Summary
Complex traits and diseases can be influenced by both genetics and environment. However, given the large number of environmental stimuli and power challenges for gene-by-environment testing, it remains a critical challenge to identify and prioritize specific disease-relevant environmental exposures. We propose a framework for leveraging signals from transcriptional responses to environmental perturbations to identify disease-relevant perturbations that can modulate genetic risk for complex traits and inform the functions of genetic variants associated with complex traits. We perturbed human skeletal-muscle-, fat-, and liver-relevant cell lines with 21 perturbations affecting insulin resistance, glucose homeostasis, and metabolic regulation in humans and identified thousands of environmentally responsive genes. By combining these data with GWASs from 31 distinct polygenic traits, we show that the heritability of multiple traits is enriched in regions surrounding genes responsive to specific perturbations and, further, that environmentally responsive genes are enriched for associations with specific diseases and phenotypes from the GWAS Catalog. Overall, we demonstrate the advantages of large-scale characterization of transcriptional changes in diversely stimulated and pathologically relevant cells to identify disease-relevant perturbations.
Keywords: colocalization, gene expression, gene-by-environment interactions, eQTL, GWAS
Introduction
Genome-wide association studies (GWASs) have identified thousands of genetic variants associated with complex diseases and traits.1 The majority of these variants fall into non-coding regions of the genome and, as a result, their mechanism of action remains largely unknown.2 In recent years, researchers have gained an increasingly clear picture of which parts of the genome are active in a range of tissues and cell types.3, 4, 5, 6 Integrating such information with results from GWASs has identified cell types, tissues, and regulatory elements relevant to specific diseases and phenotypes and moved the field toward a mechanistic understanding of GWAS hits.7, 8, 9 In addition, genomic colocalization and transcriptome-wide association studies combining results from GWASs and expression quantitative trait locus (eQTL) studies have identified candidate causal genes and their mechanisms of action.10, 11, 12
Despite these advances, a modest fraction of GWAS-associated variants and eQTLs colocalize for any trait13,14 providing the perspective that many disease-relevant effects are modulated by yet-to-be-discovered environmental factors. To address this challenge, multiple studies have mapped eQTLs in vitro that are responsive to the environment.15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26 For example, the Immune Variation Project identified eQTLs in human CD4+ T lymphocytes with different effects across distinct immune states.17 These previously unknown, immune-state-specific eQTLs were enriched for autoimmune-disease-associated variants, underscoring the importance of exploring contexts beyond tissues and cell types to reveal the specificity of genetic associations. Although there is mounting evidence that the environment modulates genetic effects, GWASs and eQTL studies rarely measure and test for genetic interactions with environmental exposures. This is, in part, due to the difficulty of identifying and collecting information on the most relevant environmental exposures in GWAS cohorts and performing eQTL studies in contexts that are relevant for the specific trait or disease.
In this study, we extend the current understanding of inherited variation in complex traits by implementing a framework to model signals from transcriptional responses to environmental perturbations to catalog and prioritize disease-relevant environments that can modulate genetic risk for complex traits and inform the functions of genetic variants and genes associated with complex traits. Specifically, we first assessed environmental effects on gene expression levels in three metabolic human cell lines by performing RNA sequencing (RNA-seq) in muscle-, fat-, and liver-relevant cell lines treated with 21 different environmental perturbations related to aspects of glucose and insulin metabolism, kinase inhibitors, inflammation, fatty acid metabolism, etc. (n = 234 samples). We identified thousands of environmentally responsive genes underlying disease-associated response pathways and characterized the specificity and sharing of these effects across perturbations and cell lines. Next, to identify disease-relevant perturbations, we coupled our gene expression data with GWAS summary statistics of 31 complex traits and diseases as well as associations from the GWAS Catalog. We confirmed several well-established environmental-phenotype associations, e.g., the role of TGF-β1 on asthma,27 and provided additional evidence for recent and less well-understood associations, e.g., the role of leptin on major depressive disorder.28 Last, to further illustrate how perturbation experiments inform the functions of complex trait-associated variants, we integrate our perturbation data with genomic colocalization studies and show that the effects of these perturbations in the relevant tissues identify context-specific molecular mechanisms of GWAS hits for diverse cardiometabolic traits.
This resource characterizes the dynamic transcriptional landscape in metabolic tissues and provides a framework for identification and prioritization of disease-relevant perturbations and disentanglement of the complex gene-environment interactions that determine disease susceptibility, which is particularly relevant for complex traits such as insulin resistance (IR), diabetes, and obesity.
Material and methods
Cell culturing and perturbations
Experiments were conducted with human skeletal muscle29,30 (HMCL-7304 myocytes; provided by Institute of Child Health, University College London), fat31 (terminally differentiated Simpson-Golabi-Behmel syndrome [SGBS] pre-adipocytes; provided by Dr. Martin Wabitsch, Ulm University, Ulm, Germany), and liver32 (HepG2; ATCC) cell lines. SGBS and HMCL-7304 cells were differentiated as described previously.30,31 Each cell line was starved for 6 h in Eagle's minimal essential medium (EMEM) with no fetal bovine serum (FBS) (HepG2), Dulbecco's modified Eagle's medium (DMEM)/F12 supplemented with pan/bio and penicillin/streptomycin but no FBS (SGBS adipocytes), or HMCL growth medium (PromoCell) without supplements. For the glucose condition DMEM, no glucose medium (Thermo) was used. After starvation and washing with PBS, the cells were exposed for 2 h to one of the 21 perturbations shown in Figure 1A in triplicate for each cell-line-perturbation combination. We selected a stimulation window of 2 h to allow enough time for transcriptional changes to occur and, at the same time, to minimize potential secondary responses that are not direct transcriptional effects of the selected perturbations, as previously assayed for insulin in the liver and skeletal muscle cells from mice.33 In addition, we selected the concentrations of use, shown in Data S1, on the basis of consensus research of available literature, particularly in the cells of interest. Last, we prepared triplicate control samples for glucose (no glucose medium) and four sets of triplicate control samples (no stimulation) for all other perturbations in each cell line, resulting in a sample size of 234.
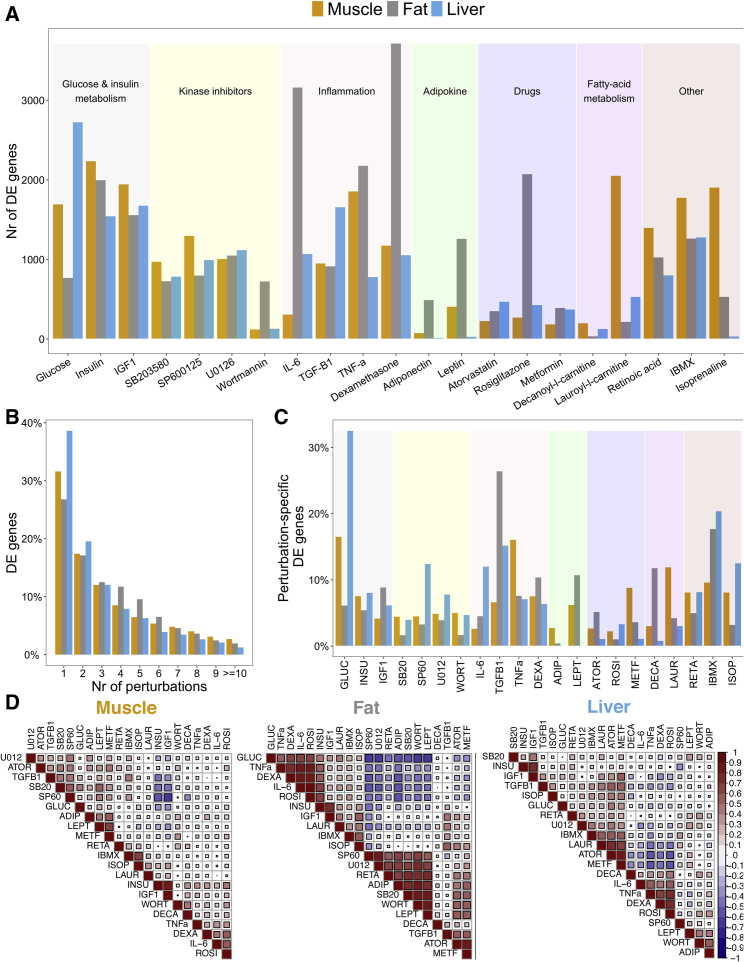
Figure 1.
Perturbations induce large-scale changes in gene expression in muscle, fat, and liver
(A) Number of DE genes for each perturbation in each cell line (FDR < 5%). See Data S2, S3, and S4 for extended DE summary statistics.
(B) Proportion of DE genes that change in response to up to ten perturbations in each cell line. See Data S4 for extended results on sharing of DE genes.
(C) Proportion of perturbation-specific DE genes, i.e., genes that change in response to a single perturbation, for each perturbation and within each cell line.
(D) Correlation of DE patterns between different perturbations within each cell line. Each square is Spearman’s correlation between the DE test statistic of a pair of perturbations across all genes.
DE, differentially expressed; FDR, false discovery rate.
RNA isolation and sequencing
After stimulation, the cells were washed with PBS and collected in PureLink RNA extraction lysis buffer supplemented with 1% 2-mercaptoethanol, flash-frozen in dry ice, and stored at −80°C. RNA extraction was performed with the PureLink RNA Mini kit (Thermo). 260/280 and RNA integrity number (RIN) values were assessed before sequencing for sample purity and integrity. Library preparation was performed at Novogene company. Liver samples were sequenced in HiSeq 4000 (Illumina), and fat and muscle were sequenced in Novaseq 6000 (Illumina) at 150 bp paired-end reads.
RNA-seq quality control
Reference genome (hg19) and gene model annotation (GRCh37.p13) files were downloaded from the UCSC Genome Browser website directly. Indexes of the reference genome were built with STAR,34 and paired-end clean reads were aligned to the reference genome via STAR (v.2.6.0; with default option mismatch = 10). Bam files were filtered for uniquely mapped reads, sorted, and indexed via SAMtools35 (v.1.4.1). We used HTSeq36 (v.0.11.0) to count the read numbers mapped to each gene (with option -m union).
