Abstract
Alzheimer's disease (AD) is a progressive and deleterious neurodegenerative disease, strongly affecting the cognitive functions and memory of seniors worldwide. Around 58% of the affected patients live in low and middle-income countries, with estimates of increasing deaths caused by AD in the coming decade. AD is a multifactor pathology. Mitochondrial function declines in AD brain and is currently emerging as a hallmark of this disease. It has been considered as one of the intracellular processes severely compromised in AD. Many mitochondrial parameters decline already during aging; mitochondrial efficiency for energy production, reactive oxygen species (ROS) metabolism and the de novo synthesis of pyrimidines, to reach an extensive functional failure, concomitant with the onset of neurodegenerative conditions. Besides its impact on cognitive functions, AD is characterized by loss of synapses, extracellular amyloid plaques composed of the amyloid-β peptide (Aβ), and intracellular aggregates of hyperphosphorylated Tau protein, accompanied by drastic sleep disorders, sensory function alterations and pain sensitization. Unfortunately, till date, effective management of AD-related disorders and early, non-invasive AD diagnostic markers are yet to be found. MicroRNAs (miRNAs) are small non-coding nucleic acids that regulate key signaling pathway(s) in various disease conditions. About 70% of experimentally detectable miRNAs are expressed in the brain where they regulate neurite outgrowth, dendritic spine morphology, and synaptic plasticity. Increasing studies suggest that miRNAs are intimately involved in synaptic function and specific signals during memory formation. This has been the pivotal key for considering miRNAs crucial molecules to be studied in AD. MicroRNAs dysfunctions are increasingly acknowledged as a pivotal contributor in AD via deregulating genes involved in AD pathogenesis. Moreover, miRNAs have been proved to control pain sensitization processes and regulate circadian clock system that affects the sleep process. Interestingly, the differential expression of miRNA panels implies their emerging potential as diagnostic AD biomarkers. In this review, we will present an updated analysis of miRNAs role in regulating signaling processes that are involved in AD-related pathologies. We will discuss the current challenges against wider use of miRNAs and the future promising capabilities of miRNAs as diagnostic and therapeutic means for better management of AD.
Keywords: Alzheimer's, mitochondria, microRNAs, synaptic plasticity, sleep disorder, pain
Introduction
Alzheimer's: A Peculiar Case of Brain Disease
“Alois Alzheimer,” The German Bavarian psychiatrist, and neurologist was the first to report Alzheimer's disease (AD) in 1906 as “A peculiar severe disease of the cerebral cortex” (Hippius and Neundörfer, 2003). Today, Alzheimer's is acknowledged as a common neurodegenerative disease affecting elderly population. It is the most common form of dementia and may account to 60–70% of cases (Plassman et al., 2007), with increasing numbers of people getting AD every year, especially in middle and low-income countries (Alzheimer's Disease Facts and Figures, 2020).
AD is an irreversible, progressive brain disorder and the most common cause of dementia among older people. It is heterogeneous in every aspect, such as the relation between the presence of plaques and tangles of Aβ and Tau in the brain, clinical symptoms, and genetic background that causes memory loss, language problems, and impulsive or unpredictable behavior. Tau tangles block the transport of nutrients and other essential molecules inside neurons. Recently, it was found that Tau signal emerges first in the rhinal cortex independently of Aβ. Tau pathology begins focally but expands catastrophically under the influence of Aβ pathology to mediate neurodegeneration and cognitive decline. Subsequent Tau elevation in the temporal neocortex is associated with age, Aβ, and APOE status (Sanchez et al., 2021). This indicates a loss of connection between the nerve cells, or neurons, in the brain. At the earlier stages of AD, patient's daily routine is impacted due to disruptions in the entorhinal cortex and hippocampus that affects memory, executive cognition, and visuospatial awareness. Meanwhile, during later AD stages, personality, behavior, and language impairments arise due to escalating damage in frontal, temporal, and parietal lobes. Such drastic damages are associated with continuous decline in independence, culminating in patients' complete dependence on their caregivers by the latest stages of the disease. Consequently, AD is one of the most debilitating disorders as it impacts both patients and families on mental, psychological, and socio-economic aspects.
Alzheimer's: What We Know So Far
Scientists do not yet fully understand the exact causes of AD. AD is a multifactorial disorder in which genetic and environmental risk factors interact to increase the rate of normal aging. It is so tightly associated with old age that there was a speculation it is a normal part of aging (Masters et al., 2015). Various causes probably contribute to AD etiology including a combination of age-related changes in the brain; brain proteins fail to function normally and the presence of toxic oligomeric species of Amyloid peptides (Aβ) and Tau within the AD brain. Recent data confirms the view that such species can propagate and spread within neural circuits, disrupting the work of brain cells (neurons) and triggering a series of toxic events (Chen and Mobley, 2019). Also, mitochondrial dysfunction, a compromised blood brain barrier, immune system dysfunction, and infectious agents probably contribute to the etiology of AD (Fulop et al., 2021). AD is a progressive neurologic disorder that causes the brain to shrink (atrophy) and brain cells to die, culminating into continuous decline in thinking, behavioral and social skills that affect a person's ability to function independently. Clinical manifestations, which are insidious in onset, include memory loss and cognitive decline (Scheltens et al., 2016). The most accepted theory is that AD is caused by misfolded proteins and the culprit in this misfolding is Aβ peptides, that aggregate or clump, killing brain cells and giving rise to the symptoms of memory loss and reduced cognition.
Current research identifies three stages of AD: preclinical Alzheimer's disease, mild cognitive impairment (MCI) due to Alzheimer's disease, and dementia due to Alzheimer's disease (Sperling et al., 2011). In the last two stages, symptoms are present, but to varying degrees. Early signs include difficulty remembering recent events or conversations. As the disease progresses, memory impairments worsen, and other symptoms develop.
People with MCI due to Alzheimer's disease have biomarker evidence of an Alzheimer's-related brain change (for example, elevated levels of Aβ) and greater cognitive decline than expected for their age, but this decline does not significantly interfere with everyday activities (Roberts and Knopman, 2013). In MCI, changes in thinking abilities may be noticeable to family members and friends but may not be noticeable to others.
Sadly till date, there is no cure for AD. Currently, the only approved drugs for AD merely alleviate some of the symptoms -partially and temporarily- but do not stop the disease from progressing.
Alzheimer's Hallmarks: Aβ and Phosphorylated Tau
The neuropathology of AD is characterized by an abnormal build-up of extracellular amyloid-β (Aβ) peptide as neuritic plaques, pathological extracellular aggregates formed around a core of Aβ and are a hallmark of AD (Stratmann et al., 2016), accompanied by intracellular hyperphosphorylated (p)-Tau fibrils which accumulate as neurofibrillary tangles (NFTs) within neurons (Stratmann et al., 2016). Amyloid peptide (Aβ) is derived from the amyloid precursor protein (APP), α-secretase (“normal” cleavage), or β-secretase (“abnormal” cleavage) cleaves APP, and a second cleavage of the β-secretase product, by γ-secretase, cleaves APP further to produce Aβ (Scheuner et al., 1996; Di Carlo et al., 2012). Depending on the cleavage site by γ-secretase, Aβ40 or Aβ42 are produced. Aβ40 is the most common form, while the 42-amino-acid–long fragment, Aβ42, is less abundant and more associated with AD. The proportion of Aβ40 to Aβ42 is important in AD because Aβ42 is far more prone to oligomerize and form fibrils than Aβ40 peptide (Sun et al., 2015). Recently, Alexandra Grubman's group isolated amyloid plaque-containing (using labeling with methoxy-XO4, XO4+) and non-containing (XO4−) microglia from an AD mouse model. Transcriptomics analysis identified different transcriptional trajectories in aging and AD mice. Where XO4+ microglial transcriptomes demonstrated dysregulated expression of genes associated with late onset AD (Grubman et al., 2021). Recent genome-wide association studies have established that the majority of AD risk loci are found in or near genes that are highly and sometimes uniquely expressed in microglia, the resident macrophages of the central nervous system. This leads to the concept of microglia being critically involved in the early steps of the disease and identified them as important potential therapeutic targets. Changes in microglial morphology, the resident macrophages of the central nervous system, and signaling is also evident in AD brains, contributing to the pathology (Hemonnot et al., 2019).
The neuropathological changes of AD brain, based on brain imaging studies, show slow and progressive cerebral atrophy, where the frontal and temporal cortices often have enlarged sulcal spaces with atrophy of the gyri, while primary motor and somatosensory cortices most often appear unaffected (Perl, 2010). Neuropathological studies in combination of MRI approaches also showed early pathological alterations to the locus coeruleus (LC), a tiny nucleus in the brainstem and the principal site of noradrenaline synthesis. The LC undergoes significant neuronal loss in AD, with postmortem studies showing as much as 80% reduction in cell number in people with AD compared to age-matched controls associated with tauopathy in AD (German et al., 1992; Hoogendijk et al., 1995; Beardmore et al., 2021). Another macroscopic feature commonly observed in AD is the loss of neuromelanin pigmentation in the locus coeruleus with age and is proposed to be toxic and inflammatory when released into the extracellular environment (Serrano-Pozo et al., 2011).
Another characteristic feature in AD pathology is the neurofibrillary tangles of hyperphosphorylated microtubule-associated protein (Tau). Microtubules (MTs) are hollow cylinders composed of parallel protofilaments of α and β tubulin subunits. Tau is a neuronal microtubule associated protein whose main biological functions are to promote microtubule self-assembly by tubulin and to stabilize those already formed. MTs dynamics are regulated by Tau proteins that stabilize or destabilize them (van der Vaart et al., 2009) and protect MTs against depolymerization by decreasing the dissociation of tubulin at both MT ends, resulting in an increased growth rate and decreased catastrophe frequency (Trinczek et al., 1995). Disruption of microtubules, as are observed in patients with AD, interrupts axonal transport which prevents vesicles and organelles from reaching the synapses. These result in the slow and steady deterioration of the synapses and retrograde degeneration of the neurons.
Genes of AD
Multiple studies reported different genes to be involved in AD development. Although twin studies support the existence of a genetic component in late onset Alzheimer's disease (LOAD), no one particular causative gene has been identified yet. Familial AD is mainly associated with mutations in the Aβ precursor protein (APP) gene and presenilin genes PSEN1 and PSEN2 that are responsible for γ-secretase cleavage of APP. Still, Familial AD comprises only the minor subclass of AD. Apolipoprotein E (APOE) on chromosome 19 is another polymorphic protein and the allele APOE4 is the strongest genetic risk factor for Sporadic AD (Giri et al., 2016). As the strong affinity of APOE for Aβ affects its production, hydrolysis, and elimination (Reitz and Mayeux, 2014). Yet, although around 80% of LOAD is associated with APOE, apolipoprotein E (APOE)-4 allele confers only a 20% risk for developing the disease. Similarly, increasing evidence confirms that Sporadic AD has a more underlying complex etiology that includes both genetic and environmental factors (Bekris et al., 2010).
The most successful approach to identifying the genetic architecture of AD is the genome-wide association studies (GWAS) which identified and confirmed 19 genome-wide-significant common variant signals in addition to APOE. Together with GWAS, Whole Exome Sequencing (WES), and Whole Genome Sequencing (WGS) defined no fewer than 20 additional genes whose variants contribute to increased risk of AD (Jun et al., 2017; Liu et al., 2017). Kunkle et al. (2019), confirmed 20 previous LOAD risk loci and identified five new genome-wide loci (IQCK, ACE, ADAM10, ADAMTS1, and WWOX), two of which (ADAM10 and ACE) were identified in a recent genome-wide association (GWAS)-by-familial-proxy of AD or dementia. Fine-mapping of the human leukocyte antigen (HLA) region confirms the neurological and immune-mediated disease haplotype HLA-DR15 as a risk factor for LOAD (Kunkle et al., 2019). Other implicated genes are Clusterin (CLU), Sortilin-related receptor-1 (SORL1), ATP-binding cassette subfamily A member 7 (ABCA7), Bridging integrator 1 (BIN1), phosphatidylinositol binding clathrin assembly protein (PICALM), CD2 associated protein (CD2AP), Complement component (3b/4b) receptor 1 (CR1), CD33, triggering receptor expressed on myeloid cells 2 (TREM2), and phospholipase D3 (PLD3) (Karch and Goate, 2015). These variants define possible contributions in AD from genes that regulate endocytosis, inflammation and the brain's innate immune system, and cholesterol/sterol metabolism (Karch and Goate, 2015).
Alzheimer's Pathophysiology: Multiple Hypotheses
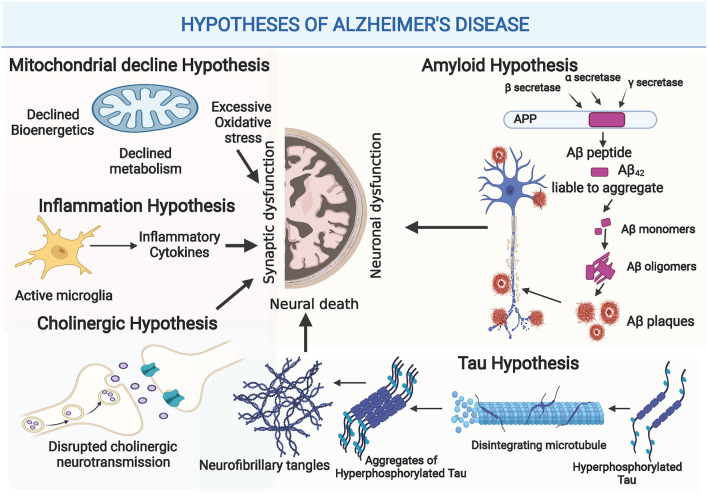
The complexity of AD led to the generation of multiple hypotheses, in trials to unravel AD pathogenesis. Illustration of the different hypotheses of AD development and progression is shown in Figure 1.
Figure 1.
Illustration of the different hypotheses of Alzheimer's Disease (AD) Development and Progression: Amyloid hypothesis: as disturbed secretase enzymes increase production of Aβ42 eventually forming Aβ plaques that impact synaptic functionality, and neuronal dysfunction. Tau Hypothesis: Where increased production of hyperphosphorylated tau provokes disintegration of microtubules and accumulation of hyperphosphorylated tau fibrils causing neuronal death. Cholinergic hypothesis: where disturbed Acetylcholine (ACH) impacts synaptic function. Inflammation hypothesis: where increased release of inflammatory modulators provoke exaggerated immune response that disrupts synaptic functions and plasticity. Mitochondrial hypothesis: where disrupted mitochondrial functions causes glucose hypometabolism, poor ATP production and increased production of ROS, eventually causing synaptic dysfunction. (Created with BioRender.com).
Aβ Cascade Hypothesis
For long, Aβ has been seen as the main causative agent in AD pathology that is followed by neurofibrillary tangles, vascular damage and neuronal loss as direct consequences of Aβ deposition (Murphy and LeVine, 2010). Where Aβ deposits in the hippocampus and basal segment, provoking Aβ to form insoluble aggregates, and inducing mitochondrial damage (Lustbader et al., 2004), and synaptic dysfunction (Hunt and Castillo, 2012). This cascade of events is followed by microglia and astrocytes activation, which arise inflammation and oxidative stress, causing synaptic loss and neuronal death (Fan et al., 2020). Recent studies show that elevated levels of Aβ plaque do not always correlate with magnified AD pathology. Whereas, increasing evidence reveals that Aβ oligomers (Aβ Prions) might be more neurotoxic as they are easily transmitted and released in the synapses (Mucke and Selkoe, 2012; Amin and Harris, 2021). Consequently, and considering the accumulated body of AD research, the majority of data still supports Aβ role as the principal initiator of AD complicated pathology in the early stages. Yet obviously, Aβ is not the only contributor to AD late stages.
Tau Hyperphosphorylation Hypothesis
Tau is mainly found in neuronal axons of the brain. It maintains microtubule structure, synaptic structure, function (Kimura et al., 2014) and regulates neuronal signaling. Tau is also a phosphoprotein that depends on protein kinase and protein phosphatase activities. Hyperphosphorylated Tau in AD patients' brains causes configuration changes and loss of tubulin polymerization capacity (Grundke-Iqbal et al., 1986) that result in defective microtubule functioning. Moreover, increased levels of cytosolic Tau induce polymerization of phosphorylated Tau to form NFTs (Iqbal et al., 2010), that contribute to reduced synapses numbers, and cell dysfunction (Callahan et al., 1999). Tau hyperphosphorylation is positively correlated with the pathological severity of AD and is apparently more detrimental to cognitive impairment than Aβ (Mocanu et al., 2008).
Cholinergic Hypothesis
The cholinergic hypothesis suggests that disruptions of acetylcholine-containing neurons contribute to the cognitive decline in Alzheimer's disease (AD). This hypothesis is supported by the fact that severity of dementia in AD, is positively correlated with the extent of cholinergic loss (Francis et al., 1985). Furthermore, multiple reports imply that cholinergic loss may be an early sign of cognitive decline in AD, and can therefore have a more crucial role in Aβ depositions, Tau phosphorylation, and neuroplasticity (Terry and Buccafusco, 2003; Hampel et al., 2018). Currently, inhibitors of acetylcholinesterase are widely used in AD management, and they show some tangible results concerning symptomatic improvement in AD patients.
Inflammation Hypothesis
Neuroinflammation is a hallmark of AD and increasing evidence shows that microglia is a central player in AD. As in early AD stage, microglia, TREM2 and complement system are responsible for synaptic disruptions (Paolicelli et al., 2011; Hong et al., 2016). Meanwhile, as the disease progresses, reactive microglia and astrocytes surround amyloid plaques and secrete numerous pro-inflammatory cytokines that drastically impact synaptic functions and neuroplasticity over the course of AD (Du et al., 2018).
Oxidative Stress Hypothesis
Likewise, oxidative stress is a significant player in AD pathogenesis. Normally, the brain utilizes more oxygen than other tissues and undergoes mitochondrial respiration, which increases the potential for ROS exposure. AD is highly associated with cellular oxidative stress that contributes to increased protein oxidation, glycol oxidation, lipid peroxidation and Aβ accumulation. In line, advanced glycation end products (AGE) and malondialdehyde have been detected in the neuro tangles and senile plaques of AD (Markesbery, 1997). Moreover, multiple studies show that Aβ is capable of generating free radicals that mediate neuron degeneration and death (Cheignon et al., 2017).
Mitochondrial Damage and Glucose Hypometabolism Hypothesis
Increasing body of evidence implies that mitochondrial impairments play fundamental roles in AD pathology. Healthy mitochondria support neuronal activity, provide proper energy supply to neurons by regulating glucose metabolism, and minimizing oxidative damage. In the neurons, mitochondria are vital for biosynthesis of iron and heme. They are also involved in presynaptic transmission, and they regulate calcium concentration during signal transduction (Picone et al., 2014).
Emerging reports spot glucose hypometabolism as an early pathogenic event in preclinical stages of AD concurrently with cognitive and functional decline. Using fluoro-2-deoxyglucose positron-emission tomography (FDG-PET), reveal that glucose hypometabolism is consistently detected in hippocampus and cortex of AD brain compared to normal individuals. In line, impaired mitochondrial bioenergetics, increased oxidative stress and disrupted mitochondrial genome are consistent features of mitochondrial abnormalities in AD and are interconnected to amplify the debilitating pathologies of AD (Wang W. et al., 2020).
Till date, it is not totally clear whether any of these characteristic deficits is the primary initiator or just a contributor in the contemporary multifactorial AD pathology. A challenge confirmed by the little therapeutic success achieved against AD so far. Consequently, we obviously need new approaches for AD diagnosis at its earliest stages before neuronal damage becomes irreversibly established. It is also crucial to concurrently target multiple axes in AD pathologies for more tangible therapeutic feasibility.
MicroRNAs: Origin, Maturation, and Role
MicroRNAs production excels with formation of a long primary transcript (pri-miRNA) via RNA polymerase II. Inside the nucleus, pri-miRNAs are cleaved by Drosha protein and DiGeorge Syndrome Critical region 8 (DGCR8) protein which dimerize together to form a functional microprocessor complex. These dimerized proteins cleave pri-miRNAs into precursor miRNAs (pre-miRNA), which are then transported to the cytoplasm and digested by Dicer and TAR RNA binding proteins (TRBP) to release a double-stranded miRNA duplex. Helicase enzyme unwinds the duplex to form mature miRNA strands. One of these strands is usually degenerated, while the other associates with Ago2 protein to form miRNA-induced Silencing Complex (miRISC). Each mature miRNA contains a sequence of 7 or 8 nucleotides that binds to its complementary region(s) on target mRNAs. Mature miRNAs bind to the 5′- or 3′-untranslated regions (UTR) of target mRNAs and rarely, both strands can serve as mature functional miRNA. Generally, functional miRNAs induce gene silencing using two different mechanisms, depending on the complementarity between the miRNA and its mRNA target (Amakiri et al., 2019; Kou et al., 2020). If mature miRNA binds perfectly to the complementary regions of the target mRNA, it induces mRNA degradation via de-adenylation, cap removal, and exonucleolytic digestion of mRNA. Meanwhile, if miRNA binds with imperfect complementarity to target mRNA, it causes a translation block by repression of translation during the initial phase or the elongation phase. Alternatively, miRNAs can repress translation by inducing premature ribosome detachment (Silvestro et al., 2019). One functional miRNA can interact with hundreds of target mRNAs to exert various levels of regulatory effects, and a single mRNA can be targeted by several miRNAs as well.
