Abstract
The brain consists of three major cell types: neurons and two glial cell types (astrocytes and oligodendrocytes). Although they are generated from common multipotent neural stem/precursor cells (NS/PCs), embryonic NS/PCs cannot generate all of the cell types at the beginning of brain development. NS/PCs first undergo extensive self‐renewal to expand their pools, and then acquire the potential to produce neurons, followed by glial cells. Astrocytes are the most frequently found cell type in the central nervous system (CNS), and play important roles in brain development and functions. Although it has been shown that nuclear factor IA (Nfia) is a pivotal transcription factor for conferring gliogenic potential on neurogenic NS/PCs by sequestering DNA methyltransferase 1 (Dnmt1) from astrocyte‐specific genes, direct targets of Nfia that participate in astrocytic differentiation have yet to be completely identified. Here we show that SRY‐box transcription factor 8 (Sox8) is a direct target gene of Nfia at the initiation of the gliogenic phase. We found that expression of Sox8 augmented leukemia inhibitory factor (LIF)‐induced astrocytic differentiation, while Sox8 knockdown inhibited Nfia‐enhanced astrocytic differentiation of NS/PCs. In contrast to Nfia, Sox8 did not induce DNA demethylation of an astrocyte‐specific marker gene, glial fibrillary acidic protein (Gfap), but instead associated with LIF downstream transcription factor STAT3 through transcriptional coactivator p300, explaining how Sox8 expression further facilitated LIF‐induced Gfap expression. Taken together, these results suggest that Sox8 is a crucial Nfia downstream transcription factor for the astrocytic differentiation of NS/PCs in the developing brain.
Keywords: astrocyte, brain development, neural stem/precursor cells, Nfia, Sox8
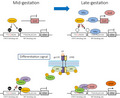
Dnmt1 maintains DNA methylation on the promoter of astrocytic genes such as Gfap in mid‐gestational NS/PCs, precluding their differentiation into astrocytes even if STAT3 is activated by the astrocyte‐inducing cytokine LIF (left). In late‐gestation, Nfia induces Sox8 expression, and sequesters Dnmt1 from astrocytic gene promoters, leading to their demethylation. As a result, NS/PCs now acquire the potential of astrocyte differentiation: upon LIF stimulation, STAT3 can associates with the astrocytic gene promoter together with p300 and Sox8 to efficiently activate the target gene.
Abbreviations
- BMP
bone morphogenetic protein
- CBP
Creb binding protein
- ChIP
chromatin immunoprecipitation
- DMEM
Dulbecco's Modified Eagle Medium
- Dnmt1
DNA methyltransferase 1
- GFAP
glial fibrillary acidic protein
- GFP
green fluorescent protein
- HBSS
Hank's balanced salt solution
- LIF
leukemia inhibitory factor
- Nfia
nuclear factor IA
- NS/PCs
neural stem/precursor cells
- Sox8
SRY‐box transcription factor 8
- STAT3
signal transducer and activator of transcription 3
1. INTRODUCTION
The brain develops rapidly over the course of embryonic stages, during which various cells are generated and integrated into not only simultaneously but also in an orderly manner. Neural stem/precursor cells (NS/PCs) are the main source of brain cells, and have abilities to self‐renew and to generate neurons and two types of glial cells (astrocytes and oligodendrocytes). 1 In early gestation, however, they divide symmetrically to expand their own pool, and gradually change their mode of division into an asymmetric mode to produce other cell types. In mid‐gestation, they start to produce neurons, and then generate glial cells from late gestation until the perinatal period. 2 This sequential acquisition of differentiation potential is essential for generating a balanced number of each cell type at the right timing to produce the elaborate brain structure.
Astrocytes are abundant in the entire central nervous system (CNS), executing various physiological functions including physical support of the neuronal network, promotion of synaptogenesis, regulation of the extracellular ion balance and neurotransmission, and support of the formation of the blood–brain barrier. 3 , 4 , 5 Therefore, disturbance of astrocyte development leads to brain dysfunctions such as neurological and psychiatric disorders. 6
Extracellular factors secreted from neurons and meningeal cells in the NS/PCs niche are known to participate in astrogenesis. 7 , 8 , 9 , 10 For example, members of the interleukin (IL)‐6 family of cytokines such as leukemia inhibitory factor (LIF), ciliary neurotrophic factor, and cardiotrophin‐1 induce astrocytic differentiation of NS/PCs through activation of the gp130‐janus kinase‐signal transducer and activator of transcription 3 (STAT3) signaling pathway. 7 , 11 , 12 The signal of bone morphogenetic proteins (BMPs), which belong to the transforming growth factor β superfamily, is transduced through the downstream pathway‐restricted transcription factors Smad1, 5 and 8. Furthermore, it has been shown that STAT3 and Smad1 form a complex bridged by transcriptional coactivator p300/Creb‐binding protein (CBP), and synergistically induce astrocytic differentiation of late‐gestational NS/PCs. 13 However, mid‐gestational NS/PCs do not differentiate into astrocytes even if STAT 14 and/or Smad1 15 are activated. 16
Notch signaling is involved in the fate switching of NS/PCs from neurogenic to gliogenic. 17 Notch ligands are expressed in neuroblasts and immature neurons, which are produced from NS/PCs, and activate Notch signaling in adjacent remaining NS/PCs. 18 Upon activation, Notch intracellular domain is cleaved and translocated into the nucleus, and forms a complex with recombinant signal sequencing‐binding protein Jκ to transcribe its target genes, such as hes family bHLH transcription factor 1/5 and hairy/enhancer‐of‐split related with YRPW motif 1/2, and inhibit neuronal differentiation via repression of proneural genes. 18 , 19 , 20 Notch signaling also induces the expression of nuclear factor IA (Nfia), a member of a family of CCAAT box element‐binding transcription factors that is essential for specification and differentiation of NS/PCs into astrocytes. 21 In spinal cord development, Nfia expression leads to generation of astrocytic precursors, while its loss results in excess neurogenesis at the expense of astrogenesis. 22 A previous study showed that decreases in the expression of an astrocyte‐specific marker, glial fibrillary acidic protein (GFAP), in the cortex and in the number of forebrain midline glia were observed in Nfia‐deficient mice. 23 It has also been reported that forced expression of Nfia in mouse telencephalon induces precocious astrocytic differentiation. 21
DNA methylation is one of the major epigenetic modifications that affects gene expression, and the maintenance DNA methyltransferase 1 (Dnmt1) reproduces 5‐methylcytocine on newly synthesized DNA during replication. 24 Using NS/PCs from Sox2‐EGFP transgenic mouse, which express EGFP under the control of a NS/PC marker gene Sox2 promoter, we performed genome‐wide analyses and demonstrated that the NFI‐consensus sequence was most enriched within regions whose DNA methylation level decreased from mid‐ to late‐gestation. 25 In addition, we previously reported that Nfia induces DNA demethylation of astrocyte‐specific gene promoters, including that of Gfap, through the induction of Dnmt1 dissociation from the promoters. 21 In fact, conditional deletion of Dnmt1 in NS/PCs causes accelerated demethylation of astrocytic genes, resulting in precocious astrocytic differentiation of NS/PCs in the developing brain. 26
Although Nfia has been characterized as being involved in the induction of DNA demethylation of astrocytic genes, it remains far from clear which Nfia downstream molecules play a critical role at the onset of astrogenesis. Therefore, in this study, we sought to identify Nfia‐target genes using genome‐wide chromatin immunoprecipitation sequencing (ChIP‐seq) and RNA‐seq analyses, and found that among identified Nfia targets, SRY‐box transcription factor 8 (Sox8) promotes astrogenesis of NS/PCs.
2. MATERIALS AND METHODS
All experiments were conducted in compliance with the ARRIVE guidelines.
2.1. Cell culture
Mice were treated according to the guidelines of Kyushu University. We obtained embryonic NS/PCs from time‐pregnant ICR mice as previously described. 14 In brief, telencephalons from embryonic day 11.5 (E11.5) mice were triturated in Hank's balanced salt solution (HBSS) (Sigma‐Aldrich, Cat# H2387) by gentle pipetting. After centrifugation, the cell pellet was resuspended in N2‐supplemented Dulbecco's Modified Eagle Medium (DMEM) /F‐12 (Thermo Fisher Scientific, Cat# 12400024), containing 10 ng/ml recombinant human basic fibroblast growth factor (PeproTech, Cat# 100‐18B) and plated on dishes precoated with poly‐L‐ornithine (Sigma‐Aldrich, Cat# P3655) /fibronectin (Sigma‐Aldrich, Cat# F1141).
