Abstract
Background/Aim:
Helicobacter pylori infects approximately 50% of individuals worldwide. Successful H. pylori eradication is associated with reduced risk of gastric cancer and peptic ulcer disease, among other conditions. We hypothesized that host genetic determinants, especially those affecting gastric pH, might contribute to eradication therapy failure, particularly when treatment adherence and antibiotic susceptibility are confirmed. We aimed to conduct a meta-analysis of host genetic variants associated with H. pylori eradication failure.
Methods:
We searched the literature for studies comparing post-treatment H. pylori eradication failure vs success (outcome) according to host genetic polymorphisms (exposure). Reference groups were defined according to genotypes (or corresponding phenotypes) hypothesized to be associated with successful eradication. We pooled estimates using a random-effects model, and performed comprehensive sensitivity analyses.
Results:
We analyzed 57 studies from 11 countries; the vast majority analyzed CYP2C19 polymorphisms. Among individuals prescribed eradication regimens with PPIs predominantly CYP2C19-metabolized, enhanced vs poor metabolizer phenotypes were associated with a 2.52-fold significantly higher likelihood of eradication failure, and 4.44-fold significantly higher likelihood when treatment adherence and H. pylori clarithromycin susceptibility (if relevant) were confirmed. There was no association between CYP2C19 variants and eradication failure if PPIs less metabolized by or which bypass CYP2C19 metabolism were used. IL-1B polymorphisms that are vs. are not associated with less gastric acid suppression were associated with 1.72-fold significantly higher likelihood of eradication failure. There was no association between MDR1 polymorphisms and H. pylori eradication failure. The certainty of evidence was moderate.
Conclusion:
Based on meta-analysis, we identified host genetic polymorphisms significantly associated with H. pylori eradication failure; host genetics might underlie eradication failure among treatment-adherent individuals with confirmed H. pylori antibiotic susceptibility.
Keywords: antibiotic resistance, pharmacogenetics, genome wide association studies, precision medicine, stomach neoplasm
Lay Summary
Failure of Helicobacter pylori eradication treatment is most commonly due to patient nonadherence and H. pylori antibiotic resistance. The findings from this meta-analysis demonstrate that host genetic polymorphisms, particularly those affecting gastric pH, are also significantly associated with H. pylori eradication treatment failure, especially when treatment adherence and antibiotic susceptibility are confirmed.
Introduction
Helicobacter pylori (H. pylori) infects approximately 50% of the global population and remains the most prevalent bacterial infection worldwide.1 H. pylori is a human carcinogen that is responsible for an estimated 5.5% of all malignancies and 80–90% of the global burden of gastric cancer.2,3 H. pylori also causes peptic ulcer disease (PUD), nonulcer dyspepsia, and other diseases that incur significant cost, morbidity, and, in some cases, even mortality. 4,5 Accordingly, international guidelines recommend H. pylori eradication when diagnosed.6–11
Initial H. pylori eradication therapy entails a combination of antibiotics and gastric acid suppression therapy (i.e. proton-pump inhibitor, PPI) for a set duration of time. Longer treatment duration, ideally 10–14 days, is generally associated with higher eradication rates compared to shorter durations.6–10,12 Failure to eradicate H. pylori places patients at ongoing risk of complications related to persistent infection, and also exposes patients to risks associated with additional antibiotics and high-dose acid suppression, since eradication failure is treated with repeated courses of alternative antimicrobial therapy.12 The broader downstream consequences such as cost5 and promoting antibiotic resistance are also very relevant.7,13,14
H. pylori antibiotic resistance and treatment regimen nonadherence among patients are two common reasons for H. pylori eradication failure. However, eradication failure still occurs despite confirmed antibiotic susceptibility and patient adherence.12 Identifying other etiologies for eradication failure might advance individualized treatment selection and ideally improve eradication success rates. Cytochrome P450 2C19 (CYP2C19) is the enzyme responsible for the primary metabolism of many drugs, including certain PPIs (namely, first-generation PPIs). CYP2C19 polymorphisms may be categorized phenotypically based on whether drug metabolism is normal, ultra-rapid, or poor, with the latter group demonstrating the highest plasma drug concentrations.15 One meta-analysis of 16 randomized controlled trials (RCTs) from Asian-Pacific countries analyzed the association between CYP2C19 phenotypes and H. pylori eradication failure among treatment-naïve individuals.16 Based on that meta-analysis, enhanced metabolizers had higher rates of eradication failure compared to normal or poor metabolizers. In addition to a restricted geographic population, another limitation of the study was that other factors known to impact eradication rates, such as patient adherence and antibiotic susceptibility, were not accounted for in the analysis. Furthermore, no other host genes were analyzed.
The primary objective of this study was to conduct a systematic review with comprehensive meta-analysis of studies that evaluated host genetic variants associated with H. pylori eradication failure. We hypothesized that specific host genetic variants, primarily those affecting gastric acid suppression, are associated with H. pylori eradication failure. We aimed to extend the clinical relevance of this analysis by conducting comprehensive sensitivity analyses, including restricting analyses based on PPI type, antibiotic susceptibility testing, confirmed treatment adherence, or longer therapeutic duration.
METHODS
This study was conducted according to the guidelines detailed in the Cochrane Handbook29 and the Preferred Items for Systematic Reviews and Meta-Analyses (PRISMA) statement.30 The protocol was registered in PROSPERO (https://www.crd.york.ac.uk/prospero/) and assigned the designation “CRD42020213464”.
Search strategy
In consultation with a biomedical librarian, we searched the biomedical databases PubMed, Embase, and Medline from their inception through December 20, 2020. We also searched the National Human Genome Research Institute European Bioinformatics Institute Catalog, which details all genome-wide association studies (GWAS) since 2008. Keywords included: “genome wide association study”, “GWAS”, “gene*”, “polymorphism”, “SNP”, “CYP2C19”, “Helicobacter pylori”, “H. pylori”, “eradication”, among others. The search details are provided in the Supplemental Methods. The search was limited to human studies. We did not apply language restrictions to our initial search.
Inclusion and exclusion criteria, data abstraction, exposure/outcome definitions
All studies were eligible for inclusion if they were comparative study designs with distinct comparisons of post-treatment H. pylori eradication failure versus success according to individual host genetic polymorphisms. Additional inclusion criteria are listed in the Supplemental Methods. Eligibility assessment was carried out independently by SCS and AT, with discrepancies resolved by a third investigator (NN).
Data were abstracted according to a standardized data collection sheet that was created specifically for this study; these data elements are listed in the Supplementary Methods. The total number of individuals with confirmed eradication failure versus success (primary outcome) was recorded. Eradication failure was defined as a persistent positive non-serological H. pylori test following a course of an approved anti-H. pylori treatment regimen, with any nuances or deviations from this definition noted on a per-study basis (see Table 1). The number of individuals with the variant versus reference genetic polymorphism (primary exposure, see definitions below) for each of the above groups (eradication failure versus success) was recorded. Study-specific effect estimates (e.g. odds ratios, OR) along with 95% confidence intervals (95% CI) and covariate adjustment were recorded, if available.
Table 1.
Details of included studies
| Author | Year | Country | Base Publication Study Design* | Total N (male, female) | Mean Age (SD) | Population Details | Gene | Method of H. pylori Testing | Therapy Duration | PPI/Dosage | Antibiotics Used | Antibiotic Resistance Tested | CLR Resistance | Medication Adherence | Overall Eradication Success13/26; 49/57 | Significant Association Between Gene and Eradication | Other Factors Associated with Eradication |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Tanigawara | 1999 | Japan | Prospective Cohort | 83 M vs. F not stated |