Around 70% of miRNAs are found in the human brain, where miRNAs are responsible for regulating synaptic functions, neurotransmitter release, and neuronal development. In the last few years, the significance of balanced miRNAs expression proved to be crucial for proper functionality and homeostasis in the body. The differential expression of miRNAs and/or single nucleotide polymorphism (SNP) of miRNAs are implicated in multiple diseases (Reddy et al., 2017). Likewise, the dysregulation of miRNAs emerged as a key contributor in AD pathology, as it leads to altered protein expressions and impairment of the complicated signaling network balance in the brain. The next section will cover the main updated findings concerning miRNAs role in AD pathogenesis and its progressive events.
Micrornas and AD Cognitive Impairment
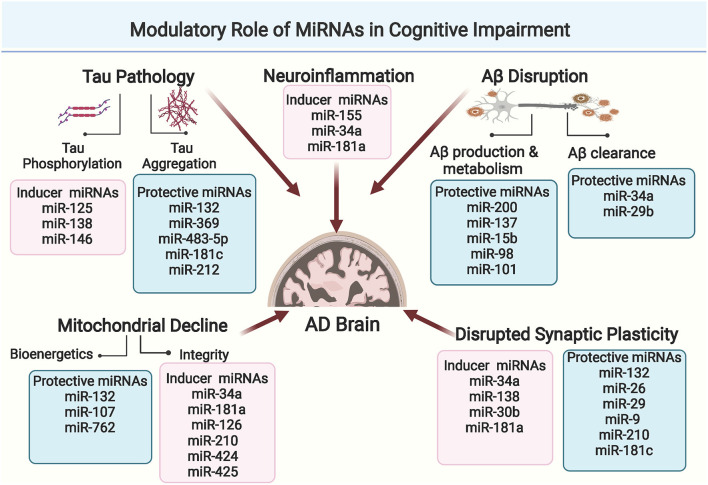
Cognition refers to the brain's ability to think, learn, remember, and process information. The loss of neuronal connections in AD brain is basically attributed to disrupted signaling pathways that affect both synaptic plasticity and dendritic functions, the two crucial controllers of cognitive processes. On the molecular level, updated studies show that Aβ and Tau pathologies cause progressive axonal degeneration and drastic downstream impairments in the synaptic processes (Pereira et al., 2021). Moreover, Aβ and Tau aggregates induce exacerbated immune microglial response, as well as disrupted astrocytes functionality, that eventually contribute to cognitive decline in AD (Fakhoury, 2018). Over the past few years, miRNAs have emerged as significant regulators of Aβ and Tau metabolism, glial functionality, and synaptic plasticity. Simultaneously, increasing studies report the drastic impact of miRNA dysregulations on cognitive functions in AD, through targeting key genes and activity-mediated protein synthesis at the synaptic level. Comprehensive illustration of the modulatory role of miRNAs in AD-related cognitive impairment is presented in Figure 2.
Figure 2.
Comprehensive illustration of the modulatory role of miRNAs in cognitive impairment in AD through affecting Aβ and Tau metabolism, mitochondrial functionality, neuroinflammation or synaptic plasticity. (Created with BioRender.com).
MicroRNAs and Aβ Pathway Disruptions
Although amyloid deposition alone is not able to produce full AD-pathology, studies that used Aβ PET in cognitively normal elderly individuals, mild cognitive impairment (MCI) and AD patients found significant relationships between cognitive deficits and increased brain fibrillar Aβ (Wang F. et al., 2015). MicroRNAs are closely related to the synaptic dysfunction induced by abnormal Aβ metabolism. Increasing body of work shows that cognitive impairment caused by Aβ can be restored by manipulation of miRNAs, which strongly supports the belief that disrupted miRNA expressions are critical in cognitive impairment in AD patients (Weldon Furr et al., 2019). MicroRNA dysregulations are repeatedly reported in association with key genes that regulate Aβ synthesis, cleavage, and clearance. Table 1 illustrates an updated comprehensive summary of the human studies showing differentially expressed miRNAs in AD samples and their relation to Aβ pathway regulations.
Table 1.
Disturbed miRNAs reported in human AD studies and their relation to Aβ pathways.
| miRNA | Sample | Regulation | Relevant pathway | References |
|---|---|---|---|---|
| miR-15a miR-15 b |
Serum | Downregulation | Downregulates BACE1 expression |
Satoh et al., 2015; Lang et al., 2016; Li and Wang, 2018 |
| miR-29a | Brain CSF Serum |
Down Up Down |
BACE1 | Hébert et al., 2008; Geekiyanage et al., 2012; Müller et al., 2016 |
| miR-29b | Blood | Down | BACE1 | Satoh et al., 2015 |
| miR-29c | Brain | Down | BACE1 | Lei et al., 2015 |
| miR-20b | Brain | Down | APP | Nunez-Iglesias et al., 2010 |
| miR-21-5p | Brain | Up | APP | Giuliani et al., 2021 |
| miR-16 | CSF | Down | APP | Müller et al., 2014 |
| miR-9 | Brain | Down | APP | Hébert et al., 2008 |
| miR-30c | Brain | Down | APP | Cogswell et al., 2008 |
| miR-101a-3p | Brain | Down | APP | Hébert et al., 2008 |
| miR-188 | Brain, Serum | Down | APP | Kou et al., 2020 |
| miR-106a | Brain Serum |
Down Up |
APP | Wang et al., 2011; Cheng et al., 2015 |
| miR-106b | Brain Whole blood |
Down Up |
APP | Hébert et al., 2008; Cheng et al., 2015 |
| miR-138-5p | Plasma Exosomes | Up | APP | Lugli et al., 2015 |
| miR-138 | Brain, CSF | Variant | APP | Boscher et al., 2019 |
| miR-135b | Blood | Down | BACE1 | Zhang et al., 2016 |
| miR-124 | CSF | Down | APP | Burgos et al., 2014 |
| miR-155 | Peripheral Blood mononuclear cells | Upregulated | Inhibits Aβ catabolism | Guedes et al., 2016 |
| miR-34 | Peripheral Blood mononuclear cells | Upregulated | Aβ clearance | Basavaraju and de Lencastre, 2016 |
| miR-181b | Peripheral Blood mononuclear cells | Upregulated | Aβ clearance | Kumar and Reddy, 2016 |
| miR-181c | Serum | Downregulated | APP | Geekiyanage et al., 2012 |
| miR-186 | Brain | Downregulated | BACE1 | Ben Halima et al., 2016 |
| miR-346 | Human Neuron Enriched culture | Upregulated | APP | Long et al., 2019 |
| miR-338 | Hippocampus | Downregulated | APP | Qian et al., 2019 |
| miR-200 | Brain | Upregulated | SIRT1, Aβ accumulation, Apoptosis | Zhang et al., 2017 |
Regarding Aβ metabolism, several miRNAs including miR-9, miR-29, miR-135, and miR-186 are significant regulators of Beta-Secretase 1 (BACE1) enzyme levels which is central in Aβ generation (Wang et al., 2019).
MiR-200-3p particularly has been grabbing increased attention for its role in Aβ pathology in AD. MiR-200-3p is repressed in AD animal and cell models. Mechanistically, miR-200-3p modulates translocation of BACE1 enzyme and ribosomal protein S6 kinase B1 (S6K1), hence it suppresses cell apoptosis, decreases Aβ1-42 and Tau phosphorylation in cell experiments (Samadian et al., 2021). To evaluate the effect of miRNA-200b/c in vivo, Tg2576 mice were treated with miRNA-200b/c by intracerebroventricular injection. This experiment confirmed that upregulating miR-200 reduced secretion of Aβ. Moreover, the treated mice were relieved of memory impairments induced by intracerebroventricular injection of oligomeric Aβ. They also demonstrated proper spatial learning, suggesting that miRNA-200b and miRNA-200c are potential therapeutic targets in AD (Higaki et al., 2018). These data are strongly supported by clinical studies that showed decreased miR-200b in serum and cerebrospinal fluid of AD patients, compared to healthy subjects (Silvestro et al., 2019).
Recent studies have recently shown that miR-137 and miR-15 b can reverse the neurotoxicity induced by Aβ abnormal metabolism in animal and cell lines (He et al., 2017; Li and Wang, 2018). In the updated pilot study of Vergallo et al. (2021), the protective anti-Aβ effect of miR-15b is reported in asymptomatic at-risk population for AD, as there were significant associations between plasma concentrations of miR-15b, with core neuroimaging biomarkers of AD pathophysiology in the hippocampus (Vergallo et al., 2021).
Several miRNAs as miR-98 and miR-124 modulate Aβ production via notch signaling pathway. Specifically, miR-98 suppresses Amyloid accumulation as it inhibits HEY2 protein levels which inactivates the notch signaling pathway responsible for Aβ production (Amakiri et al., 2019).
Furthermore, miRNAs that directly target APP, confirm the role of miRNAs in AD pathogenesis. Downregulated levels of miR-101 are reported in AD brain and are consistent with in vitro studies where inhibition of miR-101 increased APP levels (Siedlecki-Wullich et al., 2021). In vitro and in vivo studies also show that downregulation of miR-137 determines an increase in Ca2+ levels and a reduction of Aβ1-40 and Aβ1-42. These results indicate that an increase in miR-137 could cause a decrease in Ca2+ levels in neurons, improving neuronal dysfunctions of AD (Davare and Hell, 2003).
Another approach of AD Aβ pathology is the Aβ clearance from the brain to the circulation. Aβ clearance from the brain requires adequate balance of Aβ phagocytosis, glymphatic clearance and healthy system of ABC transporters ABCB1 and ABCG2. Recently, miR-34a and miR-29b have been found to interfere with at least three pathways of Aβ clearance (Weldon Furr et al., 2019). In adult mammalian brain, miR-34a and miR-29b are highly expressed and have been implicated in a range of neurodevelopmental and neuropathological processes. Both miR-34a and miR-29b are dysregulated in brain and serum samples of AD patients (Madadi et al., 2019).
Taken together, these cumulative studies show that AD disease can disrupt miRNA coordinated expression. Simultaneously, miRNA altered expression contributes to Progressive AD pathogenesis through disrupting key genes in Aβ pathway. Unleashing the mechanism of microRNA Aβ regulating pathways, can identify novel therapeutic targets for better AD management.
MicroRNAs and Tau Pathology in AD
In the last few years, abnormal phosphorylated Tau has proved to be detrimental in cognitive decline (Di et al., 2016). Moreover, extracellular soluble Tau oligomers have been recognized as a possible cause of memory loss and synaptic dysfunction (Biundo et al., 2018). Experimental AD studies helped to identify several miRNAs that are linked to Taupathy in AD. Among the most prominent miRNAs, miR-125b, and miR-138 are upregulated in AD and have been shown to induce Tau hyper phosphorylation and tangling in neuronal cultures. Subsequently their upregulation disrupts associative learning and cognition in AD mice models (Banzhaf-Strathmann et al., 2014; Wang X. et al., 2015). MiR-125 upregulation in AD promotes Tau hyperphosphorylation through activating Mitogen-activated protein kinase (MAPK) kinases, most likely by down-regulating its target phosphatases genes: DUSP6 and PPP1CA. Whereas, direct hippocampal delivery of miR-125b mimic improved learning, memory and inhibited Tau phosphorylation and expression of DUSP6, and PPP1CA in C57BL/6 mice (Banzhaf-Strathmann et al., 2014). Other miRNAs are reported to modulate Tau affinity for microtubule, regulate the maintenance of microtubule network, and affect Tau aggregation/deposition in NFTs (Siedlecki-Wullich et al., 2021). MiR-22-3p affects Tau phosphorylation through regulating Sirtuin 1 SIRT1 gene. Meanwhile, miR132-3p regulates Tau phosphorylation via PTBP2 gene and Tau splicing via modulating MeCP2 and PTEN genes (Praticò, 2020). In 3xTg mice, loss of miR-132 increased total and phosphorylated Tau levels and provoked Tau aggregation. Consistently, restoring miR-132 to normal levels improved Tau pathology and long-term memory (Smith et al., 2015). Similarly, miR146a-5p is reported to regulate Tau phosphorylation via ROCK1 gene.
Clinically and in support of these translational experiments, miR-125, miR-138, miR-146 and miR- 132 are significantly dysregulated in the cerebrospinal fluid of AD patients (Galimberti et al., 2014; Lee et al., 2016; Wei et al., 2020). MiR-369 is one of the most recently studied miRNAs. Knocking out miR-369 in 3xTg AD mice aggravated cognitive impairment and promoted hyperphosphorylation of Tau, through upregulating kinases Fyn and serine/threonine-protein kinase 2 (SRPK2) as the upstream molecules. Meanwhile, Restoring miR-369 reversed the hyperphosphorylation of Tau and downregulated Fyn and SRPK2, implying the possible therapeutic potential of miR-369 in AD (Yao et al., 2020).
Similarly, miR-483-5p is recently reported to regulate ERK1 and ERK2 kinases at both mRNA and protein levels, resulting in reduced phosphorylation of Tau protein associated with Tau neurofibrillary pathology in AD. Taking these observations together, suggests the neuroprotective action of miR-483-5p in AD pathology (Nagaraj et al., 2021).
Besides Tau phosphorylation, miRNAs have a key role in Tau clearance. Where some post-translational modifications of Tau inhibit Tau ubiquitin binding which promote Tau aggregation. Moreover, acetylation at specific sites of Tau provokes autophosphorylation, and aggregation. This acetylation process is dependent on the balance between acetyltransferase p300 (an acetylase) and sirtuin 1 (SIRT1, a deacetylase). Recent studies demonstrated that SIRT1 gene could be directly inhibited by miR-9, miR-212, and miR-181c to imply their potential role in Tau regulation and consequent AD events (Zhao et al., 2017; Praticò, 2020).
MicroRNAs and Synaptic Plasticity in AD
Dendrites and dendritic spines are the loci of long-term synaptic plasticity that facilitates cognitive processes such as learning and memory. On the cellular level, synaptic plasticity is mediated by structural changes and functionality of dendritic spines. The dendritic spines have specialized subdomains that contain scaffolding proteins, signal transduction molecules, ion channels, and cytoskeleton components which collectively regulate spine morphology, synaptic transmission, and plasticity. Assembly and remodeling of neuronal circuit are generally affected by alterations in density and properties of ionic channels and proteins of the dendritic spines (Bosch and Hayashi, 2012; Reza-Zaldivar et al., 2020).
Growing evidence confirms that alterations of spine morphology and dendritic spine (DS) loss are correlated with AD cognitive decline even before neuronal loss (Dorostkar et al., 2015). Furthermore, the characteristic Aβ and Tau pathologies in AD, suppress synaptic plasticity, which simultaneously provokes changes in dendritic morphology, synaptic maturation and synaptic loss (Pereira et al., 2021). Moreover, updated postmortem examination of AD brains shows that cognitive impairment correlates with synaptic loss better than the number of extracellular plaques or NFTs in AD, making synaptic failure a hallmark of AD (Kumar and Reddy, 2020).
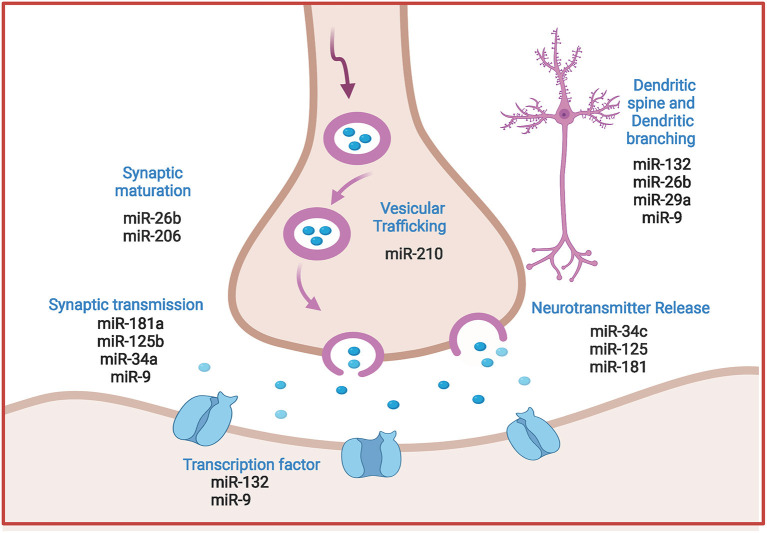
Interestingly, microRNAs are now established as principal regulators of synaptic plasticity during neuronal circuit formation and integration. Moreover, changes in neuronal microRNA expression contribute to synaptic function modification via modulating dendritic spine morphology and/ or regulating local protein translation to synaptic transmission. These mechanisms are proved determinant for both synapse formation and synaptic plasticity (Reza-Zaldivar et al., 2020). Figure 3 spots the prominent regulatory role of miRNAs in synaptic plasticity.
Figure 3.
Comprehensive illustration of the regulatory role of prominent miRNAs in synaptic plasticity in AD. MicroRNAs regulate vesicle formation and trafficking, ACH release and neurotransmission, post synaptic transcription processes and dendrites morphology. (Created with BioRender.com).
Multiple studies reported miRNAs that contribute to impaired synaptic plasticity in AD. Table 2 illustrates an updated summary of the miRNAs involved in regulating target genes of synaptic functions, morphology and dendritic spine alterations in AD studies. Among the interesting miRNAs, miR-34a overexpression is found to impact synaptic functionality and cognitive decline in AD mice models (Sarkar et al., 2019). On the molecular level, miR-34a targets SIRT1, CREB and BDNF genes that have multiple roles in AD progression via increasing Tau Phosphorylation, altering spine morphology and spine functions. Loss of miR-101 in hippocampal neurons was found to cause cognitive decline and modulation of AD-related genes in mice. Where miR-101 knockdown in the hippocampus of C57BL/6 J mice showed AMPK hyperphosphorylation, upregulation of miR-101 target genes associated with AD such as APP, and Rab5 and overproduction of Aβ42 levels (Barbato et al., 2020).
Table 2.
MicroRNAs involved in modulating key target genes of synaptic plasticity in AD.
| miRNA | Target gene | Function | References |
|---|---|---|---|
| miR-34a |
GRIN2B Syt-1 Stx-1A |
Synaptic transmission reduced dendritic trees and alteration of DS morphology and function | Agostini et al., 2011; Xu et al., 2018; Sarkar et al., 2019 |
| miR-210 | NPTX1 AMPAR | Recruitment/clustering, synaptic transmission | Pulkkinen et al., 2008 |
| miR-125b |
GRIN2A EPHA4 |
Synaptic transmissionFormation of long and narrow DS filopodia-like, with a subsequently synaptic transmission weakening | Alsharafi et al., 2016 Edbauer et al., 2010 |
| miR-181a | GRIA2 | Synaptic transmission | Rodriguez-Ortiz et al., 2020 |
| miRNA-574 | Nrn1 | DS stabilization leading to cognitive impairment | Li F. et al., 2015 |
| miRNA-29a/b | Arpc3 | Reducing the mushroom-shaped DS formation, and DS head enlargement, a fundamental step in synaptic maturation | Lippi et al., 2011 |
| miRNA-135 | CPLX1 CPLX2 | Impairment of the postsynaptic exocytosis of AMPA receptors leading to DS shrinkage | Hu et al., 2014 |
| miRNA-124 | CREB | Serotonin-induced long-term synaptic plasticity | Rajasethupathy et al., 2009 |
| miR-128 | STIM2 | Negative modulator of intracellular calcium | Deng et al., 2020 |
| miR-92, miR-137, miR-501 | GRIA1 | Synaptic transmission | Letellier et al., 2014; Hu et al., 2015; Olde Loohuis et al., 2015 |
| miR-9 | REST | Synaptogenesis, synaptic plasticity, and structuralRemodeling | Giusti et al., 2014 |
| miR-132, miR-134 |
CREB1 MMP-9 BDNF |
Transcription factors involved in synaptic plasticity Alterations in DS morphology and maturation |
Mellios et al., 2011; Jasińska et al., 2016 |
| miRNA-206 | BDNF | Decreasing DS density | Lee et al., 2012 |
| miRNA-218 | GRIA2 | Increasing the amplitude of synaptic currents and formation of thin DS | Rocchi et al., 2019 |
| miRNA-30b | EphB2, SIRT1, GRIA2 | Reduced basal synaptic transmission, impaired spatial learning and memory retention and DS density reduction | Song et al., 2019 |
Similarly, overexpression of hippocampus miR-30b disrupts basal synaptic transmission, and reduces DS density, eventually leading to declined learning and memory (Song et al., 2019). This is accomplished as miR-30b modifies SIRT1 expression which regulates Tau phosphorylation. Interestingly miR-30b also targets EphB2 that has a protective role against Aβ oligomers accumulation and disruption of glutamate receptors that are directly linked to synaptic plasticity and cognitive processes (Cissé et al., 2011). Another crucial target of miR-30b is the GRIA2 gene, the predominant excitatory neurotransmitter receptors in mammalian brain (Siedlecki-Wullich et al., 2021), and an increasingly reported linker between Aβ clearance and synaptic disruptions in AD models (Hettinger et al., 2018).
