Abstract
p130cas (Cas) is a docking protein that contains an SH3 domain and multiple tyrosine residues. p130cas is located at focal adhesions, is tyrosine phosphorylated in response to integrin stimulation, and is thought to transmit signals, via c-Crk and other proteins, for the remodeling of actin stress fibers and cell movement. In a search for the ligands of the SH3 domain of p130cas by far-Western screening, we cloned a novel protein named CIZ (for Cas-interacting zinc finger protein). CIZ consists of the following: a putative leucine zipper; a serine/threonine-rich region; a proline-rich sequence; five, six, or eight Krüppel-type C2H2 zinc fingers; and the glutamine-alanine repeat. CIZ binds Cas in cells and is located in the nucleus and at focal adhesions. We showed that CIZ is a nucleocytoplasmic shuttling protein, by using the transient interspecies heterokaryon formation assay. In order to search for the targets of CIZ in nucleus, we determined the DNA binding consensus of CIZ as (G/C)AAAAA(A) by cyclic amplification and selection of targets analysis. The consensus-like sequences are found in several promoters of matrix metalloproteinases (MMPs), which are the enzymes used to degrade the extracellular matrix proteins. CIZ binds to a consensus-like sequence in the MMP-1 (collagenase) promoter. Overexpression of CIZ upregulates the transcriptions from MMP-1, MMP-3 (stromelysin), and MMP-7 (matrilysin) promoters, and this transactivation was enhanced in the presence of Cas. Furthermore, the stable overexpression of CIZ promoted the production of MMP-7 in culture medium. In summary, CIZ, a novel zinc finger protein, binds Cas, is a nucleocytoplasmic shuttling protein, and regulates the expression of MMPs.
p130cas (Cas) is a docking protein, initially identified as a major tyrosine-phosphorylated protein in cells transformed by v-Crk or v-Src (42, 43). Cas consists of the N-terminal SH3 domain, the substrate domain, the Src binding domain, and the other regions. The substrate domain includes the repeated SH2 binding motifs for Crk, and the Src binding domain has the tyrosine residue and the proline-rich sequence that are the binding sites for Src SH2 and SH3 domains, respectively (32, 42). Subsequently, Cas was shown to be tyrosine phosphorylated in response to integrin stimulation (35, 54) and was localized mostly in the cytoplasm and partly at focal adhesions (12, 31, 37). The tyrosine-phosphorylated Cas binds c-Crk, forming a Cas-Crk complex (53), and the Cas-Crk complex leads to the activation of Rac-JNK pathway (7, 18). Nck also binds tyrosine-phosphorylated Cas, and part of ERK2 activation by integrin stimulation is shown to be mediated by the phosphorylation of Cas (44). Overexpression of Cas is reported to promote cell motility, depending on its association with FAK and c-Crk (5, 19), and the targeting of Cas revealed that Cas is essential for assembling actin filaments and for forming long actin fibers (13). Furthermore, Cas-mediated actin reorganization is critical to anchorage independence in cell transformation by activated Src (13). These results suggest that Cas constitutes the signal transduction pathway from cell attachment to cytoskeletal changes and to cell movements.
The SH3 domain of Cas is known to bind to focal adhesion-associated tyrosine kinase, FAK (12, 38), and has been shown to be necessary for the localization of Cas to focal adhesions (31). However, Cas was localized at focal adhesions in FAK−/− cells (31), and the association between Cas and FAK was reported to be dependent on the presence of Src (44). Therefore, we assumed that FAK is not the sole ligand of the Cas SH3 domain and that the other ligands would function upstream or downstream of Cas. After we began to search for the binding partners of Cas SH3 by far-Western blotting, PTP-1B (26), PTP-PEST (9), PYK2/CAKβ/RAFTK/CADTK (3), C3G (16), and CMS/CD2AP (17) were reported to be the ligands of the Cas SH3 domain.
In this paper, we identified another novel ligand of the Cas SH3 domain and named it CIZ (for Cas-interacting zinc finger protein). CIZ is shown to bind Cas in cells, is located at focal adhesions and in the nucleus, and shuttles in and out of the nucleus. We determined that the DNA binding consensus of CIZ and the CIZ-binding consensus-like sequences existed in promoters of several matrix metalloproteinases (MMPs), which are metal (zinc)-dependent proteases that can digest various extracellular matrix proteins or cell surface proteins. We showed that overexpression of CIZ upregulates the expression of MMP-1, -3, and -7.
MATERIALS AND METHODS
Far-Western screening.
The cDNA for Cas SH3 was subcloned into pGEX2TK (Pharmacia) to make pGEX2TK-CasSH3. The cDNA library from rat 3Y1 cells (42) was inserted into λgt11 and amplified. The Hybond-C extra membranes (Amersham) soaked in 10 mM isopropyl-β-d-thiogalactopyranoside (IPTG) were overlaid on phage plaques for 4 h and were blocked in Tris-buffered saline–Tween 20 (TBST) with 5% skim milk. The membranes were incubated with 0.75 μg of isotope-labelled glutathione S-transferase (GST)-Cas SH3 for 2 h at 25°C, washed, and autoradiographed. Eight positive clones were obtained. Four out of eight positive clones were CIZ. The cDNA of CIZ was labeled and was used as a probe to further screen for alternative forms of CIZ.
Production of antibody against bacterially expressed CIZ.
A clone corresponding to the region from the 140th to the 260th amino acid residue of CIZ8 was inserted into pGEX-1 (Pharmacia) to make pGEX-1-CIZ. The rabbit antiserum raised against GST-CIZ is named anti-CIZ.
Plasmid construction of CIZ-expressing vectors.
The cDNA for FLAG tag or hemagglutinin (HA) tag was added to the 3′ terminus of the coding sequence of CIZ cDNA. The cDNAs for CIZ-FLAG, CIZ-HA, or wild-type CIZ of various alternative forms were inserted into the pSSRαbsr vector (51). To make the mPR mutant of CIZ, the APPKPPR sequence was mutated to AAAKPPR by in vitro mutagenesis. To make the dZF mutant of CIZ, SalI sites were introduced to just upstream (around 879 bp) and downstream (around 1,587 bp) of the cDNA for the zinc finger domain, by in vitro mutagenesis, and the SalI fragment was cut out.
Immunoprecipitation and immunoblotting.
COS-7 cells were transfected with expression plasmids by the DEAE-dextran method as described previously (32). After lysis in radioimmunoprecipitation assay (RIPA) buffer (32), 2 mg of lysates was immunoprecipitated with 5 μl of anti-FLAG M2 antibody (Kodak), anti-Cas2 antibody (42), or preimmune serum. The sources of GST fusion proteins and the GST pulldown assay were described elsewhere (32). GST fusion proteins were incubated with 400 μg of CIZ-overexpressing COS-7 lysates. LC-540 cells were lysed in RIPA buffer, and 20 mg was immunoprecipitated with 15 μl of anti-Cas2, anti-CIZ, or preimmune serum. Immunoblottings were performed with anti-Cas2 (1:2,500), anti-CIZ (1:2,500), or 10 μg of anti-proMMP-7 (141-7B2) per ml (Fuji Chemical Industries) and detected with ProtoBlot apparatus (Promega). In some cases, immunoblottings were performed with anti-FLAG (1:400) or anti-Cas2 (1:50,000) and detected with the ECL enhanced chemiluminescence system (Amersham).
Immunofluorescence.
Cells were grown on uncoated coverslips (Matsunami) for 16 to 24 h. In the leptomycin B (LMB) treatment, 3Y1 cells were subsequently incubated in the presence of 10 ng of LMB per ml for 24 h. The cells were stained as described previously (31). Primary antibodies were used at the following dilutions after they were absorbed with purified GST or GST-CIZ: 1/400 for anti-CIZ and 1/200 for the antivinculin mouse monoclonal antibody anti-hVIN-1. LMB is a generous gift from M. Yoshida (University of Tokyo).
Cell fusion.
NIH 3T3 cells were transfected with 1 μg of pSSRαbsr-CIZ-HA by using SuperFect (Qiagen). After 2 days, the cells were resuspended and were mixed with resuspended HeLa cells. The pellet after centrifugation was resuspended in 50% polyethylene glycol 6000 for 2 min at 25°C. Subsequently, the cells were washed and replated in Dulbecco's modified Eagle's medium with 10% fetal calf serum and 20 μg of cycloheximide per ml on coverslips for 48 h. Staining was performed first with 1:1,000 anti-HA.11 (BAbCO) and then with 1:100 fluorescein isothiocyanate-conjugated anti-rabbit antibody (Jackson) together with 5 μg of Hoechst 33258 per ml (Sigma). The samples were observed with a Nikon ECLIPSE E800.
CASTing analysis.
