Abstract
Obesity has reached epidemic proportions globally. Although modern adoption of a sedentary lifestyle coupled with energy-dense nutrition is considered to be the main cause of obesity epidemic, genetic preposition contributes significantly to the imbalanced energy metabolism in obesity. However, the variants of genetic loci identified from large-scale genetic studies do not appear to fully explain the rapid increase in obesity epidemic in the last four to five decades. Recent advancements of next-generation sequencing technologies and studies of tissue-specific effects of epigenetic factors in metabolic organs have significantly advanced our understanding of epigenetic regulation of energy metabolism in obesity. The epigenome, including DNA methylation, histone modifications, and RNA-mediated processes, is characterized as mitotically or meiotically heritable changes in gene function without alteration of DNA sequence. Importantly, epigenetic modifications are reversible. Therefore, comprehensively understanding the landscape of epigenetic regulation of energy metabolism could unravel novel molecular targets for obesity treatment. In this review, we summarize the current knowledge on the roles of DNA methylation, histone modifications such as methylation and acetylation, and RNA-mediated processes in regulating energy metabolism. We also discuss the effects of lifestyle modifications and therapeutic agents on epigenetic regulation of energy metabolism in obesity.
Keywords: obesity, epigenetics, energy metabolism, treatment
Introduction
The rising prevalence of obesity and related metabolic diseases has become a major public health concern worldwide (Engin, 2017). Excess weight confers significant risk for developing numerous chronic disorders, including type 2 diabetes (T2DM), cardiovascular diseases, cancer, and aging (Lennon et al., 2016; Shapses et al., 2017; Yang et al., 2018; Oh et al., 2019). Studies from ongoing coronavirus disease-19 (COVID-19) pandemic indicate that obesity and its related disorders are also the major risk factors for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection (Abu-Farha et al., 2020). When these patients have COVID-19, the disease is usually more severe, and the mortality rate is higher comparing to patients without these conditions (Abdi et al., 2020; O'Rourke and Lumeng, 2021). It is therefore critically important to elucidate the underlying mechanisms and identify the therapeutic targets for obesity.
Genetic preposition contributes significantly to the obesity epidemic as the heritability estimates for body mass index (BMI) are up to 40%–70% (Yang et al., 2015; Young et al., 2018). Large-scale genetic studies such as genome-wide association analysis (GWAS) have identified several hundred genetic loci where sequence variations are statistically associated with the BMI at the population level. However, these associations collectively explain <3%–5% of the variation in adult BMI (Locke et al., 2015; Yengo et al., 2018). Moreover, most obesity-predisposing gene variants are not associated with weight loss or regain via interactions with lifestyle (Locke et al., 2015; Yengo et al., 2018). Consistently, only few specific gene(s) and pathway(s) have been identified to play a causative role in the development of obesity (Rohde et al., 2019). Therefore, despite the known genetic predisposition of obesity, a large portion of the heritability remains unexplained.
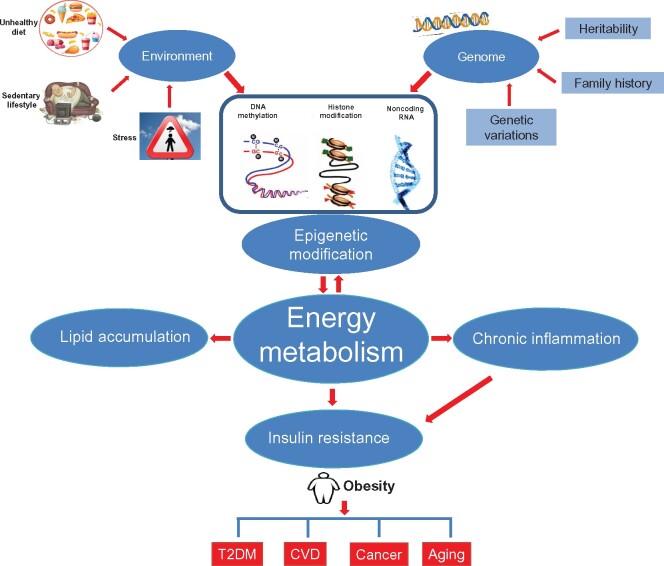
In recent years, advancements of next-generation sequencing technologies have significantly advanced epigenetics research, which allows scientists to look beyond genomic DNA and study the roles of epigenomic machinery in the development of obesity. Epigenome, including DNA methylation, histone modifications, chromatin remodeling, RNA methylation, and noncoding RNAs (ncRNAs), is characterized as mitotically or meiotically heritable changes in gene function that take place in the absence of a change in the DNA sequence (Pagiatakis et al., 2021). In contrast to the genome, which is largely static, the epigenome is much more dynamic and displays variation across cell types (Loh et al., 2019). Epigenetic modifications are crucial for several key biological processes, including cell differentiation, imprinting, and inactivation of the X chromosome (Pagiatakis et al., 2021). Mounting evidence indicates that obesity may arise from the complex interaction of environmental (‘obesogenic’) changes and the epigenome (Ling and Ronn, 2019). It has been proposed that the epigenome may represent the flexible interface of gene–environment interactions and could help explain the ‘missing heritability’ for obesity (van Dijk et al., 2015; Figure 1). In this review, we summarize the roles of epigenetic regulation in energy metabolism and provide an overview of the complex and bidirectional interplay between epigenetics and energy imbalance and consequent obesity.
Figure 1.
Relationship between environment, genome, and epigenetic modification in obesity. Genetic preposition and environmental factors such as sedentary lifestyle and energy-dense nutrition are established etiology for the development of obesity, which confers a higher risk for T2DM, cardiovascular diseases, cancer, and aging. Both genetic proposition and environmental factors may alter epigenetic modifications, including DNA methylation, histone modification, and ncRNA, to modulate energy metabolism in obesity. Obesity may in turn affect epigenetic modifications to regulate energy metabolism.
Epigenetic regulation in energy metabolism
Obesity prevalence is rapidly increased over the past 40–50 years. The duration is not long enough to generate significant amount of new DNA variants that cause obesity. It is conceivable that epigenetic regulation, which is dynamic and enables control over gene expression for the cell to respond to various signaling pathways and environmental stimuli, is a prime candidate mechanism to explain the so-called ‘developmental programming’ of body weight regulation (Waterland, 2014). The whole-body energy homeostasis is maintained by the balance between energy intake/absorption and energy expenditure/loss. When energy intake/absorption are higher than energy expenditure/loss, as in the modern adoption of a sedentary lifestyle coupled with energy-dense nutrition (Hochberg, 2018), the excessive energy would be stored in adipose tissue, leading to obesity. Many genes are activated or reduced to regulate energy metabolism, and epigenetic factors are the primary mechanisms for modulating gene expression. Accordingly, there has recently been extraordinary interest in the roles of epigenetic modifications in modulating energy metabolism.
DNA methylation
To date, DNA methylation, at either genome-wide or site-specific level, is by far the most extensively studied epigenetic mark for human diseases (Tzika et al., 2018). DNA methylation takes place on a cytosine, mainly in CG context or the so-called CpG sites and to a less extent in non-CG context. The methylation of CpG in promoter regions is often associated with transcriptional silencing achieved by repressing the binding of transcription factors or by recruiting 5-methylcytosine (5mC)-binding proteins (MBD1, MBD2, and MBD3) that initiate the formation of compact heterochromatin in part by interacting with histone-modifying enzymes (Samblas et al., 2019). The development of Illumina Infinium HumanMethylation450 (450K) array allows ‘epigenetic-wide association studies (EWAS)’ to identify methylated loci that are associated with obesity. The landscape of DNA methylation associated with obesity and its related disorders in humans has been summarized in the excellent review articles published recently (Ling and Ronn, 2019; Loh et al., 2019; Hyun and Jung, 2020). These human studies reveal several genes with altered DNA methylation that are involved in energy metabolism. For example, CpG promoter methylation of peroxisome proliferator-activated receptor gamma (PPARγ) coactivator-1alpha (PGC-1α), a transcriptional coactivator for mitochondrial biogenesis, is increased in subcutaneous adipose tissue (SAT) of obese women and in the muscle of monozygotic twins discordant for T2DM (Ribel-Madsen et al., 2012; Arner et al., 2015). Adiponectin, an adipokine that regulates systemic energy expenditure and insulin sensitivity, is reduced in adipose tissue in obesity (Liu and Liu, 2014). DNA methylation levels at the adiponectin gene locus in SAT was positively associated with BMI and waist girth in severely obese patients (Houde et al., 2015). Pro-opiomelanocortin (POMC) gene in hypothalamus controls food intake and energy expenditure. Methylation in a variably methylated region in POMC gene in postmortem human laser-microdissected melanocyte-stimulating hormone-positive neurons, is strongly associated with individual BMI (Kuhnen et al., 2016). It has to be noted that many studies of DNA methylation in obesity used DNA from blood samples (Rohde et al., 2019). The significance of blood DNA methylation of metabolism-related genes is unclear (Aslibekyan et al., 2015). Furthermore, the commonly used 450K arrays for EWAS cover ∼3% of >28 million CpG sites in the human genome (Stirzaker et al., 2014; Ling and Ronn, 2019). The whole-genome bisulfite sequencing (WGBS) has been developed and has the capacity to detect ∼95% of all CpG sites in the human genome. However, WGBS is technically challenging and not yet widely used (Stirzaker et al., 2014; Ling and Ronn, 2019). It is conceivable that much more DNA methylation will be identified to be associated with energy metabolism in obesity when the WGBS technologies are improved. Finally, the successful generation of epigenetic disease model mice by targeted demethylation of the epigenome using the CRISPR system will help differentiate the cause vs. consequence of the DNA methylation identified in obesity (Horii et al., 2020).
DNA methylation is dynamically regulated by DNA methyltransferases (DNMTs) and DNA demethylases, the ten–eleven translocation (TET) proteins (Horvath and Raj, 2018). In mammals, five family members of the DNMTs, DNMT1, DNMT2, DNMT3a, DNMT3b, and DNMT3L, have been characterized, but only DNMT1, DNMT3a, and DNMT3b possess DNMT activity. DNMT1 is responsible for maintaining DNA methylation, while DNMT3a and DNMT3b are referred as de novo DNMTs as they can establish new DNA methylation. Removal of DNA methylation is also an active process mediated by three TET proteins (TET1, TET2, and TET3). Both DNMTs and TETs have been shown to regulate metabolic genes at the cellular level. Recent reports on tissue-specific knockout of DNMTs and TETs provide novel insights into the roles of DNMTs and TETs in energy metabolism in obesity (Table 1).
Table 1.
Histone modifications involved in energy metabolism.
| Gene IDs | Model | Tissues/cells | Target genes | Modifications | Brief phenotypes | References |
|---|---|---|---|---|---|---|
| GCN5 | Knockout | Skeletal muscle | PPARγ, PGC-1α | Acetylation | Decrease fatty acid oxidation and facilitate brown adipogenesis and beige adipocyte differentiation | Gerhart-Hines et al. (2007); Jin et al. (2014); Kawabe et al. (2019) |
| HDAC1/2 | Knockdown | White adipocytes | CK2, UCP1, PGC-1α | Acetylation, phosphorylation | Inhibit thermogenesis | Shinoda et al. (2015); Kim et al. (2019) |
| HDAC3 | Knockout | Hepatocytes, white adipocytes | GPAM, REV-ERBa, NCoR, PGC-1α, PPARα/γ, UCP1 | Acetylation | Increase lipogenesis, decrease fatty acid oxidation, and impose a futile cycle of fatty acid utilization and synthesis | Feng et al. (2011); Sun et al. (2012); Ferrari et al. (2017) |
| HDAC3 | Knockout | Brown adipocytes | UCP1, PGC-1α, OXPHOS | Acetylation | Decrease capacity for thermogenesis in BAT | Emmett et al. (2017) |
| HDAC5/9 | Knockdown | Liver | PPARα, IL-6 | Acetylation, phosphorylation | Induce hepatic fatty acid oxidation and increase energy expenditure and adaptive thermogenesis | Chatterjee et al. (2014); Qiu et al. (2018) |
| HDAC6/10 | Knockout | Adipocytes | CIDEC | Acetylation | Increase fat accumulation and reduce insulin sensitivity | Qian et al. (2017) |
| HDAC11 | Knockout | Liver, adipocytes | UCP1 | Acetylation | Induce adiponectin‒AMPK signaling-mediated TG accumulation and promote thermogenic function | Sun et al. (2018) |
| CBP, p300 | Knockout | Adipocytes | UCP1 | Acetylation | Induce severe lipodystrophy along with marked hepatic steatosis, hyperglycemia, and hyperlipidemia | Namwanje et al. (2019) |
| HDAC6 | Knockout | Adipocytes | CIDEC | Acetylation | Increase fat storage and contribute to the development of obesity | Qian et al. (2017); Lieber et al. (2019) |
| LSD1 | Activate | Adipocytes | FAD, C/EBP, H3K4me1, H3K4me2 | Methylation | Promote oxidative metabolism and energy expenditure | Duteil et al. (2014) |
| EHMT1/2 | Knockout | Adipocytes | H3K9me2, H3K9me3 | Methylation | Reduce adaptive thermogenesis, obesity, and systemic insulin resistance | Ohno et al. (2013); Harms et al. (2014) |
| PRDM16 | Knockout | Brown adipocytes | IRF-E, ISRE | Methylation | Induce brown fatlike characteristics | Kissig et al. (2017) |
| JMJD1A | Knockout | Brown adipocytes | UCP1, PPARGC1A, PDK4, PCK1, ADRB1 | Methylation | Decrease heat generation in BAT and oxygen consumption | Tateishi et al. (2009); Abe et al. (2015) |
| JMJD1A | Activate | Adipocytes | H3K9me2 | Phosphorylation, methylation | Promote beige adipogenesis | Abe et al. (2018) |
| EZH2 | Knockout | Adipocytes | APOE | Methylation | Increase lipid uptake in adipocytes | Yiew et al. (2019) |
| JMJD3 | Activate | Adipocytes | UCP1, Cidea, H3K27me3 | Methylation | Induce brown adipogenesis | Pan et al. (2015) |
| KMT5A, KMT5B, KMT5C | Knockout | Adipocytes | PPARγ, TRP53, H4K20me1, H3K27me3 | Methylation | Impair thermogenic program and susceptible to HFD-induced obesity | Zhao et al. (2020) |
| KDM6B | Activate | White adipocytes | H3K27me3, H3K4me3 | Methylation | Facilitate the browning of iWAT | Pan et al. (2015) |
| PARP1 | Knockout | Adipocytes | C/EBPβ, HPF1, PPARγ, Fabp4 | ADP-ribosylation | Induce the formation of mature adipocytes | Luo et al. (2017) |
| OGT | Knockout | Breast cancer cells | AMPK, SREBP1 | O-GlcNAcylation | Induce decreases in lipids | Sodi et al. (2018) |
| PTM | Activate | BMDMs | Lactoyl-CoA, P300 | Lactylation | Promote M1 macrophage polarization and elevate intracellular lactate amounts | Zhang et al. (2019a) |
Brown adipose tissue (BAT) is characterized by high expression of the uncoupling protein-1 (UCP1), which allows the production of heat through non-shivering thermogenesis. Brown adipocytes share the same developmental origin with the skeletal muscle. PR domain containing 16 (PRDM16) determines brown adipocyte fate, while myoblast determination protein 1 (MYOD1) controls muscle development (Kajimura et al., 2015; Rui, 2017). A recent new study shows that PRDM16 recruits DNMT1 to the MYOD1 promoter, causing MYOD1 promoter hypermethylation and suppressing its expression (Li et al., 2020). Brown adipocytes with DNMT1 deficiency driven by UCP1-Cre lose the BAT identity and express higher levels of myogenic genes. Due to the reduced BAT activity, mice with DNMT1 deficiency in BAT exhibit reduced energy expenditure, increased body weight and adiposity on chow diet, and susceptibility to diet-induced obesity (DIO) when fed a high-fat diet (HFD) (Li et al., 2020). It is generally believed that DNMT3 establishes DNA methylation and DNMT1 maintains it. However, recent data also suggest that DNMT1 may coordinate with DNMT3a and DNMT3b to regulate de novo DNA methylation (Okano et al., 1999; Fatemi et al., 2002; Kim et al., 2002; FeNg et al., 2010; Jeltsch and Jurkowska, 2014). It is thus not surprising that BAT-specific DNMT3A knockout mice exhibit similar metabolic phenotype as the BAT-specific DNMT1 knockout mice (Li et al., 2020). Interestingly, human exome sequencing studies have recently identified de novo mutations in DNMT3A in individuals with autism spectrum disorder (Sanders et al., 2015; Feliciano et al., 2019; Satterstrom et al., 2019). Furthermore, mice with whole-body DNMT3A haploinsufficiency show heavier body weight in mature adults, in addition to the autism phenotype (ChrisTian et al., 2020). These results indicate that DNMT3A may play an important role in regulating body weight. Of note, a separate adipocyte-specific DNMT3A knockout mice driven by adiponectin-Cre exhibit no changes in body weight and energy metabolism but insulin resistance due to suppressed fibroblast growth factor-21 (FGF21) expression (You et al., 2017).
