ABSTRACT
Staphylococcus aureus sequence type (ST) 72, the predominant community-associated methicillin-resistant Staphylococcus aureus (CA-MRSA) lineage in South Korea, has emerged as a major cause of bloodstream infection in hospital settings. However, relatively little information is available regarding the genomic characteristics and dissemination of ST72. Here, we characterized the whole-genome sequence of 24 ST72 isolates from China, along with 83 ST72 genomes from global sources. Of these 107 ST72 isolates, 63 were MRSA and 44 were methicillin-susceptible S. aureus (MSSA). Phylogenetic analysis revealed four distinct clades (A, B, C, and D), of which clade D contained only MSSA isolates. By characterizing the evolutionary dynamics of the ST72 lineage, we found that the MRSA from China might not have developed from the MSSA in China. Furthermore, we observed both international transmission of ST72 isolates and interregional transmission within China. The distributions of the SCCmec and spa types of isolates differed among clades. Additionally, in silico analyses revealed that the distributions of resistance genes, virulence genes, and mobile genetic elements (MGEs) also differed among isolates of the four clades. This was especially true for clade D isolates, which had the lowest level of antimicrobial resistance and had obtained specific virulence genes such as tsst-1 by acquisition of specific MGEs. Notably, ST72 MRSA isolates were more antibiotic resistant than ST72 MSSA isolates, but comparably virulent. Our findings provide insight into the potential transmission and genotypic features of ST72 clones across the globe.
IMPORTANCE Understanding the evolution and dissemination of community-genotype ST72 Staphylococcus aureus isolates is important, as isolates of this lineage have rapidly spread into hospital settings and caused serious health issues. In this study, we first carried out genome-wide analysis of 107 global ST72 isolates to characterize the evolution and genetic diversity of the ST72 lineage. We found that the MSSA lineage in China might have evolved independently from the MRSA isolates from China, and that ST72 isolates have the potential to undergo both international transmission and interregional transmission within China. The diversity of isolates correlated with distinct acquisitions of SCCmec elements, antibiotic resistance genes, virulence genes, and mobile genetic elements. The comprehensive information on the ST72 lineage emerging from this study will enable improved therapeutic approaches and rapid molecular diagnosis.
KEYWORDS: Staphylococcus aureus, sequence type 72, evolutionary dynamics, transmission, antimicrobial resistance, virulence, mobile genetic elements
INTRODUCTION
Staphylococcus aureus is an important human pathogen that causes serious hospital- and community-acquired infections worldwide. It asymptomatically colonizes human skin and nasal membranes in about one-third of the human population, and may account for infections with outcomes that range from mild skin lesions to severe diseases, such as necrotizing pneumonia, septicemia, endocarditis, and toxic shock syndrome (1, 2). Until the mid-1990s, methicillin-resistant S. aureus (MRSA) was reported to be solely hospital acquired (HA-MRSA) (3); however, in the late 1990s, it began to be increasingly identified in community-acquired infections (CA-MRSA) (4). Although HA-MRSA tends to infect elderly patients with underlying health issues (5), CA-MRSA tends to infect healthy younger individuals without identified risk factors, potentially causing fatal sepsis and severe pneumonia (6).
Unlike HA-MRSA isolates, CA-MRSA isolates are generally less resistant to antibiotics and often carry the prophage genes lukS/F-PV, encoding Panton-Valentine leukocidin (PVL) toxin, which might have a role in promoting soft tissue and skin infections (7); however, several CA-MRSA clones are PVL-negative, including the CA-MRSA clone ST72 (8). In addition, CA-MRSA harbors type IV or V staphylococcal cassette chromosome mec (SCCmec) elements that are smaller than the type I, II, or III SCCmec elements carried by HA-MRSA (7, 9). The extraordinary success of CA-MRSA isolates is owing to the combination of two factors, i.e., the low fitness cost of methicillin resistance and elevated virulence (10). Distinct CA-MRSA lineages dominate in different geographical regions, e.g., USA300 in the United States and ST59 in Asia (11, 12).
CA-MRSA ST72 is the most widespread CA-MRSA genotype in South Korea (13). The first report on CA-MRSA in South Korea revealed that ST72 accounted for 35% of all CA-MRSA isolates (14). Since the late 2000s, ST72 MRSA has been introduced into hospitals and has become a major cause of bloodstream infection (BSI) (15). Joo et al. (16) reported that ST72 isolates are tolerant of stressful environments, including desiccation and hypotonic solutions, which could promote their survival in health care facilities. Moreover, although ST72 is relatively susceptible to non-β-lactams, several isolates have undergone rapid evolution of antimicrobial resistance, and some have been identified as vancomycin-intermediate isolates (17, 18). Although isolation of ST72 isolates has been reported sporadically in other countries such as China (19), Japan (18), and Spain (20), no previous study has provided detailed information on the genomic features of the ST72 lineage on a global scale.
We analyzed the genomic features of 24 ST72 strains isolated in China between 2014 and 2019. To characterize the genotypic features and potential transmission of various ST72 isolates on a global scale, 83 additional ST72 isolates with available geographic information were selected for comparison (Table S1 in the supplemental material). We then performed phylogenetic analysis to reconstruct the evolutionary dynamics of the ST72 lineage. We also analyzed the resistance genes, virulence genes, and mobile genetic elements (MGEs) of ST72 isolates. These genomic analyses clarify the genomic differences, evolutionary relationships, and variations in antibiotic resistance and virulence potential in the ST72 lineage.
Information on ST72 isolates analyzed in this study. Download Table S1, DOCX file, 0.04 MB (41.3KB, docx) .
Copyright © 2021 Zhou et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
RESULTS
Geographic distribution of the S. aureus ST72 isolates.
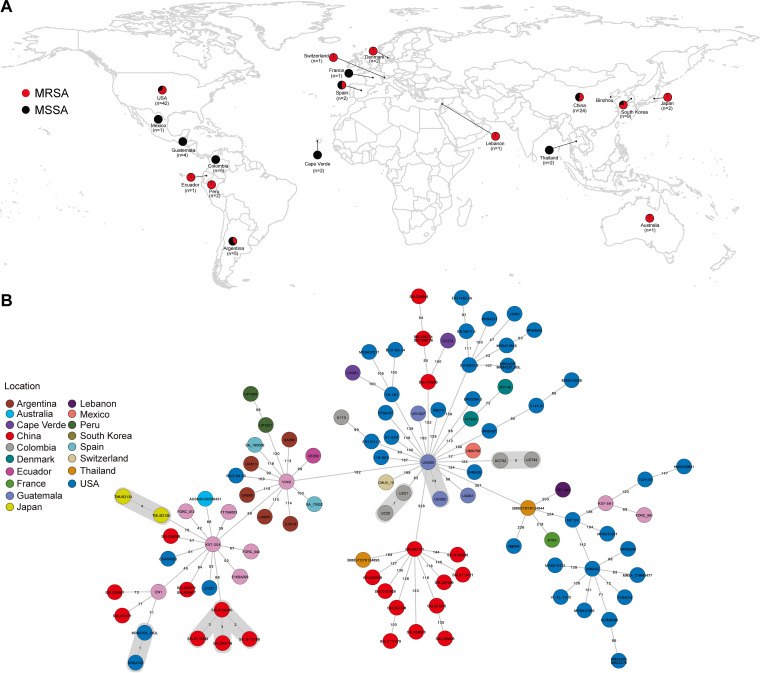
In this study, we analyzed 107 ST72 S. aureus isolates sampled between 2003 and 2019 (24 from our collection and 83 from the NCBI database), spanning 18 different countries in Asia, Europe, America, Africa, and Oceania (Fig. 1A). The top three countries with the highest numbers of ST72 isolates were the United States (39.25% of the total, n = 42), China (22.43%, n = 24), and South Korea (8.41%, n = 9). Of the 107 ST72 isolates, 63 were methicillin resistant and 44 were methicillin-susceptible S. aureus (MSSA). All isolates from China in our collection (13 MRSA and 11 MSSA) emerged in provinces in close geographic proximity, with the exception of Heilongjiang (Fig. S1A), suggesting potential transmission of ST72 isolates between geographic areas.
FIG 1.
Relationships among ST72 isolates in this study. (A) Geographical distribution of 107 ST72 S. aureus isolates. Red and black dots indicate MRSA and MSSA isolates, respectively. The map was created using R package ggplot2 (https://github.com/tidyverse/ggplot2). (B) MST of ST72 isolates. Each circle represents a ST72 isolate, with colors assigned according to geographic origin. The numbers on the connecting lines illustrate the number of SNPs in a pairwise comparison. The transmission cluster is marked with gray based on the threshold of 23 SNPs.
