Abstract
The global epidemic caused by the coronavirus severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) has resulted in the infection of over 200 million people. To extend the knowledge of interactions between SARS-CoV-2 and humans, we systematically investigate the interactome of 29 viral proteins in human cells by using an antibody-based TurboID assay. In total, 1,388 high-confidence human proximal proteins with biotinylated sites are identified. Notably, we find that SARS-CoV-2 manipulates the antiviral and immune responses. We validate that the membrane protein ITGB1 associates angiotensin-converting enzyme 2 (ACE2) to mediate SARS-CoV-2 entry. Moreover, we reveal that SARS-CoV-2 proteins inhibit activation of the interferon pathway through the mitochondrial protein mitochondrial antiviral-signaling protein (MAVS) and the methyltransferase SET domain containing 2, histone lysine methyltransferase (SETD2). We propose 111 potential drugs for the clinical treatment of coronavirus disease 2019 (COVID-19) and identify three compounds that significantly inhibit the replication of SARS-CoV-2. The proximity labeling map of SARS-CoV-2 and humans provides a resource for elucidating the mechanisms of viral infection and developing drugs for COVID-19 treatment.
Keywords: SARS-CoV-2, COVID-19, TurboID, proximity labeling, interactome
Graphical abstract
Zhang et al. use a proximity labeling technology to identify human proximal proteins of SARS-CoV-2. They show that SARS-CoV-2 manipulates key cellular processes in antiviral and immune responses. They provide a resource for elucidating the mechanisms of SARS-CoV-2 infection and developing drugs for COVID-19 treatment.
Introduction
The emergence of the severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) has resulted in extensive global transmission in more than 210 countries and territories (Li et al., 2020; Rickard et al., 2014; Wu et al., 2020; Zhou et al., 2020; Zhu et al., 2020). Infection with SARS-CoV-2 results in coronavirus disease 2019 (COVID-19), a mild to severe respiratory illness with excessive inflammation (Wiersinga et al., 2020). SARS-CoV-2 is an enveloped, positive-sense, single-stranded RNA beta-coronavirus. The genome size of this virus is 29.9 kb, and it encodes 14 open reading frames (ORFs) (Wu et al., 2020). The suggested original and intermediate hosts of SARS-CoV-2 include bats and pangolins (Lam et al., 2020; Lu et al., 2020; Zhou et al., 2020). However, the transmission route of SARS-CoV-2 remains controversial. Compared with other family members of Coronaviridae, SARS-CoV-1 and Middle East respiratory syndrome coronavirus (MERS-CoV), SARS-CoV-2 has higher morbidity and lower mortality (Yi et al., 2020). Despite intensive research, there is still limited knowledge about the interactions between SARS-CoV-2 and its host. As a result, specific antiviral drugs are not available to prevent or treat COVID-19. Information about the interactions of SARS-CoV-2 and humans is urgently needed to reveal the molecular mechanisms of COVID-19 and for the development of drugs for clinical treatment.
The interactome of SARS-CoV-2 and its host has been investigated by affinity purification coupled with mass spectrometry (AP-MS) (Gordon et al., 2020b; Stukalov et al., 2021). These resources are useful for understanding the molecular mechanisms of SARS-CoV-2 infection and the clinical progression of COVID-19. Dozens of drugs have been found to inhibit the replication of the virus, and in vitro and clinical trials are still ongoing. To identify therapeutic targets, additional strategies are still urgently required to dissect virus-host interactions. Recently, a proximity labeling method, TurboID, was developed to covalently label neighbors of a target protein with biotin within 10 min (Branon et al., 2018; Doerr, 2018). Then, the biotinylated proteins or peptides were enriched and identified by mass spectrometry (Udeshi et al., 2017). This approach allowed the investigation of interacting proteins under harsh lysis conditions while retaining weak interactions. We aimed to extend the knowledge of the interactions of SARS-CoV-2 and humans by generating a global proximity labeling map. Consequently, we revealed the basic mechanisms of viral infection and proposed potential drugs for the clinical treatment of COVID-19.
Results
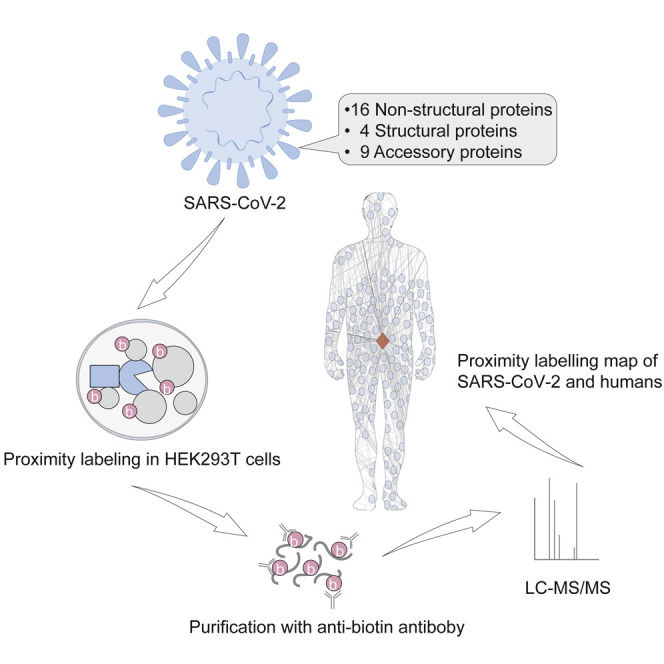
Strategy for the expression and proximity labeling of SARS-CoV-2 proteins
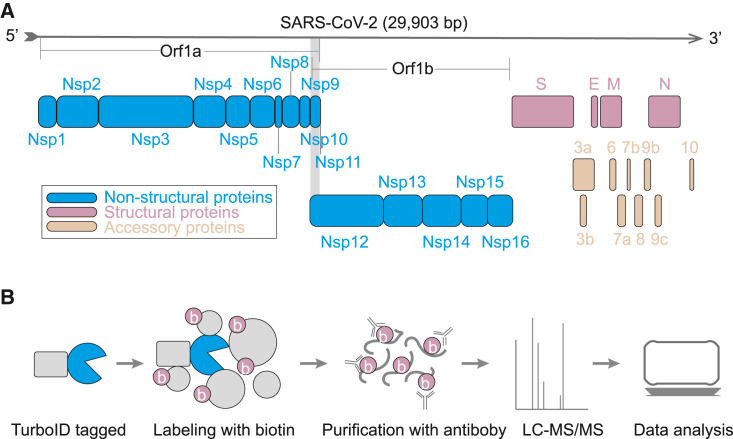
The genome RNA of SARS-CoV-2 encodes 14 putative ORFs (Kim et al., 2020). Three kinds of proteins are encoded by the viral genome: non-structural proteins, structural proteins, and accessory proteins (Figure 1 A). The non-structural replicase proteins (NSP1–16) are encoded by a polyprotein, Orf1a/Orf1b, at the amino terminus of the genome. The polyprotein is proteolytically processed into 16 non-structural proteins. Four structural proteins are expressed at the carboxyl terminus, including the spike (S), envelope (E), membrane (M), and nucleocapsid (N) proteins. The other part of the viral genome encodes nine accessory proteins.
Figure 1.
Flowchart of the proximity labeling of SARS-CoV-2 proteins
(A) The proteins encoded by the SARS-CoV-2 genome.
(B) The workflow of the TurboID assay.
To proximity label the proteins of SARS-CoV-2, all of the viral genes were cloned into mammalian expression vectors in frame with a promiscuous biotin ligase gene (TurboID) (Figure 1B; Table S1A). Additionally, five truncations were fused with TurboID, including the core domain (CD), N terminus and C terminus of NSP3; the receptor binding domain (RBD); and the S1 protein of the S protein. These cassettes were expressed in SARS-CoV-2-permissive HEK293T cells. Then, the proximal proteins were labeled with biotin for 10 min. The cells were harvested and digested with trypsin. The biotinylated peptides were enriched with a biotin antibody and identified by liquid chromatography with tandem mass spectrometry (LC-MS/MS). To obtain high-confidence interactors, three sequential steps were used to remove non-specifically binding proteins. First, we scored the interactors with the SANITexpress algorithm (Teo et al., 2014) by using the negative control samples. The negative controls were the samples that transfected with the TurboID vector. Second, we performed data mining to determine the proteins that have a higher propensity to be contaminants. We found that these proteins were detected in more than 50% of the experiments when choosing the fifth percentile as an arbitrary cutoff. To exclude these non-specifically co-purified proteins, the top fifth percentile proteins (Table S1B) ranked by the occurrence rate were removed from each of the identified proteins, the biotinylated proteins, and the CRAPome contamination proteins (Mellacheruvu et al., 2013), among which more than half of the viral proteins co-purified with these proteins. Finally, only proteins with biotinylated sites and enriched by 2-fold greater than the control were considered candidate proximal proteins of the viral proteins.
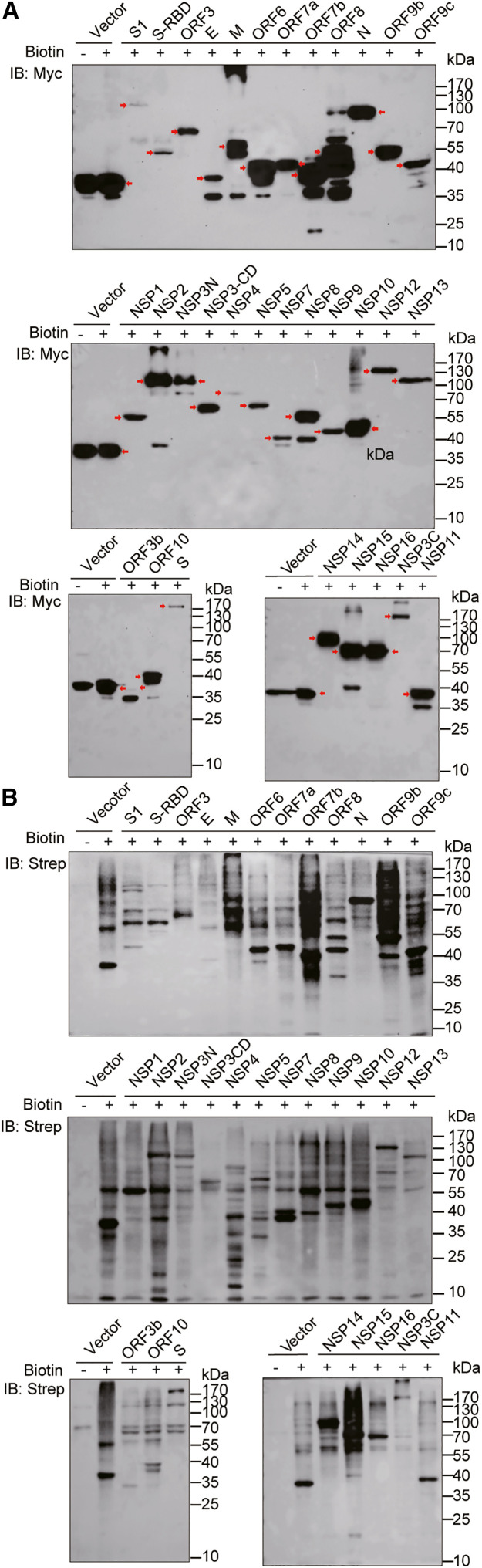
The expression of the TurboID-tagged viral proteins in HEK293T cells was detected by western blot assays using an anti-Myc antibody (Figure 2 A). A total of 32 of 33 constructs were expressed and had appropriate protein size. Only the expression of NSP6 was not detected and it was excluded from further analysis. Then, we verified the biotinylation of the proximal proteins using HRP-conjugated streptavidin (Strep) (Figure 2B). The distinct smear bands of various viral proteins represent the proximal proteins that were biotinylated by biotin ligase. As expected, significant and distinctive smear bands of the biotinylated proteins were observed by western blot analysis, indicating that the TurboID-tagged viral proteins effectively biotinylated proximal proteins of the targets.
Figure 2.
Expression and biotinylation of proximal proteins of SARS-CoV-2 proteins
(A) The expression of viral proteins in HEK293T cells was detected by the anti-Myc antibody. The red arrowheads indicate the expression of target proteins.
(B) The biotinylation of proximal proteins in HEK293T cells by SARS-CoV-2 proteins was detected by HRP-conjugated Strep.