Unavailable | HP positive gastritis of peptic ulcers | CYP2C19 | culture, histology, UBT | 7 days | OMZ 20 BID | AMX 500 QID; AMX 500 QID, CLR 200 QID | no | n/a | unavailable | 13/26; 49/57 | yes | none |
| Dojo | 2001 | Japan | RCT | 43, 43; 40, 38 | 42.4 (6.6); 44.1 (6.6) | HP positive with chronic gastritis | CYP2C19 | UBT | 7 days | AMX 750 BID, CLR 400 BID | AMX 750 BID, CLR 400 BID | no | n/a | unavailable | 70/86 PP (70/89 ITT); 65/78 PP (65/81 ITT) | no | none |
| Furuta | 2001 | Japan | ?RCT | 232, 29 PP | 49.0 (9.7) PP | HP positive with GU, DU or gastritis | CYP2C19 | culture, histology, RUT, UBT | 7 days | AMX 500 TID, CLR 200 TID | AMX 500 TID, CLR 200 TID | yes (CLR: 33/261) | 12.64% | >80% considered compliant, all in PP analysis | 226/261 PP (226/271 ITT) | yes | CLR resistance |
| Hokari | 2001 | Japan | RCT | 20, 10; 23, 8; 22, 9 | 51.9 (9.5); 51.2 (10.6); 47.0 (12.1) | HP positive with GU | CYP2C19 | culture, RUT, UBT | AMX 750 BID, CLR 200 BID | RAL/10 daily; RAL/10 BID; RAL/20 BID | AMX 750 BID, CLR 200 BID | yes (CLR: 4/98) | 4.08% | unavailable | 25/30 APT (25/30 PP); 24/31 APT (24/30 PP); 28/31 APT (27/30 PP) | no | none |
| Miyoshi | 2001 | Japan | RCT | 90, 8; 87, 14 | 49 (8.1); 50.6 (8.3) | HP positive with PUD | CYP2C19 | culture, RUT, UBT | 14 days | OMZ/20 BID, RAL/10 BID | AMX 500 TID | yes, but only to exclude abx resistance | n/a | unavailable | 65/98 ITT (65/93 APT, 65/89 PP); 63/101 ITT (63/90 APT, 63/85 PP) | no | none |
| Inaba | 2002 | Japan | RCT | 48, 11; 47, 13; 47, 17 | 56 (20–83); 52 (21–76); 55 (27–75) | HP positive with PUD | CYP2C19 | culture, histology, RUT, UBT | 7 days | OMZ/20 BID; LNZ/30 BID; RAL/20 BID | AMX 500 TID, CLR 200 TID | no | n/a | 100%; 100%; 98.40% | 49/59 ITT (49/58 PP); 52/60 ITT (52/58 PP); 49/64 ITT (49/63 PP) | yes, only for RAL groups | none |
| Isomoto | 2003 | Japan | RCT | 43, 18 | 60.7 (42.5–88.5) | HP positive | CYP2C19 | HP IgG, RUT, culture, histology, UBT | 7 days | LNZ 30 BID | AMX 750 BID, CLR 200 BID | yes (CLR: 4/61) | 6.56% | unavailable | 49/61 ITT (49/58 PP) | no | CLR resistance |
| Isomoto* | 2003 | Japan | RCT | 43, 20; 36, 24 | 47.7 (20–72); 48.6 (20–72) | HP positive after failed LNZ 30 BID, AMX 750 BID, CLR 200 BID for 7d | CYP2C19 | UBT | 14 days; 7 days | RAL/20 BID; RAL/10 BID | AMX 1000 BID; AMX 750 BID, MTZ 250 BID | yes (CLR: 45/72; MTZ 20/72) | 62.50% | 1 patient removed from each group for <80% compliance for PP analysis | 37/63 ITT (37/56 PP); 49/60 ITT (49/56 PP) | yes | none |
| Kawabata | 2003 | Japan | RCT | 138, 49 | 51.8 (20–78) | HP positive with peptic ulcers | CYP2C19 | culture, histology, UBT | 7 days (pts with active ulcers on f/u were given 4 more weeks of PPI) | RAL/10 BID; LNZ/30 BID | AMX 750 BID, CLR 400 BID | yes (CLR: 21/173) | 12.14% | unavailable | 75/100 ITT (75/93 PP); 60/87 ITT (60/80 PP) | yes | CLR resistance |
| Lee | 2003 | Korea | Prospective Cohort | 85, 31 | 48 (13) | HP positive with GU or DU | CYP2C19 | RUT, UBT | 7 days | RAL/10 BID | AMX 1000 BID, CLR 500 BID | no | n/a | unavailable | 98/116 | no | age |
| Matsuo | 2003 | Japan | Prospective Cohort | 78, 64 | Median 60 (range 40–69) | HP positive, outpatients | Le, Se; IL1A; IL-1β−31; IL1RN; MPO | HP IgG | 7 days | LNZ 40 daily | AMX 1500 TID, CLR 400 BID | no | n/a | unavailable | 87/142 | Le only | none |
| Miki | 2003 | Japan | RCT | 33, 14; 38, 8; 34, 11 (Total ITT: 113, 32) | 47.9 (13.5); 48.7 (12.7); 49.1 (20.1) (Total ITT - 48.9 (17.3)) | HP positive gastritis or peptic ulcers | CYP2C19 | culture, histology, RUT, UBT | 7 days | LNZ/30 BID; RAL/10 BID; RAL/20 BID | AMX 1000 BID, | yes (CLR: 16/138, MTZ: 11/138) | 11.59% | unavailable | 39/49 ITT (39/47 PP); 4¼8 ITT (4¼6 PP); 40/48 ITT (40/45 PP); Total - 120/145 ITT (120/138 PP) | no | CLR resistance |
| Sapone | 2003 | Italy | Prospective Cohort | 66, 77 | 53 (13.74) | HP positive with dyspepsia | CYP2C19; CYP3A4 | culture, histology, RUT | 7 days | OMZ 20 BID | AMX 1000 BID, CLR 500 BID | no | n/a | unavailable | 93/143 | yes | none |
| Furuta | 2004 | Japan | Prospective Cohort | 299, 51 | 49.0 (10.0) | HP positive with gastritis or ulcers | CYP2C19; IL-1β | histology, culture, RUT, UBT | 7 days | OMZ/20 BID; LNZ/30 BID | AMX 500 TID, CLR 200 TID | yes (CLR: 48/336) | 14.29% | unavailable | 293/350 ITT (293/336 PP) | yes | CLR resistance |
| Schwab | 2004 | Germany | Prospective Cohort | 52, 79 | 56 (15) combined | HP positive, outpatient and inpatient | CYP2C19 | UBT | 5 days | LNZ/30 BID | AMX 1000 BID, CLR 250 BID, MTZ 400 BID (for 3 or 5 days, groups combined) | yes (MTZ: 11/31, CLR: 2/31) | 6.45% | unavailable | 113/131 | yes | none |
| Furuta | 2005 | Japan | Prospective Cohort | 122, 19 | Median 51 (17–78) | HP positive with ulcer or gastritis | CYP2C19 | culture, RUT, UBT | 7 days (GU and DU continued LNZ 30 daily for 5–7 wks) | LNZ/30 BID | AMX 500 TID, CLR 200 TID | yes (CLR: 29/141) | 20.57% | unavailable | 110/139 PP (110/141 ITT) | yes | CLR resistance |
| Gawrońska-Szklarz | 2005 | Poland | Prospective Cohort | 30, 40 | 49.9 (14.5) | HP positive patients, some with ulcers, others with family hx of gastric cancer as indications for treatment | CYP2C19; MDR1 | histology, RUT, UBT | 7 days (PPI continued daily for 4 weeks) | OMZ/20 BID; PPZ/40 BID | AMX 1000 BID, CLR 500 BID; AMX 1000 BID, MTZ 500 BID | no | n/a | unavailable | 34/70 | yes | none |
| Kuwayama | 2005 | Japan | RCT | 91, 22; 82, 30 | 54.2 (11.3); 51.2 (12.8) | HP positive with healed peptic ulcers | CYP2C19 | culture, RUT, UBT | 7 days | OMZ/20 BID | AMX 750 BID, CLR 400 BID; AMX 1000 BID, CLR 500 BID | yes (CLR: 34/225) | 15.11% | unavailable | 89/113 ITT (87/109 PP); 93/112 ITT (88/106 PP) | no | CLR resistance |
| Okudaira | 2005 | Japan | RCT | 87 only male; 86 only male | 43.8 (1.0); 44.0 (1.0) | HP positive males with chronic gastritis, GU or DU | CYP2C19 | culture, RUT, UBT | 7 days | LNZ/30 BID | AMX 750 BID, CLR 200 BID | no | n/a | unavailable | 68/87 PP (68/89 ITT); 68/86 PP (75/88 ITT) | yes | none |
| Sheu | 2005 | Taiwan | RCT | 50, 50; 49, 51 | 41.6; 41.8 | HP positive patients with dyspepsia | CYP2C19 | histology, culture, UBT | 7 days | OMZ/20 BID; EMZ/40 BID | AMX 1000 BID, CLR 500 BID | yes (CLR: 10/139) | 7.19% | 85% “full compliance”, 10% moderate >4d, 5% poor <5d; 83% “full compliance”, 12% moderate >4d, 5% poor <5d | 79/100 ITT (79/93 PP); 86/100 ITT (86/92 PP) | yes | none |
| Ishida | 2006 | Japan | Prospective Cohort | 33, 34 | unavailable, but most 50–69, but included some younger than 30 | HP positive | CYP2C19; IL-1β; TNF-A | UBT or anti-HP antibody, confirme d with UBT | 7 days | LNZ/30 BID | AMX 750 BID, CLR 200 BID | no | n/a | unavailable | 57/67 | yes | none |
| Kurzawski | 2006 | Poland | Prospective Cohort | 58, 67 | 50.9 (14.4) | HP positive with peptic ulcers | CYP2C19 | histology, RUT, UBT | 7 days | PPZ/40 BID | AMX 1000 BID, MTZ 500 BID | no | n/a | unavailable | 78/125 | no | none |
| Miehlke | 2006 | Germa ny | RCT | 25, 48; 29, 43 | 50 (18–71); 48 (23–77) | HP positive with failed eradication, resistance to CLR and MTZ and indication for treatment | CYP2C19 | histology, culture, UBT | 7 days; 14 days | EMZ/20 BID; OMZ/40 BID | AMX 1000 BID, Rifabutin 150 BID; AMX 1000 TID | yes (AMX: 0/145, Rifabutin: 2/145, CLR: unavailable) | n/a | <80% adherence were removed from PP analysis | 54/73 ITT (54/69 PP); 50/72 ITT (50/67 PP) | no | none |
| Sugimoto | 2006 | Japan | Prospective Cohort | 283, 77 | 50.4 (1.6) | HP positive, sensitive to CLR, with GU, DU or gastritis | CYP2C19; IL-1β; IL-1RN; TNFa; IL-10 | RUT, serologic testing, culture, UBT | 7 days | OMZ/20 BID; LNZ/30 BID; RAL/10 BID | AMX 750 BID, CLR 400 BID | yes (unavailable) | n/a | <80% excluded from study | 301/360 ITT (301/349 PP) | yes, only CYP2C19 and IL-1β−511 | none |
| Furuta | 2007 | Japan | Prospective Cohort | 237, 84 | 53.2 (14.3) | HP positive gastritis, GU or DU | CYP2C19; MDR1 | histology, RUT, UBT | 7 days | LNZ/30 BID | AMX 750 BID, CLR 200 BID | yes (CLR: 97/321) | 30.22% | unavailable | 239/321 ITT (239/313 PP) | yes | CLR resistance |
| Furuta* | 2007 | Japan | RCT | 100, 50 | 60 (17–89) | HP positive with gastritis, GU, DU or GDU | CYP2C19 | RUT, UBT | 7 days | LNZ/30 BID | AMX 750 BID, CLR 400 BID | yes (CLR: 44/150) | 29.33% | unavailable | 105/150 ITT (105/144 PP) | yes | CLR resistance |
