ABSTRACT
Host-adapted microorganisms are generally assumed to have evolved from free-living, environmental microorganisms, as examples of the reverse process are rare. In the phylum Gammaproteobacteria, family Moraxellaceae, the genus Psychrobacter includes strains from a broad ecological distribution including animal bodies as well as sea ice and other nonhost environments. To elucidate the relationship between these ecological niches and Psychrobacter’s evolutionary history, we performed tandem genomic analyses with phenotyping of 85 Psychrobacter accessions. Phylogenomic analysis of the family Moraxellaceae reveals that basal members of the Psychrobacter clade are Moraxella spp., a group of often-pathogenic organisms. Psychrobacter exhibited two broad growth patterns in our phenotypic screen: one group that we called the “flexible ecotype” (FE) had the ability to grow between 4 and 37°C, and the other, which we called the “restricted ecotype” (RE), could grow between 4 and 25°C. The FE group includes phylogenetically basal strains, and FE strains exhibit increased transposon copy numbers, smaller genomes, and a higher likelihood to be bile salt resistant. The RE group contains only phylogenetically derived strains and has increased proportions of lipid metabolism and biofilm formation genes, functions that are adaptive to cold stress. In a 16S rRNA gene survey of polar bear fecal samples, we detect both FE and RE strains, but in in vivo colonizations of gnotobiotic mice, only FE strains persist. Our results indicate the ability to grow at 37°C, seemingly necessary for mammalian gut colonization, is an ancestral trait for Psychrobacter, which likely evolved from a pathobiont.
IMPORTANCE Host-associated microbes are generally assumed to have evolved from free-living ones. The evolutionary transition of microbes in the opposite direction, from host associated toward free living, has been predicted based on phylogenetic data but not studied in depth. Here, we provide evidence that the genus Psychrobacter, particularly well known for inhabiting low-temperature, high-salt environments such as sea ice, permafrost soils, and frozen foodstuffs, has evolved from a mammalian-associated ancestor. We show that some Psychrobacter strains retain seemingly ancestral genomic and phenotypic traits that correspond with host association while others have diverged to psychrotrophic or psychrophilic lifestyles.
KEYWORDS: genomics, phylogeny, Psychrobacter, psychrophiles
INTRODUCTION
Association with vertebrate hosts was shown to be the largest factor driving differences in the 16S rRNA diversity of microbiomes sampled globally (1, 2). Recent analysis of metagenome-assembled genomes from multiple habitats shows that many are either enriched in animal hosts or the environment but generally not both (3). Specialization to host association can also be seen at higher taxonomic levels, indicating that whole lineages may have diverged once animal hosts were first successfully colonized. For instance, within the Bacteroidetes, the taxa that are mammalian gut associated are derived from phylogenetically basal clades that include free-living and invertebrate-associated taxa (4). These patterns of distribution imply that specialization to the warm animal host habitat is mostly incompatible with fitness in other environments. The specific diet of the host, the chemical environment of the gut, competition with an extremely dense surrounding microbial community, and direct selection by the host immune system all likely contribute to the restricted taxonomy of gut-associated microbes (5).
One line of evidence for the adaptive evolution of gut microbes is the emergence of specialized functions, such as the ability to bind and degrade host-specific nutrient sources like mucin (6), or resistance to bile acids and their salt conjugates (7). Additionally, many gut-associated commensal taxa lack significant environmental reservoirs, due to the divergence in selective pressures between guts and nongut environments (8). In contrast, many gastrointestinal pathogens such as Vibrio cholerae (9) or Yersinia enterocolitica (10) have high fitness within the gut and outside the gut, typically in soil or water. One phylogenetic analysis has indicated that the evolutionary transition from a free-living lifestyle to a host-associated (including pathogenic) lifestyle is more common than the opposite (11).
Species of Psychrobacter have been recovered through culture-based and sequence-based methods from a range of animal microbiomes, including the respiratory blow of marine mammals (12); marine mammal skin (13) and guts (14–16); the throats and guts of birds (17, 18) and fish (19); and many nonhost environments such as seawater (20), sea ice (21), marine sediment (22), glacial ice (23), and permafrost soil (24). Some Psychrobacter species are capable of causing disease in mammalian hosts (25, 26); however, Psychrobacter infections are very rare, and the factors leading to infection are unclear. Psychrobacter’s wide range of environmental sources and adaptation to cold temperatures raise the question whether Psychrobacter is truly associated with mammalian hosts, or if it is an allochthonous member of mammalian microbiomes. A comparative genomics analysis of 26 Psychrobacter spp. and metadata gleaned from public sources revealed differences in cold adaptation of protein coding sequences between warm-host-associated strains and derived marine and terrestrial strains (27). Furthermore, warm-adapted strains were basal in the rpoB gene phylogeny, suggesting that Psychrobacter evolved from a mesophilic ancestor. These observations make Psychrobacter an interesting candidate to assess how phenotype maps onto phylogeny and source of isolation and to probe into the evolutionary history of a genus with a wide habitat range.
Here, we investigated the evolutionary history of the genus Psychrobacter, a group of closely related bacteria with a broad environmental distribution. We generated 85 genomes of Psychrobacter accessions, which we phenotyped for growth under different temperature and salt conditions. We used a collection of wild polar bear feces collected from across the Canadian Arctic and assessed the presence of Psychrobacter ecotypes as a function of bear diet. Finally, we conducted tests with a subset of strains for colonization of the mammal gut using germfree mice. Our phylogenomic results confirm a mesophilic ancestry for Psychrobacter and a common ancestor with the genus Moraxella. Our phenotyping revealed that overall, Psychrobacter tolerates a wide range of salinity, but growth at 37°C divided the accessions into two ecotypes: those that retained the ability to grow at warm temperatures and can colonize mammalian hosts (flexible ecotype [FE]) and those that lost the ability to grow at warmer temperatures and are instead restricted to colder temperatures (restricted ecotype [RE]). A genomic analysis of the two ecotypes showed genome reduction in the FE strains with high transposon copy numbers, and increased biofilm formation capability in RE strains, as well as higher proportions of lipid metabolism genes. We showed that Psychrobacter strains that are basal are FE but that FE strains are also phylogenetically interspersed with RE strains, indicating either readaptation to the animal host or retention of the basal traits. Although both FE and RE strains were detected in the feces of wild polar bears, of the subset of Psychrobacter strains tested, only FE strains could colonize the germfree mouse gut. Together, our results indicate the genus Psychrobacter evolved from a pathobiont ancestor, having largely lost its ability to associate with animals in its adaptation to psychrophilic, nonhost environments.
RESULTS
Psychrobacter forms a clade whose basal members are of the Moraxella genus.
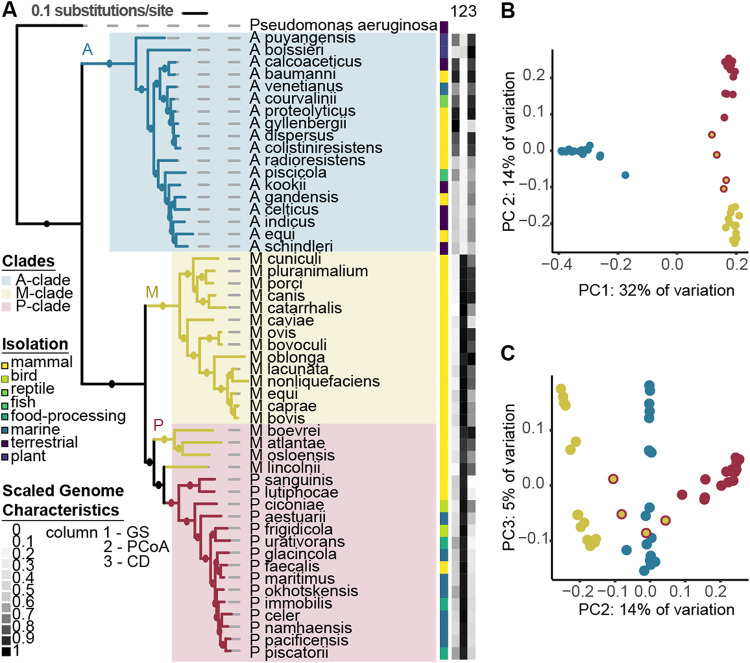
To explore the evolutionary history of Psychrobacter, we built a phylogeny based on 400 conserved marker genes using publicly available whole genomes derived from cultured isolates of 51 species from the Moraxellaceae family. We included 18 species each of Acinetobacter and Moraxella obtained from NCBI and 15 Psychrobacter genomes that we generated in this study (see Table S1 in the supplemental material). For all genomes, we incorporated phenotypic data collected from previously published type-strain research. The 51 species formed 3 distinct clades, each with robust bootstrap support. The Acinetobacter clade consists uniquely of Acinetobacter species (labeled A in Fig. 1A). The Acinetobacter clade is a sister taxon to the Moraxella (M) clade, consisting entirely of Moraxella species, and to the Psychrobacter (P) clade, which contains all of the Psychrobacter species, as well as 4 Moraxella species—M. boevrei, M. atlantae, M. osloensis, and M. lincolnii—that are basal.
FIG 1.
Genomic and phenotypic diversity in the family Moraxellaceae. (A) The Moraxellaceae family phylogeny was constructed with 51 diverse Moraxellaceae genomes using the software PhyloPhlAn, which constructs a phylogeny using FastTree with 1,000 bootstraps, refined by RAxML under the PROTCATG model. Amino acid sequences from 400 marker genes were used in the alignment. Branches with bootstrap support greater than 70% are represented by filled circles. The scale bar represents the average amino acid substitutions per site. Pseudomonas aeruginosa was used as an outgroup. Clades are highlighted in colored blocks, and branches are colored by genus. Isolation source is depicted in a color strip, along with a heatmap of scaled notable genome characteristics that differ between the genera, with 0 representing the smallest value present and 1 the largest value (P. aeruginosa not included). GS, genome size, ranging between 1.8 Mb and 4.5 Mb. PCoA, PC1 values from a PCoA based on gene presence/absence data. CD, genome coding density, ranging from 80% to 89%. Growth temperature range data were collected from type strain publications. (B) PC1 and PC2 of a PCoA of a binary matrix of gene presence/absence for 51 species of Moraxellaceae, explaining 32% and 14% of the variation, respectively. Each genome is represented by one point, colored by genus. The P-clade Moraxella spp. are represented by yellow points with red outlines. (C) PC2 and PC3, explaining 14% and 5% of the variation, respectively.
Moraxellaceae genomics. Eighteen species each of Acinetobacter and Moraxella were chosen based on availability of information on isolation source and cultivation temperatures, and their genomes were downloaded from NCBI. Fifteen additional Psychrobacter accessions were chosen for the family-level analysis based on phylogenetic breadth. Download Table S1, XLSX file, 0.01 MB (13.7KB, xlsx) .
Copyright © 2021 Welter et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Consistent with the topology of the phylogeny, a principal-coordinate analysis (PCoA) of gene presence/absence data shows that the greatest variation within the family (i.e., principal coordinate [PC] 1) is the separation between the A clade and the M and P clades, which are grouped (Fig. 1). Genes annotated with very diverse functions, falling under almost every Cluster of Orthologous Groups (COG) category, strongly contribute to the separation between clades according to envfit analysis (Fig. 1B; Table S2). The P-clade Moraxella spp. fall between the P-clade Psychrobacter and the M-clade Moraxella when visualizing PC1 and -2 (Fig. 1B) as well as PC2 and -3 (Fig. 1C).
COG separation of PCoA. An envfit analysis was performed to identify functions that might be correlated with separation of accessions along a gene presence/absence principal-component analysis, for a PCoA at the Moraxellaceae family level as well as the Psychrobacter genus level. For every COG category, the r2, indicating the strength of the correlation; the P value, indicating the significance of the correlation; and the Benjamini-Hochberg-corrected P value are reported. Download Table S2, XLSX file, 0.01 MB (9KB, xlsx) .
Copyright © 2021 Welter et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Despite their close phylogenetic relationship and high similarity in gene presence/absence, Psychrobacter and Moraxella have different genomic properties. When examined by genus rather than clade, Psychrobacter species have an average genome size of 3.12 ± 0.27 Mb, while the average of Moraxella is 2.41 ± 0.28 Mb (pairwise Wilcoxon rank sum test [WRS], P value = 8e−07). Moraxella species have an average coding density of 86.4% ± 1.33%, which is significantly higher than the Psychrobacter species average of 82.7% ± 1.33% WRS, P value = 1e−5). Despite being phylogenetically more related to Psychrobacter species than to the other Moraxella species, the P-clade Moraxella strains are significantly different from Psychrobacter species for both genome size and coding density (pairwise WRS, P values = 0.004) while not significantly different from the M-clade Moraxella spp. (pairwise WRS, P values = 0.9).
Psychrobacter ranges of growth temperatures differ from those of Moraxella.
