Abstract
A transcriptional repressor complex encoded by two essential genes, YDR1 and BUR6, was isolated from Saccharomyces cerevisiae and shown to be the functional counterpart of the human repressor complex Dr1-DRAP1. To elucidate the mechanism of repression by this complex, altered forms of Ydr1 and Bur6 were studied in vitro and in vivo. Deletion of the C-terminal 41 amino acids of Ydr1 resulted in loss of repressor activity and a growth defect, suggesting that the C-terminal domain of Ydr1 functions as a potent transcriptional repressor. A screen for extragenic suppressors of a cold-sensitive ydr1 (ydr1cs) mutant led to the identification of recessive mutations in the SIN4 gene, which encodes a component of the SRB-MED complex. The sin4 alleles suppressed not only ydr1cs mutations but also bur6cs mutations. In contrast, deletion of the gal11 gene, whose product is also a member of the SRB-MED complex, failed to suppress ydr1cs and bur6cs mutations, indicating that suppression is not due to general defects in the SRB-MED complex. Moreover, one of the sin4 alleles, but not the sin4 deletion, was found to specifically suppress the inviability of a ydr1 deletion, demonstrating that the essential function of Ydr1 becomes dispensable in a sin4 mutant background. Biochemical analysis of the SRB-MED complex from the sin4 suppressor strain revealed a structurally distinct form of the SRB-MED complex that lacks a subset of mediator subunits. These results define a delicate balance between positive and negative regulators of transcription operating through the Ydr1-Bur6 repressor complex.
Regulation of transcription in eukaryotes requires an intricate network of both positive and negative factors to maintain optimal expression of target genes (16; for details, see http://www.wi.mit.edu/young/expression.html). The identification of an additional class of regulators referred to as coactivators and corepressors underscores the complexity of transcriptional regulatory networks in eukaryotes (for reviews, see references 12 and 13). These factors, some of which are components of multiprotein complexes including RNA polymerase II (RNAPII) (the so-called “RNAPII holoenzyme” [33]), provide specific interaction sites for positive and negative regulators (14).
A large number of proteins that negatively regulate transcription have been described (for reviews, see references 15, 21, and 30). One family of repressors includes proteins that are tethered to promoters by interacting with sequence-specific DNA binding proteins and/or components of the basal transcription machinery. These include, among others, Tup1-Ssn6 (22), Mot1 (1), Sin3 (2), and Dr1-DRAP1 (17). A repressor complex from the yeast Saccharomyces cerevisiae which is encoded by two essential genes (YDR1 and BUR6) was identified as the functional counterpart of the human Dr1-DRAP1 (NC2) complex (9, 10, 24, 36). The transcriptional repressor activity of this yeast complex was demonstrated in vitro by biochemical studies and in vivo by genetic studies (9, 10, 24, 36). Amino acid sequence comparison of yeast, drosophila, and human Dr1 polypeptides revealed two highly conserved domains located at the N and C termini of the protein. The C-terminal domain of human Dr1 includes a transferable transcriptional repressor domain (45). Dr1-DRAP1 heterodimer formation occurs via the N-terminal domains of both proteins. These domains contain a histone fold motif that is crucial for DRAP1-mediated enhancement of transcriptional repression by Dr1 (11, 23, 31). Mutations in the histone fold motifs of Ydr1 or Bur6 were found to inhibit repressor function and to suppress an srb4 temperature-sensitive (ts−) phenotype (9, 26).
Characterization of the RNAPII holoenzyme has identified mechanisms by which this complex mediates transcriptional regulation. The RNAPII holoenzyme from S. cerevisiae consists of core RNAPII, a set of general transcription factors, Srb proteins (SRB), mediator proteins (MED), and several other polypeptides identified previously as both positive and negative transcriptional regulators (33, 34). Taken together, the results of several studies suggest the presence of modular subcomplexes that associate with RNAPII to mediate the response to physiological or developmental cues from specific transcription factors (for a review, see reference 13). One of these subcomplexes, the Gal11 subcomplex, contains the Gal11, Sin4, Med3 (Hrs1), and Med2 proteins and interacts physically with the Rgr1 protein (27). SIN4 (TSF3) was identified as a negative regulator of HO transcription (19) and has been implicated in the transcriptional activation and repression of a broad spectrum of genes (6, 7, 18, 19, 20). It has been suggested that the Sin4-containing Gal11 module functions as an input port for signals from a subset of gene-specific transcriptional regulators (14).
Nearly all of what we know regarding the functions of Dr1-DRAP1 has been gleaned from in vitro studies. To investigate the role of the Ydr1-Bur6 repressor complex in vivo, we have isolated and characterized extragenic suppressors of the cold-sensitive (cs−) phenotype of a ydr1 mutant. Here we report the identification of a sin4 allele as a suppressor of ydr1cs. These results define a genetic relationship between positive and negative transcriptional regulators. We also describe a possible mechanism of suppression in terms of the subunit composition of the RNAPII holoenzyme, emphasizing the importance of the delicate balance between positive and negative transcriptional regulators.
MATERIALS AND METHODS
Preparation of recombinant Ydr1 and Bur6 polypeptides.
Full-length Ydr1 (FL-Ydr1), FL-Bur6, and two truncated Ydr1 polypeptides were expressed in Escherichia coli using the pET21-a plasmid and purified through a Ni-nitrilotriacetic acid (NTA) column (Qiagen) under denaturing conditions. After mixing of FL-Ydr1 and two truncated versions of Ydr1 with FL-Bur6 in an equal molar ratio, renaturation was carried out overnight at room temperature in buffer G (30 mM Tris-HCl [pH 7.5], 150 mM KCl, 10% glycerol, 5 mM dithiothreitol). The Ydr1-Bur6 heterodimer was further purified by S-200 gel filtration chromatography.
In vitro transcription and I.P.
Transcription reactions were carried out using highly purified preparations of human transcription factors (24). Immunoprecipitation (I.P.) experiments were performed as described previously (24).
Genetic manipulation.
The yeast strains used in this study are listed in Table 1. Details of the strains and plasmid constructions are available upon request. Strain DY1717 was a generous gift from David Stillman, and strain MCY2253 was from Marian Carlson. DY1717 was described previously (18). Yeast media were prepared as previously described (41). Yeast transformations were performed by a lithium acetate procedure (39). The plasmid shuffle method was performed as previously described (3), using 5-fluoroorotic acid (FOA).
TABLE 1.