For all subsequent analyses, we focused only on expressed genes, i.e., genes that have median expression counts above 10 in at least one of the conditions (perturbation or control) within each cell line, i.e., 17,660, 17,140, and 16,722 genes for fat, muscle, and liver cells, respectively. As a measure of quality control, we looked at several RNA-seq technical metrics (Figures S1–S3), e.g., RNA integrity number, percent guanine-cytosine (GC) content, percent of uniquely mapped reads, etc. One sample (TGF-β1 in fat) was dropped because of failing these quality control metrics. All remaining samples had RIN above 8, at least 16 million reads, GC content, an average of 36% of reads marked as PCR duplicates, at least 84% of their reads mapped uniquely, and an average of 95%, 4%, and 98% exon, intron, and transcript overlapping reads, respectively (Figure S1). Moreover, for all samples, their median Spearman expression correlation (D-statistic) with other samples was at least 0.96 (Figure S1). We used principal-component analysis (PCA) to identify gene expression outliers. After removing the low-quality sample mentioned above and adjusting for major components of variability (see below), no outliers are present based on the first two principal components (Figure S3).
Identifying major components of variability in RNA-seq data
We identified and adjust for major components of gene expression variability by computing the percent of gene expression variance for each gene explained by technical and biological variables via the linear mixed models implemented in the R package variancePartition37 (Figure S2). Prior to computing the percent of variance explained, we variance stabilized and log2-transformed the expression of each gene within cell lines by using the R package DESeq2. Then, we centered and scaled each gene to have zero mean and unit variance. For each gene expressed in each cell line, we used a linear mixed model with the effect of medium, number of cells plated, plate number, sequencing batch, cell collection, differentiation, RNA extraction, starvation, and treatment date as random and the effects of all other variables as fixed. Sequencing batch and number of cells plated only differed and were modeled for liver samples, while differentiation date only differed and was modeled for fat and muscle samples. We correct all subsequent analyses for all variables that explain, on average, more than 1% of expression variability in either cell line, i.e., percent GC content, percent exon overlapping reads, RNA concentration, percent of reads marked as PCR duplicates, RNA 260/280 ratio, RIN, and percent uniquely mapped reads. For analyses done in liver cells, we also correct for sequencing batch. The number of cells plated, plate number, and cell collection, starvation, differentiation, and RNA extraction date are highly correlated with the treatment and could act as confounders of the treatment effect. To account for this, for liver cells we matched treated and untreated samples by the number of cells plated, collection and starvation date, and plate number (for most but not all treatments, see Data S1). Because of this matching, we could not correct our analyses for RNA extraction date because it was collinear with treatment status within each treated-untreated pair of samples. For fat and muscle, we matched treated and untreated samples by differentiation date and, within that, by RNA extraction date, cell collection, and starvation date, and plate number (for most but not all treatments, see Data S1). To adjust for all other variables, we include them in the model when testing for differential expression by treatment.
Principal-component analysis
To identify gene expression outliers, we run principal-component analysis (PCA) within cell lines (Figure S3). Prior to applying PCA, we variance stabilized and log2-transformed the expression of each gene within cell lines by using the R package DESeq2.38 Then, we centered and scaled each gene to have zero mean and unit variance. We also applied PCA to expression data corrected for all major components of expression variability, as defined in the previous section. After removing the outlier sample mentioned above and after we regress out all major components of expression variability, we do not see any outliers based on the two first principal components.
Differential expression analyses
We characterize transcriptional responses to each perturbation in each cell line by using the negative binomial models implemented in the R package DESeq2,38 adjusting for major technical components of expression variability identified in the last section. To account for multiple testing across cell lines, perturbations, and genes, we use the hierarchical error control strategies implemented in the R package TreeBH39 with cell line, genes, and treatments in levels 1, 2, and 3, respectively. This hierarchical procedure adjusts for all the tests performed and allows us to make statements about the differential expression at the gene, gene-perturbation, and gene-perturbation-cell-line level. We call a gene perturbation specific within a cell line if the gene is differentially expressed (DE) in that specific perturbation but not in any other perturbation in that cell line (false discovery rate [FDR] < 5% at each level). A gene is assumed perturbation and cell line specific if the gene is DE in that specific perturbation but not in any other perturbation in that or the other cell lines or that specific perturbation in the other cell lines (FDR < 5% at each level). To assess agreement between our DE results and external studies listed in Table S1, we extracted lists of DE genes from each study and computed the proportion of those DE genes that validated in our study either by using the p1 statistic40 (when the list was large enough to allow this) or by computing the proportion of those DE genes that were also DE in our study at 5% FDR.
Correlation and hierarchical clustering of the transcriptional response to perturbations
We computed correlation and performed hierarchical cluster analysis of the transcriptional response to perturbations in each cell line by using the test statistics from the DE analyses for all genes that were significant (FDR < 5%) in at least one perturbation and cell line. We used the R package corrplot41 to get a graphical display of the correlation matrix and hierarchical clustering of the perturbations.
Enrichment analyses for biological pathways
We performed over-representation analysis42 by using the R package clusterProfiler43 and pathways from the ConsensusPathDB database.44 We adjust for multiple testing within each perturbation and cell line by using the Benjamini-Hochberg45 procedure.
Linkage disequilibrium score regression analysis
We downloaded the baseline model linkage disequilibrium (LD) scores, regression weights, and allele frequencies from Kundaje et al.3 Annotations for each perturbation and cell line were built with the pipeline described on the LD score regression wiki and according to Finucane et al.9 Specifically, for each of the 63 combinations of 21 perturbations and three cell lines, we add 100 kb windows (default LDSCreg threshold) on either side of the transcribed region of each DE gene in that combination to construct a genome annotation corresponding to that perturbation-cell-line combination. Due to its unusual genetic architecture and LD pattern, we excluded the human leukocyte antigen (HLA) region from all analyses. Z scores for the significance of the estimated total heritability for each trait were computed as h2/se(h2), where h2 and se(h2) are the SNP-based heritability estimated and standard errors from LDSCreg. Z scores and p values for the significance of the partitioned and conditional heritability for each trait-perturbation-cell-type combination were obtained via the option --h2-cts flag. We adjust for multiple testing within each trait and cell line by using the Benjamini-Hochberg procedure.
Enrichment for diseases and traits in the GWAS Catalog
We downloaded the entire GWAS Catalog (v.1.0.2) with Experimental Factor Ontology (EFO) annotations, including the parent category of each trait, from the European Bioinformatics Institute (EBI) (see web resources). We assume the “MAPPED GENE”, i.e., the gene mapped to the strongest SNP as reported in the GWAS Catalog, is the GWAS gene. For the enrichment of groups of GWAS traits, i.e., EFO parent terms, we only keep the unique GWAS genes reported across all traits within each EFO parent term. We excluded all results annotated with “other measurement,” “other disease,” and “other trait” EFO parent terms as well as duplicated entries. For the enrichment of specific traits, we only test traits with at least 100 reported associated genes. To test for the significance of the enrichment, we used Fisher’s exact test. For each perturbation-cell-line combination, we use an equal number of non-DE genes matched for length and median gene expression by using the R package optmatch.46 We adjust for multiple testing across all (parent) traits, cell lines, and perturbations by using the Benjamini-Hochberg procedure.
Colocalization analysis of GWAS and eQTL effects and combination with DE signal
We performed colocalization analysis by using our custom integration of the FINEMAP47 and eCAVIAR11 methods. For each GWAS and eQTL overlap (GWAS and eQTL p < 5 × 10−8 for at least one SNP in each), we narrowed our summary statistics to the set of SNPs tested for association with both the given GWAS trait and the given QTL trait and removed all sites containing less than ten SNPs after this filter. Using the full 1000 Genomes dataset from phase 3 as a reference population,48 we estimated LD between every pair of SNPs. We then ran FINEMAP independently on the GWAS and the eQTL summary statistics to obtain posterior probabilities of causality for each of the remaining SNPs. Because the canonical colocalization posterior probability (CLPP) score described in the eCAVIAR method is highly conservative in regions with densely profiled, high-LD SNPs, we use the following LD-modified CLPP score, CLPPmod = gi ej LDij, where N is the total number of variants at the locus, gi is the probability that the ith variant is the causal variant for the GWAS trait, ej is the probability that the jth variant is the causal variant for the eQTL trait, and LDij is the estimated LD (r2) between the ith and the jth variant in a reference population (we use 1000 Genomes Phase 3 data here). CLPPmod represents the sum of causal probabilities across all pairs of GWAS-eQTL variants at the locus, and each pair’s contribution to the final score is weighted by the LD between these two variants. Like the original CLPP score, CLPPmod takes values between 0 and 1, and high values indicate higher colocalization probabilities. Subsequent visual inspection of juxtaposed GWAS and eQTL LocusCompare plots at high- and low-CLPPmod-score loci confirmed that our LD-modified CLPP score detects true colocalized loci but without disproportionately penalizing high-LD loci.
To test whether the genes DE in at least one of our perturbations and cell lines are enriched for candidate causal IR genes for at least one IR-related trait and GTEx tissue, we used Fisher’s exact test. Candidate causal IR genes, denoted as “High P(Causal),” are defined as genes with CLPPmod above 40%, which corresponds to the 80th CLPPmod percentile. To test for the significance of the difference in median CLPPmod between DE and non-DE genes for each combination of perturbation and cell line, we used the two-sample Wilcoxon rank-sum test. We adjust for multiple testing by using the Benjamini-Hochberg procedure.
