Abstract
The inhibitor of kappa B alpha (IκBα) protein is able to shuttle between the cytoplasm and the nucleus. We have utilized a combination of in vivo and in vitro approaches to provide mechanistic insight into nucleocytoplasmic shuttling by IκBα. IκBα contains multiple functional domains that contribute to shuttling of IκBα between the cytoplasm and the nucleus. Nuclear import of IκBα is mediated by the central ankyrin repeat domain. Similar to previously described nuclear import pathways, nuclear import of IκBα is temperature and ATP dependent and is blocked by a dominant-negative mutant of importin β. However, in contrast to classical nuclear import pathways, nuclear import of IκBα is independent of soluble cytosolic factors and is not blocked by the dominant-negative RanQ69L protein. Nuclear export of IκBα is mediated by an N-terminal nuclear export sequence. Nuclear export of IκBα requires the CRM1 nuclear export receptor and is blocked by the dominant-negative RanQ69L protein. Our results are consistent with a model in which nuclear import of IκBα is mediated through direct interactions with components of the nuclear pore complex, while nuclear export of IκBα is mediated via a CRM1-dependent pathway.
The transport of proteins between the nucleus and the cytoplasm is an essential activity of eukaryotic cells. Protein transport across the double lipid bilayer of the nuclear membrane occurs through large macromolecular complexes termed nuclear pore complexes (NPCs). Transport is generally dependent upon specific cis-acting signals within the cargo protein and the corresponding trans-acting factors that mediate the translocation of the protein cargo through the NPC (for reviews, see references 8 and 30). The best-characterized transport pathway is utilized by proteins that contain a classical nuclear localization signal (NLS). The prototypic NLS present in the simian virus 40 (SV40) large T protein contains a short stretch of basic residues that are critically required for nuclear localization (15, 25). NLS-bearing proteins are bound by the heterodimeric importin α-β complex (7, 16–18, 31, 36), which mediates docking of the receptor-cargo complex at the cytoplasmic face of the NPC. Translocation of the receptor-cargo complex through the NPC is poorly understood, but is thought to involve direct interactions between the importin β subunit and specific components of the NPC (37, 38). Nuclear import is terminated in the nucleus by RanGTP-induced dissociation of the receptor-cargo complex, releasing the NLS-bearing cargo into the nucleus (21, 38). The importin α and β proteins are subsequently recycled back to the cytoplasm to participate in a second round of nuclear import (29).
The export of proteins from the nucleus is accomplished by an analogous mechanism. Proteins that are destined to be exported from the nucleus typically contain nuclear export sequences (NESs) comprised of short clusters of leucine or other hydrophobic residues (12, 47). NES-bearing proteins are recognized by the nuclear export receptor, CRM1, in a RanGTP-dependent manner (4, 13, 14, 33, 45). Although the details of nuclear export are poorly understood, the NES-bearing cargo-CRM1 receptor complex is thought to dock at the nuclear side of the NPC, followed by translocation and dissociation at the cytoplasmic face of the NPC.
A critical aspect of both nuclear import and nuclear export is the asymmetric distribution of RanGTP and RanGDP between the nucleus and the cytoplasm. The GTPase-activating protein for Ran, RanGAP, is located at the cytoplasmic face of the NPC, while the guanine nucleotide exchange factor for Ran, RCC1, is tightly associated with chromatin (19, 34). This differential distribution of RanGAP and RCC1 between the cytoplasm and the nucleus has led to the prediction that Ran will be in the GTP-bound form in the nucleus, while the GDP-bound form will predominate in the cytoplasm. Experimental perturbation of the RanGTP-RanGDP gradient inhibits both nuclear import and nuclear export. For example, the GTP-bound form of the dominant-negative RanQ69L protein inhibits NLS-dependent nuclear import in digitonin-permeabilized cells, presumably by inhibiting binding of an NLS-bearing protein to the importin α-β receptor complex (21, 38). Likewise, depletion of nuclear RanGTP levels inhibits NES-dependent nuclear export, presumably by preventing a NES-bearing protein from binding to CRM1 in a manner that is competent for transport (4, 39).
Transport of proteins between the nucleus and the cytoplasm provides an effective mechanism for regulation of gene expression. A striking example of how gene expression can be regulated at the level of protein transport between the nucleus and the cytoplasm is provided by the NF-κB/Rel family of transcription factors and their inhibitory IκB proteins (reviewed in reference 5). For example, the IκBα protein is able to both inhibit nuclear import of NF-κB/Rel proteins and direct the export of NF-κB/Rel proteins from the nucleus (2, 3, 20, 22, 43). The ability of IκBα to act both in the cytoplasm and in the nucleus requires that IκBα itself travel through the NPC. The second ankyrin repeat of IκBα is critically required for nuclear localization of IκBα and is able to functionally substitute for a classical NLS (42). In this report, we have utilized a combination of in vitro and in vivo approaches to provide mechanistic insight into nuclear shuttling of IκBα. Our results indicate that nuclear import of IκBα is accomplished by a Ran-independent mechanism, while nuclear export of IκBα requires the Ran-dependent CRM1 nuclear export receptor.
MATERIALS AND METHODS
Construction of recombinant DNA molecules.
The IκBα clones used in this study were derived from the avian IκBα cDNA clone isolated by Davis et al. (9) and constructed by standard techniques (44). To construct the GST (glutathione S-transferase)-IκBα expression vector, an EcoRI fragment containing 69 bp of 5′ nontranslated sequence, the entire 954 bp of the IκBα open reading frame, and 762 bp of 3′ nontranslated sequence was cloned into the EcoRI site of pGEX-2T in the proper orientation for expression. The GST-IκBα-ΔN69 expression vector contains an IκBα insert that lacks the first 69 codons of the IκBα open reading frame. The GST-IκBα-ΔC51 expression vector contains an IκBα insert that contains a termination codon at codon 268 (41). The GST-IκBα-ΔAnk2 expression vector contains a deletion which removes codons 98 to 142 (42). The GST-IκBα-ARD (ankyrin repeat domain) expression vector contains codons 70 to 254 of IκBα cloned into the SmaI site of pGEX3X. The GFP (green fluorescent protein)-IκBα expression vectors were constructed by inserting the various IκBα fragments from the GST-based vectors into the GFP-C3 eukaryote expression vector (Clontech). The GST-NLS expression vector contains an oligonucleotide which encodes an NLS derived from the SV40 large T protein inserted into the SmaI site of pGEX1. The GST-M9 expression vector contains an oligonucleotide which encodes the M9 nuclear import sequence from the hnRNP A1 protein cloned into the SmaI site of pGEX1 (35). The pQE32-derived expression vectors for wild-type Ran and the RanQ69L protein and the pQE60-derived expression vector for the importin β(45–462) protein were obtained from Dirk Gorlich (University of Heidelberg). A pET-based expression vector for the importin β-binding domain of importin α (IBB) was obtained from Steve Adam (Northwestern University). A pQE32-derived expression vector for RanBP1 was obtained from Iain Mattaj's laboratory (EMBL, Heidelberg, Germany).
Expression and purification of recombinant proteins.
The recombinant GST fusion proteins were expressed in Escherichia coli strain BL21(DE3). Cultures were grown to an optical density at 600 nm of 0.4 and induced with 0.2 mM isopropyl thiogalactoside (IPTG) (Sigma) for 4 h at 20°C. The bacterial cell pellets were resuspended in ice-cold phosphate-buffered saline (PBS [pH 7.4]) containing 0.1% Triton X-100; 0.2 mM dithiothreitol (DTT); 1 mM phenylmethylsulfonyl fluoride; and 1 μg (each) of antipain, aprotinin, leupeptin, pepstatin, and soybean trypsin-chymotrypsin inhibitor per ml. Lysis of the cell pellets was conducted by brief sonication on ice. The lysates were cleared by centrifugation at 14,000 × g for 15 min at 4°C and the soluble GST fusion proteins were bound to glutathione-agarose beads (Sigma) for 30 min at room temperature. The glutathione-agarose beads were extensively washed with ice-cold PBS (pH 7.4), and the recombinant GST fusion proteins were eluted with 10 mM reduced glutathione in 50 mM Tris-Cl (pH 8.0) and dialyzed against transport buffer (20 mM HEPES (pH 7.3), 110 mM potassium acetate, 5 mM sodium acetate, 2 mM magnesium acetate, 1 mM EGTA).
To label the proteins with fluorescein, the recombinant GST fusion proteins were first dialyzed against labeling buffer (20 mM sodium phosphate buffer [pH 7.2], 150 mM sodium chloride) at 4°C overnight. Fluorescein 5′-maleimide (Pierce Chemical Co.) was added at an equimolar ratio, and the mixtures were incubated for 2 h on ice. The reactions were quenched by the addition of 50 mM β-mercaptoethanol. The labeled proteins were equilibrated against transport buffer by using Centricon-3 (Amicon) columns.
