Abstract
Biomolecular phase separation in the formation of membraneless organelles and biomolecular condensates has recently gained tremendous attention due to the importance of these assemblies in physiology, disease, and engineering applications. Understanding and directing biomolecular phase separation requires a multi-scale view of the biophysical properties of these phases. Yet, many classic tools to characterize biomolecular properties do not apply in these condensed phases. Here, we discuss insights obtained from spectroscopic methods, in particular NMR spectroscopy, in understanding the molecular and atomic interactions that underlie the formation of protein-rich condensates. We also review approaches closely coupling NMR data with computational methods especially coarse-grained and all-atom molecular simulations, which provide insight into molecular features of phase separation. Finally, we point to future developments, particularly visualizing biophysical properties of condensates in cells.
Introduction
Biomolecular phase separation has generated tremendous interest recently, having been found or attributed to play a role in an ever-growing list of biological processes [1,2]. Consequently, the importance of understanding the biophysical basis of phase separation is now clear [3]. A repeating theme in work linking phase separation to cellular function is: 1) the identification of key components contributing to the formation of a particular membraneless organelle or phase separated structures in cells and organisms using the tools of cell biology, and 2) demonstrating that biomolecular condensates can be minimally reconstituted in vitro using one or a few of these critical components. This reconstitution provides the important opportunity to analyze in detail the components and interactions that give rise to phase separation. Yet, unusual biophysical properties common to many phase separated condensates, including component density, sample heterogeneity, and disorder – preclude application of many common biophysical approaches to understand the structural and mechanistic details of phase separation.
Integrative biophysical tools to study biomolecular phase separation
Liquid-liquid phase separation of biomolecules requires molecules that can form multiple simultaneous contacts (necessary to stabilize a network to define a condensed phase) that lack rigid long-range order (otherwise the assemblies would be solid) [4]. Hence, disordered proteins and domains are often important contributors to phase separation as both mediators of phase separation or simply as linkers between folded domains that mediate the contacts. As liquids, these condensed phases are not directly amenable to x-ray crystallography or single-particle cryoelectron microscopy. Therefore, both solution and solid-state NMR spectroscopies have emerged as important techniques to probe the structural details of phase separation with atomic or residue-by-residue resolution [5]. Yet, NMR experiments report on average behavior, hence details on heterogeneous populations and ensembles are difficult to interrogate directly. Furthermore, because NMR experiments are uniquely sensitive probes of molecular motions, the magnitude and timescales of rotational and conformational changes impact NMR spectra – but these same features complicate quantitative interpretation. Such motions can obscure highly dynamic or static conformers or regions, depending on the particular NMR technique. Therefore, complementary approaches including molecular simulation and other spectroscopies can provide frameworks for interpreting the NMR data.
Specifically, all-atom molecular dynamics (MD) simulations using physics-based models have emerged as an essential tool in linking the laboratory measurements with molecular and atomic details with high spatiotemporal resolution [6]. Critical to using MD simulations is validation of the model (e.g. protein and water force field) for this new class of systems that are distinct from the state (folded protein and even dilute disordered proteins) and sequence compositions for which these models were originally parameterized. An explosion of activity in the last decade in protein force field refinement, new simulation algorithms[7], and the availability of modern computer hardware to generate microsecond-long trajectories[8] has made feasible near quantitative agreement of simulated properties with experimental observables such as chemical shifts, relaxation order parameters, and radius of gyration, among others. Applying these approaches to proteins that phase separate, residue-level NMR data on low-complexity (LC) disordered proteins that are implicated in cellular phase separation was used recently to assess and tune the backbone dihedral potential parameters for polar residues to balance the propensity of helical and extended structures [9*]. Going forward, these protein models will help develop a molecular mechanistic understanding of atomic interactions[10] and other important questions in condensates such as water/ion distribution and dynamics[11**]. These approaches are also used to interpret the NMR spectroscopy data on protein structure, interactions, and dynamics in dilute and dense protein phases when experimental observations alone cannot distinguish between different possible scenarios.
In situ molecular spectroscopy of phase separated condensates is an area where NMR spectroscopy is challenged due to the inherently heterogeneous nature of the sample (e.g. condensates within a homogenous bulk phase). Several solution state NMR studies of condensates have been performed with “macro-droplets” [12], while others have explicitly tested and demonstrated the correspondence between these samples and two-phase systems [13–15]. (We note that the interpretation that our previous work demonstrating direct observation of chemical shift differences in the condensed phase is due to interaction with glass [15] is not consistent with the quantification of peak intensity and spectral analysis we and others performed.) Still, spatially or spectroscopically probing droplets and particularly droplet interfaces is not straightforward. Optical spectroscopies, such as vibrational or fluorescence spectroscopy, offer an alternative set of approaches. Combined with imaging platforms to spatially resolve condensate phases in situ, they can provide fingerprints of molecular structural and chemical interactions with sub-micron spatial resolution. Below, we focus on vibrational spectroscopies (Fourier transform infrared (FTIR) spectroscopy and Raman spectroscopy) and fluorescence spectroscopy (fluorescence correlation spectroscopy (FCS)) that report on the intrinsic physical chemical environment and contacts in the phases.
Following on the success of previous work combining complementary techniques to study disordered protein conformational properties[16], here we review biophysical questions in phase separation, focusing on proteins, and integrative approaches to revealing the molecular details of phase separation (Figure 1).
Figure 1: Integrative approach to biophysical characterization of phase separation.
