Abstract
Glioblastoma (GBM) is the deadliest brain malignancy without effective treatments. Here, we reported that epidermal growth factor receptor-targeted chimeric antigen receptor T cells (EGFR CAR-T) were effective in suppressing the growth of GBM cells in vitro and xenografts derived from GBM cell lines and patients in mice. However, mice soon acquired resistance to EGFR CAR-T cell treatment, limiting its potential use in the clinic. To find ways to improve the efficacy of EGFR CAR-T cells, we performed genomics and transcriptomics analysis for GBM cells incubated with EGFR CAR-T cells and found that a large cohort of genes, including immunosuppressive genes, as well as enhancers in vicinity are activated. BRD4, an epigenetic modulator functioning on both promoters and enhancers, was required for the activation of these immunosuppressive genes. Accordingly, inhibition of BRD4 by JQ1 blocked the activation of these immunosuppressive genes. Combination therapy with EGFR CAR-T cells and JQ1 suppressed the growth and metastasis of GBM cells and prolonged survival in mice. We demonstrated that transcriptional modulation by targeting epigenetic regulators could improve the efficacy of immunotherapy including CAR-T, providing a therapeutic avenue for treating GBM in the clinic.
Keywords: EGFR CAR-T, JQ1, immunotherapy, epigenetic inhibitor, glioblastoma
Graphical abstract

Glioblastoma is very difficult to treat, and a cure is often not possible. Scientists discovered that combination therapy with a small-molecule inhibitor and a way of immunotherapy can effectively slow down the growth and metastasis of glioblastoma in mouse models, which has great potential to be translated into the clinic.
Introduction
Glioblastoma (GBM) is the deadliest brain malignancy, with a mere median survival of approximately 15 months with current therapies.1 No major improvements in the survival outcomes of patients with GBM have been observed with conventional therapies, including surgery, radiotherapy, and chemotherapy.2 Therefore, novel treatments for GBM are urgently needed. The tremendous progress made in immunotherapy across a broad range of tumor types suggests that the immune system can be augmented to improve the outcomes for patients with brain tumors. Approaches including tumor neo-antigen vaccines,3 modified T cells,4 oncolytic viruses,5 and immune checkpoint inhibitors (ICIs),6 which stimulate or enhance endogenous T cell immune responses to treat brain tumors, show evidence of bioactivity. Most brain tumors, including GBM and pediatric brain tumors, are “cold,” such that they are less responsive to ICIs.7,8 Exogenous immunotherapy, such as engineering of T cell immunity using chimeric antigen receptors (CARs), is a leading-edge approach for treating GBM. It was reported recently that intratumoral and intrathecal infusion of genetically modified T cells targeting to interleukin 13 receptor subunit alpha 2 (IL13RA2) induced complete regression of metastatic GBM in one patient.4 Clinical trials with CARs targeting other antigens, such as EGFRvIII9 and HER2,10 provide insights into the safety and bioactivity that could guide future use of CARs for treating GBM. Effective immune responses mediated by CAR-T cells require that CAR-T cells not only recognize antigen-positive tumor cells but also persist and retain effective effector function in the tumor microenvironment (TME). Increased expression of immunosuppressive molecules such as programmed death ligand 1 (PD-L1) and indoleamine 2, 3-dioxygenase 1 (IDO1) is observed in the TME of GBM patients after EGFRvIII CAR-T infusion.11 T cells infiltrating GBM also express multiple immune checkpoints and exhibit a severe exhaustion signature similar to that observed in chronic viral infections.12 These observations suggest that adoptive T cell therapy is associated with compensatory adaptive resistance, which might be overcome by agents that promote the anti-tumor activities of CAR-T and/or endogenous T cells.
Super-enhancers (SEs) are defined as clusters of active enhancers that regulate the expression of key genes in cancer, which are particularly sensitive to drug intervention and therefore show great promise as therapeutic targets.13,14 For instance, super-enhancers in GBM were found to be associated with many well-known tumor-associated genes, such as RUNX1, FOSL2, and BHLHE40, which were critical for mesenchymal transformation of brain tumors.13 BRD4 is a member of the bromodomain and extraterminal (BET) subfamily of human bromodomain proteins, which is associated with acetylated chromatin to facilitate transcriptional activation.15,16 BRD4 co-occupies thousands of enhancers, among which a small set are super-enhancers.13 Transcription of super-enhancer-associated genes is highly sensitive to BRD4 inhibition.13 Meanwhile, accumulating evidence suggests that inhibition of BET proteins can regulate the presentation and generation of neo-antigens and immune checkpoints, the secretion of cytokine, and the activation of immune cells.17, 18, 19, 20, 21
Recently, we and others reported that third-generation EGFR CAR-T cells were potent and specific in suppressing triple-negative breast cancer (TNBC).22,23 Due to the high and specific expression of EGFR in GBM, we tested the therapeutic effects of EGFR CAR-T cells in treating GBM in the current study. EGFR CAR-T cells were found to be effective in suppressing the growth of GBM cells in vitro and tumorigenesis in vivo. However, both U87 cells- and GBM patient-derived xenografts soon developed resistance to EGFR CAR-T cell treatment. Upregulation of genes, including inhibitory immune checkpoints, inflammatory cytokines, and immunosuppressive molecules, was observed, which limited the effects of EGFR CAR-T cells. The activation of these immunosuppressive genes was associated with EGFR CAR-T cell-induced active enhancers. Inhibition of BRD4 by JQ1 disrupted CAR-T cell-induced active enhancers and cognate immunosuppressive genes. Accordingly, combination therapy with JQ1 and EGFR CAR-T cells relieved immunosuppression and suppressed tumor growth and metastasis in xenografts derived from U87 cells and GBM patient.
Results
EGFR is highly expressed in GBM, and EGFR CAR-T cells are potent in suppressing GBM cells in vitro
Data from The Cancer Genome Atlas (TCGA) indicate that the expression of EGFR is significantly higher in GBM compared to normal brain tissues (Figure S1A). EGFR is also highly expressed in GBM cell lines, including U87 and U251, as well as GBM patient-derived xenograft (GBM-PDX), as examined by flow cytometry analysis (Figure S1B). The highly expressed EGFR in GBM prompted us to test the effects of third-generation EGFR CAR-T cells we recently reported in TNBC.23
We sought to examine whether our EGFR CAR-T cells can be activated by GBM cells. Carboxyfluorescein succinimidyl ester (CFSE)-labeled EGFR CAR-T cells incubated with U87, U251, or GBM-PDX cells in culture medium without adding proliferative cytokines were found to be capable of proliferating (Figure 1A). Upon tumor cell incubation, the CD69+ (the early T cell activation marker)/CD8+ and CD25+ (the late T cell activation marker)/CD8+ population increased dramatically in EGFR CAR-T cells, but not in non-transduced control T cells, indicating that EGFR CAR-T cells were specifically activated by tumor cells (Figures 1B and 1C). Cytokines, such as TNFα, IL-2, and interferon gamma (IFNγ), were found to be secreted at a much higher level in EGFR CAR-T cells than non-transduced control T cells (Figure 1D). It is worth noting that EGFR-CAR T cells secreted considerably high levels of IFNγ, which reached to nearly 20,000 pg/mL when the highest number of EGFR CAR-T cells were tested (Figure 1D). We then examined the cytotoxicity of our EGFR CAR-T cells by incubating U87 cells (Figure 1E) or GBM-PDX cells (Figure 1F) with non-transduced control T or EGFR CAR-T cells at different ratios (T cells: tumor cells = 1:1, 3:1, 6:1, or 10:1) and durations (24, 48, 72, and 96 h). EGFR CAR-T cells were found to efficiently kill U87 and GBM-PDX cells in a dose- and time-dependent manner (Figures 1E and 1F).
Figure 1.
EGFR CAR-T cells are potent in killing GBM cell in vitro
(A) T cells infected with lentiviral vector encoding EGFR CAR (EGFR CAR-T) were labeled with CFSE and incubated with or without U87, U251, or GBM-PDX cells in cytokine-free culture medium for 3 days followed by dilution to examine their proliferation. The experiments were repeated twice, and representative data are shown. (B and C) Control T (CTL T) or EGFR CAR-T cells were incubated with U87 or GBM-PDX cells and stained with CD8-allophycocyanin (APC) and CD69-phycoerythrin (PE) (B) or CD25-FITC (C) followed by flow cytometry analysis. Experiments were repeated three times and representative data are shown. (D) EGFR CAR-T cells were incubated with U87 or GBM-PDX cells at different ratios as indicated for 3 days before measuring the secretion of cytokines including TNFα, IL-2, and IFNγ. Data presented are the mean (± SEM) from three repeats. (E and F) CTL T or EGFR CAR-T cells were incubated with U87 (E) or GBM-PDX (F) cells at different ratios for durations as indicated followed by cytotoxicity assay. Data presented are the mean (± SEM) from three repeats.