DNA methylation can be removed by three TET family enzymes (TET1–3) that have different expression patterns and genomic localizations (Wu and Zhang, 2017). Adipose-selective TET1 knockout mice generated by using Fabp4-Cre improve cold tolerance, increase energy expenditure, and protect against DIO and insulin resistance (Damal Villivalam et al., 2020). For unknown reasons, adiponectin-Cre was not sufficient to delete TET1 in adipocytes. Interestingly, TET1 suppresses thermogenic gene transcription in a DNA demethylase-independent manner. Rather, TET1 coordinates with histone deacetylase 1 (HDAC1) to mediate the epigenetic changes to suppress thermogenic gene transcription (Damal Villivalam et al., 2020). TET2 expression is reduced in DIO. In mature adipocytes, TET2 facilitates the transcriptional activity of PPARγ and insulin-sensitizing efficacy of PPARγ agonist by sustaining DNA binding of PPARγ at certain target loci (Bian et al., 2018). Although TET2 knockout in metabolic organs has not been reported, TET2 loss-of-function-driven clonal hematopoiesis aggravates age- and obesity-related insulin resistance in mice. This metabolic dysfunction is paralleled by increased expression of the pro-inflammatory cytokine IL-1β in white adipose tissue (WAT) (Fuster et al., 2020).
Histone modification
Histones are well-conserved proteins that organize and package DNA into chromatin (Hammond et al., 2017). Histones are prone to various post-translational modifications such as acetylation, methylation, phosphorylation, adenosine diphosphate (ADP) ribosylation, O-GlcNAcylation, and lactylation (Hammond et al., 2017). Modifications to the histone protein tails change the chromatin structure to regulate enhancer and promoter activities. In metabolic organs, histone modifications play key roles in regulating metabolic genes in response to environmental cues.
Histone acetylation
Histone acetylation is associated with a more open chromatin configuration that is permissive for transcription, while histone deacetylation is usually associated with condensed and compacted chromatin causing transcriptional repression (Kaelin and McKnight, 2013). Histone acetylation and deacetylation are catalyzed by histone acetyltransferases (HATs) and HDACs, respectively. HATs catalyze the transfer of the acetyl group from acetyl-CoA onto a lysine residue, which can be reversed by HDACs (Kaelin and McKnight, 2013).
HATs can be classified into three major families: the general control non-derepressible 5 (GCN5)-related N-acetyltransferases (GNAT), Moz–Ybf2/Sas3–Sas2–Tip60 (MYST), and p300/cyclic adenosine monophosphate-response element-binding protein-binding protein (CBP) (Berndsen and Denu, 2008). GCN5, also known as lysine acetyltransferase 2A (KAT2A), has been shown to play a major role in regulating the expression of metabolic genes through acetylating not only histone but also PGC-1α. For example, overexpression of GCN5 in C2C12 myotubes leads to repression of PGC-1α-mediated induction of mitochondrial and fatty acid metabolism genes (Gerhart-Hines et al., 2007; Mutlu and Puigserver, 2021). Interestingly, however, muscle-specific knockout of GCN5 using the creatine kinase promoter Cre does not alter whole-body energy expenditure as well as skeletal muscle mitochondrial abundance (Dent et al., 2017). The underlying mechanism for the discrepancy between in vitro and in vivo studies is unclear. It is possible that P300/CBP-associated factor (PCAF, also known as KAT2B), which shares ∼73% homology with the GCN5 gene, is sufficient to compensate the effects of GCN5 deletion (Dent et al., 2017). GCN5 has also been shown to facilitate brown adipogenesis and beige adipocyte differentiation in vitro (Jin et al., 2014; Kawabe et al., 2019). The in vivo roles of adipose GCN5 in regulating energy metabolism remain to be investigated.
CBP and p300 are functionally related KAT3 family members. In addition to their HAT activity, CPB and p300 interact with at least 400 proteins, thereby acting as hubs in gene regulatory networks (Bedford et al., 2010). The initial evidence for the importance of CBP in regulating energy homeostasis is from the study of CBP heterozygous-null mice, which show improved DIO and glucose intolerance (Yamauchi et al., 2002). Furthermore, disrupting the CH1 domain, which is important for protein interactions in the CBP and p300, results in lean mice with improved insulin sensitivity and glucose metabolism (Bedford et al., 2011). However, defining the organ-specific effects of CBP and p300 has not been conclusive. Liver CBP knockout in adult mice using the AAV-medicated Cre does not alter glucose levels (Bedford et al., 2011). Muscle-specific loss of either p300 or CBP, either in germline or in adulthood, does not impact energy expenditure, glucose tolerance, or skeletal muscle insulin action (Martins et al., 2019). Adipose-specific CBP and p300 knockout mice are recently reported (Namwanje et al., 2019). CBP knockout mice exhibit marked brown remodeling of inguinal WAT (iWAT) but not epididymal WAT (eWAT) after cold exposure, although loss of CBP is insufficient to impact body weight or glucose tolerance. In contrast, ablation of p300 in adipose tissues has minimal effects on fat remodeling and adiposity. Double knockout of CBP and p300 causes severe lipodystrophy along with marked hepatic steatosis, hyperglycemia, and hyperlipidemia. One difference between CBP heterozygous-null mice and tissue-specific knockout mice could be the gene deletion time (Namwanje et al., 2019). Conditional knockout is achieved by Cre recombinase driven by tissue-specific promoter, which is activated after the initiation of differentiation. It is possible that CBP and p300 are essential for cellular development rather than have direct effects on metabolic genes involved in energy expenditure.
HDACs are categorized into four different classes. Class I (HDAC1–3 and HDAC8) are located in the nucleus and predominantly act as HDACs. Class IIa (HDAC4, HDAC5, HDAC7, and HDAC9) and class IIb (HDAC6 and HDAC10) shuttle from cytoplasm to nucleus and modify chromatin in a signal-responsive manner. Class III HDACs are sirtuins (Sirt1–7) that use nicotinamide adenine dinucleotide (NAD+) as a cofactor (Li and Sun, 2019). HDAC11 is the only member in the class IV. The critical roles of sirtuins in regulating energy metabolism have been well documented (Canto et al., 2015; Covarrubias et al., 2021). Here, we will focus on other classes of HDACs.
The link between HDACs and energy metabolism was initially studied using HDAC inhibitors. Treatment of obese mice with sodium butyrate, an HDAC pan-inhibitor, markedly reduces adiposity with increased brown fat thermogenesis and oxidative fibers in the muscle (Gao et al., 2009). Further studies show that class I but not class II HDAC inhibitors enhance whole-body energy expenditure through increased mitochondrial biogenesis in the skeletal muscle and adipose tissues (Galmozzi et al., 2013). Li et al. (2016) used short interfering RNA to knockdown all 11 HDACs in cultured brown adipocytes to investigate which HDAC is involved in cellular energy metabolism. It was found that only HDAC1 deletion markedly enhances thermogenic gene expression (Li et al., 2016). Mechanistically, HDAC1 knockdown increases acetylation and decreases methylation of histone H3 lysine 27 (H3K27), thereby switching the transcriptional repressive state to the active state at the promoters of thermogenic genes such as UCP1 and PGC-1α (Li et al., 2016). It has to be noted that HDAC1 and HDAC2 have redundant effects on cardiac and skeletal muscle functions (Montgomery et al., 2007; Moresi et al., 2012). For example, single deletion of either HDAC1 or HDAC2 does not evoke a phenotype whereas cardiac-specific deletion of both genes results in neonatal lethality, accompanied by cardiac arrhythmias and dilated cardiomyopathy (Montgomery et al., 2007). Future studies of adipose HDAC1 and HDAC2 knockout mice will provide insights into the in vivo roles of these two class I HDACs in regulating systemic energy metabolism.
HDAC3 has been extensively studied for its roles in regulating energy metabolism. BAT-specific knockout of HDAC3 using UCP1-Cre depletes UCP1 and reduces mitochondrial and oxidative phosphorylation genes required for non-shivering thermogenesis and cold survival (Emmett et al., 2017; Emmett and Lazar, 2019). Interestingly, a separate HDAC3 knockout mice driven by adiponectin-Cre exhibit normal BAT identity but enhanced browning of white fat (Ferrari et al., 2017). Of note, neither adipose HDAC3 knockout mouse line shows body weight changes even with HFD challenges. The significance of HDAC3 in regulating energy metabolism is also manifested by liver and muscle HDAC3 knockout mice. Deletion of HDAC3 in adult hepatocytes causes continuous lipid synthesis, leading to severe hepatosteatosis, yet these mice are more sensitive to insulin, perhaps owing to the limited availability of gluconeogenic substrates (Feng et al., 2011; Sun et al., 2012). By contrast, mice with a muscle-specific deletion of HDAC3 are insulin resistant, yet have greater endurance and resistance to muscle fatigue than wild-type mice, as the HDAC3-deficient muscle undergoes a fuel source switch favoring protein catabolism and lipid oxidation over glucose utilization (Hong et al., 2017). In addition to modulating histone acetylation such as H3K27ac and H3K9ac, HDAC3 also forms complexes with many transcriptional regulators such as REV-ERBa, NCoR, ERRα, and PGC-1α, which may contribute to the different roles of HDAC3 in regulating substrate metabolism in different metabolic organs (Emmett and Lazar, 2019).
Although inhibitors of class II HDACs do not show metabolic phenotype (Galmozzi et al., 2013), studies of genetic mouse models suggest that these HDACs may also play a role in regulating energy, lipid, and glucose metabolism. For example, overexpression of HDAC4 in adipocytes leads to beige adipocyte expansion and reduced adiposity (PauLo et al., 2018). Global knockout of HDAC5 mice exhibit increased food intake and obesity when fed HFD due to the enhanced STAT3 acetylation, which impairs the leptin signaling in hypothalamus (Kabra et al., 2016). Interestingly, HDAC5 knockout animals demonstrate normal glucose and insulin tolerance despite elevated body adiposity and increased hepatic fat deposition owing to H3K9ac-mediated interleukin-6 production in the muscle (Kabra et al., 2016; Klymenko et al., 2020). Finally, HDAC9 knockout mice show upregulated expression of beige adipocyte marker genes, particularly during an HFD, in association with increased energy expenditure and adaptive thermogenesis (Chatterjee et al., 2014). Protein levels of class IIb HDACs (HDAC6 and HDAC10) are markedly reduced in both WAT and BAT of HFD-fed obese mice (Qian et al., 2017). Mice with an adipose-specific depletion of HDAC6 display increased fat storage due to enhanced acetylation of cell death-inducing DFFA-like effector C (CIDEC) (Qian et al., 2017). HDAC6 may also affect energy metabolism by altering intestinal microbiota. Feeding HDAC6-deficient mice with HFD causes depletion in representatives of the S24-7 family and Lactobacillus but enrichment with Bacteroides and Parabacteroides, changes of which contribute to the development of obesity (Lieber et al., 2019).
HDAC11 is the only member of class IV HDAC. HDAC11 deletion enhances brown and beige fat activity, leading to enhanced energy expenditure and leanness (Bagchi et al., 2018; Sun et al., 2018). HDAC11 is also highly expressed in the skeleton muscle. HDAC11 deficiency induces a glycolytic to oxidative muscle fiber switch in vivo, providing a mechanistic explanation for the improved muscle strength and fatigue resistance (Hurtado et al., 2021).
Overall, extensive studies of HATs and HDACs have emphasized the crucial role of histone acetylation and deacetylation in bridging epigenetic, transcriptional, and signaling phenomena to metabolism in obesity (Figure 2). Interestingly, it does not appear to have a clear pattern for the different classes of HDACs in regulating energy metabolism (Table 1). Substrate specificity, tissue distribution, and interacting partners may determine their roles in regulating energy metabolism. The selectivity, on the other hand, may provide an opportunity to identify inhibitors and activators of HDACs for treatment of obesity.
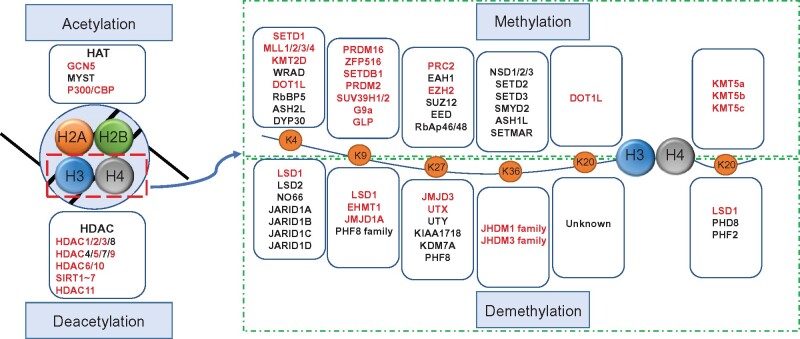
Figure 2.