The Ran proteins, RanBP1, the importin β(45–462) protein, and the IBB protein were expressed as His-tagged proteins and purified by metal-chelate affinity chromatography (Invitrogen). RanBP1, importin β(45–462), and the IBB protein were dialyzed against transport buffer prior to use. The purified Ran proteins were dialyzed against Ran loading buffer (10 mM HEPES [pH 7.3], 160 mM potassium acetate, 5 mM magnesium acetate, 1 mM DTT) at 4°C overnight. The dialyzed Ran proteins were incubated with 1 mM GTP in the presence of 15 mM EDTA for 60 min at room temperature. Magnesium chloride was added to a final concentration of 30 mM. The loading reactions were performed immediately prior to the import reactions, and the Ran proteins were placed on ice during the permeabilization step. A mock loading sample in which the respective Ran protein was left out of the loading reaction was included to ensure that the nucleotide loading conditions did not affect the nuclear import reactions.
Each of the recombinant proteins was examined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). All proteins were greater than 90% pure, as determined by Coomassie blue staining of the respective SDS-PAGE gels. Protein concentrations were determined by Bradford assays (Bio-Rad) in accordance with the instructions provided by the manufacturer.
In vitro nuclear import assay.
HeLa cells were grown in Dulbecco's modified Eagle's medium (low glucose) supplemented with 10% fetal bovine serum (FBS) in a 37°C, 5% CO2 incubator. Approximately 16 h prior to the transport assays, 6.5 × 105 HeLa cells were plated onto 35 mm-diameter plates containing glass coverslips. The in vitro nuclear transport assays were conducted essentially as described in reference 1. In brief, cells on coverslips were permeabilized with 50 μg of digitonin per ml (Calbiochem) in transport buffer for 5 min on ice. The transport reactions were typically conducted for 30 min at 30°C, except where noted. A standard 50-μl transport reaction contained an energy-regenerating system (1 mM ATP, 0.1 mM GTP, 5 mM creatine phosphate, 20 U of creatine phosphokinase per ml), protease inhibitor mix, 2 mM DTT, and 15 μl of rabbit reticulocyte lysate (50 mg of total protein per ml). The unlabeled import substrates were added to a final concentration of 100 μg/ml, while the fluorescein-labeled import substrates were added to a final concentration of 50 μg/ml. For some samples, HeLa cells growing on coverslips were pretreated with 10 nM leptomycin B for 30 min prior to digitonin permeabilization, and 10 nM leptomycin B was included throughout the 30-min time course of the transport reactions. For the energy-dependence experiments, the reticulocyte lysates were pretreated with 25 U of apyrase per ml for 30 min prior to the import reactions, which were performed in the absence of ATP, creatine phosphate, and creatine phosphokinase.
The competition experiments were carried out at room temperature with a 100-fold molar excess of unlabeled protein relative to the fluorescein-labeled protein, as indicated in the figures and legends. For the importin β(45–462) blocking experiments, the digitonin-permeabilized cells were preincubated for 10 min with 5 μg of the recombinant importin β(45–462) protein at room temperature in transport buffer before initiation of the import reaction. The Ran-dependence experiments were performed in the presence of 5 μg of the respective Ran protein preloaded with GTP. The IBB competition experiment was performed in the presence of 15 μg of the IBB polypeptide. The RanBP1 experiments were carried out in the presence of 15 μg of RanBP1.
Indirect immunofluorescence assays were conducted on coverslips as previously described (42). The primary antibody was an anti-GST monoclonal antibody (Santa Cruz Biotechnology) used at a concentration of 1:100 in PBS (pH 7.4) containing 10% FBS. Anti-mouse fluorescein isothiocyanate (FITC)-conjugated secondary antibody was used at a concentration of 1:100 in PBS (pH 7.4) containing 10% FBS. The coverslips were mounted onto glass slides with Mowiol containing 2.5% DABCO (Sigma). Pictures shown in the figures were taken with a ×40 oil immersion lens on a Nikon Optiphot-2 equipped with a Nikon UFX-IIA 35-mm camera. Unless otherwise noted, equivalent exposure time periods were used for all panels shown in the same figure. The negatives were scanned into Photoshop 3.0 (Adobe) and compiled into the figures shown. All panels shown in the same figure were treated identically during developing and compilation.
Confocal laser scanning microscopy was performed with a ×60 oil-immersion lens on a Nikon Diaphot microscope equipped with a Bio-Rad MRC-600 laser. The z-sections were captured as TIFF files by using CoSMOS software. The images were compiled with Photoshop 3.0 and treated identically during figure construction.
HeLa cell transfections.
HeLa cells were purchased from American Type Culture Collection and cultured in Dulbecco's modified Eagle's medium containing 10% FBS. HeLa cells were transfected with the GFP-IκBα expression plasmids by using Fugene 6 according to the manufacturor's instructions (Boehringer Mannheim). Expression of the GFP-IκBα fusion proteins was confirmed by immunoblot analysis. Localization of the GFP-IκBα fusion proteins was determined by indirect immunofluorescence with an anti-GFP antibody (Chemicon).
RESULTS
Nuclear shuttling of IκBα in digitonin-permeabilized HeLa cells.
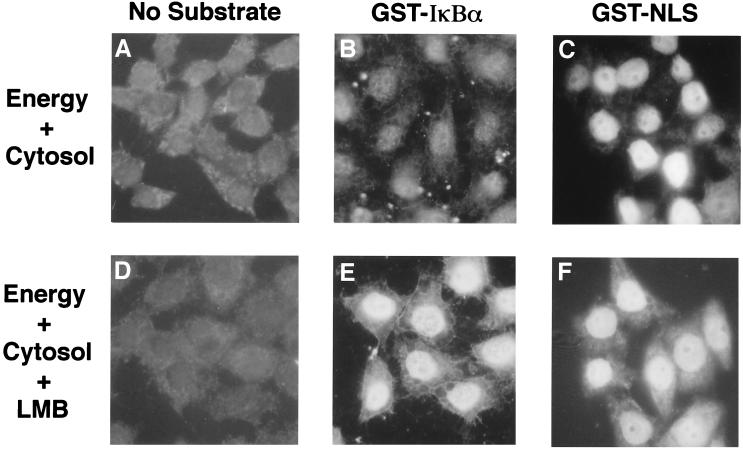
A digitonin-permeabilized cell assay was used to characterize nuclear import of IκBα. IκBα was expressed as a GST fusion protein in E. coli, and the ability of the GST-IκBα fusion protein to accumulate in the nuclei of digitonin-permeabilized HeLa cells was monitored by indirect immunofluorescence with a monoclonal antibody directed against GST. The NLS from the SV40 large T protein was fused to GST (GST-NLS) to serve as a positive control for nuclear import. Nuclear staining was not observed in the absence of an import substrate (Fig. 1A). In the presence of reticulocyte lysate and an energy-regenerating system, GST-NLS was efficiently imported into the nucleus (Fig. 1C). Under these conditions, neither GST nor a GST fusion protein containing a mutant NLS was imported into the nucleus (data not shown).
FIG. 1.
Nuclear accumulation of IκBα in digitonin-permeabilized cells is leptomycin B sensitive. HeLa cells were either left untreated (A to C) or were treated with 10 nM leptomycin B (LMB [D to F]) for 30 min prior to permeabilization with digitonin. Digitonin-permeabilized HeLa cells were incubated for 30 min at 30°C in the presence of reticulocyte lysate and an energy-regenerating system. Leptomycin B (10 nM) was included in the import reaction for the samples shown in panels D to F. The cells were incubated in the absence of import substrate (A and D) and in the presence of either GST-IκBα (B and E) or GST-NLS (C and F). The cellular localization of the import substrates was visualized by indirect immunofluorescence with a monoclonal antibody directed against GST.
In contrast to GST-NLS, the GST-IκBα protein did not efficiently accumulate in the nucleus in a standard nuclear import reaction (Fig. 1B). The inability of IκBα to localize to the nucleus in vitro was surprising, because either endogenous or ectopically expressed IκBα can readily be detected in the nucleus in vivo (2, 3, 42). Furthermore, Turpin et al. have recently reported that IκBα is actively imported into the nuclei of digitonin-permeabilized HeLa cells (46). Because IκBα contains a canonical NES and is able to direct the nuclear export of NF-κB/Rel proteins (3, 42), we asked if inhibition of nuclear export would enable nuclear accumulation of IκBα in vitro. HeLa cells were pretreated with 10 nM leptomycin B, a specific inhibitor of the nuclear export receptor CRM1 (13, 14, 33, 45), for 30 min prior to permeabilization, and 10 nM leptomycin was included throughout the 30-min time course of the import reaction. This regimen of leptomycin B treatment markedly increased the accumulation of GST-IκBα in the nuclei of digitonin-permeabilized HeLa cells (compare panels B and E of Fig. 1). Leptomycin B treatment had no effect on nuclear accumulation of GST-NLS (Fig. 1F). Nuclear accumulation of an IκBα protein containing a short N-terminal hexahistidine tag was also markedly increased by leptomycin B treatment (data not shown).
To further characterize nuclear accumulation of IκBα in digitonin-permeabilized cells, increasing amounts of FITC-labeled GST-IκBα protein were added to in vitro import reactions in the absence or presence of leptomycin B. Nuclear accumulation of IκBα was markedly enhanced by leptomycin B at 2.5 and 5.0 μg of input IκBα protein. However, nuclear accumulation of IκBα was leptomycin B independent at 10.0 and 20.0 μg of input IκBα protein (data not shown). This concentration range at which nuclear accumulation of GST-IκBα became leptomycin B independent was highly reproducible in multiple experiments (data not shown). It is likely that the addition of excess IκBα protein titrates out one or more factors that are limiting for nuclear export of IκBα.
