Significance
Glucocorticoids are widely prescribed antiinflammatory drugs that suppress inflammatory gene expression through activation of the glucocorticoid receptor (GR); however, the mechanisms of GR-mediated repression are not fully understood. We show here that bromodomain containing 9 (BRD9), a component of the SWI/SNF chromatin remodeling complex, modulates GR activity in bone marrow-derived macrophages. Loss of BRD9 activity in macrophages attenuated the response to liposaccharides and interferon inflammatory stimuli. Notably, BRD9 inhibition synergized with the synthetic glucocorticoid dexamethasone to suppress the inflammatory response in macrophages. Our data support a model in which BRD9 restricts GR binding at a subset of inflammatory related genes, and suggests that BRD9 inhibitors may increase the efficacy of glucocorticoids.
Keywords: inflammation, macrophages, bromodomain containing 9 (BRD9)
Abstract
In macrophages, homeostatic and immune signals induce distinct sets of transcriptional responses, defining cellular identity and functional states. The activity of lineage-specific and signal-induced transcription factors are regulated by chromatin accessibility and other epigenetic modulators. Glucocorticoids are potent antiinflammatory drugs; however, the mechanisms by which they selectively attenuate inflammatory genes are not yet understood. Acting through the glucocorticoid receptor (GR), glucocorticoids directly repress inflammatory responses at transcriptional and epigenetic levels in macrophages. A major unanswered question relates to the sequence of events that result in the formation of repressive regions. In this study, we identify bromodomain containing 9 (BRD9), a component of SWI/SNF chromatin remodeling complex, as a modulator of glucocorticoid responses in macrophages. Inhibition, degradation, or genetic depletion of BRD9 in bone marrow-derived macrophages significantly attenuated their responses to both liposaccharides and interferon inflammatory stimuli. Notably, BRD9-regulated genes extensively overlap with those regulated by the synthetic glucocorticoid dexamethasone. Pharmacologic inhibition of BRD9 potentiated the antiinflammatory responses of dexamethasone, while the genetic deletion of BRD9 in macrophages reduced high-fat diet-induced adipose inflammation. Mechanistically, BRD9 colocalized at a subset of GR genomic binding sites, and depletion of BRD9 enhanced GR occupancy primarily at inflammatory-related genes to potentiate GR-induced repression. Collectively, these findings establish BRD9 as a genomic antagonist of GR at inflammatory-related genes in macrophages, and reveal a potential for BRD9 inhibitors to increase the therapeutic efficacies of glucocorticoids.
Macrophages have key roles in innate immunity, regulating tissue homeostasis, inflammatory responses, pathogen elimination, and tissue repair (1). Their biological roles have been well characterized in the context of inflammation, metabolic diseases, cancer, and beyond (2, 3). Functional maturation of macrophages requires polarization, an activation process induced by a myriad of environmental cues depending on specific physiological and pathological processes (4). In a classic activation model, Toll-like receptor (TLR) agonists and interferons (IFNs) induce an inflammatory signature of macrophages, often simplified as the M1 macrophage (4). In obesity, metabolic stress leads to a shift of adipose tissue resident macrophages to the proinflammatory M1 phenotype. This shift contributes to elevated whole-body inflammation (2). A key feature of macrophages is their dynamic response to environmental challenges at transcriptional, epigenetic, and functional levels. Polarization of macrophages includes abrupt changes at the transcriptional level which is orchestrated by a variety of transcription factors (TFs) (1, 5, 6). Correspondingly, the epigenome also undergoes dynamic reprogramming during activation and repression involving changes in DNA methylation, chromatin accessibility, histone modification, and three-dimensional nuclear architecture (7). Therefore, characterization of the transcriptional and epigenetic changes during macrophage activation will provide mechanistic insight into the transition between homeostatic and pathophysiologic states.
Bromodomain containing 9 (BRD9) belongs to the large family of bromodomain proteins, known for their ability to recognize acetyl lysine and their diverse roles in physiology and oncology (8). BRD9 is a key component of the SWI/SNF (BAF) chromatin remodeling complex (9). Recent studies revealed the critical role of BRD9 as a key component of the nonconical BAF (ncBAF) complex (10–12) in cancer cell proliferation (13–16), pluripotency (10), and inflammatory responses (17), making it a favorable target for therapeutic development. Our previous work demonstrated that in cytokine stressed β-cells, inhibition of BRD9, together with the activation of the vitamin D receptor (VDR), is able to reverse genome-wide inflammatory responses to limit β-cell failure (17). Mechanistically, inhibition of BRD9 increased the antiinflammatory function of VDR through modulating VDR and BRD9 interaction (17). However, the role of BRD9 in mediating inflammatory responses in other cell types, especially in innate immunity, has not been characterized.
Glucocorticoids are potent immunosuppressive drugs that have been used in clinic for more than 60 y. Acting through the glucocorticoid receptor (GR), endogenous glucocorticoids control inflammatory responses of macrophages, dendritic cells, and other types of innate immune cells (18). Synthetic and pharmacological grade glucocorticoids, such as dexamethasone (Dex), are widely used immunosuppressants that function by repressing macrophages M1 activation (19). Mechanistically, ligand-bound GR is known to directly suppress a large number of proinflammatory targets (18). This is achieved either through directly binding to glucocorticoid response elements (GREs) (20, 21), tethering to inflammatory mediators such as AP1, STAT, or C/EBP (22), or through binding to composite sites, including the GR half-site and the binding motif of another TF (22, 23). Chromatin remodelers, such as the SWI/SNF (BAF) complex (24–26), are known to be critical for GR binding since open chromatin is a prerequisite for GR binding (27). However, whether BRD9, a critical component of the SWI/SNF complex, modulates GR transcription, has not been explored.
Here we show that BRD9 is essential for activation of bone marrow–derived macrophages (BMDMs). Pharmacological inhibition, degradation, or genetic deletion of BRD9 all consistently reduce liposaccharides (LPSs) and IFN-γ–induced inflammatory responses in BMDMs. The suppression of inflammation by BRD9 inhibitors resembles the glucocorticoid action on macrophages. Surprisingly, inhibition or degradation of BRD9 is able to further repress a subset of critical inflammation-related GR targets. Mechanistically, degradation of BRD9 increases GR occupancy at a subset of its binding sites, resulting in the additional repression of its targets. Therefore, we conclude that BRD9 is a critical regulator of inflammatory stimulation and glucocorticoid responses in macrophages.
Results
BRD9 Is Essential for BMDM Activation and Survival.
