Abstract
There is a growing global concern regarding the rise of antimicrobial resistance among Ureaplasma spp. isolates. However, studies on the antimicrobial susceptibility profiles, resistance mechanisms, and clonality of Ureaplasma spp. clinical isolates are still limited and cover only some geographic regions. Firstly, Ureaplasma species from the urogenital tracts of patients in Shanghai, China, were isolated by using the culture medium (A8 and 10B broth), and identified the genotype by polymerase chain reaction (PCR). Secondly, the antimicrobial susceptibility tests were determined by using broth microdilution assay. Then, the resistance genetic determinants to fluoroquinolones (FQs), macrolides, and tetracyclines were investigated through PCR/DNA sequencing. Finally, the molecular epidemiology of Ureaplasma species was studied by multilocus sequence typing (MLST). Among 258 isolates, Ureaplasma parvum (UPA) and Ureaplasma urealyticum (UUR) were found in 226 (87.60%) and 32 (12.40%) isolates, respectively. The minimum inhibitory concentrations (MICs) of 258 Ureaplasma spp. strains ranged from 0.015 to 64μg/ml for all 11 kinds of antimicrobials. Regardless of species, the isolates were most sensitive to AZI (1.94%), JOS (3.49%), and CLA (4.23%). Among them, there were 39 (15.12%) multidrug-resistant (MDR) strains, including 32 UPA isolates. The resistance rates of UPA to CIP (91.59%), and ROX (36.28%) were significantly higher than those of UUR. Twenty six FQ-resistant isolates had amino acid substitutions in gyrA and in parC (Ser83Leu). Mutations were detected in genes encoding ribosomal proteins L4 (Thr84Ile) and L22 (Ser81Pro) in macrolide-resistant isolates. Tet(M) was found in four UPA isolates. These mutations were mainly found in UPA isolates. Sequence type 1 (ST1) was the predominant ST, which contained 18 isolates. In conclusion, this study showed a higher resistance rate (especially to ROX and CIP), higher substitution rate, and higher MDR rate among UPA strains. The most active antimicrobial agents were AZI, JOS, and CLA. Identifying UPA or UUR in clinical isolates could help clinicians to choose appropriate drugs for treatment. The main resistance mechanisms may involve gene substitution of Ser83Leu in parC and Ser81Pro in L22. ST1 was the predominant ST of Ureaplasma isolates with MDR to FQs and macrolides in Shanghai, China.
Keywords: Ureaplasma spp., drug resistance, urogenital tract samples, multilocus sequence typing, resistance mechanism, multidrug resistance
Introduction
Ureaplasma spp. are among the best characterized mycoplasmal bacteria because they are tightly associated with the urogenital tract pathology in humans (Beeton and Spiller, 2017). Owing to their small genome and limited metabolic and biosynthetic capacities, Ureaplasma spp. are dependent on an exogenous supply of amino acids and require a close association with host cells for survival. As a result, Ureaplasma spp. infections have emerged as a clinical issue in a large group of patients with various diseases, such as nonchlamydial and nongonococcal urethritis (Beeton et al., 2019), prostatitis, cervicitis, as well as pelvic inflammatory disease (Hartmann, 2009). To date, reports have indicated that Ureaplasma spp. could be related to adverse neonatal and pregnancy outcomes, including miscarriage and meningitis in newborns (Waites, et al., 2005; Capoccia et al., 2013; Sprong et al., 2020), fatal hyperammonemia in adults (Bharat et al., 2015).
Ureaplasma spp. have no cell wall and belong to the class Mollicutes; they are characterized by small cell size and genome (Hartmann, 2009). Based on molecular analyses, human Ureaplasma spp. are now divided into Ureaplasma parvum (UPA) and Ureaplasma urealyticum (UUR), which contain at least 14 serovars: UPA (serovars 1, 3, 6, and 14) and UUR (serovars 2, 4, 5, and 7–13; Robertson et al., 2002).
Because Ureaplasma spp. lack cell wall and folic acid synthesis capacity, they are naturally insusceptible to β-lactams, sulfonamides, and trimethoprim (Beeton and Spiller, 2017). However, they are generally considered susceptible to tetracyclines, fluoroquinolones (FQs), and macrolides, which interfere with bacterial protein synthesis or DNA replication (Sweeney et al., 2016). In 2011, the Clinical and Laboratory Standards Institute (CLSI) issued the document M43-A: a standard method for performing and validating in vitro susceptibility tests for clinical Ureaplasma spp. (Clinical and Laboratory Standards Institute, 2011). CLSI M43-A highlights the requirement for standardized media (10B broth or A8 agar). The document lists only five kinds of selected antimicrobials with breakpoints. However, most laboratories lack the capability to cultivate, isolate, and purify fastidious bacteria such as Ureaplasma spp. Only a few studies have characterized antimicrobial resistance levels of Ureaplasma spp. in China and other countries (Zhang et al., 2014a; Fernández et al., 2016).
We have recently reported the relationship between Ureaplasma spp. infection, male infertility, and semen quality, and determined the minimum inhibitory concentrations (MICs) of some antimicrobials for Ureaplasma spp. (Zhou et al., 2018b; Wang et al., 2019). Although some studies have evaluated acquired resistance against several of the above-mentioned agents, the detailed mechanisms underlying the resistance and clonality of Ureaplasma spp. are still scarce and are limited to several geographic regions (Pereyre et al., 2007; Beeton et al., 2009; Leli et al., 2012; De Francesco et al., 2013; Schneider et al., 2015; Fernández et al., 2016). Therefore, the aim of this study was to study the genotype, the antimicrobial susceptibility; corresponding molecular resistance mechanisms; multidrug resistance (MDR) and sequence type (ST) distribution and clonality of large-scale clinical Ureaplasma spp. isolated from the urogenital tracts of patients in Shanghai, China.
Materials and Methods
Clinical Specimens and Species Identification
The study was approved by the Ethics Committee of Shanghai Crops Hospital of Chinese People’s Armed Police (No. 20121206). Clinical specimens were collected from the urogenital tracts of patients (33 men and 225 women; medium age, 28.40years; age range, 19–54years) between 2013 and 2014 in three departments (i.e., departments of dermatology, gynecology and obstetrics, and infertility) of the Shanghai Crops Hospital of Chinese People’s Armed Police. The specimens were collected by picking up endocervical cells or vaginal secretion using sterile swabs in women and from urethral secretions or semen in men.