Relationships among 24 ST72 isolates from China in this study. (A) Geographical distribution of 24 ST72 S. aureus isolates. Red and black dots indicate MRSA and MSSA isolates, respectively. The map was created using R package ggplot2 (https://github.com/tidyverse/ggplot2). (B) Phylogenetic tree of 24 ST72 isolates. Tips are colored by province of isolation. (C) MST of 24 ST72 isolates. Each circle represents a ST72 isolate, with colors assigned according to geographic origin. The numbers on the connecting lines illustrate the number of SNPs in a pairwise comparison. The transmission cluster is marked with gray based on the threshold of 23 SNPs. ND, not determined. Download FIG S1, PDF file, 0.3 MB (362.4KB, pdf) .
Copyright © 2021 Zhou et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Phylogenomic analysis of the ST72 lineage.
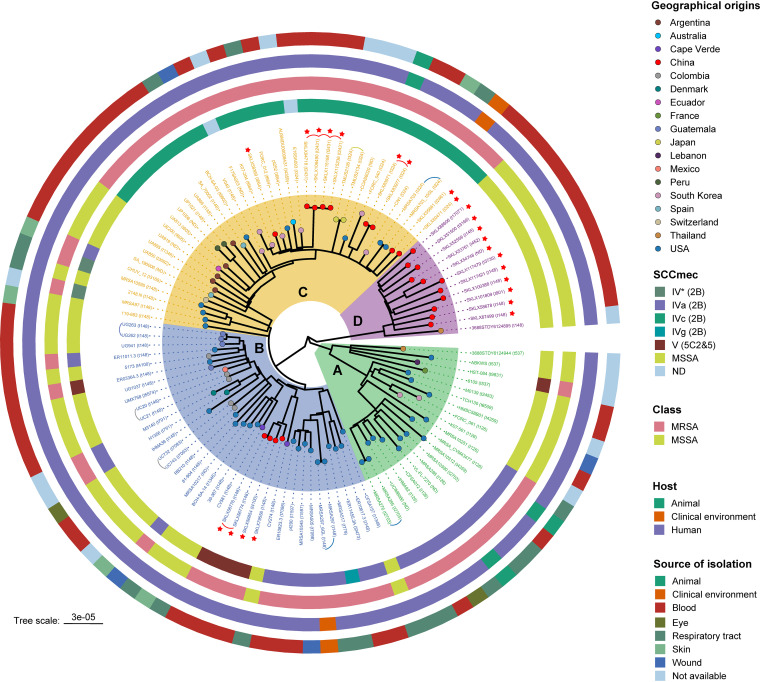
To analyze the evolutionary position and genetic diversity of the ST72 isolates examined in this study, we determined single nucleotide polymorphisms (SNPs) within the core genome among all 107 ST72 isolates. Phylogenomic analysis based on 6,345 core SNPs revealed a diverse population structure containing four distinct clades, A through D. The paired SNP distances of isolates between four clades varied from 182 to 488 (median, 368; average, 336), suggesting that the isolates from different clades were distantly related (Fig. 1B). As shown in Fig. 2, clades A, B, and C contained both MRSA and MSSA isolates, whereas clade D contained only MSSA isolates from China (n = 11) and Thailand (n = 1). In addition, the isolates collected from China and the United States were distributed across three clades of the tree, indicating that isolates from these two countries had greater variability in the core genomic regions (Fig. 2).
FIG 2.
Phylogenetic structure of 107 ST72 isolates. Clades A, B, C, and D are marked by background color fill. Tips are colored according to country of isolation. Data on the SCCmec type, MRSA/MSSA classification, host type, and source of isolation are mapped on the tree (from inner to outer circle). Red asterisks indicate the isolates obtained from our collection. Curved lines indicate isolates belonging to same transmission. The spa types of each isolate are shown in parentheses near the isolate name. ND, not determined.
Notably, when we mapped the geographic origins of the ST72 lineage onto the phylogenetic tree, we found no evidence for an association between neighboring branches and their locations, regardless of clade (except clade D). By contrast, some isolates clustered with others isolated from different countries, with paired SNP distances ranging from 47 to 488 (median, 265; average, 288). In particular, the four ST72 MRSA isolates collected from the Affiliated Hospital of Binzhou Medical College in 2019 (Binzhou, China) clustered closely with the geographically close isolate E16SA093 from South Korea (isolated in 2016) from clade C (Fig. 2), with paired SNP distance varying from 95 to 98. In addition, using 23 SNPs as the maximum pairwise distance of isolates from the same transmission (21), we identified 10 independent small-scale transmissions across the tree (except clade D) that occurred in the United States, China, Japan, Switzerland, and Guatemala (Fig. 1B and Fig. 2). Of these, two transmission events occurred between patients and the clinical environment in two U.S. hospitals (22).
We also carried out a phylogenetic analysis of 24 ST72 isolates from China. The results revealed no significant boundary existed between isolates from different provinces of China. For instance, isolate SKLX60417, collected from Jiangsu province, was closely related to isolate SKLX55661, collected from Zhejiang province (Fig. S1C). Of note, the paired SNP distances of isolates between these provinces varied from 87 to 452 (median, 365; average, 301) (Fig. S1B).
Time scale of the emergence of global ST72 isolates.
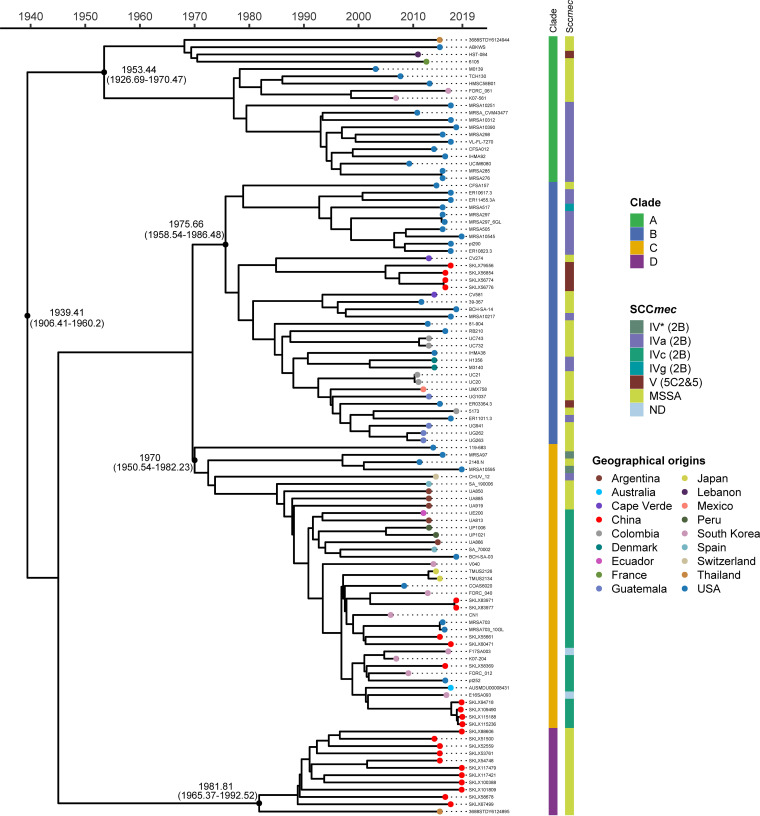
To further assess the time origin of the ST72 lineage, we used a Bayesian approach to explore the temporal spread of the ST72 isolates. The results revealed that the median time of divergence of the ST72 lineage from its most recent common ancestor (MRCA) was around 1939.41 (95% highest posterior density [HPD] interval, 1906.41 to 1960.2). However, the divergence of the four clades in the ST72 lineage occurred more recently; the MRCA of clade A isolates probably emerged in 1953.44, clade B in 1975.66, clade C in 1970, and clade D in 1981.81 (Fig. 3). This finding indicates that the expansion of ST72 occurred at least several decades after its emergence. In addition, the core genome of the ST72 isolates has accumulated variation at a rate of 1.3 × 10−6 substitutions per nucleotide site per year (95% HPD interval, 8.87 × 10−7 to 1.72 × 10−6), similar to the rate of other S. aureus lineages (23, 24).
FIG 3.
Time-based tree of the 107 ST72 isolates. Characteristics of isolates are shown on the right, including clades, SCCmec type, and geographical origin (mapped on the tips). Selected divergence time and 95% HPD intervals are shown at the nodes.
SCCmec acquisition and spa typing of ST72 isolates.