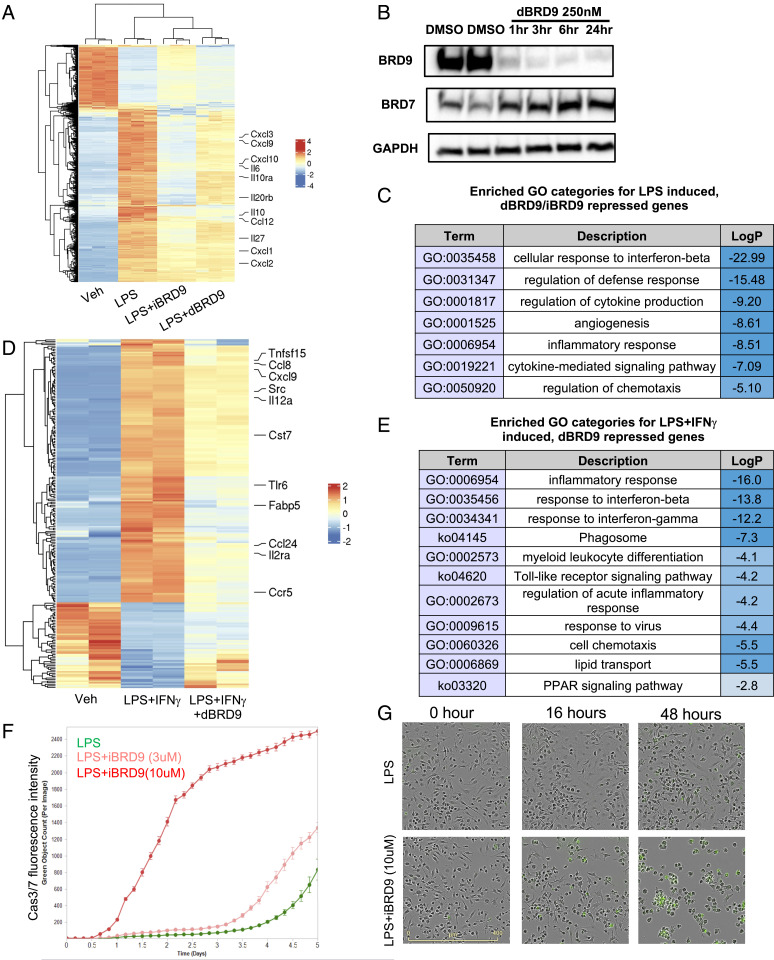
Our previous results suggest that iBRD9, a BRD9 inhibitor, is able to suppress the interleukin (IL) 1β- and IFN-γ–induced inflammatory responses in β-cells, obesity, and in type 2 diabetes (T2D) (17). In addition to islet inflammation, it has been well documented that in T2D tissue resident macrophages adopt proinflammatory M1-type signatures, causing a significant increase of whole-body inflammation (2). To determine whether inhibition of BRD9 is able to suppress inflammatory responses in macrophages, we used a classic BMDM activation model. As expected, activation of monocytes by LPS induced widespread transcriptional changes within 3 h (Fig. 1A). Interestingly, addition of iBRD9 (3 μM) strongly suppressed a large number of LPS-induced genes. We also tested a proteolysis-targeting chimera (PROTAC) degrader of BRD9, dBRD9 (28), which selectively degrades BRD9 in 3 h without cross-reacting to BRD7, the closest bromodomain ortholog to BRD9 (Fig. 1B). Similar to iBRD9, dBRD9 treatment significantly attenuated LPS-induced genes (Fig. 1A). Since the recognition ligand in dBRD9 is structurally distinct from the BRD9 inhibitor BI-7273, these results strongly suggest that selective pharmacologic targeting of BRD9 is sufficient to attenuate LPS-induced transcriptional changes in macrophages (28).
Fig. 1.
BRD9 is essential for macrophages activation and survival. (A and D) Heatmaps showing that LPS-induced, iBRD9/dBRD9-repressed (A), and LPS+IFN-g-induced, dBRD9-repressed genes (D) from RNA-seq. (B) Western blotting showing dBRD9 deplete BRD9 at protein level without affecting BRD7 stability. (C) GO suggest that iBRD9/dBRD9 inhibited genes are associated with inflammatory response. dBRD9 repress LPS+IFN-γ–induced M1 polarization, shown by RNA-seq. (E) GO of dBRD9 repressed genes in M1 polarization. (F) Long-term treatment of iBRD9 induces significant monocyte apoptosis during LPS-induced activation in vitro. (G) Representative images showing that increased cell apoptosis during BMDM activation after 2 d of iBRD9 treatment.
The subset of LPS-induced genes repressed by dBRD9/iBRD9 included many proinflammatory cytokines, chemokines, and effectors (Cxcl3, Cxcl9, Cxcl10, Il6, Il10, Cxcl1, Cxcl2, Ccl12, and so forth) (Fig. 1A), and was enriched for inflammation-related gene categories in gene ontology (GO) analysis (Fig. 1C), suggesting that BRD9 is critical for the inflammatory responses in macrophages. Classic M1 polarization of BMDM is driven by both the TLR4 agonist LPS and IFN-γ (4). In order to determine whether BRD9 is also involved in the type II IFN responses in macrophages, we treated the bone marrow-derived monocytes with LPS and IFN-γ, with or without dBRD9. RNA-sequencing (RNA-seq) showed that, similar to its capacity of partially blocking LPS-induced inflammatory transcriptional changes, dBRD9 is also able to reverse IFN-γ–induced IFN responses (Fig. 1 D and E). Interestingly, a number of dBRD9-attenuated genes are related to lipid transport and peroxisome proliferator–activated receptor (PPAR) signaling, features of mature macrophages, especially in obesity (Fig. 1 C and D). To determine whether the lipid loading capacity of macrophages is also compromised, we stressed the cells with acetylated LDL (acLDL) and measured lipid loading by Nile red staining. iBRD9 was able to reduce the lipid loading (SI Appendix, Fig. S1). Together, these results support a role for BRD9 in macrophage M1 polarization and activation. Pharmacologic inhibition or degradation of BRD9 attenuates proinflammatory gene expression, compromising the function of mature M1 macrophages. To determine long-term effects of BRD9 inhibition on monocyte/BMDM survival, we used time-lapsed imaging of caspase-3/7 to monitor apoptosis (Fig. 1F). Significant monocyte apoptosis was observed 24 h after exposure to LPS and iBRD9 (10 μM) (Fig. 1 F and G). A lower dose of iBRD9 (3 μM) also resulted in cell apoptosis, though at a much later time point (3 d) (Fig. 1F). These results suggest that long-term inhibition of BRD9 in LPS stimulated monocytes induces apoptosis.
Genetic Loss of BRD9 Compromised the M1 Activation of BMDM.