Nuclear extract was prepared from CIZ-FLAG-transfected COS-7 cells as described previously (24). The oligomers used were RANDOM (GCGTCGACAAGCTTTCTAGANNNNNNNNNNNNNNNNGAATTCGGATCCCTCGAGCG), FORWARD (GCGTCGACAAGCTTTCTAGA), and REVERSE (CGCTCGAGGGATCCGAATTC). RANDOM was annealed with 3 M excess of REVERSE, and the double-stranded DNA (dsDNA) was synthesized with the Klenow fragment at room temperature for 3 h. This dsDNA was mixed with 4 μl of nuclear extract and 16 μl of lysis buffer (24) and then was incubated for 20 min at 25°C. The protein-DNA complex was immunoprecipitated with anti-FLAG, and the DNA was extracted by adding 200 μl of extraction buffer (5 mM EDTA, 0.5% sodium dodecyl sulfate, 100 mM NaOAc, 50 mM Tris [pH 8.0]). With the ethanol-precipitated DNA, 15 cycles of PCR with the FORWARD and REVERSE primers, consisting of 1 min at 94°C, 1 min at 62°C, and 1 min at 72°C per cycle, were performed. In the following rounds of cyclic amplification and selection of targets (CASTing), 1 μl of PCR product was used as the starting material. After five rounds of CASTing, the final PCR product was cut with SalI and EcoRI, subcloned into pBluescript SK(−), and sequenced.
EMSA.
The electrophoretic mobility shift assay (EMSA) was done as described previously (59). The labeled oligonucleotide was prepared by filling in the −320 to −305 region of the human MMP-1 promoter with [α-32P]dCTP (Amersham). For competition experiments, unlabeled double-stranded oligonucleotide or the mutant double-stranded oligonucleotide (CCTGTGTCAGAGAGA) was added prior to the addition of the labeled oligonucleotides. For the antiserum supershift experiment, the antiserum was added, and this mixture was then incubated for 15 min on ice prior to the addition of the labeled oligonucleotides.
Luciferase assay and reporter plasmids.
Four tandem copies of the sequence corresponding to the −320 to −303 region of the human MMP-1 promoter or the same region with mutation (CCTGTGTCAGAGAGACC) were fused to tk-Luc (48) to generate p(CIZBC)4-tk-Luc or p(mCIZBC)4-tk-Luc, respectively. The −517/+63CAT5 plasmid, which includes the human MMP-1 promoter, is a generous gift from Peter Angel and Hans Rahmsdorf. pXP2-MMP-1 was constructed by inserting the −517 to +63 portion of the human MMP-1 promoter into pXP2 (27). For pXP2-mMMP-1, the −320 to −305 region was converted to (CCTTTGTCGACAAGA) by in vitro mutagenesis. The human MMP-7 promoter and the rat MMP-3 promoter are kind gifts from Lynn Matrisian. pXP2-MMP-7 was constructed by inserting the −301 to +35 region of the human MMP-7 promoter (30) into pXP2. pXP2-MMP-3 was constructed by inserting the −754 to −1 region of the rat MMP-3 (8) into pXP2. NIH 3T3 cells or Cas−/− primary fibroblasts (13) were transfected with SuperFect (Qiagen) with reporter plasmids, expression plasmids, and pCMV-βgal (TROPIX). After 48 h, cells were harvested for the luciferase assay. Each value was adjusted for each β-galactosidase activity. Experiments were performed three times, and the results were essentially reproducible.
Nucleotide sequence accession number.
The CIZ cDNA sequence has been deposited in the DDBJ, EMBL, and GenBank databases under accession no. AB019281.
RESULTS
Cloning and sequence analysis of CIZ.
Isotope-labeled GST-Cas SH3 fusion protein was used as a probe for far-Western blotting to screen a λgt11 cDNA expression library from rat 3Y1 cells. Approximately 106 clones were screened, and we obtained 8 positive clones. Of the eight positive clones, one clone was PTP-PEST, and another clone was C3G, which are known as Cas SH3-binding proteins (9, 16). Four other clones encoded the same zinc finger protein, and two of them encoded the full-length cDNA. We named this zinc finger protein CIZ.
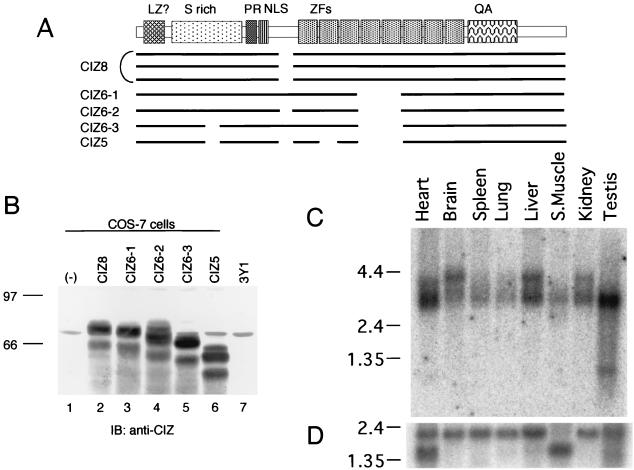
The same library was screened with the obtained full-length cDNA clone as a hybridization probe to search for the other alternatively spliced forms of cDNA. Five strongly positive clones were obtained, and they encoded the full-length cDNA of different alternatives. Three of the seven full-length clones carried the sequence shown in Fig. 1A. The protein encoded by these clones has eight zinc fingers, and we designated this alternative form as CIZ8 (Fig. 2A). Three other clones had six zinc fingers with or without the insertions indicated in Fig. 1B and 2A. These alternative forms were designated as CIZ6-1, CIZ6-2 and CIZ6-3, respectively (Fig. 2A). One clone (CIZ5) encoded the five-zinc-finger form (Fig. 2A). The longest form of CIZ (CIZ8) has 579 amino acid residues, a calculated molecular mass of 63 kDa, and a pI of 9.26. The five alternative forms were expressed in COS-7 cells and were detected with anti-CIZ antibody (Fig. 2B). Because CIZ6-1 (Fig. 2B, lane 3) had approximately the same mobility as the native 70-kDa band from 3Y1 lysate (lane 7) among the cloned species, we utilized CIZ6-1 in the following experiments.
FIG. 1.
Nucleotide and predicted amino acid sequences of rat CIZ cDNA. (A) Nucleotide sequences and predicted amino acid sequences (single-letter code) of rat CIZ cDNA are presented. Shown in lowercase letters are noncoding regions. Shown in italic type are the leucine or isoleucine residues, which appear every six or seven amino acid residues in the N-terminal portion. Shown in boldface type are the serine residues, which are rich in the following region. Overbarred residues are the proline-rich sequence, which was shown to be the Cas SH3 binding site in this paper. Underlined residues are the eight zinc fingers, which are numbered underneath. The boxed nucleotides are the possible CAG repeat portion of CIZ cDNA. <> indicates the residues that are missing in the alternative forms CIZ6-3 and CIZ5. ∗ indicates the insertion point of the residues shown in Fig. 1B, in the alternative forms CIZ6-1 and CIZ6-3. () indicates the residues that are missing in the alternative form CIZ5. {} indicates the residues that are missing in the alternative forms CIZ 6-1, CIZ6-2, CIZ6-3, and CIZ5. (B) Inserted nucleotide sequence and predicted amino acid sequences, inserted at the ∗ point of panel A, in the alternative forms CIZ6-3 and CIZ5.
FIG. 2.
Alternative splicing forms and tissue distribution of CIZ. (A) Schematic diagram indicating the relationship among the clones obtained, including five alternative forms. LZ, leucine zipper; PR, proline rich; NLS, nuclear localization signal; ZFs, zinc fingers; QA, glutamine-alanine repeat. (B) Immunoblotting (IB) with anti-CIZ of the CIZ alternatives expressed in COS-7 cells and of the native CIZ in 3Y1 cells. The lanes are the lysates of mock (lane 1)-, CIZ8 (lane 2)-, CIZ6-1 (lane 3)-, CIZ6-2 (lane 4)-, CIZ6-3 (lane 5)-, and CIZ5 (lane 6)-transfected COS-7 cells and 3Y1 cell lysate (lane 7). (C) An RNA blot (Clontech) containing poly(A)+ RNA from the indicated rat tissues (2 μg/lane) was hybridized with a labeled CIZ partial cDNA probe (RsaI fragment corresponding to 234 to 481 bp). The positions of 9.5-, 7.5-, 4.4-, 2.4-, and 1.35-kb markers are shown on the left. (D) As a control, a blot with the cDNA of human β-actin is shown.