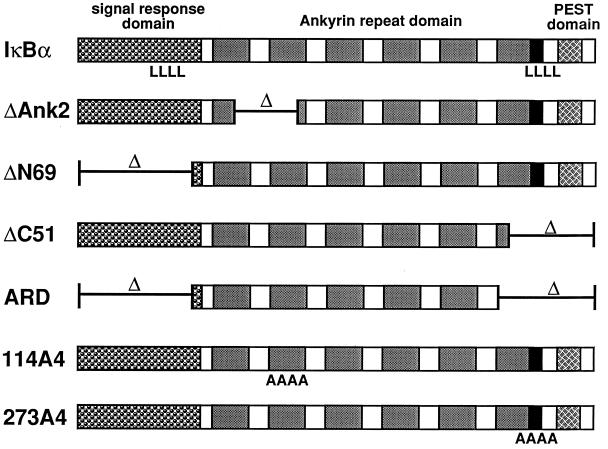
IκBα contains multiple functional domains that contribute to rapid shuttling between the cytoplasm and the nucleus.
To define regions within IκBα that specify nuclear import and nuclear export, we constructed a series of mutant GST-IκBα fusion proteins (Fig. 2). These mutant proteins were expressed in E. coli, purified with glutathione-agarose, and assayed for their ability to accumulate in the nucleus of digitonin-permeabilized HeLa cells in the absence and presence of leptomycin B (Fig. 3). To ensure that nuclear export of IκBα was not saturated by excess input protein, the ability of leptomycin B to increase nuclear accumulation of the mutant IκBα proteins was measured at 2.5 μg of input protein.
FIG. 2.
Domain organization of IκBα. IκBα contains an N-terminal signal response domain, a central ARD comprised of six ankyrin repeats, and a C-terminal PEST domain. The location of two canonical leucine-rich NESs (amino acids 45 to 55 and 273 to 283) in IκBα is indicated (LLLL). The structure of the mutant IκBα proteins used in these experiments is diagrammed.
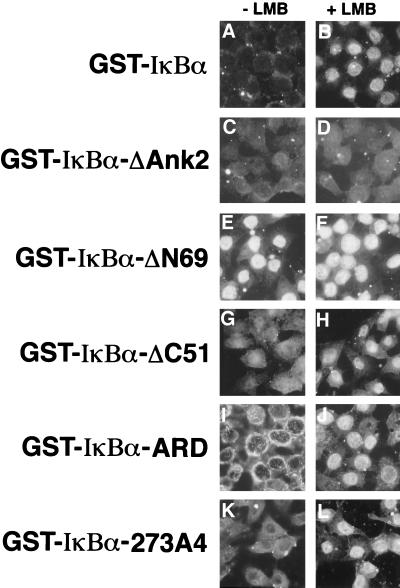
FIG. 3.
Definition of nuclear import and NESs in IκBα. Nuclear import reactions were performed with the indicated protein substrates in either the absence (A, C, E, G, I, and K) or presence of 10 nM leptomycin B (B, D, F, H, J, and L). The cellular localization of the import substrates was determined by indirect immunofluorescence with either a monoclonal antibody directed against GST or by direct fluorescence with FITC-labeled proteins. Parallel experiments with both detection methods were done for all protein samples, with the exception of the GST-IκBα-ΔC251 protein, which was only detected by direct fluorescence.
Consistent with our previous demonstration that the second ankyrin repeat of IκBα is critically required for nuclear localization of IκBα in transfected cells, a mutant IκBα protein that contained a deletion of the second ankyrin repeat (GST-IκBα-ΔAnk2; Fig. 3C and D) did not accumulate in the nucleus of digitonin-permeabilized HeLa cells, in either the absence or presence of leptomycin B. Likewise, a mutant IκBα protein that contained alanine residues in place of four hydrophobic residues within the second ankyrin repeat was defective for nuclear import (GST-IκBα-114A4 [data not shown]). These mutant IκBα proteins were unable to accumulate in the nucleus of digitonin-permeabilized cells even when 10 μg of input IκBα protein was added to the import reactions (data not shown).
IκBα contains a canonical leucine-rich NES located in its C terminus (amino acids 273 to 283). Several reports have suggested that this NES is required for IκBα-mediated nuclear export of NF-kB/Rel proteins (3, 42). However, a C-terminal-truncated GST-IκBα fusion protein, which lacked the C-terminal 51 amino acids of IκBα (GST-IκBα-ΔC51), remained leptomycin B dependent for nuclear accumulation (Fig. 3G and H). Likewise, a mutant full-length GST-IκBα protein containing four leucine-to-alanine substitutions within the canonical leucine-rich NES of IκBα was also leptomycin B dependent for nuclear accumulation (GST-IκBα-273A4; Fig. 3K and L). That these IκBα mutants remained leptomycin B dependent for nuclear accumulation despite deletion or mutation of the C-terminal NES indicates the presence of additional functional NES(s) in IκBα.
A mutant IκBα protein that lacked the N-terminal 69 amino acids of IκBα (GST-IκBα-ΔN69) was able to accumulate in the nucleus in a leptomycin B-independent manner (Fig. 3E and F). The ability of the GST-IκBα-ΔN69 protein to accumulate in the nucleus in a leptomycin B-independent manner was not due to saturation of IκBα export by excess protein, because significant leptomycin B-independent nuclear accumulation was observed at 1 μg of input FITC-labeled GST-IκBα-ΔN69 protein (data not shown). That removal of the N-terminal 69 amino acids of IκBα enabled leptomycin B-independent nuclear accumulation of IκBα suggests the presence of a functional nuclear export sequence within the N-terminal domain of IκBα. Indeed, a recent report has suggested that amino acids 45 to 55 of IκBα comprise a leucine-rich NES (24).
A GST fusion protein containing just the ARD of IκBα (GST-IκBα-ARD) was competent for nuclear import (Fig. 3I and J), consistent with our previous demonstration that the nuclear localization of IκBα in transfected cells is mediated by one or more nuclear import sequences within the ARD (42). Surprisingly, the GST-IκBα-ARD protein was leptomycin B dependent for nuclear accumulation despite the absence of both the N-terminal and C-terminal NESs. The leptomycin B-dependent nuclear accumulation of the GST-IκBα-ARD protein suggests that, under the conditions of the in vitro import reaction, the ARD contains an additional nuclear export sequence.
Nuclear shuttling of IκBα in transfected HeLa cells.
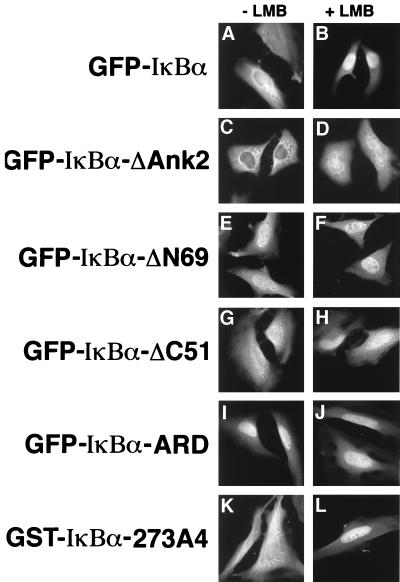
The failure of the C-terminal NES of IκBα to contribute to nuclear export of IκBα in digitonin-permeabilized HeLa cells was surprising, because several reports have indicated that this canonical leucine-rich NES is required for IκBα-mediated nuclear export of NF-κB/Rel proteins (3, 42). To further characterize nuclear shuttling of IκBα in vivo, we constructed fusion proteins between the GFP and the various IκBα proteins. These fusion proteins were expressed in HeLa cells, and the ability of leptomycin B to alter the nuclear-cytoplasmic distribution of fusion proteins was measured (Fig. 4 and Table 1).
FIG. 4.
Nuclear shuttling of GFP-IκBα chimeric proteins in transfected HeLa cells. HeLa cells transfected with expression vectors for the indicated GFP-IκBα fusion proteins were left untreated or were treated with leptomycin B for 1 h. The cellular localization of the fusion proteins was detected by indirect immunofluorescence with a chicken antibody raised against GFP. More than 200 cells were examined for each fusion protein (see Table 1 for quantitation), and representative cells are shown.
TABLE 1.
Localization of GFP-IκBα fusion proteinsa
| Protein | % of cells staining positive
|
|||||
|---|---|---|---|---|---|---|
| −LMB
|
+LMB
|
|||||
| N | N/C | C | N | N/C | C | |
| GFP | 4 | 74 | 22 | 3 | 76 | 20 |
| GFP-IκBα | 2 | 62 | 36 | 92 | 8 | 0 |
| GFP-IκBα-ΔANK2 | 1 | 11 | 88 | 53 | 46 | 1 |
| GFP-IκBα-114A4 | 1 | 11 | 88 | 20 | 78 | 2 |
| GFP-IκBα-ΔN69 | 61 | 39 | 0 | 95 | 5 | 0 |
| GFP-IκBα-ΔC51 | 2 | 71 | 27 | 54 | 46 | 0 |
| GFP-IκBα-273A4 | 3 | 64 | 33 | 90 | 9 | 1 |
| GFP-IκBα-ARD | 69 | 31 | 0 | 89 | 11 | 0 |
The cellular localization of the indicated GFP-IκBα fusion proteins was determined by indirect immunofluorescence in transfected HeLa cells. HeLa cell cultures transfected in parallel were either left untreated (−LMB) or were treated with 10 mM leptomycin B for 1 h (+LMB). The transfected cells were scored as having predominantly nuclear (N), whole-cell (N/C), or cytoplasmic (C) staining. A total of 200 cells were examined for each fusion protein.
