Summary
Currently favored models for meiotic recombination posit that both noncrossover and crossover recombination are initiated by DNA double strand breaks but form by different mechanisms: noncrossovers by synthesis dependent strand annealing; and crossovers by formation and resolution of double Holliday junctions centered around the break. This dual mechanism hypothesis predicts different hybrid DNA patterns in noncrossover and crossover recombinants. We show that these predictions are not upheld, by mapping with unprecedented resolution parental strand contributions to recombinants at a model locus. Instead, break repair in both noncrossovers and crossovers involves synthesis-dependent strand annealing, often with multiple rounds of strand invasion. Crossover-specific double Holliday junction formation occurs via processes involving branch migration as an integral feature, that can be separated from repair of the break itself. These findings reveal meiotic recombination to be a highly dynamic process and prompt a new view of the relationship between crossover and noncrossover recombination.
Keywords: meiosis, homologous recombination, gene conversion, branch migration, crossover, noncrossover, strand invasion, end extension, Saccharomyces
Graphical Abstract
eTOC blurb
Ahuja et al. analyze parental strand contributions to meiotic recombinants and conclude that the same break-repair mechanism is used during noncrossover and crossover formation. They also conclude that crossovers, but not noncrossovers, are produced by the resolution of double Holliday junctions whose formation involves branch migration as an integral feature.
INTRODUCTION
Meiosis produces haploid gametes from a replicated diploid genome by two rounds of chromosome segregation. The first (meiosis I) separates homologous chromosomes of different parental origin (homologs). Homologous recombination is crucial to this process. It promotes homolog recognition and pairing (Weiner and Kleckner, 1994), and connects homologs with crossovers (COs) to ensure proper meiosis I segregation (Buonomo et al., 2000; Hassold and Hunt, 2001; Wang et al., 2017). Meiotic recombination is initiated by DNA double strand breaks (DSBs) that are formed at high levels (de Massy, 2013) by the conserved Spo11 protein and associated proteins (Yadav and Claeys Bouuaert, 2021). DSBs are resected 5’ to 3’ to form ~1kb ssDNA overhangs, which then invade an intact template and form a displacement loop (D-loop), followed by extension by DNA synthesis of the free 3’ end in the D-loop.
The currently favored model for meiotic recombination (Figure 1A), here called the dual-mechanism model, derives mainly from studies in budding yeast. It suggests that noncrossover (NCO) and CO recombination diverge soon after D-loops form (Allers and Lichten, 2001a; Resnick, 1976). NCOs are thought to be formed by synthesis-dependent strand annealing (SDSA), where helicase-mediated displacement frees an extended invading strand to anneal with the other resected DSB end (Allers and Lichten, 2001a; McMahill et al., 2007; Nassif et al., 1994; Resnick, 1976). In contrast, COs are thought to form by a process called, for historical reasons, double-strand break repair (DSBR; Szostak et al., 1983). During DSBR, inherently unstable D-loops are stabilized by conserved ZMM proteins and are converted to a more stable 3-armed structure called a single end intermediate (SEI; Hunter and Kleckner, 2001), which captures the second DSB end to form a four-arm double Holliday junction (dHJ) intermediate (Börner et al., 2004; Hunter and Kleckner, 2001; Lynn et al., 2007; Pyatnitskaya et al., 2019; Schwacha and Kleckner, 1995). The ZMM proteins include complexes (Zip2-Zip4-Spo16 and Msh4-Msh5) that bind branched DNA (De Muyt et al., 2018; Snowden et al., 2004), the Mer3/Hfm1 3’-5’ DNA helicase, thought to extend D-loops (Mazina et al., 2004), and E3 ligases (Zip3 in budding yeast, RNF212 and HEI10 in others) that promote ZMM protein accumulation at CO-designated sites and degradation elsewhere (Ahuja et al., 2017; Cheng et al., 2006; Qiao et al., 2014; Rao et al., 2017). Most ZMM-dependent dHJs are resolved as COs by the meiosis-specific MutLγ (Mlh1/Mlh3)-Exo1 resolvase (Allers and Lichten, 2001a; Argueso et al., 2004; Cannavo et al., 2020; Keelagher et al., 2011; Kulkarni et al., 2020; Zakharyevich et al., 2010; Zakharyevich et al., 2012). A minor fraction (~10-20%) are thought to be resolved as both COs and NCOs by nucleases active in both mitosis and meiosis (De Muyt et al., 2012; Holloway et al., 2008; Kaur et al., 2015; Kohl and Sekelsky, 2013; Zakharyevich et al., 2012). In addition, ~20% of meiotic recombination intermediates contain 3 or 4 chromosomes; these can be produced when the two DSB ends invade different chromatids or by repair of two independent DSBs (Figure S1A; Oh et al., 2007). Finally, while most NCOs are thought to be formed by SDSA, helicases and topisomerases can take apart dHJs and form NCOs (Figure S1B) in a process called dissolution (Dayani et al., 2011; Gilbertson and Stahl, 1996; Nasmyth, 1982; Wu and Hickson, 2003).
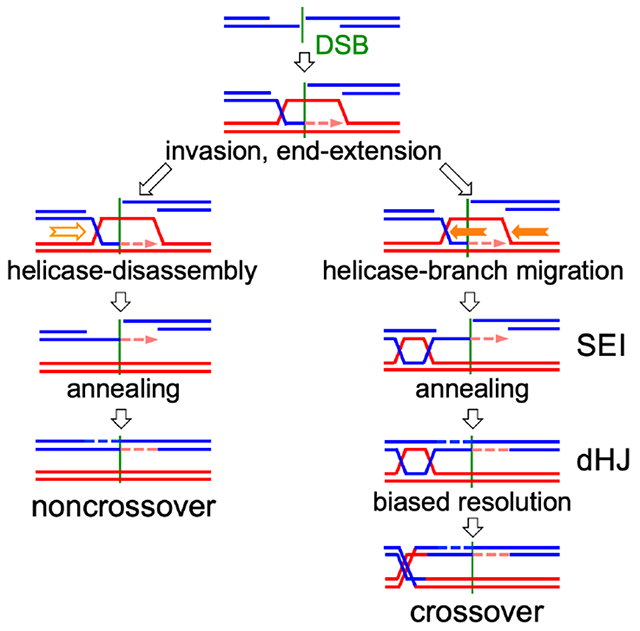
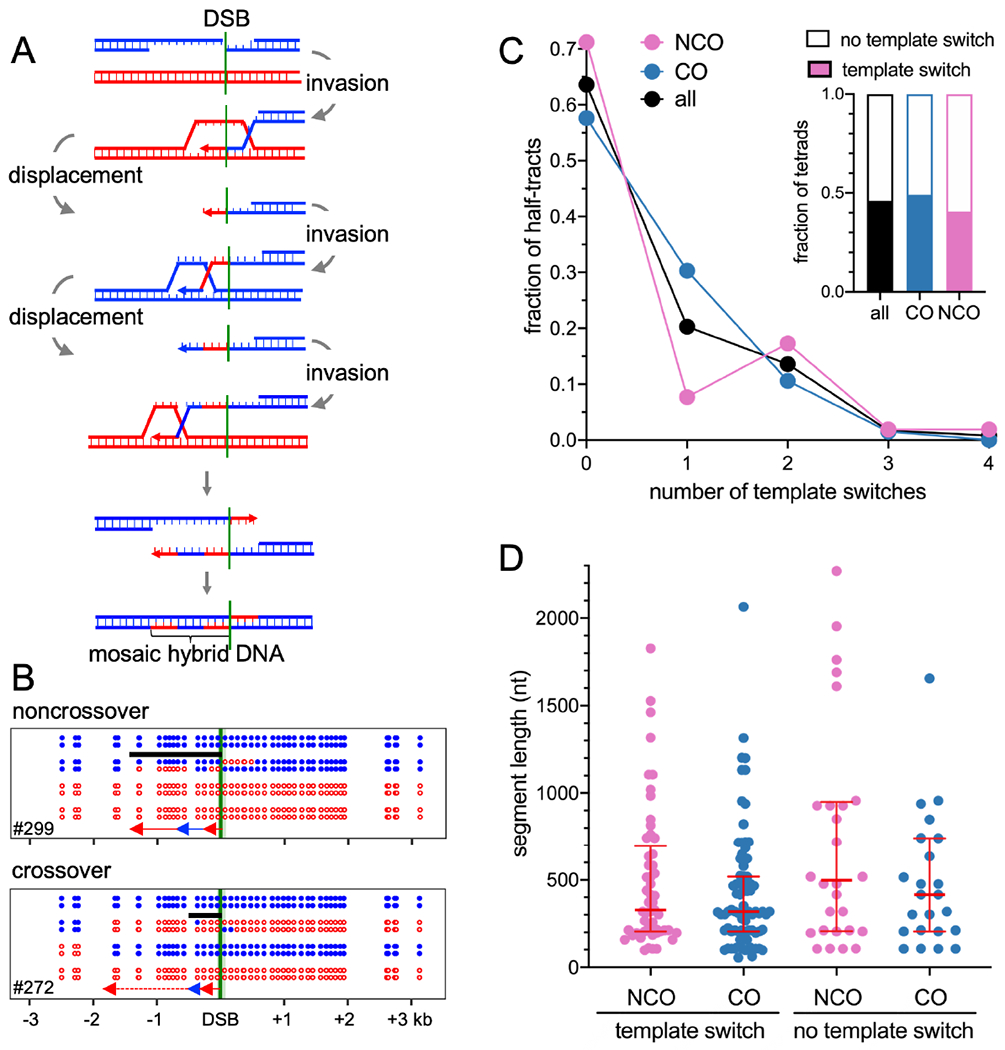
Figure 1. Current “dual-mechanism” model and marker segregation in tetrads.
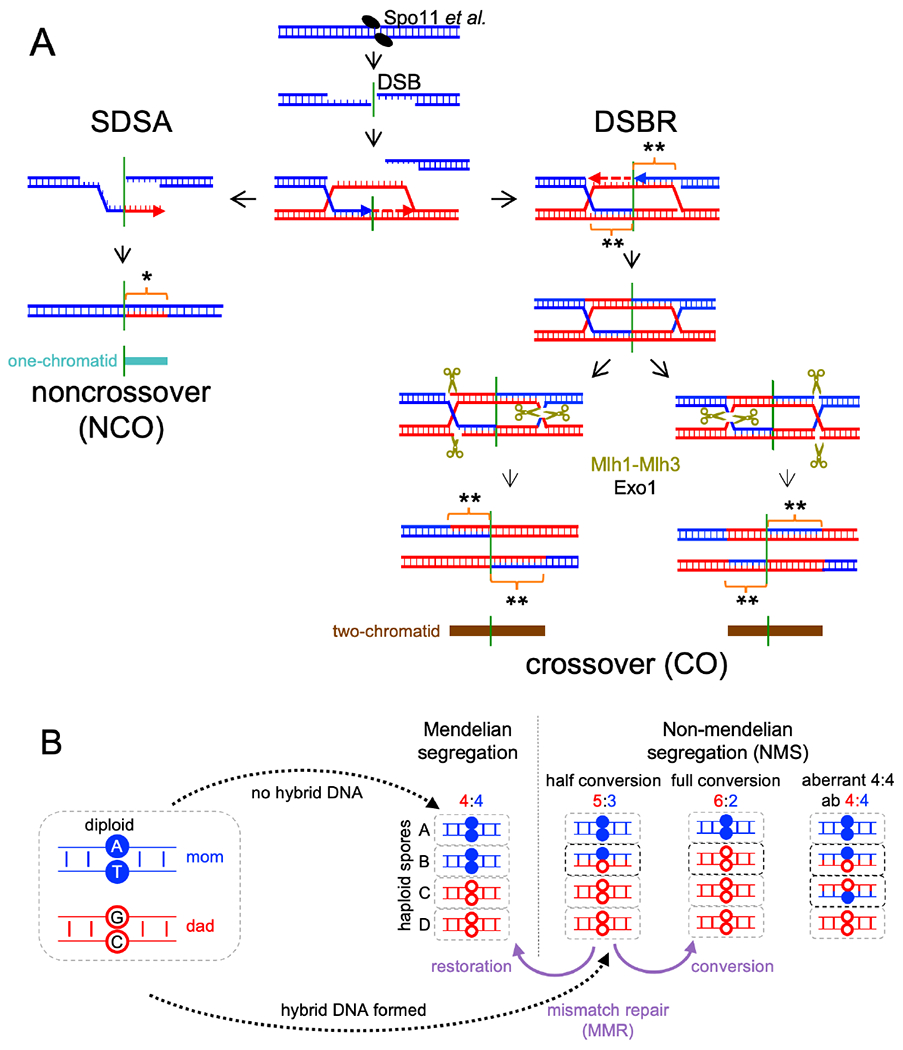
(A) Resection creates a 3’ ssDNA end that invades a homologous duplex and is extended by synthesis (arrow). SDSA – the invasion/extension strand is displaced and forms a NCO. DSBR – the second DSB end is captured and extended to form a double Holliday junction (dHJ) that resolves to form COs. Hybrid DNA in NCOs contains one “old” and one newly synthesized strand (*), and is continuous, on one chromatid, and to one side of the DSB (turquoise). COs contain hybrid DNA in two tracts (brown) that each contain two “old” parental strands (**), with one tract on each chromatid in opposite directions from the DSB. See also Figure S1.
(B) If no hybrid DNA forms, markers segregate in a 4:4 ratio. Asymmetric hybrid DNA results in a 5:3 ratio (half conversion). Correction by MMR produces either 6:2 (full conversion) or 4:4 (restoration). Symmetric hybrid DNA produces an aberrant 4:4 ratio.
Dual-mechanism model intermediates contain DNA where the two strands are from different parents (Figure 1A), here called hybrid DNA. If homologs harbor different alleles, the resulting hybrid DNA will contain mismatches, and here will be called heteroduplex. The mismatches present in heteroduplex are detected genetically as departures from Mendelian segregation (4:4, based on the eight DNA strands present) in haploid meiotic products (Figure 1B). Heteroduplex on only one homolog (asymmetric heteroduplex) produces 5:3 or 3:5 marker segregation (half conversion) if left unrepaired, while heteroduplex on both homologs (symmetric heteroduplex) produces aberrant 4:4 (ab4:4) marker segregation. Mismatches in heteroduplex are recognized by mismatch recognition complexes (Msh2-Msh6 and Msh2-Msh3) and are corrected by the removal and resynthesis of strands over large regions, producing full conversions (6:2 or 2:6) or restoration to parental 4:4 ratios (Iyer et al., 2006; Spies and Fishel, 2015). Mismatch recognition also can trigger heteroduplex rejection, where helicases disassemble recombination intermediates that contain heteroduplex (Spies and Fishel, 2015). Deleting MSH2 inactivates both heteroduplex rejection and MMR and thus preserves heteroduplex patterns, allowing inference of parental strand contributions from genotypes of spores in tetrads (Boris and Haber, 1987; Hunter and Borts, 1997; Martini et al., 2011).
