An image-based machine learning approach finds mechanisms underlying variable activation of NF-κB pathway in single cells.
Abstract
Individual cells are heterogeneous when responding to environmental cues. Under an external signal, certain cells activate gene regulatory pathways, while others completely ignore that signal. Mechanisms underlying cellular heterogeneity are often inaccessible because experiments needed to study molecular states destroy the very states that we need to examine. Here, we developed an image-based support vector machine learning model to uncover variables controlling activation of the immune pathway nuclear factor κB (NF-κB). Computer vision analysis predicts the identity of cells that will respond to cytokine stimulation and shows that activation is predetermined by minute amounts of “leaky” NF-κB (p65:p50) localization to the nucleus. Mechanistic modeling revealed that the ratio of NF-κB to inhibitor of NF-κB predetermines leakiness and activation probability of cells. While cells transition between molecular states, they maintain their overall probabilities for NF-κB activation. Our results demonstrate how computer vision can find mechanisms behind heterogeneous single-cell activation under proinflammatory stimuli.
INTRODUCTION
Individual cells show unpredictable and highly variable responses in a wide range of contexts, from immune signaling to transcriptional activation and to drug response (1–7). For example, following stimulation with signaling molecules, a portion of cells in a population will strongly activate inflammatory response pathways such as nuclear factor κB (NF-κB) and downstream transcription factors, while others will completely ignore the stimulus and will not activate (8). Determining the sources of cellular variability is of immense importance for fundamentally understanding gene regulation, signaling, immunity, and development, as well as in predicting variable drug response and tolerance (9–12). Despite previous demonstration of heterogeneous signaling responses in a wide range of contexts (8, 13, 14) such as development, immune response, infection, and cancer, it remains difficult to explore the molecular and cellular mechanisms that drive variable behavior in single cells, and the underlying causes of cellular variability are unknown in many contexts.
Here, we study the NF-κB system to investigate the underlying sources of cellular heterogeneity using a computer vision approach applied to microfluidic live-cell imaging experiments. NF-κB is a key transcriptional pathway that is activated by many signaling molecules involved in immunity (15, 16) and controls the expression of hundreds of proinflammatory and cell fate genes (17). Dysregulation of NF-κB is implicated under many physiological conditions including infection, autoimmunity, and cancer (15, 18). Live-cell imaging and single-cell analysis have shown that NF-κB activation is highly variable in single mammalian cells. When cells are stimulated with signaling molecules [i.e., tumor necrosis factor (TNF) or lipopolysaccharide], some of them show complete cytoplasm-to-nucleus translocation of the p65 transcription factor (the hallmark of pathway activation), while others ignore the stimulus and show no translocation and no NF-κB target gene expression (Fig. 1A). The fraction of cells that respond to a stimulus (signaling input) increases in a dose-dependent manner (19), and despite this seemingly noisy single-cell behavior, NF-κB nevertheless manages to mount specific responses to different signaling molecules, taking into account their concentrations and temporal ordering, and regulates gene expression in an input-specific manner (3, 13). Despite the plethora of research uncovering response relationships to input conditions, it is still unclear as to what underlying molecular components or mechanisms control variability in NF-κB pathway activation alongside the input. The accurate functioning of signaling pathways such as NF-κB is crucial to maintaining healthy immunity and immune development, and integration of internal cell state and external stimuli is important to understand the functional consequences of noise in the system (Fig. 1B). Despite several studies that substantially contributed to our understanding of cellular variability and noise (1, 20–24), the ability of NF-κB and other signaling pathways to accurately interpret complex environmental signals and create specific gene expression responses is mostly unexplained.
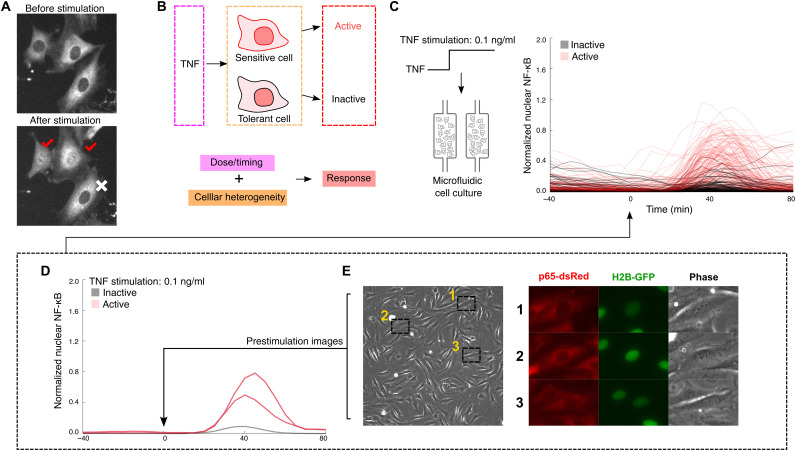
Fig. 1. Prestimulation cell images can be used to generate a predictive model for NF-κB activation.
(A) Under TNF stimulation, a fraction of individual cells persists in an unactivated state and do not show nuclear NF-κB localization, while others activate and NF-κB transcription factor p65 accumulates in the nucleus. Example images show activated cells, indicated with red check marks. (B) Analysis of single cells under constant dose of TNF shows variable single-cell activation in the population: A given cell may or may not respond to the TNF stimulus. It is unclear whether single-cell variability is due to purely stochastic processes (i.e., if a given cell can randomly achieve activated state) or whether it is deterministic where only sensitive cells activate under the TNF input. (C) We use microfluidic cell culture to stimulate cells with TNF signals and image the nuclear localization of NF-κB over time in single cells. Analysis of individual cells reveals NF-κB localization traces [TNF stimulation (0.1 ng/ml) shown on the right; stimulation starts at t = 0]. (D) Single-cell traces reveal heterogeneous activation and subpopulations of active and inactive cells. (E) We record prestimulation images of mouse 3T3 fibroblast cells that express p65-dsRed and histone 2B (H2B)–green fluorescent protein (GFP) reporters and feed them into our machine learning pipeline.
The challenge in understanding the cause of heterogeneous responses in signaling and gene regulation is a classic observer effect problem: We can only identify the responding (i.e., activated) cells after we stimulate the cell population with signaling molecules, which will inevitably perturb the cellular states that we wish to examine. Many signaling pathways, including NF-κB, contain multiple feedbacks that, upon exposure to an external signal, rapidly change the molecular composition of the pathway (25), which fundamentally limits the accuracy and power of repeat stimulation experiments (8, 26). This presents a fundamental difficulty in determining how preexisting cell-to-cell differences affect the probability of any given cell to respond to a stimulus, especially in short-term cell states that arise from systemic changes in negative and positive feedback regulation. One way to circumvent this problem is by use of mathematical modeling (27, 28). Modeling of pathway dynamics can reveal important insights and general patterns of noisy events and can offer many plausible explanations for cellular heterogeneity (29) but is bound by the underlying assumptions of the models or requires monitoring multiple components of the system for validation (30). Because of these experimental and theoretical limitations, molecular mechanisms underlying important cellular behaviors such as variable drug response, digital pathway activation, and signal tolerance are limited and often unclear (8, 31–33).
