Abstract
Scope
This study investigates the mechanism of action and functional effects of coffee extracts in colonic cells, on intestinal stem cell growth and inhibition of DSS-induced intestinal barrier damage in mice.
Methods and Results
Aqueous coffee extracts induced Ah receptor (AhR) -responsive CYP1A1, CYP1B1 and UGT1A1 gene expression in colon-derived Caco2 and YAMC cells. Tissue-specific AhR knockout (AhRf/f x Lgr5-GFP-CreERT2 x Villin-Cre), wild-type (Lgr5-CreERT2 x Villin-Cre) mice were sources of stem cell enriched organoids and both coffee extracts and norharman, an AhR-active component of these extracts inhibited stem cell growth. Coffee extracts also inhibited DSS-induced damage to intestinal barrier function and DSS-induced mucosal inflammatory genes such as IL-6 and TGF-β1 in wild-type (AhR+/+) but not AhR−/− mice. In contrast, coffee did not exhibit protective effects in intestinal-specific AhR knockout mice. Coffee extracts also enhanced overall formation of AhR-active microbial metabolites.
Conclusions
In colon-derived cells and in the mouse intestine, coffee induced several AhR-dependent responses including gene expression, inhibition of intestinal stem cell-enriched organoid growth and inhibition of DSS-induced intestinal barrier damage. We conclude that the anti-inflammatory effects of coffee in the intestine are due, in part, to activation of AhR signaling.
Keywords: Coffee, AhR, CYP1A1, colonic stem cells, anti-inflammatory
Graphical Abstract
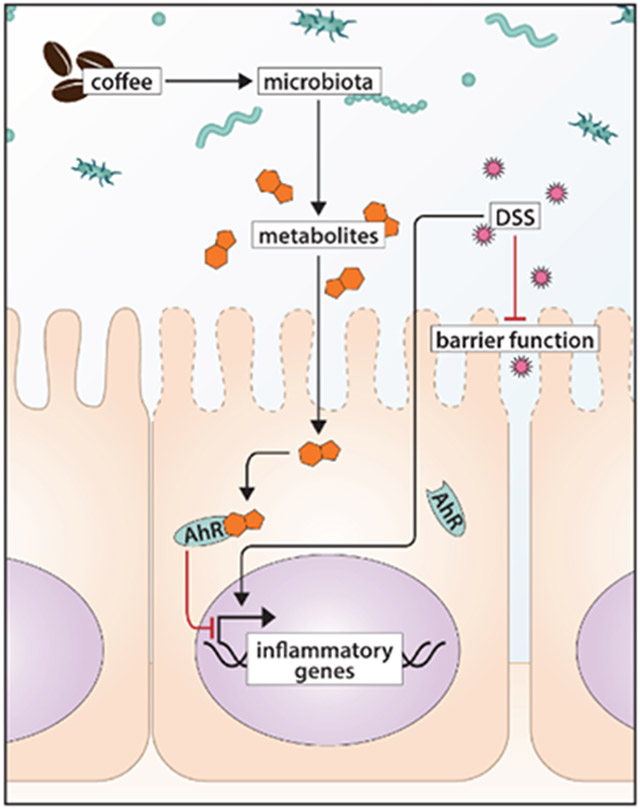
Summary diagram highlighting the AhR-dependent ability of coffee extract to suppress DSS-induced damage, intestinal barrier function and DSS-induced mucosal inflammatory genes. Coffee extracts activated Ah receptor (AhR) responsive genes in colon-derived cell lines. These same extracts and norharman, an AhR-active compound in coffee also inhibited growth in stem cell enriched organoids and enhanced intestinal barrier function in DSS-induced mice. Moreover, the effects of coffee extracts on stem cells and barrier function were AhR-dependent.
1. Introduction
Coffee is the most widely consumed beverage and worldwide consumption is over 160,000,000 x 60 kg bags. It is estimated that in the United States per capita consumption of coffee is 115.2 L annually (1,2). Several studies report an inverse association between coffee drinking and mortality and multiple adverse human health effects including Parkinson’s disease, metabolic diseases, and several cancers (3-11). For example a population-based study of 387,494 coffee drinkers in the United Kingdom showed that coffee drinking was inversely associated with mortality from all causes compared to non-coffee drinkers (3) and similar results were observed for individuals participating in the prospective Nurses’ Health Study and Health Professionals Follow-up Study (4). Coffee drinkers had a lower incidence of liver, ovarian, thyroid and endometrial cancers and melanoma (10) and meta-analysis of prospective cohort studies also showed a lower incidence of colon cancer (11). Coffee consumption has also been associated with protective effects against liver fibrosis and non-alcoholic fatty liver disease in humans (12) and this has also been observed in laboratory animal studies (13,14). In addition to the lower risk of colon cancer observed in some studies (11-15), coffee consumption protects against inflammatory bowel disease (IBD) (16) and a recent study reported that coffee consumption enhanced gut recovery in gynecological patients after surgery (17). Many of the same associations were similar for caffeinated and decaffeinated coffee, suggesting that the effects are not due to caffeine (18). Overall, these data link coffee consumption to lower risk of various health outcomes. However, the biological/signaling pathways mediating these responses are unclear. Interestingly, Hang et al (19) recently investigated the associations of total, caffeinated, and decaffeinated coffee consumption with plasma biomarkers of metabolic and inflammatory pathways in two cohorts of 15,551 women (Nurses’ Health Study) and 7,397 men (Health Professionals Follow-Up Study). Their seminal findings demonstrate that coffee consumption is associated with a favorable profile of plasma biomarkers of metabolic and inflammatory pathways.
Roasted coffee extracts contain up to 1000 different compounds which include structurally diverse aromatic and aliphatic aldehydes, alcohols, ketones, acids, polyphenolics and heterocyclic compounds including caffeine, and the health effects of coffee are due to the activity of this complex mixture. Many of the health benefits observed for coffee are similar to those reported for the aryl hydrocarbon receptor (AhR) and its ligands which include structurally diverse phytochemicals (20,21). For example, the AhR plays an important role in maintaining gut health and protecting against intestinal cancers in animal models and AhR ligands ameliorate intestinal inflammation and tumor formation (22-25) Moreover, loss of the AhR in mice results in hepatic fibrosis (26,27) and this parallels the protective effects of coffee on hepatic damage (12-14). These similarities between coffee and the AhR and its ligands coupled with a previous study showing that several different coffee extracts induced AhR-responsive genes (28) prompted us to investigate coffee extracts as potential AhR ligands in colon derived cell lines, and also determine their effects on colon epithelial stem cells, induced gut inflammation and the microbiome and its metabolites.
2. Experimental Section
2.1. Cell lines and chemicals
Caco2 cells were obtained from the American Type Culture Collection (ATCC, Manassas VA) and YAMC cells were kindly donated by Dr. Robert Whitehead (Vanderbilt University). The corresponding knockout Caco2-AhRKO and YAMC-AhRKO cell lines were generated by CRISPR/Cas9 as described (29,30). Caco2 cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 20% fetal bovine serum (FBS), 1 x MEM non-essential amino acid solution (Gibco), and 1 x antibiotic/antimycotic reagent (Sigma-Aldrich, St. Louis, MO) at 37°C in the presence of 5% CO2. YAMC cells were cultured in RPMI1640 media supplemented with 5% FBS, 1% glutamine, 0.1% ITS (Corning) and 10U IFNγ (Sigma-Aldrich) at 33°C in 5% CO2. The β-carboline alkaloids norharman, harmine hydrochloride, harmane, harmaline hydrochloride dehydrate, 1-methyl-2, 3, 4, 9-tetrahydro-1H-β-carboline-1-carboxylic acid sodium butyrate, sodium propionate and sodium acetate were purchased from Sigma-Aldrich. TCDD (99%) was synthesized in the Safe laboratory and roasted coffee (Seattle’s Best #4) was purchased locally.
2.2. Coffee extracts
Commercially available ground roasted or unroasted coffee (60 g) was stirred with 150 mL boiling water for 8-10 min and then filtered to give the aqueous coffee extract (22). An aliquot (20 mL) of the coffee extract was extracted with chloroform (2 x 20 mL), the chloroform extract was dried, concentrated and an aliquot was dissolved in DMSO at a concentration equivalent to the water extract. In addition, an aliquot of the chloroform extract was separated by semi-quantitative thin layer chromatography (55/45 hexane/acetone as solvent) into 3 fractions, top, middle (caffeine) and bottom. Gravimetric analysis indicated that % (by weight) distribution of the chloroform extract in the (top, middle (caffeine) and bottom bands was 25, 68 and 7%, respectively.
2.3. Quantitative real time PCR
After treatment of cells with coffee extracts (aqueous or chloroform) or individual compounds for 24 h, RNA was extracted and converted into cDNA using High Capacity RNA-cDNA kit (Applied Biosystem, Foster City, CA). PCR reactions were determined for three different experiments from each treatment group as previously described (29,30) and results are compared to control values. Sequences of the primers used for real time PCR are summarized in Supplemental Table S1. TBP was used to normalize TaqMan assays and GAPDH was the normalizer for SYBR green assays. The aqueous coffee extracts were added directly to the culture media. The chloroform extracts, the TLC fractions (Top, Bottom, Caffeine) other individual compounds and TCDD were dissolved in DMSO and added to the culture media (< 0.1% DMSO).
