ABSTRACT
Dolosigranulum pigrum is positively associated with indicators of health in multiple epidemiological studies of human nasal microbiota. Knowledge of the basic biology of D. pigrum is a prerequisite for evaluating its potential for future therapeutic use; however, such data are very limited. To gain insight into D. pigrum’s chromosomal structure, pangenome, and genomic stability, we compared the genomes of 28 D. pigrum strains that were collected across 20 years. Phylogenomic analysis showed closely related strains circulating over this period and closure of 19 genomes revealed highly conserved chromosomal synteny. Gene clusters involved in the mobilome and in defense against mobile genetic elements (MGEs) were enriched in the accessory genome versus the core genome. A systematic analysis for MGEs identified the first candidate D. pigrum prophage and insertion sequence. A systematic analysis for genetic elements that limit the spread of MGEs, including restriction modification (RM), CRISPR-Cas, and deity-named defense systems, revealed strain-level diversity in host defense systems that localized to specific genomic sites, including one RM system hot spot. Analysis of CRISPR spacers pointed to a wealth of MGEs against which D. pigrum defends itself. These results reveal a role for horizontal gene transfer and mobile genetic elements in strain diversification while highlighting that in D. pigrum this occurs within the context of a highly stable chromosomal organization protected by a variety of defense mechanisms.
IMPORTANCE Dolosigranulum pigrum is a candidate beneficial bacterium with potential for future therapeutic use. This is based on its positive associations with characteristics of health in multiple studies of human nasal microbiota across the span of human life. For example, high levels of D. pigrum nasal colonization in adults predicts the absence of Staphylococcus aureus nasal colonization. Also, D. pigrum nasal colonization in young children is associated with healthy control groups in studies of middle ear infections. Our analysis of 28 genomes revealed a remarkable stability of D. pigrum strains colonizing people in the United States across a 20-year span. We subsequently identified factors that can influence this stability, including genomic stability, phage predators, the role of MGEs in strain-level variation, and defenses against MGEs. Finally, these D. pigrum strains also lacked predicted virulence factors. Overall, these findings add additional support to the potential for D. pigrum as a therapeutic bacterium.
KEYWORDS: Dolosigranulum pigrum, nasal microbiota, pangenome, methylome, restriction modification, CRISPR, mobile genetic elements
INTRODUCTION
Evidence points to a prominent role for the benign nasal bacterium Dolosigranulum pigrum in structuring nasal microbiota beneficial to human health (1–30; reviewed in references 31, 32, 33, 34, 35, and 36). Individuals whose nasal microbiota is dominated by D. pigrum are less likely to be colonized by nasal pathobionts and are therefore at lower risk of invasive infections due to these microbes. For example, D. pigrum is inversely associated with Staphylococcus aureus in adult nostrils (5, 16, 28, 37). Also, the level of maternal D. pigrum is inversely associated with infant acquisition of S. aureus (38); in a small study, neonates who do not acquire S. aureus have a higher relative abundance of D. pigrum (39). During in vitro growth, D. pigrum inhibits S. aureus on agar medium, but not the reverse (28), suggesting D. pigrum might directly antagonize S. aureus in vivo. In addition, D. pigrum and nasal Corynebacterium species are frequently present in pediatric nasal microbiota when Streptococcus pneumoniae is absent (1, 8). Together, D. pigrum and Corynebacterium pseudodiphtheriticum robustly inhibit S. pneumoniae in vitro compared to either organism alone (28). As illustrated by these examples, nasal microbiota with higher levels of D. pigrum—usually alongside Corynebacterium—are often associated with health. Young infants with prolonged high levels of D. pigrum and Corynebacterium exhibit greater stability of their nasal microbiota and fewer respiratory tract infections (3, 4, 6, 11, 21). Also, higher levels of nasal D. pigrum and Corynebacterium are more common in healthy children than in children with pneumonia (12) or those with otitis media (1, 2, 15, 30).
In stark contrast to the steadily increasing data in support of D. pigrum as a candidate beneficial bacterium (40), there is a dearth of information about the basic biology of this Gram-positive organism, including the organization and stability of its genome. Ideally, bacterial strains with therapeutic potential display a reliably stable genome structure and have the capacity to resist horizontal gene transfer (HGT), since the latter might lead to unanticipated effects. The stability of bacterial genomes reflects a balance between competing factors, including invasion by mobile genetic elements (MGEs) and systems that defend against MGEs. MGEs play a key role in strain variation through acquisition and distribution of genes in the accessory genome. Analysis of the pangenome of multiple strains identifies core and soft-core gene clusters (GCs) common to all, or almost all, of the strains, respectively, and GCs present in smaller subsets of strains, which constitute the accessory genome (41, 42). Although accessory genes may result from gene loss, many are thought to be acquired via HGT. Counterbalancing this are key systems for defense against MGEs. These include well-described restriction modification systems, CRISPR-Cas systems, and the more recently identified, deity-named defense systems (43). Restriction modification (RM) systems distinguish intracellular DNA as self or nonself by virtue of specific methyl modifications within short linear sequences that allow for destruction of inappropriately methylated nonself DNA by endonuclease activity; the various RM systems are classified into types I, II, III, and IV. There are also other variations of DNA modification-based defense (44, 45). CRISPR-Cas systems mediate defense using a multistep process. Small fragments of foreign nucleic acids are first recognized as non-self and incorporated into the host genome between short DNA repeats, known as a CRISPR array. Subsequently, these fragments, now spacers within the array, are used as RNA guiding molecules for an endonuclease complex that recognizes and destroys DNA containing these sequences (46). The more recently identified deity-named defense systems consist of a set of 10 disparate antiphage/plasmid mechanisms that are often found clustered next to known defense genes (RM and CRISPR-Cas) (43) within defense islands (47) of bacterial genomes. Although deity-named defense systems have been shown to be active and limit phage/plasmid spread, their exact underlying modes of action remain to be deciphered. Collectively, these systems can protect bacteria from infection by phages and invasion by other MGEs, including plasmids and transposable elements, thus limiting the introduction of new genes and maintaining genomic stability.
Comparing genomic content and chromosomal organization of D. pigrum strains collected 20 years apart, and mostly in the United States, we identified the following characteristics: (i) highly similar strains circulating across 20 years; (ii) stable chromosomal synteny across the phylogeny; (iii) the first predicted D. pigrum prophage and insertion sequence; and (iv) a diverse collection of RM, deity-named defense and CRISPR-Cas systems incorporated at conserved chromosomal insertion sites across strains. Together, these reveal a stable synteny and a high-level of sequence conservation within the D. pigrum core genome, along with an open pangenome and active defense against HGT.
RESULTS
Detection of highly similar Dolosigranulum pigrum strains over a 20-year span.
To identify genomic shifts in D. pigrum strains currently circulating in human nasal microbiota compared to strains from approximately 20 years ago, we collected 17 new nostril isolates of D. pigrum from volunteers in 2017 and 2018 and sequenced the genomes of these isolates using SMRTSeq (PacBio), fully circularizing 14 (Table 1). We compared these 17 new genomes to 11 described genomes (28), 9 of which are from strains collected in the late 1990s (48). This refined existing and uncovered new information about the basic genomic characteristics of D. pigrum (see Table S1 in the supplemental material).
TABLE 1.
Source information for the 28 D. pigrum strains and quality description for the 17 newly SMRT-sequenced closed genomesa
| Original strain name | Internal reference | Yr isolated | Human body site | Location | Age (yrs) | NCBI assembly ID | Source or reference | Realigned bases (%)b | Coverage (fold) |
|---|---|---|---|---|---|---|---|---|---|
| ATCC 51524 | NA | 1988 | Spinal cord | UK | ? | GCF_000245815.1 | 135 | ||
| KPL1914 | KPL1914 | 2010 | Nostril | MA | Adult | GCA_003263915.2 | 28 | ||
| CDC39-95 | KPL1922 | 1995 | NP | CN | 3 | GCF_003264145.1 | 48 | ||
| CDC2949-98 | KPL1930 | 1998 | NP | AZ | ? | GCF_003264135.1 | 48 | ||
| CDC4294-98 | KPL1931 | 1998 | Blood | SC | <1 | GCF_003264085.1 | 48 | ||
| CDC4420-98 | KPL1932 | 1998 | Blood | TN | 11 | GCF_003264065.1 | 48 | ||
| CDC4545-98 | KPL1933 | 1998 | NP | AZ | ? | GCF_003264045.1 | 48 | ||
| CDC4709-98 | KPL1934 | 1998 | Eye | GA | <1 | GCA_003264015.2 | 48 | ||
| CDC4199-99 | KPL1937 | 1999 | Blood | GA | ∼2 | GCF_003264005.1 | 48 | ||
| CDC4791-99 | KPL1938 | 1999 | NP | AZ | ? | GCF_003263975.1 | 48 | ||
| CDC4792-99 | KPL1939 | 1999 | NP | AZ | ? | GCF_003263965.1 | 48 | ||
| KPL3033 | KPL3033 | 2018 | Nostril | MA | 18–30 | GCA_017655925.1 | This study | 92.61* | 498 |
| KPL3043 | KPL3043 | 2018 | Nostril | MA | 7–12 | GCA_017655905.1 | This study | 92.40* | 582 |
| KPL3050 | KPL3050 | 2018 | Nostril | MA | 31–60 | GCA_017655885.1 | This study | 92.11* | 475 |
| KPL3052 | KPL3052 | 2018 | Nostril | MA | 3–6 | GCA_017655865.1 | This study | 92.15* | 382 |
| KPL3065 | KPL3065 | 2018 | Nostril | MA | 7–12 | GCA_017655845.1 | This study | 91.73* | 460 |
| KPL3069 | KPL3069 | 2018 | Nostril | MA | 7–12 | GCA_017655825.1 | This study | 88.13* | 372 |
| KPL3070 | KPL3070 | 2018 | Nostril | MA | 31–60 | GCA_017655785.1 | This study | 91.85* | 271 |
| KPL3077 | KPL3077 | 2018 | Nostril | MA | 7–12 | GCA_017655765.1 | This study | 91.60 | 351 |
| KPL3084 | KPL3084 | 2018 | Nostril | MA | 31–60 | GCA_017655745.1 | This study | 90.24* | 433 |
| KPL3086 | KPL3086 | 2018 | Nostril | MA | <3 | GCA_017655725.1 | This study | 91.30* | 342 |
| KPL3090 | KPL3090 | 2018 | Nostril | MA | 7–12 | GCA_017655685.1 | This study | 90.72* | 423 |
| KPL3246 | KPL3246 | 2018 | Nostril | MA | 7–12 | GCA_017655805.1 | This study | 92.47* | 578 |
| KPL3250 | KPL3250 | 2018 | Nostril | MA | 7–12 | GCA_017655665.1 | This study | 92.63* | 501 |
| KPL3256 | KPL3256 | 2018 | Nostril | MA | 7–12 | GCA_017655645.1 | This study | 92.84 | 530 |
| KPL3264 | KPL3264 | 2018 | Nostril | MA | 7–12 | GCA_017655705.1 | This study | 87.61 | 342 |
| KPL3274 | KPL3274 | 2018 | Nostril | MA | 7–12 | GCA_017655945.1 | This study | 87.41* | 574 |
| KPL3911 | KPL3911 | 2017 | Nostril | MA | <3 | GCA_017655965.1 | This study | 87.13* | 595 |
NA, not applicable; NP, nasopharynx.
Percent realigned bases (from Realignment to Draft Assembly). *, circularized genome.
Pangenome contribution of all 28 D. pigrum genomes. Download Table S1, PDF file, 0.04 MB (29.5KB, pdf) .
Copyright © 2021 Flores Ramos et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
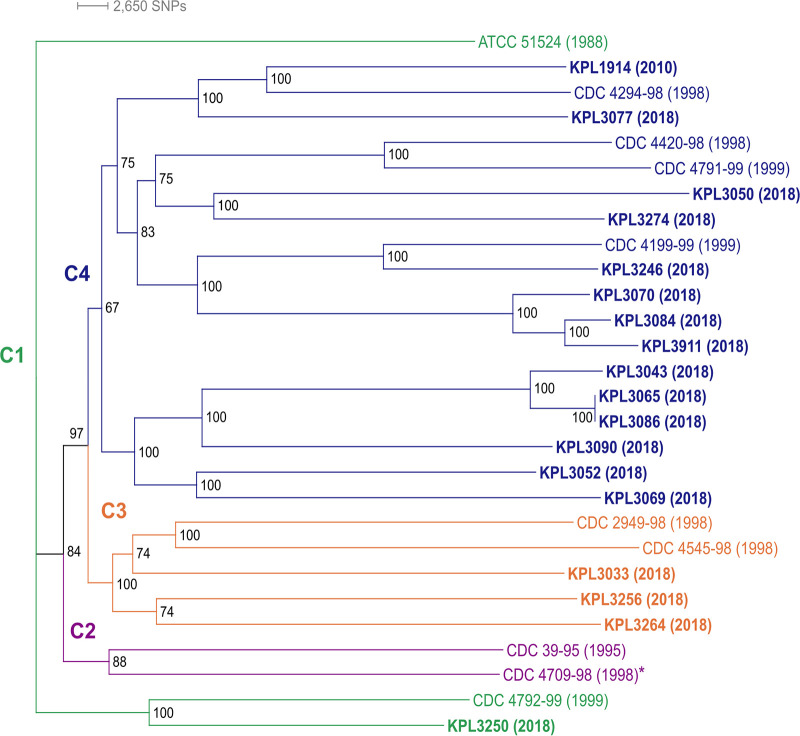
To assess the similarity of these 28 D. pigrum strains, we generated a phylogenomic tree based on 1,102 single-copy core GCs (Fig. 1). Some of the terminal clades include strains collected during different decades. The average number of pairwise single nucleotide polymorphisms (SNPs) among isolates collected approximately 20 years apart was similar to that among isolates collected recently (21,754 versus 20,834) (see Table S2A). Thus, closely related strains of D. pigrum have circulated among people in the United States over a span of time that has an upper bound of 20 years and a lower bound of 8 to 13 years. (This lower bound allows for the possibility that the recent isolates were stably acquired in infancy since most of the 2018 strains were from children in the 7- to 12-year age range.) Alloiococcus otitis (49) is the closest genome-sequenced bacterium to D. pigrum in 16S rRNA gene phylogenies. A. otitis ATCC 51267 shared 789 core GCs with the D. pigrum strains (see Fig. S1A). Using these 789 core GCs, we constructed a phylogenomic tree with A. otitis as an outgroup (see Fig. S1B and C). In contrast to the D. pigrum-only phylogeny (Fig. 1), the phylogeny including A. otitis displayed poor support for many of the branches within the D. pigrum clade. This is likely due to the reduced number of SNPs among D. pigrum strains when using only the 789 GCs shared with A. otitis (see Table S2A). Therefore, we based subsequent inferences on the D. pigrum-only phylogeny.
FIG 1.
Dolosigranulum pigrum strains collected 20 years apart are phylogenetically similar. This maximum-likelihood core-gene-based phylogeny shows recently collected strains (bold), mostly from 2018, and strains collected before 2000 intermingled in three of the four distinct clades (clades C1 to C4 are color coded, and the year of collection is in parentheses). Strains separated by 18 to 19 years grouped together in terminal clades: KPL1914 and CDC4294-98, KPL3246 and CDC4199-99, and KPL3250 and CDC4792-99. The genomes of strains in boldface plus strain CDC4709-98 (asterisk) are closed. Strains KPL3065 and KPL3086 were collected from two different individuals and have almost identical genomes, differing by just 4 core SNPs and 6 gene clusters (4 and 2 in KPL3086 and KPL3065, respectively). We created this unrooted phylogeny using the concatenated alignment of 1102 conservative single-copy core GCs (see Fig. S3A), a GTR+F+R3 substitution model of evolution, 553 maximum-likelihood searches, and 1,000 ultrafast bootstraps with IQ-Tree v.1.
Pairwise SNP analysis and MASH analysis. Download Table S2, XLSX file, 0.06 MB (59.2KB, xlsx) .