2.2. ChIP assay
Cells were fixed with 1% formaldehyde‐containing medium for 10 min and quenched with 125 mM glycine for 10 min at RT. The cells were washed with PBS, and harvested using a cell scraper. After centrifugation, the cell pellet was suspended in buffer 1 (140 mM NaCl, 1 mM EDTA, 50 mM HEPES‐KOH (pH 7.5), 0.5% NP‐40, 10% glycerol, 0.25% Triton X‐100, protease inhibitor) and kept at 4℃ for 10 min. After centrifugation, the supernatant was removed and the pellet was suspended in buffer 2 (20 mM Tris‐HCl (pH 7.5), 200 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, protease inhibitor) and kept at 4℃ for 10 min. After centrifugation, the supernatant was again removed and the pellet was suspended in buffer 3 (20 mM Tris‐HCl (pH 7.5), 150 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, 1% Triton X‐100, 0.1% Na‐deoxycholate, 0.1% SDS, protease inhibitor) and kept at 4℃ for 10 min. After incubation at 4℃ for 10 min, the lysates were sonicated with a Sonifier (Branson). After centrifugation, the supernatant was collected, and 10% of its volume was used for input.
For ChIP, 50 μL of Dynabeads M‐280 sheep anti‐mouse IgG (Thermo Fisher, Cat# DB11202), with magnetic property, were washed and pre‐incubated with primary antibody in suspension with rotation at 4℃ overnight. After incubation, beads were washed and resuspended in buffer 3. Sonicated chromatin was added to the suspension and incubated with rotation at 4℃ overnight. The immunoprecipitated samples were washed once with buffer 3, twice with buffer 4 (20 mM Tris‐ HCl (pH 7.5), 500 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, 1% Triton X‐100, 0.1% Na‐deoxycholate, 0.1% SDS), three times with buffer 5 (1 mM EDTA, 50 mM HEPES‐KOH (pH 7.4), 1% NP‐40, 0.25 M LiCl, 0.5% Na‐deoxycholate) and once with TE50 buffer (50 mM Tris‐HCl (pH 8.0), 10 mM EDTA). Immunoprecipitated and input samples were eluted in elution buffer (TE50 buffer, 1% SDS) at 65℃ overnight. Samples were supplemented with TE buffer (10 mM Tris‐HCl (pH 8.0), 1 mM EDTA), and treated with RNase A (Nacalai Tesque, Cat# 30100–31) at 37℃ for 1 h, followed by proteinase K (Roche, Cat# 3115879001) at 55℃ for 2 h. DNA was purified according to the conventional PCI method. For ChIP‐qPCR and ChIP‐seq, the following antibodies were used: mouse anti‐FLAG antibody (Sigma‐Aldrich, Cat# F3165, RRID:AB_259529), mouse anti‐STAT3 antibody (Cell Signaling Technology, Cat# 9139, RRID:AB_331757) and IgG (Santa Cruz Biotechnology, Cat# sc‐2025, RRID:AB_737182). For ChIP‐qPCR, following primers: 5’‐ CCCCAGGACCTCCTTTTGTG‐3’ (forward), 5’‐ CAGTACAAGCTCCCAGCTCA‐3’ (reverse) were used to amplify the Gfap promoter region.
2.3. ChIP‐seq and data analysis
Sequencing libraries were made according to instructions in the NEBNext Ultra II DNA Library Prep Kit for Illumina (New England Biolabs, Cat# E7645S). Obtained reads were trimmed based on read length and read quality using the FASTX‐Toolkit (http://hannonlab.cshl.edu/fastx_toolkit/, RRID:SCR_005534) and aligned with the mouse genome (UCSC mm10) using Bowtie (v1). (http://bowtie‐bio.sourceforge.net/index.shtml, RRID:SCR_005476). All redundant reads were removed from further analysis. SAM files were converted to the BAM format using SAMtools (https://www.htslib.org/, RRID:SCR_002105). Peak calling for Nfia was performed using MACS (v1.4). (https://github.com/macs3‐project/MACS, RRID:SCR_013291) with default parameters and the IgG library as control.
To obtain enriched sequences, 100‐bp sequences surrounding the summits of the Nfia peak with p‐value <1e‐10 were analyzed using MEME (v5.3.0) (http://meme‐suite.org/, RRID:SCR_001783). To investigate the biological meaning of Nfia binding sequences, Genomic Region Enrichment of Annotation Tool (GREAT) (v3) (http://great.stanford.edu/great/public‐3.0.0/html/, RRID:SCR_005807) was used.
2.4. RNA sequencing and data analysis
E11.5 NS/PCs were infected with retroviruses engineered to express green fluorescent protein (GFP) alone (control), or together with Nfia, cultured in the presence of bFGF for 3 days, and then sorted with GFP fluorescence. Total RNA was extracted using Sepasol®‐RNA I Super G (Nacalai Tesque, Cat# 09379–55), and then mRNA was further isolated with NEBNext Poly(A) mRNA Magnetic Isolation Module (New England Biolabs, Cat# E7490) according to the manufacturer's standard procedure. Purified mRNAs were supplied for library construction using the NEBNext Ultra Directional RNA library Prep Kit for Illumina (New England and Biolabs, Cat# E7420) according to the manufacturer's protocols. cDNA libraries were size‐selected with AMPure XP beads (Beckman Coulter, Cat# A63880). cDNA library quality was checked using Agilent 2100 Bioanalyser (Agilent Technologies). RNA sequencing was performed with 50‐bp single‐end sequencing using an Illumina Hiseq 2500. Obtained reads were processed with the FASTX tool kit to remove short (<30 bp) and low‐quality (quality score <20) reads, followed by trimming of the adaptor sequence. Processed sequence reads were aligned to mm10 using TopHat (http://ccb.jhu.edu/software/tophat/index.shtml, RRID:SCR_013035). Cuffdiff (http://cole‐trapnell‐lab.github.io/cufflinks/, RRID:SCR_001647), a program in Cufflinks (http://cole‐trapnell‐lab.github.io/cufflinks/cuffmerge/, SCR_014597), was used for differential gene expression analysis. To identify Nfia‐target genes, we used RPKM values and determined the genes with 1.5‐fold higher expression in the Nfia group, compared to the control group. In this procedure, genes with low expression (RPKM <5) were excluded from the analysis.
2.5. Immunocytochemistry
Cultured cells were fixed with 4% formaldehyde and washed with PBS. After treatment with blocking buffer (3% FBS and 0.3% Triton X‐100 in PBS) at RT for 30 min, cells were incubated at RT for 2 h with combinations of the following primary antibodies: chick anti‐GFP (Aves Labs, Cat# GFP‐1010, RRID:AB_2307313), rabbit anti‐RFP (MBL International, Cat# PM005, RRID:AB_591279), and mouse anti‐GFAP (Sigma‐Aldrich, Cat# G3893, RRID:AB_477010). Samples were washed with PBS, and then incubated with secondary antibody. Nuclei were stained using Hoechst 33258 (Nacalai Tesque, Cat# 04928–92). Fluorescence images were acquired using a Leica DMI6000 B microscope (Leica Biosystems) with a 20x objective lens.
2.6. Cloning
We amplified cDNAs of individual Nfia‐target genes using specific primer sets (Table 1) and cDNA derived from E14 mouse telencephalon by PCR using KOD ‐Plus‐ (TOYOBO, Cat# KOD‐201), according to the manufacturer's instruction. The PCR products of each Nfia‐target gene were subcloned into the retrovirus (pMY) or lentivirus plasmids (FUW) using restriction enzymes listed in Table 1.
TABLE 1.