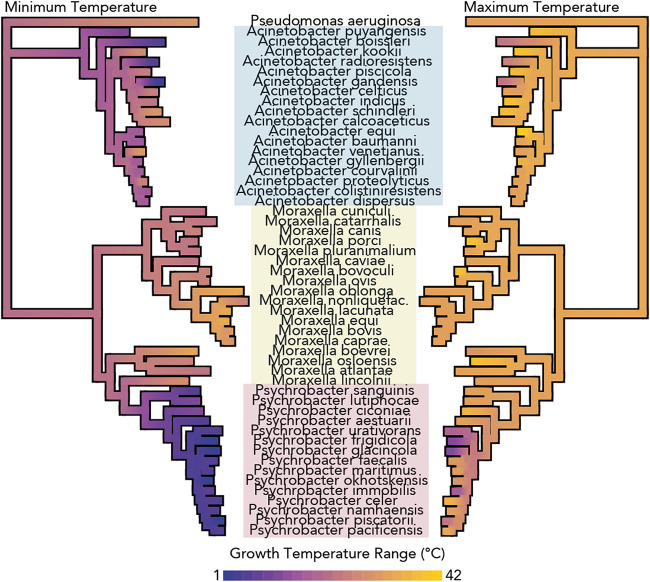
To examine the phenotypic behaviors of the Moraxellaceae family, we applied continuous trait mapping of the ranges of temperatures at which species from the Moraxellaceae family can grow (16, 17, 20, 21, 26, 28–68) onto the previously generated phylogeny (Fig. 2). The Psychrobacter spp. included here exhibit a broad range of growth temperatures (0 to 38°C), but several strains, such as P. frigidicola and P. glacincola, are psychrophilic (restricted to growth below 20°C), which is a phenotype that is not seen elsewhere in the family. Psychrobacter spp. have lower minimum growth temperatures than the included Moraxella spp. from either the P or M clades (pairwise WRS, P value = 5e−06), which have a narrow range of temperatures at which they can grow (between 22°C and 40°C). In contrast to the minimum growth temperatures of the family, there is little variation in the maximum growth temperatures, except for the notable exceptions of several Psychrobacter spp. that are entirely restricted to low temperatures.
FIG 2.
Psychrophily is observed only in Psychrobacter spp. among the family Moraxellaceae. Continuous trait mapping growth temperature ranges of 51 species from the Moraxellaceae family, taken from type strain publications. Values at nodes are imputed by maximum likelihood analysis. The phylogeny was constructed by marker-gene analysis including 400 genes, as in Fig. 1. Genera are indicated with colored boxes.
As with their genomic properties, P-clade Moraxella strains have more similar phenotypes compared to M-clade Moraxella strains than to Psychrobacter strains. Review of type strain descriptions revealed that Psychrobacter strains are consistently urease positive, nitrate reducing, salt tolerant, and nonfastidious, whereas P-clade Moraxella strains are inconsistent with their urease- and nitrate-reducing phenotypes, are sensitive to high salt concentrations, and are often nutritionally fastidious—they have complex growth requirements (in particular, often blood or bile for growth) (16, 17, 26, 32, 35, 43, 44, 59).
Few Psychrobacter strains can grow at 37°C.
To explore Psychrobacter phenotypic diversity, we established a strain collection of 85 Psychrobacter accessions isolated from diverse locations and ecological sources (Table S3). To characterize Psychrobacter phenotypes under a number of different conditions, we assessed the strain collection for resistance to bile salts as well as ability to grow under 24 different conditions: two different media combined with four different salt concentrations and with three different temperatures (Materials and Methods).
Psychrobacter cultivation. Summary of information from strain catalogues about the cultivation media and temperatures, isolation information, previously published genomes, and phenotyping batch of each of the 92 Psychrobacter strains initially included in our strain collection, with notes about the 7 strains that were ultimately not used for the analysis. Download Table S3, XLSX file, 0.02 MB (17.1KB, xlsx) .
Copyright © 2021 Welter et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
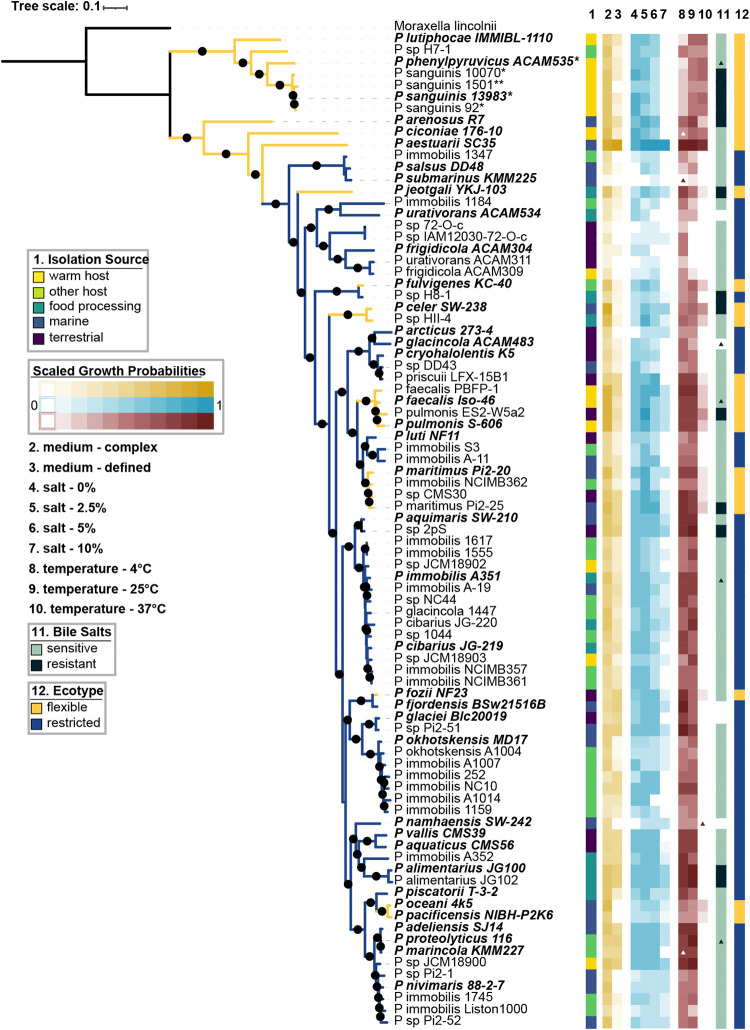
We calculated growth probabilities, or the fraction of growth-positive conditions out of total conditions tested, for every strain and given variable of the growth curve screen and compared them along with the bile resistance data across the phylogeny and by isolation source (Fig. 3). We generated a robust genus-level phylogeny for Psychrobacter using 400 conserved marker genes, with M. lincolnii as an outgroup. In agreement with single marker gene trees generated using rpoB sequences (27) and 16S rRNA gene sequences (69) as well as the P-clade structure of the Moraxellaceae family tree generated in this study, we observed a phylogenetically basal group of strains mostly isolated from mammals, and a phylogenetically derived group isolated from mixed sources. Across the entire phylogeny, closely related strains have similar growth probabilities (Pagel’s λ ranging from 0.78 to 0.97, all corrected P values < 1e−3). We observed that most Psychrobacter strains are tolerant of a wide variety of temperatures between 4 and 25°C and of salt concentrations between 0 and 5%; more than 90% of all strains can grow under these conditions. However, only 54% of the tested accessions can grow at 10% added salt, and only 31% can grow at 37°C.
FIG 3.
Psychrobacter phenotypic and phylogenomic diversity. The Psychrobacter phylogeny was constructed using FastTree with 1,000 bootstraps, refined by RAxML under the PROTCATG model. Amino acid sequences from 400 marker genes were used in the alignment. Branches with bootstrap support greater than 70% are represented by filled circles. The scale bar represents the average amino acid substitutions per site. M. lincolnii was used as an outgroup. Type strain isolate names are indicated in bold and italicized type. Strains indicated with * next to their name exhibited growth defects in liquid media and were tested on solid agar media instead. Strains indicated with ** exhibited growth defects on solid and liquid media and were tested on solid media supplemented with 0.1% Tween 80. Isolation source is depicted in column 1 as a color strip. Columns 2 to 10 represent the growth probabilities of each strain for each condition; medium complexity is represented in yellow, salt concentration is represented in blue, and temperature is represented in red. Type strain data support our temperature data except where indicated—colored triangles show conditions under which we expected growth but did not observe it, while white triangles represent conditions under which we observed growth that we did not expect. Bile salt resistance is shown in column 11, with black triangles indicating strains where we expected resistance but observed sensitivity, and green triangles representing the opposite. The “ecotype” is shown in column 12 in a color strip and in the color of the branches.
Since we were interested in the possibility of Psychrobacter interacting with warm-bodied hosts, we divided the strains into two ecotypes: the “flexible ecotype” (FE) corresponds to strains that could grow at 37°C, and the “restricted ecotype” (RE) corresponds to strains that could not grow at 37°C. FE strains are psychrotrophic (mesophilic organisms with a low minimum growth temperature but an optimal growth temperature above 15°C), and RE strains are either psychrotrophs or true psychrophiles (unable to grow at temperatures higher than 20°C).
Notably, the basal clade of the Psychrobacter-only tree (Fig. 3) consists solely of FE strains, while the rest of the phylogeny is made up both FE and RE strains. Furthermore, the basal FE strains have higher growth probabilities at 37°C than do other FE strains (Pagel’s λ = 0.89, P value = 2e−5). Frequencies of RE and FE strains vary significantly across sources of isolation: the FE group is significantly enriched in strains derived from mammalian sources, and the RE group is significantly enriched in strains derived from other hosts, food, and marine and terrestrial sources (χ2 test, 83% of P values adjusted for group size < 0.05). Nonetheless, both FE and RE contain strains from other environments, including mammalian-derived strains within the RE group.
FE strains show higher growth probabilities in complex media than in defined media, while RE strains showed little difference between the two (WRS, Wilcoxon test statistic [W] = 1.13e3, P value = 0.0005, phylogenetic P value = 0.002). FE strains also have higher growth probabilities at low to medium salt concentrations (WRS, all P values < 0.05, phylogenetic P values < 0.4), though there is no difference between FE and RE strains at 10% salt (WRS, W = 811.5, P value = 0.7, phylogenetic P value < 0.4). Finally, FE strains are more likely to be resistant to bile salts than RE strains (χ2 test, 93% of P values adjusted for group size < 0.05).
FE and RE Psychrobacter spp. have functional and structural genomic differences.
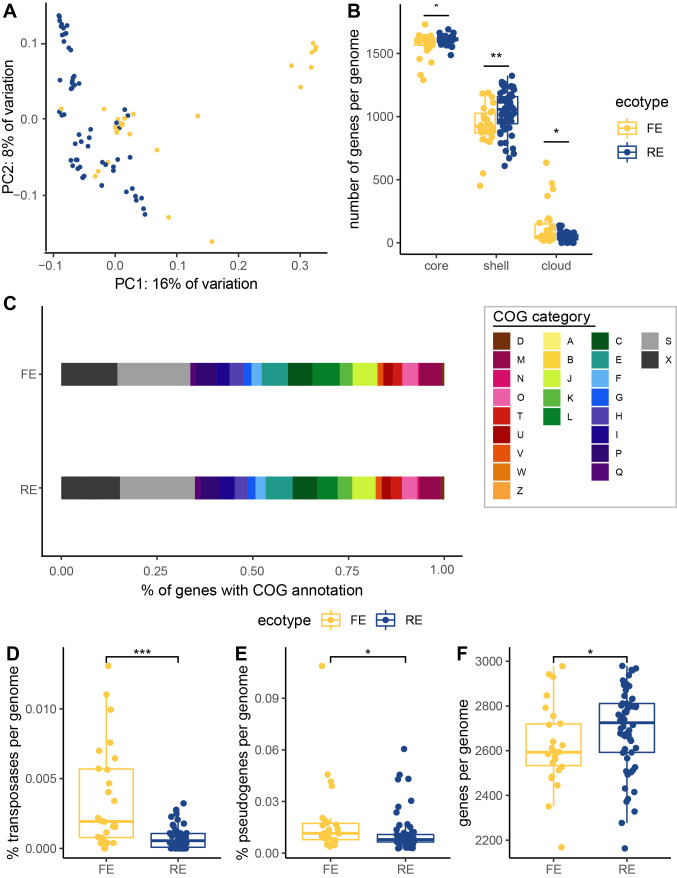
When exploring gene presence/absence via PCoA, accessions from the basal FE-only subclade cluster closely together, indicating similar gene content. In agreement with the phylogeny, these basal FE accessions are separated from the other accessions on PC1, while the derived FE and RE accessions are more scattered, indicating more diverse gene content (Fig. 4A). According to an envfit analysis, the separation is most strongly driven by genes from the COG categories T, signal transduction (r2 = 0.65, P value = 0.001); U, trafficking and secretion (r2 = 0.63, P value = 0.001); P, inorganic ion transport and metabolism (r2 = 0.56, P value = 0.001), and X, unassigned or no homologs in the COG database (r2 = 0.51, P value = 0.001) (Table S2).