The secretions were inoculated directly on the selective solid culture medium (A8) and the liquid culture medium at 37°C in an atmosphere of 5% CO2; the results were observed after 48 and 72h. Liquefied semen was inoculated in the broth medium (100μl) and on the selective agar medium in parallel (20μl). Due to an increase in pH caused by Ureaplasma spp. growth, a phenol red indicator was added to media to display a color change. A Leitz Orthoplan microscope was used for direct examination of colonies at 100× magnification. Colonies of Ureaplasma spp. were identified presumptively by their characteristic echinus appearance (brown and tiny) on A8 agar in the presence of the Mn indicator (Viscardi, 2010).
Isolation and Purification
A single Ureaplasma spp. colony was placed into 10B broth media under microscope for purification at 37°C. After the color of the liquid medium had changed into light pink or peach after approximately 16–18h of incubation, the liquid medium was measured and adjusted to pH 7.0; then, the purified strains were identified by PCR and stored in multiple aliquots at −80°C in fetal bovine serum in strains bank of Shanghai Crops Hospital. The selective medium agar (A8) and the liquid medium (10B broth) kits and reference strains (ATCC 33175 and ATCC 15488) were provided by Zhongaisheng Hebei Bioscience Technology Inc. (Xingtai, China).
For Ureaplasma spp. samples, genomic DNA extraction was performed using a Takara DNA Minikit (Shiga, Japan). These samples were reconfirmed by amplification of the Ureaplasma spp.-specific urease gene (a 430-bp DNA product) using U4 and U5 primers. Detection of 284 Ureaplasma spp. isolates was done using DNA sequences of the UM-1 primers (UMS125 and UMA226; Blanchard et al., 1993; Teng et al., 1994; Cunningham et al., 2013) and/or 16S-rRNA gene primers UMS 57-UMA 222 for the detection of UPA (327bp), as well as UMS 170-UMA 263 for the detection of UUR (476bp; Kong et al., 2000; Xie et al., 2009).
Antimicrobial Susceptibility Testing
In this study, 11 antimicrobials were used, including macrolides [erythromycin (ERY), azithromycin (AZI), roxithromycin (ROX), clarithromycin (CLA), josamycin (JOS)], FQs [levofloxacin (LEV), moxifloxacin (MXF), ciprofloxacin (CIP)], and tetracyclines [tetracycline (TET), doxycycline (DOX), and minocycline (MIN)]. All agents (Sigma-Aldrich, St. Louis, MO, United States) were obtained in a powdered form of known purity and were diluted in accordance with their respective manufacturer’s instructions. The MICs were determined in duplicate by broth microdilution performed in 96-well microtiter plates in accordance with the Clinical and Laboratory Standards (CLSI) guidelines and previous reports (Kechagia et al., 2008; Clinical and Laboratory Standards Institute, 2011) using a range of antimicrobial concentrations from 0.0625 to 128μg/ml in 10B broth (Zhong ai sheng). American Type Culture Collection (ATCC) 33,175 (Ureaplasma urealyticum) was used as a quality control strain in each broth microdilution assay.
Briefly, we first quantified these organisms for use in MIC determination; then, the frozen culture aliquots were thawed and enriched, where one part was used for colony counting and the rest was stored at 4°C for 48h for MIC. Next, all the antimicrobials were added to 96-well plates in duplicate. The 96-well plates were placed in a biosafety cabinet, dried at room temperature overnight, and then stored at 4°C for use. Then, the microdilution plates were incubated in ambient air at 37°C until the color change of the positive growth control due to the phenol red pH indicator after approximately 16–24h of incubation. The MIC was then determined as the lowest concentration of an antimicrobial agent that did not show any color change at the time of the growth control’s color change. The MIC50 and MIC90 values were recorded.
Analysis of Macrolide, FQ and Tetracycline Resistance Mechanisms
Among FQ-resistant isolates (n=31; MIC≥32μg/ml), the quinolone resistance-determining regions (QRDRs) of the gyrA, gyrB, parC, and parE genes were studied by PCR/DNA sequencing. Primers were designed for amplification, and PCR-amplified DNA was sequenced in both directions to verify the existence by previously described methods (Bébéar et al., 1999, 2000). The tet(M) gene was assessed by PCR using Blanchard et al. (1992) primers and cycling conditions described previously in a total of 20 tetracycline-resistant isolates (15 UPA and five UUR isolates) and 59 tetracycline-susceptible isolates (Blanchard et al., 1992). For all macrolide-resistant isolates (n=59, 51 UPA and eight UUR isolates), PCR amplification and sequencing were performed for both 23S rRNA alleles and the genes encoding ribosomal proteins L4 and L22 (Pereyre et al., 2007; Beeton et al., 2009; Fernández et al., 2016).
The acquired sequences were analyzed using DNAStar Lasergene 7.1 and were translated into protein sequences by blastx in NCBI. Nucleotide mutations in the 23S rRNA alleles as well as amino acid substitutions in L4 and L22, gyrA, gyrB, parC, and parE were identified by comparison with wild-type sequences, i.e., Ureaplasma parvum serovar 3 str. ATCC 700970 (GenBank accession numbers AF222894), UPA ATCC 700970, Ureaplasma urealyticum serovar 10 str. ATCC 33699 (GenBank accession numbers CP001184; Pereyre et al., 2007; Schneider et al., 2015; Fernández et al., 2016), and UPA serovar 3 str. SV3F4 (GenBank accession numbers AP 014584; Wu et al., 2014).
Multilocus Sequence Typing
Multilocus sequence typing (MLST; involving the ftsH, rpl22, valS, and thrS genes) was performed on a selection of 59 isolates (51 UPA and eight UUR isolates) that were resistant to FQs and macrolides as well as partially susceptible to tetracyclines (Fernández et al., 2016). Population structures of 59 Ureaplasma spp. isolates were predicted by eBURST. All the STs were analyzed.
Statistical Analysis
Data analysis was performed using SPSS, version 21. The Chi-square test was used for comparison of qualitative variables. p<0.05 was considered statistically significant.
Results
Species Identification and Prevalence of Resistance in Ureaplasma spp.
Of the 258 tested specimens, 226 (87.60%) were only positive for UPA and 32 (12.40%) were only positive for UUR (Table 1). There was no significant difference in the distribution of Ureaplasma spp. between the two genders (p>0.05). More than 40% of all the Ureaplasma spp. were isolated from patients aged 19–25years. There was no significant difference in the distribution of UPA and UUR isolates among the age groups (p>0.05; Table 2).
