Abstract
Using a genetic screen, we isolated three TATA-binding protein (TBP) mutants that increase transcription from promoters that are repressed by the Cyc8-Tup1 or Sin3-Rpd3 corepressors or that lack an enhancer element, but not from an equivalently weak promoter with a mutated TATA element. Increased transcription is observed when the TBP mutants are expressed at low levels in the presence of wild-type TBP. These TBP mutants are unable to support cell viability, and they are toxic in strains lacking Rpd3 histone deacetylase or when expressed at higher levels. Although these mutants do not detectably bind TATA elements in vitro, genetic and chromatin immunoprecipitation experiments indicate that they act directly at promoters and do not increase transcription by titration of a negative regulatory factor(s). The TBP mutants are mildly defective for associating with promoters responding to moderate or strong activators; in addition, they are severely defective for RNA polymerase (Pol) III but not Pol I transcription. These results suggest that, with respect to Pol II transcription, the TBP mutants specifically increase expression from core promoters. Biochemical analysis indicates that the TBP mutants are unaffected for TFIID complex formation, dimerization, and interactions with either the general negative regulator NC2 or the N-terminal inhibitory domain of TAF130. We speculate that these TBP mutants have an unusual structure that allows them to preferentially access TATA elements in chromatin templates. These TBP mutants define a criterion by which promoters repressed by Cyc8-Tup1 or Sin3-Rpd3 resemble enhancerless, but not TATA-defective, promoters; hence, they support the idea that these corepressors inhibit the function of activator proteins rather than the Pol II machinery.
Core promoters are composed of TATA and initiator elements, and they are sufficient for the RNA polymerase II (Pol II) machinery to accurately initiate transcription in vitro (47, 51). Biochemical fractionation and purification indicates that the TATA-binding protein (TBP), general transcription factors (TFIIB, -E, -F, and -H), and Pol II represent the minimal set of proteins that are sufficient for core promoter function. In unfractionated extracts and presumably in cells, TBP is a subunit of a multiprotein complex (TFIID) that contains approximately 10 associated factors (TAFs) (5), and Pol II can be found as a large holoenzyme that contains general transcription factors and a variety of Srb and mediator proteins (32, 36). Under certain experimental conditions, TFIID and the Pol II holoenzyme are sufficient to mediate high levels of basal transcription from core promoters that are not stimulated by activator proteins bound upstream of the core promoter (17). Under other conditions, such as low DNA concentrations or nucleosomal templates, basal transcription is inefficient and activators are required to achieve the maximal level.
In vivo, core promoters support very low levels of transcription, and activators bound upstream (or downstream) of the core promoter are required for physiologically significant levels of transcription (56). It is unclear whether the low levels of transcription observed from core promoters reflect basal transcription as defined in vitro or the effect of cryptic activators bound in the general vicinity of the core promoter. In Saccharomyces cerevisiae, there is considerable genetic evidence that activators function predominantly by recruiting the Pol II machinery to promoters and that association of TBP with TATA elements is a particularly important step (27, 49, 57). Direct physical analysis by protein-DNA cross-linking in vivo indicates that, in the vast majority of cases tested, TBP does not associate with core promoters in the absence of a functional activator (40, 44). TBP mutants that weaken TATA element binding (1, 41) or TFIIA association (4, 53) can selectively affect the response to certain activators. Such selective effects on activator-dependent transcription might be related to the observation in vitro that the TATA element is particularly important for supporting multiple rounds of transcription from a preinitiation complex (50, 63).
There are a number of reasons, not mutually exclusive, why core promoters are extremely inefficient for transcription in vivo. First, as TBP (and hence TFIID) is virtually unable to bind to TATA elements in the context of nucleosomal templates (20), activator-dependent modifications of chromatin structure might be required for TBP to initiate assembly of the preinitiation complex. Second, TBP interacts with general negative regulators, such as Mot1 (2), NC2 (16, 30), the N-terminal domain of TAF130 (35), and perhaps the Not-Ccr4 complex (9, 43), and these inhibitors might block TBP action at core promoters in vivo. Third, TBP forms homodimers at near-physiological concentrations, and dimer dissociation can be rate limiting for binding TATA elements in vitro (8, 59). Fourth, at some promoters, transcription is repressed by the action of gene-specific corepressors that are recruited to target promoters by DNA-binding repressors. Yeast corepressors include the Sin3-Rpd3 histone deacetylase complex (24), which functions by creating a highly localized domain of deacetylated nucleosomes and hence repressed chromatin (26, 52), and the Cyc8-Tup1 corepressor (28, 60), which functions by an unknown mechanism.
The original goal of this work was to investigate the potential role of TBP in the mechanism of Cyc8-Tup1 repression. Here, we isolate TBP mutants that overcome Cyc8-Tup1 repression and show that they directly increase transcription from a variety of enhancerless or repressed promoters containing a functional core promoter region but not from promoters that respond to moderate or strong activators. These mutants fail to bind TATA elements in vitro, but they appear to be normal in TFIID complex formation, dimerization, and interaction with the TAF130 inhibitory domain. In some, but not all, respects, these mutants resemble a TBP mutant that selectively increases transcription from weak promoters (3). These mutants provide information about why core promoters are very inefficient in vivo, and we propose a model to account for their mechanism of action.
MATERIALS AND METHODS
Isolation of TBP mutants.
JGY100, the strain used for the initial mutant isolation, is a derivative of BYΔ2 (11) in which the chromosomal copy of TBP is deleted and wild-type TBP is expressed from its natural promoter on a URA3 centromeric plasmid (YCplac33). The strain also contains a SUC2-HIS3 reporter gene at the normal HIS3 locus in which the SUC2 promoter (residues −693 to −1 relative to the ATG) is fused to the ATG initiation codon of HIS3. JGY100 was separately transformed with six mutant TBP libraries (in the TRP1 centromeric vector YCplac22) generated by regional codon randomization (13), and approximately 5,000 colonies from each library were screened on minimal medium containing 2% glucose and lacking uracil, tryptophan, and histidine. Plasmids were rescued from colonies passing this genetic selection and retransformed into JGY100 to confirm their abilities to increase expression from the SUC2-HIS3 reporter.
Phenotypic analyses.
The abilities of the TBP mutants to support cell growth were assayed by plasmid shuffling (11). Cell toxicity conferred by the TBP mutants was assessed by introducing centromeric (YCplac22) or multicopy (YEplac112) plasmids into JGY100 or a derivative containing an rpd3 deletion (24); toxicity was inferred from the inability to obtain transformants.
To determine the transcriptional requirements for increased expression from the SUC2-HIS3 reporter, we analyzed derivatives of the TBP mutants described here that also contain the following well-characterized mutations of TBP: N2-1 (K138T and Y139A), which severely reduces the interaction with TFIIA but selectively blocks the response to certain activators (53); V161A, which affects the DNA-binding surface and TATA element binding but selectively affects transcriptional activation (41); the double substitution E186A and E188, which blocks the interaction with TFIIB and generally reduces transcription (42).
To assay promoter specificity, wild-type and mutant TBPs were introduced into the following strains: derivatives of FT4 containing his3 promoters in which Rap1, Ace1, or no activator binding site was upstream of the TATA and initiator regions (21); yML2, which contains a his3 promoter lacking the normal upstream region and containing a mutated (TGTAAA; the underlined base is mutated from the consensus) TATA element (41), in the presence or absence of a plasmid expressing TBPm3, an altered-specificity derivative (55); and a derivative of FT5 generated by Jutta Deckert containing a his3 allele subject to repression by Sin3-Rpd3 histone deacetylase in which two URS1 elements from the IME2 promoter are upstream of the intact CYC1 promoter region (24). The resulting strains were analyzed for his3 expression by their growth in the presence of aminotriazole (AT).
For the artificial-recruitment experiments, the region encoding TBP in YCp91-LexA-TBP (54) was replaced by comparable regions encoding the mutant TBPs. DNAs expressing the LexA-TBP derivatives were introduced into strain FT4 containing JK103, a multicopy URA3 plasmid with four LexA operators upstream of the GAL1 TATA element and lacZ structural gene (60). The resulting strains, grown to an A600 of 0.8 in glucose medium containing 0.6% Casamino Acids and lacking tryptophan and uracil, and chloroform-permeabilized cells were assayed for β-galactosidase activity. All values shown represent averages of at least two experiments performed with four independent transformants; the error is ±20%.
S1 analyses.
