Abstract
The recently emerged coronavirus disease 2019 (COVID-19) has rapidly evolved into a pandemic with over 10 million infections and over 500 thousand deaths. There are currently no effective therapies or vaccines available to protect against this coronavirus infection. In this review, we discuss potential therapeutic options for COVID-19 based on the available information from previous research on severe acute respiratory syndrome (SARS) and Middle East respiratory syndrome (MERS). Substantial efforts are underway to discover new therapeutic agents for COVID-19, including the repurposing of existing agents and the development of novel agents that specifically target SARS-coronavirus 2 (SARS-CoV-2) or host factors. Through the screening of compound libraries, various classes of drugs, such as ribavirin, remdesivir, lopinavir/ritonavir, and hydroxychloroquine have been identified as potential therapeutic candidates against COVID-19. Novel antiviral drugs for SARS-coronavirus 2 are being developed to target viral enzymes or functional proteins, as well as host factors or cell signaling pathways.
Keywords: COVID-19, SARS-CoV-2, anti-virals
Introduction
Coronaviruses (CoVs) belong to the Orthocoronavirinae subfamily, Coronaviridae family, and Nidovirales order, and are the largest enveloped positive-sense single-stranded RNA viruses that can infect humans, livestock, bats, and many other wild animals.1,2 Currently, there are only seven CoVs that can infect humans and cause respiratory diseases. The four human CoVs, HCoV-229E, HCoV-NL63, HCoV-OC43, and HKU1, can only cause mild upper respiratory tract infections. Severe acute respiratory syndrome CoV2 (SARS-CoV-2), which was first detected in Wuhan, China, at the end of 2019, and SARS-CoV as well as Middle East respiratory syndrome CoV (MERS-CoV), which emerged in 2002 and 2012, respectively, can infect the lower respiratory tract and cause severe pneumonia in humans.3,4
SARS-CoV-2 belongs to the genus Betacoronavirus and its genome is about 30 kb, with 10 open reading frames. Viral infection is initiated from the surface spike glycoprotein, which binds to angiotensin-converting enzyme 2 (ACE2) to enter the cells, similar as the mechanism of SARS-CoV.5 Upon entry into the cells, the viral polyproteins, pp1a and pp1ab, are translated to produce non-structural proteins for the formation of the replicase-transcriptase complex, which is responsible for transcription of the subgenomic RNAs and replication of the viral genome. The structural proteins are synthesized and assembled to form mature virions. Ultimately, the new virions are released from the cells by exocytosis (Figure 1).
Figure 1.
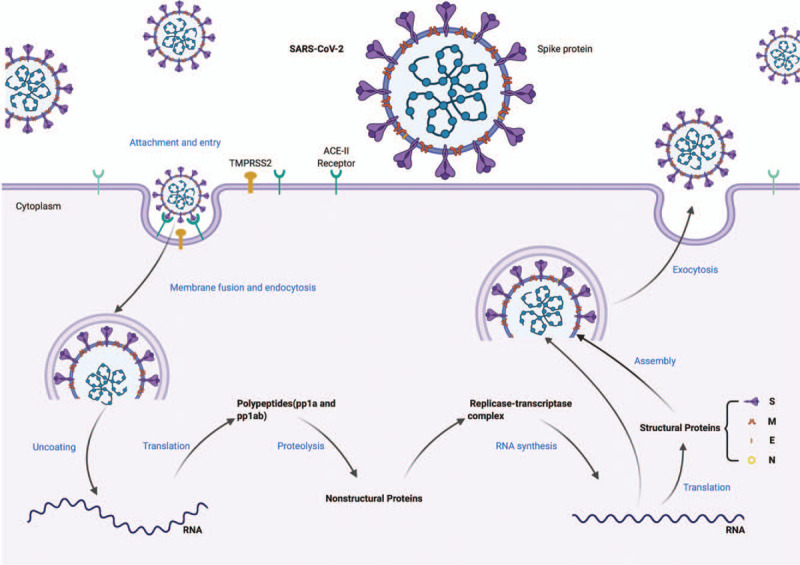
Replication cycle of SARS-CoV-2. SARS-CoV-2: severe acute respiratory syndrome-coronavirus 2.
The novel respiratory disease caused by SARS-CoV-2 was named officially coronavirus disease 2019 (COVID-19) by the World Health Organization. A recent mathematical modeling research showed that the doubling time of the number of infected persons in Wuhan was 2.3–3.3 days and the effective reproductive number (R) was 5.7, which indicates that its transmissibility is much higher than that of SARS.6 The virus spread so quickly that it was declared a global pandemic by the World Health Organization. As of July 1, 2020, there are globally over 10.3 million reported infections and more than 506,000 deaths.7 Clinically, some patients may be asymptomatic, or develop symptoms that range from mild to severe. Respiratory symptoms such as fever, cough, myalgia or fatigue, pneumonia, dyspnea, or hypoxemia are the most commonly reported clinical manifestation. Some cases worsen and present with serious complications, including acute respiratory distress syndrome (ARDS), septic shock, metabolic acidosis, coagulation dysfunction, or even death.8,9 The lack of effective antiviral treatment as well as the high morbidity rate associated with this disease highlights the need for the discovery of novel drugs. In this review, we discuss the discovery and the development of potential therapeutic options for COVID-19 based on the available information from previous researches on SARS and MERS.
