Abstract
Simple Summary
The scoliid wasps are larval parasitoids of scarabaeoid beetles and pollinators of various plants and therefore are important in biological control and pollination. However, some species of these wasps are extremely morphologically similar and difficult to identify. In this study, we used an integrative approach of species delimitation, combining morphology with molecular data, to investigate the species of Scoliidae from southern China. On the basis of both morphological and molecular evidence, we recognized 22 morphospecies of 9 genera in two tribes, among which one undescribed cryptic species of the polytypic species Solia (Discolia) superciliaris Saussure, 1864, five newly recorded species and one pending subspecies were discovered. Our study indicates that such an integrative approach is a potent tool in the effort to tackle the taxonomic challenges in Scoliidae, and even in other diverse groups of Aculeata, of which sexual dimorphism and cryptic species are common.
Abstract
Species of the family Scoliidae are larval parasitoids of scarabaeoid beetles and pollinators of various plants. Despite their great importance in pest biological control and plant pollination, the taxonomy and systematics of these parasitoids are far from clear. Some species of the family are extremely morphologically similar and difficult to identify, especially in males. In this study, an integrative taxonomic approach, combining morphology and molecular data, was used to discriminate the species of Scoliidae from southern China. In total, 52 COI sequences belonging to 22 morphospecies of 9 genera in two tribes were obtained. The COI sequences worked well for the identification of all the studied species, with intraspecific genetic distances generally less than 2%, while interspecific distances ranged between 5.3% and 20.8%. The delimitations of the problematic species and subspecies of Scolia and Megacampsomeris are well solved by COI sequences, suggesting that DNA barcoding could be a useful identification tool for Scoliidae. Based on both morphological and molecular evidence, we discovered one undescribed cryptic species of the polytypic species Solia (Discolia) superciliaris Saussure, 1864, five newly recorded species, i.e., Scolia (Discolia) sikkimensis Bingham, 1896, Sericocampsomeris flavomaculata Gupta and Jonathan, 1989, Megacampsomeris asiatica (Saussure, 1858), Megacampsomeris pulchrivestita (Cameron, 1902) and Megacampsomeris shillongensis (Betrem, 1928) and one pending subspecies of Scolia (Discolia) watanabei (Matsumura, 1912) from China. Our study indicates that such an integrative approach, combing both molecular and morphological evidence, is a potent tool to tackle the taxonomic challenges in the family Scoliidae, or even, in other diverse groups of Aculeata, of which sexual dimorphism and cryptic species are common.
Keywords: biological control, DNA barcoding, genitalia, morphology, pollinator, species delimitation
1. Introduction
Scoliid wasps (Hymenoptera, Scoliidae), commonly known as digger wasps or hairy wasps, form a medium-sized family with approximately 560 species worldwide. These wasps attack the mature larvae of scarabaeoid beetles, which are pests of agriculture and forestry, and some species have been considered as important biological control agents [1,2,3,4,5]. Some species have been imported and released in some countries, such as Australia, the Philippines and the United States, for the control of scarabaeoid larvae [6,7], at least one species—Micromeriella marginella modesta (Smith, 1855)—has been introduced and successfully established in Florida in the United States [7]. Females search for the location of scarabaeoid larva by vibratory signals, then they sting, paralyze and lay a single egg on the surface of their prey. The hatched scoliid larva develops externally and feeds on the tissues of the scarabaeoid grub mostly in the soil, though some develop in decaying wood or in rotten vegetation [8,9]. Laboratory observations have indicated that the life cycle of Radumeris tasmaniensis (Saussure, 1854) and Radumeris radula (Fabricius, 1775) lasts from 40 to 60 days, depending on season and species [2]. Scoliid adults of both sexes commonly visit the flowers of various plants, and some are found to be important pollinators [10,11,12,13,14]. Males of some scoliid wasps have been recorded as pseudocopulating with orchids. At least one species—Pygodasis bistrimaculata (Lepeletier)—may be the only pollinator of the terrestrial orchid Geoblasta penicillata (Chloraeinae) via pseudocopulation from subtropical South America [15]. Scolia nobilitata Fabricius, 1805 is the most common flower visitors in the buckwheat fields in north-central Florida [14]. Studies from Japan have suggested that scoliid wasps may play an important role in pollinating coastal plants in the grassland zone [11,12,13].
Despite their great importance, in the past few decades there have been few studies on this group from China since the rather comprehensive revisions of the Chinese Scoliidae made by Betrem [16,17]. Recently, Liu et al. [18] listed 1 subfamily (Scoliinae), 2 tribes (Campsomerini and Scoliini), 11 genera and 52 extant species in China, which provided a clearer view of the Chinese fauna. However, due to so many (overlapping) subspecies and varieties, many scoliid subspecies have been recognized merely on the basis of subtle color variations. In some cases, these subspecies are not represented by true geographically distinct populations and the boundaries between species or subspecies are often unclear. Furthermore, sexual dimorphism in Scoliidae is extreme; females and males of a single species often differ significantly, especially for species in the tribe Campsomerini. Males of the same species may vary significantly in size and color pattern and can be confused with closely related species. Further development of these important parasitoids as biological control agents against scarabaeoid pests and as pollinators in crop cultivation requires an accurate and sufficient identification of the species.
DNA barcoding (the partial sequencing of the gene cytochrome c oxidase 1, COI) has become an important species identification tool for insects [19,20]. The importance of the DNA sequences in the taxonomy and evolutionary relationships of Scoliidae or stinging wasps have been recognized by some researchers [21,22]. However, there is a serious lack of a DNA barcode database for the Chinese fauna of Scoliidae. To fill this gap, we started a research campaign with the objective of collecting specimens of Scoliidae across China in order to identify the species by morphological characters and to build a DNA database for these important parasitoids. By doing this, we aimed to test the morphological species delimitation of scoliid wasps from southern China using DNA barcoding methods.