Results
Transcriptome map of 21 perturbations across human skeletal muscle, fat, and liver cell lines
We generated a transcriptome map of multiple chemical and environmental perturbations in well-established human skeletal muscle, fat, and liver cell lines (n = 234 samples). Specifically, we studied 21 environmental perturbations covering multiple aspects of glucose and insulin metabolism, inflammation, and fatty acid metabolism and including both low-density lipoprotein (LDL)-lowering and anti-diabetic drugs (Figure 1 and Data S1). For each perturbation and cell line and matched controls, we conducted assays in triplicate and applied differential expression analysis. We observed that most perturbations induced broad gene expression changes in at least one cell line at FDR < 5% (Figures 1A and S4A, Data S2, S3, and S4). Several perturbations induced broad changes across all cell lines; for example, insulin and IGF1 altered the gene expression of 1,500–2,000 genes in each cell line. Other perturbations had broad changes only in specific cell lines. For example, IL-6, lauroyl-l-carnitine, and glucose had more pronounced effects in fat, muscle, and liver, respectively, impacting the expression of 3,161, 2,051, and 2,724 genes. A limited number of studies have examined the effect of some of the perturbations in cell lines similar to the ones considered here. Despite several differences in study design, e.g., exposure time or concentration of use, we see a high agreement (56.48%–89.69%) in the list of DE genes (Table S1).
Despite the broad effects for each perturbation, multiple DE genes showed perturbation-specific effects within each cell line, highlighting a unique molecular response to each perturbation. We observed 1,883 genes in muscle, 1,813 genes in fat, and 2,231 genes in the liver altered by only a single perturbation in their respective cell lines (Figure 1B and Data S5). The largest proportions of perturbation-specific DE genes were found in glucose-stimulated liver cell lines and TGF-β1-stimulated fat cell lines. For these perturbations, 32.6% and 26.4% of DE genes were not altered by any of the other 20 perturbations in the same cell line (Figure 1C). By further stratifying across these cell lines, we identified 627, 742, and 808 genes that were both perturbation- and cell line-specific DE genes in muscle, fat, and liver (FDR < 5%; Figure S4B and Data S5). Glucose-stimulated liver cells also provided the largest amount of perturbation- and cell-line-specific DE genes; 9.8% of DE genes were not altered by any of the other 20 perturbations in any cell line or by glucose stimulation in fat or muscle.
To identify the relationships between perturbations on the basis of their overall transcriptional responses, we assessed the correlation of DE genes between each pair of perturbations within the same cell line (Figures 1D). The correlation of the effect of some perturbations was similar across cell lines, e.g., the effects of insulin and IGF1 were positively correlated in all three cell lines, i.e., Spearman’s r = 0.88, 0.76, and 0.71 in muscle, fat, and liver, respectively. The relationship of other perturbations, however, was dependent on the cellular context, e.g., while the effects of glucose and wortmannin were moderately correlated in fat (Spearman’s r = −0.63), their correlation in muscle and liver was low (Spearman’s r = 0.02 and 0.2, respectively).
To explore the shared and specific pathways altered by each perturbation, we performed enrichment analysis of DE genes in annotated pathways from ConsensusPathDB44 (Data S6, S7, and S8). Our analysis highlighted multiple shared pathways across perturbations and cell lines related to PI3K-AKT-mTOR, MAPK, adipogenesis, and TGF-β signaling (Figure S4C). We also observed several differences in pathway enrichments; for example, pathways related to FOXA2 and FOXA3 transcription factor networks had greater enrichment across several perturbations in the liver than in muscle and fat, transcriptional regulation by RUNX2 had greater enrichment in muscle than in liver and fat, and chromatin organization and remodeling pathways had greater enrichment in fat than in liver and muscle. In addition, for genes affected by multiple perturbations, we saw strong enrichment pathways related to insulin signaling and resistance.
Combined, our concurrent assessment of multiple metabolically relevant perturbations across cell lines highlights the relationships between complex cell-specific molecular mechanisms and provides a genome-wide map of genes and signaling pathways with potential environmental contributions to complex disease susceptibility.
Prioritizing complex disease-relevant environmental perturbations
To measure the relevance of diverse environmental perturbations in complex diseases, we analyzed our transcriptome data together with GWAS summary statistics for 31 diseases and complex traits broadly related to multiple cardiometabolic, psychiatric, autoimmune, and reproductive traits, as well as hematological measurements (Figures 2 and S5; Data S9). We hypothesized that environmental perturbations impact disease through the same genes that confer susceptibility to the trait. To this end, for each of the 21 perturbations across the three cell lines, we used stratified LD score regression9 (LDSCreg) to test whether disease heritability, i.e., the proportion of phenotypic variance determined by genotypic variance, is enriched in regions surrounding DE genes for that perturbation and cell line, adjusting for heritability explained by a baseline model of genetic architecture9 and by regions surrounding genes expressed in the specific cell line.
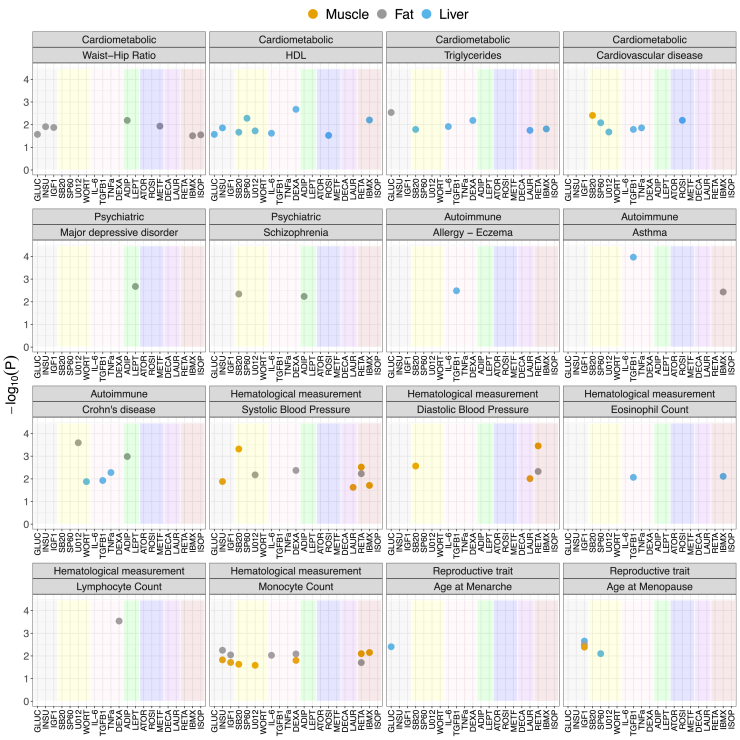
Figure 2.
Prioritizing complex disease-relevant environmental perturbations via heritability enrichment analysis
Heritability enrichment results for each complex trait. Each point represents a perturbation-cell-line combination that passes the FDR < 10% cut-off. The y axis represents the −log10(p value) of heritability enrichment, the x axis indicates perturbation, the color of the point indicates cell line, and the shading color within each panel indicates the perturbation category from Figure 1A. Numerical results are reported in Data S9.
For 26 of the 31 traits tested, the SNP-based heritability estimate was sufficiently large to partition reliably with LDSCreg, i.e., heritability Z score ≥ 7 (Data S9). In 19 of these traits, at least one perturbation in at least one cell line was enriched for heritability (FDR < 10%; Figure 2). Several of the enrichments recapitulate important known biology. For example, among cardiometabolic traits, high-density lipoprotein (HDL) and triglyceride levels were enriched for dexamethasone (p = 2.10 × 10−3 and p = 6.53 × 10−3), a corticosteroid known to induce dyslipidemia,49,50 and cardiovascular disease was enriched for rosiglitazone (p = 6.44 × 10−3), an antidiabetic drug shown to increase risk of cardiovascular disease.51 In addition, these enrichments were often manifested through a single specific relevant cell line. For example, waist-hip ratio (WHR) heritability was enriched for genes whose expression is modified by perturbations in fat, while triglyceride and HDL level heritability were enriched for genes whose expression is modified by perturbations in the liver.
Several notable examples were also observed for other tested traits. For psychiatric disorders, leptin, a hormone produced and secreted by white adipose tissue that is associated with antidepressant-like actions,28,52,53 was enriched for heritability of major depressive disorder (MIM: 608516) via its effect in fat cell lines (p = 2.11 × 10−3). In addition, adiponectin, plasma levels of which appear to be altered in neurological disorders with metabolic and inflammatory components,54, 55, 56 was enriched for heritability of schizophrenia (MIM: 181500) (p = 5.81 × 10−3). For tested autoimmune diseases, TGF-β1, an immune-suppressive cytokine dysregulated in the intestines of inflammatory bowel disease (MIM: 266600) affected individuals,57 was enriched for heritability of Crohn disease (p = 1.18 × 10−2), as well as heritability of allergy (MIM: 607154), eczema (MIM: 603165), and asthma (MIM: 600807)27,58, 59, 60 (p = 1.08 × 10−4), three diseases with shared genetic origin.61 Several perturbations were also enriched for the heritability of hematological measurements; for example, dexamethasone, a synthetic glucocorticoid known to deplete peripheral blood lymphocytes and impact immune response,62 was enriched for heritability of lymphocyte count (p = 2.88 × 10−4). Lastly, for reproductive traits, glucose was enriched for heritability of age at menarche (MIM: 610873)—older age at menarche is associated with reduced risk of glucose metabolism disorder63—while IGF1, whose serum levels rapidly decrease after menopause,64 was enriched for heritability of age at menopause (MIM: 300488) (p = 2.21 × 10−3).
Identifying environmental perturbations impacting GWAS-significant loci
Beyond the broad polygenic impact of the tested perturbations and to analyze a larger number of traits, we sought to prioritize the subset of perturbations that were enriched for impact on GWAS-significant loci in specific complex diseases. We tested for enrichment of DE genes for cis-SNPs associated with diseases and phenotypes in the GWAS Catalog.65 Because many traits had a small number of associations, we first tested for enrichment within groups of similar traits, as defined in the GWAS Catalog (Figure 3, Data S10).