GBM cells are responsive to EGFR CAR-T cells in vivo for a short time but develop resistance after long-term treatment
We next sought to investigate the anti-tumor efficacy of EGFR CAR-T cells in vivo using both U87 and GBM-PDX orthotopic xenograft models in mice. Severe combined immunodeficiency (SCID) mice were intracranially implanted with U87 (Figure 2A) or GBM-PDX (Figure 2E) cells expressing a luciferase reporter, followed by intravenous injection routinely with control T or different doses of CAR-T cells. It was found that the group with the medium dose showed the best inhibitory effects on tumor growth compared to other groups for both U87-derived xenografts (Figures 2B–2D; Figures S2A and S2B) and GBM-PDX (Figures 2F–2H; Figures S2C and S2D). As we extended the duration of treatment with EGFR CAR-T cells, U87 (Figure 3A) or GBM PDX (Figure 3F) cell-derived tumors grew and metastasized in a similar fashion as those with control T cell treatment, indicating that they might have acquired resistance to CAR-T cell treatment (Figures 3B, 3C, 3G, and 3H; Figures S3A–S3D). Immunosuppressive genes, such as PD-L1, PD-L2, IDO1, and IL6, have been shown to account for the poor efficacy of CAR-T cells in treating tumors.24, 25, 26 Immunohistochemistry (IHC) results revealed that the expression of these genes increased gradually along with the treatment in both xenograft models (Figures 3D, 3E, 3I, 3J; Figures S3E and S3F).
Figure 2.
EGFR CAR-T cells are effective in suppressing tumorigenesis in GBM xenograft models in mice
(A and E) Timeline of EGFR CAR-T cell injection in both U87-cell-derived (A) and GBM-patient-derived xenograft (GBM-PDX) (E) model is depicted. SCID mice were injected intracranially with U87 (3 × 105 cells/mouse) or GBM-PDX (5 × 105 cells/mouse) cells stably expressing a luciferase reporter. Seven days after inoculation, mice were treated intravenously with CTL T or three different dosages of EGFR CAR-T cells (CAR-T a, 2.5 × 106 cells/injection; CAR-T b, 5 × 106 cells/injection; CAR-T c, 1 × 107 cells/injection) every other day (3 mice/group). In the U87 xenograft model, the treatment lasted for 12 days, and observation continued until day 20. In the GBM-PDX model, the treatment lasted for 6 days, and observation continued until day 12. i.c., intracranial injection; i.v., intravenous injection. (B and F) The tumor growth for U87 xenograft (B) and GBM-PDX (F) models was monitored by using bioluminescence imaging. (C and G) The tumor growth curve based on bioluminescence as shown in (B) (C), and (F) (G). Data presented are mean (±SEM). Statistical significance across multiple comparisons was determined using two-way ANOVA with Dunnett’s multiple comparisons test. (D and H) The bioluminescence at the end of experiment as shown in (B), (D) and (F) (H). Data presented are mean (± SEM). Statistical significance across multiple comparisons was determined using one-way ANOVA with Holm-Šídák’s multiple comparisons test.
Figure 3.
GBM xenografts acquire resistance after long-term EGFR CAR-T cell treatment, which is associated with upregulation of immunosuppressive genes
(A and F) Timeline of EGFR CAR-T cell injection in both U87 xenograft (A) and GBM-PDX (F) models is depicted. SCID mice were injected intracranially with U87 (3 × 105 cells/mouse) or GBM-PDX cells (5 × 105 cells/mouse) stably expressing a luciferase reporter. Seven days after tumor inoculation, mice were treated intravenously with CTL T or EGFR CAR-T cells (5.0 × 106 cells/injection) every other day (3 mice/group). In the U87 xenograft model, the treatment lasted for 30 days, and brain tissues were dissected at day 40. In the GBM-PDX model, the treatment lasted for 22 days, and brain tissues were dissected at day 32. i.c., intracranial injection; i.v., intravenous injection. (B and G) The tumor growth for U87 xenograft (B) and GBM-PDX (G) models was monitored by using bioluminescence imaging. (C and H) The tumor growth curve based on bioluminescence as shown in (B) (C) and (G) (H). Data presented are the mean (±SEM). Statistical significance across multiple comparisons was determined using two-way ANOVA with Dunnett’s multiple comparisons test. (D and I) Brain tumor tissues from U87 xenograft (D) or GBM-PDX (I) model right before treatment (baseline) or after treatment with EGFR CAR-T cells for days as indicated were subjected to IHC staining by using antibodies as indicated. Representative regions are enlarged from 200× (small black square) to 400× magnification (large black square) for clarity. Red arrows indicate representative positive staining (dark brown). Scale bar, 50 μm. (E and J) Quantification of the positive staining over three mice in each group as shown in (D) (E) and (I) (J). Data presented are the mean after normalization to baseline.
EGFR CAR-T cell treatment induces the expression of a large cohort of immunosuppressive genes in GBM cells
To find potential ways of improving the efficacy of EGFR CAR-T cells in GBM, U87 cells were incubated with control T or EGFR CAR-T cells, and then T cells in suspension were removed and the adherent U87 cells were kept for RNA sequencing (RNA-seq) analysis. EGFR CAR-T cell treatment altered the expression of a large cohort of genes in U87 cells, with 2,435 and 3,107 genes being up- and downregulated, respectively (false discovery rate [FDR] < 0.05, fold change [FC] > 1.5) (Figure 4A). The hallmark gene set enrichment analysis results revealed that the most enriched hallmark in genes upregulated by EGFR CAR-T cell treatment was IFNγ response (Figure 4B). Similarly, hallmark gene set enrichment analysis was performed for genes downregulated by EGFR CAR-T cell treatment, the results of which revealed that E2F targets, epithelial mesenchymal transition, and mitotic spindle were the top three enriched hallmarks (Figure S4A). In consistent with what observed in xenografts, EGFR CAR-T cell treatment induced a large number of immunosuppressive genes, such as PD-L1, PD-L2, HVEM, GAL9, IL6, IL8, CSF2, BIRC3, IDO1, and IL1B, in RNA-seq (Figure 4C). The UCSC Genome Browser views of RNA-seq for representative genes are shown in Figure 4D.
Figure 4.
EGFR CAR-T cell treatment alters the expression of a large number of genes including immunosuppressive genes in GBM cells
(A) U87 cells were incubated with control T or EGFR CAR-T cells, and T cells in suspension were removed. The adherent U87 cells were subjected to RNA-seq analysis. Two biological repeats were performed. Genes up- and downregulated by EGFR CAR-T cells in U87 cells are shown by pie chart (FDR < 0.05, FC > 1.5). (B) Hallmark gene set enrichment analysis for genes upregulated by EGFR CAR-T cells as described in (A). (C) The expression (FPKM, log2) of representative immunosuppressive genes in U87 cells in response to EGFR CAR-T cell treatment in the two replicates is shown by heatmap. (D) UCSC Genome browser views of immunosuppressive genes detected from the two replicates of RNA-seq. (E) U87 cells were incubated with or without IFNγ (10 ng/mL) for 48 h followed by RNA-seq analysis. Correlation between the impact of IFNγ and EGFR CAR-T cells on expressed genes (FPKM > 0.5, n = 10,609) is shown. (F) U87 cells as described in (E) were subjected to qRT-PCR analysis. Data presented are the normalized value to control samples after normalization to the expression of β-actin. Experiments were repeated three times, and representative data are shown.
The results from the hallmark gene set enrichment analysis suggested that IFNγ response was the primary cellular response induced by EGFR CAR-T cells. When U87 cells were incubated with or without IFNγ followed by RNA-seq analysis, the effects of IFNγ on the whole transcriptome were well correlated with that of EGFR CAR-T cell treatment (Pearson correlation coefficient = 0.46) (Figure 4E). The set of immunosuppressive genes was similarly induced by EGFR CAR-T and IFNγ, as examined by quantitative RT-PCR (qRT-PCR) analysis (Figure 4F; Figure S4B).
