PubMed] [Google Scholar]
- 83.Yang A, Kaghad M, Wang Y, Gillett E, Fleming M D, Dotsch V, Andrews N C, Caput D, McKeon F. p63, a p53 homolog at 3q27-29, encodes multiple products with transactivating, death-inducing, and dominant-negative activities. Mol Cell. 1998;2:305–316. doi: 10.1016/s1097-2765(00)80275-0. [DOI] [PubMed] [Google Scholar]
- 84.Yang A, Schweitzer R, Sun D, Kaghad M, Walker N, Bronson R T, Tabin C, Sharpe A, Caput D, Crum C, McKeon F. p63 is essential for regenerative proliferation in limb, craniofacial and epithelial development. Nature. 1999;398:714–718. doi: 10.1038/19539. [DOI] [PubMed] [Google Scholar]
- 85.Yao T P, Ku G, Zhou N, Scully R, Livingston D M. The nuclear hormone receptor coactivator SRC-1 is a specific target of p300. Proc Natl Acad Sci USA. 1996;93:10626–10631. doi: 10.1073/pnas.93.20.10626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Yao T P, Oh S P, Fuchs M, Zhou N D, Ch'ng L E, Newsome D, Bronson R T, Li E, Livingston D M, Eckner R. Gene dosage-dependent embryonic development and proliferation defects in mice lacking the transcriptional integrator p300. Cell. 1998;93:361–372. doi: 10.1016/s0092-8674(00)81165-4. [DOI] [PubMed] [Google Scholar]
- 87.Yuan W, Condorelli G, Caruso M, Feisani A, Giordano A. Human p300 protein is a coactivator for the transcription factor MyoD. J Biol Chem. 1996;271:9009–9013. doi: 10.1074/jbc.271.15.9009. [DOI] [PubMed] [Google Scholar]
- 88.Yuan Z M, Huang Y, Ishiko T, Nakada S, Utsugisawa T, Shioya H, Utsugisawa Y, Yokoyama K, Weichselbaum R, Shi Y, Kufe D. Role for p300 in stabilization of p53 in the response to DNA damage. J Biol Chem. 1999;274:1883–1886. doi: 10.1074/jbc.274.4.1883. [DOI] [PubMed] [Google Scholar]
- 89.Yuan Z M, Shioya H, Ishiko T, Sun X, Gu J, Huang Y Y, Lu H, Kharbanda S, Weichselbaum R, Kufe D. p73 is regulated by tyrosine kinase c-Abl in the apoptotic response to DNA damage. Nature. 1999;399:814–817. doi: 10.1038/21704. [DOI] [PubMed] [Google Scholar]
- 90.Zeng X Y, Levine A J, Lu H. Non-p53 p53RE binding protein, a human transcription factor functionally analogous to p53. Proc Natl Acad Sci USA. 1998;95:6681–6686. doi: 10.1073/pnas.95.12.6681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zeng X Y, Chen L H, Jost C A, Maya R, Keller D, Wang X, Kaelin W C, Jr, Oren M, Chen J D, Lu H. MDM2 suppresses p73 function without promoting p73 degradation. Mol Cell Biol. 1999;19:3257–3266. doi: 10.1128/mcb.19.5.3257. [DOI] [PMC free article] [PubMed] [Google Scholar]