Primers used for cloning
| Primer | Experiment | Target | Direction | Sequence (5'−3’) | Restriction enzyme |
|---|---|---|---|---|---|
| Cloning | Olig1 (pMY) | Fw | GCGCGGATCCATGTACTATGCGATTTCCCA | BamHI | |
| Rv | GCGCTACGTATCACTTGGAGAACTGGG | SnaBI | |||
| Ets1 (pMY) | Fw | GCGCGGATCCATGAAGGCGGCCGTCGATCTCAA | BamHI | ||
| Rv | ATATTACGTACTAGTCAGCATCCGGCTTTA | SnaBI | |||
| Olig2 (pMY) | Fw | GCGCGAATTCATGGACTCGGACGCCAGCCT | EcoRI | ||
| Rv | GCGCGAATTCTCACTTGGCGTCGGAGG | EcoRI | |||
| Sox8 (pMY) | Fw | GCGCGGATCCATGCTGGACATGAGTGAGGC | BamHI | ||
| Rv | TATAGCGGCCGCTCAGGGTCGGGTCAGGGTGG | NotI | |||
| Nr3c1 (pMY) | Fw | GCGCGGATCCATGGACTCCAAAGAATCCTTAGC | BamHI | ||
| Rv | GCGCTACGTATCATTTCTGATGAAACAGAA | SnaBI | |||
| Sox8 (FUW) | Fw | GCGCGGATCCATGCTGGACATGAGTGAGGC | BamHI | ||
| Rv | ATATGTTAACTCAGGGTCGGGTCAGGG | HpaI | |||
| Nfia (FUW) | Fw | GCGCTCTAGAATGTATTCTCCGCTCTGTCTC | XbaI | ||
| Rv | GCGCGTTAACTTATCCCAGGTACCAGG | HpaI |
2.7. Virus preparation
To express genes in and trace the fate of NS/PCs, the NS/PCs were infected with retroviruses. We used the retrovirus vector pMY‐IRES‐GFP or tdTomato. Retroviruses were prepared as described previously. 27 , 28 In brief, Plat‐E packaging cells were transfected with retroviral vector using polyethylenimine (Polysciences, Cat# 24765). On the following day, culture medium was replaced with N2‐supplemented DMEM/F12. After 24 h, medium was collected and centrifuged, and retroviruses were re‐suspended in fresh medium.
To knock down Sox8, NS/PCs were infected with lentivirus engineered to express shRNA interfering with Sox8. We used the lentiviral vector pLLX, which had been modified to express EGFP together with a puromycin resistance gene under the ubiquitin‐C promoter. For lentivirus preparation, HEK293 T cells were transfected with pLLX together with the lentivirus constructs pCMV‐VSV‐G‐RSV‐Rev and pCAG‐HIVgp, and one day after, the medium was replaced with N2‐supplemented DMEM/F12 and the cells were cultured for 48 h before the supernatant was collected. The sequences of shRNA for mouse Sox8 and Sox9 were as follows: shSox8#1, 5′‐CGAGTTTTGCATCTCAGTAGA‐3′; shSox8#2, 5′‐GCAGAGAGTAGAATTTAAGAC‐3′; shSox9, 5′‐CGTCAATGAGTT TGACCAATA‐3′.
2.8. RT‐qPCR
Total RNAs were extracted with Sepasol‐RNA I Super G (Nacalai Tesque, Cat# 09379–97). Total RNAs were subjected to reverse transcription using a SuperScript VILO cDNA Synthesis Kit (Thermo Fisher Scientific, Cat# 11755250). Obtained cDNAs were analyzed by quantitative real‐time PCR (qRT‐PCR) with the StepOne Real‐Time PCR System (Applied Biosystems) using a KAPA SYBR FAST qPCR Kit (NIPPON Genetics, Cat# KK4602). Expression levels of each gene were normalized to Actb and calculated relative to the control. Primers were as follows: Actb, 5′‐GGGGTGTTGAAGGTCTCAAA‐3′ (forward), 5′‐TGTTACCAACTGGGACGACA‐3′ (reverse); Gfap, 5′‐AAAACAAG GCGCTGGCAGCTGAACTGAA‐3′ (forward), 5′‐TTGTTCTCTGCCTC CAGCCTCAGGTTGGT‐3′ (reverse); Sox8, 5′‐ CAAAGCTGATGCAG TGCTCG‐3′ (forward), 5′‐ TTGCTTGCTTCCATTGCTCG‐3′ (reverse);.
2.9. Bisulfite sequencing
Using a FACSAriaSORP (Becton Dickinson), NS/PCs were sorted based on GFP fluorescence. Genomic DNAs were extracted from sorted cells, and then treated with sodium bisulfite using a Methylamp DNA Modification Kit (Epigentek, Cat# P‐1001). The region of the Gfap promoter was amplified by PCR using the following primers: 5’‐ GATTTGGTAGTGTTGAGTTGGTTAGT‐3’ (forward), 5’‐ CCTACCTTCCTCTACCCATACTTAAA‐3’ (reverse). The PCR products were cloned into pGEM‐T easy vector (Promega, Cat# A1360), and randomly selected clones were sequenced.
2.10. Western blotting
To express proteins of interest, we transfected HEK293 T cells with the constructs, pcDNA3‐6xMyc‐Stat3, HA‐Sox8, Flag‐Sox8, and pCMV5‐HA‐p300. Two days after transfection, HEK293 T cells were lysed in lysis buffer (10 mM Tris‐HCl (pH 7.4), 150 mM NaCl, 0.5% (v/v) NP‐40, Protease Inhibitor Cocktail; Nacalai Tesque), using a Sonifier (Branson). Lysates were immunoprecipitated with 1 μg of anti‐Flag antibody (Sigma‐Aldrich, Cat# F3165, RRID:AB_259529) and anti‐HA antibody (MBL International, M180‐3, AB_10951811) and 30 μl of magnetic Dynabeads M‐280 sheep anti‐mouse IgG. Lysates and immunoprecipitated samples were subjected to sodium dodecyl sulfate–polyacrylamide gel electrophoresis and immunoblotted with antibodies against anti‐Flag, anti‐HA, and anti‐Myc antibody (MBL International, Cat# M047‐3, RRID:AB_591112). Detection was performed using the ECL detection system (GE Healthcare, RPN2232).
2.11. Statistical analysis
Statistical analyses were performed using one‐way ANOVA with Tukey's HSD test. Differences were considered statistically significant if the probability value was below 0.05. Data represent mean with standard error.
2.12. Data availability
The RNA‐seq and ChIP‐seq data obtained in this study are available in NCBI GEO datasets, under accession number GSE164503.
2.13. Nomenclature of Targets and Ligands
Key protein targets and ligands in this article are hyperlinked to corresponding entries in http://www.guidetopharmacology.org, the common portal for data from the IUPHAR/BPS Guide to PHARMACOLOGY, 29 and are permanently archived in the Concise Guide to PHARMACOLOGY 2019/20. 30
3. RESULTS
3.1. Identification of Nfia‐binding sites in the mid‐gestational NS/PCs
Several studies have shown that Nfia is a pivotal transcription factor for NS/PCs to acquire gliogenic potential in development, 21 , 22 yet its precise mechanisms remain to be fully elucidated. To clarify the mechanism, we first attempted to identify Nfia‐target genes. We employed a genome‐wide strategy whereby we expressed Flag‐tagged Nfia in NS/PCs derived from E11.5 forebrain, and performed ChIP with Flag antibody, followed by high‐throughput DNA sequencing (ChIP‐seq). We obtained 16,941 Nfia peaks, including those found around glia‐specific genes such as Gfap, Aqp4, and Aldh1 l1 (Figure 1A) as reported previously. 21 De novo motif discovery analysis revealed that the most highly enriched sequence (Figure 1B) was similar to previously reported NFIs consensus sequence [TTGGC(N)5GCCAA], 31 which validates the quality of our ChIP‐seq. Functional annotation of Nfia‐bound sequences using GREAT highlighted the terms involving negative regulation of neurogenesis and glial cell differentiation (Figure 1C), further supporting the function of Nfia in the acquisition of gliogenic potential by NS/PCs in mid‐gestation.
FIGURE 1.