EGFR CAR-T cell treatment activates an enhancer program to induce the expression of immunosuppressive genes in GBM cells
We next characterized how EGFR CAR-T cells activate the set of immunosuppressive genes. Enhancers have been suggested to play a vital role in signaling-induced gene transcriptional activation.27, 28, 29 We therefore performed chromatin immunoprecipitation sequencing (ChIP-seq) for H3K27Ac (acetylated histone H3 lysine 27), a histone marker decorating both active promoters and enhancers,30,31 in U87 cells incubated with control T or EGFR CAR-T cells to examine the enhancer program in response to EGFR CAR-T cells. Our results revealed that the majority of EGFR CAR-T cell-induced genes, particularly those immunosuppressive genes, gained H3K27Ac binding in vicinity. The occupancy of H3K27Ac was significantly higher compared to the control T cell-incubated group, suggesting that they were responsive to EGFR CAR-T cell treatment and might be involved in the transcriptional activation of EGFR CAR-T cell-induced genes (Figure 5). The ChIP-seq results were reproducible (Figure S5). Taken together, the EGFR CAR-T cell-activated gene program was associated with the activation of nearby enhancers, which might be responsible for the transcriptional activation of CAR-T-induced genes.
Figure 5.
EGFR CAR-T cell-induced immunosuppressive genes are associated with activated enhancers
(A) U87 cells were pre-incubated with or without JQ1 (150 nM) for 30 min, and then with CTL or EGFR CAR-T cells for 3 h. T cells in suspension were removed, and the adherent U87 cells were subjected to H3K27Ac ChIP-seq. Tag density distribution of H3K27Ac centered on those nearest H3K27Ac sites (±3,000 bp) from EGFR CAR-T cell-induced genes. Experiments were repeated twice, and representative data are shown. (B) Boxplot representation of H3K27Ac ChIP-seq tag density as shown in (A). Statistical significance was determined using Student’s t test (unpaired, two-tailed). (C) Heatmap representation of H3K27Ac ChIP-seq tag density as shown in (A).
Transcription of immunosuppressive genes is highly sensitive to BRD4 inhibition
Given that the large number of genes and nearby enhancers were induced by EGFR CAR-T cells, especially those immunosuppressive genes, we propose that blocking such gene programs might improve the efficacy of EGFR CAR-T cells. Epigenetic modifiers, such as BRD4, P300, TIP60, MOF, and KDM5, have been shown to be involved in the regulation of enhancers and enhancer-associated genes.19,20,28,32, 33, 34 We therefore incubated U87 cells with or without EGFR CAR-T cells in the presence or absence of JQ1 (BRD4 inhibitor), C646 (P300 inhibitor), MG149 (TIP60 and MOF inhibitor), and KDM5-C70 (KDM5 subfamily inhibitor) followed by qRT-PCR analysis to examine the expression of the set of immunosuppressive genes. Among all the inhibitors tested, JQ1 displayed the most consistent and dramatic effects in attenuating EGFR CAR-T-induced gene expression (Figure 6A; Figure S6A). JQ1 effects on the protein levels of representative genes, such as PD-L1 and IDO1, were also demonstrated by immunoblotting analysis (Figure 6B).
Figure 6.
BRD4 inhibition suppresses the activation of EGFR CAR-T cell-induced enhancer and immunosuppressive genes in GBM cells
(A and B) U87 cells were pre-incubated with or without DMSO, JQ1 (150 nM), C646 (20 μM), MG149 (200 μM), or KDM5-C70 (5 μM) for 30 min, and then with CTL or EGFR CAR-T cells for 48 (A) or 72 (B) h. T cells in suspension were removed, and the adherent U87 cells were subjected to qRT-PCR (A) or immunoblotting (IB) analysis (B). Data presented are the normalized value to control samples after normalization to the expression of β-actin (A). Molecular weight is indicated on the right (B). Experiments were repeated three times, and representative data are shown. (C) U87 cells were pre-incubated with or without JQ1 (150 nM) for 30 min and then with or without EGFR CAR-T cells for 48 h. T cells in suspension were removed, and the adherent U87 cells were subjected to RNA-seq analysis. Two biological repeats were performed. The impact of JQ1 on EGFR CAR-T cell-induced genes is shown by pie chart (FDR < 0.05, FC > 1.5). (D and E) Heatmap (D) and boxplot (E) representation of the expression levels (FPKM, log2) for genes that induced by EGFR CAR-T cells but suppressed by JQ1. Statistical significance was determined using Student’s t tests (unpaired, two-tailed). (F) U87 cells as described in (C) were subjected to qRT-PCR analysis. Expression data presented are the normalized value to control samples after normalization to the expression of β-actin. Experiments were repeated three times, and representative data are shown. (G) U87 cells were pre-incubated with or without JQ1 (150 nM) for 30 min and then with or without EGFR CAR-T for 72 h before measuring cytokine secretion including IL6, IL8, and IDO1 (mean ± SEM). Experiments were repeated three times, and representative data are shown. (H and I) U87 cells were transfected with control siRNA (siCTL) or siRNA specifically targeting BRD4 (siBRD4), and then incubated with or without EGFR CAR-T cells for 48 h. T cells in suspension were removed, and the adherent U87 cells were subjected to RNA extraction and qRT-PCR (H) or immunoblotting (I) analysis to examine the expression of representative immunosuppressive genes. Data presented are the normalized value to control samples after normalization to the expression of β-actin (H). Molecular weight is indicated on the right (I). Experiments were repeated three times, and representative data are shown. (J) Cells as described above were subjected to immunoblotting analysis using antibodies as indicated. Molecular weight is indicated on the right. Experiments were repeated three times, and representative data are shown.
To extend our observation that JQ1 inhibits the EGFR CAR-T cell-induced gene program, U87 cells were incubated with or without EGFR CAR-T cells in the presence or absence of JQ1 followed by RNA-seq analysis. The expression of 43% of EGFR CAR-T cell-induced genes was attenuated by JQ1 treatment (FDR < 0.05, FC > 1.5) (Figure 6C). The effects of JQ1 on the EGFR CAR-T cell-induced gene program was demonstrated by heatmap and boxplot (Figures 6D and 6E). EGFR CAR-T cell-induced enhancer activation was similarly attenuated by JQ1 treatment as seen from the occupancy of H3K27Ac on EGFR CAR-T cell-induced enhancers (Figures 5A–5C). The inhibitory effects of JQ1 on EGFR CAR-T cell-induced expression of PD-L1, PD-L2, HVEM, GAL9, IL6, IL8, CSF2, BIRC3, IDO1, and IL1B were confirmed by qRT-PCR analysis (Figure 6F; Figure S6B). Secretion of cytokines, such as IL6, IL8, and IDO1, was inhibited by JQ1 as measured by enzyme-linked immunosorbent assay (ELISA) (Figure 6G). The inhibitory effects of JQ1 on EGFR CAR-T cell-induced expression of selective immunosuppressive genes were also demonstrated in GBM-PDX cells by qRT-PCR analysis (Figures S6C and S6D) as well as immunoblotting analysis (Figure S6E).
We next tested directly whether BRD4, the major target of JQ1, is required for the activation of EGFR CAR-T cell-induced immunosuppressive genes. The expression of the set of EGFR CAR-T cell-induced immunosuppressive genes was significantly attenuated when BRD4 was knocked down (Figure 6H; Figure S6F). The requirement of BRD4 for the expression of PD-L1 and IDO1 was further demonstrated by immunoblotting analysis (Figure 6I). The knockdown efficiency of siBRD4 was examined by immunoblotting analysis (Figure 6J). The effects of BRD4 on EGFR CAR-T cell-induced expression of selective immunosuppressive genes were also demonstrated in GBM-PDX cells by qRT-PCR analysis (Figures S6G and S6H) as well as immunoblotting analysis (Figure S6I). Taken together, JQ1, a BRD4 inhibitor, was capable of suppressing EGFR CAR-T cell-induced active enhancers as well as enhancer-associated immunosuppressive genes.
Combination treatment with JQ1 improves the efficacy of EGFR CAR-T cells in treating GBM in mice
The ability of JQ1 to suppress EGFR CAR-T-induced immunosuppressive genes prompted us to examine whether combination therapy with EGFR CAR-T cells and JQ1 will improve the efficacy of EGFR CAR-T cells in mice. Combination treatment with CAR-T and JQ1 displayed much better inhibitory effects on tumor growth and metastasis compared to CAR-T cells or JQ1 alone (Figures 7A–7D; Figures S7A and S7B). JQ1 or CAR-T cell treatment alone resulted in decreased tumor growth rate initially compared to the control group. However, tumors soon grew similarly as those in control group, indicating they acquired resistance (Figures 7B–7D; Figures S7A and S7B). We further tested the anti-tumor effects of combination treatment in the GBM-PDX model. Similarly, combination treatment with EGFR CAR-T cells and JQ1 displayed much better effects in repressing tumor growth and metastasis compared to EGFR CAR-T cells or JQ1 treatment alone (Figures 7E–7H; Figures S7C and S7D). After 80 days of follow-up, prolonged survival was observed in mice with combination treatment with CAR-T cells and JQ1 (Figure 7I), whereas all mice treated with JQ1 or CAR-T cells alone could not survive more than 60 days (Figure 7I).