Yeast strains used in this study
| Strain | Genotype |
|---|---|
| YMH200 | MATa ura3-52 leu2-7 his3-11 trp1-1 ade2-1 ydr1Δ::HIS3 [CEN-URA3-YDR1] |
| YMH201 | MATα ura3-52 leu2-7 his3-11 trp1-1 ade2-1 ydr1Δ::HIS3 [CEN-URA3-YDR1] |
| YMH202 | MATα ura3-52 leu2-7 his3-11 trp1-1 ade2-1 bur6Δ::HIS3 [CEN-URA3-BUR6] |
| YMH203 | MATα ura3-52 leu2-7 his3-11 trp1-1 ade2-1 bur6Δ::HIS3 [CEN-URA3-BUR6] |
| YSK013a | MATa ydr1Δ::HIS3 [CEN-URA3-YDR1] [CEN-TRP1-YDR1] |
| YSK014a | MATa ydr1Δ::HIS3 [CEN-URA3--YDR1] [CEN-TRP1] |
| YSK025a | MATα ydr1Δ::HIS3 [CEN–LEU2–ydr1-11] |
| YSK027a | MATα ydr1Δ::HIS3 [CEN–LEU2–ydr1-13] |
| YSK028a | MATα ydr1Δ::HIS3 [CEN–LEU2–ydr1-16] |
| YSK033b | MATα bur6Δ::HIS3 [CEN–LEU2–bur6-42] |
| YSK034b | MATα bur6Δ::HIS3 [CEN–LEU2–bur6-43] |
| YSK035b | MATα bur6Δ::HIS3 [CEN–LEU2–bur6-47] |
| YSK041a | MATα ydr1Δ::HIS3 [CEN–LEU2–ydr1-13] sin4-42 |
| YSK042a | MATα ydr1Δ::HIS3 [CEN–LEU2–ydr1-13] sin4-54 |
| YSK043a | MATα ydr1Δ::HIS3 [CEN–LEU2–ydr1-13] sin4-61 |
| YSK044a | MATα ydr1Δ::HIS3 [CEN–LEU2–ydr1-13] sin4-121 |
| YSK045a | MATα ydr1Δ::HIS3 [CEN–LEU2–ydr1-13] sin4-168 |
| YSK054a | MATα ydr1Δ::HIS3 [CEN-TRP1-MET25] |
| YSK055a | MATα ydr1Δ::HIS3 [CEN-TRP1-MET25-YDR1] |
| YSK056a | MATα ydr1Δ::HIS3 [CEN-TRP1-MET25-ydr1C130] |
| YSK057a | MATα ydr1Δ::HIS3 [CEN-TRP1-MET25-ydr1C105] |
| YSK069a | MATα ydr1Δ::HIS3 [CEN–LEU2–ydr1-13] sin4-54 [CEN-URA3-SIN4] |
| YSK070a | MATα ydr1Δ::HIS3 [CEN–LEU2–ydr1-13] sin4-168 [CEN-URA3-SIN4] |
| YSK071a | MATα ydr1Δ::HIS3 [CEN–LEU2–ydr1-13] sin4-54 [CEN-URA3-YTP1] |
| YSK072a | MATα ydr1Δ::HIS3 [CEN–LEU2–ydr1-13] sin4-168 [CEN-URA3-YTP1] |
| YSK075a | MATa ydr1Δ::HIS3 [CEN-TRP1-YDR1] SIN4::[SIN4-URA3] |
| DY1717 | MATα ade2-1 his3-11 leu2-7 lys2 trp1-1 ura3-52 sin4Δ::TRP1 |
| MCY2253 | MATα ade2-101 ura3-52 spt13-10Δ::TnLUK |
| YSK084a | MATa ydr1Δ::HIS3 [CEN-URA3-YDR1] sin4Δ::TRP1 |
| YSK085a | MATa ydr1Δ::HIS3 [CEN–LEU2–ydr1-11] sin4Δ::TRP1 |
| YSK086a | MATa ydr1Δ::HIS3 [CEN–LEU2–ydr1-13] sin4Δ::TRP1 |
| YSK087a | MATa ydr1Δ::HIS3 [CEN–LEU2–ydr1-16] sin4Δ::TRP1 |
| YSK093b | MATa bur6Δ::HIS3 [CEN-URA3-BUR6] sin4Δ::TRP1 |
| YSK095b | MATa bur6Δ::HIS3 [CEN–LEU–bur6-42] sin4Δ::TRP1 |
| YSK096b | MATa bur6Δ::HIS3 [CEN–LEU2–bur6-43] sin4Δ::TRP1 |
| YSK097b | MATa bur6Δ::HIS3 [CEN–LEU2–bur6-47] sin4Δ::TRP1 |
| YSK103c | MATα ydr1Δ::HIS3 [CEN–LEU2–ydr1-11] |
| YSK104c | MATα ydr1Δ::HIS3 [CEN–LEU2–ydr1-13] |
| YSK105c | MATα ydr1Δ::HIS3 [CEN–LEU–ydr1-16] |
| YSK108c | MATa bur6Δ::HIS3 [CEN–LEU2–bur6-42] |
| YSK109c | MATa bur6Δ::HIS3 [CEN–LEU2–bur6-43] |
| YSK110c | MATa bur6Δ::HIS3 [CEN–LEU2–bur6-47] |
| YSK124a | MATα ydr1Δ::HIS3 [CEN-URA3-YDR1] sin4-168 |
| YSK125a | MATα ydr1Δ::HIS3 [CEN-URA3-YDR1] sin4-54 |
| YSK126a | MATα ydr1Δ::HIS3 [CEN-URA3-YDR1] [CEN-TRP1-YDR1] sin4-54 |
| YSK127a | MATα ydr1Δ::HIS3 [CEN-URA3-YDR1] [CEN-TRP1] sin4-54 |
| YSK128a | MATα ydr1Δ::HIS3 [CEN-URA3-YDR1] [CEN-TRP1-YDR1] sin4-168 |
| YSK129a | MATα ydr1Δ::HIS3 [CEN-URA3-YDR1] [CEN-TRP1] sin4-168 |
| YSK130b | MATa bur6Δ::HIS3 [CEN-URA3-BUR6] sin4-54 |
| YSK131b | MATa bur6Δ::HIS3 [CEN-URA3-BUR6] sin4-168 |
| YSK132b | MATa bur6Δ::HIS3 [CEN-URA3-BUR6] [CEN-TRP1-BUR6] sin4-54 |
| YSK133b | MATa bur6Δ::HIS3 [CEN-URA3-BUR6] [CEN-TRP1] sin4-54 |
| YSK134b | MATa bur6Δ::HIS3 [CEN-URA3-BUR6] [CEN-TRP1-BUR6] sin4-168 |
| YSK135b | MATa bur6Δ::HIS3 [CEN-URA3-BUR6] [CEN-TRP1] sin4-168 |
| YSK140a | MATa ydr1Δ::HIS3 sin4-54 [CEN-TRP1-SIN4] |
| YSK141a | MATα ydr1Δ::HIS3 sin4-54 [CEN-TRP1] |
| VM02 | MATα ade2 leu2 ura3 HA-6His-tagged SRB5 |
| YSK149 | MATα ade2 leu2 ura3 HA-6His-tagged SRB5 sin4-54 |
Derived from YMH200 or YMH201.
Derived from YMH202 or YMH203.
Derived from MCY2253.
Isolation of conditional mutations by error-prone PCR was done as described elsewhere (32), with the following modifications. The gapped plasmids for both ydr1 and bur6 were constructed by removing most of the open reading frames by restriction digestion. For efficient mutagenesis, the mutagenizing deoxyribose nucleoside triphosphate concentration was less than 30 μM. The FOA-resistant candidates were tested for the cs− phenotype by growth at 11°C for 14 days. The sequences of the mutant alleles were determined by sequencing the plasmid DNA isolated from mutant cells by standard methods (43).
To establish allelism between the ydr1 suppressors and the cloned SIN4 gene, a URA3-tagged ydr1 deletion strain (YSK075) was constructed by transforming YSK013 with the linearized YIp-SIN4-URA3 construct and selecting for Ura+ transformants. A diploid strain was generated by mating YSK075 with each suppressor strain, followed by sporulation and tetrad analysis using standard procedures (42). The suppressor was scored as ts− in the ydr1 background, whereas SIN4 was scored by the Ura+ phenotype.
Suppressor alleles were cloned from genomic DNA by gap repair (35) and sequenced.
Suppressor screening.
Strain YSK027 (ydr1cs) was grown overnight at 30°C in 10 ml of yeast extract-peptone-dextrose (YPD) medium, plated on YPD medium at a cell density of 106, and incubated at 11°C for 14 to 21 days. Spontaneous revertants were obtained at a frequency of 10−6. Each colony was purified by subcloning and rescored at 11°C. Five cs+ revertants (YSK041 to YSK045) were subsequently found to be ts−. To determine whether the mutations in the revertants were dominant or recessive, the five ts− revertants were crossed with YMH200 and the resulting diploid strains were scored for cold and temperature sensitivity.
In order to clone the suppressor gene from the revertants, a yeast genomic DNA library (37) obtained from the American Type Culture Collection was introduced into each suppressor strain and Ura+ transformants were selected and scored for complementation of ts− at 37°C. Library DNAs were isolated from ts+ transformants, and sequenced. DNA manipulations and PCR amplifications were performed as previously described (38).
Purification of SRB-MED complexes.
Strain VM02 (His-tagged SRB5) was a generous gift from Rick Young. Large-scale cultures of yeast strains VM02 and YSK149 were grown in the fermentation facility at the Waksman Institute. Yeast whole-cell extracts were prepared as previously described (40). The SRB-MED complex was purified from 1 kg of cells of each strain using Bio-Rex70 (Bio-Rad), DEAE-Sephacel (Pharmacia), Bio-Gel-HTP hydroxyapatite (Bio-Rad), and MonoQ HR 10/10 (Pharmacia) as described previously (25). The 1 M potassium acetate eluent from the MonoQ column was dialyzed against buffer I (20 mM Tris-acetate [pH 7.9], 10% glycerol, 10 mM imidazole, 0.02% NP-40, 5 μM β-mercaptoethanol) and loaded onto a Ni-NTA column. After extensive washing with buffer I containing 0.8 M potassium acetate, the bound material was eluted with a buffer containing 400 mM imidazole and subjected to Western blot analysis.
