SUMMARY
The suprachiasmatic nucleus (SCN) is the master circadian pacemaker in mammals and is entrained by environmental light. However, the molecular basis of the response of the SCN to light is not fully understood. We used RNA/Chromatin Immunoprecipitation/single-nucleus sequencing with circadian behavioral assays to identify mouse SCN cell types and to explore their responses to light. We identified three peptidergic cell types that responded to light in the SCN: arginine vasopressin (AVP), vasoactive intestinal peptide (VIP), and cholecystokinin (CCK). In each cell type, light-responsive subgroups were enriched for expression of Neuronal PAS Domain Protein 4 (NPAS4) target genes. Further, mice lacking Npas4 had a longer circadian period in constant conditions, a damped phase response curve to light, and reduced light-induced gene expression in the SCN. Together, our data indicate that NPAS4 is necessary for normal transcriptional responses to light in the SCN and is critical for photic phase-shifting of circadian behavior.
Keywords: suprachiasmatic nucleus, NPAS4, light simulation, single-nucleus RNA sequencing
eTOC blurb:
In this study, Xu et al. investigate the transcriptional response of the suprachiasmatic nucleus to light. They show that the light-inducible gene, Npas4, is involved in mediating the transcriptional response of SCN neurons. Furthermore, loss of Npas4 alters circadian rhythms in mice, suggesting a role for Npas4 in photic entrainment.
INTRODUCTION
Circadian rhythms are 24-hour endogenous physiological and behavioral osciilations that are entrained by environmental light-dark cycles (Golombek and Rosenstein, 2010; Pittendrigh and Daan, 1976). In mammals, the hypothalamic suprachiasmatic nucleus (SCN) is the master circadian pacemaker that coordinates circadian rhythms throughout the body (Hastings et al., 2018; Morin, 2013; Ralph et al., 1990; Welsh et al., 2010). The SCN consists of a heterogeneous cluster of approximately 10,000 neurons that are classically subdivided into a dorsomedial “shell” and a ventrolateral “core” (Morin, 2013; Park et al., 2016; Welsh et al., 2010). The neurons in the core of the SCN receive light information from intrinsically photosensitive retinal ganglion cells (ipRGCs) via the retinohypothalamic tract, and these photic inputs are sufficient to set the phase of SCN neuron oscillations (Guler et al., 2008; Hattar et al., 2002; Jones et al., 2018; Lazzerini Ospri et al., 2017; LeGates et al., 2014). In addition, other SCN cell types may also respond to light (Fernandez et al., 2016).
The molecular response of the SCN to light has been previously characterized en masse (Allen et al., 2017), and includes the induction of immediate early genes (IEGs) and signaling cascades involved in cAMP response element-binding protein (CREB) phosphorylation (Doi et al., 2007; Ginty et al., 1993; Hatori et al., 2014; Kornhauser et al., 1990, 1992; Morris et al., 1998; Porterfield et al., 2007; Tischkau et al., 2003). While these previous studies were largely limited to examining bulk SCN tissues, recent experiments in cortical tissues have paved the way towards understanding cell-type specific IEG and late-response gene responses to visual stimuli (Lin et al., 2008; Majidi et al., 2019; Mardinly et al., 2016; Spiegel et al., 2014) including the activity-induced responses of non-neuronal cells (Gibson et al., 2014; Hrvatin et al., 2018; Lacoste et al., 2014; Wang et al., 2018). These findings led us to hypothesize that the SCN may have similarly heterogenous responses to light stimulation.
Thus, in the current study we investigated the effects of light on the SCN by examining IEG expression at several time points after different durations of light exposure to examine the overall kinetics of light-induced genes. Given the cellular heterogeneity within the SCN (Park et al., 2016; Wen et al., 2020), we used single-nucleus RNA sequencing to measure gene expression in different SCN cell populations after acute light exposure to provide us with a better understanding of how these subpopulations respond to light. Our transcriptomic results revealed differential patterns of acute and sustained IEG expression after exposure to light, as well as remarkable cell-type specificity in patterns of light-induced genes. Moreover, when we performed motif analysis of the light-induced gene enhancers and promoters, we found an enrichment for NPAS4 binding sites. Npas4 is an immediate early gene encoding a bHLH-PAS-domain family member protein whose expression in the visual cortex is induced by light (Lin et al., 2008), but whose function has not been explored within the SCN. We further show that loss of Npas4 alters photic entrainment and circadian behavior in mice, suggesting that it may play an important role in mediating the hypothalamic response to light.
RESULTS
The SCN has diverse transcriptomic responses to light
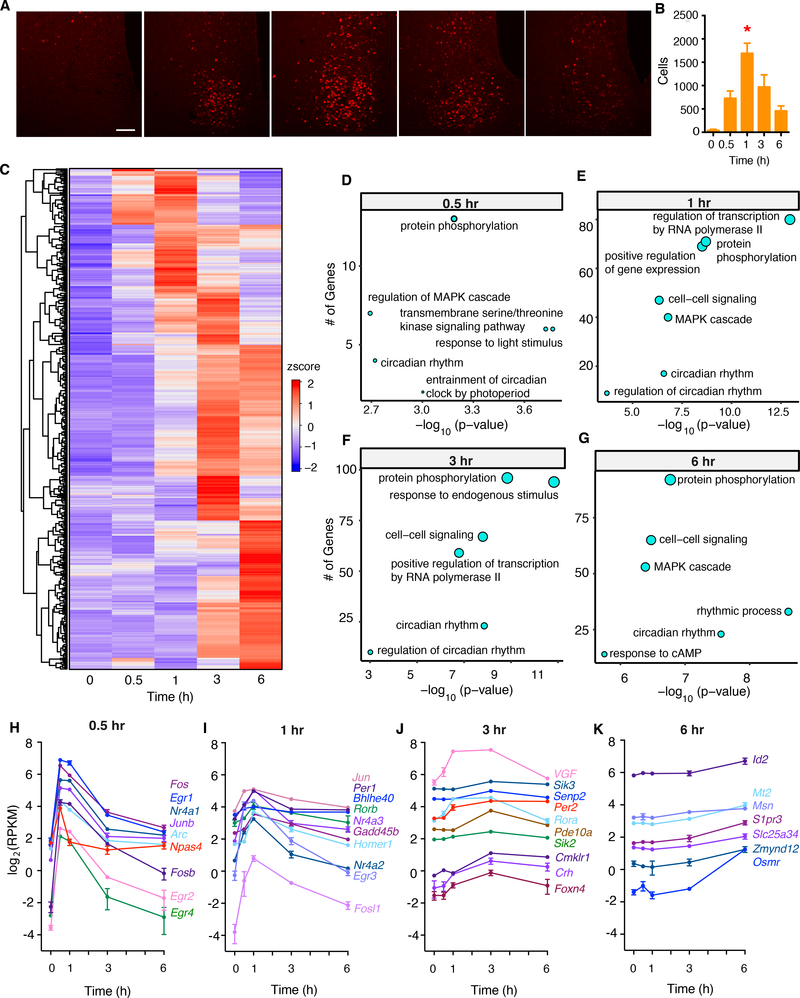
To profile transcriptomic changes in the SCN both acutely (after short light exposures) and in more sustained light, we performed bulk RNA sequencing on whole mouse SCN after different durations of light stimulation (0.5 hr, 1 hr, 3 hr, and 6 hr). To determine the responses to light of pulses that cause phase shifts of circadian activity rhythms in C57BL/6J mice, male mice were reared in the dark and exposed to light at circadian time (CT) 17, a phase that induces the maximal delays (Vitaterna et al., 2006). To validate our experimental design, we performed immunohistochemical analysis of FOS expression in the SCN after different durations of light stimulation. Elevated FOS expression was observed as early as 0.5 hr after light exposure and peaked at 1 hr post-exposure (Figure 1A and 1B).
Fig 1. Light exposure drives diverse transcriptomic changes in the SCN.
(A) Representative images for immunohistochemical analysis of FOS (red) in the SCN of wildtype mice after 0.5-, 1-, 3-, and 6-hour periods of light stimulation beginning at CT17. Scale bar, 50 μm.
(B) Quantification of FOS- immunoreactive cells in the SCN after 0.5-, 1-, 3-, and 6-hr periods of light stimulation. Mean ± s.e.m. are shown. (n = 4 for each time condition, * p < 0.05, one-way ANOVA, adjusted with Tukey’s post hoc test)
(C) Heatmap of light-induced genes grouped by time of peak expression.
(D-G) Pathway analysis of light-induced genes at different timepoints.
(H-K) Representative light-induced genes that peaked at 0.5-, 1-, 3-, or 6-hr periods of light stimulation. All examples were significantly induced genes as assessed by differential gene expression analysis (see Table S1).
RNA sequencing of whole SCN revealed that various durations of light exposure induced different patterns of gene expression (FDR < 0.05, abs(log2(Fold Change)) ≥ 0.3) (Figure 1C, H–K, Table S1). We found different clusters of genes that peaked at 0.5, 1, 3 and 6 hours of light exposure, respectively (Figure 1H). Gene ontology (GO) term enrichment analysis revealed that genes involved in the entrainment of the circadian clock by photoperiod were enriched in the acute responding group (Figure 1D), whereas other circadian genes had more sustained induction (Figure 1E–G). As expected, Fos and Per1 had transient responses, while Per2 showed a delayed but sustained induction (Vitaterna et al., 2006) (Figure 1H and 1I). Moreover, most classical IEGs in the SCN, except for Bdnf (Figure S1A), responded to light stimulation acutely (Figure 1H), consistent with previous reports (Hatori et al., 2014; Porterfield et al., 2007). Genes that were reduced after light exposure are shown in Figure S1B–I.
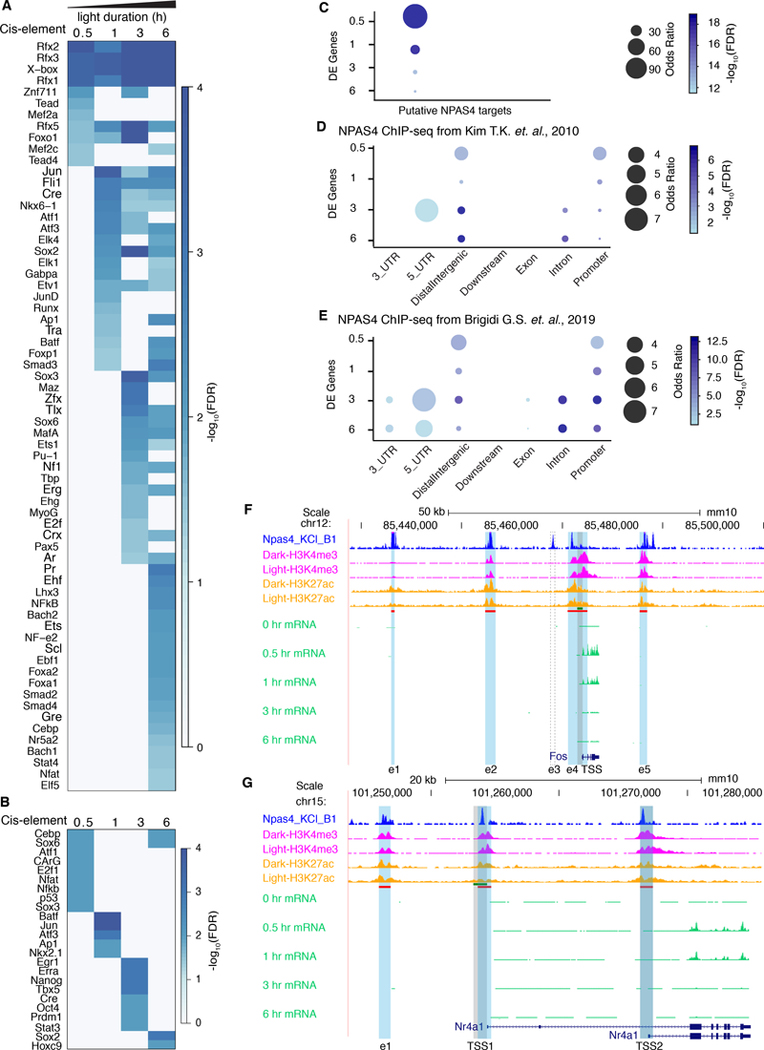
Npas4, which is involved in activity-dependent regulation of excitatory-inhibitory balance (Spiegel et al., 2014), displayed an acute response in the SCN peaking at 0.5 hr (Figure 1H) similar to its response to light in visual cortex (Lin et al., 2008). Since NPAS4 binding sites are located at Fos enhancer sites in cultured cortical neurons (Kim et al., 2010), we wondered whether NPAS4 orchestrates the light-regulated transcriptional response in SCN neurons. To find active enhancers in the SCN, we performed ChIP-seq with acetylated histone 3 lysine 27 (H3K27ac), a chromatin modification highly correlated with active enhancers (Creyghton et al., 2010) in SCN tissue with and without 1-hour light stimulation. While there were H3K27ac sites in the regulatory regions of IEGs, the binding occupancy remained stable in these regions after a 1-hr light pulse (Figure S2A–C), which is in contrast to our previous findings in the liver (Koike et al., 2012), but suggests that the SCN has persistent basal levels of H3K27ac and a more open chromatin state than the mouse liver. Motif analysis of H3K27ac enhancer sites within the region 100kb upstream of the transcription start site (TSS) of light-induced genes, ordered by the earliest time point of induction, revealed a temporal cascade of overrepresented cis-regulatory sequences in both enhancers and promoters (Figure 2A, B). As expected, at the 0.5 hr time point, motifs including Cebp, Atf1, CArG, and Nfat were significantly enriched in promoter regions (Figure 2B). CArG is a well-known motif that shares sequence similarity with the Serum Response Element, which regulates the transcriptional activation of serum-induced genes such as Fos (Greenberg et al., 1987; Santoro and Walsh, 1991). Further analysis using two independent NPAS4 ChIP-seq datasets (Brigidi et al., 2019; Kim et al., 2010) and an independent list of putative NPAS4 targets defined by ChIP-seq and RNA-seq (Bloodgood et al., 2013) revealed significant enrichment for NPAS4 binding at light-induced gene enhancer sites (Figure 2C, D, E). Genome browser views of NPAS4 and H3K27ac enhancer sites for two representative light-induced IEGs, Fos and Per1 are shown in Figure 2F and G, as well as for, Nr1a1 and Per2 in Figure S2D–E. In the Fos locus, 4 out of 5 enhancers previously identified to be activity regulated in cortical neurons (Kim et al., 2010) were also active in the SCN; whereas, enhancer, e3, had no H3K27ac signal in the SCN. In the Nr1a1 locus, NPAS4 is enriched at an upstream enhancer (e1) and at two alternative promoter sites (TSS1 and TSS2) of the gene (Figure S2D).
