Abstract
Processing of intron-encoded box C/D small nucleolar RNAs (snoRNAs) in metazoans through both the splicing-dependent and -independent pathways requires the conserved core motif formed by boxes C and D and the adjoining 5′-3′-terminal stem. By comparative analysis, we found that five out of six intron-encoded box C/D snoRNAs in yeast do not possess a canonical terminal stem. Instead, complementary regions within the flanking host intron sequences have been identified in all these cases. Here we show that these sequences are essential for processing of U18 and snR38 snoRNAs and that they compensate for the lack of a canonical terminal stem. We also show that the Rnt1p endonuclease, previously shown to be required for the processing of many snoRNAs encoded by monocistronic or polycistronic transcriptional units, is not required for U18 processing. Our results suggest a role of the complementary sequences in the early recognition of intronic snoRNA substrates and point out the importance of base pairing in favoring the communication between boxes C and D at the level of pre-snoRNA molecules for efficient assembly with snoRNP-specific factors.
The nucleolar maturation of eukaryotic rRNA is assisted by a large population of small nucleolar ribonucleoprotein particles (snoRNPs), consisting of a specific small nucleolar RNA (snoRNA) and a set of associated proteins (reviewed in references 43, 44, and 50). Few snoRNPs are required for nucleolytic processing steps of the pre-rRNA, whereas most of them guide the site-specific 2′-O-ribose methylation or pseudouridylation of rRNAs (reviewed in references 43, 44, and 50). These modifications are directed by two distinct classes of snoRNAs defined on the basis of conserved sequence and structural elements. The strict conservation of these elements makes them likely to function as binding sites for proteins which are common components of the snoRNPs in each class. Methylation guide snoRNAs belong to the box C/D class and contain conserved boxes C and D near their 5′ and 3′ ends, respectively, which are frequently brought together by short complementary sequences located in close proximity to the boxes (2, 3). Box C/D snoRNAs may also contain imperfect copies of the C and D boxes, referred to as boxes C′ and D′ (22, 46). The methylation guide snoRNAs direct 2′-O-ribose methylation by base pairing with the rRNA for 10 to 21 nucleotides (nt) immediately 5′ of box D and/or D′. The residue targeted for methylation invariably pairs with the fifth nucleotide upstream of box D or D′ (21, 32). Pseudouridylation guide RNAs belong to the box H/ACA class defined by an evolutionarily conserved “hairpin-hinge-hairpin-tail” secondary structure and by the conserved box H within the hinge region and box ACA 3 nt from the 3′ end of the snoRNA (4, 5, 16). Box H/ACA snoRNAs direct pseudouridylation by forming two short base-pairing interactions with rRNA sequences that flank the target uridine, leaving this residue unpaired within a pseudouridylation pocket located 14 to 17 nucleotides upstream of box H or ACA (15, 31).
snoRNAs of both classes are synthesized by different expression strategies depending on their genomic arrangement (reviewed in references 44 and 50). Most yeast snoRNAs and a few vertebrate ones are derived from independent transcription units. In yeast and plants, multiple different snoRNAs can be generated from polycistronic transcripts by endonucleolytic cleavage within spacer regions and subsequent maturation by exonucleases (11, 12, 25, 35, 39). The large majority of vertebrate snoRNAs and seven yeast ones, six belonging to the C/D box family and one belonging to the H/ACA box family, are intron encoded. These snoRNAs are present in genes encoding proteins involved in ribosome assembly or in nucleolar processes (28) and, in a few cases, in noncoding host genes belonging to the 5′-terminal oligopyrimidine gene family (50). This peculiar genomic location strongly suggests a form of coregulation of the host genes with the snoRNAs. Intron-encoded snoRNAs can be matured via a major splicing-dependent pathway and a secondary splicing-independent one. In the splicing-dependent pathway, exonucleases digest the flanking sequences of the debranched host intron and produce the correct 5′ and 3′ ends of the snoRNA (8, 9, 20, 26, 33, 35, 45–47). The splicing-independent pathway involves endonucleolytic cleavages within the host intron followed by exonucleolytic maturation, similar to processing of polycistronic pre-snoRNAs (6, 7, 33, 35, 47).
Regardless of the genomic arrangement, correct processing and accumulation of both box C/D and box H/ACA snoRNAs appear to depend on conserved structural elements located within their coding regions. Box H/ACA snoRNAs depend on both their signature secondary structure and the H and ACA boxes (4, 5, 16). Similarly, the processing and stability of box C/D snoRNAs both depend on the “terminal core motif” formed by the C and D boxes and the adjoining 5′-3′-terminal stem (7, 8, 19, 25, 48, 49, 51). These conserved structural elements act as assembly sites for class-specific snoRNP factors which protect pre-snoRNA molecules from exonucleolytic digestion, thus helping to specify the ends of mature snoRNAs. However, processing of polycistronic snoRNAs clearly depends also on the double-stranded structural elements within the spacer regions directing endonucleolytic cleavage by RNase III in yeast (11, 12, 39). Also, processing of several intron-encoded snoRNAs has been inferred to be influenced by context effects of neighboring intronic sequences (5, 6, 38), in one case through modulation of alternative interactions of the pre-snoRNA molecule with trans-acting factors (42). A correlation between the length of the host intron and the efficiency of the splicing-independent release of intronic snoRNAs in yeast has been described (33). Taken together, these observations suggest that processing of intron-encoded snoRNAs may depend not only on conserved structural features of the snoRNAs but also on nonconserved elements located within the host introns. So far, however, the contribution of nonconserved intronic elements to the processing of intron-encoded snoRNAs has not been investigated extensively.
In this study, we have identified two non-conserved complementary sequences within the host intron of the box C/D U18 snoRNA in the yeast Saccharomyces cerevisiae. By mutational analysis and the use of yeast strains carrying mutations in key processing enzymes, we show that host intron complementarity is essential for U18 processing. Our results indicate that such base-pairing interaction provides the structural information required for the initial recognition of the pre-snoRNA substrates, thus allowing their subsequent conversion into mature U18 molecules. Results from a comparative analysis suggest a general role of nonconserved complementary sequences in contributing to the processing efficiency of yeast intronic box C/D snoRNAs lacking a canonical terminal stem. Additionally, we show that the Rnt1p endonuclease, previously implicated in the processing of monocistronic and polycistronic snoRNAs, is not required for U18 processing.
MATERIALS AND METHODS
Strains and media.
Growth and handling of S. cerevisiae were done by standard techniques. The following strains were used: CH1462 (MATα ade2 ade3 leu2 ura3 his3 can1) (23), rat1-1 (MATα leu2-Δ1 ura3-52 his3-Δ200 rat1-1) (strain DAH18) (1), dbr1-Δ (MATα trp1-Δ1 his3-Δ200 leu2-Δ1 ura3-167 Δdbr1::HIS3) (strain KC99) (13), RNT1 (MATa his3 lys2 leu2-3,112 trp1 ura3-52 pep4 prb1 prc1 rnt1::HIS3 pRS316-RNT1), and rnt1-Δ (MATa his3 lys2 leu2-3,112 trp1 ura3-52 pep4 prb1 prc1 rnt1::HIS3) (both the RNT1 and rnt1-Δ strains were kindly provided by M. Ares [10]). For induction experiments, galactose at a final concentration of 2% was directly added to cultures grown in nonrepressive medium containing 2% raffinose and 0.1% glucose. The exonuclease-deficient strain rat1-1 was grown at 23°C until it reached mid-log phase and was then shifted to 37°C for 2 h prior to galactose induction.
Plasmid construction.