The dual-mechanism model predicts that NCOs formed by SDSA should be one-sided, with a single continuous hybrid DNA tract that starts at the DSB site, while COs formed by DSBR should be two-sided, with two continuous hybrid DNA tracts that start at the DSB and extend in opposite directions (Figure 1A). These predictions are challenged by observation of the following:
Frequent two-sided NCOs, with heteroduplex on both sides of the DSB on a single chromatid (Hoffmann and Borts, 2005; Jessop et al., 2005; Marsolier-Kergoat et al., 2018; Martini et al., 2011; McMahill et al., 2007; Porter et al., 1993).
Mosaic heteroduplex, where heteroduplex tracts alternate with parental duplex (Crown et al., 2014; Marsolier-Kergoat et al., 2018; Martini et al., 2011). This can be produced by switching between chromatids as repair templates (McVey et al., 2004; Smith et al., 2007; Yeadon et al., 2001).
Symmetrical hybrid DNA, detected as aberrant 4:4 (ab4:4) marker segregation (Figure 1B, Figure S1C). This can be formed by branch migration, where Holliday junctions move by exchanging base pairs, creating hybrid DNA on both recombining chromatids (Hamza et al., 1981; Holliday, 1964).
Full conversion in mismatch correction-deficient cells (Williamson et al., 1985) or with markers that form poorly-recognized mismatches (Getz et al., 2008; Lichten et al., 1990; Nag et al., 1989).
Many of these observations were made in studies that lacked the high density of heterozygous markers and knowledge of initiating DSB locations necessary for detailed mechanistic understanding. Recent studies used msh2Δ mutants to preserve heteroduplex in hybrid budding yeast diploids with marker densities of about one per 140-170 nucleotides, to provide compelling evidence for two-sided NCOs, template switching, branch migration and MMR-independent full conversion (Cooper et al., 2018; Marsolier-Kergoat et al., 2018; Martini et al., 2011; Oke et al., 2014). However, it remains uncertain the extent to which these noncanonical events can be generalized, in part because full analysis was precluded by uneven marker distributions and by the broad width and close spacing of many DSB hotspots (Pan et al., 2011).
To address this issue, we created a recombination reporter interval with uniformly spaced polymorphic markers and a single, narrow DSB hotspot, and mapped the origin, extent and structure of recombinants with unprecedented resolution. We find that many of the noncanonical recombination events described above occur at high frequencies during meiosis. Based on these findings, we suggest a novel mechanism for NCO and CO formation and differentiation.
RESULTS
To study meiotic recombination at high resolution, we modified a recombination reporter interval that contains the URA3 and ARG4 genes and a single DSB hotspot (his4::URA3-ARG4; Figure 2A; Jessop et al., 2005). The modified locus (his4::URA3-ARG4-SNPs; Figure 2A, File S1.2) contains sequence changes every 96 ± 29 bp (mean ± S.D.) for 1 kb to the left of the DSB hotspot and 2 kb to the right, with a lower SNP density for another 1.5 kb to the left and 1.1 kb to the right. The changes preserve wild-type coding and create XmnI site polymorphisms for molecular CO scoring. Flanking drug resistance markers allow genetic CO scoring.
Figure 2. Characteristics of the interval studied.
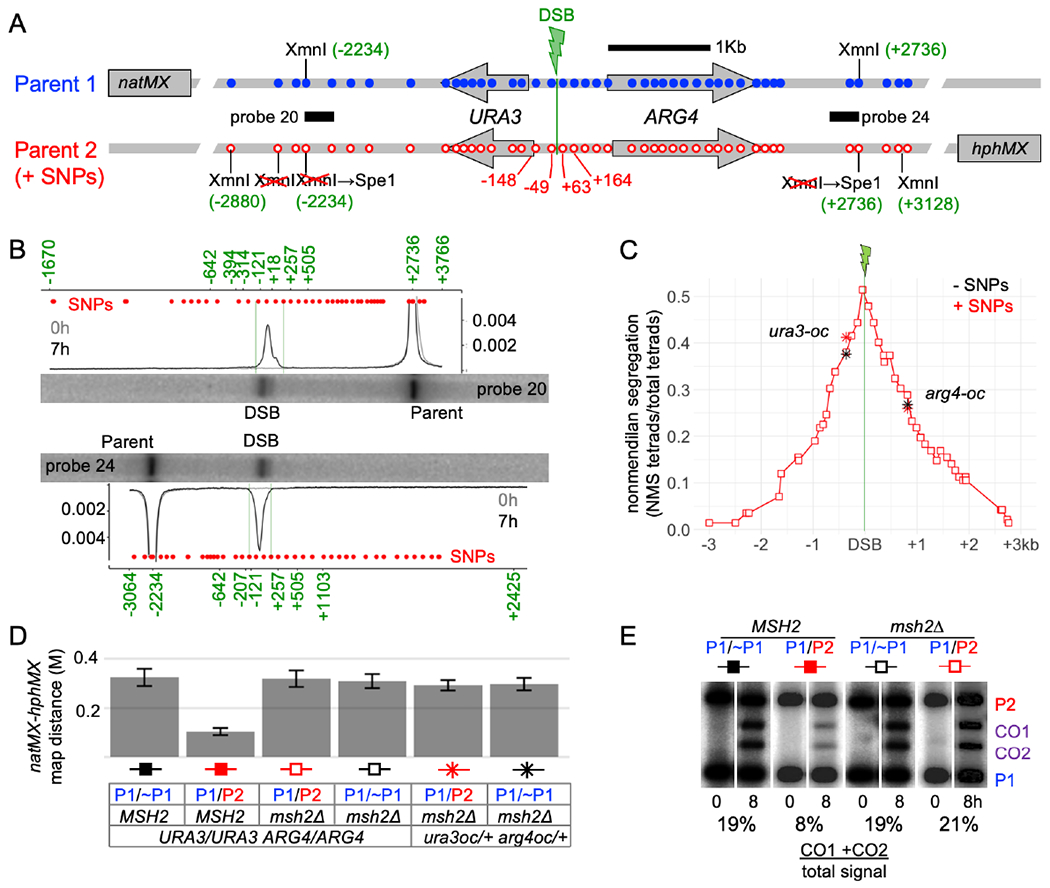
(A) The URA3-ARG4 recombination interval, showing polymorphic markers (blue—wild type; red–polymorphisms). Marker locations are relative to the DSB centroid (File S1.2).
(B) DSBs are tightly focused. Southern blots of XmnI+SpeI digests of meiotic DNA from a resection-deficient rad50S strain (probes in A). Plots are signal/total lane signal. Red dots—marker locations; green numbers—size standards (Figure S2; high resolution gels in Figure S2C, D). Full gel images available at doi:10.17632/j2jpj3p29f.1
(C) Nonmendelian segregation (NMS). Red squares—NMS determined by sequencing. Black stars—NMS for ura3-oc (−364) and arg4-oc (+819) in strains lacking polymorphisms from −2234 to +2736. Red stars—NMS for the same mutants in strains heterozygous for the full polymorphism set.
(D) Heterozygosity reduces COs in the natMX:hphMX interval in wild type but not in msh2Δ. Map distances (Morgans) from tetrad analyses; P1 and P2 are as in panel A; ~P1 lacks sequence polymorphisms from −2234 to +2736 (Figure S2A). Error bars—standard error.
(E) Heterozygosity reduces COs in MSH2 but not in msh2Δ. XmnI digests of DNA from cells before (0h) and after meiosis (8h), probed with ARG4 sequences. Superfluous lanes have been removed; full gel images available at doi:10.17632/j2jpj3p29f.1
Meiotic DSBs form in this interval in 17.5% of chromatids (Figure 2B) and are tightly focused, with 84% of DSBs in a 112 nt region between the two central markers (−49 nt and +63 nt relative to the DSB centroid). All DSBs detected in the interval are within a 312 nt region flanked by the next two markers (−148 to +164, Figure 2B, Figure S2B).
Marker segregation in 142 tetrads from a MMR-deficient msh2Δ strain was scored by high-throughput sequencing of amplicons from spore colony DNA (File S1.3, File S2). Non-mendelian segregations (NMS), ¾ of which were half-conversions, occurred in 77% of tetrads (File S1.4). Markers adjacent to the DSB centroid showed maximal NMS (51% and 48%, Figure 2C, File S1.4), which decayed exponentially with distance (median NMS ~780 nt; Figure S2E). To determine if the multiple polymorphisms affect recombination outcomes, we measured COs and NMS for ura3 and arg4 ochre mutations, in strains where the central interval was either completely homozygous (Figure S2A) or heterozygous for the full set of polymorphisms. While heterozygous polymorphisms reduced COs ~3-fold in MSH2 strains, the same high polymorphism density did not alter recombination outcomes in msh2Δ strains, (Fig. 2C–E, File S1.5 and S1.6).
39% (55/142) of all tetrads contained a CO involving two chromatids, and 23% (32/142) contained a single NCO, most of which (28/32) involved conversion of one chromatid. 15% of tetrads (22/142) contained events involving three or four chromatids. About half of these (10/22) must have involved DSBs on more than one chromatid (for example tetrad 15, File S2), while the remainder (12/22) could have been produced by more than one DSB or by resolution of a multichromatid JM initiated by a single DSB (Figure S1A, File S1.1). Because of this uncertainty, we focused on the tetrads that appeared to come from a single DSB.
Similar patterns of end invasion and extension among noncrossovers and crossovers.
The dual-mechanism model predicts that NCOs contain hybrid DNA on one side of the initiating DSB, while COs will contain hybrid DNA on both sides (Figure 1A). The heteroduplex patterns observed do not support this prediction (Figure 3). Instead, NCOs and COs have remarkably similar heteroduplex patterns: about 1/3 of both are one-sided; and 2/3 are two-sided (Figure 3D). This similarity between NCOs and COs extends to total NMS tract lengths (Figure 3E) and to half-tract lengths (distance from DSB to NMS tract end; Figures 3F; File S1.7).
Figure 3. Similar patterns of end invasion and extension in NCOs and COs.
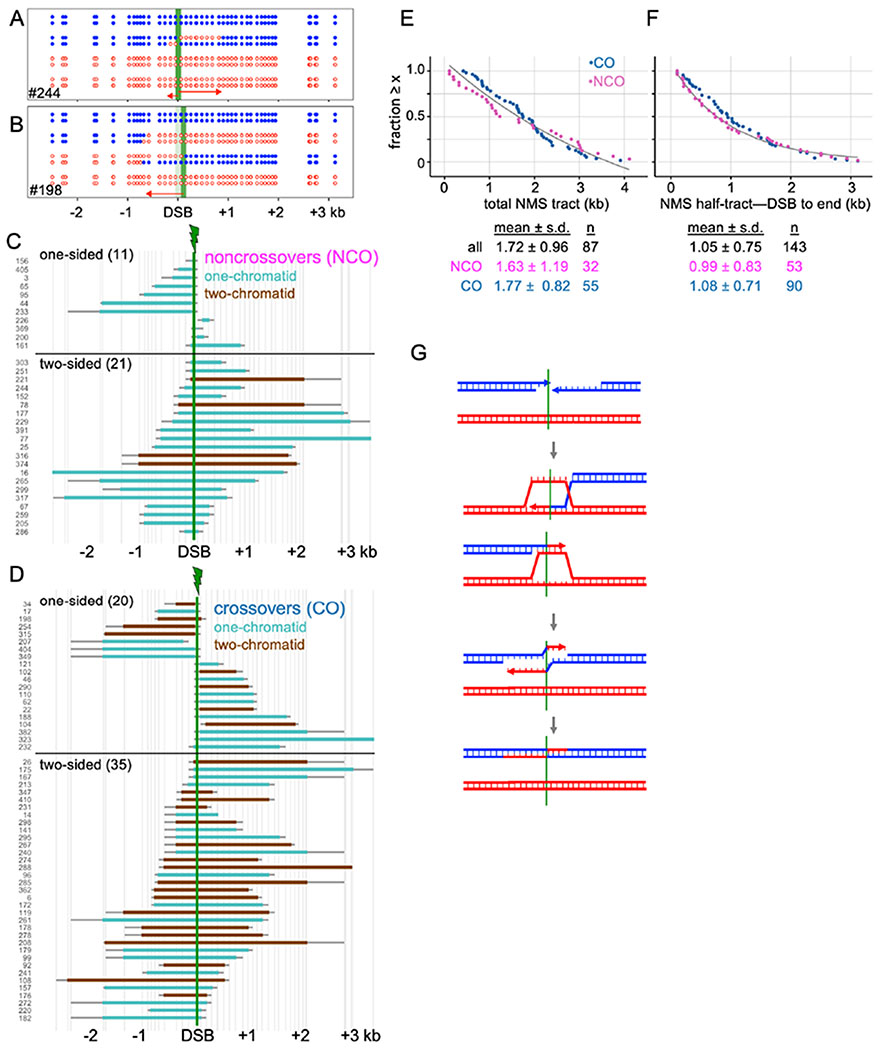
(A) NCO tetrad with two-sided NMS.
(B) CO tetrad with one-sided NMS.
(C) NMS tracts in NCOs. Turquoise—heteroduplex on one chromatid; brown—heteroduplex on two chromatids (see Figure 1A). Vertical axis-tetrad identifiers; vertical lines—marker positions. Thick colored bars and thin gray bars indicate minimum and maximum NMS tracts, respectively.
(D) NMS tracts in COs; color code as in C.
(E) Rank order plot of NMS tract lengths for crossovers (blue) and noncrossovers (purple), calculated using the average between minimum and maximum NMS tracts. Black line—exponential decay curve fit to all events.
(F) NMS half-tract lengths, distance from initiating DSB (midpoint between closest flanking markers) to NMS tract end (midpoint between the last converted and the first unconverted marker); colors as in E.