Copyright © 2021 Flores Ramos et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
A core-gene phylogenomic tree including Alloiococcus otitis ATCC 51267 as an outgroup has poor resolution. (A) The 789 GCs shared between the BDBH, COG triangle, and OMCL algorithms were used to determine the single-copy GCs shared between the 28 D. pigrum genomes and the A. otitis outgroup using GET_HOMOLOGUES v. 3.1.4. (B) The phylogenomic tree with ancestral distance displayed. (C) The phylogenomic tree as a cladogram. A. otitis is the most closely related genome-sequenced taxon to D. pigrum based on 16S rRNA gene sequence in both the eHOMD and the Living Tree Project (P. Yarza, M. Richter, J. Peplies, J. Euzeby, R. Amann, K. H. Schleifer, W. Ludwig, F. O. Glöckner, R. Rosselló-Móra. Syst Appl Microbiol. 31:241–250, 2008, [doi: 10.1016/j.syapm.2008.07.001]; P. Yilmaz, L.W. Parfrey, P. Yarza, J. Gerken, E. Pruesse, C. Quast, T. Schweer, J. Peplies, W. Ludwig, F. O. Glöckner. Nucleic Acids Res. 42:D643–D648, 2014 [doi: 10.1093/nar/gkt1209]). We generated a phylogenomic tree based on 789 concatenated core GCs shared among A. otitis ATCC 51267 (NZ_JH992957) and the 28 D. pigrum genomes with IQ-Tree using a GTR+F+R6 substitution model (BIC value 5743936.9087), 300 ML searches (ML = −2868528.1268) and 1,000 ultrafast rapid Bootstraps. There is a deep branch separating A. otitis from all the D. pigrum strains. However, many of the ancestral branches within the D. pigrum clade are poorly supported. Repeated construction resulted in phylogenies with poorly supported branching likely due to poor SNP resolution (see Table S1A), suggesting the need for a better outgroup for D. pigrum. Download FIG S1, PDF file, 0.2 MB (215.7KB, pdf) .
Copyright © 2021 Flores Ramos et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The conservative core genome of D. pigrum has 1102 GCs with a very high degree of nucleotide conservation among strains and the accessory genome size varies but the functions are similar among the 28 genomes. (A) We used the intersection of the BDBH, COG triangle, and OMCL algorithm results to determine a conservative core genome of 1102 single-copy GCs using the 28 D. pigrum genomes on GET_HOMOLOGUES v. 3.1.4. (B) We determined the GCs present in the core (n = 28), soft core (28 > n ≥ 26), shell (26 > n ≥ 3), and cloud (n ≤ 2) using the results from the OMCL and COG triangle algorithms. Of the 3700 total GCs, 30.6% are core (1,134/3,700), 5.41% are soft core (200/3,700), 22.8% are shell (845/3700), and 41.1% are cloud (1,521/3,700). (C) All paired strains share above 97.58% average nucleotide identity. We based this on the average nucleotide identity of the homologues determined using the OMCL algorithm. (D) The D. pigrum Anvi’o pangenome is similar to the GET_HOMOLOGUES pangenome. The biggest difference between the GET_HOMOLOGUES- and Anvi’o-defined pangenome was the migration of many of the GET_HOMOLOGUES cloud GCs to the Anvi’o multicopy core, causing the pangenome size to drop from 3,700 to 2,905 GCs. Occupancy of the GCs among the 28 genomes is displayed with strains collected in 2018 and 2017 (blue), strains from the CDC (pink), the ATCC strain (gold), and the strain collected in 2010 (green). Strains are arranged based on the phylogeny from Fig. 1 (right). With Anvi’o, 44.7% of the GCs (1,298/2,905) were in the core, 3.1% (90/2,905) in the soft core (green), 27.7% (805/2,905) in the shell (blue), and 24.5% (712/2,905) in the cloud (purple). A closer inspection of the core also showed 38.2% (1111/2,905) of the GCs are in the conservative single-copy (SC) core (red) and 6.4% (187/2,905) are in the multicopy (MC) core (dark red). The interactive Anvi’o pangenome can also be found on our GitHub (https://github.com/KLemonLab/DpiMGE_Manuscript/blob/master/SupplementalMethods_Anvio.md). (E) The accessory genome size varies but the functions are similar among the 28 D. pigrum genomes. Using the functional annotations of the accessory GCs identified by Anvi’o (defined as “shell” plus “cloud” bins), we found that most of the accessory GCs among all of our genomes either had no annotation (“unclassified,” 53.6%), had an ambiguous COG categorization (“ambiguous”, 6.6%) or belonged to the uninformative S or R COG category (“uninformative,” 2.6%). Only 37.2% of the GCs in the accessory genome had informative COG annotations (colored categories). (F) Of the informative GCs, there was a similar proportional distribution of accessory functions among all 28 genomes with a large fraction dedicated to “defense mechanisms” (olive) and “carbohydrate transport and metabolism” (khaki). The size of the accessory genomes varied among the strains with CDC4294-98 and KPL3090 having the largest accessory size compared to KPL3065 and KPL3086 with the smallest. Horizontal bars indicate clades: clade 1, green; clade 2, purple; clade 3, orange; and clade 4, blue. Download FIG S3, PDF file, 1.1 MB (1.1MB, pdf) .
Copyright © 2021 Flores Ramos et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The chromosome of D. pigrum exhibits conserved synteny across a phylogeny spanning 20 years.
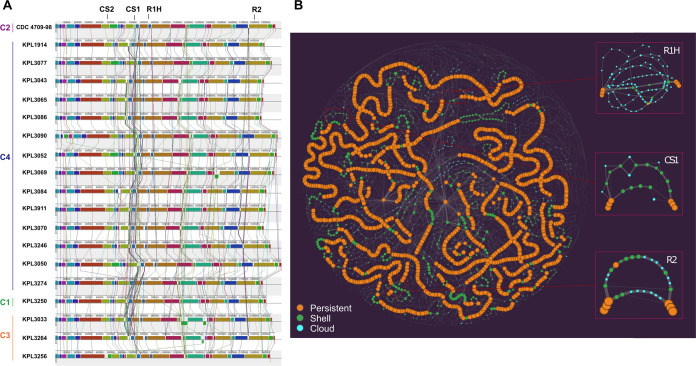
Based on the observed similarity of circulating strains over time, we hypothesized there would be a high-level of genomic stability across the D. pigrum phylogeny. To test this, we compared chromosomal synteny across the four major clades in the D. pigrum phylogeny using 19 strains with closed genome sequences (highlighted in bold or with an asterisk [*] in Fig. 1), including representative strains collected in 1998, 2010, 2017, and 2018. A MAUVE alignment (50, 51) of these 19 genomes starting at the dnaA gene revealed a remarkable conservation of the overall chromosomal structure with no visible shifts in the position of large blocks of sequence (Fig. 2A). Dispersed among these blocks are regions with higher numbers of insertions and deletions (indels) (Fig. 2A; see also Fig. S2A).
FIG 2.
D. pigrum displays conserved chromosomal synteny. (A) A MAUVE alignment of 19 closed D. pigrum genomes, with representatives from the four major clades in Fig. 1, shows a conserved order of chromosomal blocks across the phylogeny of strains collected 20 years apart. Vertical bars represent clades: clade 1, green; clade 2, purple; clade 3, orange; and clade 4, blue. CS1 and CS2 designate the CRISPR-Cas sites (Fig. 8), R1H represents the RM system insertional hot spot and R2 represents the site containing either a type II m5C RM system or a type IV restriction system (Fig. 7). (B) This PPanGGOLiN partitioned pangenome graph displays the overall genomic diversity of the 28 D. pigrum genomes. Each graph node corresponds to a GC; the node size is proportional to the total number of genes in a given cluster, and the node color represents the PPanGGOLiN assignment of GCs to the partitions: persistent (orange), shell (green), and cloud (blue). Edges connect nodes that are adjacent in the genomic context and their thickness is proportional to the number of genomes sharing that neighboring connection. The insets on the right depict subgraphs for sites R1H, CS1, and R2 showing several branches corresponding to multiple alternative shell and cloud paths. These sites with higher genomic diversity are surrounded by longer regions with conserved synteny, i.e., long stretches of consecutive persistent nodes (GCs). The static image depicted here was created with the Gephi software (https://gephi.org) (133) using the ForceAtlas2 algorithm (134) with the following parameters: scaling = 20,000, stronger gravity = true, gravity = 6.0, LinLog mode = true, and edge weight influence = 2.0.
A 100-kb region with a higher frequency of indels and a second region with a higher frequency of phage and plasmid integration events. (A) Closer inspection of positions ∼650000 to ∼750000 in the MAUVE alignment showed the presence of three sites containing sequencing heterogeneity (HS for heterogeneity sites) bounded by >10 kb blocks of homology. The first site (HS1), between abgT (p-aminobenzoyl-glutamate transport protein) and kynB (kynurenine formamidase), contained a PTS (mannitol) system for metabolism. However, the extent of completeness of this PTS system varied among the D. pigrum isolates and was absent in KPL3274 and KPL3256. The next site (a few kb downstream of HS1), between yxdM (ABC transporter permease protein YxdM) and ghrA (glyoxylate/hydroxypyruvate reductase A), contained one of the two possible CRISPR-Cas systems, with all genomes having at least one type (CS1; Fig. 8A and B). Finally, the third site (HS2), between nth (endonuclease III) and clsA_1 (cardiolipin synthase A), variably harbored two types of aspartate and threonine synthase genes. One type in KPL3033 and KPL3052; none in KPL1914, KPL3077, and KPL3274; and a variation of the second type among the remaining genomes. Also indicated on this MAUVE alignment are the second of the two CRISPR-Cas insertion sites (CS2) and the location of the pUB110 insertion, which is in a region with a higher frequency of phage and plasmid integration events. Vertical bars represent clades: clade 1, green; clade 2, purple; clade 3, orange; and clade 4, blue. (B) No autonomous plasmids were detected among the 28 genomes using the Gram-positive plasmid database, PlasmidFinder. However, as illustrated with this map from D. pigrum KPL3043, we detected a sequence match to pUB110 (GenBank ID NC_001384.1), a plasmid originally isolated from S. aureus, integrated in four of the D. pigrum isolates (KPL3043, KPL3065/KPL3086 in clade 4 and CDC4709-98 in clade 2; Fig. 1). This included a perfect match (708/708 bp) to the repB gene (blue) of pUB110, which was located adjacent to three other genes (cyan) that are also present in pUB110 and predicted to encode a kanamycin nucleotidyltransferase ANT(4′)-lb (knt), a bleomycin resistance protein (ble), and a plasmid recombination protein (pre-2). Inspection of the region adjacent to the genes from pUB110, at position ∼1.45 Mb in the MAUVE alignment (A), showed evidence of plasmid elements present in 11 of the other closed D. pigrum genomes, suggesting this region has a higher frequency of phage and plasmid integration events. For example, a 73% (288/391 bp) match to a different (non-pUB110) repB gene present in plasmids of other closely related Firmicutes including Lactobacillus plantarum (KU707950) (X. Ma, J. Li, Y. Xiong, Z. Zhai, F. Ren, Y. Hao. Curr Microbiol. 73:820–826, 2016 [doi: 10.1007/s00284-016-1124-7]) and Lactobacillus curvatus (CP031007) (R.M. Prechtl, https://www.researchgate.net/publication/337103133_Formation_and_structure_of_exopolysaccharides_of_meat_starter_cultures). A subset of the genomes, including six from clade 4 (KPL3077, KPL3090, KPL3069, KPL3084, KPL3911, and KPL3070) and two from clade 3 (KPL3264 and KPL3256), had an ltrA gene (group II reverse transcriptase/maturase) at this location. Download FIG S2, PDF file, 1.0 MB (1MB, pdf) .
Copyright © 2021 Flores Ramos et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
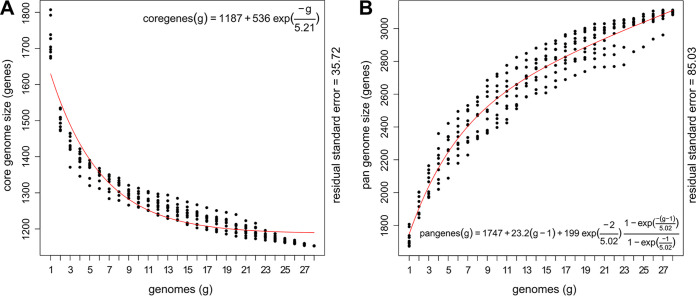
D. pigrum has a core genome that has leveled off, an open pangenome, and a high degree of conservation at the amino acid and nucleotide level.
Analysis of all 28 D. pigrum genomes revealed a conservative core of 1,102 single-copy GCs, as defined by the intersection of results from three algorithms, including bidirectional best hits (BDBH) (see Fig. S3A). A core of 1,134 GCs was defined by the intersection of two algorithms when BDBH was excluded (see Table S1 and Fig. S3B). The D. pigrum core genome has leveled off in size (Fig. 3A). Meanwhile, the pangenome continued to increase, with each additional genome (Fig. 3B) reaching 3,700 GCs (see Fig. S3B); of these, 30.6% (1,134/3,700) are core. The average number of coding sequences (CDS) per genome was 1,765 and, on average, the core constituted ∼64% (1,134/1,765) of the CDS in each individual genome (see Table S1). These results from GET_HOMOLOGUES (42) generally agreed with those from Anvi’o (52, 53), allowing us to leverage Anvi’o for additional analyses. In the Anvi’o-derived single-copy core (38.2%; Fig. S3D), 89.4% (993/1,111) of the GCs had a functional homogeneity index score ≥0.98, indicating a high degree of conservation at the amino acid level. This fits with an average nucleotide identity (ANI) over 97.58% for all 28 genomes (see Fig. S3C), matching earlier findings with 11 strain genomes (28). Moreover, two sets of three recently collected strains each shared over 99% ANI, as well as similar accessory Clusters of Orthologous Group (COG) annotations (see Fig. S3E). This revealed highly similar strains in the nasal microbiota of different individuals in Massachusetts. Of these, two strains collected from different people were nearly identical, differing by just 4 core SNPs and 6 GCs (4 and 2 in KPL3086 and KPL3065, respectively) with a MASH-distance of 3.10E-05 (P = 0; Table S2B). (Henceforth, we refer to these two strains as KPL3065/KPL3086.) In contrast KPL3086 and KPL3043, which are in that same distal clade in Fig. 1, have a MASH distance of 0.0045 (P = 0).
FIG 3.
The D. pigrum core genome levels off, and the pangenome remains open. (A and B) The D. pigrum core (n = 28) genome started to level off after 17 genomes, as predicted using a Tettelin curve fit model (red line) (A), whereas, with 28 genomes, the pangenome continued to increase in gene clusters with each additional genome (B). D. pigrum core (A) and pangenome (B) size estimations were based on 10 random genome samplings (represented by black dots) using the OMCL algorithm defined gene clusters in GET_HOMOLOGUES v.3.1.4.
The D. pigrum accessory genome is enriched for gene clusters involved in mobilome and host defense.
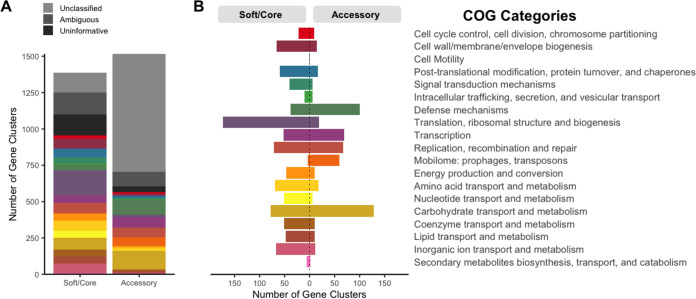
Of the 49,412 individual genes identified across the 28 genomes, 63.8% (31,501/49,412) had informative calls to a single functional COG annotation (i.e., their assignment corresponds to a single COG category other than S or R) (54, 55) (Fig. 4A). Using Anvi’o, we observed that GCs involved in mobilome, in defense mechanisms, and in carbohydrate transport and metabolism were overrepresented in the accessory compared to the core genome (Fig. 4B). GCs classified to these three COG categories accounted for 3.9, 6.6, and 8.5% of the D. pigrum accessory genome, respectively. The proportion of accessory functions was similar among all strains, but the sizes of their accessory genomes varied (see Fig. S3E and F). Because genome stability is relevant to suitability of a candidate beneficial microbe for therapeutic use, we focused subsequent analysis on the predicted mobilome and defense mechanisms.
FIG 4.
The accessory genome of D. pigrum has functional enrichment for defense mechanisms, mobilome, and carbohydrate transport and metabolism genes. (A) Of the total 49,412 individual genes identified across the 28 analyzed genomes, up to 8,242 genes (16.7%) lacked a COG annotation, 5,221 (10.6%) had an ambiguous COG category annotation (more than one COG category), and 4,448 (9.0%) had an uninformative annotation (belonging to the S or R COG category). At the gene cluster (GC) level, only 37.2% of the 1,517 GCs present in the accessory genome had an informative COG assignment compared to 68.7% of the 1,388 GCs in the soft/core. (B) The number of GCs present in the accessory genome was severalfold higher than in the soft/core for the following informative COG assignments (colored categories): defense mechanisms (olive, 2.60-fold), mobilome: prophages, transposons (orange, 14.88-fold), and carbohydrate transport and metabolism (khaki, 1.66-fold). This was determined using the COG functional annotations defined in our Anvi’o analysis of the soft/core (“core” and “soft core” bins) versus accessory (“shell” and “cloud” bins). Since many GCs have individual genes with distinct COG annotations each individual gene was counted as 1/x, with x being the number of genes in each GC.