To study the molecular mechanisms behind variable NF-κB activation in single cells, we adopted an image-based machine learning approach to predicting which individual cells will activate the NF-κB pathway in response to an inflammatory stimulus. On many occasions, machine learning algorithms have been shown to exceed human decision-making on complex game problems (34, 35). Machine learning has also previously been used to extract pathophysiological outcome predictions from images of tissue and inform classification of differentiation, disease state, and infection (36, 37). By imaging living cell morphologies before, during, and after chemical stimulation, we were able to use the cell image before stimulation to predict whether that cell will activate the NF-κB pathway. We developed an image-based support vector machine (SVM) to predict outcomes of chemical stimulation in individual cells (see the Supplementary Materials). Our computer vision–based method classifies cells into responding and nonresponding groups based solely on the unperturbed cell’s image and is able to predict which cells will respond to or ignore TNF stimulation with 79.4% accuracy. As this prediction is performed on cells that are not chemically stimulated, our approach allows studying how the cell’s unperturbed state differs between responding versus nonresponding cells.
RESULTS
Machine learning predicts single-cell NF-κB activation from prestimulation images
To develop a predictive machine learning model for NF-κB activation in individual cells, we first performed live-cell stimulation experiments to generate a reference dataset. We used a high-throughput microfluidic cell culture platform (38) to chemically stimulate and quantitatively measure NF-κB response in cultured mouse fibroblast cells (Fig. 1, C to E). These cells express p65-dsRed and histone 2B (H2B)–green fluorescent protein (GFP) fluorescent reporters (8) to track NF-κB nuclear translocation in real time. In activated cells, cytoplasmic NF-κB (p65) rapidly moves into the nucleus in a digital fashion (8). Cells were first imaged unperturbed for 1.5 hours, stimulated using the automated microfluidic system with TNF (0.1 ng/ml), and monitored for 6 hours. Custom image analysis software was used to track the nuclear localization of p65 and assign a label to each cell (activated versus not activated) (38).
To determine the characteristics of single cells that are responsible for activation, we then assigned representative features for single-cell images and analyzed predictor correlations (see the Supplementary Materials) on our defined cell features (Fig. 2A) in a single dosage dataset (fig. S1). We also expanded the experimental dataset from a single TNF dose to a range of doses and reported their individual accuracies (0.005 to 5 ng/ml; n = 3456; fig. S2A). Analyzed together using the SVM algorithm, the total accuracy is 79.4%. We also tested an alternate cell activation criterion using different single-cell p65 trace patterns as categories (fig. S3A) on a single dose (0.05 ng/ml) and found that while some trace patterns are able to be classified by the model, this does not generally hold and has lower accuracy than binary classification (fig. S3B). For our binary classifier labeling, we use a mean nuclear p65 peak height threshold of 500 (nuclear p65peak − nuclear p65t=0) (fig. S4A). We also tested with different machine learning models (table S1) and using different thresholding values for activation based on peak height and chose our threshold to balance the sample number and prediction accuracy of activated cells. While both convolutional neural network (CNN) and tree classifiers are able to classify activation more accurately than the SVM, feature analysis for both CNN and tree classifiers is typically inaccessible or difficult. Overall, we can accurately predict individual cells’ TNF response from their prestimulation images, which strongly suggests the existence of deterministic causes underlying NF-κB response in single cells.
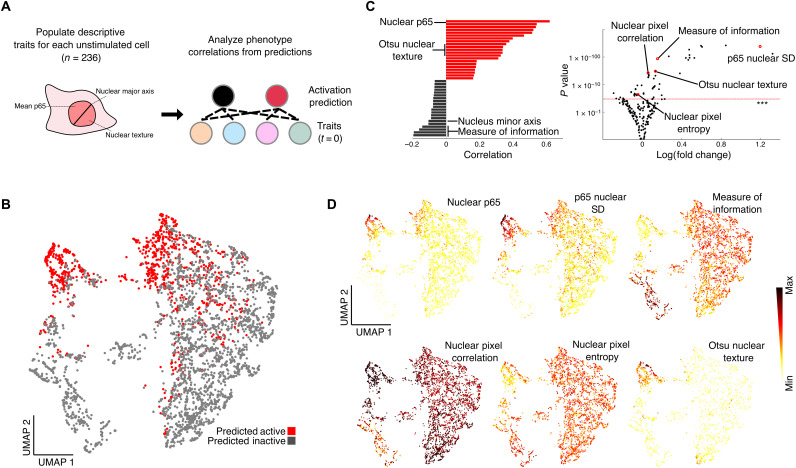
Fig. 2. Machine learning predicts NF-κB activation in single cells from prestimulation images.
(A) We generate descriptive traits for each single-cell image and analyze correlations between prestimulation traits and NF-κB activation probability. (B) Using a subset of highly predictive features (n = 8), uniform manifold approximation and projection (UMAP) corroborates the clustering of a highly predictive fraction of cells that are TNF sensitive and TNF resistant. The cells indicated with red dots have high probability of activating NF-κB under TNF stimulation. (C) We determined the top feature of activation probability to be the mean nuclear fluorescence of the NF-κB (p65-dsRed) signal in the nucleus before any exposure to TNF. The plot shows the correlation of traits to single-cell activation probability, with log(fold change) of traits from predicted active cells to predicted inactive cells on the x axis and significance of difference on the y axis determined by t test. Other highly predictive features include the SD of nuclear p65, mean nuclear phase intensity, major axis length and a texture feature describing information measure of correlation in the nucleus, and aggregative image features such as Otsu dimension, and Segmentation-based Fractal Texture Analysis (SFTA) (***P = 0.001). (D) UMAP plots of several highly predictive features determining NF-κB activation in single cells.
Leaky NF-κB localization is the primary predictor of single-cell activation
To understand the characteristics that underlie activation, we then identified a subset of highly predictive features that correlate with the cell’s likelihood to become activated, including the basal (prestimulus) level and SD of nuclear NF-κB and several texture features (Fig. 2B). Using these features, cells can be visualized by uniform manifold approximation and projection (UMAP), which present “TNF-sensitive” and “TNF-resistant” cell regions (Fig. 2C). Each point on the UMAP projection represents a cell that occupies a particular state with different amplitudes for the image features that we previously determined. The UMAP plot shows cells segregated through variables unrelated to activation; we found that features relating to size and sum of protein are responsible for creating this divide (fig. 4B). Analysis of the contribution of individual cell morphology features to the prediction revealed that most of the variation in single-cell predictions is explained by a single feature, the nuclear p65-dsRed levels before stimulation (r = 0.62), which showed a significant difference between TNF-sensitive and TNF-resistant cells.
While nuclear p65 was the most significant, we also explore the relationship between different features to see the effect of coupling on our variables: Despite the very significant P values for many features, often times, many features are coupled in nonlinear ways. While a large amount of variance can be explained using nuclear NF-κB, additional features increase the prediction for different cell states (fig. S5, A and B). The combination of p65, H2B, and phase features can all be used in one decision tree to isolate a subpopulation of inactive cells (fig. S5D). To understand some of the underlying processing occurring within the algorithm, we looked into correlations in this subpopulation. Dissecting this highly coupled subpopulation from fig. S5D, in turn, showed remarkable correlation with many features that gave high P values but did not have high fold changes.