FIG 4.
Structural differences between the genomes of FE and RE Psychrobacter strains. (A) The first two axes of a PCoA of a gene presence-absence matrix of all of the included accessions, colored by ecotype. (B) Pangenome categories. (C) Proportions of the average genome of each ecotype devoted to each cluster of orthologous genes (COG) category. Cellular processing and signaling: [D] cell cycle control cell division, chromosome partitioning; [M] cell wall/membrane/envelope biogenesis; [N] cell motility; [O] posttranslational modification, protein turnover, and chaperones; [T] signal transduction mechanisms; [U] intracellular trafficking, secretion, and vesicular transport; [V] defense mechanisms; [W] extracellular structures; [Y] nuclear structure; [Z] cytoskeleton. Information storage and processing: [A] RNA processing and modification; [B] chromatin structure and dynamics; [J] translation, ribosomal structure and biogenesis; [K] transcription; [L] replication, recombination, and repair. Metabolism: [C] energy production and conversion; [E] amino acid transport and metabolism; [F] nucleotide transport and metabolism; [G] carbohydrate transport and metabolism; [H] coenzyme transport and metabolism; [I] lipid transport and metabolism; [P] inorganic ion transport and metabolism; [Q] secondary metabolites biosynthesis, transport, and catabolism. Poorly characterized: [S] function unknown, [X] not in COG database. (D) Percent of genes that are transposases. (E) Percent of genes per genome that are predicted to be pseudogenes. (F) Number of genes per genome. Means were compared using the Wilcoxon rank sum test. *, P value less than 0.05; **, P < 0.005; ***, P < 0.0005.
The average Psychrobacter genome carries 2,763 genes, 1,598 of which are core (present in 90 to 100% of strains), 1,005 are shell (present in 2 or more strains but fewer than 90%), and 70 are cloud (present in only one strain). The average FE genome has significantly fewer core (WRS, W = 531.5, P value = 0.02, phylogenetic P value = 0.3) and shell (WRS, W = 462, P value = 0.003, phylogenetic P value = 0.4) genes but significantly more cloud genes (WRS, W = 1014, P value = 0.02, phylogenetic P value = 0.2) than the average RE genome (Fig. 4B).
In terms of functional differentiation between the ecotypes, RE strains’ genomes have a higher proportion of “W” (extracellular structure) genes (WRS, W = 275, adjusted P value = 0.00006, phylogenetic P value = 0.04), “I” (lipid transport and metabolism) genes (WRS, W = 437, adjusted P value = 0.01, phylogenetic P value = 0.2), and “S” (function unknown) genes (WRS, W = 492, adjusted P value = 0.04, phylogenetic P value = 0.4). FE strains’ genomes have a higher proportion of “P” (inorganic ion transport and metabolism) genes (WRS, W = 1.09e3, adjusted P value = 0.01, phylogenetic P value = 0.8). They also have a significantly higher proportion of COG category “L” (replication-, recombination-, and repair-related) genes (WRS, W = 1.10e3, adjusted P value = 0.01, phylogenetic P value = 0.3) (Fig. 4C) and, in particular, have higher copy numbers of transposons (WRS, W = 1.08e3, P value = 0.003, phylogenetic P value = 0.1) than do RE strains’ genomes (Fig. 4C and E).
Increased transposon activity can lead to interruption and decay of functional protein-encoding genes, leading to an increase in pseudogenes (70). Given the higher number of transposons in FE strains, we next examined the number of predicted pseudogenes between the ecotypes. FE genomes are predicted to have a higher proportion of pseudogenes than RE strains (WRS, W = 997, P value = 0.03, phylogenetic P value = 0.6) (Fig. 4C). Finally, we compared the average numbers of genes per genome between the ecotypes, as bacterial genomes are known to strongly select against the accumulation of pseudogenes resulting in gene deletions (71), and found that FE strains have significantly fewer genes than RE strains (WRS, W = 547, P value = 0.05, phylogenetic P value = 0.6).
We looked for specific gene clusters differentiating the FE and RE ecotypes while controlling for population structure by performing a microbial pangenome-wide association analysis with the R package treeWAS (72). This returned four gene clusters that significantly correlated with ecotype, each of which is enriched in RE strains compared to FE strains (Table 1). The gene with the most significant enrichment is annotated as a transcriptional regulator in the TetR/AcrR family with the KEGG KOs of K16137 and K19335, which is predicted to be either nemR or bdcR. Other significant results include lpdA, dihydrolipoamine dehydrogenase related to amino acid metabolism (KEGG KO K00382/383), a hypothetical protein with the IG domain, and a hypothetical protein with no homologs in eggNOG database and no predicted annotation. Each of these genes has paralogs that are not significantly enriched in one ecotype over the other.
TABLE 1.
Gene clusters detected as significantly associated with Psychrobacter ecotype based on a pangenome-wide association analysisa
| Gene cluster | Paralogsb | Predicted gene name | KEGG KO | COG category | eggNOG annotation | No. of FE strains | No. of RE strains | % FE strains | % RE strains |
|---|---|---|---|---|---|---|---|---|---|
| GC00000020 | GC00000020_1 | merA | K00382, K00383, K00520 | C | Mercuric reductase | 0 | 1 | 0 | 2 |
| GC00000020 | GC00000020_2 | merA | K00382, K00383, K00520 | C | Mercuric reductase | 4 | 4 | 15 | 7 |
| GC00000020 | GC00000020_3 | lpd | K00382 | C | Dihydrolipoamide dehydrogenase | 2 | 1 | 8 | 2 |
| GC00000020 | GC00000020_4 | lpd | K00382 | C | Dihydrolipoamide dehydrogenase | 1 | 0 | 4 | 0 |
| GC00000020 | GC00000020_5 | lpd | K00382 | C | Dihydrolipoamide dehydrogenase | 25 | 59 | 96 | 100 |
| GC00000020 | GC00000020_6*** | lpdA | K00382, K00383 | C | Dihydrolipoamide dehydrogenase | 9 | 53 | 35 | 90 |
| GC00000020 | GC00000020_7 | K00383 | C | Pyridine nucleotide-disulfide oxidoreductase | 0 | 3 | 0 | 5 | |
| GC00000020 | GC00000020_8 | sthA | K00322, K00382, K17883 | C | Conversion of NADPH to NADH | 26 | 59 | 100 | 100 |
| GC00000020 | GC00000020_9 | gor | K00383 | O | Glutathione reductase | 23 | 46 | 88 | 78 |
| GC00000020 | GC00000020_10 | lpdG | K00382 | C | Dihydrolipoyl dehydrogenase | 26 | 59 | 100 | 100 |
| GC00001343 | GC00001343_1 | K16137 | K | TetR family transcriptional regulator | 0 | 1 | 0 | 2 | |
| GC00001343 | GC00001343_2 | K16137 | K | Transcriptional regulator | 0 | 1 | 0 | 2 | |
| GC00001343 | GC00001343_3 | K16137 | K | TetR family transcriptional regulator | 3 | 0 | 12 | 0 | |
| GC00001343 | GC00001343_4 | K16137, K19335 | K | Transcriptional regulator TetR family | 1 | 0 | 4 | 0 | |
| GC00001343 | GC00001343_5 | K16137, K19335 | K | Transcriptional regulator TetR family | 5 | 16 | 19 | 27 | |
| GC00001343 | GC00001343_6*** | K16137, K19335 | K | Transcriptional regulator TetR family | 5 | 52 | 19 | 88 | |
| GC00001837 | GC00001837_1*** | S | Ig domain protein group 1 domain protein | 1 | 11 | 4 | 19 | ||
| GC00001837 | GC00001837_r1_1 | S | Ig domain protein group 1 domain protein | 1 | 0 | 4 | 0 | ||
| GC00001837 | GC00001837_r1_r1_1 | S | Ig domain protein group 1 domain protein | 12 | 26 | 46 | 44 | ||
| GC00001839 | GC00001839_1 | X | 1 | 0 | 4 | 0 | |||
| GC00001839 | GC00001839_r1_1*** | X | 6 | 47 | 23 | 80 | |||
Gene cluster refers to a group of protein-coding gene sequences with distant sequence homology which may be either orthologs or paralogs, while paralog refers to subsets within the gene cluster identified as exact gene matches.
***, the paralogs identified as significantly associated with ecotype by the pangenome-wide association software treeWAS.
Polar bear feces collected from the Arctic ice have a high abundance of Psychrobacter.
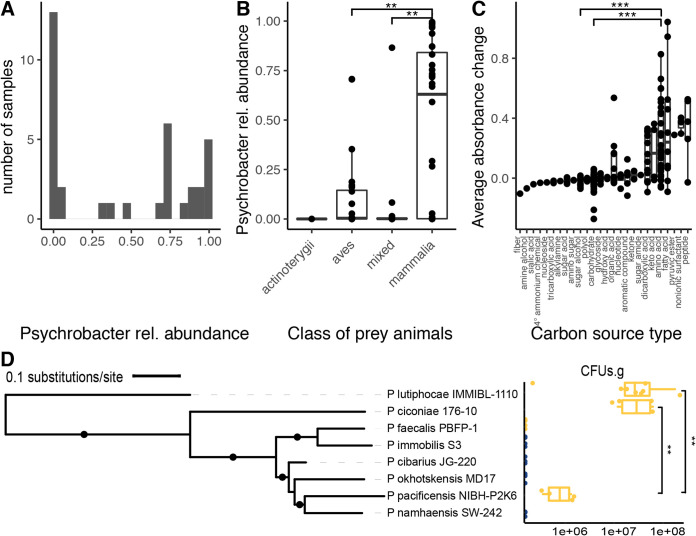
Psychrobacter spp. have been reported previously in the skin, respiratory, and gut microbiomes of several marine mammals, including whales, porpoises, seals, and sea lions; Psychrobacter presence may stem from the surrounding seawater. Polar bears, on the other hand, are marine mammals that do not spend as much time swimming. We surveyed the gut microbial diversity of 86 polar bear fecal samples, 76 wild and 10 captive, by 16S rRNA gene sequencing. Sequence variant clustering of the 16S rRNA gene sequences reveals that Psychrobacter is detectable in 88% of the samples (Fig. 5A). The large majority of Psychrobacter sequences were assigned to unclassified Psychrobacter spp., which are prevalent in 87% of samples with a mean abundance of 22%. We also detected RE strain P. immobilis in 49% of samples with a mean abundance of 3%, and FE strain P. pulmonis in 8% of samples with a mean abundance of 0.5%. Polar bear diet significantly impacts the abundance of Psychrobacter spp. (Kruskal-Wallis χ2 = 13.5, df = 3, P value = 0.004); we found that polar bears feeding on mammalian prey, including seals and reindeer, had significantly higher abundances of unclassified Psychrobacter spp. than polar bears feeding on avian prey or mixed diets (pairwise Wilcoxon rank sum test, adjusted P values < 0.05) (Fig. 5B). However, diet data are confounded with other metadata, as location (Kruskal-Wallis χ2 = 8.6, df = 4, P value = 0.0009) and year (Kruskal-Wallis χ2 = 16.6, df = 5, P value = 0.005) of sample collection also significantly impact unclassified Psychrobacter abundance. Captive status does not significantly impact abundance, but there is a trend of wild bear samples having higher unclassified Psychrobacter abundance than samples from captive bears (WRS, W = 222, P value = 0.08).
FIG 5.
Psychrobacter strains occur and persist in the mammalian gut. (A) Histogram of relative abundance at the genus level of 16S rRNA gene sequence variants (SVs), clustered at 100% identity, of polar bear fecal samples. (B) Psychrobacter relative abundance in comparison to taxonomy of prey consumed by polar bears. (C) The average change in OD600 of 19 Psychrobacter accessions grown on 190 different substrates as sole carbon sources, compared across different classes of compounds. (D) CFU per gram of cecal contents of gnotobiotic mice is shown in comparison to accession phylogeny. FE strains are shown in yellow, while RE strains are shown in blue. The phylogeny was constructed as described in the legend to Fig. 3. Branches with bootstrap support higher than 70% are indicated with a filled circle. Means were compared using the Wilcoxon rank sum test. *, P value less than 0.05; **, P < 0.005; ***, P < 0.0005.