D. pigrum hosts distinct integrated phage elements, insertional elements, and a group II intron.
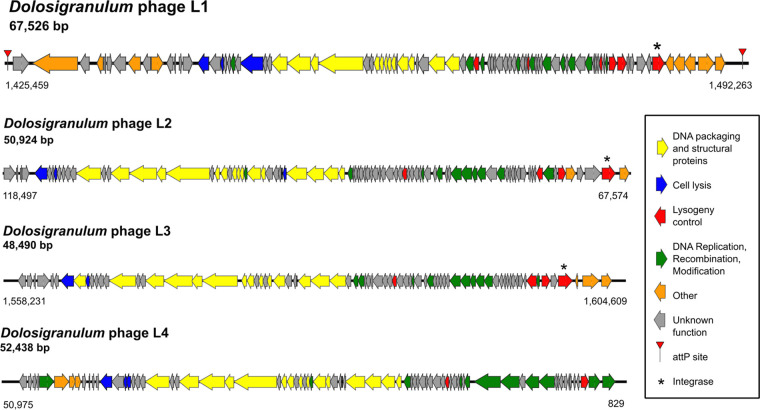
Of the total GCs in the pangenome, 2.2% were predicted to be part of the mobilome. MGEs can negatively affect genome stability and can positively affect strain diversification. Therefore, we systematically searched for various types of MGEs, including phage elements, plasmids, and insertional elements that interact with D. pigrum. First, using the Phage Tool Enhanced Release (PHASTER) database (56, 57), we identified four distinct, and mostly intact, integrated phage elements, i.e., prophages (Fig. 5). We gave these the provisional names Dolosigranulum phage L1 through L4. All four were in the size range common for Firmicutes phages and had a life cycle-specific organization of its CDS with lytic and lysogenic genes separated (Fig. 5) (58–60). Predicted prophage L1 from D. pigrum KPL3069 was the most intact with two attachment (attP) sites and an intact integrase most similar to that of the Streptococcus prophage 315.2 (NC_004585; E value 7.85e-69) (61). Prophages L2 and L3 from D. pigrum KPL3090 also had intact integrases, with similarity to other streptococcal phages, but lacked distinguishable attP sites. Beyond these similarities, other CDS from L1 to L4 displayed few and dissimilar matches to known phage elements (see Text S1), indicating that D. pigrum hosts a distinct set of lysogenic phage that are expected to have a limited host range.
FIG 5.
D. pigrum has an intact prophage. Map of the four predicted prophages: Dolosigranulum phage L1 from KPL3069, L4 from KPL3256, and L2 and L3 from KPL3090. The most complete prophage was L1 from KPL3069 with an intact integrase and two attP sites. All of the putative phages exhibited a typical life cycle-specific organization, with lytic genes on one side and lysogenic genes on the other. We detected phage elements using the PHASTER database on 8 November 2018.
Supplemental analysis of genetic elements and defense systems. Download Text S1, PDF file, 0.10 MB (98.6KB, pdf) .
Copyright © 2021 Flores Ramos et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Second, using the Gram-positive plasmid database PlasmidFinder (62), we detected no autonomous plasmids. However, a nearly complete fragment of the S. aureus plasmid pUB110 is integrated in the chromosome of four strains and includes a gene encoding kanamycin resistance (see Fig. S2B). This prompted a systematic search for antibiotic resistance genes using the Comprehensive Antibiotic Resistance Database in the Resistance Gene Identifier (CARD-RGI) (63, 64). Of the 28 genomes, 6 are predicted to encode antibiotic resistance genes for erythromycin and/or kanamycin, which are located within a CRISPR array or the integrated plasmid, respectively (see Text S1).
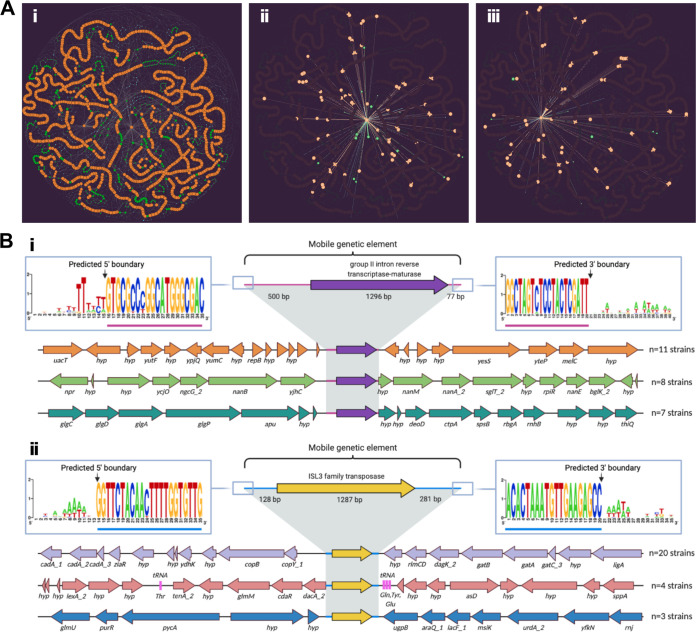
Third, we identified GCs predicted to be either transposases (eight) or integrases (five) using a multistep approach (see Table S3). Transposases are thought to function both as detrimental, selfish genetic elements that can disrupt important genes and as diversifying agents that can provide benefit to host cells through gene activation or rearrangements (65, 66). Among the 26 genomes containing at least one transposase CDS, the mean was 4.42 (median, 3.5), with a maximum of 13 per genome. Transposases were more prevalent and abundant than integrases (see Table S3). One of the predicted transposases was the GC containing the third largest number of sequences. This is consistent with reports that genes encoding transposases are the most prevalent protein-encoding genes detected across the tree of life when accounting for both ubiquity and abundance (67). We detected 74 intact instances of this most common transposase, an ISL3 family transposase with similarity to ISSau8, across 22 of the D. pigrum genomes with a mean (median) of 3.36 (2) and a maximum of 11 copies per genome (GC_00000003; Table S3). As shown on the PPanGGOLin graph (Fig. 6Ai), this transposase is inserted at multiple different sites within and across the genomes (Fig. 6Aiii; see also Table S3). The most common of these is likely the ancestral insertion site (Fig. 6Bii). The absence of a cotraveling CDS is consistent with this ISL3 family transposase being part of an insertional sequence (IS). According to standards for IS nomenclature, we propose the name ISDpi1 (66).
FIG 6.
D. pigrum genomes host a few highly prevalent MGEs. (Ai) On the PPanGGOLiN partitioned pangenome graph for the 28 D. pigrum genomes, we highlight the neighboring connections for the persistent GC of a predicted group II intron reverse transcriptase-maturase (Aii; purple in panel Bi) and a predicted ISL3 family transposase (Aiii; yellow in panel Bii). Each graph node corresponds to a GC; node size is proportional to the total number of genes in a given cluster; and node color represents PPanGGOLiN assignment of GCs to the partitions: persistent (orange), shell (green), and cloud (blue). Edges connect nodes that are adjacent in the genomic context and their thickness is proportional to the number of genomes sharing that neighboring connection. In panels Aii and Aiii, only the adjacent neighboring nodes and edges for each of the depicted GCs are contrast colored against the background pangenome graph. (B) Most common genomic neighborhoods for the predicted group II intron reverse transcriptase-maturase (Bi) and the ISL3 family transposase (Bii). OCTAPUS (https://github.com/FredHutch/octapus) identified the chromosomal coordinates of each MGE integration event in individual strains, and groupings of colocated genes residing within the same neighborhood structure across strains were visualized using Clinker (https://github.com/gamcil/clinker). ClustalOmega alignments of flanking regions across groupings revealed predicted terminal sequence boundaries (consistent 5′–3′ sequences across integration events) for each MGE. The three most common genomic loci for each MGE were rendered using BioRender.
Predicted D. pigrum transposases, integrases and group II intron. Download Table S3, PDF file, 0.2 MB (124.1KB, pdf) .
Copyright © 2021 Flores Ramos et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Fourth, the PPanGGOLin graph (68) revealed insertion of a predicted group II intron reverse transcriptase-maturase at multiple sites across multiple D. pigrum genomes (Fig. 6Aii and Bi; see also Table S3). Group II introns are MGEs commonly found in bacterial genomes that consist of a catalytic RNA and an intron-encoded protein that assists in splicing and mobility (69). Like transposases, group II introns can play both detrimental and beneficial roles within their host. We detected this intron-encoding GC in all 28 genomes with a mean (median) of 4.7 (3.5) and range of 1 to 14 copies per genome. This GC contained the highest number of individual gene sequences of any GC with 132 (GC_00000001; Table S3). It is most closely related to the bacterial class C intron-encoded protein from La.re.I1 in Lactobacillus reuteri with 44% identity and 65% similarity over 419 amino acids (70). These data are consistent with an intact bacterial reverse transcriptase/maturase expected to facilitate splicing and mobility of the group II intron (69).
A systematic search identifies multiples types of defense systems to protect D. pigrum from MGEs.
The enrichment for defense mechanisms in the accessory genome of D. pigrum is combined with the relative paucity of plasmids and prophages among D. pigrum genomes. Based on this, we performed a systematic search of the pangenome for known bacterial host defense systems, including RM, deity-named defense, and CRISPR-Cas systems.
D. pigrum harbors a diverse collection of RM systems.
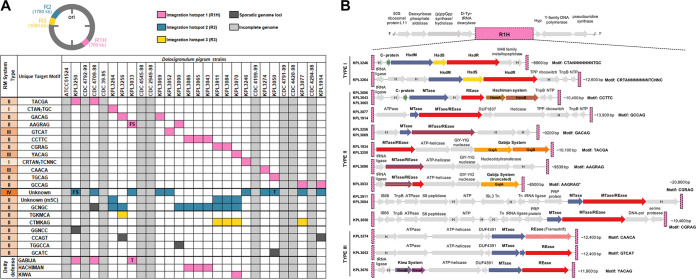
In bacteria, individual RM systems can differ with respect to target sequence, active site architecture, and reaction mechanisms, but all recognize the methylation status of target sequences on incoming DNA and degrade inappropriately methylated (non-self) DNA. Type I to III systems largely recognize and digest a target sequence when it lacks the appropriate methyl group. In contrast, type IV systems, which lack a methyltransferase, are composed of a methyl-dependent restriction endonuclease (REase) that cuts a target sequence when it contains a specific methyl modification. RM systems and their recognition sequences are often strain specific. Therefore, we characterized and compared the repertoire of RM systems present in each of the 19 D. pigrum strains sequenced via SMRTseq, defining the methylome of each strain using SMRTseq kinetics (Basemod analysis) and predicting the recognition sequences of each system via REBASE analysis (71) (Fig. 7A; see also Table S4 and Text S1). Most of the modifications detected were m6A with only one m4C being found. There were several genes coding for m5C enzymes, but their products are not usually detected by the PacBio software. Only one positive m5C enzyme was identified. Among the RM systems, most were type II, although half the strains had a type IV enzyme of unknown specificity. The type I to III systems were associated with 19 individual target recognition motifs identified by methylome analysis (Fig. 7A; see also Table S4 and Text S1).
FIG 7.
D. pigrum hosts a diverse collection of restriction-modification (RM) systems at three distinct loci. (A) Conserved methyl-modifications associated with RM defense systems of D. pigrum strains. White and light gray cells indicate that a modified motif was not detected or no SMRTseq data were available for a specific strain, respectively. Colored cells indicate that a motif was detected and the approximate genomic loci of the RM system responsible across strains are indicated by pink (R1H), blue (R2), or yellow (R3) cells. Sporadic occurrences of RM systems that do not appear conserved in more than a single strain are indicated by dark gray cells. (B) The organization of gene clusters within RM system integration site 1 hot spot (R1H), which harbors a diverse collection of RM systems, including type I (n = 2), type II (n = 7), and type III (n = 3), in addition to other mobile elements/transposons systems, including Hachiman, Gabija, and Kiwa defenses. R1H is flanked upstream by region containing genes for (p)ppGpp synthase/hydrolase and d-Tyr-tRNA (Tyr) deacylase proteins, and downstream by a region with genes for Y-family DNA polymerase and an rRNA pseudouridine synthase protein. Hypothetical genes are indicated by gray arrows labeled with an “H.”
RM systems. Download Table S4, XLSX file, 0.02 MB (18.9KB, xlsx) .
Copyright © 2021 Flores Ramos et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The D. pigrum type IV RM system is inversely related to a specific m5C-associated type II system.
We noted an inverse relationship between the presence of the D. pigrum type IV REase and a specific m5C-associated type II RM system that modified the second cytosine residue within the motif GCNGC (Fig. 7A). This inverse relationship was found to be interdependent between strains based upon a Fisher exact test (P = 0.0055). The type II m5C system was present in nine D. pigrum genomes that lacked the type IV REase. Conversely, the type II m5C system was absent in eight strains that contained the type IV REase. Type IV REase that target m5C-modified motifs have the potential to limit the spread of RM systems that utilize m5C modifications. The D. pigrum type IV REase appears related (99% coverage/43% identity) to S. aureus SauUSI, a modified cytosine restriction system targeting S5mCNGS (either m5C or 5hmC), where S is C or G. Based on the inverse relationship of the type IV and type II m5C systems, this strongly indicates that the D. pigrum type IV system targets m5C containing sequences, including GCNGC, GGNCC, and potentially the recognition sequence of the other m5C enzyme, M.Dpi3264ORF6935P.
The type IV and specific m5C-associated type II RM systems are present at the same integration site.
To decipher the basis for the inverse relationship between these two RM systems, we sought to determine where each was incorporated in the D. pigrum genomes. In 18 of the 19 strains, the type IV REase or the m5C-associated type II system are inserted into the same genetic locus, dubbed R2 (Fig. 2B; see also Fig. S4A). In the one strain that carried both the type IV REase and the m5C-associated type II system, CDC4709-98, the type IV is present at R2, whereas the m5C system is integrated at an unrelated locus downstream from a tRNA-Leu site. MGEs that carry similar integrases tend to integrate at the same sites in the chromosome, but in most strains we did not observe any integrase or additional genes cooccurring with the RM systems at this site.
The organization of GCs within RM system integration sites R2 and R3. (A) Organization of GCs within RM system integration site 2 (R2), which primarily encodes either a type II m5C-associated RM system or a type IV restriction system. R2 is flanked upstream by a region containing genes for a YacP-like NYN domain protein and an S8 family serine peptidase type protein, and downstream by genes for a DNA-3-methyladenine glycosylase protein, a hypothetical protein, and a copper-translocating P-type ATPase. (B) The organization of GCs within RM system integration site 3 (R3), which encodes two different type II RM systems. Arrows represent the direction of translation and the relative sizes of ORFs. Putative control proteins are highlighted in green; HsdR and HsdM are highlighted in red and blue, respectively; and fused hsdR/M ORFs are both red and blue. Proteins not identified as part of the RM system or those with currently unknown function are shown in gray. Download FIG S4, PDF file, 0.2 MB (229.2KB, pdf) .
Copyright © 2021 Flores Ramos et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Many D. pigrum RM systems compete for an integration hot spot.
Extending our analysis, we identified a genomic locus with an unexpectedly high frequency of variable genes across all 28 genomes. We dubbed this site RM system integration site 1 hot spot (R1H), because it harbors a diverse collection with 12 different RM systems spanning types I, II, and III across strains (Fig. 2B and Fig. 7B). Cooccurring with these RM systems in R1H, we also identified three of the antiphage deity-named defense systems: Hachiman, Gabija, and Kiwa present across seven strains (Fig. 7B). A third RM system integration site (R3) contained two different type II systems, along with an IS66 transposase family of genes (see Fig. S4B), consistent with the known association of defense systems and MGEs (72).