RESULTS
The C-terminal conserved domain of Ydr1 is critical for repression and growth.
Comparison of the human (17) and yeast (24) Dr1 proteins revealed a conserved 12-amino-acid sequence near the C termini (Fig. 1). This region of human Dr1 functions as a transferable repression domain in vitro and in transfected cells (44). In order to further characterize this region of Ydr1, we performed studies with FL-Ydr1 and two truncated Ydr1 derivatives in vitro and in vivo (Fig. 1). In the in vitro experiments, we tested the ability of the truncated Ydr1 polypeptides to heterodimerize with Bur6, bind to the yeast TATA binding protein (TBP), and repress transcription in a reconstituted human system (Fig. 2A to C). These activities were correlated with the ability of the two derivatives to support cell viability (Fig. 2D). The two Ydr1 derivatives are C-terminal truncations that retain the N-terminal 130 amino acids (C130) or 105 amino acids (C105) (Fig. 1). C130 and C105 are able to form heterodimers with FL-Bur6 in vitro, as determined by co-I.P. experiments (Fig. 2A). These two forms of Ydr1 also retain the ability to interact with TBP, although the affinity of the Ydr1-TBP interaction appears to be diminished relative to that of FL-Ydr1 (Fig. 2B and data not shown). The C130 protein retains the ability to repress transcription; however, C105 fails to repress transcription (Fig. 2C). The C130 derivative fully supports cell growth in the absence of normal Ydr1, whereas the C105 derivative is nearly inviable (Fig. 2D; summarized in Fig. 3). These results are consistent with the previous characterization of human Dr1 which demonstrated that the transferable repressor domain includes residues that have been deleted from C105 but are present in C130 (45). Thus, the essential function of Ydr1 in vivo correlates with its ability to repress transcription in vitro, rather than its interaction with Bur6.
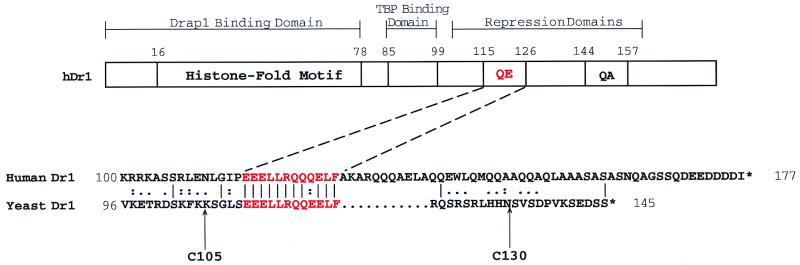
FIG. 1.
Sequence alignment of the C termini of human and yeast Dr1 polypeptides. The amino acid numbers are shown at the ends of the sequences. A conserved 12-amino-acid sequence is in red. Two truncated Ydr1 derivatives are indicated by arrows at the bottom of the yeast Dr1 sequence. A schematic representation of the human Dr1 polypeptide is shown at the top. QE, QE-rich domain; QA, QA-rich domain. For details, see reference 45.
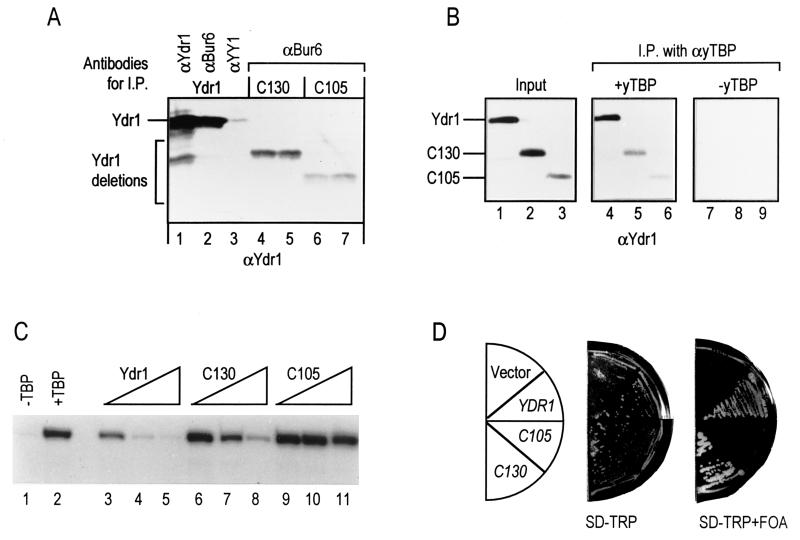
FIG. 2.
The C-terminal conserved domain of Ydr1 is critical for transcriptional repression and cell growth. (A) The ability of Ydr1 derivatives to form a heterodimer with Bur6 was examined by co-I.P. Ydr1, FL-Ydr1 (lanes 1 to 3); C130 and C105, two truncated forms of Ydr1 (lanes 4 and 5 and 6 and 7, respectively). Lanes 4 and 5 are duplicates of C130 under the same conditions; lanes 6 and 7 are the same as for C105. Antibodies for I.P. are indicated at the top. α-Ydr1, antibody against Ydr1; α-Bur6, antibody against Bur6; α-YY1, antibody against YY1 (control). The antibody used for Western blot analysis is indicated at the bottom. (B) The interaction between yeast TBP and the Ydr1-Bur6 complex was monitored in a co-I.P. experiment (24). α-yTBP antibody-beads were preincubated with (+yTBP, lanes 4 to 6) or without (−yTBP, lanes 7 to 9) yeast TBP, and then each Ydr1-Bur6 heterodimer was added. Input, 1/10 of each Ydr1-Bur6 heterodimer that was added to α-yTBP-beads (lanes 1 to 3). The polyclonal antibody against Ydr1 recognizes C105, yet its affinity for this truncated polypeptide is approximately 2.5-fold less than its affinity for FL and C130 polypeptides. This differential affinity of the antibodies is important in the interpretation of the results shown in lanes 5 and 6. The overall decrease in binding of C130 and C105 to TBP remains constant at approximately 30% of the binding observed with the FL-Ydr1 polypeptide. (C) Transcriptional repressor activity was measured in vitro using yeast TBP and a highly purified human transcription system. Ydr1, FL-Ydr1 (lanes 3 to 5); C130 and C105, C-terminal truncation of Ydr1 to amino acids 130 (lanes 6 to 8) and 105 (lanes 9 to 11), respectively. (D) Each truncated form of the ydr1 gene in the CEN plasmid was transformed into a YDR1 plasmid shuffle strain. The viability of each transformant was examined on a plate containing FOA (SD-TRP+FOA). The genotypes of the strains are shown in the semicircular diagram on the left.
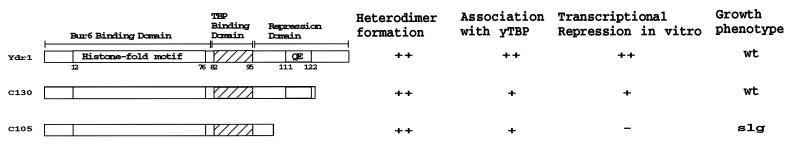
FIG. 3.
Summary of the characterization of Ydr1 derivatives. Ydr1 truncations are represented schematically by bars. The various biochemical activities of Ydr1 were measured in vitro using Ydr1-Bur6 heterodimers, and the effects on growth are summarized at the right. TBP Binding Domain, putative TBP binding domain; Repression Domain, assigned repression domain based on human Dr1 studies (44); QE, QE-rich domain; wt, wild-type growth; slg, slow growth. ++, same as FL-Ydr1; +, slightly reduced; −, significantly reduced.
Isolation of suppressors of ydr1-13.