Modifiers for histone acetylation and methylation. Histone acetylation and deacetylation are catalyzed by HATs and HDACs, respectively. Extensive studies of HATs and HDACs have emphasized the crucial role of histone acetylation and deacetylation in bridging epigenetic, transcriptional, and signaling phenomena to metabolism in obesity. Histone methylations are dynamically regulated by HMTs and HDMs. Histone methylation occurs on basic residues lysine and arginine and confers active or inhibitory transcription, depending on their location and methylation status. Red color indicates the modifiers participated in energy metabolism.
Histone methylation
Histone methylation occurs on basic residues lysine and arginine that can be mono- (me1), di- (me2), or tri-methylated (me3) on their ɛ-amine group. Different from histone acetylation, which is associated with active chromatin states, histone methylations may confer either active or repressive transcription depending on their positions and methylation states (Figure 2). For example, H3K4, H3K36, and H3K79 methylations are markers of active transcription, whereas H3K9, H3K27, and H4K20 methylations are associated with suppressive transcription. These histone methylations are dynamically regulated by histone methyltransferases (HMTs, ‘writers’) and histone demethylases (HDMs, ‘erasers’) (Han et al., 2019). HMTs, except the H3K79 methyltransferase DOT1, contain a defined SET-domain with an intrinsic histone lysine-specific methyltransferase activity. Most methyltransferases show a strong preference for specific sites. For example, the SETD1/mixed-lineage leukemia (MLL) family methylates H3K4 gene-activating marks, whereas polycomb repressive complex 2 (PRC2) promotes H3K27 methylation, a repressive mark. Histone methylation was considered stable until the report of lysine-specific demethylase 1 (LSD1) in 2004 (Shi et al., 2004). LSD1 primarily demethylates H3K4 monomethylation/dimethylation (H3K4me1/me2) and may also remove methylation from H3K9me1/me2. Subsequently, 18 HDMs containing Jumonji domain with selective substrate specificity were identified (Han et al., 2019). It is important to note that one specific lysine residue of histone protein may be modulated by multiple HMTs and HDMs. Therefore, the alteration of histone methylation occupancy on target gene promoters/enhancers is locus-specific and context-dependent. The dynamics of histone methylation regulated by HMTs and HDMs provide innovative tools to reprogram histone methylation and gene expression profiles. Several HMT and HDM inhibitors are currently in clinical trials for cancer treatment (Cheng et al., 2019). Multiple HMTs and HDMs have also been shown to regulate energy metabolism.
H3K4 can be methylated by six members of the MLL family, among which MLL4/KMT2D is associated with Kabuki syndrome, a rare genetic disease. Mutations of MLL4/KMT2D in Kabuki syndrome exhibit endocrine system-related conditions such as premature thelarche, precocious puberty, diabetes insipidus, thyroid dysfunction, obesity, and growth hormone deficiency (Ng et al., 2010; Moon et al., 2018). Interestingly, however, MLL4 heterozygous mice exhibit low body fat mass, improved hepatic steatosis, enhanced glucose tolerance, and insulin sensitivity. MLL4-regulated PPARγ2 activity and bile acids are thought to be involved in the metabolic phenotype in the MLL4 heterozygous mice (Kim et al., 2015, 2016). LSD1 is the first identified HDM that selectively demethylates H3K4me1 and H3K4me2 through a flavin adenosine dinucleotide (FAD)-dependent oxidative reaction (Shi et al., 2004). LSD1 may also act on mono- and di-methylated H3K9. Several lines of adipose LSD1 knockout mice have been reported. It appears that LSD1 is essential for brown and beige fat development via regulating H3K4 and H3K9 methylation as well as interacting with transcription factors such as PRDM16 and zinc finger protein 516 (ZFP516) (Duteil et al., 2016, 2017; Sambeat et al., 2016; Zeng et al., 2016; Wang et al., 2020a).
Notable among the histone methylation modifiers, dynamics of H3K9 methylation have been highly related to energy metabolism. The methyltransferases that methylate H3K9 include euchromatic histone–lysine N-methyltransferase 1 (EHMT1, also known as G9a-like protein, GLP), EHMT2 (also known as G9a), SETDB1, PRDM2, SUV39H1, and SUV39H2 (Krishnan et al., 2011). EHMT1 may form a complex with PRDM16 to control brown adipose cell fate by depositing the suppressive H3K9me2 and H3K9me3, leading to muscle-selective gene suppression and brown fat gene activation (Ohno et al., 2013). Adipose-specific deletion of EHMT1 leads to a marked reduction of BAT-mediated adaptive thermogenesis, obesity, and systemic insulin resistance (Ohno et al., 2013; Harms et al., 2014). EHMT2 is functionally related to EHMT1. EHMT2 knockout driven by aP2-Cre also increases adiposity with elevated insulin and leptin levels, although the mechanism is thought to be EHMT2-mediated adipogenesis (Wang et al., 2013). On the other hand, recently reported muscle EHMT2 knockout female mice are resistant to HFD-induced obesity and hepatic steatosis (Zhang et al., 2020). The liver deletion of EHMT2 results in aggravated lipopolysaccharide-induced peroxidation and proinflammatory response (Lu et al., 2019). Therefore, H3K9 methyltransferases regulate energy metabolism and adiposity in different metabolic organs in the context-dependent fashion. The H3K9 demethylase Jumonji domain-containing protein 1A (JMJD1A, also known as JHDM2A or KDM3A) is induced after β-adrenergic stimulation. JMJD1A regulates adipose thermogenesis through two mechanisms. Under acute stimulation, JMJD1A is phosphorylated by protein kinase A, which facilitates long-range chromatin interactions and target gene activation. Interestingly, this is independent of H3K9 methylation (Abe et al., 2015). Under chronic adrenergic stimulation, JMJD1A erases the suppressive H3K9me2 modification on beige-selective gene promoters in iWAT to maintain sustained activation of thermogenic genes in response to chronic cold exposure (Abe et al., 2018). JMJD1A deficiency results in obesity due to reduced thermogenesis (Tateishi et al., 2009).
Methylation of H3K27 marks suppressive gene transcription. Enhancer of zeste homolog 2 (EZH2), the functional enzymatic component of the PRC2, promotes H3K27me3, while JMJD3 (also known as KDM6B) and ubiquitously transcribed X-chromosome tetratricopeptide repeat protein (UTX) specifically demethylate H3K27. Adipocyte-specific EZH2 knockout mice, generated by crossing EZH2-floxed mice to adiponectin-Cre mice, display significantly increased body weight, adipose tissue mass, and adipocyte cell size, without changes in glucose or insulin tolerance. The increased adiposity is not caused by altered energy expenditure but by increased ApoE-mediated lipid uptake in adipocytes (Yiew et al., 2019). JMJD3 demethylates suppressive H3K27me3 to activate BAT-selective genes and promote development of beige adipocytes (Pan et al., 2015). JMJD3 may also form a complex with SIRT1 and regulates fatty acid oxidation in the liver (Seok et al., 2018). Similarly, UTX demethylase positively regulates brown adipocyte thermogenic program through coordinated control of demethylating H3K27me3 and acetylating H3K27, switching the transcriptional repressive state to the transcriptional active state at the promoters of PRDM16, UCP1, and PGC-1α. UTX deficiency in brown fat promotes HFD-induced obesity (Zha et al., 2015; Li et al., 2020). Therefore, demethylation of suppressive H3K27me3 in metabolic organs generally causes increased energy metabolism.
Methylation of suppressive H4K20 is catalyzed by three distinct lysine methyltransferase enzymes, KMT5a (also known as SETD8, SET8, and PR-Set7), KMT5b (SUV420H1), and KMT5c (SUV420H2). Double knockout of Kmt5B and Kmt5C in early mesenchymal precursor cells using Myf5-Cre increases BAT metabolic activity and enhances browning of WAT, ultimately counteracting DIO (Pedrotti et al., 2019). Activation of PPARγ is reported to be the underlying mechanism for the improved metabolic phenotype in the Kmt5B and Kmt5C double-knockout mice (Pedrotti et al., 2019). Interestingly, however, a more recent study shows that KMT5c but not KMT5a or KMT5b is essential for thermogenic gene expression in fat cells (Zhao et al., 2020). Mice with adipose-specific Kmt5c knockout driven by Adipoq-Cre exhibit reduced adipose thermogenesis and are susceptible to obesity when fed a HFD. Mechanistically, the increased p53 expression from removal of suppressive H3K20me3 in the p53 promoter with Kmt5c knockout is involved in the activation of thermogenic genes (Zhao et al., 2020). Different Cre lines (Myf5 vs. adiponectin) that delete Kmt5c at different stages of adipocyte differentiation may contribute to the discrepancy of the roles of Kmt5c in regulating adipose thermogenesis.
It is worth noting that histone methylation and acetylation are dynamically interactive. For example, H3K27ac distinguishes active enhancers from inactive/poised enhancer elements containing H3K4me1 (Creyghton et al., 2010). The enhancers with H3K4me1 have been shown to serve a priming role during development and in response to environmental cues (Ghisletti et al., 2010; Calo and Wysocka, 2013). In warm beige adipocytes, H3K4me1 signals are increased in the enhancer regions of thermogenetic genes such as UCP1 and CPT1B, indicating that warm beige adipocytes retain an epigenomic memory from prior cold exposure at a small, but key, subset of cis-elements (Roh et al., 2018).
Other histone modifications
In addition to the extensively studied acetylation and methylation, histone proteins are subjected to other post-translational modifications such as phosphorylation, succinylation, malonylation, sumoylation, ADP-ribosylation, O-GlcNAcylation, and lactylation. Quantitative proteomic analysis using the mass spectrometry-based label-free and chemical stable isotope labeling has identified 170 histone marks in the liver of HFD-induced obese mice (Nie et al., 2017). Some of these modifications may be involved in adipocyte function. For example, ADP-ribosylation of glutamate (Glu35) and the subsequent reduction of H2B-Ser36 phosphorylation inhibit adipocyte differentiation (Huang et al., 2020). H2B O-GlcNAcylation at Ser112 may be involved in brown adipogenesis (Cao et al., 2021). Overall, the roles of these histone modifications in regulating energy metabolism remain to be elucidated.
RNA methylation
More than 150 types of RNA modifications have been identified and N6-methyladenine (m6A) is the most abundant internal modification in messenger RNA (mRNA) and ncRNA (Wei et al., 2017). m6A is post-transcriptionally assembled by m6A methyltransferase complexes (‘writers’) composed of methyltransferase-like 3 (METTL3), METTL14, and Wilms’ tumor 1-associating protein (WTAP). METTL3 levels are decreased in BAT of HFD-induced obese and ob/ob mice (Pan et al., 2020). BAT-specific METTL3 knockout mice show reduced energy expenditure and are predisposed to HFD-induced obesity and metabolic syndrome. Mechanistically, deletion of METTL3 decreases m6A modification and the expression of BAT-specific mRNA, including PRDM16, PPARγ, and UCP1 (Pan et al., 2020). On the other hand, WTAP heterozygous knockout mice are protected from DIO with elevated energy metabolism and improved insulin sensitivity. WTAP knockdown induces cell cycle arrest and impairs adipogenesis by suppressing cyclin A2 during mitotic clonal expansion (Kobayashi et al., 2018). Future studies of tissue-specific knockout of WTAP will be necessary to distinguish the developmental effects vs. direct effects of m6A modification on metabolic gene expression.
m6A modifications can be removed by m6A demethylases (‘erasers’), such as fat mass- and obesity-associated (FTO) and α-ketoglutarate (α-KG)-dependent dioxygenase alkB homolog 5 (ALKBH5) (Boccaletto et al., 2018). FTO is the first identified m6A demethylase on mRNA (Jia et al., 2011). In the large-scale GWAS studies, variants in the FTO gene are strongly associated with BMI and obesity, suggesting an important role of FTO in energy metabolism (Dina et al., 2007; Frayling et al., 2007; Scott et al., 2007; Scuteri et al., 2007; Thorleifsson et al., 2009). Interestingly, a CpG site in the first intron of the FTO gene shows hypomethylation, suggesting FTO per se can also undergo epigenetic regulation (Toperoff et al., 2012). Ubiquitous overexpression of FTO leads to a marked increase in food intake, body weight, and fat accumulation in mice, while whole-body knockout of FTO reduces adiposity and DIO accompanied with increased energy expenditure and systemic sympathetic activation (Fischer et al., 2009; Church et al., 2010; Gao et al., 2010; McMurray et al., 2013). FTO is highly expressed in hypothalamus (Gerken et al., 2007). Mice with brain-specific deletion of FTO display significantly higher metabolic rates and reduced body weight (Gao et al., 2010). FTO is also expressed in peripheral metabolic organs including the adipose tissue, liver, and skeletal muscle (Gerken et al., 2007). In human WAT, FTO expression is inversely correlated with BMI (Kloting et al., 2008) and is downregulated during adipocyte differentiation (Tews et al., 2011). FTO-deficient mice exhibit an increase of UCP1 expression in WAT, indicating browning of WAT (Tews et al., 2013). By contrast, hepatocyte-specific FTO depletion fails to affect body weight, fat mass, glucose metabolism, and key parameters of energy expenditure, indicating the dispensable role of hepatic FTO for the control of energy homeostasis (Mittenbuhler et al., 2020). Entacapone, a catechol-O-methyltransferase inhibitor used in the treatment of Parkinson’s disease, is identified as an inhibitor of FTO. Treatment of obese mice with entacapone improves body weight regulation and glucose tolerance and increases adipose thermogenesis owing to decreased FTO-catalyzed m6A demethylation of FOXO1 mRNA (Peng et al., 2019). Expression of FTO is also upregulated in the skeletal muscle of obese mice and therefore demethylates the methylated mRNA by removing m6A (Wu et al., 2017). However, FTO appears to be essential to skeletal muscle development (WaNg et al., 2017b), and the physiological role of muscle FTO in regulating energy metabolism remains unclear. Another m6A demethylases ALKBH5 can be activated by hypoxia resulting in decreased m6A level of mRNA (Wang et al., 2020b). Activation of ALKBH5 in hypoxia is crucial for the adaption to hypoxia for efficient energy generation, since silencing ALKBH5 blocks cellular ATP production (Wang et al., 2020b). The effects of ALKBH5 on systemic energy metabolism in obesity remain to be elucidated.
NcRNA
NcRNAs including microRNAs (miRNAs), long noncoding RNAs (lncRNAs), and circular RNAs (circRNAs) are emerged to be important epigenetic regulators in many physiological processes, including energy metabolism (Marchese et al., 2017).
miRNAs are short molecules (19–22 nucleotides) that bind to the 3′ untranslated regions (3′ UTR) of mRNAs causing translational repression or mRNA degradation (Gharanei et al., 2020). Omics approaches have identified a variety of differentially expressed miRNAs in metabolic organs in obesity, and some of them are functional in regulating energy metabolism (Dumortier et al., 2013; Kunej et al., 2013; Landrier et al., 2019; Table 2). miRNAs may also be secreted from metabolic organs and serve as endocrine factors to regulate systemic energy expenditure (Ji and Guo, 2019). Notably, adipose tissue-specific knockout of the miRNA-processing enzyme Dicer exhibits a substantial decrease in circulating exosomal miRNAs, leading to increased FGF21 expression in the liver (Thomou et al., 2017).
