Summary
Human Accelerated Regions (HARs) are the fastest-evolving regions of the human genome and many are hypothesized to function as regulatory elements that drive human-specific gene regulatory programs. We interrogate the in vitro enhancer activity and in vivo epigenetic landscape of >3,100 HARs during human neurodevelopment, demonstrating that many HARs appear to act as neurodevelopmental enhancers and that sequence divergence at HARs has largely augmented their neuronal enhancer activity. Furthermore, we demonstrate PPP1R17 as a putative HAR-regulated gene that has undergone remarkable rewiring of its cell-type and developmental expression patterns between non-primates and primates and between non-human primates and humans. Finally, we show that PPP1R17 slows neural progenitor cell-cycle progression– paralleling the cell cycle length increase predominantly seen in primate and especially human neurodevelopment. Overall, our findings establish HARs as key components in rewiring human-specific neurodevelopmental gene regulatory programs, and provide an integrated resource to study enhancer activity of specific HARs.
eTOC Blurb
Human Accelerated Regions (HARs) are the fastest-evolving regions of the human genome. Through interrogating the in vitro enhancer activity and in vivo epigenetic landscapes of >3,100 HARs during human neurodevelopment, we demonstrate that nearly half of all HARs act as neurodevelopmental enhancers. Further we demonstrate PPP1R17 as a putative HAR-regulated gene likely involved in human-specific cortical neurodevelopment.
Introduction
The rapid evolutionary expansion of the human cerebral cortex is considered a vital step that enabled human-specific cognitive function. The increased size of the human brain is associated with a greatly prolonged period of corticogenesis (from about 7 days in mice to >130 days in humans) and increased capacity for neural progenitor cell (NPC) proliferation before neuronal differentiation (Lui et al., 2011; Pollen et al., 2015). Paradoxically, NPC cell cycle length is also lengthened in humans. Whereas mouse NPC cell cycle length ranges from 8-26.5 hours, largely by variation of G1 phase (Arai et al., 2011; Takahashi et al., 1995), NPC cell cycles in embryonic non-human primate can be up to 8 fold longer at 22-63 hours (Betizeau et al., 2013; Kornack and Rakic, 1998), and NPCs in humans show similarly long cell cycle length (Pollen et al., 2015; Subramanian et al., 2017). This lengthening of both cell cycle and neurogenic interval inevitably forces a substantially slowed overall pace of neurological development, generally referred to as a form of heterchrony, or neoteny (Geschwind and Rakic, 2013), which has dramatic implications for maternal and social behavior.
While the genetic basis underlying the evolutionary changes affecting the human brain remain largely unknown (Dehay et al., 2015; Lewitus et al., 2014; Lui et al., 2011; Namba and Huttner, 2017; Sousa et al., 2017), recent comparative genomic studies have identified over 3,100 DNA sequences with potential human-specific functions that have been hypothesized to play a role in this process. These regions, collectively termed human accelerated regions (HARs), are the fastest evolving regions of the human genome (Bird et al., 2007; Bush and Lahn, 2008; Gittelman et al., 2015; Hubisz and Pollard, 2014; Lindblad-Toh et al., 2011; Pollard et al., 2006; Prabhakar et al., 2008). HARs are defined as genomic elements that are highly conserved across non-human mammals, suggesting that they perform essential functions, but also show rapid sequence changes selectively in the human lineage. HARs are mostly noncoding and are enriched for transcription factor binding motifs, suggesting that many HARs potentially function as human-specific regulatory elements that coordinate precise spatiotemporal patterns of gene expression (Capra et al., 2013; Hubisz and Pollard, 2014). Furthermore, prior studies have uncovered some HARs that have putative enhancer activity in human neuronal tissue, with the enhancer activity of some HARs differing from that of their chimpanzee orthologs (Boyd et al., 2015; Capra et al., 2013; Franchini and Pollard, 2017; Kamm et al., 2013; Uebbing et al., 2021), raising the prospect that some HARs may contribute to human-specific brain development.
Since the particular pattern of nucleotide substitution that defines a HAR can arise by many mechanisms—including loss of function, biased gene conversion, or evolutionary drift (Pollard et al., 2006)—a critical challenge remains to define which HAR sequences represent true drivers of bona fide evolutionary changes. Prior studies using Massively Parallel Reporter Assays (MPRA) have attempted to address this problem (Uebbing et al., 2021), but have been limited both in terms of the number of HARs assessed, as well as the inherent size limitations of synthetic oligo-based MPRA assays. For example, synthetic oligo-based MPRA assays are often limited to interrogating intact elements that are smaller than 150bp in length (Uebbing et al., 2021), which represents only a small fraction of all HARs (Figure 1A).
Figure 1. Rewiring of the cis-regulatory enhancer activity of HAR elements in the human-lineage.
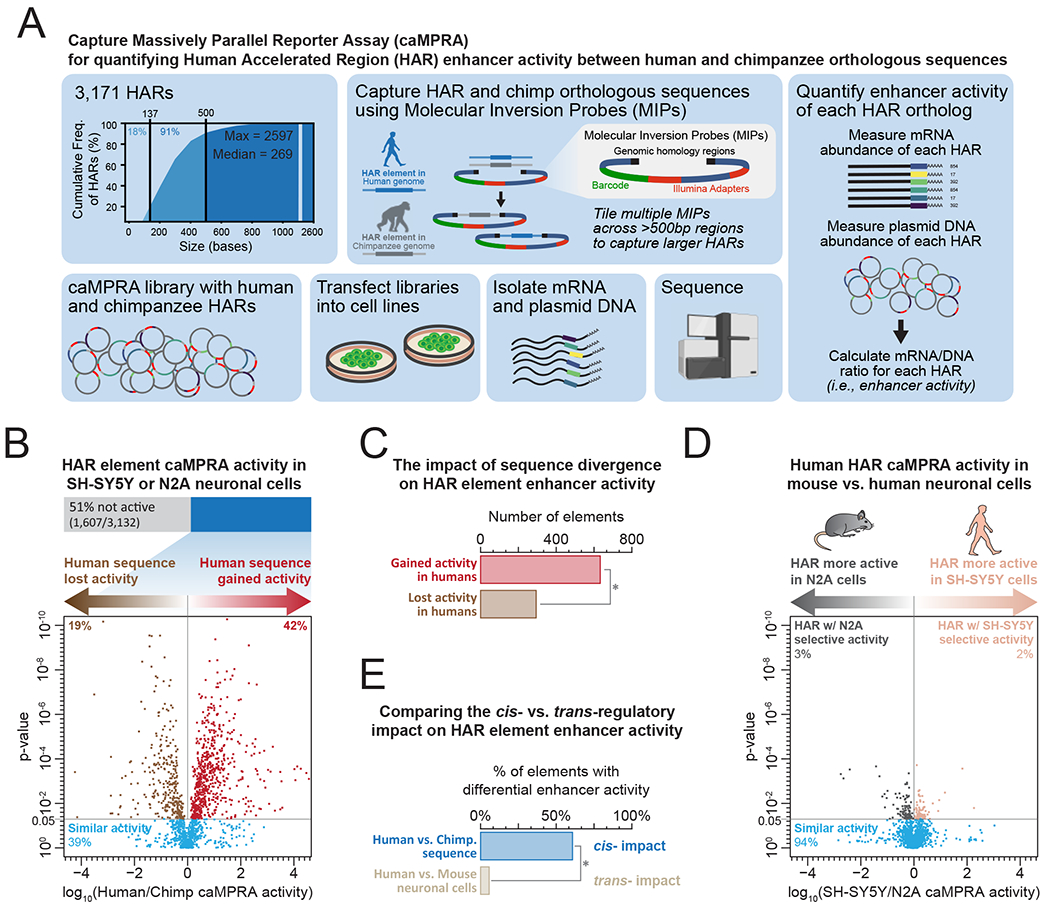
(A) Schematic describing captureMPRA assay.
(B) caMPRA activity of all HARs and their chimpanzee orthologs in human neural cells. P-value calculated using two-sided paired T-test.
(C) Bar graph showing the number of HAR elements with increased or reduced activity of the human versus chimpanzee ortholog. * signifies p-value <0.001 (2-proportions z-test)
(D) caMPRA activity of all HARs and their chimpanzee orthologs in human neural cells versus mouse neural cells. P-value calculated using two-sided paired T-test.
(E) Bar graph showing the percentage of HAR elements with altered activity of the human versus chimpanzee ortholog versus the percentage of HAR elements with altered activity in human versus mouse neural cells. * signifies p-value <0.001 (2-proportions z-test)
To systematically identify the subset of HARs most likely to be contributing to recent evolution of the human cerebral cortex, we analyzed the in vitro and in vivo gene regulatory activity of over 3,100 HARs across multiple human and mouse cell types and tissues. To overcome the size limitations of standard MPRA assays, we developed and applied an approach called CaptureMPRA, which leverages barcoded molecular inversion probes (MIPs) to capture target sequences of ~500bp in length, thereby enabling us to interrogate entire HAR elements and their surrounding DNA on individual probes. We interrogated human-chimp differences in HAR enhancer function using this CaptureMPRA approach, and integrated this data with in vivo histone marks and chromatin accessibility at HARs in human fetal neural cells to identify HARs with putative regulatory function in guiding human-specific brain development. Finally, we developed an easily searchable online resource (the HARHub) consisting of this data and published datasets of common and rare human HAR sequence variation (Doan et al., 2016) to serve as a resource for the scientific community. Overall, our findings define many HARs that play key roles in neuronal gene regulatory programs, with nearly half of all HARs showing reproducible chromatin accessibility and enhancer activity in neural cells and tissue.
Results
HAR elements demonstrate pervasive neuronal enhancer activity in vitro
To globally assess the regulatory potential of HARs in human neurodevelopment in a high-throughput manner, we first developed a new Massively Parallel Reporter Assay (MPRA) which we refer to as CaptureMPRA (caMPRA). CaptureMPRA leverages barcoded molecular inversion probes (MIPs) to capture target sequences, in this case HARs, and then simultaneously evaluates enhancer activity at these sequences in a single in vitro experiment (Figure 1A). By using MIPs to capture target sequences of ~500bp in length (Data S1), CaptureMPRA overcomes much of the length restrictions associated with the standard MPRA design, enabling the systematic evaluation of HARs, which are on average 269 bp in length. We then applied the caMPRA approach to all 3,171 reported HARs (Bird et al., 2007; Bush and Lahn, 2008; Gittelman et al., 2015; Hubisz and Pollard, 2014; Lindblad-Toh et al., 2011; Pollard et al., 2006; Prabhakar et al., 2008), as well as their paired chimpanzee orthologs.
Using 10,690 barcoded MIPs targeting flanking regions surrounding 3,171 HARs (Data S1), or tiling HARs >500bp in length, we successfully captured 3,132 HARs and their paired chimpanzee orthologs from human and chimpanzee genomic DNA, respectively. To test the enhancer activity of these elements, we used two well-described neural cell lines, one human (SH-SY5Y) and one mouse (Neuro2A/N2A), to model human as well as non-human neural developmental activity in vitro (Figure S1). Overall, 49% of the elements demonstrated reproducible and significant enhancer activity in human neural cells (Figure 1B and Data S2), demonstrating the pervasive neural precursor-like enhancer activity of these HARs. Furthermore, HARs with enhancer activity were enriched for binding motifs of transcriptional regulators known to play vital roles in cortical neurogenesis (RBPJ, TBR2 [EOMES], PAX6, ATOH1, and TEAD) (Figure S2A), suggesting that many HARs act as neurodevelopmental enhancers.
Human-specific rewiring of the cis-regulatory architecture of HARs
To identify whether human-specific sequence divergence at HAR elements is associated with altered neural enhancer activity, we compared the enhancer activity of HAR elements to their chimpanzee orthologs (Figure 1B). Overall, 61% of the HARs with caMPRA enhancer activity showed significant alterations in enhancer activity between the human and chimpanzee orthologs. Of note, human-specific sequence divergence appears to be more commonly associated with increased enhancer activity, with 68% of elements showing differential enhancer activity exhibiting greater activity for the human than chimpanzee ortholog (Figure 1C). This indicates extensive evolutionary rewiring of the cis-regulatory activity of HAR elements in the human lineage.
To disentangle the contribution of cis- versus trans-regulatory effects on the rewiring of HAR element enhancer activity, we also compared the enhancer activity of each element between the human (SH-SY5Y) and mouse (Neuro2a/N2A) neural cell lines (Figure 1D). This revealed that over 94% of elements showed similar enhancer activity in human and mouse cells, indicating that differences in the neural trans-regulatory compartment between human and mouse do not significantly alter the overall enhancer activity of each HAR element. Together, these findings demonstrate that the human-specific rewiring of HAR element enhancer activity is predominantly driven by cis-regulatory variation (Figure 1E).
