Abstract
Even though neurons are the main drivers of information processing in the brain and spinal cord, other cell types are important to mediate adequate flow of information. These include electrically passive glial cells such as microglia and astrocytes, which recently emerged as active partners facilitating proper signal transduction. In disease, these cells undergo pathophysiological changes that propel disease progression and change synaptic connections and signal transmission. In the healthy brain, astrocytic processes contact pre- and postsynaptic structures. These processes can be nanoscopic, and therefore only electron microscopy has been able to reveal their structure and morphology. However, electron microscopy is not suitable in revealing dynamic changes, and it is labour- and time-intensive. The dawn of super-resolution microscopy, techniques that ‘break’ the diffraction limit of conventional light microscopy, over the last decades has enabled researchers to reveal the nanoscopic synaptic environment. In this review, we highlight and discuss recent advances in our understanding of the nano-world of the so-called tripartite synapses, the relationship between pre- and postsynapse as well as astrocytic processes. Overall, novel super-resolution microscopy methods are needed to fully illuminate the intimate relationship between glia and neuronal cells that underlies signal transduction in the brain and that might be affected in diseases such as Alzheimer’s disease and epilepsy.
Keywords: astrocytes, super-resolution microscopy, synapses, tripartite synapses
Introduction
Information processing in the central nervous system occurs mainly at synapses, the connection points between neurons. An intimate relationship between neurons and glial cells underlies proper synaptic function. Astroglia in particular play essential roles in information processing. These cells are not only needed for structural support but astroglia also fulfil several other indispensable functions. These include clean-up of brain debris, digestion of dead neurons and pruning of synapses [1] as well as being the drivers of the glymphatic system, the major waste clearing system in the brain [2–4]. Moreover, related to synaptic function, astrocytes express many powerful neurotransmitter and ion channels as well as transporters. These allow them to sustain homeostasis in the brain through processes such as glutamate uptake [5,6] and potassium buffering [7,8]. Moreover, astrocytes release gliotransmitters that directly impact synaptic function [9,10]. These processes are dysregulated in many disorders, including Alzheimer’s disease and epilepsy [11,12].
In the brain, astrocytes adopt a sponge-like morphology, featuring a few stem processes and thousands of thin protrusions that emanate from these stem processes [13,14]. While these fine protrusions permeate the extracellular space, there is minimal overlap between territories formed by individual cells [15,16]. This is surprising as astrocytes are interconnected through gap junctions, forming a syncytium that tiles the brain [15,16]. Although astrocytes are electrically passive, they signal through Ca2+ waves and through the aforementioned syncytium [17–19]. Through this calcium signalling, the cells are able to integrate and transmit physiological signals among neuronal and glial cells [14,20,21].
The fine protrusions can either form astrocytic endfeet and encircle endothelial cells as part of the neurovascular unit of the blood–brain barrier or they can enwrap excitatory synapses throughout the brain. These processes are often called perisynaptic astrocytic processes (PAPs) and form part of the so called tripartite synapse, together with pre- and postsynaptic structures [22–25]. Depending on the brain region and the physiological state of the animal this synapse coverage varies [for reviews see 13,26]. There is a constant molecular exchange between glial cells and neurons, shaping synaptic transmission and regulating use-dependent plasticity, in the healthy brain as well as in pathological states [9,20,26–29].
Due to the nanoscopic nature (their diameter can often be smaller than the diffraction limit of light) of the PAPs, researchers have had to rely on electron microscopy (EM) to visualise and investigate tripartite synapses [14,30,31]. EM revealed the intricate relationship between neuronal, synaptic structures and astrocytes processes (e.g. reviewed in [13]). It was also shown using EM that PAPs preferentially approach thin spines over larger spines [32].
PAP coverage of synapses depends on several factors and differs depending on the physiological state, local neuronal activity, synaptic plasticity, or certain behaviours [33–39].
Confocal microscopy, which is diffraction limited, has been used to explore the dynamic nature of PAPs in real time [40–43]. The dynamic nature of PAPs has also been shown with super-resolution microscopy approaches (see below).
In this review, we will provide an overview and discussion of recent developments in tripartite synapse imaging using super-resolution methods. We will also provide a short introduction into different super-resolution methods and recent advances in revealing the nanostructure of neuronal synapses. However, for further information we point the reader to some excellent, recent reviews on super-resolution microscopy and also its use in neuroscience research [44–53].
Super-resolution methods
As mentioned above, confocal microscopy (as well as other types of light microscopy) is diffraction limited, meaning that discerning individual molecules can be difficult. EM, which has traditionally been used in order to image nanoscopic molecules, comes with several disadvantages, including its exclusive use for fixed tissue specimens. In addition, it is labour and time intensive, and correlational comparisons between different samples are difficult to attain and interpret. This is also true for some of the super-resolution methods described below.
Super resolution imaging allows the user to image beyond the diffraction limit of light while using traditional sample immunostaining methods associated with fluorescent light microscopy. Several super resolution methods have been developed, each with its own advantages and disadvantages. However, the discussion of these is beyond the scope of this review and the authors again point to some excellent reviews of this topic [44–53]. Most widely used methods include stimulated-emission depletion (STED) microscopy [54], structured illumination microscopy (SIM) [55], single molecule localisation microscopy (SMLM) [56–58] and expansion microscopy (ExM) [59,60]. Most of the super-resolution imaging is usually performed on fixed specimens. However, STED and SIM are suited for live cell and live tissue imaging (see below). Moreover, some SMLM adaptations such as single particle tracking PALM (sptPALM) and universal point accumulation for imaging in nanoscale topography (uPAINT) can be used to track single-molecule trajectories at very high densities, for short periods of time [46].
STED microscopy relies on a second, doughnut-shaped excitation beam which depletes excitation at the periphery of the primary excitation beam, hence narrowing the emission spot and allowing resolution beyond the diffraction limit [54]. One of the advantages of using STED is that there is no need for further computational processing, as is the case in other super-resolution microscopy methods. This reduces the risk of artifact generation. However, as this method requires high laser intensity, it runs the risk of bleaching the sample as well as causing physical damage to live cells [49,50,53].
SIM relies on bar code-like patterns which are shifted and rotated in the excitation path [55]. This creates high-frequency information which can be used to reconstruct a super-resolved image. As mentioned, SIM is particularly well suited for imagine live samples. However, as multiple images need to be taken at high intensities, this method runs the risk of damaging live samples, similar to STED. SIM can be expensive to set up, as it requires a dedicated microscope [53].
SMLM encompasses several different techniques that all rely on a similar molecular mechanism - pinpointing the point source of fluorescence (which can be one fluorophore/molecule) in each imaging cycle through repeated stochastic excitation of only a small, sparsely distributed subset of fluorescent molecules [58]. After acquiring several thousand frames a super-resolved image can be reconstructed. SMLM includes photo-activated localisation microscopy (PALM) and stochastic optical reconstruction microscopy (STORM) [56,57]. These images can be taken with a regular fluorescent microscope and there are many free software options available in order to process these images, making it one of the more easily accessible super-resolution microscopy methods [50]. However, due to the processing of these images, it can be prone to artifact generation [61].
ExM uses literal expansion of the tissue by a factor great enough to separate individual molecules of interest beyond the diffraction limit, followed by imaging with conventional microscopes such as confocal microscopy [59,60]. This method is relatively inexpensive and easy to implement. However, it is only suitable to be used on fixed samples [62].
Super-resolution imaging of neuronal synaptic structures
Several advances have been made in our understanding of neuronal synapses using super-resolution microscopy (for recent reviews see [45–48,52]).
Especially STED microscopy has been employed to analyse the composition of presynaptic structures and vesicle dynamics within, both in vivo and in vitro [63–67], showing for example that about 75 densely packed plasma membrane soluble N-ethylmaleimide-sensitive factor attachment protein receptor (SNARE) proteins form clusters of ∼60 nm diameter in PC12 cells [68]. SMLM was later used to investigate the nanoclustering behaviour of individual molecular players at the presynapse in vivo and in vitro [69–72].