Strains containing SUC2-HIS3, Rap1-HIS3, or Ace1-HIS3 alleles were grown in glucose medium containing 0.6% Casamino Acids and lacking tryptophan and uracil (and 400 μM copper sulfate in the case of the Ace1-HIS3 allele) to an A600 of 0.8 or 1.0. The RNA levels of the various genes were determined by the S1 nuclease protection assay using 20 to 40 μg of RNA per sample and oligonucleotide probes (22). The sequences of the oligonucleotide probes for HIS3 and DED1 (7), tRNAw (12), and CMD1 and PGK1 (45) have been described previously. The sequence of the SUC2 probe is GGGAGCGATAGCAATGGG T TGATC T TCCCAAT TAG TCAAATCATCGGAAG TAGCATGGCCCCTTTTGT. RNA levels for all genes transcribed by Pol II are normalized to the levels of tRNAw.
Analysis of TBP levels and TFIID complex integrity.
The URA3 centromeric vector YCplac22 expressing the triple-hemagglutinin (HA)-tagged derivative of TBP (HA3-TBP) from its natural promoter has been described previously (40); the comparably tagged version of the ΔN69 derivative was generated by replacing BamHI-XbaI fragments. Cells containing these DNAs (and the YCplac22 control) were grown in 2% glucose medium containing Casamino Acids lacking tryptophan to an A600 of 1. Cell extracts were made by glass bead disruption (45), and 50 μg of protein from each sample was electrophoretically separated on a sodium dodecyl sulfate (SDS)–10% polyacrylamide gel, transferred to nitrocellulose, and then probed with a polyclonal TBP antibody. Endogenous (untagged) TBP and the HA3-tagged TBP derivatives were simultaneously visualized by the alkaline phosphatase technique (Stratagene) upon development.
Immunoprecipitations (1-ml reaction volume) were performed by incubating the protein samples of interest with 50 μl of protein A-Sepharose beads containing monoclonal antibody (12CA5) to the HA-1 epitope (15) for 16 h at 4°C in buffer A (20 mM HEPES [pH 7.9], 150 mM NaCl, 5 mM MgCl2, 10% glycerol). To normalize the levels of the HA3-tagged TBPs, we analyzed 1 mg of protein from cells expressing HA3-TBP and 4 mg of protein from cells expressing HA3-TBP-ΔN69; 4 mg of protein was also used for the control cells that do not express an HA3-tagged protein. The beads were washed five times in buffer A, and the associated proteins were eluted by boiling and then resolved by SDS-polyacrylamide gel electrophoresis. Western blots using antibodies specific for TBP and TAFs were developed by the alkaline phosphatase method.
Chromatin immunoprecipitation.
To compare promoter occupancy of wild-type TBP and the ΔN69 derivative, it was necessary to express the HA3-tagged derivatives at comparable levels. We therefore placed HA3-TBP under the control of a modified version of a copper-inducible promoter (34) generated by Zarmik Moqtaderi and utilized appropriate copper concentrations. JGY100 cells expressing HA3-tagged TBP or TBP-ΔN69 were grown to an A600 of 0.8 in glucose medium containing Casamino Acids and 35 μM copper sulfate and lacking tryptophan and were cross-linked with 1% formaldehyde for 20 min. Cross-linked protein-DNA complexes were isolated, immunoprecipitated with monoclonal antibody to the HA-1 epitope (12CA5), and de-cross-linked as previously described (40). PCR mixtures contained 1 μM primers, 0.1 mM deoxynucleoside triphosphates, 0.1 mCi of [α-32P]dATP (specific activity, 3,000 Ci/mmol)/ml, and various dilutions of either immunoprecipitated or input DNAs in a total volume of 10 μl. Samples were heated to 94°C for 90 s, followed by 26 cycles of 94°C for 30 s, 55°C for 30 s, and 1 min at 72°C. After the completion of the last cycle, the samples were incubated for an additional 5 min at 72°C. The PCR products were resolved by electrophoresis through 10% polyacrylamide gels and quantitated with a Fuji phosphorimager. Units of TBP occupancy are relative to the value for the tRNACAA promoter in a strain containing HA3-TBP that has been assigned a value of 100 and corresponds to complete (or near-complete) occupancy (40).
Purification of TBPs.
Hexahistidine-tagged derivatives of wild-type and mutant TBPs were generated by subcloning NdeI-BamHI PCR fragments into the corresponding sites of pET15b (41). These TBP derivatives were expressed in Escherichia coli BL21(DE3) by induction with 1 mM isopropyl β-d-thiogalactopyranoside for 2.5 h and the cells were resuspended in 8 ml of buffer T (0.5 M KCl, 20 mM Tris [pH 7.9], 1 mM EDTA, 10% glycerol, 1 mM phenylmethylsulfonyl fluoride) per liter of culture volume. After sonication, cell debris was removed by centrifugation, and the supernatant was diluted in buffer T to a final KCl concentration of 100 mM. The supernatant was loaded on an SP-Sepharose column equilibrated in buffer T (0.1 M KCl), washed with buffer T (0.1 M KCl), and eluted in a single step with buffer T (0.5 M KCl). The peak fractions were pooled, and the proteins were purified by Ni-nitrilotriacetic acid (NTA)-agarose chromatography. TBP purity was estimated at 80 to 95%.
DNA-binding assays.
Wild-type and mutant TBPs were incubated in 10-μl reaction mixtures containing 0.7× buffer T, 6 μg of bovine serum albumin, 100 ng of poly(dG · dC), 7 mM magnesium acetate, and a 0.5 nM concentration of a 32P-labeled DNA fragment containing the adenovirus 2 E1B TATA box. Following incubation at 30°C for 30 min, the reaction products were electrophoretically separated through 5% polyacrylamide gels in Tris-glycine-EDTA buffer containing 4 mM MgCl2.
Dimerization experiments.
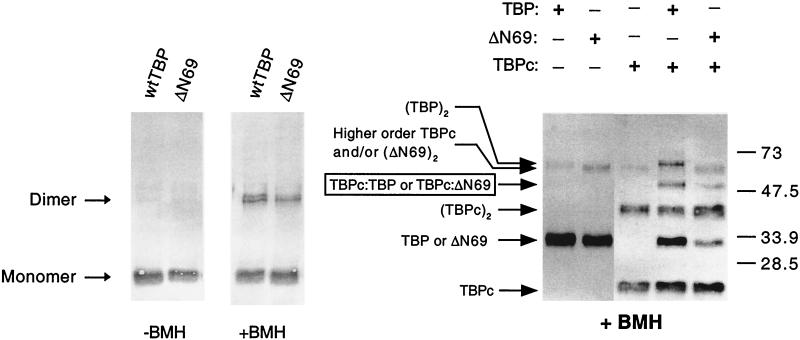
Dimerization was assayed by cross-linking with bismaleimidohexane (BMH; Pierce) essentially as described previously (59). To assay homodimerization, purified TBP and TBP-ΔN69 were separately incubated at concentrations ranging from 1 to 100 nM for 30 min at 30°C. After being cross-linked with 0.1 mM BMH for 30 s, the reaction was quenched by the addition of PSB, a buffer containing β-mercaptoethanol. To assay heterodimerization, the procedure was essentially the same except that 1 μM TBPc (the core domain of TBP, which contains the 180 C-terminal amino acids; provided by Jim Geiger) was incubated with 100 nM TBP or TBP-ΔN69. In both cases, monomers and dimers were separated by SDS–10% polyacrylamide gel electrophoresis and visualized by Western blotting with polyclonal TBP antibodies.
TBP interaction assays.
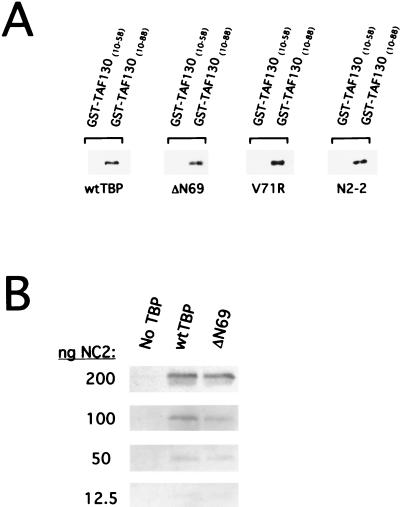
To assay the interaction of wild-type and mutant TBPs with the N-terminal inhibitory domain of TAF130, we expressed glutathione S-transferase (GST)-TAF130(10–58) and GST-TAF130(10–88) in E. coli JS5 using plasmids provided by Tetsuro Kokubo (35) and purified the GST fusion proteins from soluble extracts by glutathione-Sepharose chromatography. For the binding reactions (final volume, 100 μl), 30 pmol of GST proteins was incubated at 4°C for 30 min with 30 pmol of TBP or TBP-ΔN69 in buffer N (100 mM KCl, 20 mM Tris [pH 7.5], 12.5 mM MgCl2, 10% glycerol, 1 mM dithiothreitol) as described previously (35). Ten microliters of glutathione-Sepharose was added, and the samples were incubated an additional 30 min at 4°C. The beads were washed four times with buffer N, and the complexes were eluted by boiling and then analyzed by Western blotting with antibodies to TBP.