Plasmids pGALU18wt and pGALU18Cbs (47) were used as starting material to generate pGALU18wt/ΔS, pGALU18wt/RS, pGALU18wt/5′-3′S, pGALU18Cbs/ΔS, pGALU18Cbs/RS, and pGALU18Cbs/5′-3′S. All the mutations were introduced by PCR with Pfu polymerase (Stratagene). The PCR product obtained with oligonucleotides Δstem5′/F and E3′ was digested with HpaI and EcoRI and subsequently introduced into the corresponding sites of pGALU18 to obtain the ΔS derivatives. The PCR product obtained with oligonucleotides Rstem/long and E3′ was used for a second-step PCR with the oligonucleotide SK primer. The resulting fragment was digested with SpeI and EcoRI and then inserted into the corresponding sites of pGALU18 to obtain the RS derivatives. The same strategy was used to generate the 5′-3′S constructs, the first PCR being performed with oligonucleotides 5′-3′stem and E3′. Oligonucleotides U24HG/F and U24HG/B were used to amplify the full-length BEL1 gene in genomic DNA from the CH1462 strain. The PCR product was cloned into the SmaI site of the Bluescript KS vector to obtain pBSU24HG. A two-step amplification PCR strategy (40) was performed on this plasmid to introduce mutations in the intron of the BEL1 gene. Oligonucleotide U24HG-S, U24HG-H, 24S3/F, 24S3/B, 24S5/B, 24S5wt/F, or 24S5m/F was used to replace large regions of the U24 host intron sequence. The PCR products were digested with SmaI and HindIII and inserted into the corresponding sites of plasmid p416GAL1 (29) to obtain pGALU24wt-Stem and pGALU24m-Stem, respectively. The strategy described above was also used to amplify the full-length TEF4 gene, to introduce mutations in the intron of the gene, and to introduce a tag of 10 nt into the 3′ sequence of snR38. Oligonucleotides 38HG/F, 38HG-E/B, 38ΔS/F, 38ΔS/B, α38, and 38tag were used, and the PCR products were digested with BamHI and EcoRI and inserted into the corresponding sites of plasmid p416GAL1 to obtain pGAL38wt-tag and pGAL38ΔS-tag, respectively. All clones have been sequenced.
Oligonucleotides.
The sequences of the oligonucleotides used for the different cloning steps are as follows (5′-3′): Skprimer, CGCTCTAGAACTAGTGGATC; Δstem5′/F, TTGATTATTACTATACTTTTTTTCGCTTATGTG; Rstem/long, ATAGCACAGAGCAGAGTTAGTAATAATCAAATCTGTTATTTTTTTTT CC; 5′-3′stem, CGCTTTATCGAATGATGA; U24HG/F, ATGGCATCTAACGAAGTTTTAG; U24HG/B, TTAGTTAGCAGTCATAAC; 24HG-S, AGCCCGGGATGGCATCTAACGAAGTTTTAG; 24HG-H, ACAAGCTTTTAGTTAGCAGTCATAACTTGCC; 24S5wt/F, TCGAGTTAACTAATAATGATGGATTTGTGTATGCCATTCAAATGATGT; 24S5m/F, TCGAGTTAACTAATAATGATGGATTTGTGTATGCATGGCAAATGATGT; 24S5/B, AAATCCATCATTATTAGTTAACTCGATTGTCATCATATTCTATCATGG; 24S3/F, TACTCTATCATTATTAGTTATCGTTATGTCAAAATGGAAAC; 24S3/B, TAACGATAACTAATAATGATAGAGTAATGCTAAACCATTCATCAG; 38HG/F, CGGGATCCATGTCCCAAGGTACTTTA; 38HG-E/B, CGGAATTCCTTGTTGTATGGAATCAAACC; 38ΔS/F, GGCACGAGTAAAAAGAAGCTTTCATAATGATGAAA; 38ΔS/B, GCTTCTTTTTACTCGTGCCAAATAAACGAACGGG; 38TAG, TGTCTGAATGGGTAATAATAGTTAACGAGAGTATACTTGATATTTGTATTTCTGA; and α38, TATTACCCATTCAGACAGGG. The oligonucleotides used for RNA analyses by Northern hybridization or primer extension are as follows: anti-tag, TGCGGACTGCCTGGATGCCG; E3′, GCAAGCTTGTTGAACCATCTGAA; αU24, TCAGAGATCTTGGTGATAAT; 38 antitag, AATATCAAGTATACTCTC; 5.8S, TTTCGCTGCGTTCTTCATC; U5, CCTGTTTCTATGGAGACAACACCCGGATGGTTCTG; U3, GAAGAGTCAAAGAGTGAC; and αsnR54, GTTCTCTACAAGATCGTTTG.
RNA analysis.
RNA was extracted from exponentially growing cultures of S. cerevisiae by the hot-phenol method as previously described (47). Routinely, RNA concentrations were calibrated by absorbance at 260 nm and by ethidium bromide staining on formaldehyde gels and normalized by hybridization with U5 and U3 small nuclear RNA (snRNA) oligonucleotides. Primer extension analyses were performed as previously described (47); for the experiments in Fig. 3, a 10-fold molar excess of cold primer was included to allow a semiquantitative assessment of mRNA levels. For Northern blot analyses, typically 5 μg of total RNAs was electrophoresed on 6% polyacrylamide–7 M urea gels and electrotransferred to Amersham Hybond-N+ filters in 0.5× Tris-borate-EDTA (TBE) buffer for 3 h at 380 mA and 4°C. All hybridizations were carried out as previously described (46). Oligonucleotides (10 pmol) were routinely 5′ end labeled with 30 μCi of [γ-32P]ATP.
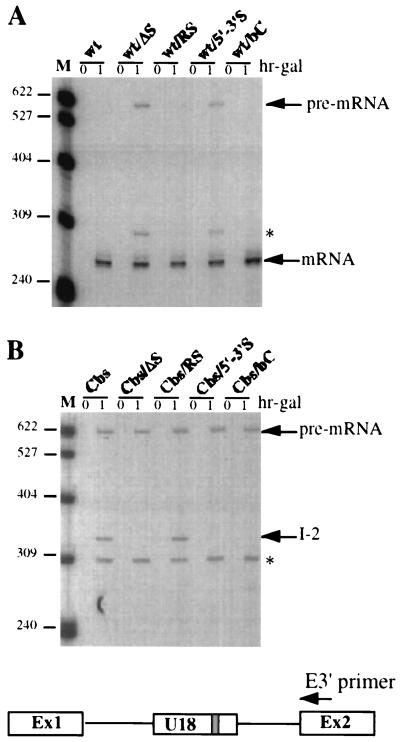
FIG. 3.
Analysis of mRNA production from the different U18 host pre-mRNAs. Total RNA extracted from strain CH1462 transformed with the indicated wt (A) and Cbs (B) constructs before or 1 h after galactose induction (hr-gal) was analyzed by primer extension with oligonucleotide E3′ complementary to the downstream exon of the plasmid-borne EFB1 gene (see Materials and Methods). A schematic representation of the EFB1 pre-mRNA is shown below, together with the position of the oligonucleotide used for the primer extension. The different elongation products are indicated on the side. The asterisk marks the position of a nonspecific stop within the U18 coding sequence, which is observed upon elongation of full-length pre-mRNA. Lanes M: pBR322 plasmid DNA, MspI digested.
Sequence analysis.
Analysis of snoRNA host intron features was performed on host introns whose sequences were available in databases. The presence of inverted repeats was investigated using the Compare software of the MacMolly program. The significance of the external inverted repeats was evaluated based on (i) the number of consecutive base pairs (≥8 nt; maximum, one mismatch) and (ii) the position with respect to the snoRNA coding sequence (contained within 40 nt from both the 5′ and 3′ ends, and not overlapping to the 5′ splice site and branchpoint regions). The potential intramolecular base-pairing interactions have been also checked by computer folding using the Mfold package (52).
RESULTS
Features of the U18 host intron: a poor canonical stem and a potential self-complementarity.
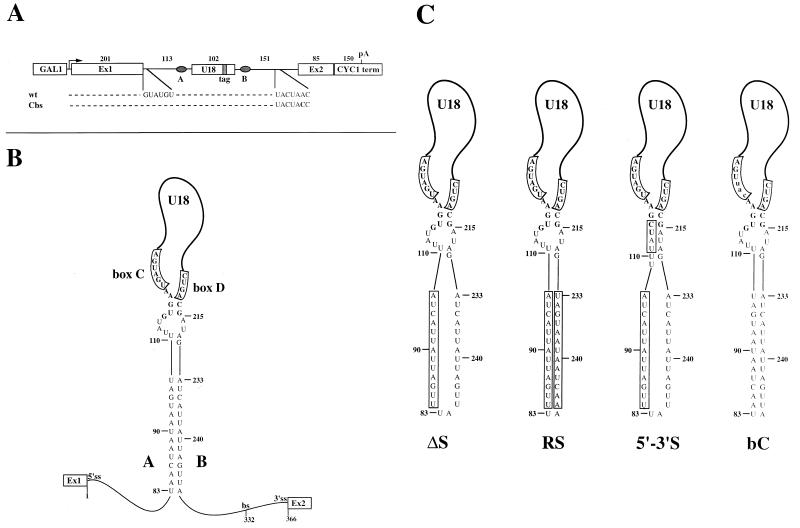
In S. cerevisiae, the U18 snoRNA is encoded within the single intron of the EFB1 gene and belongs to the box C/D class of methylation guide snoRNAs. This class is defined by the terminal core motif formed by the conserved boxes C and D and the terminal 5′-3′ stem (2, 3). This motif is required for both snoRNA processing and nucleolar localization (41, 44). Sequence analysis revealed that the yeast U18 terminal stem is very weak, being only 2 bp long (one being a G · U pair [Fig. 1B]). In addition, the adjacent intronic sequences do not display any potential to form continuously base-paired interactions. Instead, inspection of the host intron sequences, further upstream and downstream, revealed two 14-nt complementary sequences surrounding the U18 coding region (A and B in Fig. 1A). These sequence elements have the potential to form an intramolecular base-pairing interaction (Fig. 1B) (hereafter termed the external stem), as also predicted by computer folding of the U18 host intron by utilizing the MFold package (52).