Potential therapeutic options for COVID-19
Currently, the major therapeutic measures for COVID-19 are supportive care and prevention of complications, such as ARDS, organ failure, and secondary nosocomial infections. Substantial efforts are underway to discover new therapeutic agents for COVID-19. These investigations can be divided into two categories, which include the screening of compound libraries for currently available agents that may have antiviral effect on COVID-19 and the development of novel agents that specifically target SARS-CoV-2 or host factors.
Currently available agents
It would typically take several years to develop a new drug for COVID-19 and obtain approval for clinical use. Scientists have focused on the potential to repurposing existing antiviral agents that are approved or being developed for the treatment of other viral diseases, as they have known pharmacokinetic and pharmacodynamic properties, dosages, and side effects. Many compounds have strong inhibitory effects against CoVs in cell culture or in animal models, but the results of in vitro and animal experiments do not necessarily translate into efficacy in humans. Through rapid high-throughput screening, various classes of drugs that have been identified as potential candidates for the treatment of COVID-19, which include: (a) directly acting antivirals; (b) host-targeting antivirals; (c) immune-modulatory or immune-suppressive treatments; and (d) biologics targeting the virus.
Directly acting antivirals
Ribavirin, a broad-spectrum antiviral drug, exhibits moderate anti-viral activity against SARS-CoV and MERS-CoV infections in vitro.10,11 However, other studies also found that ribavirin has no significant effective inhibitory effect on the replication of SARS-CoV and causes severe adverse events such as hemolysis and reduced hemoglobin concentration.12,13 Given its toxicity, the use of ribavirin to treat COVID-19 should be considered with serious caution.
Remdesivir is a nucleotide analog RNA-dependent RNA polymerase (RdRp) inhibitor initially developed as a delayed RNA-chain terminator against Ebola virus and Marburg viruses, however, it has not been approved for marketing anywhere.14 Remdesivir has “broad-spectrum” anti-coronavirus activity due to its effectiveness against many human and zoonotic CoVs in vitro, including HCoV-NL63, HCoV-OC43, HCoV-229E, mouse hepatitis virus, SARS-CoV, MERS-CoV, and SARS-CoV-2.15–17 The first COVID-19 patient treated with intravenous remdesivir in the USA showed improved condition without obvious adverse effect.18 One recent cohort study observed 68% clinical improvement in severe COVID-19 patients treated with remdesivir on a compassionate-use basis. However, adverse events, including increased levels of hepatic enzymes, diarrhea, rash, renal impairment, and hypotension, were reported in 60% of the patients.19 In addition, it is too early to conclude on the efficacy of remdesivir against COVID-19. There are two ongoing phase III randomized, double-blind clinical trials in China to evaluate the efficacy and safety of intravenous remdesivir therapy in patients with COVID-19 (NCT04252664 and NCT04257656).
HIV-1 protease inhibitors, such as lopinavir/ritonavir, darunavir/cobicistat, emtricitabine/tenofovir, azvudine, saquinavir, and nelfinavir are potential drug candidates for the treatment of COVID-19. Nelfinavir strongly inhibited the replication of the SARS-CoV in vitro.20 By using an in silico approach, lopinavir/ritonavir and saquinavir produced strong interactions with the active site of SARS-CoV-2 3C-like protease (3CLpro).21 An open-label study suggested that lopinavir/ritonavir plus ribavirin appears to be associated with improved clinical outcomes for SARS patients.22 Besides, successful cases of MERS treatment with a triple combination therapy of lopinavir/ritonavir, ribavirin, and interferon-alpha 2a have been reported.23–25 However, the efficacy of lopinavir/ritonavir is unconvincing because of the concomitant use with ribavirin and/or interferon. Recently, the use of lopinavir/ritonavir treatment in a randomized, controlled, open-label trial involving 199 patients with COVID-19 showed no significant benefit in either overall mortality or reduction of viral loads beyond standard care.26 Currently, there are other multiple ongoing studies exploring the use of lopinavir/ritonavir for the treatment of COVID-19.