To assay the interaction of wild-type and mutant TBPs with the general negative factor NC2, various amounts of recombinant NC2 (generously provided by Rick Young) were incubated with 200 ng of either His-tagged TBP or His-tagged TBP-ΔN69 in 400 μl of buffer C (20 mM Tris [pH 8.0], 100 mM NaCl, 0.1% Nonidet P-40, 10% glycerol, 5 mM imidazole) and 10 μl Ni-NTA-agarose for 1 h at room temperature. Samples were washed three times with buffer C, followed by two washes with buffer C lacking imidazole. After elution by boiling, NC2 proteins were visualized by Western blotting using an anti-NC2 polyclonal antiserum.
RESULTS
TBP mutants that overcome repression by the Cyc8-Tup1 complex.
The Cyc8-Tup1 corepressor complex is recruited to selected sets of promoters by specific DNA-binding proteins (28, 60). It has been suggested that Cyc8-Tup1 represses transcription by directly affecting the Pol II machinery (18, 38, 61) or by altering chromatin structure (10, 19), but the mechanism is unknown. The original goal of this work was to isolate TBP mutants that overcome repression by the Cyc8-Tup1 corepressor. We devised a genetic selection in which the HIS3 structural gene is fused to the SUC2 promoter, which is repressed by glucose in a manner dependent on Cyc8-Tup1. A strain containing an integrated copy of this SUC2-HIS3 allele requires histidine for growth in repressing conditions (2% glucose) but not in nonrepressing conditions (0.05% glucose).
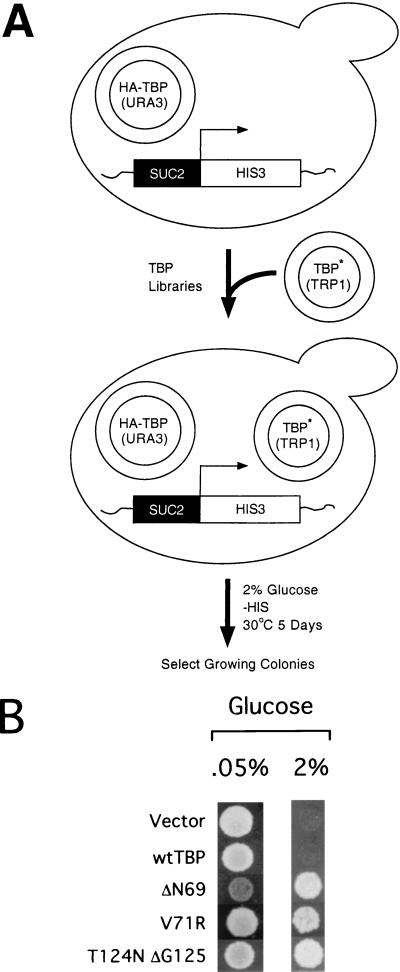
Six complex yet highly compact TBP libraries representing about half of the protein-coding region (13) were introduced into a strain carrying the integrated SUC2-HIS3 allele and expressing wild-type TBP from a URA3 centromeric plasmid (Fig. 1A). In principle, this approach should allow for the isolation of TBP mutants that are dominant or recessive (provided that they can support cell growth as the sole source of TBP after loss of the URA3 plasmid). Of 20,000 initial transformants, four reproducibly grow under the stringent selection conditions (Fig. 1B). Two mutants contain a deletion of Asn69 (ΔN69), a residue that forms two hydrogen bonds with bases T4 and A5 of the bottom strand of the TATA element (29, 31). Although single-amino-acid deletions are generally very rare, they are expected here due to the codon-based mutagenesis (13) used to generate the TBP libraries. A third mutant contains a change of valine 71 to arginine (V71R), while the fourth mutant consists of a deletion of glycine 125 and a conversion of threonine 124 to asparagine. Strikingly, residues 69, 71, 124, and 125 define a small region at the DNA-binding interface (29, 31).
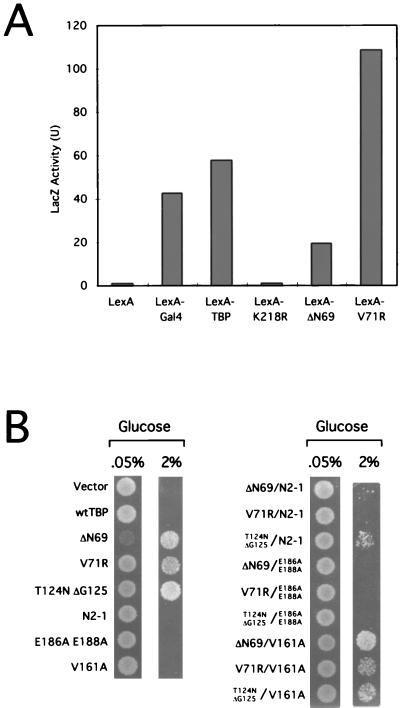
FIG. 1.
Genetic selection of TBP mutants that overcome Cyc8-Tup1 repression. (A) Selection scheme. Strain JGY100, which contains the HIS3 structural gene driven by the SUC2 promoter, was transformed with TBP mutant libraries, and colonies were selected for increased HIS3 expression by growth in the absence of histidine. (B) Growth of 104 cells of JGY100 harboring the indicated TBP derivatives on −Ura, −Trp, and −His plates containing either 0.05 or 2% glucose. wt, wild type. Asterisks indicate mutated TBPs in libraries.
TBP mutants are dominant and toxic for cell growth.
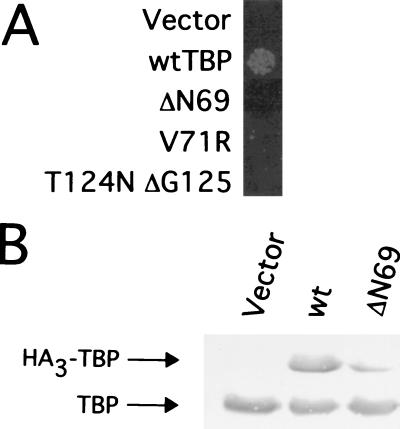
As determined by a plasmid shuffle assay, the TBP mutants are unable to support cell growth when present as the sole source of TBP (Fig. 2A). This indicates that the TBP mutants are dominant over wild-type TBP with respect to their ability to overcome Cyc8-Tup1 repression of the SUC2 promoter. Unexpectedly, the mutant proteins are expressed at 5 to 20% of the wild-type TBP level, depending on the individual mutant and the strain background (Fig. 2B and unpublished). As the proteins are all expressed from the TBP promoter, the low level of the mutant TBPs is probably due to protein instability.
FIG. 2.
TBP mutants are unable to support cell viability. (A) Growth of 104 cells of JGY100 containing the indicated TBP derivatives on medium containing 1 mg of 5-fluoroorotic acid/ml, conditions that require the elimination of the URA3-marked plasmid containing the wild-type (wt) TBP allele. (B) Western blot on cells containing YCplac22 plasmids expressing HA3-TBP, HA3-TBP-ΔN69, or no protein (vector only) using antibody to TBP. The positions of endogenous (untagged) TBP and the HA3-tagged TBPs (encoded by the YCplac22 vector) are indicated by arrows.
The relatively low levels of the TBP mutant proteins do not account for their failure to support cell growth. On the contrary, the TBP mutants are toxic for cell growth, because introduction of multicopy plasmids expressing these mutants results in barely visible microcolonies after prolonged incubation on plates. This toxicity is not observed when the TBP mutants are expressed from centromeric plasmids or when wild-type TBP is expressed from a multicopy plasmid. Thus, in the presence of wild-type TBP, relatively low levels of the mutant proteins are sufficient to override Cyc8-Tup1 repression and are important to prevent toxicity. Interestingly, strains lacking Rpd3 histone deacetylase are hypersensitive to the toxic effects of the mutant TBPs because they are unable to be transformed by centromeric plasmids expressing the mutant TBPs.
TBP mutants function at enhancerless or repressed promoters containing strong TATA elements.