Table 1.
Distribution of Ureaplasma spp. species isolated from 258 cases between female and male.
| UPA (N %) | UUR (N %) | Total (N %) | |
|---|---|---|---|
| Female | 196 (87.11%) | 29 (12.89%) | 225 (87.21%) |
| male | 30 (90.91%) | 3 (9.09%) | 33 (12.79%) |
| Total | 226 (87.60%) | 32 (12.40%) | 258 (100%) |
UPA, Ureaplasma parvum; UUR, Ureaplasma urealyticum.
Table 2.
The distribution of Ureaplasma spp. of 258 cases from the urogenital tracts of patients among different age groups.
| Age group (range, years) | UPA (N %) | UUR (N %) | Total (N %) |
|---|---|---|---|
| 19–25 | 98 (88.29%) | 13 (11.71%) | 111 (43.02%) |
| 26–30 | 63 (86.30%) | 10 (13.70%) | 73 (28.29%) |
| 31–35 | 31 (93.94%) | 2 (6.06%) | 33 (14.44%) |
| >35 | 34 (82.93%) | 7 (17.07%) | 41 (16.55%) |
| Total | 226 (87.60%) | 32 (12.40%) | 258 (100%) |
UPA, Ureaplasma parvum; UUR, Ureaplasma urealyticum.
The detailed results for the MIC range, MIC50 and MIC90 values, and the antimicrobial resistance of Ureaplasma spp. (UPA and UUR) are shown in Table 3. The MICs of 258 Ureaplasma spp. strains ranged from 0.015 to 64μg/ml for all 11 kinds of antimicrobials in vitro. Among them, ciprofloxacin had the least effective activity (MIC50 of 16μg/ml and MIC90 of 64μg/ml) against Ureaplasma spp. CLA was the most active antimicrobial with an MIC50 of 0.125μg/ml and MIC90 of 0.5μg/ml. ERY was the least effective among macrolides (MIC50 of 2μg/ml and MIC90 of 16μg/ml). DOX was the most active tetracycline, with MIC50 of 0.25μg/ml and MIC90 of 2μg/ml.
Table 3.
Antimicrobial in vitro resistance profiles of 258 clinical Ureaplasma spp. isolates.
| ERY N (%) |
AZI N (%) |
ROX N (%) |
CLA N (%) |
JOS N (%) |
LEV N (%) |
MXF N (%) |
CIP N (%) |
TET N (%) |
DOX N (%) |
MIN N (%) |
|
|---|---|---|---|---|---|---|---|---|---|---|---|
| UPA | 39 (17.26) | 4 (1.77) | 82 (36.28) | 9 (3.98) | 6 (2.65) | 167 (73.89) | 61 (27.00) | 207 (91.59) | 39 (17.2) | 22 (9.73) | 18 (7.96) |
| UUR | 6 (18.75) | 1 (3.2) | 3 (9.37) | 3 (9.37) | 3 (9.37) | 21 (65.63) | 11 (34.37) | 25 (78.12) | 6 (18.75) | 7 (21.87) | 6 (18.75) |
| p value | 0.806 | - | 0.002 | - | - | 0.395 | 0.403 | 0.027 | 0.806 | 0.042 | - |
| Total | 45 (17.44) | 5 (1.94) | 85 (32.95) | 12 (4.23) | 9 (3.49) | 188 (72.87) | 72 (27.91) | 232 (89.92) | 45 (17.44) | 29 (11.24) | 24 (9.30) |
| MIC range | 0.03–64 | 0.015–16 | 0.03–64 | 0.015–16 | 0.015–16 | 0.03–64 | 0.03–64 | 0.03–64 | 0.015–16 | 0.015–16 | 0.015–16 |
| MIC50 | 2 | 1 | 1 | 0.125 | 0.125 | 8 | 1 | 16 | 0.5 | 0.25 | 0.5 |
| MIC90 | 16 | 4 | 8 | 0.5 | 1 | 64 | 16 | 64 | 8 | 2 | 4 |
According to the CLSI, the top three drug resistance rates of Ureaplasma spp. were those to CIP (89.92%), LEV (72.87%), and ROX (32.95%). Among them, the drug resistance rates of UPA were as follows: CIP (91.59%), and ROX (36.28%); they were significantly higher than those of UUR. The most active antimicrobial agents were AZI (1.94%), JOS (3.49%), CLA (4.23%), and MIN (9.30%). Of the 258 Ureaplasma spp. strains, there were 77(29.84%) isolates uniformly resistant to ROX and CIP, and 39(15.12%) MDR isolates; both of them were dominated by UPA strains (Table 4).
Table 4.
Occurrence of ROX-CIP and MDR among Ureaplasma spp.
| Ureaplasma spp. (N %) | ROX-CIP | MDR | Total |
|---|---|---|---|
| UPA | 67 (29.65%) | 32 (14.16%) | 226 (87.60%) |
| UUR | 10 (31.25%) | 7 (21.87%) | 32 (12.40%) |
| Total | 77 (29.84%) | 39 (15.12%) | 258 (100%) |
ROX-CIP: both resistant to roxithromycin (ROX) and ciprofloxacin (CIP). MDR, multi-drug resistance: resistant to at least one agent in≥3 antimicrobial categories: macrolides, tetracyclines, and fluoroquinolones. UPA, Ureaplasma parvum; UUR, Ureaplasma urealyticum.
Molecular Characterization of Antimicrobial Resistance
In the comparison with the relative DNA and protein sequences of reference strains, none of the isolates possessed mutations in 23S rRNA. A Thr84Ile mutation was detected in the ribosomal protein L4 of one roxithromycin-resistant UPA isolate. A Ser81Pro mutation was detected in the ribosomal protein L22 of seven Ureaplasma spp. isolates (five roxithromycin-resistant UPA isolates, one erythromycin-resistant UPA isolate, and an UPA isolate resistant to roxithromycin and erythromycin); the latter also had another mutation, Lys91Asn in the protein L22.
