Abstract
The phylogenetic position and taxonomic status of Rhynchocyclidae (Aves: Passeriformes) have been the subject of debate since their first description. In most models, Rhynchocyclidae represents a subfamily-level taxon placed within the Tyrant Flycatchers (Tyrannidae). Considering that this classification does not include cytotaxonomic characters, we tested the hypothesis that the chromosome organization of Rhynchocyclidae members differs from that of Tyrannidae. Hence, we selected two species, Tolmomyias sulphurescens, and Pitangus sulphuratus, representing Rhynchocyclidae and Tyrannidae, respectively. Results revealed a diploid number (2n) of 60 in T. sulphurescens and 2n = 80 in P. sulphuratus, indicating significant chromosomal differences. Chromosome mapping of Gallus gallus (GGA) and Taeniopygia guttata bacterial artificial chromosome (BAC) corresponding to chromosomes GGA1-28 (except 16) revealed that the genome evolution of T. sulphurescens involved extensive chromosome fusions of macrochromosomes and microchromosomes. On the other hand, P. sulphuratus retained the ancestral pattern of organization of macrochromosomes (except the centric fission involving GGA1) and microchromosomes. In conclusion, comparing our results with previous studies in Tyrant Flycatchers and allies indicates that P. sulphuratus has similar karyotypes to other Tyrannidae members. However, T. sulphurescens does not resemble the Tyrannidae family, reinforcing family status to the clade named Rhynchocyclidae.
Keywords: phylogenetic relationships, chromosomal rearrangements, cytotaxonomy, passerines, tyrant flycatchers
1. Introduction
The phylogenetic position and taxonomic status of the flycatcher lineage named Rhynchocyclidae (Aves: Passeriformes) have been debated since their proposition. In most classifications, it represents a subfamily placed within the Tyrant Flycatchers (Tyrannidae), composing the most diverse Neotropical family of suboscine passerines [1]. Tyrannidae “lato sensu” exhibits high degrees of morphological, ecological, and behavioral diversity, drawing the attention of several phylogenetic studies [1,2,3,4,5]. However, some aspects of their relationships and classification remain controversial. In a recent study of a massive dataset resulting in a complete super-tree of the Tyranni passerines (“suboscines”), Tyrannidae and Rhynchocyclidae were recovered as monophyletic and well-supported sister clades which diverged 21.8 million years ago (Mya) [6]. In addition, Onychorhynchidae, Oxyruncidae, Pipritidae, Platyrinchidae, and Tachurisidae were also well-supported as separated families [6].
Most of the Tyrant Flycatchers and allies have their diploid number (2n) close to 80. However, an interesting 2n variation has been found, ranging from 2n = 60 in Platyrinchus mystaceus (Platyrinchidae) to 2n = 84 in Cnemotriccus fuscatus (Tyrannidae: Fluvicolinae) (Table 1). The typical 2n of Tyrant Flycatchers and allies represents the most frequent finding in other Passeriformes and the whole Aves. According to Degrandi et al. [7], approximately 61% of avian species have a karyotype description of 2n between 76 and 82 chromosomes. Considering that the putative avian ancestral karyotype (PAK) had 2n = 80 [8], the deviation from this diploid number resulted from different chromosomal rearrangements. Usually, the decrease or increase of the diploid number can result from fusion or fission events, respectively [9]. While in some orders, the 2n was increased, such as in Piciformes (up to more than 100 chromosomes) [10], decreased, such as in Psittaciformes [11,12], in Passeriformes, the ancestral 2n is conserved in most species [7]. Unfortunately, due to the poor quality of G-banding in macrochromosomes and the small size of microchromosomes, classical cytogenetic techniques have provided limited information concerning the process of karyotypic evolution in birds.
Table 1.
Available cytogenetic data for species of Tyrant Flycatchers and related families (classification according to the relationships found by Harvey et al. [6]).
| Species | 2n | Family | Reference |
|---|---|---|---|
| Platyrinchus mystaceus | 2n = 60 | Platyrinchidae | [13] |
| Elaenia parvirostris | 2n = 78 | Tyrannidae | [13] |
| Elaenia spectabilis | 2n = 80 | Tyrannidae | [14] |
| Serpophaga subcristata | 2n = 82 | Tyrannidae | [15] |
| Pitangus sulphuratus | 2n = 80 | Tyrannidae | [15] |
| Tyrannus melancholicus | 2n = 78 | Tyrannidae | [13] |
| Tyrannus savana | 2n = 78 | Tyrannidae | [13] |
| Myiarchus ferox | 2n = 76 | Tyrannidae | [13] |
| Knipolegus cyanirostris | 2n = 78 | Tyrannidae | [16] |
| Satrapa icterophrys | 2n = 82 | Tyrannidae | [15] |
| Cnemotriccus fuscatus | 2n = 84 | Tyrannidae | [13] |
| Empidonax alnorum | 2n = 82 | Tyrannidae | [17] |
| Empidonax flaviventris | 2n = 82 | Tyrannidae | [17] |
| Empidonax hammondii | 2n = 82 | Tyrannidae | [17] |
| Empidonax minimus | 2n = 82 | Tyrannidae | [17] |
| Empidonax traillii | 2n = 82 | Tyrannidae | [17] |
Fortunately, the use of fluorescence in situ hybridization (FISH) can overcome these limitations. Experiments of comparative chromosome painting with several sets of probes have been performed in different bird lineages to clarify the chromosomal rearrangements involved in the reorganization of avian karyotypes [7,8,18]. Among them, the most widely used sets were from Gallus gallus (GGA) and Leucopternis albicollis [7,8,18,19].
