Abstract
Background: The present study investigated the antifungal activity and mode of action of four Olea europaea leaf extracts, Thymus vulgaris essential oil (EO), and Boswellia carteri EO against Fusarium oxysporum. Methods: Fusarium oxysporum lactucae was detected with the internal transcribed spacer (ITS) region. The chemical compositions of chloroform and dichloromethane extracts of O. europaea leaves and T. vulgaris EO were analyzed using GC-MS analysis. In addition, a molecular docking analysis was used to identify the expected ligands of these extracts against eleven F. oxysporum proteins. Results: The nucleotide sequence of the F. oxysporum lactucae isolate was deposited in GenBank with Accession No. MT249304.1. The T. vulgaris EO, chloroform, dichloromethane and ethanol efficiently inhibited the growth at concentrations of 75.5 and 37.75 mg/mL, whereas ethyl acetate, and B. carteri EO did not exhibit antifungal activity. The GC-MS analysis revealed that the major and most vital compounds of the T. vulgaris EO, chloroform, and dichloromethane were thymol, carvacrol, tetratriacontane, and palmitic acid. Moreover, molecular modeling revealed the activity of these compounds against F. oxysporum. Conclusions: Chloroform, dichloromethane and ethanol, olive leaf extract, and T. vulgaris EO showed a strong effect against F. oxysporum. Consequently, this represents an appropriate natural source of biological compounds for use in healthcare. In addition, homology modeling and docking analysis are the best analyses for clarifying the mechanisms of antifungal activity.
Keywords: Olea europaea leaf extracts, Fusarium oxysporum, Thymus vulgaris, internal transcribed spacer (ITS), molecular docking
1. Introduction
Lactuca sativa (Lettuce) is one of the most commonly used crops in the leafy vegetable group, and it belongs to the family of Asteraceae [1]. Lettuce is a vital dietary vegetable, and it has some health benefits as a source of vitamin C, phenolic compounds, and fiber [2]. Overall, lettuce is susceptible to various diseases, such as viral and fungal infections, including powdery mildew, fungus, and bacterial wilt.
In this respect, the search for potent and selective inhibitors has received particular attention because of the increase in antifungal resistance, which has become one of the greatest challenges for global health, food security, and development. In particular, in the agricultural field, F. oxysporum forma sp. Lettuce (F.O.L.) represents the most serious agent of all pathologies involved in lettuce cultivation, especially in North Africa [3]. F. oxysporum forma sp. Lettuce (F.O.L.) is the most important and common fungal pathogen in lettuce plants, and it causes wilting and yield reduction. It remains with a specific host; it increases the yellowing of the leaves and wilting, and it affects the vascular system of the lettuce plant [4]. Unfortunately, to this day, no curative treatment for this fungus exists, except for some limited approaches, such as the disinfection of the soil or the propagation and use of resistant varieties, which remain the first approaches to reducing the impact of this disease [5].
Synthetic fungicides are considered successful tools, and there is no doubt that they have been used for many years in traditional agricultural systems. However, the use of synthetic fungicides has now been reduced because of their adverse effects on humans and the environment. The need to look for and evaluate alternative solutions that are environmentally friendly, available, and affordable for smallholder farmers is important for these crop protection challenges [6].
Nowadays, much attention is required in order to study plant-based ingredients and essential oils for their broad range of biological activities, such as their antiviral, anti-inflammatory, antifungal, and antibacterial properties [7,8]. Essential oils (EOs) are considered essential sources of biologically active compounds, i.e., fungicidal, insecticidal, antibacterial, nematocidal, and herbicidal compounds [8,9]. T. vulgaris includes several species of aromatic herbaceous plants. Moreover, its oil can be used externally as an antiseptic for fungal infections. Thymol is part of a naturally occurring class of compounds identified as biocides, which have strong antimicrobial traits when used alone or with other biocides, such as carvacrol [10]. O. europaea leaf extracts contain phenolic compounds that are effective against bacteria, mycoplasma, and yeasts [11]. Phenolic composites are recognized for their inhibitory activity against bacteria, fungi, and viruses [12]. An O. europaea leaf extract was found to inhibit fungi and Gram-positive bacteria [13].
Recently, the antifungal mechanisms of action of EOs were identified by determining the ergosterol content of the plasma membranes of fungi [14]. Ergosterol is the main sterol component of the fungal cell membrane, and it is responsible for cell function and integrity. Ergosterol is found in almost all fungi, and it is widely used as an indicator of fungal biomass. Therefore, the essential oils of conventionally studied plants were identified as essential instruments in the formulation of plant-based preservatives against aflatoxin and mold contamination, and also as protection against the destructive effects of free radicals [15].
On the other hand, due to inadequate information on the pathogenesis of the F. oxysporum fungus, several computational methods can be applied to better explain its mechanism of action. In this context, molecular docking analysis remains one of the most important tools, as it can provide atomistic insight into molecular recognition by predicting the ability of a molecule to bind to the active site of a protein. In our case, eleven F. oxysporum proteins were determined as targets for our docking studies. F. oxysporum (RHO1) has an essential role in maintaining the hyphal architecture and virulence of Fusarium spp., and it is also responsible for the regulation of the post-translational activity of glucan synthase, which inhibits its recognition by the host. F. oxysporum (XlnR) is responsible for the regulation of plant-cell-wall-degrading enzyme expression. Moreover, Fmk1 from F. oxysporum (MAPK) is a downstream transcription factor and has a vital role in causing invasive hyphal growth and plant infection [16,17].
No previous experiments have studied the effects of chloroform and dichloromethane extracts of O. europaea leaves or their modes of action against F. oxysporum forma species lactucae through molecular docking analysis. Therefore, this study aims to isolate and identify the purified F. oxysporum forma species lactucae using the ITS sequence of the conserved ribosomal DNA. Additionally, a preliminary screening and an evaluation of the phytochemical composition of the antifungal activities of some plant extracts, i.e., T. vulgaris, Boswellia frankincense, and O. europaea leaves, were used to test the effectiveness against the isolated F. oxysporum forma species lactucae. Moreover, a molecular docking analysis was performed for the expected antifungal ligands of these extracts in homology models that were constructed for the eleven F. oxysporum proteins in order to gain better insight into the ligand–protein binding interactions that prevent the infection process of Fusarium.
2. Results
2.1. Morphological Characterization, Isolation, and Pathogenicity Tests
The results of the morphological characterization and isolation of F. oxysporum f. sp. lactucae are presented in Figure 1a,b. The results reveal that the lettuce plants showed wilting symptoms as the result of a single, pure colony in potato dextrose agar (PDA) media. Moreover, the chlamydospores seemed to have an elliptical and spherical shape and were established only in a short, peripheral chain that reached 19.7 − 19.2 × 21.3 − 19.9 μm. A F. oxysporum isolate was able to infect the lettuce plants. It caused a disease incidence of 100% and a disease severity of 94.44%, but the control plants remained healthy. The initial symptoms appeared as yellowing on one side of the outer leaves, which then turned brown (Figure 1a). As the disease progressed, the plants wilted and died. After uprooting the infected plants from the soil and making a longitudinal section of the roots, discoloration could be noticed in the vascular area of the crown (Figure 1b). Based on the morphological characterization and isolation, the results reveal that the characteristics correspond to F. oxysporum infection.
Figure 1.
Symptoms of Fusarium wilting in lettuce under artificial infection symptoms on the margin of the outer leaves (a) and brown discoloration in the crown area (b).
2.2. Molecular Characterization and Phylogenetic Tree of the Purified F. oxysporum Isolate
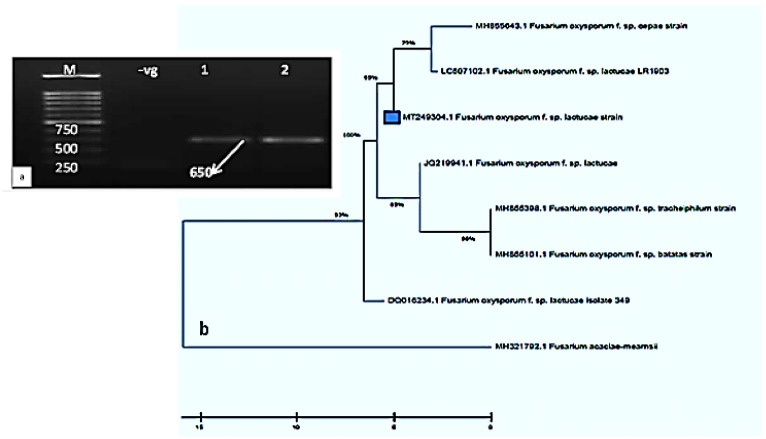
The molecular identification and phylogenetic analyses of the purified F. oxysporum lactucae isolate were carried out (Figure 2a,b). The PCR product of the ITS sequence was detected at 650 bp, as shown in Figure 2a. The sequence analysis of the fungal isolate showed 100% similarity to the F. oxysporum sp. lactucae ITS sequence, and it was deposited in the Gene Bank under the accession number MT249304.1. In addition, the phylogenetic analysis confirmed that it had the highest similarity to the F. oxysporum lactucae isolate Lux (MT249304.1). The results of the phylogenetic tree analysis revealed that the closeness of the genetic similarity between the studied F. oxysporum isolate and others from around the world was mainly with strains of F. oxysporum with different accession numbers (MH855643.1, MH855398.1, DQ016234.1, MH855101.1, MH321792.1, JQ219941.1, and LC507102.1) in the database of the GenBank (Figure 2b). Therefore, F. oxysporum was recognized as the causal mediator of the Fusarium wilt in lettuce in Egypt.