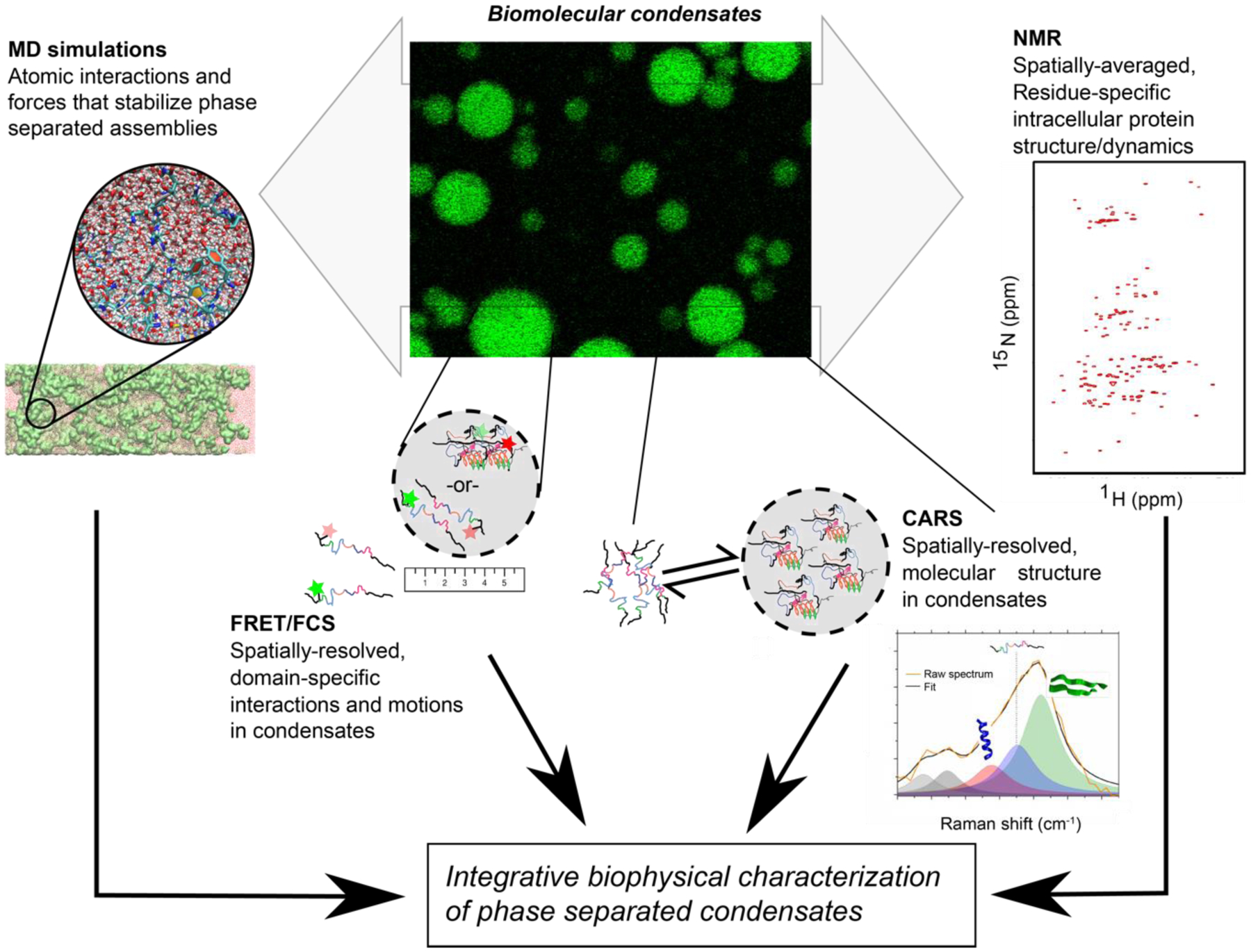
Integration of information from NMR (or EPR) and optical and vibrational spectroscopies combined with insight from molecular simulations are a powerful combination to probe structure, interactions, and molecular motions in phase separated biomolecular condensates.
Probing Conformational Properties and Structure
NMR spectroscopy:
An essential question in the biophysical characterization of phase separation is the structure of the components both in the dispersed and condensed phases. Many phase separating proteins contain sequence-repetitive low complexity domains (LCs) that are predicted to be disordered but also prone to assembly [17]. Therefore, it is not surprising that solution NMR spectroscopy (Figure 1), which provides a residue-by-residue picture of the dynamic ensemble of conformations including the secondary structure of these proteins, have shown that these domains are disordered when in the dispersed phase (i.e. not phase separated). Based on pioneering earlier work showing the formation of amyloid “hydrogels” (Box 1) from tagged forms of these domains and the correlative impact of mutations on amyloid formation and, later, phase separation[18–20], some studies have claimed that these disordered domains phase separate primarily due to β-sheet contacts. Yet, solution NMR studies directly observing the liquid condensed phases have found no support for assertions of increased β-sheet ordering. On the contrary, despite different sequence compositions and driving forces for phase separation, the LC domains of FUS, DDX4, hnRNPA2, and elastin-like peptides all retain predominant disorder within liquid condensed phases [12–14*,21]. Similarly, the acidic disordered domains of nucleophosmin (NPM1) remain disordered even in the context of a viscoelastic condensed phase formed with an arginine-rich peptide that contributes to nucleolar formation [22**]. We note that solution NMR is not sensitive to slowly moving species or minority populations. For our work, we have overcome this challenge for FUS LC with exchange-based NMR and coherent Raman scattering (CRS) to probe for conformational exchange and slow-moving states, which further confirmed no evidence of structured conformations in the liquid condensed phase [14]. Still for some proteins including FUS, it is clear that gradual conversion of liquid phases into more static structures can involve β-sheet formation depending on the conditions and missense mutations[23**]. Additionally, electron paramagnetic resonance (EPR) of site-specifically labeled samples has recently been applied to the question of disordered domain structure in droplets to probe chain motions and dimensions upon phase separation[15].
Box 1: Phase separation, gels, “hydrogels”.
It is important to distinguish between the rapid phase separation into liquid droplets from the slow conversion of liquid phases to “hydrogels”, which has been seen for FUS LC, nucleoporin FGs, hnRNPA2 LC. In phase separation, condensed phase droplets – with varying viscosities – forms, depleting the dispersed solution of the biopolymer components. For NMR samples of condensed phases, 15 to 50 ml of solution is often created in order to generate <500 μl of >200 mg/ml (up to 25 mM) condensed phase [5,21,22**]. By contrast, amyloid-based “hydrogels” are formed gradually (over days) and uniformly (the entire sample changes from a liquid to a hydrogel, phase separation is not required) from samples of ~mM concentration of proteins (that are often tagged to prevent avid phase separation). We surmise that the addition of solubility tags modifies the self-assembly process, resulting in different final states. Taking this into account, it is possible that additional semantic confusion regarding phase separation, “hydrogels”, and “gels” arises for the following reasons: 1) liquid condensed phases are sometimes referred to as “gels” as they are loosely-held networks that can achieve a percolated state, 2) very high concentration samples of liquid forming components (i.e. beyond the high concentration arm of the binodal) undergo “gelation without phase separation” in this sense that protein-dense liquids are “gels” [4], 3) some proteins with disordered regions phase separate into viscoelastic condensed phases that do not readily flow (i.e. they “gel” according to a different common usage of the word) based on the conditions, temperature, and valency/strength of interactions[12,22] yet are distinct from amyloid hydrogels, and 4) some liquid-liquid phase separated condensates “age” into static and even amyloid structures that may also be referred to as “gels” [24]. These distinctions are important because, although the same residues may contribute to both phase separation and amyloid formation, the physical features and hence molecular structures are different.