(G) Double SDSA. Both DSB ends invade a homolog, extend, are displaced, and anneal to form a two-sided NCO. Simultaneous invasion of two homologs is shown here, but invasion could occur sequentially and could involve a single chromatid.
Two-sided NCOs can be produced by a process called double SDSA, where the two DSB ends both invade, extend, and are displaced before annealing with each other (Figure 3G). Apparent one-sided COs might be two-sided events where one hybrid DNA tract (for example, the tract formed by strand invasion, see Figure 1A) is less than ~100 nt, and thus might not include a marker. If every one-sided CO contains a second “invisible” hybrid DNA tract, then 18% of CO NMS tracts are expected to be <100 nt. However, extrapolation of observed NMS half-tract lengths predicts that only ~8% should be ≤ 100nt (Figure S3B). Thus, it is likely that many COs scored as one-sided do not contain “invisible” hybrid DNA on one side of the DSB.
Template switching is common among both crossovers and noncrossovers
It has been suggested that meiotic recombination involves multiple rounds of invasion, D-loop disassembly and re-invasion (De Muyt et al., 2012; Kaur et al., 2015). If switching between the homolog and sister chromatids occurs during this process, mosaic hybrid DNA will be produced (Figure 4A). NMS tracts with at least one half-conversion (5:3 or 3:5) segment and at least one segment with two or more markers in a parental (4:4) configuration were scored as mosaic (Figure 4B). Single marker 4:4 segregations were not counted, as they can also be produced by MSH2-independent short-patch repair (Coic et al., 2000; Crown et al., 2014; Fleck et al., 1999). This suggestion is supported by the observation that NCOs with 4:4 and 6:2 single-marker segregations inside heteroduplex tracts occur at similar frequencies (File S1.12), but NCOs with 4:4 segregation for two or more adjacent markers greatly exceed 6:2s (File S1.14). Other marker segregation patterns (full conversion and symmetrical heteroduplex) that mostly occur in COs also were not counted; these are discussed below.
Figure 4. Template switching is common in both COs and NCOs.
(A) Multiple rounds of invasion, extension, and displacement can form mosaic hybrid DNA.
(B) Examples of mosaic heteroduplex among NCOs and COs. Black bars–minimum extent of mosaic heteroduplex. The NCO first invaded the homolog, while the CO first invaded the sister chromatid.
(C) Frequent template switching in COs and NCOs. Inset—fraction of tetrads with template switching. Tetrads where interpretation was uncertain are counted as “no template switch”. Main graph—number of template switches per NMS half-tract.
(D) Length of segments in NMS half-tracts with or without template switching. Red lines-median and quartiles. None of the distributions are significantly different from the others (p>0.05, Mann-Whitney test).
NCOs and COs displayed similar levels of mosaic heteroduplex (in ~0.4-0.5 of tetrads; Figure 4C, File S1.4) and similar frequencies of template-switching (mean of 0.6 ± 0.1 switches/half-tract ± S.E.M.; Figure 4C, File S1.9) and switch segment lengths (median of 327 and 319 nt/segment; Figure 4D). Non-mosaic NMS tract lengths also were similar in NCOs and COs (Figure 4D).
The DSB-adjacent segment in mosaic heteroduplex indicates which chromatid was first invaded (4:4, sister chromatid; 5:3, homolog; Figure 4B). About 1/3 of DSB-adjacent segments were parental (20/64 in NCOs, 45/110 in COs, File S1.9). This indicates that DSB ends invade the one sister and two homolog chromatids with equal likelihoods, as was previously suggested (Goldfarb and Lichten, 2010; Oh et al., 2007).
The presence of mosaic heteroduplex in about half of both COs and NCOs indicates that template switching is frequent during meiotic recombination. This is a minimum estimate, as genetically invisible template switches—for example, from one homolog chromatid to the other, or to the sister chromatid as a final switch—would not have been detected.
In summary, both DSB ends invade and extend in a majority of both NCOs and COs, and a similar fraction of NCOs and COs undergo template switching (Figure S4), consistent with the suggestion that both NCOs and COs form hybrid DNA by similar mechanisms.
Branch migration is frequent among crossovers
dHJ resolution produces a transition from hybrid DNA to parental sequences. A central feature of the DSBR model is that all hybrid DNA should be between the two resolution points (Figure 1A). Consequently, hybrid DNA should be on one side of the DSB on one chromatid, to other side of the DSB on the other (Figure 1A).
The vast majority of COs did not conform to these predictions (Figure 5, File S1.10). In half of COs (28/55), both exchange points were on the same side of the DSB (Figure 5A, D), and there was separate heteroduplex on a single chromosome that often spanned the DSB (example in Figure 5A). In addition, CO exchange points were often separated from DSB-adjacent heteroduplex by two or more markers in a parental configuration (Figure 5A, Figure S5A). In some of these COs (7/28), a stretch of symmetrical heteroduplex (hybrid DNA on both chromatids; aberrant 4:4 segregation) was present between the two exchange points (Figure 5A, File S1.10). Detection of such heteroduplex was limited by larger inter-marker intervals distant from the DSB (Figure 2A), and by the destruction of CO-adjacent heteroduplex by strand processing, which is discussed below.
Figure 5 – Branch migration is frequent among crossovers.
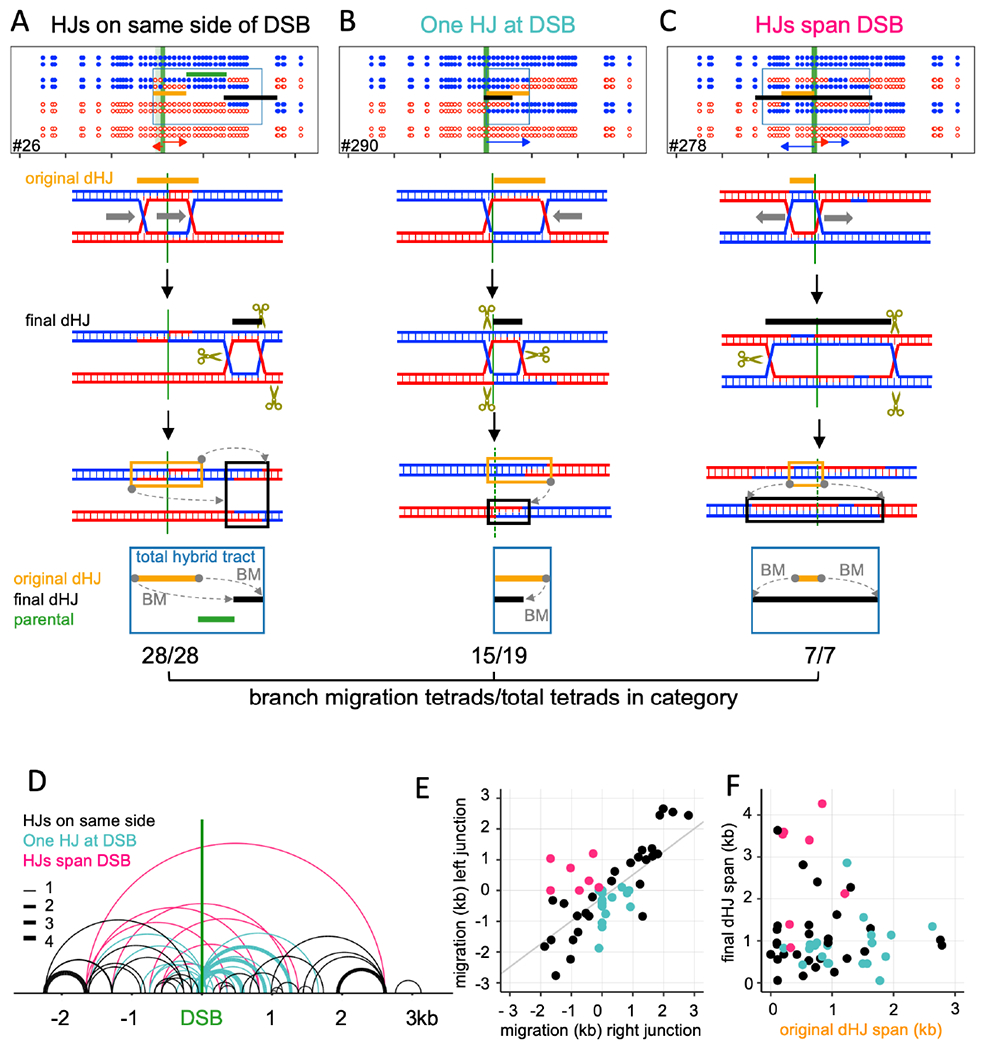
Top—COs classified with regards to final HJ locations (crossover points). Middle – An example for each category. Bottom – proposed branch migration mechanism producing the final marker pattern. Tan– inferred original HJs; black –final HJs; green –parental sequences between original and final dHJs; blue boxes—total hybrid DNA. Grey arrows (BM) – direction and extent of branch migration. The fraction of each category displaying branch migration is given below each category.
(A) Both final HJs are on the same side of the DSB interval. All display branch migration.
(B) One HJ is in the DSB interval, and the other to one side. 15/19 display branch migration.
(C) HJs are on opposite sides of the DSB interval. All display branch migration. In this example, both junctions have moved outward.
(D) Final dHJ locations. Arcs connect the two HJs. Colors correspond to categories in A-C; line thickness indicates the number of events.
(E) Distance and direction of migration by the two HJs. Negative and positive values denote left-and rightward migration; colors as in D.
(F) original and final distance between HJs (dHJ span); colors as in D.
These noncanonical heteroduplex patterns can be produced by branch migration (Holliday, 1964). HJ movement through parental duplex creates symmetrical hybrid DNA (ab4:4). Codirectional movement of the second HJ in a dHJ restores this to the original 4:4 configuration, while retaining a patch of symmetrical hybrid DNA between the two junctions (Figure S1Ci, Figure 5A). Migration of a single junction through a region of hybrid DNA transfers hybrid DNA from one chromatid to the other (Figure S1Cii). This can result in a CO where hybrid DNA switches from one homolog to the other (Figure S1Ciia). These patterns were prominent among the COs where both HJs are on one side of the DSB (Figure 5A; File S1.10).
Signals of branch migration also were present in COs where the final HJs were on opposite sides of the DSB (Figure 5C, D; 7/7), and in COs where one final HJ was inside the DSB region (Figure 5B, D; 15/19). In total, the vast majority (87%) of COs displayed hallmarks of branch migration. In contrast, a much smaller fraction of NCOs (14%, File S1.10) showed evidence of branch migration, consistent with most NCOs not involving dHJ formation and resolution.
In many tetrads, initial junction positions can be inferred from heteroduplex patterns, by assuming that the initial intermediate was a dHJ. This allows calculation of the direction and distance each HJ migrated (Figure 5E, Figure S5A–C, File S1.10). In some cases only one junction moved; in others, the two junctions moved in opposite directions, or both moved in the same direction but for different distances (Figure 5E). As a result, initial and final inter-HJ distances often differed from each other (Figure 5F, Figure S5D).
Crossovers display resolution-associated strand processing
Crossovers frequently contained full conversions (6:2 segregation). These normally would be produced by mismatch repair (Figure 1B) but are not expected in msh2Δ strains, where standard mismatch repair is absent. As with template switching, only full conversions of two or more adjacent markers were tabulated, since single-marker full conversion can be produced by MSH2-independent short-patch repair (Coic et al., 2000; Crown et al., 2014; Fleck et al., 1999). Full conversion was prominent among crossovers (19/55 crossovers, Figure 6A, Figure S6A, File S1.11), rare among noncrossovers (1/32; Figure S6A), and was associated with potential HJ resolution sites (Figure 6A, File S1.11, File S2). This might be expected if removal and resynthesis of one heteroduplex strand initiated at HJ resolution-associated nicks (Figure 6; Kulkarni et al., 2020; Marsolier-Kergoat et al., 2018). Another six tetrads had full conversion of a single marker at a resolution point, and thus are potential candidates for resolution-associated conversion (Figure S6A). Full conversion also can be produced by repair of a dsDNA gap (Johnson et al., 2019; Prieler et al., 2021; Szostak et al., 1983; Figure S1D), but most 6:2 tracts had at least one end outside of the DSB region, and frequently were separated from the DSB region by markers in a parental configuration (File S1.10, Figure S6D). It is therefore unlikely that most are products of double strand gap repair (see Discussion). Seven other COs contained other signals of resolution-associated strand processing (Figure 6B, Figure S6A, File S1.11), including tetrads where symmetrical heteroduplex was converted to asymmetrical heteroduplex. Processing tracts were short (median ~507 nt for multiple marker events; Figure 6C), so it is possible that some strand processing events were not detected.
Figure 6 – Crossovers display resolution-associated strand processing.
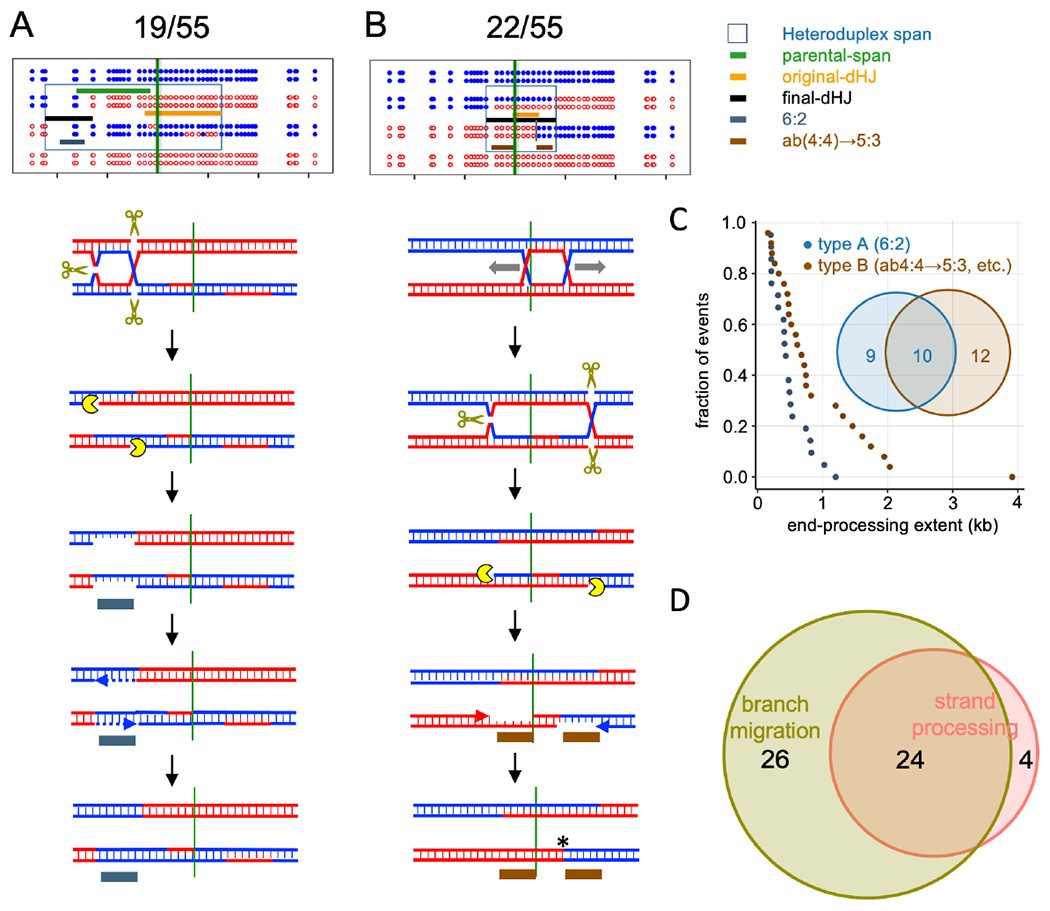
(A) CO where end-processing produces 6:2 segregation. dHJ resolution produces nicks that are converted to ssDNA gaps; repair synthesis results in full conversion (Stahl and Foss, 2010).