Systematic proximity labeling map of SARS-CoV-2 proteins
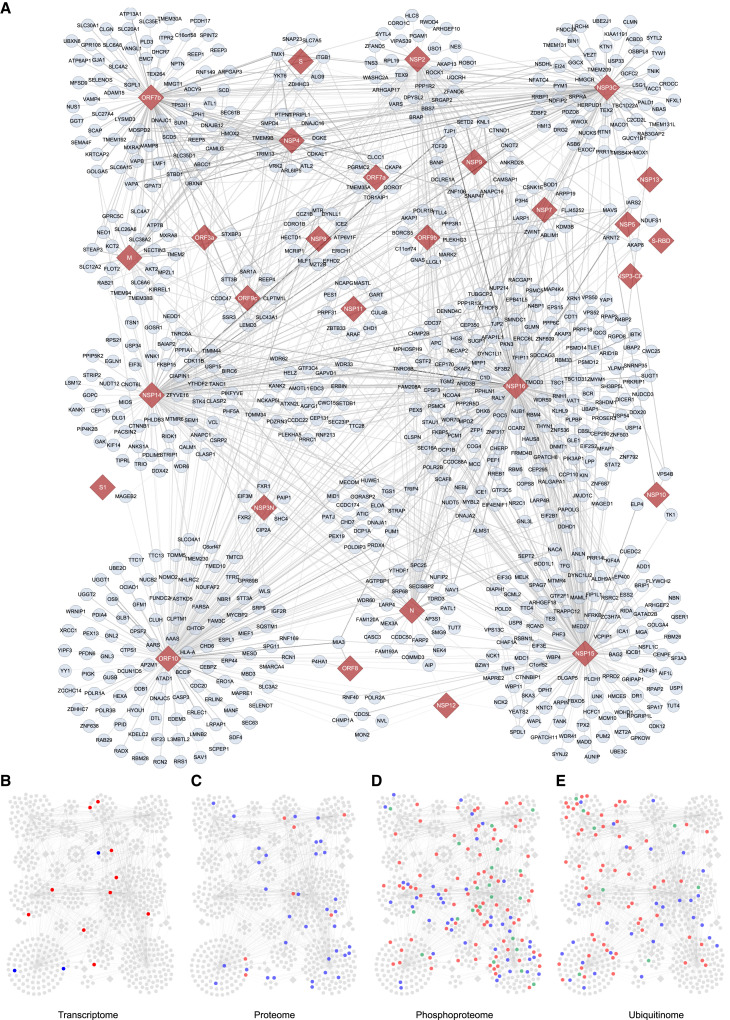
Proximity labeled samples of SARS-CoV-2 with three replicates were identified by mass spectrometry. Approximately 29% (∼21,000 proteins) of the identified proteins contained 1 to 39 biotinylated sites in the original data (Table S2A), which shows that the anti-biotin antibody effectively enriched biotinylated peptides. After filtering, we finally obtained 1,388 interactions of SARS-CoV-2 and human proteins through the TurboID assay (Figure 3 ; Table S2B; Data S1). Among the proximal proteins, we found that 90.7% of hits (1,259 proteins) emerged in all three replicates, and the other 9.3% hits (129 proteins) emerged in two replicates. The Pearson's correlation coefficient of biological replicates of each protein sample ranged from 0.72 to 0.98 (Figure S1). The number of interactions ranged from 1 to 264 for each viral protein (Table S2C). No interaction was obtained for the following viral proteins: E, NSP1, ORF3b, and ORF6.
Figure 3.
Proximity labeling map of SARS-CoV-2
(A) The 1,388 identified high-confidence human proximal proteins (blue circle nodes) for the SARS-CoV-2 proteins (red diamond nodes). The thickness of the edges is proportional to the ratio values of biotinylated sites in the sample and control.
(B–E) The proximal proteins were mapped to altered proteins of the transcriptome (B), proteome (C), phosphoproteome (D) and ubiquitinome (E). Red circle nodes, upregulated proteins; blue circle nodes, downregulated proteins; green circle nodes, dynamically regulated proteins; gray diamond nodes, viral proteins.
To define the function of the viral proteins, we classified the proximal proteins for each protein by Gene Ontology analysis (Figures S2A–S2C; Tables S3A–S3C). As expected, the significantly enriched proteins among the viral proteins were involved in the regulation of viral processes (NSP15, NSP3C, NSP7, and ORF10), Golgi vesicle transport (NSP14, NSP15, NSP16, NSP3C, and ORF7b), and RNA splicing (NSP15 and NSP16) (Figure S2A). NSP3C specifically enriched proteins involved in autophagy. The proximal proteins of ORF7b participate in the regulation of steroid metabolic processes. These proteins tended to be localized in the ER(NSP3C, ORF10), spindle (N, NSP2, NSP14, NSP15, NSP16, and ORF10), ribonucleoprotein granules (N, NSP2, NSP3N, NSP8, NSP14, NSP15, NSP16, and ORF10), and cell-cell junctions (M and NSP14) (Figure S2B). The various subcellular localizations of the proximal proteins suggest that the viral proteins affect different functions of the host. Of interest, each of the viral proteins had specific molecular functions (Figure S2C). The M protein enriches proximal proteins of SNAP receptor activity and active transmembrane transporter activity. The N protein tends to interact with proteins involved in ribonucleoprotein complex binding. The identified proteins of NSP2, NSP3C, NSP14, NSP15, NSP16, ORF3a, and ORF10 function in cadherin binding. The binding partners of these viral proteins might be useful for elucidating their biological processes, molecular functions, and cellular localization during SARS-CoV-2 infection. To further investigate the signaling pathways related to SARS-CoV-2 infection, the enriched pathways were analyzed (https://reactome.org) (Figure S2D; Table S3D). We identified numerous proteins that are reportedly related with SARS-CoV-2 infection, such as interferon (IFN), NF-κB, vascular endothelial growth factor (VEGF), and mRNA splicing-related proteins. Interestingly, we revealed that the Hippo and Wingless-INT (WNT)/β-catenin pathways were also targeted by SARS-CoV-2. The functional roles of these pathways in SARS-CoV-2 infection remain to be investigated (Figure S2D and Table S3D).
Integration analysis of the proximity labeling map and multi-omics data of SARS-CoV-2
We compared our data with previous interactome data of SARS-CoV-2 obtained by traditional AP-MS (Gordon et al., 2020b; Stukalov et al., 2021). Although we used almost the same set of viral proteins, differential results were obtained for each of the viral proteins (Figures S3A and S3B). The overlap rates were low for the three datasets, and only 57 interactions were shared by three groups, of which 36 interactions were commonly identified by this study. Unexpectedly, the two datasets from traditional AP-MS shared only 21 interactions, possibly due to the different cell lines and analysis strategies used. We used antibody-based technology to enrich biotinylated peptides in the TurboID assay and three stringent steps to filter non-specific proteins. The results of this study are complementary to those of the previous two studies, which suggests the necessity of using different methods to comprehensively elucidate the interactome of SARS-CoV-2 and its host.
To further investigate the functions of the proximal proteins, we mapped them to those identified by multi-omics studies after SARS-CoV-2 infection (Stukalov et al., 2021), including the transcriptome, proteome, phosphoproteome, and ubiquitinome. These proteins might be directly regulated by viral proteins. A total of 245 interactors were significantly altered after SARS-CoV-2 infection (Figures 3 and S4A–S4D; Table S4). As expected, only a few proteins (13 proteins) mapped to the transcriptome, which suggests that the proximal proteins tend to be regulated by direct physical interactions rather than at the transcriptional level. At the protein level, most of the proximal proteins were downregulated (35 of 42 proteins), which implies that their related biological processes are negatively regulated by viral proteins. We found that most of the proximal proteins were changed at the levels of phosphorylation (140 proteins) and ubiquitination (96 proteins). For example, the phosphorylation levels of several members of the Hippo pathway were altered, such as AMOTL1, TJP1, TJP2, and YAP1, which might enhance the activation of Yes-associated protein (YAP) and lead to a decrease in the level of IFNs. The ubiquitination level of a few components of the WNT/β-catenin pathway was altered after viral infection, including CTNNB1, CTNND1, PSMD12, and RNF213. At the transcriptome and proteome levels, the altered proximal proteins were consistently upregulated or downregulated during SARS-CoV-2 infection. However, the levels of phosphorylation and ubiquitination of the proximal proteins had different patterns during viral infection, which suggests that the interacting proteins are stringently regulated by posttranslational modification during the viral life cycle.
ITGB1 associates with hACE2 to mediate SARS-CoV-2 entry
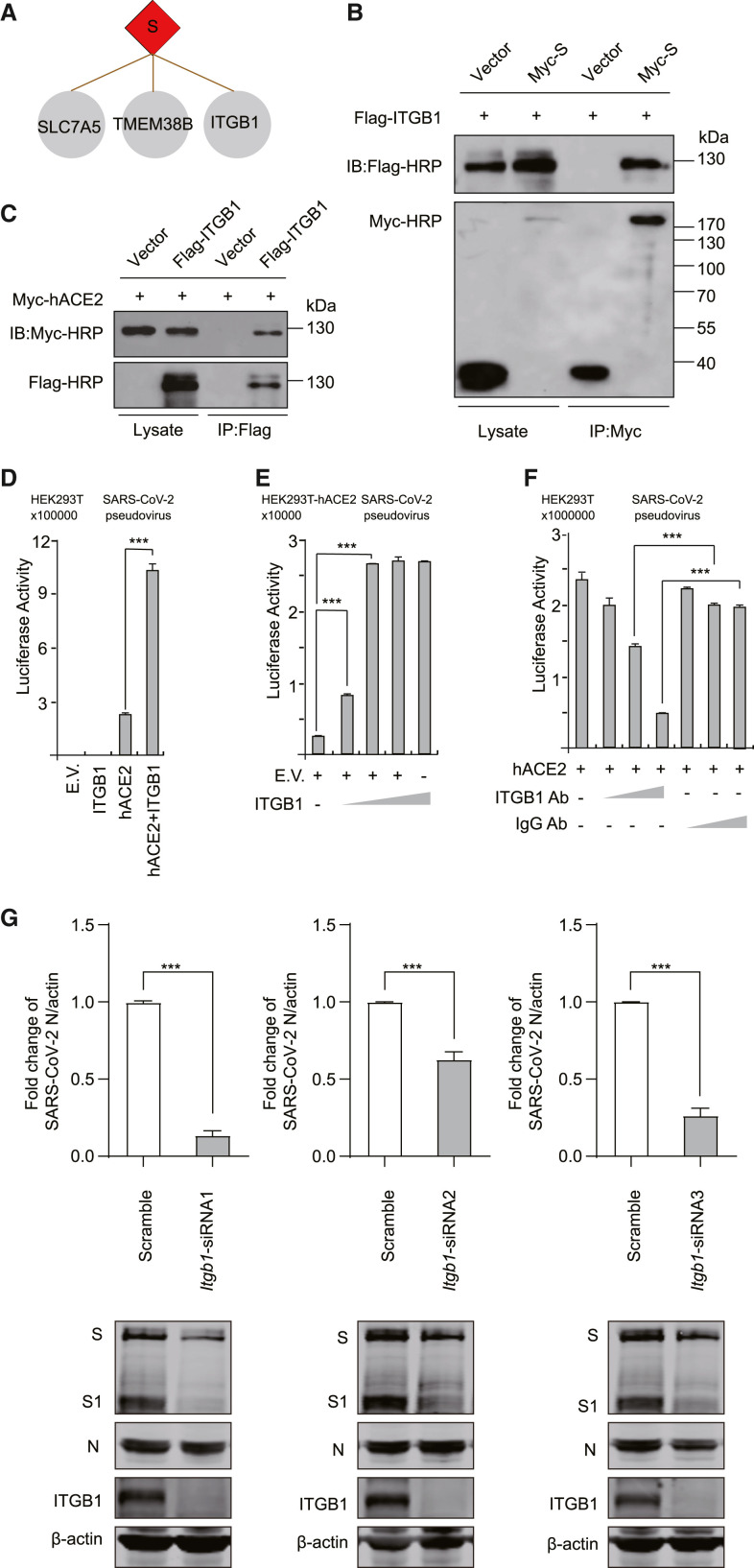
The S protein of SARS-CoV-2 forms a spike on the virion surface and is crucial for viral binding, fusion, and entry. The S protein binds to the host receptor, human angiotensin-converting enzyme 2 (hACE2), to enter cells (Walls et al., 2020; Zhou et al., 2020). Although the proximity labeling assay tends to identify intracellular proteins of targets, we still found three membrane proteins that interacted with the S protein: integrin subunit beta 1 (ITGB1), transmembrane protein 38B (TMEM38B), and Solute carrier family 7 member 5 (SLC7A5) (Figure 4 A). Of note, ITGB1 is an entry receptor of many viruses, including human echoviruses 1 and 8, cytomegalovirus, Epstein-Barr virus, human parvovirus B1, human rotavirus, and mammalian reovirus (Bergelson et al., 1993; Feire et al., 2010; Graham et al., 2003; Maginnis et al., 2006; Weigel-Kelley et al., 2003; Xiao et al., 2008). The functional role of ITGB1 in SARS-CoV-2 entry remains elusive.