STED as well as SMLM have been used to image dendritic spines and postsynaptic structures in cultured cells and brain slices [73–79]. Recently, STED has been employed to illuminate spine dynamics in living mice [80–86]. These developments now make it possible to image up to three labels in the cortex of living mice [85], and to chronically illuminate the same super-resolved structures over a time course of up to one month [86]. Besides revealing more macroscopic structures of spines and the underlying cytoskeletal structures, super-resolution microscopy has been used to further our understanding of the nanoscale organisation of the postsynaptic density (PSD).
Seminal work published in 2010 provided a detailed three-dimensional map of the nanoscale structure of excitatory synapses in fixed murine brain sections using SMLM [87]. The group also compared activity-dependent changes in the organisation of N-methyl-D-aspartate receptors (NMDARs) and α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid receptors (AMPARs) across the mouse olfactory bulb [87].
Using different super-resolution microscopy techniques, it was shown that AMPARs are packed in nanoscopic clusters in cultured cells [88–90]. These clusters are smaller than the PSD (0-4 AMPAR nanodomains per PSD, with an average size of ∼80 nm, and 20–25 AMPARs per nanodomain), and their number depends on the size of the PSD. The researchers also found that PSD95 molecules show clustering behaviour in nanodomains of ∼150 nm diameter within each PSD in mouse brain tissue and in cultured cells [88–92].
Likewise, the nanoscale clustering of the scaffolding protein gephyrin as well as of glycine and gamma-aminobutyric acid (GABA) type A receptors at inhibitory synapses has been investigated [93–95]. Using quantitative 3D-PALM, Specht and colleagues measured that 40–500 gephyrin molecules are packed at a density of about 5000 molecules/µm2 in cultured spinal cord neurons, and that in situ about three times as many gephyrin molecules are packed more densely [94].
Recent beautiful work combined super-resolution microscopy imaging with plasticity induction and revealed a trans-synaptic organisation of scaffolding molecules both at excitatory and at inhibitory synapses in organised tissue and in cultured cells [92,93,95–97]. These groups found that presynaptic proteins such as RIM1/2 form nanodomains in similar ways to postsynaptic scaffolding proteins PSD95 and gephyrin. These nanodomains are aligned in nanocolumns across the synaptic cleft, which remain intact after plasticity induction.
Super-resolution imaging of tripartite synapses
As discussed above, astrocyte processes and especially PAPs display nanoscopic features. Early super-resolution studies did not focus on the relationship between synapses and astrocytic molecules but rather on the subcellular localisation and clustering behaviour of individual proteins of interest. For example, Verkman and colleagues investigated the clustering behaviour of aquaporin 4 and of the inwardly rectifying potassium channel Kir4.1 using SMLM techniques in cultured cells and in brain sections [98–100]. Moreover, thin astroglial processes have been illuminated by expressing genetically-encoded fluorescent proteins and by labelling glutamine synthetase and S100β in cultured cells and brain sections [34,101–105].
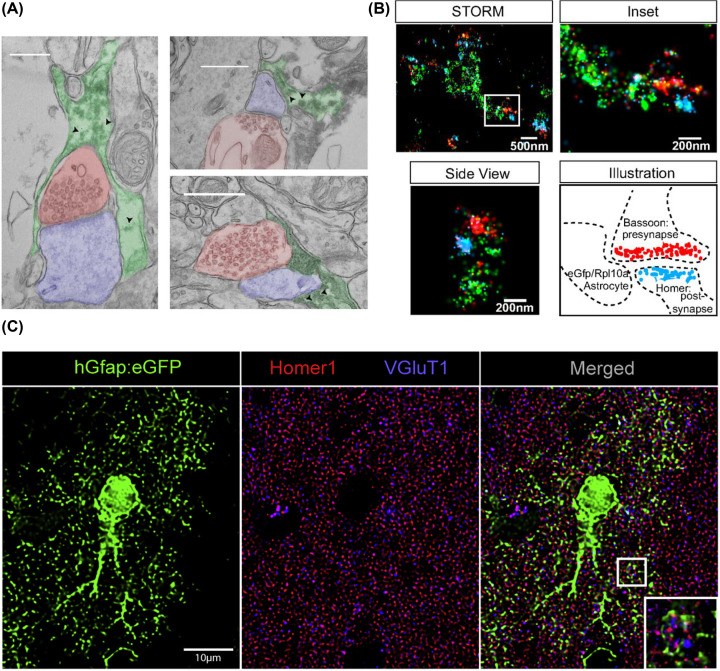
Recent studies have investigated translation events that occur locally in astrocytic processes in the vicinity of synapses [106–108]. EM and SMLM have been used to reveal the presence of Rpl10a, a component of the 60S ribosomal subunit, in close apposition to synapses in mouse brain sections [106] (Figure 1A,B). Local translation in astrocytic processes has been implicated in fear conditioning [107], and its impairment might contribute to the development of amyotrophic lateral sclerosis [109]. Here, researchers used STED and confocal microscopy to visualise astrocyte processes in the vicinity of excitatory synapses in mouse brain slices [107] (Figure 1C). The presence of mRNA and local translation has also been shown to occur in astrocytic endfeet surrounding blood vessels in the brain [110].
Figure 1. EM, SMLM and STED images visualising astrocyte processes in the vicinity of excitatory synapses.
(A) EM image of diaminobenzidine-labelled EGFP/RPL10A (arrowheads) in astrocyte processes (green) near cortical synapses (axon = red and PSD = blue); scale bars = 500 nm; modified from [106] with permission. (B) SMLM image showing an EGFP/RPL10A (green) filled astrocyte process near a synapse (presynaptic bassoon (red) and postsynaptic Homer (blue); inset of box on left, and side view is a 90° rotation of a second synapse; modified from [106] with permission. (C) Deconvoluted confocal microscopy image of an astrocyte (hGfap-eGFP, green), presynaptic vGluT1 (blue) and postsynaptic Homer1 (red); the magnified area shows the STED image for vGluT1 and Homer1 merged with deconvoluted confocal image for eGFP; scale bar = 10 µm; modified from [107] with permission.
Using STED and SIM, researchers have shown that neuronal activity increased connexin 30 expression and localisation in perisynaptic processes in hippocampal astrocytes in mouse brain slices [111]. The same group recently showed that astrocytes close the critical period for visual plasticity via developmental upregulation of connexin 30 which in turn inhibits expression of the extracellular matrix degrading enzyme matrix metalloproteinase 9 [112].
In a recent beautiful publication, researchers have used a plethora of techniques including an in vivo chemico-genetic approach that applies a cell-surface fragment complementation strategy, Split-TurboID and STED discovering that neuronal cell adhesion molecule (NRCAM) is expressed in cortical astrocytes where it localises to PAPs and restricts their neuropil infiltration [113]. Astrocytic NRCAM interacts with neuronal NRCAM coupled to gephyrin at inhibitory postsynapses, and its deletion reduces the number of inhibitory synapses and decreases inhibitory synaptic function with minimal effects on glutamatergic synaptic density or overall excitation in mice [113].
Recently, researchers visualised bestrophin-1 (Best1) expression at tripartite synapses using lattice SIM and found that, in brain tissue from wild-type mice, astrocytic Best1 localises closer to glutamatergic synapses than to GABAergic synapses [114]. In APP/PS1 mice (Alzheimer’s disease model) tissue, however, Best1 resides further away from glutamatergic synapses and closer to GABAergic synapses [114].
In a recent preprint, Südhof and colleagues used SMLM in mouse brain sections to decipher the role of astrocytic neurexin-1 in synapse function and maturation [115]. The authors showed that neurons as well as astrocytes express neurexin-1, which is organised in discrete nanoclusters at excitatory synapses. Distinct heparan sulphate modifications and alternative splicing lead to different ligand specificities and enables compartment-specific neurexin-1 signalling. Even though deletion of neurexin-1 from either astrocytes or neurons did not have an effect on synapse numbers, the authors found that neuronal neurexin-1 is required for NMDAR-mediated synaptic responses, and that astrocytic neurexin-1 is needed for silent synapse maturation, AMPAR recruitment and long-term potentiation [115].