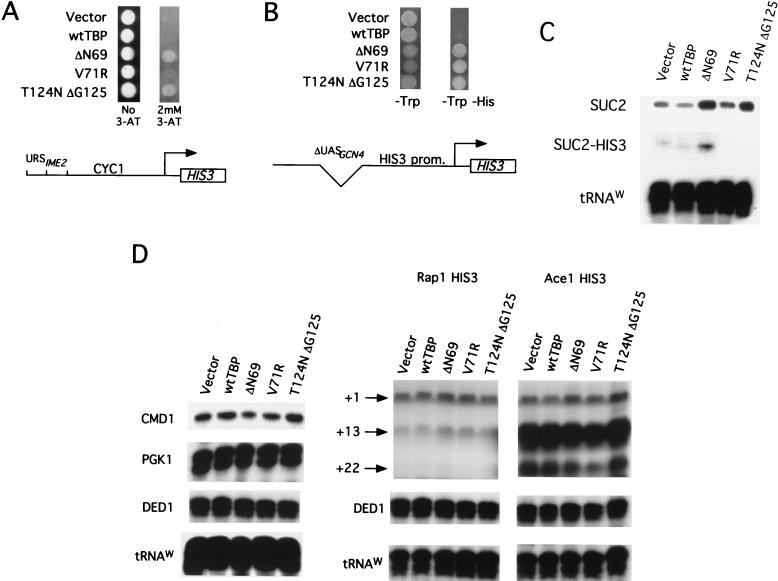
To address whether the effect of the TBP mutants is specific to Cyc8-Tup1-repressed promoters, we asked whether the mutants can also increase expression from other repressed or weak promoters that are unaffected by Cyc8-Tup1. In all cases, these promoters were fused to the HIS3 structural gene, and expression was monitored by growth in the presence of AT, a competitive inhibitor of the HIS3 gene product. As shown in Fig. 3A, the TBP mutants confer increased AT resistance and hence HIS3 expression in a strain containing a CYC1 promoter derivative repressed by Ume6 and the Sin3-Rpd3 histone deacetylase complex (24). Similarly, all three TBP mutants increase expression from a his3 promoter derivative containing the TATA and initiator elements but lacking all known upstream elements (Fig. 3B). Finally, transcriptional analysis indicates that the TBP mutants increase SUC2 or SUC2-HIS3 RNA levels approximately threefold (Fig. 3C). Thus, the TBP mutants can increase expression from three different weak promoters (SUC2, CYC1, and HIS3) that either lack a functional enhancer or are repressed by the Cyc8-Tup1 or Sin3-Rpd3 corepressor complexes.
FIG. 3.
TBP mutants specifically increase expression from enhancerless or repressed promoters. (A) Analysis of a promoter repressed by Sin3-Rpd3 histone deacetylase in which two copies of URS1 are located upstream of the CYC1 promoter and HIS3 structural gene. Strains containing the indicated TBP derivatives were examined for growth in the presence or absence of 2 mM AT. (B) Analysis of a his3 promoter derivative lacking all elements upstream of the TATA region. Strains containing the indicated TBP derivatives were examined for growth in the absence of histidine. (C) S1 protection analysis showing increased SUC2 and SUC2-HIS3 RNA levels in strains containing the indicated TBP derivatives; RNA levels were normalized to the tRNAw control. SUC2-HIS3 levels were not measured for the V71R and T124NΔG125 strains. (D) S1 analysis showing that TBP mutants do not increase transcription from moderate or strongly activated promoters. Strains containing his3 core promoters activated by Rap1 or Ace1 and the indicated TBP derivatives were assayed for transcription of HIS3 (+1, +13, and +22 transcripts are indicated), CMD1, PGK1, DED1, and tRNAw. wt, wild type.
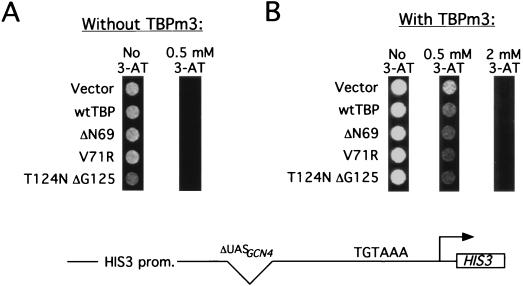
Although the TBP mutants increase expression from the his3 promoter derivative lacking upstream elements, they do not confer increased AT resistance (data not shown) or his3 transcription (Fig. 3D) from stronger his3 promoters containing binding sites for moderate (Rap1) or strong (Ace1) activators. Similarly, the TBP mutants do not cause increased transcription from natural yeast promoters that are moderately or highly active (CMD1 and PGK1, respectively [Fig. 3D]). Finally, the TBP mutants do not indiscriminately increase transcription from all weak promoters, as they have no effect on a basal his3 promoter containing a mutated TATA element (Fig. 4A), even though his3 expression from this promoter is comparable to that from the other weak promoters examined. Taken together, these results indicate that the TBP mutants specifically increase transcription from enhancerless or repressed promoters containing a functional core promoter region.
FIG. 4.
TBP mutants do not increase transcription from a HIS3 promoter with a mutated TATA element. yML2 containing YCplac33 (A) or YCplac33-TBPm3 (B) was transformed with the indicated TBP derivatives and incubated on medium containing various concentrations of AT. wt, wild type.
TBP mutants act directly at affected promoters.
Because the properties of the TBP mutants are examined in the presence of wild-type TBP, there are two classes of mechanisms to account for their ability to increase transcription from weak promoters. In one model, the TBP mutants increase transcription by acting directly at the affected promoters. In the alternative model, the TBP mutants titrate out a negative factor, thereby permitting wild-type TBP to stimulate transcription at the affected promoters. The observation of increased transcription when the mutant TBPs are expressed at only 5 to 20% of the wild-type level strongly argues against a titration model, because titration of a negative factor typically requires overexpression. Furthermore, Fig. 4B indicates that mutant TBPs do not increase transcription mediated by an altered-specificity derivative of TBP that functions on promoters containing appropriately mutated TATA elements (55). However, to provide better evidence that the TBP mutants act directly at promoters in vivo, we carried out the following two experiments.
First, we performed an artificial-recruitment experiment based on the observation that a LexA-TBP hybrid protein can activate transcription from a promoter containing LexA operators upstream of a core promoter (6, 54). In these and related experiments (33, 62), transcriptional activation requires the TBP moiety of the fusion protein to interact with the TATA element. As shown in Fig. 5A, LexA fusions to the ΔN69 or V71R derivative of TBP stimulate transcription to an extent roughly comparable to that of the wild-type LexA-TBP hybrid protein. Thus, the TBP mutants are transcriptionally competent when artificially recruited to promoters via a heterologous DNA-binding domain.
FIG. 5.
Transcriptional properties of the TBP mutants. (A) Artificial-recruitment assay. The indicated LexA-TBP derivatives were tested for their abilities to activate transcription from a promoter containing four LexA binding sites upstream of the GAL1 promoter and lacZ structural gene. LexA-T124N ΔG125 was not tested because the fusion protein is unstable in vivo. (B) Double-mutant analysis. Mutations that greatly diminish binding to TFIIA (N2-1), TFIIB (E186A and E188A), or TATA elements (V161A) were introduced into the context of the TBP mutants described here. Growth of 104 JGY100 cells containing the resulting single and double mutants on medium lacking histidine in the presence of either 0.05 or 2% glucose is shown.
Second, we examined whether mutations that block transcription in the context of wild-type TBP also abolish the ability of the TBP derivatives described here to stimulate expression of the SUC2-HIS3 promoter (Fig. 5B). The TBP mutants lose the ability to increase expression from the SUC2-HIS3 promoter when they are combined with a double substitution (E186A and E188A) on the TFIIB interaction surface that blocks transcription (42). In contrast, expression from SUC2-HIS3 is only partially affected when the TBP mutants are combined with the N2-1 (K138T and Y139A) allele, which severely weakens the TBP-TFIIA interaction (53), and it is unaffected when they are combined with the V161A mutation, which weakens the TBP-TATA interaction (41). The N2-1 and V161A derivatives of wild-type TBP support cell growth and do not generally affect the transcription of most promoters, although they have weakened responses to strong activators (41, 53). Thus, there is an excellent correlation between transcriptional activity in the context of wild-type TBP and in the context of the TBP derivatives described here.
Association of TBP mutants with promoter sequences in vivo.
Although the TBP mutants display increased expression from core promoters, they have no detectable effect on weakly or strongly activated promoters. One possibility is that the TBP mutants are truly defective in the response to activators. Alternatively, the TBP mutants may be comparable to wild-type TBP in mediating transcriptional activation but do not confer an observable phenotype because the mutant proteins represent a small proportion of the total TBP levels.
To address this issue and to avoid the contributions of wild-type TBP to transcription levels, we epitope tagged the ΔN69 mutant of TBP and directly measured its association with promoters in vivo using chromatin immunoprecipitation (40, 44). As a control, we expressed comparable levels of epitope-tagged wild-type TBP from a copper-inducible promoter. In this way, the epitope-tagged derivatives could be specifically monitored and directly compared, even though they were present at approximately 10% of the level of the untagged wild-type TBP in the same strain (Fig. 6A).