Chromosome painting studies using macrochromosomes probes from these species have been applied only in four Tyrant Flycatcher species, Elaenia spectabilis (2n = 80), Serpophaga subcristata (2n = 82), Pitangus sulphuratus (2 = 80), and Satrapa icterophrys (2n = 82) [14,15]. These studies have revealed the fission of chicken chromosome 1 in all these species, which can be considered as a candidate synapomorphy for Passeriformes since it was found in all Passeriformes studied so far [7,18]. In addition, S. icterophrys has fission in chicken chromosome 2 [15]. Compared to PAK [8], it is likely that the karyotype evolution of Tyrant Flycatchers involved mostly fissions events. However, only the macrochromosomes have been analyzed in this lineage.
Although macrochromosomes represent approximately 77% of the average avian genome size, microchromosomes contain around 50% of the avian genes [20,21,22]. Despite the importance of microchromosomes, their organization was studied in few avian orders, and interchromosomal rearrangements involving them have been found only in few orders [12,23,24,25,26]. Among the Passeriformes, only five species have been investigated: four oscine members, Taeniopygia guttata, Turdus merula, Serinus canaria, and Sicalis flaveola, and one suboscines member, Willisornis vidua [24,27,28]. No evidence of interchromosomal rearrangements involving the microchromosomes was observed in these oscine species [24,28]. On the other hand, a fusion involving G. gallus chromosome pairs 5 and 17 was observed in W. vidua [28]. Apart from this, the organization of the microchromosomes in Tyrannidae and Rhynchocyclidae flycatchers remains virtually unknown, as in most birds. In the latter, even the macrochromosomes organization has not been explored. Therefore, further studies must search for chromosome signatures to understand the phylogenetic relationship and chromosome organization in this group.
This study aimed to compare the chromosome organization of members belonging to Rhynchocyclidae and Tyrannidae to verify if cytotaxonomic characters corroborate the family-level status of Rhynchocyclidae. With this in mind, we selected Tolmomyias sulphurescens as a representative member of Rhynchocyclidae and P. sulphuratus from Tyrannidae. T. sulphurescens was selected randomly from the Rhynchocyclidae members and the P. sulphuratus was selected because it has a typical karyotype for Tyrannidae members, as indicated on previously study [15]. Our results indicated considerable chromosomal differences between both species, and the comparison with previous studies in Tyrant Flycatchers and allies reinforces that T. sulphurescens does not resemble the family Tyrannidae.
2. Materials and Methods
2.1. Specimens and Chromosome Preparation
Two male individuals of T. sulphurescens (from Porto Vera Cruz city, Rio Grande do Sul State, Brazil) and one male of P. sulphuratus (from São Gabriel city, Rio Grande do Sul State, Brazil) were used in this study. The animals were captured in their natural environment using mist nests (permissions 026/2012 and 018/2014—CEUA/Universidade Federal do Pampa, and SISBIO 33860-3—ICMBio). From each individual, skin biopsies were used to establish fibroblast cell culture, according to Furo et al. [11]. The chromosome preparations were obtained by standard arrest with colcemid (1 h), hypotonic treatment with 0.075 M KCl (15 min), and cell fixation in methanol–acetic acid (3:1).
2.2. Giemsa Staining
Chromosome morphology and diploid numbers (2n) were determined based on the analysis of at least 30 stained metaphases (5% Giemsa in phosphate buffer pH 6.8 for 5 min) from each individual. Karyotypes were arranged according to chromosome size and morphology following Guerra [29].
2.3. Fluorescence In Situ Hybridization (FISH)
Two G. gallus or T. guttata Bacterial artificial chromosome (BAC) probes corresponding to each pair GGA1-28 (except GGA16) were selected and positioned as close as possible to the end of each chromosome arms and applied to metaphases of T. sulphurescens (Table S1). In P. sulphuratus, only BAC probes for microchromosomes GGA11-28 (except GGA16) were used because the macrochromosomes have been previously published by Rodrigues et al. [15] (Table S1). The GGA16 was not tested in both species because there are no BAC probes available for this chromosome. Most of the BAC probes were chosen from G. gallus, however, for some chromosomes, the T. guttata probes give stronger signals than G. gallus ones in Passeriformes species. In this case, we chose T. guttata probes. The preparation of probes and hybridization were performed following O’Connor et al. [24]. At least 15 metaphase spreads per individual and for each probe were analyzed to confirm the FISH results. The chromosomes were counterstained with DAPI (blue), and the BAC probes were labeled with (Texas Red) (red) or FITC (green).
Although we used BAC probes from G. gallus and T. guttata, all karyotype comparisons were performed with the chicken karyotype, since it has a similar karyotype to the ancestral avian lineage (Palaeognathae) [30] and is the reference in cytogenetics and genetics studies.