On the clinical side, miR-138 is highly expressed in the dendrites of hippocampal neurons and it acts to regulate dendritic spine size and structure. Functional screening demonstrates that Acyl Protein Thioesterase1 (APT1)-induced palmitoylation of G protein α13 (Gα13) is important for the regulatory function of miR-138 during dendritic spine development. Whereas, high level of miR-138 significantly reduces APT1 level which leads to dendritic spine shrinkage and concomitant reduction in synaptic transmission (Siegel et al., 2009). Recently a panel of studies focusing on miR-138 function during the process of learning and memory showed a close association with local plasticity-related protein synthesis. Examination of human postmortem brain tissue showed expression of miR-138 and decapping mRNA 1B (DCP1B) in hippocampus and frontal cortex. Furthermore, it was found that a human memory-associated single nucleotide polymorphism could interfere with miR-138 binding to the transcripts of DCP1B, implying that miR-138 is a strong modulator of human memory performance (Ye et al., 2016).
Contrarily, some miRNAs have protective effects against synaptic disruptions in AD. For instance, miR-132 inhibits extra synaptic gene Matrix metallopeptidase 9 (MMP-9), whose overexpression promotes formation of immature DS. Consequently, MMP-9 inhibition by miR-132 supports DS head widening that potentiates synaptic plasticity (Jasińska et al., 2016). In line, a recent study showed that upregulating miR-132 by an enriched environment, enhanced hippocampal synaptic plasticity and prevented DS impairments induced by Aβ oligomers (Wei et al., 2020). Furthermore, miR-26a and miR-384-5p have been found as significant regulators of dendritic spine growth and targeting endogenous ribosomal S6 kinase 3 (RSK3). Inhibition of miR-26a is reported to attenuate neurite outgrowth and neuronal morphogenesis (Gu et al., 2015).
Collectively, these data spot the significant role of miRNAs as key players in synaptic plasticity and the impact of miRNAs dysregulations on synaptic homeostasis and functionality in AD pathology. Getting deeper understanding of miRNAs and their targets concerning synaptic modeling in AD may provide new approach to earlier diagnosis and therapeutic management of AD cognitive pathology.
MicroRNAs and Glia Cells Role in AD-Cognitive Impairment
Microglia are brain-resident myeloid cells that mediate innate immune responses in the CNS. Under normal conditions, microglia exist in a “resting” state where they “monitor” the surrounding microenvironment and maintain brain homeostasis via synapse organization, removal of debris by phagocytosis and release of neurotrophic factors (Fan and Pang, 2017). Meanwhile, activation of microglia is accompanied by morphological changes that permit motility and phagocytosis. Microglia can differentiate into either M1 (pro-inflammatory) or M2 (anti-inflammatory) phenotypes depending on the provoking signals. M2 microglia release anti-inflammatory and protective cytokines such as IL-10, TGF-β, IL-4, and IL-13, which promote repair (Guedes et al., 2013). Whereas, M1 microglia release inflammatory mediators such as ROS, MMP-9 and pro-inflammatory cytokines such as TNFα, IL-6 and IL-1β.The balance between these different microglial phenotypic states promotes inflammation or tissue repair and influences the progression of neuroinflammatory disorders (Guedes et al., 2014).
Currently, “Microglia” is an increasing hot topic in AD research, and cumulative studies spot the multifaceted role of microglia as beneficial or detrimental in AD. Recent genome-wide association studies confirm that most of AD risk loci are present in or close to genes that are highly and/or uniquely expressed in microglia. This strongly implies the significant involvement of microglia in early steps of AD (Hemonnot et al., 2019). Among the well-known genes are Cd33 and TREM2 which are linked to Aβ phagocytosis and regulation of microglial inflammatory interaction with Tau tangles (Onuska, 2020).
Additionally, the CX3CR1 receptor is predominantly expressed in microglia. Its ligand CX3CL1 is constitutively expressed by neurons, and it helps maintaining microglia in a resting state. CX3CL1-CX3CR1 is a critical signaling pathway that is disrupted in neurodegenerative conditions and is associated with a strong microglial toxicity (Keren-Shaul et al., 2017). The involvement of the CX3CL1/CX3CR1 signaling pathway in AD is confirmed by an elevated plasma concentration of CX3CL1 in AD patients compared to healthy control subjects.
Other interesting contributors are the complement proteins C1q and CR3 (Veerhuis et al., 2011) which are highly produced in the microglia as crucial factors in synapse pruning and are highly present in CSF of AD patients with emerging evidence about their role in Aβ pathogenesis (Fatoba et al., 2021). Furthermore, Numerous studies demonstrate that microglial Aβ phagocytosis contributes to degeneration by triggering NLR family pyrin domain containing 3 receptor (NLRP3) and lysosomal cathepsin-B that subsequently releases IL-1β and disrupts autophagosome degradation.
Growing body of evidence shows that miRNAs dysregulation impacts microglial hyper-activation, neuroinflammation, and alters macrophage polarization in the brain. Mechanisms that are closely implicated in AD pathology. Table 3 illustrates updated data about miRNAs that regulate key genes in the microglia.
Table 3.
MicroRNAs and their key target genes in the microglia.
| miRNA | Target Gene | Gene function | References |
|---|---|---|---|
| miR-124 | C/EBP-α | Microglial Polarization to M2 | Ponomarev et al., 2011 |
| miR-155 | C-MAf, CCM1, NOX2, NOX4 | Microglial polarization to M2, microglial activation and increased count | Guo et al., 2019 |
| miR-9 | NF-kB | Microglial activation | Yao et al., 2014 |
| miR-204 | SIRT1 | Repression of microglial activation | Li L. et al., 2015 |
| miR-125b | NF-kB | Contributing in increased TNF-α release | Parisi et al., 2013 |
| miR-34a | TREM2 | Aβ42 aggregation and accumulation and microglial activation | Alexandrov et al., 2013 |
Concerning AD studies, miR-34a is reported as a major target of TREM2, a microglial receptor that mediates Aβ42 clearance via phagocytosis in the CNS. Simultaneously, multiple clinical reports showed miR-34a level is dysregulated in AD patients (Bhattacharjee et al., 2016).
In line, both miR-155 and miR-146a are upregulated in the CNS during AD and both regulate the excessive inflammatory signaling observed in AD disease course (Su et al., 2016). MiR-155 is now established as a crucial pro-inflammatory factor in microglia, as it represses suppressor of cytokine signaling 1 gene (SOCS-1). Increased miR-155 expression was recently reported in 3xTg AD mice brains (Guedes et al., 2014), together with enhanced microglial activation. Meanwhile, knockdown of miR-155, induced SOCS-1 expression and led to downregulation of iNOS and nitric oxide production.
MiR-146 is another multi-faceted miRNA that is implicated in the pathogenesis of AD (Jayadev et al., 2013). In microglia, miR-146 is reported to target Presenilin 2 (PS2), a membrane associated protease that regulates proinflammatory microglial behavior (Wang and Wang, 2018).
Astrocytes are another class of glial cells that affect inflammatory response in the CNS. In healthy conditions, astrocytes regulate neuronal metabolism, synaptogenesis, intracellular calcium levels and interact with neuronal signaling (Vasile et al., 2017). Under pathological conditions, astrocytes participate in shaping the CNS response to stress and disease. Where neuroinflammation can be either promoted or restricted by astrocytes through release of pro-inflammatory or anti-inflammatory molecules, leukocyte recruitment and forming functional barriers for CNS parenchyma (Sofroniew, 2015).
In the brain of AD patients, the inflammatory response aggravates astrocytes number, volume and activity (Meraz-Rios et al., 2013). Interestingly, IL-1β, IL-6, and transforming growth factor-β (TGF-β) are upregulated before Aβ aggregation and Tau hyperphosphorylation. These Inflammatory factors activate astrocytes to over express BACE1 enzyme and produce excessive amounts of Aβ proteins (Blasko et al., 2000). In turn accumulated Aβ provokes astrocytes to release more cytokines as TNF-α, a crucial factor in AD-related cognitive impairment (Veeraraghavalu et al., 2014). Additionally, astrocyte dysfunction leads to a decrease in Aβ uptake and clearance (Rolyan et al., 2011). Moreover, astrocytes have been recently reported to promote Tau lesions and accelerate NFTs formation (Birch et al., 2014; Yang et al., 2019).
Increasing reports spot the disruption of miRNA expression in astrocytes during neuroinflammation and neurodegenerative processes accompanying AD. Among the characteristic miRNAs is miR-146 a. Cui et al. (2010) found that miR-146a was upregulated in human astrocytes when exposed to Aβ. The study found that miR-146a mediated down-regulation of interleukin-1 receptor-associated kinase-1 (IRAK-1). IRAK-I is coupled to NF-κB extensive sustained inflammatory response of astrocytes and downstream Toll like receptors proteins. Therefore, NF-κB inhibitors and miR-146a can present a treatment strategy against excessive immune response to Aβ in the brain (Cui et al., 2010).
Glutamate release, reuptake, and recycling are tightly regulated by astrocytes at tripartite synapses. Glutamate overload can trigger neuronal and synaptic loss (Marttinen et al., 2018). Glial glutamate transporter 1 (GLT-1) contributes to clearance and regulation of glutamate at synaptic clefts. In a 3xTg- AD mouse model, increased levels of miR-181a downregulated synaptic proteins related to GLT-1, impacting the plasticity of glutamatergic synapses in astrocytes, and implying its key mediating role in synapses plasticity (Zumkehr et al., 2015). Generally, miR-181 family regulates neuroinflammatory signaling in astrocytes, and miR-181 family has been reported to be upregulated in AD mouse model, causing impaired synaptic plasticity through targeting SIRT-1.
MiR-155 is another significant astrocyte modulator, as it affects astrocytes density during inflammation. Furthermore, increased astrocytes levels of miR-155 were shown to target SOCS1, a negative regulator of the inflammatory gene response, in Aβ-treated astrocytes, causing prolonged expression of inflammatory cytokines (Guedes et al., 2014).
MicroRNAs and Mitochondrial Damage
A large body of research shows enormous mitochondria alterations in the brains of AD patients. Interestingly, mitochondrial alterations have been consistently observed before the clinical onset of AD (Swerdlow, 2018). As a result, mitochondrial dysfunction is now a hot topic in AD for its possible role in the earlier progression of the disease.
One important feature of mitochondrial alterations in AD is the impaired mitochondrial bioenergetic machinery. Glucose hypometabolism in AD brain is strongly linked to impaired oxidative phosphorylation. Moreover, it is closely correlated to impaired levels of blood thiamine diphosphate (TDP), a crucial coenzyme of pyruvate dehydrogenase and α-ketoglutarate dehydrogenase (KGDHC) enzymes allocated in the Krebs cycle (Sang et al., 2018).
Furthermore, Redox proteomics studies found that many antioxidant enzymes that are allocated in the mitochondria, including glutathione-S-transferase Mu, peroxiredoxin 6, GSH and ATP synthase are oxidized in AD, which might compromise their functions by increasing oxidative stress conditions that prevail in AD affected brain regions (Swomley and Allan Butterfield, 2015). Interestingly, levels of oxidized nucleic acids in mtDNA are reported to be significantly elevated in preclinical AD, again stressing mitochondrial abnormalities as an early event of AD progression (Wang W. et al., 2020). Additionally, emerging studies demonstrated impaired base-excision repair (BER) activity in both AD and MCI patients (Lillenes et al., 2016), suggesting significant contribution of replication error to increased mtDNA mutations in AD.
Only recently, a subset of microRNAs is found to be localized to human mitochondria (mitomiRs) and while mitomiRs functions are still far from being completely explored, recent findings relate mitomiRs to neurodegenerative diseases, including Alzheimer's. MiR-107 is one of the recently discovered mitomirs. It regulates oxidative abilities of mitochondria and downregulated miR-107 was found to decrease mitochondrial volume, cristae and mitochondrial membrane potential. Interestingly, decreased plasma levels of miR-107 correlated with abnormal cortical anatomy, common to AD patients, while injecting miR-107 mimic reversed spatial memory impairment, decreased phosphorylated Tau levels, and Aβ neurotoxicity (Shu et al., 2018; John et al., 2020).
MitomiR-34a was recently found to affect mitochondrial metabolism contributing to declining spatial memory (Sarkar et al., 2016). In support to the significant potential of mitomiRs in AD, miR-181a is recently reported to affect mitochondrial glucose metabolism and increase mitochondrial dysfunction, while clinically it is upregulated in MCI patients' plasma and has been reported as a promising early diagnostic marker to predict progression to AD (Ansari et al., 2019).
Table 4 illustrates updates about the recently discovered mitomiRs and their potential role in AD progression.
Table 4.
Mitochondrial miRNAs in relation to mitochondrial dysfunctions.
| miRNA | Role | Altered level | References |
|---|---|---|---|
| miR-34a | Allocated in the mitochondria and Promotes apoptosis and dysfunction of mitochondria | Upregulated | Sarkar et al., 2019 |
| miR-107 | Decreased mitochondrial functions and morphological changes | Downregulated | Rech et al., 2019 |
| miR-126 | Reduces aerobic respiration | Upregulated | Tomasetti et al., 2014 |
| miR 23a/23b | Affects mitochondrial biogenesis | Downregulated | Tang, 2016 |
| miR-132 | Reduces aerobic respiration | Downregulated | Weinberg et al., 2015 |
| miR-181a | Localized in mitochondria. Promotes apoptosis and dysfunction of mitochondria | Upregulated | Giuliani et al., 2018 |
| miR-762 | Reduces ATP production | Downregulated | Yan et al., 2019 |
| miR-210 | Clinical marker for MCI and AD Targets mitochondrial iron sulfur cluster homolog |
Upregulated | Siedlecki-Wullich et al., 2019 |
| miR-424 | Suppression of ATP levels and mitochondrial integrity | Upregulated | Duarte et al., 2014 |
| miR-425 | Mitochondrial dysfunction and increased ROS production | Upregulated | Hu Y.-B. et al., 2019 |
AD, Sleep Disorders and Circadian Rhythm
Sleep: The Complex Process of Circadian Rhythm in Action
Circadian rhythms are 24 h cycles that maintain homeostasis in different body tissues. Circadian rhythm is controlled by the suprachiasmatic nucleus (SCN) which is found in the hypothalamus, and it synchronizes multiple functions as sleep/wake cycle, metabolism, thermoregulation, and hormonal regulation. This molecular clockwork involves genetically encoded autoregulatory feedback loops that provide a 24-h period of circadian oscillation (Park et al., 2020).
The core loop of this molecular clock is driven by a heterodimeric transcriptional activator that is composed of two clock genes: circadian locomotor output cycle kaput (CLOCK) and brain–muscle–arnt-like protein 1 (BMAL1). These heterodimers accelerate E-box-mediated transcription and increase gene expression of negative regulators; [Periods (PERs: PER1, PER2, and PER3) and Cryptochromes (CRYs: CRY1 and CRY2)] and circadian output genes. Expressed and dimerized PER: CRY represses the transcriptional activity of CLOCK:BMAL1, hence, downregulate their own gene transcription. It takes 24 h to complete such loop cycle and the accurate generation of 24 h cycles is regulated by post-translational modifications that includes phosphorylation, ubiquitination, and acetylation. Majorly, the phosphorylation of PER proteins by casein kinase Iε (CKI ε) and glycogen synthase kinase-3β (GSK-3 β) promotes PER nuclear translocation, thereby provide proper completion of the cycle (Eide et al., 2005).
The SCN also contains gamma-aminobutyric acid (GABA) and arginine vasopressin (AVP) neurons that send inhibitory signals to the paraventricular nucleus of the hypothalamus (Reghunandanan and Reghunandanan, 2006). This activates melatonin secretion by the pineal gland and when it binds to MT1 and MT2 receptors, it inhibits the firing of the SCN. Hence, melatonin promotes sleep and resets the circadian pacemaker (Aulinas, 2000).
AD and Sleep Disturbances
Circadian rhythms that regulate sleep gradually weaken with aging causing disturbances in sleep quality and cognitive alterations. However, circadian rhythms are markedly disturbed in AD all through the disease course. Day-time agitation, night insomnia, restlessness and sun-downing are among the characteristic changes observed in AD and they worsen with AD progression to affect around 42–52% of AD patients. Such sleep changes are majorly attributed to disruptions in the precise cascade of circadian rhythm (Todd, 2020).
AD is often associated with changes in the physiological parameters of sleep that include decrease in total sleep time and efficiency, prolonged sleep time stage 1 and stage 2 sleep, lesser time in deeper sleep, increased REM sleep latency and decreased REM sleep, together with decreased density of eye movement activity (Weldemichael and Grossberg, 2010).
Another major disruption of AD-sleep disturbance is the “Sundowning.” This condition refers to a delirium-like status usually occurring at late-afternoon and till dawn. Behavioral components of sundowning can include loud vocalization, wandering, physical aggression, maladaptive physical behaviors, and overall agitation. The prevalence of sundowning in AD ranges from 12 to 25%. The increased frequency for AD agitation at night imply that these chronobiologic changes are affected by disruptions in “the timing” of physiological events (Weldemichael and Grossberg, 2010).
Causes and Deeper Look to miRNAs Role
The main reason for the sleep-wake disrupted cycle in AD is related to alterations in the suprachiasmatic nucleus (SCN) and melatonin secretion. Moreover, expression of MT1 receptors is decreased in the SCN of AD patients resulting in reduced melatonin production and disappearance of melatonin rhythm (Todd, 2020). Till date, the molecular mechanisms underlying these disturbances are still not fully resolved which leave a tight margin for effective therapeutic intervention. Interestingly, collected data from both human and animal studies show that sleep disturbances are not only a consequence of AD progression, but may also precede AD symptoms onset and may contribute to AD pathology through affecting Tau and Aβ deposition and clearance from the brain (Musiek et al., 2015). However, despite the high frequency of sleep disturbances during AD course, there is an obvious lack of data that specifically discusses the molecular basis of AD-sleep disorders, apart from other AD-disruptions.
In a recent study, deletion of the master gene BMAL1 abrogated all circadian functions, leading to complete loss of day-night rhythmicity of sleep (Musiek et al., 2015). Simultaneously, sleep deprivation was found to change the expression of clock genes and BMAL1/CLOCK heterodimers binding, thus altering clock function.
Growing evidence now confirm that mature miRNAs are crucial for the fine-tuning of circadian rhythm regulations that also include sleep (Table 5 enlists some of the miRNAs reported to regulate Circadian Rhythm in different tissues).
Table 5.
MicroRNAs regulating core clock components.
| miRNA | Affected clock component | References |
|---|---|---|
| miR-449a | PER gene | Hansen et al., 2011 |
| miR-103 | Chen et al., 2019 | |
| miR-125a-3p | Ma et al., 2020 | |
| miR-192 | Garufi et al., 2016 | |
| miR-194 | Nagel et al., 2009 | |
| miR-24 | Oyama et al., 2017 | |
| miR-29a/b/c | ||
| miR-30a | ||
| miR-34a | Ma et al., 2020 | |
| miR-211 | Ebox, NPAS2, CLOCK Genes | |
| miR-107 | ||
| miR-124 | ||
| miR-141 | Na et al., 2009 | |
| miR-17-5p | Gao et al., 2016 | |
| miR-182-5p | Ma et al., 2020 | |
| miR-19b | Na et al., 2009 | |
| miR-199b-5p | Yuan et al., 2020 | |
| miR-206 | Garufi et al., 2016 | |
| miR-10 | BMAL1 Gene | Zheng et al., 2021 |
| miR-135b | Zheng et al., 2021 | |
| miR-142-3p | Tan et al., 2012; Shende et al., 2013 | |
| miR-155 | Shende et al., 2011 | |
| miR-211 | Ma et al., 2020 | |
| miR-27b-3p | ||
| miR-494 | Shende et al., 2011 | |
| miR-106b | CRY Genes | Zheng et al., 2018 |
| miR-181a,d | Na et al., 2009 | |
| miR-185 | Lee et al., 2013 | |
| miR-340 | Zheng et al., 2018 | |
| miR-219 | ROR output genes | Ma et al., 2020 |
| miR-142-3p | Shende et al., 2013 | |
| miR-125a-3p | (CKI ε) and (GSK-3 β) | Zheng et al., 2018 |
| miR-181a | RORα Genes | Zheng et al., 2018 |
| miR-27a-3p | Zheng et al., 2018 | |
| miR183-5p | Dambal et al., 2015 | |
| miR-450-5p | Zheng et al., 2018 | |
| miR-19b | Linnstaedt et al., 2020 | |
| miR-503-5p | Zheng et al., 2018 | |
| miR-126a-3p | E4BP4, DBP Genes | Cheng et al., 2007 |
Moreover, although differential expression of multiple miRNA panels has been detected in AD, it is of high interest that consensus miRNAs that regulate key genes in AD pathogenesis, are also involved in sleep-circadian disorders. For example, miR-219 is reported to be overexpressed in postmortem brain tissues of AD patients and interestingly, miR-219 regulates Tau phosphorylation and targets GSK-3 which is vital for phosphorylating PER genes (Kinoshita et al., 2020). Moreover, miR-219 modulates CLOCK-BMAL1 complex. Similarly, miR-132 modulates GSK-3 and Tau Phosphorylation and it is downregulated in AD neurons (El Fatimy et al., 2018).