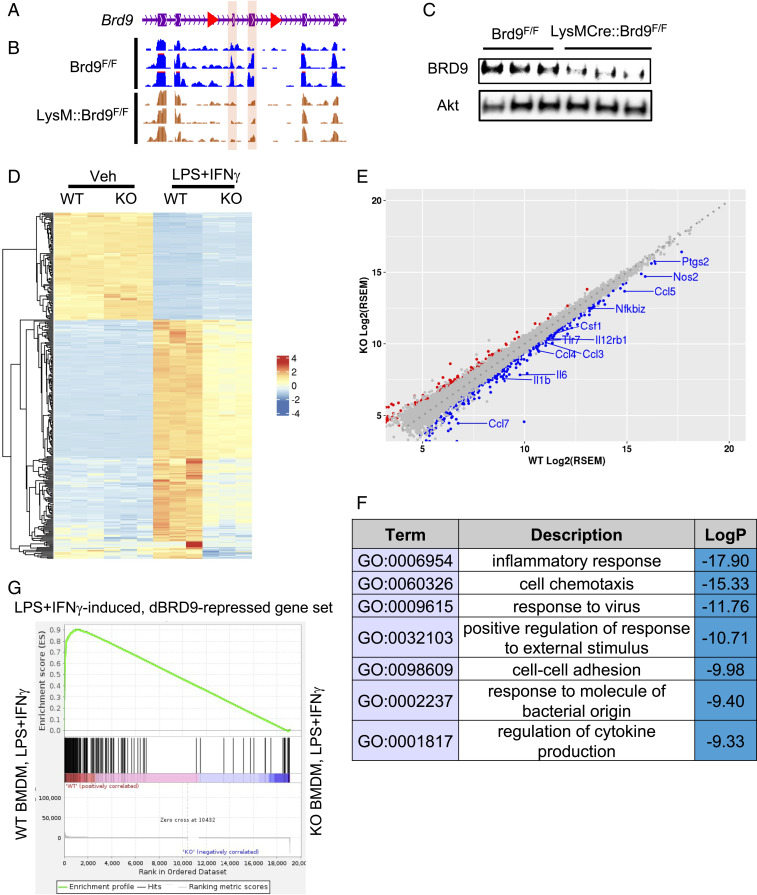
To further define the role of BRD9 in macrophage biology, we generated a genetic knockout (KO) model of BRD9 in the macrophage lineage by crossing a novel BRD9F/F line (Fig. 2A) with LysM-Cre mice. LysM-Cre induced deletion of exon 4 and 5 of BRD9, which was confirmed by comparing RNA-seq results from LysM-Cre;Brd9F/F BMDMs with the control Brd9F/F BMDMs (Fig. 2B). The deletion of these exons generates a premature stop codon and, thereby, decreased BRD9 protein level in LysM-Cre; Brd9F/F BMDMs (Fig. 2C). To determine whether deletion of BRD9 negatively affects M1 polarization of macrophages, we harvested the LysM-Cre;Brd9F/F (herein referred to as KO) and control Brd9F/F (herein referred to as WT) BMDM from littermates. After in vitro expansion, LPS and IFN-γ were added to induce M1 polarization. Loss of BRD9 attenuated the transcriptional changes induced by LPS+IFN-γ (Fig. 2D). Compared to activated WT BMDMs, 217 and 311 genes were significantly up- and down-regulated, respectively, in KO cells (Fig. 2E). Importantly, the genes repressed in BRD9 KO BMDMs were enriched in viral/IFN responses, cytokine signaling, and NF-κB signaling (Fig. 2F), including a number of key inflammatory effectors (e.g., Il6, Il1b, Ccl4, Csf1, Nfkbiz, Ccl7, Nos2) (Fig. 2E). This suggests that KO BMDMs have reduced inflammatory responses during the M1 polarization, similar to BMDMs treated with iBRD9 or dBRD9. Indeed, gene set enrichment analysis (GSEA) revealed significant overlap of repressed genes between the pharmacologic (iBRD9 and dBRD9) and genetic loss-of-function models, with the majority of dBRD9-repressed genes expressed at lower levels in KO BMDMs (Fig. 2G). Therefore, the BRD9 KO BMDMs are less inflammatory upon M1 polarization, similar to dBRD9/iBRD9 treated cells.
Fig. 2.
Conditional KO of BRD9 compromises macrophage inflammatory responses. (A and B) Design of the Brd9F/F line. Red: loxP site. Purple background: exon 4 and 5. Genomic track of RNA-seq results shows the deletion of exons of Brd9. RNA-seq reads on exon 4 and 5 (shown in purple) show significant decrease upon LysM-Cre induced deletion. (C) Western blotting demonstrated that BRD9 can be effectively depleted in LysM-Cre; Brd9F/F macrophages. (D) Heatmap of RNA-seq showing that the transcriptional responses to LPS+IFN-γ–induced M1 activation are compromised in LysM-Cre;Brd9F/F macrophages. (E) Differentially expressed genes in LPS-treated Brd9F/F and LPS-treated LysM-Cre;Brd9F/F macrophages. Red: significant up-regulated genes in KO. Blue: significant repressed genes in KO. (F) GO of RNA-seq showed down-regulated genes in LPS+IFN-γ–treated LysM-Cre;Brd9F/F are enriched in inflammatory associated pathways. (G) GSEA enrichment plot for dBRD9-repressed genes in RNA-seq of Brd9 WT vs. KO BMDM.
To determine the role of BRD9 in directing macrophage function in vivo, we challenged BRD9 WT and KO mice with a high-fat diet to induce inflammatory M1 macrophage expansion in adipose tissue (2). H&E staining of epididymal adipose tissue showed fewer crown-like structures in BRD9 KO compared to control WT mice (SI Appendix, Fig. S2 A and B), indicative of reduced adipose inflammation. Consistent with this, serum levels of proinflammatory cytokines IL-12, IL-9, IFN-γ, and the chemokine MIP-1b (CCL4) were reduced in KO mice (SI Appendix, Fig. S2C), suggesting that systemic inflammation caused by obesity is reduced in KO mice. Compared to WT mice, the BRD9 KO mice under high-fat diet have comparable fasting glucose, glucose tolerance, and insulin tolerance (SI Appendix, Fig. S3 A–E). The KO mice also gained slightly more weight (SI Appendix, Fig. S3F). Together, the data suggests that loss of BRD9 in macrophages is sufficient to reduce adipose inflammation and circulating inflammatory cytokines, but not sufficient to reverse the glucose intolerance in the context of obesity.
Pharmacological Inhibition of BRD9 Synergizes with Dex to Repress Inflammatory Responses.