Figure 4.
ITGB1 mediates the entry of SARS-CoV-2
(A) The binding partners of the S protein.
(B) Validation of the interactions of ITGB1 and S by co-immunoprecipitation assay. HEK293T cells were co-transfected with ITGB1 and S, immunoprecipited by Myc antibody, and detected by Myc or Flag antibodies.
(C) Validation of the interactions of ITGB1 and hACE2 by co-immunoprecipitation assay. HEK293T cells were co-transfected with ITGB1 and hACE2, immunoprecipited by Flag antibody, and detected by Myc or Flag antibodies.
(D) HEK293T cells were transfected with ITGB1 and hACE2. Then, the cells were infected with SARS-CoV-2 pseudovirus.
(E) HEK293T-hACE2 cells were transfected with gradually increased ITGB1. Then, the cells were infected with SARS-CoV-2 pseudovirus.
(F) HEK293T cells were transfected with hACE2. Then, the cells were infected with SARS-CoV-2 pseudovirus and treated with anti-ITGB1 antibody or scramble antibody.
(G) HeLa-hACE2 cells were transfected with scramble or ITGB1-specific siRNA oligos as indicated. The cells were infected with SARS-CoV-2. Total RNA was analyzed using real-time quantitative PCR (RT-qPCR) to determine the expression levels of the N protein. The cell lysates were analyzed using western blotting as indicated. β-Actin was used as a loading control.
∗p < 0.05, ∗∗∗p < 0.001 (two-tailed Student's t test), means + SD, n = 3. Data are representative of two (B and C) or three (D–G) independent experiments. E.V., empty vector.
To investigate the functional roles of ITGB1 in SARS-CoV-2 entry, we confirmed the interaction of ITGB1 and the S protein by co-immunoprecipitation assay (Figure 4B). Meanwhile, we found that ITGB1 was associated with hACE2 (Figure 4C). We identified that co-expression of ITGB1 with hACE2 significantly enhanced the entry of SARS-CoV-2 pseudovirus (Figure 4D). ITGB1 alone did not mediate this process. Furthermore, we found that ITGB1 dose dependently enhanced the entry of SARS-CoV-2 pseudovirus into HEK293T-hACE2 cells (Figure 4E). Next, the entry process of the SARS-CoV-2 pseudovirus was attenuated by an ITGB1 antibody in a dose-dependent manner (Figure 4F). Moreover, we knocked down the expression of ITGB1 in HeLa-hACE2 cells by RNAi assay (Figure 4G). As expected, ITGB1 expression was significantly reduced by three different small interfering RNAs (siRNAs) (Figure 4G). Then, the cells were infected with SARS-CoV-2. Total RNA or cell lysates were harvested at 24 h post infection. In contrast to the scramble siRNA group, knockdown of ITGB1 significantly reduced the replication of SARS-CoV-2 (Figure 4G). These results indicate that ITGB1 is a co-factor for SARS-CoV-2 entry.
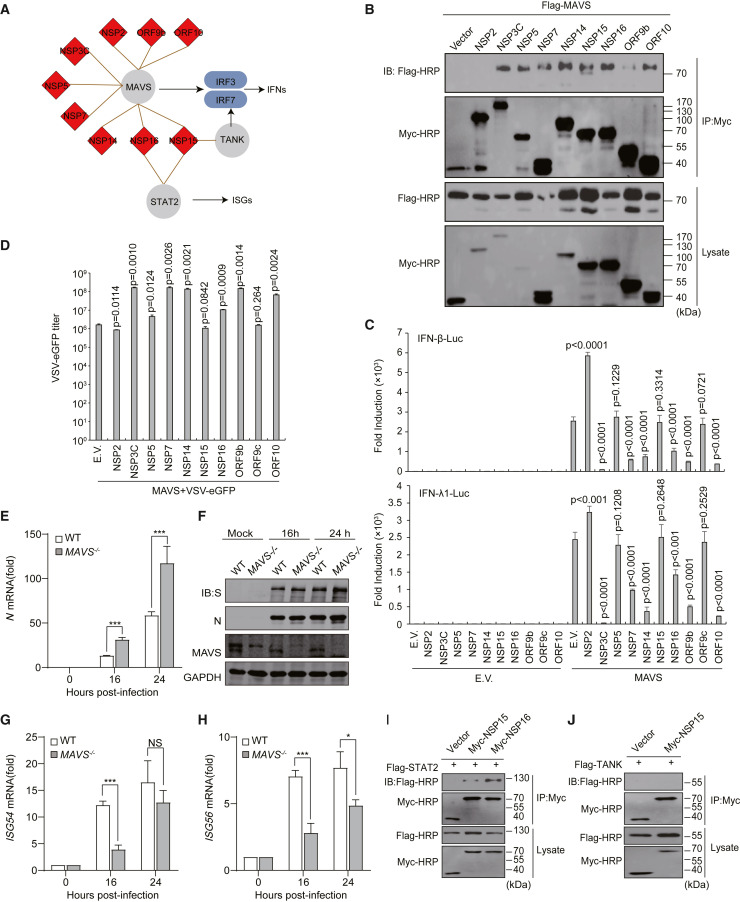
SARS-CoV-2 proteins inhibit MAVS-mediated IFN pathway
The innate immune system is crucial for the development of COVID-19 (Mangalmurti and Hunter, 2020). Non-resolving inflammation results in a cytokine storm, which is closely related to increasing severity of COVID-19 and poor clinical outcomes. It has been reported that SARS-CoV-2 regulates the immune response pathway (Mangalmurti and Hunter, 2020; Vabret et al., 2020). We found that several proteins involved in immune responses were targeted by SARS-CoV-2 proteins (Figure 5 A). A few viral proteins targeted the mitochondrial antiviral-signaling protein (MAVS) pathway, including NSP2, NSP3C, NSP5, NSP7, NSP14, NSP15, NSP16, ORF9b, and ORF10, suggesting that SARS-CoV-2 employs multiple proteins to inhibit the antiviral immunity of the host.
Figure 5.
Viral proteins interfere with the interferon pathway
(A) Innate immune pathways are targeted by viral proteins.
(B) Validation of the interactions of viral proteins and MAVS by co-immunoprecipitation assays.
(C) The effects of the SARS-CoV-2 viral proteins on MAVS-induced activation of IFN-β and IFN-λ1 luciferase reporters. The indicated vectors were co-transfected into HEK293T cells. E.V., empty vector.
(D) The effects of the SARS-CoV-2 viral proteins on MAVS-induced antiviral immunity against VSV-eGFP. The indicated vectors were co-transfected into HEK293T cells. VSV-eGFP (multiplicity of infection [MOI] = 0.001) was used to infect the cells. The culture supernatant was collected to determine the titers of extracellular VSV-eGFP using plaque assays.
(E) HeLa-hACE2 WT and HeLa-hACE2 MAVS−/− cells were infected with SARS-CoV-2. Total RNA was extracted at the indicated times and reverse transcribed to cDNA. The SARS-CoV-2 genome expression levels were measured by RT-qPCR.
(F) The cells were infected as in (E). The cell lysates were harvested at the indicated times and the protein expression levels were detected by western blot.
(G and H) The cells were treated as in (E), and ISG54 and ISG56 expression levels were measured by RT-qPCR.
(I and J) The indicated vectors were transfected into HEK293T cells, followed by immunoprecipitation with indicated antibody.
∗p < 0.05, ∗∗∗p < 0.001; NS, not significant (two-tailed Student's t test), means + SD, n = 3. Data are representative of two (B, I, and J) or three (C–H) independent experiments.
The main chymotrypsin-like protease (NSP5) and papain-like protease (NSP3) are essential for the processing of viral polyproteins. NSP3 cleaves ubiquitinated or ISGylated (ISG15 modification) proteins to inhibit antiviral immune responses (Swaim et al., 2020). Inhibition of NSP3 activates the antiviral IFN pathway and reduces viral particle release from infected cells. NSP5 and NSP3 are promising therapeutic targets for antiviral drugs. IFNs are the most important components of antiviral defense. To our knowledge, IFNs are suppressed in patients with moderate COVID-19 but are sustained at higher levels in patients with severe disease (Vabret et al., 2020). SARS-CoV-2 blocks the IFN pathway to interfere with host antiviral innate immunity. Moreover, we found that NSP15 and NSP16 bind to signal transducer and activator of transcription 2 (STAT2) (Figure 5A), which may inhibit the activation of downstream expression of interferon-stimulated genes (ISGs). It is likely that these viral proteins might lead to a dysfunctional immune response.
The aforementioned results strongly suggest that multiple viral proteins target antiviral signaling pathways through the mitochondrial protein MAVS. To further validate this finding, we investigated the interaction of viral proteins and MAVS by co-immunoprecipitation assays in mammalian cells (Figure 5B). With the exception of NSP2, the other eight viral proteins showed obvious associations with MAVS, including NSP3C, NSP5, NSP7, NSP14, NSP15, NSP16, ORF9b, and ORF10 (Figure 5B, upper panel). MAVS is a crucial protein for virus-triggered IFN signaling pathways (Seth et al., 2005). MAVS activates the transcription factors IRF3 and IRF7 (interferon regulatory factors 3 and 7) to regulate the expression of IFNs (Figure 5A).
To further validate that these viral proteins affect this pathway through MAVS, we co-transfected the viral proteins with MAVS and tested the activation of IFN-β and IFN-λ1 reporters. As expected, none of the viral proteins alone affected the activation of IFNs (Figure 5C). The expression of MAVS significantly activated IFN reporter genes. Six viral proteins blocked the activation of the IFN reporter, including NCP3C, NSP7, NSP14, NSP16, ORF9b, and ORF10. In contrast, the expression of NSP2 led to a significant increase in IFNs, but the reasons for this result remain elusive. NSP5 and NSP15 did not affect the activation of IFN reporters. ORF9c was used as a negative control. Activation of the MAVS signaling pathway contributes to antiviral immunity. Moreover, we investigated the replication of vesicular stomatitis virus-enhanced green fluorescent protein (VSV-eGFP), which is extremely sensitive to IFNs and is commonly used to study the effect of IFNs on viral replication, via co-transfection of MAVS and viral proteins (Figure 5D). Consistent with previous results, the replication of VSV-eGFP was increased by NSP3C, NSP7, NSP14, NSP16, ORF9b, and ORF10, and NSP2 decreased the viral titer of VSV-eGFP. Although NSP5 did not affect the reporter genes of IFNs (Figure 5C), it significantly increased the replication of VSV-eGFP (Figure 5D). NSP15 and ORF9c did not affect the replication of VSV-eGFP (Figure 5D). To further confirm the role of MAVS during infection, HeLa-hACE2 wild-type (WT) and HeLa-hACE2 MAVS −/− cells were infected with SARS-CoV-2 (Figures 5E–5H). Our results show that SARS-CoV-2 replication was increased in MAVS −/− cells at the RNA (Figure 5E) and protein levels compared to HeLa-hACE2 WT cells (Figure 5F). The induced expression of downstream ISGs, including ISG54 and ISG56, was significantly reduced in HeLa-hACE2 MAVS −/− cells after SARS-CoV-2 infection (Figures 5G and 5H). Additionally, we validated that NSP15 and NSP16 were associated with STAT2 (Figure 5I). However, the binding of NSP15 and TRAF family member associated NF-κB activator (TANK) was undetectable (Figure 5J).