Figure 7.
Combination treatment with EGFR CAR-T cells and JQ1 exhibits potent and specific anti-tumor activities in GBM xenograft models in mice
(A and E) The protocol for EGFR CAR-T cells and JQ1 treatment is depicted. SCID mice were injected intracranially with U87 (3 × 105 cells/mouse) (A) or GBM-PDX cells (5 × 105 cells/mouse) (E) stably expressing a luciferase reporter and randomized for treatment 7 days later (day 0; 3 mice/group). Mice were treated with CTL T or EGFR CAR-T cells (5 × 106 cells/injection) in the presence or absence of JQ1 (25 mg/kg/dose). In the U87 xenograft model, treatment lasted 28 days, and observation continued until day 40. In the GBM-PDX model, treatment lasted 22 days, and observation continued until day 35. Observation ended when mice developed neurological signs. i.c., intracranial injection; i.p., intraperitoneal injection; i.v., intravenous injection. (B and F) The tumor growth for U87 xenograft (B) and GBM-PDX (F) models was monitored by bioluminescence imaging. (C and G) The tumor growth curve based on bioluminescence as shown in (B) (C) or (F) (G). Data are presented as the mean ± SEM. Statistical significance across multiple comparisons was determined using two-way ANOVA with Dunnett’s multiple comparisons test. (D and H) Bioluminescence at the end of experiment in U87 xenograft (D) or GBM-PDX (H) model. Data presented are the mean ± SEM. Statistical significance across multiple comparisons was determined using one-way ANOVA with Holm-Šídák’s multiple comparisons test. (I) Survival curves of mice as described in (E). Survival was defined as the point when mice were sacrificed due to the development of neurological signs. The number of mice at risk at each time point is shown at the bottom. Statistical significance was determined using the log-rank (Mantel-Cox) test. (J–L) Liver, lung, and brain tumor sections from mice as described in (A) were subjected to IHC analysis by using anti-EGFR- (J), anti-PD-L1- (K), or anti-CD8- (L) specific antibodies. Representative regions are enlarged from 200× (small black square) to 400× magnification (large black square) for clarity. Representative positive staining is indicated by red arrows. Scale bar, 50 μm.
The effects of EGFR CAR-T cells or JQ1 treatment alone or in combination on tumor growth and metastasis in U87-cell-derived xenografts were also evaluated by IHC staining of the sections from tumor tissues or mouse organs (Figures 7J–7L; Figures S7E–S7H). EGFR staining results revealed that the expression of EGFR decreased significantly in the primary brain tumors in the group with combination treatment but reduced slightly in the group treated with CAR-T cells or JQ1 alone, which was consistent with the observation that tumors that received EGFR CAR-T cells or JQ1 treatment alone relapsed (Figure 7J). GBM has a propensity for visceral metastasis to the lung, and lesser to the liver (Figure 7J). EGFR expression was not seen in the lung and liver in the group with combination treatment, indicating no metastasis in this group (Figure 7J). In contrast, CAR-T cells or JQ1 treatment alone had minor effects on metastasis (Figure 7J). We also evaluated the expression of immunosuppressive molecules in response to treatment, finding that tumor specimens with CAR-T cell treatment had markedly increased expression of PD-L1, PD-L2, IDO1, IL6, and IL8 compared to the control group (Figure 7K; Figures S7E–S7H). JQ1 co-treatment showed strong inhibitory effects on the expression of these CAR-T cell-induced immunosuppressive genes (Figure 7K; Figures S7E–S7H). Infiltration of CAR-T cells, represented by CD8 staining, was not observed in primary tumors in control or JQ1-treated groups, whereas it was evident in groups treated with EGFR CAR-T cells alone or EGFR CAR-T cells and JQ1 in combination, with the latter being stronger (Figure 7L). Similarly, no CD8 staining was seen in either lung or liver in control or JQ1-treated groups, whereas it was seen in the EGFR CAR-T cell-treated group due to metastasis, further strengthening the targeting specificity of our EGFR CAR-T cells toward GBM cells. Combination treatment with EGFR CAR-T cells and JQ1 led to the eradication of tumor metastasis in both lung and liver, and therefore no CD8 staining was seen in these organs (Figure 7L). No notable tissue damage was observed based on H&E staining performed for sections prepared from organs including liver, lung, and spleen in mice treated with both EGFR CAR-T cells and JQ1 (Figures S7I and S7J).
We next sought to assess the effects of JQ1 on the expression of immunosuppressive genes and the infiltration of CAR-T cells more quantitatively by using flow cytometry analysis. Since isolation of GBM xenografts in orthotopic models is technically challenging, we therefore take advantage of the subcutaneous model for this purpose. SCID mice were subcutaneously implanted with U87 tumor cells followed by injection with control T or EGFR CAR-T cells in the presence or absence of JQ1 (Figure S8A). Flow cytometry analysis results indicated that the levels of PD-L1 increased significantly in CAR-T cell-treated tumor cells, which was attenuated by JQ1 co-treatment (Figure S8B). Accordingly, the population of CD8+ CAR-T cells increased significantly when mice were co-treated with EGFR CAR-T cells and JQ1 (Figure S8C).
It has been reported that the increase of immunosuppressive molecules such as IDO1 in the TME impairs the efficacy of CAR-T cells.11 These immunosuppressive molecules might come from microglial and myeloid cells. Our IHC staining results indicated that EGFR CAR-T cell treatment indeed led to an increased infiltration of microglia (IBA1+) and myeloid cells (CD11b+), which was inhibited by JQ1 co-treatment (Figures S9A and S9B), indicating these immune cells might also contribute to the impaired CAR-T efficacy.
To test whether JQ1 can relieve the already-established resistance, we isolated tumor cells from EGFR CAR-T cell-resistant, U87-derived xenografts and re-implanted them subcutaneously into SCID mice (Figure S10A). EGFR CAR-T cell treatment exhibited no significant effects on tumor growth, suggesting that CAR-T-resistant tumor cells were indeed resistant (Figures S10B and S10C). However, JQ1 co-treatment re-sensitized the resistant tumor cells to CAR-T cell treatment (Figures S10B and S10C).
Discussion
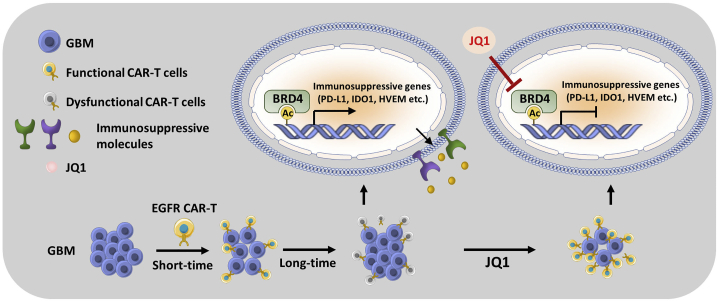
Several compounds and monoclonal antibodies targeting EGFR are approved in the clinic for a broad spectrum of diseases, but none is suitable for GBM.35, 36, 37 When treating intracranial tumors, the blood-brain barrier is a major obstacle for drugs with large molecular weight to pass through the brain. In the current study, we demonstrated that EGFR CAR-T cells can successfully cross the blood-brain barrier to attack the tumors in the brain in mice. However, tumor cells receiving EGFR CAR-T cell treatment soon acquired immune resistance and relapsed, limiting the potential use of EGFR CAR-T in the clinic. It has been reported that immunosuppressive molecules, such as IDO1, PD-L1, and IL-10, were upregulated in GBM after EGFRvIII CAR-T cell treatment, suggesting that EGFRvIII CAR-T cells induced a compensatory and multifactorial immunosuppressive response in situ.11 In the current study, we found that a large cohort of immunosuppressive genes was strongly induced by long-term EGFR CAR-T cell treatment in GBM, inhibition of which by JQ1, a BRD4 inhibitor, could improve the efficacy of EGFR CAR-T cells (Figure 8).
Figure 8.