The comparison between the QRDR DNA sequences of reference strains and the QRDRs of FQ-resistant isolates (n=31; MIC≥32μg/ml) revealed no mutations in gyrB or parE in any of the isolates. However, gyrA and parC quinolone resistance-associated mutations were found. One FQ-resistant UPA isolate showing a levofloxacin MIC of 32μg/ml harbored a Val120Phe mutation in gyrA, but no QRDR mutations were found in gyrA of FQ-resistant UUR isolates. The most frequent mutation was Ser83Leu in parC, which was present in 22 (88%) and three (50%) of the FQ-resistant UPA and UUR isolates, respectively. Among 32 MDR UPA isolates, there were 14 cases of mutation in parC S83L (Ser83Leu); 2 isolates with Ser83Leu (Ser81Pro) and 1 isolates with Tet(M) Screening of the tet(M) gene, which is associated with tetracycline resistance, was performed among 79 isolates. Two tetracycline-resistant UPA isolates were positive for this gene, but two tetracycline-susceptible UPA isolates also harbored this gene.
Multilocus Sequence Typing
Among the 59 Ureaplasma spp. isolates resistant to FQs and macrolides, the ftsH gene and thrS gene, with the highest discriminatory power, had 10 alleles and 8 alleles, respectively, while the rpL22 gene and valS gene had four and three alleles, respectively.
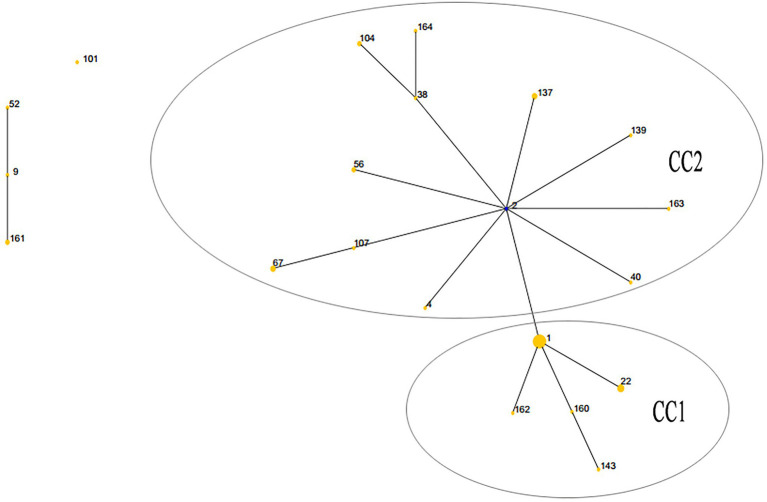
The lineage of the 21 STs in 59 strains was analyzed, and two major clonal complexes (CC1 and CC2) were revealed by the eBURST package. ST2 was the predicted ancestor, highlighted in the blue circle, while the other STs are displayed in the yellow circle. The size of the circle reflects the number of isolates in the relevant ST, and STs are represented as numbers. Twenty one STs appeared among the 59 clinical isolates. ST1 was the predominant ST, which contained 18 UPA isolates. CC1 contained the vast majority of 17 STs in 44 isolates, while the remaining four STs (ST9, ST52, ST101, and ST161) were singletons (Figure 1).
Figure 1.
Population structures of 59 Ureaplasma spp. isolates resistant to quinolones and macrolides predicted by eBURST. CC, clonal complex.
Discussion
In recent years, many questions regarding the role of Ureaplasma spp. as human pathogens, therapeutic strategies, and the antimicrobial susceptibility of individual species remain open. In this study, we examined the antimicrobial susceptibility, resistance mechanisms, and ST distribution of contemporary 258 clinical Ureaplasma spp. isolated from 2013 to 2014 in Shanghai, China. Few previous studies have focused on the individual UPA and UUR distributions in clinical Ureaplasma spp. samples (Kawai et al., 2015; Schneider et al., 2015; Yang et al., 2020). In our study, the positive rate of UPA among clinical Ureaplasma spp. isolates of the urogenital tract was 87.60%, similar to a recent estimate obtained by Fernández et al. (2016) in the United States. Studies of the frequencies of individual species may be useful for further analyses of their clinical characteristics.
The continuous increase in the prevalence of Ureaplasma spp. raised questions regarding their antimicrobial resistance (Zeng et al., 2016; Zhou et al., 2018a). Here, we analyzed the susceptibility of Ureaplasma spp. to 11 kinds of antimicrobial drugs. As expected, UPA and UUR had low resistance to most macrolides and tetracyclines, while their resistance to LEV (72.87%) was very high. While the resistance to macrolides is less than 5% in most countries, our study showed a higher rate of ERY (17.44%) and ROX (32.95%) resistance among clinical strains from Shanghai. Additionally, our results significantly differed from those of previous reports. For instance, Fernández et al. have reported a lower resistance rate to LEV (6%) and no macrolide resistance among the 250 clinical isolates in the United States (Fernández et al., 2016). Some domestic studies of Ureaplasma spp. strains indicated a 20.09% resistance rate to LEV (Zhu et al., 2012), less than 10% to macrolides (Song et al., 2014), and 76.9% to LEV (Song et al., 2015). In addition, Ureaplasma spp. exhibited high resistance to CIP (100%) and to ERY (100% in Tunisia and 80% in South Africa; Redelinghuys et al., 2014; Boujemaa et al., 2020).
Higher resistance rates to ERY, ROX, and LEV may be attributed to the following four factors. First, our study population (87.21% women with border age distribution) may have had higher antimicrobial exposure (especially to macrolides and FQs) and higher resistance levels. Kobayashi et al. found that each age group over 30years had significantly higher odds of receiving FQs for treatment compared with women between 18 and 29years (Kobayashi et al., 2016). However, in Europe and the United States, the macrolide resistance rates of M. pneumoniae are substantially lower than those in Asia (Waites et al., 2017). Second, these overall figures indicate that resistance rates vary significantly among countries; in some instances, there are even dramatic differences within the same country, which may be related to the different strategy or preference of antimicrobials usage in different areas (Yang et al., 2020). Third, another explanation for this variation may be related to the shortcomings of different Mycoplasma testing kits and the reference broth microdilution method currently used to estimate the antimicrobial susceptibility of Ureaplasma spp. Schneider et al. reported that there was conflicting results from the IST 2 kit and standard broth microdilution was observed for CIP and AZI (Schneider et al., 2015). Namely, the Mycoplasma IST2 assay identifies a number of false positives and does not conform to the approved CLSI guidelines (Beeton et al., 2009, 2015). The accuracy of some reports is still questionable since there are no standardized methods for in vitro susceptibility testing and no MIC interpretive breakpoints were designated before 2011. Of note, acidic pH and incubation time affect erythromycin MICs (Kenny and Cartwright, 1993).