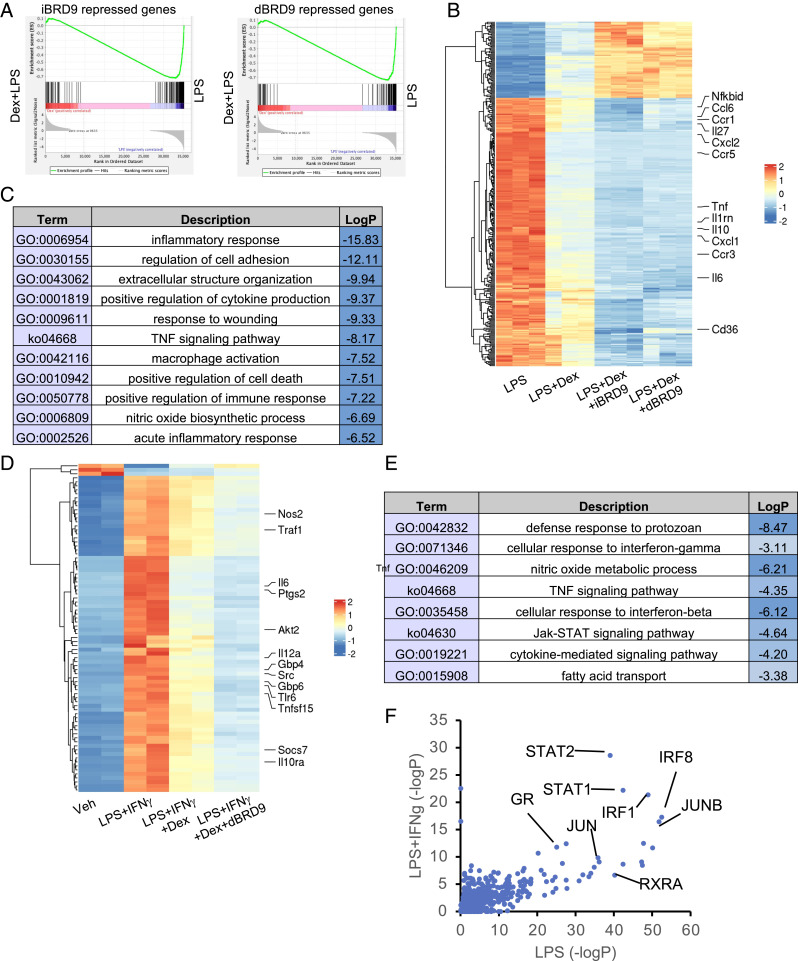
The GR plays a key role in modulating inflammation, and the synthetic ligand Dex is a powerful antiinflammatory agent in macrophages. We previously showed that BRD9 directly binds to the VDR through the conserved C-terminal extension of the VDR DNA binding domain (17), which is also present in GR (29). Previous studies have also demonstrated that GR binds to the SWI/SNF complex component Brg1 (30), further suggesting that the BRD9-containing SWI/SNF complexes may collaborate with the GR. Since both BRD9 inhibitor/degraders and glucocorticoids are able to suppress inflammatory responses in macrophage activation, we investigated whether there is significant overlap between BRD9-regulated and GR-regulated genes. The data show that under either LPS or LPS+IFN-γ stimuli, approximately half of the dBRD9/iBRD9 repressed genes and a third of the up-regulated genes were similarly regulated by treatment with the GR agonist Dex (SI Appendix, Fig. S4A). GO analysis of corepressed genes revealed viral response, inflammatory response, cytokine production (SI Appendix, Fig. S4B), as well as genes essential for macrophage activation. GSEA analysis further confirmed the similarities between iBRD9/dBRD9-repressed genes and Dex-repressed genes (Fig. 3A). In contrast, the iBRD9/dBRD9/Dex coactivated gene sets lacked any obvious pattern of GO (SI Appendix, Fig. S4B). These results suggest that the iBRD9/dBRD9 and Dex commonly repressed gene set comprises a distinct and functionally connected group of targets in macrophage activation.
Fig. 3.
BRD9 inhibitor or degrader synergize with Dex to repress macrophage inflammatory responses. (A) GSEA enrichment plot showing the similarity between RNA-seq of LPS-induced, iBRD9/dBRD9 BMDM, and LPS-induced, Dex-repressed BMDM. (B) Heatmap of genes synergistically repressed or activated by iBRD9/dBRD9 and Dex in LPS-stimulated BMDM. (C) Enriched GO categories. (D) Heatmap of genes synergistically repressed or activated by dBRD9 and Dex in LPS+IFN-γ activated BMDM. (E) Enriched GO categories. (F) LISA analysis of potential enriched upstream regulator of LPS/LPS+IFN-γ induced, Dex-dBRD9 repressed genes.
The independence of BRD9 and GR repression of inflammatory-related genes suggested that BRD9 inhibition may synergize with Dex in attenuating the macrophage inflammatory response. To explore this hypothesis, we compared the global transcriptional changes induced by Dex, Dex+dBRD9, and Dex+iBRD9 in LPS-stimulated BMDMs. Based on previous literature, 1 μm Dex was used to achieve maximum in vitro effects (31, 32). As expected, Dex induced profound transcriptional changes in BMDM. Notably, addition of iBRD9 (3 μM) or dBRD9 (250 nM) significantly enhanced Dex-mediated transcriptional changes (Fig. 3B and SI Appendix, Fig. S5A, categories A and C). Moreover, the Dex-dBRD9 synergistic gene signature was enriched in inflammatory responses categories (Fig. 3C). This ability of dBRD9 to potentiate Dex-mediated gene repression was replicated in LPS+IFN-γ–stimulated macrophages (Fig. 3D), where the enriched functional categories included defense responses, tumor necrosis factor (TNF) signaling, type II IFN responses, and lipid transport function (Fig. 3E). These findings indicate that combined treatment with Dex and dBRD9 or iBRD9 cooperatively inhibits BMDM M1 polarization. In contrast, while a subset of the genes up-regulated by Dex treatment were further up-regulated by dBRD9 (SI Appendix, Fig. S5A, category C), the synergy of Dex-dBRD9 induced genes was less related to inflammation (SI Appendix, Fig. S5B, category C).
The finding that Dex and dBRD9/iBRD9 cooperatively repressed genes are highly enriched in macrophage inflammatory responses and function led to the hypothesis of possible common regulators. To explore this notion, potential regulators of the Dex+dBRD9 synergistically regulated genes in LPS or LPS+IFN-γ stimulation were predicted using epigenetic Landscape In Silico deletion Analysis (LISA) (33). In addition to GR, predicted regulators included known macrophage regulators IRFs, JUN, and STATs (Fig. 3F), TFs known to colocalize with GR (Fig. 3F). This was further confirmed by another curated database, TRRUST (34) (SI Appendix, Fig. S5C). These findings implicate that the synergistically regulated genes are direct targets of GR, AP-1, IRFs, or other core stimulus-responsive TFs (SRTF) (1).
BRD9 Depletion Reprograms the GR Cistrome to Potentiate GR-Mediated Responses.