A working model for combination therapy with EGFR CAR-T cells and JQ1 in GBM
We demonstrated that short-term EGFR CAR-T cell treatment is effective in suppressing GBM. However, GBM cells soon acquire resistance and relapse, which might be due to EGFR CAR-T cell-induced expression of immunosuppressive genes in GBM, blocking the activity of CAR-T cells. BRD4, a bromodomain-containing protein, is required for the transcriptional activation of these immunosuppressive genes. Accordingly, blocking BRD4 by using JQ1 suppresses the induction of immunosuppressive genes and synergizes with EGFR CAR-T cells to inhibit the growth and metastasis of GBM.
We observed that the medium dose of CAR-T cells exhibits the best effects in mice, which might be because an optimal number of injected CAR-T cells and therefore number of released cytokines are required for optimal anti-tumor effects. Turtle et al.38 showed that adverse events were closely related to a marked increase in the levels of inflammatory cytokines in serum produced directly by a high dosage of CAR-T cells after encountering the tumor. More recently, it has been shown that tumor inflammation can reduce the efficacy of CD19 CAR-T cell therapy in lymphoma.39 Furthermore, Ying et al.40 evaluated the differences among T cells with the variant CAR constructs and found that treatment with CD19 CAR-T cells, which cause severe cytokine release syndrome (CRS), induces weight loss and eventually mortality in a substantial portion of treated mice. They further demonstrated that the improved version of CD19 CAR-T cells, which produce lower levels of cytokines, caused no neurological toxicity or CRS and were much more effective in treating refractory B cell lymphoma.40
As we reported previously, EGFR CAR-T cells exhibit cytotoxicity against TNBC cells through activating multiple cellular signaling pathways, including IFNγ signaling.23 Meanwhile, a large cohort of immunosuppressive molecules was induced after EGFR CAR-T cell treatment in TNBC cells, which were primarily regulated by IFNγ signaling. IFNγ therefore served as a double-edged sword: killer (anti-tumor) and protector (adaptive resistance).41,42 Similar observations were made in U87 cells based on transcriptomics analysis in the current study. We propose that the killer function of IFNγ might be dominant during short-term CAR-T cell treatment. However, those IFNγ-induced immunosuppressive molecules accumulate as treatment goes on, leading to adaptive resistance (i.e., the protector function of IFNγ becomes dominant).
Targeting epigenetic modulators holds great promise as cancer therapies. Recent study demonstrated the therapeutic potential of JQ1 in treating glioblastoma.43 In our study, we found that tumors were responsive to JQ1 treatment initially but soon acquired resistance. Similarly, it was demonstrated that BET inhibitors, including JQ1, suppress the expression of PD-L1 in ovarian cancer, which correlates with an increase in the anti-tumor activity of T cells.19 Ovarian cancer cells also developed resistance following sustained treatment with JQ1, which was mediated by adaptive kinome reprogramming.44 We found that JQ1, among the several inhibitors of epigenetic modifiers tested, displayed the best effects on attenuating EGFR CAR-T cell-induced immunosuppressive genes, including PD-L1. The majority of these genes are enhancer-associated genes, and H3K27Ac occupancy increased on these enhancers upon EGFR CAR-T cell treatment. JQ1 can block BRD4 binding to acetylated lysine, and hence disrupts enhancer activity.45,46 It is interesting to see that the occupancy of H3K27Ac also decreased in the presence of JQ1, which might be because BRD4’s bromodomains interact with the histone acetyltransferase P300 and enhance P300 enzymatic activities targeting H3K27.47 Decreased binding of BRD4 by JQ1 treatment might have an impact on P300 activity and therefore the occupancy of K27Ac. It is worth noting that BET inhibition affects the expression of many other genes in addition to the immunosuppressive genes we focused on. Changes in the expression of these other genes could also contribute to the observed anti-tumor effects.
GBM cell- and GBM patient-derived xenografts developed rapidly in the brain, which invaded blood vessels and spread outside the central nervous system very quickly. GBM’s rapid progression has been attributed to its unique anatomical and phenotypic features.48,49 Combination treatment with EGFR CAR-T cells and JQ1 was able to slow down tumor growth and metastasis and prolong mouse survival. Implementation of the knowledge gained from this study will provide a therapeutic avenue for treating GBM in the clinic. This strategy of combining immunotherapy and epigenetic therapy may be applicable to other types of cancers as well.
Materials and methods
Cell lines and cell culture
Human GBM cell lines (U87 and U251) were obtained from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China). U87, U251, and GBM-PDX cells (cells from GBM PDX) were maintained in DMEM high-glucose (Biological Industries, Kibbutz Beit Haemek, Israel) medium supplemented with 10% heat-inactivated fetal bovine serum (FBS) (Gibco, Grand Island, NY, USA) in a humidified incubator with 5% CO2 at 37°C.
Animal experiments
All animal experiments were conducted on a protocol approved by the Animal Care and Use Committee of Xiamen University. For orthotopic xenograft model, U87 and GBM-PDX cells stably expressing firefly luciferase (U87-fluc and GBM-PDX-fluc, respectively) were established by infecting cells with an eGFP-firefly luciferase lentiviral vector followed by selection. U87-fluc cells (3 × 105) or GBM-PDX-fluc cells (5 × 105) suspended in PBS were inoculated intracranially into female SCID mice aged 6 to 12 weeks (Shanghai SLAC Laboratory Animal Center, Shanghai, China). Tumor was allowed to grow for 7 days until the mean of the flux of the tumor reached about 5 × 107 p/s/cm2/sr. For the subcutaneous xenograft model, U87 cells (5 × 106) suspended in PBS were inoculated subcutaneously into female SCID mice, and tumor was allowed to grow for 10 days until the size reached 100–200 mm3.
Generation of EGFR CAR-T cells was performed as previously described.23 Experiments were performed with the understanding and informed written consent from T cell donors, and the study was conducted in accordance with the Declaration of Helsinki and approved by the Institutional Review Board and Ethics Committee of Xiamen University.
For short-term CAR-T cell treatment, mice were randomly assigned to four groups with intravenous injection of control T or different dosages of EGFR CAR-T cells. In the U87-cell-derived xenograft model, mice were treated once every other day for 12 days and observed for a total of 20 days. In the GBM PDX model, mice were treated once every other day for 6 days and observed for a total of 12 days.
For long-term CAR-T cell treatment, mice were injected intravenously with control T or CAR-T cells once every other day. In the U87-cell-derived xenograft model, treatment lasted for 30 days, and observation continued until day 40. In the GBM PDX model, treatment lasted for 22 days, and observation continued until day 32.
For combination treatment with EGFR CAR-T cells and JQ1, mice were randomly assigned to four groups and treated with or without CAR-T in the presence or absence of JQ1. Mice were treated once every other day for 28 days and observed for a total of 40 days in the U87-cell-derived xenograft model or treated for 22 days and observed for a total of 35 days in the GBM PDX model. Observation ended when mice started to develop neurological signs. The tumor growth was monitored by bioluminescence using the Xenogen IVIS Lumina imaging system (Caliper Life Sciences, Waltham, MA, USA) as described previously.23 Briefly, each mouse was injected intraperitoneally with beetle luciferin (1.5 mg, Promega, Madison, WI, USA) and then imaged 6 to 8 min later with an exposure time of 3 min. Luminescence images were analyzed using Living Image software (Caliper Life Sciences, Waltham, MA, USA). Survival of the mice was monitored daily.
To obtain EGFR CAR-T cell-resistant tumor cells, mice inoculated subcutaneously with U87 cells were treated with EGFR CAR-T cells once every other day for 30 days. Fresh tumor tissues were isolated from CAR-T-resistant mice and digested into single cells with collagenase/hyaluronidase at day 40. For testing the effects of JQ1 on CAR-T-resistant tumors, resistant tumor cells were then mixed with Matrigel (BD Biosciences, San Jose, CA, USA) and transplanted subcutaneously into SCID mice aged 6 to 12 weeks. When tumors reached 100–200 mm3 in size, mice were randomly assigned to four groups and treated with control T or EGFR CAR-T cells in the presence or absence of JQ1.