To elucidate the effect that host dietary nutrition may have on Psychrobacter growth, we tested the maximum change in absorbance for 190 different carbon sources by a subset of Psychrobacter accessions including 9 FE and 10 RE strains (Fig. 5C). All Psychrobacter spp. reached significantly higher optical densities at 600 nm (OD600) growing on amino acid carbon sources compared to carbohydrates or sugar alcohols (pairwise WRS, adjusted P values < 0.05). Psychrobacter spp. reached the highest OD600 growing on fatty acids, surfactants, and peptides, but there was no significant difference between FE and RE strains’ changes in absorbance (WRS, W = 1.55e6, P value = 0.08), though there is a slight trend of RE strains having overall higher changes in absorbance.
Psychrobacter strain survival in gnotobiotic mice.
To assess the survivorship of Psychrobacter in a mammalian gut, we tested 8 accessions for persistence in the gastrointestinal tracts of germfree mice (Fig. 5D). Choosing for phylogenetic breadth, we tested 4 FE strains (P. ciconiae, P. faecalis PBFP-1, P. lutiphocae, and P. pacificensis) and 4 RE strains (P. cibarius JG-220, P. immobilis S3, P. namhaensis, and P. okhotskensis MD17). Of the four FE strains tested, three were able to persist in the mice, while the FE strain P. faecalis PBFP-1 and none of the RE strains were detectable after 3 weeks. Phylogenetic relatedness does not correlate with ability to colonize, as FE strain P. pacificensis was able to persist, while closely related RE strains P. namhaensis and P. okhotskensis were not. Phylogenetic placement does correlate with colonization density, however, as the two most basal strains tested, P. lutiphocae and P. ciconiae, colonize at significantly higher densities than the most derived strain that was successful, P. pacificensis (pairwise WRS, both adjusted P values = 0.0007).
DISCUSSION
The phylogenomic and phenotypic characterization of the Psychrobacter genus indicates a common ancestor with Moraxella, all of which are restricted to growth at higher temperatures. Furthermore, the most basal members of the Psychrobacter clade are Moraxella species and species of Psychrobacter that can grow at 37°C, unlike most of the derived Psychrobacter species. Our extensive phenotyping indicated that members of the Psychrobacter genus grow at a wide range of salinities and temperatures, but distinguishing strains by their ability to grow at 37°C results in two groups of strains with differences in response to rich versus defined media, bile salt resistance, and genomic characteristics. Our analysis of a collection of wild polar bear species indicates both RE and FE strains are present; however, tests in germfree mice support the notion that only FE strains can colonize the mammal gut, whereas RE strains may be allochthonous members. Together with previous reports, this work indicates the genus Psychrobacter is a lineage descended from pathobionts, some of which have evolved to inhabit the colder environments of their warm-bodied hosts.
Our results corroborate those of Bakermans, who used the isolation source of Psychrobacter as a proxy for temperature adaptation to conclude the genus has a mesophilic ancestor (27). By assessing growth under the same 24 conditions for 85 strains, we remove any ambiguity that can stem from whether an isolate can indeed grow at the temperature of its source of isolation. This is particularly important given that several strains isolated from mammals proved to be RE and that many of the FE strains came from seawater or other relatively cold environments. We also expanded upon the results of Bakermans by including phylogenetically aware statistics. In any population, identifying traits that are shared due to ecological convergence versus those that are shared due to common ancestry is a well-recognized problem (73); this problem is particularly striking in the genus Psychrobacter, where not only is there phenotypic divergence between the more phylogenetically basal strains and the derived strains, but as evidenced by the gene presence/absence PCoA, there is a large difference in gene content based on phylogeny.
We found that a transcriptional repressor, either NemR or BdcR, is enriched in RE strains’ genomes compared to FE strains. NemR plays a role in electrophile sensing (74), while BdcR plays a role in cyclic-di-GMP sensing (75). Both relate to increased biofilm formation when expressed. Similarly, we found a predicted Ig-domain protein, which is thought to be an outer membrane protein with a role in adhesion (76), enriched in RE strains. Adhesion and biofilm formation can increase during cold stress (77), indicating that it may be adaptive for psychrophiles or psychrotrophs to have increased biofilm formation capability.
In addition to specific genes being enriched in RE strains, we found broader functional differences between the ecotypes. FE strains had higher proportions of COG category “L” (replication) and “P” (inorganic transport and metabolism) genes. The higher proportion of “P” category genes could point to FE strains’ association with hosts, as free iron availability is rather low within host bodies (78). The higher “L” proportion seems to be related to higher transposon copy numbers in FE strains, which could point to population bottlenecks relating to host association (79). RE strains, on the other hand, showed increased proportions of COG category “W” (extracellular structures), “S” (function unknown), and “I” (lipid metabolism) genes. The increase in the proportion of “W” genes is unlikely to have a large impact on phenotypic differences between the ecotypes, since there are very few “W” genes in genomes of either RE or FE strains. It is difficult to comment on the increased proportion of “S” genes, since their function is unknown or poorly understood. The increased proportion of “I” category genes, relating to lipid transport and metabolism, could point to increased flexibility in membrane lipids for RE strains, which is a well-characterized adaptation to cold temperatures (80).
The three of four FE strains tested were able to colonize germfree mice, whereas the RE strains could not, indicating that growth at 37°C may be necessary (although not sufficient) to colonize mammals. Opportunistic infections in mammals caused by Psychrobacter strains are limited to P. sanguinis, P. phenylpyruvicus, P. faecalis, and P. pulmonis (25), which are all FE strains but do not all belong to the basal clade. Our results suggest that the FE strains maintain an ancestral ability to grow at mammalian body temperatures and colonize mammalian host bodies, while RE strains have adapted toward psychrophilic or psychrotrophic lifestyles.
Psychrobacter’s sister taxon Moraxella in particular is commonly isolated from host mucosal tissues and exhibits the reduced genome size and nutritional fastidiousness common to many host-dependent organisms. Moraxella contains species that are frequently associated with human respiratory infections, primarily M. catarrhalis (81), as well as livestock conjunctivitis, for example, M. bovis or M. equi (36). Since they are commonly found in healthy individuals and can cause disease in healthy individuals (82), Moraxella bacteria are best categorized as pathobionts and not dedicated or opportunistic pathogens. Several species of Moraxella appear basally in the P clade of the Moraxellaceae family-level phylogeny, suggesting that Psychrobacter evolved from a “Moraxella-like” ancestor. This is supported by the fact that both phylogenetically basal and derived Psychrobacter strains carry genes related to virulence functions and that many of the basal Psychrobacter strains exhibit growth defects in liquid culture, similar to the fastidiousness of Moraxella.
Despite clear phenotypic differentiation, Psychrobacter and Moraxella have similar genomic content, although Psychrobacter genomes are larger. A psychrophile emerging from an apparently mesophilic background through widespread horizontal gene transfer has been suggested before in the genus Psychroflexus (83), though the study was limited to comparing only two genomes. In fact, it has been suggested before that this is Psychrobacter’s evolutionary trajectory (27), and although many Psychrobacter trees are constructed using a Moraxella outgroup, Psychrobacter’s potential pathobiont origin has not been explored in detail. Horizontal gene transfer would explain Psychrobacter’s larger genome size compared to Moraxella despite lower coding density, as many newly acquired horizontally transferred genes are expected to be inactivated and pruned by the recipient genome (84, 85).
Our results indicate that the history of the genus Psychrobacter is that of an ancestral pathobiont or pathogen, some of the descendants of which broadened their ecological distribution, resulting in an attenuation of pathogenicity. The emergence of a psychrotroph—a remarkable generalist—from a background of a more specialized pathobiont or pathogen showcases the remarkable adaptability of bacteria, and particularly Proteobacteria, to their environments.
MATERIALS AND METHODS
Moraxellaceae family genomics and phenotypic data.
The Moraxellaceae family includes 3 well-characterized genera: Moraxella, Acinetobacter, and Psychrobacter. To build a family-level phylogeny, we downloaded genomes of 18 species of Acinetobacter and 18 species of Moraxella from the National Center for Biotechnology Information (NCBI) genome database (86) (June 2020) (see Table S2 in the supplemental material). We also included 15 Psychrobacter genomes generated in this study (described below). We determined the phylogenetic relationship between genomes using the whole-genome marker gene analysis software PhyloPhlAn (87) (v2.0, diversity = “medium.” set to “accurate”) and determined genome quality and summary characteristics using CheckM (88) (v1.0.18) and Prokka (89) (v1.14.6, kingdom = “bacteria”). The Moraxellaceae phylogenetic tree was visualized and annotated using the interactive Tree of Life (iTOL) web interface (90) (v5.6.1).
We analyzed the Moraxellaceae pangenome using the PanX pipeline (91) (v1.6.0, core gene cutoff = 0.9). The input genomes used by PanX were initially annotated using Prokka. After their assignment into orthologous clusters using MCL (v14.137), we reannotated gene clusters using eggNOG-Mapper (92) (v1.0.3). We explored genome content by calculating a distance matrix using the Jaccard metric through the R package ecodist (93) (v0.3.0) from a binary gene presence-absence table, followed by dimensional reduction of that distance matrix through principal-coordinate decomposition (PCoA) with the cmdscale function from the R package stats. We investigated variables contributing to the separation of the PCoA using the envfit function from the R package vegan (94) (v2.5-6); we tested whether genes significantly contributing to separation were “core” (genes present in 90% to 100% of strains), “shell” (genes present in greater than two strains, but in fewer than 90%), or “cloud” (genes present in only one strain) and if general gene function—summarized by Cluster of Orthologous Groups (COG) category (95)—contributed to the separation. We collected growth temperature range data from type strain publications (16, 17, 20, 21, 26, 28–68). We used the R package phytools (96) (v0.7-47) to map the temperature ranges onto the phylogeny.
Psychrobacter strains.
We obtained 92 isolates of Psychrobacter from strain catalogues for phenotypic and genotypic characterization. These represent 38 validly published species of Psychrobacter as well as unclassified strains, all isolated from a wide variety of geographical locations and diverse environmental and host samples. All strains were purchased and maintained in compliance with the Nagoya Protocol on Access to Genetic Resources and the Fair and Equitable Sharing of Benefits Arising from their Utilization to the Convention on Biological Diversity. For a full list of accessions and their catalogue, isolation, and cultivation information, see Table S3. Unless mentioned, we grew accessions as recommended by the strain catalogue (medium and temperature) from which they were purchased.
Psychrobacter carbon utilization assay.
To inform the design of the minimal medium used in the phenotypic screen described below, we chose a subset of 19 Psychrobacter strains (P. adeliensis, P. aestuarii, P. aquaticus, P. arenosus, P. celer, P. cibarius JG-219, P. ciconiae, P. cryohalolentis, P. faecalis PBFP-1, P. fozii, P. fulvigenes, P. glacincola ACAM483, P. luti, P. lutiphocae, P. maritimus Pi2-25, P. okhotskensis MD17, P. pacificensis, P. piscatorii, and P. urativorans ACAM534) to evaluate for their ability to grow on 190 different substrates as their sole carbon source. We utilized Biolog plates PM1 and PM2 (Biolog Incorporated, Hayward, CA, USA) following a slightly modified protocol.
Briefly, we streaked out strains on agar plates of their preferred medium incubated at their preferred temperature (Table S3). We scraped cells from these plates and resuspended them at a final optical density at 600 nm (OD600) of 0.07 in the Biolog inoculation fluid (PM IF-0a GN/GP, 1.2×) diluted to 1× with sterile water. We inoculated 100 μl of cell suspension into each well of the PM1 and PM2 plates, mixing well to resuspend the carbon sources. All strains were grown under aerobic conditions at their preferred growth temperature (Table S3). Growth was monitored by measuring OD600 for 14 days. We calculated total change in absorbance by subtracting the blank wells from the substrate wells and then taking the difference between the absorbances at t = 14 days and t = 0 (inoculation time point). We then averaged the total change in absorbance for all strains for every carbon source and assigned each carbon source to a “family” of compounds.
All strains reached a significantly higher max OD600 in amino acid substrates than in other carbon sources. l-Glutamate was chosen as the carbon source for the defined medium described in our phenotypic screens below, as none of the strains tested failed to grow using it. Some of the other amino acids allowed strains to grow to a larger change in OD600 but failed to allow all strains to grow.
Psychrobacter phenotypic screen.
For the 85 Psychrobacter accessions that passed genome quality control (QC) (see Table S4), we tested their ability to grow under 24 different conditions, a growth condition being a combination of a medium (complex or defined), salt concentration (0, 2.5, 5, or 10% NaCl) and incubation temperature (4, 25, or 37°C).
Sequencing summary. Summary of the total number of short and long reads, as well as quality control information provided by CheckM, such as completeness, contamination, genome size, and scaffold information, for all 92 Psychrobacter accessions included in our initial strain collection. Some strains were removed from further analysis due to poor-quality genomes. Download Table S4, XLSX file, 0.02 MB (17.5KB, xlsx) .