| Kuwayama | 2007 | Japan | RCT | 82, 37; 83, 26; 82, 34; 84, 31 | 49.6 (12.8); 51.9 (12.8); 48.4 (13.3); 52.3 (12.0) | HP positive with endoscopically confirmed open GU or DU more than or equal to 5mm diameter | CYP2C19 | Histology, culture, UBT | 7 days | RAL/10 BID; RAL/20 BID | AMX 750 BID, CLR 200 BID; AMX 750 BID, CLR 400 BID (two groups of abx regimens for each PPI dosage group) | yes (CLR: 30/60) | 50.00% | >90% | 409/459 ITT (38–29 PP) Total | ?unclear if statistically significant | none |
| Suzuki | 2007 | Japan | Prospective Cohort | 78, 64 | Median 60 (40–69) | HP positive | CYP2C19 | serology HP IgG, culture | Not available | LNZ/30 DAILY | AMX 500 TID, CLR 200 BID | no | n/a | 93% | 87/142 | no | none |
| Kang | 2008 | Korea | Prospective Cohort | 206, 121 | 55 (12.6) | HP positive undergoing EGD | CYP2C19 | Culture, RUT, UBT | 7 days | PPZ/40 BID, EMZ/20 BID | AMX 1000 BID, CLR 500 BID | yes (CLR: 7/52) | 13.46% | >90% considered compliant | 157/190; 121/137 | yes | CLR resistance |
| Miehlke | 2008 | Germa ny | Prospective Cohort | 36, 67 | 52 (21–79) | HP positive with failed prior treatment and resistance to MTZ and CLR | CYP2C19 | Histology, culture, UBT | 7 days | EMZ/40 DAILY | MFX 400 DAILY, Rifabutin 300 Daily | yes (MFX: 2/16; Rifabutin: 5/16 - only available for those that failed) | n/a | unavailable | 80/103 ITT (80/103 PP) | no | none |
| Oh | 2009 | Korea | Prospective Cohort | 111, 99 | 52.7 (11.4) | HP positive with gastric ulcers, duodenal ulcers or functional dyspepsia | CYP2C19; MDR1 | histology, RUT, UBT | 7 days | PPZ/40 BID | AMX 1000 BID, CLR 500 BID | yes (CLR: 1¼5) | 24.44% | unavailable | 174/210 | no | CLR resistance |
| Yang | 2009 | Taiwan | RCT | 33, 15 | 51.5 | HP positive ulcers or gastritis | CYP2C19 | histology, culture, UBT | 7 days | RAL/20 BID | AMX 1000 BID, CLR 500 BID (but 3 groups with abx at various times over course o 7 days) | yes (but not available) | n/a | 98.30% | 36/47 PP | CLR resistance | |
| Gawrońska-Szklarz | 2010 | Poland | Prospective Cohort | 65, 74 | 51 (14.4) | HP positive with peptic ulcers | PPZ/40 BID | histology, RUT, UBT | 7 days (PPI BID--> 5 weeks daily) | PPZ/40 BID | AMX 1000 BID, MTZ 500 BID | no | n/a | unavailable | 103/139 | ? p = 0.4–0.99 | Peptic Ulcer Disease |
| Jinda | 2010 | Japan | Prospective Cohort | 22 M vs. F not stated | unavailable | HP positive suspected of having HP positive gastric ulcers | CYP2C19 | UBT | unavailabl e | unavaila ble | unavaila ble | yes (CAM (CLR and AMX): 6/32) | 18.75% | unavailable | 17/22 | none | |
| Kuo | 2010 | Taiwan | RCT | 41, 55; 38, 56 | 50.57 (12.23); 50.78 (10.78) | HP positive dyspepsia after failed eradication (EMZ 40 BID, CLR 500 BID, AMX 1000 BID) | CYP2C19 | UBT, rapid urease, culture, histology | 7 days | EMZ/40 BID; RAL/20 BID | Tetracycline 500 QID, MTZ 250 QID | yes (MTZ: 53/93; AMX: 4/93; CLR: 62/93; LVX: 21/93) | 66.67% | 97.76% | 70/96 ITT (70/93 PP); 74/94 ITT (74/87 PP) | yes | none |
| Lay | 2010 | China | Prospective Cohort | 71, 24 | 46.5 (6.0) | HP positive with cirrhosis and active peptic ulcers | CYP2C19 | HP DNA, histology, RUT, UBT (2/3) | 14 days | RAL/20 BID | AMX 1000 BID, CLR 500 BID | no | n/a | 92.10% | 86/95 | yes | adherence |
| Lee | 2010 | Hong Kong | RCT | 36, 68; 32, 68 | 46.1 (10.1); 47.6 (9.1) | HP positive with epigastric symptoms and NUD | CYP2C19 | histology, RUT, UBT | 7 days | EMZ/20 BID; RAL/20 BID | AMX 1000 BID, CLR 500 BID | yes (CLR: 21/193; AMP: 0/193; LFX: 22/127; Tetracycline : 0/193; MTZ 151/193) | 10.88% | >90% | 88/104 ITT (88/99 PP); 77/100 ITT (77/92 PP) | no | CLR resistance |
| Lee* | 2010 | Korea | RCT | 276, 187 (combine d PPI groups) | 57 (range 20–83) | HP positive with PUD, post–endoscopic mucosal resection of gastric cancer or adenoma, MALT lymphoma, non-ulcer dyspepsia or gastric polyps | CYP2C19 | UBT, histology | 7 days | LNZ/30 BID; RAL/20 BID | AMX 1000 BID, CLR 500 BID | no | n/a | unavailable | 185/234 PP? (185/260 ITT?); 163/229 PP? (229/261 ITT?) | no | none |
| Pan | 2010 | China | RCT | 28, 33; 30, 32; 27, 34 | 43.9 (14.3); 46.7 (13.4); 43.8 (13.9) | HP positive undergoing EGD | CYP2C19 | histology, RUT, UBT | 7 days | EMZ/20 BID; EMZ/40 BID; RAL/10 BID | LFX 500 DAILY, AMX 1000 BID | no | n/a | 98.4%; 96.80%; 100% | 52/61 ITT (52/60 PP); 54/62 ITT (54/60 PP); 46/61 ITT (46/61 PP) | no | none |
| Liou | 2011 | Taiwan | Prospective Cohort | 79, 63 | 53.7 (14.3) | HP positive failed first line triple tx (CLR, AMX, PPI) | CYP2C19 | UBT, RUT, histology | 10 days | EMZ 40 | AMX 1000 BID 1–5, MTZ 500 BID 6–10, LFX 250 BID 6–10 | yes | 51.90% | unavailable | 135/142 ITT (133/138 PP) | no | antibiotic resistance |
| Wu | 2011 | Taiwan | RCT | 30, 28; 30, 32 | 54.3 (11); 53.6 (11.7) | HP positive after failed first line tx (PPI, CLR 500 BID, AMX 1000 BID) | CYP2C19 | RUT, histology , culture, UBT | 7 days | EMZ 40 | “tetracycl ine” 500 QID, AMX 500 QID; “tetracycl ine” 500 QID, MTZ 250 QID | yes (not separat ed by drug group) | n/a | 97% | 36/58 ITT (35/55 PP); 50/62 ITT (49/59 PP) | no | none |
| Hong | 2016 | China | RCT | 121, 66 (EMZ); 106, 81 (OMZ) | 39.2 (11.9); 41.1 (11.7) | HP positive with duodenal ulcers | CYP2C19; IL-1β | HP culture and UBT | 10 days | EMZ/20 BID, OMZ/20 BID | AMX 1000 BID, CLR 500 BID, MTZ 400 BID | yes (CLR: 52/374; MTZ: 218/374; AMX: 0/374; CLRR/MTZ-R: 33/374; CLRR/MTZ-S: 19/374; CLRS/MTZ-R: 185/374) | 13.90% | 87% | 168/187 ITT (166/174 PP) | yes | compliance, abx resistance |
| Nabinger | 2016 | Brazil | RCT | 27, 121 | 46.08 (12.24) | HP positive with functional dyspepsia | CYP2C19 | RUT and histopath ological examinat ion | 10 days | OMZ/20 BID | AMX 1000 BID, CLR 500 BID | no | n/a | 94.60% | 129/148 | no | none |
| Okimoto | 2016 | Japan | RCT | 56, 52 (EMZ); 54, 57 (RAL) | 57.6 (13.4); 57.1 (13.6) | HP positive | CYP2C19 | RUT, UBT | 7 days | EMZ/20 BID, RAL/20 BID | AMX 750 BID, CLR 200 BID | yes (CLR: 85/210; MTZ: 8/210; AMP: 0/210; LFX: 115/210) | 40.48% | >90% | 69/108 ITT (69/92 PP); 65/111 ITT (65/91 PP) | no | abx resistance |
| Ormeci | 2016 | Turkey | Prospective Cohort | 82, 118 | 40 (13) | HP positive with NUD on EGD | CYP2C19 | RUT, stool antigen | 14 days | PPZ/40 BID, RAL/40 BID | AMX 1000 BID, CLR 500 BID | no | n/a | 100% | 74/104; 63/96 | yes | none |
| Phiphatpattha--maamphan | 2016 | Thailan d | RCT | 17, 33 (14d); 16, 34 (10d) | 53.6 (29–70) (14d); 48.5 (22–69) (10d) | HP positive with dyspepsia | CYP2C19; IL-1β; IL-1RN | RUT, HP culture, UBT | 14 days, 10 days | RAL/20 BID | AMX 500 QID, CLR 1000 DAILY; AMX 1000 BID 1–5, CLR 1000 DAILY 6–10, MTZ 400 TID 6–10 | yes (CLR: 8/54; MTZ: 24/54; LFX: 13/54) | 14.81% | >90% defined as adherent | 43/50 ITT (4–9 PP); 47/50 ITT (47/48 PP) | no | none |
| Song | 2016 | China | Prospective Cohort | 71, 61 | 44.4 (14.2) | HP positive dyspepsia resistant to non-bismuth quadruple therapy | CYP2C19 | UBT | 14 days | EMZ 20 | AMX 1000 BID, LFX 500 DAILY | yes (AMX: 6/72; CLR: 40/72; MTZ: 53/72; LFX: 26/72) | 55.56% | 96.1% (defined by >80%) | 97/132 ITT (95/121 PP) | no | abx resistance |
| Karaca | 2017 | Turkey | Prospective Cohort | 45, 60 (89 pts lost to follow up, no info) | 42.0 (1.73) | HP positive with PUD or gastritis | CYP2C19; CYP3A4; MDR1 | histopath ological examination, and “HP antigen” | 14 days | PPZ/40 BID | AMX 1000 BID for 1–7, MTZ 500 TID 8–14, “tetracycl ine” 500 QID 8–14 | no | n/a | unavailable | 87/105 | no, no, no | none |
| Lin | 2017 | China | RCT | 77, 83 | 45.2 (12.6) | HP positive with dyspepsia | CYP2C19 | RUT, UBT | 10 days | OMZ/20 BID, RAL/10 BID | AMX 1000 BID, LFX 500 BID | no | n/a | unavailable | 61/80 ITT (61/78 PP), 68/80 ITT (68/77 PP) | yes, no | none |
| Chang | 2018 | Korea | Prospective Cohort | 89, 101 | 54.4 (10.2) | HP positive with chronic gastritis | CYP2C19 | RUT or HP DNA in gastric mucosa | 7 days | PPZ/una vailable | AMX 1000 BID, CLR 500 BID | yes (CLR: 33/190) | 36.67% | >85% considered compliant | 130/203 ITT (130/190 PP) | no | female sex, Clarithrom ycin resistance |
| Lin | 2018 | Taiwan | Prospective Cohort | 48, 37 | 52.04 | HP positive outpatients | CYP2C19; IL-1β | RUT, histologic examinat ion and culture, UBT | 14 days | RAL/20 BID | AMX 1000 BID day 1–14, CLR 500 BID 8–14, MTZ 500 BID 8–14 | yes (AMX: 0/65; CLR: 8/65; MTZ: 26/65) | 12.31% | 3 nonadherent were excluded | 79/85 | no, no | none |
| Rech | 2018 | Brazil | RCT | 27, 121 | 46.08 (12.24) | HP positive with functional dyspepsia | IL-1β | histologic staining, RUT | 10 days | OMZ/20 BID | AMX 1000 BID, CLR 500 BID | no | n/a | 94.60% | 129/148 | yes | none |
| Shimoyama | 2018 | Japan | RCT | 90, 99 | unavailable | HP positive getting EGD | CYP2C19 | RUT, UBT, stool antigen test or serum or urine antibody | 7 days | EMZ/20 BID, RAL/20 BID | AMX 750 BID, CLR 200 BID | no | n/a | unavailable | 75/99 ITT (75/94 PP), 71/100 ITT (71/95 PP) | no | lower pepsinoge n I:II ratio |