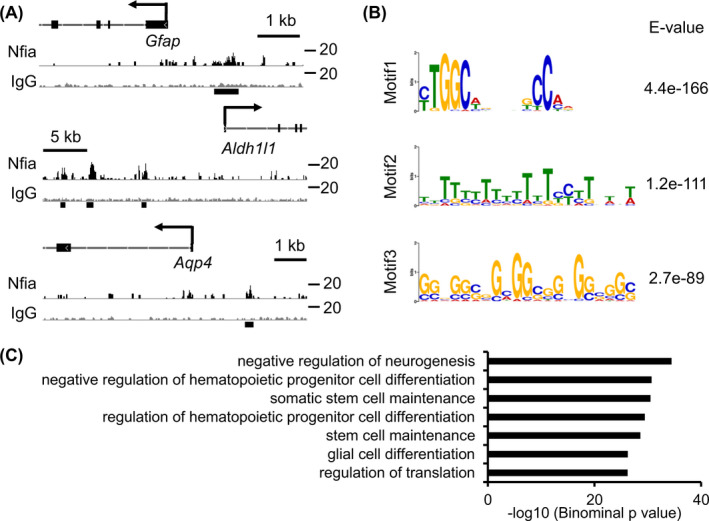
Nfia binds to glia‐specific genes in mid‐gestational NS/PCs. (A) Representative alignment of Nfia ChIP‐seq (black) and IgG control (gray) data around Gfap, Aqp4, and Aldh1 l1 loci. E11.5 NS/PCs were infected with retroviruses engineered to express Flag‐tagged Nfia, cultured for 3 days in the presence of bFGF, and then subjected to ChIP‐seq. Black lines at the bottom show Nfia peaks annotated by MACS. (B) A sequence logo of the top 3‐scoring motif discovered by MEME. (C) GO biological processes of Nfia binding genes in mid‐gestational NS/PCs
3.2. Identification of Nfia‐target genes in mid‐gestational NS/PCs
To search for genes whose expression is affected by Nfia, we expressed Nfia in NS/PCs derived from E11.5 forebrain, and performed RNA‐seq analysis. RNA‐seq revealed 616 up‐regulated and 724 down‐regulated genes with 1.5‐fold change by Nfia‐forced expression (Figure 2A). To identify Nfia direct target genes, we then combined ChIP‐seq and RNA‐seq data. We defined Nfia‐associated genes as genes that have Nfia‐binding within 5 kb from their TSS, and 120 of these genes showed overlap with up‐regulated genes (Figure 2B). Among the 120 genes, we further focused on genes encoding transcription factors because transcription factors are known to play an important role in fate‐specification and cell identity of cells (Figure 2B). The selected 18 Nfia‐associated transcription factor encoding genes are listed with fold change values in Figure 2C, and we picked the top 5 genes (Olig1, Ets1, Olig2, Sox8, and Nr3c1) in the order of descending fold change (Figure 2D and E) for further investigation.
FIGURE 2.
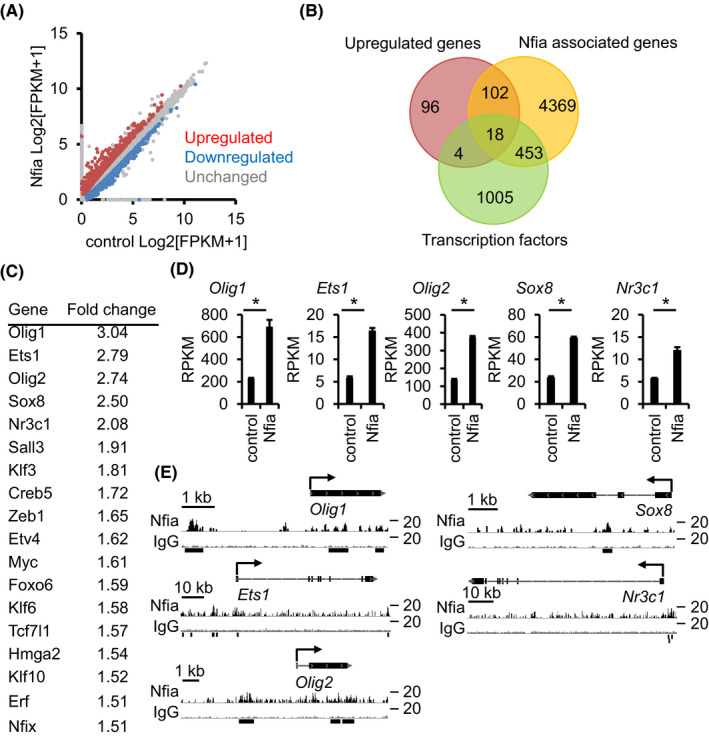
Identification of Nfia‐target genes. (A) Scatter plot showing gene expression in control and Nfia‐expressing NS/PCs. E11.5 NS/PCs were infected with Nfia, and cultured for 3 days in the presence of bFGF. Infected cells were sorted based on GFP fluorescence and then subjected to RNA‐seq. Upregulated (red) and downregulated (blue) genes (with at least 1.5‐fold change of expression) and unchanged genes (gray) were plotted. (B) Venn diagram showing the overlap among genes upregulated by Nfia‐overexpression, Nfia‐associated genes (Nfia binding within 5 kb from TSS), and transcription factors. The number of genes is indicated. (C) Fold‐change ranking of 18 genes identified in (B). (D) Expression of top 5 genes of (C) in control and Nfia‐expressing NS/PCs derived from RNA‐seq data. n = 3, *q < .05 by a program in Cufflinks. (E) Alignment of Nfia ChIP‐seq (black) and IgG control (gray) data around gene loci identified in (B). Black lines at the bottom show Nfia peaks annotated by MACS
3.3. Sox8 promotes Nfia‐primed astrocytic differentiation of NS/PCs
To examine the effects of the five transcription factors on astrocytic differentiation, we infected E11.5 NS/PCs with retroviruses engineered to express Nfia linked with GFP and the protein product of each candidate gene linked with tdTomato, cultured the infected cells for 2 days, and then stimulated them with LIF for 4 days to induce astrocytic differentiation. Among the five transcription factors, the expression of only Sox8 significantly induced GFAP‐positive astrocytes (Figure 3A and B). Combined expression of Nfia together with its target genes showed that Olig1, Ets1, and Nr3c1 did not enhance Nfia‐induced astrocytic differentiation, whereas Olig2 suppressed it, as reported previously. 32 In contrast to these 4 Nfia downstream transcription factors, Sox8 actually further promoted astrocytic differentiation induced by Nfia alone. Consistent with this fact, RT‐qPCR analysis recapitulated Sox8 enhancement of Gfap expression in Nfia‐overexpressing NS/PCs (Figure 3C). These results prompted us to examine whether Sox8 was required for Nfia‐induced astrocytic differentiation. We designed shRNAs for Sox8 (shSox8) and engineered shRNA‐containing lentiviruses to perform knockdown experiments, and confirmed the reduction of Sox8 mRNA in NS/PCs infected with these lentiviruses (Figure 3D). When we suppressed Sox8 expression in astrocytic differentiation experiments in the presence of LIF, Nfia‐induced GFAP‐positive astrocytic differentiation was inhibited (Figure 3E and F). These results indicated that Sox8 acts as an essential Nfia target gene to promote astrogenesis in mid‐gestational NS/PCs.
FIGURE 3.