Copyright © 2021 Welter et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
We used lysogeny broth (LB) as a complex medium with high nutrient availability and a variation of M9 minimal medium (MM) as a defined medium with lower nutrient availability. We prepared LB following the manufacturer’s instructions except by various concentrations of sodium chloride (NaCl) (see below) and autoclaving it at 121°C for 20 min to ensure sterility. We prepared MM by adding components in the following final concentrations, followed by filter sterilization: 33.7 mM Na2HPO4, 22.0 mM KH2PO4, 9.35 mM NH4Cl, 0.4% l-glutamic acid, 1 mM MgSO4, 0.3 mM CaCl2, 1× ATCC trace vitamins solution, and 1× ATCC trace minerals solution, pH 7 (adjusted with KOH). We added NaCl to each “base medium” to 1 of the 4 following concentrations of NaCl: low, with 0%; medium, with 2.5%; medium-high, with 5%; and high, with 10%. We grew cultures at 4°C, representing cold environments; 25°C, representing mesophilic environments; and 37°C, representing mammalian host body temperature.
We randomly assigned Psychrobacter accessions to blocks of 10 strains to be tested simultaneously (several accessions were included in multiple blocks; see Table S2). We first grew strains to saturation, washed them, diluted them to an optical density at 600 nm (OD600) of 0.3 in sterile phosphate-buffered saline (PBS), and inoculated them in 100 μl of medium with a final ratio of 1:1,000. We used 96-well plates, with 10 inocula in 5 replicates and 10 uninoculated medium wells per plate; all periphery wells were filled with water to reduce edge effects. Plates were incubated at each temperature to reflect all 24 growth conditions. We measured the OD600 (Spark plate reader; Tecan, Zürich, Switzerland) every 8 h during the first week of incubation and then every 24 h and let cultures grow until stationary phase (3 to 12 weeks).
We used the same culture dilutions from the growth assays as inocula for bile resistance assays. We spotted 10 μl of diluted culture onto both tryptic soy agar (TSA) and TSA amended with 0.5% (wt/vol) bile salt (Difco bile salts no. 3 from BD Biosciences, San Jose, CA, USA) and incubated the plates at the strains’ preferred temperatures (Table S2). We incubated the plates for 2 weeks and then scored strains for growth on bile compared to the no-bile control as either “sensitive” if no growth or “resistant” for growth similar to the control. For several strains, no growth was observed on the control plate, and these were marked as “ND” or “no data.”
P. phenylpyruvicus and P. sanguinis exhibited the inability to grow in liquid culture under the conditions tested. Hence, we first streaked 5 strains of these 2 species on Columbian blood agar which we incubated for 3 days at 37°C, and then we washed and diluted them as for the liquid cultures. We spotted dilutions onto LB or MM medium with 3% agar, supplemented with the tested salt concentrations as above, and incubated them at 4°C, 25°C, and 37°C. P. sanguinis strain 1501 was unable to grow on any of the base media, so we repeated the same process with medium supplemented with 0.1% Tween 80. We scored growth for all plates after 2 weeks.
For 2 accessions, we could not confirm the purity of the cultures used in the phenotypic screen and subsequently removed them from analysis. They were removed from subsequent genomic analysis as well.
Growth probabilities.
We scored each replicate as either “growth positive,” meaning the accession grew, or else “growth negative,” if the replicate never reached a maximum OD600 of 0.15 over the course of the experiment. For each strain and condition (medium, salt, and temperature), we calculated a growth probability, which corresponds to the median value of growth positivity/negativity of all replicates for that condition.
For the type strains included in the phenotypic screen, we cross-checked growth probabilities at 4°C and 37°C with the published type strain descriptions where the original publications were clear about conditions tested. For the majority of strains, the type strain data and our data are in agreement; however, there are discrepancies between the type strain publications and our own data for the following strains: P. ciconiae, P. fozii, P. marincola, P. namhaensis, and P. submarinus. In some cases, we observed growth under temperatures where growth was previously unobserved; P. ciconiae and P. marincola have not before been reported to grow at 4°C, and P. fozii has not been reported to grow at 37°C. In other cases, we observed no growth in temperatures that have previously been reported to support growth; P. submarinus is expected to grow at 4°C and P. namhaensis is expected to grow at 37°C. The conditions tested in the type strain publications often differ from the conditions used in our phenotypic screen.
Genome sequencing, assembly, and annotation.
We extracted genomic DNA from cultures grown in their preferred conditions using the Gentra Puregene tissue kit (Qiagen, Valencia, CA, USA). We initially sequenced samples using the MiSeq 2 × 250-bp and the HiSeq 2 × 150-bp paired-end read technology (Illumina, San Diego, CA, USA) as previously described (83). We constructed libraries using the Nextera DNA sample preparation kit (Illumina) with modifications: we sheared 1 ng of DNA with in-house-generated Tn5 transposase and then amplified and barcoded it with custom primers for 7 to 14 cycles. We pooled samples and size selected using magnetic beads for MiSeq libraries or BluePippin (Sage Science, Beverly, MA, USA) for HiSeq libraries. After dilution to 4 nM for MiSeq and 2.5 nM for HiSeq, we stored libraries at −20°C until sequencing.
After sequencing and demultiplexing, we validated raw reads with fqtools (v2.0) and deduplicated them with Clumpify (v37.78, dedupe = t, dupedist = 40 for HiSeq/2500 for MiSeq, optical = t). BBDuk (v37.78) and Skewer (v0.2.2) were used to remove sequencing adapters and filter reads (minimum read length = 100 bp, minimum PHRED quality score = 25). At multiple steps throughout, we used FastQC (v0.11.7) and MultiQC (v1.7) to monitor quality. After quality control, reads were assembled de novo. First, we subsampled reads using seqtk (v1.3, number of subsampled reads per sample = 1,000,000), normalized by bbnorm (v37.78, target = 50, k = 31, minkmers = 15, prefilter = t), and then assembled with SPAdes (v3.12.8, cov_cutoff = off, set to “careful.” minimum scaffold length = 500 bp), followed by refinement with Pilon (v1.22, chunksize = 10,000,000). Finally, we assessed the assemblies for quality using CheckM. We assigned taxonomy using Sourmash (v2.0.0a4, scaled = 10,000, k = 31) and GTDB-Tk (v1.0.2, min_perc_aa = 10) and annotated assemblies using Prokka.
For genomes that were particularly fragmented (having greater than 250 contigs), we performed additional long-read sequencing (Table S4). We constructed Oxford Nanopore libraries using the ligation sequencing and native barcode ligation kits (Oxford Nanopore, Oxford, United Kingdom). We sequenced the libraries using the MinION system run with software MinKNOW (Oxford Nanopore). We basecalled and demultiplexed reads using ont-Guppy (v3.2.4) and used Porechop (97) (v0.2.4, adapter threshold - 90, minimum PHRED quality score = 8, minimum read length = 500 bp) to ensure that the adapters were removed, along with poor-quality reads. We generated summary statistics and summary plots and removed lambda phage reads using NanoPack (98) (nanocomp v1.11.3, nanofilt v2.7.1, nanoget v1.14.0, nanolyse v1.1.3, nanomath v0.23.3, nanoplot v1.31.0, nanostat v1.2.1). After quality control, we combined the long reads with the short (generated by the MiSeq and HiSeq libraries described above) for hybrid assembly. We assembled and analyzed the hybrid assemblies in largely the same way as the short-read assemblies described above, however, replacing SPAdes with Unicycler (99) (v0.4.8, minimum contig length = 500 bp, mode = normal).
We followed the quality control cutoffs suggested by CheckM and removed two genomes for having contamination higher than 5% and one for having completion less than 90%. We removed two additional genomes as the taxonomic classification was outside the Psychrobacter genus. These accessions were removed from genomic and phenotypic analysis. All further analyses use the 85 accessions which passed all quality control (QC) measures. For a full description of sequencing for each accession, see Table S4.
We annotated genomes with Prokka and eggNOG-Mapper. A phylogeny of the accessions was generated using PhyloPhlAn, with Moraxella lincolnii as an outgroup. Again, the phylogeny was visualized and annotated using iTOL. We used PanX to analyze the Psychrobacter pangenome and R to explore gene presence-absence data as described above with the Moraxellaceae family. Pseudogenes were predicted using the DFAST core workflow (100). We used treeWAS (72) to perform a pangenome-wide association study relating data collected in the phenotypic screen to genomic data.
Microbiome diversity of polar bear feces.
We collected 86 polar bear fecal samples from several regions in Canada, including samples from 10 captive bears, fed various diets or fasted, and 76 samples from an unknown number of wild bears, feeding on unknown diets. The feces from the captive bears were forwarded from institutions within Canada and did not require permitting for their passage to Queen’s University. The captive samples comprised 5 fecal samples from a single bear sequentially fed various diets of Arctic char, harp seal, and a “zoo diet” at the Polar Bear Habitat in Cochrane during 2010; 2 samples from each of 2 bears held at the Metro Toronto Zoo, fed a consistent “zoo diet” during 2010; and a single sample from a bear held at the Churchill Polar Bear Holding Facility in Churchill during 2010, where the bears are given only water until release. We collected all wild bear feces from M’Clintock Channel and Hudson Strait in Nunavut in accordance with permits prior to shipping to Queen’s University: we collected 24 samples from M’Clintock Channel under Wildlife Research permits issued in 2007, 2008, 2009, 2010, and 2011 to Peter Van Coeverden de Groot, and 9 samples from Hudson Strait collected in 2011 under a Wildlife Research permit to Grant Gilchrest (Environment Canada). We collected 43 wild samples from the Wapusk National Park in Manitoba from 2007 to 2010 under a Canada Parks permit to Robert Rockwell (American Museum of Natural History).
We confirmed that samples collected from wild bears originated from bears by sequencing a cytochrome b gene fragment using metabarcoding approaches based on Ion Torrent (Ion Torrent Systems Inc., Gilford, NH, USA) and 454 pyrosequencing (454 Life Sciences, Branford, CT, USA) next-generation sequencing technologies (101). We also used this method to analyze what prey animals the polar bears had been feeding on at the time of sample deposition. We visually inspected the samples for confirmation on the dietary analysis. We extracted DNA from the fecal samples using the DNeasy blood and tissue kit (Qiagen) and characterized the gut bacterial community by amplification and sequencing of the V4 region of the 16S rRNA gene as described previously (102). We used QIIME (103) (v2, DADA2 for quality control, F read trimmed to 200 bp, R read trimmed to 110 bp, samples rarefied to 20,000 sequences) for sequence processing and Silva (138 SSURef NR99 515F/806R) to assign taxonomy.
Isolation of Psychrobacter sp. from polar bear feces.
Two wild polar bear fecal samples were combined and diluted to 1 mg/ml in PBS. We plated the solution onto LB agar plus 6% NaCl and incubated it at 14°C for 10 days. A single colony grew and was identified as Psychrobacter faecalis by colony PCR and Sanger sequencing of the full-length 16S rRNA gene (described below). This isolate is designated P. faecalis PBFP-1 and is included in the phenotypic screen described above and the genomic analysis.
Gnotobiotic mouse colonizations.
We selected 8 accessions of Psychrobacter—P. cibarius JG-220, P. ciconiae, P. faecalis PBFP-1, P. immobilis S3, P. lutiphocae, P. okhotskensis MD17, P. namhaensis, and P. pacificensis—as inocula for gnotobiotic mouse studies based on their derivation sources and phylogenetic breadth. We grew each accession under its preferred conditions (Table S3) to saturation, spun it at 10°C at 2,500 rpm for 25 min, washed it with sterile PBS, resuspended it in 15% (vol/vol) glycerol in PBS, and flash froze it in liquid N2. Inoculum samples were stored at −80°C until the mouse experiments were performed by animal caretakers as follows. For P. ciconiae, P. faecalis, P. namhaensis, and P. pacificensis, experiments were performed by Taconic Biosciences (Rensselaer, NY, USA) staff, while for P. cibarius, P. immobilis, and P. okhotskensis, experiments were performed by Max Planck Institute for Developmental Biology (MPIDB) animal caretakers. P. lutiphocae was included in both Taconic and MPIDB experiments.
Five- to 6-week-old germfree male C57BL/6J mice were orally inoculated with approximately 107 CFU (n = 4 per Psychrobacter accession, n = 8 for P. lutiphocae). Mice were cohoused (Taconic) or single-housed (MPIDB) in sterile cages (IsoCage P; Tecniplast) and provided autoclaved water and sterile chow (NIH31M) ad libitum. Three weeks postcolonization, mice were sacrificed via CO2 asphyxiation (Taconic) or CO2 asphyxiation followed by cervical dislocation (MPIDB), after which cecal contents were immediately collected, flash frozen, and stored at −80°C prior to use. The Taconic experiments were performed in compliance with Taconic’s IACUC, and the MPIDB experiments were approved by and performed in accordance with the local animal welfare authority’s legal requirements.