| Jiang | 2005 | China | Unavailable | 160 (82 OMZ, 78 RAL) M vs. F not stated | Unavailable | HP positive; no prior treatment | CYP2C19 | Modality not stated | 7–14 days (exact duration not specified) | OMZ, RAL (dose and freq. not stated) | AMX, CLR (dose not stated) | unavailable | unavailable | unavailable | 63/82 (OMZ); 67/78 (RAL) | Yes for EM vs. PM and EM vs. IM; not IM vs. PM (OMZ vs RAL not provided) | Full text not available^ |
| Chen | 2012 | China | Unavailable | 101 M vs. F not stated | Unavailable | HP positive; no prior treatment; GU or DU | MDR1 | UBT | 7–14 days (exact duration not specified) | EMZ, OMZ (dose and freq. not stated) | AMX, CLR (dose not stated) | unavailable | unavailable | unavailable | 83/101 (EMZ vs OMZ not provided) | No | Full text not available^ |
| Zhang | 2013 | China | Unavailable | 106 M vs. F not stated | Unavailable | HP positive; no prior treatment; GU | MDR1 | UBT | 7–14 days (exact duration not specified) | EMZ, OMZ (dose and freq. not stated) | AMX, CLR (dose not stated) | unavailable | unavailable | unavailable | 86/106 (EMZ vs OMZ not provided) | Yes | Full text not available^ |
| Shi | 2014 | China | Unavailable | 80 M vs. F not stated | Unavailable | HP positive; no prior treatment; GU | MDR1 | RUT | 7–14 days (exact duration not specified) | LNZ (dose and freq. not stated) | AMX, CLR (dose not stated) | unavailable | unavailable | unavailable | 50/80 | Yes | Full text not available^ |
Abbreviations: Helicobacter pylori, HP; per protocol, PP; intention to treat, ITT; randomized control trial, RCT; amoxicillin, AMX; clarithromycin, CLR
metronidazole, MTX; levofloxacin, LFX; rapid urease test, RUT; urea breath test, UBT; esomeprazole, EMZ; omeprazole, OMZ; pantoprazole, PPZ; lansoprazole, LNZ; rabeprazole, RAL; twice per day, BID; three times per day
Note: no studies randomized according to genetic polymorphism, which is the primary exposure for this meta-analysis; as such, for the purpose of this meta-analysis, all studies can be considered as prospective cohort studies.
Raw data were abstracted from two published meta-analyses, Li et al. and Zhao et al., but the full texts were not able to be accessed (see main text for details)
Statistical Approach
The reference groups (“wild-type”) were defined a priori and selected according to the genotypes (or corresponding phenotypes) hypothesized to be associated with successful eradication based on corresponding mechanisms of action, and supported by the literature (see Supplemental Methods). The designations “variant” and “wild-type” were used for analytic purposes, although it is acknowledged the genetic polymorphisms identified are considered normal human variants. In general, these designations mirrored the non-reference/reference groups for the included studies. For example, in studies analyzing CYP2C19, the genetic polymorphisms of CYP2C19 that correspond to the enhanced drug metabolizer phenotype (i.e. ultra-rapid metabolizer, URM; rapid metabolizer, RM; extensive metabolizer, EM) were considered the “variant” phenotypes, while those corresponding to slow metabolizer phenotypes (i.e. intermediate metabolizer, IM; poor metabolizer, PM) were considered “wild-type” phenotypes. For the primary analyses, PM only was the reference group; secondary analyses were conducted where PM+/−IM was the reference group since, in some studies, it was not possible to separate individuals as PM only or IM only. The genotype/phenotype reference groups for each gene identified are provided in Supplemental Table 1, and as footnotes in the respective data output tables.
Meta-analysis of aggregate patient data was conducted by combining the ORs of individual studies into a pooled OR using a random-effects model. We first performed a meta-analysis of all eligible studies for each host gene identified (e.g. CYP2C19, IL-1B) to calculate the pooled OR of the association between the genetic variant or corresponding phenotype and eradication failure. We then performed the following separate meta-analyses using a step-wise approach based on a priori hypotheses and clinical relevance to eradication failure. First, we stratified CYP2C19 variants by the type of PPI used in the eradication regimen. For this analysis, the primary categorization was determined by level of PPI metabolism via CYP2C19. We categorized omeprazole (OMZ), pantoprazole (PPZ), lansoprazole (LNZ) as PPIs predominantly metabolized by CYP2C19, and esomeprazole (EMZ) and rabeprazole (RAL) as PPIs non- or minimally-metabolized by CYP2C19.17 Second, we repeated the above stratification of CYP2C19 variants by PPI type, but restricted to the subgroup of studies where clarithromycin resistance was ≤15% or H. pylori clarithromycin susceptibility was confirmed (if clarithromycin-based regimens were used).6 Third, we again repeated the above stratification of CYP2C19 variants by PPI type, but separated according to geographic areas, given the ethnic differences in prevalence of CYP2C19 polymorphisms.18,19
To evaluate the effect of confirmed treatment adherence and treatment duration (≤7 days vs. >7 days) we performed meta-regression. A secondary analysis evaluated the dose and frequency of PPI. Higher or more frequent PPI is postulated to achieve adequate gastric acid suppression even in the setting of an enhanced metabolizer phenotype. PPI dosing for eradication therapy was categorized as “standard dose”, “high dose”, or “low dose”. “Standard dose” for H. pylori eradication was defined as PPI dosed two times per day at the standard dose according to the World Health Organization (WHO) (e.g. OMZ 20mg, PPZ 40mg, LNZ 30mg, EMZ 20mg, RBZ 20mg).20 Higher than standard dosages twice daily or more frequent than twice daily dosages6 were considered “high-dose”, while lower or less frequent dosing (e.g. OMZ 20mg daily) were considered “low-dose” PPI regimens for H. pylori eradication treatment.6,12
Compared to studies of CYP2C19, there were limited studies for other genotypes identified, which precluded robust sensitivity analyses. However, where possible, the pooled OR of the association between the genetic variant or corresponding phenotype (compared to reference group) and H. pylori eradication failure was calculated.
Heterogeneity, Publication Bias, and Quality Assessment
Heterogeneity was estimated with the Chi-squared test; P-value < .15 was used as the cut-off classifying a study as high heterogeneity. We also calculated I2 test statistics to quantify heterogeneity estimates, where <30%, 30–59%, 60–75%, and >75% were the thresholds for low, moderate, substantial, and considerable heterogeneity, respectively.21 AT and SCS performed quality and risk of bias assessment independently using the Quality in Prognosis Studies (QUIPS) tool for all studies, since the exposure of interest (genetic variant or phenotype) is essentially a prognostic factor for the outcome, eradication success vs failure. The QUIPS tool, which is recommended by the Cochrane Prognosis Methods Group for risk of bias assessment in prognostic factor studies, uses 6 domains scored on a 3-grade scale (high, moderate, low risk of bias).22,23 We then evaluated the overall quality of the evidence from individual studies using the Grading of Recommendations, Assessment, Development, and Evaluations (GRADE) system.24 Ultimately, the quality of evidence ratings represent an assessment as to how further research might impact the pooled effect estimates.24
All analyses were performed using Comprehensive Meta-Analysis (CMA) Software (version 2.0; Biostat, Englewood, NJ)
RESULTS
Search Results
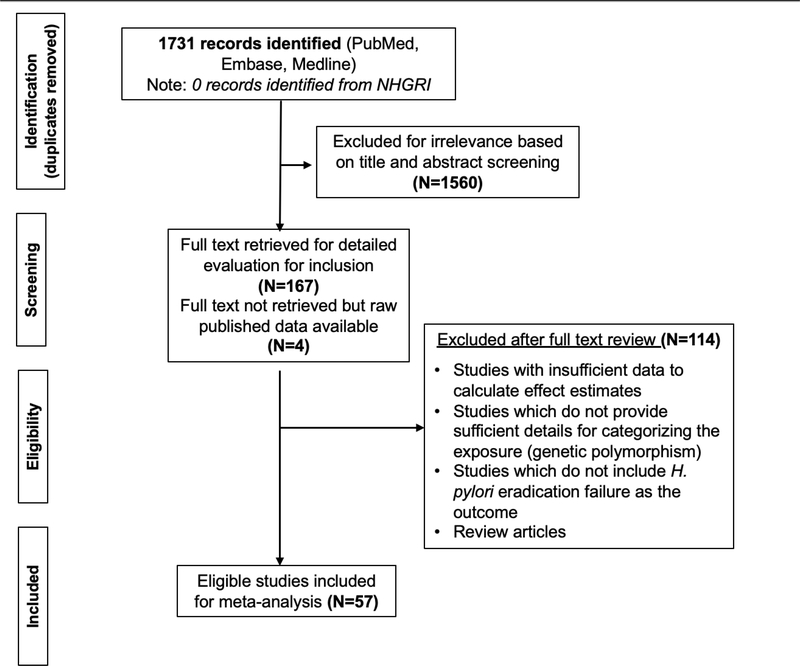
The initial search string yielded 1731 unique studies after removal of duplicates (Figure 1). From this, 1560 studies were excluded based on title/abstract screening. Of the remaining 171 studies, 167 full texts were available to review for eligibility criteria. Raw data were accessible for the other 4 studies25–28 from 2 published meta-analyses29,30; however, the full texts of these articles were not able to be retrieved despite attempts by the biomedical librarian to access the articles, nor through attempts to contact the authors directly. One-hundred twelve studies were removed for having insufficient data for calculating effect estimates; for not including genetic polymorphisms as the exposure or for providing insufficient details to appropriately categorize exposure groups as reference vs non-reference groups; and for not reporting on H. pylori eradication failure as the outcome. Two review articles were also excluded. In the search of NHGRI, we did not identify any GWAS or similarly comprehensive whole genomic studies that met inclusion criteria. The characteristics of the 57 studies included for meta-analysis are detailed in Table 1 and Supplemental Table 2.