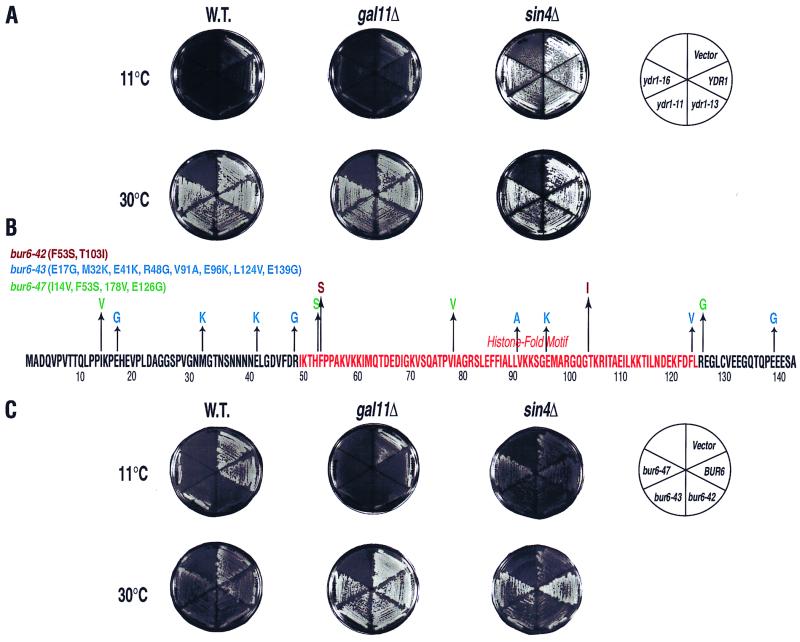
In order to extend our understanding of Ydr1 function in vivo, we sought to define the genetic relationship between Ydr1 and other transcriptional regulatory proteins. As an initial step toward this goal, we generated cs− ydr1 mutants by PCR-mediated mutagenesis. Some of the cs− mutations mapped within the 25-amino-acid C-terminal conserved domain of Ydr1 (Fig. 4A). One mutant (YSK027) encodes replacement of phenylalanine with serine at position 106 (ydr1-13; F106S) and exhibits a marked cs− phenotype at 11°C (Fig. 4B; ydr1-13 SIN4). From a total of 108 cells plated, ∼100 independent, spontaneous cs+ revertants were selected on rich medium at 11°C (YSK041 to YSK045; Table 1). In an effort to identify potential pleiotropic phenotypes associated with these suppressors, cs+ revertants were screened for ts− growth defects at 37°C. Five revertants exhibited distinct ts− phenotypes and were further characterized (Fig. 4B and data not shown).
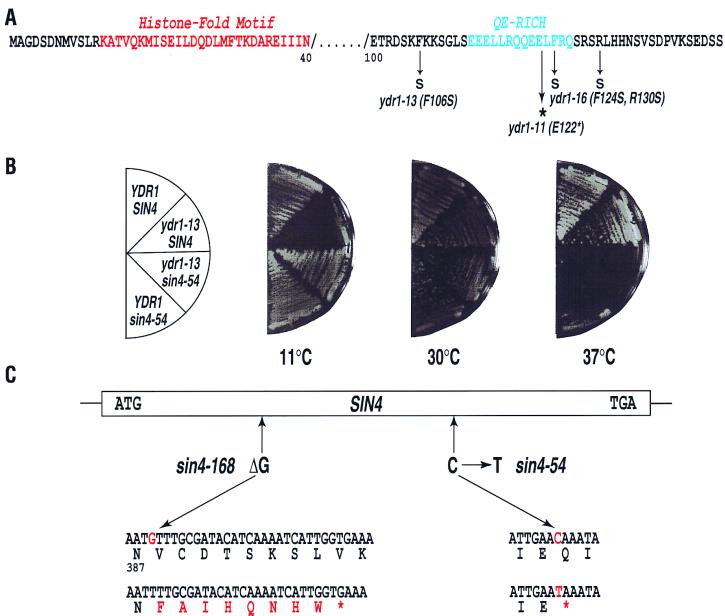
FIG. 4.
Suppression of the cs− phenotype of the ydr1 mutant allele by recessive mutation in the SIN4 gene. (A) Point mutations of cs− ydr1 mutant alleles. The histone fold motif (in red) and the conserved QE-rich domain (in blue) are indicated on the Ydr1 amino acid sequence. Amino acid substitutions for each mutation are indicated below the sequence. (B) Four different isogenic strains (genotypes are indicated on the left) were compared for the ability to grow at various temperatures on YPD medium. (C) Sequence analyses of two different sin4 suppressor mutations. Genomic DNA segments containing the sin4 alleles were retrieved from each suppressor strain by the gap repair method. The amino acid sequences changed by the sin4 mutations are indicated at the bottom in red.
The five cs+ ts− revertants were crossed with strain YMH200 (ydr1Δ::HIS3 [YDR1-URA3]). The resulting diploid strains were cured of the YDR1-URA3 plasmid and subsequently scored for the cs− and ts− phenotypes. All five diploid strains were phenotypically cs− and ts+, indicating that the revertant phenotypes were due to recessive mutations (data not shown). Plasmid-borne YDR1 was unable to complement the ts− phenotype of the haploid revertants, indicating that the suppressors are not allelic to YDR1. We tentatively designated this suppressor gene(s) scd, for suppressor of cold-sensitive ydr1-13.
The allele specificity of each suppressor was determined by plasmid shuffle using two other ydr1 cs− alleles. The ydr1-11 allele is the result of a nonsense mutation, encoding a truncated form of Ydr1 at position 122, whereas ydr1-16 encodes F124S and R130S replacements (Fig. 4A). These alleles were introduced into strains YMH201 (ydr1Δ::HIS3 SCD [YDR1-URA3]) and YSK125 (ydr1Δ::HIS3 scd [YDR1-URA3]), and the wild-type YDR1 gene was counterselected on FOA medium. The resulting strains were then scored for growth at 11°C. All three ydr1 mutants grew well in the scd background, compared to the marked cs− phenotype in the SCD background. Thus, suppression by the scd allele of strain YSK125 is not specific to the ydr1-13 allele, but instead, scd is able to suppress the cs− growth phenotype associated with three different ydr1 alleles.
Identification of sin4 as a suppressor of ydr1-13.
Since all scd suppressor strains were isolated as spontaneous cs+ revertants, we made the tentative assumption that the ts− phenotype of these revertants is a pleiotropic phenotype associated with the scd mutations. Accordingly, we exploited the ts− phenotype to clone the wild-type allele. Strain YSK042 (ydr1-13 scd1) was transformed with the YCp50 library and selected for Ura+ transformants at 37°C. Several Ura+ ts+ transformants were obtained. Plasmid DNA was isolated from one transformant and reintroduced into YSK042. In this case, all of the scored transformants were ts+, indicating that the ts+ phenotype is due to plasmid DNA rather than strain reversion. Restriction analysis of this clone identified a 5.65-kb insert. Sequence analysis using primers that annealed immediately adjacent to the insert identified the YTP1 and SIN4 genes. Both genes were cloned individually into CEN plasmids and reintroduced into both YSK042 and YSK045. Plasmid DNA containing the SIN4 gene (YSK069 and YSK070], but not YTP1 (YSK071 and YSK072), fully complemented the ts− growth defect. Furthermore, all ts+ transformants were cs−, demonstrating that SIN4 complements both the pleiotropic ts− and suppressor cs+ phenotypes.
Allelism between the scd1 suppressor and SIN4 was tested with two different suppressor strains (YSK042 and YSK043). YSK042 and YSK043 (ydr1-13 scd) were crossed with strain YSK075 (ydr1-13 SIN4-URA3), which contains the wild-type SIN4 gene tagged with URA3. Diploid strains were sporulated and dissected, and the resulting progeny were scored for uracil auxotrophy and temperature sensitivity at 37°C. Among 20 tetrads derived from the YSK042 × YSK075 cross, the Ura+:Ura− and ts+:ts− phenotypes segregated 2:2. Moreover, all Ura+ segregants were ts+ and all Ura− segregants were ts−. Similar results were obtained with the YSK043 × YSK075 cross (data not shown). Thus, the ts− phenotype conferred by the scd1 mutation segregates opposite to SIN4, thereby establishing that scd1 is allelic to SIN4. We now refer to the scd1 suppressor as sin4-54.
The cloned wild-type SIN4 gene was introduced into each of the other four scd suppressor strains. The resulting transformants were all ts+, indicating that wild-type SIN4 complements all scd suppressors. Thus, each of the other four scd suppressors is likely to be allelic to SIN4. Moreover, as defined below, SIN4 DNA was cloned from two of the five suppressors (sin4-54 and sin4-168) and found to contain specific mutations.
Characterization of sin4 suppressors.
Genomic DNA encompassing the sin4-54 and sin4-168 alleles was cloned by gap repair from strains YSK042 and YSK045, respectively. Sequence analyses showed that both alleles encode truncated forms of Sin4: sin4-54 contains a nonsense mutation at codon 672, whereas sin4-168 contains a frameshift mutation at codon 388 (Fig. 4C).