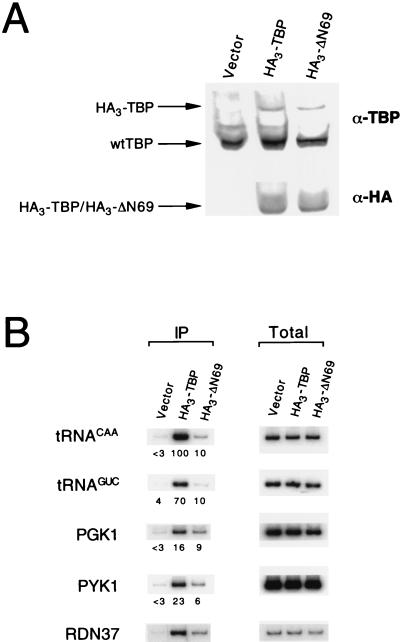
FIG. 6.
TBP occupancy at promoters in vivo. (A) Protein levels in cells expressing HA3-tagged TBP derivatives as determined by Western blotting with antibodies to TBP (α-TBP) or HA-1 epitope (12CA5) (α-HA). Levels of HA3-TBP and HA3-TBP-ΔN69 were equalized by placing HA3-TBP under the control of a copper-regulable promoter and growing cells at an appropriate concentration of copper. wt, wild types. (B) Chromatin immunoprecipitation. Strains containing the indicated HA3-tagged TBP derivatives were treated with formaldehyde, and cross-linked protein-DNA complexes were immunoprecipitated with the 12CA5 monoclonal antibody to the HA-1 epitope. DNAs recovered from protein-DNA complexes prior to (Total) or after immunoprecipitation (IP) were analyzed by PCR with promoter-specific oligonucleotides. Units of TBP occupancy (40) are indicated under the corresponding panels and are relative to the value for the tRNACAA promoter in a strain containing HA3-TBP, which was assigned a value of 100.
The Δ69 derivative of TBP associates with both the PGK1 and PYK1 promoters, although with slightly reduced (two- to threefold) efficiency in comparison to that of wild-type TBP (Fig. 6B). As the level of TBP occupancy is very strongly correlated with transcriptional activity in vivo (40, 44), this suggests that the mutant protein is mildly defective for transcription from strong promoters. The mutant protein shows a similarly mild defect at the ribosomal DNA promoter, which is transcribed by Pol I. The ΔN69 derivative binds two Pol III promoters with markedly reduced efficiency, a result that is consistent with its inability to complement a TBP mutant that is defective for Pol III transcription (data not shown). We could not assess TBP occupancy at the very weak promoters that are stimulated by the ΔN69 derivative because TBP binding at these promoters cannot be detected above the background (40). Nevertheless, the fact the ΔN69 derivative can directly associate with promoters in vivo, together with the artificial-recruitment and genetic experiments described in the previous section, provides compelling evidence that the TBP mutants increase transcription from weak promoters via direct binding to core promoter elements.
The ΔN69 derivative of TBP forms a normal TFIID complex in vivo.
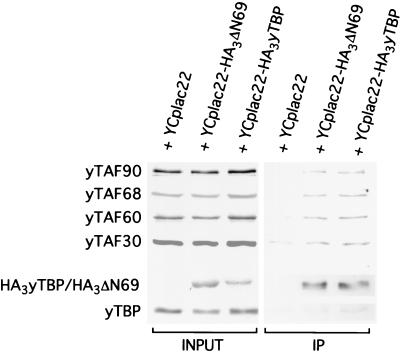
One possible model for the increased activity on core promoters is that the TBP mutant proteins form TFIID complexes that are different in composition from those formed by wild-type TBP. To address this question, we immunoprecipitated epitope-tagged versions of the wild type and the ΔN69 derivative of TBP and compared the relative levels of associated TAFs (Fig. 7). When normalized for levels of TBP, the wild-type and mutant proteins were indistinguishable in their associations with TAF90, TAF68, TAF60, and TAF30. Therefore, it appears that the ΔN69 mutant TFIID complexes are structurally similar to wild-type TFIID, although subtle changes in composition or conformation cannot be excluded.
FIG. 7.
TBP mutants form normal TFIID complexes. YCplac22 plasmids expressing the indicated HA3-tagged TBP derivatives were immunoprecipitated with antibodies to the HA-1 epitope. Samples corresponding to input or immunoprecipitated (IP) material were analyzed by Western blotting using antibodies to the indicated TAFs as well as antibodies against the HA-1 epitope or TBP. wt, wild type.
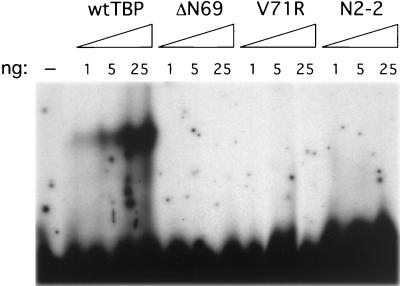
TBP mutants are defective for TATA element binding in vitro.
In accordance with the mutations mapping to the DNA-binding surface, TBP mutant proteins purified from E. coli are unable to bind a consensus TATA element in vitro, even at high concentrations (Fig. 8). The lack of TATA element binding is not due to misfolding or general inactivity of the TBP mutant proteins, because these proteins are fully functional for other biochemical activities (see below). The lack of TATA element binding in vitro does not conflict with our conclusion that the TBP mutants directly associate with TATA elements in vivo. In particular, there are a number of TBP derivatives that do not detectably associate with TATA elements in vitro yet are fully competent to support yeast cell viability and transcription in vivo (1, 41). This apparent discrepancy is probably due to the fact that TBP association with TATA elements in vivo is stabilized by TAFs and other components of the Pol II machinery (40, 44) and hence is not simply due to its inherent DNA-binding affinity.
FIG. 8.
TBP mutants are defective for TATA element binding in vitro. The indicated amounts of wild-type (wt) and mutant TBP derivatives were incubated with a double-stranded oligonucleotide containing a consensus TATA element from the adenovirus 2 E1B promoter, and TBP-DNA complexes were resolved on a 5% Tris-glycine-EDTA polyacrylamide gel containing 4 mM MgCl2.
TBP mutants are unaffected in dimerization, interaction with NC2, or interaction with the inhibitory domain of TAF130.
The DNA-binding surface of TBP is also important for dimerization (46), for interaction with the N-terminal inhibitory domain of TAF130 (35), and presumably for interaction with NC2, a general negative regulator of TBP function (16, 30). In vitro, TBP forms homodimers at concentrations similar to those found in yeast nuclei, and dimer dissociation can be rate-limiting for binding TATA elements (8, 59). If the TBP mutants have a dimerization defect, they might have a higher concentration of active TBP (and presumably TFIID) monomers, with the end result being an increase in transcription. Alternatively, a defect in interaction with the TAF130 inhibitory domain or with NC2 might liberate the DNA-binding surface in the context of TFIID and lead to increased TATA element binding.
As assayed by cross-linking (Fig. 9 and data not shown), wild-type and mutant TBP derivatives are indistinguishable for homodimerization at concentrations ranging from 10-fold lower to 10-fold higher than the apparent Kd (8). In addition, we could not detect any differences in heterodimerization when we incubated the wild-type and ΔN69 TBP derivatives with the wild-type TBP core domain. Finally, using affinity chromatography, wild-type and mutant TBPs are indistinguishable in their abilities to interact with the N-terminal inhibitory domain of TAF130 (Fig. 10A) or with NC2 (Fig. 10B). Thus, our TBP mutants are not defective for dimerization or for interaction with TAF130 or NC2 in vitro and are unlikely to be defective for these functions in vivo.
FIG. 9.
Dimerization properties of TBP and TBP-ΔN69 are indistinguishable. To examine homodimerization (left), TBP derivatives were incubated in the presence (+) or absence (−) of the BMH cross-linker, and the resulting products were analyzed by Western blotting. The protein concentration (100 nM) is approximately 10-fold higher than the observed Kd for TBP in solution. The same procedure was repeated at a range of concentrations from 10-fold lower to 10-fold higher than the Kd, with no differences observed between wild-type (wt) TBP and any of the mutant proteins. To examine heterodimerization (right), TBPc, a proteolytic fragment containing the conserved core of wild-type TBP, was incubated at a 10-fold molar excess over TBP or TBP-ΔN69 (100 nM) in order to approximate the in vivo expression levels. The band intensities of TBPc monomers and dimers are much lower than expected from the amount of protein added because the TBP antibody primarily recognizes epitopes in the nonconserved N terminus, which is not present in TBPc.
FIG. 10.
TBP mutants interact normally with the N-terminal inhibitory domain of TAF130 and the general negative regulator NC2. (A) GST fusions to regions of TAF130 that contain (10–88) or lack (10–58) the N-terminal inhibitory region were incubated with the indicated TBP derivatives. After GST pulldown, the associated proteins were analyzed by Western blotting with TBP antibodies. (B) The indicated amounts of recombinant NC2 were incubated with 200 ng of histidine-tagged TBP or TBP-ΔN69 in the presence of Ni-NTA-agarose. The amount of NC2 retained by the TBP derivatives was analyzed by Western blotting using antibodies to NC2. wt, wild type.