SCCmec typing identified only two SCCmec types (types IV [2B] and V [5C2&5]) among 63 ST72 MRSA isolates. The most common was type IV (2B), which comprised 55 isolates (90.16%, 55/61), whereas type V (5C2&5) comprised 6 isolates (9.84%, 6/61) (Fig. 2). SCCmec type IV (2B) could be further divided into three subtypes: IVa (2B), IVc (2B), and IVg (2B). This finding indicates acquisition of different SCCmec elements during the development of methicillin resistance in the ST72 MSSA lineage. Notably, most clade A MRSA isolates harbored SCCmec type IVa (2B) (91.67%, 11/12), and only one carried type V (5C2&5); clade B MRSA isolates carried three SCCmec types, including type IVa (2B) (66.67%, 12/18), IVg (2B) (5.56%, 1/18), and V (5C2&5) (27.28%, 5/18). Type IVc (2B) was only detected in the majority of clade C MRSA isolates (84.85%, 28/33), and none of the isolates within this clade carried type V (5C2&5) (Fig. 2).
The diversity of spa types within all 107 ST72 isolates was confirmed by genotypic spa typing. Twenty-nine spa types were identified, of which the top three types were spa t148 (31.78%, 34/107), t324 (9.35%, 10/107), and t126 (6.54%, 7/107). In addition, we noted that isolates within different clades exhibited considerable diversity of spa types (Fig. 2). Notably, in this regard, the spa types assigned to isolates of clade D were similar to each other and all belonged to a single spa clonal complex (spa-CC324), suggesting that this spa type (spa-CC324) is characteristic of the ST72 clade D MSSA lineage.
Distributions of AMR genes in the ST72 lineage.
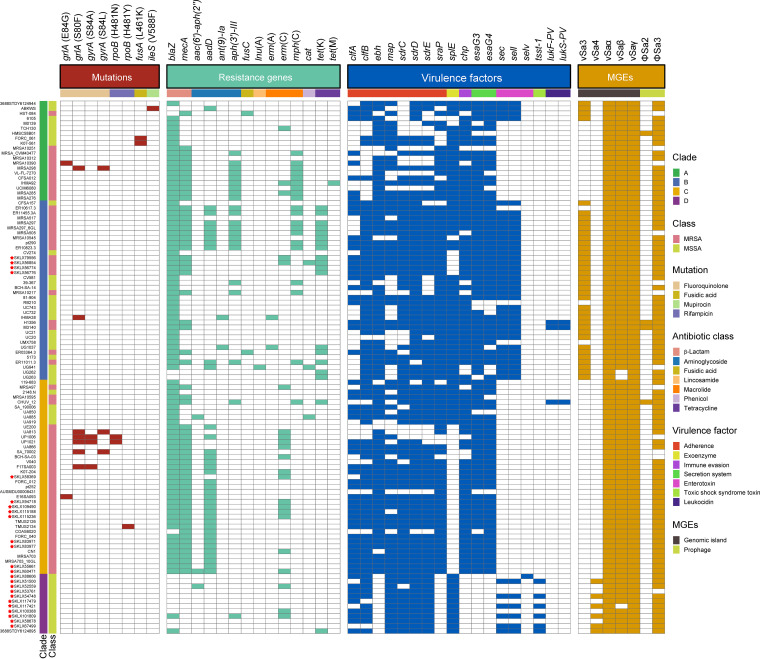
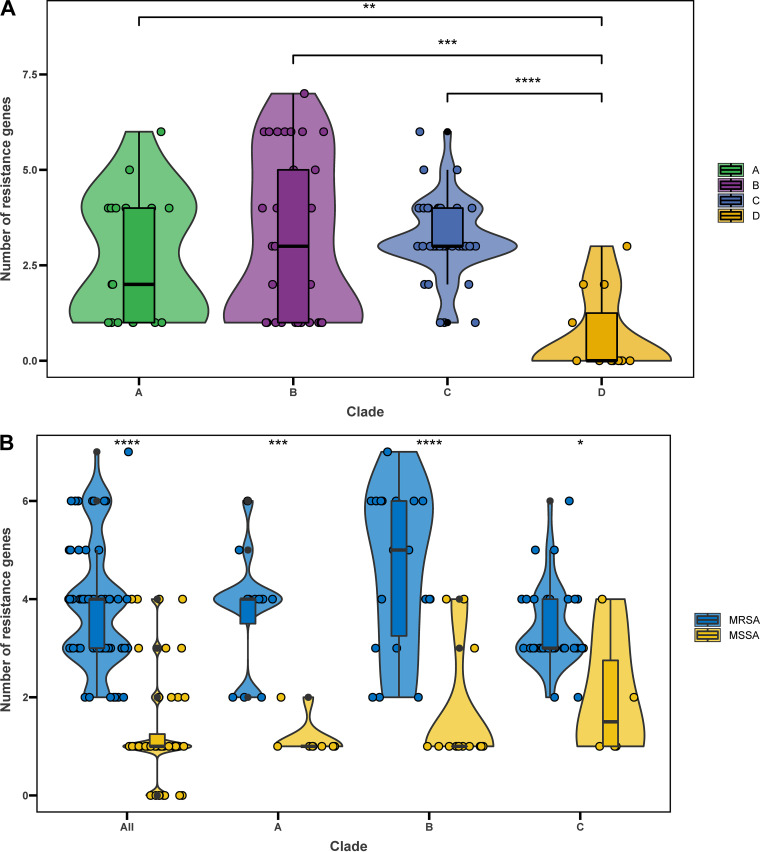
Given the clinical importance of antimicrobial resistance (AMR) in S. aureus, we compared the distributions of AMR genes in ST72 isolates from different clades. A total of 14 AMR genes associated with seven classes of antimicrobial agents (β-lactam, aminoglycoside, fusidic acid, lincosamide, macrolide, phenicol, and tetracycline) were present in isolates from the four clades; 45% (9/20, clade A), 47.2% (17/36, clade B), 28.2% (11/39, clade C), and 8.33% (1/12, clade D) of isolates from the corresponding clades were multidrug resistant (Fig. 4). Moreover, isolates from clades A, B, and C had significantly more AMR genes than those from clade D (P < 0.01) (Fig. 5A). As shown in Fig. 4, the distributions of some AMR genes differed among isolates within the four clades, indicating that isolates of different clades tended to acquire specific antibiotic resistance genes. Notably, detection of gene mutations corresponding to AMR revealed a low frequency of such events in all clades of the ST72 lineage (Fig. 4).
FIG 4.
Distributions of mutations, AMR genes, non-core-virulence genes, and MGEs in the ST72 lineage. Squares, which are colored by trait category, represent the presence of the trait examined. Red asterisks indicate the isolates obtained from our collection.
FIG 5.
AMR genes in ST72 isolates. (A) Number of acquired AMR genes per isolate in different clades. (B) Number of acquired AMR genes per isolate in MRSA and MSSA isolates from different clades. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
In each clade, MRSA isolates had more AMR genes than MSSA isolates (P < 0.05) (Fig. 5B). Furthermore, blaZ, aadD, aph(3′)-III, erm(C), and mph(C) were detected in more MRSA than MSSA isolates (P < 0.05) (Table 1), indicating that MRSA isolates exhibit a higher degree of AMR than MSSA isolates.
TABLE 1.
Distribution of antibiotic resistance and virulence genes between ST72 MRSA and ST72 MSSA isolates
| Category | Gene | No. (%) of isolates |
||
|---|---|---|---|---|
| MRSA (n = 63) | MSSA (n = 44) | P value | ||
| Antibiotic resistance | blaZ | 61 (96.83) | 31 (70.45%) | <0.001 |
| mecA | 63 (100) | 0 (0) | <0.001 | |
| aadD | 35 (55.56) | 2 (4.55) | <0.001 | |
| aph(3′)-III | 20 (31.75) | 2 (4.55) | 0.001 | |
| erm(C) | 18 (28.57) | 4 (9.09) | 0.04 | |
| mph(C) | 19 (30.16) | 1 (2.27) | <0.001 | |
| Virulence | ebh | 57 (90.48) | 28 (63.64) | 0.03 |
| sraP | 59 (93.65) | 25 (56.82) | <0.001 | |
| splE | 28 (44.44) | 41 (93.18) | <0.001 | |
| esaG3 | 60 (95.24) | 26 (59.09) | <0.001 | |
| esaG4 | 58 (92.06) | 24 (54.55) | <0.001 | |
Antibiotic susceptibilities of the ST72 lineage from China.
We also profiled the in vitro susceptibility of the 24 ST72 isolates from China to 16 common used antibiotics (Table 2). The presence of drug-resistance genes in these isolates was consistent with their antimicrobial susceptibility profiles, highlighting genome sequencing as a potential tool to assist clinicians with the treatment of complicated S. aureus infections.