The nature of the sin4 alleles suggested that suppression is due to loss of Sin4 function. We tested this possibility by determining whether a sin4 deletion would also suppress the ydr1 mutations. We constructed a ydr1 plasmid shuffle strain using sin4Δ::TRP1 deletion strain DY1717 (18). The ydr1-11, ydr1-13, and ydr1-16 alleles were introduced into the resulting strain, YSK084, and the wild-type YDR1 plasmid was counterselected on FOA medium. In contrast to the ydr1 SIN4 strains (YSK025, YSK027, and YSK028), all three ydr1 sin4Δ strains (YSK085, YSK086, and YSK087) grew well at 11°C (Fig. 5A), indicating that the deletion of SIN4 is sufficient to suppress all three ydr1 cs mutations.
FIG. 5.
Suppression of cs− ydr1 and bur6 mutants by the sin4 deletion mutation but not by the gal11 deletion mutation. (A) Different ydr1 mutant alleles in plasmid DNA were transformed into either a sin4 deletion strain (sin4Δ), a gal11 deletion strain (gal11Δ), or the wild-type strain (W.T.). The cs− phenotype of each strain was examined by streaking onto a YPD plate and incubation at either 30 or 11°C. The genotypes of the strains are shown in the circular diagram. Vector, strain YMH200 containing wild-type YDR1 and an empty LEU2 marker plasmid (control). (B) Point mutations of the bur6 cs− mutant alleles. The histone fold motif is in red. Amino acid substitutions for each mutation are indicated as follows: bur6-42, brown; bur6-43, blue; bur6-47, green. (C) Different bur6 mutant alleles in plasmid DNA were transformed into either a sin4 deletion strain, a gal11 deletion strain, or the wild-type strain. The cs− phenotype of each strain was examined by streaking onto a YPD plate and incubation at either 30 or 11°C. The genotypes of the strains are shown in the circular diagram. Vector, strain YMH203 containing wild-type BUR6 and an empty LEU2 marker plasmid (control).
Suppression of bur6 alleles by sin4 mutations.
Because of the functional relationship between Ydr1 and Bur6, we analyzed the potential effects of the sin4 alleles on bur6 mutations. Three bur6 alleles were generated by error-prone PCR as described above for ydr1 alleles. The bur6-42, bur6-43, and bur6-47 alleles of strains YSK033, YSK034, and YSK035 each encode two or more amino acid replacements (Fig. 5B) and confer cs− growth defects (Fig. 5C). The sin4-54 and sin4-168 mutations suppress the cs− phenotypes of all three bur6 alleles (data not shown). Furthermore, the sin4Δ deletion fully restored growth at 11°C in all three bur6 backgrounds (YSK095, YSK096, and YSK097) (Fig. 5C). These results, in combination with the ydr1 sin4 data, indicate that deletion of SIN4 compensates for diminished Ydr1-Bur6 function.
Neither ydr1 nor bur6 alleles are suppressed by gal11.
The SIN4 gene was identified originally as a negative regulator of HO transcription (19) and was later recovered in a screen for genes that relieve repression of the GAL1 and GAL10 genes (6). It was proposed that SIN4 plays both positive and negative roles in transcriptional regulation (20). More recently, Sin4 was identified as a component of the SRB-MED transcriptional regulatory complex that associates with RNAPII to form the so-called holoenzyme complex (29).
Gal11 is another component of the SRB-MED complex and is found in a subcomplex with Sin4 (27, 29). Moreover, sin4 and gal11 mutants have certain genetic similarities, including transcriptional defects (5, 8, 20). This prompted us to examine whether a mutation in the GAL11 gene can suppress the ydr1 and bur6 alleles. The YDR1 and BUR6 plasmid shuffle systems were set up in a gal11Δ deletion strain and transformed individually with each of the three distinct cs− ydr1 and bur6 alleles described above to generate strains YSK103, YSK104, and YSK105 (ydr1 gal11), and YSK108, YSK109, and YSK110 (bur6 gal11). Surprisingly, none of the ydr1 or bur6 alleles were suppressed by gal11Δ deletion (Fig. 5). Because we observed no genetic suppression of ydr1cs or bur6cs by gal11Δ, we conclude that suppression of ydr1cs and bur6cs is not common to these two components of the Gal11 subcomplex but is a specific feature of sin4.
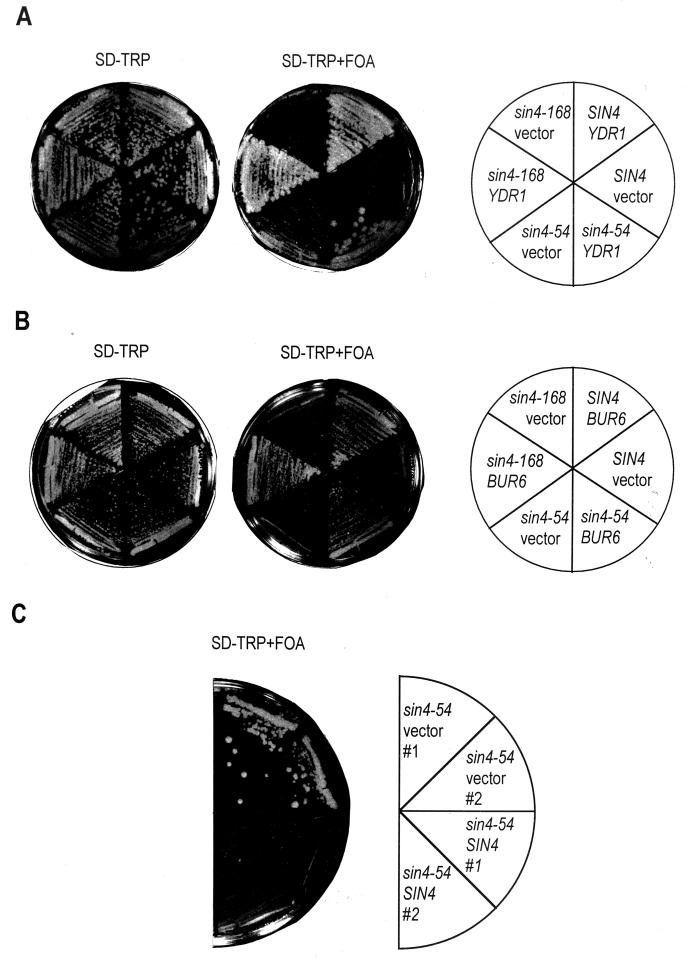
The sin4-54 allele renders YDR1, but not BUR6, dispensable for cell viability.
The genetic relationship between ydr1 or bur6 and sin4 defined in this study suggested that the activities of the Ydr1-Bur6 repressor and the Sin4 component of the SRB-MED complex exist in a delicate balance to regulate gene expression. This prompted us to examine whether YDR1 and BUR6, both essential genes in a wild-type background, are dispensable for cell viability in sin4 genetic backgrounds. It is conceivable that loss of Ydr1-Bur6 function eliminates an essential balance between positive and negative transcriptional regulatory mechanisms, with this balance apparently restored by the loss of Sin4 function.
We tested this possibility by investigating if the sin4 suppressors render YDR1 dispensable for cell viability. This was done by a YDR1 plasmid shuffle assay in the wild-type SIN4 (YSK013 and YSK014), sin4-54 (YSK126 and YSK127), and sin4-168 (YSK128 and YSK129) genetic backgrounds. Results are shown in Fig. 6A. When cured of the YDR1-URA3 plasmid, the wild-type strain failed to grow on FOA medium, consistent with YDR1's being an essential gene. However, the sin4-54 strain grew in the absence of YDR1, albeit more slowly than the wild-type SIN4 strain. To confirm that viability of the ydr1Δ sin4-54 mutant is due to sin4-54, rather than to an undefined mutation, we investigated if plasmid-borne SIN4 would render the double mutant inviable. Accordingly, the YSK125 (ydr1Δ [YDR1-URA3] sin4-54) strain was transformed with plasmid DNA containing either wild-type SIN4 (YSK140) or the vector alone (YSK141). Accordingly, plasmid-borne SIN4 will compensate the recessive sin4-54 allele. As expected, the strain containing plasmid DNA alone remained viable on the FOA medium but the strain containing SIN4 became sensitive to FOA (Fig. 6C). Thus, complementation of sin4-54 by wild-type SIN4 restored the essential Ydr1 requirement. This effect is specific to the sin4-54 allele, because sin4-168 and sin4Δ (data not shown) failed to grow when cured of YDR1. These results demonstrate that sin4-54 alone is able to suppress the inviability of ydr1Δ. We conclude that the sin4-54 suppressor mutation is able to bypass the essential requirement for YDR1 in vivo. Nonetheless, the ydr1Δ sin4-54 strain grew more slowly than the YDR1 sin4-54 strain (Fig. 6A) and a growth difference was not observed in the ydr1-13 sin4-54 strain relative to the YDR1 sin4-54 strain (Fig. 4B). This difference is due to the partial retention of Ydr1 function encoded by the partially functional ydr1-13 allele. Therefore, although sin4-54 renders YDR1 dispensable for cell viability, the normal rate of cell growth remains Ydr1 dependent. Interestingly, this effect is specific to YDR1 since neither sin4-54, sin4-168, nor sin4Δ rendered BUR6 dispensable in a similar assay (Fig. 6B). The inability of either suppressor to render BUR6 dispensable suggests that Bur6 affects another cellular process(es) independent of Ydr1.