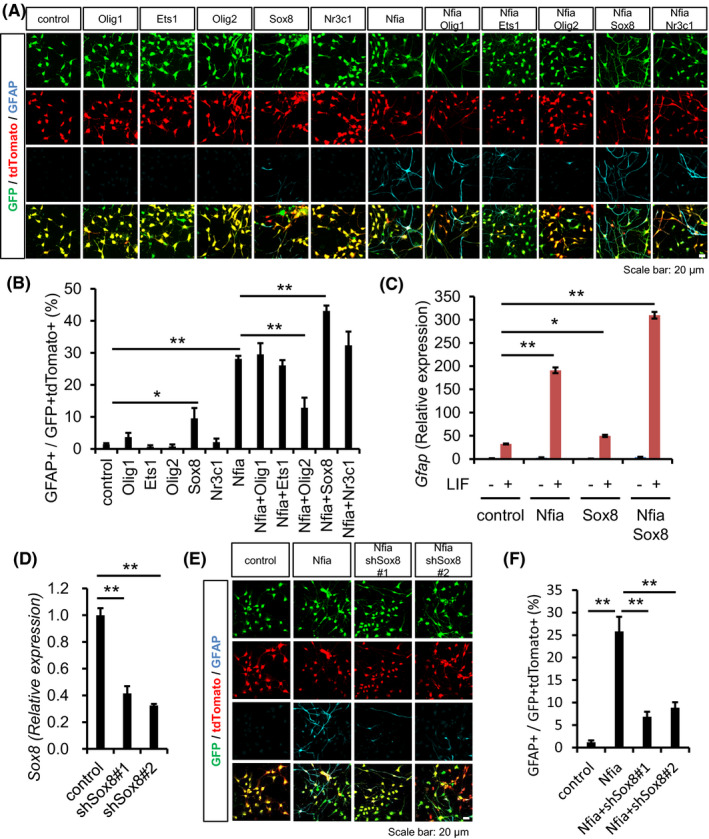
Sox8 enhances Nfia‐promoted astrocytic differentiation. (A) E11.5 NS/PCs were infected with retroviruses engineered to express GFP alone or together with Nfia, and tdTomato alone or together with Nfia‐target genes, cultured for 2 days in the presence of bFGF, and then stimulated with LIF (50 ng/ml) for a further 4 days to induce astrocytic differentiation. The cells were stained with antibodies against GFP (green), tdTomato (red), and GFAP (cyan). Scale bar =20 µm. (B) Quantification of GFAP‐positive astrocytes in GFP and tdTomato double‐positive cells. Beside Nfia, only Sox8 promoted astrocytic differentiation, and combined expression of Nfia and Sox8 further enhanced GFAP expression. n = 3–11, *p < .05, **p < .01 by ANOVA with one‐way ANOVA with Tukey's HSD test. (C) RT‐qPCR analysis of Gfap expression in NS/PCs. Combined expression of Nfia and Sox8 further enhanced Gfap expression compared to single expression of Nfia after treatment with LIF (50 ng/ml) for 24 hr. Relative expression levels to that of control without LIF are indicated. n = 3, *p < .05, **p < .01 by ANOVA with one‐way ANOVA with Tukey's HSD test. (D) RT‐qPCR analysis of Sox8 expression in NS/PCs. shSox8 significantly reduced Sox8 expression. n = 3, **p < .01 by ANOVA with one‐way ANOVA with Tukey's HSD test. (E) Representative images of immunostaining for Sox8‐knockdown NS/PCs. The cells were infected with viruses engineered to express GFP alone or together with shSox8, and tdTomato alone or together with Nfia, and stained with antibodies against GFP (green), tdTomato (red), and GFAP (cyan). Scale bar =20 µm. (F) Quantification of GFAP‐positive astrocytes in GFP and tdTomato double‐positive cells. Sox8 knockdown suppressed Nfia‐induced astrocytic differentiation. n = 3, **p < .01 by ANOVA with one‐way ANOVA with Tukey's HSD test
3.4. Sox8 does not affect DNA methylation of the Gfap promoter but interacts with STAT3 via p300 to promote astrocytic differentiation of NS/PCs
Next, we sought to investigate how Sox8 facilitates astrocytic differentiation of NS/PCs in the presence of LIF. Since both Sox8 alone and simultaneous expression with Nfia could promote astrocytic differentiation, we first hypothesized that similarly to Nfia, Sox8 induces DNA demethylation of the Gfap promoter in mid‐gestational NS/PCs, because LIF‐downstream STAT3 is incapable of binding to its methylated binding elements of target genes to activate them. 21 We then found that Sox8 expression did not alter the methylation status of the promoter compared with the control, whereas Nfia reduced it (Figure 4A and B), consistent with the previous study. 21 Consistently with the DNA methylation status, we found that Nfia is sufficient for STAT3 binding to the Gfap promoter; however, Sox8 knockdown did not affect the Nfia‐facilitated STAT3 association with the promoter (Figure 4C). These results led us to propose another idea to explain how Sox8 enhances Nfia‐potentiated astrocytic differentiation of NS/PCs in the presence of LIF.
FIGURE 4.
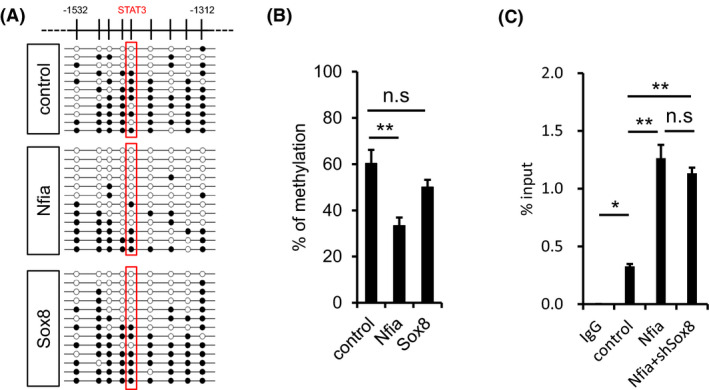
Sox8 participates in neither DNA demethylation nor STAT3 binding to Gfap promoter. (A) Bisulfite sequencing for the CpG sites of Gfap promoter around STAT3 binding site. E11.5 NS/PCs were infected with retroviruses engineered to express GFP alone or together with NFIA, and cultured for 3 days in the presence of bFGF. Infected cells were sorted according to GFP fluorescence and subjected to bisulfite sequencing. STAT3 binding site is indicated by red box. Closed and open circles indicate methylated and unmethylated CpG sites, respectively. (B) Average methylation frequency of nine CpG sites within the Gfap promoter, including STAT3 binding site. n = 4–5, **p < .01 by ANOVA with one‐way ANOVA with Tukey's HSD test. (C) Enrichment of STAT3 on Gfap promoter around STAT3 binding site. E11.5 NS/PCs were infected with lentivirus, and cultured in the presence of bFGF for 3 days. The cells were stimulated with LIF for 30 min, cross‐linked and immunoprecipitated with anti‐STAT3 antibody. Mouse IgG was used as a control antibody. Samples were assessed by qPCR using a primer set to amplify the Gfap promoter around the STAT3 binding site. n = 3, *p < .05, **p < .01 by ANOVA with one‐way ANOVA with Tukey's HSD test
It is known that transcription factors often associate with other transcription function factors to cooperatively promote their target gene expression. 33 , 34 , 35 As such, we decided to check whether Sox8 interacts with STAT3 to enhance the promoter activity of Gfap. To this end, we expressed Sox8 and STAT3 in HEK293 T cells and performed a co‐immunoprecipitation analysis (Figure 5A). Contrary to our expectation, an association between STAT3 and Sox8 was not detected (Figure 5A). Since bridging of distinct transcription factors via by transcriptional co‐activators such as p300 and CBP has been shown, 13 , 15 , 36 next we investigated the possibility that p300 mediates complex formation of Sox8 and STAT3 and found that this is the case. Sox8 interacted with p300 (Figure 5B), and interacted with STAT3 only in the presence of p300 (Figure 5C). Taking the previous finding that STAT3 associates with p300 13 into account, these results reveal the formation of a complex of Sox8 and STAT3 bridged by p300. Altogether, our findings show that Nfia induces DNA demethylation of the Gfap promoter and Sox8 expression, and upon stimulation with astrocyte‐inducing factors such as LIF, Sox8 forms a complex with STAT3 through p300, leading to the enhanced transcriptional activity of Gfap (Figure 5D).
FIGURE 5.