TABLE 2.
Antimicrobial susceptibility of 24 ST72 isolates from Chinaa
| Strain | ERY | CLI | OXA | PEN | SXT | TCY | VAN | RIF | CIP | LVX | MFX | GEN | AMK | TGC | LNZ | DAP |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SKLX58369 | R | R | R | R | S | S | S | S | S | S | S | S | S | S | S | S |
| SKLX83971 | S | S | R | R | S | S | S | S | S | S | S | S | R | S | S | S |
| SKLX83977 | S | S | R | R | S | S | S | S | S | S | S | S | R | S | S | S |
| SKLX60471 | R | R | R | R | S | S | S | S | S | S | S | S | R | S | S | S |
| SKLX94718 | R | R | R | R | S | S | S | S | S | S | S | S | R | S | S | S |
| SKLX109490 | R | R | R | R | S | S | S | S | S | S | S | S | R | S | S | S |
| SKLX115188 | R | R | R | R | S | S | S | S | S | S | S | S | R | S | S | S |
| SKLX115236 | R | R | R | R | S | S | S | S | S | S | S | S | R | S | S | S |
| SKLX55661 | S | S | R | R | S | S | S | S | S | S | S | S | R | S | S | S |
| SKLX56774 | S | S | R | R | S | R | S | S | S | S | S | S | S | S | S | S |
| SKLX56776 | S | S | R | R | S | R | S | S | S | S | S | S | S | S | S | S |
| SKLX56854 | R | R | R | R | S | R | S | S | S | S | S | S | S | S | S | S |
| SKLX79556 | R | R | R | R | S | R | S | S | S | S | S | S | S | S | S | S |
| SKLX101809 | R | R | S | R | S | S | S | S | S | S | S | S | R | S | S | S |
| SKLX58678 | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S |
| SKLX67499 | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S |
| SKLX88606 | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S |
| SKLX100388 | R | R | S | S | S | S | S | S | S | S | S | S | S | S | S | S |
| SKLX117421 | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S |
| SKLX117479 | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S |
| SKLX54748 | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S |
| SKLX52559 | R | R | S | S | S | S | S | S | S | S | S | S | S | S | S | S |
| SKLX51500 | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S |
| SKLX53761 | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S |
R, resistant; S, susceptible; ERY, erythromycin; CLI, clindamycin; OXA, oxacillin; PEN, penicillin G; SXT, trimethoprim-sulfamethoxazole; TCY, tetracycline; VAN, vancomycin; RIF, rifampin; CIP, ciprofloxacin; LVX, levofloxacin; MFX, moxifloxacin; GEN, gentamicin; AMK, amikacin; TGC, tigecycline; LNZ, linezolid; DAP, daptomycin.
Presence of virulence genes in the ST72 lineage.
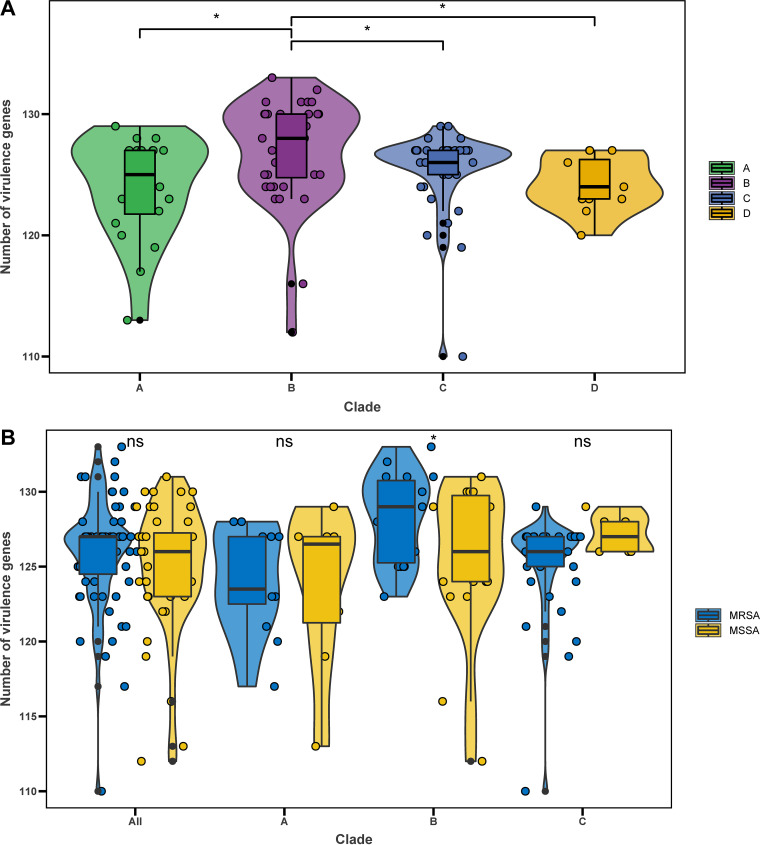
We next performed a targeted analysis of all known virulence genes in S. aureus to assess the pathogenic potential of isolates within the four clades. We identified a total of 120 virulence genes from 107 ST72 isolates. The number of virulence genes was significantly higher in isolates of clade B than in isolates of any other clade (all P < 0.05) (Fig. 6A). A series of genes associated with adhesion, exoenzyme, immune evasion, the type VII secretion system, hemolysin (including hla), iron uptake, phenol-soluble modulins (PSMs), exotoxin (set), exfoliative toxin (eta), leukocidin (lukD and lukE), and enterotoxin (seg, sei, selm, seln, selo, and selu2) were widely distributed among isolates of each clade (≥85% isolates per clade) (Table S2), suggesting that these genes are the core virulence genes of the ST72 lineage. Moreover, these core virulence genes accounted for about 85% (102/120) of all virulence genes detected in ST72 isolates.
FIG 6.
Virulence genes in ST72 isolates. (A) Number of virulence genes per isolate in different clades. (B) Number of virulence genes per isolate in MRSA and MSSA isolates from different clades. *, P < 0.05; ns, no significant difference between groups.
Distribution of core virulence genes in the ST72 lineage (≥85% isolates per clade). Download Table S2, DOCX file, 0.02 MB (23.6KB, docx) .
Copyright © 2021 Zhou et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Furthermore, with the exception of core virulence genes, the distributions of some other virulence genes differed among isolates of each clade (Fig. 4). In particular, the genes encoding Panton-Valentine leukocidins (lukS-PV and lukF-PV) were only identified in three isolates of clade B and clade C, whereas the enterotoxin gene selv and toxic shock syndrome toxin gene tsst-1 were only detected in clade D isolates.
The number of virulence genes did not significantly differ between MRSA and MSSA isolates (median 127 versus 126, P > 0.05) (Fig. 6B). Further, except for ebh, sraP, splE, esaG3, and esaG4 (P < 0.05) (Table 2), the abundance of other virulence genes did not significantly differ between MRSA and MSSA isolates. Notably, however, for isolates within clade B, MRSA isolates contained significantly more virulence genes than MSSA isolates (median 129 versus 126, P < 0.05) (Fig. 6B).
In contrast to the epidemic CA-MRSA USA300 (ST8) from the United States and ST59 from Asia, all ST72 isolates lacked virulence genes associated with exoenzyme (vWbp, coa, and splE), immune evasion (cap5ACEFGIJKNO, cap8HIJK, and capFGLMP), and enterotoxin (selq, selk and seb) (Fig. S2). The absence of these genes might decrease the invasion potential of ST72 relative to USA300 and ST59.
Network diagram of all virulence genes (VGs) identified in sequence types ST72, ST8 (USA300), and ST59 (M013). “Shared” indicates VGs are present in all STs, “Accessory” indicates VGs are present in two STs, and “ST-specific” indicates VGs are present in a specific ST. Download FIG S2, PDF file, 0.7 MB (752.8KB, pdf) .
Copyright © 2021 Zhou et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Pathogenicity islands and prophages in ST72 isolates.