Several other anti-viral drugs are also being investigated, including favipiravir, umifenovir (Arbidol), triazavirin, baloxavir, and marboxil, which are predominantly designed against various influenza subtypes, as well as danoprevir/ritonavir, sofosbuvir/ledipasvir, and sofosbuvir/daclatasvir, which are typically used in management of hepatitis C virus infections.27 A small-sample clinical study showed that danoprevir/ritonavir was able to improve the clinical symptoms and reduce the viral loads in COVID-19 patients.28
Host-targeting antivirals
Hydroxychloroquine and chloroquine, which are traditional antimalarial drugs, have exhibited effective inhibitory effect against the replication of CoVs in vitro, including SARS-CoV, MERS-CoV, and SARS-CoV-2.17,29,30 By increasing endosomal pH and disturbing the glycosylation of cell surface receptors, these medications provide an important defense against viral entry and replication.31 A recent research proposed that hydroxychloroquine and chloroquine may bind to gangliosides with high affinity, thus preventing SARS-CoV-2 from binding with the ACE-2 receptor.32 Some small studies and one subjective report have reported the effectiveness of hydroxychloroquine or chloroquine for the prevention and treatment of COVID-19. The beneficial outcome of hydroxychloroquine plus azithromycin for the treatment of COVID-19 patients has also been demonstrated.33 However, the lack of randomization and blinding, and the small sample size make these data unconvincing. A notable concern is the adverse events potentially linked to the use of hydroxychloroquine or chloroquine and azithromycin in patients with COVID-19, which include QT interval prolongation and arrhythmia. A recent study reported that cohorts of COVID-19 patients treated with chloroquine/hydroxychloroquine ± azithromycin all experienced QT interval prolongation.34 In addition, patients receiving hydroxychloroquine plus azithromycin were more likely to have cardiac arrest.35
Drugs that can regulate neurotransmitters, such as chlorpromazine, are anti-MERS-CoV and anti-SARS-CoV agents, and their effects are mediated by the ability to inhibit clathrin-mediated endocytosis.36 The male sex appears to have a higher mortality rate for COVID-19 according to epidemiologic data in several countries. China, South Korea, and Italy reported that 73%, 59%, and 70%, respectively, of the COVID-19 deaths were men.37–39 Treatment with estrogen seems to be an ideal prevention and therapeutic strategy against COVID-19, as it was able to alleviate inflammatory reactions and decrease viral titers in animal experiments.40
Other agents, such as dipyridamole (an anti-coagulation agent), leflunomide, and teriflunomide (dihydroorotate dehydrogenase inhibitors), are also potential therapeutic candidates for COVID-19, as they have been able to inhibit SARS-CoV-2 replication in cell culture models.41–43 Anti-tumor agents that act as inhibitors of poly-ADP-ribose polymerase 1, such as CVL218, which is currently in a Phase I clinical trial, exhibited effective inhibitory activity against SARS-CoV-2 replication, without obvious cytopathic effects in both animal models and cell culture.44
Immune-modulatory or immune-suppressive therapies
Interferons can help to restore innate immune responses in host cells.45 They have also been found to be potent replication inhibitors of SARS-CoV and MERS-CoV.46,47 The combination of interferon with other antivirals, such as ribavirin and/or lopinavir-ritonavir have been used to treat patients with SARS and MERS, as well as COVID-19. Some case studies have reported that the combination of interferon-α and ribavirin with lopinavir/ritonavir resulted in improved outcomes.23–25 Thus, the use of interferon in combination with other effective antivirals should be evaluated in clinical trials.
The immunosuppressive drug Cyclosporine A displays broad-spectrum antiviral activity by binding to cellular cyclophilins, which play an important role in viral infection. It has been reported that Cyclosporine A can block the replication of CoVs of all genera, including SARS-CoV and avian infectious bronchitis virus.48
The severity and outcome of COVID-19 might be associated with an excessive inflammatory response “cytokine storm.” Immune-modulatory or immune-suppressive agents such as corticosteroids, intravenous immunoglobulin, and interleukin-6 antagonists are potential therapeutic candidates for COVID-19.49 In addition, meplazumab (anti-CD147) inhibited viral entry, which indicates that it could efficiently improve clinical and virologic outcomes in COVID-19 patients.50 Moreover, immune stimulation, such as with the use of an anti-PD-1 antibody (camrelizumab), recombinant interleukin-2, and recombinant human granulocyte colony-stimulating factor, is being investigated.27
Biological therapies
The plasma of recovered COVID-19 patients may contain high-titer neutralizing antibodies. Convalescent plasma therapy has been successfully used in the treatment of SARS and MERS, and hence, it might be a promising treatment option for COVID-19 patients.51,52 One clinical study performed with 10 patients, showed the clinical benefit of convalescent plasma therapy.53 Besides, mesenchymal stem cells might relief the symptoms of ARDS in COVID-19 patients through its immunomodulatory and tissue repair effects.
Novel agents
In addition to the aforementioned potential antiviral therapeutics, the development of novel antiviral drugs that specifically target SARS-CoV-2 and host-dependent factors should also be considered. Theoretically, these drugs would exhibit better anti-CoV activity in vitro and/or in vivo; however, these drugs first need to be evaluated in animal studies and human trials and this might take several years.
Novel anti-viral drugs for CoVs are mainly targeted against viral enzymes or functional proteins to prevent the synthesis and replication of RNA. In addition, they target structural proteins to block the virus from binding to the host cell surface receptors, or inhibit the virus's self-assembly process and virulence factors to restore the host's innate immunity.