3. Results
3.1. Karyotype Description
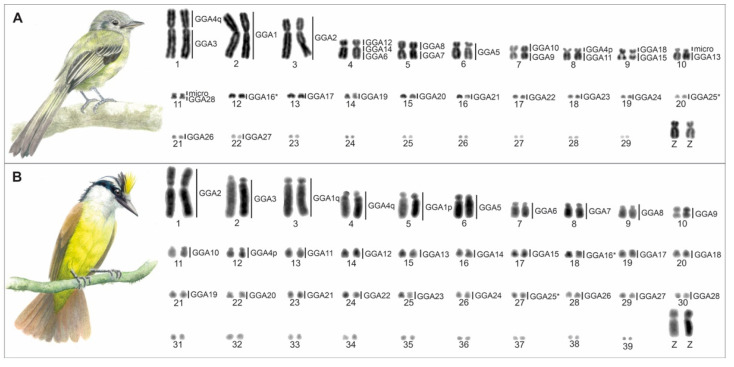
The flycatchers analyzed here showed distinct karyotypes. T. sulphurescens had a lower diploid number (2n = 60), consisting of 11 macrochromosomes, including the sex chromosomes, and 19 microchromosomes (Figure 1A). On the other hand, P. sulphuratus had a typical avian diploid number (2n = 80), consisting of 12 macrochromosomes, including the sex chromosomes, and 28 microchromosomes (Figure 1B). The Z chromosome is a submetacentric in both species.
Figure 1.
Conventionally stained complete karyotypes of Tolmomyias sulphurescens with 2n = 60 (A) and Pitangus sulphuratus with 2n = 80 (B) showing homologies to Gallus gallus (right). The homologies of P. sulphuratus macrochromosomes were based on Rodrigues et al. [15].
3.2. Fluorescence In Situ Hybridization (FISH) Experiments
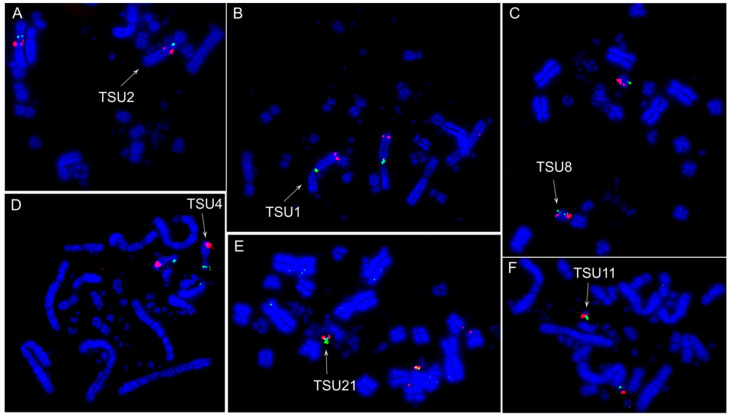
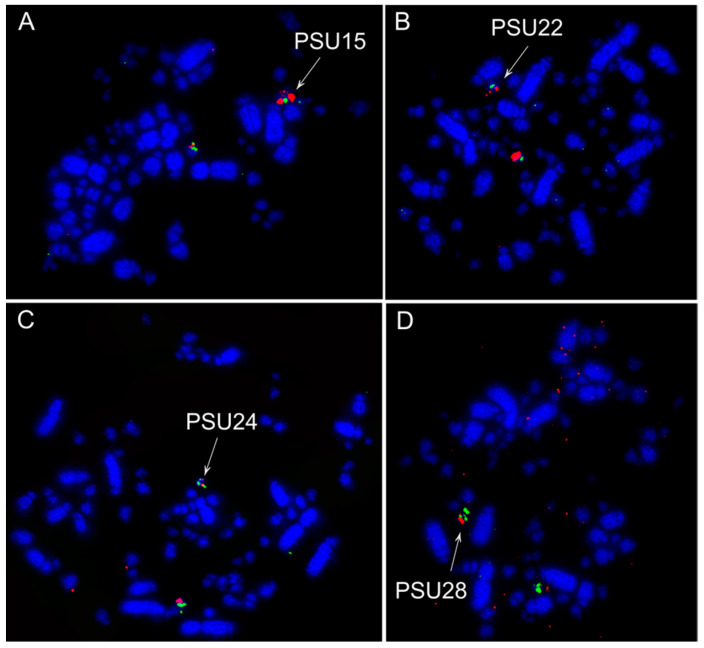
The hybridizations of BAC probes from G. gallus chromosome 1–28 (except 16) revealed extensive chromosome rearrangements in T. sulphurescens. Out of 27 chromosomes tested, only 12 chromosomes (GGA1, 2, 5, 17, and 19–27) were not involved in interchromosomal rearrangements (Figure 1A and Figure 2). The following associations were observed in T. sulphurescens: GGA3/4q (TSU 1), GGA4p/11 (TSU 8), GGA6/14/12 (TSU 4), GGA7/8 (TSU 5), GGA9/10 (TSU 7), GGA15/18 (TSU 9), GGA13/micro (TSU 10); GGA28/micro (TSU 11). It was clear that G. gallus chromosomes 13 and 28 are fused with other elements, because the signals were observed in larger chromosomes, if we compare with the sizes of G. gallus chromosomes 13 and 28. Probably, one of the microchromosome pairs not used in our analysis (GGA16, 29–38) fused with the GGA13 and 28 to originate the TSU10 and 11, respectively. On the other hand, there was no evidence of rearrangements involving microchromosomes in P. sulphuratus (Figure 1B and Figure 3).