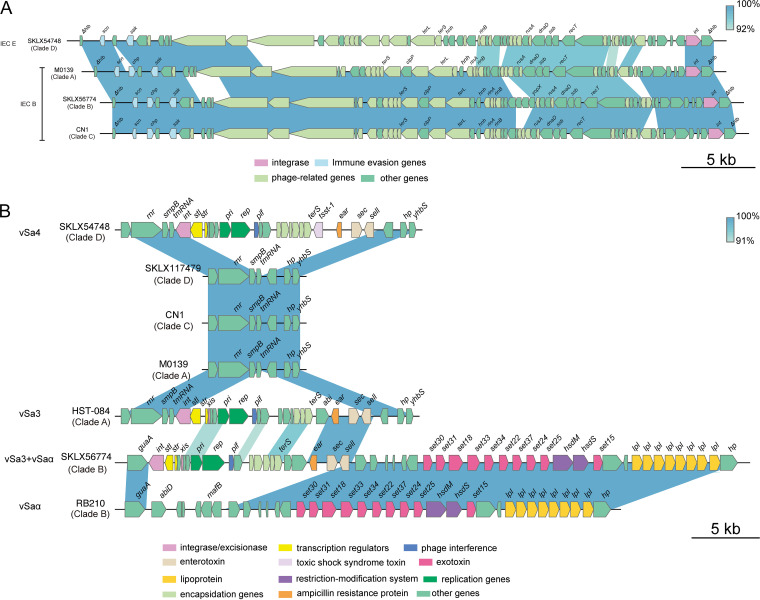
We next examined the distribution of pathogenicity islands and prophages in ST72 isolates because they are significant components of the S. aureus genome that might contribute to the evolution of virulence. The results revealed that most of the isolates from each clade (>85%) harbored three pathogenicity islands, vSaα, vSaβ, and vSaγ, as well as one prophage, ΦSa3 (Fig. 4). However, ΦSa3 of clade D isolates harbored immune evasion cluster (IEC) type E (IEC E, encoding scn-sak), whereas ΦSa3 of isolates within all other clades carried IEC B (encoding scn-chp-sak) (Fig. 7A). The vSa3 pathogenicity island, which encodes enterotoxin genes sec and sell, was mainly detected in clade B isolates (91.67%, 33/36) and a few clade A isolates (20%, 4/20); however, between the two clades, vSa3 shared a small number of homologous genes, and vSa3 carried by clade B isolates was integrated into vSaα by recombination, forming a mosaic structure. This finding implies that the vSa3 pathogenicity islands in clade A and clade B isolates are unlikely to have evolved from a common ancestor (Fig. 7B). The vSa4 pathogenicity island, encoding sec, sell, and tsst-1, was only found in half of the clade D isolates (6/12).
FIG 7.
Comparison of ΦSa3 and pathogenicity islands between ST72 isolates in each clade. (A) Comparison of ΦSa3 between ST72 isolates in each clade. (B) Comparison of pathogenicity islands between ST72 isolates in each clade. Genes are represented by arrowed boxes and colored based on gene function classification.
In all isolates of clade D, we identified an unknown prophage of 37,817 bp that was integrated immediately downstream of the glutamine synthetase gene glnA and flanked by a pair of 57-bp perfect direct repeats (Fig. S3A). The prophage contained 48 open reading frames (ORFs) arranged in a modular fashion; the genome consisted of genes involved in lysis, lysogeny, DNA replication and metabolism, capsid formation and assembly, and head and tail morphogenesis. A BLAST search against the NCBI nucleotide database indicated that this prophage resembled those found in nine S. aureus chromosomes (>98% identity and ≥80% coverage). However, even the two prophages (harbored by S. aureus NRS152 and S. aureus XQ, respectively) that shared the highest sequence similarities with this unknown prophage, only covered 87% of its entire genome. The phylogenetic relationship indicated a single ancestral acquisition of the unknown prophage in clade D isolates (Fig. S3B).
Comparison of the unknown prophage with similar prophages. (A) Circular representation of the unknown prophage and comparative genomic analysis with other similar prophages. Circle 1 (from outer to inner circle) shows the genes encoded on the unknown prophage; the colors of genes are assigned based on gene function classification. Circles 2 to 10 refer to homologous regions of S. aureus 93b_S9 (CP010952.1), 27 (CP022717.1), 78 (CP022682.1), UP_426 (CP047797.1), UP_551 (CP047794.1), 128 (CP022897.1), 422 (CP022898.1), XQ (CP013137.1), and NRS153 (CP026067.1) relative to the unknown prophage. Circles 11 and 12 represent GC content and GC skew of the unknown prophage, respectively. Direct repeats are indicated by arrows. (B) Phylogenetic analysis of the unknown prophage with other similar prophages. Download FIG S3, PDF file, 1.9 MB (1.9MB, pdf) .
Copyright © 2021 Zhou et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Genetic changes differentiating isolates of each clade.
To obtain insights into the SNP-based evolutionary analysis of the ST72 population, we searched for mutations that could differentiate the ST72 isolates of distinct clades. We identified a total of 40, 4, and 98 SNPs associated with clades A, B, and D, respectively, but no clade-specific SNPs within clade C isolates (Table S3). Strikingly, five clade A-specific nonsynonymous SNPs were located in genes involved in bacterial adhesion (atl, clfA, fnbA, and icaB) and capsule (capD), and seven clade D-specific nonsynonymous SNPs were related to bacterial adhesion (sasH), aurolysin (aur), capsule (capD and capI), exotoxin (set), and iron uptake (isdA and sfaD).
Clade-specific SNPs as identified by Scoary. Download TABLE S3 DOCX file, 0.03 MB (33.4KB, docx) .
Copyright © 2021 Zhou et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
DISCUSSION
The community-genotype S. aureus ST72 lineage, which is predominant in South Korea (13), has emerged and gradually increased in hospital settings, similar to other hospital-genotype isolates, such as ST5 and ST239 in Asia. Although ST72 isolates have rarely been reported in other countries (18–20), the 107 ST72 isolates analyzed in this study covered 18 countries on five different continents and were isolated between 2003 and 2019. The most frequently observed isolates were from the United States (n = 42), where isolation of ST72 isolates has not been previously reported, followed by China (n = 24), which has reported only sporadic isolation of ST72 isolates (19). Notably, ST72 is the trilocus variant of ST8, while ST8 is the most epidemic clone in the United States. The results of biogeography analysis indicated that the United States may be the more probable ancestral origin of ST72 isolates from which these clones began to diversify (Fig. S4), but this could also be the result of sampling bias. These findings suggest that the potential dissemination of ST72 clones around the world has been underestimated, and that large-scale monitoring will be necessary.
Phylogeographic analysis of global ST72 isolates based on the Bayesian binary MCMC method in RASP v4.2. Download FIG S4, PDF file, 0.4 MB (442.7KB, pdf) .
Copyright © 2021 Zhou et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
A recent study by Jian et al. (25) reconstructed the phylogeny of ST5 strains isolated from an affiliated tertiary hospital in China between 2008 and 2018. Those authors reported that ST5 MSSA isolates evolved independently in a manner distinct from ST5 MRSA isolates. Similarly, using phylogenetic reconstruction in conjunction with temporal information of the ST72 isolates, we found that all MSSA isolates from China formed a separate clade (clade D) in the tree with an MSSA isolate from Thailand, suggesting the ST72 MRSA and MSSA isolates of China might have evolved independently, i.e., the ST72 MRSA clones in China did not evolve from the susceptible isolates. However, our phylogenetic analysis also revealed that in some other countries, including the United States, several human-adapted ST72 MRSA isolates were much more closely related to human-adapted MSSA isolates, suggesting they may have evolved from the latter by acquisition of SCCmec elements (Fig. 2).
In addition, we identified potential international transmission of ST72 isolates, as some isolates from different countries clustered closely with each other. However, the paired SNP distances of isolates between countries surpassed the 23 SNPs suggested by Uhlemann et al. (21) as the maximum pairwise distance of isolates from direct transmission, implying that the epidemic links between clustered isolates from different countries may not be recent and direct; rather, these isolates might have undergone local expansion. Notably, our analysis of ancestral area reconstruction revealed that ST72-SCCmec IVc (2B) isolates from China in this study might have originated from South Korea (Fig. S4); in particular, four ST72-SCCmec IVc (2B) MRSA isolates from the Affiliated Hospital of Binzhou Medical College (Binzhou, China) were most closely genetically related to isolate E16SA093 from South Korea (Fig. 2, Fig. 3, and Fig. S4). Given the relatively close relationship between isolates from Binzhou and South Korea (paired SNP distance: 95 to 98) and the small geographical distance between the two locations (Fig. 1A), we speculate that international transmission of ST72 isolates might have occurred between South Korea and China. Similarly, our analysis of ST72 isolates from China showed that although the paired SNP distances of isolates from different provinces varied from 87 to 452 (Fig. S1B), the possibility of interregional dissemination in China remained due to the small geographical distances among these provinces (with the exception of Heilongjiang province).