FIG. 1.
Structures of EFB1 episomal constructs expressing the tagged U18 snoRNA. (A) Diagrammatic representation of the pGALU18wt plasmid, containing the transcriptional unit of the EFB1 gene between the GAL1 promoter and the CYC1 terminator. The Cbs construct is a mutant derivative which carries an A-to-C (shown in bold) substitution of the branch nucleotide, which abolishes splicing. The dark box within the U18 snoRNA coding region represents the tag sequence (47). Complementary sequences A and B are indicated. The lengths of the different portions of the constructs are indicated in nucleotides. pA indicates the polyadenylation site within the CYC1 terminator (term). (B) Representation of the U18 host intron secondary structure. The putative interaction between A and B sequences is shown along with the U18 box C/D motif formed by the C and D boxes and the adjoining 5′-3′-terminal stem. Nucleotides belonging to the U18 coding sequence are shown in bold. Numbering starts from the first nucleotide of the intron, and the positions of the 5′ splice site (5′ss), branch site (bs), and 3′ splice site (3′ss) are indicated. (C) Representation of the putative secondary structures of the different mutants used in this study; mutations are boxed. ΔS disrupts A-B pairing by inverting sequence A; RS restores pairing by inverting sequence B to make it complementary to the inverted A sequence; 5′-3′S lengthens the terminal stem complementarity; bC carries a mutation (lowercase letters) within conserved box C.
Host intron complementarity is required for snoRNA accumulation.
To understand in detail the role of the external stem and of the terminal stem sequences in U18 snoRNA processing, we undertook their mutational analysis. To investigate the role of the putative interaction between sequences A and B, we introduced mutations that altered the pairing within this region. Mutation ΔS (Fig. 1C) disrupts the pairing by inverting the A sequence so that no base pairing is possible and no new pairing potential with other sequences is created; mutation RS (Fig. 1C) restores pairing by inverting the B sequence to make it complementary to the inverted A sequence.
To distinguish the role played by the terminal stem from that of the potential external stem formed by A-B pairing in the U18 processing pathways, we generated mutant 5′-3′S (Fig. 1C) in the context of the ΔS mutation. This mutant has a 5′-3′-terminal stem 6 bp long, obtained by replacing the natural G · U pair with a C · G pair and by introducing an additional 4 bp.
Finally, the bC mutant (Fig. 1C) (47), carrying substitutions in the conserved box C that completely impair U18 processing, was also included as a negative control in the set of mutations analyzed during the course of this work.
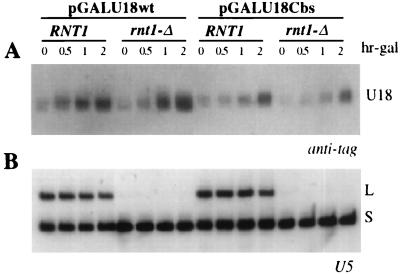
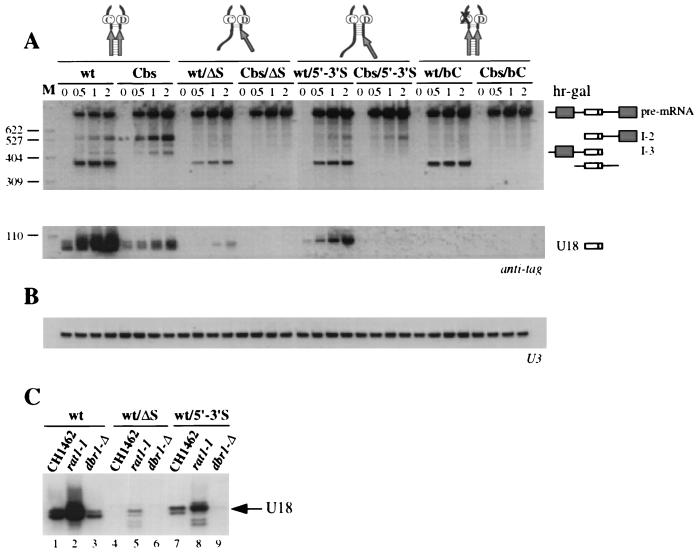
To test their effect on snoRNA accumulation, each mutation was introduced into plasmid pGALU18wt and into its derivative pGALU18Cbs, carrying a mutation of the branch nucleotide which abolishes splicing (Fig. 1A) (47). The comparison between the levels of snoRNA accumulated from these two precursors allows us to distinguish the contribution of the splicing-dependent pathway from that of the splicing-independent pathway to U18 snoRNA production (47). Each plasmid was transformed into the wild-type recipient strain CH1462 (23), and expression of the episomal EFB1 gene was induced by galactose addition. Aliquots were harvested at different times after transcriptional induction, and the accumulation of tagged U18 was assessed by Northern analysis (Fig. 2A).
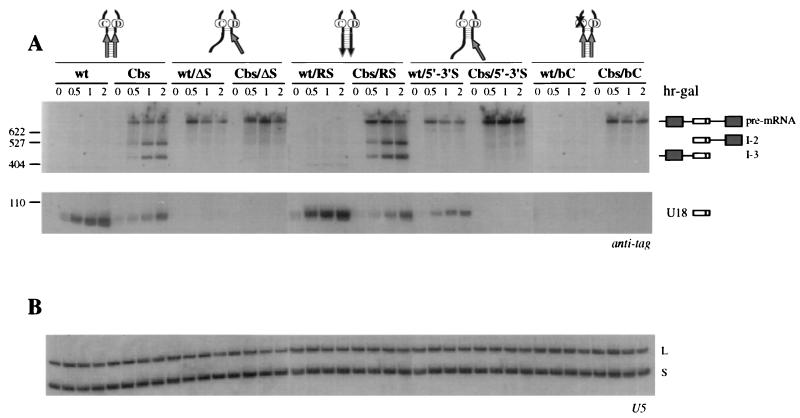
FIG. 2.
(A) Analysis of U18 processing in a wt strain. Total RNA was extracted from strain CH1462 carrying the indicated wt or Cbs constructs (schematically represented above the lanes) at the indicated times of galactose induction (expressed in hours above the lanes; hr-gal). Total RNA (5 μg) was resolved on a 6% acrylamide–7 M urea gel, electroblotted onto a nylon membrane, and hybridized with the anti-tag oligonucleotide. The different processing products are diagrammed on the side (exons are depicted as dark boxes, U18 is shown as an open box, and intronic sequences and the 5′ untranslated region are shown as lines). (B) Control hybridization to U5 snRNA. L and S indicate the long and short forms of U5 snRNA, respectively. The oligonucleotide used is indicated below each panel. Lane M: pBR322 plasmid DNA, MspI digested; molecular sizes are indicated on the left in nucleotides.
As previously described (47), production of U18 snoRNA from the Cbs precursor represented about 30% of the amount of snoRNA released from the wild-type (wt) precursor (Fig. 2A, compare wt and Cbs lanes) and led to accumulation of the I-2 and I-3 processing intermediates (Cbs lanes). Strikingly, elimination of the complementarity between sequences A and B completely abolished U18 snoRNA accumulation from both the wt/ΔS and the Cbs/ΔS precursors (wt/ΔS and Cbs/ΔS lanes). Also, the ΔS mutation triggered the accumulation of unspliced wt pre-mRNA (wt/ΔS lanes) and led to the disappearance of the I-2 and I-3 intermediates (Cbs/ΔS lanes). Restoring the complementarity in mutant RS fully recovered the tagged U18 processing from both the wt/RS and the Cbs/RS precursors (wt/RS and Cbs/RS lanes). The reason for the apparent higher efficiency of the processing pathway in the context of RS mutants was not investigated further. We conclude that complementarity within the U18 host intron is required for accumulation of the snoRNA from both the splicing-dependent and the splicing-independent pathways.