2. Material and Methods
2.1. Collection and Identification
This work is based on specimens of Scoliidae collected by sweeping net and mostly Malaise traps (MT) set up in a few provinces of southern China. Specimens were identified using the keys of Liu et al. [23] All the studied specimens are deposited in the Museum of Biology at Sun Yat-sen University, Guangzhou, China (SYSBM). Multifocal images were made using a Nikon SMZ25 microscope with a Nikon DS-Ri 2 digital camera system. Images were then post-processed with Adobe Photoshop CS6 Extended.
2.2. DNA Extraction, Amplification, and Sequencing
In total, 52 specimens of 22 morphospecies were used for DNA barcoding analysis (Table 1, detailed collecting data of the sequenced see Table S1). Both female and male specimens were selected for a species when such specimens were available. Genomic DNA were extracted from the right mid leg of each specimen using the DNeasy Blood amd Tissue Kit (Qiagen, Hilden, Germany), following the manufacturer’s protocols. Following the extraction, the “barcode” region of the mitochondrial cytochrome oxidase subunit 1 (COI) was amplified using the LCO1490/HCO2198 primer pair [24]. Polymerase chain reactions (PCRs) were performed using Tks Gflex™ DNA Polymerase (Takara, Shiga, Japan) and conducted in a T100™ Thermal Cycler (Bio-Rad, CA, USA). Thermocycling conditions were: an initial denaturing step at 94 °C for 5 min, followed by 35 cycles of 94 °C for 30s, 50 °C for 30 s, 72 °C for 30s and an additional extension at 72 °C for 5 min. Amplicons were directly sequenced in both directions with forward and reverse primers on an Applied Biosystems (ABI) 3730XL by Guangzhou Tianyi Huiyuan Gene Technology Co., Ltd. (Guangzhou, China). Chromatograms were assembled with Geneious 11.0.3. All sequences generated from this study are deposited in GenBank (accession numbers see Table 1). All residual DNAs are archived (−30 °C) in the molecular laboratory of SYSBM, Guangzhou, China, and are available for further study upon request.
Table 1.
Sequenced taxa and GenBank accession numbers.
| Code | Species | Sex | GenBank Accession No. |
|---|---|---|---|
| SCAU3043675 | Austroscolia ruficeps (Smith) | male | MZ753501 |
| SCAU3043670 | Carinoscolia vittifrons (Sichel) | male | MZ753502 |
| SCAU3043671 | Carinoscolia vittifrons (Sichel) | female | MZ753503 |
| SCAU3043672 | Carinoscolia vittifrons (Sichel) | female | MZ753504 |
| SCAU3043673 | Carinoscolia vittifrons (Sichel) | male | MZ753505 |
| En-418584 | Carinoscolia vittifrons (Sichel) | female | MZ821648 |
| SCAU3043661 | Liacos erythrosoma (Burmeister) | female | MZ753506 |
| SCAU3043662 | Liacos erythrosoma (Burmeister) | male | MZ753507 |
| SCAU3048016 | Liacos erythrosoma (Burmeister) | male | MZ753508 |
| SCAU3043656 | Megacampsomeris asiatica (Saussure) | male | MZ753509 |
| SCAU3043660 | Megacampsomeris binghami (Betrem) | male | MZ753510 |
| SCAU3048011 | Megacampsomeris binghami (Betrem) | male | MZ753511 |
| SCAU3043653 | Megacampsomeris farrenwhitei (Betrem) | female | MZ753512 |
| SCAU3043654 | Megacampsomeris farrenwhitei (Betrem) | male | MZ753513 |
| SCAU3043658 | Megacampsomeris farrenwhitei (Betrem) | male | MZ753514 |
| SCAU3043659 | Megacampsomeris farrenwhitei (Betrem) | male | MZ753515 |
| SCAU3043686 | Megacampsomeris farrenwhitei (Betrem) | female | MZ753517 |
| SCAU3048009 | Megacampsomeris farrenwhitei (Betrem) | male | MZ753518 |
| SCAU3048012 | Megacampsomeris farrenwhitei (Betrem) | male | MZ753519 |
| SCAU3043657 | Megacampsomeris prismatica (Smith) | female | MZ753520 |
| SCAU3048015 | Megacampsomeris prismatica (Smith) | male | MZ753521 |
| NZ4760 | Megacampsomeris pulchrivestita (Cameron) | male | MZ753522 |
| SCAU3043655 | Megacampsomeris shillongensis (Betrem) | male | MZ753523 |
| SCAU3043665 | Megascolia (Regiscolia) azurea (Christ) | female | MZ753524 |
| SCAU3043666 | Megascolia (Regiscolia) azurea (Christ) | male | MZ753525 |
| SCAU3043667 | Micromeriella marginella (Klug) | female | MZ753526 |
| SCAU3043668 | Micromeriella marginella (Klug) | male | MZ753527 |
| SCAU3043685 | Phalerimeris phalerata (Saussure) | male | MZ753516 |
| NZ4762 | Phalerimeris phalerata (Saussure) | female | MZ753528 |
| NZ4792 | Phalerimeris phalerata (Saussure) | male | MZ753529 |