Figure 2.
PCR product and phylogenetic tree of the Fusarium oxysporum f. sp. lactucae isolate. (a) PCR product of the isolate. M = Molecular marker, –vg = Negative control, (1–2) = F. oxysporum samples. (b) Phylogenetic tree based on the ITS gene sequences of the isolates.
2.3. Biological Activity
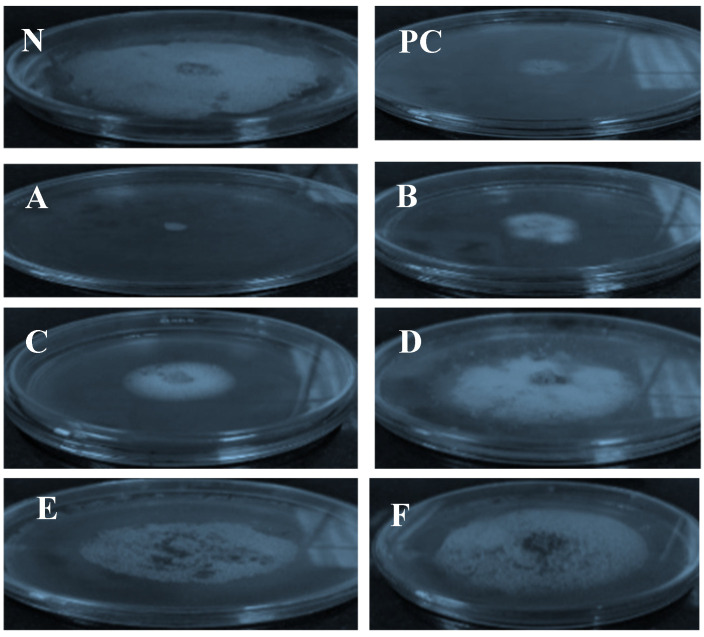
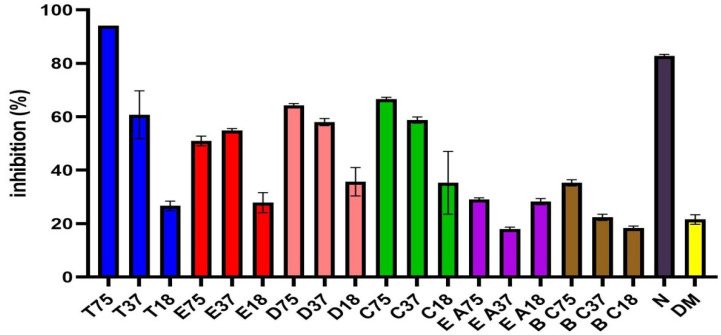
The antifungal properties of the ethanol, dichloromethane, chloroform, and ethyl acetate from O. europaea leaf extracts, and the oils of T. Vulgaris and B. carteri against F. oxysporum f. sp. lactucae were evaluated in this investigation, as shown in Figure 3 and Figure 4 and Table 1. The results revealed that the T. Vulgaris essential oil and the dichloromethane, chloroform and ethanol extracts of O. europaea leaf extract leaves efficiently inhibited the mycelial growth of the F. oxysporum f. sp. lactuca compared to control treatment, whereas the ethyl acetate extracts did not exhibit antifungal effects at the tested concentration. The data illustrated in Figure 4 indicate that all the tested plant extracts except ethyl acetate extract significantly decreased the mycelial growth of the pathogenic fungi compared to the control treatment. The oil of T. Vulgaris showed the maximum inhibition percentage (94.11) at the tested concentration (75.5 mg/mL), against the fungus. Moreover, the chloroform, dichloromethane and ethanol extracts exhibited the maximum inhibition (67.058, 64.71 and 50.59 mm, respectively) at the tested concentration (75.5 mg/mL).
Figure 3.
Effects of T. vulgaris oil (A), chloroform (B), dichloromethane (C), ethanol (D) and ethyl acetate extracts (E) of O. europaea leaves and B. carteri (F) on mycelium linear growth of. F. oxysporum f. sp. lactucae after six days of cultivating the fungus on PDA medium at 25 °C. NC: negative control (DMSO), PC: positive control (Nystatin).
Figure 4.
Mycelial growth % inhibition with different concentrations of T: thyme oil, E: ethanol, D: dichloromethane, C: chloroform, EA: ethyl acetate, B C: B. carteri, N: nystatin, DM: DMSO.
Table 1.
IC50 of different plant extracts against F. oxysporum.
| Plant Extract | IC50 (mg/mL) |
|---|---|
| Thyme oil | 28.99 (µL/ml) |
| Ethanol extract | 49.60 |
| Dichloromethane extract | 32.51 |
| Chloroform extract | 31.35 |
| Ethyl acetate extract | 521.9 |
| B. carteri | 176.2 |
2.4. GC-MS Analysis
A post-silylation GC-MS analysis was employed to profile the primary metabolites. The analysis of the silylated dichloromethane and chloroform extracts O. europaea leaves led to the detection of 39 and 54 metabolites, respectively (Table 2 and Table 3). The identified components belonged to different classes, such as sugars, organic and amino acids, fatty acids, flavonoids, and low-molecular-weight or nonpolar secondary metabolites that were exemplified in alkaloids and steroids.
Table 2.
Composition of the dichloromethane extract from the O. europaea leaves according to the GC-MS analysis.
| Compound No. | RI | Identified Compound | MWT | %Similarity | Chemical Class | Area% |
|---|---|---|---|---|---|---|
| 1 | 788 | 1,2-Bis(trimethylsiloxy)ethane | 206 | 75 | Alkane | 1.3 |
| 2 | 905 | Ethanimidic acid, N-(trimethylsilyl)-, trimethylsilyl ester | 203 | 93 | Acid | 2.2 |
| 3 | 915 | Propanoic acid, 2-[(trimethylsilyl)oxy]-, trimethylsilyl ester | 234 | 96 | Acid | 1.8 |
| 4 | 1078 | Butanoic acid, 4-[(trimethylsilyl)oxy]-, trimethylsilyl ester | 248 | 89 | Acid | 1.6 |
| 5 | 1086 | Cyclohexanone, 3,3,5-trimethyl- | 140 | 77 | Ketone | 0.4 |
| 6 | 1108 | Glycerol, 3TMS derivative | 308 | 96 | Alcohol | 6.9 |
| 7 | 1163 | Picolinic acid, TMS derivative | 195 | 95 | Acid | 2.2 |
| 8 | 1192 | Octanoic acid, trimethylsilyl ester | 216 | 75 | Fatty acid | 0.2 |
| 9 | 1252 | 2-(2-Butoxyethoxy)ethoxy-trimethylsilane | 254 | 98 | Alcohol | 0.9 |
| 10 | 1338 | 3,6,9,12-Tetraoxa-2,13-disilatetradecane, 2,2,13,13-tetramethyl- | 294 | 70 | Alcohol | 0.4 |
| 11 | 1406 | Octanoic acid, TBDMS | 258 | 76 | Fatty acid | 0.2 |
| 12 | 1612 | Hexadecane | 226 | 95 | Alkane | 0.4 |
| 13 | 1664 | 8,10-Dioxaheptadecane | 244 | 87 | Alkane | 3.8 |
| 14 | 1685 | L-Proline, 2TBDMS derivative | 343 | 73 | Amino acid | 0.9 |
| 15 | 1699 | L-Leucine, 2TBDMS derivative | 359 | 87 | Amino acid | 3.6 |
| 16 | 1699 | Isoleucine, 2TBDMS derivative | 359 | 89 | Amino acid | 1.3 |
| 17 | 1769 | Myristic acid | 228 | 93 | Fatty acid | 0.4 |
| 1788 | Tetradecanoic acid, trimethylsilyl ester | 300 | 85 | Fatty acid | 0.4 | |
| 19 | 1878 | Hexadecenoic acid, methyl ester | 270 | 87 | Fatty acid | 0.7 |
| 20 | 1916 | Mandelic acid di(tert-butyldimethylsilyl)- | 380 | 91 | Acid | 3.4 |
| 21 | 1968 | Palmitic acid | 256 | 92 | Fatty acid | 11.6 |
| 22 | 1987 | Hexadecenoic acid, trimethylsilyl ester | 328 | 77 | Fatty acid | 5.8 |
| 23 | 2009 | Eicosane | 282 | 97 | Alkane | 0.4 |
| 24 | 2037 | α.-D-(+)-Talopyranose, pentakis(trimethylsilyl) ether | 540 | 85 | Sugar | 0.4 |
| 25 | 2066 | Trimethylsilyl ether of glucitol | 614 | 70 | Sugar | 1.6 |
| 26 | 2081 | 7,9-Di-tert-butyl-1-oxaspiro(4,5)deca-6,9-diene-2,8-dione | 276 | 88 | Lactone | 0.9 |
| 27 | 2186 | Octadecanoic acid, trimethylsilyl ester | 356 | 85 | Fatty acid | 3.8 |
| 28 | 2194 | Oleic acid, trimethylsilyl ester | 354 | 86 | Fatty acid | 2.5 |
| 29 | 2581 | Hexadecanoic acid, 2,3-bis[(trimethylsilyl)oxy]propyl ester | 474 | 94 | Fatty acid | 1.3 |
| 30 | 2780 | Glycerol monostearate, 2TMS derivative | 502 | 85 | Alcohol | 0.4 |
| 31 | 2789 | β.-Sitosterol trimethylsilyl ether | 486 | 87 | Sterol | 1.1 |
| 32 | 2900 | Nonacosane | 408 | 95 | Alkane | 0.4 |
| 33 | 2914 | Squalene | 410 | 86 | Organic compound | 0.4 |
| 34 | 2931 | α.-Amyrin, TMS derivative | 498 | 96 | Triterpene | 0.9 |
| 35 | 3343 | Spirosol-5-en-3-ol, 28-acetyl-, acetate, (3.β.,22.α.,25R)- | 497 | 97 | Alcohol | 7.2 |
| 36 | 3401 | Tetratriacontane | 478 | 95 | Alkane | 1.6 |
| 37 | 3410 | Stigmasta-3,5-dien-7-one | 410 | 85 | Sterol | 0.7 |
| 38 | 3600 | Hexatriacontane | 506 | 95 | Alkane | 3.8 |
| 39 | 3986 | Propanoic acid, 3,3′-thiobis-, ditetradecyl ester | 570 | 70 | Acid | 2.7 |
Table 3.