Viral enzymes
Initial analyses of genomic sequences of SARS-CoV-2 showed that four significant functional proteins, 3CLpro, papain-like protease (PLpro), RdRp, and helicase are highly conserved in SARS-CoV-2, MERS-CoV, and SARS-CoV, especially in the functional region.54 The primary function of the coronaviral proteases PLpro and 3CLpro is to process two large viral polyproteins, pp1a and pp1ab, which is required for the release and mutation of non-structural proteins.55,56 In addition, PLpro also exhibits the ability to counteract host innate immunity through deISGylation and deglycosylation to block the production of important cytokines.57,58 Various kinds of SARS-CoV PLpro inhibitors have been identified, including natural products, thiopurine compounds, naphthalene-based inhibitors, zinc ion, and zinc conjugate inhibitors, but none has been approved by FDA for clinical practice.55 Disulfiram, initially developed for the treatment of alcohol dependence, has been reported to inhibit the PLpro of MERS and SARS in vitro, but clinical studies are lacking.59 In addition, computational screening also showed that a series of existing molecules, such as ribavirin, chloramphenicol, and levodropropizine, may have high binding affinity to PLpro, which suggests their potential use in the treatment of COVID-19.60 Recently, the crystal structure of 3CLpro of SARS-CoV-2 has been reported and new α-ketoamide inhibitors have been designed as anti-CoV agents.61 Through virtual screening, numerous classes of 3CLpro inhibitors have been reported. For example, ledipasvir and velpatasvir are particularly promising therapeutic candidates against COVID-19, with minimal side effects.62
RdRp can catalyze viral RNA synthesis from an RNA template, which makes it an attractive antiviral target. Nucleoside analogs can inhibit the activity of RdRp in a broad spectrum of RNA viruses. Recent computer modeling showed that ribavirin, remdesivir, sofosbuvir, galidesivir, IDX-184, and tenofovir can tightly bind to the RdRp of SARS-CoV-2, which suggests their potential ability to combat SARS-CoV-2.63 Nonetheless, the use of nucleoside analogs to treat COVID-19 should be closely monitored, because resistance to nucleoside analogs due to mutations in RdRp has been reported for many other RNA viruses. Small interfering RNA (siRNA) molecules have been used to inhibit the replication of SARS-CoV in vitro, and hence, siRNAs that target SARS-CoV-2 RdRp could serve as another potent treatment option for COVID-19.64
Helicase is one of the most important CoV replication enzymes, which can separate double-stranded nucleic acid into single strands using ATP generated from the hydrolysis of nucleoside triphosphate. Several chemical inhibitors, such as Bananins, 5-hydroxychromone derivatives, and triazole derivatives, can inhibit the activities of ATPase and helicase in SARS-CoV and MERS-CoV infections in cell culture experiments.65,66 However, the toxicity associated with the inhibition of cellular ATPases or kinases limits the development of these compounds. Two novel inhibitors, SSYA10-001 and aryl diketoacids, appear more promising as they can inhibit helicase in a broad range of CoVs without affecting the activity of ATPase.67,68 More animal model experiments are needed to evaluate their efficacy and safety.
Viral nucleic acids
Drugs targeting nucleic acids generally have broad-spectrum activity against various types of viruses. Mycophenolate mofetil is an inhibitor of the synthesis of inosine monophosphate dehydrogenase and guanine monophosphate, which is showing potent antiviral activity against MERS-CoV.69 DNAzymes or ribozymes targeting the viral genome of SARS have been shown to effectively inhibit viral replication.70 A small-compound inhibitor, K22, which specifically targets the synthesis of membrane-bound coronaviral RNA and the formation of double membrane vesicles, was able to inhibit a broad range of CoVs, including MERS-CoV.71
Structural proteins
Coronavirus spike glycoprotein consists of two functional subunits (S1 and S2); it is responsible for cell receptor binding, tissue tropism, and pathogenesis of viral infections. It is the primary antigenic target in the design of therapeutics and vaccines. The S1 subunit is further divided into the receptor-binding domain (RBD) and the N-terminal domain, which play essential roles in virus-cell receptor binding.5 A variety of neutralizing antibodies (nAbs) that target S1-RBD and S1-N-terminal domain are induced to inhibit viral infections.72 The RBD of SARS-CoV-2 is closely related to that of SARS-CoV, and hence, the potential cross-neutralizing effects of SARS-CoV nAbs against SARS-CoV-2 infection could be investigated.73 CR3022, the SARS-CoV RBD-specific monoclonal antibody, is able to bind to the SARS-CoV-2 RBD by recognizing a conserved epitope that does not overlap with the ACE2 binding site.74 Similarly, it has been elucidated that the human 47D11 antibody can neutralize both SARS-CoV-2 and SARS-CoV in VeroE6 cells.75 In addition, SARS-CoV RBD-specific polyclonal antibodies are cross-reactive with the SARS-CoV-2 RBD to neutralize SARS-CoV-2 infections in vitro.76 Sera obtained from convalescent SARS patients or animals are also able to specifically cross-neutralize SARS-CoV-2.77 These findings open avenues for the development of SARS-CoV nAbs as candidate therapeutics or vaccines to treat or prevent SARS-CoV-2. Cocktails comprising antiviral antibodies targeting the different epitopes of the spike protein are another promising therapeutic strategy to cope with SARS-CoV-2 and its escape-mutant strains. The S2 subunit can mediate the virus-cell fusion, and it contains an N-terminal fusion peptide, heptad repeat 1 (HR1) and HR2 domains, a transmembrane domain and a cytoplasmic domain. EK1C4, an inhibitor of coronavirus fusion, targets the HR1 domain of the S2 subunit and exhibits a strong inhibitory effect against SARS-CoV-2 in vitro and in vivo.78 Another class of anti-CoV agents that target the spike glycoprotein comprises the carbohydrate-binding agents, which do not inhibit virus-cell attachment, but rather affect viral entry at a post-binding stage.79