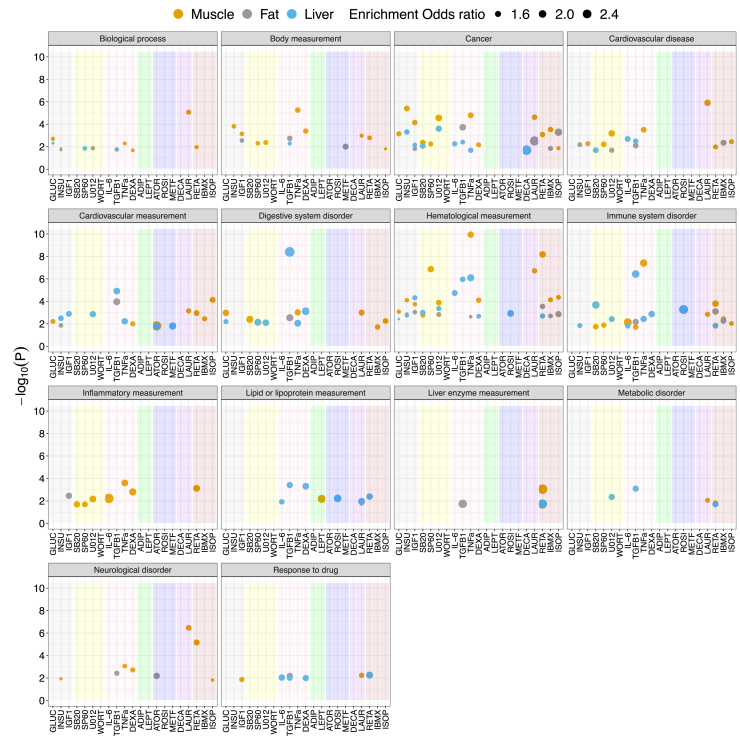
Figure 3.
Identifying environmental perturbations impacting significant GWAS loci
GWAS enrichment results for each group of complex traits from the GWAS Catalog. Each point represents a perturbation-cell-line combination that passes the FDR < 10% cut-off; the color of the point indicates the cell line and the shading color within each panel indicates the perturbation category from Figure 1A. The y axis represents the −log10(p value) of Fisher’s exact test and the size indicates the odds ratio for the enrichment of GWAS hits of each group of traits from the GWAS Catalog. Numerical results are reported in Data S10. Results for specific traits, rather than groups of traits, are displayed in Figure S5.
We observed significant enrichment for at least one perturbation and cell line across all 14 groups of complex diseases and traits tested (FDR < 10%). For example, genes responsive to the effect of rosiglitazone, an insulin sensitizer known to affect plasma lipid levels,66 were enriched within GWAS-significant hits for lipid or lipoprotein measurements (odds ratio [OR] = 2.00 and p = 5.98 × 10−3). In addition, genes responsive to the effect of retinoic acid, a metabolite of vitamin A that is synthesized in the liver and whose signaling dysregulation contributes to hepatic disease,67 were enriched within GWAS hits for liver enzyme measurements (ORMuscle = 2.71 and pMuscle = 9.06 × 10−4; ORLiver = 2.60 and pLiver = 1.8 × 10−2). Moreover, atorvastatin and metformin, two perturbations with highly correlated DE signals (Figure 1D) known to reduce cardiovascular morbidity,68, 69, 70, 71, 72 were both enriched within GWAS hits for cardiovascular measurements (ORATOR-Liver = 1.75 and pATOR-Liver = 1.58 × 10−2; ORATOR-Muscle = 2.53 and pATOR-Muscle = 1.52 × 10−2; ORMETF-Liver = 1.86 and pMETF-Liver = 1.56 × 10−2). In line with the LDSC regression-based enrichment for Crohn disease, we observed that genes responsive to TGF-β1 were enriched within GWAS-significant hits for digestive system disorders (OR = 2.7 and p = 3.96 × 10−9).
More generally, we observed that GWAS hits for immune system disorders or inflammatory measurements were enriched in genes responsive to the effect of inflammatory perturbations, e.g., ORTNFa = 1.92 and 1.77 and pTNFa = 3.84 × 10−8 and 2.54 × 10−8 for immune system disorders and inflammatory measurements, respectively. Neurological disorders were also enriched for inflammatory perturbations, although to a lesser extent (e.g., ORTNFa = 1.35 and pTNFa = 8.60 × 10−4). Associations with lipid or lipoprotein measurements and drug metabolism traits were enriched in genes responsive to several perturbations via the liver, where most drug metabolism occurs,73 and associations with body measurements were enriched via muscle.
For traits with a large number of GWAS hits, i.e., traits with at least 100 reported associated loci, we tested enrichments directly (Figure S5). In 14 of the 152 complex diseases and traits tested, we observed significant enrichment for at least one perturbation and cell line (FDR < 10%). For example, genes in muscle cells that were responsive to the effect of isoprenaline, a beta-adrenergic agonist with effects on cardiac muscle,74 were enriched within GWAS-significant hits for cardiovascular disease (OR = 2.24 and p = 2.56 × 10−4). In addition, consistent with the LDSCreg-based enrichment of dexamethasone for HDL heritability in the liver, genes responsive to dexamethasone in the liver were enriched within GWAS-significant associations for total cholesterol levels (OR = 3.08 and p = 1.94 × 10−4). Lastly, genes responsive to IGF1 in the liver were enriched within significant associations for birth weight (OR = 3.86 and p = 5.48 × 10−5), consistent with prior observations of negative correlation between IGF1 levels and birth weight.75,76
Environmental perturbations harbor causal genes and help inform their functions
A major challenge with GWAS data in isolation is identifying causal disease genes. Here, we assessed whether combining GWASs with relevant environmental perturbations helped to identify or reinforce causal disease genes and to inform on their molecular functions. Because many of our perturbations were related to cardiometabolic traits, including IR, obesity, and type 2 diabetes (T2D [MIM: 125853]), we tested whether genes affected by our panel of perturbations harbored candidate causal genes underlying loci for seven cardiometabolic traits. To assess this, we integrated our perturbation data with results from genomic colocalization analyses of GWAS loci for these seven traits and GTEx eQTLs in visceral and subcutaneous adipose, skeletal muscle, and liver tissues.5 We observed that genes with a transcriptional response to at least one of our environmental perturbations are enriched among the candidate causal genes, i.e., genes with high LD-modified posterior colocalization probability (CLPPmod), for cardiometabolic traits (Figure 4A; OR = 1.40, Fisher’s exact test p value = 5.33 × 10−4). We next assessed whether DE genes for specific perturbation-cell-line combinations were more likely to be causal compared with non-DE genes (Figure 4B). Genes responsive to isoprenaline, SP600125 (a c-Jun N-terminal kinase inhibitor that plays an essential role in TLR-mediated inflammatory responses), and TNFa in fat had significantly higher median CLPPsmod (FDR < 10%) compared to non-DE genes (Wilcoxon test; pISOP = 8.7 × 10−4, pSP60 = 6.0 × 10−3, and pTNFa = 6.0 × 10−3).
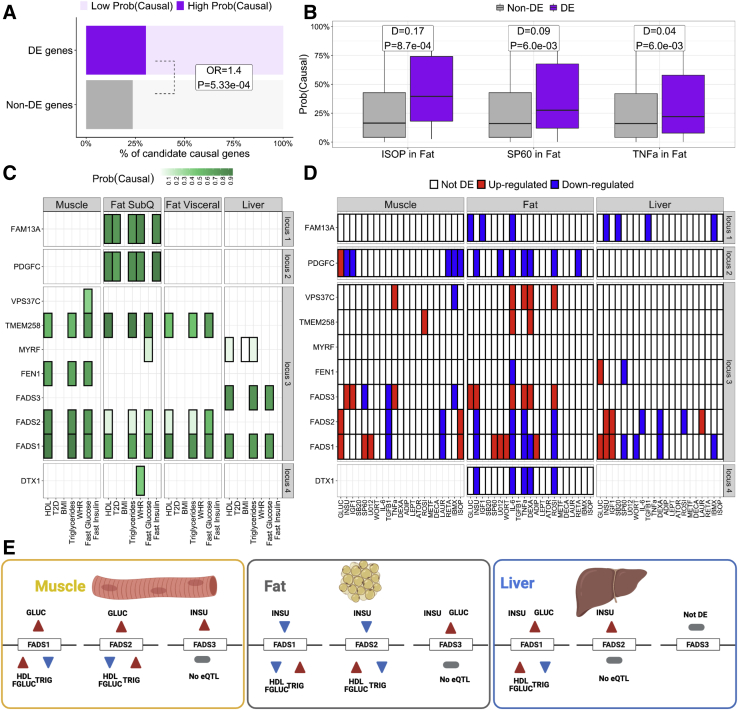
Figure 4.
Environmental perturbations can help inform the functionality of causal genes underlying cardiometabolic trait loci
(A) Percent of causal genes (High Prob(Causal)) underlying cardiometabolic traits loci that are DE (purple) or not DE (gray) in at least one perturbation and cell line. OR/P: odds ratio and Fisher’s exact test p value for the enrichment of DE genes among causal genes compared to non-DE genes.
(B) Perturbation-cell-line combinations with a significant (FDR < 10%) difference in median (D) CLPPmod between DE and non-DE genes, according to the two-samples Wilcoxon rank-sum test.
(C and D) Examples of loci for which intersecting the effects of perturbations (C) with the colocalization results (D) helps inform the functionality of candidate causal genes. The color indicates CLPPmod (C) or DE direction (D). White boxes with crosses indicate that the gene was not tested for colocalization or DE.
(E) Effect of glucose and insulin in the expression of the three FADS genes and the effect of the expression of these genes on HDL, fasting glucose (FGLUC), and triglycerides (TRIG), the three traits for which FADS genes colocalize. The color of the triangles indicates either the effect of the perturbation on the gene (red, upregulation; blue, downregulation) and the effect that up-/downregulation of the gene has on the phenotype (red, increased phenotype, blue, decreased phenotype). DE, differential expression; CLPPmod, LD-adjusted colocalization posterior probability; FDR, false discovery rate.