Composition of the chloroform extract from the O. europaea leaves according to the GC-MS analysis.
| Compound No. | RI | Identified Compound | Chemical Class | MWT | %Similarity | Area% |
|---|---|---|---|---|---|---|
| 1 | 788 | 1,2-Bis(trimethylsiloxy)ethane | Alkane | 206 | 84 | 0.9 |
| 2 | 905 | Ethanimidic acid, N-(trimethylsilyl)-, trimethylsilyl ester | Acid | 203 | 95 | 1.2 |
| 3 | 915 | Propanoic acid, 2-[(trimethylsilyl)oxy]-, trimethylsilyl ester | Acid | 234 | 95 | 0.5 |
| 4 | 993 | Hexanoic acid, TMS derivative | Acid | 188 | 78 | 0.7 |
| 5 | 1078 | Butanoic acid, 4-[(trimethylsilyl)oxy]-, trimethylsilyl ester | acid | 248 | 89 | 1.4 |
| 6 | 1086 | Cyclohexanone, 3,3,5-trimethyl- | Ketone | 140 | 75 | 0.2 |
| 7 | 1108 | Glycerol, 3TMS derivative | Alcohol | 308 | 95 | 7.3 |
| 8 | 1163 | Picolinic acid, TMS derivative | Acid | 195 | 96 | 2.1 |
| 9 | 1192 | Octanoic acid, trimethylsilyl ester | Fatty acid | 216 | 73 | 0.2 |
| 10 | 1252 | 2-(2-Butoxyethoxy)ethoxy-trimethylsilane | Alcohol | 254 | 97 | 0.9 |
| 11 | 1338 | 3,6,9,12-Tetraoxa-2,13-disilatetradecane, 2,2,13,13-tetramethyl- | Alcohol | 294 | 70 | 0.2 |
| 12 | 1406 | Octanoic acid, TBDMS | Fatty acid | 258 | 72 | 0.2 |
| 13 | 1437 | 3,6,10,13-Tetraoxa-2,14-disilapentadecane, 2,2,14,14-tetramethyl- | Alcohol | 308 | 70 | 0.2 |
| 14 | 1585 | Quinoline | Alkaloid | 197 | 73 | 0.7 |
| 15 | 1612 | Hexadecane | alkane | 226 | 70 | 0.5 |
| 16 | 1664 | 8,10-Dioxaheptadecane | Alkane | 244 | 89 | 3.1 |
| 17 | 1682 | Hexadecane, 7,9-dimethyl- | Alkane | 254 | 90 | 0.2 |
| 18 | 1685 | L-Proline, 2TBDMS derivative | Amino acid | 343 | 71 | 0.7 |
| 19 | 1692 | β.-D-(+)-Xylopyranose, 4TMS derivative | Sugar | 438 | 86 | 0.2 |
| 20 | 1699 | L-Leucine, 2TBDMS derivative | Amino acid | 359 | 90 | 0.9 |
| 21 | 1699 | Isoleucine, 2TBDMS derivative | Amino acid | 359 | 88 | 0.2 |
| 22 | 1766 | Sebacic acid, 2TMS derivative | Acid | 346 | 80 | 0.9 |
| 23 | 1769 | Myristic acid | Fatty acid | 228 | 92 | 0.2 |
| 24 | 1788 | Tetradecanoic acid, trimethylsilyl ester | Fatty acid | 300 | 76 | 0.9 |
| 25 | 1878 | Hexadecanoic acid, methyl ester | Fatty acid | 270 | 94 | 0.5 |
| 26 | 1888 | n-Pentadecanoic acid, trimethylsilyl ester | Fatty acid | 314 | 85 | 0.5 |
| 27 | 1916 | Mandelic acid di(tert-butyldimethylsilyl)- | Acid | 380 | 94 | 2.8 |
| 28 | 1968 | Palmitic acid | Fatty acid | 256 | 92 | 9.2 |
| 29 | 1987 | Hexadecanoic acid, trimethylsilyl ester | Fatty acid | 328 | 80 | 8.5 |
| 30 | 2009 | Eicosane | Alkane | 282 | 96 | 0.2 |
| 31 | 2037 | α.-D-(+)-Talopyranose, pentakis(trimethylsilyl) ether | Sugar | 540 | 87 | 0.7 |
| 32 | 2066 | Trimethylsilyl ether of glucitol | Sugar | 614 | 92 | 1.9 |
| 33 | 2067 | Heptadecanoic acid | Fatty acid | 270 | 75 | 0.9 |
| 34 | 2081 | 7,9-Di-tert-butyl-1-oxaspiro(4,5)deca-6,9-diene-2,8-dione | Lactone | 276 | 90 | 0.5 |
| 35 | 2087 | Heptadecanoic acid, TMS derivative | Fatty acid | 342 | 85 | 0.5 |
| 36 | 2186 | Octadecanoic acid, trimethylsilyl ester | Fatty acid | 356 | 75 | 5.0 |
| 37 | 2194 | Oleic acid, trimethylsilyl ester | Fatty acid | 354 | 75 | 5.2 |
| 38 | 2210 | α.-Linolenic acid, TMS derivative | Fatty acid | 350 | 90 | 0.2 |
| 39 | 2581 | Hexadecanoic acid, 2,3-bis[(trimethylsilyl)oxy]propyl ester | Fatty acid | 474 | 85 | 2.8 |
| 40 | 2780 | Glycerol monostearate, 2TMS derivative | Alcohol | 502 | 87 | 0.5 |
| 41 | 2788 | 1-Monooleoylglycerol, 2TMS derivative | Alcohol | 500 | 70 | 0.2 |
| 42 | 2789 | β.-Sitosterol trimethylsilyl ether | Sterol | 486 | 71 | 1.2 |
| 43 | 2796 | 1-Monolinolein, 2TMS derivative | Fatty acid | 498 | 91 | 14.2 |
| 44 | 2900 | Nonacosane | Alkane | 408 | 89 | 0.2 |
| 45 | 2914 | Squalene | Organic compound | 410 | 95 | 0.5 |
| 46 | 2931 | α.-Amyrin, TMS derivative | Triterpene | 498 | 96 | 1.2 |
| 47 | 2955 | Ergometrinine, 2TMS derivative | Alkaloid | 469 | 70 | 0.5 |
| 48 | 3049 | Tris(4-bromophenyl) amine | Amine | 479 | 86 | 0.2 |
| 49 | 3228 | Catechin (2R-E)-, 5TMS derivative | Flavonoid | 650 | 72 | 0.9 |
| 50 | 3343 | Spirosol-5-en-3-ol, 28-acetyl-, acetate, (3.β.,22.α.,25R)- | Alcohol | 497 | 96 | 6.6 |
| 51 | 3401 | Tetratriacontane | Alkane | 478 | 96 | 0.5 |
| 52 | 3410 | Stigmasta-3,5-dien-7-one | Sterol | 410 | 75 | 0.7 |
| 53 | 3600 | Hexatriacontane | Alkane | 506 | 95 | 2.8 |
| 54 | 3986 | Propanoic acid, 3,3′-thiobis-, ditetradecyl ester | Acid | 570 | 75 | 2.8 |
The separated oil was yellow with a spicy aromatic odor. The yield was 2.7% (w/w), and the refractive index was 1.4894. Twenty-four components were identified in the T. vulgaris essential oil at different percentages (Table 4). The GC-MS analysis of the oil revealed that the major components were thymol (41.85%), o-cymene (11.76%), gamma-terpinene (10.85), and carvacrol (3.61%). These four significant constituents represented 68.07% of all components.