While we have focused on disordered domains until now, folded domains, especially those that oligomerize, can also contribute to scaffolds for phase separation[25]. Thus, a logical question is if the structure of folded domains changes due to phase separation? The NPM1 contains a folded, pentamerizing N-terminal domain that facilitates phase separation of NPM1’s acidic disordered domains with R-rich motif peptides and RNA [26]. After phase separation, the NPM1 pentamers remain spaced as seen by neutron scattering. Solid-state NMR of phase separated of NPM1 showed that the folded domain of NPM retains the same structural fingerprint (i.e. folded pentamer) in a condensed phase [22]. Can phase separation influence folded conformations? The Dcp1/Dcp2 mRNA decapping complex can phase separate with the enhancer of decapping 3 (Edc3) to form mRNA processing body and speed mRNA decapping [27]. In these phases, the LSm domain of Edc3, which interacts with Dcp2, remains folded but is dynamically tethered via an RNA-binding disordered region to the dimerization domain[28]. Recent findings suggest that Dcp1/Dcp2 form an inactive conformation inhibiting decapping when at high concentration in condensed phases but co-condensation of Dcp1 and Dcp2 with Edc3 activates decapping within the phase-separated compartment by inducing a conformation change in the catalytic domain[29*]. Curiously, Edc3 is dimeric and hence in this model interactions with multiple copies of Dcp2 replace the autoinhibitory multivalent Dcp2 self-interactions, effectively “rewiring” the interactions that stabilize condensation.
Vibrational spectroscopy:
Though missing the atomic resolution of NMR, vibrational spectroscopy also reports on molecular structure and interactions with the added advantage that these features can be quantified in samples with heterogenous structure and motions. Linear FTIR has been used to follow pressure-induced self-assembly of γD-crystallin with crowding agents [30] and thermally-triggered phase separation of fatty-acid-modified elastin-like polypeptides [31]. Specifically, FTIR was used demonstrate the changes in solute-protein and protein-protein interactions along the process of phase separation and material maturation. Coherent, two-dimensional FTIR (2D IR), which can resolve overlapping peaks and correlations of molecular vibrations over femto- to picosecond timescales (often considered an optical analog of 2D NMR), was recently applied to study LLPS of dipeptide repeats of proline-arginine (PR) chains [32]. In this study, PR20 chains phase separated into droplets when crowded with PEG and phase separated PR20 displayed spectral signatures consistent with backbone configurations associated with polyproline I and II helices compared to a random coil state when in the dispersed phase.
Imaging spectroscopies such as atomic force microscopy IR (AFM-IR) [33**] and coherent Raman scattering (CRS) [34] can discriminate between the continuous phase and protein-rich phase separated droplets. AFM-IR offers the promise of molecular spectroscopy with ~50 nm spatial resolution but is challenging in water; CRS does not have limitations in water but offers only optical (~300 nm) resolution. St George-Hyslop and Knowles used AFM-IR to show that methylation state of FUS changes the thermodynamic state, average protein secondary structure, and mechanics of FUS condensates [35]. Our own studies, using CRS, have recently shown that various promiscuous interactions stabilize liquid FUS LC droplets, which on average showed the same overall secondary structure as dispersed FUS LC [14*]. As complementary methods to NMR, vibrational spectroscopies are useful methods to study heterogeneous phase separation samples in situ.
Molecular simulations:
Rauscher and Pomes conducted one of the first all-atom simulations of the liquid-like state of a short elastin-like peptide[36], highlighting conformational disorder within the assembled state consistent with experiment [13]. In this case, hydrophobic interactions were found to contribute to intermolecular contacts of hydrated peptides within the protein-rich phase. Some groups have also used coarse-grained (CG) models to access longer timescales including the process of phase separation as well as examine partitioning and surface properties [37,38]. To complement these approaches, we recently used our CG model based protein-rich phase configurations to generate fully atomistic models with explicit solvent/ions and probed structure and motions inside the condensed phase and on the droplet interface[11**] (Figure 3). These recent investigations suggest that these simulations can provide meaningful information that is difficult to access by experiment is available on protein structure, contacts, and motions. To realize the potential of this new frontier, accessing longer timescales in order to ensure adequate sampling of states and observe the full range of structural transitions will require further advances in both hardware and software.
Figure 3: All atom simulation of phase separation.