Although nuclear p65 before stimulation was the most predictive feature for cell activation, nuclear texture features and H2b channel features are also highly predictive components of the classifier. To investigate whether other variables were acting as proxies for nuclear p65, we look to see whether we could substitute phase image alone for biological markers in the other channels (accuracy, 73%; fig. S5C and table S1). While some of the features did show high correlation (Otsu: r = 0.600), many other features had much lower correlations (fig. S6). The clustering of correlations shows that there may be multiple underlying biological features that are being represented. For example, H2b feature sensitivity may be indicative of exogenous factors such as nuclear import/export (39) and cell cycle (40) playing a role in NF-κB activation probability. Using gray level co-occurrence matrix features from the phase channel, we used metrics such as entropy and measure of information that reflect different features of adjacent image pixels to determine whether nuclear texture was predictive of NF-κB activation. The mapping of these various texture features onto our UMAP space (Fig. 2D) show that highly activated cells have more heterogeneous nuclear morphology with low pixel correlation and that resistant cells have more homogenous nuclei with high pixel correlation. These features are suggestive of chromatin openness (41–44) and epigenetic factors having a potential relationship to NF-κB activation.
Cells transition between multiple states but maintain their overall propensity for pathway activation
To further characterize the unperturbed cell states, we look at the time window leading up to activation. By using multiple prestimulation images, we have the unique opportunity to look at the trajectories of individual cells in the UMAP space over time and see the short-term state transitions of a given cell before exposure to TNF (Fig. 3A). Unexpectedly, we find that the resting (unstimulated) cells do not occupy a fixed parameter state and instead show dynamic transitions between different states. That is, important state features such as nuclear p65 level and nuclear texture (indicative of chromatin availability) actively fluctuate in unstimulated cells. Plotting several features over time for single cells reveals coordinated changes in nuclear texture and nuclear NF-κB (p65) before stimulation (Fig. 3B) and is further observed by plotting the covariance between variables (fig. S6). As cells change over time, the image features that are changing are often correlated. This could suggest that there is compensation from some features to others, as cells dynamically change state. While there are often changes in state features during these transitions, the overall prediction score for NF-κB activation for these single cells does not change significantly (Fig. 3C). The cells maintained their overall propensity for NF-κB activation under TNF stimulation while actively transitioning between multiple cellular states. This result indicates that cellular sensitivity toward cytokine stimulation may be stable despite the fluctuations present in our features and within the cells before stimulation. To look at how stable the prediction score is at earlier time points, we look at single-cell autocorrelation of the prediction score. The correlation remains high even 35 min before stimulation (r = 0.78, n = 1109 cells, pairwise Pearson’s correlation) (Fig. 3D). While 35 min may not be enough to capture the regime of long-term state (e.g., cell cycle), it captures the relevant time scales of the NF-κB system state right before stimulation, as cells experience changes in pathway protein composition even at steady state (25). This indicates that while the cells do make transitions on the UMAP space to different states, they tend to stay within the same grouping with respect to activation prediction.
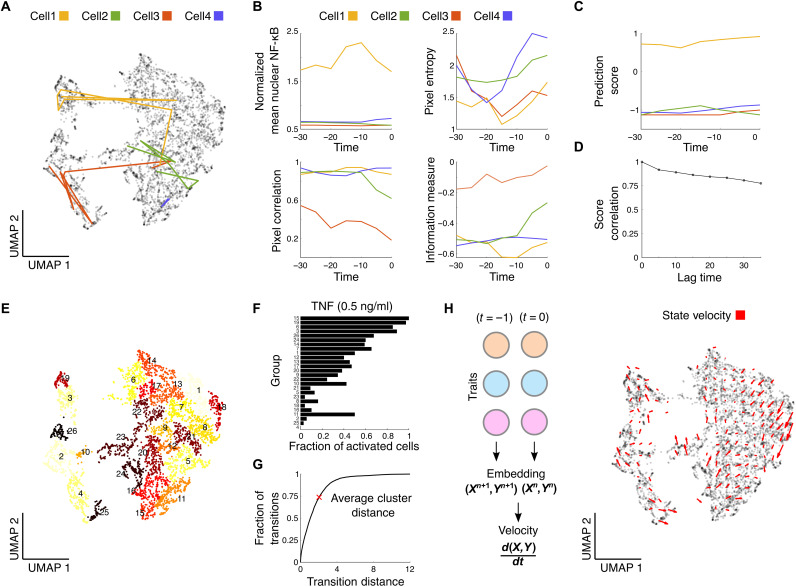
Fig. 3. Resting cells dynamically transition between different states but maintain their overall NF-κB activation probability.
Imaging of single cells before, during, and after stimulation enables analysis of single-cell state stability. (A) We use selected cellular features across multiple time points to analyze the trajectory of single cells across the latent UMAP space before stimulation. Colored lines show the temporal trajectories of four example cells on the UMAP plot before stimulation with TNF. Cells transition between various points before TNF stimulation. (B) Individual cells show coordinated changes in state features while transitioning. We show the dynamic changes in the level of various predictive features for these cells. Time is given in minutes before stimulation (stimulation starts at t = 0). (C) While cells transition between different points with different transition distances, their probability to activate NF-κB (given by the prediction score of our algorithm) remains the same. (D) Autocorrelation of the prediction score among single cells across prestimulation time points remains stable 35 min before stimulation (r = 0.78, n = 1109 cells, pairwise Pearson’s correlation). (E) Single cells are clustered in the UMAP space by local adjacency into communities to look for activation differences by state (color indicates group). (F) Analyzing end point data after TNF stimulation by individual state clusters shows different fractions of activated cells for different states. There is activation similarity in adjacent clusters, and (G) 73.73% of all cell transitions are within or to an adjacent cluster. Shown is the cumulative distribution function of state transitions by transition distance (red X indicates average distance between clusters). (H) Aggregated trajectories reveal a state velocity depicting activation score movement across the population.
To confirm that the cells maintain their activation probability through stimulation, we clustered all single cells to look for differences between states after TNF is introduced (Fig. 3E and see the Supplementary Materials). Analysis of the activation fraction (i.e., the population fraction of cells activating NF-κB) after TNF stimulation of the different state clusters reveals that the different states indeed have different activation fractions (Fig. 3F). Adjacent cell clusters have similar activation probabilities, and 73.71% of all transitions happened within or to an adjacent state cluster (Fig. 3G). That is, while cells transition between various parameter states, they maintain their overall sensitivity toward TNF stimulation. We also looked at how dosage plays a part in the magnitude and fraction of activation. While the magnitude of activation followed similar behavior across states (fig. S7B), the fraction of activated cells varies across dose for different states (fig. S7C). While some cells remain persistently sensitive or resistant, there are other cells that have their activation profiles dictated by TNF dose. This supports the hypothesis that preexisting cell features govern activation or resistance to TNF input.
To see whether there was regularity in transitions, we looked at average cellular transitions across the UMAP space. Aggregating single-cell trajectories across the UMAP space can be used to understand the state velocity across the population (Fig. 3H). We find that the average transition of single cells follows generalized patterns and indicates that while the transitions might appear non-uniform at the single-cell level, there are average population level movements.