| NZ4832 | Phalerimeris phalerata (Saussure) | male | MZ753530 |
| NZ4835 | Phalerimeris phalerata (Saussure) | female | MZ753531 |
| SCAU3043669 | Phalerimeris phalerata (Saussure) | male | MZ753532 |
| SCAU3048017 | Phalerimeris phalerata (Saussure) | male | MZ753533 |
| En-418587 | Phalerimeris phalerata (Saussure) | female | MZ821649 |
| SCAU3043683 | Scolia (Discolia) affinis Guérin-Méneville | male | MZ753534 |
| SCAU3043684 | Scolia (Discolia) binotata Fabricius | female | MZ753535 |
| SCAU3043682 | Scolia (Discolia) clypeata Sickman | female | MZ753536 |
| SCAU3043680 | Scolia (Discolia) laeviceps Smith | male | MZ753537 |
| SCAU3043681 | Scolia (Discolia) nobilis Saussure | male | MZ753538 |
| SCAU3048008 | Scolia (Discolia) sikkimensis Bingham | male | MZ753539 |
| SCAU3043676 | Scolia (Discolia) superciliaris Saussure | female | MZ753540 |
| SCAU3043677 | Scolia (Discolia) superciliaris Saussure | male | MZ753541 |
| SCAU3043678 | Scolia (Discolia) superciliaris Saussure | male | MZ753542 |
| SCAU3048010 | Scolia (Discolia) superciliaris Saussure | male | MZ753543 |
| SCAU3048013 | Scolia (Discolia) superciliaris Saussure | male | MZ753544 |
| SCAU3043679 | Scolia (Discolia) watanabei (Matsumura) | male | MZ753545 |
| En-418585 | Scolia (Discolia) watanabei (Matsumura) | male | MZ821650 |
| SCAU3043674 | Scolia sp. | male | MZ753546 |
| SCAU3048014 | Scolia sp. | male | MZ753547 |
| SCAU3043664 | Sericocampsomeris flavomacula Gupta and Jonathan | male | MZ753549 |
| En-418591 | Sericocampsomeris flavomacula Gupta and Jonathan | male | MZ821651 |
2.3. Sequence Analysis and Molecular Species Delimitation
All sequences were blasted in the BOLD (Barcode of Life Database, http://www.barcodinglife.org/index.php/IDS_OpenIdEngine) and GenBank (Accessed on: 9 August 2021). Sequences were aligned using MAFFT v7.470 by the G-INS-I strategy [25]. Genetic Kimura-2 parameter (K2P) distances within and between species were calculated in MEGA 7 with pairwise deletion for gaps [26].
Two methods, the Automatic Barcode Gap Discovery (ABGD) and the updated Poisson tree processes model (bPTP), were tested for molecular species delimitation. ABGD is a distance-based method and it sorts the sequences into hypothetical species by partitioning and comparing the difference between sequences to identify a “barcode gap” [27]. The ABGD analysis was performed on the web interface (https://bioinfo.mnhn.fr/abi/public/abgd/abgdweb.html. Accessed on: 10 August 2021), using the default priors, Pmin = 0.001, Pmax = 0.1, Steps 10, and with barcode relative gap width = 1.00. bPTP is an updated version of the original PTP with Bayesian posterior probability, which tests species boundaries on non-ultrametric phylogenetic trees by detecting significant differences in the number of substitutions between species and within species [28]. For bPTP analysis, a Maximum Likelihood (ML) tree was generated in RAxML v8.2.10 under the GTRGAMMA evolutionary model and performed on the bPTP web server (https://species.h-its.org/ptp/. Accessed on: 10 August 2021), with default parameters. The COI sequences of Tiphia minuta Linden (Hymenoptera: Tiphiidae; GenBank: JN299217) and Vespa velutina Lepeletier (Hymenoptera: Vespidae; GenBank: JKY224073) were selected as the outgroups because of their close relationships to Scoliidae [29].
3. Results and Discussion
The present study generated 52 COI sequences with an average of 678 bp. Voucher specimens of these 52 sequences were subjected to further morphological examination, and 22 species belonging to 9 genera were recognized (Table 1). When blasted in BOLD and GenBank databases, the sequences of the two Scolia watanabei specimens received close matches with Scolia oculata (Matsumura) (over 97%) and Scolia hirta (Schrank) (over 96%). Other species had no close matching sequences in BOLD or NCBI.
The K2P distances (Tables S2 and S3) showed a larger intergroup than intragroup distance for the COI sequences. The intraspecific pairwise distances were generally less than 2%, with an exception for S. watanabei (3.1%) and Scolia superciliaris (0–4%), both of which consist of two subspecies among the studied specimens. The interspecific pairwise distance ranged from 5.3% to 20.8%.