DISCUSSION
TBP mutants that directly and specifically increase transcription from core promoters.
In the presence of wild-type TBP, the mutants described here increase transcription of the following unrelated promoters: the SUC2 promoter under conditions of repression by Cyc8-Tup1, a modified CYC1 promoter under conditions of repression by the Sin3-Rpd3 histone deacetylase complex, and the core HIS3 promoter lacking all known upstream elements. In contrast, the TBP mutants do not increase transcription of a comparably weak promoter containing a mutated TATA element nor do they affect transcription of his3 promoter derivatives under conditions of modest or strong activation. In fact, chromatin immunoprecipitation experiments suggest that the ΔN69 derivative is mildly defective for transcription from moderate or strong promoters. Taken together, these results strongly suggest that the TBP mutants specifically increase transcription from core promoters under conditions where activators are either not present or functionally inhibited (either directly or indirectly) by transcriptional corepressors recruited to the promoters.
Several independent lines of evidence provide a compelling argument that the TBP mutants act directly at the affected promoters and do not function by titrating out a negative factor that permits wild-type TBP to function at the affected promoters. First, the TBP mutants are transcriptionally competent when artificially recruited to promoters via a heterologous DNA-binding domain. Second, mutational analysis reveals a correlation between transcriptional activity in the context of wild-type TBP and in the context of the TBP derivatives. Third, it is very difficult to explain how low-level expression of the TBP mutants could titrate out or sequester a negative factor to permit wild-type TBP to have increased function at core promoters, particularly since this phenotype is not observed upon overexpression of wild-type TBP. Fourth, because overexpression of the TBP mutants, but not wild-type TBP, is toxic for cell growth, any titration model would require that the mutants have a much greater affinity for the inhibitor than wild-type TBP; this seems unlikely given the radical nature of the mutations. Instead, the toxicity caused by overproduction of the TBP mutants is likely to reflect inappropriate expression of natural promoters that contain nonfunctional or repressed enhancers under the conditions examined. Fifth, chromatin immunoprecipitation experiments indicate that the ΔN69 derivative can directly associate with promoters in vivo.
Similarities to and differences from previously described TBP mutants that selectively increase transcription from weak promoters.
Blair and Cullen have described a TBP mutant that increases transcription from several weak promoters while having no significant effect on stronger promoters (3). The mutant contains an amino acid substitution, N69S, that affects the same residue that is deleted in one of our mutants (ΔN69). Furthermore, the N69S derivative is similar to our mutants in that it stimulates transcription when artificially recruited to promoters. The analysis of the N69S derivative was insufficient to determine conclusively whether it functioned directly at the affected promoters or titrated out negative factors.
Despite the similarities between the N69S mutant and the TBP derivatives described here, there are significant differences. First and most important, N69S does not pass the genetic selection used to isolate the TBP mutants described here, indicating that it is unable to increase transcription from a promoter repressed by Cyc8-Tup1. Similarly, our genetic selection yielded two independent mutants with the ΔN69 allele, which occurs very infrequently in the TBP mutant library, but did not yield any amino acid substitutions in N69, which represent approximately 5% of the TBP derivatives in the library (13). Second, N69S efficiently binds TATA elements in vitro whereas the TBP derivatives described here show no detectable DNA-binding activity. Other biochemical properties of the N69S derivative were not examined, thereby preventing comparison to the TBP derivatives described here. Third, increased transcription by our TBP mutants requires a functional TATA element, whereas this is not the case for the N69S derivative.
After our experimental work was completed, but just prior to submission of this report, additional TBP mutants that increase activator-independent transcription were described (23). Several of these TBP mutants have amino acid substitutions at N69, and one of them is identical to the V71R derivative described here. However, in apparent contradiction to our results, these TBP mutants were said to be defective for heterodimerization with wild-type TBP. This conclusion was based solely on a GST pulldown assay in which wild-type and mutant TBPs were assessed for their abilities to interact with the C-terminal core domain of TBP that was immobilized on glutathione agarose beads. This GST pulldown assay does not strictly measure heterodimerization (it monitors any form of TBP-TBP association), and the extended incubation and washing times make the assay very nonlinear with respect to Kd and are likely to magnify a subtle difference between wild-type and mutant TBPs. In addition, the assay is complicated by the dimeric nature of the GST moiety itself and the fact that experiments were performed at 4°C, conditions that favor TBP multimerization (48).
In contrast, our dimerization assays are specific in that the levels of TBP monomers and dimers were directly monitored on gels. In addition, the relative monomer and dimer levels were measured during a period of only 30 s, which represents the time of cross-linking. Finally, it is particularly important to note that the TBP-ΔN69 derivative showed no defect in homodimerization. For assessing the quality of the dimerization interface, homodimerzation is a much more stringent assay than heterodimerization because, with respect to wild-type TBP homodimers, mutant homodimers have two significant structural perturbations whereas heterodimers have only one. Indeed, in many examples of dimeric proteins (e.g., leucine zippers), mutant homodimers are virtually always more defective than mutant–wild-type heterodimers. Thus, although we cannot exclude very subtle effects, our results strongly argue that dimerization of the ΔN69 derivative is unaffected.
Potential molecular mechanisms for increased transcription by the TBP mutants.
Biochemical analysis indicates that the ΔN69 derivative is unimpaired in homodimerization, heterodimerization with wild-type TBP, interaction with NC2, and interaction with the N-terminal inhibitory domain of TAF130. In addition, the ΔN69 derivative appears to form normal TFIID complexes, although subtle differences in composition or conformation cannot be excluded. Although the other two TBP mutants were not tested for these biochemical properties, we suspect that they will behave in a manner similar to the ΔN69 derivative, given that the three TBP mutants are virtually indistinguishable by all other criteria examined. Lastly, the mutations all map to the DNA-binding surface, strongly arguing that the TBP mutants are not affected directly in their abilities to interact with TFIIA or TFIIB. Thus, it seems unlikely that the transcriptional phenotypes conferred in vivo by the ΔN69 (and presumably the other) TBP mutants are due to defects in any of the above interactions.
In considering potential mechanisms, the key observation is that the TBP mutants selectively increase transcription from core promoters. This selectivity argues against (although it does not disprove) models in which the mutations simply increase the concentration of active TBP by blocking the interaction with negative regulators that sequester TBP into inactive states. Instead, it is more likely that increased core promoter function is due to a special property of the TBP mutants, particularly since this phenotype is observed even when the mutant proteins are expressed at levels considerably below that of wild-type TBP in the same cells.
It is believed that the inactivity of core promoters in vivo is due to the repressive effects of chromatin (58). In particular, nucleosomes severely inhibit TBP binding to TATA elements because the DNA bending required for a TBP-TATA complex is structurally incompatible with DNA wrapped around nucleosomes (20). We therefore speculate that the TBP mutants might have an altered structure such that they can form TBP-TATA complexes in the context of simple (i.e., unaltered) nucleosomal templates more readily than wild-type TBP. Consistent with this idea, our TBP mutants have radically altered DNA-binding surfaces (e.g., deletions of amino acids that contact DNA) that will significantly alter the structure of the TBP-TATA complex, and the TBP-TATA complex involving the N69S derivative has reduced electrophoretic mobility suggestive of altered DNA bending (3).
In principle, this special property of the TBP mutants could account for increased function at core promoters. On the other hand, at moderately or strongly activated promoters, the chromatin structure is likely to be modified via activator-dependent recruitment of nucleosome remodeling (e.g., Swi-Snf) or histone-modifying (e.g., SAGA) activities (14, 37, 39, 58), thereby rendering this special property less important. In fact, the DNA-binding defect of these TBP mutants might actually result in a competitive disadvantage with wild-type TBP for access to moderate or strong promoters.
Inferences about the mechanism of repression by the Cyc8-Tup1 and Sin3-Rpd3 corepressors.
The Cyc8-Tup1 and Sin3-Rpd3 corepressors are recruited to specific promoters by DNA-binding repressor proteins, whereupon they inhibit transcription (24, 28, 60). In the case of the Sin3-Rpd3 histone deacetylase complex, repression is mediated by localized histone deacetylation over a range of 1 to 2 nucleosomes around the site of recruitment (25, 52). However, it is unknown whether these recruited corepressors repress transcription by inhibiting the function of activators or by blocking the activity of the core Pol II machinery. Cyc8-Tup1 can weakly inhibit basal transcription on purified DNA templates in vitro (18), but the magnitude of this effect is far below that observed for Cyc8-Tup1 repression in vivo.