D. pigrum encodes subtype II-A and I-E CRISPR-Cas systems.
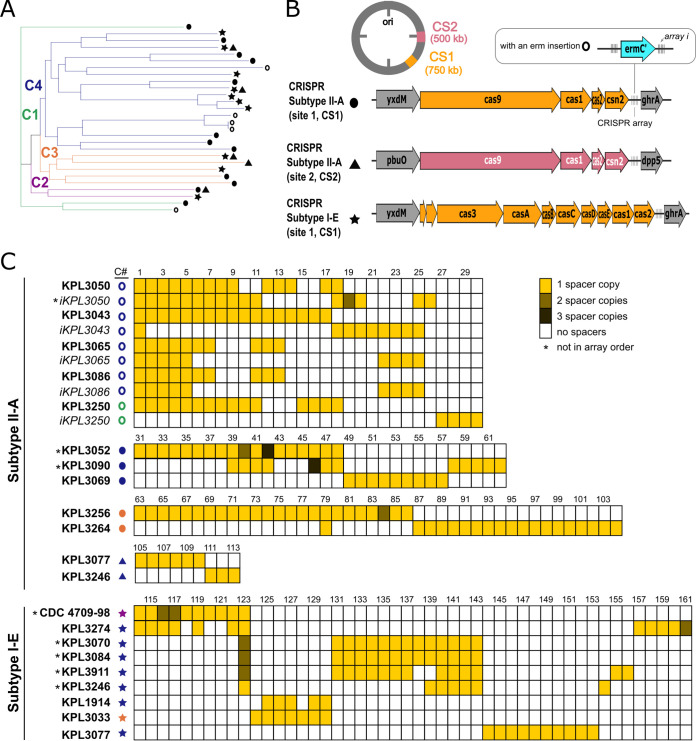
CRISPR-Cas systems provide adaptive/acquired defense (immunity) against MGEs (46). All of the complete D. pigrum genomes encoded at least one subtype II-A or I-E CRISPR-Cas system (Fig. 8A; see also Table S5A), based on the CRISPRDetect database (73). Of the 32 CRISPR-Cas systems detected, 22 are subtype II-A, which is mostly found in Firmicutes (74) and is the predominant CRISPR-Cas system among Lactobacillus spp. (75). Subtypes II-A (circles, Fig. 8A) and I-E (stars, Fig. 8A) CRISPR-Cas systems were generally intermixed within the four major clades, although several distal clades harbored only one type. A single genomic locus (CS1) contained either a subtype II-A or a subtype I-E CRISPR-Cas system in all 19 closed genomes (Fig. 2B and Fig. 8A and B). A second CRISPR-Cas system (triangles, Fig. 8A) was found at a second location (CS2) in 4 of these 19 genomes, from three of the four clades (Fig. 8B).
FIG 8.
D. pigrum encodes subtype I-E and II-A CRISPR-Cas systems with a large but sparsely shared history of MGE invasion. (A) CRISPR-Cas subtype II-A (circles and triangles) and I-E systems (stars) were intermixed among strains in all four clades, with type II-A being most common (see Table S5A). Two distal clades had only a subtype II-A system (KPL3043, KPL3065/KPL3086, KPL3090, KPL3052, and KPL3069) or a subtype I-E system (KPL3070, KPL3084, and KPL391). Three genomes (KPL3077, KPL3246, and CDC2949-98) have both types of system, with each at a different locus. (B) The most common location, CRISPR-Cas system insertion site (CS1), is between the ABC transporter permease protein (yxdM) and the glyxyolytate/hydroxypyruvate reductase A (ghrA) genes. However, subtype II-A systems are also found in between the guanine/hydoxanthine permease (pbuO; NCS2 family permease) and dipeptidyl-peptidase 5 (dpp5; S9 family peptidase) genes at CRISPR-Cas insertion site 2 (CS2). Five of the strains with a subtype II-A system in CS1 had a predicted rRNA adenine N-6-methyltransferase (ermC′) gene integrated in their CRISPR arrays (open circles) (C) Representation of the spacers (see Table S5B and Text S1 in the supplemental material) found among the different CRIPSR systems in the 19 closed genomes. We found 161 unique spacers, less than one-third of which were homologous to phages and plasmids found among other Firmicutes. Strains KPL3050, KPL3250, KPL3065/KP3086, and KPL3043 shared the most spacers among the subtype II-A CRISPR-Cas system, with the distal clade of KPL3043 and KPL3065/KPL3086 sharing 15 spacers. The distal clade with KPL3070, 3084, and 3911 shared the most spacers (12) among the subtype I-E system. CRISPR-Cas systems and spacers hits were determined using the CRISPRdetect and CRISPRtarget database on 16 February 2019, while shared spacers were determined using CRIPSRCompar on 18 March 2019.
CRISPR-Cas systems. Download Table S5, XLSX file, 0.02 MB (24.1KB, xlsx) .
Copyright © 2021 Flores Ramos et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
D. pigrum CRISPR-Cas spacers point to undiscovered D. pigrum MGEs.
Each of the 19 closed genomes included at least one complete CRISPR array. (As expected, most of the arrays were incomplete in the unclosed genomes.) Examining the CRISPR arrays in the 19 closed genomes revealed two key findings. First, the spacer sequences predict the existence of a diversity of undiscovered D. pigrum phages and plasmids with mean numbers of spacers per array of 13 (median, 12.5) for subtype II-A and 11.1 (median, 12) for subtype I-E (see Table S5A). Second, spacer sequences show a sparsely shared history of exposure to many MGEs (Fig. 8C; see also Table S5). Only 60 of the 161 unique identified spacers were shared by more than one strain (Fig. 8C). The exceptions to this limited shared history were two distal clades with shorter branch lengths within Clade 4, which shared 15 and 12 spacers, respectively. Of these 27 spacers, 9 had similarity to known MGEs (see Text S1). A few other shared spacers were scattered among D. pigrum strains outside these two distal clades. For example, D. pigrum KPL3033 (clade 3) and KPL1914 (clade 4) shared five spacers (Fig. 8C), one of which matched to the Clostridium phiCDHM19 phage (LK985322; spacer 129) (76). These shared spacers suggest strains within the host-range of specific MGEs. Spacer similarity to known MGEs indicated prior D. pigrum exposure to phage and plasmid elements that might be related to those found in other genera of Firmicutes, e.g., Clostridium, Lactococcus, Streptococcus, Staphylococcus, and Enterococcus. However, only 46/161 spacers had significant matches (match score >15) to previously identified MGEs and none had matches to any of the predicted prophage from Fig. 5, indicating that that D. pigrum CRISPR-Cas systems likely target a variety of yet-to-be-identified host-specific D. pigrum plasmids and phages.
DISCUSSION
Multiple recent studies of the composition of human nasal microbiota identify D. pigrum as a candidate beneficial bacterium (1–30). Our systematic analysis of 28 D. pigrum strain genomes, including 19 complete and closed genomes, reveals a phylogeny in which strains collected 20 years apart intermingled in clades and showed remarkable stability in genome structure (Fig. 1 and 2). Many of the older D. pigrum strains were collected in the context of human disease (48), making it unclear whether these strains were contributors to disease, bystanders, or contaminants. In a previous analysis, we detected no virulence factors in the genomes of nine of these older strains from Laclaire and Facklam (48), consistent with D. pigrum having a commensal or mutualistic relationship with humans (28). Adding further support for this, we detected no virulence factors in any of our newer strains here (see Text S1), which were all isolated from healthy volunteers. Plus, many of the older strains are closely related in the phylogeny with these recent healthy-donor-derived strains (Fig. 1). These findings are consistent with there being only a few isolated reports of D. pigrum growth in samples from different types of infections (77–82). Of these, the repeated detection of D. pigrum alone in keratitis/keratoconjunctivitis raises the possibility that some strains might be rare causes of eye surface infection (83–86). We recommend future genome sequencing of ophthalmic infection isolates to ascertain whether and how these vary from currently sequenced avirulent strains.
Our results show that strain-level variation in D. pigrum is driven by gene gain/loss in variable regions located between large blocks of syntenic DNA (Fig. 2). This pattern is consistent with the findings of Oliveira et al. for the chromosomal structures of 80 different bacterial species (87). Furthermore, D. pigrum core GCs exhibit very high conservation of nucleotide sequences (≥97.5%), and the 19 closed genomes show the order of syntenic blocks of core genes is conserved (Fig. 2A). D. pigrum has an average genome size of 1.93 Mb (median, 1.91 Mb) (see Table S1) with an open pangenome (Fig. 3B). About 64% of each D. pigrum strain genome consists of core CDS, whereas only about 30% of the D. pigrum pangenome consists of core GCs. This is similar to the percentage of core genes in the pangenomes of other colonizers of the human upper respiratory tract, such as Staphylococcus aureus (36%) and Streptococcus pyogenes (37%) (88).
HGT, much of it likely mediated by MGEs, plays an important role in strain diversification in free-living bacteria. However, a systematic search identified few such elements per genome among D. pigrum strains. In terms of MGEs that commonly mediate HGT, we detected no autonomous plasmids. However, we identified one complete and three partial predicted prophages (Fig. 5) among 27 distinct strain genomes (2 of the 28 genomes were almost identical). To our knowledge, the predicted complete prophage (L1) is the first phage element identified in D. pigrum. The disparate nature of these candidate prophages compared to those in current databases is consistent with D. pigrum having its own specific pool of yet-to-be-identified phage predators, consistent with the strain-level specificity of many known phages. This is further supported by the scarce homology of the phage spacers in the CRISPR arrays to those available in the databases. However, some D. pigrum prophages might share a distant common ancestor with streptococcal phages, as almost one fifth L1’s and at least one third of L2’s (77/202) and L3’s (51/187) predicted genes shared the most similarities to Streptococcus phage genes (see Text S1). Based on our findings, we predict that phage elements targeting D. pigrum have a narrow host range, consistent with patterns exhibited by other Firmicutes-targeting phages, such as those targeting Listeria and Clostridium difficile (58, 76). The identification of D. pigrum prophages creates the opportunity for future work to systemically query nasal metagenomic data sets for these and other D. pigrum MGEs, as well as for CRISPR spacers.
In terms of MGEs that commonly move within genomes, D. pigrum genomes host a group II intron and most also host a small number of predicted transposases and/or integrases (see Table S3). Once present in a genome, IS movement can lead to phenotypic variation among closely related strains through disruption of open reading frames (ORFs) or changes in transcription due to insertion in or adjacent to promoters (65).
The small number of MGEs identified might be related to the multiple defense mechanisms present in each D. pigrum genome. RM systems are ubiquitous in bacteria and present in ∼90% of genomes (71). They play a key role in protecting bacterial genomes from HGT, including MGEs, and maintaining genome stability. The variety of RM systems within and among D. pigrum genomes is consistent with this role. To our knowledge, this is the first report of a strongly inverse relationship between an m5C-targeting type IV REase and an m5C-associated type II system within the same chromosomal locus. A similar relationship was described previously for two antagonistic type II systems in Streptococcus pneumoniae, where strains possess either DpnI (which cleaves only modified Gm6ATC) or DpnII (which cleaves only unmodified GATC) (89). It remains unclear whether the inverse relationship observed between the two D. pigrum systems results from competition for an integration site within a D. pigrum genome (R2; Fig. 7) or whether the type II system’s m5C-modified target motif is incompatible with the type IV REase. Determination of the exact underlying mechanism for this type IV/type II relationship warrants future investigation and has implications for other bacterial genomes.
CRISPR-Cas systems are another common bacterial defense system that maintain genomic stability. In a recent analysis of complete genomes from 4010 bacterial species in NCBI RefSeq, 39% encode cas clusters (74). Several characteristics of the predicted D. pigrum CRISPR-Cas systems suggest these are active. First, the preservation of repeats and spacers along with all of the core Cas gene suggests active systems, since inactive systems often show evidence of degeneration in terms of inconsistent repeat/spacer lengths (75). Second, the diversity of spacers among D. pigrum strains supports the likelihood of activity (90). D. pigrum belongs to the order Lactobacillales in the phylum Firmicutes. Similar to our observations in D. pigrum (Fig. 8), among 171 Lactobacillus species, when multiple CRISPR-Cas systems are present in a single genome these are most often a subtype I-E and subtype II-A, and these two subtypes predominate among type I and II systems in Lactobacillus (75). More broadly, there is a positive association between subtype I-E and subtype II-A systems within the phylum Firmicutes (74). Within Lactobacillus, type I systems contain the longest arrays (average 27 spacers) (75), and we see something similar among the D. pigrum strains. Of the spacers with matches to known plasmid and phage elements in the GenBank-Phage, Refseq-Plasmid, and IMGVR databases in CRISPRTarget, almost half of the identified spacers corresponded to plasmid elements. Subtype II-A systems in Lactobacillus actively transcribe and encode spacers that provide resistance against plasmid uptake based on plasmid interference assays in which an exogenous plasmid is engineered to contain endogenous spacer sequences (75, 91). This defense mechanism might explain the lack of autonomous plasmids in D. pigrum strain genomes to date.
The majority of D. pigrum CRISPR spacers lack homology to known MGEs. This is consistent with a large-scale analysis of bacterial and archaeal genomes in which only 1% to 19% of spacers (global average ∼7%) in genomes match known MGEs, mostly phages and plasmids and uncommonly to self. Also, spacers without a match share basic sequence properties with MGE-matching spacers pointing to species-specific MGEs as the source for CRISPR spacers (92). In this context, our findings indicate D. pigrum strains defend themselves against a wealth of yet-to-be-identified D. pigrum-specific MGEs. Some of these MGEs might be key to developing a system for genetic engineering of D. pigrum.
Like other pangenomic studies, this one has both general and species-specific limitations. First, the open pangenome indicates that the accessory gene space of D. pigrum remains to be more completely assessed through sequencing strains beyond the 28 investigated here. All but 1 of these 28 strains were collected in North America, so a next step is genome sequencing D. pigrum isolates from human volunteers from diverse geographic settings on other continents. Second, many more isolates would need to be collected over time to generate a comprehensive analysis of D. pigrum strain circulation in humans across the United States, and beyond. Third, this is a systematic computational prediction of genome defense systems and MGEs. The next step is experimental verification of the function of these computationally predicted entities, which underscores the need for a system to genetically engineer D. pigrum. Fourth, in this study, we systematically identified known genomic elements that can affect bacterial genomic stability. This leaves a large proportion of D. pigrum’s accessory genome to be explored in future work.
In conclusion, a growing number of studies point to D. pigrum as a candidate beneficial bacterium with the potential for future therapeutic use to manage the composition of human nasal microbiota to prevent disease and promote health (40). One standard for bacterial strains for use in humans, either in foods, the food chain or therapeutics, is the absence of antimicrobial resistance (AMR) genes against clinically useful antibiotics (93). A prior report of 27 D. pigrum strains shows all are susceptible to clinically used antibiotics with the exception of frequent resistance to erythromycin (48). Consistent with this, only 6 of the 17 new D. pigrum genomes reported here encode AMR genes with predicted resistance to erythromycin and/or kanamycin (see Text S1). This confirms the broad antimicrobial susceptibility of D. pigrum. Further supporting its safety, we detected no virulence factors in these 28 genomes. Moreover, this pangenomic analysis of 28 D. pigrum isolates collected over the span of 20 years revealed remarkable stability in both strain circulation and chromosomal structure. Consistent with this stability, we detected relatively few MGEs in each genome; however, each genome hosted a variety of defense systems for protection against MGEs, and HGT in general. The antibiotic susceptibility, genomic stability, capacity for defense against HGT, and lack of known virulence factors described here all support the safety of D. pigrum as a candidate for use in clinical trials to determine its potential for therapeutic use.
MATERIALS AND METHODS
Collection of new D. pigrum strains.
We collected strains of D. pigrum from children and adults using supervised self-sampling of the nostrils with sterile swabs at scientific outreach events in Massachusetts in April 2017 and April 2018 under a protocol approved by the Forsyth Institutional Review Board (FIRB 17-02). All adults provided informed consent. A parent/guardian provided informed consent for children (<18 years old), and all children ≥5 years provided assent. (Self-sampling by children was considered evidence of assent.) Briefly, participants rubbed a sterile rayon swab (BBL, Franklin Lakes, NJ) around the surface of one nasal vestibule (nostril) for 20 s, and then we immediately inoculated this onto BBL Columbia colistin-nalidixic acid agar with 5% sheep’s blood (CNA blood agar). After 48 h of incubation at 37°C in a 5% CO2 enriched atmosphere, each CNA blood agar plate was examined, and colonies with a morphology typical for D. pigrum were selected for purification. Purified isolates were verified to be D. pigrum by 16S rRNA gene colony PCR (GoTaq Green; Promega, Madison, WI) using the primers 27F and 1492R and Sanger sequencing from primer 27F (Macrogen USA, Cambridge, MA).
Genomic DNA extraction.
All D. pigrum strains were cultured from frozen stocks on CNA blood agar plates at 37°C with 5% CO2 for 48 h. For each strain, cells from eight plates were harvested with a sterile cotton swab (Puritan, Guilford, ME) and resuspended in 1 ml of sterile 1× phosphate-buffered saline (PBS; Fisher, Waltham, MA). Then, 10 100-μl resuspensions were spread and grown on 47-mm, 0.22-μm-pore-size polycarbonate membranes (EMD Millipore, Burlington, MA) atop CNA blood agar plates at 37°C with 5% CO2 for 24 h. Three membranes were resuspended in 20 ml of TES buffer (20 mM Tris-HCl, 1 M [pH 8.0]; 50 mM EDTA; filter sterilized) and normalized to an optical density at 600 nm of 1.0 ± 0.02. Half the resuspension was spun down at 5,000 rpm (2,935 × g) for 10 min at 4°C. The genomic DNA was extracted using a Lucigen Masterpure (Epicentre, Middleton, WI) Gram-positive DNA purification kit according to the manufacturer’s instructions with the following modifications: we increased the amount of Ready-Lyse lysozyme added per preparation to 2.5 μl and deleted the bead-beating step. The extracted genomic DNA was assessed for quantity using a Qubit per manufacturer instructions, for quality on a 0.5% agarose gel, and for purity by measuring 260/280 and 260/230 ratios on a NanoDrop spectrophotometer.