Recent work utilising STED imaging showed that in spinal cord sections 56% of synapses (PSD95 and vGluT2 or vGluT1 pairs) are association with GLT-1 (the major astrocytic glutamate transporter), 30% with glial fibrillary acidic protein and 14% with phosphorylated ezrin [116]. PSDs associated with an astrocytic protein are larger in size and fluorescent intensity. Furthermore, PSDs that comprised more than one PSD95 nanocluster are more likely to be part of a tripartite synapse, and tripartite nanoclusters appear brighter and hence more enriched in PSD95 [116].
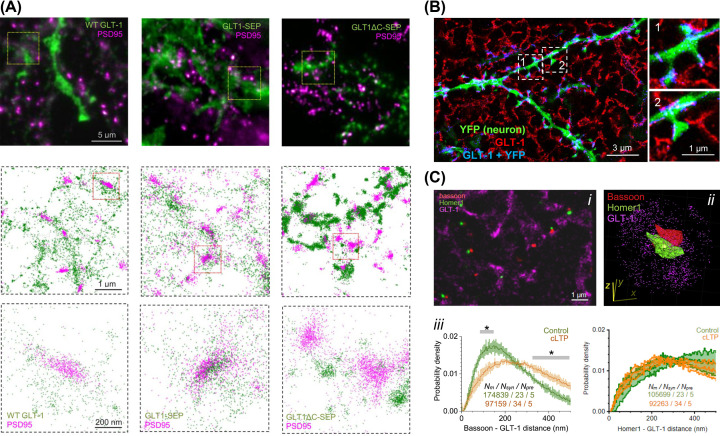
In a recent publication, we used fluorescence recovery after photobleaching in organotypic rat hippocampal sections to demonstrate that 70–75% of GLT-1 molecules dwell on the astrocyte surface, recycling with a lifetime of ∼22 s [117]. Using SMLM in cultured rat hippocampal cultures, we showed that GLT-1 surface expression and clustering behaviour relies on its C-terminus and its deletion accelerates GLT1 membrane turnover (Figure 2A) [117].
Figure 2. SMLM and ExM images highlighting GLT-1 distribution around synapses.
(A) Wide-field fluorescent images highlighting GLT-1 (green) and PSD95 (magenta) in mixed hippocampal astroglia-neuron cultures (top row). SMLM images showing individual labelled GLT-1 (green) and PSD95 (magenta) localisations (middle row depicting yellow squares in top row; bottom row depicting red squares in the middle row); modified from [117] with permission. (B) ExM image of dendrite and spines of a CA1 pyramidal neuron (green, YFP) and surrounding GLT-1 (red) labelling; with regions of ‘colocalisation’ highlighted in blue; modified from [119] with permission. (C) SMLM images highlighting localisations of presynaptic bassoon (red), postsynaptic Homer 1 (green) and astrocytic GLT-1 (magenta) (i and ii). Nearest-neighbour distances (probability density, mean ± SEM) between GLT-1 and bassoon or Homer1 (in control tissue (green) and ∼30 min after chemical long term potentiation (cLTP) induction (brown)) (iii); sample size: Nm, inter-molecular distances; Nsyn, synapses; Npre, slices; SEM relates to Npre = 5; ∗P<0.05 (grey segments, significant difference), modified from [34] with permission.
Recent attempts to study astrocyte processes and glutamate uptake have utilised ExM of mouse brain sections [118,119]. The authors showed that larger spines had reduced glutamate uptake efficiency which correlated with greater GLT-1 levels (Figure 2B) [119].
To further analyse the distribution of glutamate transporters surrounding excitatory synapses, we used a 3D SMLM protocol of brain sections [34,120,121]. Through a series of experiments involving optical glutamate sensors, patch clamp electrophysiology, sensory stimulation and super-resolution microscopy, we showed that induction of long-term potentiation leads to withdrawal of PAPs from excitatory synapses which in turn boosts glutamate escape and inter-synaptic cross-talk (Figure 2C) [34].
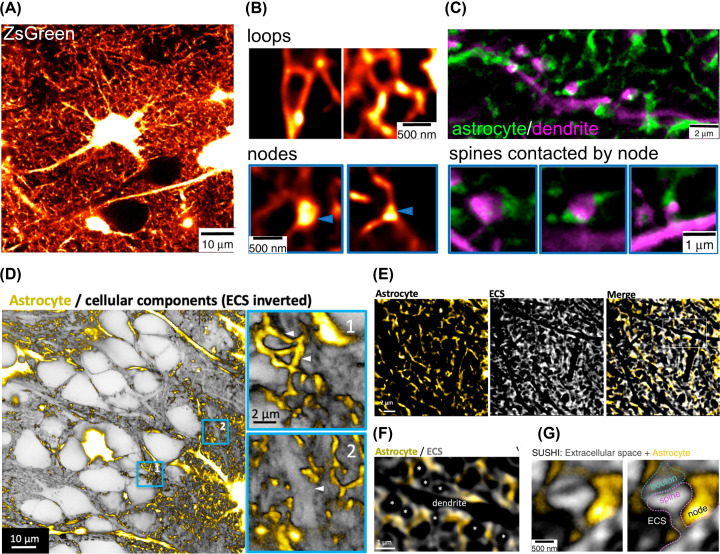
As mentioned above, astroglia show diverse calcium signalling [21]. In a recent beautiful study, Nägerl and colleagues investigated the nanoscopic structure of PAPs and their localised Ca2+ transients using STED microscopy in living organotypic brain slices (Figure 3A–C) [105]. The researchers found that thin astrocytic processes are organised in a fine reticular network, exhibiting nodes, shafts and ring-like structures. The majority of Ca2+ transients takes place in nodes, and 55% of spines are in close apposition to at least one node. Node size as well as the size of the Ca2+ transients were positively correlated to spine size, highlighting the possibility that there are different astrocyte compartments handling different modes of signalling at node versus shaft tripartite synapses [105].
Figure 3. STED images depicting nanoscopic astrocyte structures in living organotypic slices.
(A) Confocal image of ZsGreen expressing astrocytes, highlighting the entire cell morphology; modified from [105] with permission. (B) STED images of astrocyte spongiform morphology, revealing specialised process structures – loops and nodes, modified from [105] with permission. (C) STED images of astrocyte (green) and dendrites (magenta), highlighting astrocytic nodes as the main contact points of spines; modified from [105] with permission. (D) STED image of astrocytes (gold) and inverted signal of extracellular space (ECS), highlighting astrocytic processes penetrating the neuropil; modified from [104] with permission. (E) STED images of positively labelled astrocyte (gold) and stained ECS (grey), modified from [104] with permission. (F) STED image of astrocyte (gold) and ECS (grey) depicting white rectangle in (E), highlighting a negative imprint of a putative dendrite and spines (*) (black), modified from [104] with permission. (G) STED image of tripartite synapse with astrocytic node in close apposition to spine, modified from [105] with permission.
In a second study, Nägerl and colleagues used ‘super-resolution shadow imaging’ (SUSHI), a technique, in which the extracellular space is fluorescently labelled and cellular structures appear as dark shadows (Figure 3D–G) [104,105,122]. The researchers saw that the aforementioned ring-like structures that are formed by the reticular network of astrocytic processes, encircle spines and areas of interstitial fluid (Figure 3D–G) [104]. After osmotic challenge, the astrocyte structures swell, increasing the interface between astrocytic and other cellular structures but decreasing the size of interstitial pools [104].
Concluding remarks
Super-resolution microscopy has helped further our understanding of neuronal and glial cell morphology and molecular dynamics. Most findings were acquired in fixed cell culture or brain section preparations. However, it is known that cultured cells differ from their in vivo counterparts. This is especially true for astroglia as their morphology and expression profiles are very different in culture and in intact tissue. Additionally, PAPs appear to be very sensitive to classical tissue fixation protocols, with a significant difference of PAP-positive synapses found in the adult murine neocortex when comparing chemical and cryo fixation (chemical 62% versus cryo: 34%) [123]. Adopting milder fixation methods and the development of improved in vivo imaging platforms will help evaluating the true morphology of astrocytes and hence tripartite synapses. We are only just beginning to fully explore the potential of super-resolution microscopy technology and its future capabilities, especially in the field of diagnostics.