Figure 1.
PRISMA diagram of study inclusion
Characteristics of Studies
The majority of studies were from Asian-Pacific countries (24 Japan, 6 Taiwan, 10 China, 5 Korea, 1 Thailand), followed by Europe (3 Germany, 3 Poland, 1 Italy), the Middle East (2 Turkey) and South America (2 Brazil) (Table 1). There were 28 RCTs and 25 prospective cohort studies (base study design), which included a total of 9616 individuals. Of note, no studies randomized according to genetic polymorphism, the primary exposure for this meta-analysis; as such, for the purpose of this meta-analysis, all studies may be considered as prospective cohort studies.
Nearly all studies analyzed CYP2C19 polymorphisms (52/57, 91.2%). Fewer studies analyzed other genetic polymorphisms (9 IL-1B; 8 MDR; 3 IL-1RN; 2 CYP3A4; 2 TNFα). The majority of studies (41/57, 71.9%) included clarithromycin-based regimens and regimens with amoxicillin included at least 1.5g in divided doses (unable to confirm dosages in the 4 studies without full text access, all of which used clarithromycin-amoxicillin-PPI regimens25–28); although the duration and use of other antibiotic regimens varied (Table 1). Seven studies31–37 analyzed eradication outcomes in refractory H. pylori, defined as failure of at least one prior therapy12; notably, the PPI used in each of these 7 studies to treat refractory H. pylori was either EMZ or RAL. Of the studies where adherence rates were reported (40.3%, 23/57), all reported at least 80% adherence; adherence rates were not explicitly stated in the remaining included studies. Four of the included studies38–41 either defined eradication failure based a significant threshold quantitative decline in H. pylori IgG serology38,39, a definition which is now obsolete42, or included some participants based on positive serological testing only, rather than confirming active infection with a non-serological test.40,41 Notably, there was no change in the overall conclusion based on sensitivity analyses excluding these studies, except for the IL-1B-31 analysis in which the studies by Matsuo et al. and Ishida et al. were removed, as detailed below. Of the four studies in which only raw data were available and the full texts could not be accessed25–28, no information was available regarding duration of treatment, clarithromycin-sensitivity, adherence, drug dosages, nor other study-specific details except for the information delineated in Table 1. These studies were therefore only able to be included in the overall CYP2C19 analysis described below. The three studies of MDR1 variants included only patients with peptic ulcer disease.25–27 We performed sensitivity analyses (data not shown) removing each of these studies from the respective analyses, and there were no meaningful differences in the effect estimates.
Association of CYP2C19 metabolizer phenotypes with eradication failure
Overall.
The enhanced metabolizer CYP2C19 phenotypes (i.e. variant phenotypes, URM/RM/EM) were associated with a pooled 82% higher likelihood (OR 1.82; 95% CI, 1.52–2.19, P < .001) of eradication failure compared to the wild-type PM(+/−IM) phenotype(s) (Supplemental Figure 1; n=52 studies). There was moderate heterogeneity across the studies (I2 52.9%, P < .001). The heterogeneity was at least in part attributable to study design and study population characteristics (e.g. different PPI types and dosages, duration of therapy, clarithromycin resistance level, adherence) as well as reference group (PM+/−IM vs. PM only).
Visualization of the funnel plot demonstrated mild publication bias for the primary outcome of interest (eradication failure) (Supplemental Figure 2); however, Egger’s p-statistic did not confirm statistically significant publication bias (P = .18).
By PPI type and PPI dose.
Based on meta-analysis restricted to studies using PPIs predominantly metabolized by CYP2C19 (i.e. OMZ, PPZ, LNZ) only (n=29 studies), enhanced metabolizer phenotypes (i.e. URM/RM/EM, variant group) were associated with a 2.52-fold (95% CI, 1.78–3.55, P < .001) higher likelihood compared to PM only phenotype (I2 35.7%, P = .15) (Figure 2A), and 2.14-fold (95% CI, 1.65–2.76; P < .001) significantly higher likelihood of eradication failure when compared to PM+/−IM phenotypes as the wild-type/reference group (Supplemental Figure 3A; n=34 studies).
Figure 2A–2C. (CYP2C19: PPI type).
Meta-analysis of studies analyzing the association between enhanced metabolizer CYP2C19 phenotypes (i.e. URM/RM/EM, variant group) vs. PM only phenotype (wild-type/reference) and H. pylori eradication failure, when including only studies/study arms using OMZ, PPZ, LNZ (Figure 2A; low heterogeneity, I2 21.7%, P=0.15) and EMZ (Figure 2B; low heterogeneity, I2 0%, P=0.87) or RAL (Figure 2C; low heterogeneity, I2 9.8%, P=0.34)
Based on meta-analysis of studies or study arms where the PPI type was EMZ (n= 9 studies) or RAL (n=18 studies), there was no association between CYP2C19 metabolizer phenotypes and the likelihood of eradication failure when PM only was the reference group (Figure 2B, 2C). If the reference group was PM+/−IM, EMZ continued to demonstrate no association with eradication failure (Supplemental Figure 3B; n=12 studies), but for RAL, there was an increased odds of H. pylori eradication failure among individuals with enhanced metabolizer phenotypes compared to PM+/−IM phenotypes (OR 1.29; 95% CI, 1.01–1.63; P = .04) (Supplemental Figure 3C; n=21 studies). This association was no longer significant when restricted to studies where clarithromycin susceptibility and high adherence to treatment were confirmed (see below). There was no significant heterogeneity for these analyses.
When excluding studies that used only daily dosing of PPI or which did not specify the PPI dose in the H. pylori eradication regimen6, the URM/RM/EM genetic variants were associated with a 2.18-fold (95% CI, 1.65–2.88, P < .001) significantly higher likelihood of eradication failure compared to PM/IM genetic variants (n=30 studies), and a 2.74-fold (95% CI, 1.87–4.03, P < .001) higher likelihood compared to PM only genetic variants (n=26 studies). There was no significant heterogeneity (I2 20.1%, P = .18) (Supplemental Figures 4A–4B).
By clarithromycin susceptibility, duration of therapy, and adherence.
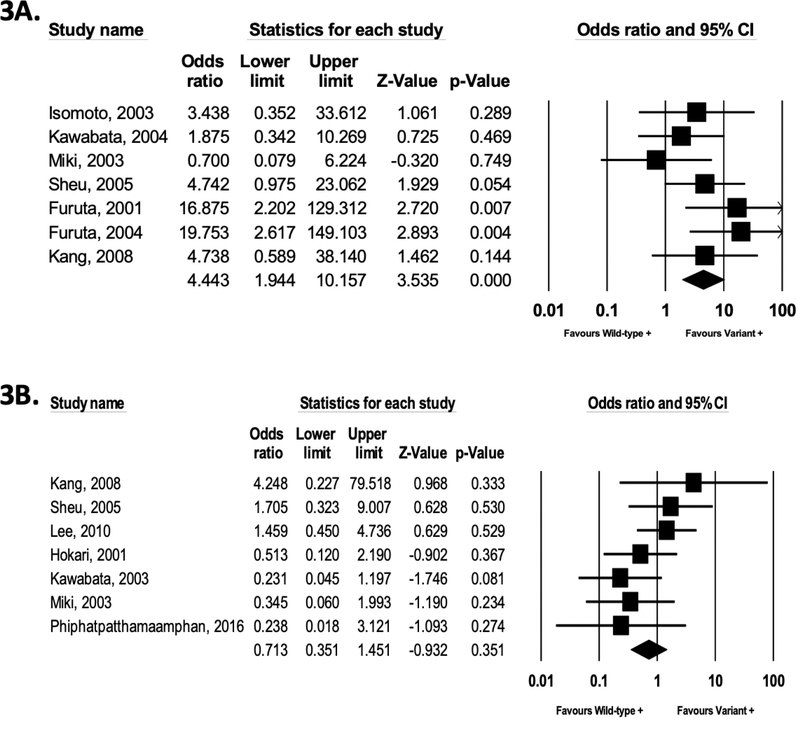
We then conducted a meta-analysis limited to studies and study arms which specifically reported clarithromycin resistance ≤15% or confirmed clarithromycin susceptibility if clarithromycin-based regimens were used. When all PPI types were analyzed together, there was no association between CYP2C19 metabolizer phenotypes and likelihood of H. pylori eradication failure, irrespective of whether PM (OR 1.69; 95% CI, 0.82–3.49; P = .15; n=14 studies) or PM+/−IM (OR 1.41; 95% CI, 0.89–2.23; P = .14; n=18 studies) were the wild-type/reference groups, Supplemental Figure 5A–5B). However, among users of OMZ, LNZ, and PPZ-containing clarithromycin-based regimens where clarithromycin resistance was ≤15% or susceptibility was confirmed, the enhanced metabolizer phenotypes were associated with a 4.44-fold (95% CI, 1.94–10.2, P < .001, Figure 3A; n=7 studies) and 2.16-fold (95% CI, 1.06–4.39; P = .03, Supplemental Figure 5C; n=9 studies) significantly higher likelihood of eradication failure compared to the PM only and PM+/−IM slow-metabolizer reference phenotypes, respectively. There was no association between CYP2C19 metabolizer phenotypes and likelihood of H. pylori eradication among users of EMZ-or RAL-containing regimens irrespective of whether the reference group was PM only (Figure 3B; n=7 studies) or PM+/−IM (Supplemental Figure 5D; n=9 studies).
Figure 3A–3B. (CYP2C19: PPI type, clarithromycin susceptibility).
Meta-analysis of studies analyzing the association between enhanced metabolizer CYP2C19 phenotypes (i.e. URM/RM/EM, variant group) vs. PM only phenotype (wild-type/reference) and H. pylori eradication failure, when including studies where clarithromycin susceptibility was confirmed, or resistance rates were confirmed ≤15% if clarithromycin-based regimens were used. Figure 3A: studies/study arms using OMZ, PPZ, LNZ (low heterogeneity, I2 20.4%, P=0.27); Figure 3B: studies/study arms using EMZ or RAL (low heterogeneity, I2 17.3%, P=0.29).