To determine the bacterial colonization density in the mouse gastrointestinal tract, we serially diluted 2 aliquots per mouse of cecal material (50 mg each) and incubated them on plates under their preferred conditions (Table S3) for 3 to 5 days. If no colonies were observed, we spread an inoculating loop of the undiluted aliquot onto brain heart infusion agar and incubated it at 37°C for 2 weeks. For samples where we again observed no colonies, we categorized these Psychrobacter accessions as “nonpersistent.” For the samples that did show colony growth, we confirmed the identity of the colonies as Psychrobacter using Sanger sequencing of the full-length 16S rRNA gene as described below.
16S rRNA gene Sanger sequencing.
We prepared “colony” PCR mixtures following the protocol for Phusion high-fidelity polymerase (New England Biolabs, Ipswich, MA, USA), swirling a colony of interest in the reaction mixture as a substitute for the DNA template, and using the 27F and 1391R universal full-length 16S rRNA gene primers (104) for amplification. The reaction mixtures were incubated on the Mastercycler Pro S thermocycler (Eppendorf, Hamburg, Germany) following the Phusion protocol, using a touchdown program for the annealing temperature (dropping the annealing temperature at a rate of 1°C per cycle from 70°C to 55°C, then annealing at 55°C for 15 cycles). We cleaned the products using the DNA Clean & Concentrator-25 kit (Zymo Research, Irvine, CA, USA) and checked the concentration using the DS 11+ spectrophotometer (DeNovix, Wilmington, DE, USA). We used the cleaned DNA products as the templates for the Sanger sequencing reaction, following the ABI Prism BigDye Terminator cycle sequencing kit (ThermoFisher Scientific, Waltham, MA, USA) protocol with the 27F primer. The sequencing was performed using the 3730xl DNA analyzer (ThermoFisher). Upon sequencing completion, we performed a BLAST search (105) of the sample sequences against the NCBI nonredundant nucleotide database. We examined the top 10 hits to confirm the sample identity.
Statistical analysis.
We performed all data processing and statistical analysis using R (106) (v3.6.2) or Python (v3.6.10). We compared means between groups using a Kruskal-Wallis test followed by a pairwise Wilcoxon rank sum when more than two groups were compared. We tested differences in frequencies between groups with more than 10 observations with a χ2 test, repeated 100 times with down-sampling in order to correct for sampling sizes between groups. We measured phylogenetic signal using a log-likelihood ratio test on Pagel’s λ (Pagel’s λ fitted using the phylosig function from the phytools R package [96] [v0.7-47] and the null hypothesis being λ = 0). When applicable, we tested for the confounding of phylogeny with our groups of interest using the aov.phylo function from the R package geiger (107) (v2.0.7). We adjusted all P values for multiple comparisons with the Benjamini-Hochberg (BH) correction method.
Data availability.
Raw sequences for the Psychrobacter genome sequencing and polar bear feces 16S rRNA gene sequencing, as well as assembled Psychrobacter genomes, are available in the European Nucleotide Archive under the accession no. PRJEB40380. Annotated Psychrobacter genomes are available at ftp://ftp.tue.mpg.de/pub/ebio/dwelter. Raw data, R notebooks, and Python scripts for the analyses are available at https://github.com/dkwelter/Welter_et_al_2020.
ACKNOWLEDGMENTS
This work was supported by the Max Planck Society. The Nunavut field work was supported by funds from the Nunavut Wildlife Management Board (NWMB), the Nunavut General Monitoring Plan (NGMP), the National Science and Engineering Research Council (NSERC), and Environment Canada (Gov. of Canada).
We thank Jacobo de la Cuesta-Zuluaga, Sara Di Rienzi, Hagay Enav, Angela Poole, Jessica Sutter, Taichi Suzuki, and William Walters for discussions regarding project design and analysis and Andrea Belkacemi, Ilja Bezrukov, Pablo Carbonell, Silke Dauser, Julia Hildebrandt, and Christa Lanz for their advice on and assistance with sequencing. We also thank Jürgen Berger and Katharina Hipp for performing electron microscopy and Ariadne Papatheodorou for her assistance with the bile sensitivity assays. We thank Markus Dyck and Patricia Morin for providing the Polar Bear Habitat samples, Maria Frank for samples from the Metro Toronto Zoo, Daryll Hedman and Manitoba Conservation for the sample from the Polar Bear Holding Facility in Churchill, and Sam Iverson for samples from Hudson Strait. The collection of Nunavut samples would not have been possible without collaboration of colleagues at the Gjoa Haven Hunters and Trappers Association and their Traditional Ecological Knowledge relating to polar bears. We also thank Marie Pages and Maxime Galan for assistance with cytochrome b barcode sequencing.
We have no competing interests to declare.
Contributor Information
Ruth E. Ley, Email: rley@tuebingen.mpg.de.
Jack A. Gilbert, University of California San Diego
REFERENCES
- 1.Ley RE, Lozupone CA, Hamady M, Knight R, Gordon JI. 2008. Worlds within worlds: evolution of the vertebrate gut microbiota. Nat Rev Microbiol 6:776–788. doi: 10.1038/nrmicro1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Lozupone CA, Turnbaugh PJ, Fierer N, Knight R. 2011. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci U S A 108(Suppl 1):4516–4522. doi: 10.1073/pnas.1000080107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Youngblut ND, de la Cuesta-Zuluaga J, Reischer GH, Dauser S, Schuster N, Walzer C, Stalder G, Farnleitner AH, Ley RE. 2020. Large scale metagenome assembly reveals novel animal-associated microbial genomes, biosynthetic gene clusters, and other genetic diversity. mSystems 5:e01045-20. doi: 10.1128/mSystems.01045-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bäckhed F, Ley RE, Sonnenburg JL, Peterson DA, Gordon JI. 2005. Host-bacterial mutualism in the human intestine. Science 307:1915–1920. doi: 10.1126/science.1104816. [DOI] [PubMed] [Google Scholar]
- 5.Ley RE, Peterson DA, Gordon JI. 2006. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell 124:837–848. doi: 10.1016/j.cell.2006.02.017. [DOI] [PubMed] [Google Scholar]
- 6.Salyers AA, Vercellotti JR, West SE, Wilkins TD. 1977. Fermentation of mucin and plant polysaccharides by strains of Bacteroides from the human colon. Appl Environ Microbiol 33:319–322. doi: 10.1128/AEM.33.2.319-322.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ruiz L, Margolles A, Sánchez B. 2013. Bile resistance mechanisms in Lactobacillus and Bifidobacterium. Front Microbiol 4:396. doi: 10.3389/fmicb.2013.00396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Browne HP, Neville BA, Forster SC, Lawley TD. 2017. Transmission of the gut microbiota: spreading of health. Nat Rev Microbiol 15:531–543. doi: 10.1038/nrmicro.2017.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sakib SN, Reddi G, Almagro-Moreno S. 2018. Environmental role of pathogenic traits in Vibrio cholerae. J Bacteriol 200:e00795-17. doi: 10.1128/JB.00795-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wren BW. 2003. The yersiniae–a model genus to study the rapid evolution of bacterial pathogens. Nat Rev Microbiol 1:55–64. doi: 10.1038/nrmicro730. [DOI] [PubMed] [Google Scholar]
- 11.Sachs JL, Skophammer RG, Regus JU. 2011. Evolutionary transitions in bacterial symbiosis. Proc Natl Acad Sci U S A 108(Suppl 2):10800–10807. doi: 10.1073/pnas.1100304108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Apprill A, Miller CA, Moore MJ, Durban JW, Fearnbach H, Barrett-Lennard LG. 2017. Extensive core microbiome in drone-captured whale blow supports a framework for health monitoring. mSystems 2:e00119-17. doi: 10.1128/mSystems.00119-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Apprill A, Robbins J, Eren AM, Pack AA, Reveillaud J, Mattila D, Moore M, Niemeyer M, Moore KMT, Mincer TJ. 2014. Humpback whale populations share a core skin bacterial community: towards a health index for marine mammals? PLoS One 9:e90785. doi: 10.1371/journal.pone.0090785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kudo T, Kidera A, Kida M, Kawauchi A, Shimizu R, Nakahara T, Zhang X, Yamada A, Amano M, Hamada Y, Taniyama S, Arakawa O, Yoshida A, Oshima K, Suda W, Kuwahara H, Nogi Y, Kitamura K, Yuki M, Iida T, Moriya S, Inoue T, Hongoh Y, Hattori M, Ohkuma M. 2014. Draft genome sequences of Psychrobacter strains JCM 18900, JCM 18901, JCM 18902, and JCM 18903, isolated preferentially from frozen aquatic organisms. Genome Announc 2:e00280-14. doi: 10.1128/genomeA.00280-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Banks JC, Craig Cary S, Hogg ID. 2014. Isolated faecal bacterial communities found for Weddell seals, Leptonychotes weddellii, at White Island, McMurdo Sound, Antarctica. Polar Biol 37:1857–1864. doi: 10.1007/s00300-014-1567-x. [DOI] [Google Scholar]
- 16.Yassin AF, Busse H-J. 2009. Psychrobacter lutiphocae sp. nov., isolated from the faeces of a seal. Int J Syst Evol Microbiol 59:2049–2053. doi: 10.1099/ijs.0.008706-0. [DOI] [PubMed] [Google Scholar]
- 17.Kämpfer P, Jerzak L, Wilharm G, Golke J, Busse H-J, Glaeser SP. 2015. Psychrobacter ciconiae sp. nov., isolated from white storks (Ciconia ciconia). Int J Syst Evol Microbiol 65:772–777. doi: 10.1099/ijs.0.000013. [DOI] [PubMed] [Google Scholar]
- 18.Kämpfer P, Glaeser SP, Irgang R, Fernández-Negrete G, Poblete-Morales M, Fuentes-Messina D, Cortez-San Martín M, Avendaño-Herrera R. 2020. Psychrobacter pygoscelis sp. nov. isolated from the penguin Pygoscelis papua. Int J Syst Evol Microbiol 70:211–219. doi: 10.1099/ijsem.0.003739. [DOI] [PubMed] [Google Scholar]
- 19.Svanevik CS, Lunestad BT. 2011. Characterisation of the microbiota of Atlantic mackerel (Scomber scombrus). Int J Food Microbiol 151:164–170. doi: 10.1016/j.ijfoodmicro.2011.08.016. [DOI] [PubMed] [Google Scholar]
- 20.Yoon J-H, Lee C-H, Kang S-J, Oh T-K. 2005. Psychrobacter celer sp. nov., isolated from sea water of the South Sea in Korea. Int J Syst Evol Microbiol 55:1885–1890. doi: 10.1099/ijs.0.63682-0. [DOI] [PubMed] [Google Scholar]
- 21.Bowman JP, Nichols DS, McMeekin TA. 1997. Psychrobacter glacincola sp. nov., a halotolerant, psychrophilic bacterium isolated from Antarctic sea ice. Syst Appl Microbiol 20:209–215. doi: 10.1016/S0723-2020(97)80067-7. [DOI] [Google Scholar]
- 22.Matsuyama H, Minami H, Sakaki T, Kasahara H, Watanabe A, Onoda T, Hirota K, Yumoto I. 2015. Psychrobacter oceani sp. nov., isolated from marine sediment. Int J Syst Evol Microbiol 65:1450–1455. doi: 10.1099/ijs.0.000118. [DOI] [PubMed] [Google Scholar]
- 23.Zeng Y-X, Yu Y, Liu Y, Li H-R. 2016. Psychrobacter glaciei sp. nov., isolated from the ice core of an Arctic glacier. Int J Syst Evol Microbiol 66:1792–1798. doi: 10.1099/ijsem.0.000939. [DOI] [PubMed] [Google Scholar]
- 24.Bakermans C, Ayala-del-Río HL, Ponder MA, Vishnivetskaya T, Gilichinsky D, Thomashow MF, Tiedje JM. 2006. Psychrobacter cryohalolentis sp. nov. and Psychrobacter arcticus sp. nov., isolated from Siberian permafrost. Int J Syst Evol Microbiol 56:1285–1291. doi: 10.1099/ijs.0.64043-0. [DOI] [PubMed] [Google Scholar]