As mentioned above, recent developments using STED microscopy have made in vivo nanoscopy possible. Further adaptations and the integration of adaptive optics [124] will improve image resolution and depth penetration, making imaging of highly dynamic and densely packed structures in living brain tissue more feasible. Implementations of adaptive optics have already shown promising results using in vivo two-photon excitation imaging [125–127], STED [128,129] and SMLM [130].
Astrocytes found in higher primates are larger and more complex, and show unique functions and different abilities to cope with stressors and disease responses when compared with murine astrocytes [131–134]. Super-resolution microscopy techniques will be essential in understanding the differences on a molecular level.
It is well documented that glial cells undergo drastic changes under pathological conditions [135]. Super-resolution imaging has already been applied for analysis of models of neurological and neurodegenerative diseases [136–139]. Additionally, it has also recently been utilised in human Alzheimer’s and Parkinson’s disease post-mortem tissue as a diagnostic tool [140,141].
Hence, the use of super-resolution will be a valuable means for both clinicians and researchers in deciphering the exact pathophysiological changes occurring in and around synapses, and in evaluating novel therapeutic options.
Acknowledgements
The authors thank Mr Trevor Tendai Shoniwa for help with initial literature search.
Abbreviations
- AMPAR
α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid receptors
- BEST1
bestrophin-1
- EM
electron microscopy
- ExM
expansion microscopy
- GABA
gamma-aminobutyric acid
- NMDAR
N-methyl-D-aspartate receptors
- NRCAM
neuronal cell adhesion molecule
- PALM
photo-activated localisation microscopy
- PAP
perisynaptic astrocytic process
- PSD
postsynaptic density
- SIM
structured illumination microscopy
- SMLM
single-molecule localisation microscopy
- SNARE
soluble N-ethylmaleimide-sensitive factor attachment protein receptor
- sptPALM
single particle tracking PALM
- STED
stimulated-emission depletion
- STORM
stochastic optical reconstruction microscopy
- SUSHI
super-resolution shadow imaging
- uPAINT
universal point accumulation for imaging in nanoscale topography
Data Availability
No data were used for this review article.
Competing Interests
The authors declare that there are no competing interests associated with the manuscript.
Funding
J.P.H. is supported by a Royal Irish Academy Charlemont Grant and Enterprise Ireland (EI) for European Research Council (ERC) Proposal-Preparation-Support.
References
- 1.Lee J.-H., Kim J., Noh S., Lee H., Lee S.Y., Mun J.Y.et al. (2020) Astrocytes phagocytose adult hippocampal synapses for circuit homeostasis. Nature 590, 612–617 10.1038/s41586-020-03060-3 [DOI] [PubMed] [Google Scholar]
- 2.Jessen N.A., Munk A.S.F., Lundgaard I. and Nedergaard M. (2015) The glymphatic system: a beginner's guide. Neurochem. Res. 40, 2583–2599 10.1007/s11064-015-1581-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Iliff J.J., Chen M.J., Plog B.A., Zeppenfeld D.M., Soltero M., Yang L.et al. (2014) Impairment of glymphatic pathway function promotes tau pathology after traumatic brain injury. J. Neurosci. 34, 16180–16193 10.1523/JNEUROSCI.3020-14.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mestre H., Hablitz L.M., Xavier A.L.R., Feng W., Zou W., Pu T.et al. (2018) Aquaporin-4-dependent glymphatic solute transport in the rodent brain. Elife 7, 10.7554/eLife.40070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bergles D.E. and Jahr C.E. (1997) Synaptic activation of glutamate transporters in hippocampal astrocytes. Neuron 19, 1297–1308 10.1016/S0896-6273(00)80420-1 [DOI] [PubMed] [Google Scholar]
- 6.Diamond J.S. (2001) Neuronal glutamate transporters limit activation of NMDA receptors by neurotransmitter spillover on CA1 pyramidal cells. J. Neurosci. 21, 8328–8338 10.1523/JNEUROSCI.21-21-08328.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hertz L. (1965) Possible role of neuroglia: a potassium-mediated neuronal - neuroglial - neuronal impulse transmission system. Nature 206, 1091–1094 10.1038/2061091a0 [DOI] [PubMed] [Google Scholar]
- 8.Orkand R.K., Nicholls J.G. and Kuffler S.W. (1966) Effect of nerve impulses on the membrane potential of glial cells in the central nervous system of amphibia. J. Neurophysiol. 29, 788–806 10.1152/jn.1966.29.4.788 [DOI] [PubMed] [Google Scholar]
- 9.Araque A., Carmignoto G., Haydon P.G., Oliet S.H.R., Robitaille R. and Volterra A. (2014) Gliotransmitters travel in time and space. Neuron 81, 728–739 10.1016/j.neuron.2014.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lalo U., Rasooli-Nejad S., Bogdanov A., More L., Koh W., Muller J.et al. (2021) Synergy between vesicular and non-vesicular gliotransmission regulates synaptic plasticity and working memory. BioRxiv, 10.1101/2021.03.25.437028 [DOI] [Google Scholar]
- 11.Dossi E., Vasile F. and Rouach N. (2017) Human astrocytes in the diseased brain. Brain Res. Bull. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Seifert G., Schilling K. and Steinhäuser C. (2006) Astrocyte dysfunction in neurological disorders: a molecular perspective. Nat. Rev. Neurosci. 7, 194–206 10.1038/nrn1870 [DOI] [PubMed] [Google Scholar]
- 13.Heller J.P. and Rusakov D.A. (2015) Morphological plasticity of astroglia: understanding synaptic microenvironment. Glia 63, 2133–2151 10.1002/glia.22821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rusakov D.A. and Stewart M.G. (2021) Synaptic environment and extrasynaptic glutamate signals: the quest continues. Neuropharmacology 195, 108688 10.1016/j.neuropharm.2021.108688 [DOI] [PubMed] [Google Scholar]
- 15.Bushong E.A., Martone M.E. and Ellisman M.H. (2004) Maturation of astrocyte morphology and the establishment of astrocyte domains during postnatal hippocampal development. Int. J. Dev. Neurosci. 22, 73–86 10.1016/j.ijdevneu.2003.12.008 [DOI] [PubMed] [Google Scholar]
- 16.Halassa M.M., Fellin T., Takano H., Dong J.-H. and Haydon P.G. (2007) Synaptic islands defined by the territory of a single astrocyte. J. Neurosci. 27, 6473–6477 10.1523/JNEUROSCI.1419-07.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cornell-Bell A.H., Finkbeiner S.M., Cooper M.S. and Smith S.J. (1990) Glutamate induces calcium waves in cultured astrocytes: Long-range glial signaling. Science (80-) 247, 470–473 10.1126/science.1967852 [DOI] [PubMed] [Google Scholar]
- 18.Nedergaard M. (1994) Direct signaling from astrocytes to neurons in cultures of mammalian brain cells. Science (80-) 263, 1768–1771 10.1126/science.8134839 [DOI] [PubMed] [Google Scholar]
- 19.Parpura V., Basarsky T.A., Liu F., Jeftinija K., Jeftinija S. and Haydon P.G. (1994) Glutamate-mediated astrocyte-neuron signalling. Nature 369, 744–747 10.1038/369744a0 [DOI] [PubMed] [Google Scholar]
- 20.Bazargani N. and Attwell D. (2016) Astrocyte calcium signaling: the third wave. Nat. Neurosci. 19, 182–189 10.1038/nn.4201 [DOI] [PubMed] [Google Scholar]
- 21.Lia A., Henriques V.J., Zonta M., Chiavegato A., Carmignoto G., Gómez-Gonzalo M.et al. (2021) Calcium signals in astrocyte microdomains, a decade of great advances. Front. Cell Neurosci. 15, 177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Halassa M.M., Fellin T. and Haydon P.G. (2007) The tripartite synapse: roles for gliotransmission in health and disease. Trends Mol. Med. 13, 54–63 10.1016/j.molmed.2006.12.005 [DOI] [PubMed] [Google Scholar]