Fig 2. Transcriptional regulation of light-induced genes in the SCN may be mediated by NPAS4.
(A-B) Sequence motif analysis of H3K27ac sites in newly induced genes. Induced genes were grouped into different time points based on the earliest time point at which their mRNA was significantly upregulated (log2(FC) ≥ 0.3, FDR < 0.05). Sequence motif analysis was carried out on the consensus peak set between dark and light stimulated conditions using HOMER (Heinz et al., 2010).
(A) HOMER motif analysis of H3K27ac sites in enhancer regions of newly upregulated genes at different durations of light stimulation duration.
(B) HOMER motif analysis of H3K27ac sites in promoter regions of newly upregulated genes at different durations of light stimulation.
(C-E) Enrichment analysis of NPAS4 ChIP-seq and putative NPAS4 target genes with light-induced differentially expressed genes.
(C) Enrichment analysis of putative NPAS4 target genes (Bloodgood et al., 2013) with light-induced differentially expression genes.
(D) Enrichment analysis of NPAS4 ChIP-seq (Kim et al., 2010) with light-induced differentially expression genes.
(E) Enrichment analysis of NPAS4 ChIP-seq (Brigidi et al., 2019) with light-induced differentially expression genes.
(F-G) UCSC genome browser views of Fos and Per1. NPAS4 ChIP-seq (Kim et al., 2010) is shown in dark blue. H3K4me3, H3K27ac ChIP-seq, and light pulse bulk RNA-seq are shown as indicated. The bed file of the promoter (−1kb - +100 bp) is shown in green while the bed file of H3K27ac sites is shown in red. The H3K27ac sites for Fos are shaded blue. The e3 enhancer of Fos is missing in SCN cells and its putative location is highlighted with a dotted box. The promoter region for Fos is highlighted in shaded grey. Per1 has 3 alternative transcriptional start sites (TSS) that correspond with the nucleosome-free H3K27ac regions within the broad H3K27ac peak that spans all three TSS. CLOCK:BMAL1 occupancy peaks at the locations of TSS1 and TSS3 in Per1 (Koike et al., 2012).
Of the core circadian clock genes in mammals, Per1 and Per2 mediate transcriptional inputs to the circadian clock mechanism for resetting and entrainment (Lowrey and Takahashi, 2011; Takahashi, 2017). In the Per1 locus, there was strong enrichment for H3K27ac and H3K4me3 at three alternative transcription start sites (TSS1-TSS3) corresponding with NPAS4 occupancy in this region. In the Per2 locus, there was enrichment for NPAS4 in the promoter region. Therefore, in addition to CREB-mediated signaling (Tischkau et al., 2003; Travnickova-Bendova et al., 2002), NPAS4 is a likely upstream regulator Per1/Per2 gene expression. These results support the hypothesis that NPAS4 is involved in the regulation of light-induced gene expression in the SCN.
Light-responsive cellular sub-populations in the SCN
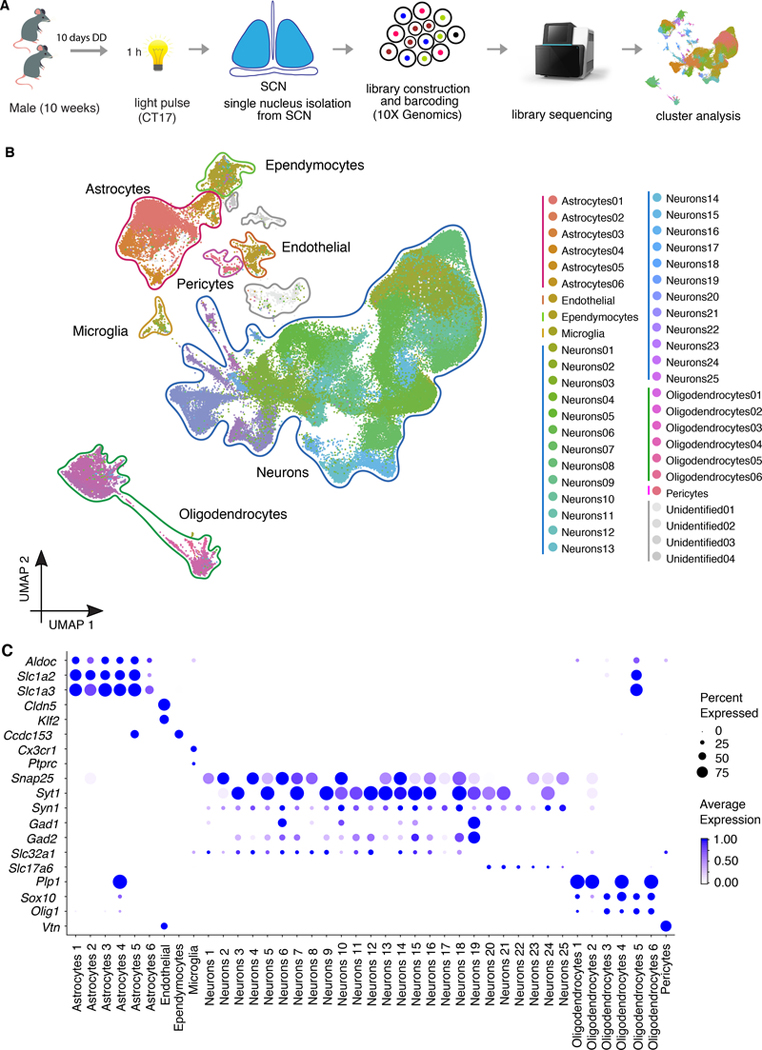
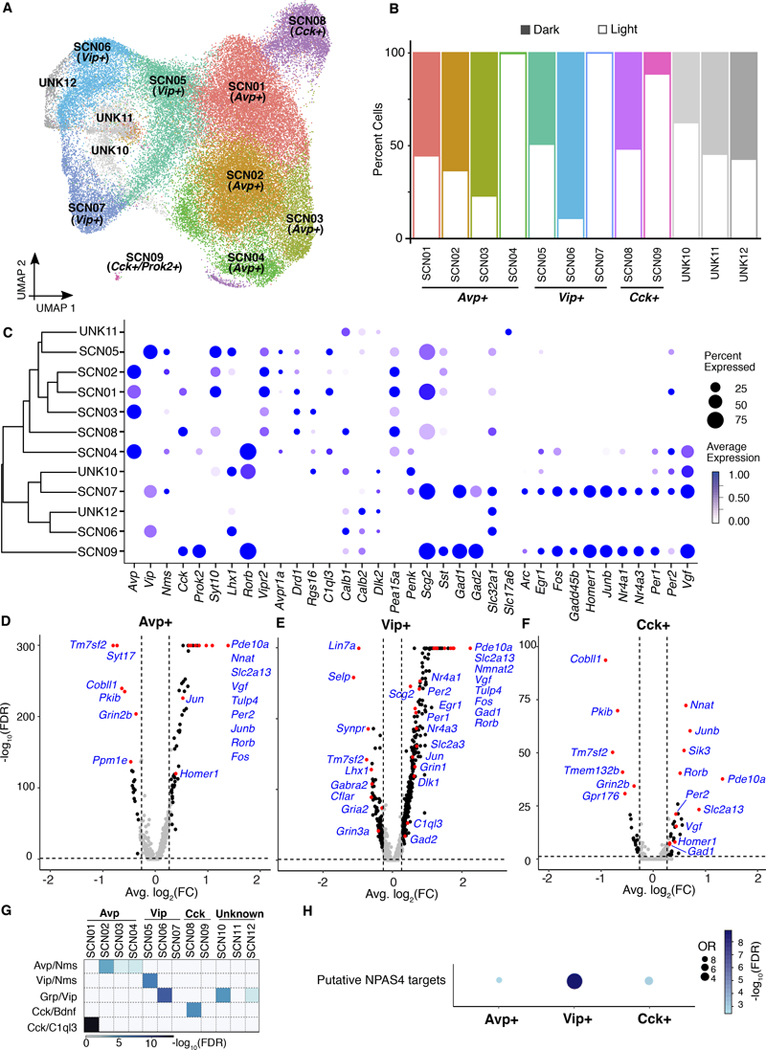
SCN neurons are heterogeneous in their responses to light (Allen et al., 2017; Park et al., 2016; Wen et al., 2020). Therefore, to understand the full spectrum of light-responsive cellular subtypes in the SCN, we adopted methods developed to study activity-induced single-cell RNA-seq gene expression (Hrvatin et al., 2018) and performed single-nucleus RNA sequencing of SCN tissue with and without 1-hr light stimulation (Figure 3A, Figure S3). From a total of five dark and five light stimulated SCN 10X Genomics libraries, 144,806 nuclei were sequenced, of which 144,582 survived quality filters and the average number of detected genes per nucleus was about 1100. From these data, we identified 45 clusters (Figure 3B) that were grouped into seven distinct cell types (neurons; oligodendrocytes; astrocytes; endothelial cells, ependymocytes, microglia, and pericytes) based on known cell identity markers (Figure 3C, Figure S4). Each cell type contained a similar proportion of cells from both light and dark libraries, suggesting that the main source of variation defining cell types was not light responsiveness, but rather other features of cellular identity (Figure S3D). These cluster identities were consistent with three recently published single-cell datasets from the mouse arcuate nucleus of the hypothalamus and median eminence (Campbell et al., 2017), the mouse visual cortex (Hrvatin et al., 2018), and the SCN (Wen et al., 2020)(Figure S4A, B, C). In the case of the SCN, the Wen et al. study used Drop-seq single-cell RNA-seq (43,846 cells from 12 time points during two circadian cycles) as compared to the single-nucleus RNA-seq with 10X Genomics technology used here (144,582 nuclei from dark and light samples at CT17).
Fig 3. Single-nucleus RNA-seq reveals the full spectrum of cell profiles in the SCN.
Experimental workflow of the 1-hr light pulse SCN single nuclei RNA-seq.
(A) UMAP plot of 144,582 nuclei, colored according to density clustering and annotated according to known cell types (clustering resolution = 0.8).
(B) Dot plots showing the relative expression of marker genes for major cell types. Size of the dot reflects the percentage of cells expressing a gene of interest and color intensity reflects the average expression (darker= more highly expressed).
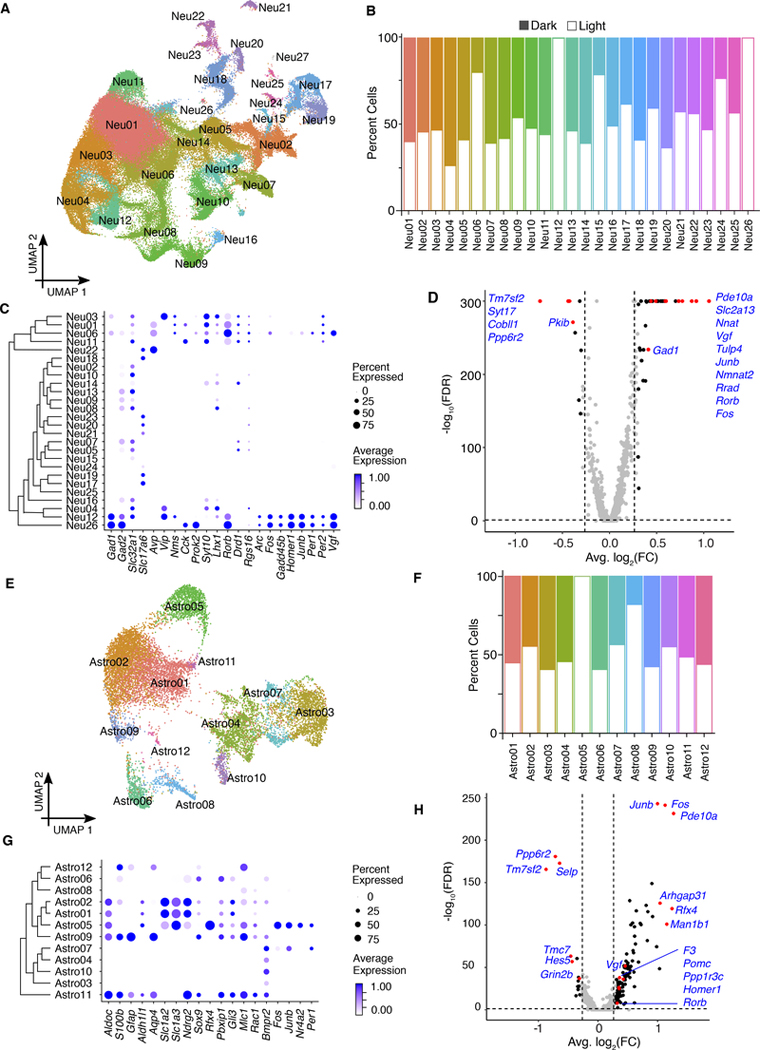
Based on known cell identity markers (Figure 3C), we selected 25 neuronal and 6 astrocyte clusters and re-clustered them separately (Figure 4A and 4E). While most of the neuronal clusters contained similar numbers of cells from dark and light libraries, 2 clusters contained almost entirely light-responsive cells (clusters 12, and 26), and 3 clusters contained > 75% light-responsive cells (clusters 6, 15 and 24) (Figure 4B). Surprisingly, of the astrocyte clusters (12,720 cells in total), there was also one cluster (Astro05) containing nearly all light-responsive cells (Figure 4F). These strongly “light-responsive cell clusters,” were enriched with IEGs (Figure 4C and 4G), providing internal validation to our approach. Differential expression (DE) analysis comparing the light and dark libraries within each cell type revealed remarkably different lists of DE genes between neurons and astrocytes, suggesting a cell-type specific response to light (Figure 4D and 4H, Table S2).