Furthermore, mutations, biofilm formation, and the distribution pattern of species or serovars of Ureaplasma spp. can also lead to this phenomenon. The top three drug resistance rates of Ureaplasma spp. were those to CIP, LEV, and ROX. Among CIP and ROX, the drug resistance rates of UPA were significantly higher than those of UUR. The drug resistance rates to AZI, CLA, JOS, and MIN were very low (less than 10%) regardless of Ureaplasma spp.
The relationship and the molecular mechanism between the antimicrobial susceptibility and genotype of Ureaplasma spp. have seldom been reported (Song, 2019). Valentine-King and Brown (2017) reported a trend to higher MIC of UUR compared with UPA. The resistance rates were different for different kinds of antimicrobial between UUR and UPA (Fernández et al., 2016). Therefore, analyses of resistance rates of Ureaplasma spp. require more data to properly explain and interpret the observed trends.
In this study, we also studied Ureaplasma spp. resistance mechanisms to the above-mentioned drugs. As described previously, FQ resistance has been attributed to mutations in gyrA, gyrB, S83L, S83W, and S84P mutations in the parC genes and parE genes (Pereyre et al., 2007; Beeton et al., 2015; Kawai et al., 2015; Schneider et al., 2015; Fernández et al., 2016; Valentine-King and Brown, 2017; Meygret et al., 2018). A sequence analysis of the QRDRs revealed that the resistance mechanism of FQ-resistant isolates involves mutations at position 248 (C-T) of parC, which leads to a substitution of Ser83 by Leu, followed by a Val120Phe substitution, which was mostly found in UPA isolates. As in previous studies, Beeton et al. (2009) also found that about 80% of isolates were UUR, and the ParC Ser83Leu mutation was largely found in all FQ-resistant Ureaplasma spp. We found that three FQ-resistant UPA isolates and three FQ-resistant UUR isolates among 31 Ureaplasma spp. isolates with high MICs (≥32μg/ml) for LEV, MXF and CIP did not harbor QRDR mutations, suggesting the existence of other undescribed resistance mechanisms in these isolates.
Macrolide resistance has been related to mutations in domain V of 23S rRNA, ribosomal proteins L4 and L22, similar to the mechanism underlying Mycoplasma pneumoniae resistance (Schneider et al., 2015; Song et al., 2015, 2019). The macrolide resistance of Ureaplasma spp. is still a controversial issue. The A2066G (A to G) mutation and C2243N (T or C) mutation in the 23S rRNA may be associated with Ureaplasma spp. macrolide resistance in the Chinese population (Meng et al., 2008; Song et al., 2019). We analyzed the aforementioned resistance traits for 59 Ureaplasma spp. isolates showing drug resistance to macrolides; while none of the isolates possessed mutations in the 23S rRNA, we found a mutation in the L4 protein gene and two mutations of the L22 protein gene in eight macrolide-resistant UPA isolates. Yang et al. (2020) reported double alterations (A121S and T141I) or a single mutation (D66N) in L22 within five strains, including one sample harboring an S21A substitution in L4 at the same time. In addition, Pereyre et al. detected two mutations (Lys91Asn and Ala94Asp) in the ribosomal protein L22 and three mutations (Thr70Lys, Gly71Val, and Trp65Arg) in the ribosomal protein L4 (Pereyre et al., 2007); the Lys91Asn mutation in L22 of an erythromycin-resistant UPA isolate was also found in one UPA isolate resistant to erythromycin and roxithromycin and harboring another mutation, Ser81Pro, in our study. The most resistance mechanism to macrolides included the gene mutations in L4 and L22 ribosomal proteins.
The presence of tet(M) has been reported in both tetracycline-susceptible and -resistant Ureaplasma spp. isolates. In this study, the prevalence of tet(M) gene (5.1%, 4/79) was lower than in other reports (Kenny and Cartwright, 1993; Teng et al., 1994; Pereyre et al., 2007; Kotrotsiou et al., 2015). We also found that the tet(M) gene may not be a specific indicator of tetracycline-resistant Ureaplasma spp. isolates.
In this study, uniform resistance to ROX and CIP was the most frequently observed pattern (29.84%) among Ureaplasma spp. isolates; an elevated MDR rate among Ureaplasma spp. (15.12%) was detected, particularly among UPA isolates. Zhu et al. (2016) reported only 1.08% MDR of Ureaplasma spp. in Nanjing, China, while Boujemaa et al. found an elevated MDR rate (37.62%) with 60.71% of UUR strains and 28.76% of UPA being MDR (Boujemaa et al., 2020). To the best of our knowledge, this is the first report of a higher MDR rate of Ureaplasma spp. isolates in Shanghai, China.
To study the molecular epidemiology and population structure of Ureaplasma spp., an MLST approach based on four housekeeping genes (ftsH, rpL22, valS, and thrS) was first developed by Zhang et al. (2014b) who used 14 serovar reference strains and 269 clinical strains. ST1 and ST22 were the predominant STs and contained 68 and 70 isolates, respectively. Zhang et al. (2014a) also found that isolates of CC1 were UPA and those of CC2 were UUR. CC2 was found more often in symptomatic patients with vaginitis, tubal obstruction, and cervicitis. In addition, two recent studies evaluated the molecular epidemiology of clinical Ureaplasma spp. isolates in the United States and Switzerland, making reference to the research of Zhang et al.; these studies suggested the association of molecular epidemiology of Ureaplasma spp. isolates with pathogenicity or spread of antimicrobial resistance (Schneider et al., 2015; Fernández et al., 2016). We found that ST1 (18 UPA isolates) was the predominant ST among 59 MDR isolates, followed by ST22 (5 UPA isolates). Among 18 ST1 UPA isolates, there was higher MDR rate (8 cases, 44.44%) and the mutations found as the following: parC gene (4 cases), GyrA/parC (one case), and L22: S81P (one case). ST1 was the main clone both in Hangzhou (Zhang et al., 2014a) and Shanghai, which may hypothesize that ST1 can be associated with spread of the urogenital tracts of different populations or the antimicrobial resistance. MLST is a well-accepted way for illustrating the diversity and population structure of different bacterial species compared to any other molecular subtyping method. As the strain numbers of STs were very limited, it was difficult to draw any decisive conclusions on any possible correlations between STs and pathologies or molecular resistance mechanisms of Ureaplasma spp.