FIG. 6.
sin4 suppressor allele sin4-54 makes YDR1, but not BUR6, dispensable for cell viability. The genotypes of the strains are shown in the circular diagram. (A) Cell viability of ydr1 deletion strains in different sin4 allele backgrounds was determined by scoring growth on a plate containing FOA (SD-TRP+FOA). Each strain (SIN4, sin4-54, or sin4-168) was transformed either with the vector containing wild-type YDR1 (SIN4-YDR1, sin4-54–YDR1, or sin4-168–YDR1) or the vector alone (SIN4-vector, sin4-54–vector, or sin4-168–vector) to generate a pair of isogenic strains. (B) Cell viability of a bur6 deletion strain in different sin4 allele backgrounds was determined as described for panel A, except that BUR6 was substituted for YDR1. (C) The specificity of lethality overcome by the sin4-54 allele was demonstrated by converting an FOA-resistant ydr1-negative strain (sin4-54–vector; two independent isolates are indicated as #1 and #2) into an FOA-sensitive strain by introducing wild-type SIN4 (sin4-54–SIN4; two independent isolates are indicated as #1 and #2).
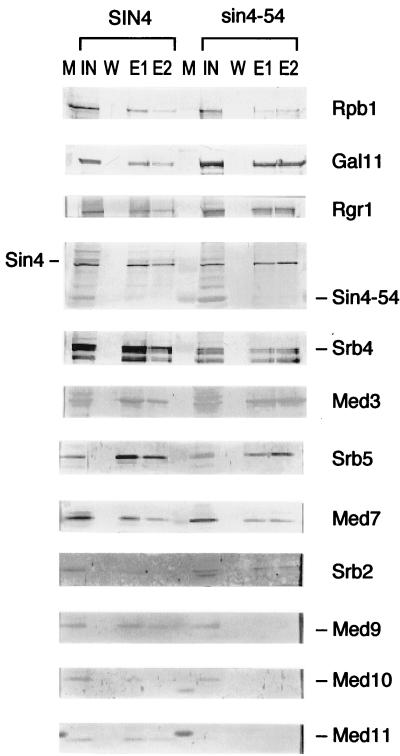
The sin4-54 suppressor alters the composition of the SRB-MED complex.
In order to elucidate both the specific effect of the sin4-54 mutation and the possible mechanism of sin4-mediated suppression, we characterized the effect of sin4 suppressor mutations on the composition of the SRB-MED complex. Deletion of SIN4 causes the loss of specific subunits from the SRB-MED complex (28, 29). Therefore, SRB-MED complexes were purified from both a wild-type SIN4 strain (VM02) and the sin4-54 suppressor strain (YSK149) and their subunit compositions were compared. We observed two interesting features of the complex isolated from the sin4-54 suppressor strain. First, Western blot analysis (Fig. 7) showed a significant reduction in the amounts of the Med9, Med10, and Med11 polypeptides. The loss of specific subunits of the SRB-MED complex suggests that the sin4 suppressor mutation weakens the association of a unique set of mediator proteins and causes a defect in transcriptional activation by certain activators in vivo. Secondly, in contrast to the effect of the total deletion of sin4, which causes the loss of all subunits of the Gal11 module except Rgr1 (29), the SRB-MED complex from the sin4-54 strain retained all of the subunits of the Gal11 module (Med2 was not determined). On the basis of these results, we conclude that sin4-54 has a unique effect on the SRB-MED complex, possibly affecting the interaction of the Gal11 subcomplex containing the Sin4 subunit with the subcomplex containing the Med9, Med10, and Med11 subunits.
FIG. 7.
The composition of the SRB-MED complex from the sin4 suppressor strain differs from that of the complex isolated from the wild-type strain. Western blot analyses of Ni-NTA column-purified SRB-MED complexes isolated from the wild-type SIN4 strain (VM02) or the sin4-54 suppressor strain (YSK149). An isogenic sin4-54 suppressor strain was generated by recombination-mediated truncation of the C terminus of the SIN4 gene using an SRB5-tagged strain (VM02). M, molecular size marker; IN, input; W, wash; E1 and E2, elution fractions (first and second lanes). The FL-Sin4 polypeptide from the wild-type strain is indicated at the left. The truncated Sin4-54 polypeptide from the sin4-54 strain is indicated at the right.
DISCUSSION
We have exploited the power of yeast genetics to further define the role of the Ydr1-Bur6 transcriptional repressor in vivo. A previously unrecognized relationship between the Ydr1-Bur6 complex and the Sin4 component of the SRB-MED complex was uncovered. These results underscore the positive role of Sin4 in transcription and define a delicate balance between positive and negative effectors of gene regulation in vivo.
We began the studies described here by analyzing the in vivo and in vitro functions of the C terminus of Ydr1, which in the human polypeptide contains a transferable repressor domain. This domain is conserved in the yeast Dr1 polypeptide. From these analyses, important implications were derived. First, deletion of the repression domain (C105) did not affect the ability of Ydr1 to interact with Bur6. This truncated polypeptide also interacted with TBP, although with approximately 30% efficiency compared to the wild-type Ydr1 polypeptide. This reduction in TBP binding is consistent with previous studies demonstrating that the C terminus of human Dr1 (residues 101 to 165) contains a low-affinity TBP binding site (46). This truncated form of Ydr1 completely lost its ability to repress transcription in vitro. Another C-terminal truncation which left the repression domain intact (C130) was capable of repressing transcription in vitro, although its ability to interact with TBP was affected to approximately the same extent as with C105. We conclude that the ability of Ydr1 to interact with TBP, while necessary and important for repression, is not sufficient but also requires the highly conserved QE-rich domain (Fig. 4). Second, the inability of C105 to repress transcription correlates with the extremely slow growth phenotype. In contrast to previous studies (10), the C-terminal domain of Ydr1, which is distinct from the histone fold motif and the TBP binding domain, is essential not only for transcriptional repression but also for normal cell growth.
We expanded the truncation analyses by introducing specific substitutions within the C terminus of Ydr1. The discovery of cs− phenotypes associated with several ydr1 mutations in the repressor domain led us to postulate that the defects caused by these mutations may be linked directly to the observed transcriptional defects. Suppressor mutations that compensate for these specific defects were expected to identify gene products that function in transcriptional activation. This expectation was borne out by the identification of sin4 alleles as suppressors of ydr1 mutations. Our analysis indicates that suppression does not involve a direct protein interaction between Sin4 and Ydr1-Bur6. This interpretation does not imply that the sin4-mediated suppression is nonspecific. The observation that the gal11 deletion was not able to suppress the cs− ydr1 mutations indicates that suppression by the sin4 mutant alleles resulted not from the disruption of the Gal11 subcomplex but from specific alterations of the Sin4 molecule. Importantly, a correlation between Ydr1-Bur6 and the RNAPII holoenzyme established in these studies is in agreement with previous findings of Young and coworkers (9, 26). Their studies, analyzing an allele of the SRB4 gene, srb4-138, which encodes another component of the SRB-MED complex, identified alleles of YDR1 and BUR6, as well as other negative regulators, including NOT1 and MOT1. Taken together, these studies point to a delicate balance between positive and negative regulators of transcription and indicate that loss of one function can be compensated for by loss of a reciprocal function in vivo.
Although the physiological role of Sin4 is not fully understood, its function has been correlated with both positive and negative regulation of gene expression. Based on the general repressive effect of Ydr1-Bur6 on transcription, our results suggest that the positive regulatory function of Sin4 has broad effects on gene expression. These results do not exclude the participation of Sin4 in transcriptional repression but suggest that negative regulation is a secondary or even indirect function of Sin4.