MiR-125a, miR-125b, miR-146a have high diagnostic potential to predict AD progression, where they modulate Tau hyperphosphorylation (Nagaraj et al., 2019), inflammatory responses and autophagy in microglia and astrocytes, while regulating PER genes in the circadian clock (Lee et al., 2016; Liang et al., 2021). MiR-125 also regulates the cholinergic functions via modulating CLOCK gene. Meanwhile miR-146a is associated with short sleep (Davis et al., 2007; Karabulut et al., 2019) and shows rhythmic expression. It is worth mentioning that miR-146a shows remarkable potential as a diagnostic AD biomarker (Siedlecki-Wullich et al., 2019). Differential expression of miR-34a has also been detected in the brains and blood of AD patients, where it regulates genes involved in memory formation, amyloid precursor protein metabolism and Tau phosphorylation (Sarkar et al., 2016). Intersecting with these functions, miR-34a also regulates PER1 and PER2 genes. MiR-29 family members are established as potential indicators of AD status. In line, miR-29a, b, c are also involved in regulating PER genes, β-secretase (BACE1) mRNA and Aβ accumulation (Müller et al., 2016). Similarly, miR-107 is down regulated in the temporal cortex and plasma of AD patients and it targets both CLOCK gene and BACE1 expression.
MiR-155 is among the most well-studied microRNAs in AD-neuroinflammation. The high expression level of miR-155 in 3xTg AD animal model is accompanied with hyperactivation of microglia and astrocytes, to trigger inflammatory mediators. Moreover, miR-155 contributes to AD through activating different T cell functions during inflammation. Clinically, miR-155 is upregulated in human AD brains and it aggravates neuroinflammation (Kou et al., 2020). Similarly, miR-181 family impaired levels are also repeatedly reported in AD and are linked to accumulated plaque formation in the temporal complex and regulation of inflammation cytokine TNF-α and IL-6 (Kou et al., 2020). Both miR-155 and miR-181 have been linked to sleep disorders in AD as well as other neurodegenerative disorders (Slota and Booth, 2019).
Despite the scarcity of studies exploring miRNAs in AD- associated sleep disorders, the previously mentioned data strongly suggests the association between miRNAs dysregulations and sleep disorders together with other AD molecular disturbances. Clarifying this interconnected network with more studies, might unravel the molecular basis of sleep disturbances in AD and provide novel approaches for better and earlier management of AD.
Pain: the Underestimated Companion in Alzheimer's Disease?
Alzheimer's is often co-morbid with chronic pain, where chronic pain prevalence is around 45.8% in AD (van Kooten et al., 2016). Pain that results from damage of body tissues is classified as nociceptive pain, while pain resulting from direct consequence of a lesion or disease that affects the somatosensory system” is classified as neuropathic pain (NP) (Merskey et al., 1994). Over the course of AD, AD patients can experience both nociceptive and neuropathic pain. And while, peripheral neuropathic lesions do not always cause chronic pain, central neuropathic pain is often chronic (Husebo et al., 2016).
Due to their inability to communicate and express their needs, pain feeling is often overlooked in AD patients. However, pain is more prevalent with severe dementia (Rajkumar et al., 2017), and its intensity is positively correlated with dementia severity (Whitlock et al., 2017). Impaired pain perception that is mediated by central nervous system is crucial in chronic pain. Simultaneously, central sensitization of pain processing pathways impacts cognition and emotional processing. These pathological interactions imply the presence of deeper link between chronic pain and AD progressiveness.
The “locus coeruleus” (LC) modulates pain and is a key player in chronic pain processing (Taylor and Westlund, 2017). It also innervates most brain areas and is the principal site of norepinephrine (NE) synthesis and neurotransmission in the CNS. The mechanistic processing underlying chronic pain is a complex issue that still needs to be resolved, nevertheless when experienced with AD.
Increasing evidence now shows that chronic pain and AD share disrupted function and structure of the LC. Moreover, both human and animal reports show that chronic pain induces microglial activation and neuroinflammation in hippocampus, anterior cingulate cortex, amygdala, nucleus accumbens, thalamus, and sensory cortex (Cao et al., 2019). This is associated with elevated release of inflammatory cytokines; TNF-α, IL-6, IL-1β that trigger disruptions in synaptic remodeling, brain connectivity and network function. Interestingly, microglial activation and neuroinflammation are found to precede cognitive decline in AD patients, implying its participation in aggravating AD disease (Fan et al., 2017). The chronic activation of microglia induces synaptic loss, and this occurs in both AD and chronic pain. Moreover, the prolonged exposure to amyloid depositions, has also been found to activate microglia, resulting in excessive secretion of synaptic-toxic cytokines, and tau accumulation that eventually cause synaptic loss and neuronal death (Cao et al., 2019). Besides microglial activation, disrupted autophagy process has been increasingly reported as a significant contributor to both chronic pain and AD (Yin et al., 2018).
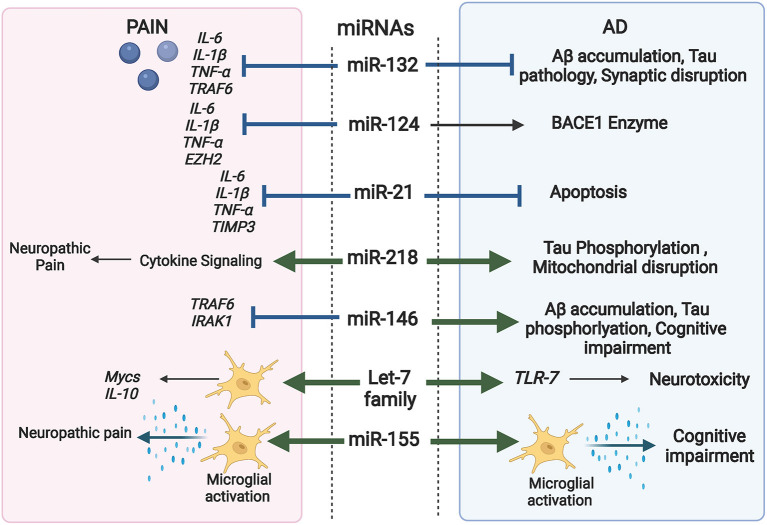
It is established that miRNAs are key players in modulating macromolecular complexes in neurons, glia, and immune cells. They regulate signals interconnecting neuro-immune network in the pain pathway and are crucial modulators of inflammation and autophagy pathways; two major factors in AD progression as well as chronic pain (López-González et al., 2017; Bernaus et al., 2020). As a result, miRNAs are now considered as significant “master switches” in chronic pain and AD. Figure 4 highlights the overlapping role of some miRNAs in AD and pain.
Figure 4.
Comprehensive illustration of the overlapping role of some miRNAs in both AD and Pain and their targeted genes. (Created with BioRender.com).
For instance, miR-132 participates in regulating inflammation and is a negative regulator of the inflammatory response in PC12 cells. IL-1β, IL-6, TNF-α and TNF receptor associated factor 6 (TRAF6) are potential targets of miR-132 (Kou et al., 2020). Recently upregulation of miR-132 in WBCs of patients were associated with chronic neuropathic pain (Leinders et al., 2016). Interestingly, miR-132 is significantly reduced in the brains of AD patients and deletion of miR-132 in mice hastened Aβ accumulation, and tau pathology via modulating the synaptic proteins (Xu N. et al., 2019). In line, the down regulation of miR-132 was found to inhibit the level of hippocampal acetylcholinesterase (AChE), impacting both cognitive function and synaptic plasticity. The downstream molecules responsible for miR-132 actions involve both p250 GTPase Activating Protein (p250GAP) and Methyl CpG-Binding Protein 2 (MeCP2) (Ye et al., 2016).
MiR-155 is one of the most prominent miRNAs that are differentially expressed in AD serum and brain. Recent reports confirm that miR-155 has significant impact on development of pain and pain hypersensitivity, where up-regulation of miR-155 is accompanied by enhanced activation of microglia and consequent production of inflammatory mediators (Kou et al., 2020). Moreover, miR-155 is a significant regulator of neuropathic pain via targeting serum and glucocorticoid regulated protein kinase 3 (SGK3), an important protein involved in phosphorylation cascades (Liu et al., 2015).
Similarly, Let-7 is critical for maintaining microglial function in inflammation-mediated injury (Roush and Slack, 2008). Let-7a inhibits the expression of inflammatory cytokines via activation of apoptosis signal-regulating kinase 1 (ASK1), IL-10 and Mycs in microglia (Song and Lee, 2015). Meanwhile, let-7 miRNAs are found to be differentially and specifically released in CSF of AD patients (Derkow et al., 2018). MiR-124 is another brain-enriched miRNA involved in the regulation of neural development. Increasing reports spot its role as a remarkable alleviator of neuropathic pain, via inhibiting (IL-6, IL-1β, and TNF-α) protein expressions and direct targeting of Enhancer of zeste homolog 2 EZH2 gene (Zhang et al., 2019). Moreover, miR-124 regulates BACE1 enzyme and is found to decrease gradually with AD progression (An et al., 2017). Most importantly, updated studies are now spotting miR-124 as a therapeutic target for its role in modulating inflammation in the central nervous system and brain injuries (Xu et al., 2021).
MiR-21 is another interesting miRNA regulator of pain that is increasingly reported as a significant contributor against AD progressive events. MiR-21 can inhibit cell apoptosis induced by Aβ1−42 via modulating cell death protein 4 (PDCD4)/phosphatidylinositol 3-kinase PI3K/AKT/GSK-3β pathway in the CNS (Feng et al., 2018). Meanwhile circulating miR-21-5p was significantly upregulated in the plasma of AD patients and was negatively correlated to cognitive impairment (Giuliani et al., 2021). Simultaneously, disturbed level of miR-21 was observed in diverse neuropathic pain models (Zhong et al., 2019) where it targets metalloproteinase-3 (TIMP3) and chemokines C-C motif ligand 1 (CCL1) that consequently evoke cytokine production of TNF-α, IL-1β, and IL-6 and aggravate neuroinflammation.
MiR-146a-5p is reported to modulate immune response and reduce inflammation by targeting both TRAF6 and interleukin-1 receptor-associated kinase 1 (IRAK1) IRAK1 in macrophages, and astrocytes. Lu et al. (2015) reported the protective effect of miR-146a against SNL-induced neuropathic pain by suppressing TRAF6 signaling in the spinal cord (Lu et al., 2015). Meanwhile, multiple studies linked miR-146a-5p with cognitive deterioration, where its upregulation in the CNS is associated with increased expression of Aβ, Taup38, and Reactive oxygen species (ROS) through targeting MAPK signaling (Alexandrov et al., 2014).
One of the emerging and promising miRNAs in both AD pathology and chronic pain is miR-218. Recently miR-218 upregulation was found to contribute to AD progression by enhancing Tau phosphorylation and disrupting mitochondrial respiratory chain through modulating Wnt signaling pathway (Gugliandolo et al., 2020; Wu et al., 2020). Meanwhile, downregulation of miR-218 was proved effective in suppressing central neuropathic pain via regulating cytokine signaling (Li and Zhao, 2016).
Elucidating the regulatory role of miRNAs on pain sensitization, neuropathic pain and the interplay with cognition and behavior alterations in AD, can unleash new resolutions on the pathophysiology of chronic pain in AD. Considering the common role of miRNAs in regulating chronic pain and its possible contribution in worsening cognitive impairment, miRNAs might also provide a prognostic tool to predict susceptible AD patients.
MicroRNAs as Diagnostic Biomarkers of AD
Current AD diagnostic markers and methods are applicable in the late stages of AD. Ultimately, they can be classified into (1) Neuropsychological tests: which are cognitive assessments used to quantitatively assess the degree of cognitive impairment and its progression over time. This method, however, has limited specificity and sensitivity and is majorly affected by patients' educational levels. (2) Neuroimaging examination: such as Magnetic resonance imaging (MRI) and fluorodeoxyglucose (FDG)-positron emission tomography (PET) that monitor the pathological and functional alterations before severe appearance of cognitive impairment (Calvillo and Irimia, 2020). However, this method is hugely limited by the high cost.
Recent trials reported the high diagnostic potential of Amyloid and Phosphorylated Tau /Aβ ratio in CSF samples. Yet again, this technique is highly invasive and requires well-trained personnel for sample acquisitions. The detection of neuro filament light chains (NFL) in biological samples is emerging as good neuronal biomarkers, however, techniques and kits for reliable detection are still limited, and more research studies are needed to explore their ability to differentiate between different neurodegenerative diseases (Gaetani et al., 2019).
In the previous decade, the reported differential expressions of miRNAs in animal models of AD have opened the field to unleash the potential of miRNAs as promising diagnostic biomarkers for multiple neurodegenerative diseases. Moreover, continuous reports show that specific miRNAs are detected in the biofluids of AD patients with different levels from normal controls, along with their correlation to AD observed pathological and cognitive changes (Wei et al., 2020). A point that can be optimized to monitor AD progression. Furthermore, circulating miRNAs that are collected from serum or plasma resist environmental degradation and can provide a cheaper and less-invasive diagnostic means, compared to neuroimaging and CSF examinations. Ongoing research continuously reveals the remarkable diagnostic potential of miRNAs in AD. Table 6 presents an updated summary for different miRNAs that have shown promising potential as AD diagnostic markers.
Table 6.
MicroRNAs with diagnostic potential in human AD studies.
| miRNA | Sample | Expression in AD | Studied cohort | Specificity/sensitivity | References |
|---|---|---|---|---|---|
| miRNA-483-5p | Plasma | Increased | 20 MCI 20 AD 20 Healthy controls |
95, 90%85, 90% | Sabry et al., 2020 |
| miR-26a | Serum | Decreased | 121 AD, 48 HC | 57, 85% | Guo et al., 2017 |
| miR-34a | Plasma CSF |
Increased Decreased |
25 AD, 27 HC 10 AD, 10 HC |
74, 84% | Cosín-Tomás et al., 2017 |
| miR-125b | Serum | Increased | 105 AD, 155 HC | 68, 80% | Tan et al., 2014a |
| miR-181a | Serum | Decreased | 121 AD, 86 HC | 73, 72% | Ansari et al., 2019 |
| miR-181c | Serum | Decreased | 150 HC, 105 AD | 64, 75% | Manzano-Crespo et al., 2019 |
| miR-206 | Serum | Decreased | 66 MCI, 33 HC | 100, 70% | Xie et al., 2015 |
| miR-342-3p | Serum | Decreased | 158 AD, 155 HC | 70, 81 | Tan et al., 2014b |
| miR-34c | Blood mononuclear cells | Increased | 110 AD, 123 HC | 74, 84% | Bhatnagar et al., 2014 |
| miR-133b | Serum | Decreased | 98 AD, 105 HC | 74.3, 90% | Yang et al., 2019 |
| miR-433a | Serum | Decreased | 32 AD, 12 HC | 0.82 AUC | Wang and Zhang, 2020 |
| miR-103 | Plasma | Decreased | 120 AD, 120 PD, 120 HC |
84, 80% | Wang J. et al., 2020 |
| miR-106 | Serum | Decreased | 56 AD, 50 HC | 62, 94% | Madadi et al., 2020 |
| miR-9 | Serum | Decreased | 36 AD female patients 38 HC females | Not available | Souza et al., 2020 |
| miR-29cmiR-19b | Serum | Decreased | 45 AD, 40HC | Not available | Wu et al., 2017 |
| miR-455-3p | Serum | Increased | 11 AD, 18 MCI, 20 Healthy | AUC 79% | Kumar et al., 2017 |
| miR-let-7b | CSF | Increased | 41 memory complaints, 36 MCI, 17 AD patients | Addition to total and p- tau panel led to improved AUC to 91.6% | Liu et al., 2018 |
Spotting the most prominent miRNAs, recent studies have investigated the predictive potential of miRNAs in longitudinal studies over time. In an updated study, plasma miR-206 level stands out as a good prognostic biomarker to monitor MCI progression to AD over a period of 5 years (Kenny et al., 2019). In the same context, Ansari et al. (2019) showed the ability of miR-181a and miR-146a blood levels to predict whether MCI would progress to AD or remain stable after 2 years monitoring (Ansari et al., 2019).
Specific single miRNAs have shown consistent differentially expressed levels in various AD samples. For instance, Persistent miR-26b upregulation is reported in both serum and whole blood AD samples (Galimberti et al., 2014). Yang et al. (2018), recently spotted the diagnostic ability of exosomal miR-384, as its expression in serum of AD and non-AD patients differ significantly. Furthermore, serum level of exosomal miR-384 showed potent differential diagnostic ability for AD and Parkinson's dementia, as well as for AD and vascular dementia, with sensitivity/specificity indices of 97.2/100% and 99.1/100%, respectively (Yang et al., 2018).
One of the most emerging and promising miRNAs in AD diagnosis is miR-455-3p. Where serum miR-455-3p is upregulated in AD patients as compared to both MCI subjects and healthy controls. Interestingly, this finding is also confirmed in fibroblast cells, postmortem AD brains examination at different Braak stages (Kumar et al., 2017; Kumar and Reddy, 2018), as well as in CSF samples of sporadic AD subjects (Kumar and Reddy, 2021). Analyzing the mechanistic effects of miR-455-3p in AD showed that APP is a validated target of miR-455-3p. Whereas, elevated levels of miR-455-3p help to alleviate Aβ toxicity, improve mitochondrial dynamics and synaptic activity (Kumar and Reddy, 2019).
Another important concept is the utilization of miRNA panels of multiple miRNAs as a signature for AD. This can provide higher accuracy, specificity, and sensitivity. In this context, miR-181c, miR-92a-3p, and miR-210-3p showed remarkable ability to differentiate between AD and healthy controls. Moreover, this signature panel showed promising results in predicting MCI progression to AD after monitoring patients for 1 to 11 years (Siedlecki-Wullich et al., 2019). Additionally, a serum 9-miNRA signature panel including; miR-26a-5p, hsa-miR-181c-3p, hsa-miR-126-5p, hsa-miR-22-3p, hsa-miR-148b-5p, hsa-miR-106b-3p, hsa-miR-6119-5p, hsa-miR-1246, and hsa-miR-660-5p was recently reported to differentiate between AD and healthy controls with an AUC ROC reaching 85%, in a study that comprised one of the biggest cohorts of AD patients (Guo et al., 2017).
Although, miRNAs possess the properties of good diagnostic tools, miRNA use is still faced by several limitations. Namely, the parameters used for groups classification, subjects' inclusion and exclusion criteria, miRNAs extraction and quantification methods and reliable reference genes, vary widely among different labs. This is why finding a method to standardize miRNAs isolation and quantification protocols, as well as recruiting wider and more diverse populations can provide deeper knowledge about miRNA diagnostic potential.
The Therapeutic Potential of miRNAs in AD
The cumulative knowledge on miRNAs functions, strongly implies their potential as an emerging therapeutic possibility for multiple AD pathologies. Generally, therapeutic intervention using miRNAs can take three approaches: (1) Using natural or synthetic compounds to modulate the expression of miRNAs (2) inhibiting the function of a particularly targeted miRNA using complementary single stranded antisense oligonucleotide (ASO). (3) Readjusting the expression of the targeted miRNA using miRNA mimics. So far, several trials have been conducted to adjust miRNA expression in AD animal models as well as cell lines.
Numerous studies used natural or synthetic compounds to readjust miRNAs expressions. Among the most characteristic trials, Osthole is reported to modulate the expression of miR-9 and miR 101a-3p in APP/PS1 mice, SHSY-5Y cells as well as neural stem cells. Consequently, this resulted in decreased cellular apoptosis, improved cell growth, improved learning and memory capacities, together with prevention of Aβ aggregation (Li et al., 2017). Berberine was also reported to remodulate the expression of miR-188 in BV2 cells, resulting in remarkable inhibition of Apoptosis (Chen et al., 2020). The Chinese herbal Tiaoxin recipe was recently reported to inhibit miR-34a expression in APPswe/PS1ΔE9 mouse AD model leading to decreased Aβ aggregation (Hu Y.-R. et al., 2019). A combination of resveratrol and curcumin was also successfully reported to readjust the expression of cortical let-7c in rats and PC-12 cells, which led to significant reduction of neuroinflammation along with readjustment of β-secretase and APP expressions (Zaky et al., 2017). In the same context, Sun et al. (2020), recently reported the ability of Dexmedetomidine to upregulate miR-129 in NIH Swiss mice, which led to cognitive improvement through targeting YAP1 gene (Sun et al., 2020).