Table 2.
NcRNAs involved in energy metabolism.
| Gene IDs | Model | Tissues/cells | Target genes | Brief phenotype | References |
|---|---|---|---|---|---|
| miR26a/b | Overexpress | 3T3-L1 | PTEN, FAS, PPARγ, C/EBPα, ADAM17 | Promote adipocyte differentiation and increase lipid accumulation | Karbiener et al. (2014); Li et al. (2017) |
| miR455 | Overexpress | Adipocytes | RIP140, PPARγ, PGC-1α, AMPK | Stimulate browning and mitochondrial respiration | Hu et al. (2015); Zhang et al. (2015) |
| miR30b/c | Overexpress | Adipocytes | UCP1, RIP140, CIDEA | Increase thermogenic gene expression and mitochondrial respiration | Hu et al. (2015) |
| miR193, miR328 | Overexpress | Muscle | BACE1 | Promote myogenic and inhibit brown fat commitment | Oliverio et al. (2016) |
| miR32 | Overexpress | Adipocytes | TOB1, FGF21, UCP1 | Increase BAT thermogenesis and promote WAT browning | Ng et al. (2017) |
| miR182, miR203 | Knockout | Adipocytes | DGCR8 | Induce brown fat dysfunction and cold intolerance | Kim et al. (2014) |
| miR133 | Knockdown | Adipocytes | PRDM16 | Promote differentiation of precursors from BAT and SAT to mature brown adipocytes and lead to increased mitochondrial activity | Trajkovski et al. (2012) |
| miR129 | Overexpress | Adipocytes | IGF2, EGR1, UCP1 | Induce energy expenditure and thermogenesis | Pahlavani et al. (2018) |
| miR155 | Knockout | Adipocytes | C/EBP | Increase BAT function and browning of WAT | Chen et al. (2013) |
| miR106b-93 | Knockdown | Adipocytes | UCP1, SIRT7 | Induce the expression of brown fat-specific genes and promote the accumulation of lipid-droplet in differentiating brown adipocytes | Wu et al. (2013); Cioffi et al. (2015) |
| miR34a | Overexpress | Adipocytes | FGFR1, FGF21, PGC-1α | Inhibit beige and brown fat formation | Fu et al. (2014) |
| miR27b | Knockdown | Adipocytes | PGC-1α, PRDM16, PPARα, PPARγ, CREB | Improve browning capacity and insulin sensitivity | Sun and Trajkovski (2014); Yu et al. (2019) |
| lncRNA H19 | Overexpress | Brown adipocytes | MBD1 | Promote oxidative metabolism and mitochondrial respiration | Schmidt et al. (2018) |
| lncBATE1, lncBATE10 | Knockout | Adipocytes | CIDEA, C/EBPα, PDRM16 PPARα, UCP1 | Reduce oxygen consumption and lower mitochondrial content | Alvarez-Dominguez et al. (2015); Bai et al. (2017) |
| lnc-Lep | Knockdown | Adipocytes | PPARγ, Adipoq | Increase fat mass with reduced plasma leptin | Lo et al. (2018); Dallner et al. (2019) |
| circSAMD4A | Knockdown | Adipocytes | miR-138-5p, ZNH2 | Increase energy expenditure and suppress preadipocyte differentiation | Liu et al. (2020) |
| circ133 | Knockdown | SGC7901, 3T3L1 | miR133, PRDM16, UCP1 | Accelerate the glucose consumption ratio and promote the oxygen consumption rate of adipocytes | Zhang et al. (2019b) |
| circNrxn2 | Overexpress | Adipocytes | miR103, PPARγ, FGF10 | Promote WAT browning through increasing M2 macrophage polarization | Zhang et al. (2019c) |
LncRNAs are conventionally defined as a transcript longer than 200 nucleotides in length lacking protein-coding capability (Kung et al., 2013). Accumulating evidence suggests that lncRNAs play important functional roles in modulating the transcription and translation of energy metabolism-related genes (Tan et al., 2020). Several BAT- and WAT-specific lncRNAs have been identified via de novo reconstruction of transcriptomes or meta-analysis of published datasets (Alvarez-Dominguez et al., 2015; Bai et al., 2017; Xiong et al., 2018). For example, comprehensive transcriptome study by RNA sequencing in adipocytes isolated from interscapular BAT, iWAT, and eWAT in DIO mice revealed a set of obesity-dysregulated lncRNAs. The most prominent lncRNAs is lnc-Lep, which is transcribed from an enhancer region upstream of leptin (Lo et al., 2018). Functional studies indicate that lnc-Lep is essential for adipogenesis and required for the maintenance of adipose leptin expression. DIO mice lacking lnc-Lep show increased fat mass with reduced plasma leptin levels and lose weight after leptin treatment. Importantly, large-scale genetic studies of humans reveal a significant association of single-nucleotide polymorphisms in the region of human lnc-Lep with lower plasma leptin levels and obesity (Dallner et al., 2019). Among those so-called BAT-enriched lncRNAs (lncBATEs), lncBATE1 and lncBATE10 are located in an intergenic locus targeted by CCAAT/enhancer binding protein α (C/EBPα), C/EBPβ, and PPARγ (Alvarez-Dominguez et al., 2015; Bai et al., 2017). Knockdown of these lncBATE1 or lncBATE10 results in significant downregulation of BAT marker genes including CIDEA, C/EBPβ, PGC-1α, PRDM16, PPARα, and UCP1, with reduced mitochondrial content and oxygen consumption, indicating impaired BAT thermogenesis (Alvarez-Dominguez et al., 2015; Bai et al., 2017). Moreover, these two lncBATEs are also required for browning of iWAT (Alvarez-Dominguez et al., 2015; Bai et al., 2017). Another important lncRNA involved in energy metabolism is encoded by the maternally imprinted gene H19. H19 is upregulated in BAT under cold exposure and decreased in BAT of DIO mice (Schmidt et al., 2018). Ubiquitous overexpression of H19 enhances BAT thermogenesis, increases energy expenditure, and prevents DIO, whereas fat H19 loss sensitizes toward HFD weight gains. Mechanistically, lncRNA H19 forms a complex with the DNA methyltransferase MBD1 and recruits suppressive H3K9me3 to maintain quiescence of obesity-predisposing paternally expressed genes in BAT (Schmidt et al., 2018). Finally, recent studies also suggest that lncRNAs can act as competitive endogenous RNA (ceRNA) by binding to miRNAs, hence inhibiting miRNA activity and regulating mRNA expression (Salmena et al., 2011; Hu et al., 2020). However, the precise roles of the ceRNA–miRNA network in regulating energy metabolism need to be further studied.
CircRNAs are covalently closed single-stranded RNA rings generated from a process known as back-splicing or head-to-tail circle splicing, which involves joining of a splice donor to an upstream splice acceptor of precursor mRNA. The biological functions of circRNAs remain to be fully elucidated, but have been shown to act as potent miRNA sponges via competition with miRNA/mRNA binding (Chen, 2016). Emerging evidence suggests that circRNAs may play an important role in regulating adipose function and energy metabolism. Differentially expressed circRNAs in the adipose tissue from obese and lean subjects have been reported using circRNA microarrays. Among these, circSAMD4A can act as a miRNA sponge by interacting with miR-138-5p. Knockdown of circSAMD4A inhibits adipocyte differentiation (Liu et al., 2020). Several other circRNAs such as ciRS-133 (circRNA sponge for miR-133) and circNrxn2 (miR-103 sponge) may promote WAT browning (Zhang et al., 2019b, c). Not all circRNAs regulate adipose function through miRNA sponges. Deep sequencing of visceral and subcutaneous fat identifies thousands of adipose circRNAs, many of which are dynamically regulated during adipogenesis and obesity. Among the regulated circRNAs, circArhgap5-2 is required for adipogenesis, not through sponging miRNAs (Arcinas et al., 2019). The understanding of circRNA biology is at a very early stage. CircRNAs are stable compared to linear RNAs. Future studies focusing on their roles in energy metabolism may help provide novel therapeutic strategy for treating obesity.
Metabolites as cofactors for the epigenetic machinery
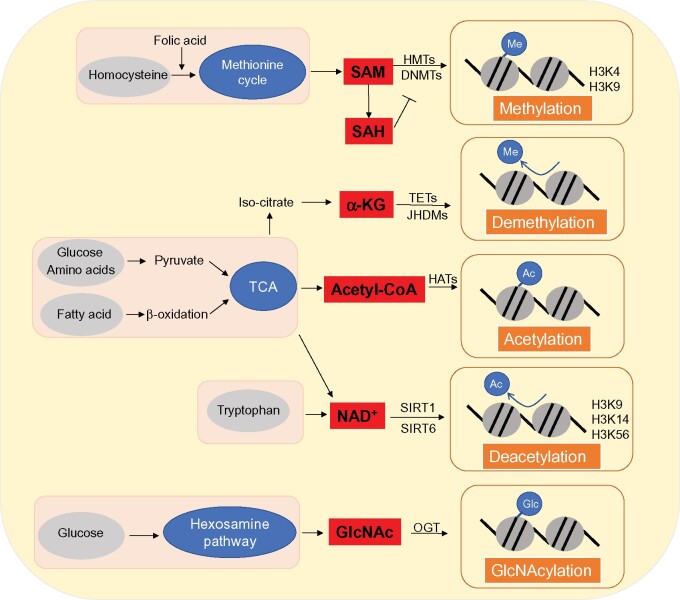
The activity of epigenetic factors is regulated at multiple levels including transcription, translation, and post-translational modifications. It is increasingly recognized that small-molecule metabolites such as acetyl-CoA, S-adenosylmethionine (SAM), NAD+, FAD, and α-KG serve as essential cofactors to modulate epigenetic factor activity. These metabolites are therefore regarded as metabolic sensors for programing pathway network of energy metabolism (Etchegaray and Mostoslavsky, 2016; Yang et al., 2018; Figure 3).
Figure 3.
Metabolites as cofactors of the epigenetic machinery. The activity of epigenetic factors is regulated at multiple levels including transcription, translation, and post-translational modifications. It is increasingly recognized that small-molecule metabolites such as acetyl-CoA, SAM, NAD+, FAD, and α-KG serve as essential cofactors to modulate epigenetic factor activity. These metabolites are therefore regarded as metabolic sensors for programing pathway network of energy metabolism. Metabolic cofactors are produced in respective metabolic pathways, which participate in epigenetic modification processes through enzymes. Epigenetic modifications can be assessed by detecting relevant metabolic cofactors, which can be interfered with by targeting the regulation of metabolic cofactor expression.
SAM
SAM is the universal methyl donor to both DNMT and HMT enzymes. SAM is synthesized from the condensation of methionine and ATP via methionine adenosyltransferase. A positive correlation between SAM levels, BMI, and adiposity mass has been reported (Elshorbagy et al., 2016). Tissue SAM levels are therefore modulated by the availability of methionine, an essential amino acid in one-carbon metabolism. Methionine restriction rapidly reduces adiposity and improves insulin sensitivity and fatty liver in mice (Malloy et al., 2006; Stone et al., 2014; Wanders et al., 2017). Although the underlying mechanisms for the beneficial metabolic effects of methionine deficiency remain to be fully elucidated, methionine deficiency drastically reduces liver SAM levels and H3K4 trimethylation (Mentch et al., 2015). In adipose tissue, SAM-regulated histone methylation may have a positive effect on systemic energy metabolism. Nicotinamide N-methyltransferase (NNMT), which is highly expressed in adipose tissue, catalyzes the methylation of nicotinamide using SAM as a methyl donor. NNMT knockdown in adipose tissue elevates SAM levels and increases H3K4 methylation, which in turn enhances the key enzymes in the polyamine flux to increase systemic energy metabolism (Kraus et al., 2014). The one-carbon metabolism and SAM production may also affect DNA methylation reprograming during mammalian development, in which genomes of germ cells and embryo undergo two waves of global demethylation and remethylation (Li et al., 2018). It has been shown that maternal one-carbon metabolism may affect the offspring energy metabolism and development of obesity, although the underlying mechanisms remain elusive (Mabasa et al., 2020).
α-KG
α-KG is an intermediate metabolite generated in the tricarboxylic acid cycle from isocitrate dehydrogenase (IDH)-mediated isocitrate conversion. α-KG can also be replenished from glutamine anaplerosis. In addition to its roles as a metabolic substrate in the cytosol and mitochondria, α-KG can also enter the nucleus and serves as a cofactor for TET-mediated DNA demethylation and JHDM family HDM activity to modify epigenetic marks (Teperino et al., 2010; Yun et al., 2012). During early brown adipogenesis, the cellular α-KG levels are profoundly increased and required for active DNA demethylation of the PRDM16 promoter (Yang et al., 2016a). A recent study also shows that IDH1-mediated α-KG modulates trimethylation of H3K4 in the promoters of genes associated with brown adipogenesis (Kang et al., 2020). Furthermore, knockdown of SIRT5 reduces intracellular α-KG concentration, leading to elevated suppressive H3K9me2 and H3K9me3 abundance at promoter regions of PPARγ and PRDM16 in adipocytes (Shuai et al., 2019). Finally, dietary α-KG supplement has been shown to promote beige adipogenesis and prevent obesity in middle-aged mice (Tian et al., 2020). These results provide strong evidence that α-KG can be an attractive therapeutic agent for obesity treatment by modulating epigenetic factor activity in brown and beige adipocytes.
Acetyl-CoA
Acetyl-CoA is a central metabolite positioned at the crossroads of carbohydrate, fat acid, and amino acid metabolism. Acetyl-CoA is also a substrate for protein acetylation including HATs (Pietrocola et al., 2015; Etchegaray and Mostoslavsky, 2016). In yeast, stem cells, and cancer cells, HATs can be regulated by the availability of acetyl-CoA, leading to global acetylation changes of histone protein. Acetyl-CoA does not modify HATs ‘non-specifically’, because different HATs have varying Kd for acetyl-CoA (Wellen et al., 2009; Cai et al., 2011; Lee et al., 2014; Moussaieff et al., 2015). Acetyl-CoA levels have been reported to be altered in the adipose tissue and liver (Dharuri et al., 2014; Perry et al., 2015; Carrer et al., 2017). However, it is challenging to determine whether the altered acetyl-CoA regulates histone acetylation and gene expression, because acetyl-CoA is modulated by multiple inputs, outputs, and inter-organ crosstalk (Yang et al., 2018).