Table 4.
Composition of the T. Vulgaris essential oil extract according to the GC-MS analysis.
| Compound No. | RI | Identified Compounds | %Similarity | Area (%) |
|---|---|---|---|---|
| 1 | 930 | α-Thujene | 97 | 1.4 |
| 2 | 948 | α-Pinene | 97 | 0.8 |
| 3 | 952 | Camphene | 97 | 0.4 |
| 4 | 975 | Sabinen | 97 | 0.1 |
| 5 | 979 | β-Pinene | 98 | 0.3 |
| 6 | 991 | β-Myrcene | 96 | 1.6 |
| 7 | 1009 | α-Phellandrene | 94 | 0.3 |
| 8 | 1013 | δ-3-Carene | 95 | 0.1 |
| 9 | 1018 | α-terpinene | 97 | 1.5 |
| 10 | 1020 | o-Cymene | 97 | 11.8 |
| 11 | 1059 | p-Cineole | 96 | 1.8 |
| 12 | 1062 | γ-Terpinene | 97 | 10.9 |
| 13 | 1109 | 2-p-Menthen-1-ol | 96 | 1.0 |
| 14 | 1112 | Linalool | 97 | 1.3 |
| 15 | 1138 | Borneol | 97 | 1.0 |
| 16 | 1177 | 4-Terpineol | 96 | 0.4 |
| 17 | 1231 | p-Cymene | 96 | 7.6 |
| 18 | 1262 | Thymol | 97 | 41.9 |
| 19 | 1298 | carvacrol | 96 | 3.6 |
| 20 | 1464 | Caryophyllene | 98 | 1.2 |
| 21 | 1515 | Germacrene D | 93 | 0.7 |
| 22 | 1524 | δ-Cadinene | 92 | 0.1 |
| 23 | 1581 | Caryophyllene oxide | 94 | 0.3 |
| 24 | 1757 | Humulane-1,6-dien-3-ol | 93 | 0.2 |
2.5. Effects of Plant Phytochemical Extracts on the Virulence Proteins of F. oxysporum According to Docking Analysis
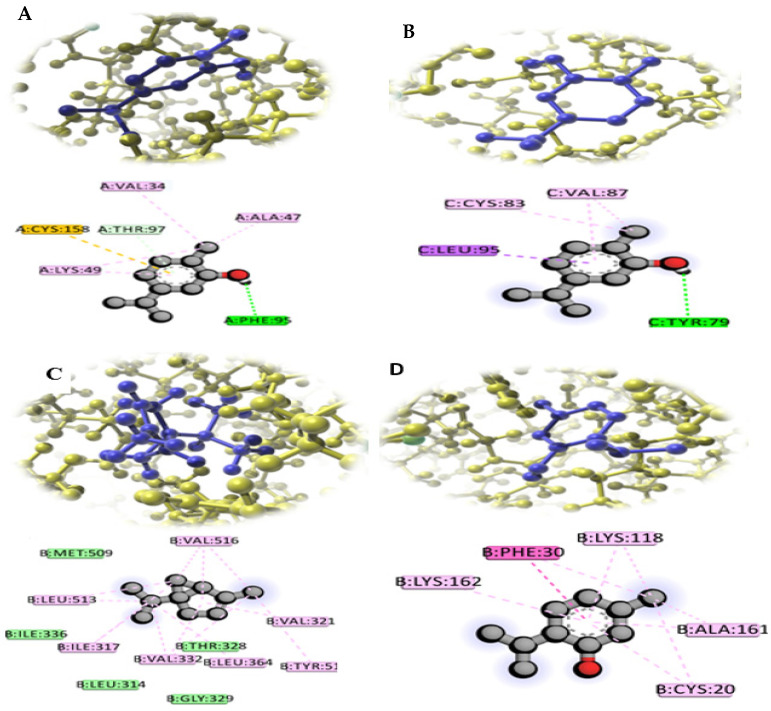
The eleven F. oxysporum proteins that were studied in relation to the active compounds present in T. vulgaris, B. carteri essential oil, and the O.europaea leaf extracts were docked as shown in Table 5, Figure 5 and Figure 6. This study was conducted to identify the ligands expected to block their activities and, hence, to gain a better clarification of their mode of action in controlling the pathogenicity of F. oxysporum f. sp. lactucae. These eleven vital proteins, i.e., AreA, MeaB, Fmk1, Ste7, Set12, Sge1, Xin R, Hog1, PacC, Mkk12, and Rho1, which have vital virulence pathways, were modeled. These proteins were the most important candidates in the virulence of F. oxysporum, as shown in Figure 4. In these connections, carvacrol, α-thujene, and thymol compounds bound with the active sites of these proteins with a binding affinity that ranged from −4.1 to −6.9 kcal/mol. Moreover, the carvacrol ligand had the highest score and bound with Hog1 (A) and PacC (B), while α–Thujene and thymol bound with Mkk12 (C) and Rho1 (D), respectively, as shown in Figure 5. It is worth mentioning that all of these compounds were present in the T. vulgaris plant extract. On the other hand, the nonacosane and tetratriacontane ligands bound with the FMK1, SET7, SEG1, and Rho1 proteins with a binding energy that varied from −5.2 to −5.9 kcal/mol. However, these compounds were found in chloroform extract of O. europaea leaves (Table 5). Hexadecenoic acid, palmitic acid, tetratetracontane, and Stigmast-5-en-3-ol (3á,24S) from the dichloromethane extract of the O. europaea leaves bound with the FMK1, SET7, SEG1, and Rho1 proteins with a binding energy ranging from −4.9 to −5.2 kcal/mol (Table 5). The model showed the docking between the virulence proteins of F. oxysporum and the ligands from the studied and tested plant extracts in order to explore the mechanisms of the binding of the selected proteins in an attempt to understand their role in the inhibition of the F. oxysporum pathogen.
Table 5.
Effects of plant phytochemical compounds, essential oils, and extracts on the F. oxysporum virulence proteins according to the docking analysis.