Figure 2.
Representative cross-species hybridization results using G. gallus (CH261) and T. guttata (TGMCBA) BAC probes on Tolmomyias sulphurescens metaphases. (A) G. gallus macrochromosome 1 CH261-36B5 (green) and CH261-118M1 (red); (B) G. gallus macrochromosome 3 TGMCBA-295P5 (green) and CH261-169K18 (red); (C) G. gallus macrochromosome 4 CH261-83E1 (green) and G. gallus microchromosome 11 CH261-121N21 (red); (D) G. gallus microchromosome 12 CH261-60P3 (green), and G. gallus macrochromosome 6 CH261-49F3 (red); (E) G. gallus microchromosome 26 CH261-186M13 (green) and CH261-170L23 (red); (F) G. gallus microchromosome 28 CH261-64A15 (green) and CH261-72A10 (red).
Figure 3.
Representative cross-species hybridization results using G. gallus (CH261) and T. guttata (TGMCBA) BAC probes on Pitangus sulphuratus metaphases. (A) G. gallus microchromosome 13 CH261-115I12 (green) and TGMCBA-321B13 (red); (B) G. gallus microchromosome 20 TGMCBA-250E3 (green) and TGMCBA-341F20 (red); (C) G. gallus microchromosome 22 CH261-40J9 (green) and CH261-18G17 (red); (D) G. gallus microchromosome 26 CH261-186M13 (green) and CH261-170L23 (red).
4. Discussion
Passerines usually show a 2n close to 80 chromosomes, however, Tyrant Flycatchers and allies have a remarkable variation, from 2n = 60 in P. mystaceus (Platyrinchidae) to 2n = 84 in C. fuscatus (Tyrannidae) [13]. To the best of our knowledge, the karyotype of T. sulphurescens (2n = 60) is described here for the first time, and the karyotype of P. sulphuratus (2n = 80) agrees with its recent description [15]. Hence, our results reinforce the chromosomal variation observed previously among Tyrant Flycatchers and allies.
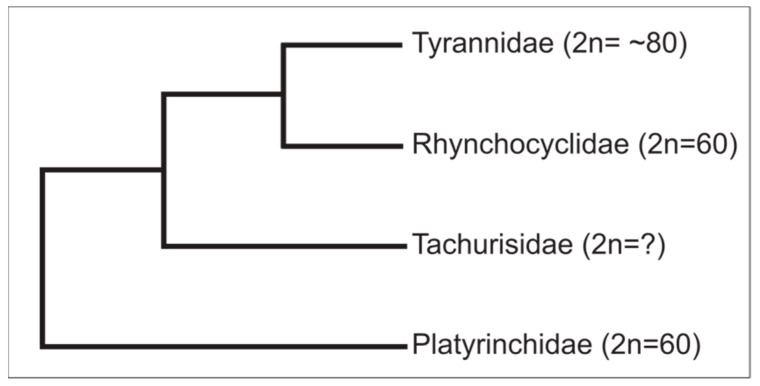
Low diploid numbers, such as seen in T. sulphurescens and P. mystaceus, both with 60 chromosomes, are rare among Passeriformes [7]. This may indicate a common ancestor in these species. According to the phylogenetic relationships found by Harvey et al. [6], Rhynchocyclidae and Tyrannidae are sister groups to Tachurisidae, and Platyrinchidae is the sister group to the former families (Figure 4). Tachurisidae represents a monotypic family, with Tachuris rubrigastra as the unique member. Despite the fact that there are no cytogenetic studies in this species, and considering the phylogenetic relationships proposed by Harvey et al. [6], a parsimony-based view would predict it has a low diploid number, similar to T. sulphurescens and P. mystaceus. Alternatively, the diploid number found in T. sulphurescens (Rhynchocyclidae) and P. mystaceus (Platyrinchidae) may be a result of independent karyotype reorganization. However, considering that low diploid number is rare in birds, especially Passeriformes, and both species have similar chromosomal morphology, this alternative seems not to be parsimoniously supported.
Figure 4.
Chromosomal data plotted on a phylogenetic tree adapted from Harvey et al. [6]. The diploid numbers (2n) for Rhynchocyclidae and Platyrinchidae are based in the data obtained to Tolmomyias sulphurescens (present study) and Platyrinchus mystaceus [13]. The 2n for Tyrannidae is considered as ~80 because most of the species karyotyped so far in this family have approximately 80 chromosomes (Table 1).
Unfortunately, few species of Tyrant Flycatchers and allies have been karyotyped (Table 1). Among the Rhynchocyclidae members, the first karyotype description is from T. sulphurescens (present study). However, Gunski et al. [13] described in their paper unpublished data that Corythopis delalandi, another Rhynchocyclidae member, has a similar karyotype to P. mystaceus, and consequently, similar to T. sulphurescens. These observations indicate that low diploid number may be a common feature among the Rhynchocyclidae members.