HARs demonstrate in vivo enhancer features during human brain development
To determine whether HAR elements demonstrate features of in vivo enhancer activity in the brain, we profiled the chromatin state at HAR elements across diverse developmental cell types and tissues using DNaseI hypersensitivity mapping (DNaseI-seq), as well as chromatin immunoprecipitation (ChIP-seq) for enhancer associated chromatin marks. Using fetal and adult DNaseI-seq data collected as part of the Roadmap Epigenomics Consortium (Roadmap Epigenomics Consortium et al., 2015), we observed that 78% of HARs demonstrated chromatin accessibility in at least one of the human cell or tissue types tested, with 56% demonstrating chromatin accessibility in human brain tissue (Figures 2A and 2B and Data S3). Of note, HAR elements were preferentially accessible in fetal as opposed to adult human brain tissue (Figure 2C and S2B), further implicating HARs as neurodevelopmental regulatory elements involved in human brain development.
Figure 2. HARs are enriched for human neurodevelopmental enhancer elements.
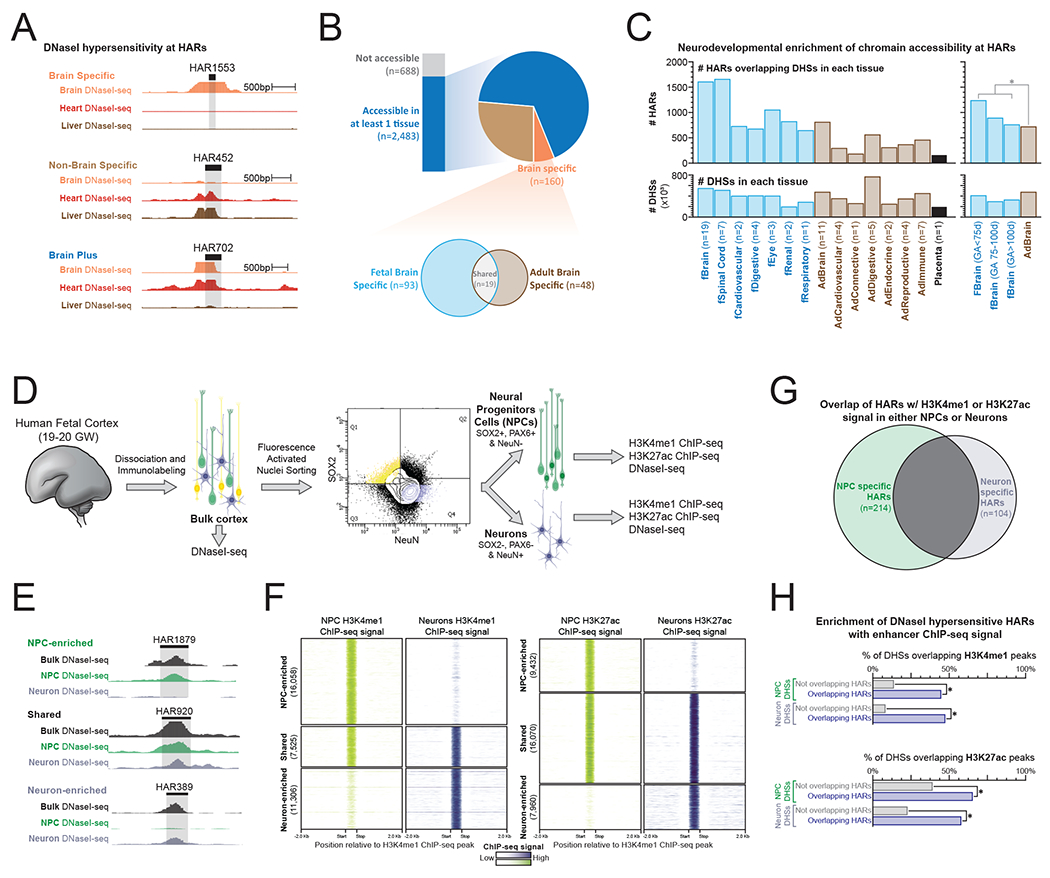
(A) Genomic loci demonstrating tissue-specific DNaseI-seq chromatin accessibility at three separate HAR elements.
(B) (top) The proportion of HAR elements that demonstrate DNaseI hypersensitivity within 79 diverse human tissues based on whether the element is DNaseI hypersensitive in brain tissues only, brain tissues plus other tissues, or non-brain tissues only. (bottom) For HAR elements that are DNaseI hypersensitive in brain tissues only, shown is a Venn diagram of the number of elements that are DNaseI hypersensitive in fetal versus adult brain tissue.
(C) Bar graphs displaying (top) the number of HAR elements overlapping DHSs in each tissue as well as (bottom) the total number of DHSs in each tissue. Fetal tissues are labeled in blue, whereas adult tissues are labeled in brown. * signifies p-value <0.001 (2-proportions z-test)
(D) Schematic for fluorescence-activated nuclear sorting (FANS) of human fetal brain tissue.
(E-F) Genomic loci and density plots demonstrating FANS tissue-specific (E) DNaseI-seq chromatin accessibility and (F) H3K4me1 and H3K27ac ChIP-seq signal at HAR elements.
(G) Venn diagram demonstrating the overlap of HARs with H3K4me1 or H3K27ac signal in either NPCs or Neurons.
(H) Bar plots showing the percentage of NPC and Neuron DHSs that overlap with H3K4me1 or H3K27ac ChIP-seq peaks based on whether the DHS is also a HAR. * signifies p-value <0.001 (2-proportions z-test)
To test the potential for HARs to behave as cell type-specific neurodevelopmental enhancers, we isolated neural progenitor cells (NPCs) and maturing neurons from human fetal brain tissue (gestational week [GW] 19-20) using fluorescence-activated nuclear sorting (FANS) and probed for chromatin features associated with enhancer activity (i.e., H3K4me1, H3K27ac, and chromatin accessibility) (Figure 2D and Figure S3A) (Creyghton et al., 2010). Specifically, nuclei from GW 19-20 human frontal cortex were isolated and labeled using PAX6, SOX2 and NeuN fluorescently-tagged antibodies. Labeled nuclei were subsequently sorted to selectively purify NPCs (SOX2+, PAX6+, NeuN−) and maturing neurons (SOX2−, PAX6−, NeuN+), and these purified populations were then subjected to ChIP-seq and DNaseI-seq. 504 HARs displayed chromatin features consistent with in vivo enhancer activity in GW19-20 NPCs or maturing neurons (evidenced by overlap with H3K4me1 and/or H3K27ac ChIP-seq peaks) (Figures 2G). Notably, HAR elements overlapping DNaseI hypersensitivity sites (DHSs) were significantly enriched for these enhancer marks when compared with other DHSs in NPCs or neurons (Figure 2H). This further demonstrates the importance of HARs as active enhancer elements during human brain development.
The majority of H3K4me1 and H3K27ac ChIP-seq peaks were unique to either NPCs or maturing neurons (Figures 2E–F and Data S4 and Data S5), indicating a substantial change in the regulatory DNA landscape during neuronal differentiation, with only 37% of H3K4me1- or H3K27ac-marked HARs overlapping these chromatin marks in both NPCs and neurons (Figure 2G). Of note, whereas NPC-selective enhancer-marked HARs were enriched for transcription factor (TFs) elements central for cerebral cortical patterning and NPC maintenance, such as LHX family proteins (Hébert and Fishell, 2008; Mangale et al., 2008; Monuki et al., 2001), neuron-selective enhancer-marked HARs were enriched for TF elements implicated in cell fate determination and cell differentiation such as RFX proteins (Figure S3B and S3C). Together, these findings implicate a substantial portion of HARs as regulatory elements involved in human-specific neuronal development.
PPP1R17 is a putative HAR-regulated neurodevelopmental gene
We next integrated our caMPRA, DNaseI-seq, and ChIP-seq HAR data to identify a core set of HARs representing likely neurodevelopmental enhancers that might be contributing to the rapid evolutionary expansion of the human cerebral cortex. Overall, this approach identified 210 HARs with both in vitro and in vivo features consistent with neurodevelopmental enhancer activity (Figure 3A and Data S6). Overlapping this set of 210 putative neurodevelopmental enhancer HARs with chromatin conformation data (HiC) from human fetal cortex (Won et al., 2016) revealed that these 210 HARs were significantly enriched for having long range chromatin interactions with target genes in fetal cortex (Figure S4A), providing further evidence for the in vivo enhancer activity of these HARs in human neurodevelopment.
Figure 3. PPP1R17 is a putative HAR-regulated neurodevelopmental gene.
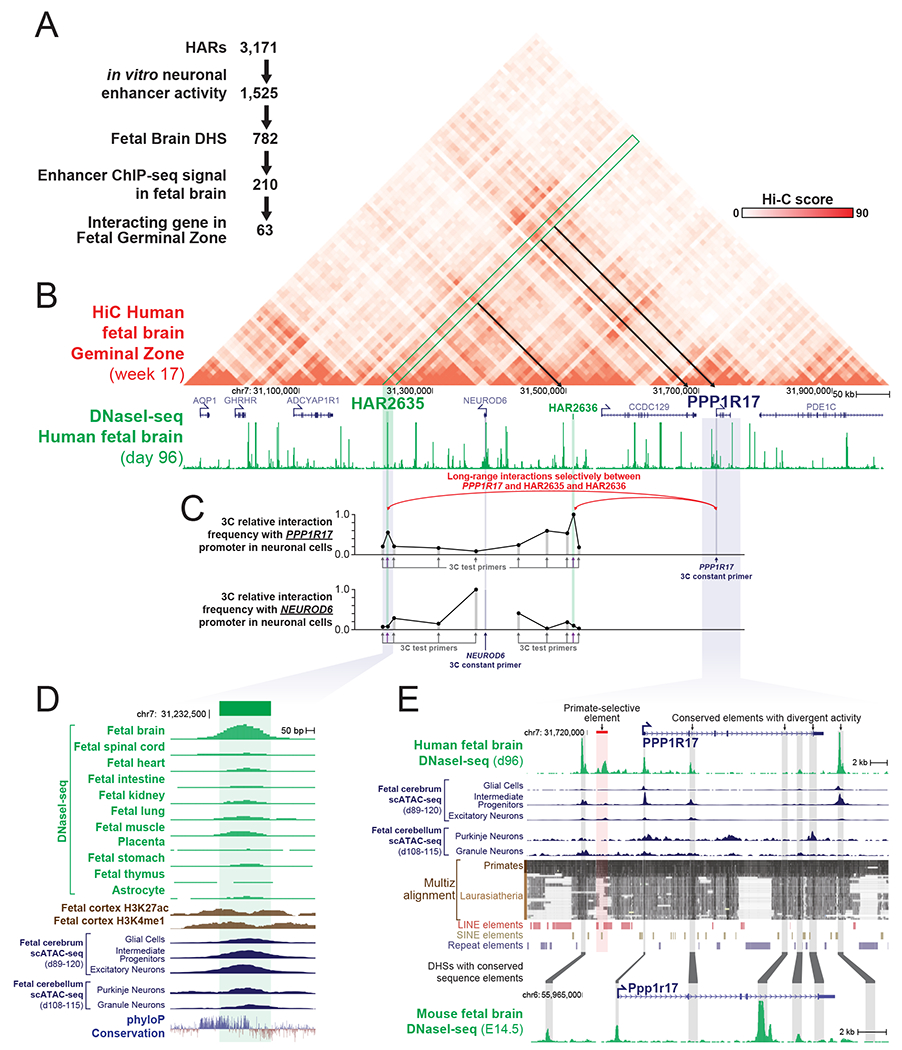
(A) Integration of caMPRA activity, chromatin accessibility, histone modification datasets and long-range chromatin interaction data defines 63 HARs as likely contributors to human brain evolution.
(B) hg19 HiC and DNaseI-seq data at the locus surrounding HAR2635, which is one of the HARs identified in panel A. HAR2635 interacting loci are indicated by the green box and black arrows.
(C) Chromatin Confirmation Capture (3C) signal in SH-SY5Y human neural cells demonstrating long-range chromatin contacts between the PPP1R17 promoter and HAR2635 and HAR3636. Data is maximum signal normalized.
(D) Genomic locus of HAR2635 showing evolutionary conservation and DNaseI-seq signal and single cell ATAC-seq signal across multiple human fetal tissues and adult astrocytes.
(E) Genomic locus of the HAR2635-interacting gene PPP1R17 showing human fetal brain DNaseI-seq signal and single cell ATAC-seq signal in addition to evolutionary conserved elements and the DNaseI-seq signal of contiguous sequence in mouse fetal brain. Red box indicates primate-selective regulatory element. Contiguous sequence elements between human and mouse are indicated by grey boxes.