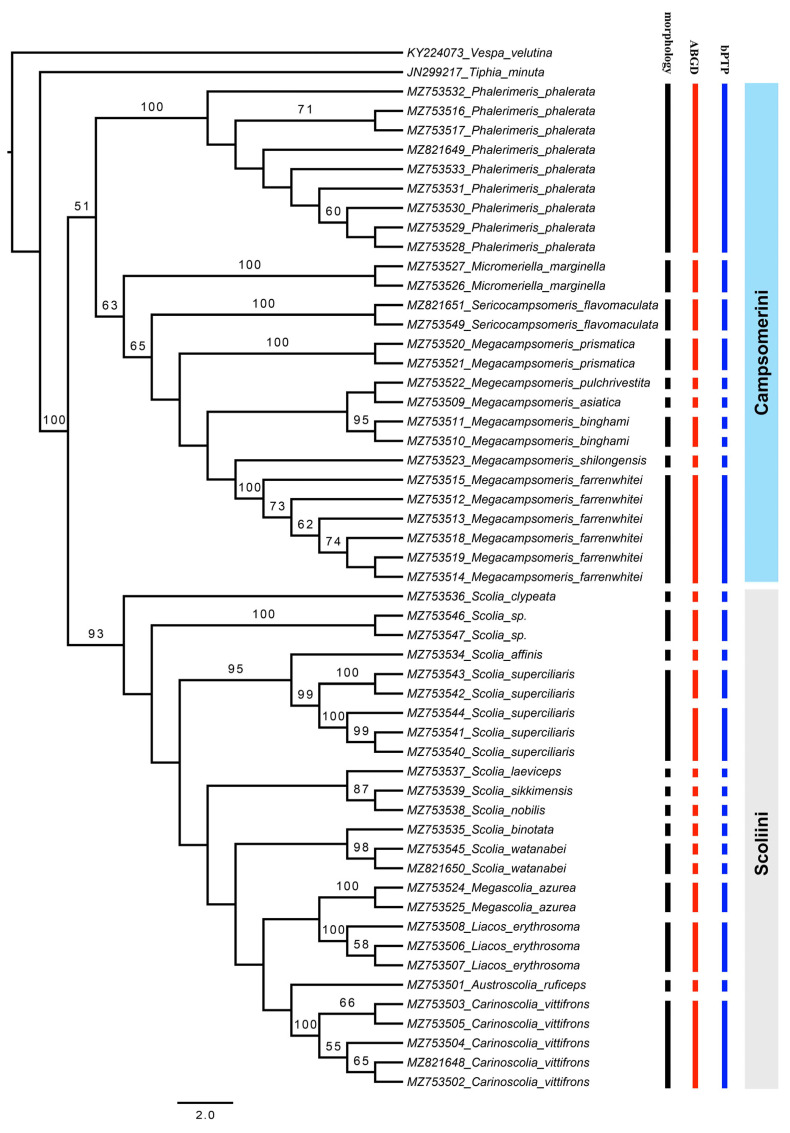
The ABGD analyses returned a total of 24 groups at a priori genetic distance thresholds of 0.002–0.036. The two subspecies of S. watanabei and S. superciliaris were assigned as putative species. After removing the outgroups, the bPTP analysis based on the ML tree delimited 25 putative species. Besides the two subspecies of S. watanabei and S. superciliaris, Megacampsomeris binghami was also recovered and consists of two putative species. The delimitations of all other species are congruent with the morphological identification results in both ABGD and bPTP methods (Figure 1).
Figure 1.
Maximum likelihood tree based on COI and results of species delimitation of three methods, only values > 50 for bootstrap are labeled.
Although the main goal of this study was not to address the phylogenetic relationships within the subfamily Scoliinae, our molecular data seem to support the tribal relationships proposed by Day et al. [30] (Figure 1) Tribe Scoliini, including genera Austroscolia (Figure 2A), Carinoscolia (Figure 2B,C), Liacos (Figure 2D,E), Megascolia (Figure 2F and Figure 3A), and Scolia (Figure 3B–F, Figure 4A–F and Figure 5A), form a highly (bootstrap = 93) supported clade. While Campsomerini, including genera Megacampsomeris (Figure 5B–F and Figure 6A–C), Micromeriella (Figure 6D,E), Phalerimeris (Figure 6F,G), and Sericocampsomeris (Figure 6H), form a relatively low (bootstrap = 51) supported clade.
Figure 2.
Habitus, lateral view. (A) Austroscolia ruficeps (Smith, 1855), male (SCAU3043675); (B) Carinoscolia vittifrons (Sichel, 1864), male (SCAU3043670); (C) Carinoscolia vittifrons (Sichel, 1864), female (SCAU3043672); (D) Liacos erythrosoma (Burmeister, 1853), male (SCAU3043662); (E) Liacos erythrosoma (Burmeister, 1853), female (SCAU3043661); (F) Megascolia azurea (Christ, 1791), male (SCAU3043666).
Figure 3.
Habitus, lateral view. (A) Megascolia azurea (Christ, 1791), female (SCAU3043665); (B) Scolia affinis Guérin-Méneville, 1845, male (SCAU3043683); (C) Scolia binotata Betrem, 1928, female (SCAU3043684); (D) Scolia clypeata Sickman, 1894, female (SCAU3043682); (E) Scolia laeviceps Smith, 1855, male (SCAU3043680); (F) Scolia (Discolia) nobilis Saussure, 1858, male (SCAU3043681).
Figure 4.
Habitus, lateral view. (A) Scolia sikkimensis Bingham, 1896, male (SCAU3048008); (B) Scolia superciliaris sauteri Betrem, 1928, male (SCAU3043677); (C) Scolia superciliaris sauteri Betrem, 1928, female (SCAU3043676); (D) Scolia superciliaris superciliaris Saussure, 1864, male (SCAU3048010); (E) Scolia watanabei Smith, 1855, male (SCAU3043679); (F) Scolia watanabei Smith, 1855, male (En-418585).
Figure 5.
Habitus, lateral view. (A) Scolia sp., male (SCAU3043674); (B) Megacampsomeris asiatica (Saussure, 1859), male (SCAU3043656); (C) Megacampsomeris binghami (Betrem, 1928), male (SCAU3043660); (D) Megacampsomeris farrenwhitei Betrem, 1928, male (SCAU3043654); (E) Megacampsomeris farrenwhitei Betrem, 1928, female (SCAU3043653); (F) Megacampsomeris prismatica (Smith, 1855), male (SCAU3048015).
Figure 6.
Habitus. (A) Megacampsomeris prismatica (Smith, 1855), female (SCAU3043653); (B) Megacampsomeris pulchrivestita (Cameron, 1902), male (NZ4760); (C) Megacampsomeris shillongensis Betrem, 1928, male (SCAU3043655); (D) Micromeriella marginella (Klug, 1810), male (SCAU3043668); (E) Micromeriella marginella (Klug, 1810), female (SCAU3043667); (F) Phalerimeris phalerata (Saussure,1858), male (SCAU3043685); (G) Phalerimeris phalerata (Saussure,1858), female (SCAU3043686); (H) Sericocampsomeris flavomacula Gupta & Jonathan, 1989, male (SCAU3043664).