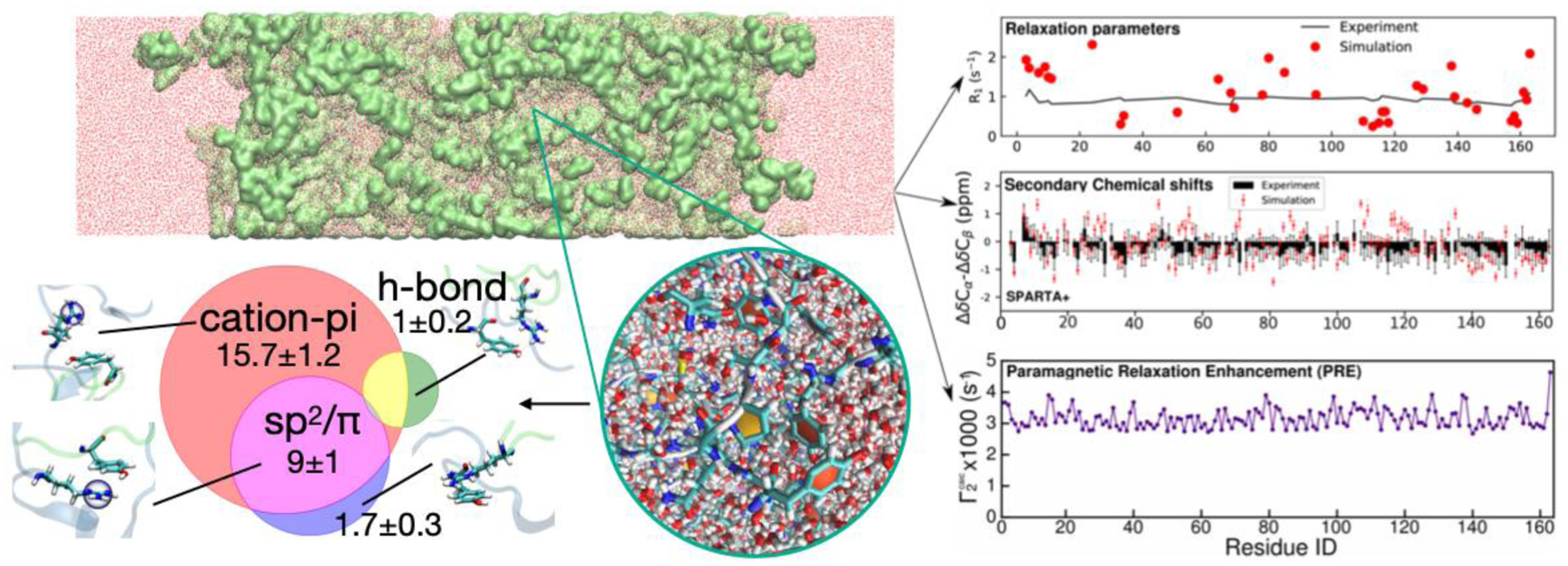
Creation and simulation of a fully atomistic “slabs” (top left) by conversion of coarse-grained models of phase separation enables atomistic analysis of contact modes (bottom left), molecular structure and solvent/ion properties (bottom center), and predicted NMR observables (right).
Molecular interactions in condensed phases
One of the central goals of many studies integrating experimental and computational results is to identify the fundamental rules (aka “molecular grammar”) based on atomic interactions that dictate the sequence-dependent phase separation of proteins [39–41]. Although perhaps starting from different initial hypotheses regarding the primary interactions (Is the dominant contribution cation-π vs. sp2/ π interactions to phase separation over non-specific hydrophobic effect and hydrogen bonding?), many studies have arrived at similar conclusions about the role of certain amino acids in promoting the phase separation, leading to some confusion about several important issues. Most importantly, are few residue types solely responsible for driving the phase separation as expected for associative polymers or do most amino acids in the sequence contribute to the phase separation to varying degrees (primary, secondary, tertiary, etc. drivers)[39,42,43]? Mutagenesis experiments, changing one residue type at a time in the low complexity domains have highlighted tyrosine and arginine as particularly important in polar-rich sequence phase separation [40,44,45]. Yet, other residues including glutamine/serine [14,46] and charged residues [21,44,47] contribute to phase separation in some systems while even hydrophobicity plays a role in some contexts [48]. In our view, together these approaches have generated consequential insights about the primary drivers of phase separation (e.g. Tyr and Arg over Phe and Lys for many polar-rich domains in vertebrates) and the role of other stabilizing interactions, e.g., involving residues such as glutamine and threonine in the case of FUS LC and stabilization of small helical motifs in case of TDP-43 CTD[49].
Still, these approaches cannot easily show what ensemble of contacts these residues types make. Therefore, NMR experiments that directly interrogate contacts have played an important role. In the dispersed phase, the contacts contributing to phase separation may form transient weak intramolecular interactions due to the repetitive nature of the sequences. Using this approach, the contribution of aromatic (tyrosine to phenylalanine) contacts was directly observed for hnRNPA1 LC using nuclear Overhauser effect experiments, where amino acids in proximity can be detected even if the interaction is transient [45]. Going further, in condensed phase samples where the density of proteins is high and hence provides higher signal to noise, a full network of contacts between many different residue types has been observed for several disordered domains often using a combination of mixing peptides with distinct isotopic labeling patterns and heteronuclear edited (and filtered) NMR experiments[13,14,21,50]. These observations are in accordance with both fully atomistic and coarse-grained simulations demonstrating a broad array of contacts both in initial contacts mimicking the interactions leading to phase separation and within condensed phases[51]. Therefore, these insights have informed efforts to understands the role of disease-associated mutations and post-translational modifications on phase separation[52–54].
It is important to note that although residue-residue contributions are directly observed by NMR and validated by mutagenesis, it is not trivial to parse out the energetic balance of specific and overlapping interaction modes that these residues make such as cation-π, sp2/π, hydrophobicity, and hydrogen bonding using available experimental techniques and computational models[55]. On the experimental side, it will be beneficial to conduct phase separation studies using non-natural amino acids to more precisely perturb the interaction modes in hope of disentangling their contributions, including further work such as the impact of fluorination of aromatic residues on phase separation[56]. On the simulation side, all-atom simulation studies could be complemented with more complex and polarizable models[57] or ab initio models that could more accurately represent, for example, explicit π-interactions and the behavior of surface water to provide confidence that they capture the interaction modes and to further probe the unusual chemical environment created by phase separation[58].
Molecular motions in condensed phases
Phase separated condensates contain high concentrations of biomolecules, and hence, it is not a surprise that local and global motions of molecules are distinct in the condensed phase compared to studies in bulk. NMR has been extensively used to demonstrate that local motions on the ns/ps timescale are slowed but the chain remains highly mobile in liquid condensed phases of disordered proteins[12–14,21,59**]. Curiously, signatures of slower timescale (ms) conformational transitions have been observed for condensed phases of DDX4, yet the origin of these remain unknown[60]. Translational protein diffusion as measured by diffusion NMR is slowed by orders of magnitude[14*,21], consistent with results from fluorescent recovery after photobleaching readily accessible via microscopy and consistent with a dense (percolated) phase[4].