| Protein/X Y Z | Ligand Name | Pubchem ID | Types of Bond | Score |
|---|---|---|---|---|
| FMK1 x = 27.4555 y = 1.5129 z = 40.778 |
Carvacrol | 10364 | H bond pi-sigma pi-alyl |
−6.4 |
| Thymol | 6989 | −6.3 | ||
| α-Thujene | 17868 | −5.7 | ||
| Hexatriacontane | 12412 | −6 | ||
| Nonacosane | 12409 | −5.9 | ||
| Palmitic acid (Hexadecenoic acid) | 985 | −5.1 | ||
| Tetratriacontane | 5282743 | −5.7 | ||
| Stigmasta-3,5-dien-7-one | 26519 | −5.8 | ||
| SET7 x = 47.3052 y = −15.6381 z = −5.1291 |
α-Thujene | 17868 | Hbond carbon Hbond pi-sulfur pi-Alkyl |
−5.6 |
| Carvacrol | 10364 | −6.1 | ||
| Thymol | 6989 | −6.9 | ||
| Hexatriacontane | 12412 | −6.5 | ||
| Nonacosane | 12409 | −5.9 | ||
| Palmitic acid | 985 | −5 | ||
| (Hexadecenoic acid) | 5282743 | −5.2 | ||
| Tetratriacontane | 26519 | −5.8 | ||
| Stigmasta-3,5-dien-7-one | 12444466 | −4.2 | ||
| SET12 x = 37.1282 y = 38.5316 z = 37.5226 |
α-Thujene | 17868 | H bond carbonHbond pi-sulfur pi-donor Hydrogen |
−3.6 |
| Carvacrol | 10364 | −4.1 | ||
| Thymol | 6989 | −4.1 | ||
| AreA x = 37.1282 y = 38.5316 z = 37.5226 |
α-Thujene | 17868 | H bond carbonHbond pi-sulfur pi-donor Hydrogen |
−3.6 |
| Carvacrol | 10364 | −4.1 | ||
| Thymol | 6989 | −4.1 | ||
| AreA x = 36.9175 y = −94.8871 z = 8.6668 |
α-Thujene | 17868 | Hbond Pi-pi-T-shaped |
4.3 |
| Carvacrol | 10364 | −4.8 | ||
| Thymol | 6989 | −4.6 | ||
| MeaB x = 16.164 y = 0.1875 z = −2.9961 |
Carvacrol | 10364 | carbon H pi-sigma pi-sulfur |
−3.9 |
| Thymol | 6989 | −4.3 | ||
| α-Thujene | 17868 | −4.1 | ||
| Rho1 x =94.6544 y =35.8305 z = 26.0388 |
α-Thujene | 17868 | Pi-pi-T-shaped pi-Alkyl |
−5.2 |
| Carvacrol | 10364 | −5.5 | ||
| Thymol | 6989 | −6.5 | ||
| Hexatriacontane | 12412 | −5.8 | ||
| Nonacosane | 12409 | −5.4 | ||
| Palmitic acid | 985 | −5.2 | ||
| (Hexadecenoic acid) | 5282743 | −5.2 | ||
| Tetratriacontane | 26519 | −5.7 | ||
| Stigmasta-3,5-dien-7-one | 12444466 | −5.1 | ||
| MKK1,2 x = 61.2779 y = −19.0646 z = 15.7687 |
α-Thujene | 17868 | Van der Waals Alkyl pi-Alkyl |
−6.8 |
| Carvacrol | 10364 | −6.1 | ||
| Thymol | 6989 | −5.8 | ||
| SEG1 x = 6.2314 y = 35.6614 z =101.8374 |
α-Thujene | 17868 | Conventional Hbond pi-sulfur pi-pi stacked |
−4.1 |
| Carvacrol | 10364 | −4.9 | ||
| Thymol | 6989 | −5.2 | ||
| Hexatriacontane | 12412 | −5.1 | ||
| Nonacosane | 12409 | −5.4 | ||
| Palmitic acid | 985 | −5.1 | ||
| (Hexadecenoic acid) | 5282743 | −4.9 | ||
| Tetratriacontane | 26519 | −4.8 | ||
| Stigmasta-3,5-dien-7-one | 12444466 | −4.7 | ||
| XINR x = −0.4654 y = −1.4131 z = −3.89 |
Thymol | 6989 | pi-donor H bond pi-Alkyl carbon H bond |
−4.2 |
| α-Thujene | 17868 | −4.1 | ||
| PACC x = 5.5317 y = 8.243 z = 49.2927 |
α-Thujene | 17868 | H bond pi-Alkyl pi-sigma |
−4.2 |
| Carvacrol | 10364 | −5.2 | ||
| Thymol | 6989 | −4.5 | ||
| Hog1 x = 26.713 y = 0.6519 z = 29.6776 |
Carvacrol | 10364 | H bond pi-sigma pi-sulfur pi-Alkyl, Alkyl |
−6.3 |
| Thymol | 6989 | −5.7 | ||
| α-Thujene | 17868 | −5.8 |
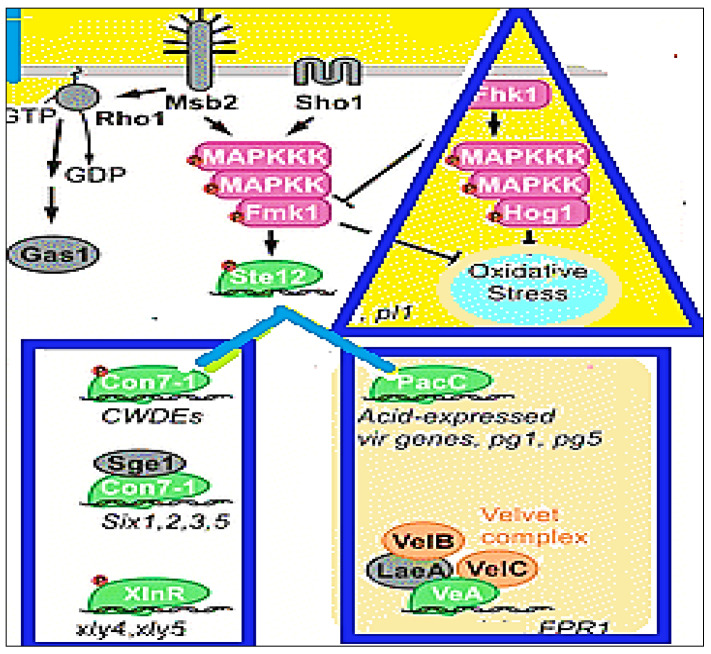
Figure 5.
The pathway of the major protein kinases and transcription factors in the infection process of F. oxysporum [18].
Figure 6.
The 2D and 3D interaction diagrams of the binding of carvacrol with Hog1 (A) and PacC (B), the binding of α-thujene with Mkk12 (C) and Thymol, and the binding of Rho1 (D) with Fusarium virulence proteins.
3. Discussion
Fusarium oxysporum is a huge complex of species of plant and human pathogens that attack a wide array of species in a host-specific manner. Fusarium is a plant disease that exists in the soil. It penetrates into plants and causes losses crop yield and production [19]. Therefore, in this study, experiments were designed to isolate and identify F. oxysporum sp. lactucae. In addition, the biological activities of T. vulgaris, B. carteri and O. europaea leaves against the studied and isolated F. oxysporum were evaluated. Then, the investigation of their modes of action in preventing the development of the Fusarium infection process and in controlling the disease was assessed by using molecular docking analysis. As a perfect, sensitive, fast, and specific means of fungal identification and detection, many authors have established molecular methods as an alternative approach to the conventional procedures used in fungal identification. The internal transcribed spacer (ITS) region of the ribosomal DNA is highly variable within the genus Fusarium. In addition, the use of polymerase chain reaction (PCR) with primers targeted to this region for the detection and identification of Fusarium species with molecular methods was summarized by [20,21]. In this investigation, based on the morphological, cultural, pathogenic, and molecular results, the fungus was identified using the IT’S sequence of the ribosomal DNA, as F. oxysporum has the accession number MT249304.1. These results are in agreement with those described by [22], as they used the same method of using the ITS sequence of the ribosomal DNA to identify the differences among the species of the genus Fusarium. The results of the PCR identification of the ITS sequence successfully categorized the studied fungus as F. oxysporum; the fungus was not capable of infecting non-lettuce hosts. All of these data agree with the findings of [23]. They stated that Fusarium yielded isolates from lettuce plants that were particularly virulent in lettuce hosts. Therefore, it could be concluded that the fungus in the experiment was F. oxysporum f. sp. lactucae. According to the available research and our knowledge, Fusarium wilt in lettuce caused by F. oxysporum f. sp. lactucae was not previously documented in Egypt. Further surveys and in-depth investigations should be considered.
In this respect, the antifungal potentials of ethanol, dichloromethane, chloroform, and ethyl acetate extracts from O. europaea leaves, as well as those of the oil of T. vulgaris, were checked with respect to F. oxysporum. The results of the preliminary screening and evaluation revealed that the T. vulgaris essential oil and dichloromethane chloroform and ethanol extracts from O. europaea leaves efficiently inhibited the growth of the tested fungus with variable potency, whereas the ethyl acetate extract did not exhibit an antifungal effect at the tested concentration (75.5, 37.75 and 18. 875 mg/mL). These results are in line with those of other studies in which phenols extracted from O. europaea leaves showed antifungal activity [24]. In this respect, T. vulgaris has antimicrobial potential against pathogenic microorganisms [25]. The effectiveness of thyme EO against food-related bacteria and fungi was tested. The synergistic, antagonistic, and additive effects of the components of EOs require further research in order to elucidate the mechanisms underlying their biological activity and to access new and natural antiseptics that are applicable to the pharmaceutical and food industries [26].
The dichloromethane and chloroform extracts from O.europaea leaves effectively inhibited F. oxysporum f. sp. lactucae with zones of inhibition of 34.83 and 23.25 mm, respectively. The extracts under investigation were potent and natural antifungal drugs, which is in agreement with the studies of [27,28]. Metabolite profiling provided insights into the mediation of the metabolites in O.europaea leaf extracts for their effects as an initial step in establishing an assessment of the quality of the extracts.
In the current study, T. vulgaris essential oil significantly inhibited F. oxysporum f. sp. lactucae with a maximum inhibition percentage (94.11) at the tested concentration (75.5 mg/mL. The results of our research are in agreement with those of prior studies that showed the powerful antifungal activity of T. vulgaris oil [29]. Our results confirm that T. vulgaris can be used as a potent natural agent against foodborne pathogens and in the protection of valuable crops [30]. The principal components, thymol and carvacrol, play a vital role in the antifungal activity of the oil [31]. Therefore, they can serve as markers for the T. vulgaris essential oil.
The potent antifungal activity of the extracts and essential oil under investigation is promising in response to previous studies that encouraged natural drugs as pesticidal, antimicrobial, and food-preservative alternatives to chemical agents [32,33].