(B) CO where end-processing converts ab(4:4) to 5:3 segregation, resulting in three apparent CO points. Colored indicators as in Figure 5, with the following additions: grey—6:2 tract produced by end processing; brown—5:3 tract produced by end processing. The fraction of COs in each category is listed.
(C) Rank order plots of strand processing tract lengths in types A and B. Only tracts of 2 or more markers were tabulated; curves including single-marker events are in Figure S6B. Median tract lengths: type A—412 nt; type B—679 nt. Inset Venn diagram--tetrads with type A, type B, or both types of strand processing.
(D) Number of CO tetrads displaying branch migration, end processing, or both. One tetrad was too complicated to be scored and is not included here.
Taken together, about half of COs display signals of resolution-associated strand processing, and the vast majority of these also display hallmarks of branch migration (Figure 6D). The overlap between template switching, branch migration and resolution associated strand processing often produced COs with highly complex parental marker segregation patterns (File S2). In all, 54/55 COs displayed evidence for one or more of these processes, a remarkable departure from the predictions of the DSBR model.
DISCUSSION
Since the dual-mechanism model was first proposed (Allers and Lichten, 2001a), most studies of meiotic recombination have been interpreted in its context. Recent reports documenting incompatible heteroduplex patterns challenge this model (Marsolier-Kergoat et al., 2018; Martini et al.,2011; Peterson et al., 2020), but leave open the question of the degree to which such events are typical of meiotic recombination. The recombination reporter we developed, with close, regularly spaced markers and a single tightly focused DSB hotspot, has allowed characterization of individual events with unparalleled precision, to show that these “non-standard” processes are the rule. Below, we discuss the implications of these findings, including a model for meiotic recombination that accounts for features shared and different between NCOs and COs.
Similar end invasion, extension and mosaic heteroduplex patterns in NCOs and COs suggest a common mechanism
We find that NCOs and COs display similar patterns of heteroduplex, both with regards to one- versus two-sided events (Figure 3) and mosaic heteroduplex (Figure 4)—a substantial majority (70-80%) involve homolog invasion by both DSB ends (two-sided events), multiple invasions by a single DSB end (template switching), or both (Figure S4). Thus, multiple rounds of repair template invasion and disengagement are the rule, rather than the exception, during all types of meiotic recombination.
We first discuss these findings in the context of NCOs. Simple SDSA produces a single, one-sided hybrid DNA tract, but 2/3 of NCOs contained heteroduplex on both sides of the DSB, most (17/21) with heteroduplex on a single chromatid and a switch in heteroduplex phase at the DSB (Figure 3B, File S2). It is likely that these were produced by double SDSA (Figure 3G). Thus, at least 2/3 of NCOs involved invasion by both DSB ends, and this fraction is likely greater, as two-ended invasions where one end invaded a sister chromatid would have been scored as one-sided.
A substantial fraction of NCO tetrads (~40%) contained mosaic heteroduplex (Figure 4C, File S1.10). While mosaic heteroduplex can be produced by Msh2-independent repair, the absence of full conversion segments inside NMS tracts (File S1.13, S1.14) indicates that this is unlikely. We therefore conclude that mosaic heteroduplex is produced by template switching (Figure 4A). As is noted above, many template switching events might not have been detected, making it likely that the majority of NCOs undergo multiple rounds of invasion, disassembly, and re-invasion.
Other studies report that two-sided NCOs are a minority class (Gilbertson and Stahl, 1996; Hoffmann and Borts, 2005; Jessop et al., 2005; Merker et al., 2003; Porter et al., 1993), and report mosaic heteroduplex frequencies lower than those we observe (Marsolier-Kergoat et al., 2018; Martini et al., 2011). This disparity may be due to uncertainty regarding DSB locations and/or limits on the ability to detect hybrid DNA, due to low or non-uniform marker densities (c.f. Marsolier-Kergoat et al., 2018).
Remarkably, COs displayed heteroduplex patterns similar to those in NCOs. A similar fraction of COs and NCOs contained heteroduplex on only one side of the initiating DSB (Figure 3C,D), and about half of COs with two-sided heteroduplex contain all heteroduplex on a single chromatid (Figure 3D). Neither outcome is predicted by the DSBR model. In addition, COs and NCOs had similar heteroduplex tract length distributions (Figure 3E, F), a finding not expected in the dual mechanism model, in light of its different mechanisms for hybrid DNA formation.
COs also contained mosaic heteroduplex at levels and with patterns comparable to those in NCOs (Figure 4C, D). Since mosaic hybrid DNA is formed by multiple rounds of invasion and end-primed synthesis (Figure 4A), the mosaic heteroduplex found in COs must be formed by annealing between an “old” and a “new” strand. This runs counter to the DSBR model, where all hybrid DNA in dHJs, and thus in COs, contains two “old” DNA strands (Figure 1A). For the same reason, it is unlikely that NCOs, at least those that contain mosaic heteroduplex, are formed by dHJ dissolution.
Our finding that 1/3 of mosaic heteroduplex involves an initial invasion of the sister (File S1.9) challenges the view, largely based on studies of dHJs in budding yeast (Schwacha and Kleckner, 1994), that inter-sister recombination is disfavored during meiosis. While it is clear that meiotic recombination does not have the strong preference for inter-sister events seen in mitotic cells (Bzymek et al., 2010; Kadyk and Hartwell, 1992; Lichten and Haber, 1989), our data support previous conclusions that DSB ends have similar likelihoods, on a per-chromatid basis, of engaging sister and homolog chromatids (Goldfarb and Lichten, 2010). Frequent engagement of the sister chromatid during meiosis has also been proposed for mouse and fission yeast (Cole et al., 2014; Cromie et al., 2006), and thus may be a feature common to meiotic recombination in all organisms. The observed bias towards interhomolog dHJs may instead reflect the preferential disassembly of inter-sister recombination intermediates (De Muyt et al., 2012; Goldfarb and Lichten, 2010; Kaur et al., 2015; Oh et al., 2007; Sandhu et al., 2020; Tang et al., 2015).
In summary, the high frequency of mosaic heteroduplex in both COs and NCOs, combined with other features common to NCOs and COs, suggests that both NCOs and COs form hybrid DNA by a common mechanism that need not involve dHJ formation.
Branch migration is intrinsic to crossover formation
While the above-discussed processes are common to all meiotic recombination, others appear to be CO-specific. The most prominent of these, branch migration, occured in the vast majority of COs but only a small fraction of NCOs (Figure 5; File S1.1 and S1.10). About half of COs (28/55) contained both final junctions on the same side of the DSB (Figure 5A, D). These two junctions often were separated from the DSB by a region of 4:4 marker segregation, and they often flanked symmetrical heteroduplex (Figure 5, Figure S5A). These configurations can be produced by branch migration of both junctions in the same direction (Allers and Lichten, 2001b; Marsolier-Kergoat et al., 2018; Figure S1C), but are incompatible with mechanisms that involve continuous DNA synthesis, such as D-loop or bubble migration (Ferguson and Holloman, 1996; Hoffmann and Borts, 2005).
The near-ubiquity of branch migration suggests that it is an essential step in CO formation. Even the canonical DSBR model requires branch migration to allow ligation of the nicks present at cross-strand junctions (Figure S7A; Szostak et al., 1983). However, the extent of junction movement we observe (median of at least 0.66 kb; Figure S5D) is considerably greater than that required for ligation, indicating that it serves other functions.
To account for these findings, we suggest that during both NCO and CO formation, early intermediates are engaged by helicases at or soon after strand invasion (Allers and Lichten, 2001b; De Muyt et al., 2012; Lao et al., 2008; Figure 7). An intermediate that is not CO-designated undergoes helicase-mediated disassembly, followed either by additional rounds of invasion and extension, or by annealing with the other DSB end to form a NCO (De Muyt et al., 2012). CO-designation of an intermediate occurs when branched structures are stabilized by ZMM proteins, most likely the Zip2-Zip4-Spo16 heterotrimer (De Muyt et al., 2018), and helicase activity is redirected towards branch migration. This confers further stability by converting D-loops into structures with two crossed-strand junctions without a need for second end capture or ligation (Figure S7B; Lao et al., 2008; Oh et al., 2007). Branch migration also re-exposes ssDNA ends, like those liberated by D-loop disassembly in SDSA, that can participate in further rounds of strand invasion and extension before they anneal with the other DSB end and create a four-armed intermediate (Figure 7, Figure S7B–D). Thus, CO formation involves two mechanistically distinct and potentially uncoupled processes, one that forms dHJs, and the other that forms DSB-associated heteroduplex (Figure 7, Figure S7). We refer to this model, first proposed by Lao et al. (2008) with features from previous studies (Allers and Lichten, 2001b), as the disassembly/migration-annealing (D/M-A) model.
Figure 7 – Disassembly/migration-annealing model for meiotic recombination.
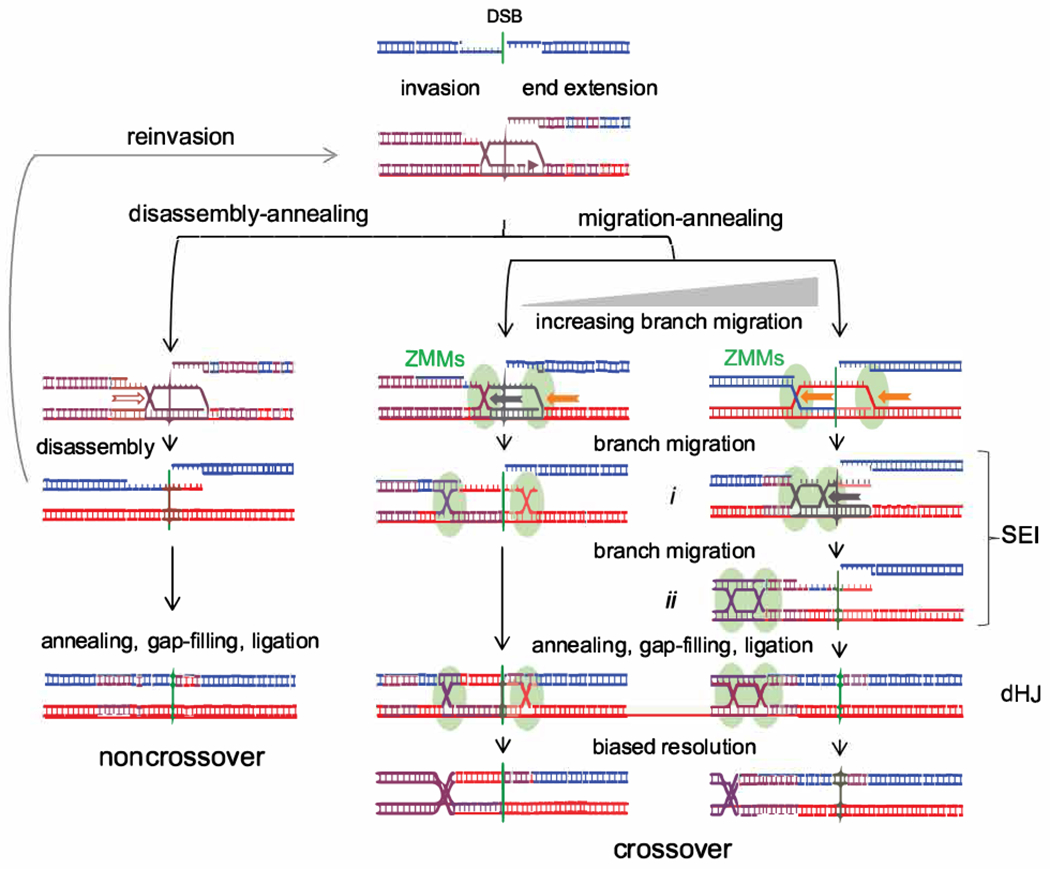
Left–helicase-mediated branch migration (hollow arrow) disassembles a D-loop, followed either by reinvasion or by end annealing to form a NCO. Center, right–CO formation. D-loop branches are captured by ZMM proteins and undergo helicase-mediated branch migration (solid arrows). Branch migration creates a three-arm single-end intermediate (SEI; Hunter and Kleckner, 2001) with a topologically closed bubble (i) or a dHJ (ii). Migration is drawn as unidirectional but may well be reversible. Annealing with the other DSB end and further processing produces a four-armed dHJ that is resolved as a CO. End reinvasion before annealing is also possible but is not drawn here. Center–limited migration can produce a DSBR-like intermediate. Migration after annealing is also possible (Figure S7A), as are different hybrid DNA configurations relative to the CO and DSB (Figure S7B). This model incorporates features of previous models (Allers and Lichten, 2001a; b; Lao et al., 2008).
While current data are difficult to reconcile with the dual mechanism model, the D/M-A model accommodates the following:
Because both NCOs and COs involve annealing between an “old” strand exposed by resection and a “new” strand formed by end-primed synthesis, NCOs and COs should display similar patterns of heteroduplex (c.f. Figures 3, 4), and mutants that affect heteroduplex tract lengths should affect NCOs and COs similarly (c.f. Duroc et al., 2017; Vernekar et al., 2021).