Fluorescence spectroscopies, specifically FCS and FRET, have a long history in biophysical measurements. Unique from NMR or vibrational spectroscopy, these methods do not directly probe molecular interactions in protein, using fluorophores as handles to measure fluorophore environment, motions or fluorophore-fluorophore distances; however, FCS and FRET offer unparalleled single-molecule sensitivity and exquisite spatial selectivity. Here, we focus on FCS as it is detected as a traditional spectroscopic measurement as a measure of molecular motion in condensed phases. FCS was used to show how RNA-FUS interactions modified the tracer mobility of condensed phases, identifying two populations of FUS upon RNA-FUS interaction[61]. Similarly, the effect of RNA concentration on morphology of hollow phase-separated protamine condensates and the mobility in the rim regions was also probed by FCS. The rim, where protein and RNA co-condense, showed significantly reduced dynamics compared to the lumen [62]. In an alternative system, FCS was used to probe protein-protein interactions between FG nucleoporins (Nups) and nuclear transport receptors, identifying changes in Nup mobility with nuclear transport receptor addition [63]. Finally, as FCS provides a measurement of mobility, it has also been used to quantify the microrheological properties in LAF-1 condensates formed with different macromolecular constituents [64]. The versatility and relative ease of FCS make it a very attractive method for physical-chemical analysis of phase separation, and one that provides highly complementary data for phase field models of condensate dynamics[65].
Conclusion
Taken together, magnetic resonance and optical spectroscopies combined with molecular simulation offer a complementary set of tools to probe the biophysical details of phase separation with atomistic and molecular resolution. As more complex condensates are reconstituted and in order to realize the ultimate goal of probing the structure, interactions, and molecular motions in condensates in living cells, cooperation between these techniques becomes even more important.
Acknowledgement
This work was supported in part by the National Institutes of Health R01NS116176 (N.L.F. and J.M.), R01GM120537 (J.M.), National Science Foundation grants DMR2004796 (J.M), the SPP2191 of the Deutsche Forschungsgemeinschaft (DFG) PA2526/3-1 (S.H.P.), the Human Frontier in Science Program (RGP0045/2018 to N.L.F. and S.H.P.). We thank Vinald Francis for designing Figure 2.
Figure 2. NMR spectroscopy of phase separation.
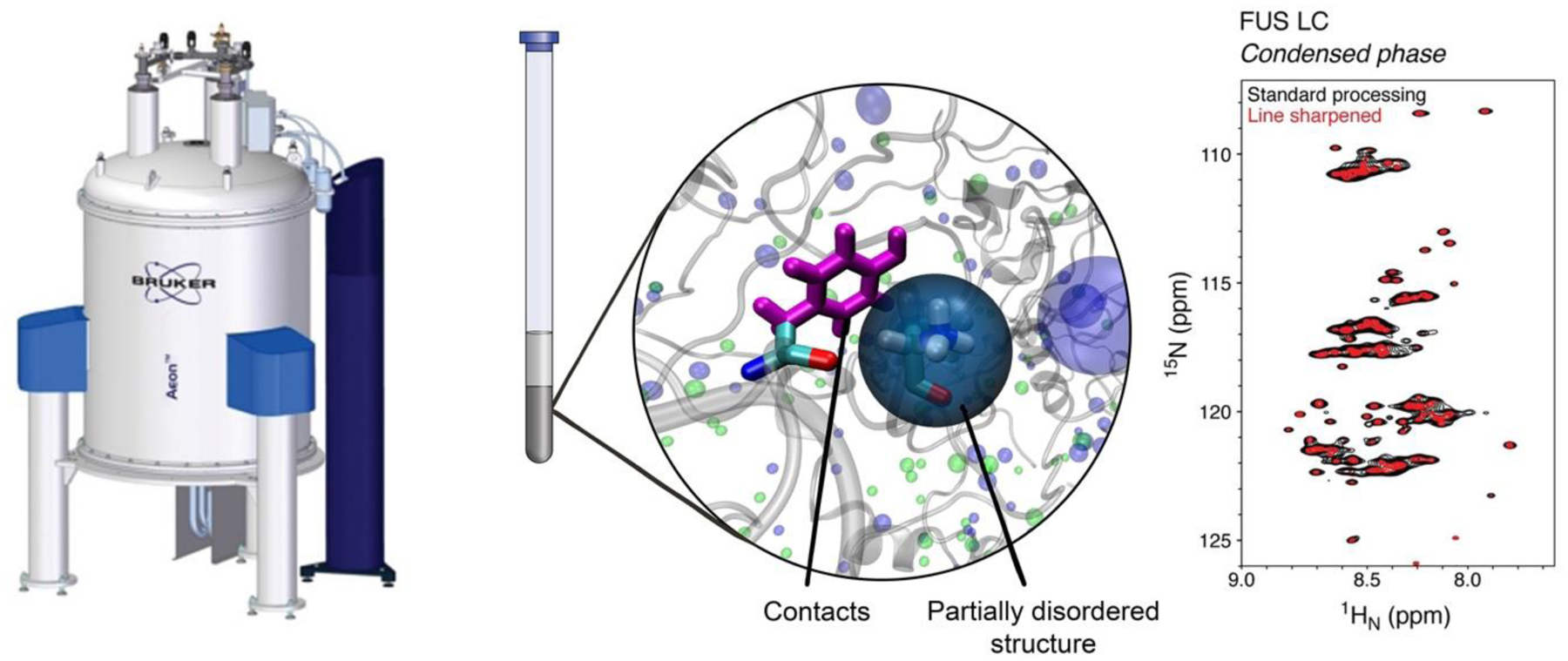
NMR spectroscopy of samples where the condensed phase (gray, center) fills the observation (coil) volume enables direct interrogation of structure and disorder in proteins and their contacts (center) with residue-by-residue resolution (right). Image concept by Vinald Francis.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Shin Y, Brangwynne CP: Liquid phase condensation in cell physiology and disease. Science 2017, 357. [DOI] [PubMed] [Google Scholar]
- 2.Lyon AS, Peeples WB, Rosen MK: A framework for understanding the functions of biomolecular condensates across scales. Nat Rev Mol Cell Biol 2021, 22:215–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mitrea DM, Chandra B, Ferrolino MC, Gibbs EB, Tolbert M, White MR, Kriwacki RW: Methods for Physical Characterization of Phase-Separated Bodies and Membraneless Organelles. J Mol Biol 2018, 430:4773–4805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Harmon TS, Holehouse AS, Rosen MK, Pappu RV: Intrinsically disordered linkers determine the interplay between phase separation and gelation in multivalent proteins. Elife 2017, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Murthy AC, Fawzi NL: The (un)structural biology of biomolecular liquid-liquid phase separation using NMR spectroscopy. J Biol Chem 2020, 295:2375–2384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hollingsworth SA, Dror RO: Molecular Dynamics Simulation for All. Neuron 2018, 99:1129–1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shea JE, Best RB, Mittal J: Physics-based computational and theoretical approaches to intrinsically disordered proteins. Curr Opin Struct Biol 2021, 67:219–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shaw DE, Grossman JP, Bank JA, Batson B, Butts JA, Chao JC, Deneroff MM, Dror RO, Even A, Fenton CH, et al. : Anton 2: Raising the Bar for Performance and Programmability in a Special-Purpose Molecular Dynamics Supercomputer. In SC ‘14: Proceedings of the International Conference for High Performance Computing, Networking, Storage and Analysis 16–21 Nov. 2014: 2014:41–53. [Google Scholar]
- 9*.Tang WS, Fawzi NL, Mittal J: Refining All-Atom Protein Force Fields for Polar-Rich, Prion-like, Low-Complexity Intrinsically Disordered Proteins. J Phys Chem B 2020, 124:9505–9512. [DOI] [PMC free article] [PubMed] [Google Scholar]; This work found that serine, threonine, and glutamine are overly helical in some forcefields when applied to polar-rich disordered domains. By tweaking the backbone torsion angle parameters for these three residue types, these domains can be simulated accurately while still correctly representing partial structure present.