Although T. sulphurescens and P. sulphuratus have a similar number of macrochromosomes, 11 and 12 pairs, respectively, they differ substantially in the number of microchromosomes, 22 pairs in T. sulphurescens and 28 in P. sulphuratus, highlighting the role of chromosome rearrangements involving microchromosomes in T. sulphurescens. In fact, our molecular cytogenetic results revealed that several microchromosomes and macrochromosomes were involved in fusion events in T. sulphurescens. At the same time, no evidence of this type of rearrangement was found in P. sulphuratus. Fusion involving microchromosomes are rare events in birds and have been found extensively in few avian orders [24,25,26]. Here, we also demonstrated extensively fusion involving microchromosomes in Passeriformes, e.g., T. sulphurescens. Recent studies demonstrated that four songbirds, Taeniopygia guttata, Turdus merula, Serinus canaria, and Sicalis flaveola [24,28], and here P. sulphuratus, have the ancestral pattern of microchromosome organization. Hence, interchromosomal rearrangements involving microchromosomes represent an unusual feature of T. sulphurescens and probably also in closely related species, e.g., Rhynchocyclidae, Tachurisidae, and Platyrinchidae (Figure 4).
Considering the high chromosomal differences between T. sulphurescens and P. sulphuratus, our data indicate that, at least from the cytogenetic point of view, these species belong to different lineages. Hence, our findings reinforce the diagnosability and recognition of the Rhynchocyclidae family [2,4,5,6,31,32,33,34].
A recent study found evidence that the microchromosomes GGA10, GGA13, and GGA14 are more prone to interchromosomal rearrangements than others. Moreover, only GGA10 was supported with statistical significance after adjusting the number of tests performed [25]. Here, we reinforce these findings since, in T. sulphurescens, these microchromosomes were also involved in fusion events (Figure 1A). In addition, we reinforce that the microchromosomes GGA22, GGA24, GGA26, and GGA27 seem not prone to interchromosomal rearrangements.
The most unexpected finding in T. sulphurescens was that the G. gallus chromosome 1 (GGA1) probe hybridizes in only one pair: up to now, all Passeriformes analyzed showed centric fission in this chromosome [18]. Hence, two hypotheses may be highlighted: (1) the GGA1 as an entire chromosome represents a plesiomorphic (ancestral) character retained in T. sulphurescens, or (2) T. sulphurescens had the fusion of GGA1p and GGA1q, restoring the ancestral character. Considering that the centric fission in this chromosome was found in all Passeriformes and Psittaciformes (Passeriformes sister group) species previously studied [7,18], it is likely that the second hypothesis is more plausible.
Together with previous studies, our results indicate that the karyotype of Tyrannidae evolved with few interchromosomal rearrangements. On the other hand, these rearrangements are likely to be the most frequent events in Rhynchocyclidae when compared to PAK [8]. In general, chromosomal rearrangements occur in breakpoint regions, usually associated with genomic features, including transposable elements, and conserved noncoding elements [35,36]. Hence, the presence of these genomic features in Rhynchocyclidae members and not in Tyrannidae members might facilitate the extensive chromosome reorganization in the former.
The Passeriformes order represents more than half of all living birds and displays great diversity in richness within subgroups, morphological and ecological diversification [37,38]. Interestingly, the rise of this great diversity was not accompanied by a high rate of interchromosomal rearrangements (e.g., fissions and fusions) [7,18], except in T. sulphurescens (present study). This may indicate that, in general, the maintenance of the ancestral pattern of karyotype in Passeriformes was crucial to the successful diversification seen in this clade. However, it remains unclear why extensive chromosome rearrangements evolved in some avian lineages, such as in T. sulphurescens and probably in other closely related species, while other Passeriformes retained the ancestral pattern of karyotype organization (~80 chromosomes). Moreover, it is known that Passeriformes underwent a high number of intrachromosomal rearrangements, such as paracentric and pericentric inversions [39,40].
In conclusion, our results indicate that the chromosome evolution of T. sulphurescens involved extensive chromosome fusions of macrochromosomes and microchromosomes, while P. sulphuratus retained the ancestral pattern of organization of macrochromosomes and microchromosomes, except for the fission of G. gallus chromosome 1. The comparison of our results with previous studies in Tyrant Flycatchers and allies indicates that the karyotype of P. sulphuratus is similar to other Tyrannidae members, however, T. sulphurescens does not resemble the Tyrannidae family, reinforcing the status of the family to Rhynchocyclidae. The high chromosomal differences observed in Tyrant Flycatchers and allies make these birds an ideal model to investigate the role of chromosomal rearrangements in speciation and to detect what contributed to chromosomal rearrangements Rhynchocyclidae, but not in Tyrannidae members.
Acknowledgments
Authors would like to thank all colleagues from the “Grupo de Pesquisa Diversidade Genética Animal” from Universidade Federal do Pampa and the “Laboratório de Cultura de Tecidos e Citogenética” from Instituto Evandro Chagas (Ananindeua, PA, Brazil) for technical and institutional support. We are grateful to Alex Pinheiro de Araújo for the illustration of the Tolmomyias sulphurescens and Pitangus sulphuratus used in Figure 1.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/cells10102650/s1, Table S1: List of BAC applied to Tolmomyias sulphurescens (TSU) and Pitangus sulphuratus (PSU).