3.1. Tribe Scoliini
Of the five genera within Scoliini, their interrelationships are still not clear because the branch supports among the genera are generally low (boostap < 50). Studies have found that COI has a low signal for phylogenetic analysis above the genus level [31]. Morphologically, the interrelationships within Scoliini are also unclear, for example, genera such as Carinoscolia, Megascolia and Liacos share many similarities and were treated as subgenera of Scolia in the past [18]. However, each species (some are represented by multiple specimens) recovered on the tree are clearly separated from all neighboring species and are identical to the number of species identified based on morphological characters, suggesting that the species in question can be identified unambiguously by DNA barcoding.
Eight determined and one undetermined species of Scolia were obtained in this study. Scolia (Discolia) sikkimensis Bingham (Figure 6C), which has been reported from Bhutan, India and Nepal [32], is newly recorded for China. The male of this species can be recognized by the presence of a distinct antero-median tubercle on the first abdominal tergite [32], as well as by the large degree of yellow maculation including the obvious markings on mesoscutum, mesopleurum, scutellum, propodeum and femura.
Scolia (Discolia) watanabei (Matsumura, 1912) is a polytypic species that is widely distributed in China, India, Japan and Myanmar [18]. The male of this species is easily confused with the males of S. oculata (Matsumura) and S. hirta (Schrank). This is also revealed in the blast results, that S. watanabei received close matches with S. oculata (over 97%) and S. hirta (over 96%) when blasted in the BOLD and GenBank databases. However, there are some characters that can be of diagnostic value: head with variable yellow patterns (all black in S. oculata and S. hirta), propodeum with larger and denser punctures and vestitures on mesosoma are golden (whitish or blackish in S. oculata and S. hirta). Three subspecies of S. watanabei, viz. watanabei, pekingensis, and kempi, have been reported and the main distinctive features between these subspecies are the variations in body markings, which were also recognized in a pending subspecies from Japan [33]. Two subspecies of S. watanabei were assigned as putative species from our ABGD and bPTP analyses. The male specimen from Hainan (SCAU3043679, Figure 4E) can be assigned to pekingensis for its color patterns (but subvertex without yellow transversal linear macula). The male specimen from Yunnan (En-418585, Figure 4F) differs from pekingensis, mainly in color patterns: Yellow maculae on clypeus, frons, scapula, fore legs and terga are larger; mid and hind femora mostly reddish. We also observed some differences between the male genitalia between this specimens and pekingensis: the squama of this specimen is polished (Figure 7A), but in pekingensis, the dorsal side of squama has some short and sparse hairs (Figure 7B). Considering the morphological distinctiveness and a relatively high genetic distance (3.1%), the specimens from Yunnan may represent a new subspecies or species. However, we need more morphological and molecular data from specimens, especially the females of the three known subspecies and the pending subspecies to confirm its identity.
Figure 7.
Male genitalia. (A) Scolia watanabei Smith, 1855 (En-418585), dorsal view; (B) Scolia watanabei Smith, 1855 (SCAU3043679), dorsal view; (C) Scolia superciliaris sauteri Betrem, 1928, dorsal view (SCAU3043677); (D) Scolia superciliaris superciliaris Saussure, 1864, dorsal view (SCAU3048010); (E) Scolia sp., (SCAU3043674) dorsal view; (F) Scolia sp., (SCAU3043674) ventral view; (G) Megacampsomeris shillongensis Betrem, 1928 (SCAU3043655), penis valve, lateral view; (H) Megacampsomeris prismatica (Smith, 1855) (SCAU3048015), penis valve, lateral view.
Solia superciliaris is also a polytypic species that is widely distributed in China, India, Japan, Myanmar, Nepal, Thailand and Vietnam [18]. It is one of the most abundant species collected from southern China during this campaign. Betrem [17] recorded three subspecies, viz. superciliaris, sauteri and staudingeri. These subspecies are distinguished from each other mainly by the extent of the reddish and black color of the antennae, especially in males [31]. In this study, we found two typical subspecies (Figure 4B–D), namely sauteri and superciliaris, which can be easily distinguished by their red (SCAU3043676, SCAU3043677 and SCAU3048013) and black (SCAU3043678, SCAU3048010) antennae, respectively. In addition, vestiture is predominantly black on mesosoma and metasoma in subspecies sauteri, while it is white in subspecies superciliaris. The male genitalia also show similar differences, as found in the two subspecies of S. watanabei mentioned above: the squama of sauteri has short hairs (Figure 7C) but are polished in superciliaris (Figure 7D). These two subspecies were also supported by the ABGD and bPTP analyses (Figure 1).
Two undetermined male specimens (SCAU3043674 (Figure 5A), SCAU3048014) from Hainan are extremely similar to the males of S. superciliaris superciliaris. Only some subtle differences can be found: an ocelli part reddish yellow (black in S. superciliaris superciliaris); vestiture predominantly black on mesoscutum and metasoma (whitish in S. superciliaris superciliaris); punctures on propodeum and scutellum are coarser and denser, interspaces between punctures are sometimes smaller than the puncture diameter (finer and sparser, interspaces between punctures always distinctly bigger than the puncture diameter in S. superciliaris superciliaris); and metasoma iridescent with a coppery sheen (less shiny in S. superciliaris superciliaris). Male genitalia, however, are distinctly different from the two subspecies of S. superciliaris mentioned above: hairs on paramere are thicker and black and paramere truncated apically (Figure 7E,F), although the sparse and short hairs present on squama are similar to S. superciliaris superciliaris sauteri. Plus, the COI sequences provide strong evidence that these are different species, with a 7.7–8.6% genetic distance between the two species. This genetic distance matches the level of genetic divergence of mtDNA between animal species (8–17%) [34]. This indicates that cryptic species may exist in Scolia and male genitalia and DNA barcoding are useful in recognizing these species.