Computational modeling could be used to determine exceptional information in order to understand the mechanisms of the modes of action of the antifungal molecules that inhibit the fungal infection process. The molecular docking approach was used to predict the molecules that could bind specifically to the protein active sites that are responsible for the fungal infection process [34]. In the present study, the docking of the active molecules of the studied plant extracts with eleven essential proteins involved in the development pathway of F. oxysporum was evaluated. In this respect, the results of the molecular docking analysis showed that the carvacrol, α-thujene, and thymol compounds in Thymus vulgaris essential oil bound with all of these F. oxysporum proteins and conferred pathogenicity, whereas hexadecenoic acid, palmitic acid, tetratetracontane, and stigmast-5-en-3-ol (3á,24S) could bind with the FMK1, SET7, SEG1, and Rho1 proteins; these represent the compositions of the dichloromethane and chloroform extracts from O. europaea leaves. Hence, this leads to the inhibition of the pathogenicity of F. oxysporum. These proteins are vital elements of the pathway of a transduction signal that controls numerous F. oxysporum infection processes [33]. Several studies have explained the importance of specific pathogenicity proteins, e.g., FMK1, SET7, SET12, AreA, MeaB, Rho1, MKK1,2, SEG1, XINR, and Hog, in various pathways that are responsible for the virulence of F. oxysporum and that have a role in disease control [18,35]. These results are in agreement with those of [35], as they stated that molecular docking analyses were performed to clarify the antifungal effectiveness of the most and least active compounds against the Fgb1 and Fophy fungal proteins. This study indicates that dichloromethane, chloroform, and O. europaea leaves may be considered as the most important sources of antifungal compounds.
4. Materials and Methods
4.1. Morphological Characterization and Isolation of Fusarium oxysporum f. sp. lactucae
Fusarium isolate samples were collected from lettuce plants at the Faculty of Agriculture, Cairo University, Giza, Egypt. The stems of the infected samples were sterilized by filling them with 2% sodium hypochlorite solution for 4 min. They were then exhaustively washed with dH2O. Then, the samples were cut into 4 pieces of 5 mm of diseased tissue, which was transferred onto potato dextrose agar (PDA) media. The antibiotic streptomycin was added to the PDA media to decrease contamination resulting from bacterial growth. The fungal culture was incubated at 26 ± 2 °C and was periodically checked. The fungal growth was identified and purified for characterization and usage in the following experiments. A single spore of the fungal isolates was grown for 11 days on a PDA medium for morphological identification. The culture properties were detected from 11 to 15 days in the PDA cultures. The microscopic features of the chlamydospores and conidia were also detected by following the methods of previous reports [36,37].
4.2. Pathogenicity Test
A test of the pathogenicity of the F. oxysporum isolates was carried out at the greenhouse of the Plant Pathology Department, Faculty of Agriculture, Cairo University, Giza, Egypt. The pathogenicity of the fungal isolates was assessed in lactucae seedlings at the true leaf stage. The roots were filled with 60 mL of a suspension of conidia for three minutes. Then, the seedlings were transferred into sterilized soil in plastic pots and incubated under greenhouse conditions. A final determination of disease development was made after 20–30 days.
4.3. Molecular Identification of Fusarium oxysporum Isolates
4.3.1. Genomic Extraction from Fungal DNA
The single, pure cultures of fungus were grown in a medium of potato dextrose broth (PDB) in darkness for 7 days at 25 °C. Mycelia were collected through purification with filter paper, then harvested in 0.85% NaCl saline solution. The collected mycelia were used directly for DNA isolation with the Gene JET Genomic DNA Purification Kit (Thermo Scientific, Lithuania, USA). The DNA yield and purity were checked using both a Nanodrop spectrophotometer and agarose gel electrophoresis.
4.3.2. Identification of F. oxysporum Isolate through ITS Gene Sequencing
The internal transcribed spacer (ITS) region was identified with the universal primers of the ITS1 (5′-CTTGGTCATTTAGAGGAAGTAA-3′) and ITS4 (5′ TCCTCCGCTTATTGATATGC-3′) sequences. The amplification step was performed using a thermal cycler for PCR (Bio-Rad T100, Hercules, CA, USA). The PCR products were amplified through agarose gel electrophoresis and by using a gel extraction kit for purification; then, the purified PCR products were sent for sequencing by Macrogen (Seoul, Korea).
4.3.3. Sequencing and Bioinformatic Analyses
The conserved sequence of the ITS gene of the studied fungus isolate’s genome was similar to the ITS sequences in the database, as established through the use of the Basic Local Alignment Search Tool (BLAST), which is found on the website of the NCBI (https://blast.ncbi.nlm.nih.gov, 2020). Then, this part of the F. oxysporum f. sp. lactucae sequence was compared with similar sequences of strains of F. oxysporum in the NCBI database; then, a phylogenetic tree was constructed by using the MEGA 6 software program (https://mega.software.informer.com/6.0/, 2020). The phylogenetic analysis was performed by using the maximum likelihood tree method. The tree distance was calculated using the maximum composite likelihood method.
4.4. Plant Extract Preparation
4.4.1. Materials
The T. vulgaris and O. europaea leaves used in this work were obtained from the Medicinal, Aromatic, and Poisonous Plant Experimental Station, Faculty of Pharmacy, Cairo University, as well as the Department of Medicinal and Aromatic Plants, Faculty of Agriculture, Cairo University, Egypt.
All solvents used were of LC/MS grade and were purchased from J. T. Baker (The Netherlands).
All other chemicals and standards were purchased from Sigma Aldrich (St. Louis, MO, USA).
4.4.2. Preparation of O. europaea Leaf Extracts
The O. europaea leaves were dried at room temperature (26 ± 2 °C) for two weeks and ground to a fine powder. The ground leaves (1 g) were extracted with 10 mL of high-purity ethanol, dichloromethane, chloroform, and ethyl acetate solvents. Whatman filter paper no. 1 was used to filtrate the extracts, and then a rotary evaporator was used to concentrate the extracts at 40 °C by using 50 mL centrifuge tubes. The extracts were dried in a glassy desiccator; then, the residues of the powder were stored for analysis. Extraction was carried out three times with the same volume of solvent added repeatedly.
4.4.3. Preparation of Essential Oil (Thymus vulgaris)
The fresh leaves were collected and subjected to hydrodistillation in a Clavenger apparatus for 5 h. according to the procedure described in the Egyptian Pharmacoepia [38].
4.5. Antifungal Activity of Plant Extracts and Essential Oils In Vitro
The antifungal activity of the ethanol, dichloromethane, chloroform, and ethyl acetate extracts from O. europaea leaves, as well as the oil of T. vulgaris and B. carteri were examined in vitro by using a poisoned food technique [39]. Each treatment was separately dissolved in DEMSO (1:1 v/v) to prepare stock of 75.5, 37.75 and 18.875 mg/mL for extracts and µL/mL for oils were added to 100 mL a sterile Erlenmeyer flask containing 60 mL cooled molten PDA, then rotated manually to disperse the solutions. We dispensed 20 milliliters of the medium into sterile Petri dishes (9 cm in diameter). The medium was allowed to solidify at room temperature. Agar discs taken from the margins of fresh and pure culture were aseptically inoculated at the center of the Petri plates. Control plates (with DMSO only) were inoculated following the same procedure. The fungicide nystatin (100,000 units/mL) was used as a positive control. We tested with 200 microliters in PDA medium. The nystatin was purchased from Sigma Aldrich (St. Louis, MO, USA). The tests were performed in triplicate, and all plates were incubated at 25 °C. We measured the fuserium radial growth after 6 days of incubation at 25 °C. All experiments were performed in triplicate for each treatment. The average of the two orthogonal diameters was measured when fungal mycelium covered one plate in control treatment.
4.6. GC/MS Analysis of Essential Oil (Thymus vulgaris)
The mass spectra were recorded with a Shimadzu GCMS-QP2010 (Tokyo, Japan) equipped with a split–splitless injector and Rtx-5MS column (30 m × 0.25 mm i.d. × 0.25 µm film thickness) (Restek, Bellefonte, PA, USA). The capillary column was attached to a quadrupole mass spectrometer (SSQ 7000; Thermo-Finnigan, Bremen, Germany). The initial temperature of the column was set to 45 °C for 2 min and programmed to 300 °C at a rate of 5 °C/min; then, it was kept constant at 300 °C for 5 min. The injector temperature was 250 °C. The flow rate of the carrier gas (helium) was 1.41 mL/min. The mass spectra were recorded according to the following conditions: filament emission current (equipment current), 60 mA; ionization voltage, 70 eV; ion source, 200 °C. Diluted samples (1% v/v) were injected via the split mode (split ratio: 1:15) [33].
Sample Sialylation for GC/MS Analysis
For the analysis of primary metabolites in different samples, a derivatization step was performed before the analysis, as described in Farag et al. (2015) [40]. Briefly, the extract (50 μL) was mixed with 100 μL of N-methyl-N-(trimethylsilyl)-trifluoroacetamide (MSTFA) and incubated at 60 °C for 45 min. Samples were equilibrated at 28 °C and analyzed using a Shimadzu model QP-5000 GC-MS mass spectrometer (Kyoto, Japan).