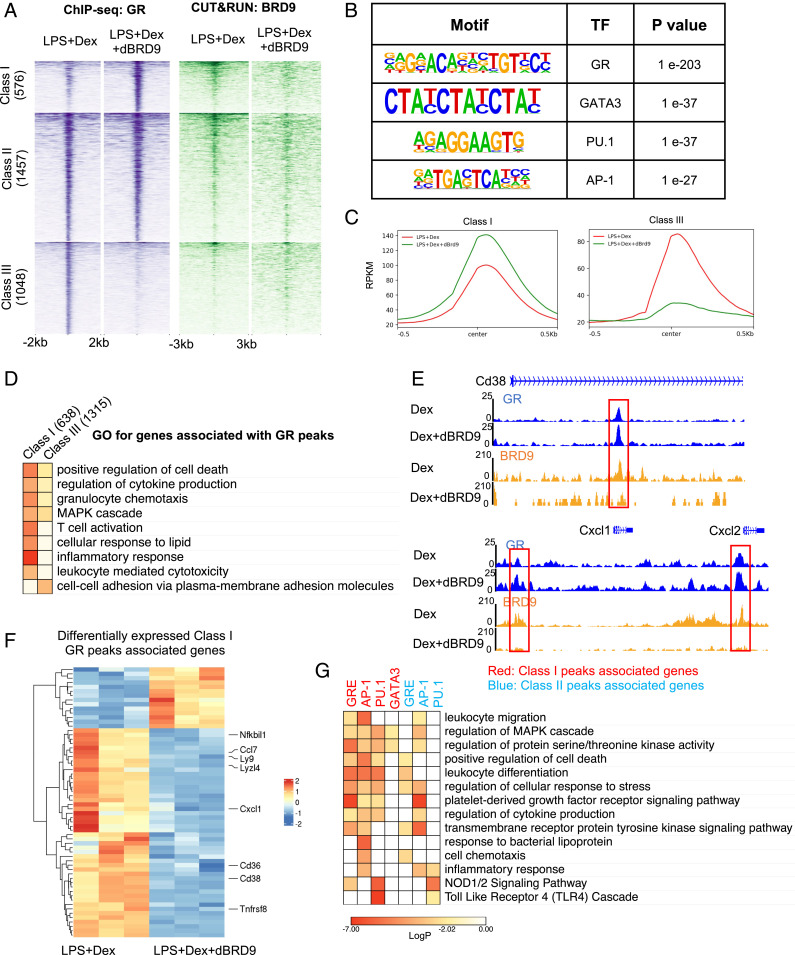
As the synergistically regulated genes include many direct GR targets, we explored the possibility that dBRD9 impacts the binding of GR to the chromatin. To identify the genome-wide changes in the GR cistrome, we performed chromatin immunoprecipitation-sequencing (ChIP-seq) studies to map the GR binding sites in LPS-induced Dex and Dex+dBRD9-treated RAW264.7 cells, a mouse macrophage cell line. GR binding sites were classified as: class I: ∼20% of peaks with enhanced GR binding with dBRD9 (>1.5-fold); class II: ∼50% of peaks with comparable binding with or without dBRD9; and class III: ∼30% of peaks with reduced GR binding in the presence of dBRD9 (>1.5-fold) (Fig. 4 A and C). Motif analysis by Homer identified that the top enriched motifs in our GR ChIP-seq include GRE, PU.1, GATA3, and AP-1 response elements (Fig. 4B), consistent with previous GR ChIP-seq studies (31).
Fig. 4.
Depletion of BRD9 reprogram GR occupancy to potentiate the GR-mediated antiinflammatory responses. (A, Left) Heatmap of GR binding sites defined in ChIP-seq in RAW264.7 cells. GR peaks are group in three categories, based on GR binding changes with the presence of dBRD9. (Right) CUT&RUN of BRD9 binding intensity around GR peaks. (B) Motif analysis showing enrichment of TF binding motifs in all GR peaks. (C) Average binding intensity of group I peaks and group III peaks. (D) GO of genes associated with class I and III GR peaks. (E) Examples of loci with increased GR binding when dBRD9 is present. (F) Heatmap of class I GR peaks associated genes that are differentially expressed between LPS+Dex and LPS+Dex+dBRD9. (G) GO of genes associated with GR peaks containing GRE, PU.1, AP-1, or GATA3 binding motifs.
In parallel studies, we determined the BRD9 cistrome in the absence and presence of dBRD9 by CUT&RUN (35). As expected, dBRD9 strongly reduced BRD9 binding to chromatin (SI Appendix, Fig. S6A). BRD9 binding sites were enriched in AP-1, IRFs, and PU.1 response elements, as well as the GRE motif, suggesting a cooccupancy with the TFs, and consistent with the results from gene-expression studies (SI Appendix, Fig. S6B). Most importantly, when examining the GR binding sites, we found BRD9 colocalized with GR at class I and II sites (Fig. 4A), suggesting that BRD9 removal facilitates GR binding at a subset of their common binding sites. In contrast, BRD9 binding at class III GR binding sites was not observed (Fig. 4A), suggesting that the loss of GR binding upon BRD9 degradation is a secondary effect. Motif enrichment analysis showed that class I/II GR peaks are highly enriched in GR, AP-1, and other known GR-associating TF motifs, whereas only minimal enrichment of the same motifs in class III GR peaks was found (SI Appendix, Fig. S6C). Using GREAT (36), we identified the genes associated with class I and class III peaks. Although the number of class III peaks and associating genes was greater than that of class I peaks, inflammatory responses and key macrophage effectors, such as Cd38 and Cxcl1, were enriched in the class I-associating genes (Fig. 4 D and E). To functionally associate changes in GR binding to downstream transcriptional responses, we identified the subset of class I-associating genes that are differentially expressed between LPS+Dex and LPS+Dex+dBRD9 from RNA-seq results. With an increase in GR binding, a majority of the differentially expressed genes are further repressed by the addition of dBRD9 (Fig. 4F). Many of the down-regulated genes are essential in regulating macrophage activation and function, including Nfkbil1, Ccl7, and Cxcl1. These findings support a mechanism in which a reduction in BRD9 enhances the suppression by Dex through increasing GR occupancy at a subset of its direct targets.
It is generally agreed that GR acts either by directly binding to its targets, binding together with other TFs on composite sites, or indirectly through “tethering” to other TFs (18). To determine whether the Dex+dBRD9 synergy preferentially acts through one of these models, GO analyses were performed on genes associated with GR peaks that contain GR, AP-1, PU-1, or GATA3 motifs. GR, AP-1, and PU-1 motif-containing GR peaks were all significantly associated with macrophage activation and function (Fig. 4G), suggesting that both directly bound and tethered GR peaks can be influenced by dBRD9. Interestingly, this enrichment of GO categories was predominantly seen in class I but not class II peaks (Fig. 4G), further supporting our notion that degradation of dBRD9 increases GR binding at a subset of essential macrophage genes.