In total, 63 HARs demonstrated neurodevelopmental enhancer activity in addition to long range chromatin interactions with a target gene in human fetal cortex (Figure 3A). Of these, the interaction between HAR2635 and PPP1R17 stood out, as the PPP1R17 topologically associated domain (TAD) contains a PPP1R17 promoter-proximal primate-selective regulatory element as well as two HARs with strong fetal brain long range chromatin interactions with the PPP1R17 gene promoter (Figures 3B–E and Figure S4). This suggests extensive rewiring of PPP1R17 expression during both non-human primate and human evolution. Targeted 3C interaction analysis confirmed the selective interaction of the promoter of PPP1R17 with both HAR2635 and HAR2636 in cultured neural cells (Figure 3C), suggesting that these two sequences likely play a role in the regulation of PPP1R17 neural expression. In contrast, the nearby gene NEUROD6, which is highly expressed in postmitotic cortical neurons but not in progenitor cells (Bormuth et al., 2013; Han et al., 2018), does not show a strong long-range chromatin interaction with these HARs. Furthermore, while little is known about PPP1R17, it is highly expressed in NPCs in the developing human cortex (Noctor et al., 2004; Pollen et al., 2015; Zhong et al., 2018). It is also a reported repressor of protein phosphatase complexes 1 (PP1) and 2A (PP2A) (Endo, 2012), which are critical for the G1→S transition during the cell cycle – a key step regulating neural progenitor cell cycle length (Wlodarchak and Xing, 2016). This prompted us to further examine the pattern of expression of PPP1R17 in neurodevelopment as well as in other cell types across both non-human primate and human evolution.
Primate-specific gain of PPP1R17 expression in the developing cerebral cortex
To define the cell-type specificity and regional distribution of PPP1R17 expression across mammalian evolution, we examined PPP1R17 expression in different regions of the developing brain in mouse, ferret, rhesus macaque, and human, thereby comparing its expression patterns between lissencephalic mammals, gyrencephalic mammals and primates. PPP1R17 is highly expressed in adult cerebellar Purkinje cells across all four of these mammals (Figure 4A–B), in a pattern that appears to be highly evolutionarily conserved. In contrast to this conserved pattern of cerebellar expression, PPP1R17 shows highly divergent expression in the developing cerebral cortex, where it is highly expressed in primates but not detected in ferret or mice at comparable ages (Figure 4C and Figure S5A). PPP1R17 expression in the prenatal macaque and human cortex is localized to the outer and inner subventricular zones (oSVZ and iSVZ) (Figure 4C) and is excluded from the ventricular zone and cortical plate. PPP1R17 expression co-localizes with TBR2, a marker of intermediate progenitor cells, and KI67, a marker of dividing cells (Figure 4C and S5B). In contrast, PPP1R17 is not expressed in the neocortex in mice or ferrets during development (Figure 4C). This gain of expression in the cortical SVZ in primates parallels several mouse-human differences in chromatin accessibility surrounding the PPP1R17 gene, including the gain of a primate specific element, as well as altered fetal brain chromatin accessibility at several surrounding conserved elements (Figure 3E).
Figure 4. Primate-specific gain of PPP1R17 expression in the developing cortex.
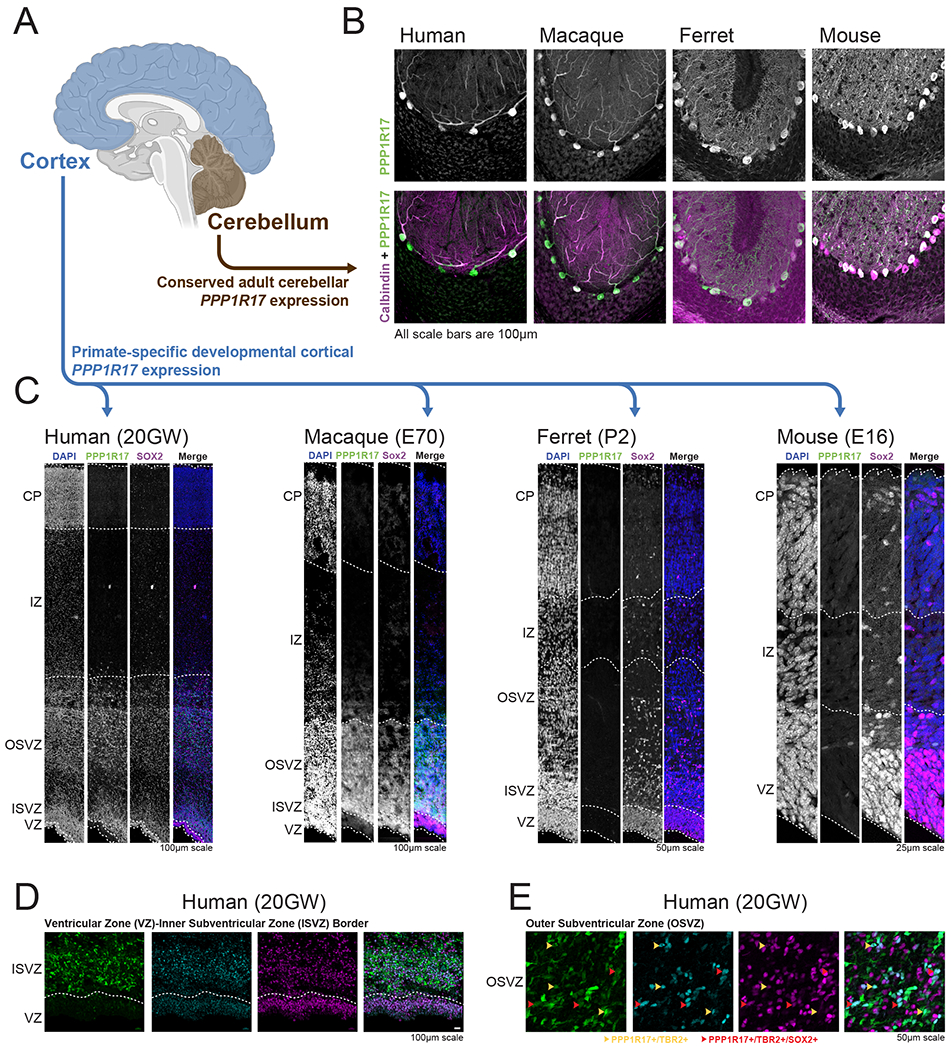
(A) Diagram of brain regions stained in this figure.
(B) Staining of human, macaque, ferret and mouse cerebellum for both PPP1R17 (green) and the Purkinje cell marker Calbindin (pink), demonstrates conserved cerebellar expression of PPP1R17 across these four mammals.
(C) Staining of human, macaque, ferret and mouse developing cortex with PPP1R17 and SOX2 demonstrates cortical expression of PPP1R17 selectively within the two primate species, which appears to be localized to both the outer and inner subventricular cortical germinal zones. VZ, ventricular zone; ISVZ, inner subventricular zone; OFL, outer fiber layer; SP, subplate; CP, cortical plate.
(D-E) Staining of human fetal cortex with PPP1R17, SOX2 and TBR2 demonstrating (D) PPP1R17 expression localized to the subventricular zones, and (E) PPP1R17, SOX2, and TBR2 colocalization.
Human-specific divergence in PPP1R17 cell-type distribution
In addition to primate-non-primate changes in early cortical expression, comparison of RNA-seq data (Zhu et al., 2018) on different regions of the macaque and human brains at various stages of fetal and adult life revealed additional, striking human-macaque differences in PPP1R17 expression (Figure 5A and Figure S6A), suggesting ongoing changes in expression within primates that we confirmed with immunofluorescence analysis (Figure 5B). Specifically, RNA-seq analysis showed highly concordant PPP1R17 expression in adult cerebellum and in fetal cortical NPCs between macaque and humans (Figure 5A and Figure S6A). However, developing cortical expression of PPP1R17 was restricted to fetal development in humans, and closely matched the time course of expression of TBR2. In contrast, in macaques, cortical expression of PPP1R17 was maintained well after neurogenesis ceased, and even well into adulthood, and hence diverged from the pattern of TBR2 expression (Figure 5A and Figure S6A). Notably, the late cortical expression of PPP1R17 in adult macaques is driven not by expression in NPCs or neurons, but rather by expression of PPP1R17 in cortical astrocytes (Figure 5B), indicating a further divergence in the cell-type expression pattern of PPP1R17 in the primate lineage. Together, these results demonstrate a primate-specific innovation in PPP1R17 expression in NPCs, especially intermediate progenitor cells, which coincides with an insertion of a new regulatory element into the promoter-proximal region of PPP1R17. Further changes in the cell-type and developmental expression patterns of PPP1R17 between macaque and human cortex also coincide with sequence changes at two HAR elements that connect to PPP1R17.
Figure 5. Human-specific divergence in PPP1R17 cell-type distribution.
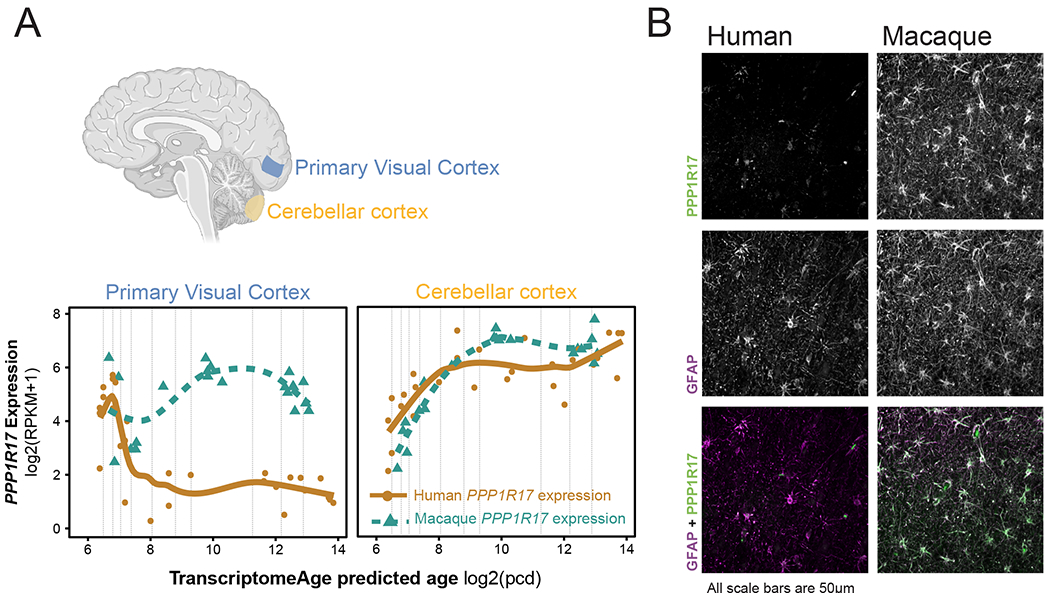
(A) (top) Diagram of brain regions evaluated by RNA-sequencing at various developmental time stages in human and macaque brain development from fetal to adult. (Bottom) PPP1R17 expression in the cerebellum and cortex in macaque and humans as a function of developmental age demonstrates persistent expression of PPP1R17 selectively in macaque well into adulthood.
(B) Staining of human and macaque adult neocortex for both PPP1R17 (green) and the astrocyte marker GFAP (purple) demonstrates predominant expression of PPP1R17 in cortical astrocytes in adult macaque.
PPP1R17 regulates neural progenitor cell-cycle progression
To assess the putative role of PPP1R17 in cell cycle regulation, we evaluated the impact of PPP1R17 misexpression on mouse primary cortical neurospheres. Neurospheres electroporated with a PPP1R17 construct were 36% smaller than those electroporated with a GFP construct (2-way ANOVA, p=0.0003), with no increase in cell death (Figure 6A), implicating PPP1R17 as a regulator of NPC proliferation. To test whether this effect was at least partially mediated through the G1→S transition, we next misexpressed PPP1R17 in mouse neural cells that genetically encoded a G1 phase cell-cycle indicator (Sakaue-Sawano et al., 2008). PPP1R17-misexpressing cells divided at half the speed of control cells (PPP1R17 cells = 68.2 hours vs GFP = 32.7 hours, p<0.0001), and demonstrated a prolonged G1 phase in comparison to GFP controls (48.5% PPP1R17 vs 33.8% GFP, p<0.0001) (Figure 6B and Figure S6B). Together, these results implicate PPP1R17 as a regulator of the G1→S transition and NPC cell cycle progression. Collectively, these results present a potential role for PPP1R17 in regulating neural progenitor cell-cycle progression. These findings parallel the increase in cell-cycle length, neurogenesis duration, and heterochrony, or slowed development, predominately seen in the primate and human lineages.
Figure 6. PPP1R17 regulates cell proliferation and G1 phase in vitro.
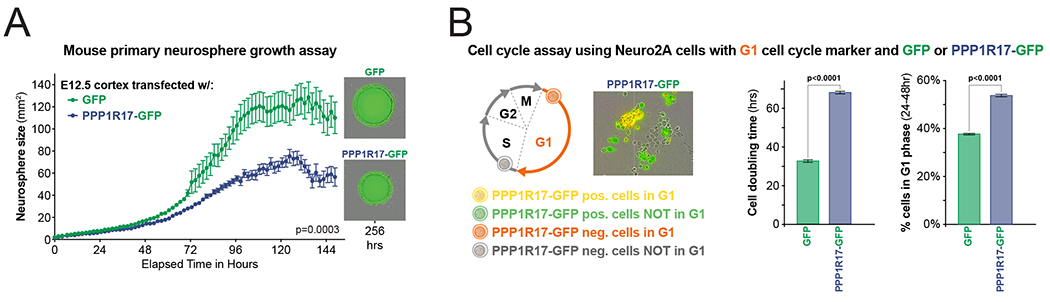
(A) Mouse primary neurosphere growth assay using cells transfected with either a GFP or PPP1R17 construct exposes a growth restriction for PPP1R17-expressing neurospheres, which were 36% smaller than GFP controls (2-way ANOVA, p=0.0003). Error bars = SEM. Individual values represent wells containing neurospheres imaged at each timepoint, n = 48 GFP- and n = 37 PPP1R17-transfected wells.