The sequence around the initiation codon (Fig. 1A) conformed to Kozak's consensus (20), and stop codons appear upstream of the initiation codon in all frames. In the N-terminal portion, leucine or isoleucine appears every six to seven residues (shown in italic in Fig. 1A). Although we do not have evidence that this portion takes the α-helix form, it is possible that this part constitutes the leucine zipper. In the following region, there appear to be plenty of serine and threonine residues. The APPKPPR sequence from the 186th amino acid residue conforms to the Cas SH3 binding consensus XXPXKPX (16) and resembles the Cas SH3 binding site of FAK (APPKPSR) (38), that of PTP-PEST (PPPKPPR) (9), and that of C3G (APPKPPLP) (16). Just downstream of this exists a possible nuclear localization signal, RKKKR.
The most prominent characteristics of CIZ are the five, six, or eight Krüppel-type C2H2 zinc fingers (Fig. 2A). This type of zinc finger is often seen in the DNA-binding motif of a transcription factor. The glutamine-alanine repeat and polyglutamines lie in the C-terminal region. The glutamine-alanine repeat is found in several transcriptional modulators, such as GAL11 and SSN6 in Saccharomyces cerevisiae, Zeste in Drosophila melanogaster, and CA150 in humans (46). Polyglutamines are often seen in the activation domain of transcription factors and have a capacity to activate transcription (10). These features, together with the existence of a possible nuclear localization signal, suggest that CIZ could act as a transcription factor.
The homology search by BLAST revealed that partial sequences of the putative human homologue of CIZ are reported (28, 36). They cloned the partial cDNA corresponding to the C-terminal portion of CIZ in their search for CAG repeat protein from a human brain cDNA library (28) or from a human testis cDNA library (36). In contrast to six glutamines of rat CIZ we cloned, the partial sequence they reported has 15 consecutive glutamines, resulting in a longer CAG repeat. They located the gene at human chromosome 12p12 D12S328–D12S89 (28) or at 12p13 (36).
Tissue distribution of CIZ transcripts.
Northern blot analysis of mRNA from a variety of rat tissues was performed (Fig. 2C). The 3-kb band corresponding to the cDNA of CIZ was observed universally, but the levels of expression in testis, heart, liver, kidney, and brain were high. Another 4-kb band was detected in brain, liver, and kidney. Although the tissue distribution of Cas is ubiquitous, the level of expression of Cas is rather high in testis, kidney, and brain (42), and Cas plays an important role in heart differentiation (13). Therefore, the tissue distribution of CIZ is compatible with the possible interaction of CIZ with Cas.
CIZ binds specifically to the Cas SH3 domain.
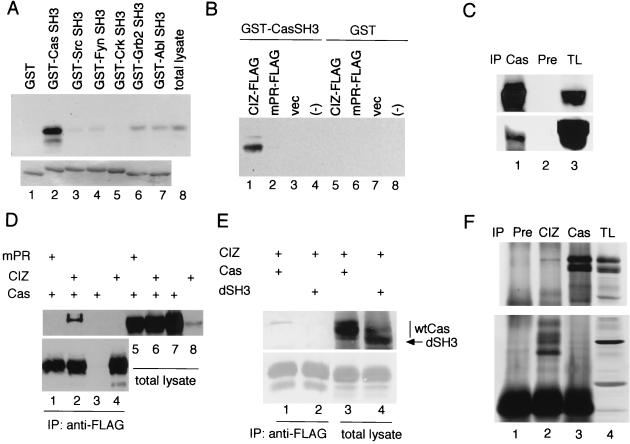
The SH3 domains often bind nonspecifically to proline-rich sequences. Therefore we investigated the association of CIZ with a panel of GST fusion proteins of various SH3 domains. FLAG-tagged CIZ (CIZ-FLAG) expressed in COS-7 cells was reacted with immobilized GST (Fig. 3A, lane 1), GST-Cas SH3 (lane 2), GST-Src SH3 (lane 3), GST-Fyn SH3 (lane 4), GST-Crk SH3 (lane 5), GST-Ash/Grb2 SH3 (lane 6), and GST-Abl SH3 (lane 7). In contrast to the strong binding of CIZ to Cas SH3 (Fig. 3A, lane 2), the association of CIZ with the other SH3s was only residual, demonstrating the specificity of the association of CIZ with Cas SH3. The mutation in the APPKPPR sequence (mPR) abolished the binding of CIZ to Cas SH3 (Fig. 3B, lanes 1, 2), strongly suggesting that this sequence was the binding site for Cas SH3.
FIG. 3.
Association of CIZ with Cas. (A) FLAG-tagged CIZ expressed in COS-7 cells was precipitated with GST (lane 1), GST-Cas SH3 (lane 2), GST-Src SH3 (lane 3), GST-Fyn SH3 (lane 4), GST-Crk SH3 (lane 5), GST-Ash/Grb2 SH3 (lane 6), and GST-Abl SH3 (lane 7) and was immunoblotted with anti-FLAG (upper row). The samples were electrophoresed and stained with Coomassie blue (bottom row). Total lysates of COS-7 cells expressing CIZ-FLAG were also immunoblotted (lane 8). (B) FLAG-tagged CIZ (lanes 1 and 5), mPR mutant of CIZ-FLAG (lanes 2 and 6), and mock (lanes 3 and 7)-transfected COS-7 cell lysates were incubated with GST-Cas SH3 (lanes 1, 2, and 3) or with GST (lanes 5, 6, and 7) and immunoblotted with anti-FLAG. Lanes 4 and 8 contained GST-Cas SH3 or GST alone, respectively. vec, vector. (C) CIZ-FLAG was expressed in COS-7 cells, and the lysates were immunoprecipitated (IP) with anti-Cas2 (lane 1) or with preimmune (Pre) serum (lane 2). The total cellular lysates (TL) are shown in lane 3. The samples were immunoblotted with anti-Cas2 in the upper row and immunoblotted with anti-FLAG in the bottom row. (D) Association between CIZ and Cas expressed in COS-7 cells. CIZ-FLAG (lanes 2 and 6) or mPR-FLAG (lanes 1 and 5) was expressed in COS-7 cells together with Cas. Cas alone (lanes 3 and 7) and CIZ-FLAG alone (lanes 4 and 8) were also expressed. In lanes 1 to 4, the lysates were immunoprecipitated with anti-FLAG. In the upper row, the filter was immunoblotted with anti-Cas2. In the bottom row, the same immunoprecipitant was immunoblotted with anti-FLAG. (E) The wild-type Cas (wtCas) (lanes 1 and 3) and the mutant of Cas that lacks the SH3 domain (lanes 2 and 4 [dSH3]) were expressed together with FLAG-tagged CIZ. The lysates were immunoprecipitated with anti-FLAG antibody and immunoblotted with anti-Cas2 (upper row) or anti-CIZ (lower row). (F) The lysates of LC-540 cells were immunoprecipitated with preimmune serum (lane 1), anti-CIZ (lane 2), and anti-Cas2 (lane 3). In the upper row, the filter was immunoblotted with anti-Cas2, and in the lower row, the filter was immunoblotted with anti-CIZ.
Association of CIZ with Cas.
We investigated the association of CIZ with Cas by overexpressing CIZ-FLAG and Cas in COS-7 cells. CIZ-FLAG was detected in the anti-Cas immunoprecipitants (Fig. 3C, lane 1). Oppositely, we detected Cas in the anti-FLAG immunoprecipitants (Fig. 3D, lane 2), whereas we could not detect Cas in the immunoprecipitants when we overexpressed mPR-FLAG and Cas (lane 1), showing again that the APPKPPR sequence is necessary for the association of Cas with CIZ. The mutant of Cas that lacks SH3 domain was not detected in anti-FLAG immunoprecipitants (Fig. 3E, lane 1). In Fig. 3D and E, the slower band corresponding to the B form of Cas (42) was detected in the immunoprecipitants by anti-FLAG antibody. We further checked the endogenous association of CIZ with Cas in a rat Leydig cell testicular tumor cell line, LC-540. Like Fig. 3D and E, the slower band corresponding to the B form of Cas (42) was detected in the immunoprecipitants by anti-CIZ antibody (Fig. 3F, lane 2), and this band was not detected in the preimmune serum precipitants (Fig. 3F, lane 1). The presence of CIZ in the reciprocal immunoprecipitation was not clear (Fig. 3F, lane 3). Neither the treatment with cytochalasin D nor the loss of cell attachment affected the binding between CIZ and Cas (data not shown), suggesting that the association between CIZ and Cas is constitutive.
CIZ localizes to focal adhesions and nucleus.