Fig 4. Neuronal and astrocyte clusters in the SCN show heterogeneous responses to light.
(A) UMAP plot of neuronal clusters, colored according to density clustering and annotated according to known cell identity markers (clustering resolution = 0.5).
(B) Cell distribution for each cluster. Open bars, cells from light sample libraries; filled bars, cells from dark sample libraries.
(C) Dot plots for marker genes for major neuronal types. Size of the dot reflects the percentage of cells expressed gene of interest and color intensity reflects the average expression (darker = more highly expressed).
(D) Differential gene expression between light clusters and control clusters. Significantly changed genes are defined log2(FC) ≥ 0.3, FDR < 0.05. Red dots indicate genes of interest (labeled).
(E) UMAP plot of astrocytes clusters, colored according to density clustering and annotated according to known cell identity markers. (clustering resolution = 0.5).
(F) Cell distribution for each cluster. Open bars, cells from light sample libraries; filled bars, cells from dark sample libraries.
(G) Dot plots showing the relative expression of marker genes for major astrocytes types. Size of the dot reflects the percent of cells expressed gene of interest and the color intensity reflects the average expression level from the cells in which the gene was detected.
(H) Differential gene expression between light clusters and control clusters. Significantly changed genes are defined as log2(FC) ≥ 0.3, FDR < 0.05. Red dots indicate genes of interest (labeled).
To further refine the clustering, we next investigated the expression of known SCN genes for inclusion and non-SCN genes for exclusion within the neuronal clusters and identified 14 potential SCN clusters (Figure S5). Clusters were selected based on canonical SCN neuronal marker genes such as Vip, Avp, Nms, Cck, Prok2, Syt10, Lhx1, Rorb, Vipr2, Avpr1a, Drd1, Rgs16, C1q13, Calb1, Calb2, Dlk2, Pea15a, Penk, Scg2, Sst, Gad1, Gad2, Slc32a1, Slc17a6, and Vgf. The clusters that showed high expression of canonical markers genes were used for subclustering. In total, reclustering resulted in 12 peptidergic SCN clusters, including Avp-expressing cells (clusters 1–4), Vip-expressing cells (clusters 5–7), Cck-expressing cells (clusters 8 and 9) and clusters that were not clearly identifiable, but that had similarities with VIP neurons (clusters 10–12; Figure 5A). Importantly, neuronal subclusters were stable across multiple arbitrary resolutions with minimal nuclei intermixing (Figure S5B).
Fig 5. SCN neuropeptide cell clusters can be further refined by their response to light.
(A) Spectral UMAP plot of neuronal clusters, colored according to density clustering and annotated according to known cell identity markers (clustering resolution = 0.5).
(B) Cell distributions for each cluster. Open bars, cells from light sample libraries; filled bars, cells from dark sample libraries.
(C) Dot plots showing the relative expression of marker genes for major SCN neuronal cell types. Size of the dot reflects the percentage of cells expressing a gene of interest and the color intensity reflects the average expression (darker = more highly expressed.
(D-F) Differential gene expression after light stimulation in (D) Avp−, (E) Vip−, and (F) Cck-expressing cells. Significantly changed genes are defined as log2(FC) ≥ 0.3, FDR < 0.05.
(G) Heatmap shows the correlation between SCN cell types discovered in a dataset from mouse SCN (Wen et al., 2020) and this study. Fisher’s exact test and the color reflects the adjusted p value.
(H) Enrichment of putative NPAS4 targets in three light-responsive, peptidergic clusters.
Because immediate early genes are indicative of light responsiveness (Kornhauser et al., 1996; Porterfield and Mintz, 2009), we used the differentially expressed IEGs to define a subgroup of light-responsive neuronal subtypes that are part of the initial peptidergic SCN clusters (Figure 5B–C, Table S3).
To validate these results, the expression of Fos, Egr, and Per1 were examined in different light-responsive clusters by fluorescence in situ hybridization (FISH) using RNAscope®. Since the Cck FISH signal was low, we instead used Prok2 as a marker for cluster 9 (Cck+/Prok2+) (Figure S5C–E). DE analysis comparing light libraries and dark libraries within each reclustered cell type again revealed remarkably different lists of DE genes among Avp+, Vip+ and Cck+ cells, suggesting a peptidergic neuron cell type-specific response to light (Figure 5D - 5F, Table S4). Further, these cluster identities were consistent with a recently published SCN single-cell dataset (Wen et al., 2020)(Figure 5G). Interestingly, there was significant enrichment of putative NPAS4 targets in light-responsive clusters, with the Vip+ cluster being the strongest, followed by Cck+ and Avp+ clusters (Figure 5H). These data support the idea that NPAS4 regulates gene expression in all light-responsive peptidergic SCN neurons.
Loss of Npas4 in the SCN causes changes in circadian behavior
Although NPAS4 is expressed in multiple cell types in the cortex, it shows remarkable cell-type specific functions: in glutamatergic neurons, NPAS4 promotes GABAergic synapses and in GABAergic interneurons it increases glutamatergic synapses (Bloodgood et al., 2013; Lin et al., 2008; Spiegel et al., 2014). SCN neurons also exhibit excitatory or inhibitory characteristics, depending on circadian phase or SCN subregion, although a majority of SCN neurons are GABAergic (Albers et al., 2017; Hastings et al., 2018; Kalsbeek et al., 2006). Because we found that Npas4 expression was induced in the SCN by light, and we saw significant enrichment of genes potentially regulated by NPAS4 in light responsive SCN neuronal clusters, we hypothesized that Npas4 is involved in mediating the response of SCN circuits to light.
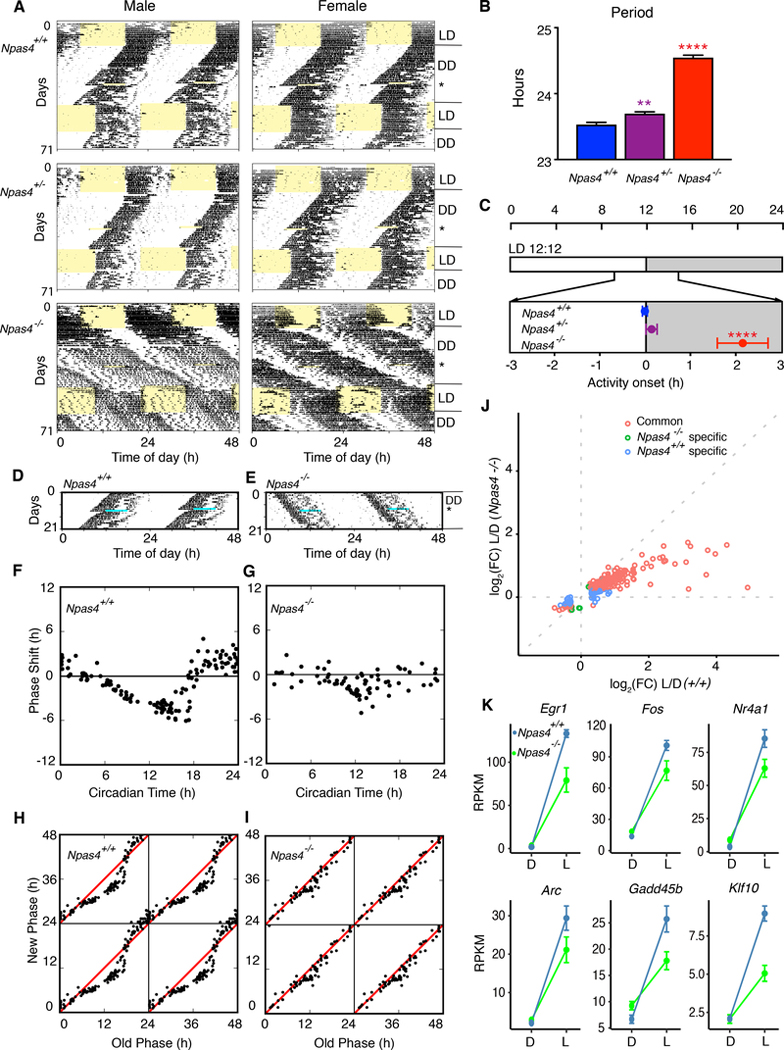
To investigate the function of NPAS4 in the behavioral response to photic stimulation, we examined the circadian locomotor activity rhythms in littermate Npas4+/+, Npas4+/−, Npas4−/− mice (Lin et al., 2008). The Npas4+/+ and Npas4+/− mice of both sexes exhibited normal circadian behavior in LD12:12 and constant darkness (DD). However, Npas4+/− and Npas4−/− mice of both sexes had a significantly longer circadian period in constant darkness with Npas4−/− mice having a mean period greater than 24.5 hrs (Figure 6A and 6B). Npas4−/− mice had a significantly delayed phase angle of entrainment to LD than either Npas4+/+ or Npas4+/− mice (Figure 6C). Notably, the entrainment of some Npas4−/− mice was unstable (Figure 6A). In addition, the magnitude of the phase shifts caused by the light pulses was reduced in Npas4−/− mice (Figure 6A, 6D and 6E). A detailed phase response curve (Figure 6F and 6G) and phase transition curve (Figure 6H and 6I) to 6-h light pulses showed that Npas4−/− mice had a significantly reduced response to light (** p= 2.22e-16, nonparametric two-way ANOVA with interaction using ARTool [Aligned Rank Transform for nonparametric factorial ANOVAs]). To further validate the function of Npas4 in the SCN, we crossed Npas4fx/fx mice with Camk2a::iCreBAC mice (CamiCre) (Casanova et al., 2001; Izumo et al., 2014) to generate forebrain-specific Npas4 null mice (cKO). Although Camk2a is thought to be an excitatory neuronal marker, it is highly expressed in SCN neurons which are GABAergic. Circadian behavior of the CamiCre driver mice has been previously reported and is indistinguishable from wildtype mice (Izumo et al., 2014). Npas4 cKO mice expressed persistent circadian locomotor rhythms in DD compared to Npas4fx/fx and CamiCre mice; however, they had a longer circadian period in DD, similar to Npas4−/− mice (Figure S6), suggesting that the long period phenotype is due to a neuron-specific function of NPAS4.
Fig 6. Mice lacking Npas4 display abnormal circadian entrainment, reduced phase response to light and long circadian period.
(A) Representative actograms of male and female Npas4+/+, Npas4+/− and Npas4−/− mice. Records are double plotted, and each horizontal line represents 48 hrs. of activity.
(B) Period of each genotype in constant darkness (Npas4+/+, n = 32, Npas4+/−, n = 32, Npas4−/−, n = 27). Mean ± s.e.m. are shown. (**** p < 0.0001, ** p < 0.01, one-way ANOVA, adjusted with Tukey’s post hoc test)
(C) Phase of entrainment of each genotype (only mice entrained to LD were included in analyses; Npas4+/+, n = 31, Npas4+/−, n = 26, Npas4−/−, n = 10). Mean ± s.e.m. are shown. (**** p < 0.0001, one-way ANOVA, adjusted with Tukey’s post hoc test)
(D-E) Activity records of representative Npas4+/+ (D) and Npas4−/− (E) mice given a 6-h light pulse (indicated by light blue stripe) at CT17. Records are double-plotted and each horizontal line represents 48 hrs. of activity.
(F-G) Representative phase-response curves to 6-h light pulses in Npas4+/+ (n = 25) (F) and Npas4−/− (n = 21) mice (G). The x axis displays the circadian time (CT) at the beginning of the light pulse. The y axis shows the phase shift induced by the light pulse. (**** p < 0.01, nonparametric two-way ANOVA (Mack-Skillings test)
(H-I) Phase-transition curves to the light pulses in Npas4+/+ (H) and Npas4−/− (I) mice. Data from F and G are re-plotted and the x axis shows the phase (CT) at the beginning of the light pulse and it is determined from the preceding free run. The y axis indicates the extrapolated phase (CT) of the light pulse and it is calculated from the reset activity rhythm.