2.4. Chromatin immunoprecipitation (ChIP) assay
The ChIP assay was performed using ChIP-IT Express Magnetic Chromatin Immunoprecipitation kit (Active Motif, Carlsbad, CA) according to the manufacturer’s protocol. Caco2 cells were treated with various compounds for 2 h and the cell workup and assay procedures for determining interactions of the AhR on the DRE region of the human CYP1A1 gene promoter were carried out as previously described (29,30). The human CYP1A1 primers were 5’-TCA ATC AAG AGG CGC GAA CCT C-3’ (sense), and 5’-CTA CAG CCT ACC AGG ACT CG-3’ (anti-sense), and then amplified by targeting a 203-bp region of human CYP1A1 promoter which contained the AhR binding sequences. PCR products were resolved on a 2% agarose gel in the presence of ethidium bromide.
2.5. Mouse models
Animals were housed under conventional conditions, adhering to the guidelines approved by the Institutional Animal Care and Use Committee at Texas A&M University. Constitutive male and female intestine-specific AhR knockout mice (AhRf/f x Lgr5-GFP-CreERT2 x Villin-Cre) and littermate AhR wild-type controls (Lgr5-GFP-CreERT2 x Villin-Cre) 8-20 wk of age (31) used for organoid growth experiments were maintained on an AIN-76A semi-purified diet (Research Diets, D12450B), fed ad libitum and housed on a 12 h light-dark cycle. Villin-Cre mice expressed constitutively active Cre across the entire intestinal epithelium under control of the Villin promoter. AhRf/f mice contained loxP sites bordering exon 2 of the AhR gene, with recombination resulting in generation of a premature stop codon at codon 29. Stem cell targeted Lgr5-GFP-CreERT2 mice expressed GFP in crypt base columnar cells in the intestine under control of the Lgr5 promoter (32). Whole body AhR knockout (AhR−/−) and wild-type littermate mice were also utilized (originally described by Schmidt et al (33)). Intestine-specific AhR knockout mice (villin Cre targeted) used for DSS-induced inflammation experiments were 8-18 wk of age and maintained on Research Diets #D10001. All mouse tails were genotyped and liver and intestine samples phenotyped with respect to AhR mRNA and protein expression (see Supplemental Methods for details).
2.6. Single cell isolation and cell sorting
Colon crypts were isolated as previously described (34). Crypt suspensions were dissociated into individual cells with 0.25% trypsin-EDTA containing 200 U/mL DNase and filtered through a 40-μm mesh. GFP-expressing stem cells were collected using Bio-Rad S3e Cell Sorter, with dead cells excluded by propidium iodide staining. Sorted cells were seeded (500 cells) onto 30 μl Matrigel containing 1 μM Jagged-1 (AnaSpec) in a flat bottom 24-well plate. Following Matrigel polymerization, cells were overlaid with 400 μL crypt culture medium containing Advanced DMEM/F12 (Gibco) supplemented with 2 mM glutamax, penicillin/streptomycin, 10 mM HEPES, 50 ng ml−1 EGF (LifeTechnologies), 100 ng mL−1 Noggin (Peprotech) or 0.2 μM LDN-193189 (Stemgent), 10% R-spondin conditioned medium, 1 μM N-acetyl-1-cysteine (Sigma), 1X N2 (LifeTechnologies), 1X B27 (Life Technologies) and 50% Wnt conditioned medium, supplemented with 10 μM Y-27632 (Sigma Y0503), 1 μM Jagged-1 and 2.5 μM CHIR99021 (Stemgent, 04-0004). Y-27632, Jagged-1 and CHIR99021 were withdrawn from crypt culture medium 2 d after plating. The treatments were included in the media starting at initial plating and replaced every 36 h. Treatments were DMSO as vehicle control, 5 or 10 μM norharman, and 1x, 2.5x or 5x coffee extract which is equivalent to the aqueous extract from 3.4, 8.5 and 17 μg of coffee beans, respectively. Organoid size was quantified on day 6 of culture using a Keyence BZ-X710 Microscope and software. Organoid forming efficiency was calculated as the percent of stem cells that formed organoids with diameter > 30 μm.
2.7. Fecal metabolite analysis
Mice were treated by oral gavage with 100 μL (volume 10% of body weight) coffee extract (equivalent to an extract of 34 mg of coffee beans) administered every 8 h for 4 d. Fecal samples were obtained after the 4 d treatment period. Samples were kept on ice during extractions. Metabolites were extracted from fecal pellets with chloroform/methanol/water as previously described (35). Untargeted liquid chromatography high resolution accurate mass spectrometry (LC-HRAM) analysis was performed on a Q Exactive Plus orbitrap mass spectrometer (Thermo Scientific, Waltham, MA) coupled to a binary pump HPLC (UltiMate 3000, Thermo Scientific). Full MS spectra were obtained in the positive mode at 70,000 resolution (200 m/z) with a scan range of 50-750 m/z. Full MS followed by ddMS2 scans were obtained at 35,000 resolution (MS1) and 17,500 resolution (MS2) with a 1.5 m/z isolation window and a stepped NCE (20, 40, 60). Samples were maintained at 4 °C before injection. The injection volume was 10 μL. Chromatographic separation was achieved on a Synergi Fusion 4μm, 150 mm x 2 mm reverse phase column (Phenomenex, Torrance, CA) maintained at 30 °C using a solvent gradient method. Solvent A was water (0.1% formic acid). Solvent B was methanol (0.1% formic acid). The gradient method used was 0-5 min (10% B to 40% B), 5-7 min (40% B to 95% B), 7-9 min (95% B), 9-9.1 min (95% B to 10% B), 9.1-13 min (10% B). The flow rate was 0.4 mL min-1. Sample acquisition was performed Xcalibur (Thermo Scientific). Data analysis was performed with Compound Discoverer 3.1 (Thermo Scientific).
Deuterated indole-3-acetic acid was used as a labeled internal quality control standard and data were analyzed using the Progenesis QI software (Waters) and the Human Metabolome Database (HMDB) and the Kegg databases. Raw abundance data were normalized to fecal sample weights and statistical analysis was performed using Kaleida Graph (Synergy).
2.8. Effects of coffee extracts on DSS-induced inflammation
Whole body AhR knockout mice (AhR−/−) (4-6 mice) and wild-type controls (10-15 mice), intestine-specific (villin Cre) AhR knockout and wildtype mice (10-12 and 10-13 mice respectively), were acclimated on Research Diets #D10001 for 2 wk with 12 h light/dark cycle before receiving aqueous coffee extract (1% of body weight, equivalent to the extract of ~70 mg of coffee on average) by gavage 3 times per day (~7 am, 2 pm and 9 pm) for the remainder of each experiment. Distal colon sections were assessed for inflammation and injury by a board-certified pathologist (M.E.H.) and scored on a scale of 0-3 as previously described (36).
2.9. Statistics
The in vitro studies were repeated at least 3 times and results are expressed as means ± SD and organoid results are expressed as means ± SE. Statistical significance for both in vitro and organoid studies were analysis of variance (ANOVA) or by a two-tailed unpaired students t test. For LC-MS, raw abundance data were normalized to fecal weights and statistically analyzed using the Mann-Whitney test at a significance level of p<0.05. Barrier function, histology scores and mucosal mRNA data were analyzed by 1-way ANOVA and Fisher’s LSD test for comparison between groups.
3. Results
The AhR agonist activities of coffee extracts were determined in Caco2 colon cancer cells, YAMC mouse colonocytes and their corresponding AhR knockout cells (Caco2-AhRKO and YAMC-AhRKO) previously generated by silencing of the AhR by CRISPR/Cas9 (29,30). The AhR agonist activities were determined using CYP1A1, CYP1B1 and UGT1A1 as Ah-responsive genes and 2,3,7,8-TCDD as an AhR ligand reference standard. Roasted coffee was extracted with boiling water as previously described (28) and an aliquot of the aqueous extract was extracted with chloroform and effects of equivalent amounts of water and chloroform extracts on AhR activity were investigated. The concentrations used (0.5 and 1.0 %) refer to the % of the crude coffee extract in the culture media (e.g.: 10 μl of coffee extract/ml media = 1.0%). In wild-type Caco2 cells both aqueous and chloroform extracts induced CYP1A1 (Fig. 1A), CYP1B1 (Fig. 1B) and UGTA1 (Fig. 1C) whereas no significant induction of these genes was observed in Caco2-AhRKO cells. There was some variability in the magnitude of the response by the chloroform and water extracts for induction of CYP1B1 and the coffee extracts induced 40-60% (CYP1A1) to ≥ 100% (UGT1A1) of the responses observed for 10 nM TCDD. The chloroform extracts induced CYP1A1 (Fig. 2A) and CYP1B1 (Fig. 2B) but only minimally induced UGTA1 (Fig. 2C) in YAMC cells whereas the water extracts were inactive as inducers of these responses. In contrast, the effects of TCDD as an inducer of CYP1A1 and CYP1B1 were similar in YAMC and Caco2 cells, however, in the former cell line only minimal induction of UGT1A1 was observed for TCDD and no induction was observed in YAMC-AhRKO cells. Thus, YAMC cells were less responsive to induction of AhR-responsive genes following incubation with the coffee extracts. The data also suggests that some cell-type specific inhibitors of Ah-responsive gene expression in YAMCs are expressed in the aqueous but not chloroform extracts of roasted coffee.