To explore how perturbation experiments can inform the function of candidate causal genes underlying cardiometabolic loci, we intersected the DE patterns in each cell line and perturbation with the colocalization patterns in the matched tissue. We illustrate four such examples (Figures 4C–4E): three loci in which a single gene showed high CLPPmod and one locus with more complex colocalization patterns (five out of seven genes in the locus showing high CLPPmod).
Results from the colocalization analysis associated FAM13A (MIM: 613299) genetic variants in subcutaneous fat with several traits of interest, i.e., HDL, T2D, triglycerides, WHR, and fasting insulin (Figure 4C, locus 1). We recently described the role of FAM13A in adipocyte differentiation and the contribution to body fat distribution.77 The DE patterns of FAM13A in our perturbation experiment (Figure 4D, locus 1) not only reinforce the role of FAM13A in adipose tissue but also suggest an additional metabolic function in the liver not captured by the colocalization results. The role of FAM13A in the regulation of hepatic glucose and lipid metabolism was recently confirmed by Lin et al.78 Another candidate gene, PDGFC (MIM: 608452), shows an identical colocalization pattern to FAM13A (Figure 4C, locus 2) and the perturbation data also support its importance in the adipose tissue (Figure 4D, locus 2). However, the perturbation data identify an additional role of PDGFC in skeletal muscle in contrast with the role of FAM13A in the liver.
Another complementary example is illustrated by the colocalization for DTX1 (MIM: 602582), which is specifically associated with WHR and subcutaneous fat (Figure 4C, locus 4) and whose expression is regulated by insulin, IL-6, TNF-a, dexamethasone, and rosiglitazone in mature human adipocytes (Figure 4D, locus 4).
Finally, genetic variants in the FADS locus have been associated with HDL cholesterol, triglyceride levels, fasting glucose, and T2D79, 80, 81 and our colocalization analysis was consistent with these observations (Figure 4C, locus 3). However, the high amount of LD, the gene density, and the pleiotropy of FADS genes have challenged the dissection of individual gene effects. Particularly informative is the case of FADS1 (MIM: 606148), FADS2 (MIM: 606149), and FADS3 (MIM: 606150), for which the DE patterns for glucose and insulin (Figure 4D, locus 3) point, among others, to a fine-tuned cell-line- and perturbation-specific regulation of the FADS locus in the context of metabolic homeostasis (Figure 4E).
Together these results highlight the importance of perturbation experiments to contextualize GWAS associations and results from genomic colocalization analyses.
Discussion
We have profiled transcriptional responses to multiple environmental perturbations to identify disease-relevant perturbations modulating genetic risk for complex traits and to inform the functionality of causal genes. By combining gene expression data with GWAS summary statistics of complex traits, we show that the heritability of multiple complex traits is enriched in regions surrounding genes responsive to particular sets of perturbation-cell-line combinations. We confirmed several well-established associations, e.g., the role of TGF-β1 on asthma, and provided additional evidence for recent and less-well-understood associations, e.g., the role of leptin on major depressive disorder. In addition, beyond the broad polygenic impacts of the tested perturbations, we were able to prioritize the subset of perturbations that are enriched for their impact on GWAS-significant loci in specific groups of complex diseases. We observed that environmentally responsive genes are enriched for cis-SNPs associated with a broad spectrum of diseases and phenotypes from the GWAS Catalog. Further, by integrating gene expression data with information from genomic colocalization studies, we showed that environmentally responsive genes are enriched for candidate causal genes for cardiometabolic traits and that the effects of these perturbations in the relevant tissues further suggest context-specific molecular mechanisms of GWAS hits for cardiometabolic traits.
2Our approach interrogated multiple cell lines and perturbations, but comparable applications will be limited by the specific cell lines and environmental perturbations assayed and the exposure time and concentrations selected. Here, we chose the concentrations of use on the basis of an extensive literature search and the exposure time to reduce the likelihood of secondary regulatory mechanisms. Our data demonstrate that, for most perturbations, there is a notable transcriptional response after a 2 h stimulation window. This suggests that longer stimulation times should be avoided, e.g., the typical 24 h stimulation period, if the purpose of the experimental setting is to describe genes that are likely to be directly regulated by the perturbations assayed. Further, the use of cell lines provides the opportunity for conducting well-controlled perturbation experiments; however, it is unknown the degree to which all findings would generalize to primary cells. Perturbation experiments on well-defined primary cells from multiple individuals might better mimic the disease-associated environment, but, until such experiments become more feasible, measuring expression in cell lines with tightly controlled cellular environments provides a more tractable setting to study gene-environment interactions.20,26 For some of the diseases we considered, the studied cell lines might not represent the cell type or tissue through which disease is manifested. However, because we observed that cell lines can share transcriptional responses (Figure 1D), our study design has shown that we can identify important perturbations without the causal cell type being examined.
To perform heritability enrichment analyses, we build annotations for each perturbation by using a fixed (100 kb size) window around each DE gene, which might lead to wide estimates of regulatory territories and decrease power to fine-map disease-relevant contexts. As perturbed chromatin immunoprecipitation sequencing (ChIP-seq) experiments become available for these perturbations and cell lines they can help further narrow down the regulatory regions for these environments. In addition, because fine-mapping summary statistics are not available for many of the traits deposited in the GWAS Catalog, we used the closest genes as proxy of the causal gene. While the closest gene is the most likely causal gene in only about 50% of examined loci, we do not expect this to disproportionately affect DE versus non-DE genes and upwardly bias our enrichment results. Furthermore, because most perturbations affect a small number of genes, the LDSC regression analysis for the enrichment of DE genes for trait heritability is based on partitions that cover a very small part of the genome. All these factors can substantially reduce power to identify disease-relevant contexts, and we thus chose to use a 10% FDR threshold and highlight several positive control examples. We illustrate the impact of the FDR threshold on the enrichment for GWAS-significant loci in Figures S6 and S7. Last, our genomic colocalization analyses are based on eQTLs mapped in GTEx tissues from healthy donors and might miss the effect of regulatory variants that manifest uniquely in the presence of the perturbation environment. Although we do not expect this to upwardly bias the enrichment of DE genes for likely causal genes, it might mask the effects of eQTLs for perturbations that are less likely to be present in healthy donors, e.g., dexamethasone, and reduce the power to detect enrichment of DE genes in candidate causal genes responsive to these perturbations.
In conclusion, we demonstrate the advantages of large-scale characterization of transcriptional changes in diversely stimulated and pathologically relevant cells to identify disease-relevant perturbations that modulate genetic risk for complex traits. We also provide a broad resource of the dynamic transcriptional landscape in metabolic tissues. To our knowledge, this is the largest and most complete study of transcriptional effects of metabolically relevant perturbations in human fat, liver, and skeletal muscle cell lines. In addition, we show that integrating GWAS and eQTL results with perturbation experiments can inform the function of candidate causal genes and prioritize genes and environmental stimuli for follow-up experiments. Combined, this work demonstrates how integrating differential expression, eQTL, and GWAS data can inform genetic and environmental components of complex disease mechanisms.
Acknowledgments
We thank Erik Ingelsson, Noah Zaitlen, Paivi Pajukanta, and Aldon J. Lusis for helpful comments and suggestions that helped to improve the quality of our manuscript. J.K. is funded by NIH R01 DK116750, R01 DK120565, R01 DK106236, R01 DK107437, P30DK116074, and ADA 1-19-JDF-108. S.B.M. is supported by NIH grants R01AG066490, U01HG009431, R01HL142015, R01HG008150, and U01HG009080. T.Q. is supported by NIH grants R01 HL139478, R01 HL145708, R01 HL134817, R01 HL151535, and R01 HL156846 and a Human Cell Atlas grant from the Chan Zuckerberg Foundation. M.G.D. is supported by the National Science Foundation Graduate Research Fellowship.
Declaration of interests
S.B.M. is on the SAB for Myome.
Published: September 27, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.ajhg.2021.08.014.
Contributor Information
Brunilda Balliu, Email: bballiu@ucla.edu.
Joshua W. Knowles, Email: knowlej@stanford.edu.
Data and code availability
The data discussed in this publication have been deposited in NCBI’s Gene Expression Omnibus (GEO) and are accessible through GEO series accession number GEO: GSE179347. The code and processed data used for these analyses are available at https://github.com/BrunildaBalliu/PerturbationGxE.
Web resources
Supplemental information
The first sheet lists the perturbations used, their abbreviations and concentration, and the category in which they belong, e.g., adipokine. The second sheet lists all the experimental parameters, e.g., medium used, RNA extraction date, etc. The third sheet lists the control samples used for DE analysis for each perturbation.
Columns indicate the cell, perturbation, Ensembl gene ID, HUGO gene name, the average expression of the gene in the control cells (baseMean), the log2 fold change estimate (log2FoldChange, negative = downregulated by perturbation) and standard error (lfcSE) in perturbed cells, the DESeq2 test statistic (test_stat) and p value. The last column (significant_5prcFDR) indicates if the gene was differentially expressed (FDR<5%) in the perturbed cells in the particular perturbation and cell after adjusting for the number of tests performed across all perturbations and cell lines.
Columns indicate the cell, perturbation, Ensembl gene ID, HUGO gene name, the average expression of the gene in the control cells (baseMean), the log2 fold change estimate (log2FoldChange, negative = downregulated by perturbation) and standard error (lfcSE) in perturbed cells, the DESeq2 test statistic (test_stat) and p value. The last column (significant_5prcFDR) indicates if the gene was differentially expressed (FDR<5%) in the perturbed cells in the particular perturbation and cell after adjusting for the number of tests performed across all perturbations and cell lines.
Columns indicate the cell, perturbation, Ensembl gene ID, HUGO gene name, the average expression of the gene in the control cells (baseMean), the log2 fold change estimate (log2FoldChange, negative = downregulated by perturbation) and standard error (lfcSE) in perturbed cells, the DESeq2 test statistic (test_stat) and p value. The last column (significant_5prcFDR) indicates if the gene was differentially expressed (FDR<5%) in the perturbed cells in the particular perturbation and cell after adjusting for the number of tests performed across all perturbations and cell lines.