NAD+
NAD+ serves as a cofactor for several nuclear proteins, notably class III HDAC sirtuins and poly(ADP-ribose) polymerase 1 (PARP1). Sirtuin deacetylases Sirt1, Sirt6, and Sirt7 are predominantly located in the nucleus, among which Sirt1 plays a major role in regulating mitochondrial function, mainly by modulating the acetylation of PGC-1α, the master regulator of mitochondrial biogenesis and function. Sirt1 may also modify histone acetylation such as H3K9ac and H4K16ac to regulate target gene expression (Canto et al., 2015; Verdin, 2015; Ryu et al., 2018). SIRT6 may deacetylate H3K9ac and SIRT6 overexpression increases male longevity with gene regulation in a similar way as mice following a caloric restriction diet (Kanfi et al., 2012). SIRT7 deacetylates GA binding protein transcription factor β (GABPβ), thereby enabling it to form the transcriptionally active GABPα/GABPβ heterotetramer to increase the expression of mitochondrial genes, especially the ones encoding mitochondrial ribosome proteins (Ryu et al., 2014). SIRT7-mediated deacetylation of H3K18ac may epigenetically facilitate the effects of SIRT7 on regulating glucose and lipid metabolism (Shin et al., 2013). It is conceivable that NAD+-regulated energy metabolism is the results of synergistic effects of sirtuin-targeted histone and non-histone acetylation.
Lifestyle modifications in the epigenetics of obesity
Epigenetic modifications are highly dynamic in response to environmental factors such as diet and exercise. Although most studies are correlational, several intervention studies have also been performed to dissect the impact of lifestyle modifications on the human epigenome in obesity.
Diet and epigenetics
Different dietary patterns, nutrients, and food components have been related to epigenetic processes that may contribute to the susceptibility of obesity (Ideraabdullah and Zeisel, 2018; Castellano-Castillo et al., 2020). In the pioneer studies of Dutch famine during the 1944–1945 winter at the end of World War II, individuals exposed to famine during gestation develop metabolic syndrome including obesity and hypercholesterolemia in adulthood (de Rooij et al., 2007). Similar studies of the Chinese Great Famine (1959–1961) show that early-life exposure to severe famine is associated with excessive risk of dyslipidemia (WaNg et al., 2017a). A genome-wide exploration of CpG methylation of whole-blood DNA identifies six CpGs that are associated with BMI and triglycerides. The serine/threonine protein kinase Pim-3, a gene involved in energy metabolism, contributes to 13.4% of the association between famine exposure and BMI (Tobi et al., 2018). As one-carbon metabolism depends on the dietary methyl donors, DNA methylation can be influenced by choline, methionine, betaine, and folate (Ducker and Rabinowitz, 2017). Maternal intake of these methyl-group donors in the periconception period is associated with DNA methylation in genes related to growth (insulin like growth factor 2, IGF2), metabolism (retinoid X receptor-α, RXRA), and appetite control (leptin) (Pauwels et al., 2017).
Interestingly, prenatal overnutrition and an obese maternal environment are also associated with DNA methylation changes in genes related to metabolic diseases in the offspring (Liu et al., 2014; Soubry et al., 2015). Both saturated fatty acids or polyunsaturated fatty acids overfeeding could increase the global degree of DNA methylation in adipose tissue of young, healthy adults in parallel with their body weight increase (Perfilyev et al., 2017). Therefore, dietary factors may alter the epigenome especially DNA methylation, although the causative roles in the pathogenesis of obesity deserve further investigation.
Dietary supplements such as resveratrol, nicotinamide riboside, and curcumin have been shown to improve energy metabolism partially through epigenetic mechanisms. Resveratrol has received great attentions because it is a component of plant-based foods especially red wine. Resveratrol may activate SIRT1 and modify histone protein (Fernandes et al., 2017; Lin et al., 2020). Although the beneficial effects of resveratrol on metabolism are convincing in cell culture and animal studies, human clinical trials have yielded mixed results (Bitterman and Chung, 2015). Nicotinamide riboside, as an NAD+ precursor, may activate multiple sirtuins. Consistently, nicotinamide riboside is effective in reducing obesity and glucose levels in animals. An 8-week randomized, double-blind, placebo-controlled clinical trial shows that nicotinamide riboside is tolerable and increases whole-blood NAD+ levels up to 142% (Conze et al., 2019). However, large clinical trials are required to determine whether nicotinamide riboside is effective in treating metabolic diseases in humans. Finally, intake of curcumin, a polyphenolic compound in turmeric, is correlated with reduced BMI and body weight in patients with metabolic syndrome (Akbari et al., 2019). Epigenetic mechanisms for the curcumin’s effects include regulation of DNA methylation, histone modifications, and miRNA expression (Tian et al., 2017; Hassan et al., 2019).
Physical activity and epigenetics
Physical activity and exercise may alter epigenetic signatures, especially DNA methylation as described in a recent systemic review (Barron-Cabrera et al., 2019). For example, individuals with physical activity of 26–30 min/day reportedly have a significantly higher level of global genomic DNA methylation compared to those with physical activity ≤10 min/day (Zhang et al., 2011). Excise increases muscle mitochondrial contents partially due to increased PGC-1α expression. Both acute and chronic exercises have been shown to reduce promoter PGC-1α methylation (Barres et al., 2012; Bajpeyi et al., 2017). In addition, chronic exercise intervention for 6 months has also been shown to alter DNA methylation in >7000 genes in adipose tissue, one-third with differential mRNA expression including HDAC4 (Ronn et al., 2013). Since PGC-1α and HDAC4 are established to play important roles in energy metabolism, altered DNA methylation of these genes may contribute to the beneficial metabolic effects of physical activity and exercise.
Therapeutic agents targeting epigenetics
The dynamics of the histone, DNA, or RNA modifications provide an opportunity to alter epigenetic factor activity for obesity treatment. Both DNMT and HDAC inhibitors are emerging treatments for cancer. Some of the inhibitors have been tested in animals for potential obesity and diabetes treatment.
Two inhibitors of DNMT, 5-azacytidine and 5-aza-2′-deoxycytidine (5-azadC), can incorporate into the DNA and trap DNMTs, resulting in DNA demethylation (WoNg et al., 2017). Inhibition of DNA methylation by 5-azacytidine at early stage of differentiation suppresses adipogenesis, while inhibition of DNA methylation at late stage of differentiation promotes lipogenesis and adipocyte phenotype (Yang et al., 2016b). In ob/ob mice, inhibiting DNA methylation by 5-azadC ameliorates obesity-induced inflammation and insulin resistance by promoting adipose tissue macrophage polarization (Wang et al., 2016).
Sodium butyrate, an HDAC pan-inhibitor, has been shown to alleviate HFD-induced obesity (Gao et al., 2009; den Besten et al., 2015; Hong et al., 2016). A subsequent study shows that class I but not class II HDAC inhibitors enhance whole-body energy expenditure through increased mitochondrial biogenesis in the skeletal muscle and adipose tissues (Galmozzi et al., 2013). Interestingly, however, the class IIa HDAC inhibitor Scriptaid is shown to increase energy expenditure without altering body weight due to increased food intake. Scriptaid enhances skeletal muscle insulin action and cardiac function in obese mice (Gaur et al., 2016). Administration of the HDAC3 inhibitor HD-75 may also improve insulin sensitivity via upregulating PPARγ acetylation in a mouse model of HFD-induced obesity (Jiang et al., 2014).
Targeting specific miRNAs that regulate energy metabolism using gain- and loss-of-function tools such as miRNA mimics, antagonists, and inhibitors may potentially be a strategy for obesity treatment. However, the major obstacle is side effects and toxicity. For example, miR-34a knockout mice are susceptible to DIO (Lavery et al., 2016). MRX34, a liposomal miR-34a mimic initially developed for treating hepatocellular carcinoma and hematological malignancies, might have beneficial effects on obesity. However, MRX34 caused severe immune-mediated adverse effects leading to early termination of the phase I clinical trial for cancer treatment (Beg et al., 2017). miR-122, the most abundant miRNA in the liver, promotes hepatic lipogenesis (Long et al., 2019). Miravirsen is an antisense oligonucleotide drug inhibiting miR-122 for hepatitis C treatment (Israelow et al., 2014). Miravirsen has also been shown to improve liver steatosis and reduce cholesterol levels (Esau et al., 2006).
Concluding remarks and perspectives
Rapid development in epigenomic technology and the increasing body of epigenomic data offer unprecedented opportunities to delineate how the interplay between genetic, environmental, and epigenetic components regulates energy metabolism in obesity and T2DM. Signatures of epigenetic markers including DNA methylation, histone modification, and ncRNAs are found to be associated with obesity. There is also substantial evidence demonstrating that epigenetic mechanisms play causal roles in the development of obesity and T2DM. Identification of metabolites as regulators of epigenetic factors such as HATs, HMTs, and sirtuins makes it possible to design small-molecule drugs to modulate epigenetic factor activity for obesity and T2DM treatment. The advancement of our understanding in epigenetic regulation of energy metabolism also represents several challenges. Many epigenetic regulators are important for development. It is less clear from studies using knockout mouse models whether the effects of the epigenetic factors on energy metabolism are developmental or physiological. Studying knockout in adult mice using inducible cre recombinase will avoid secondary effects of development and provide significant insights into the physiological roles of epigenetic factors in regulating energy metabolism. Additionally, the epigenetic factors such as HATs and HMTs often form large regulatory complexes. For drug development, it is important to understand whether the effects of histone modifiers on energy metabolism are caused by altered enzyme activity or disruption of the regulatory complexes. Furthermore, epigenetic factors regulate a broad spectrum of target genes, often in a tissue-specific manner. Understanding the mechanisms by which epigenetic factors regulate metabolic gene expression is essential to selectively target the epigenetic machinery for obesity treatment while avoiding adverse effects. Finally, studies need to elucidate how life styles such as diet and exercise alter epigenetic factor activity and how the altered epigenetic factors regulate energy metabolism at the molecular, cellular, organ, and whole-body levels. It is also key to determine which observations in animal models can be translated to humans. Despite these challenges, ultimately, in the postgenomic era, the integration of genomic and epigenomic landscapes will not only deepen our understandings of molecular mechanisms for energy metabolism, but may also lead to the identification of novel strategies for treating obesity and T2DM.
Supplementary material
Supplementary material (list of abbreviations) is available at Journal of Molecular Cell Biology online.
Funding
The study was supported by grants from the National Institutes of Health (NIH; R01 DK121146) to Q.Y., the National Natural Science Foundation of China (81970218 and 81770440 to X.L., 81970217 and 81700331 to W.G.), the Jiangsu Province Health Development Project with Science and Education (QNRC2016857 to W.G.), the Six One Project of Jiangsu Province (LGY2018100 to W.G.), and the Six Talent Peaks Project of Jiangsu Province (WSN-175 to W.G.).
Conflict of interest: none declared.