Figure 1.
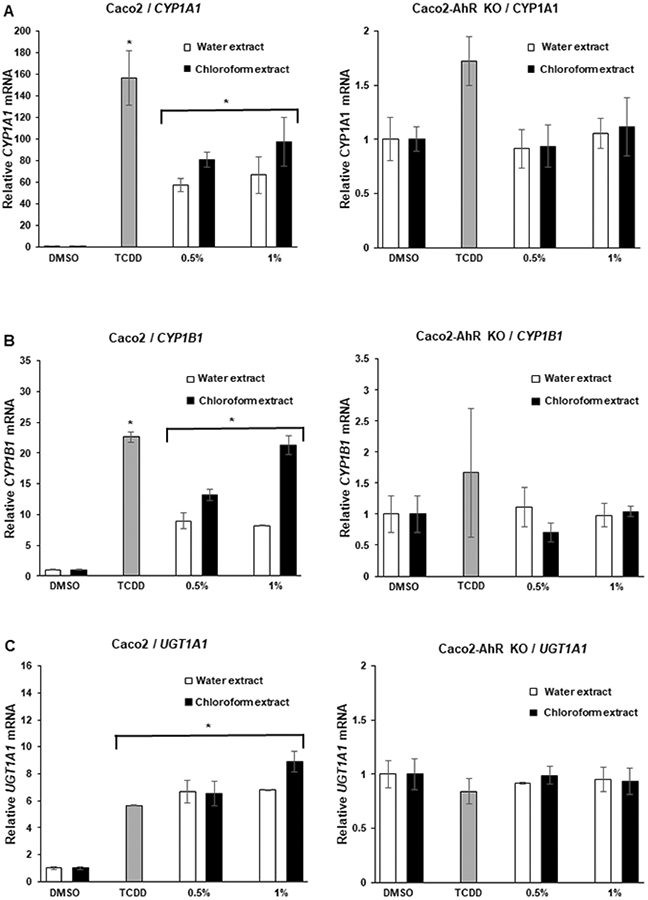
Coffee induces AhR-responsive genes in Caco2 cells. Caco2 wild-type and AhR knockout (Caco2-AhRKO) cells were treated with DMSO, 10 nM TCDD, water or chloroform extracts of coffee for 24 h and changes in expression of (A) CYP1A1, CYP1B1, (B) UGT1A1, (C) mRNA were determined by real time PCR. Results are expressed as means ± SE for at least 3 separate determinations per treatment group and significant (p<0.05) induction is indicated (*).
Figure 2.
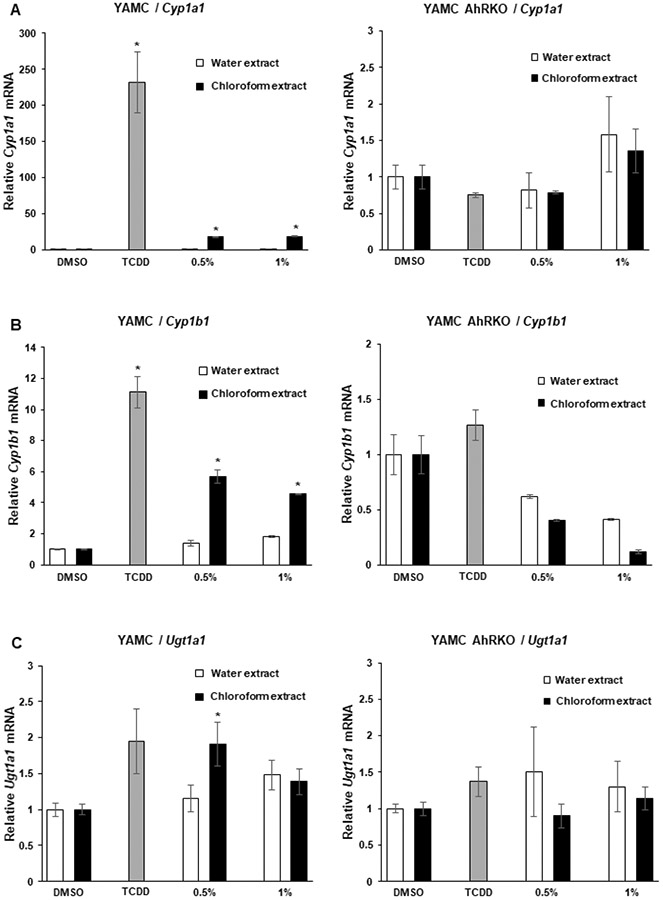
Coffee induces AhR-responsiveness in YAMC cells. YAMC wild-type and YAMC-AhRKO cells were treated with DMSO, 10 nM TCDD, water or chloroform extracts of coffee for 24 h and changes in expression of (A) CYP1A1 CYP1B1, (B) UGT1A1, (C) mRNA were determined by real time PCR. Results are expressed as means ± SE for at least 3 separate determinations per treatment group and significant (p<0.05) induction is indicated (*).
The response- and cell type-dependent differences in the AhR activity of aqueous and chloroform coffee extracts and TCDD are consistent with their activity as selective AhR modulators (SAhRMs), which have previously been observed for other structurally diverse AhR microbial metabolites (29, 30). We subsequently used the more responsive Caco2 cell line to compare the activity of roasted vs unroasted (green) coffee and the results (Supplemental Figure 1) show that unroasted coffee extracts did not induce AhR-responsive genes. In addition, we also separated the chloroform extracts by thin-layer chromatography to characterize and quantitate by weight the caffeine band (middle, 68%) less polar (top, 25%) and more polar (bottom, 7%) bands. The AhR activities of caffeine and the more and less polar bands were also determined in Caco2 cells and induction of CYP1A1, CYP1B1 and UGT1A1 was observed for both bands but not caffeine (Supplemental Figure 2) indicating the presence of multiple AhR-active compounds with different polarities.
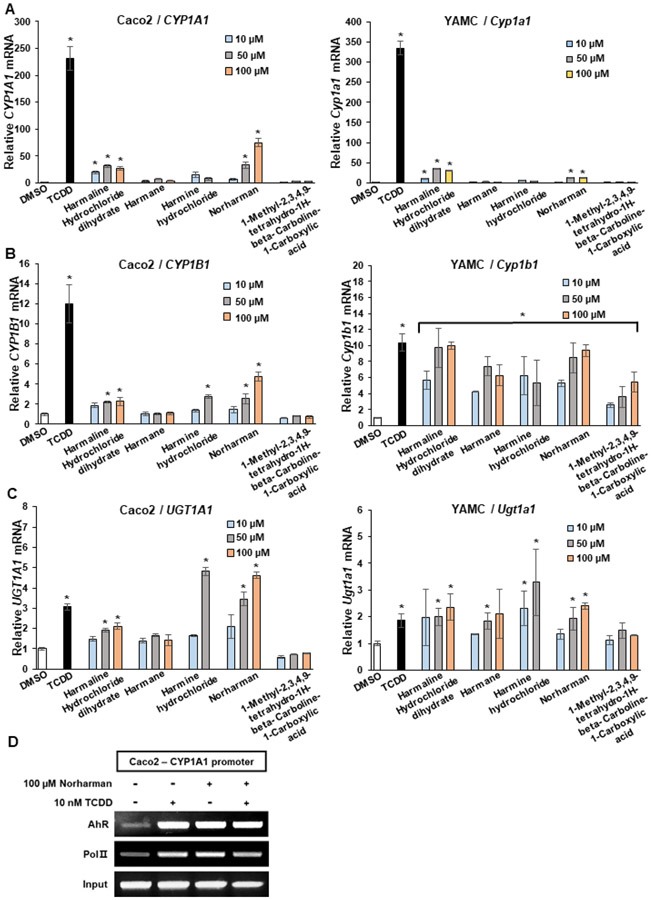
Harman and norharman are β-carboline alkaloids previously detected in brewed coffee (2) and there are prior reports indicating that these compounds are AhR-active. Results illustrated in Figure 3A show that norharman, harman, harmaline and harmine hydrochloride (10-100 μM) induced CYP1A1 in Caco2 and YAMC cells and their induction activity was more potent in Caco2 than YAMC cells. Norharman, harmine and harmaline hydrochloride induced CYP1B1 in Caco2 cells (< 40% of TCDD), whereas all the β-carboline derivatives induced CYP1B1 in YAMC cells and their response was > 50 to 100% of that observed for TCDD (Fig. 3B). Similar results were observed for induction of UGT1A1 in YAMC cells (Fig. 3C). In Caco2 cells both harmine hydrochloride and norharman were more potent than TCDD as inducers of UGT1A1 (Fig. 3C). We also examined the effects of TCDD and norharman treatment of Caco2 cells in a ChIP assay and both compounds enhanced AhR binding to the DRE region of the CYP1A1 promoter and increased RNA polymerase II (pol II) interactions with the promoter (Fig. 3D). The band intensities induced by norharman and TCDD were similar even though the former compound was a weaker inducer of CYP1A1. These results confirm the AhR activity of some β-carbolines present in coffee extracts and ongoing studies are using LC fractionation techniques to identify other AhR-active compounds in the aqueous coffee extracts.