Interestingly, extension of the terminal stem was able to partially rescue accumulation of U18 from the wt precursor only (Fig. 2A, wt/5′-3′S lanes), whereas the splicing-independent release of the snoRNA from the Cbs precursor was still impaired (Cbs/5′-3′S lanes). We conclude that the splicing-dependent release of U18 becomes insensitive to host intron self-complementarity when the snoRNA is provided with a strong terminal stem motif. Results with the ΔS and 5′-3′S mutants also rule out the possibility that an alternative folding of U18 neighboring sequences may supply the function of a terminal stem.
As expected, processing of the U18 snoRNA was totally absent in the presence of mutations within the conserved box C (Fig. 2A, wt/bC and Cbs/bC lanes), similar to what was observed with ΔS mutants. Interestingly, with respect to wt/ΔS mutations, the wt/bC mutant did not accumulate any unspliced pre-mRNA (wt/bC lanes) (see Discussion).
The intramolecular base pairing does not affect mRNA production.
Large yeast introns generally contain inverted repeats whose interaction serves to reduce the effective distance between the donor site and the branch point to a length similar to that found in small introns (34). Several studies showed that disruption of these intron helices has detrimental effects on splicing efficiency, probably inhibiting early steps of spliceosome assembly and leading to a substantial decrease in mRNA production (14, 17, 27, 30). The complementary elements described so far are usually located in close proximity to the 5′ splice site and the branch point regions. Instead, the A and B sequences in the U18 host intron are much more internally located, close to the snoRNA coding region (Fig. 1B). Nevertheless, when complementarity was disrupted, unspliced pre-mRNA accumulated (Fig. 2A, wt/ΔS and wt/5′-3′S lanes).
To assess the role of A-B pairing in the in vivo splicing phenotype of the U18 host pre-mRNA, we analyzed the mRNA released from the different precursors by primer extension analysis with a primer specific for the plasmid-borne EFB1 gene (Fig. 3). Production of mRNA from the wt/ΔS and wt/5′-3′S pre-mRNAs was not affected by the loss of the intramolecular interaction (Fig. 3A, wt/ΔS and wt/5′-3′S lanes), resulting in levels almost identical to mRNA levels released from wt, wt/RS, and wt/bC pre-mRNAs where pairing was not altered (Fig. 3A). The above results were also confirmed by Northern analysis (data not shown). Primer extension analysis of the corresponding Cbs constructs showed that no mRNA accumulated in all cases, confirming their splicing-deficient phenotype (Fig. 3B). In addition, the loss of the signal corresponding to the I-2 intermediate 5′ end confirmed the processing-deficient phenotype of the Cbs/ΔS, Cbs/5′-3′S, and Cbs/bC constructs (Fig. 3B, Cbs/ΔS, Cbs/5′-3′S, and Cbs/bC lanes). These data indicate that base pairing between sequences A and B in the U18 host intron is essential for the snoRNA processing pathway whereas, apparently, it does not affect mRNA accumulation. Nevertheless, we cannot exclude the possibility that disruption of the external stem also affects splicing, probably as the result of loosening of the pre-mRNA secondary structure (see below and Discussion).
Splicing-independent release of the U18 snoRNA relies on formation of the external stem.
A yeast strain lacking the lariat debranching enzyme (Dbr1p) has been previously used as a tool to determine the contribution of the splicing-independent pathway to the release of intronic snoRNAs (33, 35). Indeed, loss of debranching activity in dbr1-Δ cells does not affect endonuclease-mediated release of intronic snoRNAs. Those analyses showed that in dbr1-Δ cells, up to 30% of U18 is yielded as a mature snoRNA, the remainder being trapped within the lariat (references 33 and 35 and data not shown). This was consistent with levels of mature U18 snoRNA released from the Cbs precursor through the splicing-independent processing pathway (Fig. 2A) (47).
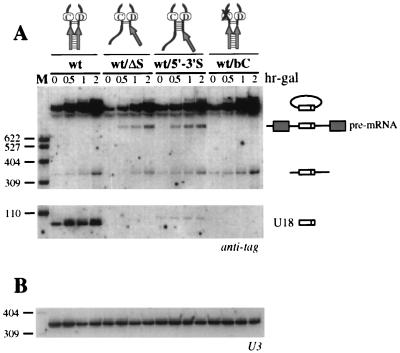
To further define the role of the external and terminal stems within the U18 host intron, we analyzed the release of tagged U18 in the dbr1-Δ strain from several of the precursors described above. As expected, no mature U18 snoRNA accumulated from the wt/ΔS and wt/bC precursors (Fig. 4, wt/ΔS and wt/bC lanes), since, as shown, the ΔS and bC mutations completely prevent accumulation of mature U18. A faint U18 snoRNA signal was detected in cells carrying the wt/5′-3′S construct (wt/5′-3′S lanes). This small amount of mature snoRNA very probably arises from exonucleolytic digestion of the minimal amounts of randomly linearized intron observed in dbr1-Δ cells (33). The apparent lower mobility of this U18 band is also consistent with exonucleolytic trimming up to the base of the extended terminal stem (see also Fig. 2A). Indeed, Cbs precursors did not produce any U18 in the dbr1-Δ strain (data not shown), similar to what is shown in Fig. 2A, confirming that the splicing-independent release of U18 from the Cbs/5′-3′S precursor is impaired. We conclude that the splicing-independent (i.e., Dbr1p-independent) release of U18 snoRNA requires the formation of the external stem within the precursor molecule. A decrease in the amount of circular intron was also observed upon disruption of the external stem (Fig. 4, wt/ΔS and wt/5′-3′S lanes), again suggesting a secondary effect of this structural element on splicing efficiency (see Discussion).
FIG. 4.
(A) Analysis of U18 processing in the debranching-deficient strain dbr1-Δ. Total RNA was extracted from strain dbr1-Δ carrying the indicated wt constructs (schematically represented above the lanes) at the indicated times of galactose induction (expressed in hours above the lanes; hr-gal). Total RNA (5 μg) was resolved on a 6% acrylamide–7 M urea gel, electroblotted onto a nylon membrane, and hybridized with the anti-tag oligonucleotide. The different processing products are diagrammed on the side, as in Fig. 2. The lariat species is depicted as a circle lacking the extension from the branch point to the 3′ splice site, since this is the major form found in strain dbr1-Δ (13). (B) Control hybridization to U3 snRNA. The oligonucleotide used is indicated below each panel. Lanes M: pBR322 plasmid DNA, MspI digested.
The Rnt1p endonuclease is not involved in U18 snoRNA processing.
Rnt1p, the yeast homolog of bacterial RNase III, possesses double-stranded endonucleolytic activity and was shown to be a key enzyme in the processing of several snoRNAs encoded by monocistronic or polycistronic transcriptional units (11, 12). Most Rnt1p cleavage sites were shown to fall within potentially double-stranded regions closed by AGNN tetraloops (11). Even if tetraloops could not be identified in the close vicinity of the U18 external stem, the strict requirement of this element in the splicing-independent release of U18 prompted us to test the involvement of Rnt1p in this pathway. The wt and Cbs constructs were transformed into the isogenic RNT1 and rnt1-Δ strains (10), and after galactose induction, the accumulation of tagged U18 was analyzed by Northern hybridization. Figure 5 shows that similar amounts of U18 were produced from the wt construct in both the RNT1 and rnt1-Δ strains (compare wt lanes in each indicated strain). Importantly, the splicing-independent production of U18 snoRNA from the Cbs precursor was found to be unaffected by the rnt1-Δ mutation. The control hybridization with the U5 snRNA probe indicates the inactivation of Rnt1p: in agreement with previous work (10), only the shorter form of U5 snRNA accumulated in the mutant strain. We conclude that the external stem is not a target for the endonucleolytic activity of Rnt1p and that this enzyme is not involved in U18 snoRNA processing. These results suggest that another as yet unidentified nuclease may be responsible for the endonucleolytic release of U18 from its host pre-mRNA.