Other structural proteins, such as membrane, envelope, and nucleocapsid proteins, and some essential accessory proteins of virion assembly are also potential antiviral targets. siRNAs could be designed to knock-down these proteins, and their antiviral effects have been demonstrated in SARS-CoV infection.80 Alternatively, an increasing number of anti-bacterial, anti-viral, anti-tumor, anti-asthmatic, and anti-inflammatory agents have shown relatively good binding affinity for these targets.60
Host-dependent targets
Host factors involved in the viral life cycle have been identified as potential targets of anti-CoV agents. SARS-CoV-2 utilizes ACE2 for viral entry in a way similar to SARS-CoV. N-(2-aminoethyl)-1-aziridine-ethanamine, a small-molecule inhibitor, can inhibit the catalytic activity of virus-cell fusion mediated by ACE2 and spike protein in vitro.81 Synthetic peptides derived from critical segments of ACE2 also exhibited anti-viral activity against SARS-CoV infections.82 The entry of SARS-CoV-2 into host cells is through endocytosis, which involves PIKfyve, TPC2, and cathepsin L.83 The inhibitors of these proteases are potential anti-viral candidates. The endosomal cysteine proteases cathepsin B and L and the cellular serine protease TMPRSS2 are also employed by SARS-CoV-2 to mediate the cell surface entry pathway. Camostat mesylate, a serine protease inhibitor, which is active against TMPRSS2, combined with E-64d, an inhibitor of cathepsin B and L, displayed full inhibition of SARS-CoV-2 infection in lung cells.77 Recent studies found that SARS-CoV-2 spike glycoprotein harbors a peculiar furin-like cleavage site at the boundary between the S1 and S2 subunits, which distinguishes the virus from SARS-CoV and SARS-related CoVs.5,84 The use of decanoyl-Arg-Val-Lys-Arg-chloromethylketone to inhibit furin has been shown to block MERS-CoV entry into the host cell in vitro.85 This suggests that the inhibition of furin-like enzymes may be a potential antiviral strategy. In addition, another study showed that the interferon-inducible gene, lymphocyte antigen 6 complex locus E, potently restricts multiple CoV infections by interfering with virus-cell fusion, including SARS-CoV-2, SARS-CoV, and MERS-CoV.86
Host cell signaling pathways in relation to coronavirus infection have also been identified as potential targets for anti-CoV treatment. Phosphoinositol 3-kinase/serine-threonine kinase/mammalian target of rapamycin (mTOR) signaling responses play important roles in MERS-CoV infection.87 Recent research showed that the mTOR pathway in conjunction with AMP activated protein kinase may help to control cell injury, oxidative stress, mitochondrial dysfunction, and the onset of hyperinflammation, which are significantly associated with COVID-19.88 Therefore, targeting the mTOR pathway is a promising antiviral strategy that can be explored for the development of therapeutics for COVID-19. Considering that CoVs are capable of hijacking type I interferon antiviral responses through structural and nonstructural proteins, the flagellin-TLR5 axis could provide a potential avenue to restore host innate immunity to fight against COVID-19.89
Outlook
The development of effective novel antiviral agents may take several months or years. The best way to deal with COVID-19 is the control of the source of infection through early detection, diagnosis, reporting, isolation, and supportive treatments. In addition, publishing of epidemic information should be controlled to avoid unnecessary panic. Looking ahead, the most feasible approach against COVID-19 is the repurposing of existing antiviral agents and the evaluation of their efficacy in clinical trials. In the long term, there is an urgent need for the development of novel broad-spectrum antiviral agents and vaccines against a wide range of CoVs.
References
- [1].Ge XY, Li JL, Yang XL, et al. Isolation and characterization of a bat SARS-like coronavirus that uses the ACE2 receptor. Nature 2013;503(7477):535–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Chen Y, Guo DY. Molecular mechanisms of coronavirus RNA capping and methylation. Virol Sin 2016;31(1):3–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Zhu N, Zhang DY, Wang WL, et al. A novel coronavirus from patients with pneumonia in China, 2019. New Engl J Med 2020;382(8):727–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Su S, Wong G, Shi WF, et al. Epidemiology, genetic recombination, and pathogenesis of coronaviruses. Trends Microbiol 2016;24(6):490–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Walls AC, Park YJ, Tortorici MA, Wall A, McGuire AT, Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell 2020;181(2):281–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Sanche S, Lin YT, Xu C, Romero-Severson E, Hengartner N, Ke R. High contagiousness and rapid spread of severe acute respiratory syndrome coronavirus 2. Emerg Infect Dis 2020;26(7):1470–1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Han Y, Yin Y, Dai X, et al. Widespread use of high-dose ceftriaxone therapy for uncomplicated gonorrhea without reported ceftriaxone treatment failure: results from 5 years of multicenter surveillance data in China. Clin Infect Dis 2020;70(1):99–105. [DOI] [PubMed] [Google Scholar]
- [8].Huang C, Wang Y, Li X, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020;395(10223):497–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Guan W-J, Ni Z-Y, Hu Y, et al. Clinical characteristics of coronavirus disease 2019 in China. N Engl J Med 2020;382(18):1708–1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Morgenstern B, Michaelis M, Baer PC, Doerr HW, Cinatl J, Jr. Ribavirin and interferon-beta synergistically inhibit SARS-associated coronavirus replication in animal and human cell lines. Biochem Biophys Res Commun 2005;326(4):905–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Omrani AS, Saad MM, Baig K, et al. Ribavirin and interferon alfa-2a for severe Middle East respiratory syndrome coronavirus infection: a retrospective cohort study. Lancet Infect Dis 2014;14(11):1090–1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Tan EL, Ooi EE, Lin CY, et al. Inhibition of SARS coronavirus infection in vitro with clinically approved antiviral drugs. Emerg Infect Dis 2004;10(4):581–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Booth CM, Matukas LM, Tomlinson GA, et al. Clinical features and short-term outcomes of 144 patients with SARS in the greater Toronto area. JAMA 2003;289(21):2801–2809. [DOI] [PubMed] [Google Scholar]