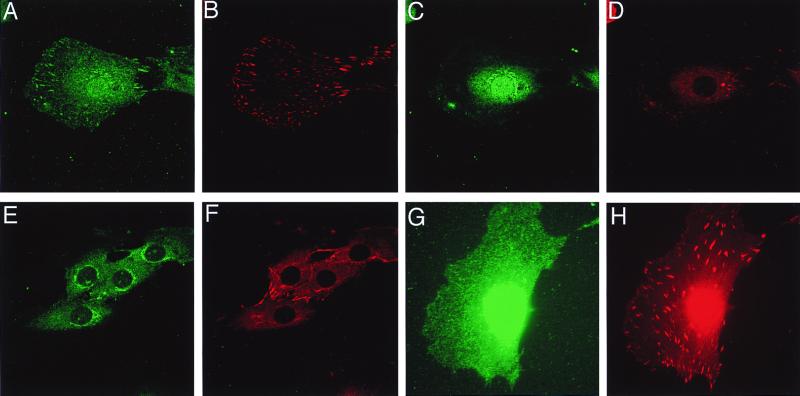
In order to see the colocalization of CIZ with Cas, we investigated the subcellular localization of CIZ. Because anti-Cas and anti-CIZ are both made in rabbits, costaining was carried out by using a mouse monoclonal antivinculin antibody, anti-hVIN-1 (Sigma). Vinculin was shown to colocalize with Cas at focal adhesions in our previous report (31). The anti-CIZ antibody stained focal adhesions (Fig. 4A), which are visualized by antivinculin antibody (Fig. 4B). Preclearing of anti-CIZ antibody with GST-CIZ, to which this antibody was raised, abrogated the staining at focal adhesions (data not shown). Another section of the same cell revealed that anti-CIZ stained the nucleus and perinuclear region (Fig. 4C). However, the perinuclear staining was also seen in the staining of GST-CIZ-precleared antibody (Fig. 4E) and was deemed to be nonspecific. The localization of CIZ in nucleus is compatible with the fact that CIZ has a putative nuclear localization signal. These results show that CIZ localizes to focal adhesions and nucleus. To further characterize the localization of CIZ, we viewed the cells under several conditions. Adhesion on poly-l-lysine or culture in serum-free medium abrogated the formation of focal adhesions together with the localization of CIZ to focal adhesions (data not shown). Stimulation with lysophosphatidic acid in serum-free medium induced the formation of focal adhesions and the localization of CIZ to focal adhesions (data not shown). Furthermore, CIZ does not localize at newly forming focal adhesions just after plating on fibronectin and the staining at the focal adhesion gradually becomes visible (data not shown). Therefore, CIZ seems to be localized at stably formed focal adhesions. The localization of CIZ in the nucleus, on the contrary, was constantly observed, as far as we examined.
FIG. 4.
Subcellular localization of CIZ. (A to F) 3Y1 cells were grown overnight on the uncoated coverslips. (G and H) After overnight culture, 3Y1 cells were treated with LMB (10 ng/ml) for 24 h. Immunofluorescence was performed with anti-CIZ (1:400) (A, C, and G) precleared by GST or with anti-CIZ precleared by GST-CIZ (E), together with antivinculin antibody (B, D, F, and H) as described previously (31). Panels A, B, G, and H show the sections at the height of focal adhesions. Panels C, D, E, and F show the sections at the height of the nucleus.
CIZ has two sequences (amino acids 52 to 62 and 84 to 94) that contain multiple hydrophobic residues and resemble the consensus nuclear export signal (NES) sequence, although they do not completely match it (34, 47, 49, 55). Therefore, we checked the effect of LMB (33), which is known to inhibit nuclear export (21, 22), on the subcellular localization of CIZ. LMB treatment abrogated the localization of CIZ to focal adhesions (Fig. 4G and H), demonstrating that CIZ can translocate from the nucleus to focal adhesions. The localization to focal adhesions of CIZ was seen in the absence of LMB under the same conditions (Fig. 4A and data not shown).
CIZ shuttles in and out of the nucleus.
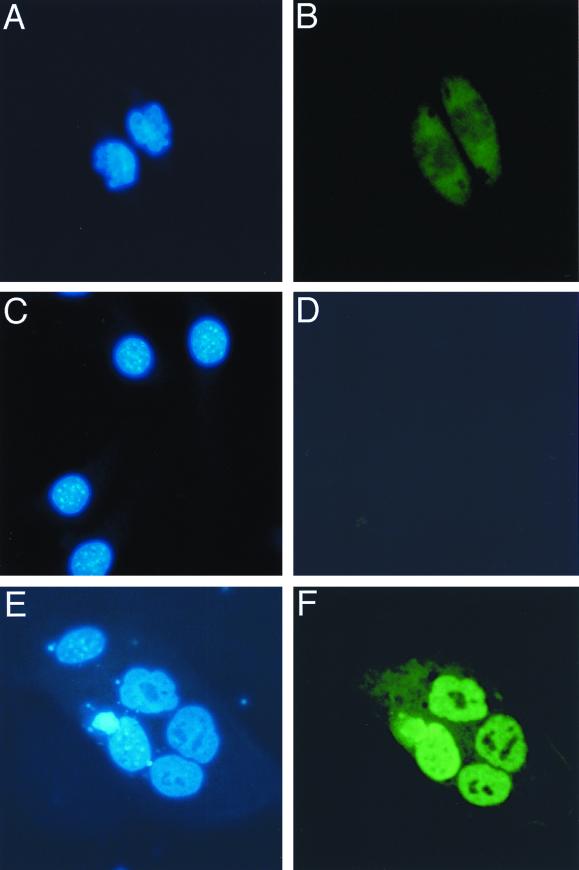
Subsequently, we examined whether CIZ really shuttles into and out of nucleus by using the cell fusion assay (4). NIH 3T3 cells transiently expressing HA-tagged CIZ were fused with HeLa cells and were stained with anti-HA antibody and Hoechst 33258 dye. The nuclear staining of mouse cells (NIH 3T3) by Hoechst 33258 is spotty (Fig. 5C), and that of human cells (HeLa) is diffuse (Fig. 5A). The staining pattern of the fused cell showed the features of the heterokaryon of NIH 3T3 cells and HeLa cells (Fig. 6E). If CIZ does not shuttle, anti-HA staining should be restricted only to mouse cells, because we blocked the protein synthesis by cycloheximide. Anti-HA antibody stained both mouse and human nuclei, suggesting CIZ shuttles between the nucleus and cytoplasm (Fig. 5F). Anti-HA antibody did not stain nontransfected mouse and human nuclei (Fig. 5B and D).
FIG. 5.
Nuclear-cytoplasmic shuttling of CIZ. (A) Nontransfected HeLa cells were stained with Hoechst 33258 dye. (B) The same cells as in panel A stained with anti-HA. (C) Nontransfected NIH 3T3 cells were stained with Hoechst 33258. (D) The same cells as in panel C stained with anti-HA. (E) A heterokaryon of HeLa cells, CIZ-HA-transfected NIH 3T3 cells, and a nearby NIH 3T3 cell were stained with Hoechst 33258. (F) The same cells as in panel E stained with anti-HA.
FIG. 6.
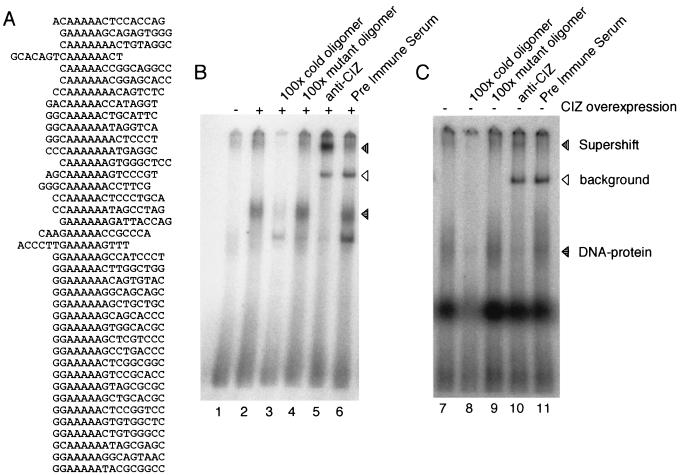
CIZ has DNA-binding consensus sequences. (A) Results of CASTing described in Materials and Methods. (B) Overexpressed CIZ binds to the CCTTTTTCAAAAAGA sequence of the human MMP-1 promoter. EMSA was performed with the nuclear extract of CIZ-FLAG (lanes 2 to 5)-transfected cells and with that of mock (lane 1)-transfected COS-7 cells. The horizontally lined arrowhead indicates the DNA-protein complex (lanes 2, 4, and 6). The supershifted band is indicated by the vertically lined arrowhead (lane 5). The background band caused by immunoglobulin was indicated by the open arrowhead (lanes 5 and 6). (C) Endogeneous CIZ is included in the DNA-protein complex (lane 7). The supershift band was observed in panel B (lane 10).