FIG. 5.
Analysis of U18 processing in the RNT1 and rnt1-Δ strains. Total RNA was extracted from strains RNT1 and rnt1-Δ carrying the indicated wt or Cbs constructs at the indicated times of galactose induction (expressed in hours above the lanes; hr-gal). Total RNA (5 μg) was resolved on a 6% acrylamide–7 M urea gel, electroblotted onto a nylon membrane, and hybridized with the anti-tag oligonucleotide. U18 snoRNA is indicated on the side. (B) Control hybridization to U5 snRNA: L and S indicate the long and short forms, respectively, of U5 snRNA (10).
Inhibition of the 5′→3′ exonuclease Rat1p partially restores accumulation of U18 snoRNA from an intron lacking the external stem.
The role played by the external and terminal stems on the accumulation of U18 snoRNA was also investigated in the rat1-1 yeast strain (1), which carries a thermosensitive allele of the gene encoding the nuclear 5′→3′ exonuclease Rat1p. We reasoned that loss of snoRNA accumulation in the absence of intramolecular base pairing could result from improper folding of the U18 coding region, probably leading to its inefficient assembly with snoRNP components and in turn to its exonucleolytic degradation. In other words, the production of U18 may be the result of a competition between efficient assembly of a snoRNP particle and turnover of the pre-snoRNA molecule. If this holds true, it would be expected that stabilization of U18 precursors in the absence of Rat1p activity would allow more time for reaching the appropriate conformation and allow the assembly into a stable snoRNP particle.
Expression of the different U18 precursors was induced after transfer of rat1-1 cells to the restrictive temperature of 37°C for 2 h, and RNA was subjected to Northern analysis (Fig. 6A). As previously reported (47), inhibition of Rat1p activity led to decreased decay rates and increased accumulation levels of pre-mRNAs and linearized lariat without impairing U18 processing (Fig. 6A, wt and Cbs lanes). This indicates that in the absence of 5′→3′ exonucleolytic processing, endonucleolytic cleavage(s) allows the formation of a mature U18 5′ end as described previously (47).
FIG. 6.
(A) Analysis of U18 processing in the 5′→3′ exonuclease-deficient strain rat1-1. Total RNA was extracted from strain rat1-1 carrying the indicated wt or Cbs constructs (schematically represented above the lanes) at the indicated times of galactose induction (expressed in hours above the lanes; hr-gal). Strain rat1-1 was shifted to the restrictive temperature of 37°C for 2 h before addition of galactose. Total RNA (5 μg) was resolved on a 6% acrylamide–7 M urea gel, electroblotted onto a nylon membrane, and hybridized with the anti-tag oligonucleotide. The different processing products are diagrammed on the side, as in Fig. 2. (B) Control hybridization to U3 snRNA. The oligonucleotide used is indicated below each panel. Lanes M: pBR322 plasmid DNA, MspI digested. (C) Levels of accumulation of U18 in the wt (CH1462; lanes 1, 4, and 7), rat1-1 (lanes 2, 5, and 8), and dbr1-Δ (lanes 3, 6, and 9) strains. Total RNAs extracted from the different strains transformed with the indicated constructs 1 h after galactose induction were analyzed by primer extension with the anti-tag oligonucleotide. Strain rat1-1 was shifted to the restrictive temperature of 37°C for 2 h before the addition of galactose.
Interestingly, U18 snoRNA was accumulated in rat1-1 cells also from the wt/ΔS precursor, although at low levels (Fig. 6A, wt/ΔS lanes). In contrast, the splicing-independent U18 snoRNA processing from the cognate Cbs/ΔS precursor was still completely inhibited in this strain (Cbs/ΔS lanes). We conclude that in the rat1-1 strain, processing of U18 in the absence of the external stem can be rescued only from the released intron.
Similar to what was observed in wt cells, in rat1-1 cells the 5′-3′S mutation allowed the accumulation of U18 only through the splicing-dependent processing pathway (Fig. 6A, compare wt/5′-3′S and Cbs/5′-3′S lanes). Also in agreement with results obtained with wt cells, both the wt/bC and Cbs/bC precursors were unable to produce mature U18 snoRNA (wt/bC and Cbs/bC lanes).
Interestingly, when we compared the levels of accumulation of the tagged U18 snoRNA released from the different wt constructs in the wt (Fig. 6C, lanes 1, 4, and 7), rat1-1 (lanes 2, 5, and 8), and dbr1-Δ strains (lanes 3, 6, and 9), we reproducibly found that U18 snoRNA accumulation was increased when the nuclear 5′→3′ exonuclease Rat1p was inhibited. Taken together, these data strongly support our view of a role of the external stem in assisting correct and productive folding of the U18 coding region, thus preventing its exonucleolytic degradation.
The external stem present in most yeast box C/D snoRNA host introns is required for processing.
We next asked whether the presence of an external stem surrounding the snoRNA coding region was a peculiar feature of the U18 host intron or a more general feature of introns hosting box C/D snoRNAs. For this reason, we searched for potential intramolecular base pairing within known yeast host intron sequences. The results of this analysis are presented in Table 1 (see also Materials and Methods). Interestingly, five out of six yeast host introns, including the U18 one, display complementary sequences (≥10 nt) in close proximity to the snoRNA coding region. Importantly, the corresponding 5 snoRNAs all lack a canonical terminal stem (5′->>>NR[boxC]---[boxD]<<<-3′) (2, 3). It is also interesting that all five of these snoRNAs display partial insensitivity to the dbr1-Δ mutation, indicating the existence of a processing pathway independent of splicing.
TABLE 1.
Presence of inverted repeats within yeast box C/D snoRNA host introns
| Host intron | Intron size (bp) | snoRNA | Length (bp) of:
|
Dbr1p-independent releasec | |
|---|---|---|---|---|---|
| Terminal stema | External stemb | ||||
| EFB1 | 366 | U18 | 2 | 14 | + |
| BEL1 | 273 | U24 | 8 | − | − |
| TEF4 | 326 | snR38 | − | 11 | + |
| YL8A-II | 467 | snR39 | − | 15* | + |
| YM9958.06c | 408 | snR54 | − | 10 | + |
| YL8B-II | 407 | snR59 | − | 12 | + |
Includes base pairs extending within the precursor molecule. −, absence of a canonical terminal stem.
−, absence of an external stem (see Materials and Methods); *, presence of a mismatch. Note that the snR39 external stem sequences are clearly distinct from those of the complementary elements which positively affect splicing efficiency of the host intron (18).
Dbr1p-independent release indicates whether an intronic snoRNA is accumulated in a dbr1-Δ strain above background levels (reference 33 and our unpublished data), indicating the existence of an endonucleolytic pathway independent from splicing.
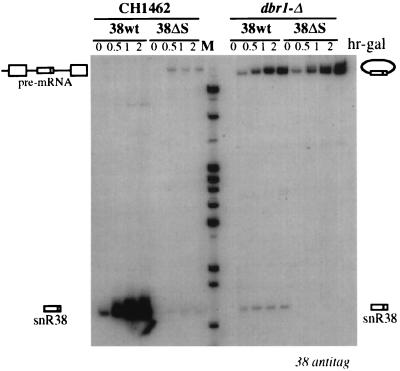
To verify whether the external stems of other yeast box C/D snoRNA host introns are also involved in processing events, we analyzed the effect of external stem disruption on the accumulation of a tagged snR38 snoRNA. snR38 also belongs to the class of snoRNAs devoid of the terminal stem structure (Table 1) (2). To this end, plasmids pGAL38wt-tag and pGAL38ΔS-tag (carrying the host wild-type TEF4 gene and a mutant derivative lacking the external stem, respectively [see Materials and Methods]) were transformed in the wt CH1462 and mutant dbr1-Δ strains. Expression of the pGAL38wt-tag and pGAL38ΔS-tag constructs was induced by galactose addition, and the accumulation of tagged snR38 was assessed by Northern analysis (Fig. 7). Similarly to U18 snoRNA, disruption of the external stem drastically impaired snR38 processing in the CH1462 strain and completely inhibited the release of snR38 in the dbr1-Δ strain (Fig. 7, compare 38wt and 38ΔS lanes in each indicated strain). These results complement those obtained by sequence comparison and strongly suggest a functional conservation of intronic processing elements in yeast box C/D snoRNA host introns.