- 10.Paloni M, Bailly R, Ciandrini L, Barducci A: Unraveling Molecular Interactions in Liquid-Liquid Phase Separation of Disordered Proteins by Atomistic Simulations. J Phys Chem B 2020, 124:9009–9016. [DOI] [PubMed] [Google Scholar]
- 11**.Zheng W, Dignon GL, Jovic N, Xu, Regy RM, Fawzi NL, Kim YC, Best RB, Mittal J: Molecular Details of Protein Condensates Probed by Microsecond Long Atomistic Simulations. J Phys Chem B 2020, 124:11671–11679. [DOI] [PMC free article] [PubMed] [Google Scholar]; Simulations of condensed phases of intrinsically disordered proteins with atomic resolution shows that proteins remain mobile and interact via a combination of hydrophobic interactions, hydrogen bonds, salt bridges, and π–π and cation–π interactions. Additionally, these simulations can probe features difficult for current experiment, demonstrating that salt ion partitioning within the condensed phase depends on the protein charge distribution.
- 12.Burke KA, Janke AM, Rhine CL, Fawzi NL: Residue-by-Residue View of In Vitro FUS Granules that Bind the C-Terminal Domain of RNA Polymerase II. Mol Cell 2015, 60:231–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reichheld SE, Muiznieks LD, Keeley FW, Sharpe S: Direct observation of structure and dynamics during phase separation of an elastomeric protein. Proc Natl Acad Sci U S A 2017, 114:E4408–E4415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14*.Murthy AC, Dignon GL, Kan Y, Zerze GH, Parekh SH, Mittal J, Fawzi NL: Molecular interactions underlying liquid-liquid phase separation of the FUS low-complexity domain. Nat Struct Mol Biol 2019, 26:637–648. [DOI] [PMC free article] [PubMed] [Google Scholar]; We used NMR, Raman spectroscopy, and simulation to show a diverse network of contacts involving many residue types in phase separated states of FUS LC.
- 15.Emmanouilidis L, Esteban-Hofer L, Damberger FF, de Vries T, Nguyen CKX, Ibanez LF, Mergenthal S, Klotzsch E, Yulikov M, Jeschke G, et al. : NMR and EPR reveal a compaction of the RNA-binding protein FUS upon droplet formation. Nat Chem Biol 2021. [DOI] [PubMed] [Google Scholar]
- 16.Gomes GW, Krzeminski M, Namini A, Martin EW, Mittag T, Head-Gordon T, Forman-Kay JD, Gradinaru CC: Conformational Ensembles of an Intrinsically Disordered Protein Consistent with NMR, SAXS, and Single-Molecule FRET. J Am Chem Soc 2020, 142:15697–15710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.King OD, Gitler AD, Shorter J: The tip of the iceberg: RNA-binding proteins with prion-like domains in neurodegenerative disease. Brain Res 2012, 1462:61–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kato M, Han TW, Xie S, Shi K, Du X, Wu LC, Mirzaei H, Goldsmith EJ, Longgood J, Pei J, et al. : Cell-free formation of RNA granules: low complexity sequence domains form dynamic fibers within hydrogels. Cell 2012, 149:753–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Murray DT, Kato M, Lin Y, Thurber KR, Hung I, McKnight SL, Tycko R: Structure of FUS Protein Fibrils and Its Relevance to Self-Assembly and Phase Separation of Low-Complexity Domains. Cell 2017, 171:615–627 e616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lin Y, Zhou X, Kato M, Liu D, Ghaemmaghami S, Tu BP, McKnight SL: Redox-mediated regulation of an evolutionarily conserved cross-beta structure formed by the TDP43 low complexity domain. Proc Natl Acad Sci U S A 2020, 117:28727–28734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brady JP, Farber PJ, Sekhar A, Lin YH, Huang R, Bah A, Nott TJ, Chan HS, Baldwin AJ, Forman-Kay JD, et al. : Structural and hydrodynamic properties of an intrinsically disordered region of a germ cell-specific protein on phase separation. Proc Natl Acad Sci U S A 2017, 114:E8194–E8203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22**.Gibbs E, Perrone B, Hassan A, Kummerle R, Kriwacki R: NPM1 exhibits structural and dynamic heterogeneity upon phase separation with the p14ARF tumor suppressor. J Magn Reson 2020, 310:106646. [DOI] [PMC free article] [PubMed] [Google Scholar]; Solid-state NMR demonstrates that NPM1 has distinct disordered, mobile and ordered, rigid parts after phase separation into viscoelastic droplets.