The continuous advances in drug delivery and biotechnology help in the rapid progress toward specific miRNA targeting and delivery. Interestingly, a recent study that used engineered exosomes to deliver miR-29 in a rat model of induced Aβ pathology, showed significant improvement of memory deficits (Jahangard et al., 2020). Meanwhile, photoactivation of targeted miRNAs, miRNAs sponges that sequester a specific miRNA and inclusion of miRNAs in labeled liposomes or cubosomes to cross the blood brain barrier are all emerging innovative means that can be optimized in the near future for miRNA delivery (López-González et al., 2017).
Discussion and Outline
Current Hopes, Challenges, and Future Perspectives
There is no doubt miRNAs provide a novel opportunity to tackle AD disease, either through their potential as earlier diagnostic markers or through modulating their expression to reinstate the relevant AD target genes.
One interesting point is that along our research, numerous miRNAs apparently act as multi-faceted modulators of AD-interconnected signaling pathways. A point that can be advantageous in re-adjusting the different impaired pathways affected by miRNAs dysregulations. Furthermore, despite their nature as epigenetic modulators that can influenced by environmental and ethnic variations, multiple miRNA panels were repeatedly reported in different AD studies of variable regions and populations. For instance, miRNAs including but not restricted to; miR-155, miR-146a, miR-34a, miR-29, miR-132, miR-483-5p, and miR-181 are reported as differentially expressed in multiple AD samples of Chinese, American, European populations (Cheng et al., 2015; Wei et al., 2020; Huaying et al., 2021; Siedlecki-Wullich et al., 2021), as well as emerging studies from Africa and middle eastern regions (Sabry et al., 2020). At the same time, these miRNAs have also been introduced as crucial modulators of Aβ and Tau pathologies, inflammatory, mitochondrial, and synaptic dysfunctions that accompany AD progression.
An emerging hot topic in AD pathology is the role of mitochondrial miRNAs in mitochondrial dysfunction and mitophagy that are highly likely to precede actual AD neuronal damage (John et al., 2020). And while recent studies show miR-7, miR-155, miR-210 and miR-125 as crucial in mitochondrial impairments accompanying cancer (Ortega et al., 2020), we are still scratching the surface concerning mitomiRs in “inflamm-aging “axis and AD.
Another hot topic in AD dilemma is the promising role of lactate level in astrocytes and its possible effect on reinstating cognitive decline of AD. Updated studies spot the role of miRNAs; miR-34a (Sarkar et al., 2016), exosomal miR-137 (Thomas et al., 2020) and miR-124-3p (Xu S.-Y. et al., 2019) in controlling astrocytes lactate shuttle in different brain disorders and the consequent impact of adjusting their levels on cognitive improvement. Further future studies can definitely elucidate more facts about miRNAs role in astrocyte metabolism during AD and unravel ways to use this knowledge.
That being said, it is inevitable to state that till date, miRNAs bench studies are still faced by several obstacles that need to be resolved before being capable of efficiently serving AD clinical applications.
For a beginning, till now, there are limitations in comparing miRNA studies due to the basal differences in AD stages of the recruited patients and unclear specificity to AD. Moreover, communication difficulties with AD patients and their caregivers, comprise a major challenge by itself to recruit bigger populations of AD patients. To improve comparability in miRNAs studies, classifying the recruited patients in comparable groups according to their disease stages, followed by correlating their pathology with their miRNA's profiles can be more reliable.
Another challenge is the scarcity of data originating from middle and low-income countries and the relatively small populations size in AD clinical studies that majorly originate from well-developed countries. This can be resolved by enhancing the means of reaching out to affected communities, performing more longitudinal studies, together with utilizing advanced screening techniques, and raising awareness about the critical significance of participating in AD studies.
On the technical level, an important challenge in miRNAs studies, is the wide variability in sample collection, storage, standardization against reference controls, and analytical protocols (Mushtaq et al., 2016). Multi-center comparisons and universal standardization of the used techniques would to a large extent improve miRNAs reliability.
Indeed, efficient use of miRNAs for AD diagnosis, still needs to be preceded by precise standardization of sampling and quantification protocols and proper staging of AD. It might also be useful to rely on multiple panels of miRNA or combinations of miRNAs together with other fluid biomarkers. Progresses in this aspect are continuously accomplished. Interestingly, some registered clinical trials are already running for example: (NCT03388242) that is using a combination of microRNAs and proteins with different expression patterns to distinguish between normal control, MCI and AD patients (Zuo, 2019).
Therapeutically, miRNAs use is challenged by some issues. Firstly, miRNAs target multiple genes and hence, manipulating one miRNA can result in unwanted effects on other sides (Siedlecki-Wullich et al., 2021). However, advances in in silico analysis and neuroinformatics can help face this problem. Besides, conducting more studies from diverse populations can contribute to getting more precise data that guide researchers to the proper beginning concerning their targeted population. Moreover, identification of the individual roles of specific miRNAs, as well as the collective role of multiple miRNAs in AD should be explored.
Another challenge is the difficulty of efficient miRNA delivery to the brain. However, the increasing advances in nanotechnology (Abdel-Mageed et al., 2021) and targeted delivery of biological materials have been successfully reported in some AD animal models.
Interestingly, miR-155 is already entering a clinical phase trial as a therapeutic compound registered by ©Miragen Therapeutics against Amyotrophic lateral sclerosis (Chakraborty et al., 2021). We believe, the near future can witness similar clinical successes to face the challenging pathologies of AD.
Author Contributions
NA and FN drafted the manuscript. AZ and AB performed critical editing. NA, MA, and FN participated in constructive outline, discussions, and editing. All authors read and approved the final manuscript.
Funding
This work was partially funded by the Science and Technology Development Fund (STDF), Egypt (Grant Number 39436) Youth Research Project.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Glossary
Abbreviations
- BACE1
Beta-Secretase 1
- HEY2
Hairy/Enhancer-Of-Split Related With YRPW Motif Protein 2
- APP
Amyloid beta precursor protein
- MAPK
Mitogen-activated protein kinase
- DUSP6
Dual specificity phosphatase 6
- PPP1CA
Protein Phosphatase 1 Catalytic Subunit Alpha
- PTBP2
Polypyrimidine Tract Binding Protein 2
- MeCP2
Methyl-CpG Binding Protein 2
- PTEN
Phosphatase And Tensin Homolog
- Fyn
Proto-oncogene tyrosine-protein kinase Fyn
- ERK1
extracellular signal-regulated kinase 1
- ERK2
extracellular signal-regulated kinase 2
- CREB
cAMP response element-binding protein
- BDNF
Brain-derived neurotrophic factor
- EphB2
Ephrin type-B receptor 2
- GRIA2
Glutamate ionotropic receptor AMPA type subunit 2
- GRIN2B
Glutamate Ionotropic Receptor NMDA Type Subunit 2B
- Syt-1
Synaptotagmin1
- Syt-1A
Syntaxin 1A
- NPTX1
Neuronal Pentraxin 1
- AMPAR
α-amino-3-hydroxy-5-methyl-4-isoxazolepropionicacid receptor
- GRIN2A
Glutamate Ionotropic Receptor NMDA Type Subunit 2A
- EPHA4
Ephrin type-A receptor 4
- Nrn1
Neuritin 1
- Arpc3
Actin Related Protein 2/3 Complex Subunit 3
- CPLX1
Complexin 1
- CPLX2
Complexin2
- STIM2
Stromal Interaction Molecule 2
- REST
RE1 Silencing Transcription Factor
- CX3CL1
C-X3-C Motif Chemokine Ligand 1
- CX3CR1
C-X3-C Motif Chemokine Receptor 1
- NPAS2
Neuronal PAS Domain Protein 2
- DBP
D-Box Binding PAR BZIP Transcription Factor
- Mycs
MYC Proto-Oncogene
- YAP1
Yes1 Associated Transcriptional Regulator
- C-MAf
C musculoaponeurotic fibrosarcoma
- CCM1
Cerebral cavernous malformations 1
- NOX2
NADPH oxidase 2
- NOX4
NADPH oxidase 4
- C/EBP-α
CCAAT/enhancer-binding protein alpha
- TREM2
Triggering receptor expressed on myeloid cells 2.
References
- 2020 Alzheimer's Disease Facts and Figures. (2020) 2020 Alzheimer's Disease Facts and figures. Alzheimer's Dementia J. Alzheimer's Assoc. 16 391–460. 10.1002/alz.12068. [Google Scholar]
- Abdel-Mageed H. M., AbuelEzz N. Z., Radwan R. A., Mohamed S. A. (2021). Nanoparticles in nanomedicine: a comprehensive updated review on current status, challenges and emerging opportunities. J. Microencapsul. 38, 414–436. 10.1080/02652048.2021.1942275 [DOI] [PubMed] [Google Scholar]
- Agostini M., Tucci P., Steinert J. R., Shalom-Feuerstein R., Rouleau M., Aberdam D., et al. (2011). MicroRNA-34a regulates neurite outgrowth, spinal morphology, and function. Proc. Natl. Acad. Sci. U.S.A. 108, 21099–21104. 10.1073/pnas.1112063108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandrov P. N., Dua P., Lukiw W. J. (2014). Up-Regulation of miRNA-146a in progressive, age-related inflammatory neurodegenerative disorders of the human CNS. Front. Neurol. 5:181. 10.3389/fneur.2014.00181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandrov P. N., Zhao Y., Jones B. M., Bhattacharjee S., Lukiw W. J. (2013). Expression of the phagocytosis-essential protein TREM2 is down-regulated by an aluminum-induced miRNA-34a in a murine microglial cell line. J. Inorg. Biochem. 128, 267–269. 10.1016/j.jinorgbio.2013.05.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alsharafi W. A., Xiao B., Li J. (2016). MicroRNA-139-5p negatively regulates NR2A-containing NMDA receptor in the rat pilocarpine model and patients with temporal lobe epilepsy. Epilepsia 57, 1931–1940. 10.1111/epi.13568 [DOI] [PubMed] [Google Scholar]
- Amakiri N., Kubosumi A., Tran J., Reddy P. H. (2019). Amyloid beta and MicroRNAs in Alzheimer's disease. Front. Neurosci. 13:430. 10.3389/fnins.2019.00430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amin L., Harris D. A. (2021). Aβ receptors specifically recognize molecular features displayed by fibril ends and neurotoxic oligomers. Nat. Commun. 12:3451. 10.1038/s41467-021-23507-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- An F., Gong G., Wang Y., Bian M., Yu L., Wei C. (2017). MiR-124 acts as a target for Alzheimer's disease by regulating BACE1. Oncotarget 8, 114065–114071. 10.18632/oncotarget.23119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ansari A., Maffioletti E., Milanesi E., Marizzoni M., Frisoni G. B., Blin O., et al. (2019). MiR-146a and miR-181a are involved in the progression of mild cognitive impairment to Alzheimer's disease. Neurobiol. Aging 82, 102–109. 10.1016/j.neurobiolaging.2019.06.005 [DOI] [PubMed] [Google Scholar]
- Aulinas A. (2000). Physiology of the pineal gland and melatonin, in Endotext, eds K. R. Feingold, B. Anawalt, A. Boyce, G. Chrousos, W. W. de Herder, K. Dhatariya (MDText.com, Inc). Available online at: http://www.ncbi.nlm.nih.gov/books/NBK550972/
- Banzhaf-Strathmann J., Benito E., May S., Arzberger T., Tahirovic S., Kretzschmar H., et al. (2014). MicroRNA-125b induces tau hyperphosphorylation and cognitive deficits in Alzheimer's disease. EMBO J. 33, 1667–1680. 10.15252/embj.201387576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbato C., Giacovazzo G., Albiero F., Scardigli R., Scopa C., Ciotti M. T., et al. (2020). Cognitive decline and modulation of Alzheimer's disease-related genes after inhibition of MicroRNA-101 in mouse hippocampal neurons. Mol. Neurobiol. 57, 3183–3194. 10.1007/s12035-020-01957-8 [DOI] [PubMed] [Google Scholar]
- Basavaraju M., de Lencastre A. (2016). Alzheimer's disease: presence and role of microRNAs. Biomol. Concepts 7, 241–252. 10.1515/bmc-2016-0014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beardmore R., Hou R., Darekar A., Holmes C., Boche D. (2021). The locus coeruleus in aging and Alzheimer's disease: a post-mortem and brain imaging review. J. Alzheimer's Dis. 83, 5–22. 10.3233/JAD-210191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekris L. M., Yu C.-E., Bird T. D., Tsuang D. W. (2010). Genetics of Alzheimer disease. J. Geriatr. Psychiatry Neurol. 23, 213–227. 10.1177/0891988710383571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben Halima S., Siegel G., Rajendran L. (2016). miR-186 in Alzheimer's disease: a big hope for a small RNA? J. Neurochem. 137, 308–311. 10.1111/jnc.13573 [DOI] [PubMed] [Google Scholar]
- Bernaus A., Blanco S., Sevilla A. (2020). Glia crosstalk in neuroinflammatory diseases. Front. Cell. Neurosci. 14:209. 10.3389/fncel.2020.00209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatnagar S., Chertkow H., Schipper H. M., Yuan Z., Shetty V., Jenkins S., et al. (2014). Increased microRNA-34c abundance in Alzheimer's disease circulating blood plasma. Front. Mol. Neurosci. 7:2. 10.3389/fnmol.2014.00002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharjee S., Zhao Y., Dua P., Rogaev E. I., Lukiw W. J. (2016). MicroRNA-34a-mediated down-regulation of the microglial-enriched triggering receptor and phagocytosis-sensor TREM2 in age-related macular degeneration. PLoS ONE 11:e0150211. 10.1371/journal.pone.0150211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birch A. M., Katsouri L., Sastre M. (2014). Modulation of inflammation in transgenic models of Alzheimer's disease. J. Neuroinflamm. 11:25. 10.1186/1742-2094-11-25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biundo F., Del Prete D., Zhang H., Arancio O., D'Adamio L. (2018). A role for tau in learning, memory and synaptic plasticity. Sci. Rep. 8:3184. 10.1038/s41598-018-21596-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blasko I., Veerhuis R., Stampfer-Kountchev M., Saurwein-Teissl M., Eikelenboom P., Grubeck-Loebenstein B. (2000). Costimulatory effects of interferon-gamma and interleukin-1beta or tumor necrosis factor alpha on the synthesis of Abeta1-40 and Abeta1-42 by human astrocytes. Neurobiol. Dis. 7(6 Pt B), 682–689. 10.1006/nbdi.2000.0321 [DOI] [PubMed] [Google Scholar]
- Bosch M., Hayashi Y. (2012). Structural plasticity of dendritic spines. Curr. Opin. Neurobiol. 22, 383–388. 10.1016/j.conb.2011.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boscher E., Husson T., Quenez O., Laquerrière A., Marguet F., Cassinari K., et al. (2019). Copy number variants in miR-138 as a potential risk factor for early-onset Alzheimer's disease. J. Alzheimer's Dis. 68, 1243–1255. 10.3233/JAD-180940 [DOI] [PubMed] [Google Scholar]
- Burgos K., Malenica I., Metpally R., Courtright A., Rakela B., Beach T., et al. (2014). Profiles of extracellular miRNA in cerebrospinal fluid and serum from patients with Alzheimer's and Parkinson's diseases correlate with disease status and features of pathology. PLoS ONE. 9:e94839. 10.1371/journal.pone.0094839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callahan L. M., Vaules W. A., Coleman P. D. (1999). Quantitative decrease in synaptophysin message expression and increase in cathepsin D message expression in Alzheimer disease neurons containing neurofibrillary tangles. J. Neuropathol. Exp. Neurol. 58, 275–287. 10.1097/00005072-199903000-00007 [DOI] [PubMed] [Google Scholar]
- Calvillo M., Irimia A. (2020). Neuroimaging and psychometric assessment of mild cognitive impairment after traumatic brain injury. Front. Psychol. 11:1423. 10.3389/fpsyg.2020.01423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao S., Fisher D. W., Yu T., Dong H. (2019). The link between chronic pain and Alzheimer's disease. J. Neuroinflamm. 16, 204. 10.1186/s12974-019-1608-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty C., Sharma A. R., Sharma G., Lee S.-S. (2021). Therapeutic advances of miRNAs: a preclinical and clinical update. J. Adv. Res. 28, 127–138. 10.1016/j.jare.2020.08.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheignon C., Tomas M., Bonnefont-Rousselot D., Faller P., Hureau C., Collin F. (2017). Oxidative stress and the amyloid beta peptide in Alzheimer's disease. Redox Biol. 14, 450–464. 10.1016/j.redox.2017.10.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Zhang B., Yang L., Bai Y.-G., Song J.-B., Ge Y.-L., et al. (2019). BMAL1 disrupted intrinsic diurnal oscillation in rat cerebrovascular contractility of simulated microgravity rats by altering circadian regulation of miR-103/CaV1.2 signal pathway. Int. J. Mol. Sci. 20:3947. 10.3390/ijms20163947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M., Li L., Liu C., Song L. (2020). Berberine attenuates Aβ-induced neuronal damage through regulating miR-188/NOS1 in Alzheimer's disease. Mol. Cell. Biochem. 474, 285–294. 10.1007/s11010-020-03852-1 [DOI] [PubMed] [Google Scholar]
- Chen X. Q., Mobley W. C. (2019). Alzheimer disease pathogenesis: insights from molecular and cellular biology studies of oligomeric Aβ and tau species. Front. Neurosci. 13:659. 10.3389/fnins.2019.00659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng H.-Y. M., Papp J. W., Varlamova O., Dziema H., Russell B., Curfman J. P., et al. (2007). MicroRNA modulation of circadian-clock period and entrainment. Neuron 54, 813–829. 10.1016/j.neuron.2007.05.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng L., Doecke J. D., Sharples R. A., Villemagne V. L., Fowler C. J., Rembach A., et al. (2015). Prognostic serum miRNA biomarkers associated with Alzheimer's disease shows concordance with neuropsychological and neuroimaging assessment. Mol. Psychiatry 20, 1188–1196. 10.1038/mp.2014.127 [DOI] [PubMed] [Google Scholar]
- Cissé M., Halabisky B., Harris J., Devidze N., Dubal D. B., Sun B., et al. (2011). Reversing EphB2 depletion rescues cognitive functions in Alzheimer model. Nature 469, 47–52. 10.1038/nature09635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cogswell J. P., Ward J., Taylor I. A., Waters M., Shi Y., Cannon B., et al. (2008). Identification of miRNA changes in Alzheimer's disease brain and CSF yields putative biomarkers and insights into disease pathways. J. Alzheimer's Dis. 14, 27–41. 10.3233/JAD-2008-14103 [DOI] [PubMed] [Google Scholar]
- Cosín-Tomás M., Antonell A., Lladó A., Alcolea D., Fortea J., Ezquerra M., et al. (2017). Plasma miR-34a-5p and miR-545-3p as early biomarkers of Alzheimer's disease: potential and LImitations. Mol. Neurobiol. 54, 5550–5562. 10.1007/s12035-016-0088-8 [DOI] [PubMed] [Google Scholar]
- Cui J. G., Li Y. Y., Zhao Y., Bhattacharjee S., Lukiw W. J. (2010). Differential regulation of interleukin-1 receptor-associated kinase-1 (IRAK-1) and IRAK-2 by microRNA-146a and NF-kappaB in stressed human astroglial cells and in Alzheimer disease. J. Biol. Chem. 285, 38951–38960. 10.1074/jbc.M110.178848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dambal S., Shah M., Mihelich B., Nonn L. (2015). The microRNA-183 cluster: the family that plays together stays together. Nucleic Acids Res. 43, 7173–7188. 10.1093/nar/gkv703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davare M. A., Hell J. W. (2003). Increased phosphorylation of the neuronal L-type Ca2+ channel Cav1.2 during aging. Proc. Natl. Acad. Sci. U.S.A. 100, 16018–16023. 10.1073/pnas.2236970100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis C. J., Bohnet S. G., Meyerson J. M., Krueger J. M. (2007). Sleep loss changes microRNA levels in the brain: a possible mechanism for state-dependent translational regulation. Neurosci. Lett. 422, 68–73. 10.1016/j.neulet.2007.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng M., Zhang Q., Wu Z., Ma T., He A., Zhang T., et al. (2020). Mossy cell synaptic dysfunction causes memory imprecision via miR-128 inhibition of STIM2 in Alzheimer's disease mouse model. Aging Cell 19:e13144. 10.1111/acel.13144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derkow K., Rössling R., Schipke C., Krüger C., Bauer J., Fähling M., et al. (2018). Distinct expression of the neurotoxic microRNA family let-7 in the cerebrospinal fluid of patients with Alzheimer's disease. PLoS ONE 13:e0200602. 10.1371/journal.pone.0200602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Carlo M., Giacomazza D., San Biagio P. L. (2012). Alzheimer's disease: biological aspects, therapeutic perspectives and diagnostic tools. J. Phys. Condens. Matt. Inst. Phys. J. 24:244102. 10.1088/0953-8984/24/24/244102 [DOI] [PubMed] [Google Scholar]