Figure 3.
β-Carboline analogs serve as AhR agonists. Caco2 and YAMC cells were treated with DMSO, 10 nM TCDD, water or chloroform extracts of coffee for 24 h and changes in expression of (A) CYP1A1, CYP1B1, (B) UGT1A1, (C) mRNA were determined by real time PCR. (D) Caco2 cells were treated with DMSO, 10 nM TCDD and 100 μM norharman alone or in combination. Interactions of the AhR and pol II with the CYP1A1 promoter were determined in a ChIP assay. Results (A-C) are expressed as means ± SE for at least 3 separate determinations per treatment group and significant (p<0.05) induction is indicated (*).
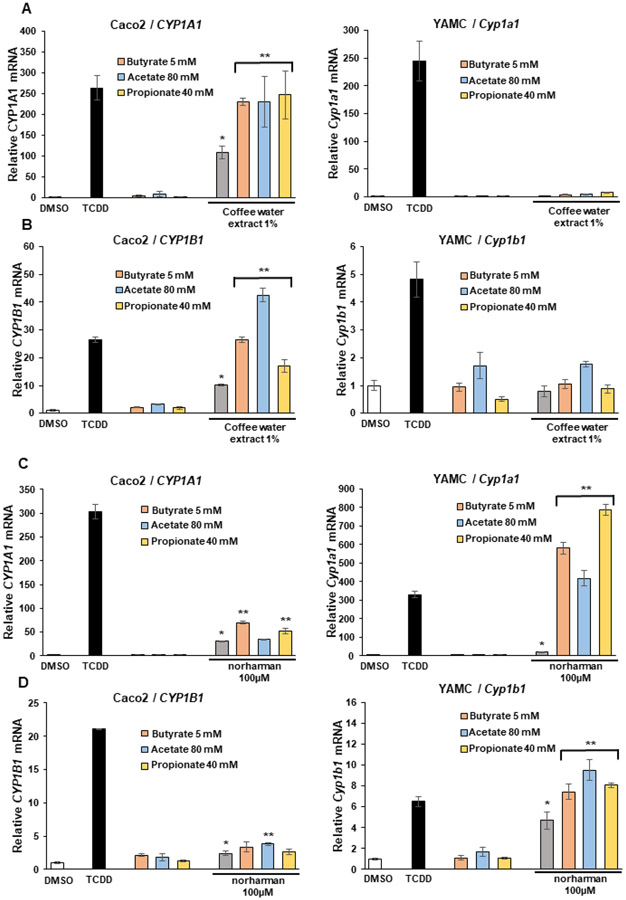
Our previous studies show that short chain fatty acids (SCFAs) e.g., butyrate, propionate and acetate, enhance AhR ligand induced CYP1A1 and CYP1B1 expression in Caco2 and YAMC cells (30,37). This could be due, in part, to their activity as histone deacetylase inhibitors (29) and it has been reported that butyrate binds and activates the AhR (38). Results in Figure 4A show that all three SCFAs enhance coffee-induced CYP1A1 in Caco2 but not in YAMC cells (Fig. 4A). SCFAs also enhanced coffee induced CYP1B1 in Caco2 but not in YAMC cells (Fig. 2B). Butyrate and propionate also enhanced norharman-induced CYP1A1 (Fig. 4C) and acetate enhanced induction of CYP1B1 (Fig. 4D) in Caco2 cells whereas all three SCFAs increased induction of CYP1A1 and CYP1B1 by norharman in YAMC cells. Thus, the pattern of induction of CYP1A1 and CYP1B1 by norharman and coffee and enhancement by SCFAs differed and was dependent on the response, the individual SCFA and cell context, and future studies will examine these interactions using inflammation endpoints.
Figure 4.
Interactions of coffee extracts and β-carboline analogs with SCFAs. Caco2 and YAMC cells were treated with DMSO, 10 nM TCDD, SCFAs alone and SCFAs in combination with (A, B) coffee extracts or (C, D) norharman and induction of (A, C) CYP1A1 and (B, D) CYP1B1 were determined by real time PCR. Results are expressed as means ± SE for at least 3 separate determinations per treatment group and significant (p<0.05) induction (*) and significantly enhanced induction after co-treatment with SCFAs is indicated (**).
The AhR functions in multiple cell types in the intestine to protect against infection, inflammation and cancer (24,39,40) and intestinal stem cells significantly contribute to these responses (24, 31). Therefore, we investigated the effects of coffee extracts on organoids derived from pooled intestinal stem cells from wild-type mice and mice in which intestinal AhR was silenced (VillinCreAhRFl/Fl) and fluorescence-activated cell sorting (FACS) was used to sort stem cells (defined as GFPhigh) on GFP expression (31). The coffee extract and norharman significantly decreased growth of organoids derived from wild type (AhR+/+) mice but not in AhR knockout mice (Fig. 5A) and this is consistent with previous studies demonstrating a role for the AhR and its ligands as inhibitors of intestinal stem cell expansion (24, 31). Figures 5B and 5C illustrate the inhibition of organoid growth by coffee extracts (5X) and 10 μM norharman. Loss of the AhR increased organoid forming efficiency, however, neither coffee or norharman affected this response (Fig. 5D). Thus, coffee extracts and one of the AhR-active components of coffee (norharman) restrict intestinal stem cell proliferation and this response was AhR-dependent.
Figure 5.
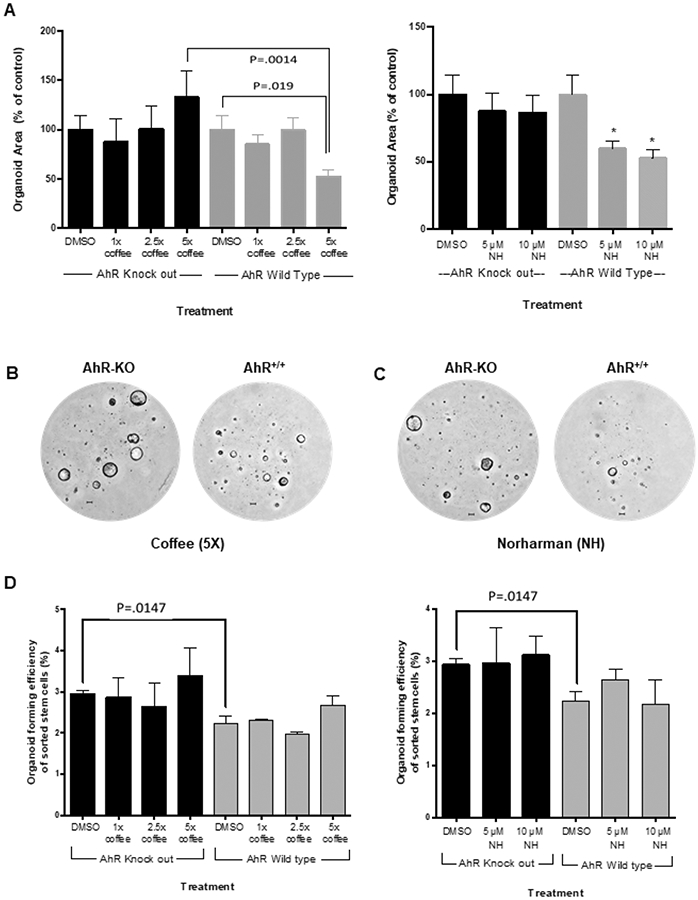
Effects of coffee extracts and norharman on colon stem cell derived organoid growth and organoid forming efficiency. (A) Effects of coffee and norharman on intestinal stem cell enriched organoid growth. Pooled stem cells from three mice were plated in Matrigel and treated with DMSO, coffee extracts or norharman every 36 h for 6 d and total organoids were quantified in the different treatment groups with the solvent control (DMSO) set at 100%. Results are expressed as means ± SE for 3 separate experiments for each treatment group and significant (p < 0.05) inhibition by coffee extracts or norharman is indicated (*). AhR knockout and AhR+/+-derived organoids growing in Matrigel and treated with (B) 5x coffee extracts and (C) 10 μM norharman are illustrated. (D) Effects of coffee extracts and norharman on organoid forming efficiencies. The percentage of sorted stem cells that develop into organoids was determined using the same treatment groups and growth conditions as indicated in (A). Results are expressed as means ± SE for at least three replicate determinations for each treatment group and significant differences were observed in solvent (DMSO) control organoids derived from wild-type and AhR-intestinal knockout mice.