The wild-type GFP-IκBα protein was distributed throughout both the nucleus and the cytoplasm in transfected HeLa cells (Fig. 4A). Treatment of transfected HeLa cells with leptomycin B for 1 h resulted in a marked redistribution of the wild-type GFP-IκBα protein to the nucleus (Fig. 4B). In contrast, leptomycin B treatment had no effect on the predominantly cytoplasmic localization of GFP (Table 1). The increased nuclear localization of GFP-IκBα in the presence of leptomycin B indicates that the GFP-IκBα protein shuttles between the nucleus and the cytoplasm in transfected HeLa cells.
The ability of the mutant GFP-IκBα proteins to shuttle between the nucleus and the cytoplasm in a leptomycin B-dependent manner was also determined. As expected, the GFP-IκBα-ΔAnk2 protein and the GFP-IκBα-114A4 protein were predominantly cytoplasmic in the absence of leptomycin B (Fig. 4C and data not shown). Treatment of the transfected HeLa cells with leptomycin B resulted in increased nuclear localization of the mutant GFP-IκBα proteins (Fig. 4D and data not shown). That leptomycin B treatment increased the extent to which these mutant proteins accumulated in the nucleus suggests that IκBα contains additional weak nuclear import sequences, likely within one or more of the remaining intact ankyrin repeats (24, 42).
The GFP-IκBα-ΔC51 protein and the GFP-IκBα-273A4 protein were distributed between the nucleus and the cytoplasm in the absence of leptomycin B, and treatment of the transfected cells with leptomycin B markedly increased the nuclear localization of these proteins (Fig. 4G, H, K, and L). These results are consistent with the shuttling phenotype of these proteins in digitonin-permeabilized cells and demonstrate that both in vitro and in vivo, the C-terminal NES of IκBα is not required for nuclear export of IκBα.
The GFP-IκBα-ΔN69 protein displayed increased nuclear localization in the absence of leptomycin B, consistent with the ability of this protein to accumulate in the nucleus of digitonin-permeabilized cells in the absence of leptomycin B (Fig. 4E). Leptomycin B treatment increased the percentage of GFP-IκBα-ΔN69-positive cells that displayed predominantly nuclear staining (Table 1).
In contrast to the results obtained in vitro, a GFP fusion protein containing just the ARD of IκBα also displayed significant nuclear accumulation in the absence of leptomycin B (Fig. 4I). Leptomycin B treatment increased the percent of GFP-IκBα-ARD-positive cells that displayed predominantly nuclear staining (Table 1), suggesting that this protein, which lacks both of the previously described NESs in IκBα, is still able to shuttle between the nucleus and the cytoplasm.
Nuclear import of IκBα is energy and temperature dependent and is blocked by a dominant-negative importin β protein.
To determine whether nuclear accumulation of IκBα reflects active nuclear transport or passive diffusion through the NPC, cytosolic extracts were pretreated with apyrase to deplete the extracts of high-energy phosphate compounds. The ability of the energy-depleted extracts to support nuclear import of GST-IκBα or GST-NLS in the absence of an energy-regenerating system was determined. Nuclear import of both GST-IκBα (Fig. 5D) and GST-NLS (Fig. 5H) was inhibited when the cytosolic extracts were pretreated with apyrase. Similar results were obtained when hexokinase and glucose were used to deplete the energy pools present in the cytosolic extracts (data not shown).
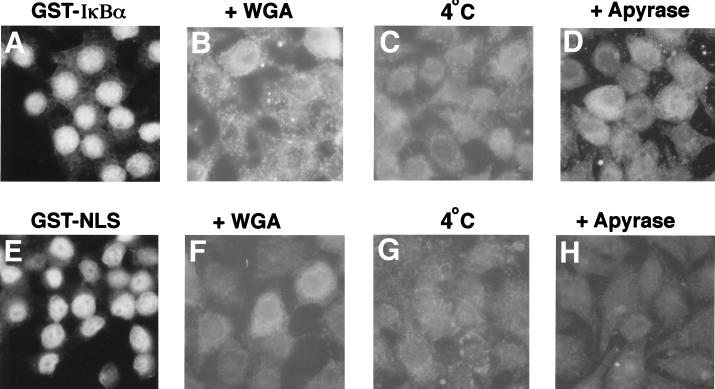
FIG. 5.
Nuclear import of IκBα is energy and temperature dependent. Nuclear import reactions into digitonin-permeabilized HeLa cells were performed with either GST-IκBα (A to D) or GST-NLS (E to H). Leptomycin B (10 nM) was included in the nuclear import reactions with GST-IκBα (A to C). The import reactions were carried out under standard conditions (A and E), in the presence of WGA (B and F), on ice (C and G), or in the presence of 25 U of apyrase per ml (D and H). The cellular localization of the import substrates was determined by indirect immunofluorescence with a monoclonal antibody directed against GST. Panels A and E were from equivalent exposure settings. Panels B to D and F to H were exposed for 10 to 20 times longer than panels A and E.
To determine the temperature dependence of IκBα nuclear import, the import reactions were performed at 4°C instead of at 30°C. Nuclear import of both GST-IκBα (Fig. 5C) and GST-NLS (Fig. 5G) was markedly reduced when the import reactions were conducted at 4°C. Nuclear import of IκBα was restored when the import reactions were shifted back to 30°C (data not shown).
The effect of wheat germ agglutinin (WGA), a lectin which binds to N-acetyl-d-glucosamine residues present on many nucleoporins, was examined. Addition of WGA significantly inhibited nuclear accumulation of both GST-IκBα and GST-NLS (Fig. 5B and F).
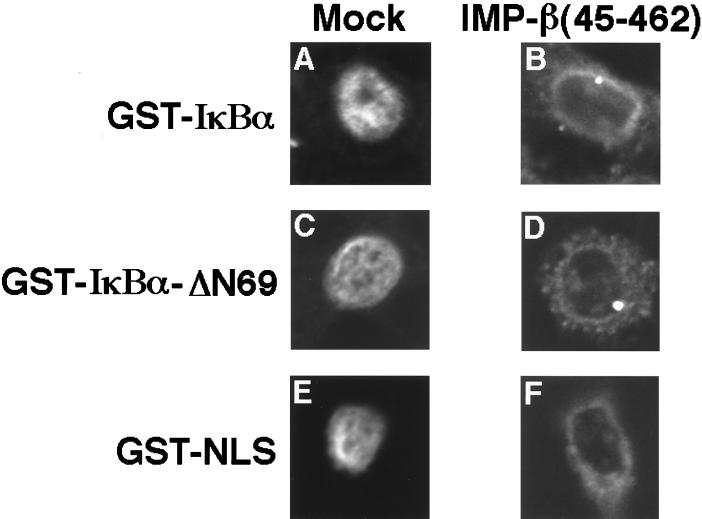
The ability of a dominant-negative importin β protein, importin β(45–462), to perturb nuclear import was also determined. The importin β(45–462) protein has been demonstrated to block multiple nuclear transport pathways, presumably by binding irreversibly to specific components of the NPC (28). Digitonin-permeabilized HeLa cells were preincubated with the mutant importin β(45–462) protein, and leptomycin B-dependent nuclear accumulation of IκBα was examined by confocal laser scanning microscopy. Consistent with previous results (27), the importin β(45–462) protein completely blocked nuclear import of GST-NLS (Fig. 6E and F). Similarly, preincubation of HeLa cells with the importin β(45–462) protein inhibited entry of both GST-IκBα (Fig. 6A and B) and of the N-terminal-truncated derivative, GST-IκBα-ΔN69 (Fig. 6C and D). The IκBα proteins, like GST-NLS, accumulated at the nuclear envelope in the presence of the importin β(45–462) protein (Fig. 6B, D, and F).
FIG. 6.
Binding of IκBα to the nuclear membrane in the presence of a dominant-negative importin β protein. Nuclear import reactions using the indicated proteins were performed in the absence (A, C, and E) or presence (B, D, and F) of the dominant-negative importin β(45–462) protein. Leptomycin B (10 nM) was included in the import reactions shown in panels A and B. The images shown are representative of more than 50 cells that were examined for each import substrate. The import substrates were visualized by indirect immunofluorescence by confocal laser scanning microscopy.
Taken together, these results demonstrate that nuclear accumulation of IκBα does not occur by passive diffusion through open channels of the NPC. Rather, nuclear import of IκBα is an energy-dependent and temperature-sensitive process that requires specific components of the NPC.
Rate-limiting factors for nuclear import of IκBα are not lost during the digitonin-permeabilization procedure.