3.2. Tribe Campsomerini
Of the four genera within Campsomerini, the generic status seems to be supported by the phylogenetic analysis of the COI sequences, as the branches representing each genus are clearly separated from each other (Figure 1). Genera Phalerimeris Betrem, Micromeriella Betrem, and Sericocampsomeris Betrem previously were all elevated from subgenera of Campsomeris [18]. In this study, these genera are all represented by one species but with multiple specimens collected from various localities and form their own clusters that are clearly separated from each other on the ML tree.
Sericocampsomeris flavomacula Gupta and Jonathan, 1989 (Figure 6H) was previously reported from India and Nepal, and here we record this species from China for the first time. This species can be distinguished from the two Sericocamposomeris species previously recorded in China by its large extent of reddish-yellow maculation on the first to fifth terga and its predominantly white vestiture.
Genus Megacampsomeris Betrem, however, is diverse and six species are included in this study. Males of Megacampsomeris are extremely morphologically similar to each other. However, the results from the ABGD and bPTP analyses are generally consistent with the results based on morphological identification.
Three species of Megacampsomeris, i.e., Megacampsomeris asiatica (Saussure, 1858), Megacampsomeris pulchrivestita (Cameron, 1902) and Megacampsomeris shillongensis (Betrem, 1928), are newly recorded in China. These three species are all widely distributed in the Oriental region [16,32]. The male of M. asiatica (Figure 5B) can be differentiated from other species by having the integument of the head and mesosoma entirely black and the vestiture distinctly white on mesosoma; the male of M. pulchrivestita (Figure 6B) can be recognized by its clypeus yellow, vestiture reddish yellow on mesosoma, fringes of terga white and short, narrow bands on terga, and completely black cutellum; while the male of M. shillongensis (Figure 6C) can be separated from other species by their yellow clypeus, largely yellow femora on the outer surface, apical bands on terga broad, golden vestiture, other than the black on the fifth to last abdominal segments, and the penis valve of male genitalia are less elongate apicad (Figure 7G) when compared with its congener M. prismatica (Figure 7H).
In some cases, DNA barcoding could enhance morphological species delimitation by assisting in discovering useful diagnostic morphological characters. Morphologically, the males of Megacampsomeris prismatica (Smith, 1855) and Megacampsomeris farrenwhitei (Betrem, 1928) are easily confused with each other. These two species are sympatric and commonly occur in southern China, but the males have never been thoroughly described. According to the literature [16,23,32], the most distinctive character for males of these two species could be the yellow marking on the hind femur (M. prismatica is all black while M. farrenwhitei is with yellow stripe beneath the hind femur). However, the male specimens of M. prismatica from Japan were found to be variable in maculation, sometimes even with a yellow stripe beneath the hind femur [35]. Females (Figure 5E and Figure 6A) of these species are easily distinguished by the punctuation on scutellum, coloration of wings and median grove of frons [23]. In this study, males of these two species were assigned to the respective species based on their identities to the female COI sequences. After such assignment, the intraspecific genetic distances of the two species are not more than 0.3%, while the interspecific genetic distance between these two species ranged between 13% to 13.5%. Further morphological examination indicates that the yellow marking on the hind femur of M. farrenwhitei is variable from presentation beneath the hind femur (e.g., SCAU3043654, SCAU3043659) to being entirely absent (e.g., SCAU3043658). Therefore, the yellow marking on the hind femur is not a valid characteristic by which to differentiate the males of these two species. However, based on the molecular species delimitation, we found that some characters (Figure 8A–F) might be of diagnostic value: vestiture on mesosoma is denser and more golden in M. prismatica, while it is sparser and more reddish in M. farrenwhitei; the mid subhorizontal portion of propodeum, in the lateral view, slopes rather steeply, making a sharper angle with the vertical portion in M. prismatica, while it is more gradual, making an obtuse angle in M. farrenwhitei; yellow bands on terga are wider (nearly 1/3 of mid length) in M. prismatica while they are narrower (about 1/4 of mid length) in M. farrenwhitei; and pubescence thicker on paramere and volsella is more acutely dentated in M. prismatica, while pubescence is sparser and volsella is more obtusely dentated in M. farrenwhitei. This example indicates that DNA barcoding is useful to recognize these problematic species in Scoliidae.
Figure 8.
(A,C,E1,E2): Male of Megacampsomeris prismatica (Smith, 1855) (SCAU3048015); (B,D,F1,F2): Megacampsomeris farrenwhitei Betrem, 1928 (SCAU3043654). (A,B). mesopleurum, lateral view; (C,D). metasoma, dorsal view; (E1,F1). paramere of male genita; (E2,F2). volsella of male genita.
The two specimens of Megacampsomeris binghami (Betrem, 1928) were recovered as two putative species by the bPTP analysis. However, the pairwise distance between these two specimens was 1.9% and few morphological differences were observed. This relatively high genetic variation is probably due to their distant geographic distribution, as one specimen (SCAU3048011) was collected in Guangdong and the other (SCAU3043660, Figure 5C) in the mountainous area of Yunnan. Besides, studies have shown that the bPTP methods tend to oversplit species [36].