- Di J., Cohen L. S., Corbo C. P., Phillips G. R., El Idrissi A., Alonso A. D. (2016). Abnormal tau induces cognitive impairment through two different mechanisms: synaptic dysfunction and neuronal loss. Sci. Rep. 6:20833. 10.1038/srep20833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorostkar M. M., Zou C., Blazquez-Llorca L., Herms J. (2015). Analyzing dendritic spine pathology in Alzheimer's disease: problems and opportunities. Acta Neuropathol. 130, 1–19. 10.1007/s00401-015-1449-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du X., Wang X., Geng M. (2018). Alzheimer's disease hypothesis and related therapies. Transl. Neurodegener. 7:2. 10.1186/s40035-018-0107-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duarte F. V., Palmeira C. M., Rolo A. P. (2014). The Role of microRNAs in mitochondria: small players acting wide. Genes 5, 865–886. 10.3390/genes5040865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edbauer D., Neilson J. R., Foster K. A., Wang C.-F., Seeburg D. P., Batterton M. N., et al. (2010). Regulation of synaptic structure and function by FMRP-associated microRNAs miR-125b and miR-132. Neuron 65, 373–384. 10.1016/j.neuron.2010.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eide E. J., Woolf M. F., Kang H., Woolf P., Hurst W., Camacho F., et al. (2005). Control of mammalian circadian rhythm by CKIepsilon-regulated proteasome-mediated PER2 degradation. Mol. Cell. Biol. 25, 2795–2807. 10.1128/MCB.25.7.2795-2807.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Fatimy R., Li S., Chen Z., Mushannen T., Gongala S., Wei Z., et al. (2018). MicroRNA-132 provides neuroprotection for tauopathies via multiple signaling pathways. Acta Neuropathol. 136, 537–555. 10.1007/s00401-018-1880-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fakhoury M. (2018). Microglia and astrocytes in alzheimer's disease: implications for therapy. Curr. Neuropharmacol. 16, 508–518. 10.2174/1570159X15666170720095240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan L., Mao C., Hu X., Zhang S., Yang Z., Hu Z., et al. (2020). New insights into the pathogenesis of Alzheimer's disease. Front. Neurol. 10:1312. 10.3389/fneur.2019.01312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan L. W., Pang Y. (2017). Dysregulation of neurogenesis by neuroinflammation: key differences in neurodevelopmental and neurological disorders. Neural Regen. Res. 12, 366–371. 10.4103/1673-5374.202926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Z., Brooks D. J., Okello A., Edison P. (2017). An early and late peak in microglial activation in Alzheimer's disease trajectory. Brain J. Neurol. 140, 792–803. 10.1093/brain/aww349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatoba O., Itokazu T., Yamashita T. (2021). Complement cascade functions during brain development and neurodegeneration. FEBS J. 10.1111/febs.15772. [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- Feng M.-G., Liu C.-F., Chen L., Feng W.-B., Liu M., Hai H., et al. (2018). MiR-21 attenuates apoptosis-triggered by amyloid-β via modulating PDCD4/ PI3K/AKT/GSK-3β pathway in SH-SY5Y cells. Biomed. Pharmacother. 101, 1003–1007. 10.1016/j.biopha.2018.02.043 [DOI] [PubMed] [Google Scholar]
- Francis P. T., Palmer A. M., Sims N. R., Bowen D. M., Davison A. N., Esiri M. M., et al. (1985). Neurochemical studies of early-onset Alzheimer's disease. Possible influence on treatment. N. Engl. J. Med. 313, 7–11. 10.1056/NEJM198507043130102 [DOI] [PubMed] [Google Scholar]
- Fulop T., Tripathi S., Rodrigues S., Desroches M., Bunt T., Eiser A., et al. (2021). Targeting impaired antimicrobial immunity in the brain for the treatment of Alzheimer's disease. Neuropsychiatr. Dis. Treat. 17, 1311–1339. 10.2147/NDT.S264910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaetani L., Blennow K., Calabresi P., Di Filippo M., Parnetti L., Zetterberg H. (2019). Neurofilament light chain as a biomarker in neurological disorders. J. Neurol. Neurosurg. Psychiatr. 90, 870–881. 10.1136/jnnp-2018-320106 [DOI] [PubMed] [Google Scholar]
- Galimberti D., Villa C., Fenoglio C., Serpente M., Ghezzi L., Cioffi S. M. G., et al. (2014). Circulating miRNAs as potential biomarkers in Alzheimer's disease. J. Alzheimer's Dis. 42, 1261–1267. 10.3233/JAD-140756 [DOI] [PubMed] [Google Scholar]
- Gao Q., Zhou L., Yang S.-Y., Cao J.-M. (2016). A novel role of microRNA 17-5p in the modulation of circadian rhythm. Sci. Rep. 6:30070. 10.1038/srep30070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garufi C., Giacomini E., Torsello A., Sperduti I., Melucci E., Mottolese M., et al. (2016). Gender effects of single nucleotide polymorphisms and miRNAs targeting clock-genes in metastatic colorectal cancer patients (mCRC). Sci. Rep. 6:34006. 10.1038/srep34006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geekiyanage H., Jicha G. A., Nelson P. T., Chan C. (2012). Blood serum miRNA: non-invasive biomarkers for Alzheimer's disease. Exp. Neurol. 235, 491–496. 10.1016/j.expneurol.2011.11.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- German D. C., Manaye K. F., White C. L., 3rd, Woodward D. J., McIntire D. D., Smith W. K., Kalaria R. N., et al. (1992). Disease-specific patterns of locus coeruleus cell loss. Ann. Neurol. 32, 667–676. 10.1002/ana.410320510 [DOI] [PubMed] [Google Scholar]
- Giri M. Zhang M. and L.ü, Y. (2016). Genes associated with Alzheimer's disease: an overview and current status. Clin. Interv. Aging 11, 665–681. 10.2147/CIA.S105769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giuliani A., Cirilli I., Prattichizzo F., Mensà E., Fulgenzi G., Sabbatinelli J., et al. (2018). The mitomiR/Bcl-2 axis affects mitochondrial function and autophagic vacuole formation in senescent endothelial cells. Aging 10, 2855–2873. 10.18632/aging.101591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giuliani A., Gaetani S., Sorgentoni G., Agarbati S., Laggetta M., Matacchione G., et al. (2021). Circulating inflamma-miRs as potential biomarkers of cognitive impairment in patients affected by Alzheimer's disease. Front. Aging Neurosci. 13:647015. 10.3389/fnagi.2021.647015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giusti S. A., Vogl A. M., Brockmann M. M., Vercelli C. A., Rein M. L., Trümbach D., et al. (2014). MicroRNA-9 controls dendritic development by targeting REST. ELife 3:e02755. 10.7554/eLife.02755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grubman A., Choo X. Y., Chew G., Ouyang J. F., Sun G., Croft N. P., et al. (2021). Transcriptional signature in microglia associated with Aβ plaque phagocytosis. Nat. Commun. 12:3015. 10.1038/s41467-021-23111-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grundke-Iqbal I., Iqbal K., Tung Y. C., Quinlan M., Wisniewski H. M., Binder L. I. (1986). Abnormal phosphorylation of the microtubule-associated protein tau (tau) in Alzheimer cytoskeletal pathology. Proc. Natl. Acad. Sci. U.S.A. 83, 4913–4917. 10.1073/pnas.83.13.4913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Q., Yu D., Hu Z., Liu X., Yang Y., Luo Y., et al. (2015). MiR-26a and miR-384-5p are required for LTP maintenance and spine enlargement. Nat. Commun. 6:6789. 10.1038/ncomms7789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guedes J., Cardoso A. L. C., Pedroso de Lima M. C. (2013). Involvement of microRNA in microglia-mediated immune response. Clin. Dev. Immunol. 2013:186872. 10.1155/2013/186872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guedes J. R., Custódia C. M., Silva R. J., de Almeida L. P., Pedroso de Lima M. C., Cardoso A. L. (2014). Early MiR-155 upregulation contributes to neuroinflammation in Alzheimer's disease triple transgenic mouse model. Hum. Mol. Genet. 23, 6286–6301. 10.1093/hmg/ddu348 [DOI] [PubMed] [Google Scholar]
- Guedes J. R., Santana I., Cunha C., Duro D., Almeida M. R., Cardoso A. M., et al. (2016). MicroRNA deregulation and chemotaxis and phagocytosis impairment in Alzheimer's disease. Alzheimer's Dementia 3, 7–17. 10.1016/j.dadm.2015.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gugliandolo A., Chiricosta L., Boccardi V., Mecocci P., Bramanti P., Mazzon E. (2020). MicroRNAs modulate the pathogenesis of Alzheimer's disease: an in silico analysis in the human brain. Genes 11, E983. 10.3390/genes11090983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo R., Fan G., Zhang J., Wu C., Du Y., Ye H., et al. (2017). A 9-microRNA signature in serum serves as a noninvasive biomarker in early diagnosis of Alzheimer's disease. J. Alzheimer's Dis. 60, 1365–1377. 10.3233/JAD-170343 [DOI] [PubMed] [Google Scholar]
- Guo Y., Hong W., Wang X., Zhang P., Körner H., Tu J., et al. (2019). MicroRNAs in microglia: how do micrornas affect activation, inflammation, polarization of microglia and mediate the interaction between microglia and glioma? Front. Mol. Neurosci. 12:125. 10.3389/fnmol.2019.00125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampel H., Mesulam M.-M., Cuello A. C., Farlow M. R., Giacobini E., Grossberg G. T., et al. (2018). The cholinergic system in the pathophysiology and treatment of Alzheimer's disease. Brain 141, 1917–1933. 10.1093/brain/awy132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen K. F., Sakamoto K., Obrietan K. (2011). MicroRNAs: a potential interface between the circadian clock and human health. Genome Med. 3:10. 10.1186/gm224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He D., Tan J., Zhang J. (2017). MiR-137 attenuates Aβ-induced neurotoxicity through inactivation of NF-κB pathway by targeting TNFAIP1 in Neuro2a cells. Biochem. Biophys. Res. Commun. 490, 941–947. 10.1016/j.bbrc.2017.06.144 [DOI] [PubMed] [Google Scholar]
- Hébert S. S., Horré K., Nicolaï L., Papadopoulou A. S., Mandemakers W., Silahtaroglu A. N., et al. (2008). Loss of microRNA cluster miR-29a/b-1 in sporadic Alzheimer's disease correlates with increased BACE1/beta-secretase expression. Proc. Natl. Acad. Sci. U.S.A. 105, 6415–6420. 10.1073/pnas.0710263105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemonnot A. L., Hua J., Ulmann L., Hirbec H. (2019). Microglia in Alzheimer disease: well-known targets and new opportunities. Front. Aging Neurosci. 11:233. 10.3389/fnagi.2019.00233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hettinger J. C., Lee H., Bu G., Holtzman D. M., Cirrito J. R. (2018). AMPA-ergic regulation of amyloid-β levels in an Alzheimer's disease mouse model. Mol. Neurodegener. 13:22. 10.1186/s13024-018-0256-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higaki S., Muramatsu M., Matsuda A., Matsumoto K., Satoh J.-I., Michikawa M., et al. (2018). Defensive effect of microRNA-200b/c against amyloid-beta peptide-induced toxicity in Alzheimer's disease models. PLoS ONE 13:e0196929. 10.1371/journal.pone.0196929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hippius H., Neundörfer G. (2003). The discovery of Alzheimer's disease. Dial. Clin. Neurosci. 5, 101–108. 10.31887/DCNS.2003.5.1/hhippius [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong S., Dissing-Olesen L., Stevens B. (2016). New insights on the role of microglia in synaptic pruning in health and disease. Curr. Opin. Neurobiol. 36, 128–134. 10.1016/j.conb.2015.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoogendijk W. J., Pool C. W., Troost D., van Zwieten E., Swaab D. F. (1995). Image analyser-assisted morphometry of the locus coeruleus in Alzheimer's disease, Parkinson's disease and amyotrophic lateral sclerosis. Brain 118 (Pt. 1), 131–143. 10.1093/brain/118.1.131 [DOI] [PubMed] [Google Scholar]
- Hu Y.-B., Zhang Y.-F., Wang H., Ren R.-J., Cui H.-L., Huang W.-Y., et al. (2019). MiR-425 deficiency promotes necroptosis and dopaminergic neurodegeneration in Parkinson's disease. Cell Death Dis. 10:589. 10.1038/s41419-019-1809-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y.-R., Xing S.-L., Chen C., Shen D.-Z., Chen J.-L. (2019). Tiaoxin recipe, a Chinese herbal formula, inhibits microRNA-34a expression in the APPswe/PS1ΔE9 mouse model of Alzheimer's disease. J. Integr. Med. 17, 404–409. 10.1016/j.joim.2019.09.002 [DOI] [PubMed] [Google Scholar]
- Hu Z., Yu D., Gu Q., Yang Y., Tu K., Zhu J., et al. (2014). MiR-191 and miR-135 are required for long-lasting spine remodelling associated with synaptic long-term depression. Nat. Commun. 5:3263. 10.1038/ncomms4263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z., Zhao J., Hu T., Luo Y., Zhu J., Li Z. (2015). MiR-501-3p mediates the activity-dependent regulation of the expression of AMPA receptor subunit GluA1. J. Cell Biol. 208, 949–959. 10.1083/jcb.201404092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huaying C., Xing J., Luya J., Linhui N., Di S., Xianjun D. (2021). A signature of five long non-coding rnas for predicting the prognosis of Alzheimer's disease based on competing endogenous RNA networks. Front. Aging Neurosci. 12:598606. 10.3389/fnagi.2020.598606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt D. L., Castillo P. E. (2012). Synaptic plasticity of NMDA receptors: mechanisms and functional implications. Curr. Opin. Neurobiol. 22, 496–508. 10.1016/j.conb.2012.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husebo B. S., Achterberg W., Flo E. (2016). Identifying and managing pain in people with Alzheimer's disease and other types of dementia: a systematic review. CNS Drugs 30, 481–497. 10.1007/s40263-016-0342-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iqbal K., Liu F., Gong C.-X., Grundke-Iqbal I. (2010). Tau in Alzheimer disease and related tauopathies. Curr. Alzheimer Res. 7, 656–664. 10.2174/156720510793611592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahangard Y., Monfared H., Moradi A., Zare M., Mirnajafi-Zadeh J., Mowla S. J. (2020). Therapeutic effects of transplanted exosomes containing miR-29b to a rat model of Alzheimer's disease. Front. Neurosci. 14:564. 10.3389/fnins.2020.00564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasińska M., Miłek J., Cymerman I. A., Łeski S., Kaczmarek L., Dziembowska M. (2016). MiR-132 regulates dendritic spine structure by direct targeting of matrix metalloproteinase 9 mRNA. Mol. Neurobiol. 53, 4701–4712. 10.1007/s12035-015-9383-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayadev S., Case A., Alajajian B., Eastman A. J., Moller T., Garden G. A. (2013). Presenilin 2 influences miR146 level and activity in microglia. J. Neurochem. 127, 592–599. 10.1111/jnc.12400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- John A., Kubosumi A., Reddy P. H. (2020). Mitochondrial microRNAs in aging and neurodegenerative diseases. Cells 9:1345. 10.3390/cells9061345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jun G. R., Chung J., Mez J., Barber R., Beecham G. W., Bennett D. A., et al. (2017). Transethnic genome-wide scan identifies novel Alzheimer's disease loci. Alzheimer's Dementia 13, 727–738. 10.1016/j.jalz.2016.12.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karabulut S., Korkmaz Bayramov K., Bayramov R., Ozdemir F., Topaloglu T., Ergen E., et al. (2019). Effects of post-learning REM sleep deprivation on hippocampal plasticity-related genes and microRNA in mice. Behav. Brain Res. 361, 7–13. 10.1016/j.bbr.2018.12.045 [DOI] [PubMed] [Google Scholar]
- Karch C. M., Goate A. M. (2015). Alzheimer's disease risk genes and mechanisms of disease pathogenesis. Biol. Psychiatry 77, 43–51. 10.1016/j.biopsych.2014.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenny A., McArdle H., Calero M., Rabano A., Madden S. F., Adamson K., et al. (2019). Elevated plasma microRNA-206 levels predict cognitive decline and progression to dementia from mild cognitive impairment. Biomolecules 9:E734. 10.3390/biom9110734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keren-Shaul H., Spinrad A., Weiner A., Matcovitch-Natan O., Dvir-Szternfeld R., Ulland T. K., et al. (2017). A unique microglia type associated with restricting development of alzheimer's disease. Cell. 169, 1276–1290. 10.1016/j.cell.2017.05.018 [DOI] [PubMed] [Google Scholar]
- Kimura T., Whitcomb D. J., Jo J., Regan P., Piers T., Heo S., et al. (2014). Microtubule-associated protein tau is essential for long-term depression in the hippocampus. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 369:20130144. 10.1098/rstb.2013.0144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita C., Okamoto Y., Aoyama K., Nakaki T. (2020). MicroRNA: a key player for the interplay of circadian rhythm abnormalities, sleep disorders and neurodegenerative diseases. Clocks Sleep 2, 282–307. 10.3390/clockssleep2030022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kou X., Chen D., Chen N. (2020). The regulation of microRNAs in Alzheimer's disease. Front. Neurol. 11:288. 10.3389/fneur.2020.00288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Reddy P. H. (2016). Are circulating microRNAs peripheral biomarkers for Alzheimer's disease? Biochim. Biophys. Acta. 1862, 1617–1627. 10.1016/j.bbadis.2016.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Reddy P. H. (2018). MicroRNA-455-3p as a potential biomarker for Alzheimer's disease: an update. Front. Aging Neurosci. 10:41. 10.3389/fnagi.2018.00041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Reddy P. H. (2019). A new discovery of MicroRNA-455-3p in Alzheimer's disease. J. Alzheimer's Dis. 72, S117–S130. 10.3233/JAD-190583 [DOI] [PubMed] [Google Scholar]
- Kumar S., Reddy P. H. (2020). The role of synaptic microRNAs in Alzheimer's disease. Biochim. Biophys. Acta Mol. Basis Dis. 1866:165937. 10.1016/j.bbadis.2020.165937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Reddy P. H. (2021). Elevated levels of MicroRNA-455-3p in the cerebrospinal fluid of Alzheimer's patients: a potential biomarker for Alzheimer's disease. Biochim. Biophys. Acta Mol. Basis Dis. 1867:166052. 10.1016/j.bbadis.2020.166052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Vijayan M., Reddy P. H. (2017). MicroRNA-455-3p as a potential peripheral biomarker for Alzheimer's disease. Hum. Mol. Gen. 26, 3808–3822. 10.1093/hmg/ddx267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkle B. W., Grenier-Boley B., Sims R., Bis J. C., Damotte V., Naj A. C., et al. (2019). Genetic and environmental risk in AD/defining genetic, polygenic and environmental risk for Alzheimer's disease consortium (GERAD/PERADES), genetic meta-analysis of diagnosed Alzheimer's disease identifies new risk loci and implicates Aβ, tau, immunity and lipid processing. Nat. Genet. 51, 414–430. 10.1038/s41588-019-0358-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang A., Grether-Beck S., Singh M., Kuck F., Jakob S., Kefalas A., et al. (2016). MicroRNA-15b regulates mitochondrial ROS production and the senescence-associated secretory phenotype through sirtuin 4/SIRT4. Aging 8, 484–505. 10.18632/aging.100905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H.-M., Kim T. S., Jo E.-K. (2016). MiR-146 and miR-125 in the regulation of innate immunity and inflammation. BMB Rep. 49, 311–318. 10.5483/BMBRep.2016.49.6.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K.-H., Kim S.-H., Lee H.-R., Kim W., Kim D.-Y., Shin J.-C., et al. (2013). MicroRNA-185 oscillation controls circadian amplitude of mouse Cryptochrome 1 via translational regulation. Mol. Biol. Cell. 24, 2248–2255. 10.1091/mbc.e12-12-0849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S.-T., Chu K., Jung K.-H., Kim J. H., Huh J.-Y., Yoon H., et al. (2012). MiR-206 regulates brain-derived neurotrophic factor in Alzheimer disease model. Ann. Neurol. 72, 269–277. 10.1002/ana.23588 [DOI] [PubMed] [Google Scholar]