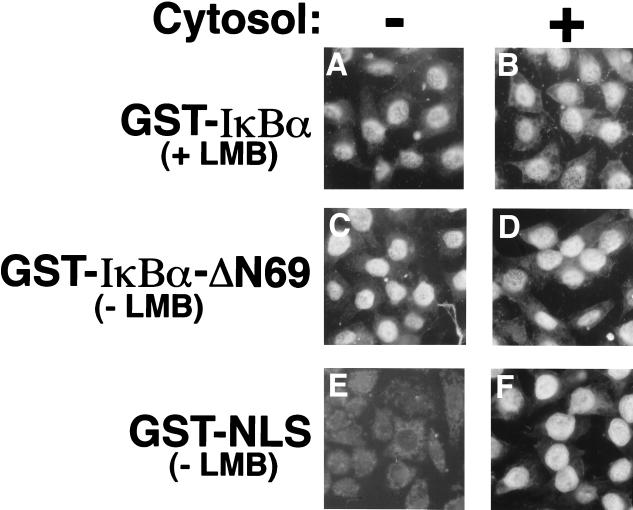
To investigate the dependence of IκBα import on soluble protein factors, the ability of GST-IκBα to accumulate in the nuclei of HeLa cells in the absence of exogenously added cytosol was determined. Nuclear import of GST-NLS was strictly dependent on the presence of exogenously added cytosol (Fig. 7E and F). In contrast, GST-IκBα readily accumulated in the nuclei of HeLa cells, even when cytosol was not added to the import reactions (Fig. 7A and B). Leptomycin B was included in the nuclear import reactions for GST-IκBα. However, the ability of GST-IκBα to accumulate in the nucleus in the absence of exogenously added soluble factors was not an artifact resulting from the inclusion of leptomycin B in the import reaction, because the GST-IκBα-ΔN69 protein efficiently accumulated in HeLa cell nuclei in a cytosol-independent manner in the absence of leptomycin B (Fig. 7C and D).
FIG. 7.
Nuclear import of IκBα is independent of exogenous cytosol. Nuclear import reactions using the indicated proteins were carried out either in the absence of reticulocyte lysate (A, C, and E) or in the presence of 100 μg of reticulocyte lysate. Leptomycin B (LMB) was included in the nuclear import reactions using GST-IκBα (A and B). The cellular localization of the import substrates was determined by indirect immunofluorescence with a monoclonal antibody directed against GST.
Nuclear import of IκBα is not blocked by saturation of the NLS-dependent or M9-dependent import pathways.
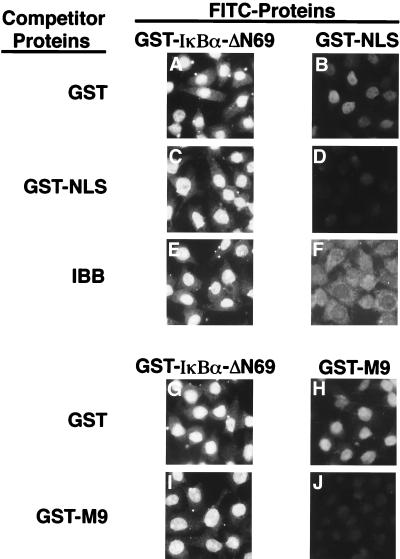
Nuclear import of proteins that utilize known nuclear import pathways, such as the NLS-dependent pathway or the M9-dependent pathway, can be inhibited by saturation of the respective import receptor (30). As anticipated, we found that a 100-fold molar excess of GST-NLS (Fig. 8D) or a 7-fold molar excess of IBB (Fig. 8F) markedly reduced nuclear import of FITC-labeled GST-NLS. Likewise, a 100-fold molar excess of GST-M9 significantly inhibited nuclear import of FITC-labeled GST-M9 (Fig. 8J). However, nuclear import of FITC-labeled GST-IκBα-ΔN69 was not inhibited by the addition of a 100-fold molar excess of GST (Fig. 8A), GST-NLS (Fig. 8C), a 7-fold molar excess of IBB (Fig. 8E), or a 100-fold molar excess of GST-M9 (Fig. 8I). Similar results were obtained with the full-length GST-IκBα protein when the nuclear import reactions were carried out in the presence of leptomycin B (data not shown). Taken together, these results indicate that nuclear import of GST-IκBα is independent of either the NLS-dependent or the M9-dependent nuclear import pathway.
FIG. 8.
Nuclear import of IκBα is independent of NLS- and M9-mediated nuclear import pathways. Nuclear import reactions with fluorescein-labeled GST-IκBα-ΔN69 (A, C, E, G, and I), fluorescein-labeled GST-NLS (B, D, and F), or fluorescein-labeled GST-M9 (H and J) were performed. The import reactions were performed in the presence of a 100-fold molar excess of unlabeled GST (A, B, G, and H), GST-NLS (C and D), or GST-M9 (I and J). For the import reactions shown in panels E and F, the IBB was included at a final concentration of 30 μM. The cellular localization of the import substrates was determined by direct fluorescence.
IκBα utilizes a high-throughput nuclear import pathway.
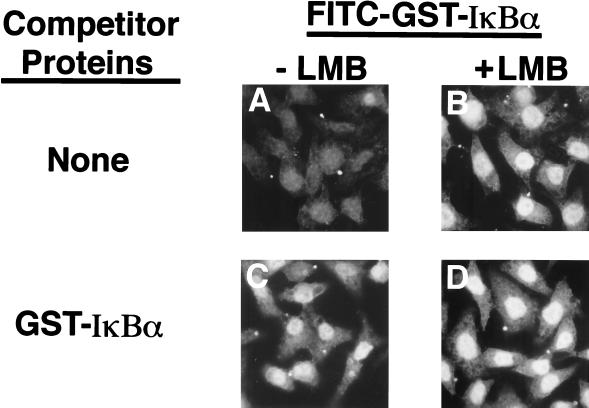
The saturability of both nuclear import and nuclear export of IκBα was determined by the ability of FITC-labeled GST-IκBα to accumulate in the nuclei of digitonin-permeabilized HeLa cells following addition of unlabeled GST-IκBα. These experiments were performed with limiting amounts of FITC-GST-IκBα protein to ensure that nuclear export was not titrated out simply by an excess of the FITC-labeled protein. Although FITC-labeled GST-IκBα does not efficiently accumulate in the nucleus in the absence of leptomycin B (Fig. 9A), addition of a 100-fold molar excess of unlabeled GST-IκBα resulted in a significant nuclear accumulation of FITC-labeled GST-IκBα (Fig. 9B), consistent with the notion that nuclear export of IκBα requires one or more rate-limiting factors that can be titrated out with an excess of IκBα protein.
FIG. 9.
High capacity of the IκBα nuclear import pathway. Nuclear import reactions with FITC-GST-IκBα were performed in the absence (A and C) or presence (B and D) of 10 nM leptomycin B (LMB). The import reactions were performed in the absence (A and B) or presence (C and D) of a 100-fold molar excess of unlabeled GST-IκBα. The cellular localization of the import substrates was determined by direct fluorescence.
Surprisingly, a 100-fold molar excess of unlabeled GST-IκBα did not inhibit the nuclear accumulation of FITC-labeled GST-IκBα in the presence of leptomycin B (compare Fig. 9C and D). The inability of an excess of GST-IκBα to inhibit the nuclear import of FITC-labeled GST-IκBα is not simply a consequence of the N-terminal GST tag, because addition of a 100-fold molar excess of a His-tagged IκBα protein was also unable to inhibit the nuclear accumulation of FITC-labeled GST-IκBα (data not shown). Furthermore, addition of a 100-fold molar excess of unlabeled GST-IκBα-ΔN69 was also unable to inhibit nuclear import of FITC-labeled GST-IκBα-ΔN69 (data not shown). Taken together, these results indicate that nuclear import of IκBα is not blocked by a 100-fold molar excess of specific competitor.
Nuclear import of IκBα is independent of GTP hydrolysis by Ran.
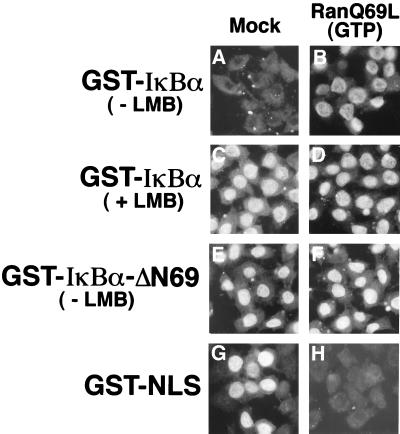
The directionality of classical nuclear import and nuclear export pathways is determined by the differential distribution of the GTP-bound and GDP-bound forms of Ran between the nucleus and the cytoplasm (21). To determine if nuclear shuttling of IκBα is sensitive to perturbation of the asymmetric Ran-nucleotide gradient, nuclear accumulation of GST-IκBα was determined in the presence of the dominant-negative mutant RanQ69L protein bound to GTP. RanQ69L-GTP blocked nuclear import of GST-NLS (compare Fig. 10G and H) and GST-M9 (data not shown). In contrast, RanQ69L-GTP did not inhibit nuclear import of GST-IκBα in the presence of leptomycin B (compare Fig. 10C and D) or of GST-IκBα-ΔN69 in the absence of leptomycin B (compare Fig. 10E and F). Consistent with the results obtained with the RanQ69L protein, addition of the wild-type Ran protein preloaded with GTPγS inhibited nuclear import of GST-NLS but did not inhibit nuclear import of either GST-IκBα or GST-IκBα-ΔN69 (data not shown). Furthermore, while addition of GTPγS or 5′-guanylylimdodiphosphate (GMP-PNP) completely inhibited nuclear import of GST-NLS and GST-M9 (data not shown), neither GTPγS nor GMP-PNP blocked nuclear import of GST-IκBα or GST-IκBα-ΔN69 (data not shown).