(J) Gene expression changes in the SCN from Npas4+/+ (n = 3) and Npas4−/− (n = 3) mice after a 1-hour light pulse. Red circles indicate differentially expressed genes in both Npas4+/+, and Npas4−/−, green circles indicate differentially expressed genes in Npas4−/− only, and blue circles indicate differentially expressed genes in Npas4+/+ only.
(K) Examples of changes in gene expression with and without light pulse from Npas4+/+, and Npas4−/− SCNs. Blue lines indicate gene expression in Npas4+/+ and green lines indicate gene expression in Npas4−/−.
Finally, we performed bulk RNA-seq on Npas4+/+ and Npas4−/− SCN after 1 hr light stimulation at CT17 in dark-reared mice. In the Npas4−/− SCN, 235 out of 320 light-induced genes showed reduced responses (Figure 6J and 6K, Table S5) compared to Npas4+/+, suggesting that the loss of Npas4 underlies gene expression changes in response to light stimulation in the majority of light-induced genes and results in a reduced amplitude phase response curve to light.
DISCUSSION
Here, using bulk RNA-sequencing and single-nucleus RNA-sequencing of SCN, we found that cells within the SCN have remarkably diverse responses to light. Our investigation of the time course of light-responsive gene expression in bulk SCN samples revealed that, in addition to an immediate early gene (IEG) response (Porterfield et al., 2007), there are more sustained changes in gene expression. Importantly, Npas4, a light responsive IEG that has been studied in the visual cortex, is among the IEGs with acute induction in the SCN. Further, we found that mice lacking Npas4 had longer circadian period and reduced response to light, providing evidence for a functional role of NPAS4 in both the entraining effects of light as well as the kinetics of the circadian pacemaker.
Our behavioral findings from Npas4 deficient mice are reminiscent of Fos deficient mice, which have normal circadian period but take longer to entrain to light shifts, also suggesting an important role for this IEG in the SCN (Honrado et al., 1996). Notably, Npas4 deficient mice do not have gross health problems seen in Fos deficient mice (Lin et al., 2008). However, studies of other IEG knockouts, such as EGR1, suggest that not all IEGs are required for a light response (Kilduff et al., 1998). Of the genetic models used to study circadian behavior, loss of Cry2, Rorb, Fbxl3, or CK1ε can all cause period lengthening to varying degrees (Etchegaray et al., 2009; Meng et al., 2008; Siepka et al., 2007). In addition, knockout of CK1d from AVP neurons in the SCN lengthens circadian period (Mieda et al., 2016). Interestingly, Npas4 deficient mice had increased basal expression of Per1 (log2(FC) = 0.34, FDR = 5×10−4) and Per2 (log2(FC) = 0.38, FDR = 3×10−4) (Table S5). Transgenic rats with overexpression of Per1 have a ~0.6–1.0-h longer circadian period than wild-type siblings in both activity and body temperature rhythms (Numano et al., 2006); thus, it is possible that the behavioral effects of Npas4 deficiency could be due to changes in the negative feedback elements of the core circadian clock.
In mice, Npas4 induction occurs in response to membrane depolarization (Ramamoorthi et al., 2011) via L-type voltage gated channels during the formation of inhibitory synapses (Lin et al., 2008), and also in hippocampal glutamatergic neurons after long-term-potentiation (Ploski et al., 2010). Within the hippocampus, NPAS4 promotes the formation of inhibitory somatostatin synapses on pyramidal neurons while also enhancing excitatory synapses on those same inhibitory neurons (Bloodgood et al., 2013; Spiegel et al., 2014). Recent studies have shown that action potentials trigger transcription of Npas4 in the nucleus, whereas excitatory postsynaptic potentials (EPSPs) induce local translation of dendritic Npas4 mRNAs. These different Npas4 sources also form different heterodimers, whereupon rapidly translated NPAS4 associates with aryl hydrocarbon receptor nuclear translocator 1 (ARNT1), while newly transcribed NPAS4 associates with ARNT2. These two different heterodimers have different transcriptional targets: NPAS4/ARNT1 bind to intra- and inter-genic loci while NPAS4/ARNT2 bind to promoters (Brigidi et al., 2019). ARNT2 appears to bind to promoters and recruit the nuclear receptor corepressor 2 (NCoR2) repressor complex prior to activity-dependent stimulation. However, NPAS4 activation/binding disrupts the promoter NCoR2 repressor complex and recruits NCoR2 to enhancers (Sharma et al., 2019). NPAS4 heterodimers recognize E-box-like response elements in the regulatory regions of target genes and DNA binding specificity is directed by ARNT1/ARNT2 (Hao et al., 2011). One known target of NPAS4 is Bdnf (Lin et al., 2008; Spiegel et al., 2014), but we see no evidence that Bdnf follows a pattern of expression indicating its induction by light in the SCN. In addition, by comparing light-induced genes in the visual cortex (Hrvatin et al., 2018) and SCN Vip+ neurons, Cck+ neurons, and astrocytes, there is no overlap in induced genes except for Fos. These findings suggest that activity-induced gene expression cascades are cell-type specific.
A previous study used high-throughput qPCR to examine gene expression in ~350 microdissected SCN neurons from mice exposed to a 1-hour light pulse at Zeitgeber time (ZT) 14 or kept in constant darkness (Park et al., 2016). The goal of that study was to identify distinct neuronal SCN cell types and investigate different activity states, and the authors concluded that traditionally used neuropeptide markers (e.g., AVP and VIP) did not accurately reflect neuronal states within the SCN. In agreement with their findings, we show that within SCN neuropeptide clusters there were subgroups of cells that were responsive to light. However, the light induced IEG list from AVP and VIP clusters are quite distinct, with VIP having a much larger list. Our interpretation of our data is that responsiveness to light, itself, is not the defining characteristic of SCN subpopulations. Rather, the distinct gene expression signatures that are induced by light may give clues to the functional organization of the SCN. A more recent single-cell RNA sequencing study (Wen et al., 2020) examined gene expression in the SCN over 12 circadian timepoints and uncovered 16 neuronal subtypes and 5 SCN neuron subtypes. These subtypes were examined in the SCN of mice after 1 h of light exposure at CT17 and then categorized from most to least light-responsive (as assessed by IEG induction): Grp+/Vip+, Vip+/Nms+, Avp+/Nms+, Cck+/C1ql3+, and Cck+/Bdnf+, which is largely consistent with our data. While Wen and colleagues showed detailed expression patterns of circadian genes throughout the SCN, the focus of our own study was not on the spatiotemporal expression of these markers. Given the role of NPAS4 in the SCN, it will be of interest in future studies to compare the anatomical distribution of NPAS4+ cells relative to the light-responsive peptidergic neurons in the SCN. Because only subsets of the neuropeptide cell clusters were light responsive, double-labeling with IEGs will likely be necessary to define these light-responsive neuropeptide subclasses.
In addition to neuropeptides, GABA and glutamate signaling are essential for SCN function. Glutamate is the primary neurotransmitter input from the retinohypothalamic tract (Welsh et al., 2010). In addition, glutamate released by astrocytes has been shown to be sufficient to maintain SCN rhythms (Brancaccio et al., 2019; Brancaccio et al., 2017). Of note, although our focus was on neuronal clusters, we did observe light-responsive astrocyte clusters, suggesting that astrocytes may also be involved in light responses in the SCN. GABA signaling in the SCN can be excitatory or inhibitory in different contexts (Myung et al., 2015), and is also critical for circadian output rhythms (Ono et al., 2019). Acute activation of GABA receptors prior to light exposure blocks the ability of light pulses to phase shift the SCN (Albers et al., 2017). In our single-nucleus analysis, all AVP clusters (light and dark) expressed glutamate receptor subunits (for example, Grin2b), with very little expression of GABA receptors (Gabra1–6). The three AVP non-light responsive clusters express Vipr2, suggesting they can respond directly to VIP signaling. Interestingly, cluster 4 was light responsive but did not express Vipr2, suggesting that it may be directly activated by retinohypothalamic projections. A recent report found that retinal ganglion cells not only communicate light signals to the SCN by the release of glutamate, but also via the release of GABA, and around 59% of VIP+ neurons in the SCN receive monosynaptic, GABAergic ipRGC input (Sonoda et al., 2020). Our gene expression analysis in VIP+ clusters shows Gabra2 and 4 expression in one of the VIP+ non-light activated clusters (cluster 6). Cluster 6 also contains the highest number of dark cells among all SCN clusters, suggesting a potential inhibitory signal which may be coming from GABAergic retinal projections.
In conclusion, we report on an essential role for NPAS4 in mediating the response of the SCN to light. Moreover, we show that the transcriptional response to light varies significantly with the duration of light exposure and is not the same in every SCN neuropeptidergic cell-type, suggesting that there are diverse transcriptional responses in both neuronal and non-neuronal cells in the SCN. Taken together, our findings suggest that our knowledge of the SCN response to light may still be open to much revision. The use of single-nucleus techniques to study the SCN will continue to provide insights into its complexity.
STAR METHODS
RESOURCE AVAILABILITY
Lead Contact:
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Joseph Takahashi (joseph.takahashi@utsouthwestern.edu).
Materials Availability:
This study did not generate unique reagents.
Data and Code Availability:
RNA-seq, Chip-seq, and Single nuclei-seq data have been deposited at GEO and are publicly available as of the date of publication. The DOI is listed in the key resources table.
All original code has been deposited at Github and is publicly available as of the date of publication. DOIs are listed in the key resources table.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Goat anti c-Fos | Santa Cruz Biotechnology | Cat# sc-52-G; RRID: AB_2629503 |
| Donkey anti-Goat IgG (H+L) Cross-Adsorbed Secondary Antibody, Alexa Fluor 546 | Thermo Fisher Scientific | Cat# A-11056; RRID: AB_2534103 |
| Rabbit polyclonal anti-H3K4me3 | Diagenode | Cat# pAb-003-050; RRID: AB_2616052 |
| Rabbit polyclonal anti-H3K27ac | Diagenode | Cat# C15410196; RRID: AB_2637079 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| TO-PRO-3 Iodide (642/661) | Thermofisher | Cat# T3605 |
| cOmplete mini protease Inhibitor cocktail | Roche (Sigma) | Cat# 11836153001 |
| PhosSTOP | Roche (Sigma) | Cat# 4906845001 |
| Tetrodotoxin 1 mg | Tocris (FISHER) | Cat# 1078 |
| DL-AP5 Sodium salt 50 mg | Tocris (FISHER) | Cat# 36-935-0R |
| Triptolide | Sigma | Cat# 645900-1 MG |
| Actinomycin D | Sigma | Cat# A1410-2MG |
| Critical Commercial Assays | ||
| RNAscope® Fluorescent Multiplex Kit v2 | ACD bio / Bio-Techne | Cat# 323100 |
| RNAscope® Target Retrieval Reagents | ACD bio / Bio-Techne | Cat# 322000 |
| TSA Plus Fluorescein Evaluation Kit | Perkin Elmer | NEL741E001KT |
| TrueMicroChIP | Diagenode | Cat. No. C01010130 (AB-002-0016) |
| DynabeadTM mRNA DIRECTTM Micro kit | Ambion | Cat# 61021 |
| Chromium™ Single Cell 3’ Library & Gel Bead Kit v2 | 10x Genomics | Cat# PN-120237 |
| Deposited Data | ||
| Chip-seq, RNA-seq, Single nuclei-seq | This paper | GEO: GSE148252 |
| MM10/GRCm38p6 for Mouse | UCSC genome browser | http://genome.ucsc.edu |
| NPAS4 ChIP-seq | (Kim et al., 2010) | GEO: GSE21161 |
| NPAS4 ChIP-seq | (Brigidi et al., 2019) | GEO: GSE127793 |
| Experimental Models: Organisms/Strains | ||
| Mouse: Npas4−/− | M.E. Greenberg, Harvard Medical School, Boston MA | Lin et al., 2008 |
| Mouse: Npas4fx/fx | M.E. Greenberg, Harvard Medical School, Boston MA | Lin et al., 2008 |
| Mouse: Camk2a::iCreBAC | (Casanova et al., 2001) | MGI: 2181426 |
| Oligonucleotides | ||
| Primer: Npas4−/−, WT forward: TGCTGAGAGGGTCTTTCTATGCG; WT reverse: TCCAGGTAGTGCTGCCACAATG; Null forward: AAAGACCCCAACGAGAAGCG; Null reverse: GCAAGTAAAACCTCTACAAATGTGG | (Lin et al., 2008) | N/A |
| Primer: Npas4fx/fx, forward: CCCTGCCCTTCTAATCAGAC; reverse: GGCATTGTTCTTTCTGTCTCC | (Lin et al., 2008) | N/A |
| Primer: Camk2a::iCreBAC, forward: TCTGATGAAGTCAGGAAGAACC; reverse: GAGATGTCCTTCACTCTGATTC | (Casanova et al., 2001) | N/A |
| Software and Algorithms | ||