The three-arm intermediates formed by branch migration (Figure 7i and ii) correspond to an observed structure, called an SEI (Hunter and Kleckner, 2001). SEIs are as ZMM-dependent as four-arm dHJs (Börner et al., 2004), and the two intermediates accumulate to similar extents in resolution-defective ndt80 mutants (Shinohara et al., 2008).
The suggestion that NCO formation and the SEI to dHJ transition occur by annealing (Figure 7) is consistent with the strong reduction in NCOs and in four-arm dHJs, but not in SEIs, seen strand annealing-defective rad52Δ mutants (Lao et al., 2008).
dHJ migration leaves sequences between initial and final dHJ positions in a parental configuration (Figure 7), consistent with previous findings that ~1/4 to ~1/3 of COs display parental marker segregation between the CO and the initiating DSB (Allers and Lichten, 2001b; Jessop et al., 2005).
Spontaneous dHJ migration is expected to be slow in vivo, especially in the presence of mismatches (Panyutin and Hsieh, 1993; 1994). The finding that branch migration can cover substantial distances (Figure 5) suggests that it is actively driven. Two leading candidates for this function are the conserved meiosis-specific Mer3/Hfm1 helicase, a ZMM protein that is required for normal dHJ and CO formation (Börner et al., 2004; Mercier et al., 2005; Pezza et al., 2006; Wang et al., 2009), and the Bloom helicase (Sgs1 in budding yeast), part of a complex with Top3 and Rmi1. Sgs1 is best known for its anti-CO activity (De Muyt et al., 2012; Ira et al., 2003) but also is required for ZMM-dependent COs (De Muyt et al., 2012; Zakharyevich et al., 2012). It is possible that Sgs1, perhaps in a post-translationally modified form (Bhagwat et al., 2021; Grigaitis et al., 2020; Wild et al., 2019), can drive branch migration during meiosis. Other helicases (Mph1, Srs2 and Pif1) participate in certain aspects of meiotic recombination in budding yeast, but mutants lacking each of these helicases show normal or near-normal CO levels (Hunt et al., 2019; Sandhu et al., 2020; Vernekar et al., 2021).
Intermediates with two cross-strand junctions, including dHJs, are topologically constrained (Kaur et al., 2015), and movement of one junction will drive similar movement of the other, thus preserving the original inter-junction distance. Since the two junctions appear to have moved differently in most COs (Figure 5E, F), relief of topological constraints, either by topoisomerase activity or by junction-nicking during resolution, would be required. In addition, distances between final dHJ positions inferred from strand switch locations in COs (median of 0.5-0.7 kb, up to ~4kb; Figure 5D, F; Figure S5A) are for the most part greater than inter-junction distances (~250 nt) in meiotic dHJs detected by electron microscopy (Bell and Byers, 1983; Oh et al., 2007; Oh et al., 2008). These observations, which remain to be accounted for, may reflect junction movement during DNA isolation and/or sample preparation in EM studies, or may reflect the possibility that, if the two HJs are not resolved at the same time, one junction can move after the other is resolved.
It is also worth noting that DSB-induced mitotic recombination in budding yeast has several features shared with those reported for meiotic recombination, including a mix of one-sided and two-sided events, frequent template-switching, and conversion tracts similar in length to those seen in meiosis (Hum and Jinks-Robertson, 2017; Hum and Jinks-Robertson, 2018; Hum and Jinks-Robertson, 2019; Mitchel et al., 2010). However, products of branch migration, seen in majority of meiotic COs, are present only in a minority of mitotic COs (Hum and Jinks-Robertson, 2018), consistent with the suggestion that meiosis-specific protein complexes and processes create a context that promotes active branch migration.
In summary, the finding that branch migration is near-ubiquitous during meiotic CO formation prompts us to suggest that helicase-driven branch migration is a central feature of meiotic dHJ formation. In the future, it will be of interest to test this model by identifying mutants and DNA sequence features that affect branch migration and determining their impact on meiotic recombination.
Crossovers frequently undergo resolution-associated strand processing.
COs also displayed frequent full conversion of multiple markers (Figure 6). Since the strains used here are MMR-deficient, it is likely that this involves removal and resynthesis of DNA strands without regards to their heteroduplex content. Such processes include the repair of gaps formed by two nearby DSBs (Johnson et al., 2019; Prieler et al., 2021), “short patch” repair (Coic et al., 2000; Crown et al., 2014; Fleck et al., 1999) initiating at spontaneous DNA lesions, and strand removal and resynthesis at nicks formed by HJ resolution (Cannavo et al., 2020; Kulkarni et al., 2020; Marsolier-Kergoat et al., 2018). Both gap-repair and short-patch repair should occur at similar levels in NCOs and COs, but the events we observed were highly skewed toward COs, making it likely that they involve dHJ resolution by the MutLγ protein complex. Consistent with this, CO-associated full conversions of 2 or more markers are strongly reduced in mlh1Δ, mlh3Δ and exo1Δ mutants (Marsolier-Kergoat et al., 2018; see also Getz et al., 2008).
We envision three ways this might occur. Strand removal might result from multiple rounds of PCNA-directed strand-specific nicking by Mlh1-Mlh3 (Kulkarni et al., 2020), by synthesis and strand-displacement by DNA polymerase recruited to resolution sites (“nick-translation”; Marsolier-Kergoat et al., 2018), or by the activity of Exol, which is associated with MutLγ. It will be of interest, in the future, to determine the activities responsible for the resolution-associated strand processing.
Recent studies suggest that a substantial fraction (~20%) of DSBs involve multiple cuts that form gaps whose repair could result in 6:2 marker segregation (Johnson et al., 2021; Prieler et al., 2021). Consistent with this, we find that 11% of COs and 12% of NCOs contain full conversions in or adjacent to the ~300 nt DSB region (File S1.1). These are a minor fraction (~11 %) of total full conversions among COs, indicating that most of the 6:2 marker segregations we observed are not due to gap-filling.
Concluding remarks
We have shown that the NCOs and COs that form during budding yeast meiosis display similar patterns of break repair-associated heteroduplex, while COs, and not NCOs, frequently display hallmarks of HJ branch migration and resolution-associated strand processing. Based on these findings, we have proposed the disassembly/migration-annealing model for meiotic recombination, where most hybrid DNA in both COs and NCOs is formed by processes akin to SDSA, and where branch migration is an integral step in CO formation. This model can account for many features of meiotic recombination in budding yeast and can also account for features of previous genetic studies in fungi (Fincham, 1974; Hamza et al., 1987; Rossignol et al., 1984; Theivendirarajah and Whitehouse, 1983). The extent to which it accurately describes events in other organisms remains to be determined. While data from other organisms are limited, studies of mouse meiosis highlight indicate differences with budding yeast, including substantially shorter NMS tracts, especially among NCOs, different NMS tract spectra in NCOs and COs, and a notable deficit of CO products with heteroduplex on both sides of the initiating DSB (Cole et al., 2014; Cole et al., 2010; Peterson et al., 2020). Despite these differences, features of mammalian data, including COs and NCO tracts remote from the initiating DSB (Cole et al., 2014; Cole et al., 2010; Peterson et al., 2020), are reminiscent of the outcomes of template switching and branch migration seen in budding yeast. It will be of considerable interest, as more high-resolution data become available, to compare parental strand contributions to meiotic recombinants in different organisms and to determine which recombination processes are shared, and which are different.
Limitations of study
The defined locus used allowed us to map recombination events at high resolution and to gain considerable insight into recombination mechanisms. It should be kept in mind that this is a single locus, and it remains to be determined if the same mechanisms apply throughout the budding yeast genome. Doing so may be difficult, since the close spacing of DSB hotspots in many parts of the yeast genome complicates analysis. While we have shown that the presence of multiple heterozygous polymorphisms does not substantially alter recombination in msh2Δ strains, we cannot exclude subtle effects due to unknown Msh2-indedpendent mechanisms. This study characterized the end-products of recombination. Therefore, both the nature and order of intermediate steps are hypothetical, and confirmation will require molecular analyses of recombination intermediates. In addition, our current conclusions are limited by the number of recombinants analyzed and the density of markers used. The current data have allowed deduction of general mechanistic principles, but as always, the characterization of additional recombinants, especially in recombination mutants, will lead to further insights.
STAR METHODS TEXT
RESOURCE AVAILABILITY
Lead contact:
Further information and requests for resources should be directed to and will be fulfilled by the lead contact, Michael Lichten (michael.lichten@nih.gov)
Materials availability
All unique reagents generated in this study are available from the Lead Contact with a completed Material Transfer Agreement.
Data and code availability
Sequencing data are deposited at the Sequence Read Archive (https://www.ncbi.nlm.nih.gov/sra) under BioProject ID: PRJNA721091.
Underlying data for all figures are in File S1, a Microsoft Excel workbook. References to File S1 in the text use the following format: File S1.x, where x is the worksheet number. Plots of individual tetrad genotypes are in File S2. Sequences of the wild-type and SNP-containing recombination intervals are in Files S3 and S4, respectively. Full Southern blot images are deposited at Mendeley Data, doi: 10.17632/j2jpj3p29f.1
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
All strains are SK1 derivatives (Kane and Roth, 1974); genotypes are listed in Supplementary Table 1.15. Drug resistance-marked deletions and insertions were created as described (Longtine et al., 1998). The previously described his4::URA3-ARG4 hotspot (Parent 1 in Figure 2A; Jessop et al., 2005) was modified to form his4::URA3-ARG4-SNPs (Parent 2 in Figure 1A) using synthetic DNA fragments and Gibson assembly. Sixty percent of the markers are G to C changes (File S1.2), and all preserve URA3 and ARG4 coding information. Sequences contained in the insert locus were deleted from the endogenous URA3 and ARG4 loci. Ochre mutations (ura3-K54oc and arg4-E93oc) were created by site-directed-mutagenesis. All constructs were confirmed by sequencing PCR amplicons. Sequences of the URA3-ARG4 and URA3-ARG4-SNPs inserts are in File S3 and File S4, respectively.
METHOD DETAILS
Sporulation, DNA preparation, and tetrad dissection
Sporulation was as described (Kaur et al., 2018), with the addition of quick mating to avoid accumulation of recessive lethal mutations in msh2Δ diploids. For liquid sporulation, 5 colonies (56h at 30°C on YPD agar plates; 2% glucose, 2% Bacto Peptone, 1% Bacto Yeast Extract, 2% Bacto agar) each from MATa and MATα parents were mixed in 20 μL of YPD broth, spotted on a YPD plate and incubated at 30°C for 7h. The patch was resuspended in 4 mL YPD broth containing 300μg/mL hygromycin B (Gibco) and aerated at 30°C for 12 h, cells were pelleted and resuspended in 4 mL YPD broth containing 100 μg/mL nourseothricin sulfate (Neta Scientific) and aerated at 30°C for 12 h. This culture was then used to initiate sporulation as described in detail by Kaur et al. (2018). Briefly, an inoculum in pre-sporulation medium (1% Bacto™ peptone, 0.5% Bacto™y east extract, 1% potassium acetate, 0.17% Bacto™ yeast nitrogen base without amino acids, 1% ammonium sulfate, 0.5% potassium hydrogen phthalate, adjusted to pH 5.5 with potassium hydroxide) was grown overnight with vigorous aeration to near-saturation. Cells were washed in 1% potassium acetate, transferred to sporulation medium (1% potassium acetate, 0.001% polypropylene glycol 2000, supplemented as required by the specific auxotrophies of the strain being used) and incubated at 30°C with vigorous aeration. DNA preparation was as described in detail by Ahuja and Börner (2011) but without crosslinking. Briefly, spheroplasted cells were lysed with SDS and proteins were digested with proteinase K, followed by removal of remaining proteins and SDS by precipitation with 1.2 M potassium acetate, followed by ethanol precipitation of nucleic acids from the resulting supernatant. Samples were treated with RNase A overnight at 4°C, followed by phenol-chloroform-isoamyl alcohol extraction, ethanol precipitation and resuspension in 10 mM Tris-HCl 1 mM EDTA.
Most tetrads (n=124) used for sequencing were from a 24 hr liquid sporulation culture and were dissected onto YPD agar plates. The rest were from quick matings as above, but cells were directly patched from YPD agar mating plates to 1% potassium acetate, 2% Bacto agar plates and were dissected after at least 2 days incubation at 30°C. For analysis of strains heterozygous for ura3-K54oc and arg4-E93oc alleles, tetrads were dissected onto YPD plates but with 4% glucose and 2.2% Bacto agar, which allowed more reliable detection of sectored colonies after replica plating.
Internal size standards
Size standards for DSB mapping were created by digesting a 0h sample from strain MJL3901 (File S1.16) as follows. Marker 1 – 2μg samples were digested with StuI, EcoRV, XbaI, NdeI, PstI, BamHI, or SacI in independent tubes. After phenol:chloroform extraction and isopropanol precipitation, equal amounts of each digest were mixed and digested with SpeI-HF, with 500 ng loaded per lane. Marker 2 – As above, but independent NdeI, DdeI, PstI and PvuII digest were mixed and digested with XcmI and HaeIII before loading.
Southern blots of agarose gels
For low resolution DSB gels (Figure 2B), 500 ng of DNA digested with SpeI-HF and XmnI was displayed on a 0.6% agarose gel along with 500ng of Marker-1. For molecular analysis of crossovers (Figure 2E) 500ng of XmnI-digested DNA was displayed on a 0.6% agarose gel. Gels were transferred to Amersham Hybond-XL nylon membrane using the Vacugene XL system, as described (Oh et al., 2009). Probes (listed in File S1.16) were prepared using a Roche Hi-Prime kit using manufacturer’s instructions.
Polyacrylamide gels
28 cm x 16cm gels were poured as per manufacturer’s recommendation (Urea Gel System – National Diagnostics) and were prerun 20 min at 40W to bring the gel to 55°C. Samples contained 750ng DNA in 45% formamide, 4mM EDTA, 0.5mM Orange-G and were heated for 5 minutes at 95°C before loading. Marker-2 was used as a size standard. Gels were transferred to membranes using a BioRad Semi-dry transfer cell at 400mA for 1h, which were then treated with 0.5M NaOH, 1M NaCl for 10 mins, rinsed with 1X SSC, and UV-crosslinked at 120,000 μJoules/cm2. Radioactive probes were prepared by PCR amplification. The template for probe 27 was amplified from yeast genomic DNA, while probe 28 sequence was synthesized to include M13 forward and M13 reverse primer sequences and was labeled by amplification with those primers.