- 23**.Berkeley RF, Kashefi M, Debelouchina GT: Real-Time Observation of Structure and Dynamics during the Liquid-to-Solid Transition of FUS LC. Biophys J 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]; This work uses solid-state NMR to watch over time how β-sheet transition can occur in condensed phases of FUS LC and how a disease mutation speeds the process.
- 24.Patel A, Lee Hyun O, Jawerth L, Maharana S, Jahnel M, Hein Marco Y, Stoynov S, Mahamid J, Saha S, Franzmann Titus M, et al. : A Liquid-to-Solid Phase Transition of the ALS Protein FUS Accelerated by Disease Mutation. Cell 2015, 162:1066–1077. [DOI] [PubMed] [Google Scholar]
- 25.Shin Y, Berry J, Pannucci N, Haataja MP, Toettcher JE, Brangwynne CP: Spatiotemporal Control of Intracellular Phase Transitions Using Light-Activated optoDroplets. Cell 2017, 168:159–171 e114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mitrea DM, Cika JA, Guy CS, Ban D, Banerjee PR, Stanley CB, Nourse A, Deniz AA, Kriwacki RW: Nucleophosmin integrates within the nucleolus via multi-modal interactions with proteins displaying R-rich linear motifs and rRNA. Elife 2016, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schutz S, Noldeke ER, Sprangers R: A synergistic network of interactions promotes the formation of in vitro processing bodies and protects mRNA against decapping. Nucleic Acids Res 2017, 45:6911–6922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Damman R, Schutz S, Luo Y, Weingarth M, Sprangers R, Baldus M: Atomic-level insight into mRNA processing bodies by combining solid and solution-state NMR spectroscopy. Nat Commun 2019, 10:4536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29*.Tibble RW, Depaix A, Kowalska J, Jemielity J, Gross JD: Biomolecular condensates amplify mRNA decapping by biasing enzyme conformation. Nat Chem Biol 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]; Decapping complex Dcp1/Dcp2 changes conformation depending on the presence of condensate binding partner Edc3 and exchanges homo- for hetero-interactions with dimeric Edc3.
- 30.Cinar H, Winter R: The effects of cosolutes and crowding on the kinetics of protein condensate formation based on liquid-liquid phase separation: a pressure-jump relaxation study. Sci Rep 2020, 10:17245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mozhdehi D, Luginbuhl KM, Simon JR, Dzuricky M, Berger R, Varol HS, Huang FC, Buehne KL, Mayne NR, Weitzhandler I, et al. : Genetically encoded lipid-polypeptide hybrid biomaterials that exhibit temperature-triggered hierarchical self-assembly. Nat Chem 2018, 10:496–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Edun DN, Flanagan MR, Serrano AL: Does liquid–liquid phase separation drive peptide folding? Chemical Science 2021, 12:2474–2479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33**.Shen Y, Ruggeri FS, Vigolo D, Kamada A, Qamar S, Levin A, Iserman C, Alberti S, George-Hyslop PS, Knowles TPJ: Biomolecular condensates undergo a generic shear-mediated liquid-to-solid transition. Nat Nanotechnol 2020, 15:841–847. [DOI] [PMC free article] [PubMed] [Google Scholar]; The authors show that a small amount of shear can induce fiber formation in otherwise non-fibrous proteins. They use AFM-IR to show that droplets and fibers from the same proteins show unique secondary structures with fibers showing substantial β-sheet character.
- 34.Lau HK, Paul A, Sidhu I, Li L, Sabanayagam CR, Parekh SH, Kiick KL: Microstructured Elastomer-PEG Hydrogels via Kinetic Capture of Aqueous Liquid-Liquid Phase Separation. Adv Sci (Weinh) 2018, 5:1701010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Qamar S, Wang G, Randle SJ, Ruggeri FS, Varela JA, Lin JQ, Phillips EC, Miyashita A, Williams D, Strohl F, et al. : FUS Phase Separation Is Modulated by a Molecular Chaperone and Methylation of Arginine Cation-pi Interactions. Cell 2018, 173:720–734 e715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rauscher S, Pomes R: The liquid structure of elastin. Elife 2017, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dignon GL, Zheng W, Kim YC, Best RB, Mittal J: Sequence determinants of protein phase behavior from a coarse-grained model. PLoS Comput Biol 2018, 14:e1005941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Benayad Z, von Bulow S, Stelzl LS, Hummer G: Simulation of FUS Protein Condensates with an Adapted Coarse-Grained Model. J Chem Theory Comput 2021, 17:525–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Choi JM, Holehouse AS, Pappu RV: Physical Principles Underlying the Complex Biology of Intracellular Phase Transitions. Annu Rev Biophys 2020, 49:107–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang J, Choi JM, Holehouse AS, Lee HO, Zhang X, Jahnel M, Maharana S, Lemaitre R, Pozniakovsky A, Drechsel D, et al. : A Molecular Grammar Governing the Driving Forces for Phase Separation of Prion-like RNA Binding Proteins. Cell 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dignon GL, Best RB, Mittal J: Biomolecular Phase Separation: From Molecular Driving Forces to Macroscopic Properties. Annu Rev Phys Chem 2020, 71:53–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rubinstein M, Semenov AN: Dynamics of Entangled Solutions of Associating Polymers. Macromolecules 2001, 34:1058–1068. [Google Scholar]
- 43.Semenov AN, Rubinstein M: Thermoreversible Gelation in Solutions of Associative Polymers. 1. Statics. Macromolecules 1998, 31:1373–1385. [Google Scholar]