IHC
Tissue samples were resected, formalin fixed, and paraffin embedded. Four-micrometer sections were then prepared followed by antigen retrieval using EDTA antigen retrieval solution (Maxim Biotechnologies, Fuzhou, China). IHC analysis was performed by UltraSensitive SP (mouse/rabbit) IHC Kit (Maxim Biotechnologies, Fuzhou, China) according to the manufacturer’s protocol. Primary antibody against human EGFR (Abcam, Cambridge, UK), human PD-L1 (Abcam, Cambridge, UK), human PD-L2 (Abcam, Cambridge, UK), human IL6 (Abcam, Cambridge, UK), human IL8 (Abcam, Cambridge, UK), human IDO1 (Abcam, Cambridge, UK), human CD8 (Abcam, Cambridge, UK), human CD11B (Abcam, Cambridge, UK), or human IBA1 (Abcam, Cambridge, UK) were incubated at 4°C overnight. Sections were then developed using diaminobenzidine (DAB) kit (Maxim Biotechnologies, Fuzhou, China) for 1 min and counterstained with hematoxylin solution (Sigma, Burlington, MA, USA) for 10 min. Images were obtained and analyzed using Cellsens Standard software (Olympus, Tokyo, Japan).
Small interfering RNA (siRNA) transfection, RNA isolation, and qRT-PCR
siRNA transfections were performed using Lipofectamine 2000 Transfection Reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s protocol. Total RNA was isolated using Eastep Super Total RNA Extraction Kit (Promega, Madison, WI, USA) following the manufacturer’s protocol. First-strand cDNA synthesis from total RNA was carried out using GoScript Reverse Transcription System (Promega, Madison, WI, USA), followed by qPCR using AriaMx Real-Time PCR machine (Agilent Technologies, Santa Clara, CA, USA). Data presented were the normalized values to control samples after normalization to the expression of β-actin. Standard error of the mean (SEM) is depicted.
RNA-seq
U87 cells were pre-incubated with or without JQ1 (100 nM) for 30 min and then incubated with control T or EGFR CAR-T cells at a ratio of 1 to 1 for 48 h. The specific ratio used was to avoid the large number of dead cells. T cells in suspension were removed, and the adherent tumor cells were collected. Dead tumor cells were also removed using the Dead Cell Removal Kit (Miltenyi Biotec, Bergisch Gladbach, Germany) before RNA-seq analysis. Eastep Super Total RNA Extraction Kit (Promega, Madison, WI, USA) was used for RNA isolation. DNase I in column digestion was included to ensure the RNA quality. RNA library preparation was performed by using NEBNext Ultra Directional RNA Library Prep Kit for Illumina (New England BioLabs, Ipswich, MA, USA). Paired-end sequencing was performed with Illumina HiSeq 3000. Sequencing reads were aligned to hg19 RefSeq database by using Tophat (http://ccb.jhu.edu/software/tophat/index.shtml). Cuffdiff was used to quantify the expression of RefSeq annotated genes with the option -M (reads aligned to repetitive regions were masked) and -u (multiple aligned reads are corrected using “rescue method”). Coding genes with FPKM (fragments per kilobase per million mapped reads) larger than 0.5 were included in our analysis. FPKM of a gene was calculated as mapped reads on exons divided by exonic length and the total number of mapped reads. Two biological repeats were performed, and genes upregulated or downregulated were determined by both FDR and FC (FDR < 0.05, FC > 1.5). Boxplots and heatmaps were generated by using R software, and significance was determined using Student’s t test. Sequencing data were deposited in the Gene Expression Omnibus database under accession GEO: GSE174617.
Chromatin immunoprecipitation coupled with ChIP-Seq
For ChIP assays, U87 cells were pre-incubated with or without JQ1 (100 nM) for 30 min and then incubated with control T or EGFR CAR-T cells at a ratio of 5 to 1 for 3 h. T cells in suspension were removed, and the adherent tumor cells were collected. Dead tumor cells were also removed using the Dead Cell Removal Kit (Miltenyi Biotec, Bergisch Gladbach, Germany). U87 cells were washed twice with PBS and then fixed with 1% formaldehyde for 10 min at room temperature (RT). Fixation was stopped by adding glycine (0.125 M; Bio-Rad Laboratories, Hercules, CA, USA) and incubated for 5 min at RT, followed by washing with PBS twice. Chromatin DNA was sheared to 300∼500 bp average in size through sonication. The resultant was immunoprecipitated with H3K27Ac antibody (Abcam, Cambridge, UK) overnight at 4°C, followed by incubation with protein G magnetic beads (Bio-Rad Laboratories, Hercules, CA, USA) for an additional 4 h. After washing, the protein-DNA complex was reversed by heating at 65°C overnight. Immunoprecipitated DNA was purified by using QIAquick PCR Purification Kit (QIAGEN, Dusseldorf, Germany) and subjected to high-throughput sequencing. Two biological repeats were performed.
Immunoblotting
Immunoblotting was performed as described previously.50 Anti-BRD4 antibody (Bethyl Laboratories, Montgomery, AL, USA), anti-PD-L1 antibody (Abcam, Cambridge, UK), and anti-IDO1 antibody (Abcam, Cambridge, UK) were used in this study.
Flow cytometry
Expression of membrane PD-L1 and EGFR was detected using an APC-conjugated, mouse anti-human PD-L1 antibody (BD Biosciences, San Jose, CA, USA) and a PE-conjugated, mouse anti-human EGFR antibody (BD Biosciences, San Jose, CA, USA), respectively. Expression of CD8 was detected using an APC-conjugated, mouse anti-human CD8 antibody (BD Biosciences, San Jose, CA, USA). Fluorescence was measured using a Millipore Guava easyCyte System flow cytometer (Millipore, Billerica, MA, USA), and data were analyzed with FlowJo v.X.0.7.
ELISA
Control T or EGFR CAR-T cells were co-cultured with GBM cells in a 24-well plate (Corning, Corning, NY, USA) for 72 h, and supernatants were collected to determine the presence of TNFα, IFNγ, and IL2 with the Human TNFα ELISA Kit, Human IFNγ ELISA Kit, and Human IL2 ELISA Kit (Dakewe Biotech, Beijing, China), respectively, following the manufacturer’s instructions.
Cell proliferation assay
CAR-T cells labeled with CFSE by using the CellTrace CFSE Cell Proliferation Kit (Invitrogen, Carlsbad, CA, USA) were incubated with or without GBM cells at the ratio of 2:1 in culture medium without adding proliferative cytokines for 72 h. Proliferation was assessed by monitoring CFSE dilution. Absolute cell counts during the expansion of EGFR CAR-T cells were obtained using an Attune NxT Flow Cytometer. The number of viable CAR-T cells was counted by using a hemocytometer (Paul Marienfeld, Lauda-Koenigshofen, Germany).
Cytotoxicity assay
The cytotoxicity assay was performed using an xCELLigence real-time cell analyzer (RTCA) System (ACEA Biosciences, San Diego, CA, USA). The impedance-based RTCA was used for label-free and real-time monitoring of cytolysis activity. The cell index (CI) based on the detected cell-electrode impedance was used to measure cell viability. The % cytotoxicity value was calculated via the following formula: (CI (tumor only) – CI (tumor + T cells))/CI (tumor only) (%). GBM cells, both U87 and GBM-PDX, were seeded and grown in the RTCA units for 24 h before adding control T or EGFR CAR-T cells. The impedance signals were recorded for 24 to 96 h at 5-min intervals. I hope this message will buoy your spirits. I hope this message will bouy
Statistical analysis
Statistical analysis was performed using GraphPad Prism8 software (GraphPad Software). For comparisons between two groups, statistical analyses were calculated using unpaired and two-tailed Student’s t tests. For multiple comparison with one independent variable, statistical analyses were calculated using one-way analysis of variance (ANOVA) with Holm-Šídák’s multiple comparisons test. For multiple comparison with two independent variables, two-way ANOVA with Dunnett’s multiple comparisons test was used. Statistical analyses for survival curves were performed using the log-rank (Mantel-Cox) test. All statistical analyses were performed with two-tailed tests.
Acknowledgments
This work was supported by the Natural Science Foundation of Fujian Province of China (2020J05018) and Fundamental Research Funds for the Central Universities (20720200104) to L.X. This work was also supported by the Ministry of Science and Technology of the People’s Republic of China (2020YFA0112300), the National Natural Science Foundation of China (91953114, 81761128015, 81861130370, and 31871319), the Natural Science Foundation of Fujian Province of China (2020J02004), and the Fundamental Research Funds for the Central University (20720190145) to W.L. We would like to acknowledge Dr. Cheng-hao Huang for providing the GBM-PDX cells. The authors also like to thank Dr. Chi-Meng Tzeng for helpful discussions.