Supplementary Material
References
- Abdi A., Jalilian M., Sarbarzeh P.A., et al. (2020). Diabetes and COVID-19: a systematic review on the current evidences. Diabetes Res. Clin. Pract. 166, 108347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abe Y., Fujiwara Y., Takahashi H., et al. (2018). Histone demethylase JMJD1A coordinates acute and chronic adaptation to cold stress via thermogenic phospho-switch. Nat. Commun. 9, 1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abe Y., Rozqie R., Matsumura Y., et al. (2015). JMJD1A is a signal-sensing scaffold that regulates acute chromatin dynamics via SWI/SNF association for thermogenesis. Nat. Commun. 6, 7052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abu-Farha M., Al-Mulla F., Thanaraj T.A., et al. (2020). Impact of diabetes in patients diagnosed with COVID-19. Front. Immunol. 11, 576818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akbari M., Lankarani K.B., Tabrizi R., et al. (2019). The effects of curcumin on weight loss among patients with metabolic syndrome and related disorders: a systematic review and meta-analysis of randomized controlled trials. Front. Pharmacol. 10, 649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez-Dominguez J.R., Bai Z., Xu D., et al. (2015). De novo reconstruction of adipose tissue transcriptomes reveals long non-coding RNA regulators of brown adipocyte development. Cell Metab. 21, 764–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arcinas C., Tan W., Fang W., et al. (2019). Adipose circular RNAs exhibit dynamic regulation in obesity and functional role in adipogenesis. Nat. Metab. 1, 688–703. [DOI] [PubMed] [Google Scholar]
- Arner P., Sinha I., Thorell A., et al. (2015). The epigenetic signature of subcutaneous fat cells is linked to altered expression of genes implicated in lipid metabolism in obese women. Clin. Epigenet. 7, 93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aslibekyan S., Demerath E.W., Mendelson M., et al. (2015). Epigenome-wide study identifies novel methylation loci associated with body mass index and waist circumference. Obesity 23, 1493–1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagchi R.A., Ferguson B.S., Stratton M.S., et al. (2018). HDAC11 suppresses the thermogenic program of adipose tissue via BRD2. JCI Insight 3, e120159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai Z., Chai X.R., Yoon M.J., et al. (2017). Dynamic transcriptome changes during adipose tissue energy expenditure reveal critical roles for long noncoding RNA regulators. PLoS Biol. 15, e2002176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajpeyi S., Covington J.D., Taylor E.M., et al. (2017). Skeletal muscle PGC1α −1 nucleosome position and −260 nt DNA methylation determine exercise response and prevent ectopic lipid accumulation in men. Endocrinology 158, 2190–2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barres R., Yan J., Egan B., et al. (2012). Acute exercise remodels promoter methylation in human skeletal muscle. Cell Metab. 15, 405–411. [DOI] [PubMed] [Google Scholar]
- Barron-Cabrera E., Ramos-Lopez O., Gonzalez-Becerra K., et al. (2019). Epigenetic modifications as outcomes of exercise interventions related to specific metabolic alterations: a systematic review. Lifestyle Genom. 12, 25–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedford D.C., Kasper L.H., Fukuyama T., et al. (2010). Target gene context influences the transcriptional requirement for the KAT3 family of CBP and p300 histone acetyltransferases. Epigenetics 5, 9–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedford D.C., Kasper L.H., Wang R., et al. (2011). Disrupting the CH1 domain structure in the acetyltransferases CBP and p300 results in lean mice with increased metabolic control. Cell Metab. 14, 219–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beg M.S., Brenner A.J., Sachdev J., et al. (2017). Phase I study of MRX34, a liposomal miR-34a mimic, administered twice weekly in patients with advanced solid tumors. Invest. New Drugs 35, 180–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berndsen C.E., Denu J.M. (2008). Catalysis and substrate selection by histone/protein lysine acetyltransferases. Curr. Opin. Struct. Biol. 18, 682–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bian F., Ma X., Villivalam S.D., et al. (2018). TET2 facilitates PPARγ agonist-mediated gene regulation and insulin sensitization in adipocytes. Metabolism 89, 39–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bitterman J.L., Chung J.H. (2015). Metabolic effects of resveratrol: addressing the controversies. Cell. Mol. Life Sci. 72, 1473–1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boccaletto P., Machnicka M.A., Purta E., et al. (2018). MODOMICS: a database of RNA modification pathways. 2017 update. Nucleic Acids Res. 46, D303–D307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai L., Sutter B.M., Li B., et al. (2011). Acetyl-CoA induces cell growth and proliferation by promoting the acetylation of histones at growth genes. Mol. Cell 42, 426–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calo E., Wysocka J. (2013). Modification of enhancer chromatin: what, how, and why? Mol. Cell 49, 825–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canto C., Menzies K.J., Auwerx J. (2015). NAD+ metabolism and the control of energy homeostasis: a balancing act between mitochondria and the nucleus. Cell Metab. 22, 31–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y., Liu X., Zhao J., et al. (2021). AMPKα1 regulates Idh2 transcription through H2B O-GlcNAcylation during brown adipogenesis. Acta Biochim. Biophys. Sin. 53, 121–118. [DOI] [PubMed] [Google Scholar]
- Carrer A., Parris J.L., Trefely S., et al. (2017). Impact of a high-fat diet on tissue acyl-CoA and histone acetylation levels. J. Biol. Chem. 292, 3312–3322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellano-Castillo D., Ramos-Molina B., Cardona F., et al. (2020). Epigenetic regulation of white adipose tissue in the onset of obesity and metabolic diseases. Obes. Rev. 21, e13054. [DOI] [PubMed] [Google Scholar]
- Chatterjee T.K., Basford J.E., Knoll E., et al. (2014). HDAC9 knockout mice are protected from adipose tissue dysfunction and systemic metabolic disease during high-fat feeding. Diabetes 63, 176–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L.L. (2016). The biogenesis and emerging roles of circular RNAs. Nat. Rev. Mol. Cell Biol. 17, 205–211. [DOI] [PubMed] [Google Scholar]
- Chen Y., Siegel F., Kipschull S., et al. (2013). miR-155 regulates differentiation of brown and beige adipocytes via a bistable circuit. Nat. Commun. 4, 1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y., He C., Wang M., et al. (2019). Targeting epigenetic regulators for cancer therapy: mechanisms and advances in clinical trials. Signal Transduct. Target. Ther. 4, 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christian D.L., Wu D.Y., Martin J.R., et al. (2020). DNMT3A haploinsufficiency results in behavioral deficits and global epigenomic dysregulation shared across neurodevelopmental disorders. Cell Rep. 33, 108416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Church C., Moir L., McMurray F., et al. (2010). Overexpression of Fto leads to increased food intake and results in obesity. Nat. Genet. 42, 1086–1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cioffi M., Vallespinos-Serrano M., Trabulo S.M., et al. (2015). MiR-93 controls adiposity via inhibition of Sirt7 and Tbx3. Cell Rep. 12, 1594–1605. [DOI] [PubMed] [Google Scholar]
- Conze D., Brenner C., Kruger C.L. (2019). Safety and metabolism of long-term administration of NIAGEN (nicotinamide riboside chloride) in a randomized, double-blind, placebo-controlled clinical trial of healthy overweight adults. Sci. Rep. 9, 9772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covarrubias A.J., Perrone R., Grozio A., et al. (2021). NAD+ metabolism and its roles in cellular processes during ageing. Nat. Rev. Mol. Cell Biol. 22, 119–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creyghton M.P., Cheng A.W., Welstead G.G., et al. (2010). Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc. Natl Acad. Sci. USA 107, 21931–21936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dallner O.S., Marinis J.M., Lu Y.H., et al. (2019). Dysregulation of a long noncoding RNA reduces leptin leading to a leptin-responsive form of obesity. Nat. Med. 25, 507–516. [DOI] [PubMed] [Google Scholar]
- Damal Villivalam S., You D., Kim J., et al. (2020). TET1 is a beige adipocyte-selective epigenetic suppressor of thermogenesis. Nat. Commun. 11, 4313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Rooij S.R., Painter R.C., Holleman F., et al. (2007). The metabolic syndrome in adults prenatally exposed to the Dutch famine. Am. J. Clin. Nutr. 86, 1219–1224. [DOI] [PubMed] [Google Scholar]
- den Besten G., Bleeker A., Gerding A., et al. (2015). Short-chain fatty acids protect against high-fat diet-induced obesity via a PPARγ-dependent switch from lipogenesis to fat oxidation. Diabetes 64, 2398–2408. [DOI] [PubMed] [Google Scholar]
- Dent J.R., Martins V.F., Svensson K., et al. (2017). Muscle-specific knockout of general control of amino acid synthesis 5 (GCN5) does not enhance basal or endurance exercise-induced mitochondrial adaptation. Mol. Metab. 6, 1574–1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dharuri H., t Hoen P.A., van Klinken J.B., et al. (2014). Downregulation of the acetyl-CoA metabolic network in adipose tissue of obese diabetic individuals and recovery after weight loss. Diabetologia 57, 2384–2392. [DOI] [PubMed] [Google Scholar]
- Dina C., Meyre D., Gallina S., et al. (2007). Variation in FTO contributes to childhood obesity and severe adult obesity. Nat. Genet. 39, 724–726. [DOI] [PubMed] [Google Scholar]
- Ducker G.S., Rabinowitz J.D. (2017). One-carbon metabolism in health and disease. Cell Metab. 25, 27–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumortier O., Hinault C., Van Obberghen E. (2013). MicroRNAs and metabolism crosstalk in energy homeostasis. Cell Metab. 18, 312–324. [DOI] [PubMed] [Google Scholar]
- Duteil D., Metzger E., Willmann D., et al. (2014). LSD1 promotes oxidative metabolism of white adipose tissue. Nat. Commun. 5, 4093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duteil D., Tosic M., Lausecker F., et al. (2016). Lsd1 ablation triggers metabolic reprogramming of brown adipose tissue. Cell Rep. 17, 1008–1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duteil D., Tosic M., Willmann D., et al. (2017). Lsd1 prevents age-programed loss of beige adipocytes. Proc. Natl Acad. Sci. USA 114, 5265–5270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elshorbagy A.K., Jerneren F., Samocha-Bonet D., et al. (2016). Serum S-adenosylmethionine, but not methionine, increases in response to overfeeding in humans. Nutr. Diabetes 6, e192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emmett M.J., Lazar M.A. (2019). Integrative regulation of physiology by histone deacetylase 3. Nat. Rev. Mol. Cell Biol. 20, 102–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emmett M.J., Lim H.W., Jager J., et al. (2017). Histone deacetylase 3 prepares brown adipose tissue for acute thermogenic challenge. Nature 546, 544–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engin A. (2017). The definition and prevalence of obesity and metabolic syndrome. Adv. Exp. Med. Biol. 960, 1–17. [DOI] [PubMed] [Google Scholar]
- Esau C., Davis S., Murray S.F., et al. (2006). miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab. 3, 87–98. [DOI] [PubMed] [Google Scholar]
- Etchegaray J.P., Mostoslavsky R. (2016). Interplay between metabolism and epigenetics: a nuclear adaptation to environmental changes. Mol. Cell 62, 695–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatemi M., Hermann A., Gowher H., et al. (2002). Dnmt3a and Dnmt1 functionally cooperate during de novo methylation of DNA. Eur. J. Biochem. 269, 4981–4984. [DOI] [PubMed] [Google Scholar]
- Feliciano P., Zhou X., Astrovskaya I., et al. (2019). Exome sequencing of 457 autism families recruited online provides evidence for autism risk genes. NPJ Genom. Med. 4, 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng D., Liu T., Sun Z., et al. (2011). A circadian rhythm orchestrated by histone deacetylase 3 controls hepatic lipid metabolism. Science 331, 1315–1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J., Zhou Y., Campbell S.L., et al. (2010). Dnmt1 and Dnmt3a maintain DNA methylation and regulate synaptic function in adult forebrain neurons. Nat. Neurosci. 13, 423–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes G.F.S., Silva G.D.B., Pavan A.R., et al. (2017). Epigenetic regulatory mechanisms induced by resveratrol. Nutrients 9, 1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari A., Longo R., Fiorino E., et al. (2017). HDAC3 is a molecular brake of the metabolic switch supporting white adipose tissue browning. Nat. Commun. 8, 93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer J., Koch L., Emmerling C., et al. (2009). Inactivation of the Fto gene protects from obesity. Nature 458, 894–898. [DOI] [PubMed] [Google Scholar]
- Frayling T.M., Timpson N.J., Weedon M.N., et al. (2007). A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science 316, 889–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu T., Seok S., Choi S., et al. (2014). MicroRNA 34a inhibits beige and brown fat formation in obesity in part by suppressing adipocyte fibroblast growth factor 21 signaling and SIRT1 function. Mol. Cell. Biol. 34, 4130–4142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuster J.J., Zuriaga M.A., Zorita V., et al. (2020). TET2-loss-of-function-driven clonal hematopoiesis exacerbates experimental insulin resistance in aging and obesity. Cell Rep. 33, 108326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galmozzi A., Mitro N., Ferrari A., et al. (2013). Inhibition of class I histone deacetylases unveils a mitochondrial signature and enhances oxidative metabolism in skeletal muscle and adipose tissue. Diabetes 62, 732–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X., Shin Y.H., Li M., et al. (2010). The fat mass and obesity associated gene FTO functions in the brain to regulate postnatal growth in mice. PLoS One 5, e14005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Z., Yin J., Zhang J., et al. (2009). Butyrate improves insulin sensitivity and increases energy expenditure in mice. Diabetes 58, 1509–1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaur V., Connor T., Sanigorski A., et al. (2016). Disruption of the class IIa HDAC corepressor complex increases energy expenditure and lipid oxidation. Cell Rep. 16, 2802–2810. [DOI] [PubMed] [Google Scholar]
- Gerhart-Hines Z., Rodgers J.T., Bare O., et al. (2007). Metabolic control of muscle mitochondrial function and fatty acid oxidation through SIRT1/PGC-1α. EMBO J. 26, 1913–1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerken T., Girard C.A., Tung Y.C., et al. (2007). The obesity-associated FTO gene encodes a 2-oxoglutarate-dependent nucleic acid demethylase. Science 318, 1469–1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gharanei S., Shabir K., Brown J.E., et al. (2020). Regulatory microRNAs in brown, brite and white adipose tissue. Cells 9, 2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghisletti S., Barozzi I., Mietton F., et al. (2010). Identification and characterization of enhancers controlling the inflammatory gene expression program in macrophages. Immunity 32, 317–328. [DOI] [PubMed] [Google Scholar]
- Hammond C.M., Stromme C.B., Huang H., et al. (2017). Histone chaperone networks shaping chromatin function. Nat. Rev. Mol. Cell Biol. 18, 141–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han D., Huang M., Wang T., et al. (2019). Lysine methylation of transcription factors in cancer. Cell Death Dis. 10, 290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harms M.J., Ishibashi J., Wang W., et al. (2014). Prdm16 is required for the maintenance of brown adipocyte identity and function in adult mice. Cell Metab. 19, 593–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan F.U., Rehman M.S., Khan M.S., et al. (2019). Curcumin as an alternative epigenetic modulator: mechanism of action and potential effects. Front. Genet. 10, 514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochberg Z. (2018). An evolutionary perspective on the obesity epidemic. Trends Endocrinol. Metab. 29, 819–826. [DOI] [PubMed] [Google Scholar]
- Hong J., Jia Y., Pan S., et al. (2016). Butyrate alleviates high fat diet-induced obesity through activation of adiponectin-mediated pathway and stimulation of mitochondrial function in the skeletal muscle of mice. Oncotarget 7, 56071–56082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong S., Zhou W., Fang B., et al. (2017). Dissociation of muscle insulin sensitivity from exercise endurance in mice by HDAC3 depletion. Nat. Med. 23, 223–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horii T., Morita S., Hino S., et al. (2020). Successful generation of epigenetic disease model mice by targeted demethylation of the epigenome. Genome Biol. 21, 77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath S., Raj K. (2018). DNA methylation-based biomarkers and the epigenetic clock theory of ageing. Nat. Rev. Genet. 19, 371–384. [DOI] [PubMed] [Google Scholar]