Mechanistic modeling shows that inhibitor of NF-κB/p65 ratio drives p65 leakiness and activation probability of a given cell
Our machine learning analysis revealed that leaky (prestimulation) p65 localization to be the main predictor of single-cell activation of NF-κB. There are many different NF-κB regulators that can influence p65 nuclear localization in resting unstimulated cells. This “leakiness” has been attributed to mechanisms of nucleocytoplasmic shuttling of inhibitor of NF-κB (IκB) and NF-κB (p65) in resting cells (39, 45, 46), and observations that basal nuclear NF-κB abundances have important biological consequences on pathway activation have been elucidated (47, 48). However, the NF-κB pathway is robust to environmental fluctuations and noise, and many built-in negative feedback mechanisms (Fig. 4A) prevent spontaneous activation (nuclear import of p65) in the absence of stimulation. Nevertheless, our imaging data reveal that many cells show minute amounts of “leaky” nuclear p65 (localization without stimulus) (fig. S8), which has been observed in cells (39) but not thoroughly investigated regarding activation. We find that this small but significant prestimulus p65 localization predetermines the sensitivity of the mouse fibroblast cells in responding to upcoming TNF challenges.
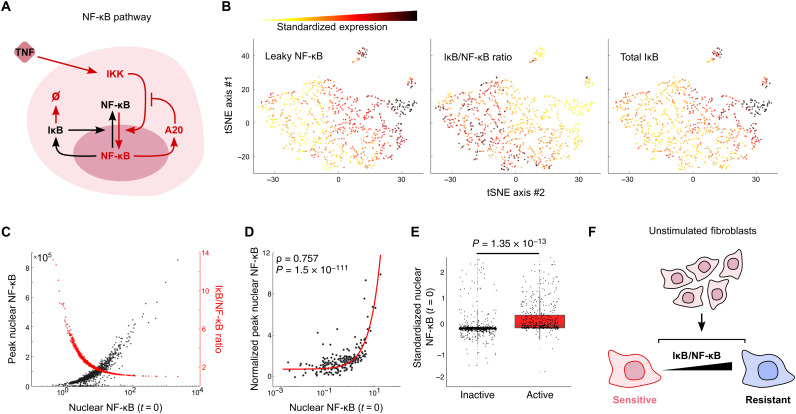
Fig. 4. Simulations suggest that leaky NF-κB localization and overall NF-κB response is predetermined by the ratio of IκB to NF-κB proteins in single cells.
(A) Simplified schematic of the NF-κB pathway used in simulations. IκB provides negative feedback to the pathway, preventing NF-κB nuclear localization in unstimulated cells. IKK, IκB kinase. (B) t-distributed stochastic neighbor embedding (tSNE) mapping of simulated single cells on NF-κB pathway protein levels shows anticorrelated nuclear NF-κB and IκB/NF-κB ratio in single cells. (C) Simulations predict a correlation between nuclear NF-κB at t = 0 and NF-κB peak height after stimulation and, consequently, an inverse correlation between NF-κB at t = 0 and the IκB/NF-κB ratio. (D) Activated live single cells confirm the prediction and show significant correlation between nuclear NF-κB at t = 0 and NF-κB peak height after stimulation. (E) Prestimulation NF-κB nuclear fluorescence accounts for a high degree of variance, and activated live single cells have a significantly higher level than inactive cells. (F) Simulations suggest that increasing IκB/NF-κB ratio makes cells more resistant to activation under TNF stimulation.
To understand the molecular mechanism behind p65 nuclear leakiness and, specifically, how it could lead to increased TNF sensitivity and NF-κB activation probability, we explored a mathematical model of NF-κB pathway in single cells (8, 14). This model accurately reproduces the experimentally measured NF-κB dynamics in a wide range of conditions in the mouse fibroblast cells used in our study (1, 6, 11, 12). We performed simulations and generated thousands of dynamic traces that describe nuclear NF-κB (p65) localization in single cells stimulated with TNF. We then analyzed the molecular composition of each simulated cell and its NF-κB activation profile. We found that variability in levels of IκB, the main inhibitor of NF-κB (p65) keeping it in the cytoplasm, explains the observed NF-κB leakiness. In addition to binding to p65 and keeping it in the cytoplasm in unstimulated cells, IκB acts as a dynamic negative feedback regulator of the pathway since IκB is a direct target gene of NF-κB. IκB is produced when NF-κB pathway is activated and p65 enters the nucleus. Note that the leaky nuclear p65 that we observe is correlated to total p65 present in the cell. Because many of these variables are entangled in the pathway, changes in one protein typically reflect changes across many proteins in the pathway.
Using dynamic simulations, we perturbed the IκB/NF-κB ratio in single cells before TNF stimulation and determined the resulting likelihood of pathway activation for single cells (n = 1000) (fig. S9B). We find a major difference in cellular activation probability and peak NF-κB amplitude for different IκB levels (fig. S9C). Counterintuitively, cells with high initial IκB levels require a smaller TNF dose to achieve NF-κB activation, whereas cells with low initial IκB levels are very resistant to any TNF dose. This unexpected finding is explained by the facts that the probability of activation depends on the IκB/NF-κB ratio and not the total level of IκB and that the IκB/NF-κB ratio is anticorrelated to the total IκB level (Fig. 4B and fig. S9A). Expression differences between activators and repressors have the capability of producing large variance in phenotype differences similar to what we see with NF-κB activation (49–51). Our simulations suggest that a preexisting imbalance in the NF-κB negative feedback is responsible for increased TNF sensitivity in single cells and that the activation probability of individual cells is predetermined by the molecular ratio of IκB to NF-κB in the cell (Fig. 4C).
Experiments show that IκB/NF-κB ratio is the main determinant of cellular activation probability
To validate our modeling predictions experimentally, we analyzed live single cells by time-lapse microscopy and immunofluorescence. First, we found that those cells that responded to TNF stimulus had total nuclear NF-κB (p65) at t = 0 significantly correlated with the NF-κB peak height after stimulation (P = 1.5 × 10−111; Fig. 4D) and, on average, show a fivefold higher level of leaky nuclear p65 before stimulation than those that did not respond (P = 1.35 × 10−13; Fig. 4E). Note that the low level of nuclear leakiness that we observe is only about 12% of the total fluorescence in a given cell and is far below what is seen during activation. This small but significant difference shows that the regulation of the steady state (i.e., prestimulus) NF-κB localization is an important determinant of NF-κB activation and demonstrates the power of using computer vision to analyze single-cell responses.
To experimentally validate our hypothesis that the IκB/NF-κB ratio is driving activation probability from the modeling, we stained unstimulated cells for IκB-α protein expression and analyzed the relationship between IκB and NF-κB in individual cells using immunofluorescence (Fig. 5A). We segmented the nuclear and cytoplasmic compartments and found that, as predicted by our simulations, there is a significant inverse correlation between the IκB/NF-κB ratio and leaky nuclear p65 localization (ρ = −0.26, P = 2.7 × 10−11; Fig. 5c), which is the main feature that determines cell activation upon TNF stimulation.