Whole-genome sequencing, assembly, and annotation.
Single molecule, real-time sequencing (SMRTseq) was carried out on a PacBio Sequel Instrument (Pacific Biosciences; Menlo Park, CA) with V2.1 chemistry, following standard SMRTbell template preparation protocols for base modification detection. Genomic DNA samples (5 to 10 μg) were sheared to an average size of 20 kbp via G-tube (Covaris, Woburn, MA), end repaired, and ligated to hairpin barcoded adapters prior to sequencing. Sequencing reads were processed using the Pacific Biosciences SMRTlink pipeline (https://smrtflow.readthedocs.io/en/latest/smrtlink_system_high_level_arch.html) according to the HGAP version 4.0 assembly tool standard protocol. Single contigs generated through HGAP were also processed through Circlator version 1.5.5 using default settings to assign the start site of each sequence to dnaA (94). All genomes were annotated with the NCBI’s Prokaryotic Genome Annotation Pipeline (PGAP) (95, 96) and uploaded to the NCBI database (accession numbers CP040408 to CP040424).
Determination of the conservative core genome and the pangenome sizes.
All the genomes were annotated with Prokka version 1.13.0 (97) prior to identification of the conservative core genome with GET_HOMOLOGUES version 3.1.4. (42, 98) using the cluster intersection (compare_clusters.pl; blastp) result of three algorithms: bidirectional best-hits (BDBH), cluster of orthologs (COG) triangles (99), and Markov Cluster Algorithm OrthoMCL (OMCL) (100). The nucleotide level clustering for each of these algorithms was calculated with the get_homologues.pl script and the following parameters: -a CDS, -A, -t 28, -c, -R, and either -G for COG, -M for OMCL, or no flag for BDBH. To obtain the nucleotide instead of the protein outputs, blastn instead of blastp was used to report clusters (parameter –a CDS).
The pangenome was established using the OMCL and COG triangle algorithm with –t 0 parameter to get all possible clusters when running get_homologues.pl. The total clusters from the OMCL and COG pangenomes were then used by compare_clusters.pl with the –m flag to create a pangenome matrix tab file. The cloud, shell, soft core, and core genome of the isolates were then determined using the parse_pangenome_matrix.pl script in GET_HOMOLOGUES using the -s flag and the pangenome matrix tab file. The average nucleotide identity and genome composition analysis were also implemented (using the –A and –c parameters, respectively, in get_homologues.pl). For the genome composition analysis, which shows how many new CDS are added to the pangenome per new genome addition, the conservative default parameters and a random seed (–R) of 1234 was selected.
Phylogenomic tree construction.
A core gene alignment was created for phylogenetic analysis using the nucleotide sequences from the conservative single-copy core GCs (n = 1,102) identified with GET_HOMOLOGUES. These GCs were aligned with MAFFT version 7.245 (101) using default settings, renamed to match the isolate’s strain name, and concatenated into an MSA file through the catfasta2phyml.pl script using the concatenate (–concatenate) and fasta (-f) parameters (copyright 2010-2018 Johan Nylander). The core gene multiple sequence alignment was converted into a phylip file format with Seaview version 4.7 (102). An unrooted phylogenetic tree of the conservative single-copy core (Fig. 1) was generated using this phylip file and IQ-Tree version 1.6.9. (103). The ModelFinder function in IQ-Tree identified the GTR+F+ as the appropriate substitution model for tree construction (BIC value 5597954.8128) (104). Using this model, 553 maximum-likelihood searches with 1,000 ultrafast rapid Bootstraps (105) were used to generate the final maximum likelihood tree (ML = −2854949.911). A clade was defined as a monophyletic group of strains sharing a well-supported ancestral node. SNP pairwise distance in the rooted and unrooted tree were determined using the “harrietr” R package (https://cran.r-project.org/web/packages/harrietr/README.html) applied with an in-house script. Pairwise MASH-distances were calculated for all D. pigrum strains using the implementation of the MASH algorithm (106) in the PanACoTA pipeline (107). Code and data files for this part of the analysis are available online (https://github.com/KLemonLab/DpiMGE_Manuscript/blob/master/SupplementalMethods_PhylogeneticDistances.md).
Synteny analysis.
We performed a whole-genome sequence alignment on all closed genomes using progressive Mauve in Mauve version 2.4.0.r4736 with its default settings (50, 51). For the five genomes that we were unable to circularize, we manually fixed the start site to dnaA and added NNNNNNNNN to the region concatenating the ends of the contigs to mark it as a region of uncertainty in the synteny alignment. Manual curating was done with SnapGene version 4.2.11 GUI platform (SnapGene software from GSL Biotech).
Functional analysis of the pangenome using Anvi’o.
All genomes were reannotated with an updated Prokka version (1.14.6) (97) with default parameters, including gene recognition and translation initiation site identification with Prodigal (108). The pangenome was analyzed using Anvi’o version 7 (52, 53). We followed the pangenome workflow to import Prokka annotated genomes into Anvi’o (http://merenlab.org/2017/05/18/working-with-prokka/), followed by the addition of functional COG annotations using the anvi-run-ncbi-cogs program with the –sensitive flag (runs sensitive version of DIAMOND [109]) and the 2020 updated COG20 database (110, 111). KEGG/KOfam (112, 113), and Pfam (114) annotations were also added to each genome .db file, as well as hmm-hits (115). The pangenome was calculated with the anvi-pan-genome program (flags: –minbit 0.5, –mcl-inflation 10, and –use-ncbi-blast) using blastp search (116), muscle alignment (117), “minbit heuristic” (118) to filter weak hits, and the MCL algorithm (119). The functional and geometric homogeneity index, and the rest of the information shown in Fig. S3D were calculated following the standard Anvi’o pangenomic pipeline (http://merenlab.org/2016/11/08/pangenomics-v2). The core (n = 28), soft core (28 > n ≥ 26), shell (26 > n ≥ 3), and cloud (n ≤ 2) annotations from GET_HOMOLOGUES were added to the Anvi’o pangenomic database using the interactive interface. We defined the accessory as GCs present in ≤25 genomes and core as GCs present in ≥26 genomes. The output of this Anvi’o pangenomic analysis and the code used to generate it are available online (https://github.com/KLemonLab/DpiMGE_Manuscript/blob/master/SupplementalMethods_Anvio.md). We used the summary file we exported from the Anvi’o pangenomic analysis to generate the functional enrichment plots shown in Fig. 4 and in Fig. S3E and F using an in-house R script (https://github.com/KLemonLab/DpiMGE_Manuscript/blob/master/SupplementalMethods_COGs.md) to wrangle and extract information on the informative COG20 annotated gene clusters (120, 121).
PPanGGOLiN analysis.
Gene clustering and annotation data were exported from the Anvi’o output and imported into PPanGGOLiN version 1.1.141 (Partitioned PanGenome Graph Of Linked Neighbors) (68) to create a partitioned pangenome graph (PPG) that assigned GCs to the “persistent,” “shell,” and “cloud” partitions. Regions of genome plasticity (RGPs) and spots of insertion were predicted (122), and subgraphs of the hot spots of interest were generated by providing the sequence of the flanking proteins in a fasta file. The output of the PPanGGOLiN analysis and the code used to generate it are available online (https://github.com/KLemonLab/DpiMGE_Manuscript/blob/master/SupplementalMethods_PPanGGOLiN.md). The subgraphs represented as inserts on Fig. 2B were obtained with the command “ppanggolin align -p pangenome.h5 –getinfo –draw_related –proteins” using the amino acid sequences for the proteins upstream and downstream of each spot of interest. Since PPanGGOLiN does not currently allow creation of subgraphs using GCs imported from external clustering methods, the pangenome was run again using the default PPanGGOLiN workflow with MMseqs2 clustering (default settings: –identity 0.8, –coverage 0.8, and –defrag).
Characterization of MGEs.
We searched all genomes for phage elements using the PHASTER database and web server (http://phaster.ca) on 8 November 2018 (56, 57). We took the “intact” phage elements as defined by a phage score of >90 and queried their ORFs using blastp to manually reannotate their phage genes in the SnapGene GUI.
We searched for plasmid elements in all genomes using the PlasmidFinder 2.0 database and GUI interface (https://cge.cbs.dtu.dk/services/PlasmidFinder/) on 13 November 2018 using the default parameters (62). For strains with hits for a plasmid element, ORFs 1,000 kb upstream and downstream of the element were queried through blastp. Manual gene reannotation was performed using the SnapGene GUI platform.
The summary file exported from the Anvi’o pangenomic analysis (see above) was also used for the identification of MGEs on the Prokka, COG20, Pfam, and KOfam annotations. We identified 23 GCs as coding for putative transposases. GC alignments were visually inspected in AliView (123), and full-length representative sequences were selected for Pfam search at the Pfam batch sequence search/HMMER website (114, 124). We identified eight GCs with complete (≥80% coverage) Pfam Transposase (tnp) domains as true predicted transposases and five GCs with complete (≥80% coverage) Pfam rve domains as integrases. We used Operon ConTextulization Across Prokaryotes to Uncover Synteny (OCTAPUS; https://github.com/FredHutch/octapus) to identify the gene neighborhoods in which the selected transposases and integrases were located across all 28 D. pigrum genomes (see Table S3). The approach used by OCTAPUS is to search for a set of defined query genes across a collection of reference genomes by translated amino acid alignment and then to summarize the results by their physical colocation and organization. In this way, operon structure can be identified as the consistent colocation of a set of genes across multiple genomes in the same relative orientation (including both position and strand). The groups of genes identified with OCTAPUS at minimum percent identity 85% and minimum coverage 80% were visualized using clinker (https://github.com/gamcil/clinker) (125), and summary data provided in (see Table S3) were calculated using the matrixStats package (https://github.com/HenrikBengtsson/matrixStats). Detailed methods for this part of the analysis, as well as relevant files, are available online (https://github.com/KLemonLab/DpiMGE_Manuscript/blob/master/SupplementalMethods_MGEs.md).
We similarly used OCTAPUS to identify the gene neighborhood of the group II intron identified with Anvi’o and PPanGGOLiN (GC_00000001). Using Pfam, we confirmed two predicted domains in a sequence from D. pigrum KPL3250 in GC_00000001: a reverse transcriptase and a maturase. The best hit in a blastx search with this same sequence against the Bacterial Group II Intron Database was to the bacterial class C intron-encoded protein from La.re.I1 in Lactobacillus reuteri with 44% identity and 65% similarity over 419 amino acids (70).
Base modification analysis and prediction of restriction-modification systems.
For methylome analysis, interpulse durations were measured and processed for all pulses aligned to each position in the reference sequence. We used Pacific Biosciences’ SMRTanalysis v8, which uses an in silico kinetic reference and a t-te st-based kinetic score detection of modified base positions, to identify modified positions (126).
We identified RM systems using SMRTseq data, as previously described (127), using the SEQWARE computer resource, a BLAST-based software module in combination with the curated restriction enzyme database (REBASE; http://rebase.neb.com/rebase/rebase.html) (71). Prediction was supported by sequence similarity, presence, and order of predictive functional motifs, plus the known genomic context and characteristics of empirically characterized RM system genes within REBASE. This facilitated reliable assignment of candidate methyltransferase genes to each modified motif based on their RM type.
Detection of 5-methylcytosine.
For D. pigrum CDC4709-98 (aka KPL1934), the presence of 5-methylcytosine in the predicted methylation motif GCNGC was assessed as previously described (127). Briefly, gDNA harvested with a Masterpure Complete DNA/RNA purification kit was bisulfite treated using an EpiMark bisulfite conversion kit (NEB, Ipswich, MA)—both according to manufacturer’s instructions, except for a final elution volume of 20 μl in the EpiMark kit. We then selected two genomic regions: each ≤700 bp containing ≥4 GCNGC motifs. We PCR amplified each region from 1 μl of the converted gDNA using TaKaRa EpiTaq HS for bisulfite-treated DNA (TaKaRa Bio USA, Mountain View, CA) according to the manufacturer’s instructions with the primers designed by MethPrimer: oKL732 (5′-AAGTTTATTTTTTTGAGTTTGTTG-3′), oKL733 (5′-TACCCATAAAATTATCACCTTC-3′), oKL734 (5′-ATTGATTTAGTAATTTTTTTGGAATAT-3′), and oKL735 (5′-TAAATAACTCTACAAAAAACTCAACTTACC-3′). After amplicon purification with a QIAquick PCR purification kit (final elution, 40 μl; Qiagen; Germantown, MD), we used Sanger sequencing (Macrogen, USA) of each PCR product to detect cytosine methylation within the predicted motif. Additional m5C-based modified motif analysis was carried out for Dolosigranulum pigrum KPL3250 using MFRE-Seq, as previously described (128).
Prediction of CRISPR-Cas systems.
CRISPR cas genes were detected using the CRISPRFinder (https://crispr.i2bc.paris-saclay.fr/Server/) (129), and the array elements downstream from these genes were found using CRISPRDetect software (http://crispr.otago.ac.nz/CRISPRDetect/predict_crispr_array.html) (73). The spacers identified using CRISPRDetect were queried through databases of possible phage targets in the GenBank-Phage, Refseq-Plasmid, and IMGVR databases with CRISPRtarget (http://crispr.otago.ac.nz/CRISPRTarget/crispr_analysis.html) (73, 130), keeping hits with a cutoff score greater than 14. These spacers were also queried against the predicted L1 to L4 prophages through CRISPRtarget using a cutoff score >0. All gene and array element searches were completed on the webserver on 16 February 2019—with the exception of the spacers’ query through the L1 to L4 prophages on 27 July 2021—using the default parameters. We also queried the genomes through CRISPRdb and CRISPRCompar (https://crispr.i2bc.paris-saclay.fr) website on 18 March 2019 to identify and annotate spacers shared among the different strains, keeping hits with scores higher than 15 to indicate similarity (129, 131, 132).
Data availability.
All genomes are available from the NCBI. Table 1 lists the accession numbers for each D. pigrum strain genome used in this study.
ACKNOWLEDGMENTS
We are deeply grateful to the participants who donated nostril swab samples at a 2017 and 2018 science festival. Their contribution was critical to expanding our knowledge of D. pigrum. We thank colleagues and lab members who provided invaluable assistance at both outreach events, in particular Javier Fernandez Juarez, Kerry Maguire, Pallavi Murugkar, Pooja Balani, Sowmya Balasubramanian, Fan Zhu, Andy Kempczynski, Andrew Collins, Brian Klein, and Megan Lambert. For critical logistical support, we are grateful to Genevieve Holmes. We also thank Melinda M. Pettigrew and Yong Kong for advice on genome analysis, as well as Tsute (George) Chen, Daniel Utter, Edoardo Pasolli, Nicola Segata, and Michael Wollenberg for their computational and phylogenetic advice over the course of the project. We thank members of the Johnston Lab, the KLemon Lab, and the Starr-Johnston-Dewhirst-Lemon joint lab meeting for critique and suggestions.
Author contributions: Conceptualization: K.P.L., S.D.B., and C.D.J. Methodology: S.F.R., S.D.B., I.F.E., C.D.J., C.A.S., S.L.C., S.M.E., T.H.T., D.S.J., S.M., and R.J.R. Strain isolation: S.M.E., S.F.R., W.G., S.D.B., L.B., and I.F.E. Investigation: S.F.R., C.A.S., S.L.C., S.D.B., S.M.E., S.M., C.D.J., and I.F.E. Interpretation of data: S.F.R., K.P.L., C.D.J., I.F.E., S.D.B., C.A.S., S.L.C., S.M.E., D.S.J., S.M., and R.J.R. Visualization: S.F.R., C.D.J., and I.F.E. Wrote original draft: S.F.R., K.P.L., C.D.J., and I.F.E. Editing and review: S.F.R., I.F.E., C.D.J., K.P.L., S.D.B., R.J.R., S.M.E., T.H.T., C.A.S., and S.M. All authors read and approved the final manuscript. Supervision: K.P.L. and C.D.J. Funding acquisition: C.D.J. and K.P.L.