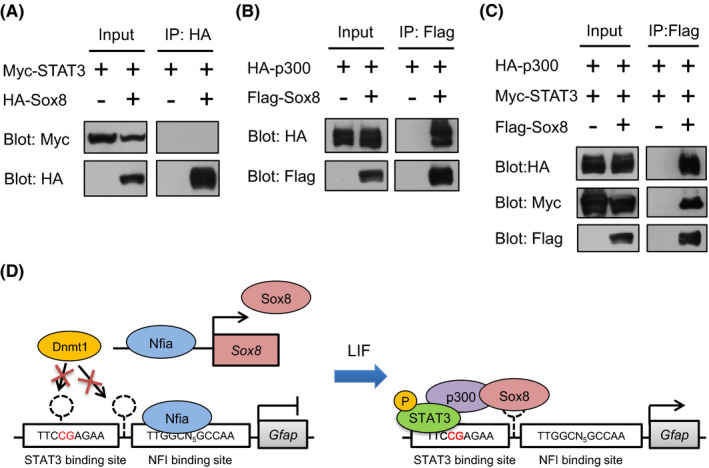
Sox8 interacts with STAT3 bridged by p300. (A) Sox8 does not interact with STAT3 by itself. HA‐Sox8‐expressing construct was transfected into HEK293 T cells with Myc‐STAT3. Lysates were immunoprecipitated with anti‐HA antibody, followed by western blotting with anti‐HA or anti‐Myc antibodies. In input, all tagged‐proteins were detected, but STAT3 was not included in Sox8‐immunoprecipitated complex. (B) Sox8 interacts with p300. Flag‐Sox8‐expressing construct was transfected into HEK293 T cells with HA‐STAT3. Lysates were immunoprecipitated with anti‐Flag antibody, followed by western blotting with anti‐Flag or anti‐HA antibodies. (C) Sox8 interacts with p300 and STAT3. Flag‐Sox8‐expressing construct was transfected into HEK293 T cells with Myc‐STAT3 and HA‐p300. Lysates were immunoprecipitated with anti‐Flag antibody, followed by western blotting with anti‐Flag, anti‐Myc, or anti‐HA antibodies. STAT3 and p300 were included in Sox8 immunoprecipitated complex. (D) Schematic model for the enhancement of astrocytic differentiation by Sox8. Nfia induces Sox8 expression and DNA demethylation by obstructing Dnmt1 binding on the Gfap promoter. Upon LIF stimulation, Sox8 forms transcriptional activation complex with p300 and STAT3 to induce Gfap expression and astrocytic differentiation
4. DISCUSSION
Nfia is a key transcription factor for glial specification and astrocytic differentiation in the developing spinal cord and cortex. 21 , 37 Unlike Nfia‐target genes in the spinal cord, Nfia‐target genes in NS/PCs derived from developmental cortex have not been well characterized. In the current study, we performed ChIP‐seq and RNA‐seq in mid‐gestational NS/PCs that do not yet have the potential to differentiate into astrocytes, and thereby identified Nfia‐direct target genes. To identify the key transcription factor encoding genes promoting astrocytic differentiation downstream of Nfia, we expressed Olig1, Ets1, Olig2, Sox8, and Nr3c1 and found that only Sox8 expression induced astrocytic differentiation, and its combined expression with Nfia further promoted LIF‐induced astrocytic differentiation of mid‐gestational NS/PCs (Figure 3A and B). To dissect the mechanism by which Sox8 promoted Nfia‐primed astrocytic differentiation, we checked whether Sox8 induces DNA demethylation of the Gfap promoter, since DNA demethylation is prerequisite for STAT3 binding to the promoter during differentiation. The results showed that, in contrast to Nfia, which was sufficient to induce DNA demethylation and STAT3 binding to the promoter, Sox8 was able to affect neither of them (Figure 4A and B). Finally, we found that Sox8 interacts with STAT3 bridged by p300, suggesting that this complex formation may enhance the transcriptional activity of Gfap (Figure 5).
Among Nfia‐target genes, we found both genes that were upregulated and genes that were downregulated by Nfia expression (Figure 2A), which probably reflects the possession of both transactivation and repression domains by Nfia. 31 The upregulated genes included Olig1 and Olig2, which are involved in oligodendrocyte differentiation and maturation. 38 In spinal cord development, it has been shown that Nfia deficiency impairs oligodendrocyte differentiation and maturation, and thus it is conceivable that Nfia is required for the expression of Olig2 at the onset of oligodendrogenesis. 22 Olig2 inhibits Nfia function in initiating astrocytic differentiation, and the balance between Nfia and Olig2 is important for defining astrocytic and oligodendrocytic lineage specifications. 22 In cortical development, we have previously shown that Nfia potentiates the expression of Olig1, which is also involved in oligodendrocyte differentiation and maturation, via inducing its promoter DNA demethylation. 39 These findings indicate that Nfia functions in the intersection of astrocytic and oligodendrocytic lineage specifications. Although we focused on upregulated genes in this study, it would be intriguing to examine Nfia‐downregulated genes, including Id4, Dlx family genes (Dlx1, 2, 5, and 6), and Fezf2 (Figure S1). Id4‐deficient mice show compromised proliferation of NS/PCs and precocious neurogenesis. 40 In Dlx1/2 double mutant mice, most cortical interneurons derived from basal ganglia are lost. 41 Simultaneous deletion of Dlx5 and 6 in mice reduces cortical interneurons. 42 Fezf2 promotes neurogenesis by repressing Hes5, a negative regulator of proneural gene Neurog2 in forebrain development. 43 Together with our results in the present study, these findings support the notion that Nfia maintains NS/PCs and represses neuronal differentiation, contributing to the accomplishment of timely‐ordered forebrain development.
Sox8 knockdown was able to greatly suppress Nfia‐enhanced astrocytic differentiation of NS/PCs in response to LIF stimulation, indicating the importance of Sox8 in astrocytic differentiation (Figure 3E and F). It has been reported that Nfia interacts with Sox9, another member of the SoxE family to which Sox8 and Sox10 belong, 44 and that Nfia and Sox9 cooperatively regulate genes implicated in gliogenesis initiation in chick spinal cord development. 37 In fact, Sox9‐deficient mice show dramatic reduction of astrocytes in the spinal cord. 45 Actually, Sox9 knockdown also reduced Nfia‐induced astrocytic differentiation in mid‐gestational NS/PCs treated with LIF (Figure S2A and B), suggesting the redundant function between Sox8 and Sox9 with regard to astrocytic differentiation. However, deletion of Sox8 alone in mice does not affect the oligodendrocyte production, but exacerbates the defect caused by loss of Sox9, 46 implying their non‐reciprocal functional redundancy in the spinal cord. In our RNA‐seq analysis, Sox9 expression was only slightly decreased rather than increased by Nfia expression (Figure S2C). Furthermore, single expression of Sox9 did not induce astrocytic differentiation, and even combined expression of Sox9 with Nfia failed to further promote Nfia‐induced astrocytic differentiation as Sox8 did in the presence of LIF (Figure S2D and E), suggesting that Sox8 was more effective in promoting astrocytic differentiation of NS/PCs than Sox9. In accord with this idea, Nfia expression increases in NS/PCs from E11 to E14, followed by upregulation of Sox8, as shown in our previous microarray data of embryonic NS/PCs (Figure S3). 25 Sox9 expression is already high in neurogenic E11 NS/PCs and is maintained at a similar level over time (Figure S3).
DNA methylation often prevents binding of transcription factors to their target sequence in the genome, 14 , 47 , 48 and is thus deeply involved in cell fate determination. Although Sox8 promoted astrocytic differentiation, Sox8 did not induce demethylation of the Gfap promoter (Figure 4A and B). This suggests that Nfia is important for conferring gliogenic potential on NS/PCs via DNA demethylation, and Sox8 plays an executive role for astrocytic differentiation in response to astrocyte‐inducing signals. In addition to being activated by the IL‐6 family of cytokines such as LIF, the Gfap promoter is also activated by BMPs and retinoic acid (RA). 13 , 49 NS/PCs in the developing brain receive these signals from surrounding neurons and meninges, which cover and protect the brain, and have recently been shown to act as a niche where NS/PCs extend their radial processes and their basal end‐feet attach. 9 , 10 These extracellular factors synergistically induce promoter activity. 8 Since Sox8 interacts with p300, which also bridges with Smad1 upon BMP stimulation, Sox8 may also be involved in the enhancement of BMP signaling.
As described previously, Nfia is induced by activation of the Notch signal by its ligands from nearby neuroblasts during mid‐ to late‐gestation, and this results in the DNA demethylation of astrocytic genes, linking alteration of extracellular signals to epigenetic change of NS/PCs toward gliogenesis. Here we revealed that one of the Nfia downstream factors, Sox8, promotes astrocytic differentiation in concert with STAT3, but one can easily imagine that as yet unelucidated mechanisms underlie astrogenesis in multiple aspects of brain development. Since impairment of astrogenesis during development is associated with a variety of neurological disorders such as epilepsy and autism, 6 full understanding of the mechanisms of astrogenesis in the developing brain would be a keystone for the development of strategies for treatment of such neurological disorders. Moreover a growing body of evidence is accumulating to demonstrate that NFIA expression is correlated with prognosis of glioma patients, 50 , 51 , 52 raising a possibility that Sox8 is involved in regulating growth and invasion of glioma. Although further investigation is warranted, Sox8 could be a novel target for treatments of glioma.