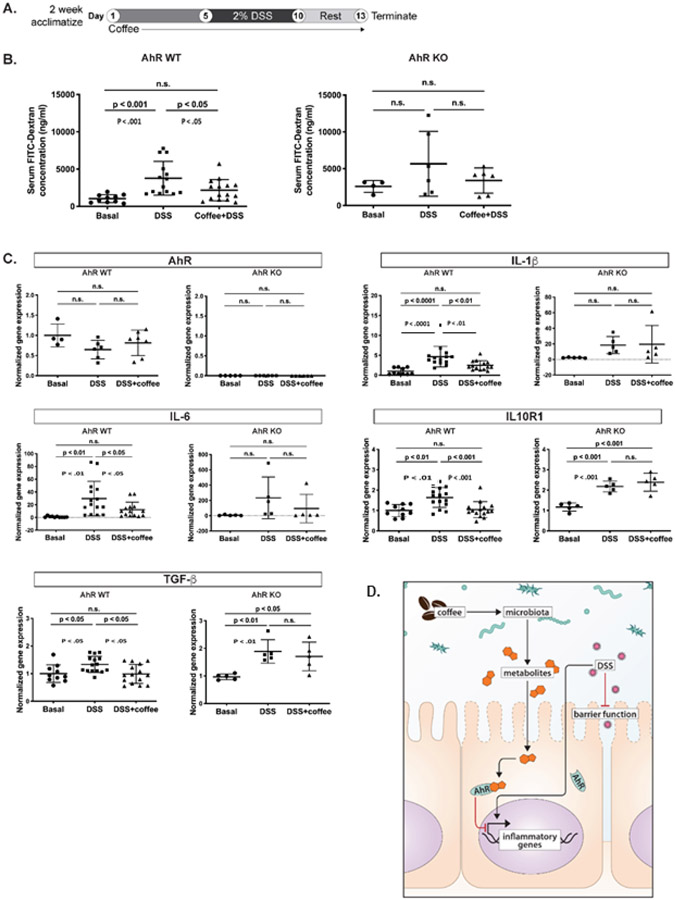
In vivo studies using a DSS-induced inflammation model in AhR wild type and AhR whole body knock out (AhR−/−) mice (Fig. 6A) showed that the coffee extract significantly reversed the effects of DSS-induced barrier permeability in AhR-WT but not AhR-KO as demonstrated by inhibition of serum levels of FITC-dextran (Fig. 6B). In addition, we also examined effects of coffee extract on inhibition of DSS-induced gene expression in intestinal mucosa and observed that coffee extracts inhibited DSS-induced IL-6, IL-1β, TGF-β1 and IL-10R1 mRNA levels in AhR-WT but not AhR-KO (Figs. 6C). DSS-induced effects on mucosal IFNγ, IL-10, IL-10R2 and Muc2 mRNA were not affected by coffee in wild-type or AhR-KO mice, nor were the AhR responsive genes Cyp1a1, Cyp1b1 or Ugt1a1 (Supplemental Figs. 3A-3G). However, the beneficial effects of coffee on barrier integrity and gene expression were not correlated with clinical markers of inflammation, where DSS induced inflammation only in the AhR KO model (Supplemental Fig. 4A&B). Coffee did not affect DSS-induced body weight changes in wild-type and AhR-KO mice (Supplemental Fig. 4D). Effects of coffee and DSS on colon length and liver weight were similar in wild-type and AhR-KO mice (Supplemental Fig. 4E&F). Using a similar protocol, we also examined the effects of coffee extracts on control and Villin Cre intestinal AhR knockout mice and observed no AhR-dependent protective effects (Supplemental Table S2). A summary diagram highlighting the AhR-dependent ability of coffee extract to suppress DSS-induced damage to intestinal barrier function and DSS-induced mucosal inflammatory genes is shown in Figure 6D.
Figure 6.
Coffee extracts inhibit DSS-induced intestinal responses in wild-type and AhR whole body knockout mice. (A) Overall treatment protocol for the experiment. (B) Barrier function was determined by accumulation of FITC-dextran into serum as outlined in the Materials and Methods. Effects of coffee on DSS-induced mucosal IL-6 and IL-1β, TGF-β and IL10R1. (C) mRNA levels were determined as outlined in the Methods. Results are expressed as means ± SD for at least five determinations per treatment group and significance as indicated in the figure. (D) Summary diagram highlighting the AhR-dependent ability of coffee extract to suppress DSS-induced damage, intestinal barrier function and DSS-induced mucosal inflammatory genes.
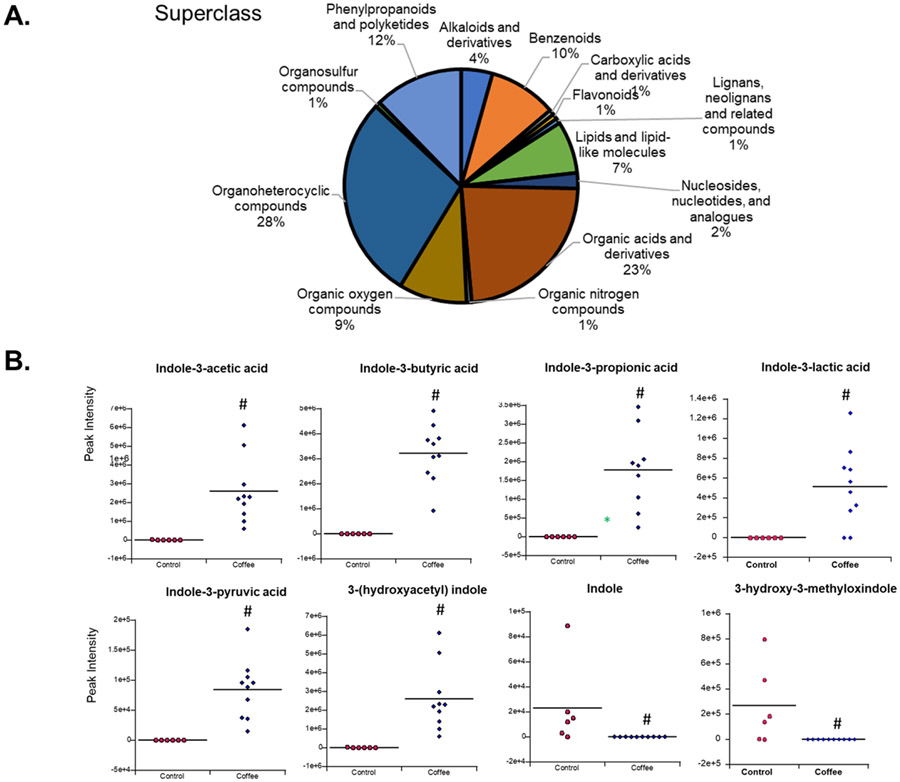
We also investigated the effects of coffee extracts on microbial-derived tryptophan metabolites and other indole derivatives. Coffee extracts and fecal pellets from control mice and mice receiving coffee extracts by oral gavage for 4 d were analyzed using LC-MS. Several hundred metabolites were detected in coffee extracts as previously reported (2). Supplemental Table S3 summarizes the microbial-derived tryptophan/indole-derived metabolites that were present only in coffee extracts and those that are present fecal material from control mice but are not detected in coffee extracts. Figure 7A shows the various classes of organic compounds identified in coffee extracts by LC-MS analysis. Figure 7B shows the relative abundances of indole derivatives including indole, 3-hydroxy-3-methyloxindole, indole-3-propionic acid, indole-3-butyric acid, indole-3-pyruvic acid, indole-3-lactic acid, indole-3-acetic acid, and 3-(hydroxyacetyl)indole.
Figure 7.
Coffee-induced changes in microbial metabolites. (A) LC-MS analysis of aqueous coffee extracts and separation into chemical classes. B. Fecal levels of several tryptophan/indole – derived metabolites that were significantly (p<0.05) altered (#) in the coffee treated group compared to controls.
There was a statistically significant increase in the abundance of several AhR-active indole-3-acids in feces from mice in the coffee group relative to control mice. Among these, previous studies reported the AhR activity of indole-3-acetic and pyruvic acids (41, 42) and the latter compound also induced CYP1A1 and CYP1B1 in both Caco2 and YAMC cells (Supplemental Figure 5) whereas indole-3-acetic acid was a weak inducer of only CYP1A1 in Caco2 cells. We also observed that administration of coffee extracts decreased fecal abundance of indole and 3-hydroxy-3-methyloxy-indole. Microbial populations in this study were relatively unchanged by coffee administration indicating that coffee extracts primarily modulate the relative amounts of specific tryptophan metabolites and current studies are focused on the relative levels of these compounds and their AhR activity.