The reciprocal relationship between Ydr1-Bur6 and Sin4 is not subtle. This is most evident in the ability of sin4-54 to render the otherwise essential YDR1 gene dispensable for cell viability. This is a dramatic effect but is not without precedent. For example, in a study by Zhao et al. (46), a mutation in the SML1 gene allowed cell growth in the absence of two otherwise essential checkpoint genes, MEC1 and RAD53. The authors suggested that Mec1 and Rad53 may be no longer required if there is a defect in an inhibitory function of the Sml1 protein. Our findings may be related to those of Zhao et al. and have two important implications. First, the defect caused by the ydr1 deletion is likely confined to a transcriptional defect. Importantly, only the sin4-54 allele, but not sin4-168 or the deletion of sin4, was capable of negating the YDR1 requirement for cell viability. Second, the C terminus of the Sin4 polypeptide (amino acids 672 to 974, those deleted in sin4-54) may have an as yet uncharacterized role(s) in vivo. Truncation of the C terminus of Sin4 does not appear to expose a cryptic transcriptional repression domain because sin4-54 did not repress lacZ expression under the control of different promoters (data not shown). Moreover, complete deletion of SIN4 will also suppress Ydr1 cs alleles. Rather, the removal of this particular region of Sin4 appears to alter the subunit composition of the RNAPII holoenzyme in a specific manner. While deletion of SIN4 causes the dissociation of most of the Gal11 subcomplex from the RNAPII holoenzyme (29), the sin4-54 mutation gave rise to the loss of a subset of mediator components, specifically, Med9, Med10, and Med11. Med9 and Med10 specifically mediate transcription signals from the BAS1-BAS2, Gcn4, and Gal4 transcriptional activators, respectively (14).
It is also important to recognize that whereas sin4-54, sin4-168, or the deletion of sin4 can suppress the cs− phenotype associated with ydr1 and bur6, sin4-54 renders YDR1, but not BUR6, dispensable for cell viability. This observation is likely related to our previous studies with the mammalian Dr1-DRAP1 repressor complex. In those studies, we analyzed the expression pattern of Dr1 and DRAP1 in the developing mouse and found that all cells expressed Dr1, yet only highly differentiated cells expressed DRAP1 (31; R. Iratni and D. Reinberg, unpublished data). In light of these findings, we proposed that there are at least two mechanisms by which Dr1 represses transcription: a low-affinity mechanism which is independent of DRAP1 (Bur6) and a high-affinity mode dependent on DRAP1. Recent studies mapped residues in TBP that define the interaction with the Ydr1-Bur6 complex (4). These residues are clustered in a previously undefined domain of TBP adjacent to the transcription factor IIB (TFIIB) binding site. The TBP mutational analysis provides a clear explanation of previous findings demonstrating that the association of Dr1 with TBP prevented the entry of TFIIB into the transcription complex. However, the mutational analysis of TBP does not provide a logical explanation for the finding that TFIIA can overcome Dr1-mediated repression of transcription. TFIIA and TFIIB-Dr1 bind to diametrically opposed surfaces of TBP. In light of these findings, we recently proposed that Ydr1-Bur6 either induces a conformational change in TBP that affects TFIIA binding or alters the structure of the TBP-DNA complex (30). Dr1-mediated repression is likely to involve two mechanisms, one directly blocking the assembly of TFIIB, the other inducing a TBP conformational change that alters TFIIA binding. This latter mechanism may be dependent on the presence of DRAP1 (Bur6), yet this speculation does not provide an explanation for the findings described above with the sin4-54 allele, which allows viability in the absence of YDR1 but not in the absence of BUR6. The most logical interpretation of this finding is that Bur6 has functions that are independent of Ydr1.
ACKNOWLEDGMENTS
We thank David Stillman for the sin4 deletion strain, Marian Carlson for the gal11 deletion strain, and Richard Young for the srb5-tagged strain and antibodies against Srb2, Srb4, and Srb5. We also thank Young-Joon Kim for antibodies against Rgr1, Sin4, Med9, Med10, and Med11; Roger Kornberg for antibodies against Med3 and Med7; and Mark Ptashne for antibodies against Gal11.
This work was supported by grants from the NIH (GM48518) and the Howard Hughes Medical Institute to D.R. and by a grant from the NIH (GM39484) to M.H.
REFERENCES
- 1.Auble D T, Hansen K E, Mueller C G, Lane W S, Thorner J, Hahn S. Mot1, a global repressor of RNA polymerase II transcription, inhibits TBP binding to DNA by an ATP-dependent mechanism. Genes Dev. 1994;8:1920–1934. doi: 10.1101/gad.8.16.1920. [DOI] [PubMed] [Google Scholar]
- 2.Ayer D E, Lawrence Q A, Eisenman R N. Mad-Max transcriptional repression is mediated by ternary complex formation with mammalian homologs of yeast repressor Sin3. Cell. 1995;80:767–776. doi: 10.1016/0092-8674(95)90355-0. [DOI] [PubMed] [Google Scholar]
- 3.Boeke J D, Trueheart J, Natsoulis G, Fink G R. 5-Fluoroorotic acid as a selective agent in yeast molecular genetics. Methods Enzymol. 1987;154:164–175. doi: 10.1016/0076-6879(87)54076-9. [DOI] [PubMed] [Google Scholar]
- 4.Cang Y, Auble D T, Prelich G. A new regulatory domain on the TATA-binding protein. EMBO J. 1999;18:6662–6671. doi: 10.1093/emboj/18.23.6662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carlson M. Genetics of transcriptional regulation in yeast: connections to the RNA polymerase II CTD. Annu Rev Cell Dev Biol. 1997;13:1–23. doi: 10.1146/annurev.cellbio.13.1.1. [DOI] [PubMed] [Google Scholar]
- 6.Chen S, West R W, Jr, Johnson S L, Gans H, Kruger B, Ma J. TSF3, a global regulatory protein that silences transcription of yeast GAL genes, also mediates repression by alpha 2 repressor and is identical to SIN4. Mol Cell Biol. 1993;13:831–840. doi: 10.1128/mcb.13.2.831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Covitz P A, Song W, Mitchell A P. Requirement for RGR1 and SIN4 in RME1-dependent repression in Saccharomyces cerevisiae. Genetics. 1994;138:577–586. doi: 10.1093/genetics/138.3.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fassler J S, Gray W, Lee J P, Yu G Y, Gingerich G. The Saccharomyces cerevisiae SPT14 gene is essential for normal expression of the yeast transposon, Ty, as well as for expression of the HIS4 gene and several genes in the mating pathway. Mol Gen Genet. 1991;230:310–320. doi: 10.1007/BF00290682. [DOI] [PubMed] [Google Scholar]
- 9.Gadbois E L, Chao D M, Reese J C, Green M R, Young R A. Functional antagonism between RNA polymerase II holoenzyme and global negative regulator NC2 in vivo. Proc Natl Acad Sci USA. 1997;94:3145–3150. doi: 10.1073/pnas.94.7.3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goppelt A, Meisterernst M. Characterization of the basal inhibitor of class II transcription NC2 from Saccharomyces cerevisiae. Nucleic Acids Res. 1996;24:4450–4455. doi: 10.1093/nar/24.22.4450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goppelt A, Stelzer G, Lottspeich F, Melsterernst M. A mechanism for repression of class II gene transcription through specific binding of NC2 to TBP-promoter complexes via heterodimeric histone fold domains. EMBO J. 1996;15:3105–3116. [PMC free article] [PubMed] [Google Scholar]
- 12.Guarente L. Transcriptional coactivators in yeast and beyond. Trends Biochem Sci. 1995;20:517–521. doi: 10.1016/s0968-0004(00)89120-3. [DOI] [PubMed] [Google Scholar]