Author Contributions
Conceptualization, R.K., T.R.O.d.F. and D.K.G.; methodology, R.K., M.S.d.S., A.D.V.G., R.J.G., and R.E.O.; validation, R.K., I.F. and M.S.d.S.; formal analysis, R.K.; investigation, R.K., I.F. and M.S.d.S.; resources, E.H.C.d.O., A.D.V.G., R.J.G., T.R.O.d.F. and D.K.G.; data curation, R.K. and M.S.d.S.; writing—original draft preparation, R.K. and I.F.; writing—review and editing, R.K., E.H.C.d.O., R.E.O.; T.R.O.d.F. and D.K.G.; visualization, R.K., M.S.d.S.; supervision, T.R.O.d.F. and D.K.G.; project administration, T.R.O.d.F. and D.K.G.; funding acquisition, R.K., E.H.C.d.O., A.D.V.G., R.J.G., T.R.O.d.F. and D.K.G. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq, Proc. PDJ 151056/2020-0 to Rafael Kretschmer), Fundação de Amparo a Pesquisa do Estado do Rio Grande do Sul (FAPERGS/PRONEX 16/2551- 00000485-4 to Thales Renato Ochotorena de Freitas) and the Biotechnology and Biological Sciences Research Council UK (BB/K008226/1 to Darren K. Griffin).
Institutional Review Board Statement
The biological material was obtained from individuals captured in their natural environment using mist nets following permissions from Sistema de Autorização e Informação em Biodiversidade (SISBIO 33860-3—ICMBio). The experiments followed protocols approved by the ethics committee from Universidade Federal do Pampa (026/2012 and 018/2014).
Data Availability Statement
All the data supporting our findings are contained within the manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Fitzpatrick J.W. Family Tyrannidae (Tyrant-Flycatchers) In: del Hoyo J., Elliott A., Christie D.A., editors. Handbook of the Birds of the World, Cotingas to Pipits and Wagtails. Volume 9. Lynx Edicions; Barcelona, Spain: 2004. pp. 170–462. [Google Scholar]
- 2.Tello J.G., Bates J.M. Molecular phylogenetics of the tody-tyrant and flatbill assemblage of tyrant flycatchers (Tyrannidae) Auk. 2007;124:134–154. doi: 10.1093/auk/124.1.134. [DOI] [Google Scholar]
- 3.Rheindt F.E., Norman J.A., Christidis L. Phylogenetic relationships of tyrant-flycatchers (Aves: Tyrannidae), with an emphasis on the elaeniine assemblage. Mol. Phylogenetics Evol. 2008;46:88–101. doi: 10.1016/j.ympev.2007.09.011. [DOI] [PubMed] [Google Scholar]
- 4.Tello J.G., Moyle R.G., Marchese D.J., Cracraft J. Phylogeny and phylogenetic classification of the tyrant flycatchers, cotingas, manakins, and their allies (Aves: Tyrannides) Cladistics. 2009;25:429–467. doi: 10.1111/j.1096-0031.2009.00254.x. [DOI] [PubMed] [Google Scholar]
- 5.Ohlson J.I., Irestedt M., Ericson P.G.P., Fjeldsa J. Phylogeny and classification of the New World suboscines (Aves, Passeriformes) Zootaxa. 2013;3613:1–35. doi: 10.11646/zootaxa.3613.1.1. [DOI] [PubMed] [Google Scholar]
- 6.Harvey M.G., Bravo G.A., Claramunt S., Cuervo A.M., Derryberry G.E., Battilana J., Seeholzer G.F., McKay J.S., O’Meara B.C., Faircloth B.C., et al. The evolution of a tropical biodiversity hotspot. Science. 2020;370:1343–1348. doi: 10.1126/science.aaz6970. [DOI] [PubMed] [Google Scholar]
- 7.Degrandi T.M., Barcelos S.A., Costa A.L., Garnero A.d.V., Hass I., Gunski R.J. Introducing the Bird Chromosome Database: An overview of cytogenetic studies on birds. Cytogenet. Genome Res. 2020;160:199–205. doi: 10.1159/000507768. [DOI] [PubMed] [Google Scholar]
- 8.Griffin D.K., Robertson L.B.W., Tempest H.G., Skinner B.M. The evolution of the avian genome as revealed by comparative molecular cytogenetic. Cytogenet. Genome Res. 2007;117:64–77. doi: 10.1159/000103166. [DOI] [PubMed] [Google Scholar]
- 9.White M.J.D. Animal Cytology and Evolution. 3rd ed. Cambridge University Press; Cambridge, UK: 1973. [Google Scholar]
- 10.Kretschmer R., Furo I.O., Cioffi M.B., Gunski R.J., Garnero A.d.V., O’Brien P.C.M., Ferguson-Smith M.A., de Freitas T.R.O., de Oliveira E.H.C. Extensive chromosomal fissions and repetitive DNA accumulation shaped the atypical karyotypes of two Ramphastidae (Aves: Piciformes) species. Biol. J. Linn. Soc. 2020;130:839–849. doi: 10.1093/biolinnean/blaa086. [DOI] [Google Scholar]
- 11.Furo I.O., Kretschmer R., dos Santos M.S., de Lima C.A.C., Gunski R.J., O’Brien P.C.M., Ferguson-Smith M.A., Cioffi M.B., de Oliveira E.H.C. Chromosomal mapping of repetitive DNAs in Myiopsitta monachus and Amazona aestiva (Psittaciformes, Psittacidae) with emphasis on the sex chromosomes. Cytogenet. Genome Res. 2017;151:151–160. doi: 10.1159/000464458. [DOI] [PubMed] [Google Scholar]