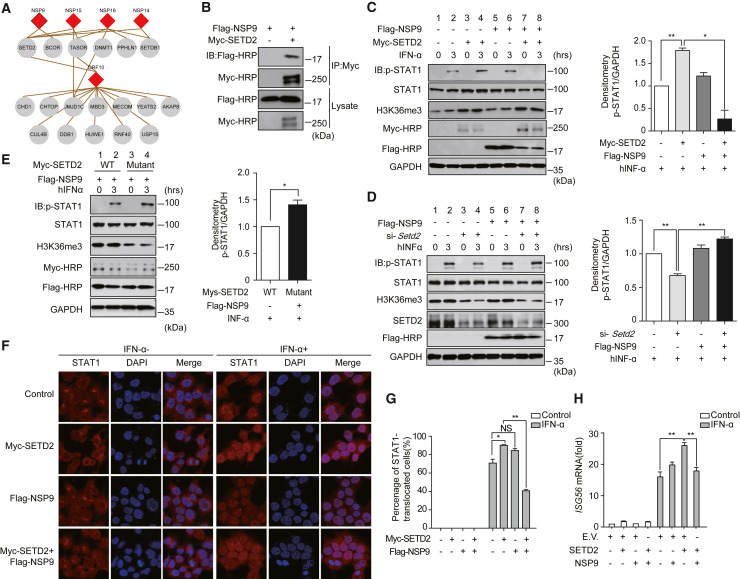
NSP9 targets the methyltransferase SETD2 to block IFN-STAT1 signaling
Epigenetic regulators are essential for viral infection. We identified that a few viral proteins targeted these molecules, including NSP9, NSP14, NSP15, NSP16, and ORF10 (Figure 6 A). NSP9 interacts with SET domain containing 2, histone lysine methyltransferase (SETD2) and BCL6 corepressor (BCOR). SETD2 also binds to NSP14, NSP15, NSP16, and ORF10, suggesting that these viral proteins might directly affect histone modifications. Transcription activation suppressor (TASOR) and periphilin 1 (PPHLN1) are components of the human silencing hub (HUSH) complex, which mediates epigenetic repression (Tchasovnikarova et al., 2015), and they were found to be partners of NSP16. The HUSH complex is recruited to genomic loci rich in H3K9me3, and it maintains transcriptional silencing by promoting the recruitment of SET domain bifurcated histone lysine methyltransferase 1 (SETDB1). SETDB1 was identified to associate with NSP14 and NSP16. Additionally, we found that DNA methyltransferase 1 (DNMT1) interacts with NSP16, which indicates that NSP16 may have a functional role in controlling DNA methylation. Our results showed that ORF10 associates with a few epigenetic regulators, including chromodomain helicase DNA binding protein 1 (CHD1), chromatin target of PRMT1 (CHTOP), and Jumonji domain containing 1C (JMJD1C). Several ubiquitination-related epigenetic proteins were enriched by ORF10, including cullin 4B (CUL4B), damage-specific DNA binding protein 1 (DDB1), HECT, UBA, and WWE domain containing E3 ubiquitin protein ligase 1 (HUWE1), ring finger protein 40 (RNF40), and ubiquitin-specific peptidase 15 (USP15). These results strongly suggest that SARS-CoV-2 might regulate host gene expression through epigenetic regulation.
Figure 6.
NSP9 inhibits the phosphorylation and nuclear translocation of STAT1 in a SETD2-dependent manner
(A) Epigenetic regulators associated with viral proteins.
(B) Validation of the interactions of NSP9 and SETD2 by co-immunoprecipitation assay.
(C) HEK293T cells were treated with hIFN-α (10 ng/mL). Phosphorylated STAT1, SETD2, and H3K36me3 in HEK293T cells were detected by the indicated antibodies. GAPDH was used as a loading control. Quantitative analysis of p-STAT1 is shown.
(D) p-STAT1, STAT1, H3K36me3, and SETD2 were detected in HEK293T cells. The cells were transfected with si-Control or si-Setd2 together with empty vector or FLAG-NSP9 and stimulated as in (C). Quantitative analysis of p-STAT1 is shown.
(E) p-STAT1, STAT1, and H3K36me3 were detected in HEK293T cells co-transfected with Myc-tagged SETD2 or mutated SETD2 and FLAG-NSP9, followed by hIFN-α treatment as in (C). Quantitative analysis of p-STAT1 is shown.
(F) The nuclear translocation of STAT1 was visualized by confocal microscopy. HEK293T cells were transfected with the indicated vectors for 24 h, followed by hIFN-α (10 ng/mL) treatment for 30 min or untreated. Scale bars, 50 μm.
(G) Quantitation of the nuclear translocation of STAT1.
(H) HEK293T cells were transfected with the indicated plasmids. hIFN-α was used to stimulate the activation of the JAK-STAT pathway. The cells were harvested for RNA isolation. The induction of ISG56 was evaluated by RT-qPCR. GAPDH was used as an internal control. The fold changes were quantified by the 2−ΔΔCt method.
∗p < 0.05, ∗∗p < 0.001; NS, not significant (two-tailed Student's t test), means +SD, n = 2 (C–E) or n = 3 (G and H). Data are representative of two (B–E) or three (F–H) independent experiments. E.V., empty vector.
The epigenetic regulator SETD2 is essential for the amplification of IFN signaling through methylation of signal transducer and activator of transcription 1 (STAT1) (Chen et al., 2017). We confirmed the binding of NSP9 and SETD2 by a co-immunoprecipitation assay (Figure 6B). Consistent with a previous report (Chen et al., 2017), overexpression of SETD2 significantly increased the level of STAT1 phosphorylation (Figure 6C, lane 4). We identified that NSP9 significantly attenuated STAT1 phosphorylation mediated by SETD2 (Figure 6C, lane 8). Moreover, knockdown of SETD2 attenuated the inhibition of STAT1 phosphorylation by NSP9 (Figure 6D, lane 8). Furthermore, we used a catalytically inactive mutant of SETD2, the R1625G mutant, which leads to a complete loss of histone H3 Lys-36 trimethylation (H3K36me3). We showed that the SETD2 mutant lost the ability to repress the level of STAT1 phosphorylation when co-transfected with NSP9 (Figure 6E, lane 4). Additionally, we found that NSP9 inhibited the nuclear translocation of STAT1 in a SETD2-dependent manner (Figures 6F and 6G). The expression of ISG56 induced by human IFN-α (hIFN-α) through the JAK-STAT pathway was also suppressed by NSP9 (Figure 6H). These results indicate that NSP9 blocks antiviral immunity through SETD2-dependent inhibition of STAT1.
NSP14 and NSP16 block the IFN signaling through the Hippo pathway
We found that the Hippo pathway is targeted by the viral proteins NSP14 and NSP16 (Figure S5A). YAP is a negative regulator of innate immunity against various viruses (Wang et al., 2017). Activation of YAP inhibits the expression of IFNs. The tight junction proteins 1 and 2 (TJP1 and TJP2) are regulators of YAP activation. Angiomotin-like 1 (AMOTL1) interacts with multiple tight junction proteins and reduces YAP activity (Yu and Guan, 2013). Serine/threonine kinase 4 (STK4, also known as MST1) is a kinase that phosphorylates and inhibits YAP1. The association of YAP regulators with NSP14 and NSP16 suggests that these viral proteins might block the expression of IFNs through the Hippo signaling pathway.
We confirmed the interaction of NSP14, NSP16, and TJP1 (Figures S5B and S5C). However, the binding of NSP14 and STK4, or NSP16 and YAP1, was undetectable. Xu and co-workers described that YAP/TAZ interacts with TANK-binding kinase 1 (TBK1) and abolishes virus-induced TBK1 activation (Zhang et al., 2017). Interferon regulatory factor 3 (IRF3) is phosphorylated by TBK1 to induce IFN transcription. Thus, we examined the activation of IRF3. The results showed that NSP14 and NSP16 impaired the phosphorylation of IRF3 induced by TBK1 (Figure S5D). The expression of the downstream target genes IFN-β and ISG1 was significantly repressed by NSP14 and NSP16 (Figures S5E and S5F). We then stimulated HEK293T cells with Sendai virus (SeV). The expression of IFN-β and ISG56 was significantly downregulated by NSP14 and NSP16 in cells stimulated with SeV (Figures S5G and S5H). Taken together, these results suggest that NSP14 and NSP16 negatively regulate IFN-β signaling partially via the Hippo pathway.
Potential targets revealed by the SARS-CoV-2 proximity labeling map and drug repurposing
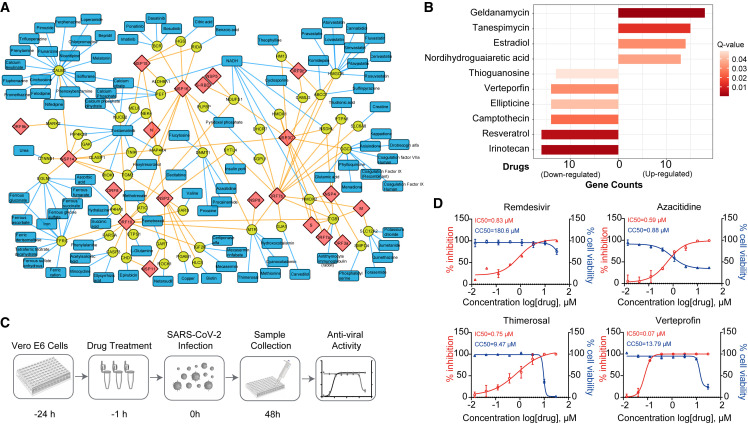
To identify the potential targets and already approved drugs for the therapeutic treatment of COVID-19, we assessed the proximal proteins of SARS-CoV-2 using the DrugBank database (Wishart et al., 2018). We obtained 70 proximal proteins targeted by 248 drugs (Table S5A), among which 101 drugs have already been approved by the Food and Drug Administration (FDA) (Figure 7 A). Several human proteins are targeted by dozens of drugs, including Calmodulin 1 (CALM 1), transferrin receptor (TFRC), and HMG-CoA reductase (HMGCR). CALM1, which is a member of the EF-hand calcium-binding protein family, binds to 21 approved drugs. Cholesterol metabolism-related proteins, including HMGCR, are enriched by the viral protein ORF7b, which indicates the potential roles of metabolic drugs in clinical testing. For example, lovastatin inhibits the lipid metabolism of the host. HMGCR is a rate-limiting enzyme in cholesterol synthesis and is targeted by statins. Consistent with this result, statins have anti-inflammatory effects, and the use of these drugs reduces the risk of mortality of COVID-19 patients (Zhang et al., 2020).
Figure 7.
Potential targets and the drug network revealed by the SARS-CoV-2 proximity labeling map
(A) The proximal proteins (yellow circle nodes) are targeted by approved drugs (blue rectangle nodes). The viral proteins of SARS-CoV-2 are shown as red diamond nodes.
(B) Ten drugs that target multiple proximal proteins obtained from the enrichment analysis by connectivity map. The length of the bar indicates the number of proximal proteins that are up- or downregulated by the drugs. The gradient red color represents the Q values (Benjamini-Hochberg adjusted p value).
(C) Schematic diagram of the antiviral assays by the selected compounds.
(D) Vero E6 cells were seeded before infection. For IC50 determination, the cells were pre-treated with drugs for 1 h at gradient concentrations. After 48 h, supernatants were harvested for RNA extraction. Then, the viral N mRNA was quantified by RT-qPCR and the inhibition ratio was calculated. Cell viability was evaluated by using a CCK8 kit. Red, percentage of inhibition; blue, cell viability.
Data are shown as means + SD of four independent experiments (n = 4) (D).
To systematically investigate the drugs that affected the proximal proteins, we mapped the proximal proteins of SARS-CoV-2 to the connectivity map (Lamb et al., 2006). Ten drugs were identified to significantly up-/downregulate the expression of multiple proximal proteins (Figure 7B; Table S5B). Among these drugs, estradiol significantly alleviates SARS-CoV-2 infection (Breithaupt-Faloppa et al., 2020). Currently, remdesivir is the only drug that has been approved by the FDA for treating COVID-19.
To validate the effects of the drugs during SARS-CoV-2 infection, we randomly selected 12 compounds from the list (Tables S5A and S5B) and tested their antiviral efficacy against SARS-CoV-2 in Vero E6 cells (Figures 7C and S6; Table S5C). Remdesivir was used as a positive control. We found that three of them significantly inhibited the replication of SARS-CoV-2, including azacitidine, thimerosal, and verteporfin (Figure 7D). Azacitidine has been used to treat cancer and has shown certain toxicity to cells, while the selective index of thimerosal and verteporfin in Vero E6 cells was greater than 10 (Figure 7D). Verteporfin significantly inhibited the replication of SARS-CoV-2 at a very low concentration (half maximal inhibitory concentration [IC50] = 0.07 μM).
Discussion
The interactome of SARS-CoV-2 and its host is essential to understand the fundamental processes of viral infection. Complementary to traditional AP-MS methods, biotin-ligase-based proximity labeling technology is a powerful tool to investigate proximal protein interactions in living cells and organisms (Branon et al., 2018; Doerr, 2018; Kim et al., 2016; Larochelle et al., 2019; Roux et al., 2012; Udeshi et al., 2017). Here, we constructed a proximity labeling map of SARS-CoV-2 and humans, which is a useful resource for antiviral drug screening and performing basic research on viral infection. We found significant enrichment of low-abundance proximal proteins compared with that with the traditional AP-MS method (p = 0.0065 and 0.0338 respectively, Student's t test; Figure S7A). As expected, the dataset generated in this study is complementary to two interactome datasets obtained previously using a different approach (Gordon et al., 2020b; Stukalov et al., 2021).