The TBP mutants increase transcription from promoters that are repressed by the Cyc8-Tup1 or Sin3-Rpd3 corepressors or that lack an enhancer element but not from an equivalently weak promoter with a mutated TATA element. Thus, the TBP mutants define a criterion by which promoters repressed by Cyc8-Tup1 or Sin3-Rpd3 histone deacetylase are similar to enhancerless promoters but distinct from TATA-defective promoters. While this consideration does not exclude direct inhibition of the basic Pol II machinery, it supports the idea that Cyc8-Tup1 and Sin3-Rpd3 repress transcription primarily by inhibiting the function of activator proteins.
ACKNOWLEDGMENTS
We thank Jutta Deckert for generating and providing the strain containing the Sin3-Rpd3-repressed his3 promoter, Jim Geiger for the TBP core domain, Michael Green for TAF antibodies, Marie Keaveney for LexA hybrid proteins, Tetsuro Kokubo for strains expressing the GST-TAF130 derivatives, Zarmik Moqtaderi for the copper-inducible promoter used to regulated TBP levels, and Rick Young for purified NC2 and antibodies to TBP and NC2. We also thank Laurent Kuras, Peter Kosa, and Mario Mencia for technical advice and fruitful discussions.
This work was supported by grants to K.S. from the National Institutes of Health (GM30186 and GM53720).
REFERENCES
- 1.Arndt K M, Ricupero-Hovasse S, Winston F. TBP mutants defective in activated transcription in vivo. EMBO J. 1995;14:1490–1497. doi: 10.1002/j.1460-2075.1995.tb07135.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Auble D T, Hansen K E, Mueller C G F, Lane W S, Thorner J, Hahn S. Mot1, a global repressor of RNA polymerase II transcription, inhibits TBP binding to DNA by an ATP-dependent mechanism. Genes Dev. 1994;8:1920–1934. doi: 10.1101/gad.8.16.1920. [DOI] [PubMed] [Google Scholar]
- 3.Blair W S, Cullen B R. A yeast TATA-binding protein mutant that selectively enhances gene expression from weak RNA polymerase II promoters. Mol Cell Biol. 1997;17:2888–2896. doi: 10.1128/mcb.17.5.2888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bryant G O, Martel L S, Burley S K, Berk A J. Radical mutations reveal TATA-box binding protein surfaces required for activated transcription in vivo. Genes Dev. 1996;10:2491–2504. doi: 10.1101/gad.10.19.2491. [DOI] [PubMed] [Google Scholar]
- 5.Burley S K, Roeder R G. Biochemistry and structural biology of transcription factor IID (TFIID) Annu Rev Biochem. 1996;65:769–799. doi: 10.1146/annurev.bi.65.070196.004005. [DOI] [PubMed] [Google Scholar]
- 6.Chatterjee S, Struhl K. Connecting a promoter-bound protein to the TATA-binding protein overrides the need for a transcriptional activation region. Nature. 1995;374:820–822. doi: 10.1038/374820a0. [DOI] [PubMed] [Google Scholar]
- 7.Chen W, Struhl K. Saturation mutagenesis of a yeast his3 TATA element: genetic evidence for a specific TATA-binding protein. Proc Natl Acad Sci USA. 1988;85:2691–2695. doi: 10.1073/pnas.85.8.2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coleman R A, Pugh B F. Slow dimer dissociation of the TATA binding protein dictates the kinetics of DNA binding. Proc Natl Acad Sci USA. 1997;94:7221–7226. doi: 10.1073/pnas.94.14.7221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Collart M A, Struhl K. NOT1(CDC39), NOT2(CDC36), NOT3, and NOT4 encode a global negative regulator of transcription that differentially affects TATA-element utilization. Genes Dev. 1994;8:525–537. doi: 10.1101/gad.8.5.525. [DOI] [PubMed] [Google Scholar]
- 10.Cooper J P, Roth S Y, Simpson R T. The global transcriptional regulators, SSN6 and TUP1, play distinct roles in the establishment of a repressive chromatin structure. Genes Dev. 1994;8:1400–1410. doi: 10.1101/gad.8.12.1400. [DOI] [PubMed] [Google Scholar]
- 11.Cormack B P, Strubin M, Ponticelli A S, Struhl K. Functional differences between yeast and human TFIID are localized to the highly conserved region. Cell. 1991;65:341–348. doi: 10.1016/0092-8674(91)90167-w. [DOI] [PubMed] [Google Scholar]
- 12.Cormack B P, Struhl K. The TATA-binding protein is required for transcription by all three nuclear RNA polymerases in yeast cells. Cell. 1992;69:685–696. doi: 10.1016/0092-8674(92)90232-2. [DOI] [PubMed] [Google Scholar]
- 13.Cormack B P, Struhl K. Regional codon randomization: defining a TATA-binding protein surface required for RNA polymerase III transcription. Science. 1993;262:244–248. doi: 10.1126/science.8211143. [DOI] [PubMed] [Google Scholar]
- 14.Cosma M P, Tanaka T, Nasmyth K. Ordered recruitment of transcription and chromatin remodeling factors to a cell cycle- and developmentally regulated promoter. Cell. 1999;97:299–311. doi: 10.1016/s0092-8674(00)80740-0. [DOI] [PubMed] [Google Scholar]
- 15.Field J, Nikawa J-I, Broek D, MacDonald B, Rodgers L, Wilson I A, Lerner R A, Wigler M. Purification of a RAS-responsive adenylyl cyclase complex from Saccharomyces cerevisiae by use of an epitope addition method. Mol Cell Biol. 1988;8:2159–2165. doi: 10.1128/mcb.8.5.2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gadbois E L, Chao D M, Reese J C, Green M R, Young R A. Functional antagonism between RNA polymerase II holoenzyme and global negative regulator NC2 in vivo. Proc Natl Acad Sci USA. 1997;94:3145–3150. doi: 10.1073/pnas.94.7.3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gaudreau L, Adam M, Ptashne M. Activation of transcription in vitro by recruitment of the yeast RNA polymerase II holoenzyme. Mol Cell. 1998;1:913–916. doi: 10.1016/s1097-2765(00)80090-8. [DOI] [PubMed] [Google Scholar]
- 18.Herschbach B M, Arnaud M B, Johnson A D. Transcriptional repression directed by the yeast a2 protein in vitro. Nature. 1994;370:309–311. doi: 10.1038/370309a0. [DOI] [PubMed] [Google Scholar]
- 19.Huang L, Zhang W, Roth S Y. Amino termini of histones H3 and H4 are required for a1-a2 repression in yeast. Mol Cell Biol. 1997;17:6555–6562. doi: 10.1128/mcb.17.11.6555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Imbalzano A N, Kwon H, Green M R, Kingston R E. Facilitated binding of TATA-binding protein to nucleosomal DNA. Nature. 1994;370:481–485. doi: 10.1038/370481a0. [DOI] [PubMed] [Google Scholar]
- 21.Iyer V, Struhl K. Mechanism of differential utilization of the his3 TR and TC TATA elements. Mol Cell Biol. 1995;15:7059–7066. doi: 10.1128/mcb.15.12.7059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Iyer V, Struhl K. Absolute mRNA levels and transcriptional initiation rates in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1996;93:5208–5212. doi: 10.1073/pnas.93.11.5208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jackson-Fisher A J, Chitikila C, Mitra M, Pugh B F. A role for TBP dimerization in preventing unregulated gene expression. Mol Cell. 1999;3:717–727. doi: 10.1016/s1097-2765(01)80004-6. [DOI] [PubMed] [Google Scholar]
- 24.Kadosh D, Struhl K. Repression by Ume6 involves recruitment of a complex containing Sin3 corepressor and Rpd3 histone deacetylase to target promoters. Cell. 1997;89:365–371. doi: 10.1016/s0092-8674(00)80217-2. [DOI] [PubMed] [Google Scholar]
- 25.Kadosh D, Struhl K. Histone deacetylase activity of Rpd3 is important for transcriptional repression in vivo. Genes Dev. 1998;12:797–805. doi: 10.1101/gad.12.6.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kadosh D, Struhl K. Targeted recruitment of the Sin3-Rpd3 histone deacetylase complex generates a highly localized domain of repressed chromatin in vivo. Mol Cell Biol. 1998;18:5121–5127. doi: 10.1128/mcb.18.9.5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Keaveney M, Struhl K. Activator-mediated recruitment of the RNA polymerase II machinery is the predominant mechanism for transcriptional activation in yeast. Mol Cell. 1998;1:917–924. doi: 10.1016/s1097-2765(00)80091-x. [DOI] [PubMed] [Google Scholar]