- [14].Warren TK, Jordan R, Lo MK, et al. Therapeutic efficacy of the small molecule GS-5734 against Ebola virus in rhesus monkeys. Nature 2016;531(7594):381–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Sheahan TP, Sims AC, Graham RL, et al. Broad-spectrum antiviral GS-5734 inhibits both epidemic and zoonotic coronaviruses. Sci Transl Med 2017;9(396):eaal3653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Brown AJ, Won JJ, Graham RL, et al. Broad spectrum antiviral remdesivir inhibits human endemic and zoonotic deltacoronaviruses with a highly divergent RNA dependent RNA polymerase. Antiviral Res 2019;169:104541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Wang M, Cao R, Zhang L, et al. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res 2020;30(3):269–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Holshue ML, DeBolt C, Lindquist S, et al. First case of 2019 novel coronavirus in the United States. N Engla J Med 2020;382(10):929–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Grein J, Ohmagari N, Shin D, et al. Compassionate use of remdesivir for patients with severe COVID-19. N Engl J Med 2020;382(24):2327–2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Yamamoto N, Yang R, Yoshinaka Y, et al. HIV protease inhibitor nelfinavir inhibits replication of SARS-associated coronavirus. Biochem Biophys Res Commun 2004;318(3):719–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Ortega JT, Serrano ML, Pujol FH, Rangel HR. Unrevealing sequence and structural features of novel coronavirus using approaches: the main protease as molecular target. EXCLI J 2020;19:400–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Chu CM, Cheng VCC, Hung IFN, et al. Role of lopinavir/ritonavir in the treatment of SARS: initial virological and clinical findings. Thorax 2004;59(3):252–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Kim UJ, Won E-J, Kee S-J, Jung S-I, Jang H-C. Combination therapy with lopinavir/ritonavir, ribavirin and interferon-α for Middle East respiratory syndrome. Antivir Ther (Lond) 2016;21(5):455–459. [DOI] [PubMed] [Google Scholar]
- [24].Spanakis N, Tsiodras S, Haagmans BL, et al. Virological and serological analysis of a recent Middle East respiratory syndrome coronavirus infection case on a triple combination antiviral regimen. Int J Antimicrob Agents 2014;44(6):528–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Min CK, Cheon S, Ha NY, et al. Comparative and kinetic analysis of viral shedding and immunological responses in MERS patients representing a broad spectrum of disease severity. Sci Rep 2016;6:25359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Cao B, Wang Y, Wen D, et al. A trial of lopinavir-ritonavir in adults hospitalized with severe COVID-19. N Engl J Med 2020;382(19):1787–1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Lythgoe PM. Ongoing clinical trials for the management of the COVID-19 pandemic. Trends Pharmacol Sci 2020;41(6):363–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Chen H, Zhang Z, Wang L, et al. First clinical study using HCV protease inhibitor danoprevir to treat naïve and experienced COVID-19 patients. medRxiv 2020;doi:10.1101/2020.03.22.20034041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Savarino A, Boelaert JR, Cassone A, Majori G, Cauda R. Effects of chloroquine on viral infections: an old drug against today's diseases? Lancet Infect Dis 2003;3(11):722–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Colson P, Rolain J-M, Raoult D. Chloroquine for the 2019 novel coronavirus SARS-CoV-2. Int J Antimicrob Agents 2020;55(3):105923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Vincent MJ, Bergeron E, Benjannet S, et al. Chloroquine is a potent inhibitor of SARS coronavirus infection and spread. Virol J 2005;2:69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Yazdany J, Kim AHJ. Use of hydroxychloroquine and chloroquine during the COVID-19 pandemic: what every clinician should know. Ann Intern Med 2020;172(11):754–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Gautret P, Lagier J-C, Parola P, et al. Hydroxychloroquine and azithromycin as a treatment of COVID-19: results of an open-label non-randomized clinical trial. Int J Antimicrob Agents 2020;105949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Saleh M, Gabriels J, Chang D, et al. Effect of chloroquine, hydroxychloroquine and azithromycin on the corrected QT interval in patients with SARS-CoV-2 infection. Circ Arrhythm Electrophysiol 2020;13(6):e008662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Rosenberg ES, Dufort EM, Udo T, et al. Association of treatment with hydroxychloroquine or azithromycin with in-hospital mortality in patients with COVID-19 in New York state. JAMA 2020;323(24):2493–2502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Plaze M, Attali D, Petit AC, et al. Repurposing of chlorpromazine in COVID-19 treatment: the reCoVery study. Encephale 2020;46(3S):S35–S39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Chen T, Wu D, Chen H, et al. Clinical characteristics of 113 deceased patients with coronavirus disease 2019: retrospective study. BMJ 2020;368:m1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Korean Society of Infectious Diseases, Korean Society of Pediatric Infectious Diseases, Korean Society of Epidemiology, Korean Society for Antimicrobial Therapy, Korean Society for Healthcare-associated Infection Control, Prevention, Korea Centers for Disease Control, Prevention Report on the epidemiological features of coronavirus disease 2019 (COVID-19) outbreak in the Republic of Korea from January 19 to March 2, 2020. J Korean Med Sci 2020;35(10):e112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].La Vignera S, Cannarella R, Condorelli RA, Torre F, Aversa A, Calogero AE. Sex-specific SARS-CoV-2 mortality: among hormone-modulated ACE2 expression, risk of venous thromboembolism and hypovitaminosis D. Int J Mol Sci 2020;21(8):2948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Suba Z. Prevention and therapy of COVID-19 via exogenous estrogen treatment for both male and female patients. J Pharm Pharm Sci 2020;23(1):75–85. [DOI] [PubMed] [Google Scholar]