4. Discussion
The AhR was initially identified as the intracellular receptor that mediated the toxicities induced by TCDD and structurally related halogenated aromatics (41). However, subsequent and ongoing studies show that the AhR also binds structurally diverse compounds including pharmaceuticals, endogenous biologicals, and health-promoting phytochemicals and microbial metabolites including several tryptophan metabolites (29,30,37,42-46). Whole body and cell-type specific AhR knockout mouse models demonstrate multiple endogenous functions for the AhR including a role for the AhR in protecting against gut infection, barrier function, inflammation and tumorigenesis. These AhR-dependent responses involve multiple cell types including intestinal epithelial cells and stem cells, innate lymphoid cells, Treg immune cells and intraepithelial lymphocytes (22,31,45-50). Moreover, AhR ligands including indole-3-carbinol, tryptophan metabolites and broccoli extracts protect against intestinal inflammation and cancer in mouse models (39-43). Since there are several reports that coffee consumption in humans is also associated with protective effects in terms of gut health (13-17), we investigated the Ah-responsiveness of coffee and some of its components in colon-derived human Caco2 and YAMC cells. In addition, we also investigated the AhR-mediated impact of coffee extracts on intestinal stem cell mediated growth, DSS-induced intestinal inflammation and changes in the microbiome and microbial metabolites.
We show that the AhR activity of coffee was due to the roasting process. In human Caco2 cells, boiling water extracts of coffee and a chloroform extract of the aqueous extract induced the expression of Ah-responsive CYP1A1, CYP1B1 AND UGT1A1 genes in wild-type but not AhR-silenced Caco2 cells. UGT1A1 was only minimally induced in mouse YAMC cells and in this cell line we observed that the chloroform but not the aqueous extract induced CYP1A1 and CYP1B1. The Ah-responsiveness of coffee extracts was substantially higher in Caco2 than in YAMC cells, however, the enhanced loss of inducibility by the aqueous extract in YAMC cells was surprising and may be due to some cell (YAMC)-specific inhibitory factors in the aqueous extract that we are currently investigating.
Our initial LC-MS screening of individual compounds in coffee extracts focused on substituted aromatic and heteroaromatics and this included both harman and norharman which have previously been identified in water extracts of roasted coffee (1,2). A previous study on activation of an Ah-responsive reported gene assay (DRE-luc) in mouse hepatoma cells showed that among a series of structurally-related β-carbolines (including harman and norharman) only harman-1,2,3,4-tetrahydro-3-carboxylic acid induced activity (51). In contrast, our results show that norharman induced CYP1A1, CYP1B1 and UGT1A1 in Caco2 and YAMC cells and other β-carbolines also exhibited some AhR activity, particularly for induction of CYP1B1 in YAMC cells. Previously, we have reported that SCFAs enhance Ah-responsiveness of AhR ligands and this includes induction of CYP1A1 (30,37). These interactions are significant in terms of their potential to enhance AhR-dependent gut health, particularly since intestinal CYP1A1 expression is important in the formation of AhR-active microbial metabolites (52,53). Butyrate, propionate and acetate significantly enhance induction of CYP1A1 (Caco2 and YAMC) and CYP1B1 (Caco2) by coffee extracts and this microbial metabolite-AhR interaction potentially impacts host metabolism (54). In contrast, the SCFAs potently enhanced induction of CYP1A1 and CYP1B1 in YAMC cells by 100 μM norharman whereas minimal and SCFA-selective effects were observed in Caco2 cells. These differences in gene specific Ah-responsiveness between coffee extracts and norharman suggest that norharman may not be a major contributor to the induction of drug-metabolizing enzymes by coffee extracts.
The AhR plays an important role in intestinal stem cells and their functions on the maintenance of intestinal epithelial homeostasis, protection from inflammation and tumor development. It was recently reported that intestinal AhR expression decreased intestinal stem cell proliferation thereby inhibiting cell transformation and development of colon cancer (24,25,31). Both coffee extracts and norharman also inhibited intestinal stem cell-derived organoid growth in AhR-expressing but not in AhR-silenced stem cells and this further demonstrates the AhR activity of coffee in an important mediator of intestinal function. We also observed that coffee inhibited DSS-induced damage to barrier function, as well as IL-6, IL-1β, TGF-β1 and IL10R1 mRNA levels, however, these responses were not observed in AhR-KO mice. Although these genes exhibit variable functions, the induction of the pro-inflammatory IL-6 gene by DSS and inhibition by coffee extracts is consistent with the effects of coffee on barrier function damage. In contrast, coffee extracts did not affect DSS-induced inflammation in intestine-specific AhR knock out mice (Supplemental Table S2) and this contrasts with their ex-vivo effects on intestinal stem cells. These data suggest that AhR-mediated responses in tissues other than the intestine contribute to the protection against DSS-induced intestinal inflammation. These results are consistent with previous studies in dysregulated CYP1A1 and CARD9 mice which exhibit altered metabolism of microbial metabolites into AhR active compounds and reduced effects on intestinal damage (53,55). This indicates that the whole-body loss of AhR and possibly the loss of this receptor in liver may compromise the metabolism and subsequent formation of coffee-induced AhR-active microbial metabolites which, in turn are required for protection from intestinal inflammation. This would be observed in whole body but not intestinal-specific AhR-KO mice.
There is evidence that coffee consumption can affect microbial metabolites and the microbiome and this included a human study which measured a total of 115 metabolites associated with coffee intake (14,56-58). Interestingly, these investigators did not distinguish between compounds present in the coffee and coffee-induced changes in microbial metabolite levels but observed a coffee-like signature. In our study, we have identified tryptophan/indole-derived compounds that are present in coffee and fecal extracts and also those that are elevated in fecal extracts relative to the coffee extract. Interestingly, coffee consumption differentially affected levels of tryptophan/indole-derived microbial metabolites and this includes enhanced levels of indole-3-pyruvic acid which is a highly active AhR agonist. This is noteworthy, because indole-3-pyruvic acid inhibits colitis in a mouse model (42) and this may contribute to the coffee-induced anti-inflammatory activities.
In summary, human studies on coffee consumption indicate that among botanical extracts, consumption of roasted coffee extracts is associated with a wide range of beneficial health effects including enhanced gut resilience. However, mechanisms that link consumption of coffee to positive health outcomes are lacking and have been addressed, in part, in this study. Our results demonstrate that roasted coffee extracts exhibit AhR activity and induce an AhR-dependent decrease in intestinal stem cells in an organoid cell culture model and protect against DSS-induced intestinal barrier damage (Fig. 6D). These results demonstrate a role for the AhR in mediating some coffee-induced protective responses in the gut but do not exclude a role for other AhR-independent pathways induced by coffee. Currently, we are investigating other AhR-dependent health benefits linked to coffee consumption and other classes of AhR-active compounds present in aqueous coffee extracts.
Supplementary Material
Acknowledgements
Funding was provided by Texas AgriLife Research, the Sid Kyle Chair Endowment, the Allen Endowed Chair in Nutrition and Chronic Disease Prevention, the Cancer Prevention Research Institute of Texas (RP160589), and the National Institutes of Health (P30-ES029067, R01-CA202697, R01-AT010282 and R35-CA197707).
Abbreviations:
- AhR
aryl hydrocarbon receptor
- ANOVA
analysis of variance
- ATCC
American Type Culture Collection
- ChIP
chromatin immunoprecipitation
- CYP
cytochrome P450
- DMEM
Dulbecco’s modified Eagle’s medium
- FACS
fluorescence-activated cell sorting
- FBS
fetal bovine serum
- HMDB
Human Metabolome Database
- SCFA
short chain fatty acids
- TCDD
2,3,7,8-tetrachlorodibenzo-p-dioxin
- UGT
uridine diphosphate glucuronosyl transferase
- YAMC
young adult mouse colonocyte
Footnotes
Disclosure of Conflicts of Interest
There are no conflicts of interest to declare.