In contrast to HA S. aureus, CA S. aureus has been categorized as susceptible to non-β-lactam antibiotics (26). However, among all isolates of the ST72 lineage analyzed in this study, we identified 14 AMR genes classified into seven antimicrobial classes. Nearly half of the isolates of clades A (45%) and B (47.2%) exhibited multidrug resistance (Fig. 3); hence, we speculate that isolates within these two clades are more likely to spread and thrive in hospitals or originate from hospital settings. Overall, we detected significantly more antibiotic-resistance genes among MRSA than MSSA isolates (Fig. 5B); this could explain why clade D isolates, which were all MSSA isolates, had a low rate of genotypic multidrug resistance. Strikingly, unlike other pandemic S. aureus clones, such as ST22 EMRSA-15, the success of which could be partially attributed to mutations within resistance genes (27), we identified a low mutation frequency in resistance genes carried by isolates of any clade of the ST72 lineage (Fig. 3). This finding indicates that gene mutations play only a limited role during the development of resistance in the ST72 lineage.
Virulence is an important determinant of whether isolates can induce clinical infections. Of the 120 virulence genes identified from all isolates, 102 were members of the core virulence genes, implying they might play a role in host adaption and evolution of virulence in the ST72 lineage. CA-ST72 isolates generally do not have PVL-encoding genes (16). Although three ST72 isolates in the current study were PVL positive and harbored PVL-encoding genes in ΦSa2 PVL prophages (2 intact), they were located within separate clades (clade B and clade C) (Fig. 2), indicating independent acquisition of PVL-encoding genes. Interestingly, the distribution of some virulence genes in clade D isolates was associated with acquisition of specific MGEs. In contrast to isolates of the other clades, clade D isolates exclusively harbored IEC E lacking chp in the ΦSa3 prophage, the vSa4 pathogenicity island containing tsst-1, and a novel 38-kb prophage encoding genes of unknown function (Fig. 6). This finding indicates that these MGEs could be used as markers to distinguish isolates of clade D from those of the other clades, and further supports the notion that the MSSA isolates from China (accounting for 91.67% of clade D isolates) evolved independently. Furthermore, we identified several clade-specific nonsynonymous SNPs associated with virulence genes, suggesting that these mutations might affect the expression of the bacterial virulence.
It is a common notion that MSSA lineages always carry a wider array of virulence genes than MRSA lineages, and that this difference is due to the fitness cost associated with methicillin resistance (28). However, we found that the number of virulence genes did not significantly differ between ST72 MRSA and MSSA isolates (median 127 versus 126, P > 0.05), and the distributions of nearly all virulence genes (115/120) did not significantly differ between the two populations. Indeed, MRSA isolates of clade B had more virulence genes than MSSA isolates (median 129 versus 126, P < 0.05) (Fig. 4B). It has been suggested that the transcription of the agr operon is repressed by the expression of β-lactam resistance (Bla/Mec system) in MRSA isolates, which prevents the production of core genome-encoded toxins (including PSMs and alpha-toxin) regulated by this operon (29). However, Chen et al. (8) showed that for the PVL-negative CA-MRSA ST72, phenol-soluble modulins (PSMs) and the global virulence regulator Agr play a key role in lysis of neutrophils and erythrocytes, while alpha-toxin and Agr strongly affect in vivo virulence, suggesting that the toxin regulator Agr is not significantly repressed in ST72 MRSA. As expected, nearly all ST72 isolates (97.2%, 104/107) in this study carried both PSMs and alpha-toxin (Fig. 4). Moreover, Otto reported that both expression of core genome-encoded toxins (including PSMs and alpha-toxin) and acquisition of virulence-associated genes could contribute to the evolution of CA-MRSA virulence (7). Hence, based on the results of our analysis of virulence genes, we speculate that the virulence potential of ST72 MRSA isolates is comparable to that of MSSA predecessors, and that this virulence potential is associated with expression of core genome-encoded toxins with no loss or even gain of related virulence genes that could contribute to virulence in ST72 MRSA isolates.
This study has some limitations. First, the number of ST72 isolates collected was not large enough. Second, the lack of detailed epidemiological information may have impacted the analysis of the epidemiology of ST72. Third, the findings of comparisons between isolates should be interpreted with caution, because these might be the result of differences in sampling sites and are subject to a statistical caveat, i.e., the presence and absence of several antibiotic resistance or virulence genes could not be considered independent since they are linked on the same MGEs. Further experimental evidence is needed to prove that ST72 MRSA and MSSA isolates have similar virulence.
In conclusion, our data provide important insight into the phylogenetic relationship, transmission, antibiotic resistance, and pathogenicity of the ST72 lineage on a global scale. Our results reveal that, in China, all MSSA isolates might have evolved independently of MRSA isolates from that nation. We demonstrated that ST72 isolates have the potential for international transmission as well as interregional transmission within China. Moreover, the MSSA isolates of clade D exhibited lower antibiotic resistance than isolates of the other clades, and tended to gain specific MGEs encoding specific virulence genes, such as tsst-1. Importantly, our findings also indicate that ST72 MRSA isolates exhibited higher antibiotic resistance than ST72 MSSA isolates, but were comparably virulent. More genomic data and experimental verification are needed to predict the epidemic potential of the ST72 lineage in countries other than South Korea.
MATERIALS AND METHODS
Collection of bacterial isolates.
From January 2014 to December 2019, we isolated 2,769 S. aureus strains from patients with bloodstream infections (BSI) in 18 provinces and municipalities in China. Identification of the tested strains was performed using biochemical methods and matrix-assisted laser desorption–time of flight mass spectrometry (MALDI-TOF MS) (Bruker, Bremen, Germany). Multilocus sequence typing of all 2,769 S. aureus isolates was performed by Sanger sequencing of seven housekeeping genes (including arcC, aroE, gmk, glpF, tpi, yqiL, and pta), after which the sequences of the housekeeping genes were submitted to the S. aureus typing database for the assignment of ST type (https://pubmlst.org/bigsdb?db=pubmlst_saureus_seqdef). Ultimately, we obtained 24 ST72 isolates among all S. aureus isolates (Table S1).
Antimicrobial susceptibility testing.
The antimicrobial susceptibilities to 16 antimicrobial agents (oxacillin, penicillin G, sulfamethoxazole and trimethoprim, ciprofloxacin, levofloxacin, moxifloxacin, gentamicin, amikacin, erythromycin, clindamycin, tetracycline, tigecycline, rifampin, vancomycin, linezolid, and daptomycin) were determined using the agar dilution method according to standard guidelines recommended by the Clinical and Laboratory Standards Institute (CLSI, 2020). S. aureus ATCC 25923 and ATCC 29213 were used as reference strains for quality control.
Whole-genome sequencing and genomic analysis of ST72 isolates.
Genomic DNA of S. aureus ST72 isolates was extracted using the Ezup Column Bacteria Genomic DNA purification kit (Sangon Biotech, Shanghai, China). Sequencing of the ST72 genomes was performed on a HiSeq X 10-PE150 platform (Illumina, San Diego, CA, USA). After adaptor trimming and quality filtering, the processed Illumina reads were assembled using SPAdes v3.14.1 (30) with default settings. SCCmec and spa typing were performed using the web-based SCCmecFinder (https://cge.cbs.dtu.dk/services/SCCmecFinder) and SpaFinder (https://cge.cbs.dtu.dk/services/spatyper), respectively. Potential open reading frames (ORFs) were predicted and annotated using Prokka v1.14.5 (31). Additional genomes of 83 ST72 isolates with available geographic information were downloaded from the NCBI genome database for comparison (Table S1).
Phylogenetic analysis and Bayesian evolutionary analysis.
Snippy v4.6.0 (https://github.com/tseemann/snippy) was used to perform reference-based mapping and identify SNPs in the ST72 core genome, with the CN1 genome (GenBank accession no. CP003979) used as a reference. Gubbins v2.4.1 (32) was used to detect the recombined regions within the core genome and remove putative recombined regions. A phylogenetic tree was constructed using the core single nucleotide polymorphisms (SNPs) from the recombination-free core-genome alignment with RAxML using a GTR model and gamma correction (1,000 bootstrap replications) (33). The ancestral geographic origins of ST72 isolates were analyzed using the Bayesian binary MCMC method in RASP v4.2 (34) with 10 parallel chains of 50 million cycles. The output from Gubbins, together with the isolation dates of the isolates, was used as the input for BactDating v1.1 (35) to obtain a dated phylogeny based on Bayesian inference. The Markov chain Monte Carlo chain lengths were run for 100 million cycles, and the effective sample sizes of the model parameters mu, sigma, and alpha were all >200. The minimum spanning trees (MSTs) based on SNP data were generated using PHYLOViZ v2.0 (https://online.phyloviz.net/index).
Identification of antimicrobial-resistance genes, virulence genes, pathogenicity islands, and prophages.