Provided as a separate excel file, one sheet for each cell line, with the first column indicating the HUGO gene name and the next 21 columns being an indicator variable for the gene being DE (0 = not DE, 1 = DE upregulated after perturbation, and −1 = DE downregulated after perturbation) between samples treated with perturbation and untreated samples. The last column (Nr_Pert_DE) shows the number of perturbations in which a gene is DE. If Nr_Pert_DE = 1 and ADIP = +1/-1 but all other columns are zero, then the gene adiponectin-specific, i.e., it is only DE in adiponectin but no other perturbations in that cell line. The last two sheets provide DE info summary across perturbations and cells. In the first sheet (deGene_by_perturbation), each cell indicates if the gene is differentially expressed in a particular perturbation in at least one of the three cell lines (FDR < 5%). In the second sheet (deGene_by_cell), each cell indicates if the gene is differentially expressed for the cell line in at least one of the perturbations (FDR < 5%).
The columns indicate pathway name, gene ratio, BgRatio, p value, BH-adjusted p value, and q-value from the over-representation analysis as well as the geneID and number of DE genes in the pathway.
The columns indicate pathway name, gene ratio, BgRatio, p value, BH-adjusted p value, and q-value from the over-representation analysis as well as the geneID and number of DE genes in the pathway.
The columns indicate pathway name, gene ratio, BgRatio, p value, BH-adjusted p value, and q-value from the over-representation analysis as well as the geneID and number of DE genes in the pathway.
Provided as a separate excel file; the first sheet lists a summary of the studies used and LDSCreg-based estimates of their SNP-based heritability (h2) and h2 z-score. The second sheet lists detailed LDSCreg results for each triplet of trait, cell line, and perturbation.
Provided as a separate excel file; the first sheet list results from the analysis of groups of related traits (Parent_Trait), as defined in the GWAS Catalog, while the second list results for specific traits. Each row corresponds to a trait or group of traits - cell line - perturbation combination. The columns indicate the specific trait (second sheet only), parent trait, cell line, and perturbation combination tested as well as the OR of enrichment with the 95% CI, the Fisher’s exact test p value for the significance of the enrichment as well as the BH-adjusted p value. The last four columns contain the number of genes that were “neither DE nor GWAS genes,” “DE but not GWAS genes,” “GWAS but not DE genes,” and “both GWAS and DE genes.”
References
- 1.MacArthur J., Bowler E., Cerezo M., Gil L., Hall P., Hastings E., Junkins H., McMahon A., Milano A., Morales J. The new NHGRI-EBI Catalog of published genome-wide association studies (GWAS Catalog) Nucleic Acids Res. 2017;45(D1):D896–D901. doi: 10.1093/nar/gkw1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Maurano M.T., Humbert R., Rynes E., Thurman R.E., Haugen E., Wang H., Reynolds A.P., Sandstrom R., Qu H., Brody J. Systematic localization of common disease-associated variation in regulatory DNA. Science. 2012;337:1190–1195. doi: 10.1126/science.1222794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kundaje A., Meuleman W., Ernst J., Bilenky M., Yen A., Heravi-Moussavi A., Kheradpour P., Zhang Z., Wang J., Ziller M.J. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–330. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moore J.E., Purcaro M.J., Pratt H.E., Epstein C.B., Shoresh N., Adrian J., Kawli T., Davis C.A., Dobin A., Kaul R. Expanded encyclopaedias of DNA elements in the human and mouse genomes. Nature. 2020;583:699–710. doi: 10.1038/s41586-020-2493-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.GTEx Consortium The GTEx Consortium atlas of genetic regulatory effects across human tissues. Science. 2020;369:1318–1330. doi: 10.1126/science.aaz1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boix C.A., James B.T., Park Y.P., Meuleman W., Kellis M. Regulatory genomic circuitry of human disease loci by integrative epigenomics. Nature. 2021;590:300–307. doi: 10.1038/s41586-020-03145-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gusev A., Lee S.H., Trynka G., Finucane H., Vilhjálmsson B.J., Xu H., Zang C., Ripke S., Bulik-Sullivan B., Stahl E. Partitioning heritability of regulatory and cell-type-specific variants across 11 common diseases. Am. J. Hum. Genet. 2014;95:535–552. doi: 10.1016/j.ajhg.2014.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Finucane H.K., Bulik-Sullivan B., Gusev A., Trynka G., Reshef Y., Loh P.R., Anttila V., Xu H., Zang C., Farh K. Partitioning heritability by functional annotation using genome-wide association summary statistics. Nat. Genet. 2015;47:1228–1235. doi: 10.1038/ng.3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Finucane H.K., Reshef Y.A., Anttila V., Slowikowski K., Gusev A., Byrnes A., Gazal S., Loh P.R., Lareau C., Shoresh N. Heritability enrichment of specifically expressed genes identifies disease-relevant tissues and cell types. Nat. Genet. 2018;50:621–629. doi: 10.1038/s41588-018-0081-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Giambartolomei C., Vukcevic D., Schadt E.E., Franke L., Hingorani A.D., Wallace C., Plagnol V. Bayesian test for colocalisation between pairs of genetic association studies using summary statistics. PLoS Genet. 2014;10:e1004383. doi: 10.1371/journal.pgen.1004383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hormozdiari F., van de Bunt M., Segrè A.V., Li X., Joo J.W.J., Bilow M., Sul J.H., Sankararaman S., Pasaniuc B., Eskin E. Colocalization of GWAS and eQTL Signals Detects Target Genes. Am. J. Hum. Genet. 2016;99:1245–1260. doi: 10.1016/j.ajhg.2016.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gusev A., Ko A., Shi H., Bhatia G., Chung W., Penninx B.W., Jansen R., de Geus E.J., Boomsma D.I., Wright F.A. Integrative approaches for large-scale transcriptome-wide association studies. Nat. Genet. 2016;48:245–252. doi: 10.1038/ng.3506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chun S., Casparino A., Patsopoulos N.A., Croteau-Chonka D.C., Raby B.A., De Jager P.L., Sunyaev S.R., Cotsapas C. Limited statistical evidence for shared genetic effects of eQTLs and autoimmune-disease-associated loci in three major immune-cell types. Nat. Genet. 2017;49:600–605. doi: 10.1038/ng.3795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Umans B.D., Battle A., Gilad Y. Where Are the Disease-Associated eQTLs? Trends Genet. 2021;37:109–124. doi: 10.1016/j.tig.2020.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maranville J.C., Luca F., Richards A.L., Wen X., Witonsky D.B., Baxter S., Stephens M., Di Rienzo A. Interactions between glucocorticoid treatment and cis-regulatory polymorphisms contribute to cellular response phenotypes. PLoS Genet. 2011;7:e1002162. doi: 10.1371/journal.pgen.1002162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee M.N., Ye C., Villani A.C., Raj T., Li W., Eisenhaure T.M., Imboywa S.H., Chipendo P.I., Ran F.A., Slowikowski K. Common genetic variants modulate pathogen-sensing responses in human dendritic cells. Science. 2014;343:1246980. doi: 10.1126/science.1246980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ye C.J., Feng T., Kwon H.K., Raj T., Wilson M.T., Asinovski N., McCabe C., Lee M.H., Frohlich I., Paik H.I. Intersection of population variation and autoimmunity genetics in human T cell activation. Science. 2014;345:1254665. doi: 10.1126/science.1254665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fairfax B.P., Humburg P., Makino S., Naranbhai V., Wong D., Lau E., Jostins L., Plant K., Andrews R., McGee C., Knight J.C. Innate immune activity conditions the effect of regulatory variants upon monocyte gene expression. Science. 2014;343:1246949. doi: 10.1126/science.1246949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Quach H., Rotival M., Pothlichet J., Loh Y.E., Dannemann M., Zidane N., Laval G., Patin E., Harmant C., Lopez M. Genetic Adaptation and Neandertal Admixture Shaped the Immune System of Human Populations. Cell. 2016;167:643–656.e17. doi: 10.1016/j.cell.2016.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moyerbrailean G.A., Richards A.L., Kurtz D., Kalita C.A., Davis G.O., Harvey C.T., Alazizi A., Watza D., Sorokin Y., Hauff N. High-throughput allele-specific expression across 250 environmental conditions. Genome Res. 2016;26:1627–1638. doi: 10.1101/gr.209759.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nédélec Y., Sanz J., Baharian G., Szpiech Z.A., Pacis A., Dumaine A., Grenier J.C., Freiman A., Sams A.J., Hebert S. Genetic Ancestry and Natural Selection Drive Population Differences in Immune Responses to Pathogens. Cell. 2016;167:657–669.e21. doi: 10.1016/j.cell.2016.09.025. [DOI] [PubMed] [Google Scholar]