FIG. 7.
Analysis of snR38 accumulation in the wt CH1462 and the mutant dbr1-Δ strains. Total RNA was extracted from strains CH1462 (left) and dbr1-Δ (right) carrying the pGAL38wt-tag and pGAL38ΔS-tag constructs at the indicated times of galactose induction (expressed in hours above the lanes; hr-gal). Total RNA (5 μg) was resolved on a 6% acrylamide–7 M urea gel, electroblotted onto a nylon membrane, and hybridized with the 38 anti-tag oligonucleotide. The different processing products are diagrammed. Lane M: pBR322 plasmid DNA, MspI digested.
An exception to this arrangement is represented by the BEL1 intron hosting U24, where no external stem could be found: instead, the U24 snoRNA can form a strong 8-bp canonical terminal stem. Interestingly, this is the only yeast box C/D snoRNA whose biosynthesis is entirely dependent on the debranching activity (Table 1) (33). When the terminal stem of U24 was reduced to 2 bp, as in the case in U18, processing could be restored when an external complementarity of 14 nt was introduced in the flanking intron sequences (data not shown).
In contrast to the arrangement found in yeast, most human box C/D snoRNAs form a canonical 5′-3′-terminal stem and their host introns do not display long external complementary sequences (data not shown). In a few cases (U30, U37, U39, and U44), where the sequences neighboring the C and D boxes do not produce canonical terminal stems, surrounding complementary sequences within the host introns can be found, suggesting that they may act as functional equivalents of the yeast external stems (data not shown).
DISCUSSION
The biosynthetic pathway of intron-encoded snoRNAs, in both yeast and higher eukaryotes, has been studied in detail. These analyses established that intronic snoRNAs can be matured via two pathways. The major pathway is dependent on the linearization of the spliced lariat by a debranching activity and subsequent exonucleolytic digestion of the flanking sequences to produce the correct 5′ and 3′ termini (8, 9, 20, 26, 33, 35, 45, 46, 47). The minor pathway relies on endonucleolytic cleavage of unspliced host pre-mRNA, producing processing intermediates that are further matured by exonucleases (6, 33, 35, 37, 47). Correct processing from both pathways and stable accumulation of mature box C/D snoRNAs are dependent upon a terminal core motif formed by the conserved C and D boxes and the short RNA helix structure (≥4 bp) found in the close vicinity of these elements (7, 8, 19, 25, 48, 49, 51). Since this motif is common to all eukaryotic box C/D snoRNAs, independent of their genomic arrangement, it is assumed that it represents a recognition signal for snoRNP core proteins. The assembled snoRNP complex protects the snoRNA from exonucleolytic digestion, thus providing the information to specify the ends of the mature molecule. Interestingly, these are the same requirements for nucleolar targeting of this class of snoRNAs (24, 41), suggesting that processing, stability, and trafficking are highly interdependent.
Our results show that processing of yeast intronic box C/D snoRNAs lacking a canonical terminal stem must be assisted by elements external to the mature snoRNA. This has been shown for U18 and snR38 snoRNAs, whose host introns contain complementary sequences outside the snoRNA coding region. The role of the external stem has been extensively studied in the U18 snoRNA. Disruption of this complementarity leads to loss of U18 processing from both the splicing-dependent and the splicing-independent pathways. While the release of U18 from the pre-mRNA uniquely relies on the presence of the external stem, processing from the released intron can be restored if a canonical terminal stem is provided. This implies that the box C/D motif of yeast U18 is not sufficient to direct processing of the snoRNA. Thus, it appears that the lack of a canonical terminal stem is compensated for by structural elements within the host intron. Very likely, the external stem mimics the function of the terminal stem by helping the initial assembly with box C/D snoRNP factors, thereby preventing progression of exonucleases within the snoRNA coding region. Consistently, inhibition of the nuclear 5′→3′ exonuclease Rat1p allows a partial recovery of U18 snoRNA release from the spliced intron lacking the external stem. This is a clear indication that one essential function of the external stem in the splicing-dependent U18 processing pathway is to provide the structural information needed for the assembly of a snoRNP complex (41). Since natural yeast U18 snoRNA lacks a 5′-3′-terminal stem, it follows that the stable association of U18 snoRNP core proteins depends entirely on conserved C and D boxes. We suggest a model in which the accumulation of U18 snoRNA is the result of a competition between the exonucleolytic degradation of the linearized host intron and the efficiency of U18 snoRNP assembly (Fig. 8). Indeed, inhibition of Rat1p activity leads to increased accumulation levels of U18 snoRNA.
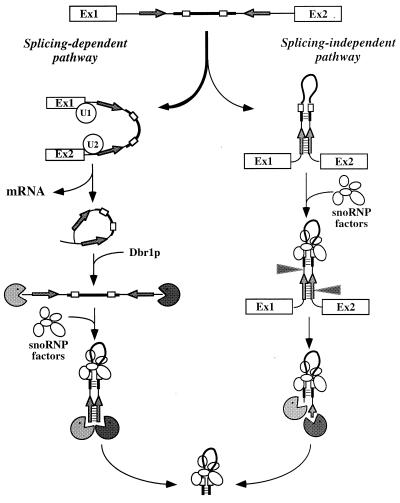
FIG. 8.
Model for intron-encoded box C/D snoRNA processing in yeast. This model applies to the intronic snoRNAs lacking a canonical terminal stem. The host intron displays nonconserved complementary sequences (gray inverted arrows). The box C/D snoRNA is depicted as a thick line, and its conserved boxes C and D are depicted as small open boxes. The host pre-mRNA mainly undergoes splicing (symbolized by the association with U1 and U2 snRNPs), releasing mRNA and the lariat. This is quickly linearized by Dbr1p, producing free 5′ and 3′ ends readily attacked by exonucleases (represented as “Pacmen”; the nuclear 5′→3′ exonuclease is Rat1p). Concomitantly with or immediately following linearization, formation of the external stem directs proper folding of the pre-snoRNA molecule (base pairing interactions are depicted by horizontal bars), allowing association of snoRNP factors with boxes C and D. Nevertheless, it cannot be excluded that snoRNP assembly may begin at some previous steps during the splicing process. This assembly protects the pre-snoRNA from exonuclease digestion and specifies the snoRNA mature ends. In the concurrent splicing-independent pathway, whose existence is evidenced by the partial insensitivity to the dbr1-Δ mutation, folding of newly synthesized host pre-mRNA molecules induced by the external stem ensures their recognition as pre-snoRNA substrates. Association with snoRNP factors is proposed to direct the endonucleolytic cleavages of the snoRNA-flanking sequences (arrowheads; their position is based on earlier results with U18 snoRNA [47]).
Our comparative analysis of host introns suggests that this situation is common in yeast, where five box C/D snoRNAs lack a canonical terminal stem whereas their host introns possess external complementary regions. Our results with snR38 indicate that such an arrangement probably reflects a conserved function of the external stems in directing the processing of yeast box C/D snoRNAs. In contrast, human box C/D snoRNAs generally share the presence of a consensus terminal motif (44, 50). In a few cases, external complementary sequences may reinforce and/or compensate for suboptimal terminal interactions. This suggests some functional interchangeability of the external and terminal stems. Indeed, yeast U24, which has an optimal terminal stem similar to human snoRNAs, can be efficiently processed from a mutated host intron if deprived of its terminal stem and provided with intronic external complementarity (data not shown).
Disruption of the external stem in the U18 host intron also induces an increase of pre-mRNA accumulation. It is largely documented that complementary sequences within large yeast introns positively influence splicing efficiency, contributing to early spliceosome assembly (14, 17, 18, 27, 30, 34, 36). Nevertheless, even if EFB1 pre-mRNA accumulates, this increase does not result in any manifest change in the level of mRNA, indicating that splicing is not limiting in the production of EFB1 mRNA (36). It is then more likely that the pre-mRNA accumulation reflects the defect of U18 processing, since the splicing-independent processing pathway completely depends on the extended base pairing provided by the external stem and cannot be supported by the shorter interaction of the terminal stem. This strict dependence on the external stem raises the possibility that this element is required for the endonucleolytic cleavage of pre-mRNA, possibly by Rnt1p, the yeast homolog of bacterial RNase III. However, when the wt and Cbs constructs are expressed in mutant yeast strains lacking Rnt1p activity, neither the splicing-dependent nor the splicing-independent U18 processing pathway is affected, also consistent with previous observations (11). It is then more likely that the external stem is required for another purpose or as a target for an unidentified endonuclease.