| Code for data pre-processing, clustering, differential gene expression analysis, and peak calling | (Berto et al., 2021) and this paper | https://doi.org/zenodo.5111722 |
| Code for the Wavelet Analysis | This paper | https://doi.org/10.5281/zenodo.5106445 |
| BWA (v0.7.16a) | (Li and Durbin, 2009) | https://github.com/lh3/bwa/releases |
| SAMtools (v1.3.1) | (Li et al., 2009) | http://samtools.sourceforge.net/ |
| SPP | (Kharchenko et al., 2008) | https://github.com/hms-dbmi/spp |
| Picard | http://broadinstitute.github.io/picard/ | |
| MACS2 (v2.1.1) | (Zhang et al., 2008) | https://github.com/macs3-project/MACS/tree/master/MACS2 |
| STAR (v2.7.1a) | (Dobin et al., 2013) | https://github.com/easybuilders/easybuild-easyconfigs/pull/8388 |
| RseqQC | (Wang et al., 2012) | https://github.com/MonashBioinformaticsPlatform/RSeQC |
| Htseq (v0.9.1) | (Anders et al., 2015) | https://github.com/simon-anders/htseq |
| ToppGene | (Chen et al., 2009) | https://toppgene.cchmc.org/ |
| bedtools (v2.25.0) | Quinlan and Hall, 2010 | https://bedtools.readthedocs.io/en/latest/ |
| Chipseeker | (Yu et al., 2015) | https://www.bioconductor.org/packages/release/bioc/html/ChIPseeker.html |
| HOMER (v4.9.1) | (Heinz et al., 2010) | http://homer.ucsd.edu/homer/ |
| CellRanger v3.0.1 | 10x Genomics | https://support.10xgenomics.com/single-cell-gene-expression/software/overview/welcome |
| FASTQC v0.11.5 | Babraham Bioinformatics | https://www.bioinformatics.babraham.ac.uk/projects/fastqc |
| UMI-tools v0.5.4 | (Kulkarni et al., 2019);(Smith et al., 2017) | https://github.com/CGATOxford/UMI-tools/releases |
| Subread package (v1.6.2) | (Liao et al., 2014) | http://subread.sourceforge.net/ |
| Seurat v3 R analysis pipeline | (Butler et al., 2018) | https://satijalab.org/seurat/ |
| Uniform Manifold Approximation and Projection (UMAP) | (Becht et al., 2018); (Kulkarni et al., 2019) | https://umap-learn.readthedocs.io/en/latest/ |
| ClockLab | Actimetrics | https://www.actimetrics.com/products/clocklab/ |
| ARTool | (Wobbrock et al., 2011) | http://depts.washington.edu/acelab/proj/art/index.html |
| Prism | GraphPad Inc. | https://www.graphpad.com/scientific-software/prism/ |
| Other | ||
| smFISH probe: Mm-Per1-C2 | ACD bio / Bio-Techne | Cat#438751-C2 |
| smFISH probe: Mm-Fos-C2 | ACD bio / Bio-Techne | Cat#316921-C2 |
| smFISH probe: Egr1-C2 | ACD bio / Bio-Techne | Cat#423371-C2 |
| smFISH probe: Mm-Avp | ACD bio / Bio-Techne | Cat#401391 |
| smFISH probe: Mm-Vip-C3 | ACD bio / Bio-Techne | Cat#415961-C3 |
| smFISH probe: Mm-Gfap-C3 | ACD bio / Bio-Techne | Cat#313211-C3 |
| smFISH probe: Mm-Map2-C3 | ACD bio / Bio-Techne | Cat#431151-C3 |
| smFISH probe: Mm-Cck-C3 | ACD bio / Bio-Techne | Cat#402271-C3 |
| smFISH probe: Mm-Prok2 | ACD bio / Bio-Techne | Cat#447941 |
EXPERIMENTAL MODEL AND SUBJECT DETAILS
All mice were maintained at the University of Texas Southwestern Medical Center in accordance with Institutional Animal Care and Use Committee (IACUC) guidelines (APN: 2015–100925). Mice were supplied with food and water ad lib under 12-h light:12-h dark cycle with controlled temperature and humidity. For studies involving wildtype mice, 7–8 week-old C57BL/6J males were used. For studies including genetically manipulated mice, 7–8 week-old males and females were used. Npas4fx/fx mice and Npas4−/− were generous gifts from Dr. Michael E. Greenberg (Lin et al., 2008). Npas4−/− (C57BL/6N, N6 backcross) were maintained by heterozygous intercross Npas4−/+ littermates and produced all experimental mice: Npas4+/+(wildtype control), Npas4+/− (heterozygote) and Npas4−/− (homozygous null). Camk2a::iCreBAC (CamiCre) (MGI:2181426) were kindly provided by Dr. G. Schutz (Casanova et al., 2001). Cre genotyping was performed as previously described (Izumo et al., 2014). Npas4fx/fx mice (C57BL/6N backcross) were crossed to CamiCre to produce Npas4fx/fx, and CamiCre+; Npas4fx /fx (Npas4 cKO) mice. The genotyping for Npas4−/− was performed as previously described (Lin et al., 2008).
METHOD DETAILS
FOS immunohistochemistry and cell counting
Mice with and without light exposure were anesthetized with 10 μl Euthasol at the end of the light stimulation. Mice were transcardially perfused with 0.1 M phosphate saline buffer (PBS, pH7.4) followed by 4% paraformaldehyde in 0.1 M PBS (PFA, pH 7.4). The brains were post-fixed in 4% PFA at 4°C overnight and then cryoprotected in 30% sucrose in PBS. The brains were embedded in Tissue-Tek® OCT (optimum cutting temperature) solution, sectioned at 40 μm on a cryostat (Leica CM3050) and processed with routine immunohistochemistry. In brief, the free-floating slices were rinsed with PBS twice and incubated with blocking buffer (10% donkey serum, Jackson ImmuneResearch, 0.3% Triton X-100 in PBS) for one hour at room temperature (RT) to reduce nonspecific antibody binding and increase antibody penetration. The sections were then incubated with primary antibody (goat anti c-Fos, 1:50, SC-52, Santa Cruz) in blocking buffer at RT for 2 days. The brain sections were washed 3 times with PBS and incubated with secondary antibody (Donkey anti Goat Alexa 546, 1:1000, Cat# A-11056, Thermofisher Scientific) in 10% Donkey serum, PBS for 1 hr. After 3 PBS washes, the sections were incubated with TO-PRO-3 Iodide (642/661) (1:100, Cat no. T3605, Thermofisher Scientific) in PBS for 1 hr at RT. The sections were then rinsed with PBS and mounted with Prolong Gold Anti-Fade Reagent with DAPI on fluorescence-free glass slides (Thermofisher). Confocal fluorescence images were acquired on a LSM550 microscope (Zeiss) using a 20X objective. The Z stack was done at 1.5 μm each slice, step size 4 μm. The FOS immunoreactive cells with identifiable nuclear staining (by TOPRO3) were counted from each z slice and 7–8 sections (40 μm apart) from each mouse (n = 4 for each time point) were counted. The experimenter was blinded to the section identity.
RNA in situ hybridization
At the end of a 1-hr light pulse at CT17, mice were euthanized by cervical dislocation and enucleated to prevent photic signaling to the SCN. A 0.5 cm thick coronal brain block containing the SCN was embedded in OCT and flash frozen in dry-ice cooled isopentane and then stored at −80°C. Within 3 days, the brain block was sectioned at 12 μm thickness on a cryostat (Leica CM3050) and mounted on Superfrost Plus Microscope Slides (ThermoFisher Scientific). RNAscope® in situ hybridizations were performed on the sections containing the SCN following the manufacturer’s instructions with RNAscope® Multiplex Fluorescent Reagent Kit v2 assay for fresh frozen sample (Cat No. 323100, Advanced Cell Diagnostics). The sections were imaged with a Zeiss LSM880 confocal microscope at 20X magnification. The probes used in the study are RNAscope® Probe -Mm-Per1-C2 (438751-C2), RNAscope® Probe -Mm-Fos-C2 (316921-C2), RNAscope® Probe- Mm-Egr1-C2 (423371-C2), RNAscope® Probe- Mm-Avp (Cat No. 401391), RNAscope® Probe- Mm-Vip-C3 (415961-C3), RNAscope® Probe- Mm-Gfap-C3 (313211-C3), RNAscope® Probe- Mm-Map2-C3 (431151-C3), RNAscope® Probe- Mm-Cck-C3 (402271-C3) and RNAscope® Probe- Mm-Prok2 (447941).
RNA-seq library preparation and sequencing
At the end of behavior tests, mice were euthanized by cervical dislocation and enucleated to prevent photic signaling to the SCN. Pairs of SCN were dissected on ice and then snap frozen in liquid nitrogen. The samples were stored under −80°C and processed within a week. For each sample, we pooled 6 pairs of SCNs. For each time condition, there were 2–4 replicates. The mRNA was directly isolated from SCN tissue using a Dynabead™ mRNA DIRECT™ Micro kit (Ambion) according to the manufacturer’s instructions. All mRNA samples were examined for quantity and quality by NanoDrop and Bioanalyzer 2100 (Agilent). The libraries were constructed following TruSeq Stranded mRNA Sample preparation guide (Illumina) and single end sequencing was performed on a HiSeq 2500 platform (Illumina) at the McDermott Sequencing Core at UT Southwestern.
ChIP-seq library preparation and sequencing
At the end of behavior tests, mice were euthanized by cervical dislocation and enucleated to prevent photic signaling to the SCN. Pairs of SCN were dissected out and fixed in freshly made 0.5 ml 1% Formaldehyde containing 100 mM NaCl, 1mM EDTA, 0.5 mM EGTA, and 50 mM HEPES, pH 8.0 for 8 mins at RT. Crosslinking was stopped by adding Glycine to reach a final 125 mM concentration for 5 mins at RT. The fixed SCN was washed twice in 0.8 ml ice-cold PBS containing protease inhibitor (cOmplete, mini protease Inhibitor cocktail, Cat#11836153001, Roche), 1 mM PMSF, 10 mM NaF, and PhosSTOP (Cat# 4906845001, Roche). In the end, the SCN was stored under −80°C and processed within a week. Each ChIP-seq library was made from two pairs of SCN. Libraries were constructed following a TrueMicroChIP-kit (Cat. No. C01010130, Diagenode) according to the manufacturer’s instructions. In brief, the tissue was homogenized in lysis buffer (10 mM EDTA, pH 8.0, 50 mM Tris-HCl, pH8.0, 1% SDS, cOmplete and PhosSTOP) on ice. DNA fragmentation was performed in a Snap-Cap microTUBE with AFA fiber with an S2 (Covaris) at 4°C (4 cycles to reach 100–600bp fragment range). The targeted DNA was immunoprecipitated with 1 μg H3K4me3 or H3K27ac antibodies (C15410003–50, C15410196, Diagenode). The amount of immunoprecipitated DNA was measured with Quant-iT PicoGreen dsDNA assay kit (Thermofisher). Single-end sequencing was performed on a HiSeq 2500 platform (Illumina) at the McDermott Sequencing Core at UT Southwestern.
ChIP-seq mapping and quality checks
FASTQ-files for ChIP-seq were processed following official ENCODE guidelines (https://github.com/ENCODE-DCC/chip-seq-pipeline2). H3K27ac/H3K4me3 reads were aligned to the mouse mm10 reference genome using BWA (v0.7.16a) (Li and Durbin, 2009). Reads were filtered for MAPQ > 10 and duplicates were removed using SAMtools (v1.3.1) (Li et al., 2009). Quality metrices were inferred using SPP (Kharchenko et al., 2008) and Picard (http://broadinstitute.github.io/picard/). MACS2 (v2.1.1) (Zhang et al., 2008) was used to identify significant peaks using input DNA without ChIP as reference.
Single-nucleus sequencing library preparation
We adopted methods developed to study activity-induced single-cell RNA-seq gene expression (Hrvatin et al., 2018). At the end of behavior tests, mice were anesthetized with 10 μl Euthasol and enucleated. Mice were transcardially perfused for 5 min with 5 ml ice-cold choline solution (per liter, 2.1 g NaHCO3, 2.16 g glucose, 0.172 g NaH2PO4·H2O, 7.5 mM MgCl2·6H2O, 2.5 mM KCl, 10 mM HEPES, 15.36 g choline chloride, 2.3 g ascorbic acid and 0.34 g pyruvic acid) which contained a small-molecule cocktail including 1 μM TTX, 100 μM AP-V, 10 μM triptolide and 5 μg actinomycin D per milliliter (Hrvatin et al., 2018). Pairs of SCN were dissected on ice and then snap frozen in liquid nitrogen. The samples were stored under −80°C and processed within a week. Nuclei were extracted following a modified protocol based on CG00055 SamplePrep Demonstrated Protocol – Mouse Neural Tissue RevA (10X Genomics). 10X Genomics libraries were constructed following CG000075 Chromium Single Cell 3 V2 Ref Cards Rev C (Cat No.PN-120237,10X Genomics). We performed 5 replicates for each light condition (dark and 1-hr light stimulation) and 10 libraries were prepared and sequenced in total.
QUANTIFICATION AND STATISTICAL ANALYSIS
For statistical comparisons apart from sequencing data, an unpaired Student’s t test was applied to compare the mean between two independent groups. One-way ANOVA followed by Tukey’s multiple comparisons was used to compare multiple independent groups. **** p < 0.0001, *** p < 0.001, ** p < 0.01, * p < 0.05. Data were analyzed using Prism (GraphPad Software).
RNA-seq mapping and quantification
Reads were aligned to the mouse mm10 reference genome using STAR (v2.7.1a) (Dobin et al., 2013). For each sample, a BAM file including mapped and unmapped reads that spanned splice junctions was produced. Secondary alignment and multi-mapped reads were further removed using in-house scripts. Only uniquely mapped reads were retained for further analyses. Quality control metrics were performed using RseqQC (Wang et al., 2012) using the m10 gene model provided. These steps include number of reads after multiple-step filtering, ribosomal RNA reads depletion, and defining reads mapped to exons, UTRs, and intronic regions. Picard tool was implemented to refine the QC metrics (http://broadinstitute.github.io/picard/). Genecode annotation for mm10 (version M21) was used as reference alignment annotation and downstream quantification. Gene level expression was calculated using Htseq (v0.9.1) (Anders et al., 2015) using intersection-strict mode by exon. Counts were calculated based on protein-coding genes from the annotation file.