References
- 1.IARC. (2018) Drinking coffee, mate and very hot beverages. in IARC Monographs On The Evaluation Of Carcinogenic Risks To Humans. pp 37–84 [Google Scholar]
- 2.Farah A (2012) Coffee Constituents. in Coffee: Emerging Health Effects and Disease Prevention, First Edition Chu Yi-Fang (Chu, Y.-F. ed.). John Wiley and Sons; pp 21–58 [Google Scholar]
- 3.Loftfield E, Cornelis MC, Caporaso N, Yu K, Sinha R, and Freedman N (2018) Association of Coffee Drinking With Mortality by Genetic Variation in Caffeine Metabolism: Findings From the UK Biobank. JAMA Intern Med 178, 1086–1097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ding M, Satija A, Bhupathiraju SN, Hu Y, Sun Q, Han JL, Lopez-Garcia E, Willett W, van Dam RM, and Hu FB (2015) Association of Coffee Consumption With Total and Cause-Specific Mortality in 3 Large Prospective Cohorts. Circulation 132, 2305–2315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Freedman ND (2012) Association of Coffee Drinking with Total and Cause-Specific Mortality (vol 366, pg 1891, 2012). New Engl J Med 367, 285–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Crippa A, Discacciati A, Larsson SC, Wolk A, and Orsini N (2014) Coffee Consumption and Mortality From All Causes, Cardiovascular Disease, and Cancer: A Dose-Response Meta-Analysis. Am J Epidemiol 180, 763–775 [DOI] [PubMed] [Google Scholar]
- 7.Tran A, Zhang CY, and Chuanhai C (2015) The Role of Coffee in the Therapy of Parkinson's Disease. Journal of Alzheimers Disease & Parkinsonism 5, 1000203 [Google Scholar]
- 8.Ding M, Bhupathiraju SN, Satija A, van Dam RM, and Hu FB (2014) Long-term coffee consumption and risk of cardiovascular disease: a systematic review and a dose-response meta-analysis of prospective cohort studies. Circulation 129, 643–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Park SY, Freedman ND, Haiman CA, Le Marchand L, Wilkens LR, and Setiawan VW (2018) Prospective Study of Coffee Consumption and Cancer Incidence in Non-White Populations. Cancer Epidemiol Biomarkers Prev 27, 928–935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gapstur SM, Anderson RL, Campbell PT, Jacobs EJ, Hartman TJ, Hildebrand JS, Wang Y, and McCullough ML (2017) Associations of Coffee Drinking and Cancer Mortality in the Cancer Prevention Study-II. Cancer Epidemiol Biomarkers Prev 26, 1477–1486 [DOI] [PubMed] [Google Scholar]
- 11.Gan Y, Wu J, Zhang S, Li L, Cao S, Mkandawire N, Ji K, Herath C, Gao C, Xu H, Zhou Y, Song X, Chen S, Chen Y, Yang T, Li J, Qiao Y, Hu S, Yin X, and Lu Z (2017) Association of coffee consumption with risk of colorectal cancer: a meta-analysis of prospective cohort studies. Oncotarget 8, 18699–18711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marventano S, Salomone F, Godos J, Pluchinotta F, Del Rio D, Mistretta A, and Grosso G (2016) Coffee and tea consumption in relation with non-alcoholic fatty liver and metabolic syndrome: A systematic review and meta-analysis of observational studies. Clin Nutr 35, 1269–1281 [DOI] [PubMed] [Google Scholar]
- 13.Watanabe S, Takahashi T, Ogawa H, Uehara H, Tsunematsu T, Baba H, Morimoto Y, and Tsuneyama K (2017) Daily Coffee Intake Inhibits Pancreatic Beta Cell Damage and Nonalcoholic Steatohepatitis in a Mouse Model of Spontaneous Metabolic Syndrome, Tsumura-Suzuki Obese Diabetic Mice. Metab Syndr Relat Disord 15, 170–177 [DOI] [PubMed] [Google Scholar]
- 14.Nishitsuji K, Watanabe S, Xiao J, Nagatomo R, Ogawa H, Tsunematsu T, Umemoto H, Morimoto Y, Akatsu H, Inoue K, and Tsuneyama K (2018) Effect of coffee or coffee components on gut microbiome and short-chain fatty acids in a mouse model of metabolic syndrome. Sci. Rep 8, 16173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kashino I, Akter S, Mizoue T, Sawada N, Kotemori A, Matsuo K, Oze I, Ito H, Naito M, Nakayama T, Kitamura Y, Tamakoshi A, Tsuji I, Sugawara Y, Inoue M, Nagata C, Sadakane A, Tanaka K, Tsugane S, Shimazu T, Research Group for the, D., and Evaluation of Cancer Prevention Strategies in, J. (2018) Coffee drinking and colorectal cancer and its subsites: A pooled analysis of 8 cohort studies in Japan. Int J Cancer 143, 307–316 [DOI] [PubMed] [Google Scholar]
- 16.Ng SC, Tang W, Leong RW, Chen M, Ko Y, Studd C, Niewiadomski O, Bell S, Kamm MA, de Silva HJ, Kasturiratne A, Senanayake YU, Ooi CJ, Ling KL, Ong D, Goh KL, Hilmi I, Ouyang Q, Wang YF, Hu P, Zhu Z, Zeng Z, Wu K, Wang X, Xia B, Li J, Pisespongsa P, Manatsathit S, Aniwan S, Simadibrata M, Abdullah M, Tsang SW, Wong TC, Hui AJ, Chow CM, Yu HH, Li MF, Ng KK, Ching J, Wu JC, Chan FK, Sung JJ, Asia-Pacific C. s., and Colitis Epidemiology Study, A. G. (2015) Environmental risk factors in inflammatory bowel disease: a population-based case-control study in Asia-Pacific. Gut 64, 1063–1071 [DOI] [PubMed] [Google Scholar]
- 17.Gungorduk K, Ozdemir IA, Gungorduk O, Gulseren V, Gokcu M, and Sanci M (2017) Effects of coffee consumption on gut recovery after surgery of gynecological cancer patients: a randomized controlled trial. Am J Obstet Gynecol 216, 145 e141–145 e147 [DOI] [PubMed] [Google Scholar]
- 18.El-Sohemy A (2019) Coffee and health: what we still don't know. Am J Clin Nutr 109, 489–490 [DOI] [PubMed] [Google Scholar]
- 19.Hang D, Kvaerner AS, Ma W, Hu Y, Tabung FK, Nan H, Hu Z, Shen H, Mucci LA, Chan AT, Giovannucci EL, and Song M (2019) Coffee consumption and plasma biomarkers of metabolic and inflammatory pathways in US health professionals. Am J Clin Nutr 109, 635–647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Safe S, Jayaraman A, and Chapkin RS (2020) Ah receptor ligands and their impacts on gut resilience: structure-activity effects. Critical reviews in toxicology 50, 463–473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Denison MS, and Faber SC (2017) And Now for Something Completely Different: Diversity in Ligand-Dependent Activation of Ah Receptor Responses. Curr Opin Toxicol 2, 124–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kawajiri K, Kobayashi Y, Ohtake F, Ikuta T, Matsushima Y, Mimura J, Pettersson S, Pollenz RS, Sakaki T, Hirokawa T, Akiyama T, Kurosumi M, Poellinger L, Kato S, and Fujii-Kuriyama Y (2009) Aryl hydrocarbon receptor suppresses intestinal carcinogenesis in ApcMin/+ mice with natural ligands. Proc Natl Acad Sci USA 106, 13481–13486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Diaz-Diaz CJ, Ronnekleiv-Kelly SM, Nukaya M, Geiger PG, Balbo S, Dator R, Megna BW, Carney PR, Bradfield CA, and Kennedy GD (2016) The Aryl Hydrocarbon Receptor is a Repressor of Inflammation-associated Colorectal Tumorigenesis in Mouse. Ann Surg 264, 429–436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Metidji A, Omenetti S, Crotta S, Li Y, Nye E, Ross E, Li V, Maradana MR, Schiering C, and Stockinger B (2018) The Environmental Sensor AHR Protects from Inflammatory Damage by Maintaining Intestinal Stem Cell Homeostasis and Barrier Integrity. Immunity 49, 353–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Han H, Davidson LA, Hensel M, Yoon G, Landrock K, Allred C, Jayaraman A, Ivanov I, Safe SH, and Chapkin RS (2021) Loss of Aryl Hydrocarbon Receptor Promotes Colon Tumorigenesis in Apc(S580/+); Kras(G12D/+) Mice. Molecular cancer research : MCR 19, 771–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wada T, Sunaga H, Miyata K, Shirasaki H, Uchiyama Y, and Shimba S (2016) Aryl Hydrocarbon Receptor Plays Protective Roles against High Fat Diet (HFD)-induced Hepatic Steatosis and the Subsequent Lipotoxicity via Direct Transcriptional Regulation of Socs3 Gene Expression. J Biol Chem 291, 7004–7016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Abbott BD, Schmid JE, Pitt JA, Buckalew AR, Wood CR, Held GA, and Diliberto JJ (1999) Adverse reproductive outcomes in the transgenic Ah receptor-deficient mouse. Toxicol. Appl. Pharmacol 155, 62–70 [DOI] [PubMed] [Google Scholar]