- 25.Deschaght P, Janssens M, Vaneechoutte M, Wauters G. 2012. Psychrobacter isolates of human origin, other than Psychrobacter phenylpyruvicus, are predominantly Psychrobacter faecalis and Psychrobacter pulmonis, with emended description of P faecalis. Int J Syst Evol Microbiol 62:671–674. doi: 10.1099/ijs.0.032631-0. [DOI] [PubMed] [Google Scholar]
- 26.Wirth SE, Ayala-del-Río HL, Cole JA, Kohlerschmidt DJ, Musser KA, Sepúlveda-Torres LDC, Thompson LM, Wolfgang WJ. 2012. Psychrobacter sanguinis sp. nov., recovered from four clinical specimens over a 4-year period. Int J Syst Evol Microbiol 62:49–54. doi: 10.1099/ijs.0.029058-0. [DOI] [PubMed] [Google Scholar]
- 27.Bakermans C. 2018. Adaptations to marine versus terrestrial low temperature environments as revealed by comparative genomic analyses of the genus Psychrobacter. FEMS Microbiol Ecol 94(7). doi: 10.1093/femsec/fiy102. [DOI] [PubMed] [Google Scholar]
- 28.Vela AI, Sánchez-Porro C, Aragón V, Olvera A, Domínguez L, Ventosa A, Fernández-Garayzábal JF. 2010. Moraxella porci sp. nov., isolated from pigs. Int J Syst Evol Microbiol 60:2446–2450. doi: 10.1099/ijs.0.016626-0. [DOI] [PubMed] [Google Scholar]
- 29.Vela AI, Arroyo E, Aragón V, Sánchez-Porro C, Latre MV, Cerdà-Cuéllar M, Ventosa A, Domínguez L, Fernández-Garayzábal JF. 2009. Moraxella pluranimalium sp. nov., isolated from animal specimens. Int J Syst Evol Microbiol 59:671–674. doi: 10.1099/ijs.0.006205-0. [DOI] [PubMed] [Google Scholar]
- 30.Lindqvist K. 1960. A Neisseria species associated with infections keratoconjunctivitis of sheep Neisseria ovis nov. spec. J Infect Dis 106:162–165. doi: 10.1093/infdis/106.2.162. [DOI] [PubMed] [Google Scholar]
- 31.Xie C-H, Yokota A. 2005. Transfer of the misnamed [Alysiella] sp. IAM 14971 (=ATCC 29468) to the genus Moraxella as Moraxella oblonga sp. nov. Int J Syst Evol Microbiol 55:331–334. doi: 10.1099/ijs.0.63250-0. [DOI] [PubMed] [Google Scholar]
- 32.Bövre K, Henriksen SD. 1967. A new Moraxella species, Moraxella osloensis, and a revised description of Moraxella nonliquefaciens. Int J Syst Evol Microbiol 17:127–135. [Google Scholar]
- 33.Catlin BW. 1970. Transfer of the organism named Neisseria Catarrhalis to Branhamella gen. nov. Int J Syst Bacteriol 20:155–159. doi: 10.1099/00207713-20-2-155. [DOI] [Google Scholar]
- 34.Eyre JW. 1900. A clinical and bacteriological study of diplo-bacillary conjunctivitis. J Pathol 6:1–13. doi: 10.1002/path.1700060102. [DOI] [Google Scholar]
- 35.Vandamme P, Gillis M, Vancanneyt l.M, Hoste l.B, Kersters K, Falsen E. 1993. Moraxella lincolnii sp. nov., Isolated from the human respiratory tract, and reevaluation of the taxonomic position of Moraxella osloensis. Int J Syst Bacteriol 43:474–481. doi: 10.1099/00207713-43-3-474. [DOI] [PubMed] [Google Scholar]
- 36.Hughes DE, Pugh GW, Jr.. 1970. Isolation and description of a Moraxella from horses with conjunctivitis. Am J Vet Res 31:457–462. [PubMed] [Google Scholar]
- 37.Berger U. 1962. Über das Vorkommen von Neisserien bei einigen Tieren. Z Hyg Infektionskr 148:445–457. doi: 10.1007/BF02156782. [DOI] [Google Scholar]
- 38.Pelczar MJ, Jr. 1953. Neisseria caviae nov spec. J Bacteriol 65:744. doi: 10.1128/JB.65.6.744-744.1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kodjo A, Tønjum T, Richard Y, Bøvre K. 1995. Moraxella caprae sp. nov., a new member of the classical Moraxellae with very close affinity to Moraxella bovis. Int J Syst Bacteriol 45:467–471. doi: 10.1099/00207713-45-3-467. [DOI] [PubMed] [Google Scholar]
- 40.Jannes G, Vaneechoutte M, Lannoo M, Gillis M, Vancanneyt M, Vandamme P, Verschraegen G, Van Heuverswyn H, Rossau R. 1993. Polyphasic taxonomy leading to the proposal of Moraxella canis sp. nov. for Moraxella catarrhalis-like strains. Int J Syst Bacteriol 43:438–449. doi: 10.1099/00207713-43-3-438. [DOI] [PubMed] [Google Scholar]
- 41.Angelos JA, Spinks PQ, Ball LM, George LW. 2007. Moraxella bovoculi sp. nov., isolated from calves with infectious bovine keratoconjunctivitis. Int J Syst Evol Microbiol 57:789–795. doi: 10.1099/ijs.0.64333-0. [DOI] [PubMed] [Google Scholar]
- 42.Henriksen SD. 1971. Designation of a neotype strain for Moraxella bovis (Hauduroy et al.) Murray. Int J Syst Evol Microbiol 21:28. doi: 10.1099/00207713-21-1-28. [DOI] [Google Scholar]
- 43.Kodjo A, Richard Y, Tønjum T. 1997. Moraxella boevrei sp. nov., a new Moraxella species found in goats. Int J Syst Bacteriol 47:115–121. doi: 10.1099/00207713-47-1-115. [DOI] [PubMed] [Google Scholar]
- 44.Bovre K, Fuglesang JE, Hagen N, Jantzen E, Froholm LO. 1976. Moraxella atlantae sp. nov. and its distinction from Moraxella phenylpyrouvica. Int J Syst Bacteriol 26:511–521. doi: 10.1099/00207713-26-4-511. [DOI] [Google Scholar]
- 45.Vaneechoutte M, Nemec A, Musílek M, van der Reijden TJK, van den Barselaar M, Tjernberg I, Calame W, Fani R, De Baere T, Dijkshoorn L. 2009. Description of Acinetobacter venetianus ex Di Cello et al. 1997 sp. nov. Int J Syst Evol Microbiol 59:1376–1381. doi: 10.1099/ijs.0.003541-0. [DOI] [PubMed] [Google Scholar]
- 46.Nemec A, De Baere T, Tjernberg I, Vaneechoutte M, van der Reijden TJ, Dijkshoorn L. 2001. Acinetobacter ursingii sp. nov. and Acinetobacter schindleri sp. nov., isolated from human clinical specimens. Int J Syst Evol Microbiol 51:1891–1899. doi: 10.1099/00207713-51-5-1891. [DOI] [PubMed] [Google Scholar]
- 47.Nishimura Y, Ino T, Iizuka H. 1988. Acinetobacter radioresistens sp. nov. isolated from cotton and soil. Int J Syst Evol Microbiol 38:209–211. doi: 10.1099/00207713-38-2-209. [DOI] [Google Scholar]
- 48.Li Y, Piao C-G, Ma Y-C, He W, Wang H-M, Chang J-P, Guo L-M, Wang X-Z, Xie S-J, Guo M-W. 2013. Acinetobacter puyangensis sp. nov., isolated from the healthy and diseased part of Populus xeuramericana canker bark. Int J Syst Evol Microbiol 63:2963–2969. doi: 10.1099/ijs.0.047274-0. [DOI] [PubMed] [Google Scholar]
- 49.Liu Y, Rao Q, Tu J, Zhang J, Huang M, Hu B, Lin Q, Luo T. 2018. Acinetobacter piscicola sp. nov., isolated from diseased farmed Murray cod (Maccullochella peelii peelii). Int J Syst Evol Microbiol 68:905–910. doi: 10.1099/ijsem.0.002608. [DOI] [PubMed] [Google Scholar]
- 50.Choi JY, Ko G, Jheong W, Huys G, Seifert H, Dijkshoorn L, Ko KS. 2013. Acinetobacter kookii sp. nov., isolated from soil. Int J Syst Evol Microbiol 63:4402–4406. doi: 10.1099/ijs.0.047969-0. [DOI] [PubMed] [Google Scholar]
- 51.Malhotra J, Anand S, Jindal S, Rajagopal R, Lal R. 2012. Acinetobacter indicus sp. nov., isolated from a hexachlorocyclohexane dump site. Int J Syst Evol Microbiol 62:2883–2890. doi: 10.1099/ijs.0.037721-0. [DOI] [PubMed] [Google Scholar]
- 52.Nemec A, Musílek M, Maixnerová M, De Baere T, van der Reijden TJK, Vaneechoutte M, Dijkshoorn L. 2009. Acinetobacter beijerinckii sp. nov. and Acinetobacter gyllenbergii sp. nov., haemolytic organisms isolated from humans. Int J Syst Evol Microbiol 59:118–124. doi: 10.1099/ijs.0.001230-0. [DOI] [PubMed] [Google Scholar]
- 53.Smet A, Cools P, Krizova L, Maixnerova M, Sedo O, Haesebrouck F, Kempf M, Nemec A, Vaneechoutte M. 2014. Acinetobacter gandensis sp. nov. isolated from horse and cattle. Int J Syst Evol Microbiol 64:4007–4015. doi: 10.1099/ijs.0.068791-0. [DOI] [PubMed] [Google Scholar]
- 54.Nemec A, Radolfova-Krizova L, Maixnerova M, Vrestiakova E, Jezek P, Sedo O. 2016. Taxonomy of haemolytic and/or proteolytic strains of the genus Acinetobacter with the proposal of Acinetobacter courvalinii sp. nov. (genomic species 14 sensu Bouvet & Jeanjean), Acinetobacter dispersus sp. nov. (genomic species 17), Acinetobacter modestus sp. nov., Acinetobacter proteolyticus sp. nov. and Acinetobacter vivianii sp. nov. Int J Syst Evol Microbiol 66:1673–1685. doi: 10.1099/ijsem.0.000932. [DOI] [PubMed] [Google Scholar]
- 55.Nemec A, Radolfova-Krizova L, Maixnerova M, Sedo O. 2017. Acinetobacter colistiniresistens sp. nov. (formerly genomic species 13 sensu Bouvet and Jeanjean and genomic species 14 sensu Tjernberg and Ursing), isolated from human infections and characterized by intrinsic resistance to polymyxins. Int J Syst Evol Microbiol 67:2134–2141. doi: 10.1099/ijsem.0.001903. [DOI] [PubMed] [Google Scholar]
- 56.Radolfova-Krizova L, Maixnerova M, Nemec A. 2016. Acinetobacter celticus sp. nov., a psychrotolerant species widespread in natural soil and water ecosystems. Int J Syst Evol Microbiol 66:5392–5398. doi: 10.1099/ijsem.0.001526. [DOI] [PubMed] [Google Scholar]
- 57.Álvarez-Pérez S, Lievens B, Jacquemyn H, Herrera CM. 2013. Acinetobacter nectaris sp. nov. and Acinetobacter boissieri sp. nov., isolated from floral nectar of wild Mediterranean insect-pollinated plants. Int J Syst Evol Microbiol 63:1532–1539. doi: 10.1099/ijs.0.043489-0. [DOI] [PubMed] [Google Scholar]
- 58.Bouvet PJM, Grimont PAD. 1986. Taxonomy of the genus Acinetobacter with the recognition of Acinetobacter baumannii sp. nov., Acinetobacter haemolyticus sp. nov., Acinetobacter johnsonii sp. nov. and Acinetobacter junii sp. nov. and emended descriptions of Acinetobacter calcoaceticus and Acinetobacter lwoffii. Int J Syst Bacteriol 36:228–240. doi: 10.1099/00207713-36-2-228. [DOI] [Google Scholar]
- 59.Baik KS, Park SC, Lim CH, Lee KH, Jeon DY, Kim CM, Seong CN. 2010. Psychrobacter aestuarii sp. nov., isolated from a tidal flat sediment. Int J Syst Evol Microbiol 60:1631–1636. doi: 10.1099/ijs.0.016782-0. [DOI] [PubMed] [Google Scholar]
- 60.Bowman JP, Cavanagh J, Austin JJ, Sanderson K. 1996. Novel Psychrobacter species from Antarctic ornithogenic soils. Int J Syst Bacteriol 46:841–848. doi: 10.1099/00207713-46-4-841. [DOI] [PubMed] [Google Scholar]
- 61.Kämpfer P, Albrecht A, Buczolits S, Busse H-J. 2002. Psychrobacter faecalis sp. nov., a new species from a bioaerosol originating from pigeon faeces. Syst Appl Microbiol 25:31–36. doi: 10.1078/0723-2020-00099. [DOI] [PubMed] [Google Scholar]
- 62.Romanenko LA, Lysenko AM, Rohde M, Mikhailov VV, Stackebrandt E. 2004. Psychrobacter maritimus sp. nov. and Psychrobacter arenosus sp. nov., isolated from coastal sea ice and sediments of the Sea of Japan. Int J Syst Evol Microbiol 54:1741–1745. doi: 10.1099/ijs.0.63096-0. [DOI] [PubMed] [Google Scholar]
- 63.Yumoto I, Hirota K, Sogabe Y, Nodasaka Y, Yokota Y, Hoshino T. 2003. Psychrobacter okhotskensis sp. nov., a lipase-producing facultative psychrophile isolated from the coast of the Okhotsk Sea. Int J Syst Evol Microbiol 53:1985–1989. doi: 10.1099/ijs.0.02686-0. [DOI] [PubMed] [Google Scholar]
- 64.Juni E, Heym GA. 1986. Psychrobacter immobilis gen. nov., sp. nov.: genospecies composed of Gram-negative, aerobic, oxidase-positive coccobacilli. Int J Syst Evol Microbiol 36:388–391. [Google Scholar]
- 65.Yoon J-H, Lee C-H, Yeo S-H, Oh T-K. 2005. Psychrobacter aquimaris sp. nov. and Psychrobacter namhaensis sp. nov., isolated from sea water of the South Sea in Korea. Int J Syst Evol Microbiol 55:1007–1013. doi: 10.1099/ijs.0.63464-0. [DOI] [PubMed] [Google Scholar]
- 66.Maruyama A, Honda D, Yamamoto H, Kitamura K, Higashihara T. 2000. Phylogenetic analysis of psychrophilic bacteria isolated from the Japan Trench, including a description of the deep-sea species Psychrobacter pacificensis sp. nov. Int J Syst Evol Microbiol 50(Pt 2):835–846. doi: 10.1099/00207713-50-2-835. [DOI] [PubMed] [Google Scholar]