In conclusion, using a large number of clinical isolates, which were mainly obtained from women, this study addressed the lack of data in China related to the antimicrobial susceptibility of Ureaplasma spp. The prevalence of antimicrobial resistance in Ureaplasma spp., especially that to ERY, ROX, and LEV, is markedly higher in China than in European countries. The most active antimicrobial agents were AZI, JOS, and CLA. Identifying UPA or UUR in clinical isolates could help clinicians to choose appropriate drugs for treatment. For UPA in particular, ERY, ROX, and FQs should be used with caution due to the relatively high rate of MDR. ST1 was the predominant ST of Ureaplasma isolates with MDR to FQs and macrolides in Shanghai, China. The main resistance mechanisms may involve gene substitution of Ser83Leu in parC and Ser81Pro in L22, in addition to other, undescribed mechanisms. One of the limitations of this study is that it did not provide sufficient data to better understand the epidemiology of Ureaplasma spp. in China. Future work should extend our investigations to address the resistance mechanisms related to the spread of antimicrobial resistance, particularly MDR strains, and the difference in drug resistance in different Ureaplasma spp.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://www.ncbi.nlm.nih.gov/genbank/MZ700119-MZ700176 for ftsH, MZ710758 – MZ710815 for valS, MZ710816 – MZ710874 for rpL22, MZ710875 – MZ710931 for thrS.
Ethics Statement
The studies involving human participants were reviewed and approved by the Ethics Committee of Shanghai Crops Hospital of Chinese People’s Armed Police. Written informed consent for participation was not required for this study in accordance with the national legislation and the institutional requirements.
Author Contributions
HM, JZ, and YZ contributed to conception and design of the study. HM, XZ, XS, and YZ performed the experimental studies and analysis. HM, XZ, and YZ wrote the section of the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was financially supported by the Key Programs of Science and Technology Commission Foundation of Changning District, Shanghai (CNKW2016Z05), the Jiangxi Provincial Health Technology Project (20202114), the National Natural Science Foundation of China (NSFC81370047), and Natural Science Foundation of Shanghai (13ZR1449700).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank LetPub (www.letpub.com) for its linguistic assistance during the preparation of this manuscript.
References
- Bébéar C. M., Grau O., Charron A., Renaudin H., Gruson D., Bébéar C. (2000). Cloning and nucleotide sequence of the DNA gyrase (gyrA) gene from Mycoplasma hominis and characterization of quinolone-resistant mutants selected in vitro with trovafloxacin. Antimicrob. Agents Chemother. 44, 2719–2727. doi: 10.1128/AAC.44.10.2719-2727.2000, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bébéar C. M., Renaudin J., Charron A., Renaudin H., de Barbeyrac B., Schaeverbeke T., et al. (1999). Mutations in the gyrA, parC, and parE genes associated with fluoroquinolone resistance in clinical isolates of Mycoplasma hominis. Antimicrob. Agents Chemother. 43, 954–956. doi: 10.1128/AAC.43.4.954, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beeton M. L., Chalker V. J., Jones L. C., Maxwell N. C., Spiller O. B. (2015). Antibiotic resistance among clinical Ureaplasma isolates recovered from neonates in England and Wales between 2007 and 2013. Antimicrob. Agents Chemother. 60, 52–56. doi: 10.1128/AAC.00889-15, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beeton M. L., Chalker V. J., Maxwell N. C., Kotecha S., Spiller O. B. (2009). Concurrent titration and determination of antibiotic resistance in Ureaplasma species with identification of novel point mutations in genes associated with resistance. Antimicrob. Agents Chemother. 53, 2020–2027. doi: 10.1128/AAC.01349-08, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beeton M. L., Payne M. S., Jones L. (2019). The role of Ureaplasma spp. in the development of nongonococcal urethritis and infertility among men. Clin. Microbiol. Rev. 32:e00137. doi: 10.1128/CMR.00137-18, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beeton M. L., Spiller O. B. (2017). Antibiotic resistance among Ureaplasma species isolates: cause for concern? J. Antimicrob. Chemother. 72, 330–337. doi: 10.1093/jac/dkw425 [DOI] [PubMed] [Google Scholar]
- Bharat A., Cunningham S. A., Scott Budinger G. R., Kreisel D., DeWet C. J., Gelman A. E., et al. (2015). Disseminated Ureaplasma infection as a cause of fatal hyperammonemia in humans. Sci. Transl. Med. 7:258.re3. doi: 10.1126/scitranslmed.aaa8419, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchard A., Crabb D. M., Dybvig K., Duffy L. B., Cassell G. H. (1992). Rapid detection of tetM in mycoplasma hominis and Ureaplasma urealyticum by PCR: tetM confers resistance to tetracycline but not necessarily to doxycycline. FEMS Microbiol. Lett. 95, 277–281. doi: 10.1016/0378-1097(92)90442-q, PMID: [DOI] [PubMed] [Google Scholar]
- Blanchard A., Hentschel J., Duffy L., Baldus K., Cassell G. H. (1993). Detection of Ureaplasma urealyticum by polymerase chain reaction in the urogenital tract of adults, in amniotic fluid, and in the respiratory tract of newborns. Clin. Infect. Dis. 17(Suppl. 1), S148–S153. doi: 10.1093/clinids/17.supplement_1.s148, PMID: [DOI] [PubMed] [Google Scholar]
- Boujemaa S., Mlik B., Ben Allaya A., Mardassi H., Ben Abdelmoumen Mardassi B. (2020). Spread of multidrug resistance among Ureaplasma serovars, Tunisia. Antimicrob. Resist. Infect. Control 9:19. doi: 10.1186/s13756-020-0681-5, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capoccia R., Greub G., Baud D. (2013). Ureaplasma urealyticum, mycoplasma hominis and adverse pregnancy outcomes. Curr. Opin. Infect. Dis. 26, 231–240. doi: 10.1097/QCO.0b013e328360db58, PMID: [DOI] [PubMed] [Google Scholar]