- 12.Furo I.O., Kretschmer R., O’Brien P.C.M., Pereira J., Garnero A.d.V., Gunski R.J., O’Connor R.E., Griffin D.K., Gomes A.J.B., Ferguson-Smith M.A., et al. Chromosomal evolution in the phylogenetic context in Neotropical Psittacidae with emphasis on a species with high karyotypic reorganization (Myiopsitta monachus) Front. Genet. 2020;11:721. doi: 10.3389/fgene.2020.00721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gunski R.J., Cabanne G.S., Ledesma M.A., Garnero A.d.V. Analisis cariotipico de siete especies de Tiranidos (Tyrannidae) El Hornero. 2000;15:103–109. [Google Scholar]
- 14.Kretschmer R., de Oliveira E.H.C., dos Santos M.S., Furo I.O., O’Brien P.C.M., Ferguson-Smith M.A., Garnero A.d.V., Gunski R.J. Chromosome mapping of the large elaenia (Elaenia spectabilis): Evidence for a cytogenetic signature for passeriform birds? Biol. J. Linn. Soc. 2015;115:391–398. doi: 10.1111/bij.12504. [DOI] [Google Scholar]
- 15.Rodrigues B.S., Kretschmer R., Gunski R.J., Garnero A.d.V., O’Brien P.C.M., Ferguson-Smith M., de Oliveira E.H.C. Chromosome Painting in Tyrant Flycatchers Confirms a Set of Inversions Shared by Oscines and Suboscines (Aves, Passeriformes) Cytogenet. Genome Res. 2017;153:205–212. doi: 10.1159/000486975. [DOI] [PubMed] [Google Scholar]
- 16.de Lucca E.J., Chamma L. Estudo do complemento cromossômico de 11 espécies de aves das ordens Columbiformes, Passeriformes e Tinamiformes. Braz. J. Med. Biol. Res. 1977;10:97–105. [PubMed] [Google Scholar]
- 17.Shields G.F., Barlow J.C., James R.D. Karyotypes of five species of Empidonax flycatchers. Wilson Bull. 1987;99:169–174. [Google Scholar]
- 18.Kretschmer R., Ferguson-Smith M.A., de Oliveira E.H.C. Karyotype evolution in birds: From conventional staining to chromosome painting. Genes. 2018;9:181. doi: 10.3390/genes9040181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.de Oliveira E.H.C., Tagliarini M.M., Rissino J.D., Pieczarka J.C., Nagamachi C.Y., O’Brien P.C.M., Ferguson-Smith M.A. Reciprocal chromosome painting between white hawk (Leucopternis albicollis) and chicken reveals extensive fusions and fissions during karyotype evolution of Accipitridae (Aves, Falconiformes) Chromosome Res. 2010;18:349–355. doi: 10.1007/s10577-010-9117-z. [DOI] [PubMed] [Google Scholar]
- 20.Smith J., Paton I.R., Bruley C.K., Windsor D., Burt D.W., Ponce de Leon F.A., Burke D. Integration of the genetic and physical maps of the chicken macrochromosomes. Anim. Genet. 2000;31:20–27. doi: 10.1046/j.1365-2052.2000.00549.x. [DOI] [PubMed] [Google Scholar]
- 21.Burt D.W. Origin and evolution of avian microchromosomes. Cytogenet. Genome Res. 2002;96:97–112. doi: 10.1159/000063018. [DOI] [PubMed] [Google Scholar]
- 22.Warren W.C., Hillier L.W., Tomlinson C., Minx P., Kremitzki M., Graves T., Markovic C., Bouk N., Pruitt K.D., Thibaud-Nissen F., et al. A new chicken genome assembly provides insight into avian genome structure. G3 Genes Genom. Genet. 2017;7:109–117. doi: 10.1534/g3.116.035923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Joseph S., O’Connor R.E., Al Mutery A.F., Watson M., Larkin D.M., Griffin D.K. Chromosome level genome assembly and comparative genomics between three falcon species reveals an unusual pattern of genome organisation. Diversity. 2018;10:113. doi: 10.3390/d10040113. [DOI] [Google Scholar]
- 24.O’Connor R.E., Kiazim L., Skinner B., Fonseka G., Joseph S., Jennings R., Larkin D.M., Griffin D.K. Patterns of microchromosome organization remain highly conserved throughout avian evolution. Chromosoma. 2019;128:21–29. doi: 10.1007/s00412-018-0685-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kretschmer R., de Souza M.S., Furo I.O., Gunski R.J., Garnero A.d.V., de Freitas T.R.O., de Oliveira E.H.C., O’Connor R.E., Griffin D.K. Interspecies Chromosome Mapping in Caprimulgiformes, Piciformes, Suliformes, and Trogoniformes (Aves): Cytogenomic Insight into Microchromosome Organization and Karyotype Evolution in Birds. Cells. 