Limitations of the study
We used individual viral proteins rather than virulent virus to investigate the protein-protein interaction network of SARS-CoV-2 and humans. The overlap rate is low for the dataset of this study and other reports that used AP-MS technology (Gordon et al., 2020b; Stukalov et al., 2021). Due to the differences of technologies, cellular background, experimental procedures, and data processing, the lack of overlap from different studies is common. Gingras and co-workers compared the proximity labeling technology with AP-MS (Lambert et al., 2015). They found two approaches permitted the recovery of biologically meaningful interactions. Similar results are shown in another study (Liu et al., 2018). No interactions were obtained for the viral proteins E, NSP1, ORF3b, and ORF6. The TurboID tag might interfere with the interactions of these proteins. As shown previously (Gordon et al., 2020b; Stukalov et al., 2021), these viral proteins have significant fewer interactions on average. In this study, proteins proximal to viral proteins were covalently biotinylated by a promiscuous biotin ligase. The biotinylated peptides were captured by an anti-biotin antibody. Although we believe that most non-specific co-purified proteins were removed from the proximal dataset, they may still contain false-positives and should be validated by other methods. It should be noted that the proximal proteins might not have direct physical interactions with the viral proteins.
During the preparation of this manuscript, three related preprints were posted on bioRxiv (Laurent et al., 2020; Samavarchi-Tehrani et al., 2020; St-Germain et al., 2020). All of these papers used a Strep-based BioID assay to enrich the proximal proteins of SARS-CoV-2. Due to the high affinity of biotin and Strep, it is difficult to identify specific sites of biotinylated proteins, which is an important parameter to evaluate the confidence of the proximal proteins. In this study, we used an antibody against biotin to enrich the biotinylated peptides, and only those significantly enriched proteins with biotinylated sites were considered high-confidence proximal proteins of SARS-CoV-2. Approximately 21,100 of the identified proteins contained biotinylated sites in the original data (Figure S7B), while other reports lacked this information. We used a stringent strategy to filter the non-specifically binding proteins (Figure S7B), such as ribosomal proteins. Of the 1,388 high-confidence proximal proteins in this study, 1,092 proteins with biotinylated sites were not covered by the other three papers (Laurent et al., 2020; Samavarchi-Tehrani et al., 2020; St-Germain et al., 2020) (Figure S7C), which indicates the technical advantages of the antibody-based TurboID assay in identifying proximal proteins. We recovered 21.3% of proteins identified by the other three studies (Figure S7C). This dataset with high confidence is useful for the fields of medical research and clinical medicine of SARS-CoV-2.
SARS-CoV-2 requires host cellular factors for its entry, fusion, release, and replication. Mapping the virus-host interactome provides an effective way to elucidate the molecular mechanisms of SARS-CoV-2 infection. ITGB1, a binding partner of the structural protein S, is an entry receptor of other viruses, and we found that it mediates the entry of SARS-CoV-2. This suggests that the combination of anti-ITGB1 and anti-S antibody treatment might effectively block the replication of SARS-CoV-2. We also confirmed that multiple viral proteins bind to MAVS and inhibit the expression of IFNs. To identify viral proteins that inhibit IFN production, three groups systematically screened the proteins of SARS-CoV-2 (Lei et al., 2020; Xia et al., 2020; Yuen et al., 2020). Consistent with these results, we found that NSP3 and NSP14 block the type I IFN (IFN-I) induction (Figure 5). It has been reported that ORF9b blocks IFN-I responses by targeting TOM70 or NEMO (Gordon et al., 2020a; Jiang et al., 2020; Wu et al., 2021a). NSP7, NSP16, and ORF10 are regulators of IFN-I production that were identified by this study. Moreover, we revealed that these viral proteins impair IFN signaling through physical interaction with MAVS and in a MAVS-dependent manner.
The virus controls gene expression via epigenetic regulation and contributes to non-resolving inflammation. We validated that NSP9 associates with STED2 to block the JAK-STAT pathway. It is likely that the perturbation of epigenetics by SARS-CoV-2 is crucial for its life cycle. Meanwhile, our data suggested that the Hippo pathway is a potential target for therapeutic intervention. We found that these pathways are modulated by the viral proteins NSP14 and NSP16. This pathway might play important roles during viral infection. On the other hand, it is critical to validated the aforementioned proximal proteins of SARS-CoV-2 under physiological conditions, such as in SARS-CoV-2-infected cells or animals.
Furthermore, we proposed 70 potential targets, 101 drugs (FDA approved), and 10 compounds that target multiple proximal proteins for drug repurposing from the proximity labeling map of SARS-CoV-2. Of note, the deprivation of some endogenous metabolites might improve the symptoms of COVID-19. Currently, the treatment strategies for COVID-19 under study include antiviral agents, immunomodulators, antibodies, and adjunctive therapies. However, only limited strategies have been approved for use in clinical practice. A number of vaccines and approved drugs are being developed and are undergoing clinical trials against SARS-CoV-2. The proximity labeling map of SARS-CoV-2 should be valuable for designing effective antiviral drugs. Our data imply that immunomodulators may ameliorate severe COVID-19 and may be able to be used as antiviral agents, such as statins and estradiol. We also found that three compounds have significant antiviral activity against SARS-CoV-2 in Vero cells. These findings strongly support efforts to test these drugs against SARS-CoV-2 infection in clinical trials.
Significance
Currently, the interactome of SARS-CoV-2 and humans has been globally investigated by AP-MS. However, the overlap of these studies is very low even using similar technology. The knowledge of the interactions of SARS-CoV-2 with its host remains limited. We systematically investigated the interactome of 29 viral proteins in human cells by using an enzyme-catalyzed proximity labeling technology, TurboID. A total of 1,388 high-confidence human proximal proteins were identified as interactors of SARS-CoV-2 proteins. Notably, we found that SARS-CoV-2 manipulates key cellular processes involved in the antiviral and immune responses: (1) the membrane protein ITGB1 mediates the entry of SARS-CoV-2; (2) SARS-CoV-2 inhibits the activation of the interferon pathway through the mitochondrial protein MAVS; (3) NSP9 blocks antiviral immunity through the methyltransferase SETD2; (4) NSP14 and NSP16 block the IFN signaling through the Hippo pathway. We discovered three compounds significantly inhibit the replication of SARS-CoV-2: azacitidine, thimerosal, and verteporfin. We hope that this resource sheds light on the basic research into SARS-CoV-2 infection and the development of clinical drugs for COVID-19.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| anti-biotin | ImmuneChem | Cat#ICP0615; RRID:AB_2893170 |
| anti-S | Sino Biological Inc | Cat#40591-T62; RRID:AB_2893171 |
| anti-N | Sino Biological Inc | Cat#40143-mm08; RRID:AB_2827978 |
| anti-Myc | MBL Life Science | Cat#M047-3; RRID:AB_591112 |
| anti-Flag | MBL Life Science | Cat#M185-3L; RRID:AB_11123930 |
| anti-Flag-HRP | Sigma-Aldrich | Cat#A8592; RRID:AB_439702 |
| anti-β-actin | Sigma-Aldrich | Cat#A5441; RRID:AB_476744 |
| anti-p-STAT1-Y701 | Abcam | Cat#ab29045; RRID:AB_778096 |
| anti-H3K36me3 | Cell Signaling Technology | Cat#4909; RRID:AB_1950412 |
| anti-MAVS | Santa Cruz Biotechnology | Cat#SC-166583; RRID:AB_2012300 |
| anti-ITGB1 | Santa Cruz Biotechnology | Cat#SC-374429; RRID:AB_11012020 |
| anti-Myc-HRP | Santa Cruz Biotechnology | Cat#SC-40 HRP; RRID:AB_2857941 |
| anti-STAT1 | Beyotime Biotechnology | Cat#AF2512; RRID:AB_2893172 |
| anti-IRF3 | Beyotime Biotechnology | Cat#AF2485; RRID:AB_2893173 |
| anti-p-IRF3-S386 | Beyotime Biotechnology | Cat#AF1594; RRID:AB_2893174 |
| anti-SETD2 | Abclonal | Cat#A3194; RRID:AB_2764980 |
| anti-GAPDH | Proteintech | Cat#60004-1-Ig; RRID:AB_2107436 |
| anti-rabbit IgG conjugated HRP | R&D Systems | Cat#HAF008; RRID:AB_357235 |
| anti- mouse IgG conjugated HRP | R&D Systems | Cat#HAF007; RRID:AB_357234 |
| Alexa fluor® 594-conjugated affinipure goat anti-rabbit IgG (H+L) | ImmunoResearch Laboratories Inc | Cat#111-585-003; RRID:AB_2338059 |
| Bacterial and virus strains | ||
| DH5α | Tsingke Biotechnology Co. | Cat#TSV-A07 |
| SeV | (Zheng et al., 2020) | PMID: 33372174 |
| SARS-CoV-2 | Peking Union Medical College | IPBCAMS-WH-01/2019 |
| Chemicals, peptides, and recombinant proteins | ||
| Human IFN-alpha 1 | Cell Signaling Technology | Cat#8927 |
| Azacitidine | Selleck | Cat#S1782 |
| Camptothecin | Selleck | Cat#S1288 |
| Dasatinib | Selleck | Cat#S1021 |
| Glycyrrhizic acid | Selleck | Cat#S2302 |
| Melatonin | Selleck | Cat#S1204 |
| Methotrexate | Selleck | Cat#S1210 |
| Nicardipine | Selleck | Cat#S5255 |
| Pemetrexed | Selleck | Cat#S1135 |
| Sulfinpyrazone | Selleck | Cat#S4628 |
| Streptavidin-HRP | Abcam | Cat#ab7403 |
| Thimerosal | Selleck | Cat#S3646 |
| Torasemide | Selleck | Cat#S1698 |
| Verteporfin | Selleck | Cat#S1786 |
| Critical commercial assays | ||
| Dual-Luciferase Reporter Assay Kit | Vazyme | DL101-01 |
| Cell Counting Kit | Yeasen | 40203ES60 |
| Deposited data | ||
| PaxDB | (Wang et al., 2015) | PMID: 25656970 |
| Multilevel proteomics data of SARS-CoV-2 | (Stukalov et al., 2021) | PMID: 33845483 |
| Raw data of mass spectrometry | This study | PXD022086 |
| Experimental models: Cell lines | ||
| HEK293T | ATCC | Cat#CRL-11268 |
| HEK293T-hACE2 | (Wu et al., 2021b) | PMID: 34117209 |
| HeLa-hACE2 cells | This study | N/A |
| MAVS-knockout HeLa-hACE2 cells | This study | N/A |
| Vero | ATCC | CCL-81 |
| Oligonucleotides | ||
| See Table S6 for oligonucleotides | ||
| Recombinant DNA | ||
| pcDNA3.1-myc-TurboID | This study | N/A |
| pcDNA6B-TurboID-myc | This study | N/A |
| pCAG-TurboID-myc | This study | N/A |
| pCDH-CMV-MCS- EF1α-copGFP | System Biosciences | Cat#CD511B-1 |
| pLP1 | Thermofisher | Cat#K4975-00 |
| pLP2 | Thermofisher | Cat#K4975-00 |
| pRK5-Myc-TBK1 | (Meng et al., 2016) | PMID: 27125670 |
| Software and algorithms | ||
| Clusterprofiler | (Yu et al., 2012) | http://www.bioconductor.org/packages/release/bioc/html/clusterProfiler.html |
| Enrichr | (Kuleshov et al., 2016) | https://maayanlab.cloud/Enrichr/ |
| Mascot | Matrix Science | http://www.matrixscience.com/ |
| PANDA | (Chang et al., 2019) | https://sourceforge.net/projects/panda-tools/ |
| Pepdistiller | (Li et al., 2012) | http://sourceforge.net/projects/pepdistiller |
| Perl | Perl Core Team | http://www.perl.org |
| R | R Core Team | http://www.r-project.org |
| ReactomePA | (Yu and He, 2016) | http://www.bioconductor.org/packages/release/bioc/html/ReactomePA.html |
| REVIGO | (Supek et al., 2011) | http://revigo.irb.hr/ |
| SAINTexpress | (Teo et al., 2014) | https://sourceforge.net/projects/saint-apms/files/ |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Jian Wang (wangjian@bmi.ac.cn).
Materials availability
Correspondence and requests for materials should be addressed to the Lead Contact.
Experimental model and subject details
Cell culture and transfection
HEK293T, HEK293T-hACE2, HeLa-hACE2 cells, MAVS-knockout HeLa-hACE2, and Vero cells were cultured in Dulbecco's Modified Eagle’s Medium (plus 10% foetal bovine serum, supplemented with 1% penicillin-streptomycin) and maintained in a humidified 5% CO2 incubator at 37°C. Cells were tested on Jun 25, 2020, using the Mycoplasma Detection Kit and were negative: B/A ratio < 1 (no detected mycoplasma).