- 22.Knowles D.A., Davis J.R., Edgington H., Raj A., Favé M.J., Zhu X., Potash J.B., Weissman M.M., Shi J., Levinson D.F. Allele-specific expression reveals interactions between genetic variation and environment. Nat. Methods. 2017;14:699–702. doi: 10.1038/nmeth.4298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim-Hellmuth S., Bechheim M., Pütz B., Mohammadi P., Nédélec Y., Giangreco N., Becker J., Kaiser V., Fricker N., Beier E. Genetic regulatory effects modified by immune activation contribute to autoimmune disease associations. Nat. Commun. 2017;8:266. doi: 10.1038/s41467-017-00366-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Calderon D., Nguyen M.L.T., Mezger A., Kathiria A., Müller F., Nguyen V., Lescano N., Wu B., Trombetta J., Ribado J.V. Landscape of stimulation-responsive chromatin across diverse human immune cells. Nat. Genet. 2019;51:1494–1505. doi: 10.1038/s41588-019-0505-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alasoo K., Rodrigues J., Danesh J., Freitag D.F., Paul D.S., Gaffney D.J. Genetic effects on promoter usage are highly context-specific and contribute to complex traits. eLife. 2019;8:e41673. doi: 10.7554/eLife.41673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Findley A.S., Monziani A., Richards A.L., Rhodes K., Ward M.C., Kalita S.A., Alazizi A., Pazokitoroudi A., Sankararaman S., Wen X. Functional dynamic genetic effects on gene regulation are specific to particular cell types and environmental conditions. eLife. 2021;10:e67077. doi: 10.7554/eLife.67077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li H., Romieu I., Wu H., Sienra-Monge J.J., Ramírez-Aguilar M., del Río-Navarro B.E., del Lara-Sánchez I.C., Kistner E.O., Gjessing H.K., London S.J. Genetic polymorphisms in transforming growth factor beta-1 (TGFB1) and childhood asthma and atopy. Hum. Genet. 2007;121:529–538. doi: 10.1007/s00439-007-0337-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ge T., Fan J., Yang W., Cui R., Li B. Leptin in depression: a potential therapeutic target. Cell Death Dis. 2018;9:1096. doi: 10.1038/s41419-018-1129-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rokach O., Ullrich N.D., Rausch M., Mouly V., Zhou H., Muntoni F., Zorzato F., Treves S. Establishment of a human skeletal muscle-derived cell line: biochemical, cellular and electrophysiological characterization. Biochem. J. 2013;455:169–177. doi: 10.1042/BJ20130698. [DOI] [PubMed] [Google Scholar]
- 30.Carcamo-Orive I., Henrion M.Y.R., Zhu K., Beckmann N.D., Cundiff P., Moein S., Zhang Z., Alamprese M., D’Souza S.L., Wabitsch M. Predictive network modeling in human induced pluripotent stem cells identifies key driver genes for insulin responsiveness. PLoS Comput. Biol. 2020;16:e1008491. doi: 10.1371/journal.pcbi.1008491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fischer-Posovszky P., Newell F.S., Wabitsch M., Tornqvist H.E. Human SGBS cells - a unique tool for studies of human fat cell biology. Obes. Facts. 2008;1:184–189. doi: 10.1159/000145784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Aden D.P., Fogel A., Plotkin S., Damjanov I., Knowles B.B. Controlled synthesis of HBsAg in a differentiated human liver carcinoma-derived cell line. Nature. 1979;282:615–616. doi: 10.1038/282615a0. [DOI] [PubMed] [Google Scholar]
- 33.Batista T.M., Garcia-Martin R., Cai W., Konishi M., O’Neill B.T., Sakaguchi M., Kim J.H., Jung D.Y., Kim J.K., Kahn C.R. Multi-dimensional Transcriptional Remodeling by Physiological Insulin In Vivo. Cell Rep. 2019;26:3429–3443.e3. doi: 10.1016/j.celrep.2019.02.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., 1000 Genome Project Data Processing Subgroup The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Anders S., Pyl P.T., Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hoffman G.E., Schadt E.E. variancePartition: interpreting drivers of variation in complex gene expression studies. BMC Bioinformatics. 2016;17:483. doi: 10.1186/s12859-016-1323-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bogomolov W., Peterson C.B., Benjamini Y., Sabatti C. Hypotheses on a tree: new error rates and testing strategies. Biometrika. 2020 doi: 10.1093/biomet/asaa086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Storey J.D., Tibshirani R. Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. USA. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wei T., Simko V. 2021. R package 'corrplot': Visualization of a Correlation Matrix. (Version 0.90)https://github.com/taiyun/corrplot [Google Scholar]
- 42.Boyle E.I., Weng S., Gollub J., Jin H., Botstein D., Cherry J.M., Sherlock G. GO:TermFinder--open source software for accessing Gene Ontology information and finding significantly enriched Gene Ontology terms associated with a list of genes. Bioinformatics. 2004;20:3710–3715. doi: 10.1093/bioinformatics/bth456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yu G., Wang L.-G., Han Y., He Q.-Y. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kamburov A., Wierling C., Lehrach H., Herwig R. ConsensusPathDB--a database for integrating human functional interaction networks. Nucleic Acids Res. 2009;37:D623–D628. doi: 10.1093/nar/gkn698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Benjamini Y., Heller R. Screening for partial conjunction hypotheses. Biometrics. 2008;64:1215–1222. doi: 10.1111/j.1541-0420.2007.00984.x. [DOI] [PubMed] [Google Scholar]
- 46.Hansen B.B., Fredrickson M., Buckner J., Errickson J., Rauh R., Solenberger P. 2018. optmatch: Functions for Optimal Matching.https://github.com/markmfredrickson/optmatch [Google Scholar]
- 47.Benner C., Spencer C.C., Havulinna A.S., Salomaa V., Ripatti S., Pirinen M. FINEMAP: efficient variable selection using summary data from genome-wide association studies. Bioinformatics. 2016;32:1493–1501. doi: 10.1093/bioinformatics/btw018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Auton A., Brooks L.D., Durbin R.M., Garrison E.P., Kang H.M., Korbel J.O., Marchini J.L., McCarthy S., McVean G.A., Abecasis G.R., 1000 Genomes Project Consortium A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Krupková M., Sedová L., Liska F., Krenová D., Kren V., Seda O. Pharmacogenetic interaction between dexamethasone and Cd36-deficient segment of spontaneously hypertensive rat chromosome 4 affects triacylglycerol and cholesterol distribution into lipoprotein fractions. Lipids Health Dis. 2010;9:38. doi: 10.1186/1476-511X-9-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Arab Dolatabadi A., Mahboubi M. A study of the influence of dexamethasone on lipid profile and enzyme lactate dehydrogenase. J. Med. Life. 2015;8:72–76. [PMC free article] [PubMed] [Google Scholar]
- 51.Wallach J.D., Wang K., Zhang A.D., Cheng D., Grossetta Nardini H.K., Lin H., Bracken M.B., Desai M., Krumholz H.M., Ross J.S. Updating insights into rosiglitazone and cardiovascular risk through shared data: individual patient and summary level meta-analyses. BMJ. 2020;368:l7078. doi: 10.1136/bmj.l7078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Westling S., Ahrén B., Träskman-Bendz L., Westrin A. Low CSF leptin in female suicide attempters with major depression. J. Affect. Disord. 2004;81:41–48. doi: 10.1016/j.jad.2003.07.002. [DOI] [PubMed] [Google Scholar]
- 53.Milaneschi Y., Lamers F., Bot M., Drent M.L., Penninx B.W.J.H. Leptin Dysregulation Is Specifically Associated With Major Depression With Atypical Features: Evidence for a Mechanism Connecting Obesity and Depression. Biol. Psychiatry. 2017;81:807–814. doi: 10.1016/j.biopsych.2015.10.023. [DOI] [PubMed] [Google Scholar]
- 54.Song X., Fan X., Song X., Zhang J., Zhang W., Li X., Gao J., Harrington A., Ziedonis D., Lv L. Elevated levels of adiponectin and other cytokines in drug naïve, first episode schizophrenia patients with normal weight. Schizophr. Res. 2013;150:269–273. doi: 10.1016/j.schres.2013.07.044. [DOI] [PubMed] [Google Scholar]
- 55.Bartoli F., Lax A., Crocamo C., Clerici M., Carrà G. Plasma adiponectin levels in schizophrenia and role of second-generation antipsychotics: a meta-analysis. Psychoneuroendocrinology. 2015;56:179–189. doi: 10.1016/j.psyneuen.2015.03.012. [DOI] [PubMed] [Google Scholar]
- 56.Bloemer J., Pinky P.D., Govindarajulu M., Hong H., Judd R., Amin R.H., Moore T., Dhanasekaran M., Reed M.N., Suppiramaniam V. Role of Adiponectin in Central Nervous System Disorders. Neural Plast. 2018;2018:4593530. doi: 10.1155/2018/4593530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ihara S., Hirata Y., Koike K. TGF-β in inflammatory bowel disease: a key regulator of immune cells, epithelium, and the intestinal microbiota. J. Gastroenterol. 2017;52:777–787. doi: 10.1007/s00535-017-1350-1. [DOI] [PubMed] [Google Scholar]
- 58.Wortley M.A., Bonvini S.J. Transforming Growth Factor-β1: A Novel Cause of Resistance to Bronchodilators in Asthma? Am. J. Respir. Cell Mol. Biol. 2019;61:134–135. doi: 10.1165/rcmb.2019-0020ED. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Duvernelle C., Freund V., Frossard N. Transforming growth factor-beta and its role in asthma. Pulm. Pharmacol. Ther. 2003;16:181–196. doi: 10.1016/S1094-5539(03)00051-8. [DOI] [PubMed] [Google Scholar]
- 60.Yao Y.S., Chang W.W., He L.P., Jin Y.L., Li C.P. An updated meta-analysis of transforming growth factor-β1 gene: Three well-characterized polymorphisms with asthma. Hum. Immunol. 2016;77:1291–1299. doi: 10.1016/j.humimm.2016.09.011. [DOI] [PubMed] [Google Scholar]
- 61.Ferreira M.A., Vonk J.M., Baurecht H., Marenholz I., Tian C., Hoffman J.D., Helmer Q., Tillander A., Ullemar V., van Dongen J. Shared genetic origin of asthma, hay fever and eczema elucidates allergic disease biology. Nat. Genet. 2017;49:1752–1757. doi: 10.1038/ng.3985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Parrillo J.E., Fauci A.S. Mechanisms of corticosteroid action on lymphocyte subpopulations. III. Differential effects of dexamethasone administration on subpopulations of effector cells mediating cellular cytotoxicity in man. Clin. Exp. Immunol. 1978;31:116–125. [PMC free article] [PubMed] [Google Scholar]
- 63.Ren Y., Zou H., Zhang D., Han C., Hu D. Relationship between age at menarche and risk of glucose metabolism disorder: a systematic review and dose-response meta-analysis. Menopause. 2020;27:818–826. doi: 10.1097/GME.0000000000001529. [DOI] [PubMed] [Google Scholar]
- 64.Nasu M., Sugimoto T., Chihara M., Hiraumi M., Kurimoto F., Chihara K. Effect of natural menopause on serum levels of IGF-I and IGF-binding proteins: relationship with bone mineral density and lipid metabolism in perimenopausal women. Eur. J. Endocrinol. 1997;136:608–616. doi: 10.1530/eje.0.1360608. [DOI] [PubMed] [Google Scholar]