- Clinical and Laboratory Standards Institute (2011). Methods for Antimicrobial Susceptibility Testing for Human Mycoplasmas; Approved Guideline. CLSI document M43-A. Wayne, PA: Clinical and Laboratory Standards Institute. [PubMed] [Google Scholar]
- Cunningham S. A., Mandrekar J. N., Rosenblatt J. E., Patel R. (2013). Rapid PCR detection of Mycoplasma hominis, Ureaplasma urealyticum, and Ureaplasma parvum. Int. J. Bacteriol. 2013, 1–7. doi: 10.1155/2013/168742, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Francesco M. A., Caracciolo S., Bonfanti C., Manca N. (2013). Incidence and antibiotic susceptibility of Mycoplasma hominis and Ureaplasma urealyticum isolated in Brescia, Italy, over 7 years. J. Infect. Chemother. 19, 621–627. doi: 10.1007/s10156-012-0527-z, PMID: [DOI] [PubMed] [Google Scholar]
- Fernández J., Karau M. J., Cunningham S. A., Greenwood-Quaintance K. E., Patela R. (2016). Antimicrobial susceptibility and clonality of clinical Ureaplasma isolates in the United States. Antimicrob. Agents Chemother. 60, 4793–4798. doi: 10.1128/AAC.00671-16, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann M. (2009). Genital mycoplasmas. J. Dtsch. Dermatol. Ges. 7, 371–377. doi: 10.1111/j.1610-0387.2008.06965.x, PMID: [DOI] [PubMed] [Google Scholar]
- Kawai Y., Nakura Y., Wakimoto T., Nomiyama M., Tokuda T., Takayanagi T., et al. (2015). In vitro activity of five quinolones and analysis of the quinolone resistance-determining regions of gyrA, gyrB, parC, and parE in Ureaplasma parvum and Ureaplasma urealyticum clinical isolates from perinatal patients in Japan. Antimicrob. Agents Chemother. 59, 2358–2364. doi: 10.1128/AAC.04262-14, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kechagia N., Bersimis S., Chatzipanagiotou S. (2008). Incidence and antimicrobial susceptibilities of genital mycoplasmas in outpatient women with clinical vaginitis in Athens, Greece. J. Antimicrob. Chemother. 62, 122–125. doi: 10.1093/jac/dkn158, PMID: [DOI] [PubMed] [Google Scholar]
- Kenny G. E., Cartwright F. D. (1993). Effect of pH, inoculum size, and incubation time on the susceptibility of Ureaplasma urealyticum to erythromycin in vitro. Clin. Infect. Dis. 17(Suppl. 1), S215–S218. doi: 10.1093/clinids/17.supplement_1.s215 [DOI] [PubMed] [Google Scholar]
- Kobayashi M., Shapiro D. J., Hersh A. L., Sanchez G. V., Hicks L. A. (2016). Outpatient antibiotic prescribing practices for uncomplicated urinary tract infection in women in the United States, 2002–2011. Open. Forum. Infect. Dis. 3:ofw159. doi: 10.1093/ofifid/ofw159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong F., Ma Z., James G., Gordon S., Gilbert G. L. (2000). Species identification and subtyping of Ureaplasma parvum and Ureaplasma urealyticum using PCR-based assays. J. Clin. Microbiol. 38, 1175–1179. doi: 10.1128/JCM.38.3.1175-1179.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotrotsiou T., Exindari M., Diza E., Gioula G., Melidou A., Malisiovas N. (2015). Detection of the tetM resistance determinant among phenotypically sensitive Ureaplasma species by a novel real-time PCR method. Diagn. Microbiol. Infect. Dis. 81, 85–88. doi: 10.1016/j.diagmicrobio.2014.10.013 [DOI] [PubMed] [Google Scholar]
- Leli C., Mencacci A., Bombaci J. C., D’Alò F., Farinelli S., Vitali M., et al. (2012). Prevalence and antimicrobial susceptibility of Ureaplasma urealyticum and Mycoplasma hominis in a population of Italian and immigrant outpatients. Infez. Med. 20, 82–87. PMID: [PubMed] [Google Scholar]
- Meng D. Y., Xue W. C., Ma X. B., Wang L. (2008). Transition mutations in 23S rRNA account for acquired resistance to macrolides in Ureaplasma urealyticum. Microb. Drug Resist. 14, 183–186. doi: 10.1089/mdr.2008.0817, PMID: [DOI] [PubMed] [Google Scholar]
- Meygret A., Le Roy C., Renaudin H., Bébéar C., Pereyre S. (2018). Tetracycline and fluoroquinolone resistance in clinical Ureaplasma spp. and Mycoplasma hominis isolates in France between 2010 and 2015. J. Antimicrob. Chemother. 73, 2696–2703. doi: 10.1093/jac/dky238 [DOI] [PubMed] [Google Scholar]
- Pereyre S., Métifiot M., Cazanave C., Renaudin H., Charron A., Bébéar C., et al. (2007). Characterisation of in vitro-selected mutants of Ureaplasma parvum resistant to macrolides and related antibiotics. Int. J. Antimicrob. Agents 29, 207–211. doi: 10.1016/j.ijantimicag.2006.09.008, PMID: [DOI] [PubMed] [Google Scholar]
- Redelinghuys M. J., Ehlers M. M., Dreyer A. W., Lombaard H. A., Kock M. M. (2014). Antimicrobial susceptibility patterns of Ureaplasma species and mycoplasma hominis in pregnant women. BMC Infect. Dis. 14:171. doi: 10.1186/1471-2334-14-171, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson J. A., Stemke G. W., Davis J. W., Jr., Harasawa R., Thirkell D., Kong F., et al. (2002). Proposal of Ureaplasma parvum sp. nov. and emended description of Ureaplasma urealyticum (Shepard et al. 1974). Int. J. Syst. Evol. Microbiol. 52, 587–597. doi: 10.1099/00207713-52-2-587 [DOI] [PubMed] [Google Scholar]
- Schneider S. C., Tinguely R., Droz S., Hilty M., Dona V., Bodmer T., et al. (2015). Antibiotic susceptibility and sequence type distribution of Ureaplasma species isolated from genital samples in Switzerland. Antimicrob. Agents Chemother. 59, 6026–6031. doi: 10.1128/AAC.00895-15, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song T., Huang J., Liu Z., Zhang Y., Kong Y., Ruan Z. (2019). Antibiotic susceptibilities and genetic variations in macrolide resistance genes of Ureaplasma spp. isolated in China. New Microbiol. 42, 225–227. PMID: [PubMed] [Google Scholar]