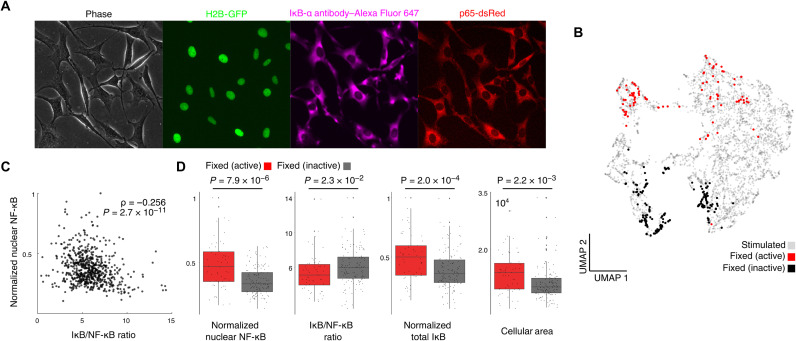
Fig. 5. Single-cell activation is largely predetermined by the NF-κB/IκB ratio.
We validated our machine learning and modeling predictions in immunofluorescence staining experiments. (A) Image of unperturbed 3T3 cells stained with IκB-α with fluorescent p65 and H2B reporters. (B) High-scoring fixed cells are mapped onto UMAP. (C) There is a significant inverse correlation between leaky nuclear NF-κB localization and IκB/NF-κB ratio. (D) IκB/NF-κB ratio is significantly correlated with nuclear NF-κB state at t = 0 in predicted fixed cells, and there is a significant difference between IκB/NF-κB ratio, leaky nuclear NF-κB localization, total IκB, and cellular area.
Next, we mapped the fixed-and-stained cells onto our UMAP visualization of the previous live imaging data (fig. S2B), allowing us to infer IκB localization on our previous experiments for high-scoring cells (Fig. 5B). We find that there is indeed a significant difference in nuclear NF-κB, IκB/NF-κB ratio, total IκB, and cellular area, validating our prediction that IκB/NF-κB ratio is driving activation probability in NF-κB (Fig. 5D).
DISCUSSION
The underlying molecular sources of noisy and variable cellular responses can be challenging to study because of a fundamental difficulty of analyzing cellular states without disturbing them. Here, we adopted an image-based machine learning approach to predicting the identity of cells that will respond to a biochemical signal and experimentally investigated them. Overall, our demonstration of machine learning in the identification and exploration of noisy NF-κB activation enables cellular analysis through a prospective lens. We supplemented our computer vision approach with additional single-cell experiments and mechanistic modeling to study how cellular states affect the probability of heterogeneous signaling in immune regulation.
Our study revealed several molecular determinants of cellular states that lead to variable signaling responses in the NF-κB pathway. Previously, there has been conjecture as to how NF-κB signaling is regulated, and metrics such as cell cycle (40) stage and basal NF-κB levels (39) have previously been implicated. In addition to this, we find that the cell-to-cell variability in NF-κB activation can largely be explained by a preexisting difference in the ratio of the NF-κB and its inhibitor, IκB. In cells that are not stimulated by a ligand, the p65 (NF-κB) transcription factor is bound by its inhibitor IκB and is kept in the cytoplasm. When the cells are exposed to a ligand, IκB degrades and NF-κB transcription factors such as p65 rapidly translocate to the nucleus and regulate inflammatory genes. These genes are expressed in a ligand-specific manner and have important functions in immunity, pathogen-host interactions, and development of adaptive immune cells. Therefore, studying which cells would show NF-κB translocation (the hallmark of NF-κB pathway activation) is important to understanding of cellular information processing and cellular variability during immunity.
We find that the cells with a lower IκB/NF-κB protein ratio show small amounts of translocation to the nucleus even without ligand stimulation, which we term leakiness. We also found that leaky cells that have a low IκB/NF-κB ratio are more sensitive to TNF stimulation and readily respond when exposed to this signal. It remains an open question as to what leads to this variation and leakiness in the IκB/NF-κB ratio: One intriguing possibility is that epigenetic variance in genes encoding for NF-κB network components enforce the variable activation chance that we observe akin to certain modes of drug tolerance (32, 52). These epigenetic changes could perhaps explain the observed predicative power of nuclear texture features that we measured (Fig. 2D), and we plan to address this possibility in follow-up studies.
Another possible explanation for our observations is that cells that are less sensitive to TNF (i.e., those that are not leaky) do not have a functioning NF-κB network. However, in our experiments, the cells that show leakiness and those that are not leaky all activate and show complete NF-κB translocation at sufficiently high TNF doses, which shows that they have intact IκB kinase/NF-κB network and transport mechanisms. On the other hand, our pathway simulations showed that simple changes in the protein abundance of p65 and IκB are sufficient to observe leakiness and increased sensitivity to TNF. These protein abundance changes can happen because of a variety of factors, including the possible epigenetic factors that we discussed above, or cells being exposed to previous signals that change their transcriptional states.
Through dynamical imaging of cells before and after stimulation, we found that resting (unstimulated) cells mostly maintain their sensitivity state at various time points, indicating the stable nature of the underlying NF-κB architecture in response to TNF. However, while cells maintain their overall sensitivity toward future TNF stimulation, these cells actively sample various molecular states. We find that most of the NF-κB response variability is explainable, but the cells do undergo spontaneous state transitions, which is a plausible explanation for the NF-κB activation variability that cannot be explained by our algorithm alone. While the NF-κB system has many components to help maintain its sensitivity toward chemical signals, there is still an element of stochastic state fluctuations that result in creating diversity in single-cell and, ultimately, population behavior. This phenotypic diversity was shown to be advantageous for a cell population when responding to rapidly changing environmental conditions. Cytokines such as TNF often activate multiple signaling pathways, and stratified state-based activation could provide a way to achieve specific responses for specific cell states at different dose ranges.
Our findings suggest an alternate characterization for signaling variability in mammalian signal transduction and offer the interesting possibility that activation heterogeneity is a proxy for cell state that a cell can interpret and act on (53). The prevalence of these state niches is further evidence that cell populations are well equipped at differentiating diverse and dynamic signals through cellular specialization even in an equilibrium population. The matching of activation probability, heterogeneity in the IκB/NF-κB ratio, and nuclear morphology is indicative of regulation mechanisms in place for decoding different signaling patterns for a variety of different cellular states.
MATERIALS AND METHODS
Experimental model and subject details
Knockout p65−/− mouse 3T3 fibroblasts were engineered with p65-dsRed under the native p65 promotor (8), and a minimum fluorescence clone was selected to represent endogenous expression of NF-κB and the pathway dynamics. A ubiquitin promotor–driven H2B-GFP cassette provides a nuclear marker for image processing.
Microfluidic cell culture
Cell culture chambers are made of polydimethylsiloxane and coated with fibronectin (FC010-10MG) overnight. Cells are seeded at a constant density of ~20,000 cells/cm2. Cells are taken at 100% confluence by incubating with 0.25% trypsin-EDTA (25200-056) for 5 min before loading and are cultured for 5 hours before stimulation. Cells were cultured using standard conditions for cell culture (5% CO2 and 37°C) and maintained using an incubation chamber during imaging. TNF-α (PMC3014_3671982503) was diluted in FluoroBrite Dulbecco’s modified Eagle’s medium (A1896701) with 2× GlutaMAX (35050061), penicillin/streptomycin (15140-122), and fetal bovine serum (16140071) for stimulation of NIH 3T3 cells. Vials of stimulation media were pressurized at 5 psi, kept on ice, and connected to the chip via microbore tubing (PEEK, VICI). The microfluidic device is mounted on the microscope.