In studies or study arms where clarithromycin resistance was confirmed ≤15% (or clarithromycin susceptibility confirmed) and adherence >80% was explicitly reported, meta-regression revealed shorter duration of therapy (≤7 days vs >7 days) was associated with higher likelihood of eradication failure in those with URM/RM/EM vs. PM+/−IM genetic variants, but only for PPIs predominantly metabolized by CYP2C19 (P = .009; n=4 studies) and not those which bypass or are minimally metabolized by CYP2C19 (P = .87; n=6 studies) (Supplemental Figures 6A–6B). Based on meta-regression analysis, there was no difference in effect estimates between studies where adherence rates were explicitly reported as >80% versus not reported (P = .87, data not shown).
By geography.
Overall, there were not marked geographic differences in the association between CYP2C19 polymorphisms and eradication failure (Supplemental Figures 7A–7D). When OMZ, LNZ, or PPZ-containing regimens were used, those with URM/RM/EM vs. PM/IM genetic variants had a statistically significant 2.06-fold higher (95% CI, 1.52–2.80, P < .001) and 2.34-fold higher (95% CI, 1.52–3.69; P < .001) likelihood of eradication failure in studies from the Asia-Pacific region and Europe/South America/Middle East region, respectively. Of note, Europe, South America, and the Middle East region were also analyzed separately, without marked differences in observations; accordingly, because of the small number of studies, these were combined as ‘non-Asian-Pacific countries’. There was no association among EMZ and RAL users based on studies from Asian-Pacific countries. Only three studies of EMZ or RAL outside of the Asia-Pacific region were available for meta-analysis, two of which were German studies of refractory H. pylori (rifabutin-based regimens). Other than country of origin, no studies provided more granular breakdown by race or ethnicity.
Association of IL-1B and other variants with eradication failure
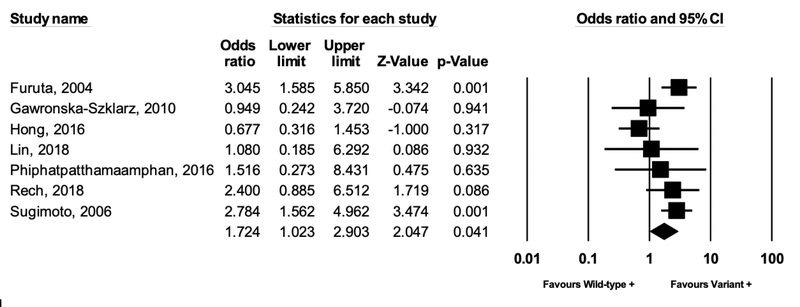
Nine studies analyzed polymorphisms of the IL-1B gene, which encodes the cytokine IL-1B, a potent gastric acid suppressor. One study by Hong et al. separated IL-1B-31 and IL-1B-511 polymorphisms, but it was unclear if individuals overlapped in these groups; thus, separate analyses were conducted. Compared to the TC or TT genotype (higher IL-1B expression, more potent acid suppression), the CC genotype was associated with a 1.72-fold (95% CI: 1.02–2.90, P = .04) significantly higher likelihood of eradication failure (Figure 4) when excluding the two studies that, in at least some of the study participants, either defined eradication failure based on the now obsolete definition of H. pylori IgG serological decline39 or included some participants based on serological testing only40. Additional IL-1B analyses, including sensitivity analyses based on clarithromycin resistance, are provided in the Supplementary Material (Supplemental Figures 8A–8C).
Figure 4 (IL-1B).
Meta-analysis of studies analyzing the association between IL-1B polymorphisms and H. pylori eradication treatment failure. Compared to the TC or TT genotype (higher IL-1B expression, more potent acid suppression), the CC genotype was associated with a 1.72-fold significantly higher likelihood of eradication failure (moderate, statistically significant heterogeneity I2 51.5%, P=0.05). Note, this analysis excludes the two studies by Matsuo et al. and Ishida et al., and includes only the IL-1B-31 genetic polymorphism from the Hong et al. study to avoid potential duplicates (see text for full details and supplemental material for additional IL-1B analyses)
There was no association between MDR1 polymorphisms and H. pylori eradication failure based on meta-analysis of 8 studies (OR 1.07; 95% CI, 0.64–1.79, P = .81) (Supplemental Figure 9A). As noted above, raw data from three of these studies25–27 were available via a previously published meta-analysis29, but the full texts were not available. Sensitivity analysis removing these three studies still yielded a null association (OR 1.08; 95% CI, 0.65–1.80, P = .81), but improved heterogeneity (Supplemental Figure 9B), as did removing only the small study Shi et al. (Supplemental Figure 9C).
There was no association between polymorphisms of CYP3A4 and H. pylori eradication failure, although there were too few studies for robust analyses for the latter (Supplemental Material, Supplemental Figure 10) The study by Sugimoto et al. which analyzed CYP2C19, IL-1B, IL-1RN, TNFα, and IL-10 had insufficient data to perform meta-analysis of the association between IL-1RN, TNFα, IL-10 and eradication failure.
Quality and Risk of Bias Assessment
The results of risk of bias (QUIPS tool) and quality of evidence (GRADE) assessments are provided in the Supplemental Tables 3 and 4, along with detailed rationale for the designated scores (Supplemental Material). Most studies were scored “low” to “low-moderate” risk of bias on the QUIPS tool assessment, with three studies scoring “moderate-high” based on study confounding43, study participation44, study attrition44,45, or outcome measurement45 (Supplemental Table 3). The quality of evidence based on detailed GRADE assessment was scored as “moderate” (Supplemental Table 5).
Discussion
Based on a meta-analysis of 57 studies representing 11 countries globally, we identified host genetic variants that are significantly associated with H. pylori eradication failure, with insufficient gastric acid suppression a presumed underlying mechanism. Specifically, use of eradication regimens containing PPIs predominantly metabolized by CYP2C19 (OMZ, PPZ, LNZ) in individuals with enhanced vs poor metabolizer phenotypes was associated with a 2.52-fold significantly higher likelihood of H. pylori eradication failure; this increased to 4.44-fold significantly higher among individuals where treatment adherence and H. pylori clarithromycin susceptibility (if clarithromycin regimen is used) were confirmed. Prolonged treatment duration (>7 days) and higher than twice daily standard dose OMZ, PPZ, or LNZ might improve eradication rates in enhanced metabolizers, but more data are needed. As hypothesized, there was no significant association between CYP2C19 metabolizer phenotypes and eradication failure when more potent PPIs that are less metabolized by (i.e. EMZ) or bypass metabolism by (i.e. RAL) CYP2C19 were used. Furthermore, IL-1B polymorphisms that are associated with less gastric acid suppression due to lower IL-1B expression were associated with 1.72-fold significantly higher likelihood of eradication failure, compared to those with genotypes associated with higher IL-1B expression. Notably, the quality of the evidence informing the effect estimates presented in this meta-analysis was rated as moderate certainty, meaning that the true effects are probably close to the estimated effects.24 Collectively, these findings support the critical, albeit often overlooked, role that achieving adequate gastric acid suppression plays in successful H. pylori eradication. Our search yielded few other host genetic polymorphisms that have been analyzed for their association with H. pylori eradication failure, and this remains an area of needed research. Meta-analyses of other genetic polymorphisms individually, including CYP3A4 and MDR1, demonstrated null associations with H. pylori eradication outcomes, but were somewhat limited due to relatively sparse literature.
In 2017, the WHO designated H. pylori eradication failure as an immediate priority that needed to be addressed given the substantial public health and economic implications. The WHO agenda, similar to other efforts focused on improving H. pylori eradication rates, is predominantly focused on H. pylori antibiotic resistance as the mechanism underlying eradication failure.14,46,47 However, the data presented here suggest that failure to achieve sufficient gastric acid suppression is likewise an important etiology of H. pylori eradication failure, especially among individuals with confirmed treatment adherence who are colonized by H. pylori strains with in vitro susceptibility to the prescribed antibiotic regimen.
Achieving gastric pH >6–7 is fundamental to successful H. pylori eradication for multiple reasons. H. pylori’s log phase, the phase in which bacteria are most susceptible to antibiotics, occurs above pH 6–7.12,48 This is also the pH at which certain antibiotics, including clarithromycin and amoxicillin, are most stable and bioavailable.48,49 Both intrinsic host and microbial factors mediate gastric acid suppression to varying degrees. Perhaps the strongest mediator of gastric acid suppression is pharmacologic acid suppression. The effectiveness of PPIs with respect to gastric acid suppression is determined by their plasma concentration, which in turn depends on their absorption (e.g. MDR1, which encodes a PPI efflux transporter; and non-genetic factors such as concomitant food consumption) and metabolism.17 CYP2C19 accounts for >80–90% of the primary metabolism of first-generation PPIs (OMZ, LNZ, PPZ) and dexlansoprazole.17,50 Decreases in gastric pH towards normal gastric pH (pH<2) during the therapeutic period, as might occur with use of these PPIs in individuals with enhanced metabolizer phenotypes, reduces the likelihood of successful H. pylori eradication. Too few studies analyzed H. pylori eradication outcomes when higher than standard twice daily dosing of first-generation PPIs were used, but individual studies do suggest higher success rates in enhanced metabolizers when higher dosages or frequencies are used. Further investigation is also needed to determine whether knowledge of CYP2C19 phenotype might additionally help tailor antibiotic dosage, frequency and overall therapeutic duration in individuals with PM phenotypes, since they may not need as intensive regimens as enhanced metabolizers. Apart from CYP2C19 variants, few other host genetic determinants have been implicated in H. pylori eradication failure, and to a varying degree, with most appearing to also modulate gastric pH. For example, IL-1B and TNFα are inflammatory cytokines, but their primary role in eradication failure appears to be through modulation of gastric pH. The role of genetic polymorphisms in genes for other inflammatory cytokines (e.g. IL-1RN, IL-10) 39,51,52 with respect to H. pylori eradication is not established and, as confirmed by our systematic review, there are far fewer data compared to CYP2C19 polymorphisms.