CIZ binds to the (G/C)AAAAA(A) nucleotide sequence.
CIZ has many characteristics of a transcription factor, such as Krüppel-type zinc fingers, the glutamine-rich region, and the nuclear localization signal, and we thus expected that CIZ could have a DNA-binding capacity. In order to determine the consensus binding sequences of CIZ, we performed the CASTing analysis (58). The nuclear extract from COS-7 cells transfected with CIZ-FLAG was mixed with random oligomers and was incubated. After immunoprecipitation by anti-FLAG, the bound oligomer was extracted and was amplified by PCR. The amplified oligomer was again incubated with the CIZ-FLAG-expressing nuclear extract. This process was repeated five times, and the oligomer obtained was subcloned into pBlueScript and was sequenced. We obtained the results shown in Fig. 6A. The consensus sequence of the obtained oligonucleotides was (G/C)AAAAA(A).
CIZ binds to the CCTTTTTCAAAAAGA sequence of the human MMP-1 promoter.
A homology search revealed that the human MMP-1 (collagenase) promoter includes the CIZ-binding consensus sequence CCTTTTTCAAAAAGA, which resides from −320 to −305 (2). In order to check if this sequence binds CIZ, we performed the EMSA. The nuclear extract of CIZ-FLAG- or mock-transfected COS-7 cells was mixed with the isotope-labeled oligomer and was electrophoresed. DNA-protein complex was detected with CIZ-overexpressing COS-7 cells (Fig. 6B, lane 2) but not with mock-transfected cells (lane 1). DNA-protein complex was abolished when competed with cold oligonucleotides (lane 3), but not by mutant oligonucleotides (lane 4). Anti-CIZ clearly supershifted the DNA-protein complex, showing that this complex includes CIZ (lane 5). The nuclear extract of LC-540 also formed a DNA-protein complex (Fig. 6C, lane 7) that was supershifted by anti-CIZ (lane 10). The DNA-protein complex was abolished when competed with cold oligonucleotides (lane 8), but not by mutant oligonucleotides (lane 9). These results indicate that endogenous CIZ is included in the DNA-protein complex.
CIZ upregulates the transcription from MMP promoters.
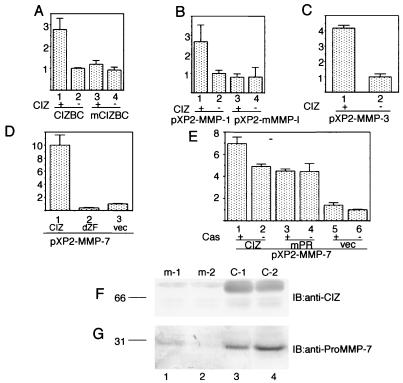
To determine whether CIZ enhances transcription through interaction with this sequence, we cotransfected NIH 3T3 cells with the expression vector of CIZ and the reporter constructs containing four tandem copies of this sequence linked to the tk basal promoter, designated (CIZBC)4-tk. Luciferase activity driven by this (CIZBC)4-tk promoter increased by 2.5- to 3-fold (Fig. 7A, lanes 1 and 2) with the expression of CIZ, while the chimeric promoter carrying mutations, designated (mCIZBC)4-tk, exhibited no increase in the luciferase activity (lanes 3 and 4). These results indicate that CIZ can act through this sequence to enhance transcription. Next, we addressed whether CIZ is capable of transactivating the native MMP-1 promoter. NIH 3T3 cells were cotransfected with the expression vector of CIZ and the −517/+63 MMP-1 promoter fused to the luciferase reporter. Luciferase activity driven by the MMP-1 promoter also increased by 2.5- to 3-fold (Fig. 7B, lanes 1 and 2). In contrast, CIZ had no effect on the luciferase activity from the MMP-1 promoter that was mutated at the CIZ binding consensus sequence (lanes 3 and 4). These results show that the CIZ transactivates the MMP-1 promoter through this sequence. The consensus sequence, (G/C)AAAAA(A), obtained in the CASTing of CIZ, is observed in the promoter regions of the other members of MMPs, such as MMP-3 (stromelysin) and MMP-7 (matrilysin). Unlike MMP-1, which has only one CIZ-binding consensus sequence, promoter regions of MMP-3 and of MMP-7 have plenty of the consensus sequences. NIH 3T3 cells were cotransfected with the expression vector of CIZ and the MMP-3 or MMP-7 promoter fused to the luciferase reporter. The luciferase activities increased by 4-fold in the MMP-3 promoter (Fig. 7C) and 10-fold in the MMP-7 promoter (Fig. 7D) by the overexpression of CIZ. In order to see whether the transactivation of MMP promoters by CIZ is direct, we generated a mutant, dZF, that lacks the zinc finger domain. This mutant had no transactivation activity, but repressed the basal level of transcription of the MMP-7 promoter (Fig. 7D), showing that the transactivation by CIZ is not mediated by other proteins but is direct. The repression by the dZF mutant was also observed with the MMP-1 and -3 promoters (data not shown).
FIG. 7.
CIZ upregulates the expression of MMPs. (A) CIZ transactivates the artificial promoter consisting of four tandem repeats of CIZ-binding consensus sequences in the human MMP-1 promoter. (B to D) CIZ upregulates the transcription from the human MMP-1 promoter mediated by the CIZ-binding consensus sequence (B) from the MMP-3 promoter (C) and from the MMP-7 promoter (D). vec, vector. The effect of Cas expression on the transactivation by CIZ was checked in Cas−/− fibroblasts (E). In these experiments, NIH 3T3 cells or Cas−/− fibroblasts were transfected with each expression vector, together with the reporter construct and β-galactosidase expression vector. Values are the means of three experiments, with bars representing the standard deviations. (F and G) Overproduction of MMP-7 in HT1080 cells stably expressing CIZ. (F) Total lysates of m-1 (lane 1), m-2 (lane 2), C-1 (lane-3), and C-2 (lane-4) cells were immunoblotted (IB) with anti-CIZ. (G) The conditioned media of m-1 (lane 1), m-2 (lane 2), C-1 (lane 3), and C-2 (lane 4) cells were immunoblotted with anti-proMMP-7.
Cas slightly enhances transactivation by CIZ.
In order to see the effect of Cas on transactivation by CIZ, we transiently expressed Cas in Cas−/− fibroblasts (13) and checked the transactivation of the MMP-7 promoter by CIZ. CIZ increased the transcription from MMP-7 promoter by four- to fivefold (Fig. 7E) in Cas−/− fibroblasts, indicating that Cas is not absolutely necessary for this transactivation. However, repeated experiments revealed that the coexpression of Cas in Cas−/− fibroblasts slightly (1.4- to 2-fold) enhanced the transactivation of the MMP-7 promoter by CIZ (Fig. 7E and data not shown). The mPR mutant of CIZ, which does not bind Cas, also increased the luciferase activities from the MMP-7 promoter by four- to fivefold, but simultaneous expression of Cas did not enhance the transactivation (Fig. 7E). This result suggests that the association of Cas with CIZ enhances the transactivation activity of CIZ. In NIH 3T3 cells, which express the endogenous Cas, the enhancement by the coexpression of Cas was not apparent (data not shown).
Secretion of MMP-7 is enhanced in HT1080 cells stably expressing CIZ.
Finally, we investigated the secretion of MMP in the cultured medium of HT1080 cells stably expressing CIZ. We checked the proMMP-7 level in the conditioned medium of HT1080 cells overexpressing CIZ, because the enhancement of the luciferase assay from the MMP-7 promoter by CIZ was larger than those from the other MMP promoters investigated. Two mock-transfected HT1080 cell lines, m-1 and m-2 (Fig. 7F, lanes 1 and 2), and two CIZ-expressing HT1080 cell lines, C-1 and C-2 (lanes 3 and 4), were cultured in serum-free Dulbecco's modified Eagle's medium with 100 ng of 12-O-tetradecanoylphorbol-13-acetate (TPA) for 3 days. TPA was added to enhance the basal production of MMP-7 above the detection level. The supernatants were concentrated 20-fold with ULTRAFREE-MC (Millipore), electrophoresed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and immunoblotted with anti-proMMP-7. The supernatants of C-1 and C-2 contained more proMMP-7 than those of m-1 and m-2 (Fig. 7G). This result shows that overexpression of CIZ upregulated the secretion of MMP-7.