- 28.Keleher C A, Redd M J, Schultz J, Carlson M, Johnson A D. Ssn6-Tup1 is a general repressor of transcription in yeast. Cell. 1992;68:709–719. doi: 10.1016/0092-8674(92)90146-4. [DOI] [PubMed] [Google Scholar]
- 29.Kim J L, Nikolov D B, Burley S K. Co-crystal structure of TBP recognizing the minor groove of a TATA element. Nature. 1993;365:520–527. doi: 10.1038/365520a0. [DOI] [PubMed] [Google Scholar]
- 30.Kim S, Na J G, Hampsey M, Reinberg D. The Dr1/DRAP1 heterodimer is a global repressor of transcription in vivo. Proc Natl Acad Sci USA. 1997;94:820–825. doi: 10.1073/pnas.94.3.820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim Y, Geiger J H, Hahn S, Sigler P B. Crystal structure of a yeast TBP-TATA box complex. Nature. 1993;365:512–520. doi: 10.1038/365512a0. [DOI] [PubMed] [Google Scholar]
- 32.Kim Y-J, Bjorklund S, Li Y, Sayre M H, Kornberg R D. A multiprotein mediator of transcriptional activation and its interaction with the C-terminal repeat domain of RNA polymerase II. Cell. 1994;77:599–608. doi: 10.1016/0092-8674(94)90221-6. [DOI] [PubMed] [Google Scholar]
- 33.Klages N, Strubin M. Stimulation of RNA polymerase II transcription initiation by recruitment of TBP in vivo. Nature. 1995;374:822–823. doi: 10.1038/374822a0. [DOI] [PubMed] [Google Scholar]
- 34.Klein C, Struhl K. Increased recruitment of TATA-binding protein to the promoter by transcriptional activation domains in vivo. Science. 1994;266:280–282. doi: 10.1126/science.7939664. [DOI] [PubMed] [Google Scholar]
- 35.Kokubo T, Swanson M J, Nishikawa J I, Hinnebusch A G, Nakatani Y. The yeast TAF145 inhibitory domain and TFIIA competitively bind to TATA-binding protein. Mol Cell Biol. 1998;18:1003–1012. doi: 10.1128/mcb.18.2.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Koleske A J, Young R A. An RNA polymerase II holoenzyme responsive to activators. Nature. 1994;368:466–469. doi: 10.1038/368466a0. [DOI] [PubMed] [Google Scholar]
- 37.Krebs J E, Kuo M-H, Allis C D, Peterson C L. Cell cycle-regulated histone acetylation required for expression of the yeast HO gene. Genes Dev. 1999;13:1412–1421. doi: 10.1101/gad.13.11.1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kuchin S, Yeghiayan P, Carlson M. Cyclin-dependent protein kinase and cyclin homologs SSN3 and SSN8 contribute to transcriptional control in yeast. Proc Natl Acad Sci USA. 1995;92:4006–4010. doi: 10.1073/pnas.92.9.4006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kuo M-H, Zhou J, Jambeck P, Churchill M E A, Allis C D. Histone acetyltransferase activity of yeast Gcn5p is required for the activation of target genes in vivo. Genes Dev. 1998;12:627–639. doi: 10.1101/gad.12.5.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kuras L, Struhl K. Binding of TBP to promoters in vivo is stimulated by activators and requires Pol II holoenzyme. Nature. 1999;389:609–612. doi: 10.1038/21239. [DOI] [PubMed] [Google Scholar]
- 41.Lee M, Struhl K. Mutations on the DNA-binding surface of TBP can specifically impair the response to acidic activators in vivo. Mol Cell Biol. 1995;15:5461–5469. doi: 10.1128/mcb.15.10.5461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee M, Struhl K. A severely defective TATA-binding protein-TFIIB interaction does not preclude transcriptional activation in vivo. Mol Cell Biol. 1997;17:1336–1345. doi: 10.1128/mcb.17.3.1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee T I, Wyrick J J, Koh S S, Jennings E G, Gadbois E L, Young R A. Interplay of positive and negative regulators in transcription initiation by RNA polymerase II holoenzyme. Mol Cell Biol. 1998;18:4455–4462. doi: 10.1128/mcb.18.8.4455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li X-L, Virbasius A, Zhu X, Green M R. Enhancement of TBP binding by activators and general transcription factors. Nature. 1999;389:605–609. doi: 10.1038/21232. [DOI] [PubMed] [Google Scholar]
- 45.Moqtaderi Z, Keaveney M, Struhl K. The histone H3-like TAF is broadly required for transcription in yeast. Mol Cell. 1998;2:675–682. doi: 10.1016/s1097-2765(00)80165-3. [DOI] [PubMed] [Google Scholar]
- 46.Nikolov D B, Hu S-H, Lin J, Gasch A, Hoffman A, Horikoshi M, Chua N-H, Roeder R G, Burley S K. Crystal structure of TFIID TATA-box binding protein. Nature. 1992;360:40–46. doi: 10.1038/360040a0. [DOI] [PubMed] [Google Scholar]
- 47.Orphanides G, Lagrange T, Reinberg D. The general initiation factors of RNA polymerase II. Genes Dev. 1996;10:2657–2683. doi: 10.1101/gad.10.21.2657. [DOI] [PubMed] [Google Scholar]
- 48.Perez-Howard G M, Weil P A, Beechem J M. Yeast TATA binding protein interaction with DNA: fluorescence determination of oligomeric state, equilibrium binding, on-rate, and dissociation kinetics. Biochemistry. 1995;34:8005–8017. doi: 10.1021/bi00025a006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ptashne M, Gann A. Transcriptional activation by recruitment. Nature. 1997;386:569–577. doi: 10.1038/386569a0. [DOI] [PubMed] [Google Scholar]
- 50.Ranish J A, Yudkovsky N, Hahn S. Intermediates in formation and activity of the RNA polymerase II preinitiation complex: holoenzyme recruitment and a postrecruitment role for the TATA box and TFIIB. Genes Dev. 1999;13:49–63. doi: 10.1101/gad.13.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Roeder R G. The role of general initiation factors in transcription by RNA polymerase II. Trends Biochem Sci. 1996;21:327–335. [PubMed] [Google Scholar]
- 52.Rundlett S E, Carmen A A, Suka N, Turner B M, Grunstein M. Transcriptional repression by UME6 involves deacetylation of lysine 5 of histone H4 by RPD3. Nature. 1998;392:831–835. doi: 10.1038/33952. [DOI] [PubMed] [Google Scholar]
- 53.Stargell L A, Struhl K. The TBP-TFIIA interaction in the response to acidic activators in vivo. Science. 1995;269:75–78. doi: 10.1126/science.7604282. [DOI] [PubMed] [Google Scholar]
- 54.Stargell L A, Struhl K. A new class of activation-defective TATA-binding protein mutants: evidence for two steps of transcriptional activation in vivo. Mol Cell Biol. 1996;16:4456–4464. doi: 10.1128/mcb.16.8.4456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Strubin M, Struhl K. Yeast TFIID with altered DNA-binding specificity for TATA elements. Cell. 1992;68:721–730. doi: 10.1016/0092-8674(92)90147-5. [DOI] [PubMed] [Google Scholar]
- 56.Struhl K. Yeast transcriptional regulatory mechanisms. Annu Rev Genet. 1995;29:651–674. doi: 10.1146/annurev.ge.29.120195.003251. [DOI] [PubMed] [Google Scholar]
- 57.Struhl K. Chromatin structure and RNA polymerase II connection: implications for transcription. Cell. 1996;84:179–182. doi: 10.1016/s0092-8674(00)80970-8. [DOI] [PubMed] [Google Scholar]
- 58.Struhl K. Fundamentally different logic of gene expression in eukaryotes and prokaryotes. Cell. 1999;98:1–4. doi: 10.1016/S0092-8674(00)80599-1. [DOI] [PubMed] [Google Scholar]
- 59.Taggart A K P, Pugh B F. Dimerization of TFIID when not bound to DNA. Science. 1996;272:1331–1333. doi: 10.1126/science.272.5266.1331. [DOI] [PubMed] [Google Scholar]
- 60.Tzamarias D, Struhl K. Functional dissection of the yeast Cyc8-Tup1 transcriptional corepressor complex. Nature. 1994;369:758–761. doi: 10.1038/369758a0. [DOI] [PubMed] [Google Scholar]
- 61.Wahi M, Johnson A D. Identification of genes required for a2 repression in Saccharomyces cerevisiae. Genetics. 1995;140:79–90. doi: 10.1093/genetics/140.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Xiao H, Friesen J D, Lis J T. Recruiting TATA-binding protein to a promoter: transcriptional activation without an upstream activator. Mol Cell Biol. 1995;15:5757–5761. doi: 10.1128/mcb.15.10.5757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yean D, Gralla J. Transcription reinitiation rate: a special role for the TATA box. Mol Cell Biol. 1997;17:3809–3816. doi: 10.1128/mcb.17.7.3809. [DOI] [PMC free article] [PubMed] [Google Scholar]