We image using Nikon Eclipse Ti2 microscope to capture both phase and fluorescence images of cultured cells at a 20× magnification. We use a Hamamatsu ORCA-Flash4.0 V3 Digital CMOS Camera (C13440) to capture an image every 5 min for the duration of the experiment. Custom MATLAB scripts were used for image processing. NF-κB activation was quantified as mean nuclear fluorescence intensity after background correction. For peak analysis, data were smoothed and normalized (MATLAB functions smooth and z-score), followed by peak detection. Activation label is crafted from binary threshold for nuclear p65.
Fixed-cell immunofluorescence
Fibroblasts were seeded in microfluidic chamber and allowed to attach. Cells were rinsed with a 1-min wash of phosphate-buffered saline (PBS) and fixed using a 4% paraformaldehyde solution at 5 hours after attachment. Cells were fixed for 10 min at room temperature and blocked and permeabilized with a 10% bovine serum album (BSA) and 0.5% Triton X-100 solution in PBS for 1 hour. Cells were then incubated with primary antibodies in a staining solution (2% BSA and 0.1% Triton X-100 in PBS) for another 1 hour at 37°C. Cells were then washed again with PBS and incubated with secondary antibodies in staining solution for 1 hour at 37°C. Primary antibody used was a rabbit polyclonal to IκB-α (ab7217). Secondary antibodies were goat polyclonal secondary antibody to rabbit immunoglobulin G H&L (Alexa Fluor 647) (G378361).
For the image analysis of fixed cells, we used cell images taken at the time point immediately preceding fixation (5 min). We then analyzed fixed images and used cell coordinates to match up the fixation analysis with the image analysis that goes into the machine learning pipeline. The images taken immediately before fixation are cells in an unperturbed state similar to prestimulation cells in the dosage experiments. We did not have to do anything in the way of specifically transforming/aligning either to map with each other in UMAP.
Data processing
Predicting the activation of cells using microscopy images can be formulated as an object classification problem. Data processing details are as follows: We first use minimum-maximum normalization to scale the pixel intensity into the range of [0, 1]. Then, we crop each cell as a 64 by 64 image patch centered around the cell nucleus. We split the dataset into 10 folds and set up cross-validation experiment to evaluate our model. In each run, eight folds are used for training, one fold is used for validation, and the last fold is used for evaluation.
Extraction of texture features and SVM model
Hand-crafted feature extraction was performed during nuclear segmentation and for each tracked single cell. The number of all extracted parameters is 236 and is shown in table S2. Texture extraction was performed using several custom MATLAB scripts. SVM model was run using cubic kernel with an overall accuracy of 73%. Fitting was performed with 10-fold cross-validation with 10% dropout on a dataset with [0.005, 0.05, 0.5, 5 ng/ml] TNF input images (n = 3456) and an independent dataset with TNF (0.1 ng/ml) input images (n = 2113) before stimulation (fig. S4). Dose feature is represented as a categorical variable in the analysis. Feature creation code is provided at https://github.com/parthivapatel/ML_cellim.git.
Validation with additional models
We validate and report the accuracies using other classifiers as well (table S1). All models use the same fivefold cross-validation split. The training fold is split into train validation to grid search the best hyperparameters. Last, we report the average performance over the test folds. The models have the following hyperparameter settings.
Logistic regression
We use logistic regression with liblinear solver and L2 regularization and optimize the best C values (for regularization) on the validation folds.
SVM with rbf kernel
We tune this SVM with “rbf” kernel to optimize different C values and gamma values (for kernel).
SVM with polynomial kernel
This SVM uses a polynomial kernel. The hyperparameters are the C value and the degree of the polynomial kernel.
Random forest
We choose to use 100 decision trees as the base estimators for the random forest classifier.
AdaBoost
This model has the same number of estimators with the random forest. The base estimator is a decision tree classifier with a maximum depth of 1.
Convolutional neural network
There are two convolutional layers, each followed by max-pooling layers. After flattening the output of the second max-pooling layer, two fully connected layers are applied with a tanh activation function. Dropout layers follow both dense layers with a dropout ratio of 0.5 during training. The last layer is a sigmoid function to calculate the probability of two classes. We used the Keras library (https://keras.io/) for the CNN implementation. We used TensorFlow (www.tensorflow.org) as the backend for Keras. We used scikit-learn (https://scikit-learn.org/) for the other classifiers.
State velocity
Individual cells at time 0 were mapped onto UMAP using parameters (nearest neighbors = 12, Chebyshev distance metric, minimum distance = 0.1) across multiple doses. Multiple additional prestimulation time points were then embedded onto the UMAP projection using nearest-neighbor embedding. UMAP embedding was turned into a grid of (i, j) components and morphological velocity was approximated using , where is the vector of displacement of a prestimulation single cell mapped onto UMAP between t and t − 1 in the grid (i, j) for all cells. The average displacement vector across all cells is shown in the state velocity.
Clustering
Clustering in Fig. 3 was performed using Louvain community detection derived from created adjacency matrix from the nearest-neighbor descent algorithm on cell database with hand-crafted features. Single cells move across Louvain partitions between time points at a frequency of less than 73.71%. Clustering for cell traces used in fig. S4 was performed through k-means clustering using a k = 6.
Statistical tests
Figure 2C uses two-sample t test to evaluate P values with n = 1028 for positive predicted and n = 2428 for negative predicted. The red line shows significance level of P = 0.001. Figure 4E uses two-sample t test to evaluate P value with n = 1028 for positive predicted and n = 2428 for negative predicted.
Figure 5D uses two-sample t test to evaluate P value on high-scoring prediction fixed cells with n = 45 for positive predicted and n = 74 for negative predicted. Misclassification error was minimized using high-scoring cells only.
Mathematical modeling of NF-κB pathway
Using a hybrid stochastic deterministic model of the NF-κB pathway, published previously (8, 14), we simulated 1000 single cells exposed to 20 TNF concentrations. The hybrid model based on Gillespie algorithm uses verified intrinsic noise (8, 14) in TNF receptor-ligand binding and in transcription of IκB-α and A20, which form the main negative feedback loops leading to oscillations.
Simulation was performed at 100-s time steps with 10-hour equilibrium period from initial conditions, 6 m of a single TNF pulse, and then 2 hours of evolution. Analysis was performed on first peak of resulting cell traces of nuclear NF-κB. Simulated single cells at t = 0 can be found in table S3 for reference TNF concentrations.
See table S4 for abbreviations and table S5 for parameters. NF-κB and TNF receptor are distributed in a lognormal distribution with means of 105 and 104 molecules and parameters μ and σ (, −1/4) and (, −1). Ordinary differential equations for the model are listed below.