(B) Cell cycle assay using Neuro2A cells with G1 cell cycle marker in addition to either a GFP or PPP1R17 construct demonstrates both (middle) a longer doubling time (PPP1R17 cells = 68.2 hours vs GFP = 32.7 hours, p<0.0001) and (right) a prolonged G1 phase for PPP1R17-expressing Neuro2A cells (48.5% PPP1R17 vs 33.8% GFP, p<0.0001, 2-way ANOVA). Total number of cells and percent of cells expressing both the G1 marker and either GFP or PPP1R17-GFP were quantitated at 24h and 48h from 4 images per well, for each well containing transfected Neuro2A cells (n=13 GFP-transfected wells, n=13 PPP1R17-transfected wells). Error bars = SEM.
An integrated resource for interrogating HAR function in humans
We have collated the data generated in this paper into an easily searchable online resource through the UCSC Genome Browser (the HARHub: http://genome.ucsc.edu/cgi-bin/hgTracks?db=hg38&hubUrl=https://allendiscoverycenter-harhub.s3.us-east-2.amazonaws.com/HAR_hub/hub.txt) (Figure S7). In addition to the caMPRA and epigenomic datasets (DNaseI hypersensitivity, H3K27ac, and H3K4me1) generated herein, this resource also contains previously published datasets of common and rare human HAR sequence variation obtained through targeted sequencing of HARs in the Homozygosity Mapping Collaborative for Autism (HMCA) (Doan et al., 2016). In addition, we also included traditional oligo-based MPRA results on a subset of HARs (Uebbing et al., 2021). This integrated HAR enhancer hub provides a comprehensive resource for guiding future investigations into the function of HARs on the evolution of the human brain.
Discussion
Through interrogating the epigenetic landscape of >3,100 HARs during human neurodevelopment, we demonstrate that nearly half of all HARs act as neurodevelopmental enhancers and that the cis-regulatory architecture of these HARs has undergone extensive human-specific rewiring. Moreover, we demonstrate that human-specific sequence changes within HARs largely augment their enhancer activity within neuronal cells – further establishing HARs as human-specific neurodevelopmental enhancers. By leveraging caMPRA to evaluate the enhancer activity of entire intact elements for >90% of HARs, our study represents the most thorough evaluation of HAR elements to-date. This is in contrast with traditional oligo-based MPRA assays that are often limited to insert sizes of <137bp in length (Uebbing et al., 2021), which represents less than 20% of intact HAR elements, as the average HAR element is 269 bp in length.
The striking conservation within the trans-regulatory activity of HARs mirrors prior studies (Stergachis et al., 2014) and demonstrates that our assessment of enhancer activity using mouse and human neural cell lines is likely capturing the majority of HARs that act as neurodevelopmental enhancers. Furthermore, by combining our caMPRA data with chromatin data from the developing fetal brain, we establish that a substantial portion of HARs act as both in vitro and in vivo enhancers and that overall HARs are enriched for being enhancers in the developing human brain. Together this firmly establishes that a subset of HARs represent genomic entities driving human-specific neurodevelopmental gene regulatory programs. In addition to their neural enhancer activity, many HARs also demonstrate evidence of enhancer activity in multiple other tissue types, indicating that HARs may also play a role in modulating human-specific gene regulatory programs for other organ systems. Of note, 11% of HARs lacked any evidence of in vitro or in vivo enhancer activity in any of the cell types analyzed (Data S3 and Data S6), indicating that these HARs may have gene regulatory element-independent functions, if they are functional at all.
Using our neurodevelopmental HAR enhancer data, we identified PPP1R17 as a putative HAR-regulated gene that appears to play a central role within the primate cortical germinal zones. PPP1R17 has undergone extensive rewiring of its cell-type and developmental expression patterns in the primate and human cortex, which appears to be potentially mediated by HARs as well as a primate-selective regulatory element, although further studies evaluating the mechanistic role of these elements in PPP1R17 gene regulation are likely needed. Notably, the primate-selective regulatory element directly upstream of PPP1R17 encompasses several recently-evolved retrotransposons, which has been proposed as the most common mechanism for generating primate-specific regulatory elements (Jacques et al., 2013).
PPP1R17 cortical expression is restricted to progenitor cells within the oSVZ and iSVZ in developing cerebral cortex of both humans and macaque. These germinal zones are the primary neurogenic niche in human cortical development and have undergone a dramatic expansion in size during primate evolution (Lui et al., 2011; Reillo et al., 2011) due to the augmentation of the transit-amplifying capacity of NPCs. Specifically, whereas rodent NPCs usually divide once (Haubensak et al., 2004; De La Torre-Ubieta et al., 2016; Noctor et al., 2004) or only a few times (Mihalas and Hevner, 2018) to produce two postmitotic neurons, human NPCs go through numerous additional rounds of symmetric division before terminal differentiation (Hansen et al., 2010; Pollen et al., 2015; Sessa et al., 2008). Together, our findings suggest that PPP1R17 may play a complex role not only in slowing cell cycle length, but also in augmenting transit-amplification and delaying neuronal differentiation– a process central to the increase in neuronal production in primate and human corticogenesis (Bystron et al., 2008). Although ferrets show longer cell cycle length than mice (Turrero García et al., 2016), we do not find PPP1R17 expression in their NPCs, suggesting that while PPP1R17 expression is sufficient to elongate cell cycle length, other mechanisms may also contribute.
Although humans and macaque both demonstrate a gain of PPP1R17 expression within cortical neural progenitor cells, humans and macaque differ in their expression of PPP1R17 within the astrocyte lineage. Specifically, whereas macaque demonstrates robust PPP1R17 expression in astrocytes throughout development, humans appear to lack significant astrocyte PPP1R17 expression postnatally. In addition, humans also lack chromatin accessibility at HAR2635 within astrocytes (Figure 3D), suggesting that this HAR may play a role in guiding the cell-specificity of PPP1R17 expression in humans. It remains unclear if this difference in astrocyte PPP1R17 expression has a phenotypic effect, as our understanding of differences in astrocyte neurobiology between primates is still in its infancy.
While we show that PPP1R17 slows neural progenitor cell-cycle progression, and while the lengthening of neural progenitor cell cycles is a well-documented feature of primates with larger brains, the evolutionary origin and presumed advantage of the remarkably extended neurogenetic period in primates remains enigmatic. Longer cell cycles are thought to reduce the rate of somatic mutation or maintain DNA quality-control, and may facilitate the formation of the remarkable diversity of neuronal types that constitute the human brain (Boldog et al., 2018; Hodge et al., 2019), but they come at the potential cost of increasing heterochrony, a longer period of infantile helplessness, and the requirement for longer maternal care and changes in societal structure, suggesting complex and balancing forces. The deepening investigation of HARs and other evolutionarily dynamic genomic sequences may increasingly enlighten these complex evolutionary changes underlying distinct human brain cytoarchitecture and cognitive capacity.
STAR★METHODS
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Christopher A. Walsh (christopher.walsh@childrens.harvard.edu).
Materials availability
All unique/stable reagents generated in this study are available from the Lead Contact with a completed Materials Transfer Agreement.
Data and code availability
HAR data have been deposited at GEO accession GSE180714. Processed ChIP-seq and DNaseI-seq and HAR data are available at http://genome.ucsc.edu/cgi-bin/hgTracks?db=hg38&hubUrl=https://allendiscoverycenter-harhub.s3.us-east-2.amazonaws.com/HAR_hub/hub.txt. Accession numbers are listed in the key resources table. All the data is available from the lead contact upon request.
All the code is available from the lead contact upon request.
Any additional information required to reanalyze the data reported in this paper is available
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| PerCP-Cy5.5-conjugated anti-PAX6 | BD Biosciences | Cat#561462, Clone O18-1330 |
| V450-conjugated anti-SOX2 | BD Biosciences | Cat#561469, Clone O30-678 |
| Alexa Fluor 488-conjugated anti-NeuN | EMD Millipore | Product No. FCMAB317PE, Clone A60 |
| Anti-H3K27ac | Abcam | Prod#ab4729 |
| Anti-H3K4me1 | Abcam | Prod#ab176877 |
| Goat anti-SOX2 (1:200) | Santa Cruz | Cat#sc-170320 |
| Chicken anti-TBR2 (1:200) | Millipore | Cat#AB15894 |
| Rabbit anti-TBR2 (1:200) | Abcam | Prod#ab23345 |
| Rabbit anti-PAX6 (1:200) | Covance | Cat#PRB-278P |
| Mouse anti-HOPX (1:200) | Santa Cruz | Cat. No.. sc-30216 |
| Rabbit anti-PPP1R17 (1:200) | Novus | Cat#NBP2-13800 |
| Mouse anti-PPP1R17 (1:200) | Novus | Cat#H00010842-B01P |
| Mouse anti-SATB2 (1:200) | Abcam | Prod#ab51502 |
| Rabbit anti-Ki67 (1:200) | Abcam | Prod#ab15580 |
| Mouse anti-PCNA | Millipore | Cat#AB93501 |
| Mouse anti-GFAP | Sigma | Cat#G3893 |
| Rat anti-GFAP | Thermo Fisher | Cat#13-0300 |
| Alexa Fluor 488 Anti-Chicken Secondary Antibody | Thermo Fisher | Cat#A11039 |
| Alexa Fluor 594 Anti-Mouse Secondary Antibody | Thermo Fisher | Cat#A11008 |
| Alexa Fluor 594 Anti-Rabbit Secondary Antibody | Thermo Fisher | Cat#A11012 |
| Alexa Fluor 647 Anti-Rat Secondary Antibody | Thermo Fisher | Cat#A21247 |
| DAPI (4′,6-Diamidino-2-Phenylindole, Dihydrochloride) | Thermo Fisher | Cat#D1306 |
| Hoechst 33342 | Thermo Fisher | Cat#62249 |
| Normal Goat Serum | Gibco | Cat#16210064-100ml |
| Bovine Serum Albumin | Sigma | Cat#A7906-100G |
| Fluoromount-G | Southern Biotech | Cat#0100-01 |
| Bacterial and Virus Strains | ||
| One Shot TOP10 Chemically Competent E. coli | Thermo Fisher | Cat#C404006 |
| Biological Samples | ||
| Human brain tissue | This paper | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Formaldehyde | Fisher Scientific | Cat#F79P-4 |
| Glycine | Sigma | Cat#G7126-5KG |
| Triton X-100 | Sigma | Cat#T9284-500ML |
| Paraformaldehyde | Electron Microscopy Sciences | Cat#15710 |
| Fetal bovine serum | Atlanta Bio | Cat#S11150 |
| Dulbecco’s Modified Eagle Medium | Thermo Fisher | Cat#MT10013CV |
| Dulbecco’s Modified Eagle Medium/F12 | Thermo Fisher | Cat#11330032 |
| Penicillin-Streptomycin | Thermo Fisher | Cat#15140122 |
| AMPure XP beads | Beckman-Coulter | Cat#A63881 |
| Retrievagen A | BD Biosciences | Cat#550524 |
| Lysis Buffer | This paper | N/A |
| Wash buffer | This paper | N/A |
| Sonication buffer | This paper | N/A |
| AsiSI restriction enzyme | NEB | Cat#R0630 |
| DNaseI | Sigma-Aldrich | Cat#D4527 |
| MboI | NEB | Cat#R0147 |
| T4 DNA Ligase | Thermo Fisher | Cat#EL0011 |
| Critical Commercial Assays | ||
| Lipofectamine LTX reagent | Thermo Fisher | Cat#A12621 |
| TRIzol reagent | Invitrogen | Cat#15596018 |
| NEBNext Single Cell/Low Input RNA Library Prep Kit for Illumina | New England BioLabs | Cat#E6420S |
| Protein A Dynabeads | Thermo Fisher | Cat#10002D |
| Ovation Ultralow V2 kit | Nugen | Cat#0344-32 |
| Neural Tissue Dissociation Kit (T) | Miltenyi Biotec | Cat#130-093-231 |
| Deposited Data | ||
| Raw and analyzed data | This paper | GSE180714 |
| Human reference genome build GRCh38/hg38 | Genome Reference Consortium | https://www.ncbi.nlm.nih.gov/assembly/GCF_000001405.26/ |
| Human, chimpanzee, and macaque brain transcriptome data | Sousa et al., 2018 | N/A |
| ENCODE Project and Roadmap Epigenomics Consortium | ENCODE Project Consortium, 2012; Roadmap Epigenomics Consortium et al., 2015 | http://www.roadmapepigenomics.org/ |
| UCSC-tools and genome browser | Kent et al., 2002 | http://genome.ucsc.edu/ |
| Pathway and gene set overdispersion analysis (PAGODA) | Fan et al., 2016 | N/A |
| Single cell ATAC-seq data | Domcke et al., 2020 | https://descartes.brotmanbaty.org/ |
| Experimental Models: Cell Lines | ||
| Neuro2A | ATCC | Cat#CCL-131 |
| SH-SY5Y cell | ATCC | Cat#CRL-2266 |
| Experimental Models: Organisms/Strains | ||
| CD1 Mice | Charles River | Crl:CD1(ICR) |
| Ferrets | Marshall Bioresources | N/A |
| Macaques | Yale University School of Medicine | N/A |
| Oligonucleotides | ||
| CustomArray pool | CustomArray, Bothell, WA | http://www.customarrayinc.com/oligos_main.htm |
| Primers for 3C interaction assay, see Supplemental Data Table S6 | This paper | N/A |
| Recombinant DNA | ||
| Human genomic DNA | Promega | Cat#G1521 |
| Chimpanzee DNA | Coriell | Cat#NS03489 |
| pMPRA1 vector | Melnikov et al., 2014 | Addgene 49349 |
| pMPRA_donor2 | Melnikov et al., 2014 | Addgene 49353 |
| pFucci-G1 Orange | Medical & Biological Laboratories Co., LTD. | Cat#AM-V9003M |
| PBCAG-eGFP | Gift from Joseph LoTurco, Chen et al., 2012 | Addgene 40973 |
| pCyl50 | Gift from Jae Yong Han, Lee et al, 2016 | N/A |
| pCAG-PB-GFP | This paper | N/A |
| pCAG-PPP1R17-PB-GFP | This paper | N/A |
| Software and Algorithms | ||
| MIPgen | Shendure Lab, University of Washington | http://shendurelab.github.io/MIPGEN/ |
| R language | R Core Team, 2019 The R Project for Statistical Computing | https://www.r-project.org/ |
| Python language, version 2.7 | Python Software Foundation | http://www.python.org |
| GraphPad Prism 8 | GraphPad Software Inc., La Jolla, CA, USA | https://www.graphpad.com/scientific-software/prism/ |
| Fiji | Schindelin et al., 2012 | https://imagej.net/ImageJ |
| ImageJ | Schneider et al., 2012 | https://imagej.net/ImageJ |
| IncuCyte Zoom | Essen Bioscience | IncuCyte S3 2018B |
| HOMER | Heinz et al., 2010 | http://homer.ucsd.edu/homer/ |
| Other | ||
| HARHub | This paper | http://genome.ucsc.edu/cgi-bin/hgTracks?db=hg38&hubUrl=https://allendiscoverycenter-harhub.s3.us-east-2.amazonaws.com/HAR_hub/hub.txt |
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Cell lines
Neuro2A/N2A cells (ATCC, CCL-131) were grown in 10% fetal bovine serum, Dulbecco’s Modified Eagle Medium, and 1% Penicillin-Streptomycin. SH-SY5Y cell (ATCC, CRL-2266) medium was 10% fetal bovine serum, Dulbecco’s Modified Eagle Medium/F12, and 1% Penicillin-Streptomycin. Both cell lines were maintained in a 5% CO2 incubator at 37°C.