Our findings are congruent with an earlier meta-analysis analyzing the association between CYP2C19 polymorphisms and H. pylori eradication failure. However, that analysis was limited to data from 16 randomized controlled trials in Asian-Pacific countries (lower prevalence of enhanced metabolizer phenotypes versus non-Asian populations) of clarithromycin-based triple therapy among treatment-naïve patients only. The analysis also did not account for other factors that impact the likelihood of H. pylori eradication, including clarithromycin resistance and patient adherence to treatment. In light of these limitations, the authors acknowledged the unclear clinical implications of their findings. In the present study, we conducted comprehensive sensitivity analyses to overcome previous limitations, including analyses based on geography. Notably, compared to Asian ethnic groups, Europeans are more likely to carry the CYP2C19*17 allele, which is associated with the ultrarapid metabolizer phenotype (*17/*17 homozygous).18,53 The lack of North American studies remains a clinically relevant knowledge gap that warrants attention. Indeed, US-population based studies demonstrate a high population prevalence of enhanced metabolizer phenotypes, approximately 60–70%, in US non-Hispanic whites, non-Hispanic blacks, and Hispanics, whereas Asian Americans have the highest prevalence of the poor metabolizer phenotype.17,19 Recognizing the high prevalence of CYP2C19 enhanced metabolizers in US populations, other fields, including cardiology, have used commercially available CYP2C19 polymorphism testing to guide dual antiplatelet therapy selection, specifically clopidogrel. Current pharmacogenetics guidelines provide recommendations on PPI dosing in H. pylori eradication when the CYP2C19 phenotype is known, but they do not provide recommendations on pre-treatment CYP2C19 polymorphism testing when CYP2C19 phenotype is not known.54 Current international H. pylori guidelines also do not comment on CYP2C19 testing, and this remains an area in need of focused investigation especially in regions with racially and ethnically diverse populations, such as the US.
When considering the high prevalence of enhanced metabolizers in the US, the availability of CYP2C19 polymorphism testing, the findings of this meta-analysis, the potential for substantial reduction in H. pylori eradication failure and the significant downstream health and economic implications, it is clear that well-designed interventional studies comparing H. pylori eradication outcomes in empiric vs. genetic polymorphism-guided selection of acid suppressive therapy are warranted. The positioning of potassium-competitive acid blockers (P-CABs) is also worth considering, particularly since vonoprazan, which is not currently approved outside of certain Asian-Pacific and South/Central American countries/regions, is actively being investigated for use in the US and Europe for conditions necessitating gastric acid suppression. Similar to PPIs, P-CABs act on the parietal cell H+, K+-ATPase transporter of the parietal cell; but, in contrast to PPIs, P-CABs instead compete with K+ ions. Data suggest that P-CABs produce more rapid and more potent gastric acid suppression vs comparable dosages of PPIs. The vast majority of studies of P-CABs in H. pylori eradication failure were conducted in Japan and may not be generalizable to other populations. Whether P-CABs are superior to PPI-based H. pylori eradication treatment in patients with normal or slow CYP2C19 metabolizer phenotypes or superior to PPIs the bypass or are only partially metabolized by CYP2C19 warrants focused investigation, especially in non-Asian-Pacific populations, along with cost valuation. Notably, the one Phase III study being conducted in the US and Europe (NCT04167670), with results anticipated late 2021, compares two vonoprazan-based regimens and one lansoprazole-based regimen, but does not include a rabeprazole or esomeprazole-based arm. P-CABs are primarily metabolized by CYP3A4/5 instead of CYP2C19, and genetic determinants still appear to be relevant. To this end, CYP3A4/5 variants have been associated with H. pylori eradication failure in vonoprazan regimens55, further underscoring the potential role of genetic polymorphism-guided H. pylori eradication treatment.
Strengths of this meta-analysis include the large size and detailed data abstraction that enabled comprehensive, clinically relevant and hypothesis-driven subgroup analyses, as well as comprehensive appraisal of the quality of the evidence, including use of the GRADE system. This is the first analysis to also investigate other genetic determinants of H. pylori eradication failure. We systematically explored reasons for observed heterogeneity, such as analyzing PMs only as the reference group as opposed to combined PMs+/−IMs. While there was heterogeneity related to treatment regimen and duration, we separately analyzed studies with longer durations and confirmed clarithromycin susceptibility. Notwithstanding, there are notable limitations, the majority of which are also limitations of the individual studies. We are unable to account for the variable effect of gene-environment and gene-gene interactions—for example, the interaction between CYP2C19 polymorphisms and smoking or gender56, and perhaps a compounded effect of CYP2C19 variants and IL-1B variants. We are also unable to account for the interaction between H. pylori strain-specific determinants and other host genetic polymorphisms, which might affect eradication outcomes. Studies of genetic polymorphisms are subject to intrinsic publication/reporting bias; while we observed mild publication bias on funnel plot visualization, this was not statistically significant.
In conclusion, based on a systematic review with meta-analysis we identified host genetic variants that are significantly associated with H. pylori eradication failure, with the predominant underlying biological mechanism most plausibly being insufficient sustained gastric acid suppression. The largest body of data support CYP2C19 variants; importantly, there is already a paved pathway for clinical implementation, since CYP2C19 polymorphism testing is available in clinical practice settings and can be expanded upon. Ideally, H. pylori eradication therapy will eventually move towards incorporating both genetic and non-genetic factors—such as age, smoking, prior antibiotic exposure—into a clinical decision-making tool that helps individualize primary H. pylori eradication therapy in order to maximize primary eradication rates and patient safety, hand in hand with minimizing patient harm and cost/waste (e.g. excess antibiotics). Given the very high prevalence of enhanced metabolizers across many racial and ethnic groups in the US, comparative trials investigating empiric vs. polymorphism-guided alteration of acid suppressive medications, as well as the cost implications, in both naïve and refractory H. pylori infection are needed before any formal recommendations can be offered, unless the CYP2C19 phenotype is already known. Even small incremental improvements in H. pylori eradication rates would likely translate to substantial collateral health, economic, and societal benefits.
Supplementary Material
What You Need to Know.
Background and Context:
Helicobacter pylori eradication treatment failure is associated with several negative downstream clinical and collateral economic consequences. Patient adherence to treatment and H. pylori antibiotic susceptibility are key determinants of successful eradication. However, the impact of host genetics have not been thoroughly investigated.
New Findings:
Based on a meta-analysis of 57 studies with over 9600 patients treated for H. pylori, we identified host genetic variants significantly associated with eradication treatment failure, ranging from 1.7 to 4.4-fold significantly higher, with insufficient gastric acid suppression likely a primary driver.
Limitations:
None of the cohorts provided detailed information on race/ethnicity, although most populations were from the Asian-Pacific region. We are unable to account for the variable effect of gene-environment and gene-gene interactions. Publication and reporting biases are also relevant.
Impact:
These findings may be used to individualize H. pylori eradication treatment by incorporating host genetic factors into treatment regimen selection, particularly with respect to ensuring adequate gastric acid suppression. However, comparative trials investigating empiric vs. polymorphism-guided H. pylori eradication treatment are needed before any formal recommendations can be offered.
Grant support:
SCS is funded by the Agency for Healthcare Research (AHRQ) and Quality and Patient-Centered Outcomes Research Institute (PCORI) under award number K12 HS026395, a 2019 American Gastroenterological Association Research Scholar Award, and Veterans Affairs Career Development Award under award number ICX002027A01. Additional funding support for co-authors include the National Institutes of Health (NIH), National Cancer Institute (NCI): R01 DK58587, R01 CA 77955, P01 116087 (RP). NN receives funding through a McMaster University Department of Medicine Internal Career Award. The content is solely the responsibility of the listed authors and does not necessarily represent the official views of the funding agencies listed.
Footnotes
Disclosures/Conflict of interest statement: The authors have no potential conflicts (financial, professional, nor personal) that are relevant to this manuscript. This work has not been previously presented.
Writing assistance: None
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hooi JKY, Lai WY, Ng WK, et al. Global Prevalence of Helicobacter pylori Infection: Systematic Review and Meta-Analysis. Gastroenterology 2017; 153: 420–9. [DOI] [PubMed] [Google Scholar]
- 2.Plummer M, Franceschi S, Vignat J, Forman D, de Martel C. Global burden of gastric cancer attributable to Helicobacter pylori. Int J Cancer 2015; 136: 487–90. [DOI] [PubMed] [Google Scholar]
- 3.Stomach Cancer - Cancer Stat Facts. https://seer.cancer.gov/statfacts/html/stomach.html (accessed Feb 12, 2019).
- 4.WHO | Disease burden and mortality estimates. https://www.who.int/healthinfo/global_burden_disease/estimates/en/index1.html (accessed Feb 16, 2019).
- 5.Peery AF, Crockett SD, Murphy CC, et al. Burden and cost of gastrointestinal, liver, and pancreatic diseases in the united states: update 2018. Gastroenterology 2019; 156: 254–272.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chey WD, Leontiadis GI, Howden CW, Moss SF. ACG Clinical Guideline: Treatment of Helicobacter pylori Infection. Am J Gastroenterol 2017; 112: 212–39. [DOI] [PubMed] [Google Scholar]
- 7.Kato M, Ota H, Okuda M, et al. Guidelines for the management of Helicobacter pylori infection in Japan: 2016 Revised Edition. Helicobacter 2019; 24: e12597. [DOI] [PubMed] [Google Scholar]
- 8.Fallone CA, Moss SF, Malfertheiner P. Reconciliation of Recent Helicobacter pylori Treatment Guidelines in a Time of Increasing Resistance to Antibiotics. Gastroenterology 2019; 157: 44–53. [DOI] [PubMed] [Google Scholar]
- 9.Fallone CA, Chiba N, van Zanten SV, et al. The Toronto Consensus for the Treatment of Helicobacter pylori Infection in Adults. Gastroenterology 2016; 151: 51–69.e14. [DOI] [PubMed] [Google Scholar]
- 10.Malfertheiner P, Megraud F, O’Morain CA, et al. Management of Helicobacter pylori infection-the Maastricht V/Florence Consensus Report. Gut 2017; 66: 6–30. [DOI] [PubMed] [Google Scholar]
- 11.Sugano K, Tack J, Kuipers EJ, et al. Kyoto global consensus report on Helicobacter pylori gastritis. Gut 2015; 64: 1353–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shah SC, Iyer PG, Moss SF. AGA Clinical Practice Update on The Management of Refractory Helicobacter pylori Infection: Expert Review. Gastroenterology 2021; published online Jan 29. DOI: 10.1053/j.gastro.2020.11.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fock KM, Talley N, Moayyedi P, et al. Asia-Pacific consensus guidelines on gastric cancer prevention. J Gastroenterol Hepatol 2008; 23: 351–65. [DOI] [PubMed] [Google Scholar]
- 14.Savoldi A, Carrara E, Graham DY, Conti M, Tacconelli E. Prevalence of Antibiotic Resistance in Helicobacter pylori: A Systematic Review and Meta-analysis in World Health Organization Regions. Gastroenterology 2018; 155: 1372–1382.e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hagymási K, Müllner K, Herszényi L, Tulassay Z. Update on the pharmacogenomics of proton pump inhibitors. Pharmacogenomics 2011; 12: 873–88. [DOI] [PubMed] [Google Scholar]
- 16.Tang H-L, Li Y, Hu Y-F, Xie H-G, Zhai S-D. Effects of CYP2C19 loss-of-function variants on the eradication of H. pylori infection in patients treated with proton pump inhibitor-based triple therapy regimens: a meta-analysis of randomized clinical trials. PLoS One 2013; 8: e62162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.El Rouby N, Lima JJ, Johnson JA. Proton pump inhibitors: from CYP2C19 pharmacogenetics to precision medicine. Expert Opin Drug Metab Toxicol 2018; 14: 447–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Petrović J, Pešić V, Lauschke VM. Frequencies of clinically important CYP2C19 and CYP2D6 alleles are graded across Europe. Eur J Hum Genet 2020; 28: 88–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martis S, Peter I, Hulot JS, Kornreich R, Desnick RJ, Scott SA. Multi-ethnic distribution of clinically relevant CYP2C genotypes and haplotypes. Pharmacogenomics J 2013; 13: 369–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.WHOCC - ATC/DDD Index. https://www.whocc.no/atc_ddd_index/ (accessed March 14, 2021).