Discussion
As a critical component of the ncBAF complex, the biological roles of BRD9 largely depend on the collaborating TFs and the specific context (10, 17, 37). In macrophages, most pattern recognition receptors induce inflammatory responses through a few SRTFs, integrating upstream signals at the level of the chromatin (1). A major unanswered question relates to the sequence of events that result in the formation and ultimate function of these complexes. A master regulator of antiinflammatory response in macrophages, GR orchestrates gene repression through a complex combinatorial mechanism involving multiple cofactors and TFs (18, 31, 38). We demonstrate that the combinatorial effects of GR and BRD9 binding regulate specific gene sets ultimately responsible for both cell identity and cell-type–specific responses in a signal-dependent fashion. Accordingly, depletion of BRD9 led to down-regulation of known STATs, AP-1, and IRFs targets, suggesting that it plays an essential role in SRTF-mediated transcription. Importantly, depletion/inhibition of BRD9 reversed LPS (TLR2/4 agonist) and IFN-γ (type II IFN) induced transcriptional responses, highlighting its previously unrecognized role in macrophage activation.
The occupancy of GR on chromatin occurs in the context of the neighboring chromatin landscape, especially by the accessibility to DNA. It has been shown that GR binding is predefined by chromatin accessibility when the ligand is present (27). However, the codependency of GR transcriptional and functional activities on the BRD9-BAF remodeling machinery has not been previously defined. Our data indicate that BRD9 colocalizes with GR at a subset of GR peaks (class I and II), and that depletion of BRD9 redistributes GR binding, with binding increased at class I peak and decreased at class III peaks. Interestingly, while BRD9 is located at class I and II GR sites, there is no significant colocalization of BRD9 and GR at the class III site. This suggests the interesting notion that in the absence of BRD9, GR binding at class III sites redistributes to class I sites. The motif enrichment and GO analyses showed that class I/II GR binding sites are closely associated with the inflammatory response gene network, whereas the class III GR sites are associated with cell–cell adhesion and MAPK signaling. These findings support a model in which BRD9 limits GR binding to a subset of inflammation response genes, and that inhibition/depletion of BRD9 allows the redistribution of GR to these class I sites, resulting in enhanced antiinflammatory responses while losing other GR-downstream responses related to cellular metabolism and cell–cell adhesion. The detailed mechanisms of how depletion of BRD9 results in this redistribution remains unclear and an interesting topic for future studies. Synthetic glucocorticoids are widely used in the clinic. However, the adverse effects of glucocorticoids, especially in inducing insulin resistance and dysregulated glucose metabolism, present a challenge in their clinic use. Interestingly, our studies suggest that the increased efficacy achieved in the presence of BRD9 inhibitor/degrader may effectively lower the required doses of steroids.
In conclusion, these results reveal an unexpected function of BRD9 in addition to regulating chromatin accessibility, and suggests that the influence of BRD9 to GR binding is context-dependent. Future exploration of tissue-, time-, and context-dependent GR regulation, shaped by specific chromatin remodelers, will lead to a deeper understanding of steroid biology and potentially the generation of safer glucocorticoid therapies.
Methods
BMDM Isolation.
BMDMs were isolated similarly as described previously (39). Briefly, bone marrow from 6- to 12-wk-old male animals. After lysis with AKC lysis buffer, cells were plated in Petri dishes, cultured with media containing RPMI, 20% FBS, 30% L-929 conditional medium, and 1% penicillin/streptomycin or antibiotic-antimycotic. Macrophages were digested with versine and plated with macrophage serum free medium.
For activation of macrophages, cells were treated with vehicle, Dex (1 μM), iBRD9 (3 or 10 μM), dBRD9 (250 nM), or combination for overnight, followed by stimulation of LPS (100 ng/mL; Sigma) for 3 h before being harvested for RNA-seq.
Animals.
All mice were housed in a 12-h light, 12-h dark cycle. Mouse experiments were approved by the Institutional Animal Care and Use Committee (IACUC) at the Salk Institute and IACUC at Mayo Clinic Arizona. Brd9F/F mice are generated by Biocytogen. LoxP sites were inserted in by CRISPR-mediated homologous recombination in C57Bl6 background. LysM-Cre mice were from Jackson Laboratory (cat. no. 004781). C57bl6 mice were from Jackson Laboratory (cat. no. 000664). For high-fat diet treatment, mice were fed with 60% fat diet paste (Bio-Serv S1850) for at least 16 wk before serum or tissue were collected.
Immunoprecipitation.
Briefly, cells were lysed using ice-cold lysis buffer (Hepes 20 mM pH 8, EDTA 0.2 mM, NaCl 0.3 mM, Nonidet P-40 0.5%, 15% glycerol) for 15 min, and after centrifuging, the supernatant was collected. For Flag-tagged BRD9, immunoprecipitations were performed using Flag-magnetic beads (Sigma, M8823) for 1.5 h at 4 °C. For other IPs, primary antibodies were incubated with lysate for 2 h followed by Protein A magnetic beads (Life Technologies, 10001D). After three washes with wash buffer (Hepes 10 mM pH 8, EDTA 0.2 mM, NaCl 0.3 mM, Nonidet P-40 0.1%, 15% glycerol), the beads were boiled with NuPage LDS sample buffer and the lysate were stored at −20 °C.
ChIP-Seq.
RAW264.7 cells were pretreated with vehicle, Dex (1 μM), or Dex+dBRD9 (250 nM) for overnight and then treated with LPS (100 ng/mL) for 3 h. Next, 20 million cells were harvested for each ChIP assay. The experimental procedure for ChIP was as previously described (40). Briefly, after fixation, nuclei from were isolated, lysed, and sheared with a Covaris Ultrasonicator ME220 to yield DNA fragment sizes of 200 to 1,000 bp followed by IP. Antibody used for ChIP-seq was GR (Cell Signaling, cat. no. 12041).
RNA-Seq.
RNA were extracted using TRIzol. Samples were then processed by Novogene Co. As described previously (41), a total amount of 1 µg RNA per sample was used as input material for the RNA sample preparations. Sequencing libraries were generated using NEBNext Ultra TM RNA Library Prep Kit for llumina (New England Biolabs) following the manufacturer’s recommendations and index codes were added to attribute sequences to each sample. Briefly, mRNA was purified from total RNA using poly-T oligo-attached magnetic beads. Fragmentation was carried out using divalent cations under elevated temperature in NEBNext First Strand Synthesis Reaction Buffer (5×). First-strand cDNA was synthesized using a random hexamer primer and M-MuLV Reverse Transcriptase (RNase H-). Second-strand cDNA synthesis was subsequently performed using DNA Polymerase I and RNase H. The remaining overhangs were converted into blunt ends via exonuclease/polymerase activities. After adenylation of 3′ ends of DNA fragments, NEBNext Adaptor with a hairpin loop structure were ligated to prepare for hybridization. In order to select cDNA fragments of preferentially 150∼200 bp in length, the library fragments were purified with AMPure XP system (Beckman Coulter). Then, 3 µL USER Enzyme (New England Biolabs) was used with size-selected, adaptor-ligated cDNA at 37 °C for 15 min followed by 5 min at 95 °C before PCR. Then PCR was performed with Phusion High-Fidelity DNA polymerase, Universal PCR primers and Index (X) Primer. At last, PCR products were purified (AMPure XP system) and library quality was assessed on the Agilent Bioanalyzer 2100 system.