DISCUSSION
In this paper, we report the molecular cloning of the cDNA encoding a novel zinc finger protein, CIZ. It was identified by far-Western screening of the rat 3Y1 cDNA library with GST-Cas SH3 fusion protein as a probe. The amino acid sequence of CIZ revealed that CIZ has a proline-rich sequence, which was subsequently shown to be the binding site for the Cas SH3 domain. The binding is specific to Cas SH3, and this sequence conforms to the Cas SH3-binding consensus (16). CIZ binds to Cas in cells and colocalized with Cas at focal adhesions. These results suggest that CIZ binds Cas at focal adhesions, at least when they are formed. However, as often happens with binding through SH3 domains, the binding of CIZ with Cas persists even after the focal adhesions are destroyed.
Subsequently, we showed that CIZ is localized both at focal adhesions and in the nucleus and that CIZ is a nucleocytoplasmic shuttling protein. Although the decisive data are lacking, these facts might imply that CIZ shuttles between focal adhesions and the nucleus. As far as we know, there are two other proteins that shuttle between focal adhesions and the nucleus, c-Abl (25, 47) and zyxin (34). Both proteins are reported to have a functional NES. As described above, two sequences of CIZ, amino acids 52 to 62 and 84 to 94, resemble the consensus NES sequence (34, 47, 49, 55), and these sequences could be the candidates for NESs. In fact, CIZ was localized exclusively to the nucleus and was not localized at focal adhesions when the cells were treated with LMB. However, the predominant localization of CIZ in the nucleus, especially when overexpressed, might indicate that the possible NES is weak or that some modification might be necessary for the NES of CIZ to become functional. Otherwise, the putative NES is usually masked by the binding partner of CIZ or by the possible oligomerization of CIZ.
The localization of Cas to focal adhesions is dependent on the Cas SH3 domain (31). The binding partners of the Cas SH3 domain, which are responsible for Cas localization to focal adhesions, are thought to be FAK or PYK2/CAKβ/RAFTK/CADTK (31, 50). These argue against the possibility that CIZ, which binds the Cas SH3 domain, is recruited to focal adhesions by binding with Cas. In contrast to c-Abl, which is known to move transiently from nucleus to focal adhesions upon cell attachment (25), CIZ does not localize at newly forming focal adhesions, but only at a later stage of the formation of focal adhesions. In addition, we could not detect the stimuli that abolish nuclear staining by anti-CIZ antibody. Abrogation of the localization of CIZ to focal adhesions by treatment with LMB indicates that CIZ molecules at focal adhesions are derived from the nucleus. These facts suggest that CIZ usually exists in the nucleus and is recruited out of the nucleus when cells form stable focal adhesions and stress fibers. We also showed that CIZ can translocate from cytoplasm to nucleus (Fig. 5). We can speculate that CIZ goes back to nucleus, not when focal adhesions are newly formed, but when focal adhesions are removed.
The other features of CIZ are its zinc fingers, glutamine-alanine repeat, glutamine-rich region, and nuclear localization signal. These features suggest that CIZ is a transcription factor. Therefore, we performed CASTing analysis to find the consensus DNA-binding sequence of CIZ. The homology search of consensus CIZ-binding sequences, (G/C)AAAAA, revealed that several MMPs have sequences similar to this consensus in their promoter regions. In fact, CIZ binds to the CCTTTTCAAAAAGA sequence of the human MMP-1 promoter and upregulates the transcription from the MMP-1, MMP-3, and MMP-7 promoters. We also showed that overexpression of CIZ enhances the secretion of proMMP-7 in the culture medium.
MMPs are reported to be upregulated by the treatment with TPA, epidermal growth factor, tumor necrosis factor, interleukin-1 (IL-1), and various other stimuli (2, 8, 29, 30, 39). Integrin stimulation, disruption of cytoskeleton, and activation of Rac-1 are also shown to increase the expression of MMP-1 (1, 14, 15, 23, 40, 56). At the transcriptional level, MMPs are known to be regulated by the AP-1 complex (2, 8, 39), by Ets transcription factors (57), and by Tcf (6). The binding of CIZ to the promoter region of MMP-1 and the transactivation of the MMP promoters by the overexpression of CIZ indicate that CIZ is another transcription factor that regulates MMP promoters. The dominant-negative form of CIZ, if it exists, would be useful for elucidating the stimulation in which CIZ is involved. Although the mutant lacking the zinc finger domain of CIZ slightly lowers the basal transcription level of MMP promoters, it did not abrogate the transactivation by the overexpression of CIZ (data not shown). Therefore, this mutant was not necessarily the dominant-negative form of CIZ. Another method of detecting the stimuli in which CIZ is involved is induction of a DNA-protein complex in the EMSA. Although we detected the stable DNA binding of CIZ, neither induction nor enhancement of the DNA-binding activity of CIZ was observed. A search for the stimulation which enhances or reduces the DNA-binding activity of CIZ would be necessary to find the signalling pathway in which CIZ has an important role.
In this report, we could show the significance of Cas-CIZ interaction only in the slight enhancement of transactivation in Cas−/− cells, and CIZ could still transactivate even without Cas. This, together with the fact that Cas is not localized in the nucleus (12, 31), argues against the idea that Cas acts as a direct cofactor for CIZ in the nucleus. Rather, Cas would act upstream of the signalling that modulates the transactivation by CIZ. Cas is reported to transmit signals from integrins (35, 44, 54). Downstream of Cas are the actin bundle formation (13), cell movement (5, 19), and activation of Rac-1 (7, 18). Although direct signaling from Cas to MMPs is not known, Rac-1, which lies downstream of tyrosine-phosphorylated Cas (7, 18), is reported to increase MMP-1 through an NF-κB–IL-1 loop (15). MMPs are also reported to be regulated by integrin stimulations (14, 23, 40, 41, 56). Furthermore, transformation by v-Src or v-Crk, which is known to enhance the tyrosine phosphorylation of Cas (42), is also reported to elevate the secretion of MMPs (11, 52). Cell motility and the secretion of MMPs are important steps in tissue morphogenesis, angiogenesis, and tumor invasion and metastasis. We can speculate that the CIZ-MMP signaling might be one of the various pathways downstream of the integrin-Cas signaling. However, at present, we have failed to show the involvement of CIZ in the integrin-Cas signaling.
Finally, another prominent feature of the CIZ cDNA is the presence of a CAG repeat. The abnormal elongation of CAG repeats and the accumulation of elongated polyglutamines in the nucleus of neurons are reported to be the cause of several neurodegenerative disorders (45). As described above, CIZ is localized in the nucleus (Fig. 4) and is expressed in the brain (Fig. 2C). Therefore, CIZ could be a candidate protein that causes a neurodegenerative disorder, and investigations in this direction should also be of interest.
ACKNOWLEDGMENTS
We thank M. Yoshida (University of Tokyo), T. Takenawa (University of Tokyo), T. Yamamoto (University of Tokyo), and B. J. Mayer (Harvard Medical School) for generously providing the leptomycin B, GST-Ash/Grb2 SH3, GST-Fyn SH3, and GST-Abl SH3, respectively. We appreciate P. Angel (German Cancer Research Center) and H. Rahmsdorf (Kernforschungszentrum Karlsruhe) for kindly providing the human MMP-1 promoter-encoding plasmid. We also appreciate L. Matrisian (Vanderbilt University) for providing the human MMP-7 promoter and the rat MMP-3 promoter. We thank K. Chida (University of Tokyo) for helpful advice.
This work was supported in part by Fellowships in Cancer Research of the Japan Society for the Promotion of Science for Young Scientists.