- 67.Yumoto I, Hirota K, Kimoto H, Nodasaka Y, Matsuyama H, Yoshimune K. 2010. Psychrobacter piscatorii sp. nov., a psychrotolerant bacterium exhibiting high catalase activity isolated from an oxidative environment. Int J Syst Evol Microbiol 60:205–208. doi: 10.1099/ijs.0.010959-0. [DOI] [PubMed] [Google Scholar]
- 68.Poppel MT, Skiebe E, Laue M, Bergmann H, Ebersberger I, Garn T, Fruth A, Baumgardt S, Busse H-J, Wilharm G. 2016. Acinetobacter equi sp. nov., isolated from horse faeces. Int J Syst Evol Microbiol 66:881–888. doi: 10.1099/ijsem.0.000806. [DOI] [PubMed] [Google Scholar]
- 69.Dziewit L, Cegielski A, Romaniuk K, Uhrynowski W, Szych A, Niesiobedzki P, Zmuda-Baranowska MJ, Zdanowski MK, Bartosik D. 2013. Plasmid diversity in arctic strains of Psychrobacter spp. Extremophiles 17:433–444. doi: 10.1007/s00792-013-0521-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bobay L-M, Ochman H. 2017. The evolution of bacterial genome architecture. Front Genet 8:72. doi: 10.3389/fgene.2017.00072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kuo C-H, Ochman H. 2009. Deletional bias across the three domains of life. Genome Biol Evol 1:145–152. doi: 10.1093/gbe/evp016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Collins C, Didelot X. 2018. A phylogenetic method to perform genome-wide association studies in microbes that accounts for population structure and recombination. PLoS Comput Biol 14:e1005958. doi: 10.1371/journal.pcbi.1005958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Stone GN, Nee S, Felsenstein J. 2011. Controlling for non-independence in comparative analysis of patterns across populations within species. Philos Trans R Soc Lond B Biol Sci 366:1410–1424. doi: 10.1098/rstb.2010.0311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Subhadra B, Surendran S, Kim DH, Woo K, Oh MH, Choi CH. 2019. The transcription factor NemR is an electrophile-sensing regulator important for the detoxification of reactive electrophiles in Acinetobacter nosocomialis. Res Microbiol 170:123–130. doi: 10.1016/j.resmic.2019.02.001. [DOI] [PubMed] [Google Scholar]
- 75.Ma Q, Yang Z, Pu M, Peti W, Wood TK. 2011. Engineering a novel c-di-GMP-binding protein for biofilm dispersal. Environ Microbiol 13:631–642. doi: 10.1111/j.1462-2920.2010.02368.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bodelón G, Palomino C, Fernández LÁ. 2013. Immunoglobulin domains in Escherichia coli and other enterobacteria: from pathogenesis to applications in antibody technologies. FEMS Microbiol Rev 37:204–250. doi: 10.1111/j.1574-6976.2012.00347.x. [DOI] [PubMed] [Google Scholar]
- 77.Lee B-H, Hébraud M, Bernardi T. 2017. Increased adhesion of Listeria monocytogenes strains to abiotic surfaces under cold stress. Front Microbiol 8:2221. doi: 10.3389/fmicb.2017.02221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bullen JJ, Rogers HJ, Griffiths E. 1978. Role of iron in bacterial infection. Curr Top Microbiol Immunol 80:1–35. [DOI] [PubMed] [Google Scholar]
- 79.Hendry TA, Freed LL, Fader D, Fenolio D, Sutton TT, Lopez JV. 2018. Ongoing transposon-mediated genome reduction in the luminous bacterial symbionts of deep-sea ceratioid anglerfishes. mBio 9:e01033-18. doi: 10.1128/mBio.01033-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Collins T, Margesin R. 2019. Psychrophilic lifestyles: mechanisms of adaptation and biotechnological tools. Appl Microbiol Biotechnol 103:2857–2871. doi: 10.1007/s00253-019-09659-5. [DOI] [PubMed] [Google Scholar]
- 81.Perez Vidakovics ML, Riesbeck K. 2009. Virulence mechanisms of Moraxella in the pathogenesis of infection. Curr Opin Infect Dis 22:279–285. doi: 10.1097/qco.0b013e3283298e4e. [DOI] [PubMed] [Google Scholar]
- 82.Juni E, Bøvre K. 2015. Moraxella, p 1–17. In Whitman WB, Rainey F, Kämpfer P, Trujillo M, Chun J, DeVos P, Hedlund B, Dedysh S (ed), Bergey’s manual of systematics of archaea and bacteria. John Wiley & Sons, Ltd, Chichester, United Kingdom. [Google Scholar]
- 83.Karasov TL, Almario J, Friedemann C, Ding W, Giolai M, Heavens D, Kersten S, Lundberg DS, Neumann M, Regalado J, Neher RA, Kemen E, Weigel D. 2018. Arabidopsis thaliana and Pseudomonas pathogens exhibit stable associations over evolutionary timescales. Cell Host Microbe 24:168–179.e4. doi: 10.1016/j.chom.2018.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Feng S, Powell SM, Wilson R, Bowman JP. 2014. Extensive gene acquisition in the extremely psychrophilic bacterial species Psychroflexus torquis and the link to sea-ice ecosystem specialism. Genome Biol Evol 6:133–148. doi: 10.1093/gbe/evt209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Liu Y, Harrison PM, Kunin V, Gerstein M. 2004. Comprehensive analysis of pseudogenes in prokaryotes: widespread gene decay and failure of putative horizontally transferred genes. Genome Biol 5:R64. doi: 10.1186/gb-2004-5-9-r64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.NCBI Resource Coordinators. 2018. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res 46:D8–D13. doi: 10.1093/nar/gkx1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Segata N, Börnigen D, Morgan XC, Huttenhower C. 2013. PhyloPhlAn is a new method for improved phylogenetic and taxonomic placement of microbes. Nat Commun 4:2304. doi: 10.1038/ncomms3304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW. 2015. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res 25:1043–1055. doi: 10.1101/gr.186072.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 90.Letunic I, Bork P. 2007. Interactive Tree Of Life (iTOL): an online tool for phylogenetic tree display and annotation. Bioinformatics 23:127–128. doi: 10.1093/bioinformatics/btl529. [DOI] [PubMed] [Google Scholar]
- 91.Ding W, Baumdicker F, Neher RA. 2018. panX: pan-genome analysis and exploration. Nucleic Acids Res 46:e5. doi: 10.1093/nar/gkx977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Huerta-Cepas J, Forslund K, Coelho LP, Szklarczyk D, Jensen LJ, von Mering C, Bork P. 2017. Fast genome-wide functional annotation through orthology assignment by eggNOG-Mapper. Mol Biol Evol 34:2115–2122. doi: 10.1093/molbev/msx148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Goslee SC, Urban DL. 2007. The ecodist package for dissimilarity-based analysis of ecological data. J Stat Softw 22:1–19. [Google Scholar]
- 94.Dixon P. 2003. VEGAN, a package of R functions for community ecology. J Veg Sci 14:927–930. doi: 10.1111/j.1654-1103.2003.tb02228.x. [DOI] [Google Scholar]
- 95.Tatusov RL, Galperin MY, Natale DA, Koonin EV. 2000. The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res 28:33–36. doi: 10.1093/nar/28.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Revell LJ. 2012. phytools: an R package for phylogenetic comparative biology (and other things). Methods Ecol Evol 3:217–223. doi: 10.1111/j.2041-210X.2011.00169.x. [DOI] [Google Scholar]
- 97.Wick R. 2018. Porechop: adapter trimmer for Oxford Nanopore reads. https://github.com/rrwick/Porechop.
- 98.De Coster W, D’Hert S, Schultz DT, Cruts M, Van Broeckhoven C. 2018. NanoPack: visualizing and processing long-read sequencing data. Bioinformatics 34:2666–2669. doi: 10.1093/bioinformatics/bty149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wick RR, Judd LM, Gorrie CL, Holt KE. 2017. Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput Biol 13:e1005595. doi: 10.1371/journal.pcbi.1005595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Tanizawa Y, Fujisawa T, Nakamura Y. 2018. DFAST: a flexible prokaryotic genome annotation pipeline for faster genome publication. Bioinformatics 34:1037–1039. doi: 10.1093/bioinformatics/btx713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Galan M, Pagès M, Cosson J-F. 2012. Next-generation sequencing for rodent barcoding: species identification from fresh, degraded and environmental samples. PLoS One 7:e48374. doi: 10.1371/journal.pone.0048374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Goodrich JK, Waters JL, Poole AC, Sutter JL, Koren O, Blekhman R, Beaumont M, Van Treuren W, Knight R, Bell JT, Spector TD, Clark AG, Ley RE. 2014. Human genetics shape the gut microbiome. Cell 159:789–799. doi: 10.1016/j.cell.2014.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Hodkinson B, Lutzoni F. 2009. 16S ribosomal DNA. http://lutzonilab.org/16s-ribosomal-dna/.
- 105.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 106.R Development Core Team. 2011. R: a language and environment for statistical computing. R Project for Statistical Computing, Vienna, Austria. [Google Scholar]
- 107.Pennell MW, Eastman JM, Slater GJ, Brown JW, Uyeda JC, FitzJohn RG, Alfaro ME, Harmon LJ. 2014. geiger v2.0: an expanded suite of methods for fitting macroevolutionary models to phylogenetic trees. Bioinformatics 30:2216–2218. doi: 10.1093/bioinformatics/btu181. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Moraxellaceae genomics. Eighteen species each of Acinetobacter and Moraxella were chosen based on availability of information on isolation source and cultivation temperatures, and their genomes were downloaded from NCBI. Fifteen additional Psychrobacter accessions were chosen for the family-level analysis based on phylogenetic breadth. Download Table S1, XLSX file, 0.01 MB (13.7KB, xlsx) .
Copyright © 2021 Welter et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
COG separation of PCoA. An envfit analysis was performed to identify functions that might be correlated with separation of accessions along a gene presence/absence principal-component analysis, for a PCoA at the Moraxellaceae family level as well as the Psychrobacter genus level. For every COG category, the r2, indicating the strength of the correlation; the P value, indicating the significance of the correlation; and the Benjamini-Hochberg-corrected P value are reported. Download Table S2, XLSX file, 0.01 MB (9KB, xlsx) .
Copyright © 2021 Welter et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Psychrobacter cultivation. Summary of information from strain catalogues about the cultivation media and temperatures, isolation information, previously published genomes, and phenotyping batch of each of the 92 Psychrobacter strains initially included in our strain collection, with notes about the 7 strains that were ultimately not used for the analysis. Download Table S3, XLSX file, 0.02 MB (17.1KB, xlsx) .
Copyright © 2021 Welter et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Sequencing summary. Summary of the total number of short and long reads, as well as quality control information provided by CheckM, such as completeness, contamination, genome size, and scaffold information, for all 92 Psychrobacter accessions included in our initial strain collection. Some strains were removed from further analysis due to poor-quality genomes. Download Table S4, XLSX file, 0.02 MB (17.5KB, xlsx) .
Copyright © 2021 Welter et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Data Availability Statement
Raw sequences for the Psychrobacter genome sequencing and polar bear feces 16S rRNA gene sequencing, as well as assembled Psychrobacter genomes, are available in the European Nucleotide Archive under the accession no. PRJEB40380. Annotated Psychrobacter genomes are available at ftp://ftp.tue.mpg.de/pub/ebio/dwelter. Raw data, R notebooks, and Python scripts for the analyses are available at https://github.com/dkwelter/Welter_et_al_2020.