CUT&RUN.
RAW264.7 cells were pretreated with vehicle, Dex (1 μM), or Dex+dBRD9 (250 nM) overnight and then treated with LPS (100 ng/mL) for 3 h. Next, 0.1 million cells were used for each CUT&RUN. The experimental procedure for CUN&RUN was previously described (42). Antibody used for CUT&RUN was BRD9 (Active Motif, cat. no. 61537).
RNA-Seq Analysis.
Raw data (raw reads) of FASTQ format were firstly processed through fastp. In this step, clean data (clean reads) were obtained by removing reads containing adapter and poly-N sequences and reads with low quality from raw data. At the same time, Q20, Q30, and GC content of the clean data were calculated. All the downstream analyses were based on the clean data with high quality. Paired-end clean reads were aligned to the reference genome using the Spliced Transcripts Alignment to a Reference (STAR) software. Fragments per kilobase of transcript per million mapped reads of each gene was calculated based on the length of the gene and reads count mapped to this gene. Differential expression analysis between two conditions/groups (three biological replicates per condition) was performed using DESeq2 R package. DESeq2 provides statistical routines for determining differential expression in digital gene expression data using a model based on the negative binomial distribution. The resulting P values were adjusted using the Benjamini and Hochberg approach for controlling the false-discovery rate. Genes with an adjusted P < 0.05 and fold-change > 1.5 found by DESeq2 were assigned as differentially expressed.
ChIP-Seq Analysis.
Sequencing was performed using Illumina technology to generate paired-end reads for each sample. Sequence data quality was determined using FastQC software. Bowtie2 (v2.3.3.1) (–very-sensitive) was used to map ChIP-seq reads to the mouse reference genome GRCm38. Duplicate reads were filtered out using the MarkDuplicate function from Picard tools v.2.17.0 (http://broadinstitute.github.io/picard/). Reads per kilobase and million mapped read (RPKM)-normalized bigWig files were generated with bamCoverage from deepTools v3.3.2. ChIP-Seq peaks were called using findPeaks within Homer using default parameters for TF (-style factor). De novo and known motif analyses were carried out using the findMotifsGenome.pl module of the Homer package with the “‐size given” option.
CUT&RUN Analysis.
Paired-end reads were trimmed and quality-filtered using cutadapt (v2.8). Trimmed qualified reads were aligned to mouse reference genome GRCm38 using bowtie2 (v2.3.3.1) with options:–end-to-end–very-sensitive–no-mixed–no-discordant–phred33 -I 10 -X 700. Only reads that were uniquely mapped were retained for further analysis. PCR-duplicated reads were removed using “MarkDuplicates.jar” from Picard tools v.2.17.0. RPKM-normalized bigWig files were generated with bamCoverage from deepTools v3.3.2. Heatmaps were generated using computeMatrix and plotHeatmap from deepTools v3.3.2. CUT&RUN peaks were called using findPeaks within Homer using default parameters for TF (-style factor). The identified peaks for BRD9 were further screened against “mouse blacklisted” genomic regions, mitochondrial DNA, and pseudochromosomes. Known motif finding on identified peaks was done by Homer findMotifsGenome.pl using parameter “-size given.”
H&E Staining.
H&E staining was performed by University of California San Diego pathology core and Mayo Clinic pathology core.
Incucyte Imaging.
BMDM were cultured in 96 wells. Vehicle, iBRD9, or LPS were added together with caspase 3/7 green dye for apoptosis (cat. no. 4440). Live imaging was carried out in Incucyte S3 Live Cell Analysis system. Brightfield and GFP images were collected every four hours for 3 d. Quantitative analysis of caspase 3/7+ objects were done using Incucyte base software.
Bioplex.
Mouse serum from HFD treated BRD9 WT and BRD9 KO mice were used for Bioplex mouse 23-plex cytokine assay.
Western Blotting.
Cells were washed with ice-cold PBS and lysed using ice-cold lysis buffer (Hepes 20 mM pH 8, EDTA 0.2 mM, NaCl 0.3 mM, Nonidet P-40 0.5%, 15% glycerol) and mixture protease inhibitor (Sigma-Aldrich, 124469). Protein concentrations in the extracts were measured using Bradford and were made equal in different samples with the extraction reagent. Lysates were mixed with sample loading buffer and denatured at 95 °C for 10 min. Samples were separated by SDS/PAGE, transferred onto PVDF membrane. Immunoblot analysis was performed with BRD9 antibody (Active Motif, cat. no. 61537) and secondary anti-rabbit antibodies conjugated to horseradish peroxidase.
Supplementary Material
Acknowledgments
We thank Y. Dai for technical support, and E. Ong and C. Brondos for administrative support. This work was supported by grants from the NIH (HL088093 and HL105278); the Glenn Foundation for Medical Research; the Leona M. and Harry B. Helmsley Charitable Trust (#2017PG-MED001); Ipsen/Biomeasure; California Institute for Regenerative Medicine; the Ellison Medical Foundation (to R.M.E); and by NIH DK120808 (to Z.W.). R.M.E is a NOMIS Foundation Distinguished Scientist and Scholar at the Salk Institute. Research reported in this publication was supported by the National Institute of Diabetes and Digestive and Kidney Diseases of the NIH under Award R01DK120480, the National Institute of Diabetes and Digestive and Kidney Diseases of the NIH under Award R01DK057978, and the National Institute of Environmental Health Sciences of the NIH under Award P42ES010337. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2109517118/-/DCSupplemental.
Data Availability
RNA-Seq data have been deposited in the National Center for Biotechnology Information Sequence Read Archive (SRA), https://www.ncbi.nlm.nih.gov/sra (BioProject PRJNA731887). ChIP-seq and CUT&RUN datasets has been deposited in Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE175585).