Mice
All procedures were reviewed and approved by the Institutional Animal Care and Use Committee (IACUC) at Boston Children’s Hospital (BCH). Timed pregnant CD1 mice (Mus musculus) were obtained from Charles River Laboratories in advance of in utero electroporation procedures after approximately 9-11 days of gestation. Mice were housed before and after procedures at BCH and maintained according to BCH and NIH animal research guidelines.
Ferrets
Wild-type ferrets (Mustela putorius furo) were obtained from Marshall Bioresources, with both males and females included in this study. No association between animal sex and findings was found. All animal work conducted as part of this study was in compliance with the NIH Principles for the Use of Animals, the NIH Guide for the Care and Use of Laboratory Animals, and the Animal Welfare Act, and all work was reviewed and approved by the Institutional Animal Care and Use Committee (IACUC) at Boston Children’s Hospital (BCH).
Human and non-human primate tissue specimens and processing
Research performed on samples of human origin was conducted according to protocols approved under expedited category 5 with waiver of consent (45 CFR 46.110) by the institutional review boards of Beth Israel Deaconess Medical Center, Boston Children’s Hospital, and Yale University School of Medicine. Fetal brain tissue was received after release from clinical pathology, with a maximum post-mortem interval of 4 h. Cases with known anomalies were excluded. Gestational ages (weeks 19, 20, and 23) were determined using fetal foot length. One subject was female, and the sex of other subjects was unknown. Tissue was transported in HBSS medium on ice to the laboratory for research processing.
Rhesus macaque (Macaca mulatta) brain samples were collected postmortem from both adult (11 years-old) and fetal (E70) specimens. All experiments using non-human primates were carried out in accordance with a protocol approved by Yale University’s Committee on Animal Research and NIH guidelines.
METHOD DETAILS
CaptureMPRA library design, cloning, and transfection
Molecular Inversion Probes (MIPs) targeting flanking regions of all HAR sequences were designed using the MIPgen program (Boyle et al., 2014) and manufactured as a pool by CustomArray (Bothell, WA). Target genomic sequences from both human (Promega, G1521) and chimpanzee DNA (Coriell, NS03489) were captured as previously described (Cantsilieris et al., 2017) and purified using Agencourt AMPure XP beads at 0.9X concentration (Beckman-Coulter, A63881). The MPRA assay then was conducted as previously described (Melnikov et al., 2014). Briefly, the captures were cloned into modified pMPRA1 vector (Addgene, 49349). The only modification to the pMPRA1 vector was the addition of an AsiSI restriction site used for inserting our captured DNA fragments. The luciferase gene and minimal promoter (pMPRA_donor2, Addgene, 49353) was then cloned into the pMPRA1_Capture construct pool.
The transfection of the MPRA libraries was performed with five replicates per condition, and the experiment was conducted in triplicate. The cells (N2A and SH-SY5Y) were grown as adherent cultures for 24hr in 15 cm plates. Then, 20ug of MPRA constructs and 4ug of a fluorescent reporter construct per plate were transfected into cells with Lipofectamine LTX reagent (Thermo Fisher, A12621). The cells were checked for fluorescent reporter expression at 24hr and harvested 48hr post-transfection. mRNA was isolated using TRIzol reagent (Invitrogen, 15596018). Resulting Tag-seq amplified cDNA was deep sequenced using the Illumina Hi-Seq 2500 with 150-bp paired-end reads.
Fluorescence-Activated Nuclear Sorting (FANS)
Nuclear preparation and sorting were performed as previously described (Lodato et al., 2015; Thomsen et al., 2016). Nuclei were labeled in aliquots of 500 μl with antibodies ((PerCP-Cy5.5-conjugated anti-PAX6 (O18-1330; BD Biosciences), V450-conjugated anti-SOX2 (O30-678; BD Biosciences), Alexa Fluor 488-conjugated anti-NeuN (A60; EMD Millipore)) for 30 min at 4°C before FANS. Pools of 750,000 to 1,000,000 nuclei were collected per condition for downstream sequencing experiments unless otherwise noted.
RNA-Sequencing of FANS nuclei
1000 nuclei from each cell population were sorted in triplicate into a 96 well plate and cDNA libraries were immediately prepared using NEBNext Single Cell/Low Input RNA Library Prep Kit for Illumina according to manufacturer’s instructions at 0.5X volume. Sorted nuclei were subjected to 18 cycles of PCR amplification following the reverse transcription step. Quality of amplified cDNA was assessed using Agilent 2100 Bioanalyzer (Agilent Technologies) Illumina-compatible cDNA libraries were subsequently prepared according to manufacturer’s instructions at 0.5X volume with 20 ng of full-length amplified cDNA and 8 cycles of PCR enrichment. Prior to sequencing, library quality was assessed using the Agilent 2100 Bioanalyzer. 150 PE reads were generated using the Illumina HiSeqX Ten and analyzed as described below.
Chromatin Immunoprecipitation (ChIP)
For ChIP of histone modifications, sorted nuclei were cross-linked via addition of 1% formaldehyde for 10 min at room temperature and quenched by the addition of 0.125 M glycine for 5 min at room temperature. Nuclei were lysed by 10 minute incubation in lysis buffer 1 (in mM: 100 HEPES-NaOH pH 7.5, 280 NaCl, 2 EDTA, 2 EGTA, 0.5% Triton X-100, 1% NP-40, 20% Glycerol) followed by washing in buffer containing 10 mM Tris-HCl pH 8.0, 200 mM NaCl. Chromatin was sheared using a Bioruptor (Diagenode) on high power mode for 50 cycles with 30 sec pulses in sonication buffer (in mM: 10 Tris-HCl pH 8.0, 100 NaCl, 1 EDTA, 0.5 EGTA, 0.1% Na-Deoxycholate, 0.5% N-Lauroylsarcosine). For H3K27ac ChIP, 10 mM sodium butyrate (Fisher NC9851678) was added to all buffers.
Following sonication, chromatin was supplemented with 1% Triton and was incubated overnight with the following antibodies coupled to Protein A Dynabeads (Thermo Fisher, 10002D): H3K27ac (Abcam, 4729), H3K4me1 (Abcam, 176877). ChIPs were performed in three biological replicates for each cell population. Libraries were generated using the Ovation Ultralow V2 kit (Nugen, 0344-32) according to the manufacturer’s instructions and PCR amplified for 13-16 cycles, depending on antibody. Library quality was assessed using the Agilent 2100 Bioanalyzer (Agilent Technologies). Seventy-five bp reads were generated on the Illumina Nextseq 500 and subsequently analyzed with our standardized ChIP-seq data analysis pipeline (below).
Chromatin Conformation Capture (3C)
3C PCR primers (Supplemental Data Table S7) were designed according to a previous 3C method (Naumova et al., 2012). To obtain an interaction profile of the genomic region spanning HAR2635, HAR2636, and the PPP1R17 promoter, we designed several test primers at and around each HAR and a single anchor primer within the promoter region of PPP1R17.
3C targeting the PPP1R17 locus was performed in SH-SY5Y cells. Cells were cross-linked in 1% methanol-free formaldehyde for 10 minutes at room temperature (RT) followed by addition of glycine to a final concentration of 125 mM. Cells were incubated for 20 minutes in lysis buffer (10 mM Tris-HCl pH 8, 10 mM NaCl, 0.2% Triton X-100) on ice, followed by dounce homogenization. Nuclei were isolated by centrifugation and treated with 0.3% SDS followed by addition of 2% Triton X-100 for 1 hour at 37C each. Cross-linked chromatin was digested with 100U of MboI restriction enzyme (NEB, R0147) at 37C overnight, followed by ligation under dilute conditions (2 ng/uL) using 75U of T4 DNA ligase (Thermo Fisher, EL0011) for 4 hours at 16C followed by 30 minutes at RT. Chromatin was reverse cross-linked via addition of Proteinase K. Purified 3C templates were amplified under standard PCR conditions. Quantification of interaction products were analyzed via agarose gel using ImageJ (Schneider et al., 2012), and relative interaction frequency was calculated from the relative intensity of each band.
Immunohistochemistry
Mouse brain tissue was fixed overnight at 4°C in 4% PFA. Fixed macaque and mouse tissue were sectioned at 50 um using a Vibratome. Tissue was permeabilized and blocked in 3% BSA and 0.3% Triton X-100 in PBS for mouse and ferret tissue, and 5% Normal Donkey Serum with 1% BSA for macaque and human tissue. Primary antibodies were diluted in blocking buffer and incubated overnight 1-2 nights at 4°C. Primary antibodies included goat anti-SOX2 (1:200, Santa Cruz sc-170320 and 1:150 R+D Systems AF2018), chicken (1:200; Millipore AB15894) and rabbit (1:200; abcam ab23345) anti-TBR2 (1:500 Abcam ab183991), rabbit anti-PAX6 (1:200; Covance PRB-278P, 1:200 Biolegend 901301, mouse anti-HOPX (1:200; Santa Cruz sc-30216), rabbit (1:200; Novus NBP2-13800) and mouse (1:200; Novus H00010842-B01P) anti-PPP1R17, mouse anti-SATB2 (1:200; abcam ab51502), rabbit anti-Ki67 (1:200; abcam ab15580, and 1:200 Invitrogen 14-5698-2), rabbit anti-TBR1 (1:500, abcam ab31940), and mouse anti-PCNA (1:200; Millipore AB93501). Sections were then stained with Alexa secondary antibodies and DAPI. Ferret and human brain sections were processed as previously described (Johnson et al., 2015, 2018). Imaging was performed using a Zeiss LSM 700 confocal microscope.
In Utero electroporation (IUE)
In utero electroporations were performed as described previously (Saito et al., 2018; Szczurkowska et al., 2016; Zhang et al., 2016) using pregnant mouse dams (embryonic age 12.5-16.5) under a Boston Children’s Hospital Institutional Animal Care and Use Committee (IACUC)-approved protocol. Embryos were injected with approximately 2μL of a 1μg/μL mixture of plasmid DNA, delivered using a pulled glass micropipette inserted into the lateral ventricle of the brain. Subsequently, 50-volt electric pulses (5 total, 1.1 second interval, 50ms duration) were passed through the brain over each injection site using paddle electrodes. Embryos were allowed to continue developing normally for 2-4 days after electroporation.