Fast reactions
Reporter is transcribed cooperatively
Slow reactions
Stochastic functions for receptors, A20, IκB, and reporter genes
Acknowledgments
Funding: This work was supported by NIH grants R01GM128042 and R01GM127527 to S.T. P.L. was supported by NSF CAREER Award #1350337.
Author contributions: P.P. did microfluidic experiments. P.L. and P.P. ran machine learning pipeline. P.P. analyzed the experimental data with assistance from N.D. M.B. and S.T. supervised the work. All authors wrote the manuscript.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. The source code and image dataset can be accessed at https://github.com/parthivapatel/ML_cellim.git (doi: 10.5281/zenodo.5361956).
Supplementary Materials
This PDF file includes:
Figs. S1 to S9
Legends for tables S1 to S5
Other Supplementary Material for this manuscript includes the following:
Tables S1 to S5
REFERENCES AND NOTES
- 1.Elowitz M. B., Levine A. J., Siggia E. D., Swain P. S., Stochastic gene expression in a single cell. Science 297, 1183–1186 (2002). [DOI] [PubMed] [Google Scholar]
- 2.Ben-David U., Siranosian B., Ha G., Tang H., Oren Y., Hinohara K., Strathdee C. A., Dempster J., Lyons N. J., Burns R., Nag A., Kugener G., Cimini B., Tsvetkov P., Maruvka Y. E., O’Rourke R., Garrity A., Tubelli A. A., Bandopadhayay P., Tsherniak A., Vazquez F., Wong B., Birger C., Ghandi M., Thorner A. R., Bittker J. A., Meyerson M., Getz G., Beroukhim R., Golub T. R., Genetic and transcriptional evolution alters cancer cell line drug response. Nature 560, 325–330 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kellogg R. A., Tian C., Etzrodt M., Tay S., Cellular decision making by non-integrative processing of TLR inputs. Cell Rep. 19, 125–135 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chakraborty A. K., Das J., Zikherman J., Yang M., Govern C. C., Ho M., Weiss A., Roose J., Molecular origin and functional consequences of digital signaling and hysteresis during Ras activation in lymphocytes. Sci. Signal. 2, pt2 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Das J., Ho M., Zikherman J., Govern C., Yang M., Weiss A., Chakraborty A. K., Roose J. P., Digital signaling and hysteresis characterize ras activation in lymphoid cells. Cell 136, 337–351 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Drayman N., Patel P., Vistain L., Tay S., HSV-1 single-cell analysis reveals the activation of anti-viral and developmental programs in distinct sub-populations. eLife 8, e46339 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hoffmann A., Levchenko A., Scott M. L., Baltimore D., The IκB-NF-κB signaling module: Temporal control and selective gene activation. Science 298, 1241–1245 (2002). [DOI] [PubMed] [Google Scholar]
- 8.Tay S., Hughey J. J., Lee T. K., Lipniacki T., Quake S. R., Covert M. W., Single-cell NF-κB dynamics reveal digital activation and analogue information processing. Nature 466, 267–271 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Avraham R., Haseley N., Brown D., Penaranda C., Jijon H. B., Trombetta J. J., Satija R., Shalek A. K., Xavier R. J., Regev A., Hung D. T., Pathogen cell-to-cell variability drives heterogeneity in host immune responses. Cell 162, 1309–1321 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee M.-C. W., Lopez-Diaz F. J., Khan S. Y., Tariq M. A., Dayn Y., Vaske C. J., Radenbaugh A. J., Kim H. J., Emerson B. M., Pourmand N., Single-cell analyses of transcriptional heterogeneity during drug tolerance transition in cancer cells by RNA sequencing. Proc. Natl. Acad. Sci. U.S.A. 111, E4726–E4735 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Otsuki L., Brand A. H., Cell cycle heterogeneity directs the timing of neural stem cell activation from quiescence. Science 360, 99–102 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sharma S. V., Lee D. Y., Li B., Quinlan M. P., Takahashi F., Maheswaran S., McDermott U., Azizian N., Zou L., Fischbach M. A., Wong K.-K., Brandstetter K., Wittner B., Ramaswamy S., Classon M., Settleman J., A chromatin-mediated reversible drug-tolerant state in cancer cell subpopulations. Cell 141, 69–80 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kellogg R. A., Tian C., Lipniacki T., Quake S. R., Tay S., Digital signaling decouples activation probability and population heterogeneity. eLife 4, e08931 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kellogg R. A., Tay S., Noise facilitates transcriptional control under dynamic inputs. Cell 160, 381–392 (2015). [DOI] [PubMed] [Google Scholar]
- 15.Lawrence T., The nuclear factor NF-κB pathway in inflammation. Cold Spring Harb. Perspect. Biol. 1, a001651 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hoffmann A., Baltimore D., Circuitry of nuclear factor κB signaling. Immunol. Rev. 210, 171–186 (2006). [DOI] [PubMed] [Google Scholar]
- 17.Zhang Q., Lenardo M. J., Baltimore D., 30 years of NF-κB: A blossoming of relevance to human pathobiology. Cell 168, 37–57 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Perkins N. D., The diverse and complex roles of NF-κB subunits in cancer. Nat. Rev. Cancer 12, 121–132 (2012). [DOI] [PubMed] [Google Scholar]
- 19.Zhang Q., Gupta S., Schipper D. L., Kowalczyk G. J., Mancini A. E., Faeder J. R., Lee R. E. C., NF-κB dynamics discriminate between TNF doses in single cells. Cell Syst. 5, 638–645.e5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Keshelava A., Solis G. P., Hersch M., Koval A., Kryuchkov M., Bergmann S., Katanaev V. L., High capacity in G protein-coupled receptor signaling. Nat. Commun. 9, 876 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Snijder B., Sacher R., Rämö P., Damm E. M., Liberali P., Pelkmans L., Population context determines cell-to-cell variability in endocytosis and virus infection. Nature 461, 520–523 (2009). [DOI] [PubMed] [Google Scholar]
- 22.Raser J. M., O’Shea E. K., Noise in gene expression: Origins, consequences, and control. Science 309, 2010–2013 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Newman J. R. S., Ghaemmaghami S., Ihmels J., Breslow D. K., Noble M., DeRisi J. L., Weissman J. S., Single-cell proteomic analysis of S. cerevisiae reveals the architecture of biological noise. Nature 441, 840–846 (2006). [DOI] [PubMed] [Google Scholar]
- 24.Losick R., Desplan C., Stochasticity and cell fate. Science 320, 65–68 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Scott M. L., Fujita T., Liou H. C., Nolan C. P., Baltimore D., The p65 subunit of NF-κB regulates IκB by two distinct mechanisms. Genes Dev. 7, 1266–1276 (1993). [DOI] [PubMed] [Google Scholar]