Southern hybridization
Blots were probed as described (Allers and Lichten, 2001b), except that blots of polyacrylamide gels were hybridized with probe at 58°C to account for shorter probes. Blots were washed sequentially with – 2X SSPE, 0.1% SDS; 0.2X SSPE, 0.5% SDS and 0.2X SSPE, 0.5% SDS respectively. Following hybridization, membranes were exposed to phosphor-imager-screens and scanned using the Typhoon phosphoimager.
DNA preparation and sequencing
Entire spore colonies were inoculated into wells of deep-well 96 well plates (USA Scientific, 1896-2110) with 500 μL of YPD broth/well and were incubated overnight at 30° with aeration. 400 μl of each culture was used to prepare DNA, and the remainder was archived at −80°C. Cells were centrifuged at 3220xG for 2 min, the supernatant was removed, and cells were resuspended in 0.2 ml of 10mM NaPO4 pH 7.2, 10 mM EDTA, 0.1% ß-mercaptoethanol, 100 μg/ml Zymolyase T100 (MP Biomedicals). Cells were incubated 30 min at 37°C with tap-mixing every 5 min. Spheroplasts were centrifuged 10 min, 300 x g, the supernatant was removed, and spheroplasts were resuspended in 180 μl ATL buffer (Qiagen) with 0.5 mg/mL proteinase-K (Roche) and incubated at 65°C for 30 min, followed by addition of 5 μl of 4 μg/mL DNase-free RNase (Roche). Samples were loaded onto 96-well Qiacube-HT® columns and DNA was purified using a Blood & Tissue kit (Qiagen). After quantification, 1ng of DNA was used to PCR amplify the entire 6.865 kb recombination interval using the following primers: ACGGCACCACTATAAACCCG and GTGGGCTAAAGAACGCGAAC, and Q5 DNA polymerase (New England Biolabs) as recommended by the manufacturer, except that an extension time of 1 min/kb was used to avoid partial synthesis and priming by partial synthesis products. Most of the samples were sequenced on a MiSeq sequencer (Illumina) after barcoding with a Nextera-384 barcoding kit (Illumina), resulting in ~20 million reads/run that were up to 300 bp in length, with a typical coverage of ~300-400 unique (deduplicated) reads/nucleotide/sample. The remaining samples were amplified from spore colony DNA using barcodes from (Guo et al., 2015) attached the outer primers listed above, and the entire region was sequenced on a Pacbio RS II instrument.
For Illumina sequencing, individual spore DNA sequences were aligned to the recombination interval sequence (Li and Durbin, 2010) followed by extraction of read base composition at each marker position, initially using a custom script and later using the sam2tsv function of jvarkit (Lindenbaum, 2016). For reads with more than one base at a marker position (i.e from a colony with heteroduplex), strand phase was determined manually from overlapping but non-conflicting reads and SNP abundance plots; in cases where phase was ambiguous, interval DNA from a single colony struck from archived samples was sequenced, initially by Illumina and later by PacBio sequencing (File S1.3). PacBio reads were demultiplexed using the locate function of seqkit (Shen et al., 2016), sample reads were aligned using minimap2 (Li, 2018), base composition per nucleotide was determined using the sam2tsv function of the jvarkit as above, and phase was determined manually from individual full-length reads.
QUANTIFICATION AND STATISTICAL ANALYSIS
Radioactive signal on Southern blots was quantified by phosphorimaging (Typhoon FLA-9500) and analyzed using Image Gauge v4.22 (FujiFilm). Statistical and other data analysis was performed using Graphpad Prism and R (https://www.R-project.org/). Plots of individual tetrad genotypes were constructed using the R packages tidyverse and ggplot2 (Wickham, 2009; Wickham et al., 2019). For tetrad analysis, map distances were calculated as described (Perkins, 1949), implemented at https://elizabethhousworth.com/StahlLabOnlineTools/.
Supplementary Material
File S2. HTML file with an illustration of each tetrad, related to Figures 2–6.
Individual tetrads can be displayed using the drop-down menu in the upper left hand corner. COs are listed first, followed by NCOs and then complex tetrads.
Supplemental Figures. Figures S1–S7.
File S1. Microsoft Excel workbook containing data underlying analysis, related to Figures 2–6.
- Cover sheet. Summary of the analysis of all tetrads. See also File S2 for a graphic depiction of each tetrad.
- Marker locations. Sequence and location of markers used, along with non-Mendelian segregation numbers and frequencies at each marker location.
- Sequencing data. Strand haplotypes for each marker location derived from sequencing.
- Fig2CD-ochres. Non-Mendelian segregation and crossovers for ura3 and arg4 point mutants in diploids where polymorphisms are present or absent.
- Fig2D-genetics. Tetrad data underlying map distances in the natMX-hygMX interval.
- Fig3CDEF tracts DSBs. Length and location of heteroduplex tracts and half-tracts.
- Fig3EF tract stats. Fit of heteroduplex tract and half tract lengths to an exponential decay model.
- Fig4 template switching. Length, location and lemplate used for each segment in mosaic heteroduplex.
- Fig5 dHJs. Location and dimensions of total heteroduplex tracts, inferred original and final dHJs, and parental segments between final dHJ and DSB.
- Fig6 SP description. Location dimensions and type of strand processing tracts.
- Point events. Single marker conversion events.
- Fig6 suppl segs corrected. NMS segments with point events removed.
- Yeast strains.
- Oligos and probes.
File S3. Genbank-format annotated sequence of the wild-type recombination interval, related to STAR Methods.
File S4. Genbank-format annotated sequence of the SNP-containing recombination interval, related to STAR Methods.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Bacterial and virus strains | ||
| Biological samples | ||
| Chemicals, peptides, and recombinant proteins | ||
| Proteinase K | Sigma Aldrich | 3115879001 |
| RNAse A | Sigma Aldrich | 11119915001 |
| Zymolyase | MP Biomedicals | 08320932 |
| XmnI | New England Biolabs | R0194L |
| SpeI-HF | New England Biolabs | R3133L |
| XcmI | New England Biolabs | R0533L |
| HaeIII | New England Biolabs | R0108L |
| NdeI | New England Biolabs | R0111S |
| DdeI | New England Biolabs | R0175S |
| PstI | New England Biolabs | R0140S |
| PvuII | New England Biolabs | R0151S |
| StuI | New England Biolabs | R0187S |
| BamHI | New England Biolabs | R3136S |
| SacI | New England Biolabs | R0156S |
| EcoRV | New England Biolabs | R0195S |
| Seakem GTG Agarose | Fisher Scientific | BMA50070 |
| Amersham Hybond-N+ nylon membrane | Fisher Scientific | 45000927 |
| Amersham Hybond-XL nylon membrane | Fisher Scientific | 45001148 |
| [a-32P]dCTP | Perkin Elmer | BLU013Z250UC |
| DNA, MB-grade from fish sperm | Sigma Aldrich | 11467140001 |
| Taq DNA Polymerase | NEB | M0273S |
| Q5® High-Fidelity DNA Polymerase | NEB | M0491L |
| Formamide | Sigma Aldrich | F7503-100ML |
| Urea Gel system | National Diagnostics | EC-833 |
| 20X-SSPE | Corning | 46-021-CM |
| 20X-SSC | KD medical | RFG-3240 |
| Nonfat dry milk | Nestle | Carnation |
| SDS | Sigma | 74255 |
| Deoxynucleotide (dNTP) Solution Set | New England Biolabs | N0446S |
| Isopropanol | Sigma-Aldrich | 190764 |
| Pottasium acetate | Sigma-Aldrich | 236497 |
| Glucose | Sigma-Aldrich | G5767 |
| Bacto Peptone | Becton, Dickinson | 211677 |
| Bacto Yeast Extract | Becton, Dickinson | 212750 |
| Bacto Agar | Becton, Dickinson | 214010 |
| Polypropylene glycol 2000 | Sigma Aldrich | 202339 |
| Nourseothricin sulfate | Neta Scientific | CAYM-16227 |
| Hygromycin B | Gibco | 10687010 |
| Critical commercial assays | ||
| Agencourt beads | Beckman Coulter Inc | A63881 |
| Qubit | Fisher Scientfic | Q32853 |
| HiPrime | Sigma Aldrich | 11585592001 |
| NexteraXT | Illumina | FC-131-2001, FC-131-2002, FC-131-2003, FC-131-2004 |
| SMRTbell template prep kit 1.0 | Pacific Biosciences | 100-259-100 |
| Sequel binding kit 3.0 | Pacific Biosciences | 101-500-400 |
| QIAamp 96 DNA QIAcube HT Kit (5) | QIAGEN | 51331 |
| Deposited data | ||
| Sequencing data | Sequence Read Archive | PRJNA721091 |
| Full Southern blot images | Mendeley Data | doi:10.17632/j2jpj3p29f.1 |
| Experimental models: Cell lines | ||
| Experimental models: Organisms/strains | ||
| Saccharomyces cerevisiae | This study | Listed in File S1 sheet 15 |
| Oligonucleotides | ||
| Primers for probe preparation | IDTdna | Listed in File S1 sheet 16 |
| Primers for amplification of hotspot for sequencing | IDTdna | Listed in File S1 sheet 16 |
| Recombinant DNA | ||
| Software and algorithms | ||
| Image Gauge v4.22 | Fujifilm | N/A |
| R (version 3.6.2) | R Project | https://www.r-project.org/ |
| Prism v9.1.0 | GraphPad Software | |
| Other | ||
| Stratalinker crosslinker | Stratagene | 1800 |
| Typhoon phosphorimager | GE | FLA 9500 |
| VacuGene XL gel transfer apparatus | GE | N/A |
| Trans-Blot® SD Semi-Dry Transfer Cell | BioRad | 1703940 |
| Gel electrophoresis thermometer | C.B.S. Scientific | EGT-100-10 |
| PAGE gel apparatus | C.B.S. Scientific | ASG-250 |
| Sub Cell GT Cell Tanks and Lids | BioRad | 1704401 |
Highlights.