- Lei X., Lei L., Zhang Z., Zhang Z., Cheng Y. (2015). Downregulated miR-29c correlates with increased BACE1 expression in sporadic Alzheimer's disease. Int. J. Clin. Exp. Pathol. 8, 1565–1574. [PMC free article] [PubMed] [Google Scholar]
- Leinders M., Üçeyler N., Pritchard R. A., Sommer C., Sorkin L. S. (2016). Increased miR-132-3p expression is associated with chronic neuropathic pain. Exp. Neurol. 283(Pt A), 276–286. 10.1016/j.expneurol.2016.06.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letellier M., Elramah S., Mondin M., Soula A., Penn A., Choquet D., et al. (2014). MiR-92a regulates expression of synaptic GluA1-containing AMPA receptors during homeostatic scaling. Nat. Neurosci. 17, 1040–1042. 10.1038/nn.3762 [DOI] [PubMed] [Google Scholar]
- Li F., Wei G., Bai Y., Li Y., Huang F., Lin J., et al. (2015). MicroRNA-574 is involved in cognitive impairment in 5-month-old APP/PS1 mice through regulation of neuritin. Brain Res. 1627, 177–188. 10.1016/j.brainres.2015.09.022 [DOI] [PubMed] [Google Scholar]
- Li J., Wang H. (2018). MiR-15b reduces amyloid-β accumulation in SH-SY5Y cell line through targetting NF-κB signaling and BACE1. Biosci. Rep. 38:BSR20180051. 10.1042/BSR20180051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Sun Q., Li Y., Yang Y., Yang Y., Chang T., et al. (2015). Overexpression of SIRT1 induced by resveratrol and inhibitor of miR-204 suppresses activation and proliferation of microglia. J. Mol. Neurosci. 56, 858–867. 10.1007/s12031-015-0526-5 [DOI] [PubMed] [Google Scholar]
- Li L., Zhao G. (2016). Downregulation of microRNA-218 relieves neuropathic pain by regulating suppressor of cytokine signaling 3. Int. J. Mol. Med. 37, 851–858. 10.3892/ijmm.2016.2455 [DOI] [PubMed] [Google Scholar]
- Li S.-H., Gao P., Wang L.-T., Yan Y.-H., Xia Y., Song J., et al. (2017). osthole stimulated neural stem cells differentiation into neurons in an Alzheimer's disease cell model via upregulation of MicroRNA-9 and rescued the functional impairment of hippocampal neurons in APP/PS1 transgenic mice. Front. Neurosci. 11:340. 10.3389/fnins.2017.00340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang C., Zou T., Zhang M., Fan W., Zhang T., Jiang Y., et al. (2021). MicroRNA-146a switches microglial phenotypes to resist the pathological processes and cognitive degradation of Alzheimer's disease. Theranostics 11, 4103–4121. 10.7150/thno.53418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lillenes M. S., Rabano A., Støen M., Riaz T., Misaghian D., Møllersen L., et al. (2016). Altered DNA base excision repair profile in brain tissue and blood in Alzheimer's disease. Mol. Brain 9:61. 10.1186/s13041-016-0237-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linnstaedt S. D., Rueckeis C. A., Riker K. D., Pan Y., Wu A., Yu S., et al. (2020). MicroRNA-19b predicts widespread pain and posttraumatic stress symptom risk in a sex-dependent manner following trauma exposure. Pain 161, 47–60. 10.1097/j.pain.0000000000001709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippi G., Steinert J. R., Marczylo E. L., D'Oro S., Fiore R., Forsythe I. D., et al. (2011). Targeting of the Arpc3 actin nucleation factor by miR-29a/b regulates dendritic spine morphology. J. Cell Biol. 194, 889–904. 10.1083/jcb.201103006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J. Z. Erlich Y. and Pickrell J. K. (2017). Case-control association mapping by proxy using family history of disease. Nat. Genet. 49, 325–331. 10.1038/ng.3766 [DOI] [PubMed] [Google Scholar]
- Liu S., Zhu B., Sun Y., Xie X. (2015). MiR-155 modulates the progression of neuropathic pain through targeting SGK3. Int. J. Clin. Exp. Pathol. 8, 14374–14382. [PMC free article] [PubMed] [Google Scholar]
- Liu Y., He X., Li Y., Wang T. (2018). Cerebrospinal fluid CD4+ T lymphocyte-derived miRNA-let-7b can enhances the diagnostic performance of Alzheimer's disease biomarkers. Biochem. Biophys. Res. Commun. 495, 1144–1150. 10.1016/j.bbrc.2017.11.122 [DOI] [PubMed] [Google Scholar]
- Long J. M., Maloney B., Rogers J. T., Lahiri D. K. (2019). Novel upregulation of amyloid-β precursor protein (APP) by microRNA-346 via targeting of APP mRNA 5'-untranslated region: implications in Alzheimer's disease. Mol. Psychiatry 24, 345–363. 10.1038/s41380-018-0266-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-González M. J., Landry M., Favereaux A. (2017). MicroRNA and chronic pain: from mechanisms to therapeutic potential. Pharmacol. Ther. 180, 1–15. 10.1016/j.pharmthera.2017.06.001 [DOI] [PubMed] [Google Scholar]
- Lu Y., Cao D.-L., Jiang B.-C., Yang T., Gao Y.-J. (2015). MicroRNA-146a-5p attenuates neuropathic pain via suppressing TRAF6 signaling in the spinal cord. Brain Behav. Immun. 49, 119–129. 10.1016/j.bbi.2015.04.018 [DOI] [PubMed] [Google Scholar]
- Lugli G., Cohen A. M., Bennett D. A., Shah R. C., Fields C. J., Hernandez A. G., et al. (2015). Plasma exosomal miRNAs in persons with and without Alzheimer disease: altered expression and prospects for biomarkers. PLoS ONE 10:e0139233. 10.1371/journal.pone.0139233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lustbader J. W., Cirilli M., Lin C., Xu H. W., Takuma K., Wang N., et al. (2004). ABAD directly links Abeta to mitochondrial toxicity in Alzheimer's disease. Science 304, 448–452. 10.1126/science.1091230 [DOI] [PubMed] [Google Scholar]
- Ma Q., Mo G., Tan Y. (2020). Micro RNAs and the biological clock: a target for diseases associated with a loss of circadian regulation. Afr. Health Sci. 20, 1887–1894. 10.4314/ahs.v20i4.46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madadi S., Saidijam M., Yavari B., Soleimani M. (2020). Downregulation of serum miR-106b: a potential biomarker for Alzheimer disease. Arch. Physiol. Biochem. 1–5. 10.1080/13813455.2020.1734842 [DOI] [PubMed] [Google Scholar]
- Madadi S., Schwarzenbach H., Saidijam M., Mahjub R., Soleimani M. (2019). Potential microRNA-related targets in clearance pathways of amyloid-β: novel therapeutic approach for the treatment of Alzheimer's disease. Cell Biosci. 9, 91. 10.1186/s13578-019-0354-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manzano-Crespo M., Atienza M., Cantero J. L. (2019). Lower serum expression of miR-181c-5p is associated with increased plasma levels of amyloid-beta 1–40 and cerebral vulnerability in normal aging. Transl. Neurodegener. 8:34. 10.1186/s40035-019-0174-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markesbery W. R. (1997). Oxidative stress hypothesis in Alzheimer's disease. Free Radic. Biol. Med. 23, 134–147. 10.1016/S0891-5849(96)00629-6 [DOI] [PubMed] [Google Scholar]
- Marttinen M., Takalo M., Natunen T., Wittrahm R., Gabbouj S., Kemppainen S., et al. (2018). Molecular mechanisms of synaptotoxicity and neuroinflammation in Alzheimer's disease. Front. Neurosci. 12:963. 10.3389/fnins.2018.00963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masters C. L., Bateman R., Blennow K., Rowe C. C., Sperling R. A., Cummings J. L. (2015). Alzheimer's disease. Nat. Rev. Dis. Primers 1:15056. 10.1038/nrdp.2015.56 [DOI] [PubMed] [Google Scholar]
- Mellios N., Sugihara H., Castro J., Banerjee A., Le C., Kumar A., et al. (2011). MiR-132, an experience-dependent microRNA, is essential for visual cortex plasticity. Nat. Neurosci. 14, 1240–1242. 10.1038/nn.2909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meraz-Rios M. A., Toral-Rios D., Franco-Bocanegra D., Villeda-Hernandez J., Campos-Pena V. (2013). Inflammatory process in Alzheimer's Disease. Front. Integr. Neurosci. 7:59. 10.3389/fnint.2013.00059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merskey H. Bogduk N. International Association for the Study of Pain (1994). Classification of Chronic Pain: Descriptions of Chronic Pain Syndromes and Definitions of Pain Terms. Seattle, WA: IASP Press. [Google Scholar]
- Mocanu M.-M., Nissen A., Eckermann K., Khlistunova I., Biernat J., Drexler D., et al. (2008). The potential for beta-structure in the repeat domain of tau protein determines aggregation, synaptic decay, neuronal loss, and coassembly with endogenous Tau in inducible mouse models of tauopathy. J. Neurosci. 28, 737–748. 10.1523/JNEUROSCI.2824-07.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mucke L., Selkoe D. J. (2012). Neurotoxicity of amyloid β-protein: synaptic and network dysfunction. Cold Spring Harb. Perspect. Med. 2:a006338. 10.1101/cshperspect.a006338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller M., Jäkel L., Bruinsma I. B., Claassen J. A., Kuiperij H. B., Verbeek M. M. (2016). MicroRNA-29a is a candidate biomarker for Alzheimer's disease in cell-free cerebrospinal fluid. Mol. Neurobiol. 53, 2894–2899. 10.1007/s12035-015-9156-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller M., Kuiperij H. B., Claassen J. A., Küsters B., Verbeek M. M. (2014). MicroRNAs in Alzheimer's disease: differential expression in hippocampus and cell-free cerebrospinal fluid. Neurobiol. Aging 35, 152–158. 10.1016/j.neurobiolaging.2013.07.005 [DOI] [PubMed] [Google Scholar]
- Murphy M. P., LeVine H. (2010). Alzheimer's disease and the β-amyloid peptide. J. Alzheimer's Dis. 19:311. 10.3233/JAD-2010-1221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mushtaq G., Greig N. H., Shaik M. M., Tamargo I. A., Kamal M. A. (2016). MiRNAs as circulating biomarkers for Alzheimer's disease and Parkinson's disease. Med. Chem. 12, 217–225. 10.2174/1573406411666151030112140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musiek E. S., Xiong D. D., Holtzman D. M. (2015). Sleep, circadian rhythms, and the pathogenesis of Alzheimer Disease. Exp. Mol. Med. 47:e148. 10.1038/emm.2014.121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Na Y.-J., Sung J. H., Lee S. C., Lee Y.-J., Choi Y. J., Park W.-Y., et al. (2009). Comprehensive analysis of microRNA-mRNA co-expression in circadian rhythm. Exp. Mol. Med. 41, 638–647. 10.3858/emm.2009.41.9.070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaraj S., Want A., Laskowska-Kaszub K., Fesiuk A., Vaz S., Logarinho E., et al. (2021). Candidate Alzheimer's disease biomarker miR-483-5p lowers TAU phosphorylation by direct ERK1/2 repression. Int. J. Mol. Sci. 22:3653. 10.3390/ijms22073653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaraj S., Zoltowska K. M., Laskowska-Kaszub K., Wojda U. (2019). MicroRNA diagnostic panel for Alzheimer's disease and epigenetic trade-off between neurodegeneration and cancer. Ageing Res. Rev. 49, 125–143. 10.1016/j.arr.2018.10.008 [DOI] [PubMed] [Google Scholar]
- Nagel R., Clijsters L., Agami R. (2009). The miRNA-192/194 cluster regulates the period gene family and the circadian clock. FEBS J. 276, 5447–5455. 10.1111/j.1742-4658.2009.07229.x [DOI] [PubMed] [Google Scholar]
- Nunez-Iglesias J., Liu C.-C., Morgan T. E., Finch C. E., Zhou X. J. (2010). Joint genome-wide profiling of miRNA and mRNA expression in Alzheimer's disease cortex reveals altered miRNA regulation. PLoS ONE 5:e8898. 10.1371/journal.pone.0008898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olde Loohuis N. F. M., Ba W., Stoerchel P. H., Kos A., Jager A., Schratt G., et al. (2015). MicroRNA-137 controls AMPA-receptor-mediated transmission and mGluR-dependent LTD. Cell Rep. 11, 1876–1884. 10.1016/j.celrep.2015.05.040 [DOI] [PubMed] [Google Scholar]
- Onuska K. M. (2020). The dual role of microglia in the progression of Alzheimer's disease. J. Neurosci. 40, 1608–1610. 10.1523/JNEUROSCI.2594-19.2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortega M. A., Fraile-Martínez O., Guijarro L. G., Casanova C., Coca S., Álvarez-Mon M., et al. (2020). The regulatory role of mitochondrial MicroRNAs (MitomiRs) in breast cancer: translational implications present and future. Cancers 12:2443. 10.3390/cancers12092443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oyama Y., Bartman C. M., Gile J., Eckle T. (2017). Circadian MicroRNAs in cardioprotection. Curr. Pharm. Des. 23, 3723–3730. 10.2174/1381612823666170707165319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paolicelli R. C., Bolasco G., Pagani F., Maggi L., Scianni M., Panzanelli P., et al. (2011). Synaptic pruning by microglia is necessary for normal brain development. Science 333, 1456–1458. 10.1126/science.1202529 [DOI] [PubMed] [Google Scholar]
- Parisi C., Arisi I., D'Ambrosi N., Storti A. E., Brandi R., D'Onofrio M., et al. (2013). Dysregulated microRNAs in amyotrophic lateral sclerosis microglia modulate genes linked to neuroinflammation. Cell Death Dis. 4:e959. 10.1038/cddis.2013.491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park I., Kim D., Kim J., Jang S., Choi M., Choe H. K., et al. (2020). MicroRNA-25 as a novel modulator of circadian Period2 gene oscillation. Exp. Mol. Med. 52, 1614–1626. 10.1038/s12276-020-00496-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira J. B., Janelidze S., Ossenkoppele R., Kvartsberg H., Brinkmalm A., Mattsson-Carlgren N., et al. (2021). Untangling the association of amyloid-β and tau with synaptic and axonal loss in Alzheimer's disease. Brain 144, 310–324. 10.1093/brain/awaa395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perl D. P. (2010). Neuropathology of Alzheimer's disease. Mount Sinai J. Med. 77, 32–42. 10.1002/msj.20157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picone P., Nuzzo D., Caruana L., Scafidi V., Di Carlo M. (2014). Mitochondrial dysfunction: different routes to Alzheimer's disease therapy. Oxid. Med. Cell. Longev. 2014:e780179. 10.1155/2014/780179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plassman B. L., Langa K. M., Fisher G. G., Heeringa S. G., Weir D. R., Ofstedal M. B., et al. (2007). Prevalence of dementia in the United States: the aging, demographics, and memory study. Neuroepidemiology 29, 125–132. 10.1159/000109998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponomarev E. D., Veremeyko T., Barteneva N., Krichevsky A. M., Weiner H. L. (2011). MicroRNA-124 promotes microglia quiescence and suppresses EAE by deactivating macrophages via the C/EBP-α-PU.1 pathway. Nat. Med. 17, 64–70. 10.1038/nm.2266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Praticò D. (2020). The functional role of microRNAs in the pathogenesis of tauopathy. Cells 9:2262. 10.3390/cells9102262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulkkinen K., Malm T., Turunen M., Koistinaho J., Ylä-Herttuala S. (2008). Hypoxia induces microRNA miR-210 in vitro and in vivo: ephrin-A3 and neuronal pentraxin 1 are potentially regulated by miR-210. FEBS Lett. 582, 2397–2401. 10.1016/j.febslet.2008.05.048 [DOI] [PubMed] [Google Scholar]
- Qian Q., Zhang J., He F.-P., Bao W.-X., Zheng T.-T., Zhou D.-M., et al. (2019). Down-regulated expression of microRNA-338-5p contributes to neuropathology in Alzheimer's disease. FASEB J. 33, 4404–4417. 10.1096/fj.201801846R [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajasethupathy P., Fiumara F., Sheridan R., Betel D., Puthanveettil S. V., Russo J. J., et al. (2009). Characterization of small RNAs in Aplysia reveals a role for miR-124 in constraining synaptic plasticity through CREB. Neuron 63, 803–817. 10.1016/j.neuron.2009.05.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajkumar A. P., Ballard C., Fossey J., Orrell M., Moniz-Cook E., Woods R. T., et al. (2017). Epidemiology of pain in people with dementia living in care homes: longitudinal course, prevalence, and treatment implications. J. Am. Med. Direct. Assoc. 18, 453.e1–453.e6. 10.1016/j.jamda.2017.01.024 [DOI] [PubMed] [Google Scholar]
- Rech M., Kuhn A. R., Lumens J., Carai P., van Leeuwen R., Verhesen W., et al. (2019). AntagomiR-103 and−107 treatment affects cardiac function and metabolism. Mol. Ther. Nucleic Acids 14, 424–437. 10.1016/j.omtn.2018.12.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy P. H., Tonk S., Kumar S., Vijayan M., Kandimalla R., Kuruva C. S. (2017). A Critical evaluation of neuroprotective and neurodegenerative MicroRNAs in Alzheimer's disease. Biochem. Biophys. Res. Commun. 483, 1156–1165. 10.1016/j.bbrc.2016.08.067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reghunandanan V., Reghunandanan R. (2006). Neurotransmitters of the suprachiasmatic nuclei. J. Circadian Rhythms. 4:2. 10.1186/1740-3391-4-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reitz C., Mayeux R. (2014). Alzheimer disease: epidemiology, diagnostic criteria, risk factors and biomarkers. Biochem. Pharmacol. 88, 640–651. 10.1016/j.bcp.2013.12.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reza-Zaldivar E. E., Hernández-Sápiens M. A., Minjarez B., Gómez-Pinedo U., Sánchez-González V. J., Márquez-Aguirre A. L., et al. (2020). Dendritic spine and synaptic plasticity in Alzheimer's disease: a focus on MicroRNA. Front. Cell Dev. Biol. 8:255. 10.3389/fcell.2020.00255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts R., Knopman D. S. (2013). Classification and epidemiology of MCI. Clin. Geriatr. Med. 29, 753–772. 10.1016/j.cger.2013.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocchi A., Moretti D., Lignani G., Colombo E., Scholz-Starke J., Baldelli P., et al. (2019). Neurite-enriched MicroRNA-218 stimulates translation of the GluA2 subunit and increases excitatory synaptic strength. Mol. Neurobiol. 56, 5701–5714. 10.1007/s12035-019-1492-7 [DOI] [PubMed] [Google Scholar]
- Rodriguez-Ortiz C. J., Prieto G. A., Martini A. C., Forner S., Trujillo-Estrada L., LaFerla F. M., et al. (2020). MiR-181a negatively modulates synaptic plasticity in hippocampal cultures and its inhibition rescues memory deficits in a mouse model of Alzheimer's disease. Aging Cell 19:e13118. 10.1111/acel.13118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolyan H., Feike A. C., Upadhaya A. R., Waha A., Van Dooren T., Haass C., et al. (2011). Amyloid-β protein modulates the perivascular clearance of neuronal apolipoprotein E in mouse models of Alzheimer's disease. J. Neural Transmiss. 118, 699–712. 10.1007/s00702-010-0572-7 [DOI] [PubMed] [Google Scholar]
- Roush S., Slack F. J. (2008). The let-7 family of microRNAs. Trends Cell Biol. 18, 505–516. 10.1016/j.tcb.2008.07.007 [DOI] [PubMed] [Google Scholar]
- Sabry R., El Sharkawy R. E., Gad N. M. (2020). MiRNA−483-5p as a potential noninvasive biomarker for early detection of Alzheimer's disease. Egyptian J. Immunol. 27, 59–72. [PubMed] [Google Scholar]
- Samadian M., Gholipour M., Hajiesmaeili M., Taheri M., Ghafouri-Fard S. (2021). The eminent role of microRNAs in the pathogenesis of Alzheimer's disease. Front. Aging Neurosci. 13:641080. 10.3389/fnagi.2021.641080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez J. S., Becker J. A., Jacobs H. I. L., Hanseeuw B. J., Jiang S., Schultz A. P., et al. (2021). The cortical origin and initial spread of medial temporal tauopathy in Alzheimer's disease assessed with positron emission tomography. Sci. Transl. Med. 13:eabc0655. 10.1126/scitranslmed.abc0655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sang S., Pan X., Chen Z., Zeng F., Pan S., Liu H., et al. (2018). Thiamine diphosphate reduction strongly correlates with brain glucose hypometabolism in Alzheimer's disease, whereas amyloid deposition does not. Alzheimer's Res. Ther. 10:26. 10.1186/s13195-018-0354-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar S., Engler-Chiurazzi E. B., Cavendish J. Z., Povroznik J. M., Russell A. E., Quintana D. D., et al. (2019). Over-expression of miR-34a induces rapid cognitive impairment and Alzheimer's disease-like pathology. Brain Res. 1721:146327. 10.1016/j.brainres.2019.146327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar S., Jun S., Rellick S., Quintana D. D., Cavendish J. Z., Simpkins J. W. (2016). Expression of MicroRNA-34a in Alzheimer's disease brain targets genes linked to synaptic plasticity, energy metabolism, and resting state network activity. Brain Res. 1646, 139–151. 10.1016/j.brainres.2016.05.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh J.-I., Kino Y., Niida S. (2015). MicroRNA-seq data analysis pipeline to identify blood biomarkers for Alzheimer's disease from public data. Biomark. Insights 10, 21–31. 10.4137/BMI.S25132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheltens P., Blennow K., Breteler M. M., de Strooper B., Frisoni G. B., Salloway S., et al. (2016). Alzheimer's disease. Lancet 388, 505–517. 10.1016/S0140-6736(15)01124-1 [DOI] [PubMed] [Google Scholar]