- Houde A.A., Legare C., Biron S., et al. (2015). Leptin and adiponectin DNA methylation levels in adipose tissues and blood cells are associated with BMI, waist girth and LDL‒cholesterol levels in severely obese men and women. BMC Med. Genet. 16, 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu F., Wang M., Xiao T., et al. (2015). miR-30 promotes thermogenesis and the development of beige fat by targeting RIP140. Diabetes 64, 2056–2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W., Ding Y., Wang S., et al. (2020). The construction and analysis of the aberrant lncRNA–miRNA–mRNA network in adipose tissue from type 2 diabetes individuals with obesity. J. Diabetes Res. 2020, 3980742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang D., Camacho C.V., Setlem R., et al. (2020). Functional interplay between histone H2B ADP-ribosylation and phosphorylation controls adipogenesis. Mol. Cell 79, 934–949.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurtado E., Nunez-Alvarez Y., Munoz M., et al. (2021). HDAC11 is a novel regulator of fatty acid oxidative metabolism in skeletal muscle. FEBS J. 288, 902–919. [DOI] [PubMed] [Google Scholar]
- Hyun J., Jung Y. (2020). DNA methylation in nonalcoholic fatty liver disease. Int. J. Mol. Sci. 21, 8138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ideraabdullah F.Y., Zeisel S.H. (2018). Dietary modulation of the epigenome. Physiol. Rev. 98, 667–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Israelow B., Mullokandov G., Agudo J., et al. (2014). Hepatitis C virus genetics affects miR-122 requirements and response to miR-122 inhibitors. Nat. Commun. 5, 5408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeltsch A., Jurkowska R.Z. (2014). New concepts in DNA methylation. Trends Biochem. Sci. 39, 310–318. [DOI] [PubMed] [Google Scholar]
- Ji C., Guo X. (2019). The clinical potential of circulating microRNAs in obesity. Nat. Rev. Endocrinol. 15, 731–743. [DOI] [PubMed] [Google Scholar]
- Jia G., Fu Y., Zhao X., et al. (2011). N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol. 7, 885–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X., Ye X., Guo W., et al. (2014). Inhibition of HDAC3 promotes ligand-independent PPARγ activation by protein acetylation. J. Mol. Endocrinol. 53, 191–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Q., Wang C., Kuang X., et al. (2014). Gcn5 and PCAF regulate PPARγ and Prdm16 expression to facilitate brown adipogenesis. Mol. Cell. Biol. 34, 3746–3753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabra D.G., Pfuhlmann K., Garcia-Caceres C., et al. (2016). Hypothalamic leptin action is mediated by histone deacetylase 5. Nat. Commun. 7, 10782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaelin W.G., Jr, and , McKnight S.L. (2013). Influence of metabolism on epigenetics and disease. Cell 153, 56–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajimura S., Spiegelman B.M., Seale P. (2015). Brown and beige fat: physiological roles beyond heat generation. Cell Metab. 22, 546–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanfi Y., Naiman S., Amir G., et al. (2012). The sirtuin SIRT6 regulates lifespan in male mice. Nature 483, 218–221. [DOI] [PubMed] [Google Scholar]
- Kang H.S., Lee J.H., Oh K.J., et al. (2020). IDH1-dependent α-KG regulates brown fat differentiation and function by modulating histone methylation. Metabolism 105, 154173. [DOI] [PubMed] [Google Scholar]
- Karbiener M., Pisani D.F., Frontini A., et al. (2014). MicroRNA-26 family is required for human adipogenesis and drives characteristics of brown adipocytes. Stem Cells 32, 1578–1590. [DOI] [PubMed] [Google Scholar]
- Kawabe Y., Mori J., Morimoto H., et al. (2019). ACE2 exerts anti-obesity effect via stimulating brown adipose tissue and induction of browning in white adipose tissue. Am. J. Physiol. Endocrinol. Metab. 317, E1140–E1149. [DOI] [PubMed] [Google Scholar]
- Kim D.H., Kim J., Kwon J.S., et al. (2016). Critical roles of the histone methyltransferase MLL4/KMT2D in murine hepatic steatosis directed by ABL1 and PPARγ2. Cell Rep. 17, 1671–1682. [DOI] [PubMed] [Google Scholar]
- Kim D.H., Rhee J.C., Yeo S., et al. (2015). Crucial roles of mixed-lineage leukemia 3 and 4 as epigenetic switches of the hepatic circadian clock controlling bile acid homeostasis in mice. Hepatology 61, 1012–1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim G.D., Ni J., Kelesoglu N., et al. (2002). Co-operation and communication between the human maintenance and de novo DNA (cytosine-5) methyltransferases. EMBO J. 21, 4183–4195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H.J., Cho H., Alexander R., et al. (2014). MicroRNAs are required for the feature maintenance and differentiation of brown adipocytes. Diabetes 63, 4045–4056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H.J., Kim D.Y., Cheon H.G. (2019). Clomiphene promotes browning of white adipocytes via casein kinase-2 inhibition. Eur. J. Pharmacol. 861, 172596. [DOI] [PubMed] [Google Scholar]
- Kissig M., Ishibashi J., Harms M.J., et al. (2017). PRDM16 represses the type I interferon response in adipocytes to promote mitochondrial and thermogenic programing. EMBO J. 36, 1528–1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloting N., Schleinitz D., Ruschke K., et al. (2008). Inverse relationship between obesity and FTO gene expression in visceral adipose tissue in humans. Diabetologia 51, 641–647. [DOI] [PubMed] [Google Scholar]
- Klymenko O., Brecklinghaus T., Dille M., et al. (2020). Histone deacetylase 5 regulates interleukin 6 secretion and insulin action in skeletal muscle. Mol. Metab. 42, 101062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi M., Ohsugi M., Sasako T., et al. (2018). The RNA methyltransferase complex of WTAP, METTL3, and METTL14 regulates mitotic clonal expansion in adipogenesis. Mol. Cell. Biol. 38, e00116–-18.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraus D., Yang Q., Kong D., et al. (2014). Nicotinamide N-methyltransferase knockdown protects against diet-induced obesity. Nature 508, 258–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan S., Horowitz S., Trievel R.C. (2011). Structure and function of histone H3 lysine 9 methyltransferases and demethylases. Chembiochem 12, 254–263. [DOI] [PubMed] [Google Scholar]
- Kuhnen P., Handke D., Waterland R.A., et al. (2016). Interindividual variation in DNA methylation at a putative POMC metastable epiallele is associated with obesity. Cell Metab. 24, 502–509. [DOI] [PubMed] [Google Scholar]
- Kunej T., Jevsinek Skok D., Zorc M., et al. (2013). Obesity gene atlas in mammals. J. Genomics 1, 45–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kung J.T., Colognori D., Lee J.T. (2013). Long noncoding RNAs: past, present, and future. Genetics 193, 651–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landrier J.F., Derghal A., Mounien L. (2019). MicroRNAs in obesity and related metabolic disorders. Cells 8, 859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavery C.A., Kurowska-Stolarska M., Holmes W.M., et al. (2016). miR-34a–/– mice are susceptible to diet-induced obesity. Obesity 24, 1741–1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.V., Carrer A., Shah S., et al. (2014). Akt-dependent metabolic reprogramming regulates tumor cell histone acetylation. Cell Metab. 20, 306–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lennon H., Sperrin M., Badrick E., et al. (2016). The obesity paradox in cancer: a review. Curr. Oncol. Rep. 18, 56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C., Fan Y., Li G., et al. (2018). DNA methylation reprogramming of functional elements during mammalian embryonic development. Cell Discov. 4, 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F., Jing J., Movahed M., et al. (2020). Epigenetic interaction between UTX and DNMT1 regulates diet-induced myogenic remodeling in brown fat. bioRxiv, 10.1101/2020.08.05.238923 [DOI] [PMC free article] [PubMed]
- Li F., Wu R., Cui X., et al. (2016). Histone deacetylase 1 (HDAC1) negatively regulates thermogenic program in brown adipocytes via coordinated regulation of histone H3 lysine 27 (H3K27) deacetylation and methylation. J. Biol. Chem. 291, 4523–4536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G., Ning C., Ma Y., et al. (2017). miR-26b promotes 3T3-L1 adipocyte differentiation through targeting PTEN. DNA Cell Biol. 36, 672–681. [DOI] [PubMed] [Google Scholar]
- Li W., Sun Z. (2019). Mechanism of action for HDAC inhibitors-insights from omics approaches. Int. J. Mol. Sci. 20, 1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieber A.D., Beier U.H., Xiao H., et al. (2019). Loss of HDAC6 alters gut microbiota and worsens obesity. FASEB J. 33, 1098–1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C., Li H., Liu J., et al. (2020). Arginine hypomethylation-mediated proteasomal degradation of histone H4-an early biomarker of cellular senescence. Cell Death Differ. 27, 2697–2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling C., Ronn T. (2019). Epigenetics in human obesity and type 2 diabetes. Cell Metab. 29, 1028–1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M., Liu F. (2014). Regulation of adiponectin multimerization, signaling and function. Best Pract. Res. Clin. Endocrinol. Metab. 28, 25–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Chen Q., Tsai H.J., et al. (2014). Maternal preconception body mass index and offspring cord blood DNA methylation: exploration of early life origins of disease. Environ. Mol. Mutagen. 55, 223–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Liu H., Li Y., et al. (2020). Circular RNA SAMD4A controls adipogenesis in obesity through the miR-138-5p/EZH2 axis. Theranostics 10, 4705–4719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo K.A., Huang S., Walet A.C.E., et al. (2018). Adipocyte long-noncoding RNA transcriptome analysis of obese mice identified Lnc-leptin, which regulates leptin. Diabetes 67, 1045–1056. [DOI] [PubMed] [Google Scholar]
- Locke A.E., Kahali B., Berndt S.I., et al. (2015). Genetic studies of body mass index yield new insights for obesity biology. Nature 518, 197–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh M., Zhou L., Ng H.K., et al. (2019). Epigenetic disturbances in obesity and diabetes: epidemiological and functional insights. Mol. Metab. 27S, S33–S41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long J.K., Dai W., Zheng Y.W., et al. (2019). miR-122 promotes hepatic lipogenesis via inhibiting the LKB1/AMPK pathway by targeting Sirt1 in non-alcoholic fatty liver disease. Mol. Med. 25, 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H., Lei X., Zhang Q. (2019). Liver-specific knockout of histone methyltransferase G9a impairs liver maturation and dysregulates inflammatory, cytoprotective, and drug-processing genes. Xenobiotica 49, 740–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo X., Ryu K.W., Kim D.S., et al. (2017). PARP-1 controls the adipogenic transcriptional program by PARylating C/EBPβ and modulating its transcriptional activity. Mol. Cell 65, 260–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mabasa L., Samodien E., Sangweni N.F., et al. (2020). In utero one-carbon metabolism interplay and metabolic syndrome in cardiovascular disease risk reduction. Mol. Nutr. Food Res. 64, e1900377. [DOI] [PubMed] [Google Scholar]
- Malloy V.L., Krajcik R.A., Bailey S.J., et al. (2006). Methionine restriction decreases visceral fat mass and preserves insulin action in aging male Fischer 344 rats independent of energy restriction. Aging Cell 5, 305–314. [DOI] [PubMed] [Google Scholar]
- Marchese F.P., Raimondi I., Huarte M. (2017). The multidimensional mechanisms of long noncoding RNA function. Genome Biol. 18, 206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins V.F., Dent J.R., Svensson K., et al. (2019). Germline or inducible knockout of p300 or CBP in skeletal muscle does not alter insulin sensitivity. Am. J. Physiol. Endocrinol. Metab. 316, E1024–E1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMurray F., Church C.D., Larder R., et al. (2013). Adult onset global loss of the fto gene alters body composition and metabolism in the mouse. PLoS Genet. 9, e1003166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mentch S.J., Mehrmohamadi M., Huang L., et al. (2015). Histone methylation dynamics and gene regulation occur through the sensing of one-carbon metabolism. Cell Metab. 22, 861–873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittenbuhler M.J., Saedler K., Nolte H., et al. (2020). Hepatic FTO is dispensable for the regulation of metabolism but counteracts HCC development in vivo. Mol. Metab. 42, 101085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery R.L., Davis C.A., Potthoff M.J., et al. (2007). Histone deacetylases 1 and 2 redundantly regulate cardiac morphogenesis, growth, and contractility. Genes Dev. 21, 1790–1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon J.E., Lee S.J., Ko C.W. (2018). A de novo KMT2D mutation in a girl with Kabuki syndrome associated with endocrine symptoms: a case report. BMC Med. Genet. 19, 102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moresi V., Carrer M., Grueter C.E., et al. (2012). Histone deacetylases 1 and 2 regulate autophagy flux and skeletal muscle homeostasis in mice. Proc. Natl Acad. Sci. USA 109, 1649–1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moussaieff A., Rouleau M., Kitsberg D., et al. (2015). Glycolysis-mediated changes in acetyl-CoA and histone acetylation control the early differentiation of embryonic stem cells. Cell Metab. 21, 392–402. [DOI] [PubMed] [Google Scholar]
- Mutlu B., Puigserver P. (2021). GCN5 acetyltransferase in cellular energetic and metabolic processes. Biochim. Biophys. Acta Gene Regul. Mech. 1864, 194626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namwanje M., Liu L., Chan M., et al. (2019). The depot-specific and essential roles of CBP/p300 in regulating adipose plasticity. J. Endocrinol. 240, 257–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng R., Hussain N.A., Zhang Q., et al. (2017). miRNA-32 drives brown fat thermogenesis and trans-activates subcutaneous white fat browning in mice. Cell Rep. 19, 1229–1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng S.B., Bigham A.W., Buckingham K.J., et al. (2010). Exome sequencing identifies MLL2 mutations as a cause of Kabuki syndrome. Nat. Genet. 42, 790–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie L., Shuai L., Zhu M., et al. (2017). The landscape of histone modifications in a high-fat diet-induced obese (DIO) mouse model. Mol. Cell. Proteomics 16, 1324–1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Rourke R.W., Lumeng C.N. (2021). Pathways to severe COVID-19 for people with obesity. Obesity 29, 645–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh A., Okazaki R., Sam F., et al. (2019). Heart failure with preserved ejection fraction and adipose tissue: a story of two tales. Front. Cardiovasc. Med. 6, 110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno H., Shinoda K., Ohyama K., et al. (2013). EHMT1 controls brown adipose cell fate and thermogenesis through the PRDM16 complex. Nature 504, 163–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okano M., Bell D.W., Haber D.A., et al. (1999). DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell 99, 247–257. [DOI] [PubMed] [Google Scholar]
- Oliverio M., Schmidt E., Mauer J., et al. (2016). Dicer1‒miR-328‒Bace1 signalling controls brown adipose tissue differentiation and function. Nat. Cell Biol. 18, 328–336. [DOI] [PubMed] [Google Scholar]
- Pagiatakis C., Musolino E., Gornati R., et al. (2021). Epigenetics of aging and disease: a brief overview. Aging Clin. Exp. Res. 33, 737–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pahlavani M., Wijayatunga N.N., Kalupahana N.S., et al. (2018). Transcriptomic and microRNA analyses of gene networks regulated by eicosapentaenoic acid in brown adipose tissue of diet-induced obese mice. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 1863, 1523–1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan D., Huang L., Zhu L.J., et al. (2015). Jmjd3-mediated H3K27me3 dynamics orchestrate brown fat development and regulate white fat plasticity. Dev. Cell 35, 568–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Y., Guan H., Zhou S., et al. (2020). Initial CT findings and temporal changes in patients with the novel coronavirus pneumonia (2019-nCoV): a study of 63 patients in Wuhan, China. Eur. Radiol. 30, 3306–3309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulo E., Wu D., Hecker P., et al. (2018). Adipocyte HDAC4 activation leads to beige adipocyte expansion and reduced adiposity. J. Endocrinol. 239, 153–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauwels S., Ghosh M., Duca R.C., et al. (2017). Maternal intake of methyl-group donors affects DNA methylation of metabolic genes in infants. Clin. Epigenet. 9, 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedrotti S., Caccia R., Neguembor M.V., et al. (2019). The Suv420h histone methyltransferases regulate PPAR-γ and energy expenditure in response to environmental stimuli. Sci. Adv. 5, eaav1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng S., Xiao W., Ju D., et al. (2019). Identification of entacapone as a chemical inhibitor of FTO mediating metabolic regulation through FOXO1. Sci. Transl. Med. 11, eaau7116. [DOI] [PubMed] [Google Scholar]