- 26.Sumit M., Jovic A., Neubig R. R., Takayama S., Linderman J. J., A two-pulse cellular stimulation test elucidates variability and mechanisms in signaling pathways. Biophys. J. 116, 962–973 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu H., Zhang F., Mishra S. K., Zhou S., Zheng J., Knowledge-guided fuzzy logic modeling to infer cellular signaling networks from proteomic data. Sci. Rep. 6, 35652 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cheong R., Hoffmann A., Levchenko A., Understanding NF-κB signaling via mathematical modeling. Mol. Syst. Biol. 4, 192 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Perlman Z. E., Slack M. D., Feng Y., Mitchison T. J., Wu L. F., Altschuler S. J., Multidimensional drug profiling by automated microscopy. Science 306, 1194–1198 (2004). [DOI] [PubMed] [Google Scholar]
- 30.Sachs K., Perez O., Pe’er D., Lauffenburger D. A., Nolan G. P., Causal protein-signaling networks derived from multiparameter single-cell data. Science 308, 523–529 (2005). [DOI] [PubMed] [Google Scholar]
- 31.Satija R., Shalek A. K., Heterogeneity in immune responses: From populations to single cells. Trends Immunol. 35, 219–229 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shaffer S. M., Dunagin M. C., Torborg S. R., Torre E. A., Emert B., Krepler C., Beqiri M., Sproesser K., Brafford P. A., Xiao M., Eggan E., Anastopoulos I. N., Vargas-Garcia C. A., Singh A., Nathanson K. L., Herlyn M., Raj A., Rare cell variability and drug-induced reprogramming as a mode of cancer drug resistance. Nature 546, 431–435 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Raj A., van Oudenaarden A., Nature, nurture, or chance: Stochastic gene expression and its consequences. Cell 135, 216–226 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Silver D., Schrittwieser J., Simonyan K., Antonoglou I., Huang A., Guez A., Hubert T., Baker L., Lai M., Bolton A., Chen Y., Lillicrap T., Hui F., Sifre L., van den Driessche G., Graepel T., Hassabis D., Mastering the game of Go without human knowledge. Nature 550, 354–359 (2017). [DOI] [PubMed] [Google Scholar]
- 35.Campbell M., Hoane A. J., Hsu F., Deep Blue. Artif. Intell. 134, 57–83 (2002). [Google Scholar]
- 36.Buggenthin F., Buettner F., Hoppe P. S., Endele M., Kroiss M., Strasser M., Schwarzfischer M., Loeffler D., Kokkaliaris K. D., Hilsenbeck O., Schroeder T., Theis F. J., Marr C., Prospective identification of hematopoietic lineage choice by deep learning. Nat. Methods 14, 403–406 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.De Fauw J., Ledsam J. R., Romera-Paredes B., Nikolov S., Tomasev N., Blackwell S., Askham H., Glorot X., O’Donoghue B., Visentin D., van den Driessche G., Lakshminarayanan B., Meyer C., Mackinder F., Bouton S., Ayoub K., Chopra R., King D., Karthikesalingam A., Hughes C. O., Raine R., Hughes J., Sim D. A., Egan C., Tufail A., Montgomery H., Hassabis D., Rees G., Back T., Khaw P. T., Suleyman M., Cornebise J., Keane P. A., Ronneberger O., Clinically applicable deep learning for diagnosis and referral in retinal disease. Nat. Med. 24, 1342–1350 (2018). [DOI] [PubMed] [Google Scholar]
- 38.Kellogg R. A., Gómez-Sjöberg R., Leyrat A. A., Tay S., High-throughput microfluidic single-cell analysis pipeline for studies of signaling dynamics. Nat. Protoc. 9, 1713–1726 (2014). [DOI] [PubMed] [Google Scholar]
- 39.Shrum C. K., Defrancisco D., Meffert M. K., Stimulated nuclear translocation of NF-κB and shuttling differentially depend on dynein and the dynactin complex. Proc. Natl. Acad. Sci. U.S.A. 106, 2647–2652 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ankers J. M., Awais R., Jones N. A., Boyd J., Ryan S., Adamson A. D., Harper C. V., Bridge L., Spiller D. G., Jackson D. A., Paszek P., Sée V., White M. R. H., Dynamic NF-κb and E2F interactions control the priority and timing of inflammatory signalling and cell proliferation. eLife 5, e10473 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Murata S. I., Mochizuki K., Nakazawa T., Kondo T., Nakamura N., Yamashita H., Urata Y., Ashihara T., Katoh R., Detection of underlying characteristics of nuclear chromatin patterns of thyroid tumor cells using texture and factor analyses. Cytometry 49, 91–95 (2002). [DOI] [PubMed] [Google Scholar]
- 42.Palcic B., Nuclear texture: Can it be used as a surrogate endpoint biomarker? J. Cell. Biochem. Suppl. 19, 40–46 (1994). [PubMed] [Google Scholar]
- 43.Doudkine A., Macaulay C., Poulin N., Palcic B., Nuclear texture measurements in image cytometry. Pathologica 87, 286–299 (1995). [PubMed] [Google Scholar]
- 44.Danielsen H. E., Kildal W., Sudbø J., Digital image analysis in pathology—Exemplified in prostatic cancer. Tidsskr. Nor. Laegeforen. 120, 479–488 (2000). [PubMed] [Google Scholar]
- 45.Mikenberg I., Widera D., Kaus A., Kaltschmidt B., Kaltschmidt C., Transcription factor NF-κB is transported to the nucleus via cytoplasmic dynein/dynactin motor complex in hippocampal neurons. PLOS ONE 2, e589 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bauer N. C., Doetsch P. W., Corbett A. H., Mechanisms regulating protein localization. Traffic 16, 1039–1061 (2015). [DOI] [PubMed] [Google Scholar]
- 47.Lee R. E. C., Walker S. R., Savery K., Frank D. A., Gaudet S., Fold change of nuclear NF-κB determines TNF-induced transcription in single cells. Mol. Cell 53, 867–879 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bagaev A. V., Garaeva A. Y., Lebedeva E. S., Pichugin A. V., Ataullakhanov R. I., Ataullakhanov F. I., Elevated pre-activation basal level of nuclear NF-κB in native macrophages accelerates LPS-induced translocation of cytosolic NF-κB into the cell nucleus. Sci. Rep. 9, 4563 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rotem E., Loinger A., Ronin I., Levin-Reisman I., Gabay C., Shoresh N., Biham O., Balaban N. Q., Regulation of phenotypic variability by a threshold-based mechanism underlies bacterial persistence. Proc. Natl. Acad. Sci. U.S.A. 107, 12541–12546 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Buchler N. E., Cross F. R., Protein sequestration generates a flexible ultrasensitive response in a genetic network. Mol. Syst. Biol. 5, 272 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Son M., Kaspar J., Ahn S. J., Burne R. A., Hagen S. J., Threshold regulation and stochasticity from the MecA/ClpCP proteolytic system in Streptococcus mutans competence. Mol. Microbiol. 110, 914–930 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Brock A., Chang H., Huang S., Non-genetic heterogeneity—A mutation-independent driving force for the somatic evolution of tumours. Nat. Rev. Genet. 10, 336–342 (2009). [DOI] [PubMed] [Google Scholar]
- 53.Torre E. A., Arai E., Bayatpour S., Jiang C. L., Beck L. E., Emert B. L., Shaffer S. M., Mellis I. A., Fane M. E., Alicea G. M., Budinich K. A., Weeraratna A. T., Shi J., Raj A., Genetic screening for single-cell variability modulators driving therapy resistance. Nat. Genet. 53, 76–85 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S9
Legends for tables S1 to S5
Tables S1 to S5