Meiotic recombination often involves template switching between sister and homolog
Crossovers and noncrossovers both involve synthesis-dependent strand annealing
Branch migration drives crossover-specific double Holliday junction formation
Holliday junction resolution is associated with strand removal and resynthesis
ACKNOWLEDGEMENTS
We thank Eric Alani, Julia Cooper, Nancy Hollingsworth, Neil Hunter, Sue Jinks-Robertson, Alexander Kelly, Bertrand Llorente and Matthew Neale for discussions and comments on this manuscript, and the Center for Cancer Research Sequencing Facility for high-throughput sequencing. This work used resources at the NIH HPC Biowulf cluster (http://hpc.nih.gov) and is supported by the Intramural Research Program of the NIH through the Center for Cancer Research at the National Cancer Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Ahuja JS, and Börner GV (2011). Analysis of meiotic recombination intermediates by two-dimensional gel electrophoresis. Methods Mol. Biol 745, 99–116. 10.1007/978-1-61779-129-1_7. [DOI] [PubMed] [Google Scholar]
- Ahuja JS, Sandhu R, Mainpal R, Lawson C, Henley H, Hunt PA, Yanowitz JL, and Börner GV (2017). Control of meiotic pairing and recombination by chromosomally tethered 26S proteasome. Science 355, 408–411. 10.1126/science.aaf4778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allers T, and Lichten M (2001a). Differential timing and control of noncrossover and crossover recombination during meiosis. Cell 106, 47–57. 10.1016/s0092-8674(01)00416-0. [DOI] [PubMed] [Google Scholar]
- Allers T, and Lichten M (2001b). Intermediates of yeast meiotic recombination contain heteroduplex DNA. Mol. Cell 8, 225–231. 10.1016/s1097-2765(01)00280-5. [DOI] [PubMed] [Google Scholar]
- Argueso JL, Wanat J, Gemici Z, and Alani E (2004). Competing crossover pathways act during meiosis in Saccharomyces cerevisiae. Genetics 168, 1805–1816. 10.1534/genetics.104.032912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell LR, and Byers B (1983). Homologous association of chromosomal DNA during yeast meiosis. Cold Spring Harbor Symp. Quant. Biol 47 Pt 2, 829–840. 10.1101/sqb.1983.047.01.095 [DOI] [PubMed] [Google Scholar]
- Bhagwat NR, Owens SN, Ito M, Boinapalli JV, Poa P, Ditzel A, Kopparapu S, Mahalawat M, Davies OR, Collins SR, et al. (2021). SUMO is a pervasive regulator of meiosis. eLife 10, e57720. 10.7554/eLife.57720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Börner GV, Kleckner N, and Hunter N (2004). Crossover/noncrossover differentiation, synaptonemal complex formation, and regulatory surveillance at the leptotene/zygotene transition of meiosis. Cell 117, 29–45. 10.1016/s0092-8674(04)00292-2. [DOI] [PubMed] [Google Scholar]
- Borts RH, and Haber JE (1987). Meiotic recombination in yeast: alteration by multiple heterozygosities. Science 237, 1459–1465. 10.1126/science.2820060. [DOI] [PubMed] [Google Scholar]
- Buonomo SB, Clyne RK, Fuchs J, Loidl J, Uhlmann F, and Nasmyth K (2000). Disjunction of homologous chromosomes in meiosis I depends on proteolytic cleavage of the meiotic cohesin Rec8 by separin. Cell 103, 387–398. 10.1016/s0092-8674(00)00131-8. [DOI] [PubMed] [Google Scholar]
- Bzymek M, Thayer NH, Oh SD, Kleckner N, and Hunter N (2010). Double Holliday junctions are intermediates of DNA break repair. Nature 464, 937–941. 10.1038/nature08868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannavo E, Sanchez A, Anand R, Ranjha L, Hugener J, Adam C, Acharya A, Weyland N, Aran-Guiu X, Charbonnier J-B, et al. (2020). Regulation of the MLH1-MLH3 endonuclease in meiosis. Nature 586, 618–622. 10.1038/s41586-020-2592-2. [DOI] [PubMed] [Google Scholar]
- Cheng C-H, Lo Y-H, Liang S-S, Ti S-C, Lin F-M, Yeh C-H, Huang H-Y, and Wang T-F (2006). SUMO modifications control assembly of synaptonemal complex and polycomplex in meiosis of Saccharomyces cerevisiae. Genes Dev. 20, 2067–2081. 10.1101/gad.1430406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coic E, Gluck L, and Fabre F (2000). Evidence for short-patch mismatch repair in Saccharomyces cerevisiae. EMBO J 19, 3408–3417. 10.1093/emboj/19.13.3408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole F, Baudat F, Grey C, Keeney S, de Massy B, and Jasin M (2014). Mouse tetrad analysis provides insights into recombination mechanisms and hotspot evolutionary dynamics. Nat. Genet 46, 1072–1080. 10.1038/ng.3068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole F, Keeney S, and Jasin M (2010). Comprehensive, fine-scale dissection of homologous recombination outcomes at a hot spot in mouse meiosis. Mol. Cell 39, 700–710. 10.1016/j.molcel.2010.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper TJ, Crawford MR, Hunt LJ, Marsolier-Kergoat M-C, Llorente B, and Neale MJ (2018). Mismatch repair impedes meiotic crossover interference. bioRxiv 480418. 10.1101/480418. [DOI] [Google Scholar]
- Cromie GA, Hyppa RW, Taylor AF, Zakharyevich K, Hunter N, and Smith GR (2006). Single Holliday junctions are intermediates of meiotic recombination. Cell 127, 1167–1178. 10.1016/j.cell.2006.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crown KN, McMahan S, and Sekelsky J (2014). Eliminating both canonical and short-patch mismatch repair in Drosophila melanogaster suggests a new meiotic recombination model. PLoS Genet. 10, e1004583. 10.1371/journal.pgen.1004583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dayani Y, Simchen G, and Lichten M (2011). Meiotic recombination intermediates are resolved with minimal crossover formation during return-to-growth, an analogue of the mitotic cell cycle. PLoS Genet. 7, e1002083. 10.1371/journal.pgen.1002083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Massy B (2003). Distribution of meiotic recombination sites. Trends Genet 19, 514–522. 10.1016/S0168-9525(03)00201-4. [DOI] [PubMed] [Google Scholar]
- de Massy B (2013). Initiation of meiotic recombination: how and where? Conservation and specificities among eukaryotes. Annu. Rev. Genet 47, 563–599. 10.1146/annurev-genet-110711-155423. [DOI] [PubMed] [Google Scholar]
- De Muyt A, Jessop L, Kolar E, Sourirajan A, Chen J, Dayani Y, and Lichten M (2012). BLM helicase ortholog Sgs1 is a central regulator of meiotic recombination intermediate metabolism. Mol. Cell 46, 43–53. 10.1016/j.molcel.2012.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Muyt A, Pyatnitskaya A, Andrani J, Ranjha L, Ramus C, Laureau R, Fernandez-Vega A, Holoch D, Girard E, Govin J, et al. (2018). A meiotic XPF-ERCC1-like complex recognizes joint molecule recombination intermediates to promote crossover formation. Genes Dev. 32, 283–296. 10.1101/gad.308510.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duroc Y, Kumar R, Ranjha L, Adam C, Gurois R, Md Muntaz K, Marsolier-Kergoat MC, Dingli F, Laureau R, Loew D, et al. (2017). Concerted action of the MutLβ heterodimer and Mer3 helicase regulates the global extent of meiotic gene conversion. Elife 6, e21900. 10.7554/eLife.21900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson DO, and Holloman WK (1996). Recombinational repair of gaps in DNA is asymmetric in Ustilago maydis and can be explained by a migrating D-loop model. Proc. Natl. Acad. Sci. U.S.A 93, 5419–5424. 10.1073/pnas.93.11.5419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fincham J (1974). Negative interference and the use of flanking markers in fine-structure mapping in fungi. Heredity 33, 116–121. 10.1038/hdy.1974.75 [DOI] [Google Scholar]
- Fleck O, Lehmann E, Schr P, and Kohli J (1999). Involvement of nucleotide-excision repair in msh2 pms1-independent mismatch repair. Nat. Genet 21, 314–317. 10.1038/6838. [DOI] [PubMed] [Google Scholar]
- Getz TJ, Banse SA, Young LS, Banse AV, Swanson J, Wang GM, Browne BL, Foss HM, and Stahl FW (2008). Reduced mismatch repair of heteroduplexes reveals “non”-interfering crossing over in wild-type Saccharomyces cerevisiae. Genetics 178, 1251–1269. 10.1534/genetics.106.067603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbertson LA, and Stahl FW (1996). A test of the double-strand break repair model for meiotic recombination in Saccharomyces cerevisiae. Genetics 144, 27–41. 10.1093/genetics/144.1.27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldfarb T, and Lichten M (2010). Frequent and efficient use of the sister chromatid for DNA double-strand break repair during budding yeast meiosis. PLoS Biol 8, e1000520. 10.1371/journal.pbio.1000520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigaitis R, Ranjha L, Wild P, Kasaciunaite K, Ceppi I, Kissling V, Henggeler A, Susperregui A, Peter M, Seidel R, et al. (2020). Phosphorylation of the RecQ helicase Sgs1/BLM controls its DNA unwinding activity during meiosis and mitosis. Dev. Cell 53, 706–723 e705. 10.1016/j.devcel.2020.05.016. [DOI] [PubMed] [Google Scholar]
- Guo X, Lehner K, O’Connell K, Zhang J, Dave SS, and Jinks-Robertson S (2015). SMRT sequencing for parallel analysis of multiple targets and accurate SNP phasing. G3 5, 2801–2808. 10.1534/g3.115.023317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamza H, Haedens V, Mekki-Berrada A, and Rossignol JL (1981). Hybrid DNA formation during meiotic recombination. Proc. Natl. Acad. Sci. U. S. A 78, 7648–7651. 10.1073/pnas.78.12.7648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamza H, Nicolas A, and Rossignol J (1987). Large heterologies impose their gene conversion pattern onto closely linked point mutations. Genetics 116, 45–53. 10.1093/genetics/116.1.45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassold T, and Hunt P (2001). To err (meiotically) is human: the genesis of human aneuploidy. Nat. Rev. Genet 2, 280–291. 10.1038/35066065. [DOI] [PubMed] [Google Scholar]
- Hoffmann ER, and Borts RH (2005). Trans events associated with crossovers are revealed in the absence of mismatch repair genes in Saccharomyces cerevisiae. Genetics 169, 1305–1310. 10.1534/genetics.104.033407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holliday R (1964). A mechanism for gene conversion in fungi. Genet. Res 5, 282–304. 10.1017/S0016672300001233. [DOI] [PubMed] [Google Scholar]
- Holloway JK, Booth J, Edelmann W, McGowan CH, and Cohen PE (2008). MUS81 generates a subset of MLH1-MLH3-independent crossovers in mammalian meiosis. PLoS Genet. 4, e1000186. 10.1371/journal.pgen.1000186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hum YF, and Jinks-Robertson S (2017). Mitotic gene conversion tracts associated with repair of a defined double-strand break in Saccharomyces cerevisiae. Genetics 207, 115–128. 10.1534/genetics.117.300057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hum YF, and Jinks-Robertson S (2018). DNA strand-exchange patterns associated with double-strand break-induced and spontaneous mitotic crossovers in Saccharomyces cerevisiae. PLoS Genet. 14, e1007302. 10.1371/journal.pgen.1007302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hum YF, and Jinks-Robertson S (2019). Mismatch recognition and subsequent processing have distinct effects on mitotic recombination intermediates and outcomes in yeast. Nucleic Acids Res 47, 4554–4568. 10.1093/nar/gkz126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt LJ, Ahmed EA, Kaur H, Ahuja JS, Hulme L, Chou T-C, Lichten M, and Goldman ASH (2019). S. cerevisiae Srs2 helicase ensures normal recombination intermediate metabolism during meiosis and prevents accumulation of Rad51 aggregates. Chromosoma 218, 249–265. 10.1007/s00412-019-00705-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter N, and Borts RH (1997). Mlh1 is unique among mismatch repair proteins in its ability to promote crossing-over during meiosis. Genes Dev. 11, 1573–1582. 10.1101/gad.11.12.1573. [DOI] [PubMed] [Google Scholar]
- Hunter N, and Kleckner N (2001). The single-end invasion: an asymmetric intermediate at the double-strand break to double-Holliday junction transition of meiotic recombination. Cell 106, 59–70. 10.1016/s0092-8674(01)00430-5. [DOI] [PubMed] [Google Scholar]
- Ira G, Malkova A, Liberi G, Foiani M, and Haber JE (2003). Srs2 and Sgs1-Top3 suppress crossovers during double-strand break repair in yeast. Cell 115, 401–411. 10.1016/s0092-8674(03)00886-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer RR, Pluciennik A, Burdett V, and Modrich PL (2006). DNA mismatch repair: functions and mechanisms. Chem. Rev 106, 302–323. 10.1021/cr0404794. [DOI] [PubMed] [Google Scholar]
- Jessop L, Allers T, and Lichten M (2005). Infrequent co-conversion of markers flanking a meiotic recombination initiation site in Saccharomyces cerevisiae. Genetics 169, 1353–1367. 10.1534/genetics.104.036509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson D, Crawford M, Cooper T, Bouuaert CC, Keeney S, Llorente B, Garcia V, and Neale MJ (2021). Concerted cutting by Spo11 illuminates DNA break mechanisms and initiates gap repair during meiosis. Nature 594, 572–576. 10.1038/s41586-021-03389-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadyk LC, and Hartwell LH (1992). Sister chromatids are preferred over homologs as substrates for recombinational repair in Saccharomyces cerevisiae. Genetics 132, 387–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kane SM, and Roth R (1974). Carbohydrate metabolism during ascospore development in yeast. J Bacteriol. 118, 8–14. 10.1128/JB.118.1.8-14.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur H, Ahuja JS, and Lichten M (2018). Methods for controlled protein depletion to study protein function during meiosis. Methods Enzymol. 601, 331–357. 10.1016/bs.mie.2017.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur H, De Muyt A, and Lichten M (2015). Top3-Rmi1 DNA single-strand decatenase is integral to the formation and resolution of meiotic recombination intermediates. Mol. Cell 57, 583–594. 10.1016/j.molcel.2015.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keelagher RE, Cotton VE, Goldman ASH, and Borts RH (2011). Separable roles for Exonuclease I in meiotic DNA double-strand break repair. DNA Repair 10, 126–137. 10.1016/j.dnarep.2010.09.024. [DOI] [PubMed] [Google Scholar]
- Kohl KP, and Sekelsky J (2013). Meiotic and mitotic recombination in meiosis. Genetics 194, 327–334. 10.1534/genetics.113.150581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkarni DS, Owens SN, Honda M, Ito M, Yang Y, Corrigan MW, Chen L, Quan AL, and Hunter N (2020). PCNA activates the MutLγ endonuclease to promote meiotic crossing over. Nature 586, 623–627. 10.1038/s41586-020-2645-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lao JP, Oh SD, Shinohara M, Shinohara A, and Hunter N (2008). Rad52 promotes postinvasion steps of meiotic double-strand-break repair. Mol. Cell 29, 517–524. 10.1016/j.molcel.2007.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H (2018). Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics 34, 3094–3100. 10.1093/bioinformatics/bty191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, and Durbin R (2010). Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 26, 589–595. 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichten M, Goyon C, Schultes NP, Treco D, Szostak JW, Haber JE, and Nicolas A (1990). Detection of heteroduplex DNA molecules among the products of Saccharomyces cerevisiae meiosis. Proc. Natl. Acad. Sci. U.S.A 87, 7653–7657. 10.1073/pnas.87.19.7653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichten M, and Haber JE (1989). Position effects in ectopic and allelic mitotic recombination in Saccharomyces cerevisiae. Genetics 123, 261–268. 10.1093/genetics/123.2.261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindenbaum P (2016). JVarkit: java-based utilities for bioinformatics. https://github.com/lindenb/jvarkit.