- Scheuner D., Eckman C., Jensen M., Song X., Citron M., Suzuki N., et al. (1996). Secreted amyloid beta-protein similar to that in the senile plaques of Alzheimer's disease is increased in vivo by the presenilin 1 and 2 and APP mutations linked to familial Alzheimer's disease. Nat. Med. 2, 864–870. 10.1038/nm0896-864 [DOI] [PubMed] [Google Scholar]
- Serrano-Pozo A., Frosch M. P., Masliah E., Hyman B. T. (2011). Neuropathological alterations in Alzheimer disease. Cold Spring Harbor Perspect. Med. 1:a006189. 10.1101/cshperspect.a006189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shende V. R., Goldrick M. M., Ramani S., Earnest D. J. (2011). Expression and rhythmic modulation of circulating MicroRNAs targeting the clock gene Bmal1 in mice. PLoS ONE 6:e22586. 10.1371/journal.pone.0022586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shende V. R., Neuendorff N., Earnest D. J. (2013). Role of miR-142-3p in the post-transcriptional regulation of the clock gene Bmal1 in the mouse SCN. PLoS ONE 8:e65300. 10.1371/journal.pone.0065300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu B., Zhang X., Du G., Fu Q., Huang L. (2018). MicroRNA-107 prevents amyloid-β-induced neurotoxicity and memory impairment in mice. Int. J. Mol. Med. 41, 1665–1672. 10.3892/ijmm.2017.3339 [DOI] [PubMed] [Google Scholar]
- Siedlecki-Wullich D., Català-Solsona J., Fábregas C., Hernández I., Clarimon J., Lleó A., et al. (2019). Altered microRNAs related to synaptic function as potential plasma biomarkers for Alzheimer's disease. Alzheimer's Res. Ther. 11:46. 10.1186/s13195-019-0501-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siedlecki-Wullich D., Miñano-Molina A. J., Rodríguez-Álvarez J. (2021). microRNAs as early biomarkers of Alzheimer's disease: a synaptic perspective. Cells 10:113. 10.3390/cells10010113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel G., Obernosterer G., Fiore R., Oehmen M., Bicker S., Christensen M., et al. (2009). A functional screen implicates microRNA-138-dependent regulation of the depalmitoylation enzyme APT1 in dendritic spine morphogenesis. Nat. Cell Biol. 11, 705–716. 10.1038/ncb1876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silvestro S., Bramanti P., Mazzon E. (2019). Role of miRNAs in Alzheimer's disease and possible fields of application. Int. J. Mol. Sci. 20:3979. 10.3390/ijms20163979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slota J. A., Booth S. A. (2019). MicroRNAs in neuroinflammation: implications in disease pathogenesis, biomarker discovery and therapeutic applications. Non Coding RNA 5:35. 10.3390/ncrna5020035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith P. Y., Hernandez-Rapp J., Jolivette F., Lecours C., Bisht K., Goupil C., et al. (2015). MiR-132/212 deficiency impairs tau metabolism and promotes pathological aggregation in vivo. Hum. Mol. Genet. 24, 6721–6735. 10.1093/hmg/ddv377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sofroniew M. V. (2015). Astrocyte barriers to neurotoxic inflammation. Nat. Rev. Neurosci. 16, 249–263. 10.1038/nrn3898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J., Lee J. E. (2015). ASK1 modulates the expression of microRNA Let7A in microglia under high glucose in vitro condition. Front. Cell. Neurosci. 9:198. 10.3389/fncel.2015.00198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y., Hu M., Zhang J., Teng Z.-Q., Chen C. (2019). A novel mechanism of synaptic and cognitive impairments mediated via microRNA-30b in Alzheimer's disease. EBioMedicine 39, 409–421. 10.1016/j.ebiom.2018.11.059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souza V. C., Morais G. S., Henriques A. D., Machado-Silva W., Perez D. I. V., Brito C. J., et al. (2020). Whole-blood levels of MicroRNA-9 are decreased in patients with late-onset Alzheimer disease. Am. J. Alzheimer's Dis. Other Dementias® 35, 1–9. 10.1177/1533317520911573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperling R. A., Aisen P. S., Beckett L. A., Bennett D. A., Craft S., Fagan A. M., et al. (2011). Toward defining the preclinical stages of Alzheimer's disease: recommendations from the National Institute on Aging-Alzheimer's Association workgroups on diagnostic guidelines for Alzheimer's disease. Alzheimer's Dementia 7, 280–292. 10.1016/j.jalz.2011.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stratmann K., Heinsen H., Korf H. W., Del Turco D., Ghebremedhin E., Seidel K., et al. (2016). Precortical phase of Alzheimer's Disease (AD)-related tau cytoskeletal pathology. Brain Pathol. 26, 371–386. 10.1111/bpa.12289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su W., Aloi M. S., Garden G. A. (2016). MicroRNAs Mediating CNS inflammation: small regulators with powerful potential. Brain Behav. Immunity 52, 1–8. 10.1016/j.bbi.2015.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun W., Zhao J., Li C. (2020). Dexmedetomidine provides protection against hippocampal neuron apoptosis and cognitive impairment in mice with Alzheimer's disease by mediating the miR-129/YAP1/JAG1 Axis. Mol. Neurobiol. 57, 5044–5055. 10.1007/s12035-020-02069-z [DOI] [PubMed] [Google Scholar]
- Sun X., Chen W. D., Wang Y. D. (2015). β-Amyloid: the key peptide in the pathogenesis of Alzheimer's disease. Front. Pharmacol. 6:221. 10.3389/fphar.2015.00221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swerdlow R. H. (2018). Mitochondria and mitochondrial cascades in Alzheimer's Disease. J. Alzheimer's Dis. 62, 1403–1416. 10.3233/JAD-170585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swomley A. M., Allan Butterfield D. (2015). Oxidative stress in Alzheimer disease and mild cognitive impairment: evidence from human data provided by redox proteomics. Arch. Toxicol. 89, 1669–1680. 10.1007/s00204-015-1556-z [DOI] [PubMed] [Google Scholar]
- Tan L., Yu J.-T., Liu Q.-Y., Tan M.-S., Zhang W., Hu N., et al. (2014a). Circulating miR-125b as a biomarker of Alzheimer's disease. J. Neurol. Sci. 336, 52–56. 10.1016/j.jns.2013.10.002 [DOI] [PubMed] [Google Scholar]
- Tan L., Yu J.-T., Tan M.-S., Liu Q.-Y., Wang H.-F., Zhang W., et al. (2014b). Genome-wide serum microRNA expression profiling identifies serum biomarkers for Alzheimer's disease. J. Alzheimer's Dis. 40, 1017–1027. 10.3233/JAD-132144 [DOI] [PubMed] [Google Scholar]
- Tan X., Zhang P., Zhou L., Yin B., Pan H., Peng X. (2012). Clock-controlled mir-142-3p can target its activator, Bmal1. BMC Mol. Biol. 13:27. 10.1186/1471-2199-13-27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang B. L. (2016). Sirt1 and the mitochondria. Mol. Cells 39, 87–95. 10.14348/molcells.2016.2318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor B. K., Westlund K. N. (2017). The noradrenergic locus coeruleus as a chronic pain generator. J. Neurosci. Res. 95, 1336–1346. 10.1002/jnr.23956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terry A. V., Buccafusco J. J. (2003). The cholinergic hypothesis of age and Alzheimer's disease-related cognitive deficits: recent challenges and their implications for novel drug development. J. Pharmacol. Exp. Ther. 306, 821–827. 10.1124/jpet.102.041616 [DOI] [PubMed] [Google Scholar]
- Thomas L., Florio T., Perez-Castro C. (2020). Extracellular vesicles loaded miRNAs as potential modulators shared between glioblastoma, and Parkinson's and Alzheimer's diseases. Front. Cell. Neurosci. 14:34. 10.3389/fncel.2020.590034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todd W. D. (2020). Potential pathways for circadian dysfunction and sundowning-related behavioral aggression in Alzheimer's disease and related dementias. Front. Neurosci. 14:910. 10.3389/fnins.2020.00910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomasetti M., Nocchi L., Staffolani S., Manzella N., Amati M., Goodwin J., et al. (2014). MicroRNA-126 suppresses mesothelioma malignancy by targeting IRS1 and interfering with the mitochondrial function. Antioxid. Redox Signal. 21, 2109–2125. 10.1089/ars.2013.5215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinczek B., Biernat J., Baumann K., Mandelkow E. M., Mandelkow E. (1995). Domains of tau protein, differential phosphorylation, and dynamic instability of microtubules. Mol. Biol. Cell 6, 1887–1902. 10.1091/mbc.6.12.1887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Vaart B., Akhmanova A., Straube A. (2009). Regulation of microtubule dynamic instability. Biochem. Soc. Transac. 37(Pt. 5), 1007–1013. 10.1042/BST0371007 [DOI] [PubMed] [Google Scholar]
- van Kooten J., Binnekade T. T., van der Wouden J. C., Stek M. L., Scherder E. J., Husebø B. S. (2016). A review of pain prevalence in alzheimer's, vascular, frontotemporal and lewy body dementias. Dementia Geriatr. Cogn. Disord. 41, 220–232. 10.1159/000444791 [DOI] [PubMed] [Google Scholar]
- Vasile F., Dossi E., Rouach N. (2017). Human astrocytes: structure and functions in the healthy brain. Brain Struct. Funct. 222, 2017–2029. 10.1007/s00429-017-1383-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veeraraghavalu K., Zhang C., Zhang X., Tanzi R. E., Sisodia S. S. (2014). Age-dependent, non-cell-autonomous deposition of amyloid from synthesis of β-amyloid by cells other than excitatory neurons. J. Neurosci. 34, 3668–3673. 10.1523/JNEUROSCI.5079-13.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veerhuis R., Nielsen H. M., Tenner A. J. (2011). Complement in the brain. Mol. Immunol. 48, 1592–1603. 10.1016/j.molimm.2011.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vergallo A., Lista S., Zhao Y., Lemercier P., Teipel S. J., Potier M.-C., et al. (2021). MiRNA-15b and miRNA-125b are associated with regional Aβ-PET and FDG-PET uptake in cognitively normal individuals with subjective memory complaints. Transl. Psychiatry 11, 1–11. 10.1038/s41398-020-01184-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F., Gordon B. A., Ryman D. C., Ma S., Xiong C., Hassenstab J., et al. (2015). Cerebral amyloidosis associated with cognitive decline in autosomal dominant Alzheimer disease. Neurology 85, 790–798. 10.1212/WNL.0000000000001903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Chen C., Zhang Y. (2020). An investigation of microRNA-103 and microRNA-107 as potential blood-based biomarkers for disease risk and progression of Alzheimer's disease. J. Clin. Lab. Anal. 34:e23006. 10.1002/jcla.23006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M., Qin L., Tang B. (2019). MicroRNAs in Alzheimer's disease. Front. Genet. 10:153. 10.3389/fgene.2019.00153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R., Zhang J. (2020). Clinical significance of miR-433 in the diagnosis of Alzheimer's disease and its effect on Aβ-induced neurotoxicity by regulating JAK2. Exp. Gerontol. 141:111080. 10.1016/j.exger.2020.111080 [DOI] [PubMed] [Google Scholar]
- Wang W., Zhao F., Ma X., Perry G., Zhu X. (2020). Mitochondria dysfunction in the pathogenesis of Alzheimer's disease: Recent advances. Mol. Neurodegener. 15:30. 10.1186/s13024-020-00376-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W.-X., Huang Q., Hu Y., Stromberg A. J., Nelson P. T. (2011). Patterns of microRNA expression in normal and early Alzheimer's disease human temporal cortex: white matter versus gray matter. Acta Neuropathol. 121, 193–205. 10.1007/s00401-010-0756-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Tan L., Lu Y., Peng J., Zhu Y., Zhang Y., et al. (2015). MicroRNA-138 promotes tau phosphorylation by targeting retinoic acid receptor alpha. FEBS Lett. 589, 726–729. 10.1016/j.febslet.2015.02.001 [DOI] [PubMed] [Google Scholar]
- Wang X. H., Wang T. L. (2018). MicroRNAs of Microglia: wrestling with Central Nervous System Disease. Neural Regener. Res. 13, 2067–2072. 10.4103/1673-5374.241444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei W., Wang Z.-Y., Ma L.-N., Zhang T.-T., Cao Y., Li H. (2020). MicroRNAs in alzheimer's disease: function and potential applications as diagnostic biomarkers. Front. Mol. Neurosci. 13:160. 10.3389/fnmol.2020.00160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg R. B., Mufson E. J., Counts S. E. (2015). Evidence for a neuroprotective microRNA pathway in amnestic mild cognitive impairment. Front. Neurosci. 9:430. 10.3389/fnins.2015.00430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weldemichael D. A., Grossberg G. T. (2010). Circadian rhythm disturbances in patients with Alzheimer's disease: a review. Int. J. Alzheimer's Dis. 2010:e716453. 10.4061/2010/716453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weldon Furr J., Morales-Scheihing D., Manwani B., Lee J., McCullough L. D. (2019). Cerebral amyloid angiopathy, Alzheimer's disease and MicroRNA: MiRNA as diagnostic biomarkers and potential therapeutic targets. Neuromol. Med. 21, 369–390. 10.1007/s12017-019-08568-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitlock E. L., Diaz-Ramirez L. G., Glymour M. M., Boscardin W. J., Covinsky K. E., Smith A. K. (2017). Association between persistent pain and memory decline and dementia in a longitudinal cohort of elders. JAMA Intern. Med. 177, 1146–1153. 10.1001/jamainternmed.2017.1622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H. Z. Y., Thalamuthu A., Cheng L., Fowler C., Masters C. L., Sachdev P., et al. (2020). Differential blood miRNA expression in brain amyloid imaging-defined Alzheimer's disease and controls. Alzheimer's Res. Ther. 12:59. 10.1186/s13195-020-00627-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., Xu J., Xu J., Cheng J., Jiao D., Zhou C., et al. (2017). Lower serum levels of miR-29c-3p and miR-19b-3p as biomarkers for Alzheimer's disease. Tohoku J. Exp. Med. 242, 129–136. 10.1620/tjem.242.129 [DOI] [PubMed] [Google Scholar]
- Xie B., Zhou H., Zhang R., Song M., Yu L., Wang L., et al. (2015). Serum miR-206 and miR-132 as potential circulating biomarkers for mild cognitive impairment. J. Alzheimer's Dis. 45, 721–731. 10.3233/JAD-142847 [DOI] [PubMed] [Google Scholar]
- Xu J., Zheng Y., Wang L., Liu Y., Wang X., Li Y., et al. (2021). miR-124: a promising therapeutic target for central nervous system injuries and diseases. Cell. Mol. Neurobiol. 10.1007/s10571-021-01091-6. [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu N., Li A.-D., Ji L.-L., Ye Y., Wang Z.-Y., Tong L. (2019). MiR-132 regulates the expression of synaptic proteins in APP/PS1 transgenic mice through C1q. Eur. J. Histochem. 63:3008. 10.4081/ejh.2019.3008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu S.-Y., Jiang X.-L., Liu Q., Xu J., Huang J., Gan S.-W., et al. (2019). Role of rno-miR-124-3p in regulating MCT1 expression in rat brain after permanent focal cerebral ischemia. Genes Dis. 6, 398–406. 10.1016/j.gendis.2019.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y., Chen P., Wang X., Yao J., Zhuang S. (2018). MiR-34a deficiency in APP/PS1 mice promotes cognitive function by increasing synaptic plasticity via AMPA and NMDA receptors. Neurosci. Lett. 670, 94–104. 10.1016/j.neulet.2018.01.045 [DOI] [PubMed] [Google Scholar]
- Yan K., An T., Zhai M., Huang Y., Wang Q., Wang Y., et al. (2019). Mitochondrial miR-762 regulates apoptosis and myocardial infarction by impairing ND2. Cell Death Dis. 10, 1–16. 10.1038/s41419-019-1734-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q., Zhao Q., Yin Y. (2019). MiR-133b is a potential diagnostic biomarker for Alzheimer's disease and has a neuroprotective role. Exp. Ther. Med. 18, 2711–2718. 10.3892/etm.2019.7855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang T. T., Liu C. G., Gao S. C., Zhang Y., Wang P. C. (2018). The serum exosome derived microRNA-135a,−193b, and−384 were potential Alzheimer's disease biomarkers. Biomed. Environ. Sci. 31, 87–96. 10.3967/bes2018.011 [DOI] [PubMed] [Google Scholar]
- Yao H., Ma R., Yang L., Hu G., Chen X., Duan M., et al. (2014). MiR-9 promotes microglial activation by targeting MCPIP1. Nat. Commun. 5:4386. 10.1038/ncomms5386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao X., Xian X., Fang M., Fan S., Li W. (2020). Loss of miR-369 promotes tau phosphorylation by targeting the fyn and serine/threonine-protein kinase 2 signaling pathways in Alzheimer's Disease mice. Front. Aging Neurosci. 11:365. 10.3389/fnagi.2019.00365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Y., Xu H., Su X., He X. (2016). Role of microRNA in governing synaptic plasticity. Neural Plast. 2016:e4959523. 10.1155/2016/4959523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y., Yi M.-H., Kim D. W. (2018). Impaired autophagy of GABAergic interneurons in neuropathic pain. Pain Res. Manag. 2018:e9185368. 10.1155/2018/9185368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan P., Yang T., Mu J., Zhao J., Yang Y., Yan Z., et al. (2020). Circadian clock gene NPAS2 promotes reprogramming of glucose metabolism in hepatocellular carcinoma cells. Cancer Lett. 469, 498–509. 10.1016/j.canlet.2019.11.024 [DOI] [PubMed] [Google Scholar]
- Zaky A., Bassiouny A., Farghaly M., El-Sabaa B. M. (2017). A combination of resveratrol and curcumin is effective against aluminum chloride-induced neuroinflammation in rats. J. Alzheimer's Dis. 60, S221–S235. 10.3233/JAD-161115 [DOI] [PubMed] [Google Scholar]
- Zhang Q.-S., Liu W., Lu G.-X. (2017). MiR-200a-3p promotes b-Amyloid-induced neuronal apoptosis through down-regulation of SIRT1 in Alzheimer's disease. J. Biosci. 42, 397–404. 10.1007/s12038-017-9698-1 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Liu H.-L., An L.-J., Li L., Wei M., Ge D.-J., et al. (2019). MiR-124-3p attenuates neuropathic pain induced by chronic sciatic nerve injury in rats via targeting EZH2. J. Cell. Biochem. 120, 5747–5755. 10.1002/jcb.27861 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Xing H., Guo S., Zheng Z., Wang H., Xu D. (2016). MicroRNA-135b has a neuroprotective role via targeting of β-site APP-cleaving enzyme 1. Exp. Ther. Med. 12, 809–814. 10.3892/etm.2016.3366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Yue D., Zhou Y., Jia L., Wang H., Guo M., et al. (2017). The role of MicroRNAs in Aβ deposition and tau phosphorylation in Alzheimer's disease. Front. Neurol. 8:342. 10.3389/fneur.2017.00342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng K., Hu F., Zhou Y., Zhang J., Zheng J., Lai C., et al. (2021). MiR-135a-5p mediates memory and synaptic impairments via the Rock2/Adducin1 signaling pathway in a mouse model of Alzheimer's disease. Nat. Commun. 12:1903. 10.1038/s41467-021-22196-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X., Wu K., Liao S., Pan Y., Sun Y., Chen X., et al. (2018). MicroRNA-transcription factor network analysis reveals miRNAs cooperatively suppress RORA in oral squamous cell carcinoma. Oncogenesis 7, 1–18. 10.1038/s41389-018-0089-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong L., Xiao W., Wang F., Liu J., Zhi L.-J. (2019). MiR-21-5p inhibits neuropathic pain development via directly targeting C-C motif ligand 1 and tissue inhibitor of metalloproteinase-3. J. Cell. Biochem. 120, 16614–16623. 10.1002/jcb.28920 [DOI] [PubMed] [Google Scholar]
- Zumkehr J., Rodriguez-Ortiz C. J., Cheng D., Kieu Z., Wai T., Hawkins C., et al. (2015). Ceftriaxone ameliorates tau pathology and cognitive decline via restoration of glial glutamate transporter in a mouse model of Alzheimer's disease. Neurobiol. Aging 36, 2260–2271. 10.1016/j.neurobiolaging.2015.04.005 [DOI] [PubMed] [Google Scholar]
- Zuo Z. (2019). Screen and Verification of Biomarkers for Early Detection of Alzheimer's Disease (Clinical Trial Registration No. NCT03388242). clinicaltrials.gov. Available online at: https://clinicaltrials.gov/ct2/show/NCT03388242