- 23.Perea G., Navarrete M. and Araque A. (2009) Tripartite synapses: astrocytes process and control synaptic information. Trends Neurosci. 32, 421–431 10.1016/j.tins.2009.05.001 [DOI] [PubMed] [Google Scholar]
- 24.Araque A., Parpura V., Sanzgiri R.P. and Haydon P.G. (1999) Tripartite synapses: glia, the unacknowledged partner. Trends Neurosci. 22, 208–215 10.1016/S0166-2236(98)01349-6 [DOI] [PubMed] [Google Scholar]
- 25.Haydon P.G. (2001) GLIA: listening and talking to the synapse. Nat. Rev. Neurosci. 2, 185–193 10.1038/35058528 [DOI] [PubMed] [Google Scholar]
- 26.Zhou B., Zuo Y.Y.-X. and Jiang R.-T.R. (2019) Astrocyte morphology: Diversity, plasticity, and role in neurological diseases. CNS Neurosci. Ther. 25, 10.1111/cns.13123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sancho L., Contreras M. and Allen N.J. (2021) Glia as sculptors of synaptic plasticity. Neurosci. Res. 167, 17–29 10.1016/j.neures.2020.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Durkee C.A. and Araque A. (2019) Diversity and specificity of astrocyte-neuron communication. Neuroscience 396, 73–78 10.1016/j.neuroscience.2018.11.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim Y.S., Choi J. and Yoon B.-E. (2020) Neuron-Glia interactions in neurodevelopmental disorders. Cells 9, 2176 10.3390/cells9102176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kiyoshi C.M., Aten S., Arzola E.P., Patterson J.A., Taylor A.T., Du Y.et al. (2020) Ultrastructural view of astrocyte-astrocyte and astrocyte-synapse contacts within the hippocampus. BioRxiv, 10.1101/2020.10.28.358200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Aboufares El Alaoui A., Jackson M., Fabri M., de Vivo L. and Bellesi M. (2021) Characterization of subcellular organelles in cortical perisynaptic astrocytes. Front Cell Neurosci. 14, 492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Medvedev N., Popov V., Henneberger C., Kraev I., Rusakov D.A. and Stewart M.G. (2014) Glia selectively approach synapses on thin dendritic spines. Philos. Trans. R. Soc. Lond. B Biol. Sci. 369, 20140047, 10.1098/rstb.2014.0047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wenzel J., Lammert G., Meyer U. and Krug M. (1991) The influence of long-term potentiation on the spatial relationship between astrocyte processes and potentiated synapses in the dentate gyrus neuropil of rat brain. Brain Res. 560, 122–131 10.1016/0006-8993(91)91222-M [DOI] [PubMed] [Google Scholar]
- 34.Henneberger C., Bard L., Panatier A., Reynolds J.P., Kopach O., Medvedev N.I.et al. (2020) LTP induction boosts glutamate spillover by driving withdrawal of perisynaptic astroglia. Neuron 108, 10.1016/j.neuron.2020.08.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jones T.A. and Greenough W.T. (1996) Ultrastructural evidence for increased contact between astrocytes and synapses in rats reared in a complex environment. Neurobiol. Learn. Mem. 65, 48–56 10.1006/nlme.1996.0005 [DOI] [PubMed] [Google Scholar]
- 36.Oliet S.H., Piet R. and Poulain D.A. (2001) Control of glutamate clearance and synaptic efficacy by glial coverage of neurons. Science 292, 923–926 10.1126/science.1059162 [DOI] [PubMed] [Google Scholar]
- 37.Lushnikova I., Skibo G., Muller D. and Nikonenko I. (2009) Synaptic potentiation induces increased glial coverage of excitatory synapses in CA1 hippocampus. Hippocampus 19, 753–762 10.1002/hipo.20551 [DOI] [PubMed] [Google Scholar]
- 38.Bernardinelli Y., Muller D. and Nikonenko I. (2014) Astrocyte-synapse structural plasticity. Neural Plast. 2014, 232105 10.1155/2014/232105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ostroff L.E., Manzur M.K., Cain C.K. and Ledoux J.E. (2014) Synapses lacking astrocyte appear in the amygdala during consolidation of pavlovian threat conditioning. J. Comp. Neurol. 522, 2152–2163 10.1002/cne.23523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hirrlinger J., Hülsmann S. and Kirchhoff F. (2004) Astroglial processes show spontaneous motility at active synaptic terminals in situ. Eur. J. Neurosci. 20, 2235–2239 10.1111/j.1460-9568.2004.03689.x [DOI] [PubMed] [Google Scholar]
- 41.Haber M., Zhou L. and Murai K.K. (2006) Cooperative astrocyte and dendritic spine dynamics at hippocampal excitatory synapses. J. Neurosci. 26, 8881–8891 10.1523/JNEUROSCI.1302-06.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bernardinelli Y., Randall J., Janett E., Nikonenko I., König S., Jones E.V.V.et al. (2014) Activity-dependent structural plasticity of perisynaptic astrocytic domains promotes excitatory synapse stability. Curr. Biol. 24, 1679–1688 10.1016/j.cub.2014.06.025 [DOI] [PubMed] [Google Scholar]
- 43.Perez-Alvarez A., Navarrete M., Covelo A., Martin E.D. and Araque A. (2014) Structural and functional plasticity of astrocyte processes and dendritic spine interactions. J. Neurosci. 34, 12738–12744 10.1523/JNEUROSCI.2401-14.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Valli J., Garcia-Burgos A., Rooney L.M., de Melo e Oliveira B.V., Duncan R.R. and Rickman C. (2021) Seeing beyond the limit: A guide to choosing the right super-resolution microscopy technique. J. Biol. Chem. 297, 100791 10.1016/j.jbc.2021.100791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gallagher B.R. and Zhao Y. (2021) Expansion microscopy: a powerful nanoscale imaging tool for neuroscientists. Neurobiol. Dis. 154, 10.1016/j.nbd.2021.105362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Choquet D., Sainlos M. and Sibarita J.-B. (2021) Advanced imaging and labelling methods to decipher brain cell organization and function. Nat. Rev. Neurosci. 22, 237–255 10.1038/s41583-021-00441-z [DOI] [PubMed] [Google Scholar]
- 47.Carvalhais L.G., Martinho V.C., Ferreiro E. and Pinheiro P.S. (2021) Unraveling the nanoscopic organization and function of central mammalian presynapses with super-resolution microscopy. Front. Neurosci. 0, 1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kashiwagi Y. and Okabe S. (2021) Imaging of spine synapses using super-resolution microscopy. Anat. Sci. Int. 96, 343–358 10.1007/s12565-021-00603-0 [DOI] [PubMed] [Google Scholar]
- 49.Möckl L. and Moerner W.E. (2020) Super-resolution microscopy with single molecules in biology and beyond-essentials, current trends, and future challenges. J. Am. Chem. Soc. 142, 17828–17844 10.1021/jacs.0c08178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jacquemet G., Carisey A.F., Hamidi H., Henriques R. and Leterrier C. (2020) The cell biologist's guide to super-resolution microscopy. J. Cell Sci. 133, 10.1242/jcs.240713 [DOI] [PubMed] [Google Scholar]
- 51.Schermelleh L., Ferrand A., Huser T., Eggeling C., Sauer M., Biehlmaier O.et al. (2019) Super-resolution microscopy demystified. Nat. Cell Biol. 21, 72–84 10.1038/s41556-018-0251-8 [DOI] [PubMed] [Google Scholar]
- 52.Werner C., Sauer M. and Geis C. (2021) Super-resolving microscopy in neuroscience. Chem. Rev. 10.1021/acs.chemrev.0c01174 [DOI] [PubMed] [Google Scholar]
- 53.Vangindertael J., Camacho R., Sempels W., Mizuno H., Dedecker P. and Janssen K.P.F.P.F. (2018) An introduction to optical super-resolution microscopy for the adventurous biologist. Methods Appl. Fluoresc. 6, 022003 10.1088/2050-6120/aaae0c [DOI] [PubMed] [Google Scholar]