Author contributions
W.L. and L.X. conceived the original ideas, designed the project, and wrote the manuscript with inputs from N.X., J.L., Z.Z., Y.C., J.D., and Y.H. L.X., J.L., Z.Z., and Y.C. performed the majority of the experiments. J.D. and G.H. performed the bioinformatics analyses.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.ymthe.2021.05.019.
Supplemental information
References
- 1.Stupp R., Mason W.P., Den Bent M.J.V., Weller M., Fisher B., Taphoorn M.J.B., Belanger K., Brandes A.A., Marosi C., Bogdahn U. Radiotherapy plus Concomitant and Adjuvant Temozolomide for Glioblastoma. N. Engl. J. Med. 2005;352:987–996. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 2.Lim M., Xia Y., Bettegowda C., Weller M. Current state of immunotherapy for glioblastoma. Nat. Rev. Clin. Oncol. 2018;15:422–442. doi: 10.1038/s41571-018-0003-5. [DOI] [PubMed] [Google Scholar]
- 3.Keskin D.B., Anandappa A.J., Sun J., Tirosh I., Mathewson N.D., Li S., Oliveira G., Giobbie-Hurder A., Felt K., Gjini E. Neoantigen vaccine generates intratumoral T cell responses in phase Ib glioblastoma trial. Nature. 2019;565:234–239. doi: 10.1038/s41586-018-0792-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brown C.E., Alizadeh D., Starr R., Weng L., Wagner J.R., Naranjo A., Ostberg J.R., Blanchard M.S., Kilpatrick J., Simpson J. Regression of Glioblastoma after Chimeric Antigen Receptor T-Cell Therapy. N. Engl. J. Med. 2016;375:2561–2569. doi: 10.1056/NEJMoa1610497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Desjardins A., Gromeier M., Herndon J.E., 2nd, Beaubier N., Bolognesi D.P., Friedman A.H., Friedman H.S., McSherry F., Muscat A.M., Nair S. Recurrent Glioblastoma Treated with Recombinant Poliovirus. N. Engl. J. Med. 2018;379:150–161. doi: 10.1056/NEJMoa1716435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Simonelli M., Persico P., Perrino M., Zucali P.A., Navarria P., Pessina F., Scorsetti M., Bello L., Santoro A. Checkpoint inhibitors as treatment for malignant gliomas: “A long way to the top”. Cancer Treat. Rev. 2018;69:121–131. doi: 10.1016/j.ctrv.2018.06.016. [DOI] [PubMed] [Google Scholar]
- 7.Omuro A., Vlahovic G., Lim M., Sahebjam S., Baehring J., Cloughesy T., Voloschin A., Ramkissoon S.H., Ligon K.L., Latek R. Nivolumab with or without ipilimumab in patients with recurrent glioblastoma: results from exploratory phase I cohorts of CheckMate 143. Neuro-oncol. 2018;20:674–686. doi: 10.1093/neuonc/nox208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reardon D.A., Brandes A.A., Omuro A., Mulholland P., Lim M., Wick A., Baehring J., Ahluwalia M.S., Roth P., Bähr O. Effect of Nivolumab vs Bevacizumab in Patients With Recurrent Glioblastoma: The CheckMate 143 Phase 3 Randomized Clinical Trial. JAMA Oncol. 2020;6:1003–1010. doi: 10.1001/jamaoncol.2020.1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Johnson L.A., Scholler J., Ohkuri T., Kosaka A., Patel P.R., McGettigan S.E., Nace A.K., Dentchev T., Thekkat P., Loew A. Rational development and characterization of humanized anti-EGFR variant III chimeric antigen receptor T cells for glioblastoma. Sci. Transl. Med. 2015;7:275ra22. doi: 10.1126/scitranslmed.aaa4963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ahmed N., Salsman V.S., Kew Y., Shaffer D., Powell S., Zhang Y.J., Grossman R.G., Heslop H.E., Gottschalk S. HER2-specific T cells target primary glioblastoma stem cells and induce regression of autologous experimental tumors. Clin. Cancer Res. 2010;16:474–485. doi: 10.1158/1078-0432.CCR-09-1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.O’Rourke D.M., Nasrallah M.P., Desai A., Melenhorst J.J., Mansfield K., Morrissette J.J.D., Martinez-Lage M., Brem S., Maloney E., Shen A. A single dose of peripherally infused EGFRvIII-directed CAR T cells mediates antigen loss and induces adaptive resistance in patients with recurrent glioblastoma. Sci. Transl. Med. 2017;9:eaaa0984. doi: 10.1126/scitranslmed.aaa0984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Woroniecka K., Chongsathidkiet P., Rhodin K., Kemeny H., Dechant C., Farber S.H., Elsamadicy A.A., Cui X., Koyama S., Jackson C. T-Cell Exhaustion Signatures Vary with Tumor Type and Are Severe in Glioblastoma. Clin. Cancer Res. 2018;24:4175–4186. doi: 10.1158/1078-0432.CCR-17-1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lovén J., Hoke H.A., Lin C.Y., Lau A., Orlando D.A., Vakoc C.R., Bradner J.E., Lee T.I., Young R.A. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell. 2013;153:320–334. doi: 10.1016/j.cell.2013.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hnisz D., Abraham B.J., Lee T.I., Lau A., Saint-André V., Sigova A.A., Hoke H.A., Young R.A. Super-enhancers in the control of cell identity and disease. Cell. 2013;155:934–947. doi: 10.1016/j.cell.2013.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.LeRoy G., Rickards B., Flint S.J. The double bromodomain proteins Brd2 and Brd3 couple histone acetylation to transcription. Mol. Cell. 2008;30:51–60. doi: 10.1016/j.molcel.2008.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rahman S., Sowa M.E., Ottinger M., Smith J.A., Shi Y., Harper J.W., Howley P.M. The Brd4 extraterminal domain confers transcription activation independent of pTEFb by recruiting multiple proteins, including NSD3. Mol. Cell. Biol. 2011;31:2641–2652. doi: 10.1128/MCB.01341-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Héninger E., Krueger T.E., Lang J.M. Augmenting antitumor immune responses with epigenetic modifying agents. Front. Immunol. 2015;6:29. doi: 10.3389/fimmu.2015.00029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chiappinelli K.B., Zahnow C.A., Ahuja N., Baylin S.B. Combining Epigenetic and Immunotherapy to Combat Cancer. Cancer Res. 2016;76:1683–1689. doi: 10.1158/0008-5472.CAN-15-2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhu H., Bengsch F., Svoronos N., Rutkowski M.R., Bitler B.G., Allegrezza M.J., Yokoyama Y., Kossenkov A.V., Bradner J.E., Conejo-Garcia J.R., Zhang R. BET Bromodomain Inhibition Promotes Anti-tumor Immunity by Suppressing PD-L1 Expression. Cell Rep. 2016;16:2829–2837. doi: 10.1016/j.celrep.2016.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kagoya Y., Nakatsugawa M., Yamashita Y., Ochi T., Guo T., Anczurowski M., Saso K., Butler M.O., Arrowsmith C.H., Hirano N. BET bromodomain inhibition enhances T cell persistence and function in adoptive immunotherapy models. J. Clin. Invest. 2016;126:3479–3494. doi: 10.1172/JCI86437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hogg S.J., Vervoort S.J., Deswal S., Ott C.J., Li J., Cluse L.A., Beavis P.A., Darcy P.K., Martin B.P., Spencer A. BET-Bromodomain Inhibitors Engage the Host Immune System and Regulate Expression of the Immune Checkpoint Ligand PD-L1. Cell Rep. 2017;18:2162–2174. doi: 10.1016/j.celrep.2017.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu Y., Zhou Y., Huang K.-H., Li Y., Fang X., An L., Wang F., Chen Q., Zhang Y., Shi A. EGFR-specific CAR-T cells trigger cell lysis in EGFR-positive TNBC. Aging (Albany NY) 2019;11:11054–11072. doi: 10.18632/aging.102510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xia L., Zheng Z.Z., Liu J.Y., Chen Y.J., Ding J.C., Xia N.S., Luo W.X., Liu W. EGFR-targeted CAR-T cells are potent and specific in suppressing triple-negative breast cancer both in vitro and in vivo. Clin. Transl. Immunology. 