- 44.Schuster BS, Dignon GL, Tang WS, Kelley FM, Ranganath AK, Jahnke CN, Simpkins AG, Regy RM, Hammer DA, Good MC, et al. : Identifying sequence perturbations to an intrinsically disordered protein that determine its phase-separation behavior. Proc Natl Acad Sci U S A 2020, 117:11421–11431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Martin EW, Holehouse AS, Peran I, Farag M, Incicco JJ, Bremer A, Grace CR, Soranno A, Pappu RV, Mittag T: Valence and patterning of aromatic residues determine the phase behavior of prion-like domains. Science 2020, 367:694–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dzuricky M, Rogers BA, Shahid A, Cremer PS, Chilkoti A: De novo engineering of intracellular condensates using artificial disordered proteins. Nat Chem 2020, 12:814–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bremer A, Farag M, Borcherds WM, Peran I, Martin EW, Pappu RV, Mittag T: Deciphering how naturally occurring sequence features impact the phase behaviors of disordered prion-like domains. bioRxiv 2021:2021.2001.2001.425046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Krainer G, Welsh TJ, Joseph JA, Espinosa JR, Wittmann S, de Csillery E, Sridhar A, Toprakcioglu Z, Gudiskyte G, Czekalska MA, et al. : Reentrant liquid condensate phase of proteins is stabilized by hydrophobic and non-ionic interactions. Nat Commun 2021, 12:1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Conicella AE, Dignon GL, Zerze GH, Schmidt HB, D’Ordine AM, Kim YC, Rohatgi R, Ayala YM, Mittal J, Fawzi NL: TDP-43 alpha-helical structure tunes liquid-liquid phase separation and function. Proc Natl Acad Sci U S A 2020, 117:5883–5894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Garcia Garcia C, Patkar SS, Jovic N, Mittal J, Kiick KL: Alteration of Microstructure in Biopolymeric Hydrogels via Compositional Modification of Resilin-Like Polypeptides. ACS Biomater Sci Eng 2021. [DOI] [PubMed] [Google Scholar]
- 51.Dignon GL, Zheng W, Mittal J: Simulation methods for liquid-liquid phase separation of disordered proteins. Curr Opin Chem Eng 2019, 23:92–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Perdikari TM, Jovic N, Dignon GL, Kim YC, Fawzi NL, Mittal J: A coarse-grained model for position-specific effects of post-translational modifications on disordered protein phase separation. Biophys J 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ukmar-Godec T, Hutten S, Grieshop MP, Rezaei-Ghaleh N, Cima-Omori MS, Biernat J, Mandelkow E, Soding J, Dormann D, Zweckstetter M: Lysine/RNA-interactions drive and regulate biomolecular condensation. Nat Commun 2019, 10:2909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tsang B, Arsenault J, Vernon RM, Lin H, Sonenberg N, Wang LY, Bah A, Forman-Kay JD: Phosphoregulated FMRP phase separation models activity-dependent translation through bidirectional control of mRNA granule formation. Proc Natl Acad Sci U S A 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Das S, Lin YH, Vernon RM, Forman-Kay JD, Chan HS: Comparative roles of charge, pi, and hydrophobic interactions in sequence-dependent phase separation of intrinsically disordered proteins. Proc Natl Acad Sci U S A 2020, 117:28795–28805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nott TJ, Petsalaki E, Farber P, Jervis D, Fussner E, Plochowietz A, Craggs TD, Bazett-Jones DP, Pawson T, Forman-Kay JD, et al. : Phase transition of a disordered nuage protein generates environmentally responsive membraneless organelles. Mol Cell 2015, 57:936–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Das AK, Demerdash ON, Head-Gordon T: Improvements to the AMOEBA Force Field by Introducing Anisotropic Atomic Polarizability of the Water Molecule. J Chem Theory Comput 2018, 14:6722–6733. [DOI] [PubMed] [Google Scholar]
- 58.Nott TJ, Craggs TD, Baldwin AJ: Membraneless organelles can melt nucleic acid duplexes and act as biomolecular filters. Nat Chem 2016, 8:569–575. [DOI] [PubMed] [Google Scholar]
- 59**.Kim TH, Tsang B, Vernon RM, Sonenberg N, Kay LE, Forman-Kay JD: Phospho-dependent phase separation of FMRP and CAPRIN1 recapitulates regulation of translation and deadenylation. Science 2019, 365:825–829. [DOI] [PubMed] [Google Scholar]; This work showed how phosphorylation alters specific sequence interactions between translational regulators CAPRIN1 and FMRP. Highlights are direct observation of a condensed phase containing both components and complementary functional assays to monitor in vitro deadenylation and translation.
- 60.Yuwen T, Brady JP, Kay LE: Probing Conformational Exchange in Weakly Interacting, Slowly Exchanging Protein Systems via Off-Resonance R1rho Experiments: Application to Studies of Protein Phase Separation. J Am Chem Soc 2018, 140:2115–2126. [DOI] [PubMed] [Google Scholar]
- 61.Maharana S, Wang J, Papadopoulos DK, Richter D, Pozniakovsky A, Poser I, Bickle M, Rizk S, Guillen-Boixet J, Franzmann TM, et al. : RNA buffers the phase separation behavior of prion-like RNA binding proteins. Science 2018, 360:918–921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62**.Alshareedah I, Moosa MM, Raju M, Potoyan DA, Banerjee PR: Phase transition of RNA-protein complexes into ordered hollow condensates. Proc Natl Acad Sci U S A 2020, 117:15650–15658. [DOI] [PMC free article] [PubMed] [Google Scholar]; This work shows how addition of RNA to a condensate forming protein can reshape the morphology of the condensates forming hollow shell-like structures. FCS confirmed that the rim area, with concentrated proteins and RNA, showed substantially less mobility compared to the lumen.
- 63.Milles S, Mercadante D, Aramburu IV, Jensen MR, Banterle N, Koehler C, Tyagi S, Clarke J, Shammas SL, Blackledge M, et al. : Plasticity of an ultrafast interaction between nucleoporins and nuclear transport receptors. Cell 2015, 163:734–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wei MT, Elbaum-Garfinkle S, Holehouse AS, Chen CC, Feric M, Arnold CB, Priestley RD, Pappu RV, Brangwynne CP: Phase behaviour of disordered proteins underlying low density and high permeability of liquid organelles. Nat Chem 2017, 9:1118–1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Muñoz-Gil G, Romero-Aristizabal C, Mateos N, de Llobet Cucalon LI, Beato M, Lewenstein M, Garcia-Parajo MF, Torreno-Pina JA: Phase separation of tunable biomolecular condensates predicted by an interacting particle model. bioRxiv 2020:2020.2009.2009.289876. [Google Scholar]