FIG. 10.
Nuclear import of IκBα is not inhibited by a dominant-negative Ran protein. Nuclear import reactions with the indicated substrate proteins were performed in the absence (A, B, and E to H) or presence (C and D) of 10 nM leptomycin B (LMB). Parallel import reactions were performed in the absence (A, C, E, and G) or presence (B, D, F, and H) of 5 μg of the RanQ69L protein preloaded with GTP. Localization of the import substrates was determined by indirect immunofluorescence with a monoclonal antibody directed against GST.
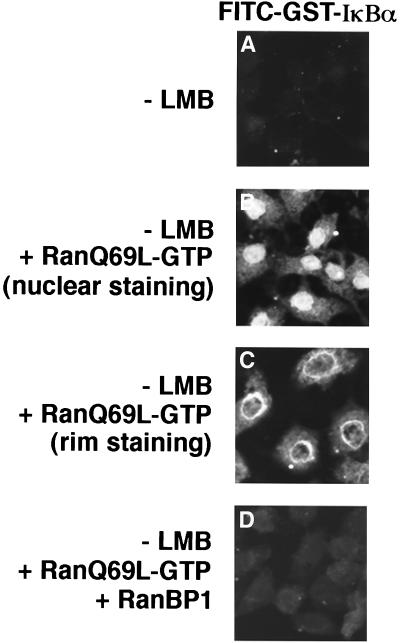
The ability of the RanQ69L-GTP mutant protein to perturb nuclear export of IκBα was also determined. Nuclear import reactions were performed in the presence of the RanQ69L-GTP mutant protein, but in the absence of leptomycin B. Analysis by confocal laser scanning microscopy demonstrated that the inclusion of RanQ69L-GTP in the import reaction allowed leptomycin B-independent nuclear accumulation of GST-IκBα (compare Fig. 11A and B). Nuclear rim accumulation of IκBα was observed in 30 to 50% of the cells (Fig. 11C), consistent with the notion that RanQ69L-GTP interfered with release of IκBα from CRM1 at nuclear pore sites involved in the termination of nuclear export. We hypothesized that the mutant RanQ69L-GTP protein interfered with termination of the nuclear export reaction by titrating out a specific factor(s) necessary for disassembly of the CRM1-cargo complex. The Ran-binding protein, RanBP1, has been suggested to play a critical role in the disassembly of the receptor-Ran-GTP complex (6). Addition of RanBP1 restored nuclear export of IκBα in the presence of RanQ69L-GTP (Fig. 11D). Thus, the ability of the RanQ69L-GTP mutant protein to inhibit nuclear export of IκBα may be due to sequestration of endogenous RanBP1.
FIG. 11.
RanQ69L-GTP inhibits nuclear export of IκBα through sequestration of RanBP1. Nuclear import reactions were performed with 2.5 μg of FITC-GST-IκBα in the absence of leptomycin B (LMB). RanQ69L-GTP (5 μg) was added to the import reactions shown in panels B to D, and RanBP1 (15 μg) was added to the import reaction shown in panel D. Localization of FITC-GST-IκBα was determined by direct fluorescence with confocal laser scanning microscopy. A single z-section of representative cells is shown in each panel.
DISCUSSION
In the present study, we have utilized both in vitro and in vivo approaches to understand how IκBα travels between the cytoplasm and the nucleus. The results of our experiments lead to three important conclusions: first, that IκBα rapidly shuttles between the cytoplasm and the nucleus; second, that IκBα contains multiple domains that specify nuclear import and nuclear export; and third, that nuclear import of IκBα is mediated by an import pathway that is mechanistically distinct from classical nuclear import pathways.
Nuclear shuttling of IκBα.
We find that leptomycin B markedly increased nuclear accumulation of IκBα in both digitonin-permeabilized cells and in transiently transfected HeLa cells. Our finding that nuclear accumulation of IκBα was increased by leptomycin B in digitonin-permeabilized cells is in apparent contrast to a recent report in which nuclear accumulation of IκBα in digitonin-permeabilized cells was observed in the absence of leptomycin B treatment (46). However, a careful titration of input FITC-GST-IκBα protein revealed that leptomycin B-independent nuclear accumulation of IκBα was observed at high levels of input protein, suggesting that one or more limiting export factors were titrated out by an excess of IκBα. It has been established that rate-limiting nuclear export factors can be lost during permeabilization with digitonin (10, 26). It is likely that subtle differences in the conditions used for permeabilization resulted in more complete loss of CRM1 in the experiments reported by Turpin et al. (46), such that their digitonin-permeabilized cells were no longer competent for nuclear export of IκBα.
The ability of leptomycin B to increase nuclear accumulation of IκBα in both digitonin-permeabilized cells and transiently transfected cells is consistent with several recent reports that leptomycin B treatment markedly increases nuclear accumulation of either ectopically expressed or endogenous IκBα (24, 40). Taken together, the available experimental evidence strongly supports a model in which IκBα rapidly shuttles between the nucleus and the cytoplasm.
IκBα contains multiple cis-acting sequences for nuclear import and export.
The distribution of IκBα between the nucleus and the cytoplasm can be altered by specific mutations with IκBα (24, 42). For example, we have previously demonstrated that mutations within the second ankyrin repeat of IκBα resulted in a marked relocalization of ectopically expressed IκBα from the nucleus to the cytoplasm (42). Consistent with this result, we find that the integrity of the second ankyrin repeat is required for nuclear import of IκBα in digitonin-permeabilized cells. However, mutation or deletion of the second ankyrin repeat does not completely abolish nuclear import of IκBα, because leptomycin B increased nuclear accumulation of the GFP-IκBα-ΔAnk2 protein in transfected HeLa cells. Likewise, Hope and coworkers found that GFP-IκBα constructs lacking the second ankyrin repeat were able to accumulate in the nucleus in a leptomycin B-dependent manner (24). In our previous work, we found that the second ankyrin repeat was able to direct nuclear localization of a heterologous cytoplasmic protein (42). The other ankyrin repeats of IκBα also possess this nuclear import function (42). Taken together, the data support a model in which IκBα contains multiple cis-acting nuclear import sequences within its ARD. Because the steady-state distribution of IκBα between the nucleus and the cytoplasm is the consequence of rapid shuttling between the nucleus and the cytoplasm, mutation of any individual nuclear import sequence will decrease the rate of nuclear import of IκBα, resulting in a net increase in cytoplasmic IκBα.
IκBα contains several leucine-rich sequences that resemble canonical NESs, one located in the N-terminal domain, amino acids 45 to 55, and a second located in the C-terminal domain, amino acids 273 to 283. We find that deletion of the N-terminal 69 amino acids of IκBα enables leptomycin B-independent nuclear accumulation of the GST-IκBα-ΔN69 fusion protein in digitonin-permeabilized cells and of the GFP-IκBα-ΔN69 fusion protein in transiently transfected HeLa cells. Likewise, Hope and coworkers have demonstrated that mutation of two hydrophobic residues in this N-terminal domain abolished nuclear shuttling of a GST-IκBα fusion protein following microinjection into multinucleated 3T3 cells (24). In these experimental situations, the mutant IκBα proteins are present in a large excess relative to the endogenous NF-κB proteins. Hence, it is likely that these assays only measure nuclear export of free IκBα (i.e., IκBα proteins that are not present in a complex with endogenous NF-κB/Rel proteins). Taken together, these results indicate that the N-terminal NES of IκBα is a major determinant for nuclear export of free IκBα.
IκBα also contains a leucine-rich NES-like sequence in its C terminus. Deletion or mutation of this C-terminal NES-like sequence has no effect on nuclear export of the IκBα proteins in all of these experimental assays described above. However, several reports have indicated that mutations within this NES-like sequence reduce or eliminate the ability of IκBα to mediate nuclear export of NF-κB/Rel proteins. In one report, the ability of IκBα to export either p50 or p65 from the nucleus of Xenopus oocytes was significantly reduced by alanine substitutions within this C-terminal NES (3). We have previously demonstrated that mutations within the C-terminal NES significantly reduced the ability of IκBα to mediate nuclear export of the v-Rel oncoprotein (43). We suggest that differential usage of the N-terminal and C-terminal NESs of IκBα is determined by the absence or presence of specific NF-κB/Rel proteins complexed with IκBα.
Surprisingly, we find that the GST-IκBα-ARD fusion protein is leptomycin B dependent for nuclear accumulation in digitonin-permeabilized cells. Minimally, this result indicates that the ARD of IκBα, when placed in a context independent of N-terminal and C-terminal flanking sequences of IκBα, is competent for CRM1-dependent nuclear export. Does the ARD contribute to nuclear export of full-length IκBα in intact cells? In transfected HeLa cells, nuclear accumulation of the GFP-IκBα-ARD fusion protein is not strictly dependent upon inhibition of nuclear export, although nuclear localization of the GFP-IκBα-ARD fusion protein is enhanced by leptomycin B. Furthermore, the results of Hope and coworkers, in which the mutation of the N-terminal NES of IκBα is sufficient to abolish nuclear export of a bacterially expressed GST-IκBα fusion protein following microinjection into one nucleus of multinucleated 3T3 cells, would suggest the IκBα ARD does not contribute to nuclear export of full-length IκBα (24). It will be important to define the residues within the ARD that mediate nuclear export and to determine the importance of the ARD to nuclear shuttling of the full-length IκBα protein.