- 21.Higgins JPT, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. BMJ 2003; 327: 557–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hayden JA, van der Windt DA, Cartwright JL, Côté P, Bombardier C. Assessing bias in studies of prognostic factors. Ann Intern Med 2013; 158: 280–6. [DOI] [PubMed] [Google Scholar]
- 23.Welcome | Cochrane Prognosis. https://methods.cochrane.org/prognosis/welcome (accessed July 1, 2021).
- 24.Guyatt GH, Oxman AD, Vist GE, et al. GRADE: an emerging consensus on rating quality of evidence and strength of recommendations. BMJ 2008; 336: 924–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen D, Sun Y, Wu Z. Effects of CYP2C19 and MDR1polymorphism on the eradication rate of Helicobacter pylori infection inesomeprazole-based triple therapy. J Pract Med 2012; 28: 2880–1. [Google Scholar]
- 26.Zhang Y, Sun Y, Zhou X. Influence of multidrug resistance gene 1 C3435T genetic polymorphism on the eradication of gastric ulcer with Helicobacter pylori infection. Chin J Postgrad Med 2013; 36: 4–6. [Google Scholar]
- 27.Shi C, Lixian W. Influence of MDR1 C3435T polymorphism on theeradication of Helicobacter pylori in patiens with gastric ulcer. . Chin J Gerontol 2014; 34: 6955–6. [Google Scholar]
- 28.Jiang YJ, Li YY, Nie YQ, Wang H, Sha WH. Effect of cytochrome P450 2C19 genetic polymorphisms on efficacy of rabeprazole or omeprazole-based triple therapy for eradication of Helicobacter pylori. . Chin J Dig 2005; 25: 458–61. [Google Scholar]
- 29.Li M, Li T, Guo S, Liang H, Jiang D. The effect of MDR1 C3435T polymorphism on the eradication rate of H. pylori infection in PPI-based triple therapy: A meta-analysis. Medicine 2017; 96: e6489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhao F, Wang J, Yang Y, et al. Effect of CYP2C19 genetic polymorphisms on the efficacy of proton pump inhibitor-based triple therapy for Helicobacter pylori eradication: a meta-analysis. Helicobacter 2008; 13: 532–41. [DOI] [PubMed] [Google Scholar]
- 31.Song Z, Zhou L, Zhang J, He L, Bai P, Xue Y. Levofloxacin, bismuth, amoxicillin and esomeprazole as second-line Helicobacter pylori therapy after failure of non-bismuth quadruple therapy. Dig Liver Dis 2016; 48: 506–11. [DOI] [PubMed] [Google Scholar]
- 32.Liou J-M, Chen C-C, Chen M-J, et al. Empirical modified sequential therapy containing levofloxacin and high-dose esomeprazole in second-line therapy for Helicobacter pylori infection: a multicentre clinical trial. J Antimicrob Chemother 2011; 66: 1847–52. [DOI] [PubMed] [Google Scholar]
- 33.Miehlke S, Schneider-Brachert W, Kirsch C, et al. One-week once-daily triple therapy with esomeprazole, moxifloxacin, and rifabutin for eradication of persistent Helicobacter pylori resistant to both metronidazole and clarithromycin. Helicobacter 2008; 13: 69–74. [DOI] [PubMed] [Google Scholar]
- 34.Miehlke S, Hansky K, Schneider-Brachert W, et al. Randomized trial of rifabutin-based triple therapy and high-dose dual therapy for rescue treatment of Helicobacter pylori resistant to both metronidazole and clarithromycin. Aliment Pharmacol Ther 2006; 24: 395–403. [DOI] [PubMed] [Google Scholar]
- 35.Kuo C-H, Wang SSW, Hsu W-H, et al. Rabeprazole can overcome the impact of CYP2C19 polymorphism on quadruple therapy. Helicobacter 2010; 15: 265–72. [DOI] [PubMed] [Google Scholar]
- 36.Wu D-C, Hsu P-I, Tseng H-H, et al. Helicobacter pylori infection: a randomized, controlled study comparing 2 rescue therapies after failure of standard triple therapies. Medicine 2011; 90: 180–5. [DOI] [PubMed] [Google Scholar]
- 37.Isomoto H, Inoue K, Furusu H, et al. High-dose rabeprazole-amoxicillin versus rabeprazole-amoxicillin-metronidazole as second-line treatment after failure of the Japanese standard regimen for Helicobacter pylori infection. Aliment Pharmacol Ther 2003; 18: 101–7. [DOI] [PubMed] [Google Scholar]
- 38.Suzuki T, Matsuo K, Sawaki A, et al. Influence of smoking and CYP2C19 genotypes on H. pylori eradication success. Epidemiol Infect 2007; 135: 171–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Matsuo K, Hamajima N, Ikehara Y, et al. Smoking and polymorphisms of fucosyltransferase gene Le affect success of H. pylori eradication with lansoprazole, amoxicillin, and clarithromycin. Epidemiol Infect 2003; 130: 227–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ishida Y, Goto Y, Kondo T, et al. Eradication rate of Helicobacter pylori according to genotypes of CYP2C19, IL-1B, and TNF-A. Int J Med Sci 2006; 3: 135–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shimoyama T, Chinda D, Sawada Y, et al. Randomized Trial Comparing Esomeprazole and Rabeprazole in First-line Eradication Therapy for Helicobacter pylori Infection based on the Serum Levels of Pepsinogens. Intern Med 2017; 56: 1621–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Marchildon P, Balaban DH, Sue M, et al. Usefulness of serological IgG antibody determinations for confirming eradication of Helicobacter pylori infection. Am J Gastroenterol 1999; 94: 2105–8. [DOI] [PubMed] [Google Scholar]
- 43.Hokari K, Sugiyama T, Kato M, et al. Efficacy of triple therapy with rabeprazole for Helicobacter pylori infection and CYP2C19 genetic polymorphism. Aliment Pharmacol Ther 2001; 15: 1479–84. [DOI] [PubMed] [Google Scholar]
- 44.Jinda S, Nakatani K, Nishioka J, et al. Personalized treatment in the eradication therapy for Helicobacter pylori. Int J Mol Med 2011; 27: 255–61. [DOI] [PubMed] [Google Scholar]
- 45.Karaca RO, Kalkisim S, Altinbas A, et al. Effects of Genetic Polymorphisms of Cytochrome P450 Enzymes and MDR1 Transporter on Pantoprazole Metabolism and Helicobacter pylori Eradication. Basic Clin Pharmacol Toxicol 2017; 120: 199–206. [DOI] [PubMed] [Google Scholar]
- 46.Meyer JM, Silliman NP, Wang W, et al. Risk factors for Helicobacter pylori resistance in the United States: the surveillance of H. pylori antimicrobial resistance partnership (SHARP) study, 1993–1999. Ann Intern Med 2002; 136: 13–24. [DOI] [PubMed] [Google Scholar]
- 47.Tacconelli E, Carrara E, Savoldi A, et al. Discovery, research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect Dis 2018; 18: 318–27. [DOI] [PubMed] [Google Scholar]
- 48.Furuta T, Graham DY. Pharmacologic aspects of eradication therapy for Helicobacter pylori Infection. Gastroenterol Clin North Am 2010; 39: 465–80. [DOI] [PubMed] [Google Scholar]
- 49.Erah PO, Goddard AF, Barrett DA, Shaw PN, Spiller RC. The stability of amoxycillin, clarithromycin and metronidazole in gastric juice: relevance to the treatment of Helicobacter pylori infection. J Antimicrob Chemother 1997; 39: 5–12. [DOI] [PubMed] [Google Scholar]
- 50.Gawrońska-Szklarz B, Siuda A, Kurzawski M, Bielicki D, Marlicz W, Droździk M. Effects of CYP2C19, MDR1, and interleukin 1-B gene variants on the eradication rate of Helicobacter pylori infection by triple therapy with pantoprazole, amoxicillin, and metronidazole. Eur J Clin Pharmacol 2010; 66: 681–7. [DOI] [PubMed] [Google Scholar]
- 51.Sugimoto M, Furuta T, Yamaoka Y. Influence of inflammatory cytokine polymorphisms on eradication rates of Helicobacter pylori. J Gastroenterol Hepatol 2009; 24: 1725–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wolfe MM, Nompleggi DJ. Cytokine inhibition of gastric acid secretion--a little goes a long way. Gastroenterology 1992; 102: 2177–8. [DOI] [PubMed] [Google Scholar]
- 53.Strom CM, Goos D, Crossley B, et al. Testing for variants in CYP2C19: population frequencies and testing experience in a clinical laboratory. Genet Med 2012; 14: 95–100. [DOI] [PubMed] [Google Scholar]
- 54.Lima JJ, Thomas CD, Barbarino J, et al. Clinical pharmacogenetics implementation consortium (CPIC) guideline for CYP2C19 and proton pump inhibitor dosing. Clin Pharmacol Ther 2020; published online Aug 8. DOI: 10.1002/cpt.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sugimoto M, Ban H, Hira D, et al. Letter: CYP3A4/5 genotype status and outcome of vonoprazan-containing Helicobacter pylori eradication therapy in Japan. Aliment Pharmacol Ther 2017; 45: 1009–10. [DOI] [PubMed] [Google Scholar]
- 56.Chang YW, Ko WJ, Oh CH, et al. Clarithromycin resistance and female gender affect Helicobacter pylori eradication failure in chronic gastritis. Korean J Intern Med 2019; 34: 1022–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.