This study was supported by the National Institutes of Health through the National Institute of General Medical Sciences (grant R01 GM117174 to K.P.L.) and through the National Institute of Dental and Craniofacial Research (grant R01DE027850 to C.D.J.) and by the Forsyth Institute through a Stimulus Pilot Grant (to K.P.L. and C.D.J.). Addition funding was from the Swiss National Science Foundation and Swiss Foundation for Grants in Biology and Medicine (grant P3SMP3_155315 to S.D.B.); by the Novartis Foundation for Medical-Biological Research (grant 16B065 to S.D.B.); and by the Promedica Foundation (grant 1449/M to S.D.B.). R.J.R. works for New England Biolabs, a company that sells research reagents, including restriction enzymes and DNA methyltransferases, to the scientific community. Funders had no role in the preparation of the manuscript or decision to publish.
Contributor Information
Christopher D. Johnston, Email: johnston@fredhutch.org.
Katherine P. Lemon, Email: katherine.lemon@bcm.edu.
Nicola Segata, University of Trento.
REFERENCES
- 1.Laufer AS, Metlay JP, Gent JF, Fennie KP, Kong Y, Pettigrew MM. 2011. Microbial communities of the upper respiratory tract and otitis media in children. mBio 2:e00245-10. doi: 10.1128/mBio.00245-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pettigrew MM, Laufer AS, Gent JF, Kong Y, Fennie KP, Metlay JP. 2012. Upper respiratory tract microbial communities, acute otitis media pathogens, and antibiotic use in healthy and sick children. Appl Environ Microbiol 78:6262–6270. doi: 10.1128/AEM.01051-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Biesbroek G, Bosch AA, Wang X, Keijser BJ, Veenhoven RH, Sanders EA, Bogaert D. 2014. The impact of breastfeeding on nasopharyngeal microbial communities in infants. Am J Respir Crit Care Med 190:140612135546007–140612135546308. doi: 10.1164/rccm.201401-0073OC. [DOI] [PubMed] [Google Scholar]
- 4.Biesbroek G, Tsivtsivadze E, Sanders EA, Montijn R, Veenhoven RH, Keijser BJ, Bogaert D. 2014. Early respiratory microbiota composition determines bacterial succession patterns and respiratory health in children. Am J Respir Crit Care Med 190:1283–1292. doi: 10.1164/rccm.201407-1240OC. [DOI] [PubMed] [Google Scholar]
- 5.Liu CM, Price LB, Hungate BA, Abraham AG, Larsen LA, Christensen K, Stegger M, Skov R, Andersen PS. 2015. Staphylococcus aureus and the ecology of the nasal microbiome. Sci Adv 1:e1400216. doi: 10.1126/sciadv.1400216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Teo SM, Mok D, Pham K, Kusel M, Serralha M, Troy N, Holt BJ, Hales BJ, Walker ML, Hollams E, Bochkov YA, Grindle K, Johnston SL, Gern JE, Sly PD, Holt PG, Holt KE, Inouye M. 2015. The infant nasopharyngeal microbiome impacts severity of lower respiratory infection and risk of asthma development. Cell Host Microbe 17:704–715. doi: 10.1016/j.chom.2015.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bosch A, Levin E, van Houten MA, Hasrat R, Kalkman G, Biesbroek G, de Steenhuijsen Piters WAA, de Groot PCM, Pernet P, Keijser BJF, Sanders EAM, Bogaert D. 2016. Development of upper respiratory tract microbiota in infancy is affected by mode of delivery. EBioMedicine 9:336–345. doi: 10.1016/j.ebiom.2016.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bomar L, Brugger SD, Yost BH, Davies SS, Lemon KP. 2016. Corynebacterium accolens releases antipneumococcal free fatty acids from human nostril and skin surface triacylglycerols. mBio 7:e01725-15–e01715. doi: 10.1128/mBio.01725-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang M, Wang R, Liao Y, Buijs MJ, Li J. 2016. Profiling of oral and nasal microbiome in children with cleft palate. Cleft Palate Craniofac J 53:332–338. doi: 10.1597/14-162. [DOI] [PubMed] [Google Scholar]
- 10.Salter SJ, Turner C, Watthanaworawit W, de Goffau MC, Wagner J, Parkhill J, Bentley SD, Goldblatt D, Nosten F, Turner P. 2017. A longitudinal study of the infant nasopharyngeal microbiota: the effects of age, illness and antibiotic use in a cohort of South East Asian children. PLoS Negl Trop Dis 11:e0005975. doi: 10.1371/journal.pntd.0005975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bosch A, de Steenhuijsen Piters WAA, van Houten MA, Chu M, Biesbroek G, Kool J, Pernet P, de Groot PCM, Eijkemans MJC, Keijser BJF, Sanders EAM, Bogaert D. 2017. Maturation of the infant respiratory microbiota, environmental drivers, and health consequences. a prospective cohort study. Am J Respir Crit Care Med 196:1582–1590. doi: 10.1164/rccm.201703-0554OC. [DOI] [PubMed] [Google Scholar]
- 12.Kelly MS, Surette MG, Smieja M, Pernica JM, Rossi L, Luinstra K, Steenhoff AP, Feemster KA, Goldfarb DM, Arscott-Mills T, Boiditswe S, Rulaganyang I, Muthoga C, Gaofiwe L, Mazhani T, Rawls JF, Cunningham CK, Shah SS, Seed PC. 2017. The nasopharyngeal microbiota of children with respiratory infections in Botswana. Pediatr Infect Dis J 36:e211–e218. doi: 10.1097/INF.0000000000001607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hasegawa K, Linnemann RW, Mansbach JM, Ajami NJ, Espinola JA, Petrosino JF, Piedra PA, Stevenson MD, Sullivan AF, Thompson AD, Camargo CA, Jr.. 2017. Nasal airway microbiota profile and severe bronchiolitis in infants: a case-control study. Pediatr Infect Dis J 36:1044–1051. doi: 10.1097/INF.0000000000001500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Langevin S, Pichon M, Smith E, Morrison J, Bent Z, Green R, Barker K, Solberg O, Gillet Y, Javouhey E, Lina B, Katze MG, Josset L. 2017. Early nasopharyngeal microbial signature associated with severe influenza in children: a retrospective pilot study. J Gen Virol 98:2425–2437. doi: 10.1099/jgv.0.000920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lappan R, Imbrogno K, Sikazwe C, Anderson D, Mok D, Coates H, Vijayasekaran S, Bumbak P, Blyth CC, Jamieson SE, Peacock CS. 2018. A microbiome case-control study of recurrent acute otitis media identified potentially protective bacterial genera. BMC Microbiol 18:13. doi: 10.1186/s12866-018-1154-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Escapa IF, Chen T, Huang Y, Gajare P, Dewhirst FE, Lemon KP. 2018. New insights into human nostril microbiome from the Expanded Human Oral Microbiome Database (eHOMD): a resource for the microbiome of the human aerodigestive tract. mSystems 3:e00187-18. doi: 10.1128/mSystems.00187-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wen Z, Xie G, Zhou Q, Qiu C, Li J, Hu Q, Dai W, Li D, Zheng Y, Wen F. 2018. Distinct nasopharyngeal and oropharyngeal microbiota of children with influenza A virus compared with healthy children. Biomed Res Int 2018:6362716. doi: 10.1155/2018/6362716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Copeland E, Leonard K, Carney R, Kong J, Forer M, Naidoo Y, Oliver BGG, Seymour JR, Woodcock S, Burke CM, Stow NW. 2018. Chronic rhinosinusitis: potential role of microbial dysbiosis and recommendations for sampling sites. Front Cell Infect Microbiol 8:57. doi: 10.3389/fcimb.2018.00057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Toivonen L, Hasegawa K, Waris M, Ajami NJ, Petrosino JF, Camargo CA, Jr, Peltola V. 2019. Early nasal microbiota and acute respiratory infections during the first years of life. Thorax 74:592–599. doi: 10.1136/thoraxjnl-2018-212629. [DOI] [PubMed] [Google Scholar]
- 20.Camelo-Castillo A, Henares D, Brotons P, Galiana A, Rodriguez JC, Mira A, Munoz-Almagro C. 2019. Nasopharyngeal microbiota in children with invasive pneumococcal disease: identification of bacteria with potential disease-promoting and protective effects. Front Microbiol 10:11. doi: 10.3389/fmicb.2019.00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Man WH, Clerc M, de Steenhuijsen Piters WAA, van Houten MA, Chu M, Kool J, Keijser BJF, Sanders EAM, Bogaert D. 2019. Loss of microbial topography between oral and nasopharyngeal microbiota and development of respiratory infections early in life. Am J Respir Crit Care Med 15:PA4995–770. doi: 10.1183/13993003.congress-2019.PA4995. [DOI] [PubMed] [Google Scholar]
- 22.Man WH, van Houten MA, Merelle ME, Vlieger AM, Chu M, Jansen NJG, Sanders EAM, Bogaert D. 2019. Bacterial and viral respiratory tract microbiota and host characteristics in children with lower respiratory tract infections: a matched case-control study. Lancet Respir Med 7:417–426. doi: 10.1016/S2213-2600(18)30449-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Man WH, van Dongen TMA, Venekamp RP, Pluimakers VG, Chu M, van Houten MA, Sanders EAM, Schilder AGM, Bogaert D. 2019. Respiratory microbiota predicts clinical disease course of acute otorrhea in children with tympanostomy tubes. Pediatr Infect Dis J 38:e116–e125. doi: 10.1097/INF.0000000000002215. [DOI] [PubMed] [Google Scholar]
- 24.Gan W, Yang F, Tang Y, Zhou D, Qing D, Hu J, Liu S, Liu F, Meng J. 2019. The difference in nasal bacterial microbiome diversity between chronic rhinosinusitis patients with polyps and a control population. Int Forum Allergy Rhinol 9:582–592. doi: 10.1002/alr.22297. [DOI] [PubMed] [Google Scholar]
- 25.de Steenhuijsen Piters WAA, Jochems SP, Mitsi E, Rylance J, Pojar S, Nikolaou E, German EL, Holloway M, Carniel BF, Chu M, Arp K, Sanders EAM, Ferreira DM, Bogaert D. 2019. Interaction between the nasal microbiota and Streptococcus pneumoniae in the context of live-attenuated influenza vaccine. Nat Commun 10:2981. doi: 10.1038/s41467-019-10814-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.De Boeck I, Wittouck S, Martens K, Claes J, Jorissen M, Steelant B, van den Broek MFL, Seys SF, Hellings PW, Vanderveken OM, Lebeer S. 2019. Anterior nares diversity and pathobionts represent sinus microbiome in chronic rhinosinusitis. mSphere 4:e00532-19. doi: 10.1128/mSphere.00532-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Man WH, Scheltema NM, Clerc M, van Houten MA, Nibbelke EE, Achten NB, Arp K, Sanders EAM, Bont LJ, Bogaert D. 2020. Infant respiratory syncytial virus prophylaxis and nasopharyngeal microbiota until 6 years of life: a subanalysis of the MAKI randomised controlled trial. Lancet Respir Med 8:1022–1031. doi: 10.1016/S2213-2600(19)30470-9. [DOI] [PubMed] [Google Scholar]
- 28.Brugger SD, Eslami SM, Pettigrew MM, Escapa IF, Henke MT, Kong Y, Lemon KP. 2020. Dolosigranulum pigrum cooperation and competition in human nasal microbiota. mSphere 5:e00852-20. doi: 10.1128/mSphere.00852-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ortiz Moyano R, Raya Tonetti F, Tomokiyo M, Kanmani P, Vizoso-Pinto MG, Kim H, Quilodran-Vega S, Melnikov V, Alvarez S, Takahashi H, Kurata S, Kitazawa H, Villena J. 2020. The ability of respiratory commensal bacteria to beneficially modulate the lung innate immune response is a strain dependent characteristic. Microorganisms 8:727. doi: 10.3390/microorganisms8050727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Coleman A, Bialasiewicz S, Marsh RL, Grahn Hakansson E, Cottrell K, Wood A, Jayasundara N, Ware RS, Zaugg J, Sidjabat HE, Adams J, Ferguson J, Brown M, Roos K, Cervin A. 2021. Upper respiratory microbiota in relation to ear and nose health among Australian Aboriginal and Torres Strait Islander children. J Pediatric Infect Dis Soc 10:468–476. doi: 10.1093/jpids/piaa141. [DOI] [PubMed] [Google Scholar]
- 31.Brugger SD, Bomar L, Lemon KP. 2016. Commensal-pathogen interactions along the human nasal passages. PLoS Pathog 12:e1005633. doi: 10.1371/journal.ppat.1005633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Krismer B, Weidenmaier C, Zipperer A, Peschel A. 2017. The commensal lifestyle of Staphylococcus aureus and its interactions with the nasal microbiota. Nat Rev Microbiol 15:675–687. doi: 10.1038/nrmicro.2017.104. [DOI] [PubMed] [Google Scholar]
- 33.Man WH, de Steenhuijsen Piters WA, Bogaert D. 2017. The microbiota of the respiratory tract: gatekeeper to respiratory health. Nat Rev Microbiol 15:259–270. doi: 10.1038/nrmicro.2017.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bomar L, Brugger SD, Lemon KP. 2018. Bacterial microbiota of the nasal passages across the span of human life. Curr Opin Microbiol 41:8–14. doi: 10.1016/j.mib.2017.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Esposito S, Principi N. 2018. Impact of nasopharyngeal microbiota on the development of respiratory tract diseases. Eur J Clin Microbiol Infect Dis 37:1–7. doi: 10.1007/s10096-017-3076-7. [DOI] [PubMed] [Google Scholar]
- 36.Mittal R, Sanchez-Luege SV, Wagner SM, Yan D, Liu XZ. 2019. Recent perspectives on gene-microbe interactions determining predisposition to otitis media. Front Genet 10:1230. doi: 10.3389/fgene.2019.01230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yan M, Pamp SJ, Fukuyama J, Hwang PH, Cho DY, Holmes S, Relman DA. 2013. Nasal microenvironments and interspecific interactions influence nasal microbiota complexity and S. aureus carriage. Cell Host Microbe 14:631–640. doi: 10.1016/j.chom.2013.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Accorsi EK, Franzosa EA, Hsu T, Joice Cordy R, Maayan-Metzger A, Jaber H, Reiss-Mandel A, Kline M, DuLong C, Lipsitch M, Regev-Yochay G, Huttenhower C. 2020. Determinants of Staphylococcus aureus carriage in the developing infant nasal microbiome. Genome Biol 21:301. doi: 10.1186/s13059-020-02209-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Khamash DF, Mongodin EF, White JR, Voskertchian A, Hittle L, Colantuoni E, Milstone AM. 2019. The association between the developing nasal microbiota of hospitalized neonates and Staphylococcus aureus colonization. Open Forum Infect Dis 6:ofz062. doi: 10.1093/ofid/ofz062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.De Boeck I, Spacova I, Vanderveken OM, Lebeer S. 2021. Lactic acid bacteria as probiotics for the nose? Microb Biotechnol 14:859–869. doi: 10.1111/1751-7915.13759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tettelin H, Masignani V, Cieslewicz MJ, Donati C, Medini D, Ward NL, Angiuoli SV, Crabtree J, Jones AL, Durkin AS, Deboy RT, Davidsen TM, Mora M, Scarselli M, Margarit y Ros I, Peterson JD, Hauser CR, Sundaram JP, Nelson WC, Madupu R, Brinkac LM, Dodson RJ, Rosovitz MJ, Sullivan SA, Daugherty SC, Haft DH, Selengut J, Gwinn ML, Zhou L, Zafar N, Khouri H, Radune D, Dimitrov G, Watkins K, O’Connor KJ, Smith S, Utterback TR, White O, Rubens CE, Grandi G, Madoff LC, Kasper DL, Telford JL, Wessels MR, Rappuoli R, Fraser CM. 2005. Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: implications for the microbial “pan-genome.” Proc Natl Acad Sci USA 102:13950–13955. doi: 10.1073/pnas.0506758102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Contreras-Moreira B, Vinuesa P. 2013. GET_HOMOLOGUES, a versatile software package for scalable and robust microbial pangenome analysis. Appl Environ Microbiol 79:7696–7701. doi: 10.1128/AEM.02411-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Doron S, Melamed S, Ofir G, Leavitt A, Lopatina A, Keren M, Amitai G, Sorek R. 2018. Systematic discovery of antiphage defense systems in the microbial pangenome. Science 359:eaar4120. doi: 10.1126/science.aar4120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Goldfarb T, Sberro H, Weinstock E, Cohen O, Doron S, Charpak-Amikam Y, Afik S, Ofir G, Sorek R. 2015. BREX is a novel phage resistance system widespread in microbial genomes. EMBO J 34:169–183. doi: 10.15252/embj.201489455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang L, Jiang S, Deng Z, Dedon PC, Chen S. 2019. DNA phosphorothioate modification-a new multi-functional epigenetic system in bacteria. FEMS Microbiol Rev 43:109–122. doi: 10.1093/femsre/fuy036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Horvath P, Barrangou R. 2010. CRISPR/Cas, the immune system of bacteria and archaea. Science 327:167–170. doi: 10.1126/science.1179555. [DOI] [PubMed] [Google Scholar]