Differential gene expression analysis
Low expressed genes were filtered using a per time-point approach with RPKM >= 0.5 in all samples in one or the other time-point as a filter to keep genes. Differential expression was performed in R using DESeq2 (Love et al., 2014). We estimated log2 fold changes and P-values. P-values were adjusted for multiple comparisons using a Benjamini-Hochberg correction (FDR). Differentially expressed genes were defined as those with FDR < 0.05 and abs(log2(fold change)) ≥ 0.3. A fold change cutoff was used to increase the stringency of the results in line with previous publications (Araujo et al., 2015; Harrington et al., 2020). Visualizations were done in R. For light stimulation in wildtype mice, each condition with different duration of light stimulation was compared to the CT 17 dark control. We also performed comparisons between CT17 and CT23 dark controls to detect differentially expressed genes that changed as a result of circadian phase and not due to the 6 hr light exposure. See Table S1.
Functional enrichment
The functional annotation of differentially expressed and co-expressed genes was performed using ToppGene (Chen et al., 2009). We used GO and KEGG databases. Pathways containing between 5 and 2000 genes were retained. A Benjamini-Hochberg FDR was applied as a correction for multiple comparisons. Significant categories were filtered for FDR < 0.05.
Gene set enrichment
Gene set enrichment was performed using a Fisher’s exact test in R with the following parameters: alternative = “greater”, conf.level = 0.95. We reported odds ratios (OR) and Benjamini-Hochberg adjusted p-value (FDR).
Enhancer-promoter calling and motif analysis
Consensus peaks between Light and Dark H3K27ac/H3K4me3 were calculated using bedtools (v2.25.0) (Quinlan and Hall, 2010) with -f 0.2 as parameter. Peak annotation and visualizations were done in R using Chipseeker library (Yu et al., 2015). Active promoters were identified by annotating consensus H3K27ac/H3K4me3 peaks within 3000 bp to the nearest transcription start sites (TSS). Active enhancers were identified by overlapping consensus H3K27ac/H3K4me3 peaks with +/− 100kb window of the genes of interest not including the promoter regions. HOMER (v4.9.1) (Heinz et al., 2010) was used for motif analysis using findMotifsGenome.pl. We assessed de novo/known motif discovery on both promoter/enhancer specific peaks.
Independent NPAS4 ChIP-seq analysis
Previously published NPAS4 ChIP-seq datasets were downloaded from GEO with accession numbers GSE21161 and GSE127793 (Brigidi et al., 2019; Kim et al., 2010). Rat NPAS4 peaks were lifted to mm10 using liftOver. Peak annotation was performed in R using Chipseeker library (Yu et al., 2015). Enrichment analysis was performed using Fisher exact test as described below.
10X Genomics single-nucleus sequencing analysis
Raw sequencing data were acquired from the North Texas Genome Center at the University of Texas at Arlington and McDermott NDA Next Generation Sequencing Core at University of Texas Southwestern Medical Center in the form of binary base call (BCL) files from both NOVAseq and NEXTseq platforms. BCL files were then de-multiplexed with the 10X Genomics i7 index (used during library preparation) using Illumina’s bcl2fastq v2.19.1 and mkfastq command from 10X Genomics CellRanger v3.0.1 tools. Extracted paired-end fastq files (26 bp long R1 - cell barcode and UMI sequence information, 124 bp long R2 - transcript sequence information) were checked for read quality using FASTQC v0.11.5 [FastQC, Babraham Bioinformatics, URL: https://www.bioinformatics.babraham.ac.uk/projects/fastqc]. R1 reads were then used to estimate and identify real cells using whitelist command from UMI-tools v0.5.4 (Kulkarni et al., 2019; Smith et al., 2017). A whitelist of cell-barcodes (putative real cells) and R2 fastq files were later used to extract reads corresponding to real cells only (excluding sequence information representing empty beads, doublets, low quality/degrading cells, etc.) using extract command from UMI-tools v0.5.4 (Smith et al., 2017). This step also appends the cell-barcode and UMI sequence information from R1 to read names in R2 fastq file. Extracted R2 reads were then aligned to reference genome (MM10/GRCm38p6 for Mouse) from UCSC genome browser (Kent et al., 2002) using STAR aligner v2.5.2b (Dobin et al., 2013) allowing up to 5 mismatches. Uniquely mapped reads were then assigned to genes (exons + intron) using featureCounts program from Subread package (v1.6.2) (Liao et al., 2014). Assigned reads sorted and indexed using Samtools v1.6 (Li et al., 2009) were then used to generate raw expression UMI count tables using count command from UMI-tools v0.5.4 (Smith et al., 2017) program. This raw expression matrix contains cells as rows and genes as columns and can be further used for downstream analysis such as normalization, clustering, differentially expressed genes, etc. Around 40% reads were assigned and the mean of assigned reads per cell is 20,732. In total, 144,806 cells were sequenced, of which 144,582 survived quality filters and the average number of detected genes per cell was about 1100.
Clustering analysis
Raw single-nucleus RNA-seq UMI count data for a total of 144,806 detected cells was used for clustering analysis using Seurat v3 R analysis pipeline (Butler et al., 2018) [https://satijalab.org/seurat/]. First, cells with more than 10,000 molecules (nUMI per cell) and cells with more than 10% mitochondrial content were filtered out to discard potential doublets and degrading cells. Also, genes from mitochondrial and Y chromosomes were removed. This dataset referred to as primary filtered dataset has 144,582 cells. Post filtering, the raw UMI counts from primary filtered dataset were used for log-normalization and scaled using a factor of 10,000 and regressed to covariates such as number of UMI per cells, percent mitochondrial content per cell and batch as described in Seurat analysis pipeline (Butler et al., 2018) [https://satijalab.org/seurat/]. Further identifying top 2000 variable genes, data was used to calculate principal components (PCs). Using Jackstraw analysis, 50 statistically significant PCs were used to identify clusters within the data using original Louvain algorithm as described in Seurat analysis pipeline (Butler et al., 2018) [https://satijalab.org/seurat/] followed by visualizing the clusters with Uniform Manifold Approximation and Projection (UMAP) (Becht et al., 2018; Kulkarni et al., 2019) in two dimensions. Genes enriched in each cluster against rest of the cells (FDR < 0.05 and log2(fold change) ≥ 0.3) were identified as described in Seurat analysis pipeline (Butler et al., 2018) [https://satijalab.org/seurat/]. Further genes corresponding to each cluster were used to identify the cell-type by correlating to genes expressed in single-cell data already published (Campbell et al., 2017; Hrvatin et al., 2018; Wen et al., 2020). Cell-types were assigned to clusters based on statistically significant enrichment of gene sets using Fisher exact test.
“Pseudobulk” differential gene expression analysis
For differentially expressed genes, cells corresponding to samples with or without light stimulation were grouped within each cellular population of SCN neurons (such as Avp+ or Vip+ or Cck+). Genes with altered expression upon light stimulation were then identified within each SCN neuronal population accounting for averaged expression differences. Wilcoxon test from the Seurat v3 (Butler et al., 2018) analysis pipeline was used to identify significant gene expression changes (FDR < 0.05 and log2(FC) ≥ 0.3) within each category SCN neurons instead of identified cellular clusters.
Analysis of circadian behavior
Circadian period in constant darkness (DD) or constant light (LL) was estimated using the Chi-square periodogram in ClockLab (ActiMetrics, IL) over 10-day intervals. Phase shifts in response to a 6-hr light pulse were determined from the steady-state phase of activity onsets for 14 days preceding and 14 days after the light pulse. Because the locomotor activity records and activity onsets of Npas4−/− mice were more variable than wild-type mice, we used a wavelet-based method to estimate phase (Leise et al., 2013). Activity onsets were determined by maximal overlap discrete wavelet transform (MODWT) based on a Matlab (Mathworks) code created by T.L. Leise (Leise et al., 2013). Using 4-tap Daubechies mother wavelet (MODWT), 15-min sampled actogram data were decomposed into 7 details (D1 ~ D7) associated with particular period ranges (e.g., D3: 2 ~ 4 h) and an approximation representing all other remaining coarse scales. An onset time on a particular day was defined as one of several local peaks in a D3 detail time series around the actual onset. The detected position of onset was dependent on the percentage of sensitivity, which controls the threshold for local peak detection. Because the amplitude of the actogram was not constant during the long period of recording time (~ 5 months per mouse), different values of sensitivity were applied during the time intervals before and after each light pulse within the same actogram. The optimal value of sensitivity was determined as one that gave the smallest product of two root mean square error (RMSE) values from the two lines which fit onsets before and after each light pulse. With this procedure, the first three days after the light pulse were skipped (due to transients and the unstable nature of the activity right after the light pulse) to improve the fitting score. Moreover, outliers (points more than two standard deviations from the fitting lines) were automatically suppressed by assigning them lower weights using iteratively reweighted least squares (“fitlm” function with robust linear regression option in Matlab).
The phase of the light pulse was calculated from the circadian period (slope of the fitting line) and the time difference between CT 12 (phase of activity onset) and the beginning of the light pulse. The magnitude of the phase shift was determined as the time difference between the intersection points of the two lines with the day of the light pulse. Phase response curves (PRCs) to light were plotted with data meeting two conditions: 1) two R-squared values of the line fitting onsets before and after the light pulses were both greater than 0.5, and 2) two RMSE values of the line fitting onsets before and after the light pulses were both smaller than 3.0 h. The total number of light pulses plotted was 195 (126 for Npas4+/+ and 69 for Npas4−/− mice). Raw actogram data from all mice and the source code for the time series analysis with MODWT are available in https://github.com/Takahashi-Lab-UTSW/Actogram-Wavelet-Analysis.
For statistical comparisons between Npas4+/+ and Npas4−/− PRCs, the data on the graphs were grouped every CT 3 hours (CT0, CT3, CT6, …, CT21) separately in Npas4+/+ and Npas4−/−, with CT0 representing the data between CT 22.5 and CT 1.5. To assess the effects of CT and genotype on PRCs, data were analyzed with a nonparametric two-way ANOVA with interaction using ARTool (Wobbrock et al., 2011). To make a complete block design, the PRC was smoothed by the kernel density estimation with Gaussian kernel function and bandwidth 1 (Härdle, 1990). Missing data were imputed by uniform random numbers ranging from minimum and maximum values of the smoothed curve within each CT group. Aligned Rank Transform (ART) nonparametric ANOVA analysis revealed a significant interaction between CT and genotype (p-value = 2.2×10−16). To determine the nature of the interaction, pairwise comparison with interaction (R phia package) showed a significant effect of genotype on the phaseshift (p < 0.05) over most of the pairs except CT0 - CT3/ CT21, CT6 - CT3/18, and CT12 - CT9/CT15. R code for the ART analysis and the results are available in the above github repository.
Supplementary Material
Table S1. Differentially expressed gene list from light pulse time course. Related to Figure 1.
Table S2. Differentially expressed gene lists between cells from light and dark conditions by cell type. Related to Figure 4.
Table S3. Gene enrichment in 12 SCN clusters. Related to Figure 5.
Table S4. Differentially expressed gene list between cells from light and dark conditions by peptidergic cell type. Related to Figure 5.
Table S5. Differentially expressed gene list between Npas4+/+ and Npas4−/− after a 1-hr light pulse. Related to Figure 6.
Highlights:
Light activates diverse patterns of gene expression in the SCN of the hypothalamus.
Light-responsive genes in the SCN show enrichment for NPAS4 regulatory elements.
Light-responsive SCN neurons also shown increased expression of NPAS4 target genes.
Mice lacking Npas4 have reduced phase shifting to light and longer circadian rhythms.
ACKNOWLEDGMENTS
We thank Gokhul Kilaru for bioinformatic support. We thank the University of Texas Southwestern Medical Center Neuroscience Imaging Core Facility and the McDermott Center for Human Growth and Development Next Generation Sequencing Core. We also thank the North Texas Genome Center at the University of Texas at Arlington. This research was supported by grants from the NIH (NS106657) and Howard Hughes Medical Institute to J.S.T; as well as the James S. McDonnell Foundation 21st Century Science Initiative in Understanding Human Cognition - Scholar Award (220020467), the Chan Zuckerberg Initiative, an advised fund of Silicon Valley Community Foundation (HCA-A-1704-01747), and grants from the NIH (DC014702, MH102603) to G.K.