Plasmid cloning
Control (pCAG-PB-GFP) and human PPP1R17 (pCAG-PPPlR17-PB-GFP) expression plasmids were cloned from PBCAG-eGFP (Addgene 49073), a generous gift from Joseph LoTurco (Chen and LoTurco, 2012). A P2A linker was inserted downstream of the eGFP coding region, and for the PPP1R17-expression plasmid, a 430bp region from the human exon 3 sequence was inserted, upstream of the rabbit beta-globin polyadenylation signal and the piggyBac 3’ terminal repeat sequence. Sequences were verified by Sanger sequencing.
Primary neurosphere growth assay
First, in utero electroporations were performed as described above with mice aged E12.5 and a mixture of control (pCAG-PB-GFP) or PPP1R17 (pCAG-PPPlR17-PB-GFP) plasmid and the PiggyBac transposase system to allow for genomic integration (Wu et al., 2006). Embryos were harvested after 48 hours in utero. The cortex of the embryos then were minced and dissociated with using the Miltenyi Biotec Neural Tissue Dissociation Kit, and the top 10% of GFP+ cells were sorted using Fluorescence Activated Cell Sorting (FACS). Cells were plated at 500 cells/well on a 96-well u-bottomed plate. Neurospheres were grown for 6-10 days in vitro, and images were collected by the IncuCyte imaging system (Essen Instruments, Ann Arbor, MI, USA) every 2 hours. Analysis was subsequently performed using IncuCyte Zoom software (Essen Bioscience) and GraphPad Prism (GraphPad Software Inc., La Jolla, CA, USA) from n = 48 GFP control and n = 37 PPP1R17-GFP wells containing neurospheres.
Quantification and Statistical Analysis
CaptureMPRA data analysis
To identify HAR barcodes, the sequence immediately preceding the adapter sequence was isolated, and the occurrence of each barcode counted. To compare to the reference 10,690 barcodes, each barcode’s Levenstein distance was calculated with respect to all reference barcodes using the stringdist package, and the minimum value was taken for each barcode. Multiple probes were summarized by taking the mean of each probe. Data was normalized to account for effective logFC differences across uneven library sizes by using the Trimmed Means of M normalization as implemented in the edgeR package. The RNA matrix was created by dividing each sample’s RNA value by the respective plasmid DNA. The gplots package is used to create the heatmap on the log2 data. HARs with significant enhancer activity were identified using a one-sided one-sample T-test of the three biological replicates to identify HARs with log2 mRNA/DNA>0. This T-test was performed independently using the human and chimpanzee orthologs as well as the data from N2A and SH-SY5Y cells, and those with a p-value <0.05 in at least one of these samples were considered to have significant enhancer activity. False discovery rates were also identified by shuffling the MPRA data between HARs and samples and recomputing the p-values, to identify the proportion of enhancers identified at a given p-value threshold that may be the result of a falsely identified enhancer. A two-sided paired T-test was used to identify HARs with significantly different log2(mRNA/DNA) ratios between the human and chimpanzee orthologs.
Transcription factor binding motif analyses
Transcription factor binding motifs for all HAR sequences were performed using HOMER (Heinz et al., 2010) with the basic configurations: database version v4.4, significance p < 0.01. Enrichment of motifs was determined using options: randomly generated sequences and HAR input regions. ConTra v3 (Kreft et al., 2017) was used for visualization of TFBS changes across species with stringency: core = 0.95, similarity matrix = 0.85.
RNA Sequencing, alignment and DGE Analysis
FASTA and Homo_sapiens.GRCh38.93.gtf annotation files were retrieved from Ensembl and STAR version 2.5.2b was used to generate a genomic index from hg38 genome release 93 (--sjdbOverhang 149). Using default settings in STAR, bam files were generated for each sample independently and a gene count matrix was created using featureCounts (-p –s0) for paired-end, unstranded RNA-seq data. Raw gene counts for technical replicates were then summed together prior to normalization of biological replicates using the Median by Ratio method implemented by DESeq2 (version 1.20.0). Box plots for genes of interest were generated with ggplot2 using gene counts after normalization of the Median by Ratio method. Normalized biological replicates were then analyzed to discover differentially expressed genes using DESeq2. Minimum requirements for a DGE was defined as a gene possessing abs(log2FoldChange) > 0.58 and a P-value < 0.05 after adjusting P-values using the Benjamini-Hochberg method.
DHS resources and analysis
Human sorted nuclei were subjected to DNaseI digestion and high-throughput sequencing, following previous methods (John et al., 2011). Additional DHS data sets were downloaded and analyzed from the ENCODE Project and Roadmap Epigenomics Consortiums (ENCODE Project Consortium, 2012; Roadmap Epigenomics Consortium et al., 2015). The number and proportion of all HARs that fell within DNase-I-sensitive regions (Hot Spots) and DHSs (150 bp peaks) were identified as were calculated as previously described (John et al., 2011). In addition, the maximum DNaseI density over each HAR was calculated using normalized DNaseI density track sets and bedops (Neph et al., 2012).
ChIP Sequencing, alignment, and genome browser track generation
All experiments were sequenced on the Nextseq 500 (Illumina). Seventy-five base pair single-end reads were obtained for all ChIP-seq datasets. All samples were aligned to the hg38 genome using default parameters for the Subread alignment software (subread-1.4.6-p3, (Liao et al., 2013)) after quality trimming with Trimmomatic v0.33 (Bolger et al., 2014) with the following command: java -jar trimmomatic-0.33.jar SE -threads 1 -phred33 [FASTQ_FILE] ILLUMINACLIP:[ADAPTER_FILE]:2:30:10 LEADINGS:5 TRAILING:5 SLIDINGWINDOW:4:20 MINLEN:45. Truseq adapters were trimmed out in ChIP-seq experiments.
To generate UCSC genome browser tracks for ChIP-seq data, all aligned bam files for each replicate of a given experiment were pooled and converted to BED format with bedtools bamtobed. The 75 base pair reads were extended in the 3’ direction to 200 basepairs (bp) (average fragment length for ChIP-seq experiments as measured by bioAnalyzer) with the bedtools slop command using the following parameters: −1 0 -r 125 -s. Published hg38 ChIP-seq blacklisted regions (ENCODE Project Consortium, 2012) were filtered out using the following command: bedops –not-element-of 1 [BLACKLIST_BED], The filtered BED files were converted to coverageBED format using the bedtools genomecov command with the following options: -scale [NORM_FACTOR to scale each library to 20M reads] –bg. Finally, bedGraphToBigWig (UCSC-tools) was used to generate the bigWIG files displayed on browser tracks throughout the manuscript.
Hi-C data was visualized using the 3D Genome Browser (Wang et al., 2018). Specifically, both Fetal_Brain_GZ/Won2016-raw and IMR90/Lieberman-raw were visualized at 10kb resolution with a scale of 0 to 90.
ChIP-seq peak calling
To identify sites with enriched ChIP-seq signal (peaks), we applied the IDR pipeline (Li et al., 2011) using the MACS2 peak calling algorithm (Zhang et al., 2008) with the following parameters: --nomodel --extsize 200 --keep-dup all. An IDR threshold of .01 was used for self-consistency, true replicate, and pooled-consistency analyses. The ‘optThresh’ cutoff was then used to obtain a final set of high-confidence, reproducible ChIP-seq peaks for each factor or chromatin modification at each stimulus condition tested.
ChIP-Seq peaks across all conditions and ChIP-ed factors were unioned to generate the comprehensive set of putative regulatory elements assessed in this study using the following command: bedops --everything [INPUT PEAKS] | bedtools sort -i - | bedtools merge -i - -c 4,5 -o collapse,collapse > [OUTPUT FILE], This peak set was then intersected with HARs using bedtools intersect.
Gene ontology analysis
For HARs overlapping ChIP peaks, bedtools (bedtools closest) was used to determine the closest gene. Functional enrichment with gene ontology was implemented with the goseq package, setting an Benjamini-Hochberg FDR of 0.05. Fold change matrices were calculated using the gtools implementation of fold change.
Human single cell RNA-sequencing and ATAC-seq
Pseudotime course analysis of previously published RNA-seq data was executed using single-cell differential expression analysis (Kharchenko et al., 2014) and pathway and gene set overdispersion analysis (PAGODA) (Fan et al., 2016). Single cell analyses for Figure S6A were performed using previously published data from human neocortical development during mid-gestation (Polioudakis et al., 2019). Single cell ATAC-seq data for Figure 3 was obtained from https://descartes.brotmanbaty.org/ (Domcke et al., 2020). The track labeled “Glial cells” in Figure 3E corresponds to the “Astrocyte” cell population in Domcke et al. Similarly, the track labeled “Intermediate Progenitors” corresponds to the “Cerebrum Unknown.3” cell population in Domcke et al based on the selective chromatin accessibility of several intermediate progenitor cell markers in this cluster, including EOMES.
Cell cycle assay
To obtain doubling time and quantification of time spent in G1, Neuro2A cells were seeded at low density (300-1000 cells/well) in a 96-well plate format after chemical transfection by Lipofectamine with two plasmids: pFucci-G1 Orange (Sakaue-Sawano et al., 2008), and either the control (CAG-GFP) or PPP1R17 misexpression vector (CAG-PPP1R17-GFP). Cells were imaged every 1-4 hours for 2-4 days at 20x magnification using the IncuCyte imaging system (Essen Instruments, Ann Arbor, MI, USA). Analysis of time-lapsed images was performed using 4 regions of interest per well (n=13 wells each for GFP control and PPP1R17-GFP transfected cells) using IncuCyte Zoom software (Essen Bioscience) and GraphPad Prism (GraphPad Software Inc., La Jolla, CA, USA). Numbers of cells expressing both pFucci-G1 Orange (channel), and GFP control or PPP1R17-expression plasmids were quantified at each timepoint recorded. Doubling time was calculated using cells that were GFP-positive in both conditions.
Supplementary Material
Data S1 | Capture MPRA probe information, Related to Figure 1
Data S2 | Capture MPRA enrichment information, Related to Figure 1.
Data S3 | DNaseI datasets used in this study, Related to Figure 2.
Data S4 | RNA-seq analysis of sorted fetal brain nuclei, Related to Figure 2.
Data S5 | ChIP-seq and DNaseI-seq analysis of fetal brain nuclei, Related to Figure 2.
Data S6 | Integtation of in vivo and in vitro enhancer activities of HAR elements, Related to Figure 3.
Data S7 | Primers used for 3C experiments, Related to Figure 3.
Highlights.
Comprehensive in vitro and in vivo epigenomic profile of >3,100 HARs
Nearly half of all HARs have features of neurodevelopmental enhancers
Extensive human-specific rewiring of the cis-regulatory architecture of HARs
Primate- and human-specific change in PPP1R17 developmental expression patterns
Acknowledgments
We are grateful to Kristina Holton and Research Commuting at Harvard Medical School for computational assistance and computing resources, including the Orchestra computing cluster (partially provided through NCRR 1S10RR028832-01). We thank John Daley and the Dana Farber Cancer Institute Flow Cytometry Core, the Boston Children’s Hospital Molecular Genetics Core Facility, and the Boston Children’s Hospital Intellectual and Developmental Disabilities Research Centers (IDDRC) Human Neuron Core, U54HD090255, for technical help. Figure 4C was created in part with BioRender.com.
This work was supported by R01NS032457, R01NS115965, T32MH020017, T32GM007753, K00CA222750, R01 NS095654, U01 MH116488, P50 MH106934, the Good Ventures Life Sciences Research Fellowship. This research was supported by the Allen Discovery Center program, a Paul G. Allen Frontiers Group advised program of the Paul G. Allen Family Foundation. A.B.S was supported by NIHs grant GM007748 (NIGMS) and OD029630, and holds a Career Award for Medical Scientists from the Burroughs Wellcome Fund. E.M.D. was supported by the Surpina and Panos Eumekian Bio Fund Fellowship. R.N.D. was supported by the Simons Foundation Autism Research Initiative Bridge to Independence Award. X.Q. was funded by Simons Center for the Social Brain Postdoctoral Fellowship. C.A.W. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interests
The authors declare no competing interests.