- [41].Liu X, Li Z, Liu S, et al. Therapeutic effects of dipyridamole on COVID-19 patients with coagulation dysfunction. bioRxiv 2020;doi:10.1101/2020.02.27.20027557. [Google Scholar]
- [42].Matsuyama S, Kawase M, Nao N, et al. The inhaled corticosteroid ciclesonide blocks coronavirus RNA replication by targeting viral NSP15. bioRxiv 2020;doi:10.1101/2020.03.11.987016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Xiong R, Zhang L, Li S, et al. Novel and potent inhibitors targeting DHODH, a rate-limiting enzymein de novo pyrimidine biosynthesis, are broad-spectrum antiviral against RNA viruses including newly emerged coronavirus SARS-CoV-2. bioRxiv 2020;doi:10.1101/2020.03.11.983056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Ge Y, Tian T, Huang S, et al. A data-driven drug repositioning framework discovered a potential therapeutic agent targeting COVID-19. bioRxiv 2020;doi:10.1101/2020.03.11.986836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Kuri T, Zhang XN, Habjan M, et al. Interferon priming enables cells to partially overturn the SARS coronavirus-induced block in innate immune activation. J Gen Virol 2009;90(Pt 11):2686–2694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Cinatl J, Morgenstern B, Bauer G, Chandra P, Rabenau H, Doerr HW. Treatment of SARS with human interferons (vol 362, pg 293, 2003). Lancet 2003;362(9385):748–1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Mustafa S, Balkhy H, Gabere MN. Current treatment options and the role of peptides as potential therapeutic components for Middle East Respiratory Syndrome (MERS): a review. J Infect Public Heal 2018;11(1):9–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Pfefferle S, Schopf J, Kogl M, et al. The SARS-coronavirus-host interactome: identification of cyclophilins as target for pan-coronavirus inhibitors. PLoS Pathog 2011;7(10):e1002331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Tufan A, Avanoğlu Güler A, Matucci-Cerinic M. COVID-19, immune system response, hyperinflammation and repurposing antirheumatic drugs. Turk J Med Sci 2020;50(SI-1):620–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Bian H, Zheng Z-H, Wei D, et al. Meplazumab treats COVID-19 pneumonia: an open-labelled, concurrent controlled add-on clinical trial. bioRxiv 2020;doi:10.1101/2020.03.21.20040691. [Google Scholar]
- [51].Cheng Y, Wong R, Soo YOY, et al. Use of convalescent plasma therapy in SARS patients in Hong Kong. Eur J Clin Microbiol Infect Dis 2005;24(1):44–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Ko J-H, Seok H, Cho SY, et al. Challenges of convalescent plasma infusion therapy in Middle East respiratory coronavirus infection: a single centre experience. Antivir Ther (Lond) 2018;23(7):617–622. [DOI] [PubMed] [Google Scholar]
- [53].Duan K, Liu B, Li C, et al. The feasibility of convalescent plasma therapy in severe COVID- 19 patients: a pilot study. medRxiv 2020;doi:10.1101/2020.03.16.20036145. [Google Scholar]
- [54].Morse JS, Lalonde T, Xu S, Liu WR. Learning from the past: possible urgent prevention and treatment options for severe acute respiratory infections caused by 2019-nCoV. Chembiochem 2020;21(5):730–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Baez-Santos YM, St John SE, Mesecar AD. The SARS-coronavirus papain-like protease: Structure, function and inhibition by designed antiviral compounds. Antivir Res 2015;115:21–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Chen Y, Liu QY, Guo DY. Emerging coronaviruses: genome structure, replication, and pathogenesis. J Med Virol 2020;92(4):418–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Sun L, Xing YL, Chen XJ, et al. Coronavirus papain-like proteases negatively regulate antiviral innate immune response through disruption of STING-mediated signaling. Plos One 2012;7(2):e30802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Mielech AM, Kilianski A, Baez-Santos YM, Mesecar AD, Baker SC. MERS-CoV papain-like protease has deISGylating and deubiquitinating activities. Virology 2014;450:64–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Lin MH, Moses DC, Hsieh CH, et al. Disulfiram can inhibit MERS and SARS coronavirus papain-like proteases via different modes. Antivir Res 2018;150:155–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Wu C, Liu Y, Yang Y, et al. Analysis of therapeutic targets for SARS-CoV-2 and discovery of potential drugs by computational methods. Acta Pharmaceutica Sinica B 2020;10(5):766–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Zhang L, Lin D, Sun X, Rox K, Hilgenfeld R. X-ray structure of main protease of the novel coronavirus SARS-CoV-2 enables design of a-ketoamide inhibitors. bioRxiv 2020;doi:10.1101/2020.02.17.952879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Chen YW, Yiu C-PB, Wong K-Y. Prediction of the SARS-CoV-2 (2019-nCoV) 3C-like protease (3CL) structure: virtual screening reveals velpatasvir, ledipasvir, and other drug repurposing candidates. F1000Research 2020;9:129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Elfiky AA. Ribavirin, remdesivir, sofosbuvir, galidesivir, and tenofovir against SARS-CoV-2 RNA dependent RNA polymerase (RdRp): a molecular docking study. Life Sci 2020;117592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Lu AL, Zhang HQ, Zhang XY, et al. Attenuation of SARS coronavirus by a short hairpin RNA expression plasmid targeting RNA-dependent RNA polymerase. Virology 2004;324(1):84–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Kim MK, Yu MS, Park HR, et al. 2,6-Bis-arylmethyloxy-5-hydroxychromones with antiviral activity against both hepatitis C virus (HCV) and SARS-associated coronavirus (SCV). Eur J Med Chem 2011;46(11):5698–5704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Zaher NH, Mostafa MI, Altaher AY. Design, synthesis and molecular docking of novel triazole derivatives as potential CoV helicase inhibitors. Acta Pharmaceut 2020;70(2):145–159. [DOI] [PubMed] [Google Scholar]