Approximately five million HEK293T or HEK293T-hACE2 cells were plated per 10-cm dish and transfected with indicated constructs after 16-20 h. PEI (Polysciences, PA) was formulated to a concentration of 1 μg/ml and used for transfection of the plasmids. After incubating DNA and PEI at a 1:5 (M/V) ratio in serum-free medium for 20 min, the transfection mix was added to the culture plate. For knockdown assay, HeLa-hACE2 cells were transfected with Lipofectamine RNAiMax (Thermo Fisher Scientific, MA) according to the manufacturer’s instructions.
Gene cloning and constructs
The DNA sequences of 29 ORFs encoded by the SARS-CoV-2 Wuhan-Hu-1 strain (GenBank: NC_045512.2) were synthesized (General Biol, China) and cloned into the pcDNA3.1 or pcDNA6B vector with the promiscuous BirA mutant (TurboID) and Myc tags by using a Seamless Cloning Kit (Beyotime Biotechnology, China). Among them, the DNA sequences of NSP3C, NSP14, S, ORF3b, and ORF10 were codon optimized to ensure their expression in HEK293T cells. To ensure their expression, some genes of SARS-CoV-2 were further cloned into the pCAG vector with TurboID and Myc tags. All of the constructs are available upon request.
Method details
Proximity labelling with TurboID assays
After transfection with TurboID-tagged constructs for 24 h, HEK293T cells were added with biotin (50 μM) for 10 minutes. The cells were washed twice with PBS at 4°C. Then, the cells were re-suspended with lysis buffer containing 8 M urea and 50 mM triethylammonium bicarbonate (TEABC), followed by sonication (30 W, 2 min pulses at 50% duty cycle). The protein concentration was measured by the BCA (bicinchoninic acid) assay. The expression of viral proteins and biotinylation of proximal proteins were detected by western blot analysis.
Approximately 6 mg protein was used for each of the three biological replicates. The samples were reduced by incubation with 20 mM DTT for 30 min at 56°C. Then, the samples were transferred to an Amicon Ultra-0.5 Centrifugal Filter Unit (Millipore, WI) and washed with 200 μL UA buffer (8 M urea, 100 mM Tris-HCl pH 8.5), and alkylation was performed by adding 100 μL 50 mM IAA for 30 min in the dark at room temperature. After washing with UA buffer and 50 mM TEABC, the proteins were digested with trypsin (Promega, WI) at 37°C overnight. The mixture of peptides was collected by centrifugation, eluted with 50 mM TEABC, and dried by vacuum centrifugation.
The peptides were resuspended in 1 mL of 50 mM MOPS pH 7.2, 10 mM sodium phosphate, and 50 mM NaCl (IAP buffer). The anti-biotin antibody was washed 3 times with IAP buffer and then added to the peptide mixture. The samples were incubated at 4°C in a rotating mixer overnight. The antibody beads were washed 4 times with ice-cold PBS. The biotinylated peptides were eluted with 50 μL 0.15% trifluoroacetic acid (TFA, Sigma-Aldrich, WI) twice. The eluted peptides were dried by vacuum centrifugation after desalting using StageTips.
Mass spectrometry analysis
Peptides were reconstituted in 8 μl 0.1% formic acid (FA) and analysed by using a Q-Exactive HF-X coupled to an UltiMat 3000 RSLCnano system (Thermo Fisher Scientific, MA). Briefly, 6 μl of the sample was loaded onto a silica-based nano-ESI column (360 μm OD × 150 μm ID) with a tip (3∼5 μM) packed with 15 cm of ReproSil-Pur 120 C18-AQ 1.9 μm beads. The samples were analysed using an 88 min LC-MS/MS method. The loading pump solvent was 0.1% FA (for the first 10 min), while NC_Pump (for the next 78 min) solvent A was 0.1% FA and solvent B was 80% acetonitrile/0.1% FA. For all the eluted peptides, we used the following gradient profile: (min:%B) 0:6; 10:6 (the first 10 min equated gradient with the loading pump solvent at a 0.3 μl/min mobile phase flow rate, the rest of the steps used a 0.6 μl/min mobile phase flow rate); 15:10; 70:30; 80:40; 80.1:95; 85:95; 85.1:6; and 88:6.
Full MS scan ranges of 350 to 1550 m/z with a resolution of 120,000, automatic gain control (AGC) target of 3 × 106, and a maximum ion time (max. IT) of 20 ms were used. The top 25 most intense ions from each MS1 scan were measured to MS2 at a range resolution of 200 to 2000 m/z, with a resolution of 15,000, AGC target of 2 × 104, fixed first mass of 110 m/z, minimum AGC target of 1 × 103, intensity threshold of 3.3 × 104, and max.IT of 30 ms. An isolation window of 1.6 m/z, an isolation offset of 0.0 m/z, and a normalized collision energy (NCE) of 27 was used. The dynamic exclusion time was set to 15 s, the FT master scan was set to preview mode and monoisotopic precursor selection was enabled. Charge exclusions were unassigned, 1, 8, or >8.
Mass spectrometry data processing
The MS data were searched against the Uniprot Swiss-Prot Human database (v20180725) with the addition of TurboID-tagged SARS-CoV-2 proteins by Mascot software (v2.6.0). The search parameters were as follows: the proteolytic enzyme was trypsin with a maximum of missed cleavages of 3; the peptide charge was set to 2+, 3+ and 4+; the peptide error tolerance was 10 ppm; the MS/MS error tolerance was 0.02 Da; and carbamidomethylation of cysteine (+57.02146 Da) as a fixed modification and biotinylation of lysine or N-terminal (+226.07759 Da) were the variable modifications. The peptide and protein false discovery rates were set to 0.01 by using Pepdistiller (Li et al., 2012) and PANDA (Chang et al., 2019), respectively. The cut-off ion score was set to 10. A three-step strategy was applied to identify high-confidence proximal proteins. First, the identified proteins were scored by SAINTexpress (Teo et al., 2014) with a cut-off value ≥ 0.6. Second, the top 5th percentile proteins ranked by occurrence rate were removed from the identified proteins (this study), the biotinylated proteins (this study), and the CRAPome contamination proteins (BirA-Flag as epitope tag). Third, the proteins were filtered by using the ratio value of biotinylated sites in the sample and control. The cut-off value was set to 2. The network of high-confidence human proximal proteins was visualized by Cytoscape (v3.7.1) (Shannon et al., 2003).
Co-immunoprecipitation and western blot assays
HEK293T cells were transfected with indicated combination of the plasmids. At least 24 h after transfection, the cells were harvested and washed with PBS (Phosphate buffered saline). The cells were lysed in lysis buffer (50 mM Tris-HCl pH 7.5, 100 mM NaCl, 1.5 mM MgCl2, 0.2% (v/v) NP-40, 5% (v/v) glycerol, protease inhibitor cocktail (Sigma-Aldrich, St. Louis, MO)) and sonicated (4°C, 50 W, 5 sec on, 5 sec off, 2 min). The samples were centrifuged at 12,000 g for 10 min. The protein A/G magnetic beads (MedChemExpress, NJ) were incubated with a Myc antibody (Medical and biological Lab, Japan) for 2 h at 4°C, followed by incubation with the supernatants of the samples overnight at 4°C. The beads were then washed three times in the lysis buffer. An aliquot (50 μl) of lysis buffer and the same amount of 2x Laemmli sample buffer were added to the beads. After boiling for 10 minutes at 100°C, a fraction of the input lysate and the immunoprecipitation samples were analysed by western blot analysis.
The protein samples were separated by SDS-PAGE, followed by immunoblot analyses with the indicated antibodies and detection with the ECL (Enhanced chemiluminescent) substrate (Thermo Fisher Scientific, MA).
Dual-luciferase reporter assays
To determine whether SARS-CoV-2 proteins that associate with MAVS can affect the activity of MAVS-induced type I and III IFN production, dual-luciferase reporter assays were performed. In brief, HEK293T cells, approximately 0.5 × 105 per well in 48-well plates, were co-transfected with a combination of the IFN-β-Luc or IFN-λ1-Luc plasmid and the MAVS plasmid and SARS-CoV-2 genes or alone. 36 h later, the cells were lysed to assess the luciferase activity using the Dual-Luciferase Reporter Assay Kit (Vazyme, China) by a Centro XS3 LB 960 microplate luminometer (Berthold Technologies, Germany). The ratio of firefly luciferase activity to Renilla luciferase activity served as the relative luciferase activity.
Pseudovirus production
Lentivirus-based SARS-CoV-2 S pseudoviruses were produced by cotransfection of the pCAG-SARS-CoV-2 SΔC19-FLAG plasmid, the lentiviral packaging plasmids pLP1 (expressing gag and pol) and pLP2 (expressing rev), and the transfer plasmid pCDH-CMV-MCS-EF1α-copGFP (encoding a luciferase reporter and a GFP marker) into HEK293T cells according to the manufacturer’s instructions (ViraPower™ Lentiviral Expression Systems, Invitrogen, USA). 6 h after transfection, the medium was replaced with fresh DMEM containing 10% FBS. 48 to 60 h after transfection, culture supernatants were harvested. After cell debris was removed via a brief centrifugation, the crude virus was concentrated by filtering through 50-kDa ultrafiltration membranes (Millipore, Germany) and was then stored at −80°C. To transduce target cells, SARS-CoV-2 S pseudovirus was diluted in DMEM containing 10 μg/mL polybrene and incubated with target cells for 12 h; subsequently, fresh DMEM containing 10% FBS was added. 48 to 60 h post inoculation, cells were observed via immunofluorescence microscopy to visualize the GFP-positive cells or harvested for the measurement of firefly luciferase activity as described above to determine the transduction efficiency.
Viral infection and plaque assays
Sendai virus (SeV) and VSV-enhanced green fluorescent protein (eGFP) was used to infect HEK293T and Vero-E6 cells as described previously (Wang et al., 2018). Briefly, before infection, the target cells were washed three times using prewarmed serum-free DMEM medium at 37°C; then, the virus diluted in serum-free DMEM was added to the target cells and incubated for 30 min. Subsequently, the virus-medium complexes were replaced with DMEM containing 10% FBS.
The titre of VSV-eGFP was assessed by plaque assays as described previously (Wang et al., 2018). Vero cells at approximately 100% confluency cultured in 24-well plates were infected with serial dilutions of VSV-eGFP. After a 30-min incubation, the medium was discarded, and DMEM containing 0.5% agar and 2% FBS was overlaid. When the agar turned solid, the cells were cultured for another 20 h. Then, the cells were fixed with a 1:1 methanol-ethanol mixture, after which the solid agarose-medium mix was removed. The cells were visualized by staining with 0.05% crystal violet. Then, the plaques on the monolayer were counted to calculate the virus titre.
RNA interference
Negative control small interfering RNA (siRNA) and specific siRNA were synthesized by Genepharma Corp (Shanghai, China). For knockdown, cells were plated and transfected with 50 nM specific siRNA oligos or non-targeting control siRNA using Lipofectamine RNAiMax (Thermo Fisher Scientific, MA) according to the manufacturer’s instructions. The cells were used for real-time PCR, western blot, and viral infection after treating with siRNA for 48 h. The siRNA sequences are as follows: ITGB1 siRNA1: 5’-GAACAGAUCUGAUGAAUGATT-3’; ITGB1 siRNA2: 5’-GAUCAUUGAUGCAUACAAUTT-3’; ITGB1 siRNA3: 5’- GUGGUUUCGAUGCCAUCAUTT-3’; si-Setd2: 5’-GCUCAGAGUUAACGUUUGA-3’.
SARS-CoV-2 infection
HeLa-hACE2 cells were plated and infected with SARS-CoV-2 at a multiplicity of infection (MOI) of 0.5 and were collected for RNA isolation or cell lysates 24 h post-infection. All experiment with SARS-CoV-2 virus were performed in the BSL-3 laboratory.
RNA extraction and RT-qPCR assays
For the test of IFN-β and ISG15, total RNA was extracted from cells using the Trizol reagent (Thermo Fisher Scientific, MA) according to the manufacturer’s instructions. For RT-qPCR analysis, cDNA was generated with ReverTra® qPCR RT Master Mix with gDNA Remover (TOYOBO, FSQ-301, Japan) and analyzed by quantitative real-time PCR using the KAPA SYBR® FAST qPCR Master Mix (2×) (Kapa Biosystems, Sigma-Aldrich, WI). The primer sequences are as follows: (1) Human IFN-β forward: 5′- CCAACAAGTGTCTCCTCCAAAT-3′; (2) Human IFN-β reverse: 5′- AATCTCCTCAGGGATGTCAAAGT-3′; (3) Human ISG15 forward: 5′- GGACAAATGCGACGAACC-3′; (4) Human ISG15 reverse: 5′- CCCGCTCACTTGCTGCTT-3′; (5) Human GAPDH forward: 5′- AGGGCTGCTTTTAACTCTGGT-3′; (6) Human GAPDH reverse: 5′- CCCCACTTGATTTTGGAGGGA-3′.