References
- 1.Glass C. K., Natoli G., Molecular control of activation and priming in macrophages. Nat. Immunol. 17, 26–33 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Reilly S. M., Saltiel A. R., Adapting to obesity with adipose tissue inflammation. Nat. Rev. Endocrinol. 13, 633–643 (2017). [DOI] [PubMed] [Google Scholar]
- 3.Ying W., Fu W., Lee Y. S., Olefsky J. M., The role of macrophages in obesity-associated islet inflammation and β-cell abnormalities. Nat. Rev. Endocrinol. 16, 81–90 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Murray P. J., Macrophage polarization. Annu. Rev. Physiol. 79, 541–566 (2017). [DOI] [PubMed] [Google Scholar]
- 5.Platanitis E., Decker T., Regulatory networks involving STATs, IRFs, and NFκB in inflammation. Front. Immunol. 9, 2542 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smale S. T., Natoli G., Transcriptional control of inflammatory responses. Cold Spring Harb. Perspect. Biol. 6, a016261 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kuznetsova T., Prange K. H. M., Glass C. K., de Winther M. P. J., Transcriptional and epigenetic regulation of macrophages in atherosclerosis. Nat. Rev. Cardiol. 17, 216–228 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Filippakopoulos P., Knapp S., Targeting bromodomains: Epigenetic readers of lysine acetylation. Nat. Rev. Drug Discov. 13, 337–356 (2014). [DOI] [PubMed] [Google Scholar]
- 9.Kadoch C., et al., Proteomic and bioinformatic analysis of mammalian SWI/SNF complexes identifies extensive roles in human malignancy. Nat. Genet. 45, 592–601 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gatchalian J., et al., A non-canonical BRD9-containing BAF chromatin remodeling complex regulates naive pluripotency in mouse embryonic stem cells. Nat. Commun. 9, 5139 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mashtalir N., et al., Modular organization and assembly of SWI/SNF family chromatin remodeling complexes. Cell 175, 1272–1288.e20 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Alpsoy A., Dykhuizen E. C., Glioma tumor suppressor candidate region gene 1 (GLTSCR1) and its paralog GLTSCR1-like form SWI/SNF chromatin remodeling subcomplexes. J. Biol. Chem. 293, 3892–3903 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alpsoy A., et al., BRD9 is a critical regulator of androgen receptor signaling and prostate cancer progression. Cancer Res. 81, 820–833 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang X., et al., BRD9 defines a SWI/SNF sub-complex and constitutes a specific vulnerability in malignant rhabdoid tumors. Nat. Commun. 10, 1881 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brien G. L., et al., Targeted degradation of BRD9 reverses oncogenic gene expression in synovial sarcoma. eLife 7, e41305 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hohmann A. F., et al., Sensitivity and engineered resistance of myeloid leukemia cells to BRD9 inhibition. Nat. Chem. Biol. 12, 672–679 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wei Z., et al., Vitamin D switches BAF complexes to protect β cells. Cell 173, 1135–1149.e15 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sacta M. A., Chinenov Y., Rogatsky I., Glucocorticoid signaling: An update from a genomic perspective. Annu. Rev. Physiol. 78, 155–180 (2016). [DOI] [PubMed] [Google Scholar]
- 19.Cain D. W., Cidlowski J. A., Immune regulation by glucocorticoids. Nat. Rev. Immunol. 17, 233–247 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Strähle U., Klock G., Schütz G., A DNA sequence of 15 base pairs is sufficient to mediate both glucocorticoid and progesterone induction of gene expression. Proc. Natl. Acad. Sci. U.S.A. 84, 7871–7875 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Surjit M., et al., Widespread negative response elements mediate direct repression by agonist-liganded glucocorticoid receptor. Cell 145, 224–241 (2011). [DOI] [PubMed] [Google Scholar]
- 22.Ratman D., et al., How glucocorticoid receptors modulate the activity of other transcription factors: A scope beyond tethering. Mol. Cell. Endocrinol. 380, 41–54 (2013). [DOI] [PubMed] [Google Scholar]
- 23.Diamond M. I., Miner J. N., Yoshinaga S. K., Yamamoto K. R., Transcription factor interactions: Selectors of positive or negative regulation from a single DNA element. Science 249, 1266–1272 (1990). [DOI] [PubMed] [Google Scholar]
- 24.Nagaich A. K., Walker D. A., Wolford R., Hager G. L., Rapid periodic binding and displacement of the glucocorticoid receptor during chromatin remodeling. Mol. Cell 14, 163–174 (2004). [DOI] [PubMed] [Google Scholar]
- 25.Fletcher T. M., et al., ATP-dependent mobilization of the glucocorticoid receptor during chromatin remodeling. Mol. Cell. Biol. 22, 3255–3263 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Murphy D. J., Hardy S., Engel D. A., Human SWI-SNF component BRG1 represses transcription of the c-fos gene. Mol. Cell. Biol. 19, 2724–2733 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.John S., et al., Chromatin accessibility pre-determines glucocorticoid receptor binding patterns. Nat. Genet. 43, 264–268 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Remillard D., et al., Degradation of the BAF complex factor BRD9 by heterobifunctional ligands. Angew. Chem. Int. Ed. Engl. 56, 5738–5743 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang S., Liang X., Danielsen M., Role of the C terminus of the glucocorticoid receptor in hormone binding and agonist/antagonist discrimination. Mol. Endocrinol. 10, 24–34 (1996). [DOI] [PubMed] [Google Scholar]
- 30.John S., et al., Interaction of the glucocorticoid receptor with the chromatin landscape. Mol. Cell 29, 611–624 (2008). [DOI] [PubMed] [Google Scholar]
- 31.Uhlenhaut N. H., et al., Insights into negative regulation by the glucocorticoid receptor from genome-wide profiling of inflammatory cistromes. Mol. Cell 49, 158–171 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Greulich F., Wierer M., Mechtidou A., Gonzalez-Garcia O., Uhlenhaut N. H., The glucocorticoid receptor recruits the COMPASS complex to regulate inflammatory transcription at macrophage enhancers. Cell Rep. 34, 108742 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Qin Q., et al., Lisa: Inferring transcriptional regulators through integrative modeling of public chromatin accessibility and ChIP-seq data. Genome Biol. 21, 32 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Han H., et al., TRRUST v2: An expanded reference database of human and mouse transcriptional regulatory interactions. Nucleic Acids Res. 46 (D1), D380–D386 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Skene P. J., Henikoff S., An efficient targeted nuclease strategy for high-resolution mapping of DNA binding sites. eLife 6, e21856 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McLean C. Y., et al., GREAT improves functional interpretation of cis-regulatory regions. Nat. Biotechnol. 28, 495–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Loo C. S., et al., A genome-wide CRISPR screen reveals a role for the non-canonical nucleosome-remodeling BAF complex in Foxp3 expression and regulatory T cell function. Immunity 53, 143–157.e8 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lamia K. A., et al., Cryptochromes mediate rhythmic repression of the glucocorticoid receptor. Nature 480, 552–556 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang X., Goncalves R., Mosser D. M., “The isolation and characterization of murine macrophages” in Current Protocols in Immunology (John Wiley & Sons, 2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Barish G. D., et al., Bcl-6 and NF-kappaB cistromes mediate opposing regulation of the innate immune response. Genes Dev. 24, 2760–2765 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li R.-J., et al., De novo assembly and characterization of the fruit transcriptome of Idesia polycarpa reveals candidate genes for lipid biosynthesis. Front Plant Sci 7, 801 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Meers M. P., Bryson T. D., Henikoff J. G., Henikoff S., Improved CUT&RUN chromatin profiling tools. Elife, e46314 (2019). [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-Seq data have been deposited in the National Center for Biotechnology Information Sequence Read Archive (SRA), https://www.ncbi.nlm.nih.gov/sra (BioProject PRJNA731887). ChIP-seq and CUT&RUN datasets has been deposited in Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE175585).