Antimicrobial-resistance (AMR) genes and virulence genes were detected using ResFinder v4.0 (https://cge.cbs.dtu.dk/services/ResFinder/) and VFanalyzer (http://www.mgc.ac.cn/cgi-bin/VFs/v5/main.cgi?func=VFanalyzer), respectively. Pathogenicity islands and prophages were identified using BLAST (https://blast.ncbi.nlm.nih.gov/Blast.cgi), with sequences of pathogenicity islands and prophages from S. aureus as the queries.
Detection of clade-specific SNPs.
The SNPs associated with isolates of each clade were detected using Scoary v1.6.16 (36).
Statistical analysis.
Wilcoxon rank-sum test and chi-square test or Fisher’s exact test were used for statistical analysis, and P < 0.05 was considered as statistically significant. All data were analyzed using the R software (https://www.r-project.org/).
Data availability.
The whole-genome sequences of 24 ST72 S. aureus isolates have been deposited in the GenBank database under BioProject accession no. PRJNA735348.
ACKNOWLEDGMENTS
We thank Jinru Ji, Chaoqun Ying, and Yunhui Fang for the technical support and quality control.
This study was supported by The National Key Research and Development Program of China (grant number 2017YFC1200203) and the National Natural Science Foundation of China (grant number 81971984).
Ethical approval for this study was submitted and approved by the Ethics Committee of the First Affiliated Hospital of Zhejiang University. Informed consent was not needed since this project was a retrospective study and all bacterial samples from patients were for anonymously scientific use.
Contributor Information
Yonghong Xiao, Email: xiaoyonghong@zju.edu.cn.
Suleyman Yildirim, Istanbul Medipol University School of Medicine.
REFERENCES
- 1.Otto M. 2010. Staphylococcus colonization of the skin and antimicrobial peptides. Expert Rev Dermatol 5:183–195. doi: 10.1586/edm.10.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lowy FD. 1998. Staphylococcus aureus infections. N Engl J Med 339:520–532. doi: 10.1056/NEJM199808203390806. [DOI] [PubMed] [Google Scholar]
- 3.Thurlow LR, Joshi GS, Richardson AR. 2012. Virulence strategies of the dominant USA300 lineage of community-associated methicillin-resistant Staphylococcus aureus (CA-MRSA). FEMS Immunol Med Microbiol 65:5–22. doi: 10.1111/j.1574-695X.2012.00937.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Centers for Disease Control and Prevention (CDC). 1999. Four pediatric deaths from community-acquired methicillin-resistant Staphylococcus aureus—Minnesota and North Dakota, 1997–1999. MMWR Morbidity and Mortality Wkly Report 48:707–710. [PubMed] [Google Scholar]
- 5.Naimi TS, LeDell KH, Como-Sabetti K, Borchardt SM, Boxrud DJ, Etienne J, Johnson SK, Vandenesch F, Fridkin S, O'Boyle C, Danila RN, Lynfield R. 2003. Comparison of community- and health care-associated methicillin-resistant Staphylococcus aureus infection. JAMA 290:2976–2984. doi: 10.1001/jama.290.22.2976. [DOI] [PubMed] [Google Scholar]
- 6.Herold BC, Immergluck LC, Maranan MC, Lauderdale DS, Gaskin RE, Boyle-Vavra S, Leitch CD, Daum RS. 1998. Community-acquired methicillin-resistant Staphylococcus aureus in children with no identified predisposing risk. JAMA 279:593–598. doi: 10.1001/jama.279.8.593. [DOI] [PubMed] [Google Scholar]
- 7.Otto M. 2013. Community-associated MRSA: what makes them special? Int J Med Microbiol 303:324–330. doi: 10.1016/j.ijmm.2013.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen Y, Yeh AJ, Cheung GY, Villaruz AE, Tan VY, Joo H-S, Chatterjee SS, Yu Y, Otto M. 2015. Basis of virulence in a Panton-Valentine leukocidin-negative community-associated methicillin-resistant Staphylococcus aureus strain. J Infect Dis 211:472–480. doi: 10.1093/infdis/jiu462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baba T, Takeuchi F, Kuroda M, Yuzawa H, Aoki K-i, Oguchi A, Nagai Y, Iwama N, Asano K, Naimi T, Kuroda H, Cui L, Yamamoto K, Hiramatsu K. 2002. Genome and virulence determinants of high virulence community-acquired MRSA. Lancet 359:1819–1827. doi: 10.1016/S0140-6736(02)08713-5. [DOI] [PubMed] [Google Scholar]
- 10.Otto M. 2010. Basis of virulence in community-associated methicillin-resistant Staphylococcus aureus. Annu Rev Microbiol 64:143–162. doi: 10.1146/annurev.micro.112408.134309. [DOI] [PubMed] [Google Scholar]
- 11.Kobayashi SD, Malachowa N, Whitney AR, Braughton KR, Gardner DJ, Long D, Wardenburg JB, Schneewind O, Otto M, DeLeo FR. 2011. Comparative analysis of USA300 virulence determinants in a rabbit model of skin and soft tissue infection. J Infect Dis 204:937–941. doi: 10.1093/infdis/jir441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li M, Dai Y, Zhu Y, Fu C-L, Tan VY, Wang Y, Wang X, Hong X, Liu Q, Li T, Qin J, Ma X, Fang J, Otto M. 2016. Virulence determinants associated with the Asian community-associated methicillin-resistant Staphylococcus aureus lineage ST59. Sci Rep 6:27899. doi: 10.1038/srep27899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Park K-H, Chong YP, Kim S-H, Lee S-O, Choi S-H, Lee MS, Jeong J-Y, Woo JH, Kim YS. 2015. Community-associated MRSA strain ST72-SCCmecIV causing bloodstream infections: clinical outcomes and bacterial virulence factors. J Antimicrob Chemother 70:1185–1192. doi: 10.1093/jac/dku475. [DOI] [PubMed] [Google Scholar]
- 14.Kim ES, Song JS, Lee HJ, Choe PG, Park KH, Cho JH, Park WB, Kim S-H, Bang J-H, Kim D-M, Park KU, Shin S, Lee MS, Choi HJ, Kim NJ, Kim E-C, Oh M-D, Kim HB, Choe KW. 2007. A survey of community-associated methicillin-resistant Staphylococcus aureus in Korea. J Antimicrob Chemother 60:1108–1114. doi: 10.1093/jac/dkm309. [DOI] [PubMed] [Google Scholar]
- 15.Park SH, Park C, Yoo J-H, Choi S-M, Choi J-H, Shin H-H, Lee D-G, Lee S, Kim J, Choi SE, Kwon Y-M, Shin W-S. 2009. Emergence of community-associated methicillin-resistant Staphylococcus aureus strains as a cause of healthcare-associated bloodstream infections in Korea. Infect Control Hosp Epidemiol 30:146–155. doi: 10.1086/593953. [DOI] [PubMed] [Google Scholar]
- 16.Joo E-J, Choi J-Y, Chung DR, Song J-H, Ko KS. 2016. Characteristics of the community-genotype sequence type 72 methicillin-resistant Staphylococcus aureus isolates that underlie their persistence in hospitals. J Microbiol 54:445–450. doi: 10.1007/s12275-016-6157-x. [DOI] [PubMed] [Google Scholar]
- 17.Chung DR, Baek JY, Kim HA, Lim MH, Kim SH, Ko KS, Kang C-I, Peck KR, Lee NY, Song J-H. 2012. First report of vancomycin-intermediate resistance in sequence type 72 community genotype methicillin-resistant Staphylococcus aureus. J Clin Microbiol 50:2513–2514. doi: 10.1128/JCM.00590-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tsukimori A, Nakamura I, Okamura S, Sato A, Fukushima S, Mizuno Y, Yamaguchi T, Matsumoto T. 2014. First case report of vancomycin-intermediate sequence type 72 Staphylococcus aureus with nonsusceptibility to daptomycin. BMC Infect Dis 14:459. doi: 10.1186/1471-2334-14-459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang X, Zhao J, Wang Y, Wu J, Wang X, Wang Y, Zhang Y, Li H. 2021. Molecular epidemiology of methicillin-resistant Staphylococcus aureus in hospitalized patients in eastern Heilongjiang Province, China. Infect Drug Resist 14:1635–1643. doi: 10.2147/IDR.S307856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Potel C, Rey S, Otero S, Rubio J, Álvarez M. 2016. Molecular characterization and clonal diversity of meticillin-resistant Staphylococcus aureus isolated from the community in Spain: emergence of clone sequence type 72. J Hosp Infect 93:382–385. doi: 10.1016/j.jhin.2016.03.012. [DOI] [PubMed] [Google Scholar]