2021;10:826. doi: 10.3390/cells10040826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kretschmer R., Gunski R.J., Garnero A.d.V., de Freitas T.R.O., Toma G.A., Cioffi M.B., de Oliveira E.H.C., O’Connor R.E., Griffin D.K. Chromosomal analysis in Crotophaga ani (Aves, Cuculiformes) reveals extensive genomic reorganization and an unusual Z-autosome Robertsonian translocation. Cells. 2021;10:4. doi: 10.3390/cells10010004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ribas T.F.A., Pieczarka J.C., Griffin D.K., Kiazim L.G., Nagamachi C.Y., O.´Brien P.C.M., Ferguson-Smith M.A., Yang F., Aleixo A., O’Connor R.E. Analysis of multiple chromosomal rearrangements in the genome of Willisornis vidua using BAC-FISH and chromosome painting on a supposed conserved karyotype. BMC Evol. Biol. 2021;21:34. doi: 10.1186/s12862-021-01768-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kretschmer R., Rodrigues B.S., Barcellos A.S., Costa A.L., Cioffi M.B., Garnero A.d.V., Gunski R.J., de Oliveira E.H.C., Griffin D.K. Karyotype Evolution and Genomic Organization of Repetitive DNAs in the Saffron Finch, Sicalis flaveola (Passeriformes, Aves) Animals. 2021;11:1456. doi: 10.3390/ani11051456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guerra M.S. Reviewing the chromosome nomenclature of Levan et al. Rev. Bras. Genética. 1986;4:741–743. [Google Scholar]
- 30.Romanov M.N., Farré M., Lithgow P.E., Fowler K.E., Skinner B.M., O’Connor R., Fonseka G., Backström N., Matsuda Y., Nishida C., et al. Reconstruction of gross avian genome structure, organization and evolution suggests that the chicken lineage most closely resembles the dinosaur avian ancestor. BMC Genom. 2014;15:1060. doi: 10.1186/1471-2164-15-1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lanyon W.E. A phylogeny of the flatbill and tody-tyrant assemblage of tyrant flycatchers. Am. Mus. Novit. 1988;2923:1–41. [Google Scholar]
- 32.Ericson P.G., Zuccon D., Ohlson J.I., Johansson U.S., Alvarenga H., Prum R.O. Higher-level phylogeny and morphological evolution of tyrant flycatchers, cotingas, manakins, and their allies (Aves: Tyrannida) Mol. Phylogenetics Evol. 2006;40:471–483. doi: 10.1016/j.ympev.2006.03.031. [DOI] [PubMed] [Google Scholar]
- 33.Ohlson J., Fjeldsa J., Ericson P.G. Tyrant flycatchers coming out in the open: Phylogeny and ecological radiation of Tyrannidae (Aves, Passeriformes) Zool. Scr. 2008;37:315–335. doi: 10.1111/j.1463-6409.2008.00325.x. [DOI] [Google Scholar]
- 34.Lopes L.E., Chaves A.V., de Aquino M.M., Silveira L.F., dos Santos F.R. The striking polyphyly of Suiriri: Convergent evolution and social mimicry in two cryptic Neotropical birds. J. Zool. Syst. Evol. Res. 2018;56:270–279. doi: 10.1111/jzs.12200. [DOI] [Google Scholar]
- 35.Zhang J., Yu C., Krishnaswamy L., Peterson T. Transposable elements as catalysts for chromosome rearrangements. Methods Mol. Biol. 2011;701:315–326. doi: 10.1007/978-1-61737-957-4_18. [DOI] [PubMed] [Google Scholar]
- 36.Farré M., Narayan J., Slavov G.T., Damas J., Auvil L., Li C., Jarvis E.D., Burt D.W., Griffin D.K., Larkin D.M. Novel in-sights into chromosome evolution in birds, archosaurs, and reptiles. Genome Biol. Evol. 2016;8:2442–2451. doi: 10.1093/gbe/evw166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Johansson U.S., Fjeldså J., Bowie R.C.K. Phylogenetic relationships within Passerida (Aves: Passeriformes): A review and a new molecular phylogeny based on three nuclear intron markers. Mol. Phylogenetics Evol. 2008;48:858–876. doi: 10.1016/j.ympev.2008.05.029. [DOI] [PubMed] [Google Scholar]
- 38.Gill F., Donsker D., Rasmussen P. IOC World Bird List. IOC; London, UK: 2020. Version 10.1. [Google Scholar]
- 39.Volker M., Backstrom N., Skinner B.M., Langley E.J., Bunzey S.K., Ellegren H., Griffin D.K. Copy number variation, chromosome rearrangement, and their association with recombination during avian evolution. Genome Res. 2010;20:503–511. doi: 10.1101/gr.103663.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Warren W.C., Clayton D.F., Ellegren H., Arnold A.P., Hillier L.W., Künstner A., Searle S., White S., Vilella A.J., Fairley S., et al. The genome of a songbird. Nature. 2010;464:757–762. doi: 10.1038/nature08819. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the data supporting our findings are contained within the manuscript.