- Perfilyev A., Dahlman I., Gillberg L., et al. (2017). Impact of polyunsaturated and saturated fat overfeeding on the DNA-methylation pattern in human adipose tissue: a randomized controlled trial. Am. J. Clin. Nutr. 105, 991–1000. [DOI] [PubMed] [Google Scholar]
- Perry R.J., Camporez J.G., Kursawe R., et al. (2015). Hepatic acetyl CoA links adipose tissue inflammation to hepatic insulin resistance and type 2 diabetes. Cell 160, 745–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietrocola F., Galluzzi L., Bravo-San Pedro J.M., et al. (2015). Acetyl coenzyme A: a central metabolite and second messenger. Cell Metab. 21, 805–821. [DOI] [PubMed] [Google Scholar]
- Qian H., Chen Y., Nian Z., et al. (2017). HDAC6-mediated acetylation of lipid droplet-binding protein CIDEC regulates fat-induced lipid storage. J. Clin. Invest. 127, 1353–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu X., Li J., Lv S., et al. (2018). HDAC5 integrates ER stress and fasting signals to regulate hepatic fatty acid oxidation. J. Lipid Res. 59, 330–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribel-Madsen R., Fraga M.F., Jacobsen S., et al. (2012). Genome-wide analysis of DNA methylation differences in muscle and fat from monozygotic twins discordant for type 2 diabetes. PLoS One 7, e51302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roh H.C., Tsai L.T.Y., Shao M., et al. (2018). Warming induces significant reprogramming of beige, but not brown, adipocyte cellular identity. Cell Metab. 27, 1121–1137.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohde K., Keller M., la Cour Poulsen L., et al. (2019). Genetics and epigenetics in obesity. Metabolism 92, 37–50. [DOI] [PubMed] [Google Scholar]
- Ronn T., Volkov P., Davegardh C., et al. (2013). A six months exercise intervention influences the genome-wide DNA methylation pattern in human adipose tissue. PLoS Genet. 9, e1003572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rui L. (2017). Brown and beige adipose tissues in health and disease. Compr. Physiol. 7, 1281–1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu D., Jo Y.S., Lo Sasso G., et al. (2014). A SIRT7-dependent acetylation switch of GABPβ1 controls mitochondrial function. Cell Metab. 20, 856–869. [DOI] [PubMed] [Google Scholar]
- Ryu K.W., Nandu T., Kim J., et al. (2018). Metabolic regulation of transcription through compartmentalized NAD+ biosynthesis. Science 360, eaan5780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmena L., Poliseno L., Tay Y., et al. (2011). A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell 146, 353–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambeat A., Gulyaeva O., Dempersmier J., et al. (2016). LSD1 interacts with Zfp516 to promote UCP1 transcription and brown fat program. Cell Rep. 15, 2536–2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samblas M., Milagro F.I., Martinez A. (2019). DNA methylation markers in obesity, metabolic syndrome, and weight loss. Epigenetics 14, 421–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders S.J., He X., Willsey A.J., et al. (2015). Insights into autism spectrum disorder genomic architecture and biology from 71 risk loci. Neuron 87, 1215–1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satterstrom F.K., Walters R.K., Singh T., et al. (2019). Autism spectrum disorder and attention deficit hyperactivity disorder have a similar burden of rare protein-truncating variants. Nat. Neurosci. 22, 1961–1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt E., Dhaouadi I., Gaziano I., et al. (2018). LincRNA H19 protects from dietary obesity by constraining expression of monoallelic genes in brown fat. Nat. Commun. 9, 3622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott L.J., Mohlke K.L., Bonnycastle L.L., et al. (2007). A genome-wide association study of type 2 diabetes in Finns detects multiple susceptibility variants. Science 316, 1341–1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scuteri A., Sanna S., Chen W.M., et al. (2007). Genome-wide association scan shows genetic variants in the FTO gene are associated with obesity-related traits. PLoS Genet. 3, e115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seok S., Kim Y.C., Byun S., et al. (2018). Fasting-induced JMJD3 histone demethylase epigenetically activates mitochondrial fatty acid β-oxidation. J. Clin. Invest. 128, 3144–3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapses S.A., Pop L.C., Wang Y. (2017). Obesity is a concern for bone health with aging. Nutr. Res. 39, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y., Lan F., Matson C., et al. (2004). Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell 119, 941–953. [DOI] [PubMed] [Google Scholar]
- Shin J., He M., Liu Y., et al. (2013). SIRT7 represses Myc activity to suppress ER stress and prevent fatty liver disease. Cell Rep. 5, 654–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinoda K., Ohyama K., Hasegawa Y., et al. (2015). Phosphoproteomics identifies CK2 as a negative regulator of beige adipocyte thermogenesis and energy expenditure. Cell Metab. 22, 997–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuai L., Zhang L.N., Li B.H., et al. (2019). SIRT5 regulates brown adipocyte differentiation and browning of subcutaneous white adipose tissue. Diabetes 68, 1449–1461. [DOI] [PubMed] [Google Scholar]
- Sodi V.L., Bacigalupa Z.A., Ferrer C.M., et al. (2018). Nutrient sensor O-GlcNAc transferase controls cancer lipid metabolism via SREBP-1 regulation. Oncogene 37, 924–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soubry A., Murphy S.K., Wang F., et al. (2015). Newborns of obese parents have altered DNA methylation patterns at imprinted genes. Int. J. Obes. 39, 650–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stirzaker C., Taberlay P.C., Statham A.L., et al. (2014). Mining cancer methylomes: prospects and challenges. Trends Genet. 30, 75–84. [DOI] [PubMed] [Google Scholar]
- Stone K.P., Wanders D., Orgeron M., et al. (2014). Mechanisms of increased in vivo insulin sensitivity by dietary methionine restriction in mice. Diabetes 63, 3721–3733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L., Marin de Evsikova C., Bian K., et al. (2018). Programming and regulation of metabolic homeostasis by HDAC11. EBioMedicine 33, 157–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L., Trajkovski M. (2014). MiR-27 orchestrates the transcriptional regulation of brown adipogenesis. Metabolism 63, 272–282. [DOI] [PubMed] [Google Scholar]
- Sun Z., Miller R.A., Patel R.T., et al. (2012). Hepatic Hdac3 promotes gluconeogenesis by repressing lipid synthesis and sequestration. Nat. Med. 18, 934–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Y.T., Lin J.F., Li T., et al. (2020). LncRNA-mediated posttranslational modifications and reprogramming of energy metabolism in cancer. Cancer Commun. 41, 109–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tateishi K., Okada Y., Kallin E.M., et al. (2009). Role of Jhdm2a in regulating metabolic gene expression and obesity resistance. Nature 458, 757–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teperino R., Schoonjans K., Auwerx J. (2010). Histone methyl transferases and demethylases; can they link metabolism and transcription? Cell Metab. 12, 321–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tews D., Fischer-Posovszky P., Fromme T., et al. (2013). FTO deficiency induces UCP-1 expression and mitochondrial uncoupling in adipocytes. Endocrinology 154, 3141–3151. [DOI] [PubMed] [Google Scholar]
- Tews D., Fischer-Posovszky P., Wabitsch M. (2011). Regulation of FTO and FTM expression during human preadipocyte differentiation. Horm Metab. Res. 43, 17–21. [DOI] [PubMed] [Google Scholar]
- Thomou T., Mori M.A., Dreyfuss J.M., et al. (2017). Adipose-derived circulating miRNAs regulate gene expression in other tissues. Nature 542, 450–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorleifsson G., Walters G.B., Gudbjartsson D.F., et al. (2009). Genome-wide association yields new sequence variants at seven loci that associate with measures of obesity. Nat. Genet. 41, 18–24. [DOI] [PubMed] [Google Scholar]
- Tian L., Song Z., Shao W., et al. (2017). Curcumin represses mouse 3T3-L1 cell adipogenic differentiation via inhibiting miR-17-5p and stimulating the Wnt signalling pathway effector Tcf7l2. Cell Death Dis. 8, e2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Q., Zhao J., Yang Q., et al. (2020). Dietary α-ketoglutarate promotes beige adipogenesis and prevents obesity in middle-aged mice. Aging Cell 19, e13059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobi E.W., Slieker R.C., Luijk R., et al. (2018). DNA methylation as a mediator of the association between prenatal adversity and risk factors for metabolic disease in adulthood. Sci. Adv. 4, eaao4364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toperoff G., Aran D., Kark J.D., et al. (2012). Genome-wide survey reveals predisposing diabetes type 2-related DNA methylation variations in human peripheral blood. Hum. Mol. Genet. 21, 371–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trajkovski M., Ahmed K., Esau C.C., et al. (2012). MyomiR-133 regulates brown fat differentiation through Prdm16. Nat. Cell Biol. 14, 1330–1335. [DOI] [PubMed] [Google Scholar]
- Tzika E., Dreker T., Imhof A. (2018). Epigenetics and metabolism in health and disease. Front. Genet. 9, 361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dijk S.J., Tellam R.L., Morrison J.L., et al. (2015). Recent developments on the role of epigenetics in obesity and metabolic disease. Clin. Epigenet. 7, 66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdin E. (2015). NAD+ in aging, metabolism, and neurodegeneration. Science 350, 1208–1213. [DOI] [PubMed] [Google Scholar]
- Wanders D., Forney L.A., Stone K.P., et al. (2017). FGF21 mediates the thermogenic and insulin-sensitizing effects of dietary methionine restriction but not its effects on hepatic lipid metabolism. Diabetes 66, 858–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Liu C.D., Li H.F., et al. (2020a). LSD1 mediates microbial metabolite butyrate-induced thermogenesis in brown and white adipose tissue. Metabolism 102, 154011. [DOI] [PubMed] [Google Scholar]
- Wang L., Xu S., Lee J.E., et al. (2013). Histone H3K9 methyltransferase G9a represses PPARγ expression and adipogenesis. EMBO J. 32, 45–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang N., Wang X., Li Q., et al. (2017a). The famine exposure in early life and metabolic syndrome in adulthood. Clin. Nutr. 36, 253–259. [DOI] [PubMed] [Google Scholar]
- Wang X., Cao Q., Yu L., et al. (2016). Epigenetic regulation of macrophage polarization and inflammation by DNA methylation in obesity. JCI Insight 1, e87748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Huang N., Yang M., et al. (2017b). FTO is required for myogenesis by positively regulating mTOR‒PGC-1α pathway-mediated mitochondria biogenesis. Cell Death Dis. 8, e2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y.J., Yang B., Lai Q., et al. (2020b). Reprogramming of m6A epitranscriptome is crucial for shaping of transcriptome and proteome in response to hypoxia. RNA Biol. 18, 131–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterland R.A. (2014). Epigenetic mechanisms affecting regulation of energy balance: many questions, few answers. Annu. Rev. Nutr. 34, 337–355. [DOI] [PubMed] [Google Scholar]
- Wei W., Ji X., Guo X., et al. (2017). Regulatory role of N6-methyladenosine (m6A) methylation in RNA processing and human diseases. J. Cell. Biochem. 118, 2534–2543. [DOI] [PubMed] [Google Scholar]
- Wellen K.E., Hatzivassiliou G., Sachdeva U.M., et al. (2009). ATP-citrate lyase links cellular metabolism to histone acetylation. Science 324, 1076–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong C.C., Qian Y., Yu J. (2017). Interplay between epigenetics and metabolism in oncogenesis: mechanisms and therapeutic approaches. Oncogene 36, 3359–3374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu W., Feng J., Jiang D., et al. (2017). AMPK regulates lipid accumulation in skeletal muscle cells through FTO-dependent demethylation of N6-methyladenosine. Sci. Rep. 7, 41606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X., Zhang Y. (2017). TET-mediated active DNA demethylation: mechanism, function and beyond. Nat. Rev. Genet. 18, 517–534. [DOI] [PubMed] [Google Scholar]
- Wu Y., Zuo J., Zhang Y., et al. (2013). Identification of miR-106b-93 as a negative regulator of brown adipocyte differentiation. Biochem. Biophys. Res. Commun. 438, 575–580. [DOI] [PubMed] [Google Scholar]
- Xiong Y., Yue F., Jia Z., et al. (2018). A novel brown adipocyte-enriched long non-coding RNA that is required for brown adipocyte differentiation and sufficient to drive thermogenic gene program in white adipocytes. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 1863, 409–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamauchi T., Oike Y., Kamon J., et al. (2002). Increased insulin sensitivity despite lipodystrophy in Crebbp heterozygous mice. Nat. Genet. 30, 221–226. [DOI] [PubMed] [Google Scholar]
- Yang J., Bakshi A., Zhu Z., et al. (2015). Genome-wide genetic homogeneity between sexes and populations for human height and body mass index. Hum. Mol. Genet. 24, 7445–7449. [DOI] [PubMed] [Google Scholar]
- Yang Q., Liang X., Sun X., et al. (2016a). AMPK/α-Ketoglutarate axis dynamically mediates DNA demethylation in the Prdm16 promoter and brown adipogenesis. Cell Metab. 24, 542–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q., Vijayakumar A., Kahn B.B. (2018). Metabolites as regulators of insulin sensitivity and metabolism. Nat. Rev. Mol. Cell Biol. 19, 654–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X., Wu R., Shan W., et al. (2016b). DNA methylation biphasically regulates 3T3-L1 preadipocyte differentiation. Mol. Endocrinol. 30, 677–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yengo L., Sidorenko J., Kemper K.E., et al. (2018). Meta-analysis of genome-wide association studies for height and body mass index in approximately 700000 individuals of European ancestry. Hum. Mol. Genet. 27, 3641–3649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yiew N.K.H., Greenway C., Zarzour A., et al. (2019). Enhancer of zeste homolog 2 (EZH2) regulates adipocyte lipid metabolism independent of adipogenic differentiation: role of apolipoprotein E. J. Biol. Chem. 294, 8577–8591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You D., Nilsson E., Tenen D.E., et al. (2017). Dnmt3a is an epigenetic mediator of adipose insulin resistance. eLife 6, e30766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young R.L., Lumsden A.L., Martin A.M., et al. (2018). Augmented capacity for peripheral serotonin release in human obesity. Int. J. Obes. 42, 1880–1889. [DOI] [PubMed] [Google Scholar]
- Yu J., Lv Y., Wang F., et al. (2019). MiR-27b-3p inhibition enhances browning of epididymal fat in high-fat diet induced obese mice. Front. Endocrinol. 10, 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun J., Song S.H., Park J., et al. (2012). Gene silencing of EREG mediated by DNA methylation and histone modification in human gastric cancers. Lab. Invest. 92, 1033–1044. [DOI] [PubMed] [Google Scholar]
- Zeng X., Jedrychowski M.P., Chen Y., et al. (2016). Lysine-specific demethylase 1 promotes brown adipose tissue thermogenesis via repressing glucocorticoid activation. Genes Dev. 30, 1822–1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zha L., Li F., Wu R., et al. (2015). The histone demethylase UTX promotes brown adipocyte thermogenic program via coordinated regulation of H3K27 demethylation and acetylation. J. Biol. Chem. 290, 25151–25163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D., Tang Z., Huang H., et al. (2019a). Metabolic regulation of gene expression by histone lactylation. Nature 574, 575–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F.F., Cardarelli R., Carroll J., et al. (2011). Significant differences in global genomic DNA methylation by gender and race/ethnicity in peripheral blood. Epigenetics 6, 623–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Guan M., Townsend K.L., et al. (2015). MicroRNA-455 regulates brown adipogenesis via a novel HIF1an‒AMPK‒PGC1α signaling network. EMBO Rep. 16, 1378–1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Zhu L., Bai M., et al. (2019b). Exosomal circRNA derived from gastric tumor promotes white adipose browning by targeting the miR-133/PRDM16 pathway. Int. J. Cancer 144, 2501–2515. [DOI] [PubMed] [Google Scholar]
- Zhang T., Zhang Z., Xia T., et al. (2019c). circNrxn2 promoted WAT browning via sponging miR-103 to relieve its inhibition of FGF10 in HFD mice. Mol. Ther. Nucleic Acids 17, 551–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Yang D., Yuan Y., et al. (2020). Muscular G9a regulates muscle–liver–fat axis by musclin under overnutrition in female mice. Diabetes 69, 2642–2654. [DOI] [PubMed] [Google Scholar]
- Zhao Q., Zhang Z., Rong W., et al. (2020). KMT5c modulates adipocyte thermogenesis by regulating Trp53 expression. Proc. Natl Acad. Sci. USA 117, 22413–22422. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.