- 13.Hampsey M, Reinberg D. RNA polymerase II as a control panel for multiple coactivator complexes. Curr Opin Genet Dev. 1999;9:132–139. doi: 10.1016/S0959-437X(99)80020-3. [DOI] [PubMed] [Google Scholar]
- 14.Han S J, Lee Y C, Gim B S, Ryu G H, Park S J, Lane W S, Kim Y J. Activator-specific requirement of yeast mediator proteins for RNA polymerase II transcriptional activation. Mol Cell Biol. 1999;19:979–988. doi: 10.1128/mcb.19.2.979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hanna-Rose W, Hansen U. Active repression mechanisms of eukaryotic transcription repressors. Trends Genet. 1996;12:229–234. doi: 10.1016/0168-9525(96)10022-6. [DOI] [PubMed] [Google Scholar]
- 16.Holstege F C, Jennings E G, Wyrick J J, Lee T I, Hengartner C J, Green M R, Golub T R, Lander E S, Young R A. Dissecting the regulatory circuitry of a eukaryotic genome. Cell. 1998;95:717–728. doi: 10.1016/s0092-8674(00)81641-4. [DOI] [PubMed] [Google Scholar]
- 17.Inostroza J A, Mermelstein F H, Ha I, Lane W S, Reinberg D. Dr1, a TATA-binding protein-associated phosphoprotein and inhibitor of class II gene transcription. Cell. 1992;70:477–489. doi: 10.1016/0092-8674(92)90172-9. [DOI] [PubMed] [Google Scholar]
- 18.Jiang Y W, Dohrmann P R, Stillman D J. Genetic and physical interactions between yeast RGR1 and SIN4 in chromatin organization and transcriptional regulation. Genetics. 1995;140:47–54. doi: 10.1093/genetics/140.1.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jiang Y W, Stillman D J. Involvement of the SIN4 global transcriptional regulator in the chromatin structure of Saccharomyces cerevisiae. Mol Cell Biol. 1992;12:4503–4514. doi: 10.1128/mcb.12.10.4503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jiang Y W, Stillman D J. Regulation of HIS4 expression by the Saccharomyces cerevisiae SIN4 transcriptional regulator. Genetics. 1995;140:103–114. doi: 10.1093/genetics/140.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johnson A D. The price of repression. Cell. 1995;81:655–658. doi: 10.1016/0092-8674(95)90524-3. [DOI] [PubMed] [Google Scholar]
- 22.Keleher C A, Redd M J, Schultz J, Carlson M, Johnson A D. Ssn6-Tup1 is a general repressor of transcription in yeast. Cell. 1992;68:709–719. doi: 10.1016/0092-8674(92)90146-4. [DOI] [PubMed] [Google Scholar]
- 23.Kim J, Parvin J D, Shykind B M, Sharp P A. A negative cofactor containing Dr1/p19 modulates transcription with TFIIA in a promoter-specific fashion. J Biol Chem. 1996;271:18405–18412. doi: 10.1074/jbc.271.31.18405. [DOI] [PubMed] [Google Scholar]
- 24.Kim S, Na J G, Hampsey M, Reinberg D. The Dr1/DRAP1 heterodimer is a global repressor of transcription in vivo. Proc Natl Acad Sci USA. 1997;94:820–825. doi: 10.1073/pnas.94.3.820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim Y J, Bjorklund S, Li Y, Sayre M H, Kornberg R D. A multiprotein mediator of transcriptional activation and its interaction with the C-terminal repeat domain of RNA polymerase II. Cell. 1994;77:599–608. doi: 10.1016/0092-8674(94)90221-6. [DOI] [PubMed] [Google Scholar]
- 26.Lee T I, Wyrick J J, Koh S S, Jennings E G, Gadbols E L, Young R A. Interplay of positive and negative regulators in transcription initiation by RNA polymerase II holoenzyme. Mol Cell Biol. 1998;18:4455–4462. doi: 10.1128/mcb.18.8.4455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee Y C, Kim Y J. Requirement for a functional interaction between mediator components Med6 and Srb4 in RNA polymerase II transcription. Mol Cell Biol. 1998;18:5364–5370. doi: 10.1128/mcb.18.9.5364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee Y C, Park J M, Min S, Han S J, Kim Y J. An activator binding module of yeast RNA polymerase II holoenzyme. Mol Cell Biol. 1999;19:2967–2976. doi: 10.1128/mcb.19.4.2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li Y, Bjorklund S, Jiang Y W, Kim Y J, Lane W S, Stillman D J, Kornberg R D. Yeast global transcriptional regulators Sin4 and Rgr1 are components of mediator complex/RNA polymerase II holoenzyme. Proc Natl Acad Sci USA. 1995;92:10864–10868. doi: 10.1073/pnas.92.24.10864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maldonado E, Hampsey M, Reinberg D. Repression: targeting the heart of the matter. Cell. 1999;99:1–4. doi: 10.1016/s0092-8674(00)81533-0. [DOI] [PubMed] [Google Scholar]
- 31.Mermelstein F, Yeung K, Cao J, Inostroza J A, Erdjument-Bromage H, Eagelson K, Landsman D, Levitt P, Tempst P, Reinberg D. Requirement of a corepressor for Dr1-mediated repression of transcription. Genes Dev. 1996;10:1033–1048. doi: 10.1101/gad.10.8.1033. [DOI] [PubMed] [Google Scholar]
- 32.Muhlrad D, Hunter R, Parker R. A rapid method for localized mutagenesis of yeast genes. Yeast. 1992;8:79–82. doi: 10.1002/yea.320080202. [DOI] [PubMed] [Google Scholar]
- 33.Myer V E, Young R A. RNA polymerase II holoenzymes and subcomplexes. J Biol Chem. 1998;273:27757–27760. doi: 10.1074/jbc.273.43.27757. [DOI] [PubMed] [Google Scholar]
- 34.Myers L C, Gustafsson C M, Bushnell D A, Lul M, Erdjument-Bromage H, Tempst P, Kornberg R D. The Med proteins of yeast and their function through the RNA polymerase II carboxy-terminal domain. Genes Dev. 1998;12:45–54. doi: 10.1101/gad.12.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Orr-Weaver T L, Szostak J W, Rothstein R J. Genetic applications of yeast transformation with linear and gapped plasmids. Methods Enzymol. 1983;101:228–245. doi: 10.1016/0076-6879(83)01017-4. [DOI] [PubMed] [Google Scholar]
- 36.Prelich G. Saccharomyces cerevisiae BUR6 encodes a DRAP1/NC2α homolog that has both positive and negative roles in transcription in vivo. Mol Cell Biol. 1997;17:2057–2065. doi: 10.1128/mcb.17.4.2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rose M D, Novick P, Thomas J H, Botstein D, Fink G R. A Saccharomyces cerevisiae genomic plasmid bank based on a centromere-containing shuttle vector. Gene. 1987;60:237–243. doi: 10.1016/0378-1119(87)90232-0. [DOI] [PubMed] [Google Scholar]
- 38.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 39.Schiestl R H, Gietz R D. High efficiency transformation of intact yeast cells using single stranded nucleic acids as a carrier. Curr Genet. 1989;16:339–346. doi: 10.1007/BF00340712. [DOI] [PubMed] [Google Scholar]
- 40.Schultz M C, Choe S Y, Reeder R H. Specific initiation by RNA polymerase I in a whole-cell extract from yeast. Proc Natl Acad Sci USA. 1991;88:1004–1008. doi: 10.1073/pnas.88.3.1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sherman F. Getting started with yeast. Methods Enzymol. 1991;194:3–21. doi: 10.1016/0076-6879(91)94004-v. [DOI] [PubMed] [Google Scholar]
- 42.Sherman F, Hicks J. Micromanipulation and dissection of asci. Methods Enzymol. 1991;194:21–37. doi: 10.1016/0076-6879(91)94005-w. [DOI] [PubMed] [Google Scholar]
- 43.Ward A C. Single-step purification of shuttle vectors from yeast for high frequency back-transformation into E. coli. Nucleic Acids Res. 1990;18:5319. doi: 10.1093/nar/18.17.5319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yeung K, Inostroza J A, Mermelstein F H, Kannabiran C, Reinberg D. Structure-function analysis of the TBP-binding protein Dr1 reveals a mechanism for repression of class II gene transcription. Genes Dev. 1994;8:2097–2109. doi: 10.1101/gad.8.17.2097. [DOI] [PubMed] [Google Scholar]
- 45.Yeung K, Kim S, Reinberg D. Functional dissection of a human Dr1-DRAP1 repressor complex. Mol Cell Biol. 1997;17:36–45. doi: 10.1128/mcb.17.1.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhao X, Muller E G, Rothstein R. A suppressor of two essential checkpoint genes identifies a novel protein that negatively affects dNTP pools. Mol Cell. 1998;2:329–340. doi: 10.1016/s1097-2765(00)80277-4. [DOI] [PubMed] [Google Scholar]