- Longtine MS, McKenzie A 3rd, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, and Pringle JR (1998). Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14, 953–961. . [DOI] [PubMed] [Google Scholar]
- Lynn A, Soucek R, and Brner GV (2007). ZMM proteins during meiosis: crossover artists at work. Chromosome Res. 15, 591–605. 10.1007/s10577-007-1150-1. [DOI] [PubMed] [Google Scholar]
- Marsolier-Kergoat M-C, Khan MM, Schott J, Zhu X, and Llorente B (2018). Mechanistic view and genetic control of DNA recombination during meiosis. Mol. Cell 70, 9–20. 10.1016/j.molcel.2018.02.032. [DOI] [PubMed] [Google Scholar]
- Martini E, Borde V, Legendre M, Audic S, Regnault B, Soubigou G, Dujon B, and Llorente B (2011). Genome-wide analysis of heteroduplex DNA in mismatch repair-deficient yeast cells reveals novel properties of meiotic recombination pathways. PLoS Genet. 7, e1002305. 10.1371/journal.pgen.1002305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazina OM, Mazin AV, Nakagawa T, Kolodner RD, and Kowalczykowski SC (2004). Saccharomyces cerevisiae Mer3 helicase stimulates 3′-5′ heteroduplex extension by Rad51; implications for crossover control in meiotic recombination. Cell 117, 47–56. 10.1016/S0092-8674(04)00294-6. [DOI] [PubMed] [Google Scholar]
- McMahill MS, Sham CW, and Bishop DK (2007). Synthesis-dependent strand annealing in meiosis. PLoS Biol. 5, e299. 10.1371/journal.pbio.0050299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McVey M, Adams M, Staeva-Vieira E, and Sekelsky JJ (2004). Evidence for multiple cycles of strand invasion during repair of double-strand gaps in Drosophila. Genetics 167, 699–705. 10.1534/genetics.103.025411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercier R, Jolivet S, Vezon D, Huppe E, Chelysheva L, Giovanni M, Nogue F, Doutriaux MP, Horlow C, Grelon M, and Mezard C (2005). Two meiotic crossover classes cohabit in Arabidopsis: one is dependent on MER3, whereas the other one is not. Curr. Biol 15, 692–701. 10.1016/j.cub.2005.02.056. [DOI] [PubMed] [Google Scholar]
- Merker JD, Dominska M, and Petes TD (2003). Patterns of heteroduplex formation associated with the initiation of meiotic recombination in the yeast Saccharomyces cerevisiae. Genetics 165, 47–63. 10.1093/genetics/165.1.47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchel K, Zhang H, Welz-Voegele C, and Jinks-Robertson S (2010). Molecular structures of crossover and noncrossover intermediates during gap repair in yeast: implications for recombination. Mol. Cell 38, 211–222. 10.1016/j.molcel.2010.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nag DK, White MA, and Petes TD (1989). Palindromic sequences in heteroduplex DNA inhibit mismatch repair in yeast. Nature 340, 318–320. 10.1038/340318a0. [DOI] [PubMed] [Google Scholar]
- Nasmyth KA (1982). Molecular genetics of yeast mating type. Annu. Rev. Genet 16, 439–500. 10.1146/annurev.ge.16.120182.002255. [DOI] [PubMed] [Google Scholar]
- Nassif N, Penney J, Pal S, Engels WR, and Gloor GB (1994). Efficient copying of nonhomologous sequences from ectopic sites via P-element-induced gap repair. Mol. Cell. Biol 14, 1613–1625. 10.1128/mcb.14.3.1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh SD, Jessop L, Lao JP, Allers T, Lichten M, and Hunter N (2009). Stabilization and electrophoretic analysis of meiotic recombination intermediates in Saccharomyces cerevisiae. Methods Molec. Biol 557, 209–234. 10.1007/978-1-59745-527-5. [DOI] [PubMed] [Google Scholar]
- Oh SD, Lao JP, Hwang PY-H, Taylor AF, Smith GR, and Hunter N (2007). BLM ortholog, Sgs1, prevents aberrant crossing-over by suppressing formation of multichromatid joint molecules. Cell 130, 259–272. 10.1016/j.cell.2007.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh SD, Lao JP, Taylor AF, Smith GR, and Hunter N (2008). RecQ helicase, Sgs1, and XPF family endonuclease, Mus81-Mms4, resolve aberrant joint molecules during meiotic recombination. Mol. Cell 31, 324–336. 10.1016/j.molcel.2008.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oke A, Anderson CM, Yam P, and Fung JC (2014). Controlling meiotic recombinational repair - specifying the roles of ZMMs, Sgs1 and Mus81/Mms4 in crossover formation. PLoS Genet. 10, e1004690. 10.1371/journal.pgen.1004690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan J, Sasaki M, Kniewel R, Murakami H, Blitzblau HG, Tischfield SE, Zhu X, Neale MJ, Jasin M, Socci ND, et al. (2011). A hierarchical combination of factors shapes the genome-wide topography of yeast meiotic recombination initiation. Cell 144, 719–731. 10.1016/j.cell.2011.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panyutin IG, and Hsieh P (1993). Formation of a single base mismatch impedes spontaneous DNA branch migration. J. Mol. Biol 230, 413–424. 10.1006/jmbi.1993.1159. [DOI] [PubMed] [Google Scholar]
- Panyutin IG, and Hsieh P (1994). The kinetics of spontaneous DNA branch migration. Proc. Natl. Acad. Sci. U.S.A 91, 2021–2025. 10.1073/pnas.91.6.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins DD (1949). Biochemical mutants in the smut fungus Ustilago maydis. Genetics 34, 607–626. 10.1093/genetics/34.5.607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson SE, Keeney S, and Jasin M (2020). Mechanistic insight into crossing over during mouse meiosis. Mol. Cell 78, 1252–1263. 10.1016/j.molcel.2020.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pezza RJ, Petukhova GV, Ghirlando R, and Camerini-Otero RD (2006). Molecular activities of meiosis-specific proteins Hop2, Mnd1, and the Hop2-Mnd1 complex. J. Biol. Chem 281, 18426–18434. 10.1074/jbc.M601073200. [DOI] [PubMed] [Google Scholar]
- Porter SE, White MA, and Petes TD (1993). Genetic evidence that the meiotic recombination hotspot at the HIS4 locus of Saccharomyces cerevisiae does not represent a site for a symmetrically processed double-strand break. Genetics 134, 5–19. 10.1093/genetics/134.1.5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prieler S, Chen D, Huang L, Mayrhofer E, Zsótér S, Vesely M, Mbogning J, and Klein F (2021). Spo11 generates chromosomal gaps through concerted cuts at sites of topological stress. Nature 594, 577–582. 10.1038/s41586-021-03632-x [DOI] [PubMed] [Google Scholar]
- Pyatnitskaya A, Borde V, and De Muyt A (2019). Crossing and zipping: molecular duties of the ZMM proteins in meiosis. Chromosoma 128, 181–198. 10.1007/s00412-019-00714-8. [DOI] [PubMed] [Google Scholar]
- Qiao H, Prasada Rao HBD, Yang Y, Fong JH, Cloutier JM, Deacon DC, Nagel KE, Swartz RK, Strong E, Holloway JK, et al. (2014). Antagonistic roles of ubiquitin ligase HEI10 and SUMO ligase RNF212 regulate meiotic recombination. Nat. Genet 46, 194–199. 10.1038/ng.2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao HBDP, Qiao H, Bhatt SK, Bailey LRJ, Tran HD, Bourne SL, Qiu W, Deshpande A, Sharma AN, Beebout CJ, et al. (2017). A SUMO-ubiquitin relay recruits proteasomes to chromosome axes to regulate meiotic recombination. Science 355, 403–407. 10.1126/science.aaf6407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resnick MA (1976). The repair of double-strand breaks in DNA; a model involving recombination. J. Theor. Biol 59, 97–106. 10.1016/S0022-5193(76)80025-2. [DOI] [PubMed] [Google Scholar]
- Rossignol JL, Nicolas A, Hamza H, and Langin T (1984). Origins of gene conversion and reciprocal exchange in Ascobolus. Cold Spring Harbor Symp. Quant. Biol 49, 13–21. 10.1101/sqb.1984.049.01.004. [DOI] [PubMed] [Google Scholar]
- Sandhu R, Monge Neria F, Monge Neria J, Chen X, Hollingsworth NM, and Borner GV (2020). DNA helicase Mph1FANCM ensures meiotic recombination between parental chromosomes by dissociating precocious displacement loops. Dev. Cell 53, 458–472. 10.1016/j.devcel.2020.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwacha A, and Kleckner N (1994). Identification of joint molecules that form frequently between homologs but rarely between sister chromatids during yeast meiosis. Cell 76, 51–63. 10.1016/0092-8674(94)90172-4. [DOI] [PubMed] [Google Scholar]
- Schwacha A, and Kleckner N (1995). Identification of double Holliday junctions as intermediates in meiotic recombination. Cell 83, 783–791. 10.1016/0092-8674(95)90191-4. [DOI] [PubMed] [Google Scholar]
- Shen W, Le S, Li Y, and Hu F (2016). SeqKit: A cross-platform and ultrafast toolkit for FASTA/Q file manipulation. PLoS One 11, e0163962. 10.1371/journal.pone.0163962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinohara M, Oh SD, Hunter N, and Shinohara A (2008). Crossover assurance and crossover interference are distinctly regulated by the ZMM proteins during yeast meiosis. Nat. Genet 40, 299–309. 10.1038/ng.83. [DOI] [PubMed] [Google Scholar]
- Smith CE, Llorente B, and Symington LS (2007). Template switching during break-induced replication. Nature 447, 102–105. 10.1038/nature05723. [DOI] [PubMed] [Google Scholar]
- Snowden T, Acharya S, Butz C, Berardini M, and Fishel R (2004). hMSH4-hMSH5 recognizes Holliday junctions and forms a meiosis-specific sliding clamp that embraces homologous chromosomes. Mol. Cell 15, 437–451. 10.1016/j.molcel.2004.06.040. [DOI] [PubMed] [Google Scholar]
- Spies M, and Fishel R (2015). Mismatch repair during homologous and homeologous recombination. Cold Spring Harb. Perspect. Biol 7, a022657. 10.1101/cshperspect.a022657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl FW, and Foss HM (2010). A two-pathway analysis of meiotic crossing over and gene conversion in Saccharomyces cerevisiae. Genetics 186, 515–536. 10.1534/genetics.110.121194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szostak JW, Orr-Weaver TL, Rothstein RJ, and Stahl FW (1983). The double-strand-break repair model for recombination. Cell 33, 25–35. 10.1016/0092-8674(83)90331-8. [DOI] [PubMed] [Google Scholar]
- Tang S, Wu MKY, Zhang R, and Hunter N (2015). Pervasive and essential roles of the Top3-Rmi1 decatenase orchestrate recombination and facilitate chromosome segregation in meiosis. Mol. Cell 57, 607–621. 10.1016/j.molcel.2015.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theivendirarajah K, and Whitehouse HL (1983). Further evidence that aberrant segregation and crossing over in Sordaria brevicollis may be discrete, though associated, events. Mol. Gen. Genet 190, 432–437. 10.1007/BF00331073. [DOI] [PubMed] [Google Scholar]
- Vernekar DV, Reginato G, Adam C, Ranjha L, Dingli F, Marsolier M-C, Loew D, Guérois R, Llorente B, Cejka P, and Borde V (2021). The Pif1 helicase is actively inhibited during meiotic recombination which restrains gene conversion tract length. Nucleic Acids Res. 49, 4522–4533. 10.1093/nar/gkab232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Tang D, Wang M, Lu J, Yu H, Liu J, Qian B, Gong Z, Wang X, Chen J, et al. (2009). MER3 is required for normal meiotic crossover formation, but not for presynaptic alignment in rice. J. Cell Sci 122, 2055–2063. 10.1242/jcs.049080. [DOI] [PubMed] [Google Scholar]
- Wang S, Hassold T, Hunt P, White MA, Zickler D, Kleckner N, and Zhang L (2017). Inefficient crossover maturation underlies elevated aneuploidy in human female meiosis. Cell 168, 977–989. 10.1016/j.cell.2017.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner BM, and Kleckner N (1994). Chromosome pairing via multiple interstitial interactions before and during meiosis in yeast. Cell 77, 977–991. 10.1016/0092-8674(94)90438-3. [DOI] [PubMed] [Google Scholar]
- Wickham H (2009). ggplot2: Elegant Graphics for Data Analysis (Springer Science & Business Media; ). 10.1007/978-0-387-98141-3. [DOI] [Google Scholar]
- Wickham H, Averick M, Bryan J, Chang W, D’Agostino McGowan L, François R, Grolemund G, Hayes A, Henry L, Hester J, et al. (2019). Welcome to the tidyverse. J. Open Source Software 4, 1686. 10.21105/joss.01686. [DOI] [Google Scholar]
- Wild P, Susperregui A, Piazza I, Drig C, Oke A, Arter M, Yamaguchi M, Hilditch AT, Vuina K, Chan KC, et al. (2019). Network rewiring of homologous recombination enzymes during mitotic proliferation and meiosis. Mol. Cell 75, 859–874. 10.1016/j.molcel.2019.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson MS, Game JC, and Fogel S (1985). Meiotic gene conversion mutants in Saccharomyces cerevisiae. I. Isolation and characterization of pms1-1 and pms1-2. Genetics 110, 609–646. 10.1093/genetics/110.4.609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, and Hickson ID (2003). The Bloom’s syndrome helicase suppresses crossing over during homologous recombination. Nature 426, 870–874. 10.1038/nature02253. [DOI] [PubMed] [Google Scholar]
- Yadav VK, and Claeys Bouuaert C (2021). Mechanism and control of meiotic DNA double-strand break formation in S. cerevisiae. Front. Cell Dev. Biol 9, 642737. 10.3389/fcell.2021.642737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeadon PJ, Rasmussen JP, and Catcheside DEA (2001). Recombination events in Neurospora crassa may cross a translocation breakpoint by a template-switching mechanism. Genetics 159, 571–579. 10.1093/genetics/159.2.571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakharyevich K, Ma Y, Tang S, Hwang PY-H, Boiteux S, and Hunter N (2010). Temporally and biochemically distinct activities of Exo1 during meiosis: double-strand break resection and resolution of double Holliday junctions. Mol. Cell 40, 1001–1015. 10.1016/j.molcel.2010.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakharyevich K, Tang S, Ma Y, and Hunter N (2012). Delineation of joint molecule resolution pathways in meiosis identifies a crossover-specific resolvase. Cell 149, 334–347. 10.1016/j.cell.2012.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
File S2. HTML file with an illustration of each tetrad, related to Figures 2–6.
Individual tetrads can be displayed using the drop-down menu in the upper left hand corner. COs are listed first, followed by NCOs and then complex tetrads.
Supplemental Figures. Figures S1–S7.
File S1. Microsoft Excel workbook containing data underlying analysis, related to Figures 2–6.
- Cover sheet. Summary of the analysis of all tetrads. See also File S2 for a graphic depiction of each tetrad.
- Marker locations. Sequence and location of markers used, along with non-Mendelian segregation numbers and frequencies at each marker location.
- Sequencing data. Strand haplotypes for each marker location derived from sequencing.
- Fig2CD-ochres. Non-Mendelian segregation and crossovers for ura3 and arg4 point mutants in diploids where polymorphisms are present or absent.
- Fig2D-genetics. Tetrad data underlying map distances in the natMX-hygMX interval.
- Fig3CDEF tracts DSBs. Length and location of heteroduplex tracts and half-tracts.
- Fig3EF tract stats. Fit of heteroduplex tract and half tract lengths to an exponential decay model.
- Fig4 template switching. Length, location and lemplate used for each segment in mosaic heteroduplex.
- Fig5 dHJs. Location and dimensions of total heteroduplex tracts, inferred original and final dHJs, and parental segments between final dHJ and DSB.
- Fig6 SP description. Location dimensions and type of strand processing tracts.
- Point events. Single marker conversion events.
- Fig6 suppl segs corrected. NMS segments with point events removed.
- Yeast strains.
- Oligos and probes.
File S3. Genbank-format annotated sequence of the wild-type recombination interval, related to STAR Methods.
File S4. Genbank-format annotated sequence of the SNP-containing recombination interval, related to STAR Methods.
Data Availability Statement
Sequencing data are deposited at the Sequence Read Archive (https://www.ncbi.nlm.nih.gov/sra) under BioProject ID: PRJNA721091.
Underlying data for all figures are in File S1, a Microsoft Excel workbook. References to File S1 in the text use the following format: File S1.x, where x is the worksheet number. Plots of individual tetrad genotypes are in File S2. Sequences of the wild-type and SNP-containing recombination intervals are in Files S3 and S4, respectively. Full Southern blot images are deposited at Mendeley Data, doi: 10.17632/j2jpj3p29f.1
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.