For the test of ISG54 and ISG56 after SARS-CoV-2 infection, total RNA was extracted by using TRIzol reagent and reverse transcript to cDNA by M-MLV Reverse Transcriptase (Promega, WI). The SARS-CoV-2 genome, ISG54 and ISG56 expression levels were measured by RT-qPCR using TB Green Premix Ex (Takara Bio, Japan). The primer sequences are as follows: (1) Human ISG54 forward: 5′- CTGCAACCATGAGTGAGAA-3′; (2) Human ISG54 reverse: 5′- CCTTTGAGGTGCTTTAGATAG-3′; (3) Human ISG56 forward: 5′- TACAGCAACCATGAGTACAA′; (4) Human ISG56 reverse: 5′- TCAGGTGTTTCACATAGGC -3′; (5) Human GAPDH forward: 5′- CGGAGTCAACGGATTTGGTCGTA′; (6) Human GAPDH reverse: 5′- AGCCTTCTCCATGGTGGTGAAGAC -3′; (7) Human β-actin forward: 5′- GGATGCAGAAGGAGATCACTG′; (8) Human β-actin reverse: 5′- CAAGTACTCCGTGTGGATCG -3′.
Anti SARS-CoV-2 infection assays by the selected compounds
Vero E6 cells were seeded in 96-well plates one day before infection. For IC50 determination, cells were pre-treated with drugs for 1 h with each drug at concentrations 0.013, 0.041, 0.123, 0.370, 1.111, 3.333, 10, 30 μM. The cells were subjected to viability assay or infected with SARS-CoV-2 at MOI of 0.1. After 48 h, supernatants were harvested for RNA extraction. Then the viral N mRNA was quantified by quantitative PCR. The inhibition ratio was obtained by dividing the viral copy number in drug-treated samples by those in the vehicle control samples. Cell viability was evaluated by using a CCK8 kit (Yeasen, China) according to the manufacturer’s instructions. The IC50 and CC50 were calculated. The selectivity index was calculated using the following formula: SI=CC50/IC50.
Fluorescence microscopy
The cells were cultured on Glass Bottom Cell Culture Dishes (Nest Scientific, China). Then, the culture medium was discarded and cells were washed with PBS for one time. Next, cells were fixed with 4% paraformaldehyde for 10 min. After that, the cells were washed with PBS, and then permeabilized in 0.2% Triton-PBS for 10-15 min on ice. After washing with PBS for 3 times, cells were blocked in PBST-BSA (PBS with 0.1%Tween-20, 3% BSA) for 1 h, and then incubated with STAT1 primary antibodies (Beyotime Biotechnology, China) diluted in PBST (PBS with 0.1% Tween-20) overnight. The cells were washed and followed by a fluorescently labeled secondary antibody (Alexa Fluor® 594-conjugated AffiniPure Goat anti-Rabbit IgG (H+L), Jackson ImmunoResearch, PA). The confocal images were examined using a microscope (LSM 880 + ELYRA S1, ZEISS, Germany) equipped with 100×/1.40 NA oil objectives.
Gene ontology and pathway over-representation analysis
The enriched terms according Gene Ontology analysis were analysed by clusterProfiler (Yu et al., 2012) with the default parameters. GO terms with Q-values less than 0.05 and more than 2 genes were considered to be significantly enriched. Redundant GO terms were removed by REViGO (Supek et al., 2011), using the following parameters: allowed similarity, 0.5; background database, Homo sapiens; and semantic similarity measure used, SimRel. The enriched Reactome pathways were analysed by ReactomePA (Yu and He, 2016).
Integration analysis of the proximity labelling map with multi-omics data
The data of the transcriptome, proteome, ubiquitinome, phosphoproteome and interactome were obtained from references (Gordon et al., 2020b; Stukalov et al., 2021). Proteins with a significantly changed abundance at more than one time point were selected. Conflicting results from the DDA (data-dependent acquisition) and DIA (data-independent acquisition) data were removed. The networks were visualized by Cytoscape software.
Analysis of the potential drugs and drug targets
Information from DrugBank (Wishart et al., 2018) was collected by the R package dbdataset (https://github.com/MohammedFCIS/dbdataset). The potential drugs and drug targets were mapped to the proximal proteins of SARS-CoV-2. The related information was collected, including type, group, category, actions and PMID (PubMed ID). The network of approved drugs and their targets was visualized by Cytoscape. Moreover, to identify drugs that regulate the expression of the proximal proteins, we mapped the proximal proteins to the Connectivity Map (Lamb et al., 2006) using Enrichr webtools (Kuleshov et al., 2016). An adjusted p value less than 0.05 was considered significant.
Quantification and statistical analysis
Unless otherwise specified, all data plotting and statistical analyses were performed using R 4.1.0 (R Core Team), and the error bars represent the SD. The statistical significance of comparisons between two groups was analyzed with two-tailed Student’s t-test. Only p values of 0.05 or lower were considered statistically significant, and ∗p < 0.05, ∗∗p < 0.01, ∗∗∗∗p < 0.001. Statistical details of experiments can be found in the figure legends.
Acknowledgments
This research was funded by grants from the National Key Research and Development Program (2017YFA0505700 to Jian Wang; 2020YFA0707600 to X.L.); from the National Key Lab of Proteomics (SKLP-K201805, SKLP-K201804 to Jian Wang); from the Key Research and Development Program of Shandong Province (2020CXGC011305 to P.-H.W); from the Natural Science Foundation of China (82101856 to P.-H.W); from the Natural Science Foundation of Shandong Province (ZR2020QC085 to P.-H.W.), and from the Natural Science Foundation of Jiangsu Province (BK20200225 to P.-H.W.).
Author contributions
P.-H.W. and Jian Wang conceived the project. J.Z. and M.-W.Z. cloned all of the viral genes of SARS-CoV-2. Yuehui Zhang, L.S., C.J., and Yanan Zhao performed TurboID assay. C.J., Yuchen Liu, and Jian Wang performed the bioinformatics analyses. Yuehui Zhang, X.Z., C.T., J.Z., Yujiao Liu, Q.M., and L.L. carried out the co-immunoprecipitation assay and functional validation experiments. X.L., W.W., and X.X. performed the assays related to SARS-CoV-2 virus. JianWang, P.-H.W., F.H., Jianwei Wang, and C.T. supervised the work. All of the authors contributed to writing and approving the manuscript. The vectors of TBK1 were kindly provided by Dr. Pinglong Xu from Zhejiang University.
Declaration of interests
The authors declare no competing interests.
Published: October 20, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.chembiol.2021.10.008.
Supplemental information
Data and code availability
All proteomics raw data have been deposited to the ProteomeXchange Consortium via the iProX (Ma et al., 2019) partner repository with the dataset identifier ProteomeXchange: PXD022086. All data reported in this paper will be shared by the lead contact upon request. This paper does not report original code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Bergelson J.M., St John N., Kawaguchi S., Chan M., Stubdal H., Modlin J., Finberg R.W. Infection by echoviruses 1 and 8 depends on the alpha 2 subunit of human VLA-2. J. Virol. 1993;67:6847–6852. doi: 10.1128/jvi.67.11.6847-6852.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branon T.C., Bosch J.A., Sanchez A.D., Udeshi N.D., Svinkina T., Carr S.A., Feldman J.L., Perrimon N., Ting A.Y. Efficient proximity labeling in living cells and organisms with TurboID. Nat. Biotechnol. 2018;36:880–887. doi: 10.1038/nbt.4201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breithaupt-Faloppa A.C., Correia C.J., Prado C.M., Stilhano R.S., Ureshino R.P., Moreira L.F.P. 17beta-Estradiol, a potential ally to alleviate SARS-CoV-2 infection. Clinics (Sao Paulo) 2020;75:e1980. doi: 10.6061/clinics/2020/e1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C., Li M., Guo C., Ding Y., Xu K., Han M., He F., Zhu Y. PANDA: a comprehensive and flexible tool for quantitative proteomics data analysis. Bioinformatics. 2019;35:898–900. doi: 10.1093/bioinformatics/bty727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K., Liu J., Liu S., Xia M., Zhang X., Han D., Jiang Y., Wang C., Cao X. Methyltransferase SETD2-mediated methylation of STAT1 is critical for interferon antiviral activity. Cell. 2017;170:492–506 e414. doi: 10.1016/j.cell.2017.06.042. [DOI] [PubMed] [Google Scholar]
- Doerr A. Proximity labeling with TurboID. Nat. Methods. 2018;15:764. doi: 10.1038/s41592-018-0158-0. [DOI] [PubMed] [Google Scholar]
- Feire A.L., Roy R.M., Manley K., Compton T. The glycoprotein B disintegrin-like domain binds beta 1 integrin to mediate cytomegalovirus entry. J. Virol. 2010;84:10026–10037. doi: 10.1128/JVI.00710-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon D.E., Hiatt J., Bouhaddou M., Rezelj V.V., Ulferts S., Braberg H., Jureka A.S., Obernier K., Guo J.Z., Batra J., et al. Comparative host-coronavirus protein interaction networks reveal pan-viral disease mechanisms. Science. 2020;370:eabe9403. doi: 10.1126/science.abe9403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon D.E., Jang G.M., Bouhaddou M., Xu J., Obernier K., White K.M., O'Meara M.J., Rezelj V.V., Guo J.Z., Swaney D.L., et al. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020;583:459–468. doi: 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham K.L., Halasz P., Tan Y., Hewish M.J., Takada Y., Mackow E.R., Robinson M.K., Coulson B.S. Integrin-using rotaviruses bind alpha2beta1 integrin alpha2 I domain via VP4 DGE sequence and recognize alphaXbeta2 and alphaVbeta3 by using VP7 during cell entry. J. Virol. 2003;77:9969–9978. doi: 10.1128/JVI.77.18.9969-9978.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H.W., Zhang H.N., Meng Q.F., Xie J., Li Y., Chen H., Zheng Y.X., Wang X.N., Qi H., Zhang J., et al. SARS-CoV-2 Orf9b suppresses type I interferon responses by targeting TOM70. Cell. Mol. Immunol. 2020;17:998–1000. doi: 10.1038/s41423-020-0514-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D., Lee J.Y., Yang J.S., Kim J.W., Kim V.N., Chang H. The architecture of SARS-CoV-2 transcriptome. Cell. 2020;181:914–921 e910. doi: 10.1016/j.cell.2020.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D.I., Jensen S.C., Noble K.A., Kc B., Roux K.H., Motamedchaboki K., Roux K.J. An improved smaller biotin ligase for BioID proximity labeling. Mol. Biol. Cell. 2016;27:1188–1196. doi: 10.1091/mbc.E15-12-0844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuleshov M.V., Jones M.R., Rouillard A.D., Fernandez N.F., Duan Q., Wang Z., Koplev S., Jenkins S.L., Jagodnik K.M., Lachmann A., et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016;44:W90–W97. doi: 10.1093/nar/gkw377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam T.T., Jia N., Zhang Y.W., Shum M.H., Jiang J.F., Zhu H.C., Tong Y.G., Shi Y.X., Ni X.B., Liao Y.S., et al. Identifying SARS-CoV-2-related coronaviruses in Malayan pangolins. Nature. 2020;583:282–285. doi: 10.1038/s41586-020-2169-0. [DOI] [PubMed] [Google Scholar]
- Lamb J., Crawford E.D., Peck D., Modell J.W., Blat I.C., Wrobel M.J., Lerner J., Brunet J.P., Subramanian A., Ross K.N., et al. The Connectivity Map: using gene-expression signatures to connect small molecules, genes, and disease. Science. 2006;313:1929–1935. doi: 10.1126/science.1132939. [DOI] [PubMed] [Google Scholar]
- Lambert J.P., Tucholska M., Go C., Knight J.D., Gingras A.C. Proximity biotinylation and affinity purification are complementary approaches for the interactome mapping of chromatin-associated protein complexes. J. Proteomics. 2015;118:81–94. doi: 10.1016/j.jprot.2014.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larochelle M., Bergeron D., Arcand B., Bachand F. Proximity-dependent biotinylation mediated by TurboID to identify protein-protein interaction networks in yeast. J. Cell Sci. 2019;132:jcs232249. doi: 10.1242/jcs.232249. [DOI] [PubMed] [Google Scholar]
- Laurent E.M.N., Sofianatos Y., Komarova A., Gimeno J.-P., Tehrani P.S., Kim D.-K., Abdouni H., Duhamel M., Cassonnet P., Knapp J.J., et al. Global BioID-based SARS-CoV-2 proteins proximal interactome unveils novel ties between viral polypeptides and host factors involved in multiple COVID19-associated mechanisms. bioRxiv. 2020 doi: 10.1101/2020.08.28.272955. [DOI] [Google Scholar]