DISCLOSURE
The authors have no conflicts of interest to declare.
AUTHORS’ CONTRIBUTIONS
JT, SK, TI, and KN contributed to study concept and design. JT contributed to acquisition of data. JT, TI, and TS contributed to resources. JT, SK, and KN contributed to analysis and interpretation of data. JT contributed to drafting of the manuscript and statistical analysis. SK and KN contributed to critical revision of the manuscript for important intellectual content. KI contributed to study supervision.
Supporting information
Fig S1‐S3
ACKNOWLEDGEMENTS
The authors thank Dr. T. Matsuda for valuable discussions and helpful comments; Dr. T. Kitamura for pMY vector and Plat‐E cells; Z. Zhou and M. E. Greenberg for pLLX vector, and Fred H. Gage for Sox2‐EGFP transgenic mice; Y. Nakagawa for excellent secretarial assistance; and E. Nakajima for proofreading the manuscript. The authors thank the Research Support Center, Kyushu University Graduate School of Medical Sciences.
Funding information
This work was supported by MEXT JSPS KAKENHI (JP16H06527 and JP16 K21734 to KN; JP26710003 and JP20 K06875 to SK; JP18 J11751 to JT), and The Naito Foundation for SK.
Contributor Information
Sayako Katada, Email: sakatada@scb.med.kyushu-u.ac.jp.
Kinichi Nakashima, Email: kin1@scb.med.kyushu-u.ac.jp.
REFERENCES
- 1. Taupin P, Gage FH. Adult neurogenesis and neural stem cells of the central nervous system in mammals. J Neurosci Res. 2002;69(6):745‐749. 10.1002/jnr.10378 [DOI] [PubMed] [Google Scholar]
- 2. Hirabayashi Y, Gotoh Y. Stage‐dependent fate determination of neural precursor cells in mouse forebrain. Neurosci Res. 2005;51(4):331‐336. 10.1016/j.neures.2005.01.004 [DOI] [PubMed] [Google Scholar]
- 3. Azevedo FAC, Carvalho LRB, Grinberg LT, et al. Equal numbers of neuronal and nonneuronal cells make the human brain an isometrically scaled‐up primate brain. J Comp Neurol. 2009;513(5):532‐541. 10.1002/cne.21974 [DOI] [PubMed] [Google Scholar]
- 4. Araque A, Parpura V, Sanzgiri RP, Haydon PG. Tripartite synapses: glia, the unacknowledged partner. Trends Neurosci. 1999;22(5):208‐215. 10.1016/S0166-2236(98)01349-6 [DOI] [PubMed] [Google Scholar]
- 5. Nedergaard M, Ransom B, Goldman SA. New roles for astrocytes: redefining the functional architecture of the brain. Trends Neurosci. 2003;26(10):523‐530. 10.1016/j.tins.2003.08.008 [DOI] [PubMed] [Google Scholar]
- 6. Molofsky AV, Krenick R, Ullian E, et al. Astrocytes and disease: a neurodevelopmental perspective. Genes Dev. 2012;26(9):891‐907. 10.1101/gad.188326.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Barnabé‐Heider F, Wasylnka JA, Fernandes KJL, et al. Evidence that embryonic neurons regulate the onset of cortical gliogenesis via cardiotrophin‐1. Neuron. 2005;48(2):253‐265. 10.1016/j.neuron.2005.08.037 [DOI] [PubMed] [Google Scholar]
- 8. Kawamura Y, Katada S, Noguchi H, et al. Synergistic induction of astrocytic differentiation by factors secreted from meninges in the mouse developing brain. FEBS Lett. 2017;591(22):3709‐3720. 10.1002/1873-3468.12881 [DOI] [PubMed] [Google Scholar]
- 9. Decimo I, Fumagalli G, Berton V, Krampera M, Bifari F. Meninges: from protective membrane to stem cell niche. Am J Stem Cells. 2012;1(2):92‐105. [PMC free article] [PubMed] [Google Scholar]
- 10. Sockanathan S, Gaiano N. Meninges play a RAdical Role in Embryonic Neural Stem Cell Regulation. Cell Stem Cell. 2009;5(5):455‐456. 10.1016/j.stem.2009.10.010 [DOI] [PubMed] [Google Scholar]
- 11. Bonni A, Sun Y, Nadal‐Vicens M, et al. Regulation of gliogenesis in the central nervous system by the JAK‐STAT signaling pathway. Science. 1997;278(5337):477‐483. 10.1126/science.278.5337.477 [DOI] [PubMed] [Google Scholar]
- 12. Nakashima K, Wiese S, Yanagisawa M, et al. Developmental requirement of gp130 signaling in neuronal survival and astrocyte differentiation. J Neurosci. 1999;19(13):5429‐5434. 10.1523/jneurosci.19-13-05429.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Nakashima K, Yanagisawa M, Arakawa H, et al. Synergistic signaling in fetal brain by STAT3‐Smad1 complex bridged by p300. Science. 1999;284(5413):479‐482. 10.1126/science.284.5413.479 [DOI] [PubMed] [Google Scholar]
- 14. Takizawa T, Nakashima K, Namihira M, et al. DNA Methylation is a critical cell‐intrinsic determinant of astrocyte differentiation in the fetal brain. Dev Cell. 2001;1(6):749‐758. 10.1016/S1534-5807(01)00101-0 [DOI] [PubMed] [Google Scholar]
- 15. Sun YI, Nadal‐Vicens M, Misono S, et al. Neurogenin promotes neurogenesis and inhibits glial differentiation by independent mechanisms. Cell. 2001;104(3):365‐376. 10.1016/S0092-8674(01)00224-0 [DOI] [PubMed] [Google Scholar]
- 16. Namihira M, Nakashima K. Mechanisms of astrocytogenesis in the mammalian brain. Curr Opin Neurobiol. 2013;23(6):921‐927. 10.1016/j.conb.2013.06.002 [DOI] [PubMed] [Google Scholar]
- 17. Ge W, Martinowich K, Wu X, et al. Notch signaling promotes astrogliogenesis via direct CSL‐mediated glial gene activation. J Neurosci Res. 2002;69(6):848‐860. 10.1002/jnr.10364. [DOI] [PubMed] [Google Scholar]
- 18. Kageyama R, Shimojo H, Imayoshi I. Dynamic expression and roles of Hes factors in neural development. Cell Tissue Res. 2015;359(1):125‐133. 10.1007/s00441-014-1888-7 [DOI] [PubMed] [Google Scholar]
- 19. Ables JL, Breunig JJ, Eisch AJ, Rakic P. Not(ch) just development: notch signalling in the adult brain. Nat Rev Neurosci. 2011;12(5):269‐283. 10.1038/nrn3024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sakamoto M, Hirata H, Ohtsuka T, Bessho Y, Kageyama R. The basic helix‐loop‐helix genes Hesr1/Hey1 and Hesr2/Hey2 regulate maintenance of neural precursor cells in the brain. J Biol Chem. 2003;278(45):44808‐44815. 10.1074/jbc.M300448200 [DOI] [PubMed] [Google Scholar]
- 21. Namihira M, Kohyama J, Semi K, et al. Committed neuronal precursors confer astrocytic potential on residual neural precursor cells. Dev Cell. 2009;16(2):245‐255. 10.1016/j.devcel.2008.12.014 [DOI] [PubMed] [Google Scholar]
- 22. Deneen B, Ho R, Lukaszewicz A, Hochstim CJ, Gronostajski RM, Anderson DJ. The transcription factor NFIA controls the onset of gliogenesis in the developing spinal cord. Neuron. 2006;52(6):953‐968. 10.1016/j.neuron.2006.11.019 [DOI] [PubMed] [Google Scholar]
- 23. Shu T, Butz KG, Plachez C, Gronostajski RM, Richards LJ. Abnormal development of forebrain midline glia and commissural projections in Nfia knock‐out mice. J Neurosci. 2003;23(1):203‐212. 10.1523/jneurosci.23-01-00203.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Meng HX, Hackett JA, Nestor C, et al. Apoptosis and DNA methylation. Cancers (Basel). 2011;3(2):1798‐1820. 10.3390/cancers3021798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sanosaka T, Imamura T, Hamazaki N, et al. DNA methylome analysis identifies transcription factor‐based epigenomic signatures of multilineage competence in neural stem/progenitor cells. Cell Rep. 2017;20(12):2992‐3003. 10.1016/j.celrep.2017.08.086 [DOI] [PubMed] [Google Scholar]