The silylated derivatives were separated on an Rtx-5MS column, and all injections were performed in the 1:15 split mode. The quadrupole mass spectrometer was operated in an electron ionization mode at 70 eV. The scan range was set to 50–650 m/z.
4.7. Molecular Docking Analysis
In this investigation, the molecular docking of the tested phytochemical compounds extracted from T. vulgaris and O. europaea leaves with eleven essential proteins involved in F. oxsyporum virulence, i.e., AreA, MeaB, Fmk1, Ste7, Set12, Sge1, Xin R, Hog1, PacC, Mkk12, and Rho1, was studied. The molecular docking analysis was performed using the SAMSON 2020 software (https://www.samson-connect.net/) to determine the interactions between the target virulence proteins of Fusarium and the ligand structures of the tested plant extract compounds and to identify the direct effects of these compounds on the inhibition of Fusarium. The sequence of each protein was downloaded from NCBI (https://www.ncbi.nlm.nih.gov/) in the FASTA format to build binding models with a 3D structure by using the TASSER server (https://zhanglab.ccmb.med.umich.edu/I-TASSER/). The SWISSMODLE server (https://swissmodel.expasy.org/) was used for the construction of the 3D proteins. The affinity minimization was performed using the 3DREFINE server (http://sysbio.rnet.missouri.edu/3Drefine/index.html). In pre-docking, all water molecules and ligands were deleted, and the hydrogen atoms were added to the target proteins. On the other hand, the ligands were downloaded from PubChem (https://pubchem.ncbi.nlm.nih.gov/) in the SDF format, and then the openbabel software (http://openbabel.org/wiki/Main_Page) was used for the conversion into the MOL2 format. The interactions of the Fusarium proteins were built into the models with the ligand structures of the ethanol, dichloromethane, chloroform, and ethyl acetate extracts of O. europaea leaves, as well as the oil of T. vulgaris. The docking of the proteins with the tested compounds was performed with the aid of the SAMSON 2020 software. The calculations of the free binding energies were performed by using the scoring function of AutoDock Vina as an element in its script. Following an exhaustive search, 100 poses were examined, and the best-scoring poses were selected to compute the binding energies of the ligands. In addition, Discovery Studio (https://www.discngine.com/discovery-studio) was used for the 2D structures of the ligands.
4.8. Statistical Data Interpretation
Data analysis and graphs were made using the GraphPad Prism Version 9 program. The data are expressed as an arithmetic mean, standard deviation and 95 percent confidence interval for the IC50 parameter. IC50 was calculated as a concentration of the tested compound which decreases the mycelial growth by half between the base and maximum. p values less than or equal 0.05 were considered statistically significant [41].
5. Conclusions
The molecular identification of F. oxysporium, which causes Fusarium wilt in lactucae plants in Egypt, was achieved. In this investigation, the dichloromethane chloroform and ethanol extracts of O. europaea leaves and the T. vulgaris essential oil showed strong effects against F. oxysporum. The antifungal screening against F. oxysporum f. sp. lactucae (F.o.L) verified that the main active ingredients of these extracts displayed considerable antifungal activity. In this respect, the results of the molecular docking analysis showed that the carvacrol, α-thujene, and thymol compounds in Thymus vulgaris essential oil bound specifically with eleven pathogenic proteins in F. oxysporium, whereas hexadecenoic acid, palmitic acid, tetratetracontane, and stigmast-5-en-3-ol (3á,24S) bound with the FMK1, SET7, SEG1, and Rho1 proteins. In particular, the chloroform extract of O. europaea leaves was selected as a source of antifungal substances for use against F. oxysporum, and it could provide a new lead in the pursuit of new biological sources of agrochemical candidates.
Acknowledgments
We thank Nour El-Houda A. Reyad, Marwa Abd-Elateef Zayton of the Plant Pathology Department, Faculty of Agriculture, Cairo University, Giza, Egypt for the help with the practical part of the study of plant pathology, and we also thank Salma Nour El-Deen of the GMO lab research team in the Cairo University Research Park (CURP) and the team members of the Crop Technology Research Department, Food Technology Research Institute, Agricultural Research Center for their technical support.
Author Contributions
H.S.O. conceived of the presented idea and wrote the manuscript, verified the analytical methods, encouraged the investigation of specific aspects, supervised the findings of this work, discussed the results, and contributed to the final manuscript. H.S.O. designed and performed the molecular identification experiment and molecular docking analysis and wrote that section. S.N.A.E.-R. conceived of the presented idea. S.N.A.E.-R. reviewed the final manuscript. S.N.A.E.-R. and S.M.A. carried out the extraction of the plant extracts. N.E.-H.A.R. Contributed the results of the morphological characterization and provided the initial source of Fusarium oxysporum f. sp. lactucae. M.S.S. prepared the essential oil and performed the sialylation step and sample preparation for the GC/MS analysis. M.S.S. and S.N.A.E.-R. performed the GC/MS analysis. M.S.S. identified the components of both the essential oil and the silylated extracts and wrote that section. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Fusarium oxysporum f. sp. lactucae from plant pathology department, Faculty of Agriculture, Cairo University, Giza, Egypt.
Conflicts of Interest
The authors declare no conflict of interest.
Sample Availability
Samples of the compounds are not available from the authors.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Singh R.J., Lebeda A., Ryder E., Grube R., Dolezalová I., KrÍstková E. Lettuce (Asteraceae; Lactuca spp.) Veg. Crops. 2006;3:377–472. doi: 10.1201/9781420009569.ch9. [DOI] [Google Scholar]
- 2.Mulabagal V., Ngouajio M., Nair A., Zhang Y., Gottumukkala A.L., Nair M.G. In vitro evaluation of red and green lettuce (Lactuca sativa) for functional food properties. Food Chem. 2010;118:300–306. doi: 10.1016/j.foodchem.2009.04.119. [DOI] [Google Scholar]
- 3.Matheron M.E., Gullino M.L. Fusarium wilts of lettuce and other salad crops. In: Gullino M.L., Katan J., Garibaldi A., editors. Fusarium Wilts of Greenhouse Vegetable and Ornamental Crops. APS Press; St. Paul, MN, USA: 2012. pp. 175–183. [Google Scholar]
- 4.Gilardi G., Ortega S.F., Van Rijswick P.C.J., Ortu G., Gullino M.L., Garibaldi A. A new race of Fusarium oxysporum f. sp. lactucae of lettuce. Plant Pathol. 2016;66:677–688. doi: 10.1111/ppa.12616. [DOI] [Google Scholar]
- 5.Gordon T.R. Fusarium oxysporum and the Fusarium Wilt Syndrome. Annu. Rev. Phytopathol. 2017;55:23–39. doi: 10.1146/annurev-phyto-080615-095919. [DOI] [PubMed] [Google Scholar]
- 6.Feliziani E., Romanazzi G. Preharvest application of synthetic fungicides and alternative treatments to control postharvest decay of fruit. Stewart Postharvest Rev. 2013;9:1–6. [Google Scholar]
- 7.Lichtenstein E.P. Insecticides Occurring Naturally in Crops. Adv. Chem. 1966;1:34–38. doi: 10.1021/ba-1966-0053.ch004. [DOI] [Google Scholar]
- 8.Turek C., Stintzing F.C. Stability of Essential Oils: A Review. Compr. Rev. Food Sci. Food Saf. 2013;12:40–53. doi: 10.1111/1541-4337.12006. [DOI] [Google Scholar]
- 9.Raveau R., Fontaine J., Sahraoui A.L.-H. Essential Oils as Potential Alternative Biocontrol Products against Plant Pathogens and Weeds: A Review. Foods. 2020;9:365. doi: 10.3390/foods9030365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ahmad A., Khan A., Yousuf S., Khan L.A., Manzoor N. Proton translocating ATPase mediated fungicidal activity of eugenol and thymol. Fitoterapia. 2010;81:1157–1162. doi: 10.1016/j.fitote.2010.07.020. [DOI] [PubMed] [Google Scholar]