In light of the proposed role of complementarity in assisting the correct folding of a pre-snoRNA molecule (41), the extended base pairing required for the splicing-independent processing correlates with the larger size of the pre-mRNA molecule, as opposed to that of the intron, in which 6 bp is sufficient to promote the release of U18. By analogy to splicing, we propose that the external complementary sequences of the U18 and snR38 host introns and presumably of the other three large yeast introns hosting box C/D snoRNAs positively influences processing efficiency. This contribution is exerted through base pairing and favors the communication between the 5′ and 3′ termini of the snoRNA coding regions. Proper folding brings together the conserved C and D boxes for interactions with the trans-acting factors that initially recognize the pre-snoRNA substrate (Fig. 8). In addition to the primary role on processing, the intramolecular interactions would enhance the splicing efficiency, thus promoting efficient conversion of newly synthesized pre-mRNAs into mature products (i.e., mRNAs and snoRNAs) and contributing to the overall yeast fitness. One interesting issue of future studies will be to determine how box C/D snoRNP proteins and/or early splicing factors modulate the initial recognition of a host pre-mRNA molecule, thus committing it to a specific maturation pathway.
ACKNOWLEDGMENTS
We thank Manny Ares for the generous gift of the RNT1 and rnt1-Δ strains. We thank members of our laboratory, in particular, Alessandro Fatica and Corinna Giorgi, for many helpful discussions. We thank Ida Ruberti and Giorgio Morelli for suggestions on sequence analysis. We also thank Massimo Arceci and Genesio Ricci for technical help.
T.V. was the recipient of an Istituto Pasteur-Fondazione Cenci Bolognetti fellowship. F.C. was the recipient of a fellowship from Fondazione Adriano Buzzati-Traverso. This work was partially supported by grants from MURST-CNR Biotechnology Program L.95/95 from PRIN 40% of MURST and from CNR Target Project on Biotechnology.
REFERENCES
- 1.Amberg D C, Goldstein A L, Cole C N. Isolation and characterization of RAT1: an essential gene of Saccharomyces cerevisiae required for the efficient nucleocytoplasmic trafficking of mRNA. Genes Dev. 1992;6:1173–1189. doi: 10.1101/gad.6.7.1173. [DOI] [PubMed] [Google Scholar]
- 2.Bachellerie J-P, Michot B, Nicoloso M, Balakin A, Ni J, Fournier M J. Antisense snoRNAs: a family of nucleolar RNAs with long complementarities to rRNA. Trends Biochem Sci. 1995;20:261–264. doi: 10.1016/s0968-0004(00)89039-8. [DOI] [PubMed] [Google Scholar]
- 3.Bachellerie J-P, Cavaillé J. Guiding ribose methylation of rRNA. Trends Biochem Sci. 1997;22:257–261. doi: 10.1016/s0968-0004(97)01057-8. [DOI] [PubMed] [Google Scholar]
- 4.Balakin A G, Smith L, Fournier M J. The RNA world of the nucleolus: two major families of small RNAs defined by different box elements with related functions. Cell. 1996;86:823–834. doi: 10.1016/s0092-8674(00)80156-7. [DOI] [PubMed] [Google Scholar]
- 5.Bortolin M-L, Ganot P, Kiss T. Elements essential for accumulation and function of small nucleolar RNAs directing site-specific pseudouridylation of ribosomal RNAs. EMBO J. 1999;18:457–469. doi: 10.1093/emboj/18.2.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Caffarelli E, Arese M, Santoro B, Fragapane P, Bozzoni I. In vitro study of the processing of the intron-encoded U16 small nucleolar RNA in Xenopus laevis. Mol Cell Biol. 1994;14:2966–2974. doi: 10.1128/mcb.14.5.2966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Caffarelli E, Fatica A, Prislei S, De Gregorio E, Fragapane P, Bozzoni I. Processing of the intron-encoded U16 and U18 snoRNAs: the conserved C and D boxes control both the processing reaction and the stability of the mature snoRNA. EMBO J. 1996;15:1121–1131. [PMC free article] [PubMed] [Google Scholar]
- 8.Cavaillé J, Bachellerie J-P. Processing of fibrillarin-associated snoRNAs from pre-mRNA introns: an exonucleolytic process exclusively directed by the common stem-box terminal structure. Biochimie. 1996;78:443–456. doi: 10.1016/0300-9084(96)84751-1. [DOI] [PubMed] [Google Scholar]
- 9.Cecconi F, Mariottini P, Amaldi F. The Xenopus intron-encoded U17 snoRNA is produced by exonucleolytic processing of its precursor in oocytes. Nucleic Acids Res. 1995;23:4670–4676. doi: 10.1093/nar/23.22.4670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chanfreau G, Abou Elela S, Ares M, Jr, Guthrie C. Alternative 3′-end processing of U5 snRNA by RNaseIII. Genes Dev. 1997;11:2741–2751. doi: 10.1101/gad.11.20.2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chanfreau G, Legrain P, Jacquier A. Yeast RNase III as a key processing enzyme in small nucleolar RNAs metabolism. J Mol Biol. 1998;284:975–988. doi: 10.1006/jmbi.1998.2237. [DOI] [PubMed] [Google Scholar]
- 12.Chanfreau G, Rotondo G, Legrain P, Jacquier A. Processing of a dicistronic small nucleolar RNA precursor by the RNA endonuclease Rnt1. EMBO J. 1998;17:3726–3737. doi: 10.1093/emboj/17.13.3726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chapman K B, Boeke J D. Isolation and characterization of the gene encoding yeast debranching enzyme. Cell. 1991;65:483–492. doi: 10.1016/0092-8674(91)90466-c. [DOI] [PubMed] [Google Scholar]
- 14.Charpentier B, Rosbash M. Intramolecular structure in yeast introns aids the early steps of in vitro spliceosome assembly. RNA. 1996;2:509–522. [PMC free article] [PubMed] [Google Scholar]
- 15.Ganot P, Bortolin M-L, Kiss T. Site-specific pseudouridine formation in preribosomal RNA is guided by small nucleolar RNAs. Cell. 1997;89:799–809. doi: 10.1016/s0092-8674(00)80263-9. [DOI] [PubMed] [Google Scholar]
- 16.Ganot J-P, Caizergues-Ferrer M, Kiss T. The family of box ACA small nucleolar RNAs is defined by an evolutionarily conserved secondary structure and ubiquitous sequence elements essential for RNA accumulation. Genes Dev. 1997;11:941–956. doi: 10.1101/gad.11.7.941. [DOI] [PubMed] [Google Scholar]
- 17.Goguel V, Rosbash M. Splice site choice and splicing efficiency are positively influenced by pre-mRNA intramolecular base pairing in yeast. Cell. 1993;72:893–901. doi: 10.1016/0092-8674(93)90578-e. [DOI] [PubMed] [Google Scholar]
- 18.Howe K J, Ares M., Jr Intron self-complementarity enforces exon inclusion in a yeast pre-mRNA. Proc Natl Acad Sci USA. 1997;94:12467–12472. doi: 10.1073/pnas.94.23.12467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang G M, Jarmolowski A, Struck J C, Fournier M J. Accumulation of U14 small nuclear RNA in Saccharomyces cerevisiae requires box C, box D, and a 5′,3′-terminal stem. Mol Cell Biol. 1992;12:4456–4463. doi: 10.1128/mcb.12.10.4456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kiss T, Filipowicz W. Exonucleolytic processing of small nucleolar RNAs from pre-mRNA introns. Genes Dev. 1995;9:1411–1424. doi: 10.1101/gad.9.11.1411. [DOI] [PubMed] [Google Scholar]
- 21.Kiss-László Z, Henry Y, Bachellerie J-P, Caizergues-Ferrer M, Kiss T. Site-specific ribose methylation of preribosomal RNA: a novel function for small nucleolar RNAs. Cell. 1996;85:1077–1088. doi: 10.1016/s0092-8674(00)81308-2. [DOI] [PubMed] [Google Scholar]