- 54.Klar T.A., Jakobs S., Dyba M., Egner A. and Hell S.W. (2000) Fluorescence microscopy with diffraction resolution barrier broken by stimulated emission. Proc. Natl. Acad. Sci. 97, 8206–8210 10.1073/pnas.97.15.8206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gustafsson M.G.L. (2000) Surpassing the lateral resolution limit by a factor of two using structured illumination microscopy. SHORT COMMUNICATION. J. Microsc. 198, 82–87 10.1046/j.1365-2818.2000.00710.x [DOI] [PubMed] [Google Scholar]
- 56.Betzig E., Patterson G.H., Sougrat R., Lindwasser O.W., Olenych S., Bonifacino J.S.et al. (2006) Imaging intracellular fluorescent proteins at nanometer resolution. Science 313, 1642–1645 10.1126/science.1127344 [DOI] [PubMed] [Google Scholar]
- 57.Rust M.J., Bates M. and Zhuang X. (2006) Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM). Nat. Methods 3, 793–795 10.1038/nmeth929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lelek M., Gyparaki M.T., Beliu G., Schueder F., Griffié J., Manley S.et al. (2021) Single-molecule localization microscopy. Nat. Rev. Methods Prim. 1, 1–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chozinski T.J., Halpern A.R., Okawa H., Kim H.-J., Tremel G.J., Wong R.O.L.et al. (2016) Expansion microscopy with conventional antibodies and fluorescent proteins. Nat. Methods 13, 10.1038/nmeth.3833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chen F., Tillberg P.W. and Boyden E.S. (2015) Expansion microscopy. Science (80-) 347, 543–548 10.1126/science.1260088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Culley S., Albrecht D., Jacobs C., Pereira P.M., Leterrier C., Mercer J.et al. (2018) Quantitative mapping and minimization of super-resolution optical imaging artifacts. Nat. Methods 15, 10.1038/nmeth.4605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chen Z., Jalabi W., Hu W., Park H.-J., Gale J.T., Kidd G.J.et al. (2014) Microglial displacement of inhibitory synapses provides neuroprotection in the adult brain. Nat. Commun. 5, 76–83 10.1038/ncomms5486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Willig K.I., Rizzoli S.O., Westphal V., Jahn R. and Hell S.W. (2006) STED microscopy reveals that synaptotagmin remains clustered after synaptic vesicle exocytosis. Nature 440, 935–939 10.1038/nature04592 [DOI] [PubMed] [Google Scholar]
- 64.Westphal V., Rizzoli S.O., Lauterbach M.A., Kamin D., Jahn R. and Hell S.W. (2008) Video-rate far-field optical nanoscopy dissects synaptic vesicle movement. Science (80-) 320, 246–249 10.1126/science.1154228 [DOI] [PubMed] [Google Scholar]
- 65.Nishimune H., Badawi Y., Mori S., Shigemoto K. and Dragunow M. (2016) Dual-color STED microscopy reveals a sandwich structure of Bassoon and Piccolo in active zones of adult and aged mice. Sci. Rep. 6, 27935 10.1038/srep27935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hua Y., Sinha R., Thiel C.S., Schmidt R., Hüve J., Martens H.et al. (2011) A readily retrievable pool of synaptic vesicles. Nat. Neurosci. 14, 833–839 10.1038/nn.2838 [DOI] [PubMed] [Google Scholar]
- 67.De Rossi P., Nomura T., Andrew R., Masse N., Sampathkumar V., Musial T.et al. (2020) Neuronal BIN1 regulates presynaptic neurotransmitter release and memory consolidation. Cell Reports 30, 3520.e7–3535.e7 10.1016/j.celrep.2020.02.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sieber J.J., Willig K.I., Kutzner C., Gerding-Reimers C., Harke B., Donnert G.et al. (2007) Anatomy and dynamics of a supramolecular membrane protein cluster. Science (80-) 317, 1072–1076 10.1126/science.1141727 [DOI] [PubMed] [Google Scholar]
- 69.Sakamoto H., Ariyoshi T., Kimpara N., Sugao K., Taiko I., Takikawa K.et al. (2018) Synaptic weight set by Munc13-1 supramolecular assemblies. Nat. Neurosci. 21, 41–49 10.1038/s41593-017-0041-9 [DOI] [PubMed] [Google Scholar]
- 70.Kusch V., Bornschein G., Loreth D., Bank J., Jordan J., Baur D.et al. (2018) Munc13-3 is required for the developmental localization of Ca2+ channels to active zones and the nanopositioning of Cav2.1 near release Sensors. Cell Rep. 22, 1965–1973 10.1016/j.celrep.2018.02.010 [DOI] [PubMed] [Google Scholar]
- 71.Bar-On D., Wolter S., van de Linde S., Heilemann M., Nudelman G., Nachliel E.et al. (2012) Super-resolution imaging reveals the internal architecture of nano-sized syntaxin clusters *. J. Biol. Chem. 287, 27158–27167 10.1074/jbc.M112.353250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Schmidt H., Brachtendorf S., Arendt O., Hallermann S., Ishiyama S., Bornschein G.et al. (2013) Nanodomain coupling at an excitatory cortical synapse. Curr. Biol. 23, 244–249 10.1016/j.cub.2012.12.007 [DOI] [PubMed] [Google Scholar]
- 73.Tønnesen J., Katona G., Rózsa B. and Nägerl U.V. (2014) Spine neck plasticity regulates compartmentalization of synapses. Nat. Neurosci. 17, 678–685 10.1038/nn.3682 [DOI] [PubMed] [Google Scholar]
- 74.Bethge P., Chéreau R., Avignone E., Marsicano G. and Nägerl U.V. (2013) Two-photon excitation STED microscopy in two colors in acute brain slices. Biophys. J. 104, 778–785 10.1016/j.bpj.2012.12.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Izeddin I., Specht C.G., Lelek M., Darzacq X., Triller A., Zimmer C.et al. (2011) Super-resolution dynamic imaging of dendritic spines using a low-affinity photoconvertible actin probe. PloS ONE 6, e15611 10.1371/journal.pone.0015611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Grotjohann T., Testa I., Leutenegger M., Bock H., Urban N.T., Lavoie-Cardinal F.et al. (2011) Diffraction-unlimited all-optical imaging and writing with a photochromic GFP. Nature 478, 204–208 10.1038/nature10497 [DOI] [PubMed] [Google Scholar]
- 77.Hoze N., Nair D., Hosy E., Sieben C., Manley S., Herrmann A.et al. (2012) Heterogeneity of AMPA receptor trafficking and molecular interactions revealed by superresolution analysis of live cell imaging. Proc. Natl. Acad. Sci. U. S. A. 109, 17052–17057 10.1073/pnas.1204589109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Shim S.-H., Xia C., Zhong G., Babcock H.P., Vaughan J.C., Huang B.et al. (2012) Super-resolution fluorescence imaging of organelles in live cells with photoswitchable membrane probes. Proc. Natl. Acad. Sci. 109, 13978–13983 10.1073/pnas.1201882109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Helm M.S., Dankovich T.M., Mandad S., Rammner B., Jähne S., Salimi V.et al. (2021) A large-scale nanoscopy and biochemistry analysis of postsynaptic dendritic spines. Nat. Neurosci. 24, 1–12 10.1038/s41593-021-00874-w [DOI] [PubMed] [Google Scholar]
- 80.Berning S., Willig K.I., Steffens H., Dibaj P. and Hell S.W. (2012) Nanoscopy in a living mouse brain. Science 335, 551 10.1126/science.1215369 [DOI] [PubMed] [Google Scholar]
- 81.Pfeiffer T., Poll S., Bancelin S., Angibaud J., Inavalli V.K., Keppler K.et al. (2018) Chronic 2P-STED imaging reveals high turnover of dendritic spines in the hippocampus in vivo. Elife 7, e34700 10.7554/eLife.34700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Masch J.-M., Steffens H., Fischer J., Engelhardt J., Hubrich J., Keller-Findeisen J.et al. (2018) Robust nanoscopy of a synaptic protein in living mice by organic-fluorophore labeling. Proc. Natl. Acad. Sci. 201807104 10.1073/pnas.1807104115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wegner W., Mott A.C., Grant S.G.N., Steffens H. and Willig K.I. (2018) In vivo STED microscopy visualizes PSD95 sub-structures and morphological changes over several hours in the mouse visual cortex. Sci. Rep. 8, 219 10.1038/s41598-017-18640-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Calovi S., Soria F.N. and Tønnesen J. (2021) Super-resolution STED microscopy in live brain tissue. Neurobiol. Dis. 156, 105420 10.1016/j.nbd.2021.105420 [DOI] [PubMed] [Google Scholar]