Nuclear import of IκBα is accomplished by a receptor-independent mechanism.
We find that nuclear import of IκBα does not require soluble factors that are lost during the digitonin permeabilization step. In contrast, both the NLS-dependent and M9-dependent nuclear import pathways require soluble transport factors that are lost during the digitonin permeabilization step and must be added back in order to reconstitute nuclear import. That nuclear import of IκBα is not dependent upon exogenously supplied factors suggests that IκBα utilizes a nuclear import pathway that is distinct from the well-characterized NLS-dependent and M9-dependent nuclear import pathways. In support of this notion, saturation of either the NLS-dependent or the M9-dependent pathway blocked nuclear import of FITC-labeled GST proteins containing the homologous NLSs, but did not inhibit nuclear import of IκBα.
Taken alone, the observation that nuclear import of IκBα does not require replenishment of soluble factors does not necessarily mean that nuclear import of IκBα is independent of a soluble transport factor(s). It is likely that low levels of importin β-related transport factors remain associated with the nucleus following permeabilization with digitonin. For example, in our digitonin-permeabilized cells, we find that CRM1, an importin-β-related nuclear export receptor, is present in sufficient amounts to mediate nuclear export of IκBα. A common feature of known nuclear import and export receptors is their dependence upon the Ran GTPase for directionality of transport through the nuclear pore. We find that nuclear import of IκBα is not disrupted by perturbation of the RanGTP gradient between the nucleus and the cytoplasm. That nuclear import of IκBα is not disrupted by the addition of the dominant-negative Ran protein provides further evidence that nuclear import of IκBα is not mediated by a typical Ran-dependent importin β-related transport factor.
Our finding that nuclear import of IκBα is not disrupted by the RanQ69L protein is in contrast to the recent report by Turpin et al. that nuclear import of IκBα into digitonin-permeabilized cells is inhibited by the RanQ69L protein (46). Although the basis for this discrepancy is not clear, the RanQ69L protein was preloaded with GTP prior to the nuclear import reaction in our experiments. In contrast, the RanQ69L protein was simply added to the import reactions in the absence of bound nucleotide in the experiments reported by Turpin et al. It is possible that the presence or absence of bound nucleotide may influence the ability of the RanQ69L protein to interfere with nuclear import. Because it is likely that the wild-type Ran protein in vivo always contains a bound nucleotide (either GDP or GTP), we believe that the RanQ69L-GTP complex is a more accurate mimic of the wild-type Ran-GTP complex than is the RanQ69L protein in the absence of bound nucleotide.
A surprising aspect of IκBα nuclear import is the very high capacity of the transport system. We find that nuclear import of FITC-labeled IκBα is not blocked by a 100-fold molar excess of unlabeled IκBα. In contrast, both NLS-dependent nuclear import and M9-dependent nuclear import are blocked by a 100-fold molar excess of a specific competitor. The failure to block nuclear import of IκBα with a 100-fold molar excess of specific competitor does not simply reflect an artifactual behavior of IκBα in the in vitro assay, since nuclear export of FITC-labeled IκBα was competitively inhibited by a 100-fold molar excess of unlabeled GST-IκBα. Furthermore, nuclear import of IκBα is not accomplished by simple diffusion through the nuclear pore, because nuclear import of IκBα is temperature and ATP dependent and is blocked by a dominant-negative importin β protein. Rather, the inability of a 100-fold molar excess of unlabeled specific competitor to block nuclear import of IκBα indicates that the transport capacity of the system utilized by IκBα is not saturated by this amount of the unlabeled specific competitor protein. Our results indicate that the nuclear import system utilized by IκBα is capable of handling a very large number of molecules within the time frame of the nuclear import assay.
Taken together, our results demonstrate that nuclear import of IκBα is not accomplished via formation of a receptor-IκBα complex which can be disrupted by Ran-GTP. In this respect, the nuclear import pathway utilized by IκBα is similar to the import pathway(s) utilized by several other proteins, including two transport receptors (importin β and transportin), β-catenin, and the Vpr protein of human immunodeficiency virus (11, 23, 27, 32, 48). A plausible mechanism to account for nuclear import of these proteins is that they interact directly with components of the nuclear pore complex. For example, these proteins might interact with mobile components of the nuclear pore complex that are able to transport protein cargoes through the pore in Ran-independent manner. Alternatively, nuclear import of these proteins might involve sequential interactions with stationary components of the nuclear pore. It is not known if these proteins interact with a common subset of nuclear pore proteins or if each of these proteins interacts with a unique group of nuclear pore proteins. Although differences between these proteins with respect to saturability and energy requirements of nuclear import have been reported, it is not clear if these differences are simply due to slight differences in experimental protocols or reflect the existence of multiple pathways for transport through the NPC. Further characterization of these receptor-independent transport pathways is likely to yield important insights into the poorly understood process of protein translocation through the NPC.
ACKNOWLEDGMENTS
We thank Candace Nichol for technical assistance and David J. Pintel for a critical reading of the manuscript. We thank Dirk Gorlich, Steve Adam, Iain Mattai, and Ludwig Englmeier for reagents and advice and Minoru Yoshida for his generous gift of leptomycin B.
This work was supported by American Cancer Society grant RPG-98-097-01, by a grant from the Charlotte Geyer Foundation, and by the University of Missouri Molecular Biology Program.
REFERENCES
- 1.Adam S A, Marr R S, Gerace L. Nuclear protein import in permeabilized cells requires soluble cytoplasmic factors. J Cell Biol. 1990;111:807–816. doi: 10.1083/jcb.111.3.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arenzana-Seisdedos F, Thompson J, Rodriguez M S, Bachelerie F, Thomas D, Hay R T. Inducible nuclear expression of newly synthesized IκBα negatively regulates DNA-binding and transcriptional activities of NF-κB. Mol Cell Biol. 1995;15:2689–2696. doi: 10.1128/mcb.15.5.2689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arenzana-Seisdedos F, Turpin P, Rodriguez M, Thomas D, Hay R T, Virelizier J-L, Dargemont C. Nuclear localization of IκBα promotes active transport of NF-κB from the nucleus to the cytoplasm. J Cell Sci. 1997;110:369–378. doi: 10.1242/jcs.110.3.369. [DOI] [PubMed] [Google Scholar]
- 4.Askjaer P, Jensen T H, Nilsson J, Englmeier L, Kjems J. The specificity of the CRM1-Rev nuclear export signal interaction is mediated by RanGTP. J Biol Chem. 1998;273:33414–33422. doi: 10.1074/jbc.273.50.33414. [DOI] [PubMed] [Google Scholar]
- 5.Baldwin A S., Jr The NF-κB and IκB proteins: new discoveries and insights. Annu Rev Immunol. 1996;14:649–681. doi: 10.1146/annurev.immunol.14.1.649. [DOI] [PubMed] [Google Scholar]
- 6.Bischoff F R, Gorlich D. RanBP1 is crucial for the release of RanGTP from importin-β-related nuclear transport factors. FEBS Lett. 1997;419:249–254. doi: 10.1016/s0014-5793(97)01467-1. [DOI] [PubMed] [Google Scholar]
- 7.Chi N, Adam E, Adam S. Sequence and characterization of cytoplasmic nuclear import factor p97. J Cell Biol. 1995;130:265–274. doi: 10.1083/jcb.130.2.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davis L I. The nuclear pore complex. Annu Rev Biochem. 1995;64:865–896. doi: 10.1146/annurev.bi.64.070195.004245. [DOI] [PubMed] [Google Scholar]
- 9.Davis N, Ghosh S, Simmons D L, Tempst P, Liou H C, Baltimore D, Bose H R., Jr Rel-associated pp40: an inhibitor of the Rel family of transcription factors. Science. 1991;253:1268–1271. doi: 10.1126/science.1891714. [DOI] [PubMed] [Google Scholar]
- 10.Englmeier L, Olivo J-C, Mattaj I W. Receptor-mediated substrate translocation through the nuclear pore complex without nucleotide triphosphate hydrolysis. Curr Biol. 1999;9:30–41. doi: 10.1016/s0960-9822(99)80044-x. [DOI] [PubMed] [Google Scholar]
- 11.Fagotto F, Gluck U, Gumbiner B M. Nuclear localization signal-independent and importin/karyopherin-independent nuclear import of β-catenin. Curr Biol. 1998;8:181–190. doi: 10.1016/s0960-9822(98)70082-x. [DOI] [PubMed] [Google Scholar]