References:
- Arai Y, Pulvers JN, Haffner C, Schilling B, Nüsslein I, Calegari F, and Huttner WB (2011). Neural stem and progenitor cells shorten S-phase on commitment to neuron production. Nat. Commun 2, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betizeau M, Cortay V, Patti D, Pfister S, Gautier E, Bellemin-Ménard A, Afanassieff M, Huissoud C, Douglas RJ, Kennedy H, et al. (2013). Precursor Diversity and Complexity of Lineage Relationships in the Outer Subventricular Zone of the Primate. Neuron 80, 442–457. [DOI] [PubMed] [Google Scholar]
- Bird CP, Stranger BE, Liu M, Thomas DJ, Ingle CE, Beazley C, Miller W, Hurles ME, and Dermitzakis ET (2007). Fast-evolving noncoding sequences in the human genome. Genome Biol. 8, R118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boldog E, Bakken TE, Hodge RD, Novotny M, Aevermann BD, Baka J, Bordé S, Close JL, Diez-Fuertes F, Ding SL, et al. (2018). Transcriptomic and morphophysiological evidence for a specialized human cortical GABAergic cell type. Nat. Neurosci 21, 1185–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, and Usadel B (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bormuth I, Yan K, Yonemasu T, Gummert M, Zhang M, Wichert S, Grishina O, Pieper A, Zhang W, Goebbels S, et al. (2013). Neuronal basic helix-loop-helix proteins neurod2/6 regulate cortical commissure formation before midline interactions. J. Neurosci 33, 641–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd JL, Skove SL, Rouanet JP, Pilaz L-J, Bepler T, Gordan R, Wray GA, and Silver DL (2015). Human-chimpanzee differences in a FZD8 enhancer alter cell-cycle dynamics in the developing neocortex. Curr. Biol 25, 772–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle EA, O’Roak BJ, Martin BK, Kumar A, and Shendure J (2014). MIPgen: optimized modeling and design of molecular inversion probes for targeted resequencing. Bioinformatics 30, 2670–2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bush EC, and Lahn BT (2008). A genome-wide screen for noncoding elements important in primate evolution. BMC Evol. Biol 8, 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bystron I, Blakemore C, and Rakic P (2008). Development of the human cerebral cortex: Boulder Committee revisited. Nat. Rev. Neurosci 9, 110–122. [DOI] [PubMed] [Google Scholar]
- Cantsilieris S, Stessman HA, Shendure J, and Eichler EE (2017). Targeted Capture and High-Throughput Sequencing Using Molecular Inversion Probes (MIPs). Methods Mol. Biol 1492, 95–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capra JA, Erwin GD, McKinsey G, Rubenstein JLR, and Pollard KS (2013). Many human accelerated regions are developmental enhancers. Philos. Trans. R. Soc. Lond. B. Biol. Sci 368, 20130025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F, and LoTurco J (2012). A method for stable transgenesis of radial glia lineage in rat neocortex by piggyBac mediated transposition. J. Neurosci. Methods 207, 172–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, Hanna J, Lodato MA, Frampton GM, Sharp PA, et al. (2010). Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc. Natl. Acad. Sci 107, 21931–21936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehay C, Kennedy H, and Kosik KS (2015). The outer subventricular zone and primate-specific cortical complexification. Neuron 85, 683–694. [DOI] [PubMed] [Google Scholar]
- Doan RN, Bae B Il, Cubelos B, Chang C, Hossain AA, Al-Saad S, Mukaddes NM, Oner O, Al-Saffar M, Balkhy S, et al. (2016). Mutations in Human Accelerated Regions Disrupt Cognition and Social Behavior. Cell 167, 341–354.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domcke S, Hill AJ, Daza RM, Cao J, O’Day DR, Pliner HA, Aldinger KA, Pokholok D, Zhang F, Milbank JH, et al. (2020). A human cell atlas of fetal chromatin accessibility. Science (80-. ). 370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ENCODE Project Consortium (2012). An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo S (2012). G-substrate: The Cerebellum and Beyond. Prog. Mol. Biol. Transl. Sci 106, 381–416. [DOI] [PubMed] [Google Scholar]
- Fan J, Salathia N, Liu R, Kaeser GE, Yung YC, Herman JL, Kaper F, Fan J-B, Zhang K, Chun J, et al. (2016). Characterizing transcriptional heterogeneity through pathway and gene set overdispersion analysis. Nat. Methods 13, 241–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franchini LF, and Pollard KS (2017). Human evolution: the non-coding revolution. BMC Biol. 15, 89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geschwind DH, and Rakic P (2013). Cortical evolution: judge the brain by its cover. Neuron 80, 633–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gittelman RM, Hun E, Ay F, Madeoy J, Pennacchio L, Noble WS, Hawkins RD, and Akey JM (2015). Comprehensive identification and analysis of human accelerated regulatory DNA. Genome Res. 25, 1245–1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han S, Dennis DJ, Balakrishnan A, Dixit R, Britz O, Zinyk D, Touahri Y, Olender T, Brand M, Guillemot F, et al. (2018). A non-canonical role for the proneural gene neurog1 as a negative regulator of neocortical neurogenesis. Dev. 145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen DV, Lui JH, Parker PRL, and Kriegstein AR (2010). Neurogenic radial glia in the outer subventricular zone of human neocortex. Nature 464, 554–561. [DOI] [PubMed] [Google Scholar]
- Haubensak W, Attardo A, Denk W, and Huttner WB (2004). Neurons arise in the basal neuroepithelium of the early mammalian telencephalon: A major site of neurogenesis. Proc. Natl. Acad. Sci. U. S. A 101, 3196–3201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hébert JM, and Fishell G (2008). The genetics of early telencephalon patterning: some assembly required. Nat. Rev. Neurosci 9, 678–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, and Glass CK (2010). Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodge RD, Bakken TE, Miller JA, Smith KA, Barkan ER, Graybuck LT, Close JL, Long B, Johansen N, Penn O, et al. (2019). Conserved cell types with divergent features in human versus mouse cortex. Nature 573, 61–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubisz MJ, and Pollard KS (2014). Exploring the genesis and functions of Human Accelerated Regions sheds light on their role in human evolution. Curr. Opin. Genet. Dev 29, 15–21. [DOI] [PubMed] [Google Scholar]
- Hunter JD, and D. J. (2007). Matplotlib: A 2D Graphics Environment. Comput. Sci. Eng 9, 90–95. [Google Scholar]
- Jacques PÉ, Jeyakani J, and Bourque G (2013). The Majority of Primate-Specific Regulatory Sequences Are Derived from Transposable Elements. PLoS Genet. 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- John S, Sabo PJ, Thurman RE, Sung M-H, Biddie SC, Johnson TA, Hager GL, and Stamatoyannopoulos JA (2011). Chromatin accessibility pre-determines glucocorticoid receptor binding patterns. Nat. Genet 43, 264–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson MB, Wang PP, Atabay KD, Murphy EA, Doan RN, Hecht JL, and Walsh CA (2015). Single-cell analysis reveals transcriptional heterogeneity of neural progenitors in human cortex. Nat. Neurosci 18, 637–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson MB, Sun X, Kodani A, Borges-Monroy R, Girskis KM, Ryu SC, Wang PP, Patel K, Gonzalez DM, Woo YM, et al. (2018). Aspm knockout ferret reveals an evolutionary mechanism governing cerebral cortical size. Nature 556, 370–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamm GB, Pisciottano F, Kliger R, and Franchini LF (2013). The developmental brain gene NPAS3 contains the largest number of accelerated regulatory sequences in the human genome. Mol. Biol. Evol 30, 1088–1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kharchenko PV, Silberstein L, and Scadden DT (2014). Bayesian approach to single-cell differential expression analysis. Nat. Methods 11, 740–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornack DR, and Rakic P (1998). Changes in cell-cycle kinetics during the development and evolution of primate neocortex. Proc. Natl. Acad. Sci. U. S. A 95, 1242–1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreft Ł, Soete A, Hulpiau P, Botzki A, Saeys Y, and De Bleser P (2017). ConTra v3: a tool to identify transcription factor binding sites across species, update 2017. Nucleic Acids Res. 45, W490–W494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De La Torre-Ubieta L, Won H, Stein JL, and Geschwind DH (2016). Advancing the understanding of autism disease mechanisms through genetics. Nat. Med 22, 345–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewitus E, Kelava I, Kalinka AT, Tomancak P, and Huttner WB (2014). An adaptive threshold in mammalian neocortical evolution. PLoS Biol. 12, e1002000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Brown JB, Huang H, and Bickel PJ (2011). Measuring reproducibility of high-throughput experiments. [Google Scholar]
- Liao Y, Smyth GK, and Shi W (2013). The Subread aligner: fast, accurate and scalable read mapping by seed-and-vote. Nucleic Acids Res. 41, e108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindblad-Toh K, Garber M, Zuk O, Lin MF, Parker BJ, Washietl S, Kheradpour P, Ernst J, Jordan G, Mauceli E, et al. (2011). A high-resolution map of human evolutionary constraint using 29 mammals. Nature 478, 476–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodato MA, Woodworth MB, Lee S, Evrony GD, Mehta BK, Karger A, Lee S, Chittenden TW, D’Gama AM, Cai X, et al. (2015). Somatic mutation in single human neurons tracks developmental and transcriptional history. Science (80-. ). 350, 94–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lui JH, Hansen DV, and Kriegstein AR (2011). Development and evolution of the human neocortex. Cell 146, 18–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangale VS, Hirokawa KE, Satyaki PRV, Gokulchandran N, Chikbire S, Subramanian L, Shetty AS, Martynoga B, Paul J, Mai MV, et al. (2008). Lhx2 Selector Activity Specifies Cortical Identity and Suppresses Hippocampal Organizer Fate. Science (80-. ). 319, 304–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melnikov A, Zhang X, Rogov P, Wang L, and Mikkelsen TS (2014). Massively parallel reporter assays in cultured mammalian cells. J. Vis. Exp [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihalas AB, and Hevner RF (2018). Clonal analysis reveals laminar fate multipotency and daughter cell apoptosis of mouse cortical intermediate progenitors. Dev. 145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monuki ES, Porter FD, and Walsh CA (2001). Patterning of the dorsal telencephalon and cerebral cortex by a roof plate-Lhx2 pathway. Neuron 32, 591–604. [DOI] [PubMed] [Google Scholar]
- Namba T, and Huttner WB (2017). Neural progenitor cells and their role in the development and evolutionary expansion of the neocortex. Wiley Interdiscip. Rev. Dev. Biol 6. [DOI] [PubMed] [Google Scholar]
- Naumova N, Smith EM, Zhan Y, and Dekker J (2012). Analysis of long-range chromatin interactions using Chromosome Conformation Capture. Methods 58, 192–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neph S, Kuehn MS, Reynolds AP, Haugen E, Thurman RE, Johnson AK, Rynes E, Maurano MT, Vierstra J, Thomas S, et al. (2012). BEDOPS: high-performance genomic feature operations. Bioinformatics 28, 1919–1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noctor SC, Martinez-Cerdeño V, Ivic L, and Kriegstein AR (2004). Cortical neurons arise in symmetric and asymmetric division zones and migrate through specific phases. Nat. Neurosci 7, 136–144. [DOI] [PubMed] [Google Scholar]
- Polioudakis D, de la Torre-Ubieta L, Langerman J, Elkins AG, Shi X, Stein JL, Vuong CK, Nichterwitz S, Gevorgian M, Opland CK, et al. (2019). A Single-Cell Transcriptomic Atlas of Human Neocortical Development during Mid-gestation. Neuron 103, 785–801.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard KS, Salama SR, King B, Kern AD, Dreszer T, Katzman S, Siepel A, Pedersen JS, Bejerano G, Baertsch R, et al. (2006). Forces shaping the fastest evolving regions in the human genome. PLoS Genet. 2, e168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollen AA, Nowakowski TJ, Chen J, Retallack H, Sandoval-Espinosa C, Nicholas CR, Shuga J, Liu SJ, Oldham MC, Diaz A, et al. (2015). Molecular Identity of Human Outer Radial Glia during Cortical Development. Cell 163, 55–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prabhakar S, Visel A, Akiyama JA, Shoukry M, Lewis KD, Holt A, Plajzer-Frick I, Morrison H, Fitzpatrick DR, Afzal V, et al. (2008). Human-specific gain of function in a developmental enhancer. Science 321, 1346–1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reillo I, de Juan Romero C, García-Cabezas MÁ, and Borrell V (2011). A role for intermediate radial glia in the tangential expansion of the mammalian cerebral cortex. Cereb. Cortex 21, 1674–1694. [DOI] [PubMed] [Google Scholar]