- 85.Willig K.I., Wegner W., Müller A., Calvet-Fournier V. and Steffens H. (2021) Multi-label in vivo STED microscopy by parallelized switching of reversibly switchable fluorescent proteins. Cell Rep. 35, 109192 10.1016/j.celrep.2021.109192 [DOI] [PubMed] [Google Scholar]
- 86.Steffens H., Mott A.C., Li S., Wegner W., Švehla P., Kan V.W.Y.et al. (2021) Stable but not rigid: Chronic in vivo STED nanoscopy reveals extensive remodeling of spines, indicating multiple drivers of plasticity. Sci. Adv. 7, eabf2806 10.1126/sciadv.abf2806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Dani A., Huang B., Bergan J., Dulac C. and Zhuang X. (2010) Superresolution imaging of chemical synapses in the brain. Neuron 68, 843–856 10.1016/j.neuron.2010.11.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Fukata Y., Dimitrov A., Boncompain G., Vielemeyer O., Perez F. and Fukata M. (2013) Local palmitoylation cycles define activity-regulated postsynaptic subdomains. J. Cell Biol. 202, 145–161 10.1083/jcb.201302071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Nair D., Hosy E., Petersen J.D., Constals A., Giannone G., Choquet D.et al. (2013) Super-resolution imaging reveals that AMPA receptors inside synapses are dynamically organized in nanodomains regulated by PSD95. J. Neurosci. 33, 13204–13224 10.1523/JNEUROSCI.2381-12.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.MacGillavry H.D., Song Y., Raghavachari S. and Blanpied T.A. (2013) Nanoscale scaffolding domains within the postsynaptic density concentrate synaptic AMPA receptors. Neuron 78, 615–622 10.1016/j.neuron.2013.03.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Broadhead M.J., Horrocks M.H., Zhu F., Muresan L., Benavides-Piccione R., DeFelipe J.et al. (2016) PSD95 nanoclusters are postsynaptic building blocks in hippocampus circuits. Sci. Rep. 6, 24626 10.1038/srep24626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Tang A.-H., Chen H., Li T.P., Metzbower S.R., MacGillavry H.D. and Blanpied T.A. (2016) A trans-synaptic nanocolumn aligns neurotransmitter release to receptors. Nature 536, 210–214 10.1038/nature19058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Yang X., Corronc H.L.E, Legendre P., Triller A. and Specht C.G. (2021) Differential regulation of glycinergic and GABAergic nanocolumns at mixed inhibitory synapses. EMBO Rep. 22, e52154 10.15252/embr.202052154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Specht C., Izeddin I., Rodriguez P., ElBeheiry M., Rostaing P., Darzacq X.et al. (2013) Quantitative nanoscopy of inhibitory synapses: counting gephyrin molecules and receptor binding sites. Neuron 79, 308–321 10.1016/j.neuron.2013.05.013 [DOI] [PubMed] [Google Scholar]
- 95.Crosby K.C., Gookin S.E., Garcia J.D., Hahm K.M., Dell'acqua M.L. and Smith Correspondence K.R. (2019) Nanoscale subsynaptic domains underlie the organization of the inhibitory synapse. Cell Rep. 26, 3284–3297 10.1016/j.celrep.2019.02.070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Sinnen B.L., Bowen A.B., Forte J.S., Hiester B.G., Crosby K.C., Gibson E.S.et al. (2017) Optogenetic control of synaptic composition and function. Neuron 93, 646.e5–660.e5 10.1016/j.neuron.2016.12.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hruska M., Henderson N., Le Marchand S.J., Jafri H. and Dalva M.B. (2018) Synaptic nanomodules underlie the organization and plasticity of spine synapses. Nat. Neurosci. 21, 10.1038/s41593-018-0138-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Smith A.J. and Verkman A.S. (2015) Superresolution imaging of aquaporin-4 cluster size in antibody-stained paraffin brain sections. Biophys. J. 109, 2511–2522 10.1016/j.bpj.2015.10.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Smith C.S., Joseph N., Rieger B. and Lidke K.A. (2010) Fast, single-molecule localization that achieves theoretically minimum uncertainty. Nat. Methods 7, 373–375 10.1038/nmeth.1449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Rossi A., Moritz T.J., Ratelade J. and Verkman A.S. (2012) Super-resolution imaging of aquaporin-4 orthogonal arrays of particles in cell membranes. J. Cell Sci. 125, 4405–4412 10.1242/jcs.109603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Volterra A., Liaudet N. and Savtchouk I. (2014) Astrocyte Ca2+ signalling: an unexpected complexity. Nat. Rev. Neurosci. 15, 327–335 10.1038/nrn3725 [DOI] [PubMed] [Google Scholar]
- 102.Heller J.P., Michaluk P., Sugao K. and Rusakov D.A.D.A. (2017) Probing nano-organization of astroglia with multi-color super-resolution microscopy. J. Neurosci. Res. 95, 2159–2171 10.1002/jnr.24026 [DOI] [PubMed] [Google Scholar]
- 103.Panatier A., Arizono M. and Nägerl U.V. (2014) Dissecting tripartite synapses with STED microscopy. Philos. Trans. R. Soc. Lond. B Biol. Sci. 369, 20130597, 10.1098/rstb.2013.0597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Arizono M., Inavalli V.V.G.K., Bancelin S., Fernández‐Monreal M. and Nägerl U.V. (2021) Super‐resolution shadow imaging reveals local remodeling of astrocytic microstructures and brain extracellular space after osmotic challenge. Glia 69, 1605–1613 10.1002/glia.23995 [DOI] [PubMed] [Google Scholar]
- 105.Arizono M., Inavalli V.V.G.K., Panatier A., Pfeiffer T., Angibaud J., Levet F.et al. (2020) Structural basis of astrocytic Ca2+ signals at tripartite synapses. Nat. Commun. 11, 1906 10.1038/s41467-020-15648-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Sakers K., Lake A.M., Khazanchi R., Ouwenga R., Vasek M.J., Dani A.et al. (2017) Astrocytes locally translate transcripts in their peripheral processes. Proc. Natl. Acad. Sci. 114, E3830–E3838 10.1073/pnas.1617782114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Mazaré N., Oudart M., Moulard J., Cheung G., Tortuyaux R., Mailly P.et al. (2020) Local translation in perisynaptic astrocytic processes is specific and changes after fear conditioning. Cell Rep. 32, 108076 10.1016/j.celrep.2020.108076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Sapkota D., Kater M.S.J., Sakers K., Nygaard K.R., Liu Y., Lake A.M.et al. (2021) Activity dependent translation in astrocytes dynamically alters the proteome of the perisynaptic astrocyte process. BioRxiv, 10.1101/2020.04.08.033027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Barton S.K., Gregory J.M., Chandran S. and Turner B.J. (2019) Could an impairment in local translation of mRNAs in glia be contributing to pathogenesis in ALS? Front. Mol. Neurosci. 12, 124 10.3389/fnmol.2019.00124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Boulay A.-C., Saubaméa B., Adam N., Chasseigneaux S., Mazaré N., Gilbert A.et al. (2017) Translation in astrocyte distal processes sets molecular heterogeneity at the gliovascular interface. Cell Discov. 3, 17005 10.1038/celldisc.2017.5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ghézali G., Vasile F., Curry N., Fantham M., Cheung G., Ezan P.et al. (2020) Neuronal Activity Drives Astroglial Connexin 30 in Perisynaptic Processes and Shapes Its Functions. Cereb. Cortex 30, 753–766 10.1093/cercor/bhz123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Ribot J., Breton R., Calvo C.-F., Moulard J., Ezan P., Zapata J.et al. (2021) Astrocytes close the mouse critical period for visual plasticity. Science (80-) 373, 77–81 10.1126/science.abf5273 [DOI] [PubMed] [Google Scholar]