- 11.Friedman M. Overview of antibacterial, antitoxin, antiviral, and antifungal activities of tea flavonoids and teas. Mol. Nutr. Food Res. 2007;51:116–134. doi: 10.1002/mnfr.200600173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee-Huang S., Huang P.L., Zhang D., Lee J.W., Bao J., Sun Y., Chang Y.-T., Zhang J., Huang P. Discovery of small-molecule HIV-1 fusion and integrase inhibitors oleuropein and hydroxytyrosol: Part I. Integrase inhibition. Biochem. Biophys. Res. Commun. 2007;354:872–878. doi: 10.1016/j.bbrc.2007.01.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Soni M., Burdock G., Christian M., Bitler C., Crea R. Safety assessment of aqueous olive pulp extract as an antioxidant or antimicrobial agent in foods. Food Chem. Toxicol. 2006;44:903–915. doi: 10.1016/j.fct.2006.01.008. [DOI] [PubMed] [Google Scholar]
- 14.Tian J., Huang B., Luo X., Zeng H., Ban X., He J., Wang Y. The control of Aspergillus flavus with Cinnamomum jensenianum Hand.-Mazz essential oil and its potential use as a food preservative. Food Chem. 2012;130:520–527. doi: 10.1016/j.foodchem.2011.07.061. [DOI] [Google Scholar]
- 15.Prakash B., Kedia A., Mishra P.K., Dubey N. Plant essential oils as food preservatives to control moulds, mycotoxin contamination and oxidative deterioration of agri-food commodities—Potentials and challenges. Food Control. 2015;47:381–391. doi: 10.1016/j.foodcont.2014.07.023. [DOI] [Google Scholar]
- 16.Di Pietro A., García-Maceira F.I., Méglecz E., Roncero M.I.G. A MAP kinase of the vascular wilt fungus Fusarium oxysporum is essential for root penetration and pathogenesis. Mol. Microbiol. 2004;39:1140–1152. doi: 10.1111/j.1365-2958.2001.02307.x. [DOI] [PubMed] [Google Scholar]
- 17.Rispail N., Soanes D.M., Ant C., Czajkowski R., Grünler A., Huguet R., Perez-Nadales E., Poli A., Sartorel E., Valiante V., et al. Comparative genomics of MAP kinase and calcium–calcineurin signalling components in plant and human pathogenic fungi. Fungal Genet. Biol. 2009;46:287–298. doi: 10.1016/j.fgb.2009.01.002. [DOI] [PubMed] [Google Scholar]
- 18.Husaini A.M., Sakina A., Cambay S.R. Host–Pathogen Interaction in Fusarium oxysporum Infections: Where Do We Stand? Mol. Plant-Microbe Interact. 2018;31:889–898. doi: 10.1094/MPMI-12-17-0302-CR. [DOI] [PubMed] [Google Scholar]
- 19.Zhang Y., Ma L.-J. Deciphering Pathogenicity of Fusarium oxysporum from a Phylogenomics Perspective. Adv. Genet. 2017;100:179–209. doi: 10.1016/bs.adgen.2017.09.010. [DOI] [PubMed] [Google Scholar]
- 20.O’Donnell K., Kistler H., Cigelnik E., Ploetz R.C. Multiple evolutionary origins of the fungus causing Panama disease of banana: Concordant evidence from nuclear and mitochondrial gene genealogies. Proc. Natl. Acad. Sci. USA. 1998;95:2044–2049. doi: 10.1073/pnas.95.5.2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zarrin M., Ganj F., Faramarzi S. Analysis of the rDNA internal transcribed spacer region of the Fusarium species by polymerase chain reaction-restriction fragment length polymorphism. Biomed. Rep. 2016;4:471–474. doi: 10.3892/br.2016.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Summerell B.A., Salleh B., Leslie J.F. A Utilitarian Approach to Fusarium Identification. Plant. Dis. 2003;87:117–128. doi: 10.1094/PDIS.2003.87.2.117. [DOI] [PubMed] [Google Scholar]
- 23.Cabral C.S., Brunelli K.R., Costa H., De Noronha Fonseca M.E., Boiteux L.S., Reis A. Identification of Fusarium oxysporum f. sp. lactucae race 1 as the causal agent of lettuce wilt in Brazil. Trop. Plant. Pathol. 2014;39:197–202. doi: 10.1590/S1982-56762014000300002. [DOI] [Google Scholar]
- 24.Zorić N., Kopjar N., Kraljić K., Oršolić N., Tomić S., Kosalec I. Olive leaf extract activity against Candida albicans and C. dubliniensis—The in vitro viability study. Acta Pharm. 2016;66:411–421. doi: 10.1515/acph-2016-0033. [DOI] [PubMed] [Google Scholar]
- 25.Khan A.A., Amjad M.S. Saboon GC-MS analysis and biological activities of Thymus vulgaris and Mentha arvensis essential oil. Turk. J. Biochem. 2019;44:388–396. doi: 10.1515/tjb-2018-0258. [DOI] [Google Scholar]
- 26.Mohsenipour Z., Hassanshahian M. The inhibitory effect of Thymus vulgaris extracts on the planktonic form and biofilm structures of six human pathogenic bacteria. Avicenna J. Phytomed. 2015;5:309–318. [PMC free article] [PubMed] [Google Scholar]
- 27.Muzzalupo I., Badolati G., Chiappetta A., Picci N., Muzzalupo R. In vitro Antifungal Activity of Olive (Olea europaea) Leaf Extracts Loaded in Chitosan Nanoparticles. Front. Bioeng. Biotechnol. 2020;8:151. doi: 10.3389/fbioe.2020.00151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sircar B., Mandal S. Screening of Elaeocarpus floribundus fruit extracts for bioactive phytocomponents and antibacterial activity against food-borne bacteria. Int. J. Res. Med. Sci. 2017;5:3665–3671. doi: 10.18203/2320-6012.ijrms20173582. [DOI] [Google Scholar]
- 29.de Lira Mota K.S., de Oliveira Pereira F., De Oliveira W.A., Lima I.O., de Oliveira Lima E. Antifungal Activity of Thymus vulgaris L. Essential Oil and Its Constituent Phytochemicals against Rhizopus oryzae: Interaction with Ergosterol. Molecules. 2012;17:14418–14433. doi: 10.3390/molecules171214418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rota C., Carramiñana J.J., Burillo J., Herrera A. In Vitro Antimicrobial Activity of Essential Oils from Aromatic Plants against Selected Foodborne Pathogens. J. Food Prot. 2004;67:1252–1256. doi: 10.4315/0362-028X-67.6.1252. [DOI] [PubMed] [Google Scholar]
- 31.Zhang J., Ma S., Du S., Chen S., Sun H. Antifungal activity of thymol and carvacrol against postharvest pathogens Botrytis cinerea. J. Food Sci. Technol. 2019;56:2611–2620. doi: 10.1007/s13197-019-03747-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Omar H.S., Elsayed T.R., Reyad N.E.A., Shamkh I.M., Sedeek M.S. Gene-targeted molecular phylogeny, phytochemical analysis, antibacterial and antifungal activities of some medicinal plant species cultivated in Egypt. Phytochem. Anal. 2020;32:724–739. doi: 10.1002/pca.3018. [DOI] [PubMed] [Google Scholar]
- 33.Abdel-Kawy M.A., Michel C.G., Kirollos F.N., Hussien R.A.A., Al-Mahallawi A.M., Sedeek M.S. Chemical composition and potentiation of insecticidal and fungicidal activities of Citrus trifoliata L. fruits essential oil against Spodoptera littoralis, Fusarium oxysporum and Fusarium solani via nano-cubosomes. Nat. Prod. Res. 2019;35:2438–2443. doi: 10.1080/14786419.2019.1675063. [DOI] [PubMed] [Google Scholar]
- 34.Toubi Y., Abrigach F., Radi S., Souna F., Hakkou A., Alsayari A., Bin Muhsinah A., Mabkhot Y.N. Synthesis, Antimicrobial Screening, Homology Modeling, and Molecular Docking Studies of a New Series of Schiff Base Derivatives as Prospective Fungal Inhibitor Candidates. Molecules. 2019;24:3250. doi: 10.3390/molecules24183250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rampersad S.N. Pathogenomics and Management of Fusarium Diseases in Plants. Pathogens. 2020;9:340. doi: 10.3390/pathogens9050340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nelson P.E., Toussoun T.A., Marasas W.F.O. An Illustrated Manual for Identification. Pennsylvania State University Press; University Park, PA, USA: 1983. Fusarium Species; p. 193. [Google Scholar]
- 37.Leslie J.F., Summerell B.A. The Fusarium Laboratory Manual. Blackwell Publishing; Hoboken, NJ, USA: 2006. pp. 1–2. [Google Scholar]
- 38.Egyptian Pharmacopoeia . The English Text of Arab Republic of Egypt. 4th ed. Central Adminstration and Pharmaceutical Affairs (CAPA); Ministry of Health and Population; Cairo, Egypt: 2005. pp. 31–33. [Google Scholar]
- 39.Gupta P., Khare V., Kumar D., Ahmad A., Banerjee G., Singh M. Comparative evaluation of disc diffusion and E-test with broth micro-dilution in sus-ceptibility testing of amphotericin B, voriconazole and caspofungin against clinical Aspergillus isolates. J. Clin. Diagn. Res. 2015;9:DC04. doi: 10.7860/JCDR/2015/10467.5395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Farag M.A., Rasheed D., Kamal I.M. Volatiles and primary metabolites profiling in two Hibiscus sabdariffa (roselle) cultivars via headspace SPME—GC-MS and chemometrics. Food Res. Int. 2015;78:327–335. doi: 10.1016/j.foodres.2015.09.024. [DOI] [PubMed] [Google Scholar]
- 41.Waller W.M., Duncan D.B. A bayes rule for symmetric multiple composition problem. J. Am. Stat. Assoc. 1969;64:1487–1503. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Fusarium oxysporum f. sp. lactucae from plant pathology department, Faculty of Agriculture, Cairo University, Giza, Egypt.