- 47.Makarova KS, Wolf YI, Snir S, Koonin EV. 2011. Defense islands in bacterial and archaeal genomes and prediction of novel defense systems. J Bacteriol 193:6039–6056. doi: 10.1128/JB.05535-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Laclaire L, Facklam R. 2000. Antimicrobial susceptibility and clinical sources of Dolosigranulum pigrum cultures. Antimicrob Agents Chemother 44:2001–2003. doi: 10.1128/AAC.44.7.2001-2003.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Aguirre M, Collins MD. 1992. Phylogenetic analysis of Alloiococcus otitis gen. nov., sp. nov., an organism from human middle ear fluid. Int J Syst Bacteriol 42:79–83. doi: 10.1099/00207713-42-1-79. [DOI] [PubMed] [Google Scholar]
- 50.Darling AC, Mau B, Blattner FR, Perna NT. 2004. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res 14:1394–1403. doi: 10.1101/gr.2289704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Darling AE, Mau B, Perna NT. 2010. progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS One 5:e11147. doi: 10.1371/journal.pone.0011147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Eren AM, Esen OC, Quince C, Vineis JH, Morrison HG, Sogin ML, Delmont TO. 2015. Anvi’o: an advanced analysis and visualization platform for ‘omics data. PeerJ 3:e1319. doi: 10.7717/peerj.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Delmont TO, Eren AM. 2018. Linking pangenomes and metagenomes: the Prochlorococcus metapangenome. PeerJ 6:e4320. doi: 10.7717/peerj.4320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tatusov RL, Fedorova ND, Jackson JD, Jacobs AR, Kiryutin B, Koonin EV, Krylov DM, Mazumder R, Mekhedov SL, Nikolskaya AN, Rao BS, Smirnov S, Sverdlov AV, Vasudevan S, Wolf YI, Yin JJ, Natale DA. 2003. The COG database: an updated version includes eukaryotes. BMC Bioinformatics 4:41. doi: 10.1186/1471-2105-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Galperin MY, Makarova KS, Wolf YI, Koonin EV. 2015. Expanded microbial genome coverage and improved protein family annotation in the COG database. Nucleic Acids Res 43:D261–D269. doi: 10.1093/nar/gku1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhou Y, Liang Y, Lynch KH, Dennis JJ, Wishart DS. 2011. PHAST: a fast phage search tool. Nucleic Acids Res 39:W347–W352. doi: 10.1093/nar/gkr485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Arndt D, Grant JR, Marcu A, Sajed T, Pon A, Liang Y, Wishart DS. 2016. PHASTER: a better, faster version of the PHAST phage search tool. Nucleic Acids Res 44:W16–W21. doi: 10.1093/nar/gkw387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dorscht J, Klumpp J, Bielmann R, Schmelcher M, Born Y, Zimmer M, Calendar R, Loessner MJ. 2009. Comparative genome analysis of Listeria bacteriophages reveals extensive mosaicism, programmed translational frameshifting, and a novel prophage insertion site. J Bacteriol 191:7206–7215. doi: 10.1128/JB.01041-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zimmer M, Sattelberger E, Inman RB, Calendar R, Loessner MJ. 2003. Genome and proteome of Listeria monocytogenes phage PSA: an unusual case for programmed +1 translational frameshifting in structural protein synthesis. Mol Microbiol 50:303–317. doi: 10.1046/j.1365-2958.2003.03684.x. [DOI] [PubMed] [Google Scholar]
- 60.van Sinderen D, Karsens H, Kok J, Terpstra P, Ruiters MHJ, Venema G, Nauta A. 1996. Sequence analysis and molecular characterization of the temperate lactococcal bacteriophage r1t. Mol Microbiol 19:1343–1355. doi: 10.1111/j.1365-2958.1996.tb02478.x. [DOI] [PubMed] [Google Scholar]
- 61.Beres SB, Sylva GL, Barbian KD, Lei B, Hoff JS, Mammarella ND, Liu M-Y, Smoot JC, Porcella SF, Parkins LD, Campbell DS, Smith TM, McCormick JK, Leung DYM, Schlievert PM, Musser JM. 2002. Genome sequence of a serotype M3 strain of group A Streptococcus: phage-encoded toxins, the high-virulence phenotype, and clone emergence. Proc Natl Acad Sci USA 99:10078–10083. doi: 10.1073/pnas.152298499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Carattoli A, Zankari E, García-Fernández A, Voldby Larsen M, Lund O, Villa L, Møller Aarestrup F, Hasman H. 2014. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother 58:3895–3903. doi: 10.1128/AAC.02412-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.McArthur AG, Waglechner N, Nizam F, Yan A, Azad MA, Baylay AJ, Bhullar K, Canova MJ, De Pascale G, Ejim L, Kalan L, King AM, Koteva K, Morar M, Mulvey MR, O’Brien JS, Pawlowski AC, Piddock LJV, Spanogiannopoulos P, Sutherland AD, Tang I, Taylor PL, Thaker M, Wang W, Yan M, Yu T, Wright GD. 2013. The Comprehensive Antibiotic Resistance Database. Antimicrob Agents Chemother 57:3348–3357. doi: 10.1128/AAC.00419-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jia B, Raphenya AR, Alcock B, Waglechner N, Guo P, Tsang KK, Lago BA, Dave BM, Pereira S, Sharma AN, Doshi S, Courtot M, Lo R, Williams LE, Frye JG, Elsayegh T, Sardar D, Westman EL, Pawlowski AC, Johnson TA, Brinkman FSL, Wright GD, McArthur AG. 2017. CARD 2017: expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res 45:D566–D573. doi: 10.1093/nar/gkw1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Siguier P, Gourbeyre E, Chandler M. 2014. Bacterial insertion sequences: their genomic impact and diversity. FEMS Microbiol Rev 38:865–891. doi: 10.1111/1574-6976.12067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Siguier P, Gourbeyre E, Varani A, Ton-Hoang B, Chandler M. 2015. Everyman’s guide to bacterial insertion sequences. Microbiol Spectr 3:MDNA3. doi: 10.1128/microbiolspec.MDNA3-0030-2014. [DOI] [PubMed] [Google Scholar]
- 67.Aziz RK, Breitbart M, Edwards RA. 2010. Transposases are the most abundant, most ubiquitous genes in nature. Nucleic Acids Res 38:4207–4217. doi: 10.1093/nar/gkq140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gautreau G, Bazin A, Gachet M, Planel R, Burlot L, Dubois M, Perrin A, Medigue C, Calteau A, Cruveiller S, Matias C, Ambroise C, Rocha EPC, Vallenet D. 2020. PPanGGOLiN: depicting microbial diversity via a partitioned pangenome graph. PLoS Comput Biol 16:e1007732. doi: 10.1371/journal.pcbi.1007732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.McNeil BA, Semper C, Zimmerly S. 2016. Group II introns: versatile ribozymes and retroelements. Wiley Interdiscip Rev RNA 7:341–355. doi: 10.1002/wrna.1339. [DOI] [PubMed] [Google Scholar]
- 70.Candales MA, Duong A, Hood KS, Li T, Neufeld RA, Sun R, McNeil BA, Wu L, Jarding AM, Zimmerly S. 2012. Database for bacterial group II introns. Nucleic Acids Res 40:D187–D190. doi: 10.1093/nar/gkr1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Roberts RJ, Vincze T, Posfai J, Macelis D. 2015. REBASE: a database for DNA restriction and modification: enzymes, genes and genomes. Nucleic Acids Res 43:D298–D299. doi: 10.1093/nar/gku1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Koonin EV, Makarova KS, Wolf YI, Krupovic M. 2020. Evolutionary entanglement of mobile genetic elements and host defence systems: guns for hire. Nat Rev Genet 21:119–131. doi: 10.1038/s41576-019-0172-9. [DOI] [PubMed] [Google Scholar]
- 73.Biswas A, Staals RH, Morales SE, Fineran PC, Brown CM. 2016. CRISPRDetect: a flexible algorithm to define CRISPR arrays. BMC Genomics 17:356. doi: 10.1186/s12864-016-2627-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bernheim A, Bikard D, Touchon M, Rocha EPC. 2020. Atypical organizations and epistatic interactions of CRISPRs and Cas clusters in genomes and their mobile genetic elements. Nucleic Acids Res 48:748–760. doi: 10.1093/nar/gkz1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Crawley AB, Henriksen ED, Stout E, Brandt K, Barrangou R. 2018. Characterizing the activity of abundant, diverse and active CRISPR-Cas systems in lactobacilli. Sci Rep 8:11544. doi: 10.1038/s41598-018-29746-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hargreaves KR, Flores CO, Lawley TD, Clokie MRJ. 2014. Abundant and diverse clustered regularly interspaced short palindromic repeat spacers in Clostridium difficile strains and prophages target multiple phage types within this pathogen. mBio 5:e01045-13. doi: 10.1128/mBio.01045-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hall GS, Gordon S, Schroeder S, Smith K, Anthony K, Procop GW. 2001. Case of synovitis potentially caused by Dolosigranulum pigrum. J Clin Microbiol 39:1202–1203. doi: 10.1128/JCM.39.3.1202-1203.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hoedemaekers A, Schulin T, Tonk B, Melchers WJ, Sturm PD. 2006. Ventilator-associated pneumonia caused by Dolosigranulum pigrum. J Clin Microbiol 44:3461–3462. doi: 10.1128/JCM.01050-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lin JC, Hou SJ, Huang LU, Sun JR, Chang WK, Lu JJ. 2006. Acute cholecystitis accompanied by acute pancreatitis potentially caused by Dolosigranulum pigrum. J Clin Microbiol 44:2298–2299. doi: 10.1128/JCM.02520-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lecuyer H, Audibert J, Bobigny A, Eckert C, Janniere-Nartey C, Buu-Hoi A, Mainardi JL, Podglajen I. 2007. Dolosigranulum pigrum causing nosocomial pneumonia and septicemia. J Clin Microbiol 45:3474–3475. doi: 10.1128/JCM.01373-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Johnsen BO, Ronning EJ, Onken A, Figved W, Jenum PA. 2011. Dolosigranulum pigrum causing biomaterial-associated arthritis. APMIS 119:85–87. doi: 10.1111/j.1600-0463.2010.02697.x. [DOI] [PubMed] [Google Scholar]
- 82.Sherret J, Gajjar B, Ibrahim L, Mohamed Ahmed A, Panta UR. 2020. Dolosigranulum pigrum: predicting severity of infection. Cureus 12:e9770. doi: 10.7759/cureus.9770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sampo M, Ghazouani O, Cadiou D, Trichet E, Hoffart L, Drancourt M. 2013. Dolosigranulum pigrum keratitis: a three-case series. BMC Ophthalmol 13:31. doi: 10.1186/1471-2415-13-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Haas W, Gearinger LS, Hesje CK, Sanfilippo CM, Morris TW. 2012. Microbiological etiology and susceptibility of bacterial conjunctivitis isolates from clinical trials with ophthalmic, twice-daily besifloxacin. Adv Ther 29:442–455. doi: 10.1007/s12325-012-0023-y. [DOI] [PubMed] [Google Scholar]
- 85.Venkateswaran N, Kalsow CM, Hindman HB. 2014. Phlyctenular keratoconjunctivitis associated with Dolosigranulum pigrum. Ocul Immunol Inflamm 22:242–245. doi: 10.3109/09273948.2013.841484. [DOI] [PubMed] [Google Scholar]
- 86.Monera-Lucas CE, Tarazona-Jaimes CP, Escolano-Serrano J, Martinez-Toldos JJ. 2020. Bilateral keratitis secondary to Dolosigranulum pigrum infection in a patient with HIV infection. Enferm Infecc Microbiol Clin doi: 10.1016/j.eimc.2020.10.017. [DOI] [PubMed] [Google Scholar]
- 87.Oliveira PH, Touchon M, Cury J, Rocha EPC. 2017. The chromosomal organization of horizontal gene transfer in bacteria. Nat Commun 8:841. doi: 10.1038/s41467-017-00808-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.McInerney JO, McNally A, O’Connell MJ. 2017. Why prokaryotes have pangenomes. Nat Microbiol 2:17040. doi: 10.1038/nmicrobiol.2017.40. [DOI] [PubMed] [Google Scholar]
- 89.Lacks SA, Mannarelli BM, Springhorn SS, Greenberg B. 1986. Genetic basis of the complementary DpnI and DpnII restriction systems of S. pneumoniae: an intercellular cassette mechanism. Cell 46:993–1000. doi: 10.1016/0092-8674(86)90698-7. [DOI] [PubMed] [Google Scholar]
- 90.Bondy-Denomy J, Davidson AR. 2014. To acquire or resist: the complex biological effects of CRISPR-Cas systems. Trends Microbiol 22:218–225. doi: 10.1016/j.tim.2014.01.007. [DOI] [PubMed] [Google Scholar]
- 91.Sanozky-Dawes R, Selle K, Klaenhammer T, O’Flaherty S, Barrangou R. 2015. Occurrence and activity of a type II CRISPR-Cas system in Lactobacillus gasseri. Microbiology (Reading) 161:1752–1761. doi: 10.1099/mic.0.000129. [DOI] [PubMed] [Google Scholar]
- 92.Shmakov SA, Sitnik V, Makarova KS, Wolf YI, Severinov KV, Koonin EV. 2017. The CRISPR spacer space is dominated by sequences from species-specific mobilomes. mBio 8:e01397-17. doi: 10.1128/mBio.01397-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.EFSA Panel. 2012. Guidance on the assessment of bacterial susceptibility to antimicrobials of human and veterinary importance. EFSA J 10:2740. https://www.efsa.europa.eu/en/efsajournal/pub/2740. [Google Scholar]
- 94.Hunt M, Silva ND, Otto TD, Parkhill J, Keane JA, Harris SR. 2015. Circlator: automated circularization of genome assemblies using long sequencing reads. Genome Biol 16:294. doi: 10.1186/s13059-015-0849-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Tatusova T, DiCuccio M, Badretdin A, Chetvernin V, Nawrocki EP, Zaslavsky L, Lomsadze A, Pruitt KD, Borodovsky M, Ostell J. 2016. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res 44:6614–6624. doi: 10.1093/nar/gkw569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Haft DH, DiCuccio M, Badretdin A, Brover V, Chetvernin V, O’Neill K, Li W, Chitsaz F, Derbyshire MK, Gonzales NR, Gwadz M, Lu F, Marchler GH, Song JS, Thanki N, Yamashita RA, Zheng C, Thibaud-Nissen F, Geer LY, Marchler-Bauer A, Pruitt KD. 2018. RefSeq: an update on prokaryotic genome annotation and curation. Nucleic Acids Res 46:D851–D860. doi: 10.1093/nar/gkx1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 98.Vinuesa P, Contreras-Moreira B. 2015. Robust identification of orthologues and paralogues for microbial pan-genomics using GET_HOMOLOGUES: a case study of pIncA/C plasmids. Methods Mol Biol 1231:203–232. doi: 10.1007/978-1-4939-1720-4_14. [DOI] [PubMed] [Google Scholar]
- 99.Kristensen DM, Kannan L, Coleman MK, Wolf YI, Sorokin A, Koonin EV, Mushegian A. 2010. A low-polynomial algorithm for assembling clusters of orthologous groups from intergenomic symmetric best matches. Bioinformatics 26:1481–1487. doi: 10.1093/bioinformatics/btq229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Li L, Stoeckert CJ, Roos DS. 2003. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res 13:2178–2189. doi: 10.1101/gr.1224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Galtier N, Gouy M, Gautier C. 1996. SEAVIEW and PHYLO_WIN: two graphic tools for sequence alignment and molecular phylogeny. Comput Appl Biosci 12:543–548. doi: 10.1093/bioinformatics/12.6.543. [DOI] [PubMed] [Google Scholar]
- 103.Nguyen L-T, Schmidt HA, Von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol 32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kalyaanamoorthy S, Minh BQ, Wong TKF, Von Haeseler A, Jermiin LS. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods 14:587–589. doi: 10.1038/nmeth.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Hoang DT, Chernomor O, von Haeseler A, Minh BQ, Vinh LS. 2018. UFBoot2: improving the ultrafast bootstrap approximation. Mol Biol Evol 35:518–522. doi: 10.1093/molbev/msx281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Ondov BD, Treangen TJ, Melsted P, Mallonee AB, Bergman NH, Koren S, Phillippy AM. 2016. Mash: fast genome and metagenome distance estimation using MinHash. Genome Biol 17:132. doi: 10.1186/s13059-016-0997-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Perrin A, Rocha EPC. 2021. PanACoTA: a modular tool for massive microbial comparative genomics. NAR Genom Bioinform 3:lqaa106. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7803007/. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Hyatt D, Chen G-L, Locascio PF, Land ML, Larimer FW, Hauser LJ. 2010. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11:119. doi: 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Buchfink B, Xie C, Huson DH. 2015. Fast and sensitive protein alignment using DIAMOND. Nat Methods 12:59–60. doi: 10.1038/nmeth.3176. [DOI] [PubMed] [Google Scholar]
- 110.Tatusov RL, Koonin EV, Lipman DJ. 1997. A genomic perspective on protein families. Science 278:631–637. doi: 10.1126/science.278.5338.631. [DOI] [PubMed] [Google Scholar]