- 113.Takano T., Wallace J.T., Baldwin K.T., Purkey A.M., Uezu A., Courtland J.L.et al. (2020) Chemico-genetic discovery of astrocytic control of inhibition in vivo. Nat 588, 296–302 10.1038/s41586-020-2926-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.An H., Koh W., Kang S., Nam M.-H. and Lee C.J. (2021) Differential proximity of perisynaptic astrocytic best1 at the excitatory and inhibitory tripartite synapses in APP/PS1 and MAOB-KO mice revealed by lattice structured illumination microscopy. Exp. Neurobiol. 30, 213–221 10.5607/en21015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Trotter J.H., Dargaei Z., Sclip A., Essayan-Perez S., Liakath-Ali K., Raju K.et al. (2021) Compartment-specific neurexin nanodomains orchestrate tripartite synapse assembly. BioRxiv, 10.1101/2020.08.21.262097 [DOI] [Google Scholar]
- 116.Broadhead M.J., Bonthron C., Arcinas L., Bez S., Zhu F., Goff F.et al. (2020) Nanostructural diversity of synapses in the mammalian spinal cord. Sci. Rep. 10, 1–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Michaluk P., Heller J.P. and Rusakov D.A. (2021) Rapid recycling of glutamate transporters on the astroglial surface. Elife 10, 10.7554/eLife.64714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Minge D., Domingos C., Unichenko P., Behringer C., Pauletti A., Anders S.et al. (2021) Heterogeneity and development of fine astrocyte morphology captured by diffraction-limited microscopy. Front. Cell Neurosci. 15, 204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Herde M.K., Bohmbach K., Domingos C., Vana N., Komorowska-Müller J.A., Passlick S.et al. (2020) Local efficacy of glutamate uptake decreases with synapse size. Cell Rep. 32, 108182 10.1016/j.celrep.2020.108182 [DOI] [PubMed] [Google Scholar]
- 120.Heller J.P. and Rusakov D.A. (2017) The nanoworld of the tripartite synapse: insights from super-resolution microscopy. Front. Cell Neurosci. 11, 10.3389/fncel.2017.00374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Heller J.P., Odii T., Zheng K. and Rusakov D.A. (2020) Imaging tripartite synapses using super-resolution microscopy. Methods 174, 10.1016/j.ymeth.2019.05.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Tønnesen J., Inavalli V.V.G.K. and Nägerl U.V. (2018) Super-resolution imaging of the extracellular space in living brain tissue. Cell 172, 1108.e15–1121.e15 10.1016/j.cell.2018.02.007 [DOI] [PubMed] [Google Scholar]
- 123.Korogod N., Petersen C.C.H. and Knott G.W. (2015) Ultrastructural analysis of adult mouse neocortex comparing aldehyde perfusion with cryo fixation. Elife 4, 10.7554/eLife.05793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Booth M., Andrade D., Burke D., Patton B. and Zurauskas M. (2015) Aberrations and adaptive optics in super-resolution microscopy. Microscopy 64, 10.1093/jmicro/dfv033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Rueckel M., Mack-Bucher J.A. and Denk W. (2006) Adaptive wavefront correction in two-photon microscopy using coherence-gated wavefront sensing. Proc. Natl. Acad. Sci. 103, 17137–17142 10.1073/pnas.0604791103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Ji N., Sato T.R. and Betzig E. (2012) Characterization and adaptive optical correction of aberrations during in vivo imaging in the mouse cortex. Proc. Natl. Acad. Sci. 109, 22–27 10.1073/pnas.1109202108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Wang K., Milkie D.E., Saxena A., Engerer P., Misgeld T., Bronner M.E.et al. (2014) Rapid adaptive optical recovery of optimal resolution over large volumes. Nat. Methods 11, 625–628 10.1038/nmeth.2925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Patton B.R., Burke D., Owald D., Gould T.J., Bewersdorf J. and Booth M.J. (2016) Three-dimensional STED microscopy of aberrating tissue using dual adaptive optics. Opt. Express 24, 8862–8876 10.1364/OE.24.008862 [DOI] [PubMed] [Google Scholar]
- 129.Gould T.J., Burke D., Bewersdorf J. and Booth M.J. (2012) Adaptive optics enables 3D STED microscopy in aberrating specimens. Opt. Express 20, 20998 10.1364/OE.20.020998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Siemons M.E., Hanemaaijer N.A.K., Kole M.H.P. and Kapitein L.C. (2021) Robust adaptive optics for localization microscopy deep in complex tissue. Nat. Commun. 12, 1–9 10.1038/s41467-021-23647-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Oberheim N.A., Takano T., Han X., He W., Lin J.H.C., Wang F.et al. (2009) Uniquely hominid features of adult human astrocytes. J. Neurosci. 29, 3276–3287 10.1523/JNEUROSCI.4707-08.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Han X., Chen M., Wang F., Windrem M., Wang S., Shanz S.et al. (2013) Forebrain engraftment by human glial progenitor cells enhances synaptic plasticity and learning in adult mice. Cell Stem Cell 12, 342–353 10.1016/j.stem.2012.12.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Vasile F., Dossi E. and Rouach N. (2017) Human astrocytes: structure and functions in the healthy brain. Brain Struct. Funct. 222, 2017–2029 10.1007/s00429-017-1383-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Li J., Pan L., Pembroke W.G., Rexach J.E., Godoy M.I., Condro M.C.et al. (2021) Conservation and divergence of vulnerability and responses to stressors between human and mouse astrocytes. Nat. Commun. 12, 1–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Lana D., Ugolini F., Nosi D., Wenk G.L. and Giovannini M.G. (2021) The Emerging Role of the Interplay Among Astrocytes, Microglia, and Neurons in the Hippocampus in Health and Disease. Front. Aging Neurosci. 13, 156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Wijetunge L.S., Angibaud J., Frick A., Kind P.C. and Nagerl U.V. (2014) Stimulated Emission Depletion (STED) Microscopy Reveals Nanoscale Defects in the Developmental Trajectory of Dendritic Spine Morphogenesis in a Mouse Model of Fragile X Syndrome. J. Neurosci. 34, 6405–6412 10.1523/JNEUROSCI.5302-13.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Schoen M., Reichel J.M., Demestre M., Putz S., Deshpande D., Proepper C.et al. (2015) Super-Resolution Microscopy Reveals Presynaptic Localization of the ALS/FTD Related Protein FUS in Hippocampal Neurons. Front. Cell Neurosci. 9, 496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Ravalia A.S., Lau J., Barron J.C., Purchase S.L.M., Southwell A.L., Hayden M.R.et al. (2021) Super-resolution imaging reveals extrastriatal synaptic dysfunction in presymptomatic Huntington disease mice. Neurobiol. Dis. 152, 105293 10.1016/j.nbd.2021.105293 [DOI] [PubMed] [Google Scholar]
- 139.Padmanabhan P., Martínez-Mármol R., Xia D., Götz J. and Meunier F.A. (2019) Frontotemporal dementia mutant Tau promotes aberrant Fyn nanoclustering in hippocampal dendritic spines. Elife 8, 10.7554/eLife.45040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Moors T.E., Maat C.A., Niedieker D., Mona D., Petersen D., Timmermans-Huisman E.et al. (2021) The subcellular arrangement of alpha-synuclein proteoforms in the Parkinson's disease brain as revealed by multicolor STED microscopy. Acta Neuropathol. 142, 10.1007/s00401-021-02329-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Querol-Vilaseca M., Colom-Cadena M., Pegueroles J., Nuñez-Llaves R., Luque-Cabecerans J., Muñoz-Llahuna L.et al. (2019) Nanoscale structure of amyloid-β plaques in Alzheimer's disease. Sci. Reports 9, 1–10 10.1038/s41598-019-41443-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No data were used for this review article.