- Lei X., Dong X., Ma R., Wang W., Xiao X., Tian Z., Wang C., Wang Y., Li L., Ren L., et al. Activation and evasion of type I interferon responses by SARS-CoV-2. Nat. Commun. 2020;11:3810. doi: 10.1038/s41467-020-17665-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N., Wu S., Zhang C., Chang C., Zhang J., Ma J., Li L., Qian X., Xu P., Zhu Y., et al. PepDistiller: a quality control tool to improve the sensitivity and accuracy of peptide identifications in shotgun proteomics. Proteomics. 2012;12:1720–1725. doi: 10.1002/pmic.201100167. [DOI] [PubMed] [Google Scholar]
- Li Q., Guan X., Wu P., Wang X., Zhou L., Tong Y., Ren R., Leung K.S.M., Lau E.H.Y., Wong J.Y., et al. Early transmission dynamics in Wuhan, China, of novel coronavirus-infected pneumonia. N. Engl. J. Med. 2020;382:1199–1207. doi: 10.1056/NEJMoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Salokas K., Tamene F., Jiu Y., Weldatsadik R.G., Ohman T., Varjosalo M. An AP-MS- and BioID-compatible MAC-tag enables comprehensive mapping of protein interactions and subcellular localizations. Nat. Commun. 2018;9:1188. doi: 10.1038/s41467-018-03523-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu R., Zhao X., Li J., Niu P., Yang B., Wu H., Wang W., Song H., Huang B., Zhu N., et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395:565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J., Chen T., Wu S., Yang C., Bai M., Shu K., Li K., Zhang G., Jin Z., He F., et al. iProX: an integrated proteome resource. Nucleic Acids Res. 2019;47:D1211–D1217. doi: 10.1093/nar/gky869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maginnis M.S., Forrest J.C., Kopecky-Bromberg S.A., Dickeson S.K., Santoro S.A., Zutter M.M., Nemerow G.R., Bergelson J.M., Dermody T.S. Beta1 integrin mediates internalization of mammalian reovirus. J. Virol. 2006;80:2760–2770. doi: 10.1128/JVI.80.6.2760-2770.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangalmurti N., Hunter C.A. Cytokine storms: understanding COVID-19. Immunity. 2020;53:19–25. doi: 10.1016/j.immuni.2020.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellacheruvu D., Wright Z., Couzens A.L., Lambert J.P., St-Denis N.A., Li T., Miteva Y.V., Hauri S., Sardiu M.E., Low T.Y., et al. The CRAPome: a contaminant repository for affinity purification-mass spectrometry data. Nat. Methods. 2013;10:730–736. doi: 10.1038/nmeth.2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng F., Zhou R., Wu S., Zhang Q., Jin Q., Zhou Y., Plouffe S.W., Liu S., Song H., Xia Z., et al. Mst1 shuts off cytosolic antiviral defense through IRF3 phosphorylation. Genes Dev. 2016;30:1086–1100. doi: 10.1101/gad.277533.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rickard D.J., Sehon C.A., Kasparcova V., Kallal L.A., Haile P.A., Zeng X., Montoute M.N., Poore D.D., Li H., Wu Z., et al. Identification of selective small molecule inhibitors of the nucleotide-binding oligomerization domain 1 (NOD1) signaling pathway. PLoS One. 2014;9:e96737. doi: 10.1371/journal.pone.0096737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux K.J., Kim D.I., Raida M., Burke B. A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells. J. Cell Biol. 2012;196:801–810. doi: 10.1083/jcb.201112098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samavarchi-Tehrani P., Abdouni H., Knight J.D.R., Astori A., Samson R., Lin Z.-Y., Kim D.-K., Knapp J.J., St-Germain J., Go C.D., et al. A SARS-CoV-2 – host proximity interactome. bioRxiv. 2020 doi: 10.1101/2020.09.03.282103. [DOI] [Google Scholar]
- Seth R.B., Sun L., Ea C.K., Chen Z.J. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell. 2005;122:669–682. doi: 10.1016/j.cell.2005.08.012. [DOI] [PubMed] [Google Scholar]
- Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St-Germain J.R., Astori A., Samavarchi-Tehrani P., Abdouni H., Macwan V., Kim D.-K., Knapp J.J., Roth F.P., Gingras A.-C., Raught B. A SARS-CoV-2 BioID-based virus-host membrane protein interactome and virus peptide compendium: new proteomics resources for COVID-19 research. bioRxiv. 2020 doi: 10.1101/2020.08.28.269175. [DOI] [Google Scholar]
- Stukalov A., Girault V., Grass V., Karayel O., Bergant V., Urban C., Haas D.A., Huang Y., Oubraham L., Wang A., et al. Multilevel proteomics reveals host perturbations by SARS-CoV-2 and SARS-CoV. Nature. 2021;594:246–252. doi: 10.1038/s41586-021-03493-4. [DOI] [PubMed] [Google Scholar]
- Supek F., Bosnjak M., Skunca N., Smuc T. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS One. 2011;6:e21800. doi: 10.1371/journal.pone.0021800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swaim C.D., Canadeo L.A., Monte K.J., Khanna S., Lenschow D.J., Huibregtse J.M. Modulation of extracellular ISG15 signaling by pathogens and viral effector proteins. Cell Rep. 2020;31:107772. doi: 10.1016/j.celrep.2020.107772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tchasovnikarova I.A., Timms R.T., Matheson N.J., Wals K., Antrobus R., Gottgens B., Dougan G., Dawson M.A., Lehner P.J. GENE SILENCING. Epigenetic silencing by the HUSH complex mediates position-effect variegation in human cells. Science. 2015;348:1481–1485. doi: 10.1126/science.aaa7227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teo G., Liu G., Zhang J., Nesvizhskii A.I., Gingras A.C., Choi H. SAINTexpress: improvements and additional features in Significance Analysis of INTeractome software. J. Proteomics. 2014;100:37–43. doi: 10.1016/j.jprot.2013.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udeshi N.D., Pedram K., Svinkina T., Fereshetian S., Myers S.A., Aygun O., Krug K., Clauser K., Ryan D., Ast T., et al. Antibodies to biotin enable large-scale detection of biotinylation sites on proteins. Nat. Methods. 2017;14:1167–1170. doi: 10.1038/nmeth.4465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vabret N., Britton G.J., Gruber C., Hegde S., Kim J., Kuksin M., Levantovsky R., Malle L., Moreira A., Park M.D., et al. Immunology of COVID-19: current state of the science. Immunity. 2020;52:910–941. doi: 10.1016/j.immuni.2020.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls A.C., Park Y.J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181:281–292 e286. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M., Herrmann C.J., Simonovic M., Szklarczyk D., von Mering C. Version 4.0 of PaxDb: protein abundance data, integrated across model organisms, tissues, and cell-lines. Proteomics. 2015;15:3163–3168. doi: 10.1002/pmic.201400441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P.H., Fung S.Y., Gao W.W., Deng J.J., Cheng Y., Chaudhary V., Yuen K.S., Ho T.H., Chan C.P., Zhang Y., et al. A novel transcript isoform of STING that sequesters cGAMP and dominantly inhibits innate nucleic acid sensing. Nucleic Acids Res. 2018;46:4054–4071. doi: 10.1093/nar/gky186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Xie F., Chu F., Zhang Z., Yang B., Dai T., Gao L., Wang L., Ling L., Jia J., et al. YAP antagonizes innate antiviral immunity and is targeted for lysosomal degradation through IKKvarepsilon-mediated phosphorylation. Nat. Immunol. 2017;18:733–743. doi: 10.1038/ni.3744. [DOI] [PubMed] [Google Scholar]
- Weigel-Kelley K.A., Yoder M.C., Srivastava A. Alpha5beta1 integrin as a cellular coreceptor for human parvovirus B19: requirement of functional activation of beta1 integrin for viral entry. Blood. 2003;102:3927–3933. doi: 10.1182/blood-2003-05-1522. [DOI] [PubMed] [Google Scholar]
- Wiersinga W.J., Rhodes A., Cheng A.C., Peacock S.J., Prescott H.C. Pathophysiology, transmission, diagnosis, and treatment of coronavirus disease 2019 (COVID-19): a review. JAMA. 2020;324:782–793. doi: 10.1001/jama.2020.12839. [DOI] [PubMed] [Google Scholar]
- Wishart D.S., Feunang Y.D., Guo A.C., Lo E.J., Marcu A., Grant J.R., Sajed T., Johnson D., Li C., Sayeeda Z., et al. DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res. 2018;46:D1074–D1082. doi: 10.1093/nar/gkx1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F., Zhao S., Yu B., Chen Y.M., Wang W., Song Z.G., Hu Y., Tao Z.W., Tian J.H., Pei Y.Y., et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J., Shi Y., Pan X., Wu S., Hou R., Zhang Y., Zhong T., Tang H., Du W., Wang L., et al. SARS-CoV-2 ORF9b inhibits RIG-I-MAVS antiviral signaling by interrupting K63-linked ubiquitination of NEMO. Cell Rep. 2021;34:108761. doi: 10.1016/j.celrep.2021.108761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z., Zhang Z., Wang X., Zhang J., Ren C., Li Y., Gao L., Liang X., Wang P., Ma C. Palmitoylation of SARS-CoV-2 S protein is essential for viral infectivity. Signal. Transduct. Target. Ther. 2021;6:231. doi: 10.1038/s41392-021-00651-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia H., Cao Z., Xie X., Zhang X., Chen J.Y., Wang H., Menachery V.D., Rajsbaum R., Shi P.Y. Evasion of type I interferon by SARS-CoV-2. Cell Rep. 2020;33:108234. doi: 10.1016/j.celrep.2020.108234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao J., Palefsky J.M., Herrera R., Berline J., Tugizov S.M. The Epstein-Barr virus BMRF-2 protein facilitates virus attachment to oral epithelial cells. Virology. 2008;370:430–442. doi: 10.1016/j.virol.2007.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi Y., Lagniton P.N.P., Ye S., Li E., Xu R.H. COVID-19: what has been learned and to be learned about the novel coronavirus disease. Int. J. Biol. Sci. 2020;16:1753–1766. doi: 10.7150/ijbs.45134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F.X., Guan K.L. The Hippo pathway: regulators and regulations. Genes Dev. 2013;27:355–371. doi: 10.1101/gad.210773.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G., He Q.Y. ReactomePA: an R/Bioconductor package for reactome pathway analysis and visualization. Mol. Biosyst. 2016;12:477–479. doi: 10.1039/c5mb00663e. [DOI] [PubMed] [Google Scholar]
- Yu G., Wang L.G., Han Y., He Q.Y. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuen C.K., Lam J.Y., Wong W.M., Mak L.F., Wang X., Chu H., Cai J.P., Jin D.Y., To K.K., Chan J.F., et al. SARS-CoV-2 nsp13, nsp14, nsp15 and orf6 function as potent interferon antagonists. Emerg. Microbes Infect. 2020;9:1418–1428. doi: 10.1080/22221751.2020.1780953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., Meng F., Chen S., Plouffe S.W., Wu S., Liu S., Li X., Zhou R., Wang J., Zhao B., et al. Hippo signalling governs cytosolic nucleic acid sensing through YAP/TAZ-mediated TBK1 blockade. Nat. Cell Biol. 2017;19:362–374. doi: 10.1038/ncb3496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X.J., Qin J.J., Cheng X., Shen L., Zhao Y.C., Yuan Y., Lei F., Chen M.M., Yang H., Bai L., et al. In-hospital use of statins is associated with a reduced risk of mortality among individuals with COVID-19. Cell Metab. 2020;32:176–187 e174. doi: 10.1016/j.cmet.2020.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y., Zhuang M.W., Han L., Zhang J., Nan M.L., Zhan P., Kang D., Liu X., Gao C., Wang P.H. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) membrane (M) protein inhibits type I and III interferon production by targeting RIG-I/MDA-5 signaling. Signal. Transduct. Target. Ther. 2020;5:299. doi: 10.1038/s41392-020-00438-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L., et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., et al. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All proteomics raw data have been deposited to the ProteomeXchange Consortium via the iProX (Ma et al., 2019) partner repository with the dataset identifier ProteomeXchange: PXD022086. All data reported in this paper will be shared by the lead contact upon request. This paper does not report original code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.