REFERENCES
- 1.Aggeler J, Frisch S M, Werb Z. Changes in cell shape correlate with collagenase gene expression in rabbit synovial fibroblasts. J Cell Biol. 1984;98:1662–1671. doi: 10.1083/jcb.98.5.1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Angel P, Baumann I, Stein B, Delius H, Rahmsdorf H J, Herrlich P. 12-O-Tetradecanoyl-phorbol-13-acetate induction of the human collagenase gene is mediated by an inducible enhancer element located in the 5′-flanking region. Mol Cell Biol. 1987;7:2256–2266. doi: 10.1128/mcb.7.6.2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Astier A, Avraham H, Manie S N, Groopman J, Canty T, Avraham S, Freedman A S. The related adhesion focal tyrosine kinase is tyrosine-phosphorylated after beta1-integrin stimulation in B cells and binds to p130cas. J Biol Chem. 1997;272:228–232. doi: 10.1074/jbc.272.1.228. [DOI] [PubMed] [Google Scholar]
- 4.Borer R A, Lehner C F, Eppenberger H M, Nigg E A. Major nucleolar proteins shuttle between nucleus and cytoplasm. Cell. 1989;56:379–390. doi: 10.1016/0092-8674(89)90241-9. [DOI] [PubMed] [Google Scholar]
- 5.Cary L A, Han D C, Polte T R, Hanks S K, Guan J L. Identification of p130Cas as a mediator of focal adhesion kinase-promoted cell migration. J Cell Biol. 1998;140:211–221. doi: 10.1083/jcb.140.1.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Crawford H C, Fingleton B M, Rudolph-Owen L A, Goss K J H, Rubinfeld B, Polakis P, Matrisian L M. The metalloproteinase matrilysin is a target of b-catenin transactivation in intestinal tumors. Oncogene. 1999;18:2883–2891. doi: 10.1038/sj.onc.1202627. [DOI] [PubMed] [Google Scholar]
- 7.Dolfi F, Garcia-Guzman M, Ojaniemi M, Nakamura H, Matsuda M, Vuori K. The adaptor protein Crk connects multiple cellular stimuli to the JNK signaling pathway. Proc Natl Acad Sci USA. 1998;95:15394–15399. doi: 10.1073/pnas.95.26.15394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gaire M, Magbanua Z, McDonnell S, McNeil L, Lovett D H, Matrisian L M. Structure and expression of the human gene for the matrix metalloproteinase matrilysin. J Biol Chem. 1994;269:2032–2040. [PubMed] [Google Scholar]
- 9.Garton A J, Burnham M R, Bouton A H, Tonks N K. Association of PTP-PEST with the SH3 domain of p130Cas; a novel mechanism of protein tyrosine phosphatase substrate recognition. Oncogene. 1997;15:877–85. doi: 10.1038/sj.onc.1201279. [DOI] [PubMed] [Google Scholar]
- 10.Gerber H P, Seipel K, Georgiev O, Höfferer M, Hug M, Rusconi S, Schaffner W. Transcriptional activation modulated by homopolymeric glutamine and proline stretches. Science. 1994;263:808–811. doi: 10.1126/science.8303297. [DOI] [PubMed] [Google Scholar]
- 11.Hamaguchi M, Yamagata S, Thant A A, Xiao H, Iwata H, Mazaki T, Hanafusa H. Augmentation of metalloproteinase (gelatinase) activity secreted from Rous sarcoma virus-infected cells correlates with transforming activity of src. Oncogene. 1995;10:1037–1043. [PubMed] [Google Scholar]
- 12.Harte M T, Hildebrand J D, Burnham M R, Bouton A H, Parsons J T. p130Cas, a substrate associated with v-Src and v-Crk, localizes to focal adhesions and binds focal adhesion kinase. J Biol Chem. 1996;271:13649–13655. doi: 10.1074/jbc.271.23.13649. [DOI] [PubMed] [Google Scholar]
- 13.Honda H, Oda H, Nakamoto T, Honda Z-I, Sakai R, Suzuki T, Saito T, Nakamura K, Nakao K, Ishikawa T, Katsuki M, Yazaki Y, Hirai H. Cardiovascular anomaly, impaired actin bundling and resistance to Src-induced transformation in mice lacking p130Cas. Nat Genet. 1998;19:361–365. doi: 10.1038/1246. [DOI] [PubMed] [Google Scholar]
- 14.Huhtala P, Humphries M J, McCarthy J B, Tremble P M, Werb Z, Damsky C H. Cooperative signaling by α5β1 and α4β1 integrins regulates metalloproteinase gene expression in fibroblasts adhering to fibronectin. J Cell Biol. 1995;129:867–879. doi: 10.1083/jcb.129.3.867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kheradmand F, Werner E, Tremble P, Symons M, Werb Z. Role of Rac-1 and oxygen radicals in collagenase-1 expression induced by cell shape change. Science. 1998;280:898–902. doi: 10.1126/science.280.5365.898. [DOI] [PubMed] [Google Scholar]
- 16.Kirsch K H, Georgescu M-M, Hanafusa H. Direct binding of p130Cas to the guanine nucleotide exchange factor C3G. J Biol Chem. 1998;273:25673–25679. doi: 10.1074/jbc.273.40.25673. [DOI] [PubMed] [Google Scholar]
- 17.Kirsch K H, Georgescu M M, Ishimaru S, Hanafusa H. CMS: an adapter molecule involved in cytoskeletal rearrangements. Proc Natl Acad Sci USA. 1999;96:6211–6216. doi: 10.1073/pnas.96.11.6211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kiyokawa E, Hashimoto Y, Kobayashi S, Sugimura H, Kurata T, Matsuda M. Activation of Rac1 by a Crk SH3-binding protein, DOCK180. Genes Dev. 1998;12:3331–3336. doi: 10.1101/gad.12.21.3331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Klemke R L, Leng J, Molander R, Brooks P C, Vuori K, Cheresh D A. CAS/Crk coupling serves as a “molecular switch” for induction of cell migration. J Cell Biol. 1998;140:961–972. doi: 10.1083/jcb.140.4.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kozak M. Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Cell. 1986;44:283–292. doi: 10.1016/0092-8674(86)90762-2. [DOI] [PubMed] [Google Scholar]
- 21.Kudo N, Khochbin S, Nishi K, Kitano K, Yanagida M, Yoshida M, Horinouchi S. Molecular cloning and cell cycle-dependent expression of mammalian CRM1, a protein involved in nuclear export of proteins. J Biol Chem. 1997;272:29742–29751. doi: 10.1074/jbc.272.47.29742. [DOI] [PubMed] [Google Scholar]
- 22.Kudo N, Wolff B, Sekimoto T, Schreiner E P, Yoneda Y, Yanagida M, Horinouchi S, Yoshida M. Leptomycin B inhibition of signal-mediated nuclear export by direct binding to CRM1. Exp Cell Res. 1998;242:540–547. doi: 10.1006/excr.1998.4136. [DOI] [PubMed] [Google Scholar]
- 23.Langholz O, Röckel D, Mauch C, Kozlowska E, Bank I, Krieg T, Eckes B. Collagen and collagenase gene expression in three-dimensional collagen lattices are differentially regulated by α1β1 and α2β1 integrins. J Cell Biol. 1995;131:1903–1915. doi: 10.1083/jcb.131.6.1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lassar A B, Davis R L, Wright W E, Kadesch T, Murre C, Voronova A, Baltimore D, Weintraub H. Functional activity of myogenic HLH proteins requires hetero-oligomerization with E12/E47-like proteins in vivo. Cell. 1991;66:305–315. doi: 10.1016/0092-8674(91)90620-e. [DOI] [PubMed] [Google Scholar]
- 25.Lewis J M, Baskaran R, Taagepera S, Schwartz M A, Wang J Y J. Integrin regulation of c-Abl tyrosine kinase activity and cytoplasmic-nuclear transport. Proc Natl Acad Sci USA. 1996;93:15174–15179. doi: 10.1073/pnas.93.26.15174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu F, Hill D E, Chernoff J. Direct binding of the proline-rich region of protein tyrosine phosphatase 1B to the Src homology 3 domain of p130Cas. J Biol Chem. 1996;271:31290–31295. doi: 10.1074/jbc.271.49.31290. [DOI] [PubMed] [Google Scholar]
- 27.Luckow B, Schütz G. CAT constructions with multiple unique restriction sites for the functional analysis of eukaryotic promoters and regulatory elements. Nucleic Acids Res. 1987;15:5490. doi: 10.1093/nar/15.13.5490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Margolis R L, Abraham M R, Gatchell S B, Li S-H, Kidwai A S, Breschel T S, Stine O C, Callahan C, McInnis M G, Ross C A. cDNAs with long CAG trinucleotide repeats from human brain. Hum Genet. 1997;100:114–122. doi: 10.1007/s004390050476. [DOI] [PubMed] [Google Scholar]