- 22.Kiss-László Z, Henry Y, Kiss T. Sequence and structural elements of methylation guide snoRNAs essential for site-specific ribose methylation of pre-rRNA. EMBO J. 1998;17:797–807. doi: 10.1093/emboj/17.3.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kranz J E, Holm C. Cloning by function: an alternative approach for identifying yeast homologs of genes from other organisms. Proc Natl Acad Sci USA. 1990;87:6629–6633. doi: 10.1073/pnas.87.17.6629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lange T S, Borovjagin A, Maxwell E S, Gerbi S A. Conserved boxes C and D are essential nucleolar localisation elements of U14 and U8 snoRNAs. EMBO J. 1998;17:3176–3187. doi: 10.1093/emboj/17.11.3176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leader D J, Clark G P, Watters J, Beven A F, Shaw P J, Brown J W S. Clusters of multiple different small nucleolar RNA genes in plants are expressed as and processed from polycistronic pre-snoRNAs. EMBO J. 1997;16:5742–5751. doi: 10.1093/emboj/16.18.5742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leverette R D, Andrews M T, Maxwell E S. Mouse U14 snRNA is a processed intron of the cognate hsc70 heat shock pre-messenger RNA. Cell. 1992;71:1215–1221. doi: 10.1016/s0092-8674(05)80069-8. [DOI] [PubMed] [Google Scholar]
- 27.Libri D, Stutz F, McCarthy T, Rosbash M. RNA structural patterns and splicing: molecular basis for an RNA-based enhancer. RNA. 1995;1:425–436. [PMC free article] [PubMed] [Google Scholar]
- 28.Maxwell E S, Fournier M J. The small nucleolar RNAs. Annu Rev Biochem. 1995;35:897–934. doi: 10.1146/annurev.bi.64.070195.004341. [DOI] [PubMed] [Google Scholar]
- 29.Mumberg D, Müller R, Funk M. Regulatable promoters of Saccharomyces cerevisiae: comparison of transcriptional activity and their use for heterologous expression. Nucleic Acids Res. 1994;22:5767–5768. doi: 10.1093/nar/22.25.5767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Newman A. Specific accessory sequences in Saccharomyces cerevisiae introns control assembly of pre-mRNAs into spliceosomes. EMBO J. 1987;6:3833–3839. doi: 10.1002/j.1460-2075.1987.tb02720.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ni J, Tien A L, Fournier M J. Small nucleolar RNAs direct site-specific synthesis of pseudouridine in ribosomal RNA. Cell. 1997;89:565–573. doi: 10.1016/s0092-8674(00)80238-x. [DOI] [PubMed] [Google Scholar]
- 32.Nicoloso M, Qu L-H, Michot B, Bachellerie J-P. Intron-encoded antisense small nucleolar RNAs: the characterization of nine novel species points to their role as guides for the 2′-O-ribose methylation of rRNAs. J Mol Biol. 1996;260:178–195. doi: 10.1006/jmbi.1996.0391. [DOI] [PubMed] [Google Scholar]
- 33.Ooi S L, Samarsky D, Fournier M J, Boeke J D. Intronic snoRNA biosynthesis in Saccharomyces cerevisiae depends on the lariat-debranching enzyme: intron length effects and activity of a precursor snoRNA. RNA. 1998;4:1096–1110. doi: 10.1017/s1355838298980785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Parker R, Patterson B. Architecture of fungal introns: implications for spliceosome assembly. In: Dudock B, Inouye M, editors. Molecular biology of RNA: new perspectives. New York, N.Y: Academic Press, Inc.; 1987. pp. 133–149. [Google Scholar]
- 35.Petfalski E, Dandekar T, Henry Y, Tollervey D. Processing of the precursors to small nucleolar RNAs and rRNAs requires common components. Mol Cell Biol. 1998;18:1181–1189. doi: 10.1128/mcb.18.3.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pikielny C W, Rosbash M. mRNA splicing efficiency in yeast and the contribution of nonconserved sequences. Cell. 1985;41:119–126. doi: 10.1016/0092-8674(85)90066-2. [DOI] [PubMed] [Google Scholar]
- 37.Prislei S, Michienzi A, Presutti C, Fragapane P, Bozzoni I. Two different snoRNAs are encoded in introns of amphibian and human L1 ribosomal protein genes. Nucleic Acids Res. 1993;21:5824–5830. doi: 10.1093/nar/21.25.5824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Prislei S, Fatica A, De Gregorio E, Arese M, Fragapane P, Caffarelli E, Presutti C, Bozzoni I. Self-cleaving motifs are found in close proximity to the sites utilized for U16 snoRNA processing. Gene. 1995;163:221–226. doi: 10.1016/0378-1119(95)00344-6. [DOI] [PubMed] [Google Scholar]
- 39.Qu L-H, Henras A, Lu Y-J, Zhou H, Zhou W X, Zhu Y-Q, Zhao J, Henry Y, Caizergues-Ferrer M, Bachellerie J-P. Seven novel methylation guide small nucleolar RNAs are processed from a common polycistronic transcript by Rat1p and RNase III in yeast. Mol Cell Biol. 1999;19:1144–1158. doi: 10.1128/mcb.19.2.1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Samarsky D A, Fournier M J. Functional mapping of the U3 small nucleolar RNA from the yeast Saccharomyces cerevisiae. Mol Cell Biol. 1998;18:3431–3444. doi: 10.1128/mcb.18.6.3431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Samarsky D A, Fournier M J, Singer R H, Bertrand E. The snoRNA box C/D motif directs nucleolar targeting and also couples snoRNA synthesis and localisation. EMBO J. 1998;17:3747–3757. doi: 10.1093/emboj/17.13.3747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Santoro B, De Gregorio E, Caffarelli E, Bozzoni I. RNA-protein interactions in the nuclei of Xenopus oocytes: complex formation and processing activity on the regulatory intron of ribosomal protein gene L1. Mol Cell Biol. 1994;14:6975–6982. doi: 10.1128/mcb.14.10.6975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Smith C M, Steitz J A. Sno storm in the nucleolus: new roles for myriad small RNPs. Cell. 1997;89:669–672. doi: 10.1016/s0092-8674(00)80247-0. [DOI] [PubMed] [Google Scholar]
- 44.Tollervey D, Kiss T. Function and synthesis of small nucleolar RNAs. Curr Opin Cell Biol. 1997;9:337–342. doi: 10.1016/s0955-0674(97)80005-1. [DOI] [PubMed] [Google Scholar]
- 45.Tycowski K T, Shu M D, Steitz J A. A small nucleolar RNA is processed from an intron of the human gene encoding ribosomal protein S3. Genes Dev. 1993;6:1120–1130. doi: 10.1101/gad.7.7a.1176. [DOI] [PubMed] [Google Scholar]
- 46.Tycowski K T, Shu M D, Steitz J A. A mammalian gene with introns instead of exons generating stable RNA products. Nature. 1996;379:464–466. doi: 10.1038/379464a0. [DOI] [PubMed] [Google Scholar]
- 47.Villa T, Ceradini F, Presutti C, Bozzoni I. Processing of the intron-encoded U18 small nucleolar RNA in the yeast Saccharomyces cerevisiae relies on both exo- and endonucleolytic activities. Mol Cell Biol. 1998;18:3376–3383. doi: 10.1128/mcb.18.6.3376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Watkins N J, Leverette R D, Xia L, Andrews M T, Maxwell E S. Elements essential for processing intronic U14 snoRNA are located at the termini of the mature snoRNA sequence and include conserved nucleotide boxes C and D. RNA. 1996;2:118–133. [PMC free article] [PubMed] [Google Scholar]
- 49.Watkins N J, Newman D R, Kuhn J F, Maxwell E S. In vitro assembly of the mouse U14 snoRNP core complex and identification of a 65-kDa box C/D-binding protein. RNA. 1998;4:582–593. doi: 10.1017/s1355838298980128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Weinstein L B, Steitz J A. Guided tours: from precursor snoRNA to functional snoRNP. Curr Opin Cell Biol. 1999;11:378–384. doi: 10.1016/S0955-0674(99)80053-2. [DOI] [PubMed] [Google Scholar]
- 51.Xia L, Watkins N J, Maxwell E S. Identification of specific nucleotide sequences and structural elements required for intronic U14 snoRNA processing. RNA. 1997;3:17–26. [PMC free article] [PubMed] [Google Scholar]
- 52.Zuker M. Prediction of RNA secondary structure by energy minimization. Methods Mol Biol. 1994;25:267–294. doi: 10.1385/0-89603-276-0:267. [DOI] [PubMed] [Google Scholar]