4. Conclusions
The scoliid wasps from southern China are polymorphic and diverse. Our analyses based on both molecular data and morphology recognized 22 species belonging to 9 genera of two tribes, including 5 species newly recorded for China: S. (Discolia) sikkimensis, S. flavomaculata, M. asiatica, M. pulchrivestita and M. shillongensis. The phylogenetic tree based on the COI sequences shows perfect consistence with the tribal system of Day et al. [30] and the general classification of the Chinese Scoliidae proposed by Liu et al. [23], suggesting that the COI sequences provide a relatively accurate picture of evolutionary history. Furthermore, DNA barcoding is not only useful in discovering cryptic species, but also in delimitating morphologically similar species. Our study indicates that the integrative approach, combing both molecular and morphological evidence, is a potent tool to tackle the taxonomic challenges in the family Scoliidae, of which sexual dimorphism and cryptic species are common.
Acknowledgments
We are grateful to Longlong Chen, Wei Lin, Jianwei Li, Lang Yi, and Chunyang Xu for their help in collecting the specimens.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/insects12100892/s1, Table S1: Details of sequenced specimens, Table S2: Genetic distance of COI within species under K2P model, Table S3: Interspecific pairwise distance of Scollidae based on COI sequences (%).
Author Contributions
Conceptualization, Z.L., H.-Y.C. and S.-X.L.; Data curation and Methodology, Z.L., S.-J.Y., Y.-Y.W. and H.-Y.C.; Formal analysis and Writing—original draft, Z.L. and H.-Y.C.; Funding acquisition, Z.L., Y.-Q.P. and S.-X.L.; Investigation, S.-J.Y., Y.-Y.W., Y.-Q.P. and H.-Y.C.; Supervision, Y.-Q.P. and S.-X.L.; Writing—review and editing, Z.L., S.-J.Y., Y.-Y.W., Y.-Q.P., H.-Y.C. and S.-X.L. All authors have read and agreed to the published version of the manuscript.
Funding
This study is supported by the Program of Ministry of Science and Technology of China (2018FY100406), Biodiversity Survey and Assessment Project of the Ministry of Ecology and Environment, China (2019HJ2096001006), National Natural Science Foundation of China (32100351), the Scientific Research Fund of Hunan Provincial Education Department (20K089) and Hunan Provincial Natural Science Foundation of China (2020JJ5392).
Institutional Review Board Statement
Not applicable.
Data Availability Statement
The data of the research were deposited in the Museum of Biology at Sun Yat-sen University (SYSBM), Guangzhou, China.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Illingworth J.F. Monthly notes on grubs and other cane pests. Qld. Bur. BSES Div. Entomol. Bull. 1919;7:1–29. [Google Scholar]
- 2.Illingworth J.F. Natural enemies of sugar-cane beetles in Queensland. Bur. BSES Div. Entomol. Bull. 1921;13:1–47. [Google Scholar]
- 3.Askew R.R. Parasitic Insects. Elsevier Publishing; New York, NY, USA: 1971. pp. 1–316. [Google Scholar]
- 4.Naumann I.D. Hymenoptera (Wasps, bees, ants, sawflies) In: Naumann I.D., Carne P.B., Lawrence J.F., Nielsen E.S., Spradberry J.P., Taylor R.W., Whitten M.J., Littlejohn M.J., editors. The Insects of Australia: A Textbook for Students and Research Workers. 2nd ed. Melbourne University Press; Carlton, VIC, Australia: 1991. pp. 916–1000. [Google Scholar]
- 5.Osten T. Checkliste der Dolchwespen der Welt (Insecta: Hymenoptera, Scoliidae) Naturforsch. Ges. 2005;220:1–62. [Google Scholar]
- 6.Elliott M.G. Annotated catalogue of the Australian scoliidae (Hymenoptera) Tech. Rep. Aust. Mus. Online. 2011;22:1–17. doi: 10.3853/j.1835-4211.22.2011.1562. [DOI] [Google Scholar]
- 7.Abbate A., Campbell J., Bremer J., Kern W.H. The introduction and establishment of Campsomeris dorsata (Hymenoptera: Scoliidae) in Florida. Fla. Entomol. 2018;101:543–545. doi: 10.1653/024.101.0334. [DOI] [Google Scholar]
- 8.Clausen C.P. Entomophagous Insects. McGraw-Hill; New York, NY, USA: 1940. pp. 1–688. [Google Scholar]
- 9.O’Neill K. Solitary Wasps: Behavior and Natural History. Cornell University Press; Ithaca, NY, USA: New York, NY, USA: 2001. p. xiv + 406. [Google Scholar]
- 10.Elliott N.B. Flower-feeding activities of Campsomeris trifasciata nassauensis Bradley (Hymenoptera: Scoliidae); Proceedings of the 3rd Symposium on the Botany of the Bahamas; San Salvador, El Salvador. 6–9 June 1989; pp. 1–6. [Google Scholar]
- 11.Minagi K., Maeta Y., Kitamura K. Studies on the conservation of pollination ecosystem in sand dune 1. Pollinators and their seasonal fluctuation at the Taisya sand dune in San-in district. Bull. Hoshizaki Green Found. 2000;4:139–160. (In Japanese with English summary) [Google Scholar]
- 12.Inoue M., Endo T. Species composition of flowervisiting insect community in the coastal sand dune of Hakoishi, Kyoto Prefecture. Hum. Sci. 2006;9:39–46. (In Japanese with English summary) [Google Scholar]
- 13.Inoue M., Endo T. Spatiotemporal distribution and resource use of scoliid wasps (Hymenoptera) in coastal sand dunes. Entomol. Sci. 2006;9:359–371. doi: 10.1111/j.1479-8298.2006.00182.x. [DOI] [Google Scholar]