- [67].Adedeji AO, Singh K, Calcaterra NE, et al. Severe acute respiratory syndrome coronavirus replication inhibitor that interferes with the nucleic acid unwinding of the viral helicase. Antimicrob Agents Chemother 2012;56(9):4718–4728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Adedeji AO, Singh K, Kassim A, et al. Evaluation of SSYA10-001 as a replication inhibitor of severe acute respiratory syndrome, mouse hepatitis, and middle east respiratory syndrome coronaviruses. Antimicrob Agents Chemother 2014;58(8):4894–4898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Chan JFW, Lau SKP, To KKW, Cheng VCC, Woo PCY, Yuen K-Y. Middle East respiratory syndrome coronavirus: another zoonotic betacoronavirus causing SARS-like disease. Clin Microbiol Rev 2015;28(2):465–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Asha K, Kumar P, Sanicas M, Meseko CA, Khanna M, Kumar B. Advancements in nucleic acid based therapeutics against respiratory viral infections. J Clin Med 2018;8(1):6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Lundin A, Dijkman R, Bergström T, et al. Targeting membrane-bound viral RNA synthesis reveals potent inhibition of diverse coronaviruses including the Middle East respiratory syndrome virus. PLoS Pathog 2014;10(5):e1004166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Jiang S, Hillyer C, Du L. Neutralizing antibodies against SARS-CoV-2 and other human coronaviruses. Trends Immunol 2020;41(5):355–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Zhou P, Yang X-L, Wang X-G, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020;579(7798):270–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Tian X, Li C, Huang A, et al. Potent binding of 2019 novel coronavirus spike protein by a SARS coronavirus-specific human monoclonal antibody. Emerg Microbes Infect 2020;9(1):382–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Wang C, Li W, Drabek D, et al. A human monoclonal antibody blocking SARS-CoV-2 infection. Nat Commun 2020;11(1):2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Tai W, He L, Zhang X, et al. Characterization of the receptor-binding domain (RBD) of 2019 novel coronavirus: implication for development of RBD protein as a viral attachment inhibitor and vaccine. Cell Mol Immunol 2020;17(6):613–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Hoffmann M, Kleine-Weber H, Schroeder S, et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 2020;181(2):271–280.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Xia S, Liu M, Wang C, et al. Inhibition of SARS-CoV-2 (previously 2019-nCoV) infection by a highly potent pan-coronavirus fusion inhibitor targeting its spike protein that harbors a high capacity to mediate membrane fusion. Cell Res 2020;30(4):343–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].van der Meer FJ, de Haan CA, Schuurman NM, et al. The carbohydrate-binding plant lectins and the non-peptidic antibiotic pradimicin A target the glycans of the coronavirus envelope glycoproteins. J Antimicrob Chemother 2007;60(4):741–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Akerstrom S, Mirazimi A, Tan YJ. Inhibition of SARS-CoV replication cycle by small interference RNAs silencing specific SARS proteins, 7a/7b, 3a/3b and S. Antiviral Res 2007;73(3):219–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Huentelman MJ, Zubcevic J, Hernandez Prada JA, et al. Structure-based discovery of a novel angiotensin-converting enzyme 2 inhibitor. Hypertension 2004;44(6):903–906. [DOI] [PubMed] [Google Scholar]
- [82].Han DP, Penn-Nicholson A, Cho MW. Identification of critical determinants on ACE2 for SARS-CoV entry and development of a potent entry inhibitor. Virology 2006;350(1):15–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Ou X, Liu Y, Lei X, et al. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat Commun 2020;11(1):1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Coutard B, Valle C, de Lamballerie X, Canard B, Seidah NG, Decroly E. The spike glycoprotein of the new coronavirus 2019-nCoV contains a furin-like cleavage site absent in CoV of the same clade. Antiviral Res 2020;176:104742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Millet JK, Whittaker GR. Host cell entry of Middle East respiratory syndrome coronavirus after two-step, furin-mediated activation of the spike protein. Proc Natl Acad Sci U S A 2014;111(42):15214–15219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Pfaender S, Mar KB, Michailidis E, et al. LY6E impairs coronavirus fusion and confers immune control of viral disease. bioRxiv 2020;doi:10.1101/2020.03.05.976167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Kindrachuk J, Ork B, Hart BJ, et al. Antiviral potential of ERK/MAPK and PI3K/AKT/mTOR signaling modulation for Middle East respiratory syndrome coronavirus infection as identified by temporal kinome analysis. Antimicrob Agents Chemother 2015;59(2):1088–1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88]. Maiese K. The mechanistic target of rapamycin (mTOR): novel considerations as an antiviral treatment. Curr Neurovasc Res 2020; doi:10.2174/1567202617666200425205122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Golonka RM, Saha P, Yeoh BS, et al. Harnessing innate immunity to eliminate SARS-CoV-2 and ameliorate COVID-19 disease. Physiol Genomics 2020;52(5):217–221. [DOI] [PMC free article] [PubMed] [Google Scholar]