- 12.Fischer U, Huber J, Boelens W C, Mattaj I W, Luhrmann R. The HIV-1 Rev activation domain is a nuclear export signal that accesses an export pathway used by specific cellular RNAs. Cell. 1995;82:475–484. doi: 10.1016/0092-8674(95)90436-0. [DOI] [PubMed] [Google Scholar]
- 13.Fornerod M, Ohno M, Yoshida M, Mattaj I W. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell. 1997;90:1051–1060. doi: 10.1016/s0092-8674(00)80371-2. [DOI] [PubMed] [Google Scholar]
- 14.Fukuda M, Asano S, Nakamura T, Adachi M, Yoshida M, Yanagida M, Nishida E. CRM1 is responsible for intracellular transport mediated by the nuclear export signal. Nature (London) 1997;390:308–311. doi: 10.1038/36894. [DOI] [PubMed] [Google Scholar]
- 15.Goldfarb D S, Gariepy J, Schoolnik G, Kornberg R D. Synthetic peptides as nuclear localization signals. Nature (London) 1986;322:641–644. doi: 10.1038/322641a0. [DOI] [PubMed] [Google Scholar]
- 16.Gorlich D, Prehn S, Laskey R A, Hartmann E. Isolation of a protein that is essential for the first step of nuclear protein import. Cell. 1994;79:767–778. doi: 10.1016/0092-8674(94)90067-1. [DOI] [PubMed] [Google Scholar]
- 17.Gorlich D, Kostka S, Kraft R, Dingwall C, Laskey R, Hartmann E, Prehn S. Two different subunits of importin cooperate to recognize nuclear localization signals and bind them to the nuclear envelope. Curr Biol. 1995;5:383–392. doi: 10.1016/s0960-9822(95)00079-0. [DOI] [PubMed] [Google Scholar]
- 18.Gorlich D, Vogel G, Mills A, Hartmann E, Laskey R. Distinct functions for the two importin subunits in nuclear protein import. Nature (London) 1995;377:246–248. doi: 10.1038/377246a0. [DOI] [PubMed] [Google Scholar]
- 19.Hopper A K, Traglia H M, Dunst R W. The yeast RNA1 gene product necessary for RNA processing is located in the cytosol and is apparently excluded from the nucleus. J Cell Biol. 1990;111:309–321. doi: 10.1083/jcb.111.2.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huxford T, Huang D-B, Malek S, Ghosh G. The crystal structure of the IκBα/NF-κB complex reveals mechanisms of NF-κB inactivation. Cell. 1998;95:759–770. doi: 10.1016/s0092-8674(00)81699-2. [DOI] [PubMed] [Google Scholar]
- 21.Izaurralde E, Kutay U, von Kobbe C, Mattaj I W, Gorlich D. The asymmetric distribution of the constituents of the Ran system is essential for transport into and out of the nucleus. EMBO J. 1997;16:6535–6547. doi: 10.1093/emboj/16.21.6535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jacobs M D, Harrison S C. Structure of an IκBα/NF-κB complex. Cell. 1998;95:749–758. doi: 10.1016/s0092-8674(00)81698-0. [DOI] [PubMed] [Google Scholar]
- 23.Jenkins Y, McEntee M, Weis K, Greene W C. Characterization of HIV-1 Vpr nuclear import: analysis of signals and pathways. J Cell Biol. 1998;143:875–885. doi: 10.1083/jcb.143.4.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johnson C, Van Antwerp D, Hope T J. An N-terminal nuclear export signal is required for the nucleocytoplasmic shuttling of IκBα. EMBO J. 1999;18:6682–6693. doi: 10.1093/emboj/18.23.6682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kalderon D, Roberts B L, Richardson W D, Smith A E. A short amino acid sequence able to specify nuclear location. Cell. 1984;39:499–509. doi: 10.1016/0092-8674(84)90457-4. [DOI] [PubMed] [Google Scholar]
- 26.Kehlenbach R H, Dickmanns A, Gerace L. Nucleocytoplasmic shuttling factors including Ran and CRM1 mediate nuclear export of NFAT in vitro. J Cell Biol. 1998;141:863–874. doi: 10.1083/jcb.141.4.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kose S, Imamoto N, Tachibana T, Shimamoto T, Yoneda Y. Ran-unassisted nuclear migration of a 97-kD component of nuclear pore-targeting complex. J Cell Biol. 1997;139:841–849. doi: 10.1083/jcb.139.4.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kutay U, Izaurralde E, Bischoff F R, Mattaj I W, Gorlich D. Dominant-negative mutants of importin-β block multiple pathways of import and export through the nuclear pore complex. EMBO J. 1997;16:1153–1163. doi: 10.1093/emboj/16.6.1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kutay U, Bischoff F, Kostka S, Kraft R, Gorlich D. Export of importin α from the nucleus is mediated by a specific nuclear transport factor. Cell. 1997;90:1061–1071. doi: 10.1016/s0092-8674(00)80372-4. [DOI] [PubMed] [Google Scholar]
- 30.Mattaj I W, Englmeier L. Nucleocytoplasmic transport: the soluble phase. Annu Rev Biochem. 1998;67:265–306. doi: 10.1146/annurev.biochem.67.1.265. [DOI] [PubMed] [Google Scholar]
- 31.Moroianu J, Hijikata M, Blobel G, Radu A. Mammalian karyopherin α1β and α2β heterodimers: α1 or α2 bind nuclear localization signal and β interacts with peptide repeat containing nucleoporins. Proc Natl Acad Sci USA. 1995;92:6532–6536. doi: 10.1073/pnas.92.14.6532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nakielny S, Dreyfuss G. Import and export of the nuclear protein import receptor transportin by a mechanism independent of GTP hydrolysis. Curr Biol. 1997;8:89–95. doi: 10.1016/s0960-9822(98)70039-9. [DOI] [PubMed] [Google Scholar]
- 33.Nishi K, Yoshida M, Fujiwara D, Nishikawa M, Horinouchi S, Beppu T. Leptomycin B targets a regulatory cascade of crm1, a fission yeast nuclear protein, involved in control of higher order chromosome structure and gene expression. J Biol Chem. 1994;269:6320–6324. [PubMed] [Google Scholar]
- 34.Ohtsubo M, Okazaki H, Nishimoto T. The RCC1 protein, a regulator for the onset of chromosome condensation, locates in the nucleus and binds DNA. J Cell Biol. 1989;109:1389–1397. doi: 10.1083/jcb.109.4.1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pollard V W, Michael W M, Nakielny S, Siomi M C, Wang F, Dreyfuss G. A novel receptor-mediated nuclear protein import pathway. Cell. 1996;86:985–994. doi: 10.1016/s0092-8674(00)80173-7. [DOI] [PubMed] [Google Scholar]
- 36.Radu A, Blobel G, Moore M S. Identification of a protein complex that is required for nuclear-protein import and mediates docking of import substrate to distinct nucleoporins. Proc Natl Acad Sci USA. 1995;92:1769–1773. doi: 10.1073/pnas.92.5.1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Radu A, Moore M S, Blobel G. The peptide repeat domain of nucleoporin Nup98 functions as a docking site in transport across the nuclear pore complex. Cell. 1995;81:215–222. doi: 10.1016/0092-8674(95)90331-3. [DOI] [PubMed] [Google Scholar]
- 38.Rexach M, Blobel G. Protein import into nuclei: association and dissociation reactions involving transport substrate, transport factors and nucleoporins. Cell. 1995;83:683–692. doi: 10.1016/0092-8674(95)90181-7. [DOI] [PubMed] [Google Scholar]
- 39.Richards S A, Carey K L, Macara I G. Requirement for guanosine triphosphate-bound Ran for signal-mediated nuclear protein export. Science. 1997;276:1842–1844. doi: 10.1126/science.276.5320.1842. [DOI] [PubMed] [Google Scholar]
- 40.Rodriguez M S, Thompson J, Hay R T, Dargemont C. Nuclear retention of IκBα protects it from signal-induced degradation and inhibits nuclear factor κB transcriptional activation. J Biol Chem. 1999;274:9108–9115. doi: 10.1074/jbc.274.13.9108. [DOI] [PubMed] [Google Scholar]
- 41.Sachdev S, Rottjakob E M, Diehl J A, Hannink M. IκBα mediated inhibition of nuclear transport and DNA-binding by Rel proteins are separable functions: phosphorylation of C-terminal serine residues of IκBα is specifically required for inhibition of DNA-binding. Oncogene. 1995;11:811–823. [PubMed] [Google Scholar]
- 42.Sachdev S, Hoffman A, Hannink M. Nuclear localization of IκBα is mediated by the second ankyrin repeat: the IκBα ankyrin repeats define a novel class of cis-acting nuclear import sequences. Mol Cell Biol. 1998;18:2524–2534. doi: 10.1128/mcb.18.5.2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sachdev S, Hannink M. Loss of IκBα-mediated control over nuclear import and DNA binding enables oncogenic activation of c-Rel. Mol Cell Biol. 1998;18:5445–5456. doi: 10.1128/mcb.18.9.5445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 45.Stade K, Ford C S, Guthrie C, Weis K. Exportin 1 (Crm1p) is an essential nuclear export factor. Cell. 1997;90:1041–1050. doi: 10.1016/s0092-8674(00)80370-0. [DOI] [PubMed] [Google Scholar]
- 46.Turpin P, Hay R T, Dargemont C. Characterization of IκBα nuclear import pathway. J Biol Chem. 1999;274:6804–6812. doi: 10.1074/jbc.274.10.6804. [DOI] [PubMed] [Google Scholar]
- 47.Wen W, Meinkoth J L, Tsien R Y, Taylor S S. Identification of a signal for rapid export of proteins from the nucleus. Cell. 1995;82:463–474. doi: 10.1016/0092-8674(95)90435-2. [DOI] [PubMed] [Google Scholar]
- 48.Yokoya F, Imammoto N, Tachibana T, Yoneda Y. β-Catenin can be transported into the nucleus in a Ran-unassisted manner. Mol Biol Cell. 1999;10:1119–1131. doi: 10.1091/mbc.10.4.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]