- 28.Ishikawa T, Takahashi S, Morita K, Okinaga H and Teramoto T, (2014) Induction of AhR-mediated gene transcription by coffee. PLoS ONE 9, e102152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cheng Y, Jin UH, Davidson LA, Chapkin RS, Jayaraman A, Tamamis P, Orr A, Allred C, Denison MS, Soshilov A, Weaver E, and Safe S (2017) Editor's Highlight: Microbial-Derived 1,4-Dihydroxy-2-naphthoic Acid and Related Compounds as Aryl Hydrocarbon Receptor Agonists/Antagonists: Structure-Activity Relationships and Receptor Modeling. Toxicol Sci 155, 458–473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jin UH, Park H, Li X, Davidson LA, Allred C, Patil B, Jayaprakasha G, Orr AA, Mao L, Chapkin RS, Jayaraman A, Tamamis P, and Safe S (2018) Structure-Dependent Modulation of Aryl Hydrocarbon Receptor-Mediated Activities by Flavonoids. Toxicol Sci 164, 205–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Han H, A. D. L, Fan YY, Goldsby J, Yoon G, Jin U, Wright GA, Landrock K, Weeks BR, Wright RC, Allred CD, Jayaraman A, Ivanov I, Roper J, Safe SH and Chapkin RS. (2020) Loss of aryl hydrocarbon receptor signaling potentiates FoxM1 signaling to enhance self-renewal of colonic crypt Lgr5+ stem and progenitor cells. . EMBO J. 2020;39: e104319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Barker N, van Es JH, Kuipers J, Kujala P, van den Born M, Cozijnsen M, Haegebarth A, Korving J, Begthel H, Peters PJ, and Clevers H (2007) Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature 449, 1003–1007 [DOI] [PubMed] [Google Scholar]
- 33.Schmidt JV, Su GH, Reddy JK, Simon MC, and Bradfield CA (1996) Characterization of a murine AhR null allele: involvement of the Ah receptor in hepatic growth and development. Proc Natl Acad Sci USA 93, 6731–6736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fan YY, Davidson LA, and Chapkin RS (2016) Murine Colonic Organoid Culture System and Downstream Assay Applications. Methods Mol. Biol 2019;1576:171–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.DeLuca JA, Allred KF, Menon R, Riordan R, Weeks BR, Jayaraman A, and Allred CD (2018) Bisphenol-A alters microbiota metabolites derived from aromatic amino acids and worsens disease activity during colitis. Exp Biol Med (Maywood) 243, 864–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jia Q, Lupton JR, Smith R, Weeks BR, Callaway E, Davidson LA, Kim W, Fan YY, Yang P, Newman RA, Kang JX, McMurray DN, and Chapkin RS (2008) Reduced colitis-associated colon cancer in Fat-1 (n-3 fatty acid desaturase) transgenic mice. Cancer Res. 68, 3985–3991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jin UH, Cheng Y, Park H, Davidson LA, Callaway ES, Chapkin RS, Jayaraman A, Asante A, Allred C, Weaver EA, and Safe S (2017) Short Chain Fatty Acids Enhance Aryl Hydrocarbon (Ah) Responsiveness in Mouse Colonocytes and Caco-2 Human Colon Cancer Cells. Sci Rep 7, 10163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Marinelli L, Martin-Gallausiaux C, Bourhis JM, Beguet-Crespel F, Blottiere HM, and Lapaque N (2019) Identification of the novel role of butyrate as AhR ligand in human intestinal epithelial cells. Sci Rep 9, 643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Esser C, and Rannug A (2015) The aryl hydrocarbon receptor in barrier organ physiology, immunology, and toxicology. Pharmacol Rev 67, 259–279 [DOI] [PubMed] [Google Scholar]
- 40.Garcia-Villatoro EL, DeLuca JAA, Callaway ES, Allred KF, Davidson LA, Hensel ME, Menon R, Ivanov I, Safe SH, Jayaraman A, Chapkin RS, and Allred CD (2020) Effects of high-fat diet and intestinal aryl hydrocarbon receptor deletion on colon carcinogenesis. Am J Physiol Gastrointest Liver Physiol 318, G451–G463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Poland A, Glover E, and Kende AS (1976) Stereospecific, high affinity binding of 2,3,7,8-tetrachlorodibenzo-p-dioxin by hepatic cytosol. Evidence that the binding species is receptor for induction of aryl hydrocarbon hydroxylase. J Biol Chem 251, 4936–4946 [PubMed] [Google Scholar]
- 42.Hubbard TD, Murray IA, and Perdew GH (2015) Indole and Tryptophan Metabolism: Endogenous and Dietary Routes to Ah Receptor Activation. Drug Metab. Dispos 43, 1522–1535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Aoki R, Aoki-Yoshida A, Suzuki C, and Takayama Y (2018) Indole-3-Pyruvic Acid, an Aryl Hydrocarbon Receptor Activator, Suppresses Experimental Colitis in Mice. J. Immunol 201, 3683–3693 [DOI] [PubMed] [Google Scholar]
- 44.Denison MS, Soshilov AA, He G, DeGroot DE, and Zhao B (2011) Exactly the same but different: promiscuity and diversity in the molecular mechanisms of action of the aryl hydrocarbon (dioxin) receptor. Toxicol Sci 124, 1–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zelante T, Iannitti RG, Cunha C, De Luca A, Giovannini G, Pieraccini G, Zecchi R, D'Angelo C, Massi-Benedetti C, Fallarino F, Carvalho A, Puccetti P, and Romani L (2013) Tryptophan catabolites from microbiota engage aryl hydrocarbon receptor and balance mucosal reactivity via interleukin-22. Immunity 39, 372–385 [DOI] [PubMed] [Google Scholar]
- 46.Kiss EA, Vonarbourg C, Kopfmann S, Hobeika E, Finke D, Esser C, and Diefenbach A (2011) Natural aryl hydrocarbon receptor ligands control organogenesis of intestinal lymphoid follicles. Science 334, 1561–1565 [DOI] [PubMed] [Google Scholar]
- 47.Scott SA, Fu J, and Chang PV (2020) Microbial tryptophan metabolites regulate gut barrier function via the aryl hydrocarbon receptor. Proc. Natl. Acad. Sci. U. S. A (2020) August 11:117(32):19376–19387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Han H, A. J., Safe S and Chapkin RS. . (2021) Targeting the aryl hydrocarbon receptor in stem cells to improve the use of food as medicine. Current Stem Cell Reports (2020) 6:109–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Han H, A. J., Safe S and Chapkin. RS (2021) Diet-microbiota interactions shape aryl hydrocarbon receptor ligand production to modulate intestinal homeostasis. Annual Review of Nutrition, in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Islam J, Sato S, Watanabe K, Watanabe T, Ardiansyah, Hirahara K, Aoyama Y, Tomita S, Aso H, Komai M, and Shirakawa H (2017) Dietary tryptophan alleviates dextran sodium sulfate-induced colitis through aryl hydrocarbon receptor in mice. J Nutr Biochem 42, 43–50 [DOI] [PubMed] [Google Scholar]
- 51.Seidel SD, Li V, Winter GM, Rogers WJ, Martinez EI, and Denison MS (2000) Ah receptor-based chemical screening bioassays: application and limitations for the detection of Ah receptor agonists. Toxicol Sci 55, 107–115 [DOI] [PubMed] [Google Scholar]
- 52.Natividad JM, Agus A, Planchais J, Lamas B, Jarry AC, Martin R, Michel ML, Chong-Nguyen C, Roussel R, Straube M, Jegou S, McQuitty C, Le Gall M, da Costa G, Lecornet E, Michaudel C, Modoux M, Glodt J, Bridonneau C, Sovran B, Dupraz L, Bado A, Richard ML, Langella P, Hansel B, Launay JM, Xavier RJ, Duboc H, and Sokol H (2018) Impaired Aryl Hydrocarbon Receptor Ligand Production by the Gut Microbiota Is a Key Factor in Metabolic Syndrome. Cell Metab 28, 737–749 e734 [DOI] [PubMed] [Google Scholar]
- 53.Schiering C, Wincent E, Metidji A, Iseppon A, Li Y, Potocnik AJ, Omenetti S, Henderson CJ, Wolf CR, Nebert DW, and Stockinger B (2017) Feedback control of AHR signalling regulates intestinal immunity. Nature 542, 242–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Korecka A, Dona A, Lahiri S, Tett AJ, Al-Asmakh M, Braniste V, D'Arienzo R, Abbaspour A, Reichardt N, Fujii-Kuriyama Y, Rafter J, Narbad A, Holmes E, Nicholson J, Arulampalam V, and Pettersson S (2016) Bidirectional communication between the aryl hydrocarbon receptor (AhR) and the microbiome tunes host metabolism. Biofilms and Microbiomes 2, 16014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lamas B, Richard ML, Leducq V, Pham HP, Michel ML, Da Costa G, Bridonneau C, Jegou S, Hoffmann TW, Natividad JM, Brot L, Taleb S, Couturier-Maillard A, Nion-Larmurier I, Merabtene F, Seksik P, Bourrier A, Cosnes J, Ryffel B, Beaugerie L, Launay JM, Langella P, Xavier RJ, and Sokol H (2016) CARD9 impacts colitis by altering gut microbiota metabolism of tryptophan into aryl hydrocarbon receptor ligands. Nature medicine 22, 598–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jaquet M, Rochat I, Moulin J, Cavin C, and Bibiloni R (2009) Impact of coffee consumption on the gut microbiota: a human volunteer study. Int. J. Food Microbiol 130, 117–121 [DOI] [PubMed] [Google Scholar]
- 57.Cornelis MC, Erlund I, Michelotti GA, Herder C, Westerhuis JA, and Tuomilehto J (2018) Metabolomic response to coffee consumption: application to a three-stage clinical trial. J. Intern. Med 283, 544–557 [DOI] [PubMed] [Google Scholar]
- 58.Cowan TE, Palmnas MS, Yang J, Bomhof MR, Ardell KL, Reimer RA, Vogel HJ, and Shearer J (2014) Chronic coffee consumption in the diet-induced obese rat: impact on gut microbiota and serum metabolomics. J. Nutr. Biochem 25, 489–495 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.