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
INCLUSION AND DIVERSITY STATEMENT
We worked to ensure sex balance in the selection of non-human subjects. While citing references scientifically relevant for this work, we also actively worked to promote gender balance in our reference list. The author list of this paper includes contributors from the location where the research was conducted who participated in the data collection, design, analysis, and/or interpretation of the work.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Albers HE, Walton JC, Gamble KL, McNeill J.K.t., and Hummer DL (2017). The dynamics of GABA signaling: Revelations from the circadian pacemaker in the suprachiasmatic nucleus. Front Neuroendocrinol 44, 35–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen CN, Nitabach MN, and Colwell CS (2017). Membrane Currents, Gene Expression, and Circadian Clocks. Cold Spring Harb Perspect Biol 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, Pyl PT, and Huber W (2015). HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araujo DJ, Anderson AG, Berto S, Runnels W, Harper M, Ammanuel S, Rieger MA, Huang HC, Rajkovich K, Loerwald KW, et al. (2015). FoxP1 orchestration of ASD-relevant signaling pathways in the striatum. Genes Dev 29, 2081–2096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becht E, McInnes L, Healy J, Dutertre CA, Kwok IWH, Ng LG, Ginhoux F, and Newell EW (2018). Dimensionality reduction for visualizing single-cell data using UMAP. Nat Biotechnol. [DOI] [PubMed] [Google Scholar]
- Berto S, Fontenot MR, Seger S, Ayhan F, Caglayan E, Kulkarni A, Douglas C, Tamminga CA, Lega BC, and Konopka G (2021). Gene-expression correlates of the oscillatory signatures supporting human episodic memory encoding. Nat Neurosci 24, 554–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloodgood BL, Sharma N, Browne HA, Trepman AZ, and Greenberg ME (2013). The activity-dependent transcription factor NPAS4 regulates domain-specific inhibition. Nature 503, 121–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brancaccio M, Edwards MD, Patton AP, Smyllie NJ, Chesham JE, Maywood ES, and Hastings MH (2019). Cell-autonomous clock of astrocytes drives circadian behavior in mammals. Science 363, 187–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brancaccio M, Patton AP, Chesham JE, Maywood ES, and Hastings MH (2017). Astrocytes Control Circadian Timekeeping in the Suprachiasmatic Nucleus via Glutamatergic Signaling. Neuron 93, 1420–1435 e1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brigidi GS, Hayes MGB, Delos Santos NP, Hartzell AL, Texari L, Lin PA, Bartlett A, Ecker JR, Benner C, Heinz S, et al. (2019). Genomic Decoding of Neuronal Depolarization by Stimulus-Specific NPAS4 Heterodimers. Cell 179, 373–391 e327. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Butler A, Hoffman P, Smibert P, Papalexi E, and Satija R (2018). Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol 36, 411–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell JN, Macosko EZ, Fenselau H, Pers TH, Lyubetskaya A, Tenen D, Goldman M, Verstegen AM, Resch JM, McCarroll SA, et al. (2017). A molecular census of arcuate hypothalamus and median eminence cell types. Nat Neurosci 20, 484–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casanova E, Fehsenfeld S, Mantamadiotis T, Lemberger T, Greiner E, Stewart AF, and Schutz G (2001). A CamKIIalpha iCre BAC allows brain-specific gene inactivation. Genesis 31, 37–42. [DOI] [PubMed] [Google Scholar]
- Chen J, Bardes EE, Aronow BJ, and Jegga AG (2009). ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res 37, W305–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, Hanna J, Lodato MA, Frampton GM, Sharp PA, et al. (2010). Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci U S A 107, 21931–21936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, and Gingeras TR (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doi M, Cho S, Yujnovsky I, Hirayama J, Cermakian N, Cato AC, and Sassone-Corsi P (2007). Light-inducible and clock-controlled expression of MAP kinase phosphatase 1 in mouse central pacemaker neurons. J Biol Rhythms 22, 127–139. [DOI] [PubMed] [Google Scholar]
- Etchegaray JP, Machida KK, Noton E, Constance CM, Dallmann R, Di Napoli MN, DeBruyne JP, Lambert CM, Yu EA, Reppert SM, et al. (2009). Casein kinase 1 delta regulates the pace of the mammalian circadian clock. Mol Cell Biol 29, 3853–3866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez DC, Chang YT, Hattar S, and Chen SK (2016). Architecture of retinal projections to the central circadian pacemaker. Proc Natl Acad Sci U S A 113, 6047–6052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson EM, Purger D, Mount CW, Goldstein AK, Lin GL, Wood LS, Inema I, Miller SE, Bieri G, Zuchero JB, et al. (2014). Neuronal activity promotes oligodendrogenesis and adaptive myelination in the mammalian brain. Science 344, 1252304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginty DD, Kornhauser JM, Thompson MA, Bading H, Mayo KE, Takahashi JS, and Greenberg ME (1993). Regulation of CREB phosphorylation in the suprachiasmatic nucleus by light and a circadian clock. Science 260, 238–241. [DOI] [PubMed] [Google Scholar]
- Golombek DA, and Rosenstein RE (2010). Physiology of circadian entrainment. Physiol Rev 90, 1063–1102. [DOI] [PubMed] [Google Scholar]
- Greenberg ME, Siegfried Z, and Ziff EB (1987). Mutation of the c-fos gene dyad symmetry element inhibits serum inducibility of transcription in vivo and the nuclear regulatory factor binding in vitro. Mol Cell Biol 7, 1217–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guler AD, Ecker JL, Lall GS, Haq S, Altimus CM, Liao HW, Barnard AR, Cahill H, Badea TC, Zhao H, et al. (2008). Melanopsin cells are the principal conduits for rod-cone input to non-image-forming vision. Nature 453, 102–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao N, Whitelaw ML, Shearwin KE, Dodd IB, and Chapman-Smith A (2011). Identification of residues in the N-terminal PAS domains important for dimerization of Arnt and AhR. Nucleic Acids Res 39, 3695–3709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Härdle W (1990). Applied nonparametric regression (Cambridge England ; New York: Cambridge University Press; ). [Google Scholar]
- Harrington AJ, Bridges CM, Berto S, Blankenship K, Cho JY, Assali A, Siemsen BM, Moore HW, Tsvetkov E, Thielking A, et al. (2020). MEF2C Hypofunction in Neuronal and Neuroimmune Populations Produces MEF2C Haploinsufficiency Syndrome-like Behaviors in Mice. Biol Psychiatry 88, 488–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastings MH, Maywood ES, and Brancaccio M (2018). Generation of circadian rhythms in the suprachiasmatic nucleus. Nat Rev Neurosci 19, 453–469. [DOI] [PubMed] [Google Scholar]
- Hatori M, Gill S, Mure LS, Goulding M, O’Leary DD, and Panda S (2014). Lhx1 maintains synchrony among circadian oscillator neurons of the SCN. Elife 3, e03357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattar S, Liao HW, Takao M, Berson DM, and Yau KW (2002). Melanopsin-containing retinal ganglion cells: architecture, projections, and intrinsic photosensitivity. Science 295, 1065–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, and Glass CK (2010). Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 38, 576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honrado GI, Johnson RS, Golombek DA, Spiegelman BM, Papaioannou VE, and Ralph MR (1996). The circadian system of c-fos deficient mice. J Comp Physiol A 178, 563–570. [DOI] [PubMed] [Google Scholar]
- Hrvatin S, Hochbaum DR, Nagy MA, Cicconet M, Robertson K, Cheadle L, Zilionis R, Ratner A, Borges-Monroy R, Klein AM, et al. (2018). Single-cell analysis of experience-dependent transcriptomic states in the mouse visual cortex. Nat Neurosci 21, 120–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izumo M, Pejchal M, Schook AC, Lange RP, Walisser JA, Sato TR, Wang X, Bradfield CA, and Takahashi JS (2014). Differential effects of light and feeding on circadian organization of peripheral clocks in a forebrain Bmal1 mutant. Elife 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JR, Simon T, Lones L, and Herzog ED (2018). SCN VIP Neurons Are Essential for Normal Light-Mediated Resetting of the Circadian System. J Neurosci 38, 7986–7995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalsbeek A, Palm IF, La Fleur SE, Scheer FA, Perreau-Lenz S, Ruiter M, Kreier F, Cailotto C, and Buijs RM (2006). SCN outputs and the hypothalamic balance of life. J Biol Rhythms 21, 458–469. [DOI] [PubMed] [Google Scholar]
- Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, and Haussler D (2002). The human genome browser at UCSC. Genome Res 12, 996–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kharchenko PV, Tolstorukov MY, and Park PJ (2008). Design and analysis of ChIP-seq experiments for DNA-binding proteins. Nat Biotechnol 26, 1351–1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilduff TS, Vugrinic C, Lee SL, Milbrandt JD, Mikkelsen JD, O’Hara BF, and Heller HC (1998). Characterization of the circadian system of NGFI-A and NGFI-A/NGFI-B deficient mice. J Biol Rhythms 13, 347–357. [DOI] [PubMed] [Google Scholar]
- Kim TK, Hemberg M, Gray JM, Costa AM, Bear DM, Wu J, Harmin DA, Laptewicz M, Barbara-Haley K, Kuersten S, et al. (2010). Widespread transcription at neuronal activity-regulated enhancers. Nature 465, 182–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koike N, Yoo SH, Huang HC, Kumar V, Lee C, Kim TK, and Takahashi JS (2012). Transcriptional architecture and chromatin landscape of the core circadian clock in mammals. Science 338, 349–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornhauser JM, Mayo KE, and Takahashi JS (1996). Light, immediate-early genes, and circadian rhythms. Behav Genet 26, 221–240. [DOI] [PubMed] [Google Scholar]
- Kornhauser JM, Nelson DE, Mayo KE, and Takahashi JS (1990). Photic and circadian regulation of c-fos gene expression in the hamster suprachiasmatic nucleus. Neuron 5, 127–134. [DOI] [PubMed] [Google Scholar]
- Kornhauser JM, Nelson DE, Mayo KE, and Takahashi JS (1992). Regulation of jun-B messenger RNA and AP-1 activity by light and a circadian clock. Science 255, 1581–1584. [DOI] [PubMed] [Google Scholar]
- Kulkarni A, Anderson AG, Merullo DP, and Konopka G (2019). Beyond bulk: a review of single cell transcriptomics methodologies and applications. Curr Opin Biotechnol 58, 129–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacoste B, Comin CH, Ben-Zvi A, Kaeser PS, Xu X, Costa Lda F, and Gu C (2014). Sensory-related neural activity regulates the structure of vascular networks in the cerebral cortex. Neuron 83, 1117–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazzerini Ospri L, Prusky G, and Hattar S (2017). Mood, the Circadian System, and Melanopsin Retinal Ganglion Cells. Annu Rev Neurosci 40, 539–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeGates TA, Fernandez DC, and Hattar S (2014). Light as a central modulator of circadian rhythms, sleep and affect. Nat Rev Neurosci 15, 443–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leise TL, Indic P, Paul MJ, and Schwartz WJ (2013). Wavelet meets actogram. J Biol Rhythms 28, 62–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, and Durbin R (2009). Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, and Genome Project Data Processing, S. (2009). The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, and Shi W (2014). featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930. [DOI] [PubMed] [Google Scholar]
- Lin Y, Bloodgood BL, Hauser JL, Lapan AD, Koon AC, Kim TK, Hu LS, Malik AN, and Greenberg ME (2008). Activity-dependent regulation of inhibitory synapse development by Npas4. Nature 455, 1198–1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, and Anders S (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowrey PL, and Takahashi JS (2011). Genetics of circadian rhythms in mammalian model organisms. Advances in Genetics 74, 175–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majidi SP, Reddy NC, Moore MJ, Chen H, Yamada T, Andzelm MM, Cherry TJ, Hu LS, Greenberg ME, and Bonni A (2019). Chromatin Environment and Cellular Context Specify Compensatory Activity of Paralogous MEF2 Transcription Factors. Cell Rep 29, 2001–2015 e2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardinly AR, Spiegel I, Patrizi A, Centofante E, Bazinet JE, Tzeng CP, Mandel-Brehm C, Harmin DA, Adesnik H, Fagiolini M, et al. (2016). Sensory experience regulates cortical inhibition by inducing IGF1 in VIP neurons. Nature 531, 371–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng QJ, Logunova L, Maywood ES, Gallego M, Lebiecki J, Brown TM, Sladek M, Semikhodskii AS, Glossop NRJ, Piggins HD, et al. (2008). Setting clock speed in mammals: the CK1 epsilon tau mutation in mice accelerates circadian pacemakers by selectively destabilizing PERIOD proteins. Neuron 58, 78–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mieda M, Okamoto H, and Sakurai T (2016). Manipulating the Cellular Circadian Period of Arginine Vasopressin Neurons Alters the Behavioral Circadian Period. Curr Biol 26, 2535–2542. [DOI] [PubMed] [Google Scholar]