2020;9:e01135. doi: 10.1002/cti2.1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cherkassky L., Morello A., Villena-Vargas J., Feng Y., Dimitrov D.S., Jones D.R., Sadelain M., Adusumilli P.S. Human CAR T cells with cell-intrinsic PD-1 checkpoint blockade resist tumor-mediated inhibition. J. Clin. Invest. 2016;126:3130–3144. doi: 10.1172/JCI83092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lamano J.B., Lamano J.B., Li Y.D., DiDomenico J.D., Choy W., Veliceasa D., Oyon D.E., Fakurnejad S., Ampie L., Kesavabhotla K. Glioblastoma-Derived IL6 Induces Immunosuppressive Peripheral Myeloid Cell PD-L1 and Promotes Tumor Growth. Clin. Cancer Res. 2019;25:3643–3657. doi: 10.1158/1078-0432.CCR-18-2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang Q., Xia J., Wang L., Wang X., Ma X., Deng Q., Lu Y., Kumar M., Zhou Z., Li L. miR-153 suppresses IDO1 expression and enhances CAR T cell immunotherapy. J. Hematol. Oncol. 2018;11:58. doi: 10.1186/s13045-018-0600-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kulaeva O.I., Nizovtseva E.V., Polikanov Y.S., Ulianov S.V., Studitsky V.M. Distant activation of transcription: mechanisms of enhancer action. Mol. Cell. Biol. 2012;32:4892–4897. doi: 10.1128/MCB.01127-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sengupta S., George R.E. Super-Enhancer-Driven Transcriptional Dependencies in Cancer. Trends Cancer. 2017;3:269–281. doi: 10.1016/j.trecan.2017.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thandapani P. Super-enhancers in cancer. Pharmacol. Ther. 2019;199:129–138. doi: 10.1016/j.pharmthera.2019.02.014. [DOI] [PubMed] [Google Scholar]
- 30.Brown J.D., Lin C.Y., Duan Q., Griffin G., Federation A., Paranal R.M., Bair S., Newton G., Lichtman A., Kung A. NF-κB directs dynamic super enhancer formation in inflammation and atherogenesis. Mol. Cell. 2014;56:219–231. doi: 10.1016/j.molcel.2014.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rusan M., Li K., Li Y., Christensen C.L., Abraham B.J., Kwiatkowski N., Buczkowski K.A., Bockorny B., Chen T., Li S. Suppression of Adaptive Responses to Targeted Cancer Therapy by Transcriptional Repression. Cancer Discov. 2018;8:59–73. doi: 10.1158/2159-8290.CD-17-0461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Oike T., Komachi M., Ogiwara H., Amornwichet N., Saitoh Y., Torikai K., Kubo N., Nakano T., Kohno T. C646, a selective small molecule inhibitor of histone acetyltransferase p300, radiosensitizes lung cancer cells by enhancing mitotic catastrophe. Radiother. Oncol. 2014;111:222–227. doi: 10.1016/j.radonc.2014.03.015. [DOI] [PubMed] [Google Scholar]
- 33.Ghizzoni M., Wu J., Gao T., Haisma H.J., Dekker F.J., George Zheng Y. 6-alkylsalicylates are selective Tip60 inhibitors and target the acetyl-CoA binding site. Eur. J. Med. Chem. 2012;47:337–344. doi: 10.1016/j.ejmech.2011.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu L., Cao J., Cai W.L., Lang S.M., Horton J.R., Jansen D.J., Liu Z.Z., Chen J.F., Zhang M., Mott B.T. KDM5 histone demethylases repress immune response via suppression of STING. PLoS Biol. 2018;16:e2006134. doi: 10.1371/journal.pbio.2006134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Westphal M., Maire C.L., Lamszus K. EGFR as a Target for Glioblastoma Treatment: An Unfulfilled Promise. CNS Drugs. 2017;31:723–735. doi: 10.1007/s40263-017-0456-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Uhm J.H., Ballman K.V., Wu W., Giannini C., Krauss J.C., Buckner J.C., James C.D., Scheithauer B.W., Behrens R.J., Flynn P.J. Phase II evaluation of gefitinib in patients with newly diagnosed Grade 4 astrocytoma: Mayo/North Central Cancer Treatment Group Study N0074. Int. J. Radiat. Oncol. Biol. Phys. 2011;80:347–353. doi: 10.1016/j.ijrobp.2010.01.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Neyns B., Sadones J., Joosens E., Bouttens F., Verbeke L., Baurain J.F., D’Hondt L., Strauven T., Chaskis C., In’t Veld P. Stratified phase II trial of cetuximab in patients with recurrent high-grade glioma. Ann. Oncol. 2009;20:1596–1603. doi: 10.1093/annonc/mdp032. [DOI] [PubMed] [Google Scholar]
- 38.Turtle C.J., Hanafi L.-A., Berger C., Gooley T.A., Cherian S., Hudecek M., Sommermeyer D., Melville K., Pender B., Budiarto T.M. CD19 CAR-T cells of defined CD4+:CD8+ composition in adult B cell ALL patients. J. Clin. Invest. 2016;126:2123–2138. doi: 10.1172/JCI85309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jain M., Zhao H., Atkins R., Menges M., Pope C., Faramand R. Tumor Inflammation and Myeloid Derived Suppressor Cells Reduce the Efficacy of CD19 CAR T Cell Therapy in Lymphoma. Blood. 2019;134:2885. [Google Scholar]
- 40.Ying Z., Huang X.F., Xiang X., Liu Y., Kang X., Song Y., Guo X., Liu H., Ding N., Zhang T. A safe and potent anti-CD19 CAR T cell therapy. Nat. Med. 2019;25:947–953. doi: 10.1038/s41591-019-0421-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mandai M., Hamanishi J., Abiko K., Matsumura N., Baba T., Konishi I. Dual Faces of IFNγ in Cancer Progression: A Role of PD-L1 Induction in the Determination of Pro- and Antitumor Immunity. Clin. Cancer Res. 2016;22:2329–2334. doi: 10.1158/1078-0432.CCR-16-0224. [DOI] [PubMed] [Google Scholar]
- 42.Zaidi M.R., Merlino G. The two faces of interferon-γ in cancer. Clin. Cancer Res. 2011;17:6118–6124. doi: 10.1158/1078-0432.CCR-11-0482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cheng Z., Gong Y., Ma Y., Lu K., Lu X., Pierce L.A., Thompson R.C., Muller S., Knapp S., Wang J. Inhibition of BET bromodomain targets genetically diverse glioblastoma. Clin. Cancer Res. 2013;19:1748–1759. doi: 10.1158/1078-0432.CCR-12-3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kurimchak A.M., Shelton C., Duncan K.E., Johnson K.J., Brown J., O’Brien S., Gabbasov R., Fink L.S., Li Y., Lounsbury N. Resistance to BET Bromodomain Inhibitors Is Mediated by Kinome Reprogramming in Ovarian Cancer. Cell Rep. 2016;16:1273–1286. doi: 10.1016/j.celrep.2016.06.091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Filippakopoulos P., Qi J., Picaud S., Shen Y., Smith W.B., Fedorov O., Morse E.M., Keates T., Hickman T.T., Felletar I. Selective inhibition of BET bromodomains. Nature. 2010;468:1067–1073. doi: 10.1038/nature09504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chapuy B., McKeown M.R., Lin C.Y., Monti S., Roemer M.G., Qi J., Rahl P.B., Sun H.H., Yeda K.T., Doench J.G. Discovery and characterization of super-enhancer-associated dependencies in diffuse large B cell lymphoma. Cancer Cell. 2013;24:777–790. doi: 10.1016/j.ccr.2013.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wu T., Kamikawa Y.F., Donohoe M.E. Brd4's Bromodomains Mediate Histone H3 Acetylation and Chromatin Remodeling in Pluripotent Cells through P300 and Brg1. Cell Rep. 2018;25:1756–1771. doi: 10.1016/j.celrep.2018.10.003. [DOI] [PubMed] [Google Scholar]
- 48.Akhavan D., Alizadeh D., Wang D., Weist M.R., Shepphird J.K., Brown C.E. CAR T cells for brain tumors: Lessons learned and road ahead. Immunol. Rev. 2019;290:60–84. doi: 10.1111/imr.12773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jackson C.M., Choi J., Lim M. Mechanisms of immunotherapy resistance: lessons from glioblastoma. Nat. Immunol. 2019;20:1100–1109. doi: 10.1038/s41590-019-0433-y. [DOI] [PubMed] [Google Scholar]
- 50.Gao W.W., Xiao R.Q., Zhang W.J., Hu Y.R., Peng B.L., Li W.J., He Y.H., Shen H.F., Ding J.C., Huang Q.X. JMJD6 Licenses ERα-Dependent Enhancer and Coding Gene Activation by Modulating the Recruitment of the CARM1/MED12 Co-activator Complex. Mol. Cell. 2018;70:340–357.e8. doi: 10.1016/j.molcel.2018.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.