- 14.Campbell J.W., Irvin A., Stanley-Stahr C., Ellis J.D. Insect visitors to flowering buckwheat, Fagopyrum esculentum (Polygonales: Polygonaceae), in north–central Florida. Fla. Entomol. 2016;99:264–268. doi: 10.1653/024.099.0216. [DOI] [Google Scholar]
- 15.Ciotek L., Giorgis P., Benitez-Vieyra S., Cocucci A.A. First confirmed case of pseudocopulation in terrestrial orchids of South America: Pollination of Geoblasta pennicillata (Orchidaceae) by Campsomeris bistrimacula (Hymenoptera, Scoliidae) Flora. 2006;201:365–369. doi: 10.1016/j.flora.2005.07.012. [DOI] [Google Scholar]
- 16.Betrem J.G. Monographie der Indo-Australischen Scoliiden mit zoogeographischen Betrachtungen. Treubia. 1928;9:1–388. [Google Scholar]
- 17.Betrem J.G. Étude systematique des Scoliidae de Chine et leurs relations avec les autres groups de Scoliidae. In: Chen T.T.H., editor. Notes d’Entomologie Chinoise. Université l’Aurore; Shanghai, China: 1941. pp. 47–188. [Google Scholar]
- 18.Liu Z., van Achterberg C., He J.H., Chen X.X. A checklist of Scoliidae (Insecta: Hymenoptera) from China. Zootaxa. 2021;4966:101–126. doi: 10.11646/zootaxa.4966.2.1. [DOI] [PubMed] [Google Scholar]
- 19.Hebert P.D.N., Cywinska A., Ball S.L., Dewaard J.R. Biological identifications through DNA barcodes. Proc. R. Soc. B Biol. Sci. 2003;270:313–321. doi: 10.1098/rspb.2002.2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hebert P.D.N., Ratnasingham S., Dewaard J.R. Barcoding animal life: Cytochrome c oxidase subunit 1 divergences among closely related species. Proc. R. Soc. B Biol. Sci. 2003;270:S96–S99. doi: 10.1098/rsbl.2003.0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Branstetter M.G., Danforth B.N., Pitts J.P., Faircloth B.C., Ward P.S., Buffington M.L., Gates M.W., Kula R.R., Brady S.G. Phylogenomic insights into the evolution of stinging wasps and the origins of ants and bees. Curr. Biol. 2017;27:1019–1025. doi: 10.1016/j.cub.2017.03.027. [DOI] [PubMed] [Google Scholar]
- 22.Dowton M., Austin A.D. Molecular phylogeny of the insect order Hymenoptera: Apocritan relationships. Proc. Natl. Acad. Sci. USA. 1994;91:9911–9915. doi: 10.1073/pnas.91.21.9911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu Z., van Achterberg C., He J.H., Chen X.X., Chen H.Y. Illustrated keys to Scoliidae (Insecta, Hymenoptera, Scolioidea) from China. ZooKeys. 2021;1025:139–175. doi: 10.3897/zookeys.1025.61385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Folmer O., Black M., Hoeh W., Lutz R., Vrijenhoek R. DNA primers for amplification of mitochondrial cytochrome C oxidase subunit I from diverse metazoan invertebrates. Mol. Marine Biol. Biotechnol. 1994;3:294–299. [PubMed] [Google Scholar]
- 25.Katoh K., Standley D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kumar S., Stecher G., Tamura K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Puillandre N., Lambert A., Brouillet S., Achaz G. ABGD, automatic barcode gap discovery for primary species delimitation. Mol. Ecol. 2012;21:1864–1877. doi: 10.1111/j.1365-294X.2011.05239.x. [DOI] [PubMed] [Google Scholar]
- 28.Zhang J., Kapli P., Pavlidis P., Stamatakis A. A general species delimitation method with applications to phylogenetic placements. Bioinformatics. 2013;29:2869–2876. doi: 10.1093/bioinformatics/btt499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Peters R.S., Krogmann L., Mayer C., Donath A., Gunkel S., Meusemann K., Kozlov A., Podsiadlowski L., Petersen M., Lanfear R., et al. Evolutionary history of the Hymenoptera. Curr. Biol. 2017;27:1013–1018. doi: 10.1016/j.cub.2017.01.027. [DOI] [PubMed] [Google Scholar]
- 30.Day M.C., George R.E., David M. The most primitive Scoliidae (Hymenoptera) J. Nat. Hist. 1981;15:671–684. doi: 10.1080/00222938100770471. [DOI] [Google Scholar]
- 31.Wilson J.J. Assessing the Value of DNA Barcodes and Other Priority Gene Regions for Molecular Phylogenetics of Lepidoptera. PLoS ONE. 2010;5:e10525. doi: 10.1371/journal.pone.0010525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gupta S.K., Jonathan J.K. Fauna of India and the adjacent countries, Hymenoptera: Scoliidae. Zoological Survey of India; Kolkata, India: 2003. pp. 1–277. [Google Scholar]
- 33.Matsumoto R., Hasegawa M., Ichikawa A. Scolia watanabei, an adventive wasp newly discovered in Japan (Hymenoptera, Scoliidae, Scoliinae) Bull. Osaka Mus. Nat. Hist. 2019;73:1–5. [Google Scholar]
- 34.Ilyasov R.A., Park J., Takahashi J., Kwon H.W. Phylogenetic uniqueness of honeybee Apis cerana from the Korean peninsula inferred from the mitochondrial, nuclear, and morphological data. J. Apic. Sci. 2018;62:189–214. doi: 10.2478/jas-2018-0018. [DOI] [Google Scholar]
- 35.Tsuneki K. Studies on the scoliid wasps of eastern Asia (Hymenoptera) Etizenia. 1972;62:1–41. [Google Scholar]
- 36.Luo A.R., Chen L., Simon Y.W.H., Zhu C.D. Comparison of Methods for Molecular Species Delimitation across a Range of Speciation Scenarios. Syst. Biol. 2018;67:830–846. doi: 10.1093/sysbio/syy011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data of the research were deposited in the Museum of Biology at Sun Yat-sen University (SYSBM), Guangzhou, China.