- Morin LP (2013). Neuroanatomy of the extended circadian rhythm system. Exp Neurol 243, 4–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris ME, Viswanathan N, Kuhlman S, Davis FC, and Weitz CJ (1998). A screen for genes induced in the suprachiasmatic nucleus by light. Science 279, 1544–1547. [DOI] [PubMed] [Google Scholar]
- Myung J, Hong S, DeWoskin D, De Schutter E, Forger DB, and Takumi T (2015). GABA-mediated repulsive coupling between circadian clock neurons in the SCN encodes seasonal time. Proc Natl Acad Sci U S A 112, E3920–3929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Numano R, Yamazaki S, Umeda N, Samura T, Sujino M, Takahashi R, Ueda M, Mori A, Yamada K, Sakaki Y, et al. (2006). Constitutive expression of the Period1 gene impairs behavioral and molecular circadian rhythms. Proc Natl Acad Sci U S A 103, 3716–3721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono D, Honma KI, Yanagawa Y, Yamanaka A, and Honma S (2019). GABA in the suprachiasmatic nucleus refines circadian output rhythms in mice. Commun Biol 2, 232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J, Zhu H, O’Sullivan S, Ogunnaike BA, Weaver DR, Schwaber JS, and Vadigepalli R (2016). Single-Cell Transcriptional Analysis Reveals Novel Neuronal Phenotypes and Interaction Networks Involved in the Central Circadian Clock. Front Neurosci 10, 481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pittendrigh CS, and Daan S (1976). A Functional Analysis of Circadian Pacemakers in Nocturnal Rodents. I. The Stability and Lability of Spontaneous Frequency. J Comp Physiol 106, 223–252. [Google Scholar]
- Ploski JE, Park KW, Ping J, Monsey MS, and Schafe GE (2010). Identification of plasticity-associated genes regulated by Pavlovian fear conditioning in the lateral amygdala. J Neurochem 112, 636–650. [DOI] [PubMed] [Google Scholar]
- Porterfield VM, and Mintz EM (2009). Temporal patterns of light-induced immediate-early gene expression in the suprachiasmatic nucleus. Neurosci Lett 463, 70–73. [DOI] [PubMed] [Google Scholar]
- Porterfield VM, Piontkivska H, and Mintz EM (2007). Identification of novel light-induced genes in the suprachiasmatic nucleus. BMC Neurosci 8, 98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, and Hall IM (2010). BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralph MR, Foster RG, Davis FC, and Menaker M (1990). Transplanted suprachiasmatic nucleus determines circadian period. Science 247, 975–978. [DOI] [PubMed] [Google Scholar]
- Ramamoorthi K, Fropf R, Belfort GM, Fitzmaurice HL, McKinney RM, Neve RL, Otto T, and Lin Y (2011). Npas4 regulates a transcriptional program in CA3 required for contextual memory formation. Science 334, 1669–1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoro IM, and Walsh K (1991). Natural and synthetic DNA elements with the CArG motif differ in expression and protein-binding properties. Mol Cell Biol 11, 6296–6305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma N, Pollina EA, Nagy MA, Yap EL, DiBiase FA, Hrvatin S, Hu L, Lin C, and Greenberg ME (2019). ARNT2 Tunes Activity-Dependent Gene Expression through NCoR2-Mediated Repression and NPAS4-Mediated Activation. Neuron 102, 390–406 e399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siepka SM, Yoo SH, Park J, Song W, Kumar V, Hu Y, Lee C, and Takahashi JS (2007). Circadian mutant Overtime reveals F-box protein FBXL3 regulation of cryptochrome and period gene expression. Cell 129, 1011–1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith T, Heger A, and Sudbery I (2017). UMI-tools: modeling sequencing errors in Unique Molecular Identifiers to improve quantification accuracy. Genome Res 27, 491–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonoda T, Li JY, Hayes NW, Chan JC, Okabe Y, Belin S, Nawabi H, and Schmidt TM (2020). A noncanonical inhibitory circuit dampens behavioral sensitivity to light. Science 368, 527–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiegel I, Mardinly AR, Gabel HW, Bazinet JE, Couch CH, Tzeng CP, Harmin DA, and Greenberg ME (2014). Npas4 regulates excitatory-inhibitory balance within neural circuits through cell-type-specific gene programs. Cell 157, 1216–1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi JS (2017). Transcriptional architecture of the mammalian circadian clock. Nat Rev Genet 18, 164–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tischkau SA, Mitchell JW, Tyan SH, Buchanan GF, and Gillette MU (2003). Ca2+/cAMP response element-binding protein (CREB)-dependent activation of Per1 is required for light-induced signaling in the suprachiasmatic nucleus circadian clock. J Biol Chem 278, 718–723. [DOI] [PubMed] [Google Scholar]
- Travnickova-Bendova Z, Cermakian N, Reppert SM, and Sassone-Corsi P (2002). Bimodal regulation of mPeriod promoters by CREB-dependent signaling and CLOCK/BMAL1 activity. Proc Natl Acad Sci U S A 99, 7728–7733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitaterna MH, Ko CH, Chang AM, Buhr ED, Fruechte EM, Schook A, Antoch MP, Turek FW, and Takahashi JS (2006). The mouse Clock mutation reduces circadian pacemaker amplitude and enhances efficacy of resetting stimuli and phase-response curve amplitude. Proc Natl Acad Sci U S A 103, 9327–9332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Symul L, Yeung J, Gobet C, Sobel J, Luck S, Westermark PO, Molina N, and Naef F (2018). Circadian clock-dependent and -independent posttranscriptional regulation underlies temporal mRNA accumulation in mouse liver. Proc Natl Acad Sci U S A 115, E1916–E1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Wang S, and Li W (2012). RSeQC: quality control of RNA-seq experiments. Bioinformatics 28, 2184–2185. [DOI] [PubMed] [Google Scholar]
- Welsh DK, Takahashi JS, and Kay SA (2010). Suprachiasmatic nucleus: cell autonomy and network properties. Annu Rev Physiol 72, 551–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen S, Ma D, Zhao M, Xie L, Wu Q, Gou L, Zhu C, Fan Y, Wang H, and Yan J (2020). Spatiotemporal single-cell analysis of gene expression in the mouse suprachiasmatic nucleus. Nat Neurosci 23, 456–467. [DOI] [PubMed] [Google Scholar]
- Wobbrock JO, Findlater L, Gergle D, and Higgins JJ (2011). The aligned rank transform for nonparametric factorial analyses using only anova procedures. In Proceedings of the SIGCHI Conference on Human Factors in Computing Systems (Vancouver, BC, Canada: Association for Computing Machinery), pp. 143–146. [Google Scholar]
- Yu G, Wang LG, and He QY (2015). ChIPseeker: an R/Bioconductor package for ChIP peak annotation, comparison and visualization. Bioinformatics 31, 2382–2383. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. (2008). Model-based analysis of ChIP-Seq (MACS). Genome Biol 9, R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Differentially expressed gene list from light pulse time course. Related to Figure 1.
Table S2. Differentially expressed gene lists between cells from light and dark conditions by cell type. Related to Figure 4.
Table S3. Gene enrichment in 12 SCN clusters. Related to Figure 5.
Table S4. Differentially expressed gene list between cells from light and dark conditions by peptidergic cell type. Related to Figure 5.
Table S5. Differentially expressed gene list between Npas4+/+ and Npas4−/− after a 1-hr light pulse. Related to Figure 6.
Data Availability Statement
RNA-seq, Chip-seq, and Single nuclei-seq data have been deposited at GEO and are publicly available as of the date of publication. The DOI is listed in the key resources table.
All original code has been deposited at Github and is publicly available as of the date of publication. DOIs are listed in the key resources table.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Goat anti c-Fos | Santa Cruz Biotechnology | Cat# sc-52-G; RRID: AB_2629503 |
| Donkey anti-Goat IgG (H+L) Cross-Adsorbed Secondary Antibody, Alexa Fluor 546 | Thermo Fisher Scientific | Cat# A-11056; RRID: AB_2534103 |
| Rabbit polyclonal anti-H3K4me3 | Diagenode | Cat# pAb-003-050; RRID: AB_2616052 |
| Rabbit polyclonal anti-H3K27ac | Diagenode | Cat# C15410196; RRID: AB_2637079 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| TO-PRO-3 Iodide (642/661) | Thermofisher | Cat# T3605 |
| cOmplete mini protease Inhibitor cocktail | Roche (Sigma) | Cat# 11836153001 |
| PhosSTOP | Roche (Sigma) | Cat# 4906845001 |
| Tetrodotoxin 1 mg | Tocris (FISHER) | Cat# 1078 |
| DL-AP5 Sodium salt 50 mg | Tocris (FISHER) | Cat# 36-935-0R |
| Triptolide | Sigma | Cat# 645900-1 MG |
| Actinomycin D | Sigma | Cat# A1410-2MG |
| Critical Commercial Assays | ||
| RNAscope® Fluorescent Multiplex Kit v2 | ACD bio / Bio-Techne | Cat# 323100 |
| RNAscope® Target Retrieval Reagents | ACD bio / Bio-Techne | Cat# 322000 |
| TSA Plus Fluorescein Evaluation Kit | Perkin Elmer | NEL741E001KT |
| TrueMicroChIP | Diagenode | Cat. No. C01010130 (AB-002-0016) |
| DynabeadTM mRNA DIRECTTM Micro kit | Ambion | Cat# 61021 |
| Chromium™ Single Cell 3’ Library & Gel Bead Kit v2 | 10x Genomics | Cat# PN-120237 |
| Deposited Data | ||
| Chip-seq, RNA-seq, Single nuclei-seq | This paper | GEO: GSE148252 |
| MM10/GRCm38p6 for Mouse | UCSC genome browser | http://genome.ucsc.edu |
| NPAS4 ChIP-seq | (Kim et al., 2010) | GEO: GSE21161 |
| NPAS4 ChIP-seq | (Brigidi et al., 2019) | GEO: GSE127793 |
| Experimental Models: Organisms/Strains | ||
| Mouse: Npas4−/− | M.E. Greenberg, Harvard Medical School, Boston MA | Lin et al., 2008 |
| Mouse: Npas4fx/fx | M.E. Greenberg, Harvard Medical School, Boston MA | Lin et al., 2008 |
| Mouse: Camk2a::iCreBAC | (Casanova et al., 2001) | MGI: 2181426 |
| Oligonucleotides | ||
| Primer: Npas4−/−, WT forward: TGCTGAGAGGGTCTTTCTATGCG; WT reverse: TCCAGGTAGTGCTGCCACAATG; Null forward: AAAGACCCCAACGAGAAGCG; Null reverse: GCAAGTAAAACCTCTACAAATGTGG | (Lin et al., 2008) | N/A |
| Primer: Npas4fx/fx, forward: CCCTGCCCTTCTAATCAGAC; reverse: GGCATTGTTCTTTCTGTCTCC | (Lin et al., 2008) | N/A |
| Primer: Camk2a::iCreBAC, forward: TCTGATGAAGTCAGGAAGAACC; reverse: GAGATGTCCTTCACTCTGATTC | (Casanova et al., 2001) | N/A |
| Software and Algorithms | ||
| Code for data pre-processing, clustering, differential gene expression analysis, and peak calling | (Berto et al., 2021) and this paper | https://doi.org/zenodo.5111722 |
| Code for the Wavelet Analysis | This paper | https://doi.org/10.5281/zenodo.5106445 |
| BWA (v0.7.16a) | (Li and Durbin, 2009) | https://github.com/lh3/bwa/releases |
| SAMtools (v1.3.1) | (Li et al., 2009) | http://samtools.sourceforge.net/ |
| SPP | (Kharchenko et al., 2008) | https://github.com/hms-dbmi/spp |
| Picard | http://broadinstitute.github.io/picard/ | |
| MACS2 (v2.1.1) | (Zhang et al., 2008) | https://github.com/macs3-project/MACS/tree/master/MACS2 |
| STAR (v2.7.1a) | (Dobin et al., 2013) | https://github.com/easybuilders/easybuild-easyconfigs/pull/8388 |
| RseqQC | (Wang et al., 2012) | https://github.com/MonashBioinformaticsPlatform/RSeQC |
| Htseq (v0.9.1) | (Anders et al., 2015) | https://github.com/simon-anders/htseq |
| ToppGene | (Chen et al., 2009) | https://toppgene.cchmc.org/ |
| bedtools (v2.25.0) | Quinlan and Hall, 2010 | https://bedtools.readthedocs.io/en/latest/ |
| Chipseeker | (Yu et al., 2015) | https://www.bioconductor.org/packages/release/bioc/html/ChIPseeker.html |
| HOMER (v4.9.1) | (Heinz et al., 2010) | http://homer.ucsd.edu/homer/ |
| CellRanger v3.0.1 | 10x Genomics | https://support.10xgenomics.com/single-cell-gene-expression/software/overview/welcome |
| FASTQC v0.11.5 | Babraham Bioinformatics | https://www.bioinformatics.babraham.ac.uk/projects/fastqc |
| UMI-tools v0.5.4 | (Kulkarni et al., 2019);(Smith et al., 2017) | https://github.com/CGATOxford/UMI-tools/releases |
| Subread package (v1.6.2) | (Liao et al., 2014) | http://subread.sourceforge.net/ |
| Seurat v3 R analysis pipeline | (Butler et al., 2018) | https://satijalab.org/seurat/ |
| Uniform Manifold Approximation and Projection (UMAP) | (Becht et al., 2018); (Kulkarni et al., 2019) | https://umap-learn.readthedocs.io/en/latest/ |
| ClockLab | Actimetrics | https://www.actimetrics.com/products/clocklab/ |
| ARTool | (Wobbrock et al., 2011) | http://depts.washington.edu/acelab/proj/art/index.html |
| Prism | GraphPad Inc. | https://www.graphpad.com/scientific-software/prism/ |
| Other | ||
| smFISH probe: Mm-Per1-C2 | ACD bio / Bio-Techne | Cat#438751-C2 |
| smFISH probe: Mm-Fos-C2 | ACD bio / Bio-Techne | Cat#316921-C2 |
| smFISH probe: Egr1-C2 | ACD bio / Bio-Techne | Cat#423371-C2 |
| smFISH probe: Mm-Avp | ACD bio / Bio-Techne | Cat#401391 |
| smFISH probe: Mm-Vip-C3 | ACD bio / Bio-Techne | Cat#415961-C3 |
| smFISH probe: Mm-Gfap-C3 | ACD bio / Bio-Techne | Cat#313211-C3 |
| smFISH probe: Mm-Map2-C3 | ACD bio / Bio-Techne | Cat#431151-C3 |
| smFISH probe: Mm-Cck-C3 | ACD bio / Bio-Techne | Cat#402271-C3 |
| smFISH probe: Mm-Prok2 | ACD bio / Bio-Techne | Cat#447941 |