- 21.Uhlemann A-C, Dordel J, Knox JR, Raven KE, Parkhill J, Holden MT, Peacock SJ, Lowy FD. 2014. Molecular tracing of the emergence, diversification, and transmission of S. aureus sequence type 8 in a New York community. Proc Natl Acad Sci USA 111:6738–6743. doi: 10.1073/pnas.1401006111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.O'Hara LM, Calfee DP, Miller LG, Pineles L, Magder LS, Johnson JK, Morgan DJ, Harris AD. 2019. Optimizing contact precautions to curb the spread of antibiotic-resistant bacteria in hospitals: a multicenter cohort study to identify patient characteristics and healthcare personnel interactions associated with transmission of methicillin-resistant Staphylococcus aureus. Clin Infect Dis 69:S171–S177. doi: 10.1093/cid/ciz621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McAdam PR, Templeton KE, Edwards GF, Holden MTG, Feil EJ, Aanensen DM, Bargawi HJA, Spratt BG, Bentley SD, Parkhill J, Enright MC, Holmes A, Girvan EK, Godfrey PA, Feldgarden M, Kearns AM, Rambaut A, Robinson DA, Fitzgerald JR. 2012. Molecular tracing of the emergence, adaptation, and transmission of hospital-associated methicillin-resistant Staphylococcus aureus. Proc Natl Acad Sci USA 109:9107–9112. doi: 10.1073/pnas.1202869109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moradigaravand D, Jamrozy D, Mostowy R, Anderson A, Nickerson EK, Thaipadungpanit J, Wuthiekanun V, Limmathurotsakul D, Tandhavanant S, Wikraiphat C, Wongsuvan G, Teerawattanasook N, Jutrakul Y, Srisurat N, Chaimanee P, Eoin West T, Blane B, Parkhill J, Chantratita N, Peacock SJ. 2017. Evolution of the Staphylococcus argenteus ST2250 clone in Northeastern Thailand is linked with the acquisition of livestock-associated staphylococcal genes. mBio 8:e00802-17. doi: 10.1128/mBio.00802-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jian Y, Zhao L, Zhao N, Lv H-Y, Liu Y, He L, Liu Q, Li M. 2021. Increasing prevalence of hypervirulent ST5 methicillin susceptible Staphylococcus aureus subtype poses a serious clinical threat. Emerg Microbes Infect 10:109–154. doi: 10.1080/22221751.2020.1868950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee GC, Dallas SD, Wang Y, Olsen RJ, Lawson KA, Wilson J, Frei CR. 2017. Emerging multidrug resistance in community-associated Staphylococcus aureus involved in skin and soft tissue infections and nasal colonization. J Antimicrob Chemother 72:2461–2468. doi: 10.1093/jac/dkx200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Holden MTG, Hsu L-Y, Kurt K, Weinert LA, Mather AE, Harris SR, Strommenger B, Layer F, Witte W, de Lencastre H, Skov R, Westh H, Zemlicková H, Coombs G, Kearns AM, Hill RLR, Edgeworth J, Gould I, Gant V, Cooke J, Edwards GF, McAdam PR, Templeton KE, McCann A, Zhou Z, Castillo-Ramírez S, Feil EJ, Hudson LO, Enright MC, Balloux F, Aanensen DM, Spratt BG, Fitzgerald JR, Parkhill J, Achtman M, Bentley SD, Nübel U. 2013. A genomic portrait of the emergence, evolution, and global spread of a methicillin-resistant Staphylococcus aureus pandemic. Genome Res 23:653–664. doi: 10.1101/gr.147710.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jiménez JN, Ocampo AM, Vanegas JM, Rodríguez EA, Garcés CG, Patiño LA, Ospina S, Correa MM. 2011. Characterisation of virulence genes in methicillin susceptible and resistant Staphylococcus aureus isolates from a paediatric population in a university hospital of Medellin, Colombia. Mem Inst Oswaldo Cruz 106:980–985. doi: 10.1590/s0074-02762011000800013. [DOI] [PubMed] [Google Scholar]
- 29.Choe D, Szubin R, Dahesh S, Cho S, Nizet V, Palsson B, Cho B-K. 2018. Genome-scale analysis of Methicillin-resistant Staphylococcus aureus USA300 reveals a tradeoff between pathogenesis and drug resistance. Sci Rep 8:2215–2213. doi: 10.1038/s41598-018-20661-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Prjibelski A, Antipov D, Meleshko D, Lapidus A, Korobeynikov A. 2020. Using SPAdes de novo assembler. Curr Protoc Bioinformatics 70:e102. doi: 10.1002/cpbi.102. [DOI] [PubMed] [Google Scholar]
- 31.Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 32.Croucher NJ, Page AJ, Connor TR, Delaney AJ, Keane JA, Bentley SD, Parkhill J, Harris SR. 2015. Rapid phylogenetic analysis of large samples of recombinant bacterial whole genome sequences using Gubbins. Nucleic Acids Res 43:e15. doi: 10.1093/nar/gku1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yu Y, Blair C, He X. 2020. RASP 4: ancestral state reconstruction tool for multiple genes and characters. Mol Biol Evol 37:604–606. doi: 10.1093/molbev/msz257. [DOI] [PubMed] [Google Scholar]
- 35.Didelot X, Croucher NJ, Bentley SD, Harris SR, Wilson DJ. 2018. Bayesian inference of ancestral dates on bacterial phylogenetic trees. Nucleic Acids Res 46:e134. doi: 10.1093/nar/gky783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brynildsrud O, Bohlin J, Scheffer L, Eldholm V. 2016. Rapid scoring of genes in microbial pan-genome-wide association studies with Scoary. Genome Biol 17:238–239. doi: 10.1186/s13059-016-1108-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Information on ST72 isolates analyzed in this study. Download Table S1, DOCX file, 0.04 MB (41.3KB, docx) .
Copyright © 2021 Zhou et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Relationships among 24 ST72 isolates from China in this study. (A) Geographical distribution of 24 ST72 S. aureus isolates. Red and black dots indicate MRSA and MSSA isolates, respectively. The map was created using R package ggplot2 (https://github.com/tidyverse/ggplot2). (B) Phylogenetic tree of 24 ST72 isolates. Tips are colored by province of isolation. (C) MST of 24 ST72 isolates. Each circle represents a ST72 isolate, with colors assigned according to geographic origin. The numbers on the connecting lines illustrate the number of SNPs in a pairwise comparison. The transmission cluster is marked with gray based on the threshold of 23 SNPs. ND, not determined. Download FIG S1, PDF file, 0.3 MB (362.4KB, pdf) .
Copyright © 2021 Zhou et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Distribution of core virulence genes in the ST72 lineage (≥85% isolates per clade). Download Table S2, DOCX file, 0.02 MB (23.6KB, docx) .
Copyright © 2021 Zhou et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Network diagram of all virulence genes (VGs) identified in sequence types ST72, ST8 (USA300), and ST59 (M013). “Shared” indicates VGs are present in all STs, “Accessory” indicates VGs are present in two STs, and “ST-specific” indicates VGs are present in a specific ST. Download FIG S2, PDF file, 0.7 MB (752.8KB, pdf) .
Copyright © 2021 Zhou et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Comparison of the unknown prophage with similar prophages. (A) Circular representation of the unknown prophage and comparative genomic analysis with other similar prophages. Circle 1 (from outer to inner circle) shows the genes encoded on the unknown prophage; the colors of genes are assigned based on gene function classification. Circles 2 to 10 refer to homologous regions of S. aureus 93b_S9 (CP010952.1), 27 (CP022717.1), 78 (CP022682.1), UP_426 (CP047797.1), UP_551 (CP047794.1), 128 (CP022897.1), 422 (CP022898.1), XQ (CP013137.1), and NRS153 (CP026067.1) relative to the unknown prophage. Circles 11 and 12 represent GC content and GC skew of the unknown prophage, respectively. Direct repeats are indicated by arrows. (B) Phylogenetic analysis of the unknown prophage with other similar prophages. Download FIG S3, PDF file, 1.9 MB (1.9MB, pdf) .
Copyright © 2021 Zhou et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Clade-specific SNPs as identified by Scoary. Download TABLE S3 DOCX file, 0.03 MB (33.4KB, docx) .
Copyright © 2021 Zhou et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Phylogeographic analysis of global ST72 isolates based on the Bayesian binary MCMC method in RASP v4.2. Download FIG S4, PDF file, 0.4 MB (442.7KB, pdf) .
Copyright © 2021 Zhou et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Data Availability Statement
The whole-genome sequences of 24 ST72 S. aureus isolates have been deposited in the GenBank database under BioProject accession no. PRJNA735348.