- Roadmap Epigenomics Consortium, Kundaje A, Meuleman W, Ernst J, Bilenky M, Yen A, Heravi-Moussavi A, Kheradpour P, Zhang Z, Wang J, et al. (2015). Integrative analysis of 111 reference human epigenomes. Nature 518, 317–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K, Kawasoe R, Sasaki H, Kawaguchi A, and Miyata T (2018). Neural Progenitor Cells Undergoing Yap/Tead-Mediated Enhanced Self-Renewal Form Heterotopias More Easily in the Diencephalon than in the Telencephalon. Neurochem. Res 43, 171–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaue-Sawano A, Kurokawa H, Morimura T, Hanyu A, Hama H, Osawa H, Kashiwagi S, Fukami K, Miyata T, Miyoshi H, et al. (2008). Visualizing spatiotemporal dynamics of multicellular cell-cycle progression. Cell 132, 487–498. [DOI] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012). Fiji: An open-source platform for biological-image analysis. Nat. Methods 9, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, and Eliceiri KW (2012). NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sessa A, Mao C. an, Hadjantonakis AK, Klein WH, and Broccoli V (2008). Tbr2 Directs Conversion of Radial Glia into Basal Precursors and Guides Neuronal Amplification by Indirect Neurogenesis in the Developing Neocortex. Neuron 60, 56–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa AMM, Meyer KA, Santpere G, Gulden FO, and Sestan N (2017). Evolution of the Human Nervous System Function, Structure, and Development. Cell 170, 226–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stergachis AB, Neph S, Sandstrom R, Haugen E, Reynolds AP, Zhang M, Byron R, Canfield T, Stelhing-Sun S, Lee K, et al. (2014). Conservation of trans-acting circuitry during mammalian regulatory evolution. Nature 515, 365–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian L, Bershteyn M, Paredes MF, and Kriegstein AR (2017). Dynamic behaviour of human neuroepithelial cells in the developing forebrain. Nat. Commun 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szczurkowska J, Cwetsch AW, dal Maschio M, Ghezzi D, Ratio GM, and Cancedda L (2016). Targeted in vivo genetic manipulation of the mouse or rat brain by in utero electroporation with a triple-electrode probe. Nat. Protoc 11, 399–412. [DOI] [PubMed] [Google Scholar]
- Takahashi T, Nowakowski RS, and Caviness VS (1995). The cell cycle of the pseudostratified ventricular epithelium of the embryonic murine cerebral wall. J. Neurosci 15, 6046–6057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomsen ER, Mich JK, Yao Z, Hodge RD, Doyle AM, Jang S, Shehata SI, Nelson AM, Shapovalova NV, Levi BP, et al. (2016). Fixed single-cell transcriptomic characterization of human radial glial diversity. Nat. Methods 13, 87–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turrero García M, Chang Y, Arai Y, and Huttner WB (2016). S-phase duration is the main target of cell cycle regulation in neural progenitors of developing ferret neocortex. J. Comp. Neurol 524, 456–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uebbing S, Gockley J, Reilly SK, Kocher AA, Geller E, Gandotra N, Scharfe C, Cotney J, and Noonan JP (2021). Massively parallel discovery of human-specific substitutions that alter enhancer activity. Proc. Natl. Acad. Sci. U. S. A 118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Song F, Zhang B, Zhang L, Xu J, Kuang D, Li D, Choudhary MNK, Li Y, Hu M, et al. (2018). The 3D Genome Browser: A web-based browser for visualizing 3D genome organization and long-range chromatin interactions. Genome Biol. 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wlodarchak N, and Xing Y (2016). PP2A as a master regulator of the cell cycle. Crit. Rev. Biochem. Mol. Biol 51, 162–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Won H, de la Torre-Ubieta L, Stein JL, Parikshak NN, Huang J, Opland CK, Gandal MJ, Sutton GJ, Hormozdiari F, Lu D, et al. (2016). Chromosome conformation elucidates regulatory relationships in developing human brain. Nature 538, 523–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu SC-Y, Meir Y-JJ, Coates CJ, Handler AM, Pelczar P, Moisyadi S, and Kaminski JM (2006). piggyBac is a flexible and highly active transposon as compared to sleeping beauty, Tol2, and Mos1 in mammalian cells. Proc. Natl. Acad. Sci. U. S. A 103, 15008–15013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Chen MH, Wu X, Kodani A, Fan J, Doan R, Ozawa M, Ma J, Yoshida N, Reiter JF, et al. (2016). Cell-Type-Specific Alternative Splicing Governs Cell Fate in the Developing Cerebral Cortex. Cell 166, 1147–1162.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nussbaum C, Myers RM, Brown M, Li W, et al. (2008). Model-based Analysis of ChIP-Seq (MACS). Genome Biol. 9, R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong S, Zhang S, Fan X, Wu Q, Yan L, Dong J, Zhang H, Li L, Sun L, Pan N, et al. (2018). A single-cell RNA-seq survey of the developmental landscape of the human prefrontal cortex. Nature 555, 524–528. [DOI] [PubMed] [Google Scholar]
- Zhu Y, Sousa AMM, Gao T, Skarica M, Li M, Santpere G, Esteller-Cucala P, Juan D, Ferrández-Peral L, Gulden FO, et al. (2018). Spatiotemporal transcriptomic divergence across human and macaque brain development. Science (80-. ). 362. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data S1 | Capture MPRA probe information, Related to Figure 1
Data S2 | Capture MPRA enrichment information, Related to Figure 1.
Data S3 | DNaseI datasets used in this study, Related to Figure 2.
Data S4 | RNA-seq analysis of sorted fetal brain nuclei, Related to Figure 2.
Data S5 | ChIP-seq and DNaseI-seq analysis of fetal brain nuclei, Related to Figure 2.
Data S6 | Integtation of in vivo and in vitro enhancer activities of HAR elements, Related to Figure 3.
Data S7 | Primers used for 3C experiments, Related to Figure 3.
Data Availability Statement
HAR data have been deposited at GEO accession GSE180714. Processed ChIP-seq and DNaseI-seq and HAR data are available at http://genome.ucsc.edu/cgi-bin/hgTracks?db=hg38&hubUrl=https://allendiscoverycenter-harhub.s3.us-east-2.amazonaws.com/HAR_hub/hub.txt. Accession numbers are listed in the key resources table. All the data is available from the lead contact upon request.
All the code is available from the lead contact upon request.
Any additional information required to reanalyze the data reported in this paper is available
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| PerCP-Cy5.5-conjugated anti-PAX6 | BD Biosciences | Cat#561462, Clone O18-1330 |
| V450-conjugated anti-SOX2 | BD Biosciences | Cat#561469, Clone O30-678 |
| Alexa Fluor 488-conjugated anti-NeuN | EMD Millipore | Product No. FCMAB317PE, Clone A60 |
| Anti-H3K27ac | Abcam | Prod#ab4729 |
| Anti-H3K4me1 | Abcam | Prod#ab176877 |
| Goat anti-SOX2 (1:200) | Santa Cruz | Cat#sc-170320 |
| Chicken anti-TBR2 (1:200) | Millipore | Cat#AB15894 |
| Rabbit anti-TBR2 (1:200) | Abcam | Prod#ab23345 |
| Rabbit anti-PAX6 (1:200) | Covance | Cat#PRB-278P |
| Mouse anti-HOPX (1:200) | Santa Cruz | Cat. No.. sc-30216 |
| Rabbit anti-PPP1R17 (1:200) | Novus | Cat#NBP2-13800 |
| Mouse anti-PPP1R17 (1:200) | Novus | Cat#H00010842-B01P |
| Mouse anti-SATB2 (1:200) | Abcam | Prod#ab51502 |
| Rabbit anti-Ki67 (1:200) | Abcam | Prod#ab15580 |
| Mouse anti-PCNA | Millipore | Cat#AB93501 |
| Mouse anti-GFAP | Sigma | Cat#G3893 |
| Rat anti-GFAP | Thermo Fisher | Cat#13-0300 |
| Alexa Fluor 488 Anti-Chicken Secondary Antibody | Thermo Fisher | Cat#A11039 |
| Alexa Fluor 594 Anti-Mouse Secondary Antibody | Thermo Fisher | Cat#A11008 |
| Alexa Fluor 594 Anti-Rabbit Secondary Antibody | Thermo Fisher | Cat#A11012 |
| Alexa Fluor 647 Anti-Rat Secondary Antibody | Thermo Fisher | Cat#A21247 |
| DAPI (4′,6-Diamidino-2-Phenylindole, Dihydrochloride) | Thermo Fisher | Cat#D1306 |
| Hoechst 33342 | Thermo Fisher | Cat#62249 |
| Normal Goat Serum | Gibco | Cat#16210064-100ml |
| Bovine Serum Albumin | Sigma | Cat#A7906-100G |
| Fluoromount-G | Southern Biotech | Cat#0100-01 |
| Bacterial and Virus Strains | ||
| One Shot TOP10 Chemically Competent E. coli | Thermo Fisher | Cat#C404006 |
| Biological Samples | ||
| Human brain tissue | This paper | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Formaldehyde | Fisher Scientific | Cat#F79P-4 |
| Glycine | Sigma | Cat#G7126-5KG |
| Triton X-100 | Sigma | Cat#T9284-500ML |
| Paraformaldehyde | Electron Microscopy Sciences | Cat#15710 |
| Fetal bovine serum | Atlanta Bio | Cat#S11150 |
| Dulbecco’s Modified Eagle Medium | Thermo Fisher | Cat#MT10013CV |
| Dulbecco’s Modified Eagle Medium/F12 | Thermo Fisher | Cat#11330032 |
| Penicillin-Streptomycin | Thermo Fisher | Cat#15140122 |
| AMPure XP beads | Beckman-Coulter | Cat#A63881 |
| Retrievagen A | BD Biosciences | Cat#550524 |
| Lysis Buffer | This paper | N/A |
| Wash buffer | This paper | N/A |
| Sonication buffer | This paper | N/A |
| AsiSI restriction enzyme | NEB | Cat#R0630 |
| DNaseI | Sigma-Aldrich | Cat#D4527 |
| MboI | NEB | Cat#R0147 |
| T4 DNA Ligase | Thermo Fisher | Cat#EL0011 |
| Critical Commercial Assays | ||
| Lipofectamine LTX reagent | Thermo Fisher | Cat#A12621 |
| TRIzol reagent | Invitrogen | Cat#15596018 |
| NEBNext Single Cell/Low Input RNA Library Prep Kit for Illumina | New England BioLabs | Cat#E6420S |
| Protein A Dynabeads | Thermo Fisher | Cat#10002D |
| Ovation Ultralow V2 kit | Nugen | Cat#0344-32 |
| Neural Tissue Dissociation Kit (T) | Miltenyi Biotec | Cat#130-093-231 |
| Deposited Data | ||
| Raw and analyzed data | This paper | GSE180714 |
| Human reference genome build GRCh38/hg38 | Genome Reference Consortium | https://www.ncbi.nlm.nih.gov/assembly/GCF_000001405.26/ |
| Human, chimpanzee, and macaque brain transcriptome data | Sousa et al., 2018 | N/A |
| ENCODE Project and Roadmap Epigenomics Consortium | ENCODE Project Consortium, 2012; Roadmap Epigenomics Consortium et al., 2015 | http://www.roadmapepigenomics.org/ |
| UCSC-tools and genome browser | Kent et al., 2002 | http://genome.ucsc.edu/ |
| Pathway and gene set overdispersion analysis (PAGODA) | Fan et al., 2016 | N/A |
| Single cell ATAC-seq data | Domcke et al., 2020 | https://descartes.brotmanbaty.org/ |
| Experimental Models: Cell Lines | ||
| Neuro2A | ATCC | Cat#CCL-131 |
| SH-SY5Y cell | ATCC | Cat#CRL-2266 |
| Experimental Models: Organisms/Strains | ||
| CD1 Mice | Charles River | Crl:CD1(ICR) |
| Ferrets | Marshall Bioresources | N/A |
| Macaques | Yale University School of Medicine | N/A |
| Oligonucleotides | ||
| CustomArray pool | CustomArray, Bothell, WA | http://www.customarrayinc.com/oligos_main.htm |
| Primers for 3C interaction assay, see Supplemental Data Table S6 | This paper | N/A |
| Recombinant DNA | ||
| Human genomic DNA | Promega | Cat#G1521 |
| Chimpanzee DNA | Coriell | Cat#NS03489 |
| pMPRA1 vector | Melnikov et al., 2014 | Addgene 49349 |
| pMPRA_donor2 | Melnikov et al., 2014 | Addgene 49353 |
| pFucci-G1 Orange | Medical & Biological Laboratories Co., LTD. | Cat#AM-V9003M |
| PBCAG-eGFP | Gift from Joseph LoTurco, Chen et al., 2012 | Addgene 40973 |
| pCyl50 | Gift from Jae Yong Han, Lee et al, 2016 | N/A |
| pCAG-PB-GFP | This paper | N/A |
| pCAG-PPP1R17-PB-GFP | This paper | N/A |
| Software and Algorithms | ||
| MIPgen | Shendure Lab, University of Washington | http://shendurelab.github.io/MIPGEN/ |
| R language | R Core Team, 2019 The R Project for Statistical Computing | https://www.r-project.org/ |
| Python language, version 2.7 | Python Software Foundation | http://www.python.org |
| GraphPad Prism 8 | GraphPad Software Inc., La Jolla, CA, USA | https://www.graphpad.com/scientific-software/prism/ |
| Fiji | Schindelin et al., 2012 | https://imagej.net/ImageJ |
| ImageJ | Schneider et al., 2012 | https://imagej.net/ImageJ |
| IncuCyte Zoom | Essen Bioscience | IncuCyte S3 2018B |
| HOMER | Heinz et al., 2010 | http://homer.ucsd.edu/homer/ |
| Other | ||
| HARHub | This paper | http://genome.ucsc.edu/cgi-bin/hgTracks?db=hg38&hubUrl=https://allendiscoverycenter-harhub.s3.us-east-2.amazonaws.com/HAR_hub/hub.txt |