- 65.Buniello A., MacArthur J.A.L., Cerezo M., Harris L.W., Hayhurst J., Malangone C., McMahon A., Morales J., Mountjoy E., Sollis E. The NHGRI-EBI GWAS Catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res. 2019;47(D1):D1005–D1012. doi: 10.1093/nar/gky1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shim W.S., Do M.Y., Kim S.K., Kim H.J., Hur K.Y., Kang E.S., Ahn C.W., Lim S.K., Lee H.C., Cha B.S. The long-term effects of rosiglitazone on serum lipid concentrations and body weight. Clin. Endocrinol. (Oxf.) 2006;65:453–459. doi: 10.1111/j.1365-2265.2006.02614.x. [DOI] [PubMed] [Google Scholar]
- 67.Shirakami Y., Lee S.-A., Clugston R.D., Blaner W.S. Hepatic metabolism of retinoids and disease associations. Biochim. Biophys. Acta. 2012;1821:124–136. doi: 10.1016/j.bbalip.2011.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Arca M., Gaspardone A. Atorvastatin efficacy in the primary and secondary prevention of cardiovascular events. Drugs. 2007;67(Suppl 1):29–42. doi: 10.2165/00003495-200767001-00004. [DOI] [PubMed] [Google Scholar]
- 69.Collins R., Reith C., Emberson J., Armitage J., Baigent C., Blackwell L., Blumenthal R., Danesh J., Smith G.D., DeMets D. Interpretation of the evidence for the efficacy and safety of statin therapy. Lancet. 2016;388:2532–2561. doi: 10.1016/S0140-6736(16)31357-5. [DOI] [PubMed] [Google Scholar]
- 70.Nesti L., Natali A. Metformin effects on the heart and the cardiovascular system: A review of experimental and clinical data. Nutr. Metab. Cardiovasc. Dis. 2017;27:657–669. doi: 10.1016/j.numecd.2017.04.009. [DOI] [PubMed] [Google Scholar]
- 71.Han Y., Xie H., Liu Y., Gao P., Yang X., Shen Z. Effect of metformin on all-cause and cardiovascular mortality in patients with coronary artery diseases: a systematic review and an updated meta-analysis. Cardiovasc. Diabetol. 2019;18:96. doi: 10.1186/s12933-019-0900-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Orkaby A.R., Driver J.A., Ho Y.L., Lu B., Costa L., Honerlaw J., Galloway A., Vassy J.L., Forman D.E., Gaziano J.M. Association of Statin Use With All-Cause and Cardiovascular Mortality in US Veterans 75 Years and Older. JAMA. 2020;324:68–78. doi: 10.1001/jama.2020.7848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Remmer H. The role of theliver in drug metabolism. Am. J. Med. 1970;49:617–629. doi: 10.1016/s0002-9343(70)80129-2. [DOI] [PubMed] [Google Scholar]
- 74.Szymanski M.W., Singh D.P. StatPearls. StatPearls Publishing; 2020. Isoproterenol. [Google Scholar]
- 75.Fall C.H., Pandit A.N., Law C.M., Yajnik C.S., Clark P.M., Breier B., Osmond C., Shiell A.W., Gluckman P.D., Barker D.J. Size at birth and plasma insulin-like growth factor-1 concentrations. Arch. Dis. Child. 1995;73:287–293. doi: 10.1136/adc.73.4.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.de Jong M., Cranendonk A., Twisk J.W.R., van Weissenbruch M.M. IGF-I and relation to growth in infancy and early childhood in very-low-birth-weight infants and term born infants. PLoS ONE. 2017;12:e0171650. doi: 10.1371/journal.pone.0171650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Fathzadeh M., Li J., Rao A., Cook N., Chennamsetty I., Seldin M., Zhou X., Sangwung P., Gloudemans M.J., Keller M. FAM13A affects body fat distribution and adipocyte function. Nat. Commun. 2020;11:1465. doi: 10.1038/s41467-020-15291-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lin X., Liou Y.H., Li Y., Gong L., Li Y., Hao Y., Pham B., Xu S., Jiang Z., Li L. FAM13A Represses AMPK Activity and Regulates Hepatic Glucose and Lipid Metabolism. iScience. 2020;23:100928. doi: 10.1016/j.isci.2020.100928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Willer C.J., Schmidt E.M., Sengupta S., Peloso G.M., Gustafsson S., Kanoni S., Ganna A., Chen J., Buchkovich M.L., Mora S. Discovery and refinement of loci associated with lipid levels. Nat. Genet. 2013;45:1274–1283. doi: 10.1038/ng.2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Dupuis J., Langenberg C., Prokopenko I., Saxena R., Soranzo N., Jackson A.U., Wheeler E., Glazer N.L., Bouatia-Naji N., Gloyn A.L. New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nat. Genet. 2010;42:105–116. doi: 10.1038/ng.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Goodarzi M.O., Guo X., Cui J., Jones M.R., Haritunians T., Xiang A.H., Chen Y.D., Taylor K.D., Buchanan T.A., Hsueh W.A. Systematic evaluation of validated type 2 diabetes and glycaemic trait loci for association with insulin clearance. Diabetologia. 2013;56:1282–1290. doi: 10.1007/s00125-013-2880-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The first sheet lists the perturbations used, their abbreviations and concentration, and the category in which they belong, e.g., adipokine. The second sheet lists all the experimental parameters, e.g., medium used, RNA extraction date, etc. The third sheet lists the control samples used for DE analysis for each perturbation.
Columns indicate the cell, perturbation, Ensembl gene ID, HUGO gene name, the average expression of the gene in the control cells (baseMean), the log2 fold change estimate (log2FoldChange, negative = downregulated by perturbation) and standard error (lfcSE) in perturbed cells, the DESeq2 test statistic (test_stat) and p value. The last column (significant_5prcFDR) indicates if the gene was differentially expressed (FDR<5%) in the perturbed cells in the particular perturbation and cell after adjusting for the number of tests performed across all perturbations and cell lines.
Columns indicate the cell, perturbation, Ensembl gene ID, HUGO gene name, the average expression of the gene in the control cells (baseMean), the log2 fold change estimate (log2FoldChange, negative = downregulated by perturbation) and standard error (lfcSE) in perturbed cells, the DESeq2 test statistic (test_stat) and p value. The last column (significant_5prcFDR) indicates if the gene was differentially expressed (FDR<5%) in the perturbed cells in the particular perturbation and cell after adjusting for the number of tests performed across all perturbations and cell lines.
Columns indicate the cell, perturbation, Ensembl gene ID, HUGO gene name, the average expression of the gene in the control cells (baseMean), the log2 fold change estimate (log2FoldChange, negative = downregulated by perturbation) and standard error (lfcSE) in perturbed cells, the DESeq2 test statistic (test_stat) and p value. The last column (significant_5prcFDR) indicates if the gene was differentially expressed (FDR<5%) in the perturbed cells in the particular perturbation and cell after adjusting for the number of tests performed across all perturbations and cell lines.
Provided as a separate excel file, one sheet for each cell line, with the first column indicating the HUGO gene name and the next 21 columns being an indicator variable for the gene being DE (0 = not DE, 1 = DE upregulated after perturbation, and −1 = DE downregulated after perturbation) between samples treated with perturbation and untreated samples. The last column (Nr_Pert_DE) shows the number of perturbations in which a gene is DE. If Nr_Pert_DE = 1 and ADIP = +1/-1 but all other columns are zero, then the gene adiponectin-specific, i.e., it is only DE in adiponectin but no other perturbations in that cell line. The last two sheets provide DE info summary across perturbations and cells. In the first sheet (deGene_by_perturbation), each cell indicates if the gene is differentially expressed in a particular perturbation in at least one of the three cell lines (FDR < 5%). In the second sheet (deGene_by_cell), each cell indicates if the gene is differentially expressed for the cell line in at least one of the perturbations (FDR < 5%).
The columns indicate pathway name, gene ratio, BgRatio, p value, BH-adjusted p value, and q-value from the over-representation analysis as well as the geneID and number of DE genes in the pathway.
The columns indicate pathway name, gene ratio, BgRatio, p value, BH-adjusted p value, and q-value from the over-representation analysis as well as the geneID and number of DE genes in the pathway.
The columns indicate pathway name, gene ratio, BgRatio, p value, BH-adjusted p value, and q-value from the over-representation analysis as well as the geneID and number of DE genes in the pathway.
Provided as a separate excel file; the first sheet lists a summary of the studies used and LDSCreg-based estimates of their SNP-based heritability (h2) and h2 z-score. The second sheet lists detailed LDSCreg results for each triplet of trait, cell line, and perturbation.
Provided as a separate excel file; the first sheet list results from the analysis of groups of related traits (Parent_Trait), as defined in the GWAS Catalog, while the second list results for specific traits. Each row corresponds to a trait or group of traits - cell line - perturbation combination. The columns indicate the specific trait (second sheet only), parent trait, cell line, and perturbation combination tested as well as the OR of enrichment with the 95% CI, the Fisher’s exact test p value for the significance of the enrichment as well as the BH-adjusted p value. The last four columns contain the number of genes that were “neither DE nor GWAS genes,” “DE but not GWAS genes,” “GWAS but not DE genes,” and “both GWAS and DE genes.”
Data Availability Statement
The data discussed in this publication have been deposited in NCBI’s Gene Expression Omnibus (GEO) and are accessible through GEO series accession number GEO: GSE179347. The code and processed data used for these analyses are available at https://github.com/BrunildaBalliu/PerturbationGxE.