- 111.Galperin MY, Wolf YI, Makarova KS, Vera Alvarez R, Landsman D, Koonin EV. 2021. COG database update: focus on microbial diversity, model organisms, and widespread pathogens. Nucleic Acids Res 49:D274–D281. doi: 10.1093/nar/gkaa1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Kanehisa M, Goto S. 2000. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res 28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Kanehisa M, Sato Y, Kawashima M, Furumichi M, Tanabe M. 2016. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res 44:D457–D462. doi: 10.1093/nar/gkv1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Mistry J, Chuguransky S, Williams L, Qureshi M, Salazar GA, Sonnhammer ELL, Tosatto SCE, Paladin L, Raj S, Richardson LJ, Finn RD, Bateman A. 2021. Pfam: the protein families database in 2021. Nucleic Acids Res 49:D412–D419. doi: 10.1093/nar/gkaa913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Eddy SR. 2011. Accelerated profile HMM searches. PLoS Comput Biol 7:e1002195. doi: 10.1371/journal.pcbi.1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 117.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Benedict MN, Henriksen JR, Metcalf WW, Whitaker RJ, Price ND. 2014. ITEP: an integrated toolkit for exploration of microbial pan-genomes. BMC Genomics 15:8. doi: 10.1186/1471-2164-15-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.van Dongen S, Abreu-Goodger C. 2012. Using MCL to extract clusters from networks. In van Helden J, Toussaint A, Thieffry D (ed), Bacterial molecular networks methods in molecular biology (methods and protocols), vol 804. Springer, New York, NY. [DOI] [PubMed] [Google Scholar]
- 120.R Core Team. 2020. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. https://www.R-project.org/. [Google Scholar]
- 121.RStudio Team. 2020. RStudio: integrated development for R. RStudio/PBSPBC, Boston, MA. http://www.rstudio.com/. [Google Scholar]
- 122.Bazin A, Gautreau G, Medigue C, Vallenet D, Calteau A. 2020. panRGP: a pangenome-based method to predict genomic islands and explore their diversity. Bioinformatics 36:i651–i658. doi: 10.1093/bioinformatics/btaa792. [DOI] [PubMed] [Google Scholar]
- 123.Larsson A. 2014. AliView: a fast and lightweight alignment viewer and editor for large datasets. Bioinformatics 30:3276–3278. doi: 10.1093/bioinformatics/btu531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Potter SC, Luciani A, Eddy SR, Park Y, Lopez R, Finn RD. 2018. HMMER web server: 2018 update. Nucleic Acids Res 46:W200–W204. doi: 10.1093/nar/gky448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Gilchrist CLM, Chooi YH. 2021. Clinker and clustermap.js: automatic generation of gene cluster comparison figures. Bioinformatics doi: 10.1093/bioinformatics/btab007. [DOI] [PubMed] [Google Scholar]
- 126.Flusberg BA, Webster DR, Lee JH, Travers KJ, Olivares EC, Clark TA, Korlach J, Turner SW. 2010. Direct detection of DNA methylation during single-molecule, real-time sequencing. Nat Methods 7:461–465. doi: 10.1038/nmeth.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Johnston CD, Skeete CA, Fomenkov A, Roberts RJ, Rittling SR. 2017. Restriction-modification mediated barriers to exogenous DNA uptake and incorporation employed by Prevotella intermedia. PLoS One 12:e0185234. doi: 10.1371/journal.pone.0185234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Anton BP, Fomenkov A, Wu V, Roberts RJ. 2021. Genome-wide identification of 5-methylcytosine sites in bacterial genomes by high-throughput sequencing of MspJI restriction fragments. PLoS One 16:e0247541. doi: 10.1371/journal.pone.0247541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Grissa I, Vergnaud G, Pourcel C. 2007. CRISPRFinder: a web tool to identify clustered regularly interspaced short palindromic repeats. Nucleic Acids Res 35:W52–W57. doi: 10.1093/nar/gkm360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Biswas A, Gagnon JN, Brouns SJ, Fineran PC, Brown CM. 2013. CRISPRTarget: bioinformatic prediction and analysis of crRNA targets. RNA Biol 10:817–827. doi: 10.4161/rna.24046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Grissa I, Vergnaud G, Pourcel C. 2007. The CRISPRdb database and tools to display CRISPRs and to generate dictionaries of spacers and repeats. BMC Bioinformatics 8:172. doi: 10.1186/1471-2105-8-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Grissa I, Vergnaud G, Pourcel C. 2008. CRISPRcompar: a website to compare clustered regularly interspaced short palindromic repeats. Nucleic Acids Res 36:W145–W148. doi: 10.1093/nar/gkn228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Bastian M, Heymann S, Jacomy M. 2009. Gephi: an open source software for exploring and manipulating networks. https://gephi.org/publications/gephi-bastian-feb09.pdf.
- 134.Jacomy M, Venturini T, Heymann S, Bastian M. 2014. ForceAtlas2, a continuous graph layout algorithm for handy network visualization designed for the Gephi software. PLoS One 9:e98679. doi: 10.1371/journal.pone.0098679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Aguirre M, Morrison D, Cookson BD, Gay FW, Collins MD. 1993. Phenotypic and phylogenetic characterization of some Gemella-like organisms from human infections: description of Dolosigranulum pigrum gen. nov., sp. nov. J Appl Bacteriol 75:608–612. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Pangenome contribution of all 28 D. pigrum genomes. Download Table S1, PDF file, 0.04 MB (29.5KB, pdf) .
Copyright © 2021 Flores Ramos et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Pairwise SNP analysis and MASH analysis. Download Table S2, XLSX file, 0.06 MB (59.2KB, xlsx) .
Copyright © 2021 Flores Ramos et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
A core-gene phylogenomic tree including Alloiococcus otitis ATCC 51267 as an outgroup has poor resolution. (A) The 789 GCs shared between the BDBH, COG triangle, and OMCL algorithms were used to determine the single-copy GCs shared between the 28 D. pigrum genomes and the A. otitis outgroup using GET_HOMOLOGUES v. 3.1.4. (B) The phylogenomic tree with ancestral distance displayed. (C) The phylogenomic tree as a cladogram. A. otitis is the most closely related genome-sequenced taxon to D. pigrum based on 16S rRNA gene sequence in both the eHOMD and the Living Tree Project (P. Yarza, M. Richter, J. Peplies, J. Euzeby, R. Amann, K. H. Schleifer, W. Ludwig, F. O. Glöckner, R. Rosselló-Móra. Syst Appl Microbiol. 31:241–250, 2008, [doi: 10.1016/j.syapm.2008.07.001]; P. Yilmaz, L.W. Parfrey, P. Yarza, J. Gerken, E. Pruesse, C. Quast, T. Schweer, J. Peplies, W. Ludwig, F. O. Glöckner. Nucleic Acids Res. 42:D643–D648, 2014 [doi: 10.1093/nar/gkt1209]). We generated a phylogenomic tree based on 789 concatenated core GCs shared among A. otitis ATCC 51267 (NZ_JH992957) and the 28 D. pigrum genomes with IQ-Tree using a GTR+F+R6 substitution model (BIC value 5743936.9087), 300 ML searches (ML = −2868528.1268) and 1,000 ultrafast rapid Bootstraps. There is a deep branch separating A. otitis from all the D. pigrum strains. However, many of the ancestral branches within the D. pigrum clade are poorly supported. Repeated construction resulted in phylogenies with poorly supported branching likely due to poor SNP resolution (see Table S1A), suggesting the need for a better outgroup for D. pigrum. Download FIG S1, PDF file, 0.2 MB (215.7KB, pdf) .
Copyright © 2021 Flores Ramos et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The conservative core genome of D. pigrum has 1102 GCs with a very high degree of nucleotide conservation among strains and the accessory genome size varies but the functions are similar among the 28 genomes. (A) We used the intersection of the BDBH, COG triangle, and OMCL algorithm results to determine a conservative core genome of 1102 single-copy GCs using the 28 D. pigrum genomes on GET_HOMOLOGUES v. 3.1.4. (B) We determined the GCs present in the core (n = 28), soft core (28 > n ≥ 26), shell (26 > n ≥ 3), and cloud (n ≤ 2) using the results from the OMCL and COG triangle algorithms. Of the 3700 total GCs, 30.6% are core (1,134/3,700), 5.41% are soft core (200/3,700), 22.8% are shell (845/3700), and 41.1% are cloud (1,521/3,700). (C) All paired strains share above 97.58% average nucleotide identity. We based this on the average nucleotide identity of the homologues determined using the OMCL algorithm. (D) The D. pigrum Anvi’o pangenome is similar to the GET_HOMOLOGUES pangenome. The biggest difference between the GET_HOMOLOGUES- and Anvi’o-defined pangenome was the migration of many of the GET_HOMOLOGUES cloud GCs to the Anvi’o multicopy core, causing the pangenome size to drop from 3,700 to 2,905 GCs. Occupancy of the GCs among the 28 genomes is displayed with strains collected in 2018 and 2017 (blue), strains from the CDC (pink), the ATCC strain (gold), and the strain collected in 2010 (green). Strains are arranged based on the phylogeny from Fig. 1 (right). With Anvi’o, 44.7% of the GCs (1,298/2,905) were in the core, 3.1% (90/2,905) in the soft core (green), 27.7% (805/2,905) in the shell (blue), and 24.5% (712/2,905) in the cloud (purple). A closer inspection of the core also showed 38.2% (1111/2,905) of the GCs are in the conservative single-copy (SC) core (red) and 6.4% (187/2,905) are in the multicopy (MC) core (dark red). The interactive Anvi’o pangenome can also be found on our GitHub (https://github.com/KLemonLab/DpiMGE_Manuscript/blob/master/SupplementalMethods_Anvio.md). (E) The accessory genome size varies but the functions are similar among the 28 D. pigrum genomes. Using the functional annotations of the accessory GCs identified by Anvi’o (defined as “shell” plus “cloud” bins), we found that most of the accessory GCs among all of our genomes either had no annotation (“unclassified,” 53.6%), had an ambiguous COG categorization (“ambiguous”, 6.6%) or belonged to the uninformative S or R COG category (“uninformative,” 2.6%). Only 37.2% of the GCs in the accessory genome had informative COG annotations (colored categories). (F) Of the informative GCs, there was a similar proportional distribution of accessory functions among all 28 genomes with a large fraction dedicated to “defense mechanisms” (olive) and “carbohydrate transport and metabolism” (khaki). The size of the accessory genomes varied among the strains with CDC4294-98 and KPL3090 having the largest accessory size compared to KPL3065 and KPL3086 with the smallest. Horizontal bars indicate clades: clade 1, green; clade 2, purple; clade 3, orange; and clade 4, blue. Download FIG S3, PDF file, 1.1 MB (1.1MB, pdf) .
Copyright © 2021 Flores Ramos et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
A 100-kb region with a higher frequency of indels and a second region with a higher frequency of phage and plasmid integration events. (A) Closer inspection of positions ∼650000 to ∼750000 in the MAUVE alignment showed the presence of three sites containing sequencing heterogeneity (HS for heterogeneity sites) bounded by >10 kb blocks of homology. The first site (HS1), between abgT (p-aminobenzoyl-glutamate transport protein) and kynB (kynurenine formamidase), contained a PTS (mannitol) system for metabolism. However, the extent of completeness of this PTS system varied among the D. pigrum isolates and was absent in KPL3274 and KPL3256. The next site (a few kb downstream of HS1), between yxdM (ABC transporter permease protein YxdM) and ghrA (glyoxylate/hydroxypyruvate reductase A), contained one of the two possible CRISPR-Cas systems, with all genomes having at least one type (CS1; Fig. 8A and B). Finally, the third site (HS2), between nth (endonuclease III) and clsA_1 (cardiolipin synthase A), variably harbored two types of aspartate and threonine synthase genes. One type in KPL3033 and KPL3052; none in KPL1914, KPL3077, and KPL3274; and a variation of the second type among the remaining genomes. Also indicated on this MAUVE alignment are the second of the two CRISPR-Cas insertion sites (CS2) and the location of the pUB110 insertion, which is in a region with a higher frequency of phage and plasmid integration events. Vertical bars represent clades: clade 1, green; clade 2, purple; clade 3, orange; and clade 4, blue. (B) No autonomous plasmids were detected among the 28 genomes using the Gram-positive plasmid database, PlasmidFinder. However, as illustrated with this map from D. pigrum KPL3043, we detected a sequence match to pUB110 (GenBank ID NC_001384.1), a plasmid originally isolated from S. aureus, integrated in four of the D. pigrum isolates (KPL3043, KPL3065/KPL3086 in clade 4 and CDC4709-98 in clade 2; Fig. 1). This included a perfect match (708/708 bp) to the repB gene (blue) of pUB110, which was located adjacent to three other genes (cyan) that are also present in pUB110 and predicted to encode a kanamycin nucleotidyltransferase ANT(4′)-lb (knt), a bleomycin resistance protein (ble), and a plasmid recombination protein (pre-2). Inspection of the region adjacent to the genes from pUB110, at position ∼1.45 Mb in the MAUVE alignment (A), showed evidence of plasmid elements present in 11 of the other closed D. pigrum genomes, suggesting this region has a higher frequency of phage and plasmid integration events. For example, a 73% (288/391 bp) match to a different (non-pUB110) repB gene present in plasmids of other closely related Firmicutes including Lactobacillus plantarum (KU707950) (X. Ma, J. Li, Y. Xiong, Z. Zhai, F. Ren, Y. Hao. Curr Microbiol. 73:820–826, 2016 [doi: 10.1007/s00284-016-1124-7]) and Lactobacillus curvatus (CP031007) (R.M. Prechtl, https://www.researchgate.net/publication/337103133_Formation_and_structure_of_exopolysaccharides_of_meat_starter_cultures). A subset of the genomes, including six from clade 4 (KPL3077, KPL3090, KPL3069, KPL3084, KPL3911, and KPL3070) and two from clade 3 (KPL3264 and KPL3256), had an ltrA gene (group II reverse transcriptase/maturase) at this location. Download FIG S2, PDF file, 1.0 MB (1MB, pdf) .
Copyright © 2021 Flores Ramos et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Supplemental analysis of genetic elements and defense systems. Download Text S1, PDF file, 0.10 MB (98.6KB, pdf) .
Copyright © 2021 Flores Ramos et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Predicted D. pigrum transposases, integrases and group II intron. Download Table S3, PDF file, 0.2 MB (124.1KB, pdf) .
Copyright © 2021 Flores Ramos et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
RM systems. Download Table S4, XLSX file, 0.02 MB (18.9KB, xlsx) .
Copyright © 2021 Flores Ramos et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The organization of GCs within RM system integration sites R2 and R3. (A) Organization of GCs within RM system integration site 2 (R2), which primarily encodes either a type II m5C-associated RM system or a type IV restriction system. R2 is flanked upstream by a region containing genes for a YacP-like NYN domain protein and an S8 family serine peptidase type protein, and downstream by genes for a DNA-3-methyladenine glycosylase protein, a hypothetical protein, and a copper-translocating P-type ATPase. (B) The organization of GCs within RM system integration site 3 (R3), which encodes two different type II RM systems. Arrows represent the direction of translation and the relative sizes of ORFs. Putative control proteins are highlighted in green; HsdR and HsdM are highlighted in red and blue, respectively; and fused hsdR/M ORFs are both red and blue. Proteins not identified as part of the RM system or those with currently unknown function are shown in gray. Download FIG S4, PDF file, 0.2 MB (229.2KB, pdf) .
Copyright © 2021 Flores Ramos et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
CRISPR-Cas systems. Download Table S5, XLSX file, 0.02 MB (24.1KB, xlsx) .
Copyright © 2021 Flores Ramos et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Data Availability Statement
All genomes are available from the NCBI. Table 1 lists the accession numbers for each D. pigrum strain genome used in this study.