- 29.Matrisian L M, Bowden G T, Krieg P, Fürstenberger G, Briand J-P, Leroy P, Breathnach R. The mRNA coding for the secreted protease transin is expressed more abundantly in malignant than in benign tumors. Proc Natl Acad Sci USA. 1986;83:9413–9417. doi: 10.1073/pnas.83.24.9413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Matrisian L M, Leroy P, Ruhlmann C, Gesnel M-C, Breathnach R. Isolation of the oncogene and epidermal growth factor-induced transin gene: complex control in rat fibroblasts. Mol Cell Biol. 1986;6:1679–1686. doi: 10.1128/mcb.6.5.1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nakamoto T, Sakai R, Honda H, Ogawa S, Ueno H, Suzuki T, Aizawa S-I, Yazaki Y, Hirai H. Requirements for localization of p130cas to focal adhesions. Mol Cell Biol. 1997;17:3884–3897. doi: 10.1128/mcb.17.7.3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nakamoto T, Sakai R, Ozawa K, Yazaki Y, Hirai H. Direct binding of C-terminal region of p130Cas to SH2 and SH3 domains of Src kinase. J Biol Chem. 1996;271:8959–8965. doi: 10.1074/jbc.271.15.8959. [DOI] [PubMed] [Google Scholar]
- 33.Nishi K, Yoshida M, Fujiwara D, Nishikawa M, Horinouchi S, Beppu T. Leptomycin B targets a regulatory cascade of crm1, a fission yeast nuclear protein, involved in control of higher order chromosome structure and gene expression. J Biol Chem. 1994;269:6320–6324. [PubMed] [Google Scholar]
- 34.Nix D A, Beckerle M C. Nuclear-cytoplasmic shuttling of the focal contact protein, zyxin: a potential mechanism for communication between sites of cell adhesion and the nucleus. J Cell Biol. 1997;138:1139–1147. doi: 10.1083/jcb.138.5.1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nojima Y, Morino N, Mimura T, Hamasaki K, Furuya H, Sakai R, Sato T, Tachibana K, Morimoto C, Yazaki Y, Hirai H. Integrin-mediated cell adhesion promotes tyrosine phosphorylation of p130Cas, an Src homology 3-containing molecule having multiple Src homology 2-binding motifs. J Biol Chem. 1995;270:15398–15402. doi: 10.1074/jbc.270.25.15398. [DOI] [PubMed] [Google Scholar]
- 36.Pawlak A, Chiannikulchai N, Ansorge W, Bulle F, Weissenbach J, Gyapay G, Guellaën G. Identification and mapping of 26 human testis mRNAs containing CAG/CTG repeats. Mamm Genome. 1998;9:745–748. doi: 10.1007/s003359900856. [DOI] [PubMed] [Google Scholar]
- 37.Petch L A, Bockholt S M, Bouton A H, Parsons J T, Burridge K. Adhesion-induced tyrosine phosphorylation of the p130 SRC substrate. J Cell Sci. 1995;108:1371–1379. doi: 10.1242/jcs.108.4.1371. [DOI] [PubMed] [Google Scholar]
- 38.Polte T R, Hanks S K. Interaction between focal adhesion kinase and Crk-associated tyrosine kinase substrate p130Cas. Proc Natl Acad Sci USA. 1995;92:10678–10682. doi: 10.1073/pnas.92.23.10678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Quinones S, Saus J, Otani Y, Harris E D, Kurkinen M. Transcriptional regulation of human stromelysin. J Biol Chem. 1989;264:8339–8344. [PubMed] [Google Scholar]
- 40.Riikonen T, Westermarck J, Koivisto L, Broberg A, Kähäri V M, Heino J. Integrin α2β1 is a positive regulator of collagenase (MMP-1) and collagen α1(I) gene expression. J Biol Chem. 1995;270:13548–13552. doi: 10.1074/jbc.270.22.13548. [DOI] [PubMed] [Google Scholar]
- 41.Saarialho-Kere U K, Kovacs S O, Pentland A P, Olerud J E, Welgus H G, Parks W C. Cell-matrix interactions modulate interstitial collagenase expression by human keratinocytes actively involved in wound healing. J Clin Investig. 1993;92:2858–2866. doi: 10.1172/JCI116906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sakai R, Iwamatsu A, Hirano N, Ogawa S, Tanaka T, Mano H, Yazaki Y, Hirai H. A novel signaling molecule, p130, forms stable complexes in vivo with v-Crk and v-Src in a tyrosine phosphorylation-dependent manner. EMBO J. 1994;13:3748–3756. doi: 10.1002/j.1460-2075.1994.tb06684.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sakai R, Iwamatsu A, Hirano N, Ogawa S, Tanaka T, Nishida J, Yazaki Y, Hirai H. Characterization, partial purification, and peptide sequencing of p130, the main phosphoprotein associated with v-Crk oncoprotein. J Biol Chem. 1994;269:32740–32746. [PubMed] [Google Scholar]
- 44.Schlaepfer D D, Broome M A, Hunter T. Fibronectin-stimulated signaling from a focal adhesion kinase–c-Src complex: involvement of the Grb2, p130cas and Nck adaptor proteins. Mol Cell Biol. 1997;17:1702–1713. doi: 10.1128/mcb.17.3.1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sisodia S S. Nuclear inclusions in glutamine repeat disorders: are they pernicious, coincidental, or beneficial? Cell. 1998;95:1–4. doi: 10.1016/s0092-8674(00)81743-2. [DOI] [PubMed] [Google Scholar]
- 46.Suñé C, Hayashi T, Liu Y, Lane W S, Young R A, Garcia-Blanco M A. CA150, a nuclear protein associated with the RNA polymerase II holoenzyme, is involved in Tat-activated human immunodeficiency virus type 1 transcription. Mol Cell Biol. 1997;17:6029–6039. doi: 10.1128/mcb.17.10.6029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Taagepera S, McDonald D, Loeb J E, Whitaker L L, McElroy A K, Wang J Y J, Hope T J. Nuclear-cytoplasmic shuttling of c-Abl tyrosine kinase. Proc Natl Acad Sci USA. 1998;95:7457–7462. doi: 10.1073/pnas.95.13.7457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tanaka T, Nishida J, Mitani K, Ogawa S, Yazaki Y, Hirai H. Evi-1 raises AP-1 activity and stimulates c-fos promoter transcription with dependence on the second zinc finger domain. J Biol Chem. 1994;269:24020–24026. [PubMed] [Google Scholar]
- 49.Toyoshima F, Moriguchi T, Wada A, Fukuda M, Nishida E. Nuclear export of cyclin B1 and its possible role in the DNA damage-induced G2 checkpoint. EMBO J. 1998;17:2728–2735. doi: 10.1093/emboj/17.10.2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ueki K, Mimura T, Nakamoto T, Sasaki T, Aizawa S, Hirai H, Yano S, Naruse T, Nojima Y. Integrin-mediated signal transduction in cells lacking focal adhesion kinase p125FAK. FEBS Lett. 1998;432:197–201. doi: 10.1016/s0014-5793(98)00862-x. [DOI] [PubMed] [Google Scholar]
- 51.Ueno H, Hirano N, Kozutsumi H, Sasaki K, Tanaka T, Yazaki Y, Hirai H. An epidermal growth factor receptor-leukocyte tyrosine kinase chimeric receptor generates ligand-dependent growth signals through the Ras signaling pathway. J Biol Chem. 1995;270:20135–20142. doi: 10.1074/jbc.270.34.20135. [DOI] [PubMed] [Google Scholar]
- 52.Vincenti M P, Coon C I, White L A, Barchowsky A, Brinckerhoff C E. src-related tyrosine kinases regulate transcriptional activation of the interstitial collagenase gene, MMP-1, in interleukin-1-stimulated synovial fibroblasts. Arthritis Rheum. 1996;39:574–582. doi: 10.1002/art.1780390406. [DOI] [PubMed] [Google Scholar]
- 53.Vuori K, Hirai H, Aizawa S, Ruoslahti E. Induction of p130cas signaling complex formation upon integrin-mediated cell adhesion: a role for Src family kinases. Mol Cell Biol. 1996;16:2606–2613. doi: 10.1128/mcb.16.6.2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vuori K, Ruoslahti E. Tyrosine phosphorylation of p130Cas and cortactin accompanies integrin-mediated cell adhesion to extracellular matrix. J Biol Chem. 1995;270:22259–22262. doi: 10.1074/jbc.270.38.22259. [DOI] [PubMed] [Google Scholar]
- 55.Wada A, Fukuda M, Mishima M, Nishida E. Nuclear export of actin: a novel mechanism regulating the subcellular localization of a major cytoskeletal protein. EMBO J. 1998;17:1635–1641. doi: 10.1093/emboj/17.6.1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Werb Z, Tremble P M, Behrendtsen O, Crowley E, Damsky C H. Signal transduction through the fibronectin receptor induces collagenase and stromelysin gene expression. J Cell Biol. 1989;109:877–889. doi: 10.1083/jcb.109.2.877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Westermarck J, Seth A, Kähäri V M. Differential regulation of interstitial collagenase (MMP-1) gene expression by ETS transcription factors. Oncogene. 1997;14:2651–2660. doi: 10.1038/sj.onc.1201111. [DOI] [PubMed] [Google Scholar]
- 58.Wright W E, Binder M, Funk W. Cyclic amplification and selection of targets (CASTing) for the myogenin consensus binding site. Mol Cell Biol. 1991;11:4104–4110. doi: 10.1128/mcb.11.8.4104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yamagata T, Nishida J, Sakai R, Tanaka T, Honda H, Hirano N, Mano H, Yazaki Y, Hirai H. Of the GATA-binding proteins, only GATA-4 selectively regulates the human interleukin-5 gene promoter in interleukin-5-producing cells which express multiple GATA-binding proteins. Mol Cell Biol. 1995;15:3830–3839. doi: 10.1128/mcb.15.7.3830. [DOI] [PMC free article] [PubMed] [Google Scholar]