- 26. Fan G, Martinowich K, Chin MH, et al. DNA methylation controls the timing of astrogliogenesis through regulation of JAK‐STAT signaling. Development. 2005;132(15):3345‐3356. 10.1242/dev.01912 [DOI] [PubMed] [Google Scholar]
- 27. Kitamura T, Koshino Y, Shibata F, et al. Retrovirus‐mediated gene transfer and expression cloning: powerful tools in functional genomics. Exp Hematol. 2003;31(11):1007‐1014. 10.1016/S0301-472X(03)00260-1 [DOI] [PubMed] [Google Scholar]
- 28. Technology VT, Communication B . Plat‐E: an efficient and stable system for transient packaging of retroviruses. Gene Ther. 2000;7(12):1063‐1066. http://www.worldcat.org/title/plat‐e‐an‐efficient‐and‐stable‐system‐for‐transient‐packaging‐of‐retroviruses/oclc/360485096&referer=brief_results. [DOI] [PubMed] [Google Scholar]
- 29. Harding SD, Sharman JL, Faccenda E, et al. The IUPHAR/BPS Guide to PHARMACOLOGY in 2019: updates and expansion to encompass the new guide to IMMUNOPHARMACOLOGY. Nucleic Acids Res. 2018;46:D1091‐D1106. 10.1093/nar/gkx1121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Alexander SPH, Kelly E, Mathie A, et al. The concise guide to PHARMACOLOGY 2019/20: introduction and other protein targets. Br J Pharmacol. 2019;176(S1):S1‐S20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gronostajski RM. Roles of the NFI/CTF gene family in transcription and development. Gene. 2000;249(1–2):31‐45. 10.1016/S0378-1119(00)00140-2 [DOI] [PubMed] [Google Scholar]
- 32. Fukuda S, Kondo T, Takebayashi H, Taga T. Negative regulatory effect of an oligodendrocytic bHLH factor OLIG2 on the astrocytic differentiation pathway. Cell Death Differ. 2004;11(2):196‐202. 10.1038/sj.cdd.4401332. [DOI] [PubMed] [Google Scholar]
- 33. Gekakis N, Staknis D, Nguyen HB, et al. Role of the CLOCK protein in the mammalian circadian mechanism. Science. 1998;280(5369):1564‐1569. 10.1126/science.280.5369.1564 [DOI] [PubMed] [Google Scholar]
- 34. Helin K, Wu CL, Fattaey AR, et al. Heterodimerization of the transcription factors E2F–1 and DP‐1 leads to cooperative trans‐activation. Genes Dev. 1993;7(10):1850‐1861. 10.1101/gad.7.10.1850 [DOI] [PubMed] [Google Scholar]
- 35. Morgunova E, Taipale J. Structural perspective of cooperative transcription factor binding. Curr Opin Struct Biol. 2017;47:1‐8. 10.1016/j.sbi.2017.03.006 [DOI] [PubMed] [Google Scholar]
- 36. Zhou P, Wan X, Zou Y, Chen Z, Zhong A. Transforming growth factor beta (TGF‐β) is activated by the CtBP2‐p300‐AP1 transcriptional complex in chronic renal failure. Int J Biol Sci. 2020;16(2):204‐215. 10.7150/ijbs.38841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kang P, Lee H, Glasgow S, et al. Sox9 and NFIA coordinate a transcriptional regulatory cascade during the initiation of gliogenesis. Neuron. 2012;74(1):79‐94. 10.1016/j.neuron.2012.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Meijer DH, Kane MF, Mehta S, et al. Separated at birth? the functional and molecular divergence of OLIG1 and OLIG2. Nat Rev Neurosci. 2012;13(12):819‐831. 10.1038/nrn3386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Semi K, Sanosaka T, Namihira M, Nakashima K. Nuclear factor I/A coordinates the timing of oligodendrocyte differentiation/maturation via olig1 promoter methylation. HAYATI J Biosci. 2018;25(2):70‐78. 10.4308/hjb.25.2.70 [DOI] [Google Scholar]
- 40. Yun K, Mantani A, Garel S, Rubenstein J, Israel MA. Id4 regulates neural progenitor proliferation and differentiation in vivo. Development. 2004;131(21):5441‐5448. 10.1242/dev.01430 [DOI] [PubMed] [Google Scholar]
- 41. Anderson SA, Eisenstat DD, Shi L, Rubenstein JLR. Interneuron migration from basal forebrain to neocortex: dependence on Dlx genes. Science. 1997;278(5337):474‐476. 10.1126/science.278.5337.474 [DOI] [PubMed] [Google Scholar]
- 42. Wang Y, Dye CA, Sohal V, et al. Dlx5 and Dlx6 regulate the development of parvalbumin‐expressing cortical interneurons. J Neurosci. 2010;30(15):5334‐5345. 10.1523/JNEUROSCI.5963-09.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Shimizu T, Nakazawa M, Kani S, et al. Zinc finger genes Fezf1 and Fezf2 control neuronal differentiation by repressing Hes5 expression in the forebrain. Development. 2010;137(11):1875‐1885. 10.1242/dev.047167 [DOI] [PubMed] [Google Scholar]
- 44. Weider M, Wegner M. SoxE factors: transcriptional regulators of neural differentiation and nervous system development. Semin Cell Dev Biol. 2017;63:35‐42. 10.1016/j.semcdb.2016.08.013 [DOI] [PubMed] [Google Scholar]
- 45. Stolt CC, Lommes P, Sock E, Chaboissier MC, Schedl A, Wegner M. The Sox9 transcription factor determines glial fate choice in the developing spinal cord. Genes Dev. 2003;17(13):1677‐1689. 10.1101/gad.259003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Stolt CC, Schmitt S, Lommes P, Sock E, Wegner M. Impact of transcription factor Sox8 on oligodendrocyte specification in the mouse embryonic spinal cord. Dev Biol. 2005;281(2):309‐317. 10.1016/j.ydbio.2005.03.010. [DOI] [PubMed] [Google Scholar]
- 47. Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16(1):6‐21. 10.1101/gad.947102 [DOI] [PubMed] [Google Scholar]
- 48. Yin Y, Morgunova E, Jolma A, et al. Impact of cytosine methylation on DNA binding specificities of human transcription factors. Science. 2017;356(6337). 10.1126/science.aaj2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Asano H, Aonuma M, Sanosaka T, Kohyama J, Namihira M, Nakashima K. Astrocyte differentiation of neural precursor cells is enhanced by retinoic acid through a change in epigenetic modification. Stem Cells. 2009;27(11):2744‐2752. 10.1002/stem.176 [DOI] [PubMed] [Google Scholar]
- 50. Yu X, Wang M, Zuo J, et al. Nuclear factor I A promotes temozolomide resistance in glioblastoma via activation of nuclear factor κB pathway. Life Sci. 2019;236:116917. 10.1016/j.lfs.2019.116917 [DOI] [PubMed] [Google Scholar]
- 51. Lee JS, Xiao J, Patel P, et al. A novel tumor‐promoting role for nuclear factor IA in glioblastomas is mediated through negative regulation of p53, p21, and PAI1. Neuro Oncol. 2014;16(2):191‐203. 10.1093/neuonc/not167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Song H‐R, Gonzalez‐Gomez I, Suh GS, et al. Nuclear factor IA is expressed in astrocytomas and is associated with improved survival. Neuro Oncol. 2010;12(2):122‐132. 10.1093/neuonc/nop044 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig S1‐S3
Data Availability Statement
The RNA‐seq and ChIP‐seq data obtained in this study are available in NCBI GEO datasets, under accession number GSE164503.