- Song J., Qiao Y., Kong Y., Ruan Z., Huang J., Song T., et al. (2015). Frequent topoisomerase IV mutations associated with fluoroquinolone resistance in Ureaplasma species. J. Med. Microbiol. 64, 1315–1320. doi: 10.1099/jmm.0.000153, PMID: [DOI] [PubMed] [Google Scholar]
- Song T., Ye A., Xie X., Huang J., Ruan Z., Kong Y., et al. (2014). Epidemiological investigation and antimicrobial susceptibility analysis of Ureaplasma species and Mycoplasma hominis in outpatients with genital manifestations. J. Clin. Pathol. 67, 817–820. doi: 10.1136/jclinpath-2014-202248, PMID: [DOI] [PubMed] [Google Scholar]
- Sprong K. E., Mabenge M., Wright C. A., Govender S. (2020). Ureaplasma species and preterm birth: current perspectives. Crit. Rev. Microbiol. 46, 169–181. doi: 10.1080/1040841X.2020.1736986, PMID: [DOI] [PubMed] [Google Scholar]
- Sweeney E. L., Dando S. J., Kallapur S. G., Knox C. L. (2016). The human Ureaplasma species as causative agents of chorioamnionitis. Clin. Microbiol. Rev. 30, 349–379. doi: 10.1128/CMR.00091-16, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng L. J., Zheng X., Glass J. I., Watson H. L., Tsai J., Cassell G. H. (1994). Ureaplasma urealyticum biovar specificity and diversity are encoded in multiple-banded antigen gene. J. Clin. Microbiol. 32, 1464–1469. doi: 10.1128/jcm.32.6.1464-1469.1994, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valentine-King M. A., Brown M. B. (2017). Antibacterial resistance in Ureaplasma species and Mycoplasma hominis isolates from urine cultures in college-aged females. Antimicrob. Agents Chemother. 61, e01104–e01117. doi: 10.1128/AAC.01104-17, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viscardi R. M. (2010). Ureaplasma species: role in diseases of prematurity. Clin. Perinatol. 37, 393–409. doi: 10.1016/j.clp.2009.12.003, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waites K. B., Katz B., Schelonka R. L. (2005). Mycoplasmas and Ureaplasmas as neonatal pathogens. Clin. Microbiol. Rev. 18, 757–789. doi: 10.1128/CMR.18.4.757-789.2005, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waites K. B., Xiao L., Liu Y., Balish M. F., Atkinson T. P. (2017). Mycoplasma pneumoniae from the respiratory tract and beyond. Clin. Microbiol. Rev. 30, 747–809. doi: 10.1128/CMR.00114-116, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang N., Liu W., Zhou Y., Liu Y. (2019). In vitro activities of Nemonoxacin and other antimicrobial agents against human Mycoplasma and Ureaplasmas isolates and their defined resistance mechanisms. Front. Microbiol. 10:1890. doi: 10.3389/fmicb.2019.01890, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H. N., Nakura Y., Motooka D., Nakamura S., Nishiumi F., Ishino S., et al. (2014). Complete genome sequence of Ureaplasma parvum Serovar 3 strain SV3F4, isolated in Japan. Genome Announc. 2, e00256–e00214. doi: 10.1128/genomeA.00256-14, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X. M., Huang K. S., Li W. H. (2009). Species identification genotyping of Ureaplasma in urogenital tract of male with secondary infertility and its clinical application. J. Reprod. Med. 18, 25–28. [Google Scholar]
- Yang T., Pan L., Wu N., Wang L., Liu Z., Kong Y., et al. (2020). Antimicrobial resistance in clinical Ureaplasma spp. and Mycoplasma hominis and structural mechanisms underlying quinolone resistance. Antimicrob. Agents Chemother. 64:e02560–19. doi: 10.1128/AAC.02560-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng X. Y., Xin N., Tong X. N., Wang J. Y., Liu Z. W. (2016). Prevalence and antibiotic susceptibility of Ureaplasma urealyticum and mycoplasma hominis in Xi’an, China. Eur. J. Clin. Microbiol. Infect. Dis. 35, 1941–1947. doi: 10.1007/s10096-016-2745-2, PMID: [DOI] [PubMed] [Google Scholar]
- Zhang J., Kong Y., Feng Y., Huang J., Song T., Ruan Z., et al. (2014a). Development of a multilocus sequence typing scheme for Ureaplasma. Eur. J. Clin. Microbiol. Infect. Dis. 33, 537–544. doi: 10.1007/s10096-013-1981-y, PMID: [DOI] [PubMed] [Google Scholar]
- Zhang J., Kong Y., Ruan Z., Huang J., Song T., Song J., et al. (2014b). Correlation between Ureaplasma subgroup 2 and genitourinary tract disease outcomes revealed by an expanded multilocus sequence typing (eMLST) scheme. PLoS One 9:e104347. doi: 10.1371/journal.pone.0104347, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y. H., Ma H. X., Shi X. X., Liu Y. (2018b). Ureaplasma spp. in male infertility and its relationship with semen quality and seminal plasma components. J. Microbiol. Immunol. Infect. 51, 778–783. doi: 10.1016/j.jmii.2016.09.004, PMID: [DOI] [PubMed] [Google Scholar]
- Zhou Y. H., Ma H. X., Yang Y., Gu W. M. (2018a). Prevalence and antimicrobial resistance of Ureaplasma spp. and mycoplasma hominis isolated from semen samples of infertile men in Shanghai, China from 2011-2016. Eur. J. Clin. Microbiol. Infect. Dis. 37, 729–734. doi: 10.1007/s10096-017-3167-5, PMID: [DOI] [PubMed] [Google Scholar]
- Zhu X., Li M., Cao H., Yang X., Zhang C. (2016). Epidemiology of Ureaplasma urealyticum and Mycoplasma hominis in the semen of male outpatients with reproductive disorders. Exp. Ther. Med. 12, 1165–1170. doi: 10.3892/etm.2016.3409, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu C. T., Liu J. M., Ling Y., Dong C. L., Wu T. T., Yu X. Y., et al. (2012). Prevalence and antimicrobial susceptibility of Ureaplasma urealyticum and Mycoplasma hominis in Chinese women with genital infectious diseases. Indian J. Dermatol. Venereol. Leprol. 78, 406–407. doi: 10.4103/0378-6323.95480, PMID: [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://www.ncbi.nlm.nih.gov/genbank/MZ700119-MZ700176 for ftsH, MZ710758 – MZ710815 for valS, MZ710816 – MZ710874 for rpL22, MZ710875 – MZ710931 for thrS.